Abstract
Objective:
To evaluate the consistency of the quantitative imaging decision support (QIDSTM) tool and radiomic analysis using 594 metrics in lung carcinoma on chest CT scan.
Materials and Methods:
We included, retrospectively, 150 patients with histologically confirmed lung cancer who underwent chemotherapy and baseline and follow-ups CT scans. Using the QIDSTM platform, 3 radiologists segmented each lesion and automatically collected the longest diameter and the density mean value. Inter-observer variability, Bland Altman analysis and Spearman’s correlation coefficient were performed. QIDSTM tool consistency was assessed in terms of agreement rate in the treatment response classification. Kruskal Wallis test and the least absolute shrinkage and selection operator (LASSO) method with 10-fold cross validation were used to identify radiomic metrics correlated with lesion size change.
Results:
Good and significant correlation was obtained between the measurements of largest diameter and of density among the QIDSTM tool and the radiologists measurements. Inter-observer variability values were over 0.85. HealthMyne QIDSTM tool quantitative volumetric delineation was consistent and matched with each radiologist measurement considering the RECIST classification (80-84%) while a lower concordance among QIDSTM and the radiologists CHOI classification was observed (58-63%). Among 594 extracted metrics, significant and robust predictors of RECIST response were energy, histogram entropy and uniformity, Kurtosis, coronal long axis, longest planar diameter, surface, Neighborhood Grey-Level Different Matrix (NGLDM) dependence nonuniformity and low dependence emphasis as Volume, entropy of Log(2.5 mm), wavelet energy, deviation and root man squared.
Conclusion:
In conclusion, we demonstrated that HealthMyne quantitative volumetric delineation was consistent and that several radiomic metrics extracted by QIDSTM were significant and robust predictors of RECIST response.
Keywords: chest CT, pulmonary carcinoma, segmentation, RECIST, CHOI, radiomic
Introduction
In recent decades, clinical research and the interpretation of the results deriving from it have brought to the fore the need to build a common language internationally for the purpose of diagnosis, staging and evaluation of the effectiveness of a treatment.1
Nowadays the evaluation of the oncological therapy tumor response was conventionally provided using the Response evaluation criteria in solid tumors (RECIST) based on the longest diameter of the lesions; however, in the context of target therapies and immunotherapy, these evaluation criteria indicate the limits linked to the inability to fully highlight the effects of molecular target therapies. Conventionally, radiologists have measured tumor extent by the longest dimension on a single image rather than performing a full segmentation of the tumor volume.2
Based on these insights, several authors have proposed alternative methods by combining morphological-dimensional criteria with physiological-metabolic characteristics in order to overcome the limitations of traditional criteria. These alternative criteria allow to highlight the response to treatment by evaluating “functional” rather than purely “morphological” parameters by providing information relating to perfusion, vascularization, diffusion and metabolic properties of the tissues, which are more appropriate especially in the case of monitoring of “Target therapies.”3-9 Choi et al in 2007,10 proposed to include the reduction of tumor density in the evaluation of the response to molecular target agents as an indirect indicator of reduced angiogenesis, defining as a response criterion the reduction of 10% of the sum of the diameters and/or at least 15% reduction in density value.
Currently, in the absence of standardized and internationally validated quantitative parameters, the limit of diagnostic imaging remains linked to the subjective interpretation of the changes undergone by the treated tissue. Jaffe et al.1 in their manuscript “Quantitative Imaging in Oncology Patients” reported that 93% of oncologists think that patient management is influenced by the subjective assessment of the size of the tumor. In this scenario, the use of automatic tools for monitoring the response to cancer therapies is fundamental.
Two major categories of computer-aided segmentation techniques can be considered based on the user interaction: fully automated techniques without user input and semi-automated techniques that require user interaction. Semi-automated techniques outperform automatic approaches obtaining accurate and robust results.2,11 Additionally, image analysis techniques have been used to provide prognostic biomarkers and to assess the treatment response with ever greater accuracy in order to provide personalized therapy. In particular, radiomic analysis methods,12,13 which describe a region of interest using multiple quantitative features derived by images, have shown great potential to predict the survival in lung cancer patients.14-19
In this study we used HealthMyne® Quantitative Imaging Decision SupportTM (QIDS) platform that provides a tool through which it is possible semi-automatically to recognize and segment the target lesions identified by the radiologist, to obtain automatically the treatment response based on several radiological criteria including RECIST and CHOI criteria and to extract automatically numerous quantitative metrics.
The aim of this manuscript was the consistency evaluation of the QIDSTM platform of Healthmyne® and of the radiomic analysis in order to identify the quantitative robust metrics in treatment evaluation of lung carcinoma on chest CT scan.
Materials and Methods
Patient Selection
In this retrospective study, we selected 150 patients (median age 67 years, range 19-88 years) with histologically confirmed lung cancer who underwent chemotherapy and baseline and follow-ups CT examinations. This retrospective study was approved by the National Cancer Institute of Naples Local Ethical Comittee as a multicentric observational retrospective spontaneous study. In addition to the promoter center (National Cancer Institute—IRCCS of Naple—G. Pascale Foundation), 2 other structures (Careggi University Hospital of Florence and University Hospital of Siena) were involved. Each center included 50 patients.
Inclusion criteria: lung cancer confirmed histologically, lung nodule size ≥ 10 mm; patients undergoing both first and second line cancer treatment; baseline CT in a time window of 30 ± 6 days before the start of treatment (CT 0); the second CT performed before the second cycle of therapy (CT 1); the third CT performed at the end of the last cycle of therapy (TC 2); CT with slice thickness ≤ 3 mm; CT must have the venous phase (70-90 seconds post contrast injection). Exclusion criteria: patients undergoing radiotherapy; patients undergoing Immunotherapy.
The gold standard to assess the consistency of the software is the radiological consensus between 3 radiologists who assessed the response according to RECIST and CHOI criteria before independently blinded to each other and after in consensus.
CT Acquisition and Analysis
CT images were acquired with 3 different scanners: 2 GE scanners with 64 detectors (Optima 660, and Discovery 750 HD General Electric Healthcare, Milwaukee, USA) and a Philips CT scanner (ICT SP 128 slice, Philips, Amsterdam, Netherlands).
The scan data was 120-140 kVp, 200–600 mA, slice thickness 1.25-2.5 mm and table speed 0.938-0.984/1 mm/rotation. Contrast-enhanced CT images were acquired in the portal venous phase (start delay 70–80 s) from pelvic brim to thoracic inlet, after the intravenous injection of 2 mL/kg of a non ionic contrast material (iodine concentration ≥ 350 mg/ml), followed by 40 mL of saline solution, using a semi-automated power injector (3,5–4 mL/s flow rate). Images reconstruction was performed by using a reconstruction algorithm.
