Alternative splicing leads to diverse mature RNAs from a given gene. Here, we review current knowledge of alternative splicing triggered by a variation in environmental temperature in plants.
Keywords: Alternative splicing, ambient temperature, cold, heat, plants, stress adaptation
Abstract
Plants have evolved numerous molecular strategies to cope with perturbations in environmental temperature, and to adjust growth and physiology to limit the negative effects of extreme temperature. One of the strategies involves alternative splicing of primary transcripts to encode alternative protein products or transcript variants destined for degradation by nonsense-mediated decay. Here, we review how changes in environmental temperature—cold, heat, and moderate alterations in temperature—affect alternative splicing in plants, including crops. We present examples of the mode of action of various temperature-induced splice variants and discuss how these alternative splicing events enable favourable plant responses to altered temperatures. Finally, we point out unanswered questions that should be addressed to fully utilize the endogenous mechanisms in plants to adjust their growth to environmental temperature. We also indicate how this knowledge might be used to enhance crop productivity in the future.
Introduction
In natural environments, plants are repeatedly or continuously exposed to a wide range of temperatures. Temperature is a key factor affecting plant growth and development. Apart from the daily fluctuations in ambient temperature (which additionally changes throughout the lifetime of a plant), plants are often exposed to ‘unpredictable’ extreme temperatures, such as unusual cold or heat, during different times of the year. The current rise in average global temperature has a huge impact on plant growth and agricultural production, often leading to a decline in crop yield. Due to thermal stresses, every degree Celsius rise in temperature above current temperature may potentially decrease yield by 3–7% for major crops like wheat, rice, and maize (Zhao et al. 2017). Thus, from an agronomic point of view, it is essential to unravel how plants perceive and respond to fluctuations in temperature. As plants are sessile organisms they cannot relocate to escape a stress. Instead, to avoid or minimize the detrimental effects of stressful conditions such as high or low temperature, plants rely on mechanisms established during evolution to effectively respond and ensure survival and reproductive growth. One such mechanism is alternative splicing (AS) of precursor mRNA (pre-mRNA) in response to environmental perturbations including changes in temperature (Filichkin et al., 2015a; Shang et al., 2017; Laloum et al., 2018). Recent studies suggest that alternative pre-mRNA splicing may serve as a ‘molecular thermometer’ in the temperature-controlled adaptation of plants, allowing them to appropriately adjust transcript abundance (Capovilla et al., 2015). AS is a common phenomenon in many organisms. For example, primary transcripts of 60% of the genes in Drosophila melanogaster undergo AS (Graveley et al., 2011), and in humans, primary transcripts of more than 95% of genes are affected by differential splicing (Pan et al., 2008). In Arabidopsis, the increased utilization of next-generation sequencing technologies in recent years has shown that up to 70% of the plant multi-exon genes generate more than a single transcript by AS, and that previous technical approaches such as microarray-based analyses had underestimated the proportion of AS-affected genes (Calixto et al., 2018; Li et al., 2020a).
Although temperature regulates diverse biological processes in plants and triggers AS events, the molecular mechanisms controlling temperature-dependent AS are still poorly understood. Here, we review the current knowledge about the effects of environmental temperature (low, high, and intermediate) on AS in plants. First, we briefly describe the mechanism of splicing regulation. We then highlight examples of how AS enables favourable plant responses to altered temperatures. Finally, we point out currently unsolved questions to be addressed, and outline how this knowledge might be used to enhance crop productivity in the future.
The mechanism of alternative splicing
Constitutive splicing of pre-mRNA is controlled by a large ribonucleoprotein complex, called the spliceosome, which leads to the removal of non-coding introns to join the flanking exons and, thereby, assembles one mature transcript (Deckert et al., 2006; Moore and Proudfoot, 2009; Kornblihtt et al., 2013). The spliceosome core components are supported by the serine/arginine-rich (SR) proteins and heterogeneous nuclear ribonucleoproteins that are responsible for splice site selection by binding to cis-regulatory elements located in exons or introns, thus activating or repressing the splicing process (Reddy, 2007; Kornblihtt et al., 2013). Selection of alternative splice sites in a single type of pre-mRNA leads to AS, and thereby to the production of multiple mature mRNA isoforms (Reddy, 2007). Five basic types of AS are observed in plants depending on the selection of splice sites at the same pre-mRNA. Elimination of splice site selection generates intron retention (IR), exon skipping, or mutual exclusion of exons, while the selection of distinct splice sites result in the generation of alternative 5′ or 3′ splice sites (Fig. 1A) (Reddy et al., 2013; Posé et al., 2013). In this way, the same single pre-mRNA may undergo different AS events resulting in the formation of two or more mature transcripts encoding different protein isoforms, and these may functionally differ from one another (Nilsen and Graveley, 2010).
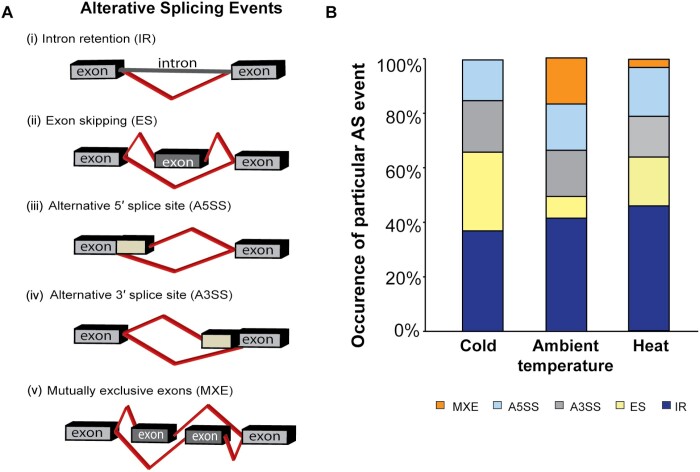
Fig. 1.
Alternative splicing (AS) events in plants. (A) Types of splicing events (see also Verhage et al., (2017)). (B) Percentage of different AS events reported to date in response to cold, changes in ambient temperature, and heat in plants. Data were extracted from PubMed Central, December 2020 (https://pubmed.ncbi.nlm.nih.gov/). A3SS, alternative 3′ splice site selection; A5SS, alternative 5′ splice site selection; ES, exon skipping; IR, intron retention; MXE, mutual exclusion of exons.
AS has the potential to change the number of protein variants (and, hence, their biochemical properties) encoded by a given genome. It can lead to the production of proteins with or without functional domains, thereby resulting in protein variants with a loss or gain of function, or to proteins with changed biochemical or cellular properties that may affect subcellular localization, stability, and function. In plants, a large number of transcripts of genes are alternatively spliced during stressful and non-stressful changes in the environment. Experimental evidence shows that those changes in response to environmental stresses can be beneficial for plants, allowing them to rapidly adjust the transcript abundance of essential genes including key regulators involved in stress response, thereby promoting stress tolerance (Fig. 1B). In plants, approximately 40% of all AS events in response to changes in temperature are due to IR, making it the most common type of AS (Fig. 1B). IR was found to be the predominant AS event in plants exposed to cold, moderate temperature changes, and heat stress, while the occurrence of other AS events varies between different temperature conditions.
Three modes of action of AS transcripts have been reported in plants in response to temperature variations (Fig. 2). The first is peptide interference by the formation of small interfering peptides (siPEPs). In this case, the alternatively spliced mRNA is translated into a truncated protein, the siPEP, which forms a non-functional heterodimer with an otherwise functional protein, which may be a transcription factor (TF) (Seo et al., 2011a; Staudt and Wenkel, 2011). The siPEPs have dimerization domains but lack other functional domains, such as DNA-binding and/or transcription regulation domains (Yun et al., 2008; Seo et al., 2011a). siPEPs can, therefore, act as competitive inhibitors of the targeted TFs (Seo et al., 2011a; Staudt and Wenkel 2011). In contrast to RNA interference (RNAi) mediated by small interfering RNAs (siRNAs), or micro-RNAs (miRNAs) that interact with mRNAs to block their translation or induce their cellular degradation, peptide interference functions at the protein level (Ramachandran and Chen 2008; Staudt and Wenkel 2011; Naqvi et al., 2012).
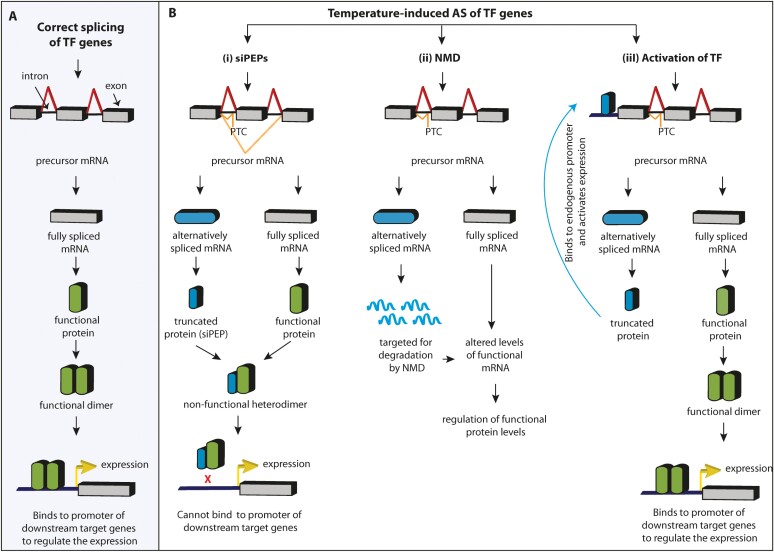
Fig. 2.
