Molecular–genetic analysis of Arabidopsis mutants revealed a novel role for abscisic acid in promoting auxin-induced somatic embryogenesis at three different levels: biosynthesis, signalling, and downstream transcription.
Keywords: ABA; Arabidopsis; 2,4-D; mature zygotic embryo; somatic embryogenesis; seed maturation; totipotency
Abstract
Somatic embryogenesis (SE) is a type of induced cell totipotency where embryos develop from vegetative tissues of the plant instead of from gamete fusion after fertilization. SE can be induced in vitro by exposing explants to growth regulators, such as the auxinic herbicide 2,4-dichlorophenoxyacetic acid (2,4-D). The plant hormone abscisic acid (ABA) has been proposed to be a downstream signalling component at the intersection between 2,4-D- and stress-induced SE, but it is not known how these pathways interact to induce cell totipotency. Here we show that 2,4-D-induced SE from the shoot apex of germinating Arabidopsis thaliana seeds is characterized by transcriptional maintenance of an ABA-dependent seed maturation pathway. Molecular–genetic analysis of Arabidopsis mutants revealed a role for ABA in promoting SE at three different levels: ABA biosynthesis, ABA receptor complex signalling, and ABA-mediated transcription, with essential roles for the ABSCISIC ACID INSENSITIVE 3 (ABI3) and ABI4 transcription factors. Our data suggest that the ability of mature Arabidopsis embryos to maintain the ABA seed maturation environment is an important first step in establishing competence for auxin-induced cell totipotency. This finding provides further support for the role of ABA in directing processes other than abiotic stress response.
Introduction
Plant embryogenesis begins at fertilization with the formation of a totipotent zygote that develops into an embryo within the confines of maternal and filial seed tissues. Embryo development proceeds through defined developmental stages that are characteristic for each plant species. In the model plant Arabidopsis thaliana, the first phase of embryo development comprises a period of cell proliferation and morphogenesis, where the basic cell types, tissues, and organs are established (Zhao et al., 2017; Tian et al., 2020a). This phase is driven in part by the plant hormone auxin, which acts as a major instructor of cell identity and patterning (Smit and Weijers, 2015; Zhao et al., 2017; Figueiredo and Köhler, 2018). Thereafter, the embryo enters the maturation phase during which cell division is reduced and storage products accumulate that are used to drive embryo growth during germination (Devic and Roscoe, 2016). During the last phase of development, the desiccation and dormancy phase, the water content of the embryo decreases and the embryo enters a quiescent state (Leprince et al., 2017). The maturation and desiccation phases of embryo development are largely controlled by the plant hormone abscisic acid (ABA) (Yan and Chen, 2017) and by a well characterized network of ABA-dependent transcription factors. Among these are the LAFL [for LEAFY COTYLEDON1 (LEC1), ABSCISIC ACID INSENSITIVE 3 (ABI3), FUSCA3 (FUS3), and LEAFY COTYLEDON2 (LEC2)] and ABI4 and ABI5 transcription factors. Mutants of these transcription factors are characterized by a reduction in storage product accumulation and/or desiccation tolerance, but also by the failure to maintain embryo identity (Brocard-Gifford et al., 2003; Carbonero et al., 2016; Devic and Roscoe, 2016; Skubacz et al., 2016; Lepiniec et al., 2018). Seed dormancy can be broken in response to specific environmental signals and by hydration of the seed. During germination, ABA levels decline to promote the transition from embryo development to seedling development (Shu et al., 2016b).
Plant cells are developmentally flexible, and many plant cells other than the zygote can develop into embryos, either naturally as part of an altered seed development programme (León-Martínez and Vielle-Calzada, 2019) or when induced in vitro (Soriano et al., 2013; Horstman et al., 2017a; Testillano, 2019). Somatic embryogenesis (SE) is a type of cell totipotency in which embryos develop from vegetative tissues of the plant (Méndez-Hernández et al., 2019; Schmidt, 2020). SE can be induced in vitro by exposing explants to exogenous plant growth regulators, usually synthetic herbicidal auxins such as 2,4-dichlorophenoxyacetic acid (2,4-D), often with an additional abiotic stress treatment (Fehér, 2015; Nic-Can et al., 2016). SE forms the basis for a number of plant breeding and biotechnology applications, including clonal propagation (Park et al., 1998; Egertsdotter et al., 2019), but is also used as a model system to understand cell fate changes, in particular in Arabidopsis (Horstman et al., 2017a). SE protocols have been developed for a wide range of Arabidopsis explants, which show different levels of competence and follow different developmental routes to somatic embryo development, including directly from the explant, indirectly through callus, and by secondary SE (Luo and Koop, 1997; Gaj, 2001; Ikeda-Iwai, 2002; Ikeda-Iwai et al., 2003; Wei et al., 2006; Kobayashi et al., 2010; Horstman et al., 2017a).
At present it is not known whether these different routes to SE represent a single pathway or multiple pathways that converge at different downstream points. Nonetheless, a general framework for somatic embryo induction has been proposed in which chromatin-modifying proteins, transcription factors, stress response, and exogenous growth regulator pathways converge at the level of endogenous hormone production and signalling to reprogramme cells to a totipotent state (Fehér, 2015; Horstman et al., 2017a; Pasternak and Dudits, 2019). Direct links between embryo repressive chromatin-modifying proteins and their downstream embryo identity transcription factor genes have been established in Arabidopsis seedlings (Jia et al., 2013; Yang et al., 2013; Chen et al., 2018), as have links between embryo identity transcription factors and endogenous hormone production (Horstman et al., 2017a; Wójcik et al., 2020; Wójcikowska et al., 2020), However, it is not clear how stress modulates SE. With the exception of Daucus carota (Kamada et al., 1989, 1993; Nishiwaki et al., 2000), stress treatments on their own are not sufficient to induce SE. Rather, abiotic stress appears to act as an enhancer of plant growth regulator-induced SE (Ikeda-Iwai et al., 2003; Gaj, 2004). In addition to its role as a developmental regulator (Nambara et al., 2010; Hong et al., 2013; Yamaguchi et al., 2018; Yoshida et al., 2019), ABA has key roles as an integrator and modulator of abiotic stress response (Vishwakarma et al., 2017). It has been suggested that an ABA stress response is an important component of competence for SE, as changes in ABA levels and ABA-related gene expression can be associated with competence for SE (Gaj et al., 2006; Su et al., 2013; Fehér, 2015; Kadokura et al., 2018). In Arabidopsis, ABA enhances 2,4-D-induced SE from otherwise non-embryogenic seedling root explants of the POLYCOMB REPRESSIVE COMPLEX 2 CURLY LEAF/SWINGER mutant (clf swn) through an unknown mechanism (Mozgová et al., 2017), and modulates auxin response and transport during 2,4-D-induced secondary SE from embryogenic callus (Su et al., 2013). It is not clear whether ABA is required during SE in its role as a developmental regulator or as a stress response modulator. Neither is it known which ABA signalling components have roles during SE.
In this study, we show that 2,4-D-induced SE from the shoot apex of germinating after-ripened Arabidopsis embryos is characterized by the maintenance of an ABA-dependent seed maturation environment. We show genetically that not only ABA, but also ABA perception, signalling, and transcriptional output are required for efficient 2,4-D-induced SE. We also show that the AUXIN RESPONSE FACTORS (ARFs) ARF10 and ARF16, which act upstream and downstream of ABI3 expression, are also required for efficient SE. These data provide a mechanistic link between 2,4-D and ABA signalling in somatic embryo induction, and suggest a developmental role for ABA in promoting plant cell totipotency.
Materials and methods
Plant materials and growth
The mutant, reporter, and overexpression lines used in this study are described in Supplementary Table S1. Primers used for cloning and genotyping are listed in Supplementary Table S2.
The 35S:PYL10 vector was made by amplifying the PYL10 protein-coding sequence from Col-0 genomic DNA and then inserting it into the pGD625 binary vector by Gateway cloning (Immink et al., 2002). The 35S:ABI3 vector was made by amplifying the ABI3 protein-coding sequence from Col-0 cDNA and then inserting it into the pH7GW2 binary vector (Karimi et al., 2002) using Gateway cloning. The pBBM:BBM-GFP-GUS construct was made using a Col-0 PCR fragment containing 4200 bp upstream of the translational start codon up to the end of the BBM coding region. This PCR fragment was cloned into the pARC175 binary vector by Gateway cloning (Karimi et al., 2002), which also contains the GFP-GUS (green fluorescent protein–β-glucuronidase) reporter and the FAST-Red (OLEO:OLE01:RFP) cassette for seed selection (Castel et al., 2019). The FAST-Red cassette was introduced into the pARC175 vector in between the XabI and SpeI restriction sites.
PYL10 CRISPR/Cas9 [clustered regularly interspaced palindromic repeats/CRISPR-associated protein 9] mutagenesis was performed by combining four guide RNAs in the pAGM4723 vector using Golden Gate cloning, as described in Wang et al. (2019).
Arabidopsis Col-0 transgenics were obtained by Agrobacterium tumefaciens-mediated floral dip transformation (Clough and Bent, 1998), except for the PYL10 crispants, which were generated by A. tumefaciens-mediated root transformation (Vergunst et al., 1998). The pyl8-1/pyl10CR and pyl8-1/pyl9/pyl10CR mutants were generated by crossing pyl8-1 and pyl8-1/pyl9 with pyl10CR, respectively.
All plants were grown in a growth chamber with 70% relative humidity at 20 °C on rock wool cubes (Grodan), which were supplemented twice a week with 1 g l–1 6.5-6-19 liquid fertilizer (Hyponex). The snrk2.2snrk2.3snrk2.6 triple mutant seedlings and plants were covered with a plastic cap to maintain a high humidity level (Fujii and Zhu, 2009). Plants were maintained under LED light (150 μmol m–2 s–1) on a 16 h light/8 h dark day/night cycle. Slight differences in plant growth conditions and age at seed harvest can affect the efficiency of somatic embryo cultures (Wu et al., 2019), therefore wild-type control and mutant lines for any given experiment were always grown and harvested at the same time. Unless otherwise indicated, siliques were harvested when they were completely brown and then dried to 30% relative humidity (Wu et al., 2019).
Somatic embryo culture
Seeds were surface sterilized with liquid bleach and then added to 30 ml of 1/2 MS-10 medium [half-strength Murashige and Skoog macro- and microelements and vitamins (Murashige and Skoog, 1962; Duchefa), 1% (w/v) sucrose, pH 5.8] supplemented with 1 μM 2,4-D (Duchefa) in 190 ml plant tissue culture containers (Greiner). Approximately 60–100 seeds per container were used. The containers were placed at 4 °C in the dark for 2 d and then placed on a shaker (130 rpm) at 25 °C on a 16 h/8 h day/night cycle (100 μmol m–2 s–1). SE efficiency and productivity were determined after 2 weeks of culture by counting, respectively, the number of seedlings that formed embryogenic tissues or bipolar somatic embryos, and the number of explants with more than two somatic embryos. For some experiments, explants were transferred after 2 weeks of culture to 1/2 MS-10 medium without 2,4-D to promote embryo elongation and thereby facilitate scoring. The results for three technical replicates (same seed batch) are shown for each experiment, and are in agreement with numerous experiments with biological replicates from independent seed batches.
For the ABA treatments, a mixture of ±ABA stereoisomers (Sigma) was dissolved in DMSO and added to the SE culture medium prior to or immediately after stratification or at the indicated time during culture, and then left in the medium for the duration of the culture. The same volume of DMSO was added to control cultures.
Gene expression analysis
Arabidopsis Col-0 seeds were surface sterilized and grown in containers as described above, with or without 1 μM 2,4-D. The seeds were stratified at 4 °C in the dark for 2 d and then grown for 2 d on a 16 h/8 h day/night cycle at 25 °C on a shaker platform at 130 rpm.
Total RNA was isolated with the Invitrap Plant Spin RNA Mini Kit (Invitek), treated with DNase I (Invitrogen), and sent to the Nottingham Arabidopsis Stock Centre (NASC, http://arabidopsis.info/) for hybridization to the Arabidopsis Affymetrix GeneChip ATH1-121501 microarrays. Three biological replicates were used for the 2,4-D and control treatments.
Raw data were analysed using R Bioconductor packages (www.bioconductor.org; Gentleman et al., 2004). The raw array data were normalized using a robust multichip average (RMA) normalization, which was carried out using the affy package (Gautier et al., 2004). Probe sets that were differentially expressed were identified with linear models generated with limma using a Benjamin and Hochberg adjustment for multiple testing [false discovery rate FDR)] for calculation of the adjusted P-values (FDR values) (Ritchie et al., 2015). Gene Ontology (GO) analysis of differentially expressed genes induced by 2,4-D was performed by using DAVID with EASE score (P-value <0.05) (Huang et al., 2009).