Clinical and Radiological Measurements
Three radiologists with different experience in reading and interpreting of chest CT (low experience ≤5 years, medium experience from 5 to 15 years and high experience > 15 years) performed the evaluation collecting the measurement of longest diameter and of the Hounsfield Unit (HU) density of the target lesions in 2D on the CT venous phase. The target lesions were selected according to their size, considering those with a larger diameter clearly visible and outlined, based on the intrinsic reliability in the measurement repeatability. In the case of primary carcinoma, only 1 target lesion were considered; in the presence of lung metastases, up to 5 target lesions were considered. The longest diameter was calculated on the axial plane. The density was measured on the region of interest (ROI) obtained by surrounding the entire lesion including both the hypervascular and necrotic parts excluding the atelectasis pulmonary parenchyma. Each radiologist classified the treatment response of the 2 follow-ups according to the RECIST 1.1 criteria.8 Objective therapeutic responses according to RECIST 1.1 are as follows: complete response (CR) is the complete target lesion disappearance; partial response (PR) is a reduction of at least 30% in tumor diameter; progressive disease (PD) is at least a 5 mm increase in tumor diameter, and percent change from nadir is at least 20% and stable disease (SD) is neither PR nor PD target lesions’ diameter has neither decreased at least 30% from baseline nor has increased at least 20% from nadir. Moreover, the response was evaluated according to the Choi criteria10: CR is disappearance of target lesion; PR is a decrease in tumor size ≥ 10% or decrease in tumor density ≥ 15% on CT; SD is neither PR nor PD, target lesions’ diameter has neither decreased at least 10% from baseline longest diameter nor has increased at least 10% from nadir and the decrease in the average of all Target lesions’ Mean HU value has also not met nor exceeded 15%; and PD is an increase in tumor size ≥ 10% from nadir and does not meet PR criteria by tumor density.
Radiologists performed the CT analysis before, blinded to each other, and then in consensus on the dedicate post processing workstations of the CT scanners and then on HealthMyne® QIDSTM platform. To reduce recall bias, all 3 readers maintained a gap of more than 2 weeks between the 2 interpretation sessions (blinded to each other and consensus assessment).
CT Post Processing With QIDSTM Tool
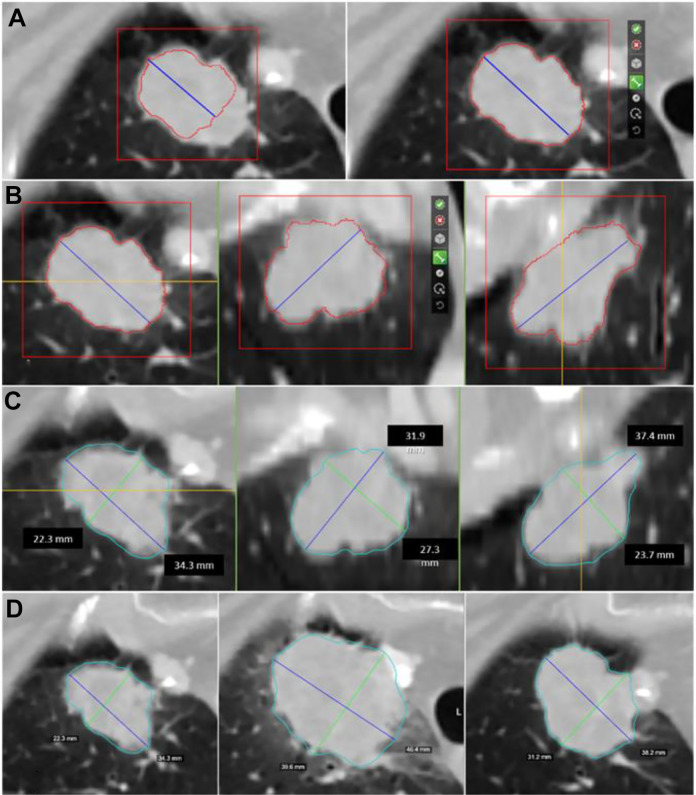
Among HealthMyne® QIDS™ functionalities, there is the capability to extract the target lesion volume implementing an interactive Rapid Precise Metrics (RPM™) algorithm with user interaction and control: the user initializes the lesion segmentation by drawing a long axis on a plane of the multiplanar reconstruction (MPR). Then a 2D segmentation updates in real-time for interactive feedback11 and then the 2D segmentation happens immediately on the other MPR planes. When the contours on a MPR plane is unsatisfactory, the user can upgrade the segmentation by either drawing long axes on this plane or using the ball tool. When the segmentation is satisfactory, the user can initiate 3D segmentation by a single click. 3D segmentation occurs quickly (approximate time = 1–2 s), and the user may examine the segmentation contours by scrolling through slices. If unsatisfied, the user can delete the segmentation or alternatively edit it using a 3D sphere tool, otherwise the user clicks a button to confirm the 3D segmentation (Figure 1).
Figure 1.
Semi-automated identification of the lesion: (A) A first step consists of the manual indication of the ROI to segment. The blue line represents the initial drag of an axis crossing the lesion manually delineated by the radiologist. As the blue line is drawn an intensity-based estimation of the lesion boundary is displayed with a red contour. On the right: the initial long axis delineated by the radiologist and the 2D contour on the axial plane. (B) Additional axes can be dragged on all the orthogonal MPR views. From left to right: the 2D contours on the axial, coronal and sagittal views of the lesion used as a starting point for the HealthMyne RPM™ algorithms. (C) HealthMyne RPM™ algorithms combine intensity gradients with statistical sampling methods for delineation of the volumetric 3D contour of the lesion (light blue contour). The blue line represents the longest long axes and the green line represents the longest short axes automatically determined leveraging the 3D delineation. From left to right: the 3D delineation of the lesion on the axial, coronal and sagittal views. (D) The 3D delineation of the lesion is automatically determined on current studies through the lesion propagation across studies. From left to right: the longest diameters of the lesion in axial plane for the diagnostic study and the 2 follow-ups.
Therefore, using the HealthMyne® QIDSTM platform, the following procedures was performed: advanced semi-automatic segmentation of target lesions identified by the radiologist including 3D outlines; propagation of regions of interest segmented through scans acquired at different times; semi-automatic identification and segmentation of new lesions; classification of response to treatment using the RECIST and CHOI criteria by means of the “Therapy Response Assessment Module “of the software (Long/Short axis are registered instantly).
Using the HealthMyne® QIDSTM platform, we recorded for each target lesion and for each time (baseline, follow-up 1 and follow-up 2) the longest diameter and the mean value of density in Hounsfield unit on the 2D slice with the longest diameter (2D density) and on the entire segmented volume (3D density).
The average elapsed time for each target lesion segmentation was collected along with the percentage of cases for which the radiologists had to make changes.
Moreover, using QIDSTM platform we extracted 594 radiomic metrics (see Appendix 1): 28 delta radiomic features considered to obtain radiological response according to RECIST and CHOI criteria (measure of change over time, percent growth, projected doubling time, and other metrics determined by comparing change in 1st and 2nd order metrics across multiple time points); 66 first Order profile features based on intensity values (statistical distribution of image value); 50 second order profile features based on lesion shape (geometric analysis of shape, volume, curvature, and volumetric lengths); 393 third order profile features based on texture (analysis of voxel sub-environments—voxel neighborhood statistical distributions—to show location-specific characteristics within a lesion or tumor) and 57 features with higher order profiles (statistical metrics after transformations and wavelet analysis).
We extracted each radiomic metrics on the lung nodules and for each feature (except that delta radiomic features) and we calculated the percentage change respect to baseline value.
Statistical Analysis
The correlation (Spearman’s correlation coefficient) between the measurements of the target lesions diameter and density provided by the QIDSTM software and the measurements provided by radiologists was calculated.
