Abstract
Iron is unique among all minerals in that humans have no regulatable excretory pathway to eliminate excess iron after it is absorbed. Iron deficiency anemia occurs when absorbed iron is not sufficient to meet body iron demands, whereas iron overload and subsequent deposition of iron in key organs occur when absorbed iron exceeds body iron demands. Over time, iron accumulation in the body can increase risk of chronic diseases, including cirrhosis, diabetes, and heart failure. To date, only ∼30% of the interindividual variability in iron absorption can be captured by iron status biomarkers or iron regulatory hormones. Much of the regulation of iron absorption may be under genetic control, but these pathways have yet to be fully elucidated. Genome-wide and candidate gene association studies have identified several genetic variants that are associated with variations in iron status, but the majority of these data were generated in European populations. The purpose of this review is to summarize genetic variants that have been associated with alterations in iron status and to highlight the influence of ethnicity on the risk of iron deficiency or overload. Using extant data in the literature, linear mixed-effects models were constructed to explore ethnic differences in iron status biomarkers. This approach found that East Asians had significantly higher concentrations of iron status indicators (serum ferritin, transferrin saturation, and hemoglobin) than Europeans, African Americans, or South Asians. African Americans exhibited significantly lower hemoglobin concentrations compared with other ethnic groups. Further studies of the genetic basis for ethnic differences in iron metabolism and on how it affects disease susceptibility among different ethnic groups are needed to inform population-specific recommendations and personalized nutrition interventions for iron-related disorders.
Keywords: iron, iron deficiency, iron overload, ethnicity, genetics, hemochromatosis, polymorphism, nutrigenomics, mutation
Introduction
Iron is an essential trace element involved in numerous metabolic processes, including oxygen transport and utilization, cellular proliferation, DNA synthesis, neurotransmitter synthesis, and energy production (1, 2). Iron is the fourth most common element in the Earth's crust (1, 3), yet iron deficiency (ID) remains prevalent, affecting an estimated 2.5 billion people worldwide (4). Iron overload (IO), on the other hand, is associated with adverse health outcomes caused by the accumulation of iron in organs, particularly the pancreas, liver, and heart. Elevated iron stores are associated with increased risk of type 2 diabetes, independent of inflammation (5, 6), and increased risk of cardiovascular disease (7, 8), liver fibrosis, and cancer (9–11). Moreover, age-related iron accumulation in the brain strongly predicts cognitive decline, motor impairment, and the development of neurodegenerative diseases (12, 13). A causal role of enhanced iron status in these diseases has been noted in animal models of IO (14, 15).
Increased attention has been focused on the genetic contributions to iron status. Population differences in the frequency of genetic variants that are associated with increased risk of iron disorders may explain varying iron status in different ethnic groups. No studies to date have pooled existing data to summarize current findings and knowledge on ethnic differences in iron status. This review summarizes data on genetic variants found to be associated with iron metabolism across different ethnic groups and statistically evaluates published data on iron status as a function of ethnicity.
Current Status of Knowledge
Iron physiology
Iron absorption
Dietary iron is ingested as heme iron (from animal-based foods) and nonheme iron (from animal- and plant-based foods). Heme iron is only minimally impacted by iron stores. The proteins involved in heme iron absorption are unique to heme iron and the pathways involved in this process continue to be elucidated (16–18). In contrast, nonheme iron absorption is tightly regulated in response to body iron demands and mutations in the proteins involved in nonheme iron absorption are associated with known iron-related diseases (16, 19, 20).
Dietary nonheme iron is ingested primarily as ferric iron (Fe3+) that must be reduced to ferrous iron (Fe2+) (by duodenal cytochrome b) before it enters the enterocyte via the divalent metal transporter 1 (DMT1). Once absorbed, Fe2+ can either be stored as ferritin, used for intracellular functions, or exported via the only known nonheme iron export protein in the body, ferroportin (FPN). Once exported, the ferroxidase hephaestin oxidizes Fe2+ to Fe3+, which can then bind to transferrin (TF) in the bloodstream (16, 20, 21). At other locations in the body, ceruloplasmin functions as a ferroxidase (22), and the placenta also expresses a unique ferroxidase (zyklopen) (23).
Systemic iron homeostasis
Transferrin-bound Fe3+ in circulation is taken up by cells using receptor-mediated endocytosis via transferrin receptor (TFR) 1 (TFR1). Cellular expression of TFR1 is highest in erythroid tissue to support erythropoiesis. Under normal circumstances, ∼30% of the iron-binding sites in the plasma TF pool are occupied. This value is referred to as transferrin saturation (TSAT) (16, 24). When TSAT exceeds 45%, iron begins to circulate free or bound to low-molecular-weight molecules (citrate, albumin), generating potentially toxic iron species known as non–transferrin-bound iron (NTBI). The pancreatic cells and hepatocytes can internalize NTBI via Zrt- and Irt-like protein 14, while cardiomyocytes are purported to internalize NTBI through L-type or T-type calcium channels (25, 26). Cellular uptake of NTBI can increase the intracellular labile iron pool, resulting in generation of reactive oxygen species that can cause oxidative damage, adversely impacting specific organs and, over time, increasing the risk of chronic diseases.
