Abstract
Importin α plays a pivotal role in the classical nuclear protein import pathway. Importin α shuttles between nucleus and cytoplasm, binds nuclear localization signal-bearing proteins, and functions as an adapter to access the importin β-dependent import pathway. In contrast to what is found for importin β, several isoforms of importin α, which can be grouped into three subfamilies, exist in higher eucaryotes. We describe here a novel member of the human family, importin α7. To analyze specific functions of the distinct importin α proteins, we recombinantly expressed and purified five human importin α’s along with importin α from Xenopus and Saccharomyces cerevisiae. Binding affinity studies showed that all importin α proteins from humans or Xenopus bind their import receptor (importin β) and their export receptor (CAS) with only marginal differences. Using an in vitro import assay based on permeabilized HeLa cells, we compared the import substrate specificities of the various importin α proteins. When the substrates were tested singly, only the import of RCC1 showed a strong preference for one family member, importin α3, whereas most of the other substrates were imported by all importin α proteins with similar efficiencies. However, strikingly different substrate preferences of the various importin α proteins were revealed when two substrates were offered simultaneously.
Heavy trafficking between the nucleus and the cytoplasm takes place in eucaryote cells. Various substrates, such as different forms of RNA and many nuclear proteins, must be exported into the cytoplasm. Other proteins, such as transcription factors and ribonucleoprotein particles, must be imported into the nucleus. Nuclear transport occurs through the nuclear pore complexes (NPCs), which are about 125 MDa in size (34). Whereas smaller molecules up to 20 to 60 kDa may passively diffuse through the NPCs into the nucleus, the import of larger substrates is generally receptor mediated (12). This import process depends on the presence of specific signal sequences within the substrate. Different types of import signals exist. One such signal consists of the so-called nuclear localization signals (NLSs), which are mainly characterized by clusters of basic amino acids, predominately lysines. Depending on the numbers of their charged clusters, the NLSs may be classified into monopartite and bipartite NLSs (8, 9).
Several soluble factors of the classical nuclear protein import pathway have been identified so far, including the small GTPase Ran/TC4 (26, 30), importin α (16, 20, 45), importin β (1, 4, 14, 19, 39), and NTF2 (31, 36), which is involved in the import and export of Ran (41). Importin α functions as an adapter molecule by binding importin β via its amino-terminally located importin β binding (IBB) domain (13, 46) and by binding NLS-bearing proteins via its two NLS binding sites in the central area (5, 18). Importin β is the transport receptor that carries the importin α-NLS complex from the cytoplasm into the nuclear side of the NPC (17). Once inside the nucleus, importin β binds to RanGTP, which is generated within the nucleus by the chromatin-bound RanGDP/GTP exchange factor RCC1. This binding of importin β to RanGTP leads to the dissociation of the import complex (15). Whereas importin β is thought to return to the cytoplasm rapidly without other soluble factors, the export of importin α is mediated by its nuclear export factor CAS, which binds to importin α preferentially in the presence of RanGTP (24). In the cytoplasm, the importins are set free for another round of import by the concerted action of RanGAP1 and RanBP1.
Only one gene coding for importin β has been identified in the organisms analyzed thus far. In contrast to what was found for importin β, several isoforms of importin α in humans have been described. These include importin α1/Rch1 (7, 45), importin α5/hSRP1 (6), importin α3/Qip1 (23, 42), importin α4/hSRP1γ (23, 32), importin α6 (23), and, here, the newly reported importin α7. Importin α7 is the human homologue of the recently identified mouse importin α-S2 (44). Based on the sequence similarity, the importin α proteins can be grouped into three subfamilies. Members of different subfamilies have about 50% sequence identity. Within one subfamily, the identity is at least 80%. Whereas several of these isoforms are also found in invertebrates, the yeast Saccharomyces cerevisiae has only one gene for importin α, SRP1. Why so many importin α isoforms exist in higher eucaryotes has not yet been definitely answered. Although there is some tissue specificity in the expression of these proteins (23, 32, 38, 44), most isoforms are expressed within the same tissues. Initial data indicate that there may be distinct substrate specificities of different importin α family members (11, 28, 33, 43). However, other data show that different importin α proteins can interact with the same substrate (37, 38). We compared all of the ubiquitously expressed human importin α proteins, Xenopus importin α2, and yeast SRP1p in their efficiencies to promote the nuclear import of different substrates. When testing only one substrate per assay, we found that most substrates (NLS-bovine serum albumin [BSA], nucleoplasmin, P/CAF, and hnRNP K) were imported by all importin α proteins with only marginal differences. The exception was the nuclear import of RCC1, which was efficient only with importin α3, not with other isoforms. If two substrates are offered at the same time, the various importin α proteins show striking differences in their substrate-specific import efficiencies.
MATERIALS AND METHODS
Isolation of importin α7 cDNA.
For isolation of importin α7, 5′ rapid amplification of cDNA ends (RACE) and 3′ RACE were performed with a HeLa Marathon Ready cDNA kit (Clontech) by using the primers AAT(C)TGTTCT(A)GCC(A)C(T)TACCC(T)TGT(C)CT, CTCTTCCGCTGGCTGGTGGTG, and TATAACAAGCCTTTATTGAGCCCT, which correspond to human cDNA clones (GenBank accession no. T08580 and U68730). Several positive clones, which harbored the proposed start codon or the proposed stop codon, were isolated and sequenced. Full-length cDNA of importin α7 was obtained by using the cDNAs of the N and C termini and primers AACCCCGGCATGCAGACCATGGCGAGCCCAGGGAAAGAC and CAATTTGGATCCTAGCTGGAAGCCCTCCATGGGGGCC. Full-length cDNA of importin α5/hSRP1 was obtained via PCR with the same HeLa cDNA kit and primers TTGCGCCCATGGCCACCCCAGGAAAAGAGAAC and GAAGCCGGATCCAAGCTGGAAACCTTCCATAGGA. The cloning of the other importin α cDNAs has been described earlier (14, 23).
Northern blotting.
Human multiple-tissue Northern blots (Clontech) were hybridized according to the manufacturer’s instructions with an [α-32P]dATP-labeled 0.5-kbp cDNA fragment from importin α7 and with a 2.0-kbp fragment of β-actin.
Generation of antibodies and immunoblotting.
