Abstract
The healthy vaginal microbiota is dominated by Lactobacillus spp., which provide an important critical line of defense against pathogens, as well as giving beneficial effects to the host. We characterized L. gasseri 1A‐TV, L. fermentum 18A‐TV, and L. crispatus 35A‐TV, from the vaginal microbiota of healthy premenopausal women, for their potential probiotic activities. The antimicrobial effects of the 3 strains and their combination against clinical urogenital bacteria were evaluated together with the activities of their metabolites produced by cell‐free supernatants (CFSs). Their beneficial properties in terms of ability to interfere with vaginal pathogens (co‐aggregation, adhesion to HeLa cells, biofilm formation) and antimicrobial activity mediated by CFSs were assessed against multidrug urogenital pathogens (S. agalactiae, E. coli, KPC‐producing K. pneumoniae, S. aureus, E. faecium VRE, E. faecalis, P. aeruginosa, P. mirabilis, P. vulgaris, C. albicans, C. glabrata). The Lactobacilli tested exhibited an extraordinary ability to interfere and co‐aggregate with urogenital pathogens, except for Candida spp., as well as to adhere to HeLa cells and to produce biofilm in the Lactobacillus combination. Lactobacillus CFSs and their combination revealed a strong bactericidal effect on the multidrug resistant indicator strains tested, except for E. faecium and E. faecalis. The antimicrobial activity was maintained after heat treatment but decreased after enzymatic treatment. All Lactobacilli showed lactic dehydrogenase activity and production of D‐ and L‐lactic acid isomers on Lactobacillus CFSs, while only 1A‐TV and 35A‐TV released hydrogen peroxide and carried helveticin J and acidocin A bacteriocins. These results suggest that they can be employed as a new vaginal probiotic formulation and bio‐therapeutic preparation against urogenital infections. Further, in vivo studies are needed to evaluate human health benefits in clinical situations.
Keywords: antimicrobial activity, Lactobacilli, MDR‐urogenital infection, supernatants, vaginal probiotics
The present study characterized a novel combination of Lactobacillus gasseri 1A‐TV, L. fermentum 18A‐TV, and L. crispatus 35A‐TV as live strains and as non‐live cell‐free supernatants showing antimicrobial activity against the most common multidrug‐resistant urogenital pathogens (Streptococcus agalactiae, Escherichia coli, KPC‐producing Klebsiella pneumoniae, Staphylococcus aureus, Enterococcus faecium VRE, Enterococcus faecalis, Pseudomonas aeruginosa, P. mirabilis, P. vulgaris, Candida albicans, C. glabrata). Our findings are promising for their use as a new valid vaginal probiotic and bio‐therapeutic formulation against multidrug‐resistant pathogens.
1. INTRODUCTION
Lactobacilli are important members of the human gastrointestinal, oral, and vaginal microbiota and are gaining great interest for their health‐promoting effects in the host both on direct interactions between cells and indirectly through their released metabolites, thus making them suitable to be used as probiotic strains (Reid et al., 2011). Over the last few years, the search for probiotic strains possessing innovative functional characteristics and formulations has been evolving and is an attractive goal in therapeutic strategies to restore the natural microbiota. Antibiotic treatment is the main approach used to fight bacterial infections (Aslam et al., 2018), but excessive and inappropriate use in both hospital and community settings has been one of the main factors of the onset of antibiotic resistance, and urogenital tract infections (UGTIs) are the most common infections in which many multidrug‐resistant (MDR) pathogenic strains are recorded due to the abuse of antibiotic therapy (Matulay et al., 2016).
Lactobacilli dominate the healthy vaginal microbiota and are considered gatekeepers of this ecosystem, maintaining a healthy state and impeding the growth of pathogens (Bautista et al., 2016; Martin, 2012; Ravel et al., 2011). Recent studies have focused on the vaginal microbiome in healthy reproductive‐aged women by 16S rRNA gene sequencing showed at least 5 community state types (CSTs), in which four were dominated by L. crispatus (CST‐I), L. gasseri (CST‐II), L. iners (CST‐III), L. jensenii (CST‐V), and only one by the microbial community (CST‐IV) composed of polymicrobial species confirming an important protection factor of the Lactobacillus population against potential pathogens associated with urogenital tract infections (UTIs) (Borges et al., 2014; Eryilmaz et al., 2018; Razzak et al., 2011; Wijgert et al., 2014). However, besides the most abundant vaginal Lactobacilli, other species have been encountered in the healthy vaginal microbiota such as L. rhamnosus, L. fermentum, L. plantarum, L. brevis, L. casei, L. delbrueckii, L. vaginalis, and L. salivarius (Dimitonova et al., 2008; Kiss et al., 2007; Pino et al., 2019; Smith & Ravel, 2017).
Perturbations of this highly regulated ecosystem occur during urogenital tract infections (UGTIs), as well as urinary tract infections (UTIs), bacterial vaginosis (BV), and during antimicrobial therapy, resulting in an even greater aberration of the microbiota and, eventually, in the extension of an infectious state (Donders et al., 2017; Eryilmaz et al., 2018; Matulay et al., 2016). Restoration of vaginal homeostasis, driven by Lactobacilli, may be accomplished through numerous mechanisms: (i) “competitive exclusion,” the first critical line of defense against local pathogens, which is the ability of bacteria to adhere to vaginal epithelial cells competing for nutrients and adhesion receptors (Liu et al., 2008), (ii) “co‐aggregation,” the assembly of microbial communities into distinct, interlinked structures (Pino et al., 2019); in addition, (iii) an intense production of antimicrobial compounds such as lactic acid, hydrogen peroxide (H2O2), bacteriocin‐like substances, and biosurfactants may inhibit pathogen growth (Petrova et al., 2015).
In vitro and in vivo studies have indicated the use of probiotics as an alternative approach for restoring healthy vaginal microbiota by interfering with potential pathogens. Although the use of live microorganisms is currently widely employed, safety issues remain a matter of debate, mainly for vulnerable subjects (Borges et al., 2014; Ravel et al., 2011; Reid et al., 2011). To overcome these issues, in the last decade, the use of non‐live microorganisms such as heat‐killed probiotics, microbial extracts, and cell‐free supernatants has been growing in interest for their applications in therapeutic strategies also considering that they can confer relevant beneficial effects (Piqué et al., 2019).
In this study, we characterized three vaginal Lactobacilli, L. gasseri 1A‐TV, L. fermentum 18A‐TV, and L. crispatus 35A‐TV from healthy vaginal microbiota for their probiotic properties mainly focusing on their antimicrobial activity against the most common MDR UGTI pathogens (Ahmed et al., 2019; Al‐Zahrani et al., 2019), considering also both adhesive properties and inhibitory substances released in their cell‐free supernatants (CFS).
2. METHODS
2.1. Sample collection and microbial growth conditions
L. gasseri 1A‐TV, L. fermentum 18A‐TV, and L. crispatus 35A‐TV were isolated from vaginal swabs taken from healthy premenopausal women without symptoms of vaginal or urinary tract infections during normal gynecological examinations for routine analyses at the Obstetrics and Gynecology Unit of the University Hospital of Catania, Italy. The authors received the strains for the subsequent analysis and their characterization. All Lactobacillus strains were grown on Man Rogosa and Sharpe (MRS) agar (Oxoid), incubated for 48 h at 37°C under anaerobic conditions, using the GasPakEZ Gas Generating Pouch Systems (BD). All Lactobacilli were taxonomically identified at the species level by amplification and sequencing of the tuf and 16S rRNA genes for accurate identification. Genomic DNA was extracted from overnight cultures of isolates in 5 ml of MRS and the tuf and16S rRNA genes were amplified. All PCR products obtained were purified using the QIAquick PCR gel extraction kit (Qiagen) and sequenced (Hütt et al., 2016; Marchisio et al., 2015; Ventura et al., 2003). Sequence analyses were performed using Gapped BLAST (Altschul et al., 1997).
The indicator strains were selected from our microbial bank at the MMARLab as having MDR profiles. The strains Streptococcus agalactiae GB022, Enterococcus faecalis EFS1, Enterococcus faecium 75 VRE (vanA‐positive), Staphylococcus aureus (MSSA) SA3, Pseudomonas aeruginosa IF1, Proteus vulgaris IF3, Proteus mirabilis IF2, Escherichia coli GM1, Klebsiella pneumoniae 340 KPC (KPC‐3 positive), Candida albicans CA312, and Candida glabrata CG2824 were used as target microorganisms for the determination of antagonistic activity (Table 1). All clinical isolates had been tested for antimicrobial susceptibility profiles according to the interpretative standard of the European Committee on Antimicrobial Susceptibility Testing 2019 (EUCAST) (2019) and INTEGRAL SYSTEM YEASTS Plus (Liofilchem®) for antimycotic resistance profile.
TABLE 1.
