SUMMARY
Large clostridial toxins (LCTs) are a family of bacterial exotoxins that infiltrate and destroy target cells. Members of the LCT family include Clostridioides difficile toxins TcdA and TcdB, Paeniclostridium sordellii toxins TcsL and TcsH, Clostridium novyi toxin TcnA, and Clostridium perfringens toxin TpeL. Since the 19th century, LCT-secreting bacteria have been isolated from the blood, organs, and wounds of diseased individuals, and LCTs have been implicated as the primary virulence factors in a variety of infections, including C. difficile infection and some cases of wound-associated gas gangrene. Clostridia express and secrete LCTs in response to various physiological signals. LCTs invade host cells by binding specific cell surface receptors, ultimately leading to internalization into acidified vesicles. Acidic pH promotes conformational changes within LCTs, which culminates in translocation of the N-terminal glycosyltransferase and cysteine protease domain across the endosomal membrane and into the cytosol, leading first to cytopathic effects and later to cytotoxic effects. The focus of this review is on the role of LCTs in infection and disease, the mechanism of LCT intoxication, with emphasis on recent structural work and toxin subtyping analysis, and the genomic discovery and characterization of LCT homologues. We provide a comprehensive review of these topics and offer our perspective on emerging questions and future research directions for this enigmatic family of toxins.
KEYWORDS: Clostridium difficile, large clostridial toxin, toxin, toxin-mediated diseases, toxin-receptor interaction
INTRODUCTION
Clostridia are a polyphyletic class of anaerobes that are prolific producers of toxins. Well-known clostridial toxins include pore-forming toxins, such as Clostridium perfringens epsilon toxin (Etx) (1), binary toxins, such as Clostridioides difficile binary toxin (CDT) (2), and large clostridial toxins (LCTs) (3). LCTs are a family of six bacterial exotoxins secreted by Gram-positive, spore-forming clostridial species. Members of the LCT family include Clostridioides difficile toxins TcdA and TcdB, Paeniclostridium sordellii toxins TcsL and TcsH, Clostridium novyi toxin TcnA, and Clostridium perfringens toxin TpeL. LCTs were first grouped together as a family of related toxins on basis of their large size (>200 kDa), similarities in primary structure (Table 1), and unusual ability to induce profound changes in cell morphology (3). Members of the LCT family have been implicated as the primary virulence factors in a variety of human and animal infections, including C. difficile infection (CDI), some cases of wound-associated gas gangrene, toxic shock syndrome, and severe soft tissue infections in injection drug users (4–7).
TABLE 1.
Sequence identity and similarity of LCT holotoxins
| LCT | % sequence identity (% similarity)a |
||||
|---|---|---|---|---|---|
| TcdA | TcdB | TcsH | TcsL | TcnA | |
| TcdB | 46 (66) | ||||
| TcsH | 77 (87) | 48 (68) | |||
| TcsL | 46 (66) | 76 (88) | 49 (70) | ||
| TcnA | 31 (51) | 30 (50) | 32 (52) | 31 (50) | |
| TpeL | 41 (61) | 39 (60) | 42 (61) | 40 (61) | 31 (51) |
Sequence identity and similarity were calculated using Water (EMBOSS) local alignment. LCTs are from the following strains: C. difficile VPI 10463, P. sordellii VPI 9048, C. novyi 19402, and C. perfringens JGS1495. Under new subtyping analysis, TcdA and TcdB from C. difficile VPI 10463 are referred to as TcdA1 and TcdB1 (138).
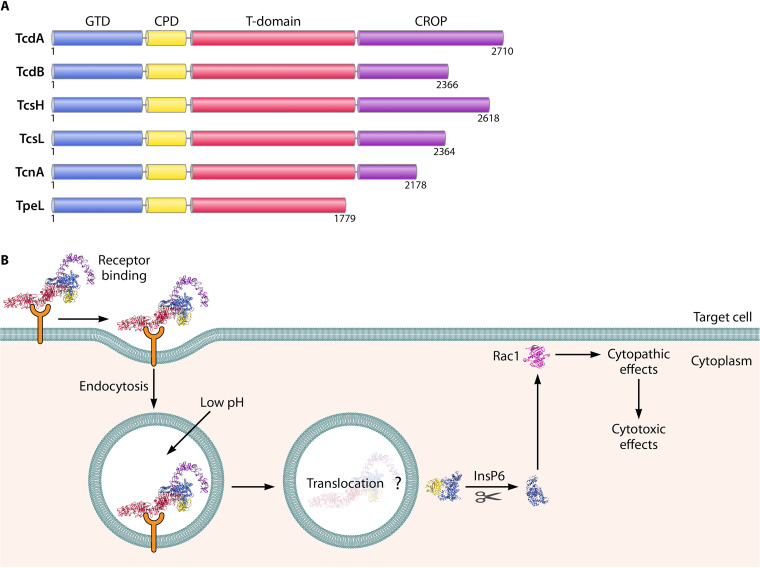
LCTs are single-chain multidomain polypeptides with similar gene organization, regulation, and overall domain architecture. Clostridia express LCTs in response to various environmental and physiological signals, enabling LCTs to infiltrate and ultimately destroy eukaryotic cells to promote bacterial infection. Symptoms of infection are thought to arise owing to the cytosolic delivery of the N-terminal glycosyltransferase domain (GTD) through concerted actions of the cysteine protease domain (CPD), the central translocation and receptor-binding domain (T domain) and combined repeating oligopeptide (CROP) (8) (Fig. 1). In brief, LCTs first bind target receptors on host cells, and become internalized into vesicles. The internalized LCT-containing vesicles subsequently become acidified. In response to acidic pH, conformational changes occur within LCTs, triggering translocation of the glycosyltransferase and cysteine protease domain across the endosomal membrane and into the cytosol. The cysteine protease domain autocatalytically cleaves and releases the glycosyltransferase from the rest of the polypeptide, freeing the glycosyltransferase to access membrane-tethered Rho and Ras guanine triphosphatases (GTPases), leading first to cytopathic effects (cell rounding) and later to cytotoxic effects (cell death).
FIG 1.
LCT domain architecture and mechanism of action. (A) Individual domains are colored as follows: glycosyltransferase domain (GTD) in blue, cysteine protease domain (CPD) in yellow, translocation and receptor-binding domain (T domain) in red, and combined repeating oligopeptide (CROP) in purple. (B) Schematic illustrating the major steps of LCT intoxication of host cells, using TcdB (PDB 6OQ5).
Here, we provide a comprehensive review of the LCT family. We detail seminal contributions spanning the 19th, 20th, and 21st centuries that have established LCTs as potent poisons, highlight recent structural work and toxin subtyping analysis to provide a thorough understanding of LCT structure, function, diagnostic, and therapeutic development, and discuss the burgeoning field of LCT homologue identification and characterization. We end this review by providing our perspective on pressing questions and pertinent future research directions.
ROLE OF LCTs IN CLOSTRIDIAL INFECTION AND DISEASE
C. difficile, TcdA, and TcdB
C. difficile was first described by I. C. Hall and E. O’Toole in 1935 (9). During their investigations of the intestinal microbiota of newborns, Hall and O’Toole isolated C. difficile from the stools of healthy infants. The researchers found that cell-free supernatants of C. difficile cultures were lethal to a variety of animals (10), suggesting the presence of secreted factors that were toxic toward animals. Hall and O’Toole named the bacterium Bacillus difficilis: Bacillus for the rod-like morphology and difficilis for the difficulty in cultivating the anaerobic bacterium. Since then, the bacterium formally known as B. difficilis has been renamed several times. First, to Clostridium difficile, then to Peptoclostridium difficile (11), and finally to Clostridioides difficile in 2016 by Lawson et al. (12).
Because C. difficile was originally isolated from healthy infants, the bacterium was considered a normal component of the intestinal flora in humans. In the 1970, J. G. Bartlett was the first to determine that C. difficile was the causative agent of C. difficile infection (CDI) (13–15). CDI is characterized by damage and injury of the colonic epithelium, with clinical symptoms ranging from diarrhea to pseudomembranous colitis and toxic megacolon (16). Bartlett’s work connected years of puzzling findings on antibiotic-associated colitis in guinea pigs and hamsters, first noted during animal studies of penicillin during World War II, and the rapid increase of antibiotic-associated colitis in hospitals during the 1950s, 60s, and 70s. In animal and human studies, Bartlett and colleagues identified undescribed toxins in the stools of infected patients (14, 15). The toxic components were later identified as TcdA, toxin C. difficile A, or simply toxin A/toxA, and TcdB, toxin C. difficile B, or simply toxin B/toxB (17–22).
CDI is the leading cause of antibiotic-associated diarrhea in the developing world (23), and in the United States alone, there are approximately a quarter of a million cases of CDI annually, resulting in 12,800 deaths and attributable health care costs of $1 billion (23). CDI is most prevalent in hospitalized elderly patients (>65 years) who have recently taken antibiotics (24). In animals, antibiotic usage has also been identified a major risk factor for CDI (25). It has been hypothesized that antibiotic usage wipes out the normal gut microflora, enabling C. difficile to colonize the gastrointestinal tract, eventually leading to toxin production (26). Although C. difficile is considered a hospital-acquired infection, the epidemiology of CDI is changing, with increasing reports of community-acquired CDI in populations without established risk factors (27). Multiple reports have also identified C. difficile in livestock and in the food chain and have suggested that the presence of C. difficile in the agricultural industry may provide a reservoir for common community-acquired CDI (25, 28).
In animal models, TcdA and TcdB recapitulate the symptoms associated with CDI, including disruption of tight junctions, epithelial cell death, and mucosal inflammation (16), and are thus believed to be the major virulence factors in infection (16) (Table 2). Furthermore, C. difficile strains lacking TcdA and TcdB are avirulent and nonpathogenic (29, 30), and TcdA and TcdB levels correlate with the severity of C. difficile infection in epidemic C. difficile strains (31, 32). It remains contentious whether TcdA or TcdB is the major virulence factor in infection. In support of TcdB as the major virulence factor, TcdA− TcdB+ clinical isolates cause CDI in in humans (33, 34). Additionally, laboratory TcdA− TcdB+ strains cause CDI in animal models (29, 30, 35), and TcdB alone is directly responsible for severe intestinal damage (36). In contrast, laboratory TcdA+ TcdB− strains are attenuated in virulence compared to TcdA+ TcdB+ strains, and TcdA alone is capable of only causing minimal intestinal damage in animal models (36). Recently, the first ever TcdA+ TcdB− clinical isolate was isolated from an individual with antibiotic-associated diarrhea (37). The existence of a TcdA+ TcdB− clinical strain suggests that TcdA may be capable of causing CDI, although it is not yet clear if TcdA can cause more severe disease symptoms. Notably, hamsters infected with the TcdA+ TcdB− strain did not recapitulate the clinical symptoms associated with human infection, suggesting potential limitations of animal models in understanding and defining the role of C. difficile toxins in human infection.
TABLE 2.
Large clostridial toxins: overview
| Organism | Toxin | Mol wt (kDa) | Biological activity | Role in infection |
|---|---|---|---|---|
| C. difficile | TcdA | 308 | Enterotoxica (269) | Primary virulence factor in TcdB− TcdA+ strains, less central role in TcdB+ TcdA+ strains |
| TcdB | 270 | Enterotoxica (270) | Primary virulence factor in TcdB+ TcdA(+/−) strains | |
| P. sordellii | TcsL | 270 | Necrotizing, edematizing (45) | Primary virulence factor in TcsL+ TcsH(+/−) strains |
| TcsH | 299 | Hemorrhaging (45) | Unknown | |
| C. novyi | TcnA | 250 | Necrotizing, edematizing (271) | Primary virulence factor in C. novyi type A and B infections |
| C. perfringens | TpeL | 206 | Unknown | May enhance virulence of C. perfringens type G strains |
P. sordellii, TcsL, and TcsH
P. sordellii was first isolated by A. Sordelli in 1922 (5). Sordelli isolated the bacterium from acute edematous human wound infections and named it Bacillus oedematic sporogenes. The name of the bacterium was derived on the basis of shared features to Bacillus oedematiens (presently known as C. novyi), which causes edema (fluid retention and swelling), and Bacillus sporogenes, which has a similar rod-like morphology. To avoid confusion with B. oedematiens and B. sporogenes, the bacterium was renamed Bacillus sordellii in 1927, in honor of Sordelli (38), and later Clostridium sordellii (39, 40). Recently, the bacterium was reclassified as a species of the genus Paeniclostridium, a new closely related genus to Clostridium (41).
TcsL, toxin C. sordellii lethal, also known as lethal toxin (LT), and TcsH, toxin C. sordellii hemorrhagic, also known as hemorrhagic toxin (HT), were first described in 1969 by Arseculeratne and colleagues as two independent toxins secreted by P. sordellii with edematizing and hemorrhagic activities (42). In 1987, Popoff purified TcsL and described TcsL as an ∼250-kDa cytotoxin that was immunologically related to TcdB (43). One year later, Martinez and Wilkins purified and characterized TcsH, describing it as an ∼300-kDa protein that was immunologically similar to TcdA (44).
P. sordellii has been implicated in a myriad of sporadic infections in both humans and animals which are characterized by a mild or completely absent inflammatory response (45). P. sordellii has been associated in wound-associated gas gangrene in humans and animals (46, 47). Gas gangrene is typified by large amounts of gas, which can form bubbles and blisters in tissue that can lead to necrotizing infection and tissue death. More recently, P. sordellii infections have been identified in deep tissue infections (most often in injection drug users), and during childbirth, abortion, and surgery (5, 48–51). P. sordellii soft tissue infections in injection drug users are characterized by a rapid onset of illness, massive edema, and, in some cases, necrotizing fasciitis, also known as flesh-eating disease (48, 49). In toxic shock syndrome, P. sordellii colonizes the genital tract and causes a rapidly fatal onset of infection. Toxic shock syndrome is characterized by several clinical features, including leukocytosis, edema, and refractory hypotension (5, 50, 51).
Most pathogenic strains of P. sordellii produce TcsL, while few produce TcsH, suggesting that TcsL has a more central role in virulence than TcsH (52–54) (Table 2). In support of TcsL as the major virulence factor in P. sordellii infections, animal studies have demonstrated that TcsL alone causes extensive tissue edema and death, particularly in the lung vascular endothelium (55, 56), and inactivation of the tcsL gene prevents mice from developing tissue edema or dying (57). Amimoto et al. have, however, shown that vaccination with toxoids of both TcsH and TcsL was required to protect guinea pigs against a P. sordellii spore challenge (58), suggesting a possible role of TcsH in virulence. In P. sordellii culture supernatants, however, TcsH has been shown to account for a marginal amount of toxicity, with ∼98% of supernatant toxicity attributable to TcsL, suggesting that TcsH may have a minimal, if any, role in infection (52).
C. novyi and TcnA
C. novyi was first isolated in the late 19th century by F. G. Novy (4). An excellent historical review of C. novyi is provided by Aronoff and Kazanjian (4). In brief, to determine the bacteriological components of food substances, Novy injected nuclein isolated from milk into rabbits. Unexpectedly, the rabbits developed septicemia with malignant edema and rapidly died. From the deceased rabbits, Novy was able to, albeit with great difficulty due to its extreme oxygen sensitivity, cultivate and isolate the bacterium responsible for infection and death, naming it Bacillus novyi in 1897. The bacterium was again isolated in 1915 from a combat wound in a soldier who later developed gas gangrene and named Bacillus oedematiens (59). In 1923, the bacterium was formally reclassified as Clostridium novyi (60).
C. novyi alpha toxin, also known as TcnA for toxin C. novyi alpha, or simply alpha toxin, was first isolated from C. novyi by Izumi et al. in 1983 and was shown to have lethal and edematizing activity in mice (61, 62). TcnA was later shown to have similar cytopathic effects and sequence homology to other members of the LCT family (3, 63, 64).
C. novyi is a rare pathogen of both animals and humans (46). Owing to its extreme oxygen sensitivity, C. novyi is very difficult to cultivate, which may contribute to the rarity in which C. novyi is implicated in infection. Like P. sordellii, C. novyi has been implicated in some cases of wound-associated gas gangrene that resulted in lethal infections of deep soft tissue (46, 47). C. novyi has also been implicated in hepatic damage and infectious necrotic hepatitis in animals, also known as black disease, due to the dark discoloration of subcutaneous tissue caused by severe congestion of blood vessels (65, 66). Infectious necrotic hepatitis is usually accompanied by subcutaneous edema, hemorrhaging, and necrotic lesions in the liver (65). More recently, C. novyi has been identified as the causative agent in severe soft tissue infections in injection drug users (67–69). C. novyi infection in injection drug users has similar clinical features as infection with P. sordellii, including leukocytosis, edema, and refractory hypotension, which can lead to necrotizing infection (67–69).
TcnA is produced by C. novyi type A and B (Table 2). C. novyi type A is associated with gas gangrene infections in humans and animals and infections in injection drug users (67–69). C. novyi type B is associated with necrotic enteritis in animals (66, 70, 71). TcnA is believed to be the major virulence factor in C. novyi type A and B infections, largely owing to its lethal and edematizing activity in vitro and in vivo in animal models (4) (Table 2). For livestock infected with necrotic hepatitis, detection of the tcnA gene has been used to confirm C. novyi type B infection (66). Interestingly, C. novyi infection in injection drug users has very similar clinical manifestations as those in patients with P. sordellii-induced toxic shock syndrome, suggesting that both diseases may be mediated by toxins with similar biochemical and biological activities (4).
C. perfringens and TpeL
C. perfringens was first isolated near the end of the 19th century by two independent research groups. For an excellent review on the history and isolation of C. perfringens, please consult Rood et al. (72). In brief, in 1891, W. H. Welch isolated C. perfringens from the blood and organs of a deceased male who had died of an aortic aneurism (73). Concurrently, M. P. Achalme isolated C. perfringens from a patient with acute articular rheumatism (74). Welch and colleagues named the bacterium Bacillus aerogenes capsulatus (aerogenes for air/gas producing and capsulatus for capsule), and Achalme named the bacterium Bacillus phlegmonis emphysematosae (phlegmonis for phlegmon, an area of acute inflammation in soft tissue, and emphysematosae for emphysema, a condition of abnormal enlargement of tissues). Since then, C. perfringens has been renamed multiple times before the formal adoption of Clostridium perfringens (perfringens for per, meaning through, and frango for burst) in the 1930 (75, 76).
C. perfringens large toxin, also known as TpeL for toxin perfringens large, is the most recently discovered LCT. TpeL was isolated from C. perfringens culture filtrate in 2007 by Amimoto et al. (77). The researchers noted that TpeL had sequence homology to other LCTs and was toxic when injected into mice.
C. perfringens has been implicated in numerous diseases in humans and animals (78). Like P. sordellii and C. novyi, C. perfringens is associated with some cases of wound-associated gas gangrene, with reports suggesting that C. perfringens is responsible for up to 90% of all clostridium-mediated gas gangrene (46, 47). In humans, C. perfringens is one of the leading causes of bacterium-mediated food poisoning (79) and is associated with antibiotic-associated diarrhea (80) and necrotizing enterocolitis, a lethal infection characterized by profound inflammation of the intestine that occurs mostly in neonates (81). In animals, particularly poultry, C. perfringens is implicated in severe and often fatal disease, including necrotic enteritis, which costs the agricultural industry in excess of two billion dollars per year in the United States alone (82). Necrotic enteritis, also known as pulpy kidney or overeating disease, is an acute enterotoxemia that has clinical manifestations that differ between animals. Clinical manifestations include enterocolitis, a soft consistency of the kidney, and encephalomalacia within the brain (83, 84).
TpeL is present in isolates of C. perfringens types B, C, and G (85–87) (Table 2). The majority of C. perfringens type B and C strains are associated with hemorrhagic and necrotic enteritis in animals and enteritis necroticans in humans, respectively (88). Chen and McClane have shown that natural production levels of TpeL in C. perfringens type C supernatants contribute to cytotoxic activity, suggesting a potential role of TpeL in type C infections (87). However, there is no direct evidence that TpeL contributes to C. perfringens type B and C infections. Strong evidence instead supports a central role for other C. perfringens toxins, including β-toxin and ε-toxin in type B infections and β-toxin and enterotoxin (CPE) in type C infections (88). C. perfringens type G strains are associated with necrotic enteritis in poultry, and there is some evidence to suggest that TpeL may contribute to virulence in type G strains (89). TpeL-positive strains are associated with a more rapid course of infection and a higher fatality rate than TpeL-negative strains (89). C. perfringens toxin NetB is, however, believed to be the major virulence factor in type G infections (90). It is yet unclear if TpeL contributes to virulence, by acting either alone or synergistically with NetB.
