Abstract
Dbs was identified initially as a transforming protein and is a member of the Dbl family of proteins (>20 mammalian members). Here we show that Dbs, like its rat homolog Ost and the closely related Dbl, exhibited guanine nucleotide exchange activity for the Rho family members RhoA and Cdc42, but not Rac1, in vitro. Dbs transforming activity was blocked by specific inhibitors of RhoA and Cdc42 function, demonstrating the importance of these small GTPases in Dbs-mediated growth deregulation. Although Dbs transformation was dependent upon the structural integrity of its pleckstrin homology (PH) domain, replacement of the PH domain with a membrane localization signal restored transforming activity. Thus, the PH domain of Dbs (but not Dbl) may be important in modulating association with the plasma membrane, where its GTPase substrates reside. Both Dbs and Dbl activate multiple signaling pathways that include activation of the Elk-1, Jun, and NF-κB transcription factors and stimulation of transcription from the cyclin D1 promoter. We found that Elk-1 and NF-κB, but not Jun, activation was necessary for Dbl and Dbs transformation. Finally, we have observed that Dbl and Dbs regulated transcription from the cyclin D1 promoter in a NF-κB-dependent manner. Previous studies have dissociated actin cytoskeletal activity from the transforming potential of RhoA and Cdc42. These observations, when taken together with those of the present study, suggest that altered gene expression, and not actin reorganization, is the critical mediator of Dbl and Rho family protein transformation.
The Dbl family of proteins is a large family of structurally related proteins that share an approximately 300-residue region of significant sequence similarity with Dbl, a transforming protein that was originally isolated from a diffuse B-cell lymphoma (reviewed in references 4 and 66). Members of this family that were discovered as transforming or invasion-inducing proteins include Vav, Ect2, Tim, Ost, Lsc, Lbc, Net, and Lfc. Other members include proteins identified as gene products of sequences that are rearranged in human diseases (BCR and FGD1) or as proteins with other catalytic functions, such as the SOS or RasGRF/CDC25 Ras guanine nucleotide exchange factors (GEFs). The deregulated expression of many Dbl family proteins causes tumorigenic growth and promotes invasion of a variety of cell types (66).
The region of sequence similarity that defines members of the Dbl family consists of a Dbl homology (DH) domain (unique to the family) arranged in tandem with a pleckstrin homology (PH) domain. The DH domains of many Dbl family proteins have been shown to serve as GEFs and activators of specific members of the Rho family of Ras-related small GTPases (reviewed in references 4 and 66). The Rho family comprises over 14 distinct family members, including RhoA, RhoB, RhoC, RhoD, RhoE, RhoG, Rac1, Rac2, Cdc42, TC10, TTF, Rnd1, and Rnd2 (reviewed in reference 73). Like Ras, Rho family proteins bind and hydrolyze GTP and cycle between biologically active GTP-bound and inactive GDP-bound forms (2). GEFs stimulate the exchange of GDP for GTP and are thus activators of Rho function. GTPase-activating proteins increase the low intrinsic rate of GTP hydrolysis, thus converting Rho proteins to the inactive state. Finally, guanine nucleotide dissociation inhibitors bind to Rho proteins and lock them into their existing nucleotide-bound state.
The PH domain is invariably located immediately COOH-terminal to the DH domain, and this invariant topography suggests a functional interdependence between the two domains. Consistent with this possibility, derivatives of Dbl family members that are truncated within either the DH or the PH domain are impaired in their transforming activity (22, 35, 51, 67–69). Although the precise role of the PH domain in regulating DH domain function remains to be clarified, present evidence supports two possible roles. First, we have shown that mutation of the PH domain of Lfc abolishes its transforming activity and that the addition of a plasma membrane-targeting sequence can restore Lfc transforming activity (69). Therefore, the PH domain may promote the translocation of Dbl family proteins to membranes, where its GTPase substrates are located. Second, several recent reports have demonstrated that an interaction between the PH domain and products of phosphoinositide 3-kinase is necessary to activate the catalytic activity of the DH domains of Vav and Sos, suggesting that the PH domain is a negative regulator of DH function (18, 40). However, the isolated DH domain of Trio was found to be more active when expressed together with the PH domain (29). Thus, a second function of the PH domain may be to serve as a positive or negative regulator of DH domain function via an intramolecular interaction.
The recent demonstrations that constitutively activated derivatives of the Rho family proteins RhoA, RhoB, RhoG, Cdc42, Rac1, and TC10 are transforming when expressed in rodent fibroblasts suggests that Dbl family members may transform cells by stimulating the activity of their GTPase substrates (25, 28, 39, 44–47). In support of this, we have observed that rodent fibroblasts that are transformed by Dbl family oncoproteins form similar foci composed of rounded, piled-up nonrefractile cells (64). This phenotype is distinct from that seen when cells are transformed with oncogenic Ras, Raf, or Src and is more similar to that of cells transformed by constitutively activated derivatives of Rac1 or RhoA. Furthermore, Dbl family protein transforming activity is associated with the activation of signaling pathways known to be mediated by their GTPase targets (64). Finally, it has been reported that coexpression of Dbl family members with dominant-inhibitory mutants of Rho family GTPases blocks their ability to transform NIH 3T3 cells (2, 3, 17, 23, 60). However, a Tiam1 mutant that lacked the DH domain could still cause invasion (16). Furthermore, although the transforming activity of FGD1 is dependent on its substrate, Cdc42, FGD1 transformation may be mediated by Cdc42-independent functions (65). Thus, Dbl family oncoprotein transforming activity may not always be completely attributable to the activation of its Rho family targets.
Although one of the immediate in vivo activities associated with Dbl family protein expression may be upregulation of Rho-related GTPases, the subsequent signaling events that lead to full cellular transformation have not yet been identified. Rho family proteins control multiple aspects of cellular behavior, including regulation of the actin cytoskeleton, transcriptional activation, and regulation of progression through the cell cycle (reviewed in reference 73), yet the relative contributions of these activities to full cellular transformation is unclear. For example, microinjection studies with Swiss 3T3 cells have shown that Cdc42 is involved in the extension of filopodia, Rac1 is involved in the formation of lamellipodia, and RhoA regulates the formation of actin stress fibers and focal adhesions. However, the recent demonstration that Rac1 mutants that are impaired in their ability to induce lamellipodium formation retain potent transforming activity in NIH 3T3 cells suggests that this activity is dispensable for transformation (63). Similarly, the actin reorganization functions of RhoA and Cdc42 have also been dissociated from their transforming functions (45, 52).
A second cellular activity that may contribute to Dbl family-mediated effects on cell growth is the ability of these proteins to stimulate transcriptional activation of specific genes. Both Dbl and Rho family members have demonstrated roles in the regulation of gene expression as measured by (i) activation of p38/Mpk2 (64, 71), an activator of the ATF-2 transcription factor; (ii) activation of the c-Jun NH2-terminal kinases (JNKs), activators of the ATF-2 and c-Jun transcription factors (8, 37, 41, 64); (iii) transcriptional activation of the serum response factor (SRF) (21, 64); (iv) activation of the NF-κB transcription factor (38, 43, 58); (v) activation of the ternary complex factor (TCF) protein Elk-1 (70); and (vi) regulation of expression from the cyclin D1 promoter (63, 64). However, only in the case of cyclin D1 expression has a good correlation been observed between a transcriptional event and the transforming activity of Dbl family members (64). Although no nuclear signaling events have been shown to be necessary to promote the transforming activity of either Dbl or Rho family members, signaling pathways that are regulated by the JNK activator SEK1 are required for transformation by Ras and other oncoproteins (7, 48, 50), and NF-κB activation is necessary for full transformation by Ras and Bcr-Abl (13, 49).
We previously identified Dbs in a retrovirus-based cDNA expression screen for transforming proteins that exhibit the ability to cause focus formation in NIH 3T3 mouse fibroblasts (68). Dbs is a member of the Dbl family and is the murine homolog of the rat protein Ost, although Dbs differs from Ost in having a COOH-terminal Src homology 3 domain and a considerably extended NH2-terminal domain (22). Dbs and Ost are most closely related to Dbl (51). Like other Dbl family members, Dbs is a potent activator of the JNK and p38 mitogen-activated protein kinases (MAPKs), can stimulate transcription from SRF and c-Jun promoter response elements, and has been shown to regulate expression from the cyclin D1 promoter (64). In this study, we show that Dbs exhibits guanine nucleotide exchange activity for RhoA and Cdc42 in vitro and that blocking the activation of these GTPases impairs Dbs transforming activity. We have also determined that the PH domain, which is essential for Dbs transforming activity, could be replaced by a plasma membrane targeting sequence. Thus, one role of the PH domain involves the translocation of Dbs to the plasma membrane. Finally, we found that Dbs and Dbl activation of Elk-1 and NF-κB, but not Jun, was required for their transforming activities. These observations suggest an important role for transcriptional activation in the regulation of Dbl family protein transforming activity.
MATERIALS AND METHODS
Molecular constructs.
The pAX142, pCTV3H, and pCTV3P mammalian expression vectors have been described previously (69). pAX142-dbl-HA1, pCTV3H-dbl-HA1, pAX142-dbs-HA6, and pCTV3H-dbs-HA6 encode transforming derivatives of the Dbl and Dbs proteins fused in frame at the NH2 terminus to an epitope from the hemagglutinin (HA) protein of influenza virus (64). pCTV3H-dbs-HA8 encodes residues 525 to 833 of the Dbs protein fused at the NH2 terminus to an HA epitope tag. It was made by replacing the FspI/BsiWI fragment of pCTV3H-dbs-HA6 with the FspI/BsiWI fragment of pCTV3H-dbs-D16 (68). pCTV3P-dbs-HA7 encodes the same fragment of Dbs as pCTV3H-dbs-HA8 (residues 525 to 833) except that it is fused at the COOH terminus to the plasma membrane-targeting sequence (GCMSCKCVLS) present at the COOH terminus of H-Ras. It was made by (i) cloning the 871-bp NsiI fragment from pCTV3-TL19-10c2 (68) into the PstI site of pBS-KS(+) (Stratagene) (pBS-TL19-10c2/S2), (ii) shuttling the EcoRV/SmaI fragment of pBS-TL19-10c2/S2 into the HpaI site of pCTV3P (69) to make pCTV3P-19-10B, and finally (iii) replacing the FspI/BsiWI fragment of pCTV3H-dbs-HA6 with the FspI/BsiWI fragment of pCTV3P-19-10B. We and others have shown that this membrane-targeting sequence can promote the membrane association of heterologous proteins (56).
