SUMMARY
Fifty years ago, David Baltimore published a brief conceptual paper delineating the classification of viruses by the routes of genome expression. The six “Baltimore classes” of viruses, with a subsequently added 7th class, became the conceptual framework for the development of virology during the next five decades. During this time, it became clear that the Baltimore classes, with relatively minor additions, indeed cover the diversity of virus genome expression schemes that also define the replication cycles. Here, we examine the status of the Baltimore classes 50 years after their advent and explore their links with the global ecology and biology of the respective viruses. We discuss an extension of the Baltimore scheme and why many logically admissible expression-replication schemes do not appear to be realized in nature. Recent phylogenomic analyses allow tracing the complex connections between the Baltimore classes and the monophyletic realms of viruses. The five classes of RNA viruses and reverse-transcribing viruses share an origin, whereas both the single-stranded DNA viruses and double-stranded DNA (dsDNA) viruses evolved on multiple independent occasions. Most of the Baltimore classes of viruses probably emerged during the earliest era of life evolution, at the stage of the primordial pool of diverse replicators, and before the advent of modern-like cells with large dsDNA genomes. The Baltimore classes remain an integral part of the conceptual foundation of biology, providing the essential structure for the logical space of information transfer processes, which is nontrivially connected with the routes of evolution of viruses and other replicators.
KEYWORDS: virus classification, virus evolution, virus realms, virus taxonomy
INTRODUCTION
In September 1971, nearly 50 years prior to this writing, David Baltimore published in Bacteriology Reviews (now Microbiology and Molecular Biology Reviews) a short paper entitled “Expression of animal virus genomes” (1) (the subject has been further discussed in Baltimore’s review articles that appeared shortly thereafter [2, 3]). This article (hereafter B71) does not report any particular discovery or theory as such. Instead, it is a simple classification of viruses (those that infect animals, according to the title, but effectively all viruses) by the routes of information transmission from the nucleic acid that is encapsidated in the virion (hereafter genome) to the mRNA, from which virus proteins are translated.
Objectively measuring the impact of a particular publication on the further development of science is an unsolved and, perhaps, unsolvable problem. Still, however imperfect and difficult to interpret, the citation history is a useful source of information. By that measure, B71 fares well, even if not overwhelmingly, having been cited a total of 988 times (Google Scholar, as of 25 June 2021). The citation dynamics of B71 (as easily traced using Google Scholar) is remarkable, with active initial citation followed by a period of neglect, with virtually no citations around 1990, and then, by resurgence in the 2010s, a striking rediscovery of a temporarily forsaken classic.
In this article commemorating the 50th anniversary of B71, we argue that, despite the relatively modest citation record and notwithstanding (or thanks to?) the simplicity of Baltimore’s scheme, this is one of the most important papers in virology ever published. It also has more general implications for the study of the evolution of life. Indeed, the importance of classification in the evolution of any science is hard to overestimate. Typically, the mature age of a scientific field is marked by the appearance of a powerful classification scheme (4–7). Suffice it to mention animal and plant systematics created by Linnaeus, Fedorov’s classification of crystals, Mendeleev’s periodic system of chemical elements, and the classification of elementary particles underlying the standard model of modern particle physics. Each of these classifications had demonstrable predictive power, and each has become the foundational framework of the respective field of science. The Baltimore classification plays an analogous foundational role in virology, even if the scope of the discipline may not be comparable to the whole of chemistry or particle physics. Moreover, viewed from a higher plane of abstraction, the Baltimore system classifies not only viruses but also the routes of biological information transfer and, in that capacity, is central to all of biology. Indeed, although not usually discussed in these terms, the Baltimore scheme, in addition to viruses, formally accommodates all cells, which, with their uniform route of information transmission, fit into one of the Baltimore classes.
The publication of B71 came in the wake of the ground-breaking discovery of the retrovirus reverse transcriptase (RT) that was made by Baltimore (8) and, simultaneously and independently, by Temin and Mizutani (9) (and rewarded by the Nobel Prize in Physiology or Medicine to Baltimore and Temin in 1975). The discovery of the RT triggered conceptual discussion of the routes of transfer of biological information and, in particular, the explicit formulation of the “Central Dogma of Molecular Biology” by Crick (10), following the conceptual framework laid out in Crick’s earlier seminal paper on translation (10, 11) (Fig. 1). In the central dogma, Crick amended the canonical scheme of information transfer from DNA to RNA to protein by allowing, in special situations such as virus replication, the back transfer from RNA to DNA, RNA replication without involvement of DNA, and even direct translation of DNA that, however, does not seem to occur in cells. By contrast, direct information transfer from protein to nucleic acid (reverse translation) was explicitly and strictly prohibited. The impossibility of the direct information flow from protein to nucleic acids is the central dogma, which, now as then, stands as a fundamental biological exclusion principle. A generalization of this principle is that all routes of information transfer between the digital information carriers (DNA and RNA) are allowed and actually occur in nature, whereas the information transfer from a digital to an analog device, that is, from nucleic acids to protein, is strictly unidirectional (12). The classification presented in B71 is an embodiment of this principle that is based on the preceding discoveries in virology, namely, virus RNA genomes and reverse transcription.
FIG 1.
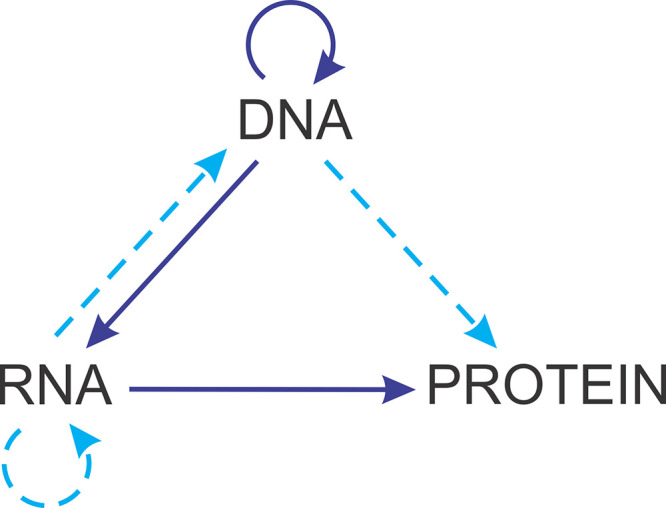
The central dogma of molecular biology. The figure is redrawn from Crick’s 1970 article. The solid arrows represent the mainstream routes of information flow, and the dashed arrows show (putative) “special routes” after Crick. Adapted from reference 10 with permission of Springer Nature.
THE BALTIMORE CLASSES OF VIRUSES
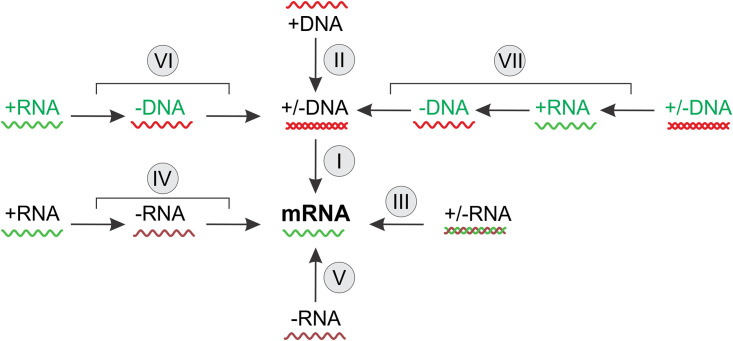
The system described in B71 consists of six classes of viruses (hereafter Baltimore classes, BCs) that are distinguished by their distinct routes of information transfer from the nucleic acid that is incorporated into virions (virus genome). These routes, evidently, reflect the chemical nature and polarity of the genome (Fig. 2).
FIG 2.
The amended scheme of the seven Baltimore classes of viruses. Shown is the transfer of genetic information between genomic nucleic acids encapsidated into virions (genomes, for short) and mRNA. This transfer is enabled by enzymatic reactions; in the case of the RNA viruses, with the exception of hepatitis delta and related viruses, these reactions are catalyzed by the virus-encoded RNA-dependent RNA polymerases, which are encapsidated into virions of viruses of BCIII and BCV, but not BCIV; reverse-transcription of RNA to DNA is catalyzed by virus-encoded reverse transcriptases, which may or may not be encapsidated into virions; replication of DNA genomes is catalyzed by virus-encoded or host-encoded DNA polymerases, which are not encapsidated. The roman numerals denote the BCs. BCVII was added to the scheme. Adapted from reference 1 with permission.
I. BCI: double-stranded (ds) DNA viruses. These are the viruses that encapsidate dsDNA and use the classical route of information transmission, the same as in all cells.
II. BCII: single-stranded (ss) DNA viruses that encapsidate ssDNA, which is then replicated and expressed via a dsDNA intermediate; Baltimore noted that all ssDNA viruses known at the time incorporated into virions either a ssDNA of the same polarity as the mRNA ([+]DNA) or ssDNA molecules of both polarities (in separate particles), but not exclusively (–)DNA.
III. BCIII: dsRNA viruses that package a dsRNA genome that has to be transcribed (transcription here being defined as mRNA synthesis, irrespective of the nature of the template) to produce the mRNA.
IV. BCIV: positive-sense (+)RNA viruses that pack into virions a ssRNA of the same polarity as the mRNA for the synthesis of virus proteins such that the genome RNA can be translated directly.
V. BCV: negative-sense (–)RNA viruses that package an RNA that is complementary to the mRNA and is transcribed to produce the latter.
VI. BCVI: reverse-transcribing RNA viruses that package a positive-sense RNA that is replicated via a DNA intermediate.
Shortly after the publication of B71, a group of viruses has been characterized, hepatitis B viruses (HBV) later named hepadnaviruses (13, 14), that qualified as BCVII, viruses that package a dsDNA genome (although one of the strands is typically incomplete) that use RT to replicate via an RNA intermediate.
BCVI and BCVII are of special interest because their status as distinct classes emphasizes that the classes reflect the actual route of information transmission and not the structure of the genome directly. Hence, viruses that encapsidate dsDNA and (+)RNA but use reverse transcription are not included in BCI and BCIV, respectively, but merit the creation of separate BCs.
BCII is another point in the same case, where the route of information transfer is a more fundamental feature of a virus than the genome structure. Small ssDNA viruses that appear to package exclusively (–)DNA have been discovered, namely, the family Anelloviridae (15, 16), as well as certain members of the family Parvoviridae, such as mink enteritis virus (17). Formally, viruses with a (–)DNA genome would qualify as a distinct BC, potentially BCVIII. However, many ssDNA viruses, such as circoviruses, geminiviruses, genomoviruses, and smacoviruses, possess ambisense genomes, in which some genes are located on one strand, and other genes are located on the complementary strand (18, 19). Furthermore, to the best of the current knowledge, viral ssDNA is never transcribed directly. Rather, the template for transcription producing the mRNA is always a dsDNA intermediate. Given the wide spread of ambisense ssDNA and the ubiquity of the dsDNA intermediate, which makes the route of information transfer effectively uniform among the ssDNA viruses, it appears that they all should be assigned to a single BC (BCII).