Only 1–2 mg of absorbed iron/d is needed to offset the typical amounts of endogenous daily iron losses. The majority of iron utilized to support erythropoiesis (20–25 mg/d) is obtained from catabolism of senescent RBCs. Several hormones are now known to be involved in the regulation of systemic iron homeostasis, including hepcidin, erythropoietin, and erythroferrone.
Hormonal control of iron physiology
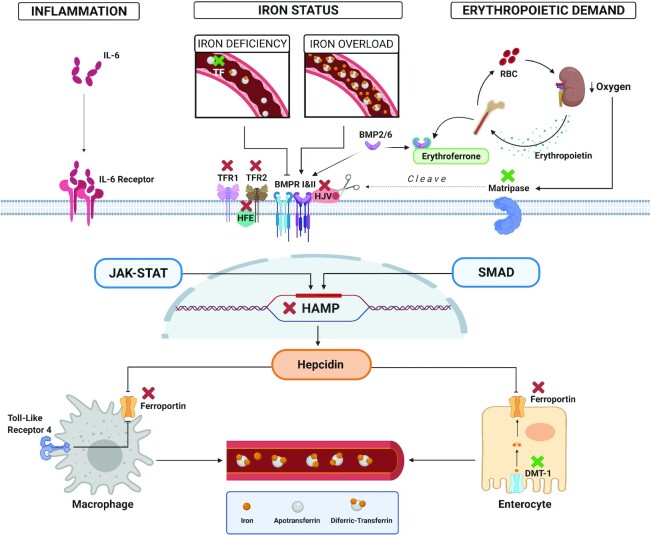
Of the 3 iron-related hormones, hepcidin is the major regulator of body iron balance. This hepatic hormone binds to FPN, thereby reducing iron export from the enterocyte and iron release from stores (27). Hepcidin production is induced by iron loading or inflammation/infection and suppressed by increased erythropoietic demand or hypoxia. The regulation of hepcidin involves multiple membrane proteins [matriptase-2 (MT-2), hemojuvelin (HJV), bone morphogenetic protein receptor (BMPR) 1 and 2 (BMPR1 and BMPR2), TFR1 and TFR2, high iron protein (HFE), and IL-6 receptor], intracellular signaling pathways [bone morphogenetic protein–small mothers against decapentaplegic (BMP-SMAD), Janus kinase–signal transducer and activator of transcription protein (JAK-STAT)], and hormones (erythropoietin, erythroferrone) (21, 28–30). Genetic mutations in these proteins are associated with iron disorders as shown in Figure 1.
FIGURE 1.
Genetic mutations in hepatic hepcidin signaling pathways. Hepcidin is regulated by infection/inflammation (left), iron status (middle), and erythropoietic demand (right). Pathogenic mutations in genes that encode proteins involved in the regulation of the hepcidin or FPN result in iron deficiency (green X) or iron overload (red X). When serum transferrin is saturated, diferric-transferrin binds to TFR1 and displaces HFE. HFE can then form a complex with TFR2 and possibly HJV to promote the BMP-SMAD hepcidin signaling pathway. Recessive mutations in the genes encoding these proteins (HFE, TFR2, HJV, hepcidin) result in decreased hepcidin production preventing hepcidin from being upregulated as iron stores accumulate leading to iron overload. Under low oxygen conditions, MT-2 cleaves HJV, generating a soluble and inactive form of this protein, resulting in the inactivation of the BMP-SMAD signaling pathway and downregulation of hepcidin transcription. Genetic mutations in MT-2 result in uninhibited hepcidin production leading to IRIDA. High erythropoietic demand results in upregulation of erythroferrone, which suppresses hepcidin production by sequestering BMP6 and inhibiting BMP-SMAD signaling. In inflammation, IL-6 binds to IL-6 receptor, stimulating the JAK-STAT pathway and upregulating hepcidin production. Dominant gain- and loss-of-function FPN1 mutations result in hepcidin resistance or in cellular iron accumulation (particularly in macrophages), respectively. Recessive mutations in DMT1 result in iron loading anemia. Likewise, recessive mutations in TF result in reduced concentrations of functional TF and iron loading anemia. The figure was created using BioRender.com. BMP, bone morphogenetic protein; BMPR, bone morphogenetic protein receptor; DMT-1, divalent metal transporter 1; FPN, ferroportin; HAMP, hepcidin, HFE, high iron protein, HJV, hemojuvelin; IRIDA, iron refractory iron deficiency anemia; JAK-STAT, Janus kinase-signal transducer and activator of transcription protein; MT-2, matriptase-2; SMAD, small mothers against decapentaplegic; TF, transferrin; TFR, transferrin receptor.