The assessment of observer variability for the 3 chest CT readings was performed by calculating the intraclass correlation coefficient.20
The consistency of the QIDSTM tool in defining the treatment radiological response was assessed in terms of agreement rate of the response (according to RECIST and CHOI criteria) respect to the single reader and compared with the radiological consensus of 3 readers.
Chi square test was applied to detect differences statistically significant among percentage values in different groups.
Kruskal Wallis test was applied to identify the radiomic features that had significant differences in median value in the groups based on RECIST response (PR, SD and PD). Moreover, the robust features were selected by the least absolute shrinkage and selection operator (LASSO) method to best predict the classification response based on RECIST response (PR, SD and PD).21 In the LASSO method, 10-fold cross-validation was used to select the optimal regularization parameter alpha, as the average of mean square error of each patient was the smallest. With the optimal alpha, features having nonzero coefficient in LASSO were considered robust predictors of RECIST response.
Median and range values were reported for significant quantitative metrics.
A value of p <0.05 was considered statistically significant.
All analyzes was performed using Matlab’s Statistics Toolbox (The Math-Works Inc., Natick, MA).
Results
Median size of lung nodules were 29.87 mm ± 26.14 mm (10-164 mm). No patient with complete response was observed in the selected cases for both RECIST and CHOI criteria; 42 patients resulted in in PR, 82 in SD and 26 in PD according to RECIST criteria while 62 patient resulted in PR, 40 in SD and 48 in PD according to CHOI criteria.
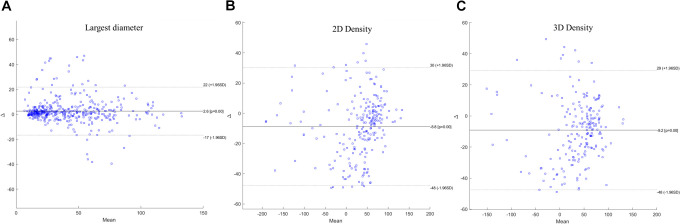
Tables 1 and 2 reports Spearman’s Correlation coefficients between the measurements of the longest diameter and of HU density of the target lesions provided by the HealthMyne (HM) QIDSTM software and the measurements provided individually by the 3 radiologists. Tables 3 and 4 reports Spearman’s Correlation coefficients between the measurements of the longest diameter and of HU density of the target lesions provided by the QIDSTM software and the measurements of radiological consensus. Good and significant correlation was obtained between the measurements of longest diameter and of density among the QIDSTM tool and the radiologists: the correlation coefficient between measurements of the longest diameter provided by radiologists compared with those provided by QIDSTM ranges from 0.82 to 0.83; the correlation coefficient between measurements of the 2D density provided by radiologists compared with those provided by QIDSTM ranges from 0.74 to 0.76; the correlation coefficient between measurements of the 3D density provided by radiologists compared with those provided by QIDSTM ranges from 0.78 to 0.79. Figure 2 reports the Bland-Altman plots for the comparison between the longest diameter provided by radiological consensus and by QIDSTM tool (a), for the comparison between 2D density provided by radiological consensus and by QIDSTM tool (b) and for the comparison between 3D density provided by radiological consensus and by QIDSTM tool (c).
Table 1.
Spearman’s Correlation Coefficients Between the Measurements of the Diameter of the Target Lesions Provided by the QIDSTM Software and the Measurements Provided Individually by the 3 Radiologists.
| Reader1 size | Reader2 size | Reader3 size | HM size | |||
|---|---|---|---|---|---|---|
| Spearman’s Correlation | Reader1 size | Correlation Coefficient | 1.00 | 0.98** | 0.99** | 0.82** |
| P value | – | 0.00 | 0.00 | 0.00 | ||
| Reader2 size | Correlation Coefficient | 0.98** | 1.00 | 0.99** | 0.82** | |
| P value | 0.00 | – | 0.00 | 0.00 | ||
| Reader3 size | Correlation Coefficient | 0.99** | 0.99** | 1.00 | 0.82** | |
| P value | 0.00 | 0.00 | – | 0.00 | ||
| HM size | Correlation Coefficient | 0.82** | 0.82** | 0.82** | 1.00 | |
| P value | 0.00 | 0.00 | 0.00 | – | ||
** The correlation is significant at the 0.01 level (2-tailed).
* The correlation is significant at 0.05 level (2-tailed).
Table 2.
Spearman’s Correlation Coefficients Between the HU Density of the Target Lesions Provided by the QIDSTM Software and the Measurements Provided Individually by the 3 Radiologists.
| Reader1 2D density | Reader2 2D density | Reader3 2D density | HM 2D density | HM 3D density | |||
|---|---|---|---|---|---|---|---|
| Spearman’s Correlation | Reader1 2D density | Correlation Coefficient | 1.00 | 0.96** | 0.98** | 0.75** | 0.79** |
| P value | – | 0.00 | 0.00 | 0.00 | 0.00 | ||
| Reader2 2D density | Correlation Coefficient | 0.96** | 1.00 | .96** | 0.74** | .78** | |
| P value | 0.00 | – | 0.00 | 0.00 | 0.00 | ||
| Reader3 2D density | Correlation Coefficient | 0.98** | 0.96** | 1.00 | 0.76** | 0.80** | |
| P value | 0.00 | 0.00 | – | 0.00 | 0.00 | ||
| HM 2D density | Correlation Coefficient | 0.75** | 0.74** | 0.76** | 1.00 | 0.96** | |
| P value | 0.00 | 0.00 | 0.00 | – | 0.00 | ||
| HM 3D density | Correlation Coefficient | 0.79** | 0.78** | 0.79** | 0.96** | 1.00 | |
| P value | 0.00 | 0.00 | 0.00 | 0.00 | – | ||
** The correlation is significant at the 0.01 level (2-tailed).
* The correlation is significant at 0.05 level (2-tailed).
Table 3.
Spearman’s Correlation Coefficients Between the Measurements of the Diameter of the Target Lesions Provided by the QIDSTM Software and the Measurements of Radiological Consensus.
| Radiological consensus size | HM size | |||
|---|---|---|---|---|
| Spearman’s Correlation | Radiological consensus size | Spearman Correlation Coefficient | 1.00 | 0.83** |
| P value | – | 0.00 | ||
| HM size | Spearman Correlation Coefficient | 0.83** | 1.00 | |
| P value | 0.00 | – | ||
** The correlation is significant at the 0.01 level (2-tailed).
* The correlation is significant at 0.05 level (2-tailed).
Table 4.
Spearman’s Correlation Coefficients Between the Measurements of HU Density of the Target Lesions Provided by the QIDSTM Software and the Measurements of Radiological Consensus.
| Radiological consensus density | HM 2D density | HM 3D density | |||
|---|---|---|---|---|---|
| Spearman’s Correlation | Radiological consensus density | Spearman Correlation Coefficient | 1.00 | 0.76** | 0.79** |
| P value | – | 0.00 | 0.00 | ||
| HM 2D density | Spearman Correlation Coefficient | 0.76** | 1.00 | 0.91** | |
| P value | 0.00 | – | 0.000 | ||
| HM 3D density | Spearman Correlation Coefficient | 0.79** | 0.91** | 1.00 | |
| P value | 0.00 | 0.00 | – | ||
Figure 2.
