Abstract
The first and second generations of the Cobas Amplicor HCV assay were compared among patients at risk of hepatitis C virus (HCV) infection. The second-generation test was found to be of greater sensitivity and of good specificity among clinical specimens containing HCV RNA of different genotypes. Finally, this new test is shown to predict the outcome of interferon therapy better.
The detection of hepatitis C virus (HCV) RNA by reverse transcription-PCR is the method of choice for monitoring HCV infection. Detection of HCV RNA by reverse transcription-PCR has been widely developed with standardized ready-to-use assays such as the Amplicor HCV test (5, 11). This test was successfully automated by using an integrated PCR system based on Cobas Amplicor technology (1, 3, 6) that fully automates all steps of PCR amplification and detection. The target HCV RNA sequence is amplified in the thermal cycler section of the Cobas analyzer with biotin-labelled primer KY78 and reverse primer KY80, which allow the amplification of a 244-bp sequence from the highly conserved 5′-untranslated region of the HCV genome (11). Following PCR amplification, biotin-labelled amplicons are specifically captured by target-specific oligonucleotide probes bound to magnetic particles and are colorimetrically detected (at 660 nM) through an enzyme-linked immunosorbent assay-like reaction. The Cobas Amplicor HCV assay, version 1.0 (CA v1.0), has been recently modified to generate a new version (CA v2.0), proposed as more reliable and of greater sensitivity. CA v2.0 uses more serum (200 μl instead of 100 μl), and nucleic acids are concentrated to one-fifth of the initial volume. These modifications would lead to an increase in the sensitivity of HCV RNA detection by an least 10-fold. In addition, a synthetic RNA containing HCV primer binding regions and a unique probe sequence is added with the lysis buffer before nucleic acid extraction from the serum. Thus, this RNA is extracted and amplified simultaneously with the clinical specimen and serves as an internal control (IC) to assure the successful recovery of nucleic acid.
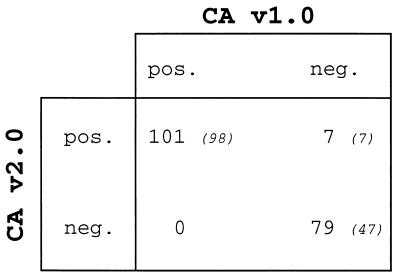
The presence of HCV RNA was investigated with both Cobas assays (CA v1.0 and CA v2.0) in 187 specimens consecutively collected in our laboratory from patients at risk of HCV infection (Fig. 1). Among these clinical specimens 152 were from HCV-seropositive patients, 10 were from children born to HCV-infected mothers, and 25 were from individuals without serological evidence of HCV infection but who either had been recently exposed to HCV or had an unexplained elevation of alanine aminotransferase levels. Each specimen was processed once with both assays, and the concordance between both assays was quite good since 180 specimens gave the same result (96.2%). Among the 152 HCV-seropositive patients, HCV RNA was detectable with both assays in sera from 98 patients and undetectable in 47 samples, and seven samples were discrepant (Fig. 1). The seven discrepant specimens which were all positive with CA v2.0 but negative with CA v1.0 were processed again in triplicate with both Cobas HCV versions (data not shown). With CA v1.0, repeat testings confirmed the seven samples as true negatives. With CA v2.0, four specimens were confirmed as positive with at least three of four positive determinations, but three specimens showed inconsistent results (one or two positive determinations of four) and finally were classified as indeterminate. Since these specimens were all from HCV-seropositive patients with documented HCV infections (three of them had increased levels of alanine aminotransferase), it is likely that these patients have a very low level of circulating HCV RNA.
FIG. 1.
Overall performance of CA v1.0 and CA v2.0. HCV RNA was detected with both Cobas assays in 187 clinical specimens consecutively collected from patients at risk of HCV infection. Data are the numbers of samples with the indicated test results. Italic numbers in parentheses are the numbers of HCV-seropositive patients in each group.
