Abstract
Background
Uveal melanoma (UVM) is the leading cause of eye-related mortality worldwide. This study aimed to explore the expression and prognostic value of matrix metalloproteinases (MMPs) in UVM.
Methods
Gene expression levels were obtained from the Gene Expression Omnibus (GEO) and Oncomine databases. Functional and pathway enrichment analyses were performed using the Metascape database. GeneMANIA was then applied to construct a protein-protein interaction network and identify the hub genes. Moreover, overall survival (OS) and disease-free survival (DFS) analysis for the hub genes was performed using the UALCAN and Gene Expression Profiling Interactive Analysis (GEPIA) online tool. Furthermore, TRRUST was used to predict the targets of the MMPs.
Results
Our results revealed that the transcriptional levels of MMP1, MMP9, MMP10, MMP11, MMP13, MMP14, and MMP17 were upregulated in UVM tissues compared to normal tissues. A protein-protein interaction (PPI) network was constructed and the top 50 hub genes were identified. The functions of MMPs and their neighboring proteins are mainly associated with ECM-receptor interaction, proteoglycans in cancer, the IL-17 signaling pathway, and microRNAs in cancer. Among the MMPs, MMP1/2/9/11/14/15/16/17/24 played significant roles in the progression of UVM from stage 3 to stage 4. We also found that the expression of MMP1, MMP2, MMP9, and MMP16 positively correlated with OS and DFS in patients with UVM. Additionally, 18 transcription factors associated with nine MMPs were identified.
Conclusions
The results of this study may provide potential biomarkers and targets for UVM. However, further studies are required to confirm these results.
Keywords: Matrix metallopeptidase, Transcriptional level, Protein-protein interaction, Prognosis, Overall survival, Disease-free survival
Background
Uveal melanoma (UVM) is the most common primary malignant tumor of the eye in adults [1]. It often occurs in people aged 50–60 years. The clinical manifestations, complications, high degree of malignancy, and poor prognosis of UVM seriously threaten the visual function and life of patients [2]. At present, the main treatment methods for UVM are enucleation, photocoagulation, local tumor resection, and external scleral application radiotherapy [1, 3]. Although these treatments have achieved a certain degree of success, there are still some problems. The most common sites of metastasis are the liver, lung, and bone. Furthermore, the mortality rate of UVM is as high as 50% [4]. Currently, the etiology and molecular mechanisms of UVM are still unclear [5, 6]. The abnormal expression of multiple genes may be involved. Therefore, finding new molecular therapeutic targets to establish early diagnosis and inhibit tumor metastasis is key to improving the survival rate of patients.
To date, 26 MMPs have been investigated, most of which are present in the human proteome [7]. MMPs can degrade various protein components in the extracellular matrix. Meanwhile, they can also destroy the histological barrier of tumor cell and affect tumor migration, invasion, metastasis, and angiogenesis [8–12]. Therefore, MMPs are considered as the main proteolytic enzymes in this process and the relation between MMPs and extracellular matrix components is interactive. According to their substrates, MMPs can be divided into six categories: (1) collagenases, including MMP1 (collagenase-1), MMP8 (collagenase-2), MMP13 (collagenase-3) with the main function of degrading collagen I, II, and III; (2) gelatinases, including MMP2 (gelatinase A) and MMP9 (gelatinase B), with the function of degrading collagen I and IV; (3) stromelysins, including MMP3 (stromelysin-1) and MMP11 (stromelysin-3); (4) MMP7. Its substrates are elastin, fibronectin, and laminin. They are necessary for the epithelization of mucosal wounds; (5) MMP12, the substrates of MMP12 are mainly type IV collagen, gelatin, fibronectin, and laminin; (6) Membrane-type MMPs (MT-MMPs), including MMP14, MMP15, MMP16, MMP17, MMP24, and MMP25. The substrates of MMP14 are type I, II, and III collagen. They can promote keratinocyte growth, airway re-epithelization, and cell migration. However, the role of the other MT-MMPs remains unclear. MMP activity is regulated at three levels: gene transcription, the activation of enzyme precursors and the effect of tissue inhibitors of metalloproteinases (TIMP).
In recent years, studies have shown that MMPs expression is associated with the occurrence and development of a variety of tumors. These molecules are important in the growth and invasion of malignant tumors and their expression has an important impact on the prognosis of patients [13–16] . However, there are few reports on the MMPs gene family role in the prognosis of UVM.
In recent years, with the development of high-throughput technology, gene chips and gene sequencing have become necessary and efficient methods for studying tumor diseases. In the era of big data, much genomic, epigenomic, and transcriptomic data are available in online databases. However, effective data mining remains lacking. This study used bioinformatic methods to detect the expression and mutation of the core MMPs, aiming to screen the genes closely related to the occurrence, development, and prognosis of UVM through clinical big data. We aimed to provide a reference for the clinical treatment of UVM, by establishing a theoretical basis for the research and development of targeted therapies for this malignancy.
Methods
Since all the data were retrieved from the online databases, we assumed that all written informed consent had been already obtained.
Malacards database
MalaCards (https://www.malacards.org) is an integrated database of human maladies and their annotations, modeled on the architecture and richness of the popular GeneCards database of human genes [17]. In this study, we aimed to identify more potential classical genes in UVM.
Oncomine database
ONCOMINE (www.oncomine.org) is a classic cancer sample database. It is a cancer microarray database and integrated data mining platform which aims to promote the discovery of genome-wide expression analysis [18, 19]. In this study, transcriptional differential expression of 17 different MMPs between different cancer tissues and their corresponding adjacent normal controls were obtained from the ONCOMINE database. Student’s t-test was used to compare the differences in transcriptional expression. Fold change and cut-off of p-value were as follows: fold change, 1.5; p value, 0.01; gene rank, 10%; data type, mRNA.
TCGA data and cBioPortal
TCGA (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) is currently the largest cancer gene information database that is comprehensive in many cancer types and multi-omics data [20]. We selected a UVM dataset (TCGA, Firehose Legacy) including 80 samples and further analyzed the expression of MMPs using cBioPortal (http://www.cbioportal.org/) [21]. Mutations, putative copy-number alterations from GISTIC, and mRNA expression z-scores relative to diploid samples (RNA Seq V2 RSEM) were included in the genomic map.
Metascape
Metascape (http://metascape.org/gp/index.html#/main/step1) is a public online analysis database used to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses [22]. In our study, Metascape was used for functional annotation and enrichment analysis of MMPs and their 50 neighboring genes and the relevant parameters were as follows: minimum overlap, 3; p value cutoff, 0.01; minimum enrichment, 1.5.
GeneMANIA
GeneMANIA (http://genemania.org/) is an online prediction website that provides information on protein-DNA, protein-protein, and genetic interactions, reactions, pathways, and protein domains of related genes [23]. In our study, GeneMANIA was used to compare the correlation between MMPs in UVM patients.
GEPIA 2
Based on tumor and normal samples from the TCGA and GTEx databases, GEPIA 2 (http://gepia2.cancer-pku.cn/#index) is an indispensable and highly cited database for gene expression analysis [24]. It can be used to perform a survival analysis of specific genes. In GEPIA 2, UVM patients were divided into low and high expression groups based on the median values of mRNA expression and validated by GEPIA 2 survival curves. The methods used were OS and DFS with a cut-off of 50%.
