Abstract
Background:
Recent studies have shown that methyltransferase-like 3, a catalytic enzyme that is predominant in the N6-methyladenosine methyltransferase system, is abnormally expressed in various types of carcinoma and is correlated with poorer prognosis. However, the clinical functions of methyltransferase-like 3 in the prognosis of tumors are not fully understood.
Methods:
We identified studies by searching PubMed, Web of Science, and MedRvix for literature (up to June 30, 2020), and collected a total of 9 studies with 1257 patients for this meta-analysis. The cancer types included gastric cancer, breast cancer, non-small cell lung cancer, bladder cancer, colorectal cancer and ovarian. We further used The Cancer Genome Atlas dataset to validate the results.
Results:
High methyltransferase-like 3 expression clearly predicted a worse outcome (high vs. low methyltransferase-like 3 expression group; hazard ratio = 2.09, 95% confidence interval 1.53–2.89, P = 0.0001). Moreover, methyltransferase-like 3 expression was associated with differentiation (moderate + poor vs. well, pooled odds ratio = 1.76, 95% confidence interval 1.32–2.35, P = 0.0001), and gender (male vs. female, pooled odds ratio = 0.73, 95% confidence interval 0.55-0.97, P = 0.029).
Conclusion:
Our results suggest that methyltransferase-like 3 upregulation is significantly associated with poor prognosis and could potentially function as a tumor biomarker in cancer prognosis.
Keywords: METTL3, prognosis, overall survival, cancer, meta-analysis, TCGA
Introduction
Cancer, which is a leading cause of disability and death globally, has become a major problem in fundamental public health worldwide.1,2 Driven by the evolution of exposure to high-risk factors and demographic change, more people are developing cancers, while the expenditure on treatment is likewise spiraling.3 Although science and technology are developing rapidly, oncotherapy is continually faced with great challenges such as poor prognosis and delayed diagnosis, owing to the lack of specific biomarkers. Hence, it is important to explore novel biomarkers and help reveal the association between the biomarker expression levels and cancer progression. In addition, it may aid in the identification of potential therapeutic targets and novel diagnostic methods for cancer.
A complicated interplay among “reader,” “eraser,” and “writer” proteins, plays an essential role in mediating N6-methyladenosine (m6A) modifications.4,5 Erasers and writers regulate the prevalence, distribution, and abundance of m6A, whereas readers modulate the gene modification functions in m6A, such as RNA stability, RNA export, mRNA splicing, and translation.6 m6A mediates a variety of RNAs, such as long non-coding RNAs (lncRNAs), eukaryotic messenger RNAs (mRNAs), and microRNAs (miRNAs). m6A is concentrated in the sites near the termination codon and 3′ untranslated region (UTR), translating in a cap-independent manner near the 5′ UTR.7
Methyltransferase-like 3 (METTL3) is a major catalytic enzyme in the m6A methyltransferase (MTase) system.8 MTase complex “writers” accomplish m6A deposition. In mammalian cells, 2 separate proteins, MT-A and MT-B, are required for m6A MTase activity on mRNA. METTL3, also known as MT-A70, is a 70 KDa S-adenosylmethionine-binding subunit of the multimeric protein MT-A,8 and belongs to the class I MTase family. METTL3 participates in all phases of the RNA M6A life-cycle, such as mRNA splicing,9 nuclear export,10 translation regulation,11 miRNA processing,12 and mRNA decay.13 Other components of “writers” help METTL3 to complete the catalytic process, which is considered the most common m6A pathway, modifying most m6A sites, especially in mRNA.14
m6A modification mediated by MTETTL3 plays a dual and essential role in the progression of human cancers, with a complex underlying mechanism involving multiple pathways and molecules. For instance, METTL3 interacts with DGCR8 and results in a reduction in the phosphatase and tensin homolog (PTEN) levels in an m6A-dependent manner, positively regulating miRNA maturation and processing.15,16 On one hand, Lin et al. revealed that m6A modification influences epithelial-mesenchymal transition (EMT) depending on the reader ELAVL1, and that METTL3 downregulation impairs EMT, both in vitro and in vivo.17 On the other hand, Lin et al. demonstrated that METTL3 knockdown upregulated the active caspase-3 and positive regulator BAX, but downregulated the negative apoptosis regulator, BCL2, indicating that the apoptosis-related pathway was activated by METTL3 knockdown.18 In addition, Li et al. discovered that SOX2, in which expression is correlated to cancer stem cell differentiation, exhibited the most persistently decreased level of m6A in METTL3-downregulation colorectal cancer cells.19 In summary, METTL3 dysregulation in diverse cancers can regulate apoptosis, EMT, and stem cell self-renewal, which were found to be essential for tumor progression.
Only a few articles focused on why the expression of METTL3 is abnormal in tumor. Current evidence indicated that METTL3 expression can be influenced by histone modification and non-coding RNAs. For instance, Wang et al. found that the activation of H3K27 acetylation mediated by P300 can lead to the upregulation of METTL3 expression in gastric cancer (GC).20 In another study, Zhang et al. proved that hypomethylation at the METTL3 promoter increased the expression of METTL3 by using a cigarette smoke condensate-induced malignant transformation model of pancreatic duct epithelial cells.21 In addition, bioinformatics analysis indicated that METTL3 mRNA might be activated by the transcription factor GFI-1,22 and further functional verification was necessary. It has also been pointed that miR-24-2 can upregulate METTL3 expression in liver cancer23 and that METTL3 expression was reduced by miR-4429 in gastric cancer.24 Additionally, other components of MTase complex (MTC) should be emphasized, especially METTL14, when exploring the m6A-related function of METTL3 in tumor. The expression of METTL3 was positively associated with the expression of METTL14, and both revealed high expression in normal breast-like and luminal A/B breast cancer.25 In brief, these studies indicated that METTL3 expression can be influenced by promoter methylation, histone modification, other MTC components and non-coding RNAs.23
The clinical function of METTL3 is controversial.26 Most tumor cell lines and tissues show increased METTL3 expression, which plays an oncogenic role in various stages of tumor progression.15 However, a few studies have indicated converse conclusions regarding METTL3 expression and its role in bladder-urothelial carcinoma (BLCA), glioblastoma multiforme (GBM), colorectal cancer (CRC), and invasive breast carcinoma (BRCA). Therefore, in this study, meta-analysis and bioinformatic analyses were conducted, to identify the possible correlation, and evaluate the association between METTL3 expression and prognosis or the clinicopathological characteristics of cancer.
Materials and Methods
Search Strategy
A standard guideline was used to perform this study for this meta-analysis of original articles.27,28 The databases PubMed, Web of Science, and MedRvix were searched. The latest time for publishing was June 30, 2020. And the searching key words were “METTL3” OR “Methyltransferase-like 3” and “Tumor” or “Carcinoma” or “Cancer” and “prognosis” or “outcome.” The search was broadened by browsing the related methods, summary, and references of retrieved articles. Notably, articles from any regions in the databases were included, as long as the conditions were satisfied. Two researchers finished the assessment of all the studies included respectively.
