Abstract
We report a simple method for tailoring the size of in-plane nanopores fabricated in thermoplastics for single-molecule sensing. The in-plane pores were fabricated via nanoimprint lithography (NIL) from resin stamps, which were generated from Si masters. We could reduce the size of the in-plane nanopores from 30 to ∼10 nm during the thermal fusion bonding (TFB) step, which places a cover plate over the imprinted polymer substrate under a controlled pressure and temperature to form the relevant nanofluidic devices. Increased pressures during TFB caused the cross-sectional area of the in-plane pore to be reduced. The in-plane nanopores prepared with different TFB pressures were utilized to detect single-λ-DNA molecules via resistive pulse sensing, which showed a higher current amplitude in devices bonded at higher pressures. Using this method, we also show the ability to tune the pore size to detect single-stranded (ss) RNA molecules and single ribonucleotide adenosine monophosphate (rAMP). However, due to the small size of the pores required for detection of the ssRNA and rAMPs, the surface charge arising from carboxylate groups generated during O2 plasma oxidation of the surfaces of the nanopores to make them wettable had to be reduced to allow translocation of coions. This was accomplished using EDC/NHS coupling chemistry and ethanolamine. This simple modification chemistry increased the event frequency from ∼1 s−1 to >136 s−1 for an ssRNA concentration of 100 nM.
Keywords: nanofluidics, nanopore, thermoplastics, thermal fusion bonding, single-molecule sensing
Graphical Abstract
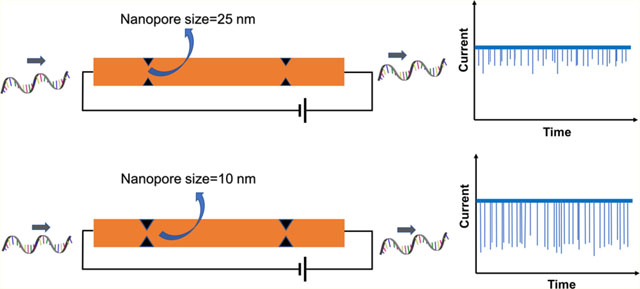
Pores with nanometer dimensions are typically fabricated in a thin membrane separating two fluid chambers.1 When an electrical bias is applied across the membrane, the resulting electric field can transport charged molecules through the pore producing signals in the transmembrane ionic current that can be used to detect single molecules via resistive pulse sensing (RPS). Among single-molecule sensors, nanopores have garnered significant interest because they allow the detection of single molecules without requiring fluorescence labeling of the target and the need for sophisticated optical equipment for transduction.2–5 These and other attractive properties have led to the development of many nanopore-based applications including detection of DNA-protein interactions,6–9 measurement of molecular forces,10,11 and nucleic acid sequencing.12,13 Although biological pores, such as α-hemolysin14 and MspA,15 have proven to be useful sensors, several disadvantages remain primarily due to their fixed size and limited stability under extreme conditions of salt, pH, temperature, and mechanical stress. As an alternative, solid-state nanopores3,16 have captured attention to address challenges associated with biological pores. Moreover, solid-state nanopores can be integrated with other micro- and nanofluidic components to form lab-on-a-chip systems.
Most solid-state nanopores have been fabricated on inorganic thin-film membranes.3,17–19 Several approaches have been demonstrated to produce small nanopores in these substrates using charged particle beams,17,18,20,21 electrical breakdown,22 and to control the size of the pores ex post facto via exposure with a defocused beam of electrons,17 ions,23 direct thermal heating,24 or focused ion beam (FIB) deposition of materials such as gold.25 Even though these methods have proven successful in the fabrication of small-diameter pores, they are generally not conducive to production at a scale and cost that will ultimately enable them to be translated for clinical applications that require disposable devices as is necessary for in vitro diagnostics.
Solid-state nanopores have also been fabricated in planar substrates (“in-plane” nanopores) embedded within a fluidic network rather than suspended on a thin membrane.26 FIB has been used to fabricate monolithic channels with micro- and nanoscale components including in-plane nanopores.26–28 In-plane pores can also be produced in series so that other measurement modalities can be realized. For example, Kondylis et al. used glass nanopore devices with two, four, and eight pores (width: 60 nm and depth: 70 nm) in series for real-time, resistive pulse analysis of viral capsids. They showed that the standard deviation of the pulse amplitude distributions of individual molecules decreased with increasing number of pores in series leading to increased measurement precision,29 while the electrophoretic mobility of virus particles was determined.30–32
Thermoplastics provide the means for both medium and high-scale manufacturing at low production costs even at the nanoscale due to a plethora of fabrication technologies, such as nanoimprint lithography (NIL) and injection molding, respectively.33,34 Additionally, due to the diverse physiochemical properties of different thermoplastics, the appropriate material can be selected according to measurement requirements.35,36 However, it has been difficult to achieve sub-20 nm structures using thermoplastics due to challenges associated with bonding a cover plate to the nanofluidic network, which can result in deformation of the patterned nanostructures. The cover plate bonding process in thermoplastic devices can use thermal fusion bonding (TFB), which bonds a thin cover plate to the nanopatterned substrate under a controlled pressure at temperatures near the glass transition temperature, Tg, of the substrate and/or cover plate. The TFB process involves motion of polymer chains between the cover plate and the substrate, which inevitably alters the dimensions of the nanostructures in the enclosed nanofluidic devices. We have shown that high process yield rates of thermoplastic nanofluidic devices with minimal deformation of nanostructures can be realized using a hybrid bonding process in which a lower Tg cover plate is thermally fusion-bonded to a higher Tg substrate.37
The ability to control the size of in-plane nanopores imprinted from the same mold allows for reduction in the development and production costs by obviating the need for FIB-milled Si masters to accommodate a particular application, for example, reducing in-plane nanopore size to sense molecules of various sizes. Several reports have demonstrated reduction of nanostructure dimensions by applying pressure to the patterned polymer substrate at elevated temperatures.38 For example, Choi et al. reduced the size of micropores in a perforated SU-8 membrane produced by NIL from 3000 nm to 300 nm.39 The same group utilized polymer reflow to reduce the nanopore size from 12 to 6 nm.34 In another report, Chou et al. described the use of a method called pressed self-perfection by liquefaction (P-SPEL), where the transiently molten thermoplastic nanostructures were pressed using a blank Si plate to achieve sub-20 nm structures.38 However, these methods have not been demonstrated to reduce the in-plane nanopore size for nanofluidic devices to sense differently sized molecules.
The transport properties of biomolecules through nanopores depend on interactions of analytes with the nanopore’s surface.36,40 Several reports have discussed the functionalization of pore surfaces to facilitate transport or other properties. For example, Martin et al. reported a method to alter the surface properties of track-etched nanopores in polycarbonate with gold by electroless deposition41 followed by chemisorption of thiols.42,43 For polyimide (PI) and polyethylene terephthalate (PET), the surface carboxyl groups created during track etching were chemically functionalized with an alkyl bromide and KF catalyst,44–46 methylation,47 or amidation.48,49 Previously, our group reported surface modification of PMMA nanochannels to generate both negatively charged and positively charged surfaces.50 A negatively charged surface was generated via O2 plasma treatment, which forms carboxyl groups on the thermoplastic surface. These surface-confined carboxyl groups were subsequently converted into positively charged surfaces by covalently attaching ethylenediamine.
In this study, we demonstrate a post fabrication method to tailor the dimensions of in-plane nanopores in enclosed nanofluidic devices using TFB, a process step needed to produce enclosed nanofluidic devices. Thermoplastic dual in-plane nanopore devices were fabricated in either a poly(methyl methacrylate) (PMMA) or cyclic olefin polymer (COP) substrate, and the O2 plasma-activated imprinted substrates and cover plates (made from COC; cyclic olefin copolymer), which were used to increase the wettability of the surfaces by the formation of surface carboxylate groups, were subjected to different bonding pressures to vary the size of the in-plane nanopores. The change in depth and width of the nanopores with bonding pressure was measured by AFM and SEM, respectively. COMSOL simulations and experimental conductance measurements further demonstrated the pore closing behavior of the nanopores with higher bonding pressures. The devices bonded at different pressures were used to analyze λ-DNA and showed an improved signal-to-noise ratio (SNR) with a lower nanopore size. Furthermore, the use of nanopore devices with different pore sizes fabricated by changing the bonding pressure during TFB was used to analyze different types of molecules, such as single-stranded (ss) RNA and ribonucleotide monophosphate (rAMP) molecules.
Moreover, to reduce coion exclusion effects leading to suboptimal event frequency, a simple surface modification step was carried out using ethanolamine on assembled devices. The high carboxyl group density generated during O2 plasma treatment prior to TFB created a high surface charge, which led to exclusion of coions passing through small nanopores. To alter the surface charge, EDC/NHS chemistry with ethanolamine was used.50 Dual in-plane nanopore devices modified with ethanolamine showed a significant increase in translocation event frequency.
MATERIALS AND METHODS
Reagents and Materials.
For a full list of reagents and materials used, please see Supporting Information.
Device Fabrication and Assembly.
Thermoplastic nanofluidic devices were fabricated and assembled as we have previously reported.33,51 A detailed explanation on device fabrication is provided in Supporting Information.
Atomic Force Microscopy.
To determine the depth of the nanopores with increasing pressure, AFM (SPM HT-9700, Shimadzu Corporation, Kyoto, Japan) analysis was carried out. The probe used for imaging was a SuperSharpSilicon tip (Nanosensors, Switzerland) with a tip radius of <2 nm, a half cone angle of 10°, an aspect ratio of 4:1 at 200 nm from the tip apex, and a frequency of 300 kHz. A dynamic scanning mode was used for imaging with a scanning frequency of 0.5 Hz. The acquired images were analyzed using SPM Manager v4.76.1 software.
