SUMMARY
Regulatory T cells (Treg cells) are critical mediators of self-tolerance, but they can also limit effective anti-tumor immunity. Although under homeostasis a small fraction of Treg cells in lymphoid organs express the putative checkpoint molecule Tim-3, this protein is expressed by a much larger proportion of tumor-infiltrating Treg cells. Using a mouse model that drives cell-type-specific inducible Tim-3 expression, we show that expression of Tim-3 by Treg cells is sufficient to drive Treg cells to a more effector-like phenotype, resulting in increases in suppressive activity, effector T cell exhaustion, and tumor growth. We also show that T-reg-cell-specific inducible deletion of Tim-3 enhances anti-tumor immunity. Enhancement of Treg cell function by Tim-3 is strongly correlated with increased expression of interleukin-10 (IL-10) and a shift to a more glycolytic metabolic phenotype. Our data demonstrate that Tim-3+ Treg cells may be a relevant therapeutic target cell type for the treatment of cancer.
In brief
Regulatory T cells (Treg cells) limit the immune response to tumors, and tumor-infiltrating Treg cells are especially suppressive. However, the mechanisms underlying enhanced Treg cell function are poorly understood. Banerjee et al. show that Tim-3 expression is linked to increased Treg cell suppressive activity, possibly through the cytokine IL-10, in mouse models and people with cancer.
Graphical Abstract
INTRODUCTION
Immune checkpoint blockade therapies like those targeting the PD-1/PD-L1 axis have resulted in dramatic gains in the treatment of some tumors, but in most settings, only 20%–30% of people respond (Ferris et al., 2016; Hamid et al., 2013; Hargadon et al., 2018; Ribas and Wolchok, 2018). The mechanisms responsible for this resistance are still not clear. One barrier to successful immunotherapy can be the presence within the tumor microenvironment of significant numbers of regulatory T cells (Treg cells) (Tanaka and Sakaguchi, 2017). The glycoprotein T cell (or transmembrane) immunoglobulin and mucin domain 3 (Tim-3) (HAVCR2) has attracted significant attention as a potential negative regulator of T cell responses, including in tumors (Anderson, 2014; Du et al., 2017). Many groups have developed Tim-3 monoclonal antibodies (mAbs), with the goal of using these antibodies for immunotherapy of solid tumors, and early-stage clinical trials are now underway (Acharya et al., 2020). This effort has mainly focused on the role of Tim-3 in regulating CD8+ cytotoxic (CTL) T cells, although Tim-3 is also constitutively expressed by several other immune cell types, including mast cells, dendritic cells, and some natural killer (NK) cells (Banerjee and Kane, 2018). At least four ligands have been described to bind to Tim-3: galectin-9; high-mobility group box 1 protein (HMGB1); phosphatidylserine; and carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1), with virtually no studies directly comparing the effects of these different ligands. Moreover, these ligands are also known to bind other receptors in addition to Tim-3 (Segawa and Nagata, 2015; Su et al., 2011; Venereau et al., 2016; Wu et al., 2014; Yang et al., 2021).
Tim-3 is also expressed by a small subset (2%–5%) of circulating Treg cells in the secondary lymphoid organs and peripheral blood. Interestingly, the proportion of Tim-3+ Treg cells is significantly higher (40%–60%) among tumor-infiltrating Treg cells (Gao et al., 2012; Jie et al., 2013; Liu et al., 2018b; Sakuishi et al., 2013; Shen et al., 2016). These studies showed that Tim3-expressing Treg cells, in contrast to paired Tim-3-negative Treg cells, express an array of genes normally associated with effector T cell function. Along with this effector Treg cell (eTreg cell) phenotype, Tim-3+ Treg cells also possess enhanced in vitro suppressive capacity. Because modulation of Tim-3 is being explored as a possible therapeutic strategy, it is important to understand the role of Tim-3 on this substantial population of Treg cells. Indeed, Tim-3 appears to identify a particularly functional Treg cell subset and could act as a signaling mediator, enforcing unique activity of the Treg cell expressing this protein (Gao et al., 2012; Gautron et al., 2014; Jie et al., 2013; Liu et al., 2018b; Sakuishi et al., 2013). Elimination or impairment of Tim-3+ Treg cells may represent an important mechanism by which mAbs targeting Tim-3 enhance anti-tumor immunity (Liu et al., 2018a). However, it is still not known whether Tim-3 expression or signaling themselves modify the phenotype or function of Treg cells on which it is expressed or whether its expression merely correlates with altered Treg cell phenotype and function.
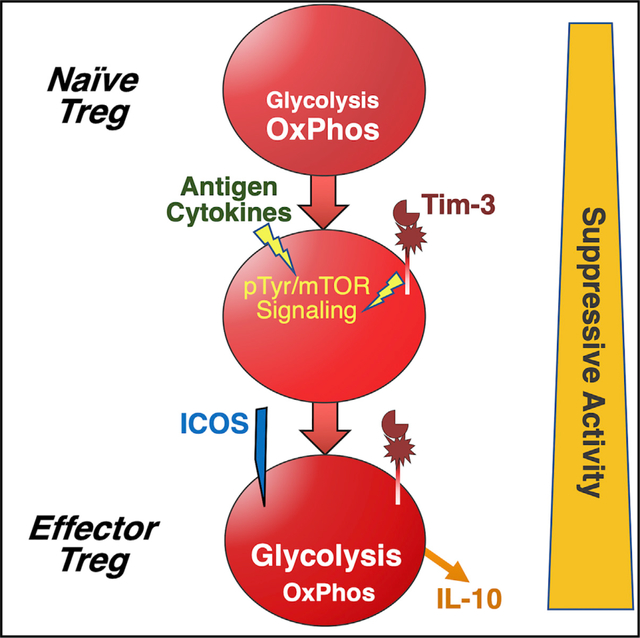
Using a mouse model that allows us to drive Tim-3 expression in a Cre-inducible fashion, we have now found that expression of Tim-3 by Treg cells is sufficient to push a significant proportion of Treg cells toward an effector-like Treg cell state, as revealed by surface protein expression and gene expression. Among the phenotypic changes we observe are enhanced gene and protein expression of interleukin-10 (IL-10) and a dramatic shift in cellular metabolism, away from oxidative phosphorylation and toward glycolysis. These changes were also accompanied by decreased signaling through the Akt/mechanistic target of rapamycin (mTOR) signaling pathway and increased signaling through the ERK mitogen-activated protein kinase (MAPK) pathway. At a functional level, Treg cells with enforced Tim-3 expression display enhanced in vitro suppressive activity. Even more striking, transplanted tumors grow more rapidly in mice with Treg-cell-specific expression of Tim-3. Finally, we generated an inducible knockout (KO) model to study the requirement for Tim-3 and show that specific elimination of Tim-3 from Treg cells results in slower tumor growth and decreased CD8+ T cell exhaustion in the tumor microenvironment. Together, our data demonstrate that Tim-3 can promote acquisition of at least part of the eTreg cell phenotype observed in Tim-3+ tumor-infiltrating Treg cells. These findings have significant implications for the further development of immunotherapy approaches targeting Tim-3 and/or Treg cells in the tumor microenvironment.
RESULTS
Differential expression of Tim-3 on Treg cells and CD8+ T cells in lymphoid compartments versus tumors
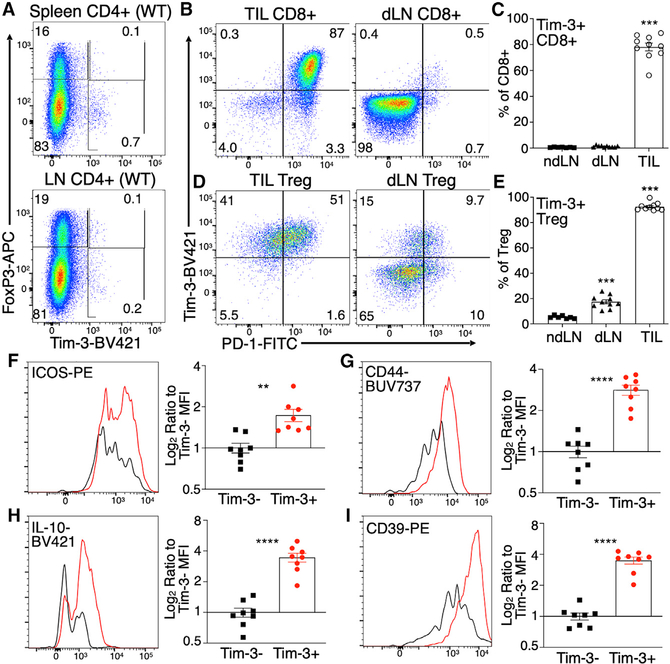
Previous reports demonstrated a large increase in the proportion of Tim-3+ Treg cells in the tumor microenvironment, relative to Treg cells in circulation. Consistent with those findings, we observe very few Tim-3+ T cells among circulating CD4+ T cells, whether expressing FoxP3 or not, from wild-type (WT) C57BL/6 mice under steady-state conditions (Figure 1A), in the spleen (top) or lymph nodes (bottom). To confirm and extend this finding, we implanted syngeneic MC38 tumors into WT C57BL/6 mice and examined the phenotype of tumor-infiltrating Treg cells and effector T cells, as well as T cells from both tumor-draining and non-draining lymph nodes. As expected, we observed a large proportion of Tim-3+ CD8+ T cells in tumors, with very few such cells in either draining or non-draining lymph nodes (Figures 1B and 1C). Most tumor CD8+Tim-3+ cells were also PD-1+, consistent with many previous reports. As we and others had shown previously, there was a significant proportion of Tim3-expressing Treg cells among the tumor-infiltrating lymphocytes (TIL), which was smaller in draining lymph nodes and smaller still in non-draining lymph nodes (Figures 1D and 1E). The presence of Tim-3+ Treg cells generally tracked with the appearance of CD8+ effector T cells bearing an exhausted (PD-1hiTim-3+) phenotype. Interestingly, we did observe a population of PD-1intTim3+ Treg cells in the draining lymph nodes (Figures 1D and 1E), which was not observed among CD8+ T cells (Figures 1B and 1C). At least one previous study reported no increase in Tim-3+ Treg cells in tumor-draining lymph nodes (Sakuishi et al., 2013), although we have reported PD-1−Tim-3+ Treg cells infiltrating the tumors of patients with head and neck cancer (Liu et al., 2018b). Although their significance is not clear, these cells may represent a precursor of the PD-1+Tim-3+ Treg cells. The enriched presence of these Tim3-expressing Treg cells in tumors raises the question of whether and how Tim-3 expression directly influences the function of Treg cells.
Figure 1. Differential expression of Tim-3 on Treg cells in lymphoid compartments and tumors of WT mice.
(A) Minimal Tim-3 expression on Treg cells and other CD4+ T cells from spleen and lymph nodes of WT C57BL/6 mice.
(B–E) Expression of Tim-3 on CD8+ T cells (B and C) and FoxP3+CD25+ Treg cells (D and E) 14 days after implantation of MC38 tumors into C57BL/6 mice. TIL, tumor-infiltrating lymphocytes; dLN, tumor-draining lymph node. Mean ± SEM; one-way ANOVA.