Assessment of iron status
Multiple biomarkers are used to represent iron status of individuals and/or populations. Ferritin is the primary intracellular iron storage protein and some is released into circulation as serum ferritin (SF) (31). Low SF concentrations are indicative of depleted iron stores. Elevated concentrations of SF can indicate excessive iron stores or inflammatory conditions since ferritin is an acute-phase protein (31). Total iron binding capacity (TIBC) indicates the total number of binding sites for iron in TF, which is the main iron transport protein in circulation. TIBC, in combination with serum iron, can be used to calculate TSAT, which reflects iron supply to tissues (31). Soluble transferrin receptor (sTfR) reflects the intensity of erythropoietic and cellular demands for iron (31). An increase in sTfR concentrations indicates an increase in cellular iron requirements or insufficient iron supply to tissues (31). In late stages of ID, hemoglobin production is compromised, which can result in anemia (31). A summary of the typical cutoffs for ID and/or IO for common iron status indicators is provided in Supplemental Table 1.
Genetic variants of iron metabolism
Tight control of iron absorption is needed to prevent ID or IO, but to date, only ∼30% of the interindividual variability in iron absorption can be captured by hepcidin or other iron status biomarkers (32–36). Marked ethnic differences in the risk of ID or IO have been known to exist, as highlighted by the CDC cutoffs used to denote anemia (37). To investigate the genetic contributions to variations in iron status between and within populations, multiple genome-wide association studies (GWASs) and candidate gene association studies have been undertaken. The following sections cover pathogenic mutations in iron-related genes identified to date that result in ID or IO, and polymorphisms within iron- and non–iron-related genes found to be associated with variations in iron traits among healthy populations. Genetic variants associated with iron status in healthy populations stratified by ethnic group are summarized (Supplemental Table 2).
Genetic variants related to IO
Genetic mutations in proteins involved in the regulation of the hepcidin-FPN axis result in aberrant expression of hepcidin or FPN (38) and can eventually lead to IO. Disorders of the hepcidin-FPN axis cause subtypes of hereditary hemochromatosis (HH), a heterogeneous genetic condition found primarily in Northern Europeans. The most common form of HH occurs from mutations in HFE, but HH can also result from mutations in HJV, hepcidin gene (HAMP), TFR2, and FPN1, which are collectively referred to as non-HFE hemochromatosis. The pattern of inheritance of pathogenic mutations in HFE, HJV, HAMP, and TFR2 is autosomal recessive, whereas in FPN1 is autosomal dominant (39). While mutations in these iron-related genes have been described as causative for HH, some single nucleotide polymorphisms (SNPs) within, or in close proximity to these genes, have been associated with iron traits in healthy populations.
HFE
HFE was named as the high iron (high Fe) or hemochromatosis protein when it was discovered in 1996 to harbor a mutation that was highly prevalent in patients with hemochromatosis (40). This major histocompatibility complex class I–like protein stimulates the intracellular BMP-SMAD hepcidin signaling pathway. Recessive mutations in HFE result in decreased hepcidin production and, thus, hepcidin is not appropriately upregulated as iron stores accumulate (41) (Figure 1). HFE-related hemochromatosis or HH type 1 is most commonly associated with 2 missense mutations, C282Y and H63D. The iron burden and clinical phenotype presented in patients with HFE-related hemochromatosis is highly variable. The classic biochemical abnormalities seen in C282Y homozygous individuals include elevated TSAT and SF and tissue IO. Although biochemical penetrance of homozygosity for this mutation is high (75–100% in males, 40–60% in females) (42–44), the clinical penetrance is much lower, affecting men at higher rates than women (2–38% and 1–10%, respectively) (45, 46). Interestingly, H63D homozygosity rarely results in clinical disease development, except when this mutation is present with C282Y, both in heterozygote states (40, 47).
The prevalence of the C282Y and H63D mutations varies among ethnic groups and is one of the best examples of ethnic differences in iron metabolism (48). The C282Y homozygosity is most prevalent (0.3–0.5%) in individuals of Northern European descent (44, 49–52). This mutation is thought to have originated in a Celtic population 60–70 generations ago (53–55). A Viking origin of this mutation has been proposed as the highest frequencies are observed in populations of Northern European descent (i.e., Viking populations) (56). The Hemochromatosis and Iron Overload Screening (HEIRS) Study reported the highest prevalence (0.44%) of C282Y homozygosity among unrelated non-Hispanic white individuals (44). The prevalence of C282Y homozygosity has been reported at even higher frequencies in specific regions of Northern Europe, particularly Ireland (1.24–1.96%) (57, 58). In addition, the highest average allele frequency in HFE has been reported among this population (10.1%) (48). The lowest prevalence of C282Y homozygosity in the HEIRS study was reported among Asians (0.000039%), followed by Pacific Islanders (0.0132%), African Americans (0.014%), Hispanics (0.027%), and Native Americans (0.11%) (44).
