Few trials of pharmacological interventions in critical care show benefit despite decades of laboratory-based and translational research dedicated to identifying mechanisms responsible for critical illness (1). This reality has prompted us to consider the limitations of our current definitions of critical illness syndromes. Syndromes such as acute respiratory distress syndrome (ARDS) and sepsis are defined based on clinical criteria that yield little insight into their underlying biology. This syndromic approach to disease classification has led to trial designs that target heterogeneous populations, which may explain why treatments have been unsuccessful.
The identification of biologically distinct subphenotypes in ARDS (2, 3), sepsis, and acute kidney injury, which have divergent outcomes and may respond differently to randomly allocated therapies (2, 4–7), may point toward an alternative approach to testing pharmacological therapies. These subphenotypes tend to be defined by biomarkers that are agnostic to syndromic diagnosis within critical illness, suggesting mechanistic pathways may be conserved across these syndromes. Interestingly, similarities between subphenotypes identified using varying methods for phenotype allocation and in different syndromes have emerged (7, 8). Indeed, ARDS subphenotypes with distinct clinical trajectories have also been identified in patients with coronavirus disease (COVID-19) (9). These findings led to the hypothesis that there may be host-response subphenotypes of critical illness, which might also be termed “treatable traits,” that are independent of syndromic definitions (Figure 1). If this hypothesis is correct, our approach to clinical trials premised on those syndromic definitions should be reevaluated.
Figure 1.
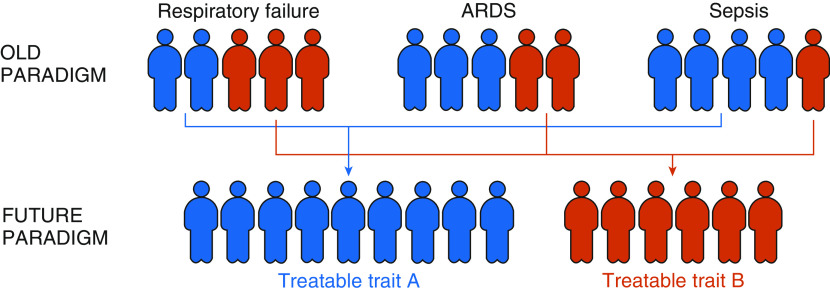
The impending transformation in critical care syndrome classification. In the old paradigm, we classify syndromes such as respiratory failure, acute respiratory distress syndrome, and sepsis based on clinical criteria that poorly inform treatments. In the future paradigm, we identify more biologically meaningful subgroup characteristics that inform disease-modifying treatment options (treatable traits). As pictured above, the data presented by Heijnen and colleagues (10) in this issue of the Journal brings us closer to identifying treatable traits. ARDS = acute respiratory distress syndrome.
In this issue of the Journal, Heijnen and colleagues (pp. 1503–1511) provide the most compelling evidence to date supporting the concept that treatable traits in critical illness are independent of syndromic diagnosis (10). In a retrospective analysis of a large cohort of mechanically ventilated patients (n = 2,499), they used previously described parsimonious models for subphenotype allocation for their reactive and uninflamed ARDS subphenotypes (cluster-derived, using IL-6, IFN-γ, ANG1/2, and PAI-1) (3) and for the hyperinflammatory and hypoinflammatory subphenotypes (latent class analysis [LCA]-derived, using IL-8, protein C, and bicarbonate) (11) to demonstrate that these classifier methods also provide prognostic enrichment in a non-ARDS population (n = 1,825). Regardless of ARDS diagnosis, the reactive and hyperinflammatory subphenotypes retained their associations with increased ICU mortality, increased 30-day mortality, and lower probability of successful extubation. In keeping with previous ARDS studies, this study demonstrates that in univariable analysis, the reactive and hyperinflammatory subphenotypes were the strongest predictors of ICU mortality in patients without ARDS(hazard ratio, 2.43; 95% confidence interval [CI], 1.90–3.11; P < 0.001; and hazard ratio, 2.54; 95% CI, 2.00–3.24; P < 0.001, respectively). These subphenotypes retained their association with mortality even when adjusted for APACHEIV score, demonstrating prognostic value independent of severity of illness.
A subset of patients (n = 719) had leukocyte gene expression profiles examined by microarray. Principal component analysis revealed that members of the same cluster-derived and LCA-derived subphenotypes grouped together regardless of the presence of ARDS, suggesting that the transcriptome is conserved within these subgroups across syndromic definitions and providing further evidence that the biology of these subphenotypes is fundamentally distinct.