Clinical information and antimicrobial profiles of indicators strains used in this study
| ID | Species | Infection disease | Source | Phenotypic resistance profile |
|---|---|---|---|---|
| GB022 | S. agalactiae | asymptomatic | Vaginal swab | AK‐TOB‐LEV‐CIP‐LNZ‐TE‐TGC‐E‐DA‐RD |
| SA3 | S. aureus | vaginitis | Vaginal swab | TOB‐AMC‐TZP‐LEV‐CIP‐E |
| EFS1 | E. faecalis | vaginitis | Vaginal swab | FOS‐IPM‐TOB‐AMC‐LNZ‐F‐SXT‐CIP |
| 75VRE | E. faecium | vaginitis | Vaginal swab | AMC‐TOB‐IPM‐TZP‐LEVCIP‐LNZ‐E‐QDA‐RD‐TEC‐VA |
| GM1 | E. coli | Symptomatic cystistic | urine | AMC‐RD‐F |
| 340KPC | K. pneumoniae | Symptomatic cystistic | urine | ETP‐MRP‐MEM‐AMC‐TZP‐C/T‐CAZ‐TGC‐RD‐F |
| IF1 | P. aeruginosa | Symptomatic cystistic | urine | IPM‐MRP‐MEM‐TOB‐AMC‐TZP‐LEV‐CIP‐RD‐ATM‐SXT |
| IF2 | P. mirabilis | Symptomatic cystistic | urine | FOS‐MRP‐MEM‐TOB‐AMC‐TZP‐C/T‐CAZ‐FEP‐CTZ‐LEV‐CIP‐TGC‐RD‐CS‐ATM‐SXT |
| IF3 | P. vulgaris | Symptomatic cystistic | urine | ETP‐MRP‐MEM‐AMC‐C/T‐TGC‐RD‐CS |
| CA312 | C. albicans | Vulvo‐vaginal candidosis | Vaginal swab | ECN‐KCA‐CLO‐MIC‐AMB*‐ITR*‐VOR*‐FLU* |
| CG2824 | C. glabrata | Vulvo‐vaginal candidosis | Vaginal swab | CLO‐MIC‐ITR‐VOR‐FLU‐NY*‐AMB*‐ECN*‐KCA* |
Abbreviations: AK, Amikacin; AMB, Amphotericin; AMC, Amoxicillin–clavulanic acid; ATM, Aztreonam; C/T, ceftolozane/tazobactam; CAZ, Ceftazidime; CIP, Ciprofloxacin; CLO, Clotrimazole; CN, Gentamicin; CS, Colistin; CTZ, Cefotaxime; CZA, Ceftazidime/avibactam; DA, Clindamycin; E, Erythromycin; ECN, Econazole; ETP, Ertapenem; F, Nitrofurantoin; FCY, Flucytosine; FEP, Cefepime; FLU, Fluconazole; FOS, Fosfomycin; IPM, Imipenem; ITR, Itraconazole; KCA, Ketoconazole; KPC, Klebsiella pneumoniae carbapenemase; LEV, Levofloxacin; LNZ, Linezolid; MIC, Miconazole; MRP, Meropenem; NY, Nystatin; QDA, Quinupristin–dalfopristin; RD, Rifampicin; SXT, Trimethoprim/Sulfamethoxazole; TE, Tetracycline; TEC, Teicoplanin; TGC, Tigecycline; TOB, Tobramycin; TZP, Piperacillin–tazobactam; VA, Vancomycin; VRE, vancomycin‐resistant enterococci; VOR, Voriconazole.
intermediate resistance.
2.2. In vitro safety assessment of Lactobacillus strains
i. Antibiotic susceptibility testing and detection of hemolytic activity.
The antibiotic susceptibility profiles of the three Lactobacilli were determined by the Kirby‐Bauer diffusion and E‐test methods on MRS agar at 37°C for 48 h under anaerobic conditions (Charteris et al., 2007). The following antibiotics were tested: penicillin, ampicillin, amoxicillin–clavulanic acid, vancomycin, gentamicin, streptomycin, tetracycline, chloramphenicol, erythromycin, clindamycin, trimethoprim–sulfamethoxazole, rifampicin, ciprofloxacin, levofloxacin, and metronidazole. The antimicrobial susceptibility profiles were analyzed according to the interpretative standard of the European Union Commission recommendations for probiotic safety (Authority EFS, 2012).
ii. The hemolytic activity of Lactobacilli was visually verified on Columbia agar base supplemented with 5% sheep and horse blood (Oxoid) after 24 h and 48 h of incubation under anaerobic conditions at 37°C(Maragkoudakis et al., 2006). Streptococcus pyogenes, strain ATCC 19615, was used as a positive control. Both experiments mentioned above were performed in triplicate.
2.3. Determination of antagonistic activity
The MDR indicator strains, S. agalactiae, E. faecalis VRE, E. faecium, S. aureus, P. aeruginosa, P. mirabilis, P. vulgaris, E. coli, KPC‐producing K. pneumoniae, C. albicans, and C. glabrata, were used for detecting the antimicrobial activity of Lactobacilli. The inhibitory activity of vaginal strains was determined by the deferred antagonism test and quantified by the agar spot test with some modifications (Santagati et al., 2012; Siroli et al., 2017). In addition, for the evaluation of Lactobacillus combination, L. gasseri 1A‐TV, L. fermentum 18A‐TV, and L. crispatus 35A‐TV were grown in MRS broth for 48 h at 37°C under anaerobic conditions, using the GasPakEZ Gas Generating Pouch System (BD, New Jersey, USA) and approximately 2 × 108 CFU/ml of each Lactobacillus culture in a 1:1:1 ratio were used. Briefly, for the deferred antagonism assay, the test strain was inoculated diametrically across MRS agar with the addition of 0.1% CaCO3 and incubated for 48 h at 37°C under anaerobic conditions, as reported before. Lactobacillus growth was stopped, and the surface of the plate was sterilized by exposure to chloroform vapors for 30 min. The broth cultures of the indicator strains, grown for 18 h at 37°C, were streaked across the Lactobacillus growth line, and the plates were incubated for 18 h at 37°C to examine the interference zones with the indicator. Lactobacillus isolates that inhibited the growth of an indicator strain were considered inhibitory for that species (Maragkoudakis et al., 2006). For the agar spot test, the Lactobacillus cultures were spotted (5 µl) on the surface of MRS agar (1.2%) (20 ml) and incubated anaerobically for 48 h at 37°C. Then, 100 µl of an overnight culture of indicator strains (approximately 107 CFU/ml) was mixed with 10 ml of BHI soft agar (0.7%) and poured over the plate in which Lactobacilli were grown. After incubation for 24 h at 37°C, the inhibition zones around Lactobacillus spots were diametrically measured and expressed as diameter >10 (+ + + +); Diameter between 6 and 10 mm (+ + +); Diameter between 3 and 6 mm (+ +); Diameter between 1 and 3 mm (+); no inhibition (−) (Siroli et al., 2017).
2.4. Auto‐aggregation and co‐aggregation assays
Auto‐aggregation assays were performed according to Kos et al. (Kos et al., 2003). The auto‐aggregation percentage is expressed as A% = 1−(At5/At0) × 100, where At5 represents the absorbance measured by a microplate reader (BioTek Synergy™ H1) at 600 nm after centrifugation at 650 × g for 2 min at time t = 5 h and At0 the absorbance at t = 0. The percentage of co‐aggregation (CoA%) was calculated according to the equation of Malik et al. (Malik et al., 2003): CoA% = ODTOT−ODS/ODTOT × 100, where the ODTOT value represents total absorbance, taken immediately after the relevant strains were paired; and ODS refers to the absorbance of the supernatant after 5 h from when the mixture was centrifuged. The statistical analysis was determined by ANOVA with Fisher's least significant difference (LSD) test, p < 0.05 (De Gregorio et al., 2014). Both tests were repeated in triplicate.
2.5. In vitro adhesion test
HeLa cells (ATCC® CCL‐2.2 TM) were grown in RPMI 1640 (Sigma‐Aldrich Inc, St. Louis, MO, USA) at 37°C with 5% CO2 supplemented with 10% (v/v) fetal bovine serum (FBS, Thermo Fisher Scientific), 1% (v/v) L‐glutamine, penicillin G (100 IU mL−1), and streptomycin (100 mg/L) (Sigma‐Aldrich). The Lactobacillus adhesion to the HeLa cell layer was performed on microscope cover glasses and expressed as percentage adherence. Briefly, Lactobacillus cultures grown anaerobically for 48 h at 37°C in MRS broth (Oxoid) were harvested by centrifugation (5000 × g for 15 min, 4°C), and the cells were washed twice with a sterile solution of 0.85% NaCl (w/v) (Sigma‐Aldrich) diluted in RPMI 1640 medium at 5 × 108 CFU/ ml and incubated with a monolayer of HeLa for 1 h at 37°C (Martín et al., 2020; Mastromarino et al., 2002). After washes, the cells were fixed with 3 ml of methanol and stained with 3 ml of Giemsa stain solution (1:20; Carlo Erba, Milan, Italy) for 30 min at room temperature. Wells were washed and dried at 30°C for 1 h. Adherent bacteria were examined microscopically by light‐microscopy DM5500 (Leica, Wetzlar, Germany) in 20 random microscopic fields to obtain Lactobacillus counts and averages. The adhesion indexes (ADI; the number of bacteria/100 HeLa cells) were expressed as strong adhesion: ADI >2500; good adhesion: good adhesion: ADI between 2500 and 500, weak adhesion between 500 and 100, no adhesion, ADI <100. Bacterial adhesion to the HeLa cell layer was also evaluated by viable counts. After incubation, supernatants were discarded and non‐adherent bacteria were removed by washing each well twice with PBS and after the detachment by 1 ml of PBS with 0.1% Triton X‐100 (Sigma‐Aldrich, USA). The viable counts of adherent lactobacilli were evaluated by CFU/ml on MRS agar plates after incubation anaerobically for 48 h at 37°C (Santagati et al., 2012).
2.6. Biofilm formation assay
Biofilm production was tested in MRS broth. Lactobacillus biofilm development was evaluated as described by Ibarreche et al. (Perez Ibarreche et al., 2014) with modifications. Briefly, 200 μl of the medium was added to each well of sterile 96‐well plates (Corning® Incorporated Life Sciences, NY, USA) and was inoculated with LAB cultures at 3 × 108 CFU/ml. The plates were incubated under anaerobiosis at 37°C for 72 h. To quantify the biofilm formation, the wells were washed 3 times with PBS, fixed for 1 h at 37°C, and then stained for 30 min with 200 μl of 2% (v/v) crystal violet. The excess dye was rinsed with sterile distilled water, and the plates were allowed to dry at room temperature. The dye that had adhered to the cells was resolubilized with 200 μl of 95% (v/v) ethanol, and the absorbance of each well was measured at 570 nm using a microplate reader (BioTek Synergy™ H1). We used L. rhamnosus GGATCC 53103 as a positive control strain as it was a good biofilm producer (Lebeer et al., 2007), and MRS medium without inoculum was included as a negative control. As a selection criterion for biofilm formation, a cutoff OD (ODc) for the test was defined as three standard deviations above the mean OD of the negative control. The strains were considered non‐biofilm producers (OD_ODc); weak biofilm producers (ODc<OD_2_ODc); moderate biofilm producers (2_ODc<OD_4_ODc); strong biofilm producers (4_ODc<OD_8_ODc); and very strong biofilm producers (8_ODc<OD). These experiments were performed in triplicate.
