Correction to: BMC Genomics 22, 545 (2021)
https://doi.org/10.1186/s12864-021-07884-9
Following publication of the original article [1], it was reported that an incorrect image was published as Fig.6. The correct Fig. 6 is included in this Correction and the original article has been corrected.
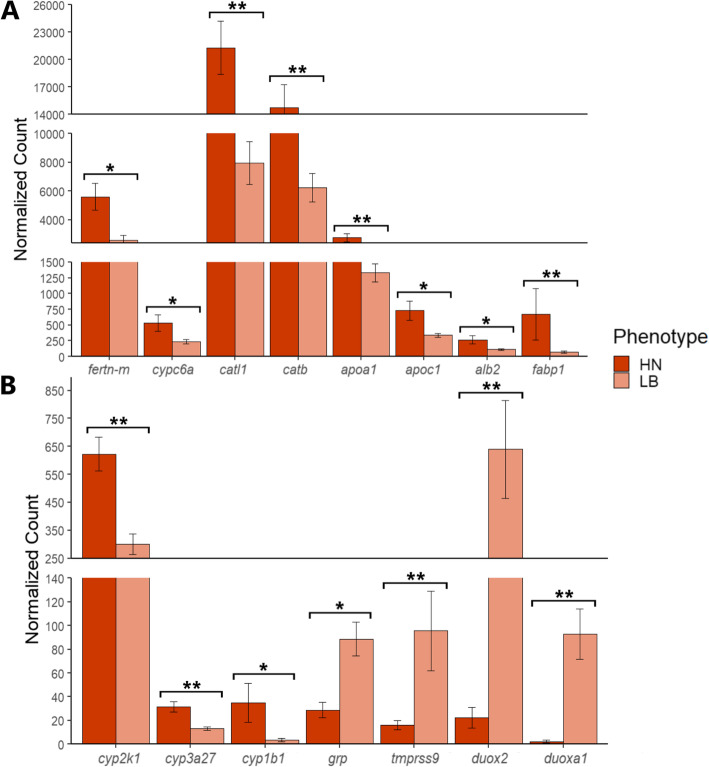
Fig. 6.
Analysis comparing HN and LB phenotypes. Subset of DEGs of interest identified in the transcriptiomes from the two library preprations. A Expression pattern of eight genes identified in the QuantSeq library: ferritin-m (fertn-m), involved in iron ion transport; cytochrome c oxidative subunit 6 A mitochondrial-like (cypc6a), involved in oxidation-reduction process; cathepsin L1 (catl1) and cathepsin B (catb) involved in apoptosis and muscle degradation; apolipoprotein A1 (apoa1), apolipoprotein C1 (apoc1), serum albumin 2 (alb2), fatty acid-binding protein 1 (fabp1) involved in lipid metabolism; B Expression pattern of seven genes identified in the TruSeq library: cyp450 gene family cyp2k1, cyp3a27 and cyp1b1, involved in oxidation-reduction process; gastrin-releasing peptide (grp) involved in regulation of feeding, and tmprss9, dual oxidase 2-like (duox2), dual oxidase maturation factor 1-like (duoxa1) involved in SNP analysis. Asterisks (* and **) indicate significant difference between the HN and LB phenotypes at P < 0.05 and P < 0.01, respectively
Contributor Information
Tomer Ventura, Email: tventura@usc.edu.au.
Abigail Elizur, Email: aelizur@usc.edu.au.
Reference
- 1.Vo TTM, Nguyen TV, Amoroso G, Ventura T, Elizur A. Deploying new generation sequencing for the study of flesh color depletion in Atlantic Salmon (Salmo salar) BMC Genomics. 2021;22(1):545. doi: 10.1186/s12864-021-07884-9. [DOI] [PMC free article] [PubMed] [Google Scholar]