These data have important implications. First, they suggest that the subphenotypes identified in ARDS to date (e.g., reactive vs. uninflamed and hyper- vs. hypoinflammatory) may be translatable to patients without ARDS. Furthermore, independently described subphenotypes have significant overlap, and analogous cluster- and LCA-derived subphenotypes (hyperinflammatory and reactive; hypoinflammatory and uninflamed) demonstrated similar blood leukocyte gene expression profiles. This latter finding suggests convergence at a common biological signal, further increasing confidence that these subphenotypes represent a generalizable and reproducible finding.
Despite the strength of this data, some limitations should be considered. The study population consisted solely of mechanically ventilated patients, and there was a bias in biomarker data availability toward those patients with definite or probable infection, who have a high risk of developing ARDS. Subphenotyping methods developed in ARDS may therefore be more translatable to this population rather than a broader population. Furthermore, classification was accomplished using parsimonious models rather than the gold-standard cluster analysis or LCA. Despite the strong concordance of these models with gold standard (AUC = 0.94; 95% CI, 0.92–0.95 for LCA-derived subphenotypes; AUC = 0.98; 95% CI, 0.97–0.99 for cluster-derived subphenotypes) (3, 11), without de novo cluster analysis or LCA, these results cannot provide statistical evidence that a two subphenotype approach is the most robust approach to subdivision in patients without ARDS.
These results imply that treatments that are found to work in ARDS-specific subphenotypes might be translatable to a broader population of patients. In a post hoc analysis of the HARP-2 clinical trial of simvastatin in ARDS, which showed no overall benefit, the LCA-derived hyperinflammatory subphenotype had improved 28-day survival with simvastatin versus placebo (5). Although prospective validation is needed, the results presented here suggest that “hyperinflammatory” patients without ARDS might also benefit from simvastatin. Unfortunately, the ability to prospectively phenotype critically ill patients on ICU admission remains a limitation to the translation of these subphenotypes into clinical trials and, ultimately, practice. Allocation of ARDS subphenotypes requires the measurement of biomarkers that are not available in routine care, limiting their clinical utility at present. A prospective multicenter study (NCT04009330) that employs a point-of-care assay for allocation of the subphenotypes is in progress and aims to address this challenge. Alternatively, a clinical variable–only classifier has recently been developed for the LCA-based subphenotypes, though prospective validation of this approach is also needed (12). Of note, Heijnen and colleagues demonstrate incomplete overlap between cluster-derived and LCA-derived subphenotypes. Notably, only 56% of patients assigned to the reactive subphenotype were also assigned to the hyperinflammatory subphenotype, whereas 44% were assigned to the hypoinflammatory group (10). Further studies of the reasons for the divergence in these two approaches are needed.
Once we can consistently and prospectively identify subphenotypes, we need a better understanding of the underlying biology to identify which therapies might be appropriate to test in each subphenotype. Leukocyte expression profiles presented here and previously (13) point toward neutrophil activation, oxidative phosphorylation, and mitochondrial dysfunction in the reactive subphenotype, with upregulation of MAPK pathways in the uninflamed group. As more advanced techniques that allow detection of novel transcripts and modifications (RNA sequencing) and transcriptomic mapping of individual cell types (single-cell RNA sequencing) become increasingly available, we hope to further advance our understanding (14, 15). Other key unanswered mechanistic questions include how the biology of subphenotypes differs beyond the circulating plasma (e.g., for lung or kidney injury), whether metagenomic sequencing can identify differences in either pathogens or the microbiome between subphenotypes, and which experimental models are best suited to test novel interventions for each subphenotype.
The novel findings presented in this issue of the Journal (10) represent important evidence in support of a new ontology of critical illness—identifying treatable traits in critical care that are independent of clinical diagnosis (Figure 1). Whether such a sea change will ultimately be usefully translated to clinical trials and clinical care remains unknown, but other fields such as oncology have embraced this possibility. Heijnen and colleagues have helped bring us one step closer to this new frontier.
Footnotes
Supported by NIH–NHLBI grant R35 HL140026 (C.S.C.).