The generation of antibodies against peptide sequences of importin α1/Rch1, importin α5/hSRP1, importin α3, and importin α4 was described previously (23). Two antibodies against the newly identified human importin α7 were raised against peptide sequences MASPGKDNYR, representing amino acids 3 to 12 of human importin α7, and PEAPMEGFQUL, representing amino acids 526 to 536. Since very similar peptides are also present in importin α6, the antibodies against importin α7 recognize recombinant importin α6 as well. Cross-reaction of the antibodies with other importin α forms was excluded by immunoblotting with the recombinantly expressed proteins. For the analysis of the tissue-specific expression of the importin α forms by immunoblotting, human protein lysates (Clontech; 25 μg per lane) were separated sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS–10% PAGE) and blotted. Detection was achieved by chemiluminescence (Du Pont).
CAS binding assay.
Ran-[γ-32P]GTP (50 pM) was preincubated in a mixture of 20 mM HEPES-NaOH (pH 7.2), 50 mM sodium acetate, 1 mM MgCl2, 0.5% hydrolyzed gelatin, 0.4% sodium azide, and either 1 μM CAS or mixtures of 1 μM CAS and the different importin α homologous proteins. After 30 min at 15°C, 40 nM Rna1p was added and the reaction was allowed to proceed for 30 s. The hydrolysis of Ran-bound GTP was determined as released [32P]phosphate. The final reaction volume was 25 μl, and the concentrations of importin α proteins are as indicated in the legends for Fig. 4 and 5.
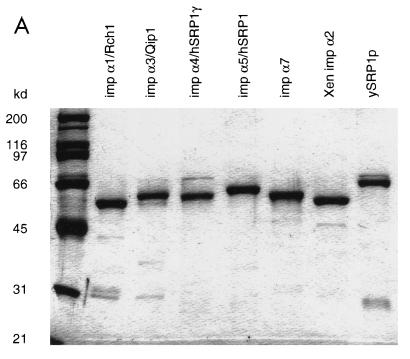
FIG. 4.
(A) Purified recombinant importin α proteins migrate between 50 and 60 kDa. Solutions (7.5 μl, 2 μM) of each recombinantly expressed and purified importin α protein were separated by SDS-PAGE and stained with Coomassie blue. imp, importin; Xen, Xenopus; ySRP1, yeast SRP1p. (B) Determination of the binding affinities of importin α1/Rch1 (RCH1), importin α5/hSRP1 (hSRP1), and importin α3 to CAS. (C) Determination of the binding affinities of importin α4, importin α7, Xenopus importin α2 (importin α Xen), and yeast SRP1p (ySrp1p) to human CAS. Ran-[γ-32P]GTP (50 pM) was preincubated either with 1 μM CAS or with mixtures of 1 μM CAS and the different importin α homologous proteins. After 30 min at 15°C, 40 nM Rna1p (the S. pombe homologue of RanGAP1) was added and the reaction was allowed to proceed for 30 s. Hydrolysis of Ran-bound GTP was determined as released [32P]phosphate.
FIG. 5.
All importin α proteins can import standard substrates with monopartite (NLS-BSA) and bipartite NLS (nucleoplasmin) in vitro. HeLa cells were grown on coverslips and permeabilized for 8 min with digitonin. Coverslips were incubated with 20 μl of import mixture for 8 min. Reactions were stopped by fixation with 4% paraformaldehyde. The coverslips were mounted and analyzed by confocal microscopy. The import reaction mixtures consisted of an energy-regenerating system, nucleoplasmin core buffer, 3 μM RanGDP, 0.2 μM Rna1p, 0.3 μM RanBP1, 0.4 μM NTF2, 1 μM importin β, a 2 μM concentration of the indicated importin α protein, and 10% reticulocyte lysate. (A) Importin α-dependent nuclear import of Texas red-labeled nucleoplasmin. α3, importin α3; α4, importin α4; α7, importin α7; Xen α2, Xenopus importin α2; ySRP1, yeast SRP1p; no α, no importin α added to the import reaction mixture. (B) Importin α-dependent nuclear import of fluorescein-labeled simian virus 40 large-T antigen coupled to NLS-BSA.
Importin β binding assay.
Equilibrium dissociation constants (KDs) of the interaction between the importin α’s and importin β were determined by surface plasmon resonance (SPR) measurements with a Biacore 2000 instrument. The different importin α isoforms were diluted to a final concentration of about 100 ng/μl and immobilized on the surface of a Pioneer F1 sensor chip (Biacore AB) by the amine-coupling method described in the Biacore manual (22). The immobilization level was 250 to 800 RU for importin α’s and for BSA, which served as a control. Unreacted normal human serum was blocked for 8 min with 1 M ethanolamin. To determine kinetic constants, sensorgrams were collected at 22°C, an 8-μl/min flow rate, and a 2.5-Hz data collection rate. Importin β (220 μl) dissolved in HBS-EP buffer (10 mM HEPES [pH 7.4], 100 mM NaCl, 1 mM EDTA, 0.05% P20; Biacore AB) was injected at different concentrations (6.25 to 200 nM) by using the KINJECT command specifying 27.5 min of association time and 6 min of dissociation time. Subsequently, 8 μl of regeneration solution (2 M NaCl in HBS-EP buffer) was injected. Sets of sensorgrams with analyte concentrations of 6.25 to 200 nM and without protein for background correction were collected. For evaluation we used BIAevaluation 3.0 software (Biacore AB). The data were fitted with the steady-state affinity model. As a reference, we subtracted the BSA and buffer control.
Recombinant expression and purification of the importin α proteins.
Full-length cDNAs were digested with NcoI/BamHI (importin α5/hSRP1, importin α3), NcoI/BglII (importin α4), and SphI/BamHI (importin α7). The particular restriction sites had been introduced by PCR primers. After ligation into expression vectors (for importin α5/hSRP1, importin α3, and importin α4, pQE60; for importin α7, pQE70; Qiagen), the resulting constructs encoding C-terminally His-tagged proteins were verified by DNA sequencing. Expression was performed in a culture of Escherichia coli XL1/pSB161 at 25°C for 4 h. Phenylmethylsulfonyl fluoride (2 mM) was added immediately before the culture was chilled on ice. After centrifugation, the bacterial pellet was resuspended in sonification buffer (50 mM Tris-HCl [pH 7.5], 200 mM NaCl, 5 mM magnesium acetate, 5% glycerol), and bacteria were lysed by sonification. The lysate was cleared by ultracentrifugation in a 50.2 Ti rotor at 50,000 rpm for 2 h. After 20 mM imidazole was added, the supernatant was loaded onto nickel agarose. Elution of the column was performed with an imidazole gradient, and peak fractions were pooled. For importin α5/hSRP1, pooled fractions were dialyzed against sonification buffer and stored at −80°C after 250 mM sucrose had been added. The other importin α protein peak fractions were loaded onto a Mono Q column and eluted in 50 mM Tris-HCl (pH 7.5)–5% glycerol by using a NaCl gradient. Peak fractions were pooled again and stored at −80°C after 250 mM sucrose had been added. The concentrations of the proteins were determined via photometry at 280 nm with the molar extinction coefficient described by Edelhoch (10). Preparation of the following proteins was described earlier: C-terminally His-tagged Xenopus importin α2, nucleoplasmin, nucleoplasmin core, human Ran, Schizosaccharomyces pombe Rna1p, murine RnaBP1, and NTF2 (25); NLS-BSA, Rch1, and yeast SRP1p (14); and importin β (17). RCC1 was expressed and purified exactly as described here for the newly identified importin α proteins. Fluorescence labeling of purified import substrates was performed with fluorescein 5′-maleimide, FLUOS, or Texas red, as described earlier (24).
