Abstract
Trophic interactions play a central role in driving microbial community assembly and function. In gut or soil ecosystems, successful inoculants are always facilitated by efficient colonization; however, the metabolite exchanges between inoculants and resident bacteria are rarely studied, particularly in the rhizosphere. Here, we used bioinformatic, genetic, transcriptomic, and metabonomic analyses to uncover syntrophic cooperation between inoculant (Bacillus velezensis SQR9) and plant-beneficial indigenous Pseudomonas stutzeri in the cucumber rhizosphere. We found that the synergistic interaction of these two species is highly environmental dependent, the emergence of syntrophic cooperation was only evident in a static nutrient-rich niche, such as pellicle biofilm in addition to the rhizosphere. Our results identified branched-chain amino acids (BCAAs) biosynthesis pathways are involved in syntrophic cooperation. Genome-scale metabolic modeling and metabolic profiling also demonstrated metabolic facilitation among the bacterial strains. In addition, biofilm matrix components from Bacillus were essential for the interaction. Importantly, the two-species consortium promoted plant growth and helped plants alleviate salt stress. In summary, we propose a mechanism in which synergic interactions between a biocontrol bacterium and a partner species promote plant health.
Subject terms: Microbial ecology, Bacteria, Biofilms, Microbial communities, Soil microbiology
Introduction
Plants host enormous diverse communities of microorganisms, the plant microbiome, which is crucial for plant health. Beneficial plant−microbiome interactions improve plant fitness through growth promotion, stress alleviation, and defense against pathogens through various mechanisms [1]. Direct stimulations are mediated through the production of phytohormones or 1-aminocyclopropane-1-carboxylate deaminases, induction of systematic resistance, and increasing nutrient acquisition through nitrogen fixation, phosphorus solubilization, and secretion of siderophores [2]. Indirect stimulation includes suppression of pathogens by antibiotic production, competition for niches within the rhizosphere, promotion of mycorrhizal functioning, and changing the rhizosphere microbial community structure [3–5]. However, the high complexity of microbiome composition makes it challenging to answer fundamental ecological questions surrounding natural microbial communities. Reductionist approaches conducted under laboratory conditions have been a promising strategy to decipher the microbial interactions in the rhizosphere microbiome along with their relevance for host health [6]. Driven by great application potential, studies focusing on synthetic community (SynCom) have emerged in the last decade providing sustainable agriculture solutions [7]. SynCom is a microbial community designed by mixing selected strains to demonstrate plant-beneficial impact. Such an approach enables detailed assessment of host and microbe characteristics under controlled, reproducible conditions.
Two fundamental principles are commonly used for designing SynComs, function- and interaction-based approaches, during which cell−cell interactions play a key role in stability and robustness [7]. Given the high complexity of metabolites in the rhizosphere, metabolite exchange possibly drives species interactions in the plant microbiome. A well-studied example of positive microbial interactions includes cross-feeding, i.e., the exchange of essential nutritional molecules [8]. Gut microbiota studies highlighted that obligate cross-feeding can significantly expand the ecological niche space of each member involved in SynComs [9]. Moreover, cross-feeding also helps maintaining the diversity and stability of natural microbial communities [10], while spatial structure, especially biofilm, is vital for cross-feeding development and its evolution [11]. However, compared with the plentiful knowledge from gut microbiota studies, relatively few studies have been published on metabolite cross-feeding in the rhizosphere. We suggest that such metabolic mutualisms could be exploited for designing robust probiotic SynComs for green agriculture.
Generally, Bacillus spp. and Pseudomonas spp. are the most extensively studied beneficial microorganisms in the rhizosphere [12, 13], several commercial products belong to these two genera are currently available for agricultural production of crops. Even though the biocontrol ability and plant-growth promotion by plant growth-promoting rhizobacteria (PGPR) have been investigated thoroughly, their impacts on the composition of the indigenous rhizosphere microbiome are still not fully explored. Microbial inoculants were shown to recruit assemblages of beneficial taxa, like Flavobacterium, Pseudomonas, Agrobacterium, and Lysobacter [14, 15], and inhibit soil−borne pathogens. It has been suggested that in addition to direct association with host plants, disease suppression, and growth promotion may also be achieved by recruitment of beneficial species that reshape soil microbiome structure and function. However, these findings were largely constrained by the microbiome sequencing approach, with limited information on the activity of the organisms behind them. We propose that the undiscovered interactions between PGPR and recruited beneficial microbes affect the functional capacities of the applied inoculants in the rhizosphere. Instead of focusing on the mono-association with plant, more attention should be paid to how inoculants modulate the structure of indigenous microbiome and how microbial interaction affect the functionality of the applied inoculants [16].
To address this important knowledge gap, we used B. velezensis SQR9 as a representative of PGPR to characterize the influence of PGPR on resident rhizosphere microbiome and study emerging microbial interactions using a simplified two-species community with target rhizosphere community bacteria. Plant beneficial Bacillus, including B. velezensis SQR9 are known for plant growth promotion, disease suppression, and enhanced salt stress tolerance [17–20]. Root colonization and plant growth-promoting properties require efficient biofilm formation on the roots. The essential B. velezensis genes to produce biofilm extracellular matrix include the epsA-O operon (encoding exopolysaccharide EPS) [21] and the tapA-sipW-tasA operon (encoding TasA protein fibers) [22]. Inoculation of corresponding bio-fertilizer to soil increased the abundance of indigenous microbial groups with reported antifungal activity, such as Lysobacter spp., which could play a keystone role in soil suppressiveness [14]. However, the exact mechanisms involved in recruitment have not been characterized in detail previously. In this study, we discovered that B. velezensis SQR9 stimulated resident rhizosphere member, P. stutzeri. These two microorganisms formed robust biofilms in vitro and on the plant root surface. Metabolic and transcriptomic analysis revealed potential cross-feeding to increase community performance. We demonstrated that the BCAAs biosynthesis pathways are involved in the syntrophic interaction of these two species. Furthermore, a dual-species consortium excelled in plant growth promotion compared to the individual species. Together, our work expanded the knowledge on complex microbial interactions that generally occur in the rhizosphere, providing guidance for rhizosphere engineering in safe and eco-friendly agriculture.
Methods and materials
Rhizosphere sample collection
This study focused on cucumber rhizosphere bacterial communities. The soil used in this study was collected from a field in Taizhou city, Jiangsu province, China (32.4555° N, 119.9229° E) in October 2017. Cucumber seeds (Jinchun 4) were purchased from Jiangsu Academy of Agricultural Sciences. The seeds were surface-sterilized and grown in ¼ Muarshige Skoog (MS) medium [23] for two weeks and then transferred to 2 kg soil pre-inoculated with 10 ml B. velezensis SQR9 suspensions (108 cells/ml). Plants were grown in a greenhouse at 30 °C, 16 h light/8 h dark. Plants grown in ordinary soil without inoculation of bacteria were set as control. After 16 days, rhizosphere soil was collected as described by Bai [24]. Each treatment had six replicates.
Strain isolations, culture conditions, and mutant construction
Bacterial strains and plasmids used in this study are listed in Table S1. B. velezensis strain SQR9 (CGMCC accession number 5808) was isolated previously from the cucumber rhizosphere and used throughout this study. Deletion mutants were generated by using a markerless deletion method described by Zhou [25]. Oligonucleotides used for PCR in this study are listed in Table S2.
To isolate cooperating bacteria, fresh cucumber rhizosphere soil inoculated with strain SQR9 was collected and suspended in PBS buffer, and vortexed vigorously. Dilution series were plated on 0.1X TSB, R2A, TYG, and M715 media [24] solidified with 1.5% agar, then incubated at 30 °C for 2−7 days. 267 colonies were picked and phylogenetically characterized by colony PCR using a universal primer set (27F and 1492R) for the 16S rRNA gene and stored at −80 °C in 25% (v/v) glycerol. Other media used in this study include M9 medium, MSgg medium and root exudate medium (REM). M9 medium was prepared from M9 minimal salts (5x, Sigma-Aldrich M6030), 2 mM MgSO4, 0.1 mM CaCl2. M9 medium was supplemented with sole carbon sources listed in Table S3. Water-soluble compounds were provided at a concentration of 10 mg/ml, other compounds were provided at the maximum amount of solubility. MSgg medium consists of 5 mM potassium phosphate, 100 mM 3-(N-morpholino) propanesulfonic acid (MOPS), 2 mM MgCl2, 700 μM CaCl2, 50 μM MnCl2, 50 μM FeCl3, 1 μM ZnCl2, 2 μM thiamine, 0.5% glycerol, 0.5% glutamate, 50 μg/ml tryptophan, 50 μg/ml phenylalanine, pH7 [26]. REM is M9 glucose medium supplemented with 0.02 mg/ml root exudates. Root exudates were collected as described by Feng [26]. REM and TSB medium are rich media, while M9 and MSgg are chemically defined minimal media.
Genome assembly and annotation
P. stutzeri XL272 was selected in the following investigation and modified with a mini-Tn7 transposon containing a DsRed marker [27]. The genome was sequenced using a PacBio RS II platform and a HiSeq 4000 platform (Illumina) at the Beijing Genomics Institute (BGI, Shenzhen, China). Four SMRT cells Zero-Mode Waveguide arrays of sequencing, were used by the PacBio platform to generate the subreads set. PacBio subreads (length < 1 kb) were removed. The program Pbdagcon (https://github.com/PacificBiosciences/pbdagcon) was used for self-correction. Draft genomic unitigs, which are uncontested groups of fragments, were assembled using the Celera Assembler against a high-quality corrected circular consensus sequence subreads set. To improve the accuracy of the genome sequences, GATK (https://www.broadinstitute.org/gatk/) and SOAP tool packages (SOAP2, SOAPsnp, SOAPindel) were used to make single-base corrections. To trace the presence of any plasmid, the filtered Illumina reads were mapped using SOAP to the bacterial plasmid database (http://www.ebi.ac.uk/genomes/plasmid.html, last accessed July 8, 2016). Raw sequencing data have been deposited to the NCBI Sequence Read Archive (SRA) database under BioProject accession number PRJNA593625. The assembled sequence of the genome was deposited at the National Center for Biotechnology Information (NCBI) under Nucleotide accession number NZ_CP046538. Genome was annotated in 2019 using PROKKA v1.14.4 [28]. The annotations were provided as supplementary Dataset S1.
Metabolic model reconstruction and interspecies metabolic exchange analysis
The CarveMe pipeline [29] was used to reconstruct the genome-scale metabolic models for B. velezensis SQR9 (NCBI Nucleotide accession number NZ_CP006890.1) and P. stuzeri XL272. In general, these models were used to simulate growth and metabolite production capabilities of corresponding species under TSB medium. The quality of the metabolic models was validated using MEMOTE [30]. The metabolic interaction potential of the dual species consortium was analyzed using SMETANA [31], this identified all likely exchanged metabolites. The metabolic model was provided as Dataset S2.