Some viruses span the boundaries between the BCs and formally can be assigned to two BCs. Thus, different members of the family Pleolipoviridae, despite having conserved genome sequences, encapsidate either ssDNA or dsDNA genomes, with some genomes being a patchwork of single-stranded and double-stranded regions (20–22). Similarly, the genomes of the ssDNA viruses in the family Bacilladnaviridae contain short dsDNA regions (23–25). Thus, technically, both pleolipoviruses and bacilladnaviruses can also be considered representatives of two different BCs, namely, BCI and BCII.
An even more notable twist to the Baltimore classification is the discovery of ambisense RNA viruses (arenaviruses and some other members of the order Bunyavirales) that possess a genome partitioned into two or three segments of RNA, respectively, one of which combines regions of positive and negative polarity (26–28). Furthermore, a group of fungal viruses, dubbed ambiviruses with nonsegmented bicistronic genomes where the RNA-dependent RNA polymerase (RdRP) is encoded on the (+) strand and the other ORF is located on the (–) strand, has been recently discovered (29). It can be argued that these viruses would formally qualify as a new BC, or else could be considered as belonging both in BCIV and BCV, although, effectively they are distinct varieties of (–)RNA and (+)RNA viruses, respectively, as discussed below.
On the whole, despite many notable discoveries of variations on the main routes of information flow in diverse viruses, after the addition of BCVII, the strikingly simple Baltimore classification has remained stable for nearly half a century.
MOLECULAR AND ECOLOGICAL CORRELATES OF THE BALTIMORE CLASSES OF VIRUSES
The ultimate value of any classification can be measured by the fraction of the variance in the classified data set it accounts for. A perfect classification (which hardly can exist in reality) would partition the data in accord with all the salient features. The BCs fare well in this regard. Indeed, the inclusion of a virus in a particular BC defines, in large part, its replication cycle, genome size, gene content, the presence or absence in the virion of key functional systems, such as the replication and transcription machineries, and other important features (Table 1).
TABLE 1.
The Baltimore classes of viruses and their key molecular and biological features
| BC | Virion nucleic acid | Genome structure | Genome size, kba | Genome segmentation | Packaging of components of replication/transcription machineries | Host range |
|---|---|---|---|---|---|---|
| I | dsDNA | Mostly linear | 5–2,500 | None | No replication, transcription in some | Bacteria and archaea, protists, animals; none in plants, rare in fungi |
| II | ssDNA | Mostly circular | 1.7–25 | Mostly nonsegmented | None | Bacteria, rare in archaea; most eukaryotes |
| III | dsRNA | linear | 4–30 | Mostly segmented | All packaged | Protists, animals, plants; one family in bacteria, none in archaea |
| IV | (+)RNA | linear | 3.5–40 | Mostly nonsegmented but many segmented | None | All eukaryotes; one class with six families in bacteria, none in archaea |
| V | (–)RNA | Mostly linear | 1.7–20 | Roughly half segmented | Nearly all packaged | Animals, plants, rare in fungi, protists; none in bacteria or archaea |
| VI | (+)RNA, RT | Linear | 5–13 | Nonsegmented | All packaged | All eukaryotes; none in bacteria or archaea |
| VII | dsDNA, RT | Circular | 3–10 | Nonsegmented | Mostly packaged | Animals, plants; unknown in protists; none in bacteria or archaea |
The information on genome size ranges is from the 10th ICTV report (109).
General patterns, some of which amount to strict rules, follow from the BC classification under biologically interpretable (albeit to different degrees) constraints. Among the most remarkable patterns associated with the BCs is the genome size distribution. All the viruses in BCII to BCVII have tiny genomes compared to the organismal genomes and replicate with high error rates. The record size in all these BCs is reached by the 30- to 40-kb coronavirus genomes (30), which, unlike all other known viruses in these BCs, encode proofreading enzymes that boost replication fidelity (31–33). Conceivably, the size of the genome in these BCs is limited by the relative chemical instability of ssRNA and structural constraints on ssDNA (potential for extensive secondary structure formation) and dsRNA (structural rigidity). The expansive BCI stands apart from the rest of the BCs in terms of the span of the genome size, which ranges between about 4 kb and 2.5 Mb, and the organization of the replication process (34). The latter can vary, arguably because the genomes of dsDNA viruses share the same physical organization with the host genomes and thus have the “choice” of exploiting cellular replication machineries or encoding their own. Thus, it appears that dsDNA that is replicated and expressed according to the classical scheme of genetic information flow is the only type of nucleic acid that provides for the maintenance and efficient replication of genomes in excess of about 50 kb.
RNA replication and reverse transcription of RNA into DNA are generally shunned by cells, with the exception of some highly specialized situations, such as telomere synthesis and RNA interference in eukaryotes. Therefore, all viruses in BCIII to BCVII that rely on these processes invariably encode an RNA-dependent RNA polymerase (RdRP) or RT, with the exception of satellite viruses that depend on other viruses for replication. Moreover, all these viruses, with the notable exceptions of BCIV and certain members of BCVII (caulimoviruses), incorporate the RdRP or RT and the rest of the enzymatic machinery required for the expression of the mRNA into the virions because these enzymes are required to produce the first viral mRNA in the infected cells. Conversely, no BCIV virus carries any enzymatic machinery in the virion because the (+)RNA genome is directly translated upon infection. The ssDNA viruses in BCII generally can afford to rely on the host enzymatic machinery for genome replication and transcription. Indeed, with the sole exception of members of the Bidnaviridae family, which encode their own protein-primed DNA polymerase (35, 36), and members of the Anelloviridae family, which encode proteins without detectable homologs, the small ssDNA viruses in BCII encode a single replicative enzyme, the endonuclease involved in the initiation of rolling circle/hairpin DNA replication (37, 38). This endonuclease has to be encoded in the virus genome because rolling circle replication is not among the normal cellular nucleic acid synthesis processes.
Although the dsDNA viruses of BCI also have the luxury of relying on the host replication and transcription systems, they encode a broad range of the repertoires of proteins involved in these processes (39). Having broken through the genome size limits affecting the rest of the BCs, many of these viruses have acquired their own replication and transcription machineries, in some cases nearly complete ones that, apparently, provide for efficient, partly autonomous genome expression and replication.
Some other correlates of the BCs are harder to interpret. For example, all RNA-containing reverse-transcribing viruses comprising BCVI contain the RT within the virions where it reverse transcribes the virion (+)RNA, although it is unclear what would preclude direct translation of this RNA as it occurs in the BCIV viruses. Baltimore noted when introducing the BCs (1) that all dsRNA viruses known at the time had segmented genomes, although it was unclear why there could not be dsRNA viruses with nonsegmented genomes. And, indeed, several years after the publication of B71, the totiviruses, a widespread group of viruses with small (about 4 kb) nonsegmented dsRNA genomes, have been discovered (40, 41). The trend remains, however, that most of the viruses in BCIII have segmented genomes, and there are no large (by RNA virus standards) nonsegmented dsRNA genomes. This trend might have to do with the rigidity of long dsRNA molecules hampering their utilization as the templates for RNA synthesis and/or packaging into virions.
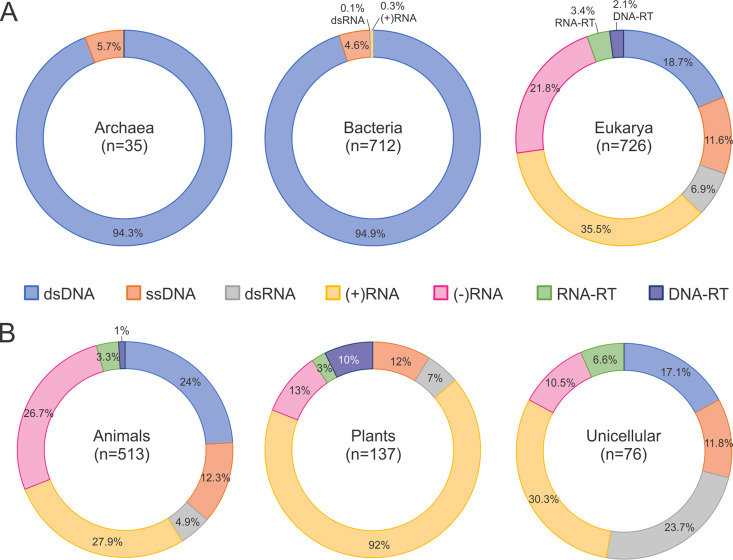
The different BCs show a nontrivial distribution of host ranges (Table 1 and Fig. 3) (42). The characterized portion of the prokaryotic virosphere is dominated by dsDNA viruses (BCI), with a substantial minority of ssDNA viruses (BCII), a low representation of (+)RNA viruses (BCIV), a single small family of dsRNA viruses (BCIII), and no known reverse-transcribing viruses (BCVI to BCVII). In contrast, well-characterized eukaryotic viromes are dominated by (+)RNA viruses (BCIV) and in some major taxa, for example, fungi, by dsRNA viruses (BCIII). The reverse-transcribing viruses of BCVI are also widespread and highly abundant, especially in animals. The dsDNA viruses of BCI, although lagging behind the (+)RNA viruses in diversity, are a major presence in various unicellular eukaryotes as well as in animals, but not in plants (Fig. 3). There are no compelling explanations for most of these notable patterns of host range across the BCs. Some biological considerations are apparent. For instance, the absence of BCI in plants can be attributed to the inability of large dsDNA molecules to spread between cells by passing through plasmodesmata (43). The dominance of BCI and, to a lesser extent, BCII in prokaryotes that contrasts the high prevalence of RNA viruses (primarily BCIV) in eukaryotes is a more general pattern, and the biological underpinnings are harder to decipher. It seems plausible that in prokaryotes, where access to the replication and transcriptional machineries is relatively unencumbered for a virus, the more efficient and versatile DNA viruses outcompete RNA viruses. In contrast, in eukaryotic cells, the nucleus presents a barrier that required special adaptations for large DNA viruses to cross. The DNA viruses either have to adapt to penetrate the nuclear envelope and replicate in the nucleus or have to acquire replication and transcription apparatus to replicate within “virus factories” in the cytosol (44, 45). Conversely, the endomembranes of eukaryotic cells seem to provide a fertile niche for the replication of RNA viruses (46, 47). With regard to RNA viruses, one might be compelled to ask why do (–)RNA and dsRNA viruses exist at all and are widespread in eukaryotes, considering the ultimate simplicity of the information transmission route of (+)RNA viruses and their obvious evolutionary success? A potential explanation linking the information transmission routes in these BCs with the host biology could be that (–)RNA and dsRNA viruses hide their replicating genomes inside capsids or nucleocapsids from the powerful systems of host innate immunity that operate in the cytoplasm of eukaryotic cells, in particular RNA interference (48, 49), the interferon response (50, 51), and other dsRNA-triggered defense pathways (52).