In vitro nuclear protein import assay.
Import assays were performed as described previously (21) based on the method described by Adam et al. (2). Briefly, HeLa cells were grown on 12-mm coverslips to 40 to 80% confluence, washed once in ice-cold phosphate-buffered saline (PBS), and permeabilized for 8 min in ice-cold 20 mM HEPES-KOH (pH 7.5)–110 mM potassium acetate–5 mM magnesium acetate–0.5 mM EGTA–250 mM sucrose–40 μg of digitonin (Sigma) per ml. Coverslips were incubated with 20 μl of import mixture for 8 min at room temperature, and reactions were stopped by fixation with 4% paraformaldehyde in PBS. After being washed in PBS and water, the coverslips were mounted with Vectorshield mounting medium (Vector) and analyzed by confocal microscopy (Leika; TCS NT). The import reaction mixtures consisted of an energy-regenerating system (0.5 mM ATP, 0.5 mM GTP, 10 mM creatine phosphate, 50 μg of creatine kinase per ml), core buffer (2 μg of nucleoplasmin core per ml, 20 mM HEPES-KOH [pH 7.5], 140 mM potassium acetate, 6 mM magnesium acetate, 250 mM sucrose), 0.5 mM EGTA, 3 μM RanGDP, 0.2 μM Rna1p, 0.3 μM RanBP1, 0.4 μM NTF2, 1 μM importin β, a 2 μM concentration of an importin α protein, and 10% reticulocyte lysate.
Nucleotide sequence accession number.
The nucleotide sequence associated with importin α7 has been assigned GenBank accession no. AF060543.
RESULTS
Cloning and analysis of the distribution of human importin α7 in tissue.
Whereas in different mammals only one importin β protein has been identified so far, both humans and mice harbor at least five different importin α proteins. The human homologues for most of the mouse proteins have been definitely determined. However, whether or not importin α-S2 (44) represents the mouse homologue of human importin α6 or the homologue of a yet-unknown human importin α protein was not clear. Since we wanted to investigate the functions of all human importin α proteins, we screened the GenBank database and identified a partial sequence of an unknown human cDNA which displayed a high degree of homology with both human importin α6 and mouse importin α-S2. We isolated the corresponding coding cDNA by PCR and found it to be 1,611 bp in length. The encoded protein (importin α7) has about 85% identity to human importin α6 and more than 99% identity to mouse importin α-S2 (Fig. 1A). Therefore, importin α7 belongs to the SRP1-like subfamily of vertebrate importin α proteins (Fig. 1B).
FIG. 1.
(A) Amino acid sequence of human importin α7. Residues in human importin α6 and mouse importin α-S2 that differ from those in human importin α7 are indicated above or below the importin α7 sequence, respectively. Epitopes used for antibody generation are underlined. (B) Alignment tree of all known importin α homologues. Tree construction was performed with the CLUSTAL program. All putative complete proteins of the GenBank and Swiss-Prot databases containing arm repeats and an IBB domain were included in the analysis. The accession numbers of the proteins are (Imp, importin): Imp α homologue (hom.), rice, AB006788; Imp α homologue A, C. eleg., gi1707027; Srp1p, yeast, Q02821; Srp1 A, S. pombe, O14063; Srp1 B, S. pombe, AL034433; Srp1 C, A. thal., O04294; Srp1 D, A. thal., AC003114; Srp1, tomato, O22478; Srp1, rice, AB004660; Srp1 A, A. thal., AF077528; Srp1 B, A. thal., Y14615; Srp1, D. mela., AF074957; Imp α6, human, O15131; Imp α7, mouse, O35345; Imp α7, human, AF060543; Srp1/Imp α5, mouse, U34228; Srp1/Imp α5, human, P52294; Imp α3, C. eleg., AF040995; Imp α3, D. mela., AF074958; Imp α3, human, O00629; Imp α3, mouse, O35343; Imp α4, human, O00505; Imp α4, mouse, O35344; OHO31/Imp α1, D. mela., A57319; pendulin/Imp α1, mouse, P52293; Rch1/Imp α1, human, P52292; Imp α2a, X. laev., P52170; Imp α2b, X. laev., P52171; Imp α homologue B, C. eleg., AF040997. C. eleg., Caenorhabditis elegans; A. thal., Arabidopsis thaliana; D. mela., Drosophila melanogaster; X. laev., Xenopus laevis; yeast, S. cerevisiae.
To determine the distribution of importin α7 in tissue, we first performed RNA analysis by Northern blotting. Transcripts of 8 kbp were detectable in human poly(A)+ RNA from almost all tissues tested (Fig. 2). However, the levels of expression differed considerably. No transcript for importin α7 was detectable in thymus, and the expression levels in lung, liver, small intestine, and colon were significantly lower than those of the other tissues tested, even though control hybridization with β-actin demonstrated that comparable amounts of RNA were loaded (1.7 and 2.0 kbp). The signal at 2.4 kbp in testis is probably due to a cross-reaction with human importin α6, which we previously reported to be expressed only in testis (23). Thus, in contrast to its most highly related isoform, importin α6, importin α7 is expressed in a variety of tissues.
FIG. 2.
Expression of importin α7 mRNA in human tissues. Human multiple-tissue Northern blots were hybridized with probes specific for importin α7 and β-actin. A suggested cross-reactive band of importin α6 was detected in testis at 2.4 kb. sk. muscle, skeletal muscle; sm. intest., small intestine; bl. leuk., peripheral blood leukocytes.
Protein expression pattern of soluble factors involved in importin-dependent protein transport.
