Abstract
Evolutionary selective pressure on lactation has resulted in milk that provides far more than simply essential nutrients, delivering a complex repertoire of agents from hormones to intact cells. Human infants are born with low barrier integrity of their gut, which means that many of the complex biopolymer components of milk enter and circulate in lymph and blood, reaching organs throughout the body. Due to this state of gut maturation, all components of milk are potentially part of the crosstalk between mother and infants. This article highlights the functions of milk's complex biopolymers, more specifically the potential role of microRNAs (miRNAs) contained in extracellular vesicles in human milk. miRNAs are key effectors in the regulation of many biological processes during early-age development, and consequently milk-sourced miRNAs must be considered to provide unique biological assets to the infant during breastfeeding. This article interprets the evidence of the potential action of human milk miRNAs on infant development, taking into account their abundance in milk based on the literature and current knowledge. Human milk miRNAs appear to influence lipid and glucose metabolism, gut maturation, neurogenesis, and immunity. We also show growing evidence that human milk miRNAs are epigenetic modulators that play a pivotal role in the regulation of tissue-specific gene expression throughout life. Furthermore, this article addresses the ongoing debate regarding the potential influence of human milk miRNAs on viral infection as a new research area. This article highlights that these bioactive molecules are now being incorporated into our overall understanding of nutrient needs for healthy infant development, preparing each individual infant to succeed as a healthy and protected adult throughout its life. In essence, miRNAs are a new language in the Rosetta stone of health that is mammalian lactation.
Keywords: human milk, exosome, miRNA, infant, health
Statement of Significance: This manuscript presents analyses that are new in the field.
Introduction
The emergence of lactation as the sole source of nourishment for neonatal mammalian infants has been central to the success of mammals (1), providing a complete system of nourishment. The composition of milk has been a singularly valuable guide for nutrition scientists to identify nutrients, their quantitative requirements, and various mechanisms that ensure their successful absorption (2, 3).
The current paradigm for nourishment is that biopolymers, including proteins, saccharides, polynucleotides, and complex lipids, are denatured by stomach acid, dispersed by bile acids, and attacked by endogenous hydrolytic enzymes. This adult model envisions a milk digestive process that rapidly leads to the release and complete absorption of monomeric amino acids, sugars, nucleotides, and lipids by the intestinal epithelia (4). However, human infants are developmentally naive, produce little gastric acid or bile acids, and express low levels of digestive enzyme activities (5). As a result, the infant is exposed to far more of the components of milk intact. Additionally, the lower barrier integrity of the infant intestine means that many of the complex biopolymer components of milk enter and circulate in lymph and blood, reaching targets throughout the infant and playing a role in establishing immunity in newborns (6). The chemical and biological examination of milk must now consider the functions of those intact and semi-intact biopolymers and the ensembles of molecules from the mammary gland in the infant.
The most innovative opportunity for milk research is to discover the targets on which milk acts to protect infants, support development, and prevent diseases proactively (7). With the goal of identifying mechanisms of “function for prevention,” multiple independent biopolymers have been selected for efficacy as ingested components towards the targets that guide the success of the mother-infant dyad.
Neonates and infants are particularly challenging as models of biological functions because all of their complex systems are actively proceeding through their development processes. Many systems (e.g., cardiovascular, respiratory, gastrointestinal) undergo significant changes at birth, and many others (such as neural systems) have not yet completed their development.
Human milk (HM) contains a complex combination of lipids, proteins, carbohydrates, and minerals that are essential for infant growth, development, and immune system (8, 9). Identifying and annotating the complex repertoire of milk components is part of the daunting scientific task. Evidence has shown the importance of milk constituents for infant development. For example, milk lipids illuminate the complexity of milk's diverse roles in infant nutrition. Their presence and abundance are the keys to diverse levels of metabolic regulation, from subcellular compartments to whole-body energy control and signaling (10). Intensive research over the last decade has highlighted oligosaccharides as having a very important role in infant development, in particular, acting on the gut microbiota (GM) (11). More recently, scientists have focused on nucleic acids present in milk. Life scientists have now recognized that cells transcribe far more RNAs than simply those encoding structural proteins.
Of special interest are microRNAs (miRNAs), which are now considered key regulators of numerous biological processes (12). They are actively secreted out of cells that synthesize them, including in milk. The discovery of miRNAs in milk in significant amounts has led the scientific milk community to focus their attention on the potential role of milk in the health of infants (13–15). In this article, we provide evidence in support of the multieffector strategy of milk and the use of miRNAs in milk as a conceptual window into the targets of those tactics. After a brief description of miRNA biogenesis and evidences of food-source miRNAs in humans, this article focuses on small noncoding RNAs (miRNAs) in human breast milk and their potential effects on infant development, finishing with the concept of their role in counteracting viral infections.
MicroRNA Biogenesis
miRNAs are small noncoding RNAs [∼22 nucleotides (nts)] transcribed by RNA polymerase II as part of “pri-miRNAs” (16, 17). Each pri-miRNA forms a hairpin that is a substrate for the Drosha-DGCR8 complex (18). Drosha has 2 RNase III domains that each cut the pri-miRNA hairpin, liberating a ∼60-nt stem-loop called a “pre-miRNA” (19). This pre-miRNA is exported to the cytoplasm via exportin 5 and RAN(RAS-related Nuclear protein)-GTP) (20–22), where Dicer, an endonuclease with 2 RNase III domains, cuts both strands near the loop to generate the miRNA duplex. Then, the miRNA duplex is loaded into an Argonaute protein with chaperone proteins (HSC70/HSP90) to expulse the second strand of the duplex (named miRNA*) to form the mature silencing complex (23). Next, the miRNA targets mRNAs by base pairing to direct their posttranscriptional repression. miRNAs control various biological processes and are thought to regulate ≥60% of genes at the posttranslational level (12, 24). miRNAs have been described as participating in the crosstalk between cells in the same organ (25). However, their isolation from extracellular vesicles (EVs; so-called exosomes) in bodily fluids (as serum) emphasizes their function in the crosstalk between organs (26, 27).
Milk miRNAs as Food-Sourced miRNAs
There is increasing evidence of the presence of plant food–sourced miRNAs in human plasma (28). As an example, miR-168a, an abundant miRNA in rice, was detected in the sera and tissues of the Chinese population. This exogenous plant miRNA could target the human LDL receptor adapter protein 1 (LDLRAP1) mRNA, inhibiting its expression in liver (29). Similarly, a negative correlation between plant miR-159 and breast cancer incidence and progression was shown in a Western population (30). Such studies suggest cross-kingdom action of dietary intake miRNAs and demonstrate that miRNAs acquired orally through food intake influence human gene expression after migration through the plasma and delivery to specific organs (31, 32). The functional influence of ingested miRNAs on organisms consuming them has already emerged as a new signaling system affecting the physiology of consumers through modulation of host gene expression profiles.
The detection of miRNAs in milk has served as a starting point for studies investigating the role of miRNAs, their transfer to and from milk and potential impact on consumers, and of course, better characterization of miRNAs in milk. Thus, miRNAs were detected in different compartments of milk, such as milk fat globules and exosomes (33–35). A comparison between different fractions of milk showed that the milk lipid fractions contained higher concentrations of miRNA compared with skim milk (33). Milk fat globules have proven to be a complex window to the miRNA repertoire of the mammary gland, with discrepancies (35, 36) due, at least in part, to the secretion mechanisms of milk fat globules (37, 38). Discouragingly, few studies are dedicated to the survival or to the effects of milk fat globule miRNA on infants.