(F–I) Expression of eTreg-cell-associated proteins on Tim-3− versus Tim-3+ TIL Treg cells.
Dot plots show representative data from individual mice from 8 to 10 mice analyzed over two independent experiments. Compiled data from all mice are shown in the column plots. Mean ± SEM; two-tailed t test.
We and others have reported some phenotypic changes associated with Tim-3 expression on Treg cells. We looked at this more completely, finding an array of markers whose expression is changed on tumor Tim3-expressing Treg cells versus tumor Treg cells that do not express Tim-3. We found that Tim-3+ TIL Treg cells express greater levels of surface inducible T cell costimulator (ICOS) (Figure 1F) and CD44 (Figure 1G), which are both markers of the more-activated phenotype of eTreg cells (Teh et al., 2015). This phenotype is also associated with enhanced Treg cell function; indeed, we noted that Tim-3+ TIL Treg cells also expressed higher levels of the suppressive cytokine IL-10 (Figure 1H) and the ectonucleotidase CD39 (Figure 1I). Thus, expression of endogenous Tim-3 by Treg cells is associated with an eTreg cell phenotype in tumors, leading us to ask whether expression of Tim-3 might be sufficient to induce or reinforce such a phenotype.
Enforced expression of Tim-3 leads to an increase in Treg cell frequency in lymphoid compartments, with no detectable changes in conventional T cells
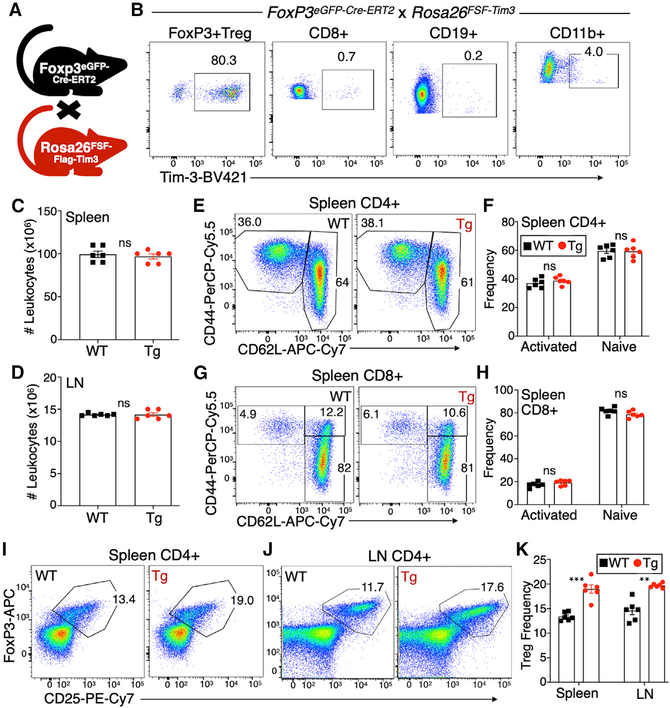
To probe the effects of Tim-3 on Treg cells in vivo, we employed a mouse model that we had previously described (Avery et al., 2018), which carries a flox-stop-flox cassette followed by a full-length murine Tim-3 cDNA, knocked into the Rosa26 locus (referred to as “FSF-Tim-3” mice). Upon expression of the Cre recombinase, cells carrying this allele express Tim-3 for the remaining life of that cell. To drive restricted but also temporally regulated expression of Tim-3, we bred the FSF-Tim-3 mice to mice with a knockin of a tamoxifen-inducible Cre (Cre-ERT2) fused to EGFP in the Foxp3 locus (Levine et al., 2014; Figure 2A). After tamoxifen administration (1 mg/mouse intraperitoneally [i.p.] on 5 successive days), females homozygous for the Cre and transgenic males had 80%–95% Tim-3+ Treg cells (Figure 2B). As expected, we did not observe nonspecific induction of Tim-3 on non-Treg cells, among a number of immune cell types tested (Figure 2B). In the results that follow below, Cre-only control animals will also be referred to as “WT” and those carrying both the Cre and FSF-Tim-3 allele as “transgenic” (Tg).
Figure 2. Increased Tim-3+ Treg cell frequency in peripheral lymphoid organs of FoxP3EGFP-Cre-ERT2 × FSF-Tim-3 mice.
(A) Depiction of the cross used to generate FoxP3EGFP-Cre-ERT2 × FSF-Tim-3 mice.
(B) Tim-3 transgene expression is restricted to CD4+ Treg cells when driven by the FoxP3-specific, tamoxifen-inducible Cre. Mice (both WT and Tg) were dosed on 5 consecutive days with tamoxifen, followed by 1 day of rest, before analysis. Shown is the analysis of splenic lymphocytes from representative animals.
(C and D) No change in the total number of splenocytes or lymph node cells isolated from WT versus Tim-3 Tg animals after tamoxifen treatment.
(E–H) Antigen-experienced and naive conventional CD4+ (E and F) and CD8+ (G and H) T cells from spleens of WT and Tim-3 Tg animals, based on CD44 and CD62L expression.
(I–K) Proportions of FoxP3+CD25+ Treg cells among CD4+ T cells in the spleen (I) and lymph nodes (J) of WT and Tim-3 Tg animals, quantified in (K).
Graphs show individual animals and mean ± SEM; two-tailed t test.
Having established that the double transgenic system drives robust, inducible, expression of Tim-3 specifically on Treg cells, we proceeded to perform detailed phenotypic and functional characterization of these animals and their T cell populations. Enforced expression of Tim-3 on Treg cells did not lead to any detectable changes in immune homeostasis under the conditions examined (standard specific-pathogen-free (SPF) housing; out to 6–8 weeks after tamoxifen administration). Specifically, we noted no changes in the total number of leukocytes recovered from the spleen or peripheral lymph nodes of these mice (Figures 2C and 2D). The proportions of conventional CD4+ and CD8+ T cells were also unchanged in either compartment after Tim-3 induction on Treg cells (Figures S2A–S2D). In addition, the frequencies of previously activated (CD44+CD62LLo) and naive (CD44−CD62LHi) CD4+ and CD8+ T cells were unchanged by Treg-cell-specific expression of Tim-3, in both the spleen (Figures 2E–2H) and lymph nodes (Figures S2E–S2H).
We next began to assess the Treg cells in these animals. Treg cells were identified by intracellular staining for FoxP3, along with surface staining for CD25, the alpha chain of the IL-2 receptor. We observed moderately higher proportions of Treg cells in spleen (Figure 2I) and peripheral lymph nodes (Figure 2J) of WT versus Tim-3 Tg animals. Analysis of multiple animals revealed a slightly higher, and statistically significant, frequency of FoxP3+ Treg cells in both spleen and lymph nodes 7 days after completion of tamoxifen (Figure 2K). Enforced expression of Tim-3 on Treg cells may therefore drive a modest expansion of these cells, at least in the short term.
Tim-3 Tg Treg cells acquire an effector-like Treg cell phenotype
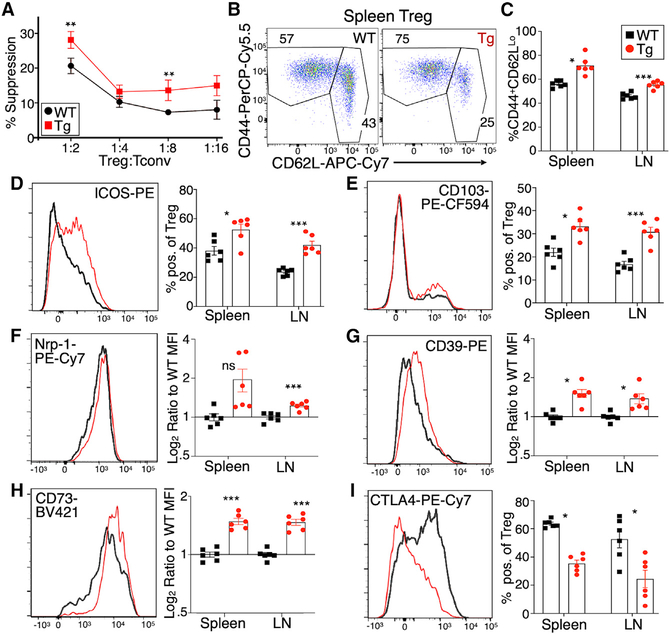
As discussed above, previous studies associated the acquisition of Tim-3 expression with enhanced Treg cell suppressive function. We tested the in vitro suppressive capacity of Tim-3-positive versus Tim-3-negative Treg cells, using a standard in vitro suppression assay. FoxP3+ cells were sorted (based on GFP expression) from FoxP3EGFP-Cre-ERT2 mice, with or without the FSF-Tim3 transgene. Treg cells from the Tim-3 transgenic mice were more active in this assay, i.e., Tim-3 Tg Treg cells suppressed the proliferation of conventional CD8 cells to a greater extent than WT Treg cells did (Figure 3A).
Figure 3. Enhanced function and effector-like phenotype of Tim-3 Tg Treg cells.
(A) In vitro suppression assay conducted with WT conventional CD8+ T cells (responders) and either WT or Tim-3 Tg Treg cells as suppressors. Responder cells were labeled with CellTrace Violet and stimulated with anti-CD3 mAb, and proliferation was assessed by flow cytometry after 72 h.
(B and C) Proportions of naive (CD44−CD62LHi) and activated (CD44+CD62LLo) Treg cells in WT and Tim-3 Tg mice.
(D–I) Flow cytometry staining for multiple surface markers of Treg cell differentiation and function. (D) ICOS, (E) CD103, (F) neuropilin 1 (Nrp1), (G) CD39, (H) CD73, and (I) CTLA-4.
Flow cytometry histograms show representative staining in splenic Treg cells. Graphs show individual animals and mean ± SEM; two-tailed t test, for both spleen and lymph node.
Enhanced suppressive activity of Treg cells under some conditions is associated with acquisition of a so-called eTreg cell phenotype (Cretney et al., 2013; Teh et al., 2015). This is typified by the enhanced expression of CD44 and ICOS, among other markers (Cretney et al., 2013; Smigiel et al., 2014). Tim-3 Tg Treg cells from both spleen and lymph nodes displayed higher baseline activation, based on increased expression of CD44 and decreased expression of CD62L (Figures 3B and 3C). We next analyzed other cell-surface proteins associated with Treg cell differentiation and function. One of the most consistently increased markers was the costimulatory molecule ICOS (Figures 3D and S3A). Induction of Tim-3 expression also altered other phenotypic aspects of Treg cells, with increased expression of markers like CD103 (Figure 3E), Nrp-1 (Figure 3F), and the ectonucleotidases CD39 and CD73 (Figures 3G and 3H), although these changes were lower in magnitude than for ICOS. Similar changes were seen on lymph node Treg cells (Figures 3 and S3A). By contrast, there were minimal or no differences in CD25, TIGIT, and Helios (Figures S3B and S3C). Surprisingly, even though we observed increased function and activation of Tim-3 Tg Treg cells, we noted a significant downregulation of CTLA-4 expression on both splenic and lymph node Treg cells (Figures 3I and S3A).
