Abstract
With the increasing maternal age and the use of assisted reproductive technology in various countries worldwide, the influence of epigenetic modification on embryonic development is increasingly notable and prominent. Epigenetic modification disorders caused by various nutritional imbalance would cause embryonic development abnormalities and even have an indelible impact on health in adulthood. In this scoping review, we summarize the main epigenetic modifications in mammals and the synergies among different epigenetic modifications, especially DNA methylation, histone acetylation, and histone methylation. We performed an in-depth analysis of the regulation of various epigenetic modifications on mammals from zygote formation to cleavage stage and blastocyst stage, and reviewed the modifications of key sites and their potential molecular mechanisms. In addition, we discuss the effects of nutrition (protein, lipids, and one-carbon metabolism) on epigenetic modification in embryos and emphasize the importance of various nutrients in embryonic development and epigenetics during pregnancy. Failures in epigenetic regulation have been implicated in mammalian and human early embryo loss and disease. With the use of reproductive technologies, it is becoming even more important to establish developmentally competent embryos. Therefore, it is essential to evaluate the extent to which embryos are sensitive to these epigenetic modifications and nutrition status. Understanding the epigenetic regulation of early embryo development will help us make better use of reproductive technologies and nutrition regulation to improve reproductive health in mammals.
Keywords: early embryo, epigenetics, DNA methylation, histone modification, imprinting, nutrition, protein, fat, blastocyst, one-carbon metabolism
Introduction
Assisted reproductive technology (ART) has been increasingly used in animals and humans to help infertility patients achieve higher pregnancy rates and birth rates (1). However, studies have shown that some rare imprinted diseases are likely related to the use of ART (2). The incidence of Beckwith-Wiedemann syndrome and macrosomia in children born via ART has increased, and abnormal epigenetic modifications of many imprinted genes, such as insulin-like growth factor 2 (Igf2) and H19, have been detected in patients (3, 4). There are also various abnormal epigenetic modifications in the early development of mammalian cloned embryos that lead to an extremely low development efficiency of cloned embryos and birth rate of cloned animals. However, by appropriately regulating epigenetic modification, early embryos can return to a normal developmental track (5–7).
In addition to affecting embryonic developmental efficiency, abnormal epigenetic modifications during embryonic development continue to affect the growth and health of infants after birth, resulting in a variety of adult diseases, such as obesity, diabetes, cardiovascular disease, neurological diseases, and behavioral diseases (8, 9). In pregnant women suffering famine during the Dutch War, changes in the pattern of fetal DNA methylation were common, and the risk of early-onset coronary artery disease was significantly increased in adult offspring. DNA methylation patterns were different among fetuses at different stages of pregnancy, and males and females responded differently to starvation (10, 11). In the umbilical stem cells of fetuses with intrauterine growth restriction, DNA methylation was altered in the promoter region of the hepatocyte nuclear factor 4 gene, a gene that is involved in the development of juvenile diabetes mellitus (12). These studies indicated that germ cells and embryonic development are accompanied by extensive epigenetic reprogramming and that nutrition plays an important role in the epigenetic modification of embryos (13, 14).
With increasing maternal age and social pressure, reproductive diseases have become increasingly prominent. Epigenetic modification–related reproductive diseases have become a research hotspot in reproductive medicine. In recent years, advances in sequencing methods have improved our understanding of the critical developmental processes in early embryos. However, at the same time, some viewpoints that contradict traditional cognition have emerged, bringing new problems and challenges in the study of epigenetic modification during early embryonic development. We repeatedly made a thorough literature search in PubMed and Web of Science, with a limitation for articles written in the English language. Search terms used were embryo, epigenetic, DNA methylation, histone modification, and nutrient. In addition, reference sections of all relevant studies or reviews were manually searched for more information. In this review, we introduce the main epigenetic modifications in mammals, summarize the latest knowledge on epigenetic modifications at early stages of embryonic development in mammals and humans, and discuss nutritional regulation that may cause epigenetic modifications in embryos. Our intention is to better understand the molecular mechanism of epigenetics by exploring various epigenetic modification changes and their nutritional regulation mechanisms during embryonic development and then apply them in reproductive medicine. This review will be helpful for nutrition researchers to better understand where and when nutritional status is potentially problematic in epigenetic regulation, which can provide new ideas for the use of ART and improve reproductive health in mammals and humans.
Current Status of Knowledge
Part 1: Epigenetic modifications
Epigenetic modifications in mammals mainly include histone modification, DNA methylation, and noncoding RNA (ncRNA) (15). Abnormal epigenetic modification can lead to a variety of diseases, which can be divided into 2 major categories: 1) the abnormal epigenetic modification of a specific gene during the development and reprogramming process, also known as epigenetic mutation, and 2) the mutations in protein-coding genes associated with epigenetic modifications, such as mutations in DNA methyltransferase (Dnmt) genes or in methyl-CpG binding protein (MeCP) genes (16). It is generally believed that the embryonic origin of adult diseases occurs mainly through histone modification and DNA methylation (17).
Histone modification
In eukaryotes, histones undergo a variety of post-transcriptional modifications, including acetylation, methylation, phosphorylation, ubiquitination, ubiquitin-like modification, and crotonylation (Figure 1). Among various histone modifications, acetylation and methylation are the most common and important (18).
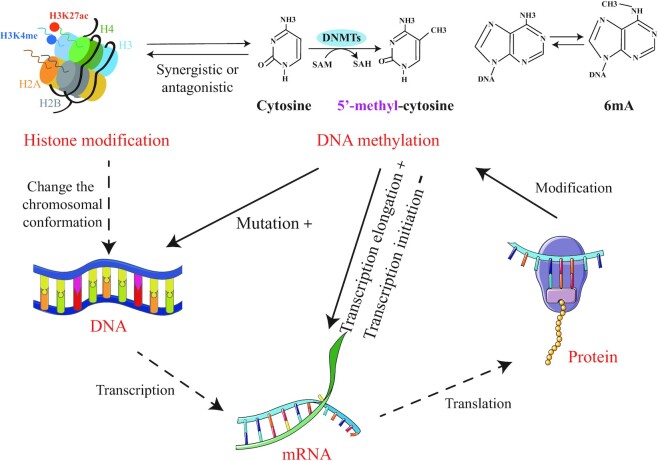
FIGURE 1.
Influence of DNA methylation and histone modification in mammals. Histone acetylation and methylation are the most common and important processes to regulate gene expression, which may change the chromosomal conformation. DNA methylation includes 5mC and 6mA, which increase the mutation of DNA, enhance transcription elongation, and inhibit transcription initiation. In addition, mRNA translates into protein that affects DNA methylation. DNMT, DNA methyltransferase; SAH, S-adenosylhomocysteine; SAM, S-adenosylmethionine.
Histone acetylation is related to gene activation, while deacetylation is related to gene silencing (Figure 1). The degree of histone acetylation is jointly determined by histone acetyltransferase (HAT) and histone deacetylase (HDAC) (19). Unlike histone acetylation, histone methylation promotes gene expression as well as inhibits gene expression, mainly depending on the residues undergoing methylation modification and the regions where the modified gene is located (20). For example, H3K8 methylation has a significant inhibitory effect on expression. However, the methylation of other residues, such as H3K4 and H3K36, is associated with active transcription (21, 22). As an important means to regulate gene expression, histone modification can affect the expression and function of downstream proteins, thereby determining the status of cells and affecting embryogenesis and development. In recent years, it has been reported that abnormal histone modification affects embryonic development. Studies have shown that the probability of fetal death from cell proliferation defects and nerve development retardation before 10.5 d of embryonic development is high in early embryos lacking HDAC1 (23). With knockout of different members of the Hat family, the embryonic development of mice is severely impaired, manifesting as a lack of specific embryonic structures derived from the chordamesoderm and paraxial mesoderm, which results in severe cranial neural tube closure defects, heart development defects, etc. Histone acetyltransferase GCN5 (GCN5) is the first histone acetylase identified. Gcn5 knockout mouse embryos can survive normally until 7.5 d, but embryo development is severely impeded in 7.5–8.5 d, and most embryos die at 10.5 d (24).
DNA methylation
DNA methylation is a very common genome modification and plays an important role in biological processes, such as regulating genome functions, maintaining chromatin structure stability, and genetic imprinting (Figure 1).
The relation between DNA methylation and certain reproductive diseases has gradually drawn increasing attention. Under normal circumstances, the CpG sites outside CpG islands in the normal genome are often methylated, thereby inhibiting gene expression. When CpG sites in CpG islands are in an unmethylated state, gene expression will be promoted. However, during tissue differentiation or early embryonic development, a small portion of CpG (6%) is methylated (25). Numerous studies have confirmed that DNA in mature sperm and oocytes is highly methylated, with almost no transcriptional activity. Hany et al. (26) proposed that DNA methylation mainly inhibits gene expression, inactivates the female X chromosome, regulates imprinted gene expression, silences transposable elements, and regulates and expresses tissue-specific genes during embryonic development. After the inhibition of Dnmt1 gene expression, the early embryonic development rate in mice significantly decreases; after Dnmt gene knockout, early embryonic development and the formation and differentiation of multiple organs are severely affected, even leading to early embryonic death (27). In human ART, the expression levels of DNMT in abnormal embryos and frozen embryos were lower than those in fresh embryos, indicating that the expression levels of DNMT in embryos may be related to abnormal embryo development (28).
Studies have shown that nearly 150 imprinted genes have been found in mice, and half of them have functions in humans (29). Most imprinted genes aggregate into clusters, which are controlled by imprinted control elements. Gene imprinting was first recognized during spermatogenesis; during zygogenesis, germ cell imprinting was erased and subsequently reconstructed during development based on characteristics such as fetal sex (30). After fertilization, the imprinted genes avoid whole-genome DNA demethylation during the reprogramming process, retain their methylation or demethylation status in the early development process, and further transfer to somatic cells (31). The low efficiency of somatic cell nuclear transfer (SCNT) and the abnormal development of animals after SCNT are both associated with the abnormal expression of imprinted genes. For example, the commonly seen “macrojugular disease” in cloned animals is closely related to the abnormal expression of important imprinted genes such as Igf2 and H19. Abnormal X chromosome inactivation is also an important cause of abnormalities in animals after SCNT. Studies have found that in the late stage of gastrula in most SCNT embryos, the 2 X chromosomes are in an activated status without proper random inactivation, leading to embryonic death (32). Knockout of the X inactive specific transcript (Xist) in mice increased the SCNT efficiency by 8–9 times (33), indicating the importance of X chromosome inactivation in the normal development of embryos after SCNT.
ncRNAs
ncRNAs can be divided into long ncRNAs (lncRNAs) and small ncRNAs, according to their size. Currently, lncRNAs, which are well studied, have important functions in regulating complex cell networks and are involved in important physiological processes such as chromosome formation and mRNA splicing.