FIG 3.
The host range distribution in the seven Baltimore classes of viruses. (A) Distribution of the BCs in the three cellular domains. The panel illustrates the dominance of dsDNA viruses in Bacteria and Archaea, which contrasts the dominance of viruses with RNA genomes in Eukarya. (B) Distribution of the BCs in eukaryotes. Each circle represents the breakdown of the virus genera (according to the ICTV taxonomy release number 35 [https://talk.ictvonline.org/files/master-species-lists/m/msl/9601]) associated with the indicated group of hosts. The number of virus genera (n) is indicated inside each circle. The BCs are denoted by the virion nucleic acid and are color coded.
Admittedly, the above are general, vague arguments, and attaining a deep understanding of the aspects of the virus-host interactions that underlie the strikingly nonuniform distribution of the BCs across the broad range of hosts requires further, extensive experimental studies. Furthermore, the possibility remains that the current host range pattern is substantially biased by different rates of discovery of viruses across the BCs; for example, significant undercounting of both ssDNA viruses of BCII (53) and (+)RNA viruses of BCIV (54, 55) in various environments has been reported.
All the caveats notwithstanding, the key message from the examination of the features of viruses that correlate with the BCs, be it molecular mechanisms or (perhaps to a lesser extent) host range, is that the assignment of a virus to a BC is highly predictive. Furthermore, the major differences between the BCs, such as the contrasting host ranges, pose fundamental challenges for further research. These are hallmarks of a productive, working classification system.
EXTENSION OF THE BALTIMORE CLASSIFICATION: THE SPACE OF LOGICAL POSSIBILITIES AND WHY IT REMAINS UNFILLED
Classification systems that capture patterns existing in nature have the remarkable capacity to identify missing elements and predict their properties. Mendeleev’s prediction of the chemical elements that were missing in his table was promptly validated and became a glorious triumph that established the periodic table as the foundational framework of chemistry (56). The prediction and discovery of a number of elementary particles, including the famous Higgs boson, was equally momentous for high energy physics (57). Could the Baltimore classification do anything similar for virology, or even beyond, by predicting the discovery of new routes of genetic information transmission or explaining why some of the logically possible routes might not be realized in any genetic systems?
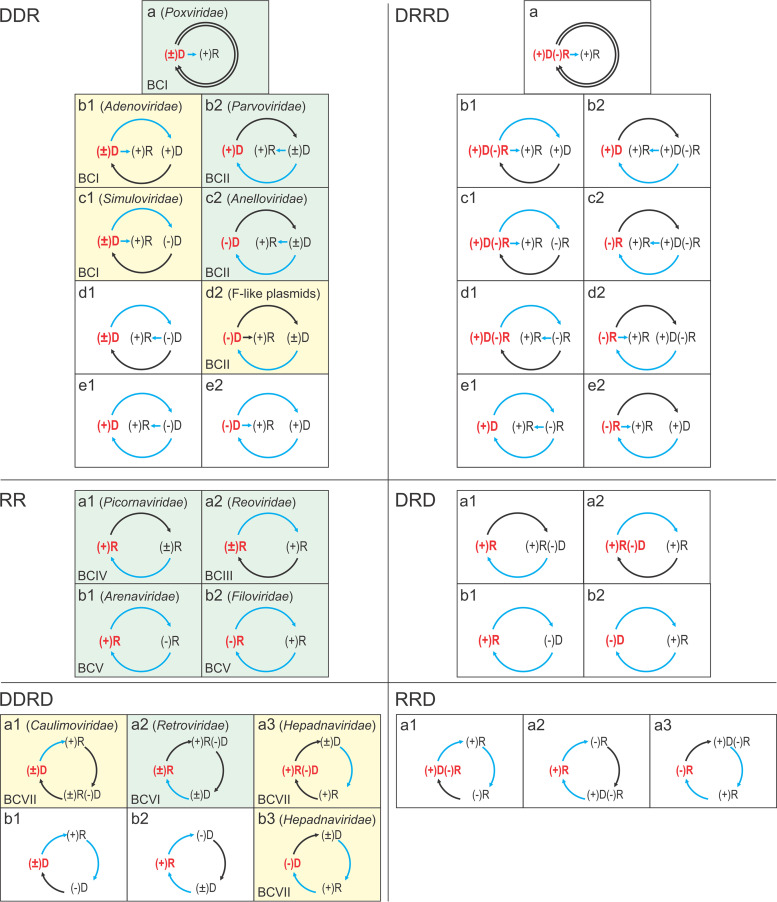
In B71, Baltimore only makes a general comment that “Viruses with similar transcriptional systems could have different replicational systems leading to the necessity to extend the class designations” (1). Three years after the publication of B71, one of us (V.I.A.), building upon this suggestion, modified and expanded the BC system (58). This classification takes the form of a complete hierarchical table of the routes of information transmission between nucleic acids that, unlike Baltimore’s original scheme that focused on mRNA formation (Fig. 2), takes into account both expression and replication of virus genomes (Fig. 4). The analysis started with the definition of genetic elements, of which there are four: (+)RNA, (–)RNA, (+)DNA, and (–)DNA. As each element can serve as the template for the synthesis of two types of complementary molecules, there are eight elementary acts of synthesis. Then, there are eight genetic units, namely, the four genetic elements and four types of double-stranded molecules made of those, including DNA-RNA hybrids. Considering the simplest cycles, in which one of the eight genetic units involved is the genome (the nucleic acid incorporated in the virion as far as viruses are concerned), there are 35 distinct cyclic graphs (evidently, replication presupposes a cycle) that represent the routes of information transfer (Fig. 4). The classification becomes naturally hierarchical, with six types defined by the genetic units that comprise the vertices of the graph [DDR, RR, etc.; here, DDR means three acts of synthesis, namely, synthesis of (+)DNA and (−)DNA on the respective complementary DNA templates and synthesis of (+)RNA on the (−)DNA template, whereas RR means two acts of synthesis, namely, synthesis of (+)RNA and (−)RNA on the respective complementary RNA templates], 17 superclasses, each with the same graph topology but different distributions of the genetic units over the vertices, and the 35 classes that correspond to unique information transmission routes (not to be confused with the BCs) (Fig. 4). By the design of this system, the classes within each superclass differ only by the nature of the genome, that is, the encapsidated nucleic acid (Fig. 4). Now, as in 1974, this table is only sparsely occupied by known genetic systems, which appears unexpected given that we are unaware of any physicochemical limitations on genetic units and acts of synthesis, and so one would suspect that all simple routes of genetic information transmission would have been explored during the nearly 4 billion years of the evolution of life. Thus, if exclusion principles exist that preclude occupation of some of the classes, these have to be uncovered (58).
FIG 4.
The extended, hierarchical version of virus classification by information transmission routes. Types DDR, DRRD, RR, DRD, DDRD, and RRD represent theoretical paths of genetic information transfer. The elementary acts of synthesis of DNA or RNA on a DNA template are denoted, respectively, as DD or DR, and the elementary acts of synthesis of DNA or RNA on an RNA template are denoted as RD and RR. Each type is further divided into superclasses, denoted with Latin letters, and the superclasses are divided into classes denoted with Arabic numerals and shown in separate boxes. In each class, the leftmost genetic unit, shown in red, represents the virion nucleic acid (genome). The occupied classes are shown by colored background, green for those known at the time of the original publication (58) and yellow for those discovered subsequently (see the text for details). The blue arrows show synthesis of single-stranded molecules on double-stranded templates (multiplicational acts of synthesis), and black arrows show synthesis of dsDNA or dsRNA on single-stranded templates (nonmultiplicational acts of synthesis). Abbreviations: D, DNA; R, RNA. For each occupied class, the corresponding BC is indicated in the bottom left corner, whereas examples of virus families that use the corresponding routes of information transfer are shown in parentheses. The examples were chosen arbitrarily. Class DDR-d2 is currently not occupied by known viruses, but the corresponding flow of information has been described for conjugative F-like plasmids. Reproduced from reference 58 with permission from Elsevier.
At the time of the publication of the table, three types, six superclasses, and eight classes were assigned to viruses (and cellular organisms, which all fit in one class, DDR-a) (58). The occupied cells in the table include all four simple, two-edge classes in type RR, that is, all RNA viruses that comprise BCIII to BCV, two classes in type DDR (BCI and BCII), and one class in type DDRD (BCVI) (Fig. 4). One more class, DDR-c2 with the virion negative-sense ssDNA, could be considered “conditionally” represented because some ssDNA viruses, specifically parvoviruses, have been known to encapsidate positive-sense and negative-sense ssDNA molecules in different particles, but none have been discovered at the time with negative-sense virion ssDNA only.
So how did the occupation of the extended classification table change over the next 47 years? A group of parvoviruses in the genus Protoparvovirus that package exclusively (–)DNA has been discovered (17, 59). Furthermore, all known members of the family Anelloviridae also have a (–)DNA genome (15, 60), so that class DDR-c2 is definitively occupied. The classes DDR-b1 and DDR-c1 are both represented by ambisense dsDNA viruses, such as corticoviruses, simuloviruses, and pleolipoviruses, that replicate by the rolling circle mechanism, a process that produces an ssDNA intermediate (21, 61, 62). Adenoviruses that have linear dsDNA genomes also squarely fit in because they replicate via an ssDNA intermediate, albeit by an entirely different mechanism that involves strand displacement by the virus-encoded protein-primed DNA polymerase (63, 64). Notably, both strands of the adenovirus genome can be individually displaced from different dsDNA molecules (63, 64), satisfying the requirements for both DDR-b1 and DDR-c1.
The class DDR-d2, which envisions direct transcription from an ssDNA template, has also been discovered in nature, albeit in plasmids rather than in viruses (65). The leading region of the ssDNA of F-like plasmids, the first to be transferred into recipient cells during conjugation, contains a promoter that functions exclusively in the single-stranded form for both transcription of the mRNA for several early proteins and initiation of the plasmid DNA replication, that is, synthesis of the second DNA strand (65, 66). Although such transcription of ssDNA, to our knowledge, has not been reported in viruses, it is worth noting that the experiment leading to the discovery of the promoter was performed with ssDNA bacteriophages, whereby the plasmid DNA was cloned into the phage genome (65), showing the principal possibility of direct transcription from a phage ssDNA. Regardless, the ssDNA entering the cell through conjugation is conceptually equivalent to the DNA encapsidated into virions and is delivered into the host cells upon virus infection. Alternatively, if the dsDNA form of the conjugative plasmid is considered genome, the case would qualify as filling class DDR-d1.