Scanning Electron Microscopy.
SEMs of the nanopore devices were acquired using an FEI VERSA 3D Dualbeam field emission/low vacuum SEM instrument. A 2 nm-thin conductive iridium layer was sputter-coated onto the devices using an EMS 150ES sputter coater before SEM imaging. All images were acquired using a 5.0 kV accelerating voltage and 8.7 mm working distance. The SEM images of the Si mold masters were collected using a Quanta 3D DualBeam FEI FIB-SEM instrument and were analyzed using the instrument’s software and Image J.
COMSOL.
Simulations were performed in COMSOL v5.5 for the dual-nanopore devices. The length of both nanopores was kept at 30 nm, but the size (width and depth) was varied to calculate the corresponding conductance. The electrolyte used was 1 M KCl with a DC applied voltage of −1 V at 293 K with the electrostatics module used to calculate the electric potential, current density, and conductance across the pores.
Conductance Measurements.
Experimental conductance measurements were performed using the dual in-plane nanopore devices made in PMMA and COP bonded at different pressures using 1 M KCl as the electrolyte. Assembled devices were filled with 50% v/v methanol/water for 5 min. The 50% methanol solution was then replaced with 1 M KCl and allowed to equilibrate for 15 min. The device was placed in a Faraday cage, and Ag/AgCl electrodes were placed in the reservoirs filled with buffer. The current was measured from −1 to 1 V in 0.2 V steps. Current data were acquired using an Axopatch Digidata 1440B instrument and analyzed using Clampfit 11.1. The current measurements corresponding to the applied voltages were measured for different devices (n ≥ 3) at each bonding pressure, and the conductance was calculated.
λ-DNA, RNA, and rNMP Translocation.
Translocation experiments were performed for λ-DNA in PMMA dual in-plane nanopore devices bonded at 110, 170, and 200 psi. Briefly, after methanol/water priming, 1× TBE buffer was introduced into the device. Finally, 100 nM λ-DNA in 1 M KCl seeded into 1× TBE was injected into the device. The devices were placed in a Faraday cage, and Ag/AgCl electrodes were immersed in the reservoirs of the device. A potential of −1 V was applied between two electrodes, and the data were acquired using the Axopatch Digidata 1440B and analyzed using Clampfit 11.1. The Wilcoxon p-test was used to calculate statistical differences of peak amplitudes used for each TFB pressure.
For RNA and rAMP translocation, in-plane nanopore devices were primed as described above and 100 nM ssRNA (60 nt) in 1× NEBuffer 3 (10 mM NaCl; 5 mM Tris-HCl; 1 mM MgCl2; 0.1 mM DTT; pH 7.9 at 25 °C) was introduced into one of the reservoirs of the device. For rAMP in-plane nanopore measurements, the same carrier buffer was used and, in some cases, different concentrations of rAMP were employed. For RNA/rAMP translocation experiments, the applied potential was increased by serially connecting a 1.5 V battery to the Axopatch circuit, which increased the applied potential to 2.5 V compared to the 1 V maximum obtainable using the Axopatch instrument. Potentials were applied using Ag/AgCl electrodes, and all data were collected at a sampling frequency of 250 kHz, a head stage configuration of β = 0.1, gain = 1, and a low pass filter of 10 kHz. The nanofluidic devices were kept inside the Faraday cage while recording current transient data. Data were collected for a period of 10 min, and Clampfit 11.1 software was used for data acquisition and analysis.
Surface Modification with Ethanolamine.
After fabrication and assembly of the dual in-plane nanopore devices and to suppress the surface charge and EOF of O2 plasma-activated surfaces, the devices were modified with ethanolamine (see Figure S1). Ethanolamine was covalently attached to surface-confined carboxylic acid groups using EDC/NHS coupling chemistry, which covalently attaches primary amine-containing molecules to carboxylated surfaces via the formation of an amide bond.52,53 PMMA substrates and COC cover plates were exposed to O2 plasma at 50 W for 1 min, which generated surface carboxylic acid groups to improve the wettability of the surface. Then, a buffered solution (0.1 M MES, pH 4.7) containing 100 mg of EDC, 10 mg of NHS, and 16 μL of ethanolamine was filled into the plasma-treated device and kept for 30 min at room temperature. After reaction, the device was washed with ultrapure water.
Statistical Analysis.
All reported data sets were compared by either the two-sided t-test or Wilcoxon signed rank test using R Studio v1.0.153 and R v3.5.1 software.
RESULTS AND DISCUSSION
Device Fabrication and Assembly.
Nanofluidic devices were fabricated in a thermoplastic using a method we have reported, which consists of making microstructures and nanostructures in Si masters followed by producing resin stamps via UV-NIL and production of the final device using thermal NIL (see Supporting Information for more details). 33,51 The in-plane nanopores were positioned at either end of a nanochannel, which was 5 μm in length and 50 nm × 50 nm in width and depth (Figure 1A). SEM of the resin stamp is shown in Figure 1B,54 and Figure 1C shows an imprinted substrate. The average height of the in-plane nanopores on the resin stamp was 30.3 ± 2.0 nm (n = 4), and the depth of the nanopores in the imprinted substrate was 29.6 ± 1.7 nm (n = 3).
Figure 1.
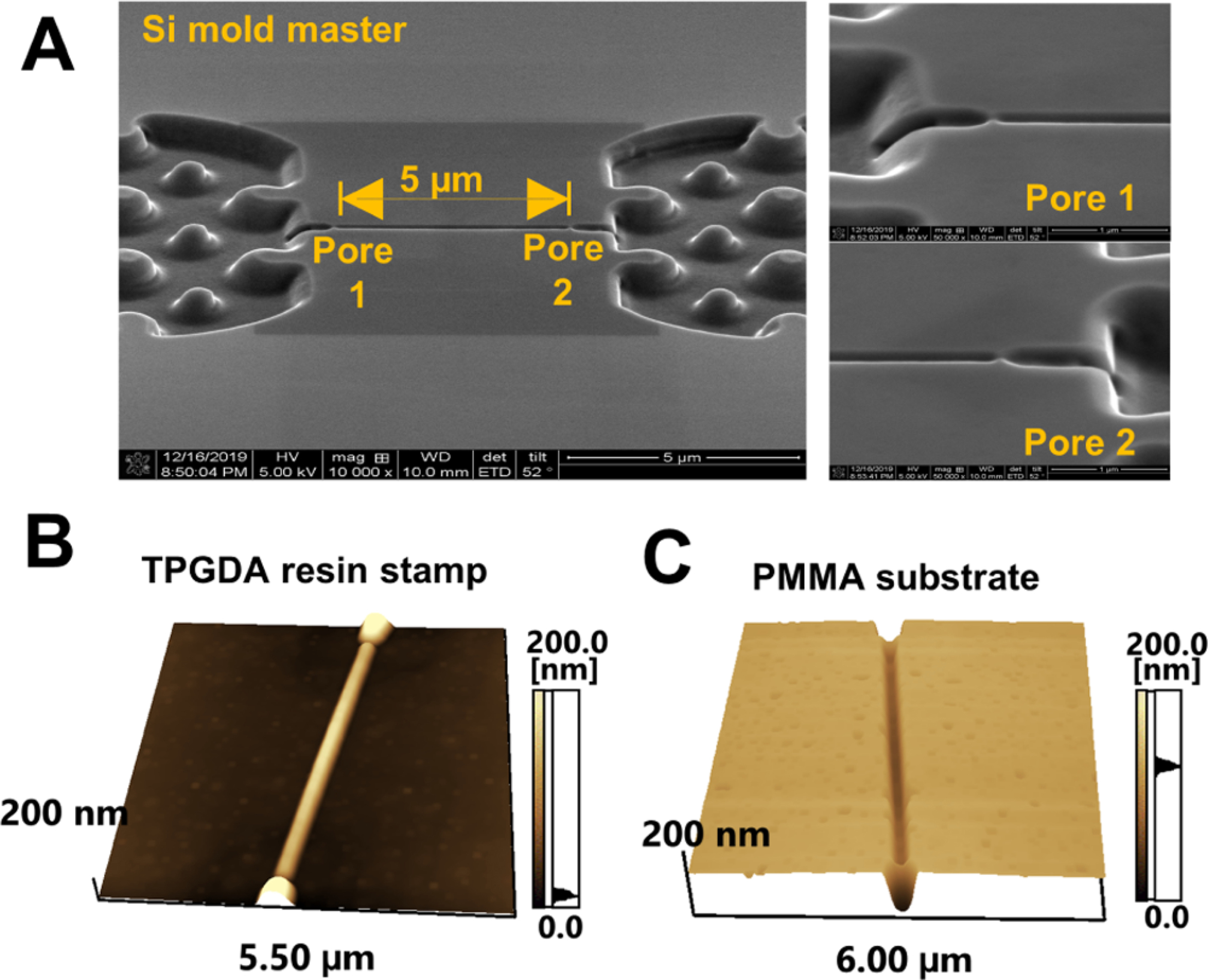
Dual in-plane nanopore device. (A) SEM image of the Si mold master. The two in-plane nanopores are 5 μm apart from each other. AFM scans of the (B) TPGDA resin stamp and (C) imprinted PMMA substrate. Tapping-mode AFM scans were acquired at 0.5 Hz scanning frequency using a high aspect ratio tip with a radius of <2 nm.
