SUMMARY
Pneumocystis species colonize mammalian lungs and cause deadly pneumonia if the immune system of the host weakens. Each species presents a specificity for a single mammalian host species. Pneumocystis jirovecii infects humans and provokes pneumonia, which is among the most frequent invasive fungal infections. The lack of in vitro culture methods for these fungi complicates their study. Recently, high-throughput sequencing technologies followed by comparative genomics have allowed a better understanding of the mechanisms involved in the sexuality of Pneumocystis organisms. The structure of their mating-type locus corresponding to a fusion of two loci, Plus and Minus, and the concomitant expression of the three mating-type genes revealed that their mode of sexual reproduction is primarily homothallism. This mode is favored by microbial pathogens and involves a single self-compatible mating type that can enter into the sexual cycle on its own. Pneumocystis sexuality is obligatory within the host’s lungs during pneumonia in adults, primary infection in children, and possibly colonization. This sexuality participates in cell proliferation, airborne transmission to new hosts, and probably antigenic variation, processes that are crucial to ensure the survival of the fungus. Thus, sexuality is central in the Pneumocystis life cycle. The obligate biotrophic parasitism with obligate sexuality of Pneumocystis is unique among fungi pathogenic to humans. Pneumocystis organisms are similar to the plant fungal obligate biotrophs that complete their entire life cycle within their hosts, including sex, and that are also difficult to grow in vitro.
KEYWORDS: obligate parasite, obligate sexuality, Pneumocystis, Taphrinomycotina, opportunistic fungi, sexuality
INTRODUCTION
Pneumocystis species are fungi that colonize the lungs of mammals (1). They belong to the subphylum Taphrinomycotina of the ascomycetes, which also includes plant pathogens and commensals. Each Pneumocystis species presents specificity for one mammalian host species, although the strictness of this characteristic remains to be understood (2, 3). Should the host’s immune system weaken, Pneumocystis organisms turn into pathogens that cause deadly pneumonia. That caused by the species infecting humans, Pneumocystis jirovecii, is among the most frequent invasive fungal infections (4), including in HIV-negative children (5). The lack of a long-term in vitro culture methods for these fungi complicates their study. Infections of rats and mice by Pneumocystis carinii and Pneumocystis murina, respectively, are used as models of the infection in humans. Pneumocystis organisms proliferate extracellularly within the lumen of the host’s lungs’ alveoli. They are biotrophic parasites that acquire nutrients from living host cells (6, 7). Whole-genome sequencing followed by comparative genomics showed that Pneumocystis species miss enzymes to carry out several biosynthetic pathways, revealing that they are obligate parasites scavenging essential compounds from their host (8–11).
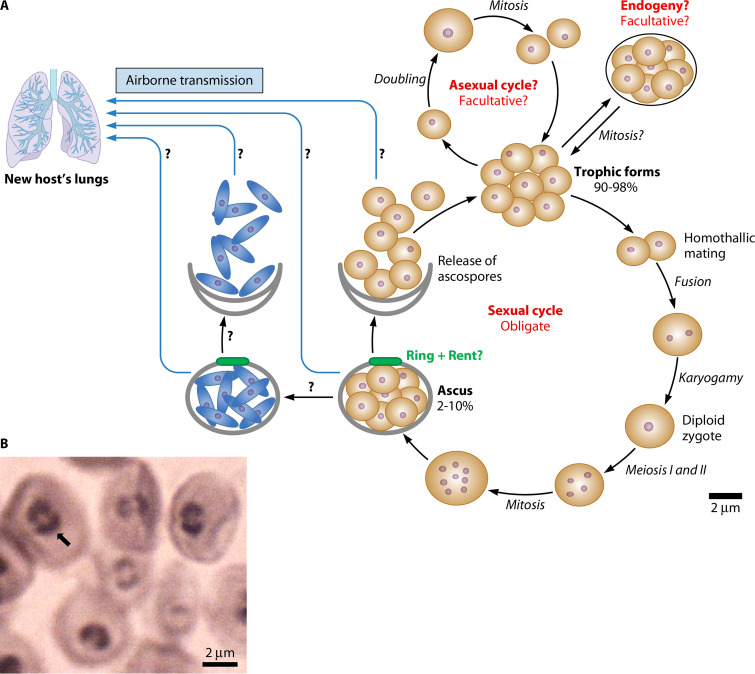
Researchers believed first that the life cycle of Pneumocystis organisms might be devoid of a sexual phase. Nevertheless, microscopic observation of synaptonemal complexes within P. carinii cells revealed that meiosis occurs (12, 13). Indeed, these structures are involved in the alignment of the homologous chromosomes and mediate crossover between them. Characterization of several genes involved in sexuality subsequently confirmed the occurrence of sex during infection (14–18). Accordingly, the life cycles proposed for Pneumocystis organisms typically include both asexual and sexual phases (19–21); the most updated one is shown in Fig. 1 (22). The haploid trophic forms would multiply asexually by binary fission and possibly by endogeny and would be involved in mating, which initiates the sexual phase. The latter culminates by the production of asci containing each of eight haploid ascospores that ensure dissemination by the air route and participate into proliferation within the host lungs. Thus, sexuality proved to be central in the Pneumocystis life cycle. Other features of the cell cycle shown in Fig. 1 are commented on the following sections.
FIG 1.
(A) Hypothetical life cycle of Pneumocystis. Violet dots represent nuclei. The question marks indicate events not supported or poorly supported by the data. Elongated ascospores that present a condensed cytoplasm are shown as blue spindle-shaped cells. No differences between the different Pneumocystis species were reported. (B) Structures like parentheses on P. jirovecii asci. The arrow points to this structure of a single ascus. It may correspond to the thickened ring observed in ascomycetes, in the center of which a rent is formed, allowing dehiscence (i.e., the release of the ascospores). Grocott’s methenamine silver staining is shown. (Panels A and B are redrawn and reproduced from Fig. 1 and 2, respectively, of reference 22 under the terms of the Creative Commons Public Domain declaration [https://creativecommons.org/publicdomain/zero/1.0].)
Early studies have assessed the occurrence of sex during the Pneumocystis cell cycle, but the process remained obscure. Recently, the mechanisms of Pneumocystis sexuality began to be unraveled thanks to whole-genome analyses, transcriptomics, and comparative genomics. In this article, I summarize the present knowledge of Pneumocystis sexuality and point to open questions.
PNEUMOCYSTIS SEX-RELATED GENES
Eighty-three putative sex-related genes have been identified in the genome of each Pneumocystis species (14–18, 23–25) (Table 1). Their detection mainly relies on their homology with genes involved in the well-characterized sexuality of the close relative Schizosaccharomyces pombe (26). These genes are potentially involved in all processes of fungal sexuality: i.e., mating signaling, cell-cell fusion, karyogamy, meiosis, and mating-type (MAT) locus silencing and switching (24, 25). However, extensive rewiring of the MAT pathways is common among fungi (27, 28), and therefore, the presence or absence of specific orthologs might be insignificant. For example, the gene mei3, which is required for entry into meiosis in S. pombe, is absent in Pneumocystis, which implies that the latter species uses another way to carry out this step. Nevertheless, the set of sex-related genes present in the Pneumocystis genome is consistent with the occurrence of the processes that are integral to sexuality (i.e., karyogamy and meiosis), whereas other sexual processes may not take place. Indeed, the genes tht1 and dmc1 have conserved function among fungi (16, 29) and thus are signatures of karyogamy and meiosis, respectively (Table 1). The structure of the Pneumocystis MAT locus, which is discussed in the following section, provides insights into this issue.
TABLE 1.
