Abstract
Diffusion MRI tractography produces massive sets of streamlines that need to be clustered into anatomically meaningful white-matter bundles. Conventional clustering techniques group streamlines based on their proximity in Euclidean space. We have developed AnatomiCuts, an unsupervised method for clustering tractography streamlines based on their neighboring anatomical structures, rather than their coordinates in Euclidean space. In this work, we show that the anatomical similarity metric used in AnatomiCuts can be extended to find corresponding clusters across subjects and across hemispheres, without inter-subject or inter-hemispheric registration. Our proposed approach enables group-wise tract cluster analysis, as well as studies of hemispheric asymmetry. We evaluate our approach on data from the pilot MGH-Harvard-USC Lifespan Human Connectome project, showing improved correspondence in tract clusters across 184 subjects aged 8–90. Our method shows up to 38% improvement in the overlap of corresponding clusters when comparing subjects with large age differences. The techniques presented here do not require registration to a template and can thus be applied to populations with large inter-subject variability, e.g., due to brain development, aging, or neurological disorders.
Keywords: Hierarchical clustering, Normalized cuts, Tractography, Diffusion MRI
1. Introduction
The human brain undergoes structural changes from birth to adulthood (Giedd et al., 1999; Sowell et al., 1999; Zuo et al., 2017). Understanding and characterizing typical brain development, maturation, and aging is crucial for early detection of neurological diseases (Dinstein et al., 2011; Mahone et al., 2011; Peters and Karlsgodt, 2015) and to inform treatment and intervention approaches (Lindenberger, 2014; Raz and Rodrigue, 2006).
Postmortem studies have found age-related differences in various morphological aspects of the brain, such as size, weight, expansion of cerebral ventricles, axon diameter, and myelin sheath thickness (Benes, 1989, 1994). MRI provides in-vivo insights into morphological changes during the lifespan, such as in gray and white matter (WM) volumes (Paus, 1999; Sullivan and Pfefferbaum, 2006; Tamnes et al., 2010; Taki et al., 2012). Changes in connectivity as measured by functional MRI (fMRI) have also been demonstrated during normal development. Young populations appear to have more efficient networks (Betzel et al., 2014), while aging increases local segregation and thus requires interaction between networks (Chan et al., 2014; Fair et al., 2009; Reuter-Lorenz, 2002; Tsang et al., 2017). Although such macrostructural functional network changes have been shown, it is still unclear whether these are accompanied by WM microstructural alterations (Baum et al., 2017; Betzel et al., 2014; Tsang et al., 2017).
White matter undergoes conspicuous growth during the early years of life and continues to develop at a slower pace into adulthood (Lebel et al., 2017; Paus, 1999). Diffusion MRI (dMRI) provides indirect measurements of the microstructural changes in WM. Neurodevelopmental studies have used diffusion tensor imaging (DTI) (Basser et al., 1994) to analyze changes in fractional anisotropy, mean, radial, and axial diffusivity, which are presumed to be markers of myelination, axonal density and coherence (Betzel et al., 2014; Lebel et al., 2008; Sullivan and Pfefferbaum, 2006; Tsang et al., 2017). Studies show that diffusion anisotropy, which may be a marker of coherence or myelination, increases in several WM bundles until young adulthood, reaching a peak during adulthood, before decreasing with age. More recent studies investigated age-related changes in other dMRI-based microstructural measures, which attempt to distinguish among the various factors that contribute to diffusion anisotropy, such as measures from diffusion kurtosis imaging (Grinberg et al., 2017; Paydar et al., 2013) or neurite orientation, dispersion and density imaging (Chang et al., 2015; Genc et al., 2017; Kodiweera et al., 2016).
White matter changes through the lifespan are non-linear and their rates differ between bundles. For example, maturation in callosal and projection pathways appear to be mostly complete by the end of adolescence, while maturation in certain fronto-temporal association pathways continues into the twenties. The most commonly used models for lifespan trajectories of WM microstructural measures are linear (Kodiweera et al., 2016; Sullivan and Pfefferbaum, 2006), quadratic (Betzel et al., 2014) and exponential (Lebel et al., 2008; Paydar et al., 2013; Tamnes et al., 2010). However, these models are not necessarily linked to any particular biological process and their accuracy may depend on the age range and target WM bundle (Lebel et al., 2017).
The study of WM development relies on the delineation of anatomically meaningful bundles. Most lifespan studies of WM microstructure to date have focused on a small set of predefined WM pathways. The majority did not use tractography to delineate these tracts, instead resorting to region of interest (ROI) volumes that were defined manually in each individual (Lebel et al., 2008; Paydar et al., 2013; Sullivan et al., 2001), obtained by growing an individual cortical parcellation into the WM (Tamnes et al., 2010), or mapped to the individual from an atlas (Chang et al., 2015; Genc et al., 2017; Grinberg et al., 2017; Kodiweera et al., 2016; Pohl et al., 2016; Tamnes et al., 2010). Alternatively, some studies performed tractography in individuals and used ROIs from an atlas to define tracts of interest (Clayden et al., 2012; Lebel et al., 2008, 2012). Any approach that relies on ROIs from an atlas requires accurate alignment of each individual to the atlas template space, which can be challenging for populations with large age ranges or other sources of morphological variability. Using priors on the anatomical neighborhood of the tracts of interest is an alternative approach that does not require perfect alignment in template space and that can be readily applied to longitudinal studies of age-related WM change (Fjell et al., 2016, 2017; Storsve et al., 2016).
Here we focus on the problem of performing exploratory, data-driven analysis of whole-brain tractography data, rather that analyzing predefined tracts of interest. This can be done by applying unsupervised clustering to the whole-brain tractography streamlines of each individual and then establishing correspondence between the clusters of different individuals. A multitude of tract clustering algorithms exist in the literature. When it comes to across-subject analysis of clusters, some follow a supervised approach, i.e., require input from experts to define a set of tracts of interest and constrain the analysis to clusters that match those tracts (Garyfallidis et al., 2018; Jin et al., 2014; Guevara et al., 2012; O’Donnell and Westin, 2007; Ros et al., 2013; Zhang et al., 2018; Ziyan et al., 2009). Purely unsupervised approaches usually register all subjects to a common template space and match clusters across subjects based on their Euclidean distance in that space (Guevara et al., 2012; Ros et al., 2013; Visser et al., 2011; Wang et al., 2011; Ziyan et al., 2009). Unsupervised approaches that do not require inter-subject registration rely either on shape descriptors of the clusters (Zhang et al., 2014) or on the anatomical regions that the clusters terminate in. The termination regions may be used in a post-processing step after unsupervised clustering (Wassermann et al., 2010) or in the similarity metric of the clustering algorithm itself (Tunç et al., 2014). The method that we present here generalizes the approach of using individual anatomical regions in the tract similarity metric, as it quantifies the similarity of streamlines and clusters based on all their anatomical neighbors, and not just their termination points.
In addition to inter-subject correspondence for group-wise tract cluster analysis, the method that we present may be used to find corresponding tract clusters between a subject’s hemispheres, which can be useful for studying laterality. When focusing on predefined tracts of interest one can readily compare the left and right counterparts of the same tract (Garic et al., 2018; Propper et al., 2010; Wu et al., 2012). When following an unsupervised approach, however, this becomes less straightforward. Prior work computed the similarity of streamlines from contralateral hemispheres by assuming that they are mirror images of each other (O’Donnell et al., 2010) or performed a nonlinear registration between hemispheres (Park et al., 2004).
In this work, we build upon our previously proposed method for unsupervised clustering of streamlines based on anatomical similarity (Siless et al., 2016, 2018). Previously, we evaluated this similarity for clustering the whole-brain tractography streamlines of a single subject, showing that it produces clusters more similar to WM bundles defined manually by a human rater, even though it does not use any manually labeled training data (Siless et al., 2018). Our similarity metric compares the histograms of neighboring anatomical structures of two streamlines for different directions. Here we take this approach a step further, showing that, once this clustering has been performed in each individual subject, an extended anatomical similarity metric with canonical directions can be used to find corresponding clusters between individuals and between hemispheres, without inter-subject or inter-hemispheric registration. We evaluate the approach on data from the MGH-Harvard-USC lifespan human connectome project (Fan et al., 2016). We compare our method with a conventional similarity metric based on Euclidean distances, which does rely on inter-subject registration, and show that our approach improves correspondence of tract clusters across subjects aged 8–90. This work expands substantially on the preliminary results presented in (Siless et al., 2017) and extends the methodology to finding corresponding clusters between hemispheres, for studies of symmetry. Our method is particularly relevant to the analysis of populations with large morphological variability, such as healthy subjects across the lifespan or subjects with neurodegenerative diseases and healthy controls.
2. Methods
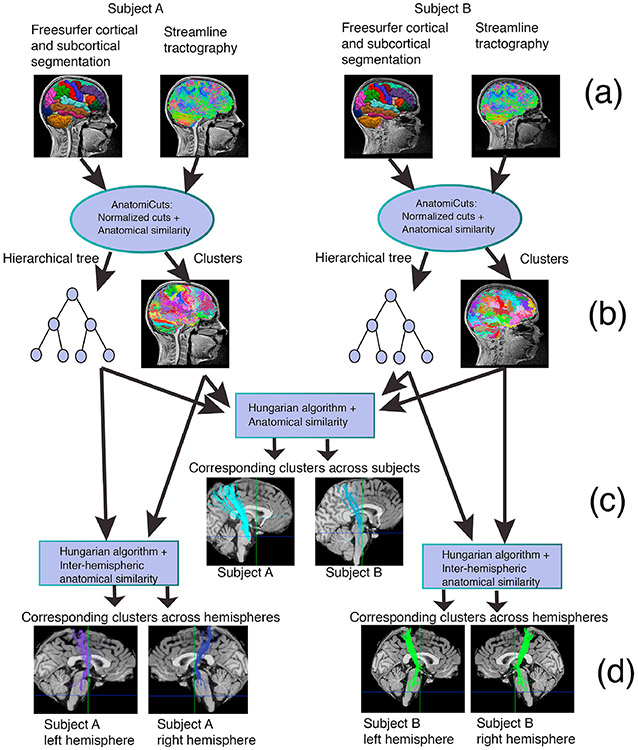
We propose an unsupervised method for finding corresponding WM structures across subjects without the need for inter-subject registration. Briefly, we group each subject’s whole-brain tractography streamlines into clusters with our recently developed algorithm for hierarchical clustering based on anatomical similarity, AnatomiCuts (Siless et al., 2018). Diffusion measures for each cluster can be extracted in each subject’s native space and compared between subjects without aligning subjects to a template. In the following we define our measure of anatomical similarity between two streamlines, as well as a more conventional one based on Euclidean distance, which we use for comparison. We then generalize these definitions to quantify similarity between two clusters of streamlines that may come from different subjects or from different hemispheres of the same subject. The procedures described below are summarized in Fig. 1.