Recent studies have found that lncRNAs are closely related to the occurrence and development of germ cells and embryos and that abnormal lncRNA functions or mutations can lead to a series of complex diseases. However, most studies on lncRNAs remain at the macroscopic analysis stage and are mainly focused on sperm research. Sperm cells contain many special RNAs, such as small interfering RNAs (siRNAs) and microRNAs (miRNAs) (34). miRNAs mainly bind to mRNA sequences to block translation, thereby inhibiting DNA expression. Maternal miRNAs are abundant in early embryos, and the expression of miRNAs is significantly increased at the 2-cell stage of mouse embryos (35). If Dicer, an important enzyme in the process of RNA interference (RNAi), is lacking during embryonic development in mice, miRNA will not function, thereby reducing the number of mouse embryonic stem cells (ESCs). This result indicates that miRNA can maintain the pluripotency of stem cells and participate in embryonic development (36). A pseudochromosome is a structure unique to haploid sperm cells. lncRNA, RNA connexin, Piwi protein interacting RNA (piRNA), mRNA, and other proteins involved in RNA regulation are co-assembled in this structure (37). During the process of epigenetic modification, RNA in sperm plays a substantial role. Due to the lack of paternal RNA in parthenogenetic mouse embryos, embryo development is hindered, and the birth mortality rate is high. Melissa et al. (38) studied the early embryonic development in parthenogenetic mice and found that parthenogenetic mice had a mortality rate as high as 80%, while the success rate was as low as 0.3%. Further studies analyzed the mechanism of lncRNAs and found that they do not function alone but require interactions with other factors, such as mRNA. Liang et al. (39) found that expression in mature sperm required specific synergistic actions between lncRNA and mRNA; otherwise, spermatids had abnormalities. However, there have been few studies on lncRNAs in human embryos, and their mechanism of action remains unclear and needs further exploration.
Interactions between epigenetic modifications
Evidence has shown that there is rare regulation by 1 epigenetic modification alone in the body and most regulation involves synergy among a variety of epigenetic modifications, in which DNA methylation, histone acetylation, and histone methylation are closely related (40).
DNA methylation and histone acetylation
Gene promoter region DNA methylation and histone deacetylation synergistically inhibit gene transcription (41). Normally, in the case of low-density CpG methylation in the promoter region, the inhibition of transcription mainly depends on the histone deacetylation pathway, while in the case of high-density CpG methylation, the inhibition of transcription mainly plays a role in DNA methylation (42) (Figure 2 ).
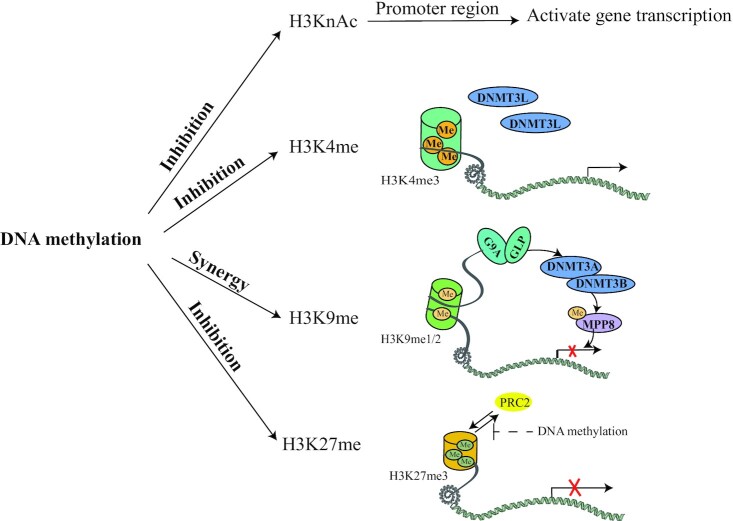
FIGURE 2.
Interactions between DNA methylation and histone modifications. Gene promoter region DNA methylation and histone deacetylation synergistically inhibit gene transcription. DNA methylation and H3K4 methylation inhibit each other. Once H3K4 is occupied by methylated groups, the methylated complex subunit DNMT3L cannot find attachment points, leading to DNA methylation failure. H3K9 dimethylation and trimethylation and DNA methylation have a synergistic effect on gene silencing mechanisms. The histone methyltransferases G9A and GLP catalyze the methylation and dimethylation of H3K9; at the same time, they can directly recruit DNMT3A and DNMT3B. PRC2 generates H3K27me3 and promotes PRC2 binding, which inhibits gene transcription. However, DNA methylation inhibits the binding of PRC2 to DNA. DNMT, DNA methyltransferase; GLP, euchromatic histone lysine methyltransferase 1; G9A, euchromatic histone lysine methyltransferase 2; MPP8, M-phase phosphoprotein 8; PRC2, Polycomb repressive complex 2.
DNA methylation and histone methylation
DNA methylation in mammals is closely related to H3K9 methylation (43–45). H3K9 methylation is catalyzed by 1 of the 5 histone lysine methyltransferase SET/SUV39 (SUV39) protein family members [SUV38H1, SUV38H2, euchromatic histone lysine methyltransferase 2 (G9A), euchromatic histone lysine methyltransferase 1 (GLP), and SET domain bifurcated histone lysine methyltransferase 1 (SETDB1)]. SUV38H1 and SUV38H2 mainly catalyze the trimethylation of H3K9, primarily acting in centromeres and the surrounding heterochromatin (46, 47). The histone methyltransferases G9A and GLP catalyze the methylation and dimethylation of H3K9 and are mainly related to gene silencing in autosomes (48). SETDB1 is an H3K9 trimethyltransferase that is responsible for the methylation of endogenous retroviruses (ERVs) and X chromosome inactivation (49, 50). H3K9 dimethylation and trimethylation and DNA methylation have a synergistic effect on gene silencing mechanisms (51), and they bind to heterochromatin protein HP1 (52, 53). Mutation studies in mouse ESCs demonstrated that the level of DNA methylation in major satellites was decreased in Suv39H1−/− Suv39H2−/− mice (54, 55). G9a knockout in mouse ESCs caused DNA hypomethylation at specific sites in the genome (56, 57). Setdb1 knockout resulted in a small loss of DNA methylation in some gene loci, including imprinted genes (50, 58). No change in H3K9 methylation was observed in Dnmt1−/− Dnmt3a −/− Dnmt3b −/− knockout mouse ESCs (59, 60). However, in human cancer cells, H3K9 methylation has been shown to be dependent on DNA methylation (61–63).
H3K27 methylation also interacts with DNA methylation. Polycomb repressive complex 2 (PRC2), a transcription repression complex, generates H3K27me3, which further promotes PRC2 binding and inhibits gene transcription. However, DNA methylation inhibits the binding of PRC2 to DNA (Figure 2). In addition, DNA methylation and H3K4 methylation also inhibit each other. Once H3K4 is occupied by methylated groups (H3K4 becomes H3K4me3), the methylated complex subunit DNMT3L cannot find attachment points, leading to DNA methylation failure.
With regard to the interaction between DNA methylation and histone modification, it is generally believed that DNA methylation and histone modification can trigger each other and that these processes are interlinked by MeCP. MeCP can bind to CpG dinucleotide, thus promoting heterochromatin formation and initiating gene silencing by recruiting histone deacetylase and histone methyltransferase (16). Histone-binding proteins or proteins that modify histones can also recruit DNMT for DNA methylation, resulting in the silencing of gene expression (64). For example, G9A and GLP can directly recruit DNMT3A and DNMT3B, causing de novo DNA methylation (65, 66).
Part 2: Epigenetic modification during embryonic development
After gamete fertilization, newly formed zygotes encounter a series of important challenges regarding cell fate and germline maintenance. First, the epigenetic modifications carried by sperm and oocytes will be reprogrammed to complete genome expression in zygotes, allowing them to enter into the developmental stage. In addition, this reprogramming process can prevent parents from transmitting inappropriate or abnormal epigenetic modifications to their offspring. Then, zygotes divide into many small cells (i.e., blastomeres). Cleavage is the first stage of embryonic development. The number of blastomeres increases with the increase of cleavage times. Afterwards, the blastomeres undergo the first cell lineage differentiation, gradually becoming the inner cell mass (ICM) or trophectoderm (TE) (67). Many epigenetic modifications jointly participate in maintaining and completing these stages (Table 1).
TABLE 1.
Epigenetic modification during embryonic development1
| Name | Functions | Influences | References |
|---|---|---|---|
| Zygote formation | |||
| TET1 | Bound to the unmethylated CpG-rich region, impeded DNMT approach to DNA | Decreased pyrimidine activity, abnormal morphology, unable to form blastocysts | (68–72) |
| TET2 | Oxidized 5hmC to 5fC and 5caC | Hematopoietic malignancies | (73, 74) |
| TET3 | Oxidized 5mC to 5hmC | Embryo lethal | (75, 76) |
| H3K27me | Changed the transcription around centromeres | Chromosome separation defects, early embryonic arrest | (77) |
| AID | Changed the deamination of cytosine, 5mC and 5hmC | Reduced demethylation level in genome | (78, 79) |
| MLL3/4 | Enhanced H3K27ac and H3K4me, activate paternal transcription regulation | — | (80, 81) |
| Cleavage stage | |||
| H2A.Zac | Activated embryonic genome | Caused defects in proliferation and differentiation of embryonic ICM, leading to the death of embryos before implantation | (82) |
| H3K9me | Transcriptional activation | Abnormal developmental and epigenetic reprogramming | (83–85) |
| KDM4A | Removed histone H3K9me2/me3, maintaining the transcription ability | Blastocyst formation | (86–89) |
| H3K27me | Regulated mammalian imprinted genes | Loss of the imprint | (90) |
| H3K4me | Caused abnormalities in embryonic genome activation | Low developmental efficiency during late stage | (91) |
| WDR82 | Reduced H3K4me3 at the transcription initiation site | Decreased blastocyst cell numbers, embryonic cell apoptosis, embryonic retardation | (92) |
| CHD1 | Caused H3K4me3 and promote mRNA transcription | Developmental disorders of the ectoderm, implanted embryos death | (93) |
| H3K36me | Affected gene transcription through the interaction between CTD and RNA polymerase II | Regulated cell proliferation | (94) |
| Blastocyst stage | |||
| DNMT1 | Maintains DNA methylation | Embryonic death | (95) |
| DNMT2 | — | Lost RNA methylase activity | (96, 97) |
| DNMT3A | Major de novo methylases | Lost imprints in both female and male germ cells, died immediately after birth | (98) |
| DNMT3B | Major de novo methylases | Developmental defects and demethylation of the satellite repeat sequence | (99) |
| DNMT3L | Promoted the catalytic activity of DNMT3A and DNMT3B | Male mice were sterile, female offspring could not develop to term | (99–102) |
| H3K27me | Transcriptional inhibition promoter of CpG islands and genomic regions | — | (103–105) |
| H3K9me | Effective inhibition of a few species-specific genes (by silencing gene promoters or enhancers) | — | (106, 107) |
| H3K4m2 | Regulated transcription | — | (108–110) |
| SETD1A | Major H3K4 methyltransferase in post-implanted embryos | Played an important role in lineage differentiation, an essential factor in gastrulation | (111) |
| MLL2 | Major H3K4 methyltransferase in pre-implantation embryos | Embryonic development defects, arrested at mid-pregnancy | (111) |
1AID, activation-induced deaminase; CHD1, chromodomain helicase DNA binding protein 1; DNMT, DNA methyltransferase; ICM, inner cell mass; KDM4A, lysine demethylase 4A; MLL, lysine methyltransferase 2A; SETD1A, SET domain containing 1A; TET, ten-eleven translocation methylcytosine dioxygenase; WDR82, WD repeat domain 82; 5mC, 5-methylcytosine; 5hmC, 5‐hydroxymethylcytosine.
Zygote formation
During the process of fertilization, female and male gametes express their epigenetic specificity, and the formed zygote is a unique cell with the ability to differentiate from a cell into any tissue. To achieve pluripotency in embryos, the germ cell genome is highly modified by transcription factors (Table 1).
