Summary
Force generation in epithelial tissues is often pulsatile, with actomyosin networks generating contractile forces before cyclically disassembling. This pulsed nature of cytoskeletal forces implies there must be ratcheting mechanisms that drive processive transformations in cell shape. Previous work has shown that force generation is coordinated with endocytic remodeling – however, how ratcheting becomes engaged at specific cell surfaces remains unclear. Here, we report that PtdIns(3,4,5)P3 is a critical lipid-based cue for ratcheting engagement. The Sbf RabGEF binds to PIP3, and disruption of PIP3 reveals a dramatic switching behavior in which medial ratcheting is activated and epithelial cells begin globally constricting apical surfaces. PIP3 enrichments are developmentally regulated, with mesodermal cells having high apical PIP3 while germband cells have higher interfacial PIP3. Finally, we show that JAK/STAT signaling constitutes a second pathway that combinatorially regulates Sbf/Rab35 recruitment. Our results elucidate a complex lipid-dependent regulatory machinery that directs ratcheting engagement in epithelial tissues.
Graphical Abstract
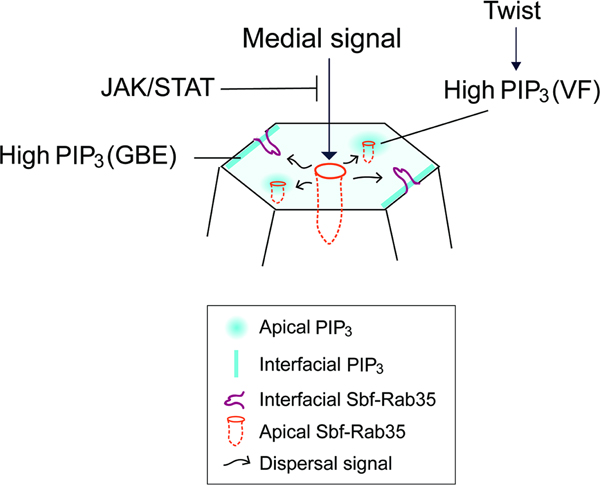
eTOC Blurb:
Miao et al. examine how PIP phospholipid cues direct contractile ratcheting engagement at specific epithelial surfaces. PtdIns(3,4,5)P3 is developmentally patterned and regulates a switching behavior in Sbf/Rab35 cell ratcheting - loss of PIP3 leads to global constriction of cell apices, while junctional enrichment of PIP3 directs interface contraction driving cell intercalation.
Introduction
Cell shaping processes use contractile force generation to drive the active contraction of specific cell surfaces that causes tissues to adopt new morphogenetic forms. This selective contraction of cell surfaces drives a diverse range of processes from tissue invagination to cell intercalation to epithelial cell extrusion and wound healing (Martin et al., 2009; Rauzi et al., 2010; Blanchard et al., 2010; David et al., 2010; Fernandez-Gonzalez et al., 2011; Sawyer et al., 2011; Michel and Dahmann, 2020). A key discovery in the last decade of work on these processes is that they are often pulsatile in nature, with highly-transient actomyosin populations that briefly apply a tensioning force to an area of the cell cortex (Mason and Martin, 2011; Levayer and Lecuit, 2012; Loerke and Blankenship, 2020). This has raised a central outstanding question – if actomyosin networks display such pulsed behaviors, how does processivity emerge from cyclic systems? To obtain lasting changes from such systems, there is a requirement for ratcheting mechanisms that gain irreversible changes out of periodic, contractile cycles.
Several potential molecular mechanisms of cell ratcheting have been discovered (Solon et al., 2009; Martin et al., 2009; Levayer et al., 2011; Munjal et al., 2015; Clément et al., 2017; Jewett et al., 2017; Cavanaugh et al., 2020), ranging from processes that direct the turnover and reformation of elastic cortical networks to pathways that rely on direct remodeling of the plasma membrane. Previous work has shown that a membranous ratchet centered on SET domain binding factor (Sbf) RabGEF and Rab35 function is a critical part of the pathway used to remodel the cell surfaces during early morphogenesis in Drosophila (Jewett et al., 2017; Miao et al., 2019). This membrane ratcheting module is deployable to specific cell surfaces – during cell intercalation, Sbf and Rab35 tubular invaginations of the plasma membrane are primarily found at planar polarized contracting interfaces, where they mediate the processive loss of AP (anterior-posterior) surfaces required for neighbor exchange through endocytic mechanisms (Jewett et al., 2017). Alternatively, during the apical constrictions that drive invagination of the ventrally-located mesoderm, Sbf and Rab35 are enriched at the apical surface, where they direct the ratcheted loss of apical surface areas (Miao et al., 2019). However, how this ratcheting activity is selectively recruited to specific cell surfaces is unknown. Although actomyosin function is required to terminate Sbf/Rab35 compartments, it is not required for Sbf and Rab35 recruitment (Jewett et al., 2017). Thus, Sbf and Rab35 recruitment is not downstream of actomyosin function, suggesting that yet-to-be-identified mechanisms are in place to control Sbf/Rab35 compartmental formation.
One potential important cue for directing the localized activities of plasma membrane-associated proteins are the lipid-signaling phosphoinositides (PIPs), which are often found in microdomains in the plasma membrane. PIPs have been shown to be potent regulators of both membrane trafficking and cytoskeletal networks (Sakisaka et al., 1997; Sechi and Wehland, 2000; Gassama-Diagne et al., 2006; Sun et al., 2007; Zhang and McCormick, 2010). The various PIP phospho-species, especially PI(4,5)P2 and PI(3,4,5)P3, can regulate the activity of a variety of endocytic-regulatory proteins, such as AP-2 and Dynamin (Czech, 2000; Gaidarov and Keen, 1999; Salim et al., 1996; Naga Prasad et al., 2002). PIPs are also implicated in controlling actin assembly and plasma membrane-cytoskeletal linkage by binding directly and tightly to at least 30 regulatory proteins (Janmey et al., 1999; Oikawa et al., 2004; Goldschmidt-Clermont et al., 1990). PI(3,4,5)P3 in particular has been deeply implicated in the regulation of membrane trafficking processes such as regulated endocytosis and exocytosis. Upregulation of PIP3 levels induces recycling of the epidermal growth factor receptor to the cell surface (Laketa et al., 2014), and PIP phospho-balance can regulate syncytial cleavage furrow lengths (Reversi et al., 2014). Further, recently published work suggests that PIPs may lay downstream of Toll receptor activity and Src activation in the germband epithelium (Tamada et al., 2021). However, how PIPs regulate the dynamic cell shape changes that occur as epithelial sheets change dimensions is unclear, and the function of PIP phospho-species during Drosophila gastrulation remains to be closely examined. Here, we identify phosphatidylinositol phosphates as providing lipid-based membrane cues for morphogenesis and cellular gastrulation dynamics in the early Drosophila embryo. We find that PI(3,4,5)P3 inhibition leads to a potent disruption of contractile cellular behaviors, and that PIP3 function controls a “constriction switch” that determines if epithelial cells enter into an apical constriction or cell interface contraction regime. This switch in contractile surfaces then determines which of two primary morphogenetic paradigms the developing tissue will follow – a primarily intercalatory process driven by neighbor exchange events, or the loss of apical areas that lead to furrow formation and tissue invagination.
Results
PtdIns(3,4,5)P3 controls the recruitment of Sbf-Rab35 compartments to specific cell surfaces
We previously identified a Sbf-Rab35 centered membrane trafficking pathway that directs the processive loss of cell surface areas. During Drosophila gastrulation, these endocytic compartments localize to either: 1) shrinking cell interfaces to drive cell intercalation, or 2) to contracting apical surfaces to promote apical constriction (Jewett et al., 2017; Miao et al., 2019). However, the pathways that regulate the engagement of Sbf-Rab35 ratcheting function at precise locations on the plasma membrane are unknown. Sbf is an interesting multidomain protein that contains a conserved PH domain (Fig. 1A), as well as a tripartite DENN domain that functions as a GEF for the Rab35 small GTPase. The presence of a PH domain could be indicative of a function for phosphoinositides (PIPs) in regulating Sbf localization and function. As a starting point to address potential Sbf interactions with PIPs, we used an in vitro lipid binding assay to assess whether Sbf binds specific PIP phosphoforms. To this end, we purified the Sbf-PH domain (amino acids 1890–1993) and performed a protein-lipid overlay assay. This revealed that Sbf-PH binds specifically to phosphatidylinositol (3,4,5)-trisphosphate (PIP3) and, at lower levels, to PtdIns(3,5)P2 (Fig. 1B). As this binding suggests a potential interaction, we assessed whether PIP3 was present at Rab35 tubular plasma membrane compartments in vivo. Rab35 compartments at cell interfaces in the intercalating germband were found to colocalize with a biosensor for PIP3, tGPH (a fusion protein composed of GFP and the PH domain of GRP1; Britton et al., 2002), although PIP3 is also found along the interfacial plasma membrane (Fig. 1C; Fig. S1A) (Britton et al., 2002).
Figure 1. PIP3 recruits Sbf-Rab35 compartments to interfaces during cell intercalation.
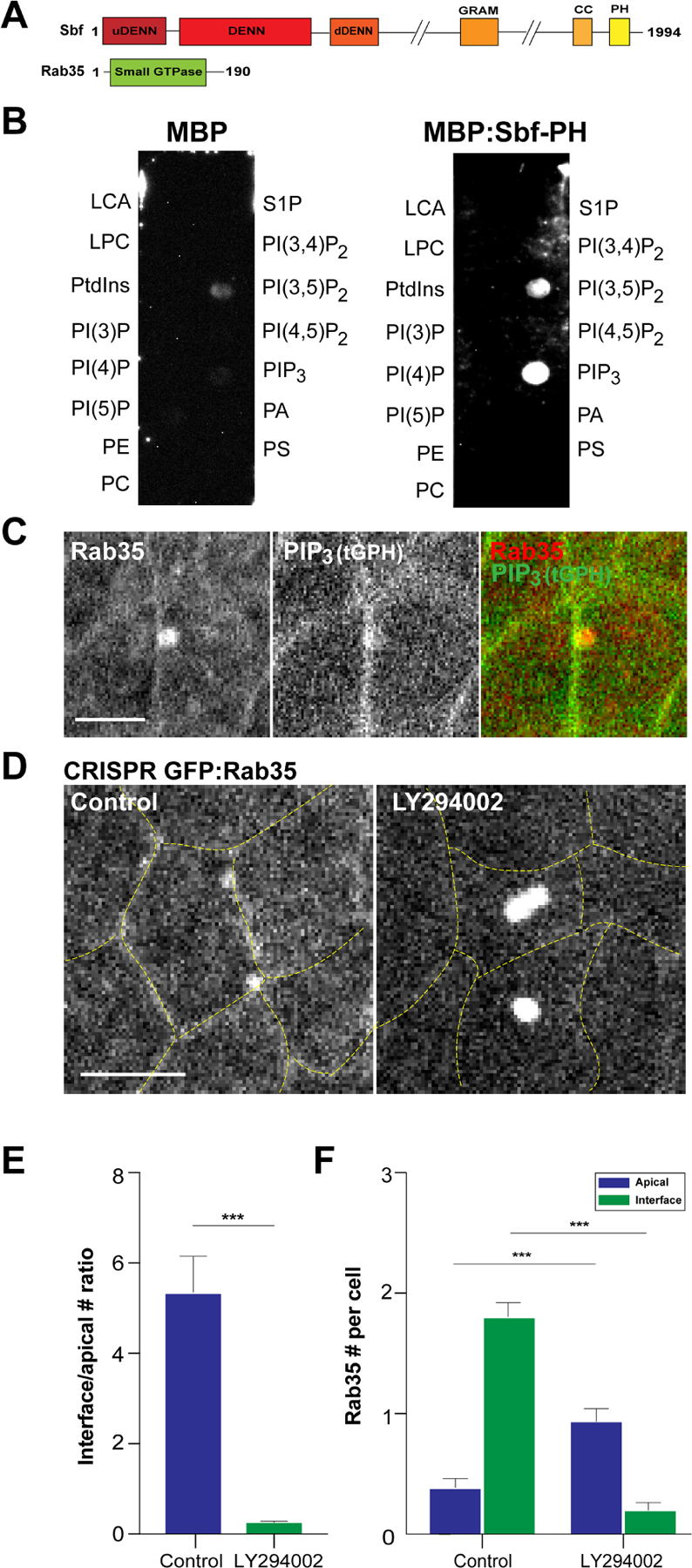
(A) Structural domains of Drosophila Sbf and Rab35 protein. (B) Protein-lipid binding assay of MBP and MBP:Sbf-PH with indicated lipids. (C) Two-color live imaging of mCh:Rab35 and PIP3 sensor (tGPH). (D) Still images of embryo expressing CRISPR GFP:Rab35 after either control or LY294002 injections. Yellow dashed lines mark cell outlines. (E) Ratio between the number of Rab35 compartments at interface or apical surfaces after control or LY294002 injections. (F) Absolute number of Rab35 compartments at interface or apical surfaces (per cell) in control or LY294002 background. n= 105 cells from 3 embryos (control) and 108 cells from 3 embryos (LY294002) in (E) and (F). Scale bars in (C) and (D) are 2.5 µm and 5 µm, respectively. Error bars indicate measured standard error; statistical significance has been calculated using Mann-Whitney U-test. ***P<0.0005.