UALCAN
UALCAN (http://ualcan.path.uab.edu/index.html) is an effective website for online analysis and mining of cancer data, mainly based on the relevant cancer data in the TCGA database [25]. In this study, UALCAN was used to analyze the mRNA expression of 17 different MMP family members in primary UVM tissues and its association with clinicopathological parameters. Student’s t-test was used to compare the difference in transcriptional expressions and statistical significance was set at p < 0.05.
TRRUST
TRRUST (https://www.grnpedia.org/trrust/) is a curated database of human and mouse transcriptional regulatory networks that contains information derived from 11,237 published articles [26]. In our study, this database was used to explore the association between MMPs and transcriptional factors.
Timer
TIMER (http://timer.cistrome.org/) is a comprehensive resource for systematical analysis of immune infiltrations across diverse cancers [27–29]. “Immune association” and “gene” module were used in this study to evaluate the correlation between MMPs and the infiltration of immune cells. R software and GSVA package were also used to draw the pictures of associations between MMPs and the immune cells [30, 31].
Results
Eight elite genes related to UVM
Using MalaCards, we identified eight elite genes related to UVM. The relationship between these elite genes and UVM comes from artificial annotation and reliable resources (Table 1).
Table 1.
8 elite genes related to uveal melanoma
| Number | Gene | Description | Category | Score |
|---|---|---|---|---|
| 1 | BAP1 | BRCA1 Associated Protein 1 | Protein coding | 764.58 |
| 2 | GNAQ | G Protein Subunit Alpha Q | Protein coding | 759.14 |
| 3 | GNA11 | G Protein Subunit Alpha 11 | Protein coding | 759.09 |
| 4 | PLCB4 | Phospholipase C Beta 4 | Protein coding | 407.24 |
| 5 | CYSLTR2 | Cysteinyl Leukotriene Receptor 2 | Protein coding | 356.9 |
| 6 | UVM1 | Melanoma, Uveal, Susceptibility To, 1 | Genetic Locus | 57.97 |
| 7 | UVM2 | Melanoma, Uveal, Susceptibility To, 2 | Genetic Locus | 57.97 |
| 8 | SF3B1 | Splicing Factor 3b Subunit 1 | Protein coding | 38.14 |
MMPs transcriptional differences in melanoma cancers and normal tissues
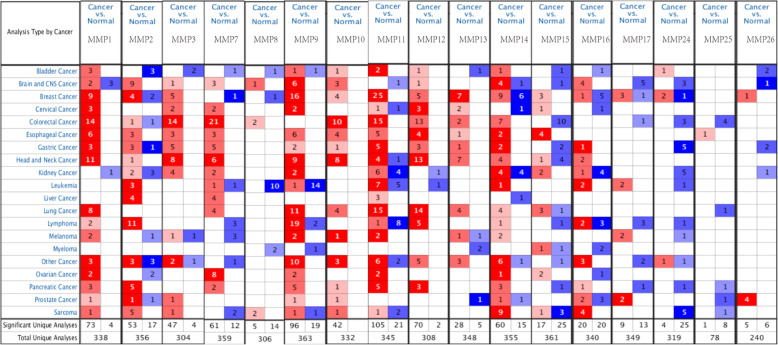
We used the ONCOMINE database to explore the expression levels of MMPs in UVM and normal tissues (Fig. 1). This analysis revealed that the transcriptional levels of MMP1, MMP9, MMP10, MMP11, MMP13, MMP14, and MMP17 were upregulated in patients with UVM, while the transcriptional levels of MMP2, MMP7, and MMP24 were lower than those in normal samples. Next, we discuss the relationship between UVM and MMP1, MMP2, MMP9, MMP11, MMP14, MMP15, MMP16, MMP17, and MMP24.
Fig. 1.
Transcriptional expression of MMPs in 20 different types of cancer diseases (ONCOMINE database). Difference of transcriptional expression was compared by students’ t-test. Cut-off of p value and fold change were as following: p value: 0.01, fold change: 1.5, gene rank: 10%, data type: mRNA
GO and KEGG explorations of MMPs in UVM patients
Through GeneMANIA analysis, we identified 50 neighboring genes with the highest frequency association with MMPs. These data indicated that CTB-96E2.2, HPX, PRG4, VTN, ASTL, MEP1B, MFAP2, MEP1A, BSPH1, ELSPBP1, IGF2R, ENDOU, TLL2, TLL1, BMP1, SEL1L, FN1, MFAP5, F12, MRC2, LY75-CD302, LY75, TIMP2, HGFAC, MRC1, TINAG, PLA2R1, ENPP2, ENPP1, IGFBP1, TIMP1, TFPI, OSCAR, SPP1, TIMP4, ENPP3, PTAFR, TIMP3, PIK3IP1, LAIR1, SUSD2, GP6, TARM1, FCAR, CRISP3, CRISP2, PROC, SDC1, VSTM1, and CRISP1 were associated with the function and pathway of MMPs in patients with UVM. The functions of MMP members and their neighboring genes were predicted using Metascape.
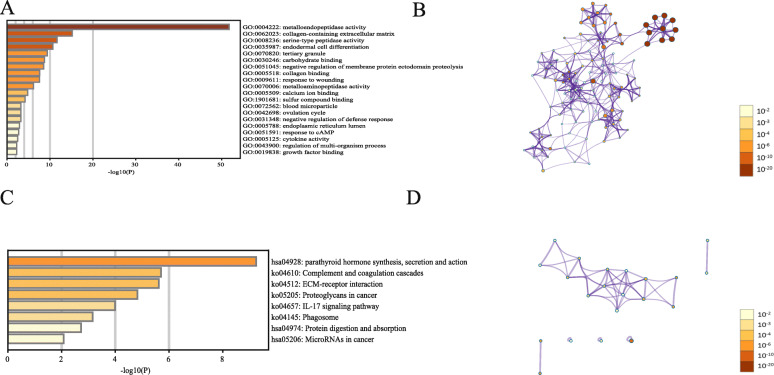
The biological process (BP) functions for these genes were primarily endodermal cell differentiation, negative regulation of membrane protein ectodomain proteolysis, response to wounding, ovulation cycle, negative regulation of defense response, response to cAMP, and regulation of multi-organism processes. The cellular components (CC) related to these genes were mainly collagen-containing extracellular matrix, tertiary granules, blood microparticles, and endoplasmic reticulum lumen. The identified molecular functions (MF) related to these genes were metalloendopeptidase activity, serine-type peptidase activity, carbohydrate binding, collagen binding, metalloaminopeptidase activity, calcium ion binding, sulfur compound binding, cytokine activity, and growth factor binding (Fig. 2A, B, Table 2).
Fig. 2.
PPI and functional enrichment analysis of MMPs in patients with UVM (Metascape). a MMPs gene ontology (GO) enriched terms, colored by p-value. b Networks of GO enriched terms colored by p value, and terms containing more genes tend to have a more significant p value. c MMPs Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment terms, colored by p-value. d Networks of KEGG enriched terms colored by p value, and terms containing more genes tend to have a more significant p value
Table 2.
