SUMMARY
True morels (Morchella spp., Morchellaceae, Ascomycota) are widely regarded as a highly prized delicacy and are of great economic and scientific value. Recently, the rapid development of cultivation technology and expansion of areas for artificial morel cultivation have propelled morel research into a hot topic. Many studies have been conducted in various aspects of morel biology, but despite this, cultivation sites still frequently report failure to fruit or only low production of fruiting bodies. Key problems include the gap between cultivation practices and basic knowledge of morel biology. In this review, in an effort to highlight the mating systems, evolution, and life cycle of morels, we summarize the current state of knowledge of morel sexual reproduction, the structure and evolution of mating-type genes, the sexual process itself, and the influence of mating-type genes on the asexual stages and conidium production. Understanding of these processes is critical for improving technology for the cultivation of morels and for scaling up their commercial production. Morel species may well be good candidates as model species for improving sexual development research in ascomycetes in the future.
KEYWORDS: mating type, heterothallism, unisexual reproduction, pseudohomothallism, spatial competition, skewed distribution, mitospore, asexual reproduction, genome analysis, evolution
INTRODUCTION
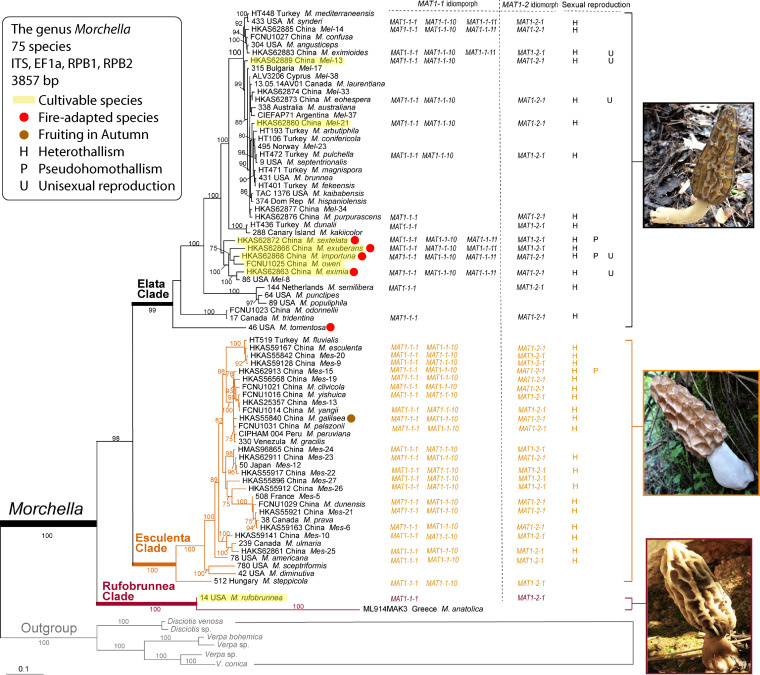
True morels (Morchella spp., Morchellaceae, Pezizomycotina, Ascomycota) are highly prized edible fungi, renowned for their aromatic and gustatory qualities. They are distinguished by a unique honeycomb-like hymenium of the hollow fruiting body and usually fruit in the spring (at least in the temperate zone), except for some autumnal species (1–3). Phenotypic plasticity and subtle morphological characters make morel species difficult to distinguish from each other (4, 5). Consequently, genealogical concordance phylogenetic species recognition (GCPSR) of multiple gene sequences (internal transcribed spacer [ITS], RPB1, RPB2, and EF1-α) is extensively used in Morchella identification (5–14). Three evolutionary clades are easily recognized: (i) the basal Rufobrunnea clade (blushing morels), (ii) the Esculenta clade (yellow morels), and (iii) the Elata clade (black morels), which also includes semifree capped morels (10). To date, 78 phylogenetically distinct species have been recognized worldwide in the genus Morchella (Fig. 1).
FIG 1.
Maximum likelihood (ML) phylogenetic analysis of 75 species in the genus Morchella based on a four-gene data set (ITS plus EF1-α plus RPB1 plus RPB2), with Verpa and Disciotis as outgroups. The species Morchella castaneae, M. vulgaris, and Morchella sp. Mes-28 were not included in this analysis because only ITS sequences for them are available from GenBank. The ML analyses were run in PhyML 3.0 using the GTR+I+G model of molecular evolution. The numbers by the nodes represent branch support >75%. Mating-type genes detected and sexual reproduction modes known in the species of Morchella are listed on the right. (Photo of M. rufobrunnea of the Rufobrunnea clade courtesy of Michael Loizides, reproduced with permission.)
The key importance of morels lies not only in their nutritional and medicinal properties (15, 16) but also in their economical and scientific value (1, 17). Morel cultivation has been a research focus worldwide for more than 100 years, starting with an original report from 1882 on their outdoor cultivation in France (18). Artificial cultivation of Morchella rufobrunnea in the United States (19), and especially of M. importuna, M. sextelata, and M. eximia in China (17), has spurred morel research worldwide. Recently, important new insights have been gained into various aspects of morel biology, including their reproductive modes (20–25), interspecific hybridization (26–28), biological and physiological characteristics (29–32), cultivation conditions (33), trophic modes (34, 35), transcriptome analysis (36), nuclear and mitochondrial genome analysis (25, 37–40), and the microbial community dynamics associated with the cultivation of morels (41, 42). The accelerated research into morels makes it highly probable that additional morel species will be successfully cultivated in the future. In 2019, the morel cultivation area in China had already reached 8,000 ha. However, typically, half of the morels in cultivation fail to fruit or exhibit a low rate of reproduction because of the low quality of strains (e.g., strain degeneration), environmental factors, management measures, and other factors. Undoubtedly, even considering the available knowledge amassed from basic and applied studies, the biology and cultivation technology for morels are both far from being fully elucidated.
In this review, we summarize the current understanding of the structure and diversification of mating-type genes, sexual reproduction, sexual process, segregation and imbalance of mating-type genes, and influence of the mating-type (MAT) locus on the asexual stages and the production of conidia in Morchella. We also present a hypothetical life cycle for morels, highlighting outstanding questions in the field. Finally, we propose morels as candidate model species to elucidate sexual development in ascomycetes.
SEXUAL REPRODUCTION IN ASCOMYCETES
More than 100,000 species of fungi have been described to date (43), exhibiting various life cycles and strong reproductive plasticity. The mechanisms of sexual and asexual reproduction in fungi are very interesting and deeply studied (44). Unlike animals with sex chromosomes, sexual reproduction in ascomycetes is under the control of a single locus, the mating type (MAT) locus (45, 46), first described over a century ago by Blakeslee (47) in Mucorineae. In fungi, the chromosomes bearing mating-type genes are usually called mating-type chromosomes or sex-related chromosomes (48, 49). The mating type locus normally has two mating types present, which by convention are indicated MAT1-1 and MAT1-2 (50, 51), also with alternative terminologies used in some species, such as mat A and mat a (e.g., in Neurospora) or mat− and mat+ (e.g., in Podospora) (52). These genes, located on the MAT locus, share little homology and are not true alleles but rather are termed “idiomorphs” (53).
The MAT genes determine the mating strategies of a fungal species (54), which include either heterothallism (self-incompatible), primary homothallism (self-compatible), or secondary homothallism (self-compatible) (50, 55). In heterothallic fungi, individuals possess either the MAT1-1 or MAT1-2 idiomorph in each haploid nucleus, and mating occurs only between partners of opposite mating types to complete the sexual life cycle (56). In contrast, primary homothallic species typically possess both mating types in each haploid nucleus (57) and are thus able to complete the sexual cycle without seeking a mate. Two mating-type idiomorphs are fused in a single genome or unlinked in a single genome but at different genomic regions (44, 56, 58).
In secondary homothallic fungi, there are two types of mechanisms, which have been described as pseudohomothallism and mating-type switching. Pseudohomothallic species usually package nuclei of opposite mating types (either mating type in each haploid nucleus) within a single multinucleus spore and are fully able to undergo independent sexual reproduction (59, 60). Mating-type switching is a “cassette model,” in which each haploid individual expresses only one mating type, even though it carries the information of both mating types (61, 62). The “silent” copies of the alternate mating type lie in a region of the genome that is transcriptionally nonactive due to the modified chromatin structure and are used during asexual reproduction to change the mating type through the transposition of information from a silent cassette to an active site (50, 62). The HO gene, vital for mating-type switching, encodes an endonuclease responsible for the initiation of switching by generating the double-stranded DNA break, which induces the gene conversion between MAT1-1 and MAT1-2 (63). For example, in Saccharomyces cerevisiae, with the active MAT locus that governs the mating type, there are two MAT cassettes, HML and HMR, in the genome, which are normally kept silent by a modified chromatin structure (64). During mitosis, the existing allele at the active MAT locus can be replaced with the opposite allele from one of the two silent MAT cassettes via gene conversion (64). Due to the ability of switching mating type, this mating strategy may appear to be outwardly homothallic, as both mating types can be produced from a single haploid individual (65).
The most recently reported and, to date, incompletely elucidated form of homothallism is unisexual reproduction, first described by Lin et al. (66), also termed monokaryotic, homokaryotic, or haploid fruiting (58, 67), same-sex mating (68), or single mating-type reproduction (62). In contrast to primary homothallism, unisexual reproduction describes the process by which a single isolate independently undergoes sexual reproduction despite having genes of only one mating type (69), with sexual processes such as meiosis and genome diploidization being involved (70, 71), similar to parthenogenesis (46, 72). Though unisexual reproduction is suggested to be derived from heterothallism via the mutation of genes involved in the initiation of sexual reproduction (69), the molecular mechanisms of unisexual reproduction are not yet fully elucidated.
In ascomycetes, MAT idiomorphs (MAT1-1 and MAT1-2) include two primary MAT genes (MAT1-1-1 and MAT1-2-1) and other secondary MAT genes, which include 11 other MAT1-1 genes (MAT1-1-2 to MAT1-1-11 plus “MAT1-1-10,” a later homonym of MAT1-1-10) and 11 other MAT1-2 genes (MAT1-2-2 to MAT1-2-12) reported from the MAT locus of various species (23, 73, 74). The two primary MAT genes, MAT1-1-1 and MAT1-2-1, have been functionally characterized in many ascomycete fungi and are typically essential for successful sexual reproduction, although their precise functions may change with species (75). In Podospora anserina, Aspergillus nidulans, and Neurospora crassa (76–79), both genes are known to be essential for fertilization, but the MAT1-1-1 gene is also essential for ascospore production in P. anserina (77).
The MAT1-1-1 and MAT1-2-1 genes encode highly conserved proteins for the alpha domain of MAT1-1 and an HMG box domain of MAT1-2, respectively, which possess DNA binding domains (51, 72, 80). The MAT proteins act as transcription factors that regulate the expression of several hundred genes related to sex, such as those involved in mate recognition of MAT1-1 and MAT1-2 identities, the pheromone and receptor systems, the conversion of vegetative mycelia into sexual tissue, cellular differentiation, and meiosis (50, 76, 81–86). In heterothallic ascomycete fungi, mate recognition is initiated by cell-specific pheromone and receptor combinations (87, 88), regulated by the MAT1-1-1 protein and the MAT1-2-1 protein (69, 89). Mating-type-specific pheromone receptors are located on the cell surface and sense the reciprocal signals (50). In homothallic ascomycete fungi, which harbor both MAT genes, both pheromone precursors are usually expressed (90). However, the discovery of unisexual reproduction has overturned the general view that the expression of two primary MAT genes is indispensable for sexual reproduction in fungi (64). The pheromone expression and response pathway of unisexual reproduction remain unclear.
The secondary MAT genes, which are not highly conserved as the primary MAT genes, usually do not have typical functional domains and have only been partially characterized (57, 75). For example, the MAT1-1-2 gene of P. anserina (91) and MAT1-1-5 gene of Sclerotinia sclerotiorum (92) are essential for producing sexual ascospores, the MAT1-1-3 gene of P. anserina is beneficial for determining the nuclear identity (91), and the MAT1-2-4 gene of S. sclerotiorum (92) and MAT1-2-10 gene of Botrytis cinerea (93) act on ascomatal development.
During the sexual process in ascomycete fungi, a key feature is that ploidy changes from diploid to haploid and back to diploid via meiosis and sporulation (45). Meiosis is integral to this process and increases diversity through recombination. Two key meiosis-specific genes, SPO11 and DMC1, are critical for meiosis (64). SPO11, which is highly conserved across eukaryotes (94), encodes the endonuclease that generates the double-strand DNA breaks that provoke meiotic recombination, and DMC1, which is important in the processing of double-strand DNA breaks for repair during homologous recombination, encodes a meiotic recombinase (45, 95). As the master regulator of meiosis, IME1 is only active during meiotic events and encodes a transcription factor that activates the expression of early meiotic genes (63, 96). Other genes with a specific function during meiosis include IME2, NDT80, RME1, SET3, etc. (63, 96). These genes, SPO11, DMC1, ME1, IME2, IME4, KAR4, NDT80, DIG1, and others, are also involved in sporulation, and their deletion severely impairs or entirely disables sporulation (66, 97, 98).
MATING-TYPE STRUCTURES IN MORCHELLA
As a new focus of intense research in the fields of biology and cultivation of edible fungi, morels and their reproductive modes are of interest not only in the context of basic questions regarding their biology, but also for the improvement of their cultivation. Several studies have reported possible reproductive modes of morel species (26, 99–101), but structures of the mating-type genes, mating system, and sexual reproduction of true morels have only recently been confirmed through the availability of the genome sequences of the species M. importuna, M. eximia, M. yangii, Morchella sp. Mes-15, and Morchella sp. Mes-21 in the genus Morchella (20–25).
With the draft genomes database of M. eximia and Morchella sp. Mes-21, Du et al. (20, 21) identified the α domain of MAT1-1-1 and the MATA_HMG-box of MAT1-2-1 from both species and then subsequently obtained the α domain and MATA_HMG-box from 34 other species in Morchella by performing PCR using specially designed primers. Based on the genomes of two M. importuna ascospore isolates with either mating type and the resultant primers, Chai et al. (22, 23) identified the MAT locus and its flanking regions of M. importuna and were able to obtain the MAT locus of Morchella sp. Mes-20 as well as MAT1-1-1 and MAT1-2-1 genes of 15 other species. With the MAT locus and its flanking regions characterized in the genomes of M. importuna (24), the genome of one MAT1-1 ascospore isolate from a yellow morel species was sequenced, with a size of approximately 56 Mb and with at least 14 chromosomes identified, by Liu et al. (25), in which the species was regarded as M. crassipes, based on the ITS sequence FJ860052.1. However, our analysis indicated that FJ860052.1 and the ITS sequence of the genome-sequenced yellow morel are identical to that of Morchella sp. Mes-20, and M. yangii, respectively. In this review, given the detailed analysis on the MAT locus and flanking regions of both M. importuna (22, 24) and Morchella sp. Mes-20 (23; X.-H. Du, L. Liu, and K. Chen, unpublished data), we focus particular attention on the mating type structures in the two species. Information about mating-type genes in other species in Morchella is also discussed later, with detailed information given in Table 1.