These results are consistent with Sbf potentially interacting with plasma membrane PIP3, so we then explored whether Sbf-Rab35 compartmental dynamics are altered when PIP3 levels are disrupted (as demonstrated by tGPH, PIP3 biosensor levels after injection with PI3K class I inhibitor, LY294002; Fig. S1B,C). We observed a remarkable change in Sbf-Rab35 compartmental behaviors after injection with LY294002. Interestingly, interface-associated Sbf-Rab35 compartments are almost completely lost, indicating PIP3 is required for Sbf-Rab35 localization to contracting cell interfaces during cell intercalation (Fig. 1D,E,F, Video1). However, Sbf-Rab35 is not simply lost from the cell surface – instead, strikingly large Sbf-Rab35 compartments form in the middle of the apical cell surface (Fig. 1D,E,F; Fig. S1D,E). These medio-apical compartments are reminiscent of the apical Rab35 tubules that form during apical constriction (Miao et al., 2019), although their formation is even more robust than in the invaginating mesoderm (Fig. S1F). These results suggest that PIP3 function may direct the relative engagement of Sbf-Rab35 compartmental function at cell interfaces versus cell apices, and also indicates the potential presence of a medio-apical signaling pathway that competes with PIP3 signaling to direct Sbf/Rab35 recruitment at specific cell surfaces.
Pi3K class I protein function represses medio-apical formation of Sbf-Rab35 compartments
As Sbf-Rab35 function has been shown to be essential for the morphogenetic movements that drive gastrulation (Jewett et al., 2017; Miao et al., 2019), we explored whether PIP function is similarly required in the Drosophila epithelium at these stages. We therefore performed a screen of the 13 different PI kinases present in the fly genome by using a collection of bioinformatically validated shRNA lines (DSRC/TRiP genome collection, https://fgr.hms.harvard.edu/trip-in-vivo-fly-rnai). We found that embryos that were compromised for the function of PI3K class I genes (Pi3K92E and Pi3K21B) displayed defects during gastrulation, although these defects are in the moderate range when scored at the level of gross tissue morphologies (Fig. 2A). Additionally, disrupting the function of the Fab1 kinase, which phosphorylates the 5’ position in PIPs, also leads to defects in early syncytial development prior to gastrulation, consistent with data from previous biochemically-driven disruptions (Reversi et al., 2014).
Figure 2. A screen of PIP kinases reveals a PIP3-dependent localization switch.
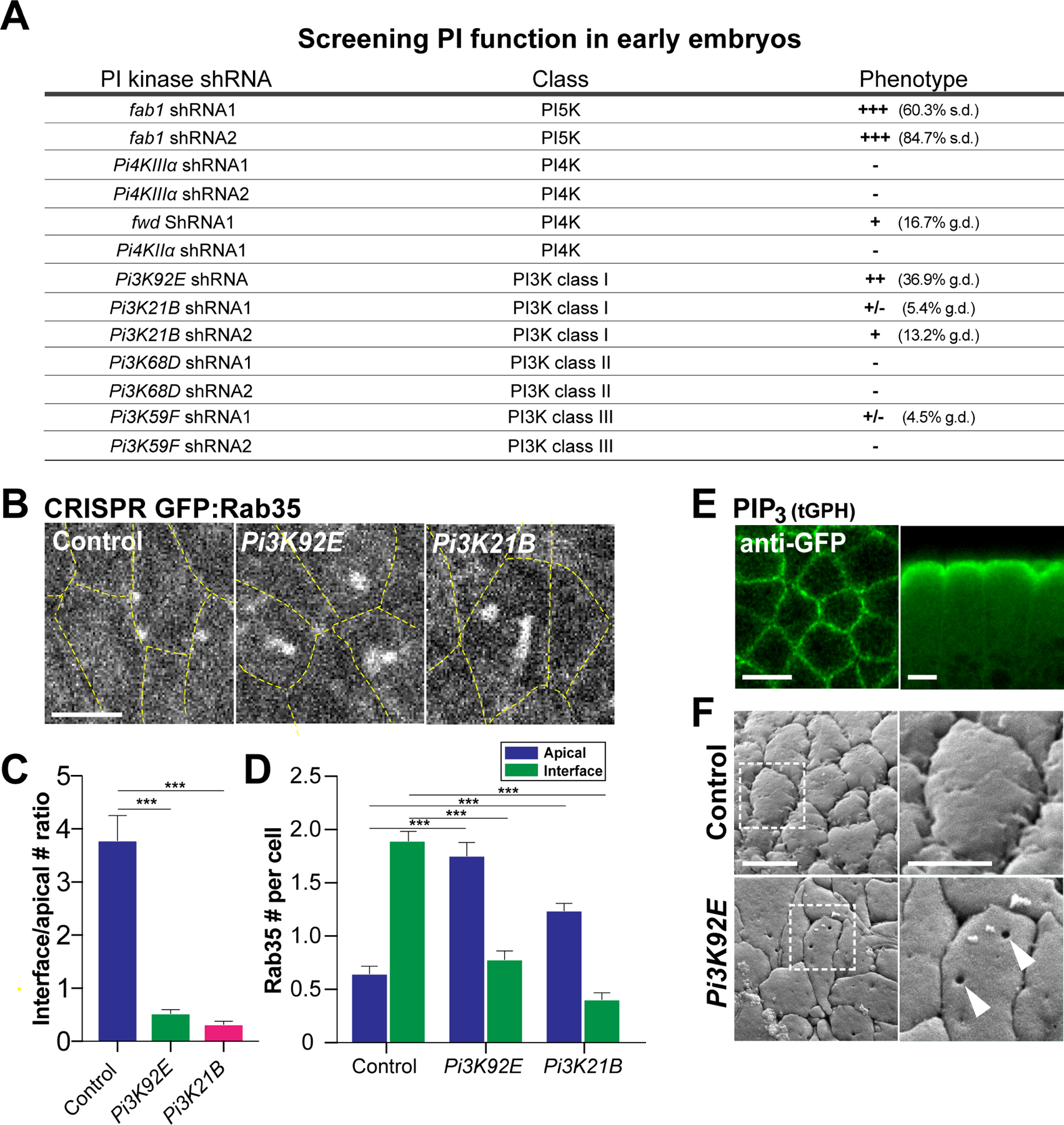
(A) shRNA screen of the genomic Drosophila PI Kinases as scored by gross tissue morphologies. n>120 embryos for each PI Kinase shRNA line. Phenotypic categories: +/−, +,++ and +++ represent 0–10%, 10%−30%, 30%−60% and 60% and above defective embryos, respectively. s.d. stands for syncytial division defects and g.d. represents gastrulation defects (typically embryos with a 50% or greater reduction in the amount of germband extension). For comparison purposes, ~95% of LY294002 injected embryos displayed gross gastrulation. (B) Images of CRISPR GFP:Rab35 in control, Pi3K92E shRNA and Pi3K21B shRNA embryos. Yellow dashed lines mark cell outlines. (C) Ratio of the number of Rab35 compartments at interfaces to apical surfaces in control, Pi3K92E shRNA and Pi3K21B shRNA cells. (D) Absolute number of Rab35 compartments at interfaces or apical surfaces (per cell) in control, Pi3K92E shRNA and Pi3K21B shRNA cells. n= 102 cells from 3 embryos (control), 105 cells from 3 embryos (Pi3K92E shRNA) and 101 cells from 3 embryos (Pi3K21B shRNA) in (C) and (D). (E) Immunostaining images of PIP3 sensor, tGPH in either planar (left) or apical-basal orientations (right). (F) SEM images of germband cells in embryos expressing control or Pi3K92E shRNA. The white dashed boxes on the left panels mark the magnified regions shown on the right. Arrowheads mark invaginated pits. Scale bars in (B) and (E) equal 5 µm. Scale bar in (F) left panel equals 5 µm and the scale bar in right panel equals 2.5 µm. Error bars indicate measured standard error; statistical significance has been calculated using Mann-Whitney U-test in (C) and (D). ***P<0.0005.
The PI3K class I proteins, Pi3K92E and Pi3K21B, like their mammalian homologues, p110 and p60, can phosphorylate phosphatidylinositol (4,5)-bisphosphate (PIP2) to generate PIP3 (Leevers et al., 1996). Indeed, PIP3 levels are significantly reduced in PI3K class I deficient embryos when examined in the gastrulating epithelium (Fig. S2A–E,H). Importantly, although the penetrance of Pi3K92E and Pi3K21B shRNAs is not high when scoring gross tissue morphologies under a dissecting scope, 100% of embryos displayed gastrulation defects at the cellular level. Similar to LY294002 injection results (Fig. 1D–F), large medio-apical Sbf-Rab35 compartments form in both Pi3K92E and Pi3K21B disrupted embryos, and there is a loss of interface-associated compartments (Fig. 2B–D). The large apical compartments fill immediately after extracellular injection of dextran, consistent with previous work demonstrating that that both interfacial and apical Rab35 compartments are tubular invaginations of the plasma membrane (Fig. S2F; Jewett et al., 2017; Miao et al., 2019). This provides further evidence that PIP3 is required to recruit Rab35 compartments to the interface during cell intercalation while also suppressing medio-apical compartment formation. This result is also consistent with the apical/junctional enrichment of PIP3 in which low levels are present at apical surfaces and higher levels are at junctional interfaces (Fig. 2E; Fig. S2D,E). Given the size and strength of the medio-apical Sbf-Rab35 compartments after PIP3 disruption, we were interested in examining the appearance of the apical surface in these backgrounds. Intriguingly, SEM imaging of the cell surface of PI3K defective embryos revealed deep pits in the plasma membrane, which likely represent the Sbf-Rab35 compartments seen in fluorescent imaging, and which are not present in control embryos (Fig 2F; Fig. S2I). These results implicate PIP3 and PI3K kinase function in controlling a switch in the recruitment of Sbf-Rab35 tubular compartments to either interfacial or medio-apical cell surfaces.
Ectopic engagement of Sbf-Rab35 ratcheting function drives aberrant apical constriction
Cells in the intercalating germband undergo apically-driven cell area oscillations in a manner similar to cells of the ventral furrow. However, in the germband these area oscillations are fully reversible and are used to drive the ratcheted contraction of cell interfaces, while in the ventral furrow apical area contractions are stabilized to produce constrictions of the apical surface (Fig. 3A). Previous results have shown that Sbf-Rab35 compartments are key to this membrane-dependent ratcheting function (Jewett et al., 2017; Miao et al., 2019). Given this, we wanted to know if the medio-apical mislocalization of Sbf-Rab35 in PIP3 disrupted embryos would be sufficient to engage ectopic apical constriction in the intercalating epithelium. In wild-type germband extending (GBE) embryos, cell size remains uniform through the process of intercalation (Fig. 3A). However, in Pi3K92E and Pi3K21B disrupted germband cells, apical cell areas become progressively more contracted, until cells disappear into ectopic furrows in the embryo surface (Fig. 3A; Fig. S3A,B). This is not due to a defect in cell adhesion, as intact adherens junctions are maintained throughout the process (Fig. S3C,D). We also examined apical surface morphologies in both control and Pi3K92E disrupted embryos using scanning electron microscopy (SEM). These images revealed that Pi3K92E disrupted cells lose their typical rounded apical “cobblestone” appearance in early GBE and possess flattened apices (Fig. 3B), a further hallmark of processes driven by apical constriction (Sweeton et al., 1991). Thus, the cellular machinery to generate apical constriction and drive the ratcheted stabilization of apical surfaces is relocalized from cell interfaces to the apical cortex when PIP3 function is compromised.