The class DDRD-a1, which corresponds to BCVII, has been claimed through the discovery of reverse-transcribing viruses containing dsDNA in the virion, hepadnaviruses and caulimoviruses (67). Notably, the hepadnavirus genomic DNA is only partially double stranded (68), so it technically could be considered to belong to both BCI and BCII, and, respectively, to both DDRD-a1 and DDRD-b3. So, these classes can be considered occupied. Furthermore, some hepadnaviruses package into virions (+)RNA-(–)DNA hybrids (69, 70), thus filling the class DDRD-a3. Therefore, in the updated information route classification table, 14 (the 6 original ones and the 8 filled here based on the current knowledge) of the 35 classes are now claimed (Fig. 4).
Why are the remaining 21 cells of the table empty? At the time of the publication of the extended classification in 1974, it seemed likely that some of these would be occupied as a result of the continuing study of the diversity of viruses that was then in its infancy, although the possibility of exclusion has been considered (58). The empty classes, which do not include any acts of synthesis not known to exist in nature, have been explicitly predicted to eventually fill with new viruses (58). These classes are DDRD-a1, DDRD-a3, DDR-b1, and DDR-c1. Indeed, viruses occupying each of these classes have been discovered, fully vindicating the prediction. The first two correspond to reverse-transcribing viruses with dsDNA genomes (BCVII), whereas DDR-b1 and DDR-c1 are both represented by dsDNA viruses (BCI) with ambisense genomes replicated by mechanisms involving single-stranded intermediates (Fig. 4).
Considering the enormous expansion of the collection of sequenced virus genomes thanks to the advances of genomics and metagenomics in the 21st century, the discovery of viruses that would fill the remaining empty classes appears now increasingly unlikely. At the very least, if any are discovered, these should be extremely rare in the biosphere. No hard exclusion principles of the type of the central dogma are likely to be at play, given that all genetic units are known to exist in nature. In part, the preponderance of unoccupied classes can be attributed to avoidance of one genetic unit; evolution seems to disfavor RNA-DNA hybrids as a genome and, to a lesser extent, as an intermediate. Conceivably, conversion of this type of molecule into dsDNA, the nucleic acid form that appears to be best suited for genome replication, thanks, partly, to its physical properties including stability and regular structure and, partly, to the opportunity to utilize the cellular replication machinery, was advantageous in the course of evolution and was fixed independently on multiple occasions.
The apparent nonexistence of the DRD-a superclass merits a special comment. In principle, its two classes correspond to the most economical mode of replication for reverse-transcribing viruses that would involve only two genetic units. The class DRD-a1 seems particularly plausible given that packaging of (+)RNA is so widespread. However, all the numerous known RNA-containing reverse-transcribing viruses belong to class DDRD-a2, that is, replicate via a dsDNA intermediate, a third genetic unit (Fig. 4) that integrates into the host genome in an essential stage of the virus replication cycle. Many viruses in this class, such as metaviruses (also known as Ty3/gypsy-like retrotransposons) (71), pseudoviruses (Ty1/copia-like retrotransposons) (72), and belpaoviruses (Bel/Pao-like retrotransposons) (67), lead a dual lifestyle as viruses and transposons. Switching between these lifestyles, known as lysis-lysogeny switch in bacteriophages (73, 74), is also typical of numerous viruses in classes DDR-a and DDR-b2 (dsDNA and [+]DNA viruses, respectively). This dual lifestyle is a bet-hedging strategy that allows the virus to alternate between horizontal spread among hosts and vertical propagation with the host, depending on the conditions (75, 76), which could be another reason why it is advantageous for viruses to include dsDNA in their replication cycle. Thus, routes of information transfer deviating from minimal complexity apparently can evolve under the pressure for optimization of the evolutionary strategy.
The discovery of the ambisense RNA genome organization in many RNA viruses of the order Bunyavirales, in which one or two of the genome segments contain two genes, one in the negative sense and the other one in the positive sense strand, formally violates an assumption implicit in both the original BC scheme and in the extended table, namely, that single-stranded genomes have the same polarity throughout. The same applies to the numerous ambisense ssDNA viruses in which different genes are transcribed from either the positive or the negative strand (77–79). Dismissing this assumption, the ambisense RNA viruses can be placed simultaneously in BCIV and BCV or in the RR-b1 and RR-b2 classes of the extended table. Ambisense ssDNA viruses belong to BCII, as discussed above, but in the extended table could be assigned to both DDR-b2 and DDR-c2. Alternatively, two new BCs and new types in the extended table could be created for the ambisense viruses. Regardless of the formal classification, the ambisense RNA viruses are associated with the (–)RNA viruses in terms of both molecular features and evolutionary origins. In some of the ambisense bunyaviruses, the RdRP is encoded in a fully negative-sense genome segment and, accordingly, is packaged in the virions. In other viruses of the Arenaviridae family, both genomic segments are ambisense, but the RdRP is encoded in the negative-sense portion of the large segment and is a virion protein as well (80). In either case, the proteins encoded in the packaged positive-sense strand are never translated directly from the virion RNA but rather from a subgenomic mRNA that is transcribed from a negative-sense intermediate (80). In agreement with these features, the ambisense bunyaviruses clearly evolved from (–)RNA viruses (see next section). With regard to the ssDNA viruses, the ambisense organization of the genome is of a lesser consequence because mRNA transcription occurs on a dsDNA intermediate in which case there is no substantial distinction between genes located on the positive-sense or the negative-sense strand.
In summary, extending the Baltimore classification of viruses into a complete classification of information transfer routes, unlike the original scheme, allows one to ask questions that are inherent to those classifications that represent the entire space of logical possibilities for the analyzed category of objects. In this sense, the original Baltimore scheme is analogous to the Linnean taxonomy, whereas the extended table more closely resembles the periodic system of chemical elements. The main question to be asked is why is a particular subset of the classes in this table occupied, whereas the rest are empty? Although attempts to address this question might not lead to major biological generalizations, they do suggest “weak” exclusion principles that merit further investigation.
THE EVOLUTIONARY STATUS OF THE BALTIMORE CLASSES: UNIFICATION VERSUS POLYPHYLY
The Baltimore classes were delineated based on the structure of the encapsidated nucleic acid and the route of information transfer from the genome to the mRNA. By design, the BCs do not expressly reflect evolutionary relationships among viruses, which were intractable in 1971. Nevertheless, a tantalizing hypothesis that might have not been explicitly stated in print, but that seemed to have been permeating the thinking on virus evolution for at least 3 decades after the publication of B71, is that the BCs were major monophyletic groups of viruses. This conjecture seemed to be implicit, for example, in the taxonomies of viruses that have been adopted by the International Committee on Taxonomy of Viruses (ICTV) or NCBI versions for decades until the recent overhaul (81), where the BCs were adopted as informal top rank taxa or even formally proposed in that capacity (82, 83).
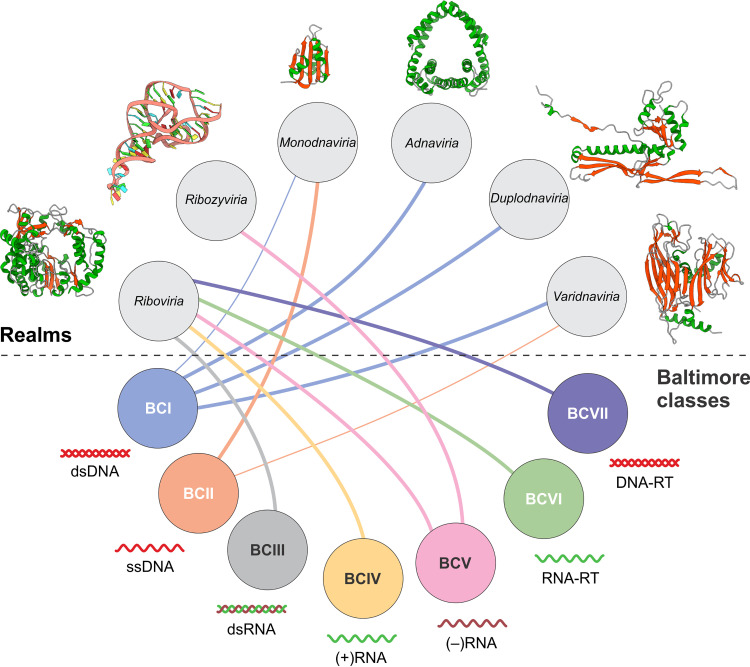
The advances of genomic and especially metagenomic sequencing in the 21st century rendered the study of virus evolution a realistic and productive enterprise so that 50 years after B71, the contours of the evolutionary landscape of the entire virosphere are becoming discernible. The recent synthesis of the evolutionary relationship among the major groups of viruses (42) reveals a highly complex relationship between the BCs and virus evolution, far removed from the simplistic identification of the BCs with the largest monophyletic taxa of viruses (Fig. 5). The comprehensive, hierarchical taxonomy of viruses reflecting the evolutionary synthesis and formally adopted by the ICTV includes four major realms (the top rank in virus taxonomy), each including an enormous diversity of viruses, and two subsequently added, much smaller realms (Fig. 5) (81). The viruses within the realms are considered monophyletic or at least connected through shared components (see below).
FIG 5.
The Baltimore classes and monophyletic realms of viruses. The connections between the BCs and the virus realms are shown by colored edges. Thick lines denote major associations, and thin lines denote exceptional cases (see the text for details). For each virus realm, a ribbon diagram of a hallmark structure is shown as follows: Riboviria, poliovirus RdRP (1RA7); Ribozyviria, HDV ribozyme (4PRF); Monodnaviria, porcine circovirus 2 rolling circle replication initiation endonuclease (5XOR); Adnaviria, major capsid protein of Sulfolobus rod-shaped virus 2 (3J9X); Duplodnaviria, major capsid protein of bacteriophage HK97 (1OHG); Varidnaviria, double jelly roll major capsid protein of bacteriophage PRD1 (1HX6). The protein structures are colored by secondary structure: α-helices, green; β-strands, red.