RhoA(WT), RhoA(19N), Cdc42(WT), and Cdc42(17N) are wild-type and dominant-inhibitory versions of their respective GTPases that have been described previously (67). cDNAs encoding wild-type p190 RhoGAP (54) and C3 transferase were provided by R. Weinberg and J. Settleman, respectively.
pAX142-sek1(WT) and pAX142-sek1(AL) encode wild-type and dominant-inhibitory versions of SEK1, respectively. They were made by removing the BamHI fragments from pEBG-GST-sek1 and pEBG-GST-sek1(AL) (53), thus cleaving off the sequence encoding the glutathione S-transferase (GST) portion; filling in the ends with T4 DNA polymerase; and cloning the sequence into the SmaI site of pAX142. pAX142-mek1(WT) and pAX142-mek1(2A) encode the sequences for wild-type and dominant-inhibitory MEK1, respectively. They were made by removing the ApaI/XhoI fragments from pCMV-mek(WT) and pCMV-mek(2A) (kindly provided by N. Ahn), filling in the ends with T4 DNA polymerase, and cloning the sequence into the SmaI site of pAX142. pAX142-FLAG-IκBα(WT) and pAX142-IκBα(SS) encode the sequences for wild-type and super-repressor versions of IκBα, respectively. These were made by excising the MluI/SmaI fragments from pCMV4-IκBα and pCMV4-IκBα(SS) (kindly provided by A. Baldwin) and cloning the sequence into pAX142 digested with MluI/SmaI. The IκBα(WT) protein is fused to the FLAG epitope sequence at its NH2 terminus.
The reporter constructs utilized in the luciferase-coupled transcriptional assays have been described previously: Gal4–Elk-1 (32) and Gal-Jun(1–223) (57) encode the Gal4 DNA-binding domain fused to the transactivation domains of Elk-1 and Jun, respectively; 5×Gal4-luc contains the luciferase gene under control of the c-fos minimal promoter that contains tandem copies of the Gal4 DNA-binding sequence (57). HIV-luc contains the luciferase gene under control of the c-fos minimal promoter that contains tandem copies of the HIV NF-κB binding sequence (14). Cyclin D1-luciferase (CD1-Luc) consists of sequences from −963 of human cyclin linked to luciferase (provided by R. Pestell) (1). pCMVnlac encodes the sequences for the β-galactosidase gene under the control of the cytomegalovirus promoter (provided by J. Samulski).
Cell culture, transfection, and transformation assays.
NIH 3T3 and BOSC23 cells were maintained in Dulbecco’s modified Eagle medium (DMEM; high glucose) supplemented with 10% calf or fetal calf serum, respectively. Primary focus formation assays were performed with NIH 3T3 cells exactly as described previously (6). Briefly, NIH 3T3 cells were transfected by calcium phosphate coprecipitation in conjunction with a glycerol shock. Focus formation was scored at 14 days. Cognate empty vectors for each construct were employed as controls. NIH 3T3 cell lines that stably express Dbs-HA6 and Dbl-HA1 were generated by retrovirus infection. Retroviral particles were generated as described previously and then used to infect NIH 3T3 cells at low density (67). Following infection, the cells were selected for 14 days in growth medium supplemented with hygromycin (200 μg/ml). Multiple drug-resistant colonies (>100) were pooled to establish cell lines for the transformation assays. The growth properties of NIH 3T3 cells expressing Dbs-HA6 or Dbl-HA1 were compared in terms of their growth rates and saturation densities on plastic and for their abilities to proliferate in a low concentration of serum (2%) or soft agar by procedures that we have described previously (6). All assays for transformation were performed in triplicate.
Membrane fractionation analyses.
Mass populations of NIH 3T3 cells stably expressing HA epitope-tagged derivatives of Dbs were washed with ice-cold PBS and resuspended in cold TSA buffer (2 mM Tris, pH 8.0, 0.14 M NaCl) for 1 h. The lysates were homogenized in TSA buffer supplemented with 0.25 M sucrose, 1 mM EDTA, 100 μg of phenylmethylsulfonyl fluoride/ml, 25 μg of leupeptin/ml, and 1 mM NaVO4, and then centrifuged at 3,000 rpm in a Beckman centrifuge to acquire total protein (200 μl of supernatant). The remaining supernatant was then centrifuged at 100,000 × g to separate it into crude cytosolic (S100) and membrane (P100) fractions. The protein concentrations of the total, cytosolic, and membrane fractions were determined with a biciuchoninic acid protein assay kit (Pierce) with 30 μg of protein for each fraction, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to Immobilon-P membranes (Millipore), and probed with anti-HA epitope antibody (BabCo).
Transient expression reporter gene assays.
For transient expression reporter gene assays, NIH 3T3 cells were transfected by calcium phosphate coprecipitation, allowed to recover for 30 h, and starved in DMEM that was supplemented with 0.5% newborn-calf serum for 14 h before lysate preparation (6, 20, 62). Analysis of luciferase expression in transiently transfected NIH 3T3 cells was performed as described previously with enhanced chemiluminescent reagents and a Monolight 2010 luminometer (Analytical Luminescence, San Diego, Calif.) (20). β-Galactosidase activity in transiently transfected NIH 3T3 cells was determined exactly as described previously (30). All assays were performed in triplicate.
Protein expression.
Protein expression in transiently transfected 293 cells, or in stably transfected NIH 3T3 cell lines, was determined by Western blot analysis as described previously (69). Protein was visualized with enhanced chemiluminescence reagents (Amersham).
GDP dissociation assays.
The cDNA sequence encoding a GST-Dbs fusion protein was prepared by inserting a fragment of the Dbs cDNA (residues 604 to 968) that encompasses both the DH and PH domains into pAX142-GST (15). A fragment encoding the GST-Dbs fusion was then excised from pAX142 at the MluI/SalI sites, blunt ended, and inserted into the SmaI site of the baculovirus transfer vector pVL1393. Spodoptera frugiperda cells (SF21) were infected with the recombinant baculovirus, and then recombinant GST-Dbs protein was collected and purified as described previously (15). Preparation of GST-Cdc24 and GST-Lbc and their expression in SF21 cells have been described previously, as has the expression of GST-GRF in E. coli (15). Cdc24 and Lbc are Dbl family proteins and are GEFs for RhoA and Cdc42 and for RhoA, respectively, whereas GRF is a GEF for Ras proteins. The GTP-binding proteins RhoA, Rac1, Cdc42, and Ras were expressed as polyhistidine-tagged fusion proteins in Escherichia coli as described previously (15).
The GDP dissociation assays were carried out by the filter binding method at 24°C as described previously (15). Briefly, the GTP-binding proteins were loaded with [3H]GDP and then incubated with control and test proteins. After 15 min, the samples were quenched with ice-cold dilution buffer containing 10 mM MgCl2, collected by filter binding, and counted to determine the relative amount of bound [3H]GDP remaining.
GDP dissociation assays were also done with a bacterially expressed form of the isolated DH domain of Dbs. A cDNA fragment encoding the Dbs DH domain (residues 623 to 832) was generated by PCR and inserted into the NcoI/XhoI sites of the pET-28a (Novagen) bacterial expression vector. The bacterial expression construct was transformed into the BL21 (DE3) E. coli strain, and protein expression was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 25°C. The recombinant protein contained a COOH-terminal polyhistidine tag and was purified from bacterial lysate on a Ni nitrilotriacetic acid agarose column (Qiagen). Bacterially expressed Tiam1 DH-PH protein was kindly provided by J. Sondek. The GDP dissociation assay with the purified Dbs DH and Tiam1 DH-PH proteins was done with recombinant GST-RhoA, GST-Rac1, and GST-Cdc42 as described above with the exception that 30-μl aliquots of the exchange reaction mixture were removed at 0, 5, 10, and 15 min and quenched with ice-cold dilution buffer.
RESULTS
Stable expression of an activated derivative of the Dbs protein in NIH 3T3 cells is sufficient to cause anchorage- and serum-independent growth.
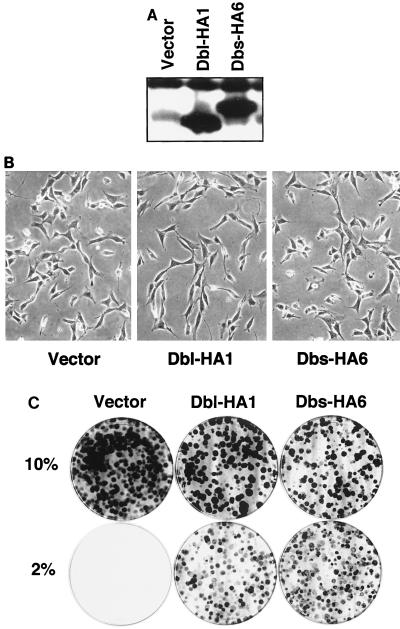
We have recently described the isolation of Dbs, a Dbl family member that is transforming in primary NIH 3T3 focus formation assays (68). Dbs-HA6 is an NH2-terminal-truncated, HA epitope-tagged derivative of Dbs (residues 525 to 1097) that contains an intact DH-PH domain module and retains full transforming activity (64). To further assess the effects of Dbs expression on the growth properties of NIH 3T3 cells, we established NIH 3T3 cell lines that stably express either Dbs-HA6 or Dbl-HA1. Dbl-HA1 is a transforming derivative of the Dbl oncoprotein (64) and was used as a positive control for transformation. NIH 3T3 cells were infected at low density with either the empty pCTV3H retroviral vector (69), pCTV3H-dbs-HA6, or pCTV3H-dbl-HA1 and then selected for 14 days in growth medium supplemented with hygromycin (200 μg/ml). Multiple drug-resistant colonies (>200) were then pooled to establish mass populations of cells that stably expressed Dbs-HA6 or Dbl-HA1 proteins at approximately equivalent levels (Fig. 1A).
FIG. 1.
Stable expression of Dbl and Dbs causes growth, but not morphologic transformation, of NIH 3T3 cells. (A) Expression of activated derivatives of Dbl and Dbs in pooled populations of hygromycin-resistant NIH 3T3 cells that were infected with pCTV3 retroviral constructs encoding the indicated protein. Protein expression was determined by Western blotting with an anti-HA epitope antibody. (B) NIH 3T3 cells that are stably overexpressing Dbl or Dbs do not show morphologic transformation compared to the empty-vector-transfected cell population. Shown are representative polyclonal populations of NIH 3T3 cells that express empty pCTV3 vector, Dbl-HA1, or Dbs-HA6. (C) NIH 3T3 cells that stably express Dbl or Dbs proliferate in low serum. Cells (103) were plated in duplicate in DMEM containing the indicated concentration of calf serum. The dishes were stained with crystal violet at 14 days to better visualize the appearance of colonies of proliferating cells.
