Abstract
Background
Cardiogenic shock (CS) is a heterogeneous syndrome with varied presentations and outcomes. We used a machine learning approach to test the hypothesis that patients with CS have distinct phenotypes at presentation, which are associated with unique clinical profiles and in‐hospital mortality.
Methods and Results
We analyzed data from 1959 patients with CS from 2 international cohorts: CSWG (Cardiogenic Shock Working Group Registry) (myocardial infarction [CSWG‐MI; n=410] and acute‐on‐chronic heart failure [CSWG‐HF; n=480]) and the DRR (Danish Retroshock MI Registry) (n=1069). Clusters of patients with CS were identified in CSWG‐MI using the consensus k means algorithm and subsequently validated in CSWG‐HF and DRR. Patients in each phenotype were further categorized by their Society of Cardiovascular Angiography and Interventions staging. The machine learning algorithms revealed 3 distinct clusters in CS: "non‐congested (I)", "cardiorenal (II)," and "cardiometabolic (III)" shock. Among the 3 cohorts (CSWG‐MI versus DDR versus CSWG‐HF), in‐hospital mortality was 21% versus 28% versus 10%, 45% versus 40% versus 32%, and 55% versus 56% versus 52% for clusters I, II, and III, respectively. The "cardiometabolic shock" cluster had the highest risk of developing stage D or E shock as well as in‐hospital mortality among the phenotypes, regardless of cause. Despite baseline differences, each cluster showed reproducible demographic, metabolic, and hemodynamic profiles across the 3 cohorts.
Conclusions
Using machine learning, we identified and validated 3 distinct CS phenotypes, with specific and reproducible associations with mortality. These phenotypes may allow for targeted patient enrollment in clinical trials and foster development of tailored treatment strategies in subsets of patients with CS.
Keywords: cardiogenic shock, clusters, heart failure, hemodynamics, machine learning, myocardial infarction, phenotypes
Subject Categories: Myocardial Infarction, Heart Failure, Machine Learning
Nonstandard Abbreviations and Acronyms
- CS
cardiogenic shock
- CSWG
Cardiogenic Shock Working Group Registry
- DRR
Danish Retroshock MI Registry
- ML
machine learning
- SCAI
Society of Cardiovascular Angiography and Interventions
Clinical Perspective
What Is New?
Using an unbiased machine learning approach, we were able to identify 3 distinct cardiogenic shock (CS) clinical phenotypes ("noncongested," "cardiorenal," and "cardiometabolic" shock) with specific characteristics and associations with outcomes.
These phenotypes were identified and validated in CS attributable to myocardial infarction as well as acute‐on‐chronic heart failure in 2 different data sets.
Our data validate the clinical assumption that hemometabolic shock is associated with a higher mortality and stress the importance of renal function, systemic congestion, and metabolic failure for CS outcomes.
What Are the Clinical Implications?
The identified phenotypes of CS may be used by clinicians in the intensive care unit or in the catheterization laboratory to quickly assess patients with CS since only 6 baseline variables were required.
This approach could improve risk stratification, particularly by defining subsets of mortality risk within the Society of Cardiovascular Angiography and Interventions shock staging system.
These data may enhance clinical trials by developing treatment strategies tailored to a shock phenotype instead of aiming for a one‐size‐fits‐all solution, thereby paving the way for more individualized health care.
Cardiogenic shock (CS) is a heterogeneous clinical syndrome with increasing incidence and high mortality.1, 2, 3 Two primary causes of CS include acute myocardial infarction (AMI‐CS) and acute‐on‐chronic heart failure (HF‐CS).3, 4 Previous clinical trials designed to reduce mortality in CS have focused on the use of temporary mechanical circulatory support devices in AMI‐CS.4 However, up until now, trials of temporary mechanical circulatory support in AMI‐CS have not shown any significant improvement in clinical outcomes.3, 5, 6, 7 In fact, there has been little impact on the 30‐day mortality associated with CS in the past 20 years, which remains between 30% and 60%.2, 3, 8, 9
One factor that has potentially limited our ability to prove benefit of new therapies through randomized studies is our inability to "characterize" patients with CS beyond cause.10 The lack of large, comprehensive, and contemporary databases for all‐cause CS has further limited our ability to develop evidence‐based therapeutic approaches, especially in HF‐CS. As a result, attempts at staging CS have been based on expert opinions and consensus.11, 12, 13, 14 To avoid complexity, some of these classification systems include only a few variables and rely on specific, although arbitrary, cutoffs that introduce bias and fail to capture the full variability of patient profiles. The recently proposed Society of Cardiovascular Angiography and Interventions (SCAI) staging provides discriminatory potential for morbidity and mortality.15, 16 It can be used to track the severity of shock over the course of a hospital stay.
Thus, a means of appropriately phenotyping patients with CS at admission remains a much‐needed critical step in the development of treatment algorithms and prospective clinical studies to improve patient outcomes. New insights to subclassify patients with CS may be gained by using an unbiased, algorithmic approach to data analysis using machine learning (ML). Clustering algorithms (a form of unsupervised ML) have been effectively used for classification and phenotyping of other clinical syndromes and diseases, including diabetes mellitus, HF with preserved ejection fraction, and sepsis.17, 18, 19, 20
The objective of this investigation was to identify and evaluate CS phenotypes in large, contemporary CS data sets. We hypothesized that the use of ML algorithms can mathematically reduce routine clinical information at the time of presentation to discrete, reproducible phenotypes of CS. These phenotypes could further our understanding of shock physiology, inform patient selection for clinical trials, and be incorporated into clinical practice as an enhancement to our approach to risk assessment.
Methods
Overview
This study involved 2 data sets and a multistep statistical approach. The CSWG (Cardiogenic Shock Working Group Registry) includes data from patients with both AMI‐CS (CSWG‐MI) and acute‐on‐chronic systolic HF (CSWG‐HF), whereas the DRR (Danish Retroshock MI Registry) gathers data on patients with AMI‐CS. The CSWG‐MI cohort data were used as a derivation cohort for the CS phenotypes using consensus k‐means clustering applied to relevant clinical variables. We then assessed phenotype reproducibility in the DRR data set with further subsequent validation in both the DRR and the CSWG‐HF cohort. We assessed the association of phenotypes with mortality and clinical profiles across both causes: AMI and HF. Last, we studied the association between ML‐derived phenotypes and in‐hospital mortality within individual SCAI stages.
