Abstract
The factors that control the development of an effective immune response to the recently emerged SARS-CoV-2 virus are poorly understood. Herein, we provide a cross-sectional analysis of the dynamics of B cell responses to SARS-CoV-2 infection in hospitalized COVID-19 patients. We observe changes in B cell subsets consistent with a robust humoral immune response, including significant expansion of plasmablasts and activated RBD specific memory B cell populations. We observe elevated titers of antibodies to SARS-CoV-2 RBD, full-length spike, and nucleoprotein over the course of infection, with higher levels of RBD-specific IgG correlating with increased serum neutralization. Depletion of RBD-specific antibodies from serum removed a major portion of neutralizing activity in most individuals. Some donors did retain significant residual neutralization activity, suggesting a potential antibody subset targeting non-RBD epitopes. Taken together, these findings are instructive for future vaccine design and monoclonal antibody strategies.
Introduction
The novel coronavirus, SARS-CoV-2, emerged in December 2019(1) and continues to take an unprecedented toll on the global population with over 2.4 million deaths reported worldwide, a half million of which have occurred in the U.S. alone.(2) Two mRNA vaccines (Pfizer-BioNTech and Moderna) are currently approved for emergency use in the United States.(3,4) In addition to these U.S.-approved SARS-CoV-2 vaccines, there are over 200 other vaccine candidates at various developmental stages from preclinical testing to approved usage outside the U.S.(5) In order to understand differences in efficacy between vaccine candidates and between immunity developed from vaccination versus natural infection, it is important to continue to explore the characteristics of protective immunity after natural infection. Discoveries concerning the generation, dynamics, and durability of natural immunity may influence discussions and decisions concerning future vaccination development and distribution efforts. Furthermore, a detailed insight into the mechanism of viral neutralization is also essential for both vaccine and monoclonal antibody-based treatment efforts, potentially influencing considerations of necessary antigen targets to achieve effective thresholds of protection.
SARS-CoV-2, a beta-coronavirus (6), shares a high level of homology to SARS-CoV(1), the coronavirus responsible for the 2002–2003 SARS epidemic. These coronaviruses have also been found to share the same host entry receptor, ACE2, which is bound by the receptor binding domain (RBD) on the spike (S) homotrimer that is present on the viral surface.(7) RBD is located within the S1 subunit of the protein and appears to be only accessible in the “open” or “up” confirmation of the trimer.(7) Given the homology between the two beta-coronaviruses, a predictable relationship between RBD binding and SARS-CoV-2 neutralization exists, and studies by us(8) and others(9–11) have clearly illustrated a strong correlation between RBD binding and viral neutralization. Like their SARS-CoV counterparts, antibodies targeting RBD appear to be an integral component of the protective immune response against SARS-CoV-2.(12) In support of this, several antibodies isolated from RBD-specific memory B cells have been characterized and shown to be potent neutralizers of SARS-CoV-2 both in vitro(11,13,14) and in vivo.(15) In addition, several groups have shown potent plasmablast responses during acute infection(16), and sizeable RBD- or spike-specific memory B cell responses(11,13–15,17) early after infection. Interestingly, potent RBD-specific neutralizing antibodies can be isolated from individuals regardless of the neutralizing serum titer(11) and follicular T cells, a critical part of germinal center reactions, are rarely RBD-specific.(17) Given the contribution of antibodies to the neutralization of SARS-CoV-2 in vivo, understanding the protective characteristics of the virus-specific B cell responses to SARS-CoV-2 infection remains crucial.
An early expansion of plasmablasts followed by the formation of a circulating antigen-specific memory B cell pool has been reported for numerous acute viral infections, including SARS-CoV-2.(11,13–17) Previous studies of SARS-CoV and SARS-CoV-2 infection have shown that patients develop severe lymphopenia with significant decreases in T cell numbers.(18,19) In contrast, B cell numbers in these individuals remain unimpacted by infection.(18,19) However, recent evidence from autopsied patients that succumbed to SARS-CoV-2 infection suggests suboptimal germinal center reactions in these patients, which could potentially contribute to short-lived and immature antibody responses to SARS-CoV-2.(16,20) Additional alterations among B cell subsets have also been reported. For example, expanded atypical memory B cells have been reported in patients with severe COVID-19.(20,21) A subset most often described in patients with autoimmunity, immunodeficiencies, or chronic viral infection, these CD27-CD21- B cells are thought to mature independently of germinal center reactions through an extrafollicular pathway.(21,22) The apparent expansion of this subset within severe COVID-19 patients raises questions concerning the nature of their role in the immune response to SARS-CoV-2 and their contribution durable antibody responses, which is a continued concern given several recent reports of SARS-CoV-2 re-infection after only a few months in individuals previously infected with SARS-CoV-2.(23,24)
Herein, we report a cross-sectional study of the dynamics of human B cell responses during acute SARS-CoV-2 infection. We show that infection induces a potent plasmablast response, and RBD-specific memory B cell responses that correlate with virus-specific serological responses. We also show, using a serum depletion approach, that RBD specific antibodies are the primary driver of viral neutralization in the majority of patients. Interestingly, a subset of the individuals examined had significant portions of their neutralizing response that appeared resistant to RBD depletion, potentially suggesting alternate mechanisms of protection outside the direct inhibition of RBD. These findings have significant implications for ongoing vaccine strategies, as well as for efforts to identify, characterize and deploy preventative and therapeutic monoclonal antibodies against SARS-CoV-2.
Methods
Study cohort
The current study draws on patient samples from hospitalized COVID-19 patients with RT-PCR confirmed SARS-CoV-2 infection at the Emory University Hospital and Emory University Hospital Midtown (n=50). While no specific criteria or demographics were used for enrollment beyond PCR confirmed SARS-CoV-2 infection, all patients were symptomatic at the time of enrollment. Specimens were collected after receiving informed consent, except for 00022371 for which a consent waiver was obtained. The clinical studies from which these samples were obtained was approved by the Emory University Institutional Review Board IRB #00000510, IRB #00045690 and IRB #00022371. For IRB #00000510 and #00045690, informed consent was obtained prior to patient participation. For #00022371, an IRB waiver was obtained allowing the use of discarded samples in the clinical laboratory at the Emory Hospital. The majority of the patients were diagnosed with severe disease (91%) and trended towards being older (median age = 58.5) and male (59%). Further details of the cohort can be found in Supplementary Table 1. Limitations of the cohort include (i) all individuals were hospitalized with the majority diagnosed with severe disease, (ii) a majority of individuals had one or more pre-existing conditions, and (iii) a relatively small sample size.
Sample preparation
Briefly, plasma and PBMC were isolated from peripheral blood collected in CPT tubes from these patients at various times after disease onset (3–57 days post-symptom onset). Briefly, CPT tubes were processed according to manufacturer’s protocol, and plasma and PBMCs separated collected separately. PBMCs were treated with ACK lysis buffer (Quality Biological #118–156-101) for 5 minutes and washed 3 times with PBS with 2% FBS before counting and analysis by flow cytometry. PBMC and plasma were frozen at −80C prior to long-term storage at −80C (plasma) or in liquid nitrogen (PBMC).
Viruses and cells
The infectious clone SARS-CoV-2 (icSARS-CoV-2) and mNG-tagged SARS-CoV-2 (icSARS-CoV-2-mNG) was kindly provided to us and previously described by Dr. Vineet Menachery (UTMB).(25) Briefly, the SARS-CoV-2 virus used was derived from infectious clone 2019-nCOV/USA_WA1/2020 and tagged with a fluorescent reporter gene (mNG) in ORF7.(25) Viral titers were determined by plaque assay on VeroE6 cells (ATCC). VeroE6 cells were cultured in complete DMEM medium consisting of 1x DMEM (Corning Cellgro), 10% FBS, 25 mM HEPES Buffer (Corning Cellgro), 2 mM L-glutamine, 1mM sodium pyruvate, 1x Non-essential Amino Acids, and 1x antibiotics. Viral stocks were titered on VeroE6 cells and stored at −80°C until use.