Effects of Tim-3 expression on Treg cell gene expression
To further explore the nature of the effects of Tim-3 induction on Treg cells, we performed RNA sequencing of Treg cells from WT (FoxP3EGFP-Cre-ERT2 alone) and Tim-3+ transgenic (FoxP3-Cre × FSF-Tim3) Treg cells. In addition, we sorted both GFPHi (FoxP3+) and GFPLo (FoxP3−/Lo) cells, and both populations were compared to WT Treg cells (Figure 4A, inset). The GFPLo population was included because we had noted that the proportion of such cells (Tim-3 Tg+FoxP3-GFPLo) increased over time, after induction of Tim-3 expression (not shown). We therefore suspected that these cells may represent a terminally differentiated subset of Treg cells driven to such a state by constitutive Tim-3 expression and/or signaling. RNA was made from the sorted populations and prepared for sequencing. Tim-3 induction led to major changes in the Treg cells’ transcript signature, as illustrated in the volcano plot in Figure 4A. Based on Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of the top 500 differentially expressed genes in the Tim-3+FoxP3hi population, major housekeeping processes in the Treg cells seem to be altered due to expression of Tim-3, including translation (ribosomal genes) and metabolism (Figures 4B, S4, and S5). Relative expression of selected genes from these modules, as well as cytokines or receptors and transcription factors, are shown in Figure 4C, where we also compared their expression in the Tim-3+GFPLo cells (see also Figures S4 and S5). Of note, the message encoding the suppressive cytokine IL-10 is very highly upregulated in the Tim-3 Tg Treg cells (>1,000-fold; p = 4.6 × 10−13; see also the volcano plot in Figure 4A). In addition, there were broad decreases in ribosomal and oxidative phosphorylation and mitochondrial genes, with a corresponding increase in genes involved in glycolysis. Consistent with acquisition of an eTreg-cell-like phenotype, we noted increased expression of transcription factors Eomes, Myc, and T-bet (Tbx21) in Tim-3 Tg Treg cells (Figure 4C).
Figure 4. Tim-3 expression drives widespread changes in Treg cell gene expression.
(A) Inset: populations of Treg cells sorted for RNA-seq analysis. Volcano plot of RNA-seq data comparing Tim-3+FoxP3hi cells from Tg animals to FoxP3+ cells from WT (Cre-only) animals is shown. Only comparisons with a p < 0.05 are shown.
(B) KEGG pathway analysis of the top 500 differentially expressed genes by fold change and false discovery rate (FDR) (either up or down), comparing FoxP3HiTim-3+ cells to WT FoxP3+ cells.
(C) Differential expression of selected genes in the indicated pathways, comparing either Tim-3+GFPHi (i.e., FoxP3Hi) or Tim-3+GFPLo cells to WT Treg cells from FoxP3-Cre-GFP mice. Mice were injected with tamoxifen at 6–8 weeks of age and analyzed between 3 and 6 weeks of age.
(D) KEGG analysis of TIM-3+ versus TIM-3− TIL Treg cells from patients with HNC, based on scRNA-seq, using the top 100 upregulated and top 100 downregulated genes.
(E) Heatmap showing relative expression of selected genes in TIM-3+ versus TIM-3− human Treg cells from the scRNA-seq dataset.
(F) Pseudotime analysis of scRNA-seq data from TIL Treg cells of patients with HNC, showing that increased HAVCR2 and IL-10 expression are associated with differentiation (arrow trajectory) of Treg cells.
(G) UMAP analysis of Treg cells from people with HNC, for HAVCR2 and IL-10.
(H) Top: definition of clusters from UMAP analysis shown in (G); bottom: regression analysis of HAVCR2 versus IL-10 expression in the various Treg cell clusters.
We previously reported that about 50% of human tumor-infiltrating Treg cells express TIM-3 and that these cells possess enhanced suppressive function, compared to Treg cells not expressing TIM-3 (Jie et al., 2013; Liu et al., 2018b). We wanted to determine whether the pathways identified as driven by Tim-3 expression by RNA sequencing (RNA-seq) analysis of our Tg mouse model would also be observed in human TIL Treg cells. Recently, we published a comprehensive study of leukocytes isolated from blood and tumors of people with head and neck cancer (HNC) (Cillo et al., 2020). Analysis of an updated scRNA-seq dataset, using the top 100 differentially expressed genes (Figure S6), revealed overlap with the mouse Tim-3 Tg Treg cell analysis. Of note, the ribosome pathway and glycolysis were two of the most significantly altered pathways (Figure 4D). Looking at individual genes associated with eTreg cells, we noted upregulation of messages encoding IL-10 and ICOS, among other genes (Figure 4E). One difference from Tim-3 Tg mouse Treg cells was the upregulation of CTLA-4 in TIM-3+ human TIL Treg cells, which contrasts with the lower expression in Tim-3 Tg mouse Treg cells. Given the abundance of IL-10 (both message and protein) in mouse Tim-3 Tg Treg cells, we examined the expression of IL-10 in human TIL Treg cells in more detail. We also wanted to assess whether TIM-3+ human TIL Treg cells represent a more differentiated state, as suggested by the analysis of Tim-3 Tg Treg cells over time in the mouse model. We performed pseudotime analysis of FOXP3-expressing cells from the single-cell RNA sequencing (scRNA-seq) dataset. As shown in Figure 4F, pseudotime progression of TIL Treg cells (indicated by the diagonal arrow) was associated with increased expression of both HAVCR2 (the gene encoding TIM-3) and IL-10. Consistent with the enrichment of Treg cells in tumors, most Treg cells in the dataset were of tumor origin, with a minor fraction from the blood (Figure 4G). Overlaying the expression of Tim-3 onto the Treg cell UMAP, we again observed a significant number of tumor-infiltrating, but not peripheral blood lymphocyte (PBL), Treg cells expressing HAVCR2 (Figure 4G), which again appeared to correlate with expression of IL-10. This same UMAP analysis revealed the presence of seven distinct clusters of Treg cells in the tumors, based on gene expression signatures (Figure 4H). When we performed linear regression analysis, we indeed found a significant correlation between HAVCR2 and IL-10 expression among these clusters (Figure 4I).
Tim-3 expression drives specific changes to Treg cell metabolism and signaling
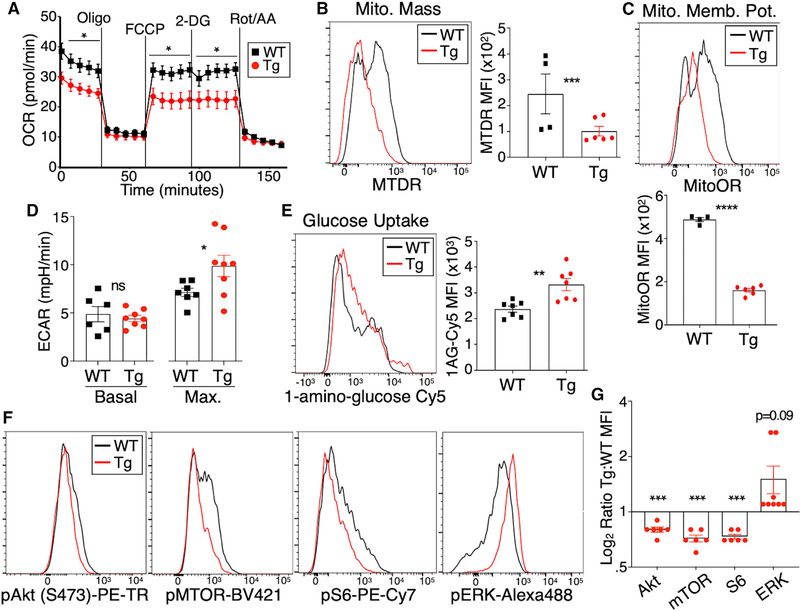
Circulating Treg cells normally derive most of their energy from oxidative phosphorylation (including lipid oxidation), with a lesser role for glycolysis, although alterations in these pathways may accompany tissue-specific pressures put on Treg cells in various compartments (Shi and Chi, 2019). RNA-seq analysis discussed above (Figures 4 and S5) suggested a shift of Treg cell metabolic pathways away from oxidative phosphorylation and toward glycolysis in Tim3-expressing Treg cells, so we examined these pathways more directly in Tim-3 Tg versus WT Treg cells. We performed Seahorse metabolic flux assays to assess oxygen consumption (a measure of oxidative phosphorylation) and extracellular acidification (a measure of glycolysis). Consistent with the gene expression data, Tim-3 Tg Treg cells were less efficient at performing oxidative phosphorylation (Figure 5A). To assess this via independent assays, we performed flow cytometry with dyes that reveal the number and functionality of mitochondria in intact cells. Overall mitochondrial mass (Figure 5B) and membrane potential (Figure 5C) were both significantly decreased in Tim-3 Tg Treg cells. Next, we performed a glucose “stress test,” suspending sorted Treg cells in glucose-free media before injecting glucose during metabolic flux analysis in the Seahorse. Using this approach, we found that Tim-3 Tg Treg cells achieved a higher maximal glycolytic capacity than WT Treg cells (Figure 5D). We also directly examined uptake of glucose in the Treg cell populations by incubation with a fluorescent glucose analog. Consistent with the Seahorse results, we found that Tim-3 Tg Treg cells took up more glucose than their WT counterparts (Figure 5E).
Figure 5. Ectopic expression of Treg cells drives shifts in Treg cell metabolism and signaling.
Tamoxifen was administered to the indicated mice on 5 successive days, followed by 2 days of rest, after which Treg cells were analyzed.
(A) Oxygen consumption rate (OCR) of cells over time, measured in Seahorse, without any prior stimulation.
(B) Flow cytometry staining of gated Treg cells with MitoTracker Deep Red (MTDR), which reflects mitochondrial mass.
(C) Flow cytometry staining of gated Treg cells with MitoTracker Orange CMTMRos, which is proportional to mitochondrial membrane potential.
(D) Glucose stress test Seahorse assay showing ECAR of sorted Treg cells assessed in glucose-free media, before (“basal”) or after (“max”) injection of glucose.
(E) Uptake of the glucose analog 1-amino-glucose-Cy5, assayed by flow cytometry of gated Treg cells.
(F and G) Flow cytometry analysis of signaling molecules in gated Treg cells from WT and Tim-3 Tg mice. Single-cell suspensions were obtained from spleen and immediately fixed in paraformaldehyde (PFA), followed by permeabilization and staining for the indicated targets. Quantitation in (G) represents pooled data from multiple experiments, normalized to WT values.
Individual data points in (A) were obtained from five biological replicates (from five mice), obtained over the course of three experiments. Other graphs show individual mice pooled from 2 to 3 experiments. Graphs show mean ± SEM, and statistical significance is based on two-tailed t test.