Role of temperature-induced alternative splicing (AS) and mode of action of different splice variants. (A) Regulation of down-stream target genes when the transcription factor (TF) transcript undergoes constitutive splicing to form a functional protein. A functional homodimer of the TF is formed that binds to the promoters of target genes to activate or repress their expression. (B) Temperature-induced AS can lead to three types of TF regulation. (i) Peptide interference by the formation of small interfering peptides (siPEPs). The alternatively spliced mRNA leads to a truncated protein that functions as an siPEP by forming a non-functional heterodimer with the functional protein; this inhibits the TF from binding to the promoters of target genes to affect their expression. (ii) Nonsense-mediated decay (NMD) of alternatively spliced transcripts. Many splice variants that contain premature termination codons (PTCs) are targeted for degradation by NMD, thereby altering the transcript levels available for translation to form the functional TF protein. (iii) Activation of the TF. AS leads to the formation of a truncated protein that has the ability to bind to the promoter of its own gene and modify its expression. Red lines represent constitutive splicing and orange lines represent temperature-induced AS.
The second mode of action is nonsense-mediated decay (NMD) in which alternatively spliced transcripts are degraded. Many AS events, in particular IR, lead to the introduction of a premature translation termination codon (PTC) in the spliced transcript, thereby limiting the amount of transcript encoding functional protein. Splice variants containing a PTC are often targeted for degradation by NMD (Kurihara et al., 2009; Rebbapragada and Lykke-Andersen 2009; Palusa and Reddy 2010). IR thus represents a molecular mechanism to down-regulate the functional output of a gene that is already actively transcribed, thereby bypassing the need for regulating its transcription, which may be a slower response. In the case of Arabidopsis it has been predicted that about 13% of the intron-containing genes are targeted by NMD (Kalyna et al., 2012), while in humans up to one-third of the transcripts generated by AS are degraded by NMD (García-Moreno and Romão, 2020).
The third mode of action is activation of a TF. In this scenario, which has been reported for heat shock transcription factor A2 (HSFA2) in Arabidopsis, AS leads to the formation of a truncated, and C-terminally modified, TF protein with an extra leucine-rich motif. The modified TF has the capacity to bind to its own promoter to activate HSFA2 transcription (Liu et al., 2013).
Alternative splicing in response to low temperature
Cold is one of the major abiotic stresses affecting growth and development in higher plants, leading to reduced crop yields (Xin and Browse, 2000). Plants differ in their tolerance to chilling (0–15 °C) and freezing (<0 °C) temperatures. Many important tropical and subtropical crop plants like rice, corn, and tomato are sensitive to chilling stress whereas temperate crops like wheat, barley, and rye are better adapted to survive freezing temperatures (Chinnusamy et al., 2007; Zhu et al., 2007). In higher plants, low temperature (4–15 °C) has a huge impact on splicing regulation (Fig. 3) (Calixto et al., 2018; Chechanovsky et al., 2019; Li et al., 2020a, b). AS in response to cold appears to occur very rapidly (in the range of minutes), and small shifts in the cold temperature range result in changes in the number of transcripts undergoing AS (Calixto et al., 2018; Gallegos, 2018). It is estimated that approximately 33% of the cold-responsive transcripts are alternatively spliced (Calixto et al., 2019). Here, we summarize recent findings that describe how low temperature induces AS in different plants.
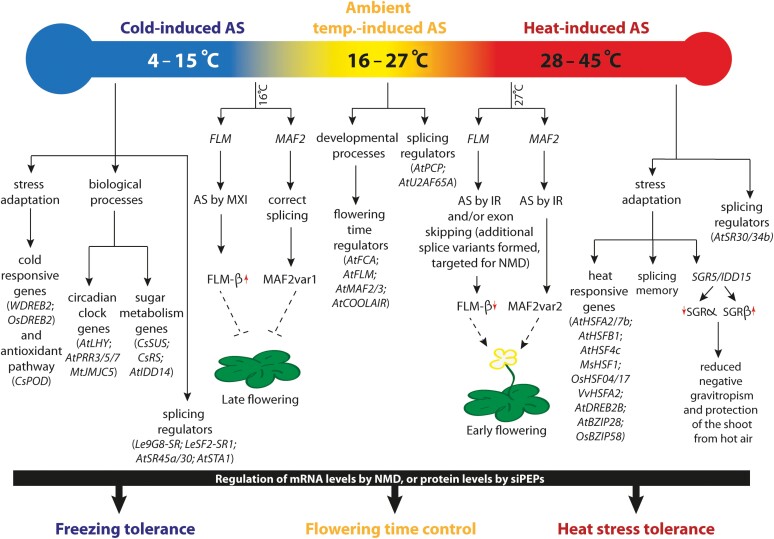
Fig. 3.
Temperature-induced alternative splicing (AS) under different temperature conditions resulting in the plant’s adaptation. Overview of the genes undergoing AS under diverse temperature regimes like cold (around 4–15 °C), changes in ambient temperature (16–27 °C), and heat stress (28–45 °C), and the resulting physiological responses. AS in response to changes in temperature (including extreme temperatures) has been shown to play an important role in improving plant performance and stress tolerance. As an example, AS of the flowering time genes FLM and MAF2 in response to a change in ambient temperature regulates the transition to flowering and reproductive growth in plants. A higher ambient temperature (27 °C) induces flowering while a lower temperature (16 °C) represses flowering. Dashed lines indicate indirect responses. The red arrows indicate up-regulation (upward arrow) or down-regulation (downward arrow). IR, intron retention; MXI, mutually exclusive incorporation; NMD, non-sense mediated decay; siPEPs, small interfering peptides.
In general, the response to cold is regulated by the CBF–COR hierarchical network with C-REPEAT/DEHYDRATION-RESPONSIVE ELEMENT BINDING FACTORS (CBF/DREBs) and their downstream targets, COLD-REGULATED (COR) genes, as the central components (Badawi et al., 2007; Mao and Chen 2012). The transcripts of DREB genes undergo AS in response to cold whereby the changes in temperature regulate the abundance of particular isoforms. For example, the wheat WDREB2 gene, a homologue of the Arabidopsis DREB2A and DREB2B genes, undergoes AS by exon skipping to produce three splice variants at 4 °C (Egawa et al., 2006). Similarly, the rice DREB2-type gene OsDREB2B is alternatively spliced into two active variants, OsDREB2B1 and OsDREB2B2, at low temperatures (Matsukura et al., 2010). In addition to DREBs, the COR genes undergo AS in response to cold. In durum wheat (Triticum durum), the transcripts of two early COR (e-COR) genes putatively encoding a ribokinase and a C3H2C3 RING-finger protein are alternatively spliced to retain a subset of introns in the mature mRNA during the cold period (Mastrangelo et al., 2005). In the tea plant (Camellia sinensis), the CsCOR gene was also found to undergo AS to form a truncated protein at low temperature; however, whether this shortened protein has a biological function has not been reported (Li et al., 2020b).
It is interesting to note that AS affects not only transcripts of genes that regulate the cold response in plants, but also a range of genes involved in plant growth and development including the circadian clock. In Arabidopsis, the expression of isoforms of many clock components, e.g. LATE ELONGATED HYPOCOTYL (LHY), PSEUDO-RESPONSE REGULATOR 3 (PRR3), PRR5, PRR7, PRR9, and TIMING OF CAB (TOC1), is controlled by AS by IR (Filichkin et al., 2015b). Similarly, cold-dependent IR of CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) leads to the formation of the two isoforms CCA1α and CCA1β. Here, the retention of the fourth intron produces the CCA1β isoform that forms a non-functional heterodimer with CCA1α and inhibits CCA1α’s activity. In this way, CCA1β acts as a siPEP. AS of the CCA1 transcript is suppressed by cold, which releases CCA1α repression and allows it to be fully functional in regulating freezing tolerance in Arabidopsis (Park et al., 2012; Seo et al., 2012). In Medicago truncatula, cold induction results in the formation of four alternatively spliced variants of MtJMJC5, which encodes a JmjC domain-containing protein, a circadian clock component (Shen et al., 2016). Also, in a commercial sugarcane variety (Saccharum hybrid, SP80-3280), the temperature- and organ-dependent AS of five clock genes, ScLHY, ScPRR37, ScPRR73, ScPRR95, and ScTOC1, was observed (Dantas et al., 2019). These findings suggest a strong correlation between temperature and AS events in circadian clock genes.
In addition to the already mentioned AS events in clock genes, transcripts of genes involved in flowering time regulation are alternatively spliced in response to prolonged cold exposure (Guan et al., 2013; Wang et al., 2019). The main mechanism of vernalization-mediated flowering requires repression of the transcription of FLOWERING LOCUS C (FLC), a MADS-box TF that acts as a negative regulator of flowering (Sheldon et al., 2000). The cold-dependent silencing of FLC is controlled by the long non-coding anti-sense RNA COLD INDUCED LONG ANTISENSE INTRAGENIC RNA (COOLAIR) produced from the FLC locus (Swiezewski et al., 2009; Sun et al., 2013). The temperature-dependent AS of COOLAIR subsequently regulates the expression of FLC through co-transcriptional coupling mechanisms (Swiezewski et al., 2009; Hornyik et al., 2010; Marquardt et al., 2014), providing plants with the ability to measure temperature and to integrate this external information to regulate flowering time.