Flow cytometry
Freshly isolated peripheral blood mononuclear cells were stained first for viability with Live/dead Yellow (ThermoFisher) and then for markers with the following monoclonal antibodies: IgA (IS11–8E10, Miltenyi), IgD (IA6–2, BD), IgG (G18–145, BD), IgM (MHM-88, Biolegend), CD3 (SK7, BD), CD4 (RPA-T4, BD), CD8 (SK1, BD), CD14 (61D3, eBioscience), CD16 (CB16, eBioscience), CD19 (SJ25C1, BD), CD20 (2H7, BD), CD27 (O323, BioLegend or M-T271, BD), CD38 (HB7, BD), and CD71 (CY1G4, BioLegend. Antigen-specific B cells were detected by staining with RBD conjugated to Alexa Fluor 488 (Protein Labeling Kit, ThermoFisher). RBD was conjugated according to manufacturer’s instructions, with the following changes: protein was labeled at a concentration of 1mg/mL, and incubated for 30 minutes without the addition of bicarbonate. After staining, PBMCs were washed and then fixed for 30 minutes using 2% paraformaldehyde (ThermoFisher). Data were acquired on a BD FACSymphony A5 and analyzed using FlowJo 10.7.1 (BD).
ELISA
ELISAs were conducted as we have previously described.(8) Recombinant RBD for this assay was generated as previously described.(8) Briefly, recombinant RBD derived from SARS-CoV-2, Wuhan-Hu-1 (GenPept:QHD43416) was cloned, expressed in an Expi293F cell system, and purified on HisTALON Superflow Cartridges.(8) Recombinant RBD, recombinant monomeric spike (obtained from the CDC), or nucleoprotein (Sinobiological, # 40588-V08B) were coated overnight at 4C on Maxisorb plates at 0.5 (NP) and 1 (RBD, Spike) μg/mL in Dulbecco’s phosphate-buffered saline (DPBS). After blocking for 2 h with 1% bovine serum albumin (BSA) in PBS containing 0.05%Tween 20 (PBS-T), serially diluted serum samples were added and incubated for 90 minutes. The bound antibodies were detected using goat anti-human isotype specific secondary antibodies conjugated to horseradish peroxidase (HRP) that were added for 60 minutes (Jackson ImmunoResearch, anti-IgG Cat#109–036-098, anti-IgM Cat#109–036-129, anti-IgA Cat#109–036-011). Plates were developed with 0.4 mg/mL o-phenylenediamine dihydrochloride (OPD) diluted in phosphate-citrate buffer pH 5.0 containing 0.012% H2O2. The reaction was stopped with 1M hydrochloric acid and the absorbance was measured at 490 nm using a spectrophotometer (BioRad). Unless noted, plates were washed 3 times with PBS-T between each step. Endpoint titers were interpolated based on a sigmoidal 4-parameter logistic where X is concentration with the baseline value for each isotype/antigen pair derived from the average plus three times the standard deviation of pre-pandemic negative control samples (n=20).
Focus Reduction Neutralization Titer assay
COVID-19 patient or healthy control plasma were incubated at 56°C for 30 min and manually diluted in duplicate in serum-free Dulbecco’s modified media and incubated with 750–1000 focus-forming units of either icSARS-CoV-2 or SARS-CoV-2-mNG virus at 37° C for 1 hour. The virus/serum mixture was added to VeroE6 cell monolayers seeded in 96-well clear or blackout plates and incubated at 37° C for 1 hour. Post incubation, the inoculum was removed and replaced with pre-warmed complete DMEM containing 0.85% methylcellulose. Plates were incubated at 37° C for 24 hours. After 24 hours, the methylcellulose overlay was removed, cells were washed three times with phosphate-buffered saline (PBS), and fixed with 2% paraformaldehyde (PFA) in PBS for 30 minutes at room temperature. For the FRNT assay, plates were washed twice with 1x PBS and 100 μl of permeabilization buffer (0.1% BSA-Saponin in PBS) (Sigma Aldrich), was added to the fixated Vero cell monolayer for 20 minutes. Cells were incubated with an anti-SARS-CoV spike protein primary antibody conjugated to biotin (CR3022-biotin) for 1 hour at room temperature, then with avidin-HRP conjugated secondary antibody for 1 hour at room temperature. Foci were visualized using True Blue HRP substrate and imaged on an ELISPOT reader (CTL). For the FRNT-mNG assay, the 2% PFA is removed and washed twice with PBS. The foci were visualized using an ELISPOT reader (CTL ImmunoSpot S6 Universal Analyzer) under a FITC channel and enumerated using Viridot. The neutralization titers were calculated as follows: 1 - (ratio of the mean number of foci in the presence of sera and foci at the highest dilution of respective sera sample). Each specimen is tested in two independent assays performed at different times. The FRNT-mNG50 titers were interpolated using a 4-parameter nonlinear regression in GraphPad Prism 8.4.3. Samples with an FRNT50 value that was below the limit of detection, are plotted at 10. For these samples, this value was used in fold reduction calculations.
Depletion of RBD specific serum antibody
RBD binding antibodies in patient sera were depleted using RBD-coupled paramagnetic beads. Recombinant SARS-CoV-2 RBD was covalently attached to paramagnetic M-270 epoxy Dynabeads using the “Dynabeads Antibody Coupling Kit” (ThermoFisher Scientific # 14311D) according to manufacturer’s instructions for labeling 60 mg of beads. Beads were prepared using 30 μg of RBD per mg of beads. After coupling, beads were suspended at a concentration of 10 mg/mL in buffer SB containing 0.02% (w/v) sodium azide for up to two weeks at 4ºC. Immediately before use, RBD-coupled beads were washed once for 5 minutes in PBS with 0.1% BSA and then resuspended in DPBS at a concentration of 30 mg/mL. Patient sera were added to beads at a ratio of 1:10 (v/v) and gently mixed for 1 hour at ambient temperature using a rotating mixer. Depleted sera were separated from beads with a magnet tube rack and transferred to a fresh tube that contained RBD coupled beads equal in amount to the first depletion, which had been separated from the storage solution. Samples were incubated again for 1 hour at ambient temperature and then magnetically separated from beads yielding RBD-depleted sera diluted 1 to 10 in DPBS. Samples were aliquoted and stored at −80ºC prior to use in binding assays or neutralization assays as described above. Endpoint binding titers for this assay were interpolated based on a sigmoidal 4-parameter logistic where X is concentration using 3x background as the baseline value. Neutralization titers were calculated as previously stated. Percent reduction was then calculated from the fold-change between the pre-depletion and post-depletion samples.
Statistics
Data were analyzed using GraphPad Prism 8.4.3. A one-way ANOVA Brown-Forsythe test or Holm-Sidak multiple-T test, as appropriate for all comparisons of cell populations and antibody titers between groups. Pearson correlation coefficients and linear regressions were applied as appropriate.
Results
Highly expanded plasmablasts and reduced memory B cell frequencies in peripheral blood of hospitalized COVID-19 patients.
