Graphical abstract
Keywords: tncRNA Toolkit, transfer RNA (tRNA)-derived non-coding RNAs (tncRNAs), tRFs, tRNA halves, Transcription regulation
Highlights
-
•
Computational pipeline for accurate identification of transfer RNA-derived non-coding RNAs.
-
•
Six major tncRNA classes with a size between 14 and 50 nt were identified by exploiting 2,448 samples from six different angiosperms.
-
•
tRNA nucleoside modifications may direct tRNA cleavage.
-
•
Conserved tncRNAs target transcripts that are involved in plant growth, development, and metabolism.
-
•
tncRNAs are expressed in a tissue-dependent, and stress-specific manner in plants.
Abstract
The emergence of distinct classes of non-coding RNAs has led to better insights into the eukaryotic gene regulatory networks. Amongst them, the existence of transfer RNA (tRNA)-derived non-coding RNAs (tncRNAs) demands exploration in the plant kingdom. We have designed a methodology to uncover the entire perspective of tncRNAome in plants. Using this pipeline, we have identified diverse tncRNAs with a size ranging from 14 to 50 nucleotides (nt) by utilizing 2448 small RNA-seq samples from six angiosperms, and studied their various features, including length, codon-usage, cleavage pattern, and modified tRNA nucleosides. Codon-dependent generation of tncRNAs suggests that the tRNA cleavage is highly specific rather than random tRNA degradation. The nucleotide composition analysis of tncRNA cleavage positions indicates that they are generated through precise endoribonucleolytic cleavage machinery. Certain nucleoside modifications detected on tncRNAs were found to be conserved across the plants, and hence may influence tRNA cleavage, as well as tncRNA functions. Pathway enrichment analysis revealed that common tncRNA targets are majorly enriched during metabolic and developmental processes. Further distinct tissue-specific tncRNA clusters highlight their role in plant development. Significant number of tncRNAs differentially expressed under abiotic and biotic stresses highlights their potential role in stress resistance. In summary, this study has developed a platform that will help in the understanding of tncRNAs and their involvement in growth, development, and response to various stresses. The workflow, software package, and results are freely available at http://nipgr.ac.in/tncRNA.
1. Introduction
The profusion of non-coding RNAs (ncRNAs) with diverse regulatory actions discovered over the last decade has been proven to be a crucial breakthrough in molecular biology. They are powerful regulators of gene expression at the epigenetic, transcriptional, and post-transcriptional levels in the living system [1]. Amongst the diverse pool of smaller ncRNAs, microRNAs (miRNAs), and small interfering RNAs (siRNAs) are the most extensively surveyed molecules [2]. They alter gene expression by binding to their target mRNAs [3]. The advancement of high-throughput sequencing technology has tremendously resolved the surveillance of the other classes of small untranslated RNAs beyond miRNAs and siRNAs [4], [5]. Amongst them, transfer RNA (tRNA)-derived non-coding RNAs (tncRNAs) have been reported in all three domains of life [6]. This distinct group of regulatory RNAs is derived from the endonucleolytic cleavage of precursor tRNAs (pre-tRNAs) or mature tRNAs [7]. tncRNAs includes well-known shorter tRNA-derived RNA fragments (∼12 to 30 nucleotides [nt]) popularly termed as tRFs [8] or tDRs [9], and longer tRNA halves or tRHs (∼30 to 40 nt) [10]. Precise tRNA cleavage leading to their production is governed by tRNA type, cell type, developmental stage, stress, and tissue [11], [12], [13]. It has been revealed that these novel molecules can originate from nuclear as well as organellar tRNAs [14], [15]. This class of small RNAs has been particularly well studied in human cancers [16], [17], [18]. In plants, the earliest report by Thompson et al. 2008 showed that processed tRNA halves were generated in Arabidopsis during oxidative stress [19]. In another study, phloem-specific tRNA fragments were shown to interfere with protein synthesis in pumpkin (Cucurbita maxima) [20]. For the first time in plants, Loss Morais and his group in 2013 showed the association of 19 nt tRFs with Argonaute under stress conditions [21]. Alves et al. extended the research and highlighted the differential accumulation of specific tRFs under abiotic stresses in Arabidopsis and rice [22]. In addition to abiotic stresses, some studies have shown links to tncRNAs generation in response to biotic stresses, e.g., in response to Apple stem grooving virus (ASGV) infection in domesticated apple [23], and Phytophthora capsici infection in black pepper [24].
Depending on the position of cleavage on mature tRNA, tRFs belong to two categories: 5′ end derived tRF-5, and 3′ end derived tRF-3 generated upon cleavage in the D and T region, respectively. Among the tRNA halves, 5′ or 3′ tRHs are produced by cleavage in the anticodon region containing 5′ or 3′ portions of mature tRNA [25]. The pre-tRNA produces 5′U-tRFs/leader tRF, and tRF-1/tsRNA/3′U-tRFs from the 5′ leader and 3′ trailer portion respectively [7], [26], [27]. By now, it has been established that tncRNAs can be generated in a DICER-dependent or independent manner in plants [28], [29]. Besides Dicer-like proteins (DCLs), members of RNases T2, namely RNS1, RNS2, and RNS3 are the key contributors to the biogenesis of tncRNAs originating from mature tRNAs in Arabidopsis [22], [30]. Noteworthy, the tncRNA generation from pre-tRNAs in plants has been understudied.
Identification of tncRNAs in small RNA-seq (sRNA-seq) datasets is a very challenging and error-prone process. During the library preparation, extensively modified tRNA nucleosides affect the reverse transcription process, resulting in truncation or misincorporation of nucleotides [31]. This, in turn, increases the probability of mismatch. However, allowing just a single mismatch to overcome sequencing errors, may lead to base misidentification, and hence causes the inflation of the false negatives. This is due to high sequence similarities among 20 major tRNA isotypes covering the whole tRNA family of isoacceptors (tRNAs with different anticodons but charged with the same amino acid), and isodecoders (tRNAs carrying the same anticodon with variations in the body sequence). Researchers have suggested different mapping strategies for tRNA-derived reads [32], [33], [34]. Some tools are available for tncRNA detection, e.g. tDRmapper [35], MINTmap [36], and tRF2Cancer [37], but they are trained and tested on human datasets, and thereby suit for human data only. As compared to humans, tncRNAs have been less explored in the plant domain. Although, exclusive plant databases, e.g. tRex [38] and PtRFdb [39], harboring information related to the transfer RNA-derived fragments (tRFs), have also been developed in the recent past, yet a convenient methodology for the accurate identification and exploration of tncRNAs is still lacking in plants.
In this study, we developed a computational methodology for accurate tncRNAs detection. We have identified a diverse pool of tncRNAs in six different angiosperms viz. Arabidopsis thaliana (model plant), Medicago truncatula (model legume), tomato (Solanum lycopersicum), chickpea (Cicer arietinum), rice (Oryza sativa), and maize (Zea mays) by exploiting publicly available sRNA-seq datasets. With the help of our in-house pipeline, we have identified, classified, and quantified tncRNAs into different subtypes based on their origin and cleavage position viz. tRF-5, tRF-3, tRF-1, leader-tRF, 5′ and 3′ tRNA halves (tRHs). As there is no single standardized nomenclature for tncRNA subtypes yet, researchers have followed the classical and most frequently used terms for tncRNA subtypes. tRNA modifications play an integral role in tRNA maturation, structural integrity, and stabilization in all domains of life [40], [41]. Recently, few studies have indicated that some specific tRNA modifications may drive programmed tRNA cleavage [42], [43]. Thereby, in addition to the tncRNA identification, we have also included information related to the modified nucleoside sites detected along the individual tncRNAs. We also scrutinized the organization of tncRNA classes in various plant tissues. Additionally, we have also cataloged differentially expressed tncRNAs under various abiotic and biotic stresses. Henceforth, by applying our methodology, this study highlights a dynamic landscape of tncRNAs. Certainly, this will help in elucidating their significance in plants.
2. Results
2.1. In silico tncRNA identification across six plants
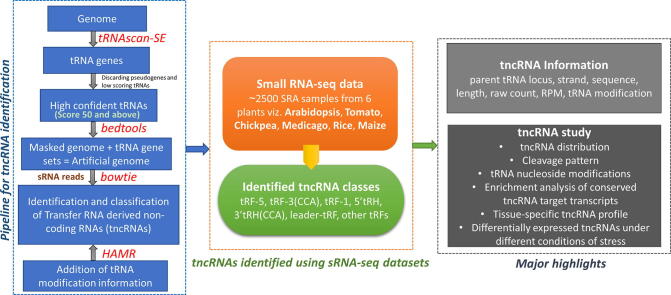
By utilizing sRNA-seq data available at Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra), from the National Center for Biotechnology Information (NCBI), we have identified diverse groups of tncRNAs from 2,448 samples in six angiosperms viz. A. thaliana (1676), S. lycopersicum (1 6 0), C. arietinum (21), M. truncatula (1 2 7), O. sativa (2 4 3), and Z. mays (2 2 1). After processing and quality control, filtered reads were provided as input to the in-house pipeline (Fig. 1A). Due to the huge number of tRNA gene copies predicted by tRNAscanSE [44], we have filtered our tRNA gene pool by eliminating pseudogenes and keeping genuine tRNAs based on score. As the score is an important indicator of the structural propensity of tRNA [44], [45], only high-quality tRNAs with a score equal to or above 50 were selected for mapping. To avoid ambiguous reads from non-tRNA regions, we created an artificial genome by masking the genuine tRNA gene region with 50 nt upstream and downstream in the genome (masked genome) and adding them as artificial chromosomes (Fig. 1A). We classified only those reads as tncRNAs that mapped exclusively to the artificial chromosomes (i.e. tRNA set consisting of mature tRNA, 50 nt leader, and 50 nt trailer). The identified tncRNAs were categorized into tRF-5, tRF-3, tRF-1, leader-tRF, 5′tRH, 3′tRH, and other tRF (Fig. 1B). The sample-wise tncRNA analysis results containing various information related to each tncRNA, i.e. tncRNA type, parent tRNA locus information, position (start–end), sequence, length, read count, RPM, nature, and position of modified nucleosides, alignment of each tncRNA on its progenitor tRNA, and the count of different tncRNA subclasses utilized in this study can be downloaded from “Data download” module at http://www.nipgr.ac.in/tncRNA. The sample number, total, and unique tncRNAs counts for individual plants have been summarised in Table 1 (the same tncRNA sequences were grouped and termed as unique).