Data Sources and Study Populations
The CSWG is a multicenter database initiated in 2016 and currently includes 16 clinical sites across the United States, contributing data on patients with CS.15 For the purpose of this analysis, all available data by the 8 initial participating sites at the time of initial presentation for patients with CS between years 2016 and 2019 were included. Patients admitted to respective hospitals’ catheterization laboratories and intensive care units were screened by the clinical coordinators (retrospectively) if they met the predefined criteria for CS. The screening processes were physician adjudicated, and any patient who met the inclusion criteria was included. The registry includes a standardized set of data elements that were predefined by principal investigators and collected retrospectively. These include patient demographics, clinical presentation, procedural factors, and hospital characteristics. Patients from the CSWG with myocardial infarction (MI) as underlying cause of shock represented our derivation cohort (CSWG‐MI), whereas those with acute‐on‐chronic HF serve as a validation cohort (CSWG‐HF). Quality assurance was achieved through monitoring at each site by the respective clinical coordinators and principal investigator. Values were centrally audited and screened by the CSWG research team for any discrepancies or major outliers and resolved with the submitting site. Data collection at each US clinical site and data sharing were approved by individual site’s institutional review boards. The need for informed consent was waived because of the retrospective deidentified nature of data collection. The CSWG is housed and analyzed by the lead operational team (E.Z., M.A., and K.T.) at Tufts Medical Center.
The DRR database is derived from the DNPR (Danish National Patient Registry), which records all patient contacts within the Danish healthcare system. For this study, baseline data on patients with AMI‐CS hospitalized between 2012 and 2017 were collected retrospectively from 2 tertiary academic facilities in Denmark (Odense University Hospital and Copenhagen University Hospital Rigshospitalet) that cover nearly two thirds of the entire Danish population.1 In all cases, data were collected from medical records after being evaluated for the diagnosis of CS. The DRR was approved by the Danish Patient Safety Authority (file number: 3‐3013‐1133/1) and the Danish Data Protection Agency (file numbers: 16/7381 and 18/23756). The need for informed consent was waived. Further information on the data sources can be found in previous publications.1, 15
Definition of CS
CS diagnosis was physician adjudicated at each site based on criteria defined by the CSWG.15 CS was defined retrospectively if at least 1 of the following 3 conditions were met: (1) a sustained episode of systolic blood pressure ≤90 mm Hg for at least 30 minutes or the need for vasoactive agents to maintain such blood pressure; (2) a cardiac index <2.2 L/min per m2 determined to be secondary to cardiac dysfunction, in the absence of hypovolemia; or (3) the use of a temporary mechanical circulatory support device for suspected CS. Only adult (aged ≥18 years) patients with a known clinical outcome and sufficiently complete data for common variables in both data sets were considered for analyses. All data were recorded at an available time point as close to index hospital admission as possible.
Data Processing and Variable Handling
To derive the phenotypes, we first assessed the candidate variables’ distribution, missingness, and correlation (Data S1). Within the CSWG cohorts, variables and patients with high proportions of missing data were removed from the derivation data set, to ensure that overall missingness did not exceed 10% for imputation (Data S1). Any remaining missing values were imputed with random forest imputation. For the validation DRR cohort, only patients with complete data for clustering were included, to exclude any bias introduced by imputation. To rule out any potential bias introduced because of the exclusion step, we performed sensitivity analyses of CSWG‐HF and DRR without removal of any patients. To reduce variable collinearity and to limit complexity and dimensionality, we used a classification algorithm to select variables from continuous clinical and laboratory data (Data S1).21 Concretely, we used a random forest classifier to identify demographic and laboratory variables that predicted in‐hospital mortality in the CSWG‐MI derivation cohort because it does not assume linear relationships between variables and removed highly correlated variables (|r|>0.6), leaving 6 variables to be included in the final analyses (Data S1).
Cluster Analyses and Validation
Clustering is an ML technique used to identify homogeneous subgroups within data, such that data points in each cluster are as similar as possible while being as different from other clusters as possible. K‐means clustering is one of the most common unsupervised algorithms that iteratively tries to partition the data set into distinct, nonoverlapping subgroups (clusters) where each data point belongs to only one group. We deployed semisupervised consensus k‐means clustering on the 6 identified variables with highest predictive value using the ConsensusClusterPlus package22 for R 3.6.0 and defined the optimal number of clusters (k) using several metrics (Data S1). We selected various methods to visualize the clusters (t‐distributed stochastic neighbor embedding plots,23 chord diagrams, and rank plots) and their characteristics which are detailed in the Supplemental Material.
For validation, first, cases in the DRR were clustered independently using the same variables and cluster algorithm as in the derivation cohort to assess external reproducibility. Phenotype characteristics were then scrutinized within and between these cohorts to assess whether a similar pattern can be seen when the 2 cohorts are clustered completely independently. Finally, cases in DRR and CSWG‐HF were assigned to a respective cluster using the centroids from the clusters in the CSWG MI derivation cohort (Data S1) to validate applicability of the clusters in external data sets and future clinical practice. Distribution, mortality, and characteristics of the clusters in the validation cohorts were compared with those in the derivation CSWG‐MI cohort.
Application of SCAI Shock Classification Scheme
To put the novel phenotypes into clinical context, we assessed the SCAI shock stages in the CSWG cohort and combined this established method of CS classification to our proposed phenotypes. Each patient was categorized into the most severe SCAI stage encountered during his/her hospital stay, using a method previously described by Thayer et al.15 Of note, the phenotypes were derived from data at the time of initial presentation, whereas the SCAI staging was denoted as the "highest stage" during hospitalization.
Statistical Analysis
P values were calculated using ANOVA, Kruskal‐Wallis test, or χ2 test. Continuous data are displayed as mean and SD or as median and interquartile ranges, depending on distribution. All statistical tests were performed using SAS Enterprise Guide 9.4 (SAS Institute Inc, Cary, NC) and Python 3.7.4 (Python Software Foundation, DE). Figures were created using Python 3.7.4 and GraphPad Prism 8.2.1 (GraphPad Software, Inc, San Diego, CA). Statistical significance threshold was P<0.05.
Results
A total of 1959 patients (410 in CSWG‐MI cohort, 1069 in DRR cohort, and 480 in CSWG‐HF cohort) were eligible for final analyses (Figure S1). Mean ages at presentation were 65±13, 66±11, and 57±14 years, whereas male sex represented 68%, 77%, and 75% patients in the CSWG‐MI, DRR, and CSWG‐HF cohorts, respectively (Table 1). The in‐hospital mortality from CS was noted to be 39% in CSWG‐MI cohort, 45% in DRR cohort, and 26% in CSWG‐HF cohort. The AMI‐CS cohorts (CSWG‐MI and DRR) were similar on various clinical variables but differed in age and treatment strategy, especially in use of temporary mechanical circulatory support (Table 1). CSWG‐HF differed from the MI cohorts on several clinical features, including younger age, higher body weight, more severe kidney and liver injury, and lower lactate levels (Table 1).
Table 1.