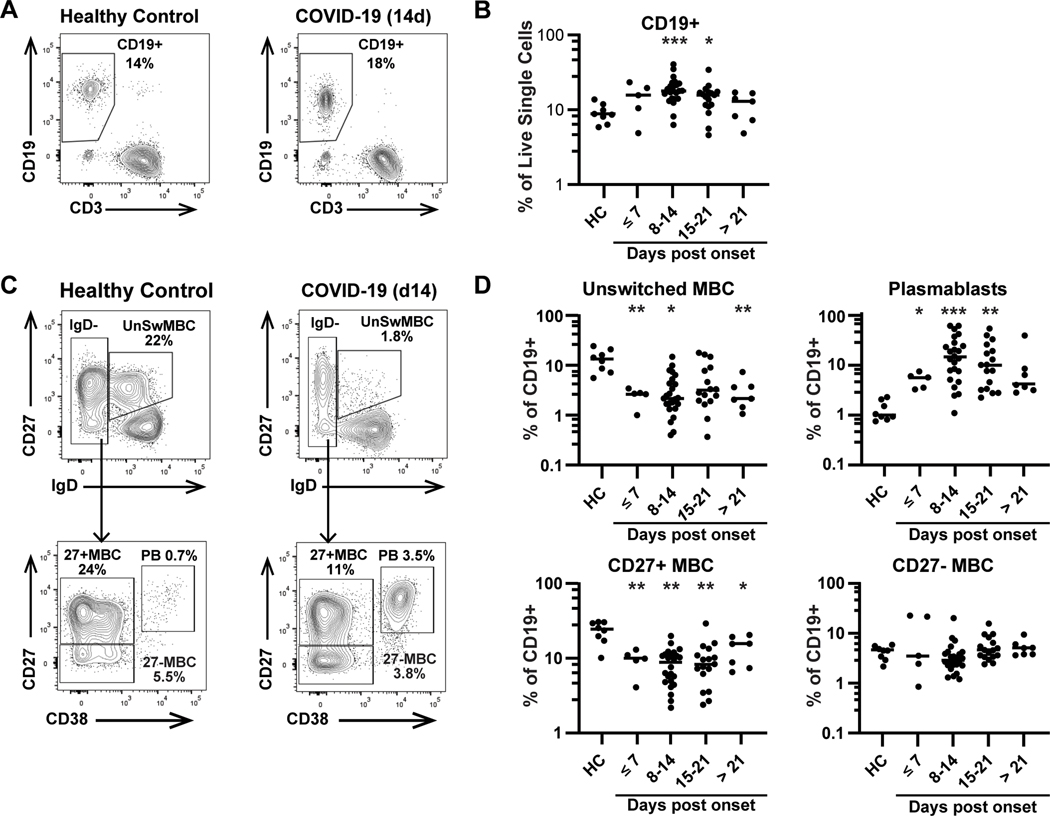
To define the dynamics of human B cell responses during acute SARS-CoV-2 infection, we assessed CD19+ cell subsets in a cross-sectional study of 46 hospitalized patients, sampled at timepoints ranging from 3 to 57 days post symptom onset (DpSO), compared to 8 healthy controls samples collected during the study period and confirmed to be SARS-CoV-2 negative by serology (Figure 1). Six of the acutely infected patients were sampled at least twice (Supplementary Table 1). We found that the overall B cell compartment (CD19+ cells) in peripheral blood was significantly increased in the second (19.1±7.54) and third (15.1±6.69) weeks after symptom onset as compared to healthy controls (9.16±2.67) (p<0.001, p=0.017). This increase is likely due to both plasmablast expansion as well as a loss of peripheral CD3+ T cells (Supplemental Fig 1), as has been previously reported.(26) Focusing on antigen-experienced B cell subsets, we analyzed both infection-induced plasmablasts and total memory B cells (MBC). We found highly expanded plasmablast responses in the majority of COVID-19 patients, rising early after symptom (<d7) onset (5.0±1.7, p=0.016), peaking at 8–14 DpSO (19.5±17.6, p<0.001), and remaining significantly increased 15–21 DpSO (14.5±14.5 p=0.006), as compared to healthy controls (1.3±0.6). In contrast, classical MBCs (CD27+/IgD- B cells) were significantly reduced, falling before 7 DpSO (HC=23.2±7.2, CVD=9.4±3.3, p=0.003) and remaining low in patients at >21 days of illness (13.4±5.8, p=0.044). CD27- MBCs were not significantly reduced in frequency at any timepoint. Additionally, unswitched memory B cells (CD27+/IgD+) were dramatically reduced ≤7 DpSO (2.6±0.9) compared to controls (14.2±6.9) (p=0.008) and remained low in patients hospitalized >21 DpSO (3.2±2.2, p=0.008). Unswitched MBC are known to exhibit reactivity similar to naïve B cells, and are able to rapidly respond to antigen.(22) The significant loss observed may be in part due to differentiation into plasmablasts, but this issue requires further study. However, both CD27+ and CD27- switched MBCs remained low after 21 DpSO, when plasmablasts were no longer significantly expanded, at least in peripheral blood. These data show an early expansion of plasmablasts that is reminiscent of other serious viral infections, such as H1N1 influenza(27), dengue(28) and Ebola(29) infection, and is likely responsible for the early SARS-CoV-2-specific antibody responses seen in these patients.
Figure 1: Acute COVID-19 patients exhibit loss of circulating memory B cells and expansion of plasmablasts.
A) CD19+ B cells are identified from live single CD14-CD16- cells in a healthy control (left) or COVID-19 (right) participant. B) Percentage of CD19+ B cells are shown for healthy controls (n=8) or hospitalized COVID-19 patients (n=46) over time, measured as days post symptom onset. Six patients contributed more than one timepoint. C) CD19+ B cells are further subsetted as unswitched memory B cells (MBC), isotype-switched CD27+ MBC and CD27- MBC, and plasmablasts (PB). Unswitched MBC are identified as CD27+IgD+, while MBC and PB are IgD- and then separated by CD27 and CD38 expression. D) Unswitched MBC, CD27+ MBC, CD27- MBC and plasmablasts are shown as % of CD19+ cells in healthy controls and COVID-19 patients. Significance is calculated by Brown-Forsythe ANOVA test. *p≤0.05, **p≤0.01, ***p≤0.001
RBD-specific memory cells appear 8–14 days after symptom onset.
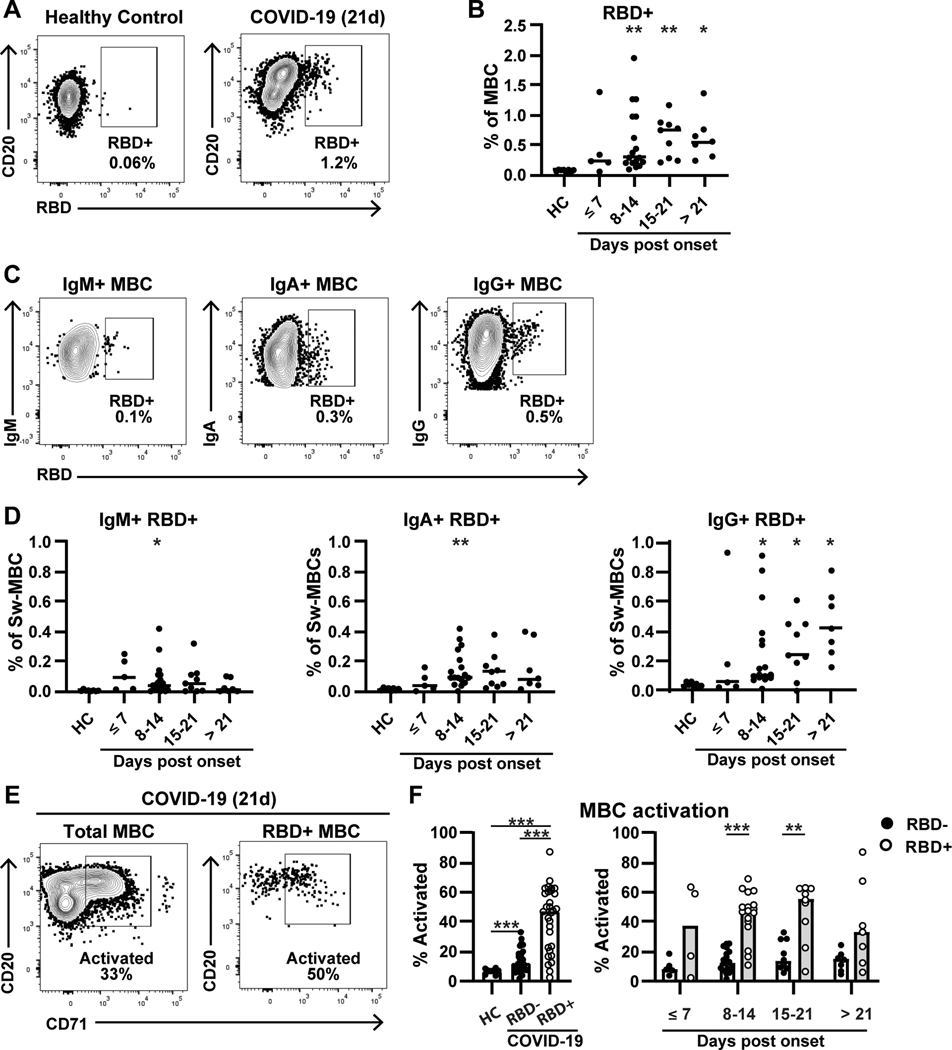
To further assess the dynamics of B cell responses to SARS-CoV-2, we analyzed antigen-specific memory B cell responses by flow cytometry, using fluorescently-labeled RBD as a probe (Figure 2A). We observed a significant expansion of RBD-specific switched MBC at 8–14 DpSO, corresponding to 0.57±0.53% of the overall MBC population, compared to the negligible background of 0.07±0.02% in the healthy controls (p=0.005) (Figure 2B). The mean frequency of RBD-specific MBCs continued to increase 15–21 DpSO (0.63±0.33%, p=0.004), and then plateaued >21 DpSO (0.63±0.37, p=0.027). Notably, in some patients, a proportion of the RBD-specific switched MBCs did not express CD27 (Supplemental Fig 2). In fact, overall 31.6±19.5% of the RBD+ cells were CD27-, and in one participant reached as high as 74%. This observation could be connected to the loss of CD27+ switched MBCs, as the frequency of CD27 expression did not differ between RBD-specific and non-specific MBCs, or a sign that some RBD+ MBCs are generated in an early extrafollicular or T-independent manner.(30) Therefore, we have reported RBD-specificity as a function of total switched MBCs (Figure 2B).