Bland-Altman plots. In (A) comparison between the longest diameter provided by radiological consensus and by QIDSTM tool; in (B) comparison between 2D density provided by radiological consensus and by QIDSTM tool; in (C) comparison between 3D density provided by radiological consensus and by QIDSTM tool.
We found that the ICC was over 0.85 among measurements provided by 3 radiologists both for longest diameter and for density value (Table 5); however a variability among the radiologists measurements determined a different treatment response classification based on RECIST and CHOI criteria (see Table 6).
Table 5.
Elapsed Time for Each Target Lesion Segmentation, User Interactions and Rate of Modified Segmentation.
|
Central nodules with regular shape N. 6 |
Central nodules with irregular shape N. 29 |
Peripheral nodules with regular shape N. 63 |
Peripheral nodules with irregular shape N. 52 |
P value * |
Noduleswith size ≤ 30 mm N. 72 |
Noduleswith size > 30 mm N. 78 |
P value * | |
|---|---|---|---|---|---|---|---|---|
| Radiologists Intraclass correlation coefficient for longest diameter | 0.99 | 0.92 | 0.93 | 0.89 | >0.05 | 0.87 | 0.98 | >0.05 |
| Radiologists Intraclass correlation coefficient for 2D density | 0.98 | 0.93 | 0.92 | 0.87 | >0.05 | 0.98 | 0.92 | >0.05 |
| HM QIDSTM Intraclass correlation coefficient for longest diameter | 0.99 | 0.98 |
0.97 |
0.95 | >0.05 | 0.94 | 0.97 | >0.05 |
| HM QIDSTM Intraclass correlation coefficient for 2D density | 0.95 | 0.93 | 0.90 | 0.86 | >0.05 | 0.90 | 0.96 | >0.05 |
| HM QIDSTM Intraclass correlation coefficient for 3D density | 0.99 | 0.99 | 0.99 | 0.98 | >0.05 | 0.97 | 0.99 | >0.05 |
| Elapsed time for each target lesion segmentation (min) | 2,4 | 2,5 | 5,3 | 7,9 | << 0.01 | 2,5 | 6,7 |
<< 0.01 |
|
User interactions and rate of modified segmentation
(N. / %) |
2 / 0.00% (0/6) | 3 / 6.90% (2/29) | 2 / 9.52% (6/63) | 4 / 38.50% (20/52) | << 0.01 | 11.10% (8/72) | 25.60% (20/78) | << 0.01 |
* P value at Chi square test.
Table 6.
Evaluation of Agreement Among Radiologists and Between Radiologists and HM QIDSTM Tool Based on RECIST and CHOI Criteria.a
| Rate (%) | RECIST response | CHOI response | P value* |
|---|---|---|---|
| Reader 1 versus Reader 2 | 94.67 | 83.00 | > 0.05 for both RECIST and CHOI |
| Reader 1 versus Reader 3 | 92.33 | 85.67 | |
| Reader 2 versus Reader 3 | 89.67 | 84.67 | |
| Reader 1 versus radiological consensus (gold standard) | 98.33 | 92.33 | > 0.05 for both RECIST and CHOI |
| Reader 2 versus radiological consensus (gold standard) | 95.67 | 89.67 | |
| Reader 3 versus radiological consensus (gold standard) | 93.33 | 92.67 | |
| HM QIDSTM versus Reader 1 | 82.33 | 57.67 | > 0.05 for both RECIST and CHOI |
| HM QIDSTM versus Reader 2 | 81.00 | 59.00 | |
| HM QIDSTM versus Reader 3 | 80.00 | 60.00 | |
| HM QIDSTM versus radiological consensus (gold standard) | 84.33 | 62.67 |
a The table reports the rate of patients with the same treatment response categorized using RECIST and CHOI criteria.
* P value at Chi square test.
HealthMynes’ quantitative volumetric delineation was consistent and matched with each individual radiologist measurement considering the RECIST classification (80-84% of agreement). Instead, a lower concordance among QIDSTM results and the radiologists was obtained considering CHOI classification (58-63%) (Table 6). For this aspect, the radiomic analysis and extracted metrics were non compared with density measurement change.
No significant difference was observed considering the different experience grade of radiologist (p value > 0.05 at Chi square test, Table 6). Moreover, no difference in the agreement rate was observed considering the radiological consensus and the QIDSTM tool measurements (p value > 0.05 at Chi square test). These results strengthen the consistency proof between the software measurements and the measurements made by the radiologists.
The average elapsed time for each target lesion segmentation was estimated to be 4.5 min; this was a weighted average computed over 6 categories of lesions: central or peripheral with regular or irregular shape and nodules with size ≤ 3 cm and > 3 cm (Table 5). The percentage of the patients with modifications of segmentation implemented by radiologists using the QIDSTM platform was of 28/150 (18.7%) of cases mainly in peripheral nodules with irregular shape (Table 5).
For all cases the software allowed to segment the target lesions using the long and short axis drawing by the radiologists and allowed to propagate automatically the lesion segmentation in the follow-up CT scan.
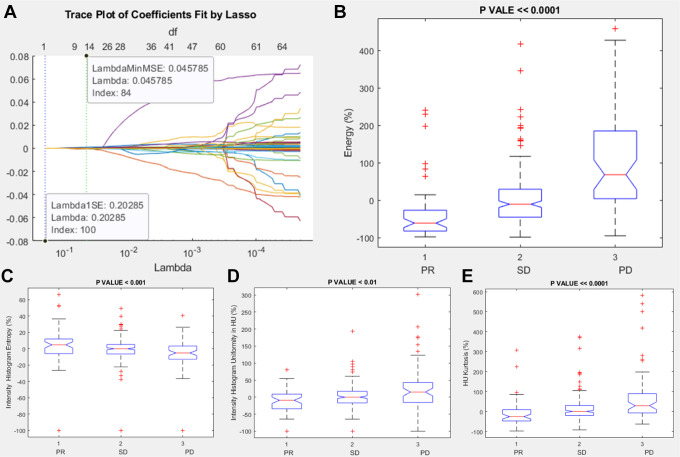
Among intensity features, the significant and robust metrics correlated to RECIST response (PR, SD or PD) were energy percentage change, intensity histogram entropy percentage change, intensity histogram uniformity percentage change, and HU Kurtosis percentage change (Table 7 and Figure 3).
Table 7.