LCT GENE ORGANIZATION AND REGULATION
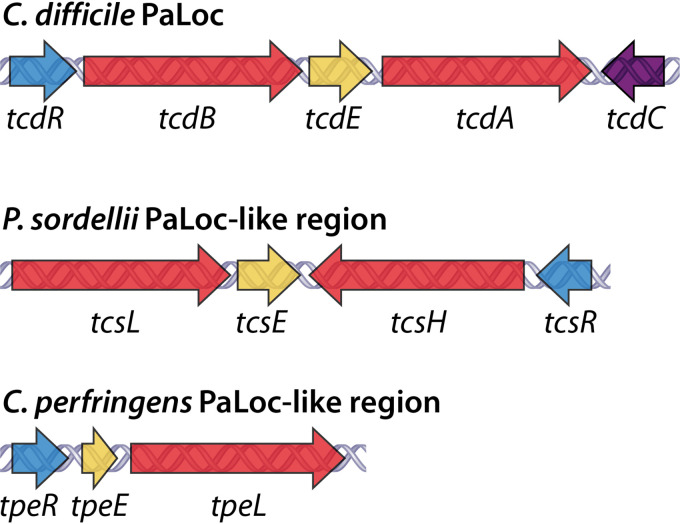
In 1996, a 19.6-kb chromosomal region termed the pathogenicity locus (PaLoc) was uncovered in C. difficile strain VPI 10463 (91). The C. difficile PaLoc contains genes for both TcdA and TcdB as well as three accessory genes: tcdR, tcdC, and tcdE (Fig. 2). The accessory gene tcdR encodes an alternative RNA polymerase sigma (σ) factor, TcdR. TcdR belongs to the σ70 family of alternative σ factors, which includes alternative σ factors of other pathogenic clostridia, such as BotR of C. botulinum and UviA of C. perfringens (92). TcdR is a positive regulator of LCT expression and is critical for the initiation of TcdA and TcdB gene expression (92). The accessory gene tcdC encodes an anti-σ factor, TcdC. TcdC negatively regulates LCT expression by directly interacting with TcdR or the TcdR-RNA polymerase holoenzyme (93, 94). TcdC may have a role in controlling TcdA and TcdB levels, as C. difficile strains with frameshift mutations or deletions of the tcdC gene have increased toxin production (31, 95). The extent to which TcdC contributes to LCT regulation remains controversial, as several studies have demonstrated that TcdC only moderately contributes to LCT expression (96, 97). The accessory gene tcdE encodes a bacteriophage holin-like protein, TcdE. Holins are bacteriophage-encoded membrane proteins that oligomerize and form holes in the host cell membrane to release progeny phage (98). Multiple reports have suggested that holin-like proteins may be responsible for release of proteins from bacteria (99–101). Based on sequence homology to holins and the lack of obvious export signatures on TcdA and TcdB, TcdE was proposed to regulate TcdA and TcdB release from C. difficile (102). In 2012, Govind and Dupuy demonstrated that TcdE facilitates the release of TcdA and TcdB without inducing cell lysis (103), providing the first experimental evidence of holin-like proteins mediating the secretion of proteins from bacteria. In addition to holin-dependent secretion, there is evidence that C. difficile can also release TcdA and TcdB by bacteriolysis mediated by the cell surface peptidoglycan hydrolase Cwp19 (104, 105).
FIG 2.
LCT gene organization. The PaLoc of C. difficile and PaLoc-like regions of P. sordellii and C. perfringens, first identified in C. difficile strain VPI 10463, P. sordellii strain VPI 9048, and C. perfringens strain ATCC 3626. LCT genes are colored red. Accessory genes are colored as follows: alternative σ factor (tcdR/tcsR/tpeR) in blue, anti-σ factor (tcdC) in purple, and holin-like protein (tcdE/tcsE/tpeE) in yellow.
The C. difficile PaLoc is located at the same chromosomal site in the majority of C. difficile strains (91, 106, 107). C. difficile strains lacking the PaLoc harbor a 75/115-bp noncoding region, are nontoxigenic, and do not cause disease (91, 108). The C. difficile PaLoc can be transferred from toxigenic to nontoxigenic strains and thus has characteristics of a mobile genetic element (109). There is considerable genetic variation in PaLocs of different C. difficile strains, including truncated and monotoxin PaLoc variants (110, 111). Genetic variations of the C. difficile PaLoc have been assessed by toxinotyping, a PCR restriction fragment length polymorphism method that distinguishes strains into 34 toxinotypes based on the PaLoc (107). Based on genetic studies of the PaLoc, it has been suggested that the bitoxin PaLoc (i.e., containing TcdA and TcdB) may have evolved from the merging of two monotoxin PaLocs (i.e., containing either TcdA or TcdB) (111).
A PaLoc-like region has been identified in strains of P. sordellii (112) and C. perfringens (113) (Fig. 2). In contrast to the chromosomally localized C. difficile PaLoc, the P. sordellii and C. perfringens PaLoc-like regions are located on conjugative plasmids (54, 85, 114, 115). Both the P. sordellii and C. perfringens PaLoc-like regions contain accessory genes that are homologous to the C. difficile genes tcdR and tcdE and encode TcdR- and TcdE-like proteins, respectively. In P. sordellii, the genes tcsR and tcsE encode TcsR and TcsE (112), and in C. perfringens, the genes tpeR and tpeE encode TpeR and TpeE (113) (Fig. 2). TcdR, TcsR, and TpeR are all members of the σ70 family of alternative σ factors (113). TcsR and TpeR have been demonstrated to regulate the expression of TcsL and TpeL, respectively (112, 113). Notably, TcdR and TcsR are functionally interchangeable, while neither TcdR or TcsR can be functionally exchanged with TpeR (113). TcdE and TcsE belong to superfamily 4 of bacteriophage holins, and TpeE belongs to the DUF2762 superfamily of bacteriophage holins (116). Recently, TpeE was demonstrated to facilitate the secretion of TpeL without cell lysis, supporting a model of holin-dependent toxin secretion in C. perfringens (117). Due to the similarities in structure and function of TcdR and TcsR and of TcdE and TcsE, it has been suggested that the C. difficile PaLoc and P. sordellii PaLoc-like region may share a recent common ancestor, while the PaLoc-like region of C. perfringens may be more divergent (113). This is consistent with C. difficile and P. sordellii both belonging to the Peptostreptococcaceae family, while C. perfringens belongs to the Clostridiaceae family.
There are few studies on tcnA gene organization. In C. novyi, the tcnA gene is phage localized (118), and toxigenic strains of C. novyi can transduce nontoxigenic strains of C. novyi to produce TcnA, indicating mobile transfer of the tcnA gene (119). It is not currently known whether the tcnA gene resides within a PaLoc-like region or how tcnA expression is regulated.
The C. difficile PaLoc and the P. sordellii and C. perfringens PaLoc-like regions are influenced by environmental, physiological, and nutrient signals. Regulation of the PaLoc has been extensively studied in C. difficile (120) and has been studied in less detail in P. sordellii and C. perfringens (113). In C. difficile, P. sordellii, and C. perfringens, LCT production follows a similar pattern of temporal expression, with increases in toxin production as bacterial cells approach the stationary phase of growth (113, 121, 122). The regulatory pathways that coordinate LCT production and bacterial growth are not well understood. Quorum sensing has been proposed to coordinate LCT production and bacterial growth, enabling the clostridia to modulate LCT production in response to bacterial cell densities (113). In C. difficile, P. sordellii, and C. perfringens, LCT production is repressed by glucose (112, 113). In C. difficile, the catabolite control protein A (CcpA) mediates the bacterium’s response to glucose, by directly and indirectly inhibiting transcription of numerous regulators (123, 124). Both P. sordellii and C. perfringens harbor CcpA homologues (113), which may mediate the glucose response in these organisms, although this has not been experimentally demonstrated. It has been proposed that TcdA and TcdB production is triggered by C. difficile in response to particular states of nutrient availability during infection and that toxin production improves nutrient availability for the bacterium, thus enabling C. difficile to persist and cause damage to the host (120). The integration of regulation, toxin production, and infection in P. sordellii and C. perfringens is not well understood.
In addition to growth conditions and glucose, the C. difficile PaLoc is regulated by a complex array of environmental and physiological factors through several global regulators, including CodY, SigD, PrdR, Rex, RstA, and Spo0A (120, 125–128). These environmental and physiological factors include temperature (129), amino acids such as proline and cysteine (130), short-chain fatty acids such as butyric acid that are present in the gut (131), subinhibitory concentrations of antibiotics (132, 133), stress responses (134), and sporulation (135, 136). Interestingly, several studies have reported that regulation of the C. difficile PaLoc is strain specific, with the PaLoc across different strains responding differently to antibiotics (120) and sporulation cues (126, 128). Furthermore, epidemic C. difficile RT027 strains encode a binary toxin locus, CdtLoc, which has been suggested to regulate PaLoc expression (137).
LCT Classification and Subtyping Analysis
With the increasing number of sequenced bacteria, LCTs are being detected in an expanding number of clostridial genomes. A key question that has emerged is whether LCT genes vary across clostridial strains, and if so, if sequence variation contributes to biological and functional differences, which may manifest in different clinical presentations of infection and disease. Recently, Mansfield et al. (138) and Shen et al. (139) and proposed a method of sequence-based subtyping of TcdA and TcdB to enable more accurate predictions of variations in toxin activity. The subtyping analysis on a larger set of C. difficile genomes by Mansfield et al. partitions TcdA and TcdB into 7 and 12 distinct subgroups, referred to as A1-7 and B1-12, respectively (138). TcdA and TcdB from C. difficile VPI 10463, which has been the reference strain since the 1980s, belong to the A1/B1 subtype, and TcdA and TcdB from epidemic strains, such as RT027, tend to cluster outside the A1/B1 subtype (138). Interestingly, TcdA variants differ mainly in the number of repeats in the C-terminal repetitive region, while TcdB has diversified through extensive homologous recombination throughout its entire sequences (138). Variations in TcdB sequence have been correlated with distinct antigenic, receptor-binding, and phenotypic properties, which will be reviewed in the upcoming sections on LCT structure and function. To the best of our knowledge, sequence variations in TcsL, TcsH, TcnA, and TpeL across different strains have not been investigated.
STRUCTURE AND FUNCTION OF LCTs
Holotoxin Structure
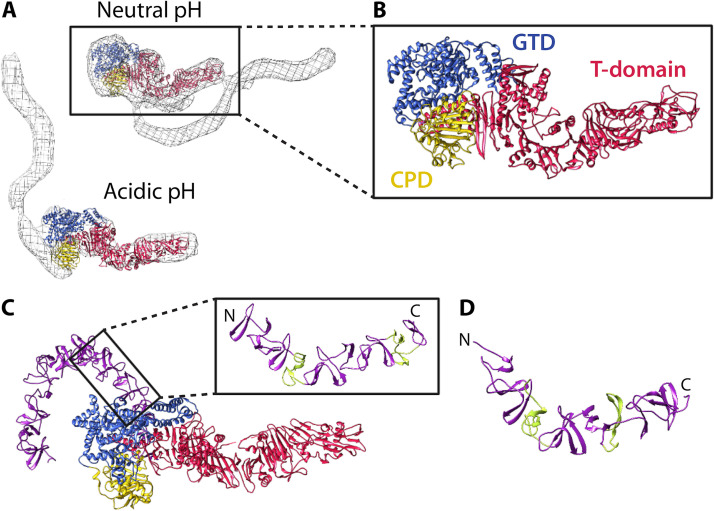
Over the years, numerous structures have been solved for individual LCT domains as well as multidomain and full-length fragments of TcdA and TcdB, providing enormous insights into LCT structure and function. In 2010, the structure of full-length TcdA was determined by negative stain electron microscopy (EM) and small-angle X-ray scattering (SAXS) (140, 141). TcdA was shown to have a bilobed organization, with a globular “head” region consisting of the glycosyltransferase and cysteine protease domain, from which the T domain extends to the opposite end of the molecule. The structure of the CROP-less TcdA later confirmed the organization of the glycosyltransferase domain, the cysteine protease domain, and the T domain and enabled refinement of structural changes at neutral and acidic pH by fitting the high-resolution TcdA structure to EM maps of the TcdA holotoxin (142) (Fig. 3A and B). At neutral pH, the TcdA CROP makes structural contacts with the T domain, and at acidic pH, the CROP extends away from the T domain (Fig. 3A and B). The X-ray structure of full-length TcdB later revealed the precise positioning of the TcdB CROP at acidic pH and the dynamism of the CROP at neutral pH (143) (Fig. 3C). At acidic pH, the TcdB CROP extends ∼130 Å from the base of the cysteine protease domain and T domain, curving around the glycosyltransferase domain like a hook in a “open” configuration (143). Although the full-length TcdB structure was solved at acidic pH, no major structural changes were observed for the glycosyltransferase, cysteine protease, and T domain compared to that at neutral pH (143). Notably, the glycosyltransferase domain and T domain were bound to nanobodies, which may have prevented the full suite of pH-mediated conformational changes.
FIG 3.
Structure and organization of the C. difficile holotoxin. (A) TcdA1–1832 (PDB 4R04) fit into the EM map of the TcdA holotoxin at neutral and acidic pH (140) using Chimera (267). The TcdA EM maps were kindly provided by Borden Lacy and are reproduced with permission. (B) Focused view of TcdA1–1832. (C) Structure of TcdB1–2366 at acidic pH (PDB 6OQ5). In the focused image, short repeats (SRs) are colored purple and long repeats (LRs) are colored green. (D) Structure of a fragment of the TcdA CROP (PDB 2G7C), with SRs colored purple and LRs colored green.
Combined Repeating Oligopeptide
The combined repeating oligopeptide (CROP) is the C-terminal domain of LCTs, except the naturally CROP-less TpeL. As the name suggests, the CROP is composed of multiple repeating units: 19- to 24-amino-acid short repeats (SRs) and 29- to 31-amino-acid long repeats (LRs). The LCT CROP is variable in sequence, sharing between 27% and 75% sequence identity among LCTs (Table 3). The LCT CROP ranges in size, from 40 kDa to 100 kDa, and correspondingly, in number of SRs and LRs (Table 4). The CROPs of TcdA and TcsH are the largest, with 30 and 33 SRs and 6 and 7 LRs, respectively, while the CROP of TcnA is the shortest, with 13 SRs and 3 LRs.
TABLE 3.
LCT CROP sequence identity and similarity
| LCT | % sequence identity (% similarity)a |
|||
|---|---|---|---|---|
| TcdA | TcdB | TcsH | TcsL | |
| TcdB | 37 (51) | |||
| TcsH | 68 (77) | 43 (57) | ||
| TcsL | 36 (50) | 75 (87) | 43 (59) | |
| TcnA | 34 (45) | 27 (40) | 39 (57) | 31 (48) |
aSequence identity and similarity were calculated using Water (EMBOSS) local alignment. LCTs are from the following strains: C. difficile VPI 10463, P. sordellii VPI 9048, and C. novyi 19402. Under new subtyping analysis, TcdA and TcdB from C. difficile VPI 10463 are referred to as TcdA1 and TcdB1 (138).
TABLE 4.
LCT CROP repeating units
| LCTa | CROP bounds | No. of short repeats | No. of long repeats | Mol wt (kDa) |
|---|---|---|---|---|
| TcdA | 1812–2710 | 33 | 7 | 102 |
| TcdB | 1814–2366 | 21 | 4 | 64 |
| TcsH | 1812–2618 | 30 | 6 | 92 |
| TcsL | 1815–2364 | 21 | 4 | 63 |
| TcnA | 1822–2178 | 13 | 3 | 41 |
aLCTs are from the following strains: C. difficile VPI 10463, P. sordellii VPI 9048, and C. novyi 19402. Under new subtyping analysis, TcdA and TcdB from C. difficile VPI 10463 are referred to as TcdA1 and TcdB1 (138).
At present, there is structural information for the TcdA and TcdB CROP (Fig. 3C and D). The SRs and LRs of the CROP consist of one and three β-hairpins, respectively, with a variable loop region of 7 to 10 amino acids for SRs and ∼18 amino acids for LRs (144, 145) (Fig. 3D). SRs pack together, forming a solenoid-like fold, and LRs introduce kinks and curvature into the CROP structure. Using the structure of the TcdA CROP fragment as a framework, Ho et al. built models of the full-length TcdA and TcdB CROPs (144). Ho et al. were the first to propose the S-shape and hook-shape structures of the TcdA and TcdB CROPs, respectively; these predictions were later supported by EM (140) and X-ray crystallography (143) (Fig. 3A, B, and C). Furthermore, work by Chen et al. has shown that the TcdB CROP has a C-terminal SR hinge region that confers conformational mobility to the CROP, enabling the CROP to adopt “open” and “closed” conformations at acidic and neutral pHs, respectively (143).
The CROP has long been assumed to mediate cellular binding and entry into host cells. Antibodies against the CROP prevent cell-surface binding and toxicity (146, 147), and recently, the TcdA CROP was shown to mediate binding to the colonic epithelium (148). Due to homology with streptococcal glycosyltransferases, the CROP was first postulated to mediate LCT attachment to cell surfaces by binding to carbohydrate moieties (20), a common strategy of bacterial and viral pathogens (149). In support of the CROP as a carbohydrate binding region, the TcdA and TcdB CROPs have been demonstrated to bind cell surface carbohydrates with low affinity (150–153). The physiological relevance of TcdA and TcdB binding to carbohydrates in the context of cellular intoxication has not been clarified. Recent work has demonstrated that the protein receptor chondroitin sulfate proteoglycan 4 (CSPG4) binds in part to the TcdB CROP (amino acids 1831 to 1850) and to the T domain (154), providing evidence that regions of the CROP are involved in receptor binding. Presently, no other protein receptors have been identified that bind to the CROP of other LCTs.
It is clear that the CROP is not the sole LCT domain that mediates cell surface binding and entry, as was originally hypothesized. Multiple receptors have been identified that bind in the LCT T domain (148, 155–159) (reviewed in the upcoming section), including a receptor for the CROP-less TpeL (160). Functional studies with TcdA and TcdB have also demonstrated that toxin entry is attenuated or not affected by truncations or complete removal of the CROP (77, 161, 162). Interestingly, several studies have demonstrated that the LCT CROP has functions apart from binding to cell surface carbohydrates and protein receptors. These functions include aiding in holotoxin folding, stabilization, and prevention of premature autoprocessing (163–165). Recently, it was shown that the TcdB CROP directly and reversibly binds intestinal bile acids, inhibiting toxin uptake and thereby intoxication (166). Bile acids prevent TcdB from binding to cell surface receptors and induce conformational changes that enable TcdB to become more resistant to proteolytic digestion (166). Notably, intestinal bile acids do not bind to the TcdA CROP or to the CROP-less TpeL. Although the role of bile acid binding to TcdB requires further study, this work suggests that bile acids may impact the timing of TcdB intoxication of cells, modulating virulence with respect to bile acid concentration in the gastrointestinal tract. It is not yet known if bile acids bind to other LCTs to modulate cellular intoxication.
Bezlotoxumab is a neutralizing antibody against TcdB that is used in the treatment of recurrent CDI (167). To neutralize TcdB, bezlotoxumab binds two epitopes in the N terminus of the TcdB CROP (146). Notably, bezlotoxumab was generated using TcdB1 antigens (from strain VPI 10463) and has exhibited reduction in neutralization efficacy against TcdB B2/4/5 subtypes from RT027, 8864, and RT078 strains (138, 168, 169). Alignment of key residues in the epitope region across all TcdB subtypes by Mansfield et al. revealed residue changes in B2/4/5 TcdB subtypes (138), providing strong evidence that the reduced efficacy of bezlotoxumab is related to sequence variations in the epitope binding region of subtypes. TcdB subtyping is clearly essential to direct the clinical use of bezlotoxumab and other emerging toxin-targeted therapies.
Translocation and Receptor-Binding Domain
The translocation and receptor-binding domain (T domain) is an ∼1,000-amino-acid domain that shares between 28% and 79% sequence identity among LCTs (Table 5; Fig. 4A). At present, there are high-resolution structures of the full-length T domains of TcdA (142) and TcdB (143, 170) and of a fragment of TcsL (158). The LCT T domain is structurally unique and, at neutral pH, is composed largely of β-sheets, with a short helical region (amino acids 956 to 1135) that extends from one end of the T domain to the other, wrapping around β-sheet structures (142) (Fig. 4A). Interestingly, although no major structural rearrangements were observed for the T domain at acidic pH, there is a loss of electron density within the helical stretch (944 to 949 and 1032 to 1047), suggesting structural flexibility of this region (143).
TABLE 5.
Sequence identity and similarity of LCT translocation and receptor-binding domains
| LCT | % sequence identity (% similarity)a |
||||
|---|---|---|---|---|---|
| TcdA | TcdB | TcsH | TcsL | TcnA | |
| TcdB | 47 (69) | ||||
| TcsH | 79 (92) | 48 (70) | |||
| TcsL | 47 (69) | 77 (88) | 48 (70) | ||
| TcnA | 28 (50) | 28 (51) | 28 (51) | 28 (50) | |
| TpeL | 36 (56) | 33 (56) | 37 (56) | 34 (56) | 28 (47) |
aSequence identity and similarity were calculated using Water (EMBOSS) local alignment. The following boundaries of the LCT T domains were used for sequence comparison: TcdA (802 to 1812), TcdB (800 to 1814), TcsH (802 to 1812), TcsL (800 to 1815), and TcnA (800 to 1822). LCTs are from the following strains: C. difficile VPI 10463, P. sordellii VPI 9048, C. novyi 19402, and C. perfringens JGS1495. Under new subtyping analysis, TcdA and TcdB from C. difficile VPI 10463 are referred to as TcdA1 and TcdB1 (138).