Conversely, the potential biological effects of milk EVs containing miRNAs have been demonstrated both in vitro (39) and in vivo (40), suggesting a role of dietary miRNAs. The stability of miRNAs, including those in milk under various harsh conditions, has led investigators to suggest that their packaging in milk EVs has the net effect of protecting them (41), thus allowing their survival in the gut intestinal tract of offspring (29). This is an important step in the delivery of dietary EVs and their cargo, as reported in human vascular endothelial cells with bovine milk (42). However, few studies have reported the lack of transfer of miRNAs from milk to mouse tissues (43, 44). The disagreement between the 2 competing hypotheses could be due to actions modulated by their packaging within “transporting vehicles,” which play an important role in their transfer and action in the consumer (33, 45). Exosomes containing miRNAs protect them against degradation and facilitate their uptake by endocytosis in bovine milk and HM (46, 47). In vitro studies have revealed the survivability and complexity of HM exosome miRNAs upon simulated gastric/pancreatic digestion (48), demonstrating the same ability as preterm milk exosomes (49), suggesting their functional and nutritive role (50). This is confirmed by the in vivo detection in different tissues (such as intestinal mucosa, spleen, liver, heart, and brain) of fluorophore-labeled synthetic miRNAs administered in milk exosomes to mice and pigs (51). Because miRNAs regulate numerous biological processes, their involvement in infant development is indubitable, and crossing through the intestinal barrier is one mechanism of this action.
The Potential Effects of Breast Milk EV miRNAs on Newborn Development
The abundance of miRNAs in milk and their persistence in milk across millennia of selective pressure argue that they play important roles in the postnatal development of mammalian neonates. Here, we performed a functional annotation of miRNAs present in human breast milk exosomes with recent advances in the understanding of their function related to infant development. Their potential action is predicted from the influence of miRNAs on metabolism, gut, neurogenesis, immunity, and epigenetics.
Strategy
We used CAB Abstract (via Ovid at UC Davis Library) and PubMed to search for articles referring to the effect of miRNA on development, avoiding those related to diseases. We focused on 5 main functions that have an important impact during the first weeks of development in a newborn just after birth when the intestine wall is thin and still under development. The thin intestine wall could more easily allow miRNA–target interactions. The keywords that were used for the search were miRNA, immunity, lipid, glycerol, glucose, gut, intestine, epithelium, neurogenesis, and neuron. We focused on metabolism (lipids and glucose), gut maturation, neurogenesis, and immunity. We searched miRNAs regulating these functions. Then, we took into account only miRNAs detected in HM exosomes (48). The abundance was also taken into account on the basis of the total of read counts calculated from Liao et al. (48) with the categories defined as grade A > 250.00, 250.00 ≥ B > 150.00, 150.00 ≥ C > 50.00, and 50.00 ≥ D > 0 sum of the total normalized read counts. We identified 32, 86,170, and 302 miRNAs in grades A, B, C, and D, respectively (Supplemental Table 1). Then, we created a list of the miRNAs that have a potential role in the selected functions based on the rank of abundance (Table 1).
TABLE 1.
Categorization of potential influence of milk microRNAs (miRNAs) on health from bibliographic analyses crossed with human milk miRNA detection1
| Function | miRNAs | Rank | Observations | Reference2 |
|---|---|---|---|---|
| Lipid metabolism | let-7f-5p, miR-148a-3p, miR-182–5p, miR-22–3p | A | AGPAT6 is regulated by some of the most highly expressed human milk cell miRNAs, and has a direct effect on the synthesis of triacylglycerol and long chain acyl-CoA | Alsaweed et al. 2016 (45) |
| let-7f-5p | A | Modulates FADS2 and involved in oleate biosynthesis | Alsaweed et al. 2016 (45) | |
| miR-33 | C | miR-33a and miR33b are intronic miRNAs located within the SREBP genes; regulate lipid metabolism in concert with their host genes | Goedeke et al. 2013 (57) | |
| miR-26a | A | Regulates insulin sensitivity and metabolism of lipids | Fu et al. 2015 (53) | |
| Glucose metabolism | miR-30a-5p | A | Controls THEM4, which is essential for the phosphorylation and synthesis of fatty acids | Alsaweed et al. 2016 (45) |
| miR-143 | B | Induced transgenic overexpression of miR-143 impairs insulin-stimulated AKT activation and glucose homeostasis | Jordan et al. 2011 (59) | |
| miR-33 | C | Cooperates with SREBP in regulating glucose metabolism by targeting PCK1 and G6PC, key regulatory enzymes of hepatic gluconeogenesis | Ramirez et al. 2013 (60) | |
| miR-26a | A | Regulates insulin sensitivity and metabolism of glucose | Fu et al. 2015 (53) | |
| miR-181b | A | Improves glucose homeostasis and insulin sensitivity by regulating endothelial function in white adipose tissue | Sun et al. 2016 (61) | |
| Gut maturation | miR-375, miR-200c | A | Both modulate epithelial function, which can influence exosomal endocytosis and thus uptake of the miRNAs | Alsaweed et al. 2016 (45) |
| miR-200b | B | Inhibits tight junction disruption of intestinal epithelial cells in vitro | Shen et al. 2017 (62) | |
| miR-21 | A | Regulates intestinal epithelial tight junction permeability | Yang et al. 2013 (63) | |
| miR-99b | A | Inhibits the gene expression of MFG-E8, known to maintain intestinal homeostasis by enhancing enterocyte migration | Wang et al. 2016 (64) | |
| miR-200 family | A–D | Critical gatekeepers of the epithelial state linked to epithelial-mesenchymal transition | Pillman et al. 2018 (65) | |
| miR-30 family | A–D | Control proliferation and differentiation of intestinal epithelial cells | Peck et al. 2016 (66) | |
| Neurogenesis | let-7 family | A–D | Neural differentiation of EC cells was accompanied by an increase in let-7 precursor processing activity | Wulczyn et al. 2007 (67) |
| miR-574 | B | Promotes neurogenesis, but reduces the neural progenitor pool | Zhang et al. 2014 (68) | |
| miR-15b | C | Inhibits cortical neural progenitor cell proliferation and promotes cell-cycle exit and neuronal differentiation | Lv et al. 2014 (69) | |
| miR-210 | C | miR-210 inhibition significantly increased neuronal survival of inflammation but reduced proliferation | Voloboueva et al. 2017 (70) | |
| miR-29b | C | Plays a pivotal role in fetal neurogenesis by regulating VDAC1 | Roshan et al. 2014 (71) | |
| Immunity | miR-223 | C | Activates proliferation of granulocytes | Johnnidis et al. 2008 (72) |
| miR-146b-5p | A | Targets signaling proteins of innate immune responses | Taganov et al. 2006 (73) | |
| miR-181a | A | Regulates inflammation responses in monocytes and macrophages in part by downregulating IL-1α | Xie et al. 2013 (74) | |
| miR-150 | C | Blocks B-cell development | Zhou et al. 2007 (75) | |
| miR-182–5p | A | Promotes T-cell–mediated immune responses | Stittrich et al. 2010 (76) | |
| miR-17, miR-92 | C, A | Regulate monocyte development as well as B- and T-cell differentiation and maturation | Mendell 2008 (77) | |
| miR-29a-3p | A | Suppresses immune responses to intracellular pathogens by targeting IFN-γ | Ma et al. 2011 (78) | |
| miR-155 | C | Regulates T- and B-cell maturation and the innate immune response | Vigorito et al. 2013 (79) |
Exosome human milk miRNAs from Liao et al. (48). Abundances of miRNAs in milk are ranked with A > 250.00, 250.00 ≥ B > 150.00, 150.00 ≥ C > 50.00, and 50.00 ≥ D > 0 sum of the total normalized read counts. AGPAT6, 1-acylglycerol-3-phosphate O-acyltransferase 6; AKT, Protein kinase; EC, embryocarcinoma; FADS2, fatty acid desaturase 2; G6PC, Glucose-6-phosphatase Catalitic subunit; MFG-E8, milk fat globule EGF and factor V/VIII domain containing; PCK1, phosphoenolpyruvate carboxykinase 1; SREBP, sterol regulatory element-binding protein; THEM4, thioesterase superfamily member 4; VDAC1, Voltage-dependent anion channel 1.