Sex-related genes identified in Pneumocystis species
| Role | Gene name (alternate) | Gene ID |
S. pombe putative ortholog |
Reference(s) | |||
|---|---|---|---|---|---|---|---|
| P. jirovecii | P. carinii | P. murina | Gene ID | Function | |||
| Mating-type locus | matMc | T551_02162 | T552_02831 | PNEG_02275 | SPBC1711.02 | Expression of M-specific genes | 22, 24, 25 |
| matPi | T551_02159 | T552_02829 | PNEG_02373 | SPMTR.02 | Expression of mei3, required for meiosis | 24, 25 | |
| matMi | Supercontig 9: 23185–23497+ | Supercontig 13: 81309–81521+ (24) or 81264–81476+ (25) | Supercontig 13: 81097–81427+ | SPBC1711.01c | Expression of mei3, required for meiosis | 24, 25 | |
| Signal transmission | ste12 (fab1) | T551_03275 | T552_00248 | PNEG_03088 | SPBC3E7.01 | Secretion of pheromones | 24 |
| mam1 | T551_03503 | T552_00261 | PNEG_03100 | SPBC25B2.02c | Export of M-pheromone | 24 | |
| mam2 | T551_00015 | T552_02343 | PNEG_03148 | SPAC11H11.04 | Response to P-pheromone | 23 – 25 | |
| map3 (ste3) | T551_02750 | T552_00176 | PNEG_03013 | SPAC3F10.10c | Response to M-pheromone | 15, 23–25 | |
| Signal transduction | ste11 | T551_02014 | T552_02504 | PNEG_02134 | SPBC32C12.02 | Expression of MAT genes | 14, 23, 24 |
| cdc42 | T551_02552 | T552_03168 | PNEG_01785 | SPAC110.03 | Development of cell polarity | 24, 25 | |
| byr1 | T551_02571 | T552_03147 | PNEG_01806 | SPAC1D4.13 | Regulation of sexual differentiation and conjugation | 24 | |
| byr2 (ste8) | T551_00958 | T552_01502 | PNEG_00717 | SPBC1D7.05 | Regulation of conjugation | 24, 25 | |
| gap1 | T551_02909 | T552_04103 | PNEG_00245 | SPBC646.12c | Regulation of cell morphogenesis during conjugation | 24 | |
| gpa1 | T551_00018 | T552_02341 | PNEG_03151 | SPBC24C6.06 | Regulation of pheromone signaling | 24 | |
| ral2 | T551_01782 | T552_03378 | PNEG_03376 | SPBC21.05c | Regulation of pheromone signaling | 24 | |
| ras1 | T551_03115 | T552_00578 | PNEG_00279 | SPAC17H9.09c | Regulation of pheromone signaling | 24 | |
| shk1 (ste20) | T551_02174 | T552_03023 | PNEG_03507 | SPBC1604.14c | Regulation of pheromone signaling | 24, 25 | |
| spk1 (fus3) | T551_02467 | T552_02142 | PNEG_03249 | SPAC31G5.09c | Regulation of pheromone signaling | 23 – 25 | |
| tim10 | T551_03530 | T552_01369 | PNEG_00015 | SPAC232.03c | Regulation of pheromone signaling | 25 | |
| git11 (ste18) | T551_00557 | T552_01736 | PNEG_02719 | SPBC215.04 | Regulation of pheromone signaling | 25 | |
| git5 (gpb1) | T551_00090 | T552_00915 | PNEG_01572 | SPBC32H8.07 | Regulation of pheromone signaling | 25 | |
| ste4 | T551_01873 | T552_01857 | PNEG_02045 | SPAC1565.04c | Regulation of pheromone signaling | 25 | |
| ste6 | T551_02045 | T552_00858 | PNEG_01634 | SPCC1442.01 | Positive regulation of pheromone signaling | 24 | |
| rgs1 | T551_01456 | T552_01691 | PNEG_02767 | SPAC23F3.12c | Negative regulation of pheromone signaling | 24, 25 | |
| scd1 | T551_02407 | T552_03331 | PNEG_03360 | SPAC16E8.09 | Regulation of cell shape | 24, 25 | |
| scd2 | T551_02328 | T552_03072 | PNEG_03558 | SPAC23H10.07 | Regulation of cell shape | 24, 25 | |
| zfs1 (moc4) | T551_01998 | T552_01981 | PNEG_01914 | SPBC1718.07c | Regulation of cell shape | 24 | |
| Signal regulation | map1 | T551_01436 | T552_02410 | PNEG_00576 | SPAC11E3.06 | Expression of P-specific genes | 24 |
| bob1 (pfd5) | T551_01168 | T552_02444 | PNEG_02075 | SPBC215.02 | Regulation of sexual differentiation | 24 | |
| cdc2 | T551_01094 | T552_00498 | PNEG_00359 | SPBC11B10.09 | Negative regulation of mitotic to meiotic cycle | 24 | |
| Mating process | cwp1 (ram2) | T551_01863 | T552_01846 | PNEG_02057 | SPAPB1A10.04c | Sexual pheromone maturation | 25 |
| kex1 | T551_02518 | T552_01067 | PNEG_01420 | SPBC16G5.09 | Sexual pheromone maturation | 25 | |
| kex2 | T551_03487 | T552_04260 | PNEG_00129 | SPAC23E12.09c | Sexual pheromone maturation | 25 | |
| dpp2 | T551_03591 | Not detected | Not detected | SPACUNK4.08 | Sexual pheromone maturation | 25 | |
| iph1 | T551_03640 | T552_02967 | PNEG_03454 | SPACUNK4.12c | Sexual pheromone maturation | 25 | |
| rce1 | T551_02384 | T552_02990 | PNEG_03476 | SPAC1687.02 | Sexual pheromone maturation | 25 | |
| ste24 | T551_02959 | T552_04168 | PNEG_02341 | SPAC3H1.05 | Sexual pheromone maturation | 25 | |
| cpp1 (ram1) | T551_02704 | T552_03212 | PNEG_01739 | SPAC17G6.04c | Sexual pheromone maturation | 25 | |
| mam4 (ste14) | T551_02897 | T552_01163 | PNEG_00234 | SPAC10F6.12c | Sexual pheromone maturation | 25 | |
| pmd1 | T551_03503 | T552_00261 | PNEG_03100 | SPCC663.03 | Sexual pheromone export | 25 | |
| Cell-cell fusion | fus1 | T551_03421 | T552_02692 | PNEG_02523 | SPAC20G4.02c | Cytoplasmic membrane fusion | 24 |
| prm1 | T551_03460 | T552_00209 | PNEG_03049 | SPAP7G5.03 | Cytoplasmic membrane fusion | 24 | |
| cfr1 | T551_03398 | T552_02419 | PNEG_00567 | SPAC6G9.12 | Cytoplasmic membrane fusion | 24 | |
| Karyogamy | tht1 (kar5) | T551_03047 | T552_03361 | PNEG_03392 | SPAC13C5.03 | Nuclear membrane fusion | 25 |
| pkl1 (kar3) | T551_01305 | T552_02382 | PNEG_02951 | SPAC3A11.14c | Nuclear membrane fusion | 25 | |
| mal3 (bim1) | T551_00265 | T552_00117 | PNEG_01219 | SPAC18G6.15 | Nuclear membrane fusion | 25 | |
| bip1 (kar2) | T551_03062 | T552_03346 | PNEG_03406 | SPAC23A12.15c | Nuclear membrane fusion | 25 | |
| Meiosis | dmc1 | T551_00146 | T552_01043 | PNEG_01443 | SPAC8E11.03c | Meiotic recombination | 16, 24, 25 |
| rad51 (rhp51) | T551_03070 | T552_03387 | PNEG_03414 | SPAC644.14c | Meiotic strand invasion and exchange | 18, 24, 25 | |
| mei2 | T551_03162 | T552_02431 | PNEG_02898 | SPAC27D7.03c | Commitment to meiosis | 17, 24 | |
| ran1 (pat1) | T551_02982 | T552_00659 | PNEG_01031 | SPBC19C2.05 | Repression of sexual conjugation | 24 | |