Whereas the cellular morphology of isolated NIH 3T3 cells stably expressing Dbs-HA6 or Dbl-HA1 retained the nonrefractile appearance and well-adherent nature of untransformed NIH 3T3 cells (Fig. 1B), the cells did exhibit significant secondary focus-forming activity, grew well in low serum (Fig. 1C), and were able to grow in an anchorage-independent fashion. Thus, Dbs-expressing NIH 3T3 cells exhibit a transformed phenotype similar to that seen for NIH 3T3 cells transformed by other members of the Dbl family as well as by activated derivatives of Rho family proteins (4, 66, 73). We also observed previously that NIH 3T3 cell lines that stably express activated derivatives of Rac1 and RhoA have a higher growth rate and grow to higher saturation densities than control cell lines (25). Consistent with this, NIH 3T3 cells that stably express Dbs-HA6 and Dbl-HA1 exhibit high levels of expression of the cell cycle protein cyclin D1 (64). However, although we observed that cell lines that stably express Dbl-HA1 and Dbs-HA6 grow to a higher saturation density than control cell lines (Table 1), we observed no differences in growth rate between experimental and control cell lines (data not shown). Thus, the differences in cyclin D1 expression that we have observed previously are not reflected in a change in the growth rate of the Dbs- and Dbl-transformed cells when they are grown under high-serum conditions (10%).
Dbs exhibits GEF activity for RhoA and Cdc42 in vitro.
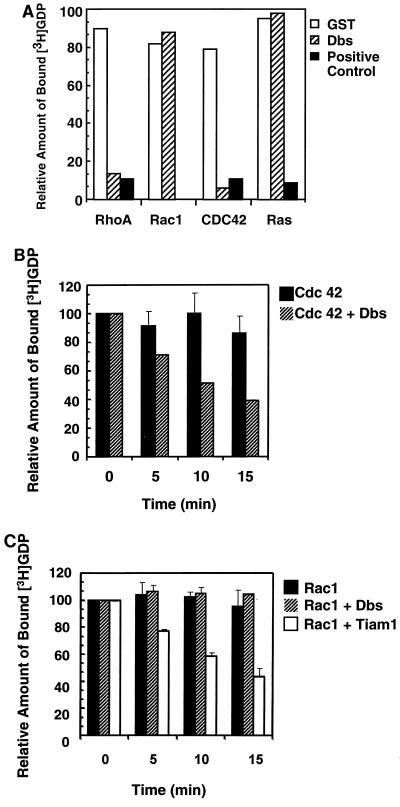
Based on numerous biochemical studies performed on members of the Dbl family, it is generally presumed that DH domain-containing proteins will possess GEF activity specific for members of the Rho family (reviewed in references 4 and 66). Although this has generally proven to be the case, there are several notable exceptions in which no GEF activity has been assigned to Dbl family members (e.g., RasGRF and Ect2) (35, 55). Dbs shows the greatest sequence similarity to Dbl and Ost, both of which have been shown to possess GEF activity for RhoA and Cdc42 but not for Rac1 in vitro (22). To determine whether Dbs shares this biochemical property with these closely related family members, we expressed and purified a GST fusion protein that contained the tandem DH-PH domains as well as some flanking sequences of Dbs. Purified GST-Dbs DH-PH was highly effective in stimulating the dissociation of [3H]GDP from bacterially expressed RhoA and Cdc42, but not Rac1 or Ras (Fig. 2A). The rate at which Dbs catalyzed GDP dissociation from RhoA and Cdc42 was equivalent to that seen with Lsc and Cdc24, respectively. This in vitro profile is similar to those that have been reported for Dbl and Ost (22), suggesting that these three structurally related proteins share common targets for their exchange activity.
FIG. 2.
Dbs exhibits GEF activity for RhoA and Cdc42 but not Rac1. (A) Dbs exhibits GEF activity for RhoA and Cdc42, but not Rac1 or Ras, in vitro. Two micrograms of the various recombinant GTP-binding proteins were preloaded with [3H]GDP and incubated with 2 μg of GST or 1 μg of GST-Dbs before termination of the reaction after 15 min. One microgram of GST-Lbc was used as a positive control for assaying stimulated dissociation of GDP from RhoA, 1 μg of GST-Cdc24 was used as a control for stimulated GDP dissociation from Cdc42, and 1 μg of GST-Ras-GRF was used as a control for stimulated GDP dissociation from Ras. (B) Time course for Cdc42 nucleotide exchange catalyzed by the isolated Dbs DH domain. Reaction mixtures containing 4 μM [3H]GDP-preloaded GST-Cdc42 and 1 μM Dbs DH domain were sampled at 0, 5, 10, and 15 min and terminated as described above. (C) The isolated Dbs DH domain fails to stimulate GDP dissociation from Rac1. Reaction mixtures containing 4 μM [3H]GDP-preloaded GST-Rac1 and 1 μM Dbs DH domain or 1 μM Tiam1 DH-PH domain were sampled at 0, 5, 10, and 15 min. The results (+ standard deviations) are the average of two independent experiments.
Since the PH domain may serve as a negative regulator of DH domain function (18, 40), we also analyzed the GEF activity of the isolated DH domain of Dbs. Bacterially expressed Dbs DH protein catalyzed GDP dissociation from Cdc42 (Fig. 2B) but not Rac1 (Fig. 2C). In contrast, bacterially expressed versions of the isolated tandem DH-PH domains of Tiam1 (Fig. 2C) or the isolated DH domain of Vav2 (not shown) catalyzed GDP dissociation from Rac1, indicating that the lack of activity seen with the Dbs DH domain was not due to inactive Rac1 protein. Thus, like Dbl, Dbs does not serve as a GEF for Rac1 in vitro.
The PH domain of Dbs regulates plasma membrane association.
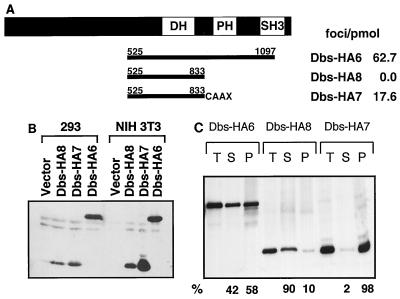
To further assess the role of the DH-PH domain module of the Dbs protein in regulating transforming activity, we generated a derivative of Dbs in which the PH domain had been replaced by the plasma membrane-targeting sequence present at the COOH terminus of H-Ras. It was shown previously that this membrane localization signal is sufficient to restore transforming activity to a PH domain-minus derivative of Lfc, but not Dbl (69, 72). Whereas the removal of the PH domain of the Dbs protein (Dbs-HA8) completely eliminated its focus-forming activity, the addition of the plasma membrane-targeting sequence (Dbs-HA7) restored potent transforming activity (Fig. 3A). The loss of transforming activity associated with Dbs-HA8 was not due to inherent instability of the protein, since good expression was observed in transiently transfected 293 cells and stably transfected NIH 3T3 cells (Fig. 3B).
FIG. 3.
Transforming activity of a PH domain-minus derivative of Dbs is restored by the addition of a plasma membrane-targeting signal from H-Ras. (A) The domain structure of the full-length Dbs protein is illustrated in the upper line (DH, Dbl homology domain; PH, pleckstrin homology domain; SH3, Src homology 3 domain), and the lines below indicate the regions of the protein included in predicted translation products of the cDNA derivatives. The numbers refer to amino acid positions in the full-length protein. The Dbs-HA7 derivative is fused to the H-Ras COOH-terminal plasma membrane-targeting sequence that includes the CAAX tetrapeptide isoprenylation signal (designated CAAX). All derivatives were fused at their NH2 termini to an HA epitope tag. The values at the right indicate the transforming activity of the different derivatives in NIH 3T3 cell focus formation assays. The data shown are representative of three independent assays done on triplicate plates. (B) Expression of HA epitope-tagged Dbs proteins. Epitope-tagged proteins were expressed transiently in 293 cells (left-hand lanes) or stably in NIH 3T3 cells (right-hand lanes) and detected by Western blotting with an anti-HA epitope antibody. (C) Subcellular localization of PH domain variants of Dbs. HA epitope-tagged proteins were expressed stably in NIH 3T3 cells. The cells were lysed and fractionated at 100,000 × g. Thirty micrograms of protein from total (T), soluble (S), and particulate (P) fractions were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Protein expression was determined with the anti-HA epitope antibody (BabCo). The relative amounts of protein in particulate and soluble fractions (%) were determined with a PhosphorImager (Molecular Dynamics).
These results suggest that the recruitment of Dbs to the plasma membrane via a PH domain-mediated mechanism is a necessary step for the activation of Dbs GEF function in vivo. To address this directly, we determined if loss of the PH domain decreased Dbs membrane association and if addition of the membrane-targeting sequence restored efficient membrane association. NIH 3T3 cells stably expressing each Dbs protein were used for fractionation analyses into crude cytosolic (S100) or particulate, membrane-containing (P100) fractions. Whereas the PH domain-containing Dbs-HA6 protein showed 60% association with the particulate fraction, deletion of the PH domain (Dbs-HA8) resulted in a predominantly cytosolic protein (Fig. 3C). Addition of the membrane-targeting sequence (Dbs-HA7) resulted in essentially complete association with the particulate fraction. Interestingly, although membrane targeting strongly restored a degree of Dbs membrane association that was greater than that seen with the counterpart with the PH domain intact, it showed a level of transforming activity that was more than threefold less. This suggests that the PH domain may have a function in addition to membrane targeting that is not restored by addition of a membrane-targeting sequence.
Dbs transforming activity is mediated by Rho family GTPases.