Furthermore, links between AS, the response to cold stress, and plant metabolism were reported. In particular, in C. sinensis, a large number of AS events occurs in transcripts associated with sugar metabolism and antioxidant pathways, such as SUCROSE SYNTHASE (CsSUS), RAFFINOSE SYNTHASE (CsRS), and PEROXIDASE (CsPOD) (Li et al., 2020b). These pathways contribute to establishing cold tolerance by protecting membrane structures, reducing oxidative damage through the induction of reactive oxygen species (ROS)-degrading enzymes, and the accumulation of sugars that can have a protective function at high concentration (Uemura and Steponkus, 2003; Sun et al., 2010; Rai et al., 2013). Similarly, in Arabidopsis, the primary transcript of INDETERMINATE DOMAIN 14 (IDD14), a regulator of starch metabolism, undergoes AS by IR to form the functional IDD14α isoform and the truncated form IDD14β (Seo et al., 2011b). IDD14α and IDD14β form a heterodimer that displays reduced binding capacity to the promoter of the QUA-QUINE STARCH (QQS) gene that regulates starch accumulation. Overexpression of IDD14α leads to retarded growth, similar to QQS overexpression. As IDD14β overexpression rescues the IDD14α overexpression phenotype, AS of the IDD14 transcript may be a cold adaptation strategy for the plant to modulate starch accumulation to withstand the cold conditions (Seo et al., 2011b). These findings suggest a possible link between carbohydrate metabolism and low-temperature signalling mediated by AS. Also in potato (Solanum tuberosum), sugar metabolism genes undergo AS. Here, cold-induced AS by exon skipping occurs in the transcripts of one of the plant’s INVERTASE genes responsible for the conversion of sucrose to glucose and fructose (Bournay et al., 1996). The accumulation of these reducing sugars in tubers stored under cold conditions leads to reduced tuber quality in many cultivars. This phenomenon is known as low-temperature sweetening (Davies et al., 1989). Here, invertase inhibitors bind to invertases to inactivate them (Hothorn et al., 2010). Interestingly, one of the most abundant forms of invertase inhibitor genes expressed in potato tubers, INH2, is regulated by AS. During cold storage of potato tubers of cultivars resistant to cold-induced sweetening, the levels of the unspliced INH2α transcript, which encodes the full-length INH2 protein, and hybrid INH2β mRNAs generated through splicing of INH2α and INH1 transcripts, were higher than in tubers of sensitive cultivars. The increase in invertase inhibitor levels in the resistant cultivars may be responsible for the suppression of acid invertase activity and sucrose breakdown, thereby leading to reduced cold-induced sweetening (Brummell et al., 2011). Also, sugar transporter genes, e.g. SUGARS WILL EVENTUALLY BE EXPORTED TRANSPORTER 17 (SWEET17) in C. sinensis undergo AS in response to cold (Yao et al., 2020). Interestingly, a metabolic or physiological reprogramming of the transcriptome as a result of AS was also observed in animals such as Caenorhabditis elegans, mouse, and human, where genes involved in pyruvate metabolism, glycolysis, and gluconeogenesis were found to be alternatively spliced (Tabrez et al., 2017; Biamonti et al., 2018).
A recent study showed that 46 of the 379 long non-coding RNAs (lncRNAs) in Arabidopsis are alternatively spliced in response to cold (of which half are also differentially expressed after cold treatment), suggesting an important regulatory mechanism involved in cold acclimation and freezing tolerance (Calixto et al., 2019). For example, the transcript of TAS1a is alternatively spliced upon exposure to cold, without a major change in gene expression level. The non-spliced TAS1a transcript, which is more abundant at normal growth temperature, contains a miR173 binding site in its intron; binding of miR173 to the non-spliced intron sequence causes the formation of transacting siRNAs (tasiRNAs). The splicing of the primary TAS1a transcript at low temperature removes its intron, thereby eliminating the miR173 binding site and leading to a reduction of the level of siRNAs. The data demonstrate that AS regulates the production of siRNAs in response to cold stress (Calixto et al., 2019). In addition, various studies provide evidence that cold induces AS in a tissue-specific manner. For example, tissue-specific cold-induced AS occurs in the wild rice Oryza longistaminata, which is tolerant to cold non-freezing temperatures, unlike Oryza sativa, which is chilling stress-sensitive. Transcriptome profiling of shoots and rhizomes of O. longistaminata subjected to chilling stress revealed that cold-induced AS was transcript-specific in these tissues. Most of the chilling-induced genes undergoing AS only in shoots are involved in photosynthesis and the regulation of gene expression, whereas those undergoing AS only in rhizomes are mainly involved in stress signal transduction. This observation suggests that tissue-specific AS may play an important role in regulating cold acclimation in O. longistaminata (Zhang et al., 2017).
In summary, compelling evidence from various studies demonstrates that a large number of AS events observed in plants in response to cold temperature are not random. In fact, those events contribute to the acquisition of cold acclimation in plants, allowing them to develop cold tolerance.
Alternative splicing induced by moderate changes in ambient temperature
In addition to low temperature, moderate changes in ambient temperature may trigger AS (Fig. 3) (Verhage et al., 2017). For example, transcripts of Arabidopsis genes encoding the RNA-binding protein PUMILIO 23 (PUM23) and three other regulatory proteins, i.e. mitochondrial EMBRYO DEFECTIVE 3114 (EMB3114), SNF1 KINASE HOMOLOG 11 (AKIN11), and FYD (Streitner et al., 2013), are subject to AS at small changes in ambient temperature (from 16 °C to 20 °C, from 20 °C to 24 °C, and from 20 °C to 16 °C) (Streitner et al., 2013).
AS is also involved in regulating flowering time in response to fluctuating ambient temperature. The temperature-dependent AS observed in the transcript of FLOWERING CONTROL LOCUS A (FCA), which encodes an RNA-binding protein involved in the biosynthesis of the flowering time regulator microRNA172 (miR172) (Macknight et al., 1997; Jung et al., 2012), is a well-known example. The FCA mRNA undergoes AS so that functional FCA protein is more abundant at 23 °C than 16 °C, which leads to an accumulation of miR172 at the higher temperature (Jung et al., 2012).
Other flowering time regulators are also controlled by ambient temperature-triggered AS, including FLOWERING LOCUS M (FLM), a MADS-domain TF (Scortecci et al., 2001; Balasubramanian et al., 2006). FLM modulates flowering time by creating a repressor complex with FLC or SHORT VEGETATIVE PHASE (SVP) proteins in a wide range of temperatures. Interestingly, the FLM transcript undergoes alternative splicing in response to changes in ambient temperature (Balasubramanian et al., 2006). FLM-β and FLM-δ are the predominant splice variants formed by the mutual exclusion of exons: while exon 2 is maintained in the FLM-β transcript, exon 3 is retained in the FLM-δ transcript (Lee et al., 2013; Posé et al., 2013). Low ambient temperature (16 °C) favours the expression of the repressive isoform FLM-β, and its expression decreases when ambient temperature increases (27 °C) (Lee et al., 2013; Posé et al., 2013). FLM-β protein forms a complex with SVP, a MADS-domain TF that actively represses flowering by binding to, for example, the promoter of the flowering time integrator SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) (Posé et al., 2013). Although initial research had indicated an important role of the FLM-δ isoform for flowering control, as a competitor of FLM-β in its interaction with SVP, follow-up research revealed a less important role of FLM-δ (Lutz et al., 2015; Sureshkumar et al., 2016; Lutz et al., 2017). Apart from FLM-β and FLM-δ, additional FLM splice variants are generated in Arabidopsis Col-0, particularly at higher ambient temperatures (Sureshkumar et al., 2016; Capovilla et al., 2017). Most of the additional splice variants harbour PTCs and are targeted for degradation by NMD, which in consequence leads to a decrease in the level of transcripts available for translation into the functional isoform FLM-β at higher ambient temperatures. Furthermore, the SVP protein is degraded at higher ambient temperatures (Lee et al., 2013). This decrease in the FLM–SVP repressor complex promotes flowering at elevated temperatures (Sureshkumar et al., 2016). To further elucidate the role of FLM-β and FLM-δ in flowering, CRISPR-Cas9-generated mutants lacking the second (FLM-∆E2, FLM-δ) or third (FLM-∆E3, FLM-β) exon of the FLM transcript were studied (Capovilla et al., 2017). A comparison of the genome-edited plants with the flm loss-of function mutant showed that expression of FLM-δ alone did not promote flowering while expression of FLM-β delayed flowering. Taken together, the results suggest that FLM-δ is unlikely to promote flowering, while temperature-induced AS regulates the levels of FLM-β, which plays a crucial role in flowering time regulation (Fig. 3).
The balance between the FLM-β and FLM-δ isoforms is regulated by splicing factors like the U2 AUXILIARY FACTORs ATU2AF65A and ATU2AF65B, the GLYCINE-RICH PROTEINs ATGRP7 and ATGRP8, and SPLICING FACTOR 1 (ATSF1) (Lee et al., 2017; Park et al., 2019; Steffen et al., 2019). Recently, the CYCLIN-DEPENDENT KINASE G2 (CDKG2)–CYCLIN L1 (CYCL1) complex has also been shown to regulate this balance and to be required for the correct processing of the alternative introns 1 and 4 in FLM pre-mRNA (Nibau et al., 2020). When the CDKG2–CYCL1 complex was absent, expression of the FLM-δ isoform increased while expression of the FLM-β variant decreased in a temperature-dependent manner, resulting in an early-flowering phenotype (Nibau et al., 2020). Interestingly, the CDKG2–CYCL1 complex also regulates the temperature-dependent splicing of another CDK, i.e. CDKG1, which regulates AS of the splicing factor ATU2AF65A transcript (Cavallari et al., 2018). ATU2AF65A undergoes AS when ambient temperature is lowered from 23 °C to 16 °C (Verhage et al., 2017). This observation indicates that apart from their primary function in the progression of the cell cycle, CDKs may play an essential role in linking temperature sensing mechanisms with the AS of specific transcripts.
Another flowering time component subject to AS is MADS AFFECTING FLOWERING 2 (MAF2), a gene closely related to FLM at the sequence level. Different AS events occur in the MAF2 primary transcript. However, only one of them leads to the formation of a functional, full-length MIKC-type MADS-domain TF that represses flowering (Rosloski et al., 2013). The expression of this splice variant is elevated at low ambient temperature (16 °C), and the protein it encodes interacts with SVP to repress flowering. Interestingly, at higher ambient temperatures (21–27 °C), the MAF2 primary transcript undergoes AS by IR to introduce a PTC (Rosloski et al., 2013). This isoform encodes a protein incapable of interacting with SVP and, consequently, to inhibit flowering. A progressive temperature-dependent early-flowering phenotype has been observed in Arabidopsis plants, which flower earlier at 27 °C than at 21 °C, and earlier at 21 °C than at 16 °C (Airoldi et al., 2015). At higher ambient temperatures, AS triggers the increased production of MAF2 and FLM isoforms, which lack the repressive function that may be responsible for the decrease in flowering repression. Similarly, also MAF3 undergoes AS in response to changes in ambient temperature (Verhage et al., 2017). Among the several AS products derived from the primary MAF3 transcript, only the transcript that includes a skipping of exon 2 shows temperature sensitivity (Verhage et al., 2017).