The H63D mutation has a broader distribution, with higher frequencies throughout Europe and other geographical locations (48). The estimated prevalence of H63D homozygosity in the HEIRS study was 2.4% in non-Hispanic whites, 1.3% in Native Americans, 1.1% in Hispanics, 0.089% in African Americans, and 0.02% in Pacific Islanders and Asians (44). The prevalence of C282Y/H63D compound heterozygosity reported in the HEIRS study was 2.0% in non-Hispanic whites, 0.77% in Native Americans, 0.33% in Hispanics, 0.071% in African Americans, 0.096% in Pacific Islanders, and 0.0055% in Asians (44).
SNPs in HFE corresponding to C282Y and H63D have been associated with iron and erythrocyte traits in healthy populations. The SNP rs1800562 that results in the C282Y variant has been associated with several iron biomarkers (59–63) and hematological parameters (64) that reflect both systemic and cellular iron homeostasis at the genome-wide level among Europeans. The SNP rs1799945 that results in the H63D variant has also been associated at the genome-wide level with various iron traits (59, 65) in Europeans. In Hispanics, both rs1800562 and rs1799945 have been shown to be associated with iron status biomarkers (66). In African Americans, only the association of rs1800562 with SF and TF seen in Europeans has been replicated (67). It is noteworthy that these variants are nearly absent in Asians and Pacific Islanders (44, 52), and no associations between these mutations and iron biomarkers have been reported in these populations (68).
HJV and HAMP
HJV encodes the hemojuvelin protein, which acts as a BMP co-receptor to regulate the expression of HAMP (21) (Figure 1). Juvenile hemochromatosis (JH) or HH type 2 is the most severe form of HH (69, 70) and can arise from pathogenic mutations in HJV (HH subtype 2a) or HAMP (HH subtype 2b) and results in cardiomyopathy, diabetes, and hypogonadism by the early 20s (69, 70). In rare instances, adult-onset HH due to HJV mutations has been observed (71, 72). Biochemical abnormalities of JH include high TSAT and marked increases in SF (70). Interestingly, a systematic review assessing the genotypic and phenotypic spectrum of HJV mutations in patients with HH reported ethnic disparities in the clinical presentation of HJV-related HH between Europeans and East Asians (73). This same review (73), in agreement with a phenotypic analysis of HH subtypes (70), found that European males and females were affected equally, while East Asian males were affected at higher rates than their female counterparts (73).
The first comparative study of the prevalence of HFE- and non–HFE-related HH reported a predicted prevalence of homozygous HJV pathogenic mutations of 1 in 5 million using available next-generation sequence (NGS) databases, with the highest predicted prevalence in South Asians (74). Homozygous HAMP pathogenic mutations were predicted to be even rarer (1 in 182 million) (74). Of note, these predictions may be restricted given that the populations were not representative of all the pathogenic mutations within the HH-related genes identified to date.
Mutations in HJV account for up to 90% of JH cases (73), and most have been identified in a single family or small populations (73). The G320V mutation is a more common HJV mutation and is restricted to European ancestry (75, 76). Other mutations have been described only within individuals of Asian or Pacific Island ancestry (73, 77), some are more common among East Asians (e.g., C321X, Q312X) (71, 78–85), and some among South Asians (e.g., G336X, G99R, P192L, L194, C80Y, A343PfsX23) (77, 86, 87). Few mutations have been described in Africans (e.g., R385X, A310G) (73, 88, 89); however, these are not restricted to African ancestry (90–93). Mutations in HAMP result in a rare form of JH and have been characterized in patients from varying geographical locations (87, 93, 94–102).
Interestingly, no polymorphisms within HJV or HAMP have been associated with variation in iron traits among healthy individuals. A candidate gene association study reported an association between the SNP rs10421768 in HAMP and hemoglobin among a Kenyan cohort (n = 628) (103), but the significance of this association disappeared after adjustment for multiple testing.
TFR2
TFR2 forms a complex with HFE and possibly HJV to promote BMP-SMAD signaling and upregulate hepcidin production (29) (Figure 1). HH type 3 results from pathogenic mutations in TFR2 and is a disease mainly of adult onset, although more severe mutations have been described in early childhood (104–106). Biochemical abnormalities include elevated TSAT and SF (24). Although the true prevalence of TFR2 mutations is unknown, homozygous TFR2 pathogenic genotypes have been estimated (using available NGS databases) to be 1 in 6 million and were predicted to be most frequent among non-Finnish European populations from the ExAC database (74).