Microbiome analysis
Total genomic DNA of soil samples was isolated using the PowerSoil DNA Isolation Kit (Mo Bio Laboratories, Inc., Carlsbad, CA, USA). Mixed universal primers targeting the V3−V4 regions of 16S rRNA gene were used to construct the DNA library for sequencing. Paired-end sequencing of bacterial amplicons was performed on the MiSeq 300 instrument (Illumina). Raw sequencing data have been deposited to the NCBI SRA database under BioProject accession number PRJNA727458. Reads were processed using the UPARSE pipeline [32]. The paired-end reads were merged using the “fastq_mergepairs” command. High-quality sequences were then selected using the “fastq_filter” command and dereplicated using the “derep_fulllength” command. The singletons were removed using USEARCH-unoise3 algorithm and chimeric sequences were removed using “uchime_ref” command. The remaining sequences were used to create amplicon sequencing variant (ASV) table. Taxonomy assignment was performed using the Ribosomal Database Project (RDP) classifier. Bray−Curtis distance-based PCoA analysis, and permutational multivariate analysis of variance (PERMANOVA) were performed based on the ASV table using the vegan R package. Welch’s t test was used to calculate the significance of differences between two treatments using STAMP [33]. ASVs whose relative abundance were higher than 0.05% in all samples were used in this analysis.
Biofilm formation assay
To observe the cocultured colony biofilm, an equal volume of B. velezensis and P. stutzeri were mixed at an OD600 of 0.0001, then 5 μl of bacteria were spotted on TSB medium solidified with 1.5% agar. The plates were incubated at 30 °C for 48 h.
To observe pellicle biofilm formation, 20 μl of the initial inoculum was cultivated in 2 ml of TSB liquid medium in a 24-well microtiter plate (Fisher Scientific). The microtiter plates were incubated statically at 30 °C for 24 h. The initial inoculum was obtained by growing the cells overnight to exponential growth in TSB medium at 30 °C, 180 rpm shaken condition, the cells were spun down and diluted to OD600 of 1 in 0.9% NaCl buffer. For coculture, the initial inoculum was prepared by mixing an equal volumes of two species. Unless otherwise specified, the initial inoculum was prepared as described here in all the experiments.
Biofilm biomass quantification assay
Pellicle was grown in six-well microtiter plates insert with 100 μm Sterile Nylon Mesh Cell Strainers (Biologix Cat#15-1100). 10 ml of TSB liquid medium and 100 μl of the initial inoculum was added. The plates were incubated for 24 h at 30 °C statically to allow the pellicle to grow on top of the nylon mesh cell strainer. The cell strainer was taken out, removed visible drops with paper, and weighed. The fresh weight was the total weight minus the weight of the nylon mesh. The dry weight was measured by drying the pellicle within the laminar hood for 24 h. Each treatment has six replicates.
Cell numbers quantification
Cell numbers in coculture were quantified under four conditions: static TSB medium, shaken TSB medium, static MSgg medium [34], static REM. For quantifying individual cell numbers in pellicle, 100 μl of the initial inoculum was grown in six-well microtiter plates (VWR) with 10 ml medium. A 100 μm sterile nylon mesh cell strainer and a Spectra Mesh Woven Filter (Fisher Scientific, Spectrum 146488) were put inside. The mesh was manually cut into 1.5 cm2 squares and autoclaved. After 48 h of pellicle development, the nylon mesh cell strainer was taken out, the inner filter was transferred to a 1.5 ml microcentrifuge tube, stored at −80 °C for following DNA or RNA extraction. This ensured equal sampling of biofilm. For quantifying individual cell numbers in shaken TSB, 40 μl of the initial inoculum was inoculated in sterile test tubes with 4 ml TSB medium and incubated at 30 °C, 180 rpm shaken condition. After 48 h, 2 ml of cell cultures were spun down and stored at −80 °C. Total DNA of biofilm formed on the mesh filter or cell pellets was extracted with E.Z.N.A. Bacterial DNA Kit (Omega Bio-tek, Inc.) according to the manufacturer’s instructions.
Alignment of the B. velezensis SQR9 and P. stutzeri XL272 genomes was conducted with Roary [35] to find different genes of the two isolates. The strain-specific primer pairs for qPCR were designed based on different gene sequences according to the guidelines of Oligo (v7). The specificity of obtained primers was checked by conventional PCRs and qPCR melt curves. Standard curves were generated using plasmids containing corresponding fragments. qPCR was performed with Applied Biosystems (ABI) Real-Time PCR Instrument. Reaction components are as follow: 7.2 μl H2O, 10 μl 2× ChamQ SYBR qPCR Master Mix (Vazyme), 0.4 μl of each primer (10 μM) and 2 μl template DNA. The PCR programs were carried out under the following conditions: 95 °C for 30 s, 40 cycles of 95 °C for 5 s, 60 °C for 45 s, followed by a standard melting curve segment. Each treatment has six replicates, and each sample was run in triplicates.
Whole-genome transcriptomic analysis and qRT-PCR validation
Total RNA was obtained from 24 h biofilms formed on Spectra Mesh Woven Filter in static TSB medium using the E.Z.N.A. bacterial RNA kit (Omega Bio-tek, Inc.), according to the instructions. Pair-end reads libraries were generated using NEBNext Ultra Directional RNA Library Prep Kit and sequenced on an HiSeq (Illumina) platform. Raw sequencing data have been deposited to the NCBI SRA database under BioProject accession number PRJNA727814. Raw reads were quality-trimmed and then mapped to reference genomes using Bowtie 2-2.2.3 software [36]. Differential expression analysis was performed using the DESeq2 R package [37]. The resulting p values of genes were adjusted using Benjamini and Hochberg’s approach for controlling the false discovery rate (FDR). Genes were assigned as differentially expressed when log2 fold change (LFC) > 2 and FDR < 0.05. For functional analysis, the protein-coding sequences were mapped with KEGG Orthology terms using EggNOG-mapper v2 [38]. P values of pathways were corrected for multiple hypothesis testing using the Benjamini and Hochberg’s approach.
Isolated RNAs were reverse transcribed into single-stranded complementary DNA (cDNA) using the PrimeScript RT reagent kit with a genomic DNA (gDNA) eraser (Toyobo). Transcript levels of ilvA, ilvC, ilvD, ilvE, ilvH, leuA, leuB, leuC, and leuD were measured by qRT-PCR using a ChamQ SYBR qPCR Master Mix (Vazyme). The recA gene was used as an internal control for B. velezensis SQR9. The rpoD gene was used as an internal control for P. stutzeri XL272. ABI Real-Time PCR Instrument was operated under the following conditions: cDNA was denatured for 10 s at 95 °C, followed by 40 cycles consisting of 5 s at 95 °C and 34 s at 60 °C. The relative expression of specific genes was calculated by using the 2−ΔΔCT method [39].
Growth curve assay
For the TSB-based growth curve assays, B. velezensis SQR9 and P. stutzeri XL272 were grown in TSB rich medium individually for 24 h, the cell cultures were spun down, then the supernatants were filter sterilized as bacterial metabolites. 2 μl of start inoculum was inoculated to 200 μl TSB medium or TSB supplemented with 10% bacterial metabolites in a 10×10 well Honeycomb Microplate. OD600 was measured every 30 min at 30 °C with Bioscreen C Automated Microbiology Growth Curve Analysis System. Each treatment has 5 replicates.
For the M9-based growth curve assays, sole carbon source was provided at the concentration listed in Table S3. B. velezensis SQR9 and P. stutzeri XL272 were grown in M9 medium with corresponding carbon source individually for 96 h. Other settings were the same as described above. Each treatment has five replicates.
Metabolic facilitation assay and metabolome analysis
Both isolates were inoculated individually in 100 ml of M9 glucose medium, then incubated at 30 °C, 180 rpm shaken condition. The consumption of glucose was measured every day using the Glucose GO Assay Kit (Sigma). As a result, B. velezensis SQR9 consumed all the glucose provided in 6 days, while P. stutzeri XL272 took 4 days. The cell cultures were spun down, then the spent medium was filter sterilized. Each isolate was inoculated in 20 ml of each other’s spent medium at 1% v/v, then incubated at 30 °C, 180 rpm shaken condition for extra 4 days. Growth was measured by OD600 and colony-forming units (CFUs) at end time point. Each treatment had six replicates.
Two rounds of extracellular metabolites were collected and analyzed by UHPLC-MS/MS. Untargeted metabolomics analysis was performed using a Vanquish UHPLC system (Thermo Fisher) coupled with an Orbitrap Q Exactive HF-X mass spectrometer (Thermo Fisher). Samples were injected into an Hyperil Gold column (100 × 2.1 mm, 1.9 μm) using a 16 min linear gradient at a flow rate of 0.2 ml/min. The eluents for the positive polarity mode were eluent A (0.1% FA in water) and eluent B (methanol). The eluents for the negative polarity mode were eluent A (5 mM ammonium acetate, pH 9.0) and eluent B (methanol). The solvent gradient was set as follows: 2% B, 1.5 min; 2−100% B, 12.0 min; 100% B, 14.0 min; 100−2% B, 14.1 min; 2% B, 16 min. Q Exactive HF-X mass spectrometer was operated in positive/negative polarity mode with spray voltage of 3.2 kV, capillary temperature of 320 °C, sheath gas flow rate of 35 arb and aux gas flow rate of 10 arb. The raw data files generated by UHPLC-MS/MS were processed using the Compound Discoverer 3.0 (CD 3.0, Thermo Fisher) to perform peak alignment, peak picking, and quantitation for each metabolite. After that, peak intensities were normalized to the total spectral intensity. The normalized data were used to predict the molecular formula based on additive ions, molecular ion peaks, and fragment ions. And then peaks were matched with the mzCloud (https://www.mzcloud.org/) and ChemSpider (http://www.chemspider.com/) database to obtain the relative quantitative results. The results were processed on the free online platform of Majorbio Cloud Platform (www.majorbio.com) and provided as Dataset S3. Each treatment has four replicates.
Chemotaxis assay
A capillary assay was performed according to the method of Ling et al [40]. Briefly, the assay setup consisted of a 200 μl pipette tip as a chamber for holding 100 μl of P. stutzeri XL272 cell suspension (around 1 × 107 cells) in PBS buffer. A 2 cm 25-gauge needle (Becton Dickinson) was used as the chemotaxis capillary and was attached to a 1 ml tuberculin syringe (Becton Dickinson) containing 200 μl of the compound (secreted by B. velezensis SQR9). After 1.5 h of incubation at room temperature, the needle syringe was removed from the bacterial suspension and the content was diluted and plated on TSB medium solidified with 1.5% agar. Accumulation of bacteria in the capillaries was calculated as the average CFUs obtained on triplicate plates, and the accumulation value was expressed as the mean of three independent capillary assays for each determination. The relative chemotaxis index (RCI) was calculated as the ratio of the accumulation value for the bacteria that entered the test capillary to the corresponding value for the negative control capillary. An RCI of 2 or greater is considered significant for this method [41].
Root colonization assay
Colonization of Arabidopsis thaliana roots was performed according to the protocol from Dragoš [42]. To access root colonizing ability of the bacterial strains, the roots were transferred into Eppendorf tubes, subjected to standard sonication protocol, and the CFU assays were performed for obtained cell suspensions. To extract CFU/mm of the root, the obtain CFU values were divided by the total length of a corresponding root. Each treatment has six replicates. The experiments were performed independently at 24 and 48 h.