There is increasing evidence that AS in response to changes in ambient temperature is not only limited to genes involved in flowering time regulation, but also affects other developmental genes. For example, the splice regulator PORCUPINE (PCP) has recently been reported to be a temperature-specific regulator of development in Arabidopsis (Capovilla et al., 2018). When pcp-1 knockout plants grown at permissive ambient temperature (23 °C) were shifted to a lower temperature (16 °C), growth was arrested and plants were rendered male sterile. When the permissive temperature was re-established, the pcp-1 loss-of-function mutant restored the wild type-like phenotype. At low ambient temperature (16 °C), pcp-1 plants showed aberrant shoot apical meristems, and altered lateral organ formation. In a proteome study, two variants of the PCP protein, PCP-α and PCP-β, which are encoded by alternatively spliced PCP transcripts, were identified (Ito et al., 2011; Capovilla et al., 2018). The constitutive overexpression of PCP-α, but not PCP-β, rescues the pcp-1 phenotype at 16 °C. The two splice variants differ from each other only in the extreme 3′ part of the protein-coding region, indicating that the C-terminal region of the PCP protein may be imperative for its function (Capovilla et al., 2018).
It is important to note that among the genes in Arabidopsis undergoing AS due to elevated ambient temperature, approximately 96% harbour a histone H3 lysine 36 tri-methylation (H3K36me3)-enriched region in their gene body. Several flowering time regulators (such as FLM, MAF2, and FCA) and circadian clock components (PRR3 and PRR7) affected by temperature-induced AS harbour this mark (Pajoro et al., 2017). In addition, lack of histone methyltransferases involved in the deposition of H3K36me3 marks led to altered AS upon a temperature shift from 16 °C to 25 °C. Mutants affected in writing, erasing, and reading the H3K36me3 mark showed altered elevated temperature-induced flowering. Therefore, the H3K36me3 mark plays an important role in regulating ambient temperature-induced AS of biological relevance. The results also demonstrate that AS and epigenetic regulation of chromatin are coupled (Pajoro et al., 2017). Evidence that AS affects the epigenetic landscapes of cells, thereby modulating gene expression, has also been obtained in mammals. For example, LYSINE-SPECIFIC DEMETHYLASE 1 has been reported to repress and activate gene expression programmes in neurons by mediating histone H3K4me1/2 and H3K9me1/2 demethylation, respectively (Laurent et al., 2015).
Heat-regulated alternative splicing
Heat stress is another abiotic stress that adversely affects plant growth and reduces crop yields. Each crop has a different threshold temperature (above which growth and development are severely affected). For example, a temperature threshold of 35 °C has been reported for rice, and 33–38 °C for maize. The susceptibility to high temperature can also depend on the stage of development of the plant. For example, the temperature threshold in wheat was observed to be 20–30 °C for vegetative growth and 15 °C for reproductive growth, whereas in tomato the temperature threshold was observed to be 37 °C for vegetative growth and 28–30 °C for reproductive growth (Janni et al., 2020). Given the current level of knowledge of AS in plants in response to high temperature, it is predicted that ~46, 46, and 55% of intron-containing genes in Arabidopsis undergo AS in response to mild, severe, or extreme/lethal changes in temperature, respectively (Ling et al., 2018). Here, we summarize recent findings that describe how heat stress induces AS in different plant species that in turn can lead to plant adaptation to higher temperatures.
The heat stress response is controlled by members of the heat shock factor (HSF) transcription factor family that activate (or repress) HEAT SHOCK PROTEIN (HSP) genes. The HSPs in turn function as molecular chaperones to protect the proteome against the negative effects of heat stress (Zhang et al., 2010; Wu et al., 2013). Interestingly, HSFs are strongly impacted by splicing regulation in the response to heat stress in plants. In the Drosophila HSF (dHSF), both heat- and cold-induced AS are observed, in which the abundance of the dHSFb isoform increases upon heat exposure, while that of the dHSFd isoform increases upon cold exposure, which may lead to the induction of different HSPs (Fujikake et al., 2005). AS of HSFs in response to cold has not yet been reported in plants. In Arabidopsis, various HSFs, e.g. HSFA2, HSFA7b, HSFB1, HSFB2a, and HSF4c, undergo extensive AS in response to heat stress by producing intron-retained splice variants (He et al., 2008; Sugio et al., 2009; Amano et al., 2011; Liu et al., 2013; Cheng et al., 2015; Jiang et al., 2017; Zhang et al., 2020). For example, at 22 °C the HSFA2 transcript is fully spliced, while at a moderately high temperature (37 °C) a 31-nucleotide-long cryptic mini-exon is derived from within the HSFA2 intron. This leads to the generation of the splice variant HSFA2-II that is further degraded by NMD due to a PTC present within the mini-exon (Sugio et al., 2009). Interestingly, during extreme heat (45 °C) a third splice variant, HSFA2-III, is generated by retaining an approximately 80-nucleotide-long 5′ region of the intron in the processed mRNA. HSFA2-III is translated into a truncated, C-terminally modified protein, S-HSFA2. The S-HSFA2 variant binds to the HSFA2 promoter, thereby creating a positive auto-regulatory loop controlling HSFA2 expression (Liu et al., 2013). Of note, alternatively spliced HSF transcripts are observed in different plant species, e.g. Medicago sativa HSF1, Potamogeton malaianus and P. perfoliatus HSFA2a2, rice (Oryza sativa) HSFA2d, maize (Zea mays) ZmHSF04 and ZmHSF17, and grape (Vitis vinifera) HSFA2 (He et al., 2008; Sugio et al., 2009; Amano et al., 2011; Liu et al., 2013; Cheng et al., 2015; Jiang et al., 2017; Zhang et al., 2020). In lily (Lilium spp.), LlHSFA3B undergoes heat-induced AS to generate splice variant LlHSFA3B-III (Wu et al., 2019). The LlHSFA3B-III protein appeared to be transcriptionally inactive in a yeast test system. A green fluorescent protein (GFP)–LlHSFA3B-III fusion protein accumulated in both the nucleus and cytoplasm. Notably, LlHSFA3B-III negatively affects the interaction of the full-length HSFA3 proteins LlHSFA3A-I and LlHSFA3B-I (Wu et al., 2019). Enhanced tolerance to both salinity stress and prolonged heat treatment at 40 °C was observed in transgenic Arabidopsis and Nicotiana benthamiana plants expressing LlHSFA3B-III, while tolerance to acute heat shock at 45 °C was reduced. Furthermore, the interaction between LlHSFA3B-III with LlHSFA3A-I was found to be essential for reducing the transactivation function of LlHSFA3A-I, thereby limiting various adverse effects of increased LlHSFA3A-I accumulation such as sensitivity to salinity and heat during prolonged heat stress (Wu et al., 2019). More importantly, the AS pattern for HSFs appears to be largely conserved between plants indicating that heat-induced AS regulation is an evolutionarily conserved phenomenon (Chang et al., 2014).
It is important to note that heat stress-induced AS not only occurs in transcripts of HSF genes, but also in transcripts of HSPs and other heat stress-inducible genes such as DREB2B and BZIP28 in Arabidopsis (Liu et al., 2013). As in the case of HSFs, retention of the first intron was observed in HSP transcripts, suggesting a conserved mechanism of AS in response to high temperature (Neves-da-Rocha et al., 2019). Interestingly, recent research suggested that STABILIZED1 (STA1), a U5-snRNP-interacting protein, acts as a high temperature-specific splicing factor involved in AS of HSP and HSF primary transcripts, i.e. HSFA3 (Kim et al., 2018). Pre-mRNA of HSFA3, whose transcription is induced by the upstream transcription factor DREB2A, is spliced with a contribution from STA1. The protein produced from its mature mRNA induces the expression of HSP genes. The findings also suggest that heat-inducible STA1 is required for the establishment of acquired, but not basal, heat stress tolerance (Kim et al., 2017, 2018).
In natural environments and in agricultural fields, plants are often subject to recurrent stress. One of the strategies to cope with recurring stress is a process called ‘priming’ through which moderately stressed plants ‘prepare’ themselves to successfully withstand a later, and often more severe, stress. The period between both stresses is called the ‘memory phase’. During this phase the specific molecular-biochemical changes induced by a priming stress are maintained to allow the plant to withstand the second stress, which otherwise would be more harmful or even deadly (Hilker et al., 2016). Some of the molecular mechanisms underlying molecular memory have been discovered in recent years, although a coherent picture is not yet available (Sedaghatmehr et al., 2016; Luo et al., 2020; Mayer and Charron, 2020).
Heat priming-induced splicing memory has been studied in Arabidopsis, where plants subjected to a priming heat stress retain the memory of constitutive splicing of transcripts of many heat-responsive genes (including HSFs and HSPs). In response to heat priming, the transcripts of HSP genes like HSP21, HSP101, HSP70.10, HSP70.6, HSP90.5, and HSP100.3 are alternatively spliced, mostly by IR (Ling et al., 2018). In primed plants, these gene transcripts undergo constitutive splicing when subjected to a second heat stress (called triggering). Unlike in primed plants, a significant splicing repression was observed in non-primed plants directly subjected to the triggering heat stress, which subsequently led to an increase in alternatively spliced isoforms of several HSF and HSP transcripts in these plants. This ‘splicing memory’ enables the primed plants to combat subsequent and otherwise lethal heat stress conditions (Ling et al., 2018). Heat-induced AS may, therefore, play an important role in the thermomemory response in plants. However, the molecular mechanisms underpinning the splicing memory are unknown at present.