DNA demethylation
After fertilization, paternal and maternal genomic DNA undergo large-scale demethylation. Ten-eleven translocation methylcytosine dioxygenase (TET) proteins are a new family of DNA modifiers involved in DNA demethylation and are divided into TET1, TET2, and TET3. TET1 binds to the unmethylated CpG-rich region mainly through the CXXC region (a protein domain of DNMTs), impedes DNMT1’s approach to DNA, and is highly expressed in blastocysts and ESCs. Tet1 knockout embryos showed abnormal morphology, decreased pyrimidine activity, and were unable to form normal blastocysts. In tetraploid complementation experiments, mouse embryos with Tet1 gene knockout were pluripotent and developed to term. These mice were able to survive and reproduce, producing smaller fetuses (68–72). TET2 is highly expressed in almost all tissues, especially in hematopoietic cells and stem cells. Tet2 gene knockout embryos had normal morphology and normal transcription factor AP activity, but reduced 5‐hydroxymethylcytosine (5hmC) level. Tet2 mutations can lead to hematopoietic malignancies (73, 74). TET3 is highly expressed in oocytes and sperm (expressed in neural stem cells, lungs, and pancreas). After intra-sperm injection of Tet3 siRNA, 5-methylcytosine (5mC) level was increased and 5hmC level was decreased. After the deletion of maternal Tet3, the level of 5mC in the paternal genome was reduced and the embryo had a higher mortality (75, 76). All of this evidence points to the importance of demethylation during embryonic development.
In addition, activation-induced deaminase (AID) is also involved in the active DNA demethylation (78). AID can change the deamination of cytosine into uracil, the deamination of 5mC into thymine, and the deamination of 5hmC into 5-hydroxymethyl uracil. Under normal conditions, early primordial germ cells (PGCs) and ESCs have the same methylation level compared with somatic cells. When AID is lacking in PGCs, the demethylation level in the genome is reduced by 20%. Meanwhile, 5mC hydroxymethylated enzymes TET1 and TET2 are also expressed in PGCs (79), indicating that 5mC may initiate demethylation through different mechanisms.
Although there have been some studies on DNA demethylation in the parental genome after fertilization, researchers have differentiated the parental genome and demethylation mechanisms. In fact, DNA demethylation in the parental genome is extremely complex, and it is likely to involve a combination of active and passive approaches, affecting the maternal and paternal genome demethylation levels to varying degrees at the whole-genome level. What are the effects of dynamic changes in the level of DNA methylation in early zygotes on cellular fate? What are the interactions between DNA demethylation and other epigenetic modifications? These questions remain unanswered.
Histone modification
The late-stage transcriptional regulation of zygotes is asymmetrical in paternal and maternal pronuclei (112). Studies have shown that the initiation of transcriptional regulation in the paternal pronucleus occurs several hours earlier than that in the maternal pronucleus (113). However, how this asymmetric transcriptional regulation is established has not been explained for decades. Studies have shown that activated histone markers, including the acetylation of H3.3 and H3K27, are preferentially deposited in the late paternal pronucleus (114). H3K4 methylation is the most representative histone activation marker and exists in the paternal pronucleus in the middle and late stages of zygotes (115). Therefore, it is speculated that paternal chromatin is more susceptible to transcriptional activation than maternal chromatin, due to less inhibition of histone markers such as H3K9 and H3K27 methylation (116).
Studies have further shown that, in zygotes, lysine methyltransferase 2C/2B (MLL3/4) can act as an enhancer, participate in histone H3K27 acetylation and H3.3-K4 methylation, and activate paternal transcription regulation (80, 81). However, early studies have also reported that the transcription in the paternal pronucleus is independent of enhancers (117). Therefore, the function of enhancers during the zygotic stage is still unclear, and more evidence is needed to demonstrate whether enhancers participate in transcription in zygotes.
Cleavage stage
After fusion of the paternal and maternal pronuclei in zygotes, the cytoplasm is rearranged, and the zygote undergoes cell division. Studies have found that the cleavage time of embryos with different qualities is inconsistent. Early cleavage can be used as a marker to determine late embryonic development potential. It can be used to identify the quality of human in vitro fertilization embryos. Embryos with early cleavage are more likely to develop to the blastocyst stage than those that undergo late cleavage (118, 119).
Histone modification
The acetylation of histones in specific regions of zygotic genes (such as H2A, H3, and H4) is very important for embryonic development. Among the core histones, H2A has the most variation and H2A.Z is the most studied. H2A.Z is the first histone variant found to be closely related to mammalian development. Although its sequence is highly conserved, it plays different roles in different organisms. In the mouse embryonic 2-cell stage, the H2A.Z acetylation level is decreased. The main activation of the embryonic genome occurs during this stage, and the lack of H2A.Z in mice causes defects in the proliferation and differentiation of the embryonic ICM, leading to the death of embryos before implantation (82). This evidence indicated that activity markers are related to the transcriptional activation of the embryonic genome.
The acetylation of different residues in H3 varies at different stages of embryonic development. The abundant expression of H3K64ac and H3K56ac can be detected during embryonic cleavage, but H3K122ac is only weakly expressed, suggesting that different histone modifications play different roles in embryo cleavage (120). In addition, the methylation of various histone residues in H3 also plays a critical role in the transcriptional activation of early embryos. During the 2-cell stage of embryonic development, H3K9me3 modification is abundant in the maternal chromosome, but this modification on the parental chromosome is removed. After entering the 4-cell stage of embryonic development, the histone modification asymmetry in the paternal and maternal genomes gradually disappears, and the modification levels of H3K27me, H3K4me, and H3K9me in each blastomere are comparable. H3K9me3 and H3K27me3 are considered as inhibitory markers of transcriptional activity, while H3K4me3 and H3K36me3 are considered as markers of transcriptional activation.
Lysine demethylase 4A (KDM4A) can remove histone H3K9me2/me3 (86–88) to prevent the accumulation of H3K9me3 on activated promoters, thereby maintaining the transcription ability (89). KDM4B is a key factor leading to 2-cell arrest in cloned embryos in mice (121). The injection of Kdm4a and Kdm4d mRNA significantly increased the blastocyst formation rate of cloned embryos (122, 123). Kdm4a not only serves as the maternal effector gene to ensure the normal development of embryos before implantation during the early stage but also maintains a suitable uterine environment conducive to embryo development and implantation by regulating cytokines such as colony stimulating factor 2 receptor subunit alpha (Csf2ra) (124).
The gene silencing of WD repeat domain 82 [Wdr82, one of the subunits of the homocysteine S-methyltransferase 1 (HMT) complex] in early-stage mouse embryos reduces H3K4me3 at the transcription initiation site of the gene for the transcription factor POU class 5 homeobox 1 (POU5F1), and this reduction is accompanied by a decrease in the number of blastocyst cells, embryonic cell apoptosis, and embryonic retardation (92). Chromodomain helicase DNA binding protein 1 (CHD1) is a member of the ATPase-dependent chromatin-remodeling factor family. It causes methylation of H3K4 and promotes mRNA transcription, which is essential for maintaining the pluripotency of mouse ESCs (125). In the process of ectoderm development, CHD1 can promote gene transcription, allowing the rapid development of the ectoderm. After the CHD1 gene is knocked out, mouse embryos stop developing on embryonic day 6.5 (E6.5) due to developmental disorders of the ectoderm, ultimately leading to the death of implanted embryos (93). Studies have shown that during early mouse embryonic development, inhibition of the Chd1 gene transcription factors POU5F1, nanog homeobox (NANOG), and caudal type homeobox 2 (CDX2) causes a sharp decrease in gene activation before embryo implantation, while the expression of upstream binding transcription factor like 1 (Hmgp1) and kruppel like factor 5 (Klf5), which regulate these transcription factors, are significantly inhibited until the blastocyst stage, and the inhibition of Klf5 expression is reversed in the morula stage. This result suggests that during the preimplantation gene activation stage, CHD1 expression is mainly influenced by the transcription factors HMGP1 and KLF5, thus controlling the development and survival of mouse embryos (93). Studies have also suggested that CHD1 affects gene expression by changing the chromosome structure. These studies all indicate that CHD1 is essential for the normal development of early mouse embryos.
Blastocyst stage
Before the implantation of mammalian embryos into the uterus, zygotes undergo multiple cleavages, and with cell polarization and densification, cells undergo asymmetric division and then differentiate into 2 different cell types: TE and ICM. Then, embryos continue to develop into a hollow spheroid blastocyst. Although the time of the first cell lineage differentiation is different due to species differences, all mammals experience this process. The role of epigenetic regulation in this differentiation is extremely complex and thus far unknown. Studies of its regulatory mechanism mostly use transgenic mouse models to identify the key regulatory factors and the in vivo localization and dynamic changes remain poorly understood.
DNA methylation
When embryos develop into blastocysts, embryonic cells undergo DNA remethylation processes, and the methylation level gradually recovers (126). Approximately 80% of the embryonic genome has established DNA methylation (94). In the process of DNA remethylation, the methylating enzyme DNMT plays a critical role. DNMT1 is an enzyme that maintains DNA methylation. In the process of DNA replication, DNA strands modified by methylation are recognized to methylate the CpG sites of methylation. During the process of gastrula formation in mice, Dnmt1 knockout leads to an extensive reduction in DNA methylation levels until embryonic death (95). DNMT2 is highly expressed in the heart and testis. Dnmt2 knockout mice had normal reproductive ability and showed a normal genomic methylation pattern, but RNA methylase activity was lost (96, 97). DNMT3A, DNMT3B, and DNMT3L are the major de novo methylases and are mainly involved in remethylation after active and passive demethylation of the parental and maternal genomes. Part of Dnmt3a knockout embryos continue to develop, but although the embryo develops to term, it dies immediately after birth due to losing imprints in both female and male germ cells (98). DNMT3B is significantly expressed at the blastocyst stage, and the knockout embryo shows many developmental defects and demethylation of the satellite repeat sequence, which occurs between embryonic days 13.5 and 16.5 and leads to the death of the embryo. DNMT3L has no methyltransferase activity, but it is an important regulatory factor, which can promote the catalytic activity of DNMT3A and DNMT3B and express specifically in the embryonic development stage. Dnmt3l knockout mice could survive, but male mice were sterile because their germ cells showed sporadic nuclear duplication and A particle reactivation in the cistern, as well as severe meiosis defects. Female offspring with this gene knocked out do not develop to term [death of the developing embryo is caused by neural tube defects, in part due to co-expression of the missing maternal imprinting gene (99–102)]. Ubiquitin like with PHD and ring finger domains 1 (NP95) can recruit DNMT1 into the replication site and is highly expressed in proliferating cells. After this gene is knocked out, the embryo develops to the gastrula and dies immediately, and the genome is demethylated on a large scale (127, 128).
Zhang et al. (129) described changes in DNA methylation during early embryonic lineage differentiation in mouse embryos using STEM-SEQ sequencing. They found that there was a large range of DNA hypomethylation regions around the homeobox A cluster (Hoxa) gene cluster and some developmental regulatory genes, such as hepatocyte nuclear factor 4 alpha (Hnf4a, endoderm marker), Pou5f1 (also called Oct4) and teratocarcinoma-derived growth factor 1 (Tdgf1, epidermal cell marker), exhibit dynamic changes in DNA methylation. Further methylation analysis of ICM and TE showed that the methylation levels in the promoter regions of most differentially expressed genes in ICM and TE cells, such as Pou5f1 and Tdgf1, were not significantly different. These 2 genes showed high levels of expression in the ICM, while the expression levels in TE cells were very low, and their promoters were not methylated (129). These results indicate that, despite the widespread presence of DNA methylation, it is not necessary for guiding early embryonic transcription processes but is a necessary condition to strengthen lineage differentiation.