Most mutations in TFR2 that result in HH type 3 have been clustered in ∼45 families worldwide (107), but some mutations have been described among unrelated individuals from different ethnic groups (105, 108–111). TFR2-related HH may be the leading cause of IO in Asians, predominantly those from Japan (112, 113). The I238M variant is present in Asians at a higher frequency (0.0192) (112); however, it is not restricted to Asian ancestry (93).
The rs7385804 SNP in TFR2 has been associated with iron biomarkers (59, 64, 65) and RBC parameters (59, 114) at genome-wide significance in healthy Europeans. The associations of rs7385804 with serum iron and TSAT have been replicated in Hispanics (66) and Chinese women (115). Another SNP within TFR2, rs7786877, has been associated with mean corpuscular volume (MCV) at the genome-wide level among individuals of European ancestry (64).
FPN
The FPN1 gene encodes FPN, which regulates cellular export of non-heme iron (116) (Figure 1). Pathogenic mutations in FPN1 can result in 2 phenotypically distinct diseases, HH type 4A (FPN disease) and HH type 4B. HH type 4B is caused by gain-of-function mutations resulting in partial or complete hepcidin resistance (117). Individuals with these mutations present with high SF, TSAT, organ IO, and progressive organ damage (117). The phenotype of HH type 4B resembles other recessive HH-causing mutations (117). Conversely, FPN disease is due to dominant loss-of-function mutations that result in impaired iron export, particularly from reticuloendothelial cells (117). The classical phenotype of FPN disease includes high SF, normal to low TSAT, low hemoglobin concentrations, and progressive iron loading (117, 118). FPN disease seems to have a milder clinical presentation as it has not been shown to cause major organ damage (119, 120), conceivably because macrophages protect against reactive oxygen species when burdened with iron (117). Not all FPN mutations have been classified based on their phenotypic presentation or pathogenicity (117, 119).
Globally, the estimated pathogenic genotype carrier rate of FPN1 mutations (using available NGS databases) is 1 in 1373 (74). The highest predicted pathogenic genotype carrier rate was reported in African Americans (0.25%), followed by Americans (0.039%), East Asians (0.033%), and non-Finnish Europeans (0.03%) (74). A systematic meta-analysis of FPN1 mutations found 31 disease-causing mutations in 161 individuals by 2010 (119), and more recently, a total of 60 variants in 359 individuals was described between 1999 and 2019 (120). The most frequently reported FPN1 mutation is the V162del loss-of-function mutation (119–126).
Few polymorphisms in FPN1 have been associated with iron traits in healthy populations. Q248H is the most common FPN1 variant among individuals of African ancestry and is present at polymorphic frequencies in African populations (2.2–13.4%) (127–131). The Q248H variant has been weakly associated with increased SF concentrations among individuals with primary IO and healthy individuals (127–129, 131–133), and this association seems to be stronger in men (132, 133). This polymorphism may confer a protective effect against ID (128, 134), anemia, and iron deficiency anemia (IDA) (134) in African children, particularly those with underlying inflammation. While conflicting (134), a protective effect against malarial infection has also been suggested. The largest GWAS on iron status conducted in healthy Europeans identified an SNP (rs744653) near FPN1 that was associated with SF and TF at genome-wide significance (59), and the association with SF was replicated in a recent candidate gene association study among Europeans (135). Last, a GWAS conducted in healthy Chinese men found an SNP (rs5742933) located in close proximity to the FPN1 gene that was associated with SF at the genome-wide level (136). However, this SNP seemed to be in weak linkage disequilibrium (R2 and D′ <0.20) with SNPs located within FPN1 (136).
Genetic variants related to ID
As with IO, several SNPs in iron-related genes have been associated with increased risk of anemia, ID, or IDA in healthy populations of various ethnicities. Hereditary disorders that result in anemia include iron-refractory iron deficiency anemia (IRIDA) and iron loading anemia.
TMPRSS6
MT-2 is produced in the liver and negatively regulates expression of hepcidin by cleaving HJV and, in turn, inactivating the BMP-SMAD pathway (137) (Figure 1). Genetic mutations in the MT-2 protein [transmembrane serine protease 6 (TMPRSS6)] can result in uninhibited hepcidin production leading to IRIDA (138–140). This disease is characterized by congenital hypochromic, microcytic anemia; low MCV and TSAT; low to normal SF concentrations; and defective iron absorption and utilization (138, 141). IRIDA is usually unresponsive to oral iron, and only partially responsive to parenteral iron treatment (138).