| srw1 (ste9) | T551_00377 | T552_01429 | PNEG_00790 | SPAC144.13c | Regulation of meiotic metaphase | 24 | |
| rec12 (spo11) | T551_00124 | T552_00948 | PNEG_01539 | SPAC17A5.11 | Initiation of meiotic recombination | 25 | |
| mcp7 (mnd1) | T551_02965 | T552_02592 | PNEG_02325 | SPAC13A11.03 | Recombinase | 25 | |
| hop1 | T551_02693 | T552_03234 | PNEG_01729 | SPBC1718.02 | Meiotic chromosome synapsis | 25 | |
| meu13 (hop2) | T551_02448 | T552_02162 | PNEG_03219 | SPAC232.15 | Meiotic chromosome pairing | 25 | |
| rec8 | T551_00216 | T552_02404 | PNEG_00582 | SPBC29A10.14 | Meiotic sister chromatid cohesion | 25 | |
| MAT locus silencing | chp2 | T551_03054 | T552_03354 | PNEG_03399 | SPBC16C6.10 | Heterochromatin assembly | 24 |
| cul4 (pcu4) | T551_01564 | T552_00671 | PNEG_01020 | SPAC3A11.08 | Heterochromatin assembly | 24 | |
| hip1 (hir1) | T551_01243 | T552_02514 | PNEG_02145 | SPBC31F10.13c | Heterochromatin assembly | 24 | |
| hpc2 | T551_02364 | T552_02145 | PNEG_03236 | SPBC947.08c | Heterochromatin assembly | 24 | |
| hrk1 | T551_01486 | T552_01661 | PNEG_02797 | SPAC23C4.03 | Heterochromatin assembly | 24 | |
| hrp3 | T551_02019 | T552_02498 | PNEG_02128 | SPAC3G6.01 | Heterochromatin assembly | 24 | |
| mit1 | T551_02073 | T552_02028 | PNEG_01867 | SPBP35G2.10 | Heterochromatin assembly | 24 | |
| pip1 (rbx1) | T551_00033 | T552_03469 | PNEG_01300 | SPAC23H4.18c | Heterochromatin assembly | 24 | |
| pob3 | T551_03410 | T552_01102 | PNEG_01383 | SPBC609.05 | Heterochromatin assembly | 24 | |
| psc3 | T551_02039 | T552_00863 | PNEG_01628 | SPAC17H9.20 | Heterochromatin assembly | 24, 25 | |
| rhp6 (ubc2) | T551_01736 | T552_02893 | PNEG_02338 | SPAC18B11.07c | Heterochromatin assembly | 24 | |
| spt6 | T551_02570 | T552_03148 | PNEG_01805 | SPAC1F7.01c | Heterochromatin assembly | 24 | |
| clr4 | T551_00331 | T552_02811 | PNEG_02256 | SPBC428.08c | Histone methyltransferase | 24 | |
| raf1 | T551_01972 | T552_01956 | PNEG_01941 | SPCC613.12c | Histone methyltransferase | ||
| lid2 | T551_00473 | T552_00980 | PNEG_01507 | SPBP19A11.06 | Histone demethylase | 24 | |
| pmt3 | T551_01148 | T552_02465 | PNEG_02095 | SPBC365.06 | Histone demethylase | 24 | |
| tra1 | T551_00450 | T552_01000 | PNEG_01486 | SPBP16F5.03c | Histone acetyltransferase | 24 | |
| clr6 | T551_00446 | T552_01004 | PNEG_01482 | SPBC36.05c | Histone deacetylase | 24 | |
| phd1 (hos2) | T551_00611 | T552_02792 | PNEG_02422 | SPAC3G9.07c | Histone deacetylase | 24 | |
| MAT locus switching | hsk1 | T551_01602 | T552_02102 | PNEG_03279 | SPBC776.12c | Imprinting | 24 |
| swi3 | T551_01272 | T552_02542 | PNEG_02174 | SPBC30D10.04 | Imprinting, conversion | 24 | |
| msh2 | T551_02326 | T552_02183 | PNEG_03198 | SPBC19G7.01c | Conversion | 24, 25 | |
| rad16 (rad1) | T551_00218 | T552_02406 | PNEG_00580 | SPCC970.01 | Conversion | 24, 25 | |
| swi1 | T551_01097 | T552_00501 | PNEG_00356 | SPBC216.06c | Conversion | 24 | |
| swi10 | T551_02136 | T552_01836 | PNEG_01115 | SPBC4F6.15c | Conversion | 24 | |
| swi5 | T551_02707 | T552_03209 | PNEG_01744 | SPBC409.03 | Switching | 24 | |
| rad22 | T551_01250 | T552_02521 | PNEG_02152 | SPAC30D11.10 | Switching | 24, 25 | |
THE MATING-TYPE LOCUS OF PNEUMOCYSTIS
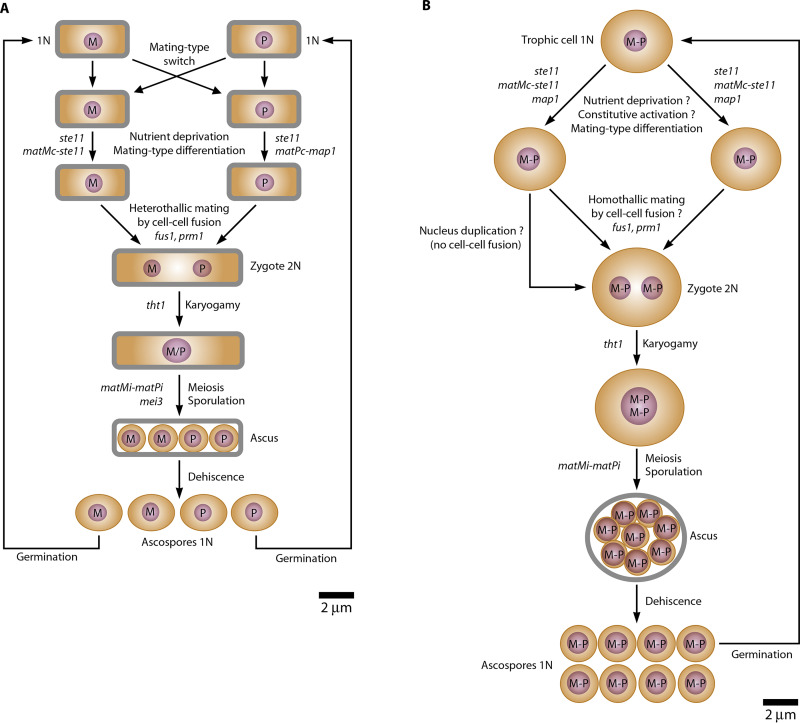
The 83 Pneumocystis sex-related genes include three putative MAT genes, the genes that govern fungal sexuality by controlling cellular mating-type identity and that are central to this process (Table 1). Taking S. pombe sexuality as a model (26) (Fig. 2A), the Pneumocystis matMc gene encodes the transcription factor with a high-mobility group DNA-binding domain responsible for the differentiation into the cellular mating type Minus (M). The Pneumocystis matPi gene encodes the transcription factor with a homeobox DNA-binding domain that functions with the cofactor matMi, which is the third Pneumocystis MAT gene identified. In S. pombe, matPi with matMi activates mei3, which derepresses meiosis in the zygote (Fig. 2A). In the absence of mei3 in Pneumocystis species, the function of matPi with matMi remains to be understood. Neither an ortholog to the S. pombe transcription factor matPc responsible for differentiation into the mating type Plus (P) was identified in Pneumocystis, nor were any other types of MAT transcription factors (24, 30). Importantly, the identity of Pneumocystis matMc was confirmed experimentally by functional complementation restoring meiosis and sporulation of the S. pombe matMc mutant (30). These observations suggest that the three MAT genes identified are sufficient to trigger sexual mating and meiosis in Pneumocystis species, but that the mechanisms involved differ from those in S. pombe.
FIG 2.
Schematic sexual cycles of Schizosaccharomyces pombe (A) and Pneumocystis species (B). The genes involved in the regulation of steps are indicated. The circles filled in violet represent the nuclei. M, minus MAT locus; P, plus MAT locus; N, number of chromosomes of a haploid set. (A) Secondary homothallism of S. pombe involving mating-type switch and heterothallic mating by cell-cell fusion. The haploid and diploid mitotic cycles by fission are not shown. P/M, active P and M MAT loci on different chromosomes. (B) Hypothetical primary homothallism of Pneumocystis species. The question marks indicate events that are not demonstrated. The occurrence or not of homothallic mating by cell-cell fusion remains to be determined. The involvement of the genes mentioned is presumed but not established. The genes matPc and mei3 are absent in Pneumocystis species (see the text). The haploid mitotic cycles shown in Fig. 1 are not shown (asexual cycle and endogeny). P-M, fused P and M MAT loci on the same chromosome.