Figure 3. Ectopic apical constriction and furrow formation in Pi3K92E shRNA embryos.
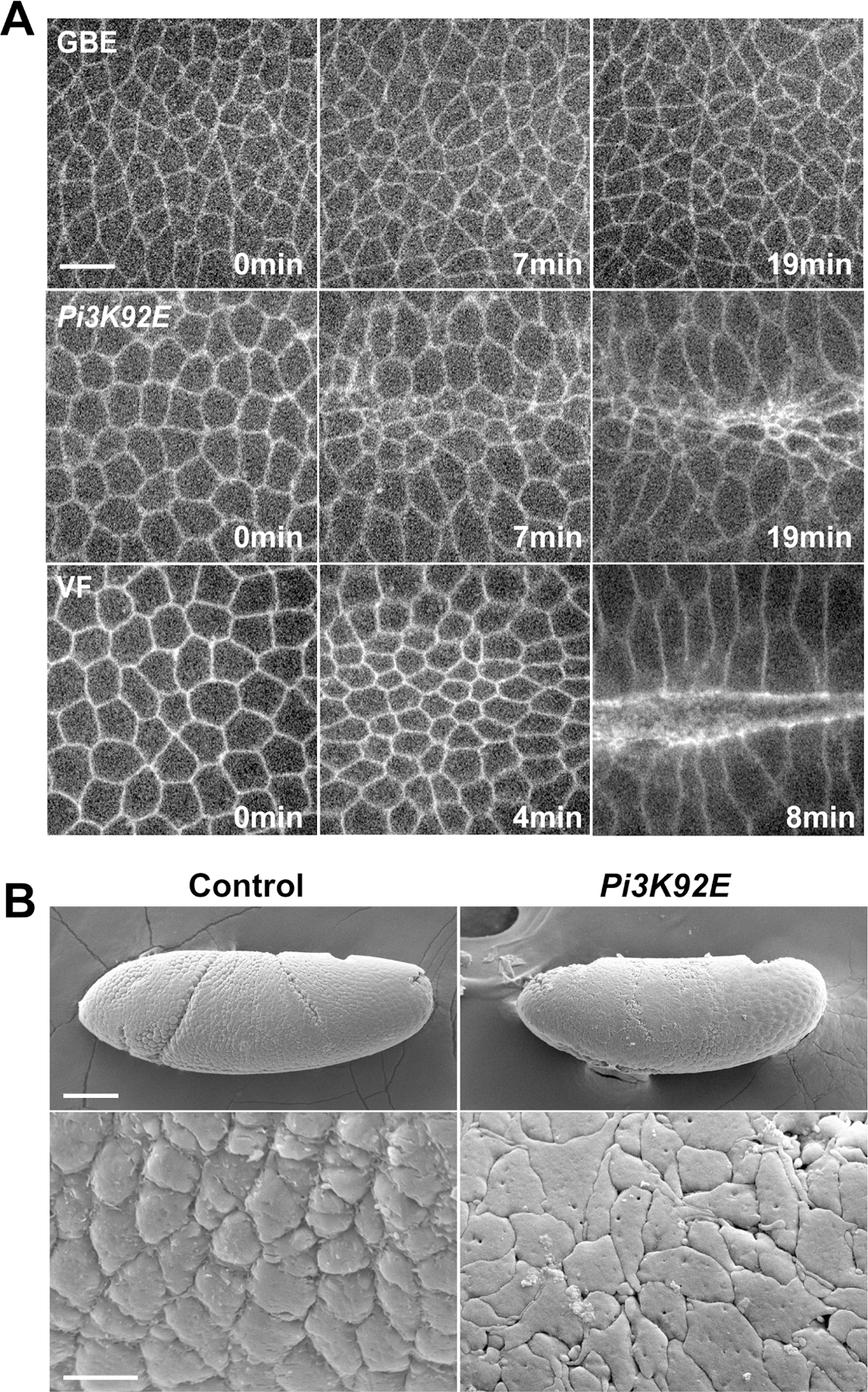
(A) Time-lapse images of embryos expressing a cell outline marker (Spider:GFP) in the germband epithelium in control and Pi3K92E background, or during wild-type ventral furrow formation. (B) SEM images of germband cells in embryos expressing control or Pi3K92E shRNA. Scale bar in (A) equals 5 µm. Scale bar in (B) top panel equals 50 µm and scale bar in (B) bottom panel equals 5 µm.
Ratcheted and rapid apical constriction occurs in germband cells after disruption of PIP3
Given the observed surface-switching behaviors in Sbf-Rab35 compartmental function, we wanted to know how individual contractile step profiles were changing to enable apical constriction (Fig. 4A). In theory, a number of possible changes could drive apical constriction – for example, asymmetric changes in contractile frequencies, amplitudes or directionality could each cause an increase in the contraction of apical surface areas. We therefore performed automated segmentation analysis on time-lapse movies with a temporal resolution of 2 seconds, and then used a sliding-window MSD-based step detection algorithm to identify periods of active contraction and expansion (see Methods). Interestingly, PIP3 disruption did not significantly change the frequency or duration of contractile steps (Fig. 4D,E). Therefore, the oscillatory machinery that initiates contractions in PIP3 disrupted embryos still resembles that of wild-type intercalating cells. However, Pi3K92E disrupted cells have an increased total contractile step rate and a greater number of individual steps that produce contractions of apical cell areas (Fig. 4B,C,F,G). This is due to steps spending less time in an expanding state (Fig. 4H) and those steps having a lower frequency of expanding individual steps that would reverse contractile gains from previous steps (Fig. 4I). This indicates that contractions are stabilized, a hallmark of a cellular ratchet, and that changes in PIP3 function regulate the processivity of contractions without altering the overall strength or frequency of contractions. Of note, it is interesting to compare the step dynamics in Pi3K92E disrupted embryos to those in the apically constricting ventral furrow – here, we see that Pi3K92E disrupted embryos begin to possess step profiles that are very similar to those of the ventral furrow, while still maintaining bulk step durations and frequencies characteristic of the germband (Fig. 4B–I). Taken together, these results indicate that PIP3 function is a key determinant which is responsible for directing Sbf and Rab35 membrane ratcheting activity to specific cell surfaces.
Figure 4. PIP3 disruption leads to a VF-like engagement of apical cell ratcheting.
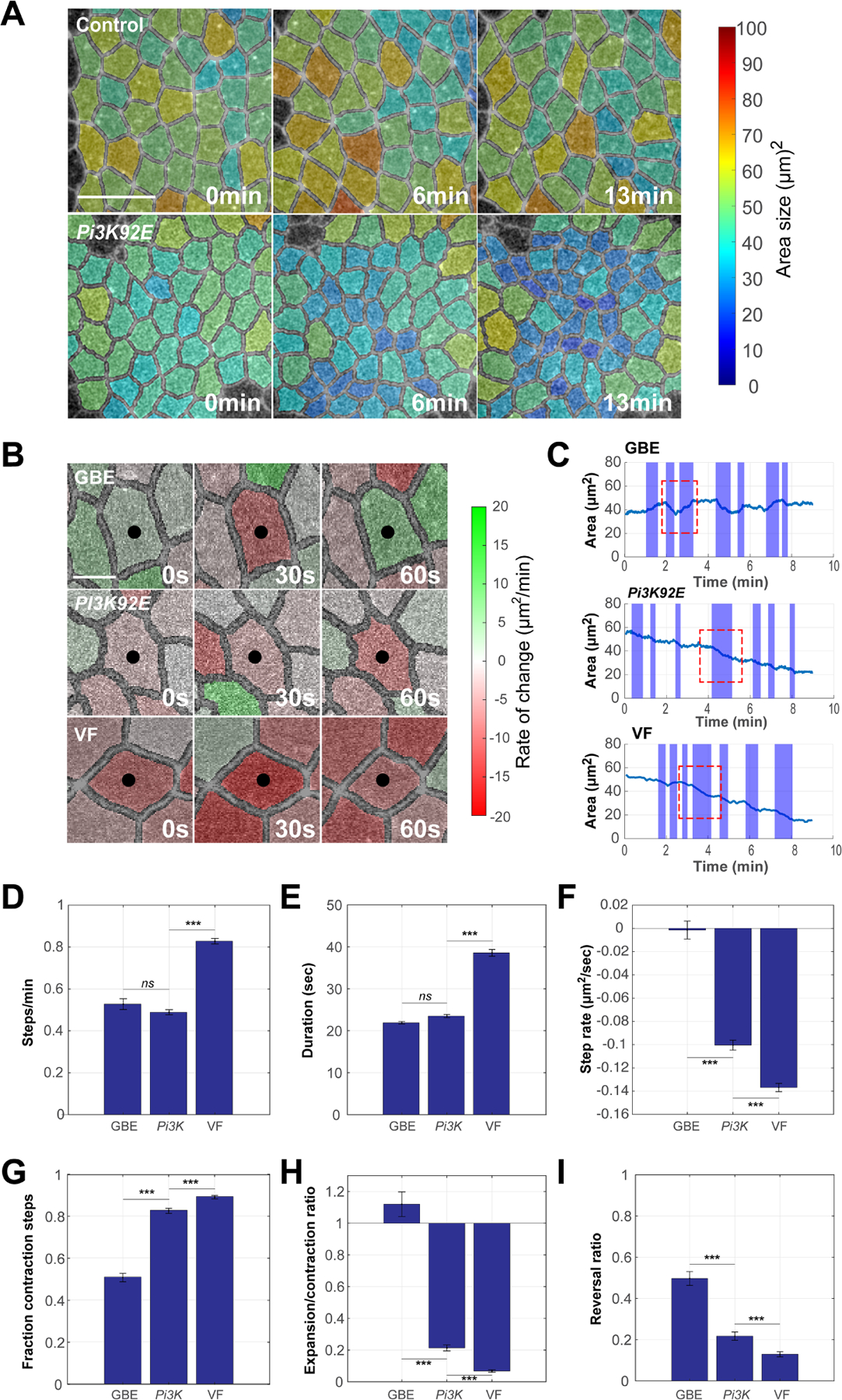
(A) Time-lapse images of embryos expressing Spider:GFP and automatically segmented for area analysis. Cell area size is color-coded (see color bar on the right) – smaller cell areas (blue colors) are observed after Pi3K disruption than in control embryos in the germband. (B) Time-lapse images of individual cells in control and Pi3K shRNA germband or wild-type ventral furrow. Expansion and constriction rates are color-coded (see color bar on the right). Red indicates constriction and green expansion. With wild-type oscillatory, GBE cells show reversible contractions in cell areas, while PI3K and ventral furrow cells show stabilized losses in cell areas. Black dots mark the central cells. (C) Automated step detection of periods of active area change (shaded area) in control and Pi3K shRNA germband, or wild-type ventral furrow cells. Red boxed regions mark examples of a reversing step in germband cells, and stabilized contractions in Pi3K and ventral furrow cells. (D,E and F) Frequency, duration, and step rate (D,E,F, respectively) of active steps in control germband, Pi3K shRNA germband, and ventral furrow cells. n= 731 steps from 3 embryos (GBE), 981 steps from 3 embryos (Pi3K92E shRNA) and 1295 steps from 3 embryos (VF) in (D), (E) and (F). Step frequency and duration remain germband-like after PIP3 disruption, but contractile rates become similar to cells of the ventral furrow. (G-I) The fraction of contractile steps (G), the ratio of expansion steps to contractile steps (H), and the likelihood of contractile steps followed by the expansion steps (I) in germband, Pi3K shRNA germband, and VF cells. VF and PIP3 disrupted cells have lower rates of reversibility than control germband cells. n= 154 cells from 3 embryos (GBE), 223 cells from 3 embryos (Pi3K92E shRNA) and 192 cells from 3 embryos (VF) in (G), (H) and (I). Scale bar equals 5 µm in (A) and 2.5 µm in (B). Error bars indicate measured standard error; statistical significance has been calculated using Kolmogorov-Smirnov test. ***P<0.0005.