Figure 2: RBD-specific memory B cells expand rapidly and exhibit high levels of activation in COVID-19 patients.
A) RBD-specific memory B cells (CD19+CD20+IgD-CD38-) are shown for a healthy control (left) or COVID-19 patient (right). B) RBD+ MBCs are shown as a percentage of total MBCs for healthy controls (n=8) and COVID-19 patients (n=34) over time. Four COVID-19 patients contribute repeat timepoints. C) Gating is shown for IgM+ (left), IgA+ (middle), and IgG+ (right) RBD+ MBCs in the COVID-19 patient shown in (A). Percentages shown are % of total MBC. D) RBD+ MBCs are shown as in (B), split into IgM+ (left), IgA+ (middle) and IgG+ (right). E) Activated B cells, gated by CD71 expression, for both total (left) and RBD+ (right) MBC. F, left) Total activation in healthy controls (n=8) and COVID-19 patients RBD- MBC (n=33) or RBD+ MBC (n=30). F, right) A comparison of activation over time between RBD- and RBD+ MBC. Significance is calculated by Brown-Forsythe ANOVA test (B, D, F left) or Holm-Sidak multiple T test (F right). *p≤0.05, **p≤0.01, ***p≤0.001
The RBD+ MBC in COVID-19 patients are primarily of the IgG isotype (Figure 2C/D). RBD-specific IgM+ MBC were only significantly expanded at 8–14 DpSO (0.08±0.11) compared to healthy controls (0.005±0.005) (p=0.04) and responses at that time were highly variable. IgA+ MBCs were also only significantly expanded at 8–14 DpSO (0.15±0.12) compared to healthy controls (0.01±0.01) (p=0.002). Though a subset of patients did have measurable RBD+ IgA+ MBCs at later timepoints, other patients did not seem to mount strong IgA+ MBC responses. In contrast, IgG+ RBD-specific MBCs were significantly expanded starting at 8–14 DpSO (0.26±0.27, p=0.02) through 15–21 DpSO (0.29±0.20, p=0.02). By 21 DpSO or later all donors were positive for IgG+ RBD-specific cells (0.45±0.23, p=0.01) (Figure 2D). Our data show that RBD-specific switched MBC arise by the second week of infection, and highlight that focusing on only CD27+ memory may exclude a sizeable percentage of the RBD-specific memory response.
RBD-specific MBCs upregulate the activation marker CD71.
Recently, activated memory B cells (ABCs) have been shown to be an important subset in several diseases, such as Ebola and influenza(29). Therefore, we assessed the expression of CD71 on the memory B cells of healthy controls and patients with acute SARS CoV-2 infection, and further compared the CD71 expression of non-RBD and RBD-binding memory B cells during disease progression (Figure 2E–F). MBC obtained from healthy controls had low frequencies of CD71+ (6.8±1.9). In contrast, the frequency of activated switched MBCs in COVID-19 patients were significantly higher (14.3±8.0) (p<0.001). The RBD-specific switched MBCs express CD71 at markedly higher frequencies (42.5±21.3) than non-RBD-specific MBCs. This difference was not only significant compared to healthy control MBC (p<0.001) but was also significantly increased relative to non-RBD-specific MBC from the same patients (p<0.001). The difference between RBD-specific and non-specific MBC was most apparent between days 8 and 14 (RBD=44.8±16.1 vs non-RBD=13.7±7.29, p<0.001) and days 14 and 21 (RBD=27.2±20.1 vs non-RBD=17.7±10.0, p=0.001). These data show that not only are RBD-specific MBCs are present early in the course of COVID-19, but that these B cells are an active part of the ongoing immune response.
Circulating IgG and IgA titers against RBD, S, and NP antigens peak 3 weeks post-symptom onset.
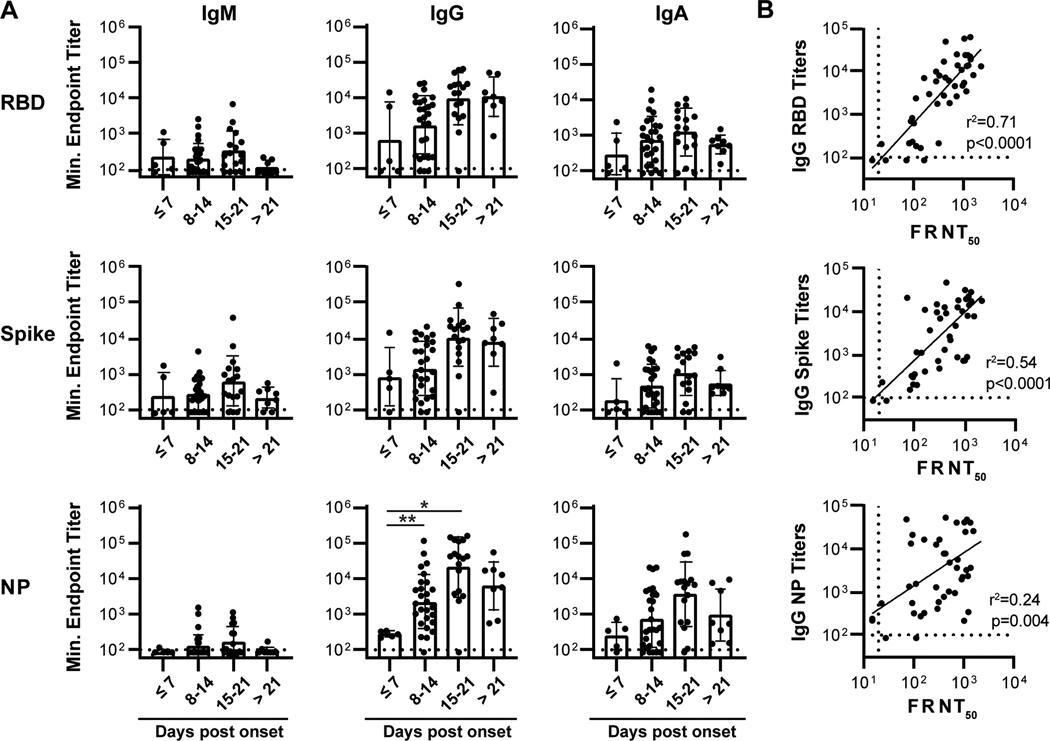
To determine the dynamics of antibody responses during infection, we measured circulating antibody titers against SARS-CoV-2 antigens by ELISA in the 46 individuals analyzed above, using recombinant RBD, monomeric spike (S), or nucleoprotein (NP). In agreement with previous reports showing that seroconversion against SARS-CoV-2 and other coronaviruses occurs within two weeks post-symptom onset(6,9,10), all but two individuals in the cohort had positive IgG and IgA titers against all three antigens by two weeks post-symptom onset. IgG and IgA titers against all antigens trended with DpSO with significant increases in antibody titer observed between the first, second, and third weeks post-symptom onset with the NP-specific IgG serum fraction (Figure 3A). Between 8–14 DpSO, 93% (25/27), 93%, (25/27), and 96% (26/27) of individuals had positive IgG titers towards RBD, S, and NP, respectively, as compared to 88% (24/27), 78% (21/27), and 81% (22/27) of individuals with positive IgA titers (Figure 3A, Supplementary Table 1). We also note that, while IgG titers against all antigens remain robust in individuals greater than a month post-symptom onset, IgA titers tended to decrease in the individuals sampled one-month post onset as compared to the early timepoints (Figure 3A). Thus, the antibody responses to SARS-COV-2 infection were dominated by IgG, even early after infection, illustrating that isotype switching occurs rapidly during the acute infection, with lower level responses of the IgM and IgA isotypes also detectable in most donors.