Greenhouse experimental design
The trial was conducted from June to August 2020, in the greenhouse of Nanjing Agricultural University. The soil used for pot experiments was collected from a field site with a history of cucumber cultivation. The field site located in Nanjing, Jiangsu Province, China, and the soil had the following properties: pH 5.62, organic matter 21.6 mg/kg, available N 157 mg/kg, available P 128 mg/kg, available K 268 mg/kg, total N 1.85 g/kg, total P 1.86 g/kg, total K 15.3 g/kg. Pots with 2 kg soil were divided into two groups. Group A: untreated soil. Group B: salt-treated soil group, the required amount of NaCl was added into the soil and stirred well to attain 3.00 g/kg salt concentration. After one-week soil incubation, one-week-old cucumber seedlings were transferred into the soil and grown for another 5 days. Then, for each group, the experiment includes four treatments: CTL, plants grown in ordinary soil without inoculation of bacteria; S, plants treated with 10 ml B. velezensis SQR9 suspensions (108 cells/ml); P, plants treated with 10 ml P. stutzeri XL272 suspensions (108 cells/ml); PS, plants treated with 5 ml B. velezensis SQR9 and 5 ml P. stutzeri XL272 suspensions, respectively (108 cells/ ml). Plants were grown for another 30 days at 30 °C, 16 h light/8 h dark. Each treatment has ten to twelve replicates.
Plant growth-promoting (PGP) traits analysis
P. stutzeri XL272 was tested for PGP traits including production of indoleacetic acid (IAA), ammonia, siderophore, and phosphate solubilization. IAA production and ammonia production were detected by the method as described in [43]. Siderophore production was detected on CAS agar [44]. Phosphate solubilization was detected on NBRIP agar containing calcium phytate or Ca3(PO4)2 [45].
Microscopy/confocal laser scanning microscopy
Fluorescent images of colonies and whole pellicle were obtained with an Axio Zoom V16 stereomicroscope (Carl Zeiss, Jena, Germany) equipped with a Zeiss CL 9000 LED light source and an AxioCam MRm monochrome camera (Carl Zeiss) and HE eGFP (excitation at 470/40 nm and emission at 525/50 nm), and HE mRFP (excitation at 572/25 nm and emission at 629/62 nm) filter sets. The exposure times for green and red fluorescence were set up to maximal possible values before reaching overexposure, using the range indicator function.
The pellicles formed in TSB medium at 24 h and root colonization at 24 h were visualized with a confocal laser scanning microscopy (CLSM, LMI-005-Leica Microsystems Confocal Microscope-SP8). Fluorescent reporter excitation was performed with the argon laser at 488 and 556 nm, the emitted fluorescence was recorded at 484−536 nm and 560−612 nm for GFP and DsRed, respectively. ImageJ software was used to obtain overlaid, artificially colored images for both stereomicroscope and CLSM.
Statistical analysis
Analysis was conducted in R 4.0.3 or GraphPad Prism 8. Figures were produced using the ggplot2 R package or GraphPad Prism 8. Detailed statistical analysis was described in the figure legends.
Results
B. velezensis SQR9 induce the enrichment of Pseudomonas spp. in the cucumber rhizosphere
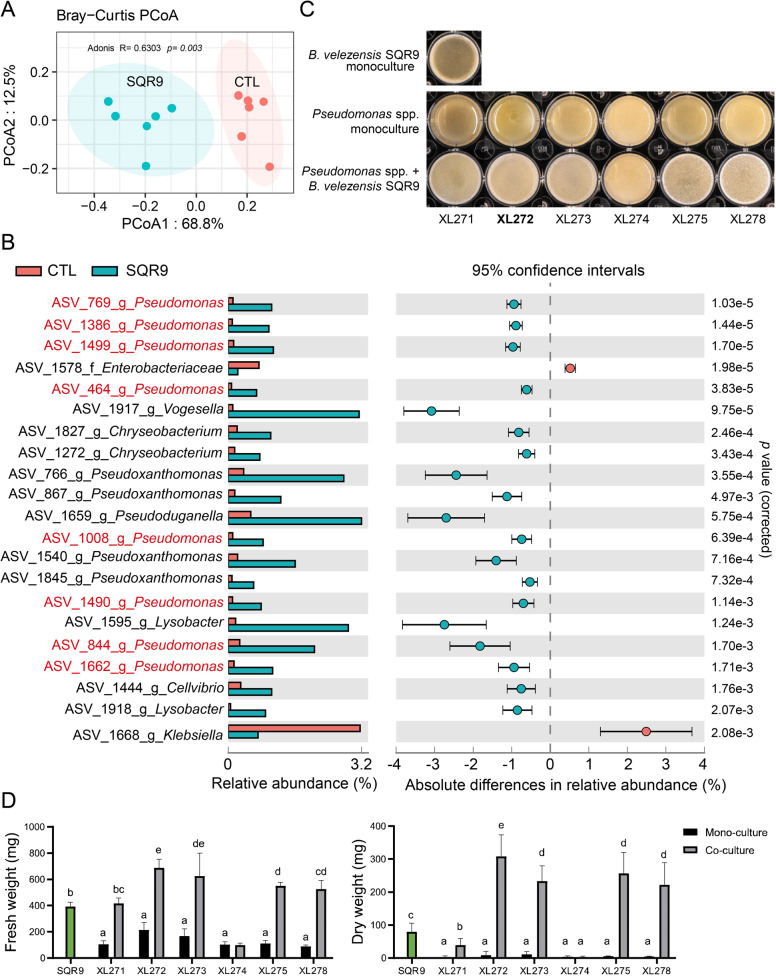
To explore the effects of B. velezensis SQR9 on rhizosphere microbiota, two-weeks-old cucumber seedlings were inoculated with strain SQR9 and the rhizosphere soil samples were collected after sixteen days. 16S rRNA amplicon sequencing was applied to compare the composition of rhizo-microbiomes of untreated control plant and plant treated with B. velezensis SQR9. Principal coordinates analysis (PCoA) was used to visualize differences in taxonomic abundance using bray-curtis distances (Fig. 1A). Untreated rhizosphere samples (CTL) were clearly separated from the B. velezensis SQR9 inoculated samples (SQR9), demonstrating that strain SQR9 had an influence on the indigenous bacterial community. To illustrate changes in the community composition and reveal the affected species at genus level, STAMP analysis was applied. In total, 21 ASVs were significantly influenced by strain SQR9 (t test; p < 0.05) (Fig. 1B). Based on the differences in relative abundance, members of the genera Pseudomonas, Vogesella, Pseudoxanthomonas, Chryseobacterium, Pseudoduganella, Lysobacter, Klebsiella, and Cellvibrio were increased after SQR9 application. Remarkably, eight out the twenty-one ASVs mapped to the Pseudomonas genus, suggesting a positive interaction with strain SQR9. Recent studies previously demonstrated that these two beneficial genera have the potential of interacting positively to enhance plant growth [46] and rescue their host plant from a sudden-wilt disease [47, 48]. We hypothesized that B. velezensis SQR9 may recruit and then synergistically interact with beneficial Pseudomonas spp. that contribute to plant growth promotion.
Fig. 1. Influence of B. velezensis SQR9 on rhizosphere microbiota, biofilm phenotype, and quantification of isolated bacteria with predicted synergism.
A Principal Coordinates Analysis (PCoA) of the rhizosphere bacteria community is plotted based on the Bray−Curtis distance metrices for taxonomical data (p < 0.01). Permutational multivariate analysis of variance (PERMANOVA) was performed using the adonis function from the vegan R package. Samples were isolated from rhizosphere of untreated control (CTL) and B. velezensis SQR9 inoculated (SQR9) plants (n = 6). B Absolute differences in relative abundance of genera between control (CTL) and B. velezensis SQR9 inoculated (SQR9) rhizosphere samples (Welch’s t test; p < 0.05). ASVs matched to Pseudomonas spp. are marked in red. Error bars represent standard deviations. C Biofilm phenotype of predicted cooperating strains. XL271-278 represents different Pseudomonas isolates. Well diameter is 15.6 mm. D Pellicle biomass quantified by fresh weight and dry weight. SQR9 represent B. velezensis SQR9, XL271-278 represent different Pseudomonas spp. Green bars represent monoculture of B. velezensis SQR9. Coculture means co-cultivated with strain SQR9. Pellicles were cultivated in TSB medium for 24 h. Data presented are the mean ± s.d. (n = 4−6). Significance test was performed using two-way ANOVA followed by Tukey’s post hoc test via Prism 8. Different letters indicate statistically significant (p < 0.05) differences.
To further characterize the potentially synergistic interaction between SQR9 and the recruited Pseudomonas spp., we isolated candidate bacteria from the rhizosphere of the B. velezensis SQR9 inoculated plants. In total, 267 bacterial isolates were obtained and phylogenetically characterized based on distinct 16S rRNA gene sequences (Fig. S1), allowing us to recover six Pseudomonas isolates, for further interaction studies. Plant beneficial bacterial consortia have been suggested to form biofilm synergistically [47, 49]. Whether Pseudomonas spp. isolates act synergistically with B. velezensis SQR9 in vitro for biofilm formation was tested. Intriguingly, five out of six Pseudomonas isolates showed enhanced biofilm phenotype in coculture as indicated by floating biofilm biomass (Fig. 1C, D). The phenotype was especially pronounced in P. stutzeri XL272 where the coculture biofilm dry weight increased more than three-fold (t test; p < 0.01) compared to the SQR9 monoculture pellicle. This suggested that the recruited Pseudomonas species have positive interactions with B. velezensis SQR9.
P. stutzeri XL272 form biofilm with B. velezensis SQR9 synergistically in rich medium but not in minimal medium
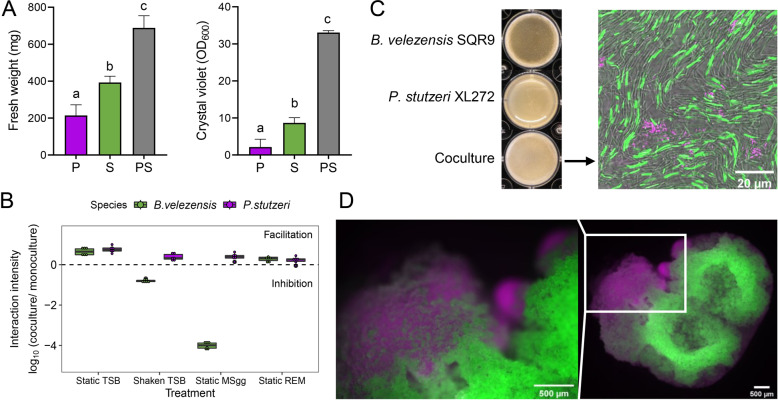
As P. stutzeri XL272 showed the highest synergy with B. velezensis SQR9 in biofilm formation, as indicated by enhanced biomass production when compared with that of single-species biofilm (Figs. 1D, 2A), this isolate was selected for further experiments. This isolate has 98.06% sequence similarity too with ASV_1490, which is one of the increased ASVs. To illustrate whether the enhanced coculture biofilm resulted from cooperation [50] or competition [51], the absolute cell numbers of the two species were quantified using qPCR. The cell numbers of both interaction partners were significantly higher in the dual-species biofilm (t test, p < 0.01) in comparison with single-species biofilms in static TSB or REM, indicative of interspecies cooperation (Fig. 2B and Figs. S2A, S2D). The distribution of these two strains were also visualized under confocal laser scanning microscope using GFP tagged B. velezensis SQR9 and DsRed tagged P. stutzeri XL272. The two strains formed distinct cell clusters and segregated within the biofilm. B. velezensis SQR9 appeared as the dominant species in the population (Fig. 2C and Fig S3), in line with qPCR quantification data (Fig S3A). In a cocultured colony, the two strains also occupied different niches, with P. stutzeri at the bottom which is oxygen-limited and nutrient-rich, and B. velezensis on the top which is oxygen-rich but nutrient-limited (Fig. 2D). Altogether these results indicate that B. velezensis and P. stutzeri cooperate in biofilm mode potentially by niche partitioning.
