Abstract
Initially recognized as a HeLa factor essential for parvovirus DNA replication, parvovirus initiation factor (PIF) is a site-specific DNA-binding complex consisting of p96 and p79 subunits. We have cloned and sequenced the human cDNAs encoding each subunit and characterized their products expressed from recombinant baculoviruses. The p96 and p79 polypeptides have 40% amino acid identity, focused particularly within a 94-residue region containing the sequence KDWK. This motif, first described for the Drosophila homeobox activator DEAF-1, identifies an emerging group of metazoan transcriptional modulators. During viral replication, PIF critically regulates the viral nickase, but in the host cell it probably modulates transcription, since each subunit is active in promoter activation assays and the complex binds to previously described regulatory elements in the tyrosine aminotransferase and transferrin receptor promoters. Within its recognition site, PIF binds coordinately to two copies of the tetranucleotide PuCGPy, which, remarkably, can be spaced from 1 to 15 nucleotides apart, a novel flexibility that we suggest may be characteristic of the KDWK family. Such tetranucleotides are common in promoter regions, particularly in activating transcription factor/cyclic AMP response element-binding protein (ATF/CREB) and E-box motifs, suggesting that PIF may modulate the transcription of many genes.
Differential gene expression is achieved by the assembly of multicomponent complexes containing precise combinations and arrangements of individual transcription factors. On certain cis-acting promoter sequences, the activity of specific factors within these complexes can be further modulated in a combinatorial fashion by site-specific regulatory proteins. One example, the Drosophila homeobox protein Deformed, binds weakly to low-complexity DNA sequences present in many homeotically regulated promoters but specifically upregulates transcription at a subset of promoters which harbor additional binding sites for the Deformed activator molecule DEAF-1 (10). The effects of such activator molecules in transient transcriptional regulation are less well documented than in homeotic control, and few candidate complexes have been identified. We have isolated and cloned a previously undescribed but widely expressed heterodimer from HeLa cells, called parvovirus initiation factor (PIF), which resembles DEAF-1 in a number of critical ways. We suggest that both are members of the same emerging family of transcriptional modulators characterized by conservation of a domain containing the signature amino acid motif KDWK.
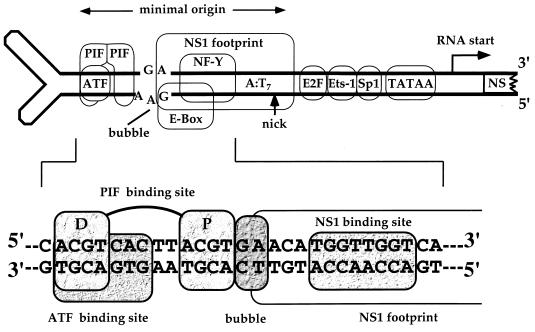
PIF was first identified as an essential ancillary factor for DNA replication initiation from the minimal left-end origin of the parvovirus minute virus of mice (MVM) (4). In this and comparable viral systems, specific cellular transcription factors stimulate in vitro initiation up to a 1,000-fold, so that successful initiation can often provide a sensitive indicator for monitoring how such factors cooperate with other proteins and interact with their DNA substrates (23, 24). The MVM origin is approximately 50 bp long (7) and contains sequences which play a dual role both in initiation and as part of the early viral transcriptional promoter (Fig. 1). MVM replicates by a rolling-hairpin mechanism which resembles prokaryotic rolling-circle replication, so that DNA synthesis is initiated by the introduction of a site-specific single-strand nick, mediated by the virally encoded nickase NS1. NS1 is itself a DNA-binding protein which recognizes an ACCA repeat motif in the origin, as shown in Fig. 1, and it is oriented in such a way that its footprint lies over the nick site (6). However, by itself, NS1 is in a catalytically inactive state. Before it can nick the origin, a cellular factor must become bound to one end of the origin sequence, at a site which overlaps a consensus activated transcription factor (ATF) binding motif. This site is known to bind ATF and is involved in regulation of the promoter during the cell cycle and in response to cellular transformation (8, 18). However, ATF/cyclic AMP response element-binding protein (CREB) family members inhibit, rather than activate, the viral replication origin, since they compete for their binding site with PIF, the authentic activator complex (3). PIF comprises related polypeptides denoted p96 and p79 and, as depicted in Fig. 1, binds coordinately to the ACGT motif within the ATF site and to an identical motif located 5 bp away, proximal to the NS1 footprint (3). In this study, we examined the remarkably flexible way in which the PIF complex interacts with its bipartite recognition sequence and showed that its individual component polypeptides are capable of activating transcription in transfected cells. Finally, we investigated the possible relationship between PIF and other previously described activities which modulate transcription at the tyrosine aminotransferase and transferrin receptor promoters.
FIG. 1.
The left end of the MVM genome. The left (3′) hairpin of the MVM genome contains both the upstream regulatory region of the P4 promoter and sequences that give rise to the left-end DNA replication origin. This origin, part of which is shown in expanded form, is a duplex copy of sequences from the top strand of the hairpin, which is formed in dimeric replication intermediates. The PIF half-sites (spaced ACGT motifs) overlapping a consensus ATF site are indicated and marked proximal (P) and distal (D) to denote their positions with respect to the NS1 binding site. The dinucleotide “bubble” sequence is a critical spacer element lying between the proximal ACGT and the NS1 binding site. The box which starts at the bubble sequence and extends rightwards toward the other end of the minimal origin indicates the sequences protected from DNase I digestion by NS1. Within this box are the NS1-binding motif, which overlaps the E box and NF-Y sites of the promoter, and the nick site, where NS1 initiates replication by nicking the DNA to liberate a base-paired 3′ nucleotide capable of priming DNA synthesis.
MATERIALS AND METHODS
Purification of HeLa PIF complexes.
Nuclear extracts from HeLa S3 cells were prepared and fractionated on Q-Sepharose and Zn2+-metal chelate Sepharose as described previously (3). The Zn2+-metal chelate Sepharose eluate was diluted fivefold in buffer A (25 mM Tris-HCl [pH 7.8], 1 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride, 0.01% Nonidet P-40, 1 mM dithiothreitol, 100 mM NaCl, 10% glycerol) supplemented with sonicated salmon sperm DNA (200 μg/ml) and 60 μg of a nonspecific, duplex oligonucleotide (SCRAM; described previously [3]) per ml and absorbed for 1 h on nonspecific DNA-Sepharose coupled with duplex SCRAM DNA. The extract was then applied serially to two 1-ml DNA-Sepharose columns coupled with a duplex copy of the TR56 oligonucleotide 5′-GATCGATCTGTCAGAGCACCTCGCGAGCGTACGTGCCTCAGGAAGTGACGCACAGC-3′, representing nucleotides −61 through −115 from the transferrin receptor promoter (19), equilibrated in buffer A. The columns were washed with 50 volumes of buffer A, and bound proteins were eluted with buffer A adjusted to 300 mM NaCl. Fractions containing PIF activity were identified by gel shift and nicking assays and were flash-frozen in liquid N2 before being stored at −80°C. Selected fractions were concentrated by ultrafiltration on Centricon 30 filters and separated by quantitative sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). PIF bands were localized by staining with Coomassie brilliant blue, excised, and submitted to the Keck Biotechnology Center for peptide sequence analysis.
cDNA cloning of PIF subunits.