The list of articles reported here is not exhaustive.
The ranking allowed evaluation of the bioavailability of each miRNA. In this Perspective article, we chose to discuss only the high- and medium-ranking miRNAs. We assume that very low concentrations of miRNA in milk are less captured by the intestinal cells and thus have less effect on the infant. However, it should be noted that considering that exosomes protect their cargoes against the intestinal environment (low pH, enzymatic activities, etc.), we cannot exclude that miRNAs in very small quantities might also have effects.
Potential of human milk miRNAs in infant lipid metabolism
An increasing number of publications report the effects of miRNAs on metabolism regulation. Such miRNAs underlying metabolic regulation were detected in HM (Table 1; Figure 1). Indeed, we identified highly expressed miRNAs in HM that were shown to target genes involved in lipid metabolism, such as miR-182-5p, miR-148a-3p, and miR-22-3p, which were among the top 25 most abundant miRNAs in HM exosomes (48). They regulate the expression of the AGPAT6 gene (coding for 1-acylglycerol-3-phosphate O-acyltransferase 6), having a direct effect on the synthesis of triacylglycerol and long-chain acyl-CoA fatty acids in cells (52). Lipid metabolism could also be regulated by miR-26a, which regulates glucose and lipid metabolism in the liver of mice fed a high-fat diet (53), and miR-30a-5p has been reported to influence fatty acid synthesis by regulating the expression of THEM4 (Thioesterase Superfamily Member 4), a member of the thioesterase superfamily (54). let-7f-5p (rank A for abundance) is a member of the large let-7 family. This family is highly conserved across species in sequence and function (55). let-7f-5p (rank A) was also identified as potentially targeting mRNA coding for AGPAT6, which is an enzyme involved in the synthesis of triacylglycerol (52), and mRNAs coding for stearoyl-coA desaturase and fatty acid desaturase 2, two enzymes catalyzing the biosynthesis of PUFAs, which play pivotal roles in many biological functions. In addition, the members of this family, let-7f, are proposed to regulate stem cells by promoting differentiation during development (56). In addition to these highly abundant miRNAs (rank A) in HM, miR-33 detected with a lower abundance (rank C) has also been reported to influence lipid metabolism. miR-33* (miR-33a and miR-33b) is located within sterol regulatory element-binding protein (SREBP) genes and is well known as a key transcription factor regulating lipogenic gene expression. mir-33* regulates host gene expression and therefore lipid metabolism (57, 58).
FIGURE 1.
Potential effects of human milk exosomal microRNAs (miRNAs) on infant development. The font size and color of each miRNA corresponds to the abundance classification. The classification was based on the total count reads reported by Liao et al. (48) with A > 250.00, 250.00 ≥ B > 150.00, 150.00 ≥ C > 50.00, and 50.00 ≥ D > 0 sum of the total normalized read counts (Supplemental Table 1). miRNAs playing a role in ≥2 functions are in bold. miR, microRNA.
Potential of human milk miRNAs in infant glucose metabolism
In addition to their action on lipid metabolism, miR-33 and miR-26a are also depicted as influencing glucose metabolism. miR-33b is reported to inhibit the expression of phosphoenolpyruvate carboxykinase and glucose 6-phosphatase, 2 key enzymes of hepatic gluconeogenesis leading to the regulation of glucose production (60). The involvement of hepatic miR-26a in glucose metabolism, lipid metabolism, and insulin signaling through the regulation of critical metabolic genes suggests miR-26a is a promising novel target for the treatment of obesity-associated metabolic syndrome (53). miR-33a and miR-143, both present in HM EVs, have been associated with glucose homeostasis and energy metabolism in a model of Per-Arnt-Sim kinase (PASK)–deficient mice fed a high-fat diet (80). The action of miR-143 on glucose homeostasis has also been identified in mice overexpressing this miRNA (59). Additionally, adipose glucose homeostasis and insulin sensitivity are reported to be regulated by miR-181b, which is abundant in HM EVs (61). All these data suggest a potentially important role of HM miRNAs in metabolism influencing the development of infants and therefore their immediate health as well as their future.
Potential of human milk miRNAs in the infant gut
An expected potential role of breast milk miRNAs is in gut maturation and function. Our analysis identified ≥4 miRNAs or families of miRNAs with high abundance and potentially influencing gut function and structure (Table 1; Figure 1). This influence is particularly important due to the permeability of the gut during the first days of life. Intestinal epithelial cells play a fundamental role in the selective absorption of nutrients. They are also a major source of immunoregulatory cytokines and a critical part of the physiological epithelial barrier (81). As mentioned above, studies have demonstrated the interaction of milk EVs and intestinal epithelial cells (14, 47), suggesting their regulation by HM miRNAs. Thus, miR-99b, which is very abundant (rank A) in milk, is known to enhance intestinal MFG-E8 (also known as milk fat globule membrane protein) and restore enterocyte migration (64). The miR-200 family, which is also abundant in HM EVs, is known to influence epithelial-mesenchymal transition (EMT), which has a key role in the structure of epithelia. The members of this family are critical gatekeepers of the epithelial state, restraining the expression of promesenchymal genes that drive EMT (65). miR-200c was shown to regulate several important signaling pathways, such as transforming growth factor β, PI3K/Akt (Phosphoinositide 3 Kinase/protein kinase), Notch, and NF-κB signaling (82). miR-200/375 are also reported to control epithelial plasticity–associated alternative splicing by repressing the RNA-binding protein Quaking (QKI) known to directly bind to and regulate alternative splicing targets (65). The miR-200/miR-375/QKI axis exerts pleiotropic effects, such as increasing cell migration and invasion. Another member of this family, miR-200b, inhibits tight junction disruption of intestinal epithelial cells in vitro (62). Similarly, intestinal epithelial barrier function is modified by miR-21 (63), which is also abundant in breast milk EVs. Gastrointestinal tract function is influenced by the microbiota. The GM plays an important role in host metabolism and therefore regulates a large number of biological processes. The GM interacts with its host, working together to maintain symbiosis (83). miRNAs have been shown to act in the intercommunication between the host and GM (83, 84). This intercommunication includes an influence of the GM on host miRNA expression but also an influence of host miRNAs on the GM (83). miRNAs can enter bacteria and thus regulate their growth (84). Because the GM is shaped by many factors, including host diet (85), we can hypothesize that dietary miRNAs could influence the GM. However, these potential effects are still unknown. In particular, the role of milk miRNAs in human neonates and their effects on GM are unidentified.