The prevalence of pathogenic mutations resulting in IRIDA is unknown (140–142). So far, 69 different TMPRSS6 mutations in 65 IRIDA families of different ethnicities have been identified (137, 141–144). Most IRIDA patients have homozygous mutations in TMPRSS6 and, thus, the mode of inheritance is considered to be recessive (143). However, heterozygous mutations may also result in a clinical phenotype that resembles IRIDA but with a milder presentation (141, 143). More recent evidence suggests that IRIDA is a highly heterogeneous disease and some patients have been found to respond to oral iron therapy (143).
Several genome-wide and candidate gene association studies conducted in healthy populations of mainly non-African origin have found common SNPs in TMPRSS6 to be associated with iron traits. The most reported SNP in TMPRSS6, rs855791 (V736A), has been associated with iron status indicators (59, 62, 145, 146), RBC parameters (59, 64, 147), and liver iron content (148) in healthy individuals of European ancestry at genome-wide significance. In addition, candidate gene studies in Europeans have replicated the association of this SNP with SF (135, 149) and serum iron (149) and have identified associations with sTfR (61) and the sTfR-to-SF ratio (150). This SNP was also associated with hemoglobin (6, 115) and iron status indicators (6, 115) in East Asians, and with serum iron in Hispanics (66). A systematic review with meta-analysis found the minor allele (A) frequency (MAF) of rs855791 to be significantly higher in Asians than in Europeans (0.55 vs. 0.42) (151). This same review showed that each A allele was associated with 0.11-g/dL lower hemoglobin concentrations, 3.71-μg/L lower SF concentrations, and 0.2-mg/L higher sTfR, and the differences in effect estimates between ethnicities were not significant (151). Significant differences in MAF between Africans and non-Africans have been reported (152). The rs855791 SNP has a significantly lower MAF in Africans (<0.1) compared with non-Africans (<0.35), yet Africans have a high prevalence of anemia (152).
The second most reported SNP in TMPRSS6 is rs4820268 (D521D) and it has been associated with iron biomarkers (60, 65, 146), and hemoglobin in Europeans (147), with SF and hemoglobin in East Asians (6), and with hemoglobin in South Asians (147). The association with serum iron was replicated in a candidate gene study of Europeans and an association with SF was identified (149). A study in Europeans found the rs4820268 GG genotype to be associated with lower serum iron, hemoglobin, MCV, and mean cell hemoglobin, and higher TF, sTfR, and sTfR:SF (65). Furthermore, a meta-analysis of 13 study populations showed that the G allele resulted in a 0.12-μg/L increase in SF concentrations among Europeans and a 3.69-μg/dL decrease among Asians (151). Analysis of 3 studies among Europeans suggested an association between the G allele and a reduction in sTfR (151). However, there was high heterogeneity of the effect of each allele on the variation in iron status parameters reported in studies among Europeans included in this meta-analysis (151). Consistent with these observations, a recent study assessing differences in allele frequencies among different populations found that South and East Asians had the highest number of iron-lowering alleles and Africans had the lowest number of low-iron risk alleles (152). Furthermore, studies in Chinese populations identified rs855791 and rs4820268 polymorphisms as genetic risk factors for developing anemia, ID, and IDA (6, 115). Several other SNPs in TMPRSS6 have been associated with RBC parameters (64, 147), and iron biomarkers (146) among different ethnic groups.
TF
Autosomal recessive mutations in TF cause severely reduced serum concentrations of functional TF and lead to hypotransferrinemia (or atransferrinemia) (Figure 1). This rare disorder is characterized by iron-deficient erythropoiesis and anemia due to insufficient iron supply to erythropoietic tissues and severe IO in non-hematopoietic organs due to low hepcidin concentrations and increased non–TF-mediated iron uptake (153, 154). Only 18 cases among 16 families worldwide have been described to date (155, 156).
While mutations that result in hypotransferrinemia are extremely rare, several SNPs in TF have been associated with iron status in populations from different ethnic origins. The rs3811647 SNP in TF has been associated with TIBC and TF in genome-wide and candidate gene association studies in Europeans (60, 63, 65, 150, 157). Other SNPs in TF have been associated with serum iron, TF, and TSAT (59). Associations of several polymorphisms in TF with TIBC found in Europeans have been replicated in Asians, Hispanics, and/or African Americans (157). In addition, a candidate gene association study in Chinese women found TF polymorphisms were significantly associated with serum iron, TF, and TSAT (115). Other SNPs in TF have been associated with TSAT and TIBC in Hispanics (66), and with TIBC in African Americans (67). Moreover, the GWAS in African Americans found the top 2 SNPs in TF to explain 11.2% of the variation in TIBC in this population (67). Interestingly, unlike other iron-related genes discussed in this section, in TF, no SNPs have been reported to be associated with hematological traits in any ethnic group.