Predicted nucleotide sequences for peptides MMDSGQIDFYQHDK from p96 and IMDSGELDFYQHDK from p79 were used to design the degenerate oligonucleotides 5′-ATGGATTCIGGICAIATIGATTTCTACCAICATGATAA-3′ and 5′-ATIATGGATGGITCIGAICTIGATTTCTACCAICATGATAA-3′, respectively. These were used simultaneously to screen a lambda gt11 HeLaS3 cDNA library (Clontech, Palo Alto, Calif.) and a Uni-Zap XR HeLa cDNA library (Stratagene, San Diego, Calif.). Probes were incubated with duplicate filters overnight at 45°C in 1× SSC (0.15 M NaCl, 0.015 M sodium citrate)–20 mM NaH2PO4–0.4% SDS containing denatured salmon sperm DNA (500 μg/ml) and 32P-labeled probe (2 × 105 cpm/ml) and washed twice for 20 min in 3× SSC–0.1% SDS at room temperature and once for 10 min at 45°C. Four clones were identified out of 3 × 106 plaques screened, all of which hybridized exclusively to the p96 probe. Inserts derived from the lambda gt11 library were excised with EcoRI and subcloned into pGem4Z (Promega, Madison, Wis.) before analysis. Those from the Uni-Zap library were excised in vivo as specified by the manufacturer. All the clones had open reading frames (ORFs) containing the probe peptide and were identical in their overlapping sequence, but only one clone contained the full p96 ORF.
Since phages encoding the p79 gene were not obtained in this screen, we cloned it by a different approach. First we amplified a 756-bp fragment from the p79 ORF from HeLa cDNA by PCR using two degenerate primers: 5′-GARYTIGAYTTYTAYCARCAYGAYAA-3′ and 5′-YTTICCIAGIACIGTIGAIGGIAGIGTIGAIACIAC-3′, based on the two longest p79 peptide sequences. The 5′ and 3′ sequences were then obtained from HeLaS3 cDNA by rapid amplification of cDNA ends (RACE), using a combined RACE-cDNA kit from Clontech and primers based on internal sequences from the 756-bp fragment. Finally, the full cDNA sequence was PCR amplified from HeLa cDNA with primers based on the extreme 5′ and 3′ sequences of the 5′ and 3′ RACE products, respectively. PCR products were gel purified and cloned into pCR2.1-TOPO (Invitrogen, Carlsbad, Calif.). Three clones were sequenced on both strands to generate a consensus sequence.
Recombinant baculoviruses.
To optimize the expression of wild-type p96 and p79, PCR fragments which started 3 nucleotides upstream of the ORF were cloned into the baculovirus transfer vector pVL1393 (Invitrogen, Carlsbad, Calif.) to generate pVL1393-p96 and pVL1393-p79, respectively. Recombinant baculoviruses were recovered, grown, and assayed as previously described (2).
Purification and analysis of recombinant proteins.
Recombinant PIF p96-p79 heterodimers were generated by coinfecting High Five insect cells at high multiplicity (∼10 PFU per cell) with both recombinant baculoviruses expressing p79 and p96. Nuclear extracts were prepared essentially as described previously (2) and purified using by fast protein liquid chromatography on a MonoQ (HR5/5) column equilibrated in 25 mM Tris-HCl (pH 7.8)–1 mM EDTA–0.1 mM phenylmethylsulfonyl fluoride–0.01% Nonidet P-40–1.0 mM dithiothreitol–10% glycerol, containing 150 mM NaCl. Bound proteins were eluted with a 40-ml linear NaCl gradient (0.05 to 0.7 M) in the same buffer.
Recombinant baculoviruses expressing human ATF1, CREB, and NS1 were expressed in Sf9 insect cells by using baculovirus vectors, and their products were purified as described previously (3).
Electrophoretic mobility shift assays (EMSA), nicking assays, DNase I protection assays, and methylation interference assays were performed as described previously (4).
Transcription vector construction and assays.
Vectors used in this study were part of a mammalian two-hybrid system purchased from Clontech. To generate pGal-p96 and pVP16-p96 vectors expressing p96 molecules with amino-terminal yeast GAL4 DNA-binding domains or herpes simplex virus VP16 transcriptional activator domains, respectively, SmaI-SalI or EcoRI-SalI fragments containing the full p96 ORF were inserted, in frame with the relevant domains, in the vectors pM and pVP16, respectively. Similarly, the full p79 ORF was cloned into the EcoRI site of pM and pVP16, resulting in the vectors pGal-p79 and pVP16-p79.
To measure transcriptional activity, expression vectors were cotransfected with pG5CAT, a vector containing five GAL4-binding sites located upstream of a minimal adenovirus E1B promoter driving a chloramphenicol acetyltransferase (CAT) reporter gene. Human 293 cells were transfected with combinations of vectors as described in the legend to Fig. 7 by using Superfect (Qiagen GmbH, Hilden, Germany) as recommended by the manufacturer. The vector pCH110 (Pharmacia, Uppsala, Sweden) expressing β-galactosidase was included in all assay mixtures. Cells were harvested 48 h after transfection, and β-galactosidase and CAT activity were measured as described previously (5) by using a dynamic liquid scintillation assay.
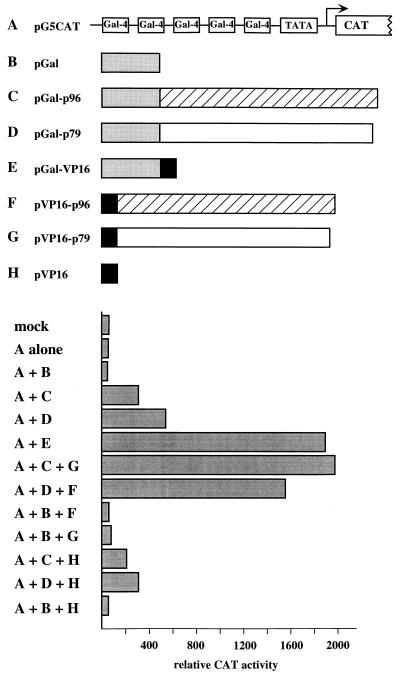
FIG. 7.
PIF activates transcription from a model reporter construct in vivo. The pG5CAT reporter plasmid contains the CAT gene driven by the adenovirus E1B minimal promoter (TATA) fused to five upstream GAL4-binding sites. N-terminal fusions of p96pif (hatched box) or p79pif (open box) to amino acids 1 to 147 of the GAL4 DNA-binding domain (shaded box) or with amino acids 411 through 455 of HSV VP16 (solid box) were constructed in vectors driven by the simian virus 40 early promoter, as described in Materials and Methods. 293 cells were mock transfected or transfected with pG5CAT alone or with various combinations of the fusion proteins, as indicated below the bar graph. A 5-μg portion of each plasmid was transfected. Transfection efficiency was monitored by measuring β-galactosidase activity generated by cotransfected pCH110, and CAT activity is represented as the mean of duplicate samples, which differed by less than 10%.
Nucleotide sequence accession number.
The sequence of p79pif and p96pif have been deposited with GenBank under accession no. AF173867 and AF173868, respectively.
RESULTS
Two polypeptides copurify with PIF activity.
The PIF complex was purified from HeLa S3 nuclear extracts (from ∼1010 cells) by the scheme depicted in Fig. 2A. For the final site-specific DNA-binding column, we initially used an oligomerized form of a 23-mer containing the PIF-binding site from the MVM origin, but were unable to release the active complex from this substrate. Instead we substituted a nonoligomerized duplex 53-mer sequence, derived from the transferrin receptor promoter, which Roberts et al. (20) had previously shown bound and eluted a factor that we believed was identical to PIF (3). Purification was monitored at each step by measuring site-specific DNA binding activity in EMSA, in the presence and absence of the duplex alternating copolymer poly(dI-dC)-poly(dI-dC), which effectively competes for PIF but not for most other DNA-binding proteins (3).