Quantitative reverse transcription–PCR (qRT–PCR) was performed using RNA isolated with a cetyltrimethylammonium bromide (CTAB)/LiCl protocol and treated with DNase (TURBO DNA-free kit; Invitrogen). cDNA synthesis was performed with the iScript cDNA synthesis kit (BioRad). qRT–PCR was performed as previously described (Horstman et al., 2017b) using the primers shown in Supplementary Table S2. Relative gene expression was calculated according to the 2−ΔΔCT method (Livak and Schmittgen, 2001) using wild-type or DMSO-treated samples as the calibrator (as indicated) and the SAND family gene (At2g28390) and the TIP41-like gene (At4g34270) (Czechowski et al., 2005) as the reference.
Histochemistry
GUS activity was determined histochemically as previously described (Soriano et al., 2014), using 1.0–2.5 mM potassium ferri- and ferrocyanide and up to 24 h incubation time. Explants and seedlings were cleared with 70% ethanol prior to imaging.
Neutral lipids were visualized by Sudan Red staining (Sudan Red 7B, Sigma) (Brundrett et al., 1991). Whole explants were incubated for 1 h in filtered Sudan Red solution (0.5% Sudan Red in 60% isopropanol) at room temperature, followed by three washes with water.
Light images were recorded as described below.
Microscopy
For confocal laser scanning microscopy, seedlings were embedded in 0.2% agarose containing 10 µM FM4-64 (Invitrogen) (de Folter et al., 2007) and imaged with a Leica SPE DM5500 upright confocal microscope using the LAS AF 1.8.2 software. GFP and FM4-64 were excited with a 488 nm and 532 nm solid-state laser, respectively, and emissions were detected at band widths of 500–530 nm and 617–655 nm, respectively.
Light images of explants from SE culture were taken with a Nikon DS-Fi1 camera mounted on a ZEISS Stemi SV 11 binocular. Images were processed with NIS-Elements D 3.2 software.
Results
2,4-D induces SE from the shoot apical meristem (SAM) of germinating seeds
Contrary to a recent report (Wang et al., 2020), embryos from mature, after-ripened Arabidopsis seeds can be readily reprogrammed from seedling development to somatic embryo development by culturing them in 2,4-D (Mordhorst et al., 1998; Thakare et al., 2008; Kobayashi et al., 2010; Wu et al., 2019; Tian et al., 2020a). Here we followed the development of somatic embryos from mature, after-ripened seed explants treated with 1 µM 2,4-D (Wu et al., 2019) by using morphological and embryo identity markers to define the major developmental steps in this process.
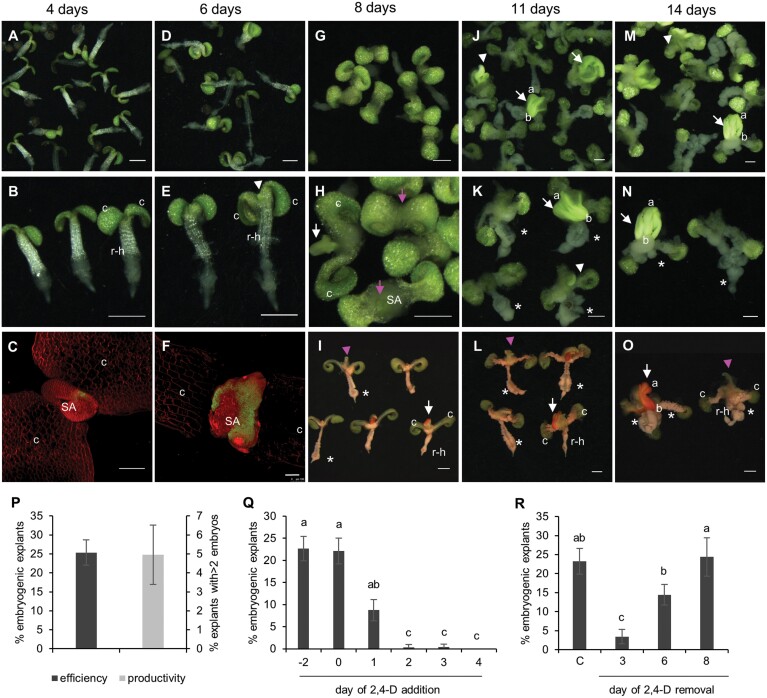
During the first 4 d of culture, the seedling cotyledons and petioles enlarged and the epidermal and cortex cells of the root elongation zone and the hypocotyl expanded and began to detach from the underlying tissue (Fig. 1A, B; Supplementary Fig. S1A). Embryogenic and non-embryogenic explants could not be distinguished morphologically on the fourth day of culture, but small patches of LEC1:LEC1-GFP embryo reporter expression could already be observed at the enlarged shoot apex of some explants (Fig. 1C). By 6 d of culture, the majority of explants had an elongated hypocotyl–root region, in which the epidermal and cortical cell layers had completely detached from the vascular cylinder above the root meristem (Fig. 1D, E). At this time, cytoplasmic dense, bright green embryogenic protrusions (Fig. 1E) (Verdeil et al., 2007; Godel-Jedrychowska et al., 2020) with LEC1–GFP expression (Fig. 1F) were observed at the shoot apex. The absence of callus at the shoot apex suggests that somatic embryos are formed directly from the shoot apex. Bipolar somatic embryos were visible at the shoot apex from day 8 of culture onward (Fig. 1G, H), but could be most clearly distinguished morphologically during days 11–14 of culture (Fig. 1J, K, M, N; Supplementary Fig. S1C). In addition to expressing LEC1–GFP, these embryogenic protrusions and bipolar somatic embryos were intensely stained by Sudan Red, a dye that stains neutral lipids including the triacylglycerols that accumulate to high levels in Arabidopsis zygotic embryos (Fig. 1I, L, O) (Brundrett et al., 1991). In non-embryogenic explants, the shoot apex either failed to develop or formed a (fused) leaf-like structure (Fig. 1I, J–O). These leaf-like structures were not stained by Sudan Red (Fig. 1I, L, O). Non-embryogenic callus developed in both embryogenic and non-embryogenic explants on the abaxial surface of the cotyledon petiole, under the shoot apex, and from the root–hypocotyl vascular cylinder (Fig. 1G–O; Supplementary Fig. S1B). SE efficiency and productivity were calculated after 14 d of culture (Fig. 1P). SE was induced in ~25% of the explants (SE efficiency) of which ~5% developed more than two bipolar embryos (SE productivity).
Fig. 1.
2,4-D-induced somatic embryogenesis (SE) from wild-type mature after-ripened embryo explants. The time of culture is indicated above the panels. (A–N) Overview of somatic embryo cultures in time. (B, E, H, K, N) Magnified images. The images are light micrographs. (C, F) LEC1:LEC1-GFP explant showing LEC1–GFP expression (green) at the shoot apex. The explants were counterstained with FM4-64 (red). The images are confocal laser scanning micrographs. (I, L, O) Sudan Red-stained explants. Sudan Red stains the bright green structures and embryos at the shoot apex, but not the ectopic leaf-like structure that develops at the shoot apex of non-embryogenic explants. The images are light micrographs. (A–O) c, cotyledon; r-h, root–hypocotyl; a, apical pole; b, basal pole; white arrowhead, embryogenic structures; white arrow, somatic embryos; pink arrowhead, leaf-like structure; pink arrow, non-embryogenic shoot apex; asterisk, callus. The scale bars are 1 mm in (A), (B), (D), (E), and (G–O), and 100 µm in (C) and (F). (P) Somatic embryogenesis efficiency (percentage of explants with embryogenic tissues and/or bipolar embryos) and productivity (percentage of explants with >2 bipolar embryos) from germinating seeds. (Q) Effect of 2,4-D addition on SE. 2,4-D was added during stratification (–2), at the start of culture (0), or at the indicated time points (1–4) after the start of cultures. (R) Effect of 2,4-D removal on SE induction. 2,4-D was added during stratification (–2) and then removed at the indicated time points by refreshing the medium. C, continuous 2,4-D treatment was used as a control. For (Q) and (R), statistically significant differences in SE efficiency were calculated using Fisher’s least significant difference test. Error bars represent the SD of three technical replicates in one experiment.
We determined the developmental window in which 2,4-D is required to induce SE in germinating embryos by adding or removing 2,4-D at different time points in culture (Fig. 1Q, R). Addition of 2,4-D during seed stratification at 4 °C or at the start of culture induced the highest SE efficiency, while adding 2,4-D at progressively later time points decreased SE efficiency, such that SE could no longer be induced when 2,4-D was added after the third day of culture (Fig. 1Q). Removal of 2,4-D after 3 d of culture dramatically decreased somatic embryo induction, while removal at later time points only had a mild effect on SE efficiency compared with continuous treatment.
These results indicate that treatment of mature after-ripened embryos with 2,4-D inhibits normal shoot apex development to promote embryogenesis, and that the developmental competence for shoot apex embryogenesis is established within 48 h of culture. These results are in contrast to previous reports showing loss of SE competence in mature embryo explants within 1 d after germination (Mozgová et al., 2017), but might reflect differences in the type of SE under study (direct versus indirect).
2,4-D maintains the seed ABA maturation pathway post-germination
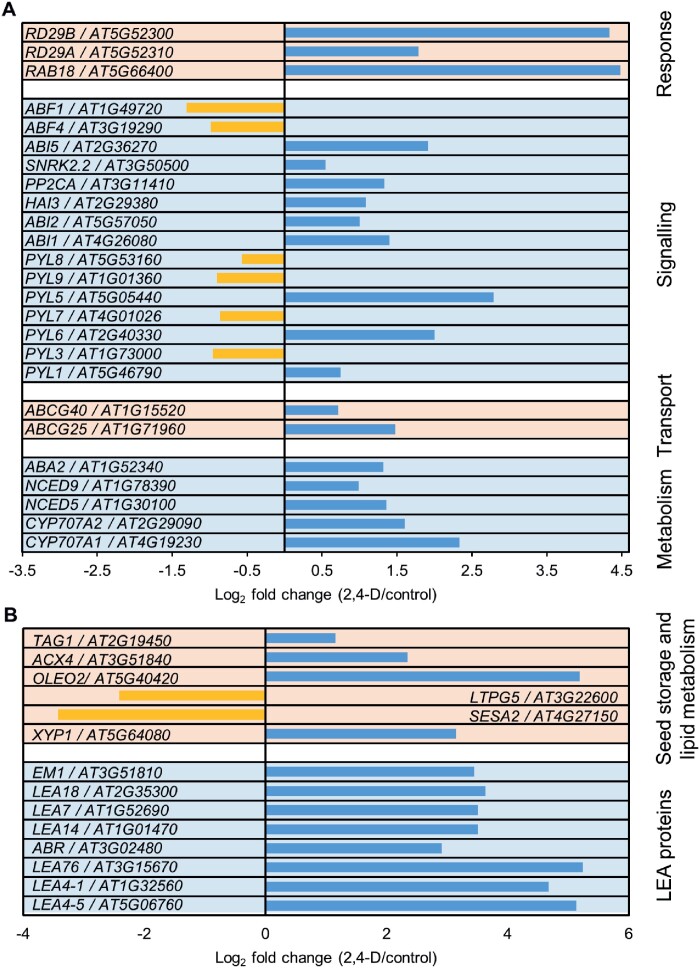
To identify the signalling pathways that are affected by 2,4-D treatment, we compared the transcriptomes of imbibed seeds cultured for 48 h in medium with or without 1 µM 2,4-D. We identified 5687 and 5300 genes that were significantly up- or down-regulated, respectively, by 2,4-D compared with the untreated control (log2 fold change >0.5 or < –0.5, FDR <0.05; see Supplementary Data Set S1). GO analysis of both up- and down-regulated genes revealed that changes in the expression of genes involved in response to cadmium ion, salt stress, cytokinin, auxin signalling, homeostasis, and response were among the most highly enriched categories (Supplementary Figs S2, S3; Supplementary Data Set S1). The 2,4-D treatment also induced statistically significant changes in expression of genes involved in ABA, dehydration, and cold stress, and seed maturation pathways (Fig. 2A, B; Supplementary Fig. S2; Supplementary Data Set S1). The expression of these seed-expressed ABA and maturation-related genes is normally down-regulated during the transition to germination (Carles et al., 2002; Lopez-Molina et al., 2002; Cadman et al., 2006; Nakashima et al., 2006; Braybrook and Harada, 2008; Yamaguchi et al., 2018), suggesting that 2,4-D treatment maintains the ABA seed maturation pathway post-germination. The differential expression of selected auxin and ABA pathway genes was confirmed by qRT–PCR analysis (Supplementary Fig. S3, Supplementary Fig. S4).
Fig. 2.
2,4-D promotes ABA-related gene expression post-germination. (A) Selection of statistically significant differentially expressed ABA-related genes. (B) Selection of statistically significant differentially expressed seed maturation genes. In (A) and (B), the gene name and Arabidopsis gene identifier (AGI), as well as the log2 fold expression change for 2,4-D-treated versus control seedlings are shown for each gene. Genes were grouped per functional category. The complete dataset can be found in Supplementary Data Set S1.