Ventral Twist function regulates PIP3 levels and ratcheting engagement
As the constriction of apical surfaces is a normal part of mesodermal ingression (Martin and Goldstein, 2014), we examined if PIP3 levels are altered in the tissue anlagen that will form the ventral furrow and drive tissue invagination. To examine this, we first imaged PIP3 biosensor levels in ventral cells. Interestingly, and in contrast to intercalating cells, PIP3 is enriched apically in cells at the beginning of ventral furrow formation and possess low PIP3 levels at cell junctions (Fig. 5A,B). In previous work, we have shown that Sbf-Rab35 compartments are active at apical surfaces during ventral furrow formation (Miao et al., 2019) – we therefore assessed whether PIP3 was present at Rab35 tubular plasma membrane compartments on ventral-apical surfaces. Indeed, Rab35 compartments in constricting ventral furrow cells were found to colocalize with PIP3 (Fig. S4A,B). These results are consistent with apically localized PIP3 redirecting the engagement of Sbf-Rab35 compartmental function to cell apices versus cell interfaces in the ventral furrow.
Figure 5. PIP3 levels and Sbf-Rab35 ratcheting are developmentally patterned.
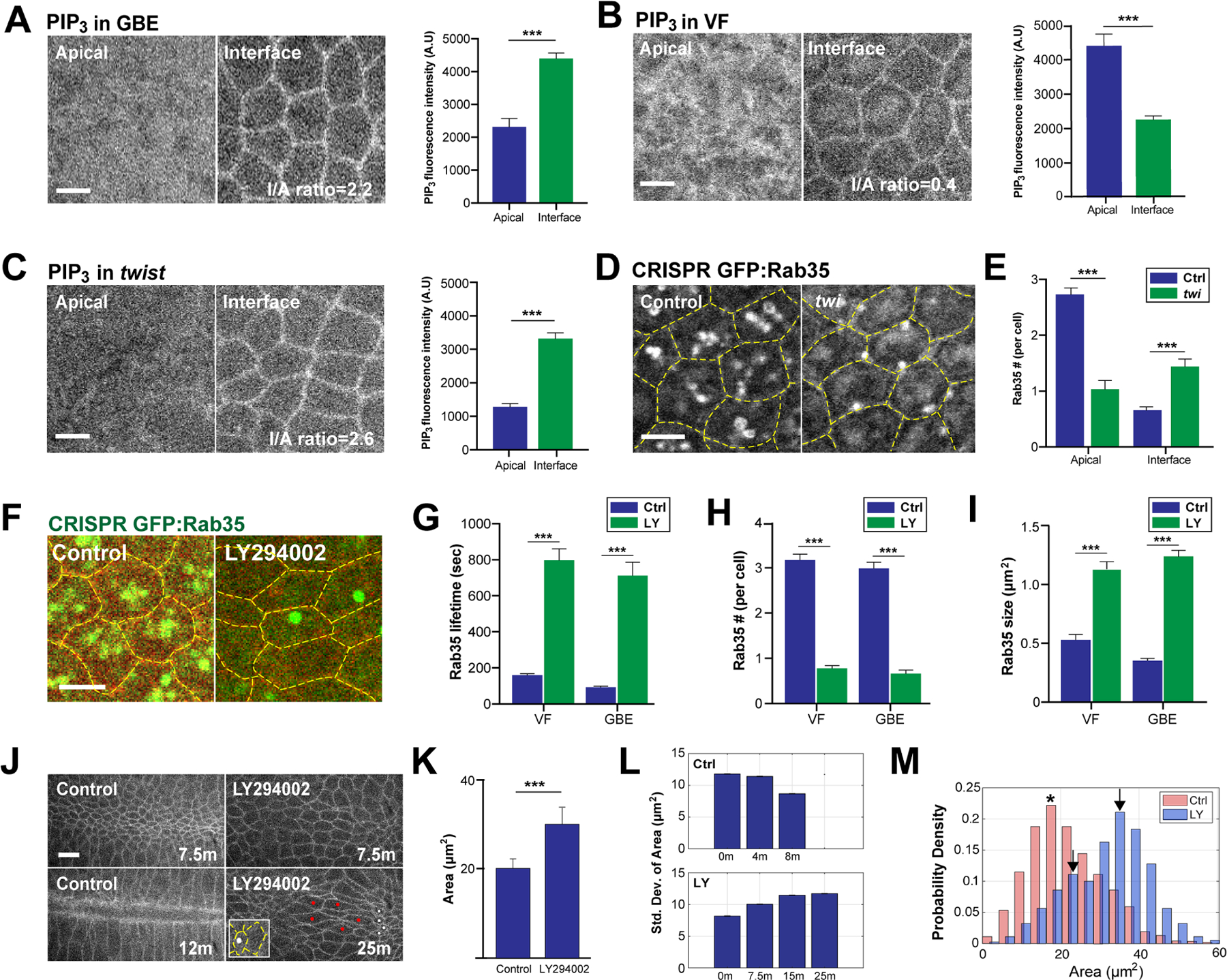
(A,B) PIP3 levels at apical surfaces and cell junctions/interfaces in intercalating cells (A) and constricting ventral furrow cells (B). Left panel: still images of PIP3 biosensor, tGPH. Right panel: quantitation of PIP3 levels at apical surfaces and cell junctions. n= 254 cells from 12 embryos (GBE) and 156 cells from 9 embryos (VF). I/A ratio is interfacial PIP3/apical PIP3 fluorescence intensity ratio. (C) PIP3 level at apical surfaces and cell junctions/interfaces of ventral furrow cells in twist mutant embryos. Left panel: still images of PIP3 biosensor, tGPH. Right panel: quantitation of PIP3 level at apical surfaces and cell junctions. n= 168 cells from 7 embryos. (D) Live imaging of CRISPR GFP:Rab35 in control and twist cells during ventral furrow formation. (E) Number of Rab35 compartments at apical surfaces and cell junctions during ventral furrow formation. n=145 cells from 3 embryos (control) and 59 cells from 3 embryos (twist). (F) Still images of CRISPR GFP:Rab35 embryo injected with either control or LY294002. The yellow dashed lines mark cell outlines. (G, H and I) Lifetime (G), number (H) and size (I) of Rab35 compartments in control and LY294002 injected germband and ventral furrow cells. Measured compartment location varied depending on predominant Rab35 localization (medio-apical region in ventral cells and LY294002 injected embryos, interface-associated in GBE cells). n= 101 compartments from 3 embryos (VF-Ctrl), 62 compartments from 3 embryos (VF-LY), 103 compartments from 3 embryos (GBE-Ctrl) and 69 compartments from 3 embryos (GBE-LY) in (G). n= 115 cells from 3 embryos (VF-Ctrl), 142 cells from 3 embryos (VF-LY), 106 cells from 3 embryos (GBE-Ctrl) and 101 cells from 3 embryos (GBE-LY) in (H). n= 109 compartments from 3 embryos (VF-Ctrl), 109 compartments from 3 embryos (VF-LY), 103 compartments from 3 embryos (GBE-Ctrl) and 109 compartments from 3 embryos (GBE-LY) in (I). (J) Time-lapse images of cells outlined with plasma membrane marker (Spider:GFP) in control and LY294002 injected background during ventral furrow formation. White dots indicate cells with small apical surfaces and red dots mark cells with large apical surfaces demonstrating heterogeneity of contraction after PIP3 disruption. Inset shows CRISPR GFP:Rab35 compartments after LY294002 injection. As ventral furrow dynamics are much slower in PIP3 disrupted embryos, a later time point is included to show maximal contraction. (K) Average area of control and LY294002 injected ventral cells at 7.5 minutes. n=250 cells from 3 embryos (control) and 357 cells from 3 embryos (LY294002). (L) Standard deviation of cell area of control (top panel) and LY294002 injected (bottom panel) cells at the indicated time points. n>114 control cells and n>215 LY294002 cells from 3 embryos. (M) Area distribution of control and LY294002 injected ventral cells at 7.5 minutes. n=250 cells from 3 embryos (control) and 357 cells from 3 embryos (LY294002). Asterisk and arrows mark the peaks in control and LY294002 injected background, respectively. Scale bars equal 5 µm; error bars indicate measured standard error. Statistical significance has been calculated using Student’s t-test in (A), (B) and (C), Kolmogorov-Smirnov test in (K) and Mann-Whitney U-test in (E), (G), (H) and (I). ***P<0.0005.
Given the observed changes in apical-to-junctional ratios of PIP3 in germband and ventral furrow cells, we next determined if this is patterned by the fate-determining transcription factors that specify ventral identities and morphogenetic behaviors. The transcription factor Twist is necessary for dorsal-ventral pattern establishment and directs processive apical constriction during ventral furrow formation (Thisse et al., 1987; Martin et al., 2009). We therefore examined if PIP3 levels are altered in twist mutant embryos. Indeed, twist mutants accumulate PIP3 predominantly at cell junctions, similar to intercalating cells in the germband epithelium (Fig. 5C). We then determined the compartmental localization of Sbf and Rab35 in ventral cells of twist mutant embryos, and observed that Sbf and Rab35 compartments switch their localization and are present at interfacial surfaces (Fig. 5D,E; Fig. S4C,D). These results provide further evidence that PIP3 function is developmentally patterned and is active in controlling a switch in the recruitment of Sbf-Rab35 tubular compartments to either interfacial or medio-apical cell surfaces.
However, the above results, while consistent with PIP3 recruitment of the Sbf RabGEF protein, pose an interesting contrast with Sbf-Rab35 compartmental dynamics in PIP3 disrupted embryos – in one case, high apical PIP3 levels correlate with a shift to apical surfaces, while in the other case a deep reduction in PIP3 levels trigger ectopic constriction and medial Sbf-Rab35 compartments. We therefore more closely examined the nature of the Sbf-Rab35 compartments formed in both control ventral furrow and PIP3 disrupted embryos. Intriguingly, time-lapse imaging revealed immediate and obvious differences in the nature of the Sbf-Rab35 compartments formed in control and PIP3 disrupted embryos (Video 2). PIP3 disruption produces large, hyperstable medial Rab35 compartments, while ventral Rab35 compartments in control embryos are shorter-lived, more numerous, and smaller (Fig. 5F–I). One interpretation of these results is that PIP3 is required to seed the dispersal of Sbf from a large, apically-centered aggregate into smaller, more dynamic compartments. Given these changes in Sbf-Rab35 compartmental behaviors, we wondered if the disruption of PIP3 in ventral furrow cells would either accelerate or interfere with apical constriction rates. We therefore examined apical area contraction during ventral furrow formation when PIP3 levels are compromised (as shown by tGPH, PIP3 biosensor levels after LY294002 injection; Fig. S4E). Interestingly, PIP3 disrupted embryos failed to form a complete furrow (Fig. 5J,K). However, contractions of apical areas in ventral epithelial cells were still observed, although these cells became increasingly disordered (Fig. 5J,L). Regions of small highly-constricted cells are present, which are juxtaposed with patches of larger cells that possess a “stretched” appearance (Fig. 5J). This is also evident in systematic measurements of cell area distributions in which two peaks are observed (Fig. 5M). It is interesting to note that Rab35 compartments are often found in the small highly-constricted cells, but are absent in larger cells (Inset in Fig. 5J; Fig. S4F). It appears that a balanced, uniform contraction of ventral furrow cells is not possible after PIP3 disruption. These results indicate that the dynamics of PIP3-driven Sbf-Rab35 compartmental formation and function must be carefully regulated, and that they are essential for uniform cell behaviors and processive apical surface constriction during ventral furrow formation.
JAK/STAT signaling regulates apical engagement of the Sbf-Rab35 ratchet
As Rab35 compartments adopt a medio-apical localization after PIP3 disruption, we wondered if a second, independent signaling pathway existed that can also direct Sbf and Rab35 localization to specific cell surfaces. Interestingly, ectopic apical constrictions in the embryonic epithelium have been observed once before in the literature – when JAK/STAT signaling is compromised, similar abnormal apical narrowing occurs (Bertet et al., 2009). Mutants in either unpaired (Upd), a Drosophila ligand of JAK/STAT signaling, or the single STAT transcriptional factor, Stat92E, have been shown to cause ectopic apical constriction in germband cells. We therefore first asked if Rab35 compartments are relocalized to the apical surface in Stat92E disrupted embryos. Indeed, there is a similar shift of Rab35 compartments to the apical surface, with an average of 2.7 apical Rab35 compartments per cell in Stat92E disrupted embryos, compared to 0.3 in wild type (Fig. 6A,B). About 83% of these compartments are filled with dextran immediately after injection, suggesting that these compartment represent infoldings of the plasma membrane as has been previously observed during germband extension as well as during ventral furrow formation (Fig. S5A,B) (Jewett et al., 2017; Miao et al., 2019).