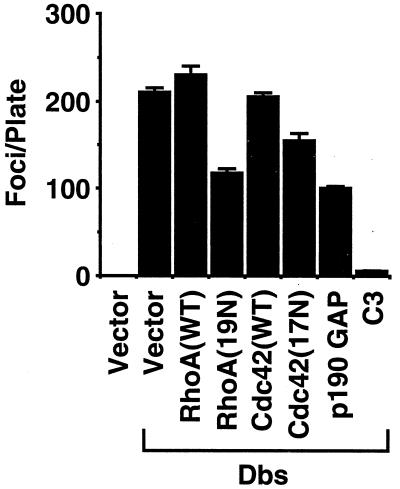
If Cdc42 and RhoA are required for Dbs transformation, then blocking the endogenous activity of these GTPases should block Dbs transforming activity. We have previously described dominant-inhibitory versions of the Cdc42 and RhoA proteins [Cdc42(17N) and RhoA(19N), respectively] that are analogous to the Ras(17N) mutant, and both have been shown to be specific inhibitors of their respective GTPases (25, 67). We determined whether the coexpression of Cdc42(17N) or RhoA(19N) could block the focus-forming activity of Dbs-HA6. Coexpression of the Dbs-HA6 protein with dominant-negative RhoA or Cdc42 alone partially inhibited transforming activity (50 and 30%, respectively), whereas no inhibition was observed with the coexpression of wild-type RhoA or Cdc42 (Fig. 4).
FIG. 4.
Suppression of Dbs-mediated transformation by p190 GAP, C3 transferase, or dominant-negative RhoA and Cdc42. NIH 3T3 cells were cotransfected with pAX142-dbs-HA6 (500 ng) and with either empty vector (500 ng) or vector containing the indicated proteins (500 ng). The appearance of foci of transformed cells was quantitated after 14 days. Clonogenic analyses showed that transfection of the C3 expression plasmid did not cause significant inhibition of cell growth: pRc-CMV (empty vector), 210 ± 16 drug-resistant colonies; pRc-CMV-C3, 176 ± 26 colonies (data are representative of two independent experiments and are the average of three plates). The data shown are representative of three independent determinations, with each determination representing the average number of foci from three dishes. The error bars indicate standard deviations.
The transformation inhibition seen with the dominant-negative RhoA and Cdc42 proteins may not be specific but instead may be a consequence of nonspecific growth inhibition. To examine this possibility, we determined the consequences of expressing the dominant-negative derivatives on the growth properties of NIH 3T3 cells. NIH 3T3 cells were transfected with equimolar amounts of either pZIP-rhoA(WT), pZIP-rhoA(19N), pZIP-cdc42(WT), or pZIP-cdc42(17N) and selected for 14 days with G418; and then the appearance of drug-resistant colonies was quantitated. No significant differences were observed between wild-type and dominant-negative derivatives with respect to the number of colonies that arose (data not shown). Thus, the inhibition of focus-forming activity that we observed could not be attributed to a nonspecific growth inhibition associated with the mutant proteins. However, we did see a growth-inhibitory activity with the Rac1(17N) dominant negative. Consequently, we could not use this mutant to verify that Rac1 is not required for Dbs transforming activity.
To further examine the role of Rho family proteins in regulating Dbs transforming activity we examined the consequences of cotransfecting Dbs-HA6 with plasmid constructs that encode cDNA sequences for p190 GAP and for C3 transferase. p190 GAP encodes GAP activity that is specific for members of the Rho family (including RhoA and Cdc42) and thus functions as a general inhibitor of Rho family function (54). C3 is an exoenzyme that has been shown to ADP-ribosylate and thus inactivate specific members of the Rho family, including RhoA, RhoB, and RhoC (but not Rac1 or Cdc42) (5). Whereas neither C3 nor p190 GAP exhibited growth inhibition when expressed in NIH 3T3 cells (see the legend to Fig. 4), both were effective in blocking the transforming activity of Dbs-HA6. These results further support the requirement for Rho family GTPases in mediating Dbs transformation. The very potent inhibition seen with C3, compared with that seen with dominant-negative RhoA, may simply reflect its more effective inhibition of RhoA function. Dominant-negative RhoA may require a very high level of expression to effectively sequester Dbs and prevent its activation of endogenous RhoA. These results further support the requirement for Rho family GTPases in mediating Dbs transformation. Additionally, they suggest that RhoA may be more important for Dbs transformation than Cdc42.
Dbs and Dbl transformation is independent of SEK1 activation.
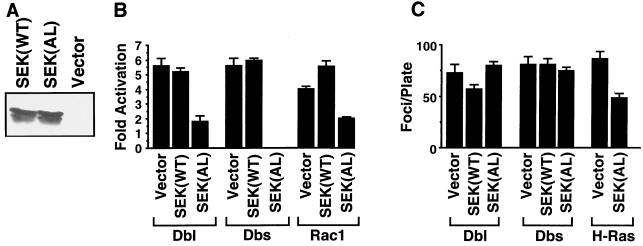
We and others have shown that Dbl family members are potent activators of JNK MAPKs and their in vivo substrate, the Jun transcription factor (8, 37, 41, 64). However, it has been observed for at least one family member (Lfc) that a derivative that is completely impaired in its transforming activity is still able to stimulate the activation of JNK-mediated signaling, suggesting that activation of this pathway is not sufficient for transformation (64). To assess the role of JNK-Jun signaling in mediating Dbs transforming activity we determined if a dominant-negative mutant of the JNK activator SEK1-JNKK (53) could block the transforming activity of Dbs and the closely related Dbl protein. Dominant-negative mutants of SEK1 have previously been shown to selectively block JNK activation by a variety of stimuli (37, 53, 61). We have constructed wild-type and dominant-inhibitory [SEK1(AL)] versions of the SEK1 protein that are expressed from the pAX142 mammalian expression vector, where expression is regulated by the EF-1α promoter (Fig. 5A). Coexpression of dominant-inhibitory SEK1(AL), but not SEK(WT), blocked Dbl and Dbs activation of Jun, indicating that Jun activation was occurring via a SEK-dependent mechanism (Fig. 5B). SEK1(AL) also blocked activation of Jun by an activated derivative of Rac1, which is in accordance with previous observations that Rac1 is a potent activator of the SEK-JNK-Jun signaling pathway (8, 37).
FIG. 5.
Activation of SEK1-mediated signaling pathways is dispensable for Dbl and Dbs transformation. (A) Expression of wild-type and dominant-inhibitory SEK1 in transiently transfected 293 cells, or in stably transfected NIH 3T3 cells, was determined with the C-20 rabbit polyclonal anti-SEK1 antibody (Santa Cruz Biotechnology). The lower band presumably corresponds to either a cleavage product or a nonspecific degradation product or is the result of translational initiation from an internal methionine. (B) Dominant-negative SEK(AL) blocks activation of c-Jun by Dbl and Dbs. NIH 3T3 cells were cotransfected with either pAX142-dbl-HA1 (500 ng), pAX142-dbs-HA6 (500 ng), or pAX142-rac1(61L) (500 ng) and vector pAX142, pAX142-sek(AL), or pAX-sek(WT) (500 ng) along with Gal4–c-Jun (500 ng), the Gal4-responsive 5×Gal4-Luc construct (2.5 μg), and pCMVnlac (0.5 μg) as an internal control for transfection efficiency and nonspecific growth inhibition. Luciferase and β-galactosidase activities were measured and expressed as fold activation relative to the level of activation seen with the empty vector control. Luciferase activity was standardized relative to β-galactosidase activity. Data shown are representative of at least three independent experiments performed on duplicate plates. (C) Dominant-negative SEK(AL) does not block focus formation by Dbl and Dbs. NIH 3T3 cells were cotransfected with pAX142-dbl-HA1 (500 ng), pAX142-dbs-HA6 (500 ng), or pAX-H-ras(61L) and either empty vector (500 ng), pAX142-sek1(WT) (500 ng), or pAX142-sek(AL) (500 ng). Foci were counted at 14 days. The data presented are representative of three independent determinations performed on triplicate plates. The error bars indicate standard deviations.
Next, we determined if SEK1(AL) inhibition of Jun activation was associated with a change in Dbl and Dbs transforming potency. As we have shown previously, coexpression of dominant-negative SEK1(AL) significantly reduced H-Ras(61L) focus-forming activity (7) (Fig. 5C). In contrast, cotransfection of Dbl or Dbs with either SEK(WT) or SEK(AL) had no effect on transformation as measured by focus formation in NIH 3T3 cells. These observations suggest that Jun and JNK activation are not necessary for Dbl and Dbs transformation, which is in accordance with our previous observations that activation of the JNK pathway and transforming activity can be genetically uncoupled in at least one Dbl family member (64). These results also suggest that Dbl and Dbs transform NIH 3T3 cells through a mechanism that is distinct from Ras transformation.
Dbl and Dbs transformation is dependent on MEK1 activation.
Whereas Dbl family members are potent activators of the JNK-mediated signaling pathway, they have been found to be poor activators of the ERK MAPKs (10, 34, 42). Consistent with this, we are unable to detect activation of ERK1 or ERK2 in 293 cells transiently transfected with expression plasmids encoding Dbl and Dbs or in NIH 3T3 cells stably expressing these two proteins (data not shown). However, there is some evidence that the highly conserved Raf-MEK-ERK signaling pathway may contribute to Dbl family protein signaling and transformation. We have observed elevated levels of activated ERK1 and ERK2 in NIH 3T3 cells stably transfected with Vav or Dbl, and we have shown that dominant-inhibitory mutants of ERK1 and ERK2 can partially inhibit Dbl transforming activity (24).