TABLE 1.
Known sexual reproduction modes and mating-type genes of species in Morchella and the approximate number of nuclei in their ascospores
| Clades | Species | MAT1-1-1 | MAT1-1-10 | MAT1-1-11 | MAT1-2-1 | Heterothallism | Pseudohomothallism | Unisexual reproduction | No. of nuclei per ascospore |
|---|---|---|---|---|---|---|---|---|---|
| Rufobrunnea clade | M. rufobrunnea | √a | Ub | U | √ | U | U | U | U |
| Elata clade | M. tridentina | √ | ×c | × | √ | √ | U | U | N ≥ 10 |
| M. semilibera | √ | × | × | √ | √ | U | U | N ≥ 6 | |
| M. sextelata | √ | √ | √ | √ | √ | √ | U | N ≥ 4 | |
| M. eximia | √ | √ | √ | √ | √ | U | √ | N ≥ 6 | |
| M. exuberans | √ | √ | √ | √ | √ | U | U | N ≥ 6 | |
| M. importuna | √ | √ | √ | √ | √ | √ | √ | N ≥ 6 | |
| Morchella sp. Mel-13 | √ | √ | × | √ | √ | U | √ | N ≥ 10 | |
| Morchella sp. Mel-14 | √ | √ | √ | √ | √ | U | U | N ≥ 6 | |
| M. eximioides | √ | √ | √ | √ | √ | U | √ | N ≥ 6 | |
| M. eohespera | √ | √ | × | √ | √ | U | √ | N ≥ 10 | |
| M. purpurascens | √ | × | × | √ | √ | U | U | N ≥ 6 | |
| Morchella sp. Mel-21 | √ | √ | × | √ | √ | U | U | N ≥ 6 | |
| M. dunalii | √ | × | × | √ | √ | U | U | N ≥ 6 | |
| M. pulchella | √ | √ | × | √ | √ | U | U | N ≥ 6 | |
| M. snyderi | √ | √ | √ | √ | √ | U | U | U | |
| Morchella sp. Mes-6 | √ | √ | /d | √ | √ | U | U | N = 2/4/6/8/≥10 | |
| Esculenta Clade | Morchella sp. Mes-9 | √ | √ | / | √ | √ | U | U | N = 2/4/6/8/≥10 |
| Morchella sp. Mes-10 | √ | √ | / | √ | √ | U | U | N ≥ 10 | |
| Morchella sp. Mes-15 | √ | √ | / | √ | √ | √ | U | N ≥ 10 | |
| Morchella sp. Mes-19 | √ | √ | / | √ | √ | U | U | N = 2/4/6/8/≥10 | |
| Morchella sp. Mes-20 | √ | √ | / | √ | √ | U | U | N = 2/4/6/8/≥10 | |
| Morchella sp. Mes-21 | √ | √ | / | √ | √ | U | U | N = 2/4/6/8/≥10 | |
| Morchella sp. Mes-22 | √ | √ | / | √ | √ | U | U | N = 2/4/6/8/≥10 | |
| Morchella sp. Mes-23 | √ | √ | / | √ | √ | U | U | N = 2/4/6/8/≥10 | |
| Morchella sp. Mes-24 | √ | √ | / | √ | U | U | U | U | |
| Morchella sp. Mes-25 | √ | √ | / | √ | √ | U | U | N ≥ 10 | |
| Morchella sp. Mes-26 | √ | √ | / | √ | √ | U | U | N ≥ 10 | |
| Morchella sp. Mes-27 | √ | √ | / | √ | U | U | U | U | |
| M. americana | √ | √ | / | √ | √ | U | U | N ≥ 10 | |
| M. clivicola | √ | √ | / | √ | √ | U | U | N ≥ 10 | |
| M. dunensis | √ | √ | / | √ | √ | U | U | N = 2/4/6/8/≥10 | |
| M. esculenta | √ | √ | / | √ | √ | U | U | N = 2/4/6/8/≥10 | |
| M. galilaea | √ | √ | / | √ | √ | U | U | N ≥ 20 | |
| M. palazonii | √ | √ | / | √ | √ | U | U | N ≥ 10 | |
| M. steppicola | √ | √ | / | √ | U | U | U | U | |
| M. yangii | √ | √ | / | √ | √ | U | U | N = 2/4/6/8/≥10 | |
| M. yishuica | √ | √ | / | √ | √ | U | U | N ≥ 10 |
√, gene or sexual reproduction mode was reported in the species.
U, gene, sexual reproduction mode, or number of nuclei per ascospore was not reported in the species so far.
×, gene was not detected in the species by performing PCR amplification.
/, analysis of MAT1-1-11 was not applied in the species.
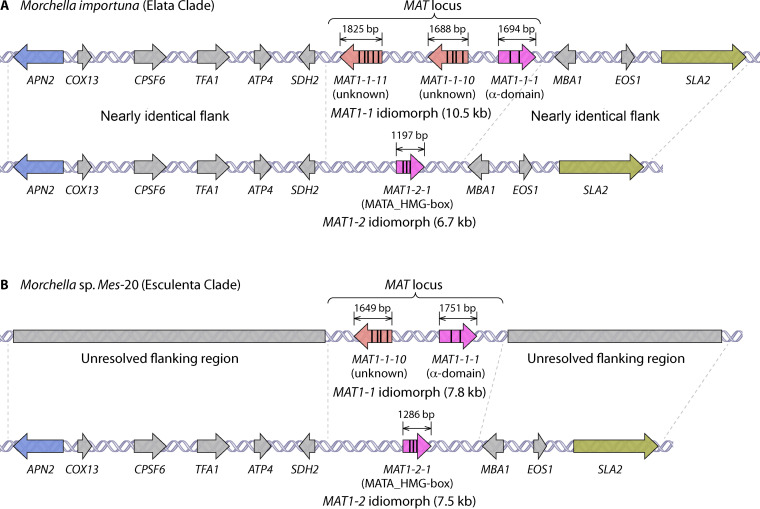
The genome of single-ascospore strains of M. importuna, with either MAT1-1 and MAT1-2 present, is reported to be approximately 46 Mb (22) or 51 Mb (24). The number and characters of chromosomes in the genome of M. importuna remain unknown. Analyses of the M. importuna genomes revealed that the lengths of MAT1-1 and MAT1-2 idiomorphs are 10.5 and 6.7 kb, respectively (22). Detailed analysis of the structure of the MAT locus showed that the MAT1-1 idiomorph contains MAT1-1-1 and two further newly described MAT genes, MAT1-1-10 and MAT1-1-11, which were first reported and named from Morchella, while MAT1-2 contains only a single MAT1-2-1 gene (22, 23) (Fig. 2A). The MAT1-1-1 gene, with a size of 1,694 bp, contains two introns and a conserved α domain; the MAT1-2-1 gene, at 1,197 bp in size, contains three introns and a conserved MATA_HMG-box. The MAT1-1-10, at 1,688 bp in size, contains four introns and encodes a hypothetical idiomorph-specific protein with 488 amino acids; the MAT1-1-11 gene, at 1,825 bp in length, contains five introns and encodes an idiomorph-specific hypothetical protein with 512 amino acids (22, 23) (Fig. 2A). The MAT genes, including MAT1-1-1, MAT1-1-10, MAT1-1-11, and MAT1-2-1, are constitutively expressed in M. importuna under laboratory conditions (22, 23). However, no further progress has been made on elucidating the molecular biological function of the MAT1-1-1, MAT1-2-1, MAT1-1-10, and MAT1-1-11 genes in M. importuna (22). It is worth noting that the MAT1-1-10 gene was not found in the genome of M. importuna by Liu et al. (24). We used the primers designed by Chai et al. (22) to perform PCR and did detect the MAT1-1-10 gene from samples of M. importuna.
FIG 2.
Structure of the MAT locus of Morchella importuna (Elata clade) (A) and Morchella sp. Mes-20 (Esculenta clade) (B). Introns are represented inside the MAT genes with vertical black lines. At the MAT locus, either a MAT1-1 or MAT1-2 idiomorph is present, containing at least a MAT1-1-1 α-domain gene or a MAT1-2-1 MATA_HMG-box gene, respectively. In addition, further genes are present in the MAT1-1 idiomorph: MAT1-1-10 and MAT1-1-11 in M. importuna (A) and MAT1-1-10 in Morchella sp. Mes-20 (B). The genes flanking the MAT idiomorphs are fairly well conserved. The flanking region of the MAT1-1 idiomorph of Morchella sp. Mes-20 has not yet been revealed. APN2 encodes an abasic endonuclease/DNA lyase; COX13 encodes the cytochrome c oxidase subunit VIa; CPSF6 encodes cleavage and polyadenylation specificity factor subunit 6; TFA1 encodes transcription initiation factor IIE subunit alpha; ATP4 encodes mitochondrial membrane ATP synthase subunit 4; SDH2 encodes succinate dehydrogenase (ubiquinone) iron-sulfur subunit; MBA1 encodes mitochondrial inner membrane-associated mitoribosome receptor; EOS1 encodes N-glycosylation protein; and SLA2 encodes a protein that binds to cortical patch actin. Distances and sizes are not drawn to scale. Mating-type structures from M. importuna and Morchella sp. Mes-20 were summarized from references 20–24 and our unpublished data.
The regions flanking the two idiomorphs are highly conserved in M. importuna. The flanking sequences of the MAT1-1 idiomorph harbor the same genes in the same syntenic orientation as those of the MAT1-2 idiomorph (22, 24). However, the pattern of flanking genes and the gene names are slightly different between Chai et al. (22) and Liu et al. (24). An extra flanking gene, EOS1, was given by Liu et al. (24) but not by Chai et al. (22). Analysis of our unpublished genomic data indicated the gene EOS1 does exist in the flanking regions of MAT locus in M. importuna. Moreover, the genes encoding the same protein are annotated with different names by Chai et al. (22) and Liu et al. (24), namely, atp-3/ATPs4, and Mba1/Mab1. By searching for the information in UniProt, a freely accessible database on proteins and their functions, the names of two genes should be ATP4 and MBA1, respectively. Taking the total information into account, the genes flanking the MAT locus of M. importuna are shown in Fig. 2A.
The genome of Morchella sp. Mes-20 generated from one MAT1-2 ascospore isolate is haploid, and the size is approximately 48 Mb (X.-H. Du, L. Liu, and K. Chen, unpublished data). The MAT1-1 idiomorph is 7.8 kb in length and harbors the MAT1-1-1 and MAT1-1-10 genes, while the MAT1-2 idiomorph is 7.5 kb in length, containing only the MAT1-2-1 gene (23). The MAT1-1-1 gene, which has a size of 1,751 bp, contains two introns and a conserved α domain; the MAT1-2-1 gene, which is 1,286 bp in size, contains three introns and a conserved MATA_HMG-box. The MAT1-1-10, with a size of 1,649 bp, contains four introns and encodes a hypothetical idiomorph-specific protein with 476 amino acids (23) (Fig. 2B). The MAT1-1-1, MAT1-2-1, and MAT1-1-10 genes are expressed in Morchella sp. Mes-20 under laboratory conditions; however, their molecular functions remain unknown (23). The flanking sequences of the MAT1-2 idiomorph of Morchella sp. Mes-20 are highly conserved and harbor the same genes as those of the MAT1-1 and MAT1-2 idiomorphs of M. importuna (22–24; X.-H. Du, L. Liu, and K. Chen, unpublished data), with the pattern shown in Fig. 2B. The flanking regions of the MAT1-1 idiomorph in Morchella sp. Mes-20 are not confirmed yet. Considering the conservative character of this area, we presume the flanking regions of its MAT1-1 idiomorph are likely to be the same as those of its MAT1-2 idiomorph.
The size and arrangement of the MAT1-2 idiomorph (7.5 kb) of Morchella sp. Mes-20 is similar to that of M. importuna (6.7 kb), while the size of the MAT1-1 idiomorph is truncated in Morchella sp. Mes-20 (7.8 kb) due to a missing MAT1-1-11 gene compared with that of M. importuna (10.5 kb) (23). Also, 66.5%, 72.1%, and 71.1% of the MAT1-2-1, MAT1-1-1, and MAT1-1-10 genes of Morchella sp. Mes-20, respectively, are identical to those of M. importuna (23). In Morchella, the MAT1-2 idiomorph is less complex than that of the MAT1-1 idiomorph.
In filamentous ascomycetes, the MAT idiomorphs usually vary in size from 1.2 to 5.7 kb (56), but in certain taxonomic groups, the size of the MAT locus can be up to 6 kb or larger due to the inclusion of additional genes (102). The MAT1-1 and MAT1-2 idiomorphs of Coccidioides are 8.1 and 9 kb in length, respectively, and were once reported to be the largest for ascomycetes (103). However, the MAT1-1 idiomorph of M. importuna, reported to be 10.5 kb in size, is much larger than that of Coccidioides and may well be the largest MAT1-1 idiomorph in filamentous ascomycetes, whereas the MAT1-2 idiomorphs of either M. importuna (6.7 kb) or Morchella sp. Mes-20 (7.5 kb) are both smaller than that of Coccidioides (22, 23, 103).
The MAT locus normally occupies a similar chromosomal position between homologs of APN2 and SLA2 in many filamentous ascomycetes (57, 104). In Sordariomycetes, homologs of APN2 and SLA2 are adjacent to each side of the MAT locus in almost all species, with an interruption only in a few species (104). In Morchella, the 5′ flanking regions of both MAT idiomorphs contain APN2, COX13, and four other predicted genes, and the 3′ flanking region houses SLA2 and two other predicted genes. However, the molecular functions of the flanking genes of the MAT1-1 and MAT1-2 idiomorphs in M. importuna and Morchella sp. Mes-20 are as yet unknown.