2.7. Assessment of in vitro antimicrobial activity of Lactobacillus cell‐free supernatants
Cell‐free supernatants (CFSs) of L. gasseri 1A‐TV, L. fermentum 18A‐TV, and L. crispatus 35A‐TV and the CFS of the Lactobacilli combination were prepared as previously reported (Parolin et al., 2015). Each Lactobacillus culture was centrifuged at 7000 × g for 30 min at 4°C, and their supernatants were filtered through a 0.2 μm membrane, and pH values were measured by a pH meter (pH50+DHS Bench pH meter). For the CFS combination, each Lactobacillus culture at 2 × 108 CFU/ml, after the filtration step, was mixed in a 1: 1:1 ratio.
The antimicrobial activity of CFSs was assayed against the indicator strains previously mentioned. The antagonism experiment was performed in a sterile 96‐well plate (Corning® Incorporated Life Sciences, NY, USA) using the indicator strains at 3 × 105 CFU/ml. In each plate, 50 μl of Lactobacillus CFS was mixed with 50 μl of each indicator of antagonist tests and of control growth, 50 μl of sterile MRS medium and 50 μl of each indicator strain were mixed. The 96‐well plates were incubated at 37°C under aerobic conditions and evaluated at 6 h and 24 h.
The results were considered by evaluating the growth inhibition of the indicator strains. The viable microbial cell counts (CFU/ml) of each indicator strain were recorded as log10 reduction of the total count of CFU/ml in the original inoculum, planting on Mueller–Hinton agar (Oxoid, Basingstoke, UK) and Sabouraud dextrose agar plates (BD). The bactericidal activity was defined as a reduction of at least 99.9% (≥3 log10) (NCCLS, 1999). This experiment was repeated in triplicate.
2.8. Evaluation of the antimicrobial activity of CFSs after pH, heat, catalase, and proteolytic enzymatic treatment
The effects of heat treatment, catalase, and proteolytic enzymatic treatments were evaluated for all CFSs. The effect of temperature was determined by exposing 5 ml of each aliquot of CFS to 70°C and 100°C for 30 min and 121°C for 15 min. The sensitivity of the CFSs to enzymatic activity was assayed by catalase (E. C.1.11.1.6) at pH 7.0 (50 mM potassium phosphate buffer), trypsin (E. C.3.4.21.4, type II), and proteinase K (E. C. 3.4.21.64) at pH 7.5 (100 mM Tris‐HCl buffer). Aliquots of the CFSs were incubated (1:1 v/v) with enzyme solutions (1 mg/ml) and their respective controls at 37°C for 2 h under aerobic conditions (Oliveira et al., 2017). After these treatments, the antibacterial activity of the CFSs was determined by antagonism experiments in 96‐well plates and expressed as total (+++), good (++), partial (+), and no inhibition (‐). The effects of pH were tested at pH 5.5, 6.5, and 7.5 adjusted by 10 N NaOH, and untreated cell‐free supernatants were used as controls. The antagonism experiments were performed in a sterile 96‐well plate (Corning® Incorporated Life Sciences) using the indicator strains at 3 × 105 CFU/ml as described above. After incubation for 6 and 24 h at 37°C, the results were estimated by the growth rates of the indicator strains measured by a turbidimetric method with Microplate Reader (BioTek Synergy™ H1) system using OD600 for bacterial strains and OD530 for Candida spp. (Yang et al., 2018). All experiments were repeated three times.
2.9. Determination of hydrogen peroxide production, lactic dehydrogenase activity, L‐ and D‐lactic acid production, and the presence of bacteriocin genes
The production of H2O2 was tested by the Eschenbach method (Eschenbach et al., 1989) using the scale previously reported by Parolin et al. (Parolin et al., 2015). All strains were scored as low (score 1 [>20 min]), medium (score 2 [10–20 min]), or high (score 3 [<10 min]) producing strains. Isolates not producing blue coloration were scored as 0. We used L. acidophilus ATCC 4356 as a positive control strain for H2O2 production. To determine the activity of lactic dehydrogenase (LDH), the cells were harvested after 48 h at 37°C under anaerobic conditions at an optical density (OD600 nm) of 1.5 and centrifuged at 10000 × g for 10 s. The cells were washed and resuspended in 2 ml of phosphate‐buffered saline (PBS; 137 mmol/L NaCl, 2.7 mM KCl, 10 mM Na2HPO4·12H2O, 1.8 mM KH2PO4, pH 7.4). The cell suspensions were ultrasonicated using a BANDELIN SONOPULS HD 2070 sonicator. The LDH activity of bacterial cell lysates from 1A‐TV, 18A‐TV, and 35A‐TV strains was determined through the kinetics of the decrease in NADH absorbance (Δmin) that was measured by a spectrophotometer (Hitachi U‐2000) at λ = 340 nm (Kasai et al., 2019). The enzyme assay was performed at 30°C, and 1 U of the enzyme was defined as the amount of enzyme that catalyzes the degradation of 1 µmol of NADH per minute (Sung et al., 2004).
The production of D‐ and L‐lactic acid produced by Lactobacillus were determined on cell‐free supernatants using a commercial assay kit (Cat. No.11112821035, R‐Biopharm) according to the manufacturer's instructions, the kit used the internal control solutions for the enzymatic determination. The lactic acid production was expressed in g/L. In both tests, lactic dehydrogenase activity, L‐ and D‐lactic acid production, we used L. rhamnosus GGATCC 53103, lactic acid producer as a control strain. The probiotic L. rhamnosus GGATCC 53103 had lactic dehydrogenase activity (46 U mg/L) and was L‐lactic acid (2.8 g/L) and D‐lactic acid (0.03 g/L) producer (Allonsius et al., 2019). The detection of bacteriocin‐encoding genes was conducted by analyzing those most frequently present in the Lactobacilli species: nisinA, nisinB, nisinF, gassericinA, gassericinT, gassericinK, gassericinE, lactacinF, helveticinJ, acidocinA, acidocinB, plantericinA, plantericinEF, and pediocinA. The primers used, designed by Vector NTI software, are listed in Table A1. PCR was performed as previously published (Santagati et al., 2012).
2.10. Statistical analysis
Statistical analyses were performed using GraphPad Prism 6 software (GraphPad Software Inc.), and results were expressed as mean ±standard deviation (SD) of 3 independent experiments. For the co‐aggregation assays, ANOVA with Fisher's least significant difference (LSD) test was used to determine significant differences (p < 0.05).
3. RESULTS
3.1. Evaluation of Lactobacillus antagonistic activity against multidrug‐resistant clinical isolates
The antagonistic activity of L. gasseri 1A‐TV, L. fermentum 18A‐TV, and L. crispatus 35A‐TV, assessed by the agar spot test, showed the best growth inhibition with diameters >10 mm (+ + + +) for E. coli GM1, S. aureus SA3, E. faecalis EFS1, S. agalactiae GB022, E. faecium 75 VRE and for K. pneumoniae 340 KPC, multidrug‐resistant pathogens frequently associated with serious infections. All three Lactobacilli antagonized C. albicans, showing inhibition zones between 6 and 10 mm (+ + +), and exerted a partial inhibition versus C. glabrata, with inhibition zones between 1 and 3 mm. They also showed good inhibition versus P. aeruginosa, P. mirabilis, and P. vulgaris with diameters between 3 and 6 mm (++). The same results were obtained with the combination (1:1:1 ratio) of the three Lactobacilli (Table 2).
TABLE 2.