Although the concept of monophyly as applied to viruses has been discussed in detail previously (42, 84), a brief comment is due here. There is not a single universally conserved gene among all viruses, so viruses as a whole are clearly polyphyletic. However, a small set of virus hallmark genes (about 20 altogether) encoding key proteins involved in virus replication and virion formation link viruses within the realms (42, 84, 85). Thus, all RNA viruses, along with all reverse-transcribing viruses, are connected through the hallmark gene encoding the homologous RdRP or RT, the only gene all these viruses share, and are accordingly unified in the realm Riboviria. This is considered evidence of the monophyly of the viruses themselves under the explicit or implicit scenario where they all evolved from a simple genetic element that encoded an ancestral RNA-templated polymerase (86). Nevertheless, it has to be kept in mind that, when addressing the origin and deep evolutionary relationships among viruses, we can only trace the evolutionary history of one or several hallmark genes. This is conceptually not different from using universal genes, such as rRNA, for the reconstruction of organismal evolution, with the difference that among viruses, even the most highly conserved hallmark genes do not cover the entire diversity of the virosphere but only the diversity within a realm.
With this understanding, we can trace the relationships between the BCs and the now established realms of viruses, in other words, the evolutionary status of the BCs (Fig. 5) (42). With the sole exception of hepatitis delta virus (HDV) and its relatives, all viruses with RNA genomes, together with the reverse-transcribing viruses with dsDNA genomes (that is, BCIII to BCVII or the RR and DDRD types in the extended table), share a common ancestry and form the realm Riboviria. However, within this assemblage that is monophyletic as a whole, the relationships among the BCs are nontrivial. Among the viruses of BCIII to BCV (type RR) that comprise the kingdom Orthornavira in the current taxonomy, (+)RNA viruses (BCIV, RR-a1) are the “primary” group in terms of the greatest diversity but, more importantly, because they are paraphyletic with respect to dsRNA viruses (BCIII, RR-a2) and (–)RNA viruses (BCV, RR-b2) (87). This means that dsRNA viruses evolved on at least two independent occasions from different branches of (+)RNA viruses, whereas (–)RNA viruses likely evolved from within dsRNA viruses (87). Thus, BCIV and BCV are each monophyletic, whereas BCIII is polyphyletic. A similar pattern is observed among reverse-transcribing viruses, which comprise the kingdom Pararnavira. The kingdom as such is monophyletic, but within it, one group of DNA-packaging viruses (caulimoviruses) originate from within the RNA-packaging viruses, whereas the other one, hepadnaviruses, is external, pointing to the polyphyly of BCVII (67).
Viruses related to HDV comprise the small realm Ribozyviria. These are unusual satellite viruses with small (about 1.7 kb), circular, negative-sense genomic RNAs that resemble a viroid and contain a distinct ribozyme involved in virus RNA maturation but, unlike viroids, encode the nucleocapsid protein (88, 89). The members of Ribozyviria do not encode any polymerase and, like viroids, are replicated by the host DNA-directed RNA polymerases (90, 91). For virion formation, all ribozyviruses require a helper virus, which is HBV in the case of HDV, but can be a different enveloped virus for other members of this realm (89, 92). Formally, these viruses belong to BCV (RR-b2), but they are clearly unrelated to the numerous viruses that comprise the bulk of this class and form a distinct clade in the RdRP tree (Fig. 5). Thus, unlike the monophyletic BCIV and BCVI but similar to BCIII and BCVII, BCV is polyphyletic.
The ssDNA viruses comprising BCII, and the realm Monodnaviria, present an unusual evolutionary history. Most of these small viruses share one homologous hallmark gene encoding the rolling circle replication endonuclease, and many members also share the single jelly roll capsid protein. Thus, the viruses in this realm are clearly evolutionarily related. However, there is strong evidence of multiple origins of monodnaviruses via recombinational merge of the endonuclease gene from different bacterial plasmids with the capsid protein genes from different (+)RNA viruses (38). This evolutionary scenario does not appear to fully fit the definitions of either monophyly or polyphyly. Furthermore, the realm Monodnaviria also includes two families of dsDNA viruses with small circular genomes, Polyomaviridae and Papillomaviridae, that apparently evolved from ssDNA viruses, possibly parvoviruses (38). Thus, in this case, one realm combines viruses from two BCs. The evolutionary provenance of anelloviruses remains unclear because no homologs of the proteins encoded by these ssDNA viruses have been so far detected. The possibility remains that this group becomes another small realm, in which case BCII might become definitively polyphyletic.
The evolutionary landscape of the dsDNA viruses in BCI is the most complex. We already mentioned the small DNA viruses of the families Polyomaviridae and Papillomaviridae that are unrelated to any other dsDNA viruses. Apart from these, there are two unrelated vast realms of dsDNA viruses that are defined primarily by the hallmark genes encoding the major capsid protein and enzymes involved in DNA packaging into the capsids. The first realm Duplodnaviria includes the enormously diverse tailed bacteriophages and archaeal viruses together with the relative herpesviruses infecting animals, and the second realm Varidnaviria consists of nontailed phages and viruses of archaea along with diverse viruses of eukaryotes, including the nucleocytoplasmic large DNA viruses (NCLDV; now phylum Nucleocytoviricota). Similar to Riboviria and Monodnaviria, Varidnaviria includes viruses from more than one BC; the vast majority have dsDNA genomes (BCI), but one family, Finnlakeviridae, includes viruses with ssDNA genomes (93, 94) and, accordingly, belongs to BCII. In addition, a recently established small realm, Adnaviria, consists of viruses infecting hyperthermophilic archaea that form rod-shaped or filamentous virions packing A-form DNA (95–97). A variety of viruses of hyperthermophilic archaea, some with odd-shaped virions and without any discernible relationship with other viruses, remain to be classified, so several additional small realms are likely to emerge (98, 99). Thus, the vast variety of dsDNA viruses that comprise BCI originate from at least four and, probably, more independent ancestors.
To summarize, the relationship between the BCs and the realms, the largest evolutionarily coherent divisions of viruses, is far from a simple one-to-one correspondence. Rather, it is a network that includes both one-to-many relationships, that is, multiple BCs combined in one realm, and many-to-one relationships, that is, multiple realms within one BC (Fig. 5). The BCs organize the information transfer routes of viruses, whereas the realms reflect evolutionary relationships traced through the hallmark genes. As such, the two classifications present complementary perspectives on the virosphere that have to be investigated jointly in order to develop a comprehensive view of virus evolution.
THE BALTIMORE CLASSES OF VIRUSES AND THE ORIGIN OF LIFE
Six of the seven BCs encompass replication-expression cycles that do not occur in cells but rather are unique to viruses, including the simplest RNA-only ones (BCIII to BCV and the RR type in the extended table) as well as those based on reverse transcription (BCVI and BCVII and the DDRD type). Thus, the hypothesis presents itself that these different replication-expression cycles are the legacy of a primordial stage in the evolution of life antedating modern-like cells, during which the uniform mode of dsDNA genome replication-expression enacted by all extant cells was not yet established. Speaking metaphorically, viruses seem to be evolution’s workshop for exploring, refining, and selecting genome replication and expression circuits.
This proposition meshes perfectly with the concept of a primordial RNA world, in which information transmission and catalysis were both performed by RNA molecules, perhaps aided by nontemplated peptides (100–103). Although the RNA world remains a hypothesis, it is effectively a logical necessity to avoid the chicken and egg paradox whereby efficient protein catalysts are required for the function of the translation system, but these only can be produced by an elaborate, high-fidelity translation machinery. The workings of the RNA world are far from being experimentally validated in full, but the growing list of reactions that can be efficiently catalyzed by ribozymes lends increasing credence to this scenario (102, 104, 105). A further, highly plausible proposition on the RNA world is that, at this stage of evolution, RNA was the only type of nucleic acid, whereas DNA, the dedicated information storage device, came to the scene later, probably after the emergence of the translation system.
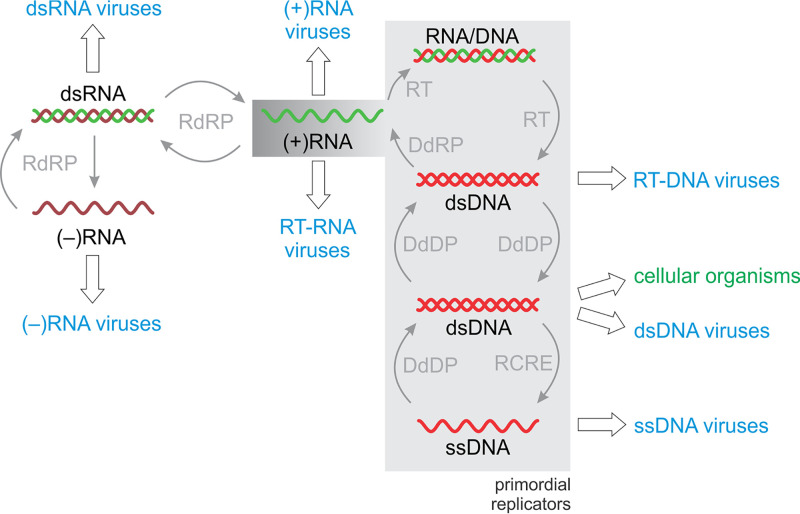
Assuming the RNA world as an essential stage of evolution that gave rise to an RNA-protein stage, it seems natural to speculate that RNA viruses are relics of that primordial era. Admittedly, however, the extant host range distribution of RNA viruses (BCIII to BCVI) does not immediately appear to bode well for this hypothesis, because the diversity of RNA viruses in eukaryotes dramatically exceeds that in prokaryotes, with (–)RNA and dsRNA viruses being (nearly) confined to eukaryotes (Fig. 3). As discussed above, this might be due to the competitive advantages enjoyed by DNA viruses in the prokaryotic intracellular environment. However, this conundrum needs to be addressed within the more general context of virus evolution. Detailed analysis of the evolutionary provenance of the key structural components of virions (primarily major capsid proteins) has led to the conclusion that the genes encoding these proteins were acquired by the emerging viruses from the hosts at different stages of the evolution of life (106). Some of these captures, notably that of the single jelly roll capsid protein that is found in an enormous variety of viruses with icosahedral capsids, most likely occurred very early, perhaps prior to the stage of the last universal cellular ancestor (LUCA) (107) but after the advent of modern-type cells with large DNA genomes. Thus, viruses sensu stricto in all likelihood did not exist during the primordial, precellular phase of the evolution of life (86). Deep analysis of the evolutionary history of virus replicative proteins leads to contrasting conclusions (86). In the discussion above, we note that the realms of viruses are evolutionarily independent. However, at a deeper level, there is a crucial unifying theme (42). The replicative enzymes of the viruses comprising all four major realms (Fig. 5) are built upon the core RNA recognition motif (RRM) domain, one of the most common nucleic acid-binding domains in nature. This profound unity suggests an evolutionary scenario in which all these modes of replication (and expression) emerged within the primordial pool of genetic elements (replicators). The replication-expression cycles most likely diversified prior to the major symmetry breaking event that led to the separation of large dsDNA replicators that became cellular genomes from the other varieties of replicators that became selfish elements and eventually gave rise to viruses (Fig. 6).