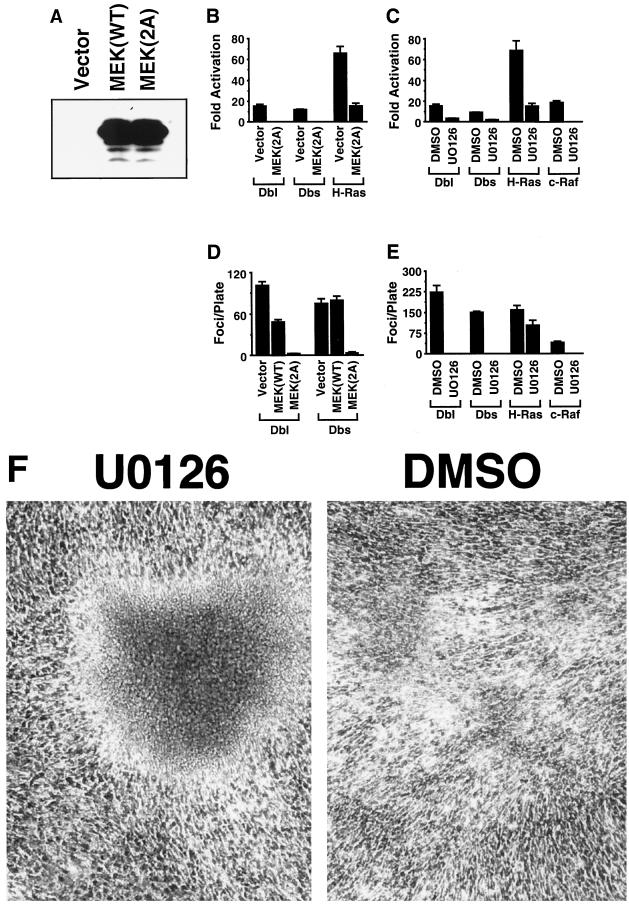
To further assess the ability of Dbl family members to stimulate the activation of the Raf-MEK-ERK signaling pathway, we examined the abilities of Dbl and Dbs to transcriptionally activate the ERK substrate Elk-1. Both Dbl and Dbs are efficient activators of Elk-1 in transcription-coupled luciferase assays, albeit at a lower level than that stimulated by activated Ras (Fig. 6B). Although Elk-1 has been shown to be activated primarily by ERKs, it is also activated by the JNK and p38 MAPKs in response to a variety of extracellular stimuli (36, 59). To determine if Dbl and Dbs activation of Elk-1 occurs through stimulation of a MEK-ERK signaling pathway, we measured Elk-1 activation by Dbl and Dbs in the presence of specific inhibitors of MEK1 and MEK2. MEK(2A) is a dominant-inhibitory derivative of MEK1 that has been shown to antagonize the activation of endogenous MEK1, whereas U0126 is a pharmacological agent that specifically inhibits MEK1 and MEK2 activity (11, 12). We have constructed wild-type and dominant-inhibitory versions of MEK1 that are expressed from the EF-1α promoter of pAX142 (Fig. 6A). Both MEK1(2A) and U0126 inhibited activation of Elk-1 by Dbl and Dbs as well as by activated derivatives of Ras and Raf (Fig. 6B and C), suggesting a common mechanism of activation of Elk-1 through MEK-mediated signaling.
FIG. 6.
Activation of MEK-mediated signaling pathways is necessary for Dbl and Dbs transformation. (A) Expression of wild-type and dominant-inhibitory MEK1 in transiently transfected 293 cells. MEK1 expression was determined with the C-18 rabbit polyclonal anti-MEK1 antibody (Santa Cruz Biotechnology). (B) Dominant-negative MEK(AL) blocks activation of Elk-1 by Dbl and Dbs. NIH 3T3 cells were cotransfected with either pAX142-dbl-HA1 (500 ng), pAX142-dbs-HA6 (500 ng), or pAX142-ras(61L) (50 ng) and vector (500 ng) or pAX142-mek1(2A) (500 ng), along with Gal4–Elk-1 (500 ng), 5×Gal4-Luc (2.5 μg), and pCMVnlac (0.5 μg). The data shown are calculated and presented as in Fig. 5B and are representative of at least three independent experiments performed on duplicate plates. (C) The MEK1/2-specific inhibitor U0126 blocks activation of Elk-1 by Dbl, Dbs, H-Ras, or c-Raf-1. NIH 3T3 cells were transfected with pAX142-dbl-HA1 (500 ng), pAX142-dbs-HA6 (500 ng), pAX142-H-ras(61L) (50 ng), or pZIP-raf(22W) (500 ng) along with Gal4–Elk-1 (500 ng), 5×Gal4-Luc (2.5 μg), and pCMVnlac (0.5 μg). U0126 (30 μM) or dimethyl sulfoxide was added directly to the growth medium 12 h prior to lysate preparation. The data shown are calculated as in Fig. 5B and are representative of three experiments performed on duplicate plates. (D) Dominant-negative MEK(2A) blocks focus formation by Dbl and Dbs. NIH 3T3 cells were transfected with pAX142-dbl-HA1 (500 ng) or pAX142-dbs-HA6 (500 ng) and either empty vector (500 ng), pAX142-mek1(WT) (500 ng), or pAX142-mek1(2A) (500 ng). Foci were counted at 14 days. The data presented are representative of three independent experiments performed on triplicate plates. (E) U0126 blocks focus formation by Dbl, Dbs, and Raf but not Ras. NIH 3T3 cells were transfected with pAX142-dbl-HA1 (500 ng), pAX142-dbs-HA6 (500 ng), pZIP-raf(22W) (500 ng), or pAX142-ras(63L) (50 ng). U0126 (30 μM) or dimethyl sulfoxide was added to the medium at 48 h posttransfection and in all subsequent medium changes. Foci were counted at 14 days. The data presented are representative of three independent experiments performed on triplicate plates. (F) Ras focus morphology is distinct when the transfected cultures are maintained in the presence of U0126.
To determine a possible role for the MEK–ERK–Elk-1 signaling pathway in Dbl and Dbs transformation, we measured Dbl and Dbs transforming potency in the presence of MEK(2A) or U0126. Although U0126 and MEK(2A) do not significantly impair growth of NIH 3T3 cells (data not shown), they are both potent inhibitors of Dbl and Dbs transforming activities when assayed in a primary focus formation assay (Fig. 6D and E). Thus, although Dbl and Dbs are relatively weak activators of MEK–ERK–Elk-1 signaling, this activation may be essential for transformation. Alternatively, since we did not detect significant ERK activation by Dbl and Dbs, it remains possible that basal ERK activity is required for Dbl and Dbs transforming activity.
Interestingly, we observed that the U0126 inhibitor had relatively little effect on the focus-forming activity of activated Ras, yet the transforming activity of Raf was completely inhibited in the same assay. Although it is not clear what the additional pathways are that are required for Ras-induced focus formation, we observed a dramatic change in the morphology of Ras-induced foci in the presence of the U0126 inhibitor such that they more closely resemble foci that are induced by activated derivatives of Rho or Dbl family members (Fig. 6F). These foci also resemble those caused by Ras effector domain mutants (37G and 40C) that are impaired in Raf activation (26).
Dbs and Dbl transformation is dependent upon NF-κB activation.
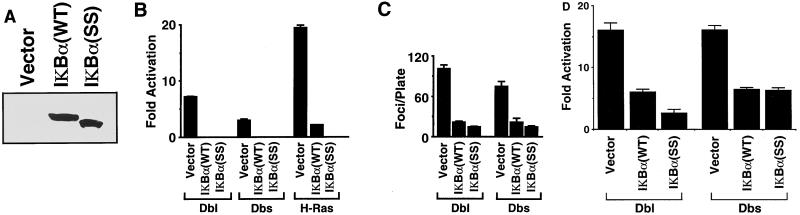
It has been reported recently that both Dbl and Rho family members can drive transcription from NF-κB-responsive elements (38, 43). Since transformation by Ras and Bcr-Abl has been shown to occur in an NF-κB-dependent manner (13, 49, 58), we wished to determine if NF-κB activation is necessary for Dbl and Dbs transforming activity. Transient transfection of NIH 3T3 cells with Dbs-HA6 and Dbl-HA1 led to a significant activation of expression of an NF-κB-dependent reporter, which is in accordance with previous observations with the Dbl and Ost proteins (Fig. 7B) (38). IκBα(SS) is a derivative of IκBα that is unable to be inducibly phosphorylated or degraded in response to stimuli and thus is a potent inhibitor of NF-κB activation. We have generated pAX142 expression vectors encoding wild-type IκBα and mutant IκBα(SS) that are expressed from the pAX142-encoded EF-1α promoter (Fig. 7A). Coexpression with IκBα(SS), as well as IκBα(WT), blocked Dbl and Dbs activation of the NF-κB-dependent reporter, indicating that NF-κB regulates the transcriptional response (Fig. 7B).
FIG. 7.
Activation of NF-κB is necessary for Dbl and Dbs transformation. (A) Expression of wild-type and super-repressor IκBα(SS) in transiently transfected 293 cells. IκBα expression was determined with the C-21 rabbit polyclonal antibody (Santa Cruz Biotechnology). The difference in mobility is attributable to the presence of a FLAG epitope tag at the NH2 terminus of the wild-type derivative. We also determined that expression of IκBα(SS) did not cause significant inhibition of growth of NIH 3T3 cells by using a clonogenic assay by drug selection of transfected NIH 3T3 cells: pCTV3 (empty vector), 311 ± 23 colonies; pCTV3-IκBα(WT), 380 ± 41 colonies; pCTV3-IκBα(SS), 349 ± 14 colonies (the data are representative of independent assays and are the average of three plates). (B) Both wild-type and super-repressor IκBα block activation of NF-κB by Dbl, Dbs, and Ras. NIH 3T3 cells were cotransfected with either pAX142-dbl-HA1 (500 ng), pAX142-dbs-HA6 (500 ng), or pAX142-H-ras(61L) (50 ng) and vector (500 ng) or pAX142-IκBα(SS) (500 ng), along with the HIV-luc construct (2.5 μg) and pCMVnlac (0.5 μg). The data shown are calculated and presented as in Fig. 5B and are representative of at least three independent experiments performed on duplicate plates. (C) Wild-type and super-repressor IκBα(SS) block focus formation by Dbl and Dbs. NIH 3T3 cells were transfected with pAX142-dbl-HA1 (500 ng) or pAX142-dbs-HA6 (500 ng) and either empty vector (500 ng), pAX142-IκBα(WT) (500 ng), or pAX142-IκBα(SS) (500 ng). Foci were counted at 14 days. The data presented are representative of three independent experiments performed on triplicate plates. (D) Dbl and Dbs activate transcription from the cyclin D1 promoter in an NF-κB-dependent manner. NIH 3T3 cells were cotransfected with either pAX142-dbl-HA1 (500 ng) or pAX142-dbs-HA6 (500 ng) and vector (500 ng), pAX142-IκBα(WT) (500 ng), or pAX142-IκBα(SS) (500 ng), along with CD1-Luc (2.5 μg) and pCMVnlac. Fold activation was calculated as described in the legend to Fig. 5B and is representative of at least three independent experiments performed on duplicate plates.
Next, we determined whether NF-κB activity is necessary for Dbl and Dbs transformation. IκBα(WT) and IκBα(SS) exhibited no significant growth inhibition when expressed alone in NIH 3T3 cells (See the legend to Fig. 7), yet they were equally effective in blocking the transforming activity of Dbl-HA1 and Dbs-HA6 (Fig. 7C). This suggests that transformation of NIH 3T3 cells by Dbl and Dbs requires NF-κB activation.