Interestingly, transcripts undergoing AS are not only directly involved in the heat stress response, but are also involved in plant growth and metabolism. For example, the zinc finger (ZF)-containing transcription factor SHOOT GRAVITROPISM 5 (SGR5; also called INDETERMINATE DOMAIN 15, IDD15), which is responsible for mediating initial events of gravitropic responses in inflorescence stems in Arabidopsis (Morita et al., 2006), undergoes AS that is accelerated by higher temperatures. IR in the SGR5 transcript produces two protein isoforms, the completely spliced SGR5α transcription factor and the truncated SGR5β form lacking the functional ZF motif. SGR5β binds to SGR5α to form a non-functional heterodimer that lacks DNA-binding activity and thus functions as an siPEP. Since this AS is enhanced at higher temperature, SGR5β protein accumulates during heat stress; this heat-induced accumulation of SGR5β enables shoots to curve away from the source of heat, thereby serving as an adaptive response developed to protect shoots from hot air when plants are growing in heat stress conditions (Kim et al., 2016).
A second example is the rice transcription factor OsbZIP58, a regulator of various starch synthesis and storage protein genes (Hakata et al., 2012; Xu et al., 2020). The loss-of-function phenotype of OsbZIP58 is characterized by reduced seed storage material, and floury and shrunken endosperms under high-temperature conditions. The OsbZIP58 primary transcript undergoes AS by IR during heat stress to generate PTCs, leading to the increased accumulation of a truncated protein, OsbZIP58β. The newly formed OsbZIP58β protein is transcriptionally less active than OsbZIP58α derived from the fully spliced transcript. The heat-induced AS of OsbZIP58 is more pronounced in heat sensitive rice varieties like japonica than in heat resistant varieties like indica. The production of the transcriptionally less active protein OsbZIP58β due to increased AS in japonica may lead to an impaired accumulation of storage materials, thereby leading to the higher heat sensitivity. The reduced AS of OsbZIP58 in less heat sensitive indica may be responsible for the heat tolerance observed during grain filling in these varieties (Xu et al., 2020).
In addition, heat stress-induced AS provides a mechanism for regulating miRNA processing in plants. An example for this has been reported in Arabidopsis, where intronic miR400 is co-transcribed together with its host gene (AT1G32583). Under conditions of elevated temperature (37 °C, 30 min–12 h), miR400 and the host gene transcript display opposite responses. While the abundance of the host gene transcript slightly decreased, the abundance of miR400 primary transcript strongly increased by 12 h compared with controls; the level of mature miR400 continuously declined. It turned out that the accumulation of miR400 was triggered by an AS in intron 1 under higher temperatures that produced a version of the transcript from which the miR400-containing intron could not be spliced out. This then blocked the efficient further processing to mature miR400 (Yan et al., 2012). Overexpression of miR400 reduces the tolerance to heat stress, although the precise molecular mode of action for this is not well known yet. However, miR400 guides PPR1 and PPR2 mRNAs, which encode pentatricopeptide repeat proteins, for cleavage, which may suggest an involvement of the two proteins in the heat stress response (Yan et al., 2012; Park et al., 2014).
In summary, AS in response to high temperature occurs in many genes involved in the heat stress response like HSFs, bZIPs, HSPs, and others. The heat-induced AS of these genes leads to favourable adaptive responses in plants as seen in the case of AS in SGR5, OsbZIP58, and the heat priming-induced splicing memory.
Temperature-induced alternative splicing of splicing regulators
In addition to the above mentioned examples, also members of the spliceosomal complex themselves undergo AS in different temperature regimes. For example, transcriptome analysis of ambient temperature-induced AS in Arabidopsis and Brassica oleracea ssp. botrytis revealed differential splicing in several classes of splicing-related genes, indicating that the whole spliceosome is affected by fluctuations in ambient temperature (Verhage et al., 2017). These findings suggest a two-step model controlling ambient temperature perception through AS: first, the splicing regulator genes undergo changes in their splicing patterns, and second, the altered splicing machinery subsequently affects the splicing of downstream genes involved in the adaptation to altered temperature (Verhage et al., 2017).
A good example of splicing-related genes affected by AS in response to a wide range of temperatures are those for SR proteins. In fact, transcripts of SR proteins undergo AS to generate approximately 100 distinct splice variants from 15 SR genes in Arabidopsis (Palusa et al., 2007). The AS of some SR genes was found to be controlled during development and with tissue specificity. The AS of the transcripts of many SR genes drastically changes in response to extreme temperatures (cold and heat). The transcripts of SR30, RS31a, RS41, RSZ33, and SCL30a undergo heat-induced AS, while the transcripts of SR33/SCL33, SR1/SR34, and RS40 undergo AS under both heat and cold conditions (Palusa et al., 2007). Heat or cold treatments also had an impact on the recruitment of splice variants of many SR genes to polysomes for translation (Palusa and Reddy, 2015). Here, the functional isoforms of SR30, SR34, and SR34a were found to be more abundant in polysome-associated RNA of heat- and cold-treated Arabidopsis seedlings. On the contrary, many isoforms associated with polysomal RNA in the control were not recruited to polysomes under heat or cold stress conditions. One splice variant of SR34 is recruited to polysomes only during heat stress. The differential AS in response to cold produces new splice variants of SCL33 that are not recruited to polysomes. These splice variants may be involved in regulating the amount of functional SCL33 mRNA. In heat-treated seedlings, the functional isoform of SCL33 was found to be more enriched (Palusa and Reddy, 2015). In Arabidopsis, cold stress mediates the repression of the active isoforms of SR45a and SR30 (Tanabe et al., 2007), whereas it leads to an elevated expression level of STABILIZED1 (STA1), which encodes a protein responsible for the correct splicing of the COLD REGULATED 15a (COR15a) gene transcript (Lee et al., 2006). In tomato (Solanum lycopersicum), the splicing factor genes Le9G8-SR and LeSF2-SR1 undergo AS at low temperature (Fung et al., 2006).
The AS of splicing regulators that leads to the formation of different isoforms and differential protein levels in response to varying temperatures may be responsible for rapid and dynamic transcriptome changes, providing plants with development plasticity.
Open questions regarding temperature-controlled alternative splicing
Rapid progress in exploring the regulatory aspects of AS in plants has been achieved recently. Yet, most studies have concentrated on the splicing events themselves and their impact on protein levels, while the impact of AS on the plant’s physiology has often been neglected. However, understanding the functional consequences of AS events for plants has an important practical relevance, in particular with respect to improving the resilience of crops towards climate-related environmental changes in the coming decades. In mammals the biological importance of AS is relatively well understood. A large number of human diseases are caused by mutations in splicing factors or other components of the spliceosome (Scotti and Swanson, 2016). In fact, the Human Genome Mutation Database predicts that more than one-third of the diseases caused by single nucleotide polymorphisms are due to mutations within splice sites or splicing elements (Krawczak et al., 2007). Interestingly, in mammals, physiological body temperature differences of only 1 °C occurring during day–night cycles lead to functionally relevant AS events that affect the levels of TATA-box binding proteins (Tbp) and, as a consequence, global gene expression (Preußner et al., 2017). Also in ectotherm insects such as D. melanogaster, temperature changes dramatically affect splicing patterns. Anduaga et al. (2019) demonstrated the presence of two thermosensitive transcript isoforms of the circadian clock gene TIMELESS (TIM). Temperature-dependent switching of the isoforms, e.g. TIM-COLD (18 °C) and TIM-MEDIUM (29 °C), mediates temperature adaptation of flies by acting as a thermometer within the circadian clock (Anduaga et al., 2019).
Temperature-induced AS also plays a functional role in conferring an adaptive benefit in single-cell eukaryotic species. For example, in Saccharomyces cerevisiae, the HSP70 nucleotide exchange factor FACTOR EXCHANGE FOR SSA1 PROTEIN 1 (FES1) undergoes AS to produce two isoforms, FES1L and FES1S. At elevated temperature (37 °C), the expression of FES1S is highly induced. The protein encoded by stress-inducible FES1S localizes to the cytosplasm and plays an important role in proteasomal degradation of cytosolic misfolded proteins, thereby contributing to proteostasis (Gowda et al., 2016).
An important question is how environmental temperature changes are registered by plants, and how monitoring such changes then leads to altered molecular responses. A recent study (Jung et al., 2020) showed that a prion-like domain present in the EARLY FLOWERING 3 (ELF3) protein serves as a thermosensor in Arabidopsis. However, it is currently unclear whether temperature sensing through the ELF3 protein, or other known thermosensory mechanisms (Dai Vu et al., 2019), controls nuclear splicing, and if so, which transcripts are actually affected. Addressing this will be an important aspect of future research.
Another issue that recently attracted increased interest tackles evolutionary conservation of temperature-triggered AS events in orthologous (and paralogous) genes in diverse plants (including crops), or—equally relevant—the diversification of AS in genes conserved between species. Addressing this will be essential in future research. In silico analysis suggests that SR proteins are evolutionarily conserved between multicellular green algae, bryophytes, and angiosperms (Melo et al., 2020). In the future, experimental evidence is required to support the function of those proteins for (alternative) splicing and to investigate how they contributed to processes that allowed plants to colonize terrestrial habitats and their adaptation to terrestrial temperatures.
As described in this review, changes in environmental temperature modify the splicing patterns of many genes. A possibly important aspect that needs further investigation is how day–night changes in environmental temperature shape the AS landscape, since in their natural environment and in the field, plants are subject to frequent day–night temperature shifts. In Arabidopsis, temperature-induced AS may be involved in the regulation of the plant circadian clock (James et al., 2012) and the circadian clock can also regulate AS (Yang et al., 2020). Recently, an increase in daytime temperature was found to affect the splicing of the SR45a transcript in maize. SR45a isoforms with higher splicing efficiencies were formed later in the day (afternoon) when the temperatures rose, while earlier in the day (morning) when the temperature is lower, SR45a isoforms less efficient in splicing were produced (Li et al., 2021). As plants are not able to control their ‘body’ temperature, the effects of day–night cycles on AS, besides the effect of abiotic stress-related environmental changes in temperature, may be significant. Analysing this in the future would be highly important, in particular for crops to improve their growth and yield potential at more severe day–night temperature regimes.
In order to further unravel the role of AS in response to temperature and its impact on agriculture in the context of climate change, it is important to acknowledge that even within a single species (e.g. in accessions, cultivars, mutants) there is considerable variation in the tolerance to temperature stress. In the future, detailed analyses will be required to relate differences in AS between species/accessions/cultivars to their actual temperature change response profiles.