Cohort Characteristics
| Characteristics | CSWG‐MI Cohort | DRR Cohort | CSWG‐HF Cohort | |||
|---|---|---|---|---|---|---|
| No. | (%) | No. | (%) | No. | (%) | |
| Nonsurvivors | 161 | 39.27 | 478 | 44.71 | 127 | 26.46 |
| Men | 279 | 68.05 | 819 | 76.61 | 362 | 75.42 |
| IABP | 249 | 60.73 | 127 | 11.88 | 200 | 41.67 |
| ECMO | 128 | 31.22 | 43 | 4.02 | 93 | 19.38 |
| Impella | 170 | 41.46 | 152 | 14.22 | 109 | 22.71 |
| Any t‐MCS | 395 | 96.34 | 288 | 26.99 | 325 | 67.71 |
| No t‐MCS | 15 | 3.66 | 799 | 73.01 | 155 | 32.29 |
| Multiple t‐MCS | 134 | 32.68 | 34 | 3.19 | 76 | 15.83 |
| Mechanical ventilation | 242 | 59.02 | Not captured | Not captured | 163 | 33.96 |
| Vasopressor/inotrope use | 318 | 77.56 | 1032 | 96.54 | 384 | 80 |
| Vasodilators | 62 | 15.12 | Not captured | Not captured | 201 | 41.88 |
| History of hypertension | 281 | 68.54 | 512 | 47.9 | 216 | 45 |
| History of CKD (any stage) | 73 | 17.8 | Not captured | Not captured | 169 | 35.21 |
| History of COPD | 24 | 5.85 | 104 | 9.73 | 49 | 10.21 |
| History of CVA/TIA | 57 | 13.9 | 85 | 7.95 | 76 | 15.83 |
| Prior HF | 92 | 22.44 | Not captured | Not captured | 363 | 75.63 |
| Prior MI | 110 | 26.83 | 152 | 14.22 | 132 | 27.5 |
| History of PCI | 138 | 33.66 | Not captured | Not captured | 94 | 19.58 |
| History of CABG | 33 | 8.05 | Not captured | Not captured | 45 | 9.38 |
| History of diabetes mellitus | 179 | 43.66 | 173 | 16.18 | 142 | 29.58 |
| History of PVD | 20 | 4.88 | 77 | 7.2 | 18 | 3.75 |
| Mean | SD | Mean | SD | Mean | SD | |
|---|---|---|---|---|---|---|
| Age, y | 65.12 | 13.27 | 66 | 11.04 | 56.87 | 14.38 |
| Weight, kg | 81.9 | 18.81 | 81.11 | 15.77 | 86.69 | 22.24 |
| Laboratory Values (Normal Range) | Mean | SD | % OOR | Mean | SD | % OOR | Mean | SD | % OOR |
|---|---|---|---|---|---|---|---|---|---|
| Sodium, mEq/L | 137.16 | 4.2 | 137.88 | 4.52 | 134.22 | 5.53 | |||
| Potassium, mEq/L (3.6–5.1) | 4.3 | 0.73 | 23.82 | 4.07 | 0.78 | 36.84 | 4.26 | 0.69 | 23.48 |
| HCO3, mEq/L (21–28) | 20.2 | 4.79 | 55.00 | 18.47 | 4.69 | 73.71 | 24.65 | 4.95 | 36.25 |
| BUN, mg/dL (6–24) | 27.83 | 16.65 | 55.40 | 24.6 | 16.14 | 33.84 | 37.86 | 22.73 | 68.00 |
| Creatine, mg/dL (0.6–1.3) | 1.61 | 1.08 | 50.64 | 1.48 | 1.07 | 44.43 | 1.84 | 1.18 | 64.77 |
| WBC, 103/mm3 (4–11) | 14.59 | 7.09 | 64.19 | 16.39 | 6.71 | 81.01 | 10.77 | 5.69 | 35.61 |
| Hemoglobin, g/dL (11–16) | 12.52 | 2.58 | 39.34 | 13.54 | 2.11 | 21.68 | 11.98 | 2.34 | 37.35 |
| Hematocrit, % (32–47) | 37.33 | 7.53 | 34.81 | 39.04 | 7.33 | 31.59 | 36.79 | 6.76 | 29.92 |
| Platelets, 103/mm3 (150–400) | 217.94 | 88.94 | 24.79 | 247.27 | 89.01 | 14.13 | 198.95 | 78.57 | 30.42 |
| ALT, U/L (<55) | 173.17 | 392.18 | 43.49 | 198.81 | 464.87 | 34.80 | 237.83 | 774.43 | 65.44 |
| Total bilirubin, mg/dL (0.2–1.1) | 0.88 | 0.55 | 23.97 | 0.77 | 0.93 | 19.15 | 1.78 | 2.05 | 46.80 |
| INR (0.9–1.3) | 1.38 | 0.47 | 67.83 | 1.27 | 0.51 | 21.12 | 1.95 | 1.18 | 37.85 |
| GFR, mL/min per 1.73 m2 (>90) | 55.37 | 26.83 | 88.49 | 58.29 | 23.55 | 89.71 | 50.48 | 26.33 | 94.30 |
| Lactate, mEq/L (0.5–2.2) | 4.54 | 3.99 | 35.46 | 5.76 | 4.37 | 81.67 | 3.94 | 4.14 | 51.30 |
| pH (7.35–7.45) | 7.28 | 0.15 | 68.20 | 7.26 | 0.13 | 82.76 | 7.35 | 0.14 | 60.38 |
| Hemodynamic Values | Mean | SD | Mean | SD | Mean | SD |
|---|---|---|---|---|---|---|
| MAP, mm Hg | 74.72 | 16.71 | 64.09 | 11.79 | 73.06 | 13.14 |
| Cardiac index, L/min per m2 | 1.88 | 0.59 | 1.91 | 0.58 | ||
| Cardiac output, L/min | 3.71 | 1.49 | 4.04 | 2.8 | ||
| CPI, W/m2 | 0.31 | 0.13 | 0.31 | 0.11 | ||
| CPO, W | 0.61 | 0.3 | 0.66 | 0.43 | ||
| LVEDD, mm | 4.79 | 0.92 | 6.5 | 1.18 | ||
| Heart rate, 1/min | 91.21 | 22.35 | 86.02 | 23.99 | 91.65 | 22.44 |
| DBP, mm Hg | 60.41 | 15.91 | 52.94 | 11.31 | 61.73 | 12.89 |
| SBP, mm Hg | 100.85 | 23.85 | 83.71 | 14.27 | 94.47 | 16.16 |
| PCWP, mm Hg | 24.39 | 9.38 | 24.12 | 8.76 | ||
| PADP, mm Hg | 23.69 | 7.71 | 26.15 | 8.45 | ||
| PASP, mm Hg | 43.84 | 13.92 | 48.87 | 14.37 | ||
| Mean PAP, mm Hg | 30.41 | 9.16 | 33.74 | 9.82 | ||
| RAP, mm Hg | 14.72 | 6.63 | 12.56 | 5.07 | 13.68 | 7.28 |
| PAPI, arbitrary units | 1.83 | 2.48 | 2.7 | 3.42 | ||
| RVSWI, mm Hg×mL/m2 | 4.67 | 2.98 | 5.9 | 3.24 |
Table displays only nonimputed data. The proportions of patients with abnormal values are calculated using the normal values from the laboratory at Tufts Medical Center and do not necessarily represent the normal ranges for each participating site. ALT indicates alanine aminotransferase; BUN, blood urea nitrogen; CABG, coronary artery bypass grafting; CKD, chronic kidney disease; COPD, chronic obstructive pulmonary disease; CPI, cardiac power index; CPO, cardiac power output; CSWG, Cardiogenic Shock Working Group Registry; CVA, cerebrovascular accident; DBP, diastolic blood pressure; DRR, Danish Retroshock MI Registry; ECMO, extracorporeal membrane oxygenation; GFR, glomerular filtration rate; HCO3, sodium bicarbonate; HF, heart failure; IABP, intra‐aortic balloon pump; INR, International Normalized Ratio; LVEDD, left ventricular end‐diastolic dimension; MAP, mean arterial pressure; MI, myocardial infarction; OOR, out of range; PADP, pulmonary artery diastolic pressure; PAP, pulmonary artery pressure; PAPI, pulmonary artery pulsatility index; PASP, pulmonary artery systolic pressure; PCI, percutaneous coronary intervention; PCWP, pulmonary capillary wedge pressure; PVD, peripheral vascular disease; RAP, right atrial pressure; RVSWI, right ventricular stroke volume index; SBP, systolic blood pressure; TIA, transient ischemic attack; t‐MCS, temporary mechanical circulatory support; and WBC, white blood cell count.