To confirm the greater sensitivity of CA v2.0, series of diluted specimens were analyzed in parallel with both assays (Table 1). Diluted specimens were prepared by diluting sera containing HCV RNA of different genotypes in a negative serum obtained from two healthy donors free of any sign of HCV infection. HCV genotypes were determined through the sequencing of the 5′ noncoding region of HCV RNA as previously described (4). The dilution factor was different for each specimen, and dilutions were undertaken until the HCV RNA detection signal became negative within each assay. As shown in Table 1, final positive dilutions of specimens were always much lower when processed with CA v2.0 than with CA v1.0. These results were independent of the viral genotype, showing that CA v2.0 works well with most of the genotypes encountered. Additionally, series of calibrated dilutions were also used in order to quantify the low limit detection level of CA v2.0 (Table 2). Calibrated dilutions were prepared by diluting a calibrated serum containing known amounts of HCV RNA 1b (Accurun HCV RNA from Boston Biomedica, Inc.), and testing was undertaken in triplicate. As shown in Table 2, samples containing approximately 100 copies of HCV RNA per ml were always positive but became negative below this limit (30 copies/ml). These results confirm the great sensitivity of CA v2.0, which is capable of detecting fewer than 100 copies of HCV RNA per ml in clinical specimens.
TABLE 1.
Detection of HCV RNA with both Cobas assays in series of diluted specimens
| HCV genotype | Last positive dilutiona (−log)
|
|
|---|---|---|
| CA v1.0 | CA v2.0 | |
| 1b | 4.0 | 6.0 |
| 1a | 4.0 | 6.0 |
| 1b | 5.0 | 6.0 |
| 3 | 3.0 | 4.8 |
| 1b | 3.0 | 4.9 |
| 3 | 1.4 | 2.8 |
| 3 | 1.4 | 2.8 |
| 4 | 0.4 | 3.3 |
| 2a/c | 0.95 | 3.3 |
Series of diluted specimens were tested until the HCV RNA signal was negative within each assay. The last dilution found positive is given in the table (log).
TABLE 2.
Determination of the low limit detection level of CA v2.0
| No. of HCV RNA copies/mla | Detection of:
|
|
|---|---|---|
| HCV RNAb (OD660) | IC | |
| 6 × 103 | Pos. (3.861) | Pos. |
| 103 | Pos. (3.556) | Pos. |
| 102 | Pos. (0.613) | Pos. |
| Pos. (1.633) | Pos. | |
| Pos. (0.498) | Pos. | |
| 30 | Neg. (0.008) | Pos. |
| Neg. (0.007) | Pos. | |
| Neg. (0.007) | Pos. | |
Series of diluted specimens containing known amounts of HCV RNA were obtained by diluting a calibrated serum (Accurun HCV RNA).
Crude readings of the optical density at 660 nm (OD660) are furnished by Cobas. Samples are positive (Pos.) with optical densities of >0.15 and negative (Neg.) below this value.
A large random screening of individuals was retrospectively conducted using 300 serum specimens collected in our virology laboratory from individuals free of human immunodeficiency virus or syphilis infection. Samples from this large group were processed with CA v2.0, and HCV RNA was detected in 11 specimens (3.6%) (data not shown). The high rate of positive samples encountered in this cohort could be explained by the fact that most specimens were from patients at risk of viral infection. Further investigations confirmed that these patients were in fact infected with HCV, since they all presented serological evidence of HCV infection. The concordance between HCV RNA detection and HCV serological status supports the findings of the good specificity of CA v2.0 in identifying HCV-infected people in large-scale screening.
Concurrently, 25 patients were selected from a larger group of patients treated with interferon (IFN) for chronic hepatitis C. These patients had received recombinant IFN-α2a (Roferon* A; Produits Roche) three times a week at a dose of 6 × 106 U for 3 months and 3 × 106 U for the following 9 months. The 25 patients were selected because of a relapse (9 patients) or a sustained response to therapy (16 patients) as defined 6 months after the cessation of therapy. Serum samples collected during and after therapy from these patients were retrospectively analyzed with CA v2.0, and the results were compared to those obtained previously with CA v1.0 (data not shown). Specimens processed with CA v2.0 were indicative of its high prognostic value, since it appeared that 1 month after the initiation of treatment, HCV RNA was detected in seven sera from the relapsers (78%) and in only four specimens from the responders (25%). In contrast, results were less informative when samples were processed with CA v1.0, since at the same time, HCV RNA was detected in only 44% of specimens from relapsers (four of nine). These observations confirmed previous data suggesting that it should be possible to predict a long-term response to IFN by identifying the presence or absence of HCV RNA a few weeks after the initiation of treatment (2, 7, 9, 10).