Thus far, several groups have investigated the distribution of different importin α isoforms in tissue (23, 32, 38, 44). In contrast, little is known about the distribution of the shuttling transport factors, importin β, CAS, and Ran in tissue. Therefore, we compared the expression levels of all known human shuttling transport factors of the classical import pathway by immunoblot analysis of protein lysates obtained from various tissues. Only NTF2, which was recently shown to transport Ran into the nucleus (41), was not included in our study. First, we generated antibodies against importin α7, CAS, and Ran. Antibodies that specifically recognize the other factors (importin α1/Rch1, importin α5/hSRP1, importin α3, importin α4, and importin β) had been obtained previously (17, 23). Antibodies for importin α7 were raised against two different peptides which correspond to the amino terminus and the carboxy terminus of the protein. Due to the high sequence similarity of the proteins, these antibodies were found to recognize recombinant importin α6 as well but do not cross-react with other importin α proteins (data not shown).
By analyzing lysates of different human tissues by immunoblotting, we detected all transport factors in the investigated tissues (Fig. 3). In agreement with the mRNA analysis, importin α7 protein was found in all tissues tested. In terms of total protein concentration, the expression levels of importin α, importin β, CAS, and Ran vary between the different tissues and were low in spleen and liver. However, these variations were not always uniform. For example, in ovary and lung, where importin α proteins are quite abundant, the expression levels of the other factors are not elevated. Testis and brain contain the highest relative amounts of CAS, but not of importin α proteins. In the heart, the Ran levels were lowest, while the other factors were present in average amounts. A reason for the lack of correlation could be that these factors are also involved in functions other than the importin α-dependent protein transport process. This is well established for Ran (29) and importin β (21), but not for CAS. In several tissues, the relative expression levels of particular importin α proteins differed from those of other importin α forms, as has been reported earlier (23). Notably, the antibodies against importin α7 and importin α4 detect several proteins in the range of 60 kDa which are not present in all tissues. In both cases, these bands are recognized by two independent antibodies raised against the extreme N terminus and C terminus, respectively (data not shown), which probably reflects the existence of modified or alternatively spliced versions of these proteins.
FIG. 3.
Expression of the shuttling transport factors of the classical nuclear import pathway in human tissues at the protein level. Twenty-five micrograms of total protein lysates was loaded per lane, separated by SDS-PAGE, and probed by immunoblotting with the antibodies against importin α1/Rch1, importin α3, importin α4, importin α5/hSRP1, importin α7, importin β, CAS, and Ran. imp, importin; sm. intest., small intestine.
Binding affinities between importin α proteins and their carriers importin β and CAS.
To investigate the properties of the various importin α’s during protein import, we recombinantly expressed and purified the five ubiquitously expressed human isoforms, as well as Xenopus importin α2 and the only yeast importin α homologue, SRP1p (Fig. 4A). We first wanted to investigate if there were differences in binding to the nuclear import factor importin β or in binding to CAS, the nuclear export factor of importin α.
To quantitate the binding affinities of the different importin α proteins to CAS, we employed the fact that the binding affinity of CAS for RanGTP is greatly enhanced in the presence of importin α proteins. Furthermore, the binding results in protection against activation of Ran/TC4 GTPase by RanGAP1 (24). We preincubated 50 pM Ran-[γ-32P]GTP either with 1 μM human CAS or with mixtures of 1 μM human CAS and the different importin α homologous proteins. After 30 min at 15°C, we added 40 nM Rna1p for 30 s and determined the hydrolysis of Ran-bound GTP as released [32P]phosphate. As can be seen in Fig. 4B, we found that importin α5/hSRP1, importin α1/Rch1, and importin α3 bind CAS with high affinity within the same range (KD < 2 nM) (Fig. 4B). The binding affinity of Xenopus importin α2 was only marginally weaker (KD > 3 nM) (Fig. 4C). However, in comparison to the human isoforms importin α7 and importin α4 (KD > 5 nM), the binding affinity of Xenopus importin α2 to CAS was even higher. Only yeast SRP1p showed a binding affinity to CAS characterized by much less efficiency (KD > 20 nM).
The binding of the different importin α proteins to importin β was analyzed with the Biacore 2000 instrument. We immobilized the recombinant importin α proteins and BSA (as a control) on sensor chips and injected various concentrations of recombinant importin β (6.25 to 200 nM). We measured the SPR response and calculated the equilibrium KD from the results. Thus, we found that all importin α proteins tested were able to bind importin β very efficiently (Table 1). The differences between the human importin α proteins for binding importin β that we detected were only marginal. The highest binding affinities to importin β were found for importins α4 and α7 (5 nM). The lowest binding affinity was detected for importin α3 (18 nM). Interestingly, Xenopus importin α2 and yeast SRP1 showed no marked differences in their binding affinities to importin β in comparison to the human isoforms (3 and 5 nM, respectively). In contrast, BSA was not able to bind importin β significantly, demonstrating that the binding of the α importins to importin β is specific.
TABLE 1.
Binding affinities of importin α proteins to importin βa
| Proteinb | KD (nM) | Dc (%) |
|---|---|---|
| α1/Rch1 | 10 | 18 |
| α3 | 18 | 16 |
| α4 | 5 | 20 |
| α5/hSRP1 | 9 | 18 |
| α7 | 5 | 33 |
| Xenopus α2 | 3 | 35 |
| Yeast SRP1p | 5 | 40 |
KDs for the interaction between the importin α’s and importin β were determined by SPR with the Biacore 2000 instrument (see Materials and Methods).
β, importin β; α1, importin α1; α3, importin α3; α4, importin α4; α5, importin α5; α7, importin α7; α2, importin α2; yeast, S. cerevisiae.
D, range of deviation.
Comparison of the import efficiencies of different importin α proteins in vitro by using standard substrates.
To analyze the specific functions of the different importin α proteins, we wanted to compare their import efficiencies in parallel by a defined in vitro nuclear import assay. First, we established that all recombinant proteins were able to import the standard substrates simian virus 40 large-T antigen–NLS–BSA and Xenopus nucleoplasmin. Import assays were generally performed in the presence of reticulocyte lysate, which improved the efficiency of protein import. In the absence of importin α, the reticulocyte lysate had no effect on the nuclear import of any substrate tested. Presently, we cannot decide whether this improvement of import efficiency by reticulocyte lysate is due to unspecific protein-protein interactions or caused by as-yet-unknown specific import factors. Fractionation of reticulocyte lysate showed that several fractions display this effect, but no fraction was as stimulating as the total lysate. Hemoglobin, the most abundant protein of reticulocyte lysate, did not enhance the import efficiency.