In Morchella, the MAT1-1-10 and MAT1-1-11 genes were first reported for the Elata clade in the MAT1-1 idiomorph of M. importuna, and the MAT1-1-10 gene was first reported in the Esculenta clade in the MAT1-1 idiomorph of Morchella sp. Mes-20 (22, 23). To confirm whether these new genes exist in other species of Morchella, we performed a facile PCR test to rapidly assess presence or absence of these genes in samples of 14 other species in the Elata clade and 21 species in the Esculenta clade, using the primers designed by Chai et al. (23) and X.-H. Du, H. Li, and B. Yuan (unpublished data). Results showed that MAT1-1-10 exists in all the tested species from the Esculenta clade. In Elata clade, besides M. importuna, both the MAT1-1-10 and MAT1-1-11 genes were also found in an additional six species but partially failed to be amplified in other species (Table 1) (X.-H. Du, H. Li, and B. Yuan, unpublished data). Future genomic studies should uncover whether these species harbor MAT1-1-11 (and MAT1-1-10). To date, of the 38 species studied from the Rufobrunnea, Elata, and Esculenta clades of Morchella, all are confirmed to have MAT1-1-1 and MAT1-2-1 genes, while seven species from the Elata clade also harbor both MAT1-1-10 and MAT1-1-11, and 22 species from the Esculenta clade contain MAT1-1-10 as well. The mating-type genes currently obtained from each species are listed in Table 1 and are shown in Fig. 1. The structures of both idiomorphs and their flanking regions of M. importuna and Morchella sp. Mes-20 are summarized in Fig. 2.
EVOLUTION OF MATING-TYPE GENES IN MORCHELLA
Although sexual reproduction in fungi is associated with dynamic evolution of sexual behaviors and mating-type-determining mechanisms, it may influence important evolutionary and ecological processes, such as speciation and adaptation (105, 106). Accordingly, the phylogenetic relationships between the MAT1-1-1 and MAT1-2-1 genes from different species of Morchella were examined to elucidate the evolution of mating-type genes in Morchella (20, 21, 23). In ascomycetes, mating-type genes have evolved more rapidly than other protein-coding genes (107, 108). That the interspecific nucleotide diversity of MAT1-1-1 and MAT1-2-1 sequences is higher than the intraspecific nucleotide diversity suggests that MAT genes evolve rapidly in Morchella (20, 21, 23). This is also consistent with the strong purifying selection against deleterious mutations in MAT genes (46, 109).
The MAT genealogies and species phylogeny of the Elata clade share similar topologies, and the two MAT genes were recommended as good candidate markers for phylogenetic inferences in this clade (20, 23). For the Esculenta clade, Du et al. (21) reported that the MAT genealogies, generated from the alpha domain of MAT1-1 and an HMG box domain of MAT1-2, and a species phylogeny of 22 yellow morel species had different topologies. The MAT genealogies reported by Du et al. (21) resolved most of the species, indicating their potential usefulness as phylogenetic markers for species delimitation. However, several species could not be distinguished. Chai et al. (23) compared the genealogies of MAT1-1-1 and MAT1-2-1 with a phylogeny of nine species generated from the combined ITS plus 28S plus RPB1 plus RPB2 plus EF1-α data set. The authors reported that either MAT gene could be used to distinguish each of the nine species, although the trees had different topologies. The highly conserved HMG and α-box domains of the mating-type genes found in Morchella (23) might partially contribute to the unresolved phylogeny of several species reported by Du et al. (21). However, the potential introgression of the two MAT idiomorphs into a species from a single ancestral source could not be excluded, and a more extensive analysis incorporating additional samples is required to resolve this question.
The evolution of MAT genes is commonly regarded as a source of reproductive isolation during speciation (80). Reproductive isolation acts to preserve the genetic integrity of species and prevents the introduction of “foreign” DNA (110). However, the phylogenies of MAT idiomorphs in the genus Morchella presented by Du et al. (20, 21) and Chai et al. (23) highlight a few well-supported topological discordances from the species trees which can be taken as an indication of potential interspecific hybridization.
In fungi, interspecific hybridization can be achieved by sexual mating or by fusion of vegetative structures (111) and acts as a major factor affecting speciation and adaptation (112, 113). In Morchella, multiple potential interspecific hybridization events have been reported between species within the Elata clade (26–28) or within the Esculenta clade (21), as well as between species of the aforementioned two clades (X.-H. Du and K. Chen, unpublished data). These interspecific hybridization events in Morchella suggest that the phylogenetic divergence of morels might have preceded the development of reproductive barriers, as has also been proposed for other fungi (114–116). Taken together, these works highlight the complex evolutionary trajectories of the reproductive genes of Morchella, which are important for the understanding of speciation and reproductive behavior in this genus.
Inbreeding has also been indicated in Morchella in several studies (26, 99, 100, 117). Typically, in heterothallic species, heterozygosity can be identified when the genetic diversity of fruiting bodies is evaluated using codominant markers because of the existence of two different parental genotypes. However, little heterozygosity has been reported from Morchella except the MAT locus, which indeed indicates the occurrence of frequent inbreeding (26, 99, 100, 117). Although heterozygosity in M. importuna, M. sextelata, M. eximia, M. exuberans, Mel-13, and Mel-21 was reported using 22 genetic markers (simple sequence repeats [SSRs]), the observed intraspecific heterozygosity was lower than expected, and the discrepancy was attributed to forces such as inbreeding (27). In this case, the parents may have been closely related (inbred), which would potentially explain why little marker segregation was found in Morchella. In heterothallic species, the recognition and response of gametes of opposite mating types is regulated by pheromones and pheromone receptors produced in a matin-type-specific manner (54). This probably means that parental fungi that are physically close and therefore more likely to be related to each other may more easily sense each other and receive pheromone signals to mate than more geographically distant individuals, ultimately resulting in the prevalence of inbreeding in morels. This is similar to the results from works in Tuber melanosporum, which suggest that two mating partners most probably have to be located sufficiently close to allow sensing each other for mating (118). However, the exact mechanisms that underpin the frequent inbreeding of morel species remain unknown.
MATING SYSTEMS IN MORCHELLA
Sex involving two mating partners, males and females with clear morphological differences, is compulsory in some eukaryotes. However, in fungi, the two mating partners are morphologically indistinguishable for most of their life cycles (51, 102). Mating types are used to define the sexual identity of a fungal individual, according to its ability to recognize and mate with different sexual cell types, and then to determine the mode of sexual reproduction (45, 56).
The availability of the complete or draft genome sequences has created the opportunity to unveil the structure of the MAT locus and mating strategies in the genus Morchella. Knowledge of MAT genes in M. eximia (20), M. importuna (22, 24), Morchella sp. Mes-21 (21), and Morchella sp. Mes-20 (23) made it possible to develop universal primers that can be used for amplification of MAT genes and sequencing of these genes from other species of this genus.
To date, in Morchella, MAT1-1-1 and MAT1-2-1 genes have been successfully amplified from one species of the Rufobrunnea clade (23), 15 species of the Elata clade (20, 22, 24), and 22 species of the Esculenta clade (21, 23) (Fig. 1, Table 1). Single-spore analyses were conducted in all of the species except Morchella sp. Mes-24, Morchella sp. Mes-27, and M. rufobrunnea, and the results revealed that the ascospores produced by each species harbor a single idiomorph with nearly equal ratios of both mating types (20–25). The results support the notion that these species are self-sterile and have a heterothallic life style (20–25). Notably, the sexual reproductive mode of M. rufobrunnea, a basal species in Morchella, remains a mystery. This needs to be investigated further to get a better understanding of reproductive modes and its evolution in Morchella.
Remarkably, as well as the heterothallism mentioned above, it has also been discovered that unisexual reproduction, the most recent known homothallism, can also, if rarely, occur in Morchella (20). In M. eximia, M. importuna, Morchella sp. Mel-13, M. eximioides, and M. eohespera, only one idiomorph was detected in the individual fruiting bodies, which suggests that unisexual reproduction (haploid fruiting) does occur in these species, without the involvement of an opposite mating type (20). That means some species in Morchella have two sexual reproduction modes, heterothallism and unisexual reproduction (homothallism). Du et al. (20) did not observe any ascospores in the hymenium at maturity in those ascocarps produced by unisexual reproduction, indicating a possible failure to complete the life cycle. Sterile fruiting bodies and a lack of ascospores have also been reported in Cordyceps militaris (119, 120). However, one or two asci with ascospores, rarely up to 13, were observed to be produced in mtA perithecia in Sordaria brevicollis (121), even though the majority were empty.
Interestingly, pseudohomothallism has recently been reported in the species M. sextelata, M. importuna, and Morchella sp. Mes-15, in which single ascospores harboring both MAT1-1 and MAT1-2 genes were found (122). One wild and one cultivated fruiting body of M. importuna were analyzed, and heterokaryon ascospores were only detected in the wild one, with a ratio of 12.1% (four heterokaryon ascospores detected from 33 investigated ascospores). The ratios of heterokaryon to total ascospores were approximately 10% and 1.2% in M. sextelata and Morchella sp. Mes-15, respectively, when single wild ascocarps were analyzed (122). The biological characters and ontogeny of the heterokaryon ascospores in Morchella remain unknown.
To date, three kinds of sexual reproductive modes have been found in Morchella: heterothallism, unisexual reproduction (homothallism), and pseudohomothallism. Mixed mating systems do exist in several species of Morchella, and heterothallism is the dominant reproductive mode. To date, M. importuna is the only species in Morchella having all three sexual reproductive modes known from the genus, and it is also the species which is most widely cultivated in China. The known sexual reproductive modes of species in Morchella are listed in Table 1. It is anticipated that more species in Morchella having mixed sexual reproduction modes could be found in the future.
DOMINANT HETEROTHALLISM WITH A PREVALENT HAPLOID LIFESTYLE
Separate sexes are not known in fungi, and most filamentous heterothallic ascomycetes mate in a hermaphroditic fashion, where the male and female roles are not associated with the MAT locus (mat A/MAT1-1 or mat a/MAT1-2), and the sexual identity of a fungal individual is defined according to its ability to recognize and mate with different sexual cell types (56, 123). Haploid strains are hermaphroditic and self-sterile, mating by donation or acceptance of nuclei from other individuals with the opposite mating type (124, 125).
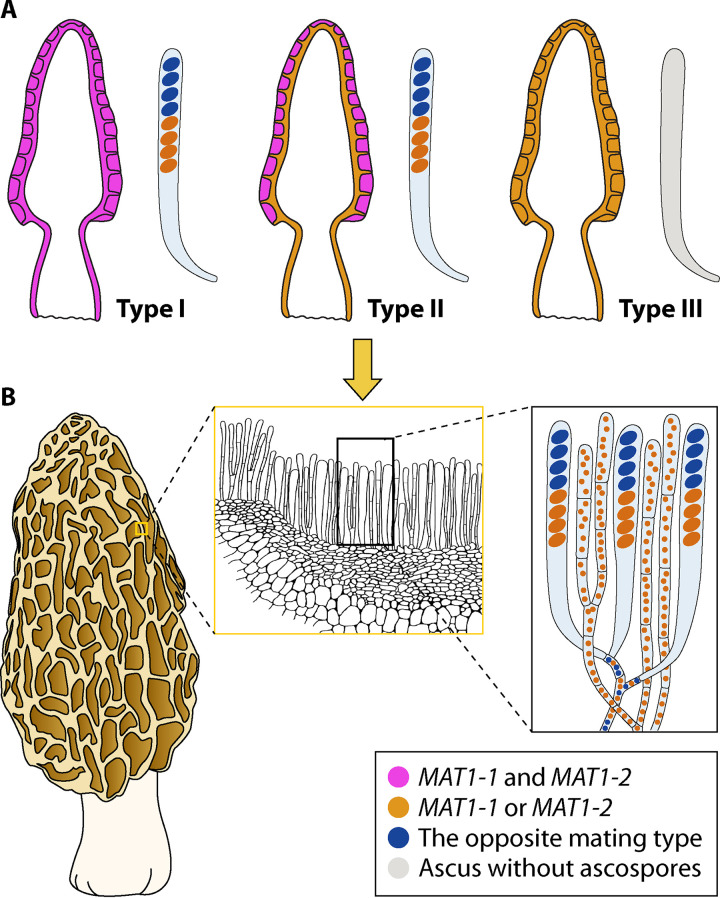
In Morchella, the genetic material in the ascocarps exhibits maternal and paternal partitioning, but the sexual identity is not genetically defined. The predominant partner (mating type) in the ascocarps of Morchella is usually defined as maternal or female, and the opposite mating type as paternal or male, as used in truffles (126–128). By defining the stipe as representing the sterile portion and the hymenium as representing the fertile portion, Du et al. (20) described three types of MAT segregation in Morchella ascocarps, namely, types I to III (Fig. 3).
FIG 3.
Schematic illustration of distribution of mating types in mature ascomata of morels. (A) Three recognized types: I (both mating types equally distributed and present in the hymenium and stipe), II (the dominant type, MAT1-1 or MAT1-2 acting female, and opposite as male, with both mating types present in the hymenium while either of them is present in the stipe), and III (only one MAT idiomorph present in the ascomata, no mating, and ascospores not formed). (B) Distribution of mating types in the hymenium of the type II morels.
In type I (Fig. 3A), the hymenium as well as the corresponding stipe of certain cultivated morel ascocarps from the species M. importuna, M. sextelata, and M. eximia harbor both MAT genes, unlike wild morels (as described below). Du et al. (20) speculated that this might be related to the available nutrition at the cultivated sites. However, given the recently observed pseudohomothallism in Morchella (122), especially in M. importuna and M. sextelata, we presume that this type of fruiting body is likely to develop from the heterokaryotic ascospore and be constructed from heterokaryotic mycelia and tissue, with both mating types found in the hymenium and stipe.
Type II (Fig. 3A) is the dominant distribution type of MAT genes observed in wild morel ascocarps. In this type, the hymenium harbors both MAT genes, but only a single mating type (either MAT1-1 or MAT1-2) is exclusively amplified in the corresponding stipe. As in other ascomycetes, including the truffle species, this implies that a haploid life cycle is prevalent (129, 130). The one mating type (either MAT1-1 or MAT1-2) is likely to contribute to the formation of the main structure of the ascocarp (the stipe and the infertile tissues of the hymenium) (Fig. 3A and B), and is defined as maternal, behaving as a “maternal” partner or a female gamete during mating and growth. The other mating type, which is only detected in the hymenium (ascospore pools), is likely to have functioned as a “paternal” partner (the male gamete) via contribution to fertilization. A haploid morel strain may be hermaphroditic, with the ability to differentiate into maternal and paternal sexual structures, but the sexual cycle is initiated only when one mating type is fertilized by cells of a different mating type, as reported in N. crassa (124, 125). In heterothallic ascomycetes, hermaphroditism is common and expected in most species and supposedly represents the ancestral state (123, 131).