In vitro inhibitory activity against UGTI pathogens (indicator strains), H2O2production, bacteriocin gene, lactic dehydrogenase activity, detection and sensitivity of the CFS antimicrobial activity to heat, catalase, and proteolytic enzymatic treatment of vaginal Lactobacilli isolates 1A‐TV, 18A‐TV, 35 A‐TV, and their combination
| Bacteriocingenes | Lactic dehydrogenase activity | H2O2production testb | Indicators strains | S. agalactiae GB022 | E. coli GM1 | K. pneumonia 340KPC | S. aureus SA3 | E. faecium 75VRE E. faecalis EFS1 | P. aeruginosa IF1 | P. vulgaris IF1 P. mirabilis IF3 | C. albicans CA312 | C. glabrata CG2824 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 A‐TVL. gasseri | Acidocin A, Helveticin J | 25.57 U mg/1 | High | Deferred agar spot assaya | + + + + | + + + + | + + + + | + + + + | + + + + | ++ | ++ | + + + | + | ||
| Untreated‐pH4.2 | +++ | +++ | +++ | +++ | + | +++ | +++ | − | − | ||||||
| Enzymesc | Trypsin | ‐ | + | + | − | − | +++ | − | − | − | |||||
| Proteinase K | + | + | + | + | − | +++ | − | − | − | ||||||
| Catalase | + | + | + | + | − | ++ | − | − | − | ||||||
| Temperaturec | 70°C, 30 min | +++ | +++ | +++ | +++ | + | +++ | +++ | − | − | |||||
| 100°C, 30 min | +++ | +++ | +++ | +++ | + | +++ | +++ | − | − | ||||||
| 121°C, 15 min | +++ | +++ | +++ | +++ | + | +++ | +++ | − | − | ||||||
| 18 A‐TV L. fermentum | No bacteriocin | 28.27 U mg/1 | 0 | Deferred agar spot assaya | + + + + | + + + + | + + + + | + + + + | + + + + | ++ | ++ | + + + | + | ||
| Untreated‐pH4.3 | +++ | ++ | +++ | +++ | + | +++ | +++ | − | − | ||||||
| Enzymesc | Trypsin | − | + | ++ | − | − | +++ | +++ | − | − | |||||
| Proteinase K | − | + | − | − | − | +++ | +++ | − | − | ||||||
| Catalase | +++ | ++ | +++ | +++ | + | +++ | +++ | − | − | ||||||
| Temperaturec | 70°C, 30 min | +++ | ++ | +++ | +++ | + | +++ | +++ | − | − | |||||
| 100°C, 30 min | +++ | ++ | +++ | +++ | + | +++ | +++ | − | − | ||||||
| 121°C, 15 min | +++ | ++ | +++ | +++ | + | +++ | +++ | − | − | ||||||
| 35 A‐TVL. crispatus | Acidocin A | 56.17 U mg/1 | Low | Deferred agar spot assaya | + + + + | + + + + | + + + + | + + + + | + + + + | ++ | ++ | + + + | + | ||
| Untreated‐pH4.8 | +++ | +++ | +++ | +++ | + | +++ | +++ | − | − | ||||||
| Enzymesc | Trypsin | − | − | − | − | − | − | − | − | − | |||||
| Proteinase K | − | − | − | − | − | +++ | − | − | − | ||||||
| Catalase | − | − | − | − | − | ++ | − | − | − | ||||||
| Temperaturec | 70°C, 30 min | +++ | ++ | ++ | ++ | + | +++ | +++ | − | − | |||||
| 100°C, 30 min | +++ | ++ | ++ | ++ | + | +++ | +++ | − | − | ||||||
| 121°C, 15 min | +++ | ++ | ++ | ++ | + | +++ | +++ | − | − | ||||||
| Lactobacilli Mix | Acidocin A, Helveticin J | N.D | N.D | Deferred agar spot assaya | + + + + | + + + + | + + + + | + + + + | + + + + | ++ | ++ | + + + | + | ||
| Untreated‐pH4.4 | +++ | +++ | +++ | +++ | + | +++ | +++ | − | − | ||||||
Interpretation criteria for the deferred agar spot test: Diameter > 10 (+ + + +); Diameter between 6 and 10 mm ( + + +); Diameter between 3 and 6 mm (+ +); Diameter between 1 and 3 mm (+); no inhibition (−).
Interpretation criteria for H2O2 production: low (score 1[>20 min]), medium (score 2[10–20 min]); high (score 3 [<10 min]), no production 0, ND: undefined.
Interpretation criteria for antagonistic activity after, heat, catalase, and proteolytic enzymatic treatment: total (+++), good (++), partial (+), and no inhibition (−).
3.2. In vitro safety assessment
L. gasseri 1A‐TV, L. fermentum 18A‐TV, and L. crispatus 35A‐TV were sensitive to penicillin G, ampicillin, amoxicillin–clavulanic acid, tetracycline, chloramphenicol, erythromycin, rifampicin, and clindamycin. Intrinsic resistance to trimethoprim–sulfamethoxazole, metronidazole, gentamicin, levofloxacin, ciprofloxacin, streptomycin, and vancomycin was confirmed, except for L. gasseri that was sensitive to vancomycin. Safety assessment tests showed that none of the tested Lactobacilli caused the complete lysis (β‐hemolysis) of erythrocytes on sheep and horse blood agar. The in vitro safety assessment of vaginal Lactobacilli isolates is given in Table A2.
3.3. Aggregation assays and biofilm formation
Aggregation properties were assayed with the auto‐aggregation and co‐aggregation tests measuring two characteristics of the strains. Auto‐aggregation can be mediated by intra‐species cellular promoting factors and cell‐wall hydrophobicity, while co‐aggregation is the ability to achieve an adequate mass by co‐aggregating other bacterial species, however, the ability of a probiotic to aggregate is a desirable property.
The auto‐aggregation rates of L. gasseri 1A‐TV, L. fermentum 18A‐TV, and L. crispatus 35A‐TV, measured after 5 h of incubation, gave the following values: 75.14% ±0.01, 79.41% ±0.01, 83.10% ±0.02, respectively. The degree of Lactobacilli co‐aggregation with S. agalactiae, E. coli. K. pneumoniae, S. aureus, E. faecium, E. faecalis, P. aeruginosa, P. vulgaris, and P. mirabilis was very high, ranging between 51.3 ± 0.02 and 83.19 ± 0.03. C. albicans and C. glabrata, despite a strong value of selective interactions versus Lactobacilli strains (83.51, 89.58%, respectively), possessed a strong auto‐aggregation property (95.1 and 96.8%, respectively).
Despite the co‐aggregation percentage of all bacterial strains being higher than self‐aggregation percentages, significant co‐aggregation (p < 0.05) was found only for S. aureus, P. aeruginosa, and Proteus spp. with 1A‐TV, S. agalactiae, E.coli, S. aureus, E. faecium, and P. aeruginosa with 18 A‐TV and 35A‐TV; in addition, 35A‐TV also showed significant co‐aggregation with K. pneumoniae (Figure 1). Regarding the biofilm production, we found different levels: weak for L. gasseri 1A‐TV, moderate for L. fermentum 18A‐TV while L. crispatus 35A‐TV was not a biofilm producer; however, the Lactobacillus combination stood out as being a strong biofilm producer.
FIGURE 1.
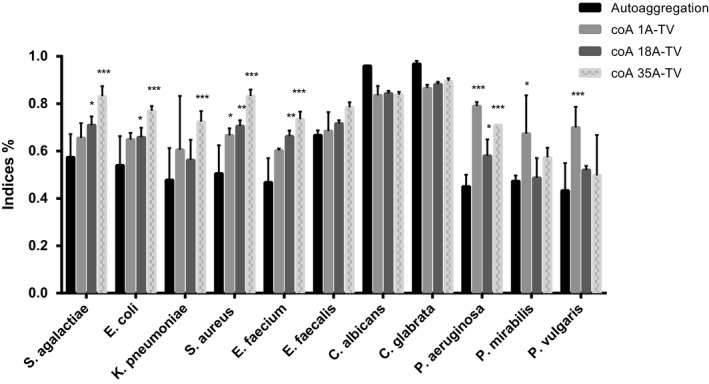
Co‐aggregation ability of Lactobacilli after 5 h incubation at room temperature in PBS (pH 7.4). Results are presented as the average of at least three independent experiments, and the error bars correspond to standard deviations. Statistical significance was evaluated by ANOVA with Fisher’s least significant difference (LSD) (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001)
3.4. Adhesion test on HeLa cells
L. gasseri 1A‐TV, L. fermentum 18A‐TV, and L. crispatus 35A‐TV were tested for their capability to adhere to HeLa cells. After being extensively washed with PBS, a significant proportion of cells from all bacterial strains remained attached to the HeLa monolayer displaying a strong adhesive phenotype, coinciding with an adhesion index (ADI) greater than 2500, as shown in Figure 2 (a, b). This adhesion in L. crispatus 35A‐TV showed an extraordinary ADI of 70000. The Lactobacillus adhesion was also tested by viable counts showing that L. crispatus 35A‐TV (6x106± 0.24 CFU/ml) L. gasseri 1A‐TV (4,5×105± 0.47 CFU/ml) and L. fermentum 18A‐TV (2,88105± 0.38 CFU/ml) displayed a good ability to adhere to HeLa cells.
FIGURE 2.
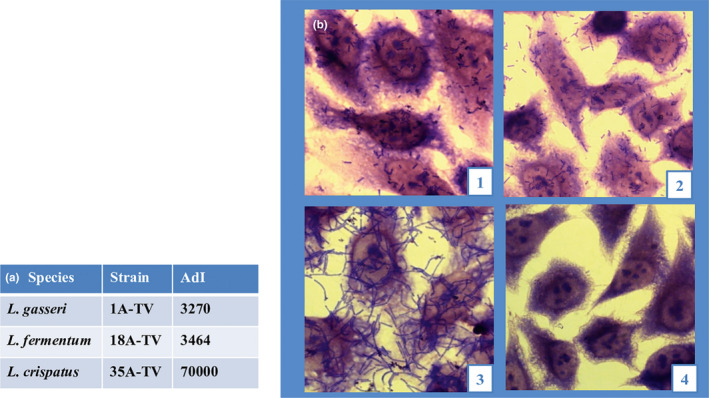
Bacterial adhesion to HeLa cell layer. (a) Adhesion indexes (ADI; the number of bacteria/100 HeLa cells). (b) Cell layers observed after Giemsa staining using light microscopy: (1) L. gasseri 1A‐TV; (2) L. fermentum 18A‐TV; (3) L. crispatus 35A‐TV; (4) Adhesion to HeLa cell monolayer as a negative control
3.5. In vitro antimicrobial activity of Lactobacilli CFSs and their sensitivity to pH, heat, catalase, and proteolytic enzymatic treatment
Cell‐free supernatants of L. gasseri 1A‐TV, L. fermentum 18A‐TV, and L. crispatus 35A‐TV, at pH 4.2, 4.3, and 4.8, respectively, were assayed for their ability to inhibit the pathogens by time‐killing tests (Figure 3 and Figure A1). After 6 and 24 h of incubation, L. gasseri 1A‐TV CFS could inhibit the growth of S. agalactiae, E. coli, K. pneumoniae, S. aureus, P. aeruginosa, P. mirabilis, and P. vulgaris exhibiting bactericidal activity. L. fermentum 18A‐TV reduced the growth by 1 log10 for S. agalactiae, E. coli, and S. aureus at T6 h, while it reduced the growth by 2 log10 for K. pneumoniae. At T24 h, 18A‐TV reduced the growth by 2 log10 for E. coli, whereas it exhibited a complete inhibition (bactericidal effect) of S. agalactiae, S. aureus, and K. pneumoniae. Furthermore, L. fermentum 18A‐TV had bactericidal activity at both T6 h and T24 h against P. aeruginosa, P. mirabilis, and P. vulgaris.