FIG 6.
The Baltimore classes and evolution of the primordial replicator pool. Shading shows the primordial replicator pool; the dark hue for (+)RNA shows that this genome structure was, most likely, the first on the route from the primordial RNA world to diverse genetic systems. Solid arrows show the inferred origins of different types of replicators, and empty block arrows show origin of viruses belonging to each of the BCs and cellular organisms. Abbreviations: DdDP, DNA-dependent DNA polymerase; DdRP, DNA-directed RNA polymerase; RdRP, RNA-dependent RNA polymerase; RCRE, rolling circle replication initiation endonuclease; RT, reverse transcriptase.
More specifically, the crucial step of “inventing” DNA as the dedicated information storage device required reverse transcription that would become possible upon the split of the primordial replicases into RdRP and RT. Notably, modern prokaryotes lack reverse-transcribing viruses, but nonviral retroelements, such as the mobile group II introns, abound (108). These elements might be direct descendants of the primordial replicators that were the first to give rise to DNA (Fig. 6).
Thus, viruses, as such, seem to have evolved at different stages of evolution when modern-type cells with large DNA genomes were already fully formed, but, in all likelihood, the first bona fide viruses predate the emergence of LUCA (107). However, the BCs and especially the extended table can be considered a general classification system for routes of replication-expression of all types of replicators, not only viruses, and in that capacity appear to be of direct relevance to the origin of life.
CONCLUDING REMARKS
Half a century is an enormous amount of time in the history of modern science. The entire scientific enterprise has changed almost beyond recognition since 1971 so that only a tiny fraction of publications from that time retain any relevance. Those that remain part of the active scientific process are only a few, but we hope and believe the discussion above presents compelling evidence that B71 is among those. Consideration of the BCs and extension of the classification of the replication-expression routes of viruses (and other replicators) trigger questions that remain unanswered and difficult to this day. The underlying causes of the contrasting host ranges of viruses from different BCs (Fig. 3) are outstanding problems but, arguably, are of fundamental interest and importance. Furthermore, the superposition of the functional classification of viruses, which is offered by the BCs, and the evolutionary classification (Fig. 5) clearly provide a deeper perspective on virus evolution than each of these complementary approaches alone.
Classification is essential for the successful development of any scientific discipline. However, it is only the first step en route to true fundamental theory. Thus, the periodic system of chemical elements is both a crowning achievement of the early phase in the evolution of chemistry and the foundation of a new era. Despite its predictive power and enormous utility, the periodic system is merely a distillation of empirical observations. Decades after its creation by Mendeleev, it became the basis first of a semiempirical theory encapsulated in the Moseley law and then of the modern theory of electron shells centered around the Pauli exclusion principle. Might it be possible to develop a general theory of replicators similarly underlying the Baltimore classification? Arguably, this is one of deepest questions faced by today’s evolutionary biology.
ACKNOWLEDGMENTS
E.V.K. was supported by the Intramural Research Program of the National Institutes of Health of the USA (National Library of Medicine). M.K. was supported by l’Agence Nationale de la Recherche (grant ANR-20-CE20-0009) and Emergence(s) project MEMREMA from Ville de Paris. V.I.A. was supported by the Russian Science Foundation grant number 20-14-00178.
Biographies

Eugene V. Koonin received his B.Sc. and Ph.D. from Moscow State University, Russia. He was a Researcher at the Institute of Poliomyelitis of the Russian Academy of Medical Sciences and a Laboratory Chief at the Institute of Microbiology of the Russian Academy of Sciences. He is currently the Leader of the Evolutionary Genomics Group and an NIH Distinguished Investigator at the NCBI at the NIH in Bethesda, Maryland, USA. He is a fellow of the American Academy of Microbiology and the American Academy of Arts and Sciences, a member of the National Academy of Sciences of the USA, a Foreign Associate of the European Molecular Biology Organization, and a Foreign Member of the Russian Academy of Sciences. He received honorary doctorates from L'Université d'Aix-Marseille in France and Wageningen University in the Netherlands and is a recipient of the National Library of Medicine Board of Regents Award, the NIH Director’s Award, and the Benjamin Franklin Award. He is the founder of the journal Biology Direct, where he served as Editor-in-Chief from 2006 to 2019. Dr. Koonin has published about 1,000 articles and the book The Logic of Chance: The Nature and Origin of Biological Evolution.

Mart Krupovic is the Head of the Archaeal Virology Unit in the Department of Microbiology at Institut Pasteur of Paris, France. He received his Ph.D. in 2010 in General Microbiology from the University of Helsinki, Finland. His current research focuses on the diversity, origins, and evolution of the virosphere as well as on the molecular mechanisms of virus-host interactions in archaea. Dr. Krupovic has published close to 200 articles and serves as an editor or on the editorial boards of Biology Direct, Research in Microbiology, Scientific Reports, Virology, and Virus Evolution. He is the Chair of the Archaeal Viruses Subcommittee of the Executive Committee of the International Committee on Virus Taxonomy (ICTV) and is the chair or a member of several ICTV study groups.

Vadim I. Agol graduated from the 1st Moscow Medical School in 1951. He received Ph.D. and D.Sc. degrees in 1954 and 1967, respectively. In 1956 he joined the Laboratory of Biochemistry of the Poliomyelitis Research Institute of the USSR Academy of Medical Sciences (now M.P. Chumakov Center for Research and Development of Immune-Biological Products of the Russian Academy of Sciences) and headed it from 1961 to 2009. In 1963, he participated in the organization of the Virology Department at the School of Biology at Moscow State University, where he was Full Professor from 1969 to 2012. In 1965, he founded the Department of Virus-Cell Interactions at the A.N. Belozersky Institute of Physical-Chemical Biology of the same university and headed it until 2019. His research is in molecular and cell biology and molecular epidemiology of picornaviruses as well as evolution of RNA viruses. He is an Honorary Scientist of Russia, an elected Corresponding Member of the Russian Academy of Sciences, and a Foreign Member of the Bulgarian Academy of Sciences. He has been awarded the Triumph Prize in Life Sciences and Medicine, the A.N. Belozersky Award of the Russian Academy of Sciences, and the Lifetime Achievement Award for Scientific Contribution of the Institute of Human Virology, University of Maryland. Dr. Agol has published about 300 articles.
REFERENCES
- 1.Baltimore D. 1971. Expression of animal virus genomes. Bacteriol Rev 35:235–241. 10.1128/br.35.3.235-241.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baltimore D. 1971. Viral genetic systems. Trans N Y Acad Sci 33:327–332. 10.1111/j.2164-0947.1971.tb02600.x. [DOI] [PubMed] [Google Scholar]
- 3.Baltimore D. 1974. The strategy of RNA viruses. Harvey Lect 70 Series: 57–74. [PubMed] [Google Scholar]
- 4.Mayr E. 1968. Theory of biological classification. Nature 220:545–548. 10.1038/220545a0. [DOI] [PubMed] [Google Scholar]
- 5.Sokal RR. 1974. Classification: purposes, principles, progress, prospects. Science 185:1115–1123. 10.1126/science.185.4157.1115. [DOI] [PubMed] [Google Scholar]
- 6.Kuhn JH, Wolf YI, Krupovic M, Zhang YZ, Maes P, Dolja VV, Koonin EV. 2019. Classify viruses - the gain is worth the pain. Nature 566:318–320. 10.1038/d41586-019-00599-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayr E. 1981. Biological classification: toward a synthesis of opposing methodologies. Science 214:510–516. 10.1126/science.214.4520.510. [DOI] [PubMed] [Google Scholar]
- 8.Baltimore D. 1970. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature 226:1209–1211. 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- 9.Temin HM, Mizutani S. 1970. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature 226:1211–1213. 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- 10.Crick F. 1970. Central dogma of molecular biology. Nature 227:561–563. 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 11.Crick FH. 1958. On protein synthesis. Symp Soc Exp Biol 12:138–163. [PubMed] [Google Scholar]
- 12.Koonin EV. 2015. Why the central dogma: on the nature of the great biological exclusion principle. Biol Direct 10:52. 10.1186/s13062-015-0084-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galibert F, Mandart E, Fitoussi F, Tiollais P, Charnay P. 1979. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature 281:646–650. 10.1038/281646a0. [DOI] [PubMed] [Google Scholar]
- 14.Kay A, Zoulim F. 2007. Hepatitis B virus genetic variability and evolution. Virus Res 127:164–176. 10.1016/j.virusres.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 15.Kaczorowska J, van der Hoek L. 2020. Human anelloviruses: diverse, omnipresent and commensal members of the virome. FEMS Microbiol Rev 44:305–313. 10.1093/femsre/fuaa007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Webb B, Rakibuzzaman A, Ramamoorthy S. 2020. Torque teno viruses in health and disease. Virus Res 285:198013. 10.1016/j.virusres.2020.198013. [DOI] [PubMed] [Google Scholar]
- 17.Kariatsumari T, Horiuchi M, Hama E, Yaguchi K, Ishigurio N, Goto H, Shinagawa M. 1991. Construction and nucleotide sequence analysis of an infectious DNA clone of the autonomous parvovirus, mink enteritis virus. J Gen Virol 72:867–875. 10.1099/0022-1317-72-4-867. [DOI] [PubMed] [Google Scholar]
- 18.Rosario K, Duffy S, Breitbart M. 2012. A field guide to eukaryotic circular single-stranded DNA viruses: insights gained from metagenomics. Arch Virol 157:1851–1871. 10.1007/s00705-012-1391-y. [DOI] [PubMed] [Google Scholar]
