Abstract
There is an urgent need to identify the cellular and molecular mechanisms responsible for severe COVID-19 that results in death. We initially performed both untargeted and targeted lipidomics as well as focused biochemical analyses of 127 plasma samples and found elevated metabolites associated with secreted phospholipase A2 (sPLA2) activity and mitochondrial dysfunction in patients with severe COVID-19. Deceased COVID-19 patients had higher levels of circulating, catalytically active sPLA2 group IIA (sPLA2-IIA), with a median value that was 9.6-fold higher than that for patients with mild disease and 5.0-fold higher than the median value for survivors of severe COVID-19. Elevated sPLA2-IIA levels paralleled several indices of COVID-19 disease severity (e.g., kidney dysfunction, hypoxia, multiple organ dysfunction). A decision tree generated by machine learning identified sPLA2-IIA levels as a central node in the stratification of patients who died from COVID-19. Random forest analysis and least absolute shrinkage and selection operator–based (LASSO-based) regression analysis additionally identified sPLA2-IIA and blood urea nitrogen (BUN) as the key variables among 80 clinical indices in predicting COVID-19 mortality. The combined PLA-BUN index performed significantly better than did either one alone. An independent cohort (n = 154) confirmed higher plasma sPLA2-IIA levels in deceased patients compared with levels in plasma from patients with severe or mild COVID-19, with the PLA-BUN index–based decision tree satisfactorily stratifying patients with mild, severe, or fatal COVID-19. With clinically tested inhibitors available, this study identifies sPLA2-IIA as a therapeutic target to reduce COVID-19 mortality.
Keywords: COVID-19, Inflammation
Keywords: Cellular immune response, Molecular pathology
Introduction
Host resistance and disease tolerance are essential to mounting a successful defense against infections such as SARS-CoV-2. Up to 80% of individuals infected with SARS-CoV-2 are asymptomatic or develop mild-to-moderate symptoms. However, others progress to severe disease with life-threatening complications requiring hospitalization and specialized medical care. Severe COVID-19 correlates with respiratory symptoms (i.e., dyspnea, hyperpnea, hypoxemia, pulmonary infiltration) and concomitant multiple organ failure with disseminated intravascular coagulation (1). Consequently, there is an urgent need to elucidate the central molecular mechanisms underlying severe and fatal COVID-19 disease to develop targeted therapeutic approaches.
Early studies suggested that the host response to COVID-19 may be associated with an excessive proinflammatory response caused by a cytokine storm syndrome (CSS) (2). However, more recent studies show that persistent CSS is uncommon (3%–4%) in severe COVID-19 disease, where high-dose steroids benefit only a small proportion of individuals with organ failure (3, 4). Mounting evidence supports the idea that immunometabolic suppression, and not CSS, compromises host immunity, leading to unrestrained viral replication and severe COVID-19 (5, 6). Even when viral burdens decrease, pathologies including tissue and organ damage often remain (7).
Lipid metabolism plays an important role in determining COVID-19 outcomes. Early lipidomic studies (8, 9) revealed that severe COVID-19 modifies the circulating lipidome, with decreases in plasma levels of phospholipids and elevated quantities of lysophospholipids (lyso-PLs), unesterified unsaturated fatty acids (UFAs), and acylcarnitines. This lipidomic pattern suggests that severe COVID-19 may be accompanied by cellular or circulating phospholipase(s) that cleave intact phospholipids from cellular and mitochondrial membranes to form lyso-PLs and UFAs. Among phospholipases, the secreted phospholipase A2 (sPLA2) family includes 12 members with highly conserved characteristics, including low molecular weight (13–17 kDa), high Ca2+ levels for catalytic activity, and the presence of histidine/aspartic acid dyads in the catalytic site (10). Elevated sPLA2 group IIA (sPLA2-IIA) levels have been associated with various clinical conditions, including sepsis and systemic bacterial infections, adult respiratory disease syndrome (ARDS), atherosclerosis, cancer, and multiple organ trauma (10). Basal levels of circulating sPLA2-IIA in healthy humans are 1–3 ng/mL; however, sPLA2-IIA plasma concentrations can reach 250–500 ng/mL during acute sepsis (11).
Here, we identified lipidomic signatures of PLA2 hydrolysis and mitochondrial dysfunction that corresponded with COVID-19 severity in 127 patient plasma samples. Marked elevations in circulating sPLA2-IIA levels mirrored disease severity, particularly in deceased COVID-19 patients. Circulating sPLA2-IIA was catalytically active and paralleled several indices of disease severity, including hyperglycemia, kidney dysfunction, hypoxia, anemia, and multiple organ dysfunction. Importantly, 3 independent machine-learning approaches identified sPLA2-IIA as the central feature in predicting survivors versus nonsurvivors in cases of severe COVID-19. Since blood urea nitrogen (BUN) was identified alongside plasma sPLA2-IIA as a key stratification feature, we evaluated a novel PLA-BUN index as a prognostic biomarker of COVID-19–related mortality. Indeed, our PLA-BUN index successfully predicted COVID-19 mortality markedly better than did either feature alone. A validation cohort (n = 154) recapitulated the increases in plasma sPLA2-IIA in deceased COVID-19 patients, with the PLA-BUN index accurately stratifying patients with severe disease and deceased patients. Collectively, our study provides evidence to support a targeted therapeutic approach using clinically available sPLA2-IIA inhibitors to reduce COVID-19 mortality.
Results
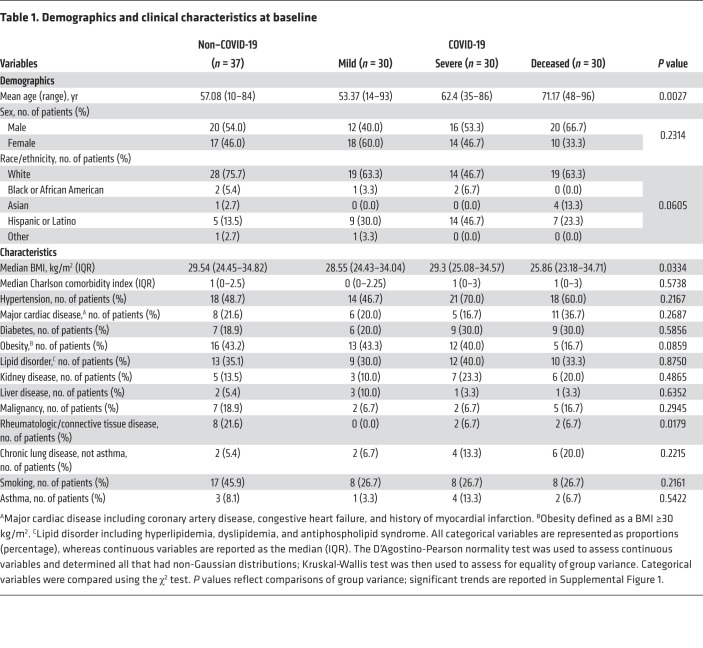
We analyzed an initial cohort of 127 patient plasma samples collected between May 2020 and July 2020 from Stony Brook University Medical Center (referred to hereafter as Stony Brook). The plasma sampling information is summarized in Supplemental Table 1 (supplemental material available online with this article; https://doi.org/10.1172/JCI149236DS1). Non–COVID-19 patients and those with mild COVID-19 had significantly shorter hospital stays than did patients with severe or fatal COVID-19. Overall, samples from patients with COVID-19 were collected at comparable time points during their hospital stay, whereas samples from non–COVID-19 patients were collected toward the end of the inpatient stay. The demographics and baseline clinical characteristics of the patients are shown in Table 1. Ages differed across groups, with deceased COVID-19 patients being older on average (Supplemental Figure 1). We noted no significant trends in BMI or obesity. The prevalence of various comorbidities was comparable across groups, except for a higher prevalence of rheumatologic disease among the non–COVID-19 patients. Patients with severe COVID-19 and deceased COVID-19 patients had presented with enhanced signs and symptoms of disease, such as an elevated national early warning score 2 (NEWS2) and 7-category ordinal scale score, pulmonary infiltration, and low oxygen saturation requiring oxygen therapy (Supplemental Table 2 and Supplemental Figure 1). Furthermore, patients with severe COVID-19 and those with fatal COVID-19 experienced more complications, such as cardiac arrests, acute kidney injury and renal failure, bacterial pneumonia, ARDS, and sepsis (Supplemental Figure 1). Patients who developed severe COVID-19 received various therapies to treat hypoxemia, ARDS, superimposed/ventilator-associated bacterial pneumonia, hyperinflammation, acute kidney injury/renal failure, and sepsis. A majority (70%–80%) of patients with severe or fatal COVID-19 received corticosteroid therapy, which has been shown to attenuate the expression of sPLA2-IIA and induce the synthesis of proteins that inhibit sPLA2-IIA activity (12–15). However, a larger number of patients received vasopressors in the deceased group, possibly because of the patients’ uncontrolled septic shock that eventually led to multiple organ failure and cardiac arrest (Supplemental Table 3).