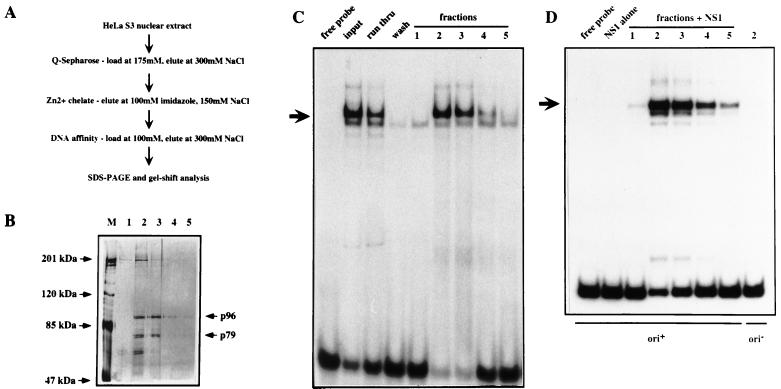
FIG. 2.
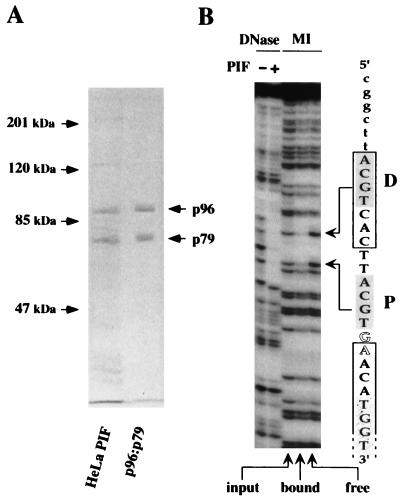
Purification and analysis of the HeLa PIF complex. (A) Fractionation scheme used to purify PIF from HeLa nuclear extracts. (B) Silver-stained SDS-PAGE gel showing fractions 1, 2, 3, 4, and 5 eluted from the site-specific DNA affinity chromatography. Lane M contains molecular mass markers. (C) EMSA analysis of fractions. The input is a PIF fraction from a Zn2+-metal chelate Sepharose column which was loaded on to the site-specific DNA affinity column; run thru is the unbound material; wash is the last column wash before elution; lanes 1, 2, 3, 4, and 5 are serial fractions obtained by elution with 300 mM NaCl. (D) Eluted fractions catalyze nicking and covalent attachment of NS1 to the minimal MVM origin. Nicking assay mixtures contained 100 ng of GST-NS1 and ATP; samples in lanes labeled 1 through 5 also received fractions eluted from the DNA affinity column. The nicking assay in the last lane was performed in the presence of fraction 2, GST-NS1, and ATP but using a control substrate (ori−), in which a single extra nucleotide inserted into the bubble sequence inactivates viral replication origin function (7).
The ability of each fraction to activate NS1, the MVM initiator endonuclease, was also tracked by using a nicking assay, in which fractions were tested for their ability to allow recombinant NS1 to introduce a site-specific, single-strand nick into the minimal MVM origin sequence (Fig. 1). These activities coeluted from the transferrin receptor DNA affinity column (Fig. 2C and D). Analysis of the elution profile on SDS-polyacrylamide gels revealed several polypeptides, two of which, with apparent molecular masses of about 96 and 79 kDa, coeluted in equimolar concentration with the peak DNA-binding and -nicking activity (Fig. 2B, lanes 2 and 3). Final estimates of protein yield versus activity suggest that there are about 2 × 104 copies each of p96 and p79 per HeLa cell.
A 60-pmol sample of purified p96 and 32.5 pmol of p79 were excised from a gel and submitted for peptide sequencing to the Keck Center for Biotechnology. Both p96 and p79 gave three peptide sequences which proved useful in the identification of cDNAs encoding them.
Isolation of PIF cDNAs.
The longest peptides obtained from each molecule were identical at 11 of 14 residues, and their sequences were used to design separate degenerate oligonucleotide probes for screening HeLa-derived lambda phage cDNA libraries. Screening ∼3 × 106 plaques gave four cDNA clones which hybridized to the p96-based probe but none that hybridized to p79. The most extensive p96 insert was 1,862 nucleotides long and contained an ORF capable of encoding a protein of 563 amino acids, with a predicted molecular mass of 61,379 Da, which contained all three of the p96 peptide sequences, as shown in Fig. 3A. The discrepancy between the migration rate of p96 on SDS-PAGE and the calculated molecular weight of the protein prompted us to check the sequence in HeLa cDNA by using 5′ RACE, but all RACE clones gave the same 5′ sequence. Although the p96 gene is not present in the standard GenBank database, the dbEST database contains many sequences related to it, derived from both human and murine tissues, representing various cell types including colon, liver, embryonic stem cells, neuroepithelium, and placenta, suggesting that p96 is expressed in a broad range of tissues. One insert, from human colon (AA148980), contains a predicted single insertion of 10 amino acids, which is suggested by genome sequencing to be the product of alternate splicing (22a).
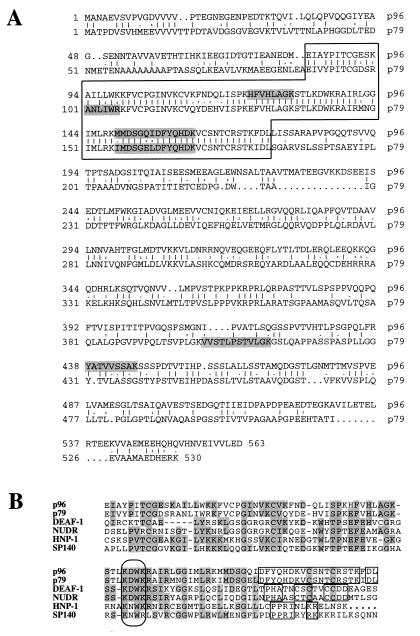
FIG. 3.
Polypeptide sequence analysis of the PIF subunits. (A) Alignment of the amino acid sequences predicted from the ORFs of cloned cDNAs encoding p79pif and p96pif, aligned by using the GCG BESTFIT program. The highly conserved region which contains the KDWK motif discussed in the text and panel B is boxed. (B) Alignment of sequences from the most highly conserved domain of human p96pif and p79pif, with individual proteins or representative members of protein groups, obtained from the GenBank database. The alignment, obtained by using the Genetics Computer Group Pile-Up program, was refined manually. Residues identical in at least half of the compared sequences are shaded. The oval box indicates the highly conserved KDWK motif, and the smaller boxes indicate extended relationships between pairs of compared molecules. Full names, GenBank accession numbers, and references for these proteins are cited in the text.
Since we were unable to locate the p79 gene in cDNA libraries, we resorted to a three-step PCR strategy, described in Materials and Methods, with HeLa cDNA as template. Cloned products included a 2,333-nucleotide insert with a single ORF, encoding a protein of 530 amino acids with a predicted molecular mass of 56,397 Da, which contained the three p79 peptide sequences (Fig. 3A). As with p96, this sequence was not present in the GenBank database at the time of its isolation, although a very similar sequence (AF059273) encoding one subunit of the rat glucocorticoid modulating element-binding protein (GMEB-2) has since appeared (26). However, p79 did show 83 to 97% identity to six partial sequences from the dbEST database, of which two were human, three were murine, and one was from the rat.
PIF polypeptides p96 and p79 are related and share a conserved KDWK domain with several known proteins.