Study Inclusion/Exclusion Criteria
The inclusion criteria of this meta-analysis were as follows: (1) The language was restricted in English and the study should be published in full paper. (2) The expression of METTL3 was measured by reverse-transcriptase polymerase chain reaction (qRT-PCR) in different tumor tissue. All the patients in the studies were separated into 2 groups based on their METTL3 expression; (3) The patients were diagnosed with cancer definitively;
The exclusion criteria of this meta-analysis were as follows: (1) Animal and cell experiments, letter, case report or review; (2) Studies without enough data required. Data must contain the following content: (1) Authors, countries and published years (2) sample sizes (3) result of METTL3 testing by qRT-PCR or immunohistochemistry (IHC). (4) HRs and the corresponding 95% confidence interval (CI) produced by multivariate analysis investigating the relationship between METTL3 and overall survival. (5) Clinical features such as age, gender, tumor size, differentiation, metastasis, and TNM stage. Among them, 1-3 are necessary items. Either 4 or 5 can satisfy the conditions.
Data Extraction
The following data of each studies was extracted as follows: (1) Authors, countries and published years (2) Ages, sample sizes, gender, METTL3 testing. (3) The followed-up time or the description of followed-up. (4) Overall survival (5) HRs and the corresponding 95% confidence interval (CI) produced by multivariate analysis investigating the relationship between METTL3 and. (6) Clinical features such as age, gender, tumor size, differentiation, metastasis, and TNM stage.
Quality Assessment
As was shown in Table S1. Tumor Marker Prognostic Studies (REMARK) guidelines were adopted for quality assessment of the included studies; The scores varied from 60% to 80%. It would be regarded as high-quality studies when score was more than 60% scores. The detailed scores were shown in Table S2.
Statistical Analysis
STATA 15.1 software was used to process the data extracted. HR and 95% CI values were collected to evaluate the relationship between METTL3 and OS. Moreover, ORs with 95% CIs were pooled to evaluate the relationship between expression of METTL3 and clinicopathological features. Subgroup analysis were performed to confirm whether different regions, sample sizes and cancer types resulted in the changes of overall results.
The heterogeneity was conducted by the I2 statistics and chi-squared Q test. A random-effects model was selected when severe heterogeneity was calculated(I2 > 40% or P < 0.05). Otherwise, a fixed-effects model would be chosen. Publication bias together with sensitivity analyses were examined to evaluate the stability by Begg’s test.
Bioinformatics Analysis
The Htseq-counts of METTL3, survival information and clinical information of patients were extracted from TCGA (https://cancergenome.nih.gov/). Differential expression analysis was conducted between normal samples and tumor samples by using edge R package. The Benjamini-Hochberg test was conducted to avoid false-positive result. Differences were considered statistically significant with |Log2fold change (FC)| cutoff >1 and P < 0.01. Considering that the patient situation has certain heterogeneity, such as the stage of cancer and taking different treatment schemes. Selection bias can be controlled when strictly enforcing the inclusion and exclusion criteria of study subjects. Therefore, Inclusion and exclusion criteria were adopted to further sort out the data. On the premise of ensuring the representativeness of the study subjects, the scope of the study was narrowed by limiting exposure or intervention factors. The inclusion and exclusion criteria were presented in the supplement material. All the patients in the studies were divided into 2 groups based on their expression of METTL3. The differences in survival time were assessed by using Kaplan–Meier survival curves and log-rank tests, using P < 0.05 as a threshold. Then clinical information was used to explore the relationships between METTL3 expression and gender, tumor metastasis, tumor stage. Due to the lack of relevant data in glioblastoma, we only carried out this work in bladder cancer, colorectal cancer and breast cancer. Patients were divided into 2 groups according to their clinical information, including gender, tumor metastasis and tumor stage. Unpaired t-test was conducted to assess the differences in METTL3 expression between 2 groups, using P < 0.05 as a threshold.
Results
Characteristics of the Selected Studies
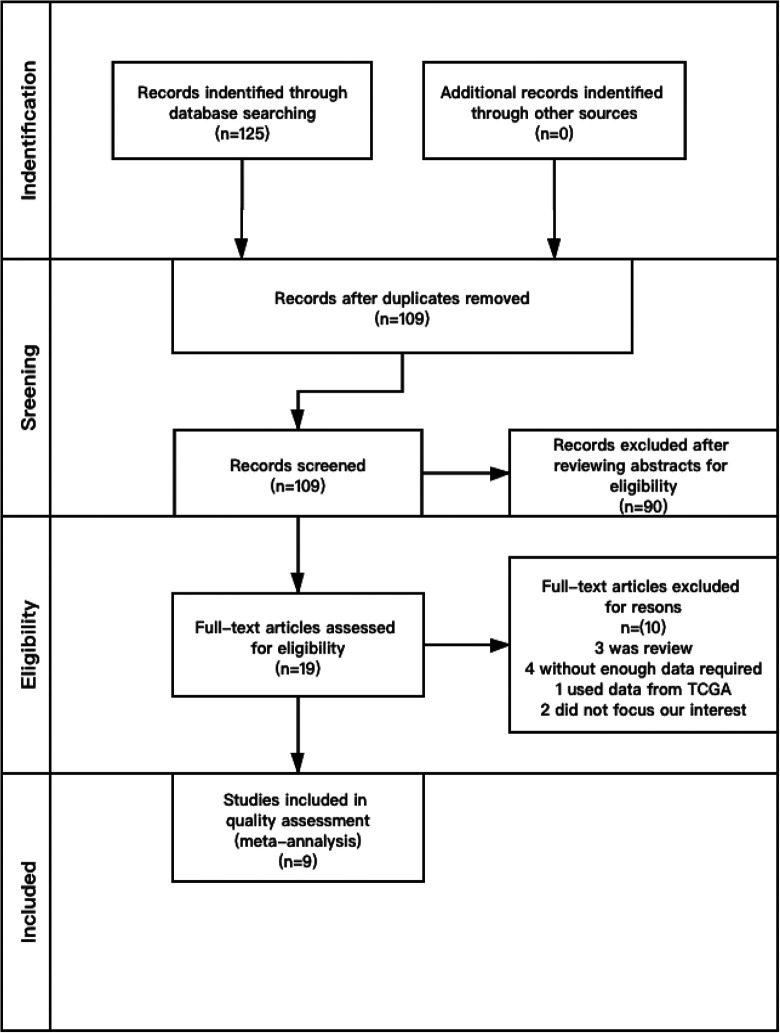
The initial search yielded a total of 125 articles. A total of 109 articles remained, after duplicates were removed. And then, the abstracts were independently and carefully reviewed by 2 researchers according to the criteria mentioned above. Disagreement was resolved by discussion between the 2 researchers until a consensus was reached. Upon assessment of the 19 full-text articles, 10 were excluded for the following reasons: 3 articles were reviews, 4 articles did not have enough required enough data, 1 article used data from The Cancer Genome Atlas (TCGA), and 2 articles did not focus on human subjects, which we were not interested in.
As shown in Table 1, 9 studies published between 2016 and 2020 were finally selected for our meta-analysis after meticulous inspection. The study selection process is illustrated in the flowchart (Figure 1). The 9 studies included for the final analysis involved 1437 patients with different cancers. The study with the largest sample size had 432 samples, and that with smallest sample size had 50 samples. The studies consisted of 6 tumor categories including bladder, colorectal, gastric, ovarian, breast, and non-small-cell lung cancer. The published year was from 2016 to 2020. Among the 9 studies, patients were separated into 2 groups based on METTL3 expression. The first group was characterized by a high level of METTL3, and the second group was compared with the former group.15,19,29-35
Table 1.