Robust Metrics Correlated to RECIST Classification.
| Robust metrics correlated to RECIST classification | Description | |
|---|---|---|
| 1st order profile metrics based on intensity values (intensity features) | energy percentage change | A measure of the magnitude of raw voxel values in an image. A greater amount of larger values implies a greater sum of the squares of these values |
| intensity histogram entropy percentage change | Entropy of discretized voxels | |
| intensity histogram uniformity percentage change | Uniformity of discretized voxels | |
| HU Kurtosis percentage change | A measure of the “peakedness” of the distribution of HU values in the ROI. A higher kurtosis implies that the mass of the distribution is concentrated toward the tail(s) rather than toward the mean. A lower kurtosis implies the reverse, that the mass of the distribution is concentrated toward a spike the mean | |
| 2nd order profile metrics based on lesion shape (morphological features) | Coronal long axis percentage change | A measure of the longest straight line that can fit entirely inside an XZ-planar slice of the 3D structure (from edge to edge, without ever leaving structure) |
| Longest planar diameter percentage change | A measure of the longest straight line that can fit entirely inside an XY-planar slice of the 3D structure (from edge to edge, without ever leaving structure) | |
| Surface percentage change | Surface area of the specified ROI of the image | |
| 3 rd order profile metrics based on texture (textural features) | NGLDM Dependence Nonuniformity by Slice percentage change | Dependence nonuniformity from merging matrices by each slice and averaging the result |
| NGLDM Low Dependence Emphasis as Volume percentage change | Low dependence emphasis from merging matrices by each slice and averaging the result | |
| Higher order features |
Entropy of Log(2.5 mm) percentage change | Entropy of 2.5D LoG transformed voxels at 2.5 mm smoothing |
| Wavelet energy percentage change | Energy of voxels under wavelet transforms with filters HHL | |
| Wavelet mean deviation percentage change | Absolute deviation from the mean of voxels under wavelet transforms with filters HHL | |
| Wavelet root man squared percentage change | Root mean squared of voxels under wavelet transforms with filters HHL |
Figure 3.
Lasso results and boxplots of robust metrics among intensity features group: in (A) is visualized the trace plot of LASSO fit. Each line represents a trace of the values for a single predictor variable. The parameters under the zero line are the redundant predictors. The dashed vertical lines represent the Lambda value with minimal mean squared error MSE (on the right), and the Lambda value with minimal mean squared error plus 1 standard deviation. The upper part of the plot shows the degrees of freedom (df), meaning the number of nonzero coefficients in the regression, as a function of Lambda. This latter value is a recommended setting for Lambda. In (B), (C), (D) and (E) were represented the boxplots of the robust metrics: energy percentage change, intensity histogram entropy percentage change, intensity histogram uniformity percentage change and HU Kurtosis percentage change.
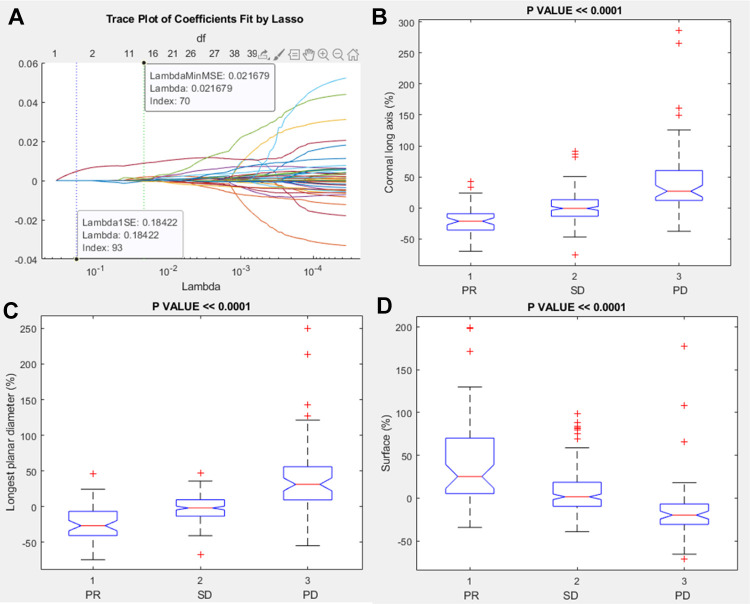
Among morphological features, the significant and robust metrics correlated to RECIST response (PR, SD or PD) were coronal long axis percentage change, longest planar diameter percentage change and surface percentage change (Table 7 and Figure 4).
Figure 4.
Lasso results and boxplots of robust metrics among morphological features group: in (A) is visualized the trace plot of LASSO fit. Each line represents a trace of the values for a single predictor variable. The parameters under the zero line are the redundant predictors. The dashed vertical lines represent the Lambda value with minimal mean squared error MSE (on the right), and the Lambda value with minimal mean squared error plus 1 standard deviation. The upper part of the plot shows the degrees of freedom (df), meaning the number of nonzero coefficients in the regression, as a function of Lambda. This latter value is a recommended setting for Lambda. In (B), (C) and (D) were represented the boxplots of the robust metrics: coronal long axis percentage change, longest planar diameter percentage change, surface percentage change.
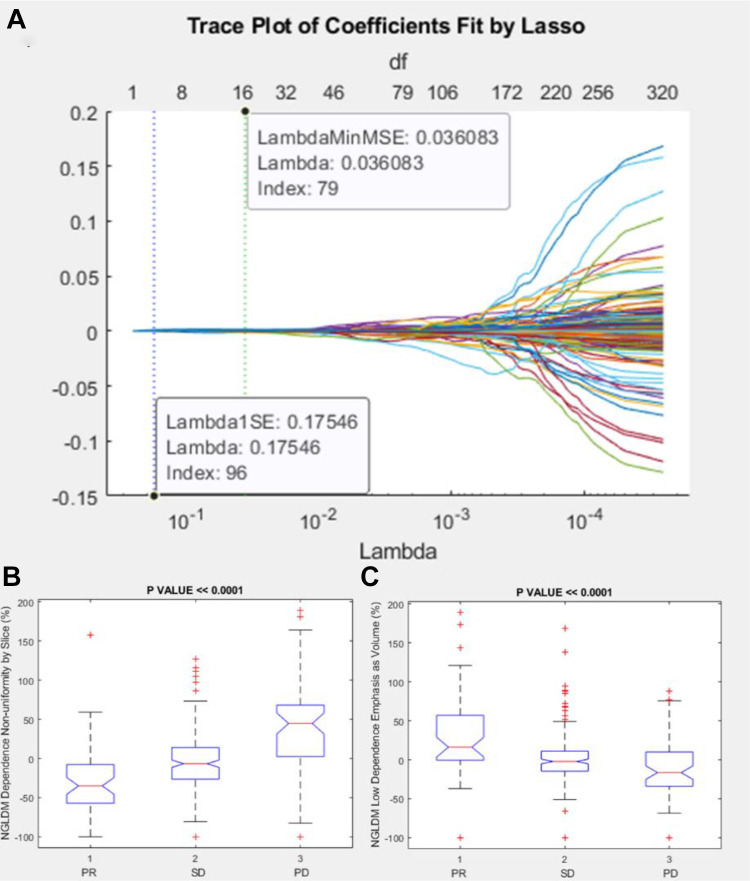
Among textural features, the significant and robust metrics correlated to RECIST response (PR, SD or PD) were Neighborhood Grey-Level Different Matrix (NGLDM) Dependence Nonuniformity by Slice percentage change and NGLDM Low Dependence Emphasis as Volume percentage change (Table 7 and Figure 5).
Figure 5.
Lasso results and boxplots of robust metrics among textural features group: in (A) is visualized the trace plot of LASSO fit. Each line represents a trace of the values for a single predictor variable. The parameters under the zero line are the redundant predictors. The dashed vertical lines represent the Lambda value with minimal mean squared error MSE (on the right), and the Lambda value with minimal mean squared error plus 1 standard deviation. The upper part of the plot shows the degrees of freedom (df), meaning the number of nonzero coefficients in the regression, as a function of Lambda. This latter value is a recommended setting for Lambda. In (B) and (C) were represented the boxplots of the robust metrics: NGLDM Dependence Nonuniformity by Slice percentage change and NGLDM Low Dependence Emphasis as Volume percentage change.