Taken together, our results show that the clinical relevance of HCV RNA detection in serum is reinforced by the use of a very sensitive test. Given the high sensitivity of the second-generation Cobas Amplicor assay the potential of contamination could be a serious concern. However, this assay contains the AmpErase enzyme, which is used to prevent the carryover contamination of previously amplified materials (8). On the other hand, the addition of an IC at the very beginning of the procedure ensures that serum specimens are correctly processed and that RNA is not lost during the extraction steps. From more than 500 processed samples, we have failed to detect the IC in six samples (1.2%) (data not shown). However, the IC was always found to be positive in these cases when specimens were processed again with a new aliquot of serum (data not shown). Therefore, CA v2.0 appears to be very well suited for the monitoring and diagnosis of HCV infection, since this standardized automated assay is shown to be very sensitive. Thus, it meets most of the required criteria for wide, large-scale screening of HCV infection or for monitoring HCV replication in infected patients.
Acknowledgments
Special thanks are due to F. Eberle (Roche Diagnostics) for providing the assays and helpful discussions. Sylvain Thyss is acknowledged for expert technical assistance.
This work was supported by the Agence Française du Sang and the Institut National de la Santé et de la Recherche Médicale (contrat AFS no. 96019).
REFERENCES
- 1.Albadalejo J, Alonso R, Antinozzi R, Bogard M, Bourgault A-M, Colucci G, Fenner T, Petersen H, Sala E, Vincelette J, Young C. Multicenter evaluation of the COBAS AMPLICOR HCV assay, an integrated PCR system for rapid detection of hepatitis C virus RNA in the diagnostic laboratory. J Clin Microbiol. 1998;36:862–865. doi: 10.1128/jcm.36.4.862-865.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis G L, Lau J Y. Factors predictive of a beneficial response to therapy of hepatitis C. Hepatology. 1997;26:122S–127S. doi: 10.1002/hep.510260721. [DOI] [PubMed] [Google Scholar]
- 3.DiDomenico N, Link H, Knobel R, Caratsch T, Weschler W, Loewy Z G, Rosenstraus M. COBAS AMPLICOR: fully automated RNA and DNA amplification and detection system for routine diagnostic PCR. Clin Chem. 1996;42:1915–1923. [PubMed] [Google Scholar]
- 4.Doglio A, Laffont C, Thyss S, Lefebvre J. Rapid genotyping of hepatitis C virus by direct cycle sequencing of PCR-amplified cDNAs and capillary electrophoresis. Res Virol. 1998;149:219–227. doi: 10.1016/s0923-2516(98)80003-4. [DOI] [PubMed] [Google Scholar]
- 5.Gerken G, Pontisso P, Roggendorf M, Grazia R M, Simmonds P, Trepo C, Zeuzem S, Colucci G. Clinical evaluation of a single reaction, diagnostic polymerase chain reaction assay for the detection of hepatitis C virus RNA. J Hepatol. 1996;24:33–37. doi: 10.1016/s0168-8278(96)80183-8. [DOI] [PubMed] [Google Scholar]
- 6.Jungkind D, DiRenzo S, Beavis K G, Silverman N S. Evaluation of automated COBAS AMPLICOR PCR system for detection of several infectious agents and its impact on laboratory management. J Clin Microbiol. 1996;34:2778–2783. doi: 10.1128/jcm.34.11.2778-2783.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kakumu S, Aiyama T, Okumura A, Iwata K, Ishikawa T, Yoshioka K. Earlier loss of hepatitis C virus RNA in interferon therapy can predict a long-term response in chronic hepatitis C. J Gastroenterol Hepatol. 1997;12:468–472. doi: 10.1111/j.1440-1746.1997.tb00468.x. [DOI] [PubMed] [Google Scholar]
- 8.Longo M C, Berninger M S, Hartley J L. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene. 1990;93:125–128. doi: 10.1016/0378-1119(90)90145-h. [DOI] [PubMed] [Google Scholar]
- 9.Tong M J, Blatt L M, McHutchison J G, Co R L, Conrad A. Prediction of response during interferon alfa 2b therapy in chronic hepatitis C patients using viral and biochemical characteristics: a comparison. Hepatology. 1997;26:1640–1645. doi: 10.1002/hep.510260637. [DOI] [PubMed] [Google Scholar]
- 10.Yamakawa Y, Sata M, Suzuki H, Tanaka K, Tanaka E, Noguchi S, Ono K, Tanikawa K. Monitoring of serum levels of HCV RNA in early phase of IFN therapy; as a predictive marker of subsequent response. Hepatogastroenterology. 1998;45:133–136. [PubMed] [Google Scholar]
- 11.Young K K Y, Resnick R M, Myers T W. Detection of hepatitis C virus RNA by a combined reverse transcription-polymerase chain reaction assay. J Clin Microbiol. 1993;31:882–886. doi: 10.1128/jcm.31.4.882-886.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]