FIGURE 3.
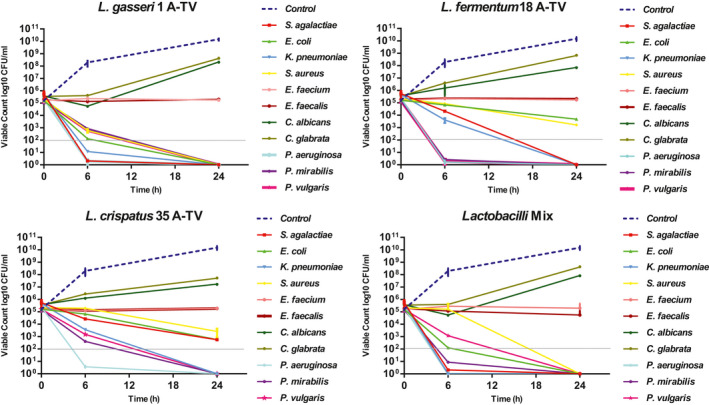
In vitro antimicrobial activity of cell‐free supernatants (CFSs) on indicator strains by time‐killing curves analysis. The gray dotted line indicates a 3‐log10 decrease in the number of CFU/ml versus the number at the baseline (bactericidal effect), while the blue dotted line indicates a general control growth
Regarding L. crispatus 35A‐TV, no effect was found for S. agalactiae, E. coli, K. pneumoniae, S. aureus, and P. vulgaris at T6 h, but switched to bactericidal at T24 h, for K. pneumoniae and P. vulgaris, whereas 35A‐TV had a bactericidal activity from T6 h against P. aeruginosa and P. mirabilis.
Moreover, all three Lactobacilli CFSs showed no antimicrobial effect versus E. faecium and E. faecalis at T6 h and T24 h, as well as both strains of Candida spp. In addition, the curve of candidal growth maintained a concentration similar to the initial inoculum (105–106 CFU/ml) also at T24 h compared to the CFS‐free curve that reached a concentration of 109–1010 CFU/ml.
Also, the inhibitory effect of the CFS combination (the three Lactobacilli together), evaluated at 6 h and 24 h, showed bactericidal activity only against S. agalactiae, E. coli, K. pneumoniae, S. aureus, P. aeruginosa, P. vulgaris, and P. mirabilis.
In Table 2, the effects of heat treatment and proteolytic enzymes on the CFS activity of each strain and their combination are shown. The treatment at 70, 100, and 121°C of the CFSs of L. gasseri 1A‐TV and L. fermentum 18A‐TV did not alter their antagonistic activity, whereas a reduction was observed only for L. crispatus 35A‐TV CFS versus E. coli, K. pneumoniae, and S. aureus. The catalase treatment decreased inhibitory activity for 1A‐TV and 35A‐TV CFSs, while no effects were observed in 18A‐TV.
Regarding proteolytic treatment, we registered the loss, or the strong reduction, of the inhibitory activity of the three singular supernatants versus S. agalactiae, E. coli, S. aureus, K. pneumoniae, E. faecalis, and E. faecium. The growth of Proteus spp. was inhibited by 18A‐TV CFS and maintained with 1A‐TV and 35A‐TV CFSs. Only for P. aeruginosa, was the bactericidal effect maintained by the three Lactobacilli CFSs after proteolytic treatment, except for L. crispatus 35A‐TV CFS after trypsin treatment.
The pH‐dependent effects on antimicrobial activity of CFSs were tested at pH 5.5, 6.5, and 7.5. by measurements of the growth rates (OD) of the indicator strains. All CFS Lactobacilli and their formulation at pH 5.5 maintained their activity up to 6 h and weakly lost their efficiency at 24 h compared to the untreated pH, while the antagonistic activity of CFS at pH 6.5 and 7.5 was lost after 6 h despite the growth of the indicator curve showed a slight decrease in slope compared to controls (Figure A2).
3.6. Determination of hydrogen peroxide production, lactic dehydrogenase activity, and bacteriocin‐encoding genes
L. gasseri 1A‐TV produced a higher quantity of hydrogen peroxide with respect to L. crispatus 35A‐TV, while L. fermentum 18A‐TV did not release this metabolite (Table 2).
The lactic dehydrogenase activity was evaluated using cell lysates, in particular, L. crispatus 35A‐TV had a specific activity of 56.17 U mg/L, higher than that observed in L. gasseri 1A‐TV and L. fermentum 18A‐TV, which were, respectively, 25.57 U mg/L and 28.27 U mg/L (Table 2). In addition, all Lactobacilli were L‐ and D‐lactic acid producers showing the production of D‐lactic acid 4.04 (g/L), 3.7 (g/L), 3.11 (g/L), and L‐lactic acid between 2.94 (g/L), 2.95 (g/L), and 3.34 (g/L) for L. gasseri 1A‐TV, L. fermentum 18A‐TV, and L. crispatus 35A‐TV, respectively.
The detection of bacteriocin‐encoding genes revealed helveticin J only in L. gasseri and acidocin A in L. gasseri and L. crispatus. L. fermentum, despite showing activity against pathogens, was negative for all genes tested (Table 2).
4. DISCUSSION
Several studies have reported beneficial effects exerted by probiotics, and it has been well demonstrated that functional properties are strain‐dependent (Borges et al., 2014). In this study, we characterized three Lactobacilli, L. gasseri 1A‐TV, L. fermentum 18A‐TV, and L. crispatus 35A‐TV isolated from the vaginal microbiota, with the activities of their metabolites produced by CFSs for their beneficial features addressed mainly to their antimicrobial activity against multidrug‐resistant clinical isolates.
In accordance with the objectives of our study, the selected Lactobacilli were tested in vitro for surface properties to determine their capability to colonize the human vagina. In vitro experiments showed their ability to adhere to HeLa cells, and this is also related to their predisposition to self‐aggregate. As is well known, adhesion and auto‐aggregation represent the determining factors for the initial development of biofilm, which is a strategy of some organisms to persist in harsh environments promoting microbial resistance to antimicrobial agents, the immune system, and stress conditions (Leccese Terraf et al., 2016). In this regard, our Lactobacilli possessed strong biofilm formation capacity when tested in combination; however, they were poor producers when tested alone. These data make us hypothesize a synergistic interspecific interaction between our Lactobacilli to optimize their living conditions. Biofilm formation is a phenomenon that can promote mucosal colonization and masking epithelial cell receptors, can exert a protective role by interfering with the growth and adhesion of pathogens (Leccese Terraf et al., 2016).
Another mechanism that promotes an exclusion/competition behavior is the ability of beneficial bacteria to co‐aggregate with pathogens (Santos et al., 2016). In this regard, our Lactobacillus strains showed a significant capability to co‐aggregate with S. agalactiae, E. coli. KPC‐producing K. pneumoniae, S. aureus E. faecium VRE and E. faecalis, P. aeruginosa, P vulgaris, and P. mirabilis. This is an important contributing factor to create a microenvironment where pathogens can be exposed to higher concentrations of inhibitory substances or metabolites such as organic acids (e.g., lactic acid) and hydrogen peroxide mainly produced by Lactobacilli strains as the dominant bacterial population in the vaginal ecosystem (Verdenelli et al., 2014).
In this study, we found that cell‐free supernatants released from three Lactobacilli as single entities, and their combination, exhibited an antagonistic effect against multidrug‐resistant clinical isolates including S. agalactiae, E. coli, KPC‐producing K. pneumoniae, S. aureus, E. faecium VRE, E. faecalis, P. aeruginosa, P. mirabilis, and P. vulgaris.
Conversely, the anti‐candida activity of the three Lactobacilli showed different behavior with the two approaches: agar diffusion and using cell‐free supernatants, which had no growth‐inhibitory activity and could maintain the candidal growth almost at the same concentration as the initial inoculum compared to the control (CFS‐free). These conflicting results could be explained by the physical state of the media; the concentration of antimicrobial substances released into the solid and liquid media, and by the environment where the substances exert their effects. Scorzoni et al. also reported that the microdilution test is more sensitive to agar diffusion in the evaluation of anti‐candida activity highlighting the need to apply different methods to evaluate in vitro antimicrobial effects of Lactobacilli (Scorzoni et al., 2007).
Notably, the CFS combination maintained the same antagonistic profile of each strain, excluding a possible interference between them.
The activity of Lactobacillus CFSs after the heat and enzymatic treatments was reduced in some cases compared with untreated CFSs hypothesizing the presence of thermostable and thermosensitive substances such as bacteriocins in the supernatants, while the neutralization treatment at pH 6.5 and 7.5 canceled antagonistic effects. These data suggested that the acid environment and antimicrobial metabolites released by our strains such as bacteriocins had synergetic action against the growth of pathogens tested, showing a better antagonistic activity. Further, several reports suggested that the pH‐induced alterations of net charge might facilitate the translocation of some bacteriocin molecules through the cell wall (Oliveira et al., 2017) and that an acid environment could interfere with the production and bactericidal activity of several bacteriocins (Yang et al., 2018). pH and lactic acid levels display a strong inverse correlation demonstrating that lactic acid is the main acidifier of the human vagina, increasing its production under hypoxic conditions (Tachedjian et al., 2017), which displays antimicrobial and anti‐inflammatory properties. In this context, all three isolated Lactobacilli can produce D‐ and L‐lactic acid that could mainly contribute to the vaginal health promotion having also anti‐inflammatory effects (Alvarez‐Olmos et al., 2004; José Aníbal Mora‐Villalobos JM‐Z, 2020). Moreover, Lactobacillus production of hydrogen peroxide as diffusible inhibitory substances could be connected to antimicrobial properties of the vaginal microbiota, representing an important nonspecific antimicrobial defense mechanism due to a highly toxic state (Kullisaar et al., 2002; Mijac et al., 2006).