A number of Arabidopsis genes have been identified that induce spontaneous SE when ectopically expressed and/or enhance 2,4-D-induced SE (Horstman et al., 2017a). We therefore examined whether any of these genes are differentially expressed within the first few days of somatic embryo induction (Supplementary Data Set S1). Surprisingly, of these genes, only PLT1, PLT2, and BBM expression was significantly up-regulated in 2-day-old 2,4-D-treated explants. However, BBM:BBM-GUS reporter analysis (Supplementary Fig. S5) showed that BBM was not expressed at the shoot apex in 2-day-old explants, but was restricted to its normal expression domain in the root meristem (Galinha et al., 2007). BBM:BBM-GUS activity was only observed in the shoot apex of embryogenic explants from 6 d of culture onward. These data suggest that expression of somatic embryo identity genes is enhanced by 2,4-D in their natural expression domain in the root, followed later by ectopic expression in the shoot meristem. The relatively late expression of those embryo identity genes in the shoot meristem suggests that other developmental changes precede expression of SE-inducing transcription factors in the shoot meristem.
Endogenous ABA is required for efficient somatic embryo induction
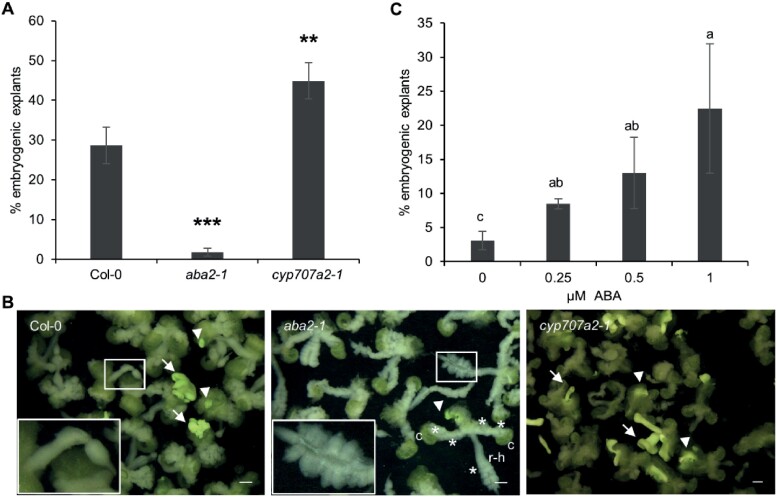
2,4-D-treated somatic embryo cultures showed up-regulation of ABA pathway genes that are normally expressed during embryo maturation and down-regulated during embryo germination (Fig. 2). ABA/ABA stress signalling has been proposed to promote SE, but the mechanism has not been well characterized. We therefore focused our subsequent analysis on the role of ABA in SE from the shoot apex of germinating embryos. Previously we showed that ABA biosynthesis is important for SE competence in fresh mature seeds (harvested from yellow siliques that are dried at 30% relative humidity and then stored at –80 °C) and aged seeds (stored at room temperature for 5 years) (Wu et al., 2019). The ABA biosynthesis mutant, aba2-1, which has reduced endogenous ABA levels (Leon-Kloosterziel et al., 1996; González-Guzmán et al., 2002), negatively affected SE efficiency in both fresh mature and aged seeds, while the cyp707a2-1 mutant, which has a higher endogenous ABA level (Kushiro et al., 2004), enhances SE efficiency (Wu et al., 2019). We obtained similar results with these mutants using mature after-ripened seed explants (Fig. 3). Compared with wild-type explants, the cyp707a2-1 mutant enhanced SE efficiency and the aba2-1 mutant reduced SE efficiency (Fig. 3A). The aba2-1 mutant also developed more callus on the cotyledon petioles, under the shoot apex, and throughout the root–hypocotyl region than wild-type explants (Fig. 3B). The reduction in SE efficiency in the aba2-1 background could be fully complemented by addition of 1 µM ABA to the culture medium (Fig. 3C). The mutant phenotypes and ABA complementation experiments indicate that endogenous ABA is required and limiting for efficient SE. However, we have shown previously that treatment of mature after-ripened seed explants with exogenous ABA slightly inhibits SE (Wu et al., 2019). Thus, although ABA levels are limiting for SE from the shoot apex, they also need to be tightly regulated to promote SE.
Fig. 3.
Endogenous ABA is required for efficient somatic embryogenesis. (A) The effect of ABA biosynthesis mutants on SE. Statistically significant differences in SE efficiency between the wild type and the mutant lines were calculated using a two-tailed Student’s t-test (**P<0.01; ***P<0.001). Error bars represent the SD of three technical replicates. (B) Light images of 14-day old explants from somatic embryo cultures of the indicated wild-type and mutant lines. The insets in the wild-type and aba2-1 panels are magnifications of the respective boxed regions, showing excessive callus formation at the root–hypocotyl region of the explant of the aba2-1 mutant. The images are light micrographs. Arrows, somatic embryos; arrowheads, embryogenic tissue; asterisks, callus; c, cotyledon; r-h, root–hypocotyl. Scale bars, 1 mm. (C) Application of exogenous ABA restores SE efficiency to wild-type Col-0 levels in the aba2-1 mutant. The Col-0 control was previously reported by Wu et al. (2019). Statistically significant differences in SE efficiency were calculated using Fisher’s least significant difference test. Error bars represent the SD of three technical replicates.
The ABA receptor complex positively regulates auxin-induced SE
Given the requirement of endogenous ABA for somatic embryo development from germinating seeds (Fig. 3; Wu et al., 2019), we focused our efforts on identifying the specific components of the ABA signalling pathway (Fig. 6) that are required for this developmental process. ABA is perceived and transduced by a ternary ABA signalling complex comprising RCAR/PYR1/PYL ABA receptors (hereafter referred to as PYLs), clade A PP2C protein phosphatases, and SnRK2 kinases (Fujii et al., 2009; Ma et al., 2009; Park et al., 2009). ABA-bound PYLs interact with and inhibit PP2Cs, which in turn promotes activation of SnRK2 kinases such as SnRK2.2/3/6. The activated SnRK2 protein kinases phosphorylate and activate various downstream substrates, including ABA-responsive transcription factors (Fujii et al., 2009; Melcher et al., 2009; Yin et al., 2009; Raghavendra et al., 2010; Soon et al., 2012; Xie et al., 2012).
Fig. 6.
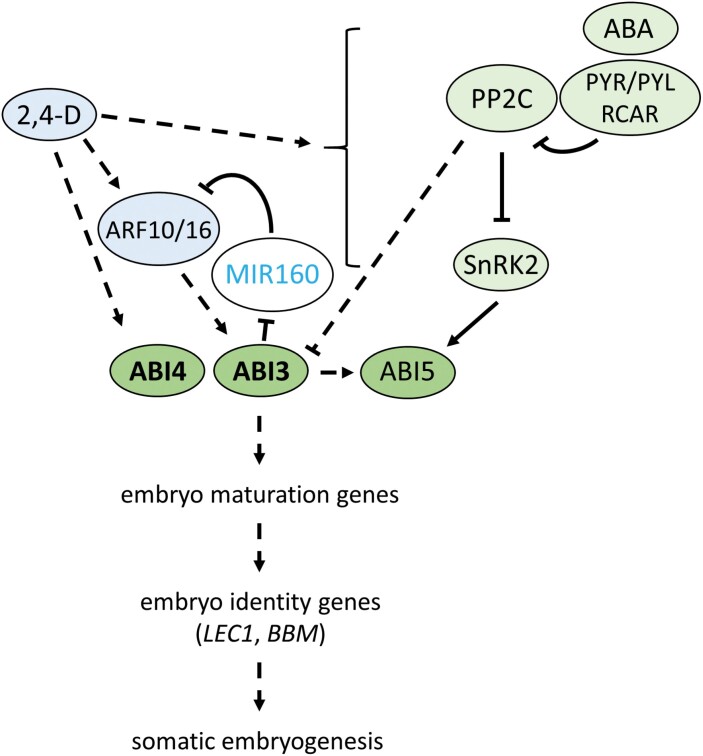
A proposed model for auxin and ABA interaction during somatic embryogenesis (SE) in Arabidopsis mature after-ripened embryos. Auxin treatment induces expression of ABA signalling genes and maintains expression of the downstream ABI3/4/5 transcription factor genes. ABA signalling and ABI5 transcription factors are required for efficient SE from germinating seeds, while ABI3 and ABI4 (bold) are essential for SE. ABA signals through the upstream components, including the receptors and the PP2Cs, to indirectly up-regulate ABI3 (Lopez-Molina et al., 2002), while core ABA signalling promotes ABI5 phosphorylation (Fujii et al., 2009; Melcher et al., 2009; Yin et al., 2009). ABI3 also regulates ARF10 and ARF16 expression through MIR160, which post-transcriptionally regulates ARF10 and ARF16 levels (Tian et al., 2020b). Dashed lines indicate indirect transcriptional regulation, while black solid lines indicate known protein-level regulation.
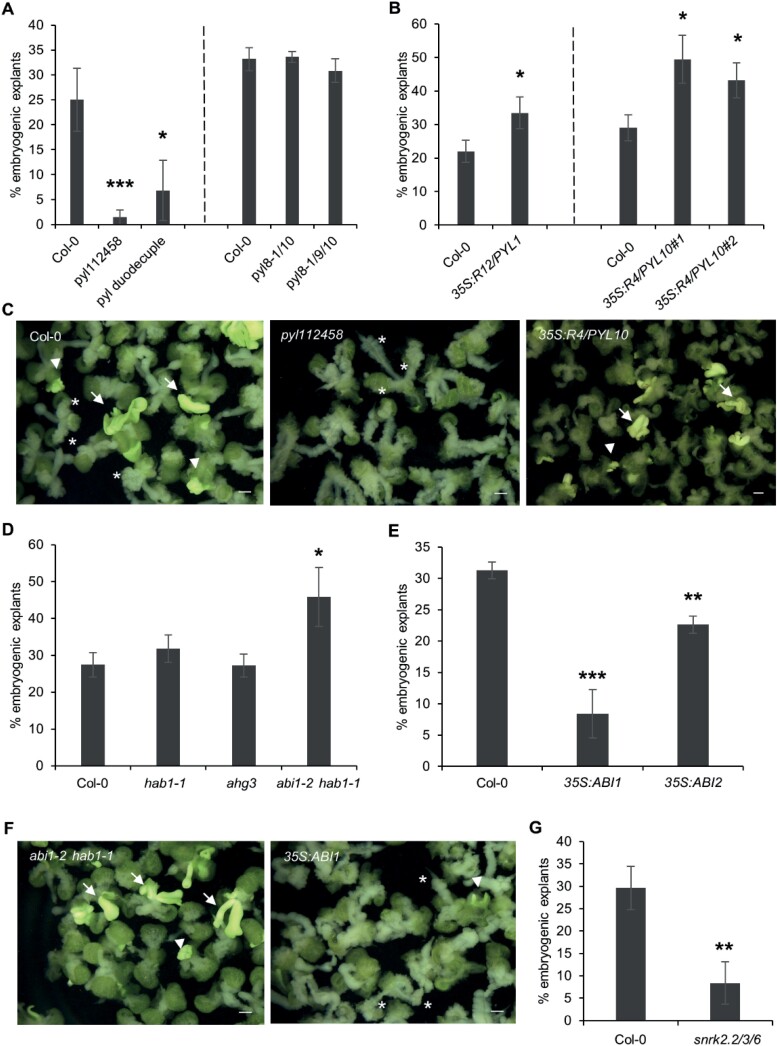
We first determined whether ABA receptors play a role in somatic embryo induction from germinating seeds. Our transcriptome analysis showed that of the 14 Arabidopsis PYL genes, seven were (differentially) expressed in 2,4-D-treated cultures, including PYL1 (up-regulated) and PYL3 (down-regulated) in subfamily III, PYL5 and PYL6 (both up-regulated) in subfamily II, and PYL7, PYL8, and PYL9 (all down-regulated) in subfamily I (Fig. 2A; Supplementary Data Set S1). PYL10, PYL11, PYL12, and PYL13 are not represented on the ATH1 microarray. Given the functional redundancy between ABA receptors (Park et al., 2009; Zhao et al., 2018), we analysed higher order RCAR/PYR1/PYL mutants for their effect on SE. ABA signalling is blocked to a large extent in the pyl112458 sextuple mutant, which carries T-DNA insertions in the PYL1, PYL2, PYL4, PYL5, and PYL8 genes and a point mutation in PYR1 (Gonzalez-Guzman et al., 2012). This sextuple mutant had a strong negative effect on somatic embryo formation (Fig. 4A, C) that could not be rescued by exogenous ABA application (Supplementary Fig. S6), suggesting that the requirement for ABA for efficient SE depends on a functional ABA receptor complex. Similarly, the pyl duodecuple mutant (pyl112458379101112), in which only one functional ABA receptor, PYL6, is a wild-type allele (Zhao et al., 2018), also had a negative effect on somatic embryo development (Fig. 4A). The phenotype of these higher order ABA receptor mutants resembled that of the aba2-1 mutant explants, in which SE efficiency was compromised and non-embryogenic callus formation was stimulated. Of the genes in the PYL7, PYL8, PYL9, PYL10 subfamily (subfamily I), loss-of-function mutants are only available for PYL8 and PYL9. We therefore made a PYL10 null mutant using CRISPR/Cas9 mutagenesis (pyl10CR) and crossed this mutant with PYL8 and PYL9 null mutants to obtain double and triple mutants (pyl8-1pyl10CR and pyl8-1pyl9pyl10CR). The pyl8-1pyl10CR and pyl8-1pyl9pyl10CR mutants did not show obvious mutant phenotypes during somatic embryo culture (Fig. 4A). In contrast to the negative effect of the loss-of-function pyl mutants on SE, overexpression of both RCAR12/PYL1 (35S:RCAR12/PYL1; Yang et al., 2016) and RCAR4/PYL10 (35S:RCAR4/PYL10) (Fig. 4B, C; Supplementary Fig. S7) slightly, but significantly, enhanced SE efficiency. Together, the data from both loss of function and overexpression of ABA receptor complex components suggest that functional ABA receptors are required for efficient SE and that enhanced basal ABA signalling promotes 2,4-D-induced SE.