The 8 clinical variables that were most associated with in‐hospital mortality were: glomerular filtration rate, serum bicarbonate, serum lactate, alanine aminotransferase, platelet count, serum creatinine, white blood cell count, and blood urea nitrogen levels (Figure S2). Of these, glomerular filtration rate, lactate, serum bicarbonate, and alanine aminotransferase were the most predictive ones (Figure S2).
Phenotypes in CS
Consensus k‐means clustering in the CSWG‐MI derivation cohort identified 3 as the optimal number (k) of clusters, based on the calculated silhouette score, cluster consensus, and other metrics (Figure 1 and Figure S3). These clusters demonstrated important differences beyond the variables used to design them, suggesting that the clustering algorithm successfully identified 3 discrete clusters of CS that were able to identify as distinct clinical phenotypes. On the basis of their clinical characteristics, we labeled the 3 phenotypes as "noncongested (I)," "cardiorenal shock (II)," and "cardiometabolic shock (III).”
Figure 1. Derivation of the clusters: consensus k‐means clustering.
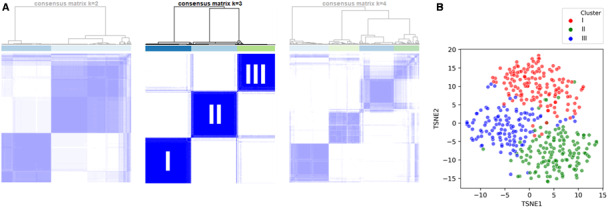
A, Representative plots illustrating the method of consensus k‐means clustering in the CSWG (Cargiogenic Shock Working Group Registry) myocardial infarction derivation cohort. Each column represents one patient, whereas each row displays the assigned clusters. Well‐defined (ie, segregated) squares indicate stable clusters. Compared with k (number of clusters)=2 and k=4, k=3 shows highest cluster stability, suggesting that 3 may be a good choice for the number of clusters. B, A t‐distributed stochastic neighbor embedding (TSNE) plot for visual representation of the clusters in a 2‐dimensional space. The algorithm uses probability estimates to calculate the similarity of data points in the high‐dimensional space (ie, identifies the “neighbors”) and then calculates the distance of these “neighbors” in a lower‐dimensional space (in this case, 2 dimensions).23 The wider the different clusters separate in the plot, the larger is the difference between them.
The 3 phenotypes differed from one another on various demographic and clinical parameters (Table 2). Radar plots were used to display the deviation of metabolic and hemodynamic values from the mean value (derived from the total derivation cohort average values) (Figure 2), and chord plots were used to reveal the sources of phenotype differences (Figure S4). The noncongested phenotype (I) exhibited lower heart rate, filling pressures (right atrial and pulmonary capillary wedge pressures), and a higher blood pressure relative to the other phenotypes. This represents a relatively stable profile of a noncongested patient with CS. In contrast, the patients in the cardiorenal shock (II) group were older, with multiple comorbidities. They exhibited a lower heart rate, elevated pulmonary arterial and pulmonary capillary wedge pressures, as well as lower glomerular filtration rate, suggesting renal involvement from shock. Last, the patients in the cardiometabolic shock (III) group exhibited elevated lactate, alanine aminotransferase, heart rate, and right atrial pressure, along with low blood pressure, cardiac power output, and index. This suggested a multiorgan involvement, featured by transaminases and lactic acidosis in a patient with CS.
Table 2.
Selection of Outstanding Characteristics of the Phenotypes
| Characteristics |
Cluster/Phenotype I “Noncongested” CS |
Cluster/Phenotype II “Cardiorenal” CS |
Cluster/Phenotype III “Cardiometabolic” CS |
|---|---|---|---|
| Mean age, y | ≈60 | ≈70 | ≈65 |
| Comorbidities | Few | DM2, CKD, hypertension… | Few |
| Blood pressure | ↓ | ↓ | ↓↓ |
| Congestion | None | Left ventricular | Right ventricular |
| Heart rate | ↔ | ↔ | ↑↑ |
| Hemoglobin | ↔ | ↓ | ↔ |
| Transaminases | ↔ | ↔ | ↑↑ |
| Lactate | ↔ or ↑ | ↓ | ↑↑ |
| Kidney function | ↔ | ↓↓ | ↓ |
CKD indicates chronic kidney disease; CS, cardiogenic shock; and DM2, type 2 diabetes mellitus.
Figure 2. Metabolic and hemodynamic profiles of the different phenotypes.