However, hydrogen peroxide production by Lactobacilli as well as L. gasseri 1A‐TV and L. crispatus 35A‐TV can be considered an additional beneficial effect for vaginal health (Antonio et al., 2005; Pendharkar et al., 2013), in vivo, conversely, under microaerobic (hypoxic) conditions such as the cervicovaginal environment, the concentration of H2O2 produced does not achieve the amount necessary to have antimicrobial activity in the vaginal environment. The low vaginal O2 levels measured in in vivo studies have been associated with little or no H2O2 in the hypoxic cervicovaginal environment (O'Hanlon et al., 2011). Therefore, these findings support an important role of lactic acids as main products at high concentrations in a hypoxic environment such as the vagina. However, H2O2 production remains an in vitro marker for beneficial vaginal properties (Tachedjian et al., 2018).
Additionally, bacteriocins are believed to contribute to the competitiveness between strains by acting against pathogenic strains; therefore, the production of bacteriocins represents an important antimicrobial factor (Soltani et al., 2020). Among our strains, L. gasseri 1A‐TV and L. crispatus 35A‐TV are producers of helveticin J and acidocin A, which is a small thermostable peptide with maximum production at pH 5, exerting antagonistic activity versus several bacterial genera, including Lactococcus, Pediococcus Staphylococcus, Enterococcus, Streptococcus, Listeria, Clostridium, and Bacillus (Kanatani et al., 1995). The stability at a high temperature of acidocin A and its bacterial targets suggested its decisive role in the antimicrobial activity exerted by the supernatants of L. gasseri 1A‐TV and L. crispatus 35A‐TV against the indicators tested, also considering that L. crispatus is considered a major determinant in the stability of the normal vaginal microbiota in women of reproductive age (Miller et al., 2016)
Our study strengthens the concept of using probiotic Lactobacillus to protect the host against MDR pathogens including E. faecium VRE and KPC‐producing Klebsiella pneumoniae, based on the antimicrobial activity of our Lactobacilli, L. gasseri 1A‐TV, L. fermentum 18A‐TV, and L. crispatus 35A‐TV and their combination, as well as their CFSs, showed clear antibacterial activity against multidrug‐resistant pathogens. Moreover, the three Lactobacilli, with some intra‐species diversity, share many probiotic features both as live and non‐live bacteria such as their released metabolites (CFSs) possessing the potential of colonizing the vaginal epithelium, producing antagonistic metabolites, and keeping their activity in different environmental conditions. Taken together, all these results support novel therapeutic strategies as a new vaginal formulation for the prevention and treatment of urogenital infections, acting on the rebalance of the vaginal microbiome.
Further experiments are planned to complete the characterization of these CFSs, with a more detailed knowledge of their metabolic profiles to better understand their nature and mode of action. Future work will also characterize the probiotic potential of these bacteria in the vaginal tract through in vivo studies.
5. CONCLUSIONS
The novel combination of L. gasseri 1A‐TV, L. fermentum 18A‐TV, and L. crispatus 35A‐TV characterized both as live strains and as non‐live CFSs in this study showed an antimicrobial activity versus the most common MDR pathogens, such as E. faecium VRE and KPC‐producing Klebsiella pneumoniae involved in UTIs, considering the limited antibiotic choice against these MDR microorganisms. In addition, we demonstrated the antimicrobial effect of their cell‐free supernatant, thanks to different substances released by the three Lactobacilli both singularly and in combination. The strong bactericidal effect on MDR isolates was also maintained in selected conditions. These results are promising for new vaginal probiotic formulations against MDR bacterial infections.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
Marina Scillato: Conceptualization (lead); Data curation (equal); Formal analysis (equal); Methodology (lead); Validation (lead); Writing‐original draft (lead); Writing‐review & editing (equal). Ambra Spitale: Conceptualization (equal); Data curation (lead); Formal analysis (lead); Methodology (lead); Supervision (lead); Validation (equal); Writing‐original draft (equal); Writing‐review & editing (lead). Gino Mongelli: Conceptualization (equal); Data curation (lead); Formal analysis (equal); Methodology (equal); Validation (equal); Writing‐original draft (equal); Writing‐review & editing (equal). GreteFrancesca Privitera: Conceptualization (supporting); Methodology (equal); Validation (supporting); Writing‐original draft (equal); Writing‐review & editing (equal). Katia Mangano: Methodology (equal); Writing‐original draft (supporting); Writing‐review & editing (equal). Antonio Cianci: Investigation (supporting); Writing‐original draft (supporting); Writing‐review & editing (equal). Stefania Stefani: Conceptualization (equal); Formal analysis (supporting); Methodology (equal); Project administration (equal); Resources (lead); Supervision (equal); Writing‐original draft (lead); Writing‐review & editing (lead). Maria Santagati: Conceptualization (lead); Data curation (lead); Formal analysis (equal); Methodology (lead); Supervision (lead); Validation (lead); Writing‐original draft (lead); Writing‐review & editing (lead).
ETHICS STATEMENT
Ethical approval is not required. Written informed consent was obtained from all participants.
ACKNOWLEDGMENTS
We wish to thank the Scientific Bureau of the University of Catania for language support. The authors would like to thank Nicola Musso and the CS B. R. I. T. for their technical assistance, and Prof. Marcella Renis and Barbara Tomasello for the LDH assay. A grant from DMG Italia supported the preparation of test materials. The 2016/2018 Department Research Plan of the University of Catania (the second line of intervention) supported the molecular test, writing, and publishing of this article.
1.
TABLE A1.
Primers designed for the detection of bacteriocin genes
| Bacteriocin | Primer name | Primer sequence (5’‐3’) |
|---|---|---|
| Nisin A | Nis A | GCGAGCATAATAAACGGCTCTGATT |
| Nis Aa | CAACCACTGAGTATCCAATCTTATACCC | |
| Nisin B | Nis B | CTCAGCTAAATGTTCTAATTGTTGCTTC |
| Nis Ba | AGCTCACAGTATGACTTAACGGGAA | |
| Nisin F | Nis F | TGGAACAGTCTGTGGTTTATTAGGAG |
| Nis Fa | TCACATTCCTCCATGCACAATCTTAA | |
| Gassericin A | GaaA | AGTATCAGTTGGTGGGTTCGTTTG |
| GaaAa | CACCAACGAGTATTCCAATAAATAGG | |
| Gassericin T | GatA | CACAATAGTGACAGGTCGTAGCACATA |
| GatAa | CCGTAGCAGCTCCTATTACAGCAT | |
| Gassericin K | MS 480 | TCCCCAACTAGTCTATCTGTTGTTCC |
| MS 481a | GCAATCAGACAGAGTACAGTTACATCTAC | |
| Lactacin F | LaF | AGGGGAATGTGACGATAATGACC |
| LaFa | TATAGCCAAAATAACCTCCTATTGCTG | |
| Elveticin J | MS 478 | TGTATGCGGGCTGGGCTGACT |
| MS 479a | AGGTTCAGGCTATGGCGATGGAA | |
| Acidocin A | Acd A | CGTAAATTGGGGTAGTGTTGC |
| Acd Aa | AGAACTCAAACGCTGCCTACA | |
| Acidocin B | Acd B | GTCCTGCTTGTGGCTTTGTT |
| Acd Ba | GCCCGTTTGATACAAGTTACCT |
Reverse primers.
TABLE A2.
In vitro safety assessment of vaginal Lactobacilli isolates
| Producer Strains | Antibiotic susceptibility profilea | Hemolytic activityb | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | AMP | VA | SXT | RD | MTZ | CN | S | TE | C | E | AMC | LEV | CIP | DA | ||
| 1A‐TV L. gasseri | S | S | S | R | S | R | R | R | S | S | S | S | R | R | S | − |
| 18A‐TV L. fermentum | S | S | R | R | S | R | R | R | S | S | S | S | R | R | S | − |
| 35A‐TV L. crispatus | S | S | R | R | S | R | R | R | S | S | S | S | R | R | S | − |
Interpretation criteria for the antibiotic susceptibility profile determined by Kirby‐Bauer: resistant (R); susceptible (S). (European Food Safety Authority, 2012). P: Penicillin G, AMP: Ampicillin, VA: Vancomycin, SXT: Cotrimoxazole, RD: Rifampicin, MTZ: Metronidazole, CN: Gentamicin, S: Streptomycin, TE: Tetracycline, C: Chloramphenicol, E: Erythromycin, AMC: Amoxicillin–clavulanic acid, CIP: Ciprofloxacin, LEV: Levofloxacin, DA: Clindamycin
Interpretation criteria for the hemolytic activity: hemolysis (+); no hemolysis (−).
FIGURE A1.
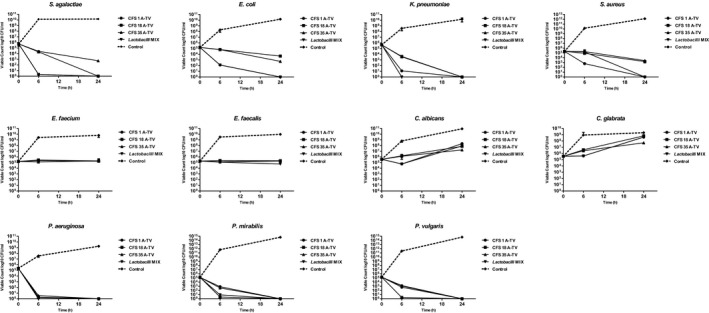
In vitro antimicrobial activity of cell‐free supernatants (CFSs) on each single indicator strain by time‐killing curve analysis. *The black dotted line indicates a growth control of each indicator strain, the solid black line with circles indicates the CFS of L. gasseri 1A‐TV, while the line with rectangles indicates the CFS of L. fermentum 18A‐TV, the line with triangles indicates the CFS of L. crispatus 35 A‐TV, and the line with triangles facing down indicates the CFS Lactobacilli mix
FIGURE A2.