Fig. 1.
Algorithm overview: Whole-brain tractography streamlines from each individual (a) are grouped into a fixed number of clusters (b) with our anatomical similarity metric, which utilizes a cortical and subcortical segmentation from Freesurfer. Clusters from different subjects are matched based on their anatomical similarity (c), which does not require inter-subject registration. Clusters are also matched between each subject’s hemispheres based on their anatomical similarity, for symmetry analysis (d).
2.1. Streamline similarity metrics
Let fi = [xi1,…, xiN] be a streamline defined as a sequence of N points , k = 1,…,N. A tractography dataset is a set of M streamlines, F = {f1,…, fM}. In the following, we assume that all streamlines have the same number of points N. This can be enforced by downsampling, which is performed commonly to make computation for clustering algorithms tractable (Garyfallidis et al., 2012; Guevara et al., 2012; O’Donnell and Westin, 2007; Siless et al., 2013, 2018; Visser et al., 2011; Wu et al., 2012).
2.1.1. Euclidean similarity
This similarity metric has been used widely for tract clustering and consists in the mean Euclidean distance between the corresponding points of two streamlines fi and fj (Garyfallidis et al., 2012; Guevara et al., 2012; Siless et al., 2013, 2018; Visser et al., 2011; Wu et al., 2012):
The ordering of points is not consistent across streamlines, i.e., it is possible for the first point of fi to be closer to the last point of fj and vice versa. It is typical to account for this by also evaluating the similarity between fi and the reversed fj. This leads to the following definition for the similarity metric:
| (1) |
where fjrev = [xjN, …, xj1].
2.1.2. Anatomical similarity
In (Siless et al., 2018) we introduced a streamline similarity metric that makes use of a whole-brain cortical and subcortical segmentation, . Each point x on a streamline is associated with a set of segmentation labels, S(x + dl(x)vl), l = 1, …, P, where dl(x) is the minimum d > 0 such that S(x + dvl) ≠ S(x). That is, for each point x, we find the nearest neighboring segmentation labels in a set of directions vl, l = 1, …, P. Here we modify the definition of those directions, to ensure that they are consistent across different subjects. Specifically, a neighborhood of P = 26 elements includes neighboring labels in the directions vl = U · el, where U ≜ [uLR uAP uSI], e ≜ [e1 e2 e3]T, e1,2,3 ∈ {−1,0,1} and we use v0 = [0,0,0] to represent the segmentation label that the streamline passes through. The columns of the matrix U are unit vectors in the left-right, anterior-posterior, and superior-inferior direction. We find these vectors as follows. We fit the mid-sagittal plane using the FreeSurfer segmentation labels of the mid-sagittal section for the corpus callosum and the third ventricle. We define uAP by projecting the line that connects the center of mass of the anterior and posterior cingulate labels onto the mid-sagittal plane. We define uSI as perpendicular to uAP within the mid-sagittal plane, and uLR as perpendicular to that plane.
For each direction l = 0,…,P, we compute a label histogram , where K is the total number of labels in the anatomical segmentation. This histogram represents the frequency with which different segmentation labels are the l-th neighbor across all points on the i-th streamline. The anatomical similarity measure between two streamlines fi and fj expresses the joint probability of their anatomical neighborhoods:
| (2) |
where ⟨·, ·⟩ is the inner product, and Li, Lj are the sets of all labels found to be neighbors of streamlines fi, fj. The normalization factor ∣Lj∩ Lj∣, which is the number of common neighbors between the two streamlines, penalizes trivial streamlines with too few neighbors.
2.1.3. Inter-hemispheric anatomical similarity
Let fi and fj be streamlines in a subject’s left and right hemisphere, respectively. Let SL(x) and SR(x) be anatomical segmentations of the left and right hemisphere, such that each structure and its contralateral counterpart (e.g., the left and right amygdala) have the same label ID. We define the inter-hemispheric anatomical similarity of streamlines fi and fj as:
| (3) |
where the neighbor histograms , are computed for the segmentations SL(x), SR(x), respectively, and, if the l-th neighbor is defined by in the left hemisphere, then it is defined by in the right.
2.2. Cluster similarity metrics
We define the Euclidean-distance similarity of two streamline clusters as the Euclidean-distance similarity of their centroid streamlines. This definition has been previously used in the literature (Guevara et al., 2012; Ros et al., 2013). The centroid streamline is the closest streamline to the average of all streamlines in a cluster. The centroids of two clusters must be in a common space to compute this similarity metric. We define the anatomical similarity of two streamline clusters by substituting the cumulative histogram of anatomical neighbors of all streamlines in each cluster in equation (2) or (3). Such histograms can be compared between two clusters even if the clusters are not in the same space.
2.3. Matching clusters across subjects
Let C be the number of streamline clusters per subject. This number can be fixed by stopping the hierarchical clustering of each subject’s tractography dataset when the hierarchical tree has C leaf nodes. Given the C × C matrix W = {wmn}, where wmn is the similarity of the m-th cluster from one subject and the n-th cluster from another, our goal is to find the permutation n1,…,nC of 1,…,C that maximizes , i.e., the matching of clusters between the two subjects that maximizes the total cluster-to-cluster similarity. We solve the problem efficiently with the Hungarian algorithm (Kuhn, 1955, 2009), which has been previously applied to tract matching (Tunç et al., 2014). A subject is chosen at random and the Hungarian algorithm is used to assign correspondence between the clusters of that subject and those of every other subject, based either on Euclidean distance or on anatomical similarity between clusters.
Note that this implementation enforces one-to-one correspondence of clusters between subjects, which may be a stringent requirement. Thus we investigate the use of inter-subject consistency, quantified by the coefficient of variation (CV), as a means for detecting outlier clusters with inconsistent similarity across subjects. For the Euclidean-distance similarity metric, these would be clusters with inconsistent alignment across subjects in template space. For our anatomical similarity metric, they would be clusters with inconsistent anatomical neighbors across subjects. Inter-subject consistency has been used previously to threshold the edges of brain networks for graph-theory analysis (Baum et al., 2018; Roberts et al., 2017).
2.4. Matching clusters across hemispheres
In this case, the Hungarian algorithm is applied to the inter-hemispheric anatomical similarities of clusters from a subject’s left and right hemispheres. Before computing the similarity matrix, every streamline that switches hemispheres is removed. Clusters with more than 20% of streamlines switching hemispheres are excluded from this computation entirely. There is no guarantee that the two hemispheres will have the same number of clusters. This, however, is not a requirement for the Hungarian algorithm and any excess clusters of low inter-hemispheric similarity will remain unmatched.
2.5. Data acquisition
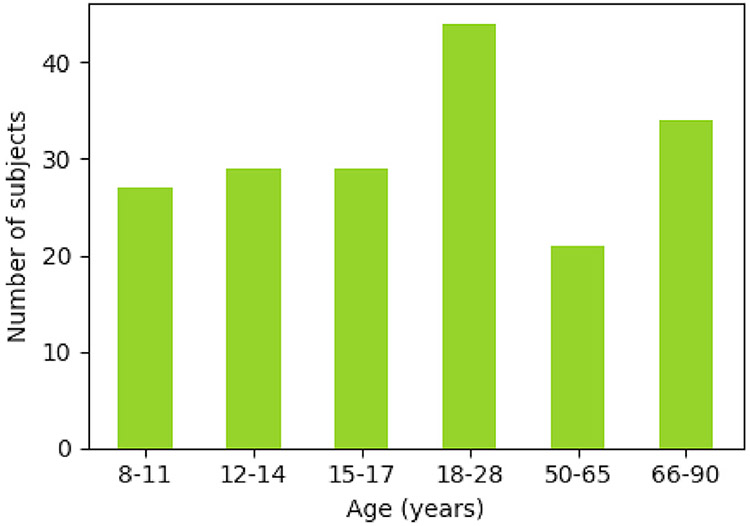
In the following experiments, we use dMRI and structural MRI (sMRI) data from 184 healthy subjects, scanned as part of the pilot MGH-Harvard-USC Lifespan Human Connectome project (Fan et al., 2016). Subjects’ ages range from 8 to 90 years as shown in Fig. 2. T1-weighted images were acquired with 3D multi-echo magnetization prepared rapid gradient echo (MEMPRAGE) (Van der Kouwe et al., 2008) at 1mm isotropic resolution with an acquisition time under 4 min. Diffusion data were acquired using a 2D spin echo echo-planar imaging (EPI) sequence with a generalized autocalibrating partial parallel acquisition (GRAPPA) factor of 3, combined with fast low angle excitation echo-planar technique with auto-calibration signal (FLEET-ACS) (Polimeni et al., 2016). A simultaneous multi-slice (SMS) factor of 2 was used, allowing for faster data acquisition. The dMRI acquisition scheme used two b-values, 2500s/mm2 and 7500s/mm2, with 60 and 180 diffusion-encoding directions, respectively. Data were acquired on the MGH 3T Skyra Connectom scanner, featuring a maximum gradient strength of 300mT/m (Setsompop et al., 2013). The T1-weighted data were processed in FreeSurfer to extract cortical parcellations and subcortical segmentations (Fischl et al., 2002, 2004). The dMRI data were corrected for eddy-current (Andersson and Sotiropoulos, 2016) and magnetic susceptibility artifacts (Andersson et al., 2003).
Fig. 2.
Age distribution of the 184 volunteers.
2.6. Data analysis
We reconstructed orientation distribution functions from the dMRI data using the generalized q-sampling imaging model (Yeh et al., 2010) and performed whole-brain, deterministic tractography using DSI Studio (Yeh et al., 2013). We seeded every voxel in the segmentation map computed by FreeSurfer. Following (Siless et al., 2018) we focused only on long-range connections, as analysis methods can be better optimized for long- and short-range connections separately (Guevara et al., 2012, 2017; Román et al., 2017; Zhang et al., 2014). We obtained a large number of long-range connections while keeping computation tractable by generating a total of 500,000 streamlines per subject and then excluding any streamlines shorter than 55 mm. This yielded between 100,000 and 150,000 streamlines per subject. For reference, (Román et al., 2017) defined short connection streamlines as shorter than 80mm, thus our criterion was less stringent. We downsampled all streamlines to N = 10 equispaced points. Previously we found that increasing N did not have enough of an impact on the clustering to justify the increased computational cost (Siless et al., 2018). The dMRI data were registered to the individual’s T1 volume by applying a boundary-based, affine registration (Greve and Fischl, 2009) to the b = 0 volumes.