The GO function enrichment analysis of MMP family members and neighboring genes (Metascape)
| GO | Category | Description | Count | % | Log10(P) | Log10(q) |
|---|---|---|---|---|---|---|
| GO:0004222 | GO Molecular Functions | metalloendopeptidase activity | 29 | 39.73 | −51.76 | −47.41 |
| GO:0062023 | GO Cellular Components | collagen-containing extracellular matrix | 17 | 23.29 | −15.20 | −11.96 |
| GO:0008236 | GO Molecular Functions | serine-type peptidase activity | 11 | 15.07 | −11.66 | −8.46 |
| GO:0035987 | GO Biological Processes | endodermal cell differentiation | 7 | 9.59 | −10.68 | −7.54 |
| GO:0070820 | GO Cellular Components | tertiary granule | 9 | 12.33 | −9.36 | −6.33 |
| GO:0030246 | GO Molecular Functions | carbohydrate binding | 10 | 13.70 | −8.74 | −5.77 |
| GO:0051045 | GO Biological Processes | negative regulation of membrane protein ectodomain proteolysis | 4 | 5.48 | −8.54 | −5.59 |
| GO:0005518 | GO Molecular Functions | collagen binding | 6 | 8.22 | −7.62 | −4.78 |
| GO:0009611 | GO Biological Processes | response to wounding | 13 | 17.81 | −7.55 | −4.73 |
| GO:0070006 | GO Molecular Functions | metalloaminopeptidase activity | 4 | 5.48 | −6.16 | −3.57 |
| GO:0005509 | GO Molecular Functions | calcium ion binding | 10 | 13.70 | −4.82 | −2.42 |
| GO:1901681 | GO Molecular Functions | sulfur compound binding | 6 | 8.22 | −4.22 | −1.89 |
| GO:0072562 | GO Cellular Components | blood microparticle | 4 | 5.48 | −3.24 | −1.00 |
| GO:0042698 | GO Biological Processes | ovulation cycle | 3 | 4.11 | −3.23 | −1.00 |
| GO:0031348 | GO Biological Processes | negative regulation of defense response | 5 | 6.85 | −3.22 | −0.99 |
| GO:0005788 | GO Cellular Components | endoplasmic reticulum lumen | 5 | 6.85 | −2.92 | −0.72 |
| GO:0051591 | GO Biological Processes | response to cAMP | 3 | 4.11 | −2.72 | −0.56 |
| GO:0005125 | GO Molecular Functions | cytokine activity | 4 | 5.48 | −2.48 | −0.34 |
| GO:0043900 | GO Biological Processes | regulation of multi-organism process | 5 | 6.85 | −2.25 | −0.12 |
| GO:0019838 | GO Molecular Functions | growth factor binding | 3 | 4.11 | −2.25 | −0.12 |
The top eight KEGG pathways for MMP family members and their neighboring genes are shown in Fig. 2C and Table 3. Among these pathways, ECM-receptor interaction, proteoglycans in cancer, IL-17 signaling pathway, and microRNAs in cancer were found to be related to carcinoma pathogenesis and metastasis (Fig. 2C, D, Table 3).
Table 3.
The KEGG function enrichment analysis of MMP family and neighboring genes (Metascape)
| GO | Category | Description | Count | % | Log10(P) | Log10(q) |
|---|---|---|---|---|---|---|
| hsa04928 | KEGG Pathway | parathyroid hormone synthesis, secretion and action | 8 | 10.96 | −9.24 | −6.34 |
| ko04610 | KEGG Pathway | Complement and coagulation cascades | 5 | 6.85 | −5.70 | −3.21 |
| ko04512 | KEGG Pathway | ECM-receptor interaction | 5 | 6.85 | −5.62 | −3.21 |
| ko05205 | KEGG Pathway | Proteoglycans in cancer | 6 | 8.22 | −4.83 | −2.70 |
| ko04657 | KEGG Pathway | IL-17 signaling pathway | 4 | 5.48 | −3.99 | −2.00 |
| ko04145 | KEGG Pathway | Phagosome | 4 | 5.48 | −3.15 | −1.36 |
| hsa04974 | KEGG Pathway | Protein digestion and absorption | 3 | 4.11 | −2.72 | −0.99 |
| hsa05206 | KEGG Pathway | MicroRNAs in cancer | 4 | 5.48 | −2.08 | −0.49 |
Genetic alteration, co-expression, and interaction analyses of MMPs in patients with UVM
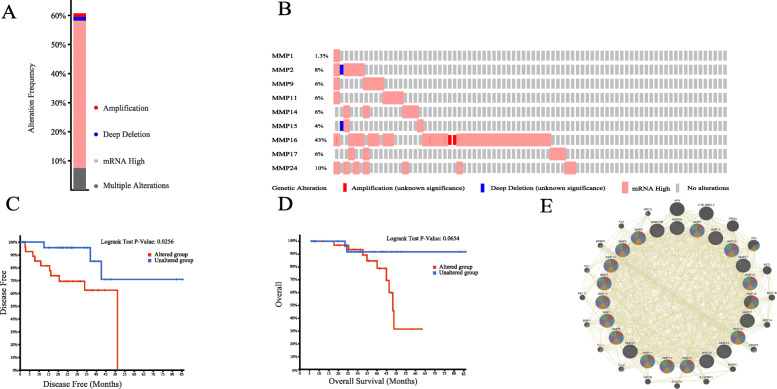
The cBioportal online tool was used to analyze the alteration frequency of MMP mutations in patients with UVM. According to the TCGA dataset, among the 80 UVM samples, MMPs were changed in 49 samples (61%) (Fig. 3A). MMP16 (43%) had the highest genetic variation rate, while MMP1 (1.3%) had the lowest. MMP2, MMP9, MMP11, MMP14, MMP15, MMP17, and MMP24 were altered in 8, 6, 6, 6, 4, 6, and 10% of the patients with UVM, respectively (Fig. 3B). The result of Kaplan-Meier plotter and log-rank test showed no significant difference in OS between the altered and unaltered groups (log-rank test p-value =0.0654), while there was a significant difference in DFS (log-rank test p-value =0.0256) (Fig. 3C, D).
Fig. 3.
Alteration frequency of MMPs family members and neighboring gene network in patients with UVM (cBioPortal and geneMANIA). a Summary of alterations in 9 MMPs family members. b OncoPrint visual summary of alteration on a query of MMPs family members. c Kaplan-Meier plots comparing OS in patients with/without MMPs family member gene alterations. d Kaplan-Meier plots comparing DFS in patients with/without MMPs family member gene alterations. e Gene-gene interaction network of different expressed MMPs. Each node represents a gene, and the size of which represents the strength of the interaction
Moreover, a gene-gene interaction (GGI) network of 23 MMPs was constructed and analyzed using the GeneMANIA database (Fig. 3E). Twenty genes surrounding the 17 MMPs were significantly associated with each other in the shared protein, prediction, co-expression, pathway, and co-localization parameters. The top five genes most associated with MMPs were HPX (hemopexin), CTB-96E2.2, PRG4 (proteoglycan 4), VTN (vitronectin), and ASTL (astacin-like metalloendopeptidase). Figure 3E shows that 95.59% of these genes had shared protein domains, 2.89% shared prediction, 0.72% shared physical interactions, and only 0.38% of these genes shared co-localization. Further functional analysis indicated that the most significant correlation was with the extracellular matrix (FDR = 2.85e-27). In addition, these genes were also correlated with extracellular structure, extracellular structure organization, collagen catabolic process, and multicellular organismal catabolic process.