Fig. 2. P. stutzeri XL272 and B. velezensis SQR9 form biofilm synergistically in certain conditions.
A Biofilm biomass quantification by fresh weight and crystal violet assay. Biofilm was cultivated in static TSB medium. P represents P. stutzeri XL272 monocultured pellicle, S represents B. velezensis SQR9 monocultured pellicle, PS represents cocultured pellicle. Bars represent the mean ± s.d. (n = 6). Different letters indicate statistically significant (p < 0.05) differences according to ANOVA, Tukey test via Prism 8. B Interaction intensity is defined as logarithmic scale of DNA copies in cocultures relative to the average DNA copies in monocultures. Interaction intensity > 1 indicates facilitation, while < 1 indicates inhibition. Bars represent the mean ± s.d. (n = 6). Static TSB and static REM (root-exudate medium) represented structured, nutrient-rich condition, shaken TSB represented unstructured, nutrient-rich condition, static MSgg represented structured, nutrient-limited condition. Cooperation only occurred under structured, nutrient-rich condition. C Biofilm formation of monoculture and coculture in TSB medium. Biofilm formed by P. stutzeri XL272 (magenta) and B. velezensis SQR9 (green) were viewed under the CLSM. Well diameter is 15.6 mm. D Colony grown on TSB agar. DsRed tagged P. stutzeri XL272 were colored in magenta, GFP tagged B. velezensis SQR9 were colored in green.
Next, we tested the role of structured environment and nutrient availability of the cooperative relationship between B. velezensis SQR9 and P. stutzeri XL272. The interaction intensity was calculated as a logarithmic value of cell numbers in coculture relative to monoculture, where value > 1 indicates facilitation, while value < 1 indicates inhibition (Fig. 2B). In the absence of spatial structure (shaking TSB medium) or under limited nutrient availability (static MSgg medium), only P. stutzeri was benefited in the coculture (Fig. 2B, and Figs. S2B, S2C). While in the static, nutrient-rich medium (TSB or REM), both species were mutually facilitated (Fig. 2B and Figs. S2A, S2D). These observations suggested that the synergism between B. velezensis and P. stutzeri is only maintained under static, nutrient-rich condition.
B. velezensis biofilm matrix EPS and TasA are required for the synergism
To understand the molecular mechanisms of cooperation between B. velezensis and P. stutzeri, we examined the contribution of B. velezensis biofilm matrix components, EPS and TasA, for the interaction. In TSB medium, B. velezensis ∆tasA mutant showed severely impaired monocultured pellicle, but not ∆epsD (Fig. 3A). To test whether these components contribute to the coculture pellicle phenotype, we mixed P. stutzeri strains with B. velezensis strains lacking either EPS or TasA. The resulting coculture pellicles appeared weaker than those formed by wildtype cocultures (Fig. 3A). The cell numbers of both B. velezensis matrix mutants, were reduced in cocultures, indicating that both EPS and TasA were necessary for efficient synergism (Fig. 3B). Meanwhile, the cell numbers of P. stutzeri were higher in all cocultures than in monoculture (Fig. 3B). We also tested the interaction of P. stutzeri with B. velezensis matrix mutants in biofilm-inducing minimal medium MSgg. Surprisingly, although both B. velezensis mutants showed impaired individual fitness in minimal medium, they benefit from the presence of P. stutzeri (Fig. S4). In conclusion, biofilm matrix components EPS and TasA are necessary for positive interspecies interaction in certain conditions.
Fig. 3. Extracellular matrix EPS and TasA are essential for mutualism in TSB-rich medium.
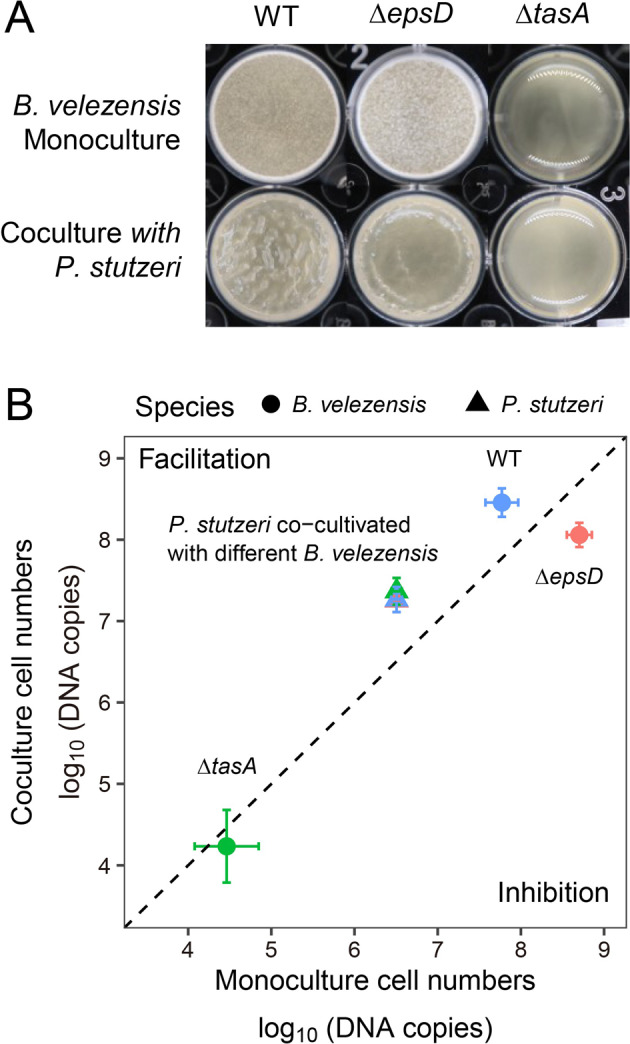
A Formation of pellicle biofilms by the mutants deficient in biosynthesis of exopolysaccharide EPS (∆epsD) and TasA protein fibers (∆tasA). Cells were incubated in TSB at 30 °C for 24 h before images were taken. Well diameter is 15.6 mm. B Cell numbers in dual-species biofilm. Circle dots represent B. velezensis, triangles represent P. stutzeri. Colors indicate B. velezensis strain genotypes in coculture: WT (blue), ∆epsD (pink), ∆tasA (green). Data presented are the mean ± s.d. (n = 6).
P. stutzeri XL272 might provide BCAAs to B. velezensis SQR9
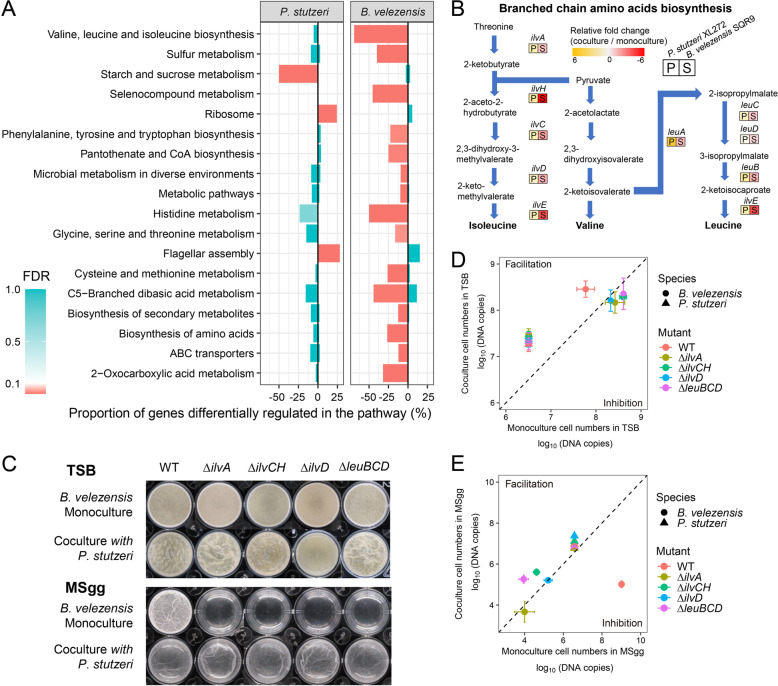
Our results above demonstrated that P. stutzeri XL272 cooperates with B. velezensis SQR9 in TSB medium during biofilm formation (Fig. 2A and Fig. S2A). To disentangle the mechanism of cooperation, the transcriptomes of both species were determined to elucidate the potential mode of actions underpinning the remarkably enhanced biomass in dual-species biofilm. In total, 345 genes of B. velezensis SQR9 and 443 genes of P. stutzeri XL272 were significantly regulated at a minimum of four-fold expression change in dual-species biofilms compared to single-species biofilms (24 h of interaction in TSB medium). Major transcriptional alterations were observed in genes related to bacterial metabolism, biosynthesis of amino acids, flagellar assembly, and ABC transporter (Fig. 4A). In the cocultures, fifteen B. velezensis SQR9 pathways were downregulated including amino acid biosynthesis and metabolism, sulfur metabolism, selenocompound metabolism, carbohydrate biosynthesis and metabolism, biosynthesis of secondary metabolites, and ABC transporter (Fig. 4A). Intriguingly, the BCAAs biosynthesis pathways were downregulated in B. velezensis SQR9. qRT-PCR assay validated that all these genes involved in this pathway were downregulated in B. velezensis SQR9 while conversely upregulated in P. stutzeri XL272 during co-culturing (Fig. 4B). This result suggested that the reduced production of BCAAs in B. velezensis SQR9 might be compensated by P. stutzeri XL272. To explore the potential metabolic exchanges between the two species, we reconstructed genome-scale metabolic models and predicted the likely exchanged metabolites (Dataset S2). The results highlighted that P. stutzeri XL272 could potentially release two of the BCAAs (L-valine and L-leucine) to B. velezensis SQR9 (Dataset S2).
Fig. 4. Transcriptional response of P. stutzeri XL272 and B. velezensis SQR9 in dual-species TSB biofilm and validation on mutants.
A KEGG pathway analysis of the genes induced (right bars) and repressed (left bars) in response to coculture in TSB. Pathways which LFC > 2, FDR < 0.05 were significantly regulated and shown in red. B The relative fold change of genes involved in the branched-chain amino acid biosynthesis pathways examined by qRT-PCR. Color represents the mean relative fold change of mRNA in coculture compared to monoculture (n = 6). C Formation of pellicle biofilms by the mutants. Cells were incubated in TSB or MSgg at 30 °C for 24 h before images were taken. D Cell numbers of dual-species biofilm in TSB medium. E Cell numbers of dual-species biofilm in MSgg medium. Circle dots represent B. velezensis, triangles represent P. stutzeri. Colors indicate B. velezensis strain genotypes in coculture: WT (pink), ∆ilvA (brown), ∆ilvCH (green), ∆ilvD (blue), ∆leuBCD (magenta). P. stutzeri XL272 promoted the growth of B. velezensis SQR9 ∆ilvD and ∆leuBCD in nutrient-limited condition. Data presented are the mean ± s.d. (n = 6).