Figure 6. JAK/STAT signaling regulates ratcheting engagement upstream of Sbf-Rab35.
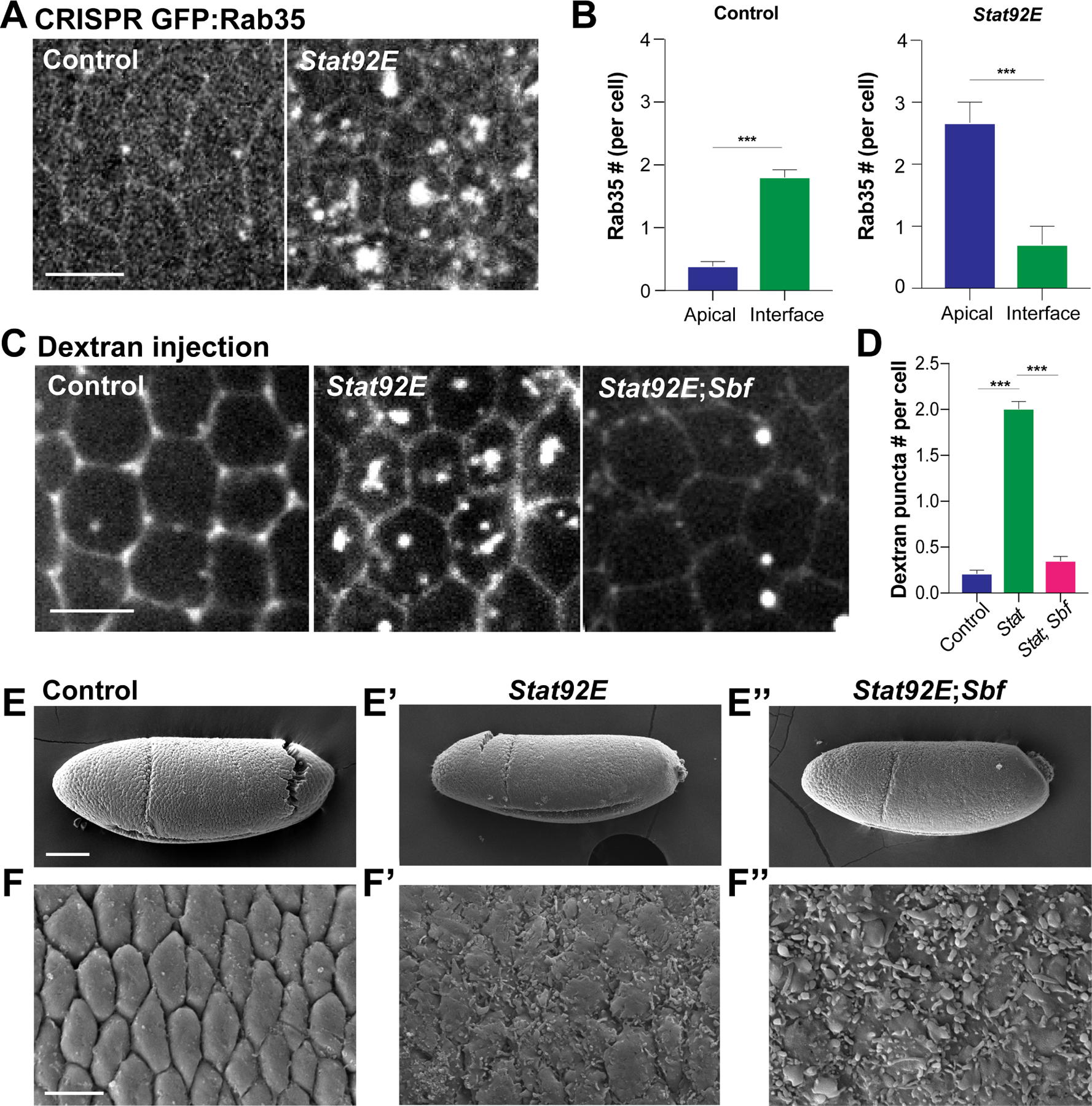
(A) Live imaging of CRISPR GFP:Rab35 in control and Stat92E shRNA cells. (B) Number of Rab35 compartments per cell at either interfaces or apical surfaces in control and Stat92E shRNA cells. n= 77 cells from 3 embryos (control) and 125 cells from 3 embryos (Stat92E). (C) Still images of control, Stat92E shRNA and Stat92E shRNA; Sbf shRNA embryos injected with dextran to extracellular space. (D) Apical dextran puncta number in control, Stat92E shRNA and Stat92E shRNA; Sbf shRNA cells. n= 114 cells from 3 embryos (control), 132 cells from 3 embryos (Stat92E shRNA), 145 cells from 3 embryos (Stat92E shRNA; Sbf shRNA). (E-F’’) Scanning electron microscopy images of germband epithelial cells in control (E,F), Stat92E shRNA (E’,F’) and Stat92E shRNA; Sbf shRNA (E’’,F’’) embryos. Scale bars in (A), (C) and (F) equal 5 µm and scale bar in (E) equals 50 µm. Error bars indicate measured standard error; statistical significance has been calculated using Mann-Whitney U-test. ***P<0.0005.
We then asked if Sbf-Rab35 ratcheting function is essential for the ectopic apical constriction triggered by compromising JAK/STAT signaling. We imaged dextran labeled plasma membrane infoldings or pits in doubly compromised Stat92E; Sbf disrupted cells and found that the number of dextran-filled surface compartments is greatly reduced (Fig. 6C,D). When the apical surfaces of these cells are examined by SEM, it is apparent that the absence of JAK/STAT signaling still induces apical flattening and deep pits, but, interestingly, the surface is filled with membrane blebs (Fig. 6E–F’’; S5C–E). This indicates that removing JAK/STAT signaling is sufficient to trigger contractile forces in the apical dome of epithelial cells, but, without Sbf-Rab35 function, the membrane ratcheting activity to remove excess apical surface is missing. Indeed, in the absence of Sbf-Rab35 function, the narrowed cell apices present in JAK/STAT disrupted embryos are no longer present, and the epithelium is rescued back to a more wild-type state (Fig. S5F). These results suggest removing JAK/STAT signaling is sufficient to trigger a switch to an apical constricting fate, which depends on Sbf-Rab35 function to achieve functional processivity.
PIP3 and JAK/STAT signaling pathways act combinatorially to control Sbf-Rab35 compartmental function
Our data demonstrates that two different pathways (PIP3 and JAK/STAT signaling) regulate the switch between apical constriction and interface contraction. We therefore wanted to know if these pathways individually converge on Sbf-Rab35 ratcheting or if one pathway is upstream of the other. To test this, we first compared individual cell behaviors during ectopic apical constriction between Pi3K92E shRNA and Stat92E shRNA embryos. In 15 mins, Pi3K92E cells become increasingly disordered, with a variety of apical cell sizes (Fig. 7A,B,C). By contrast, Stat92E cells maintain a greater uniformity during the ectopic apical constrictions (Fig. 7A,B,C). We therefore took a closer look at the apical area steps in Pi3K92E shRNA and Stat92E shRNA cells using our automated active step detection method. Stat92E shRNA cells possess similar step and reversal frequencies as Pi3K92E shRNA cells in 7.5 mins, but possess higher constriction rates that are driven by prolonged step durations, higher contractile step rates and more frequent contractile steps (Fig. S6A–F). These results suggest that the JAK/STAT signaling pathways regulate both Sbf-Rab35 ratcheting as well as the core oscillatory contraction machinery.
Figure 7. Functional independence of PIP3 and JAK/STAT signaling pathways in regulating cell ratcheting.
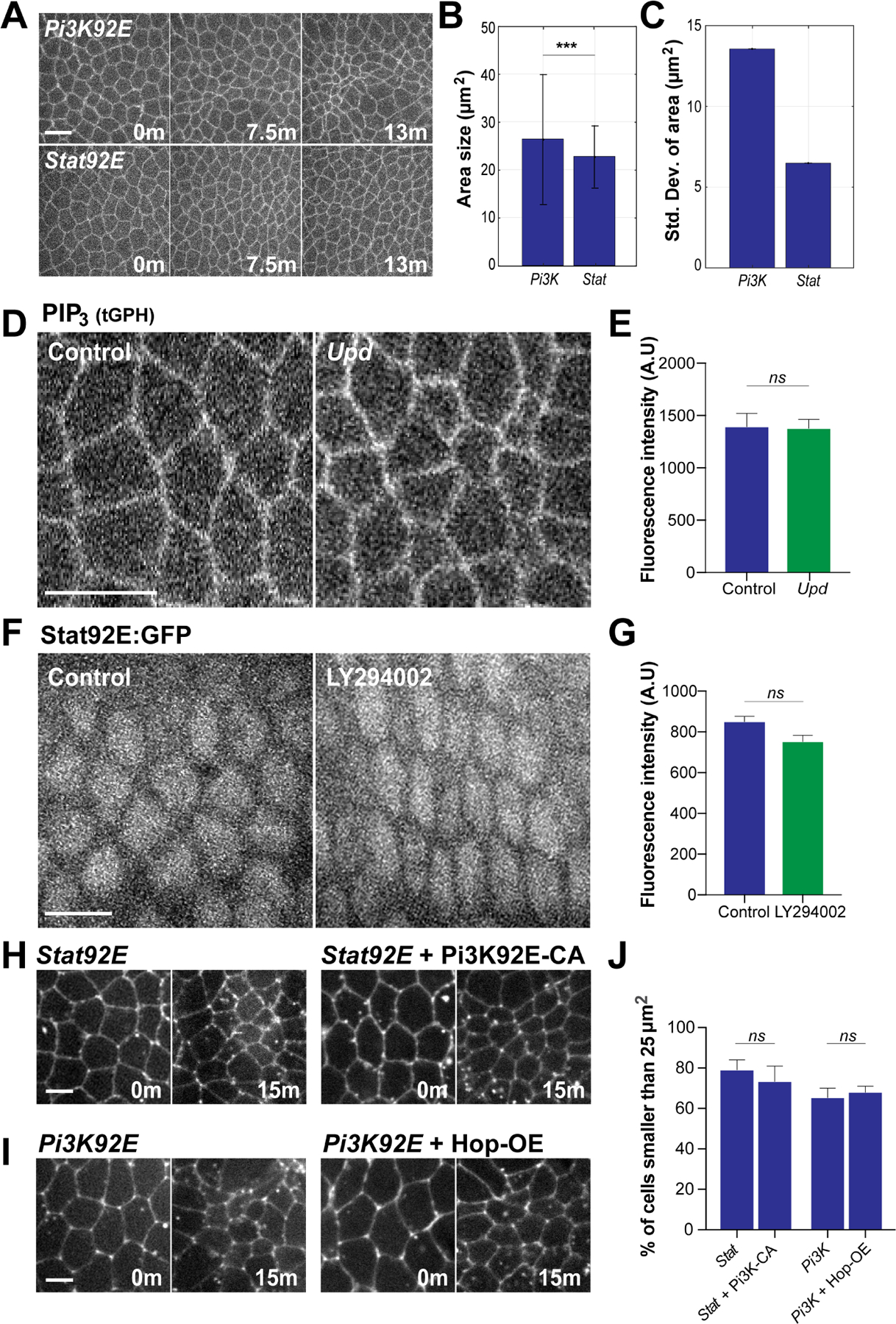
(A) Time-lapse images of embryos expressing cell outline marker (Spider:GFP) during ectopic apical constriction induced by the disruption of Pi3K92E or Stat92E. (B) The area size of Pi3K92E and Stat92E cells after 13 minutes. (C) Standard deviation of area of Pi3K92E and Stat92E cells at 13 minutes. n=489 cells from 3 embryos (Pi3K) and 407 cells from 3 embryos (Stat) in (B) and (C). (D) Images of PIP3 sensor in control and Upd mutants. (E) Fluorescence intensity of PIP3 sensor in control and Upd embryos . n= 112 cells from 4 embryos (control) and 125 cells from 4 embryos (Upd). (F) Stat92E:GFP localization and expression in control and LY294002 injected cells. (G) Fluorescence intensity of Stat92E:GFP in control and Upd embryos. n= n= 123 cells from 4 embryos (control) and 116 cells from 4 embryos (Upd). (H) Still images of embryos expressing either Stat92E shRNA or Pi3K92E constitutively active (CA); Stat92E shRNA during cell intercalation. (I) Still images of embryos expressing either Pi3K92E shRNA or Hop overexpression (OE); Pi3K92E shRNA during cell intercalation. (J) Percentage of cells smaller than 25 µm2 after 15 minutes. n= 217 cells from 3 embryos (STAT), 268 cells from 3 embryos (STAT+Pi3K-CA), 247 cells from 3 embryos (Pi3K) and 326 (Pi3K+ Hop-OE). Error bars indicate standard deviation in (B). Scale bars equal 5 µm. Error bars indicate measured standard error; statistical significance has been calculated using Kolmogorov-Smirnov test in (B) and Student’s t-test in (E), (G) and (J). ***P<0.0005.