Figure 3. RBD-binding fraction of patient serum antibody strongly correlates with neutralization capacity.
(A) ELISA endpoint titers for serum binding against SARS-CoV-2 receptor binding domain (RBD), Spike (S), and nucleoprotein (NP) recombinant protein from a cohort of acutely infected individuals (n=46). Significance is calculated by Brown-Forsythe ANOVA test. *p≤0.05, **p≤0.01 (B) IgG binding titers against RBD, Spike, and NP correlated with SARS-CoV-2 serum neutralization activity. The coefficient of determination (r2) is reported following linear regression analysis.
Almost all of the acutely infected hospitalized patients had detectable SARS-CoV-2 neutralizing antibody responses (Figure 3B) with an average reciprocal titer of 568 and a range from 23 to 2205 (Supplementary Table 1). These responses displayed a strong correlation with RBD-specific IgG antibody titers, as we have previously shown (Figure 3B).(8) Although weaker than the correlation with RBD-specific IgG titers, neutralization titers also had a positive correlation with anti-S IgG titers (Figure 3B). Finally, NP-specific IgG titers correlated quite poorly with SARS-CoV-2 neutralization (Figure 3B). Overall, this data illustrates the occurrence of a rapid and robust antibody response to multiple SARS-CoV-2 antigens in individuals with severe COVID-19.
The RBD-specific serum fraction is responsible for neutralizing activity in a majority, but not all, hospitalized COVID-19 patients.
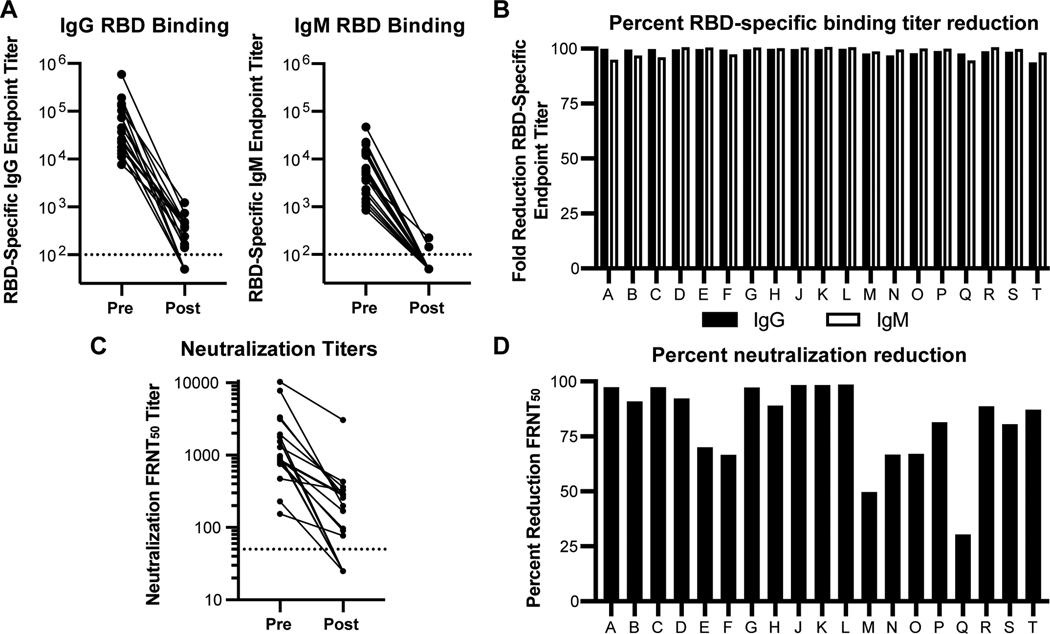
As has been previously reported, circulating titers of RBD-specific IgG correlated with time after disease onset(8) (Supplemental Fig 3) and with serum neutralizing potency (Figure 3B). Given differences in time of sampling between patients and limited clinical data, we were unable to correlate metrics of disease severity or resolution within this cohort to RBD-specific titers (Supplementary Table 1). To quantify the overall contribution of RBD-specific antibodies to SARS-CoV-2 viral neutralization, we depleted RBD binding antibodies from serum samples collected from a randomly selected subset of infected patients. To assess the effectiveness of the depletion, we determined the endpoint RBD binding titer for paired pre- and post-depletion serum samples. We found that all samples were efficiently depleted with an average percent reduction of RBD specific IgG of 98% (Figure 4B). Irrespective of the initial titer (Figure 4A/B), it is important to note that a subset of individuals had post-depletion titers that dropped below the limit of detection (Figure 4A). For these individuals, the fold reduction was estimated using half the limit of detection as a baseline value (10) and therefore may be greater than what was measurable in this assay. The pre- and post-depletion serum samples from each individual were then analyzed using a viral neutralization assay. Pre-depletion, reciprocal neutralization titers ranged from 154 to 10,270 with a median titer of 973 (Figure 4C). Post-depletion, 13 individuals had titers above baseline, and the remaining individuals had titers below the limit of detection (Figure 4C). The neutralization potency of depleted serum samples was markedly reduced in the majority of individuals assayed relative to pre-depletion control samples. Specifically, 13 of 19 of serum samples had a greater than 80% reduction in the viral neutralization titers as a consequence of depleting the RBD binding antibodies (Figure 4C&D). These results provide evidence that epitopes within the RBD are the main target of antibody-mediated viral neutralization in these individuals. In the remaining 6 individuals, 4 had >65% reduction in neutralization titer, and the remaining two individuals had 49.7% and 30.3% reductions in neutralization, respectively (Figure 4C&D). This observation indicates that these donors retained more than 30% of their neutralization activity despite RBD depletion (Figure 4D). This result indicates that over one third of the neutralizing activity in these individuals may be due to antibodies that do not bind directly to the RBD region of spike or that bind to a confirmation of RBD that is not preserved in its recombinant form. Taken together, this analysis shows that while the majority of neutralizing antibodies are RBD specific, some individuals may generate neutralizing responses that target non-RBD epitopes. These antibodies may represent an important class of immunoglobulins that could act in synergy with clinically relevant RBD-specific neutralizing antibodies or enhance protection to other coronaviruses and SARS-CoV-2 RBD escape variants.
Figure 4. SARS-CoV-2 viral neutralization activity is mediated by RBD specific antibody in a majority of COVID-19 patients.
(A) ELISA endpoint titers for SARS-CoV-2 receptor binding domain (RBD) specific IgG and IgM in sera from 19 acute COVID-19 patients before (Pre) and after (Post) depletion of RBD binding antibodies. (B) For each patient, bars represent the percent reduction in serum IgG (black) or IgM (white) RBD binding endpoint titers relative to pre-depleted samples. (C) Serum neutralization activity against SARS-CoV-2 before and after depleting RBD binding antibodies. Values represent the FRNT50 titer. (D) For each donor, the effect of reducing RBD binding serum antibodies on the viral neutralization titer is expressed as percent reduction in the FRNT50 value relative to pre-depleted samples.
Discussion
Important components of the humoral response to viral infection include not only a rapid expansion of antibody secreting cells (ASCs) to boost circulating serum titers towards the invading pathogen but also the formation of an antigen-specific memory B cell pool responsible for long-lasting protection. While multiple groups have described strong B cell responses in SARS-CoV-2 patients, and RBD specific memory B cells encoding neutralizing antibodies at convalescence, the dynamics of these responses have not been well characterized, either cross-sectionally or longitudinally. Furthermore, several recent reports(20,21) have described “dysregulated” B cell responses during severe SARS-CoV-2 infection, suggesting mechanisms that could lead to ineffective and short-lived antibody responses, as in the case of chronic viral infections such as HIV(31) and HCV.(32) A failure to develop or a later loss of germinal center structures within the lymph nodes of deceased COVID-19 patients(16) and the abundance of several extrafollicular B cell populations in severe COVID-19 patients, normally observed in autoimmune individuals (such as double negative (DN) B cells)(20), have suggested this dysregulation. In our cohort of acutely infected SARS-CoV-2 patients, we found robust infection-induced plasmablast responses and the development of RBD-specific MBCs, which exhibited greater activation than their non-RBD-specific counterparts. Taken together, this data provides evidence for robust and functional humoral response to SARS-CoV-2 infection even in the face of severe disease. However, we also observe that, within the RBD-specific MBC compartment, a significant fraction of the cells are negative for CD27, a population that has been previously described to have an extrafollicular origin.(22) This finding is line with previous reports(16,20,21) that SARS-CoV-2 infection generates an extrafollicular response and raises questions as to the contribution of these extrafollicular subsets to the robustness and durability of the immune response against SARS-CoV-2. In addition, we observe a significant decrease of the unswitched MBC population in infected individuals, which could also be suggestive of immune dysregulation. These findings clearly highlight the heterogeneity of COVID-19 as a disease and the continued need to dissect the cellular response to SARS-CoV-2 infection.