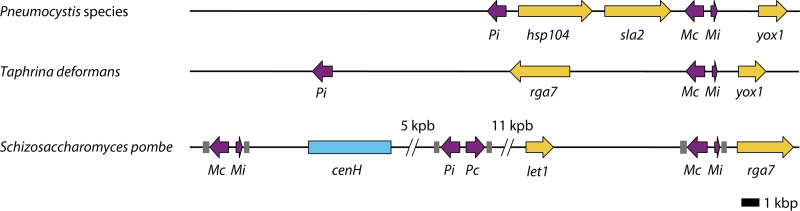
The three MAT genes identified are close to each other in the genome of each Pneumocystis species; they constitute the MAT locus (Fig. 3). An intergenic region of 100 to 300 bp separates matMc and matMi genes, which are divergently transcribed. The third MAT gene, matPi, is located approximately 8 kbp away, on the same strand as matMc. The upstream region of each MAT gene harbors a potential transcription promoter (30). The shared promoter region of matMc and matMi suggests a common regulation ensuring a tightly coordinated expression. Such an arrangement is a hallmark of most active genes that could structure chromatin and nascent mRNAs for subsequent regulation, providing fine-tuned expression (31–33). The arrangement with divergent transcription is frequent among fungal MAT loci, including in the ascomycetous models Saccharomyces cerevisiae and S. pombe (Fig. 3), as well as in basidiomycetes (34). The common regulation of matMc and matMi is likely to be ensured by the Pneumocystis ortholog to the S. pombe transcription factor Ste11. In S. pombe, this regulation relies on the single recognition motif TTTCTTTGTT, which is present close to the middle of the intergenic region (26, 35). In Pneumocystis, the recognition motif, or part of it, could be the CCTTG sequence that resembles it, which is conserved in P. jirovecii and P. carinii at the same location. The latter hypothesis is also based on the observation that this motif is duplicated in about 50% of the P. jirovecii isolates, which suggests its importance (24). In P. murina, the motif CCTGT or CCGTT might be involved. Consistent with its potential important role, ste11 is expressed during pneumonia in rats as the second most abundant transcript (23). The latter observation is also compatible with Ste11 participating in conjunction with MatMc in the activation of the M-specific genes, as observed in S. pombe.
FIG 3.
Schematic representation of MAT loci present in the subphylum Taphrinomycotina. MAT genes are shown in violet, other relevant genes in yellow, and the centromere-homologous sequence cenH in blue. The chromosomes carrying the genes are shown as black lines. The drawings are roughly to scale. The primary homothallic species Pneumocystis and Taphrina deformans each harbor a single MAT locus that includes three genes. The secondary homothallic species S. pombe harbors three MAT loci (or cassettes), each including two genes. The two MAT loci flanking cenH are silenced, whereas the third locus is active (here, for example, the M mating type). The cis-acting sequence motifs H1, H2, and H3, involved in mating-type switching and flanking each MAT locus, are shown in dark gray. The MAT locus is located on supercontigs (scaffolds) 9, 13, and 13, respectively, of the P. jirovecii, P. carinii, and P. murina genome assemblies (25). (Each scaffold presumably corresponds to one chromosome.)
Two genes are generally conserved close to the MAT locus among ascomycetes: sla2 and apn2 or dic1 (36, 37). The Pneumocystis MAT locus harbors only sla2, between matPi and matMc, together with the hsp104 gene (Fig. 3). The Pneumocystis MAT locus presents only a limited synteny with that of the other members of the Taphrinomycotina subphylum: i.e., only one gene in common with the close relative Taphrina deformans (yox1) and none in common with S. pombe (24) (Fig. 3). The MAT loci of the two latter relatives do not harbor the sla2 gene and present only one gene conserved (rga7). This lack of synteny suggests important evolutionary distances among members of the subphylum Taphrinomycotina. The gene sla2 encodes the adaptor linking actin to clathrin involved in endocytosis and the cytoskeleton. As in S. pombe, it might be essential under most conditions in Pneumocystis. This essentiality might prevent deletions and ensure the stability of the Pneumocystis MAT locus. Such a phenomenon is also postulated for the gene let1, which is present between the S. pombe active and silent MAT loci (26, 38) (Fig. 3).
The structure and content of the Pneumocystis MAT locus suggest a specific mode of sexual reproduction, which is discussed in the following section.
MODE OF SEXUAL REPRODUCTION OF PNEUMOCYSTIS SPECIES
The presence of genes involved in the differentiation of both P and M mating types (P and M genes) in the Pneumocystis MAT locus is incompatible with heterothallism because this mode of sexual reproduction implies that the P and M genes are in different genomes (39). Secondary homothallism is also unlikely because this mode involves more than one MAT locus: i.e., two or three, with one active and the other(s) silenced. Moreover, the latter mode requires genetic elements for switching the active MAT locus and silencing the others that are absent in Pneumocystis genomes (cis-acting sequence motifs of H, proximity to telomere or centromere-like repeats) (24, 30) (Fig. 3). Homothallism resulting from unidirectional mating-type switching (40) is also unlikely because (i) it requires indirect or direct repeats for the deletion and reconstitution of one MAT locus, and (ii) it implies a mixture of different MAT loci in each cell population, which is not observed in Pneumocystis. The presence of both M and P genes in the Pneumocystis MAT locus is consistent with a fusion of two MAT loci, M and P, that were present in an ancestor, followed by the loss of one MAT gene (matPc). Such a scenario has previously been proposed to account for fused MAT loci of other fungi (41). These observations suggest that the sexual mode of reproduction of Pneumocystis species is primary homothallism, the mode that involves a single self-compatible mating type that can engage on its own into the sexual phase.
The hypothesis of primary homothallism is supported by the presence of the same MAT locus in all P. jirovecii DNA samples (30). It is further supported by the finding that all three MAT genes are expressed concomitantly during pneumonia in both humans and mice (Table 2). Such coexpression is expected because expression of both P and M genes is generally required for successful initiation of the sexual cycle in primary homothallic species (39). This finding is particularly relevant in mice because infections are thought to be caused by a single P. murina strain, strongly suggesting expression from the same MAT locus. On the other hand, infections in humans are most often, if not always, polyclonal (42), which leaves the possibility that the coinfecting P. jirovecii strains may differ in the expression of their MAT genes. It must be stressed that, although primary homothallism of Pneumocystis appears almost certain, one cannot totally exclude a new mode of sexual reproduction involving previously unknown mechanisms. Indeed, fungi present a myriad of different mechanisms to trigger sexuality (39), and new ones could be discovered in the future. The definitive ascertainment of the mode of sexual reproduction of Pneumocystis organisms may require their culturing in vitro.
TABLE 2.
Results from reverse transcriptase PCR amplification of Pneumocystis transcripts from bronchoalveolar lavage fluid samples from 10 patients with Pneumocystis pneumonia and from infected mouse lungsa
| cDNA source | PCR result for: |
|||||
|---|---|---|---|---|---|---|
| β-Tubulinb |
MAT transcription factors |
Pheromone receptors |
||||
| matMc | matMi | matPi | mam2 | map3 | ||
| P. jirovecii patient no.: | ||||||
| 1 | + | + | + | + | + | + |
| 2 | − | − | − | − | − | − |
| 3 | + | − | + | − | + | − |
| 4 | + | + | + | + | + | + |
| 5 | + | + | + | + | + | + |
| 6 | + | − | − | + | + | − |
| 7 | + | + | + | + | + | + |
| 8 | + | − | − | + | − | − |
| 9 | − | − | − | − | − | − |
| 11 | + | + | + | + | − | + |
| P. murina | + | + | + | + | + | + |
+, positive PCR result; −, negative PCR result. (Adapted from reference 47.)
Amplification of the β-tubulin transcripts was used as a control (30). It assessed adequate reverse transcriptase PCR by the absence of the intron in these PCR products. This control suggested that the negative PCR results obtained in these experiments are due to RNA degradation. The latter may have occurred during the uncontrolled period between collection of the samples from the patients and their arrival in our laboratory.
Primary homothallism is also observed in other human-pathogenic fungi: e.g., Candida albicans (43) and Cryptococcus neoformans (44). Moreover, sexual systems resembling homothallism are probably used by human-pathogenic protozoans: i.e., Toxoplasma, Giardia, Trypanosoma, and Leishmania (45). This suggests that this mode of sexual reproduction is advantageous for microbial pathogens. The reason hypothesized is that it alleviates the need to find a compatible mating partner in the restricted niches of the host body, while still providing the benefits of sex (increase in genetic diversity and virulence and elimination of deleterious mutations) (45, 46).
The mechanisms involved in primary homothallism in fungi are poorly known compared to those of secondary homothallism and heterothallism. In addition, they probably vary considerably according to the fungal species and rewiring of the pathways. The mechanisms of sexuality in action in Pneumocystis species are discussed in the following section.