Table 1. Demographics and clinical characteristics at baseline.
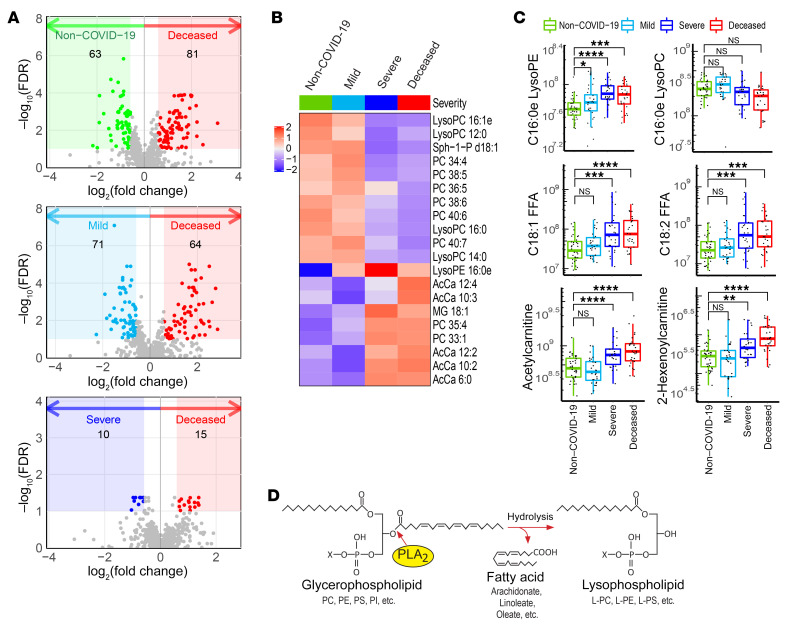
Initial studies were designed to identify lipidomic changes linked to COVID-19 outcomes. Untargeted lipidomic analysis of the plasma samples revealed that the most significant changes in the lipid profile occurred in the deceased COVID-19 patients (Figure 1A), with 181 unique molecules identified. Further analysis of the 20 most significant molecules showed enrichment in metabolites associated with acylcarnitine and phospholipid metabolism (Figure 1B). Specifically, several lysophosphatidylethanolamine (lyso-PE) molecular species typified by C16eLysoPE and UFAs such as linoleic (18:2) and oleic (18:1) acids were elevated in patients with severe COVID-19 and deceased COVID-19 patients (Figure 1C). Targeted lipidomics confirmed the untargeted analysis, revealing significant increases in major molecular species of lyso-PE and lysophosphatidylserine (lyso-PS), while showing no changes in lysophosphatidylcholine (lyso-PC) (Supplemental Figure 2). Hydrolysis of PE and PS, but not PC, to form corresponding lyso-PLs, together with mobilization of UFAs such as linoleic and oleic acids are hallmarks of catalysis by a secreted PLA2 isoform (16). Given the critical role of sPLA2-IIA in several related diseases, these data suggest that PLA2 hydrolysis (Figure 1D) may contribute to COVID-19 disease severity and mortality (10).
Figure 1. Untargeted lipidomics analysis and COVID-19 status.
Plasma samples from non–COVID-19 patients, those with mild COVID-19, those with severe COVID-19, and deceased COVID-19 patients were subjected to untargeted metabolomics analyses. Lipidome data were extracted from the metabolomics data set and analyzed. (A) Volcano plots show significant alterations in the lipidome of the deceased COVID-19 patients compared with that of the non–COVID-19 patients, patients with mild COVID-19, and patients with severe COVID-19. Colored areas highlight compounds with a FC of greater than 1.5 and a FDR of less than 0.1. (B) Heatmap of the top 20 metabolites whose abundances varied markedly across non–COVID-19 patients (Non–COVID-19), patients with mild COVID-19 (Mild), patients with severe COVID-19 (Severe), and deceased COVID-19 patients (Deceased). (C) Abundances of 2 lyso-PLs, 2 FFAs, and 2 short-chain acyl carnitines extracted from the untargeted lipid data. C16:0e lyso-PC in the upper right panel is an example of a PC-containing lysolipid that did not meet the FC and FDR criteria in A and is not a primary substrate of sPLA2-IIA. The other 5 compounds were selected from the colored regions in A (FDR <0.1) and may have resulted from the action of sPLA2-IIA. The levels in each panel were further compared using a 1-sided Wilcoxon test with Holm’s correction for multiple testing. For the box plots, the upper and lower bounds indicate the 75th (Q3) and 25th (Q1) percentiles, respectively; the line within the box indicates the median value; whiskers extend to values within 1.5 IQR (IQR, Q3–Q1) of the upper or lower bound; outlying values are shown between 1.5 and 3 IQR beyond the upper or lower bound. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. (D) Model of PLA2 reaction showing how PLA2 hydrolyzes the sn-2 position of the glycerol backbone of phospholipids to form lyso-PL and FFA products.
Patients with severe COVID-19 and deceased COVID-19 patients also showed elevations of short- and medium-chain acylcarnitines (acetyl and hexanoyl carnitines) as well as of mitochondrial DNA (mtDNA) (Figure 1C and Supplemental Figure 3B). Moreover, acetylcarnitine showed high areas under receiver operating characteristic (ROC) curves: 0.810 (95% CI, 0.694–0.925) for patients with mild versus severe COVID-19, and 0.849 (95% CI, 0.752–0.945) for those with mild versus fatal COVID-19 (Supplemental Figure 3A). Accordingly, acetylcarnitine may also be an indicator of COVID-19 severity and mortality. Circulating short-chain acylcarnitines (particularly acetylcarnitine) were recently reported as a prognostic biomarker of death during sepsis (17). Plasma concentrations of mitochondrially encoded cytochrome B (MT-CYB) and cytochrome c oxidase subunit III (MT-COX3) were also significantly elevated in deceased COVID-19 patients compared with non–COVID-19 patients and patients with mild COVID-19 (Supplemental Figure 3B). The increases in mtDNA levels we observed confirmed the results from a report linking mtDNA levels to COVID-19 severity and mortality (18). Together, these data also implicate defective fatty acid oxidation and mitochondrial dysfunction in COVID-19 severity and mortality.