Conclusion
Temperature-dependent AS plays a crucial role in integrating environmental temperature information into the molecular networks that control plant biological processes, including development and growth. From a biological point of view, it seems reasonable that plants evolved AS to allow rapid adjustments of transcript abundance to changes in temperature, thereby providing developmental plasticity as seen in the regulation of flowering time by AS in response to changes in environmental temperature. Although AS is a common phenomenon in many organisms, we are only starting to understand the possibly wide relevance of AS for stress adaptation in plants. Importantly, most of the studies on temperature-induced AS have so far been conducted under controlled laboratory conditions. However, considering the huge effect climate change has on our global ecosystem and agricultural productivity it would be tremendously important to study the role of AS in natural (outdoor) settings. Only then will we be able to fully appreciate the biological role of AS in the response to changes in environmental temperature. Biotechnological approaches including, for example, CRISPR-Cas-mediated genome editing to place mutations in splice sites in order to generate the desired temperature-responsive splicing patterns, or overexpression of splice variants and siPEPs essential for improving temperature stress tolerance, could be employed to engineer crops resistant to different temperature stresses.
Acknowledgements
We thank the Deutsche Forschungsgemeinschaft (DFG), Germany, for funding the Collaborative Research Centre 973 ‘Priming and Memory of Organismic Responses to Stress’ (www.sfb973.de), the European Union for funding the Horizon 2020 project PlantaSYST (SGA-CSA No. 739582 under FPA No. 664620), and the International Max Planck Research School ‘Primary Metabolism and Plant Growth’ (IMPRS-PMPG) for supporting SJ.
Author contributions
BM-R, JJO, and SJ: conceptualization; BM-R: funding acquisition, project administration, and resources; BM-R and JJO: supervision; BM-R, JJO, and SJ: validation; JJO and SJ: visualization; SJ: writing—original draft; BM-R, JJO, and SJ: writing—review and editing. The authors declare the absence of a potential conflict of interest.
References
- Airoldi CA, McKay M, Davies B. 2015. MAF2 is regulated by temperature-dependent splicing and represses flowering at low temperatures in parallel with FLM. PLoS One 10, e0126516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano M, Iida S, Kosuge K. 2011. Comparative studies of thermotolerance: different modes of heat acclimation between tolerant and intolerant aquatic plants of the genus Potamogeton. Annals of Botany 109, 443–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anduaga AM, Evantal N, Patop IL, Bartok O, Weiss R, Kadener S. 2019. Thermosensitive alternative splicing senses and mediates temperature adaptation in Drosophila. eLife 8, e44642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badawi M, Danyluk J, Boucho B, Houde M, Sarhan F. 2007. The CBF gene family in hexaploid wheat and its relationship to the phylogenetic complexity of cereal CBFs. Molecular Genetics and Genomics 277, 533–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Sureshkumar S, Lempe J, Weigel D. 2006. Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genetics 2, e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biamonti G, Maita L, Montecucco A. 2018. The Krebs cycle connection: reciprocal influence between alternative splicing programs and cell metabolism. Frontiers in Oncology 8, 408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournay AS, Hedley PE, Maddison A, Waugh R, Machray GC. 1996. Exon skipping induced by cold stress in a potato invertase gene transcript. Nucleic Acids Research 24, 2347–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummell DA, Chen RK, Harris JC, Zhang H, Hamiaux C, Kralicek AV, McKenzie MJ. 2011. Induction of vacuolar invertase inhibitor mRNA in potato tubers contributes to cold-induced sweetening resistance and includes spliced hybrid mRNA variants. Journal of Experimental Botany 62, 3519–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calixto CPG, Guo W, James AB, Tzioutziou NA, Entizne JC, Panter PE, Knight H, Nimmo HG, Zhang R, Brown JWS. 2018. Rapid and dynamic alternative splicing impacts the Arabidopsis cold response transcriptome. The Plant Cell 30, 1424–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calixto CPG, Tzioutziou NA, James AB, Hornyik C, Guo W, Zhang R, Nimmo HG, Brown JWS. 2019. Cold-dependent expression and alternative splicing of Arabidopsis long non-coding RNAs. Frontiers in Plant Science 10, 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capovilla G, Delhomme N, Collani S, Shutava I, Bezrukov I, Symeonidi E, de Francisco Amorim M, Laubinger S, Schmid M. 2018. PORCUPINE regulates development in response to temperature through alternative splicing. Nature Plants 4, 534–539. [DOI] [PubMed] [Google Scholar]
- Capovilla G, Pajoro A, Immink RG, Schmid M. 2015. Role of alternative pre-mRNA splicing in temperature signaling. Current Opinion in Plant Biology 27, 97–103. [DOI] [PubMed] [Google Scholar]
- Capovilla G, Symeonidi E, Wu R, Schmid M. 2017. Contribution of major FLM isoforms to temperature-dependent flowering in Arabidopsis thaliana. Journal of Experimental Botany 68, 5117–5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallari N, Nibau C, Fuchs A, Dadarou D, Barta A, Doonan JH. 2018. The cyclin-dependent kinase G group defines a thermo-sensitive alternative splicing circuit modulating the expression of Arabidopsis ATU2AF65A. The Plant Journal 94, 1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CY, Lin WD, Tu SL. 2014. Genome-wide analysis of heat-sensitive alternative splicing in Physcomitrella patens. Plant Physiology 165, 826–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chechanovsky N, Hovav R, Frenkel R, Faigenboim A, Eselson Y, Petreikov M, Moy M, Shen S, Schaffer AA. 2019. Low temperature upregulates cwp expression and modifies alternative splicing patterns, increasing the severity of cwp-induced tomato fruit cuticular microfissures. Horticulture Research 6, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Zhou Y, Liu Z, Zhang L, Song G, Guo Z, Wang W, Qu X, Zhu Y, Yang D. 2015. An alternatively spliced heat shock transcription factor, OsHSFA2dI, functions in the heat stress-induced unfolded protein response in rice. Plant Biology 17, 419–429. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu J, Zhu JK. 2007. Cold stress regulation of gene expression in plants. Trends in Plant Science 12, 444–451. [DOI] [PubMed] [Google Scholar]
- Dai Vu L, Gevaert K, De Smet I. 2019. Feeling the heat: searching for plant thermosensors. Trends in Plant Science 24, 210–219. [DOI] [PubMed] [Google Scholar]
- Dantas LLB, Calixto CPG, Dourado MM, Carneiro MS, Brown JWS, Hotta CT. 2019. Alternative splicing of circadian clock genes correlates with temperature in field-grown sugarcane. Frontiers in Plant Science 10, 1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies HV, Jefferies RA, Scobie L. 1989. Hexose accumulation in cold-stored tubers of potato (Solanum tuberosum L.): The effects of water stress. Journal of Plant Physiology 134, 471–475. [Google Scholar]
- Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, Stark H, Urlaub H, Lührmann R. 2006. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Molecular and Cellular Biology 26, 5528–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa C, Kobayashi F, Ishibashi M, Nakamura T, Nakamura C, Takumi S. 2006. Differential regulation of transcript accumulation and alternative splicing of a DREB2 homolog under abiotic stress conditions in common wheat. Genes & genetic systems 81, 77–91. [DOI] [PubMed] [Google Scholar]
- Filichkin S, Priest HD, Megraw M, Mockler TC. 2015a. Alternative splicing in plants: directing traffic at the crossroads of adaptation and environmental stress. Current Opinion in Plant Biology 24, 125–135. [DOI] [PubMed] [Google Scholar]
- Filichkin SA, Cumbie JS, Dharmawardhana P, Jaiswal P, Chang JH, Palusa SG, Reddy AS, Megraw M, Mockler TC. 2015b. Environmental stresses modulate abundance and timing of alternatively spliced circadian transcripts in Arabidopsis. Molecular Plant 8, 207–227. [DOI] [PubMed] [Google Scholar]
- Fujikake N, Nagai Y, Popiel HA, Kano H, Yamaguchi M, Toda T. 2005. Alternative splicing regulates the transcriptional activity of Drosophila heat shock transcription factor in response to heat/cold stress. FEBS Letters 579, 3842–3848. [DOI] [PubMed] [Google Scholar]
- Fung RW, Wang CY, Smith DL, Gross KC, Tao Y, Tian M. 2006. Characterization of alternative oxidase (AOX) gene expression in response to methyl salicylate and methyl jasmonate pre-treatment and low temperature in tomatoes. Journal of Plant Physiology 163, 1049–1060. [DOI] [PubMed] [Google Scholar]
- Gallegos J. 2018. Alternative splicing plays a major role in plant response to cold temperatures. The Plant Cell 30, 1378–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Moreno JF, Romão L. 2020. Perspective in alternative splicing coupled to nonsense-mediated mRNA decay. International Journal of Molecular Sciences 21, 9424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda NK, Kaimal JM, Masser AE, Kang W, Friedländer MR, Andréasson C. 2016. Cytosolic splice isoform of Hsp70 nucleotide exchange factor Fes1 is required for the degradation of misfolded proteins in yeast. Molecular Biology of the Cell 27, 1210–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, et al. 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471, 473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Q, Wu J, Zhang Y, Jiang C, Liu R, Chai C, Zhu J. 2013. A DEAD box RNA helicase is critical for pre-mRNA splicing, cold-responsive gene regulation, and cold tolerance in Arabidopsis. The Plant Cell 25, 342–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakata M, Kuroda M, Miyashita T, Yamaguchi T, Kojima M, Sakakibara H, Mitsui T, Yamakawa H. 2012. Suppression of α-amylase genes improves quality of rice grain ripened under high temperature. Plant Biotechnology Journal 10, 1110–1117. [DOI] [PubMed] [Google Scholar]