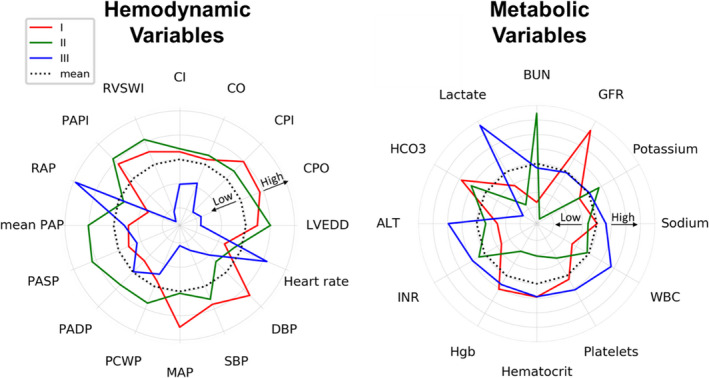
Radar plots illustrate the association of each phenotype with hemodynamic and metabolic variables in CSWG( Cargiogenic Shock Working Group Registry) myocardial infarction cohort. Data were normalized across all phenotypes to a mean of 0 and an SD of 1. The dashed black line marks the mean (0), whereas every concentric gray line signifies a 0.1‐SD difference from the overall mean. Values that were higher than the mean are drawn outside, whereas values that were lower than the mean are drawn inside the dashed line for each variable. ALT indicates alanine aminotransferase; BUN, blood urea nitrogen; CI, cardiac index; CO, cardiac output; CPI, cardiac power index; CPO, cardiac power output; DBP, diastolic blood pressure; GFR, glomerular filtration rate; HCO3, sodium bicarbonate; Hgb, hemoglobin; INR, International Normalized Ratio; LVEDD, left ventricular end‐diastolic dimension; MAP, mean arterial pressure; PADP, pulmonary artery diastolic pressure; PAP, pulmonary artery pressure; PAPI, pulmonary artery pulsatility index; PASP, pulmonary artery systolic pressure; PCWP, pulmonary capillary wedge pressure; RAP, right atrial pressure; RVSWI, right ventricular stroke work index; SBP, systolic blood pressure; and WBC, white blood cell count.
Validation of Phenotypes
First‐pass validation of the derived phenotypes was performed to test external reproducibility by using the same consensus k‐means clustering algorithm de novo in the DRR validation cohort. Using this approach, similar patterns of the clusters were identified compared with the derivation cohort (Figures S5 and S6). In the second validation procedure, we assigned patients from DRR and CSWG‐HF to the derived phenotypes using a classifier based on the cluster centroids of the derivation cohort to test for potential future clinical application (Figure S7). Both validation cohorts revealed a similar trend in mortality and comparable clinical profiles across the 3 clusters, as was seen in the independent clustering of the CSWG‐MI and DRR cohorts (Table S1 and Figure S8). These trends were consistent in all sensitivity analyses, including the imputation of all missing values in CSWG‐HF and DRR instead of removing cases with a large number of missing values (Tables S2 and S3 and Figure S9).
Association of Phenotypes With Outcomes
In‐hospital mortality rates differed among phenotypes but were similar within a given phenotype, even across cohorts. Relative to phenotype I, patients in phenotype II had a higher mortality (CSWG‐MI odds ratio [OR], 3 [95% CI, 1.8–5.1]; DRR OR, 2 [95% CI, 1.4–2.8]), whereas those in phenotype III (CSWG‐MI OR, 4.6 [95% CI, 2.7–5.1]; DRR OR, 3.5 [95% CI, 2.5–4.9]) were at highest risk of dying (Figure 3).
Figure 3. In‐hospital mortality in the 3 distinct phenotypes of cardiogenic shock (CS).
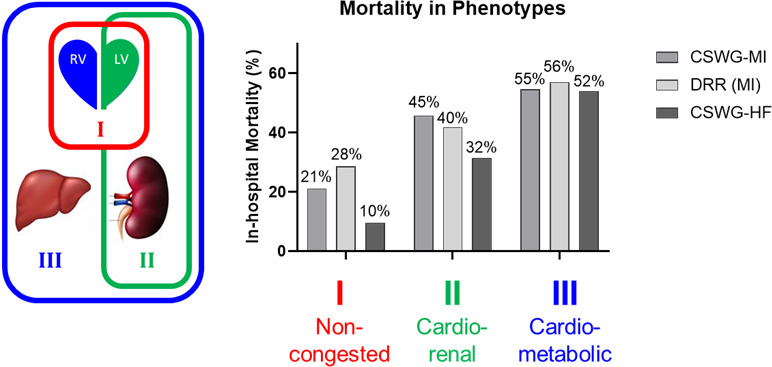
Phenotype I (noncongested), phenotype II (cardiorenal), and phenotype III (cardiometabolic) are associated with in‐hospital mortality across 2 international multicenter registries of CS attributable to acute myocardial infarction (MI) and a multicenter registry of CS attributable to acute‐on‐chronic heart failure. CSWG indicates Cardiogenic Shock Working Group Registry; and DRR, Danish Retroshock MI Registry.
Phenotypes and Shock Stages
We compared the distribution of each phenotypes’s patients with regard to their most severe SCAI shock stage encountered during the hospital stay (Figure 4). The risk of developing stage D or E shock during the hospital stay was lowest in phenotype I and highest in phenotype III for patients with both CS‐MI and CS‐HF. Within each phenotype, the SCAI staging (C‐E) further stratified mortality. Similarly, within each SCAI stage, the 3 phenotypes further stratified mortality. In a bivariate logistic regression model, both SCAI stage (C‐E) and phenotype were significant predictors (P<0.0001) of in‐hospital mortality in CSWG‐MI, CSWG‐HF, and CSWG‐MI and CSWG‐HF combined (Table S4).
Figure 4. Society for Cardiovascular Angiography and Interventions (SCAI) stages reached during hospital stay by phenotype.
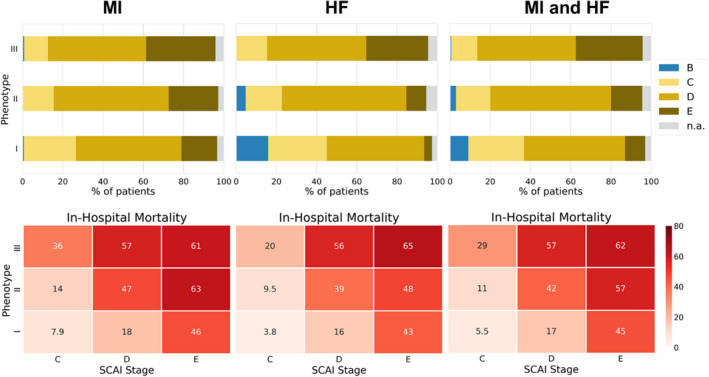
Upper panel: percentage of patients in each phenotype (I: noncongested; II: cardiorenal; III: cardiometabolic) reaching SCAI stage B, C, D, or E during their hospitalization in CSWG (Cardiogenic Shock Working Group Registry) myocardial infarction (MI) cohort, CSWG heart failure (HF) cohort, and both cohorts combined. These graphs can be read as follows: for example, a patient in admission phenotype 3 had a chance of >80% to reach SCAI stage D or E during his/her hospitalization independent of cause of shock (MI or HF). Bottom panel: in‐hospital mortality stratified by phenotype and SCAI stage in percentage for the CSWG‐MI cohort, CSWG‐HF cohort, and both cohorts combined. These graphs can be read as follows: For example, a patient with MI in admission phenotype 2 who only reached SCAI stage C had a probability of 14% for in‐hospital death; however, if the same patient reaches SCAI stage E, his/her probability to die in hospital is 63%. n.a. indicates data insufficient to assign SCAI stage.