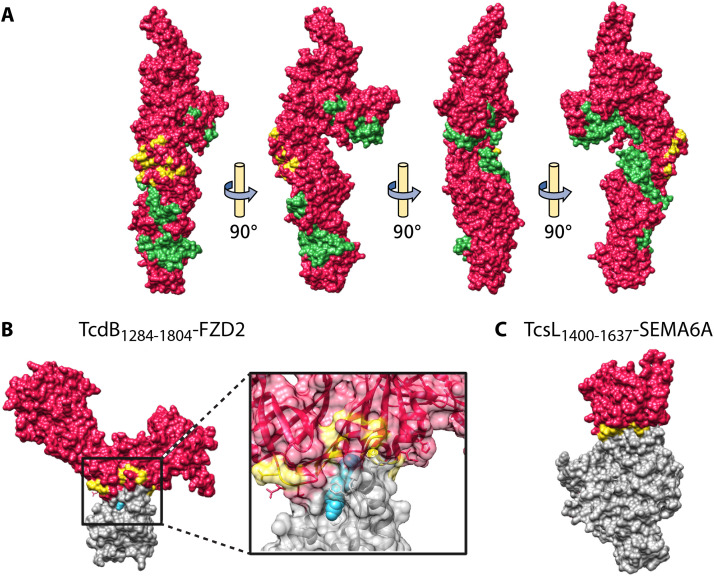
FIG 4.
Structure of the LCT T domain and LCT-receptor complexes. (A) T domain of TcdB (PDB 6OQ5), with the hydrophobic helical stretch (residues 956 to 1135) colored green and the interface residues for FZD2 binding colored yellow. (B) TcdB1284–1804-FZD2 (PDB 6C0B), with focus on the receptor-binding interface. TcdB is colored red, FZD2 interface residues on TcdB are colored yellow, FZD2 is colored gray, and palmitoleic acid (PAM) is colored blue. (C) TcsL1400–1637-SEMA6A (PDB 6WTS). TcsL is colored red, SEMA6A interface residues on TcsL are colored yellow, and SEMA6A is colored gray.
Several cell surface receptors have been identified that bind to the LCT T domain. Receptors for TcdA (148), TcdB (155–157), TcsL (158, 159), and TpeL (160) have been identified (Table 6), while the receptors for TcsH and TcnA remain more elusive. Notably, LCTs do not bind to the same receptors, and several LCTs have been demonstrated to bind multiple receptors (Table 6).
TABLE 6.
Host cell factors that mediate cell surface binding and internalization of LCTs
| Host cell factora | Binding regionb | Expression in human tissuec | PDBd |
|---|---|---|---|
| TcdA | |||
| Sulfated glycosaminoglycans (sGAGs) (148) | 1–1832 (148) | Colonic epithelium (148) | NA |
| Low-density lipoprotein receptor (LDLR) (148) | 1–1832 (148) | ND | NA |
| Low-density lipoprotein receptor-related protein-1 (LRP1) (174) | ND | ND | NA |
| TcdB | |||
| Chondroitin sulfate proteoglycan 4 (CSPG4) (155) | 1500–1850 (154) | Multinucleated intestinal subepithelial myofibroblasts (179) | NA |
| Frizzled family receptors (FZD)1/2/7 (156) | Discontinuous surface, 1433–1599 (170) | Colonic epithelium (156) | 6C0B |
| Poliovirus receptor-like protein 3 (PVRL3) (157) | 1372–1493 (162) | Colonic epithelium (157) | NA |
| TcsL | |||
| Semaphorins (SEMA)6A/6B (158, 159) | Discontinuous surface, 1433–1601 (158) | Vascular endothelium (158, 159) | 6WTS |
| TpeL | |||
| LRP1 (160) | 1335–1779 (160) | ND | NA |
aLCTs are from the following strains: C. difficile VPI 10463, P. sordellii JGS6382 and 6018, and C. perfringens JGS1495. Under new subtyping analysis, TcdA and TcdB from C. difficile VPI 10463 are referred to as TcdA1 and TcdB1 (138). Sucrase isomaltase (275) and glycoprotein 96 (Gp96) (276) have also been identified as TcdA receptors, but their lack of expression on the intestinal epithelial cells makes their physiological relevance to C. difficile pathogenesis unclear.
bBinding region indicates the minimal region demonstrated to bind to the receptor or the stretch of LCT interface residues that bind to the receptor, as determined from the LCT-receptor complex. ND, not determined if the full-length LCT was used to detect binding.
cIncludes human tissues that are physiologically relevant for infection. ND, not determined.
d NA, not available.
Sulfated glycosaminoglycans (sGAGs) and low-density lipoprotein receptor (LDLR) have been identified as host cell factors that mediate cell surface binding and entry of TcdA (148) (Table 6). Both factors were identified using genome-wide CRISPR-Cas9 screens using a truncated TcdA lacking the majority of the CROP. Biolayer interferometry confirmed that sGAGs bound directly to TcdA independent of the CROP. Binding could not be directly detected between LDLR and TcdA, suggesting that TcdA and LDLR bind weakly or require other cellular factors (148). While both sGAGs and LDLR mediate cellular binding and entry, the former were suggested to be the major attachment factors in the colonic epithelium (148). The exact role of LDLR binding in the context of TcdA intoxication remains to be clarified. Due to the major function of LDLR family receptors in mediating endocytosis (171), LDLR may facilitate endocytosis of TcdA bound to sGAGs (148). It is possible that TcdA binds to structurally similar LDLR family receptors, many of which are cell surface receptors for other pathogens (160, 172, 173). In support of TcdA binding structurally similar LDLR family members, Schöttelndreier et al. have recently provided evidence that TcdA binds to low-density lipoprotein receptor-related protein-1 (LRP1) (174).
At present, three receptors for TcdB have been identified: Wnt receptor frizzled family (FZD) FZD1/FZD2/FZD7 (156), chondroitin sulfate proteoglycan 4 (CSPG4) (155), and poliovirus receptor-like protein 3 (PVRL3/nectin 3) (157) (Table 6). Competition studies have demonstrated that FZD and CSPG4 bind independently of each other, indicating distinct nonoverlapped binding sites for these two receptors (156). It is not known whether PVRL3 binds at distinct sites or competes with binding to TcdB with FZD and CSPG4. FZD binds TcdB in the central region of the T domain, with palmitoleic acid (PAM) mediating binding of TcdB and FZD by making extensive contacts with both TcdB and FZD (170) (Table 6; Fig. 4A and B). Several reports have indicated that FZD binding varies between TcdB subtypes, with B2 and B4 subtypes having reduced affinity for FZD compared to that of B1 and B3 subtypes (175–178). Mansfield et al. have shown that the FZD binding motif is not conserved in B2/4/7/10/11 subtypes, suggesting that these subtypes may all have reduced affinity for FZD (138). The exact binding sites of CSPG4 and PVRL3 on TcdB are unknown. Functional studies have indicated that CSPG4 binds to both the T domain and the CROP (154) (Table 6) and that B3 and B4 subtypes have reduced affinity for CSPG4 binding compared to that of B1 and B2 subtypes (178). Recently, truncation and mutational analysis of TcdB revealed that the GTD and CPD contribute to CSPG4 binding, suggesting that the CSPG4 binding interface may be composed of multiple TcdB domains that converge in the holotoxin (178). PVRL3 is believed to bind to the central region of the TcdB T domain (162) (Table 6). Chung et al. have demonstrated that the B2 subtype has lower affinity for PVRL3 than B1 and suggested that the CROP may modulate PVRL3 binding, although it is not yet clear how this modulation occurs (176).
TcdB may utilize multiple receptors with different binding sites to broaden the selection of mammalian cells it can target. Both PVRL3 and FZDs are highly expressed on the surface epithelium of the human colon (156, 157), while CSPG4 is predominantly expressed in the multinucleated intestinal subepithelial myofibroblasts (ISEMFs) (179). The expression of PVRL3 and FZDs on the colonic epithelium suggests that PVRL3 and FZD might be the first receptors TcdB encounters when released into the lumen of the colon, and CSPG4 might serve as an important target to cause further tissue damage by exposing subepithelial myofibroblast cells. Notably, FZDs are receptors in the Wnt signaling pathway, an essential pathway for maintaining colonic stem cells (180). Healthy colonic stem cells constantly supply new colonic epithelial cells, which is central to colonic epithelial cell renewal and repair. TcdB competes with Wnt for binding to FZDs and, subsequently, inhibits Wnt signaling (156), suggesting that colonic stem cells are a potential target in C. difficile pathogenesis (181). Interestingly, recent work by Mileto et al. has shown that the B2 subtype can induce stem cell damage in an FZD-independent manner, suggesting the involvement of other TcdB receptors in mediating colonic epithelial damage (182). More recently, Pan et al. have also demonstrated that TcdB subtypes induce different pathological effects in mouse colonic tissue, suggesting that receptor preference can mediate colonic pathology (178).
Semaphorins 6A and 6B (SEMA6A/6B) have been identified as cellular receptors for TcsL by two independent genome-wide CRISPR-Cas9 screens (158, 159) (Table 6). SEMA6A binds to the central region of the T domain, forming a discontinuous binding interface along the T domain (158) (Table 6; Fig. 4C). Interestingly, the interface on TcsL that binds SEMA6A corresponds to the same interface on TcdB that binds FZD2, indicating that LCTs bind structurally unrelated receptors using the same receptor-binding interface (158). Furthermore, mutation of multiple residues in TcsL changes binding specificity to FZD2, suggesting that LCT receptor binding can be fine-tuned by changing key residues in the interaction surfaces (158, 159).
Intraperitoneal injection of TcsL causes major damage to lung endothelial cells, resulting in increased vascular permeability and edema in the lungs (55). Therefore, the vascular endothelium is believed to be the primary target of TcsL in vivo (55). Mice coinjected with TcsL and SEMA6A fused to an Fc fragment are protected from developing fluid edema in lung tissues, indicating that blocking SEMA6A-mediated TcsL entry into cells prevents toxin-induced symptoms (158). The physiological consequences of TcsL binding SEMA are not entirely clear. TcsL binds to SEMA6A at a position partially overlapping the functional site used by the plexin A2 cognate ligand; thus, TcsL binding to SEMA6A/6B has been suggested to interfere with semaphorin-plexin signaling in the vascular endothelium (158, 159). It is not yet known whether TcsL binding inhibits or disrupts semaphorin-plexin mediated downstream signaling pathways, many of which have key roles in controlling cell shape and movement (183).
LRP1 was identified as a receptor for TpeL using a haploid genetic screen (160) (Table 6). Functional studies have indicated that LRP1 binds the C-terminal region of TpeL, although the exact binding site of TpeL on LRP1 has not been determined (160) (Table 6). In a similar manner to that of LDLR family receptors and TcdA, LRP1 has been hypothesized as an endocytic receptor for TpeL (160).
For LCTs to gain entry into cells, LCTs must be internalized. TcdB, TcsL, and TcnA are endocytosed in a dynamin- and clathrin-dependent manner (184), while TcdA endocytosis is clathrin independent but dependent on dynamin and on the host factor protein kinase C and casein kinase substrate in neurons 2 (PACSIN2) (185). Investigation into the mechanism of LCT-mediated endocytosis and the role of cell surface receptors is ongoing. Schöttelndreier et al. have provided evidence that LRP1 contributes to TcdA internalization (174) and that FZD2/7, CSPG4, and PVRL3 do not contribute to TcdB internalization, suggesting the presence of a yet-unidentified receptor that facilitates TcdB endocytosis (186). The latter finding is in direct contrast to previous reports that CSPG4 and PVRL3 facilitate receptor-mediated endocytosis of TcdB (155, 157). To account for binding to both nonendocytic and endocytic receptors, Schöttelndreier et al. (174, 186) have proposed a model for TcdA/TcdB uptake and entry. In this model, TcdA/TcdB first bind nonendocytic receptors, enriching the cell surface. Nonendocytic receptors then associate with endocytic receptors, facilitating TcdA/TcdB binding to the endocytic receptor and internalization (174, 186). This model for TcdA/TcdB internalization requires experimental testing. It is not yet known if other LCTs exploit a similar mechanism of cellular internalization.
For LCTs to modify cytosolic factors and exert their toxic effects, LCTs must escape from internalized vesicles. Early on, it was noted that blocking endosomal acidification with small molecule inhibitors of v-ATPases such as bafilomycin prevented LCT toxicity (187–189). Furthermore, acidic pH was shown to trigger conformational changes within LCTs, leading to exposure of hydrophobic surfaces and changes to protease susceptibility (187–189). The optimal pH for the hydrophobic transition of LCTs is between pH 4.0 and 5.0 (187–189), although differences in the optimal pH have been reported for a TcdB subtype (190). The requirement of endosomal acidification for toxicity and acidic pH-induced conformational changes suggest that LCTs translocate out of endosomes in a similar manner to that of other bacterial toxins, including diphtheria toxin (191) and botulinum neurotoxin (192). These bacterial toxins have long been thought to form membrane-inserted pores in the endosomal membrane using their central translocation domains that act as conduits for translocation of toxin enzymatic domains into the cytosol. Several functional studies have shown that LCTs insert into the membrane at acidic pH, leading to the release of rubidium from preloaded cells and formation of ion-conductive pores (193, 194). TcdA has been demonstrated to require cholesterol-enriched membranes for insertion, which may indicate that LCTs preferentially insert into membranes, or regions of membranes, with a distinct lipid composition. Notably, unlike diphtheria toxin (195) and botulinum neurotoxin (196), LCTs do not form stable ion-conductive pores, instead exhibiting characteristic “flickering” electrophysiological behavior, with large conductances of up to ∼1 to 2 nS and lifetimes of several milliseconds (193, 194, 197).
Since the primary sequences of LCTs were determined, it was postulated that the ∼172-residue marginally hydrophobic stretch near the N terminus of the T domain (TcdB amino acids 956 to 1128) was involved in membrane insertion, pore formation, and translocation (20, 22, 140, 193, 194). In 2014, a site-directed loss-of-function mutagenesis screen was performed on conserved LCT residues in the hydrophobic region of TcdB (197). Highly sensitive residues were identified that were >100-fold defective in both pore formation and cellular toxicity when mutated (197). Several years later, regions of the LCT hydrophobic region were directly shown to insert into the membrane, demonstrating the membrane insertion propensity of the hydrophobic region (198). Additionally, an aspartate residue in the hydrophobic region, D1037 in TcdB, was identified as a part of a yet-incompletely described “pH sensor” for membrane insertion (198). A model for how the hydrophobic region may facilitate translocation was proposed, based largely on similarities in hydropathy to the translocation domain of diphtheria toxin and membrane insertion data of the LCT hydrophobic region (197, 198). This model posits that the LCT hydrophobic region forms a membrane-inserted pore, inserting as a “double dagger” of two α-helical hairpins in the membranes, with a nonhydrophobic inserting element located at the N-terminal edge of the hydrophobic region. Importantly, this model has not been experimentally tested and many details of translocation are not known, including the toxin oligomeric state and whether the glycosyltransferase and cysteine protease domain must unfold in order to translocate.
Recently, hundreds of LCT T domain homologues were identified, providing an unprecedented opportunity to gain insights into the elusive mechanism of translocation (199). LCT homologues share on average 18.6% amino acid identity with the TcdB T domain (199) and retain important LCT translocation features (199). Unlike the canonical LCT family, T domain homologues are found elsewhere in addition to clostridia and have variable domain architectures. Nearly 150 (∼20% of all homologues) have an upstream glycosyltransferase and cysteine protease domain, and >300 (∼40% of all homologues) have either an upstream glycosyltransferase or cysteine protease (199). The remaining 40% of homologues have an upstream sequence with no known domain annotation (199). The diversity of the upstream protein region suggests that LCT-like translocases are permissible in the types of proteins they can translocate. Several LCT T domain homologues have been functionally characterized as protein translocases, including an LCT homologue from Serratia marcescens (199) and two LCT homologues from Yersinia mollaretii (200). The S. marcescens LCT homologue has been demonstrated to translocate its upstream cysteine protease and a domain of unknown function into cells, while the LCT homologues from Yersinia mollaretii translocate their upstream enzymatic domains (a cysteine protease domain and either an ADP-ribosyltransferase or glycosyltransferase domain) into cells, which inactivate Rab proteins through ADP ribosylation and glycosylation, respectively (200).
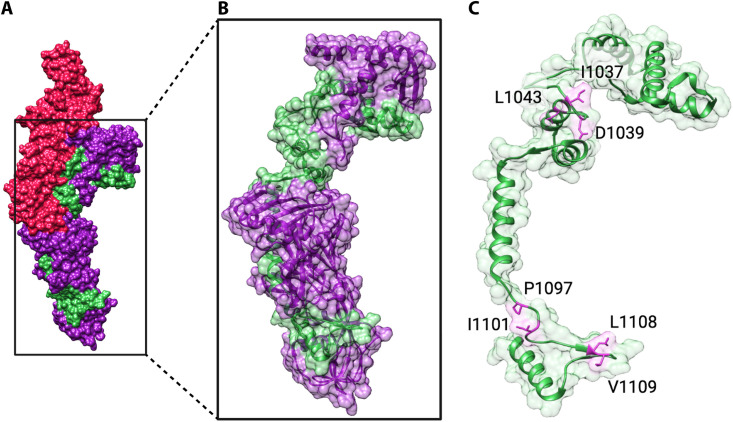
Strikingly, distant T domain homologues have the highest degree of conservation across the N terminus of the LCT T domain; this region was termed the “evolutionarily conserved translocase” (199) (Fig. 5). In TcdB, the evolutionarily conserved translocase was demonstrated to be a functional domain intertwined within receptor-binding sites of the T domain that could independently facilitate translocation of the glycosyltransferase and cysteine protease domain (199). The evolutionarily conserved translocase spans a region of the T domain that extends beyond the hydrophobic region, indicating that the hydrophobic region is not necessary and sufficient for translocation. It is not clear if the evolutionarily conserved translocase is an independently folded domain in other LCTs or LCT homologues or whether this region requires other parts of the protein for stability/solubility.
FIG 5.
Translocation features of the TcdA T domain. (A) Structure of the TcdA T domain (PDB 4R04), with the evolutionarily conserved translocase colored purple (residues 853 to 1475) and the hydrophobic region (residues 958 to 1137) colored green. (B) Focus on the evolutionarily conserved region and (C) the hydrophobic region without the rest of the T domain. Residues with important functions in translocation identified by Zhang et al. (197) are shown as pink sticks.
In a recent analysis of >8,000 tcdB genes, the majority of conserved surfaces across the entire toxin were located in the evolutionarily conserved translocase (138). Thus, the evolutionarily conserved translocase is also an attractive target for broad-spectrum therapeutics, which could target multiple TcdB subtypes, other LCTs, and LCT T domain homologues.
Cysteine Protease Domain
The LCT cysteine protease domain (CPD) autocatalytically cleaves the N-terminal glycosyltransferase from the polypeptide, resulting in the release of the glycosyltransferase into the cytosol (201, 202). The LCT cysteine protease domain shares between 33% and 86% sequence identity among LCTs (Table 7) and belongs to the C80 family of proteases (203). C80 cysteine proteases are found in bacterial pathogens, such as the multifunctional autoprocessing repeats-in-toxin (MARTX) from Vibrio cholerae (204). All LCTs have a conserved histidine, cysteine, and aspartate, which form a catalytic triad that is essential for autoprocessing (201, 205). Autoprocessing is not essential for cytotoxicity, but mutation of the catalytic triad render LCTs less toxic (201, 205–207). It has been hypothesized that autoprocessing is required for optimal activity of the LCT glycosyltransferase, most likely by improving the access of the glycosyltransferase to cellular substrates (208). Enhanced autoprocessing activity has been observed for a TcdB subtype from the epidemic C. difficile RT027, suggesting that more efficient autoprocessing may be responsible for increased toxicity of toxin subtypes (209).
TABLE 7.
Sequence identity and similarity of LCT cysteine protease domains
| LCT | % sequence identity (% similarity)a |
||||
|---|---|---|---|---|---|
| TcdA | TcdB | TcsH | TcsL | TcnA | |
| TcdB | 57 (77) | ||||
| TcsH | 86 (95) | 58 (77) | |||
| TcsL | 57 (77) | 79 (90) | 59 (79) | ||
| TcnA | 33 (57) | 35 (57) | 36 (59) | 36 (60) | |
| TpeL | 53 (71) | 51 (70) | 54 (72) | 51 (72) | 38 (60) |
aSequence identity and similarity were calculated using Water (EMBOSS) local alignment. LCTs are from the following strains: C. difficile VPI 10463, P. sordellii VPI 9048, C. novyi 19402, and C. perfringens JGS1495. Under new subtyping analysis, TcdA and TcdB from C. difficile VPI 10463 are referred to as TcdA1 and TcdB1 (138).