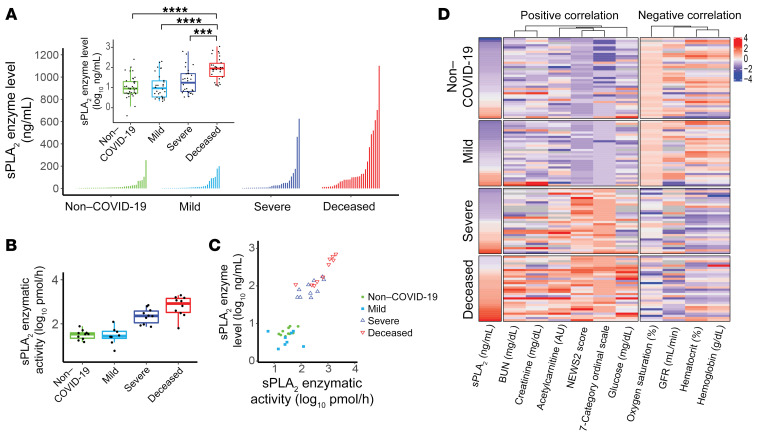
We subsequently quantified sPLA2-IIA levels in the 127 plasma samples to corroborate our lipidomic analyses. Figure 2A shows the distribution of sPLA2-IIA levels with markedly higher median values in deceased (89.3 ng/mL) and severe (17.9 ng/mL) COVID-19 patients compared with mild COVID-19 (9.3 ng/mL) and non–COVID-19 patients (8.9 ng/mL). Given that the non–COVID-19 patients in this cohort showed a higher prevalence of rheumatologic disease compared with patients with COVID-19 (Table 1 and Supplemental Figure 1), it is important to note that rheumatologic diseases significantly increase plasma sPLA2-IIA levels (19) when compared with levels in healthy control individuals (median = 0 ng/mL, IQR: 0–6.5) (20). Importantly, deceased COVID-19 patients had sPLA2-IIA levels as high as 1020 ng/mL, and their overall levels were 9.6- and 5.0-fold higher than those in patients with mild or severe COVID-19, respectively. Furthermore, we found that circulating sPLA2-IIA was catalytically active (Figure 2B), and its potential pathologic impact on organism-wide membranes was supported by a strong correlation (r2 = 0.84, P = 1.2 × 10–13) between sPLA2-IIA concentrations and enzymatic activity (Figure 2C).
Figure 2. Association between sPLA2-IIA and COVID-19 status.
(A) sPLA2-IIA levels were determined in 127 plasma samples and are shown here sorted within each group. The inset box plot compares the log-transformed data across groups and shows the medians and quartiles. Groups were compared using a 1-sided Wilcoxon test with Holm’s correction for multiple testing. ***P < 0.001 and ****P < 0.0001. Pairwise comparisons were computed from a linear model that included age and sex, and P values were adjusted for multiple comparisons. (B) sPLA2 enzymatic activity within plasma was assayed in a selected subset of samples. In the box plots in A and B, the upper and lower bounds designate the 75th (Q3) and 25th (Q1) percentiles, respectively; the line within the box indicates the median value; whiskers extend to values within 1.5 IQR (IQR, Q3–Q1) of the upper or lower bound; outlying values are shown between 1.5 and 3 IQR beyond the upper or lower bound. (C) Scatter plot shows plasma sPLA2-IIA levels versus sPLA2 activity in the selected subset of samples. Enzyme levels and activity were strongly correlated, indicating that plasma levels of sPLA2-IIA reflect the levels of active enzyme in the larger sample set. (D) A heatmap showing the significant Spearman correlations (FDR <0.05) between sPLA2-IIA and other clinical indices of disease severity. Indices that were positively or negatively correlated with sPLA2-IIA are as indicated. Indices with missing values above 25 were removed, and those with a skewness (absolute value) below 1.0 were log transformed. Index values were mean centered and scaled according to the SD. Blue to red represents low to high index values, with color intensity indicating the value magnitude (see the color scheme). Missing values are shown in gray.
Elevated levels of plasma sPLA2-IIA were significantly associated with several clinical indices (Figure 2D). Positive correlations with higher baseline NEWS2 and 7-category ordinal scale scores suggest a role for sPLA2-IIA in disease severity. The positive correlation of sPLA2-IIA with glucose levels highlights a potential link to dysregulated systemic inflammation. Accordingly, hyperglycemia is an important prognostic factor for COVID-19 and associates with a pro-oxidative/proinflammatory state (21). The positive correlations with creatinine and BUN levels (and a corresponding negative correlation with the glomerular filtration rate [GFR]) demonstrate how sPLA2-IIA levels may also reflect kidney dysfunction. Finally, the negative correlations of sPLA2 levels with hematocrit, hemoglobin levels, and oxygen saturation further support the notion that sPLA2-IIA may contribute to disease severity, including hypoxemia and multiple organ dysfunction (22).