MECHANISMS INVOLVED IN THE PRIMARY HOMOTHALLISM OF PNEUMOCYSTIS SPECIES
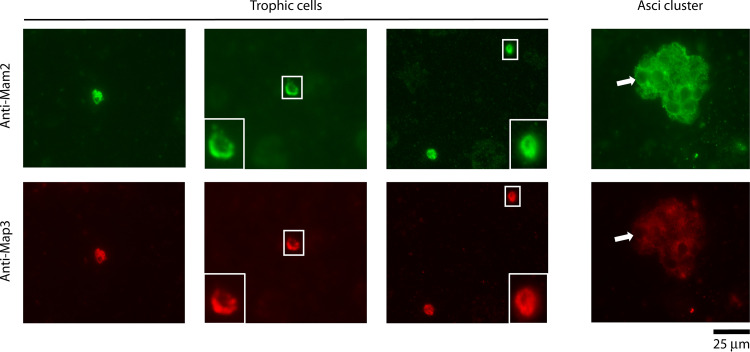
The absence of the transcription factor matPc, responsible in S. pombe for the differentiation into the cellular mating type P, suggests that Pneumocystis species are unisexual, involving a single mating type, M, as observed in other fungi (C. albicans and C. neoformans) (43, 44). However, analysis of the pheromone receptors suggested that the P. jirovecii trophic forms are of both mating types at the same time (47). Indeed, the genes mam2 and map3, encoding, respectively, the receptors M and P, are present in every Pneumocystis genome (24, 25) and are both expressed during pneumonia in rats, mice, and humans (24, 25, 47) (Table 1). The P. murina infection is again particularly relevant because it strongly suggests that both receptors are expressed from the same genome, and not from different coinfecting strains. Immunostaining revealed that P. jirovecii trophic forms expose at their surface both receptors M and P at the same time (Fig. 4). Consequently, they could excrete both pheromones M and P, but this could not be investigated so far because the encoding genes could not be identified due to their significant divergence (24, 25). In S. pombe, the transcription factor Map1 forms a heterodimer with MatPc, which is responsible for the activation of the P-specific genes (26). An ortholog of map1 is present in Pneumocystis and could be involved in the differentiation into the mating type P despite the absence of matPc.
FIG 4.
Indirect immunofluorescence microscopic analysis of Mam2 and Map3 pheromone receptors on P. jirovecii cells from a bronchoalveolar lavage fluid sample of a patient with Pneumocystis pneumonia. Shown is costaining with anti-Mam2 and anti-Map3 (pheromone receptors M and P, respectively). The presumed trophic cells in the small white squares are shown enlarged in the insets at the left or right bottom corners of the images, and a single spherical cell corresponding to an ascus within the cluster is indicated by the white arrows. A fluorescein isothiocyanate (FITC) filter (green) was used to visualize Mam2 staining (Alexa Fluor 488), and a tetramethyl rhodamine isocyanate (TRITC) filter (red) was used to visualize Map3 staining (Alexa Fluor 594). Bar, 25 μm. (Reproduced from reference 47.)
The receptors M and P present at the cell surface of Pneumocystis trophic cells could be involved in the recognition of the mating partners. Being identical, these cells might mate randomly by cell-cell fusion within host’s lung alveoli. However, this is not necessarily the case because cell-cell fusion can be replaced by nucleus duplication in homothallism, and the pheromone receptor systems play other roles in some primary homothallic fungi (48–50). For example, the close relative T. deformans does not rely on cell-cell fusion despite the fact that it harbors one pheromone receptor (24). Electron microscopy studies have presented few images of two Pneumocystis trophic cells with connected cytoplasmic membranes that could correspond to cell-cell fusion events (22). Moreover, plasmogamy during mating and cytokinesis during mitosis cannot be distinguished morphologically. Similarly, the connected nuclear membranes observed on some images may correspond to karyogamy or karyokinesis. Indeed, Pneumocystis might have a closed mitosis with the nuclear membrane present throughout the cell cycle as in most fungi, rather than an open one as in many basidiomycetes. Pneumocystis harbors potential orthologs of the S. pombe genes fus1 and prm1 (24). (Note that prm1 is also present in P. jirovecii [our unpublished data].) These genes are essential to cell-cell fusion during mating in S. pombe (51). However, they are also present in T. deformans (24). Thus, the occurrence of cell-cell fusion during Pneumocystis sexuality remains an open question.
Heteroplasmy of mitochondria has been reported in Pneumocystis based on the presence of more alleles of the mitochondrial markers than of the nuclear ones (42). Although it cannot be excluded that it results from a higher frequency of mutations in these organelles, this heteroplasmy might result from biparental inheritance of mitochondria that could be generated by cell-cell fusion during sexuality. This would fit that no studies reported cells other than the trophic cells and asci that could participate in anisogamy (22), one of the phenomena that can lead to uniparental inheritance of mitochondria. It is possible that the Pneumocystis mitochondrial heteroplasmy reflects frequent cell-cell fusions, which would be coherent with sexuality being necessary and possibly the preponderant mechanism of proliferation (22). This would imply a system of homoplasmy control weaker than that generally present in fungi (39), although persistent heteroplasmy in fungi has been reported (52).
Splicing variants corresponding to intron retention have been observed among the transcripts of the pheromone receptors (47, 53). These variants might be involved in the regulation of expression and associated with specific stages of the cell cycle. The timing of expression of the Pneumocystis sex genes is discussed in the following section.
OCCURRENCE OF PNEUMOCYSTIS SEXUAL CYCLE
The sexual cycle of many fungi, including S. cerevisiae and S. pombe, is triggered by deprivation of essential nutrients, such as a fermentable carbon source or nitrogen, or by stress (26, 54). As far as Pneumocystis is concerned, expression of MAT and other sex-related genes has been observed during pneumonia in humans, mice, and rats (15, 23, 25, 30, 47) (Table 2). It is plausible that Pneumocystis sexuality occurs when the host alveoli are filled with fungal cells (i.e., when pneumonia is overt), because this stage may correspond to an exhaustion of the nutriments as well as marked stress (23). This hypothesis would be consistent with the activation of sexuality upon treatment of P. murina with echinocandins, inhibiting growth and provoking stress (55). The sensitivity of the trophic forms to echinocandins might result from a link between asexual and sexual cycles through regulatory factors or from the presence of small amounts of 1,3-β glucan in their wall (22). However, sexuality may also take place in the lungs of colonized individuals as they are a source of P. jirovecii in a cluster of nosocomial cases (56), which implies airborne transmission by asci (Fig. 1A). The alveoli might not be filled in the latter situation because of the lower fungal load present in colonized humans (56). Thus, one cannot exclude that Pneumocystis sexuality is constitutively induced during growth—possibly by the stress resulting from the action of the host immune system. This hypothesis would fit that this sexuality might be the preponderant mechanism of proliferation. It would also be consistent with the fact that Pneumocystis sexuality proved to be obligatory within host lungs, a characteristic that is discussed in the following section.
OBLIGATE PNEUMOCYSTIS SEXUALITY WITHIN HOST LUNGS DURING PNEUMONIA
Two facts strongly suggest that Pneumocystis sexuality is obligatory within host lungs during pneumonia: the MAT and sex-related genes are concomitantly expressed, and asci are always present. Asci are observed in all pneumonia, so that their staining has been used for decades to diagnose the disease (Fig. 1B). Only a few infections with a reduced proportion of asci have been reported, and only under particular conditions: an athymic host (57), immunity reconstitution (58), and prophylaxis breakthrough (59). Asci are also observed in all primary infections that occur during the first 2 years of life (60, 61). Obligate sexuality is consistent with (i) asci and/or ascospores being the airborne infectious particles responsible for transmission between hosts because this ensures survival (62, 63) and (ii) its necessity for proliferation by the release of ascospores within the host lungs (22). On the other hand, the data gathered so far suggest that the asexual phase of proliferation by mitosis and possibly endogeny might be facultative (22) (Fig. 1A). It could be activated under certain peculiar conditions or at early stages of the infection and might be capable of latency, ensuring survival upon growth inhibition (62).
The obligate nature of Pneumocystis sexuality may also ensure the antigenic variation of these fungi, a system crucial for colonization and thus survival. Indeed, these pathogens dedicate about 8% of their genomes to a subtelomeric superfamily of genes encoding six families of major surface glycoproteins (25, 64–67). The antigenic variation relies on recombinations creating continuously new mosaic genes, as well as on mutually exclusive expression of a single gene out of approximately 80 genes of the family encoding the most abundant glycoprotein at the cell surface (family I, also named A1). Recombinations among each of the six families and the exchange of the expressed gene of family I probably occur when all subtelomeres are close to each other: i.e., when they are clustered as a “bouquet” at the nuclear membrane during the meiotic prophase (68). Thus, antigenic variation probably requires sexuality to occur.