Histone modification
When genomic DNA undergoes de novo methylation during the blastocyst stage, some regions are not affected. Studies have used mouse ESCs as a model to confirm that transcription factors and RNA polymerase complexes bind to DNA prior to de novo methylation, thereby preventing methylation at these sites. This protection likely prevents de novo methylation of the histone complex by recruiting the enzyme complex for local methylation (130).
Studies have shown that a second asymmetric histone modification will occur between ICM cells and TE cells. Compared with TE cells, ICM cells contain more transcriptional inhibition-related histone modifications. Histone methylation (such as H3K27me1/2/3) first appears in the ICM cells, but H3K27me2/3 can only be detected on the inactivated X chromosome. Compared with post-blastocyst stage, H3K27me3 is lower during the pre-blastocyst stage, indicating that H3K27me3 in differentiated cells is higher than that in dividing cells. The possible reason is that the promoter of the pluripotency gene is methylated and cannot be transcribed. In postimplanted embryos, de novo H3K27me3 targets the transcriptional silencing promoter of CpG islands and genomic regions through Polycomb inhibition complex protein (103, 104) and maintains the transcriptional inhibition of these genes (105). However, histone H3K9me2/3 levels are comparable in ICM cells and TE cells (131). After embryo implantation, there is a large amount of H3K9me2 deposition. However, this is not an essential condition for whole-genome DNA methylation; it is only necessary for the effective inhibition of a few species-specific genes (by silencing gene promoters or enhancers) (106, 107). In addition, the deposition of H3K9me2 is also not necessary for the silencing of most repeated elements (106). These findings together indicate that H3K9me2 is commonly associated with methylated DNA in implanted embryos but that its function is more specific. This may be due to the redundancy of inhibitory epigenetic markers and the possibility that these inhibitory modifications are not used as upstream transcription factors but act as enhancers of DNA transcriptional inactivation regions.
In addition, active chromatin markers, such as H3K4me3, may also play a role in transcriptional regulation during lineage differentiation. Active histone modification can enhance transcription by promoting the accessibility of transcription factors to regulatory regions. However, whether these markers are essential factors in the process of transcriptional construction or are only used to enhance the transcription process remains controversial (132). Studies have shown that the H3K4me3 level in promoter regions is related to transcription. However, in many cases, the effect of H3K4me3 removal on transcription is extremely limited (108–110), and the major H3K4 methyltransferase in preimplantation embryos is MLL2, and the major H3K4 methyltransferase in postimplanted embryos is SETD1A (111). Due to the overlapping redundancy of H3K4 methyltransferase proteins, the function of each methyltransferase and the function of H3K4me3 during embryogenesis are difficult to explain (111). Studies have shown that embryos lacking H3K4 methyltransferases MLL1 and MLL2 are arrested at midpregnancy and exhibit embryonic development defects, which may be due to abnormal expression levels of a subset of Hox genes (133, 134), indicating that MLL1/2-mediated H3K4me3 regulation is critical for these genes to maintain normal expression levels. The upregulation of SETD1A expression in embryos after implantation may play an important role in lineage differentiation and is an essential factor in gastrulation (111), indicating that SETD1A plays an important role in the establishment of transcriptional patterns in blastocysts. H3K27me3 can also coexist with H3K4me3 in the chromatin state to form a “bivalent modification” (104). A series of results showed that, from the early stage of embryonic development until implantation, this “bivalent modification” is rare. It is mainly enriched in promoters that are not methylated but express silent embryonic ectoderm development genes (135). This bivalent modification can balance the rapid activation or inhibition of development-related genes during embryonic lineage differentiation (136). Studies have confirmed that the bivalent modification of genes maintains neural crest cell migration and plasticity and chromatin accessibility during development through the Polycomb region (137). After differentiation, a decrease in H3K27me3 and an increase in H3K4me2 lead to cell-specific gene expression (138). However, further studies are needed to determine how H3K27me3 targets and regulates embryonic cell differentiation.
These studies together show that lineage differentiation processes require the participation of a variety of epigenetic modifications, especially H3K27me3 and H3K4me3. However, in most cases, the role of epigenetic modification is to strengthen lineage differentiation rather than guide it. These studies have created a new chapter in the study of the function of histone modification in embryonic development, providing answers to many pending scientific questions related to epigenetics. However, this is just the beginning, and more questions are waiting to be answered. The various modifications influence each other, forming a network. How the upstream and downstream regulatory molecules in this network play their roles and the diseases and their underlying mechanisms caused by related abnormalities are worth further investigation.
Part 3: Nutritional regulation of epigenetic modification
As early as 1997, British epidemiologist David Barker suggested that malnutrition during pregnancy is one of the important reasons for heart and metabolic disorders in early adulthood (138). The intrauterine environment has a permanent impact on body structure, function, metabolism, and the possibility of disease in adulthood. With the establishment of the concept of developmental origins of health and disease (DOHaD), Barker's hypothesis has been widely accepted and confirmed by many studies (139). During pregnancy, the lifestyle and nutrient intake of parents may greatly affect the health of the fetus through epigenetic regulation (140) (Figure 3). Nutrient concentrations not only have short- and long-term effects on metabolism, but may even stimulate memory that can be transmitted to offspring (141).
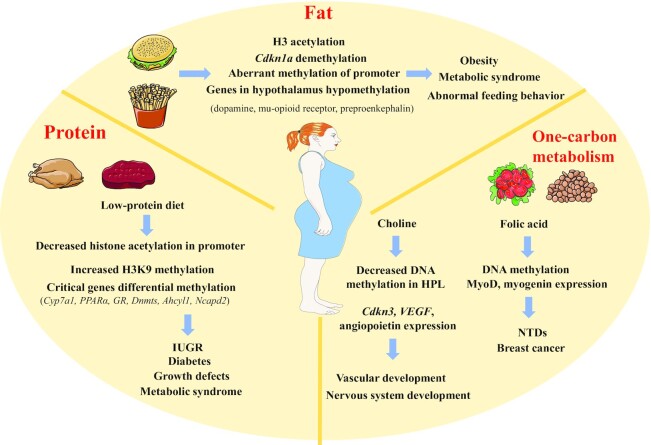
FIGURE 3.
Nutritional regulation of epigenetic modification. During pregnancy, maternal nutrition concentrations greatly influence the health of the fetus through epigenetic regulation. Fat, protein, and one-carbon metabolism can affect the epigenetics of the mother of fetuses in different ways. Ahcyl, adenosylhomocysteinase like 1; Cdkn1a, cyclin dependent kinase inhibitor 1A; Cyp7a1, cholesterol 7α-hydroxylase; DNMT, DNA methyltransferase; GR, glucocorticoid receptor; HPL, hippocampal neuroepithelial layer; IUGR, intrauterine growth restriction; Ncapd2, non-SMC condensin I complex subunit D2; NTD, neural tube defect; PPARα, peroxisome proliferator-activated receptor α; VEGF, vascular endothelial growth factor.
Protein
Protein is the material cornerstone of life and has a certain regulatory effect on fetal growth and development and the DNA methylation status of the offspring (142–144). Children born during the Dutch famine at the end of the Second World War had worse glucose tolerance than those born 1 y before the famine. A low-protein diet during pregnancy is also associated with increased incidences of diabetes and growth defects in the next generation (145). Painter et al. (146) found that protein restriction during pregnancy decreased the expression of liver cholesterol 7α-hydroxylase (Cyp7a1) in intrauterine growth-restriction mice, and this decrease was accompanied by decreased histone acetylation modification in the promoter region and increased H3K9 methylation modification. van Straten et al. (147) and Rees et al. (148) also found that when mice were fed a low-protein diet (9% casein) during pregnancy, the degree of DNA methylation in fetal liver was significantly higher than that in the control group (18% casein). Further studies showed that the expression of peroxisome proliferator-activated receptor α (PPARα) and glucocorticoid receptor (GR) significantly increased in the livers of 34-d-old pups in a low-protein diet group, while the methylation levels of the PPARα and GR genes significantly decreased (149–151). Jousse et al. (152) fed mice a low-protein diet (10% protein) throughout the entire pregnancy and lactation periods. They found that the body weight and body fat of the offspring were significantly decreased and the dietary intake significantly increased compared with those of control mice, and these changes were sustained into adulthood. The underlying reason is that the intake of a low-protein diet leads to the methylation of CpG islands in the promoter region of the leptin gene, and this specific modification further downregulates the expression levels of the leptin gene and protein. Altmann et al. (153) fed landrace sows either a high-protein or low-protein diet during pregnancy and found that the genes of key enzymes in methionine metabolic pathways [Dnmt1, Dnmt3a, Dnmt3b, betaine-homocysteine S-methyltransferase (Bhmt),methionine adenosyltransferase 2B (Mat2b), and adenosylhomocysteinase like 1 (Ahcyl1)] and chromosome morphology-related enzymes [non-SMC condensin I complex subunit D2 (Ncapd2), non-SMC condensin I complex subunit G (Ncapg), and non-SMC condensin I complex subunit H (Ncaph)] showed different degrees of differential methylation in the liver and skeletal muscle of offspring. Among those genes, the expression of DNA methyltransferases (Dnmt1, Dnmt3a, and Dnmt3b) was affected by the protein concentration, indicating that the epigenetic regulation of DNA methylation in fetal programmed growth was affected by maternal protein supply. In the livers of the offspring, the expression levels of the Ncapd2, Ncapg, and Ncaph3 genes in the protein-restriction group were significantly different from those in the normal group, while the expression levels of the Ncapd2 and Ncaph genes in skeletal muscle also showed significant differences. These results indicate that the maternal protein concentration during pregnancy changes the epigenetic markers in offspring, further affecting the gene expression process. In addition, Altmann et al. (154) showed that, compared with that in a normal-protein group, the expression level of the mitochondrial cytochrome C gene was significantly increased in the liver of piglets produced by the low-protein group, while the expression level of the cytochrome C gene in the high-protein group was only slightly increased. In 47 CpG loci in the promoter region of this gene, significantly high levels of DNA methylation occurred in the piglets in the high-protein and low-protein groups, indicating that the restriction of maternal protein concentrations during pregnancy affected the gene expression and metabolic status of offspring more significantly.
Fat
Fat is the main energy source for the body and plays a very important role in animal life activities such as growth, development, and metabolism. Aagaard-Tillery et al. (155) showed that in Japanese macaques fed a high-fat diet during pregnancy, the acetylation modification of H3 in offspring liver tissue changed, affecting the gene expression in the fetus. The results provided a molecular basis for the occurrence of adult diseases. A high-fat diet not only affects the F2 generation through the transmission of aberrant methylation of the promoter of the growth hormone secretin to offspring (156) but also affects the F3 generation (157). Dudley et al. (158) found that in rats fed a high-fat diet during pregnancy and lactation, the triglyceride content in the liver of 2-d-old offspring increased, leading to nonalcoholic fatty liver. In addition, the gene for the specific cell cycle inhibitor cyclin dependent kinase inhibitor 1A (Cdkn1a) was demethylated at a specific CpG site and the first exon region, resulting in an increase in gene expression, thereby inhibiting the division and proliferation of stem cells. Vucetic et al. (159) found that in mice fed a high-fat diet during pregnancy and lactation, 3 genes (i.e., dopamine, mu-opioid receptor, and pre-proenkephalin) in the hypothalamus of the offspring (18–24 wk old) were hypomethylated, and the expression of dopamine reabsorption transporter increased significantly. These changes not only caused abnormal feeding behavior in mice but also metabolic syndrome and an increased risk of obesity.