We further constructed B. velezensis BCAAs biosynthetic mutants: ∆ilvA, ∆ilvCH, ∆ilvD that were unable to synthesize all three BCAAs, and ∆leuBCD that was unable to synthesize leucine. All the mutants showed severe growth defects in MSgg minimal medium under both planktonic and static conditions (Fig. S5A, C). However, these strains showed increased static biofilm formation and higher planktonic biomass than wildtype in TSB, as tryptone can be hydrolyzed by B. velezensis to generate the required amino acids (Fig. S5A–C). It was expected that these auxotrophic mutants of B. velezensis SQR9 could be complemented by P. stutzeri XL272 during interaction. Consistently, part of the auxotrophic mutants, such as ∆ilvCH and ∆leuBCD showed significantly increased cell numbers (4.3 × 105 and 2.0 × 105, respectively) in cocultures with P. stutzeri XL272 compared to monocultures of each mutant (4.4 × 104 and 1.0 × 104, respectively) under nutrient-limited condition (MSgg medium) (Fig. 4E). Furthermore, we observed that BCAAs mutants of SQR9 (ilvD and ∆leuBCD) not only benefited in cocultures, but also promoted the growth of P. stutzeri (Fig. 4E). Nevertheless, when the mutants were co-cultivated with P. stutzeri in nutrient-rich TSB medium, they reached similar cell numbers like the wildtype (Fig. 4D and Fig. S5C), likely due to excess of BCAAs in the medium. All together, these analyses indicate that B. velezensis SQR9 and P. stutzeri XL272 might exchange BCAAs in certain conditions.
B. velezensis SQR9 facilitates the growth of P. stutzeri XL272 by metabolic cross-feeding
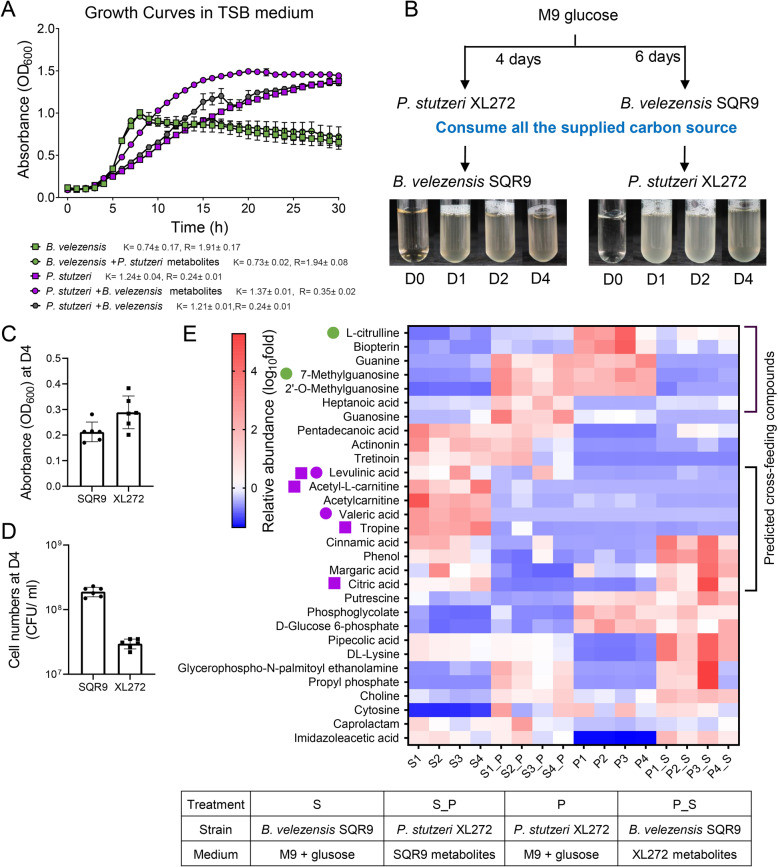
To elucidate whether metabolic facilitation is responsible for syntrophic cooperation, TSB spent medium complementation assays were performed. In support of our hypothesis, the growth of P. stutzeri XL272 was significantly enhanced by the supplementation of B. velezensis SQR9 supernatant (Fig. 5A), which indicated strong metabolic facilitation. However, the supernatant of P. stutzeri XL272 did not affect the growth of B. velezensis SQR9 under these conditions (Fig. 5A).
Fig. 5. Metabolic facilitation and chemotactic attraction stabilized cooperation.
A Growth curves of monoculture cultivated in pure TSB media (square), monoculture grown on TSB supplemented with 10% supernatant of another species (circle) and coculture (hexagon). B. velezensis SQR9 TSB spent medium facilitated the growth of P. stutzeri XL272. “K” indicates growth capacity, “R” indicates growth rate, which are calculated by growthcurver R package. B Schematic representation of the M9 spent medium experiment. Isolates were independently grown in M9 glucose medium for 6 days (B. velezensis SQR9) or 4 days (P. stutzeri XL272) till the glucose was under detection, after which cells were filtered out from the suspension. The filtrate was used as the growth medium for another isolates (see “Methods”). Both isolates were able to grow on each other isolate’s metabolic by-products. C Absorbance of OD600 at day 4 of cell cultures in panel (B). D Viable cell numbers quantified by CFU assay at day 4 of cell cultures in panel (B). E Metabolic profiles of the spent medium experiment. S indicated the spent medium of B. velezensis grown on M9 glucose, S_P indicated the spent medium of P. stutzeri grown on cell-free filtrate of B. velezensis, P indicated the spent medium of P. stutzeri grown on M9 glucose, P_S indicated the spent medium of B. velezensis grown on cell-free filtrate of P. stutzeri. Magenta circle symbol indicated P. stutzeri can grow on M9 medium with these compounds, green circle symbol indicated B. velezensis can grow on M9 medium with these compounds (see Fig. S6 for detail). Magenta square symbol indicated P. stutzeri showed chemotactic response towards these compounds (see Fig. S8 for detail).
In an alternative complementation assay, spent medium was created from M9 minimal medium with 1% glucose as the sole carbon source in which monocultures of the two isolates were previously cultivated until all the supplied carbons have been consumed, thus all the carbons present in the supernatant were originated from metabolites secreted by the respective bacterium. We found that both isolates were able to grow and multiply on the metabolic by-products present in each other’s, but not on their own spent medium (Fig. 5B–D). Untargeted UPLC-MS-based metabolomic approach was used to compare the metabolic profiles of cell culture grown in M9 medium and bacterial supernatant filtrate. The top 30 differential compounds were selected based on their relative abundance displayed in the heatmap (Fig. 5E). Compounds that were more abundant in glucose M9 spent medium of one species, while decreased after cultivation of the other species were assumed to be metabolized by the subsequently growing species. As a result, levulinic acid, acetyl-L-carnitine, acetylcarnitine, valeric acid, cinnamic acid, margaric acid, citric acid, tropine, and phenol secreted by B. velezensis SQR9 could potentially be utilized by P. stutzeri XL272 subsequently. Correspondingly, P. stutzeri XL272 can produce L-citrulline, biopterin, guanine, 7-methylguanosine, and 2′-O-methylguanosine, which could potentially be metabolized by B. velezensis SQR9. To test this hypothesis, growth curve assays on M9 medium with these individual compounds were performed. Results confirmed the metabolism of some but not all compounds largely due to the low solubility (Table S3). P. stutzeri XL272 could utilize levulinic acid and valeric acid, while B. velezensis SQR9 could utilize 7-methylguanosine and L-citrulline (Fig. S6). This suggests a strong potential for metabolic cross-feeding between the two species, which was further supported by dissimilar preference of B. velezensis SQR9 and P. stutzeri XL272 for carbon sources that are present in root exudates (Fig. S7). Additional chemotaxis assays were performed on selected individual compounds secreted by B. velezensis SQR9. Interestingly, P. stutzeri XL272 showed a strong chemotactic response towards tropine, citric acid, and acetyl-L-carnitine although these compounds cannot serve as sole carbon sources (Fig. S8). Altogether, metabolic facilitation and chemotactic attraction could explain the observed cooperation in TSB-grown co-culture biofilms and in the rhizosphere.
Performance of two species consortia in the rhizosphere
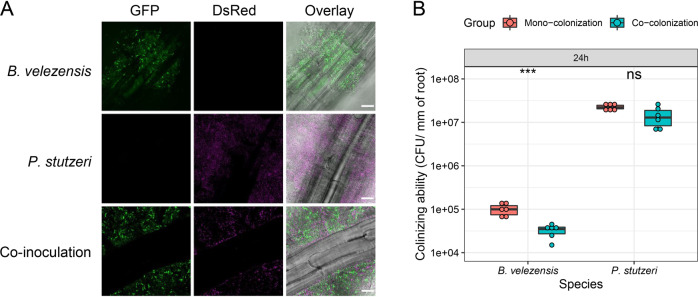
Since Pseudomonas spp. were recruited by B. velezensis SQR9 and the two isolates showed syntrophic exchange in biofilm formation in vitro, we were interested in the biological relevance of biofilm formation on plant roots. First, we monitored the colonization of these two strains on the roots of model plant, A. thaliana, which allows easy monitoring of bacterial colonization in the laboratory [42, 52]. P. stutzeri XL272 colonized the roots of A. thaliana at higher abundance than B. velezensis SQR9 using hydroponic conditions (Fig. 6 and Fig S9). Notably, these two species were able to co-colonize the A. thaliana roots (Fig. 6A). We further monitored the proportion of each strain on these root-surface biofilms. In contrast to pellicle biofilm, P. stutzeri XL272 was the dominant species in the population colonized in the A. thaliana roots (Fig. 6B and Fig. S9). B. velezensis SQR9 was inhibited by P. stutzeri XL272 in co-colonization.
Fig. 6. Co-colonization on plant roots.
A A. thaliana roots were colonized by B. velezensis SQR9 (GFP, colored green), P. stutzeri XL272 (DsRed, false-colored magenta), and mixture, visualized using CLSM. Scale bar represents 20 µm. B Colonizing ability of the bacteria was measured as CFUs per millimeter of root (n = 6) (see “Methods”). The samples were collected at 24 h. The colonizing ability of mono-colonization is compared with co-colonization. Asterisks indicate statistically significant (p < 0.01) according to unpaired student’s t test via R.