Although multicellular organisms cannot be compared easily with microbes, it is striking that mammals and plants, but only few fungi, share obligate sexuality during their life cycles (39). Pneumocystis is similar to the plant fungal obligate biotrophs that complete their entire life cycles within their hosts, including sex (6). Thus, Pneumocystis is an animal parasite resembling plant parasites that, consistently, has nutritional requirements observed in both these types of parasites (7, 10). Moreover, the reluctance of Pneumocystis organisms to sustain axenic growth in vitro is consistent with that of the fungal obligate biotrophs infecting plants (6). The Pneumocystis lifestyle differs from that of other human fungal pathogens that are necrotrophs obtaining nutriments from killed host cells with a facultative sexuality (Candida, Aspergillus, and dermatophytes).
The relationship between Pneumocystis organisms and their hosts fits the concept of “compatibility” used in the fungal plant pathogen field (6, 23). The latter consists of a relationship between an adapted biotrophic fungal pathogen and a susceptible host where there is complementation, but which may eventually lead to the development of the disease. Each Pneumocystis genome is only approximately 8 Mbp, which is among the smallest and most compact fungal genomes (69, 70). This compaction results not only from the loss of essential pathways but also from the presence of a single copy of the ribosomal DNA. Genome compaction is also observed in fungal pathogens adapted to host waxy surface of plants or fruits (69, 71). Like the surface of plants, the epithelial cells’ surface in mammalian lungs may constitute an extreme environment that imposes restrictions on the parasites, such as nutrient limitation.
The obligate sexuality of Pneumocystis organisms probably has a great importance in their evolution. The selection of the homothallic mode for this sexuality during evolution is discussed in the following section.
POSSIBLE EVOLUTION OF PNEUMOCYSTIS SEXUALITY
The ancestral mode of sexual reproduction of fungi remains to be determined: homo- versus heterothallism (39). Primary homothallism has often been found to result from recombination events between heterothallic partners (39, 72). This possibility is compatible with the structure of the Pneumocystis MAT locus because it includes both P and M genes. The species T. deformans is putatively the closest known relative of Pneumocystis (9, 10, 25) and harbors a MAT locus similar to that of Pneumocystis (24) (Fig. 3). Thus, a heterothallic ancestor may have generated these two primary homothallic genera. On the other hand, this ancestor would have also evolved into the secondary homothallic relative S. pombe by subsequent acquisition of a supplementary MAT locus as well as the switching and silencing mechanisms (72). This putative scenario suggests that the acquisition of obligate biotrophy on mammals (Pneumocystis) or plants (Taphrina) involved the selection of primary homothallic strains about 100 million years ago (73). This would be consistent with the belief that primary homothallism is advantageous for microbial pathogens. At least for Pneumocystis, acquisition of primary homothallism would have ensured maintenance of sex, which is essential for its survival. This evolutionary hypothesis could be challenged by the characterization of the MAT locus and the mode of sexual reproduction of other related fungi.
CONCLUSIONS
Pneumocystis is unique among fungi pathogenic to humans. It differs by its obligate and biotrophic parasitism, its transmissibility between host individuals, and its obligate sexuality within the host’s lungs. The latter appears essential because it would ensure proliferation, dissemination, and possibly antigenic variation, processes that are all required for the survival of the fungus. Figure 2B shows the hypothetical sexual cycle of Pneumocystis that can be derived from the observations made so far. Importantly, whether or not cell-cell fusion occurs remains to be determined. The understanding of the mechanisms of this sexuality and its implications for Pneumocystis genetic diversity and evolution deserves further study.
ACKNOWLEDGMENTS
P.H.M.’s research is presently funded by the Swiss National Science Foundation (grant 310030_ 192802). The funder had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
I warmly thank Sophie Richard for the invaluable and outstanding technical expertise she brought to our research during many years. I am indebted to Marco Pagni of the Swiss Bioinformatics Institute for review and improvements of the manuscript and for our long-standing and extremely fruitful collaboration. I also very much thank João Almeida of the UCIBIO (Applied Molecular Biosciences Unit, Universidade Nova de Lisboa) for precious analyses and inputs into our research on Pneumocystis sexuality. I also thank the reviewers for their constructive comments.
I have no conflicts of interest to declare.
Biography

Philippe M. Hauser obtained his academic degrees in Biology from the University of Lausanne, Switzerland. For his Ph.D., he studied Gram-positive bacteria. Then he did a postdoctoral training at the University of Oxford, United Kingdom, during which he studied bacterial sporulation. Since 1996, he has been the leader of a research group studying Pneumocystis infection in humans at the Centre Hospitalier Universitaire Vaudois in Lausanne. He has investigated aspects of the molecular epidemiology, drug resistance, and genomics of Pneumocystis. Presently, his research focuses on the sexuality and the antigenic variation of the fungus.
This review is dedicated to my dear wife Brigitte, who provides constant support throughout the years.
REFERENCES
- 1.Gigliotti F, Limper AH, Wright T. 2014. Pneumocystis. Cold Spring Harb Perspect Med 4:a019828. 10.1101/cshperspect.a019828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Latinne A, Bezé F, Delhaes L, Pottier M, Gantois N, Nguyen J, Blasdell K, Dei-Cas E, Morand S, Chabé M. 2018. Genetic diversity and evolution of Pneumocystis fungi infecting wild Southeast Asian murid rodents. Parasitology 145:885–900. 10.1017/S0031182017001883. [DOI] [PubMed] [Google Scholar]
- 3.Babb-Biernacki SJ, Esselstyn JA, Doyle VP. 2020. Rethinking host range in Pneumocystis. PLoS Pathog 16:e1008824. 10.1371/journal.ppat.1008824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White T. 2012. Hidden killers: human fungal infections. Sci Transl Med 4:165rv13. 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 5.Pneumonia Etiology Research for Child Health (PERCH) Study Group. 2019. Causes of severe pneumonia requiring hospital admission in children without HIV infection from Africa and Asia: the PERCH multi-country case-control study. Lancet 394:757–779. 10.1016/S0140-6736(19)30721-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cushion MT, Stringer JR. 2010. Stealth and opportunism: alternative lifestyles of species in the fungal genus Pneumocystis. Annu Rev Microbiol 64:431–452. 10.1146/annurev.micro.112408.134335. [DOI] [PubMed] [Google Scholar]
- 7.Hauser PM. 2014. Genomic insights into the fungal pathogens of the genus Pneumocystis: obligate biotrophs of humans and other mammals. PLoS Pathog 10:e1004425. 10.1371/journal.ppat.1004425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hauser PM, Burdet FX, Cissé OH, Keller L, Taffé P, Sanglard D, Pagni M. 2010. Comparative genomics suggests that the fungal pathogen Pneumocystis is an obligate parasite scavenging amino acids from its host’s lungs. PLoS One 5:e15152. 10.1371/journal.pone.0015152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cissé OH, Pagni M, Hauser PM. 2012. De novo assembly of the Pneumocystis jirovecii genome from a single bronchoalveolar lavage fluid specimen from a patient. mBio 4:e00428-12. 10.1128/mBio.00428-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cissé OH, Pagni M, Hauser PM. 2014. Comparative genomics suggests that the human pathogenic fungus Pneumocystis jirovecii acquired obligate biotrophy through gene loss. Genome Biol Evol 6:1938–1948. 10.1093/gbe/evu155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porollo A, Sesterhenn TM, Collins MS, Welge JA, Cushion MT. 2014. Comparative genomics of Pneumocystis species suggests the absence of genes for myo-inositol synthesis and reliance on inositol transport and metabolism. mBio 5:e01834-14. 10.1128/mBio.01834-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsumoto Y, Yoshida Y. 1984. Sporogony in Pneumocystis carinii: synaptonemal complexes and meiotic nuclear divisions observed in precysts. J Protozool 31:420–428. 10.1111/j.1550-7408.1984.tb02989.x. [DOI] [PubMed] [Google Scholar]