One-carbon metabolism
The methylation of cytosine in mammals requires the consumption of the one-carbon donor S-adenosylmethionine. Demethylation is carried out through Tet enzyme–mediated oxidation reactions, requiring α-ketoglutaric acid and Fe2+, and is enhanced by the micronutrient vitamin C. Therefore, changes in the concentrations of these substances may also affect the expression of downstream genes.
The effect of folic acid concentrations on the methylation level in porcine C2C12 cells was analyzed using MeDIP-chip and bisulfite sequencing. The results showed that when the concentration of folic acid changed, there were different degrees of hypermethylation upstream of the Myod transcription start site. Both the high-folic-acid group and the low-folic-acid group exhibited hypermethylation, while the normal-folic-acid-concentration group exhibited hypomethylation (160). In addition, during C2C12 differentiation, with the increase in folic acid concentration, myogenin mRNA expression increased, indicating that folic acid might affect the gene expression of Myod and myogenin through the regulation of the methylation status of the CpG locus in the gene promoter regions (160). Dietary supplementation with folic acid during pregnancy and lactation (5 mg/d) in rats significantly reduced mammary gland genomic DNA methylation in adult offspring and increased breast cancer risk (160). Folic acid deficiency during pregnancy and lactation in mice significantly reduced whole-genome DNA methylation levels in the small intestine tissue in the F1 generation, and this effect was persistent (161).
Studies have shown that maternal choline regulates the growth and development of the nervous system through DNA methylation. Low maternal dietary choline content at the embryonic stage in mice results in a significant reduction in the degree of genomic DNA methylation in the fetal hippocampal neuroepithelial layer, and the methylation level of protein kinase inhibitor (Cdkn3) genomic DNA also significantly decreases (162). A lack of choline during pregnancy also leads to reduced degrees of methylation in the vicinity of the promoter of the vascular endothelial growth factor C and angiopoietin 2 genes, and a significant decrease in the proliferation rate of vascular endothelial cells and the number of blood vessels in the hippocampus (163).
Robert et al. (164) found that exposure of agouti mice to methyl donors (such as folic acid, choline, and betaine) during pregnancy induced extensive methylation at CpG sites in embryonic genes, manifesting as yellow to brown hair changes in the offspring. The deletion of DNA methylase can cause embryonic development abnormalities. For example, mouse embryos with point mutations in the Dnmt1 catalytic center may exhibit abnormal development and even death, which may be related to decreased DNA methylation levels, disordered methylation patterns, and allelic loss (165).
The expression level of vitamin C transporters in germ cells is very high. From the beginning of pregnancy until embryonic day 13.5, vitamin C deficiency in mice caused a decreased number of germ cells, and the expression of TET1-dependent reproductive regulatory factors was defective. Starting from embryonic day 13.5, a vitamin C–containing diet was resumed and continued until delivery. The number of developing oocytes in the offspring of the vitamin C–deficient female mice was very small, and the number of implanted embryos after each adult female offspring mating was significantly reduced, and the completely failed pregnancy rate (lack of implantation sites) was very high. Even if a pregnancy is successful, the frequency of embryo resorption is very high. By comparing the DNA methyl groups in embryonic day 13.5 germ cells, it was found that two-thirds of differentially methylated regions (DMRs) were methylated after vitamin C deficiency, indicating that vitamin C is required for DNA demethylation. Hypermethylated DMRs induced by vitamin C deficiency are closely associated with ovarian development abnormalities and female infertility (166). In vitro–cultured embryonic stem cell experiments also showed that vitamin C induced DNA demethylation and the expression of a series of key reproductive genes in a TET1/TET2-dependent manner (167). Furthermore, vitamin C deficiency can also promote KDM3A/B-mediated H3K9me2 demethylation (168, 169).
Conclusions
In recent years, with rapid advances in sequencing technology, knowledge in the field of epigenetics has surged, revealing some unique epigenetic modifications involved in early embryo development. Particularly, in the field of ART, the epigenetic modification abnormalities of early embryos are becoming increasingly common. Understanding the epigenetic regulation of early embryo development is helpful for nutrition researchers to better understand where and when nutritional status is potentially problematic in epigenetic regulation. In addition, further understanding of the interaction between cellular metabolism and epigenetic marks provides new ideas for the targeted regulation and improvement of embryonic development by means of nutrition.
ACKNOWLEDGEMENTS
The authors’ responsibilities were as follows—SC: conceptualized the review, conducted the literature search, and wrote the manuscript; SQ, GY, MC, and QY: developed and formatted the tables; GW, HY, YW, SQ, and XZ: critically reviewed the manuscript; and all authors: read and approved the final manuscript.
Notes
This study was supported by the National Natural Science Foundation of China (32022080).
Author disclosures: The authors report no conflicts of interest.
Abbreviations used: AHCYl, adenosylhomocysteinase like 1; AID, activation-induced deaminase; ART, assisted reproductive technology; BHMT, betaine-homocysteine S-methyltransferase; CDKN1A, cyclin dependent kinase inhibitor 1A; CDX2, caudal type homeobox 2; CHD1, chromodomain helicase DNA binding protein 1; CSF2RA, colony stimulating factor 2 receptor subunit alpha; DMR, differentially methylated region; DNMT, DNA methyltransferase; ESC, embryonic stem cell; GCN5, histone acetyltransferase GCN5; GLP, euchromatic histone lysine methyltransferase 1; GR, glucocorticoid receptor; G9A, euchromatic histone lysine methyltransferase 2; HAT, histone acetyltransferase; HDAC, histone deacetylase; HMGP1, upstream binding transcription factor like 1; HMT, homocysteine S-methyltransferase 1; HNF4A, hepatocyte nuclear factor 4 alpha; ICM, inner cell mass; IGF2, insulin-like growth factor 2; KDM4A, lysine demethylase 4A; KLF5, kruppel like factor 5; lncRNA, long noncoding RNA; MAT2B, methionine adenosyltransferase 2B; MeCP, methyl-CpG binding protein; MLL, lysine methyltransferase 2A; miRNA, microRNA; ncRNA, noncoding RNA; NANOG, nanog homeobox; NCAPD2, non-SMC condensin I complex subunit D2; NCAPG, non-SMC condensin I complex subunit G; NCAPH, non-SMC condensin I complex subunit H; PGC, primordial germ cell; PN3, pronuclear stage 3; POU5F1, POU class 5 homeobox 1; PPARα: peroxisome proliferator-activated receptor α; PRC2, Polycomb repressive complex 2; SAH, S-adenosylhomocysteine; SAM, S-adenosylmethionine; SCNT, somatic cell nuclear transfer; SETDB1, SET domain bifurcated histone lysine methyltransferase 1; siRNA, small interfering RNA; SUV39, histone lysine methyltransferase SET/SUV39; TDGF1, teratocarcinoma-derived growth factor 1; TE, trophectoderm; TET, ten-eleven translocation methylcytosine dioxygenase; WDR82, WD repeat domain 82; 5mC, 5-methylcytosine; 5hmC, 5‐hydroxymethylcytosine.
Contributor Information
Shuang Cai, State Key Laboratory of Animal Nutrition, China Agricultural University, Beijing, China; Beijing Key Laboratory of Bio-feed Additives, China Agricultural University, Beijing, China.
Shuang Quan, State Key Laboratory of Animal Nutrition, China Agricultural University, Beijing, China; Beijing Key Laboratory of Bio-feed Additives, China Agricultural University, Beijing, China.
Guangxin Yang, State Key Laboratory of Animal Nutrition, China Agricultural University, Beijing, China; Beijing Key Laboratory of Bio-feed Additives, China Agricultural University, Beijing, China.
Meixia Chen, State Key Laboratory of Animal Nutrition, China Agricultural University, Beijing, China; Beijing Key Laboratory of Bio-feed Additives, China Agricultural University, Beijing, China.
Qianhong Ye, State Key Laboratory of Animal Nutrition, China Agricultural University, Beijing, China; Beijing Key Laboratory of Bio-feed Additives, China Agricultural University, Beijing, China.
Gang Wang, State Key Laboratory of Animal Nutrition, China Agricultural University, Beijing, China; Beijing Key Laboratory of Bio-feed Additives, China Agricultural University, Beijing, China.
Haitao Yu, State Key Laboratory of Animal Nutrition, China Agricultural University, Beijing, China; Beijing Key Laboratory of Bio-feed Additives, China Agricultural University, Beijing, China.
Yuming Wang, State Key Laboratory of Animal Nutrition, China Agricultural University, Beijing, China; Beijing Key Laboratory of Bio-feed Additives, China Agricultural University, Beijing, China.
Shiyan Qiao, State Key Laboratory of Animal Nutrition, China Agricultural University, Beijing, China; Beijing Key Laboratory of Bio-feed Additives, China Agricultural University, Beijing, China.
Xiangfang Zeng, State Key Laboratory of Animal Nutrition, China Agricultural University, Beijing, China; Beijing Key Laboratory of Bio-feed Additives, China Agricultural University, Beijing, China.