Summary of the 11 Included Articles.
| Study | Country | Tumor type |
Sample sizes | METTL3 expression High. Low | Survival information | HR | Laboratory method |
|---|---|---|---|---|---|---|---|
| Yue(2019) | China | GC | 120 | H:40 L:80 | OS | 2.794 | qRT-PCR |
| Wang(2019) | China | GC | 166 | H:83 L:83 | OS | 4.495 | qRT-PCR |
| Li(2019) | China | CRC | 432 | H: 221 L:211 | OS | 3.259 | qRT-PCR |
| Han(2019) | China | BCa | 180 | H:83 L:97 | OS | NA | qRT-PCR |
| Jin2019 | China | NSCLC | 50 | H:36 L:14 | OS | 1.3 | qRT-PCR |
| Wang(2020) | China | Breast | 60 | H:30 L:30 | OS | 1.6 | qRT-PCR |
| Deng(2019) | China | CRC | 181 | H:47 L:134 | OS | 1.908 | qRT-PCR |
| Hua(2018) | China | Ovarian | 162 | H:73 L:89 | OS | 1.755 | qRT-PCR |
| Liu(2019) | China | CRC | 86 | H:43 L:43 | OS | 2.055 | qRT-PCR |
Abbreviations: GC, gastric cancer; CRC, colorectal carcinoma; BCa, bladder cancer; H, high expreesion of METTL3; L, low expression of METTL3; NA, not associated. qRT-PCR, reverse-transcriptase polymerase chain reaction.
Figure 1.
Flow diagram of the study search and selection process in the meta-analysis.
Correlation Between METTL3 Expression and Prognosis
The correlation between METTL3 expression levels and outcomes is presented in Figure 2A. The role of METTL3 in overall survival (OS) in patients with different types of carcinoma was evaluated in the meta-analysis. Statistical analysis of all the articles revealed a correlation between OS and METTL3. Considering that I2 > 40% and heterogeneity was found in these articles, a random-effects model was selected. The pooled hazard ratios (HRs) demonstrated that higher METTL3 expression levels were associated with worse survival (high vs. low METTL3 expression group; pooled HR = 2.09, 95% confidence interval (CI) 1.53–2.89, P = 0.0001, random effect; Figure 2A). Sensitivity analysis was used to evaluate the pooled results (Figure 2B), which proved the reliability. Publication bias was evaluated using Begg’s funnel plot, and there was no obvious publication bias observed for OS (P = 0.063; Figure 2C). In summary, the results of combined data revealed that METTL3 carriers could predict worse overall survival than patients with lower METTL3 expression in various cancers, and this conclusion was reliable, excluding heterogeneity and publication bias.
Figure 2.
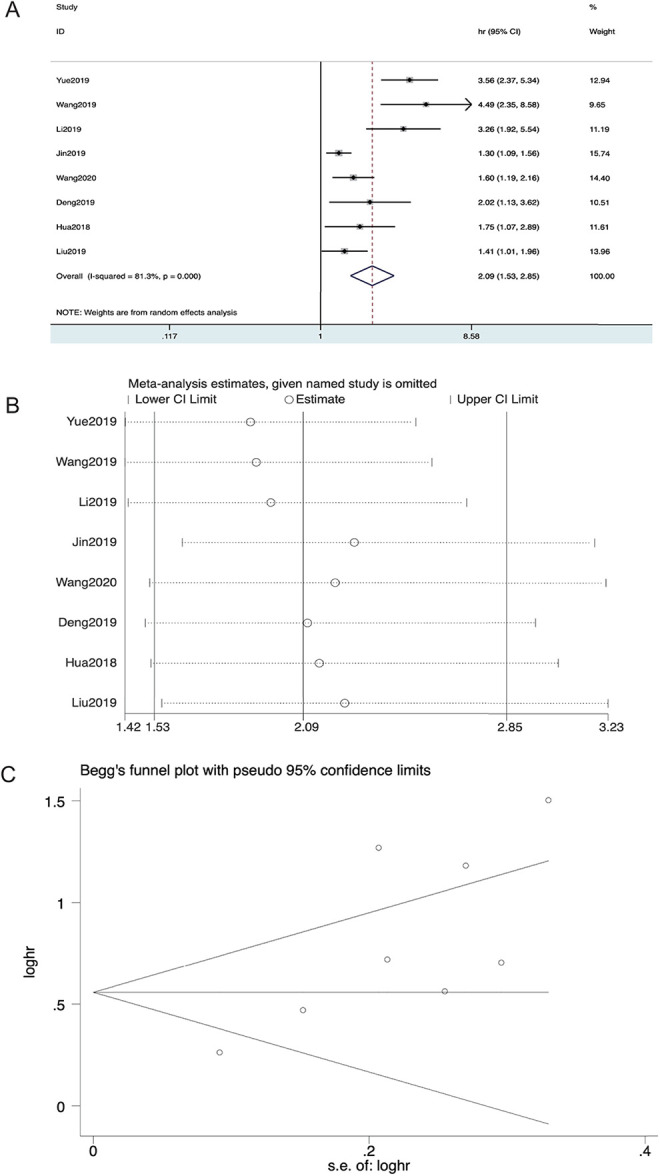
Forest plot of studies evaluating (A) the relationship between METTL3 expression and overall survival (OS) rate, (B) sensitivity analysis for OS, and (C) Begg’s publication bias plots of OS.
Subgroup analysis was performed to determine whether variations in the results were caused by the different cancer types, sample size, and regions (Table 2). Regions were analyzed for OS, suggesting that higher METTL3 expression could predict a worse survival outcome in both the Yantse River Delta (HR = 2.84, 95% CI 1.47–5.49, P = 0.002; Figure 3A) and Pearl River Delta (HR = 2.26, 95% CI 1.55–3.29, P = 0.0001; Figure 3A), respectively. Thus, this result indicated that METTL3 was differentially expressed in 2 local populations. Further analysis of the cancer types showed that higher METTL3 expression indicated poorer OS in both the CRC group (HR = 2.04, 95% CI 1.22-3.41, P = 0.007; Figure 3B), and other cancer type groups (HR = 2.15, 95% CI 1.39–3.31, P = 0.001; Figure 3B). No significant differences were found in this subgroup analysis. We stratified all the studies included into 2 groups based on the sample size, with 100 set as the cut-off value separating the studies. We found that the group of studies with a sample size over 100 had a higher HR (HR = 2.82, 95% CI 2.02–3.95, P = 0.0001; Figure 3C), and studies with a sample size less than 100 had a lower HR (HR = 1.38, 95% CI 1.20–1.59, P = 0.0001; Figure 3C). The larger sample size reduced the sampling error and increased the value of the test statistic, which demonstrated the reliability of the study.
Table 2.