We have previously observed a close correlation between transforming activity and the ability of Dbl family members to drive transcription from the cyclin D1 promoter (64). Since Dbl and Dbs transformation is dependent upon NF-κB activation, we wished to determine if activation of the cyclin D1 promoter by Dbl and Dbs is occurring in an NF-κB-dependent manner. Whereas Dbl and Dbs alone showed activation of the CD1-luc reporter construct, activation was significantly impaired in cotransfections with IκBα(SS) or IκBα(WT), indicating the dependence of the response upon NF-κB activity (Fig. 7D). These observations establish a link between NF-κB activation and upregulation of cyclin D1 and provide an explanation for the elevated levels of cyclin D1 that we have observed previously in Dbs- and Dbl-transformed cells (64). However, since coexpression of IκBα(SS) caused a complete inhibition of NF-κB activation, but only partial inhibition of cyclin D1 upregulation, Dbl and Dbs may also stimulate the cyclin D1 promoter through NF-κB-independent promoter elements.
DISCUSSION
Dbs is a Dbl family member that was isolated as a transforming protein in a screen for proteins whose expression causes focus formation in NIH 3T3 transformation assays (68). Like all Dbl family members, Dbs contains tandem DH and PH domains, both of which are required for transformation. Thus, Dbs transforming activity is likely to be mediated by its ability to cause aberrant upregulation of specific Rho family protein signaling. However, the precise targets of Dbs GEF activity, and the signaling activity important for Dbs transformation, have not been established. In the present study, we show that Dbs, like Dbl, is an activator of RhoA and Cdc42 and causes transformation in a RhoA- and Cdc42-dependent manner. Since a plasma membrane-targeting sequence can restore the loss of PH domain function and promotes Dbs transformation, the PH domain may be involved in regulating Dbs membrane association. Finally, we found that Dbs and Dbl transformation is dependent on activation of the Elk-1 and NF-κB, but not Jun, transcription factors, indicating that transcription regulation is a key mediator of Dbs and Dbl transformation. Cyclin D1 may be an important target of this regulation.
Dbs is the murine homolog of the rat Ost protein, and Dbs and Ost show the greatest amino acid identity with Dbl (19, 22). Since Dbl and Ost have been shown to be GEFs for RhoA and Cdc42, but not Rac1, we anticipated that Dbs would also serve as a GEF for these specific Rho family GTPases. Our sequence alignment analyses of the DH domains of all known Dbl family proteins show that their degree of sequence similarity correlates closely with substrate specificity (66). Consistent with these observations in vitro, we also found that Dbs transforming activity was partially inhibited by coexpression of dominant-negative mutants of RhoA and Cdc42. The incomplete nature of this inhibition may reflect the incomplete efficiency of these dominant-negative proteins in forming nonproductive complexes with Dbs. However, the fact that Dbs exhibits a potent focus-forming activity in primary focus formation assays that is far greater than that seen when GTPase-deficient mutants of RhoA and Cdc42 are coexpressed (no activity in primary focus formation assays) suggests that Dbs transforming activity may involve RhoA- and Cdc42-independent mechanisms. Alternatively, it may reflect the possibility that Dbs transformation is caused by promoting enhanced GDP-GTP cycling of RhoA and Cdc42 rather than constitutive GTP binding. This possibility is supported by the previous observation that Cdc42 may exhibit more potent transforming activity when its GDP-GTP cycling is enhanced than when it is rendered constitutively GTP bound (28).
The invariant topography of the DH and PH domains of Dbl family proteins argues that their functions are interdependent. Consistent with such a scenario, mutation of the PH domain typically causes loss of Dbl family oncoprotein transforming activity (73). Our observation that the PH domain of Dbs can be functionally replaced by a plasma membrane-targeting sequence suggests that the catalytic activity of the Dbs protein is not dependent upon the presence of a structurally intact PH domain. It further suggests that the PH domain of Dbs functions as a plasma membrane localization signal and that recruitment of the Dbs protein to the cellular membrane is a necessary step for cellular transformation. This is similar to what we have observed for Lfc (69) but distinct from Dbl, where the loss of transformation caused by deletion of the PH domain was not restored by the addition of a plasma membrane-targeting sequence (72). Thus, despite the strong structural and functional relationship between Dbs and Dbl, their respective PH domains may make distinct contributious to the regulation of DH domain function. Alternatively, a trivial explanation may simply be the difference in efficiency of membrane association achieved by the two different CAAX-containing proteins. In the present study the Dbs-CAAX chimeric protein showed over 95% association with the membrane fraction, whereas in the previous study the Dbl-CAAX chimeric proteins showed only 10 to 20% association with membranes. Even though Dbs-CAAX was more efficiently membrane targeted than its PH domain-containing counterpart (Dbs-HA6), its transforming activity was still substantially lower than that of Dbs-HA6. Perhaps more efficient membrane targeting would also restore to Dbl the transforming activity lost through the loss of PH domain function. Finally, recent studies have shown that the substrates and products of phosphoinositide 3-phosphate kinase interact with the PH domains of Vav and SOS to positively regulate the activities of their PH domains (18, 40). Even with 95% of Dbs-CAAX in the membrane fraction, its transforming activity is substantially lower than that of the PH domain-containing Dbs-HA6 protein, which showed half as much protein in the membrane fraction. Thus, the function of the PH domain is not simply promoting membrane association, and it will be important to determine whether phosphoinositide interaction with the PH domain of Dbs also serves as a regulator of DH function.
Although it has been documented that activated derivatives of Rho and Dbl family members have a common ability to activate multiple signaling pathways to the nucleus, the contribution of transcription regulation, if any, to transforming activity has not been examined. Of particular interest has been the potent stimulation of the JNK-Jun signaling pathway by many Dbl family members (8, 37, 41, 64). Although this has been shown to be an apoptotic signaling pathway in several cell systems (36), we have observed that the integrity of this pathway is necessary for full Ras-mediated transformation in NIH 3T3 cells (7). This raises the possibility that the JNK pathway represents a proliferative pathway in NIH 3T3 cells and that its activation accounts for the transforming activity of Dbl and Rho family members. However, several recent observations would argue against this model. First, whereas Rac1 and Cdc42, but not RhoA, are potent activators of JNK in NIH 3T3 cells, only activated derivatives of RhoA are transforming in primary NIH 3T3 cell focus formation assays (25). Second, we have recently observed that a derivative of the Lfc protein that is impaired in its transforming activity can fully activate the JNK signaling pathway (64). Finally, in this study we observed that inhibition of the JNK signaling pathway did not inhibit the transforming activity of the Dbl and Dbs oncoproteins. Taken together, these results suggest that signaling through JNK is neither necessary nor sufficient for transformation by Dbl or Rho family members.
Whereas Dbl and Rho family proteins are strong activators of JNK, they are relatively weak activators of the related ERK MAPKs. ERK MAPKs are components of the Ras-Raf-MEK-ERK signaling pathway that constitutes an important component of Ras-mediated mitogenic signaling (73), and inhibition of this pathway results in impairment of Ras-mediated transformation (9, 27, 31). Although it has been generally assumed that the weak activation of ERK kinases by Dbl and Rho family members would not be sufficient to account for their transforming activity, we observed previously that the transforming activity of the Vav and Dbl proteins was partially inhibited by kinase-dead mutants of ERK1 and ERK2 (24). In support of an involvement of ERK-mediated signaling in Dbl family protein transformation, we have observed that the transforming activity of the Dbl and Dbs proteins can be completely abrogated by both genetic and pharmacologic inhibitors that block this signaling pathway. Surprisingly, our results suggest an absolute requirement for at least a low level of ERK activation for transformation by these two oncoproteins.
Although previous studies with dominant-inhibitory versions of MEK have suggested that activation of this kinase is necessary for Ras-mediated focus formation in NIH 3T3 cells, our results with a specific pharmacological inhibitor of MEK activity suggest that this may not be the case. Whereas the U0126 inhibitor effectively blocked transformation by Raf, Dbl, and Dbs, it only partially blocked the focus-forming activity of an activated derivative of Ras. This suggests that a majority of the focus-forming activity associated with activated Ras is a consequence of the stimulation of Raf-independent signaling pathways. Although these Raf-independent pathways are not sensitive to the U0126 inhibitor, they are clearly responsive to the MEK(2A) dominant-inhibitory mutant, thus bringing into question the specificity of this reagent.
In addition to Jun and Elk-1, Rho and Dbl family members have also been shown to be strong activators of the NF-κB transcription factor (38, 43, 58). Although the contribution of NF-κB-regulated signaling to Rho- and Dbl-mediated transformation has not been assessed, we demonstrated recently that NF-κB supports Ras transformation of NIH 3T3 cells by protecting the transformed cells from apoptotic death (33). A second report has also demonstrated the importance of NF-κB activation for full transformation by Bcr-Abl, although the mechanism has not yet been determined (49). In this study we observed that the potency of transformation by Dbl and Dbs is also linked to NF-κB activation. Although we have not yet determined the signaling pathways that are involved in Dbs- and Dbl-mediated NF-κB activation, it has been reported that Cdc42 and Rac1, but not RhoA, activate NF-κB in a MEKK1-dependent manner (38). Our observation that Dbl and Dbs transformation is dependent upon NF-κB signaling, yet independent of SEK1-mediated signaling, suggests that either SEK1 and NF-κB are activated independently by MEKK1 or that activation of NF-κB by Dbl and Dbs is not occurring through a MEKK1-mediated signaling pathway. Finally, an NF-κB-responsive element has not been described for the cyclin D1 promoter (1). However, Pestell and colleagues have identified an NF-κB site in the cyclin D1 promoter that is required for activated Rac1 stimulation of transcription (unpublished observation). This element is most likely responsible for the NF-κB-dependent upregulation of cyclin D1 caused by Dbs.
In a recent study we observed a close correlation between the transforming activities of a panel of Dbl family members and their abilities to activate transcription from the cyclin D1 promoter (64). In addition, we have observed elevated levels of cyclin D1 in NIH 3T3 cells that are stably transformed by Dbl and Dbs (64). Our present observation that activation of the cyclin D1 promoter by Dbl and Dbs can be blocked by a specific inhibitor of NF-κB function suggests a possible direct interaction between NF-κB and cyclin D1 promoter elements and further suggests a mechanism by which the Dbl family proteins upregulate cyclin D1 levels in NIH 3T3 cells. NF-κB-dependent activation of transcription from the cyclin D1 promoter has also been observed in other situations (1a).