Fig. 4.
Signalling through the ABA receptor complex positively regulates 2,4-D-induced somatic embryogenesis. (A) Somatic embryogenesis (SE) efficiency in ABA receptor mutants. The corresponding wild-type Col-0 control is shown for each set of mutants. (B) Effect of overexpression of the RCAR12/PYL1 and RCAR4/PYL10 ABA receptor genes on SE efficiency. Independent experiments are separated by a dashed line in (A) and (B). (C) Overview of 14-day-old wild-type Col-0 and ABA receptor mutant phenotypes in somatic embryo culture. The images are light micrographs. (D) Effect of PP2C single and double mutants on SE efficiency. (E) Effect of overexpression of the PP2C genes ABI1 and ABI2 on SE efficiency. (F) Overview of 14-day-old PP2C mutant phenotypes in somatic embryo culture. The images are light micrographs. (G) Effect of SnRK2 mutants on SE efficiency. Statistically significant differences in SE efficiency between wild-type Col-0 and mutant explants were calculated using a two-tailed Student’s t-test (*P<0.05; **P<0.01; ***P<0.001). Error bars represent the SD of three technical replicates. Arrows, somatic embryos; arrowhead, embryogenic tissue; asterisks, callus. Scale bars, 1 mm.
Next, we determined whether mutants for PP2C protein phosphatases (negative ABA signalling regulators) and SnRK2 protein kinases (positive ABA signalling regulators) affect 2,4-D-induced somatic embryogenesis from germinating seeds. ABI1, ABI2, and AHG3/PP2CA are up-regulated in our 2,4-D-treated seed explants (Fig. 2A; Supplementary Data Set S1). These genes are known to play roles in ABA-mediated repression of seed germination together with the PP2C phosphatase gene HAB1 (Rubio et al., 2009). SE efficiency was not affected in the hab1-1 or ahg3 single mutants, while the higher order abi1-2 hab1-1 PP2C mutant showed enhanced SE efficiency (Fig. 4D, F). Accordingly, overexpression of either ABI1 (35S:ABI1) or ABI2 (35S:ABI2) (Wang et al., 2018) inhibited somatic embryo formation (Fig. 4E, F). Compared with wild-type explants, explants from the abi1-2 hab1-1 loss-of-function mutant did not show any abnormal phenotypes, while ABI1 overexpression explants produced more non-embryogenic callus over the entire explant (Fig. 4F).
Among the subclass III SnRK2 kinases, only SnRK2.2 was up-regulated by 2,4-D treatment (Fig. 2A; Supplementary Data Set S1). SnRK2.2, SnRK2.3, and SnRK 2.6 function redundantly in the regulation of ABA-mediated seed germination, therefore the snrk2.2snrk2.3snrk2.6 triple mutant (Fujii and Zhu, 2009) was evaluated for its effect on SE. This mutant had a strong negative effect on SE from germinating seeds (Fig. 4G). As with other positive regulators of ABA signalling, snrk mutant explants produced more non-embryogenic callus than wild-type explants (data not shown).
Together, these data indicate that signalling through the ABA receptor complex, from ABA perception to protein kinase function, is required for 2,4-D-mediated SE from germinating seeds.
Seed maturation transcription factors positively regulate auxin-induced SE
The ABI3, ABI4, and ABI5 transcription factor genes act downstream of ABA signalling and encode, respectively, B3-, APETALA2- (AP2), and basic leucine zipper- (bZIP) domain DNA-binding proteins. These ABI genes were identified based on genetic screens for mutants that are insensitive to ABA during seed germination, but were later shown to also have overlapping roles in seed maturation (Parcy et al., 1994; Carles et al., 2002; Penfield et al., 2006; Delmas et al., 2013), in abiotic stress responses during seed germination and seedling growth (Söderman et al., 2000; Lopez-Molina et al., 2001, 2002; Brocard et al., 2002), as well as other post-germination functions (Brady et al., 2003; Shkolnik-Inbar and Bar-Zvi, 2010; Wang et al., 2013). ABI3, ABI4, and ABI5 are subject to extensive transcriptional cross-regulation during seed maturation and germination (Söderman et al., 2000; Yan and Chen, 2017). A number of ABI3, ABI4, and ABI5 target genes were up-regulated in the transcriptome dataset, including the LEA genes LEA4-1, LEA76, AT5G44310, and AT4G21020, and PGIP1 and NYC1 (Fig. 2B; Supplementary Fig. S4; Supplementary Data Set S1; Reeves et al., 2011; Mönke et al., 2012; Skubacz et al., 2016) Given the expression patterns of ABA- and ABI-regulated seed maturation pathway genes, we examined the expression and function of the three ABI genes during SE. Microarray-based transcriptome analysis showed that of these three genes, only ABI5 expression was up-regulated in 2,4-D-treated mature after-ripened seeds (Fig. 2A; Supplementary Data Set S1), but qRT–PCR analysis showed that ABI3 and ABI4 expression was also up-regulated significantly at this time point (Supplementary Fig. S4).
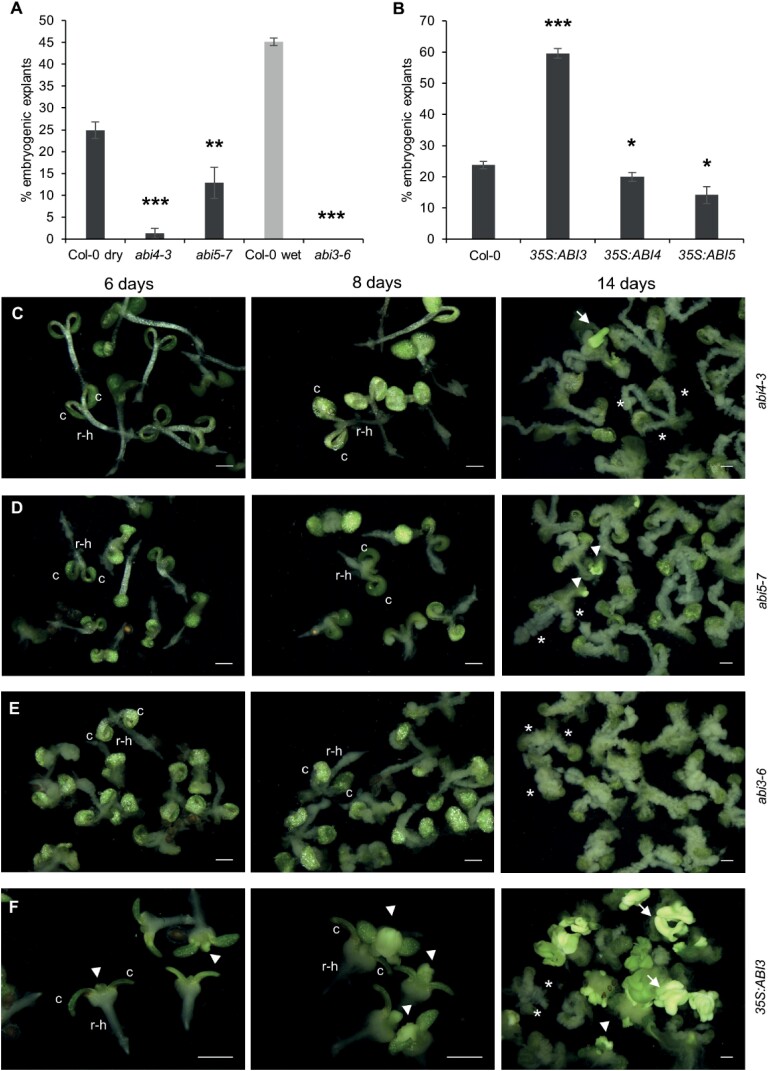
Next, we examined the effect of ABI3, ABI4, and ABI5 mutant and overexpression lines on 2,4-D-induced somatic embryo induction. abi3 null mutants are desiccation intolerant; therefore, we analysed the response of weak abi3 alleles (abi3-8, abi3-9, and abi3-10; Nambara et al., 2002) using mature, after-ripened seed explants, and the response of the abi3-6 null allele (Nambara et al., 1994) using mature wet seed explants. All abi mutants displayed a strong reduction in SE efficiency from mature seed explants, with the abi3-6 (0%) and abi4 mutants (2–5%) being most severely affected (Fig. 5A–E; Supplementary Fig. S8). Consistent with our results on the core ABA signalling complex, abi3, abi4, and abi5 explants also produced more non-embryogenic callus than wild-type explants (Fig. 5C–E). The higher SE response in the abi3-8, abi3-9, and abi3-10 mutants compared with the abi3-6 mutant might be due to, respectively, partial versus complete loss of function of the mutant alleles (Nambara et al., 2002; Delmas et al., 2013). The higher SE responses in abi5-7 compared with abi4-3 and abi3-6 might be due to functional redundancy of ABI5 with another eight ABRE-binding factors (ABFs). The number of embryogenic explants was also severely reduced in the weak abi3 mutants and in the abi4 and abi5-7 mutants.
Fig. 5.
Seed maturation transcription factors are required for efficient somatic embryogenesis (SE). (A) Effect of ABI4, ABI5, and ABI3 loss-of-function mutants on 2,4-D-induced SE. The abi3-6 mutant is desiccation intolerant, therefore mature wet seeds were analysed for this mutant, using mature wet Col-0 seeds as a control. (B) Effect of ABI3, ABI4, and ABI5 overexpression on 2,4-D-induced SE. (C–F) Overview of somatic embryo cultures in time for the indicated mutants. The images are light micrographs. The day of culture is indicated above the panels. Statistically significant differences in SE efficiency between wild-type Col-0 and mutant explants were calculated using a two-tailed Student’s t-test (*P<0.05; **P<0.01; ***P<0.001). Error bars represent the SD of three technical replicates. c, cotyledon; r-h, root–hypocotyl region; arrow, somatic embryos; arrowheads, embryogenic tissues; asterisks, callus formed on the petioles or the root–hypocotyl region. Scale bars, 1 mm.
In wild-type immature zygotic embryo explants, prolific SE takes place on the adaxial surface of the cotyledon petiole, while a single somatic embryo develops from the shoot apex (Supplementary Fig. S9A). abi3-6 immature zygotic embryo explants produced somatic embryos on the adaxial surface of the petiole, as in wild-type explants, but, unlike wild-type explants, a shoot-like structure developed from the apex instead of a somatic embryo (Supplementary Fig. S9B). These results indicate that abi3-6 immature zygotic embryo explants retain the ability to form somatic embryos from the petiole, but not from the shoot apex (Supplementary Fig. S9C), and suggest that ABI3 is required to repress shoot development from the shoot apex.
We determined whether exogenous ABA application could improve SE efficiency in the abi3, abi4, and abi5 mutant backgrounds. ABA application did not improve SE efficiency in the abi3-6, abi4-1, or abi4-2 backgrounds, but did have a positive effect on SE in the abi5-7 background (Supplementary Fig. S10), suggesting that auxin-induced SE is less dependent on ABI5 than on ABI3 and ABI4.
Next, we examined the effect of ABI3, ABI4, and ABI5 ectopic overexpression on somatic embryo formation using 35S:ABI3 (Supplementary Fig. S7), 35S:ABI4, and 35S:ABI5 lines (Brocard et al., 2002; Shu et al., 2013). ABI3 overexpression enhanced, while ABI4 and ABI5 overexpression reduced, SE efficiency in 2,4-D-treated cultures (Fig. 5B, F). 2,4-D-treated 35S:ABI3 explants had a larger embryogenic shoot apex than 2,4-D-treated wild-type explants, which was already visible after 4 d of culture compared with 6 d in wild-type explants (compare Fig. 1E and Fig. 5F). A similarly enlarged shoot apex was not observed in 2,4-D-treated ABI4 and ABI5 overexpression lines.