DMT1
DMT1 transports dietary iron into enterocytes (Figure 1) or out of intracellular endosomes. Pathogenic mutations in DMT1 result in microcytic anemia and severe organ iron loading (141). Despite having body iron excess, patients with defects in DMT1 have normal to mildly elevated SF concentrations (141). Iron loading anemia due to mutations in DMT1 is extremely rare, with only 7 patients from 6 families with homozygotes or compound heterozygotes described to date (141). Despite the importance of DMT1 in iron trafficking, few polymorphisms have been associated with iron traits. In a healthy Turkish cohort, the IVS4+44 polymorphism in DMT1 was associated with interindividual variations in serum iron (158). This same SNP was associated with up to a 4-fold increased risk of developing anemia in Italian children with celiac disease (159).
Other genetic variants related to iron metabolism
Iron-related genes
Polymorphisms in other iron-related genes involved in transport of non-heme and heme iron [TFR1, cytochrome b reductase 1 (CYBRD1), feline leukemia virus subgroup C receptor-related protein (FLVCR), 6-transmembrane epithelial antigen of prostate 3 (STEAP3), cluster of differentiation 163 (CD163)], in the regulation of cellular hepcidin signaling pathways (SMAD8, BMP2, BMP4, BMP9, BMPR1B, BMPR2, neogenin 1 (NEO1), protein convertase suntilisin/kexin type 7 (PCSK7)], or intracellular iron signaling [hypoxia-inducible factor 2ɑ (HIF2A), iron regulatory protein 1 (IRP1] have been associated with or have suggestive associations with ≥1 biomarker of iron status and/or erythrocyte phenotype in healthy populations (59, 61, 135) or act as modifiers of HH phenotype (160–162) among individuals of European descent.
Non–iron-related genes
Although most genetic variants associated with iron status indicators are within or in close proximity to iron-related genes, genetic variants of non–iron-related genes have also been identified. Among Europeans, SNPs in genes involved in lipid metabolism were shown to be associated with TF [N-acetyltransferase 2 (NAT2), aryl hydrocarbon receptor nuclear translocator–like (ARNTL), fatty acid desaturase 2 (FADS2)], SF [ɑ1-3-N-acetylgalactosaminyltransferase and ɑ1-3-galactosyltransferase (ABO), testis expressed 14 (TEX14)] (59), or serum iron [lipoprotein lipase (LPL)] (146). In Hispanics, a SNP in myelin regulatory factor (MYRF) that is in linkage disequilibrium with the FADS2 SNP identified in Europeans (59) was associated with TIBC (66). Moreover, the associations between SNPs in NAT2 with TF and in ABO with SF found in Europeans (59) were generalized to Hispanics (66). Whether such associations result from the confounding influence of variations in lipid metabolism on iron status or reveal a pleiotropic effect of lipid-related genes on iron regulation needs further investigation. Interestingly, in analysis of the UK Biobank samples, ferritin-associated SNPs in HFE and TMPRSS6 conferred significant protection against hypercholesterolemia, suggesting interplay of metabolic pathways between lipid and iron (163). Other SNPs in non–iron-related genes have been associated with iron traits, including TIBC, unsaturated iron-binding capacity, serum iron, and sTfR in Europeans (146, 157).
In African Americans, SNPs in the hepatoma-derived growth factor–like protein 1 (HDGFL1) and MAF bZIP transcription factor (MAF) have been associated with TIBC and in growth factor receptor bound protein 2–associated protein 3 (GAB3) with SF at genome-wide significance (67). Among these associations, only the significant SNP in GAB3 was replicated in Hispanics (66). GAB3 is a member of the GBR2-associated binding protein gene family and is involved in several growth factor and cytokine signaling pathways. The protein encoded by this gene is expressed in hematopoietic tissues and facilitates macrophage differentiation (164, 165).
In East Asians, an SNP in postmeiotic segregation increased 1 (PMS1) was found to be significantly associated with SF (136). PMS1 encodes a protein thought to be involved in DNA mismatch repair pathways and is expressed by various tissues, including hematopoietic tissues. Interestingly, anti-PMS1 antibodies were found in Japanese patients with aplastic anemia (10%), but none were detected in aplastic anemia patients from the United States (166). In addition, while PMS1 has not been shown to play a direct role in iron homeostasis, it is in close proximity to FPN1.