Our previous studies suggested that Tim-3 can enhance antigen receptor activation of Akt and mTOR signaling (Avery et al., 2018; Lee et al., 2011), although Tim-3 alone could activate ERK MAPK signaling, at least under some circumstances (Kataoka et al., 2021). These findings, and the above-described effects of Tim-3 on metabolism and gene expression, led us to examine the status of signaling pathways in Tim-3 Tg Treg cells. In contrast to our previous findings with Tim-3 Tg conventional T cells, which had enhanced activation of Akt/mTOR signaling (Avery et al., 2018), Tim-3 Tg Treg cells had lower levels of pAkt, pMTOR, and pS6, compared with WT Treg cells (Figures 5F and 5G). In contrast, we did note a modest, but not statistically significant, increase in the levels of pERK in the Tim-3 Tg Treg cells (Figures 5F and 5G), which was consistent with our previous findings in Tim-3 Tg conventional T cells (Kataoka et al., 2021). Altogether, our data suggest that expression of Tim-3 drives a metabolic phenotype consistent with an eTreg cell phenotype.
Ectopic Tim-3 expression on Treg cells leads to increased tumor growth and CD8+ TIL exhaustion
To define the in vivo functional competence of Treg cells with enforced Tim-3 expression, we implanted tumors into control (FoxP3EGFP-Cre-ERT2) or Tim-3 Tg mice. We chose two tumor models for their different kinetics and relative immunogenicity—B16 melanoma, which is relatively aggressive, and MC38 carcinoma, which is both more immunogenic and more responsive to checkpoint blockade (Woo et al., 2012). B16 tumors grew with typical kinetics in WT (FoxP3EGFP-Cre-ERT2 only) mice (Figures 6A and 6B), and this was not affected by administration of vehicle alone (sunflower oil). By contrast, even this aggressive tumor grew more quickly in Tim-3 Tg animals that were given tamoxifen to induce Cre-mediated recombination, and Tim-3 expression, in Treg cells (Figures 6A and 6B). We also observed an acceleration of tumor growth with a second syngeneic tumor line, MC38, which grows with somewhat slower kinetics than B16 tumors (Figures 6C and 6D). These effects of Treg-cell-specific Tim-3 expression in vivo correspond well with the shift toward a more effector-like Treg cell phenotype and enhanced suppressive function.
Figure 6. Enforced expression of Tim-3 on Treg cells drives increased tumor growth and effector T cell exhaustion.
(A–D) Growth of transplanted syngeneic tumors in WT (Cre only) versus Tim-3 Tg mice. The indicated mice were treated with tamoxifen or vehicle on 5 successive days, followed by 1 day of rest and injection of B16 (A and B) or MC38 (C and D) tumors. Growth curves for individual animals are shown (A and C), as well as average tumor growth (B and D).
(E) Expression of PD-1, Tim-3, and IL-10 (mapped in pseudo-color) in CD8+ TIL T cells of WT versus Treg-cell-specific Tim-3 Tg mice.
(F) Expression of FoxP3 and CD25 (to identify Treg cells), as well as IL-10, among CD4+ TIL of WT versus Treg-cell-specific Tim-3 Tg mice. Representative of two experiments of four mice each, with 6-week-old mice, are shown.
(G and H) Staining of IL-10 and pSTAT3 in CD8+ (G) and Treg cell (H) TIL.
All graphs show mean ± SEM, and statistical significance is based on two-tailed t test.
Treg cells employ multiple mechanisms to effect suppression of conventional T cell and myeloid cell function. One of the most critical factors in this regard, including in the tumor microenvironment, is the regulatory cytokine IL-10 (Chen et al., 2016; Yano et al., 2019). Previous studies suggested that expression of Tim-3 is linked to increased IL-10 expression on both effector and Treg cells (Gorman and Colgan, 2018; Gupta et al., 2012; Jin et al., 2010; Sakuishi et al., 2013; Zhu et al., 2015). We therefore examined the expression of IL-10 in CD8+ and FoxP3+ TIL isolated from MC38 tumors implanted into control or Tim-3 Tg mice. When we plotted the expression of IL-10 heatmapped onto flow cytometry plots for Tim-3 and PD-1, which identify the most-exhausted CD8+ T cells in the tumor microenvironment, we found that Tim-3+PD-1+ effector T cells were enriched for IL-10 expression when Tim-3 was induced on Treg cells (Figure 6E). In addition, there was a higher proportion of these PD-1+Tim-3+CD8+ T cells in tumors implanted into the Treg cell Tim-3 Tg animals. We also examined IL-10 expression in Treg cells from these animals. As shown in Figure 6F, Treg cells (FoxP3+CD25+) from Tim-3 Tg animals showed higher expression of IL-10, compared with Cre-only control animals. This is quantified in Figures 6G and 6H. We also observed statistically significant increases in both IL-10 and pSTAT3, a target of IL-10 receptor signaling, among CD8+ TIL T cells in Tim-3 Tg animals, indicative of an effect of the Tg Treg cells on effector T cells (Figure 6G). Consistent with data discussed above, we noted even larger increases in both IL-10 and pSTAT3 among TIL Treg cells from tumors in Tim-3 Tg mice (Figure 6H). Thus, constitutive expression of Tim-3 on Treg cells leads to both increased IL-10 production and sensitivity to IL-10 by Treg cells themselves and terminally exhausted CD8+ T cells.
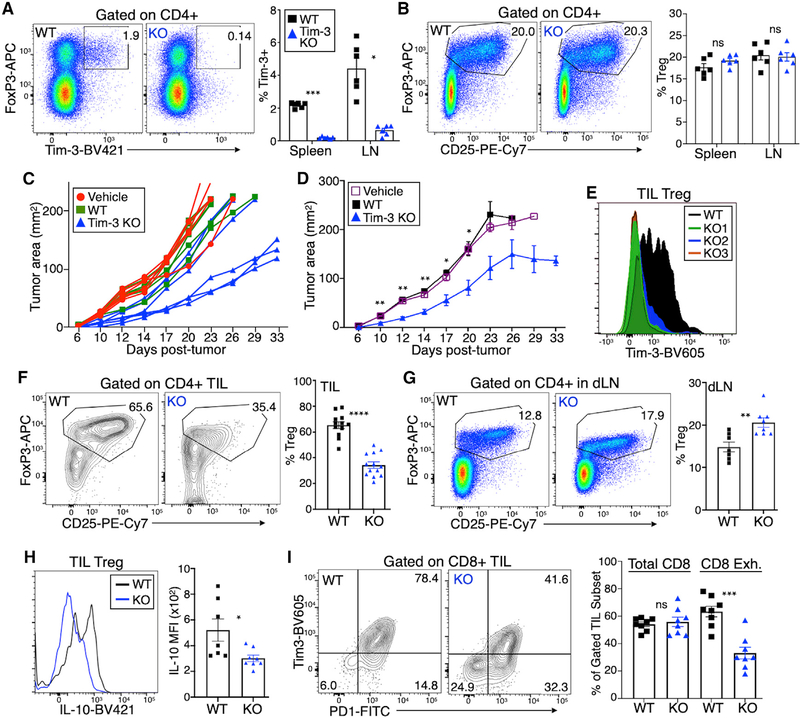
Effects of Treg-cell-specific Tim-3 deficiency on tumor growth
We next addressed whether expression of Tim-3 is necessary for establishment of a more suppressive environment in solid tumors. We generated a mouse line containing LoxP sites flanking exon 4 of the Havcr2 gene, which encodes Tim-3. We bred these mice to the FoxP3EGFP-Cre-ERT2 strain used above and treated Cre-only or Cre × floxed mice with tamoxifen. This treatment eliminated virtually all of the (relatively low) expression of Tim-3 on circulating Treg cells at steady state (Figures 7A and S7A), while maintaining Tim-3 expression in other cell lineages, including CD11b+ monocytes, about 20% of which express Tim-3 (Figure S7B). As expected, the overall proportion of peripheral Treg cells was not affected by inducible KO (iKO) of Tim-3, at least at this early time point (Figure 7B), nor were conventional CD4+ and CD8+ T cells (Figure S7C), B cells, or monocytes (Figure S7D). We implanted MC38 tumors into these mice to assess the effects of Treg-cell-specific Tim-3 deficiency on tumor growth. As shown in Figure 7C, tumors grew more slowly overall in the Tim-3 Treg cell KO mice, compared with WT (Cre-only) animals treated with tamoxifen or Cre × Tim-3 floxed animals treated with vehicle alone. Average tumor growth is shown in Figure 7D. We next examined the status of TIL at day 30 after tumor implantation. Although there was ample expression of Tim-3 on Treg cells from control mice, TIL Treg cells from Cre × Tim-3 floxed mice treated with tamoxifen expressed little if any Tim-3 (Figure 7E). The absence of Tim-3 expression on Treg cells was also associated with a decreased frequency of TIL Treg cells (Figure 7F). This may be specific to the tumor, as there was a small increase in Treg cells within tumor-draining lymph nodes (Figure 7G). In addition, TIL Treg cells from the Tim-3 iKO mice expressed less IL-10 than WT Treg cells (Figure 7H), suggesting that Tim-3 is required for the enhanced IL-10 expression of TIL Treg cells. Finally, we found that the absence of Tim-3 expression on Treg cells resulted in decreased CD8+ T cell exhaustion, as determined by lower levels of PD-1 and Tim-3 expression (Figure 7I).
Figure 7. Inducible knockout of Tim-3 in Treg cells leads to decreased tumor growth and less severe T cell exhaustion.
Tamoxifen was administered on 5 successive days to the indicated mice, followed by 1 day of rest.
(A) Analysis of splenic T cells revealed efficient deletion of Tim-3 from Treg cells.
(B) Normal overall proportion of Treg cells in lymph nodes of Tim-3 inducible KO (iKO) mice.
(C and D) Growth of MC38 tumors implanted into individual WT or Tim-3 KO mice after tamoxifen treatment (C) and average tumor growth (D).
(E) Levels of Tim-3 on TIL Treg cells from representative animals.
(F and G) Diminished proportion of Treg cells in tumors of mice with Tim-3 iKO (F); increased proportion of Treg cells in tumor-draining lymph nodes of the same animals (G).
(H) Reduced expression of IL-10 in TIL Treg cells from Tim-3 iKO mice.
(I) Reduced exhaustion phenotype (PD1+Tim-3+) of TIL CD8+ T cells, with unchanged percentage of total CD8+ T cells among all TIL T cells.
Statistical significance is based on two-tailed t test, and all graphs show mean ± SEM; symbols represent individual mice pooled from 2 to 3 experiments each.
DISCUSSION
Tim-3 has attracted significant attention as a possible target for immune checkpoint blockade therapy, based on its expression on terminally exhausted T cells. However, Tim-3 is also expressed by at least 50% of Treg cells in the tumor microenvironment, and this expression is associated with more potent Treg cell suppressive activity. Here, we set out to address whether Tim-3 itself can drive phenotypic and functional changes in Treg cells. We found that transgenic expression of Tim-3 on Treg cells is indeed sufficient to increase the proportion of circulating Treg cells with a cell-surface phenotype reminiscent of eTreg cells. These cells are also more suppressive, which also resulted in enhanced growth of syngeneic tumors. Consistent with previous studies on eTreg cells (Cretney et al., 2013; Teh et al., 2015), Tim-3 Tg Treg cells displayed higher expression of cell-surface protein like ICOS and CD44, with decreased CD62L. We also noted higher expression of proteins known to help mediate Treg cell function, including the ectonucleotidases CD39 and CD73.