Following fabrication, the ability to control the depth and width of the in-plane nanopores via TFB was examined by subjecting NIL-imprinted devices to different bonding pressures at 70 °C for 15 min and measuring the depth and width of the in-plane nanopores using AFM and SEM, respectively. For this purpose, we only treated the imprinted PMMA substrate with O2 plasma and not the COC cover plate prior to TFB to reduce the bonding strength so that the cover plate and substrate could be pulled apart without damaging the underlying structures (see Figure S2A). Previously, we reported the bond strength of PMMA/COC devices to be 0.086 ± 0.014 mJ/cm2 using the crack opening method.55 In these experiments, the bond strength between the O2 plasma-treated substrate and untreated cover plate was 0 mJ/cm2 (i.e., no crack was measured), meaning that the cover plate could be removed without material removal or deformation of the nanostructures in the substrate. A TFB temperature of 70 °C was used as it was close to the Tg of the COC 8007 cover plate. For single-molecule translocation studies and RPS, the dual in-plane nanopore devices were assembled by O2 plasma treatment of the substrate and cover plate before TFB at varying pressures (see Figure S2B) so that the bond strength was sufficient to sustain fluidic/translocation experiments. The surface roughness of the COC cover plate as determined by AFM was <1 nm, which was much smaller than the nanopore depth.
The PMMA substrate was exposed to O2 plasma to form oxygen-containing groups.35,53,56–58 In TFB, these oxygen-containing species are involved in bond formation between the substrate and cover plate.35 In addition, polymer chain scissioning can result in photofragments that are thermally mobile due to their low molecular weight. This leads to a lowering of the Tg of the polymer on the surface, making it easier for the polymer chains to fuse into the mating substrate.53 Depending on the type of polymer and the O2 plasma power used and exposure time, polymer chain scission can occur up to several molecular layers into the bulk of the polymer. Therefore, during TFB of the cover plate to the substrate, the O2 plasma-activated polymer surfaces can rearrange, leading to a change in nanostructure dimensions.
Nanopore Size Analysis.
The depth of the in-plane nanopores was measured by dynamic mode AFM at a 0.5 Hz scanning rate (Figure 2A). To measure the width of the in-plane nanopores following TFB, SEM was performed (see Figure S3). The depth of the in-plane nanopores reduced from 22.3 ± 1.4 nm (110 pressure, n = 6) to 10.2 ± 1.5 nm (200 psi, n = 4) with increasing bonding pressure used for TFB (Figure 2B). The relative width of the in-plane nanopores decreased initially with bonding pressure to 0.47 ± 0.04 (n = 4) at 130 psi compared to the imprinted device but showed no statistically different widths at higher pressures (130–200 psi, p > 0.05, see Figure 2C). SEM and AFM images did not show statistically significant changes in the nanopore length before and after TFB. However, the overall cross-sectional area of the nanopores decreased with increasing pressure, demonstrating the pore closing behavior (Figure 2D). Our in-plane nanopores in the imprinted PMMA are a U-shaped constriction attributed to the nature of the FIB milling process in the Si master.
Figure 2.
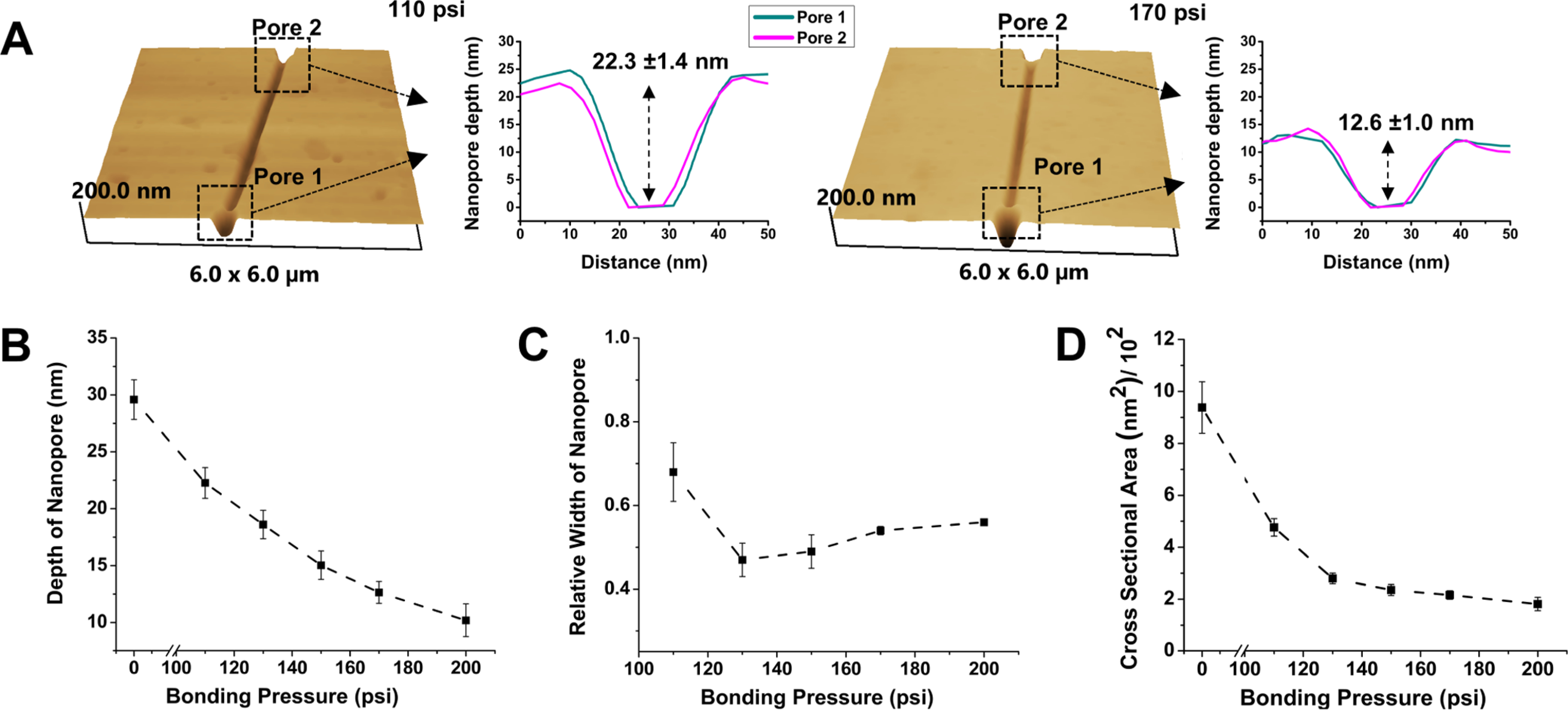
Nanopore depth and width with varying thermal fusion bonding pressure. (A) AFM scans of PMMA devices at 110 and 170 psi bonding pressures. (B) Change in the depth of the in-plane nanopores with bonding pressure. (C) Relative width of the in-plane nanopores after bonding at different pressures relative to the width of the nanopore before bonding (0 psi). There was no statistical difference in relative width from 130 to 200 psi at the 95% confidence interval (p > 0.05). (D) Cross-sectional area of the in-plane nanopore with thermal fusion bonding pressure.
The results of the pore depth (Figure 2B) and width (Figure 2C) versus the bonding pressure provided a hint on the pore closing behavior. The decrease in both the pore depth and width in the low bonding pressure range can be attributed to the fusion of polymer chains between the two mating polymers, primarily in the thin layer of the polymer subjected to O2 plasma prior to bonding and to the squeeze flow toward the nanopores. As the bonding pressure increases, the lateral squeeze flow will preferentially flow along the border of nanopores as was observed during the squeeze flow into hydrophilic nanostructures59,60 and thus further reduce the pore depth while limiting the decrease in the pore width. Consequently, the nanopore bonded at higher pressures became a shallower U shape. At the same time, the decrease in the thickness of the surface-modified thermoplastic layer further limits the squeeze flow because of increases in the Tg for polymers in the vicinity of a hydrophilic surface due to the alignment of polymer chains along the surface.61 This may account for the slight increase in the pore width at higher bonding pressures. Further increases in the bonding pressure beyond 200 psi ultimately led to collapse of the nanopores, as evidenced by the cessation of the open pore current.
COMSOL Simulations and Conductance Measurements of Devices.
We carried out COMSOL simulations of the dual in-plane nanopore sensor to estimate the change in the conductance with the change in pore size. The length of the nanopore was maintained at 30 nm, but the width was changed from 10 to 50 nm (see Figure S4A). A DC bias of −1 V was applied across the ends of the device, and 1 M KCl was used as the electrolyte. From Figure S4B, it can be seen that the majority of the potential drop occurred across the two in-plane nanopores (0.03 V in each pore) and through the 5 μm-long nanochannel. A current density graph was plotted that showed a sharp increase in current density at the nanopores due to their small dimensions (Figure S4C). The current density was integrated over the area of the nanopore to calculate the current. The conductance of the pores was then calculated using Ohm’s law and was plotted to estimate the conductance at different pore sizes. As shown in Figure S4D, a linear increase in conductance was seen as the size of the nanopore increased. The conductance values obtained from simulations and depths/widths from AFM and SEM, respectively, were used to estimate the size of the nanopores from conductance values obtained experimentally.
Experimental conductance measurements were performed using the dual in-plane nanopore devices made in PMMA and COP bonded at different pressures using 1 M KCl as the electrolyte and COC as the cover plate. The conductance was calculated from the slope of the curve for all bonding pressures and is shown in Figure S4E. The average conductances at 110 psi for PMMA and COP devices were 83 ± 29 nS (RSD 34%) and 128 ± 89 nS (RSD 69%), respectively. However, at 130 psi, the average conductance of the PMMA and COP dual in-plane nanopore devices was 23 ± 6 nS (RSD 26%) and 21 ± 16 nS (RSD 76%), respectively. These values correlate well with the conductance (∼23 nS) obtained from COMSOL for an 18 nm pore, which is the size of the pore obtained from AFM and SEM when using 130 psi TFB pressure. The conductance values showed a slight increase from 24 ± 5 nS (RSD 22%) to 33 ± 6 nS (RSD 18%) for PMMA devices bonded at 150 and 170 psi, respectively, but there was no statistical difference in the conductance values at bonding pressures >130 psi (p > 0.05). Similarly, in the case of COP devices, the conductance values decreased at 150 psi to 9 ± 7 nS (RSD 77%) but showed a statistically insignificant increase at 170 psi to 12 ± 6 nS (RSD 48%). This small increase in conductance can be correlated to the results from SEM and COMSOL that showed a <5 nm increase in pore width. Overall, both PMMA and COP devices followed a similar trend of decrease in conductance at 130 and 150 psi followed by a slight increase at 170 psi, showing the reproducibility of our approach with different substrate materials.