- 19.Krupovic M, Ghabrial SA, Jiang D, Varsani A. 2016. Genomoviridae: a new family of widespread single-stranded DNA viruses. Arch Virol 161:2633–2643. 10.1007/s00705-016-2943-3. [DOI] [PubMed] [Google Scholar]
- 20.Sencilo A, Paulin L, Kellner S, Helm M, Roine E. 2012. Related haloarchaeal pleomorphic viruses contain different genome types. Nucleic Acids Res 40:5523–5534. 10.1093/nar/gks215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pietila MK, Roine E, Sencilo A, Bamford DH, Oksanen HM. 2016. Pleolipoviridae, a newly proposed family comprising archaeal pleomorphic viruses with single-stranded or double-stranded DNA genomes. Arch Virol 161:249–256. 10.1007/s00705-015-2613-x. [DOI] [PubMed] [Google Scholar]
- 22.Mizuno CM, Prajapati B, Lucas-Staat S, Sime-Ngando T, Forterre P, Bamford DH, Prangishvili D, Krupovic M, Oksanen HM. 2019. Novel haloarchaeal viruses from Lake Retba infecting Haloferax and Halorubrum species. Environ Microbiol 21:2129–2147. 10.1111/1462-2920.14604. [DOI] [PubMed] [Google Scholar]
- 23.Tomaru Y, Toyoda K, Suzuki H, Nagumo T, Kimura K, Takao Y. 2013. New single-stranded DNA virus with a unique genomic structure that infects marine diatom Chaetoceros setoensis. Sci Rep 3:3337. 10.1038/srep03337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimura K, Tomaru Y. 2015. Discovery of two novel viruses expands the diversity of single-stranded DNA and single-stranded RNA viruses infecting a cosmopolitan marine diatom. Appl Environ Microbiol 81:1120–1131. 10.1128/AEM.02380-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kazlauskas D, Dayaram A, Kraberger S, Goldstien S, Varsani A, Krupovic M. 2017. Evolutionary history of ssDNA bacilladnaviruses features horizontal acquisition of the capsid gene from ssRNA nodaviruses. Virology 504:114–121. 10.1016/j.virol.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Auperin DD, Romanowski V, Galinski M, Bishop DH. 1984. Sequencing studies of pichinde arenavirus S RNA indicate a novel coding strategy, an ambisense viral S RNA. J Virol 52:897–904. 10.1128/JVI.52.3.897-904.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bishop DH. 1989. Infection and coding strategies of arenaviruses, phleboviruses, and nairoviruses. Rev Infect Dis 11 Suppl 4:S722–S729. 10.1093/clinids/11.supplement_4.s722. [DOI] [PubMed] [Google Scholar]
- 28.Bouloy M, Weber F. 2010. Molecular biology of Rift Valley fever virus. Open Virol J 4:8–14. 10.2174/1874357901004020008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sutela S, Forgia M, Vainio EJ, Chiapello M, Daghino S, Vallino M, Martino E, Girlanda M, Perotto S, Turina M. 2020. The virome from a collection of endomycorrhizal fungi reveals new viral taxa with unprecedented genome organization. Virus Evol 6:veaa076. 10.1093/ve/veaa076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saberi A, Gulyaeva AA, Brubacher JL, Newmark PA, Gorbalenya AE. 2018. A planarian nidovirus expands the limits of RNA genome size. PLoS Pathog 14:e1007314. 10.1371/journal.ppat.1007314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eckerle LD, Lu X, Sperry SM, Choi L, Denison MR. 2007. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J Virol 81:12135–12144. 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith EC, Case JB, Blanc H, Isakov O, Shomron N, Vignuzzi M, Denison MR. 2015. Mutations in coronavirus nonstructural protein 10 decrease virus replication fidelity. J Virol 89:6418–6426. 10.1128/JVI.00110-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robson F, Khan KS, Le TK, Paris C, Demirbag S, Barfuss P, Rocchi P, Ng WL. 2020. Coronavirus RNA proofreading: molecular basis and therapeutic targeting. Mol Cell 79:710–727. 10.1016/j.molcel.2020.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koonin EV, Yutin N. 2019. Evolution of the large nucleocytoplasmic DNA viruses of eukaryotes and convergent origins of viral gigantism. Adv Virus Res 103:167–202. 10.1016/bs.aivir.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 35.Hayakawa T, Kojima K, Nonaka K, Nakagaki M, Sahara K, Asano S, Iizuka T, Bando H. 2000. Analysis of proteins encoded in the bipartite genome of a new type of parvo-like virus isolated from silkworm - structural protein with DNA polymerase motif. Virus Res 66:101–108. 10.1016/s0168-1702(99)00129-x. [DOI] [PubMed] [Google Scholar]
- 36.Krupovic M, Koonin EV. 2014. Evolution of eukaryotic single-stranded DNA viruses of the Bidnaviridae family from genes of four other groups of widely different viruses. Sci Rep 4:5347. 10.1038/srep05347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ilyina TV, Koonin EV. 1992. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res 20:3279–3285. 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kazlauskas D, Varsani A, Koonin EV, Krupovic M. 2019. Multiple origins of prokaryotic and eukaryotic single-stranded DNA viruses from bacterial and archaeal plasmids. Nat Commun 10:3425. 10.1038/s41467-019-11433-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kazlauskas D, Krupovic M, Venclovas C. 2016. The logic of DNA replication in double-stranded DNA viruses: insights from global analysis of viral genomes. Nucleic Acids Res 44:4551–4564. 10.1093/nar/gkw322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang S, Ghabrial SA. 1996. Organization and expression of the double-stranded RNA genome of Helminthosporium victoriae 190S virus, a totivirus infecting a plant pathogenic filamentous fungus. Proc Natl Acad Sci U S A 93:12541–12546. 10.1073/pnas.93.22.12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghabrial SA. 1998. Origin, adaptation and evolutionary pathways of fungal viruses. Virus Genes 16:119–131. 10.1023/A:1007966229595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koonin EV, Dolja VV, Krupovic M, Varsani A, Wolf YI, Yutin N, Zerbini FM, Kuhn JH. 2020. Global organization and proposed megataxonomy of the virus world. Microbiol Mol Biol Rev 84:e00061-19. 10.1128/MMBR.00061-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dolja VV, Krupovic M, Koonin EV. 2020. Deep roots and splendid boughs of the global plant virome. Annu Rev Phytopathol 58:23–53. 10.1146/annurev-phyto-030320-041346. [DOI] [PubMed] [Google Scholar]
- 44.Katsafanas GC, Moss B. 2007. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe 2:221–228. 10.1016/j.chom.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu X, Meng X, Yan B, Rose L, Deng J, Xiang Y. 2012. Vaccinia virus virion membrane biogenesis protein A11 associates with viral membranes in a manner that requires the expression of another membrane biogenesis protein, A6. J Virol 86:11276–11286. 10.1128/JVI.01502-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.den Boon JA, Ahlquist P. 2010. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu Rev Microbiol 64:241–256. 10.1146/annurev.micro.112408.134012. [DOI] [PubMed] [Google Scholar]
- 47.Romero-Brey I, Bartenschlager R. 2014. Membranous replication factories induced by plus-strand RNA viruses. Viruses 6:2826–2857. 10.3390/v6072826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shabalina SA, Koonin EV. 2008. Origins and evolution of eukaryotic RNA interference. Trends Ecol Evol 23:578–587. 10.1016/j.tree.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Broecker F, Moelling K. 2019. Evolution of immune systems from viruses and transposable elements. Front Microbiol 10:51. 10.3389/fmicb.2019.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Secombes CJ, Zou J. 2017. Evolution of interferons and interferon receptors. Front Immunol 8:209. 10.3389/fimmu.2017.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watson SF, Knol LI, Witteveldt J, Macias S. 2019. Crosstalk between mammalian antiviral pathways. Noncoding RNA 5:29. 10.3390/ncrna5010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aguado LC, tenOever BR. 2018. RNase III nucleases and the evolution of antiviral systems. Bioessays 40:1700173. 10.1002/bies.201700173. [DOI] [PubMed] [Google Scholar]
- 53.Labonte JM, Suttle CA. 2013. Previously unknown and highly divergent ssDNA viruses populate the oceans. ISME J 7:2169–2177. 10.1038/ismej.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steward GF, Culley AI, Mueller JA, Wood-Charlson EM, Belcaid M, Poisson G. 2013. Are we missing half of the viruses in the ocean? ISME J 7:672–679. 10.1038/ismej.2012.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Starr EP, Nuccio EE, Pett-Ridge J, Banfield JF, Firestone MK. 2019. Metatranscriptomic reconstruction reveals RNA viruses with the potential to shape carbon cycling in soil. Proc Natl Acad Sci U S A 116:25900–25908. 10.1073/pnas.1908291116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strathern P. 2019. Mendeleev’s dream: the quest of the elements. Pegasus Books, New York, NY. [Google Scholar]
- 57.Oerter R. 2006. The theory of almost everything: the standard model, the unsung triumph of modern physics. Penguin Books, New York, NY. [Google Scholar]
- 58.Agol VI. 1974. Towards the system of viruses. Biosystems 6:113–132. 10.1016/0303-2647(74)90003-3. [DOI] [PubMed] [Google Scholar]
- 59.Cotmore SF, Agbandje-McKenna M, Canuti M, Chiorini JA, Eis-Hubinger AM, Hughes J, Mietzsch M, Modha S, Ogliastro M, Penzes JJ, Pintel DJ, Qiu J, Soderlund-Venermo M, Tattersall P, Tijssen P, ICTV Report Consortium . 2019. ICTV virus taxonomy profile: Parvoviridae. J Gen Virol 100:367–368. 10.1099/jgv.0.001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fahsbender E, Burns JM, Kim S, Kraberger S, Frankfurter G, Eilers AA, Shero MR, Beltran R, Kirkham A, McCorkell R, Berngartt RK, Male MF, Ballard G, Ainley DG, Breitbart M, Varsani A. 2017. Diverse and highly recombinant anelloviruses associated with Weddell seals in Antarctica. Virus Evol 3:vex017. 10.1093/ve/vex017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y, Chen B, Cao M, Sima L, Prangishvili D, Chen X, Krupovic M. 2018. Rolling-circle replication initiation protein of haloarchaeal sphaerolipovirus SNJ1 is homologous to bacterial transposases of the IS91 family insertion sequences. J Gen Virol 99:416–421. 10.1099/jgv.0.001009. [DOI] [PubMed] [Google Scholar]
- 62.Espejo RT, Canelo ES, Sinsheimer RL. 1971. Replication of bacteriophage PM2 deoxyribonucleic acid: a closed circular double-stranded molecule. J Mol Biol 56:597–621. 10.1016/0022-2836(71)90404-9. [DOI] [PubMed] [Google Scholar]