The deduced amino acid sequences of p96 and p79 (Fig. 3A) show an overall identity of 40 to 42% (for p96 and p79, respectively), which fluctuates along the length of the polypeptide chains but is most evident between residues 82 and 174 in p96 and residues 89 and 182 in p79, where the two chains show 80% identity. When these highly conserved sequences were used to search available databases, a group of protein and expressed sequence tag sequences were found with peptide homologies that stretched through most of the 94 residues of the conserved domain. This region includes a KDWK motif first noticed by Gross and McGinnis (10) in the Drosophila melanogaster DEAF-1 protein (U46686), which itself shows 31% identity to p96 and 28% identity to p79 over this entire region. Other KDWK domain proteins recognized in the search included two human interferon-induced nuclear phosphoproteins (L22342 and L22343) of unknown function (represented by HNP-1 in Fig. 3B), a human leukocyte-specific SP140 protein (U63420), and human lymphoid cell-specific SP100 homologues (U36499 to U36501), which are tissue-specific components of dot-like, subnuclear bodies often described as PML/SP100 bodies. The search also identified various rat and human forms (AF007165, AF068892 to AF068895, and U59659) of a protein initially named “suppressin” (12, 13). Recently a monkey cDNA encoding a very closely related molecule, dubbed NUDR (for “nuclear DEAF-1 related”) transcription factor, was identified by its ability to bind, albeit weakly, to a synthetic retinoic acid response element (AF049461) (11). Significantly, recombinant NUDR recognizes a bipartite DNA sequence in which both half-sites contain CpG dinucleotides, and it is able to enhance transcription from a promoter containing such elements, both properties described in this paper for PIF. NUDR-homologous cDNAs were also isolated from human (AF04959) and rat (AF04960) libraries (11). The alignment between the PIF polypeptides and one member from each of these groups is shown in Fig. 3B. Over this region, identity scores between PIF and these polypeptides are all about 30% and include some broadly scattered positions where sequence is absolutely conserved throughout all members of the family and others where one or two particular residues are clearly favored. Boxed sequences at the C terminus of this region are conserved in a pairwise fashion between protein classes, suggesting that they might characterize distinct subsets of the KDWK family. Multiple cosmids from Caenorhabditis elegans also have this pattern of conserved residues, including one (Z81089) which shows 40% identity to the p79 sequence across this region. Significantly, no matches were found in the genomes of prokaryotes or eukaryotic unicellular organisms, suggesting that this gene family is a feature of metazoans.
Both PIF polypeptides are acidic, with a particularly high concentration of acidic residues in their carboxy-terminal regions, which are also rich in proline, serine, and threonine, suggesting that these could be transactivation domains. When analyzed with the program COILS (ICREC Bioinformatics), sequences around residue 300 in both polypeptide chains show a 100% probability of forming coiled-coil structures, which are frequently involved in protein dimerization. No other motifs commonly associated with DNA-binding proteins were detected with programs in the Genetics Computer Group package.
During analysis of the PIF sequences, we became aware that a heterodimeric rat protein complex which binds to the glucocorticoid-modulating element (GME) of the tyrosine aminotransferase promoter (17) contained six peptides which were closely related to peptides that we subsequently identified in the human p96:p79 PIF heterodimer. Subsequently, Zeng et al. (26) used these sequences to clone a rat cDNA encoding a protein they called GMEB-2 (AF059273), which is most probably the rat homolog of the human p79 gene.
Recombinant PIF heterodimers bind to the ACGT motifs that overlap the ATF/CREB site in the viral origin.
Cloning and DNA sequencing revealed p96 and p79 to be proteins of 563 and 530 amino acids, with predicted molecular masses of 61,379 and 56,397 Da, respectively. These values are both significantly lower than expected from their mobilities measured in denaturing gels. When recombinant baculoviruses were used to coexpress p96 and p79 in insect cells, the purified products comigrated on denaturing gels with their authentic HeLa counterparts (Fig. 4A). Each of the PIF cDNAs also generated proteins with mobilities that matched those of the HeLa cell-derived proteins when translated in vitro (data not shown), indicating that each cloned cDNA contains the entire coding sequence of its respective protein. This also suggests that the discrepancies between their calculated and observed molecular weights are intrinsic properties of the polypeptide chains themselves rather than the result of posttranslational modification.
FIG. 4.
Characterization of recombinant PIF subunits expressed from separate baculovirus vectors. (A) SDS-PAGE analysis of purified recombinant PIF. DNA-affinity purified PIF from HeLaS3 cells was compared to purified recombinant PIF p96-p79, expressed in insect cells by using baculovirus vectors. The positions of the p96 and p79 subunits are indicated by arrows. (B) DNase I protection and methylation interference patterns of rPIF binding to a duplex DNA fragment containing the minimal replication origin, in which the top strand, as represented in Fig. 1, was 32P labeled at its 3′ end. The sequence through this part of the origin is indicated on the right. Lowercase letters indicate flanking vector DNA sequence. PIF half-sites are shaded, the consensus ATF site is boxed, bubble nucleotides are shown in outline, and the open-ended box indicates the start of the NS1 footprint, in which the first TGGT motif is stippled. Sequences protected by rPIF from DNase I digestion are shown in the left lanes. The right three lanes, labeled MI, show methylation interference profiles: input indicates piperidine cleavage products of the methylated DNA sample used in the binding assay; bound indicates cleaved DNA from the retarded rPIF-DNA complex; and free indicates free probe which was not shifted by the binding reaction. Methylated residues which impair rPIF binding are indicated by arrows.
Gel filtration chromatography and electrophoresis on nondenaturing polyacrylamide gels both indicate that when expressed alone, p96 and p79 can each self-associate (data not shown). However, since the HeLa PIF complex which normally activates NS1 is a heterodimer, we wished to enrich for such products among the recombinant molecules. To this end, we coexpressed p96 and p79 by dual infection of insect cells at high input multiplicity, to ensure that most cells received both viruses, and purified the products by anion-exchange chromatography. When they were expressed alone, wild-type p96 bound efficiently to this resin although wild-type p79 did not, but when they were coexpressed, p79 formed heterodimers with p96 and could be copurified. When cross-linked with glutaraldehyde, a fraction of the purified product migrated on SDS-PAGE as a homogeneous band at the position expected for the dimer but not higher-order multimers (data not shown).
Purified preparations of recombinant PIF were also tested in EMSAs in the presence of antisera specific for p96 or p79, or both. These sera were each able to supershift the complex independently, and together they shifted the complex to a position of even lower mobility (1a). Therefore we conclude that the great majority of PIF complexes are heterodimers, suggesting that this may be the preferred configuration. This recombinant complex was shown to activate the nickase of NS1 in a manner indistinguishable from that of the purified HeLa factor in Fig. 2A (4a) and was used to explore further the DNA-binding specificity of PIF. Initially, recombinant p96-p79 complexes were tested for their ability to recognize the two spaced ACGT motifs in the viral origin sequence. As shown in Fig. 4B, recombinant PIF protects the entire 13-bp region spanning these two motifs from digestion by DNase I, while methylation interference analysis highlights the critical importance of the G residues within each of the motifs, paralleling exactly our previously published observations with authentic PIF purified from HeLa cells (4).