λ-DNA Translocation through the Dual In-Plane Nanopores.
Assembled dual in-plane nanopore devices could be used to electrokinetically drive charged single molecules through the nanopores. Based on our results from COMSOL simulations and experimental conductance measurements, it was evident that the increase in bonding pressure caused a drop in conductance, which indicated reduction in pore size. To further reaffirm that the pore size indeed decreased with increasing bonding pressure, we used λ-DNA and electrokinetically translocated them through the nanopore devices under different bonding conditions to estimate the current blockage amplitudes as a function of pore size.9,62,63 Figure 3 shows the use of dual in-plane nanopore devices as a sensing platform for the detection of 48.5 Kbp λ-DNA that has a contour length of 16.5 μm. When the electrokinetically driven molecule entered the first nanopore, there was a partial current blockage creating a transient increase in the electrical resistance, which manifested itself as a negative peak (current drop) in the measured trace, as shown in the schematic of Figure 3A. Because the contour length of λ-DNA is longer than the distance between the two in-plane nanopores (5 μm), the initial drop of current was accompanied by a subsequent drop in current when the DNA coresided in the first and second in-plane nanopores. Furthermore, when the DNA exited the first nanopore, it was resident only in the second nanopore, causing a subsequent small shoulder in the current trace and eventually returning to the baseline. A current trace of multiple current transient signals over a time interval of 400 s for λ-DNA is shown in Figure 3B, demonstrating the characteristic shape of the peaks at 110 and 170 psi. The various stages of the translocation are marked in both traces to show how the shape of the current transients agrees with the aforementioned description. Although the average amplitude increased with every increase in bonding pressure, the shape of the peaks remained similar to that shown in Figure 3B, indicating that the size of the nanopore did not alter the translocation dynamics rather changed only the SNR of the signal.
Figure 3.
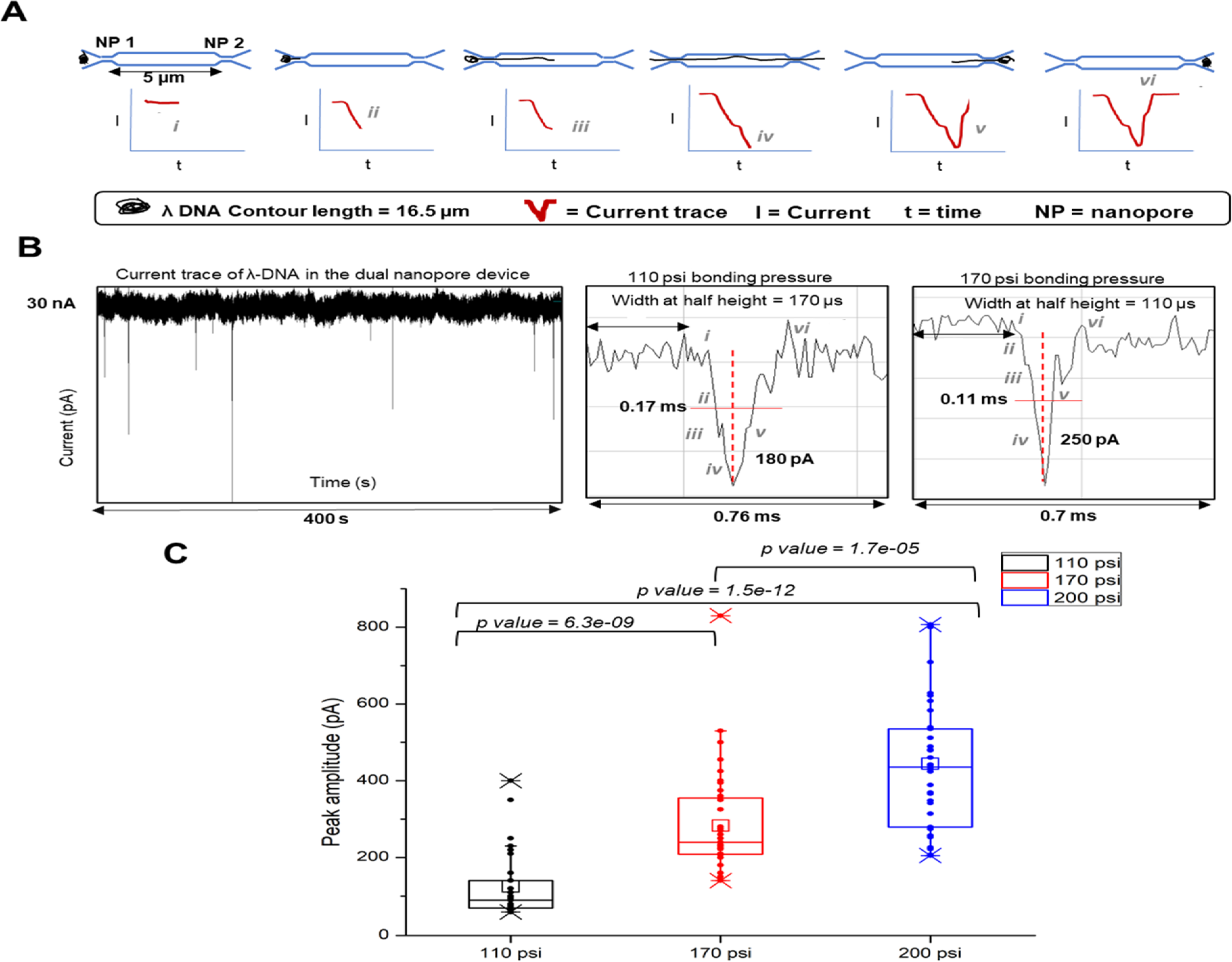
λ-DNA translocation through the dual in-plane PMMA nanopores and the ramifications of the size of the nanopore on peak amplitude. (A) Schematic of the λ-DNA translocation through the in plane dual nanopore device that gives rise to a negative peak as the DNA enters the first pore. Since the contour length of the DNA is longer than both the pores, there is a second subsequent peak when the DNA coresides in both the pores. The DNA then leaves the pores very quickly which makes the current return to the baseline. (B) Detected current transient trace typically observed in a time interval of 400 s as a result of λ-DNA translocation and magnified images of individual peak shapes at various translocation stages of the DNA through the dual nanopore at 110 and 170 psi pressure, respectively. (C) Distribution of peak amplitudes of λ-DNA at 110, 170, and 200 psi bonding pressures. The average peak amplitude increases with the increasing bonding pressure. p values calculated between each bonding pressure condition (Wilcoxon signed rank test) show statistically a significant difference at 95% confidence interval (p < 0.05).
Peak height measurements (n ≥ 120) of the current transient amplitudes of λ-DNA in devices bonded at different pressures showed differences between each bonding pressure as determined by the Wilcoxon signed rank test (Figure 3C). The peak amplitudes were collected from >10 devices for each bonding pressure as we were only able to see ∼6 events per device. The average peak amplitude of λ-DNA in devices bonded at 110 psi for 15 min was 130 pA corresponding to a pore depth of ∼22 nm and a width of ∼21 nm. Devices bonded at 170 psi for 15 min having a pore depth × width of 13 nm × 17 nm yielded a current amplitude of 280 pA, and devices bonded at 200 psi for 5 min having a depth × width of 10 nm × 18 nm yielded a current amplitude of 437 pA. There was a 3.5-fold increase in current amplitude when the pore dimensions decreased from 22 nm (depth) × 21 nm (width) to 10 nm (depth) × 18 nm (width). Devices bonded at 200 psi for 15 min served as the upper limit because the nanopore became unusable as the cover plate collapsed into the nanopore. However, for 200 psi bonding pressure, the TFB time could be reduced to 5 min to generate functional devices.
We calculated the apparent mobility (μapp) of λ-DNA based on the time interval between peaks (Δt) at time points (i) and (iv) (see Figure 3A). These time points were used because one end of λ-DNA was entering the first in-plane nanopore and that same end was then entering the second nanopore corresponding to a length of 5 μm (l) at the applied electric field strength (E). The average apparent mobility for λ-DNA was determined to be (2.57 ± 0.94) × 10−7 m2/V s.
ssRNA Translocation through O2 Plasma-Modified PMMA Dual In-Plane Nanopore Devices.
To further demonstrate the use of the dual in-plane nanopores for detection of various sized molecules, we carried out translocation experiments of 60 nt ssRNA (Rg ∼6 nm) using the dual in-plane nanopore devices bonded at 170 psi, which resulted in a pore depth × width of 12.6 ± 0.9 nm × 17.1 ± 0.2 nm. The assembled PMMA/COC dual in-plane nanopore devices were primed with 50% v/v methanol followed by introduction of 1× NEBbuffer 3 (Figure S5A in Supporting Information). Before introducing the ssRNA solution, the current was monitored to establish a baseline (Figure S5B in Supporting Information). Then, the buffer in one reservoir was replaced with 100 nM ssRNA solution and a potential (1–2.5 V) was applied.