References
- 1.Helmerhorst FM, Perquin DAM, Donker D, Keirse M. Perinatal outcome of singletons and twins after assisted conception: a systematic review of controlled studies. BMJ. 2004;328(7434):261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maher ER, Brueton LA, Bowdin SC, Luharia A, Cooper W, Cole TR, Macdonald F, Sampson JR, Barratt CL, Reik W, Hawkins MM. Beckwith-Wiedemann syndrome and assisted reproduction technology (ART). J Med Genet. 2003;40(1):62–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeBaun MR, Niemitz EL, Feinberg AP. Association of in vitro fertilization with Beckwith-Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am J Hum Genet. 2003;72(1):156–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim D, Bowdin SC, Tee L, Kirby GA, Blair E, Fryer A, Lam W, Oley C, Cole T, Brueton LAet al. Clinical and molecular genetic features of Beckwith-Wiedemann syndrome associated with assisted reproductive technologies. Hum Reprod. 2009;24:741–7. [DOI] [PubMed] [Google Scholar]
- 5.Peat JR, Reik W. Incomplete methylation reprogramming in SCNT embryos. Nat Genet. 2012;44(9):965–6. [DOI] [PubMed] [Google Scholar]
- 6.Matoba S, Liu Y, Lu F, Iwabuchi KA, Shen L, Inoue A, Zhang Y. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell. 2014;159(4):884–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dean W, Santos F, Stojkovic M, Zakhartchenko V, Walter J, Wolf E, Reik W. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc Natl Acad Sci. 2001;98(24):13734–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho S, Tang W. Techniques used in studies of epigenome dysregulation due to aberrant DNA methylation: an emphasis on fetal-based adult diseases. Reprod Toxicol. 2007;23(3):267–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenberg MVC, Glaser J, Borsos M, Marjou FE, Walter M, Teissandier A, Bourchis D. Transient transcription in the early embryo sets an epigenetic state that programs postnatal growth. Nat Genet. 2017;49(1):110–18. [DOI] [PubMed] [Google Scholar]
- 10.Tobi EW, Lumey LH, Talens RP, Kremer D, Putter H, Stein AD, Slagboom PE, Heijmans BT. DNA methylation differences after exposure to prenatal famine are common and timing-and sex-specific. Hum Mol Genet. 2009;18(21):4046–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Painter RC, de Rooij SR, Bossuyt PM, Simmers TA, Osmond C, Barker DJ, Bleker OP, Roseboom TJ. Early onset of coronary artery disease after prenatal exposure to the Dutch famine. Am J Clin Nutr. 2006;84(2):322–7. [DOI] [PubMed] [Google Scholar]
- 12.Francine E, Reid FT, Yushar DB, Melissa JF, Amit V, Nir B, John MG. Cytosine methylation dysregulation in neonates following intrauterine growth restriction. PLoS One. 2010;5:e8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241(1):172–82. [DOI] [PubMed] [Google Scholar]
- 14.Stefanie S, Simon A, Felix K, Julia A, Jörn W, Fátima S, Christian P, Bernard T, Wendy D, Wolf R. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell. 2012;48:849–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charney E. Cytoplasmic inheritance redux. Adv Child Dev Behav. 2013;44:225–55. [DOI] [PubMed] [Google Scholar]
- 16.Tatiana N, Rajarshi PG, Rachel AH, Jeffrey CH, Sergei AG, Christopher LW. MeCP2-chromatin interactions include the formation of chromatosome-like structures and are altered in mutations causing Rett syndrome. J Biol Chem. 2007;282:28237–45. [DOI] [PubMed] [Google Scholar]
- 17.Dana CD, Jennifer RW, Randy LJ. Epigenetic gene regulation: linking early developmental environment to adult disease. Reprod Toxicol. 2007;23:297–307. [DOI] [PubMed] [Google Scholar]
- 18.Lingjun S, Ji W. Epigenetic regulation in mammalian preimplantation embryo development. Reprod Biol Endocrinol. 2009;7:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu N, Li S, Wu N, Cho K. Acetylation and deacetylation in cancer stem-like cells. Oncotarget. 2017;8(51):89315–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christopher RV, Sean AM, Benjamin AO, Gerd AB. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381–9. [DOI] [PubMed] [Google Scholar]
- 21.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. [DOI] [PubMed] [Google Scholar]
- 23.Dovey OM, Foster CT, Cowley SM. Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation. Proc Natl Acad Sci. 2010;107(18):8242–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu W, Edmondson DG, Evrard YA, Wakamiya M, Behringer RR, Roth SY. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat Genet. 2000;26(2):229–32. [DOI] [PubMed] [Google Scholar]
- 25.Straussman R, Nejman D, Roberts D, Steinfeld I, Blum B, Benvenisty N, Simon I, Yakhini Z, Cedar H. Developmental programming of CpG island methylation profiles in the human genome. Nat Struct Mol Biol. 2009;16(5):564–71. [DOI] [PubMed] [Google Scholar]
- 26.Hany A, Yusuke Y, Shinichi H. Active demethylation of paternal genome in mammalian zygotes. J Reprod Dev. 2009;55:356–60. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, Ho SM. Epigenetics meets endocrinology. J Mol Endocrinol. 2011;46(1):R11–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrussa L, Van de Velde H, De Rycke M. Dynamic regulation of DNA methyltransferases in human oocytes and preimplantation embryos after assisted reproductive technologies. Mol Hum Reprod. 2014;20(9):861–74. [DOI] [PubMed] [Google Scholar]
- 29.Jo P. The role of genomic imprinting in biology and disease: an expanding view. Nat Rev Genet. 2014;15:517–40. [DOI] [PubMed] [Google Scholar]
- 30.Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;9(2):129–40. [DOI] [PubMed] [Google Scholar]
- 31.Spahn L, Barlow DP. An ICE pattern crystallizes. Nat Genet. 2003;35(1):11–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xue F, Tian XC, Du FL, Kubota C, Taneja M, Dinnyes A, Dai YP, Levine H, Pereira VL, Yang XZ. Aberrant patterns of X chromosome inactivation in bovine clones. Nat Genet. 2002;31(2):216–20. [DOI] [PubMed] [Google Scholar]
- 33.Inoue K, Kohda T, Sugimoto M, Sado T, Ogonuki N, Matoba S, Shiura H, Ikeda R, Mochida K, Fujii Tet al. Impeding Xist expression from the active X chromosome improves mouse somatic cell nuclear transfer. Science. 2010;330(6003):496–9. [DOI] [PubMed] [Google Scholar]
- 34.Toshio H. Human spermatozoal RNAs. Fertil Steril. 2012;97:275–81. [DOI] [PubMed] [Google Scholar]
- 35.Sirard MA. Factors affecting oocyte and embryo transcriptomes. Reprod Domestic Animals. 2012;47:148–55. [DOI] [PubMed] [Google Scholar]
- 36.Murchison EP, Stein P, Xuan ZY, Pan H, Zhang MQ, Schultz RM, Hannon GJ. Critical roles for Dicer in the female germline. Genes Dev. 2007;21(6):682–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliver M, Vasily VV, Frédéric C, Karin S, Aurélie L, Molly H, Ying J, Matteo DR, Kaja AW, Jorma Tet al. An atlas of chromatoid body components. RNA. 2014;20:483–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melissa RR, Wayne RJ. Epigenetics in fertilization and preimplantation embryo development. Prog Biophys Mol Biol. 2013;113:423–32. [DOI] [PubMed] [Google Scholar]
- 39.Liang M, Li WQ, Tian H, Hu T, Wang L, Lin Y, Li YL, Huang HF, Sun F. Sequential expression of long noncoding RNA as mRNA gene expression in specific stages of mouse spermatogenesis. Sci Rep. 2015;4(1):5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delcuve GP, Rastegar M, Davie JR. Epigenetic control. J Cell Physiol. 2009;219(2):243–50. [DOI] [PubMed] [Google Scholar]
- 41.Vaissière T, Sawan C, Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat Res/Rev Mutat Res. 2008;659(1-2):40–8. [DOI] [PubMed] [Google Scholar]
- 42.Michela C, Annalisa I, Gianfranco B, Nicoletta L. Molecular mechanisms of gene silencing mediated by DNA methylation. Mol Cell Biol. 2002;22:3157–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meissner A, Mikkelsen TS, Gu HC, Wernig M, Hanna J, Sivachenko A, Zhang XL, Bernstein B, Nusbaum C, Jaffe DBet al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454(7205);766–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mikkelsen TS, Ku MC, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim T, Koche RPet al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du J, Johnson LM, Jacobsen SE, Patel DJ. DNA methylation pathways and their crosstalk with histone methylation. Nat Rev Mol Cell Biol. 2015;16(9):519–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Antoine H, Stefan K, Karl M, Roderick JOS, Alwin A, Laura PB, Alexander K, Susanne O, Makoto T, Yoichi Set al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–89. [DOI] [PubMed] [Google Scholar]
- 47.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6(11):838–49. [DOI] [PubMed] [Google Scholar]
- 48.Yoichi S, Makoto T. H3K9 methyltransferase G9a and the related molecule GLP. Gene Dev. 2011;25:781–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alissa M, Anna S, Elyse R, Giancarlo B, Sanjeet P, Kathrin P. The Mbd1-Atf7ip-Setdb1 pathway contributes to the maintenance of X chromosome inactivation. Epigenet Chromatin. 2014;7:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karimi MM, Goyal P, Maksakova IA, Bilenky M, Leung D, Tang JX, Shinkai Y, Mager DL, Jones S, Hirst Met al. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell. 2011;8(6):676–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hou J, Liu L, Zhang J, Cui XH, Yan FX, Guan H, Chen YF, An XR. Epigenetic modification of histone 3 at lysine 9 in sheep zygotes and its relationship with DNA methylation. BMC Dev Biol. 2008;8(1):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410(6824):116–20. [DOI] [PubMed] [Google Scholar]
- 53.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410(6824):120–4. [DOI] [PubMed] [Google Scholar]
- 54.Arand J, Spieler D, Karius T, Branco MR, Mwilinger D, Meissner A, Jenuwein T, Xu GL, Leonhardt H, Wolf Vet al. In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases. PLos Genet. 2012;8(6):e1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bernhard L, Yoshihide U, Alwin A, Ulrich B, Laura P, Stefan K, Chen TP, Li E, Thomas J, Antoine H. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–200. [DOI] [PubMed] [Google Scholar]
- 56.Kohta I, Misa I, Masako S, Makoto T, Yoichi S, Satoshi T, John MG, Shintaro Y, Naka H, Kunio S. Genome-wide and locus-specific DNA hypomethylation in G9a deficient mouse embryonic stem cells. Genes Cells. 2007;12:1–11. [DOI] [PubMed] [Google Scholar]
- 57.Kevin BD, Irina AM, Fabio M, Danny L, Ruth A, Sandra L, Hao WY, Lucia LL, Dixie LM, Dirk Set al. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J. 2008;27:2691–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Danny L, Du TT, Ulrich W, Xie W, Young LA, Preeti G, Li YJ, Keith ES, Jin P, Lorincz MCet al. Regulation of DNA methylation turnover at LTR retrotransposons and imprinted loci by the histone methyltransferase Setdb1. Proc Natl Acad Sci USA. 2014;111:6690–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Akiko T, Tomohiro H, Yuichi K, Shin-ichiro T, Morito S, Chisa M, Kunitada S, Fuyuki I, Li E, Hiroki RUet al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006;11:805–14. [DOI] [PubMed] [Google Scholar]
- 60.Toshiyuki M, Danny L, Hiroki M, Irina AM, Hitoshi M, Hiroshi K, Makoto T, Matthew CL, Yoichi S. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–31. [DOI] [PubMed] [Google Scholar]
- 61.Bachman KE, Park BH, Rhee I, Rajagopalan H, Herman JG, Baylin SB, Kinzler KW, Vogelstein B. Histone modifications and silencing prior to DNA methylation of a tumor suppressor gene. Cancer Cell. 2003;3(1):89–95. [DOI] [PubMed] [Google Scholar]
- 62.Jesus E, Esteban B, Mario FF, Ana VG, Angeles J, Juan CS, Keith DR, Francois F, Manel E. Human DNA methyltransferase 1 is required for maintenance of the histone H3 modification pattern. J Biol Chem. 2004;279:37174–84. [DOI] [PubMed] [Google Scholar]