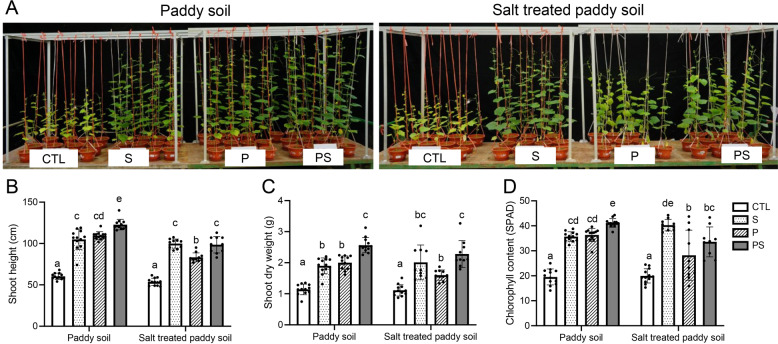
We further tested whether the two species consortium performed better than the single strains in the cucumber pot assays. Before test, indeed, results showed that both strains were able to promote the growth of cucumber plants individually and in the mixture (Fig. 7). The growth promotion ability of P. stutzeri XL272 is potentially explained by IAA, ammonia and siderophore production (Fig. S10). For B. velezensis SQR9, previous studies demonstrated that strain SQR9 can directly stimulate the cucumber growth by producing phytohormones (IAA), volatile compounds (acetoin and 2,3-butanediol), and extracellular enzymes (phytase) [53]. Remarkably, the consortium had a stronger promoting effect in paddy soil, as the dual-species consortium significantly increased the shoot height, shoot dry weight, and chlorophyll content of plants in comparison to plant inoculated with one species. In previous work, B. velezensis SQR9 was shown to enhance plant salt tolerance [19], the consortium was also tested under salt-treated paddy soil. Compared with non-inoculated control plants, P. stutzeri XL272 protects the plant against salt stress (Fig. 7). The protective effect of the consortium was higher than that of P. stutzeri XL272 alone in comparison to control plants. Collectively, these results showed that the genus Pseudomonas recruited by PGPR B. velezensis SQR9 in the rhizosphere promotes the growth of cucumber plants synergistically with B. velezensis SQR9. These results support the suggestion that the PGPR can induce the assemblage of the indigenous beneficial microbiome, leading to the promotion of plant health and resistance to salt stress.
Fig. 7. The consortium promoted plant growth and alleviated salt stress.
A Photos showing 6-week-old cucumber plants grown in normal paddy soil or salt-treated paddy soil inoculated with S (B. velezensis SQR9), P (P. stutzeri XL272) or PS (mixture of P and S). CTL represents plants grown in ordinary soil without inoculation on bacteria. B Shoot height of plants. C Shoot dry weight of plants. D Leaf chlorophyll content of plants. Bars represent the mean mean ± s.d. (n = 10−12). Significance test was performed using two-way ANOVA followed by Tukey’s post hoc test via Prism 8. Different letters indicate statistically significant (p < 0.05) differences.
Discussion
Plant genotypes and local soil environmental parameters are known to play pivotal roles in shaping the root-associated microbiota [54], while bacterial inoculants also modulate the plant microbiota. The present study highlights the importance of microbial ecological interactions in PGPR-based biocontrol. We demonstrated that a well-established biocontrol bacterium B. velezensis SQR9 alters rhizosphere microbiome, by changing the abundance of other beneficial bacteria. The recruited strains can serve as cooperative partners of SQR9 and assist in plant growth promotion.
We demonstrated that PGPR B. velezensis SQR9 can facilitate increases in the abundance of several genera with reported beneficial functions including Pseudomonas [55], Chryseobacterium [56], Lysobacter [57]. Stimulating beneficial bacteria in the rhizosphere seems to be a common property of Bacillaceae. Previously, Bacillaceae group species were reported to promote the colonization of rhizobia [58], promote the growth of rhizosphere bacterium Flavobacterium johnsoniae [59], and stimulate indigenous soil Pseudomonas populations that enhance plant disease suppression [60].
Here, we concentrated on the interaction between Bacillus and Pseudomonas genera as widely commercialized beneficial bacteria that dominate the rhizosphere microbiota [61, 62] and are more investigated at the molecular level compared with other genera. P. stutzeri XL272 was chosen as a representative to design a consortium as it shared the same region of colonization with B. velezensis SQR9 in the rhizosphere and formed dual-species biofilm synergistically in TSB and REM medium. Forming biofilm synergistically is a common trait of plant beneficial consortium [47, 49]. Rhizosphere bacteria frequently reside in multispecies biofilms, the lifestyle of multispecies biofilm facilitate the emergence of community intrinsic properties [63], such as enhanced tolerance to antimicrobial agents [64, 65], horizontal gene transfer (HGT), and sharing of public goods. In this study, the dual-species consortium not only formed synergistic biofilm in vitro (Fig. 2), but also created mixed-species biofilms on plant roots.
We further revealed that both biofilm matrix components, EPS and TasA are important for the cooperation between B. velezensis and P. stutzeri. The role of TasA in mixed-species interaction was also reported in B. subtilis-Pantoea agglomerans interaction, during which both species contributed matrix components to the coculture colony biofilm structure, with B. subtilis producing matrix protein TasA protein and P. agglomerans producing exopolysaccharides [66]. In the case of B. velezensis-P. stutzeri cocultures, matrix components were important for synergism under cooperation-promoting conditions (nutrient-rich medium), while under competition-promoting conditions (minimal medium), the loss of biofilm matrix conferred B. velezensis with a fitness advantage in the presence of P. stutzeri. We hypothesize that in minimal medium, B. velezensis might exploit the matrix components produced by P. stutzeri. Whether and how P. stutzeri complements the lacking extracellular matrix components of B. velezensis in the mixed-species biofilms needs further investigation.
Our results showed that the interaction outcome of B. velezensis with P. stutzeri is highly context-dependent. The synergism between the two species only occurred under static, nutrient-rich conditions. As static conditions allow biofilm formation, the cooperation could be facilitated by more intimate interactions between the two species, facilitating the metabolic cross-feeding, as previously demonstrated [67]. This would also be in line with aggregation-promoting matrix components being important for the cooperative interaction under this condition.
In the hydroponic root colonization assays, the environment was mixed evenly by continuous shaking of the cultures and limited plant exudates were available to bacteria, which might explain the lower colonizing cell numbers of B. velezensis in co-inoculation compared with that in mono-colonization (Fig. 6 and Fig. S7). Competition is more prevalent in free-living habitats wherein the resources are scarcer, such as the hydroponic shaking root system in this study. In contrast, the natural rhizosphere soil environments provide stages for cooperative interaction due to the local presence of diverse nutrients, root exudates, and heterogeneity. These findings are in accordance with previous studies, support the impact of spatial organization and nutrient condition on microbial interaction [68, 69]. Our experimental setup involved co-cultivation of B. velezensis and P. stutzeri in liquid medium. We documented the potential for positive interaction and cross-feeding between the two species in such in vitro system. We also highlighted the potential advantages and disadvantages of the use of rich medium (like TSB) as an in vitro model for biofilm studies. As an advantage, such a model enables systematic in-depth analysis of the mechanisms underlying bacterial interaction. Moreover, our system rules out the impact of other factors, for instance, temperature change, competition with other species, the influence of the plant, etc. This enables the downstream process to be performed separately for each species, aiding the effect of bacterial interaction. As a disadvantage, this model does not actually mimic the natural habitat, solid rhizo-sphere environment that is surrounded by a diverse microbiome. Other influence factors were overlooked in this system. It is essential to develop a rhizosphere mimicking system to replace hydroponic system and study microbial interactions in the natural habitat in follow-up research.
Interaction between members of Bacillus and Pseudomonas genera was previously reported to be dependent on abiotic conditions and species involved. In particular cases, these two genera utilize sophisticated competition strategies, such as type VI secretion system used by P. chlororaphis to attack B. subtilis colonies [70]. It was suggested that B. subtilis either produces extracellular matrix to reduce infiltration by P. chlororaphis or enters sporulation as self-defense strategies [70]. In another example, the presence of B. subtilis reduces the appearance of spontaneous mutants lacking secondary metabolite production in P. protegens [71]. In other circumstances, B. licheniformis and P. fluorescens interact positively in biofilm mode, enhancing plant growth and photosynthetic attributes [46], further supporting our findings.
So far, the role of Bacilli in recruiting beneficial bacteria was only investigated using phenotypic analysis and amplicon sequencing, with limited knowledge about interspecies interactions at the molecular level. To close this gap, we took advantage of dual RNA-seq analysis to characterize the transcriptional consequences of bacterial interactions. Transcriptome analysis revealed distinct gene expression profiles in dual-species biofilm compared with single-species biofilm. For B. velezensis SQR9, we noted that six pathways related to amino acids biosynthesis were down-regulated in the presence of P. stutzeri XL272, possibly reducing the metabolic costs related to these pathways. In accordance, the essential amino acids could partly be compensated by P. stutzeri XL272. This cross-feeding hypothesis was supported by genome-scale metabolic modeling (Dataset S2 and Fig. 8B). In synthetic microbial communities, the metabolic exchange of biosynthetically costly amino acids tends to promote strong cooperative interactions [72]. This hypothesis was further confirmed using B. velezensis SQR9 auxotrophic mutants of BCAAs biosynthesis, which could be rescued by P. stutzeri XL272 under nutrient-limiting conditions. Thus, the ability to synthesize BCAAs is important to survive under nutrient-limited condition, while dispensable when the required nutrients are provided by co-existing species or other environmental sources.
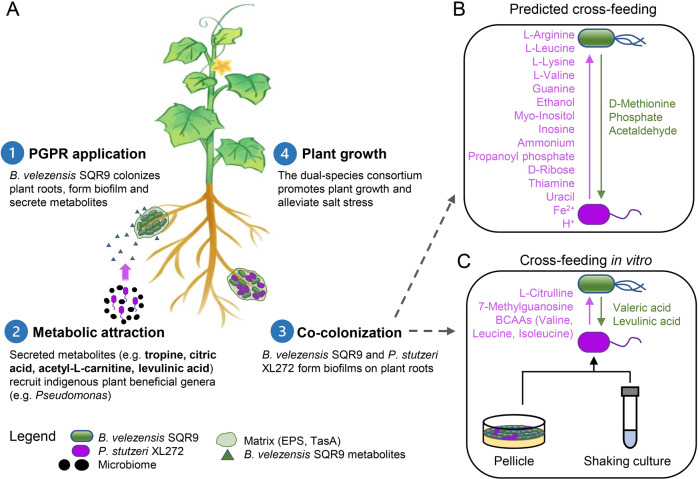
Fig. 8. Schematic diagram.
A A model illustrating the sequential events taking place in the rhizosphere after PGPR B. velezensis SQR9 application. B Cross-feeding between B. velezensis SQR9 and P. stutzeri XL272 based on genome-scale metabolic modeling (see “Methods” and Dataset S2). C Cross-feeding between B. velezensis SQR9 and P. stutzeri XL272 based on in vitro experiments.
We demonstrated that supernatants obtained using M9 minimal medium were able to support the growth of the other respective species, indicating metabolic cross-feeding, as reported previously for various mixed-species communities [10, 73–75]. Syntrophic interactions mediated by the exchange of nutrients might help to maintain the stability of the bacterial consortium. As species interactions alter the evolution of resource utilization [75, 76], it will be interesting to investigate whether the cooperation between B. velezensis SQR9 and P. stutzeri XL272 can be maintained in a longer time scale and whether the interacting partners influence the evolutionary diversification of each other.