To further examine the relationship between these two signaling pathways, we measured PIP3 levels in JAK/STAT disrupted embryos, as well as STAT92E nuclear levels in PIP3 disrupted embryos. PIP3 levels are unchanged in Upd mutant embryos, as revealed by imaging a PIP3 biosensor (Fig. 7D,E). Conversely, STAT92E nuclear levels also showed no significant changes after LY294002 injection (Fig. 7F,G). These results suggest these pathways function independently of each other – to prove this we next examined potential epistatic interactions between these pathway. Consistent with pathway independence, we found that overexpression of constitutively active PI3K does not rescue ectopic furrow formation induced by Stat92E disruption (Fig. 7H,J). Conversely, embryos that overexpress the only Drosophila JAK kinase, Hopscotch (Hop), still display ectopic constriction in Pi3K92E compromised embryos (Fig. 7I,J). These overexpression constructs are likely active, as PI3K CA can further deplete apical Rab35 puncta (Fig. S5G) and, although JAK/STAT overexpression does not further deplete apical Rab35, Hop-OE leads to developmental arrest at mid-embryogenesis. These results suggest PIP3 and JAK/STAT signaling pathways work independently to control whether apical or interfacial surfaces contract during early embryonic morphogenesis.
Discussion
The ability of epithelial cells to remodel specific cellular surfaces is central to determining cell dimensions as well as the neighbor community that they will interact with and adhere to. This ability to either grow or contract certain cell sides will determine overall cell shape, and the cumulative effects of these cell shapes changes determines tissue behaviors and morphology. By regulating the contraction of apical surfaces versus cell-cell interfaces, a tissue can drive events as diverse as furrow formation and cell ingression to cell intercalation and the intermixing of cells along the AP axis. However, it has been unclear if direct cues reside within the plasma membrane that may guide and control the engagement of contractile forces. Here, we have examined the function of plasma membrane phospholipids in recruiting Sbf-Rab35-driven ratcheting. We show that PI(3,4,5)P3 regulates a switch in ratcheting engagement – a reduction in PIP3 levels causes a reorientation of Sbf-Rab35 compartment formation to apicomedial surfaces (Model, Fig. S6G,H). This relocalization is sufficient to change the reversible oscillations in cell area that occur in the wild-type germband epithelium into a processive regime in which apical cell areas shrink and ectopic furrows are formed. Sbf, the guanine nucleotide exchange factor for Rab35, can directly bind PIP3, and PIP3 levels and sites of enrichment are differentially regulated between the germband and ingressing mesoderm to provide a differential lipid-based cue between these two tissues.
Uniformity of contractile events and extracting processivity from contractile pulses
It is interesting to note that while ectopic furrows form after PIP3 disruption, these furrows are often disorganized and lack the regular appearance of the main ventral furrow that drives the ingression of mesoderm during gastrulation. In some respects, we believe this is to be expected – if the major portion of the embryo is transformed to attempt to contract apical surfaces, then cells will be engaged in a contractile tug-of-war against each other. This condition is likely shown by the juxtaposition of small and large cells which is observed in both the germband as well as the ventral furrow after PIP3 disruption. The uneven contraction of apical surfaces may also reflect the different temporal dynamics of Sbf-Rab35 compartments after PIP3 disruption. The new medial compartments that form after PIP3 disruption are much more stable (indeed, lifetimes up to 10x longer in some measurements) and robust than those that form in either the germband or ventral furrow in control embryos. Previous work has shown the importance of contractile cycles to achieve a uniform overall contraction of the apical surfaces. For example, in circumstances where pulsatility, but not contractility, is compromised, contractile networks have been observed to tear and separate – this results in a similar loss of cell area uniformity as detected after PIP3 levels are downregulated (Jodoin et al., 2015; Mason et al., 2016). Thus, the change in Sbf-Rab35 compartment function to much more stable and longer cycles may enhance the tug-of-war element of the cell contractions previously referenced, producing “winner” cells of much smaller apical areas and “loser” cells that cannot shrink against the pulling forces of neighboring cells, and thus possess larger apical areas. We find it intriguing that the pulsatility of contraction appears to be such a fundamental element of contractile processes – pulsatility has been observed across a huge variety of contraction-driven processes ranging from wound healing to compaction of the mouse embryo to neuroblast ingression (Munro et al., 2004; Martin et al., 2009; Solon et al., 2009; Rauzi et al., 2010; Blanchard et al., 2010; David et al., 2010; Fernandez-Gonzalez and Zallen, 2011; Kim and Davidson, 2011; Sawyer et al., 2011; Maître et al., 2015; Michel and Dahmann, 2020; Mason and Martin, 2011; Levayer and Lecuit, 2012; Miao and Blankenship, 2020).
What is the nature of the signals that recruit ratcheting engagement?
Another interesting aspect of this work is that both phosphatidyl inositol phosphate species and JAK/STAT-dependent signaling control where ratcheting engagement occurs. If either of these pathways is disrupted, then a medial signal dominates and Sbf-Rab35 compartment formation occurs in a central, apical location (Model, Fig. S6G,H). Our model suggests that PIP3 and JAK/STAT signaling may provide a dispersal signal that guides the compartments from a single apico-central location to the cell periphery (in the case of germband epithelial cells) or to smaller, more dispersed apical locations (in cells of the ventral furrow). Based on the direct binding of PIP3 by the Sbf RabGEF in the PIP binding assay, this dispersal may be through a direct interaction. Our experiments did not have the resolution to determine if small PIP3 microdomains exist in the plasma membrane, or if further systems direct the formation of smaller, compartmental assemblies. On the other hand, how does JAK/STAT direct ratcheting engagement? Previous work examining the apical constrictions driven by an absence of JAK/STAT signaling implicated a repression of WASP actin networks that, when activated, may cause the enhanced recruitment of apical Myosin II populations (Bertet et al., 2009). Interestingly, this fits with our step detection measurements. In addition to the changes in Sbf-Rab35 localization, pulsed contractions are stronger and more sustained in JAK/STAT embryos than in PIP3 disrupted embryos. However, our previous results have shown that the generation of Sbf-Rab35 compartments is independent of Myosin II function (Jewett et al., 2017). Thus, from these results, we suggest that, while PIP3 directly regulates ratcheting engagement, JAK/STAT may regulate both the underlying oscillatory machinery as well as the strength of the medial signal that directs ratcheting engagement. It is interesting to note that, through the use of automated step detection measurements based on mean squared displacements, we have previously shown that the oscillatory machinery appears to be strengthened specifically in the ventral cells that will undergo processive apical constriction to form the ventral furrow (Miao et al., 2019), which suggests a commonality with the behaviors observed after JAK/STAT disruption.
Phosphatidylinositol phosphates and their function in morphogenetic processes
PIP phospho-species are attractive candidates to provide important spatial information as they are directly embedded in target membranes. PIP3 has long been implicated in distinguishing the leading edge in migrating cells, and is rapidly upregulated after cells are stimulated with chemoattractant where it promotes F-actin assembly necessary for cell crawling (Insall and Weiner, 2001; Parent et al., 1998; Meili et al., 1999; servant et al., 2000; Haugh et al.,2000). However, how PIP3 regulates gastrulation events has been relatively unstudied. A recent work has demonstrated the Pi3K92E can bind to active Toll receptors and Src signaling complexes, and is planar polarized at AP interfaces, in keeping with our finding of developmental enrichment of PIP3 in the germband, and suggesting an intriguing connection between planar positional information and PIP function (Tamada et al., 2021). PIP3 is necessary for cell migration events in the mesenchymal gastrulation movements that occur in zebrafish (Montero et al., 2003), while data from Drosophila has shown that PIP3 levels are upregulated after wounding and help cells recognize affected surfaces (Pickering et al., 2013). Disrupting PIP3 levels disrupted dorsal closure in the late embryo, where, once again, PIP3 is found at higher levels specifically at those surfaces that are driving tissue remodeling. Our work similarly finds the PIP3 levels are developmentally patterned, where they are enriched at contractile surfaces. Other work has shown that a PIP2/PIP3 balance affects actomyosin contractility during the cellularization process that creates the early embryonic epithelium through the recruitment of an actin stabilizer, bottleneck (Reversi et al.,2014). Sbf-Rab35 compartments have represented an interesting convergence point between pathways that directly regulate cell membrane remodeling and those that control cortical force generation (Dambournet et al., 2011; Jewett et al., 2017; Frémont et al., 2017, Miao et al., 2019). Going forward, it will be interesting to examine if this convergence includes similar higher-level regulation of the protein networks that have been implicated in migrating systems.
Limitations of the Study
Our work demonstrates a fundamental switch in contractile behaviors depending on the activity and localization of the PI(3,4,5)P3 lipid cue. There were several limitations to our studies: firstly, many of our functional disruptions relied on pharmacological or shRNA knockdown lines which often produce only hypomorphic disruptions. Phenotypes were confirmed with secondary shRNA lines that targeted different regions of the selected mRNA and yet produced similar defects; however, deeper disruption of these genes may produce more severe defects at these stages or earlier in development. Secondly, we used a PIP3 biosensor (tGPH-GFP) to detect PIP3 localization and levels – this is the standard in the field, but represents an indirect binder of PIP3, so future probe development may allow a better resolution of PIP3 behaviors. As we mention in the Discussion, we would be very interested to be able to better address if PIP3 microdomains exist in the plasma membrane, and if these may directly trigger site-specific Sbf/Rab35 compartmental formation, but these were not resolvable with our combination of probe and microscopy elements. Finally, gastrulation in the early Drosophila embryo shows a remarkable robustness to disruption, so additional phenotypes may be concealed by compensatory mechanisms that were not detected in our analysis.
STAR★METHODS
RESOURCE AVAILABILITY
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and fulfilled by the Lead Contact, J. Todd Blankenship (todd.blankenship@du.edu).
MATERIAL AVAILABILITY
All fly stocks and plasmid DNAs used in this study are freely available on request from the authors.
DATA AND CODE AVAILABILITY
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fly Lines and Maintenance
The flies used are listed in the Key Resources table. All the flies were maintained at 25°C. UAS transgenic flies were crossed to matαTub-Gal4VP16 67C;15 (D. St. Johnson, Gurdon Institute, Cambridge, UK) maternal driver females. shRNA lines were scored for gross tissue morphologies by transmitted light and under Halocarbon oil.