Fig. 1.
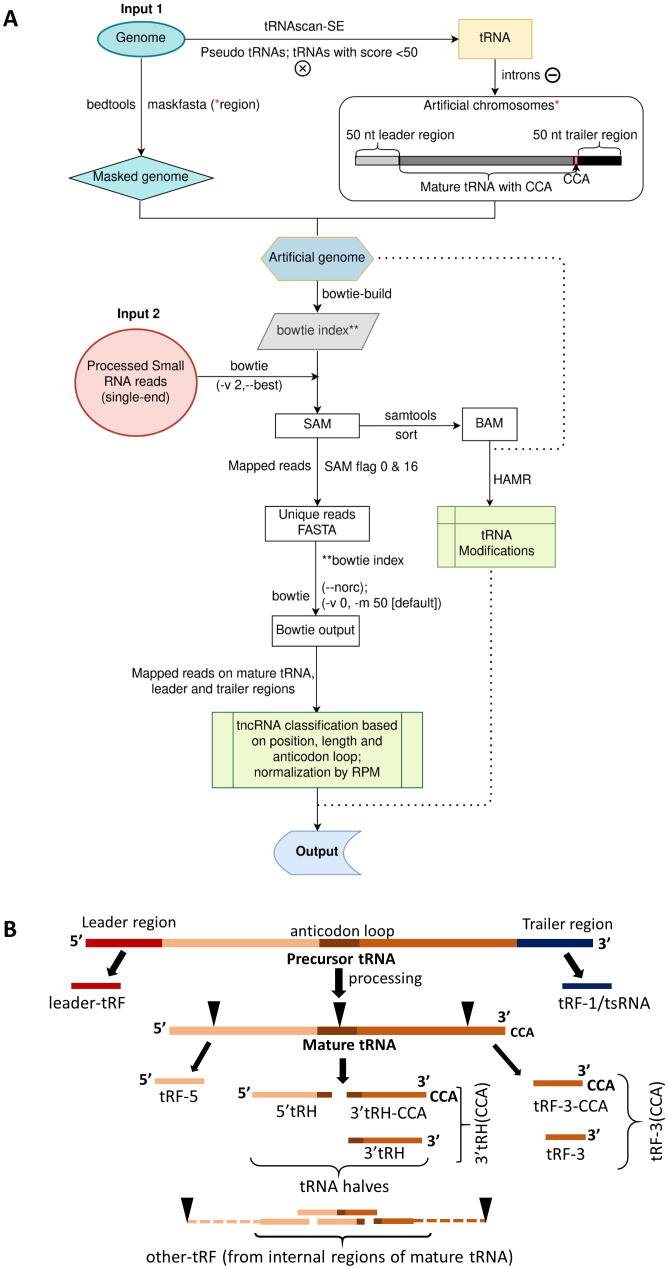
Workflow for the identification of transfer-RNA-derived non-coding RNAs (tncRNAs). A Genome and filtered reads are provided as inputs to the pipeline. For avoiding ambiguous reads from non-tRNA regions, an artificial genome is created by masking the genuine tRNA gene region with 50 nt upstream and downstream in the genome and adding them as artificial chromosomes* (*as represented in the box). Only those reads are classified as tncRNAs that are mapped exclusively to the artificial chromosomes. tncRNAs are classified based on tRNA origin, and site of cleavage. B Biogenesis of distinct tncRNA subtypes generated from the excision of pre-tRNA, and mature tRNA. pre-tRNA gives rise to leader tRF and tRF-1/tsRNA, whereas mature tRNA generates tRF-5, tRF-3 (with or without CCA), 5′tRH, 3′tRH (with or without CCA), and other tRF (derived from internal portions excluding extreme ends of mature tRNA denoted by dashed lines).
Table 1.
Summary of identified groups of transfer-RNA-derived non-coding RNAs (tncRNAs) abundance in six plants.
| Plant species | Total Samples | tncRNAs | tRF-5 | tRF-3(CCA) | tRF-1 | 5′tRH | 3′tRH(CCA) | Leader tRF | Other tRFs |
|---|---|---|---|---|---|---|---|---|---|
| Total/Unique | Total/Unique | Total/Unique | Total/Unique | Total/Unique | Total/Unique | Total/Unique | Total/Unique | ||
| A. thaliana | 1676 | 3,362,128/50619 | 760,155/2180 | 782,722/4513 | 22,505/693 | 164,074/662 | 28,835/840 | 2359/238 | 1,601,478/41502 |
| C. arietinum | 21 | 54,390/10627 | 8837/1012 | 11,763/1792 | 77/35 | 2135/306 | 778/175 | 26/12 | 30,774/7310 |
| M. truncatula | 127 | 306,836/11971 | 63,157/1182 | 82,262/1967 | 779/72 | 15,232/321 | 4372/139 | 137/32 | 140,897/8270 |
| O. sativa | 243 | 406,496/23117 | 100,828/1812 | 98,089/2787 | 1553/189 | 19,070/447 | 2595/294 | 53/26 | 184,308/17585 |
| S. lycopersicum | 160 | 265,435/23312 | 62,838/1753 | 59,833/3323 | 1878/688 | 19,298/560 | 2429/264 | 130/78 | 119,029/16664 |
| Z. mays | 221 | 428,193/18304 | 95,748/1819 | 99,510/2651 | 1959/265 | 37,768/618 | 7457/354 | 164/27 | 185,587/12584 |
2.2. tncRNA classification according to origin and site of cleavage
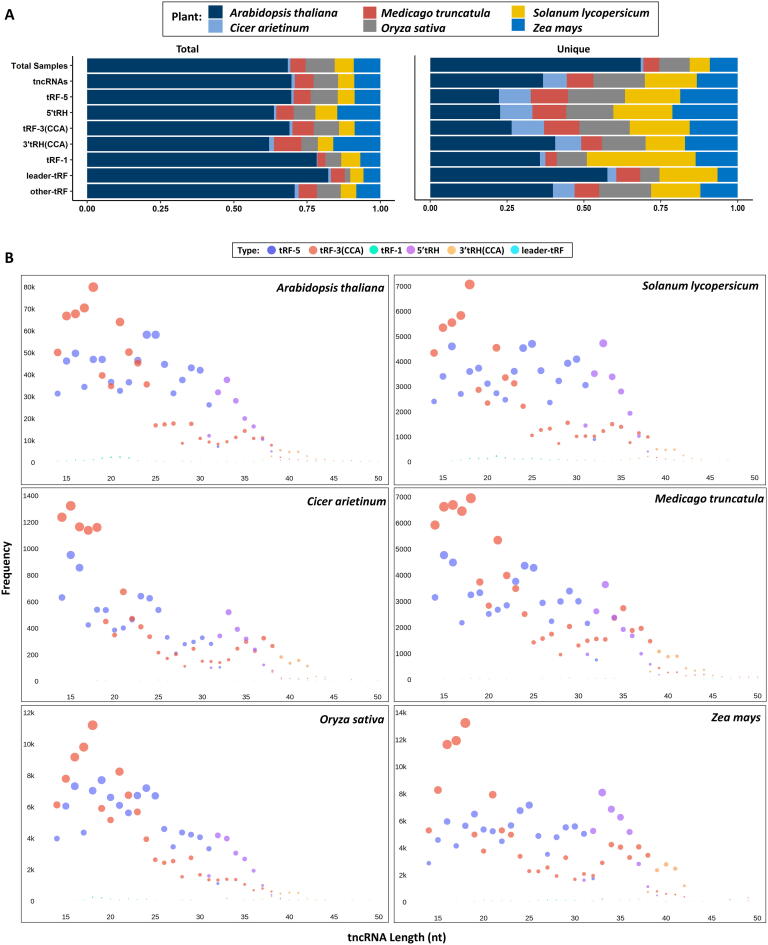
We have computed the relative percentages of sRNA-seq samples, tncRNA abundance (total and unique), and tncRNA classes (total and unique) for each plant (Fig. 2A). The total tncRNAs in Arabidopsis showed higher abundance due to greater sample size, but the relative abundance of unique sequences in Arabidopsis was fairly comparable to other plants (removing duplicate sequences from several samples reduces the bias of sample number). The unique count of reads per tncRNA category for individual samples has been provided in Supplementary sheets 1.1–1.6. The length-wise abundance of major tncRNA classes was studied in different plants (Fig. 2B). The bulk of the identified tncRNAs belonging to the tRF-5 and tRF-3 series, fall in a smaller length range up to 25 nt and 21 nt respectively. The high frequency of specific-sized tncRNAs indicates cleavage preference by the endoribonuclease machinery at a certain position in the loop/stem region. Also, more than 92% of the total identified tRF-3 sequences contained terminal end CCA in all plants. The high abundance of CCA ending fragments indicates that the terminal CCA might also play a role in the structural and functional integrity of fragments generated from the 3′ end. We also observed a remarkable fraction of fragments generated from internal portions of mature tRNA. These fragments constitute nearly 45% of the total tncRNA sequences identified in each plant (Supplementary Fig. 1A–F). Longer tRHs occupy less than 10% of the tncRNAs. Compared to 3′tRHs, the 5′tRHs were more abundant. The remaining fraction (<1%) consisted of tncRNAs generated from pre-tRNAs. The length diversity of tncRNAs indicates their prospective roles besides canonical miRNA-like gene expression regulation. The high abundance of smaller tncRNAs, e.g. 15–17 nt tRF-5 and tRF-3 sequences, need to be carefully examined as smaller fragments are usually discarded or considered to be randomly degraded fragments (Fig. 2B). The highly abundant and diverse reads belonging to tRF-5, tRF-3, as well as those derived from the internal region of tRNAs, suggest that they may have varied roles in plants whereas longer tRNA halves are generated during specific conditions (Supplementary Fig. 1A–F). As cleavage in the anticodon loop can generate both types of tRNA halves i.e. 5′ and 3′ tRHs, but it is not clear that a single cleavage in the tRNA molecule can generate functional halves simultaneously, and should be explored by further experiments.