Finally, the combined use of Bacillus and Pseudomonas inoculants provide a synergistic effect on plant health, including plant growth promotion, salt stress alleviation, as observed in our pot experiments, and increased disease suppression [60]. Inspired by their complementary colonization dynamics on plant roots [77] and positive interaction in biofilm formation, we hypothesize that the fast-growing Bacillus could serve as a pioneer to occupy the available niche in the rhizosphere at an early stage. Through metabolism, they secrete metabolic by-products that increase the viability of successor Pseudomonas. In line with this hypothesis, B. velezensis SQR9 had a broader ability of catabolizing sugars, while P. stutzeri XL272 had a stronger ability to utilize organic acids (Fig. S7). In agreement with the metabolome data, B. velezensis SQR9 converts glucose to organic acids which could be metabolized by P. stutzeri XL272. Whether such metabolic cooperation occurs in the rhizosphere environment and contributes to the synergistic plant beneficial functions, remains to be elucidated. Basically, P. stutzeri XL272 was capable of IAA, ammonia and siderophore production in the lab condition (Fig S10). However, we considered that understanding the molecular processes underlying the plant-microbe interactions of P. stutzeri XL272 is an important step to disentangle the synergy in the rhizosphere context.
In conclusion, our findings demonstrate that a PGPR recruits indigenous beneficial bacteria and can cooperatively interact with them via cross-feeding. Synergistic biofilm formation was accompanied by enhanced plant-growth-promoting and salt stress-relieving ability. Based on our findings, the conceptual ecological model was summarized in Fig. 8. In the first step, B. velezensis SQR9 is attracted by root exudates and colonizes the rhizosphere [78–80]. After establishing biofilm on plant roots, it secretes metabolites that increase the abundance of indigenous plant beneficial genera (such as Pseudomonas spp.). By forming tightly associated biofilm, they share extracellular matrix and essential metabolites that increase their fitness in the rhizosphere. As a result, the simplified community has an increased ability to promote plant growth and alleviate salt stress. Our study proposes an ecological approach for plant health using microbial inoculants with synergistic effects, in addition to being an important step towards the understanding of microbial interactions in the rhizosphere microbiome.
Supplementary information
Acknowledgements
This work was financially supported by the National Nature Science Foundation of China (31972512, 32072675, and 32072665), the Agricultural Science and Technology Innovation Program of CAAS (CAAS-ZDRW202009), the Fundamental Research Funds for the Central Universities (KYXK202009). XS was supported by a Chinese Scholarship Council fellowship. ÁTK, MLS, and AD were supported by the Danish National Research Foundation (DNRF137) for the Center for Microbial Secondary Metabolites. AD was supported by Slovenian Research Agency (N1-0177). Biofilm-related work in the group of ÁTK is supported by a DTU Alliance Strategic Partnership PhD fellowship. Funding from Novo Nordisk Foundation (grant NNFOC0055625) for the infrastructure “Imaging microbial language in biocontrol (IMLiB)” is acknowledged. Author XS is very grateful to Prof. Shen and Prof. Zhang for the strong supports during the period of outbreak epidemics of COVID-19.
Author contributions
XS, ZX, RZ, and ÁTK designed the study; XS, JX, and TT performed the experiments. XS, VHT, and MLS analyzed the data and created the figures. DZ constructed the metabolic models. XS and ZX wrote the first draft of the paper; AD, ÁTK, MLS, RZ, and QS revised the paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xinli Sun, Zhihui Xu.
Contributor Information
Zhihui Xu, Email: xzh2068@njau.edu.cn.
Ruifu Zhang, Email: rfzhang@njau.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-01125-3.
References
- 1.Leach JE, Tringe SG. Plant–microbiome interactions: from community assembly to plant health. Nat Rev Microbiol. 2020;18:607–21. doi: 10.1038/s41579-020-0412-1. [DOI] [PubMed] [Google Scholar]
- 2.Pii Y, Mimmo T, Tomasi N, Terzano R, Cesco S, Crecchio C. Microbial interactions in the rhizosphere: beneficial influences of plant growth-promoting rhizobacteria on nutrient acquisition process. A Rev Biol Fertil Soils. 2015;51:403–21. [Google Scholar]
- 3.Backer R, Rokem JS, Ilangumaran G, Lamont J, Praslickova D, Ricci E, et al. Plant growth-promoting rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front Plant Sci. 2018;871:1473–89. doi: 10.3389/fpls.2018.01473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lugtenberg B, Kamilova F. Plant-growth-promoting rhizobacteria. Annu Rev Microbiol. 2009;63:541–56. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharyya PN, Jha DK. Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J Microbiol Biotechnol. 2012;28:1327–50. doi: 10.1007/s11274-011-0979-9. [DOI] [PubMed] [Google Scholar]
- 6.Leach JE, Triplett LR, Argueso CT, Trivedi P. Communication in the phytobiome. Cell. 2017;169:587–96. doi: 10.1016/j.cell.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 7.Vorholt JA, Vogel C, Carlström CI, Müller DB. Establishing causality: opportunities of synthetic communities for plant microbiome research. Cell Host Microbe. 2017;22:142–55. doi: 10.1016/j.chom.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Douglas AE. The microbial exometabolome: ecological resource and architect of microbial communities. PTRBAE. 2020;375:20190250. doi: 10.1098/rstb.2019.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turroni F, Milani C, Duranti S, Mahony J, van Sinderen D, Ventura M. Glycan utilization and cross-feeding activities by Bifidobacteria. Trends Microbiol. 2018;26:339–50. doi: 10.1016/j.tim.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 10.Evans CR, Kempes CP, Price-Whelan A, Dietrich LEP. Metabolic heterogeneity and cross-feeding in bacterial multicellular systems. Trends Microbiol. 2020;28:732–43. doi: 10.1016/j.tim.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith NW, Shorten PR, Altermann E, Roy NC, McNabb WC. The classification and evolution of bacterial cross-feeding. Front Ecol Evol. 2019;7:153. [Google Scholar]
- 12.Santoyo G, del Orozco-Mosqueda MC, Govindappa M. Mechanisms of biocontrol and plant growth-promoting activity in soil bacterial species of Bacillus and Pseudomonas: a review. Biocontrol Sci Technol. 2012;22:855–72. [Google Scholar]
- 13.Borriss R. Use of plant-associated Bacillus strains as biofertilizers and biocontrol agents in agriculture. Bacteria in agrobiology: plant growth responses. Springer: Berlin; 2011. 41–76.
- 14.Xiong W, Guo S, Jousset A, Zhao Q, Wu H, Li R, et al. Bio-fertilizer application induces soil suppressiveness against Fusarium wilt disease by reshaping the soil microbiome. Soil Biol Biochem. 2017;114:238–47. [Google Scholar]
- 15.Qin Y, Shang Q, Zhang Y, Li P, Chai Y. Bacillus amyloliquefaciens L-S60 reforms the rhizosphere bacterial community and improves growth conditions in cucumber plug seedling. Front Microbiol. 2017;8:2620. doi: 10.3389/fmicb.2017.02620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Compant S, Samad A, Faist H, Sessitsch A. A review on the plant microbiome: ecology, functions, and emerging trends in microbial application. J Adv Res. 2019;19:29–37. doi: 10.1016/j.jare.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao Y, Zhang Z, Ling N, Yuan Y, Zheng X, Shen B, et al. Bacillus subtilis SQR9 can control Fusarium wilt in cucumber by colonizing plant roots. Biol Fertil Soils. 2011;47:495–506. [Google Scholar]
- 18.Xu Z, Shao J, Li B, Yan X, Shen Q, Zhang R. Contribution of bacillomycin D in Bacillus amyloliquefaciens SQR9 to antifungal activity and biofilm formation. Appl Environ Microbiol. 2013;79:808–15. doi: 10.1128/AEM.02645-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L, Liu Y, Wu G, Veronican Njeri K, Shen Q, Zhang N, et al. Induced maize salt tolerance by rhizosphere inoculation of Bacillus amyloliquefaciens SQR9. Physiol plant. 2016;158:34–44. doi: 10.1111/ppl.12441. [DOI] [PubMed] [Google Scholar]
- 20.Blake C, Nordgaard Christensen M, Kovács ÁT. Molecular aspects of plant growth promotion and protection by Bacillus subtilis. Mol Plant Microbe Interact. 2020;34:15–25. doi: 10.1094/MPMI-08-20-0225-CR. [DOI] [PubMed] [Google Scholar]
- 21.Al-Ali A, Deravel J, Krier F, Béchet M, Ongena M, Jacques P. Biofilm formation is determinant in tomato rhizosphere colonization by Bacillus velezensis FZB42. Environ Sci Pollut Res. 2018;25:29910–20. doi: 10.1007/s11356-017-0469-1. [DOI] [PubMed] [Google Scholar]
- 22.Xu Z, Mandic-Mulec I, Zhang H, Liu Y, Sun X, Feng H, et al. Antibiotic bacillomycin D affects iron acquisition and biofilm formation in Bacillus velezensis through a Btr-mediated FeuABC-dependent pathway. Cel Rep. 2019;29:1192–202. doi: 10.1016/j.celrep.2019.09.061. [DOI] [PubMed] [Google Scholar]
- 23.Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–97. [Google Scholar]
- 24.Bai Y, Müller DB, Srinivas G, Garrido-Oter R, Potthoff E, Rott M, et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature. 2015;528:364–9. doi: 10.1038/nature16192. [DOI] [PubMed] [Google Scholar]
- 25.Zhou C, Shi L, Ye B, Feng H, Zhang J, Zhang R, et al. pheS *, an effective host-genotype-independent counter-selectable marker for marker-free chromosome deletion in Bacillus amyloliquefaciens. Appl Microbiol Biotechnol. 2017;101:217–27. doi: 10.1007/s00253-016-7906-9. [DOI] [PubMed] [Google Scholar]
- 26.Feng H, Zhang N, Fu R, Liu Y, Krell T, Du W, et al. Recognition of dominant attractants by key chemoreceptors mediates recruitment of plant growth-promoting rhizobacteria. Environ Microbiol. 2019;21:402–15. doi: 10.1111/1462-2920.14472. [DOI] [PubMed] [Google Scholar]
- 27.Lambertsen L, Sternberg C, Molin S. Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ Microbiol. 2004;6:726–32. doi: 10.1111/j.1462-2920.2004.00605.x. [DOI] [PubMed] [Google Scholar]