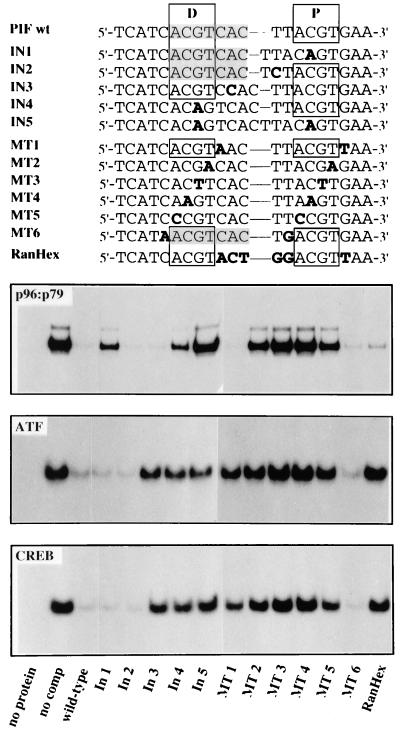
Of the two spaced ACGT motifs in the viral origin sequence (Fig. 1), site D coincides with a predicted ATF/CREB-binding site. A panel of double-stranded oligonucleotides which had insertions or mutations within and between the two ACGT motifs were then used to compare the specificities of PIF, ATF1, and CREB on this site. Initially, the mutated sequences were tested for their ability to compete for rPIF in a EMSA with a 32P-labeled wild-type sequence, designated PIF wt (Fig. 5). The mutant origin sequences In1 and In4 have a single nucleotide inserted between the C and G residues of ACGT motifs D and P, respectively. Both of these mutants competed poorly with the wild-type probe for recombinant p96-p79, even when present at 100-fold molar excess, as shown in Fig. 5, while mutant In5, which has similar insertions in both motifs, was unable to compete. In contrast, mutants In2 and In3, which have a single base inserted between the two ACGT blocks, competed as effectively for recombinant PIF as the wild-type sequence did. Thus, rPIF heterodimers bind cooperatively to the two spaced ACGT half-sites in this origin, since they are essentially unable to recognize the sequence if the CpG dinucleotides in both motifs are disrupted, and they bind very poorly to the remaining single site if only one of the two is mutant. However, insertion of a single extra nucleotide between the two half-sites had a negligible influence on binding.
FIG. 5.
Recombinant PIF heterodimers bind spaced, bipartite ACGT motifs. Sequences of double-stranded oligonucleotides, representing wild-type and mutant forms of the PIF-binding region in the MVM 3′ origin that were used as competitors in the EMSAs, are shown at the top. Positions of wild-type proximal (P) and distal (D) ACGT half-sites are boxed, and the ATF/CREB-binding site is shaded. Individually mutated nucleotides are indicated in bold type. The lower three panels compare rPIF-, rATF1-, and rCREB-binding specificities on these sequences by competitive EMSA with 100-fold molar excesses of the wild-type or mutant competitor oligonucleotide. Assay mixtures contained the same 32P-labeled PIF wild-type oligonucleotide probe and constant amounts of rPIF, rATF1, or rCREB, as indicated in each panel.
The relative importance of individual nucleotides in the half-sites was also monitored by using a series of double-stranded oligonucleotides, called MT1 to MT6, which carried transversions at a particular position in each block (Fig. 5). Transversions in residues immediately flanking the ACGT motifs had no effect on the ability of these sequences to compete for PIF, while sequences with transversion at positions 1 and 4 in each motif (A to C and T to G, respectively) competed very poorly but still showed a trace of residual binding and those with transversions in the central CpG dinucleotide were totally unable to compete. Thus, all four positions within the motif influence binding, but the central CpG dinucleotide is of primary importance. Mutant RanHex has transversions at all 5 nucleotides in the linker region between the two half-sites (and an additional transversion immediately downstream of site P). It competes very effectively for recombinant PIF, albeit slightly less well that the wild-type sequence. This indicates that the sequence of the spacer region between the tetranucleotide repeats is of minimal importance.
Although the PIF-binding site in the MVM origin shares an ACGT motif with the consensus ATF site (ACGTCAC), the binding of PIF and ATF/CREB can be distinguished readily by using the panel of oligonucleotides illustrated in Fig. 5. For example, the In1 mutation inactivates ACGT site P and so drastically impairs PIF binding, but this oligonucleotide competes efficiently for ATF1 and CREB because the insertion is outside the consensus ATF site. In contrast, mutants MT1, In3, and RanHex change nucleotides in the ATF site but outside its ACGT core, and so they compete poorly for ATF and CREB but efficiently for PIF. Thus, within this dual MVM replication origin and transcriptional promoter sequence, PIF and ATF/CREB factors use overlapping but distinct binding sites and probably compete with each other for binding in vivo.
The spacing between the two half-sites can be highly flexible.
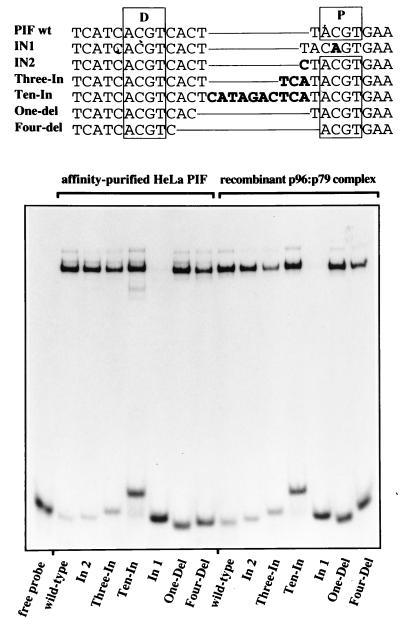
Although the PIF recognition sequence in the MVM origin involves two ACGT motifs spaced by 5 nucleotides, insertions of a single extra nucleotide between the half-sites had little effect on binding (Fig. 5). This prompted us to ask if PIF would tolerate more substantial changes in the length of this spacer element. Accordingly, we constructed a series of oligonucleotides with insertions or deletions at this position (Fig. 6), end labeled them with 32P, and compared their ability to bind to both authentic, affinity-purified HeLa PIF and the recombinant p96-p79 complex in direct binding assays. Both complexes bound sequences which had 1, 3, or 10 additional nucleotides inserted into the spacer region (Fig. 6, mutants In2, Three-In, and Ten-In, respectively), although comparing the proportion of bound to unbound probe in each reaction made it clear that as the inserts became longer the mutants bound progressively less well than wild-type molecules. For comparison, mutant In1, which has only a single intact ACGT site, failed to bind either of the protein complexes. Mutant sequences with deletions of 1 or 4 nucleotides in the normal spacer region (Fig. 6, One-del and Four-del, respectively) also bound both forms of PIF, although, once again, binding efficiency progressively diminished relative to the wild type. Thus, PIF heterodimers can bind sequences in which the spacing element varies in length from 1 to 15 nucleotides, although spacers of around 5 nucleotides may be optimal. Methylation interference data for the mutant with a 15-nucleotide spacer confirmed that PIF was binding to the two spaced ACGT motifs in this sequence, ruling out the possibility that we had created alternate binding sites (data not shown).
FIG. 6.
Both HeLa PIF and rPIF recognize flexibly spaced bipartite sites. Sequences of the probe oligonucleotides, with differently spaced ACGT motifs, that were used as substrates, are shown at the top. The proximal and distal PIF half-sites are boxed, deletions are indicated by thin lines connecting adjacent nucleotides, and inserted nucleotides are shown in bold type. The bottom panel shows direct EMSAs with 32P-labeled, double-stranded wild-type and mutant oligonucleotides and either DNA affinity-purified PIF from HeLa cells or purified recombinant p96-p79 heterodimers.
p96 and p79 function as transcriptional activators.
Although we initially identified PIF because it is an essential cofactor for viral replication initiation, in the host cell it is most likely to function as a transcriptional regulator. Because its binding site in the viral origin/promoter overlaps binding sites for other transcription factors, using this sequence to assess its effects on transcription proved problematic. To circumvent these difficulties, we fused p79 and p96 coding sequences to the yeast GAL4 DNA binding domain and cotransfected the resulting constructs into 293 cells along with pG5CAT, in which the CAT reporter gene is driven by the adenovirus EIB minimal promoter fused to a tandem array of five GAL4-binding sites (Fig. 7). In this context, both Gal-p79 and Gal-p96 activated the transcription of the reporter gene between 5- and 20-fold in several independent experiments (Fig. 7). Although these experiments indicate that p79 and p96 may individually activate transcription, it is also possible that the observed activity results from heterodimerization with an endogenous partner polypeptide in the transfected cell. Cotransfection of Gal-p79 or Gal-p96 with p96 or p79 fused to the herpes simplex virus transcriptional activator VP16 (VP16-p96 and VP16-p79, respectively) increased the activity of the GAL4 fusion constructs approximately fourfold (Fig. 7), indicating that the products of the transfected genes probably assemble into heterodimers in the transfected cell.
p96-p79 heterodimers bind to regulatory elements within the tyrosine aminotransferase and transferrin receptor promoters.