Fig. 2.
Abundance of the identified transfer-RNA-derived non-coding RNAs (tncRNAs) in different plants. A Relative distribution of samples, total and unique tncRNA, tRF-5, tRF-3(CCA), tRF-1, 5′tRH, 3′tRH(CCA), leader, and other tRFs analyzed in this study. B Size distribution of major tncRNA categories viz. tRF-5, tRF-3(CCA), tRF-1, 5′tRH, 3′tRH(CCA), and other tRFs in individual plants.
In addition to the nucleus, the tncRNAs from chloroplast and mitochondria were also observed. A considerable number of tncRNAs originated exclusively from either chloroplast or mitochondria (Supplementary sheet 1.7). For example, in Arabidopsis, 24% tRF-5 s, 33% tRF-3 s, 29% 5′tRHs, and 25% 3′tRHs were exclusively derived from organellar tRNAs. We checked the relative abundance of chloroplastic tncRNAs in green vs. non-green tissue samples. Interestingly, there was a higher percentage of chloroplastic tncRNAs in leaf samples as compared to the root samples (Supplementary Fig. 2). It can be suggested that the presence of a separate pool of organellar tncRNAs in distinct tissues may have potential functions in plants.
2.3. Cleavage of tncRNAs is highly specific and conserved amongst plants
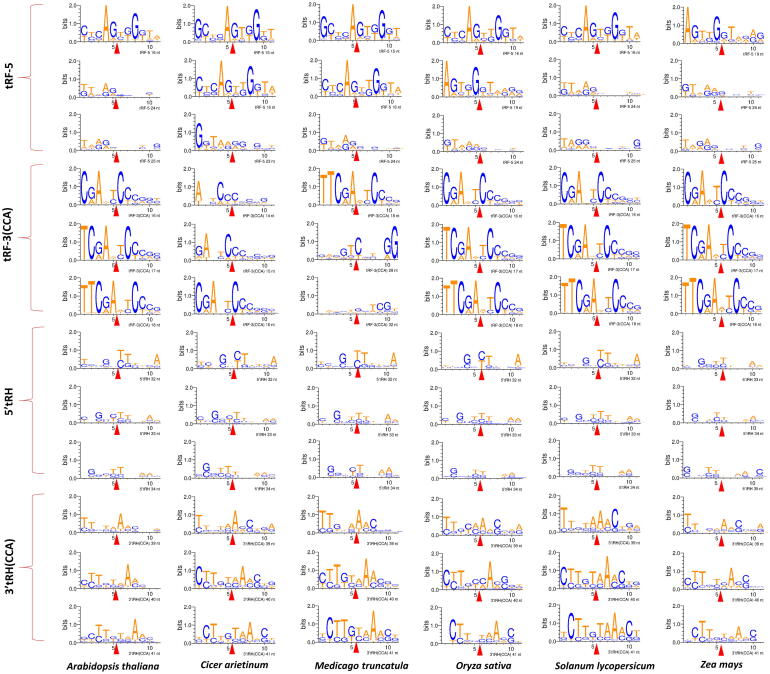
Nucleotide composition and the ratio of the constituent monomer units is an important characteristic feature for studying nucleic acids. For studying the tncRNA cleavage pattern, we looked into the nucleotide composition of tncRNA::tRNA junction for highly recurring tRF-5, tRF-3(CCA), 5′tRH, and 3′tRH(CCA) sequences (Fig. 3). A common conserved motif was observed in tRF-5::tRNA interface among all plants i.e. AG::TGG (first row in Fig. 3) in tRF-5 s ranging between 15 and 19 nt. Also, the last nucleotide in tRF-5 was observed to be A/G rich (first three rows from top in Fig. 3). In tRNA::tRF-3(CCA) junction, CGA(N)::(N)TC pattern was commonly observed across all species (fourth to sixth rows in Fig. 3). Similarly, in the majority of the 5′ tRNA halves, the breakpoint region in the middle (fifth and sixth position) is either T/C (seventh to ninth rows in Fig. 3), whereas the first nucleotide in 39 nt 3′ tRH (CCA) is A rich in all species (sixth nucleotide in the tenth rows in Fig. 3). Also, a common conserved pattern, CTTGTAAAC was observed in the majority of 40–41 nt length 3′tRHs (eleventh and twelfth rows in Fig. 3). Despite the variability due to the occurrence of anticodon triplets among various isoacceptors, we have seen common motifs at tRNA cleavage points among different plants indicating that the cleaving enzymes mostly prefer specific isoacceptors for spawning tRHs. Also, the majority of tRF-1 sequences identified in rice, maize, and tomato samples had terminal ends rich in T residues (Supplementary Fig. 3). It has been reported that G at 18th and 19th position in tRF-5 plays a crucial role in the inhibition of protein translation in both human and Arabidopsis [46], [47]. The production of tncRNAs from pre-tRNAs has not been well studied in plants. In humans, tRF-1/pre-tRF-3U usually end in a short stretch of T residues due to the release of polymerase III [6], [57]. Based on observed motifs and specific nucleotide composition of tncRNAs, it is worth speculating that the enzymatic cleavage might first recognize or prefer a particular motif located on parental tRNA to cleave precise fragments for at least a reasonable number of genuine tncRNAs.
Fig. 3.
Web logo representation depicting conserved cleavage sites for four tncRNA subclasses viz. tRF-5, tRF-3(CCA), 5′tRH, and 3′tRH(CCA). Web Logos were made using top-three recurring tncRNAs by taking 5 nt stretches from progenitor tRNA and respective tncRNA fragments viz. tRF-5::tRNA; 5′tRH::tRNA; tRNA::tRF-3(CCA);tRNA::3′tRH(CCA). To study the breakpoint and its flanking nucleotide composition, for tRF-5 and 5′tRH classes, the first five nucleotides (from left to right) represent the last five nucleotides of tRF-5/5′tRH and the remaining stretch of nucleotides is the tRNA (sixth to the eleventh nucleotide). For tRF-3(CCA) and 3′tRH(CCA) classes, the first five nucleotides (from left to right) represent progenitor tRNA sequence, and the remaining stretch represents the first sixth nucleotide of tRF-3(CCA)/3′tRH(CCA) (sixth to the eleventh nucleotide). The red arrow represents the breaking point of each tncRNA fragment from its parental tRNA. All the sequences shown are oriented in the 5′–3′ direction. G|T; T|C; A|A breakpoint was commonly found in all types of tncRNAs. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.4. Specific anticodons, not the whole tRNA repertoire are responsible for tncRNA generation
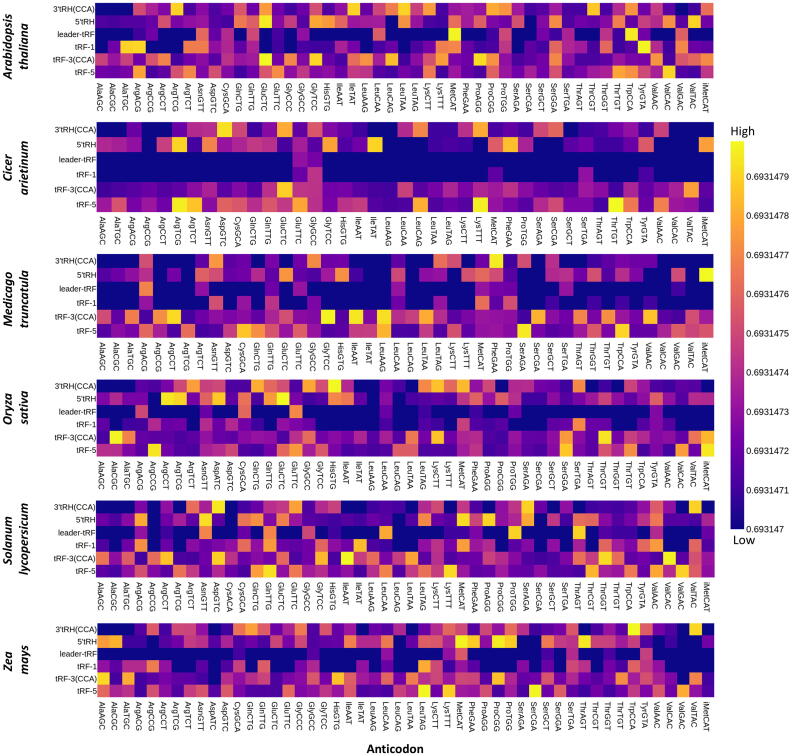
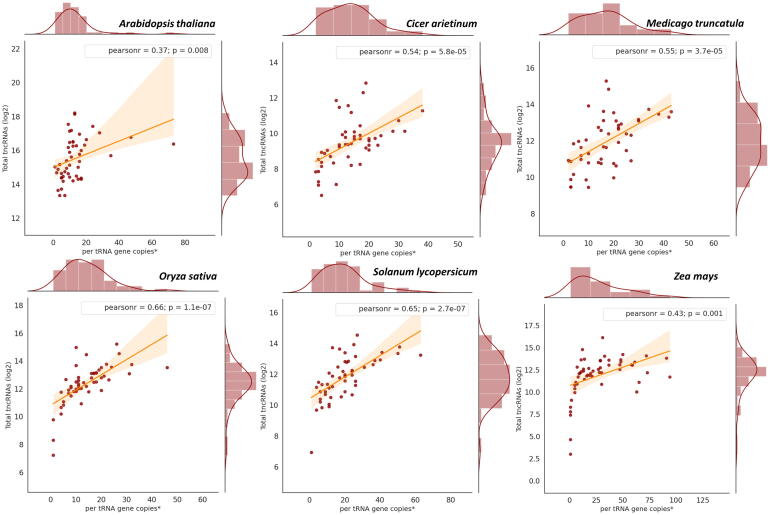
Out of the 64 triplet codons, 61 are sense codons that code for 20 amino acids. We observed that not all 61 codons generate tncRNAs. Out of these 61 codons, Arabidopsis, chickpea, and Medicago utilized 49, 41, and 44 isoacceptors respectively, while 48 isoacceptors contributed to the generation of different tncRNAs in tomato, rice, and maize (Fig. 4). Different isoacceptors contributed to the production of different tncRNA types and varied from one plant to another. Although coding for the same amino acids, there was marked variation among the isoacceptors for contributing to the tncRNA generation, e.g. in Arabidopsis, among five tRNA-Arg isoacceptors viz. ACG, CCG, CCT, TCG, and TCT, tRNA-ArgACG generated the most number of tRF-5 s. Also, many anticodons can generate more than one class of tncRNAs, e.g. GluCTC, and ProCGG gave rise to tRF-5 and tRF-3, while GluCTC, GlyTCC, and LeuTAA generated 5′tRH and 3′tRH in Arabidopsis. By comparing the number of tncRNAs generated per the number of copies for each tRNA isoacceptors, we observed that the tncRNAs generation showed a slightly positive correlation with the increasing abundance of isoacceptor copies (Fig. 5). Pearson correlation coefficient ranged from 0.37 to 0.66, which implies that the number of tncRNAs generated is not directly proportional to tRNA frequency. Still, there is a possibility that the tRNA isoacceptors with more copy numbers may contribute more to tncRNAs generation.