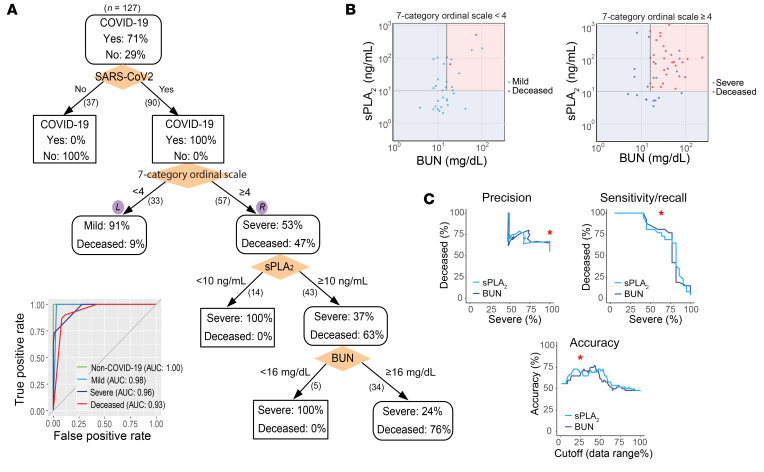
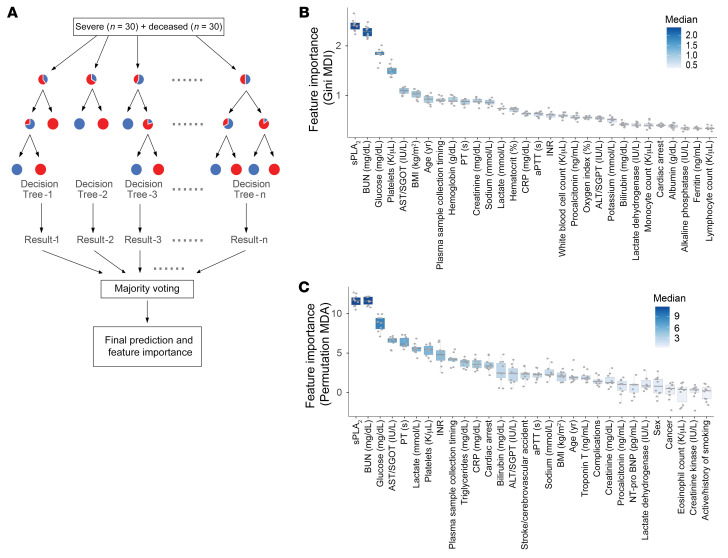
Three parallel, unbiased machine-learning analyses also identified elevated sPLA2-IIA as a critical risk factor for COVID-19 mortality. First, 80 clinical indices, including sPLA2-IIA levels, in the initial cohort of 127 patients were analyzed in a clinical decision tree model (23). A decision tree generated by recursive partitioning identified critical indices necessary to stratify the 4 patient groups with high accuracy (area under the ROC curve = 0.93–1.0, Figure 3A, inset). Patients positive for COVID-19 were stratified using the predictor “7-category ordinal scale” into “mild” and “severe or deceased,” with 91% and 100% accuracy, respectively. Surprisingly, the sPLA2-IIA level was identified as the central node that stratified survivors versus nonsurvivors in the “severe or deceased” category. Indeed, none of the patients with severe COVID-19 with sPLA2-IIA levels below 10 ng/mL died from COVID-19. In contrast, 75.4% of the patients in the “severe or deceased” category had sPLA2-IIA levels of 10 ng/mL or higher, and 63% of the patients in this subset died from this disease. Of the remaining patients with high sPLA2-IIA levels (≥10 ng/mL), all who had BUN levels below 16 mg/dL survived. Conversely, 76% of the patients with high sPLA2-IIA levels (≥10 ng/mL) and BUN levels of 16 mg/dL or higher died from the disease. The clinical decision tree developed in this study provides a framework to identify patients with COVID-19 at high risk of mortality. Of the 80 clinical indices measured, circulating levels of sPLA2-IIA emerged as the most important prognostic factor for COVID-19–related mortality. A cutoff value of 10 ng/mL or higher accurately predicted mortality in patients with severe COVID-19. Combining sPLA2-IIA and BUN levels into a PLA-BUN index (sPLA2-IIA ≥10 ng/mL and BUN ≥16 mg/dL) resulted in a parameter that predicted COVID-19 mortality more accurately than did using either feature alone (Figure 3, B and C).
Figure 3. Clinical decision tree predicting COVID-19 severity and mortality.
(A) Clinical decision tree model. Patients were classified on the basis of the indicated clinical indices (shown in orange diamonds) and boundary conditions (above the split arrows). The number of patients following each split is shown in parentheses beneath the split arrow (patients with missing index values were not included in the split). In each node, the percentages of patients in the corresponding categories are shown. The inset graph shows the area under the ROC curve, AUC, of the tree in determining each group designation (e.g., deceased vs. nondeceased patients). (B) Decision surface based on the sPLA2 and BUN boundary conditions in A. The left and right graphs show the results following application of the sPLA2 and BUN boundary conditions to the subsets of patients in these graphs (split following the 7-category ordinal scale), as indicated in A. (C) PLA-BUN index. The precision, sensitivity/recall, and accuracy in classifying patients with severe COVID-19 and deceased COVID-19 patients (7-category ordinal scale ≥4) by combining both decision boundary conditions of sPLA2 and BUN, as in B (i.e., the PLA-BUN index), are indicated with a red star in each graph, respectively. The corresponding classification results obtained by using the single index of sPLA2 (light blue curve) or BUN (dark blue curve) are shown with varying cutoff values in the corresponding data range (sPLA2, 3.4–1101.2 ng/mL; BUN, 5–242 mg/dL).
To separately assess and rank the relative importance of the 80 clinical indices (features) in predicting COVID-19 mortality, we used 2 additional machine-learning approaches. A random forest analysis ranked feature importance by removing a feature from the model and then evaluating the corresponding decrease in prediction accuracy obtained from an assembly of decision trees (n = 1000 each in 10 repeats, Figure 4A and ref. 24). This method identified sPLA2-IIA and BUN as the top 2 features, ranking significantly higher (P < 0.0001) than all other clinical indices (including age and BMI) in accurately predicting COVID-19–related mortality (Figure 4, B and C). A logistic regression model using least absolute shrinkage and selection operator (LASSO) also identified sPLA2-IIA and BUN as 2 of the 6 features selected among the 80 clinical variables for classifying patients with severe COVID-19 and deceased patients (Supplemental Table 4 and ref. 25). Supplemental Figure 4 illustrates that 3 independent machine-learning approaches found sPLA2-IIA and BUN to be key features for predicting COVID-19 mortality.
Figure 4. Feature importance ranking of clinical indices.
(A) The relative importance of the 80 clinical indices in separating the deceased patients from patients with severe COVID-19 (n = 30 each) was evaluated in a random forest analysis. In this random forest, an assembly of decision trees (n = 1000) was generated using randomly selected subsets of patients and features (clinical indices) to collectively arrive at the final model prediction (deceased vs. severe). The importance of a feature (i.e., clinical index) was evaluated by the decrease in prediction accuracy, when such a feature was excluded from the model, assessed on the basis of (B) Gini impurity following a node split (MDI) and (C) the permuted values of the feature (MDA). The feature importance was evaluated in 10 repeated random forest analyses. The top 30 features in B and C are shown (the color scheme is proportional to the importance score).
We obtained a second independent test cohort of plasma samples from patients with mild or severe COVID-19 and from deceased COVID-19 patients (n = 154) from both Banner-University Medical Center Tucson (referred to hereafter as Banner) and Stony Brook. The clinical decision tree that minimized the classification error in the first cohort was applied to this cohort. Consistently, sPLA2-IIA levels were significantly higher in the deceased patients than in those with severe or mild disease (Supplemental Figure 5A), with the PLA-BUN index–based decision tree model stratifying the patient groups (mild, severe, and deceased) with reasonably high accuracy (AUC = 0.72–0.99, Supplemental Figure 5B; also see the decision surface plot in Supplemental Figure 5C).
Given the potential for antibody-based assay cross-reactivity due to extensive homology across numerous sPLA2 isoforms, we validated the ELISA results using a proteomics assay with greater specificity. SomaScan (aptamer-based proteomics) analysis of the Banner samples validated the presence of the group IIA sPLA2 isoform in plasma from patients with severe COVID-19 and deceased COVID-19 patients compared with that from non–COVID-19 patients (Supplemental Figure 6). SomaScan data are compositional and therefore not quantitative like the ELISA-based quantitation we used for both our initial and validation cohorts.
In a subset of sequential samples (n = 46) for which each patient’s plasma was sampled at 2 time points, sPLA2-IIA levels tapered in the “late” compared with “early” samples in the mild and severe groups but remained relatively constant in the plasma from deceased patients (median sPLA2-IIA levels in the linear scale dropped by 60.2%, 62.0%, and 18.7%, respectively, in patients with mild COVID-19, patients with severe COVID-19, and deceased patients; Supplemental Figure 7). Given the limited number of sequential samples available, the differences in sPLA2-IIA levels between time points were not statistically significant for patients with mild or severe COVID-19. However, these trends are consistent with the possible role of sPLA2-IIA in COVID-19 mortality. Moreover, these data suggest that monitoring sPLA2-IIA levels in patients with severe COVID-19 may be clinically useful.