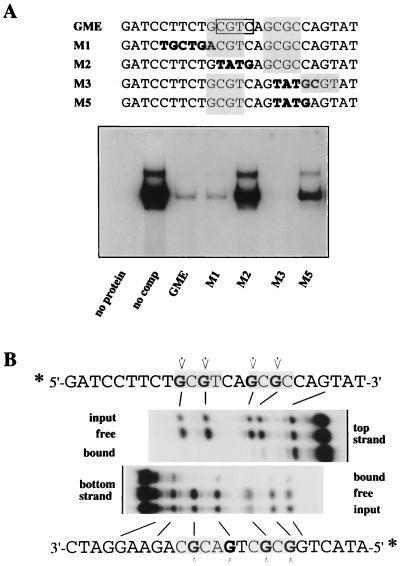
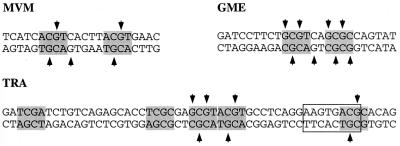
During these studies, we found two reports of other DNA-binding activities which showed similar physical characteristics. The first of these, mentioned above, is the ubiquitously expressed GMEB heterodimer, which binds to a regulatory element in the rat tyrosine aminotransferase promoter and makes it sensitive to induction by low concentrations of glucocorticoids (17). The sequences of six tryptic peptides from the rat complex all corresponded to sequences present in the human p96 and p79 genes, and subsequent cloning and DNA sequencing strongly indicate that one of the GMEB polypeptides, GMEB-2, is the rat homolog of p79pif (26). Surprisingly, the element which responds to this complex, GME (Fig. 8A), is very different from the sequence in the MVM origin but does contain two spaced CpG dinucleotides within the sequence 5′-GCGTCAGCGC-3′. When used as substrate in an EMSA, the GME sequence was recognized and shifted efficiently by recombinant p96-p79 (Fig. 8A). Moreover, mutant oligonucleotides M1 through M3 competed for rPIF in exactly the way described previously for GMEB. Thus, M1, which carries a mutation upstream of the first CpG dinucleotide, competed as well as the wild-type oligonucleotide, whereas M2, which has the first CpG mutated, was unable to compete. A significant and initially confusing observation was that oligonucleotide M3, which has the second CpG mutated, was also able to compete. Although the activity of mutant M3 had led Oshima and Simons to conclude that GMEB bound to the first CGTC motif, boxed in Fig. 8A (16), its binding is quite consistent with the flexible bipartite nature of the PIF binding site as described in this paper, since M3 contains a new CpG dinucleotide, fortuitously introduced 3 bases downstream of the mutated CpG. To test whether this new CpG dinucleotide introduced into M3 was critical in allowing recombinant PIF to bind this site, mutant M5, which resembles M3 but no longer contains the new CpG, was constructed. As predicted, this single-base substitution drastically reduced recognition of the oligonucleotide by recombinant PIF (Fig. 8A).
FIG. 8.
rPIF binds to an element in the tyrosine aminotransferase promoter. (A) EMSAs of recombinant PIF binding to a double-stranded, 32P-labeled oligonucleotide representing the wild-type GME present in the rat tyrosine aminotransferase promoter. Samples in lanes labeled GME through M5 were incubated in the presence of a 100-fold molar excess of an unlabeled wild-type (GME) or mutant (M1 to M5) oligonucleotide competitor, as indicated. (B) Methylation interference analysis of recombinant PIF binding to the rat tyrosine aminotransferase promoter element. The double-stranded wild-type GME oligonucleotide was 5′-end labeled with 32P on either the top or bottom strand, partially methylated, incubated with rPIF, and gel purified following fractionation by EMSA. Methylated residues which impair rPIF binding are indicated in boldface, and residues which interfered with GMEB binding, as determined by Oshima et al. (17), are indicated by arrows.
When methylation interference was used to detail the interaction between rPIF and GME (Fig. 8B), the interference pattern was identical to that previously described for the rat factor (17) and pinpointed the critical importance of two GCG sequences spaced 3 nucleotides apart in the binding site. By comparing this sequence with the PIF site in the viral origin, we conclude that the consensus half-site for PIF is likely to be PuCGPy, although there may be some preference for a thymidine residue in the fourth position of at least one of the boxes.
The second complex which resembles PIF is a human factor called TRAC that binds a regulatory element, TRA, in the promoter of the transferrin receptor (TR) gene, allowing it to respond to mitogen induced stimulation (19). The chromatographic profile of TRAC resembled that of PIF, and, like PIF, TRAC bound the synthetic alternating copolymer poly(dI-dC). Originally, we had been unable to elute PIF from a DNA-Sepharose column conjugated with highly oligomerized copies of its binding site, as found in the MVM origin, but were able to purify PIF activity by using DNA columns conjugated with a single long oligonucleotide containing this TR element, shown in Fig. 9, which had been shown by Roberts et al. to bind to and elute TRAC (20). These authors mapped the TRAC-binding site to the sequence AAGTGACG (boxed in Fig. 9), containing most of an isolated ACGC tetranucleotide, which lies 76 nucleotides upstream of the RNA start. However, consistent with the bipartite nature of the PIF recognition element, there is an additional ACGT half-site 13 nucleotides upstream of the first motif, and a further GCGT site immediately next to that, either or both of which might be expected to cooperate with the downstream ACGC to anchor PIF. We used methylation interference to map the PIF interaction with this oligonucleotide and found that the retarded band in the EMSA phase of the assay contained multiple species. The interference pattern obtained, diagrammed in Fig. 9, was complex and implicated all of the G residues within the upstream GCGTACGT sequence, as well as those within the downstream ACGC, without identifying any subset of residues as being absolutely critical. This suggests that some PIF heterodimers might bind to the two immediately juxtaposed half-sites in the GCGTACGT sequence while others interact with one of these sites plus the ACGC site mapped by Roberts et al. (19). This supports the notion that the consensus recognition element for PIF involves two flexibly spaced PuCGPy motifs and suggests that the PIF gene products might be involved in transcriptional modulation of the transferrin receptor. This analysis also highlights the difficulty in mapping such binding sites, where the presence of—or inadvertent mutational introduction of—additional tetranucleotide motifs provides redundant binding capacity.
FIG. 9.
PIF binding to regulatory elements within viral and cellular promoters. Comparison of the methylation interference pattern of rPIF binding to the MVM origin site, the GMEB site in the tyrosine aminotransferase promoter, and the TRA element of the TR promoter. Tetranucleotide motifs conforming to the PuCGPy consensus are shaded, and residues which, when methylated, interfere with rPIF binding are indicated by arrows.
DISCUSSION
PIF binds variably spaced CpG motifs and has probably been encountered in previous studies of transcriptional activation.