Fig. 4.
Heatmap showing the abundance of tncRNA subclasses viz. tRF-5, tRF-3(CCA), tRF-1, 5′tRH, 3′tRH(CCA), and leader tRF sourcing from each tRNA isoacceptor (specified using their respective anticodon) in six plants; The frequencies are given in read count converted to log10 values.
Fig. 5.
Correlation graph showing the total tncRNAs identified vs per tRNA gene copies in individual plants.
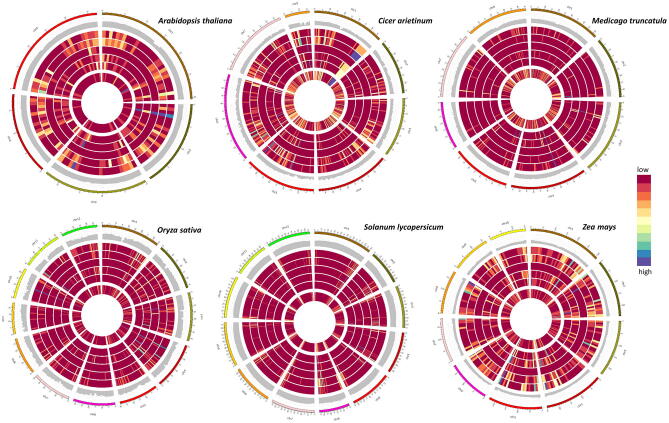
The tRNA and tncRNA generating regions were visualized on the genome (Fig. 6). Various classes of tncRNAs are generated from distinct portions of genomic tRNAs. We found many regions on the genome that showed lesser tRNA density but generated more tncRNAs and vice-versa. For example, tRNA-dense regions can be seen particularly on chromosome 1 (chr1) in Arabidopsis but most of the tncRNAs were generated from chr2. Similarly, despite low tRNA gene density, a large number of tncRNAs originated from chr4 in tomato, chr6 and 8 in chickpea, chr3 in Medicago, chr7 and 11 in rice, and chr1, 5, and 8 in maize. It can be observed that the tncRNAs are not necessarily generated from tRNA-rich regions of the genome. Interestingly, some regions generated several classes of tncRNAs, like chr2 in Arabidopsis, chr1 in tomato, chr4 in Medicago and rice, and chr5 in maize, while some regions gave rise to specific tncRNA classes. For example, more 5′ containing tncRNAs, i.e. tRF-5 and 5′tRHs, on chr3 in Arabidopsis, chr6 in tomato, chr4 in Medicago, chr11 in rice, and chr5 in maize. It is quite conceivable that some of these regions may govern rapid turn-over of tRNAs into tncRNAs. These findings give clues about the additional insights into the genomic organization of tRNA into clusters and their implications on tRNA functions. The prevalence of tRNA gene clusters in all kingdoms of life reveals various insights into evolutionary history and tRNA functions. Whether tRNA gene clusters impact the generation of tncRNAs in the living system, including plants, is worth intriguing.
Fig. 6.
Circos plots for interactive visualization of the genomic origin of various tncRNAs subclasses generated from parental tRNAs and the relative position of tRNA genes on the genome for individual plants. The relative abundance of various tncRNA subtypes and tRNA gene density with respect to the genomic location is shown by the heatmap. From the outer to the inner circle, the first ring represents the chromosomes, the next ring (second) shows the GC content of the chromosomes. The third, fourth, fifth, sixth, and seventh ring shows the genomic tRNA region producing tRF-5, tRF-3(CCA), tRF-1, 5′ tRH, and 3′tRH respectively. The innermost ring shows the tRNA gene distribution across the genome. tncRNA/tRNA-rich regions are shown in purple (highest density) and tncRNA/tRNA-poor regions are shown in the red (minimum density) spectrum. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.5. Modification may play a role in regulating the tncRNA generation and function
As tRNAs are heavily modified molecules with over 100 known modifications, we scrutinized the modifications present in our identified tncRNAs. For this, we utilized HAMR [48], a high utility tool for predicting modified nucleosides in high throughput sequencing datasets. Various modifications were detected by HAMR on tncRNAs in different plants, viz. 3-methylcytosine (m3C), pseudouridine (Y/Ψ), dihydrouridine (D), N2-methylguanosine (m2G), N2-dimethylguanosine (m22G), 1-methyl guanosine (m1G), 1-methyladenosine (m1A), 1-methylinosine (m1I), 2-methylthio-N6-isopentenyladenosine (ms2i6A), N6-isopentenyl adenosine (i6A), Threonylcarbamoyladenosine (t6A) in various isoacceptors generating tncRNAs. The visualization of various modifications on tncRNAs concerning their position on the respective consensus sequence of mature tRNA in 49 isoacceptors in Arabidopsis has been shown in Supplementary Fig. 4. The plots for modification distribution in the other five angiosperms have been provided in Supplementary Figs. 5–9. We observed that most modifications were predicted from 20th to 40th position and 50th to 60th position, i.e. containing the cleavage sites for tRF-5, tRHs, and tRF-3, indicating a high probability of tncRNA generation to be regulated by tRNA modifications. The recurring tRNA modifications in tncRNAs from Arabidopsis samples have been highlighted in Supplementary Fig. 10. It was observed that some modifications were specific for certain anticodons and were found in abundance at specific positions. For example, in the D region, Guanosine (G) was modified to m1G on GlyGCC and iMetCAT (Supplementary Fig. 10). Also, pseudouridine (Y) modification on GlyGCC tRNAs was highly abundant. In the anticodon loop region, Y and D were abundant in tncRNAs originating from GluTTC tRNAs. In the T region, m1A|m1I|ms2i6A were observed in a large number of tRNAs.
We also looked for modifications found common in all six plants selected for this study (Supplementary Fig. 11A-U). Adenosine (A) replaced with m1A|m1I| ms2i6A modifications were found in 20 isoacceptors viz. tRNA ArgACG|CCG|TCG|TCT (58th position), AspGTC (57th position), GlnTTG (57th position), GlyGCC (57th position), iMetCAT (57th position), LeuCAA (69th position), LeuCAG (66th position), LeuTAG (65th position), LysCTT (58th position), LysTTT (57th position), SerAGA|CGA|TGA (67th position), ThrTGT (57th position), TyrGTA (58th position), and ValAAC|CAC (59th position). G is modified to m1G on the T loop at 58th position in TyrGTA and 59th position in ValAAC|CAC. These modifications were prominent in the region of the T loop, which is the site of cleavage for tRF-3 generation. Also, on the 9th nucleotide, G was replaced with m1G in ArgACG, GlyGCC, and iMetCAT in all plants. Am nucleoside (2′-O-methyladenosine) is reported to be induced by salt stress in a variety of land plants, including Arabidopsis, Brachypodium, poplar, and rice [49]. 2′-O-methylguanosine (Gm), 5-methyluridine (m5U), and 5-methylcytidine (m5C) are also tightly linked to plant development [50]. Overall, the findings suggest the possibility of a very delicate cross-talk among the genetic machinery involved in tncRNA biogenesis, functioning, as well as tRNA nucleoside modification in response to plant growth, development, as well as environmental changes.