We clustered each subject’s streamlines using the normalized cuts algorithm, with our anatomical similarity and with the conventional, Euclidean-distance similarity, as in (Siless et al., 2018). For the anatomical similarity, segmentation labels came from each subject’s cortical, subcortical and white-matter segmentations computed by Freesurfer. The number of anatomical neighbors was set to P = 26, as we have previously found this to perform better than smaller neighborhoods (Siless et al., 2018). In our previous evaluations, we have found that 200 clusters outperformed smaller numbers of clusters when comparing the overlap of tract clusters with a set of WM bundles that were defined manually by a human rater (Siless et al., 2018). Hence, we obtained 200 clusters per subject with each method. We then used each of the two similarity metrics to find one-to-one correspondence of clusters between subjects with the Hungarian algorithm.
Although our anatomical similarity metric does not require inter-subject registration to find inter-subject cluster correspondence, the conventional, Euclidean-distance similarity metrics do. Clusters from different subjects must be mapped to a common template space before Euclidean distances between them can be computed. For this purpose, we performed a combined volume and surface (CVS) registration (Postelnicu et al., 2009) between each subject’s T1 and a template T1 volume. We opted to use a single-subject template, i.e., the T1 of a randomly selected subject from the young adult cohort (age 25). We mapped each individual’s clusters to this template by composing the affine registration from individual dMRI to individual T1 space and the nonlinear registration from individual T1 to template T1 space. We compared this to a more standard nonlinear inter-subject registration approach for dMRI data. We stress once again that this registration is required only for computing the Euclidean-distance similarity metric, and not our proposed anatomical similarity metric.
Finally, we fit tensors to the b = 2500 dMRI data of each individual and computed fractional anisotropy (FA) and mean/radial/axial diffusivity (MD/RD/AD) maps from the tensors.
2.7. Evaluation metrics
2.7.1. Inter-subject overlap
For each cluster, we generated a binary volume with values of 1 in the voxels that intersected the streamlines of the cluster and 0 elsewhere. We quantified the overlap for a pair of corresponding clusters from two subjects by computing their Dice coefficient (Dice, 1945; Sørensen, 1948), which is defined as for two sets A and B. For each pair of ∣Ar+∣Br subjects, we averaged the Dice coefficients of all 200 corresponding clusters. We investigated how the overlap of two subjects’ clusters changed as the age difference between subjects increased. Clusters that had been generated and matched based on anatomical similarity had to be mapped to the template space to compute their inter-subject overlap. Clusters that had been generated and matched between subjects based on Euclidean distance were already mapped to template space.
Previously, CVS registration was shown to perform better than other nonlinear methods at aligning WM bundles between subjects in a cohort with a smaller age range than ours (Zöllei et al., 2010). For comparison, we used FSL tools (Jenkinson et al., 2002, 2012) to perform nonlinear registration of each subject’s FA map to the FMRIB FA template. We compared the image difference between target and source FA images for the two inter-subject registration methods.
2.7.2. Inter-subject consistency
We quantified the variability of corresponding clusters across subjects by the coefficient of variation (CV), , where σ is the standard deviation and μ is the mean. We first computed the CVs of the Dice coefficients of corresponding clusters between pairs of subjects.
We also computed histograms of the CV of the anatomical and Euclidean-distance similarities, for different numbers of clusters per subject. The goal of this evaluation was to understand how the number of clusters can impact the inter-subject consistency of corresponding clusters, and whether the latter can be used for outlier exclusion.
2.7.3. Inter-hemispheric vs. inter-subject similarity
We expected anatomical similarity to be comparable between corresponding clusters in different hemispheres and in different subjects, as it is based on histograms of neighboring anatomical structures, and the same structures exist in both hemispheres and all subjects. Thus, we compared the mean anatomical similarity between corresponding clusters obtained with the inter-hemispheric (intra-subject) or the inter-subject Hungarian algorithm. We evaluated this for different numbers of clusters.
2.7.4. Fitting age-related changes of diffusion measures
We computed the average FA, MD, RD and AD for each cluster in the individual dMRI space. After establishing cluster correspondence across subjects, we used these measures for a cross-sectional study of microstructure changes in each of the 200 clusters across the lifespan. We fit the models that are most commonly used to study age-related WM changes in the literature: linear, quadratic and Poisson (exponential).
Linear: y = β0 + β1 · t + β2 · s
Quadratic: y = β0 + β1 · t + β2 · t2 + β3 · s
Poisson: y = β0 + β1 · t · e−β2 · t + β3 · s
Here y is a microstructural measure (FA/MD/RD/AD), t is age, and s is a discrete variable of gender. We performed least-squares estimation of βi. We used the residual errors to compare the goodness-of-fit for clusters generated with each of the similarity metrics.
2.7.5. Computation time
The computational complexity of the Hungarian algorithm is O(C3) (Jonker and Volgenant, 1987). The complexity of computing the similarity matrix is O(CD), where D is the complexity of the similarity metric, which is O(N) and O(NP) for the Euclidean and anatomical similarity, respectively. However, the Euclidean-distance similarity requires a registration step whose complexity is variable and not accounted for in the above. As explained in previous sections, our experiments used C = 200, N = 10, P = 26.
3. Results
3.1. Inter-subject overlap
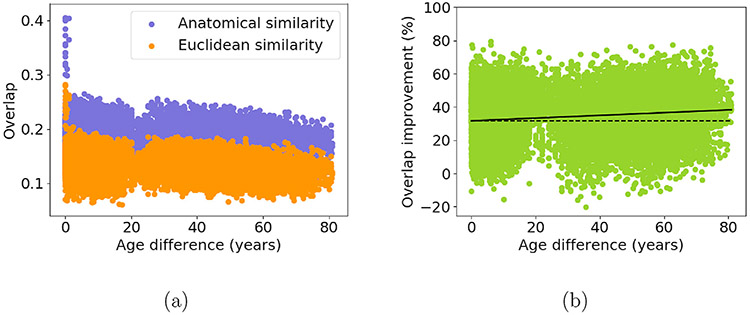
Fig. 3 shows the overlap (Dice coefficient) of corresponding clusters between pairs of subjects, as a function of the age difference between the subjects. Fig. 3(a) plots the average overlap over all 200 clusters, for clusters generated and matched across subjects with the anatomical and the Euclidean-distance similarity metric. Fig. 3(b) plots the percent change between the former and the latter. Our anatomical similarity metric yielded clusters with greater inter-subject overlap than the conventional, Euclidean-distance similarity metric, and it also led to less pronounced deterioration of the overlap as the age difference between subjects increased (p < 0.0001 based on a two-sided T-test on the linear regression of percent improvement vs. age difference). The average improvement in inter-subject cluster overlap afforded by using the anatomical over the Euclidean-distance similarity metric ranged from 31% for subjects of the same age to 38% for subjects who were 80 years apart in age.
Fig. 3.
Inter-subject overlap. For each pair of subjects we average the Dice coefficients of the 200 corresponding clusters. We plot the average Dice coefficient of corresponding clusters for each pair of subjects, for clusters obtained with the anatomical and the Euclidean-distance similarity metric (a) and the percent difference between them (b), as a function of the age difference between subjects.
In Fig. S1 we show that inter-subject mean squared differences between registered FA images were smaller with CVS registration to a single-subject T1 template composed with the mapping between T1 and diffusion space, than nonlinear registration to an average FA template. Hence we proceed with using CVS registration for the Euclidean-distance similarity in all remaining analyses.
3.2. Inter-subject consistency
Fig. 4 shows the CV of the Dice coefficients of corresponding clusters between pairs of subjects. We evaluated the effect of the subject that is chosen at random as the target for the Hungarian algorithm by repeating the CV calculation for different target subjects. The plot shows the average and standard error of these CV values. The inter-subject overlap had lower CV for clusters produced with the anatomical than the Euclidean-distance similarity metric, with few outliers. The choice of target subject had a negligible effect on this outcome. Hence we proceed with a single, randomly selected target subject for all remaining results.
Fig. 4.
Coefficient of variation of the overlap of corresponding clusters across subjects for each similarity metric. Average values and standard error bars are plotted across different subjects chosen as the target for the Hungarian algorithm.
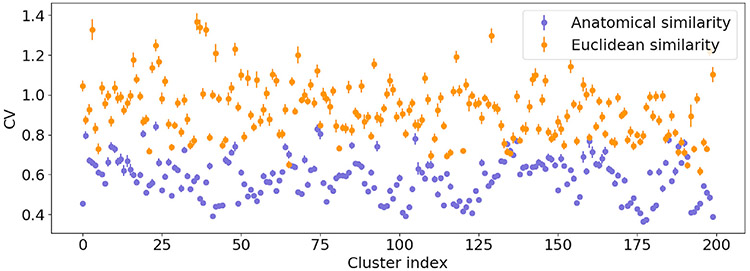
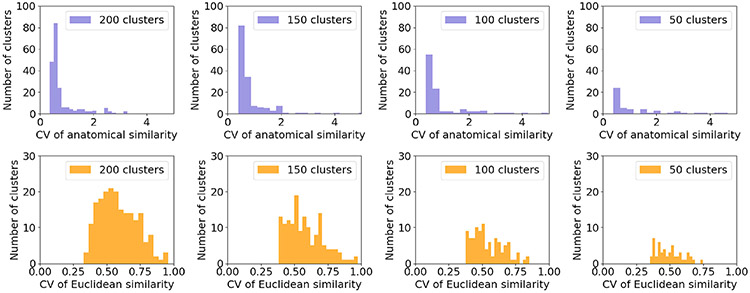
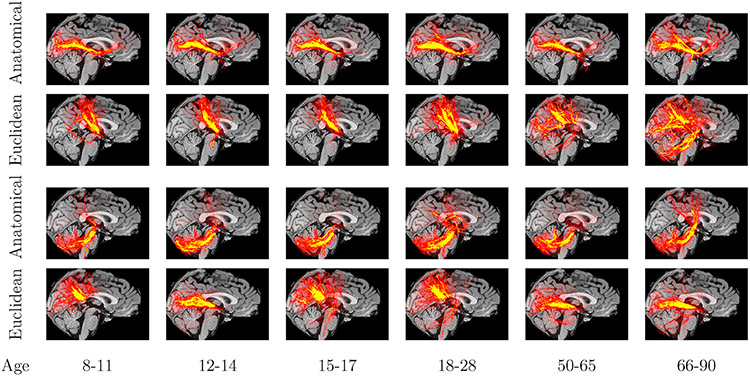
We show histograms of the CV of the similarities between corresponding clusters, for different numbers of clusters, in Fig. 5. For all numbers of clusters, the distribution of cluster CVs for the Euclidean-distance similarity did not allow the detection of outliers. Histograms for the anatomical similarity had a few easily discernible outlier clusters of high CV. In Fig. 6 we show the clusters with the highest and lowest CV of similarities. The heat maps are binary images of corresponding clusters from all subjects in an age group, summed in template space. For the anatomical similarity, the cluster with lowest CV shows a portion of the inferior longitudinal fasciculus, and the cluster with highest CV shows the corticospinal tract that decussated for some age groups but not others. This illustrates a clear differentiation, in terms of anatomical consistency, between the low-CV and high-CV clusters and confirms that it would be reasonable to exclude the latter as outliers. For the Euclidean-distance similarity metric, the cluster with lowest CV contains a portion of the thalamic radiation and the cluster with highest CV mixes portions of the inferior longitudinal fasciculus and the corpus callosum.