Expression of ICOS by Treg cells has been shown to track with expression of the immunoregulatory cytokine IL-10 (Ito et al., 2007, 2008). Indeed, the most highly upregulated gene in Tim-3 Tg Treg cells was that encoding IL-10, an important mediator of Treg cell suppression, including at barrier sites (Rubtsov et al., 2008) and in the tumor microenvironment (Sawant et al., 2019; Wei et al., 2017). We confirmed upregulation of IL-10 protein in mouse TIL and also noted that there was a tight correlation between Tim-3 and IL-10 in human TIL Treg cells. Also, in mice, the absence of Tim-3 expression, by genetic deletion in Treg cells, led to a decrease in IL-10 expression by TIL Treg cells. We also found that levels of IL-10 in CD8+ TIL T cells correlated with the expression of Treg cell IL-10. In addition, our data suggest that this enhanced IL-10 is sensed by the Treg cells themselves and the exhausted CD8+ TIL T cells, as pSTAT3, a target of IL-10r signaling, was increased in both cell types among the TIL of Tim-3 Tg mice. Interestingly, these data corroborate the findings in a previous study of patients with osteoarthritis, which noted a correlation between Tim-3 and IL-10 in peripheral blood Treg cells (Li et al., 2016). Finally, in a gene expression analysis of WT versus Tim-3 KO CD4+ T cells, one of the most highly upregulated genes was IL-10 (Gorman and Colgan, 2018). These findings suggest that IL-10 may be a general target of Tim-3 signaling.
An unexpected effect of Tim-3 expression on murine Treg cells was the downregulation of CTLA-4, especially because this occurred in the context of enhanced Treg cell suppression, and on human TIM-3+ Treg cells, CTLA-4 was actually higher (Liu et al., 2018b). RNA-seq revealed that, in mice, the effect of Tim-3 was due at least in part to a decrease in Ctla4 message. There is a precedent for such a dichotomy, as inducible KO of CTLA-4 in mice still allowed for normal Treg cell suppressive activity, both in vitro and in vivo (Paterson et al., 2015). At the time, this split phenotype was attributed to corresponding upregulation of a number of genes, including IL-10 and ICOS (Paterson et al., 2015), both of which we also observed in Tim-3 Tg. In addition, lower CTLA-4 on Tim-3+ Treg cells may promote T cell over-stimulation, leading to exhaustion, due to more sustained expression of B7 family members within tumors (Greenwald et al., 2005; Zang and Allison, 2007).
Cellular metabolism and protein translation were two of the pathways most closely linked to Treg-cell-specific expression of Tim-3. There is a paucity of literature regarding the regulation of protein translation in Treg cells. However, a recent report documented a role for ribosome biogenesis in controlling Treg cell activation and function (Zhu et al., 2019). There is an extensive literature exploring the roles of different metabolic pathways among subsets of T cells. Treg cells are thought to rely predominantly on mitochondrial metabolism, including lipid oxidation, while actively suppressing glycolysis (Shi and Chi, 2019). However, there is likely a more nuanced role for glucose metabolism in Treg cells, and indeed, glucose uptake and glycolysis are heterogeneous within Treg cells infiltrating various tissues, including tumors (Watson et al., 2021). Although peripheral Treg cells that are avid for glucose have an overall decreased Treg cell signature, they notably express elevated levels of both Tim-3 and IL-10 (Watson et al., 2021). Thus, the upregulation of glycolysis genes, along with downregulation of oxidative phosphorylation genes, in Tim3-expressing Treg cells may be due at least in part to downregulation of FoxP3 during the acquisition of an effector Treg-cell-like state. There could also be more direct effects of Tim-3 expression, because we previously reported that ectopic expression of Tim-3 in T cells modulates the mTOR pathway (Avery et al., 2018; Lee et al., 2011), which can promote glycolysis in T cells (Pollizzi and Powell, 2014). Surprisingly, Tim-3 Tg Treg cells had lower levels of pAkt, pMTOR, and pS6, suggesting that this pathway was actually downregulated in the presence of Tim-3. The reasons for this discrepancy are not clear, but it is possible that constitutive expression of Tim-3 drove desensitization of this pathway over time, because Treg cells usually maintain lower levels of Akt/mTOR signaling than effector T cells (Huynh et al., 2015). Conversely, we observed an increase in steady-state pERK in Tim-3 Tg Treg cells, consistent with previous reports that MAPK signaling may lead to an increase in glycolysis (Huynh et al., 2015).
How do the effects of Tim-3 on Treg cell biology fit within the wider context of Tim-3 function? As described above, although there are indications that Tim-3 can enhance immune cell function, other studies support a model of Tim-3 as a negative regulator of T cells in the contexts of chronic viral infection or within the tumor microenvironment (Anderson et al., 2016; Banerjee and Kane, 2018; Du et al., 2017). The ability of Tim-3 to increase Treg cell activation is consistent with our previous findings in effector T cells and, indeed in myeloid and mast cells, that a major proximal and cell-intrinsic effect of Tim-3 is to enhance phosphotyrosine-dependent signaling (Anderson et al., 2007; Avery et al., 2018; Lee et al., 2011; Phong et al., 2015). Furthermore, mAbs to Tim-3 may have either agonistic or antagonistic function, and most studies have not explicitly addressed the role of Tim-3 antibodies in blocking access to specific ligands (Banerjee and Kane, 2018; Ferris et al., 2014). The ability of Tim-3 to enhance the suppressive activity of Treg cells suggests that such antibodies could interfere with the ability of Tim-3 to drive increased Treg cell suppressive activity, indirectly promoting anti-tumor immunity.
Although our study addresses the function of Tim-3, it does not answer the question of how Treg cells acquire expression of Tim-3. There are at least two mechanisms by which this might occur. One is the recruitment and expansion of circulating or tissue Tim-3+ Treg cells to tumors. We do observe a very small fraction of Tim-3+ Treg cells in spleen and lymph nodes of normal mice and also within the peripheral blood of people with cancer (Liu et al., 2018b). We have not examined expression of Tim-3 on Treg cells from non-lymphoid tissues. However, there is evidence for increased expression of immune checkpoint molecules on tissue-resident memory T cells (Gamradt et al., 2019; Weisberg et al., 2019). TIL Treg cells may also acquire expression of Tim-3 after entry into the tumor. Although factors that regulate Tim-3 expression have not been exhaustively defined, several cytokines and transcription factors with such activity have been identified. The transcription factors T-bet and NFIL3 can apparently help drive expression of Tim-3 on effector T cells in response to interferon γ (IFN-γ) and IL-27, respectively (Anderson et al., 2010; Zhu et al., 2015). Additional factors likely remain to be revealed.
We also report here the development of a mouse model with Cre-inducible deletion of Tim-3. We bred these Tim-3 targeted mice to the same FoxP3EGFP-Cre-ERT2 line described above. Using these mice, we found that Treg-cell-specific deletion of Tim-3 led to decreased tumor growth and lower T cell exhaustion. We did not observe any obvious changes in the proportion or phenotype of Treg cells in these mice at baseline. However, there was a significant decrease in the frequency of Treg cells present in the tumors and those that were present had decreased FoxP3 and CD25 expression, suggesting that the absence of Tim-3 on this population was leading to Treg cell destabilization. While this study was under review, another group reported an anti-tumor effect of dendritic-cell-specific Tim-3 deletion but no detectable effect of Treg-cell-specific Tim-3 deletion (Dixon et al., 2021). We do not yet know the reason(s) for this discrepancy, although in the study of Dixon et al., the authors reported that they floxed exon 1, although we floxed exon 4. Ultimately, side-by-side comparison of these two KO models in the same animal facility may be required to resolve these divergent findings.
Taken together, our studies demonstrate that modulation of Tim-3 expression can have dramatic effects on Treg cell function and differentiation within tumors. Given the highly suppressive nature of Tim-3+ Treg cells, therapies that specifically target these cells may be effective for promoting anti-tumor immunity, without attendant autoimmunity.
Limitations of the study
One caveat of this study lies in the high levels of Tim-3 expression achieved in our transgenic system. We have taken this approach in part because of the aforementioned complexity in Tim-3 ligands. Based on previous studies from our group and others, we believe that such overexpression results in enhanced Tim-3 signaling (Avery et al., 2018; Lee et al., 2011, 2020). Indeed, our finding that Treg cells expressing endogenous levels of Tim-3 display a very similar phenotype to the Tim-3 Tg Treg cells supports this assertion. Finally, further investigation will be necessary to determine the precise relationship between Tim-3 signaling pathways and downstream expansion of existing eTreg cells versus de novo acquisition of an eTreg cell phenotype.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Lawrence P. Kane (lkane@pitt.edu).
Materials availability
FSF-Tim-3 and floxed Tim-3 mouse lines described in this study are available on request from the lead contact.