KEY RESOURCES TABLE
| REAGENT | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-GFP (1:1000) | Invitrogen | Cat# A-11122 RRID:AB_221569 |
| Mouse anti-Ecad (1:10) | Developmental Studies Hybridoma Bank (DSHB) | Cat# 5D3 anti-Ecad RRID:AB_528116 |
| Goat anti-mouse Alexa Fluor 488 (1:500) | Invitrogen | Cat# A-11029 RRID:AB_2534088 |
| Goat anti-Rabbit Alexa Fluor 488 (1:500) | Invitrogen | Cat# A-11008 RRID:AB_143165 |
| Alexa Fluor 647 phalloidin (1:200) | Invitrogen | Cat# A-22287 RRID:AB_2617428 |
| Mouse anti-MBP (1:200) | DSHB | Cat# 2A1 anti-MBP RRID:AB_2617428 |
| Goat anti-mouse HRP conjugate (1:10,000) | Bio-Rad | Cat# 1706516 |
| Chemicals, peptides, and recombinant proteins | ||
| Halocarbon 700 oil | Halocarbon Products | Series 700 |
| Halocarbon 27 oil | Halocarbon Products | Series 27 |
| LY294002 | Sigma-Aldrich | Cat# 440202 |
| Dextran Alexa Fluor 568 | ThermoFisher Scientific | Cat# D22912 |
| ProLong gold Mountant | ThermoFisher Scientific | Cat# P36930 |
| Critical commercial assays | ||
| Protein-lipid overlay assay | Echelon Biosciences | Cat# P-6001 |
| PH domain of Sbf synthesis | Integrated DNA Technologies | N/A |
| Experimental models: organisms/strains | ||
| tGPH | Bloomington Drosophila Stock Center (BDSC) | BDSC Cat# 8164 |
| UAS-Pi3K21B TRiP Valium 22 | BDSC | BDSC Cat# 36810 |
| UAS-Pi3K21B TRiP Valium 20 | BDSC | BDSC Cat# 38991 |
| UAS-Pi3K92E TRiP Valium 22 | BDSC | BDSC Cat# 35798 |
| UAS-Pi3K92E TRiP Valium 20 | BDSC | BDSC Cat# 61182 |
| UAS-fab1 TRiP Valium 20 | BDSC | BDSC Cat# 51782 |
| UAS-fab1 TRiP Valium 22 | BDSC | BDSC Cat# 35793 |
| UAS-Pi4KIIIα TRiP Valium 22 | BDSC | BDSC Cat# 35256 |
| UAS-Pi4KIIIα TRiP Valium 20 | BDSC | BDSC Cat# 38242 |
| UAS-fwd TRiP Valium 22 | BDSC | BDSC Cat# 35257 |
| UAS-Pi4KIIα TRiP Valium 22 | BDSC | BDSC Cat# 35278 |
| UAS-Pi3K68D TRiP Valium 20 | BDSC | BDSC Cat# 34621 |
| UAS-Pi3K68D TRiP Valium 22 | BDSC | BDSC Cat# 35265 |
| UAS-Pi3K59F TRiP Valium 20 | BDSC | BDSC Cat# 33384 |
| UAS-Pi3K59F TRiP Valium 22 | BDSC | BDSC Cat# 36056 |
| UAS-Stat92E TRiP Valium 20 | BDSC | BDSC Cat# 33637 |
| UAS-Sbf TRiP Valium 20 | BDSC | BDSC Cat# 44004 |
| Stat92E:GFP | BDSC | BDSC Cat# 38670 |
| Upd | BDSC | BDSC Cat# 4767 |
| UAS-Pi3K92E.CAAX | BDSC | BDSC Cat# 8294 |
| UAS-Hop | BDSC | BDSC Cat# 79033 |
| CRISPR GFP:Rab35 | Blankenship lab, (Jewett et al., 2017) | N/A |
| UAS:mCh:Rab35 | Blankenship lab, (Miao et al., 2019) | N/A |
| UAS:eGFP:Sbf | A. Kiger, University of California San Diego, USA, (Jean et al., 2012) | N/A |
| mCh:Sqh | A. Martin, Massachusetts Institute of Technology, USA, (Martin et al., 2009) | N/A |
| Spider:GFP | A. Debec, Paris Diderot University, France | N/A |
| Resille:GFP | A. Debec, Paris Diderot University, France | N/A |
| matαTub-Gal4VP16 67C;15 | D. St. Johnson, Gurdon Institute, UK | N/A |
| Oligonucleotides | ||
| pET-15b_Sbf_PH5’: AAACATATGATGAA- TAGAAGCTTATCGATGA- TAAGC | This study | Eurofins MWG |
| pET-15b_Sbf_PH3’: ACTCGAGTTAG- CAAACATGCCTG- CAATTTTTC | This study | Eurofins MWG |
| Recombinant DNA | ||
| pET-15b vector | C. Asensio, University of Denver | N/A |
| Software and algorithms | ||
| ImageJ | NIH | N/A |
| MATLAB | MathWorks | N/A |
| MATLAB Segmentation Code | Jewett et al., 2017; Miao et al., 2019 | N/A |
| Prism | GraphPad | N/A |
Fly stocks were maintained on standard BDSC Cornmeal Food (https://bdsc.indiana.edu/information/recipes/bloomfood.html; water, yeast, soy flour, yellow cornmeal, light malt extract (dehydrated), agar, light corn syrup, propionic acid). Embryos were collected from medium-sized collection cups on apple juice agar plates (apple juice, dextrose, agar, Nipagin) with a dab of baker’s yeast paste (dry active baker’s yeast mixed with water) on the surface of the plate. Flies were housed in pan-humidifier Percival incubators (no lighting) at 25°C and 65% humidity.
METHOD DETAILS
SEM embryos preparation
OreR, PI3K shRNA, STAT shRNA and STAT shRNA; Sbf shRNA embryos were dechorionated in 50% bleach solution for two minutes, fixed for 20 min at the interface of heptane and 25% glutaraldehyde in 50mM sodium cacodylate buffer (pH 7.4) and then post-fixed in 1% OsO4, 50 mM cacodylate buffer (pH 7.4). The embryos were dried using HMDS and imaged on a JEOL JSM-6100LA Scanning Electron Microscope at 10 kV.
Live imaging and injection
Embryos were dechorionated in 50% bleach solution and then transferred to an air-permeable membrane and covered with Halocarbon 27 oil. A CSU10b Yokogawa spinning disk confocal from Zeiss/ solamere Technologies Group with a 60x 1.4 NA objective was used to perform all time-lapse imaging. For drug injection, after dechorionation as described above, embryos were dehydrated for 15 minutes, covered with Halocarbon 700 oil and then injected with either LY294002 (Sigma, 25mM) or dextran Alexa568 (Thermo Fisher, 1mg/mL). Embryos were imaged 20 minutes after LY294002 injection or were imaged immediately after the dextran injection. LY294002 injection embryos showed reduced germband extension and variable defects in cephalic furrow formation.
Embryo fixation, immunostaining and imaging
Embryos were collected on apple juice agarose plates and then dechorionated for 2 min before fixation. The embryos were fixed for 1 hr 10 min at the interface of heptane and 4% formaldehyde in 0.1 M sodium phosphate buffer (pH 7.4). Then the embryos were manually devitellinized and stained with Rabbit anti-GFP (Invitrogen, 1:1000), mouse anti-Ecad (DSHB, 1:100). Conjugated secondary antibodies Alexa-488 (Molecular Probes, 1:500) were used. Embryos were mounted in Prolong Gold (Molecular Probes). Immunostained embryos were imaged with a confocal laser scanning microscope with 60x objective lens.
Protein preparation and protein-lipid overlay assay
The PH domain of Sbf was synthesized by Integrated DNA Technologies IDT and cloned in-frame into pET-15b vector. Proteins were induced in Escherichia coli BL21 (DE3) at 20°C for 16 hr using 1mM IPTG (sigma). The bacteria were lysed by sonication. MBP and MBP-tagged fusion proteins were purified by the incubation with amylose resin beads (New England BioLabs) and then eluted with 20mM maltose in PBS buffer. PIP strips (Echelon Biosciences, cat#P-6001) were used to analyze protein-lipid binding. 0.5 μg/mL of the proteins were incubated with the lipid membrane for 1 hr at room temperature. Mouse anti-MBP antibody (DSHB, 1:200) and anti-mouse HRP conjugated secondary antibodies (Bio-Rad, 1:10,000) were used to detect the protein. The lipid membrane was imaged on FluorChem R.
DNA subcloning
The PH domain of Sbf (amino acids 1891–1992 of Sbf-PA reference isoform) was synthesized by Integrated DNA Technologies IDT and cloned in-frame into pET-15b vector at the NdeI-XhoI sites.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification of fluorescence intensity and colocalization
Fluorescence intensity was measured using ImageJ. For apical PIP3 intensity measurement in time-lapse images, multiple square regions (605 μm2) in the most apical region of either GBE or VF cells were quantified. For the interface measurements and PIP3 intensity measurement in immunostaining images, the ROI was identified as all pixels within 2 pixels of the center of interfaces or apical surfaces. As for Stat92E intensity measurement, a circular region (~3 μm2) in was quantified in each nuclei. Colocalization between Sbf and Rab35 was performed on time-lapse images. Rab35 puncta equal or bigger than 2x2 pixels were selected. The selected Rab35 compartments were then overlaid with the opposing channel. When the overlapping region was equal or larger than 2 × 2 pixels, the relationship between two proteins was determined as “colocalized.” Average colocalization was found by performing a weighted average calculation from images collected.
Cell Segmentation
Image and data analysis were performed in Matlab. Cells were segmented using a seeded watershed algorithm and tracked in time. We measured cell area as the sum of the pixels within the contour of the watershed segmentation lines converted to square microns.
Step detection
To detect active motion steps in our vertex position trajectories we used a rolling analysis window technique adapted from Huet et al. (2006). The MSD is the customary method to classify a sub-trajectory into active, diffusive, or constrained motions based on whether the MSD curves upward, is linear, or curves downward, respectively. For periods of active motion the MSD behaves as a power law , where γ > 1. By calculating the parameter gamma along a signal using a rolling window we can identify periods of active and non-active (i.e. either diffusive or constrained) motion. We used two rolling window sizes, 21 and 27 seconds. For each time window, we fit the MSD to lags between 4 and (N-1)/2 frames where N is the odd-numbered number of points in the window. The first 3 lags were left out of the fitting because localization error leads to artifactual sub-diffusion at this short time scale lowering the value of γ. To reduce computation time, we performed linear fitting of the MSD verses r on a log-log plot. The determination of systematic from non-systematic periods is made by setting a threshold on (t) of 1. We applied a minimum duration requirement of 15 seconds because we found that positive detections below that duration didn’t represent real active periods.
Reversal likelihood
The Reversal likelihood is a measure of how many step reversals occur within a cell’s time course. It is the ratio of the number of times a contraction step is immediately followed by an expansion step to the total number of contraction steps. The Reversal likelihood can take values between 0 and 1, 0 if every step of a cell is a contraction, and 1 if every contraction is followed by an expansion.
Rate of Change color overlay images
The rate of Area change, , was taken over a timescale of ∆t = 30 seconds.
Image editing and figure preparation
Spinning disk images and laser scanning confocal images were edited with ImageJ or Photoshop and images were leveled identically between samples for optimal appearance. Besides the ventral furrow embryos, all embryos were oriented with anterior left, posterior right, dorsal up and ventral down in the figure. Ventral furrow embryos were oriented with anterior left, posterior right, ventral up and dorsal down. All the graphs are generated in Graphpad Prism. Figures were prepared and labeled in Adobe Illustrator. Error bars indicate measured standard error in all graphs besides Fig. 7B.
Repeatability
All measurements were quantified from a minimum of three embryos and represented at least two individual trials.
Supplementary Material
Live imaging of CRISPR GFP:Rab35; tdTomato:Gish (plasma membrane marker) in control and LY294002 injected embryos. Scale bar equals 5 µm.
Live imaging of CRISPR GFP:Rab35; tdTomato:Gish (plasma membrane marker) in control and LY294002 injected embryos. Scale bar equals 5 µm.
Highlights:
Phospholipid cues in the form of PIP3 direct contractile ratcheting engagement
Loss of PIP3 leads to a switching behavior in which cell apices globally constrict
PIP3 enrichment sites are developmentally patterned
JAK/STAT signaling controls a medial signal for ratcheting engagement
Acknowledgements
We thank members of the Blankenship and Loerke labs for critical reading and constructive comments on the manuscript. We thank A. Kiger (UCSD) for GFP:Sbf fly stocks. This work was supported by grants from the NIH NIGMS: R01GM127447 and R15 GM126422 to J.T.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare no competing interests.