Fig. 5.
Histograms of the CV of anatomical (top row) and Euclidean distance (bottom row) inter-subject similarities. This is shown for different numbers of clusters per subject.
Fig. 6.
Examples of clusters with low (top 2 rows) and high (bottom 2 rows) CV of inter-subject similarity. Images show heat maps of binary cluster images, summed across subjects from each age group in template space.
3.3. Inter-hemispheric vs. inter-subject similarity
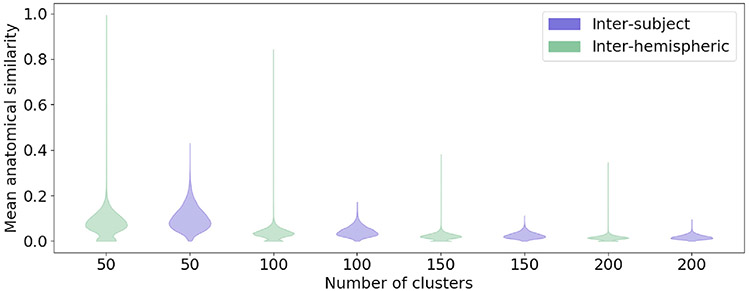
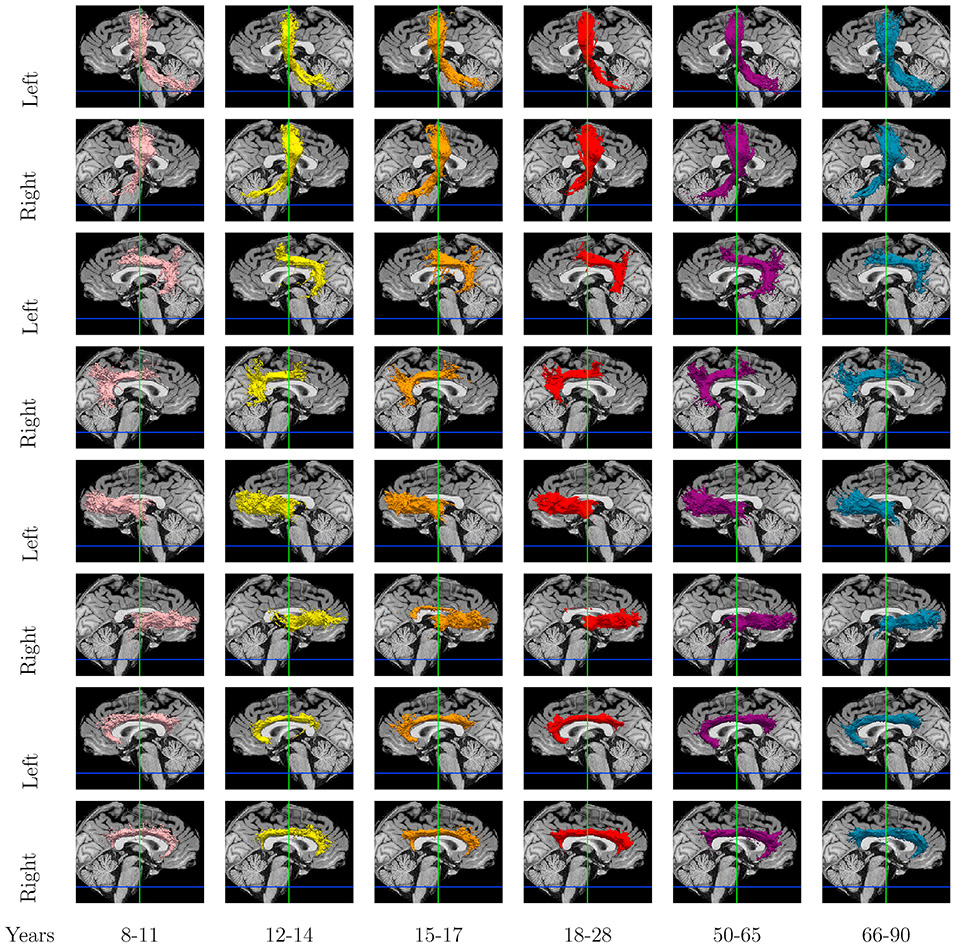
Fig. 7 shows the mean anatomical similarity between clusters, when the Hungarian algorithm is used to find inter-hemispheric and inter-subject cluster correspondence. Anatomical similarity for corresponding clusters between hemispheres and subjects is comparable. Fig. S2 shows the effect of the percentage of streamlines crossing hemispheres that clusters were allowed to have to be included in this analysis. We did not find this to affect the results noticeably. In Fig. 8 we show examples of clusters that were found to correspond between hemispheres. For display, individual tract clusters were transformed to binary images, summed across subjects by age group in template space and converted to isosurfaces at 10% of the maximum number of subjects.
Fig. 7.
Mean anatomical similarity of corresponding clusters between subjects and between hemispheres. This is shown for the hierarchical tree pruned at 50, 100, 150 and 200 clusters. Similarity values are normalized by the maximum value over all for display.
Fig. 8.
Inter-hemispheric cluster correspondence shown in alternate rows. We show isosurfaces color-coded by age group: pink (8–11), yellow (12–14), orange (15–17), red (18–28), purple (50–65), and blue (66–90).
3.4. Fitting age-related changes of diffusion measures
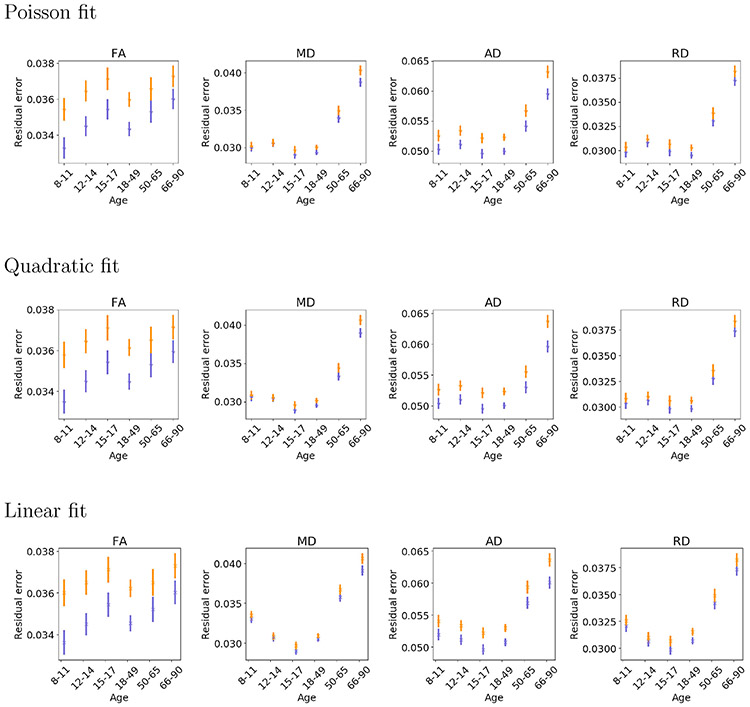
Fig. 9 shows the residual errors of the Poisson, quadratic and linear fits of FA, MD, AD, and RD vs. age, averaged over all 200 clusters. The anatomical similarity metric led to lower residual errors for all three models and all four diffusion measures. As the Poisson model had the lowest residual errors, we show examples of Poisson curves for some clusters of interest in Figs. 10 and 11. Clusters in Fig. 10 were selected based on their anatomical similarity to some of the manually labeled tracts from (Siless et al., 2018), and clusters in Fig. 11 were not included in that manually labeled set. Consistent with the literature, inter-hemispheric connections, such as the forceps major of the corpus callosum, show little to no changes in FA after age 8 (Clayden et al., 2012; Lebel et al., 2008; Pohl et al., 2016), while MD continues to decrease until early adulthood (Lebel et al., 2008). The superior longitudinal fasciculus develops until late adolescence, with FA increasing and MD decreasing until adulthood (Lebel et al., 2008; Pohl et al., 2016). This increase in FA is only seen with the anatomical similarity metric. The cingulum bundle shows the greatest changes during development, with FA increasing substantially until adulthood (Clayden et al., 2012; Lebel et al., 2008; Pohl et al., 2016). This is better captured by clusters produced with the anatomical similarity measure. In Fig. 11 we show Poisson curves of structures that have been reported on less frequently in the literature. A mid-component of the corpus callosum shows subtle increase of FA into adulthood (Pohl et al., 2016; Tamnes et al., 2010) but large decrease of MD. The FA trend is only seen in the clusters produced with the anatomical similarity metric. This subtle change would be missed if FA were averaged over the entire corpus callosum, instead of decomposing it into smaller clusters. Another structure that is analyzed less frequently is the frontal aslant tract, where only clusters obtained with the anatomical similarity metric show an FA increase in both hemispheres until early adulthood (Garic et al., 2018). Clusters produced with the Euclidean-distance similarity metric show no development in the right hemisphere.
Fig. 9.
Residual errors of Poisson, quadratic, and linear fits of the average FA, MD, AD, and RD vs. age, averaged over 200 clusters obtained with the anatomical (blue) and Euclidean-distance (orange) similarity metric. The bars represent standard deviation.