2.6. tncRNAs are conserved amongst plants
In our entire analysis, we observed that various tncRNAs were identical in all six plant species. We found a total of 252 tRF-5, 351 tRF-3, 46 5′tRH, and 12 3′tRH common sequences among all six plant species included in our analysis. The sequence conservation provides a clue to the conservation in the regulatory function of these tncRNAs. They can act as post-transcriptional regulators by complementary binding to their messenger RNA (mRNA) targets leading to mRNA degradation. Thus, conserved sequences were utilized for target prediction and pathway enrichment analysis to gain some functional insights into gene expression regulation by tncRNAs. The detailed list of conserved tRF-5 s, and tRF-3 s along with their respective target genes have been provided in Supplementary sheets 2.1 and 2.2, respectively. We observed that several target genes were associated with the molecular function, biological process, and cellular components for tRF-5 and tRF-3. The tRF-5 target genes were related to 9 distinct molecular functions, 25 types of biological processes, and six cellular components, while tRF-3 target genes were associated with 13 molecular functional categories, 26 biological activities, and seven cellular components (Supplementary Fig. 12). A large number of target genes for the tRF-5 series were involved in catalytic (7 1 1), transferase (3 3 9), kinase (1 3 8) activities (Supplementary sheet 2.3). The target genes for the tRF-3 series were mostly associated with catalytic (9 0 3), and transferase activities (4 1 6) (Supplementary sheet 2.4). For a more organized visualization of our enrichment results, we clustered our enriched genes into enrichment maps, and clusters of similar pathways representing major biological processes were generated. The tRF-5 targets were enriched in growth and development, transport and localization, and cellular metabolic processes (Supplementary Fig. 13), while the tRF-3 targets formed two distinct clusters related to developmental and cellular metabolic processes (Supplementary Fig. 14). It can be predicted that many tRF-5 and tRF-3 may act as potent regulators of the genes involved in plant development, and cellular metabolic machinery in a tissue/condition-specific manner. The top one target analysis reveals that conserved tRF sequences can bind to different transcripts in plants, but the genes targeted by the same tRF don't need to have an identical function in different plants (Supplementary sheets 2.5 and 2.6). It can be speculated that tRFs can interact with multiple genes, and may regulate various activities in the plant system.
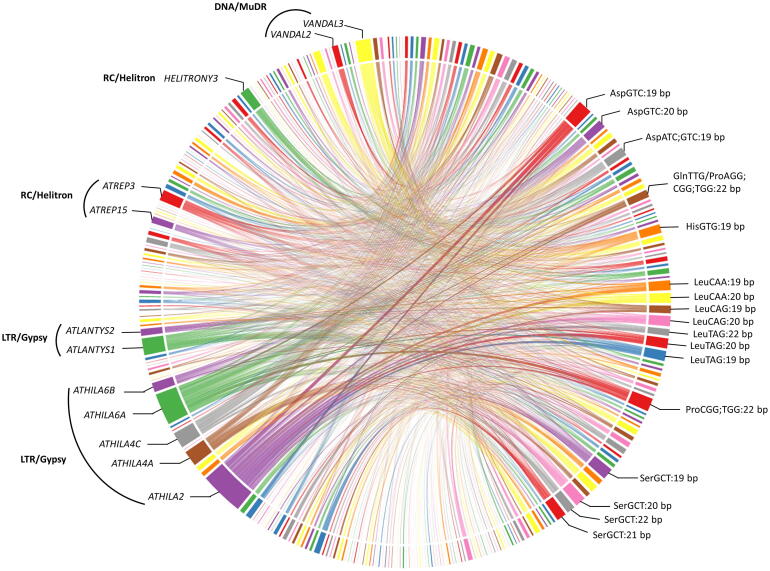
We also examined the conserved tRF-5 and tRF-3 against full transposable elements (TEs) from Arabidopsis. Many tRF-5 and tRF-3 sequences were associated with several TE transcripts (Supplementary sheets 2.7 and 2.8). A large number of tRF-5 showed association with various transposon families (Fig. 7). As compared to tRF-5 s, tRF-3 showed a weaker association with TEs. TE belonging to the LTR/Gypsy superfamily were the most frequent targets of the tRF-5 s. With this superfamily, TE from ATHILA2, ATHILA6A, ATHILA4C, ATHILA4A, ATLANTYS1, ATHILA6B, and ATLANTYS2 families were found to be associated with a large number of tRFs. Apart from them, TE superfamilies like RC/Helitron (ATREP3 and ATREP15) and DNA/Mudr (VANDAL2 and VANDAL3) also show considerable association with tRFs. Smaller length tRFs (mostly 19–22 nt) originating from AspGTC/ATC, GlnTTG, ProAGG/CGG/TGG, HisGTG, LeuCAA/TAG, and SerGCT tRNAs were associated with TEs in high numbers. tncRNAs binding with TE can bestow genome stability to plants. Interestingly, two recent reports have shown that tRFs target transposable elements (retrotransposons) in both plants and mammals, indicating their potential as epigenetic regulators [51]. For instance, in Arabidopsis, a 19 nt tRF-5 from tRNA-MetCAT cleaves its target LTR retrotransposon transcript, Athila6A [52]. Deciphering the synchronization among the tncRNA generation, TE expression, and their implications in plants under normal and stress conditions can unveil additional regulatory functions of this class of ncRNAs.
Fig. 7.
A chord diagram representing the connections between conserved tRF-5 s and their corresponding target transposable elements (TE) family and their superfamily. Most recurring TE families and associated tRF-5 have been labeled. The broader fragments reflect the greater number of associations.
2.7. tncRNAs profiling
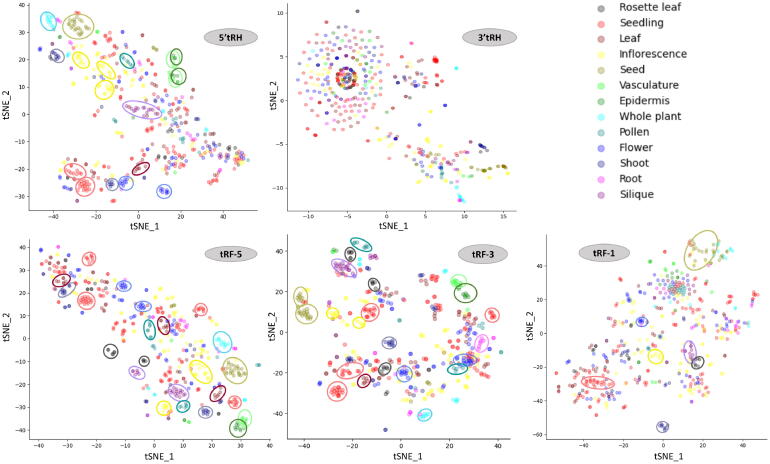
We checked the organization of tncRNA sequences in each tncRNA class for tissue samples using t-Distributed Stochastic Neighbor Embedding (tSNE). The t-SNE plots revealed numerous clusters for individual tncRNA classes in different plants. In Arabidopsis, seedlings and seed clusters were frequently seen in tncRNAs belonging to tRF-5, tRF-3, tRF-1, and 5′tRH (Fig. 8). As compared to the tRNA halves, smaller fragments particularly belonging to tRF-5 and tRF-3 classes showed enriched clustering for more than ten different tissues. It was interesting to observe that vasculature, and epidermal tissue clusters were found to be in close proximity in tRF-5, tRF-3, and 5′tRHs. Surprisingly, seedling, seed, shoot, rosette leaf, inflorescence, silique, and flower clusters were also seen in the tRF-1 series. Some clusters belonging to various tissues like leaf, anther, and seedling were also observed in rice, tomato, and maize (Supplementary Figs. 15–17). The distribution pattern of tncRNAs belonging to various tissues indicates that spatiotemporal expression of tncRNAs occurs in a tissue-specific manner at normal physiological conditions.
Fig. 8.
t-Distributed Stochastic Neighbor Embedding (t-SNE) for the visualization of the tncRNA organization in Arabidopsis tissue samples. Individual t-SNE plots were made for tRF-5, tRF-3, tRF-1, 5′tRH, and 3′tRH classes. The tncRNAs from a total of 525 samples from 13 different tissues were utilized for this analysis. Tissue clusters observed have been shown by similar color-coded dashed circles.
2.8. Differentially expressed (DE) tncRNAs
We checked the expression of tncRNAs under various abiotic and biotic stresses in Arabidopsis, tomato, rice, and maize (Supplementary sheet 3.2–3.5). Significant differences in tncRNA expression were observed in different plants under different conditions. Besides up-regulated fragments, a significant number of tncRNA were down-regulated in various samples. We observed stress, time, or tissue-dependent tncRNA expression in different plants. For instance, in Arabidopsis, only 3 tncRNAs showed dysregulation after 0.5 h of heat stress but their number rose to 42 after a time duration of 6 h. Likewise, only 25 tncRNAs were expressed during drought stress at 20% field capacity (FC), whereas 182 DE tncRNAs were detected when FC was 30% drought stress. On the other hand, less than 10 DE tncRNAs were detected in Magnaporthe oryzae infected root tissues (9 and 7 DE tncRNAs after 30 and 120 min respectively). However, the numbers were highly elevated in infected leaves as compared to root tissues (87 and 62 tncRNAs after 30 and 120 min respectively). The time-dependent difference in expression was also observed in few tissues in maize under heat stress (2 h vs 48 h). Among all the conditions studied, we found that heat exposure leads to the maximum number of differentially expressed tncRNAs in different maize tissues, both in vegetative and reproductive stages when compared with control. The highest among all with a total of 1748 tncRNAs were differentially expressed in tassel in reproductive stage upon exposure to 48 h of heat stress, among which 35 nt 5′tRH derived from GlyTCC, was significantly downregulated with a log2FC of −30. The tomato plant when subjected to Tomato Mosaic Virus (TMV) infection showed 757 DE tncRNAs, of which the majority of the fraction were contributed by 15 nt tRFs. In addition, it was noticed that DE tncRNAs in Arabidopsis were generated from HisGTG at a comparatively elevated level than others, GlnCTG in maize, GluTTC in rice, and tomato. In addition, it was noticed that most of the DE tncRNAs were generated from HisGTG in Arabidopsis, GlnCTG in maize, and GluTTC in rice as well as tomato. We also observed the upregulation of 19 nt tRF-5 (ArgCCT) under drought (30% FC) and NaCl stress in Arabidopsis. It also showed elevated expression in maize leaf subjected to heat stress. In earlier studies, it was reported that this tRF was expressed at high levels in Arabidopsis seedlings under drought conditions and cold-treated rice inflorescences [22]. The findings suggest that the accumulation of evolutionarily conserved plant tncRNAs is regulated by biotic and environmental stresses.