In filamentous fungi, heterokaryons could be formed by the fusion of somatic cells from different isolates, and somatic nonself recognition is controlled by vegetative incompatibility (78, 132–134). Recognition of a dominant haploid life cycle highlights the absence of heterokaryons during most stages of the morel life cycle, hinting at the existence of a system of vegetative incompatibility in Morchella. In ascomycetes, genetic nonself recognition systems usually prevent the formation of anastomosis between strains, likely at the prefusion level, which maintains the genetic integrity of each strain and protects resources within hyphae from exploitation by nonkin individuals during vegetative growth (132). However, it is not clear which genes play a role in vegetative incompatibility in Morchella, and the factors that regulate the transition from the vegetative to reproductive stage await to be uncovered.
Finally, in type III (Fig. 3A), only one idiomorph is detected in the fruiting body. This type of ascomata is rare and develops through unisexual reproduction from a haploid individual with only one mating type.
SPATIAL COMPETITION AND SKEWED DISTRIBUTION OF MATING TYPES
In some fungi, mating type genes not only drive sexual reproduction but also influence other functions, such as pathogenicity (135), virulence (54, 136), cellular processes (137), and cell wall integrity (138). These correlations were reported to likely cause the patchy and imbalanced distribution of two mating types at natural sites in some species, such as C. militaris (120), Cryptococcus neoformans (121), and T. melanosporum (128, 139). Even in species that reproduce asexually, an imbalanced distribution of mating types was readily observed, such as in Teratosphaeria destructans (140).
As mentioned in the section above, there are three types of ascocarps with different MAT distributions known from Morchella (20). To investigate the distribution of mating types in different populations of the same species, we focus here on the dominant type II ascocarps. In type II, the maternal partner (mating type) contributes to most parts of the ascocarps (the stipe and the infertile tissue of hymenium) (20). The stipe, haploid with either mating type, has been chosen to represent the maternal tissue and can be monitored by MAT genotyping (20). MAT1-1 was found to serve as the maternal partner overwhelmingly more often than MAT1-2, appearing in almost all investigated species of Morchella (20). In M. eximioides, a ratio of being maternal of nearly 6:1 for MAT1-1 to MAT1-2 was reported (20). Furthermore, an unexpectedly large imbalance was also reported from individual populations of M. eohespera and Morchella sp. Mel-13, in which the maternal partners of all investigated ascocarps were MAT1-1 (20). Apparently, MAT1-1 carries a strong competitive advantage acting as the maternal partner, and the frequency of its being so is skewed and unbalanced toward it. The observations suggest that MAT1-1 tends more to serve a female sexual role than MAT1-2, and a strong sexual selection on MAT1-1 exists in Morchella. Although separate sexes (male and female genders) are not known in fungi (123), these findings would suggest the mating types of Morchella are seemingly more like “sexes” than the typical (hermaphroditic) mating types.
The strong mating type competition and predominance of a single mating type in morels may reduce the opportunity for successful mating in some cases and cause inhibition of growth. It has been reported that the divergence of ecological, physiological, pathogenic, or other functional aspects of isolates having opposite mating types may affect the spatial distribution of some fungi (72, 135, 141, 142). However, the reasons underlying the skewed spatial distribution of opposite mating types in Morchella are unknown. In Morchella, MAT1-2-1 shows higher genetic diversity than MAT1-1-1 (20, 21, 23), which is likely related to potential competition and sexual selection.
IMPACT OF MATING TYPE ON THE ASEXUAL STAGE AND CONIDIAL PRODUCTION IN MORCHELLA
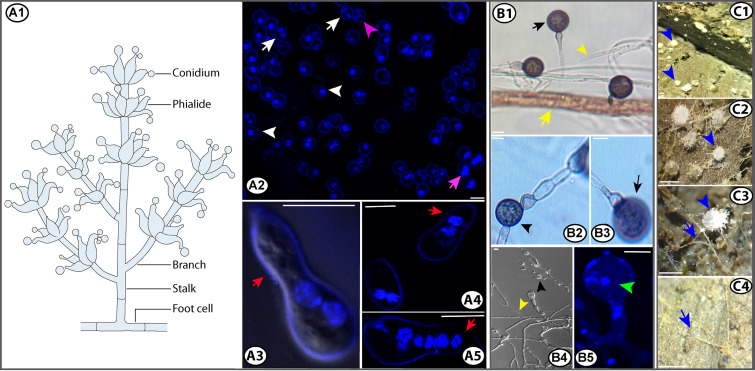
Asexual reproduction is an alternative to sexual reproduction and occurs in the majority of fungi, although the evolutionary forces governing the relationship between asexual and sexual reproductions are not well known. In Morchella, the mitospore stage (conidia) was first observed by Molliard (143, 144) and then repeatedly reported for other morel species at natural or cultivation sites (145–151). Alvarado-Castillo et al. (152) first reported the formation of chlamydospores in morels. Recently, Yuan et al. (153) successfully induced conidiation (Fig. 4A) in single-ascospore cultures of either mating type and in cross-mating cultures with opposite mating types of M. sextelata. They also observed the germination of conidia (Fig. 4A3 to A5) and chlamydospore formation from conidial hyphae (Fig. 4B1 to B5). These results support the notion that Morchella species produce two kinds of mitospores, conidia and chlamydospores.
FIG 4.
Schematic diagram of a conidiophore from Morchella, conidial nuclei, and conidial germination (A), chlamydospore formation from conidial hyphae (B), and primordia of morels and probable conidial hyphae at cultivation sites (C). Uninucleate (white arrowheads, A2), binucleate (white arrows, A2), and trinucleate conidia (purple arrowhead, A2). Nucleus mitosis in detached conidia (purple arrow, A2). Conidial germination (red arrows, A3 to A5). Terminal chlamydospores (black arrows, B1 and B3). Intercalary chlamydospores (black arrowheads, B2 and B4). Nuclei of chlamydospores (green arrowhead, B5). Much narrower conidial hyphae (yellow arrowheads, B1 and B4) than other hyphae (yellow arrow, B1). Primordia from morels (blue arrowheads, C1 to C3). Putative conidial hyphae at cultivation sites (blue arrows, C3 to C4). Bars, 5 μm (A and B) or 0.2 mm (C). (Panels A and B are modified from reference 153 with permission from the British Mycological Society.)
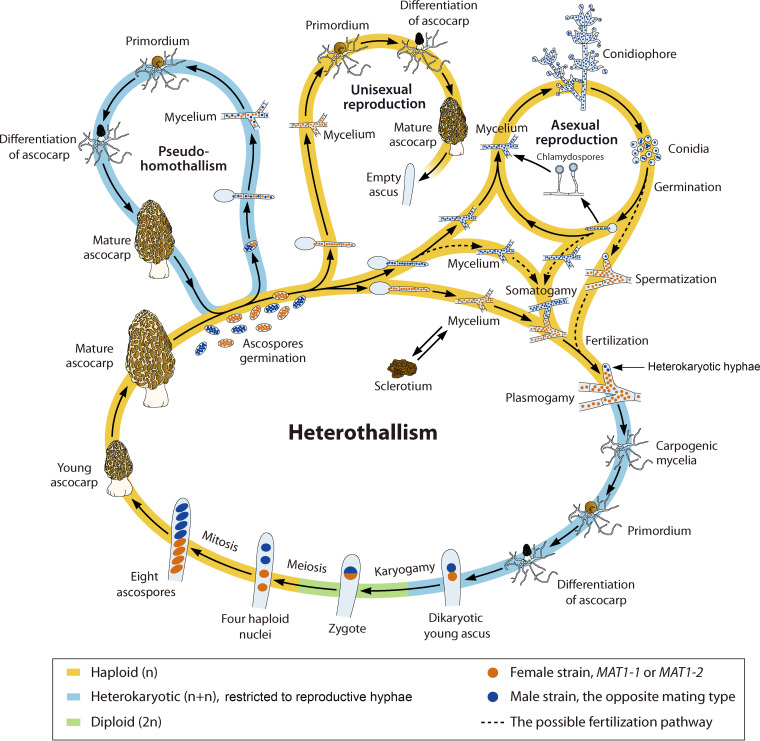
FIG 5.
Generalized life cycle of Morchella, with heterothallism, pseudohomothallism, unisexual reproduction, and asexual reproduction included. Of the known three types of ascocarps with different MAT distribution patterns (20), the ascocarps produced by heterothallism are likely to correspond to type II (Fig. 3A), needing two partners with opposite mating types to mate for sexual reproduction and being the dominant strategy. The ascocarps produced by pseudohomothallism may correspond to type I (Fig. 3A), developing from a single heterokaryotic ascospore and undergoing independent sexual reproduction. The ascocarps produced by unisexual reproduction should coincide with type III (Fig. 3A), independently undergoing sexual reproduction with only one mating type, with no ascospores observed (20). In the sexual process of heterothallism, presumably the specialized cell of the female hypha receives male nuclei from one conidium via spermatization or from a specialized cell of the male hypha via somatogamy to form a heterokaryotic cell. The heterokaryotic cell probably acts as an ascogonium and produces multinucleate ascogenous hyphae. Meiosis takes place almost immediately after karyogamy in the developing asci, followed by several postmeiotic mitoses, giving rise to eight plurinucleate ascospores. The heterokaryotic phase is short in the life cycle of morels and is restricted to the reproductive hyphae, being embedded in the homokaryotic haploid vegetative mycelium. The vegetative mycelia contribute to the main structure of the ascocarp (the nonascogenous sterile tissue), and the reproductive hyphae contribute to the ascospore pools. In asexual reproduction, mitospores (conidia and chlamydospores) are formed. The dashed lines represent hypotheses of fertilization pathways that require verification.
Intriguingly, Yuan et al. (153) demonstrated that a vegetative mycelium (either MAT1-1 or MAT1-2 mycelium) formed by germinating ascospores could produce conidia, which supports the suggestion by Volk and Leonard (151) that conidia may be formed from vegetative mycelia as shown in their diagram. Hence, the ability of either MAT1-1 or MAT1-2 mycelium-producing conidia indicates, in turn, that the mating type of conidia could be MAT1-1 or MAT1-2. In addition, the development of conidiophores in cross-mating cultures (MAT1-1:MAT1-2) was observed to occur earlier and to result in the production of more conidia than was observed in single-spore (MAT1-1 or MAT1-2) cultures (153). This suggests that cross-mating cultures produce some specific compounds that may stimulate conidiation. The asexual stage and conidial production in morels are most likely to be impacted and regulated in a mating-type-specific manner. In Ulocladium botrytis (154) and Penicillium chrysogenum (155), MAT genes were demonstrated to play essential roles in different aspects of conidiation, such as conidial size and numbers.
Conidia are commonly observed in both wild and cultivated morels but, at first, were not observed to germinate or initiate a new colony, and were once thought to function as spermatia rather than reproductive propagules (147, 156–159). Yuan et al. (153) first observed the germination of conidia and their subsequent development into mycelia in Morchella and also demonstrated that conidia are able to act as reproductive propagules rather than only as spermatia. Since a morel strain of either mating type can be hermaphroditic and have the potential to form “maternal” and “paternal” sexual structures (20), we presume that morel conidia might act as fertilizing agents (spermatia) or germinate to act as a hermaphroditic and facultative mycelium performing the function of fertilizing agents or reproductive propagules. Although hyphae derived from conidia were observed at cultivation sites and around primordia (Fig. 4C), there is still no direct evidence that shows conidia or hyphae derived from them acting as fertilizing agents. On the other hand, it remains an open question whether conidia are indispensable for morel fruiting, considering that some morel fruiting bodies are seemingly produced without conidial formation observed outwardly at the cultivation sites.
HYPOTHETICAL LIFE CYCLE OF MORCHELLA
Because of the high economic value of morels, illumination of the life cycle of Morchella species is crucial to understand their biology and advance morel cultivation technology. Over the past few decades, investigators have proposed a tentative life cycle of Morchella (4, 20, 151). Recently, several aspects of the morel mating system and asexual stages have been elucidated (20–25, 147, 152, 153), including characterization of two mating types, the identification of heterothallism, pseudohomothallism, and unisexual reproduction, the recognition of a dominant haploid life cycle, and the discovery of two kinds of mitospores. These discoveries have advanced our understanding of the morel life cycle and have led to the reinterpretation of existing data.
However, some aspects that may be related to the life cycle still remain controversial, or understudied, such as the trophic status of morels and the role of the microbiome. Based on the isotopic evidence, Hobbie et al. (35) concluded that fire-adapted morel species are saprotrophic. Current cultivation modes also support the ideas that some successfully cultivated morel species are saprotrophic. Nevertheless, mycorrhiza-like interactions have also been found in species of Morchella (160, 161). So far, there is no clear final conclusion as to the trophic status of Morchella species. In addition, little is known of the role of the microbiome in the life cycle of morels. Some studies have indicated that 29 bacterial taxa are statistically associated with the fruiting of M. sextelata (41) and that the dominating fungal community can change from the early to the cropping stage during the fruiting of M. rufobrunnea (42). In this review, in order to make a generalized hypothetical life cycle focusing on the mating systems, we consider the trophic status of morels to be saprotrophic, according to the results reported by Hobbie et al. (35), and we do not discuss the possible relationships among morels, plants, and the microbiome.
A hypothetical life cycle of morels is given in Fig. 5, with heterothallism, pseudohomothallism, unisexual reproduction, and asexual reproduction included. Of the three known types of ascocarps with different MAT distribution patterns (20), we speculate that ascocarps produced by heterothallism are likely to correspond to type II (Fig. 3A), serving as the dominant type and needing two partners to mate. Ascocarps produced by pseudohomothallism are likely to correspond to type I (Fig. 3A), resulting from a single heterokaryotic ascospore and undergoing independent sexual reproduction. We suggest that ascocarps produced by unisexual reproduction are likely to correspond to type III (Fig. 3A), independently undergoing sexual reproduction with only one mating type; although no ascospores of this type were observed (20), the possibility of rare asci with ascospores being missed cannot be entirely excluded. In asexual reproduction, mitospores (conidia and chlamydospores) are formed.
For the following hypothetical statement, we focus on heterothallism and asexual reproduction. The Morchella life cycle involves alternating haploid, heterokaryotic, and diploid phases and involves mitosis, plasmogamy, karyogamy, and meiosis. The life cycle commences with a mature fruiting body. Its ascus contains eight multinucleate ascospores (151), which are produced by meiosis and postmeiotic mitoses from a diploid nucleus formed by a karyogamy of two parental haploid (n, MAT1-1 and MAT1-2) nuclei. These ascospores are usually haploid, with either MAT1-1 or MAT1-2 (20–23). Once mature, the ascospores are released and, under suitable environmental conditions, germinate, grow, and form a haploid multikaryotic mycelium. The haploid vegetative mycelia, with either MAT1-1 or MAT1-2, are able to go into the asexual stages and produce conidiophores and conidia (153).