For the 100 nM ssRNA solution, we only observed very few translocation events even after increasing the concentration to 1 μM and the driving voltage to −2.5 V (see Figure S5C in Supporting Information). Due to the low event frequency, a large number of devices would have to be used to collect a reasonable number of events to secure meaningful statistics as was carried out for the λ-DNA data shown in Figure 3. The low event frequency could have been due to coion exclusion due to the high surface charge of the O2 plasma-modified PMMA devices, which is reported as −40 mC/cm2.50 Due to the high negative charge on the nanopore surface and partial electric double layer overlap within the nanopore, the entry of negatively charged coions such as ssRNAs is impeded. In addition, the opposing EOF also serves to reduce event frequency.
Surface Modification with Ethanolamine.
The PMMA/ COC surfaces were modified with ethanolamine using EDC/ NHS coupling chemistry (see Figure S1 in Supporting Information). Sessile drop water contact angle measurements were acquired for native, O2 plasma-activated, and ethanolamine-treated PMMA surfaces with and without EDC/NHS. The contact angle dropped from 79.2 ± 1.8° for pristine PMMA to 42.3 ± 2.7° after O2 plasma activation, indicating the generation of surface carboxyl groups (Figure S6 in Supporting Information). These values agreed with values reported in the literature.55 After amidation of the carboxyl groups with ethanolamine via EDC/NHS coupling chemistry, the sessile drop water contact angle (53.4 ± 2.6°) slightly increased compared to the plasma-treated surface. However, in the absence of the EDC/NHS coupling reagents, no change in water contact angle was seen with respect to the O2 plasma-activated surface. As evident from the water contact angle measurements, the ethanolamine-treated surfaces remained hydrophilic compared to native PMMA due to the presence of terminal hydroxyl groups. The hydrophilic nature of the ethanolamine-treated surface is helpful in consistent filling of the nanofluidic device without generating air bubbles.
To examine the molecular nature of the modified and unmodified thermoplastic surfaces, ATR—FTIR experiments were performed (Figure S7A in Supporting Information). After treatment with ethanolamine, bands at 3396 and 1635 cm−1 corresponding to the v(N—H) stretch of a primary amine and v(C=O) of an amide appeared, respectively, which confirmed the successful addition of ethanolamine to the activated surface.
The surface charge density (σs) of the ethanolamine-modified PMMA surfaces was measured using conductance plots (Figure S7B in Supporting Information).50 For ethanolamine-modified PMMA nanochannels, σs was found to be −3.8 mC/m2, which was ∼10-fold less than −40.5 mC/m2 reported for O2 plasma-activated nanochannels.50 The EOF was also measured using a current monitoring method64 and showed a value of 3.63 × 10−5 cm2/V s after ethanolamine modification, which was approximately ninefold less than the value reported for O2 plasma-activated channels (Figure S7C in Supporting Information).65
ssRNA Translocation through Ethanolamine-Modified PMMA/COC Dual In-Plane Nanopore Devices.
After ethanolamine modification, ssRNA translocation was carried out. For these experiments, a 100 nM solution of 60 nt ssRNA in 1× NEBuffer 3 was introduced into the nanopore device, which used TFB at 170 psi and a potential of −2.5 V applied across the nanopores. As shown in Figure 4, after ethanolamine treatment, the event frequency increased significantly compared to the O2 plasma-treated nanopore devices, as noted in the data shown in Figure S5C (see Supporting Information). Only a single ssRNA event (concentration = 100 nM) was observed over a span of 900 ms for the O2 plasma-treated devices, while in the case of the ethanolamine-treated PMMA/ COC device, ∼34 single-molecule events (concentration = 100 nM) were observed over a time span of 250 ms. The average dwell time of the peaks obtained for single 60 nt ssRNA events was determined to be 0.92 ± 0.38 ms. The current transient amplitudes of the ssRNA events ranged between 0.10 and 0.8 nA with an average of 311.75 ± 137.49 pA (n = 325; see Figure S8A).
Figure 4.
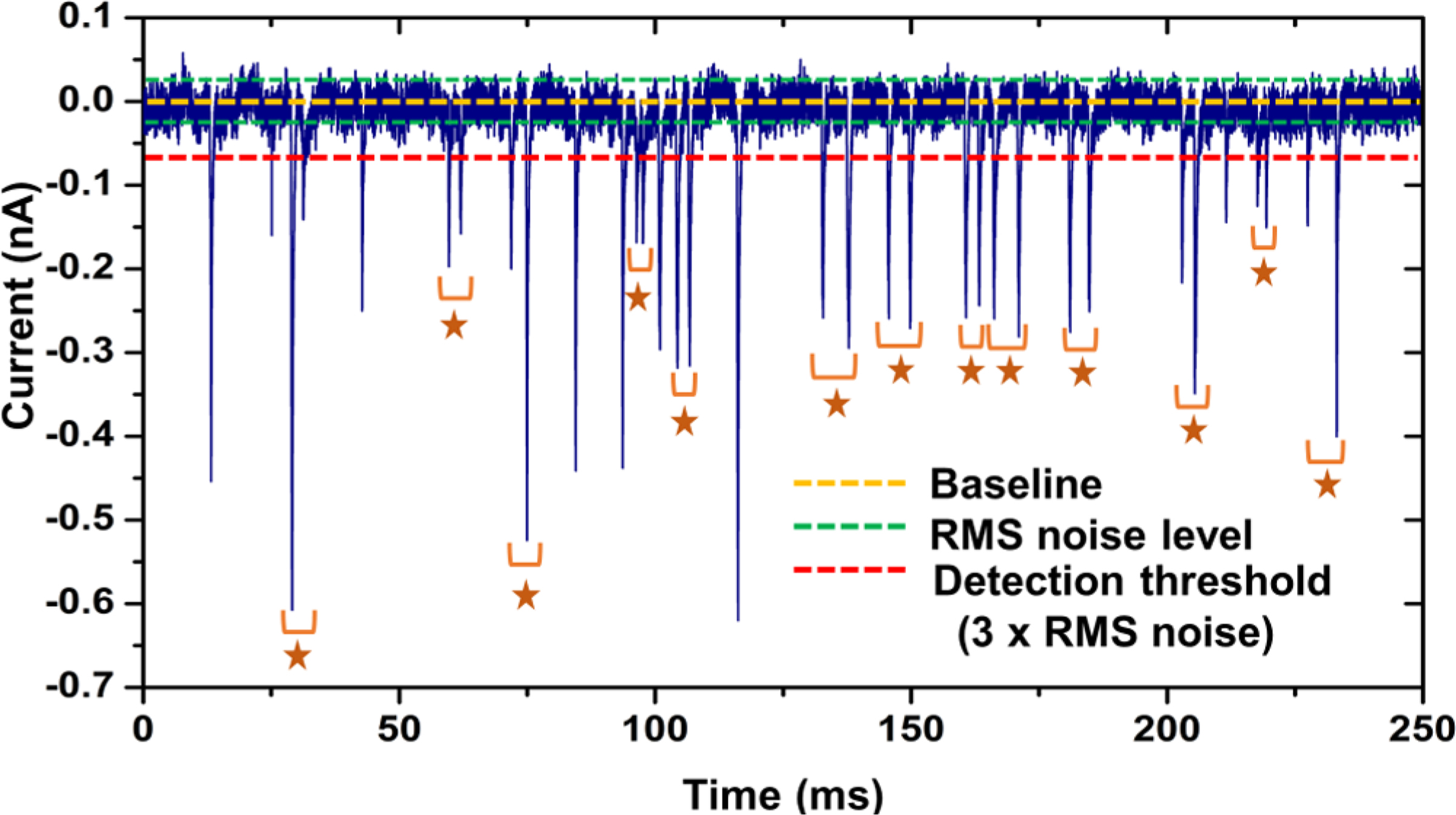
250 ms trace of the current transient amplitude signal obtained for 100 nM solution of 60 nt RNA obtained using dual in-plane nanopore devices bonded at 170 psi bonding pressure. The stars represent paired peaks which corresponded to a single RNA molecule translocating through both nanopores.
We also measured the time-of-flight (TOF) of the ssRNA, which corresponds to the time of the molecule to travel between the two pores. The TOF ranged from 1 to 4 ms with an average of 2.09 ± 0.97 ms (n = 51; see Figure S8B). An example peak pair is shown in Figure S8C in Supporting Information. In the 250 ms current trace shown in Figure 4, 76.5% (26/34) of the events corresponded to peak pairs.
rAMP Translocation through Ethanolamine-Modified PMMA/COC Dual In-Plane Nanopore Devices.
We next carried out experiments to detect single rAMP molecules using the PMMA/COC dual in-plane nanopore devices bonded at 200 psi for 5 min, which was used to create a smaller pore to accommodate the smaller size of the rAMP molecule compared to the ssRNA 60 nt molecule. Figure 5A shows the current traces for a blank, 10, 100 nM, and 1 μM solution of rAMP. With increasing concentration, a linear increase in event frequency was observed (R2 = 0.9757). An example peak pair obtained for rAMP translocation between the two pores in the series is shown in Figure 5B. The average current blockage amplitudes for rAMP was 425.89 ± 175.89 pA (n = 185; Figure 5C). The average dwell time of rAMPs within the nanopores was 0.31 ± 0.26 ms (n = 185; Figure 5D).
Figure 5.