Correlation between MMPs and clinicopathological parameters in patients with UVM
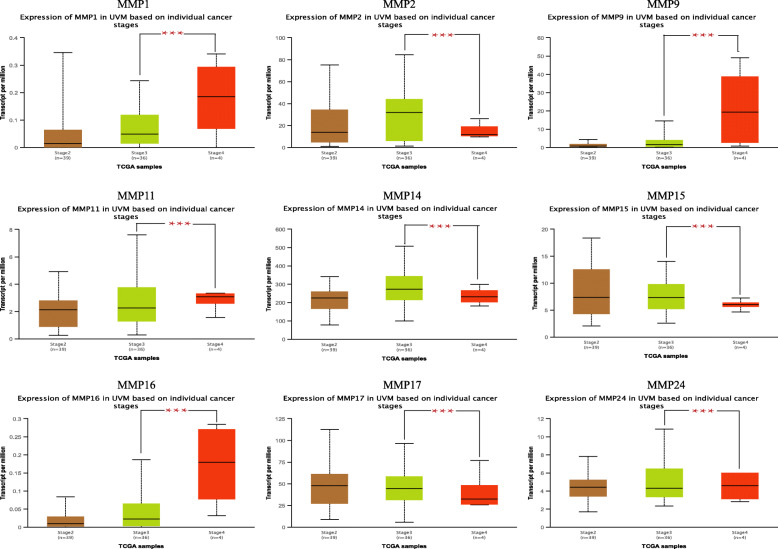
The correlation between MMPs and the pathological and histological subtypes of UVM was further assessed by UALCAN (Fig. 4). As shown in Fig. 4, for patients with UVM, all MMPs above were markedly statistically significant during the transition from stage 3 to stage 4. These data strongly suggested that these MMPs play significant roles in the progression from stage 3 to stage 4.
Fig. 4.
Correlation between 9 MMPs expression and tumor stage in UVM patients (GEPIA). The p value was set at 0.05. *** represents p < 0.001
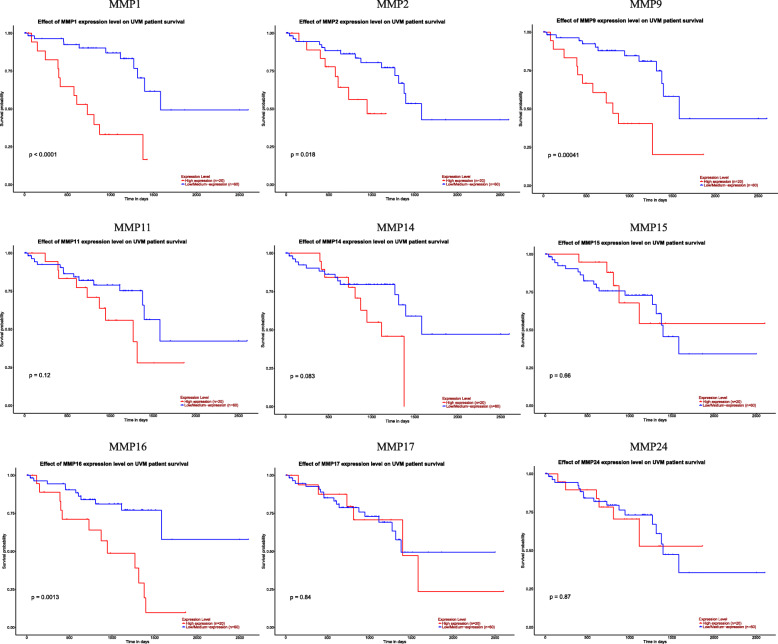
Prognostic value of mRNA expression of MMPs in patients with UVM
In addition, we also obtained survival plots of MMPs in the uveal melanoma TCGA dataset from UALCAN. Kaplan Meier plots showed the effect of MMPs on patients’ OS (Fig. 5). The mRNA expression of four MMPs was significantly associated with the prognosis of UVM. The increased mRNA levels of MMP1, MMP2, MMP9, and MMP16 were strongly associated with poor OS, while those of MMP14, MMP15, MMP17, and MMP24 were not related to OS in UVM.
Fig. 5.
The prognostic value of 9 MMPs family members in UVM patients (UALCAN). The OS survival curves comparing patients with high (red) and low (blue) MMPs family member expressions were plotted. The threshold of p value is 0.05
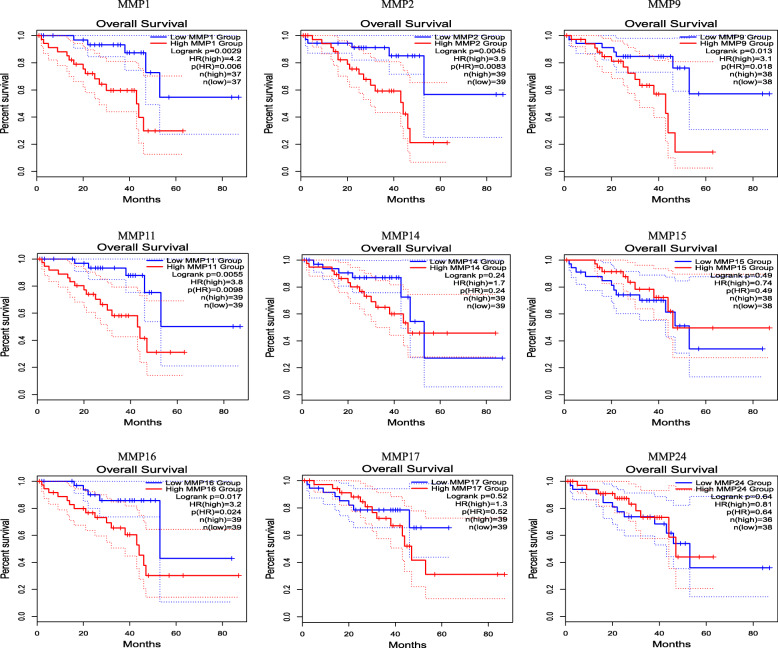
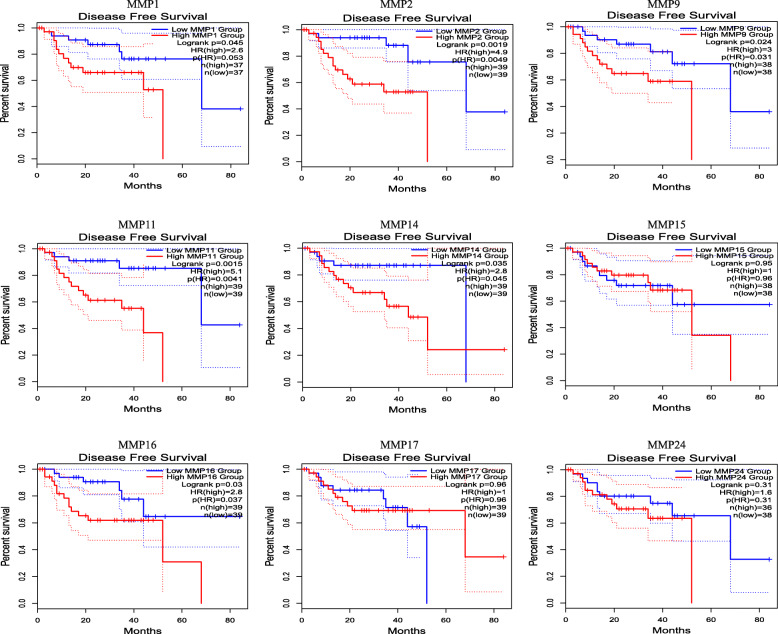
We also used the GEPIA dataset to compare the mRNA expression of MMPs to the OS and DFS of patients with UVM. DFS and OS curves are presented in Figs. 6 and 7. There were significant associations between some MMPs (MMP1, MMP2, MMP9, MMP11, and MMP16) and OS in patients, which was almost consistent with the results presented above. Moreover, there were significant associations between certain MMPs (MMP1, MMP2, MMP9, MMP11, MMP14, and MMP16) and DFS.