Discussion
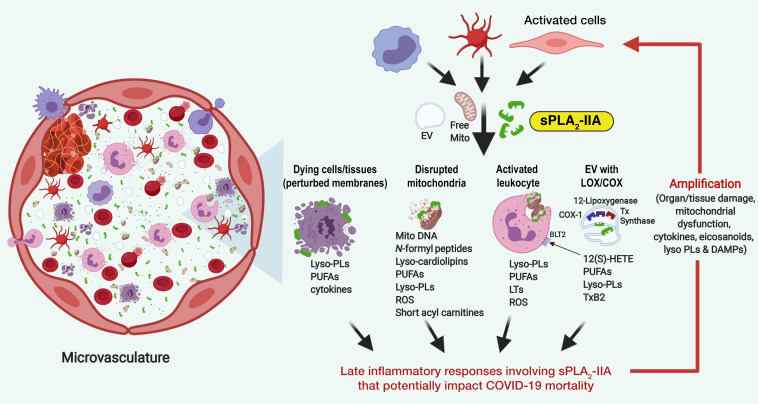
Early studies from the 1980s first described elevated circulating PLA2 activity in severe sepsis, mirroring the observations in our study (26–28). Indeed, patients who died from sepsis showed sustained and increasing trends in PLA2 activity, whereas surviving patients showed marked tapering. Although it was not possible to determine the temporal nature of sPLA2-IIA levels in these retrospective cohorts with a limited number of sequential samples, we found that deceased COVID-19 patients had persistently elevated sPLA2-IIA levels. When activated, sPLA2-IIA has direct and organism-wide pathogenic potential (10, 29–32), which may contribute to COVID-19 severity and mortality (Figure 5). During cell activation and the initiation of multiple cell death mechanisms, the anionic phospholipids PS and PE are externalized, exposing them to phospholipid hydrolysis by sPLA2-IIA (33). Hydrolysis of cellular membranes can broadly invoke tissue damage and organ dysfunction. Additionally, activated cells and damaged tissues and organs secrete extracellular mitochondria (29). Given that mitochondrial phospholipids are preferred substrates for sPLA2-IIA, our data suggest the occurrence of cell catalysis resulting in the release of mtDNA, acetylcarnitine, and several danger-associated molecular patterns (DAMPs) (34) during severe COVID-19 disease. Damaged mitochondria can then be internalized by bystander leukocytes to increase inflammatory mediators including lyso-PLs, UFAs, eicosanoids, and cytokines. sPLA2-IIA also hydrolyzes platelet-derived extracellular vesicles (EVs) to release cyclooxygenase, thromboxane synthase, and 12-lipoxygenase inflammatory eicosanoids (10). Given the known properties of sPLA2-IIA, our findings suggest that sPLA2-IIA may prolong and exacerbate tissue and organ damage during fatal COVID-19 disease.
Figure 5. Potential direct and organism-wide pathogenic mechanism of sPLA2-IIA.
Mechanisms include: (a) hydrolysis of cellular membranes that broadly invoke tissue damage and organ dysfunction; (b) hydrolysis of mitochondrial membranes leading to the release of mtDNA, acetylcarnitine, and DAMPs; (c) internalization of damaged mitochondria by bystander leukocytes to increase inflammatory mediators including lyso-PLs, UFAs, eicosanoids, and cytokines; and (d) hydrolysis of platelet-derived EVs to release eicosanoids, platelet-activating factor, and lyso-PLs.
As a multi-institutional retrospective observational study, there are limitations. Since the patients were selected on the basis of plasma sample availability, this study is subject to potential confounders and may not represent the general population. Additionally, clinical data availability was restricted to existing medical records, and there were missing values in the data set (especially in the second cohort). Moreover, temporal relationships were difficult to assess, as the plasma sampling was not standardized. Given that one of the test cohorts comprised plasma samples from 2 institutions, the patient populations may be heterogeneous. Finally, given the chaotic nature of COVID-19 management in early 2020, current standards of care may differ. Despite these limitations, this study consistently identified sPLA2-IIA as a previously unrecognized and plausible life-threatening mechanism underlying COVID-19 severity and mortality. Our study also offers a clinical blueprint for identifying those patients with COVID-19 who are at risk of death and identifies sPLA2-IIA as a potential therapeutic target. Our findings are further supported by the fact that high levels of sPLA2 also predict clinical disease severity in children, both in those in the acute phase of COVID-19 and in those who develop multisystem inflammatory syndrome (35).
Given the role of sPLA2-IIA during other forms of critical illness that are often complicated by multiple organ failure and high mortality rates, structurally diverse classes of sPLA2-IIA inhibitors have been developed (36–40). Although deemed safe for clinical use, clinical trials using a sPLA2-IIA inhibitor only improved survival in patients with sepsis when treatment was initiated within 18 hours of organ failure (39, 40). Further examination of the design of these studies revealed limitations: (a) patient selection criteria did not incorporate patient sPLA2-IIA levels and (b) circulating sPLA2-IIA levels were not reported in the studies. Therefore, inappropriate patient selection probably contributed to patient heterogeneity, resulting in negative findings. A recent study reported that, using a cutoff value of 25 ng/mL, sPLA2-IIA is highly sensitive and specific in detecting sepsis (20). Given that deceased COVID-19 patients in this study had elevated sPLA2-IIA levels (≥10 ng/mL), we propose incorporating sPLA2-IIA levels and the PLA-BUN index as prognostic clinical parameters. Our study further highlights the merit of exploring sPLA2-IIA inhibitors to reduce COVID-19–related morality.
Methods
Study design.
The study was designed according to Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) guidelines (41). COVID-19 was diagnosed by real-time reverse transcriptase PCR (RT-PCR), and COVID-19–positive patients were classified into 3 groups: (a) mild, in which patients had mild symptoms without pneumonia on imaging and were discharged from inpatient care; (b) severe, in which patients had respiratory tract or nonspecific symptoms, pneumonia confirmed on imaging, an oxygenation index below 94% on room air, and were discharged from inpatient care; and (c) deceased, in which the patients died during inpatient care. All plasma samples were collected during each patient’s hospital stay, except for the late (2nd) time points for patients with mild COVID-19 (Supplemental Figure 6). Only non–COVID-19 patients and those with mild COVID-19 with NEWS2 scores of 3 or lower were included in this study in order to exclude patients hospitalized for unrelated, possibly confounding major clinical presentations.
Sample processing and lipidomics analyses.