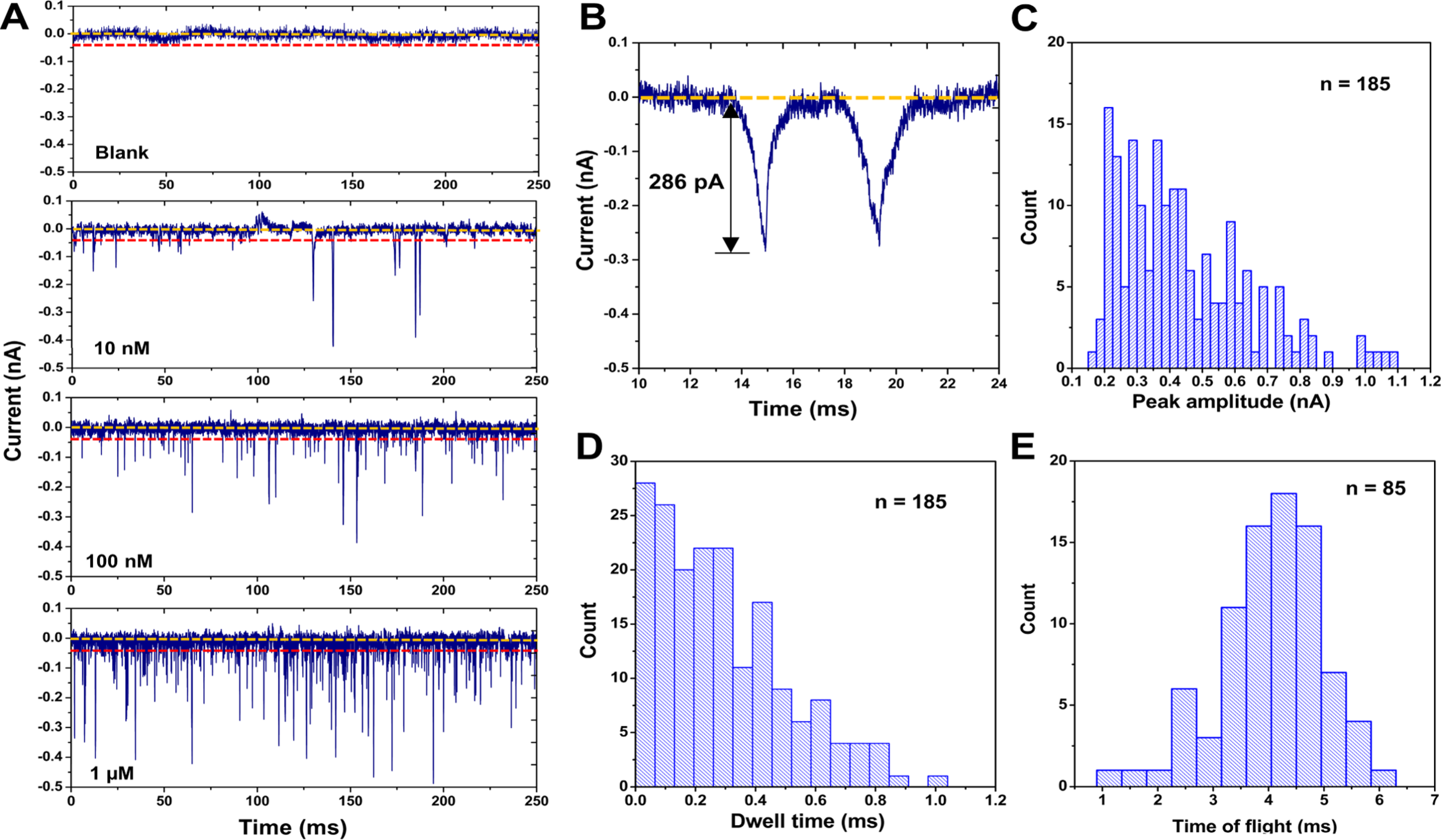
Translocation of rAMPs through dual in-plane nanopore devices assembled at 200 psi bonding pressure. (A) 250 ms current transient trace of signal amplitudes obtained for blank, 10, 100 nM, and 1 μM solutions of rAMP using dual in-plane nanopore devices. An increase in event frequency was observed with increasing concentration (R2 = 0.9757). (B) Example peak pair selected using the peak pair criteria. (C) Peak amplitude distribution of rAMP events. (D) Dwell time distribution for rAMP events. (E) TOF distribution for rAMP.
We also used the dual in-plane nanopore devices to measure the apparent electrophoretic mobility (i.e., time-of-flight, TOF) of rAMP using rigorous selection criteria.66 The first criterion was that the peak amplitude should be > 3× the RMS noise of the open pore current. The RMS noise of the open pore current of the 200 psi bonded device was found to be 19.6 pA, and therefore, only peaks with amplitudes >58.5 pA were considered as true events. The second criterion was the minimum TOF, where the travel time between the two pores (TOF) for the single molecule should be greater than the dwell time (peak width) of each peak comprising the peak pair. The third criterion was that the maximum TOF needed to be within 1.5 times the theoretical TOF. The maximum TOF (5.7 ms) was calculated using the mobility values we recently reported for ATTO-532-labeled rAMP.65 The average TOF for rAMP was 4.14 ± 0.97 ms (n = 85; Figure 5E). The percentage of peaks identified as paired events was 82, 63, and 64% for 10, 100 nM, and 1 μM rAMP solutions, respectively. For measurements performed with the rAMPs using the dual in-plane nanopore devices bonded at 110 psi, no current transients were observed (data not shown).
Unpaired peaks found in this work may be attributed to the relatively large pore size compared to that of rNMP and the irregular shape (shallow U-shape) of the in-plane nanopores, which resulted in a large variation in the peak amplitude, as seen in Figure 5. Thus, some peaks may not produce sufficient amplitudes to be selected as paired events and account for the unpaired peaks. Also, there may be cases where entry of a molecule into the first nanopore before the previous molecule left the second nanopore may lead to unpaired events, which can be seen in the decreased percentage of the paired events with increasing concentration. In previous work by Langecker et al.66 where double-stranded (ds) DNA was detected by stacked nanopores with diameters of 23 and 28 nm, 94% of detected peaks could unambiguously be assigned to the translocating dsDNA.
CONCLUSIONS
In this paper, we have shown the ability to tailor the size of an in-plane nanopore by TFB using different pressures during an assembly step already required for thermoplastic nanofluidic chip production. We noticed that the size and shape of the in-plane nanopore changed with different bonding pressures in a controlled fashion as measured using AFM and SEM. Furthermore, COMSOL simulations with varying pore widths gave the approximate conductance observed in the PMMA and COP dual in-plane nanopore devices connected using a 5 μm-long nanochannel. Furthermore, λ-DNA was electrokinetically driven by a DC voltage through the dual in-plane nanopore devices bonded at 110, 170, and 200 psi. The transient current amplitudes were seen to increase with higher TFB pressures. Therefore, thermoplastic nanofluidic devices allow for tuning the nanostructure size to accommodate a given application by simply altering device assembly conditions without requiring direct FIB milling of a new Si master. This was further demonstrated using 60 nt ssRNAs and rAMPs with devices bonded at 170 and 200 psi bonding pressure, respectively. We should note that we could produce >1000 resin stamps from a single Si master and from each resin stamp, >20 nanofluidic devices could be produced with a success rate of producing functional devices following TFB of the COC cover plate to the PMMA or COP substrate >90%.33,37,51 For high-scale production, preliminary data in our laboratories indicate that these devices can be made via injection molding, which will be the focus of a future report.
The dual in-plane nanopores with their associated fluidic network and the fact that they were fabricated in thermoplastics in a single step using NIL make it feasible to integrate this sensing technology into other nanofluidic components for the label-free identification of biomolecules. For example, we are currently developing a chip-based single-molecule exo-sequencing method, termed exonuclease time-of-flight (X-TOF).65,67–69 This method involves a solid-phase enzymatic reactor coupled to a nanoflight tube that contains dual in-plane nanopores to measure free nucleotides’ TOFs. Recently, we reported the use of solid-phase XRN1 reactions to sequentially produce rNMPs (5' → 3' direction).70 Previous work from our group also demonstrated the identification of labeled rNMPs via their molecular-dependent electrophoretic mobility (i.e., TOF) in thermoplastic nanochannels; we were able to achieve TOF identification accuracies of >99%.65
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank the NIH for funding of this work (NIBIB: P41 EB020594; NCI: P30 CA168524; and NIGMS: P20 GM130423). Finally, the authors would like to thank the Kansas University Nanofabrication Facility for assistance in making the nanofluidic devices and Dr. Prem Thapa from the Microscopy and Analytical Imaging facility at KU for helping with SEM imaging. The authors would also like to thank Collin McKinney and Matt Verber (CRITCL; UNC-Chapel Hill) for generating the current amplifier circuitry and the RPS data analysis software used in this work.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssensors.1c01359.
Reagents and materials used in this study; device fabrication; and the following figures—protocol for ethanol amine modification of PMMA/COC devices; procedure for securing depth/width of in-plane pores; SEM of in-plane pores; COMSOL simulations; RNA translocation of the O2 plasma-treated device; water contact angle results; surface characterization results; and RNA translocation results (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acssensors.1c01359
Contributor Information
Uditha S. Athapattu, Department of Chemistry, The University of Kansas, Lawrence, Kansas 66045, United States; Center of BioModular Multiscale Systems for Precision Medicine, The University of Kansas, Lawrence, Kansas 66045, United States.
Chathurika Rathnayaka, Department of Chemistry, The University of Kansas, Lawrence, Kansas 66045, United States; Center of BioModular Multiscale Systems for Precision Medicine, The University of Kansas, Lawrence, Kansas 66045, United States.
Swarnagowri Vaidyanathan, Center of BioModular Multiscale Systems for Precision Medicine and Bioengineering Program, The University of Kansas, Lawrence, Kansas 66045, United States.
Sachindra S. T. Gamage, Department of Chemistry, The University of Kansas, Lawrence, Kansas 66045, United States; Center of BioModular Multiscale Systems for Precision Medicine, The University of Kansas, Lawrence, Kansas 66045, United States.