Discussion
This is the first study using ML approaches in large, multicenter cohorts of patients with CS. Using this novel approach, we identified 3 distinct clusters of CS patient profiles in our derivation cohort of patients with MI‐CS from the CSWG. Next, we tested the reproducibility of our analysis in an independent, Danish registry of patients with MI‐CS and in patients with HF‐CS from the CSWG and identified the same 3 clusters with matching rates of in‐hospital mortality. These CS phenotypes exhibited distinct demographic, hemodynamic, and metabolic signatures and correlated with inpatient mortality. The "noncongested" (phenotype I) group represented lowest mortality, and the "cardiometabolic shock" (phenotype III) group resulted in highest mortality across all cohorts. We further identified that these 3 clusters were directly associated with mortality within individual SCAI stages. Accordingly, these findings may improve risk stratification and enable the development of treatment algorithms tailored to each phenotype of CS and inform patient selection in future clinical trials in CS.
Major discriminators of phenotype membership included glomerular filtration rate, lactate, serum bicarbonate, and alanine aminotransferase, which have all previously been associated with risk of mortality in CS.24, 25 Previously, SCAI staging was shown to have a strong correlation with mortality in a heterogeneous intensive care unit population.16 Patients with refractory shock (SCAI shock stage E) were shown to have a significantly higher in‐hospital mortality than other stages, regardless of cause.16 Identifying the clinical CS phenotypes that impact outcomes could allow for early classification into treatment groups. In our analysis, SCAI stage C had the lowest and stage E, the highest, mortality in each individual phenotype cluster. These findings support that the 3 phenotypes of CS reflect clinically relevant features that are expected to be associated with mortality. Therefore, the phenotypes seem to be compatible with the SCAI staging system and provide further support to it in guiding even more individualized therapy. Unlike the SCAI staging system, whose aim is to characterize disease severity as it evolves over the course of a hospital stay, the aim of the current analysis was the identification of CS phenotypes at the time of admission.
Although “labeling” phenotypes with 1‐dimensional titles (noncongested, cardiorenal, and cardiometabolic) may obscure the fascinating mathematical interactions of the entirety of variables defining the phenotypes,21 each phenotype was composed of strikingly different characteristics: young patients with few comorbidities, normal hematocrit, low lactate, and good kidney function in phenotype I; older patients with chronic kidney disease, diabetes mellitus, hypertension, and anemia in phenotype II; and patients with lactic acidosis, inflammation, and elevated liver enzymes in phenotype III. Hemodynamically, phenotype I was characterized by relatively high cardiac output and blood pressures; phenotype II exhibited pulmonary congestion (high wedge pressure); phenotype III displayed systemic congestion (high central venous pressure but low wedge pressure, suggesting involvement of right‐sided HF), high heart rate, and low cardiac output and arterial blood pressure. Taken together, these findings suggest that phenotype I represents the noncongested patient with high likelihood of salvage. Phenotype II, on the other hand, may involve more left‐sided HF with worsening kidney function (cardiorenal), whereas phenotype III represents worsening venous congestion, liver damage, and likelihood of multiorgan involvement. Identifying these distinct phenotypes may change the way we classify patients with CS, each of whom may benefit from a different management strategy that could be tested in prospective trials.
Reproducibility and external validity are important measurements of cluster quality.21 A strength of this study is the finding that similar phenotypes evolved when we clustered 2 completely independent AMI‐CS cohorts on the same variables, especially as these cohorts represent patients from 2 countries on different continents with different healthcare systems and treatment strategies. Future studies may validate these phenotypes globally, even further supporting the universal existence of 3 distinct groups of CS with sufficiently nonoverlapping characteristics and prognosis, such that patients can be classified on presentation. In addition, many of the phenotypes’ clinical characteristics align with existing clinical consensus, including the concepts that venous congestion and high lactate are associated with increased mortality, making their incorporation into practice more feasible.
The phenotypes of CS may also allow for the stratification of patients in clinical trials, both past and future, which could improve their design and interpretation. The inability of randomized controlled trials in CS to demonstrate significant differences in outcomes, despite technological advances and improvement in hemodynamic markers, implicates heterogeneity as a potential confounder. Previous work with ML in sepsis (another clinically heterogeneous population) has shown clinical phenotypes within large, randomized trials derive harm or benefit from an intervention, which may be opposite from the larger population when all patients are grouped together.18
Although the 3 phenotypes of CS were derived from patients with AMI‐CS, these were equally applicable to patients with HF‐CS. Despite baseline differences between the HF‐CS and AMI‐CS cohorts, the clinical presentation and mortality trends of the phenotypes were consistent, regardless of whether they were derived from patients with HF‐CS or AMI‐CS. This finding indicates applicability in both the 2 most common forms of CS.
Creating phenotypes in CS through ML has the potential to address both implicit biases and identify subgroups within large populations that may derive harm or benefit from a certain therapeutic intervention. Information gained from this type of data analysis may also impact future clinical trials. Although we found similar treatment protocols across all phenotypes in our analysis, the vast differences in mortality we report between these phenotypes stress the importance of more granularity in defining patient characteristics and phenotypes in future treatment protocols and clinical trial designs.
Limitations
Major limitations of this study include the retrospective nature of the data sources, limited number of patients and clinical variables, and the inability to assess long‐term outcomes. ML algorithms tend to perform better on larger data sets, and the sample size may limit the complexity of a cluster model. However, there is currently a lack of large, comprehensive, contemporary multicenter CS databases that represent all causes of CS. Within these limitations, we were able to demonstrate and validate reproducible, clinically relevant phenotypes in CS using ML algorithms, which were associated with mortality. Future studies may collect comprehensive data in a prospective manner and may allow for enhanced, even more nuanced examination of the CS phenotypes.