Fig. 6.
The prognostic value of 9 MMPs family members in UVM patients (GEPIA). The DFS survival curves comparing patients with high (red) and low (blue) MMPs family member expressions were plotted. The threshold of p value is 0.05
Fig. 7.
The prognostic value of 9 MMPs family members in UVM patients (GEPIA). The OS survival curves comparing patients with high (red) and low (blue) MMPs family member expressions were plotted. The threshold of p value is 0.05
Taken together, these results showed that MMP1, MMP2, MMP9 and MMP16 expression levels may be exploited as potential biomarkers for the prediction of the survival of patients with UVM.
Transcription factor targets in patients with UVM
Owing to the significant difference in the expression of MMPs in UVM vs. normal tissues, we explored related transcription factor targets of the differentially expressed MMPs using TRRUST DATABASE. MMP1, MMP2, MMP9, MMP11, MMP14, MMP15, MMP16, MMP17, and MMP24 were included in TRRUST. We found that 18 transcription factors (MAZ, ETV4, SRF, ETS2, STAT3, KLF8, RELA, NFKB1, SNAI2, NFKBIA, JUN, SP1, TWIST1, FOS, PPARG, TFAP2A, ETS1, and TP53) were associated with the regulation of MMPs (Table 4).
Table 4.
Transcription factors of the differentially expressed MMPs
| Key TF | Description | P value | FDR | Regulated genes |
|---|---|---|---|---|
| MAZ | MYC-associated zinc finger protein (purine-binding transcription factor) | 4.20E-09 | 7.55E-08 | MMP9, MMP14, MMP1 |
| ETV4 | ets variant 4 | 1.15E-07 | 1.03E-06 | MMP14, MMP1, MMP2 |
| SRF | serum response factor (c-fos serum response element-binding transcription factor) | 1.72E-07 | 1.03E-06 | MMP14, MMP2, MMP9 |
| ETS2 | v-ets erythroblastosis virus E26 oncogene homolog 2 (avian) | 3.69E-07 | 1.35E-06 | MMP1, MMP9, MMP2 |
| STAT3 | signal transducer and activator of transcription 3 (acute-phase response factor) | 3.76E-07 | 1.35E-06 | MMP9, MMP2, MMP1, MMP14 |
| KLF8 | Kruppel-like factor 8 | 5.65E-06 | 1.70E-05 | MMP14, MMP9 |
| RELA | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | 7.50E-06 | 1.73E-05 | MMP9, MMP2, MMP14, MMP1 |
| NFKB1 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | 7.70E-06 | 1.73E-05 | MMP1, MMP2, MMP9, MMP14 |
| SNAI2 | snail homolog 2 (Drosophila) | 1.83E-05 | 3.67E-05 | MMP17, MMP9 |
| NFKBIA | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | 3.44E-05 | 6.20E-05 | MMP1, MMP9 |
| JUN | jun proto-oncogene | 3.91E-05 | 6.40E-05 | MMP9, MMP2, MMP1 |
| SP1 | Sp1 transcription factor | 4.40E-05 | 6.61E-05 | MMP2, MMP9, MMP11, MMP14 |
| TWIST1 | twist basic helix-loop-helix transcription factor 1 | 0.000119 | 0.000165 | MMP2, MMP1 |
| FOS | FBJ murine osteosarcoma viral oncogene homolog | 0.000318 | 0.000409 | MMP9, MMP1 |
| PPARG | peroxisome proliferator-activated receptor gamma | 0.000427 | 0.000512 | MMP9, MMP1 |
| TFAP2A | transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) | 0.000494 | 0.000556 | MMP2, MMP9 |
| ETS1 | v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | 0.000611 | 0.000647 | MMP1, MMP9 |
| TP53 | tumor protein p53 | 0.0026 | 0.0026 | MMP1, MMP2 |
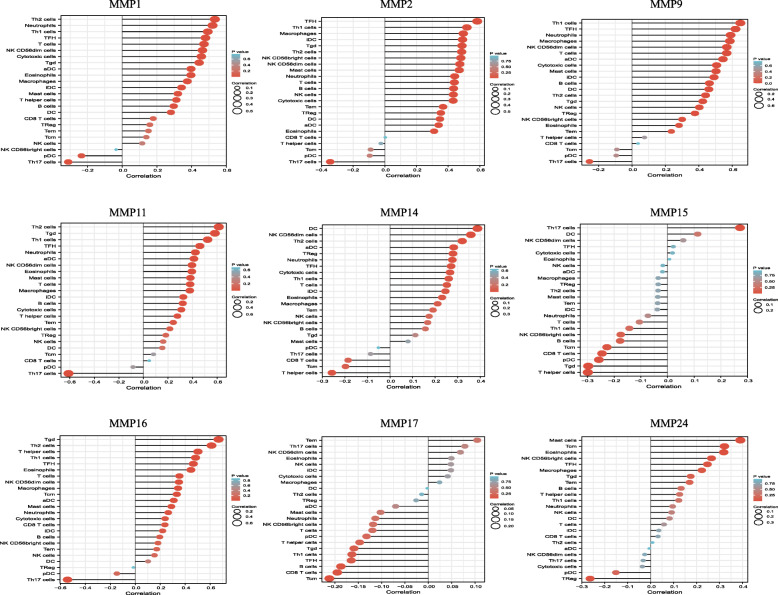
Immune cell infiltration of MMPs in patients with UVM
MMPs are involved in cancer-related inflammation and immune cell infiltration, thus affecting the clinical outcome of UVM patients. Therefore, the TIMER database was used to evaluate the comprehensive relationship between differentially expressed MMPs and immune cell infiltration. The results are presented in Fig. 8. Except for MMP15 and MMP17, seven matrix metalloproteinases (including MMP1, MMP2, MMP9, MMP11, MMP14, MMP16, MMP24) positively correlated with the infiltration of most of the immune cells. We also found that MMP15 and MMP17 negatively correlated with the infiltration of most of the immune cells.
Fig. 8.
The relationship between differentially expressed MMPs and immune cell infiltration (TIMER)
Discussion
Numerous studies have suggested that dysregulation of MMPs is involved in the development and progression of many tumors. Although some members of MMPs have been shown to play crucial roles in carcinoma, the distinct roles of MMP family members in UVM remain to be elucidated. In this study, the mutations, mRNA expression, prognostic values, and potential functions of different MMP family members in UVM were identified with bioinformatics tools.
MMP proteins are associated with the breakdown of the extracellular matrix during physiological processes and disease processes. Mutations in MMP1 are associated with chronic obstructive pulmonary disease [32]. Moreover, recent studies have shown that MMP1 is highly expressed in human oral squamous cell carcinoma and cervical squamous cell carcinoma. It can promote tumor growth and cell motility, participate in the development of human oral squamous cell carcinoma [33], and serve as a potential biomarker for cervical squamous cell carcinoma [12]. In our study, MMP1 was found to be upregulated in UVM tissues, and high expression indicated shorter OS and DFS, which was consistent with the results of previous studies.
MMP2 is a zinc-dependent enzyme that cleaves the components of the extracellular matrix and molecules. Unlike most MMP family members, activation of MMP2 occurs on the cell membrane. This enzyme can be activated extracellularly and intracellularly and is involved in multiple pathways. Mutations in this gene are associated with the Winchester syndrome [34]. Recent studies have indicated that MMP2 is highly expressed in many tumors, and its increase promotes proliferation and metastasis, and reduces tumor cell apoptosis [35–38]. In our study, MMP2 was found to be upregulated in UVM, and high expression indicated shorter OS and DFS.