Collectively, our genetic data showed that ABI3, ABI4, and ABI5 positively regulate auxin-induced SE from germinating seeds. SE efficiency was reduced in the abi5-7 mutant, even more so in the abi4-3 mutant, and was completely abolished in the abi3-6 mutant. The complete absence of embryogenic growth in the abi3-6 mutant and the positive effect of ABI3 overexpression on SE suggests that ABI3 expression is essential and limiting for somatic embryo initiation. The slight negative effect of ABI4 and ABI5 overexpression on SE efficiency might reflect additional stress signalling roles for these proteins that interfere with SE. Exogenous ABA application enhanced SE in the abi5 loss-of-function mutant, but not in the abi3 and abi4 loss-of-function mutants. Together, these data suggest that ABI3 and ABI4 are the main downstream effectors of ABA signalling during SE.
ABI3 is a key component of 2,4-D-induced SE from mature after-ripened embryos. Auxin signals through the ARFs ARF10 and ARF16 to maintain ABI3 expression and enhance ABA-mediated inhibition of seed germination (Liu et al., 2013). ABI3 also regulates ARF10 and ARF16 expression through MIR160, which post-transcriptionally regulates ARF10 and ARF16 levels (Tian et al., 2020b). Therefore, we determined whether mutations in ARF10 and ARF16 influence 2,4-D-induced SE. The efficiency of 2,4-D-induced SE was significantly reduced in the arf10/16 double mutant (Supplementary Fig. S12), suggesting that 2,4-D and ABA converge at the level of ARF10 and/or ARF16 and ABI3.
Developmental timing of ABI3 and ABI4 expression in SE culture
ABI3 and ABI4 are strong positive regulators of auxin-induced SE. We examined their expression patterns during the course of somatic embryo induction to better understand their roles in this process. ABI3 and ABI4 expression was followed during SE culture using ABI3:GUS (Ryu et al., 2014) and ABI4:GUS (Söderman et al., 2000) reporter lines (Supplementary Fig. S11). Seeds were cultured with or without 2,4-D, and explants stained for GUS activity over an 8 d period. ABI3 (Supplementary Fig. S11A) was expressed initially in the cotyledons and the hypocotyl of germinating seeds from both auxin-treated and control seedlings, with higher expression in 2,4-D-treated samples at day 2 of culture. Four days after the start of culture, ABI3 expression could no longer be detected in control seedlings, but was still present in auxin-treated explants. Later, from day 6 to day 8 of culture, ABI3:GUS expression became restricted to the cotyledons and shoot apex (Supplementary Fig. S11A). ABI4 expression (Supplementary Fig. S11B) was also gradually lost in control seedlings compared with 2,4-D-treated seedlings, although ABI4 expression declined earlier than ABI3 expression. No obvious difference in ABI3- or ABI4-driven GUS activity was found between embryogenic and non-embryogenic explants from 2,4-D-induced somatic embryo culture (Supplementary Fig. S11).
ABI3 and ABI4 are induced transiently during seed germination (https://www.bioinformatics.nl/dormancy/). Our reporter, microarray, and qPCR data therefore suggest that ABI gene expression is maintained post-germination in response to auxin treatment. The developmental window in which ABI expression normally decreases in control seedlings corresponds to the window for efficient 2,4-D-induced SE. ABI3 and ABI4 are essential for SE, but GUS reporter analysis suggests that they are not differentially regulated at the transcriptional level between embryogenic and non-embryogenic explants. The lack of difference in expression of these two genes between embryogenic and non-embryogenic explants suggests that ABI3 and ABI4 expression is regulated post-transcriptionally in these explants (Zhang et al., 2005; Finkelstein et al., 2011; Gregorio et al., 2014).
Discussion
The vast majority of SE protocols use 2,4-D as the inducer treatment, alone or in combination with an abiotic stress treatment (Gaj, 2004). 2,4-D treatment has also been shown to induce a transcriptional stress response during SE (Rai et al., 2011; Nic-Can and Loyola-Vargas, 2016; Kadokura et al., 2018). Somatic embryo induction by stress treatment alone has rarely been described (Kamada et al., 1989, 1993; Nishiwaki et al., 2000), suggesting that a stress response is in itself not sufficient for somatic embryo initiation, but rather is needed to enhance the effect of auxin treatment. The role of ABA as a core regulator of diverse plant abiotic stress responses has been well documented in many plant species (Vishwakarma et al., 2017; Cho et al., 2018). It is clear that auxin interacts with the ABA pathway during somatic embryo induction, but it is not clear whether these interactions are stress related or simply reflect developmental roles for ABA in basal signalling pathways (Yoshida et al., 2019). The observation that ABA modulates auxin response and transport during 2,4-D-induced secondary SE from embryogenic callus in Arabidopsis supports a developmental role for ABA during SE (Su et al., 2013), but it is not known which ABA signalling components regulate this response.
Here we show that 2,4-D-induced SE from mature after-ripened Arabidopsis embryos induces a transcriptional cascade that is characteristic for the ABA seed maturation pathway (Fig. 2). Genes in this pathway are normally down-regulated in mature after-ripened seeds or during germination, but their expression is maintained when imbibed seeds are cultured in 2,4-D. We show that ABA promotes and is limiting for SE at three different levels: ABA biosynthesis, ABA receptor complex signalling, and ABA-mediated transcription, and that ABI3 and ABI4 are essential players in this process (Figs 3–5). Our results suggest a novel developmental role for a basal ABA signalling pathway in modulating auxin-dependent cell fate changes in the shoot apex.
SE requires and is limited by upstream components of the ABA signalling pathway
Endogenous ABA is required for 2,4-D-induced SE from the shoot apex of germinating seeds (Fig. 3). Higher order ABA receptor mutants, where ABA signalling is blocked to a large extent (Gonzalez-Guzman et al., 2012), also show reduced SE efficiency that cannot be complemented by exogenous ABA (Figs 4, 5). Together, these data suggest that a basal ABA level is required for SE and that ABA signals through the RCAR/PYR1/PYL receptor complex to regulate 2,4-D induced SE.
Endogenous ABA is required for 2,4-D-induced (indirect) secondary SE from callus, where it is thought to modulate auxin response and transport during secondary embryo outgrowth (Su et al., 2013). In this system, immature zygotic embryos are cultured in 2,4-D to induce somatic embryo formation and then callus formation, followed by secondary SE from callus after removal of auxin from the medium. Here SE is induced by 2,4-D treatment directly from the SAM of germinating mature after-ripened embryos. Whether ABA-regulated auxin response and transport is a common component of/is required for SE systems that rely on different explants and follow different developmental pathways (primary/secondary, direct/indirect) remains to be determined by genetic analysis.
In line with the transcriptome data showing 2,4-D-induced PYL1 and PYL5 up-regulation (Fig. 2A), the higher order loss-of-function mutant (pyl112458) in subfamilies I, II, and III negatively affected SE progression (Fig. 4A), while overexpression of PYL1/RCAR12 (subfamily III) enhanced SE (Fig. 4B). Expression of the subfamily I receptor genes PYL7, PYL8, and PYL9 was down-regulated by 2,4-D treatment (Fig. 2A), but pyl8-1, pyl9, and pyl10CR mutant combinations had no effect on SE efficiency (Fig. 4A). However, we cannot rule out a (different) role for subfamily I RCAR/PYR1/PYL receptors, as genetic redundancy between PYL7 and the other subfamily I members and/or other receptor subfamilies might mask a role for these genes during SE (Park et al., 2009; Zhao et al., 2018). Although subfamily I RCAR/PYR1/PYL receptors do not appear to have a major role in SE, overexpression of one subfamily 1 receptor, PYL10, did enhance SE efficiency. The ability of 35S:PYL10 to enhance SE might therefore indicate a lack of specificity of the ABA receptors with respect to the downstream signalling pathways that are regulated during 2,4-D-induced SE.
Overexpression of the ABI1 and ABI2 PP2C protein phosphatase genes inhibited SE (Fig. 4E), while the abi1 hab1 double mutant showed enhanced SE, in line with their role as negative regulators of ABA signalling (Fig. 4D). According to the transcriptome data, expression of the ABI1, ABI2, and AHG3/PP2CA PP2C protein phosphatase genes, which are negative regulators of ABA signalling, was slightly up-regulated by 2,4-D treatment (Fig. 2A). This suggests that up-regulation of these PP2C genes after 2,4-D treatment is due to negative feedback regulation that keeps downstream ABA signalling in check (Maia et al., 2014).
Overall, we show dependence on various mediators of ABA biosynthesis and signalling to enhance 2,4-D-induced SE. However, the transcriptome data imply that the ABA transcriptional response is far more complex. For example, genes for both ABA biosynthesis (ABA2, NCED5, and NCED6) and inactivating enzymes (CYP707A1 and CYP707A2) are up-regulated after 2,4-D treatment (Fig. 2A), yet our mutant analysis showed that increased SE potential was correlated with loss of CYP707A2 activity (Fig. 3A). Our transcriptome data were obtained from whole embryos, while only a subset of the explant cells contribute either cell autonomously or non-autonomously to somatic embryo competence. In addition, the time point at which specific genes function during the course of culture needs to be taken into consideration, and can be difficult to define with mutants. Additional studies using RNAi and reporter lines for specific genes, as well as pharmacological intervention (Park et al., 2009; Nemoto et al., 2018), will help to resolve the contributions of the different signalling components to 2,4-D-induced SE.
Auxin maintains the seed maturation environment
In Arabidopsis, ABI4 and ABI5 together with LAFL genes (LEC1, ABI3, FUS3, and LEC2) control the maturation and desiccation phases of zygotic embryo development (Brocard-Gifford et al., 2003; Carbonero et al., 2016; Skubacz et al., 2016; Lepiniec et al., 2018). Mutant analysis has shown that LAFL genes also regulate other aspects of embryo development, including repression of seedling-expressed genes in the early embryo (Yamamoto et al., 2014) and promotion of suspensor and cotyledon development. In line with these functions, ectopic overexpression of these genes in seedlings confers embryo identity traits, and in the case of LEC1 and LEC2 also induces spontaneous SE (Parcy et al., 1994; Lotan et al., 1998; Stone et al., 2001, 2008; Gazzarrini et al., 2004; Braybrook et al., 2006; Horstman et al., 2017a). LAFL and ABI genes are regulated in part by larger, complex transcriptional and post-transcriptional feedback LAFL loops during seed development (Gazzarrini et al., 2004; Zhang et al., 2005; To et al., 2006; Lepiniec et al., 2018). Unlike LEC1/2 overexpression, ectopic expression of ABI genes has not been reported to induce SE, but does confer seed maturation traits such as storage product accumulation (Parcy et al., 1994; Reeves et al., 2011).
ABI4, ABI5, and LAFL gene expression begins early in embryo development and decreases or becomes restricted to a subset of tissues in germinating seeds and seedlings (Brocard et al., 2002; Kroj et al., 2003; To et al., 2006; Braybrook and Harada, 2008; Wang et al., 2010; Wind et al., 2013). Our data show that expression of ABI3, ABI4, ABI5, and other ABA signalling genes is maintained within the first 48 h of 2,4-D treatment (Fig. 2A), yet most known SE inducers or enhancers were either not differentially expressed or were down-regulated at the same time point (Supplementary Dataset S1). Three genes, BBM, PLT1, and PLT2, showed significant up-regulation after 2 d of 2,4-D treatment, but we demonstrated that at this time point, BBM is expressed in the root rather than the shoot, and that ectopic expression at the shoot apex, the site of somatic embryo initiation, occurs later, after 6 d of culture, following LEC1 expression (Supplementary Fig. S5). These data suggest a two-step mechanism for SE induction in which 2,4-D first induces an ABA seed maturation response, followed by induction of embryo identity genes such as LEC1 and BBM (Supplementary Fig. S13). 2,4-D and 2,4-D-induced maintenance of ABA-related gene expression during seed germination and beyond might be required to create a permissive transcriptional environment for expression of embryo identity genes such as LEC1 and BBM.
Polycomb Repressive Complex 2 (PRC2) proteins regulate the transition from seed development to seed germination by repressing seed dormancy and embryo maturation traits in seedlings (Mozgova et al., 2015). In Arabidopsis, shoots from 7-day-old clf swn seedlings occasionally make differentiated somatic embryos (Mozgová et al., 2017). A combined wounding and 2,4-D treatment provides the additional competence for efficient somatic embryo induction in the shoot meristem of clf swn seedlings, but is not sufficient to induce SE from root tissues. ABA response appears to be limiting in clf swn roots, as addition of exogenous ABA to 2,4-D-treated roots is sufficient to induce SE. In this study, we found that endogenous ABA is limiting and required for efficient 2,4-D-induced SE from the shoot apex of wild-type mature embryo explants. However, exogenous ABA application does not induce SE from the explant root and actually inhibits SE from the shoot apex. This suggests that the pathways leading to 2,4-D-induced SE from mature embryos and from seedlings are different.