Results of Subgroup Analyses for OS.
| Group | studies | patients | HR | P value | Heterogeneity. I2(%) | ph |
|---|---|---|---|---|---|---|
| OS | 8 | 1257 | 2.09 | 0.0001 | 81.3 | 0.0001 |
| Type of cancer | 3 | 699 | 2.04 | 0.007 | 71.6 | 0.029 |
| CRC | 5 | 558 | 2.15 | 0.001 | 86.6 | 0.0001 |
| Others | 3 | 196 | 1.38 | 0.0001 | 0.0 | 0.499 |
| Sample size | 5 | 1061 | 2.82 | 0.0001 | 51.6 | 0.083 |
| <100 | 3 | 346 | 2.84 | 0.002 | 86.0 | 0.001 |
| ≥100 | 3 | 775 | 2.26 | 0.0001 | 32.4 | 0.0357 |
| Region | ||||||
| Yantse River Delta Pearl river delta |
Abbreviation: CRC, colorectal carcinoma; GBM (Glioblastoma multiforme), BLCA (Bladder Urothelial Carcinoma), CRC (colorectal carcinoma) and BRCA (Breast invasive carcinoma).
Figure 3.
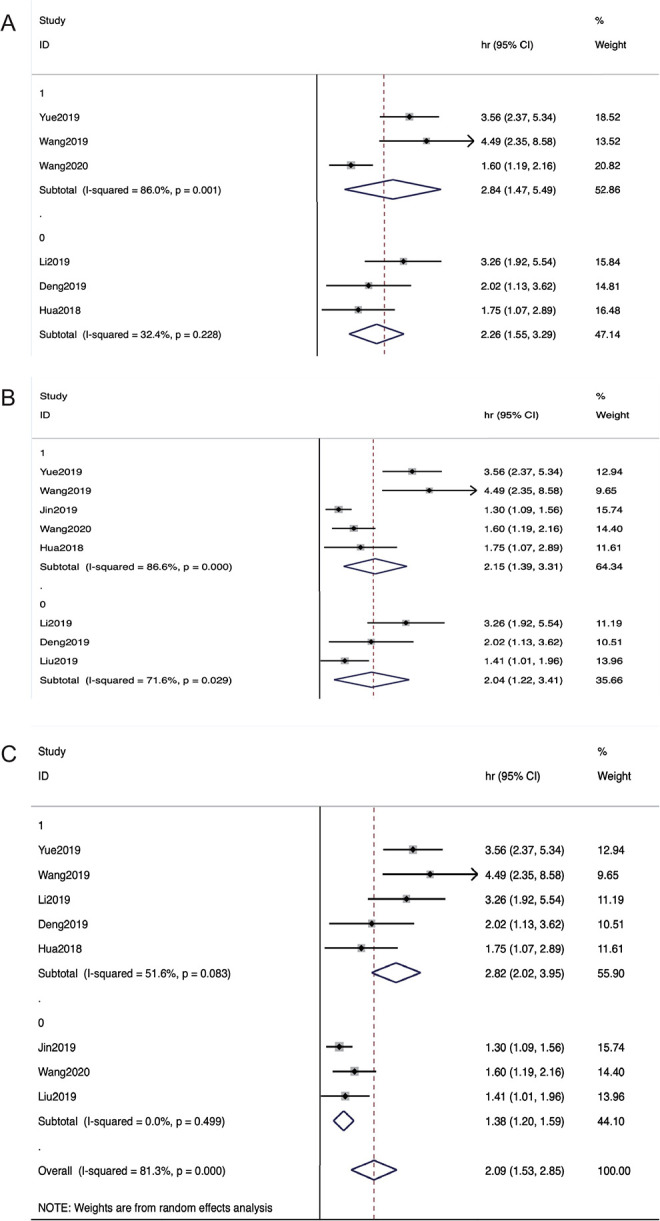
Subgroup analyses of studies evaluating (A)regions, (B)cancer types, and (C) sample sizes of OS.
Verification of the Results Using TCGA
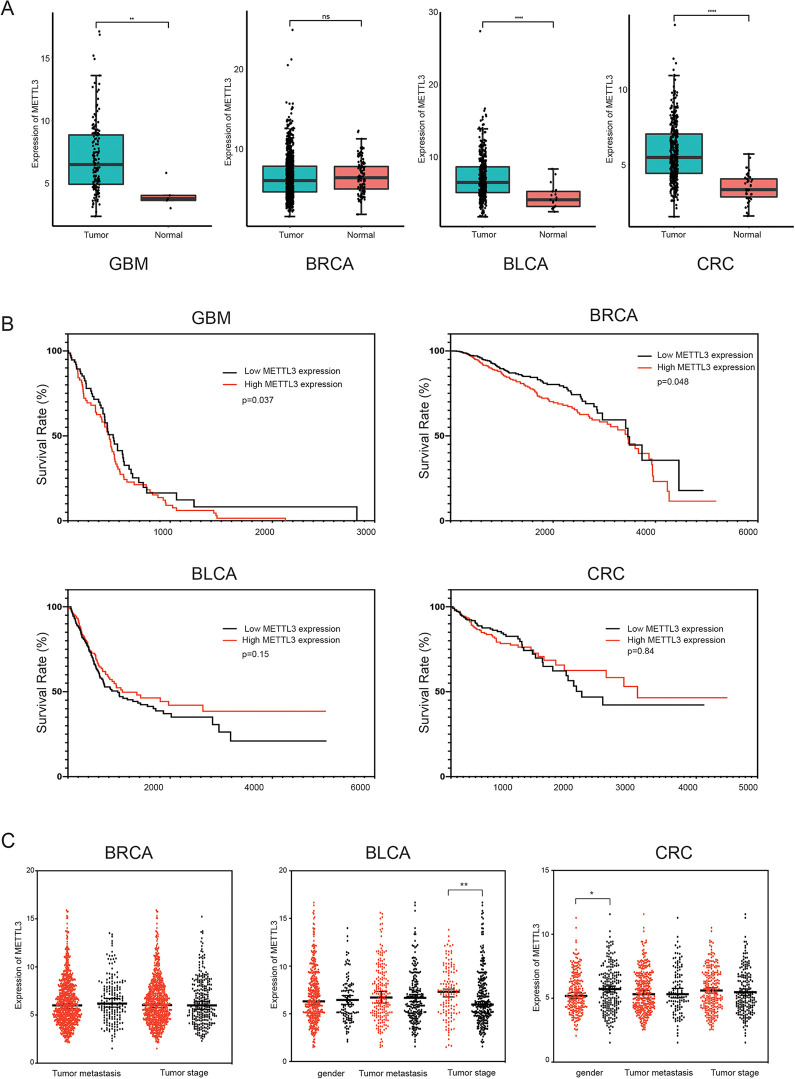
Here, insight into functional influence of METTL3 expression was gained by conducting bioinformatic analyses in various cancers. First, data from TCGA were used to evaluate METTL3 expression in 4 different cancers. As shown in Figure 4A, METTL3 was upregulated in GBM, BLCA and CRC (|Log2fold change (FC)| cutoff >1 and P<0.01). We used Kaplan-Meier curves together with the log-rank test to define the associations between METTL3 and OS in patients with different types of cancer, according to the TCGA dataset. Similar results were observed in GBM, with higher METTL3 expression levels being associated with worse survival, as compared to this meta-analysis (P = 0.037 < 0.05, log-rank; Figure 4B). Higher METTL3 expression was also correlated with better OS in BRCA (P = 0.048 < 0.05, log-rank; Figure 4B). The results revealed that METTL3 could play a role as an independent prognostic biomarker in GBM and BRCA. Unpaired t-test was conducted to assess the association between METTL3 expression and clinical characteristics in TCGA data. METTL3 expression was lower in male patients than in female patients in CRC (P = 0.022 < 0.05; Figure 4C). And patients with lower tumor grade showed higher METTL3 expression in BLCA (P = 0.007 < 0.05; Figure 4C). The results demonstrated that METTL3 expression was correlated with gender in CRC and tumor grade in BLCA.