Junseo Choi, Center of BioModular Multiscale Systems for Precision Medicine, The University of Kansas, Lawrence, Kansas 66045, United States; Mechanical & Industrial Engineering Department, Louisiana State University, Baton Rouge, Louisiana 70803, United States.
Ramin Riahipour, Center of BioModular Multiscale Systems for Precision Medicine, The University of Kansas, Lawrence, Kansas 66045, United States; Mechanical & Industrial Engineering Department, Louisiana State University, Baton Rouge, Louisiana 70803, United States.
Anishkumar Manoharan, Department of Chemistry, The University of Kansas, Lawrence, Kansas 66045, United States; Center of BioModular Multiscale Systems for Precision Medicine, The University of Kansas, Lawrence, Kansas 66045, United States.
Adam R. Hall, Wake Forest School of Medicine, Virginia Tech-Wake Forest University School of Biomedical Engineering and Sciences and Comprehensive Cancer Center, Winston-Salem, North Carolina 27101, United States.
Sunggook Park, Center of BioModular Multiscale Systems for Precision Medicine, The University of Kansas, Lawrence, Kansas 66045, United States; Mechanical & Industrial Engineering Department, Louisiana State University, Baton Rouge, Louisiana 70803, United States.
Steven A. Soper, Department of Chemistry, The University of Kansas, Lawrence, Kansas 66045, United States; Center of BioModular Multiscale Systems for Precision Medicine, Bioengineering Program, and Department of Mechanical Engineering, The University of Kansas, Lawrence, Kansas 66045, United States; KU Cancer Center, University of Kansas Medical Center, Kansas City, Kansas 66160, United States.
REFERENCES
- (1).Branton D; Deamer DW; Marziali A; Bayley H; Benner SA; Butler T; Di Ventra M; Garaj S; Hibbs A; Huang X; Jovanovich SB; Krstic PS; Lindsay S; Ling XS; Mastrangelo CH; Meller A; Oliver JS; Pershin YV; Ramsey JM; Riehn R; Soni GV; Tabard-Cossa V; Wanunu M; Wiggin M; Schloss JA The potential and challenges of nanopore sequencing. Nat. Biotechnol. 2008, 26, 1146–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Wanunu M; Sutin J; McNally B; Chow A; Meller A DNA translocation governed by interactions with solid-state nanopores. Biophys. J. 2008, 95, 4716–4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Dekker C Solid-state nanopores. Nat. Nanotechnol. 2007, 2, 209–215. [DOI] [PubMed] [Google Scholar]
- (4).Venkatesan BM; Bashir R Nanopore sensors for nucleic acid analysis. Nat. Nanotechnol. 2011, 6, 615–624. [DOI] [PubMed] [Google Scholar]
- (5).Smeets RMM; Keyser UF; Krapf D; Wu M-Y; Dekker NH; Dekker C Salt dependence of ion transport and DNA translocation through solid-state nanopores. Nano Lett. 2006, 6, 89–95. [DOI] [PubMed] [Google Scholar]
- (6).Carlsen AT; Zahid OK; Ruzicka JA; Taylor EW; Hall AR Selective detection and quantification of modified DNA with solid-state nanopores. Nano Lett. 2014, 14, 5488–5492. [DOI] [PubMed] [Google Scholar]
- (7).Kowalczyk SW; Hall AR; Dekker C Detection of local protein structures along DNA using solid-state nanopores. Nano Lett. 2010, 10, 324–328. [DOI] [PubMed] [Google Scholar]
- (8).Plesa C; Ruitenberg JW; Witteveen MJ; Dekker C Detection of individual proteins bound along DNA using solid-state nanopores. Nano Lett. 2015, 15, 3153–3158. [DOI] [PubMed] [Google Scholar]
- (9).Smeets RMM; Kowalczyk SW; Hall AR; Dekker NH; Dekker C Translocation of RecA-coated double-stranded DNA through solid-state nanopores. Nano Lett. 2009, 9, 3089–3095. [DOI] [PubMed] [Google Scholar]
- (10).Hornblower B; Coombs A; Whitaker RD; Kolomeisky A; Picone SJ; Meller A; Akeson M Single-molecule analysis of DNA-protein complexes using nanopores. Nat. Methods 2007, 4, 315–317. [DOI] [PubMed] [Google Scholar]
- (11).Tabard-Cossa V; Wiggin M; Trivedi D; Jetha NN; Dwyer JR; Marziali A Single-molecule bonds characterized by solid-state nanopore force spectroscopy. ACS Nano 2009, 3, 3009–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Cherf GM; Lieberman KR; Rashid H; Lam CE; Karplus K; Akeson M Automated forward and reverse ratcheting of DNA in a nanopore at 5-Å precision. Nat. Biotechnol. 2012, 30, 344–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Ricroch AE Assessment of GE food safety using “-omics” techniques and long-term animal feeding studies. New Biotechnol. 2013, 30, 349–354. [DOI] [PubMed] [Google Scholar]
- (14).Song L; Hobaugh MR; Shustak C; Cheley S; Bayley H; Gouaux JE Structure of Staphylococcal alpha -Hemolysin, a Heptameric Transmembrane Pore. Science 1996, 274, 1859–1865. [DOI] [PubMed] [Google Scholar]
- (15).Faller M; Niederweis M; Schulz GE The structure of a mycobacterial outer-membrane channel. Science 2004, 303, 1189–1192. [DOI] [PubMed] [Google Scholar]
- (16).Plesa C; van Loo N; Dekker C DNA nanopore translocation in glutamate solutions. Nanoscale 2015, 7, 13605–13609. [DOI] [PubMed] [Google Scholar]
- (17).Storm AJ; Chen JH; Ling XS; Zandbergen HW; Dekker C Fabrication of solid-state nanopores with single-nanometre precision. Nat. Mater. 2003, 2, 537–540. [DOI] [PubMed] [Google Scholar]
- (18).Li J; Stein D; McMullan C; Branton D; Aziz MJ; Golovchenko JA Ion-beam sculpting at nanometre length scales. Nature 2001, 412, 166–169. [DOI] [PubMed] [Google Scholar]
- (19).Wanunu M; Meller A Chemically modified solid-state nanopores. Nano Lett. 2007, 7, 1580–1585. [DOI] [PubMed] [Google Scholar]
- (20).Gierak J; Madouri A; Biance AL; Bourhis E; Patriarche G; Ulysse C; Lucot D; Lafosse X; Auvray L; Bruchhaus L Sub-5 nm FIB direct patterning of nanodevices. Microelectron. Eng. 2007, 84, 779–783. [Google Scholar]
- (21).Yang J; Ferranti DC; Stern LA; Sanford CA; Huang J; Ren Z; Qin L-C; Hall AR Rapid and precise scanning helium ion microscope milling of solid-state nanopores for biomolecule detection. Nanotechnology 2011, 22, 285310. [DOI] [PubMed] [Google Scholar]
- (22).Kwok H; Briggs K; Tabard-Cossa V Nanopore fabrication by controlled dielectric breakdown. PLoS One 2014, 9, No. e92880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Stein DM; McMullan CJ; Li J; Golovchenko JA Feedback-controlled ion beam sculpting apparatus. Rev. Sci. Instrum. 2004, 75, 900–905. [Google Scholar]
- (24).Asghar W; Ilyas A; Billo J; Iqbal S Shrinking of solid-state nanopores by direct thermal heating. Nanoscale Res. Lett. 2011, 6, 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Schiedt B; Auvray L; Bacri L; Oukhaled G; Madouri A; Bourhis E; Patriarche G; Pelta J; Jede R; Gierak J Direct FIB fabrication and integration of “single nanopore devices” for the manipulation of macromolecules. Microelectron. Eng. 2010, 87, 1300–1303. [Google Scholar]
- (26).Harms ZD; Haywood DG; Kneller AR; Selzer L; Zlotnick A; Jacobson SC Single-particle electrophoresis in nanochannels. Anal. Chem. 2015, 87, 699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Henley WH; Dennis PJ; Ramsey JM Fabrication of microfluidic devices containing patterned microwell arrays. Anal. Chem. 2012, 84, 1776–1780. [DOI] [PubMed] [Google Scholar]
- (28).Menard LD; Ramsey JM Fabrication of sub-5 nm nanochannels in insulating substrates using focused ion beam milling. Nano Lett. 2011, 11, 512–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Kondylis P; Zhou J; Harms ZD; Kneller AR; Lee LS; Zlotnick A; Jacobson SC Nanofluidic Devices with 8 Pores in Series for Real-Time, Resistive-Pulse Analysis of Hepatitis B Virus Capsid Assembly. Anal. Chem. 2017, 89, 4855–4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Harms ZD; Mogensen KB; Nunes PS; Zhou K; Hildenbrand BW; Mitra I; Tan Z; Zlotnick A; Kutter JP; Jacobson SC Nanofluidic Devices with Two Pores in Series for Resistive-Pulse Sensing of Single Virus Capsids. Anal. Chem. 2011, 83, 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Harms ZD; Selzer L; Zlotnick A; Jacobson SC Monitoring Assembly of Virus Capsids with Nanofluidic Devices. ACS Nano 2015, 9, 9087–9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Zhou K; Li L; Tan Z; Zlotnick A; Jacobson SC Characterization of Hepatitis B Virus Capsids by Resistive-Pulse Sensing. J. Am. Chem. Soc. 2011, 133, 1618–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Chantiwas R; Park S; Soper SA; Kim BC; Takayama S; Sunkara V; Hwang H; Cho Y-K Flexible fabrication and applications of polymer nanochannels and nanoslits. Chem. Soc. Rev. 2011, 40, 3677–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Choi J; Lee CC; Park S Scalable fabrication of sub-10 nm polymer nanopores for DNA analysis. Microsyst. Nanoeng. 2019, 5, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).O’Neil CE; Taylor S; Ratnayake K; Pullagurla S; Singh V; Soper SA Characterization of activated cyclic olefin copolymer: effects of ethylene/norbornene content on the physiochemical properties. Analyst 2016, 141, 6521–6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Jia Z; Choi J; Park S Surface Charge Density-Dependent DNA Capture through Polymer Planar Nanopores. ACS Appl. Mater. Interfaces 2018, 10, 40927–40937. [DOI] [PubMed] [Google Scholar]