3. Discussion
We have developed a workflow/pipeline to accurately identify genuine tncRNAs from sRNA-seq datasets. The workflow has been designed for convenient tncRNA mining and can be tailored as per the requirement. In the past two decades, various research groups have contributed to the tncRNA identification in the animal world, particularly in humans. Some databases e.g. tRex and PtRFdb have been developed in the recent past for the exploration of tncRNAs in plants. Also, an annotation pipeline, SPORTS1.0 was developed for profiling canonical small RNAs like miRNAs, piRNAs, rRNAs, and tRNA-derived sRNAs in various organisms including some plants [53]. In this study, we have reported 43,153 novel tncRNAs (Supplementary Fig. 18) by using our workflow/pipeline (i.e. tncRNA Toolkit), and tested it on previously published datasets [52]; and further compared it with SPORTS 1.0. Both pipelines (i.e. tncRNA Toolkit and SPORTS 1.0) detected the previously validated 19 nt tRF-5 (AlaAGC) in all five replicates in Arabidopsis pollen. Comparison of various features, and outputs of the tncRNA Toolkit with already existing databases (i.e. tRex and PtRFdb), and methodologies (SPORTS 1.0) reveals the clear difference in the number of reported tncRNAs because of the differences in mapping strategies, read count, read length, and read classification parameters (Supplementary sheets 4.1–4.3). SPORTS1.0 profiled canonical tncRNAs coming from mature tRNAs and categorized them in four categories based on whether they are derived from 5′ end, 3′ end, 3′ CCA end of tRNAs, or from other regions within the mature tRNA. The various tncRNA subtypes and their respective counts from different regions of mature tRNAs from both pipelines have been provided in Supplementary sheet 4.3. By comparing the overall results, it can be stated that tncRNA Toolkit identify and classify the tncRNAs into various subclasses sourcing from both mature as well as pre-tRNAs, and can detect potential tncRNAs including experimentally validated ones. Also, with tncRNA Toolkit, the read count cut-off, number of allowed mismatches, and threshold of suppressing multi-mapped alignments can be altered as per the user requirements, making this a superior tool. Moreover, in addition to the application of computational approaches for the prediction of tncRNAs, studying and validating the tncRNAs in the future will provide the opportunity for better benchmarking the tools, and pipeline using real positive datasets.
In recent years, many studies associated with eukaryotic tRNA fragments have revealed newer complexities and functions linked with the conserved tRNA molecules beyond their established role in the canonical translation process [54], [55], [56]. However, structured searches for tncRNAs have so far been restricted to their biogenesis and functional role as regulatory molecules analogous to miRNAs. The existence of tRNAs with the same anticodon sequence (isodecoders) necessitates further exploration of their role in tncRNAs biogenesis. Our observations confirm the existence of a heterogeneous pool of tncRNAs in plants. Organellar tncRNAs identified in this pool demand further attention for exploring their probable functions in the cellular milieu. In plants, the existence of chloroplastic tRNAs in addition to mitochondrial tRNAs adds another layer of intricacy in terms of their biogenesis, localization, and functions. Conservation of cleavage consensus sites in different plants, and codon-dependent tncRNA generation support that they are the product of non-random ribonucleolytic tRNA cleavage. Also, the DE tRF-1 reported for the first time in our study encourages us to commence the exploration of these fragments in planta. The identification of tncRNA targets, as well as DE tncRNAs, is just a head start to explore innumerable pathways that may be critically regulated by tncRNAs in various cellular processes. The tissue-specific organization of tncRNAs suggests their potential biological role in plants. Further validation of DE tncRNAs and their putative targets will pave the way for a better understanding of the underlying mechanism in regulating stress responses. Differences in tncRNAs expression suggest their multifarious physiological roles particularly in relevance to stress responses. The tncRNA sequences particularly in the length range of 19–25 nt have been previously studied for the Argonaute proteins (AGOs) interaction and Argonaute-immunoprecipitation (AGO-IP) libraries have been observed to be rich in tRNA fragments [21], [22]. However, in our study, we have seen the abundance of tncRNAs in other libraries as well. This indicates that AGOs may or may not associate with all tncRNAs. Apart from conventional AGO binding mediated RNA interference, many tncRNAs may be functionally independent of AGO, unlike miRNAs. Also, the expression of tncRNAs in non-AGO IPs indicates the existence of some unrevealed tncRNA regulatory mechanisms which might be involved in fine-tuning gene expression. tRNA is post-transcriptionally modified at various nucleotides, their vital role in structural integrity and the translation is well established, but the detailed mechanisms showing how these tRNA modifications affect the regulation of their cleavage remain unclear. Our study provides a future perspective for better understanding the structural and functional impact of tRNA modifications in tRNA biology.
Although the in-silico approach will help in identifying these novel molecules across a wide range of organisms, experimental validation and characterization are vital for distinguishing more and more bona fide tncRNAs. Switching from conventional RNA-seq to tRNA-seq to overcome modification biases will be advantageous for the identification of tncRNAs. To study the molecular mechanisms of tncRNAs, apart from classical molecular biology methods, advanced ribonomics [57] like cross-linking, ligation, and sequencing of hybrids (CLASH) may lay the foundation for studying tncRNA-target hybrids in plants. As tncRNAs are ubiquitous and conserved across different domains of life, our comprehensive study is believed to substantiate research of these novel molecules. Their global identification will facilitate deciphering common conserved pathways in eukaryotes and their mechanism of unexplored regulatory action. Leveraging computational power together with molecular biology techniques will augment the current understanding of the vast tncRNAomic landscape in the living system.
4. Methods
4.1. Data retrieval
Genome assembly for Arabidopsis thaliana (TAIR10.1), Solanum lycopersicum (nuclear: SL4.0; organellar: SL3.0), Cicer arietinum (nuclear: ASM33114v1; organellar: ASM33114v1, Medicago truncatula (nuclear: MedtrA17_4.0; organellar, MtrunA17r5.0-ANR), Oryza sativa (Build 4.0; organellar: IRGSP-1.0), and Zea mays (Zm-B73-REFERENCE-NAM-5.0) were downloaded, and IDs were transformed for starting with “chr”, to make them convenient for secondary analysis. Single-end sRNA-seq datasets (Illumina) available for the abovementioned plants were downloaded using NCBI SRA Toolkit (v2.10) [58].
4.2. tRNA annotation and genome pre-processing
Nuclear tRNAs were predicted by tRNAscan-SE [44] (v2.0.6) using the eukaryotes model, while organellar tRNAs were detected by using the ‘-O’ option in this tool. The tRNA pseudogenes and tRNAs with a score less than 50 were eliminated, and filtered tRNA regions with 50 base pair (bp) flank at 3′ and 5′ were masked in the reference genome by ‘maskfasta’ script of bedtools [59] (v2.29). Further, three FASTA files were prepared, 1) mature tRNA, built from filtered tRNAs after intron removal, and addition of CCA at 3′ terminus, 2) leader tRNA, 50 bp 5′ tRNA genomic region flank, and 3) trailer tRNA, 50 bp 3′ tRNA genomic region flank. These FASTA files were added to the masked genome as ‘additional chromosomes’ to create an artificial genome. The Bowtie index for the artificial genome was built by bowtie-build (bowtie v1.3) [60].
4.3. Modification site prediction
The single-end small RNA reads were processed by Trim Galore v0.6.6 (https://github.com/FelixKrueger/TrimGalore) to remove the low-quality bases and trim the adapter sequences. To permit error-tolerant and uniquely mapping reads alignments, processed reads were aligned to the artificial genome index using bowtie [60] with ‘-v2 --best’ options. The SAM output was sorted and converted to BAM by samtools [61] (v1.10). The modification sites were predicted by HAMR (v1.2) (python hamr.py IN.bam REF.fa models/euk_trna_mods.Rdata OUT ath 30 10 0.05 H4 0.01 0.05 0.05) [48], [62]. Further, modification sites only on mature tRNA and leader/trailer regions were counted.
4.4. tncRNAs identification and classification
The mapped reads from previously generated SAM files were fetched to create a FASTA file of unique reads with the count, using SAM flag 0 & 16 for single-end reads. Mapped unique reads were aligned to + ve strand only with no mismatch, and multi-mapped reads were discarded if alignment occurred greater than 50 times, using bowtie1 with “--norc -v 0 -m 50” arguments. The bowtie combinatorial arguments “--best” and “--strata” were used, which guarantees that reported singleton alignments are best in terms of the stratum. Then, the output was used to identify locus, location, length of read mapped to mature tRNA, and their 50 bp upstream and downstream flank; based on that, reads were classified into different classes of tncRNAs. The tRNA halves were classified by cleavage at the anticodon loop (2 nt + 3 nt of anticodon + 2 nt = 7 nt), and information for modification site was added for tncRNAs. Reads per million (RPM) was calculated for each tncRNA by using the formula:
Per Million Factor (PMF) = Total mapped reads/106
Reads per million (RPM) = Number of reads mapped to progenitor tRNA/ PMF
A local alignment file of tncRNA to the sequence of origin was also created for visualization using the pairwise2 biopython module [63]. The alignment score is given as per
identical = 1, non-identical = −1, gap-open = −1, gap-extend = −0.5
4.5. tRNA model for consensus sequence and structure
The FASTA files were generated for specific isoacceptor tRNAs and provided as an input to the LocARNA [64] (v1.9.2) software package with “--stockholm” option for consensus sequence study. These ‘stockholm’ format files were used to create postscript files by RNAalifold [65] (v2.4.11) for the consensus tRNA sequence. The modification sites predicted for each tRNA isoacceptor have been shown on the consensus tRNA structures.