Like other MMP family members, MMP9 is involved in disease processes such as metastasis and arthritis, as well as in the degradation of extracellular matrix components in normal processes such as reproduction and tissue remodeling. MMP9 has been found to play an essential role in cancer progression and is significantly associated with metastasis, angiogenesis, and tumor growth [39]. Dysregulation of MMP9 is involved in various diseases, such as neurological disorders [40], inflammatory diseases [41], cardiovascular diseases [16], and lung diseases [8]. In our study, MMP9 was found to be upregulated in UVM, and high expression indicated shorter OS and DFS, which was consistent with the results of previous studies.
MMP11 is a protein-coding gene, and MMP25 is an important paralog of this gene. MMP11, a zinc-dependent endopeptidase and the main protease involved in ECM degradation, plays an important role in various biological processes, such as breast cancer [13], prostate cancer [9], gastric cancer [42], and lung cancer [43], where the high expression of MMP11 is associated with metastasis and a poor clinical outcome. Furthermore, several studies have shown that MMP11 is a potential biomarker gene in breast cancer, which may facilitate the diagnosis and prognosis of breast cancer [44, 45]. In our study, MMP11 was found to be upregulated in UVM, and high expression indicated shorter OS and DFS, which was consistent with the results of previous studies.
The protein encoded by MMP14 is a member of the membrane-type MMP (MT-MMP), which contains a potential transmembrane domain, indicating that it is expressed on the cellular surface.
Studies have reported that high MMP14 expression is associated with poor prognosis and might serve as a marker of progression in muscle-invasive bladder cancer and colorectal cancer [10, 46]. Low MMP14 expression can inhibit the invasion and metastasis of breast cancer and gastric cancer cells [47, 48]. In our study, MMP14 was found to be upregulated in UVM, and high expression indicated shorter DFS, but it was not associated with OS in patients with UVM.
Like MMP14, the protein encoded by MMP15 is also a member of the MT-MMP. MMP15, also known as MT2-MMP, was first extracted from a human lung cDNA library.
It is composed of 669 amino acids encoded in 3530 bp of the gene. Proteins in this family are associated with the breakdown of the extracellular matrix in various diseases. MMP15 is highly expressed in bladder cancer and contributes to inflammation and angiogenesis in cancer cells [49]. MMP15 is also associated with malignancy, aggressiveness, and survival prognosis in human urinary bladder cancer by activating other MMPs [50]. The expression of MMP15 is significantly higher in tumors with a higher level of aggressiveness and malignancy [15]. In our study, MMP15 was not found to be upregulated in UVM and the expression of MMP15 was not associated with OS or DFS in patients with UVM.
MMT16 was once referred to as MT-MMP2 and is has been renamed as MT-MMP3 or MMP16.
MMP16 is activated when cleaved by other proteinases. Degradation of extracellular and microRNAs in cancer is an important pathway related to MMP16. MMP24 is an important paralog of the MMP16 gene. Some studies have indicated that high expression of MMP16 is associated with the proliferation, migration, and invasion of cutaneous squamous cell carcinoma cells [51], while knockdown of MMP16 could inhibit cell proliferation and invasion in chordoma cells [11]. Moreover, some variations in MMP16 have been found to be associated with different types of diseases [52, 53]. In our study, MMP16 was found to be upregulated in UVM, and high expression was associated with shorter OS and DFS.
MMP17 is a protein-coding gene that is associated with diseases such as breast cancer [14], colon cancer [54], and head and neck cancer [55]. While the expression of MMP17 has been associated with different physiological and pathological processes, the mechanisms underlying these processes remain unknown. In our study, MMP17 was not found to be upregulated in UVM and the expression of MMP17 was not associated with OS and shorter DFS in UVM.
The MMP24 protein is associated with the breakdown of the extracellular matrix in disease processes and normal physiological processes. The proteins cadherin 2 and MMP2 are substrates of this protease. MMP16 is an important paralog of this gene.
In the present study, we comprehensively assessed the expression and prognostic value of 26 MMPs in UVM. In our study, MMP24 was not found to be upregulated in UVM and the expression of MMP24 was not associated with overall survival and shorter disease-free survival in uveal melanoma patients. However, MMP24 play a significant role in the progression from stage 3 to stage 4 for UVM patients.
To date, our study is the first bioinformatic analysis to explore the association between UVM and all the members of the matrix metalloproteinase family. However, there are some limitations in our study. First, due to the limitation of conditions, there is no relevant experimental verification in our study. At the appropriate time in the future, we will carry out related experiments. Second, due to the lack of clinical data on UVM in relevant databases, such as TCGA, we could not find sufficient data for an effective analysis.
Conclusions
In conclusion, the current study suggests that the transcription levels of MMP1/9/10/11/13/14/17 are obviously upregulated in UVM and may play an important role in its occurrence and progression. MMP 1/2/9/11/14/15/16/17/24 could be used as molecular markers to elucidate the stages of UVM. In addition, abnormal expression of MMP1/2/9/11/16 was found to be related to OS in UVM, and MMP1/2/9/11/14/16 were found to be associated with DFS. Moreover, the expression of MMP1, MMP2, MMP9, and MMP16 was positively correlated with OS and DFS. We also found that most MMPs correlated with the infiltration of immune cells. Overall, our results revealed that MMP1 and MMP9 may serve as potential prognostic biomarkers and as therapeutic targets for UVM. However, further studies are needed to confirm our results and assess the clinical utility of MMPs.
Acknowledgements
The authors thank the contributors of the TCGA (https://tcga-data.nci.nih.gov/) database and GEO (http://www.ncbi.nlm.nih.gov/geo/) database and concerned authors for sharing their data on open access.
Abbreviations
- UVM
Uveal melanoma
- MMP
Matrix metalloproteinases
- GEO
Gene Expression Omnibus
- OS
Overall survival
- DFS
Disease-free survival
- GEPIA
Gene Expression Profiling Interactive Analysis
- PPI
Protein-protein interaction
- BP
Biological process
- CC
Cellular component
- MF
Molecular function
- GO
Gene ontology
- KEGG
Kyoto encyclopedia of genes and genomes
- GGI
Gene-gene interaction
- TCGA
The cancer genome atlas
Authors’ contributions
Q.P and T.Y.W conceived and designed the study. Y.Y.Z and J.H.B extracted the data and performed the analysis. Y.W.X and Y.Y.Z wrote the manuscript. Q. P and T.Y.W revised the manuscript. All authors approved the final manuscript to be published.
Funding
This work was financially supported by the National Natural Science Foundation of China (No. 81470025) and by the Shanghai Municipal Health Commission (No. ZY (2018–2020)-ZWB-1001-CPJS10) and by the Three-Year Action Plan for Promoting Clinical Skills and Clinical Innovation in Municipal Hospitals (No. SHDC2020CR5014).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the Oncomine repository, [https://www.oncomine.org/resource/main.html]; the TCGA respository, [https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga]; the Metascape respository, [http://metascape.org/gp/index.html#/main/step1]; the GeneMANIA respository, [http://genemania.org/]; GEPIA 2 respository, [http://gepia2.cancer-pku.cn/#index]; the UALCAN respository, [http://ualcan.path.uab.edu/index.html]; the TRRUST respository, [https://www.grnpedia.org/trrust/]; the TIMER respository, [http://timer.cistrome.org/].
Declarations
Ethics approval and consent to participate
Since all the data were retrieved from the online databases, we assumed that all written informed consent had been already obtained.