In the case of previously studied coronaviruses, both pandemic and endemic species, seroconversion has been reported to take place within 2–3 weeks from the time of infection.(33–35) It has now been well-documented in cohorts containing both mild and severe cases of COVID-19 that, on average, seroconversion takes place two weeks post-infection(9,10). Our serological analysis of a cohort of severe COVID-19 patients supports the findings of previously published reports with the majority of individuals exhibiting positive titers against multiple SARS-CoV-2 antigens by two weeks post symptom onset. This analysis suggests that, even in individuals with severe COVID-19, the humoral response to SARS-CoV-2 remains functional. In fact, as has been previously reported(9,11), individuals with greater disease severity tend to have higher levels of RBD-binding antibodies in circulation, suggesting a robust humoral response to infection. While this trend could be potentially due to later seroconversion in these individuals, i.e. a delayed antibody response, previous studies of SARS-CoV patients demonstrate that patients with both earlier seroconversion and, in some cases, higher antibody titers were more likely to experience severe disease.(36,37) The data presented herein supports a model in which neutralizing antibodies may be insufficient for mitigating disease progression and pathology in certain individuals. Critical questions remain as to why circulating antibody responses observed in our cohort were unable to prevent severe disease given that the serum antibodies were able to bind effectively to multiple viral antigens and potently neutralize the virus in vitro. Thus, the contribution and functional role of the humoral response in severe SARS-CoV-2 infection in vivo still needs to be elucidated.
We have previously reported a highly significant correlation between serum neutralizing potency in vitro and RBD binding titers (8), as have others.(9,11) However, recent investigations into both the cellular and serological aspects of the B cell response to SARS-CoV-2 infection have begun to raise questions about the contribution of antibodies derived against additional antigenic targets.(17) While the strongest correlation within our cohort is undoubtably between anti-RBD IgG titers and serum neutralization, significant correlations can also be found between full length S and NP antibody titers and serum neutralization. Despite the significant body of evidence now exists that supports the neutralizing potential of RBD-specific antibodies(38), it is possible that antibodies targeting epitopes outside of the RBD epitopes could also contribute to neutralization potency. To provide quantitative evidence for the role of RBD-specific antibodies in circulating serum neutralization, we employed a strategy similar to that previously published by He, et al after the SARS pandemic in 2002–2003.(39) We show that depletion of the RBD-specific serum fraction reduced the neutralizing potency of the remaining serum antibody by greater than 80% in 13 out 19 individual serum samples tested. Interestingly, the percentage of RBD-specific B cells observed within the cohort represented an exceedingly small percentage of the overall MBC population. The contrast between the small percentage of RBD-specific MBCs observed and their potent contribution to the neutralizing activity is echoed by the findings of Rogers et al., where the percentage of RBD specific antibodies derived from spike-specific MBCs was minimal and yet the RBD-specific antibodies contributed an equal number of neutralizing antibodies as their non-RBD counterparts.(40) In addition, this group also found that the non-RBD antibodies had lower neutralization than their RBD-specific counterparts and failed to provide protection in an in vivo small animal model.(40) In contrast, a subset of the donors we analyzed showed a significant residual activity after RBD depletion, such that greater than 30% of the neutralization activity remained after depletion. There could be a number of explanations for this difference. It is possible that non-RBD antibodies are most potent in a synergistic environment in which antibodies against multiple epitopes or antigens act together to elicit neutralizing responses – this hypothesis would explain why neutralization effects observed in serum are not seen when testing monoclonal antibodies. Alternatively, these donors may have initiated a response that produced potently neutralizing antibodies to non-RBD epitopes. Analysis of monoclonal antibodies derived from these donors is currently ongoing. Thus, in conclusion, while our study shows that the majority of neutralizing activity in circulating serum is driven by RBD-specific antibodies, questions remain concerning the importance and combinatorial potency of non-RBD antibodies in vivo. This finding has potential implications for vaccine design, as it appears that generation of antibodies targeting solely SARS-CoV-2 RBD are sufficient for viral neutralization in the majority of individuals assayed. Thus, vaccines containing RBD rather than FL Spike or whole virus would seem likely provide sufficient, if not greater, elicitation of SARS-CoV-2 neutralizing antibodies. It was also found that a subset of individuals possess neutralizing antibodies targeting potentially non-RBD epitopes, which could lead to the discovery of potent neutralization targets outside the RBD. However, further investigation is necessary to ascertain the targets of the neutralizing antibodies, as confirmation of RBD can be critical for the function of specific antibody subsets and thus, we cannot confirm that these antibodies do not bind to RBD in some form. Taken together, these findings serve as a platform for further exploration of the immune response to SARS-CoV-2 and will be instructive for current vaccine design and development and optimization of prophylactic and therapeutic strategies based on monoclonal antibodies
Supplementary Material
Key Points.
Increased plasmblasts/activated RBD-specific MBCs observed SARS-CoV-2 infection.
RBD/S/NP titers increase over infection; RBD titers correlate with neutralization.
RBD-specific antibody depletion greatly reduces neutralization in most individuals.
Acknowledgements
We would like to acknowledge the contribution of the staff at both the Vaccine Research Center at Emory Children’s Center and Hope Clinic for coordinating the clinical aspects of this study.
Financial Support Footnote
The research reported in this publication was supported in part by an Emory EVPHA Synergy Fund award (M.S.S. and J.W.), COVID-Catalyst-I3 Funds from the Woodruff Health Sciences Center (M.S.S), Center for Childhood Infections and Vaccines (J.W and M.S.S), Children’s Healthcare of Atlanta (J.W and M.S.S), Woodruff Health Sciences Center 2020 COVID-19 CURE Award (M.S.S), and by the National Institutes of Health (NIH) through the National Institute for Allergy and Infectious Diseases under the award numbers ORIP/OD P51OD011132 (M.S.S), 3U19AI057266-17S1 (J.W, R.A and M.S.S.), and the Infectious Diseases Clinical Research Consortium UM1AI148684 (N.R., E.J.A, J.W., M.S.S.), R00AG049092 (V.D.M) and the World Reference Center for Emerging Viruses and Arboviruses R24AI120942 (V.D.M). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Declaration of Interests
E.J.A has received personal fees from AbbVie, Pfizer, and Sanofi Pasteur for consulting, and his institution receives funds to conduct clinical research unrelated to this manuscript from MedImmune, Regeneron, PaxVax, Pfizer, GSK, Merck, Novavax, Sanofi-Pasteur, Janssen, and Micron. He also serves on a safety monitoring board for Kentucky BioProcessing, Inc.