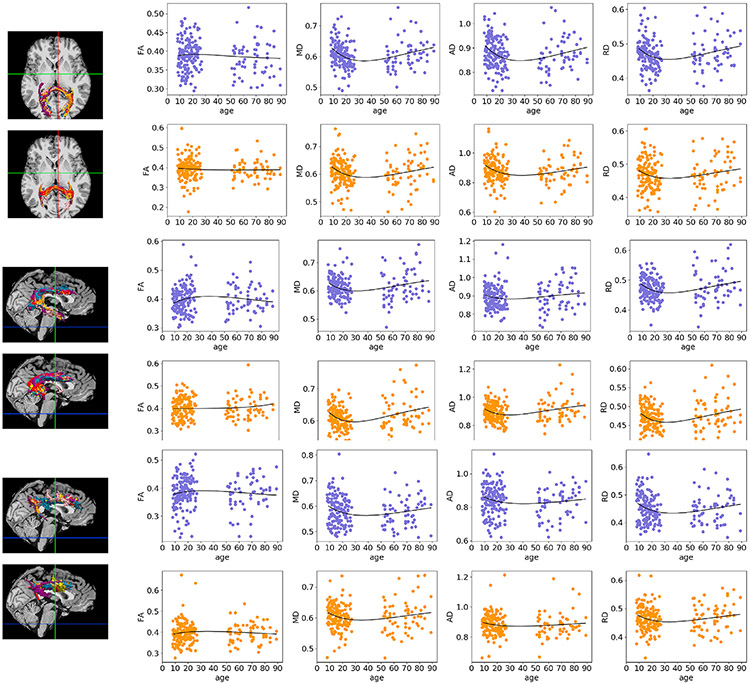
Fig. 10.
Poisson curves fit to average FA/MD/AD/RD of clusters obtained with the anatomical (blue) and the Euclidean-distance (orange) similarity metric. Results are shown for the three clusters of the left, where isosurfaces are color-coded by age group: pink (8–11), yellow (12–14), orange (15–17), red (18–28), purple (50–65), and blue (66–90). The clusters represent portions of the forceps major of the corpus callosum (top), left arcuate fasciculus (middle), and left cingulum bundle (bottom).
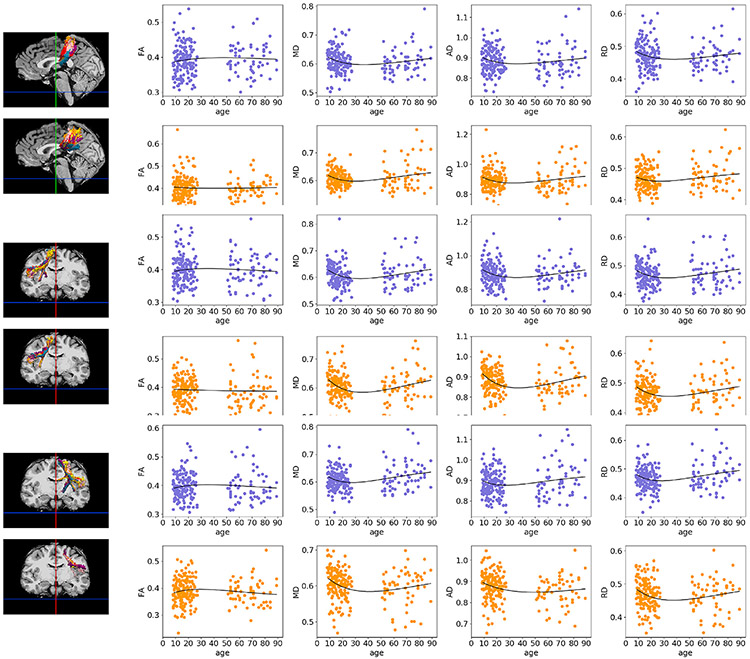
Fig. 11.
Poisson curves fit to average FA/MD/AD/RD of clusters obtained with the anatomical (blue) and the Euclidean-distance (orange) similarity metric. Results are shown for the three clusters on the left, where isosurfaces are color-coded by age group: pink (8–11), yellow (12–14), orange (15–17), red (18–28), purple (50–65), and blue (66–90). The clusters represent portions of the corpus callosum (top), right frontal aslant tract (middle), left frontal aslant tract (bottom).
Fig. S3 shows residual error plots for smaller numbers of clusters, i.e., when the hierarchical clustering tree was pruned at 50, 100 or 150 clusters. In all cases, clusters produced with the anatomical similarity metric yielded lower residual errors than those produced with the Euclidean-distance similarity metric. Fig. S4 shows Poisson, quadratic, and linear fit of diffusion measures vs. age for each individual cluster, at the 200-cluster level. These confirm that, overall, our data are able to capture expected lifespan trends, with FA increasing during development and decreasing during aging, while MD/AD/RD follow opposite trends. Fig. S5 shows the same for different numbers of total clusters, only for the Poisson model.
3.5. Computation time
Computation time for finding corresponding clusters across subjects, when each subject’s hierarchical clustering tree is pruned at different levels, is shown in Table 1 for the anatomical and the Euclidean-distance similarity metric. Times are reported for a quadcore Xeon 5472 with 3.0 GHz CPUs and 7 GB of RAM.
Table 1.
Computation times for finding inter-subject cluster correspondence.
| #clusters | Anatomical | Euclidean |
|---|---|---|
| 200 | 6:37 min | 4:38 min |
| 150 | 6:11 min | 4:27 min |
| 100 | 5:90 min | 4:20 min |
| 50 | 4:44 min | 4:18 min |
3.6. Visual evaluation
For visual evaluation, we show corresponding clusters obtained with the anatomical and Euclidean-distance similarity metrics in two individuals from each age group in Figs. S6, S7, S8. The clusters shown in the three figures represent portions of a long-range (corticospinal tract, S6), a short-range (frontal aslant tract, S7) and an inter-hemispheric (corpus callosum, Fig. S8) connection.
Fig. S9 shows clusters with high anatomical similarity with some of the manually labeled tracts from (Siless et al., 2018). Here clusters were transformed to binary images, summed over subjects in each age group in template space, and converted to isosurfaces at 10% of the maximum number of subjects. These clusters show portions of various known tracts.
4. Discussion
We present a method for comparing whole-brain tract clusters across subjects without the need for inter-subject registration. It relies on an extended version of our recently proposed, anatomical similarity metric, which groups tractography streamlines based on the similarity of their anatomical neighbors, rather than their distance in Euclidean space. Previously, we used this similarity metric to cluster whole-brain tractography streamlines in an individual subject, and evaluated it in a small group of young to middle-aged adults (Siless et al., 2018). Here we extend this approach to across-subject analysis, and evaluate it on a large cohort of subjects aged 8–90. We show that our anatomical similarity metric can be extended to match clusters across subjects without co-registering them. It leads to much greater overlap of corresponding clusters across subjects, in comparison to a conventional similarity metric based on Euclidean distance, which relies on accurate inter-subject registration. The improvement ranges from 31% for subjects of the same age to 38% for subjects who are 80 years apart in age (Fig. 3). The improved inter-subject consistency is also demonstrated by lower CV of cluster similarities across subjects (Fig. 5) and lower residual error in the fitting of lifespan trajectories of microstructural measures (Fig. 9). Our anatomical similarity metric can also be used to match clusters between hemispheres, without the need for inter-hemispheric registration (Fig. 8). This could be used for unsupervised analysis of the laterality of WM bundles, which is observed in many neurological disorders.
Unsupervised streamline tractography may produce artifactual bundles, e.g., a truncated section of a true WM pathway or merged sections of different pathways, in a manner that is inconsistent across subjects. Hence the requirement for one-to-one correspondence of clusters between subjects may be too stringent. One way to relax this requirement is by detecting clusters that are outliers in terms of inter-subject consistency. We found that the anatomical similarity metric produced a few easily discernible outliers of low consistency (high CV) (Figs. 4 and 5). These outlier clusters were predominantly due to tractography artifacts (Fig. 6), hence it would be appropriate to exclude them from a population study. Note, however, that the reverse is not necessarily true, i.e., high inter-subject consistency does not guarantee anatomical validity. It is possible for a certain tractography error to occur consistently across subjects. This is a limitation of unsupervised tractography that could only be remedied by the introduction of prior anatomical information.
It is important to note that unsupervised clustering of whole-brain tractography is a purely data-driven analysis that is not optimized for reconstructing specific named tracts from the neuroanatomical literature (e.g., the arcuate fasciculus, uncinate fasciculus, etc.). Supervised approaches, which incorporate prior information on such tracts, are specifically designed for this task. Such approaches may use ROI-based rules (Clayden et al., 2008; De Groot et al., 2013; Yeatman et al., 2012; Zhang et al., 2008) or cluster similarity metrics (Garyfallidis et al., 2018; Guevara et al., 2012; Jin et al., 2014; O’Donnell and Westin, 2007; Ros et al., 2013; Zhang et al., 2018; Ziyan et al., 2009) to identify named tracts after performing tractography; may introduce prior anatomical information in the tractography itself (Yendiki et al., 2011); or may forgo tractography entirely, performing volumetric segmentation instead (Bazin et al., 2011; Hagler et al., 2009; Ratnarajah and Qiu, 2014; Wasserthal et al., 2018). The named tracts reconstructed by these supervised algorithms represent the main highways of the brain. However, it is known from anatomical studies that these large pathways are not monolithic structures, but instead comprise many smaller bundles, each projecting to different anatomical regions (Lehman et al., 2011). These sub-bundles may be topographically organized within the large white-matter pathways, and disease effects may be specific to certain sub-bundles (Safadi et al., 2018). Therefore, statistical power to detect these effects may be reduced when diffusion measures are averaged over a large pathway. As a result, it is useful to have an anatomically meaningful way of subdividing white-matter bundles that goes beyond the large, named tracts. The unsupervised, hierarchical clustering based on anatomical similarity that we have presented here is suitable for studying the entire white matter, and can divide it into arbitrary small tract clusters, within the limits posed by the granularity of the anatomical segmentation. We have shown previously that our anatomical similarity metric leads to clusters that resemble manual dissections of named tracts more than the clusters produced by the conventional, Euclidean-distance similarity (Siless et al., 2018). Encouragingly, the unsupervised approach proposed here yielded several findings on WM microstructural development in agreement with prior studies that had used supervised tractography to study specific pathways.
Clusters that were generated and matched across subjects with our anatomical similarity metric yielded results consistent with previous findings on the development of the forceps major of the corpus callosum, the arcuate fasciculus, and the cingulum bundle (Clayden et al., 2012; Lebel et al., 2008; Pohl et al., 2016). Clusters produced by a conventional Euclidean-distance similarity metric do not show the expected increase in FA of the arcuate or as large an increase in FA of the cingulum into late adulthood. While the corpus callosum as a whole shows little development of FA into adulthood (Pohl et al., 2016), subdividing the corpus callosum based on anatomical similarity resulted in a mid-callosal cluster whose FA showed late increases. This finding would be missed if clustering were preformed based on Euclidean similarity or if FA were averaged in the entire corpus callosum. The frontal aslant tract is believed to have functional lateralization (Catani et al., 2013; Dick et al., 2019) but its developmental trajectory in terms of FA does not differ between hemispheres (Broce et al., 2015; Garic et al., 2018). In agreement with this, clustering of anatomical similarity shows an increase in FA for both hemispheres until early adulthood. However, clustering based on Euclidean similarity shows no development on the right hemisphere.