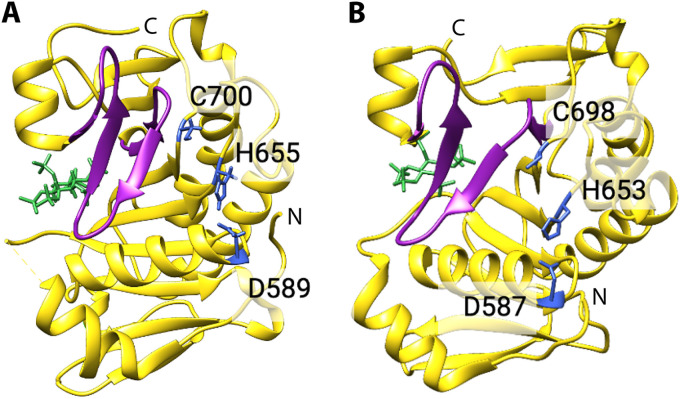
To induce autoprocessing, LCTs must bind to the cellular host factor inositol hexakisphosphate (InsP6), which is found exclusively in the cytosol of eukaryotic cells (210). The equilibrium dissociation constant (KD) values for InsP6 binding to TcdA, TcdB, TcsL, TcnA, and TpeL range from ∼2.0 μM to 9.0 μM (189, 205). The structures of the TcdA and TcdB cysteine protease domains bound to the cellular factor InsP6 were solved in 2009 and 2010, respectively (211, 212) (Fig. 6). The TcdA and TcdB cysteine protease domains have a central β-sheet flanked by a number of α-helices. Structurally, the cysteine protease domains of TcdA and TcdB are very similar to, albeit larger than, the cysteine protease domains of MARTX (213, 214), containing additional helical regions and an additional β-strand. The catalytic triad and InsP6-binding site are on opposite faces of the domain and are separated by a three-stranded β-sheet, termed a β-flap. Binding of InsP6, which is negatively charged, occurs at a basic, lysine-rich positively charged pocket. In TcdA and TcdB, InsP6 binding to the cysteine protease domain induces conformational changes to the β-flap, which in turn transduces conformational changes to the active site region (211, 215). Cleavage occurs after a conserved leucine residue and at neutral pH (205, 216), resulting in release of the glycosyltransferase domain into the cytosol, while the remainder of the toxin is localized in endosomes (217).
FIG 6.
Structural features of the C. difficile toxin cysteine protease domain. (A) TcdA cysteine protease domain (PDB 3HO6) and (B) TcdB cysteine protease domain (PDB 3PEE) bound to InsP6 (268). The β-flap is colored purple, residues of the catalytic triad are depicted as blue sticks, and InsP6 is colored green.
The mechanism of autoprocessing is highly similar among LCTs, although a few key differences have emerged (189, 205). The TcsL holotoxin requires acidic pH to bind InsP6 with high affinity, while all other LCTs preferentially bind InsP6 at neutral pH (205). Binding of TcsL to InsP6 at acidic pH is unexpected, as InsP6 is present solely in the cytosol of eukaryotic cells (210). Interestingly, the TcsL cysteine protease domain binds to InsP6 with ∼10-fold higher affinity at neutral pH than at acidic pH, suggesting that the TcsL holotoxin must undergo major conformational changes at acidic pH before binding to InsP6 at neutral pH (205). Studies have also revealed differences in sensitivity to InsP6-induced cleavage between the TcdA and TcdB holotoxins (165). TcdA is less sensitive to InsP6-induced cleavage, probably due to extensive domain interactions between the TcdA CROP and N terminus at neutral pH (164, 206). It is possible that holotoxins with similar-sized CROP domains to that of TcdA, such as TcsH, may be less sensitive to InsP6-induced cleavage, although to the best of our knowledge, this has not been investigated. Several features relating specifically to TcdA and TcdB autoprocessing activity and regulation have been reported. The histidine and cysteine of the TcdA and TcdB catalytic triad coordinate a zinc ion that is essential for autoprocessing activity (142), and the cysteine of the TcdA and TcdB catalytic triad is regulated through endogenous S-nitrosylation (218). Additionally, autoprocessing has been implicated in regulating the proinflammatory activities of TcdA and TcdB (219). It has not been demonstrated if zinc and S-nitrosylation are involved in autoprocessing activity and regulation or if autoprocessing mediates inflammatory activity of other LCTs.
Glycosyltransferase Domain
Once autocatalytically cleaved from the polypeptide and released into the cytosol, the LCT glycosyltransferase modifies Rho and Ras GTPases by glycosylation, using UDP-glucose or UDP-N-acetylglucosamine (GlcNAc) as a cosubstrate (189, 202, 220–223) (Table 8). Related glycosyltransferases have been identified in pathogenic bacteria, including Escherichia coli (NleB) (224), and species of Legionella (Lgt1, 2, 3) (225) and Photorhabdus (PaTox) (226). Numerous in vitro studies have provided evidence that LCT-mediated glycosylation of Rho and Ras GTPases is essential for cellular toxicity (189, 222, 227–229). Recently, Bilverstone et al. demonstrated that animals infected with strains of C. difficile with glycosyltransferase-defective mutations were unable to induce CDI, providing strong support that glycosyltransferase activity is essential in C. difficile disease pathogenesis (230). At high nanomolar (>1 nM) concentrations of TcdB (163), glycosylation-independent effects, including necrosis (231) and pyknosis (232), have also been reported. The physiological relevance of glycosylation-independent effects of TcdB remains unclear and requires further investigation.
TABLE 8.
Substrate specificity of LCT glycosyltransferase domainsa
| LCT | Sugar donor(s) | Transfer(s) | Strain | Target(s) |
|---|---|---|---|---|
| TcdA | UDP-glucose (220) | Glucose | VPI 10463 (A1) | RhoA/B/C, Rac1, RhoG, Cdc42, Rap1/2, H/K/N-Ras (277, 278) |
| C34 | Rho, Rac, Cdc42, Rap (279) | |||
| TcdB | UDP-glucose (220) | Glucose | VPI 10463 (B1) | RhoA/B/C, Rac1/2/3, Cdc42, RhoG (177, 277, 278, 280) |
| RT027 (B2) | RhoA/B, Rac1/2/3, Cdc42, Rap1A/2A, R-Ras (177, 277, 281) | |||
| 1470 (B3) | Rac, Cdc42, Rap, Ral, R-Ras (282) | |||
| 8864 (B4) | Rac, Cdc42, Rap, Ral, R-Ras (279, 283) | |||
| C34 (B7)b | Rho, Rac, Cdc42, Rap, Ral, R-Ras (279) | |||
| RT019 (B7) | Rac1/2/3, Cdc42, Rap2A/1B, R-Ras1/2, (H/K)-Ras (177, 281) | |||
| HMX-152 (B7) | RhoA (110) | |||
| HSJD-312 (B7) | RhoA (110) | |||
| HMX-149 (B11) | RhoA (110) | |||
| TcsL | UDP-glucose (221) | Glucose | VPI 9048 | Rac, Cdc42, Rap, Ras (221, 284) |
| IP 82 | Rac, Rap, Ral, Ras (221, 285) | |||
| 6018 | Rac1, RhoG, Rap1/2, Ral, (H/K/N)-Ras (277, 284) | |||
| TcsH | UDP-glucosec (221) | Glucose | VPI 9048 | Rho, Rac, Cdc42, Ras (221, 286) |
| TcnA | UDP-GlcNAc (222) | N-Acetylglucosamine | 590, 19402 | Rho, Rac, Cdc42 (221, 287) |
| TpeL | UDP-glucose, UDP-GlcNAcd (189, 223) | Glucose, N-acetylglucosamine | MC18 | Rac, Rap, Ral, Ras (223) |
| JGS1495 | (H/K/N)-Ras (288) |
aThis table is based in part on data from Chandrasekaran and Lacy (239). If the sequence of the C. difficile subtype was available, the TcdA or TcdB subtype is indicated in parentheses. Subtyping based on that reported by Mansfield et al. (138).
bBased on partial sequencing of the toxin gene.
cBased on conservation of key UDP-glucose binding features.
dPreferentially modifies GTPases by GlcNAcylation using UDP-GlcNAc (189).
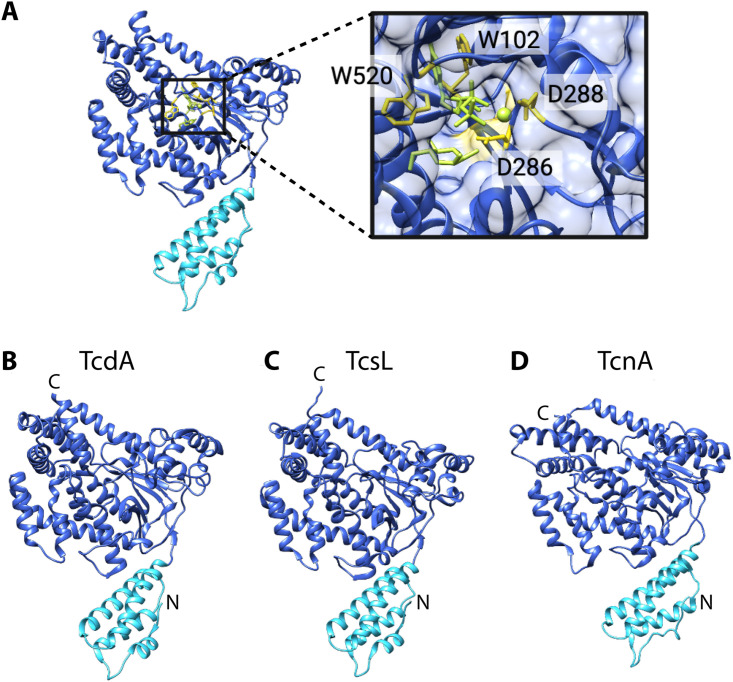
LCTs glycosylate Rho and Ras GTPases by cleaving the UDP-glucose or UDP-GlcNAc cosubstrate and transferring glucose or N-acetylglucosamine onto the conserved Thr35/37 of the target GTPase (233). LCT glycosyltransferases have conserved residues that are essential for catalysis, most notably the aspartate-X-aspartate (DXD) motif (TcdB D286/D288) and tryptophan (TcdB W102, W520) (52, 234, 235) that are important for binding and coordination of the cosubstrate and a manganese ion (189, 227, 228, 234, 236, 237) (Fig. 7A). The glycotransferase domain shares between 34% and 85% sequence identity among LCTs (Table 9), and differences in sequence have been ascribed to differences in cosubstrate and GTPase specificity. In LCTs, specificity for UDP-glucose or UDP-GlcNAc is dictated by two residues (I383 and Q385 in TcdB) (223, 238, 239). The molecular basis for GTPase specificity in LCTs has not been completely defined. Preferential targeting of Rho and Ras GTPases in TcdB and TcsL has been attributed to specific residues in the glycosyltransferase domain, such as those in α-helix 17 (residues 444 to 455) in TcdB (45).
FIG 7.
Structural features of the LCT glycosyltransferase domain. (A) TcdB glycosyltransferase domain (PDB 2BVM) in complex with the cosubstrate and a manganese ion, depicted as green sticks and a sphere, respectively. Residues important for catalytic function are depicted as yellow sticks. Structures of the (B) TcdA (PDB 3SRZ), (C) TcsL (PDB 2VKD), and (D) TcnA (PDB 2VK9) glycosyltransferase domains. The membrane localization domain (MLD) is colored aqua.
TABLE 9.
Sequence identity and similarity of LCT glycosyltransferase domains
| LCT | % sequence identity (% similarity)a |
||||
|---|---|---|---|---|---|
| TcdA | TcdB | TcsH | TcsL | TcnA | |
| TcdB | 51 (72) | ||||
| TcsH | 85 (92) | 51 (73) | |||
| TcsL | 53 (73) | 76 (88) | 55 (75) | ||
| TcnA | 34 (53) | 34 (54) | 34 (52) | 36 (54) | |
| TpeL | 46 (66) | 44 (65) | 46 (67) | 48 (66) | 34 (57) |
aSequence identity and similarity were calculated using Water (EMBOSS) local alignment. LCTs are from the following strains: C. difficile VPI 10463, P. sordellii VPI9048, C. novyi 19402, and C. perfringens JGS1495. Under new subtyping analysis, TcdA and TcdB from C. difficile VPI 10463 are referred to as TcdA1 and TcdB1 (138).
The structures of the TcdA (240, 241), TcdB (237), TcsL (242), and TcnA (242) glycosyltransferase domains have been solved by X-ray crystallography (Fig. 7). LCT glycosyltransferases belong to the GT-A family of glycosyltransferases (243), which are defined by a core α/β/α sandwich that resembles a Rossmann fold, a tertiary fold found in many nucleotide-binding proteins (244). In addition to the Rossmann-like fold, LCTs have multiple α-helical subdomains. The N-terminal subdomain (∼1 to 90 amino acids) is a membrane localization domain that targets the glycosyltransferase to the cytosolic leaflets of the cell membrane, where it can access membrane-bound Rho and Ras GTPases. The nucleotide sugar-binding pocket is formed by the edge of the β-sheet and several α-helices and is overlaid with a flexible loop that is involved in binding of the phosphate of the nucleotide sugar (245, 246). The flexible loop undergoes conformational changes upon substrate binding, defining open and closed conformations of the LCT glycosyltransferase. The TcdA and TcdB glycosyltransferase domains have been shown to interact with the TRiC/CCT chaperonin system (247), which may aid in glycosyltransferase refolding in the cytoplasm after unfolding in the acidic environment of the endosome. It is not yet known if the TcsL, TcsH, TpeL, or TcnA glycosyltransferase interacts with cytosolic chaperones.
GTPases are molecular switches that cycle between an active GTP-bound state and an inactive GDP-bound state. LCTs preferentially target GTPases in their inactive GDP-bound state (202, 233). Rho GTPases are master regulators of the actin cytoskeleton, control motile cellular processes, and are involved in cell cycle control and polarity (248). Ras GTPases are essential for assembly and function of cell-cell junctions, cell differentiation, and proliferation (249).
LCT-mediated glycosylation of Rho and Ras GTPases has been correlated with both cytopathic effects (i.e., loss of cytoskeletal structure, resulting in the characteristic cell rounding phenotype) and cytotoxic effects (i.e., apoptosis, resulting in cell death) (Tables 9 and 10). In addition to the major cytopathic and cytotoxic effects, glycosylation of Rho and Ras proteins has also been correlated with cell cycle arrest and defects in cell proliferation in TcdB and TcsL (250–252). Glycosylation-dependent cytopathic and cytotoxic effects are believed to be primarily responsible for LCT-mediated tissue damage, although the precise mechanisms underlying tissue damage and cell death are not well defined. Broadly, two types of cytopathic effects have been described for LCTs: cell rounding with protrusions radiating around rounded cells, also referred to as the “arborizing” cytopathic effect, and cell rounding without protrusions (253). TcdA, TcnA, and TcdB subtypes B1 and B2 have been reported to induce the arborizing cytopathic effect, while TcsL and TcdB subtypes B3, B4, and B7 have been reported to induce cell rounding without protrusions (110, 138, 253, 254). It is not yet clear why some LCTs induce the arborizing phenotype, although altered specificity for GTPases has been proposed (138, 253, 254).
TABLE 10.
LCT-mediated glycosylation of Rho and Ras GTPases and correlated cellular effectsa
| LCT | Cytopathic effect | Cytotoxic effect | Pyroptosis |
|---|---|---|---|
| TcdA | Rho GTPases (Rac) (289) | Rho GTPases (290) | RhoA (260) |
| TcdB | Rho GTPases (Rac) (291) | Rho GTPases (RhoA) (292) | RhoA (258) |
| TcsL | Rho GTPases (Rac) (250, 293) | Ras GTPases ([H/K/N]-Ras) (250) | ND |
| TcsH | Rho GTPases (221, 286) | Rho GTPases (221, 286) | ND |
| TcnA | Rho GTPases (221, 287) | Rho GTPases (221, 287) | ND |
| TpeL | Rho GTPases (Rac) (223) | Ras GTPases (189) | ND |
The following strains are used for each LCT: VPI10463 (TcdA, TcdB); VPI 9048 (TcsL); VPI 9048 (TcsH); 590 (TcnA); MC18 (TpeL). ND, not determined.
It is well established that TcdA and TcdB induce an inflammatory response in intestinal epithelial cells (239). In the context of CDI, inflammation may be both beneficial as a host defense mechanism for pathogen eradication and harmful if the inflammatory response is prolonged and uncontrolled (255, 256). There are conflicting reports on the requirement for glycosyltransferase activity in the TcdA- and TcdB-induced inflammatory response (229, 239, 257–259). In myeloid cells, glycosylation of RhoA by TcdA and TcdB has been correlated with the activation of the pyrin inflammasome, a multimeric protein complex that activates procascapse-1, which then activates proinflammatory cytokines, such as interleukin-1β (IL-1β) and IL-18 (258, 260, 261). Activation of the pyrin inflammasome can lead to pyroptosis, a caspase-1-dependent cell death that is highly inflammatory and characterized by cell swelling and lysis (258, 262, 263). A recent report by Saavedra et al. provided evidence that the pyrin inflammasome does not have a role in TcdA/TcdB-induced killing of mouse intestinal epithelial cells, due to the absence of pyrin expression (264). The role of TcdA/TcdB-induced activation of the pyrin inflammasome in human intestinal epithelial cells has yet to be investigated. Interestingly, TcsL has been reported to induce an anti-inflammatory response to inactivation of Ras proteins (45). To the best of our knowledge, it is not known whether TpeL, TcnA, and TcsH induce an inflammatory response.
NEWLY IDENTIFIED LCT HOMOLOGUES
The recent identification of hundreds of LCT homologues (199) has opened up a new avenue of LCT research and has provided a novel lens for understanding toxin structure and function. As detailed in this review, LCTs were first identified in context of disease and defined by their toxicity to humans at low doses. The genomics-driven identification of hundreds of LCT homologues has reversed the paradigm of LCT discovery, as bioinformatically identified homologues are identified on the basis of sequence similarity alone. It is not known if LCT homologues are involved in disease processes or what the ecological function of homologues may be.
On the basis of their frequent co-occurrence with virulence and mobile genes and presence in pathogenic bacteria, recently identified LCT homologues have been hypothesized to be putative toxins (199). Notably, the majority of recently identified LCT homologues are present in nonclostridial species that are directly pathogenic to insects (i.e., Pseudomonas, Photorhabdus, and Yersinia) or in species that, due to plant growth-promoting properties, may be pathogenic to insects or other plant pathogens, such as nematodes (199). Interestingly, as reported by Mansfield and Doxey (265) and Contreras et al. (266), numerous links to insects have also been made for botulinum neurotoxin homologues. The significance and evolutionary implications of LCT homologues—and more broadly, bacterial toxins—targeting insects and other plant pathogens is unclear. It may be suggested that human-targeted LCTs evolved from insect-targeting homologues and/or that LCTs pose a selective advantage to clostridia in the environment.
CONCLUDING REMARKS AND FUTURE DIRECTIONS
Through decades of work spanning the fields of clinical and veterinary medicine, microbiology, biochemistry, and structural biology, LCTs have emerged as some of the deadliest poisons of humans and animals. While members of the LCT family share many similar features, including sequence homology and general mechanism of action, LCTs also have distinct clinical and molecular features. Recent work has demonstrated that sequence variation in TcdA and TcdB across different C. difficile strains results in toxin subtypes with different functional and immunological activities, warranting the creation of a toxin subtyping system. C. difficile toxin subtyping is directly relevant for diagnostic and therapeutic development and should be used to guide the use of toxin-mediated treatments, such as bezlotoxumab, in the clinic. It remains to be determined whether sequence variation exists in other LCTs across clostridial strains and, if so, whether sequence variations result in toxins with different biological and functional activities.
A clearer picture of LCT-mediated infection is emerging, particularly for TcdA, TcdB, and TcsL; the roles in virulence of TcnA, TpeL, and TcsH remain less well defined. Although great strides have been made in LCT structure and function, a detailed mechanistic understanding of LCT intoxication is still elusive. Central to understanding LCT intoxication is identifying the receptors LCTs bind to mediate cell surface attachment and internalization and the GTPases LCTs target. The additional layer of complexity that also must be addressed is how the difference in receptor engagement and substrate specificity between LCTs and toxin subtypes contributes to differences in toxin-mediated pathology and disease progression. Perhaps the greatest challenge is determining how LCTs translocate their glycosyltransferase and cysteine protease domain across the endosomal membrane and into the cytosol. Based on conserved sequence features in membrane-inserting regions, it seems likely that the mechanism of translocation is highly similar among LCTs and subtypes. Additional work is required to gain a deeper and more precise understanding of the enigmatic mechanism of translocation.
Genomics has expanded the LCT family from six proteins to several hundred, thus establishing a new frontier in LCT research. Characterization and determination of the receptors that homologues engage and the intracellular substrates they modify—and by extension, the organisms they target—remain difficult. Elucidating the function of LCT homologues, particularly in their native environment, will clarify their ecological roles and potential adaptive value in bacteria. Importantly, homologues also hold clues to how LCTs evolved to become deadly human poisons and may lead us closer to identifying the ancestral LCT toxin.
ACKNOWLEDGMENTS
Figures were created with BioRender, Toronto, Canada.
Biographies

Kathleen E. Orrell, Ph.D., is a postdoctoral fellow in the Molecular Medicine Program at The Hospital for Sick Children in Toronto, Canada. She received her doctoral degree in biochemistry from the University of Toronto in 2021. Her doctoral work focused on large clostridial toxin translocation across host cell membranes. Dr. Orrell’s current work investigates structural and functional features of novel bacterial toxin effectors.

Roman A. Melnyk, Ph.D., is an Associate Professor of Biochemistry at the University of Toronto and Senior Scientist in Molecular Medicine at The Hospital for Sick Children. He received his Ph.D. in biochemistry from the University of Toronto in 2004 and then was a postdoctoral fellow at Harvard Medical School from 2004 to 2006. Following this, he was a Sr. Scientist at Merck Frosst Centre for Therapeutic Research between 2006 and 2011. His research focuses on understanding the structure and function of bacterial toxins and using this information both to develop novel small molecule antitoxins and for engineering intracellular delivery platforms.