- 63.Carvell TN, Daniel JW, Mihaela V, Felicidad AG, Joy CYL, Liang GN, Peter AJ. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2'-deoxycytidine. Cancer Res. 2002;62:6456–61. [PubMed] [Google Scholar]
- 64.Handel AE, Ebers GC, Ramagopalan SV. Epigenetics: molecular mechanisms and implications for disease. Trends Mol Med. 2010;16(1):7–16. [DOI] [PubMed] [Google Scholar]
- 65.Kokura K, Sun LD, Bedford MT, Fang J. Methyl-H3K9-binding protein MPP8 mediates E-cadherin gene silencing and promotes tumour cell motility and invasion. EMBO J. 2010;29(21):3673–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chang TQ, Sun LD, Kokura K, Horton JR, Fukuda M, Espejo A, Izumi V, Koomen JM, Bedford MT, Zhang Xet al. MPP8 mediates the interactions between DNA methyltransferase Dnmt3a and H3K9 methyltransferase GLP/G9a. Nat Commun. 2011;2(1):533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14(3):204–20. [DOI] [PubMed] [Google Scholar]
- 68.Ito S, D Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466(7310):1129–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi SW, Page DCet al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9(2):166–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aleksandra S, Sebastian B, Christine S, Fabio S, Heinrich L. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38:e181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maria EF, Omar A, Chao L, Patrick SW, Jay P, Alan S, Li YS, Neha B, Aparna V, Hugo FFet al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tahiliani M, Koh KP, Shen YH, Pastor WA, Bandukwala H, Brudno YB, Suneet, A, Lyer LM, Liu DR, Aravind Let al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumer A, Patel J, Zhao XYet al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20(1):11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Quivoron C, Couronné L, Valle VD, Lopez CK, Plo I, Wagner-Ballon O, Cruzeiro MD, Delhommeau F, Arnulf B, Stern MHet al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20(1):25–38. [DOI] [PubMed] [Google Scholar]
- 75.Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi LY, He XY, Jin Set al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477(7366):606–10. [DOI] [PubMed] [Google Scholar]
- 76.Mark W, Toshinobu N, Konstantin L, Joana MC, Valeri Z, Michele B, Julia A, Toru N, Wolf R, Jorn W. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011;2:241. [DOI] [PubMed] [Google Scholar]
- 77.Santenard A, Ziegler-Birling C, Koch M, Tora L, Bannister AJ, Torres-Padilla M. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat Cell Biol. 2010;12(9):853–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guo JJU, Su YJ, Zhong C, Ming GL, Song HJ. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145(3):423–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science. 2010;329(5987):78–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Keisuke A, Erina I, Hirofumi S, Yuki O. Paternal H3K4 methylation is required for minor zygotic gene activation and early mouse embryonic development. EMBO Rep. 2015;16:803–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hans-Martin H, Man M, Garruss AS, Liang KW, Yoh-Hei T, Kristen M, Voets O, Peter V, Ali S. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Gene Dev. 2012;26:2604–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reis E, Silva AR, Bruno C, Fleurot R, Daniel N, Archilla C, Peynot N, Lucci CM, Beaujean N, Duranthon V. Alteration of DNA demethylation dynamics by in vitro culture conditions in rabbit pre-implantation embryos. Epigenetics. 2012;7(5):440–6. [DOI] [PubMed] [Google Scholar]
- 83.Cao ZB, Li YS, Chen Z, Wang H, Zhang ML, Zhou NR, Wu RH, Ling YH, Fang FG, Li Net al. Genome-wide dynamic profiling of histone methylation during nuclear transfer-mediated porcine somatic cell reprogramming. PLoS One. 2015;10(12):e0144897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Santos F, Zakhartchenko V, Stojkovic M, Peters A, Jenuwein T, Wolf E, Reik W, Dean W. Epigenetic marking correlates with developmental potential in cloned bovine preimplantation embryos. Curr Biol. 2003;13(13):1116–21. [DOI] [PubMed] [Google Scholar]
- 85.Fu L, Zhang J, Yan F, Guan H, An X, Hou J. Abnormal histone H3K9 dimethylation but normal dimethyltransferase EHMT2 expression in cloned sheep embryos. Theriogenology. 2012;78(9):1929–38. [DOI] [PubMed] [Google Scholar]
- 86.Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T, Hansen KH, Helin K. The putative oncogene GASC1 demethylates tri-and dimethylated lysine 9 on histone H3. Nature. 2006;442(7100):307–11. [DOI] [PubMed] [Google Scholar]
- 87.Klose RJ, Yamane K, Bae Y, Zhang DZ, Erdjument-Bromage H, Tempst P, Wong JM, Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442(7100):312–16. [DOI] [PubMed] [Google Scholar]
- 88.Liu X, Wang YZ, Gao YP, Su JM, Zhang JC, Xing XP, Zhou C, Yao KZ, An QL, Zhang Y. H3K9 demethylase KDM4E is an epigenetic regulator for bovine embryonic development and a defective factor for nuclear reprogramming. Development. 2018;145(4):dev158261. [DOI] [PubMed] [Google Scholar]
- 89.Pedersen MT, Kooistra SM, Radzisheuskaya A, Laugesen A, Johansen JV, Hayward DG, Nilsson J, Agger K, Helin K. Continual removal of H3K9 promoter methylation by Jmjd2 demethylases is vital for ESC self-renewal and early development. EMBO J. 2016;35(14):1550–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Inoue A, Jiang L, Lu F, Suzuki T, Zhang Y. Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature. 2017;547(7664):419–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shao G, Ding H, Gong A. Role of histone methylation in zygotic genome activation in the preimplantation mouse embryo. In Vitro Cell Dev Biol Animal. 2008;44(3-4):115–20. [DOI] [PubMed] [Google Scholar]
- 92.Bi Y, Lv Z, Wang Y, Hai T, Huo R, Zhou ZM, Zhou ZM, Shao JH. WDR82, a key epigenetics-related factor, plays a crucial role in normal early embryonic development in mice. Biol Reprod. 2011;84(4):756–64. [DOI] [PubMed] [Google Scholar]
- 93.Guzman-Ayala M, Sachs M, Koh FM, Onodera C, Bulut-Karslioglu A, Lin C, Wong P, Nitta R, Song JS, Santos MR. Chd1 is essential for the high transcriptional output and rapid growth of the mouse epiblast. Development. 2015;142(1):118–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tamás A, András P. The constant variation: DNA methylation changes during preimplantation development. FEBS Lett. 2006;580:6521–6. [DOI] [PubMed] [Google Scholar]
- 95.Lei H, Oh SP, Okano M, Juttermann R, Goss KA, Jaenisch R, Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:173–7. [DOI] [PubMed] [Google Scholar]
- 96.Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang XY, Golic KG, Jacobsen SE, Bestor TH. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311(5759):395–8. [DOI] [PubMed] [Google Scholar]
- 97.Okano M, Xie S, Li E. Dnmt2 is not required for de novo and maintenance methylation of viral DNA in embryonic stem cells. Nucleic Acids Res. 1998;26(11):2536–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Masahiro K, Masaki O, Kenichiro H, Takashi S, Naomi T, En L, Hiroyuki S. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–3. [DOI] [PubMed] [Google Scholar]
- 99.Margot JB, Cardoso MC, Leonhardt H. Mammalian DNA methyltransferases show different subnuclear distributions. J Cell Biochem. 2001;83(3):373–9. [DOI] [PubMed] [Google Scholar]
- 100.Déborah B, Timothy HB. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–9. [DOI] [PubMed] [Google Scholar]
- 101.Kenichiro H, Masaki O, Hong L, En L. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–93. [DOI] [PubMed] [Google Scholar]
- 102.Webster KE, O'Bryan MK, Fletcher S, Crewther PE, Aapola U, Craig J, Harrison DK, Aung H, Phutikanit N, Lyle Ret al. Meiotic and epigenetic defects in Dnmt3L-knockout mouse spermatogenesis. Proc Natl Acad Sci. 2005;102(11):4068–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu XY, Wang C, Liu WQ, Li JY, Li C, Kou XC, Chen JY, Zhao YH, Gao HB, Wang Het al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature. 2016;537(7621):558–62. [DOI] [PubMed] [Google Scholar]
- 104.Zheng H, Huang B, Zhang BJ, Xiang YL, Du ZH, Xu QH, Li YY, Wang QJ, Ma J, Peng Xet al. Resetting epigenetic memory by reprogramming of histone modifications in mammals. Mol Cell. 2016;63(6):1066–79. [DOI] [PubMed] [Google Scholar]
- 105.Yang XF, Hu BQ, Hou Y, Qiao Y, Wang R, Chen YY, Qian Y, Feng S, Chen J, Liu Cet al. Silencing of developmental genes by H3K27me3 and DNA methylation reflects the discrepant plasticity of embryonic and extraembryonic lineages. Cell Res. 2018;28(5):593–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zylicz JJ, Dietmann S, Günesdogan U, Hackett JA, Cougot D, Lee C, Surani MA. Chromatin dynamics and the role of G9a in gene regulation and enhancer silencing during early mouse development. Elife. 2015;4:e09571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Auclair G, Borgel J, Sanz LA, Vallet J, Guibert S, Dumas M, Cavelier P, Girardot M, Forne T, Feil Ret al. EHMT2 directs DNA methylation for efficient gene silencing in mouse embryos. Genome Res. 2016;26(2):192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15(24):3286–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Clouaire T, Webb S, Skene P, Illingworth R, Kerr A, Andrews R, Lee J, Skalnik D, Bird A. Cfp1 integrates both CpG content and gene activity for accurate H3K4me3 deposition in embryonic stem cells. Genes Dev. 2012;26(15):1714–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Margaritis T, Oreal V, Brabers N, Maestroni L, Vitaliano-Prunier A, Benschop JJ, van Hooff S, van Leenen D, Dargemont C, Geli Vet al. Two distinct repressive mechanisms for histone 3 lysine 4 methylation through promoting 3′-end antisense transcription. PLos Genet. 2012;8(9):e1002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Anita SB, Kerstin S, Katrin N, Undine H, Giovanni C, Ashish G, Davi CT, Jun F, Andrea K, A Francis Set al. The H3K4 methyltransferase Setd1a is first required at the epiblast stage, whereas Setd1b becomes essential after gastrulation. Development. 2014;141:1022–35. [DOI] [PubMed] [Google Scholar]
- 112.Okada Y, Yamaguchi K. Epigenetic modifications and reprogramming in paternal pronucleus: sperm, preimplantation embryo, and beyond. Cell Mol Life Sci. 2017;74(11):1957–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol. 1997;181(2):296–307. [DOI] [PubMed] [Google Scholar]
- 114.Yoko H, Kazuo Y, Teruhiko W, Timothy JS, Takashi K, Toshiki T, Makoto T, Yoichi S, Hitoshi K, Naohito Net al. Tracking epigenetic histone modifications in single cells using Fab-based live endogenous modification labeling. Nucleic Acids Res. 2011;39:6475–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Konstantin L, Jörn W. Differential dynamics of histone H3 methylation at positions K4 and K9 in the mouse zygote. BMC Dev Biol. 2004;4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Santos F, Peters AH, Otte AP, Reik W, Dean W. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev Biol. 2005;280(1):225–36. [DOI] [PubMed] [Google Scholar]
- 117.Wiekowski M, Miranda M, Depamphilis ML. Requirements for promoter activity in mouse oocytes and embryos distinguish paternal pronuclei from maternal and zygotic nuclei. Dev Biol. 1993;159(1):366–78. [DOI] [PubMed] [Google Scholar]
- 118.Andres S, Christel H, Sirpa M, Anne-Maria S, Aila T, Timo T. Early cleavage predicts the viability of human embryos in elective single embryo transfer procedures. Hum Reprod. 2003;18:821–5. [DOI] [PubMed] [Google Scholar]