Two mechanisms for SE from embryo explants
In Arabidopsis, SE can be induced from both immature bent cotyledon stage embryos and embryos from mature after-ripened seeds. SE efficiency is much higher in immature zygotic explants (~80%) than in embryo explants from mature after-ripened seed (~20%). The tissue competence for SE also differs between these two explants. In mature embryo explants, somatic embryos develop from the shoot apex, while in immature zygotic embryo explants somatic embryos develop from the cotyledon petioles and only a single somatic embryo develops from the shoot apex. Thus, overall and tissue competence for SE is gradually reduced during the late maturation phase (Gaj et al., 2005; Wu et al., 2019).
The aba2, abi3, abi4, and abi5 mutants showed reduced SE from the shoot apex of mature geminating seeds, yet similar mutants show either normal or less severely reduced SE efficiency from the cotyledonary petioles of immature zygotic embryo explants (Gaj et al., 2006). In wild-type immature zygotic embryo explants, somatic embryos develop from the petioles and shoot apex, but in abi3-6 explants somatic embryos only develop from the petioles (Supplementary Fig. S9). This suggests two different mechanisms for 2,4-D-induced SE, one that operates in the cotyledonary petioles and one that operates in the shoot apex.
In immature Arabidopsis zygotic embryo explants, LEC1/2 and FUS3 are still relatively highly expressed in the cotyledons (Lotan et al., 1998; Kroj et al., 2003; To et al., 2006). SE is severely compromised in lec1, lec2, and fus3 immature zygotic embryos, and any embryos that develop, develop indirectly from callus rather than directly from the protoderm as in wild-type explants (Gaj et al., 2005). Mature seeds show no or low LEC1/2 and FUS3 expression (Lotan et al., 1998; Stone et al., 2001; Lu et al., 2010; Junker and Bäumlein, 2012), but LEC1 expression can be induced after 4 d of 2,4-D treatment. Thus, the existence of a largely intact embryo identity programme in cotyledonary petioles might be sufficient to facilitate 2,4-D-induced SE from immature zygotic embryo cotyledons, even in the absence of individual ABI genes, while reinduction of a similar state is required for SE from the shoot apex of germinating embryos. These results are in line with studies showing that ectopic expression of the LEC1, LEC2, and FUS3 seed maturation transcription factors can induce and/or enhance SE in different explants (Lowe et al., 2003; Zhang et al., 2014; Liu et al., 2018).
Somatic embryo development from the shoot apex of germinating embryos relies on ABI3, ABI4, and ABI5 (Fig. 5A). Little is known about the involvement of these genes in shoot meristem development, but roles for ABI3, ABI4, and ABI5 in the auxin-dependent control of (lateral) root meristem size/number have been described (Brady et al., 2003; Shkolnik-Inbar and Bar-Zvi, 2010; Yuan et al., 2014; Ding et al., 2015; Mu et al., 2017). Only ABI3 has been shown to have a role in development of the shoot apex. abi3 embryos show seedling-like characteristics, including premature activation of the shoot meristem and development of leaf primordia (Nambara et al., 1995; Holdsworth et al., 1999). ABI3 also promotes vegetative shoot meristem quiescence in seedlings in response to ABA and dark (Rohde et al., 1999). Meristems of dark-grown seedlings show ectopic ABI3 expression and activation of a seed storage protein gene reporter (Rohde et al., 1999), suggesting that meristem quiescence involves transdifferentiation to an embryogenic state. ABI gene expression is low/repressed in germinating embryos, allowing them to transition to vegetative growth (Lopez-Molina et al., 2001, 2002; Wind et al., 2013; Lepiniec et al., 2018), but is maintained after 2,4-D treatment (Supplementary Fig. S4) . We propose that ectopic ABI expression in germinating embryos represses vegetative shoot differentiation, which provides the developmental framework required for 2,4-D-induced totipotent cell growth. In germinating embryos, 2,4-D represses vegetative shoot meristem development in favour of somatic embryo development, while (lateral root meristem) callus formation is induced on the abaxial surface of the cotyledons and in the basal region of the explant (Supplementary Fig. S1). De novo shoot organogenesis (pluripotency) in Arabidopsis has been shown to rely on 2,4-D-induced lateral root meristem formation from pericycle cells (Che et al., 2007; Atta et al., 2009; Sugimoto et al., 2010). We showed that ABA biosynthesis mutants and mutants for positive ABA signalling components have reduced capacity for SE from the shoot apex, but increased callus formation in both the apical and basal regions of the explant (Figs 4C, F, 5C–E). ABA and ABI4 inhibit lateral root formation by reducing polar auxin transport (Shkolnik-Inbar and Bar-Zvi, 2010). Enhanced callus formation in ABA signalling mutants in 2,4-D-treated explants suggests that ABA and ABA signalling are required in these tissues to repress 2,4-D-induced lateral root formation. However, our mutant analyses show that enhanced ABA biosynthesis and signalling are not sufficient to induce SE in these tissues. Together, these data suggest that additional factors are limiting for 2,4-D,induced SE from the roots of mature embryo explants.
A narrow window for somatic embryo induction from germinating seeds
ABI3, ABI4, and ABI5 are required for efficient 2,4-D-induced SE from the shoot apex of germinating seeds (Fig. 5A). ABI3/4/5 expression in germinating embryos declines during the first 2 d of seed germination, but is extended beyond this developmental window after treatment with 2,4-D (Supplementary Fig. S4). Our 2,4-D addition and removal experiments showed that this developmental window corresponds to the time frame in which the 2,4-D treatment is most effective for somatic embryo induction (Fig. 1Q, R). Treatment of germinating seeds with ABA within a short developmental window of 60 h after stratification can reinstate a seed ABA response and seed osmotolerance; thereafter ABA application induces a vegetative ABA response (Lopez-Molina et al., 2001, 2002). ABI3 and ABI5 are both required to induce this developmental checkpoint. ABI3 expression is up-regulated by ABA application within this developmental window, but not thereafter (Lopez-Molina et al., 2002). ABI3 acts upstream of ABI5 to regulate ABI5 expression, and ectopic expression of ABI5 is sufficient to rescue the negative effect of the abi3 mutant on this developmental checkpoint (Lopez-Molina et al., 2002). Similarly, desiccation tolerance can be re-induced in a narrow developmental window during seed germination, and also relies on ABI3, ABI4, and ABI5 function (Maia et al., 2014).
A model for auxin–ABA interaction during induced cell totipotency
We propose a model (Fig. 6) in which 2,4-D promotes an ABI-mediated transcriptional cascade in germinating seeds leading to repression of vegetative meristem development in favour of somatic embryo induction. This pathway is dependent on ARF10/16 and on signalling through the core ABA signalling pathway (RCARs/PYR1/PYLs–PP2Cs–SnRK2s), as reduced/enhanced ABA signalling negatively/positively affects 2,4-D-induced SE, respectively. ABA positively regulates ABI3/4/5 expression (Lopez-Molina et al., 2001, 2002; Arroyo et al., 2003; Shkolnik-Inbar and Bar-Zvi, 2010), ABI3/4/5 protein stability/accumulation (Lopez-Molina et al., 2001, 2002; Shu et al., 2016a), and ABI5 protein phosphorylation (Lopez-Molina et al., 2001; Fujii et al., 2009; Melcher et al., 2009; Yin et al., 2009). ABI3 and ABI4 play a larger role in SE than ABI5, as increased signalling through the ABA receptor can partially restore SE efficiency in the abi5 mutant, but not in the abi3 and abi4 mutants. Auxin and ABA might interact synergistically through an ARF10/ARF16–ABI3 expression module to regulate SE, as was shown for seed germination (Liu et al., 2013). Together, this model provides a new framework for identifying additional, intersecting plant totipotency pathways, and for directing efficient SE in systems that make use of mature seed explants (Wu et al., 2019).
Supplementary data
The following supplementary data are available at JXB online.
Table S1. Plant materials.
Table S2. Primers used in this study.
Fig. S1. Development of the root hypocotyl region in 2,4-D-treated explants.
Fig. S2. GO analysis of 2,4-D differentially regulated genes.
Fig. S3. Statistically significant differentially regulated auxin pathway genes.
Fig. S4. qRT–PCR validation of differentially expressed ABA-related genes in SE culture.
Fig. S5. 2,4-D treatment induces ectopic BBM:BBM-GUS expression post-germination.
Fig. S6. The effect of ABA application on SE in ABA receptor mutant explants.
Fig. S7. 35S:PYL10 and 35S:ABI3 overexpression lines.
Fig. S8. Effect of abi3 weak alleles on 2,4-D-induced somatic embryogenesis.
Fig. S9. Effect of the abi3-6 allele on 2,4-D-induced somatic embryogenesis from immature zygotic embryo explants.
Fig. S10. Effect of ABA application on SE efficiency in abi3, abi4, and abi5-7 explants.
Fig. S11. 2,4-D treatment maintains ABI3 and ABI4 expression post-germination.
Fig. S12. ARF10 and ARF16 are required for 2,4-D-induced SE.
Dataset S1. Microarray data (all data, ABA-related genes, auxin-related genes, seed maturation-related genes, GO analysis of 2,4-D-induced DEGs, somatic embryogenesis inducer and enhancer genes).
Acknowledgements
This work was supported by a ZonMW Horizon grant to MF, and a China Scholarship Council grant to BC. We thank Erwin Grill for providing the 35S:R12/PYL1 line; Xuelu Wang for providing the 35S:ABI1 and 35S:ABI2 lines; Qi Xie for providing the 35S:ABI4 line; Ruth Finkelstein for providing the 35S:ABI5 line; and Hojin Ryu and Ildoo Hwang for providing the ABI3:GUS reporter line.
Glossary
Abbreviations
- ABA
abscisic acid
- 2,4-D
2,4-dichlorophenoxyacetic acid
- SE
somatic embryogenesis
Author contributions
BC, MF, BD, GCA, YZ, and KB: design of the experiments; BC and MF: performing experiments; BC, MF, and GWvE: data analysis; LM: providing novel material; BC and KB: writing the manuscript with input from all authors.
Data availability
All data supporting the findings of this study are available within the paper and within its supplementary data published online.