In addition, the clusters in this article are semisupervised. Operating on the assumption that variables driving mortality are clinically the most relevant variables in CS, we identified important variables using supervised ML based on mortality association before applying an unsupervised clustering algorithm (semisupervised learning). However, because the phenotypes differed in several clinical features, even beyond the variables included in the clustering algorithm, this is likely to be the case for characteristics not captured in our data sets as well, thus providing a comprehensive classification. The selected ML algorithm, (consensus) k means, is a well‐established algorithm in unsupervised learning,17, 18 particularly in medical research, but is not simply applicable to categorical data; thus, only continuous variables were used for clustering. Renal impairment had a major impact on cluster assignment but did not discriminate between acute and chronic kidney injury. Furthermore, although it may be a limitation that the calendar time frames of data collection for the CSWG and DRR cohorts were not identical, they do overlap and changes in CS treatment protocol were not significant enough to introduce any bias in our results. Although we were able to assess the maximum SCAI stage across the patients’ hospital course, based on drug and device escalation, the initial SCAI stage at admission could not be validly identified retrospectively because there was only admission data for most patients in CSWG, and for patients to be assigned SCAI stage D or E, they would have to fail to respond to initial interventions first.15 Taken together, these limitations identify the need for new, larger, prospective CS registries that account for temporal changes in patient variables and treatment to improve our understanding of the nature of these phenotypes.
Conclusions
We report that using an ML approach to define unbiased clusters in the complex and heterogeneous clinical syndrome of CS is feasible. Our analysis of multicenter cohorts of patients with CS identified 3 distinct phenotypes of CS with unique clinical profiles that are associated with different risks of in‐hospital mortality. These clusters exhibited clinically relevant differences in hemodynamic and metabolic profiles and are equally applicable to patients with CS attributable to MI or acute‐on‐chronic HF. Future studies are needed to evaluate the clinical implications of these phenotypes as they relate to prognosis and optimal treatment.
Sources of Funding
This work was supported by a NIH RO1 grant to NKK (RO1HL139785‐01) and institutional grants from Abbott Laboratories Inc (Abbott Park, IL), Abiomed Inc (Danvers, MA), Boston Scientific Inc (Minneapolis, MN), and Getinge Inc (Wayne, NJ) to Tufts Medical Center and by a grant from the Danish Heart Foundation (16‐R107‐A6576) to OKLH.
Disclosures
Dr Garan is an unpaid consultant for Abiomed Inc. Dr Hernandez‐Montfort is a consultant for Abiomed Inc (research and education). Dr Burkhoff reports an unrestricted, educational grant from Abiomed Inc to Cardiovascular Research Foundation. Dr Vorovich is a consultant and in the speakers’ bureau of Abiomed Inc. Dr Abraham is a consultant for Abbott Laboratories and Abiomed Inc. Dr Møller receives speaker honoraria and a research grant from Abiomed Inc. Dr Kapur receives consulting/speaker honoraria and institutional grant support from: Abbott Laboratories, Abiomed Inc, Boston Scientific, Edwards, Medtronic, Getinge, LivaNova, MDStart, Precardia, and Zoll. Dr Sinha is a consultant for Abiomed Inc (Critical Care Advisory Board). Dr O’Neill receives consulting/speaker honoraria from Abiomed Inc, Boston Scientific Inc, and Abbott Laboratories. None of the listed disclosures could be perceived as a competing interest for the content of this article. The remaining authors have no disclosures to report.
Supporting information
Supplementary Material for this article is available at https://www.ahajournals.org/doi/suppl/10.1161/JAHA.120.020085
For Sources of Funding and Disclosures, see page 11.
References
- 1.Helgestad OKL, Josiassen J, Hassager C, Jensen LO, Holmvang L, Sørensen A, Frydland M, Lassen AT, Udesen NLJ, Schmidt H, et al. Temporal trends in incidence and patient characteristics in cardiogenic shock following acute myocardial infarction from 2010 to 2017: a Danish cohort study. Eur J Heart Fail. 2019;21:1370–1378. DOI: 10.1002/ejhf.1566. [DOI] [PubMed] [Google Scholar]
- 2.Kolte D, Khera S, Aronow WS, Mujib M, Palaniswamy C, Sule S, Jain D, Gotsis W, Ahmed A, Frishman WH, et al. Trends in incidence, management, and outcomes of cardiogenic shock complicating ST‐elevation myocardial infarction in the United States. J Am Heart Assoc. 2014;3:e000590. DOI: 10.1161/JAHA.113.000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Diepen S , Katz JN, Albert NM, Henry TD, Jacobs AK, Kapur NK, Kilic A, Menon V, Ohman EM, Sweitzer NK, et al. Contemporary management of cardiogenic shock: a scientific statement from the American Heart Association. Circulation. 2017;136:e232–e268. DOI: 10.1161/CIR.0000000000000525. [DOI] [PubMed] [Google Scholar]
- 4.Thiele H, Ohman EM, de Waha‐Thiele S , Zeymer U, Desch S. Management of cardiogenic shock complicating myocardial infarction: an update 2019. Eur Heart J. 2019;40:2671–2683. DOI: 10.1093/eurheartj/ehz363. [DOI] [PubMed] [Google Scholar]
- 5.Thiele H, Jobs A, Ouweneel DM, Henriques JPS, Seyfarth M, Desch S, Eitel I, Poss J, Fuernau G, de Waha S . Percutaneous short‐term active mechanical support devices in cardiogenic shock: a systematic review and collaborative meta‐analysis of randomized trials. Eur Heart J. 2017;38:3523–3531. DOI: 10.1093/eurheartj/ehx363. [DOI] [PubMed] [Google Scholar]
- 6.Hochman JS, Sleeper LA, Webb JG, Sanborn TA, White HD, Talley JD, Buller CE, Jacobs AK, Slater JN, Col J, et al. Early revascularization in acute myocardial infarction complicated by cardiogenic shock: Shock Investigators: should we emergently revascularize occluded coronaries for cardiogenic shock. N Engl J Med. 1999;341:625–634. DOI: 10.1056/NEJM199908263410901. [DOI] [PubMed] [Google Scholar]
- 7.Werdan K, Gielen S, Ebelt H, Hochman JS. Mechanical circulatory support in cardiogenic shock. Eur Heart J. 2014;35:156–167. DOI: 10.1093/eurheartj/eht248. [DOI] [PubMed] [Google Scholar]