Frozen EDTA plasma samples were processed using Biosafety Level 2 conditions following the CDC Guidelines for the handling and processing of specimens associated with COVID-19. Metabolites were isolated from plasma via methanol-based extraction containing 10 μL Splash Lipidomix (Avanti Polar Lipids) and separated using reversed-phase chromatography as previously described by Najdekr et al. (43). Samples were analyzed using an ultra-high-performance liquid chromatography–electrospray ionization–tandem mass spectrometry (UHPLC–ESI–MS/MS) system (UHPLC: Thermo Horizon Vanquish Duo System from Thermo Horizon; MS: Thermo Exploris 480, both from Thermo Fisher Scientific, and separation was achieved using a Hypersil GOLD aQ UHPLC column (100 × 2.1 mm, 1.9 μm, Thermo Fisher Scientific, part no. 25302-102130) with mobile phases composed of water containing 0.1% formic acid and methanol containing 0.1% formic acid. Metabolites were eluted over a 15-minute gradient with the Exploris 480 operating in positive ion mode, at an ion transfer tube temperature of 350°C, a sheath gas of 45, an aux gas of 5, and a spray voltage of 4000. Mass spectrometric data for all samples were collected using dynamic exclusion and then aligned with pooled samples collected using the Thermo AquireX to achieve optimal metabolite identification in LipidSearch 4.0 and Thermo Compound Discoverer 2.3 software (both from Thermo Fisher Scientific).
Targeted lipidomics analysis was performed using an Agilent 1200 HPLC Tandem Thermo Quantum Ultra triple quadrupole mass spectrometer (Agilent Technologies) to quantify levels of major molecular species of lyso-PLs. C16, C18:1, C18:2, and C20:4 molecular species for lyso-PC, lyso-PE, and lyso-PS (Cayman Chemical) were used as standards and deuterated Splash Lipidomix (Avanti Polar Lipids) as internal standards. Lyso-PLs were separated using an Agilent Poroshell 120 EC-C18 1.9 μm (2.1 × 50 mm) with mobile phases composed of water containing 2 mM ammonium formate/0.1% formic acid (A) and methanol containing 1 mM ammonium formate/0.1% formic acid. Chromatographic gradient elution began at 40% A and remained there for the first minute, proceeding to 1% A at 6 minutes and remaining there for 10.5 minutes, before returning to 40% MPA over 1.5 minutes and remaining there until the end of the 20-minute run.
Determination of sPLA2-IIA concentrations.
sPLA2-IIA levels in plasma were determined by ELISA (Cayman Chemical). Plasma samples were diluted (1:20–1:800) and assayed in duplicate. sPLA2-IIA concentrations in plasma were calculated using standard curves.
Proteomics analysis.
Plasma samples from 82 Banner patients (21 non–COVID-19 patients, 30 patients with severe COVID-19, and 31 deceased COVID-19 patients) were analyzed using multiplexed SomaScan and Selex processing technology by SomaLogic. The aptamer-based SomaScan assay (44) and its performance characteristics (45, 46) have been previously described and were used for the targeted analysis of nearly 7000 proteins, one of which was sPLA2-IIA.
Enzymatic assay for sPLA2-IIA activity.
A subset of 34 patients’ samples from the initial Stony Brook cohort (n = 127) was selected for PLA2 activity analysis. sPLA2 activity was assayed by modifying techniques from Kramer and Pepinsky (47). Hydrolytic activity was determined in plasma samples from 34 patients (9 non–COVID-19 patients, 8 patients with mild COVID-19, 7 patients with severe COVID-19, and 10 deceased COVID-19 patients) representing a wide range of sPLA2-IIA levels. Assays were performed using 5 μL plasma in a final volume of 400 μL containing 50 mM Tris/NaCl, pH 8.5, with 5 mM CaCl2 and 5 nmol 3H-oleate–labeled E. coli phospholipids, and the plasma was mixture incubated for 30 minutes at 37°C. Lipids were extracted using a modified Bligh and Dyer method (48), and hydrolyzed fatty acids were separated from phospholipids using thin-layer chromatography (Silica Gel G) and a mobile phase of hexane/ether/formic acid (90:60:6, v/v/v), followed by visualization by iodine vapor relative to cold standards.
mtDNA quantification.
mtDNA was quantified in the same 34 patients’ samples used in the enzymatic assay. mtDNA was quantified by adapting methods from Scozzi et al (18). Using genes for human MT-CYB and MT-COX3, mtDNA was quantified in plasma samples from the same 34 patients (9 non–COVID-19 patients, 8 patients with mild COVID-19, 7 patients with severe COVID-19, and 10 deceased COVID-19 patients) as in the sPLA2 activity assay using an ABI 7900HT real-time PCR instrument (Applied Biosystems) in a 384-well format. Synthetic oligonucleotide copies of the MT-CYB and MT-COX3 genomic sequences (gBlock Gene Fragments from Integrated DNA Technologies) were included to generate a standard curve at 105, 104, 103, and 102 copies/μL. The following primer sequences were used: MT-CYB forward, 5′– ATGACCCCAATACGCAAAA-3′, MT-CYB reverse, 5′–CGAAGTTTCATCATGCGGAG-3′; and MT-COX3 forward, 5′–ATGACCCACCAATCACATGC-3′, MT-COX3 reverse, 5′–ATCACATGGCTAGGCCGGAG-3′.
Each diluted serum sample was compared with a control reaction of a gBlock standard, and the ΔCt was used to correct the calculated concentrations from triplicate reactions.
Statistics.
Untargeted lipidomics data were transformed, normalized, and analyzed using MetaboAnalyst 4.0. The Benjamini-Hochberg procedure was used to control the FDR, and the molecules with a FDR of 0.1 or less and an absolute log2 fold change (FC) of 1.5 or greater were considered to be significant and biologically relevant. Individual metabolites, sPLA2 levels, sPLA2 activity, and mtDNA levels were compared between groups with nonparametric Mann-Whitney Wilcoxon tests at an α level of 0.05, with an additional Holm adjustment for multiple comparisons. Spearman’s correlations between sPLA2 levels and clinical indices were computed in R. ROC curves, AUC, and CIs were generated using the R packages ROCR and pROC.
Decision tree, random forest analyses, and logistic regression analysis (LASSO).
Eighty initial clinical indices were used as input variables to build a predictive model (i.e., decision tree) by recursive partitioning, using the classification and regression trees (CART) algorithm (23) implemented in the R package RPART. The tree model identified a set of predictive features (branch conditions) that best classified the initial cohort of 127 patients into the 4 groups: non–COVID-19, patients with mild COVID-19, patients with severe COVID-19, and deceased COVID-19 patients. The tree split points were determined by the Gini index with a minimum leaf size of 10. A 10-fold cross-validation method was used to tune the tree model and evaluate its prediction accuracy. To avoid overfitting, the tree was pruned back to the smallest size, while minimizing the cross-validated error. The classification accuracy of the tree to determine each group membership (e.g., deceased vs. nondeceased) was assessed using the area under the ROC curve.
To further evaluate the relative feature importance in accurately separating patients with severe COVID-19 and deceased COVID-19 patients, a random forest analysis was performed using the R package Random Forest (24). An assembly of 1000 random decision trees was constructed in each forest, and 10 forests were constructed in replicate. The importance of a given feature (i.e., 1 of the 80 clinical indices) was assessed by the decrease of prediction accuracy when such a feature was omitted in the model, based on 2 measurement metrics: the Gini importance or mean decrease impurity (MDI) and the permutation importance or mean decrease accuracy (MDA).
A logistic regression model was built to classify patients with severe COVID-19 and deceased COVID patients using the glmnet package in R (25). The same 80 initial clinical indices used to construct the decision tree were used as the input variables; 48 indices with fewer than 10 missing values (across the total of 60 patients with severe COVID-19 and deceased COVID-19 patients in the initial cohort of 127 patients) were further reserved to construct the LASSO model, with the missing values imputed using the imputeMissing function (42). The classification accuracy of the LASSO model was determined by 10-fold cross-validation.