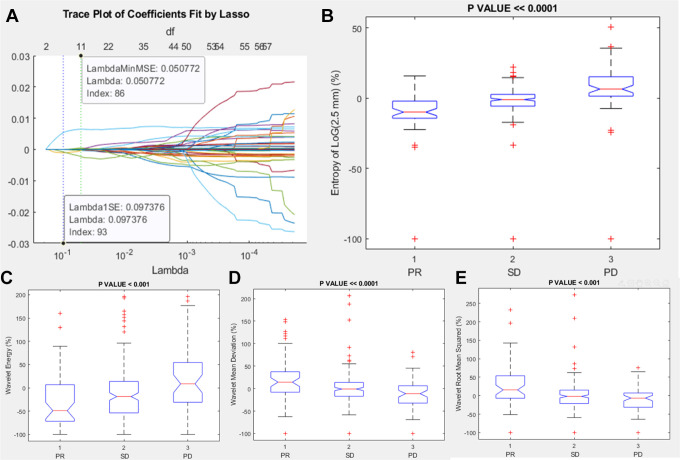
Among Higher order features, the significant and robust metrics correlated to RECIST response (PR, SD or PD) were entropy of Log(2.5 mm) percentage change, wavelet energy percentage change, wavelet mean deviation percentage change and wavelet root man squared percentage change (Table 7 and Figure 6).
Figure 6.
Lasso results and boxplots of robust metrics among higher order features group: in (A) is visualized the trace plot of LASSO fit. Each line represents a trace of the values for a single predictor variable. The parameters under the zero line are the redundant predictors. The dashed vertical lines represent the Lambda value with minimal mean squared error MSE (on the right), and the Lambda value with minimal mean squared error plus 1 standard deviation. The upper part of the plot shows the degrees of freedom (df), meaning the number of nonzero coefficients in the regression, as a function of Lambda. This latter value is a recommended setting for Lambda. In (B), (C), (D) and (E) were represented the boxplots of the robust metrics: entropy of Log(2.5 mm) percentage change, wavelet energy percentage change, wavelet mean deviation percentage change and wavelet root man squared percentage change.
Table 8 reports the median values and range for the significant and robust metrics correlated to RECIST response (PR, SD or PD). In PR group, median values of energy percentage, intensity histogram entropy percentage change, intensity histogram uniformity percentage change, HU Kurtosis percentage change, coronal long axis percentage change, longest planar diameter percentage change, surface percentage change, NGLDM Dependence Nonuniformity by Slice percentage change, NGLDM Low Dependence Emphasis as Volume percentage change, entropy of Log(2.5 mm) percentage change, wavelet energy percentage change, wavelet mean deviation percentage change and wavelet root man squared percentage change were respectively of −60,77%; 4,71%; −9,30%; −25,32%; −21,39%; −26,71%; 25,22%; −35,00%; 16,50%; −9,89%; −37,61%; 14,26%; 15,67%.
Table 8.
Median Value and Range for Significant and Robust Radiomic Features.
| Energy | Intensity histogram entropy | Intensity histogram uniformity | HU Kurtosis | Coronal long axis | Longest planar diameter | Surface | NGLDM dependence nonuniformity by slice | NGLDM low dependence emphasis as volume | Entropy of Log(2.5 mm) | Wavelet energy | Wavelet mean deviation | Wavelet root man squared | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (%) | (%) | (%) | (%) | (%) | (%) | (%) | (%) | (%) | (%) | (%) | (%) | (%) | ||
| PR | Median Value | −60.77 | 4.71 | −9.30 | −25.32 | −21.39 | −26.71 | 25.22 | −35.00 | 16.50 | −9.89 | −37.61 | 14.26 | 15.67 |
| Range | 338.03 | 166.13 | 180.10 | 404.06 | 112.28 | 120.20 | 233.24 | 258.00 | 349.00 | 115.81 | 1466.67 | 253.60 | 332.56 | |
| Minimum | −97.57 | −100.00 | −100.00 | −97.45 | −69.73 | −74.47 | −34.28 | −100.00 | −100.00 | −100.00 | −100.00 | −100.00 | −100.00 | |
| Maximum | 240.46 | 66.13 | 80.10 | 306.61 | 42.55 | 45.73 | 198.96 | 158.00 | 249.00 | 15.81 | 1366.67 | 153.60 | 232.56 | |
| SD | Median Value | −10.20 | −0.08 | −0.05 | 0.53 | −0.67 | −1.98 | 1.47 | −6.26 | −2.05 | −1.00 | −14.67 | −1.04 | −1.72 |
| Range | 1481.10 | 149.24 | 293.19 | 467.06 | 166.53 | 115.09 | 137.53 | 306.00 | 321.00 | 122.02 | 1181.48 | 306.30 | 372.91 | |
| Minimum | −98.25 | −100.00 | −100.00 | −91.94 | −75.48 | −67.57 | −39.20 | −100.00 | −100.00 | −100.00 | −100.00 | −100.00 | −100.00 | |
| Maximum | 1382.85 | 49.24 | 193.19 | 375.11 | 91.05 | 47.52 | 98.33 | 206.00 | 221.00 | 22.02 | 1081.48 | 206.30 | 272.91 | |
| PD | Median Value | 97.93 | −5.20 | 14.91 | 29.13 | 27.02 | 31.20 | −19.87 | 49.65 | −16.45 | 6.49 | 34.02 | −11.34 | −6.48 |
| Range | 5588.13 | 140.53 | 401.84 | 644.04 | 323.71 | 304.50 | 248.57 | 792.00 | 187.60 | 150.55 | 2921.96 | 180.61 | 175.62 | |
| Minimum | −94.58 | −100.00 | −100.00 | −62.72 | −37.59 | −54.79 | −71.10 | −100.00 | −100.00 | −100.00 | −100.00 | −100.00 | −100.00 | |
| Maximum | 5493.55 | 40.53 | 301.84 | 581.32 | 286.12 | 249.71 | 177.47 | 692.00 | 87.60 | 50.55 | 2821.96 | 80.61 | 75.62 | |
| Total | Median Value | −8.53 | −0.18 | 0.00 | 2.65 | 0.10 | −0.30 | 0.37 | −4.88 | −1.26 | −0.75 | −7.31 | −0.88 | −1.22 |
| Range | 5591.80 | 166.13 | 401.84 | 678.77 | 361.60 | 324.18 | 270.06 | 792.00 | 349.00 | 150.55 | 2921.96 | 306.30 | 372.91 | |
| Minimum | −98.25 | −100.00 | −100.00 | −97.45 | −75.48 | −74.47 | −71.10 | −100.00 | −100.00 | −100.00 | −100.00 | −100.00 | −100.00 | |
| Maximum | 5493.55 | 66.13 | 301.84 | 581.32 | 286.12 | 249.71 | 198.96 | 692.00 | 249.00 | 50.55 | 2821.96 | 206.30 | 272.91 | |
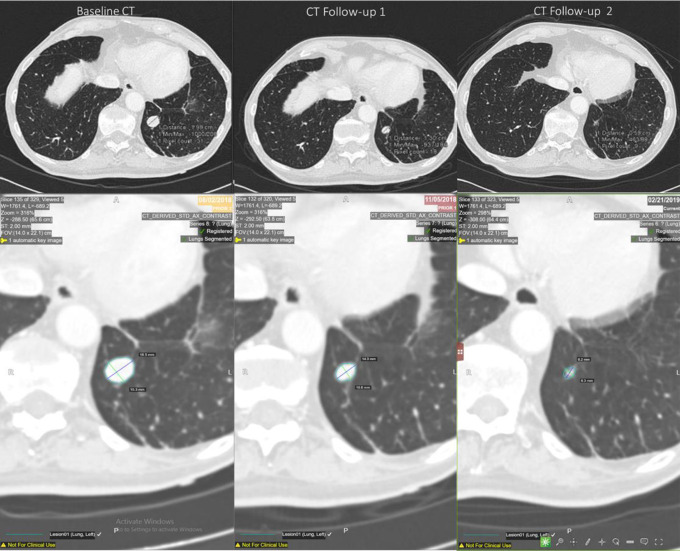
Figure 7 reports an example of segmentation using the QIDSTM tool for each time of a partial responder patient.