Consent for publication
Not applicable.
Competing interests
The authors declared no conflicts of interest in this work.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Krantz BA, Dave N, Komatsubara KM, Marr BP, Carvajal RD. Uveal melanoma: epidemiology, etiology, and treatment of primary disease. Clin Ophthalmol (Auckland, N.Z.) 2017;11:279–289. doi: 10.2147/OPTH.S89591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreoli MT, Mieler WF, Leiderman YI. Epidemiological trends in uveal melanoma. Br J Ophthalmol. 2015;99(11):1550–1553. doi: 10.1136/bjophthalmol-2015-306810. [DOI] [PubMed] [Google Scholar]
- 3.Schank TE, Hassel JC. Immunotherapies for the treatment of Uveal melanoma-history and future. Cancers. 2019;11(8):1048. doi: 10.3390/cancers11081048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh AD, Bergman L, Seregard S. Uveal melanoma: epidemiologic aspects. Ophthalmol Clin N Am. 2005;18(1):75–84. doi: 10.1016/j.ohc.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Derrien AC, Rodrigues M, Eeckhoutte A, Dayot S, Houy A, Mobuchon L, Gardrat S, Lequin D, Ballet S, Pierron G, Alsafadi S, Mariani O, el-Marjou A, Matet A, Colas C, Cassoux N, Stern MH. Germline MBD4 mutations and predisposition to Uveal melanoma. J Natl Cancer Inst. 2021;113(1):80–87. doi: 10.1093/jnci/djaa047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katopodis P, Khalifa MS, Anikin V. Molecular characteristics of uveal melanoma and intraocular tumors. Oncol Lett. 2021;21(1):9. doi: 10.3892/ol.2020.12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mondal S, Adhikari N, Banerjee S, Amin SA, Jha T. Matrix metalloproteinase-9 (MMP-9) and its inhibitors in cancer: a minireview. Eur J Med Chem. 2020;194:112260. doi: 10.1016/j.ejmech.2020.112260. [DOI] [PubMed] [Google Scholar]
- 8.Churg A, Wang R, Wang X, Onnervik PO, Thim K, Wright JL. Effect of an MMP-9/MMP-12 inhibitor on smoke-induced emphysema and airway remodelling in guinea pigs. Thorax. 2007;62(8):706–713. doi: 10.1136/thx.2006.068353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roscilli G, Cappelletti M, De Vitis C, Ciliberto G, Di Napoli A, Ruco L, Mancini R, Aurisicchio L. Circulating MMP11 and specific antibody immune response in breast and prostate cancer patients. J Transl Med. 2014;12(1):54. doi: 10.1186/1479-5876-12-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang JF, Gong YQ, He YH, Ying WW, Li XS, Zhou XF, Zhou LQ. High expression of MMP14 is associated with progression and poor short-term prognosis in muscle-invasive bladder cancer. Eur Rev Med Pharmacol Sci. 2020;24(12):6605–6615. doi: 10.26355/eurrev_202006_21646. [DOI] [PubMed] [Google Scholar]
- 11.Xu DM, Han PH, Chen L, Li TT, Yang XH, Guo R. Knockdown of MMP16 inhibits cell proliferation and invasion in chordoma in vitro. J Biol Regul Homeost Agents. 2020;34(6):2263–2270. doi: 10.23812/20-559-L. [DOI] [PubMed] [Google Scholar]
- 12.Zhao S, Yu M. Identification of MMP1 as a potential prognostic biomarker and correlating with immune infiltrates in cervical squamous cell carcinoma. DNA Cell Biol. 2020;39(2):255–272. doi: 10.1089/dna.2019.5129. [DOI] [PubMed] [Google Scholar]
- 13.Eiro N, Cid S, Fernandez B, Fraile M, Cernea A, Sanchez R, Andicoechea A, DeAndres Galiana EJ, Gonzalez LO, Fernandez-Muniz Z, et al. MMP11 expression in intratumoral inflammatory cells in breast cancer. Histopathology. 2019;75(6):916–930. doi: 10.1111/his.13956. [DOI] [PubMed] [Google Scholar]
- 14.Host L, Paye A, Detry B, Blacher S, Munaut C, Foidart JM, Seiki M, Sounni NE, Noel A. The proteolytic activity of MT4-MMP is required for its pro-angiogenic and pro-metastatic promoting effects. Int J Cancer. 2012;131(7):1537–1548. doi: 10.1002/ijc.27436. [DOI] [PubMed] [Google Scholar]
- 15.Mohammad MA, Ismael NR, Shaarawy SM, El-Merzabani MM. Prognostic value of membrane type 1 and 2 matrix metalloproteinase expression and gelatinase a activity in bladder cancer. Int J Biol Markers. 2010;25(2):69–74. doi: 10.1177/172460081002500202. [DOI] [PubMed] [Google Scholar]
- 16.Yabluchanskiy A, Ma Y, Iyer RP, Hall ME, Lindsey ML. Matrix metalloproteinase-9: many shades of function in cardiovascular disease. Physiology (Bethesda) 2013;28(6):391–403. doi: 10.1152/physiol.00029.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rappaport N, Twik M, Plaschkes I, Nudel R, Iny Stein T, Levitt J, Gershoni M, Morrey CP, Safran M, Lancet D. MalaCards: an amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res. 2017;45(D1):D877–DD87. doi: 10.1093/nar/gkw1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, Varambally S, Ghosh D, Chinnaiyan AM. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer genome atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19(1A):A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38(Web Server issue):W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han H, Cho JW, Lee S, Yun A, Kim H, Bae D, Yang S, Kim CY, Lee M, Kim E, Lee S, Kang B, Jeong D, Kim Y, Jeon HN, Jung H, Nam S, Chung M, Kim JH, Lee I. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018;46(D1):D380–D3D6. doi: 10.1093/nar/gkx1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–WW14. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–ee10. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, Jiang P, Shen H, Aster JC, Rodig S, Signoretti S, Liu JS, Liu XS. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(1):174. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14(1):7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A, Bruneval P, Fridman WH, Becker C, Pagès F, Speicher MR, Trajanoski Z, Galon J. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 32.Ostridge K, Williams N, Kim V, Bennett M, Harden S, Welch L, Bourne S, Coombs NA, Elkington PT, Staples KJ, Wilkinson TMA. Relationship between pulmonary matrix metalloproteinases and quantitative CT markers of small airways disease and emphysema in COPD. Thorax. 2016;71(2):126–132. doi: 10.1136/thoraxjnl-2015-207428. [DOI] [PubMed] [Google Scholar]
- 33.Wang C, Mao C, Lai Y, Cai Z, Chen W. MMP1 3’UTR facilitates the proliferation and migration of human oral squamous cell carcinoma by sponging miR-188-5p to up-regulate SOX4 and CDK4. Mol Cell Biochem. 2021;476(2):785–796. doi: 10.1007/s11010-020-03944-y. [DOI] [PubMed] [Google Scholar]
- 34.de Vos I, Wong ASW, Welting TJM, Coull BJ, van Steensel MAM. Multicentric osteolytic syndromes represent a phenotypic spectrum defined by defective collagen remodeling. Am J Med Genet Part A. 2019;179(8):1652–1664. doi: 10.1002/ajmg.a.61264. [DOI] [PubMed] [Google Scholar]