References
- 1.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, & Shi ZL 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. doi: 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong E, Du H, & Gardner L. 2020. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis, 20(5), 533–534. doi: 10.1016/S1473-3099(20)30120-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oliver SE, Gargano JW, Marin M, Wallace M, Curran KG, Chamberland M, McClung N, Campos-Outcalt D, Morgan RL, Mbaeyi S, Romero JR, Talbot HK, Lee GM, Bell BP, & Dooling K. 2020. The Advisory Committee on Immunization Practices’ Interim Recommendation for Use of Pfizer-BioNTech COVID-19 Vaccine - United States, December 2020. MMWR Morb Mortal Wkly Rep, 69(50), 1922–1924. doi: 10.15585/mmwr.mm6950e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oliver SE, Gargano JW, Marin M, Wallace M, Curran KG, Chamberland M, McClung N, Campos-Outcalt D, Morgan RL, Mbaeyi S, Romero JR, Talbot HK, Lee GM, Bell BP, & Dooling K. 2021. The Advisory Committee on Immunization Practices’ Interim Recommendation for Use of Moderna COVID-19 Vaccine - United States, December 2020. MMWR Morb Mortal Wkly Rep, 69(5152), 1653–1656. doi: 10.15585/mmwr.mm695152e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao J, Zhao S, Ou J, Zhang J, Lan W, Guan W, Wu X, Yan Y, Zhao W, Wu J, Chodosh J, & Zhang Q. 2020. COVID-19: Coronavirus Vaccine Development Updates. Front Immunol, 11, 602256. doi: 10.3389/fimmu.2020.602256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang AT, Garcia-Carreras B, Hitchings MDT, Yang B, Katzelnick LC, Rattigan SM, Borgert BA, Moreno CA, Solomon BD, Rodriguez-Barraquer I, Lessler J, Salje H, Burke D, Wesolowski A, & Cummings DAT 2020. A systematic review of antibody mediated immunity to coronaviruses: antibody kinetics, correlates of protection, and association of antibody responses with severity of disease. medRxiv. doi: 10.1101/2020.04.14.20065771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, & Veesler D. 2020. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell, 181(2), 281–292 e286. doi: 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suthar MS, Zimmerman M, Kauffman R, Mantus G, Linderman S, Vanderheiden A, Nyhoff L, Davis C, Adekunle S, Affer M, Sherman M, Reynolds S, Verkerke H, Alter DN, Guarner J, Bryksin J, Horwath M, Arthur C, Saakadze N, Smith GH, Edupuganti S, Scherer EM, Hellmeister K, Cheng A, Morales JA, Neish AS, Stowell SR, Frank F, Ortlund E, Anderson E, Menachery V, Rouphael N, Metha A, Stephens DS, Ahmed R, Roback J, & Wrammert J. 2020. Rapid generation of neutralizing antibody responses in COVID-19 patients. medRxiv. doi: 10.1101/2020.05.03.20084442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham NR, Whitaker AN, Strother CA, Miles AK, Grier D, McElvany BD, Bruce EA, Poynter ME, Pierce KK, Kirkpatrick BD, Stapleton RD, An G, Botten JW, Crothers JW, & Diehl SA 2020. Kinetics and Isotype Assessment of Antibodies Targeting the Spike Protein Receptor Binding Domain of SARS-CoV-2 In COVID-19 Patients as a function of Age and Biological Sex. medRxiv. doi: 10.1101/2020.07.15.20154443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iyer AS, Jones FK, Nodoushani A, Kelly M, Becker M, Slater D, Mills R, Teng E, Kamruzzaman M, Garcia-Beltran WF, Astudillo M, Yang D, Miller TE, Oliver E, Fischinger S, Atyeo C, Iafrate AJ, Calderwood SB, Lauer SA, Yu J, Li Z, Feldman J, Hauser BM, Caradonna TM, Branda JA, Turbett SE, LaRocque RC, Mellon G, Barouch DH, Schmidt AG, Azman AS, Alter G, Ryan ET, Harris JB, & Charles RC 2020. Dynamics and significance of the antibody response to SARS-CoV-2 infection. medRxiv. doi: 10.1101/2020.07.18.20155374 [DOI] [Google Scholar]
- 11.Robbiani DF, Gaebler C, Muecksch F, Lorenzi JCC, Wang Z, Cho A, Agudelo M, Barnes CO, Gazumyan A, Finkin S, Hagglof T, Oliveira TY, Viant C, Hurley A, Hoffmann HH, Millard KG, Kost RG, Cipolla M, Gordon K, Bianchini F, Chen ST, Ramos V, Patel R, Dizon J, Shimeliovich I, Mendoza P, Hartweger H, Nogueira L, Pack M, Horowitz J, Schmidt F, Weisblum Y, Michailidis E, Ashbrook AW, Waltari E, Pak JE, Huey-Tubman KE, Koranda N, Hoffman PR, West AP Jr., Rice CM, Hatziioannou T, Bjorkman PJ, Bieniasz PD, Caskey M, & Nussenzweig MC 2020. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. doi: 10.1038/s41586-020-2456-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du L, He Y, Zhou Y, Liu S, Zheng BJ, & Jiang S. 2009. The spike protein of SARS-CoV--a target for vaccine and therapeutic development. Nat Rev Microbiol, 7(3), 226–236. doi: 10.1038/nrmicro2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kreer C, Zehner M, Weber T, Ercanoglu MS, Gieselmann L, Rohde C, Halwe S, Korenkov M, Schommers P, Vanshylla K, Di Cristanziano V, Janicki H, Brinker R, Ashurov A, Krahling V, Kupke A, Cohen-Dvashi H, Koch M, Eckert JM, Lederer S, Pfeifer N, Wolf T, Vehreschild M, Wendtner C, Diskin R, Gruell H, Becker S, & Klein F. 2020. Longitudinal Isolation of Potent Near-Germline SARS-CoV-2-Neutralizing Antibodies from COVID-19 Patients. Cell. doi: 10.1016/j.cell.2020.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wan J, Xing S, Ding L, Wang Y, Gu C, Wu Y, Rong B, Li C, Wang S, Chen K, He C, Zhu D, Yuan S, Qiu C, Zhao C, Nie L, Gao Z, Jiao J, Zhang X, Wang X, Ying T, Wang H, Xie Y, Lu Y, Xu J, & Lan F. 2020. Human-IgG-Neutralizing Monoclonal Antibodies Block the SARS-CoV-2 Infection. Cell Rep, 32(3), 107918. doi: 10.1016/j.celrep.2020.107918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zost SJ, Gilchuk P, Case JB, Binshtein E, Chen RE, Nkolola JP, Schafer A, Reidy JX, Trivette A, Nargi RS, Sutton RE, Suryadevara N, Martinez DR, Williamson LE, Chen EC, Jones T, Day S, Myers L, Hassan AO, Kafai NM, Winkler ES, Fox JM, Shrihari S, Mueller BK, Meiler J, Chandrashekar A, Mercado NB, Steinhardt JJ, Ren K, Loo YM, Kallewaard NL, McCune BT, Keeler SP, Holtzman MJ, Barouch DH, Gralinski LE, Baric RS, Thackray LB, Diamond MS, Carnahan RH, & Crowe JE Jr. 2020. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. doi: 10.1038/s41586-020-2548-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaneko N, Kuo HH, Boucau J, Farmer JR, Allard-Chamard H, Mahajan VS, Piechocka-Trocha A, Lefteri K, Osborn M, Bals J, Bartsch YC, Bonheur N, Caradonna TM, Chevalier J, Chowdhury F, Diefenbach TJ, Einkauf K, Fallon J, Feldman J, Finn KK, Garcia-Broncano P, Hartana CA, Hauser BM, Jiang C, Kaplonek P, Karpell M, Koscher EC, Lian X, Liu H, Liu J, Ly NL, Michell AR, Rassadkina Y, Seiger K, Sessa L, Shin S, Singh N, Sun W, Sun X, Ticheli HJ, Waring MT, Zhu AL, Li J, Lingwood D, Schmidt AG, Lichterfeld M, Walker BD, Yu X, Padera RF, Pillai S, & Group M. 2020. The Loss of Bcl-6 Expressing T Follicular Helper Cells and the Absence of Germinal Centers in COVID-19. SSRN, 3652322. doi: 10.2139/ssrn.3652322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juno JA, Tan HX, Lee WS, Reynaldi A, Kelly HG, Wragg K, Esterbauer R, Kent HE, Batten CJ, Mordant FL, Gherardin NA, Pymm P, Dietrich MH, Scott NE, Tham WH, Godfrey DI, Subbarao K, Davenport MP, Kent SJ, & Wheatley AK 2020. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat Med. doi: 10.1038/s41591-020-0995-0 [DOI] [PubMed] [Google Scholar]