There are three pathways for conidial growth, which differ primarily in whether the conidia germinate or not. In pathway I, conidia act as spermatia, as has been suggested for other ascomycetes (55, 158, 159). In pathway II, conidia undergo vegetative growth (153) and then keep growing as reproductive propagules or act as fertilization agents in the form of hyphae. In pathway III, conidial hyphae are able to produce chlamydospores (asexual spores) (153). The chlamydospores keep dormant under adverse environmental conditions or germinate into a vegetative mycelium under suitable conditions (162). The vegetative mycelium (either MAT1-1 or MAT1-2) continues growing, intertwining, and forming sclerotia rich in lipids and polysaccharides, which appear to be adapted for survival under adverse conditions (4, 30, 151, 162). Although Morchella sclerotia are considered to be pseudosclerotia because they lack the medulla and rind (151), the term “sclerotia” is used in many studies (4, 17, 30, 151–153, 162–164). Considering the similar function of pseudosclerotia and sclerotia, despite the differences in their development and anatomy, we use the term sclerotia here for convenience. Sclerotia can germinate again into a vegetative mycelium under certain conditions (149, 151). Volk and Leonard (151) proposed that sclerotia produced by a vegetative mycelium may develop directly into fruiting bodies. This view has been questioned by Pilz et al. (4), who deem that scenario unlikely because of the haploid nature of sclerotium produced by the vegetative mycelium.
The differentiated gametangia (ascogonia and antheridia), trichogynes, and croziers observed in other ascomycetes (56) have not yet been identified in morel species (29, 151, 165, 166). According to the trend of morphological differentiation of the ascocarp in ascomycetes (167), we speculate that the morel sexual apparatus may be reduced. In Morchella, the female gametes are presumably the hyphae of the haploid vegetative mycelium (either MAT1-1 or MAT1-2), in which the specialized cell plays a role similar to that of the ascogonium. Male gametes with opposite mating types are presumably conidia acting as spermatia (147, 153, 156–159) or hyphae from a mycelium produced by germinating ascospores or conidia (131, 153, 168). The latter two fertilization possibilities occur by somatogamy, also suggested in Gäumann (169) and Kirschner (170). Meiosis takes place twice almost immediately after karyogamy in the young asci produced by multinucleate ascogenous hyphae, and the nuclei divide into four haploid nuclei, followed by one subsequent mitosis giving rise to eight nuclei (29, 151). Eight incipient ascospores with one nucleus in each spore are formed after spore delimitation (29, 151). The number of nuclei in mature ascospores and the number of subsequent mitoses occurring in incipient ascospores could change with different morel species (20, 21).
In ascomycetes, all fruiting body-forming tissues except ascospores arise from haploid, nonheterokaryotic hyphae (86). The heterokaryotic state of Morchella is most likely to be restricted to sexual reproductive cells. The reproductive/fertile hyphae are supported by and embedded in the haploid homokaryotic multinucleate vegetative mycelia, which contribute to the main structure of the ascocarp (the nonascogenous sterile tissue) having the same mating type (Fig. 3A and B), as indicated by the analysis of haploid stipes (representing the sterile tissue) of type II. The type II stipes, which have only one mating type (Fig. 3A) (20), have the same mating type as the haploid ridges (ectal excipulum; X.-H. Du et al., unpublished data). Development of morels does not begin from the base of the stipe followed by development of the hymenium and the ridges but rather has a fertilized hypha as the starting point, in which the haploid hyphae grow positively geotropically to form the stipe, and the fertilized and haploid vegetative hyphae grow negatively geotropically to form the hymenium and the ridges.
Some studies on morels suggest that mycelium containing both sterile and fertile hyphae may develop a sclerotium, which then could form carpogenic mycelium, or could germinate and form sterile and fertile mycelia (19, 152). To date, there is no direct evidence to support the notion that there are both sterile and fertile hyphae in a sclerotium. We have analyzed the mating-type genes in hundreds of morel sclerotia produced in cross-mating cultures of MAT1-1 and MAT1-2 ascospores and found that most of them harbored only one of the idiomorphs, while very few harbored both MAT1-1 and MAT1-2 (X.-H. Du, T. Li, and N. Xu, unpublished data). However, for the sclerotia with both MAT1-1 and MAT1-2, we were not able to determine whether they formed from a mycelium containing both sterile and fertile hyphae or from a compact mass of MAT1-1 and MAT1-2 haploid sterile mycelia. According to Frazer (171), sclerotia are formed from vegetative hyphae, and mating-type genes do not operate in this situation. Furthermore, Horn et al. (172) reported that most sclerotia of Aspergillus flavus are likely to originate from single strains of one mating type. Faretra et al. (173) suggested that sclerotia of heterothallic Botryotinia species could be fertilized by conidia of the opposite mating type. Sclerotia formation is widely thought to be critical for morel fructification (4, 17, 19, 30, 151, 152, 162, 164, 174); however, the role and importance of sclerotia in the morel life cycle remain unclear.
Under suitable environmental conditions, carpogenic mycelium produce primordia that continue to grow and differentiate into fruiting bodies. Nevertheless, under unfavorable conditions, primordia tend to abort, partially contributing to reduced or failed production at cultivation sites in China. The specific triggers of morel fructification are currently unknown.
In summary, we have provided a generalized hypothetical life cycle of Morchella based on the latest new findings. We hope that this tentative model will instigate discussions on the biology of Morchella and lead to improvements in morel cultivation. With the rapid acceleration of morel research, morels will become readily available and inexpensive in the near future.
MORELS AS CANDIDATE MODEL ORGANISMS IN STUDYING SEXUAL DEVELOPMENT IN ASCOMYCETES
We think morel species, especially M. importuna, M. sextelata, and M. eximia, could be good candidates as model organisms for the future study of sexual development, mating genetics, and mating genes in ascomycetes (75, 175–177).
As an economically important research subject, morels also have worldwide geographical distribution. Importantly, they are amenable both to culture in the laboratory, with rapid ascospore germination, and to artificial cultivation, both of which render them highly suitable for physiological and genetic studies. The current knowledge of mating systems of morel species suggests that they might be ideal models for the understanding of sexual development in Morchella and other ascomycetes. Furthermore, the maintenance of sexual and asexual reproduction strategies within a single organism makes morels a suitable model for understanding how sexual morphogenesis and the asexual stages are controlled and initiated in filamentous ascomycetes and for analyzing the function of mating-type genes and mating-type-regulated processes. In the near future, it might be possible to use transformation and, presumably, targeted mutation for gene replacement and for MAT deletion and shuffling in different genetic backgrounds, which will be useful in addressing various questions regarding the functions of MAT genes. Another added advantage of morels as model organisms is their production of hermaphrodite spores. These can be used to advance our understanding of the biology of sexual and asexual behavior in fungi. Finally, the sequenced genomes of morel species and the ongoing genome-sequencing projects will yield invaluable resources, particularly in facilitating the analysis of function and evolution of MAT genes and their roles on sexual development in Morchella. All these considerations support the use of morels as candidate model organisms to study various aspects of fungal biology.
CONCLUDING REMARKS
Morels are a fascinating group of fungi for studying the molecular mechanisms and evolutionary implications of sexual reproduction. To date, three kinds of sexual reproductive modes have been found in Morchella: heterothallism, pseudohomothallism, and unisexual reproduction (homothallism). Mixed mating systems exist in some Morchella species, and heterothallism is the dominant reproductive mode in this genus. Whether there are other sexual reproductive modes, and whether more species have mixed mating systems in Morchella, should be revealed by further studies in the future. Understanding of the mating systems of these economically important fungal species is a major issue that needs to be addressed for the better management of their life cycles, cultivation, and domestication.
ACKNOWLEDGMENTS
We thank Pierre-Arthur Moreau and Jean-Michael Bellanger for their assistance in obtaining French articles, Gerhard Kost for his help in obtaining German articles, Michael Loizides for his contribution of the photo of M. rufobrunnea, and Han-Yue Hu for her skilled help in preparing the illustrations, which were artistically largely improved by Patrick Lane, ScEYEnce Studios. We also thank the anonymous reviewers for their constructive comments and suggestions.
This work was financially supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (no. XDB31000000), the International Partnership Program of the Chinese Academy of Sciences (no. 151853KYSB20170026), the Chongqing Basic and Frontier Research Project (no. cstc2017jcyjAX0179), the Natural Science Foundation of Chongqing (no. cstc2018jcyjA3693), and the Science and Technology Service Network Initiative-Regional Key Project of the Chinese Academy of Sciences (KFJ-STS-QYZD-171).
We have no conflict of interest to declare.
Biographies

Xi-Hui Du received her Ph.D. from Kunming Institute of Botany, Chinese Academy of Sciences, in 2012, working with Professor Zhu L. Yang on the species diversity, phylogeny, and biogeography of Morchella. Then, she worked in the laboratory of Professor Zhu L. Yang as a research assistant from 2012 to 2016, and focused on the research of mating systems and population genetics of black morels. In 2011 and 2015, she visited the labs of Professor Kerry O’Donnell (Bacterial Foodborne Pathogens and Mycology Research Unit, NCAUR, USDA) and Professor Franck Richard (Centre d'Écologie Fonctionnelle et Évolutive, CNRS), respectively, for several months each as a visiting scholar. She is currently an Associate Professor at Chongqing Normal University, China, with research interests focusing on the mating systems, species diversity, and phylogeny of Morchella, which are critical for uncovering its life cycle and also for improving the commercial cultivation and resource utilization of morels.

Zhu L. Yang received his Ph.D. from the Universität Tübingen in Germany in 1997. He is now professor at Kunming Institute of Botany, a campus of the University of Chinese Academy of Sciences. In order to understand evolution of some selected fungi, his group has focused on taxonomy, molecular phylogeny, population genetics, and biogeography of Morchellaceae, Amanitaceae, and Boletaceae over the past 20 years. His team has collected >50,000 fungal collections in various parts of China and in many other regions of the world in the past years, which has led to the mycological herbarium of Kunming Institute of Botany (abbreviated KUN-HKAS) becoming the second largest mycological herbarium in China. He was awarded an honorary membership in the Mycological Society of America in 2019. Currently, he serves as editor-in-chief of Fungal Diversity and as an editorial board member for Fungal Biology Reviews, Mycological Progress, etc.
APPENDIX
GLOSSARY
- MAT
Mating type.
- MAT locus
Mating-type locus.
- MAT genes
Mating-type genes.
- idiomorph
Mating-type genes are located on the MAT locus and share little homology, and so they are not true alleles and are termed “idiomorphs.”
- heterothallism
Self-incompatible; individuals have either a MAT1-1 or MAT1-2 idiomorph in each haploid nucleus, and mating occurs only between partners of opposite mating types to complete the sexual life cycle.
- primary homothallism
Self-compatible; individuals have both mating types in each haploid nucleus and are able to complete the sexual cycle without seeking a mate.
- pseudohomothallism
Self-compatible; individuals package nuclei of opposite mating types (each haploid nucleus having one mating type) within a single multinucleus spore and are fully able to undergo independent sexual reproduction.
- mating-type switching
Each haploid individual expresses only one mating type, though it carries the information of both mating types, and the “silent” copies of the alternate mating type, which reside in a transcriptionally nonactive region, are used to change the mating type identity via the transposition of information from a silent cassette to an active site.
- unisexual reproduction
Also termed monokaryotic, homokaryotic, or haploid fruiting, same-sex mating or single-mating-type reproduction. It occurs when a single isolate having only one mating-type gene independently undergoes sexual reproduction, with sexual processes such as meiosis and genome diploidization being involved.
- silent cassette
Additional copies of the two opposite mating idiomorphs in the genome, used for mating-type switching. These cassettes are transcriptionally inactive because of modified chromatin structure.
- spermatium (pl. spermatia)
A nonmotile male reproductive cell.
- propagule
Any material that functions in propagating an organism to the next stage in its life cycle.
Contributor Information
Xi-Hui Du, Email: duxihuimorel@outlook.com.
Zhu L. Yang, Email: fungi@mail.kib.ac.cn.