Study approval.
This retrospective study initially analyzed 127 plasma samples from patients hospitalized at Stony Brook from January 2020 to July 2020. This study was approved by the central IRB at Stony Brook University (IRB 2020-00423). For the second independent cohort, we analyzed 154 plasma samples from patients at Stony Brook (n = 98) and Banner (n = 56), who were hospitalized from January 2020 to November 2020. The Banner study was approved by the University of Arizona Human Subjects Protection Program (IRB 2007847180).
Author contributions
FHC conceived and designed the study. JKY, RRK, MDP, JMS, AJS, YAH, and SS prepared IRB documentation, coordinated the collection of samples, and handled the logistics of sample transfers between Stony Brook University Medical Center and the University of Arizona. KL, SP, CB, and SG prepared IRB documentation, coordinated the collection of samples, and handled logistics of sample transfers between Banner – University Medical Center Tucson and the University of Arizona. JKY, SG, and MDP collected the clinical data. JMS, AJS, and QW performed untargeted and targeted metabolomics and data analysis. MCS and SS performed the sPLA2 activity assay. AJS, MMZ, and TFC performed sPLA2 ELISAs. RS performed mtDNA analysis. XW, GY, BH, LJ, QW, and HHZ conducted the statistical analysis and modeling. FHC and JMS wrote the initial draft of the manuscript. CEM, XW, JKY, MDP, YAH, AJS, CL, BH, and GY contributed to revisions and discussion.
Supplementary Material
Acknowledgments
The authors thank the Stony Brook Medicine Biobank for the procurement and distribution of plasma samples from COVID-19 and non–COVID hospitalized patients. We acknowledge the University of Arizona Data Science Institute for their help in applying machine-learning modeling. This work was supported by the following grants: NIH R01 AT008621 (to FHC); National Cancer Institute (NCI), NIH P01-CA97132 (to YAH); NIH AI136934 and AI125770 (to MDP); NIH R35 GM126922 (to CM); National Institute of Allergy and Infectious Diseases (NIAID), NIH AI135108 (to SG); Merit Review Grant, Veterans Affairs I01BX002924 (to MDP); Technology and Research Initiative Fund, BIO5 Institute (to FHC); and Cancer Center Support Grant (CCSG), NIH P30 CA023074 (to JMS).
Version 1. 08/24/2021
In-Press Preview
Version 2. 10/01/2021
Electronic publication
Funding Statement
Yasuf A. Hannun
Maurizio Del Poeta
Maurizio Del Poeta
Charles E. McCall
Floyd H. Chilton
Maurizio Del Poeta
Floyd H. Chilton
Stefano Guerra
Footnotes
Conflict of interest: Maurizio Del Poeta is a co-founder and chief scientific officer of MicroRid Technologies Inc.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(19):e149236.https://doi.org/10.1172/JCI149236.
Contributor Information
Justin M. Snider, Email: justinsnider@arizona.edu.
Jeehyun Karen You, Email: jeehyunkaren.you@stonybrook.edu.
Xia Wang, Email: xiawang@arizona.edu.
Ashley J. Snider, Email: ashleysnider@arizona.edu.
Brian Hallmark, Email: bhallmark@statlab.bio5.org.
Manja M. Zec, Email: manjazec@arizona.edu.
Michael C. Seeds, Email: mseeds@wakehealth.edu.
Susan Sergeant, Email: ssergean@wakehealth.edu.
Laurel Johnstone, Email: laureljo@arizona.edu.
Qiuming Wang, Email: qiumingwang@email.arizona.edu.
Ryan Sprissler, Email: ryans1@arizona.edu.
Tara F. Carr, Email: tcarr@deptofmed.arizona.edu.
Karen Lutrick, Email: klutrick@email.arizona.edu.
Sairam Parthasarathy, Email: spartha1@email.arizona.edu.
Christian Bime, Email: cbime@email.arizona.edu.
Chiara Luberto, Email: chiara.luberto@stonybrookmedicine.edu.
Richard R. Kew, Email: Richard.Kew@stonybrookmedicine.edu.
Yusuf A. Hannun, Email: Yusuf.Hannun@stonybrookmedicine.edu.
Stefano Guerra, Email: stefano@email.arizona.edu.
Charles E. McCall, Email: chmccall@wfubmc.edu.
Guang Yao, Email: guangyao@arizona.edu.
Maurizio Del Poeta, Email: maurizio.delpoeta@stonybrook.edu.
Floyd H. Chilton, Email: fchilton@arizona.edu.
References
- 1.Liao D, et al. Haematological characteristics and risk factors in the classification and prognosis evaluation of COVID-19: a retrospective cohort study. Lancet Haematol. 2020;7(9):e671–e678. doi: 10.1016/S2352-3026(20)30217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fajgenbaum DC, June CH. Cytokine Storm. N Engl J Med. 2020;383(23):2255–2273. doi: 10.1056/NEJMra2026131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sinha P, et al. Is a “cytokine storm” relevant to COVID-19? JAMA Intern Med. 2020;180(9):1152–1154. doi: 10.1001/jamainternmed.2020.3313. [DOI] [PubMed] [Google Scholar]
- 4.Horby P, et al. Dexamethasone in hospitalized patients with Covid-19 — Preliminary Report. N Engl J Med. 2021;384(8):693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Remy KE, et al. Severe immunosuppression and not a cytokine storm characterizes COVID-19 infections. JCI Insight. 2020;5(17):e140329. doi: 10.1172/jci.insight.140329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng HY, et al. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol Immunol. 2020;17(5):541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nienhold R, et al. Two distinct immunopathological profiles in autopsy lungs of COVID-19. Nat Commun. 2020;11(1):5086. doi: 10.1038/s41467-020-18854-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen B, et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182(1):59–72. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu D, et al. Plasma metabolomic and lipidomic alterations associated with COVID-19. Natl Sci Rev. 2020;7(7):1157–1168. doi: 10.1093/nsr/nwaa086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dore E, Boilard E. Roles of secreted phospholipase A(2) group IIA in inflammation and host defense. Biochim Biophys Acta Mol Cell Biol Lipids. 2019;1864(6):789–802. doi: 10.1016/j.bbalip.2018.08.017. [DOI] [PubMed] [Google Scholar]
- 11.van Hensbergen VP, et al. Type IIA secreted phospholipase A2 in host defense against bacterial infections. Trends Immunol. 2020;41(4):313–326. doi: 10.1016/j.it.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Nakano T, et al. Glucocorticoids suppress group II phospholipase A2 production by blocking mRNA synthesis and post-transcriptional expression. J Biol Chem. 1990;265(21):12745–12748. doi: 10.1016/S0021-9258(19)38408-X. [DOI] [PubMed] [Google Scholar]
- 13.Miele L. New weapons against inflammation: dual inhibitors of phospholipase A2 and transglutaminase. J Clin Invest. 2003;111(1):19–21. doi: 10.1172/JCI17506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kol S, et al. Glucocorticoids suppress basal (but not interleukin-1-supported) ovarian phospholipase A2 activity: evidence for glucocorticoid receptor-mediated regulation. Mol Cell Endocrinol. 1998;137(2):117–125. doi: 10.1016/S0303-7207(98)00003-3. [DOI] [PubMed] [Google Scholar]