- He ZS, Zou HS, Wang YZ, Zhu JB, Yu GQ. 2008. Maturation of the nodule-specific transcript MsHSF1c in Medicago sativa may involve interallelic trans-splicing. Genomics 92, 115–121. [DOI] [PubMed] [Google Scholar]
- Hilker M, Schwachtje J, Baier M, et al. 2016. Priming and memory of stress responses in organisms lacking a nervous system. Biological Reviews of the Cambridge Philosophical Society 91, 1118–1133. [DOI] [PubMed] [Google Scholar]
- Hornyik C, Duc C, Rataj K, Terzi LC, Simpson GG. 2010. Alternative polyadenylation of antisense RNAs and flowering time control. Biochemical Society Transactions 38, 1077–1081. [DOI] [PubMed] [Google Scholar]
- Hothorn M, Van den Ende W, Lammens W, Rybin V, Scheffzek K. 2010. Structural insights into the pH-controlled targeting of plant cell-wall invertase by a specific inhibitor protein. Proceedings of the National Academy of Sciences, USA 107, 17427–17432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J, Batth TS, Petzold CJ, Redding-Johanson AM, Mukhopadhyay A, Verboom R, Meyer EH, Millar AH, Heazlewood JL. 2011. Analysis of the Arabidopsis cytosolic proteome highlights subcellular partitioning of central plant metabolism. Journal of Proteome Research 10, 1571–1582. [DOI] [PubMed] [Google Scholar]
- James AB, Syed NH, Bordage S, Marshall J, Nimmo GA, Jenkins GI, Herzyk P, Brown JW, Nimmo HG. 2012. Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. The Plant Cell 24, 961–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janni M, Gullì M, Maestri E, Marmiroli M, Valliyodan B, Nguyen HT, Marmiroli N. 2020. Molecular and genetic bases of heat stress responses in crop plants and breeding for increased resilience and productivity. Journal of Experimental Botany 71, 3780–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Liu X, Liu C, Liu G, Li S, Wang L. 2017. Integrating omics and alternative splicing reveals insights into grape response to high temperature. Plant Physiology 173, 1502–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Barbosa AD, Hutin S, et al. 2020. A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature 585, 256–260. [DOI] [PubMed] [Google Scholar]
- Jung JH, Seo PJ, Ahn JH, Park CM. 2012. Arabidopsis RNA-binding protein FCA regulates microRNA172 processing in thermosensory flowering. The Journal of Biological Chemistry 287, 16007–16016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyna M, Simpson CG, Syed NH, et al. 2012. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Research 40, 2454–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GD, Cho YH, Lee BH, Yoo SD. 2017. STABILIZED1 modulates pre-mRNA splicing for thermotolerance. Plant Physiology 173, 2370–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GD, Yoo SD, Cho YH. 2018. STABILIZED1 as a heat stress-specific splicing factor in Arabidopsis thaliana. Plant Signaling & Behavior 13, e1432955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Ryu JY, Baek K, Park CM. 2016. High temperature attenuates the gravitropism of inflorescence stems by inducing SHOOT GRAVITROPISM 5 alternative splicing in Arabidopsis. New Phytologist 209, 265–279. [DOI] [PubMed] [Google Scholar]
- Kornblihtt AR, Schor IE, Alló M, Dujardin G, Petrillo E, Muñoz MJ. 2013. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nature Reviews. Molecular Cell Biology 14, 153–165. [DOI] [PubMed] [Google Scholar]
- Krawczak M, Thomas NS, Hundrieser B, Mort M, Wittig M, Hampe J, Cooper DN. 2007. Single base-pair substitutions in exon-intron junctions of human genes: nature, distribution, and consequences for mRNA splicing. Human Mutation 28, 150–158. [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Matsui A, Hanada K, et al. 2009. Genome-wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis. Proceedings of the National Academy of Sciences, USA 106, 2453–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloum T, Martín G, Duque P. 2018. Alternative splicing control of abiotic stress responses. Trends in Plant Science 23, 140–150. [DOI] [PubMed] [Google Scholar]
- Laurent B, Ruitu L, Murn J, et al. 2015. A specific LSD1/KDM1A isoform regulates neuronal differentiation through H3K9 demethylation. Molecular Cell 57, 957–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Kapoor A, Zhu J, Zhu JK. 2006. STABILIZED1, a stress-upregulated nuclear protein, is required for pre-mRNA splicing, mRNA turnover, and stress tolerance in Arabidopsis. The Plant Cell 18, 1736–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Ryu HS, Chung KS, Posé D, Kim S, Schmid M, Ahn JH. 2013. Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science 342, 628–632. [DOI] [PubMed] [Google Scholar]
- Lee KC, Jang YH, Kim SK, Park HY, Thu MP, Lee JH, Kim JK. 2017. RRM domain of Arabidopsis splicing factor SF1 is important for pre-mRNA splicing of a specific set of genes. Plant Cell Reports 36, 1083–1095. [DOI] [PubMed] [Google Scholar]
- Li S, Yu X, Cheng Z, Zeng C, Li W, Zhang L, Peng M. 2020a. Large-scale analysis of the cassava transcriptome reveals the impact of cold stress on alternative splicing. Journal of Experimental Botany 71, 422–434. [DOI] [PubMed] [Google Scholar]
- Li Y, Mi X, Zhao S, Zhu J, Guo R, Xia X, Liu L, Liu S, Wei C. 2020b. Comprehensive profiling of alternative splicing landscape during cold acclimation in tea plant. BMC Genomics 21, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Tang J, Bassham DC, Howell SH. 2021. Daily temperature cycles promote alternative splicing of RNAs encoding SR45a, a splicing regulator in maize. Plant Physiology, doi: 10.1093/plphys/kiab110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Y, Serrano N, Gao G, et al. 2018. Thermopriming triggers splicing memory in Arabidopsis. Journal of Experimental Botany 69, 2659–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Sun N, Liu M, Liu J, Du B, Wang X, Qi X. 2013. An autoregulatory loop controlling Arabidopsis HsfA2 expression: role of heat shock-induced alternative splicing. Plant Physiology 162, 512–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Ou Y, Li R, He Y. 2020. Maternal transmission of the epigenetic ‘memory of winter cold’ in Arabidopsis. Nature Plants 6, 1211–1218. [DOI] [PubMed] [Google Scholar]
- Lutz U, Nussbaumer T, Spannagl M, Diener J, Mayer KF, Schwechheimer C. 2017. Natural haplotypes of FLM non-coding sequences fine-tune flowering time in ambient spring temperatures in Arabidopsis. eLife 6, e22114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz U, Posé D, Pfeifer M, Gundlach H, Hagmann J, Wang C, Weigel D, Mayer KF, Schmid M, Schwechheimer C. 2015. Modulation of ambient temperature-dependent flowering in Arabidopsis thaliana by natural variation of FLOWERING LOCUS M. PLoS Genetics 11, e1005588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macknight R, Bancroft I, Page T, et al. 1997. FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell 89, 737–745. [DOI] [PubMed] [Google Scholar]
- Mao D, Chen C. 2012. Colinearity and similar expression pattern of rice DREB1s reveal their functional conservation in the cold-responsive pathway. PLoS One 7, e47275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt S, Raitskin O, Wu Z, Liu F, Sun Q, Dean C. 2014. Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Molecular Cell 54, 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrangelo AM, Belloni S, Barilli S, Ruperti B, Di Fonzo N, Stanca AM, Cattivelli L. 2005. Low temperature promotes intron retention in two e-cor genes of durum wheat. Planta 221, 705–715. [DOI] [PubMed] [Google Scholar]
- Matsukura S, Mizoi J, Yoshida T, Todaka D, Ito Y, Maruyama K, Shinozaki K, Yamaguchi-Shinozaki K. 2010. Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Molecular Genetics and Genomics 283, 185–196. [DOI] [PubMed] [Google Scholar]
- Mayer BF, Charron JB. 2020. Transcriptional memories mediate the plasticity of cold stress responses to enable morphological acclimation in Brachypodium distachyon. New Phytologist 229, 1615–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo JP, Kalyna M, Duque P. 2020. Current challenges in studying alternative splicing in plants: The case of Physcomitrella patens SR proteins. Frontiers in Plant Science 11, 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ. 2009. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136, 688–700. [DOI] [PubMed] [Google Scholar]
- Morita MT, Sakaguchi K, Kiyose S, Taira K, Kato T, Nakamura M, Tasaka M. 2006. A C2H2-type zinc finger protein, SGR5, is involved in early events of gravitropism in Arabidopsis inflorescence stems. The Plant Journal 47, 619–628. [DOI] [PubMed] [Google Scholar]
- Naqvi AR, Sarwat M, Hasan S, Roychodhury N. 2012. Biogenesis, functions and fate of plant microRNAs. Journal of Cellular Physiology 227, 3163–3168. [DOI] [PubMed] [Google Scholar]
- Neves-da-Rocha J, Bitencourt TA, de Oliveira VM, Sanches PR, Rossi A, Martinez-Rossi NM. 2019. Alternative splicing in heat shock protein transcripts as a mechanism of cell adaptation in Trichophyton rubrum. Cells 8, 1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibau C, Gallemí M, Dadarou D, Doonan JH, Cavallari N. 2019. Thermo-sensitive alternative splicing of FLOWERING LOCUS M is modulated by cyclin-dependent kinase G2. Frontiers in Plant Science 10, 1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463, 457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajoro A, Severing E, Angenent GC, Immink RGH. 2017. Histone H3 lysine 36 methylation affects temperature-induced alternative splicing and flowering in plants. Genome Biology 18, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palusa SG, Ali GS, Reddy AS. 2007. Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. The Plant Journal 49, 1091–1107. [DOI] [PubMed] [Google Scholar]
- Palusa SG, Reddy AS. 2010. Extensive coupling of alternative splicing of pre-mRNAs of serine/arginine (SR) genes with nonsense-mediated decay. New Phytologist 185, 83–89. [DOI] [PubMed] [Google Scholar]