- 28.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 29.Machado D, Andrejev S, Tramontano M, Patil KR. Fast automated reconstruction of genome-scale metabolic models for microbial species and communities. Nucleic Acids Res. 2018;46:7542–53. doi: 10.1093/nar/gky537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lieven C, Beber ME, Olivier BG, Bergmann FT, Ataman M, Babaei P, et al. MEMOTE for standardized genome-scale metabolic model testing. Nat Biotechnol. 2020;38:272–6. doi: 10.1038/s41587-020-0446-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zelezniak A, Andrejev S, Ponomarova O, Mende DR, Bork P, Patil KR. Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc Natl Acad Sci USA. 2015;112:6449–54. doi: 10.1073/pnas.1421834112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–8. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 33.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–4. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Branda SS, González-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci USA. 2001;98:11621–6. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langdon WB. Performance of genetic programming optimised Bowtie2 on genome comparison and analytic testing (GCAT) benchmarks. BioData Min. 2015;8:1–7. doi: 10.1186/s13040-014-0034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21.. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, von Mering C, et al. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol. 2017;34:2115–22. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔ C(T) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 40.Ling N, Raza W, Ma J, Huang Q, Shen Q. Identification and role of organic acids in watermelon root exudates for recruiting Paenibacillus polymyxa SQR-21 in the rhizosphere. Eur J Soil Biol. 2011;47:374–9. [Google Scholar]
- 41.Gordillo F, Chávez FP, Jerez CA. Motility and chemotaxis of Pseudomonas sp. B4 towards polychlorobiphenyls and chlorobenzoates. FEMS Microbiol Ecol. 2007;60:322–8. doi: 10.1111/j.1574-6941.2007.00293.x. [DOI] [PubMed] [Google Scholar]
- 42.Dragoš A, Kiesewalter H, Martin M, Hsu CY, Hartmann R, Wechsler T, et al. Division of labor during biofilm matrix production. Curr Biol. 2018;28:1903–.e5. doi: 10.1016/j.cub.2018.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahmad F, Ahmad I, Khan MS. Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiol Res. 2008;163:173–81. doi: 10.1016/j.micres.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 44.Lynne AM, Haarmann D, Louden BC. Use of blue agar CAS assay for siderophore detection. J Microbiol Biol Educ. 2011;12:51–53. doi: 10.1128/jmbe.v12i1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nautiyal CS. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol Lett. 1999;170:265–70. doi: 10.1111/j.1574-6968.1999.tb13383.x. [DOI] [PubMed] [Google Scholar]
- 46.Ansari FA, Ahmad I. Fluorescent Pseudomonas -FAP2 and Bacillus licheniformis interact positively in biofilm mode enhancing plant growth and photosynthetic attributes. Sci Rep. 2019;9:1–12. doi: 10.1038/s41598-019-40864-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santhanam R, Menezes RC, Grabe V, Li D, Baldwin IT, Groten K. A suite of complementary biocontrol traits allows a native consortium of root‐associated bacteria to protect their host plant from a fungal sudden‐wilt disease. Mol Ecol. 2019;28:1154–69. doi: 10.1111/mec.15012. [DOI] [PubMed] [Google Scholar]
- 48.Santhanam R, Luu VT, Weinhold A, Goldberg J, Oh Y, Baldwin IT. Native root-associated bacteria rescue a plant from a sudden-wilt disease that emerged during continuous cropping. Proc Natl Acad Sci USA. 2015;112:E5013–E5120. doi: 10.1073/pnas.1505765112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berendsen RL, Vismans G, Yu K, Song Y, de Jonge R, Burgman WP, et al. Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 2018;12:1496–507. doi: 10.1038/s41396-018-0093-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ren D, Madsen JS, Sørensen SJ, Burmølle M. High prevalence of biofilm synergy among bacterial soil isolates in cocultures indicates bacterial interspecific cooperation. ISME J. 2015;9:81–89. doi: 10.1038/ismej.2014.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oliveira NM, Martinez-Garcia E, Xavier J, Durham WM, Kolter R, Kim W, et al. Biofilm formation as a response to ecological competition. PLoS Biol. 2015;13:1–23. doi: 10.1371/journal.pbio.1002191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gallegos-Monterrosa R, Mhatre E, Kovács ÁT. Specific Bacillus subtilis 168 variants form biofilms on nutrient-rich medium. Microbiology. 2016;162:1922–32. doi: 10.1099/mic.0.000371. [DOI] [PubMed] [Google Scholar]
- 53.Shao J, Xu Z, Zhang N, Shen Q, Zhang R. Contribution of indole-3-acetic acid in the plant growth promotion by the rhizospheric strain Bacillus amyloliquefaciens SQR9. Biol Fertil Soils. 2015;51:321–30. [Google Scholar]
- 54.Stopnisek N, Shade A. Persistent microbiome members in the common bean rhizosphere: an integrated analysis of space, time, and plant genotype. ISME J. 2021;15:2708–22. doi: 10.1038/s41396-021-00955-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sivasakthi S, Usharani G, Saranraj P. Biocontrol potentiality of plant growth promoting bacteria (PGPR)—Pseudomonas fluorescens and Bacillus subtilis: a review. Afr J Agric Res. 2014;9:1265–77. [Google Scholar]
- 56.Dardanelli MS, Manyani H, González-Barroso S, Rodríguez-Carvajal MA, Gil-Serrano AM, Espuny MR, et al. Effect of the presence of the plant growth promoting rhizobacterium (PGPR) Chryseobacterium balustinum Aur9 and salt stress in the pattern of flavonoids exuded by soybean roots. Plant Soil. 2010;328:483–93. [Google Scholar]
- 57.Gómez Expósito R, Postma J, Raaijmakers JM, de Bruijn I. Diversity and activity of Lysobacter species from disease suppressive soils. Front Microbiol. 2015;6:1243. doi: 10.3389/fmicb.2015.01243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Han Q, Ma Q, Chen Y, Tian B, Xu L, Bai Y, et al. Variation in rhizosphere microbial communities and its association with the symbiotic efficiency of rhizobia in soybean. ISME J. 2020;14:1915–28. doi: 10.1038/s41396-020-0648-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peterson SB, Dunn AK, Klimowicz AK, Handelsman J. Peptidoglycan from Bacillus cereus mediates commensalism with rhizosphere bacteria from the Cytophaga-Flavobacterium group. Appl Environ Microbiol. 2006;72:5421–7. doi: 10.1128/AEM.02928-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tao C, Li R, Xiong W, Shen Z, Liu S, Wang B, et al. Bio-organic fertilizers stimulate indigenous soil Pseudomonas populations to enhance plant disease suppression. Microbiome. 2020;8:137. doi: 10.1186/s40168-020-00892-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar A, Singh J. Biofilms forming microbes: diversity and potential application in plant-microbe interaction and plant growth. Springer: Cham; 2020. 173−97.
- 62.Tabassum B, Khan A, Tariq M, Ramzan M, Iqbal Khan MS, Shahid N, et al. Bottlenecks in commercialisation and future prospects of PGPR. Appl Soil Ecol. 2017;121:102–17. [Google Scholar]
- 63.Madsen JS, Røder HL, Russel J, Sørensen H, Burmølle M, Sørensen SJ. Coexistence facilitates interspecific biofilm formation in complex microbial communities. Environ Microbiol. 2016;18:2565–74. doi: 10.1111/1462-2920.13335. [DOI] [PubMed] [Google Scholar]
- 64.Burmølle M, Webb JS, Rao D, Hansen LH, Sørensen SJ, Kjelleberg S. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl Environ Microbiol. 2006;72:3916–23. doi: 10.1128/AEM.03022-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee KWK, Periasamy S, Mukherjee M, Xie C, Kjelleberg S, Rice SA. Biofilm development and enhanced stress resistance of a model, mixed-species community biofilm. ISME J. 2014;8:894–907. doi: 10.1038/ismej.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yannarell SM, Grandchamp GM, Chen SY, Daniels KE, Shank EA. A dual-species biofilm with emergent mechanical and protective properties. J Bacteriol. 2019;201:e00670–18.. doi: 10.1128/JB.00670-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Preussger D, Giri S, Muhsal LK, Oña L, Kost C. Reciprocal fitness feedbacks promote the evolution of mutualistic cooperation. Curr Biol. 2020;30:1–11. doi: 10.1016/j.cub.2020.06.100. [DOI] [PubMed] [Google Scholar]
- 68.Nadell CD, Drescher K, Foster KR. Spatial structure, cooperation, and competition in biofilms. Nat Rev Microbiol. 2016;14:589–600. doi: 10.1038/nrmicro.2016.84. [DOI] [PubMed] [Google Scholar]
- 69.Estrela S, Sanchez-Gorostiaga A, Vila JCC, Sanchez A. Nutrient dominance governs the assembly of microbial communities in mixed nutrient environments. eLife. 2021;10:e65948. doi: 10.7554/eLife.65948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Molina-Santiago C, Pearson JR, Navarro Y, Berlanga-Clavero MV, Caraballo-Rodriguez AM, Petras D, et al. The extracellular matrix protects Bacillus subtilis colonies from Pseudomonas invasion and modulates plant co-colonization. Nat Commun. 2019;10:1919. doi: 10.1038/s41467-019-09944-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yan Q, Lopes LD, Shaffer BT, Kidarsa TA, Vining O, Philmus B, et al. Secondary metabolism and interspecific competition affect accumulation of spontaneous mutants in the GacS-GacA regulatory system in Pseudomonas protegens. mBio. 2018;9:e01845–17. doi: 10.1128/mBio.01845-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mee MT, Collins JJ, Church GM, Wang HH. Syntrophic exchange in synthetic microbial communities. Proc Natl Acad Sci USA. 2014;111:E2149–E2156. doi: 10.1073/pnas.1405641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.D’Souza G, Shitut S, Preussger D, Yousif G, Waschina S, Kost C. Ecology and evolution of metabolic cross-feeding interactions in bacteria. Nat Prod Rep. 2018;35:455–88. doi: 10.1039/c8np00009c. [DOI] [PubMed] [Google Scholar]
- 74.Goldford JE, Lu N, Bajić D, Estrela S, Tikhonov M, Sanchez-Gorostiaga A, et al. Emergent simplicity in microbial community assembly. Science. 2018;361:469–74. doi: 10.1126/science.aat1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lawrence D, Fiegna F, Behrends V, Bundy JG, Phillimore AB, Bell T, et al. Species interactions alter evolutionary responses to a novel environment. PLoS Biol. 2012;10:e1001330. doi: 10.1371/journal.pbio.1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Evans R, Beckerman AP, Wright RCT, McQueen-Mason S, Bruce NC, Brockhurst MA. Eco-evolutionary dynamics set the tempo and trajectory of metabolic evolution in multispecies communities. Curr Biol. 2020;30:1–5. doi: 10.1016/j.cub.2020.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gamez RM, Ramirez S, Montes M, Cardinale M. Complementary dynamics of banana root colonization by the plant growth-promoting rhizobacteria Bacillus amyloliquefaciens Bs006 and Pseudomonas palleroniana Ps006 at spatial and temporal scales. Micro Ecol. 2020;80:656–68. doi: 10.1007/s00248-020-01571-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Feng H, Zhang N, Fu R, Liu Y, Krell T, Du W, et al. Recognition of dominant attractants by key chemoreceptors mediates recruitment of plant growth-promoting rhizobacteria. Environ Microbiol. 2019;21:402–15. doi: 10.1111/1462-2920.14472. [DOI] [PubMed] [Google Scholar]
- 79.Feng H, Zhang N, Du W, Zhang H, Liu Y, Fu R, et al. Identification of chemotaxis compounds in root exudates and their sensing chemoreceptors in plant-growth-promoting rhizobacteria Bacillus amyloliquefaciens SQR9. Mol Plant Microbe interact. 2018;31:995–1005. doi: 10.1094/MPMI-01-18-0003-R. [DOI] [PubMed] [Google Scholar]
- 80.Xu Z, Xie J, Zhang H, Wang D, Shen Q, Zhang R. Enhanced control of plant wilt disease by a xylose-inducible degQ gene engineered into Bacillus velezensis strain SQR9XYQ. Phytopathology. 2019;109:36–43. doi: 10.1094/PHYTO-02-18-0048-R. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.