REFERENCES
- 1.Du XH, Zhao Q, Yang ZL. 2015. A review on research advances, issues, and perspectives of morels. Mycology 6:78–85. 10.1080/21501203.2015.1016561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matočec N, Kušan I, Mrvoš D, Raguzin E. 2014. The autumnal occurrence of the vernal genus Morchella (Ascomycota, Fungi). Nat Croat 23:163–177. [Google Scholar]
- 3.Taşkın H, Doğan HH, Büyükalaca S. 2015. Morchella galilaea, an autumn species from Turkey. Mycotaxon 130:215–221. 10.5248/130.215. [DOI] [Google Scholar]
- 4.Pilz D, McLain R, Alexander S, Villarreal-Ruiz L, Berch S, Wurtz TL, Parks CG, McFarlane E, Baker B, Molina R, Smith JE. 2007. Ecology and management of morels harvested from the forests of western North America. General Technical Report PNW-GTR-710. US Department of Agriculture, Forest Service, Pacific Northwest Research Station, Portland, OR. [Google Scholar]
- 5.Baroni TJ, Beug MW, Cantrell SA, Clements TA, Iturriaga T, Læssøe T, Holgado Rojas ME, Aguilar FM, Quispe MO, Lodge DJ, O'Donnell K. 2018. Four new species of Morchella from the Americas. Mycologia 110:1205–1221. 10.1080/00275514.2018.1533772. [DOI] [PubMed] [Google Scholar]
- 6.Du XH, Zhao Q, O'Donnell K, Rooney AP, Yang ZL. 2012. Multigene molecular phylogenetics reveals true morels (Morchella) are especially species-rich in China. Fungal Genet Biol 49:455–469. 10.1016/j.fgb.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Du XH, Wu DM, He GQ, Wei W, Xu N, Li TL. 2019. Six new species and two new records of Morchella in China using phylogenetic and morphological analyses. Mycologia 111:857–870. 10.1080/00275514.2019.1640012. [DOI] [PubMed] [Google Scholar]
- 8.Elliott TF, Bougher NL, O'Donnell K, Trappe JM. 2014. Morchella australiana sp. nov., an apparent Australian endemic from New South Wales and Victoria. Mycologia 106:113–118. 10.3852/13-065. [DOI] [PubMed] [Google Scholar]
- 9.Loizides M, Bellanger JM, Clowez P, Richard F, Moreau PA. 2016. Combined phylogenetic and morphological studies of true morels (Pezizales, Ascomycota) in Cyprus reveal significant diversity, including Morchella arbutiphila and M. disparilis spp. nov. Mycol Progress 15:39. 10.1007/s11557-016-1180-1. [DOI] [Google Scholar]
- 10.O'Donnell K, Rooney AP, Mills GL, Kuo M, Weber NS, Rehner SA. 2011. Phylogeny and historical biogeography of true morels (Morchella) reveals an early Cretaceous origin and high continental endemism and provincialism in the Holarctic. Fungal Genet Biol 48:252–265. 10.1016/j.fgb.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 11.Richard F, Bellanger JM, Clowez P, Hansen K, O'Donnell K, Urban A, Sauve M, Courtecuisse R, Moreau PA. 2015. True morels (Morchella, Pezizales) of Europe and North America: evolutionary relationships inferred from multilocus data and a unified taxonomy. Mycologia 107:359–382. 10.3852/14-166. [DOI] [PubMed] [Google Scholar]
- 12.Taşkin H, Büyükalaca S, Doğan HH, Rehner SA, O'Donnell K. 2010. A multigene molecular phylogenetic assessment of true morels (Morchella) in Turkey. Fungal Genet Biol 47:672–682. 10.1016/j.fgb.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 13.Taskin H, Büyükalaca S, Hansen K, O'Donnell K. 2012. Multilocus phylogenetic analysis of true morels (Morchella) reveals high levels of endemics in Turkey relative to other regions of Europe. Mycologia 104:446–461. 10.3852/11-180. [DOI] [PubMed] [Google Scholar]
- 14.Voitk A, Beug MW, O'Donnell K, Burzynski M. 2016. Two new species of true morels from Newfoundland and Labrador: cosmopolitan Morchella eohespera and parochial M. laurentiana. Mycologia 108:31–37. 10.3852/15-149. [DOI] [PubMed] [Google Scholar]
- 15.Meng X, Che C, Zhang J, Gong Z, Si M, Yang G, Cao L, Liu J. 2019. Structural characterization and immunomodulating activities of polysaccharides from a newly collected wild Morchella sextelata. Int J Biol Macromol 129:608–614. 10.1016/j.ijbiomac.2019.01.226. [DOI] [PubMed] [Google Scholar]
- 16.Tietel Z, Masaphy S. 2018. True morels (Morchella)—nutritional and phytochemical composition, health benefits and flavor: a review. Crit Rev Food Sci Nutr 58:1888–1901. 10.1080/10408398.2017.1285269. [DOI] [PubMed] [Google Scholar]
- 17.Liu Q, Ma H, Zhang Y, Dong C. 2018. Artificial cultivation of true morels: current state, issues and perspectives. Crit Rev Biotechnol 38:259–271. 10.1080/07388551.2017.1333082. [DOI] [PubMed] [Google Scholar]
- 18.Roze ME. 1882. Adherence de la base d’appareils ascospores de Morchella sur Helianthus tuberosus. Acta Bot Gall 19:166–167. [Google Scholar]
- 19.Ower R. 1982. Notes on the development of the morel ascocarp: Morchella esculenta. Mycologia 74:142–144. 10.2307/3792639. [DOI] [Google Scholar]
- 20.Du XH, Zhao Q, Xia EH, Gao LZ, Richard F, Yang ZL. 2017. Mixed-reproductive strategies, competitive mating-type distribution and life cycle of fourteen black morel species. Sci Rep 7:1493. 10.1038/s41598-017-01682-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du XH, Wu D, Kang H, Wang H, Xu N, Li T, Chen K. 2020. Heterothallism and potential hybridization events inferred for twenty-two yellow morel species. IMA Fungus 11:4. 10.1186/s43008-020-0027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chai H, Chen L, Chen W, Zhao Q, Zhang X, Su K, Zhao Y. 2017. Characterization of mating-type idiomorphs suggests that Morchella importuna, Mel-20 and M. sextelata are heterothallic. Mycol Progress 16:743–752. 10.1007/s11557-017-1309-x. [DOI] [Google Scholar]
- 23.Chai H, Chen W, Zhang X, Su K, Zhao Y. 2019. Structural variation and phylogenetic analysis of the mating-type locus in the genus Morchella. Mycologia 111:551–562. 10.1080/00275514.2019.1628553. [DOI] [PubMed] [Google Scholar]
- 24.Liu W, Chen L, Cai Y, Zhang Q, Bian Y. 2018. Opposite polarity monospore genome de novo sequencing and comparative analysis reveal the possible heterothallic life cycle of Morchella importuna. Int J Mol Sci 19:2525. 10.3390/ijms19092525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu W, Cai Y, Zhang Q, Shu F, Chen L, Ma X, Bian Y. 2020. Subchromosome-scale nuclear and complete mitochondrial genome characteristics of Morchella crassipes. Int J Mol Sci 21:483. 10.3390/ijms21020483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du XH, Zhao Q, Xu J, Yang ZL. 2016. High inbreeding, limited recombination and divergent evolutionary patterns between two sympatric morel species in China. Sci Rep 6:22434. 10.1038/srep22434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du XH, Wang H, Sun J, Xiong L, Yu J. 2019. Hybridization, characterization and transferability of SSRs in the genus Morchella. Fungal Biol 123:528–538. 10.1016/j.funbio.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 28.He P, Yu M, Wang K, Cai Y, Li B, Liu W. 2020. Interspecific hybridization between cultivated morels Morchella importuna and Morchella sextelata by PEG-induced double inactivated protoplast fusion. World J Microbiol Biotechnol 36:1–10. 10.1007/s11274-020-02835-0. [DOI] [PubMed] [Google Scholar]
- 29.He P, Wang K, Cai Y, Liu W. 2017. Live cell confocal laser imaging studies on the nuclear behavior during meiosis and ascosporogenesis in Morchella importuna under artificial cultivation. Micron 101:108–113. 10.1016/j.micron.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 30.He P, Wang K, Cai Y, Hu X, Zheng Y, Zhang J, Liu W. 2018. Involvement of autophagy and apoptosis and lipid accumulation in sclerotial morphogenesis of Morchella importuna. Micron 109:34–40. 10.1016/j.micron.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 31.Tan H, Kohler A, Miao R, Liu T, Zhang Q, Zhang B, Jiang L, Wang Y, Xie L, Tang J, Li X, Liu L, Grigoriev IV, Daum C, LaButti K, Lipzen A, Kuo A, Morin E, Drula E, Henrissat B, Wang B, Huang Z, Gan B, Peng W, Martin FM. 2019. Multi-omic analyses of exogenous nutrient bag decomposition by the black morel Morchella importuna reveal sustained carbon acquisition and transferring. Environ Microbiol 21:3909–3926. 10.1111/1462-2920.14741. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Q, Miao R, Liu T, Huang Z, Peng W, Gan B, Zhang X, Tan H. 2019. Biochemical characterization of a key laccase-like multicopper oxidase of artificially cultivable Morchella importuna provides insights into plant-litter decomposition. 3 Biotech 9:171. 10.1007/s13205-019-1688-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He S, Zhao K, Ma L, Yang J, Chang Y. 2018. Effects of different cultivation material formulas on the growth and quality of Morchella spp. Saudi J Biol Sci 25:719–723. 10.1016/j.sjbs.2017.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen X, Lv SY, Ma RX, Bian YB, Kang H. 2020. A study on the nutritional pattern of Morchella importuna. Mycosystema 39:323–334. [Google Scholar]
- 35.Hobbie EA, Rice SF, Weber NS, Smith JE. 2016. Isotopic evidence indicates saprotrophy in post-fire Morchella in Oregon and Alaska. Mycologia 108:638–645. 10.3852/15-281. [DOI] [PubMed] [Google Scholar]
- 36.Hao H, Zhang J, Wang H, Wang Q, Chen M, Juan J, Feng Z, Chen H. 2019. Comparative transcriptome analysis reveals potential fruiting body formation mechanisms in Morchella importuna. AMB Express 9:103. 10.1186/s13568-019-0831-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han M, Wang Q, Baiyintala W. 2019. The whole-genome sequence analysis of Morchella sextelata. Sci Rep 9:15376. 10.1038/s41598-019-51831-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li W, Zhang F, Gao LZ. 2020. SMRT-based mitochondrial genome of the edible mushroom Morchella conica. Mitochondrial DNA B Resour 5:3201–3202. 10.1080/23802359.2020.1810160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu W, Cai Y, Zhang Q, Chen L, Shu F, Ma X, Bian Y. 2020. The mitochondrial genome of Morchella importuna (272.2 kb) is the largest among fungi and contains numerous introns, mitochondrial non-conserved open reading frames and repetitive sequences. Int J Biol Macromol 143:373–381. 10.1016/j.ijbiomac.2019.12.056. [DOI] [PubMed] [Google Scholar]
- 40.Sa W, Shang QH, Yang T, Gao QY, Liu ML, Liang J, Li ZH. 2020. Characterization of the complete mitochondrial genome of Morchella eohespera. Mitochondrial DNA B Resour 5:3048–3049. 10.1080/23802359.2020.1797559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benucci GMN, Longley R, Zhang P, Zhao Q, Bonito G, Yu F. 2019. Microbial communities associated with the black morel Morchella sextelata cultivated in greenhouses. PeerJ 7:e7744. 10.7717/peerj.7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Longley R, Benucci GMN, Mills G, Bonito G. 2019. Fungal and bacterial community dynamics in substrates during the cultivation of morels (Morchella rufobrunnea) indoors. FEMS Microbiol Lett 366:fnz215. 10.1093/femsle/fnz215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hawksworth DL, Lücking R. 2017. Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol Spectr 5:FUNK-0052-201. 10.1128/microbiolspec.FUNK-0052-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Billiard S, López-Villavicencio M, Hood ME, Giraud T. 2012. Sex, outcrossing and mating types: unsolved questions in fungi and beyond. J Evol Biol 25:1020–1038. 10.1111/j.1420-9101.2012.02495.x. [DOI] [PubMed] [Google Scholar]
- 45.Heitman J, Sun S, James TY. 2013. Evolution of fungal sexual reproduction. Mycologia 105:1–27. 10.3852/12-253. [DOI] [PubMed] [Google Scholar]
- 46.Heitman J. 2015. Evolution of sexual reproduction: a view from the fungal kingdom supports an evolutionary epoch with sex before sexes. Fungal Biol Rev 29:108–117. 10.1016/j.fbr.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blakeslee AF. 1904. Sexual reproduction in the Mucorineae. Proc Am Acad Arts Sci 40:205–319. 10.2307/20021962. [DOI] [Google Scholar]
- 48.Branco S, Carpentier F, Rodríguez de la Vega RC, Badouin H, Snirc A, Le Prieur S, Coelho MA, de Vienne DM, Hartmann FE, Begerow D, Hood ME, Giraud T. 2018. Multiple convergent supergene evolution events in mating-type chromosomes. Nat Commun 9:2000. 10.1038/s41467-018-04380-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bazzicalupo AL, Carpentier F, Otto SP, Giraud T. 2019. Little evidence of antagonistic selection in the evolutionary strata of fungal mating-type chromosomes (Microbotryum lychnidis-dioicae). G3 (Bethesda) 9:1987–1998. 10.1534/g3.119.400242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ni M, Feretzaki M, Sun S, Wang X, Heitman J. 2011. Sex in fungi. Annu Rev Genet 45:405–430. 10.1146/annurev-genet-110410-132536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dyer P, Inderbitzin P, Debuchy R. 2016. 14. Mating-type structure, function, regulation and evolution in the Pezizomycotina, p 351–385, In Wendland J (ed), Growth, differentiation and sexuality, the mycota, 3rd ed, vol 1. Springer, Cham, Switzerland. [Google Scholar]
- 52.Turgeon BG, Yoder O. 2000. Proposed nomenclature for mating type genes of filamentous ascomycetes. Fungal Genet Biol 31:1–5. 10.1006/fgbi.2000.1227. [DOI] [PubMed] [Google Scholar]
- 53.Metzenberg RL, Glass NL. 1990. Mating type and mating strategies in Neurospora. Bioessays 12:53–59. 10.1002/bies.950120202. [DOI] [PubMed] [Google Scholar]
- 54.Kronstad J, Staben C. 1997. Mating type in filamentous fungi. Annu Rev Genet 31:245–276. 10.1146/annurev.genet.31.1.245. [DOI] [PubMed] [Google Scholar]
- 55.Nelson MA. 1996. Mating systems in ascomycetes: a romp in the sac. Trends Genet 12:69–74. 10.1016/0168-9525(96)81403-x. [DOI] [PubMed] [Google Scholar]
- 56.Coppin E, Debuchy R, Arnaise S, Picard M. 1997. Mating types and sexual development in filamentous ascomycetes. Microbiol Mol Biol Rev 61:411–428. 10.1128/mmbr.61.4.411-428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Butler G. 2007. The evolution of MAT: the ascomycetes, p 1–18. In Heitman J, Kronstad JW, Taylor JW, Casselton LA (ed), Sex in fungi: molecular determination and evolutionary implications. ASM Press, Washington, DC. [Google Scholar]