Data and code availability
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the Key resources table. This paper also analyzes existing, publicly available data. The accession numbers for these datasets are also listed in the Key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| mCD25 | BD Biosciences, BioLegend, Tonbo | Clone PC61 |
| FoxP3 | Invitrogen | Clone FJK16S |
| mCD44 | BD Biosciences, BioLegend, Tonbo | Clone IM7 |
| mCD62L | BioLegend, Tonbo | Clone MEL-14 |
| mICOS | BioLegend | Clone 398.4A |
| Ki67 | BD Biosciences | Clone B56 |
| mCTLA-4 | BioLegend | Clone UC10–4B9 |
| mCD103 | BD Biosciences | Clone M290 |
| mNrp-1 | BD Biosciences | Clone 3E12 |
| mCD39 | BioLegend | Clone Duha 59 |
| mCD73 | BioLegend | Clone TY/11.8 |
| mTim-3 | BioLegend | Clone RMT 3–23 |
| mTim-3 | R&D Systems | Clone FAB1529 |
| Flag | BioLegend | Clone L5 |
| mPD-1 | BioLegend | Clone RMP1–30 |
| mIL-10 | BioLegend | Clone JES5–16E3 |
| mCD90.2 | BD Biosciences, BioLegend | Clone 532.1 |
| mCD45.2 | BioLegend | Clone 104 |
| Helios | BioLegend | Clone 22F6 |
| mKlrg-1 | BioLegend | Clone 2F1 |
| mLy6g | Tonbo | Clone IA8 |
| mGITR | eBioscience | Clone DTA-1 |
| mCD4 | BD Biosciences, BioLegend, Tonbo | Clone RM-4–5 |
| mTIGIT | BioLegend | Clone 1G9 |
| mLAG-3 | Invitrogen | Clone C9B7W |
| mGARP | BioLegend | Clone F011–5 |
| phospho-S6 | BioLegend | Clone A17020B |
| phospho-ERK | BioLegend | Clone 4B11B69 |
| phospho-Akt | BD Biosciences | Clone M89–61 |
| phospho-MTOR | eBioscience | Clone MRRBY |
| phospho-STAT3 | BD Biosciences | Clone 4 |
| CD16/CD32 (Fc Block) | Tonbo | Clone 2.4G2 |
| mCD8a | BD Biosciences, BioLegend, Tonbo | Clone H35–17.2 |
| mCD19 | BD Biosciences, BioLegend, Tonbo | Clone ID3 |
| mCD11b | eBioscience | Clone M1/70 |
| mCD3 (purified) | BioLegend | Clone 2C11 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| CellTrace Violet | Invitrogen | Cat#C34571 |
| FoxP3 Staining Kit | eBioscience | Cat#00–5523-00 |
| GhostDye-Violet510 | Tonbo Biosciences | Cat#13–0870-T500 |
| RBC lysis buffer | eBioscience | Cat#00–4333-57 |
| Collagenase D | Roche | Cat#11088866001 |
| DNase I, type IV | Sigma-Aldrich | Cat#D5025 |
| Hyclone Fetal bovine serum | Cytiva | Cat#SH30071 |
| Tamoxifen | MP Biomedical | Cat#02156738-CF |
|
| ||
| Deposited data | ||
|
| ||
| RNA sequencing of Tim-3 Tg Treg and WT Treg | Data obtained for this study | GEO record GSE155825 |
| single-cell RNA sequencing of HNC tumors | Cillo et al. (2020) | GEO record GSE139324 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| MC38 tumor line | Dario Vignali, Univ. of Pittsburgh | RRID:CVCL_B288 |
| B16-F10 tumor line | Dario Vignali, Univ. of Pittsburgh | RRID:CVCL_0159 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: FoxP3eGFP-Cre-ERT2: Foxp3tm9(EGFP/cre/ERT2)Ayr | The Jackson Laboratory | Cat#016961 |
| Mouse: Tim-3fl/fl: B6-Havcr2fl/fl | Developed with the Pitt Department of Immunolgy Transgenic and Gene Targeting Core for this study | N/A |
| Mouse: FSF-Tim-3: B6-Rosa26flox-stop-flox-Flag-Tim-3 | Developed for the investigators by GenOway | Avery et al. (2018) |
| Mouse: C57BL/6J | The Jackson Laboratory | Cat#000664 |
|
| ||
| Software and algorithms | ||
|
| ||
| FlowJo | BD Biosciences | version 10 |
| Prism | GraphPad | version 9 |
| FastQC | Babraham Bioinformatics | version 0.11.7 |
| Cutadapt | https://cutadapt.readthedocs.io/en/stable/ | version 1.18 |
| HISAT2 | http://daehwankimlab.github.io/hisat2/main/ | version 2.1.0 |
| SAMtools | http://www.htslib.org/ | version 1.9 |
| HTSeq | https://htseq.readthedocs.io/en/master/ | version 0.11.2 |
| Bioconductor edgeR | https://bioconductor.org/packages/release/bioc/html/edgeR.html | version 3.24.1 |
| Enrichr | https://maayanlab.cloud/Enrichr/ | Kuleshov et al., 2016 |
| Scanpy | https://github.com/theislab/scanpy | Wolf et al., 2018 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All animal studies were approved by the University of Pittsburgh Institutional Animal Care and Use Committee (IACUC). C57BL/6 mice carrying a Rosa26 knockin of a flox-stop-flox cassette containing a Flag-tagged mTim-3 cDNA have been described (Avery et al., 2018). Mice with a floxed allele of Tim-3 were created by the University of Pittsburgh Department of Immunology Transgenic and Gene Targeting (TGT) core. LoxP sites were inserted on either side of exon 4 of the Havcr2 gene in C57BL/6J zygotes, using Crispr/Cas9. Disruption of this exon was predicted to result in a premature stop codon 20 bp into exon 5, before the TM domain, resulting in nonsense mediated decay. Insertion of the LoxP sites was initially confirmed by PCR and restriction digest, followed by sequencing of the targeted region. For inducible Cre-mediated expression or deletion, tamoxifen was administered at 1 mg via the i.p. route on five successive days. FoxP3eGFP-Cre-ERT2 had been backcrossed to C57BL/6 mice for at least ten generations.
For tumor experiments in WT (C57BL/6J) mice, groups consisted of both male and female mice 6–8 weeks of age at the start of the experiment. For experiments employing Tim-3 Tg or Tim-3 inducible KO mice, male mice were used, as the source of Cre was from the Foxp3 locus, which is located on the X chromosome.
Cell Lines
B16-F10 tumors were derived from a melanoma in a male C57BL/6 mouse. MC38 tumors were derived from a colon carcinoma in a female CD57BL/6 mouse. Both were obtained from Dr. Dario Vignali. The lines were not independently validated.
METHOD DETAILS
Flow cytometry
Single cell suspensions from organs and tumors were incubated with Live/Dead fixable stain and Fc Block for 20 min, followed by staining for various cell surface markers. For analytic flow cytometry, cells were captured on a BD LSR-II and BD Fortessa. Data were exported and analyzed in FlowJo. Cell sorting was carried out with a BD FACS Aria. The gating strategy used for flow cytometry analysis of spleen and lymph nodes is shown in Figure S1A. The gating strategy used for tumor analysis is shown in Figure S1B.
Samples analyzed for transcription factors and cytokines were first stained for Live/Dead staining along with Fc Block (CD16/32) followed by cell surface marker staining, fixed/permeabilized with a FoxP3 staining kit, followed by intracellular protein staining. For analysis of intracellular signaling at steady state, we prepared single-cell suspensions from the spleens of WT and Tim-3 Tg mice and immediately fixed the cells in paraformaldehyde (PFA). Cells were then permeabilized and stained with phospho-specific antibodies to various signaling intermediates.
Transplantable tumor models
B16-F10 and MC38 tumor cells were grown in RPMI1640 (for B16) or Dulbecco’s modified Eagle’s medium (DMEM; for MC38), supplemented with 10% fetal bovine serum. Cell cultures were maintained at 37°C and 5% CO2. Mice (day six after tamoxifen/oil) were injected with B16F10 melanoma (1.25 × 105 cells intradermally) or MC38 (5 × 105 cells subcutaneously). Tumors were measured every three days starting at day five, in two dimensions, using digital calipers, and plotted as tumor area (mm2).
Lymphocyte isolation from organs and tumors
Splenocytes and lymph node cells were isolated by mechanical dissociation between frosted glass slides. Spleen cells were further processed using RBC lysis buffer, followed by filtering through 70 μM nylon mesh. Single cell suspensions of splenocytes and lymph node cells were then used for downstream experiments and staining for flow cytometry purposes. TIL from transplanted tumors were isolated by roughly chopping tumors, followed by incubation with collagenase D and 0.2 mg/ml DNase I, type IV for 40 min at 37°C with constant gentle shaking in Miltneyi C-tubes, using a GentleMACS tissue dissociator (Miltenyi). Digestion was quenched using complete DMEM followed by two washes with PBS. RBC lysis was performed, and cells were filtered through 70 μM nylon mesh before resuspension and incubation with Fc Block.
In vitro suppression assay
Treg were isolated by pooling spleen and LNs together. For the suppression assay in Figure 3A, Tim-3+ Treg were isolated from a tamoxifen-induced FSF-Tim-3 x Foxp3eGFP-Cre-ERT2 mice, and control Tim-3 negative Treg were isolated similarly from a tamoxifen-treated, Cre-only mice. Treg were sorted into Tim-3+ Treg (live CD4+eGFP+CD25Hi Flag+Tim-3+) and control Tim-3− Treg (live CD4+eGFP+CD25HiTim-3−). Conventional T cells (Tconv; live CD3+CD8+) were sorted from the spleen of a C57BL/6Thy1.1 mouse as responder cells and labeled with 5 μM CellTrace Violet (Invitrogen). Whole splenocytes from 6–8 week-old female C57BL/6 mice were irradiated with 3,000 rads and used as antigen presenting cells (APCs). Responder cells and APCs were plated in triplicate at 5 × 104 cells/well; co-cultures were set up with the following ratios of control Tim-3− or Tim-3+ Treg: 1:2–1:16 Treg:Teff; 25,000–3,125 Treg (Polanczyk et al., 2005). T cell activation was achieved by adding anti-CD3 mAb at a final concentration of 1 μg/ml and cells were co-cultured in 200 μL RPMI for 72 hr. Stained cells were analyzed with a BD Fortessa flow cytometer and CellTrace Violet dilution was quantified using FlowJo. Suppression was calculated using the following formula, adapted from Collison and Vignali (2011): percent suppression = ((fraction of proliferated Tconv cells alone-fraction of proliferated Tconv cells with Treg)/(fraction of proliferated Tconv cells alone)*100).
Gene expression analysis and RNaseq pipeline
Live CD4+CD25+GFP+ from WT mice, CD4+CD25+GFP+Tim-3+FLAG+ and CD4+CD25+GFP−Tim-3+FLAG+ cells from spleens of transgenic mice were sorted on a FACSAria directly into SmartSeq low-input RNA kit lysis buffer in a 96 well plate. DNA libraries were prepared using Nextera XT Kit and RNaseq was performed on an Illumina NextSeq 500 by The Children’s Hospital of Pittsburgh Sequencing core facility.
The reverse unstranded paired-end RNA-Seq reads of mouse Treg, generated by SMART-seq HT kit, were checked for presence of adapters and high-quality bases using FastQC (v 0.11.7). These high-quality reads were trimmed for adapters using Cutadapt (v 1.18). The trimmed reads were later mapped against the Ensembl mouse reference genome (GRCm38) using the HISAT2 (v 2.1.0) mapping tool. The output file from HISAT2 was converted from SAM format to BAM format using SAMtools (v 1.9). Counts for expressed genes were generated using HT-Seq (v 0.11.2) and were output in text format. These count text files were then imported into the Bioconductor R package, edgeR (v 3.24.1). The package was used to identify the differentially expressed genes based on those having an expression count of absolute value log base 2 greater than 1 between two experimental conditions and a false discovery rate of less than 0.05. Based on this standard, between the 3 comparisons of WT Treg versus FoxP3Hi, WT Treg versus FoxP3Lo, and FoxP3Hi versus FoxP3Lo produced 5027, 5685, and 834 differentially expressed genes, respectively. Processed read data have been deposited to the Gene Expression Omnibus, record number GSE155825. KEGG analysis was performed with Enrichr (Kuleshov et al., 2016).
Analysis of single cell RNaseq data from human TIL and PBL of patients with HNC were carried out with our previously published dataset described in Cillo et al. (2020), using the Scanpy package (Wolf et al., 2018). Data are available at the Gene Expression Omnibus, record number GSE139324.
Seahorse metabolic flux assays
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR), as measures of mitochondrial respiration and glycolysis, respectively, were measured essentially as described (Scharping et al., 2016; Watson et al., 2021), except cells were not stimulated during the assay. Treg were sorted based on expression of CD4, CD25 and FoxP3-GFP. Sorted cells were plated on Cell-Tak-coated Seahorse plates (250,000 cells per well) in DMEM with 2 mM glutamine (for OCR) or glucose-free media (for ECAR). Basal ECAR and OCR were recorded for 15–30 min. Maximal and minimal rates were obtained by injecting oligomycin (2 μM), 2-DG (10 mM), glucose (10 mM) or FCCP (0.5 mM).