References
- Arroyo A, Bossi F, Finkelstein RR, León P. 2003. Three genes that affect sugar sensing (abscisic acid insensitive 4, abscisic acid insensitive 5, and constitutive triple response 1) are differentially regulated by glucose in Arabidopsis. Plant Physiology 133, 231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atta R, Laurens L, Boucheron-Dubuisson E, Guivarc’h A, Carnero E, Giraudat-Pautot V, Rech P, Chriqui D. 2009. Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. The Plant Journal 57, 626–644. [DOI] [PubMed] [Google Scholar]
- Brady SM, Sarkar SF, Bonetta D, McCourt P. 2003. The ABSCISIC ACID INSENSITIVE 3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in Arabidopsis. The Plant Journal 34, 67–75. [DOI] [PubMed] [Google Scholar]
- Braybrook SA, Harada JJ. 2008. LECs go crazy in embryo development. Trends in Plant Science 13, 624–630. [DOI] [PubMed] [Google Scholar]
- Braybrook SA, Stone SL, Park S, Bui AQ, Le BH, Fischer RL, Goldberg RB, Harada JJ. 2006. Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proceedings of the National Academy of Sciences, USA 103, 3468–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard IM, Lynch TJ, Finkelstein RR. 2002. Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiology 129, 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard-Gifford IM, Lynch TJ, Finkelstein RR. 2003. Regulatory networks in seeds integrating developmental, abscisic acid, sugar, and light signaling. Plant Physiology 131, 78–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundrett MC, Kendrick B, Peterson CA. 1991. Efficient lipid staining in plant material with Sudan Red 7B or Fluoral Yellow 088 in polyethylene glycol–glycerol. Biotechnic & Histochemistry 66, 111–116. [DOI] [PubMed] [Google Scholar]
- Cadman CSC, Toorop PE, Hilhorst HWM, Finch-Savage WE. 2006. Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. The Plant Journal 46, 805–822. [DOI] [PubMed] [Google Scholar]
- Carbonero P, Iglesias-Fernández R, Vicente-Carbajosa J. 2016. The AFL subfamily of B3 transcription factors: evolution and function in angiosperm seeds. Journal of Experimental Botany 68, erw458. [DOI] [PubMed] [Google Scholar]
- Carles C, Bies-Etheve N, Aspart L, Leon-Kloosterziel KM, Koornneef M, Echeverria M, Delseny M. 2002. Regulation of Arabidopsis thaliana Em genes: role of ABI5. The Plant Journal 30, 373–383. [DOI] [PubMed] [Google Scholar]
- Castel B, Tomlinson L, Locci F, Yang Y, Jones JDG. 2019. Optimization of T-DNA architecture for Cas9-mediated mutagenesis in Arabidopsis. PLoS One 14, e0204778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che P, Lall S, Howell SH. 2007. Developmental steps in acquiring competence for shoot development in Arabidopsis tissue culture. Planta 226, 1183–1194. [DOI] [PubMed] [Google Scholar]
- Chen N, Veerappan V, Abdelmageed H, Kang M, Allen RD. 2018. HSI2/VAL1 silences AGL15 to regulate the developmental transition from seed maturation to vegetative growth in Arabidopsis. The Plant Cell 30, 600–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SH, von Schwartzenberg K, Quatrano R. 2018. The role of abscisic acid in stress tolerance. Annual Plant Reviews 36, 282–297. [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology 139, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Folter S, Urbanus SL, van Zuijlen LG, Kaufmann K, Angenent GC. 2007. Tagging of MADS domain proteins for chromatin immunoprecipitation. BMC Plant Biology 7, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas F, Sankaranarayanan S, Deb S, Widdup E, Bournonville C, Bollier N, Northey JGB, McCourt P, Samuel MA. 2013. ABI3 controls embryo degreening through Mendel’s I locus. Proceedings of the National Academy of Sciences, USA 110, E3888–E3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devic M, Roscoe T. 2016. Seed maturation: simplification of control networks in plants. Plant Science 252, 335–346. [DOI] [PubMed] [Google Scholar]
- Ding ZJ, Yan JY, Li CX, Li GX, Wu YR, Zheng SJ. 2015. Transcription factor WRKY46 modulates the development of Arabidopsis lateral roots in osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. The Plant Journal 84, 56–69. [DOI] [PubMed] [Google Scholar]
- Egertsdotter U, Ahmad I, Clapham D. 2019. Automation and scale up of somatic embryogenesis for commercial plant production, with emphasis on conifers. Frontiers in Plant Science 10, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehér A. 2015. Somatic embryogenesis—stress-induced remodeling of plant cell fate. Biochimica et Biophysica Acta 1849, 385–402. [DOI] [PubMed] [Google Scholar]
- Figueiredo DD, Köhler C. 2018. Auxin: a molecular trigger of seed development. Genes & Development 32, 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R, Lynch T, Reeves W, Petitfils M, Mostachetti M. 2011. Accumulation of the transcription factor ABA-insensitive (ABI)4 is tightly regulated post-transcriptionally. Journal of Experimental Botany 62, 3971–3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. 2009. In vitro reconstitution of an abscisic acid signalling pathway. Nature 462, 660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Zhu JK. 2009. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proceedings of the National Academy of Sciences, USA 106, 8380–8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj MD. 2001. Direct somatic embryogenesis as a rapid and efficient system for in vitro regeneration of Arabidopsis thaliana. Plant Cell, Tissue and Organ Culture 64, 39–46. [Google Scholar]
- Gaj MD. 2004. Factors influencing somatic embryogenesis induction and plant regeneration with particular reference to Arabidopsis thaliana (L.) Heynh. Plant Growth Regulation 43, 27–47. [Google Scholar]
- Gaj MD, Trojanowska A, Ujczak A, Medrek M, Koziol A, Garbaciak B. 2006. Hormone-response mutants of Arabidopsis thaliana (L.) Heynh. impaired in somatic embryogenesis. Plant Growth Regulation 49, 183–197. [Google Scholar]
- Gaj MD, Zhang S, Harada JJ, Lemaux PG. 2005. Leafy cotyledon genes are essential for induction of somatic embryogenesis of Arabidopsis. Planta 222, 977–988. [DOI] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B. 2007. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449, 1053–1057. [DOI] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. 2004. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315. [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M, McCourt P. 2004. The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Developmental Cell 7, 373–385. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biology 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godel-Jedrychowska K, Kulinska-Lukaszek K, Horstman A, Soriano M, Li M, Malota K, Boutilier K, Kurczynska EU. 2020. Symplasmic isolation marks cell fate changes during somatic embryogenesis. Journal of Experimental Botany 71, 2612–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Guzmán M, Apostolova N, Bellés JM, Barrero JM, Piqueras P, Ponce MR, Micol JL, Serrano R, Rodríguez PL. 2002. The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. The Plant Cell 14, 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guzman M, Pizzio GA, Antoni R, et al. 2012. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. The Plant Cell 24, 2483–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorio J, Hernández-Bernal AF, Cordoba E, León P. 2014. Characterization of evolutionarily conserved motifs involved in activity and regulation of the ABA-INSENSITIVE (ABI) 4 transcription factor. Molecular Plant 7, 422–436. [DOI] [PubMed] [Google Scholar]
- Holdsworth M, Kurup S, MKibbin R. 1999. Molecular and genetic mechanisms regulating the transition from embryo development to germination. Trends in Plant Science 4, 275–280. [Google Scholar]
- Hong JH, Seah SW, Xu J. 2013. The root of ABA action in environmental stress response. Plant Cell Reports 32, 971–983. [DOI] [PubMed] [Google Scholar]
- Horstman A, Bemer M, Boutilier K. 2017a. A transcriptional view on somatic embryogenesis. Regeneration 4, 201–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstman A, Li M, Heidmann I, Weemen M, Chen B, Muiño JM, Angenent GC, Boutilier K. 2017b. The BABY BOOM transcription factor activates the LEC1–ABI3–FUS3–LEC2 network to induce somatic embryogenesis. Plant Physiology 175, 848–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Ikeda-Iwai M. 2002. Establishment of a reproducible tissue culture system for the induction of Arabidopsis somatic embryos. Journal of Experimental Botany 53, 1575–1580. [DOI] [PubMed] [Google Scholar]
- Ikeda-Iwai M, Umehara M, Satoh S, Kamada H. 2003. Stress-induced somatic embryogenesis in vegetative tissues of Arabidopsis thaliana. The Plant Journal 34, 107–114. [DOI] [PubMed] [Google Scholar]
- Immink RGH, Gadella TWJ, Ferrario S, Busscher M, Angenent GC. 2002. Analysis of MADS box protein–protein interactions in living plant cells. Proceedings of the National Academy of Sciences, USA 99, 2416–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, McCarty DR, Suzuki M. 2013. Distinct roles of LAFL network genes in promoting the embryonic seedling fate in the absence of VAL repression. Plant Physiology 163, 1293–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker A, Bäumlein H. 2012. Multifunctionality of the LEC1 transcription factor during plant development. Plant Signaling & Behavior 7, 1718–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadokura S, Sugimoto K, Tarr P, Suzuki T, Matsunaga S. 2018. Characterization of somatic embryogenesis initiated from the Arabidopsis shoot apex. Developmental Biology 442, 13–27. [DOI] [PubMed] [Google Scholar]
- Kamada H, Ishikawa K, Saga H, Harada H. 1993. Induction of somatic embryogenesis in carrot by osmotic stress. Plant Tissue Culture Letters 10, 38–44. [Google Scholar]
- Kamada H, Katsunori K, Tomohiro K, Hiroshi H. 1989. Stress induced somatic embryogenesis in carrot and its application to synthetic seed production. In Vitro Cellular & Developmental Biology 25, 1163–1166. [Google Scholar]
- Karimi M, Inzé D, Depicker A. 2002. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Nagayama Y, Higashi K, Kobayashi M. 2010. Establishment of a tissue culture system for somatic embryogenesis from germinating embryos of Arabidopsis thaliana. Plant Biotechnology 27, 359–364. [Google Scholar]
- Kroj T, Savino G, Valon C, Giraudat J, Parcy F. 2003. Regulation of storage protein gene expression in Arabidopsis. Development 130, 6065–6073. [DOI] [PubMed] [Google Scholar]
- Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E. 2004. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. The EMBO Journal 23, 1647–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JAD, Koornneef M. 1996. Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. The Plant Journal 10, 655–661. [DOI] [PubMed] [Google Scholar]
- León-Martínez G, Vielle-Calzada JP. 2019. Apomixis in flowering plants: developmental and evolutionary considerations. Current Topics in Developmental Biology 131, 565–604. [DOI] [PubMed] [Google Scholar]
- Lepiniec L, Devic M, Roscoe TJ, Bouyer D, Zhou DX, Boulard C, Baud S, Dubreucq B. 2018. Molecular and epigenetic regulations and functions of the LAFL transcriptional regulators that control seed development. Plant Reproduction 31, 291–307. [DOI] [PubMed] [Google Scholar]
- Leprince O, Pellizzaro A, Berriri S, Buitink J. 2017. Late seed maturation: drying without dying. Journal of Experimental Botany 68, 827– 841. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang H, Zhao Y, Feng Z, Li Q, Yang H-Q, Luan S, Li J, He Z-H. 2013. Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proceedings of the National Academy of Sciences, USA 110, 15485–15490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Ge X-X, Qiu W-M, Long J-M, Jia H-H, Yang W, Dutt M, Wu X-M, Guo W-W. 2018. Overexpression of the CsFUS3 gene encoding a B3 transcription factor promotes somatic embryogenesis in Citrus. Plant Science 277, 121–131. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua N-H. 2001. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proceedings of the National Academy of Sciences, USA 98, 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua N-H. 2002. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. The Plant Journal 32, 317–328. [DOI] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. 1998. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93, 1195–1205. [DOI] [PubMed] [Google Scholar]
- Lowe K, Hoerster G, Sun X, Rasco-Gaunt S, Lazerri P, Ellis S, Abbitt S, Glassman K, Gordon-Kamm B. 2003. Maize LEC1 improves transformation in both maize and wheat. In: Vasil IK, ed. Plant biotechnology 2002 and beyond. Dordrecht: Springer Netherlands, 283–284. [Google Scholar]
- Lu QS, Paz JD, Pathmanathan A, Chiu RS, Tsai AY, Gazzarrini S. 2010. The C-terminal domain of FUSCA3 negatively regulates mRNA and protein levels, and mediates sensitivity to the hormones abscisic acid and gibberellic acid in Arabidopsis. The Plant Journal 64, 100–113. [DOI] [PubMed] [Google Scholar]
- Luo Y, Koop HU. 1997. Somatic embryogenesis in cultured immature zygotic embryos and leaf protoplasts of Arabidopsis thaliana ecotypes. Planta 202, 387–396. [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. 2009. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. [DOI] [PubMed] [Google Scholar]
- Maia J, Dekkers BJ, Dolle MJ, Ligterink W, Hilhorst HW. 2014. Abscisic acid (ABA) sensitivity regulates desiccation tolerance in germinated Arabidopsis seeds. New Phytologist 203, 81–93. [DOI] [PubMed] [Google Scholar]
- Melcher K, Ng LM, Zhou XE, et al. 2009. A gate–latch–lock mechanism for hormone signalling by abscisic acid receptors. Nature 462, 602–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez-Hernández HA, Ledezma-Rodríguez M, Avilez-Montalvo RN, Juárez-Gómez YL, Skeete A, Avilez-Montalvo J, De-la-Peña C, Loyola-Vargas VM. 2019. Signaling overview of plant somatic embryogenesis. Frontiers in Plant Science 10, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mönke G, Seifert M, Keilwagen J, et al. 2012. Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon. Nucleic Acids Research 40, 8240–8254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordhorst AP, Voerman KJ, Hartog MV, Meijer EA, van Went J, Koornneef M, de Vries SC. 1998. Somatic embryogenesis in Arabidopsis thaliana is facilitated by mutations in genes repressing meristematic cell divisions. Genetics 149, 549–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozgova I, Köhler C, Hennig L. 2015. Keeping the gate closed: functions of the polycomb repressive complex PRC2 in development. The Plant Journal 83, 121–132. [DOI] [PubMed] [Google Scholar]