- 58.Lin X, Heitman J. 2007. Mechanisms of homothallism in fungi and transitions between heterothallism and homothallism, p 35–57. In Heitman J, Kronstad JW, Taylor JW, Casselton LA (ed), Sex in fungi: molecular determination and evolutionary implications. ASM Press, Washington, DC. [Google Scholar]
- 59.Whitehouse H. 1949. Heterothallism and sex in the fungi. Biol Rev Camb Philos Soc 24:411–447. 10.1111/j.1469-185x.1949.tb00582.x. [DOI] [PubMed] [Google Scholar]
- 60.Wilson AM, Wilken PM, van der Nest MA, Steenkamp ET, Wingfield MJ, Wingfield BD. 2015. Homothallism: an umbrella term for describing diverse sexual behaviours. IMA Fungus 6:207–214. 10.5598/imafungus.2015.06.01.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hicks J, Strathern JN, Klar AJ. 1979. Transposable mating type genes in Saccharomyces cerevisiae. Nature 282:478–483. 10.1038/282478a0. [DOI] [PubMed] [Google Scholar]
- 62.Nieuwenhuis BP, Immler S. 2016. The evolution of mating-type switching for reproductive assurance. Bioessays 38:1141–1149. 10.1002/bies.201600139. [DOI] [PubMed] [Google Scholar]
- 63.Usher J. 2019. The mechanisms of mating in pathogenic fungi—a plastic trait. Genes 10:831. 10.3390/genes10100831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roach KC, Feretzaki M, Sun S, Heitman J. 2014. Unisexual reproduction. Adv Genet 85:255–305. 10.1016/B978-0-12-800271-1.00005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wallen RM, Perlin MH. 2018. An overview of the function and maintenance of sexual reproduction in dikaryotic fungi. Front Microbiol 9:503. 10.3389/fmicb.2018.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin X, Hull CM, Heitman J. 2005. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature 434:1017–1021. 10.1038/nature03448. [DOI] [PubMed] [Google Scholar]
- 67.Wickes BL, Mayorga ME, Edman U, Edman JC. 1996. Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the alpha-mating type. Proc Natl Acad Sci U S A 93:7327–7331. 10.1073/pnas.93.14.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fu C, Sun S, Billmyre RB, Roach KC, Heitman J. 2015. Unisexual versus bisexual mating in Cryptococcus neoformans: consequences and biological impacts. Fungal Genet Biol 78:65–75. 10.1016/j.fgb.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilson AM, Gabriel R, Singer SW, Schuerg T, Wilken PM, Van der Nest MA, Wingfield MJ, Wingfield BD. 2021. Doing it alone: unisexual reproduction in filamentous ascomycete fungi. Fungal Biol Rev 35:1–13. 10.1016/j.fbr.2020.12.003. [DOI] [Google Scholar]
- 70.Lin X, Jackson JC, Feretzaki M, Xue C, Heitman J. 2010. Transcription factors Mat2 and Znf2 operate cellular circuits orchestrating opposite- and same-sex mating in Cryptococcus neoformans. PLoS Genet 6:e1000953. 10.1371/journal.pgen.1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang P, Perfect JR, Heitman J. 2000. The G-protein β subunit GPB1 is required for mating and haploid fruiting in Cryptococcus neoformans. Mol Cell Biol 20:352–362. 10.1128/MCB.20.1.352-362.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee SC, Ni M, Li W, Shertz C, Heitman J. 2010. The evolution of sex: a perspective from the fungal kingdom. Microbiol Mol Biol Rev 74:298–340. 10.1128/MMBR.00005-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wilken PM, Steenkamp ET, Wingfield MJ, De Beer ZW, Wingfield BD. 2017. Which MAT gene? Pezizomycotina (Ascomycota) mating-type gene nomenclature reconsidered. Fungal Biol Rev 31:199–211. 10.1016/j.fbr.2017.05.003. [DOI] [Google Scholar]
- 74.Aylward J, Havenga M, Dreyer LL, Roets F, Wingfield BD, Wingfield MJ. 2020. Genomic characterization of mating type loci and mating type distribution in two apparently asexual plantation tree pathogens. Plant Pathol 69:28–37. 10.1111/ppa.13094. [DOI] [Google Scholar]
- 75.Wilson AM, Wilken PM, van der Nest MA, Wingfield MJ, Wingfield BD. 2019. It’s all in the genes: the regulatory pathways of sexual reproduction in filamentous ascomycetes. Genes 10:330. 10.3390/genes10050330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ferreira AVB, An ZQ, Metzenberg RL, Glass NL. 1998. Characterization of mat A-2, mat A-3 and ΔmatA mating-type mutants of Neurospora crassa. Genetics 148:1069–1079. 10.1093/genetics/148.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Debuchy R, Coppin E. 1992. The mating types of Podospora anserina: functional analysis and sequence of the fertilization domains. Mol Gen Genet 233:113–121. 10.1007/BF00587568. [DOI] [PubMed] [Google Scholar]
- 78.Paoletti M, Seymour FA, Alcocer MJ, Kaur N, Calvo AM, Archer DB, Dyer PS. 2007. Mating type and the genetic basis of self-fertility in the model fungus Aspergillus nidulans. Curr Biol 17:1384–1389. 10.1016/j.cub.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 79.Staben C, Yanofsky C. 1990. Neurospora crassa a mating-type region. Proc Natl Acad Sci U S A 87:4917–4921. 10.1073/pnas.87.13.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martin SH, Steenkamp ET, Wingfield MJ, Wingfield BD. 2013. Mate-recognition and species boundaries in the ascomycetes. Fungal Divers 58:1–12. 10.1007/s13225-012-0217-2. [DOI] [Google Scholar]
- 81.Saupe S, Stenberg L, Shiu KT, Griffiths AJ, Glass NL. 1996. The molecular nature of mutations in the mt A-1 gene of the Neurospora crassa A idiomorph and their relation to mating-type function. Mol Gen Genet 250:115–122. 10.1007/BF02191831. [DOI] [PubMed] [Google Scholar]
- 82.Bobrowicz P, Pawlak R, Correa A, Bell-Pedersen D, Ebbole DJ. 2002. The Neurospora crassa pheromone precursor genes are regulated by the mating type locus and the circadian clock. Mol Microbiol 45:795–804. 10.1046/j.1365-2958.2002.03052.x. [DOI] [PubMed] [Google Scholar]
- 83.Glass NL, Grotelueschen J, Metzenberg RL. 1990. Neurospora crassa A mating-type region. Proc Natl Acad Sci U S A 87:4912–4916. 10.1073/pnas.87.13.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chang S, Staben C. 1994. Directed replacement of mt A by mt a-1 effects a mating type switch in Neurospora crassa. Genetics 138:75–81. 10.1093/genetics/138.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Klix V, Nowrousian M, Ringelberg C, Loros JJ, Dunlap JC, Pöggeler S. 2010. Functional characterization of MAT1-1-specific mating-type genes in the homothallic ascomycete Sordaria macrospora provides new insights into essential and nonessential sexual regulators. Eukaryot Cell 9:894–905. 10.1128/EC.00019-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pöggeler S, Nowrousian M, Teichert I, Beier A, Kück U. 2018. Fruiting-body development in ascomycetes, p 1–56. In Anke T, Schüffler A (ed), Physiology and genetics, the Mycota (a comprehensive treatise on fungi as experimental systems for basic and applied research), vol 15. Springer, Cham, Switzerland. [Google Scholar]
- 87.Bölker M, Kahmann R. 1993. Sexual pheromones and mating responses in fungi. Plant Cell 5:1461–1469. 10.1105/tpc.5.10.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kothe E. 1999. Mating types and pheromone recognition in the homobasidiomycete Schizophyllum commune. Fungal Genet Biol 27:146–152. 10.1006/fgbi.1999.1129. [DOI] [PubMed] [Google Scholar]
- 89.Coppin E, de Renty C, Debuchy R. 2005. The function of the coding sequences for the putative pheromone precursors in Podospora anserina is restricted to fertilization. Eukaryot Cell 4:407–420. 10.1128/EC.4.2.407-420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pöggeler S, Kück U. 2001. Identification of transcriptionally expressed pheromone receptor genes in filamentous ascomycetes. Gene 280:9–17. 10.1016/s0378-1119(01)00786-7. [DOI] [PubMed] [Google Scholar]
- 91.Arnaise S, Debuchy R, Picard M. 1997. What is a bona fide mating-type gene? Internuclear complementation of mat mutants in Podospora anserina. Mol Gen Genet 256:169–178. 10.1007/pl00008611. [DOI] [PubMed] [Google Scholar]
- 92.Doughan B, Rollins JA. 2016. Characterization of MAT gene functions in the life cycle of Sclerotinia sclerotiorum reveals a lineage-specific MAT gene functioning in apothecium morphogenesis. Fungal Biol 120:1105–1117. 10.1016/j.funbio.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 93.Rodenburg SY, Terhem RB, Veloso J, Stassen JH, van Kan JA. 2018. Functional analysis of mating type genes and transcriptome analysis during fruiting body development of Botrytis cinerea. mBio 9:e01939-17. 10.1128/mBio.01939-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Forche A, Alby K, Schaefer D, Johnson AD, Berman J, Bennett RJ. 2008. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol 6:e110. 10.1371/journal.pbio.0060110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sherwood RK, Bennett RJ. 2009. Fungal meiosis and parasexual reproduction-lessons from pathogenic yeast. Curr Opin Microbiol 12:599–607. 10.1016/j.mib.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kassir Y, Granot D, Simchen G. 1988. IME1, a positive regulator gene of meiosis in S. cerevisiae. Cell 52:853–862. 10.1016/0092-8674(88)90427-8. [DOI] [PubMed] [Google Scholar]
- 97.Feretzaki M, Heitman J. 2013. Genetic circuits that govern bisexual and unisexual reproduction in Cryptococcus neoformans. PLoS Genet 9:e1003688. 10.1371/journal.pgen.1003688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wendland J. 2020. Sporulation in Ashbya gossypii. J Fungi 6:157. 10.3390/jof6030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dalgleish H, Jacobson K. 2005. A first assessment of genetic variation among Morchella esculenta (morel) populations. J Hered 96:396–403. 10.1093/jhered/esi045. [DOI] [PubMed] [Google Scholar]
- 100.Pagliaccia D, Douhan GW, Douhan L, Peever TL, Carris LM, Kerrigan JL. 2011. Development of molecular markers and preliminary investigation of the population structure and mating system in one lineage of black morel (Morchella elata) in the Pacific Northwestern USA. Mycologia 103:969–982. 10.3852/10-384. [DOI] [PubMed] [Google Scholar]
- 101.Volk TJ, Leonard TJ. 1989. Experimental studies on the morel. I. Heterokaryon formation between monoascosporous strains of Morchella. Mycologia 81:523–531. 10.2307/3760127. [DOI] [Google Scholar]
- 102.Debuchy R, Berteaux-Lecellier V, Silar P. 2010. Mating systems and sexual morphogenesis in ascomycetes, p 501–535. In Borkovich KA, Ebbole DJ (ed), Cellular and molecular biology of filamentous fungi. ASM Press, Washington, DC. [Google Scholar]
- 103.Mandel MA, Barker BM, Kroken S, Rounsley SD, Orbach MJ. 2007. Genomic and population analyses of the mating type loci in Coccidioides species reveal evidence for sexual reproduction and gene acquisition. Eukaryot Cell 6:1189–1199. 10.1128/EC.00117-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Debuchy R, Turgeon B. 2006. Mating-type structure, evolution, and function in Euascomycetes, p 293–323. In Wendland J (ed), Growth, differentiation and sexuality, the mycota, 3rd ed, vol 1. Springer, Berlin, Germany. [Google Scholar]
- 105.Billiard S, López-Villavicencio M, Devier B, Hood ME, Fairhead C, Giraud T. 2011. Having sex, yes, but with whom? Inferences from fungi on the evolution of anisogamy and mating types. Biol Rev Camb Philos Soc 86:421–442. 10.1111/j.1469-185X.2010.00153.x. [DOI] [PubMed] [Google Scholar]
- 106.Nieuwenhuis BP, Billiard S, Vuilleumier S, Petit E, Hood ME, Giraud T. 2013. Evolution of uni- and bifactorial sexual compatibility systems in fungi. Heredity (Edinb) 111:445–455. 10.1038/hdy.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pöggeler S. 1999. Phylogenetic relationships between mating-type sequences from homothallic and heterothallic ascomycetes. Curr Genet 36:222–231. 10.1007/s002940050494. [DOI] [PubMed] [Google Scholar]
- 108.Wik L, Karlsson M, Johannesson H. 2008. The evolutionary trajectory of the mating-type (mat) genes in Neurospora relates to reproductive behavior of taxa. BMC Evol Biol 8:109–112. 10.1186/1471-2148-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Turgeon BG. 1998. Application of mating type gene technology to problems in fungal biology. Annu Rev Phytopathol 36:115–137. 10.1146/annurev.phyto.36.1.115. [DOI] [PubMed] [Google Scholar]
- 110.Orr HA. 1995. The population genetics of speciation: the evolution of hybrid incompatibilities. Genetics 139:1805–1813. 10.1093/genetics/139.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Feurtey A, Stukenbrock EH. 2018. Interspecific gene exchange as a driver of adaptive evolution in fungi. Annu Rev Microbiol 72:377–398. 10.1146/annurev-micro-090817-062753. [DOI] [PubMed] [Google Scholar]
- 112.Hubka V, Barrs V, Dudová Z, Sklenář F, Kubátová A, Matsuzawa T, Yaguchi T, Horie Y, Nováková A, Frisvad JC, Talbot JJ, Kolařík M. 2018. Unravelling species boundaries in the Aspergillus viridinutans complex (section Fumigati): opportunistic human and animal pathogens capable of interspecific hybridization. Persoonia 41:142–174. 10.3767/persoonia.2018.41.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stukenbrock EH. 2016. The role of hybridization in the evolution and emergence of new fungal plant pathogens. Phytopathology 106:104–112. 10.1094/PHYTO-08-15-0184-RVW. [DOI] [PubMed] [Google Scholar]
- 114.Dettman JR, Jacobson DJ, Turner E, Pringle A, Taylor JW. 2003. Reproductive isolation and phylogenetic divergence in Neurospora: comparing methods of species recognition in a model eukaryote. Evol 57:2721–2741. 10.1554/03-074. [DOI] [PubMed] [Google Scholar]