A typical result for the different importin α proteins is shown in Fig. 5. We found that all recombinant importin α proteins, including the hitherto-uninvestigated importin α7, were able to import both substrates. However, there were clear differences in the import efficiencies of the different importin α proteins. For NLS-BSA, the best import efficiencies were found with importin α5/hSRP1, importin α3, α7, and Xenopus importin α2. The effect of importin α4 was mildly weaker, but still stronger than that of importin α1/Rch1. The weakest import efficiency on NLS-BSA was displayed by yeast SRP1p, although the nuclear accumulation of the labeled substrate was still evident in comparison to that of the negative control, which was performed without adding any importin α protein. The pattern for nuclear import of nucleoplasmin was similar to the one for NLS-BSA. Importin α5/hSRP1 showed the strongest effect on nuclear import of nucleoplasmin, followed by importin α3, α4, and Xenopus importin α2. The import efficiencies of importin α1/Rch1 and importin α7 were mildly weaker, and that of yeast SRP1p was again the weakest. The negative control showed a typical pattern, with a few cells displaying a weak nuclear staining which is most likely due to leaky nuclear envelopes. However, most of the nuclei are clearly negative, in contrast to the results of all import assays with one of the importin α proteins added to the import reaction mixture.
Importin α-dependent nuclear import of hnRNP K, P/CAF, and RCC1.
We next analyzed the abilities of the various importin α proteins to mediate the import of three functionally completely different human proteins, namely, the shuttling RNA-binding protein hnRNP K containing a nuclear shuttling domain which confers bidirectional transport across the nuclear envelope and also a classical NLS (27), the stimulator of the Rous sarcoma virus promoter P/CAF (40), and Ran’s major GDP/GTP exchange factor, the chromatin-bound protein RCC1, which is exclusively located inside the nucleus (3, 35). First, we added the fluorescein-labeled proteins as single substrates into the import assay reaction mixtures (Fig. 6A, 7A, and 8A). We found that both hnRNP K and P/CAF were imported by most of the importin α proteins very efficiently (Fig. 6A and 7A). Similar to the results of assays testing the import of the standard substrates NLS-BSA and nucleoplasmin, the efficiencies of the import reactions differed slightly depending on the importin α protein used. Importin α1/Rch1, Xenopus importin α2, importin α3, and importin α5/hSRP1 showed the best stimulation of the nuclear import of hnRNP K, whereas importins α4, α7, and yeast SRP1p displayed a somewhat weaker effect. For P/CAF, these differences were even less pronounced. Here, the pattern after import with importin α1/Rch1 was somewhat more heterogeneous and the effects of importin α4 and yeast SRP1p were weaker than those of the other importin α isoforms. In contrast to what was found for hnRNP K and P/CAF, when RCC1 was added as the only substrate to the import reaction mixture, we found striking differences in its nuclear import depending on the importin α protein used in the import assay (Fig. 8A). While importin α3 proved to behave as a very good import factor for RCC1, the effect of importin α4 was moderate, and the other importin α proteins had hardly any effect at all.
FIG. 6.
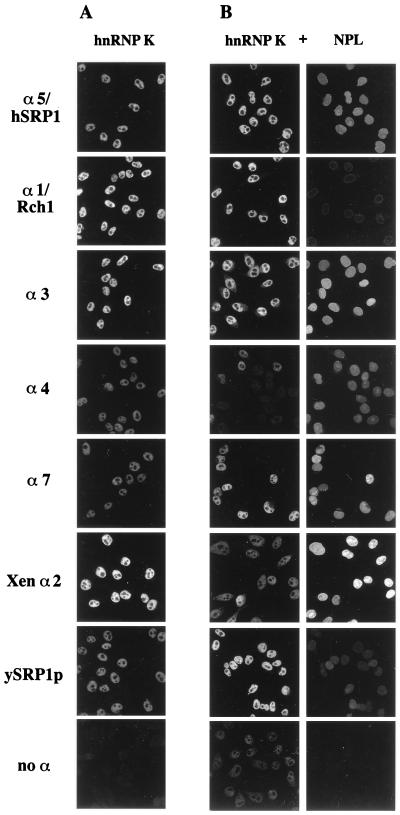
Importin α-dependent nuclear import of hnRNP K. In vitro nuclear import of fluorescein-labeled hnRNP K was performed as described above (see Materials and Methods and legend for Fig. 5) either in the absence (A) or in the presence (B, right panels) of Texas red-labeled nucleoplasmin by using the importin α proteins indicated. (B) Left panels, hnRNP K staining; right panels, nucleoplasmin (NPL) staining. α3, importin α3, α4, importin α4; α7, importin α7; Xen α2, Xenopus importin α2; ySRP1, yeast SRP1p; no α, no importin α added to the import reaction mixture.
FIG. 7.
Importin α-dependent nuclear import of P/CAF. In vitro nuclear import of fluorescein-labeled P/CAF was performed as described above (see Materials and Methods and legend for Fig. 5) either in the absence (A) or in the presence (B, right panels) of Texas Red-labeled nucleoplasmin (NPL) by using the importin α proteins indicated. (B) Left panels, P/CAF staining; right panels, nucleoplasmin staining. α3, importin α3; α4, importin α4; α7, importin α7; Xen α2, Xenopus importin α2; ySRP1, yeastSRP1p; no α, no importin α added to the import reaction mixture.
FIG. 8.
Importin α-dependent nuclear import of RCC1. In vitro nuclear import of fluorescein-labeled RCC1 was performed as described above (see Materials and Methods and legend for Fig. 5) either in the absence (A) or in the presence (B, right panels) of Texas red-labeled nucleoplasmin (NPL) by using the importin α proteins indicated. (B) Left panels, hnRNP K staining; right panels, nucleoplasmin staining. α3, importin α3; α4, importin α4; α7, importin α7; Xen α2, Xenopus importin α2; ySRP1, yeast SRP1p; no α, no importin α added to the import reaction mixture.