- 15.Kuzniatsova O. Dexamethasone inhibits secretory and cytosolic phospholipases A2 in alveolar macrophages in acute lung injury. Eur Respir J. 2013;42(Suppl 57):P3863 [Google Scholar]
- 16.Dennis EA, et al. Phospholipase A2 enzymes: physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chem Rev. 2011;111(10):6130–6185. doi: 10.1021/cr200085w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung KP, et al. Increased plasma acetylcarnitine in sepsis is associated with multiple organ dysfunction and mortality: a multicenter cohort study. Crit Care Med. 2019;47(2):210–218. doi: 10.1097/CCM.0000000000003517. [DOI] [PubMed] [Google Scholar]
- 18.Scozzi D, et al. Circulating mitochondrial DNA is an early indicator of severe illness and mortality from COVID-19. JCI Insight. 2021;6(4):e143299. doi: 10.1172/jci.insight.143299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boilard E, et al. A novel anti-inflammatory role for secretory phospholipase A2 in immune complex-mediated arthritis. EMBO Mol Med. 2010;2(5):172–187. doi: 10.1002/emmm.201000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berg E, et al. Measurement of a novel biomarker, secretory phospholipase A2 group IIA as a marker of sepsis: a pilot study. J Emerg Trauma Shock. 2018;11(2):135–139. doi: 10.4103/JETS.JETS_29_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ceriello A. Hyperglycemia and the worse prognosis of COVID-19. Why a fast blood glucose control should be mandatory. Diabetes Res Clin Pract. 2020;163:108186. doi: 10.1016/j.diabres.2020.108186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hariyanto TI, Kurniawan A. Anemia is associated with severe coronavirus disease 2019 (COVID-19) infection. Transfus Apher Sci. 2020;59(6):102926. doi: 10.1016/j.transci.2020.102926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Breiman L, et al. Classification and Regression Trees. Chapman & Hall; 1984. [Google Scholar]
- 24.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 25.Friedman J, et al. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 26.Vadas P. Elevated plasma phospholipase A2 levels: correlation with the hemodynamic and pulmonary changes in gram-negative septic shock. J Lab Clin Med. 1984;104(6):873–881. [PubMed] [Google Scholar]
- 27.Guidet B, et al. Secretory non-pancreatic phopholipase A2 in severe sepsis: relation to endotoxin, cytokines and thromboxane B2. Infection. 1996;24(2):103–108. doi: 10.1007/BF01713312. [DOI] [PubMed] [Google Scholar]
- 28.Vadas P, et al. A predictive model for the clearance of soluble phospholipase A2 during septic shock. J Lab Clin Med. 1991;118(5):471–475. [PubMed] [Google Scholar]
- 29.Boudreau LH, et al. Platelets release mitochondria serving as substrate for bactericidal group IIA-secreted phospholipase A2 to promote inflammation. Blood. 2014;124(14):2173–2183. doi: 10.1182/blood-2014-05-573543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hurt-Camejo E, et al. Phospholipase A(2) in vascular disease. Circ Res. 2001;89(4):298–304. doi: 10.1161/hh1601.095598. [DOI] [PubMed] [Google Scholar]
- 31.Chilton F. Would the real role(s) for secretory PLA2s please stand up. J Clin Invest. 1996;97(10):2161–2162. doi: 10.1172/JCI118654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murakami M, et al. Regulatory functions of phospholipase A2. Crit Rev Immunol. 2017;37(2–6):127–195. doi: 10.1615/CritRevImmunol.v37.i2-6.20. [DOI] [PubMed] [Google Scholar]
- 33.Atsumi G, et al. The perturbed membrane of cells undergoing apoptosis is susceptible to type II secretory phospholipase A2 to liberate arachidonic acid. Biochim Biophys Acta. 1997;1349(1):43–54. doi: 10.1016/s0005-2760(97)00082-9. [DOI] [PubMed] [Google Scholar]
- 34.Roh JS, Sohn DH. Damage-associated molecular patterns in inflammatory diseases. Immune Netw. 2018;18(4):e27. doi: 10.4110/in.2018.18.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuypers FA, et al. Secretory phospholipase A2 in SARS-CoV-2 infection and multisystem inflammatory syndrome in children (MIS-C) Exp Biol Med (Maywood) doi: 10.1177/15353702211028560. [published online July 13, 2021]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan TL, Goh YY. The role of group IIA secretory phospholipase A2 (sPLA2-IIA) as a biomarker for the diagnosis of sepsis and bacterial infection in adults-A systematic review. PLoS One. 2017;12(7):e0180554. doi: 10.1371/journal.pone.0180554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson BO, et al. Phospholipase A2 regulates critical inflammatory mediators of multiple organ failure. J Surg Res. 1994;56(2):199–205. doi: 10.1006/jsre.1994.1032. [DOI] [PubMed] [Google Scholar]
- 38.Corke C, et al. Circulating secretory phospholipase A2 in critical illness--the importance of the intestine. Crit Care Resusc. 2001;3(4):244–249. [PubMed] [Google Scholar]
- 39.Zeiher BG, et al. LY315920NA/S-5920, a selective inhibitor of group IIA secretory phospholipase A2, fails to improve clinical outcome for patients with severe sepsis. Crit Care Med. 2005;33(8):1741–1748. doi: 10.1097/01.CCM.0000171540.54520.69. [DOI] [PubMed] [Google Scholar]
- 40.Abraham E, et al. Efficacy and safety of LY315920Na/S-5920, a selective inhibitor of 14-kDa group IIA secretory phospholipase A2, in patients with suspected sepsis and organ failure. Crit Care Med. 2003;31(3):718–728. doi: 10.1097/01.CCM.0000053648.42884.89. [DOI] [PubMed] [Google Scholar]
- 41.von Elm E, et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Ann Intern Med. 2007;147(8):573–577. doi: 10.7326/0003-4819-147-8-200710160-00010. [DOI] [PubMed] [Google Scholar]
- 42. imputeMissings: Impute missing values in a predictive context. Version 0.0.3. Meire M, et al; 2016. Accessed July 21, 2021. https://CRAN.R-project.org/package=imputeMissings.
- 43.Najdekr L, et al. Collection of untargeted metabolomic data for mammalian urine applying HILIC and reversed phase ultra performance liquid chromatography methods coupled to a Q exactive mass spectrometer. Methods Mol Biol. 2019;1996:1–15. doi: 10.1007/978-1-4939-9488-5_1. [DOI] [PubMed] [Google Scholar]
- 44.Gold L, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010;5(12):e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim CH, et al. Stability and reproducibility of proteomic profiles measured with an aptamer-based platform. Sci Rep. 2018;8(1):8382. doi: 10.1038/s41598-018-26640-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Candia J, et al. Assessment of variability in the SOMAscan assay. Sci Rep. 2017;7(1):14248. doi: 10.1038/s41598-017-14755-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kramer RM, Pepinsky RB. Assay and purification of phospholipase A2 from human synovial fluid in rheumatoid arthritis. Methods Enzymol. 1991;197:373–381. doi: 10.1016/0076-6879(91)97163-s. [DOI] [PubMed] [Google Scholar]
- 48.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.