- 35.Li C, Deng L, Shen H, Meng Q, Qian A, Sang H, Xia J, Li X. O-6-methylguanine-DNA methyltransferase inhibits gastric carcinoma cell migration and invasion by Downregulation of matrix metalloproteinase 2. Anti Cancer Agents Med Chem. 2016;16(9):1125–1132. doi: 10.2174/1871520615666150914114455. [DOI] [PubMed] [Google Scholar]
- 36.Liu C. Pathological and prognostic significance of matrix metalloproteinase-2 expression in ovarian cancer: a meta-analysis. Clin Exp Med. 2016;16(3):375–382. doi: 10.1007/s10238-015-0369-y. [DOI] [PubMed] [Google Scholar]
- 37.Ren F, Tang R, Zhang X, Madushi WM, Luo D, Dang Y, Li Z, Wei K, Chen G. Overexpression of MMP family members functions as prognostic biomarker for breast Cancer patients: a systematic review and Meta-analysis. PLoS One. 2015;10(8):e0135544. doi: 10.1371/journal.pone.0135544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bi MC, Hose N, Xu CL, Zhang C, Sassoon J, Song E. Nonlethal levels of Zeaxanthin inhibit cell migration, invasion, and secretion of MMP-2 via NF-kappaB pathway in cultured human Uveal melanoma cells. J Ophthalmol. 2016;2016:8734309–8734308. doi: 10.1155/2016/8734309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiranjeevi P, Spurthi KM, Rani NS, Kumar GR, Aiyengar TM, Saraswati M, Srilatha G, Kumar GK, Sinha S, Kumari CS, Reddy BN, Vishnupriya S, Rani HS. Gelatinase B (−1562C/T) polymorphism in tumor progression and invasion of breast cancer. Tumour Biol. 2014;35(2):1351–1356. doi: 10.1007/s13277-013-1181-5. [DOI] [PubMed] [Google Scholar]
- 40.Tokito A, Jougasaki M. Matrix Metalloproteinases in non-neoplastic disorders. Int J Mol Sci. 2016;17(7):1178. doi: 10.3390/ijms17071178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vandooren J, Knoops S, Aldinucci Buzzo JL, Boon L, Martens E, Opdenakker G, Kolaczkowska E. Differential inhibition of activity, activation and gene expression of MMP-9 in THP-1 cells by azithromycin and minocycline versus bortezomib: a comparative study. PLoS One. 2017;12(4):e0174853. doi: 10.1371/journal.pone.0174853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Gao M, An J, Wang X, Jia Y, Xu J, Zhu J, Cui J, Li W, Xing R, Song L, Liu K, He Y, Sheng J, Qi S, Pan Y, Lu Y. Dysregulation of MiR-30a-3p/gastrin enhances tumor growth and invasion throughSTAT3/MMP11 pathway in gastric Cancer. Onco Targets Ther. 2020;13:8475–8493. doi: 10.2147/OTT.S235022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen GL, Wang SC, Huang WC, Chang WS, Tsai CW, Li HT, Shen TC, Hsia TC, Bau DT. The association of MMP-11 promoter polymorphisms with susceptibility to lung Cancer in Taiwan. Anticancer Res. 2019;39(10):5375–5380. doi: 10.21873/anticanres.13731. [DOI] [PubMed] [Google Scholar]
- 44.Fu J, Khaybullin R, Zhang Y, Xia A, Qi X. Gene expression profiling leads to discovery of correlation of matrix metalloproteinase 11 and heparanase 2 in breast cancer progression. BMC Cancer. 2015;15(1):473. doi: 10.1186/s12885-015-1410-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han J, Choi YL, Kim H, Choi JY, Lee SK, Lee JE, Choi JS, Park S, Choi JS, Kim YD, Nam SJ, Nam BH, Kwon MJ, Shin YK. MMP11 and CD2 as novel prognostic factors in hormone receptor-negative, HER2-positive breast cancer. Breast Cancer Res Treat. 2017;164(1):41–56. doi: 10.1007/s10549-017-4234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Claesson-Welsh L. How the matrix metalloproteinase MMP14 contributes to the progression of colorectal cancer. J Clin Invest. 2020;130(3):1093–1095. doi: 10.1172/JCI135239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ager EI, Kozin SV, Kirkpatrick ND, Seano G, Kodack DP, Askoxylakis V, Huang Y, Goel S, Snuderl M, Muzikansky A, et al. Blockade of MMP14 activity in murine breast carcinomas: implications for macrophages, vessels, and radiotherapy. J Natl Cancer Inst. 2015;107(4):djv017. doi: 10.1093/jnci/djv017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng L, Li D, Xiang X, Tong L, Qi M, Pu J, Huang K, Tong Q. Methyl jasmonate abolishes the migration, invasion and angiogenesis of gastric cancer cells through down-regulation of matrix metalloproteinase 14. BMC Cancer. 2013;13(1):74. doi: 10.1186/1471-2407-13-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, Zhang L, Wei N, Sun Y, Pan W, Chen Y. Silencing LINC00482 inhibits tumor-associated inflammation and angiogenesis through down-regulation of MMP-15 via FOXA1 in bladder cancer. Aging. 2020;13(2):2264–2278. doi: 10.18632/aging.202247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kudelski J, Mlynarczyk G, Darewicz B, Bruczko-Goralewska M, Romanowicz L. Dominative role of MMP-14 over MMP-15 in human urinary bladder carcinoma on the basis of its enhanced specific activity. Medicine. 2020;99(7):e19224. doi: 10.1097/MD.0000000000019224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng W, Li ZY, Zhao DL, Li XL, Liu R. microRNA-26a directly targeting MMP14 and MMP16 inhibits the Cancer cell proliferation, migration and invasion in cutaneous squamous cell carcinoma. Cancer Manag Res. 2020;12:7087–7095. doi: 10.2147/CMAR.S265775. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Morton SE, O'Hare KJM, Maha JLK, Nicolson MP, Machado L, Topless R, Merriman TR, Linscott RJ. Testing the validity of Taxonic Schizotypy using genetic and environmental risk variables. Schizophr Bull. 2017;43(3):633–643. doi: 10.1093/schbul/sbw108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu Y, Wang Y, Liu H, Shi Q, Zhu D, Amos CI, Fang S, Lee JE, Hyslop T, Li X, Han J, Wei Q. Genetic variants in the metzincin metallopeptidase family genes predict melanoma survival. Mol Carcinog. 2018;57(1):22–31. doi: 10.1002/mc.22716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nimri L, Barak H, Graeve L, Schwartz B. Restoration of caveolin-1 expression suppresses growth, membrane-type-4 metalloproteinase expression and metastasis-associated activities in colon cancer cells. Mol Carcinog. 2013;52(11):859–870. doi: 10.1002/mc.21927. [DOI] [PubMed] [Google Scholar]
- 55.Huang CH, Yang WH, Chang SY, Tai SK, Tzeng CH, Kao JY, Wu KJ, Yang MH. Regulation of membrane-type 4 matrix metalloproteinase by SLUG contributes to hypoxia-mediated metastasis. Neoplasia. 2009;11(12):1371–1382. doi: 10.1593/neo.91326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the Oncomine repository, [https://www.oncomine.org/resource/main.html]; the TCGA respository, [https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga]; the Metascape respository, [http://metascape.org/gp/index.html#/main/step1]; the GeneMANIA respository, [http://genemania.org/]; GEPIA 2 respository, [http://gepia2.cancer-pku.cn/#index]; the UALCAN respository, [http://ualcan.path.uab.edu/index.html]; the TRRUST respository, [https://www.grnpedia.org/trrust/]; the TIMER respository, [http://timer.cistrome.org/].