In living cells many substrates are present in the cytoplasm and may compete for transport into the nucleus by a particular importin α protein. Therefore, we wondered whether or not good import substrates might be efficient competitors in our import assays. Thus, we repeated the import reactions with fluorescein-labeled hnRNP K, P/CAF, and RCC1, adding in every case Texas red-labeled nucleoplasmin as well, which has been shown to be imported by all importin α proteins (Fig. 5). If hnRNP K was combined with nucleoplasmin, importins α4 and α7 had a weaker effect on the nuclear import of hnRNP K than importin α5/hSRP1, importin α3, and importin α1/Rch1 (Fig. 6B). These findings were similar to what we found with hnRNP K alone (Fig. 6A). However, in contrast to the single-substrate assay, in this two-substrate assay, yeast SRP1p was as efficient as importin α5/hSRP1 and importin α1/Rch1 in transporting hnRNP K into the cell nucleus. Surprisingly, Xenopus importin α2 now had no effect at all on the nuclear import of hnRNP K. This finding clearly demonstrates that the relative effect on nuclear import of hnRNP K by a particular importin α isoform can be decreased if another competing substrate is present. These changes were found to be even more dramatic if fluorescein-labeled P/CAF was added to the import assay mixtures together with Texas red-labeled nucleoplasmin (Fig. 7B). Only importin α3 was able to import P/CAF very efficiently in the presence of nucleoplasmin. In contrast, the effect of all other importin α proteins on the nuclear transport of P/CAF was greatly diminished (importin α5/hSRP1, importin α4, importin α7, yeast SRP1p) or even abolished (importin α1/Rch1 and Xenopus importin α2) if nucleoplasmin was added to the assay mixture. A comparison of the various patterns found for nuclear import of nucleoplasmin (Fig. 5, 6B, 7B, and 8B) indicates that good import substrates can strongly compete for each other depending on the particular importin α protein present in the assay system. Whereas, in the presence of hnRNP K, Xenopus importin α2 is the most efficient transport factor for nucleoplasmin, importin α5/hSRP1 becomes the best import-stimulating isoform for nucleoplasmin if RCC1 is added to the import reaction mixture, and the effect of Xenopus importin α2 on nucleoplasmin import is much weaker (compare Fig. 6B with Fig. 8B). It should be noted that different settings of the confocal microscope were used for single- and double-labeling experiments, i.e., one cannot directly compare the intensities between these different types of experiments. This can be seen if, e.g., one compares the negative controls in Fig. 6A and B. Within one experimental series, the settings had of course been identical for all combinations of a given substrate with the indicated import factors.
DISCUSSION
The classical nuclear protein import pathway is mediated by the α and β subunits of importin. Importin α functions as an adapter molecule by binding both importin β and the NLS-bearing import substrate. While only one importin β isoform has been found in humans thus far, six human genes for importin α exist, including that for importin α7, newly described here. Some of these importin α isoforms may even occur in different versions. We found partial cDNAs in the database corresponding to importin α6 and importin α7, which may represent alternatively spliced mRNAs. Those cDNAs would code for proteins with amino termini three amino acids shorter or longer than those published, respectively. As reported previously (23, 32, 38, 44) and now confirmed in more detail here, the relative expression levels of a particular importin α form may vary between different tissues, indicating a special demand of different cell types for specific importin α’s. Nevertheless, all human importin α proteins can be found in various tissues with the exception of importin α6, which has thus far been found only in testis. Even more striking, these proteins are expressed simultaneously in many cell lines, such as HeLa cells or human umbilical vein epithelial cells (reference 23 and data not shown).
The radiation of ancestral importin α genes into different orthologues probably occurred several times during evolution (Fig. 1B). Most of the known importin α forms, such as the six mammalian importin α’s, belong to one of three main subgroups, which differ from one another in about 50% of their amino acids. However, several other species such as C. elegans and rice possess additional importin α-like proteins differing in more than 75% of their amino acids from those isoforms of the main subgroups. Most species examined so far possess at least one protein which belongs to the SRP1-like subgroup. Humans have three different SRP1-like proteins, namely, importin α5, importin α6, and the newly reported importin α7. In contrast to the diversity found in other organisms, Schizosaccharomyces pombe has two genes coding for importin α proteins and S. cerevisiae has only one importin α gene, SRP1. This indicates that one importin α isoform alone may be sufficient to fulfill the basic requirements of a eucaryotic cell.
Although the identity of the primary sequences of the importin α isoforms including the IBB domain varies between 50 and 85%, the proteins do not differ dramatically in their interactions with their transport receptors CAS and importin β. The human importin α proteins bind CAS with a KD between 2 and 5 nM. These differences are small and could be caused by variations in the quality of the protein purification process. Whereas the binding affinity of Xenopus importin α2 turned out to be in the same range as that of the human importin α forms, yeast SRP1p showed a clearly less efficient binding affinity to CAS (KD > 20 nM). Since yeast SRP1p is less homologous to the human importin α proteins than Xenopus importin α2 is, the weaker binding affinity of yeast SRP1p to human CAS was not surprising. No binding to Crm1/exportin was observed. These results fit with recently reported data from Herold et al., who found similar binding affinities of importin α1/Rch1, importin α5/hSRP1, and importin α4 to CAS but not to Crm1 in two-hybrid studies (18). The KDs for the interactions between importin β and the various importin α forms in the Biacore assay are also in a nanomolar range (between 3 and 18 nM). These differences are unlikely to have an influence on the in vitro assay, where the proteins are present in micromolar concentrations. Again, these small differences may be caused by variations in the quality of the protein purification process. In addition, the coupling of the proteins to the sensor chips may impair the importin α forms to different extents.
The simultaneous existence of several highly divergent importin α proteins in a given cell led to the question whether they might be specialized in their efficiency to transport different nuclear proteins. Several experiments clearly support this hypothesis. Sekimoto et al. recently reported that intracellular injection of antibodies against importin α5/hSRP1, but not against importin α1/Rch1, can inhibit nuclear import of the transcription factor Stat1 (43). Fisher et al. reported that the Epstein-Barr virus protein EBNA1 interacts with importin α1/Rch1 but not with importin α5/hSRP1 in the yeast two-hybrid system (11). By pull-down assays with different NLS-BSA conjugates, Nadler et al. showed that importin α1/Rch1 and importin α5/hSRP1 share distinct binding affinities for various NLSs (33). Finally, Miyamoto et al. demonstrated that the efficiency of nuclear import of different NLS-reporter protein conjugates in vitro may depend on the importin α protein present in the assay (28). Thus, earlier studies indicated that there might exist substrate specificities for the different importin α proteins. Therefore, we compared the import activities of all known ubiquitously expressed human importin α proteins on different artificial and natural substrates by using a defined in vitro import system. For comparison we also included frog importin α2, a paralogue of human importin α1/Rch1, and yeast SRP1p in our study. Most substrates were imported with about the same efficiency by all importin α isoforms, if they are added as single substrates to the assay. Nevertheless, significant differences between the different isoforms were detectable, and the nuclear import of RCC1, a protein that is strictly localized within the nucleus, showed a very strong dependence on the presence of one particular importin α isoform (importin α3). The addition of two differently labeled substrates into one import reaction mixture clearly demonstrated that those differences are unlikely to be caused by variations in the quality of purification of the recombinant proteins. For example, the very strong effect of importin α1/Rch1 on the nuclear import of hnRNP K demonstrates that its weak import efficiency on nucleoplasmin in the same assay reaction is not due to a functionally inactive protein. Only yeast SRP1p usually showed a weak import efficiency, probably because its evolutionary distance from the human proteins fostered an impaired interaction with the mammalian substrates in our import assays.