Role in neurogenesis
To date, few highly abundant miRNAs present in breast milk EVs are known to influence neurogenesis (Table 1; Figure 1), and in vivo studies have been performed in rodents. The most abundant miRNAs belong to the let-7 family. An increase in let-7 precursor processing activity is associated with neural differentiation of cells, suggesting the role of let-7 in early developmental regulation of embryonic stem cell differentiation and neurogenesis reported in mice in vivo and in vitro (67). miR-29b is also detected in HM EVs and is reported to influence neurogenesis. Its knockdown in vivo in mice results in neural cell death (71). miR-574-5p, moderately abundant in HM EVs, is also an actor in neurogenesis. The overexpression and downregulation of this miRNA promotes and inhibits neurogenesis, respectively, in rat mesenchymal stem cells (68). Three other miRNAs (miR-15b, -132, and -210), detected in HM but in lower abundance, were described as influencing neurogenesis. As a let-7 family member, miR-15b promotes neuronal differentiation and inhibits neural progenitor proliferation in mice (69). miR-132 has been shown to exert effects within the central nervous system by improving memory (86). Conversely, miR-210 inhibition was reported to increase neuronal survival in vitro (70). These few data report the potential role of some miRNAs on neurogenesis but also underline the need to increase our knowledge on the link between miRNAs and neurogenesis and open up new avenues of investigation.
Influence on immune function development
Milk is well known to transfer immunity-related substances from mother to infants. For instance, HM contains large quantities of immunological components but also several nonspecific factors, such as lysozyme, lactoferrin, and oligosaccharides, which have antimicrobial properties (87–89). In addition, miRNAs could participate in this transfer (Table 1; Figure 1). HM is rich in immune-related miRNAs (innate and acquired), including miR-223, -146b-5p, -181a, -150, -155, -92a, and -17 (41, 90). Among them, miR-223, -146, -181a, and -155 have been detected in higher abundance in colostrum than in mature HM (91). Such abundance is in line with the role of transfer of immunity to neonates. Thus, miR-181a is reported to target the 3´-untranslated region (UTR) of IL1a mRNA, regulating inflammatory responses in monocytes and macrophages in vitro (74), whereas miR-223 regulates granulocyte function and fine-tunes the inflammatory response in mice (72). Both miRNAs act on human T cell and granulocyte cell populations as selective targets (92), suggesting that miRNAs could affect newborn immune homeostasis at early stages of life. The innate immune response is also regulated by miR-146b, which is predicted, in vitro, to base-pair the 3′-UTRs of the TNF receptor-associated factor 6 and IL-1α receptor-associated kinase 1 mRNAs encoding 2 key adapter molecules downstream of Toll-like receptors and cytokine signaling (73). miR-155 regulates T-helper-cell differentiation and participates in the development of immune cells such as B and T lymphocytes and macrophages by regulating their function and activation (93, 79). miR-150 is also reported in mice to be an effector controlling B-cell differentiation (94). These studies demonstrate a potential role of HM miRNAs in the establishment of the immune system in infants.
In addition to the potential effects of EV milk miRNAs on neonatal immune function, recent evidence demonstrates the influence of EVs during viral infections. During viral infection, host EVs can package the virus, thus giving them potential protection from the host's immune system and providing secreted entry into host cells. In this way, EVs can contribute to the spread of the virus, as suggested for COVID-19 virus infection in a review (95). However, EVs must be considered to influence various aspects of the infection process, because EVs carry miRNAs, which could affect the interactions between viruses and hosts. A burgeoning body of data suggests a complex 2-way relation between miRNAs and viruses (95). Host miRNAs can act on virus. Host miRNAs can affect RNA virus replication and pathogenesis through direct binding to the RNA virus genome or through virus-mediated changes in the host transcriptome (96). An increasing number of examples have described the influence of host miRNAs during viral infection, leading to the identification of novel mechanisms to block RNA virus replication. As described above, food-source miRNAs can act in consumer cells, and we can hypothesize that they also influence the spread of a viral infection by acting on virus replication (96). To explore this possibility, we compared the 23 miRNAs (Supplemental Table 1) already known to affect virus replication by directly binding numerous RNA virus genomes (96) regardless of the type of virus, with the miRNAs detected in EV milk (48). We observed that among those 23 miRNAs, 16 were present in EV milk (Figure 2). We identified miR-29a, miR-21, and miR-181, which can inhibit the replication of different viruses and are abundantly present in milk EVs (rank A or B). For example, miR-29a, miR-21, miR-181, miR-23, miR-28, and let-7c were reported to bind HIV, porcine reproductive and respiratory syndrome virus, infectious bursal disease virus, enterovirus 71, human T-cell leukemia virus 1, and H1N1 influenza virus, respectively. miR-23, miR-28, and let-7c or miR-150, miR-223, miR-378, miR-505, and miR-296 were also identified to act on virus replication and detected in rank A, B, or C abundance as milk exosomal miRNAs. The last 4 miRNAs (miR-142, miR-32, miR-491, miR-654) were weakly abundant. In addition, other publications reported a role of host miRNAs in viral replication, because mir-30a (97), miR-16-5p (98), and miR-103a (99) were highly expressed (rank A for abundance in HM exosomes), and miR-203 (100) had lower expression (rank C). In contrast, miR-340-5p, belonging to the rank B class in milk, has been described to enhance influenza A virus replication. The diverse roles of miRNAs at the host–virus interface must be deeply explored as well as the mechanisms of action. The effectiveness of the role of food-source miRNAs on viral replication is one of the exciting challenges of the field, particularly in the COVID-19 pandemic context. We used the same strategy to compare the milk exosomal miRNA (from reference 48) with the miRNA computationally predicted to target SARS-CoV-2 RNA (101). We identified 10 miRNAs (Figure 3A). However, their abundances reported by Liao et al. (48) are low. Further analyses of their abundance during different lactation statuses showed that 9 of them were less abundant during late lactation than during early or midlactation (Figure 3B).
FIGURE 2.
Comparison between milk exosomal microRNAs (miRNAs) taking into account their abundance [rank A (high) to D (low) abundance with A > 250.00, 250.00 ≥ B > 150.00, 150.00 ≥ C > 50.00, and 50.00 ≥ D > 0 sum of the total normalized read counts] and the list of miRNAs targeting viral RNA. Additional knowledge on the effects of host miRNAs on virus replication is indicated in the panel bottom right. 1From Liao et al. (48); 2from Trobaugh and Klimstra (96). miR, microRNA.
FIGURE 3.