References
- Bertet C, Rauzi M, and Lecuit T (2009). Repression of Wasp by JAK/STAT signalling inhibits medial actomyosin network assembly and apical cell constriction in intercalating epithelial cells. Development 136(24), 4199–4212. [DOI] [PubMed] [Google Scholar]
- Blanchard GB, Murugesu S, Adams RJ, Martinez-Arias A, and Gorfinkiel N (2010). Cytoskeletal dynamics and supracellular organisation of cell shape fluctuations during dorsal closure. Development 137(16), 2743–2752. [DOI] [PubMed] [Google Scholar]
- Blankenship JT, Backovic ST, Sanny JS, Weitz O, and Zallen JA (2006). Multicellular rosette formation links planar cell polarity to tissue morphogenesis. Dev. cell 11(4), 459–470. [DOI] [PubMed] [Google Scholar]
- Britton JS, Lockwood WK, Li L, Cohen SM, and Edgar BA (2002). Drosophila’s insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev. cell 2(2), 239–249. [DOI] [PubMed] [Google Scholar]
- Cavanaugh KE, Staddon MF, Munro E, Banerjee S, and Gardel ML (2020). RhoA mediates epithelial cell shape changes via mechanosensitive endocytosis. Dev. Cell 52(2), 152–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clément R, Dehapiot B, Collinet C, Lecuit T, and Lenne PF (2017). Viscoelastic dissipation stabilizes cell shape changes during tissue morphogenesis. Curr. Biol 27(20), 3132–3142. [DOI] [PubMed] [Google Scholar]
- Czech MP (2000). PIP2 and PIP3: complex roles at the cell surface. Cell 100(6), 603–606. [DOI] [PubMed] [Google Scholar]
- Dambournet D, Machicoane M, Chesneau L, Sachse M, Rocancourt M, El Marjou A, … and Echard A (2011). Rab35 GTPase and OCRL phosphatase remodel lipids and F-actin for successful cytokinesis. Nat. Cell Biol 13(8), 981–988. [DOI] [PubMed] [Google Scholar]
- David DJ, Tishkina A, and Harris TJ (2010). The PAR complex regulates pulsed actomyosin contractions during amnioserosa apical constriction in Drosophila. Development 137(10), 1645–1655. [DOI] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R, and Zallen JA (2011). Oscillatory behaviors and hierarchical assembly of contractile structures in intercalating cells. Phys. Boil 8(4), 045005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frémont S, Romet-Lemonne G, Houdusse A, and Echard A (2017). Emerging roles of MICAL family proteins–from actin oxidation to membrane trafficking during cytokinesi J. Cell sci 130(9), 1509–1517. [DOI] [PubMed] [Google Scholar]
- Gaidarov I, and Keen JH (1999). Phosphoinositide–AP-2 interactions required for targeting to plasma membrane clathrin-coated pits. J. Cell Biol 146(4), 755–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassama-Diagne A, Yu W, ter Beest M, Martin-Belmonte F, Kierbel A, Engel J, and Mostov K (2006). Phosphatidylinositol-3,4,5-trisphosphate regulates the formation of the basolateral plasma membrane in epithelial cells. Nat. Cell Biol 8(9), 963–70. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont PJ, Machesky LM, Baldassare JJ, and Pollard TD (1990). The actin-binding protein profilin binds to PIP2 and inhibits its hydrolysis by phospholipase C. Science 247(4950), 1575–1578. [DOI] [PubMed] [Google Scholar]
- Haugh JM, Codazzi F, Teruel M, and Meyer T (2000). Spatial sensing in fibroblasts mediated by 3′ phosphoinositides. J. Cell Biol 151(6), 1269–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet S, Karatekin E, Tran VS, Fanget I, Cribier S, and Henry JP (2006). Analysis of transient behavior in complex trajectories: application to secretory vesicle dynamics. Biophys. J 91(9), 3542–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insall RH, and Weiner OD (2001). PIP3, PIP2, and cell movement—similar messages, different meanings?. Dev. Cell 1(6), 743–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmey PA, Xian W, and Flanagan LA (1999). Controlling cytoskeleton structure by phosphoinositide–protein interactions: phosphoinositide binding protein domains and effects of lipid packing. Chem. Phys. Lipids 101(1), 93–107. [DOI] [PubMed] [Google Scholar]
- Jean S, Cox S, Schmidt EJ, Robinson FL, and Kiger A (2012). Sbf/MTMR13 coordinates PI (3) P and Rab21 regulation in endocytic control of cellular remodeling. Mol. Biol. Cell 23(14), 2723–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett CE, Vanderleest TE, Miao H, Xie Y, Madhu R, Loerke D, and Blankenship JT (2017). Planar polarized Rab35 functions as an oscillatory ratchet during cell intercalation in the Drosophila epithelium. Nat. Commun 8(1), 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jodoin JN, Coravos JS, Chanet S, Vasquez CG, Tworoger M, Kingston ER, … and Martin AC (2015). Stable force balance between epithelial cells arises from F-actin turnover. Dev. Cell 35(6), 685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, and Davidson LA (2011). Punctuated actin contractions during convergent extension and their permissive regulation by the non-canonical Wnt-signaling pathway. Journal of cell science, 124(4), 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laketa V, Zarbakhsh S, Traynor-Kaplan A, MacNamara A, Subramanian D, Putyrski M, … and Schultz C (2014). PIP3 induces the recycling of receptor tyrosine kinases. Sci. signal 7(308), ra5-ra5. [DOI] [PubMed] [Google Scholar]
- Leevers SJ, Weinkove D, MacDougall LK, Hafen E, and Waterfield MD (1996). The Drosophila phosphoinositide 3‐kinase Dp110 promotes cell growth. EMBO J 15(23), 6584–6594. [PMC free article] [PubMed] [Google Scholar]
- Levayer R, Pelissier-Monier A, and Lecuit T (2011). Spatial regulation of Dia and Myosin-II by RhoGEF2 controls initiation of E-cadherin endocytosis during epithelial morphogenesis. Nat. Cell Biol 13(5), 529–540. [DOI] [PubMed] [Google Scholar]
- Levayer R, and Lecuit T (2012). Biomechanical regulation of contractility: spatial control and dynamics. Trends Cell Biol 22(2), 61–81. [DOI] [PubMed] [Google Scholar]
- Loerke D, and Blankenship JT (2020). Viscoelastic voyages–Biophysical perspectives on cell intercalation during Drosophila gastrulation. In Seminars in Cell Dev. Biol (Vol. 100, pp. 212–222). Academic Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maître JL, Niwayama R, Turlier H, Nédélec F, and Hiiragi T (2015). Pulsatile cell-autonomous contractility drives compaction in the mouse embryo. Nat. Cell Biol 17(7), 849–855. [DOI] [PubMed] [Google Scholar]
- Martin AC, Kaschube M, and Wieschaus EF (2009). Pulsed contractions of an actin–myosin network drive apical constriction. Nature 457(7228), 495–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AC, and Goldstein B (2014). Apical constriction: themes and variations on a cellular mechanism driving morphogenesis. Development 141(10), 1987–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason FM, and Martin AC (2011). Tuning cell shape change with contractile ratchets. Curr. Opin. Genet 21(5), 671–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason FM, Xie S, Vasquez CG, Tworoger M, and Martin AC (2016). RhoA GTPase inhibition organizes contraction during epithelial morphogenesis. J. Cell Biol 214(5), 603–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meili R, Ellsworth C, Lee S, Reddy TBK, Ma H, and Firtel RA (1999). Chemoattractant‐mediated transient activation and membrane localization of Akt/PKB is required for efficient chemotaxis to cAMP in Dictyostelium. EMBO J 18(8), 2092–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao H, Vanderleest TE, Jewett CE, Loerke D, and Blankenship JT (2019). Cell ratcheting through the Sbf RabGEF directs force balancing and stepped apical constriction. J. Cell Biol 218(11), 3845–3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao H, and Blankenship JT (2020). The pulse of morphogenesis: actomyosin dynamics and regulation in epithelia. Development 147(17). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel M, and Dahmann C (2020). Tissue mechanical properties modulate cell extrusion in the Drosophila abdominal epidermis. Development 147(5). [DOI] [PubMed] [Google Scholar]
- Montero JA, Kilian B, Chan J, Bayliss PE, and Heisenberg CP (2003). Phosphoinositide 3-kinase is required for process outgrowth and cell polarization of gastrulating mesendodermal cells. Curr. Biol 13(15), 1279–1289. [DOI] [PubMed] [Google Scholar]
- Munjal A, Philippe JM, Munro E, and Lecuit T (2015). A self-organized biomechanical network drives shape changes during tissue morphogenesis. Nature 524(7565), 351–355. [DOI] [PubMed] [Google Scholar]
- Munro E, Nance J, and Priess JR (2004). Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev. Cell, 7(3), 413–424. [DOI] [PubMed] [Google Scholar]
- Naga Prasad SV, Laporte SA, Chamberlain D, Caron MG, Barak L, and Rockman HA (2002). Phosphoinositide 3-kinase regulates β2-adrenergic receptor endocytosis by AP-2 recruitment to the receptor/β-arrestin complex. J. Cell Biol 158(3), 563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa T, Yamaguchi H, Itoh T, Kato M, Ijuin T, Yamazaki D, … and Takenawa T (2004). PtdIns (3, 4, 5) P 3 binding is necessary for WAVE2-induced formation of lamellipodia. Nat. Cell Biol 6(5), 420–426. [DOI] [PubMed] [Google Scholar]
- Parent CA, Blacklock BJ, Froehlich WM, Murphy DB, and Devreotes PN (1998). G protein signaling events are activated at the leading edge of chemotactic cells. Cell 95(1), 81–91. [DOI] [PubMed] [Google Scholar]
- Pickering K, Alves-Silva J, Goberdhan D, and Millard TH (2013). Par3/Bazooka and phosphoinositides regulate actin protrusion formation during Drosophila dorsal closure and wound healing. Development 140(4), 800–809. [DOI] [PubMed] [Google Scholar]
- Rauzi M, Lenne PF, and Lecuit T (2010). Planar polarized actomyosin contractile flows control epithelial junction remodelling. Nature 468(7327), 1110–1114. [DOI] [PubMed] [Google Scholar]
- Reversi A, Loeser E, Subramanian D, Schultz C, and De Renzis S (2014). Plasma membrane phosphoinositide balance regulates cell shape during Drosophila embryo morphogenesis. J. Cell Biol 205(3), 395–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakisaka T, Itoh T, Miura K, and Takenawa T (1997). Phosphatidylinositol 4, 5-bisphosphate phosphatase regulates the rearrangement of actin filaments. Mol. Cell. Biol, 17(7), 3841–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim K, Bottomley MJ, Querfurth E, Zvelebil MJ, Gout I, Scaife R, … and Panayotou G (1996). Distinct specificity in the recognition of phosphoinositides by the pleckstrin homology domains of dynamin and Bruton’s tyrosine kinase. EMBO J 15(22), 6241–6250. [PMC free article] [PubMed] [Google Scholar]
- Sawyer JK, Choi W, Jung KC, He L, Harris NJ, and Peifer M (2011). A contractile actomyosin network linked to adherens junctions by Canoe/afadin helps drive convergent extension. Mol. Biol. Cell 22(14), 2491–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sechi AS, and Wehland J (2000). The actin cytoskeleton and plasma membrane connection: PtdIns (4, 5) P (2) influences cytoskeletal protein activity at the plasma membrane. J. Cell Sci 113(21), 3685–3695. [DOI] [PubMed] [Google Scholar]
- Servant G, Weiner OD, Herzmark P, Balla T, Sedat JW, and Bourne HR (2000). Polarization of chemoattractant receptor signaling during neutrophil chemotaxis. Science 287(5455), 1037–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solon J, Kaya-Copur A, Colombelli J, and Brunner D (2009). Pulsed forces timed by a ratchet-like mechanism drive directed tissue movement during dorsal closure. Cell 137(7), 1331–1342. [DOI] [PubMed] [Google Scholar]
- Sun Y, Carroll S, Kaksonen M, Toshima JY, and Drubin DG (2007). PtdIns (4, 5) P2 turnover is required for multiple stages during clathrin-and actin-dependent endocytic internalization. J. Cell Biol 177(2), 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeton D, Parks S, Costa M, and Wieschaus E (1991). Gastrulation in Drosophila: the formation of the ventral furrow and posterior midgut invaginations. Development 112(3), 775–789. [DOI] [PubMed] [Google Scholar]
- Tamada M, Shi J, Bourdot KS, Supriyatno S, Palmquist KH, Gutierrez-Ruiz OL, and Zallen JA (2021). Toll receptors remodel epithelia by directing planar-polarized Src and PI3K activity. Developmental Cell doi: 10.1016/j.devcel.2021.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B, Messal ME, and Perrin-Schmitt F (1987). The twist gene: isolation of a Drosophila zygotle gene necessary for the establishment of dorsoventral pattern. Nucleic Acids Res 15(8), 3439–3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, and McCormick S (2010). The regulation of vesicle trafficking by small GTPases and phospholipids during pollen tube growth. Sex. Plant Reprod 23(2), 87–93. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Live imaging of CRISPR GFP:Rab35; tdTomato:Gish (plasma membrane marker) in control and LY294002 injected embryos. Scale bar equals 5 µm.
Live imaging of CRISPR GFP:Rab35; tdTomato:Gish (plasma membrane marker) in control and LY294002 injected embryos. Scale bar equals 5 µm.
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.