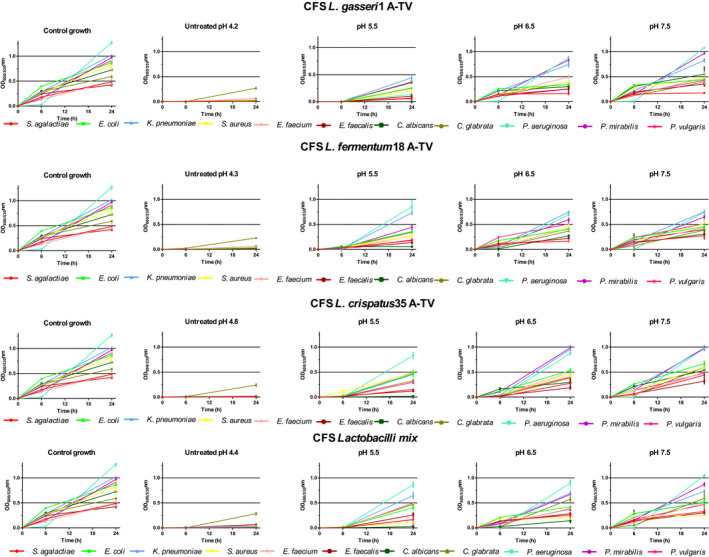
Effects of pH on CFS antimicrobial activity on indicator strain growth. Comparison of growth curves at 6 and 24h (OD = 600 nm for bacterial strains and OD = 530 for Candida spp.) of control indicator strains with respect to the growth of untreated Lactobacilli CFS, CFS adjusted to pH 5.5, 6.5, and 7.5. The results are expressed as mean ± standard deviations of values obtained from triplicate experiments
Marina Scillato and Ambra Spitale contributed equally to the work.
DATA AVAILABILITY STATEMENT
All data generated or analyzed during this study are included in this published article.
REFERENCES
- Ahmed, S. S., Shariq, A., Alsalloom, A. A., Babikir, I. H., & Alhomoud, B. N. (2019). Uropathogens and their antimicrobial resistance patterns: Relationship with urinary tract infections. International Journal of Health Sciences., 13(2), 48–55. [PMC free article] [PubMed] [Google Scholar]
- Allonsius, C. N., Vandenheuvel, D., Oerlemans, E. F. M., Petrova, M. I., Donders, G. G. G., Cos, P., Delputte, P., & Lebeer, S. (2019). Inhibition of Candida albicans morphogenesis by chitinase from Lactobacillus rhamnosus GG. Scientific Reports., 9(1), 2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997). Gapped BLAST and PSI‐BLAST: A new generation of protein database search programs. Nucleic Acids Research., 25(17), 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez‐Olmos, M. I., Barousse, M. M., Rajan, L., Van Der Pol, B. J., Fortenberry, D., Orr, D., & Fidel, P. L. (2004). Vaginal lactobacilli in adolescents: Presence and relationship to local and systemic immunity, and to bacterial vaginosis. Sexually Transmitted Diseases, 31(7), 393–400. [DOI] [PubMed] [Google Scholar]
- Al‐Zahrani, J., Al Dossari, K., Gabr, A. H., Ahmed, A. F., Al Shahrani, S. A., & Al‐Ghamdi, S. (2019). Antimicrobial resistance patterns of Uropathogens isolated from adult women with acute uncomplicated cystitis. BMC Microbiology, 19(1), 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonio, M. A., Rabe, L. K., & Hillier, S. L. (2005). Colonization of the rectum by Lactobacillus species and decreased risk of bacterial vaginosis. The Journal of Infectious Diseases., 192(3), 394–398. [DOI] [PubMed] [Google Scholar]
- Aslam, B., Wang, W., Arshad, M. I., Khurshid, M., Muzammil, S., Rasool, M. H., & Baloch, Z. (2018). Antibiotic resistance: A rundown of a global crisis. Infect Drug Resist., 11, 1645–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Authority EFS (2012). Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA Journal., 10, 1–10. [Google Scholar]
- Bautista, C. T., Wurapa, E., Sateren, W. B., Morris, S., Hollingsworth, B., & Sanchez, J. L. (2016). Bacterial vaginosis: A synthesis of the literature on etiology, prevalence, risk factors, and relationship with chlamydia and gonorrhea infections. Military Medical Research, 3, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges, S., Silva, J., & Teixeira, P. (2014). The role of lactobacilli and probiotics in maintaining vaginal health. Archives of Gynecology and Obstetrics., 289(3), 479–489. [DOI] [PubMed] [Google Scholar]
- Charteris, W. P., Kelly, P. M., Morelli, L., & Collins, J. K. (2007). Gradient diffusion antibiotic susceptibility testing of potentially probiotic lactobacilli. Journal of Food Protection., 64(12), 2007–2014. [DOI] [PubMed] [Google Scholar]
- De Gregorio, P. R., Tomas, M. S. J., Terraf, M. C. L., & Nader‐Macias, M. E. F. (2014). In vitro and in vivo effects of beneficial vaginal lactobacilli on pathogens responsible for urogenital tract infections. Journal of Medical Microbiology., 63(Pt 5), 685–696. [DOI] [PubMed] [Google Scholar]
- Dimitonova, S. P., Bakalov, B. V., Aleksandrova‐Georgieva, R. N., & Danova, S. T. (2008). Phenotypic and molecular identification of lactobacilli isolated from vaginal secretions. Journal of Microbiology, Immunology, and Infection, 41(6), 469–477. [PubMed] [Google Scholar]
- Donders, G. G. G., Bellen, G., Grinceviciene, S., Ruban, K., & Vieira‐Baptista, P. (2017). Aerobic vaginitis: No longer a stranger. Research in Microbiology., 168(9–10), 845–858. [DOI] [PubMed] [Google Scholar]
- Eryilmaz, M., Gurpinar, S. S., Palabiyik, I. M., Guriz, H., & Gerceker, D. (2018). Molecular Identification and Antimicrobial Activity of Vaginal Lactobacillus sp. Current Pharmaceutical Biotechnology., 19(15), 1241–1247. [DOI] [PubMed] [Google Scholar]
- Eschenbach, D. A., Davick, P. R., Williams, B. L., Klebanoff, S. J., Young‐Smith, K., Critchlow, C. M. et al (1989). Prevalence of hydrogen peroxide‐producing Lactobacillus species in normal women and women with bacterial vaginosis. Journal of Clinical Microbiology., 27(2), 251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EUCAST ECoAST . 2019. [Available from: http://www.eucast.org/clinical_breakpoints/.