- 18.He Z, Dong Q, Zhuang H, Song S, Peng G, Luo G, & Dwyer DE 2004. Kinetics of severe acute respiratory syndrome (SARS) coronavirus-specific antibodies in 271 laboratory-confirmed cases of SARS. Clin Diagn Lab Immunol, 11(4), 792–794. doi: 10.1128/CDLI.11.4.792-794.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang F, Nie J, Wang H, Zhao Q, Xiong Y, Deng L, Song S, Ma Z, Mo P, & Zhang Y. 2020. Characteristics of Peripheral Lymphocyte Subset Alteration in COVID-19 Pneumonia. J Infect Dis, 221(11), 1762–1769. doi: 10.1093/infdis/jiaa150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodruff M, Ramonell R, Cashman K, Nguyen D, Ley A, Kyu S, Saini A, Haddad N, Chen W, Howell JC, Ozturk T, Lee S, Estrada J, Morrison-Porter A, Derrico A, Anam F, Wu H, Le S, Jenks S, Hu W, Lee FE, & Sanz I. 2020. Critically ill SARS-CoV-2 patients display lupus-like hallmarks of extrafollicular B cell activation. medRxiv. doi: 10.1101/2020.04.29.20083717 [DOI] [Google Scholar]
- 21.Oliviero B, Varchetta S, Mele D, Mantovani S, Cerino A, Perotti CG, Ludovisi S, & Mondelli MU 2020. Expansion of atypical memory B cells is a prominent feature of COVID-19. Cell Mol Immunol. doi: 10.1038/s41423-020-00542-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor JJ, Pape KA, & Jenkins MK 2012. A germinal center-independent pathway generates unswitched memory B cells early in the primary response. J Exp Med, 209(3), 597–606. doi: 10.1084/jem.20111696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bentivegna E, Sentimentale A, Luciani M, Speranza ML, Guerritore L, & Martelletti P. 2020. New IgM seroconversion and positive RT-PCR test after exposure to the virus in recovered COVID-19 patient. J Med Virol. doi: 10.1002/jmv.26160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.To KK, Hung IF, Ip JD, Chu AW, Chan WM, Tam AR, Fong CH, Yuan S, Tsoi HW, Ng AC, Lee LL, Wan P, Tso E, To WK, Tsang D, Chan KH, Huang JD, Kok KH, Cheng VC, & Yuen KY 2020. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis. doi: 10.1093/cid/ciaa1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie X, Muruato A, Lokugamage KG, Narayanan K, Zhang X, Zou J, Liu J, Schindewolf C, Bopp NE, Aguilar PV, Plante KS, Weaver SC, Makino S, LeDuc JW, Menachery VD, & Shi PY 2020. An Infectious cDNA Clone of SARS-CoV-2. Cell Host Microbe, 27(5), 841–848 e843. doi: 10.1016/j.chom.2020.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lucas C, Wong P, Klein J, Castro TBR, Silva J, Sundaram M, Ellingson MK, Mao T, Oh JE, Israelow B, Takahashi T, Tokuyama M, Lu P, Venkataraman A, Park A, Mohanty S, Wang H, Wyllie AL, Vogels CBF, Earnest R, Lapidus S, Ott IM, Moore AJ, Muenker MC, Fournier JB, Campbell M, Odio CD, Casanovas-Massana A, Yale IT, Herbst R, Shaw AC, Medzhitov R, Schulz WL, Grubaugh ND, Dela Cruz C, Farhadian S, Ko AI, Omer SB, & Iwasaki A. 2020. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature, 584(7821), 463–469. doi: 10.1038/s41586-020-2588-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng NY, Mays I, Garman L, Helms C, James J, Air GM, Capra JD, Ahmed R, & Wilson PC 2008. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature, 453(7195), 667–671. doi: 10.1038/nature06890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wrammert J, Onlamoon N, Akondy RS, Perng GC, Polsrila K, Chandele A, Kwissa M, Pulendran B, Wilson PC, Wittawatmongkol O, Yoksan S, Angkasekwinai N, Pattanapanyasat K, Chokephaibulkit K, & Ahmed R. 2012. Rapid and massive virus-specific plasmablast responses during acute dengue virus infection in humans. J Virol, 86(6), 2911–2918. doi: 10.1128/JVI.06075-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ellebedy AH, Jackson KJ, Kissick HT, Nakaya HI, Davis CW, Roskin KM, McElroy AK, Oshansky CM, Elbein R, Thomas S, Lyon GM, Spiropoulou CF, Mehta AK, Thomas PG, Boyd SD, & Ahmed R. 2016. Defining antigen-specific plasmablast and memory B cell subsets in human blood after viral infection or vaccination. Nat Immunol, 17(10), 1226–1234. doi: 10.1038/ni.3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanz I, Wei C, Jenks SA, Cashman KS, Tipton C, Woodruff MC, Hom J, & Lee FE 2019. Challenges and Opportunities for Consistent Classification of Human B Cell and Plasma Cell Populations. Front Immunol, 10, 2458. doi: 10.3389/fimmu.2019.02458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moir S, Ho J, Malaspina A, Wang W, DiPoto AC, O’Shea MA, Roby G, Kottilil S, Arthos J, Proschan MA, Chun TW, & Fauci AS 2008. Evidence for HIV-associated B cell exhaustion in a dysfunctional memory B cell compartment in HIV-infected viremic individuals. J Exp Med, 205(8), 1797–1805. doi: 10.1084/jem.20072683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doi H, Tanoue S, & Kaplan DE 2014. Peripheral CD27-CD21- B-cells represent an exhausted lymphocyte population in hepatitis C cirrhosis. Clin Immunol, 150(2), 184–191. doi: 10.1016/j.clim.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Callow KA, Parry HF, Sergeant M, & Tyrrell DA 1990. The time course of the immune response to experimental coronavirus infection of man. Epidemiol Infect, 105(2), 435–446. doi: 10.1017/s0950268800048019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsueh PR, Huang LM, Chen PJ, Kao CL, & Yang PC 2004. Chronological evolution of IgM, IgA, IgG and neutralisation antibodies after infection with SARS-associated coronavirus. Clin Microbiol Infect, 10(12), 1062–1066. doi: 10.1111/j.1469-0691.2004.01009.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ko JH, Muller MA, Seok H, Park GE, Lee JY, Cho SY, Ha YE, Baek JY, Kim SH, Kang JM, Kim YJ, Jo IJ, Chung CR, Hahn MJ, Drosten C, Kang CI, Chung DR, Song JH, Kang ES, & Peck KR 2017. Serologic responses of 42 MERS-coronavirus-infected patients according to the disease severity. Diagn Microbiol Infect Dis, 89(2), 106–111. doi: 10.1016/j.diagmicrobio.2017.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee N, Chan PK, Ip M, Wong E, Ho J, Ho C, Cockram CS, & Hui DS 2006. Anti-SARS-CoV IgG response in relation to disease severity of severe acute respiratory syndrome. J Clin Virol, 35(2), 179–184. doi: 10.1016/j.jcv.2005.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L, Zhang F, Yu W, He T, Yu J, Yi CE, Ba L, Li W, Farzan M, Chen Z, Yuen KY, & Ho D. 2006. Antibody responses against SARS coronavirus are correlated with disease outcome of infected individuals. J Med Virol, 78(1), 1–8. doi: 10.1002/jmv.20499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ju B, Zhang Q, Ge J, Wang R, Sun J, Ge X, Yu J, Shan S, Zhou B, Song S, Tang X, Yu J, Lan J, Yuan J, Wang H, Zhao J, Zhang S, Wang Y, Shi X, Liu L, Zhao J, Wang X, Zhang Z, & Zhang L. 2020. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature, 584(7819), 115–119. doi: 10.1038/s41586-020-2380-z [DOI] [PubMed] [Google Scholar]
- 39.He Y, Zhu Q, Liu S, Zhou Y, Yang B, Li J, & Jiang S. 2005. Identification of a critical neutralization determinant of severe acute respiratory syndrome (SARS)-associated coronavirus: importance for designing SARS vaccines. Virology, 334(1), 74–82. doi: 10.1016/j.virol.2005.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rogers TF, Zhao F, Huang D, Beutler N, Burns A, He WT, Limbo O, Smith C, Song G, Woehl J, Yang L, Abbott RK, Callaghan S, Garcia E, Hurtado J, Parren M, Peng L, Ramirez S, Ricketts J, Ricciardi MJ, Rawlings SA, Wu NC, Yuan M, Smith DM, Nemazee D, Teijaro JR, Voss JE, Wilson IA, Andrabi R, Briney B, Landais E, Sok D, Jardine JG, & Burton DR 2020. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science, 369(6506), 956–963. doi: 10.1126/science.abc7520 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.