- (37).Uba FI; Hu B; Weerakoon-Ratnayake K; Oliver-Calixte N; Soper SA High process yield rates of thermoplastic nanofluidic devices using a hybrid thermal assembly technique. Lab Chip 2015, 15, 1038–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Wang Y; Liang X; Liang Y; Chou SY Sub-10-nm wide trench, line, and hole fabrication using pressed self-perfection. Nano Lett. 2008, 8, 1986–1990. [DOI] [PubMed] [Google Scholar]
- (39).Choi J; Farshchian B; Kim J; Park S Fabrication of perforated micro/nanopore membranes via a combination of nanoimprint lithography and pressed self-perfection process for size reduction. J. Nanosci. Nanotechnol. 2013, 13, 4129–4133. [DOI] [PubMed] [Google Scholar]
- (40).Siwy Z; Trofin L; Kohli P; Baker LA; Trautmann C; Martin CR Protein biosensors based on biofunctionalized conical gold nanotubes. J. Am. Chem. Soc. 2005, 127, 5000–5001. [DOI] [PubMed] [Google Scholar]
- (41).Martin CR; Nishizawa M; Jirage K; Kang M Investigations of the transport properties of gold nanotubule membranes. J. Phys. Chem. B 2001, 105, 1925–1934. [Google Scholar]
- (42).Siwy Z; Heins E; Harrell CC; Kohli P; Martin CR Conical-nanotube ion-current rectifiers: the role of surface charge. J. Am. Chem. Soc. 2004, 126, 10850–10851. [DOI] [PubMed] [Google Scholar]
- (43).Harrell CC; Kohli P; Siwy Z; Martin CR DNA—Nanotube Artificial Ion Channels. J. Am. Chem. Soc. 2004, 126, 15646–15647. [DOI] [PubMed] [Google Scholar]
- (44).Maekawa Y; Suzuki Y; Maeyama K; Yonezawa N; Yoshida M Visualization of chemical modification of pore internal surfaces using fluorescence microscopy. Chem. Lett. 2004, 33, 150–151. [Google Scholar]
- (45).Maekawa Y; Suzuki Y; Maeyama K; Yonezawa N; Yoshida M Chemical Modification of the Internal Surfaces of Cylindrical Pores of Submicrometer Size in Poly(ethylene terephthalate). Langmuir 2006, 22, 2832–2837. [DOI] [PubMed] [Google Scholar]
- (46).Li J; Maekawa Y; Yamaki T; Yoshida M Chemical Modification of a Poly(ethylene terephthalate) Surface by the Selective Alkylation of Acid Salts. Macromol. Chem. Phys. 2002, 203, 2470–2474. [Google Scholar]
- (47).Pasternak CA; Alder GM; Apel PY; Bashford CL; Edmonds DT; Korchev YE; Lev AA; Lowe G; Milovanovich M; Pitt CW; Rostovtseva TK; Zhitariuk NI Nuclear track-etched filters as model pores for biological membranes. Radiat. Meas. 1995, 25, 675–683. [Google Scholar]
- (48).Marchand-Brynaert J; Deldime M; Dupont I; Dewez J-L; Schneider Y-J Surface Functionalization of Poly(ethylene terephthalate) Film and Membrane by Controlled Wet Chemistry: Chemical Characterization of Carboxylated Surfaces. J. Colloid Interface Sci. 1995, 173, 236–244. [Google Scholar]
- (49).Papra A; Hicke H-G; Paul D Synthesis of peptides onto the surface of poly(ethylene terephthalate) particle track membranes. J. Appl. Polym. Sci. 1999, 74, 1669–1674. [Google Scholar]
- (50).Uba FI; Pullagurla SR; Sirasunthorn N; Wu J; Park S; Chantiwas R; Cho Y-K; Shin H; Soper SA Surface charge, electroosmotic flow and DNA extension in chemically modified thermoplastic nanoslits and nanochannels. Analyst 2015, 140, 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Wu J; Chantiwas R; Amirsadeghi A; Soper SA; Park S Complete plastic nanofluidic devices for DNA analysis via direct imprinting with polymer stamps. Lab Chip 2011, 11, 2984–2989. [DOI] [PubMed] [Google Scholar]
- (52).Wei S; Vaidya B; Patel AB; Soper SA; McCarley RL Photochemically Patterned Poly(methyl methacrylate) Surfaces Used in the Fabrication of Microanalytical Devices. J. Phys. Chem. B 2005, 109, 16988–16996. [DOI] [PubMed] [Google Scholar]
- (53).Jackson JM; Witek MA; Hupert ML; Brady C; Pullagurla S; Kamande J; Aufforth RD; Tignanelli CJ; Torphy RJ; Yeh JJ; Soper SA UV activation of polymeric high aspect ratio microstructures: ramifications in antibody surface loading for circulating tumor cell selection. Lab Chip 2014, 14, 106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Jia Z; Choi J; Park S Selection of UV-resins for nanostructured molds for thermal-NIL. Nanotechnology 2018, 29, 365302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Amarasekara CA; Athapattu US; Rathnayaka C; Choi J; Park S; Soper SA Open-tubular nanoelectrochromatography (OT-NEC): gel-free separation of single stranded DNAs (ssDNAs) in thermoplastic nanochannels. Electrophoresis 2020, 41, 1627–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Miller AA; Lawton EJ; Balwit JS Effect of chemical structure of vinyl polymers on crosslinking and degradation by ionizing radiation. JPoSc 1954, 14, 503–504. [Google Scholar]
- (57).Mahmood Raouf R; Abdul Wahab Z; Azowa Ibrahim N; Abidin Talib Z; Chieng B Transparent Blend of Poly-(Methylmethacrylate)/Cellulose Acetate Butyrate for the Protection from Ultraviolet. Polymers 2016, 8, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Dole M. The Radiation Chemistry of Macromolecules: Volume II; Elsevier, 2013; Vol. 2. [Google Scholar]
- (59).Jeong J-H; Choi Y-S; Shin Y-J; Lee J-J; Park K-T; Lee E-S; Lee S-R Flow behavior at the embossing stage of nanoimprint lithography. Fibers Polym. 2002, 3, 113–119. [Google Scholar]
- (60).Heyderman LJ; Schift H; David C; Gobrecht J; Schweizer T Flow behaviour of thin polymer films used for hot embossing lithography. Microelectron. Eng. 2000, 54, 229–245. [Google Scholar]
- (61).Fryer DS; Peters RD; Kim EJ; Tomaszewski JE; de Pablo JJ; Nealey PF; White CC; Wu W.-l. Dependence of the Glass Transition Temperature of Polymer Films on Interfacial Energy and Thickness. Macromolecules 2001, 34, 5627–5634. [Google Scholar]
- (62).Kowalczyk SW; Grosberg AY; Rabin Y; Dekker C Modeling the conductance and DNA blockade of solid-state nanopores. Nanotechnology 2011, 22, 315101. [DOI] [PubMed] [Google Scholar]
- (63).Li J; Gershow M; Stein D; Brandin E; Golovchenko JA DNA molecules and configurations in a solid-state nanopore microscope. Nat. Mater. 2003, 2, 611–615. [DOI] [PubMed] [Google Scholar]
- (64).Huang X; Gordon MJ; Zare RN Current-monitoring method for measuring the electroosmotic flow rate in capillary zone electrophoresis. Anal. Chem. 1988, 60, 1837–1838. [Google Scholar]
- (65).Amarasekara CA; Rathnayaka C; Athapattu US; Zhang L; Choi J; Park S; Nagel AC; Soper SA Electrokinetic identification of ribonucleotide monophosphates (rNMPs) using thermoplastic nanochannels. J. Chromatogr. A 2021, 1638, 461892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Langecker M; Pedone D; Simmel FC; Rant U Electrophoretic Time-of-Flight Measurements of Single DNA Molecules with Two Stacked Nanopores. Nano Lett. 2011, 11, 5002–5007. [DOI] [PubMed] [Google Scholar]
- (67).Novak BR; Moldovan D; Nikitopoulos DE; Soper SA Distinguishing Single DNA Nucleotides Based on Their Times of Flight Through Nanoslits: A Molecular Dynamics Simulation Study. J. Phys. Chem. B 2013, 117, 3271–3279. [DOI] [PubMed] [Google Scholar]
- (68).Oliver-Calixte NJ; Uba FI; Battle KN; Weerakoon-Ratnayake KM; Soper SA Immobilization of lambda exonuclease onto polymer micropillar arrays for the solid-phase digestion of dsDNAs. Anal. Chem. 2014, 86, 4447–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).O’Neil C; Amarasekara CA; Weerakoon-Ratnayake KM; Gross B; Jia Z; Singh V; Park S; Soper SA Electrokinetic transport properties of deoxynucleotide monophosphates (dNMPs) through thermoplastic nanochannels. Anal. Chim. Acta 2018, 1027, 67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Athapattu US; Amarasekara CA; Immel JR; Bloom S; Barany F; Nagel AC; Soper SA Solid-phase XRN1 reactions for RNA cleavage: application in single-molecule sequencing. Nucleic Acids Res. 2021, 49, No. e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