We fit three commonly used models for WM microstructural changes through the lifespan: linear, quadratic, and Poisson. Clusters that were generated and matched across subjects based on our anatomical similarity metric exhibited lower residual errors than clusters produced with a conventional, Euclidean-distance similarity metric, for all models (Fig. 9) and all levels of granularity of the hierarchical clustering (Fig. S3). The improved fitting of lifespan trajectories is likely to stem from the higher inter-subject consistency of clusters produced with our anatomical similarity metric (Fig. 4). Overall, we found lower residual errors with the Poisson model than the quadratic and linear ones. In the future, it may be worth investigating more flexible approaches to modeling lifespan trajectories, such as generalized additive models. It is worth noting that the dataset used in this study lacks subjects of ages between 30 and 50. Although the majority of development and aging is hypothesized to happen outside that range, this may be a limitation of this study. Despite this gap, all models fit here show the expected trends in anisotropy and diffusivity changes across the lifespan (Figs. S4 and S5). Given that in our previous report (Siless et al., 2018) we evaluated our methodology on data from subjects who were predominantly in the 30–50 age range, we do not expect that adding this age group here would alter our conclusions on the benefits of our anatomical similarity metric over more conventional metrics for unsupervised clustering.
5. Conclusion
This work builds upon our previously proposed method for tract clustering based on anatomical similarity, AnatomiCuts, and extends it for across-subject analyses. Specifically, we use our anatomical similarity metric to find corresponding clusters across subjects or hemispheres without the need for registration. We evaluate this approach on a large cohort of subjects aged 8–90 and we show that it yields clusters that are more consistent across subjects than those produced by a conventional, Euclidean-distance similarity metric. The improvement in inter-subject overlap of clusters increases with the age difference between subjects. Furthermore, our approach leads to lower residual errors when fitting several commonly used models for age-related changes in WM microstructure across the lifespan. We expect this method to facilitate exploratory analyses of WM microstructure in populations with large inter-subject variability.
Supplementary Material
Acknowledgements
Support for this research was provided in part by the National Institute of Biomedical Imaging and Bioengineering (R01-EB021265) and the National Institute of Mental Health (U01-MH108168; Boston Adolescent Neuroimaging of Depression and Anxiety project) and was made possible by the resources provided by Shared Instrumentation Grants 1S10RR023401, 1S10RR019307, and 1S10RR023043. Additional support was provided by the NIH Blueprint for Neuroscience Research (5U01-MH093765; part of the multi-institutional Human Connectome Project, T90DA022759/R90DA023427), and a Harvard University Mind, Brain, and Behavior Interfaculty Initiative Research Grant.
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neuroimage.2020.116703.
References
- Andersson JLR, Sotiropoulos SN, January2016. An integrated approach to correction for off-resonance effects and subject movement in diffusion mr imaging. Neuroimage 125, 1063–1078. 10.1016/j.neuroimage.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson JLR, Skare S, Ashburner J, 2003. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. Neuroimage 20 (2), 870–888. 10.1016/S1053-8119(03)00336-7. ISSN 10538119. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Mattiello J, LeBihan D, January1994. MR diffusion tensor spectroscopy and imaging. Biophys. J 66 (1), 259–267. 10.1016/s0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum GL, Ciric R, Roalf DR, Betzel RF, Moore TM, Shinohara RT, Kahn AE, Vandekar SN, Rupert PE, Quarmley M, Cook PA, Elliott MA, Ruparel K, Gur RE, Gur RC, Bassett DS, Satterthwaite TD, June2017. Modular segregation of structural brain networks supports the development of executive function in youth. Curr. Biol 27 (11), 1561–1572. 10.1016/j.cub.2017.04.051e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum GL, Roalf DR, Cook PA, Ciric R, Rosen AFG, Xia C, Elliott MA, Ruparel K, Verma R, Tunç B, Gur RC, Gur RE, Bassett DS, Satterthwaite TD, 2018. The impact of in-scanner head motion on structural connectivity derived from diffusion MRI. Neuroimage 173, 275–286. 10.1016/j.neuroimage.2018.02.041. ISSN 10959572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazin Pierre-Louis, Ye Chuyang, Bogovic John A., Shiee Navid, Reich Daniel S., Prince Jerry L., Pham Dzung L., 2011. Direct Segmentation of the Major White Matter Tracts in Diffusion Tensor Images. 10.1016/j.neuroimage.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benes FM, January1989. Myelination of cortical-hippocampal relays during late adolescence. Schizophr. Bull 15 (4), 585–593. 10.1093/schbul/15.4.585. [DOI] [PubMed] [Google Scholar]
- Benes FM, June1994. Myelination of a key relay zone in the hippocampal formation occurs in the human brain during childhood, adolescence, and adulthood. Arch. Gen. Psychiatr 51 (6), 477. 10.1001/archpsyc.1994.03950060041004. [DOI] [PubMed] [Google Scholar]
- Betzel RF, Byrge L, He Y, Goñi J, Zuo X, Sporns O, November2014. Changes in structural and functional connectivity among resting-state networks across the human lifespan. Neuroimage 102, 345–357. 10.1016/j.neuroimage.2014.07.067. [DOI] [PubMed] [Google Scholar]
- Broce I, Bernal B, Altman N, Tremblay P, Dick AS, October2015. Fiber tracking of the frontal aslant tract and subcomponents of the arcuate fasciculus in 5–8-year-olds: relation to speech and language function. Brain Lang. 149, 66–76. 10.1016/J.BANDL.2015.06.006. ISSN 0093-934X. [DOI] [PubMed] [Google Scholar]
- Catani M, Mesulam MM, Jakobsen E, Malik F, Martersteck A, Wieneke C, Thompson CK, Thiebaut de Schotten M, Dell’Acqua F, Weintraub S, Rogalski E, August2013. A novel frontal pathway underlies verbal fluency in primary progressive aphasia. Brain 136 (8), 2619–2628. 10.1093/brain/awt163. ISSN 1460-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan MY, Park DC, Savalia NK, Petersen SE, Wig GS, November2014. Decreased segregation of brain systems across the healthy adult lifespan. Proc. Natl. Acad. Sci. Unit. States Am 111 (46), E4997–E5006. 10.1073/pnas.1415122111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YS, Owen JP, Pojman NJ, Thieu T, Bukshpun P, Wakahiro MLJ, Berman JI, Roberts TPL, Nagarajan SS, Sherr EH, Mukherjee P, June2015. White matter changes of neurite density and fiber orientation dispersion during human brain maturation. PloS One 10 (6), e0123656. 10.1371/journal.pone.0123656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayden JD, Jentschke S, Munoz M, Cooper JM, Chadwick MJ, Banks T, Clark CA, Vargha-Khadem F, August2012. Normative development of white matter tracts: similarities and differences in relation to age, gender, and intelligence. Cerebr. Cortex 22 (8), 1738–1747. 10.1093/cercor/bhr243. ISSN 1047-3211. [DOI] [PubMed] [Google Scholar]
- Clayden Jonathan D., Storkey Amos J., Maniega Susana Muñoz, Bastin Mark E., 2008. Reproducibility of tract segmentation between sessions using an unsupervised modelling-based approach. Neuroimage 45, 377–385. 10.1016/j.neuroimage.2008.12.010. [DOI] [PubMed] [Google Scholar]
- De Groot Marius, Vernooij Meike W., Klein Stefan, Arfan Ikram M, Vos Frans M., Smith Stephen M., Niessen Wiro J., Andersson Jesper L.R., 2013. Improving alignment in Tract-based spatial statistics: evaluation and optimization of image registration. Neuroimage. 10.1016/j.neuroimage.2013.03.015. ISSN 10538119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dice LR, 1945. Measures of the amount of ecologic association between species. Ecology 26 (3), 297–302. 10.2307/1932409. ISSN 00129658. [DOI] [Google Scholar]
- Dick AS, Garic D, Graziano P, Tremblay P, February2019. The frontal aslant tract (FAT) and its role in speech, language and executive function. Cortex 111, 148–163. 10.1016/J.CORTEX.2018.10.015. ISSN 0010-9452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinstein I, Pierce K, Eyler L, Solso S, Malach R, Behrmann M, Courchesne E, June2011. Disrupted neural synchronization in toddlers with autism. Neuron 70 (6), 1218–1225. 10.1016/j.neuron.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair DA, Cohen AL, Power JD, Dosenbach NUF, Church JA, Miezin FM, Schlaggar BL, Petersen SE, May2009. Functional brain networks develop from a “local to distributed” organization. PLoS Comput. Biol 5 (5), e1000381 10.1371/journal.pcbi.1000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q, Witzel T, Nummenmaa A, Van Dijk KRA, Van Horn JD, Drews MK, Somerville LH, Sheridan MA, Santillana RM, Snyder J, Hedden T, Shaw EE, Hollinshead MO, Renvall V, Zanzonico R, Keil B, Cauley S, Polimeni JR, Tisdall D, Buckner RL, Wedeen VJ, Wald LL, Toga AW, Rosen BR, January2016. MGH–USC human connectome project datasets with ultra-high b-value diffusion MRI. Neuroimage 124, 1108–1114. 10.1016/j.neuroimage.2015.08.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Salat David H., Busa E, Albert M, Dieterich M, Haselgrove C, Van Der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM, 2002. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 33 (3), 341–355. 10.1016/S0896-6273(02)00569-X. ISSN 08966273. [DOI] [PubMed] [Google Scholar]
- Fischl B, Van Der Kouwe A, Destrieux C, Halgren E, Ségonne F, Salat DH, Busa E, Seidman LJ, Goldstein J, Kennedy D, Caviness V, Makris N, Rosen B, Dale AM, 2004. Automatically parcellating the human cerebral cortex. Cerebr. Cortex 14 (1), 11–22. 10.1093/cercor/bhg087. ISSN 10473211. [DOI] [PubMed] [Google Scholar]