- Hütt, P., Lapp, E., Štšepetova, J., Smidt, I., Taelma, H., Borovkova, N., Oopkaup, H., Ahelik, A., Rööp, T., Hoidmets, D., Samuel, K., Salumets, A., & Mändar, R. (2016). Characterisation of probiotic properties in human vaginal lactobacilli strains. Microb Ecol Health Dis., 27, 30484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanatani, K., Oshimura, M., & Sano, K. (1995). Isolation and characterization of acidocin A and cloning of the bacteriocin gene from Lactobacillus acidophilus. Applied and Environmental Microbiology., 61(3), 1061–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai, T., Suzuki, Y., Kouzuma, A., & Watanabe, K. (2019). Roles of d‐Lactate Dehydrogenases in the Anaerobic Growth of Shewanella oneidensis MR‐1 on Sugars. Applied and Environmental Microbiology, 85(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss, H., Kögler, B., Petricevic, L., Sauerzapf, I., Klayraung, S., Domig, K., Viernstein, H., & Kneifel, W. (2007). Vaginal Lactobacillus microbiota of healthy women in the late first trimester of pregnancy. BJOG : an International Journal of Obstetrics and Gynaecology., 114(11), 1402–1407. [DOI] [PubMed] [Google Scholar]
- Kos, B., Suskovic, J., Vukovic, S., Simpraga, M., Frece, J., & Matosic, S. (2003). Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. Journal of Applied Microbiology., 94(6), 981–987. [DOI] [PubMed] [Google Scholar]
- Kullisaar, T., Zilmer, M., Mikelsaar, M., Vihalemm, T., Annuk, H., Kairane, C. et al (2002). Two antioxidative lactobacilli strains as promising probiotics. International Journal of Food Microbiology., 72(3), 215–224. [DOI] [PubMed] [Google Scholar]
- Lebeer, S., Verhoeven, T. L. A., Perea Vélez, M., Vanderleyden, J., & De Keersmaecker, S. C. J. (2007). Impact of environmental and genetic factors on biofilm formation by the probiotic strain Lactobacillus rhamnosus GG. Applied and Environmental Microbiology., 73(21), 6768–6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leccese Terraf, M. C., Juarez Tomas, M. S., Rault, L., Le Loir, Y., Even, S., & Nader‐Macias, M. E. (2016). Biofilms of vaginal Lactobacillus reuteri CRL 1324 and Lactobacillus rhamnosus CRL 1332: Kinetics of formation and matrix characterization. Archives of Microbiology., 198(7), 689–700. [DOI] [PubMed] [Google Scholar]
- Liu, C. H., Lee, S. M., Vanlare, J. M., Kasper, D. L., & Mazmanian, S. K. (2008). Regulation of surface architecture by symbiotic bacteria mediates host colonization. Proceedings of the National Academy of Sciences of the United States of America., 105(10), 3951–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, A., Sakamoto, M., Hanazaki, S., Osawa, M., Suzuki, T., Tochigi, M., & Kakii, K. (2003). Coaggregation among nonflocculating bacteria isolated from activated sludge. Applied and Environmental Microbiology., 69(10), 6056–6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragkoudakis, P., Zoumpopoulou, G., Miaris, C., Kalantzopoulos, G., Pot, B., & Tsakalidou, E. (2006). Probiotic potential of Lactobacillus strains isolated from dairy products. International Dairy Journal., 16, 189–199. [Google Scholar]
- Marchisio, P., Santagati, M., Scillato, M., Baggi, E., Fattizzo, M., Rosazza, C. et al (2015). Streptococcus salivarius 24SMB administered by nasal spray for the prevention of acute otitis media in otitis‐prone children. European Journal of Clinical Microbiology & Infectious Diseases : Official Publication of the European Society of Clinical Microbiology., 34(12), 2377–2383. [DOI] [PubMed] [Google Scholar]
- Martín, C., Fernández‐Vega, I., Suárez, J. E., & Quirós, L. M. (2020). Adherence of Lactobacillus salivarius to HeLa cells promotes changes in the expression of the genes involved in biosynthesis of their ligands. Frontiers in Immunology., 10(3019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, D. H. (2012). The microbiota of the vagina and its influence on women's health and disease. The American Journal of the Medical Sciences., 343(1), 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastromarino, P., Brigidi, P., Macchia, S., Maggi, L., Pirovano, F., Trinchieri, V., Conte, U., & Matteuzzi, D. (2002). Characterization and selection of vaginal Lactobacillus strains for the preparation of vaginal tablets. Journal of Applied Microbiology., 93(5), 884–893. [DOI] [PubMed] [Google Scholar]
- Matulay, J. T., Mlynarczyk, C. M., & Cooper, K. L. (2016). Urinary tract infections in women: Pathogenesis, diagnosis, and management. Current Bladder Dysfunction Reports., 11(1), 53–60. [Google Scholar]
- Mijac, V. D., Dukic, S. V., Opavski, N. Z., Dukic, M. K., & Ranin, L. T. (2006). Hydrogen peroxide producing lactobacilli in women with vaginal infections. European Journal of Obstetrics, Gynecology, and Reproductive Biology., 129(1), 69–76. [DOI] [PubMed] [Google Scholar]
- Miller, E. A., Beasley, D. E., Dunn, R. R., & Archie, E. A. (2016). Lactobacilli dominance and vaginal pH: Why Is the human vaginal microbiome unique? Frontiers in Microbiology, 7:1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora‐Villalobos, J. A., Montero‐Zamora, J., Barboza, N., Rojas‐Garbanzo, C., Usaga, J., Redondo‐Solano, M., Schroedter, L., Olszewska‐Widdrat, A., & López‐Gómez, J. P. (2020). Multi‐Product Lactic Acid Bacteria Fermentations: A Review. Multidisciplinary Digital Publishing Institute. [Google Scholar]
- NCCLS . (1999). Methods for determining bactericidal activity of antimicrobial agents: Approved guideline M26‐A. National Committee for Clinical Laboratory Standards, Wayne
- O'Hanlon, D. E., Moench, T. R., & Cone, R. A. (2011). In vaginal fluid, bacteria associated with bacterial vaginosis can be suppressed with lactic acid but not hydrogen peroxide. BMC Infectious Diseases., 11, 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira, L. C., Silveira, A. M. M., Monteiro, A. S., Dos Santos, V. L., Nicoli, J. R., Azevedo, V. A. D. C., Soares, S. D. C., Dias‐Souza, M. V., & Nardi, R. M. D. (2017). In silico Prediction, in vitro antibacterial spectrum, and physicochemical properties of a putative bacteriocin produced by lactobacillus rhamnosus Strain L156.4. Frontiers in Microbiology, 8, 876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parolin, C., Marangoni, A., Laghi, L., Foschi, C., Nahui Palomino, R. A., Calonghi, N., Cevenini, R., & Vitali, B. (2015). Isolation of vaginal lactobacilli and characterization of Anti‐Candida activity. PLoS One, 10(6), e0131220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendharkar, S., Magopane, T., Larsson, P.‐G., Bruyn, G. D., Gray, G. E., Hammarström, L., & Marcotte, H. (2013). Identification and characterisation of vaginal lactobacilli from South African women. BMC Infectious Diseases, 13(1), 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez Ibarreche, M., Castellano, P., & Vignolo, G. (2014). Evaluation of anti‐Listeria meat borne lactobacillus for biofilm formation on selected abiotic surfaces. Meat Science., 96(1), 295–303. [DOI] [PubMed] [Google Scholar]
- Petrova, M. I., Lievens, E., Malik, S., Imholz, N., & Lebeer, S. (2015). Lactobacillus species as biomarkers and agents that can promote various aspects of vaginal health. Frontiers in Physiology., 6, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino, A., Bartolo, E., Caggia, C., Cianci, A., & Randazzo, C. L. (2019). Detection of vaginal lactobacilli as probiotic candidates. Scientific Reports., 9(1), 3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piqué, N., Berlanga, M., & Miñana‐Galbis, D. (2019). Health benefits of heat‐killed (Tyndallized) probiotics: An overview. International Journal of Molecular Sciences., 20(10), 2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravel, J., Gajer, P., Abdo, Z., Schneider, G. M., Koenig, S. S., McCulle, S. L., Karlebach, S., Gorle, R., Russell, J., Tacket, C. O., Brotman, R. M., Davis, C. C., Ault, K., Peralta, L., & Forney, L. J. et al (2011). Vaginal microbiome of reproductive‐age women. Proceedings of the National Academy of Sciences of the United States of America., 108(Suppl 1), 4680–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzak, M. S., Al‐Charrakh, A. H., & Al‐Greitty, B. H. (2011). Relationship between lactobacilli and opportunistic bacterial pathogens associated with vaginitis. North American Journal of Medical Sciences., 3(4), 185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid, G., Younes, J. A., Van der Mei, H. C., Gloor, G. B., Knight, R., & Busscher, H. J. (2011). Microbiota restoration: Natural and supplemented recovery of human microbial communities. Nature Reviews Microbiology., 9(1), 27–38. [DOI] [PubMed] [Google Scholar]
- Santagati, M., Scillato, M., Patane, F., Aiello, C., & Stefani, S. (2012). Bacteriocin‐producing oral streptococci and inhibition of respiratory pathogens. FEMS Immunology and Medical Microbiology., 65(1), 23–31. [DOI] [PubMed] [Google Scholar]
- Santos, C. M. A., Pires, M. C. V., Leao, T. L., Hernandez, Z. P., Rodriguez, M. L., Martins, A. K. S., Miranda, L. S., Martins, F. S., & Nicoli, J. R. (2016). Selection of lactobacillus strains as potential probiotics for vaginitis treatment. Microbiology (Reading, England)., 162(7), 1195–1207. [DOI] [PubMed] [Google Scholar]
- Scorzoni, L., Benaducci, T., Almeida, A. M. F., Silva, D. H. S., Bolzani, V. S., & Gianinni, M. J. S. M. (2007). The use of standard methodology for determination of antifungal activity of natural products against medical yeasts Candida sp and Cryptococcus sp. Brazilian Journal of Microbiology., 38, 391–397. [Google Scholar]
- Siroli, L., Patrignani, F., Serrazanetti, D. I., Parolin, C., Nahui Palomino, R. A., Vitali, B., & Lanciotti, R. (2017). Determination of antibacterial and technological properties of vaginal lactobacilli for their potential application in dairy products. Frontiers in Microbiology., 8, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S. B., & Ravel, J. (2017). The vaginal microbiota, host defence and reproductive physiology. The Journal of Physiology., 595(2), 451–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltani, S., Hammami, R., Cotter, P. D., Rebuffat, S., Said, L. B., Gaudreau, H., Bédard, F., Biron, E., Drider, D., & Fliss, I. (2020). Bacteriocins as a new generation of antimicrobials: Toxicity aspects and regulations. FEMS Microbiology Reviews, 45(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, H. G., Lee, J. H., & Shin, H. T. (2004). Screening and characterization of lactate dehydrogenase‐producing microorganism. Asian‐Australas J Anim Sci., 17(10), 1411–1416. [Google Scholar]
- Tachedjian, G., Aldunate, M., Bradshaw, C. S., & Cone, R. A. (2017). The role of lactic acid production by probiotic Lactobacillus species in vaginal health. Research in Microbiology., 168(9), 782–792. [DOI] [PubMed] [Google Scholar]
- Tachedjian, G., O'Hanlon, D. E., & Ravel, J. (2018). The implausible "in vivo" role of hydrogen peroxide as an antimicrobial factor produced by vaginal microbiota. Microbiome., 6(1), 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wijgert, J. H., Borgdorff, H., Verhelst, R., Crucitti, T., Francis, S., Verstraelen, H. et al (2014). The vaginal microbiota: What have we learned after a decade of molecular characterization? PLoS One, 9(8), e105998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura, M., Canchaya, C., Meylan, V., Klaenhammer, T. R., & Zink, R. (2003). Analysis, characterization, and loci of the tuf genes in lactobacillus and bifidobacterium species and their direct application for species identification. Applied and Environmental Microbiology., 69(11), 6908–6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdenelli, M. C., Coman, M. M., Cecchini, C., Silvi, S., Orpianesi, C., & Cresci, A. (2014). Evaluation of antipathogenic activity and adherence properties of human lactobacillus strains for vaginal formulations. Journal of Applied Microbiology., 116(5), 1297–1307. [DOI] [PubMed] [Google Scholar]
- Yang, E., Fan, L., Yan, J., Jiang, Y., Doucette, C., Fillmore, S., & Walker, B. (2018). Influence of culture media, pH and temperature on growth and bacteriocin production of bacteriocinogenic lactic acid bacteria. AMB Express., 8(1), 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