It is unlikely that differences in binding affinity between substrate and importin α are the main reason for the observed effects in the import assay. Although experiments using SPR demonstrate that RCC1 binds significantly better to importin α3 and α4 (KDs ∼9 nM) than to the other isoforms (KDs ∼18 to 30 nM) while nucleoplasmin shows no significant differences in its binding to the various importin α forms (KDs ∼4 to 8 nM) (data not shown), these differences are too small to have an influence on the in vitro import assay where substrates and importin α’s are present in micromolar concentrations.
Some of our results disagree with data obtained by other groups. Nachury et al. (32) reported that importin α4/hSRP1γ had a weaker efficiency to import NLS-BSA in vitro than importin α1/Rch1. We could not detect those differences. In contrast to Nachury et al. (32), who obtained importin α4 from HeLa cells via vaccinia virus infection and importin α1/Rch1 from E. coli, we purified all importin α proteins from the same system (E. coli), which might explain those differences. Moreover, Miyamoto et al. (28) reported that a CBP80-allophycocyanin fusion protein becomes imported in their in vitro import system by importin α1/Rch1 and by importin α5/SRP1 but not by importin α3/Qip1. In our hands, recombinant human cap binding protein is imported by all human importin α proteins tested (data not shown). This finding demonstrates that artificial NLS fusion proteins and their corresponding full-length proteins may behave quite differently in the import assay system and that testing the functional activity of the purified import factors is very important for comparison of their efficiencies. Furthermore, the source of the recombinant proteins might influence the result to some extent.
If one adds two substrates simultaneously, the preference of a particular importin α for a certain substrate can get more clear-cut. For example, if nucleoplasmin and P/CAF are added to one import reaction mixture at the same time, importin α3 is still able to import P/CAF very efficiently. This result is in clear contrast to those for the other importin α proteins, although all of them can still import nucleoplasmin. The situation for RCC1 is similar. This protein is transported efficiently as a single added substrate into the nucleus only by importin α3 and to a less efficient extent by importin α4, which is highly homologous to importin α3. The addition of nucleoplasmin as a second substrate enhances these differences between the members of the importin α3/α4 subfamily and members of the other subfamilies. This observation indicates that one level of the import efficiency regulation in the cell may be the competition between import substrates for their “preferred” importin α form. However, the results of our in vitro studies cannot predict to what extent this concurrence happens in living cells and whether or not different importin α proteins can substitute for each other in vivo.
ACKNOWLEDGMENTS
We thank E. Bürger, B. Nentwig, and A. Wittstruck for technical help and M. Wellner and D. Fiedler for sequencing. We also thank C. Maasch for assistance with the Biacore instrument, J. Francke for purifying nucleoplasmin, S. Thiel for assistance with immunoblottings, and K. Ribbeck for detailed introduction into the import assay system. The expression clone for hnRNP K was a kind gift from Dirk Ostarek. Finally, we are grateful to F. C. Luft for critical reading of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG KO 1950/1-1).
REFERENCES
- 1.Adam E J, Adam S A. Identification of cytosolic factors required for nuclear location sequence-mediated binding to the nuclear envelope. J Cell Biol. 1994;125:547–555. doi: 10.1083/jcb.125.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adam S A, Sterne-Marr R, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bischoff F R, Postingl H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature. 1991;354:80–82. doi: 10.1038/354080a0. [DOI] [PubMed] [Google Scholar]
- 4.Chi N C, Adam E J H, Adam S A. Sequence and characterization of cytoplasmic nuclear import factor p97. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- 6.Cortes P, Zheng-Sheng Y, Baltimore D. RAG-1 interacts with the repeated amino acid motif of the human homologue of the yeast protein SRP1. Proc Natl Acad Sci USA. 1994;91:7633–7637. doi: 10.1073/pnas.91.16.7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuomo C A, Kirch S A, Gyuris J, Brent R, Oettinger M A. Rch1, a protein that specifically interacts with the RAG-1 recombination-activating protein. Proc Natl Acad Sci USA. 1994;91:6156–6160. doi: 10.1073/pnas.91.13.6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 9.Dingwall C, Sharnick A V, Laskey R A. A polypeptide domain that specifies migration of nucleoplasmin into the nucleus. Cell. 1982;30:449–458. doi: 10.1016/0092-8674(82)90242-2. [DOI] [PubMed] [Google Scholar]
- 10.Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 11.Fischer N, Kremmer E, Lautscham G, Mueller-Lantzsch N, Grässer F A. Epstein-Barr virus nuclear antigen 1 forms a complex with the nuclear transporter karyopherin α2. J Biol Chem. 1997;272:3999–4005. doi: 10.1074/jbc.272.7.3999. [DOI] [PubMed] [Google Scholar]
- 12.Görlich D. Transport into and out of the cell nucleus. EMBO J. 1998;17:2721–2727. doi: 10.1093/emboj/17.10.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Görlich D, Henklein P, Laskey R A, Hartmann E. A 41 amino acid motif in importin alpha confers binding to importin beta and hence transit into the nucleus. EMBO J. 1996;15:1810–1817. [PMC free article] [PubMed] [Google Scholar]
- 14.Görlich D, Kostka S, Kraft R, Dingwall C, Laskey R A, Hartmann E, Prehn S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- 15.Görlich D, Panté N, Kutay U, Aebi U, Bischoff F R. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- 16.Görlich D, Prehn S, Laskey R A, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 17.Görlich D, Vogel F, Mills A D, Hartmann E, Laskey R A. Distinct functions for the two importin subunits in nuclear protein import. Nature. 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- 18.Herold A, Truant R, Wiegand H, Cullen B R. Determination of the functional domain organization of the importin α nuclear import factor. J Cell Biol. 1998;143:309–318. doi: 10.1083/jcb.143.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imamoto N, Shimamoto T, Kose S, Takao T, Tachibana T, Matsubae M, Sekimoto T, Shimonishi Y, Yoneda Y. The nuclear pore targeting complex binds to nuclear pores after association with a karyophile. FEBS Lett. 1995;368:415–419. doi: 10.1016/0014-5793(95)00699-a. [DOI] [PubMed] [Google Scholar]