- Fjell AM, Sneve MH, Storsve AB, Grydeland H, Yendiki A, Walhovd KB, March2016. Brain events underlying episodic memory changes in aging: a longitudinal investigation of structural and functional connectivity. Cerebr. Cortex 26 (3), 1272–1286. 10.1093/cercor/bhv102. ISSN 1047-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjell AM, Sneve MH, Grydeland H, Storsve AB, Amlien IK, Yendiki A, Walhovd KB, January2017. Relationship between structural and functional connectivity change across the adult lifespan: a longitudinal investigation. Hum. Brain Mapp 38 (1), 561–573. 10.1002/hbm.23403. ISSN 10659471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garic D, Broce I, Graziano P, Mattfeld A, Dick AS, September2018. Laterality of the frontal aslant tract (FAT) explains externalizing behaviors through its association with executive function. page e12744.Dev. Sci. 10.1111/desc.12744. ISSN 1363755X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garyfallidis E, Brett M, Correia MM, Williams GB, Nimmo-Smith I, 2012. QuickBundles, a method for tractography simplification. Front. Neurosci 6 (December), 175. 10.3389/fnins.2012.00175. ISSN 1662-453X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garyfallidis E, Côté M-A, Rheault F, Sidhu J, Hau J, Petit L, Fortin D, Cunanne S, Descoteaux M, April2018. Recognition of white matter bundles using local and global streamline-based registration and clustering. Neuroimage 170, 283–295. 10.1016/j.neuroimage.2017.07.015. [DOI] [PubMed] [Google Scholar]
- Genc S, Malpas CB, Holland SK, Beare R, Silk TJ, March2017. Neurite density index is sensitive to age related differences in the developing brain. Neuroimage 148, 373–380. 10.1016/j.neuroimage.2017.01.023. [DOI] [PubMed] [Google Scholar]
- Giedd JN, Blumenthal J, Jeffries NO, Castellanos FX, Liu H, Zijdenbos A, Paus T, Evans AC, Rapoport JL, October1999. Brain development during childhood and adolescence: a longitudinal MRI study. Nat. Neurosci 2 (10), 861–863. 10.1038/13158. ISSN 1097-6256. [DOI] [PubMed] [Google Scholar]
- Greve DN, Fischl B, October2009. Accurate and robust brain image alignment using boundary-based registration. Neuroimage 48 (1), 63–72. 10.1016/j.neuroimage.2009.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinberg F, Maximov II, Farrher E, Neuner I, Amort L, Thönneßen H, Oberwelland E, Konrad K, Jon Shah N, January2017. Diffusion kurtosis metrics as biomarkers of microstructural development: a comparative study of a group of children and a group of adults. Neuroimage 144 (12–22). 10.1016/j.neuroimage.2016.08.033. [DOI] [PubMed] [Google Scholar]
- Guevara M, Román C, Houenou J, Duclap D, Poupon C, Mangin J-F, Guevara P, February2017. Reproducibility of superficial white matter tracts using diffusion-weighted imaging tractography. Neuroimage 147, 703–725. 10.1016/j.neuroimage.2016.11.066. [DOI] [PubMed] [Google Scholar]
- Guevara P, Duclap D, Poupon C, Marrakchi-Kacem L, Fillard P, Le Bihan D, Leboyer M, Houenou J, Mangin JF, 2012. Automatic fiber bundle segmentation in massive tractography datasets using a multi-subject bundle atlas. Neuroimage 61 (4), 1083–1099. 10.1016/j.neuroimage.2012.02.071. ISSN 10538119. [DOI] [PubMed] [Google Scholar]
- Hagler Donald J., E Ahmadi Mazyar, Kuperman Joshua, Holland Dominic, Carrie R, McDonald Halgren, Eric Dale, Anders M, 2009. Automated white-matter tractography using a probabilistic diffusion tensor atlas: application to temporal lobe epilepsy. Hum. Brain Mapp 30 (5), 1535–1547. 10.1002/hbm.20619. ISSN 10659471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady M, Smith S, October2002. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 17 (2), 825–841. ISSN 1053-8119. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM, August2012. FSL. NeuroImage 62 (2), 782–790. 10.1016/j.neuroimage.2011.09.015. ISSN 10538119. [DOI] [PubMed] [Google Scholar]
- Jin Yan, Shi Yonggang, Zhan Liang, Gutman Boris A., de Zubicaray Greig I., McMahon Katie L., Wright Margaret J., Toga Arthur W., Thompson Paul M., 2014. Automatic clustering of white matter fibers in brain diffusion {MRI} with an application to genetics. Neuroimage 100, 75–90. 10.1016/j.neuroimage.2014.04.048. ISSN 1053-8119. www.sciencedirect.com/science/article/pii/S1053811914003176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonker R, Volgenant A, December1987. A shortest augmenting path algorithm for dense and sparse linear assignment problems. Computing 38 (4), 325–340. 10.1007/BF02278710. ISSN 0010-485X. [DOI] [Google Scholar]
- Kodiweera C, Alexander AL, Harezlak J, McAllister TW, Wu Y, March2016. Age effects and sex differences in human brain white matter of young to middle-aged adults: a DTI, NODDI, and q-space study. Neuroimage 128, 180–192. 10.1016/j.neuroimage.2015.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn HW, March1955. The Hungarian method for the assignment problem. Nav. Res. Logist. Q 2 (1-2), 83–97. 10.1002/nav.3800020109. ISSN 00281441. [DOI] [Google Scholar]
- Kuhn HW, November2009. The Hungarian method for the assignment problem. In: In 50 Years of Integer Programming 1958–2008. Springer Berlin Heidelberg, pp. 29–47. 10.1007/978-3-540-68279-0_2. [DOI] [Google Scholar]
- Lebel C, Walker L, Leemans A, Phillips L, Beaulieu C, April2008. Microstructural maturation of the human brain from childhood to adulthood. Neuroimage 40 (3), 1044–1055. 10.1016/j.neuroimage.2007.12.053. [DOI] [PubMed] [Google Scholar]
- Lebel C, Gee M, Camicioli R, Wieler M, Martin W, Beaulieu C, 2012. Diffusion tensor imaging of white matter tract evolution over the lifespan. Neuroimage. 10.1016/j.neuroimage.2011.11.094. ISSN 10538119. [DOI] [PubMed] [Google Scholar]
- Lebel C, Treit S, Beaulieu C, September2017. A review of diffusion MRI of typical white matter development from early childhood to young adulthood. page e3778.NMR Biomed.. 10.1002/nbm.3778. ISSN 09523480. [DOI] [PubMed] [Google Scholar]
- Lehman Julia F., Greenberg Benjamin D., Mcintyre Cameron C., Rasmussen Steve A., Haber Suzanne N., July2011. Rules ventral prefrontal cortical axons use to reach their targets: implications for diffusion tensor imaging tractography and deep brain stimulation for psychiatric illness. J. Neurosci 31 (28), 10392–10402. 10.1523/JNEUROSCI.0595-11.2011. ISSN 02706474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenberger U, October2014. Human cognitive aging: corriger la fortune? Science 346 (6209), 572–578. 10.1126/science.1254403. [DOI] [PubMed] [Google Scholar]
- Mahone EM, Crocetti D, Ranta ME, Gaddis A, Cataldo M, Slifer KJ, Denckla MB, Mostofsky SH, August2011. A preliminary neuroimaging study of preschool children with ADHD. Clin. Neuropsychol 25 (6), 1009–1028. 10.1080/13854046.2011.580784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell LJ, Westin C-F, Norton I, Whalen S, Rigolo L, Propper R, Golby AJ, 2010. The fiber laterality histogram: a new way to measure white matter asymmetry. In: MICCAI 2010. Lecture Notes in Computer Science, vol. 6362. Springer, Berlin, Heidelberg, pp. 225–232. 10.1007/978-3-642-15745-5_28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell LJ, Westin CF, 2007. Automatic tractography segmentation using a highdimensional white matter atlas. IEEE Trans. Med. Imag 26 (11), 1562–1575. 10.1109/TMI.2007.906785. ISSN 02780062. [DOI] [PubMed] [Google Scholar]
- Park H-J, Westin C-F, Kubicki M, Maier SE, Niznikiewicz M, Baer A, Frumin M, Kikinis R, Jolesz FA, McCarley RW, Shenton ME, September2004. White matter hemisphere asymmetries in healthy subjects and in schizophrenia: a diffusion tensor MRI study. Neuroimage 23 (1), 213–223. 10.1016/j.neuroimage.2004.04.036. ISSN 1053-8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paus TSAA, March1999. Structural maturation of neural pathways in children and adolescents: in vivo study. Science 283 (5409), 1908–1911. 10.1126/science.283.5409.1908. [DOI] [PubMed] [Google Scholar]
- Paydar A, Fieremans E, Nwankwo JI, Lazar M, Sheth HD, Adisetiyo V, Helpern JA, Jensen JH, Milla SS, November2013. Diffusional kurtosis imaging of the developing brain. Am. J. Neuroradiol 35 (4), 808–814. 10.3174/ajnr.a3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters BD, Karlsgodt KH, January2015. White matter development in the early stages of psychosis. Schizophr. Res 161 (1), 61–69. 10.1016/j.schres.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl KM, Sullivan EV, Rohlfing T, Chu W, Kwon D, Nichols BN, Zhang Y, Brown SA, Tapert SF, Cummins K, Thompson WK, Brumback T, Colrain IM, Baker FC, Prouty D, De Bellis MD, T Voyvodic J, Clark DB, Schirda C, Nagel BJ, Pfefferbaum A, April2016. Harmonizing DTI measurements across scanners to examine the development of white matter microstructure in 803 adolescents of the NCANDA study. Neuroimage 130, 194–213. 10.1016/j.neuroimage.2016.01.061. ISSN 1095-9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polimeni JR, Bhat H, Witzel T, Benner T, Feiweier T, Inati SJ, Renvall V, Heberlein K, Wald LL, February2016. Reducing sensitivity losses due to respiration and motion in accelerated echo planar imaging by reordering the autocalibration data acquisition. Magn. Reson. Med 75 (2), 665–679. 10.1002/mrm.25628. ISSN 07403194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postelnicu G, Zollei L, Fischl B, April2009. Combined volumetric and surface registration. IEEE Trans. Med. Imag 28 (4), 508–522. 10.1109/TMI.2008.2004426. ISSN 1558-254X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Propper RE, O’Donnell LJ, Whalen S, Tie Y, Norton IH, Suarez RO, Zollei L, Radmanesh A, Golby AJ, July2010. A combined fMRI and DTI examination of functional language lateralization and arcuate fasciculus structure: effects of degree versus direction of hand preference. Brain Cognit. 73 (2), 85–92. 10.1016/J.BANDC.2010.03.004. ISSN 0278-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnarajah Nagulan, Qiu Anqi, 2014. Multi-label Segmentation of White Matter Structures: Application to Neonatal Brains. 10.1016/j.neuroimage.2014.08.001. [DOI] [PubMed] [Google Scholar]