- 13.Peters SE, English K, Rana A, Akter S, Malik S, Warburton NC, Duckett JG. 2001. Synaptonemal complexes in the pre-cyst of Pneumocystis carinii. J Eukaryot Microbiol 10.1111/j.1550-7408.2001.tb00485.x. [DOI] [PubMed] [Google Scholar]
- 14.Vohra PK, Puri V, Kottom TJ, Limper AH, ThomasCF, Jr.. 2003. Pneumocystis carinii STE11, an HMG-box protein, is phosphorylated by the mitogen activated protein kinase PCM. Gene 312:173–179. 10.1016/s0378-1119(03)00614-0. [DOI] [PubMed] [Google Scholar]
- 15.Vohra PK, Park JG, Sanyal B, Thomas CF. 2004. Expression analysis of PCSTE3, a putative pheromone receptor from the lung pathogenic fungus Pneumocystis carinii. Biochem Biophys Res Commun 319:193–199. 10.1016/j.bbrc.2004.04.154. [DOI] [PubMed] [Google Scholar]
- 16.Kutty G, Achaz G, Maldarelli F, Varma A, Shroff R, Becker S, Fantoni G, Kovacs JA. 2010. Characterization of the meiosis-specific recombinase Dmc1 of Pneumocystis. J Infect Dis 202:1920–1929. 10.1086/657414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burgess JW, Kottom TJ, Limper AH. 2008. Pneumocystis carinii exhibits a conserved meiotic control pathway. Infect Immun 76:417–425. 10.1128/IAI.00986-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kutty G, Kovacs JA. 2007. Identification and characterization of rad51 of Pneumocystis. Gene 389:204–211. 10.1016/j.gene.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 19.Cushion MT, Ruffolo JJ, Walzer PD. 1988. Analysis of the developmental stages of Pneumocystis carinii, in vitro. Lab Invest 58:324–331. [PubMed] [Google Scholar]
- 20.Yoshida Y. 1989. Ultrastructural studies of Pneumocystis carinii. J Protozool 36:53–60. 10.1111/j.1550-7408.1989.tb02696.x. [DOI] [PubMed] [Google Scholar]
- 21.Aliouat-Denis CM, Martinez A, Aliouat EM, Pottier M, Gantois N, Dei-Cas E. 2009. The Pneumocystis life cycle. Mem Inst Oswaldo Cruz 104:419–426. 10.1590/s0074-02762009000300004. [DOI] [PubMed] [Google Scholar]
- 22.Hauser PM, Cushion MT. 2018. Is sex necessary for the proliferation and transmission of Pneumocystis? PLoS Pathog 14:e1007409. 10.1371/journal.ppat.1007409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cushion MT, Smulian AG, Slaven BE, Sesterhenn T, Arnold J, Staben C, Porollo A, Adamczak R, Meller J. 2007. Transcriptome of Pneumocystis carinii during fulminate infection: carbohydrate metabolism and the concept of a compatible parasite. PLoS One 2:e423. 10.1371/journal.pone.0000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Almeida JM, Cissé OH, Fonseca Á, Pagni M, Hauser PM. 2015. Comparative genomics suggests primary homothallism of Pneumocystis species. mBio 6:e02250-14. 10.1128/mBio.02250-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma L, Chen Z, Huang DW, Kutty G, Ishihara M, Wang H, Abouelleil A, Bishop L, Davey E, Deng R, Deng X, Fan L, Fantoni G, Fitzgerald M, Gogineni E, Goldberg JM, Handley G, Hu X, Huber C, Jiao X, Jones K, Levin JZ, Liu Y, Macdonald P, Melnikov A, Sassi M, Sherman BT, Song X, Sykes S, Tran B, Waish L, Xia Y, Yang J, Young S, Zeng Q, Zheng X, Stephens R, Nusbaum C, Birren BW, Azadi P, Lempicki RA, Cuomo CA, Kovacs JA. 2016. Genome analysis of three Pneumocystis species reveals adaptation mechanisms to life exclusively in mammalian hosts. Nat Commun 7:10740. 10.1038/ncomms10740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nielsen O, Egel R. 2007. The mat genes of Schizosaccharomyces pombe: expression, homothallic switch, and silencing, p 143–157. In Heitman J, Kronstad JW, Taylor JW, Casselton LA (ed), Sex in fungi: molecular determination and evolutionary implications. ASM Press, Washington, DC. [Google Scholar]
- 27.Reedy JL, Floyd AM, Heitman J. 2009. Mechanistic plasticity of sexual reproduction and meiosis in the Candida pathogenic species complex. Curr Biol 19:891–899. 10.1016/j.cub.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherwood RK, Scaduto CM, Torres SE, Bennett RJ. 2014. Convergent evolution of a fused sexual cycle promotes the haploid lifestyle. Nature 506:387–390. 10.1038/nature12891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu C, Heitman J. 2017. PRM1 and KAR5 function in cell-cell fusion and karyogamy to drive distinct bisexual and unisexual cycles in the Cryptococcus pathogenic species complex. PLoS Genet 13:e1007113. 10.1371/journal.pgen.1007113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richard S, Almeida JM, Cissé OH, Luraschi A, Nielsen O, Pagni M, Hauser PM. 2018. Functional and expression analyses of the Pneumocystis MAT genes suggest obligate sexuality through primary homothallism within host lungs. mBio 9:e02201-17. 10.1128/mBio.02201-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. 2008. Divergent transcription from active promoters. Science 322:1849–1851. 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seila AC, Core LJ, Lis JT, Sharp PA. 2009. Divergent transcription: a new feature of active promoters. Cell Cycle 8:2557–2564. 10.4161/cc.8.16.9305. [DOI] [PubMed] [Google Scholar]
- 33.Lepoivre C, Belhocine M, Bergon A, Griffon A, Yammine M, Vanhille L, Zacarias-Cabeza J, Garibal MA, Koch F, Maqbool MA, Fenouil R, Loriod B, Holota H, Gut M, Gut I, Imbert J, Andrau JC, Puthier D, Spicuglia S. 2013. Divergent transcription is associated with promoters of transcriptional regulators. BMC Genomics 14:914. 10.1186/1471-2164-14-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coelho MA, Bakkeren G, Sun S, Hood ME, Giraud T. 2017. Fungal sex: the basidiomycota. Microbiol Spectr 5. 10.1128/microbiolspec.FUNK-0046-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelly M, Burke J, Smith M, Klar A, Beach D. 1988. Four mating-type genes control sexual differentiation in the fission yeast. EMBO J 7:1537–1547. 10.1002/j.1460-2075.1988.tb02973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanson SJ, Wolfe KH. 2017. An evolutionary perspective on yeast mating-type switching. Genetics 206:9–32. 10.1534/genetics.117.202036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolfe KH, Butler G. 2017. Evolution of mating in the Saccharomycotina. Annu Rev Microbiol 71:197–214. 10.1146/annurev-micro-090816-093403. [DOI] [PubMed] [Google Scholar]
- 38.Michael H, Schmidt H, Fleck O, Gutz H, Liedtke C, Lorentz A, Ostermann K. 1994. The mating-type region of Schizosaccharomyces pombe contains an essential gene encoding a protein homologous to human modulators of HIV transactivation. Gene 145:205–210. 10.1016/0378-1119(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 39.Ni M, Feretzaki M, Sun S, Wang X, Heitman J. 2011. Sex in fungi. Annu Rev Genet 45:405–430. 10.1146/annurev-genet-110410-132536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yun SH, Kim HK, Lee T, Turgeon BG. 2017. Self-fertility in Chromocrea spinulosa is a consequence of direct repeat-mediated loss of MAT1-2, subsequent imbalance of nuclei differing in mating type, and recognition between unlike nuclei in a common cytoplasm. PLoS Genet 13:e1006981. 10.1371/journal.pgen.1006981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, Agrafioti I, Arnaud MB, Bates S, Brown AJ, Brunke S, Costanzo MC, Fitzpatrick DA, de Groot PW, Harris D, Hoyer LL, Hube B, Klis FM, Kodira C, Lennard N, Logue ME, Martin R, Neiman AM, Nikolaou E, Quail MA, Quinn J, Santos MC, Schmitzberger FF, Sherlock G, Shah P, Silverstein KA, Skrzypek MS, Soll D, Staggs R, Stansfield I, Stumpf MP, Sudbery PE, Srikantha T, Zeng Q, Berman J, Berriman M, Heitman J, Gow NA, Lorenz MC, Birren BW, Kellis M, Cuomo CA. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459:657–662. 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alanio A, Gits-Muselli M, Mercier-Delarue S, Dromer F, Bretagne S. 2016. Diversity of Pneumocystis jirovecii during infection revealed by ultradeep pyrosequencing. Front Microbiol 7:733. 10.3389/fmicb.2016.00733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alby K, Schaefer D, Bennett RJ. 2009. Homothallic and heterothallic mating in the opportunistic pathogen Candida albicans. Nature 460:890–893. 10.1038/nature08252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin X, Hull CM, Heitman J. 2005. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature 434:1017–1021. 10.1038/nature03448. [DOI] [PubMed] [Google Scholar]