- 115.Olarte RA, Worthington CJ, Horn BW, Moore GG, Singh R, Monacell JT, Dorner JW, Stone EA, Xie DY, Carbone I. 2015. Enhanced diversity and aflatoxigenicity in interspecific hybrids of Aspergillus flavus and Aspergillus parasiticus. Mol Ecol 24:1889–1909. 10.1111/mec.13153. [DOI] [PubMed] [Google Scholar]
- 116.Talbot JJ, Houbraken J, Frisvad JC, Samson RA, Kidd SE, Pitt J, Lindsay S, Beatty JA, Barrs VR. 2017. Discovery of Aspergillus frankstonensis sp. nov. during environmental sampling for animal and human fungal pathogens. PLoS One 12:e0181660. 10.1371/journal.pone.0181660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yoon CS, Gessner RV, Romano MA. 1990. Population genetics and systematics of the Morchella esculenta complex. Mycologia 82:227–235. 10.2307/3759851. [DOI] [Google Scholar]
- 118.Rubini A, Belfiori B, Riccioni C, Paolocci F. 2012. Genomics of Tuber melanosporum: new knowledge concerning reproductive biology, symbiosis, and aroma production, p 57–72. In Zambonelli A, Bonito GM (ed), Edible ectomycorrhizal mushrooms. Springer, Berlin, Germany. [Google Scholar]
- 119.Zhang G, Liang Y. 2013. Improvement of fruiting body production in Cordyceps militaris by molecular assessment. Arch Microbiol 195:579–585. 10.1007/s00203-013-0904-8. [DOI] [PubMed] [Google Scholar]
- 120.Zheng P, Xia Y, Xiao G, Xiong C, Hu X, Zhang S, Zheng H, Huang Y, Zhou Y, Wang S. 2012. Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional Chinese medicine. Genome Biol 12:R116. 10.1186/gb-2011-12-11-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Litvintseva AP, Kestenbaum L, Vilgalys R, Mitchell TG. 2005. Comparative analysis of environmental and clinical populations of Cryptococcus neoformans. J Clin Microbiol 43:556–564. 10.1128/JCM.43.2.556-564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu P, Ma Y, Zhao Y, Chai H, Li Y. 2021. Sexual and vegetative compatibility of single ascospores isolations in the genus Morchella. Acta Edulis Fungi 28:40–47. [Google Scholar]
- 123.Nieuwenhuis BP, Aanen D. 2012. Sexual selection in fungi. J Evol Biol 25:2397–2411. 10.1111/jeb.12017. [DOI] [PubMed] [Google Scholar]
- 124.Bistis G. 1981. Chemotropic interactions between trichogynes and conidia of opposite mating-type in Neurospora crassa. Mycologia 73:959–975. 10.2307/3759806. [DOI] [Google Scholar]
- 125.Fischer MS, Glass NL. 2019. Communicate and fuse: how filamentous fungi establish and maintain an interconnected mycelial network. Front Microbiol 10:619. 10.3389/fmicb.2019.00619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rubini A, Belfiori B, Riccioni C, Arcioni S, Martin F, Paolocci F. 2011. Tuber melanosporum: mating type distribution in a natural plantation and dynamics of strains of different mating types on the roots of nursery-inoculated host plants. New Phytol 189:723–735. 10.1111/j.1469-8137.2010.03493.x. [DOI] [PubMed] [Google Scholar]
- 127.Paolocci F, Rubini A, Riccioni C, Arcioni S. 2006. Reevaluation of the life cycle of Tuber magnatum. Appl Environ Microbiol 72:2390–2393. 10.1128/AEM.72.4.2390-2393.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Selosse MA, Taschen E, Giraud T. 2013. Do black truffles avoid sexual harassment by linking mating type and vegetative incompatibility? New Phytol 199:10–13. 10.1111/nph.12329. [DOI] [PubMed] [Google Scholar]
- 129.Riccioni C, Belfiori B, Rubini A, Passeri V, Arcioni S, Paolocci F. 2008. Tuber melanosporum outcrosses: analysis of the genetic diversity within and among its natural populations under this new scenario. New Phytol 180:466–478. 10.1111/j.1469-8137.2008.02560.x. [DOI] [PubMed] [Google Scholar]
- 130.Belfiori B, Riccioni C, Paolocci F, Rubini A. 2016. Characterization of the reproductive mode and life cycle of the whitish truffle T. borchii. Mycorrhiza 26:515–527. 10.1007/s00572-016-0689-0. [DOI] [PubMed] [Google Scholar]
- 131.Glass NL, Kuldau GA. 1992. Mating type and vegetative incompatibility in filamentous ascomycetes. Annu Rev Phytopathol 30:201–224. 10.1146/annurev.py.30.090192.001221. [DOI] [PubMed] [Google Scholar]
- 132.Glass NL, Jacobson DJ, Shiu PK. 2000. The genetics of hyphal fusion and vegetative incompatibility in filamentous ascomycete fungi. Annu Rev Genet 34:165–186. 10.1146/annurev.genet.34.1.165. [DOI] [PubMed] [Google Scholar]
- 133.Glass NL, Kaneko I. 2003. Fatal attraction: nonself recognition and heterokaryon incompatibility in filamentous fungi. Eukaryot Cell 2:1–8. 10.1128/EC.2.1.1-8.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Saupe SJ. 2000. Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiol Mol Biol Rev 64:489–502. 10.1128/MMBR.64.3.489-502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhan J, Torriani SF, McDonald BA. 2007. Significant difference in pathogenicity between MAT1–1 and MAT1–2 isolates in the wheat pathogen Mycosphaerella graminicola. Fungal Genet Biol 44:339–346. 10.1016/j.fgb.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 136.Kwon-Chung KJ, Edman JC, Wickes BL. 1992. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect Immun 60:602–605. 10.1128/iai.60.2.602-605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kunz BA, Haynes RH. 1981. Phenomenology and genetic control of mitotic recombination in yeast. Annu Rev Genet 15:57–89. 10.1146/annurev.ge.15.120181.000421. [DOI] [PubMed] [Google Scholar]
- 138.Verma I, Rohilla A, Khuller G. 1999. Alterations in macromolecular composition and cell wall integrity by ciprofloxacin in Mycobacterium smegmatis. Lett Appl Microbiol 29:113–117. 10.1046/j.1365-2672.1999.00597.x. [DOI] [PubMed] [Google Scholar]
- 139.Rubini A, Riccioni C, Belfiori B, Paolocci F. 2014. Impact of the competition between mating types on the cultivation of Tuber melanosporum: Romeo and Juliet and the matter of space and time. Mycorrhiza 24:19–27. 10.1007/s00572-013-0551-6. [DOI] [PubMed] [Google Scholar]
- 140.Havenga M, Wingfield BD, Wingfield MJ, Roets F, Dreyer LL, Tatham CT, Duong TA, Wilken PM, Chen S, Aylward J. 2020. Mating strategy and mating type distribution in six global populations of the Eucalyptus foliar pathogen Teratosphaeria destructans. Fungal Genet Biol 137:103350. 10.1016/j.fgb.2020.103350. [DOI] [PubMed] [Google Scholar]
- 141.Clarke D, Woodlee G, McClelland C, Seymour T, Wickes B. 2001. The Cryptococcus neoformans STE11α gene is similar to other fungal mitogen-activated protein kinase kinase kinase (MAPKKK) genes but is mating type specific. Mol Microbiol 40:200–213. 10.1046/j.1365-2958.2001.02375.x. [DOI] [PubMed] [Google Scholar]
- 142.Samils N, Gioti A, Karlsson M, Sun Y, Kasuga T, Bastiaans E, Wang Z, Li N, Townsend JP, Johannesson H. 2013. Sex-linked transcriptional divergence in the hermaphrodite fungus Neurospora tetrasperma. Proc Biol Sci 280:20130862. 10.1098/rspb.2013.0862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Molliard M. 1904. Mycelium et forme conidienne de la morille. C R Acad Sci 138:516–517. [Google Scholar]
- 144.Molliard M. 1904. Forme conidienne et sclerotes de Morchella esculenta Pers. Rev Gen Bot 16:209–218. [Google Scholar]
- 145.Baran J, Boroń P. 2017. Two species of true morels (the genus Morchella, Ascomycota) recorded in the Ojców National Park (south Poland). Acta Mycol 52:1094. 10.5586/am.1094. [DOI] [Google Scholar]
- 146.Brock HD. 1951. Studies on the nutrition of Morchella Esculenta Fries. Mycologia 43:402–422. 10.2307/3755649. [DOI] [Google Scholar]
- 147.Liu W, Cai Y, He P, Zhang Y, Bian Y. 2016. Morphological and structural analysis of mitospores of Morchella importuna. J Fungal Res 14:157–161. [Google Scholar]
- 148.Masaphy S. 2010. Biotechnology of morel mushrooms: successful fruiting body formation and development in a soilless system. Biotechnol Lett 32:1523–1527. 10.1007/s10529-010-0328-3. [DOI] [PubMed] [Google Scholar]
- 149.Ower RD, Mills GL, Malachowski JA. June1986. Neogen Corporation, Cultivation of Morchella. US patent US4594809A.
- 150.Paden J. 1972. Imperfect states and the taxonomy of the Pezizales. Persoonia 6:405–414. [Google Scholar]
- 151.Volk TJ, Leonard TJ. 1990. Cytology of the life-cycle of Morchella. Mycol Res 94:399–406. 10.1016/S0953-7562(09)80365-1. [DOI] [Google Scholar]
- 152.Alvarado-Castillo G, Mata G, Sangabriel-Conde W. 2014. Understanding the life cycle of morels (Morchella spp.). Rev Mex Micol 40:47–50. [Google Scholar]
- 153.Yuan BH, Li H, Liu L, Du XH. 2021. Successful induction and recognition of conidiation, conidial germination and chlamydospore formation in pure culture of Morchella. Fungal Biol 125:285–293. 10.1016/j.funbio.2020.11.005. [DOI] [PubMed] [Google Scholar]
- 154.Wang Q, Wang S, Xiong CL, James TY, Zhang XG. 2017. Mating-type genes of the anamorphic fungus Ulocladium botrytis affect both asexual sporulation and sexual reproduction. Sci Rep 7:7932. 10.1038/s41598-017-08471-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Böhm J, Hoff B, O'Gorman CM, Wolfers S, Klix V, Binger D, Zadra I, Kürnsteiner H, Pöggeler S, Dyer PS, Kück U. 2013. Sexual reproduction and mating-type-mediated strain development in the penicillin-producing fungus Penicillium chrysogenum. Proc Natl Acad Sci U S A 110:1476–1481. 10.1073/pnas.1217943110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Carris LM, Peever TL, McCotter SW. 2015. Mitospore stages of Disciotis, Gyromitra and Morchella in the inland Pacific Northwest USA. Mycologia 107:729–744. 10.3852/14-207. [DOI] [PubMed] [Google Scholar]
- 157.Clowez P, Moreau PA. 2020. Morilles de France et d'Europe. Cap Régions Editions, Noyon, France. [Google Scholar]
- 158.Doehlemann G, Ökmen B, Zhu W, Sharon A. 2017. Plant pathogenic fungi, p 701–726. In Heitman J, Howlett BJ, Crous PW, Stukenbrock EH, James TY, Gow NAR (ed), The fungal kingdom. ASM Press, Washington, DC. [Google Scholar]
- 159.Maheshwari R. 1999. Microconidia of Neurospora crassa. Fungal Genet Biol 26:1–18. 10.1006/fgbi.1998.1103. [DOI] [PubMed] [Google Scholar]
- 160.Dahlstrom J, Smith J, Weber N. 2000. Mycorrhiza-like interaction by Morchella with species of the Pinaceae in pure culture synthesis. Mycorrhiza 9:279–285. 10.1007/PL00009992. [DOI] [Google Scholar]
- 161.Buscot F. 1994. Ectomycorrhizal types and endobacteria associated with ectomycorrhizas of Morchella elata (Fr.) Boudier with Picea abies (L.) Karst. Mycorrhiza 4:223–232. 10.1007/s005720050025. [DOI] [Google Scholar]
- 162.Chen LJ, Chai HM, Chen WM, Huang XQ, Zhao YC. 2014. Gene expressing difference in sclerotial formation of Morchella conica. Indian J Microbiol 54:274–283. 10.1007/s12088-014-0445-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kanwal HK, Reddy MS. 2012. The effect of carbon and nitrogen sources on the formation of sclerotia in Morchella spp. Ann Microbiol 62:165–168. 10.1007/s13213-011-0241-6. [DOI] [Google Scholar]
- 164.Liu Q, Zhao Z, Dong H, Dong C. 2018. Reactive oxygen species induce sclerotial formation in Morchella importuna. Appl Microbiol Biotechnol 102:7997–8009. 10.1007/s00253-018-9104-4. [DOI] [PubMed] [Google Scholar]
- 165.Greiss H. 1940. Befruchtungsarten bei Morchella. Jahrbücher Für Wissenschaftliche Botanik 89:245–253. [Google Scholar]
- 166.Masaphy S. 2005. External ultrastructure of fruit body initiation in Morchella. Mycol Res 109:508–512. 10.1017/s0953756204002126. [DOI] [PubMed] [Google Scholar]
- 167.Gäumann EA. 1952. The fungi. A description of their morphological features and evolutionary development. (Translated from German by Frederic Lyle Wynd.)H. Hafner Publishing, New York, NY. [Google Scholar]
- 168.Leslie JF, Klein KK. 1996. Female fertility and mating type effects on effective population size and evolution in filamentous fungi. Genetics 144:557–567. 10.1093/genetics/144.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gäumann E. 1964. Die Pilze. Grundzüge ihrer Entwicklungsgeschichte und Morphologie. Reihe der experimentellen Biologie IV. Birkhäuser Verlag, Basel, Switzerland. [Google Scholar]
- 170.Kirschner R. 2019. Sex does not sell: the argument for using the terms “anamorph” and “teleomorph” for fungi. Mycol Progress 18:305–312. 10.1007/s11557-018-1421-6. [DOI] [Google Scholar]
- 171.Frazer N. 1996. Control of growth and patterning in the fungal fruiting structure. A case for the involvement of hormones, p 156–181. In Chiu SW, Moore D (ed), Patterns in fungal development. Cambridge University Press, Cambridge, UK. [Google Scholar]
- 172.Horn BW, Gell RM, Singh R, Sorensen RB, Carbone I. 2016. Sexual reproduction in Aspergillus flavus sclerotia: acquisition of novel alleles from soil populations and uniparental mitochondrial inheritance. PLoS One 11:e0146169. 10.1371/journal.pone.0146169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Faretra F, Antonacci E, Pollastro S. 1988. Sexual behaviour and mating system of Botryotinia fuckeliana, teleomorph of Botrytis cinerea. Microbiology 134:2543–2550. 10.1099/00221287-134-9-2543. [DOI] [Google Scholar]
- 174.Volk TJ, Leonard TJ. 1989. Physiological and environmental studies of sclerotium formation and maturation in isolates of Morchella crassipes. Appl Environ Microbiol 55:3095–3100. 10.1128/aem.55.12.3095-3100.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Teichert I, Pöggeler S, Nowrousian M. 2020. Sordaria macrospora: 25 years as a model organism for studying the molecular mechanisms of fruiting body development. Appl Microbiol Biotechnol 104:3691–3704. 10.1007/s00253-020-10504-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Teichert I, Nowrousian M, Pöggeler S, Kück U. 2014. The filamentous fungus Sordaria macrospora as a genetic model to study fruiting body development. Adv Genet 87:199–244. 10.1016/B978-0-12-800149-3.00004-4. [DOI] [PubMed] [Google Scholar]
- 177.Raju NB. 2009. Neurospora as a model fungus for studies in cytogenetics and sexual biology at Stanford. J Biosci 34:139–159. 10.1007/s12038-009-0015-5. [DOI] [PubMed] [Google Scholar]