REFERENCES
- 1.Savva CG, Clark AR, Naylor CE, Popoff MR, Moss DS, Basak AK, Titball RW, Bokori-Brown M. 2019. The pore structure of Clostridium perfringens epsilon toxin. Nat Commun 10:2641. 10.1038/s41467-019-10645-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerding DN, Johnson S, Rupnik M, Aktories K. 2014. Clostridium difficile binary toxin CDT: mechanism, epidemiology, and potential clinical importance. Gut Microbes 5:15–27. 10.4161/gmic.26854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bette P, Oksche A, Mauler F, Eichel-Streiber Cv, Popoff MR, Habermann E. 1991. A comparative biochemical, pharmacological and immunological study of Clostridium novyi alpha-toxin, C. difficile toxin B and C. sordellii lethal toxin. Toxicon 29:877–887. 10.1016/0041-0101(91)90224-F. [DOI] [PubMed] [Google Scholar]
- 4.Aronoff DM, Kazanjian PH. 2018. Historical and contemporary features of infections due to Clostridium novyi. Anaerobe 50:80–84. 10.1016/j.anaerobe.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 5.Aldape MJ, Bryant AE, Stevens DL. 2006. Clostridium sordellii infection: epidemiology, clinical findings, and current perspectives on diagnosis and treatment. Clin Infect Dis 43:1436–1446. 10.1086/508866. [DOI] [PubMed] [Google Scholar]
- 6.Shindo Y, Dobashi Y, Sakai T, Monma C, Miyatani H, Yoshida Y. 2015. Epidemiological and pathobiological profiles of Clostridium perfringens infections: review of consecutive series of 33 cases over a 13-year period. Int J Clin Exp Pathol 8:569–577. [PMC free article] [PubMed] [Google Scholar]
- 7.Popoff MR, Bouvet P. 2009. Clostridial toxins. Future Microbiol 4:1021–1064. 10.2217/fmb.09.72. [DOI] [PubMed] [Google Scholar]
- 8.Orrell KE, Zhang Z, Sugiman-Marangos SN, Melnyk RA. 2017. Clostridium difficile toxins A and B: receptors, pores, and translocation into cells. Crit Rev Biochem Mol Biol 52:461–473. 10.1080/10409238.2017.1325831. [DOI] [PubMed] [Google Scholar]
- 9.Hall IC, O’Toole E. 1935. Intestinal flora in new-born infants with a description of a new pathogenic anaerobic, Bacillus difficilis. Am J Dis Child 49:390–402. 10.1001/archpedi.1935.01970020105010. [DOI] [Google Scholar]
- 10.Snyder ML. 1937. Further studies on Bacillus difficilis (Hall and O'Toole). J Infect Dis 60:223–231. 10.1093/infdis/60.2.223. [DOI] [Google Scholar]
- 11.Yutin N, Galperin MY. 2013. A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ Microbiol 15:2631–2641. 10.1111/1462-2920.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawson PA, Citron DM, Tyrrell KL, Finegold SM. 2016. Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O'Toole 1935) Prevot 1938. Anaerobe 40:95–99. 10.1016/j.anaerobe.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 13.Bartlett JG. 1994. Clostridium difficile: history of its role as an enteric pathogen and the current state of knowledge about the organism. Clin Infect Dis 18:S265–S272. 10.1093/clinids/18.Supplement_4.S265. [DOI] [PubMed] [Google Scholar]
- 14.Bartlett JG, Onderdonk AB, Cisneros RL, Kasper DL. 1977. Clindamycin-associated colitis due to a toxin-producing species of Clostridium in hamsters. J Infect Dis 136:701–705. 10.1093/infdis/136.5.701. [DOI] [PubMed] [Google Scholar]
- 15.Bartlett JG, Chang TW, Gurwith M, Gorbach SL, Onderdonk AB. 1978. Antibiotic-associated pseudomembranous colitis due to toxin-producing clostridia. N Engl J Med 298:531–534. 10.1056/NEJM197803092981003. [DOI] [PubMed] [Google Scholar]
- 16.Smits WK, Lyras D, Lacy DB, Wilcox MH, Kuijper EJ. 2016. Clostridium difficile infection. Nat Rev Dis Primers 2:16020. 10.1038/nrdp.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor NS, Thorne GM, Bartlett JG. 1981. Comparison of two toxins produced by Clostridium difficile. Infect Immun 34:1036–1043. 10.1128/IAI.34.3.1036-1043.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.von Eichel-Streiber C, Suckau D, Wachter M, Hadding U. 1989. Cloning and characterization of overlapping DNA fragments of the toxin A gene of Clostridium difficile. J Gen Microbiol 135:55–64. 10.1099/00221287-135-1-55. [DOI] [PubMed] [Google Scholar]
- 19.Dove CH, Wang SZ, Price SB, Phelps CJ, Lyerly DM, Wilkins TD, Johnson JL. 1990. Molecular characterization of the Clostridium difficile toxin A gene. Infect Immun 58:480–488. 10.1128/IAI.58.2.480-488.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.von Eichel-Streiber C, Sauerborn M. 1990. Clostridium difficile toxin A carries a C-terminal repetitive structure homologous to the carbohydrate binding region of streptococcal glycosyltransferases. Gene 96:107–113. 10.1016/0378-1119(90)90348-u. [DOI] [PubMed] [Google Scholar]
- 21.von Eichel-Streiber C, Laufenberg-Feldmann R, Sartingen S, Schulze J, Sauerborn M. 1990. Cloning of Clostridium difficile toxin B gene and demonstration of high N-terminal homology between toxin A and B. Med Microbiol Immunol 179:271–279. 10.1007/BF00192465. [DOI] [PubMed] [Google Scholar]
- 22.von Eichel-Streiber C, Laufenberg-Feldmann R, Sartingen S, Schulze J, Sauerborn M. 1992. Comparative sequence analysis of the Clostridium difficile toxins A and B. Mol Gen Genet 233:260–268. 10.1007/BF00587587. [DOI] [PubMed] [Google Scholar]
- 23.CDC. 2020. Biggest threats and data: 2019 AR threats report. CDC, Atlanta, GA. [Google Scholar]
- 24.Jump RL. 2013. Clostridium difficile infection in older adults. Aging Health 9:403–414. 10.2217/ahe.13.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown AWW, Wilson RB. 2018. Clostridium difficile colitis and zoonotic origins-a narrative review. Gastroenterol Rep (Oxf) 6:157–166. 10.1093/gastro/goy016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaffler H, Breitruck A. 2018. Clostridium difficile - from colonization to infection. Front Microbiol 9:646. 10.3389/fmicb.2018.00646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vindigni SM, Surawicz CM. 2015. C. difficile infection: changing epidemiology and management paradigms. Clin Transl Gastroenterol 6:e99. 10.1038/ctg.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta A, Khanna S. 2014. Community-acquired Clostridium difficile infection: an increasing public health threat. Infect Drug Resist 7:63–72. 10.2147/IDR.S46780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP. 2010. The role of toxin A and toxin B in Clostridium difficile infection. Nature 467:711–713. 10.1038/nature09397. [DOI] [PubMed] [Google Scholar]
- 30.Kuehne SA, Collery MM, Kelly ML, Cartman ST, Cockayne A, Minton NP. 2014. Importance of toxin A, toxin B, and CDT in virulence of an epidemic Clostridium difficile strain. J Infect Dis 209:83–86. 10.1093/infdis/jit426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warny M, Pepin J, Fang A, Killgore G, Thompson A, Brazier J, Frost E, McDonald LC. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079–1084. 10.1016/S0140-6736(05)67420-X. [DOI] [PubMed] [Google Scholar]
- 32.Akerlund T, Svenungsson B, Lagergren A, Burman LG. 2006. Correlation of disease severity with fecal toxin levels in patients with Clostridium difficile-associated diarrhea and distribution of PCR ribotypes and toxin yields in vitro of corresponding isolates. J Clin Microbiol 44:353–358. 10.1128/JCM.44.2.353-358.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drudy D, Fanning S, Kyne L. 2007. Toxin A-negative, toxin B-positive Clostridium difficile. Int J Infect Dis 11:5–10. 10.1016/j.ijid.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 34.Rupnik M, Kato N, Grabnar M, Kato H. 2003. New types of toxin A-negative, toxin B-positive strains among Clostridium difficile isolates from Asia. J Clin Microbiol 41:1118–1125. 10.1128/jcm.41.3.1118-1125.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lyras D, O'Connor JR, Howarth PM, Sambol SP, Carter GP, Phumoonna T, Poon R, Adams V, Vedantam G, Johnson S, Gerding DN, Rood JI. 2009. Toxin B is essential for virulence of Clostridium difficile. Nature 458:1176–1179. 10.1038/nature07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carter GP, Chakravorty A, Pham Nguyen TA, Mileto S, Schreiber F, Li L, Howarth P, Clare S, Cunningham B, Sambol SP, Cheknis A, Figueroa I, Johnson S, Gerding D, Rood JI, Dougan G, Lawley TD, Lyras D. 2015. Defining the roles of TcdA and TcdB in localized gastrointestinal disease, systemic organ damage, and the host response during Clostridium difficile infections. mBio 6:e00551. 10.1128/mBio.00551-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marvaud JC, Quevedo-Torres S, Eckert C, Janoir C, Barbut F. 2019. Virulence of new variant strains of Clostridium difficile producing only toxin A or binary toxin in the hamster model. New Microbes New Infect 32:100590. 10.1016/j.nmni.2019.100590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hall IC, Scott JP. 1927. Bacillus sordellii, a cause of malignant edema in man. J Infect Dis 41:329–335. 10.1093/infdis/41.5.329. [DOI] [Google Scholar]
- 39.Humphreys F, Meleney FL. 1928. The identity of C. oedematoides and B. sordelli. Proc Soc Exp Bio Med 25:611–614. 10.3181/00379727-25-3975. [DOI] [Google Scholar]
- 40.Hall IC, Rymer MR, Jungherr E. 1929. Comparative study of Bacillus sordellii (Hall and Scott) and Clostridium oedematoides (Meleney, Humphreys, and Carp). J Infect Dis 45:42–66. 10.1093/infdis/45.1.42. [DOI] [Google Scholar]
- 41.Sasi Jyothsna TS, Tushar L, Sasikala C, Ramana CV. 2016. Paraclostridium benzoelyticum gen. nov., sp. nov., isolated from marine sediment and reclassification of Clostridium bifermentans as Paraclostridium bifermentans comb. nov. Proposal of a new genus Paeniclostridium gen. nov. to accommodate Clostridium sordellii and Clostridium ghonii. Int J Syst Evol Microbiol 66:1268–1274. 10.1099/ijsem.0.000874. [DOI] [PubMed] [Google Scholar]
- 42.Arseculeratne SN, Panabokke RG, Wijesundera S. 1969. The toxins responsible for the lesions of Clostridium sordelli gas gangrene. J Med Microbiol 2:37–53. 10.1099/00222615-2-1-37. [DOI] [PubMed] [Google Scholar]
- 43.Popoff MR. 1987. Purification and characterization of Clostridium sordellii lethal toxin and cross-reactivity with Clostridium difficile cytotoxin. Infect Immun 55:35–43. 10.1128/IAI.55.1.35-43.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez RD, Wilkins TD. 1988. Purification and characterization of Clostridium sordellii hemorrhagic toxin and cross-reactivity with Clostridium difficile toxin A (enterotoxin). Infect Immun 56:1215–1221. 10.1128/IAI.56.5.1215-1221.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Popoff MR. 2018. Clostridium difficile and Clostridium sordellii toxins, proinflammatory versus anti-inflammatory response. Toxicon 149:54–64. 10.1016/j.toxicon.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 46.Maclennan JD. 1962. The histotoxic clostridial infections of man. Bacteriol Rev 26:177–276. 10.1128/BR.26.2_Pt_1-2.177-274.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buboltz JB, Murphy-Lavoie HM. 2020. Gas gangrene. StatPearls, Treasure Island, FL. [PubMed] [Google Scholar]
- 48.Brett MM, Hood J, Brazier JS, Duerden BI, Hahne SJ. 2005. Soft tissue infections caused by spore-forming bacteria in injecting drug users in the United Kingdom. Epidemiol Infect 133:575–582. 10.1017/S0950268805003845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kimura AC, Higa JI, Levin RM, Simpson G, Vargas Y, Vugia DJ. 2004. Outbreak of necrotizing fasciitis due to Clostridium sordellii among black-tar heroin users. Clin Infect Dis 38:e87–e91. 10.1086/383471. [DOI] [PubMed] [Google Scholar]
- 50.Rorbye C, Petersen IS, Nilas L. 2000. Postpartum Clostridium sordellii infection associated with fatal toxic shock syndrome. Acta Obstet Gynecol Scand 79:1134–1135. [PubMed] [Google Scholar]
- 51.Fischer M, Bhatnagar J, Guarner J, Reagan S, Hacker JK, Van Meter SH, Poukens V, Whiteman DB, Iton A, Cheung M, Dassey DE, Shieh WJ, Zaki SR. 2005. Fatal toxic shock syndrome associated with Clostridium sordellii after medical abortion. N Engl J Med 353:2352–2360. 10.1056/NEJMoa051620. [DOI] [PubMed] [Google Scholar]
- 52.Thiele TL, Stuber TP, Hauer PJ. 2013. Detection of Clostridium sordellii strains expressing hemorrhagic toxin (TcsH) and implications for diagnostics and regulation of veterinary vaccines. Vaccine 31:5082–5087. 10.1016/j.vaccine.2013.08.065. [DOI] [PubMed] [Google Scholar]
- 53.Voth DE, Martinez OV, Ballard JD. 2006. Variations in lethal toxin and cholesterol-dependent cytolysin production correspond to differences in cytotoxicity among strains of Clostridium sordellii. FEMS Microbiol Lett 259:295–302. 10.1111/j.1574-6968.2006.00287.x. [DOI] [PubMed] [Google Scholar]
- 54.Couchman EC, Browne HP, Dunn M, Lawley TD, Songer JG, Hall V, Petrovska L, Vidor C, Awad M, Lyras D, Fairweather NF. 2015. Clostridium sordellii genome analysis reveals plasmid localized toxin genes encoded within pathogenicity loci. BMC Genomics 16:392. 10.1186/s12864-015-1613-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Geny B, Khun H, Fitting C, Zarantonelli L, Mazuet C, Cayet N, Szatanik M, Prevost MC, Cavaillon JM, Huerre M, Popoff MR. 2007. Clostridium sordellii lethal toxin kills mice by inducing a major increase in lung vascular permeability. Am J Pathol 170:1003–1017. 10.2353/ajpath.2007.060583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hao Y, Senn T, Opp JS, Young VB, Thiele T, Srinivas G, Huang SK, Aronoff DM. 2010. Lethal toxin is a critical determinant of rapid mortality in rodent models of Clostridium sordellii endometritis. Anaerobe 16:155–160. 10.1016/j.anaerobe.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carter GP, Awad MM, Hao Y, Thelen T, Bergin IL, Howarth PM, Seemann T, Rood JI, Aronoff DM, Lyras D. 2011. TcsL is an essential virulence factor in Clostridium sordellii ATCC 9714. Infect Immun 79:1025–1032. 10.1128/IAI.00968-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amimoto K, Oishi E, Yasuhar H, Sasak O, Katayama S, Kitajima T, Izumida A, Hirahara T. 2001. Protective effects of Clostridium sordellii LT and HT toxoids against challenge with spores in guinea pigs. J Vet Med Sci 63:879–883. 10.1292/jvms.63.879. [DOI] [PubMed] [Google Scholar]
- 59.Weinberg M. 1917. La gangrene gazeuse: bacteriologie, reproduction experimentale, serotherapie. Masson & Cie, Paris, France. [Google Scholar]
- 60.Bergey FH, Breed R, Hammer B, Huntoon F. 1923. Bergey's manual of determinative bacteriology. Williams and Wilkins, Baltimore, MD. [Google Scholar]
- 61.Izumi N, Kondo H, Ohishi I, Sakaguchi G. 1983. Purification and characterization of alpha-toxin of Clostridium oedematiens type A. Jpn J Med Sci Biol 36:135–146. 10.7883/yoken1952.36.135. [DOI] [PubMed] [Google Scholar]
- 62.Izumi N, Niiro M, Kondo H. 1983. Clostridium oedematiens type A toxin: the correlation between the lethal and edematizing activities. Jpn J Med Sci Biol 36:67–74. 10.7883/yoken1952.36.67. [DOI] [PubMed] [Google Scholar]
- 63.Bette P, Frevert J, Mauler F, Suttorp N, Habermann E. 1989. Pharmacological and biochemical studies of cytotoxicity of Clostridium novyi type A alpha-toxin. Infect Immun 57:2507–2513. 10.1128/IAI.57.8.2507-2513.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hofmann F, Herrmann A, Habermann E, von Eichel-Streiber C. 1995. Sequencing and analysis of the gene encoding the alpha-toxin of Clostridium novyi proves its homology to toxins A and B of Clostridium difficile. Mol Gen Genet 247:670–679. 10.1007/BF00290398. [DOI] [PubMed] [Google Scholar]
- 65.Navarro MA, Uzal FA. 2020. Pathobiology and diagnosis of clostridial hepatitis in animals. J Vet Diagn Invest 32:192–202. 10.1177/1040638719886567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nyaoke AC, Navarro MA, Beingesser J, Uzal FA. 2018. Infectious necrotic hepatitis caused by Clostridium novyi type B in a horse: case report and review of the literature. J Vet Diagn Invest 30:294–299. 10.1177/1040638717737125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McGuigan CC, Penrice GM, Gruer L, Ahmed S, Goldberg D, Black M, Salmon JE, Hood J. 2002. Lethal outbreak of infection with Clostridium novyi type A and other spore-forming organisms in Scottish injecting drug users. J Med Microbiol 51:971–977. 10.1099/0022-1317-51-11-971. [DOI] [PubMed] [Google Scholar]
- 68.Ryan JM, Paul J, Curtis S, Patel NK. 2001. Clostridium novyi infection: a fatal association with injecting drug users. Emerg Med J 18:138–139. 10.1136/emj.18.2.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Finn SP, Leen E, English L, O'Briain DS. 2003. Autopsy findings in an outbreak of severe systemic illness in heroin users following injection site inflammation: an effect of Clostridium novyi exotoxin? Arch Pathol Lab Med 127:1465–1470. 10.5858/2003-127-1465-AFIAOO. [DOI] [PubMed] [Google Scholar]
- 70.Mauricio Navarro FDQ, Uzal FA. 2016. Bacillary hemoglobinuria, p 265–274. In Uzal FA, Songer JG, Prescott JF, Popoff MR. (ed), Clostridial diseases of animals. Wiley Blackwell, Hoboken, NJ. [Google Scholar]
- 71.Jeong CG, Seo BJ, Nazki S, Jung BK, Khatun A, Yang MS, Kim SC, Noh SH, Shin JH, Kim B, Kim WI. 2020. Characterization of Clostridium novyi isolated from a sow in a sudden death case in Korea. BMC Vet Res 16:127. 10.1186/s12917-020-02349-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rood JI, Adams V, Lacey J, Lyras D, McClane BA, Melville SB, Moore RJ, Popoff MR, Sarker MR, Songer JG, Uzal FA, Van Immerseel F. 2018. Expansion of the Clostridium perfringens toxin-based typing scheme. Anaerobe 53:5–10. 10.1016/j.anaerobe.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Welch WH, Nuttall GHF. 1892. A gas-producing bacillus (Bagillus aerogenes capsulatus, nov. spec.) capable of rapid development in the blood-vessels after death. Johns Hopkins Hosp Bull 24. [Google Scholar]
- 74.Achalme MP. 1891. Examen bacteriologie d’un cas de rhumatisme articulaire aigu mort de rhumatisme cerebral. Compt Rendu Soc Biol Paris 1891:651–656. [Google Scholar]
- 75.Veillon A, Zuber A. 1898. Recherches sur quelques microbes strictement anaerobies et leur vole en pathologie. Arch Med Exp Anat Pathol 10:517–545. [Google Scholar]
- 76.Hauduroy P, Urbain A, Guillot J. 1937. Dictionnaire des bacteries pathogenes pour l'homme, les animaux et les plantes. Masson & Cie, Paris, France. [Google Scholar]
- 77.Amimoto K, Noro T, Oishi E, Shimizu M. 2007. A novel toxin homologous to large clostridial cytotoxins found in culture supernatant of Clostridium perfringens type C. Microbiology 153:1198–1206. 10.1099/mic.0.2006/002287-0. [DOI] [PubMed] [Google Scholar]
- 78.Uzal FA, Freedman JC, Shrestha A, Theoret JR, Garcia J, Awad MM, Adams V, Moore RJ, Rood JI, McClane BA. 2014. Towards an understanding of the role of Clostridium perfringens toxins in human and animal disease. Future Microbiol 9:361–377. 10.2217/fmb.13.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Grass JE, Gould LH, Mahon BE. 2013. Epidemiology of foodborne disease outbreaks caused by Clostridium perfringens, United States, 1998–2010. Foodborne Pathog Dis 10:131–136. 10.1089/fpd.2012.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brett MM, Rodhouse JC, Donovan TJ, Tebbutt GM, Hutchinson DN. 1992. Detection of Clostridium perfringens and its enterotoxin in cases of sporadic diarrhoea. J Clin Pathol 45:609–611. 10.1136/jcp.45.7.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Heida FH, van Zoonen A, Hulscher JBF, Te Kiefte BJC, Wessels R, Kooi EMW, Bos AF, Harmsen HJM, de Goffau MC. 2016. A necrotizing enterocolitis-associated gut microbiota is present in the meconium: results of a prospective study. Clin Infect Dis 62:863–870. 10.1093/cid/ciw016. [DOI] [PubMed] [Google Scholar]