QUANTIFICATION AND STATISTICAL ANALYSIS
Experiments were analyzed using Student’s t test for two-way comparisons and ANOVA for experiments involving multiple comparisons. Statistical analyses were performed with GraphPad Prism. Tumor growth curves were analyzed using t test for each time point and statistical significance was determined by the Holm-Sidak method, with alpha = 0.05. Each time point was analyzed individually, without assuming a consistent variance between samples. p values are indicated as follows: *p < 0.05, **p < 0.01 and ***p < 0.001 and ****p < 0.0001 where statistical significance was found, and all data are represented as mean ± SEM. Statistical details for specific experiments can be found within the figure legends.
Supplementary Material
Highlights.
Treg cells expressing Tim-3 display a more activated phenotype
Enforced Treg cell expression of Tim-3 increases activated T cell frequency
Enforced expression of Tim-3 on Treg cells impairs T cell responses to tumors
Deletion of Tim-3 specifically from Treg cells improves anti-tumor T cell responses
ACKNOWLEDGMENTS
We thank Shunsuke Kataoka for helpful discussions; Mia Simms, Julia Correa, and Isabella Bosco for performing mouse genotyping; Lawrence Andrews, Angela Gocher, and Elisa Ruffo for technical assistance with tumor experiments and Treg cell suppression assays; Creg Workman for input into experimental design; The Department of Immunology flow facility and Dwayne Falkner for cell sorting; Health Sciences Sequencing Core at UPMC Children’s Hospital of Pittsburgh; Rachel Cumberland for assistance with Seahorse assays; Tim Hand, Mandy McGeachy, Mark Shlomchik, and Dario Vignali for providing antibodies; and Reinhardt Hinterleitner and Marlies Meisel for advice on intracellular IL-10 staining. Support was from NIH awards CA206517 (R.L.F. and L.P.K.), AI138504 (L.P.K.), and DP2AI136598 (G.M.D.). H.N.-R. is supported by a diversity supplement to NIH award AI138504 and by T32GM008208 to the University of Pittsburgh Medical Scientist Training Program. B.M. is supported by NCI training grant T32CA082084 (PI, Vignali); additional support was from NCI SPORE CA097190.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109699.
REFERENCES
- Acharya N, Sabatos-Peyton C, and Anderson AC (2020). Tim-3 finds its place in the cancer immunotherapy landscape. J. Immunother. Cancer 8, e000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson AC (2014). Tim-3: an emerging target in the cancer immunotherapy landscape. Cancer Immunol. Res. 2, 393–398. [DOI] [PubMed] [Google Scholar]
- Anderson AC, Anderson DE, Bregoli L, Hastings WD, Kassam N, Lei C, Chandwaskar R, Karman J, Su EW, Hirashima M, et al. (2007). Promotion of tissue inflammation by the immune receptor Tim-3 expressed on innate immune cells. Science 318, 1141–1143. [DOI] [PubMed] [Google Scholar]
- Anderson AC, Lord GM, Dardalhon V, Lee DH, Sabatos-Peyton CA, Glimcher LH, and Kuchroo VK (2010). T-bet, a Th1 transcription factor regulates the expression of Tim-3. Eur. J. Immunol. 40, 859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson AC, Joller N, and Kuchroo VK (2016). Lag-3, Tim-3, and TIGIT: co-inhibitory receptors with specialized functions in immune regulation. Immunity 44, 989–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L, Filderman J, Szymczak-Workman AL, and Kane LP (2018). Tim-3 co-stimulation promotes short-lived effector T cells, restricts memory precursors, and is dispensable for T cell exhaustion. Proc. Natl. Acad. Sci. USA 115, 2455–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee H, and Kane LP (2018). Immune regulation by Tim-3. F1000Res. 7, 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Du Y, Lin X, Qian Y, Zhou T, and Huang Z (2016). CD4+CD25+ regulatory T cells in tumor immunity. Int. Immunopharmacol. 34, 244–249. [DOI] [PubMed] [Google Scholar]
- Cillo AR, Kürten CHL, Tabib T, Qi Z, Onkar S, Wang T, Liu A, Duvvuri U, Kim S, Soose RJ, et al. (2020). Immune landscape of viral- and carcinogen-driven head and neck cancer. Immunity 52, 183–199.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collison LW, and Vignali DA (2011). In vitro Treg suppression assays. Methods Mol. Biol. 707, 21–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cretney E, Kallies A, and Nutt SL (2013). Differentiation and function of Foxp3(+) effector regulatory T cells. Trends Immunol. 34, 74–80. [DOI] [PubMed] [Google Scholar]
- Dixon KO, Tabaka M, Schramm MA, Xiao S, Tang R, Dionne D, Anderson AC, Rozenblatt-Rosen O, Regev A, and Kuchroo VK (2021). TIM-3 restrains anti-tumour immunity by regulating inflammasome activation. Nature 595, 101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Yang M, Turner A, Xu C, Ferris RL, Huang J, Kane LP, and Lu B (2017). TIM-3 as a target for cancer immunotherapy and mechanisms of action. Int. J. Mol. Sci. 18, 645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris RL, Lu B, and Kane LP (2014). Too much of a good thing? Tim-3 and TCR signaling in T cell exhaustion. J. Immunol. 193, 1525–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris RL, Blumenschein G Jr., Fayette J, Guigay J, Colevas AD, Licitra L, Harrington K, Kasper S, Vokes EE, Even C, et al. (2016). Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 375, 1856–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamradt P, Laoubi L, Nosbaum A, Mutez V, Lenief V, Grande S, Redoulès D, Schmitt AM, Nicolas JF, and Vocanson M (2019). Inhibitory checkpoint receptors control CD8+ resident memory T cells to prevent skin allergy. J. Allergy Clin. Immunol. 143, 2147–2157.e9. [DOI] [PubMed] [Google Scholar]
- Gao X, Zhu Y, Li G, Huang H, Zhang G, Wang F, Sun J, Yang Q, Zhang X, and Lu B (2012). TIM-3 expression characterizes regulatory T cells in tumor tissues and is associated with lung cancer progression. PLoS ONE 7, e30676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautron AS, Dominguez-Villar M, de Marcken M, and Hafler DA (2014). Enhanced suppressor function of TIM-3+ FoxP3+ regulatory T cells. Eur. J. Immunol. 44, 2703–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman JV, and Colgan JD (2018). Acute stimulation generates Tim-3-expressing T helper type 1 CD4 T cells that persist in vivo and show enhanced effector function. Immunology 154, 418–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald RJ, Freeman GJ, and Sharpe AH (2005). The B7 family revisited. Annu. Rev. Immunol. 23, 515–548. [DOI] [PubMed] [Google Scholar]
- Gupta S, Thornley TB, Gao W, Larocca R, Turka LA, Kuchroo VK, and Strom TB (2012). Allograft rejection is restrained by short-lived TIM-3+PD-1+Foxp3+ Tregs. J. Clin. Invest. 122, 2395–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R, Wolchok JD, Hersey P, Joseph RW, Weber JS, et al. (2013). Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N. Engl. J. Med. 369, 134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargadon KM, Johnson CE, and Williams CJ (2018). Immune checkpoint blockade therapy for cancer: An overview of FDA-approved immune checkpoint inhibitors. Int. Immunopharmacol. 62, 29–39. [DOI] [PubMed] [Google Scholar]
- Huynh A, DuPage M, Priyadharshini B, Sage PT, Quiros J, Borges CM, Townamchai N, Gerriets VA, Rathmell JC, Sharpe AH, et al. (2015). Control of PI(3) kinase in Treg cells maintains homeostasis and lineage stability. Nat. Immunol. 16, 188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Yang M, Wang YH, Lande R, Gregorio J, Perng OA, Qin XF, Liu YJ, and Gilliet M (2007). Plasmacytoid dendritic cells prime IL-10-producing T regulatory cells by inducible costimulator ligand. J. Exp. Med. 204, 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Hanabuchi S, Wang YH, Park WR, Arima K, Bover L, Qin FX, Gilliet M, and Liu YJ (2008). Two functional subsets of FOXP3+ regulatory T cells in human thymus and periphery. Immunity 28, 870–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jie HB, Gildener-Leapman N, Li J, Srivastava RM, Gibson SP, Whiteside TL, and Ferris RL (2013). Intratumoral regulatory T cells upregulate immunosuppressive molecules in head and neck cancer patients. Br. J. Cancer 109, 2629–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin HT, Anderson AC, Tan WG, West EE, Ha SJ, Araki K, Freeman GJ, Kuchroo VK, and Ahmed R (2010). Cooperation of Tim-3 and PD-1 in CD8 T-cell exhaustion during chronic viral infection. Proc. Natl. Acad. Sci. USA 107, 14733–14738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka S, Manandhar P, Lee J, Workman CJ, Banerjee H, Szymczak-Workman AL, Kvorjak M, Lohmueller J, and Kane LP (2021). The costimulatory activity of Tim-3 requires Akt and MAPK signaling and its recruitment to the immune synapse. Sci. Signal. 14, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44 (W1), W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Su EW, Zhu C, Hainline S, Phuah J, Moroco JA, Smithgall TE, Kuchroo VK, and Kane LP (2011). Phosphotyrosine-dependent coupling of Tim-3 to T-cell receptor signaling pathways. Mol. Cell. Biol. 31, 3963–3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Yun SJ, Lee B, Jeong E, Yoon G, Kim K, and Park S (2020). Association of TIM-3 expression with glucose metabolism in Jurkat T cells. BMC Immunol. 21, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AG, Arvey A, Jin W, and Rudensky AY (2014). Continuous requirement for the TCR in regulatory T cell function. Nat. Immunol. 15, 1070–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wan J, Anderson W, Sun H, Zhang H, Peng X, Yu Z, Wang T, Yan X, and Smith W (2016). Downregulation of IL-10 secretion by Treg cells in osteoarthritis is associated with a reduction in Tim-3 expression. Biomed. Pharmacother. 79, 159–165. [DOI] [PubMed] [Google Scholar]