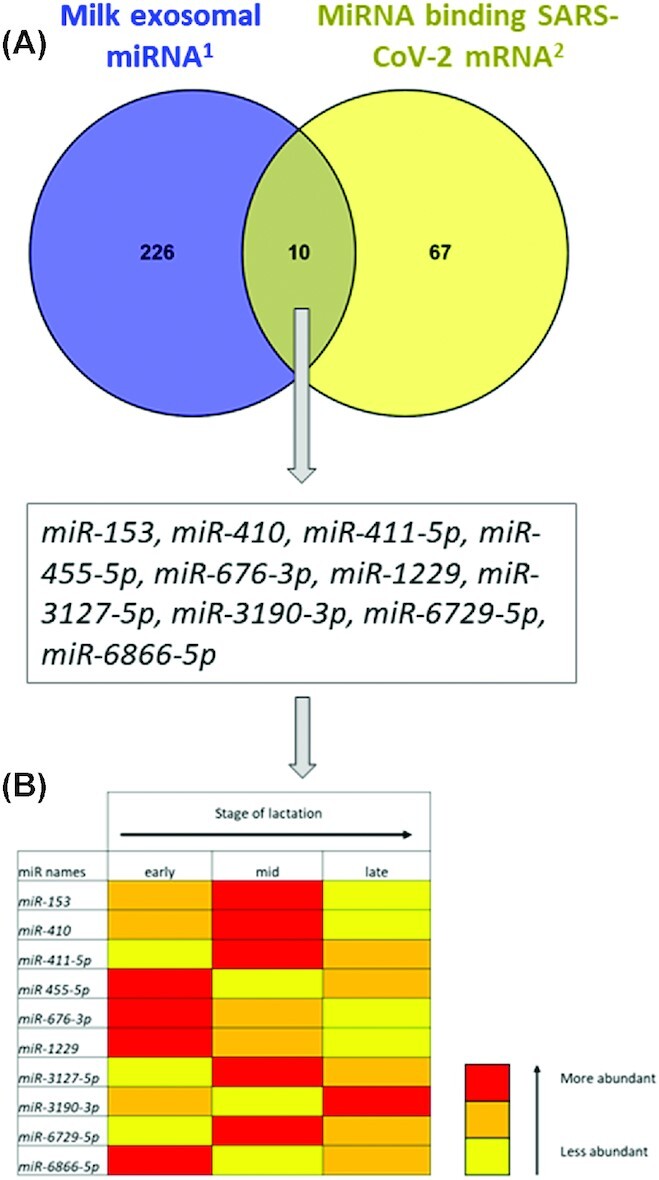
Potential relation between milk exosomal microRNAs (miRNAs) and SARS-CoV-2 virus. (A) Comparison between milk exosomal miRNAs and the list of miRNAs targeting SARS-CoV-2 RNA. (B) Abundance of common miRNAs in undigested exosomal miRNAs according to lactation status. Yellow, orange, and red colors indicate the level of abundance, with yellow being less abundant than orange than red. 1From Liao et al. (48); 2from Demirci and Adan (101). miR, microRNA.
In conclusion, based on all these observations, we can suggest that EV miRNAs from milk should be predicted to target viral RNA, including SARS-CoV-2. The determination of the conceivable action of milk miRNAs on viral replication raises the question of their possible role in protecting against viral infection. Such a role should be investigated to be considered in parallel with more conventional therapeutic strategies.
Predicted role of HM in epigenetic regulation
In addition to the diverse roles of miRNAs in neonatal development, long-term effects must be considered such as their actions on epigenetic processes. Epigenetic processes are shown to play a pivotal role in regulating tissue-specific gene expression and can induce long-term changes, which persist throughout the life course (102). There is accumulating evidence that milk is a major epigenetic modulator of gene expression in infants and therefore in adults. miRNAs were demonstrated to act on epigenetics via the expression regulation of DNA methyltransferases (DNMTs) involved in DNA methylation, which is crucial for gene expression and hallmarks of human diseases. For example, the miR-29 family targets DNMT family members, because miR-29 reverts aberrant methylation via complementarities to the 3′-UTRs of DNMT3A and DNMT3B mRNA, encoding 2 key de novo methyltransferases (103). Similarly, miR-148a-3p represses human DNMT3B gene expression (104). These 2 miRNAs reduced DNA methylation in humans. Thus, the presence of miR-29b and miR-148a-3p in HM exosomes (41) raises the question of the role of HM miRNAs in the epigenome of infants.
Conclusion
This study highlighted the predicted key role of HM miRNAs in infant development: they participate in the continuum between mother and infant. We related the predicted effects on the infant to the abundance of miRNA in milk, presented here as a ranking. This point is important to strengthen their potential influence. All these data underscore the complexity of regulation by miRNAs. Their abundance in milk must be taken into account to assess their effects on infants. There was compelling evidence showing the importance of milk as a diverse, complex, and highly functional matrix of support systems selected through evolution for newborn development. More recently, the presence of miRNAs in breast milk allowed us to answer the following question: what targets does milk act upon to improve the health of infants? We report a detailed examination of the effects of several dozen of those miRNAs involved in the control of functions related to infant development. Several miRNAs that are abundant in milk could potentially influence various functions (Figure 1). For example, miR-21 was reported to be involved in immunity and gut maturation, miR-29b could regulate epigenetics and influence neurogenesis, miR-182-5p was described to act both on metabolism and immunity, and miR-148 influenced metabolism and epigenetics. The let-7 family is represented by several members in milk and is known to be involved in multiple functions (e.g., neurogenesis, immunity, metabolism). Based on this set of evidences, in addition to the greater permeability of the gut during the first days of life, the potential role of miRNAs, especially those abundant in milk, in infant development can reasonably be suggested. These discoveries have to be incorporated into our overall understanding of nutrient needs for healthy infant development, preparing each individual infant to succeed as a healthy and protected adult throughout life. This study builds on the concept of natural selection throughout evolution for a biofluid that transfers bioactive components from mother to infant through milk for successful infant development. In addition, this study strengthens the crucial role of milk in infant development and protection using examples of the importance of multiple milk constituents. This evolutionary perspective on the protection and prevention of disease is envisioned as a complement to the history of therapeutic interventions based on the model of 1 target–1 molecule drug development. In practice, HM is a model combining efficacy and safety in natural selection that takes into account the daunting challenge of this complexity. In the most modern context, milk miRNAs can be considered in the face of viral infection and how to build scientific strategies for prevention. We propose that within the significant ongoing research to fully understand human breast milk and its targeted function, miRNAs provide an attractive path for functional discovery. However, functional redundancy across biopolymers acting upon unique targets in concert should be the goal of future strategies. This massive task of reverse engineering a bioreactor as complex as the mammary gland and a product as dynamic as milk will require new tools, models, and paradigms (105). Nonetheless, accomplishing this task will have immediate benefits on infants and mothers and provide a clear map for guiding improved health for everyone. In essence, miRNAs are a new language in the Rosetta stone of health, mammalian lactation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the NIH for the grant (no. 1R01DK124193-01).
The authors’ responsibilities were as follows—MLC: conducted the analyses to build Table 1; JBG, CL: conceptualized the article; CL: wrote the first draft; and all authors: read and approved the final manuscript.
Notes
Supported by NIH grant (no: 1R01DK124193-01).
Author disclosures: The authors report no conflicts of interest.
Supplemental Table 1 is available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/advances/.
Perspective articles allow authors to take a position on a topic of current major importance or controversy in the field of nutrition. As such, these articles could include statements based on author opinions or point of view. Opinions expressed in Perspective articles are those of the author and are not attributable to the funder(s) or the sponsor(s) or the publisher, Editor, or Editorial Board of Advances in Nutrition. Individuals with different positions on the topic of a Perspective are invited to submit their comments in the form of a Perspectives article or in a Letter to the Editor.
Abbreviations used: AGPAT6, 1-acylglycerol-3-phosphate O-acyltransferase 6; DNMT, DNA methyltransferase; EMT, epithelial-mesenchymal transition; EV, extracellular vesicle; GM, gut microbiota; HM, human milk; miRNA, microRNA; nt, nucleotide; QKI, Quaking; UTR, untranslated region.