Figure 7.
Semi-automated identification of the target lesion in baseline and follow-ups CT scans for a partial responder.
Discussions and Conclusions
The HealthMyne® Quantitative Imaging Decision SupportTM platform provides a tool through which it is possible to semi-automatically recognize and segment the target lesions identified by the radiologist, to obtain automatically the treatment response based on several radiological criteria including RECIST and CHOI criteria and to extract automatically numerous quantitative metrics useful in the evaluating and in the prediction of the treatment response.
The use of automatic tools for monitoring the response to cancer therapies is fundamental in order to obtain quantitative measurements automatically and to reduce the intra and inter observer variability. We demonstrated that a good and significant correlation was obtained between the measurements of longest diameter and of density among the QIDSTM tool and the radiologists. However, some few cases showed a variability of 40% between the QIDS tool and radiologist measurements as reported by Bland-Altman plots; this variability could be linked to the intrinsic variability of the measure of the longest diameter that influences mainly on the lesions with less size. The measurement of the longest diameter has the highest correlation and the 3D density had a greater correlation than the 2D density probably because the average density value calculated on a slice by the radiologist could be different from that automatically identified by the QIDSTM tool. However, HealthMyne quantitative volumetric delineation was consistent and matched with each individual radiologist measurement considering the RECIST classification (80-84% of concordance). A lower concordance among HM results and the radiologists was obtained considering CHOI classification (58-63%). No significant difference was observed considering the different experience grade of radiologist and no difference in the agreement rate was observed considering the radiological consensus and QIDSTM tool measurements. This as a proof of learning ease in the platform use and of consistency of the automatic tool. Moreover, for all cases the software allowed to segment the target lesion using the long and short axis drawing by the radiologists and allowed to propagate automatically the segmentation in the follow-up CT scan.
With improved consistency of lesion measurement, HealthMyne eliminates the need for user to go back and to re-measure previous lesions saving additional time and effort. The average elapsed time for each target lesion segmentation was estimated to be 4.5 min and the percentage of the patients with modifications of segmentation implemented by radiologists using the HM platform was of 18.7%. The correct segmentation was clearly linked to the localization and shape of the target lesions, in fact we reported that the prevalence of segmentation changes was in peripheral nodules with irregular shape.
Gering et al11 in their study designed an experiment to simulate the level of user interaction in semi-automatic segmentation using the HealthMyne platform mimicking the radiologist process to perform segmentation with real-time interaction.22 Clicks and drags were positioned only where needed in response to the deviation between real-time segmentation results and assumed radiologist’s goal. Results of accuracy for various levels of interaction are presented using the Dice similarity coefficient (DSC) to quantity the similarity between 2 sets of segmentations: DSC values range from 0.857-0.943.22
Moreover, another aim of the study was the identification of the robust metrics, by radiomics analysis, correlated to RECIST criteria, able to follow size change and that could be used to monitor or evaluate oncological treatments with quantitative and objective approaches, considering that RECIST criteria is influenced by radiologist measure that suffer from intra and inter observer variability.
Several studies23-28 have examined the correlations between features extracted from X-ray images and lung cancer. The first complete application of radiomic in lung cancer was reported by Aerts et al in 2014.16 In this study 1019 cancer patients were enrolled, 788 with non-small cell lung tumors and the other 231 with head-neck district tumors. 404 parameters that quantify the signal strength of the tumor, the shape, the structure and the wavelet were extracted from a single CT scan. Together with clinical information and gene expression data, a radiomic map was developed to show patient clusters with similar radiomic expression patterns. The results of this study showed that radiomic is able to identify the tumor prognostic phenotype in lung and head-neck district tumors by a single CT scan. In another study,24 583 radiomic characteristics of 127 pre-treatment lung nodules were extracted to measure the shape, intensity and heterogeneity of the nodule. The results showed satisfactory accuracy (80%) in the distinction between primary and malignant primary lung nodules with a sensitivity and specificity of 85.5% and 82.7% respectively.
Another challenge in lung cancer is predicting the response to therapeutic treatment or survival and the onset of local recurrence or distant metastasis. A radiomic model capable of effectively identify patients whose tumors do not respond to treatment would be desirable and could be used to direct patients to personalized treatments.25-27 Coraller et al.25 built a radiomic model with 635 parameters; 35 predictors of distant metastasis and 12 predictors of overall survival and in a further analysis of the response to neoadjuvant chemo-radiotherapy showed that 7 radiomic characteristics were predictive of macroscopic residual pathological disease and one characteristic was predictive of the complete pathological response; it has been shown that tumors with a more rounded shape and heterogeneous texture are more likely to have a poor response to neoadjuvant chemo-radiotherapy. Huang et al.27 reports that the “radiomic signature” is a biomarker of independent estimate of disease-free survival and that the combination of radiomic metrics with traditional staging and high clinical-pathological risk factors allows a better estimate.
These results suggest that radiomic has the potential to be used as a decision-making tool in evaluating treatment in patients with lung cancer. However, a complete analysis of the variance has shown that predictive accuracy depends on the lesion segmentation and of selection of the characteristics and of the analysis techniques, therefore suggesting that standardized methods are needed for further investigations.27 A recent paper by Langlotz et al29 noted that inter- and intra-observer variability can occur at rates as high as 37% and that diagnostic errors could play a role in up to 10% of patient deaths.
We demonstrated that different features were correlated with reduction or increase of the target lesion and then to RECIST response: among 594 extracted metrics the robust predictors of size change were energy that measures the magnitude of raw voxel values in an image; the histogram entropy and uniformity; the HU Kurtosis that measures the “peakedness” of the distribution of HU values in the ROI; coronal long axis that measures the longest straight line that can fit entirely inside an XZ-planar slice of the 3D structure; the longest planar diameter that measures the longest straight line that can fit entirely inside an XY-planar slice of the 3D structure; the surface of volume of interest; the NGLDM Dependence Nonuniformity by Slice linked to the dependence nonuniformity from merging matrices by each slice and averaging the result; the NGLDM Low Dependence Emphasis as Volume that measures the low dependence emphasis from merging matrices by each slice and averaging the result; the entropy of Log(2.5 mm); the wavelet energy; the wavelet mean deviation and the wavelet root man squared that were statistical metrics obtained after log and wavelet transformations. Among the morphological characteristics, the long coronal axis and the percentage variations of the longest planar diameter were clearly correlated to the RECIST response and are therefore less interesting than the other correlated metrics.