- 20.Imamoto N, Shimamoto T, Takao T, Tachibana T, Kose S, Matsubae M, Sekimoto T, Shimonishi Y, Yoneda Y. In vivo evidence for involvement of a 58 kDa component of nuclear pore targeting complex in nuclear protein import. EMBO J. 1995;14:3617–3626. doi: 10.1002/j.1460-2075.1995.tb00031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jäkel S, Görlich D. Importin β, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson B, Lofas S, Lindquist G. Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors. Anal Biochem. 1991;198:268–277. doi: 10.1016/0003-2697(91)90424-r. [DOI] [PubMed] [Google Scholar]
- 23.Köhler M, Ansieau S, Prehn S, Leutz A, Haller H, Hartmann E. Cloning of two novel importin-α subunits and analysis of the expression pattern of the importin-α protein family. FEBS Lett. 1997;417:104–108. doi: 10.1016/s0014-5793(97)01265-9. [DOI] [PubMed] [Google Scholar]
- 24.Kutay U, Bischoff F R, Kostka S, Kraft R, Görlich D. Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- 25.Kutay U, Izaurralde E, Bischoff F R, Mattaj I W, Görlich D. Dominant-negative mutants of importin-β block multiple pathways of import and export through the nuclear pore complex. EMBO J. 1997;16:1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melchior F, Paschal B, Evans E, Gerace L. Inhibition of nuclear protein import by nonhydrolyzable analogs of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michael W M, Eder P S, Dreyfuss G. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 1997;16:3587–3598. doi: 10.1093/emboj/16.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyamoto Y, Imamoto N, Sekimoto T, Tachibana T, Seki T, Tada S, Enomoto T, Yoneda Y. Differential modes of nuclear localization signal (NLS) recognition by three distinct classes of NLS receptors. J Biol Chem. 1997;272:26375–26381. doi: 10.1074/jbc.272.42.26375. [DOI] [PubMed] [Google Scholar]
- 29.Moore M S. Ran and nuclear transport. J Biol Chem. 1998;273:22857–22860. doi: 10.1074/jbc.273.36.22857. [DOI] [PubMed] [Google Scholar]
- 30.Moore M S, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- 31.Moore M S, Blobel G. Purification of a Ran-interacting protein that is required for protein import into the nucleus. Proc Natl Acad Sci USA. 1994;91:10212–10216. doi: 10.1073/pnas.91.21.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nachury M V, Ryder U W, Lamond A I, Weis K. Cloning and characterization of hSRP1γ, a tissue-specific nuclear transport factor. Proc Natl Acad Sci USA. 1998;95:582–587. doi: 10.1073/pnas.95.2.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nadler S G, Tritschler D, Haffar O K, Blake J, Bruce A G, Cleaveland J S. Differential expression and sequence-specific interaction of karyopherin α with nuclear localization sequences. J Biol Chem. 1997;272:4310–4315. doi: 10.1074/jbc.272.7.4310. [DOI] [PubMed] [Google Scholar]
- 34.Ohno M, Fornerod M, Mattaj I W. Nucleocytoplasmic transport: the last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- 35.Ohtsubo M, Okazaki H, Nishimoto T. The RCC1 protein, a regulator for the onset of chromosome condensation, locates in the nucleus and binds to DNA. J Cell Biol. 1989;109:1389–1397. doi: 10.1083/jcb.109.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paschal B M, Gerace L. Identification of NTF2, a cytosolic factor for nuclear import that interacts with nuclear pore protein p62. J Cell Biol. 1995;129:925–937. doi: 10.1083/jcb.129.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prieve M G, Guttridge K L, Munguia J, Waterman M L. Differential importin-α recognition and nuclear transport by nuclear localization signals within the high-mobility-group DNA binding domains of lymphoid enhancer factor 1 and T-cell factor 1. Mol Cell Biol. 1998;18:4819–4832. doi: 10.1128/mcb.18.8.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prieve M G, Guttridge K L, Munguia J, Waterman M L. The nuclear localization signal of lymphoid enhancer factor-1 is recognized by two differentially expressed Srp1-nuclear localization sequence receptor proteins. J Biol Chem. 1996;271:7654–7658. doi: 10.1074/jbc.271.13.7654. [DOI] [PubMed] [Google Scholar]
- 39.Radu A, Blobel G, Moore M S. Identification of a protein complex that is required for nuclear protein import and mediates docking of the import substrate to distinct nucleoporins. Proc Natl Acad Sci USA. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reid J L, Bannister J A, Zegerman P, Martinez-Balbás M A, Kouzarides T. E1A directly binds and regulates the P/CAF acetyltransferase. EMBO J. 1998;17:4469–4477. doi: 10.1093/emboj/17.15.4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ribbeck K, Lipowsky G, Kent H M, Stewart M, Görlich D. NTF2 mediates nuclear import of Ran. EMBO J. 1998;17:6587–6598. doi: 10.1093/emboj/17.22.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seki T, Tada S, Katada T, Enomoto T. Cloning of a cDNA encoding novel importin-α homologue, Qip1: discrimination of Qip1 and Rch1 from hSrp1 by their ability to interact with DNA helicase Q1/RecQL. Biochem Biophys Res Commun. 1997;234:7633–7637. doi: 10.1006/bbrc.1997.6535. [DOI] [PubMed] [Google Scholar]
- 43.Sekimoto T, Imamoto N, Nakajima K, Hirano T, Yoneda Y. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J. 1997;16:7067–7077. doi: 10.1093/emboj/16.23.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsuji L, Takumi T, Imamoto N, Yoneda Y. Identification of novel homologues of mouse importin α, the α subunit of the nuclear pore-targeting complex, and their tissue-specific expression. FEBS Lett. 1997;416:30–34. doi: 10.1016/s0014-5793(97)01092-2. [DOI] [PubMed] [Google Scholar]
- 45.Weis K, Mattaj I W, Lamond A I. Identification of hSRP1α as a functional receptor for nuclear localization sequences. Science. 1995;268:1049–1053. doi: 10.1126/science.7754385. [DOI] [PubMed] [Google Scholar]
- 46.Weis K, Ryder U, Lamond A I. The conserved amino terminal domain of hSRP1α is essential for nuclear protein import. EMBO J. 1996;15:1818–1825. [PMC free article] [PubMed] [Google Scholar]