- Liu JF, Wu L, Yang LL, Deng WW, Mao L, Wu H, Zhang WF, and Sun ZJ (2018a). Blockade of TIM3 relieves immunosuppression through reducing regulatory T cells in head and neck cancer. J. Exp. Clin. Cancer Res. 37, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, McMichael EL, Shayan G, Li J, Chen K, Srivastava R, Kane LP, Lu B, and Ferris RL (2018b). Novel effector phenotype of Tim-3+ regulatory T cells leads to enhanced suppressive function in head and neck cancer patients. Clin. Cancer Res. 24, 4529–4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AM, Lovitch SB, Sage PT, Juneja VR, Lee Y, Trombley JD, Arancibia-Cárcamo CV, Sobel RA, Rudensky AY, Kuchroo VK, et al. (2015). Deletion of CTLA-4 on regulatory T cells during adulthood leads to resistance to autoimmunity. J. Exp. Med. 212, 1603–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phong BL, Avery L, Sumpter TL, Gorman JV, Watkins SC, Colgan JD, and Kane LP (2015). Tim-3 enhances FcεRI-proximal signaling to modulate mast cell activation. J. Exp. Med. 212, 2289–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polanczyk MJ, Hopke C, Huan J, Vandenbark AA, and Offner H (2005). Enhanced FoxP3 expression and Treg cell function in pregnant and estrogen-treated mice. J. Neuroimmunol. 170, 85–92. [DOI] [PubMed] [Google Scholar]
- Pollizzi KN, and Powell JD (2014). Integrating canonical and metabolic signalling programmes in the regulation of T cell responses. Nat. Rev. Immunol. 14, 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas A, and Wolchok JD (2018). Cancer immunotherapy using checkpoint blockade. Science 359, 1350–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov YP, Rasmussen JP, Chi EY, Fontenot J, Castelli L, Ye X, Treuting P, Siewe L, Roers A, Henderson WR Jr., et al. (2008). Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 28, 546–558. [DOI] [PubMed] [Google Scholar]
- Sakuishi K, Ngiow SF, Sullivan JM, Teng MW, Kuchroo VK, Smyth MJ, and Anderson AC (2013). TIM3+FOXP3+ regulatory T cells are tissue-specific promoters of T-cell dysfunction in cancer. OncoImmunology 2, e23849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawant DV, Yano H, Chikina M, Zhang Q, Liao M, Liu C, Callahan DJ, Sun Z, Sun T, Tabib T, et al. (2019). Adaptive plasticity of IL-10+ and IL-35+ Treg cells cooperatively promotes tumor T cell exhaustion. Nat. Immunol. 20, 724–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharping NE, Menk AV, Moreci RS, Whetstone RD, Dadey RE, Watkins SC, Ferris RL, and Delgoffe GM (2016). The tumor microenvironment represses T cell mitochondrial biogenesis to drive intratumoral T cell metabolic insufficiency and dysfunction. Immunity 45, 374–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segawa K, and Nagata S (2015). An apoptotic ‘eat me’ signal: phosphatidylserine exposure. Trends Cell Biol. 25, 639–650. [DOI] [PubMed] [Google Scholar]
- Shen P, Yue R, Tang J, Si H, Shen L, Guo C, Zhang L, Han H, Song HK, Zhao P, et al. (2016). Preferential Tim-3 expression on Treg and CD8(+) T cells, supported by tumor-associated macrophages, is associated with worse prognosis in gastric cancer. Am. J. Transl. Res. 8, 3419–3428. [PMC free article] [PubMed] [Google Scholar]
- Shi H, and Chi H (2019). Metabolic control of Treg cell stability, plasticity, and tissue-specific heterogeneity. Front. Immunol. 10, 2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smigiel KS, Richards E, Srivastava S, Thomas KR, Dudda JC, Klonowski KD, and Campbell DJ (2014). CCR7 provides localized access to IL-2 and defines homeostatically distinct regulatory T cell subsets. J. Exp. Med. 211, 121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su EW, Bi S, and Kane LP (2011). Galectin-9 regulates T helper cell function independently of Tim-3. Glycobiology 21, 1258–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, and Sakaguchi S (2017). Regulatory T cells in cancer immunotherapy. Cell Res. 27, 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh PP, Vasanthakumar A, and Kallies A (2015). Development and function of effector regulatory T cells. Prog. Mol. Biol. Transl. Sci. 136, 155–174. [DOI] [PubMed] [Google Scholar]
- Venereau E, De Leo F, Mezzapelle R, Careccia G, Musco G, and Bianchi ME (2016). HMGB1 as biomarker and drug target. Pharmacol. Res. 111, 534–544. [DOI] [PubMed] [Google Scholar]
- Watson MJ, Vignali PDA, Mullett SJ, Overacre-Delgoffe AE, Peralta RM, Grebinoski S, Menk AV, Rittenhouse NL, DePeaux K, Whetstone RD, et al. (2021). Metabolic support of tumour-infiltrating regulatory T cells by lactic acid. Nature 591, 645–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Zhang J, Gu Q, Huang M, Zhang W, Guo J, and Zhou X (2017). Reciprocal expression of IL-35 and IL-10 defines two distinct effector Treg subsets that are required for maintenance of immune tolerance. Cell Rep. 21, 1853–1869. [DOI] [PubMed] [Google Scholar]
- Weisberg SP, Carpenter DJ, Chait M, Dogra P, Gartrell-Corrado RD, Chen AX, Campbell S, Liu W, Saraf P, Snyder ME, et al. (2019). Tissue-resident memory T cells mediate immune homeostasis in the human pancreas through the PD-1/PD-L1 pathway. Cell Rep. 29, 3916–3932.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf FA, Angerer P, and Theis FJ (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo SR, Turnis ME, Goldberg MV, Bankoti J, Selby M, Nirschl CJ, Bettini ML, Gravano DM, Vogel P, Liu CL, et al. (2012). Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Cancer Res. 72, 917–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Thalhamer T, Franca RF, Xiao S, Wang C, Hotta C, Zhu C, Hirashima M, Anderson AC, and Kuchroo VK (2014). Galectin-9-CD44 interaction enhances stability and function of adaptive regulatory T cells. Immunity 41, 270–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Sun L, Li CF, Wang YH, Yao J, Li H, Yan M, Chang WC, Hsu JM, Cha JH, et al. (2021). Galectin-9 interacts with PD-1 and TIM-3 to regulate T cell death and is a target for cancer immunotherapy. Nat. Commun. 12, 832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano H, Andrews LP, Workman CJ, and Vignali DAA (2019). Intratumoral regulatory T cells: markers, subsets and their impact on anti-tumor immunity. Immunology 157, 232–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang X, and Allison JP (2007). The B7 family and cancer therapy: costimulation and coinhibition. Clin. Cancer Res. 13, 5271–5279. [DOI] [PubMed] [Google Scholar]
- Zhu C, Sakuishi K, Xiao S, Sun Z, Zaghouani S, Gu G, Wang C, Tan DJ, Wu C, Rangachari M, et al. (2015). An IL-27/NFIL3 signalling axis drives Tim-3 and IL-10 expression and T-cell dysfunction. Nat. Commun. 6, 6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Zhang W, Guo J, Zhang X, Li L, Wang T, Yan J, Zhang F, Hou B, Gao N, et al. (2019). Noc4L-mediated ribosome biogenesis controls activation of regulatory and conventional T cells. Cell Rep. 27, 1205–1220.e4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the Key resources table. This paper also analyzes existing, publicly available data. The accession numbers for these datasets are also listed in the Key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| mCD25 | BD Biosciences, BioLegend, Tonbo | Clone PC61 |
| FoxP3 | Invitrogen | Clone FJK16S |
| mCD44 | BD Biosciences, BioLegend, Tonbo | Clone IM7 |
| mCD62L | BioLegend, Tonbo | Clone MEL-14 |
| mICOS | BioLegend | Clone 398.4A |
| Ki67 | BD Biosciences | Clone B56 |
| mCTLA-4 | BioLegend | Clone UC10–4B9 |
| mCD103 | BD Biosciences | Clone M290 |
| mNrp-1 | BD Biosciences | Clone 3E12 |
| mCD39 | BioLegend | Clone Duha 59 |
| mCD73 | BioLegend | Clone TY/11.8 |
| mTim-3 | BioLegend | Clone RMT 3–23 |
| mTim-3 | R&D Systems | Clone FAB1529 |
| Flag | BioLegend | Clone L5 |
| mPD-1 | BioLegend | Clone RMP1–30 |
| mIL-10 | BioLegend | Clone JES5–16E3 |
| mCD90.2 | BD Biosciences, BioLegend | Clone 532.1 |
| mCD45.2 | BioLegend | Clone 104 |
| Helios | BioLegend | Clone 22F6 |
| mKlrg-1 | BioLegend | Clone 2F1 |
| mLy6g | Tonbo | Clone IA8 |
| mGITR | eBioscience | Clone DTA-1 |
| mCD4 | BD Biosciences, BioLegend, Tonbo | Clone RM-4–5 |
| mTIGIT | BioLegend | Clone 1G9 |
| mLAG-3 | Invitrogen | Clone C9B7W |
| mGARP | BioLegend | Clone F011–5 |
| phospho-S6 | BioLegend | Clone A17020B |
| phospho-ERK | BioLegend | Clone 4B11B69 |
| phospho-Akt | BD Biosciences | Clone M89–61 |
| phospho-MTOR | eBioscience | Clone MRRBY |
| phospho-STAT3 | BD Biosciences | Clone 4 |
| CD16/CD32 (Fc Block) | Tonbo | Clone 2.4G2 |
| mCD8a | BD Biosciences, BioLegend, Tonbo | Clone H35–17.2 |
| mCD19 | BD Biosciences, BioLegend, Tonbo | Clone ID3 |
| mCD11b | eBioscience | Clone M1/70 |
| mCD3 (purified) | BioLegend | Clone 2C11 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| CellTrace Violet | Invitrogen | Cat#C34571 |
| FoxP3 Staining Kit | eBioscience | Cat#00–5523-00 |
| GhostDye-Violet510 | Tonbo Biosciences | Cat#13–0870-T500 |
| RBC lysis buffer | eBioscience | Cat#00–4333-57 |
| Collagenase D | Roche | Cat#11088866001 |
| DNase I, type IV | Sigma-Aldrich | Cat#D5025 |
| Hyclone Fetal bovine serum | Cytiva | Cat#SH30071 |
| Tamoxifen | MP Biomedical | Cat#02156738-CF |
|
| ||
| Deposited data | ||
|
| ||
| RNA sequencing of Tim-3 Tg Treg and WT Treg | Data obtained for this study | GEO record GSE155825 |
| single-cell RNA sequencing of HNC tumors | Cillo et al. (2020) | GEO record GSE139324 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| MC38 tumor line | Dario Vignali, Univ. of Pittsburgh | RRID:CVCL_B288 |
| B16-F10 tumor line | Dario Vignali, Univ. of Pittsburgh | RRID:CVCL_0159 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: FoxP3eGFP-Cre-ERT2: Foxp3tm9(EGFP/cre/ERT2)Ayr | The Jackson Laboratory | Cat#016961 |
| Mouse: Tim-3fl/fl: B6-Havcr2fl/fl | Developed with the Pitt Department of Immunolgy Transgenic and Gene Targeting Core for this study | N/A |
| Mouse: FSF-Tim-3: B6-Rosa26flox-stop-flox-Flag-Tim-3 | Developed for the investigators by GenOway | Avery et al. (2018) |
| Mouse: C57BL/6J | The Jackson Laboratory | Cat#000664 |
|
| ||
| Software and algorithms | ||
|
| ||
| FlowJo | BD Biosciences | version 10 |
| Prism | GraphPad | version 9 |
| FastQC | Babraham Bioinformatics | version 0.11.7 |
| Cutadapt | https://cutadapt.readthedocs.io/en/stable/ | version 1.18 |
| HISAT2 | http://daehwankimlab.github.io/hisat2/main/ | version 2.1.0 |
| SAMtools | http://www.htslib.org/ | version 1.9 |
| HTSeq | https://htseq.readthedocs.io/en/master/ | version 0.11.2 |
| Bioconductor edgeR | https://bioconductor.org/packages/release/bioc/html/edgeR.html | version 3.24.1 |
| Enrichr | https://maayanlab.cloud/Enrichr/ | Kuleshov et al., 2016 |
| Scanpy | https://github.com/theislab/scanpy | Wolf et al., 2018 |