Contributor Information
Christine Leroux, Foods for Health Institute, Department of Food Science and Technology, University of California, Davis, Davis, CA, USA; INRAE, UMR Herbivores, Université Clermont-Auvergne, VetAgro Sup, Saint Genès Champanelle, France.
Mathilde Lea Chervet, Foods for Health Institute, Department of Food Science and Technology, University of California, Davis, Davis, CA, USA.
J Bruce German, Foods for Health Institute, Department of Food Science and Technology, University of California, Davis, Davis, CA, USA.
References
- 1.Oftedal OT. The mammary gland and its origin during synapsid evolution. J Mammary Gland Biol Neoplasia. 2002;7(3):225–52. [DOI] [PubMed] [Google Scholar]
- 2.Domellof M, Lonnerdal B, Dewey KG, Cohen RJ, Hernell O. Iron, zinc, and copper concentrations in breast milk are independent of maternal mineral status. Am J Clin Nutr. 2004;79(1):111–15. [DOI] [PubMed] [Google Scholar]
- 3.Feeley RM, Eitenmiller RR, Jones JB Jr, Barnhart H. Copper, iron, and zinc contents of human milk at early stages of lactation. Am J Clin Nutr. 1983;37(3):443–8. [DOI] [PubMed] [Google Scholar]
- 4.Giromini C, Cheli F, Rebucci R, Baldi A. Invited review: dairy proteins and bioactive peptides: modeling digestion and the intestinal barrier. J Dairy Sci. 2019;102(2):929–42. [DOI] [PubMed] [Google Scholar]
- 5.Gan J, Bornhorst GM, Henrick BM, German JB. Protein digestion of baby foods: study approaches and implications for infant health. Mol Nutr Food Res. 2018;62(1):10.1002/mnfr.201700231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Westrom B, Arevalo Sureda E, Pierzynowska K, Pierzynowski SG, Perez-Cano FJ. The immature gut barrier and its importance in establishing immunity in newborn mammals. Front Immunol. 2020;11:1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ford EL, Underwood MA, German JB. Helping mom help baby: nutrition-based support for the mother-infant dyad during lactation. Front Nutr. 2020;7:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.German JB, Dillard CJ. Composition, structure and absorption of milk lipids: a source of energy, fat-soluble nutrients and bioactive molecules. Crit Rev Food Sci Nutr. 2006;46(1):57–92. [DOI] [PubMed] [Google Scholar]
- 9.Neville MC, Anderson SM, McManaman JL, Badger TM, Bunik M, Contractor N, Crume T, Dabelea D, Donovan SM, Forman Net al. Lactation and neonatal nutrition: defining and refining the critical questions. J Mammary Gland Biol Neoplasia. 2012;17(2):167–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.German JB, Gillies LA, Smilowitz JT, Zivkovic AM, Watkins SM. Lipidomics and lipid profiling in metabolomics. Curr Opin Lipidol. 2007;18(1):20–4. [DOI] [PubMed] [Google Scholar]
- 11.Smilowitz JT, Lebrilla CB, Mills DA, German JB, Freeman SL. Breast milk oligosaccharides: structure-function relationships in the neonate. Annu Rev Nutr. 2014;34(1):143–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. [DOI] [PubMed] [Google Scholar]
- 13.Leroux C, Milenkovic D, Mobuchon L, Le Guillou S, Faulconnier Y, German B, Le Provost F. Nutritional regulation of mammary mirnome: implications for human studies. In: Patel V, Preedy VReditors. Handbook of nutrition, diet, and epigenetics. Springer; 2019. p.1–17. [Google Scholar]
- 14.Zempleni J, Aguilar-Lozano A, Sadri M, Sukreet S, Manca S, Wu D, Zhou F, Mutai E. Biological activities of extracellular vesicles and their cargos from bovine and human milk in humans and implications for infants. J Nutr. 2017;147(1):3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zempleni J, Baier SR, Howard KM, Cui J.. Gene regulation by dietary microRNAs. Can J Physiol Pharmacol. 2015;93(12):1097–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10(12):1957–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21(17):4663–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen TA, Jo MH, Choi YG, Park J, Kwon SC, Hohng S, Kim VN, Woo JS. Functional anatomy of the human microprocessor. Cell. 2015;161(6):1374–87. [DOI] [PubMed] [Google Scholar]
- 19.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim Set al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–19. [DOI] [PubMed] [Google Scholar]
- 20.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10(2):185–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303(5654):95–8. [DOI] [PubMed] [Google Scholar]
- 22.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawamata T, Tomari Y., Making R. Trends Biochem Sci. 2010;35(7):368–76. [DOI] [PubMed] [Google Scholar]
- 24.Friedman RC, Farh KKH, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, Gonzalez MA, Bernad A, Sanchez-Madrid F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weber JA, Baxter DH, Zhang SL, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56(11):1733–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu H, Fan GC. Extracellular/circulating microRNAs and their potential role in cardiovascular disease. Am J Cardiovasc Dis. 2011;1(2):138–49. [PMC free article] [PubMed] [Google Scholar]
- 28.Liu YC, Chen WL, Kung WH, Huang HD. Plant miRNAs found in human circulating system provide evidences of cross kingdom RNAi. BMC Genomics. 2017;18(S2):112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Hou D, Chen X, Li D, Zhu L, Zhang Y, Li J, Bian Z, Liang X, Cai Xet al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 2012;22(1):107–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chin AR, Fong MY, Somlo G, Wu J, Swiderski P, Wu X, Wang SE. Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res. 2016;26(2):217–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaucheret H, Chupeau Y. Ingested plant miRNAs regulate gene expression in animals. Cell Res. 2012;22(1):3–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang L, Shen J, Cheng J, Fan X. MicroRNA-21 regulates intestinal epithelial tight junction permeability. Cell Biochem Funct. 2015;33(4):235–40. [DOI] [PubMed] [Google Scholar]
- 33.Alsaweed M, Hepworth AR, Lefevre C, Hartmann PE, Geddes DT, Hassiotou F. Human milk microRNA and total RNA differ depending on milk fractionation. J Cell Biochem. 2015;116(10):2397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munch EM, Harris RA, Mohammad M, Benham AL, Pejerrey SM, Showalter L, Hu M, Shope CD, Maningat PD, Gunaratne PHet al. Transcriptome profiling of microRNA by Next-Gen deep sequencing reveals known and novel miRNA species in the lipid fraction of human breast milk. PLoS One. 2013;8(2):e50564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li R, Dudemaine PL, Zhao X, Lei C, Ibeagha-Awemu EM. Comparative analysis of the miRNome of bovine milk fat, whey and cells. PLoS One. 2016;11(4):e0154129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lago Novais D, Pawlowski K, Pires J, Mobuchon L, Bes S, Martin P, Leroux C. Milk fat globules as a source of mammary microRNA. In: ADSA/ASAS (American Society of Animal Science /American Dairy Science Association) Joint Annual Meeting. Salt Lake City, USA, 2016. [Google Scholar]
- 37.Pawlowski K, Faulconnier Y, Bes S, Boby C, Pires J. Leroux C. Comparison of miRNome from cow milk fat fraction and mammary gland tissue. In: 10th ISNH Symposium, Clermont-Ferrand, France, 2018.