- Raz N, Rodrigue KM, January2006. Differential Aging of the Brain: Patterns, Cognitive Correlates and Modifiers. ISSN 01497634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter-Lorenz PA, September2002. New visions of the aging mind and brain. Trends Cognit. Sci 6 (9), 394–400. 10.1016/s1364-6613(02)01957-5. [DOI] [PubMed] [Google Scholar]
- Roberts JA, Perry A, Roberts G, Mitchell PB, Breakspear M, 2017. Consistency-based thresholding of the human connectome. Neuroimage 145, 118–129. 10.1016/j.neuroimage.2016.09.053. ISSN 10959572. [DOI] [PubMed] [Google Scholar]
- Román C, Guevara M, Valenzuela R, Figueroa M, Houenou J, Duclap D, Poupon C, Mangin J-F, Guevara P, December2017. Clustering of whole-brain white matter short association bundles using HARDI data. Front. Neuroinf 11 10.3389/fninf.2017.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros C, Güllmar D, Stenzel M, Mentzel H-J, Reichenbach JR, 2013. Atlas-guided cluster analysis of large tractography datasets. PloS One 8 (12), e83847. 10.1371/journal.pone.0083847. ISSN 1932-6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safadi Ziad, Grisot Giorgia, Jbabdi Saad, Behrens Timothy E., Heilbronner Sarah R., McLaughlin Nicole C.R., Joe Mandeville, Amelia Versace, Phillips Mary L., Lehman Julia F., Yendiki Anastasia, Haber Suzanne N., February2018. Functional segmentation of the anterior limb of the internal capsule: linking white matter abnormalities to specific connections. J. Neurosci 38 (8), 2106–2117. 10.1523/JNEUROSCI.2335-17.2017. ISSN 15292401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setsompop K, Kimmlingen R, Eberlein E, Witzel T, Cohen-Adad J, McNab JA, Keil B, Tisdall MD, Hoecht P, Dietz P, Cauley SF, Tountcheva V, Matschl V, Lenz VH, Heberlein K, Potthast A, Thein H, Van Horn J, Toga A, Schmitt F, Lehne D, Rosen BR, Wedeen V, Wald LL, 2013. Pushing the limits of in vivo diffusion MRI for the human connectome project. Neuroimage 80, 220–233. 10.1016/j.neuroimage.2013.05.078. ISSN 10538119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siless V, Medina S, Varoquaux G, Thirion B, June2013. A comparison of metrics and algorithms for fiber clustering. In: In 2013 International Workshop on Pattern Recognition in Neuroimaging, pp. 190–193. 10.1109/PRNI.2013.56. [DOI] [Google Scholar]
- Siless V, Chang K, Fischl B, Yendiki A, 2016. Hierarchical Clustering of Tractography Streamlines Based on Anatomical Similarity. Springer International Publishing, Cham, ISBN 978-3-319-46720-7, pp. 184–191. 10.1007/978-3-319-46720-7_22. [DOI] [Google Scholar]
- Siless V, Davidow JY, Nielsen J, Fan Q, Hedden T, Hollinshead M, Bustamante CV, Drews MK, Van Dijk KRA, Sheridan MA, Buckner RL, Fischl B, Somerville L, Yendiki A, 2017. Registration-free analysis of diffusion mri tractography data across subjects through the human lifespan. In: ISMRM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siless V, Chang K, Fischl B, AnatomiCuts A. Yendiki, 2018. Hierarchical clustering of tractography streamlines based on anatomical similarity. Neuroimage 166, 32–45. 10.1016/j.neuroimage.2017.10.058. ISSN 10959572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen TJ, 1948. A method of establishing groups of equal amplitude in plant sociology based on similarity of species and its application to analyses of the vegetation on Danish commons. Biol. Skr 5 (1–34). [Google Scholar]
- Sowell ER, Thompson PM, Holmes CJ, Batth R, Jernigan TL, Toga AW, 1999. Localizing age-related changes in brain structure between childhood and adolescence using statistical parametric mapping. Neuroimage. 10.1006/nimg.1999.0436. ISSN 10538119. [DOI] [PubMed] [Google Scholar]
- Storsve AB, Fjell AM, Yendiki A, Walhovd KB, June2016. Longitudinal changes in white matter tract integrity across the adult lifespan and its relation to cortical thinning. PloS One 11 (6), e0156770. 10.1371/journal.pone.0156770. ISSN 1932-6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan EV, Pfefferbaum A, January2006. Diffusion tensor imaging and aging. Neurosci. Biobehav. Rev 30 (6), 749–761. 10.1016/j.neubiorev.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Sullivan EV, Adalsteinsson E, Hedehus M, Ju C, Moseley M, Lim KO, Pfefferbaum A, January2001. Equivalent disruption of regional white matter microstructure in ageing healthy men and women. Neuroreport 12 (1), 99–104. 10.1097/00001756-200101220-00027. ISSN 0959-4965. [DOI] [PubMed] [Google Scholar]
- Taki Y, Hashizume H, Thyreau B, Sassa Y, Takeuchi H, Wu K, Kotozaki Y, Nouchi R, Asano M, Asano K, Fukuda H, Kawashima R, April2012. Linear and curvilinear correlations of brain gray matter volume and density with age using voxel-based morphometry with the akaike information criterion in 291 healthy children. Hum. Brain Mapp 34 (8), 1857–1871. 10.1002/hbm.22033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamnes CK, Østby Y, Fjell AM, Westlye LT, Due-Tønnessen P, Walhovd KB, 2010. Brain maturation in adolescence and young adulthood: regional age-related changes in cortical thickness and white matter volume and microstructure. Cerebr. Cortex 10.1093/cercor/bhp118. ISSN 10473211. [DOI] [PubMed] [Google Scholar]
- Tsang A, Lebel CA, Bray SL, Goodyear BG, Hafeez M, Sotero RC, McCreary CR, Frayne R, May2017. White matter structural connectivity is not correlated to cortical resting-state functional connectivity over the healthy adult lifespan. Front. Aging Neurosci 9 10.3389/fnagi.2017.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunç B, Parker WA, Ingalhalikar M, Verma R, November2014. Automated tract extraction via atlas based adaptive clustering. Neuroimage 102 (P2), 596–607. 10.1016/j.neuroimage.2014.08.021. ISSN 10959572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Kouwe A, Benner T, Salat DH, Fischl B, 2008. Brain morphometry with multiecho MPRAGE. Neuroimage 40 (2), 559–569. 10.1016/j.neuroimage.2007.12.025. ISSN 10538119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser E, Nijhuis EHJ, Buitelaar JK, Zwiers MP, 2011. Partition-based mass clustering of tractography streamlines. Neuroimage 54 (1), 303–312. 10.1016/j.neuroimage.2010.07.038. ISSN 10538119. [DOI] [PubMed] [Google Scholar]
- Wang X, Grimson WEL, Westin C-F, January2011. Tractography segmentation using a hierarchical Dirichlet processes mixture model. Neuroimage 54 (1), 290–302. 10.1016/j.neuroimage.2010.07.050. ISSN 1095-9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassermann D, Bloy L, Kanterakis E, Verma R, Deriche R, 2010. Unsupervised white matter fiber clustering and tract probability map generation: applications of a Gaussian process framework for white matter fibers. Neuroimage 51 (1), 228–241. 10.1016/j.neuroimage.2010.01.004. ISSN 1053-8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserthal Jakob, Neher Peter, Klaus H, Maier-Hein, December2018. TractSeg - fast and accurate white matter tract segmentation. Neuroimage 183, 239–253. 10.1016/j.neuroimage.2018.07.070. ISSN 10538119. https://linkinghub.elsevier.com/retrieve/pii/S1053811918306864. [DOI] [PubMed] [Google Scholar]
- Wu X, Xie M, Zhou J, Anderson AW, Gore JC, Ding Z, May2012. Globally optimized fiber tracking and hierarchical clustering – a unified framework. Magn. Reson. Imag 30 (4), 485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeatman JD, Dougherty RF, Myall NJ, Wandell BA, Feldman HM, 2012. Tract profiles of white matter properties: automating fiber-tract quantification. PloS One 7 (11), 49790. 10.1371/journal.pone.0049790. www.plosone.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh FC, Wedeen VJ, Tseng WYI, 2010. Generalized q-sampling imaging. IEEE Trans. Med. Imag 29 (9), 1626–1635. 10.1109/TMI.2010.2045126. ISSN 02780062. [DOI] [PubMed] [Google Scholar]
- Yeh FC, Verstynen TD, Wang Y, Fernández-Miranda JC, Tseng WYI, 2013. Deterministic diffusion fiber tracking improved by quantitative anisotropy. PloS One 8 (11). 10.1371/journal.pone.0080713. ISSN 19326203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yendiki A, Panneck P, Srinivasan P, Stevens A, Zöllei L, Augustinack J, Wang R, Salat D, Ehrlich S, Behrens T, Jbabdi S, Gollub R, Fischl B, 2011. Automated probabilistic reconstruction of white-matter pathways in Health and disease using an atlas of the underlying anatomy. Front. Neuroinf 5 (23) 10.3389/fninf.2011.00023. ISSN 1662-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wu Y, Norton I, Rigolo L, Rathi Y, Makris N, O’Donnell LJ, October2018. An anatomically curated fiber clustering white matter atlas for consistent white matter tract parcellation across the lifespan. Neuroimage 179, 429–447. 10.1016/J.NEUROIMAGE.2018.06.027. ISSN 1053-8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Chen H, Guo L, Li K, Li L, Zhang S, Shen D, Hu X, Liu T, 2014. Characterization of U-shape streamline fibers: methods and applications. Med. Image Anal 18 (5), 795–807. 10.1016/j.media.2014.04.005. ISSN 13618423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Weihong, Olivi Alessandro, Hertig Samuel J., Van Zijl Peter, Mori Susumu, 2008. Automated Fiber Tracking of Human Brain White Matter Using Diffusion Tensor Imaging. 10.1016/j.neuroimage.2008.04.241.www.mristudio.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziyan U, Sabuncu MR, Grimson WEL, Westin C-F, 2009. Consistency clustering: a robust algorithm for group-wise registration, segmentation and automatic atlas construction in diffusion MRI. Int. J. Comput. Vis 85 (3), 279–290. 10.1007/s11263-009-0217-1. ISSN 0920-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zöllei L, Stevens A, Huber K, Kakunoori S, Fischl B, May2010. Improved tractography alignment using combined volumetric and surface registration. Neuroimage 51 (1), 206–213. 10.1016/j.neuroimage.2010.01.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, He Y, Betzel RF, Colcombe S, Sporns O, Milham MP, January2017. Human connectomics across the life span. Trends Cognit. Sci 21 (1), 32–45. 10.1016/j.tics.2016.10.005.55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.