- 82.Keyburn AL, Bannam TL, Moore RJ, Rood JI. 2010. NetB, a pore-forming toxin from necrotic enteritis strains of Clostridium perfringens. Toxins (Basel) 2:1913–1927. 10.3390/toxins2071913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cooper KK, Songer JG, Uzal FA. 2013. Diagnosing clostridial enteric disease in poultry. J Vet Diagn Invest 25:314–327. 10.1177/1040638713483468. [DOI] [PubMed] [Google Scholar]
- 84.M'Sadeq SA, Wu S, Swick RA, Choct M. 2015. Towards the control of necrotic enteritis in broiler chickens with in-feed antibiotics phasing-out worldwide. Anim Nutr 1:1–11. 10.1016/j.aninu.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gurjar A, Li J, McClane BA. 2010. Characterization of toxin plasmids in Clostridium perfringens type C isolates. IAI 78:4860–4869. 10.1128/IAI.00715-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sayeed S, Li J, McClane BA. 2010. Characterization of virulence plasmid diversity among Clostridium perfringens type B isolates. Infect Immun 78:495–504. 10.1128/IAI.00838-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen J, McClane BA. 2015. Characterization of Clostridium perfringens TpeL toxin gene carriage, production, cytotoxic contributions, and trypsin sensitivity. Infect Immun 83:2369–2381. 10.1128/IAI.03136-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Freedman JC, Theoret JR, Wisniewski JA, Uzal FA, Rood JI, McClane BA. 2015. Clostridium perfringens type A-E toxin plasmids. Res Microbiol 166:264–279. 10.1016/j.resmic.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Coursodon CF, Glock RD, Moore KL, Cooper KK, Songer JG. 2012. TpeL-producing strains of Clostridium perfringens type A are highly virulent for broiler chicks. Anaerobe 18:117–121. 10.1016/j.anaerobe.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 90.Keyburn AL, Boyce JD, Vaz P, Bannam TL, Ford ME, Parker D, Di RA, Rood JI, Moore RJ. 2008. NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Pathog 4:e26. 10.1371/journal.ppat.0040026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Braun V, Hundsberger T, Leukel P, Sauerborn M, von Eichel-Streiber C. 1996. Definition of the single integration site of the pathogenicity locus in Clostridium difficile. Gene 181:29–38. 10.1016/s0378-1119(96)00398-8. [DOI] [PubMed] [Google Scholar]
- 92.Mani N, Dupuy B. 2001. Regulation of toxin synthesis in Clostridium difficile by an alternative RNA polymerase sigma factor. Proc Natl Acad Sci U S A 98:5844–5849. 10.1073/pnas.101126598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Carter GP, Douce GR, Govind R, Howarth PM, Mackin KE, Spencer J, Buckley AM, Antunes A, Kotsanas D, Jenkin GA, Dupuy B, Rood JI, Lyras D. 2011. The anti-sigma factor TcdC modulates hypervirulence in an epidemic BI/NAP1/027 clinical isolate of Clostridium difficile. PLoS Pathog 7:e1002317. 10.1371/journal.ppat.1002317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Matamouros S, England P, Dupuy B. 2007. Clostridium difficile toxin expression is inhibited by the novel regulator TcdC. Mol Microbiol 64:1274–1288. 10.1111/j.1365-2958.2007.05739.x. [DOI] [PubMed] [Google Scholar]
- 95.MacCannell DR, Louie TJ, Gregson DB, Laverdiere M, Labbe AC, Laing F, Henwick S. 2006. Molecular analysis of Clostridium difficile PCR ribotype 027 isolates from eastern and western Canada. J Clin Microbiol 44:2147–2152. 10.1128/JCM.02563-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cartman ST, Kelly ML, Heeg D, Heap JT, Minton NP. 2012. Precise manipulation of the Clostridium difficile chromosome reveals a lack of association between the tcdC genotype and toxin production. Appl Environ Microbiol 78:4683–4690. 10.1128/AEM.00249-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bakker D, Smits WK, Kuijper EJ, Corver J. 2012. TcdC does not significantly repress toxin expression in Clostridium difficile 630ΔErm. PLoS One 7:e43247. 10.1371/journal.pone.0043247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Young I, Wang I, Roof WD. 2000. Phages will out: strategies of host cell lysis. Trends Microbiol 8:120–128. 10.1016/S0966-842X(00)01705-4. [DOI] [PubMed] [Google Scholar]
- 99.Desvaux M, Hebraud M. 2006. The protein secretion systems in Listeria: inside out bacterial virulence. FEMS Microbiol Rev 30:774–805. 10.1111/j.1574-6976.2006.00035.x. [DOI] [PubMed] [Google Scholar]
- 100.Desvaux M, Khan A, Beatson SA, Scott-Tucker A, Henderson IR. 2005. Protein secretion systems in Fusobacterium nucleatum: genomic identification of type 4 piliation and complete type V pathways brings new insight into mechanisms of pathogenesis. Biochim Biophys Acta 1713:92–112. 10.1016/j.bbamem.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 101.Desvaux M, Khan A, Scott-Tucker A, Chaudhuri RR, Pallen MJ, Henderson IR. 2005. Genomic analysis of the protein secretion systems in Clostridium acetobutylicum ATCC 824. Biochim Biophys Acta 1745:223–253. 10.1016/j.bbamcr.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 102.Tan KS, Wee BY, Song KP. 2001. Evidence for holin function of tcdE gene in the pathogenicity of Clostridium difficile. J Med Microbiol 50:613–619. 10.1099/0022-1317-50-7-613. [DOI] [PubMed] [Google Scholar]
- 103.Govind R, Dupuy B. 2012. Secretion of Clostridium difficile toxins A and B requires the holin-like protein TcdE. PLoS Pathog 8:e1002727. 10.1371/journal.ppat.1002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Olling A, Seehase S, Minton NP, Tatge H, Schroter S, Kohlscheen S, Pich A, Just I, Gerhard R. 2012. Release of TcdA and TcdB from Clostridium difficile cdi 630 is not affected by functional inactivation of the tcdE gene. Microb Pathog 52:92–100. 10.1016/j.micpath.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 105.Wydau-Dematteis S, El Meouche I, Courtin P, Hamiot A, Lai-Kuen R, Saubamea B, Fenaille F, Butel MJ, Pons JL, Dupuy B, Chapot-Chartier MP, Peltier J. 2018. Cwp19 is a novel lytic transglycosylase involved in stationary-phase autolysis resulting in toxin release in Clostridium difficile. mBio 9:e00648-18. 10.1128/mBio.00648-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Janezic S, Marin M, Martin A, Rupnik M. 2015. A new type of toxin A-negative, toxin B-positive Clostridium difficile strain lacking a complete tcdA gene. J Clin Microbiol 53:692–695. 10.1128/JCM.02211-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rupnik M, Janezic S. 2016. An update on Clostridium difficile toxinotyping. J Clin Microbiol 54:13–18. 10.1128/JCM.02083-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dingle KE, Elliott B, Robinson E, Griffiths D, Eyre DW, Stoesser N, Vaughan A, Golubchik T, Fawley WN, Wilcox MH, Peto TE, Walker AS, Riley TV, Crook DW, Didelot X. 2014. Evolutionary history of the Clostridium difficile pathogenicity locus. Genome Biol Evol 6:36–52. 10.1093/gbe/evt204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brouwer MS, Roberts AP, Hussain H, Williams RJ, Allan E, Mullany P. 2013. Horizontal gene transfer converts non-toxigenic Clostridium difficile strains into toxin producers. Nat Commun 4:2601. 10.1038/ncomms3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ramirez-Vargas G, Lopez-Urena D, Badilla A, Orozco-Aguilar J, Murillo T, Rojas P, Riedel T, Overmann J, Gonzalez G, Chaves-Olarte E, Quesada-Gomez C, Rodriguez C. 2018. Novel clade C-I Clostridium difficile strains escape diagnostic tests, differ in pathogenicity potential and carry toxins on extrachromosomal elements. Sci Rep 8:13951. 10.1038/s41598-018-32390-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Monot M, Eckert C, Lemire A, Hamiot A, Dubois T, Tessier C, Dumoulard B, Hamel B, Petit A, Lalande V, Ma L, Bouchier C, Barbut F, Dupuy B. 2015. Clostridium difficile: new insights into the evolution of the pathogenicity locus. Sci Rep 5:15023. 10.1038/srep15023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sirigi Reddy AR, Girinathan BP, Zapotocny R, Govind R. 2013. Identification and characterization of Clostridium sordellii toxin gene regulator. J Bacteriol 195:4246–4254. 10.1128/JB.00711-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Carter GP, Larcombe S, Li L, Jayawardena D, Awad MM, Songer JG, Lyras D. 2014. Expression of the large clostridial toxins is controlled by conserved regulatory mechanisms. Int J Med Microbiol 304:1147–1159. 10.1016/j.ijmm.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 114.Vidor CJ, Watts TD, Adams V, Bulach D, Couchman E, Rood JI, Fairweather NF, Awad M, Lyras D. 2018. Clostridium sordellii pathogenicity locus plasmid pCS1-1 encodes a novel clostridial conjugation locus. mBio 9:e01761-17. 10.1128/mBio.01761-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Han X, Du XD, Southey L, Bulach DM, Seemann T, Yan XX, Bannam TL, Rood JI. 2015. Functional analysis of a bacitracin resistance determinant located on ICECp1, a novel Tn916-like element from a conjugative plasmid in Clostridium perfringens. Antimicrob Agents Chemother 59:6855–6865. 10.1128/AAC.01643-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Reddy BL, SaierMH, Jr.. 2013. Topological and phylogenetic analyses of bacterial holin families and superfamilies. Biochim Biophys Acta 1828:2654–2671. 10.1016/j.bbamem.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Saadat A, Melville SB. 2021. Holin-dependent secretion of the large clostridial toxin TpeL by Clostridium perfringens. J Bacteriol 203:e00580-20. 10.1128/JB.00580-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Eklund MW, Poysky FT, Peterson ME, Meyers JA. 1976. Relationship of bacteriophages to alpha toxin production in Clostridium novyi types A and B. Infect Immun 14:793–803. 10.1128/IAI.14.3.793-803.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Eklund MW, Poysky FT, Meyers JA, Pelroy GA. 1974. Interspecies conversion of Clostridium botulinum type C to Clostridium novyi type A by bacteriophage. Science 186:456–458. 10.1126/science.186.4162.456. [DOI] [PubMed] [Google Scholar]
- 120.Martin-Verstraete I, Peltier J, Dupuy B. 2016. The regulatory networks that control Clostridium difficile toxin synthesis. Toxins 8:153. 10.3390/toxins8050153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dupuy B, Sonenshein AL. 1998. Regulated transcription of Clostridium difficile toxin genes. Mol Microbiol 27:107–120. 10.1046/j.1365-2958.1998.00663.x. [DOI] [PubMed] [Google Scholar]
- 122.Hundsberger T, Braun V, Weidmann M, Leukel P, Sauerborn M, von Eichel-Streiber C. 1997. Transcription analysis of the genes tcdA-E of the pathogenicity locus of Clostridium difficile. European J Biochemistry 244:735–742. 10.1111/j.1432-1033.1997.t01-1-00735.x. [DOI] [PubMed] [Google Scholar]
- 123.Antunes A, Martin-Verstraete I, Dupuy B. 2011. CcpA-mediated repression of Clostridium difficile toxin gene expression. Mol Microbiol 79:882–899. 10.1111/j.1365-2958.2010.07495.x. [DOI] [PubMed] [Google Scholar]
- 124.Antunes A, Camiade E, Monot M, Courtois E, Barbut F, Sernova NV, Rodionov DA, Martin-Verstraete I, Dupuy B. 2012. Global transcriptional control by glucose and carbon regulator CcpA in Clostridium difficile. Nucleic Acids Res 40:10701–10718. 10.1093/nar/gks864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Edwards AN, Anjuwon-Foster BR, McBride SM. 2019. RstA is a major regulator of Clostridioides difficile toxin production and motility. mBio 10:e01991-18. 10.1128/mBio.01991-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mackin KE, Carter GP, Howarth P, Rood JI, Lyras D. 2013. Spo0A differentially regulates toxin production in evolutionarily diverse strains of Clostridium difficile. PLoS One 8:e79666. 10.1371/journal.pone.0079666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Deakin LJ, Clare S, Fagan RP, Dawson LF, Pickard DJ, West MR, Wren BW, Fairweather NF, Dougan G, Lawley TD. 2012. The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect Immun 80:2704–2711. 10.1128/IAI.00147-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Edwards AN, Krall EG, McBride SM. 2020. Strain-dependent RstA regulation of Clostridioides difficile toxin production and sporulation. J Bacteriol 202:e00586-19. 10.1128/JB.00586-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Karlsson S, Dupuy B, Mukherjee K, Norin E, Burman LG, Akerlund T. 2003. Expression of Clostridium difficile toxins A and B and their sigma factor TcdD is controlled by temperature. Infect Immun 71:1784–1793. 10.1128/iai.71.4.1784-1793.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Karlsson S, Burman LG, Akerlund T. 1999. Suppression of toxin production in Clostridium difficile VPI 10463 by amino acids. Microbiology 145:1683–1693. 10.1099/13500872-145-7-1683. [DOI] [PubMed] [Google Scholar]
- 131.Karlsson S, Lindberg A, Norin E, Burman LG, Åkerlund T. 2000. Toxins, butyric acid, and other short-chain fatty acids are coordinately expressed and down-regulated by cysteine in Clostridium difficile. Infect Immun 68:5881–5888. 10.1128/IAI.68.10.5881-5888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Honda T, Hernadez I, Katoh T, Miwatani T. 1983. Stimulation of enterotoxin production of Clostridium difficile by antibiotics. Lancet 321:655. 10.1016/S0140-6736(83)91832-9. [DOI] [PubMed] [Google Scholar]
- 133.Gerber M, Walch C, Loffler B, Tischendorf K, Reischl U, Ackermann G. 2008. Effect of sub-MIC concentrations of metronidazole, vancomycin, clindamycin and linezolid on toxin gene transcription and production in Clostridium difficile. J Med Microbiol 57:776–783. 10.1099/jmm.0.47739-0. [DOI] [PubMed] [Google Scholar]
- 134.Onderdonk AB, Lowe BR, Bartlett JG. 1979. Effect of environmental stress on Clostridium difficile toxin levels during continuous cultivation. Appl Environ Microbiol 38:637–641. 10.1128/AEM.38.4.637-641.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Saujet L, Monot M, Dupuy B, Soutourina O, Martin-Verstraete I. 2011. The key sigma factor of transition phase, SigH, controls sporulation, metabolism, and virulence factor expression in Clostridium difficile. J Bacteriol 193:3186–3196. 10.1128/JB.00272-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pettit LJ, Browne HP, Yu L, Smits WK, Fagan RP, Barquist L, Martin MJ, Goulding D, Duncan SH, Flint HJ, Dougan G, Choudhary JS, Lawley TD. 2014. Functional genomics reveals that Clostridium difficile Spo0A coordinates sporulation, virulence and metabolism. BMC Genomics 15:160. 10.1186/1471-2164-15-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lyon SA, Hutton ML, Rood JI, Cheung JK, Lyras D. 2016. CdtR regulates TcdA and TcdB production in Clostridium difficile. PLoS Pathog 12:e1005758. 10.1371/journal.ppat.1005758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mansfield MJ, Tremblay BJ, Zeng J, Wei X, Hodgins H, Worley J, Bry L, Dong M, Doxey AC. 2020. Phylogenomics of 8,839 Clostridioides difficile genomes reveals recombination-driven evolution and diversification of toxin A and B. PLoS Pathog 16:e1009181. 10.1371/journal.ppat.1009181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shen E, Zhu K, Li D, Pan Z, Luo Y, Bian Q, He L, Song X, Zhen Y, Jin D, Tao L. 2020. Subtyping analysis reveals new variants and accelerated evolution of Clostridioides difficile toxin B. Commun Biol 3:347. 10.1038/s42003-020-1078-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pruitt RN, Chambers MG, Ng KK, Ohi MD, Lacy DB. 2010. Structural organization of the functional domains of Clostridium difficile toxins A and B. Proc Natl Acad Sci U S A 107:13467–13472. 10.1073/pnas.1002199107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Albesa-Jove D, Bertrand T, Carpenter EP, Swain GV, Lim J, Zhang J, Haire LF, Vasisht N, Braun V, Lange A, von Eichel-Streiber C, Svergun DI, Fairweather NF, Brown KA. 2010. Four distinct structural domains in Clostridium difficile toxin B visualized using SAXS. J Mol Biol 396:1260–1270. 10.1016/j.jmb.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 142.Chumbler NM, Rutherford SA, Zhang Z, Farrow MA, Lisher JP, Farquhar E, Giedroc DP, Spiller BW, Melnyk RA, Lacy DB. 2016. Crystal structure of Clostridium difficile toxin A. Nat Microbiol 1:15002. 10.1038/nmicrobiol.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chen P, Lam KH, Liu Z, Mindlin FA, Chen B, Gutierrez CB, Huang L, Zhang Y, Hamza T, Feng H, Matsui T, Bowen ME, Perry K, Jin R. 2019. Structure of the full-length Clostridium difficile toxin B. Nat Struct Mol Biol 26:712–719. 10.1038/s41594-019-0268-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ho JG, Greco A, Rupnik M, Ng KK. 2005. Crystal structure of receptor-binding C-terminal repeats from Clostridium difficile toxin A. Proc Natl Acad Sci U S A 102:18373–18378. 10.1073/pnas.0506391102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Greco A, Ho JG, Lin SJ, Palcic MM, Rupnik M, Ng KK. 2006. Carbohydrate recognition by Clostridium difficile toxin A. Nat Struct Mol Biol 13:460–461. 10.1038/nsmb1084. [DOI] [PubMed] [Google Scholar]
- 146.Orth P, Xiao L, Hernandez LD, Reichert P, Sheth PR, Beaumont M, Yang X, Murgolo N, Ermakov G, DiNunzio E, Racine F, Karczewski J, Secore S, Ingram RN, Mayhood T, Strickland C, Therien AG. 2014. Mechanism of action and epitopes of Clostridium difficile toxin B-neutralizing antibody bezlotoxumab revealed by X-ray crystallography. J Biol Chem 289:18008–18021. 10.1074/jbc.M114.560748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hernandez LD, Kroh HK, Hsieh E, Yang X, Beaumont M, Sheth PR, DiNunzio E, Rutherford SA, Ohi MD, Ermakov G, Xiao L, Secore S, Karczewski J, Racine F, Mayhood T, Fischer P, Sher X, Gupta P, Lacy DB, Therien AG. 2017. Epitopes and mechanism of action of the Clostridium difficile toxin A-neutralizing antibody actoxumab. J Mol Biol 429:1030–1044. 10.1016/j.jmb.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tao L, Tian S, Zhang J, Liu Z, Robinson-McCarthy L, Miyashita SI, Breault DT, Gerhard R, Oottamasathien S, Whelan SPJ, Dong M. 2019. Sulfated glycosaminoglycans and low-density lipoprotein receptor contribute to Clostridium difficile toxin A entry into cells. Nat Microbiol 4:1760–1769. 10.1038/s41564-019-0464-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kato K, Ishiwa A. 2015. The role of carbohydrates in infection strategies of enteric pathogens. Trop Med Health 43:41–52. 10.2149/tmh.2014-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hartley-Tassell LE, Awad MM, Seib KL, Scarselli M, Savino S, Tiralongo J, Lyras D, Day CJ, Jennings MP. 2018. Lectin Activity of the TcdA and TcdB Toxins of Clostridium difficile. Infect Immun 87:e00676-18. 10.1128/IAI.00676-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Krivan HC, Clark GF, Smith DF, Wilkins TD. 1986. Cell surface binding site for Clostridium difficile enterotoxin: evidence for a glycoconjugate containing the sequence Gal alpha 1-3Gal beta 1-4GlcNAc. Infect Immun 53:573–581. 10.1128/IAI.53.3.573-581.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tucker KD, Wilkins TD. 1991. Toxin A of Clostridium difficile binds to the human carbohydrate antigens I, X, and Y. Infect Immun 59:73–78. 10.1128/IAI.59.1.73-78.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Teneberg S, Lonnroth I, Torres Lopez JF, Galili U, Halvarsson MO, Angstrom J, Karlsson KA. 1996. Molecular mimicry in the recognition of glycosphingolipids by Gal alpha 3 Gal beta 4 GlcNAc beta-binding Clostridium difficile toxin A, human natural anti alpha-galactosyl IgG and the monoclonal antibody Gal-13: characterization of a binding-active human glycosphingolipid, non-identical with the animal receptor. Glycobiology 6:599–609. 10.1093/glycob/6.6.599. [DOI] [PubMed] [Google Scholar]