- 38.Pawlowski K, Lago-Novais D, Bevilacqua C, Mobuchon L, Crapart N, Faulconnier Y, Boby C, Carvalho G, Martin P, Leroux C. Different miRNA contents between mammary epithelial cells and milk fat globules: a random or a targeted process?. Mol Biol Rep. 2020;47(10):8259–64. [DOI] [PubMed] [Google Scholar]
- 39.Pieters BC, Arntz OJ, Bennink MB, Broeren MG, van Caam AP, Koenders MI, van Lent PL, van den Berg WB, de Vries M, van der Kraan PMet al. Commercial cow milk contains physically stable extracellular vesicles expressing immunoregulatory TGF-beta. PLoS One. 2015;10(3):e0121123. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arntz OJ, Pieters BC, Oliveira MC, Broeren MG, Bennink MB, de Vries M, van Lent PL, Koenders MI, van den Berg WB, van der Kraan PMet al. Oral administration of bovine milk derived extracellular vesicles attenuates arthritis in two mouse models. Mol Nutr Food Res. 2015;59(9):1701–12. [DOI] [PubMed] [Google Scholar]
- 41.Zhou Q, Li M, Wang X, Li Q, Wang T, Zhu Q, Zhou X, Wang X, Gao X, Li X. Immune-related microRNAs are abundant in breast milk exosomes. Int J Biol Sci. 2012;8(1):118–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kusuma RJ, Manca S, Friemel T, Sukreet S, Nguyen C, Zempleni J. Human vascular endothelial cells transport foreign exosomes from cow's milk by endocytosis. Am J Physiol Cell Physiol. 2016;310(10):C800–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laubier J, Castille J, Le Guillou S, Le Provost F. No effect of an elevated miR-30b level in mouse milk on its level in pup tissues. RNA Biol. 2015;12(1):26–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Title AC, Denzler R, Stoffel M. Uptake and function studies of maternal milk-derived microRNAs. J Biol Chem. 2015;290(39):23680–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alsaweed M, Lai CT, Hartmann PE, Geddes DT, Kakulas F. Human milk cells contain numerous miRNAs that may change with milk removal and regulate multiple physiological processes. Int J Mol Sci. 2016;17(6):956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Izumi H, Kosaka N, Shimizu T, Sekine K, Ochiya T, Takase M. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. J Dairy Sci. 2012;95(9):4831–41. [DOI] [PubMed] [Google Scholar]
- 47.Zempleni J. Milk exosomes: beyond dietary microRNAs. Genes Nutr. 2017;12:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liao Y, Du X, Li J, Lonnerdal B. Human milk exosomes and their microRNAs survive digestion in vitro and are taken up by human intestinal cells. Mol Nutr Food Res. 2017;61(11). doi:10.1002/mnfr.201700082. [DOI] [PubMed] [Google Scholar]
- 49.Kahn S, Liao Y, Du X, Xu W, Li J, Lonnerdal B. Exosomal microRNAs in milk from mothers delivering preterm infants survive in vitro digestion and are taken up by human intestinal cells. Mol Nutr Food Res. 2018;62(11):e1701050. [DOI] [PubMed] [Google Scholar]
- 50.Melnik BC, Kakulas F, Geddes DT, Hartmann PE, John SM, Carrera-Bastos P, Cordain L, Schmitz G. Milk miRNAs: simple nutrients or systemic functional regulators?. Nutr Metab. 2016;13:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manca S, Upadhyaya B, Mutai E, Desaulniers AT, Cederberg RA, White BR, Zempleni J. Milk exosomes are bioavailable and distinct microRNA cargos have unique tissue distribution patterns. Sci Rep. 2018;8(1):11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alsaweed M, Lai CT, Hartmann PE, Geddes DT, Kakulas F. Human milk cells and lipids conserve numerous known and novel miRNAs, some of which are differentially expressed during lactation. PLoS One. 2016;11(4):e0152610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fu X, Dong B, Tian Y, Lefebvre P, Meng Z, Wang X, Pattou F, Han W, Wang X, Lou Fet al. MicroRNA-26a regulates insulin sensitivity and metabolism of glucose and lipids. J Clin Invest. 2015;125(6):2497–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. [DOI] [PubMed] [Google Scholar]
- 55.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18(10):505–16. [DOI] [PubMed] [Google Scholar]
- 56.Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14(9):400–9. [DOI] [PubMed] [Google Scholar]
- 57.Goedeke L, Vales-Lara FM, Fenstermaker M, Cirera-Salinas D, Chamorro-Jorganes A, Ramirez CM, Mattison JA, de Cabo R, Suarez Y, Fernandez-Hernando C. A regulatory role for microRNA 33* in controlling lipid metabolism gene expression. Mol Cell Biol. 2013;33(11):2339–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328(5985):1566–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jordan SD, Kruger M, Willmes DM, Redemann N, Wunderlich FT, Bronneke HS, Merkwirth C, Kashkar H, Olkkonen VM, Bottger Tet al. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat Cell Biol. 2011;13(4):434–46. [DOI] [PubMed] [Google Scholar]
- 60.Ramirez CM, Goedeke L, Rotllan N, Yoon JH, Cirera-Salinas D, Mattison JA, Suarez Y, de Cabo R, Gorospe M, Fernandez-Hernando C. MicroRNA 33 regulates glucose metabolism. Mol Cell Biol. 2013;33(15):2891–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun X, Lin J, Zhang Y, Kang S, Belkin N, Wara AK, Icli B, Hamburg NM, Li D, Feinberg MW. MicroRNA-181b improves glucose homeostasis and insulin sensitivity by regulating endothelial function in white adipose tissue. Circ Res. 2016;118(5):810–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen Y, Zhou M, Yan J, Gong Z, Xiao Y, Zhang C, Du P, Chen Y. miR-200b inhibits TNF-alpha-induced IL-8 secretion and tight junction disruption of intestinal epithelial cells in vitro. Am J Physiol Gastrointest Liver Physiol. 2017;312(2):G123–32. [DOI] [PubMed] [Google Scholar]
- 63.Yang Y, Ma Y, Shi C, Chen H, Zhang H, Chen N, Zhang P, Wang F, Yang J, Yang Jet al. Overexpression of miR-21 in patients with ulcerative colitis impairs intestinal epithelial barrier function through targeting the Rho GTPase RhoB. Biochem Biophys Res Commun. 2013;434(4):746–52. [DOI] [PubMed] [Google Scholar]
- 64.Wang X, Hao L, Bu HF, Scott AW, Tian K, Liu F, De Plaen IG, Liu Y, Mirkin CA, Tan XD. Spherical nucleic acid targeting microRNA-99b enhances intestinal MFG-E8 gene expression and restores enterocyte migration in lipopolysaccharide-induced septic mice. Sci Rep. 2016;6(1):31687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pillman KA, Phillips CA, Roslan S, Toubia J, Dredge BK, Bert AG, Lumb R, Neumann DP, Li X, Conn SJet al. miR-200/375 control epithelial plasticity-associated alternative splicing by repressing the RNA-binding protein Quaking. EMBO J. 2018;37(13):e99016). . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peck BC, Sincavage J, Feinstein S, Mah AT, Simmons JG, Lund PK, Sethupathy P. miR-30 family controls proliferation and differentiation of intestinal epithelial cell models by directing a broad gene expression program that includes SOX9 and the ubiquitin ligase pathway. J Biol Chem. 2016;291(31):15975–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O, Strehle M, Seiler A, Schumacher S, Nitsch R. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J. 2007;21(2):415–26. [DOI] [PubMed] [Google Scholar]