Figure 4.
Bioinformatics analysis using data from TCGA. (A)The expression levels of METTL3 in 4 kinds of cancer tissues and normal tissues in GBM, BRCA, BLCA and CRC. (B)Survival curves of METTL3 are plotted for 4 cancer types from TCGA dataset. The survival curve of patients with GBM, BRCA, BLCA and CRC. (C)The expression of METTL3 in different gender, tumor metastasis, tumor stage in BRCA, BLCA and CRC, respectively.
Correlation Between METTL3 and Clinicopathological Characteristics
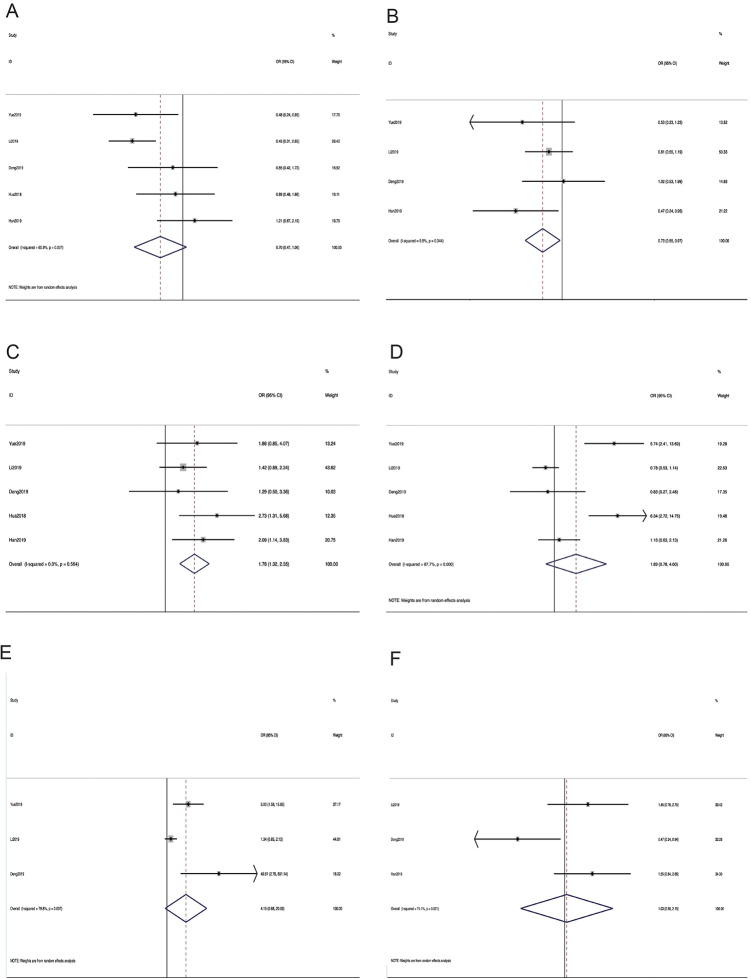
As shown in Table 3, 6 types of clinicopathological characteristics were recorded in 6 studies, and 1075 patients were included. Higher METTL3 expression correlated with poor differentiation (moderate + poor vs. well, pooled odds ratio (OR) = 1.76, 95% CI 1.32–2.35, P = 0.0001, fixed effect). The pooled ORs further revealed that METTL3 upregulation was associated with advanced tumor node metastasis (TNM) stage (III + IV vs. I + II, OR = 1.89, 95% CI 0.78-4.60, P = 0.016, random effect), and metastasis (yes vs. no, OR = 4.19, 95% CI 0.88–20.3, P = 0.007, random effect). Unfortunately, the confidence intervals of the 2 groups were greater than 1. Nevertheless, no significant differences were observed among the 2 groups for tumor size (OR = 1.03, 95% CI 0.50–2.15, P = 0.929, random effect), and age (OR = 0.70, 95% CI 0.47–1.06, P = 0.093, random effect). Pooled data also revealed that METTL3 regulation was associated with gender (OR = 0.73, 95% CI 0.55–0.97, P = 0.029, fixed effect). Therefore, our results demonstrated that higher METTL3 expression could result in substantially worse clinicopathological characteristics. (Figure 5A-F). Publication bias was examined using Begg’s funnel plot (Figure S1 A–F), and was determined for age (P = 0.487), gender (P = 0.716), differentiation (P = 0.899), metastasis (P = 0.101), TNM stage (P = 0.066), and tumor size (P = 0.806). The positive results indicated that differentiation and gender were directly related to the expression of METTL3. Though the OR values of TNM stage and metastasis were positive, the confidence intervals of 2 groups are over 1. This phenomenon suggested that the role of METTL3 in tumors is multi-directional and complicated. Studying the clinical features of METTL3 will enable us better understanding the biological function of METTL3 in various cancers.
Table 3.
The Association Between METTL3 Expression and Clinical Features.
| Clinicopathological parameters | studies | patients | OR | P value | Heterogeneity. I2(%) | ph | model |
|---|---|---|---|---|---|---|---|
| Age | 5 | 1075 | 0.70 | 0.093 | 60.9 | 0.037 | Random |
| Gender | 4 | 913 | 0.73 | 0.029 | 9.8 | 0.344 | Fixed |
| Differentiation | 5 | 1075 | 1.76 | 0.0001 | 0.0 | 0.564 | Fixed |
| TNM stage | 5 | 1075 | 1.89 | 0.016 | 87.7 | 0.0001 | Random |
| Metastasis | 3 | 733 | 4.19 | 0.007 | 79.8 | 0.0072 | Random |
| Tumor size | 3 | 793 | 1.03 | 0.929 | 74.1 | 0.021 | Random |
Figure 5.
Forest plot of studies evaluating the relationship between METTL3 expression and (A) age, (B) gender, (C)differetiation, (D) TNM stage, (E) metastasis, and (F) tumor size.
Discussion
In the majority of tumor studies, METTL3 expression has been discovered to be elevated and to function as an oncogene accompanied by m6A upregulation in tumor cell lines or tissues. The diverse pathways in tumors, which are influenced by METTL3, mostly focused on cell death resistance, cell proliferation, metastasis and invasion.36 METTL3 and m6A were upregulated in human lung cancer,8 leukemia,37 gastric cancer,17,29 osteosarcoma,38 melanoma,39 hepatocellular carcinoma,40 and ovarian carcinoma.35 METTL3 expression level was positively correlated with cancer stage and grade, demonstrating that METTL3 acts as an oncogene in these cancers.