PIF has a bipartite recognition sequence, binding coordinately to two ACGT motifs spaced 5 nucleotides apart in the MVM origin, one of which also forms the core of an ATF site. However, PIF can accommodate linker sequences which vary in size from 1 to at least 15 nucleotides. In addition, some degeneracy in the first and fourth positions of each ACGT motif is tolerated, giving a consensus binding site of PuCGPy, with thymidine residues possibly being favored in the fourth position. Since the CpG dinucleotides at the centers of each motif are absolutely essential for binding, it is not surprising that synthetic polymers of alternating dI and dC residues were found to inhibit PIF binding (3). Historically, this is likely to be significant because ACGT is the core of consensus ATF/CREB binding sites and is also present in a subset of E-box motifs, comprising binding sites for members of the upstream stimulatory factor (USF) family and complexes containing c-Myc. PIF is expressed at around 2 × 104 copies per HeLa cell, so that without widespread use of poly(dI-dC) as a nonspecific competitor, PIF-binding activity would probably have been noticed in previous analyses of sequences which contained a single ATF site or E-box but which had an additional PuCGPy tetranucleotide within 15 bp or so. The spacing criteria for PIF appear so flexible that high-affinity binding sites for it are likely to be present in many human promoters, particularly those which incorporate ATF/CREB-, USF-, and c-Myc-binding sites.
A similar poly(dI-dC)-sensitive factor with a purification profile identical to that of PIF was reported by Roberts et al. (20) in their characterization of TRAC, a factor which binds the TRA element of the TR promoter (19), stimulating increased expression of this receptor in response to proliferation. As we have reported for PIF, these authors found that TRAC copurifies with the autoantigen Ku, which will also band-shift the various target oligonucleotides used in the present study. However, Ku binding is sequence independent and relatively poly(dI-dC) resistant, and in our hands, the site-specific binding activity detected in these partially purified fractions was due exclusively to PIF (3).
The TRA element contains several variably spaced CpG repeats, and, as detailed in Fig. 9, we have found that PIF binds to various combinations of these, making definitive footprinting of any one interaction tenuous. The fact that PIF binds to this site suggests that it may well be identical to TRAC and modulate transferrin receptor levels in vivo. This in turn suggests that PIF levels themselves may change in response to proliferation, although we have not examined this possibility directly.
Likewise, human PIF heterodimers bind effectively to the GME sequence in the rat tyrosine aminotransferase promoter, and mutational analysis of the site now indicates that this response element is also bipartite, containing paired PuCGPy motifs spaced 2 nucleotides apart. As mentioned above, Zeng et al. (26) have recently identified rat cDNAs encoding a protein which corresponds closely to the human p79 gene described here, so that the two complexes are clearly homologous. Binding of the GMEB complex to the GME site in the rat tyrosine aminotransferase promoter renders it responsive to low glucocorticoid concentrations (16), probably via direct interactions with the glucocorticoid receptor. This indicates that human PIF p96-p79 may similarly regulate the human tyrosine aminotransferase promoter and could also play a more general role in the combinatorial control of cellular promoters that interact with steroid receptors.
p96 and p79 belong to an emerging family of polypeptides which share a KDWK domain.
Sequence analysis shows that p96 and p79 have calculated molecular masses of 61 and 56 kDa, respectively, and are highly related but quite distinct, with only 40 to 42% identity overall. This homology is most evident in a 94-amino-acid domain, which shows 80% identity, beginning 80 to 90 residues from the amino termini of both chains. Recent studies with progressive truncations of both PIF subunits indicate that this region contains the DNA-binding domain. Searches involving this highly conserved sequence identified a small group of nuclear proteins which show conspicuous homology throughout most of this domain but not elsewhere. Initially recognized in the Drosophila homeobox modulator DEAF-1, this domain was designated KDWK, after one highly conserved cluster of residues (10). Another transcriptionally active nuclear protein, NUDR, which also shares this motif, was recently cloned from human, monkey, and rat cDNA libraries (11), while the SP100 group of nuclear dot antigens, which have also been implicated in transcriptional regulation (25), all contain a related domain. Significantly, most of these proteins have been reported to exhibit aberrantly low migration rates when analyzed by SDS-PAGE (1, 9, 21), suggesting that the KDWK domain itself may well be responsible for this anomaly. To date, no definitive binding specificities have been published for any of these proteins, but available data appear consistent with the novel mode of DNA binding described here for PIF. It thus seems likely that these molecules and, by analogy, all members of the KDWK family may bind DNA by engaging low-complexity, flexibly spaced half-sites containing a CpG dinucleotide core.
Factors which recognize flexibly spaced half-sites are rare.
Dimeric DNA-binding proteins generally recognize both the sequence and spacing of their two half-sites, but a few exceptions to this rule are currently recognized. For example, a subfamily of nuclear receptors which bind nonsteroid ligands, including vitamin D, thyroid hormone, and retinoic acid, all recognize variously spaced copies of the same minimal hexad half-site, AGGTCA. However, to activate transcription, these all associate with a second molecule, the retinoid X receptor (RXR), and interactions within the heterodimers cause them to have precise spacing requirements, so that vitamin D receptor-RXR heterodimers bind direct repeats spaced 3 nucleotides apart, thyroid hormone receptor-RXR heterodimers require a 4-nucleotide spacing, etc., in accordance with a model called the “1-to-5 rule” (reviewed in reference 14). Thus, this flexibility is more apparent than real and actually results from the use of different combinations of binding modules.
In contrast, γδ resolvase, a site-specific recombinase encoded by transposon γδ of Escherichia coli, can accommodate substantial differences in the disposition of its half-sites. This molecule interacts with three operator sequences in a 114-bp recombination element called res, and although each operator contains two copies of the 12-bp recognition sequence arranged as inverted repeats and all three sites bind resolvase dimers equally well, the three operators differ in having 1, 4, or 10 bp inserted between the half-sites (15).
A third and more extreme example of such flexibility is provided by the MATα2 protein of yeast (22). This homeodomain transcription factor normally recognizes a sequence in which its half-sites are separated by 2.5 helical turns, but it binds with similar affinity to sequences in which the spacing is modified by insertions and deletions that range over at least 27 nucleotides, while an insertion of 100 bp could be recognized, albeit with severely impaired efficiency. MATα2 contains quite distinct dimerization and DNA-binding domains, which appear to move relatively independently of each other, so that its DNA-binding domains are largely free from constraints imposed by the dimerization module while the complex still benefits from the enhanced avidity of a bipartite target. By analogy, it seems likely that the dimerization and DNA-binding domains of at least one PIF subunit must also be spaced separately and articulate relatively freely with respect to the other, in order to accommodate the spacing flexibility described here.
Homodimers of p96 bind the MVM origin at least as efficiently as p96-p79 heterodimers do, but they are unable to activate NS1. In contrast, p79 homodimers bind origin DNA poorly but can activate the nickase (4a), suggesting that p79 may carry the domain(s) required for interaction with NS1 while p96 exerts a greater influence on substrate affinity. However, in other cell types, PIF subunits might be selectively expressed, forming homodimers, perhaps altering the response of the cell to exogenous signals for transcriptional activation and incidentally rendering it refractory to MVM replication. This scenario would provide an additional level of combinatorial control, as would the ability, currently hypothetical, of p79 or p96 to heterodimerize with other members of the KDWK family. Such subunit shuffling could operate during differentiation to generate unique complexes, each with a subtly different site specificity or avidity, able to interact with a different spectrum of transcription factors, and thus effect extensive, coordinated shifts in gene expression.
ACKNOWLEDGMENTS
We thank Christine Ticknor for help in assembling and analyzing cDNA sequences and Ulla Toftegaard and Jessica Bratton for excellent technical assistance. We are indebted to John Flory, Kathy Stone, Karl Hager, and colleagues at the Keck Biotechnology Facility.
This work was supported by U. S. Public Health service grants AI26109 and CA29303 (to P.T.) from the National Institutes of Health. J.C. was supported by grants from RVAU, the Danish Agricultural and Veterinary Research Council, and the Danish Center for Biotechnology.