- 119.Van Montfoort Aafke PA, Dumoulin John CM, Kester Arnold DM, Evers Johannes LH. Early cleavage is a valuable addition to existing embryo selection parameters: a study using single embryo transfers. Hum Reprod. 2004;19(9):2103–8. [DOI] [PubMed] [Google Scholar]
- 120.Ziegler-Birling C, Daujat S, Schneider R, Torres-Padilla M. Dynamics of histone H3 acetylation in the nucleosome core during mouse pre-implantation development. Epigenetics. 2016;11(8):553–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liu WQ, Liu XY, Wang CF, Gao YW, Gao R, Kou XC, Zhao YH, Li JY, Wu Y, Xiu WCet al. Identification of key factors conquering developmental arrest of somatic cell cloned embryos by combining embryo biopsy and single-cell sequencing. Cell Discovery. 2016;2(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chung YG, Matoba S, Liu YT, Eum JH, Lu FL, Jiang W, Lee JE, Sepilian V, Cha KY, Lee DRet al. Histone demethylase expression enhances human somatic cell nuclear transfer efficiency and promotes derivation of pluripotent stem cells. Cell Stem Cell. 2015;17(6):758–66. [DOI] [PubMed] [Google Scholar]
- 123.Matoba S, Liu YT, Lu FL, Iwabuchi KA, Shen L, Inoue A, Zhang Y. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell. 2014;159(4):884–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sankar A, Kooistra SM, Gonzalez JM, Ohlsson C, Poutanen M, Helin K. Maternal expression of the histone demethylase Kdm4a is crucial for pre-implantation development. Development. 2017;144(18):3264–77. [DOI] [PubMed] [Google Scholar]
- 125.Suzuki S, Nozawa Y, Tsukamoto S, Kaneko T, Manabe I, Imai H, Minami N. CHD1 acts via the Hmgpi pathway to regulate mouse early embryogenesis. Development. 2015;142(13):2375–84. [DOI] [PubMed] [Google Scholar]
- 126.Tang W, Ho S. Epigenetic reprogramming and imprinting in origins of disease. Rev Endocr Metab Disord. 2007;8:173–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bostick M, Kim JK, Estève P, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317(5845):1760–4. [DOI] [PubMed] [Google Scholar]
- 128.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura Ket al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450(7171):908–12. [DOI] [PubMed] [Google Scholar]
- 129.Zhang Y, Xiang YL, Yin QZ, Du ZH, Peng X, Wang QJ, Fidalgo M, Xia WK, Li YY, Zhao Zet al. Dynamic epigenomic landscapes during early lineage specification in mouse embryos. Nat Genet. 2018;50(1):96–105. [DOI] [PubMed] [Google Scholar]
- 130.Greenfield R, Tabib A, Keshet I, Moss J, Sabag O, Goren A, Cedar H. Role of transcription complexes in the formation of the basal methylation pattern in early development. Proc Natl Acad Sci. 2018;115(41):10387–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Erhardt S, Su I, Schneider R, Barton S, Bannister AJ, Perez-Burgos L, Jenuwein T, Kouzarides T, Tarakhovsky A, Surani MA. Consequences of the depletion of zygotic and embryonic enhancer of zeste 2 during preimplantation mouse development. Development. 2003;130:4235–48. [DOI] [PubMed] [Google Scholar]
- 132.Howe FS, Fischl H, Murray SC, Mellor J. Is H3K4me3 instructive for transcription activation?. Bioessays. 2017;39(1):1–12. [DOI] [PubMed] [Google Scholar]
- 133.Ernst P, Fisher JK, Avery W, Wade S, Foy D, Korsmeyer SJ. Definitive hematopoiesis requires the mixed-lineage leukemia gene. Dev Cell. 2004;6(3):437–43. [DOI] [PubMed] [Google Scholar]
- 134.Glaser S, Schaft J, Lubitz S, Vintersten K, Hoeven F, Tufteland KR, Aasland R, Anastassiadis K, Ang S, Stewart AF. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development. 2006;133:1423–32. [DOI] [PubMed] [Google Scholar]
- 135.Rugg-Gunn PJ, Cox BJ, Ralston A, Rossant J. Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo. Proc Natl Acad Sci. 2010;107(24):10783–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath Ket al. A bivalent chromatin structure marks key developmental gene in embryonic stem cells. Cell. 2006;125(2):315–26. [DOI] [PubMed] [Google Scholar]
- 137.Minoux M, Holwerda S, Vitobello A, Kitazawa T, Kohler H, Stadler MB, Rijli FM. Gene bivalency at Polycomb domains regulates cranial neural crest positional identity. Science. 2017;355(6332):eaal2913. [DOI] [PubMed] [Google Scholar]
- 138.Barker DJP. Maternal nutrition, fetal nutrition, and disease in later life. Nutrition. 1997;13(9):807–13. [DOI] [PubMed] [Google Scholar]
- 139.Wadhwa PD, Buss C, Entringer S, Swanson JM. Developmental origins of health and disease: brief history of the approach and current focus on epigenetic mechanisms. Semin Reprod Med. 2009;27(05):358–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang Y, Liu Q, Tang FC, Yan LY, Qiao J. Epigenetic regulation and risk factors during the development of human gametes and early embryos. Annu Rev Genomics Hum Genet. 2019;20(1):21–40. [DOI] [PubMed] [Google Scholar]
- 141.Bartke T, Schneider R. You are what you eat—how nutrition and metabolism shape the genome through epigenetics. Mol Metabol. 2020;38:100987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Milagro FI, Mansego ML, Miguel CD, Martinez JA. Dietary factors, epigenetic modifications and obesity outcomes: progresses and perspectives. Mol Aspects Med. 2013;34(4):782–812. [DOI] [PubMed] [Google Scholar]
- 143.Sharma U, Rando OJ. Metabolic inputs into the epigenome. Cell Metab. 2017;25(3):544–58. [DOI] [PubMed] [Google Scholar]
- 144.Adelheid S. Epigenetic inheritance and evolution: a paternal perspective on dietary influences. Prog Biophys Mol Biol. 2015;118:79–85. [DOI] [PubMed] [Google Scholar]
- 145.Zambrano E, Martinez-Samayoa PM, Bautista CJ, Deas M, Guillen L, Rodriguez-Gonzalez GL, Guzman C, Larrea F, Nathanielsz PW. Sex differences in transgenerational alterations of growth and metabolism in progeny (F2) of female offspring (F1) of rats fed a low protein diet during pregnancy and lactation. J Physiol. 2005;566(1):225–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Painter RC, de Rooij SR, Bossuyt PM, Simmers TA, Osmond C, Barker DJ, Bleker OP, Roseboom TJ. Early onset of coronary artery disease after prenatal exposure to the Dutch famine. Am J Clin Nutr. 2006;84(2):322–7. [DOI] [PubMed] [Google Scholar]
- 147.van Straten EM, Bloks VW, Huijkman NC, Baller JF, Meer HV, Lütjohann D, Kuipers F, Plosch T. The liver X-receptor gene promoter is hypermethylated in a mouse model of prenatal protein restriction. Am J Physiol Regul Integr Comp Physiol. 2010;298(2):R275–82. [DOI] [PubMed] [Google Scholar]
- 148.Rees WD, Hay SM, Brown DS, Antipatis C, Palmer RM. Maternal protein deficiency causes hypermethylation of DNA in the livers of rat fetuses. J Nutr. 2000;130(7):1821–6. [DOI] [PubMed] [Google Scholar]
- 149.Lillycrop KA, Slater-Jefferies JL, Hanson MA, Godfrey KM, Jackson AA, Burdge GC. Induction of altered epigenetic regulation of the hepatic glucocorticoid receptor in the offspring of rats fed a protein-restricted diet during pregnancy suggests that reduced DNA methyltransferase-1 expression is involved in impaired DNA methylation and changes in histone modifications. Br J Nutr. 2007;97:1064–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lillycrop KA, Phillips ES, Torrens C, Hanson MA, Jackson AA, Burdge GC. Feeding pregnant rats a protein-restricted diet persistently alters the methylation of specific cytosines in the hepatic PPARα promoter of the offspring. Br J Nutr. 2008;100(2):278–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr. 2005;135(6):1382–6. [DOI] [PubMed] [Google Scholar]
- 152.Jousse C, Parry L, Lambert Langlais S, Maurin AC, Averous J, Bruhat A, Carraro V, Tost J, Letteron P, Chen Pet al. Perinatal undernutrition affects the methylation and expression of the leptin gene in adults: implication for the understanding of metabolic syndrome. FASEB J. 2011;25(9):3271–8. [DOI] [PubMed] [Google Scholar]
- 153.Altmann S, Murani E, Schwerin M, Metges CC, Wimmers K, Ponsuksili S. Maternal dietary protein restriction and excess affects offspring gene expression and methylation of non-SMC subunits of condensin I in liver and skeletal muscle. Epigenetics. 2012;7(3):239–52. [DOI] [PubMed] [Google Scholar]
- 154.Altmann S, Murani E, Schwerin M, Metges CC, Wimmers K, Ponsuksili S. Somatic cytochrome c (CYCS) gene expression and promoter-specific DNA methylation in a porcine model of prenatal exposure to maternal dietary protein excess and restriction. Br J Nutr. 2012;107(6):791–9. [DOI] [PubMed] [Google Scholar]
- 155.Aagaard-Tillery KM, Grove K, Bishop J, Ke XR, Fu Q, McKnight R, Lane RH. Developmental origins of disease and determinants of chromatin structure: maternal diet modifies the primate fetal epigenome. J Mol Endocrinol. 2008;41(2):91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gregory AD, Tracy LB. Maternal high-fat diet promotes body length increases and insulin insensitivity in second-generation mice. Endocrinology. 2009;150:4999–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gregory AD, Tracy LB. Maternal high-fat diet effects on third-generation female body size via the paternal lineage. Endocrinology. 2011;152:2228–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Dudley KJ, Sloboda DM, Connor KL, Beltrand J, Vickers MH. Offspring of mothers fed a high fat diet display hepatic cell cycle inhibition and associated changes in gene expression and DNA methylation. PLoS One. 2011;6(7):e21662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Vucetic Z, Kimmel J, Totoki K, Hollenbeck E, Reyes TM. Maternal high-fat diet alters methylation and gene expression of dopamine and opioid-related genes. Endocrinology. 2010;15(:10):4756–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ly A, Lee H, Chen J, Sie KK, Renlund R, Medline A, Sohn K, Croxford R, Thompson LU, Kim Y. Effect of maternal and postweaning folic acid supplementation on mammary tumor risk in the offspring. Cancer Res. 2011;71(3):988–97. [DOI] [PubMed] [Google Scholar]
- 161.McKay JA, Waltham KJ, Williams EA, Mathers JC. Folate depletion during pregnancy and lactation reduces genomic DNA methylation in murine adult offspring. Genes Nutr. 2011;6(2):189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Niculescu MD, Craciunescu CN, Zeisel SH. Dietary choline deficiency alters global and gene-specific DNA methylation in the developing hippocampus of mouse fetal brains. FASEB J. 2006;20(1):43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mehedint MG, Craciunescu CN, Zeisel SH, Cousins RJ. Maternal dietary choline deficiency alters angiogenesis in fetal mouse hippocampus. Proc Natl Acad Sci. 2010;107(29):12834–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Robert AW, Randy LJ. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Shin-ichiro T, Takashi T, Chisa M, Masaki O. Major and essential role for the DNA methylation mark in mouse embryogenesis and stable association of DNMT1 with newly replicated regions. Mol Cell Biol. 2007;27:8243–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.DiTroia SP, Percharde M, Guerquin M, Wall E, Collignon E, Ebata KT, Mesh K, Mahesula S, Agathocleous M, Laird DJet al. Maternal vitamin C regulates reprogramming of DNA methylation and germline development. Nature. 2019;573(7773):271–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Blaschke K, Ebata KT, Karimi MM, Zepeda-Martínez JA, Goyal P, Mahapatra S, Tam A, Laird DJ, Hirst M, Rao Aet al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500(7461):222–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ebata KT, Mesh K, Liu S, Bilenky M, Fekete A, Acker MG, Horst M, Garcia BA, Ramalho-Santos M. Vitamin C induces specific demethylation of H3K9me2 in mouse embryonic stem cells via Kdm3a/b. Epigenetics Chromatin. 2017;10(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13(2):115–26. [DOI] [PMC free article] [PubMed] [Google Scholar]