However, some studies have yielded opposite results, even within the same cancer type, including glioblastoma, bladder cancer, colorectal cancer and breast cancer. The reasons for controversy regarding the function of METTL3 in various tumors may be explained by the diversity of METTL3 mechanisms:
First, METTL3 can deposit m6A modification on critical transcripts. Then, the m6A modification directly influence the transcription and translation of tumor suppressors and oncogenes involving most of the essential pathways such as the Wnt/β-catenin,29 P38/ERK,34 and PI3K/AKT8,18,37 pathways. Recent studies pointed that METTL3 was upregulated in colorectal cancer tissues and that this feature correlated with poor prognosis. METTL3 overexpression promoted cancer growth by stabilizing cyclin E1 (CCNE1) 41 and SOX219 mRNA in an m6A-dependent manner. However, Deng et al. showed that METTL3 functioned as a tumor suppressor by inhibiting the proliferation of colorectal cancer cell through p38/ERK pathways.34 In the nervous system, GBM exhibited increased levels of METTL3 transcripts, and tumor growth was inhibited by silencing METTL3-SOX2 axis coupled with prolonged survival of mice in vivo.42 On the contrary, in another study on the role of m6A in glioblastoma, upregulation of METTL3 suppressed stem cell self-renewal growth and was accompanied by inhibited mRNA m6A enrichment and altered mRNA expression of FTO and ADAM19.43 In BRCA, METTL3 expression was higher in tumor cells and tissues. METTL3 knockdown reduced tumor proliferation and accelerated migration together with apoptosis by targeting BCL2, indicating the oncogenic function of METTL3 in BRCA.44,45 Another study showed that METTL3 was reduced in BRCA, and MTase overexpression induced m6A upregulation and suppressed colony formation and cancer cell viability.25 Overall, METTL3 regulates the maturation, stability and degradation of mRNA in various pathways and these pathways may play converse roles in the cancers above.
Second, in addition to regulating mRNA, METTL3 also influences non-coding RNA and miRNA metabolism. For example, METTL3 downregulation significantly inhibited bladder cancer cell proliferation, invasion, and survival, both in vivo and in vitro.46 On the contrary, Han, et al. proved that METTL3 promoted the proliferation of bladder cancer by accelerating the maturation of pri-miR221/222, resulting in the reduction of PTEN. Moreover, they found that METTL3 was significantly increased in bladder cancer and correlated with poor prognosis of bladder cancer patients.15
Third, METTL3 can activate oncogene translation independently of “readers” and the activity of relevant MTase.47 By recruiting eIF3 h, METTL3 is linked to reporter mRNAs at sites near the stop codon, promoting growth and invasion of lung tumor cells.48 A recent work by Hua et al. also showed a similar mechanism, in which translation of the receptor tyrosine kinase AXL is promoted by METTL3, independent of its methyltransferase activity.35 Notably, METTL3 simply binds toward approximately 22% of the m6A sites,49 indicating a mechanism of selectivity of METTL3 to the targets for translational regulation.
Collectively, the studies mentioned above have differences in research methods, model systems, intratumoral heterogeneity and tumor tissue origin, which may explain the opposite roles of METTL3 in cancers. Therefore, more detailed and comprehensive studies are warranted to achieve a better view.
In the present meta-analysis, 9 studies with 1437 patients were included to analyze the association between METTL3 expression, clinical outcomes, and clinicopathologic characteristics. Two main conclusions were drawn from the meta-analysis of the available evidence: (1) METTL3 carriers could predict worse OS compared with patients with lower METTL3 expression in various cancers and (2) METTL3 expression was higher in females than in males, and high METTL3 expression was associated with poorer differentiation.
Further, through subgroup analysis, we found that the association between METTL3 and OS was different between the larger and the smaller sample size groups. The HR for the group with sample size over 100 was 2.82 compared with 1.38. Both heterogeneities were significantly lower than that of the combined group. The significance level was closely related to sample size. A larger sample size reduced the sampling error and increased the value of the test statistic, which demonstrated the reliability of the study. We then divided the patients into 2 groups according to the geographical locations of the study centers. The HR of the group from Pearl River Delta was 2.26 with low heterogeneity compared with the group from the Yangtze River Delta. Notably, as the 2 places are more than 1,000 km apart, it was speculated that METTL3 was differentially expressed in the 2 local populations. Future prospective studies, which aim to define the role of METTL3 in cancer outcomes, should include patients from multiple centers, and offer the METTL3 genetic test to entire study populations. Notably, the OR value of METTL3 with gender was 0.73, with a low heterogeneity, which indicated that METTL3 expression was higher in females compared to that in males. In addition, clinical data from TCGA showed that METTL3 expression was lower in male patients than in female patients in CRC. Similarly, Li et al. found that METTL3 differentially expressed in gender by using optimized consensus matrix of lung cancer tissue.50 It was also reported that METTL3 was highly expressed in female with clear cell renal cell carcinoma51 and lung cancer.52 The reasons for this discrepancy remain unknown. The OR value of METTL3 with differentiation was 1.76, with a low heterogeneity. Consistently, a recent report examining the function of METTL3 in tumor differentiation was carried out using glioma samples. METTL3 expression was found to be elevated in glioma stem-like cells and attenuated during differentiation.52 In addition, the OR values of METTL3 with tumor size, TNM stage, and metastasis were relatively high, but with high heterogeneities. This negative result revealed that more clinical samples are needed to explore the function of METTL3. In the analysis of clinical data from TCGA, patients with lower tumor grade showed higher METTL3 expression in BLCA. The potential of predicting the pathological grading of tumors was beneficial to developing METTL3 as a diagnostic biomarker in BLCA.
The HR values of groups with controversial tumors, including BLCA, CRC, and BRCA, were significantly high, indicating that METTL3 acted as an oncogene in these cancers. To validate the results, TCGA database and bioinformatic analyses were used. Our results showed that, in GBM, BLCA and CRC, the METTL3 expression was higher in tumor samples compared to that in in normal samples, indicating that METTL3 can function as a diagnostic biomarker for GBM, BLCA and CRC. Unfortunately, in BRCA, no significant differential expressions were found between the tumor and normal samples. In the Kaplan-Meier curves with log-rank analysis, patients with high METTL3 expression in GBM had shorter OS. Conversely, METTL3 expression was positively correlated with longer OS in BRCA. The results indicated that METTL3 could act as a prognostic biomarker with the potential for new therapies in GBM. As the result in BRCA was contrary to the included literature, the function of METTL3 in BRCA cannot be conclusively determined.
Based on the emerging data on the roles and the molecular mechanisms in cancer, m6A regulators have attracted growing investigation as therapeutic targets. Taketo et al. found that METTL3 may promote drug resistance in pancreatic cancer, indicating that METTL3 is a potential target for the enhancement of therapeutic efficacy.53 Currently, no specific inhibitors of METTL3 have been found except one compound. Bedi et al. reported the first study to identify the existence of 7 compounds by using high-throughput docking into METTL3 among 4000 analogs and derivatives. Only one compound has the most favorable inhibitory potency.54 However, there were several problems remaining to be addressed. Thus, other potential candidates need to be further explored. The oncogenic role of METTL3 depends on the heterodimer structure formed with METTL14 in most cancers and inhibitors to interfere the interaction between proteins will be a feasible option.55 In addition, seeking inhibitors which target METTL3 molecules upstream or downstream may be a reasonable strategy for tumor treatment. Collectively, the clinical application of targeting METTL3 is still in its infancy. With the deepening knowledge of the mechanisms, regulation and functions in various cancer, it is promising to explore METTL3 targeted therapy.