- 154.Gupta P, Zhang Z, Sugiman-Marangos SN, Tam J, Raman S, Julien JP, Kroh HK, Lacy DB, Murgolo N, Bekkari K, Therien AG, Hernandez LD, Melnyk RA. 2017. Functional defects in Clostridium difficile TcdB toxin uptake identify CSPG4 receptor-binding determinants. J Biol Chem 292:17290–17301. 10.1074/jbc.M117.806687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yuan P, Zhang H, Cai C, Zhu S, Zhou Y, Yang X, He R, Li C, Guo S, Li S, Huang T, Perez-Cordon G, Feng H, Wei W. 2015. Chondroitin sulfate proteoglycan 4 functions as the cellular receptor for Clostridium difficile toxin B. Cell Res 25:157–168. 10.1038/cr.2014.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tao L, Zhang J, Meraner P, Tovaglieri A, Wu X, Gerhard R, Zhang X, Stallcup WB, Miao J, He X, Hurdle JG, Breault DT, Brass AL, Dong M. 2016. Frizzled proteins are colonic epithelial receptors for C. difficile toxin B. Nature 538:350–355. 10.1038/nature19799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.LaFrance ME, Farrow MA, Chandrasekaran R, Sheng J, Rubin DH, Lacy DB. 2015. Identification of an epithelial cell receptor responsible for Clostridium difficile TcdB-induced cytotoxicity. Proc Natl Acad Sci U S A 112:7073–7078. 10.1073/pnas.1500791112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lee H, Beilhartz GL, Kucharska I, Raman S, Cui H, Lam MHY, Liang H, Rubinstein JL, Schramek D, Julien JP, Melnyk RA, Taipale M. 2020. Recognition of semaphorin proteins by P. sordellii lethal toxin reveals principles of receptor specificity in clostridial toxins. Cell 182:345.e16–356.e16. 10.1016/j.cell.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tian S, Liu Y, Wu H, Liu H, Zeng J, Choi MY, Chen H, Gerhard R, Dong M. 2020. Genome-wide CRISPR screen identifies semaphorin 6A and 6B as receptors for Paeniclostridium sordellii toxin TcsL. Cell Host Microbe 27:782.e7–792.e7. 10.1016/j.chom.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Schorch B, Song S, van Diemen FR, Bock HH, May P, Herz J, Brummelkamp TR, Papatheodorou P, Aktories K. 2014. LRP1 is a receptor for Clostridium perfringens TpeL toxin indicating a two-receptor model of clostridial glycosylating toxins. Proc Natl Acad Sci U S A 111:6431–6436. 10.1073/pnas.1323790111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Olling A, Goy S, Hoffmann F, Tatge H, Just I, Gerhard R. 2011. The repetitive oligopeptide sequences modulate cytopathic potency but are not crucial for cellular uptake of Clostridium difficile toxin A. PLoS One 6:e17623. 10.1371/journal.pone.0017623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Manse JS, Baldwin MR. 2015. Binding and entry of Clostridium difficile toxin B is mediated by multiple domains. FEBS Lett 589:3945–3951. 10.1016/j.febslet.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 163.Chumbler NM, Farrow MA, Lapierre LA, Franklin JL, Haslam DB, Goldenring JR, Lacy DB. 2012. Clostridium difficile toxin B causes epithelial cell necrosis through an autoprocessing-independent mechanism. PLoS Pathog 8:e1003072. 10.1371/journal.ppat.1003072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Olling A, Huls C, Goy S, Muller M, Krooss S, Rudolf I, Tatge H, Gerhard R. 2014. The combined repetitive oligopeptides of Clostridium difficile toxin A counteract premature cleavage of the glucosyl-transferase domain by stabilizing protein conformation. Toxins (Basel) 6:2162–2176. 10.3390/toxins6072162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhang Y, Hamza T, Gao S, Feng H. 2015. Masking autoprocessing of Clostridium difficile toxin A by the C-terminus combined repetitive oligo peptides. Biochem Biophys Res Commun 459:259–263. 10.1016/j.bbrc.2015.02.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Tam J, Icho S, Utama E, Orrell KE, Gomez-Biagi RF, Theriot CM, Kroh HK, Rutherford SA, Lacy DB, Melnyk RA. 2020. Intestinal bile acids directly modulate the structure and function of C. difficile TcdB toxin. Proc Natl Acad Sci U S A 117:6792–6800. 10.1073/pnas.1916965117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wilcox MH, Gerding DN, Poxton IR, Kelly C, Nathan R, Birch T, Cornely OA, Rahav G, Bouza E, Lee C, Jenkin G, Jensen W, Kim YS, Yoshida J, Gabryelski L, Pedley A, Eves K, Tipping R, Guris D, Kartsonis N, Dorr MB, MODIFY I and MODIFY II Investigators. 2017. Bezlotoxumab for prevention of recurrent Clostridium difficile infection. N Engl J Med 376:305–317. 10.1056/NEJMoa1602615. [DOI] [PubMed] [Google Scholar]
- 168.Marozsan AJ, Ma D, Nagashima KA, Kennedy BJ, Kang YK, Arrigale RR, Donovan GP, Magargal WW, Maddon PJ, Olson WC. 2012. Protection against Clostridium difficile infection with broadly neutralizing antitoxin monoclonal antibodies. J Infect Dis 206:706–713. 10.1093/infdis/jis416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hernandez LD, Racine F, Xiao L, DiNunzio E, Hairston N, Sheth PR, Murgolo NJ, Therien AG. 2015. Broad coverage of genetically diverse strains of Clostridium difficile by actoxumab and bezlotoxumab predicted by in vitro neutralization and epitope modeling. Antimicrob Agents Chemother 59:1052–1060. 10.1128/AAC.04433-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chen P, Tao L, Wang T, Zhang J, He A, Lam KH, Liu Z, He X, Perry K, Dong M, Jin R. 2018. Structural basis for recognition of frizzled proteins by Clostridium difficile toxin B. Science 360:664–669. 10.1126/science.aar1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Li Y, Cam J, Bu G. 2001. Low-density lipoprotein receptor family: endocytosis and signal transduction. Mol Neurobiol 23:53–67. 10.1385/MN:23:1:53. [DOI] [PubMed] [Google Scholar]
- 172.Nikolic J, Belot L, Raux H, Legrand P, Gaudin Y, Albertini AA. 2018. Structural basis for the recognition of LDL-receptor family members by VSV glycoprotein. Nat Commun 9:1029. 10.1038/s41467-018-03432-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Finkelshtein D, Werman A, Novick D, Barak S, Rubinstein M. 2013. LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc Natl Acad Sci U S A 110:7306–7311. 10.1073/pnas.1214441110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Schottelndreier D, Langejurgen A, Lindner R, Genth H. 2020. Low density lipoprotein receptor-related protein-1 (LRP1) is involved in the uptake of Clostridioides difficile toxin A and serves as an internalizing receptor. Front Cell Infect Microbiol 10:565465. 10.3389/fcimb.2020.565465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Peng Z, Simeon R, Mitchell SB, Zhang J, Feng H, Chen Z. 2019. Designed Ankyrin Repeat Protein (DARPin) Neutralizers of TcdB from Clostridium difficile Ribotype 027. mSphere 4:e00596-19. 10.1128/mSphere.00596-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Chung SY, Schottelndreier D, Tatge H, Fuhner V, Hust M, Beer LA, Gerhard R. 2018. The conserved Cys-2232 in Clostridioides difficile toxin B modulates receptor binding. Front Microbiol 9:2314. 10.3389/fmicb.2018.02314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lopez-Urena D, Orozco-Aguilar J, Chaves-Madrigal Y, Ramirez-Mata A, Villalobos-Jimenez A, Ost S, Quesada-Gomez C, Rodriguez C, Papatheodorou P, Chaves-Olarte E. 2019. Toxin B variants from Clostridium difficile strains VPI 10463 and NAP1/027 share similar substrate profile and cellular intoxication kinetics but use different host cell entry factors. Toxins 11:348. 10.3390/toxins11060348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Pan Z, Zhang Y, Luo J, Li D, Zhou Y, He L, Yang Q, Dong M, Tao L. 2021. Functional analyses of epidemic Clostridioides difficile toxin B variants reveal their divergence in utilizing receptors and inducing pathology. PLoS Pathog 17:e1009197. 10.1371/journal.ppat.1009197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Terada N, Ohno N, Murata S, Katoh R, Stallcup WB, Ohno S. 2006. Immunohistochemical study of NG2 chondroitin sulfate proteoglycan expression in the small and large intestines. Histochem Cell Biol 126:483–490. 10.1007/s00418-006-0184-3. [DOI] [PubMed] [Google Scholar]
- 180.Crosnier C, Stamataki D, Lewis J. 2006. Organizing cell renewal in the intestine: stem cells, signals and combinatorial control. Nat Rev Genet 7:349–359. 10.1038/nrg1840. [DOI] [PubMed] [Google Scholar]
- 181.Tao J, Abudoukelimu M, Ma YT, Yang YN, Li XM, Chen BD, Liu F, He CH, Li HY. 2016. Secreted frizzled related protein 1 protects H9C2 cells from hypoxia/re-oxygenation injury by blocking the Wnt signaling pathway. Lipids Health Dis 15:72. 10.1186/s12944-016-0240-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mileto SJ, Jarde T, Childress KO, Jensen JL, Rogers AP, Kerr G, Hutton ML, Sheedlo MJ, Bloch SC, Shupe JA, Horvay K, Flores T, Engel R, Wilkins S, McMurrick PJ, Lacy DB, Abud HE, Lyras D. 2020. Clostridioides difficile infection damages colonic stem cells via TcdB, impairing epithelial repair and recovery from disease. Proc Natl Acad Sci U S A 117:8064–8073. 10.1073/pnas.1915255117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Alto LT, Terman JR. 2017. Semaphorins and their signaling mechanisms. Methods Mol Biol 1493:1–25. 10.1007/978-1-4939-6448-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Papatheodorou P, Zamboglou C, Genisyuerek S, Guttenberg G, Aktories K. 2010. Clostridial glucosylating toxins enter cells via clathrin-mediated endocytosis. PLoS One 5:e10673. 10.1371/journal.pone.0010673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Chandrasekaran R, Kenworthy AK, Lacy DB. 2016. Clostridium difficile toxin A Undergoes clathrin-independent, PACSIN2-dependent endocytosis. PLoS Pathog 12:e1006070. 10.1371/journal.ppat.1006070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Schottelndreier D, Seeger K, Grassl GA, Winny MR, Lindner R, Genth H. 2018. Expression and (lacking) internalization of the cell surface receptors of Clostridioides difficile toxin B. Front Microbiol 9:1483. 10.3389/fmicb.2018.01483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Qa'Dan M, Spyres LM, Ballard JD. 2000. pH-induced conformational changes in Clostridium difficile toxin B. Infect Immun 68:2470–2474. 10.1128/IAI.68.5.2470-2474.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Qa'Dan M, Spyres LM, Ballard JD. 2001. pH-enhanced cytopathic effects of Clostridium sordellii lethal toxin. Infect Immun 69:5487–5493. 10.1128/iai.69.9.5487-5493.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Guttenberg G, Hornei S, Jank T, Schwan C, Lu W, Einsle O, Papatheodorou P, Aktories K. 2012. Molecular characteristics of Clostridium perfringens TpeL toxin and consequences of mono-O-GlcNAcylation of Ras in living cells. J Biol Chem 287:24929–24940. 10.1074/jbc.M112.347773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lanis JM, Heinlen LD, James JA, Ballard JD. 2013. Clostridium difficile 027/BI/NAP1 encodes a hypertoxic and antigenically variable form of TcdB. PLoS Pathog 9:e1003523. 10.1371/journal.ppat.1003523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Collier RJ. 2001. Understanding the mode of action of diphtheria toxin: a perspective on progress during the 20th century. Toxicon 39:1793–1803. 10.1016/S0041-0101(01)00165-9. [DOI] [PubMed] [Google Scholar]
- 192.Pirazzini M, Azarnia Tehran D, Leka O, Zanetti G, Rossetto O, Montecucco C. 2016. On the translocation of botulinum and tetanus neurotoxins across the membrane of acidic intracellular compartments. Biochim Biophys Acta 1858:467–474. 10.1016/j.bbamem.2015.08.014. [DOI] [PubMed] [Google Scholar]
- 193.Barth H, Pfeifer G, Hofmann F, Maier E, Benz R, Aktories K. 2001. Low pH-induced formation of ion channels by Clostridium difficile toxin B in target cells. J Biol Chem 276:10670–10676. 10.1074/jbc.M009445200. [DOI] [PubMed] [Google Scholar]
- 194.Giesemann T, Jank T, Gerhard R, Maier E, Just I, Benz R, Aktories K. 2006. Cholesterol-dependent pore formation of Clostridium difficile toxin A. J Biol Chem 281:10808–10815. 10.1074/jbc.M512720200. [DOI] [PubMed] [Google Scholar]
- 195.Donovan JJ, Simon MI, Draper RK, Montal M. 1981. Diphtheria toxin forms transmembrane channels in planar lipid bilayers. Proc Natl Acad Sci U S A 78:172–176. 10.1073/pnas.78.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Fischer A, Montal M. 2007. Single molecule detection of intermediates during botulinum neurotoxin translocation across membranes. Proc Natl Acad Sci U S A 104:10447–10452. 10.1073/pnas.0700046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhang Z, Park M, Tam J, Auger A, Beilhartz GL, Lacy DB, Melnyk RA. 2014. Translocation domain mutations affecting cellular toxicity identify the Clostridium difficile toxin B pore. Proc Natl Acad Sci U S A 111:3721–3726. 10.1073/pnas.1400680111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Orrell KE, Tellgren-Roth A, Di Bernardo M, Zhang Z, Cuviello F, Lundqvist J, von Heijne G, Nilsson I, Melnyk RA. 2018. Direct detection of membrane-inserting fragments defines the translocation pores of a family of pathogenic toxins. J Mol Biol 430:3190–3199. 10.1016/j.jmb.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 199.Orrell KE, Mansfield MJ, Doxey AC, Melnyk RA. 2020. The C. difficile toxin B membrane translocation machinery is an evolutionarily conserved protein delivery apparatus. Nat Commun 11:432. 10.1038/s41467-020-14306-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ost GS, Wirth C, Bogdanovic X, Kao WC, Schorch B, Aktories PJK, Papatheodorou P, Schwan C, Schlosser A, Jank T, Hunte C, Aktories K. 2020. Inverse control of Rab proteins by Yersinia ADP-ribosyltransferase and glycosyltransferase related to clostridial glucosylating toxins. Sci Adv 6:eaaz2094. 10.1126/sciadv.aaz2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Egerer M, Giesemann T, Jank T, Satchell KJ, Aktories K. 2007. Auto-catalytic cleavage of Clostridium difficile toxins A and B depends on cysteine protease activity. J Biol Chem 282:25314–25321. 10.1074/jbc.M703062200. [DOI] [PubMed] [Google Scholar]
- 202.Aktories K, Schwan C, Jank T. 2017. Clostridium difficile toxin biology. Annu Rev Microbiol 71:281–307. 10.1146/annurev-micro-090816-093458. [DOI] [PubMed] [Google Scholar]
- 203.Rawlings ND, Barrett AJ, Bateman A. 2010. MEROPS: the peptidase database. Nucleic Acids Res 38:D227–D233. 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Sheahan KL, Cordero CL, Satchell KJ. 2007. Autoprocessing of the Vibrio cholerae RTX toxin by the cysteine protease domain. EMBO J 26:2552–2561. 10.1038/sj.emboj.7601700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Guttenberg G, Papatheodorou P, Genisyuerek S, Lu W, Jank T, Einsle O, Aktories K. 2011. Inositol hexakisphosphate-dependent processing of Clostridium sordellii lethal toxin and Clostridium novyi alpha-toxin. J Biol Chem 286:14779–14786. 10.1074/jbc.M110.200691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kreimeyer I, Euler F, Marckscheffel A, Tatge H, Pich A, Olling A, Schwarz J, Just I, Gerhard R. 2011. Autoproteolytic cleavage mediates cytotoxicity of Clostridium difficile toxin A. Naunyn Schmiedebergs Arch Pharmacol 383:253–262. 10.1007/s00210-010-0574-x. [DOI] [PubMed] [Google Scholar]
- 207.Barroso LA, Moncrief JS, Lyerly DM, Wilkins TD. 1994. Mutagenesis of the Clostridium difficile toxin B gene and effect on cytotoxic activity. Microb Pathog 16:297–303. 10.1006/mpat.1994.1030. [DOI] [PubMed] [Google Scholar]
- 208.Shen A. 2010. Autoproteolytic activation of bacterial toxins. Toxins (Basel) 2:963–977. 10.3390/toxins2050963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lanis JM, Hightower LD, Shen A, Ballard JD. 2012. TcdB from hypervirulent Clostridium difficile exhibits increased efficiency of autoprocessing. Mol Microbiol 84:66–76. 10.1111/j.1365-2958.2012.08009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Reineke J, Tenzer S, Rupnik M, Koschinski A, Hasselmayer O, Schrattenholz A, Schild H, von Eichel-Streiber C. 2007. Autocatalytic cleavage of Clostridium difficile toxin B. Nature 446:415–419. 10.1038/nature05622. [DOI] [PubMed] [Google Scholar]
- 211.Pruitt RN, Chagot B, Cover M, Chazin WJ, Spiller B, Lacy DB. 2009. Structure-function analysis of inositol hexakisphosphate-induced autoprocessing in Clostridium difficile toxin A. J Biol Chem 284:21934–21940. 10.1074/jbc.M109.018929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Puri AW, Lupardus PJ, Deu E, Albrow VE, Garcia KC, Bogyo M, Shen A. 2010. Rational design of inhibitors and activity-based probes targeting Clostridium difficile virulence factor TcdB. Chem Biol 17:1201–1211. 10.1016/j.chembiol.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Lupardus PJ, Shen A, Bogyo M, Garcia KC. 2008. Small molecule-induced allosteric activation of the Vibrio cholerae RTX cysteine protease domain. Science 322:265–268. 10.1126/science.1162403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Prochazkova K, Shuvalova LA, Minasov G, Voburka Z, Anderson WF, Satchell KJ. 2009. Structural and molecular mechanism for autoprocessing of MARTX toxin of Vibrio cholerae at multiple sites. J Biol Chem 284:26557–26568. 10.1074/jbc.M109.025510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Shen A, Lupardus PJ, Gersch MM, Puri AW, Albrow VE, Garcia KC, Bogyo M. 2011. Defining an allosteric circuit in the cysteine protease domain of Clostridium difficile toxins. Nat Struct Mol Biol 18:364–371. 10.1038/nsmb.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Rupnik M, Pabst S, Rupnik M, von Eichel-Streiber C, Urlaub H, Soling HD. 2005. Characterization of the cleavage site and function of resulting cleavage fragments after limited proteolysis of Clostridium difficile toxin B (TcdB) by host cells. Microbiology (Reading) 151:199–208. 10.1099/mic.0.27474-0. [DOI] [PubMed] [Google Scholar]
- 217.Pfeifer G, Schirmer J, Leemhuis J, Busch C, Meyer DK, Aktories K, Barth H. 2003. Cellular uptake of Clostridium difficile toxin B. Translocation of the N-terminal catalytic domain into the cytosol of eukaryotic cells. J Biol Chem 278:44535–44541. 10.1074/jbc.M307540200. [DOI] [PubMed] [Google Scholar]
- 218.Savidge TC, Urvil P, Oezguen N, Ali K, Choudhury A, Acharya V, Pinchuk I, Torres AG, English RD, Wiktorowicz JE, Loeffelholz M, Kumar R, Shi L, Nie W, Braun W, Herman B, Hausladen A, Feng H, Stamler JS, Pothoulakis C. 2011. Host S-nitrosylation inhibits clostridial small molecule-activated glucosylating toxins. Nat Med 17:1136–1141. 10.1038/nm.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Zhang Y, Li S, Yang Z, Shi L, Yu H, Salerno-Goncalves R, Saint Fleur A, Feng H. 2018. Cysteine protease-mediated autocleavage of Clostridium difficile toxins regulates their proinflammatory activity. Cell Mol Gastroenterol Hepatol 5:611–625. 10.1016/j.jcmgh.2018.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Jank T, Giesemann T, Aktories K. 2007. Rho-glucosylating Clostridium difficile toxins A and B: new insights into structure and function. Glycobiology 17:15R–22R. 10.1093/glycob/cwm004. [DOI] [PubMed] [Google Scholar]
- 221.Genth H, Pauillac S, Schelle I, Bouvet P, Bouchier C, Varela-Chavez C, Just I, Popoff MR. 2014. Haemorrhagic toxin and lethal toxin from Clostridium sordellii strain vpi9048: molecular characterization and comparative analysis of substrate specificity of the large clostridial glucosylating toxins. Cell Microbiol 16:1706–1721. 10.1111/cmi.12321. [DOI] [PubMed] [Google Scholar]