- 45.Heitman J. 2010. Evolution of eukaryotic microbial pathogens via covert sexual reproduction. Cell Host Microbe 8:86–99. 10.1016/j.chom.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roach KC, Heitman J. 2014. Unisexual reproduction reverses Muller’s ratchet. Genetics 198:1059–1069. 10.1534/genetics.114.170472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luraschi A, Richard S, Almeida J, Pagni M, Cushion MT, Hauser PM. 2019. Expression and immunostaining analyses suggest that Pneumocystis primary homothallism involves trophic cells displaying both plus and minus pheromone receptors. mBio 10:e01145-19. 10.1128/mBio.01145-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.David-Palma M, Sampaio JP, Gonçalves P. 2016. Genetic dissection of sexual reproduction in a primary homothallic basidiomycete. PLoS Genet 12:e1006110. 10.1371/journal.pgen.1006110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mayrhofer S, Weber JM, Pöggeler S. 2006. Pheromones and pheromone receptors are required for proper sexual development in the homothallic ascomycete Sordaria macrospora. Genetics 172:1521–1533. 10.1534/genetics.105.047381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nygren K, Strandberg R, Gioti A, Karlsson M, Johannesson H. 2012. Deciphering the relationship between mating system and the molecular evolution of the pheromone and receptor genes in Neurospora. Mol Biol Evol 29:3827–3842. 10.1093/molbev/mss193. [DOI] [PubMed] [Google Scholar]
- 51.Merlini L, Dudin O, Martin SG. 2013. Mate and fuse: how yeast cells do it. Open Biol 3:30008. 10.1098/rsob.130008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barr CM, Neiman M, Taylor DR. 2005. Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol 168:39–50. 10.1111/j.1469-8137.2005.01492.x. [DOI] [PubMed] [Google Scholar]
- 53.Smulian AG, Sesterhenn T, Tanaka R, Cushion MT. 2001. The ste3 pheromone receptor gene of Pneumocystis carinii is surrounded by a cluster of signal transduction genes. Genetics 157:991–1002. 10.1093/genetics/157.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jungbluth M, Mösch H-U, Taxis C. 2012. Acetate regulation of spore formation is under the control of the Ras/cyclic AMP/protein kinase A pathway and carbon dioxide in Saccharomyces cerevisiae. Eukaryot Cell 11:1021–1032. 10.1128/EC.05240-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cushion MT, Ashbaugh A, Hendrix K, Linke MJ, Tisdale N, Sayson SG, Porollo A. 2018. Gene expression of Pneumocystis murina after treatment with anidulafungin results in strong signals for sexual reproduction, cell wall integrity, and cell cycle arrest, indicating a requirement for ascus formation for proliferation. Antimicrob Agents Chemother 62:e02513-17. 10.1128/AAC.02513-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Le Gal S, Damiani C, Rouillé A, Grall A, Tréguer L, Virmaux M, Moalic E, Quinio D, Moal MC, Berthou C, Saliou P, Le Meur Y, Totet A, Nevez G. 2012. A cluster of Pneumocystis infections among renal transplant recipients: molecular evidence of colonized patients as potential infectious sources of Pneumocystis jirovecii. Clin Infect Dis 54:e62–e71. 10.1093/cid/cir996. [DOI] [PubMed] [Google Scholar]
- 57.Matsumoto Y, Frenkel JK, Aikawa M, Yoshida Y. 1989. Proliferation of Pneumocystis trophozoites in human lymph nodes and in nude mice lungs. J Protozool 36:33S–34S. 10.1111/j.1550-7408.1989.tb02682.x. [DOI] [PubMed] [Google Scholar]
- 58.Sukura A. 1995. Trophozoite-to-cyst ratio increases during recovery from Pneumocystis carinii pneumonia in rats. APMIS 103:300–306. 10.1111/j.1699-0463.1995.tb01110.x. [DOI] [PubMed] [Google Scholar]
- 59.Tamburrini E, Mencarini P, Visconti E, De Luca A, Zolfo M, Siracusano A, Ortona E, Murri R, Antinori A. 1996. Imbalance between Pneumocystis carinii cysts and trophozoites in bronchoalveolar lavage fluid from patients with pneumocystosis receiving prophylaxis. J Med Microbiol 45:146–148. 10.1099/00222615-45-2-146. [DOI] [PubMed] [Google Scholar]
- 60.Vargas SL, Ponce CA, Gallo M, Pérez F, Astorga JF, Bustamante R, Chabé M, Durand-Joly I, Iturra P, Miller RF, Aliouat EM, Dei-Cas E. 2013. Near-universal prevalence of Pneumocystis and associated increase in mucus in the lungs of infants with sudden unexpected death. Clin Infect Dis 56:171–179. 10.1093/cid/cis870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ponce CA, Bustamante RI, Gallo M, Vargas SL. 2014. Diagnosis of the primary infection by Pneumocystis in autopsy specimens from two infants using lung impression smears (touch preps). Med Mycol Case Rep 5:28–31. 10.1016/j.mmcr.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cushion MT, Linke MJ, Ashbaugh A, Sesterhenn T, Collins MS, Lynch K, Brubaker R, Walzer PD. 2010. Echinocandin treatment of Pneumocystis pneumonia in rodent models depletes cysts leaving trophic burdens that cannot transmit the infection. PLoS One 5:e8524. 10.1371/journal.pone.0008524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martinez A, Halliez MC, Aliouat EM, Chabé M, Standaert-Vitse A, Fréalle E, Gantois N, Pottier M, Pinon A, Dei-Cas E, Aliouat-Denis CM. 2013. Growth and airborne transmission of cell-sorted life cycle stages of Pneumocystis carinii. PLoS One 8:e79958. 10.1371/journal.pone.0079958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Keely SP, Stringer JR. 2009. Complexity of the MSG gene family of Pneumocystis carinii. BMC Genomics 10:367. 10.1186/1471-2164-10-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schmid-Siegert E, Richard S, Luraschi A, Mühlethaler K, Pagni M, Hauser PM. 2017. Mechanisms of surface antigenic variation in the human pathogenic fungus Pneumocystis jirovecii. mBio 8:e01470-17. 10.1128/mBio.01470-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ma L, Chen Z, Huang DW, Cissé OH, Rothenburger JL, Latinne A, Bishop L, Blair R, Brenchley JM, Chabé M, Deng X, Hirsch V, Keesler R, Kutty G, Liu Y, Margolis D, Morand S, Pahar B, Peng L, Van Rompay KKA, Song X, Song J, Sukura A, Thapar S, Wang H, Weissenbacher-Lang C, Xu J, Lee CH, Jardine C, Lempicki RA, Cushion MT, Cuomo CA, Kovacs JA. 2020. Diversity and complexity of the large surface protein family in the compacted genomes of multiple Pneumocystis species. mBio 11:e02878-19. 10.1128/mBio.02878-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schmid-Siegert E, Richard S, Luraschi A, Mühlethaler K, Pagni M, Hauser PM. 2020. Expression pattern of the Pneumocystis jirovecii major surface glycoprotein superfamily in patients with pneumonia. J Infect Dis 223:310–318. 10.1093/infdis/jiaa342. [DOI] [PubMed] [Google Scholar]
- 68.Barry JD, Ginger ML, Burton P, McCulloch R. 2003. Why are parasite contingency genes often associated with telomeres? Int J Parasitol 33:29–45. 10.1016/s0020-7519(02)00247-3. [DOI] [PubMed] [Google Scholar]
- 69.Wang B, Liang X, Gleason ML, Hsiang T, Zhang R, Sun G. 2020. A chromosome-scale assembly of the smallest Dothideomycete genome reveals a unique genome compaction mechanism in filamentous fungi. BMC Genomics 21:321. 10.1186/s12864-020-6732-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cushion MT. 2004. Comparative genomics of Pneumocystis carinii with other protists: implications for life style. J Eukaryot Microbiol 51:30–37. 10.1111/j.1550-7408.2004.tb00158.x. [DOI] [PubMed] [Google Scholar]
- 71.Wang B, Liang X, Gleason ML, Zhang R, Sun G. 2017. Genome sequence of the ectophytic fungus Ramichloridium luteum reveals unique evolutionary adaptations to plant surface niche. BMC Genomics 18:729. 10.1186/s12864-017-4118-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Inderbitzin P, Turgeon BG. 2015. Pondering mating: Pneumocystis jirovecii, the human lung pathogen, selfs without mating type switching, in contrast to its close relative Schizosaccharomyces pombe. mBio 6:e00583-15. 10.1128/mBio.00583-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cissé OH, Hauser PM. 2018. Genomics and evolution of Pneumocystis species. Infect Genet Evol 65:308–320. 10.1016/j.meegid.2018.08.015. [DOI] [PubMed] [Google Scholar]