Originally Published in Press as DOI: 10.1164/rccm.202101-0218ED on February 10, 2021
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Santacruz CA, Pereira AJ, Celis E, Vincent JL. Which multicenter randomized controlled trials in critical care medicine have shown reduced mortality? A systematic review. Crit Care Med. 2019;47:1680–1691. doi: 10.1097/CCM.0000000000004000. [DOI] [PubMed] [Google Scholar]
- 2. Calfee CS, Delucchi K, Parsons PE, Thompson BT, Ware LB, Matthay MA, NHLBI ARDS Network. Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir Med. 2014;2:611–620. doi: 10.1016/S2213-2600(14)70097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bos LD, Schouten LR, van Vught LA, Wiewel MA, Ong DSY, Cremer O, et al. MARS consortium. Identification and validation of distinct biological phenotypes in patients with acute respiratory distress syndrome by cluster analysis. Thorax. 2017;72:876–883. doi: 10.1136/thoraxjnl-2016-209719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Famous KR, Delucchi K, Ware LB, Kangelaris KN, Liu KD, Thompson BT, et al. Acute respiratory distress syndrome subphenotypes respond differently to randomized fluid management strategy. Am J Respir Crit Care Med. 2017;195:331–338. doi: 10.1164/rccm.201603-0645OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Calfee CS, Delucchi KL, Sinha P, Matthay MA, Hackett J, Shankar-Hari M, et al. Irish Critical Care Trials Group. Acute respiratory distress syndrome subphenotypes and differential response to simvastatin: secondary analysis of a randomised controlled trial. Lancet Respir Med. 2018;6:691–698. doi: 10.1016/S2213-2600(18)30177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Antcliffe DB, Burnham KL, Al-Beidh F, Santhakumaran S, Brett SJ, Hinds CJ, et al. Transcriptomic signatures in sepsis and a differential response to steroids: from the VANISH randomized trial. Am J Respir Crit Care Med. 2019;199:980–986. doi: 10.1164/rccm.201807-1419OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reddy K, Sinha P, O’Kane CM, Gordon AC, Calfee CS, McAuley DF. Subphenotypes in critical care: translation into clinical practice. Lancet Respir Med. 2020;8:631–643. doi: 10.1016/S2213-2600(20)30124-7. [DOI] [PubMed] [Google Scholar]
- 8. Kitsios GD, Yang L, Manatakis DV, Nouraie M, Evankovich J, Bain W, et al . Host-response subphenotypes offer prognostic enrichment in patients with or at risk for acute respiratory distress syndrome. Crit Care Med. 2019;47:1724–1734. doi: 10.1097/CCM.0000000000004018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sinha P, Calfee CS, Cherian S, Brealey D, Cutler S, King C, et al. Prevalence of phenotypes of acute respiratory distress syndrome in critically ill patients with COVID-19: a prospective observational study. Lancet Respir Med. 2020;8:1209–1218. doi: 10.1016/S2213-2600(20)30366-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heijnen NFL, Hagens LA, Smit MR, Cremer OL, Ong DSY, van der Poll T, et al. MARS Consortium. Biological subphenotypes of acute respiratory distress syndrome show prognostic enrichment in mechanically ventilated patients without acute respiratory distress syndrome. Am J Respir Crit Care Med. 2021;203:1503–1511. doi: 10.1164/rccm.202006-2522OC. [DOI] [PubMed] [Google Scholar]
- 11. Sinha P, Delucchi KL, McAuley DF, O’Kane CM, Matthay MA, Calfee CS. Development and validation of parsimonious algorithms to classify acute respiratory distress syndrome phenotypes: a secondary analysis of randomised controlled trials. Lancet Respir Med. 2020;8:247–257. doi: 10.1016/S2213-2600(19)30369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sinha P, Churpek MM, Calfee CS. Machine learning classifier models can identify acute respiratory distress syndrome phenotypes using readily available clinical data. Am J Respir Crit Care Med. 2020;202:996–1004. doi: 10.1164/rccm.202002-0347OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bos LDJ, Scicluna BP, Ong DSY, Cremer O, van der Poll T, Schultz MJ. Understanding heterogeneity in biologic phenotypes of acute respiratory distress syndrome by leukocyte expression profiles. Am J Respir Crit Care Med. 2019;200:42–50. doi: 10.1164/rccm.201809-1808OC. [DOI] [PubMed] [Google Scholar]
- 14. Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stuart T, Satija R. Integrative single-cell analysis. Nat Rev Genet. 2019;20:257–272. doi: 10.1038/s41576-019-0093-7. [DOI] [PubMed] [Google Scholar]