- 68.Zhang W, Thevapriya S, Kim PJ, Yu WP, Je HS, Tan EK, Zeng L. Amyloid precursor protein regulates neurogenesis by antagonizing miR-574-5p in the developing cerebral cortex. Nat Commun. 2014;5(1):3330. [DOI] [PubMed] [Google Scholar]
- 69.Lv X, Jiang H, Liu Y, Lei X, Jiao J. MicroRNA-15b promotes neurogenesis and inhibits neural progenitor proliferation by directly repressing TET3 during early neocortical development. EMBO Rep. 2014;15(12):1305–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Voloboueva LA, Sun X, Xu L, Ouyang YB, Giffard RG. Distinct effects of miR-210 reduction on neurogenesis: increased neuronal survival of inflammation but reduced proliferation associated with mitochondrial enhancement. J Neurosci. 2017;37(11):3072–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roshan R, Shridhar S, Sarangdhar MA, Banik A, Chawla M, Garg M, Singh VP, Pillai B. Brain-specific knockdown of miR-29 results in neuronal cell death and ataxia in mice. RNA. 2014;20(8):1287–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451(7182):1125–9. [DOI] [PubMed] [Google Scholar]
- 73.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103(33):12481–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xie W, Li M, Xu N, Lv Q, Huang N, He J, Zhang Y. MiR-181a regulates inflammation responses in monocytes and macrophages. PLoS One. 2013;8(3):e58639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci U S A. 2007;104(17):7080–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stittrich AB, Haftmann C, Sgouroudis E, Kuhl AA, Hegazy AN, Panse I, Riedel R, Flossdorf M, Dong J, Fuhrmann Fet al. The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nat Immunol. 2010;11(11):1057–62. [DOI] [PubMed] [Google Scholar]
- 77.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133(2):217–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, Hua M, Li N, Yao H, Cao X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nat Immunol. 2011;12(9):861–9. [DOI] [PubMed] [Google Scholar]
- 79.Vigorito E, Kohlhaas S, Lu D, Leyland R. miR-155: an ancient regulator of the immune system. Immunol Rev. 2013;253(1):146–57. [DOI] [PubMed] [Google Scholar]
- 80.Perez-Garcia A, Dongil P, Hurtado-Carneiro V, Blazquez E, Sanz C, Alvarez E. High-fat diet alters PAS kinase regulation by fasting and feeding in liver. J Nutr Biochem. 2018;57:14–25. [DOI] [PubMed] [Google Scholar]
- 81.Eckmann L, Jung HC, Schurer-Maly C, Panja A, Morzycka-Wroblewska E, Kagnoff MF. Differential cytokine expression by human intestinal epithelial cell lines: regulated expression of interleukin 8. Gastroenterology. 1993;105(6):1689–97. [DOI] [PubMed] [Google Scholar]
- 82.Mutlu M, Raza U, Saatci O, Eyupoglu E, Yurdusev E, Sahin O. miR-200c: a versatile watchdog in cancer progression, EMT, and drug resistance. J Mol Med. 2016;94(6):629–44. [DOI] [PubMed] [Google Scholar]
- 83.Williams MR, Stedtfeld RD, Tiedje JM, Hashsham SA. MicroRNAs-based inter-domain communication between the host and members of the gut microbiome. Front Microbiol. 2017;8:1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu S, da Cunha AP, Rezende RM, Cialic R, Wei Z, Bry L, Comstock LE, Gandhi R, Weiner HL. The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe. 2016;19(1):32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3(4):213–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hansen KF, Sakamoto K, Aten S, Snider KH, Loeser J, Hesse AM, Page CE, Pelz C, Arthur JS, Impey Set al. Targeted deletion of miR-132/-212 impairs memory and alters the hippocampal transcriptome. Learning Memory. 2016;23(2):61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Goldman AS. The immune system in human milk and the developing infant. Breastfeed Med. 2007;2(4):195–204. [DOI] [PubMed] [Google Scholar]
- 88.Newburg DS, Walker WA. Protection of the neonate by the innate immune system of developing gut and of human milk. Pediatr Res. 2007;61(1):2–8. [DOI] [PubMed] [Google Scholar]
- 89.Zivkovic AM, German JB, Lebrilla CB, Mills DA. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4653–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kosaka N, Izumi H, Sekine K, Ochiya T. microRNA as a new immune-regulatory agent in breast milk. Silence. 2010;1(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Na RS, GX E, Sun W, Sun XW, Qiu XY, Chen LP, Huang YF. Expressional analysis of immune-related miRNAs in breast milk. Genet Mol Res. 2015;14(3):11371–6. [DOI] [PubMed] [Google Scholar]
- 92.Perri M, Lucente M, Cannataro R, De Luca IF, Gallelli L, Moro G, De Sarro G, Caroleo MC, Cione E. Variation in immune-related microRNAs profile in human milk amongst lactating women. MicroRNA. 2018;7(2):107–14. [DOI] [PubMed] [Google Scholar]
- 93.Baumjohann D, Ansel KM. MicroRNA-mediated regulation of T helper cell differentiation and plasticity. Nat Rev Immunol. 2013;13(9):666–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131(1):146–59. [DOI] [PubMed] [Google Scholar]
- 95.Hassanpour M, Rezaie J, Nouri M, Panahi Y. The role of extracellular vesicles in COVID-19 virus infection. Infect Genet Evol. 2020;85:104422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Trobaugh DW, Klimstra WB. MicroRNA regulation of RNA virus replication and pathogenesis. Trends Mol Med. 2017;23(1):80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ma YS, Yu F, Zhong XM, Lu GX, Cong XL, Xue SB, Xie WT, Hou LK, Pang LJ, Wu Wet al. miR-30 family reduction maintains self-renewal and promotes tumorigenesis in NSCLC-initiating cells by targeting oncogene TM4SF1. Mol Ther. 2018;26(12):2751–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zheng C, Zheng Z, Sun J, Zhang Y, Wei C, Ke X, Liu Y, Deng L, Wang H. MiR-16-5p mediates a positive feedback loop in EV71-induced apoptosis and suppresses virus replication. Sci Rep. 2017;7(1):16422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Duan X, Liu X, Li W, Holmes JA, Kruger AJ, Yang C, Li Y, Xu M, Ye H, Li Set al. MicroRNA-130a downregulates HCV replication through an atg5-dependent autophagy pathway. Cells. 2019;8(4):338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang S, Li J, Li J, Yang Y, Kang X, Li Y, Wu X, Zhu Q, Zhou Y, Hu Y. Up-regulation of microRNA-203 in influenza A virus infection inhibits viral replication by targeting DR1. Sci Rep. 2018;8(1):6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Demirci MDS, Adan A. Computational analysis of microRNA-mediated interactions in SARS-CoV-2 infection. PeerJ. 2020;8:e9369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Melnik BC, Schmitz G. Milk's role as an epigenetic regulator in health and disease. Diseases. 2017;5(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering Cet al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104(40):15805–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14(5):872–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ashworth J, Wurtmann EJ, Baliga NS. Reverse engineering systems models of regulation: discovery, prediction and mechanisms. Curr Opin Biotechnol. 2012;23(4):598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