In summary, we have observed that two members of the Dbl family, Dbl and Dbs, transform NIH 3T3 cells in a manner that is dependent upon transcriptional activation. Although Dbl and Dbs transformations are dependent upon their abilities to activate MEK- and NF-κB-regulated signaling pathways, the strong activation of JNK by Dbl and Dbs appears to be dispensable for transformation. Recent studies observed that derivatives of Rac1, RhoA, and Cdc42 that are impaired in their actin cytoskeletal reorganization activities retain full transforming activity (45, 52, 63). These observations, when taken together with those of this study, argue that nuclear signaling, and not cytoskeletal modification, is the key biological event required for the transformation of NIH 3T3 cells by Dbl and Rho family proteins. Therefore, identification of the growth-regulatory genes whose activities are regulated by NF-κB and Elk-1 will be important in defining the mechanism of Dbs and Dbl transformation.
ACKNOWLEDGMENTS
We thank A. Christine Tabaka and Jia-Sheng Yan for the synthesis of U0126, Carol Martin for technical support, and Jennifer Parrish for the preparation of figures.
This work was supported by Public Health Service grants CA42978, CA55008, CA63071 (C.J.D.), and CA77493 (I.P.W.) from the National Cancer Institute.
REFERENCES
- 1.Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell R G. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem. 1995;270:23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- 1a.Baldwin, A. Unpublished observation.
- 2.Boguski M S, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 3.Bourne H R, Sanders D A, McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990;348:125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- 4.Cerione R A, Zheng Y. The Dbl family of oncogenes. Curr Opin Cell Biol. 1996;8:216–222. doi: 10.1016/s0955-0674(96)80068-8. [DOI] [PubMed] [Google Scholar]
- 5.Chardin P, Boquet P, Madaule P, Popoff M R, Rubin E J, Gill D. The mammalian G protein rhoC is ADP-ribosylated by Clostridium botulinum exoenzyme C3 and affects actin microfilaments in Vero cells. EMBO J. 1989;8:1087–1092. doi: 10.1002/j.1460-2075.1989.tb03477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark G J, Cox A D, Graham S M, Der C J. Biological assays for Ras transformation. Methods Enzymol. 1995;255:395–412. doi: 10.1016/s0076-6879(95)55042-9. [DOI] [PubMed] [Google Scholar]
- 7.Clark G J, Westwick J K, Der C J. p120 GAP modulates Ras activation of Jun kinases and transformation. J Biol Chem. 1997;272:1677–1681. doi: 10.1074/jbc.272.3.1677. [DOI] [PubMed] [Google Scholar]
- 8.Coso O A, Chiariello M, Yu J-C, Teramoto H, Crespo P, Xu N, Miki T, Gutkind J S. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 9.Cowley S, Paterson H, Kemp P, Marshall C J. Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell. 1994;77:841–852. doi: 10.1016/0092-8674(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 10.Crespo P, Bustelo X R, Aaronson D S, Coso O A, Lopez-Barahona M, Barbacid M, Gutkind J S. Rac-1 dependent stimulation of the JNK/SAPK signaling pathway by Vav. Oncogene. 1996;13:455–460. [PubMed] [Google Scholar]
- 11.DeSilva D R, Jones E A, Favata M F, Jaffee B D, Magolda R L, Trzaskos J M, Scherle P A. Inhibition of mitogen-activated protein kinase kinase blocks T cell proliferation but does not induce or prevent anergy. J Immunol. 1998;160:4175–4181. [PubMed] [Google Scholar]
- 12.Favata M F, Horiuchi K Y, Manos E J, Daulerio A J, Stradley D A, Feeser W S, Van Dyk D E, Pitts W J, Earl R A, Hobbs F, Copeland R A, Magolda R L, Scherle P A, Trzaskos J M. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J Biol Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- 13.Finco T S, Westwick J K, Norris J L, Beg A A, Der C J, Baldwin A S., Jr Oncogenic Ha-Ras-induced signaling activates NF-κB transcriptional activity, which is required for cellular transformation. J Biol Chem. 1997;272:24113–24116. doi: 10.1074/jbc.272.39.24113. [DOI] [PubMed] [Google Scholar]
- 14.Galang C K, García-Ramírez J J, Solski P A, Westwick J K, Der C J, Neznanov N N, Oshima R G, Hauser C A. Oncogenic Neu/ErbB-2 increases Ets, AP-1 and NF-κB-dependent gene expression, and inhibiting Ets activation blocks Neu-mediated cellular transformation. J Biol Chem. 1996;271:7992–7998. doi: 10.1074/jbc.271.14.7992. [DOI] [PubMed] [Google Scholar]
- 15.Glaven J A, Whitehead I P, Nomanbhoy T, Kay R, Cerione R A. Lfc and Lsc oncoproteins represent two new guanine nucleotide exchange factors for the Rho GTP-binding protein. J Biol Chem. 1996;271:27374–27381. doi: 10.1074/jbc.271.44.27374. [DOI] [PubMed] [Google Scholar]
- 16.Habets G G M, Scholtes E H M, Zuydgeest D, van der Kammen R A, Stam J C, Berns A, Collard J G. Identification of an invasion-inducing gene, Tiam-1, that encodes a protein with homology to GDP-GTP exchangers for Rho-like proteins. Cell. 1994;77:537–549. doi: 10.1016/0092-8674(94)90216-x. [DOI] [PubMed] [Google Scholar]
- 17.Hall A. Ras-related proteins. Curr Biol. 1993;5:265–268. doi: 10.1016/0955-0674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- 18.Han J, Luby-Phelps K, Das B, Shu X, Xia Y, Mosteller R D, Krishna U M, Falck J R, White M A, Broek D. Role of substrates and products of PI 3-kinase in regulating activation of Rac-related guanosine triphosphatases by Vav. Science. 1998;279:558–560. doi: 10.1126/science.279.5350.558. [DOI] [PubMed] [Google Scholar]
- 19.Hart M J, Eva A, Evans T, Aaronson S A, Cerione R A. Catalysis of guanine nucleotide exchange on the CDC42Hs protein by the dbl oncogene product. Nature. 1991;354:311–314. doi: 10.1038/354311a0. [DOI] [PubMed] [Google Scholar]
- 20.Hauser C A, Westwick J K, Quilliam L A. Ras-mediated transcription activation: analysis by transient cotransfection assays. Methods Enzymol. 1995;255:412–426. doi: 10.1016/s0076-6879(95)55043-7. [DOI] [PubMed] [Google Scholar]
- 21.Hill C S, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1 and Cdc42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 22.Horii Y, Beeler J F, Sakaguchi K, Tachibana M, Miki T. A novel oncogene, ost, encodes a guanine nucleotide exchange factor that potentially links Rho and Rac signaling pathways. EMBO J. 1994;13:4776–4786. doi: 10.1002/j.1460-2075.1994.tb06803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kahn R A, Der C J, Bokoch G M. The Ras superfamily of GTP-binding proteins: guidelines on nomenclature. FASEB J. 1992;6:2512–2513. doi: 10.1096/fasebj.6.8.1592203. [DOI] [PubMed] [Google Scholar]
- 24.Khosravi-Far R, Chrzanowska-Wodnicka M, Solski P A, Eva A, Burridge K, Der C J. Vav and Dbl mediate transformation via Ras-independent mitogen activated protein kinase activation. Mol Cell Biol. 1994;14:6848–6857. doi: 10.1128/mcb.14.10.6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khosravi-Far R, Solski P A, Kinch M S, Burridge K, Der C J. Activation of Rac and Rho, and mitogen activated protein kinases, are required for Ras transformation. Mol Cell Biol. 1995;15:6443–6453. doi: 10.1128/mcb.15.11.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khosravi-Far R, White M A, Westwick J K, Solski P A, Chrzanowska-Wodnicka M, Van Aelst L, Wigler M H, Der C J. Oncogenic Ras activation of Raf/MAP kinase-independent pathways is sufficient to cause tumorigenic transformation. Mol Cell Biol. 1996;16:3923–3933. doi: 10.1128/mcb.16.7.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolch W, Heidecker G, Lloyd P, Rapp U R. Raf-1 protein kinase is required for growth of induced NIH/3T3 cells. Nature. 1991;349:426–428. doi: 10.1038/349426a0. [DOI] [PubMed] [Google Scholar]
- 28.Lin R, Bagrodia S, Cerione R, Manor D. A novel Cdc42Hs mutant induces cellular transformation. Curr Biol. 1997;7:794–797. doi: 10.1016/s0960-9822(06)00338-1. [DOI] [PubMed] [Google Scholar]
- 29.Liu X, Wang H, Eberstadt M, Schnuchel A, Olejniczak E T, Meadows R P, Schkeryantz J M, Janowick D A, Harlan J E, Harris E A, Staunton D E, Fesik S W. NMR structure and mutagenesis of the N-terminal Dbl homology domain of the nucleotide exchange factor Trio. Cell. 1998;95:269–277. doi: 10.1016/s0092-8674(00)81757-2. [DOI] [PubMed] [Google Scholar]
- 30.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 31.Mansour S J, Matten W T, Hermann A S, Candia J M, Rong S, Fukasawa K, Vande Woude G F, Ahn N G. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- 32.Marais R, Wynne J, Treisman R. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell. 1993;73:381–393. doi: 10.1016/0092-8674(93)90237-k. [DOI] [PubMed] [Google Scholar]
- 33.Mayo M W, Wang C-Y, Cogswell P C, Rogers-Graham K S, Lowe S W, Der C J, Baldwin A S., Jr Requirement of NF-kappaB activation to suppress p53-independent apoptosis induced by oncogenic Ras. Science. 1997;278:1812–1815. doi: 10.1126/science.278.5344.1812. [DOI] [PubMed] [Google Scholar]
- 34.Michiels F, Stam J C, Hordijk P L, van der Kammen R A, Ruuls-Van Stalle L, Feltkamp C A, Collard J G. Regulated membrane localization of TIAM1, mediated by the N-terminal pleckstrin homology domain, is required for Rac-dependent membrane ruffling and JNK activation. J Cell Biol. 1997;137:1–12. doi: 10.1083/jcb.137.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miki T, Smith C L, Long J E, Eva A, Fleming T P. Oncogene ect2 is related to regulators of small GTP-binding proteins. Nature. 1993;362:462–465. doi: 10.1038/362462a0. [DOI] [PubMed] [Google Scholar]