There were some limitations to our meta-analysis. First, all the potential studies were queried in PubMed, Web of Science, and MedRxiv. Unfortunately, all eligible studies were localized to China. Hence, our results do not appropriately represent global populations. Second, the cancer diversity might have led to an obvious bias due to different baseline characteristics. Since significant heterogeneities were discovered in the pooled outcomes, subgroup, and sensitivity analysis further proved the robustness of our results.
Conclusion
In conclusion, this is the first comprehensive study that discusses the prognostic value of METTL3 in various tumor types. The results revealed that METTL3 could be used prognostic marker for most carcinomas and emphasized its potential as a drug target, by downregulating METTL3 expression. METTL3 acts as an oncogene in BLCA, CRC and GBM according to our results. However, the functions of METTL3 in BRCA are currently controversial, and larger sample sizes and multiple centers are needed for future studies. In addition, owing to the limitations mentioned above, further studies should focus on the utility of METTL3 in cancer prognosis, diagnosis, and therapeutics.
Supplemental Material
Supplemental Material, sj-eps-1-ccx-10.1177_1073274821997455 for Prognostic Roles of N6-Methyladenosine METTL3 in Different Cancers: A System Review and Meta-Analysis by KuangZheng Liu, Yue Gao, Kai Gan, YuQing Wu, Bin Xu, LiHua Zhang and Ming Chen in Cancer Control
Supplemental Material, sj-pdf-1-ccx-10.1177_1073274821997455 for Prognostic Roles of N6-Methyladenosine METTL3 in Different Cancers: A System Review and Meta-Analysis by KuangZheng Liu, Yue Gao, Kai Gan, YuQing Wu, Bin Xu, LiHua Zhang and Ming Chen in Cancer Control
Abbreviations
- BLCA
bladder-urothelial carcinoma
- BRCA
breast carcinoma
- CI
confidence interval
- CRC
colorectal cancer
- EMT
epithelial-mesenchymal transition
- GBM
glioblastoma multiforme
- HR
hazard ratio
- lncRNA
long non-coding RNAs
- m6A
N6-methyladenosine
- METTl3
methyltransferase-like 3
- Mtase
N6-methyladenosine methyltransferase
- mRNA
messenger RNA
- miRNA
microRNA
- OR
odds ratio
- OS
overall survival
- PTEN
phosphatase and tensin homolog
- TCGA
The Cancer Genome Atlas
- TNM
tumor node metastasis
Footnotes
Author Contributions: KuangZheng Liu: Methodology, Software, Validation, Investigation, Writing—Original Draft, Supervision. YuQing Wu: Writing—Review & Editing, Visualization. Kai Gan: Data curation, Validation, Investigation. Yue Gao: Visualization, Investigation. Bin Xu: Conceptualization, Writing—Review & Editing, Funding acquisition. LiHua Zhang: Supervision. Ming Chen: Conceptualization, Funding.
Data Availability Statement: All data, models, or code generated or used during the study are available in a repository or online in accordance with funder data retention policies.
Declaration of Conflicting Interests: The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article. We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the position presented in, or the review of, the manuscript entitled, “Prognostic roles of N6-methyladenosine METTL3 in different cancers: a system review and meta-analysis.”
Ethics Statement: Ethics statement (including the committee approval number) for animal and human studiess is not applicable.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was funded by The National Natural Science Foundation of China (No. 81872089, 81370849, 81672551, 81300472, 81070592, 81202268, 81202034); Natural Science Foundation of Jiangsu Province (BK20161434, BL2013032, BK20150642 and BK2012336); Six talent peaks project in Jiangsu Province, Jiangsu Provincial Medical Innovation Team (CXTDA2017025, WSW-034); The National Key Research and Development Program of China (SQ2017YFSF090096); Jiangsu Provincial Key Research and Development Program (BE2019751); Innovative Team of Jiangsu Provincial (2017ZXKJQWO7); Jiangsu Provincial Medical Talent (ZDRCA2016080).
ORCID iDs: KuangZheng Liu  https://orcid.org/0000-0002-2810-1511
https://orcid.org/0000-0002-2810-1511
Ming Chen  https://orcid.org/0000-0001-8414-7242
https://orcid.org/0000-0001-8414-7242
Supplemental Material: Supplemental material for this article is available online.
References
- 1.Zafar SN, Siddiqui AH, Channa R, Ahmed S, Javed AA, Bafford A. Estimating the global demand and delivery of cancer surgery. J Surg. 2019;43(9):2203–2210. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. A Cancer J Clin. 2019;69(1):7–34. [DOI] [PubMed] [Google Scholar]
- 3.Wild CP, Espina C, Bauld L. et al. Cancer prevention Europe. Mol Oncol. 2019;13(3):528–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18(1):31–42. doi:10.1038/nrm.2016.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, He C. Reading RNA methylation codes through methyl-specific binding proteins. RNA Biol. 2014;11(6):669–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Y, Yin Z, Hou B, Yu M, Chen R, Jin H, Jian Z. Expression profiles and prognostic significance of RNA N6-methyladenosine-related genes in patients with hepatocellular carcinoma: evidence from independent datasets. Cancer Manag Res. 2019;11:3921–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng W, Dong X, Zhao Y. et al. Multiple functions and mechanisms underlying the role of METTL3 in human cancers. Front Oncol. 2019;9:1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei W, Huo B, Shi X. miR-600 inhibits lung cancer via downregulating the expression of METTL3. Cancer Manage Res. 2019;11;1177–1187. doi:10.2147/CMAR.S181058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haussmann IU, Bodi Z, Sanchez-Moran E. et al. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540(7632):301–304. doi:10.1038/nature 20577 [DOI] [PubMed] [Google Scholar]
- 10.Zheng G, Dahl JA, Niu Y. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi:10.1016/j.molcel.2012. 10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Zhao BS, Roundtree IA. et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161(6):1388–1399. doi:10.1016/j.cell.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519(7544):482–485. doi:10.1038/nature14281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Lu Z, Gomez A. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–120. doi:10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer KD, Jaffrey SR. Rethinking m6 a readers, writers, and erasers. Annu Rev Cell Dev Biol. 2017;33:319–342. doi:10.1146/annurev-cellbio-100616-060758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han J, Wang JZ, Yang X. et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18(1):110. 10.1186/s12943-019-1036-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Ishfaq M, Xu L, Xia C, Chen C, Li J. METTL3/m6A/miRNA-873–5p attenuated oxidative stress and apoptosis in colistin-induced kidney injury by modulating Keap1/Nrf2 pathway. Front Pharmacol. 2019;10:517. doi:10.3389/fphar.2019.00517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin X, Chai G, Wu Y. et al. RNA m6A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat Commun. 2019;10(1):2065. doi:10.1038/s41467-019-09865-9 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Lin S, Liu J, Jiang W. et al. METTL3 promotes the proliferation and mobility of gastric cancer cells. Open Med. 2019;14:25–31. doi:10.1515/med-2019-0005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li T, Hu PS, Zuo Z. et al. METTL3 facilitates tumor progression via an m6A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18(1):112. doi:10.1186/s12943-019-1038-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Q, Chen C, Ding Q. et al. METTL3-mediated m 6 a modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020;69(7):1193–1205. [DOI] [PubMed] [Google Scholar]