4.6. Target prediction, GO, and pathway enrichment
The psRNATarget [66] software was utilized for tncRNA target prediction (2017 release; default parameters). Common tncRNAs (tRF-5 and tRF-3 series; 18–25 nt long) were used as a query, while Arabidopsis cDNA and transposons sequences were used as probable targets. The protein targets were analyzed for pathway enrichment analysis using g:Profiler [67] and visualized in Cytoscape [68] (EnrichmentMap [69]and AutoAnnotate [70]).
4.7. t-Distributed Stochastic Neighbor Embedding (t-SNE) plot
t-SNE is a non-linear technique for dimensionality reduction that is particularly well suited for the visualization of high-dimensional datasets. We employed t-SNE to study datasets comprising of unique tncRNA sequences belonging to tRF-5, tRF-3, tRF-1, 5′tRH, and 3′tRH in major tissues from the Arabidopsis, rice, tomato, and maize. The tissues with at least 10 samples (Supplementary sheets 5.1–5.4) were utilized for matrix generation. A binary matrix, for presence (1) and absence (0), was created for unique tncRNA sequences from each tissue sample. From this, the plot was generated for each aforementioned class of tncRNAs.
4.8. Differential expression study
DESeq2 was utilized for the identification of differentially expressed tncRNAs under stress conditions [71]. Only those samples were selected in which at least three biological replicates for each control and treatment were present (Supplementary sheet 3.1). In DESeq2, the p-values attained by the Wald test are corrected for multiple testing using the Benjamini and Hochberg method by default. For our analysis, only those tncRNAs with a p value less than 0.05 were considered to be significant. Only significant tncRNAs were reported in expression analysis. tncRNAs with log2FC value greater than 1 (>1), and less than −1 (<−1) were considered up- and down-regulated, respectively.
5. Code and Data availability
All pipeline scripts, codes, data generated, and analyzed for each of the species are freely available at our website (URL: http://nipgr.ac.in/tncRNA). The codes and usage are also available at https://github.com/skbinfo/tncRNA-Toolkit.
Author contributions
S.Z. and A.S. designed the pipeline, wrote the codes, and performed all the analysis. N.P. contributed to the data analysis. S.Z. and SK wrote the manuscript. SK conceived the study and coordinated the project.
Funding
Grant SRG/2019/000097 from Science and Engineering Research Board (SERB), Department of Science and Technology, Government of India. Core research grant of National Institute of Plant Genome Research, India.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
S.Z. and A.S. acknowledge the Council of Scientific and Industrial Research (CSIR), India, for the Senior Research Fellowship. This study was supported by Grant SRG/2019/000097 from Science and Engineering Research Board (SERB), Department of Science and Technology, Government of India, and Grant BT/PR40146/BTIS/137/4/2020 from the Department of Biotechnology (DBT), Government of India. The authors are thankful to the Department of Biotechnology (DBT)-eLibrary Consortium, India, for providing access to e-resources. The authors thank Dr. Senjuti Sinharoy, Scientist, NIPGR, for checking the English in the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.09.021.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.P. Zhang, W. Wu, Q. Chen, and M. Chen, “Non-Coding RNAs and their Integrated Networks,” Journal of integrative bioinformatics, vol. 16, no. 3. NLM (Medline), Jul. 13, 2019, doi: 10.1515/jib-2019-0027. [DOI] [PMC free article] [PubMed]
- 2.Costa F.F. Non-coding RNAs: meet thy masters. BioEssays. 2010;32(7):599–608. doi: 10.1002/bies.200900112. [DOI] [PubMed] [Google Scholar]
- 3.Vaucheret H. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 2006;20(7):759–771. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- 4.Vickers K.C., Roteta L.A., Hucheson-Dilks H., Han L., Guo Y. Mining diverse small RNA species in the deep transcriptome. Trends Biochem Sci. 2015;40(1):4–7. doi: 10.1016/J.TIBS.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vivek A.T., Kumar S. Computational methods for annotation of plant regulatory non-coding RNAs using RNA-seq. Brief Bioinform. 2021;22(4):1–24. doi: 10.1093/BIB/BBAA322. [DOI] [PubMed] [Google Scholar]
- 6.Keam S.P., Hutvagner G. tRNA-derived fragments (tRFs): emerging new roles for an ancient RNA in the regulation of gene expression. Life (Basel, Switzerland) 2015;5(4):1638–1651. doi: 10.3390/life5041638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee Y.S., Shibata Y., Malhotra A., Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23(22):2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu Y., Lee I., Lee Y.S., Bao X. Small non-coding transfer RNA-derived RNA fragments (tRFs): Their biogenesis, function and implication in human diseases. Genomics Inform. 2015;13(4):94–101. doi: 10.5808/GI.2015.13.4.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fricker R., Brogli R., Luidalepp H., Wyss L., Fasnacht M., Joss O. A tRNA half modulates translation as stress response in Trypanosoma brucei. Nat Commun. 2019;10(1) doi: 10.1038/s41467-018-07949-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.J. M. Dhahbi, S. R. Spindler, H. Atamna, D. Boffelli, and D. I. K. Martin, “Deep Sequencing of Serum Small RNAs Identifies Patterns of 5′ tRNA Half and YRNA Fragment Expression Associated with Breast Cancer,” Biomark. Cancer, vol. 6, p. BIC.S20764, Jan. 2014, doi: 10.4137/BIC.S20764. [DOI] [PMC free article] [PubMed]
- 11.Kumar P., Anaya J., Mudunuri S.B., Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014;12(1):78. doi: 10.1186/s12915-014-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raina M., Ibba M. tRNAs as regulators of biological processes. Front Genet. 2014;5:171. doi: 10.3389/fgene.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soares A.R., Santos M. Discovery and function of transfer RNA-derived fragments and their role in disease. Wiley Interdiscip Rev RNA. 2017;8(5):e1423. doi: 10.1002/wrna.2017.8.issue-510.1002/wrna.1423. [DOI] [PubMed] [Google Scholar]
- 14.V. Cognat et al., “The nuclear and organellar tRNA-derived RNA fragment population in Arabidopsis thaliana is highly dynamic.,” Nucleic Acids Res., vol. 45, no. 6, pp. 3460–3472, Apr. 2017, doi: 10.1093/nar/gkw1122. [DOI] [PMC free article] [PubMed]
- 15.Pliatsika V., Loher P., Telonis A.G., Rigoutsos I. MINTbase: a framework for the interactive exploration of mitochondrial and nuclear tRNA fragments. Bioinformatics. 2016;32(16):2481–2489. doi: 10.1093/bioinformatics/btw194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson P., Ivanov P. tRNA fragments in human health and disease. FEBS Lett. 2014;588(23):4297–4304. doi: 10.1016/j.febslet.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X., Zhang Y., Ghareeb W.M., Lin S., Lu X., Huang Y. A comprehensive repertoire of transfer RNA-derived fragments and their regulatory networks in colorectal cancer. J Comput Biol. 2020;27(12):1644–1655. doi: 10.1089/cmb.2019.0305. [DOI] [PubMed] [Google Scholar]
- 18.Zhu L., Ge J., Li T., Shen Y., Guo J. tRNA-derived fragments and tRNA halves: the new players in cancers. Cancer Lett. 2019;452:31–37. doi: 10.1016/J.CANLET.2019.03.012. [DOI] [PubMed] [Google Scholar]
- 19.Thompson D.M., Lu C., Green P.J., Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14(10):2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang S., Sun L., Kragler F. The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol. 2009;150(1):378–387. doi: 10.1104/pp.108.134767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loss-Morais G., Waterhouse P.M., Margis R. Description of plant tRNA-derived RNA fragments (tRFs) associated with argonaute and identification of their putative targets. Biol Direct. 2013;8(1):6. doi: 10.1186/1745-6150-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alves C.S., Vicentini R., Duarte G.T., Pinoti V.F., Vincentz M., Nogueira F.T.S. Genome-wide identification and characterization of tRNA-derived RNA fragments in land plants. Plant Mol Biol. 2017;93(1–2):35–48. doi: 10.1007/s11103-016-0545-9. [DOI] [PubMed] [Google Scholar]
- 23.Visser M., Maree H.J., Rees D.J.G., Burger J.T. High-throughput sequencing reveals small RNAs involved in ASGV infection. BMC Genom. 2014;15(1):568. doi: 10.1186/1471-2164-15-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asha S., Soniya E.V. Transfer RNA derived small RNAs targeting defense responsive genes are induced during phytophthora capsici infection in black pepper (Piper nigrum L.) Front Plant Sci. 2016;7:767. doi: 10.3389/fpls.2016.00767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dhahbi J.M. 5’ tRNA halves: the next generation of immune signaling molecules. Front Immunol. 2015;6:74. doi: 10.3389/fimmu.2015.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olvedy M., Scaravilli M., Hoogstrate Y., Visakorpi T., Jenster G., Martens-Uzunova E.S. A comprehensive repertoire of tRNA-derived fragments in prostate cancer. Oncotarget. 2016;7(17):24766–24777. doi: 10.18632/oncotarget.v7i1710.18632/oncotarget.8293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pekarsky Y., Balatti V., Palamarchuk A., Rizzotto L., Veneziano D., Nigita G. Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc Natl Acad Sci U S A. 2016;113(18):5071–5076. doi: 10.1073/pnas.1604266113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haussecker D., Huang Y., Lau A., Parameswaran P., Fire A.Z., Kay M.A. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16(4):673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuscu C., Kumar P., Kiran M., Su Z., Malik A., Dutta A. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA. 2018;24(8):1093–1105. doi: 10.1261/rna.066126.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.C. Megel et al., “Plant RNases T2, but not Dicer-like proteins, are major players of tRNA-derived fragments biogenesis,” Nucleic Acids Res., vol. 47, no. 2, pp. 941–952, Jan. 2019, doi: 10.1093/nar/gky1156. [DOI] [PMC free article] [PubMed]