- Mozgová I, Muñoz-Viana R, Hennig L. 2017. PRC2 represses hormone-induced somatic embryogenesis in vegetative tissue of Arabidopsis thaliana. PLoS Genetics 13, e1006562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Y, Zou M, Sun X, He B, Xu X, Liu Y, Zhang L, Chi W. 2017. Basic pentacysteine proteins repress Abscisic Acid Insensitive4 expression via direct recruitment of the Polycomb-Repressive Complex 2 in Arabidopsis root development. Plant & Cell Physiology 58, 607–621. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. 1962. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum 15, 473–497. [Google Scholar]
- Nakashima K, Fujita Y, Katsura K, Maruyama K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. 2006. Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Molecular Biology 60, 51–68. [DOI] [PubMed] [Google Scholar]
- Nambara E, Keith K, McCourt P, Naito S. 1994. Isolation of an internal deletion mutant of the Arabidopsis thaliana ABI3 gene. Plant & Cell Physiology 35, 509–513. [PubMed] [Google Scholar]
- Nambara E, Nambara E, McCourt P, Naito S. 1995. A regulatory role for the ABI3 gene in the establishment of embryo maturation in Arabidopsis thaliana. Development 121, 629–636. [Google Scholar]
- Nambara E, Okamoto M, Tatematsu K, Yano R, Seo M, Kamiya Y. 2010. Abscisic acid and the control of seed dormancy and germination. Seed Science Research 20, 55–67. [Google Scholar]
- Nambara E, Suzuki M, Abrams S, McCarty DR, Kamiya Y, McCourt P. 2002. A screen for genes that function in abscisic acid signaling in Arabidopsis thaliana. Genetics 161, 1247–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto K, Kagawa M, Nozawa A, Hasegawa Y, Hayashi M, Imai K, Tomii K, Sawasaki T. 2018. Identification of new abscisic acid receptor agonists using a wheat cell-free based drug screening system. Scientific Reports 8, 4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nic-Can GI, Avilez-Montalvo JR, Aviles-Montalvo RN, Márquez-López RE, Mellado-Mojica E, Galaz-Ávalos RM, Loyola-Vargas VM. 2016. The relationship between stress and somatic embryogenesis. In: Loyola-Vargas V, Ochoa-Alejo N, eds. Somatic embryogenesis: fundamental aspects and applications. Cham: Springer International Publishing, 151–170. [Google Scholar]
- Nic-Can GI, Loyola-Vargas VM. 2016. The role of the auxins during somatic embryogenesis. In: Loyola-Vargas V, Ochoa-Alejo N, eds. Somatic embryogenesis: fundamental aspects and applications. Cham: Springer International Publishing, 171–182. [Google Scholar]
- Nishiwaki M, Fujino K, Koda Y, Masuda K, Kikuta Y. 2000. Somatic embryogenesis induced by the simple application of abscisic acid to carrot (Daucus carota L.) seedlings in culture. Planta 211, 756–759. [DOI] [PubMed] [Google Scholar]
- Parcy F, Valon C, Raynal M, Gaubier-Comella P, Delseny M, Giraudat J. 1994. Regulation of gene expression programs during Arabidopsis seed development: roles of the ABI3 locus and of endogenous abscisic acid. The Plant Cell 6, 1567–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, et al. 2009. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YS, Barrett JD, Bonga JM. 1998. Application of somatic embryogenesis in high-value clonal forestry: deployment, genetic control, and stability of cryopreserved clones. In Vitro Cellular & Developmental Biology - Plant 34, 231–239. [Google Scholar]
- Pasternak T, Dudits D. 2019. Epigenetic clues to better understanding of the asexual embryogenesis in planta and in vitro. Frontiers in Plant Science 10, 778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Li Y, Gilday AD, Graham S, Graham IA. 2006. Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. The Plant Cell 18, 1887–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E. 2010. ABA perception and signalling. Trends in Plant Science 15, 395–401. [DOI] [PubMed] [Google Scholar]
- Rai MK, Shekhawat NS, Harish, Gupta AK, Phulwaria M, Ram K, Jaiswal U. 2011. The role of abscisic acid in plant tissue culture: a review of recent progress. Plant Cell, Tissue and Organ Culture 106, 179–190. [Google Scholar]
- Reeves WM, Lynch TJ, Mobin R, Finkelstein RR. 2011. Direct targets of the transcription factors ABA-Insensitive(ABI)4 and ABI5 reveal synergistic action by ABI4 and several bZIP ABA response factors. Plant Molecular Biology 75, 347–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde A, Van Montagu M, Boerjan W. 1999. The ABSCISIC ACID-INSENSITIVE 3 (ABI3) gene is expressed during vegetative quiescence processes in Arabidopsis. Plant, Cell & Environment 22, 261–270. [Google Scholar]
- Rubio S, Rodrigues A, Saez A, Dizon MB, Galle A, Kim TH, Santiago J, Flexas J, Schroeder JI, Rodriguez PL. 2009. Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiology 150, 1345–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Cho H, Bae W, Hwang I. 2014. Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nature Communications 5, 4138. [DOI] [PubMed] [Google Scholar]
- Schmidt A. 2020. Controlling apomixis: shared features and distinct characteristics of gene regulation. Genes 11, 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkolnik-Inbar D, Bar-Zvi D. 2010. ABI4 mediates abscisic acid and cytokinin inhibition of lateral root formation by reducing polar auxin transport in Arabidopsis. The Plant Cell 22, 3560–3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K, Chen Q, Wu Y, et al. 2016a. ABI4 mediates antagonistic effects of abscisic acid and gibberellins at transcript and protein levels. The Plant Journal 85, 348–361. [DOI] [PubMed] [Google Scholar]
- Shu K, Liu XD, Xie Q, He ZH. 2016b. Two faces of one seed: hormonal regulation of dormancy and germination. Molecular Plant 9, 34–45. [DOI] [PubMed] [Google Scholar]
- Shu K, Zhang H, Wang S, Chen M, Wu Y, Tang S, Liu C, Feng Y, Cao X, Xie Q. 2013. ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in arabidopsis. PLoS Genetics 9, e1003577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skubacz A, Daszkowska-Golec A, Szarejko I. 2016. The role and regulation of ABI5 (ABA-Insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Frontiers in Plant Science 7, 1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit ME, Weijers D. 2015. The role of auxin signaling in early embryo pattern formation. Current Opinion in Plant Biology 28, 99–105. [DOI] [PubMed] [Google Scholar]
- Söderman EM, Brocard IM, Lynch TJ, Finkelstein RR. 2000. Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiology 124, 1752–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soon FF, Ng LM, Zhou XE, et al. 2012. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 335, 85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano M, Li H, Boutilier K. 2013. Microspore embryogenesis: establishment of embryo identity and pattern in culture. Plant Reproduction 26, 181–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano M, Li H, Jacquard C, Angenent GC, Krochko J, Offringa R, Boutilier K. 2014. Plasticity in cell division patterns and auxin transport dependency during in vitro embryogenesis in Brassica napus. The Plant Cell 26, 2568–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Braybrook SA, Paula SL, Kwong LW, Meuser J, Pelletier J, Hsieh TF, Fischer RL, Goldberg RB, Harada JJ. 2008. Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: implications for somatic embryogenesis. Proceedings of the National Academy of Sciences, USA 105, 3151–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ. 2001. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proceedings of the National Academy of Sciences, USA 98, 11806–11811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YH, Su YX, Liu YG, Zhang XS. 2013. Abscisic acid is required for somatic embryo initiation through mediating spatial auxin response in Arabidopsis. Plant Growth Regulation 69, 167–176. [Google Scholar]
- Sugimoto K, Jiao Y, Meyerowitz EM. 2010. Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Developmental Cell 18, 463–471. [DOI] [PubMed] [Google Scholar]
- Testillano PS. 2019. Microspore embryogenesis: targeting the determinant factors of stress-induced cell reprogramming for crop improvement. Journal of Experimental Botany 70, 2965–2978. [DOI] [PubMed] [Google Scholar]
- Thakare D, Tang W, Hill K, Perry SE. 2008. The MADS-domain transcriptional regulator AGAMOUS-LIKE15 promotes somatic embryo development in Arabidopsis and soybean. Plant Physiology 146, 1663–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian R, Paul P, Joshi S, Perry SE. 2020a. Genetic activity during early plant embryogenesis. The Biochemical Journal 477, 3743–3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian R, Wang F, Zheng Q, Niza VMAGE, Downie AB, Perry SE. 2020b. Direct and indirect targets of the arabidopsis seed transcription factor ABSCISIC ACID INSENSITIVE3. The Plant Journal 103, 1679–1694. [DOI] [PubMed] [Google Scholar]
- To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J, Parcy F. 2006. A network of local and redundant gene regulation governs Arabidopsis seed maturation. The Plant Cell 18, 1642–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdeil JL, Alemanno L, Niemenak N, Tranbarger TJ. 2007. Pluripotent versus totipotent plant stem cells: dependence versus autonomy? Trends in Plant Science 12, 245–252. [DOI] [PubMed] [Google Scholar]
- Vergunst AC, de Waal EC, Hooykaas PJ. 1998. Root transformation by Agrobacterium tumefaciens. Methods in Molecular Biology 82, 227–244. [DOI] [PubMed] [Google Scholar]
- Vishwakarma K, Upadhyay N, Kumar N, et al. 2017. Abscisic acid signaling and abiotic stress tolerance in plants: a review on current knowledge and future prospects. Frontiers in Plant Science 8, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zhang C, Hearn DJ, Kang IH, Punwani JA, Skaggs MI, Drews GN, Schumaker KS, Yadegari R. 2010. Identification of transcription-factor genes expressed in the Arabidopsis female gametophyte. BMC Plant Biology 10, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang FX, Shang GD, Wu LY, Xu ZG, Zhao XY, Wang JW. 2020. Chromatin accessibility dynamics and a hierarchical transcriptional regulatory network structure for plant somatic embryogenesis. Developmental Cell 54, 742–757. [DOI] [PubMed] [Google Scholar]
- Wang H, Tang J, Liu J, Hu J, Liu J, Chen Y, Cai Z, Wang X. 2018. Abscisic acid signaling inhibits brassinosteroid signaling through dampening the dephosphorylation of BIN2 by ABI1 and ABI2. Molecular Plant 11, 315–325. [DOI] [PubMed] [Google Scholar]
- Wang R, Tavano ECDR, Lammers M, Martinelli AP, Angenent GC, de Maagd RA. 2019. Re-evaluation of transcription factor function in tomato fruit development and ripening with CRISPR/Cas9-mutagenesis. Scientific Reports 9, 1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li L, Ye T, Lu Y, Chen X, Wu Y. 2013. The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. Journal of Experimental Botany 64, 675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Li XR, Sun MX. 2006. Establishment of a simple and efficient system for somatic embryo induction via ovule culture in Arabidopsis thaliana. Plant Cell Reports 25, 1275–1280. [DOI] [PubMed] [Google Scholar]
- Wind JJ, Peviani A, Snel B, Hanson J, Smeekens SC. 2013. ABI4: versatile activator and repressor. Trends in Plant Science 18, 125–132. [DOI] [PubMed] [Google Scholar]
- Wójcik AM, Wójcikowska B, Gaj MD. 2020. Current perspectives on the auxin-mediated genetic network that controls the induction of somatic embryogenesis in plants. International Journal of Molecular Sciences 21, 1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wójcikowska B, Wójcik AM, Gaj MD. 2020. Epigenetic regulation of auxin-induced somatic embryogenesis in plants. International Journal of Molecular Sciences 21, 2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Chen B, Fiers M, Wróbel-Marek J, Kodde J, Groot SPC, Angenent G, Feng H, Bentsink L, Boutilier K. 2019. Seed maturation and post-harvest ripening negatively affect arabidopsis somatic embryogenesis. Plant Cell, Tissue and Organ Culture 139, 17–27. [Google Scholar]
- Xie T, Ren R, Zhang Y, et al. 2012. Molecular mechanism for inhibition of a critical component in the Arabidopsis thaliana abscisic acid signal transduction pathways, SnRK2. 6, by protein phosphatase ABI1. Journal of Biological Chemistry 287, 794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Kamiya Y, Nambara E. 2018. Regulation of ABA and GA levels during seed development and germination in Arabidopsis. Annual Plant Reviews 27, 224–247. [Google Scholar]
- Yamamoto A, Yoshii M, Murase S, Fujita M, Kurata N, Hobo T, Kagaya Y, Takeda S, Hattori T. 2014. Cell-by-cell developmental transition from embryo to post-germination phase revealed by heterochronic gene expression and ER-body formation in Arabidopsis leafy cotyledon mutants. Plant & Cell Physiology 55, 2112–2125. [DOI] [PubMed] [Google Scholar]
- Yan A, Chen Z. 2017. The pivotal role of abscisic acid signaling during transition from seed maturation to germination. Plant Cell Reports 36, 689–703. [DOI] [PubMed] [Google Scholar]
- Yang C, Bratzel F, Hohmann N, Koch M, Turck F, Calonje M. 2013. VAL- and AtBMI1-mediated H2Aub initiate the switch from embryonic to postgerminative growth in Arabidopsis. Current Biology 23, 1324–1329. [DOI] [PubMed] [Google Scholar]
- Yang Z, Liu J, Tischer SV, Christmann A, Windisch W, Schnyder H, Grill E. 2016. Leveraging abscisic acid receptors for efficient water use in Arabidopsis. Proceedings of the National Academy of Sciences, USA 113, 6791–6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P, Fan H, Hao Q, Yuan X, Wu D, Pang Y, Yan C, Li W, Wang J, Yan N. 2009. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nature Structural & Molecular Biology 16, 1230–1236. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Christmann A, Yamaguchi-Shinozaki K, Grill E, Fernie AR. 2019. Revisiting the basal role of ABA—roles outside of stress. Trends in Plant Science 24, 625–635. [DOI] [PubMed] [Google Scholar]
- Yuan TT, Xu HH, Zhang KX, Guo TT, Lu YT. 2014. Glucose inhibits root meristem growth via ABA INSENSITIVE 5, which represses PIN1 accumulation and auxin activity in Arabidopsis. Plant, Cell & Environment 37, 1338–1350. [DOI] [PubMed] [Google Scholar]
- Zhang X, Garreton V, Chua NH. 2005. The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes & Development 19, 1532–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Clemens A, Maximova SN, Guiltinan MJ. 2014. The Theobroma cacao B3 domain transcription factor TcLEC2 plays a dual role in control of embryo development and maturation. BMC Plant Biology 14, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Begcy K, Dresselhaus T, Sun MX. 2017. Does early embryogenesis in eudicots and monocots involve the same mechanism and molecular players? Plant Physiology 173, 130–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zhang Z, Gao J, et al. 2018. Arabidopsis duodecuple mutant of PYL ABA receptors reveals PYL repression of ABA-independent SnRK2 activity. Cell Reports 23, 3340–3351.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and within its supplementary data published online.