- 36.Minden A, Karin M. Regulation and function of the JNK subgroup of MAP kinases. Biochim Biophys Acta. 1997;1333:85–104. doi: 10.1016/s0304-419x(97)00018-8. [DOI] [PubMed] [Google Scholar]
- 37.Minden A, Lin A, Claret F-X, Abo A, Karin M. Selective activation of the JNK signaling cascade and Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 38.Montaner S, Perona R, Saniger L, Lacal J C. Multiple signalling pathways lead to the activation of the nuclear factor kappaB by the Rho family of GTPases. J Biol Chem. 1998;273:12779–12785. doi: 10.1074/jbc.273.21.12779. [DOI] [PubMed] [Google Scholar]
- 39.Murphy G A, Solski P A, Jillian S A, Perez de la Ossa P, D’Eustachio P, Der C J, Rush M G. Cellular functions of TC10, a Rho family GTPase: regulation of morphology, signal transduction, and cell growth. Oncogene. 1999;15:3831–3845. doi: 10.1038/sj.onc.1202758. [DOI] [PubMed] [Google Scholar]
- 40.Nimnual A S, Yatsula B A, Bar-Sagi D. Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science. 1998;279:560–563. doi: 10.1126/science.279.5350.560. [DOI] [PubMed] [Google Scholar]
- 41.Olson M F, Ashworth A, Hall A. An essential role for Rho, Rac and Cdc42 GTPases in cell cycle progression through G1. Science. 1995;269:1270–1272. doi: 10.1126/science.7652575. [DOI] [PubMed] [Google Scholar]
- 42.Olson M F, Pasteris N G, Gorski J L, Hall A. Faciogenital dysplasia protein (FGD1) and Vav, two related proteins required for normal embryonic development, are upstream regulators of Rho GTPases. Curr Biol. 1996;6:1628–1633. doi: 10.1016/s0960-9822(02)70786-0. [DOI] [PubMed] [Google Scholar]
- 43.Perona R, Montaner S, Saniger L, Sánchez-Pérez I, Bravo R, Lacal J C. Activation of the nuclear factor-κB by Rho, CDC42, and Rac-1 proteins. Genes Dev. 1997;11:463–475. doi: 10.1101/gad.11.4.463. [DOI] [PubMed] [Google Scholar]
- 44.Prendergast G C, Khosravi-Far R, Solski P A, Kurzawa H, Lebowitz P F, Der C J. Critical role of RhoB in cell transformation by oncogenic Ras. Oncogene. 1995;10:2289–2296. [PubMed] [Google Scholar]
- 45.Qiu R-G, Abo A, McCormick F, Symons M. Cdc42 regulates anchorage-independent growth and is necessary for Ras transformation. Mol Cell Biol. 1997;17:3449–3458. doi: 10.1128/mcb.17.6.3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiu R-G, Chen J, Kirn D, McCormick F, Symons M. An essential role for Rac in Ras transformation. Nature. 1995;374:457–459. doi: 10.1038/374457a0. [DOI] [PubMed] [Google Scholar]
- 47.Qiu R-G, Chen J, McCormick F, Symons M. A role for Rho in Ras transformation. Proc Natl Acad Sci USA. 1995;92:11781–11785. doi: 10.1073/pnas.92.25.11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raitano A B, Halpern J R, Hambuch T M, Sawyers C L. The Bcr-Abl leukemia oncogene activates Jun kinase and requires Jun for transformation. Proc Natl Acad Sci USA. 1995;92:11746–11750. doi: 10.1073/pnas.92.25.11746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reuther J Y, Reuther G W, Cortez D, Pendergast A M, Baldwin A S., Jr A requirement for NF-kappaB activation in Bcr-Abl-mediated transformation. Genes Dev. 1998;12:1–14. doi: 10.1101/gad.12.7.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodrigues G A, Park M, Schlessinger J. Activation of the JNK pathway is essential for transformation by the Met oncogene. EMBO J. 1997;16:2634–2645. doi: 10.1093/emboj/16.10.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ron D, Zannini M, Lewis M, Wickner R B, Hunt L T, Graziani G, Tronick S R, Aaronson S A, Eva A. A region of Proto-dbl essential for its transforming activity shows sequence similarity to a yeast cell cycle gene, CDC24, and the human breakpoint cluster gene, bcr. New Biol. 1991;3:372–379. [PubMed] [Google Scholar]
- 52.Sahai E, Alberts A S, Treisman R. RhoA effector mutants reveal distinct effector pathways for cytoskeletal reorganization, SRF activation and transformation. EMBO J. 1998;17:1350–1361. doi: 10.1093/emboj/17.5.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanchez I, Hughes R T, Mayer B J, Yee K, Woodgett J R, Avruch J, Kyriakis J M, Zon L I. Role of SAPK/ERK kinase-1 in the stress-activated pathway regulating transcription factor Jun. Nature. 1994;372:794. doi: 10.1038/372794a0. [DOI] [PubMed] [Google Scholar]
- 54.Settleman J, Albright C F, Foster L C, Weinberg R A. Association between GTPase activators for Rho and Ras families. Nature. 1992;359:153–154. doi: 10.1038/359153a0. [DOI] [PubMed] [Google Scholar]
- 55.Shou C, Farnsworth C L, Neel B G, Feig L A. Molecular cloning of cDNAs encoding a guanine-nucleotide-releasing factor for Ras p21. Nature. 1992;358:351–354. doi: 10.1038/358351a0. [DOI] [PubMed] [Google Scholar]
- 56.Solski P A, Quilliam L A, Coats S G, Der C J, Buss J E. Targeting proteins to membranes using signal sequences for lipid modification. Methods Enzymol. 1995;250:435–454. doi: 10.1016/0076-6879(95)50089-8. [DOI] [PubMed] [Google Scholar]
- 57.Su B, Jacinto E, Hibi M, Kallunki T, Karin M, Ben-Neriah Y. JNK is involved in signal integration during costimulation of T lymphocytes. Cell. 1994;77:727–736. doi: 10.1016/0092-8674(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 58.Sulciner D J, Irani K, Yu Z-X, Ferrans V J, Goldschmidt-Clermont P, Finkel T. rac1 regulates a cytokine-stimulated, redox-dependent pathway necessary for NF-κB activation. Mol Cell Biol. 1996;16:7115–7121. doi: 10.1128/mcb.16.12.7115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Treisman R. Regulation of transcription by MAP kinase cascades. Curr Opin Cell Biol. 1996;8:205–215. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- 60.Valencia A, Chardin P, Wittinghofer A, Sander C. The ras protein family: evolutionary tree and role of conserved amino acids. Biochemistry. 1991;30:4637–4648. doi: 10.1021/bi00233a001. [DOI] [PubMed] [Google Scholar]
- 61.Verheij M, Bose R, Lin X H, Yao B, Jarvis W D, Grant S, Birrer M J, Szabo E, Zon L I, Kyriakis J M, Haimovitz-Friedman A, Fuks Z, Kolesnick R N. Requirement for ceramide-initiated SAPK/JNK signalling in stress-induced apoptosis. Nature. 1996;380:75–79. doi: 10.1038/380075a0. [DOI] [PubMed] [Google Scholar]
- 62.Westwick J K, Brenner D A. Methods for analyzing c-Jun kinase. Methods Enzymol. 1995;255:342–360. doi: 10.1016/s0076-6879(95)55037-2. [DOI] [PubMed] [Google Scholar]
- 63.Westwick J K, Lambert Q T, Clark G J, Symons M, Van Aelst L, Pestell R G, Der C J. Rac regulation of transformation, gene expression, and actin organization by multiple, PAK-independent pathways. Mol Cell Biol. 1997;17:1324–1335. doi: 10.1128/mcb.17.3.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Westwick J K, Lee R J, Lambert Q T, Symons M, Pestell R G, Der C J, Whitehead I P. Transforming potential of Dbl family proteins correlates with transcription from the cyclin D1 promoter but not with activation of Jun NH2-terminal kinase, p38/Mpk2, serum response factor or c-Jun. J Biol Chem. 1998;273:16739–16747. doi: 10.1074/jbc.273.27.16739. [DOI] [PubMed] [Google Scholar]
- 65.Whitehead I P, Abe K, Gorski J L, Der C J. CDC42 and FGD1 cause distinct signaling and transforming activities. Mol Cell Biol. 1998;18:4689–4697. doi: 10.1128/mcb.18.8.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Whitehead I P, Campbell S, Rossman K L, Der C J. Dbl family proteins. Biochem Biophys Acta. 1997;1332:F1–F23. doi: 10.1016/s0304-419x(96)00040-6. [DOI] [PubMed] [Google Scholar]
- 67.Whitehead I P, Khosravi-Far R, Kirk H, Trigo-Gonzalez G, Der C J, Kay R. Expression cloning of Isc, a novel oncogene with structural similarities to the Dbl family of guanine nucleotide exchange factors. J Biol Chem. 1996;271:18643–18650. doi: 10.1074/jbc.271.31.18643. [DOI] [PubMed] [Google Scholar]
- 68.Whitehead I P, Kirk H, Kay R. Retroviral transduction and oncogenic selection of a cDNA encoding Dbs, a homolog of the Dbl guanine nucleotide exchange factor. Oncogene. 1995;10:713–721. [PubMed] [Google Scholar]
- 69.Whitehead I P, Kirk H, Tognon C, Trigo-Gonzalez G, Kay R. Expression cloning of lfc, a novel oncogene with structural similarities to guanine nucleotide exchange factors and to the regulatory region of protein kinase C. J Biol Chem. 1995;271:18388–18395. doi: 10.1074/jbc.270.31.18388. [DOI] [PubMed] [Google Scholar]
- 70.Whitmarsh A J, Yang S-H, Su M S S, Sharrocks A D, Davis R J. Role of p38 and JNK mitogen-activated protein kinases in the activation of ternary complex factors. Mol Cell Biol. 1997;17:2360–2371. doi: 10.1128/mcb.17.5.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang S, Han J, Sells M A, Chernoff J, Knaus U G, Ulevitch R J, Bokoch G M. Rho family GTPases regulate p38 mitogen-activated protein kinase through the downstream mediator Pak1. J Biol Chem. 1995;270:23934–23936. doi: 10.1074/jbc.270.41.23934. [DOI] [PubMed] [Google Scholar]
- 72.Zheng Y, Zangrilli D, Cerione R A, Eva A. The pleckstrin homology domain mediates transformation by oncogenic Dbl through specific intracellular targeting. J Biol Chem. 1996;271:19017–19020. doi: 10.1074/jbc.271.32.19017. [DOI] [PubMed] [Google Scholar]
- 73.Zohn I E, Campbell S L, Khosravi-Far R, Rossman K L, Der C J. Rho family proteins and Ras transformation: the RHOad less traveled gets congested. Oncogene. 1998;17:1415–1438. doi: 10.1038/sj.onc.1202181. [DOI] [PubMed] [Google Scholar]