These 13 radiomic metrics have been identified as robust parameters in order to quantitatively and objectively track the reduction or increase in tumor size over time and therefore could be used to assess or predict the response to cancer treatment in the lung. Moreover, these features could be linked to other physiological-metabolic processes (tissue vascularization changes, cellular density changes, etc…), different to lesion size changes, and could be able to follow the temporal changes of these processes determining a possible major performances in the monitoring and in the evaluation of the treatments response. The future endpoint of the study is to verify whether these radiomic characteristics could follow the response to treatment even independently of a reduction in tumor size.
A limit of the study can be the absence of an analysis about the robustness of the QIDSTM measurements and of extracted metrics by varying the parameters of the CT acquisition. However, the radiomics analysis was made considering the percentage change of parameters that can be considered less dependent upon the acquisition settings.
In conclusions, we demonstrated that HealthMyne quantitative volumetric delineation was consistent and matched each individual radiologist and that several radiomic metrics extracted by QIDSTM were significant and robust predictors of RECIST response.
Footnotes
Authors’ Note: Our study was approved by Ethics Committee of the National cancer Institute of Naples Pascale Foundation (approval no. 215 of 04/03/2019). All patients provided written informed consent prior to enrollment in the study.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Vincenza Granata, MD  https://orcid.org/0000-0002-6601-3221
https://orcid.org/0000-0002-6601-3221
References
- 1.Jaffe TA, Wickersham NW, Sullivan DC. Quantitative imaging in oncology patients: part 2, oncologists’ opinions and expectations at major U.S. cancer centers. AJR Am J Roentgenol. 2010;195(1):W19–23. [DOI] [PubMed] [Google Scholar]
- 2.Gering D, Kotrotsou A, Young-Moxon B, et al. Measuring efficiency of semi-automated brain tumor segmentation by simulating user interaction. Front Comput Neurosci. 2020;14:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faivre S, Sablin MP, Dreyer C, Raymond E. Novel anticancer agents in clinical trials for well-differentiated neuroendocrine tumors. Endocrinol Metab Clin N Am. 2010; 39(4):811–826. [DOI] [PubMed] [Google Scholar]
- 4.Joensuu H, Roberts PJ, Sarlomo-Rikala M, et al. Effect of the tyrosine kinase inhibitor STI-571 in a patient with a metastatic gastrointestinal stromal tumor. N Engl J Med. 2011;344(14):1052–1056. [DOI] [PubMed] [Google Scholar]
- 5.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors. J Natl Cancer Inst. 2011;92(3):205–216. [DOI] [PubMed] [Google Scholar]
- 6.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45(2):228–247. [DOI] [PubMed] [Google Scholar]
- 7.Kang H, Lee HY, Lee KS, Kim J-H.Imaging-based tumor treatment response evaluation: review of conventional, new, and emerging concepts. Korean J Radiol. 2012;13(4):371–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lencioni R, Llovet JM. Modified RECIST (mRECIST) assessment for hepatocellular carcinoma. Semin Liver Dis. 2010;30(1):52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byrne MJ, Nowak AK. Modified RECIST criteria for assessment of response in malignant pleural mesothelioma. Ann Oncol. 2004;15(2):257–260. [DOI] [PubMed] [Google Scholar]
- 10.Choi H. Response evaluation of gastrointestinal stromal tumors. Oncologist. 2008;13(2):4–7. [DOI] [PubMed] [Google Scholar]
- 11.Gering D, Sun K, Avery A, et al. Semi-Automatic Brain Tumor Segmentation by Drawing Long Axes on Multi-Plane Reformat; in International MICCAI Brainlesion Workshop. Springer. 2018:441–455. [Google Scholar]
- 12.Fusco R, Granata V, Maio F, Sansone M, Petrillo A. Textural radiomic features and time-intensity curve data analysis by dynamic contrast-enhanced MRI for early prediction of breast cancer therapy response: preliminary data. Eur Radiol Exp. 2020;4(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fusco R, Vallone P, Filice S, et al. Radiomic features analysis by digital breast tomosynthesis and contrast-enhanced dual-energy mammography to detect malignant breast lesions. Biomed Signal Process Control. 2019;53:101568. [Google Scholar]
- 14.Parmar C, Grossmann P, Bussink J, Lambin P, Aerts HJWL. Machine learning methods for quantitative radiomic biomarkers. Sci Rep. 2015;5:13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coroller TP, Grossmann P, Hou Y, et al. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol. 2015;114(3):345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5(1):4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fave X, Zhang L, Yang J, et al. Delta-radiomics features for the prediction of patient outcomes in non-small cell lung cancer. Sci Rep. 2017;7(1):588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coroller TP, Agrawal V, Huynh E, et al. Radiomic-based pathological response prediction from primary tumors and lymph nodes in NSCLC. J Thorac Oncol. 2017;12(3):467–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bae JM, Jeong JY, Lee HY, et al. Pathologic stratification of operable lung adenocarcinoma using radiomics features extracted from dual energy CT images. Oncotarget. 2017;8(1):523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGraw KO, Wong SP. Forming inferences about some intraclass correlation coefficients. Psychol Methods. 1996;1(1):30. [Google Scholar]
- 21.Tibshirani R. The lasso method for variable selection in the cox model. Stat Med. 1997;16(4):385–395. [DOI] [PubMed] [Google Scholar]
- 22.Bakas S, Reyes M, Jakab A, et al. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. 2018. arXiv preprint arXiv:1811.02629.
- 23.Reginelli A, Capasso R, Petrillo M, et al. Looking for lepidic component inside invasive adenocarcinomas appearing as CT solid solitary pulmonary nodules (SPNs): CT morpho-densitometric features and 18-FDG PET findings. Biomed Res Int. 2019;2019:7683648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma J, Wang Q, Ren Y, Hu H, Zhao J.Automatic lung nodule classification with radiomics approach. SPIE Medical Imaging: International Society for Optics and Photonics. 2016:978906. [Google Scholar]
- 25.Coroller TP, Agrawal V, Narayan V, et al. Radiomic phenotype features predict pathological response in non-small cell lung cancer. Radiother Oncol. 2016;119(3):480–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaddad A, Desrosiers C, Toews M, Abdulkarim B. Predicting survival time of lung cancer patients using radiomic analysis. Oncotarget. 2017;8(61):104393–104407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Y, Liu Z, He L, et al. Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non-small cell lung cancer. Radiology. 2016;281(3):947–957. [DOI] [PubMed] [Google Scholar]
- 28.Mattonen SA, Palma DA, Johnson C, et al. Detection of local cancer recurrence after stereotactic ablative radiation therapy for lung cancer: physician performance versus radiomic assessment. Int J Radiat Oncol Biol Phys. 2016;94(5):1121–1128. [DOI] [PubMed] [Google Scholar]
- 29.Langlotz CP, Allen B, Erickson BJ, et al. A roadmap for foundational research on artificial intelligence in medical imaging: from the 2018 NIH/RSNA/ACR/The Academy Workshop. Radiology. 2019;291(3):781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]