- 31.Y. Motorin, S. Muller, I. Behm-Ansmant, and C. Branlant, “Identification of Modified Residues in RNAs by Reverse Transcription-Based Methods,” Methods in Enzymology, vol. 425. Academic Press, pp. 21–53, Jan. 01, 2007, doi: 10.1016/S0076-6879(07)25002-5. [DOI] [PubMed]
- 32.Telonis A.G., Loher P., Honda S., Jing Y.i., Palazzo J., Kirino Y. Dissecting tRNA-derived fragment complexities using personalized transcriptomes reveals novel fragment classes and unexpected dependencies. Oncotarget. 2015;6(28):24797–24822. doi: 10.18632/oncotarget.4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Telonis A.G., Loher P., Kirino Y., Rigoutsos I. Consequential considerations when mapping tRNA fragments. BMC Bioinf. 2016;17:123. doi: 10.1186/s12859-016-0921-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.A. Hoffmann, J. Fallmann, E. Vilardo, M. Mörl, P. F. Stadler, and F. Amman, “Accurate mapping of tRNA reads,” Bioinformatics, vol. 34, no. 7, pp. 1116–1124, Apr. 2018, doi: 10.1093/bioinformatics/btx756. [DOI] [PubMed]
- 35.S. R. Selitsky and P. Sethupathy, “tDRmapper: challenges and solutions to mapping, naming, and quantifying tRNA-derived RNAs from human small RNA-sequencing data.,” BMC Bioinformatics, vol. 16, no. 1, p. 354, Nov. 2015, doi: 10.1186/s12859-015-0800-0. [DOI] [PMC free article] [PubMed]
- 36.Loher P., Telonis A.G., Rigoutsos I. MINTmap: fast and exhaustive profiling of nuclear and mitochondrial tRNA fragments from short RNA-seq data. Sci Rep. 2017;7(1):41184. doi: 10.1038/srep41184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng L.-L., Xu W.-L., Liu S., Sun W.-J., Li J.-H., Wu J. tRF2Cancer: a web server to detect tRNA-derived small RNA fragments (tRFs) and their expression in multiple cancers. Nucleic Acids Res. 2016;44(W1):W185–W193. doi: 10.1093/nar/gkw414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.A. Thompson et al., “tRex: A Web Portal for Exploration of tRNA-Derived Fragments in Arabidopsis thaliana,” Plant Cell Physiol., vol. 59, no. 1, pp. e1–e1, Jan. 2018, doi: 10.1093/pcp/pcx173. [DOI] [PubMed]
- 39.N. Gupta, A. Singh, S. Zahra, and S. Kumar, “PtRFdb: a database for plant transfer RNA-derived fragments,” Database, vol. 2018, no. 10, pp. 63–63, 2018, [Online]. Available: http://www.nipgr.res.in/PtRFdb/. [DOI] [PMC free article] [PubMed]
- 40.Y. Motorin and M. Helm, “TRNA stabilization by modified nucleotides,” Biochemistry, vol. 49, no. 24. American Chemical Society, pp. 4934–4944, Jun. 22, 2010, doi: 10.1021/bi100408z. [DOI] [PubMed]
- 41.El Yacoubi B., Bailly M., De Crécy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu Rev Genet. 2012;46(1):69–95. doi: 10.1146/annurev-genet-110711-155641. [DOI] [PubMed] [Google Scholar]
- 42.Lyons S.M., Fay M.M., Ivanov P. The role of RNA modifications in the regulation of tRNA cleavage. FEBS Lett. 2018;592(17):2828–2844. doi: 10.1002/feb2.2018.592.issue-1710.1002/1873-3468.13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.M. Pereira, D. R. Ribeiro, M. M. Pinheiro, M. Ferreira, S. Kellner, and A. R. Soares, “m 5 U54 tRNA hypomodification by lack of TRMT2A drives the generation of tRNA-derived small RNAs,” no. January, 2021, doi: 10.20944/preprints202101.0227.v1. [DOI] [PMC free article] [PubMed]
- 44.Lowe T.M., Eddy S.R. TRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1996;25(5):955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pan T. Modifications and functional genomics of human transfer RNA. Cell Res. 2018;28(4):395–404. doi: 10.1038/s41422-018-0013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sobala A., Hutvagner G. Small RNAs derived from the 5’ end of tRNA can inhibit protein translation in human cells. RNA Biol. 2013;10(4):553–563. doi: 10.4161/rna.24285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lalande S., Merret R., Salinas-Giegé T., Drouard L. Arabidopsis tRNA-derived fragments as potential modulators of translation. RNA Biol. 2020;17(8):1137–1148. doi: 10.1080/15476286.2020.1722514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ryvkin P., Leung Y.Y., Silverman I.M., Childress M., Valladares O., Dragomir I. HAMR: High-throughput annotation of modified ribonucleotides. RNA. 2013;19(12):1684–1692. doi: 10.1261/rna.036806.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Y. Wang et al., “The 2′-O-methyladenosine nucleoside modification gene OsTRM13 positively regulates salt stress tolerance in rice,” J. Exp. Bot., vol. 68, no. 7, pp. 1479–1491, Mar. 2017, doi: 10.1093/jxb/erx061. [DOI] [PMC free article] [PubMed]
- 50.Wang Y., Pang C., Li X., Hu Z., Lv Z., Zheng B.o. Identification of tRNA nucleoside modification genes critical for stress response and development in rice and Arabidopsis. BMC Plant Biol. 2017;17(1) doi: 10.1186/s12870-017-1206-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martinez G. tRNA-derived small RNAs: new players in genome protection against retrotransposons. RNA Biol. 2018;15(2):170–175. doi: 10.1080/15476286.2017.1403000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martinez G., Choudury S.G., Slotkin R.K. tRNA-derived small RNAs target transposable element transcripts. Nucleic Acids Res. 2017;45(9):5142–5152. doi: 10.1093/nar/gkx103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi J., Ko E.A., Sanders K.M., Chen Q., Zhou T. SPORTS1.0: a tool for annotating and profiling non-coding RNAs Optimized for rRNA- and tRNA-derived small RNAs. Genom Proteom Bioinform. 2018;16(2):144–151. doi: 10.1016/J.GPB.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schimmel P. The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat Rev Mol Cell Biol. 2017;19(1):45–58. doi: 10.1038/nrm.2017.77. [DOI] [PubMed] [Google Scholar]
- 55.Y. Cao, K. Liu, Y. Xiong, C. Zhao, and L. Liu, “Increased expression of fragmented tRNA promoted neuronal necrosis,” Cell Death Dis. 2021 129, vol. 12, no. 9, pp. 1–15, Aug. 2021, doi: 10.1038/s41419-021-04108-6. [DOI] [PMC free article] [PubMed]
- 56.G. Shin, H. J. Koo, M. Seo, S.-J. V. Lee, H. G. Nam, and G. Y. Jung, “Transfer RNA-derived fragments in aging Caenorhabditis elegans originate from abundant homologous gene copies,” Sci. Reports 2021 111, vol. 11, no. 1, pp. 1–9, Jun. 2021, doi: 10.1038/s41598-021-91724-z. [DOI] [PMC free article] [PubMed]
- 57.Dziublenski M., Roff A.N., Ishmael F.T. Ribonomic approaches to identify protein-mRNA and microRNA-mRNA interactions: Implications for drug design. Drug Dev Res. 2012;73(7):406–413. doi: 10.1002/ddr.21031. [DOI] [Google Scholar]
- 58.S. Sherry et al., “NCBI SRA Toolkit Technology for Next Generation Sequence Data,” Accessed: Jul. 22, 2021. [Online]. Available: http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?view=software.
- 59.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N. The sequence alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma X., Si F., Liu X., Luan W. PRMdb: a repository of predicted RNA modifications in plants. Plant Cell Physiol. 2020;61(6):1213–1222. doi: 10.1093/PCP/PCAA042. [DOI] [PubMed] [Google Scholar]
- 63.Cock P.J.A., Antao T., Chang J.T., Chapman B.A., Cox C.J., Dalke A. Biopython: freely available python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25(11):1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Will S., Joshi T., Hofacker I.L., Stadler P.F., Backofen R. LocARNA-P: accurate boundary prediction and improved detection of structural RNAs. RNA. 2012;18(5):900–914. doi: 10.1261/rna.029041.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bernhart S.H., Hofacker I.L., Will S., Gruber A.R., Stadler P.F. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinf. 2008;9(1):474. doi: 10.1186/1471-2105-9-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dai X., Zhuang Z., Zhao P.X. PsRNATarget: a plant small RNA target analysis server (2017 release) Nucleic Acids Res. 2018;46(W1):W49–W54. doi: 10.1093/nar/gky316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.U. Raudvere et al., “g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update),” Nucleic Acids Res., no. 1, 2019, doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed]
- 68.Shannon P. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Merico D., Isserlin R., Stueker O., Emili A., Bader G.D., Ravasi T. Enrichment Map: a network-based method for gene-set enrichment visualization and interpretation. PLoS ONE. 2010;5(11):e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.M. Kucera, R. Isserlin, A. Arkhangorodsky, and G. D. Bader, “AutoAnnotate: A Cytoscape app for summarizing networks with semantic annotations,” F1000Research, vol. 5, 2016, doi: 10.12688/F1000RESEARCH.9090.1. [DOI] [PMC free article] [PubMed]
- 71.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All pipeline scripts, codes, data generated, and analyzed for each of the species are freely available at our website (URL: http://nipgr.ac.in/tncRNA). The codes and usage are also available at https://github.com/skbinfo/tncRNA-Toolkit.