- 21.Zhang J, Bai R, Li M. et al. Excessive miR-25-3p maturation via N(6)-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat Commun. 2019;10(1):1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu T, Yang S, Sui J. et al. Dysregulated N6-methyladenosine methylation writer METTL3 contributes to the proliferation and migration of gastric cancer. J Cell Physiol. 2020;235(1):548–562. [DOI] [PubMed] [Google Scholar]
- 23.Wang Q, Geng W, Guo H. et al. Emerging role of RNA methyltransferase METTL3 in gastrointestinal cancer. J Hematol Oncol. 2020;13(1):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He H, Wu W, Sun Z, Chai L. MiR-4429 prevented gastric cancer progression through targeting METTL3 to inhibit m(6)A-caused stabilization of SEC62. Biochem Biophys Res Commun. 2019;517(4):581–587. [DOI] [PubMed] [Google Scholar]
- 25.Wu L, Wu D, Ning J, Liu W, Zhang D. Changes of N6-methyladenosine modulators promote breast cancer progression. BMC Cancer. 2019;19(1):326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lobo J, Barros-Silva D, Henrique R, Jeronimo C. The emerging role of epitranscriptomics in cancer: focus on urological tumors. Genes. 2018;9:E552. doi:10.3390/genes9110552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moher D, Liberati A, Tetzlaff J, Altman DG, The PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2019;6(7):e1000097. doi:10.1371/ journal Pmed 1000097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siddaway AP, Wood AM, Hedges LV. How to do a systematic review: a best practice guide for conducting and reporting narrative reviews, meta-analyses, and meta-syntheses. Annu Rev Psychol. 2019;70:747–770. [DOI] [PubMed] [Google Scholar]
- 29.Yue B, Song C, Yang L. et al. METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol Cancer. 2019;18(1):142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiang W, Chen C, Ding Q. et al. METTL3-mediated m6A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2019;69(7):1–13. [DOI] [PubMed] [Google Scholar]
- 31.Liu X, Liu L, Dong Z. et al. Expression patterns and prognostic value of m6A-related genes in colorectal cancer. Am J Transl Res. 2019;11(7):3972–3991. [PMC free article] [PubMed] [Google Scholar]
- 32.Jin D, Guo J, Wu Y. et al. m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J Hematol Oncol. 201912(1):135. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Wang H, Xu B, Shi J. N6-methyladenosine METTL3 promotes the breast cancer progression via T targeting Bcl-2. Gene. 2020;722:144076. [DOI] [PubMed] [Google Scholar]
- 34.Deng R, Cheng Y, Ye S. et al. m6A methyltransferase METTL3 suppresses colorectal cancer proliferation and migration through p38/ERK pathways. Onco Targets Ther. 2019:124391–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hua W, Zhao Y, Jin X. et al. METTL3 promotes ovarian carcinoma growth and invasion through the regulation of AXL translation and epithelial to mesenchymal transition. Gynecol Oncol. 2018;151(2):356–365. [DOI] [PubMed] [Google Scholar]
- 36.Zeng C, Huang W, Li Y, Weng H. Roles of METTL3 in cancer: mechanisms and therapeutic targeting. J Hematol Oncol. 2020;13(1):117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vu LP, Pickering BF, Cheng Y. et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23(11):1369–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miao W, Chen J, Jia L, Ma J, Song D. The m6A methyltransferase METTL3 promotes osteosarcoma progression by regulating the m6A level of LEF1. Biochem Biophys Res Commun. 2019;516:719–725. doi:10.1016/j.bbrc.2019.06.128 [DOI] [PubMed] [Google Scholar]
- 39.Dahal U, Le K, Gupta M. RNA m6A methyltransferase METTL3 regulates invasiveness of melanoma cells by matrix metallopeptidase 2. Melanoma Res. 2019;29(4):382–389. [DOI] [PubMed] [Google Scholar]
- 40.Chen M, Wei L, Law CT. et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67(6):2254–2270. [DOI] [PubMed] [Google Scholar]
- 41.Zhu W, Si Y, Xu J, Lin Y, Wang JZ, Cao M. et al. Methyltransferase like 3 promotes colorectal cancer proliferation by stabilizing CCNE1 mRNA in an m6A-dependent manner. J Cell Mol Med. 2020;24(6):3521–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Visvanathan A, Patil V, Arora A. et al. Essential role of METTL3-mediated m6A modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018;37(4):522–533. [DOI] [PubMed] [Google Scholar]
- 43.Cui Q, Shi H, Ye P. et al. m6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18(11):2622–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang H, Xu B, Shi J. N6-methyladenosine METTL3 promotes the breast cancer progression via targeting Bcl-2. Gene. 2020;722:144076. doi:10.1016/j.gene.2019.144076 [DOI] [PubMed] [Google Scholar]
- 45.Cai X, Wang X, Cao C. et al. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7 g. Cancer Lett. 2018;415:11–19. [DOI] [PubMed] [Google Scholar]
- 46.Zhao S, Liu J, Nanga P. et al. Detailed modeling of positive selection improves detection of cancer driver genes. Nat Commun. 2019;10(1):3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62(3):335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.METTL3 Promotes mRNA translation to drive tumorigenesis. Cancer Discov. 2018;8(11):1346. doi:10.1158/2159-8290.CD-RW2018-167 [Google Scholar]
- 49.Liu J, Yue Y, Han D. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10(2):93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li N, Zhan X. Identification of pathology-specific regulators of m 6A RNA modification to optimize lung cancer management in the context of predictive, preventive, and personalized medicine. EPMA J. 2020;11(3):485–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Q, Luan J, Song L, et al. m6A RNA methylation regulators correlate with malignant progression and have potential predictive values in clear cell renal cell carcinoma. Exp Cell Res. 2020;392(1):112015. [DOI] [PubMed] [Google Scholar]
- 52.Zhu J, Wang M, Hu D. Deciphering N 6-methyladenosine-related genes signature to predict survival in lung adenocarcinoma. Biomed Res Int. 2020;2020(10):2514230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taketo K, Konno M, Asai A. et al. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int J Oncol. 2018;52(2):621–629. [DOI] [PubMed] [Google Scholar]
- 54.Bedi RK, Huang D, Eberle SA, Wiedmer L, Caflisch A, Sledz P. Small-molecule inhibitors of METTL3, the major human epitranscriptomic writer. ChemMedChem. 2020;15(9):744–748. [DOI] [PubMed] [Google Scholar]
- 55.Scott DE, Bayly AR, Abell C, Skidmore J. Small molecules, big targets: drug discovery faces the protein-protein interaction challenge. Nat Rev Drug Discov. 2016;15(8):533–550. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, sj-eps-1-ccx-10.1177_1073274821997455 for Prognostic Roles of N6-Methyladenosine METTL3 in Different Cancers: A System Review and Meta-Analysis by KuangZheng Liu, Yue Gao, Kai Gan, YuQing Wu, Bin Xu, LiHua Zhang and Ming Chen in Cancer Control
Supplemental Material, sj-pdf-1-ccx-10.1177_1073274821997455 for Prognostic Roles of N6-Methyladenosine METTL3 in Different Cancers: A System Review and Meta-Analysis by KuangZheng Liu, Yue Gao, Kai Gan, YuQing Wu, Bin Xu, LiHua Zhang and Ming Chen in Cancer Control