REFERENCES
- 1.Bloch D B, de la Monte S M, Guigaouri P, Filippov A, Bloch K D. Identification and characterization of a leukocyte-specific component of the nuclear body. J Biol Chem. 1996;271:29198–29204. doi: 10.1074/jbc.271.46.29198. [DOI] [PubMed] [Google Scholar]
- 1a.Burnett, E., J. Christensen, C. Ticknor, and P. Tattersall. Unpublished data.
- 2.Christensen J, Cotmore S F, Tattersall P. The minute virus of mice transcriptional activator protein, NS1, binds directly to the trans-activating region (tar) of the viral P38 promoter in a strictly ATP-dependent manner. J Virol. 1995;69:5422–5430. doi: 10.1128/jvi.69.9.5422-5430.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christensen J, Cotmore S F, Tattersall P. A novel cellular site-specific DNA-binding protein cooperates with the viral NS1 polypeptide to initiate parvovirus DNA replication. J Virol. 1997;71:1405–1416. doi: 10.1128/jvi.71.2.1405-1416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christensen J, Cotmore S F, Tattersall P. Parvovirus initiation factor PIF: a novel human DNA-binding factor which coordinately recognizes two ACGT motifs. J Virol. 1997;71:5733–5741. doi: 10.1128/jvi.71.8.5733-5741.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Christensen, J., S. F. Cotmore, and P. Tattersall. Unpublished data.
- 5.Christensen J, Storgaard T, Viuff B, Aasted B, Alexandersen S. Comparison of promoter activity in Aleutian mink disease parvovirus, minute virus of mice, and canine parvovirus: possible role of weak promoters in the pathogenesis of Aleutian mink disease parvovirus infection. J Virol. 1993;67:1877–1886. doi: 10.1128/jvi.67.4.1877-1886.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cotmore S F, Christensen J, Nuesch J P F, Tattersall P. The NS1 polypeptide of the murine parvovirus minute virus of mice binds to DNA sequences containing the motif [ACCA]2-3. J Virol. 1995;69:1652–1660. doi: 10.1128/jvi.69.3.1652-1660.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotmore S F, Tattersall P. An asymmetric nucleotide in the parvoviral 3′ hairpin directs segregation of a single active origin of DNA replication. EMBO J. 1994;13:4145–4152. doi: 10.1002/j.1460-2075.1994.tb06732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deleu L, Fuks F, Spitkovsky D, Horlein R, Faisst S, Rommelaere J. Opposite transcriptional effects of cyclic AMP-responsive elements in confluent or p27KIP-overexpressing cells versus serum-starved or growing cells. Mol Cell Biol. 1998;18:409–419. doi: 10.1128/mcb.18.1.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dent A L, Yewdell J, Puvion-Dutilleul F, Koken M H, de The H, Staudt L M. LYSP100-associated nuclear domains (LANDs): description of a new class of subnuclear structures and their relationship to PML nuclear bodies. Blood. 1996;88:1423–1426. [PubMed] [Google Scholar]
- 10.Gross C T, McGinnis W. DEAF-1, a novel protein that binds an essential region in a Deformed response element. EMBO J. 1996;15:1961–1970. [PMC free article] [PubMed] [Google Scholar]
- 11.Huggenvik J I, Michelson R J, Collard M W, Ziemba A J, Gurley P, Mowen K A. Characterization of a nuclear deformed epidermal autoregulatory factor-1 (DEAF-1)-related (NUDR) transcriptional regulator protein. Mol Endocrinol. 1998;12:1619–1639. doi: 10.1210/mend.12.10.0181. [DOI] [PubMed] [Google Scholar]
- 12.LeBoeuf R D, Ban E M, Green M M, Stone A S, Propst S M, Blalock J E, Tauber J D. Molecular cloning, sequence analysis, expression, and tissue distribution of suppressin, a novel suppressor of cell cycle entry. J Biol Chem. 1998;273:361–368. doi: 10.1074/jbc.273.1.361. [DOI] [PubMed] [Google Scholar]
- 13.LeBoeuf R D, Burns J N, Bost K L, Blalock J E. Isolation, purification, and partial characterization of suppressin, a novel inhibitor of cell proliferation. J Biol Chem. 1990;265:158–165. [PubMed] [Google Scholar]
- 14.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murley L L, Grindley N D F. Architecture of the gamma-delta resolvase synaptosome—oriented heterodimers identify interactions essential for synapsis and recombination. Cell. 1998;95:553–562. doi: 10.1016/s0092-8674(00)81622-0. [DOI] [PubMed] [Google Scholar]
- 16.Oshima H, Simons S S., Jr Modulation of transcription factor activity by a distant steroid modulatory element. Mol Endocrinol. 1992;6:416–428. doi: 10.1210/mend.6.3.1584217. [DOI] [PubMed] [Google Scholar]
- 17.Oshima H, Szapary D, Simons S S., Jr The factor binding to the glucocorticoid modulatory element of the tyrosine aminotransferase gene is a novel and ubiquitous heteromeric complex. J Biol Chem. 1995;270:21893–21901. doi: 10.1074/jbc.270.37.21893. [DOI] [PubMed] [Google Scholar]
- 18.Perros M, Deleu L, Vanacker J M, Kherrouche Z, Spruyt N, Faisst S, Rommelaere J. Upstream CREs participate in the basal activity of minute virus of mice promoter P4 and in its stimulation in ras-transformed cells. J Virol. 1995;69:5506–5515. doi: 10.1128/jvi.69.9.5506-5515.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts M R, Han Y, Fienberg A, Hunihan L, Ruddle F H. A DNA-binding activity, TRAC, specific for the TRA element of the transferrin receptor gene copurifies with the Ku autoantigen. Proc Natl Acad Sci USA. 1994;91:6354–6358. doi: 10.1073/pnas.91.14.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts M R, Miskimins W K, Ruddle F H. Nuclear proteins TREF1 and TREF2 bind to the transcriptional control element of the transferrin receptor gene and appear to be associated as a heterodimer. Cell Regul. 1989;1:151–164. doi: 10.1091/mbc.1.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seeler J S, Marchio A, Sitterlin D, Transy C, Dejean A. Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc Natl Acad Sci USA. 1998;95:7316–7321. doi: 10.1073/pnas.95.13.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith D L, Johnson A D. A molecular mechanism for combinatorial control in yeast: MCM1 protein sets the spacing and orientation of the homeodomains of an alpha 2 dimer. Cell. 1992;68:133–142. doi: 10.1016/0092-8674(92)90212-u. [DOI] [PubMed] [Google Scholar]
- 22a.Ticknor, C. M., and P. Tattersall. Unpublished data.
- 23.van der Vliet P. Roles of transcription factors in DNA replication. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 87–118. [Google Scholar]
- 24.van Leeuwen H C, Rensen M, van der Vliet P C. The Oct-1 POU homeodomain stabilizes the adenovirus preinitiation complex via a direct interaction with the priming protein and is displaced when the replication fork passes. J Biol Chem. 1997;272:3398–3405. doi: 10.1074/jbc.272.6.3398. [DOI] [PubMed] [Google Scholar]
- 25.Xie K, Lambie E J, Snyder M. Nuclear dot antigens may specify transcriptional domains in the nucleus. Mol Cell Biol. 1993;13:6170–6179. doi: 10.1128/mcb.13.10.6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng H W, Jackson D A, Oshima H, Simons S S. Cloning and characterization of a novel binding factor (GMEB-2) of the glucocorticoid modulatory element. J Biol Chem. 1998;273:17756–17762. doi: 10.1074/jbc.273.28.17756. [DOI] [PubMed] [Google Scholar]