- 8.Goldberg RJ, Makam RC, Yarzebski J, McManus DD, Lessard D, Gore JM. Decade‐long trends (2001‐2011) in the incidence and hospital death rates associated with the in‐hospital development of cardiogenic shock after acute myocardial infarction. Circ Cardiovasc Qual Outcomes. 2016;9:117–125. DOI: 10.1161/CIRCOUTCOMES.115.002359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becher PM, Schrage B, Sinning CR, Schmack B, Fluschnik N, Schwarzl M, Waldeyer C, Lindner D, Seiffert M, Neumann JT, et al. Venoarterial extracorporeal membrane oxygenation for cardiopulmonary support. Circulation. 2018;138:2298–2300. DOI: 10.1161/CIRCULATIONAHA.118.036691. [DOI] [PubMed] [Google Scholar]
- 10.Burkhoff D, Garan AR, Kapur NK. The SCAI cardiogenic shock staging system gets taken for a test drive. J Am Coll Cardiol. 2019;74:2129–2131. DOI: 10.1016/j.jacc.2019.08.1020. [DOI] [PubMed] [Google Scholar]
- 11.Baran DA, Grines CL, Bailey S, Burkhoff D, Hall SA, Henry TD, Hollenberg SM, Kapur NK, O'Neill W, Ornato JP, et al. SCAI clinical expert consensus statement on the classification of cardiogenic shock: this document was endorsed by the American College of Cardiology (ACC), the American Heart Association (AHA), the Society of Critical Care Medicine (SCCM), and the Society of Thoracic Surgeons (STS) in April 2019. Catheter Cardiovasc Interv. 2019;94:29–37. DOI: 10.1002/ccd.28329. [DOI] [PubMed] [Google Scholar]
- 12.Killip T, Kimball JT. Treatment of myocardial infarction in a coronary care unit. Am J Cardiol. 1967;20:457–464. DOI: 10.1016/0002-9149(67)90023-9. [DOI] [PubMed] [Google Scholar]
- 13.Diamond GA, Forrester JS. Analysis of probability as an aid in the clinical diagnosis of coronary‐artery disease. N Engl J Med. 1979;300:1350–1358. DOI: 10.1056/NEJM197906143002402. [DOI] [PubMed] [Google Scholar]
- 14.Stevenson LW, Pagani FD, Young JB, Jessup M, Miller L, Kormos RL, Naftel DC, Ulisney K, Desvigne‐Nickens P, Kirklin JK. INTERMACS profiles of advanced heart failure: the current picture. J Heart Lung Transplant. 2009;28:535–541. DOI: 10.1016/j.healun.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 15.Thayer KL, Zweck E, Ayouty M, Garan AR, Hernandez‐Montfort J, Mahr C, Morine KJ, Newman S, Jorde L, Haywood JL, et al. Invasive hemodynamic assessment and classification of in‐hospital mortality risk among patients with cardiogenic shock. Circ Heart Fail. 2020;13:e007099. DOI: 10.1161/CIRCHEARTFAILURE.120.007099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jentzer JC, van Diepen S , Barsness GW, Henry TD, Menon V, Rihal CS, Naidu SS, Baran DA. Cardiogenic shock classification to predict mortality in the cardiac intensive care unit. J Am Coll Cardiol. 2019;74:2117–2128. [DOI] [PubMed] [Google Scholar]
- 17.Ahlqvist E, Storm P, Käräjämäki A, Martinell M, Dorkhan M, Carlsson A, Vikman P, Prasad RB, Aly DM, Almgren P, et al. Novel subgroups of adult‐onset diabetes and their association with outcomes: a data‐driven cluster analysis of six variables. Lancet Diabetes Endocrinol. 2018;6:361–369. DOI: 10.1016/S2213-8587(18)30051-2. [DOI] [PubMed] [Google Scholar]
- 18.Seymour CW, Kennedy JN, Wang S, Chang C‐C, Elliott CF, Xu Z, Berry S, Clermont G, Cooper G, Gomez H, et al. Derivation, validation, and potential treatment implications of novel clinical phenotypes for sepsis. JAMA. 2019;321:2003–2017. DOI: 10.1001/jama.2019.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaharia OP, Strassburger K, Strom A, Bönhof GJ, Karusheva Y, Antoniou S, Bódis K, Markgraf DF, Burkart V, Müssig K, et al. Risk of diabetes‐associated diseases in subgroups of patients with recent‐onset diabetes: a 5‐year follow‐up study. Lancet Diabetes Endocrinol. 2019;7:684–694. DOI: 10.1016/S2213-8587(19)30187-1. [DOI] [PubMed] [Google Scholar]
- 20.Shah SJ, Katz DH, Selvaraj S, Burke MA, Yancy CW, Gheorghiade M, Bonow RO, Huang CC, Deo RC. Phenomapping for novel classification of heart failure with preserved ejection fraction. Circulation. 2015;131:269–279. DOI: 10.1161/CIRCULATIONAHA.114.010637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altman RB, Ashley EA. Using "big data" to dissect clinical heterogeneity. Circulation. 2015;131:232–233. DOI: 10.1161/CIRCULATIONAHA.114.014106. [DOI] [PubMed] [Google Scholar]
- 22.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. DOI: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Maaten L, Hinton G. Visualizing data using t‐SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 24.Pöss J, Köster J, Fuernau G, Eitel I, de Waha S , Ouarrak T, Lassus J, Harjola V‐P, Zeymer U, Thiele H, et al. Risk stratification for patients in cardiogenic shock after acute myocardial infarction. J Am Coll Cardiol. 2017;69:1913–1920. DOI: 10.1016/j.jacc.2017.02.027. [DOI] [PubMed] [Google Scholar]
- 25.Harjola V‐P, Lassus J, Sionis A, Køber L, Tarvasmäki T, Spinar J, Parissis J, Banaszewski M, Silva‐Cardoso J, Carubelli V, et al. Clinical picture and risk prediction of short‐term mortality in cardiogenic shock. Eur J Heart Fail. 2015;17:501–509. DOI: 10.1002/ejhf.260. [DOI] [PubMed] [Google Scholar]
- 26.Stekhoven DJ, Buhlmann P. Missforest–non‐parametric missing value imputation for mixed‐type data. Bioinformatics. 2012;28:112–118. DOI: 10.1093/bioinformatics/btr597. [DOI] [PubMed] [Google Scholar]
- 27.Shah AD, Bartlett JW, Carpenter J, Nicholas O, Hemingway H. Comparison of random forest and parametric imputation models for imputing missing data using MICE: a CALIBER study. Am J Epidemiol. 2014;179:764–774. DOI: 10.1093/aje/kwt312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trunk GV. A problem of dimensionality: a simple example. IEEE Trans Pattern Anal Mach Intell. 1979;PAMI‐1:306–307. DOI: 10.1109/TPAMI.1979.4766926. [DOI] [PubMed] [Google Scholar]
- 29.Dolnicar S. A review of unquestioned standards in using cluster analysis for data‐driven market segmentation, CD conference proceedings of the Australian and New Zealand marketing academy conference 2002 (ANZMAC 2002), Deakin University, Melbourne, 2‐4 December 2002. Available at: https://ro.uow.edu.au/commpapers/273.
- 30.Formann AK. Die latent‐class‐analyse: einführung in theorie und anwendung. Weinheim [W. Germany]: Beltz; 1984. Available at: http://catalog.hathitrust.org/api/volumes/oclc/16851238.html [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