- 222.Voth DE, Ballard JD. 2005. Clostridium difficile toxins: mechanism of action and role in disease. Clin Microbiol Rev 18:247–263. 10.1128/CMR.18.2.247-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Nagahama M, Ohkubo A, Oda M, Kobayashi K, Amimoto K, Miyamoto K, Sakurai J. 2011. Clostridium perfringens TpeL glycosylates the Rac and Ras subfamily proteins. Infect Immun 79:905–910. 10.1128/IAI.01019-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Gao X, Wang X, Pham TH, Feuerbacher LA, Lubos ML, Huang M, Olsen R, Mushegian A, Slawson C, Hardwidge PR. 2013. NleB, a bacterial effector with glycosyltransferase activity, targets GAPDH function to inhibit NF-kappaB activation. Cell Host Microbe 13:87–99. 10.1016/j.chom.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Belyi Y, Tabakova I, Stahl M, Aktories K. 2008. Lgt: a family of cytotoxic glucosyltransferases produced by Legionella pneumophila. J Bacteriol 190:3026–3035. 10.1128/JB.01798-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Jank T, Bogdanovic X, Wirth C, Haaf E, Spoerner M, Bohmer KE, Steinemann M, Orth JH, Kalbitzer HR, Warscheid B, Hunte C, Aktories K. 2013. A bacterial toxin catalyzing tyrosine glycosylation of Rho and deamidation of Gq and Gi proteins. Nat Struct Mol Biol 20:1273–1280. 10.1038/nsmb.2688. [DOI] [PubMed] [Google Scholar]
- 227.Teichert M, Tatge H, Schoentaube J, Just I, Gerhard R. 2006. Application of mutated Clostridium difficile toxin A for determination of glucosyltransferase-dependent effects. Infect Immun 74:6006–6010. 10.1128/IAI.00545-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Craven R, Lacy DB. 2016. Clostridium sordellii lethal-toxin autoprocessing and membrane localization activities drive GTPase glucosylation profiles in endothelial cells. mSphere 1:e00012-15. 10.1128/mSphere.00012-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Cowardin CA, Jackman BM, Noor Z, Burgess SL, Feig AL, PetriWA, Jr.. 2016. Glucosylation drives the innate inflammatory response to Clostridium difficile toxin A. Infect Immun 84:2317–2323. 10.1128/IAI.00327-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Bilverstone TW, Garland M, Cave RJ, Kelly ML, Tholen M, Bouley DM, Kaye P, Minton NP, Bogyo M, Kuehne SA, Melnyk RA. 2020. The glucosyltransferase activity of C. difficile toxin B is required for disease pathogenesis. PLoS Pathog 16:e1008852. 10.1371/journal.ppat.1008852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Farrow MA, Chumbler NM, Lapierre LA, Franklin JL, Rutherford SA, Goldenring JR, Lacy DB. 2013. Clostridium difficile toxin B-induced necrosis is mediated by the host epithelial cell NADPH oxidase complex. Proc Natl Acad Sci U S A 110:18674–18679. 10.1073/pnas.1313658110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Wohlan K, Goy S, Olling A, Srivaratharajan S, Tatge H, Genth H, Gerhard R. 2014. Pyknotic cell death induced by Clostridium difficile TcdB: chromatin condensation and nuclear blister are induced independently of the glucosyltransferase activity. Cell Microbiol 16:1678–1692. 10.1111/cmi.12317. [DOI] [PubMed] [Google Scholar]
- 233.Schirmer J, Aktories K. 2004. Large clostridial cytotoxins: cellular biology of Rho/Ras-glucosylating toxins. Biochim Biophys Acta 1673:66–74. 10.1016/j.bbagen.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 234.Busch C, Hofmann F, Selzer J, Munro S, Jeckel D, Aktories K. 1998. A common motif of eukaryotic glycosyltransferases is essential for the enzyme activity of large clostridial cytotoxins. J Biol Chem 273:19566–19572. 10.1074/jbc.273.31.19566. [DOI] [PubMed] [Google Scholar]
- 235.Wiggins CA, Munro S. 1998. Activity of the yeast MNN1 alpha-1,3-mannosyltransferase requires a motif conserved in many other families of glycosyltransferases. Proc Natl Acad Sci U S A 95:7945–7950. 10.1073/pnas.95.14.7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Busch C, Hofmann F, Gerhard R, Aktories K. 2000. Involvement of a conserved tryptophan residue in the UDP-glucose binding of large clostridial cytotoxin glycosyltransferases. J Biol Chem 275:13228–13234. 10.1074/jbc.275.18.13228. [DOI] [PubMed] [Google Scholar]
- 237.Reinert DJ, Jank T, Aktories K, Schulz GE. 2005. Structural basis for the function of Clostridium difficile toxin B. J Mol Biol 351:973–981. 10.1016/j.jmb.2005.06.071. [DOI] [PubMed] [Google Scholar]
- 238.Jank T, Reinert DJ, Giesemann T, Schulz GE, Aktories K. 2005. Change of the donor substrate specificity of Clostridium difficile toxin B by site-directed mutagenesis. J Biol Chem 280:37833–37838. 10.1074/jbc.M506836200. [DOI] [PubMed] [Google Scholar]
- 239.Chandrasekaran R, Lacy DB. 2017. The role of toxins in Clostridium difficile infection. FEMS Microbiol Rev 41:723–750. 10.1093/femsre/fux048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Pruitt RN, Chumbler NM, Rutherford SA, Farrow MA, Friedman DB, Spiller B, Lacy DB. 2012. Structural determinants of Clostridium difficile toxin A glucosyltransferase activity. J Biol Chem 287:8013–8020. 10.1074/jbc.M111.298414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.D'Urzo N, Malito E, Biancucci M, Bottomley MJ, Maione D, Scarselli M, Martinelli M. 2012. The structure of Clostridium difficile toxin A glucosyltransferase domain bound to Mn2+ and UDP provides insights into glucosyltransferase activity and product release. FEBS J 279:3085–3097. 10.1111/j.1742-4658.2012.08688.x. [DOI] [PubMed] [Google Scholar]
- 242.Ziegler MO, Jank T, Aktories K, Schulz GE. 2008. Conformational changes and reaction of clostridial glycosylating toxins. J Mol Biol 377:1346–1356. 10.1016/j.jmb.2007.12.065. [DOI] [PubMed] [Google Scholar]
- 243.Liu J, Mushegian A. 2003. Three monophyletic superfamilies account for the majority of the known glycosyltransferases. Protein Sci 12:1418–1431. 10.1110/ps.0302103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Breton C, Snajdrova L, Jeanneau C, Koca J, Imberty A. 2006. Structures and mechanisms of glycosyltransferases. Glycobiology 16:29R–37R. 10.1093/glycob/cwj016. [DOI] [PubMed] [Google Scholar]
- 245.Hofmann F, Busch C, Prepens U, Just I, Aktories K. 1997. Localization of the glucosyltransferase activity of Clostridium difficile toxin B to the N-terminal part of the holotoxin. J Biol Chem 272:11074–11078. 10.1074/jbc.272.17.11074. [DOI] [PubMed] [Google Scholar]
- 246.Jank T, Giesemann T, Aktories K. 2007. Clostridium difficile glucosyltransferase toxin B-essential amino acids for substrate binding. J Biol Chem 282:35222–35231. 10.1074/jbc.M703138200. [DOI] [PubMed] [Google Scholar]
- 247.Steinemann M, Schlosser A, Jank T, Aktories K. 2018. The chaperonin TRiC/CCT is essential for the action of bacterial glycosylating protein toxins like Clostridium difficile toxins A and B. Proc Natl Acad Sci U S A 115:9580–9585. 10.1073/pnas.1807658115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Spiering D, Hodgson L. 2011. Dynamics of the Rho-family small GTPases in actin regulation and motility. Cell Adh Migr 5:170–180. 10.4161/cam.5.2.14403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Colicelli J. 2004. Human RAS superfamily proteins and related GTPases. Sci STKE 2004:re13. 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Genth H, Just I. 2011. Functional implications of lethal toxin-catalysed glucosylation of (H/K/N)Ras and Rac1 in Clostridium sordellii-associated disease. Eur J Cell Biol 90:959–965. 10.1016/j.ejcb.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 251.May M, Schelle I, Brakebusch C, Rottner K, Genth H. 2014. Rac1-dependent recruitment of PAK2 to G2 phase centrosomes and their roles in the regulation of mitotic entry. Cell Cycle 13:2211–2221. 10.4161/cc.29279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Huelsenbeck SC, May M, Schmidt G, Genth H. 2009. Inhibition of cytokinesis by Clostridium difficile toxin B and cytotoxic necrotizing factors–reinforcing the critical role of RhoA in cytokinesis. Cell Motil Cytoskeleton 66:967–975. 10.1002/cm.20390. [DOI] [PubMed] [Google Scholar]
- 253.Popoff MR, Geny B. 2011. Rho/Ras-GTPase-dependent and -independent activity of clostridial glucosylating toxins. J Med Microbiol 60:1057–1069. 10.1099/jmm.0.029314-0. [DOI] [PubMed] [Google Scholar]
- 254.Chaves-Olarte E, Freer E, Parra A, Guzman-Verri C, Moreno E, Thelestam M. 2003. R-Ras glucosylation and transient RhoA activation determine the cytopathic effect produced by toxin B variants from toxin A-negative strains of Clostridium difficile. J Biol Chem 278:7956–7963. 10.1074/jbc.M209244200. [DOI] [PubMed] [Google Scholar]
- 255.Abt MC, Lewis BB, Caballero S, Xiong H, Carter RA, Susac B, Ling L, Leiner I, Pamer EG. 2015. Innate immune defenses mediated by two ILC subsets are critical for protection against acute Clostridium difficile infection. Cell Host Microbe 18:27–37. 10.1016/j.chom.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Yu H, Chen K, Sun Y, Carter M, Garey KW, Savidge TC, Devaraj S, Tessier ME, von Rosenvinge EC, Kelly CP, Pasetti MF, Feng H. 2017. Cytokines are markers of the Clostridium difficile-induced inflammatory response and predict disease severity. Clin Vaccine Immunol 24:e00037-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Sun X, He X, Tzipori S, Gerhard R, Feng H. 2009. Essential role of the glucosyltransferase activity in Clostridium difficile toxin-induced secretion of TNF-alpha by macrophages. Microb Pathog 46:298–305. 10.1016/j.micpath.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Xu H, Yang J, Gao W, Li L, Li P, Zhang L, Gong YN, Peng X, Xi JJ, Chen S, Wang F, Shao F. 2014. Innate immune sensing of bacterial modifications of Rho GTPases by the pyrin inflammasome. Nature 513:237–241. 10.1038/nature13449. [DOI] [PubMed] [Google Scholar]
- 259.Ng J, Hirota SA, Gross O, Li Y, Ulke-Lemee A, Potentier MS, Schenck LP, Vilaysane A, Seamone ME, Feng H, Armstrong GD, Tschopp J, Macdonald JA, Muruve DA, Beck PL. 2010. Clostridium difficile toxin-induced inflammation and intestinal injury are mediated by the inflammasome. Gastroenterology 139:542.e3–552.e3. 10.1053/j.gastro.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 260.Gao W, Yang J, Liu W, Wang Y, Shao F. 2016. Site-specific phosphorylation and microtubule dynamics control pyrin inflammasome activation. Proc Natl Acad Sci U S A 113:E4857–E4866. 10.1073/pnas.1601700113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Van Gorp H, Saavedra PH, de Vasconcelos NM, Van Opdenbosch N, Vande Walle L, Matusiak M, Prencipe G, Insalaco A, Van Hauwermeiren F, Demon D, Bogaert DJ, Dullaers M, De Baere E, Hochepied T, Dehoorne J, Vermaelen KY, Haerynck F, De Benedetti F, Lamkanfi M. 2016. Familial Mediterranean fever mutations lift the obligatory requirement for microtubules in pyrin inflammasome activation. Proc Natl Acad Sci U S A 113:14384–14389. 10.1073/pnas.1613156113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F. 2015. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526:660–665. 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 263.Russo HM, Rathkey J, Boyd-Tressler A, Katsnelson MA, Abbott DW, Dubyak GR. 2016. Active caspase-1 induces plasma membrane pores that precede pyroptotic lysis and are blocked by lanthanides. J Immunol 197:1353–1367. 10.4049/jimmunol.1600699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Saavedra PHV, Huang L, Ghazavi F, Kourula S, Vanden Berghe T, Takahashi N, Vandenabeele P, Lamkanfi M. 2018. Apoptosis of intestinal epithelial cells restricts Clostridium difficile infection in a model of pseudomembranous colitis. Nat Commun 9:4846. 10.1038/s41467-018-07386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Mansfield MJ, Doxey AC. 2018. Genomic insights into the evolution and ecology of botulinum neurotoxins. Pathog Dis 76:fty040. 10.1093/femspd/fty040. [DOI] [PubMed] [Google Scholar]
- 266.Contreras E, Masuyer G, Qureshi N, Chawla S, Dhillon HS, Lee HL, Chen J, Stenmark P, Gill SS. 2019. A neurotoxin that specifically targets Anopheles mosquitoes. Nat Commun 10:2869. 10.1038/s41467-019-10732-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 268.Crosby JR, Zhao C, Jiang C, Bai D, Katz M, Greenlee S, Kawabe H, McCaleb M, Rotin D, Guo S, Monia BP. 2017. Inhaled ENaC antisense oligonucleotide ameliorates cystic fibrosis-like lung disease in mice. J Cyst Fibros 16:671–680. 10.1016/j.jcf.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 269.Lyerly DM, Saum KE, MacDonald DK, Wilkins TD. 1985. Effects of Clostridium difficile toxins given intragastrically to animals. Infect Immun 47:349–352. 10.1128/IAI.47.2.349-352.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Savidge TC, Pan W-h, Newman P, O’Brien M, Anton PM, Pothoulakis C. 2003. Clostridium difficile toxin B is an inflammatory enterotoxin in human intestine. Gastroenterology 125:413–420. 10.1016/S0016-5085(03)00902-8. [DOI] [PubMed] [Google Scholar]
- 271.Hatheway CL. 1990. Toxigenic clostridia. Clin Microbiol Rev 3:66–98. 10.1128/cmr.3.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Di Bella S, Ascenzi P, Siarakas S, Petrosillo N, di Masi A. 2016. Clostridium difficile toxins A and B: insights into pathogenic properties and extraintestinal effects. Toxins (Basel) 8:134. 10.3390/toxins8050134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Mattila E, Arkkila P, Mattila PS, Tarkka E, Tissari P, Anttila VJ. 2013. Extraintestinal Clostridium difficile infections. Clin Infect Dis 57:e148–e153. 10.1093/cid/cit392. [DOI] [PubMed] [Google Scholar]
- 274.Urban E, Terhes G, Gajdacs M. 2020. Extraintestinal Clostridioides difficile infections: epidemiology in a university hospital in Hungary and review of the literature. Antibiotics (Basel) 9:16. 10.3390/antibiotics9010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Pothoulakis C, Gilbert RJ, Cladaras C, Castagliuolo I, Semenza G, Hitti Y, Montcrief JS, Linevsky J, Kelly CP, Nikulasson S, Desai HP, Wilkins TD, LaMont JT. 1996. Rabbit sucrase-isomaltase contains a functional intestinal receptor for Clostridium difficile toxin A. J Clin Invest 98:641–649. 10.1172/JCI118835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Na X, Kim H, Moyer MP, Pothoulakis C, LaMont JT. 2008. gp96 is a human colonocyte plasma membrane binding protein for Clostridium difficile toxin A. Infect Immun 76:2862–2871. 10.1128/IAI.00326-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Genth H, Junemann J, Lammerhirt CM, Lucke AC, Schelle I, Just I, Gerhard R, Pich A. 2018. Difference in mono-O-glucosylation of Ras subtype GTPases between toxin A and toxin B from Clostridioides difficile strain 10463 and lethal toxin from Clostridium sordellii strain 6018. Front Microbiol 9:3078. 10.3389/fmicb.2018.03078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Chaves-Olarte E, Weidmann M, Eichel-Streiber C, Thelestam M. 1997. Toxins A and B from Clostridium difficile differ with respect to enzymatic potencies, cellular substrate specificities, and surface binding to cultured cells. J Clin Invest 100:1734–1741. 10.1172/JCI119698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Mehlig M, Moos M, Braun V, Kalt B, Mahony DE, Eichel-Streiber C. 2001. Variant toxin B and a functional toxin A produced by Clostridium difficile C34. FEMS Microbiol Lett 198:171–176. 10.1111/j.1574-6968.2001.tb10638.x. [DOI] [PubMed] [Google Scholar]
- 280.Just I, Selzer J, Wilm M, von Eichel-Streiber C, Mann M, Aktories K. 1995. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature 375:500–503. 10.1038/375500a0. [DOI] [PubMed] [Google Scholar]
- 281.Quesada-Gomez C, Lopez-Urena D, Chumbler N, Kroh HK, Castro-Pena C, Rodriguez C, Orozco-Aguilar J, Gonzalez-Camacho S, Rucavado A, Guzman-Verri C, Lawley TD, Lacy DB, Chaves-Olarte E. 2016. Analysis of TcdB proteins within the hypervirulent clade 2 reveals an impact of RhoA glucosylation on Clostridium difficile proinflammatory activities. Infect Immun 84:856–865. 10.1128/IAI.01291-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Chaves-Olarte E, Low P, Freer E, Norlin T, Weidmann M, von Eichel-Streiber C, Thelestam M. 1999. A novel cytotoxin from Clostridium difficile serogroup F is a functional hybrid between two other large clostridial cytotoxins. J Biol Chem 274:11046–11052. 10.1074/jbc.274.16.11046. [DOI] [PubMed] [Google Scholar]
- 283.Muller S, von Eichel-Streiber C, Moos M. 1999. Impact of amino acids 22–27 of Rho-subfamily GTPases on glucosylation by the large clostridial cytotoxins TcsL-1522, TcdB-1470 and TcdB-8864. Eur J Biochem 266:1073–1080. 10.1046/j.1432-1327.1999.00951.x. [DOI] [PubMed] [Google Scholar]
- 284.Hofmann F, Rex G, Aktories K, Just I. 1996. The ras-related protein Ral is monoglucosylated by Clostridium sordellii lethal toxin. Biochem Biophys Res Commun 227:77–81. 10.1006/bbrc.1996.1470. [DOI] [PubMed] [Google Scholar]
- 285.Popoff MR, Chaves-Olarte E, Lemichez E, von Eichel-Streiber C, Thelestam M, Chardin P, Cussac D, Antonny B, Chavrier P, Flatau G, Giry M, de Gunzburg J, Boquet P. 1996. Ras, Rap, and Rac small GTP-binding proteins are targets for Clostridium sordellii lethal toxin glucosylation. J Biol Chem 271:10217–10224. 10.1074/jbc.271.17.10217. [DOI] [PubMed] [Google Scholar]
- 286.Genth H, Hofmann F, Selzer J, Rex G, Aktories K, Just I. 1996. Difference in protein substrate specificity between hemorrhagic toxin and lethal toxin from Clostridium sordellii. Biochem Biophys Res Commun 229:370–374. 10.1006/bbrc.1996.1812. [DOI] [PubMed] [Google Scholar]
- 287.Selzer J, Hofmann F, Rex G, Wilm M, Mann M, Just I, Aktories K. 1996. Clostridium novyi alpha-toxin-catalyzed incorporation of GlcNAc into Rho subfamily proteins. J Biol Chem 271:25173–25177. 10.1074/jbc.271.41.25173. [DOI] [PubMed] [Google Scholar]
- 288.Schorch B, Heni H, Zahaf NI, Brummer T, Mione M, Schmidt G, Papatheodorou P, Aktories K. 2018. Targeting oncogenic Ras by the Clostridium perfringens toxin TpeL. Oncotarget 9:16489–16500. 10.18632/oncotarget.24740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Just I, Wilm M, Selzer J, Rex G, von Eichel-Streiber C, Mann M, Aktories K. 1995. The enterotoxin from Clostridium difficile (ToxA) monoglucosylates the Rho proteins. J Biol Chem 270:13932–13936. 10.1074/jbc.270.23.13932. [DOI] [PubMed] [Google Scholar]
- 290.Gerhard R, Nottrott S, Schoentaube J, Tatge H, Olling A, Just I. 2008. Glucosylation of Rho GTPases by Clostridium difficile toxin A triggers apoptosis in intestinal epithelial cells. J Med Microbiol 57:765–770. 10.1099/jmm.0.47769-0. [DOI] [PubMed] [Google Scholar]
- 291.Halabi-Cabezon I, Huelsenbeck J, May M, Ladwein M, Rottner K, Just I, Genth H. 2008. Prevention of the cytopathic effect induced by Clostridium difficile toxin B by active Rac1. FEBS Lett 582:3751–3756. 10.1016/j.febslet.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 292.Hippenstiel S, Tannert-Otto S, Vollrath N, Krull M, Just I, Aktories K, von Eichel-Streiber C, Suttorp N. 1997. Glucosylation of small GTP-binding Rho proteins disrupts endothelial barrier function. Am J Physiol 272:L38–L43. 10.1152/ajplung.1997.272.1.L38. [DOI] [PubMed] [Google Scholar]
- 293.Geny B, Grassart A, Manich M, Chicanne G, Payrastre B, Sauvonnet N, Popoff MR. 2010. Rac1 inactivation by lethal toxin from Clostridium sordellii modifies focal adhesions upstream of actin depolymerization. Cell Microbiol 12:217–232. 10.1111/j.1462-5822.2009.01392.x. [DOI] [PubMed] [Google Scholar]