- 63.Charman M, Herrmann C, Weitzman MD. 2019. Viral and cellular interactions during adenovirus DNA replication. FEBS Lett 593:3531–3550. 10.1002/1873-3468.13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoeben RC, Uil TG. 2013. Adenovirus DNA replication. Cold Spring Harb Perspect Biol 5:a013003. 10.1101/cshperspect.a013003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Masai H, Arai K. 1997. Frpo: a novel single-stranded DNA promoter for transcription and for primer RNA synthesis of DNA replication. Cell 89:897–907. 10.1016/s0092-8674(00)80275-5. [DOI] [PubMed] [Google Scholar]
- 66.Bates S, Roscoe RA, Althorpe NJ, Brammar WJ, Wilkins BM. 1999. Expression of leading region genes on IncI1 plasmid ColIb-P9: genetic evidence for single-stranded DNA transcription. Microbiology (Reading) 145:2655–2662. 10.1099/00221287-145-10-2655. [DOI] [PubMed] [Google Scholar]
- 67.Krupovic M, Blomberg J, Coffin JM, Dasgupta I, Fan H, Geering AD, Gifford R, Harrach B, Hull R, Johnson W, Kreuze JF, Lindemann D, Llorens C, Lockhart B, Mayer J, Muller E, Olszewski NE, Pappu HR, Pooggin MM, Richert-Poggeler KR, Sabanadzovic S, Sanfacon H, Schoelz JE, Seal S, Stavolone L, Stoye JP, Teycheney PY, Tristem M, Koonin EV, Kuhn JH. 2018. Ortervirales: new virus order unifying five families of reverse-transcribing viruses. J Virol 92:e00515-18. 10.1128/JVI.00515-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shih C, Yang CC, Choijilsuren G, Chang CH, Liou AT. 2018. Hepatitis B virus. Trends Microbiol 26:386–387. 10.1016/j.tim.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 69.Miller RH, Tran CT, Robinson WS. 1984. Hepatitis B virus particles of plasma and liver contain viral DNA-RNA hybrid molecules. Virology 139:53–63. 10.1016/0042-6822(84)90329-5. [DOI] [PubMed] [Google Scholar]
- 70.Miller RH, Marion PL, Robinson WS. 1984. Hepatitis B viral DNA-RNA hybrid molecules in particles from infected liver are converted to viral DNA molecules during an endogenous DNA polymerase reaction. Virology 139:64–72. 10.1016/0042-6822(84)90330-1. [DOI] [PubMed] [Google Scholar]
- 71.Llorens C, Soriano B, Krupovic M, ICTV Report Consortium . 2020. ICTV virus taxonomy profile: Metaviridae. J Gen Virol 101:1131–1132. 10.1099/jgv.0.001509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Llorens C, Soriano B, Krupovic M, ICTV Report Consortium . 2021. ICTV virus taxonomy profile: Pseudoviridae. J Gen Virol 102:001605. 10.1099/jgv.0.001563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Johnson AD, Poteete AR, Lauer G, Sauer RT, Ackers GK, Ptashne M. 1981. Lambda repressor and cro–components of an efficient molecular switch. Nature 294:217–223. 10.1038/294217a0. [DOI] [PubMed] [Google Scholar]
- 74.Benler S, Koonin EV. 2020. Phage lysis-lysogeny switches and programmed cell death: Danse macabre. Bioessays 42:e2000114. 10.1002/bies.202000114. [DOI] [PubMed] [Google Scholar]
- 75.Avlund M, Dodd IB, Semsey S, Sneppen K, Krishna S. 2009. Why do phage play dice? J Virol 83:11416–11420. 10.1128/JVI.01057-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maslov S, Sneppen K. 2015. Well-temperate phage: optimal bet-hedging against local environmental collapses. Sci Rep 5:10523. 10.1038/srep10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tijssen P, Li Y, El-Far M, Szelei J, Letarte M, Zadori Z. 2003. Organization and expression strategy of the ambisense genome of densonucleosis virus of Galleria mellonella. J Virol 77:10357–10365. 10.1128/jvi.77.19.10357-10365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mankertz A, Hillenbrand B. 2002. Analysis of transcription of Porcine circovirus type 1. J Gen Virol 83:2743–2751. 10.1099/0022-1317-83-11-2743. [DOI] [PubMed] [Google Scholar]
- 79.Shivaprasad PV, Akbergenov R, Trinks D, Rajeswaran R, Veluthambi K, Hohn T, Pooggin MM. 2005. Promoters, transcripts, and regulatory proteins of mungbean yellow mosaic geminivirus. J Virol 79:8149–8163. 10.1128/JVI.79.13.8149-8163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hallam SJ, Koma T, Maruyama J, Paessler S. 2018. Review of Mammarenavirus biology and replication. Front Microbiol 9:1751. 10.3389/fmicb.2018.01751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.International Committee on Taxonomy of Viruses Executive Committee. 2020. The new scope of virus taxonomy: partitioning the virosphere into 15 hierarchical ranks. Nat Microbiol 5:668–674. 10.1038/s41564-020-0709-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bao Y, Federhen S, Leipe D, Pham V, Resenchuk S, Rozanov M, Tatusov R, Tatusova T. 2004. National Center for Biotechnology information viral genomes project. J Virol 78:7291–7298. 10.1128/JVI.78.14.7291-7298.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gorbalenya AE. 2018. Increasing the number of available ranks in virus taxonomy from five to ten and adopting the Baltimore classes as taxa at the basal rank. Arch Virol 163:2933–2936. 10.1007/s00705-018-3915-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Koonin EV, Senkevich TG, Dolja VV. 2006. The ancient virus world and evolution of cells. Biol Direct 1:29. 10.1186/1745-6150-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Iranzo J, Krupovic M, Koonin EV. 2016. The double-stranded DNA virosphere as a modular hierarchical network of gene sharing. mBio 7:e00978-16. 10.1128/mBio.00978-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krupovic M, Dolja VV, Koonin EV. 2019. Origin of viruses: primordial replicators recruiting capsids from hosts. Nat Rev Microbiol 17:449–458. 10.1038/s41579-019-0205-6. [DOI] [PubMed] [Google Scholar]
- 87.Wolf YI, Kazlauskas D, Iranzo J, Lucia-Sanz A, Kuhn JH, Krupovic M, Dolja VV, Koonin EV. 2018. Origins and evolution of the global RNA virome. mBio 9:e02329-18. 10.1128/mBio.02329-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Littlejohn M, Locarnini S, Yuen L. 2016. Origins and evolution of hepatitis B virus and hepatitis D virus. Cold Spring Harb Perspect Med 6:a021360. 10.1101/cshperspect.a021360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Iwamoto M, Shibata Y, Kawasaki J, Kojima S, Li YT, Iwami S, Muramatsu M, Wu HL, Wada K, Tomonaga K, Watashi K, Horie M. 2021. Identification of novel avian and mammalian deltaviruses provides new insights into deltavirus evolution. Virus Evol 7:veab003. 10.1093/ve/veab003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sureau C, Negro F. 2016. The hepatitis delta virus: replication and pathogenesis. J Hepatol 64:S102–S116. 10.1016/j.jhep.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 91.Goodrum G, Pelchat M. 2018. Insight into the contribution and disruption of host processes during HDV replication. Viruses 11:21. 10.3390/v11010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Szirovicza L, Hetzel U, Kipar A, Martinez-Sobrido L, Vapalahti O, Hepojoki J. 2020. Snake deltavirus utilizes envelope proteins of different viruses to generate infectious particles. mBio 11:e03250-19. 10.1128/mBio.03250-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Laanto E, Mantynen S, De Colibus L, Marjakangas J, Gillum A, Stuart DI, Ravantti JJ, Huiskonen JT, Sundberg LR. 2017. Virus found in a boreal lake links ssDNA and dsDNA viruses. Proc Natl Acad Sci U S A 114:8378–8383. 10.1073/pnas.1703834114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mäntynen S, Laanto E, Sundberg L-R, Poranen MM, Oksanen HM, ICTV Report Consortium . 2020. ICTV virus taxonomy profile: Finnlakeviridae. J Gen Virol 101:894–895. 10.1099/jgv.0.001488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang F, Baquero DP, Su Z, Osinski T, Prangishvili D, Egelman EH, Krupovic M. 2020. Structure of a filamentous virus uncovers familial ties within the archaeal virosphere. Virus Evol 6:veaa023. 10.1093/ve/veaa023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang F, Baquero DP, Beltran LC, Su Z, Osinski T, Zheng W, Prangishvili D, Krupovic M, Egelman EH. 2020. Structures of filamentous viruses infecting hyperthermophilic archaea explain DNA stabilization in extreme environments. Proc Natl Acad Sci U S A 117:19643–19652. 10.1073/pnas.2011125117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Krupovic M, Kuhn JH, Wang F, Baquero DP, Dolja VV, Egelman EH, Prangishvili D, Koonin EV. 2021. Adnaviria: a new realm for archaeal filamentous viruses with linear A-form double-stranded DNA genomes. J Virol 10.1128/JVI.00673-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Iranzo J, Koonin EV, Prangishvili D, Krupovic M. 2016. Bipartite network analysis of the archaeal virosphere: evolutionary connections between viruses and capsidless mobile elements. J Virol 90:11043–11055. 10.1128/JVI.01622-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Prangishvili D, Bamford DH, Forterre P, Iranzo J, Koonin EV, Krupovic M. 2017. The enigmatic archaeal virosphere. Nat Rev Microbiol 15:724–739. 10.1038/nrmicro.2017.125. [DOI] [PubMed] [Google Scholar]
- 100.Gilbert W. 1986. Origin of life: the RNA world. Nature 319:618. 10.1038/319618a0. [DOI] [Google Scholar]
- 101.Joyce GF. 2002. The antiquity of RNA-based evolution. Nature 418:214–221. 10.1038/418214a. [DOI] [PubMed] [Google Scholar]
- 102.Joyce GF, Szostak JW. 2018. Protocells and RNA self-replication. Cold Spring Harb Perspect Biol 10:a034801. 10.1101/cshperspect.a034801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Robertson MP, Joyce GF. 2012. The origins of the RNA world. Cold Spring Harb Perspect Biol 4:a003608. 10.1101/cshperspect.a003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lehman N. 2010. RNA in evolution. Wiley Interdiscip Rev RNA 1:202–213. 10.1002/wrna.37. [DOI] [PubMed] [Google Scholar]
- 105.Breaker RR. 2020. Imaginary ribozymes. ACS Chem Biol 15:2020–2030. 10.1021/acschembio.0c00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Krupovic M, Koonin EV. 2017. Multiple origins of viral capsid proteins from cellular ancestors. Proc Natl Acad Sci U S A 114:E2401–E2410. 10.1073/pnas.1621061114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Krupovic M, Dolja VV, Koonin EV. 2020. The LUCA and its complex virome. Nat Rev Microbiol 18:661–670. 10.1038/s41579-020-0408-x. [DOI] [PubMed] [Google Scholar]
- 108.Zimmerly S, Wu L. 2015. An unexplored diversity of reverse transcriptases in bacteria. Microbiol Spectr 3:MDNA3-0058-2014. 10.1128/microbiolspec.MDNA3-0058-2014. [DOI] [PubMed] [Google Scholar]
- 109.ICTV Report Consortium. 2021. Virus taxonomy. The ICTV report on virus classification and taxon nomenclature. https://talk.ictvonline.org/ictv-reports/ictv_online_report/. Accessed 25 June 2021.