- Palusa SG, Reddy AS. 2015. Differential recruitment of splice variants from SR pre-mRNAs to polysomes during development and in response to stresses. Plant & Cell Physiology 56, 421–427. [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature Genetics 40, 1413–1415. [DOI] [PubMed] [Google Scholar]
- Park HY, Lee HT, Lee JH, Kim JK. 2019. Arabidopsis U2AF65 regulates flowering time and the growth of pollen tubes. Frontiers in Plant Science 10, 569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MJ, Seo PJ, Park CM. 2012. CCA1 alternative splicing as a way of linking the circadian clock to temperature response in Arabidopsis. Plant Signaling & Behavior 7, 1194–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YJ, Lee HJ, Kwak KJ, Lee K, Hong SW, Kang H. 2014. MicroRNA400-guided cleavage of pentatricopeptide repeat protein mRNAs renders Arabidopsis thaliana more susceptible to pathogenic bacteria and fungi. Plant & Cell Physiology 55, 1660–1668. [DOI] [PubMed] [Google Scholar]
- Posé D, Verhage L, Ott F, Yant L, Mathieu J, Angenent GC, Immink RG, Schmid M. 2013. Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503, 414–417. [DOI] [PubMed] [Google Scholar]
- Preußner M, Goldammer G, Neumann A, Haltenhof T, Rautenstrauch P, Müller-McNicoll M, Heyd F. 2017. Body temperature cycles control rhythmic alternative splicing in mammals. Molecular Cell 67, 433–446.e4. [DOI] [PubMed] [Google Scholar]
- Rai GK, Rai NP, Rathaur S, Kumar S, Singh M. 2013. Expression of rd29A::AtDREB1A/CBF3 in tomato alleviates drought-induced oxidative stress by regulating key enzymatic and non-enzymatic antioxidants. Plant Physiology and Biochemistry 69, 90–100. [DOI] [PubMed] [Google Scholar]
- Ramachandran V, Chen X. 2008. Small RNA metabolism in Arabidopsis. Trends in Plant Science 13, 368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbapragada I, Lykke-Andersen J. 2009. Execution of nonsense-mediated mRNA decay: what defines a substrate? Current Opinion in Cell Biology 21, 394–402. [DOI] [PubMed] [Google Scholar]
- Reddy AS. 2007. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annual Review of Plant Biology 58, 267–294. [DOI] [PubMed] [Google Scholar]
- Reddy AS, Marquez Y, Kalyna M, Barta A. 2013. Complexity of the alternative splicing landscape in plants. The Plant Cell 25, 3657–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosloski SM, Singh A, Jali SS, Balasubramanian S, Weigel D, Grbic V. 2013. Functional analysis of splice variant expression of MADS AFFECTING FLOWERING 2 of Arabidopsis thaliana. Plant Molecular Biology 81, 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scortecci KC, Michaels SD, Amasino RM. 2001. Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. The Plant Journal 26, 229–236. [DOI] [PubMed] [Google Scholar]
- Scotti MM, Swanson MS. 2016. RNA mis-splicing in disease. Nature Reviews. Genetics 17, 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedaghatmehr M, Mueller-Roeber B, Balazadeh S. 2016. The plastid metalloprotease FtsH6 and small heat shock protein HSP21 jointly regulate thermomemory in Arabidopsis. Nature Communications 7, 12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Hong SY, Kim SG, Park CM. 2011a. Competitive inhibition of transcription factors by small interfering peptides. Trends in Plant Science 16, 541–549. [DOI] [PubMed] [Google Scholar]
- Seo PJ, Kim MJ, Ryu JY, Jeong EY, Park CM. 2011b. Two splice variants of the IDD14 transcription factor competitively form nonfunctional heterodimers which may regulate starch metabolism. Nature Communications 2, 303. [DOI] [PubMed] [Google Scholar]
- Seo PJ, Park MJ, Lim MH, Kim SG, Lee M, Baldwin IT, Park CM. 2012. A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. The Plant Cell 24, 2427–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang X, Cao Y, Ma L. 2017. Alternative splicing in plant genes: a means of regulating the environmental fitness of plants. International Journal of Molecular Sciences 18, 432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES. 2000. The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proceedings of the National Academy of Sciences, USA 97, 3753–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Wu X, Liu D, Song S, Liu D, Wang H. 2016. Cold-dependent alternative splicing of a Jumonji C domain-containing gene MtJMJC5 in Medicago truncatula. Biochemical and Biophysical Research Communications 474, 271–276. [DOI] [PubMed] [Google Scholar]
- Staudt AC, Wenkel S. 2011. Regulation of protein function by ‘microProteins’. EMBO Reports 12, 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen A, Elgner M, Staiger D. 2019. Regulation of flowering time by the RNA-binding proteins AtGRP7 and AtGRP8. Plant & Cell Physiology 60, 2040–2050. [DOI] [PubMed] [Google Scholar]
- Streitner C, Simpson CG, Shaw P, Danisman S, Brown JW, Staiger D. 2013. Small changes in ambient temperature affect alternative splicing in Arabidopsis thaliana. Plant Signaling & Behavior 8, e24638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio A, Dreos R, Aparicio F, Maule AJ. 2009. The cytosolic protein response as a subcomponent of the wider heat shock response in Arabidopsis. The Plant Cell 21, 642–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Csorba T, Skourti-Stathaki K, Proudfoot NJ, Dean C. 2013. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science 340, 619–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W-H, Duan M, Li F, Shu D-F, Yang S, Meng Q-W. 2010. Overexpression of tomato tAPX gene in tobacco improves tolerance to high or low temperature stress. Biologia Plantarum 54, 614–620. [Google Scholar]
- Sureshkumar S, Dent C, Seleznev A, Tasset C, Balasubramanian S. 2016. Nonsense-mediated mRNA decay modulates FLM-dependent thermosensory flowering response in Arabidopsis. Nature Plants 2, 16055. [DOI] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C. 2009. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462, 799–802. [DOI] [PubMed] [Google Scholar]
- Tabrez SS, Sharma RD, Jain V, Siddiqui AA, Mukhopadhyay A. 2017. Differential alternative splicing coupled to nonsense-mediated decay of mRNA ensures dietary restriction-induced longevity. Nature Communications 8, 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe N, Yoshimura K, Kimura A, Yabuta Y, Shigeoka S. 2007. Differential expression of alternatively spliced mRNAs of Arabidopsis SR protein homologs, atSR30 and atSR45a, in response to environmental stress. Plant & Cell Physiology 48, 1036–1049. [DOI] [PubMed] [Google Scholar]
- Uemura M, Steponkus P. 2003. Modification of the intracellular sugar content alters the incidence of freeze‐induced membrane lesions of protoplasts isolated from Arabidopsis thaliana leaves. Plant, Cell & Environment 26, 1083–1096 [Google Scholar]
- Verhage L, Severing EI, Bucher J, Lammers M, Busscher-Lange J, Bonnema G, Rodenburg N, Proveniers MC, Angenent GC, Immink RG. 2017. Splicing-related genes are alternatively spliced upon changes in ambient temperatures in plants. PLoS One 12, e0172950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X-L, Qi H-D, Lin Y, Ren Q-P, Wang Y-Y, Xiong F. 2019. RNA splicing of FLC modulates the transition to flowering. Frontiers in Plant Science 10, 1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TY, Juan YT, Hsu YH, Wu SH, Liao HT, Fung RW, Charng YY. 2013. Interplay between heat shock proteins HSP101 and HSA32 prolongs heat acclimation memory posttranscriptionally in Arabidopsis. Plant Physiology 161, 2075–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Liang J, Wang C, Ding L, Zhao X, Cao X, Xu S, Teng N, Yi M. 2019. Alternative splicing provides a mechanism to regulate LlHSFA3 function in response to heat stress in lily. Plant Physiology 181, 1651–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Z, Browse J. 2000. Cold comfort farm: the acclimation of plants to freezing temperatures. Plant, Cell & Environment 23, 893–902. [Google Scholar]
- Xu H, Li X, Zhang H, Wang L, Zhu Z, Gao J, Li C, Zhu Y. 2020. High temperature inhibits the accumulation of storage materials by inducing alternative splicing of OsbZIP58 during filling stage in rice. Plant Cell & Environment 43, 1879–1896. [DOI] [PubMed] [Google Scholar]
- Yan K, Liu P, Wu CA, Yang GD, Xu R, Guo QH, Huang JG, Zheng CC. 2012. Stress-induced alternative splicing provides a mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Molecular Cell 48, 521–531. [DOI] [PubMed] [Google Scholar]
- Yang Y, Li Y, Sancar A, Oztas O. 2020. The circadian clock shapes the Arabidopsis transcriptome by regulating alternative splicing and alternative polyadenylation. The Journal of Biological Chemistry 295, 7608–7619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao L, Ding C, Hao X, Zeng J, Yang Y, Wang X, Wang L. 2020. CsSWEET1a and CsSWEET17 mediate growth and freezing tolerance by promoting sugar transport across the plasma membrane. Plant & Cell Physiology 61, 1669–1682. [DOI] [PubMed] [Google Scholar]
- Yun J, Kim SG, Hong S, Park CM. 2008. Small interfering peptides as a novel way of transcriptional control. Plant Signaling & Behavior 3, 615–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Li G, Fu C, Duan S, Hu D, Guo X. 2020. Genome-wide identification, transcriptome analysis and alternative splicing events of Hsf family genes in maize. Scientific Reports 10, 8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JX, Wang C, Yang CY, Wang JY, Chen L, Bao XM, Zhao YX, Zhang H, Liu J. 2010. The role of Arabidopsis AtFes1A in cytosolic Hsp70 stability and abiotic stress tolerance. The Plant Journal 62, 539–548. [DOI] [PubMed] [Google Scholar]
- Zhang T, Huang L, Wang Y, Wang W, Zhao X, Zhang S, Zhang J, Hu F, Fu B, Li Z. 2017. Differential transcriptome profiling of chilling stress response between shoots and rhizomes of Oryza longistaminata using RNA sequencing. PLoS One 12, e0188625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Liu B, Piao S, et al. 2017. Temperature increase reduces global yields of major crops in four independent estimates. Proceedings of the National Academy of Sciences, USA 114, 9326–9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Dong CH, Zhu JK. 2007. Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Current Opinion in Plant Biology 10, 290–295. [DOI] [PubMed] [Google Scholar]