Ethnic differences in iron status
Iron regulation is highly conserved, but risk of iron-related disorders differs across major ethnic groups. These ethnic differences may be a consequence of adaptive changes during evolution that occurred due to limited iron availability, a condition referred to as antagonistic pleiotropy. In antagonistic pleiotropy, adaptive changes that arise from evolutionary adaptations can become deleterious in the current environment. For iron, due to its importance in physiology, strong selective pressures would have been expected to maintain adequate iron status. These adaptations needed in an iron-poor environment (i.e., mutations that increase iron absorption) may now be deleterious when iron is considerably more abundant in our food systems. Understanding the evolutionary context underlying dietary adaptation could have strong implications in precision nutrition (167).
Large epidemiological reports of ethnic differences in iron status indicators have been published over the past few decades. Several studies of North American cohorts have evaluated iron status in large groups of otherwise healthy adults. The largest epidemiological study to date that evaluated iron stores as a function of ethnicity was the HEIRS study. This study recruited 101,168 primary-care adults aged 25 y or older from the United States and Canada, and evaluated SF and TSAT as a function of ethnicity (self-reported as Hispanic, European, African American, Asian, Pacific Islander, Native American) (168). Another large epidemiological study, the Recipient Epidemiology and Donor Evaluation Study–III recruited 12,683 men and women participants of different ethnicities (self-reported as African American, Asian, white, Hispanic, and other) aged 18 y or older who had successfully donated whole blood (169). This study was designed to examine the genetic and metabolic basis of blood donor susceptibility to ID and iron-related symptoms in multiple ethnic groups. The Iron and Atherosclerosis Study (FeAST) was a prospective, randomized, controlled, single-blinded clinical trial to test whether iron reduction using phlebotomy in participants with symptomatic but stable peripheral arterial disease can effectively improve clinical outcomes. This study recruited 1277 European and African-American veterans over the age of 21 (primarily males) (170). An additional study among 1491 African-American and 31,005 European men and women compared hematologic and iron status between these 2 groups (171). Another study designed to determine the frequency of HFE mutations and its association with iron-related genotypes involved 10,198 adults (self-reported as black, Asian, white, and Hispanic) (172). Additional epidemiological data analysis examining iron status among multiple ethnic groups was conducted using the NHANES-III database involving 20,040 individuals aged 18 y or older (grouped as black, white, Hispanic, and other) (173).
Similarly, large-cohort data on iron status have been published in Europe. The population-based SUNSET (Surinamese in The Netherlands: Study on Ethnicity and Health) was a multiethnic, cross-sectional study designed to test the association between SF and the prevalence of type 2 diabetes and fasting glucose concentrations in a total of 2975 adults (174). In addition, 2 other cohort studies targeting women of reproductive age were conducted to examine ethnic differences in iron status (175, 176). Major findings from these data are summarized below by ethnicity.
Europeans
Most research to date has been conducted in Europeans, who are often used as the reference group in comparisons to other ethnic groups. Europeans have been consistently shown to have a lower risk of ID and anemia, and higher hemoglobin concentrations (171, 177–180). In addition, TSAT in Europeans has been found to be significantly higher than mean values observed in African Americans and Hispanics (179, 180).
African Americans
Lower hemoglobin concentrations are consistently reported among African Americans compared with Europeans. These differences have been noted in infancy and appear to be maintained in the elderly (171, 176, 181–184). Existing data from population groups with sample sizes ranging from 388 to 3074 indicate that the hemoglobin distribution observed among African Americans is shifted to the left by ∼0.8 g/dL (184, 185). This difference has been highlighted by the CDC and WHO to promote ethnicity-specific cutoffs for hemoglobin in the diagnosis of anemia (37, 186).
Ethnic differences in the hemoglobin distribution remain significant even after controlling for iron status (SF and TSAT) (182, 184) and/or dietary iron intake (182), suggesting that variation in hemoglobin concentration is not entirely driven by factors related to iron metabolism. Despite lower hemoglobin concentrations, African-American adults have elevated SF concentrations compared with Europeans and Hispanics (44, 170, 173, 183, 186–188). NHANES-III (173, 189), FeAST (170, 188), and HEIRS (179) data all found significantly higher SF in African Americans compared with Europeans. However, both NHANES and HEIRS (179, 180) and Li et al.’s genetic study in African Americans (67) found that African ancestry was associated with decreased concentrations of serum iron and TSAT as well as increased concentrations of sTfR. Moreover, the prevalence of ID and anemia in HEIRS was greater in African Americans than in Europeans and Asians among women of reproductive age (177, 190, 191). At present, whether the elevated SF concentrations in African Americans reflect increased systemic inflammation or elevated iron stores is unclear. African-American populations are often at increased risk for obesity compared with other ethnic groups (192), and elevated BMI is positively correlated with SF concentrations, perhaps due to adiposity-induced inflammation. The lower TSAT and hemoglobin concentrations but higher SF concentrations observed in African Americans might indicate that the mobilization of iron from stores for erythropoiesis is reduced due to some unknown genetic contributors (182).
East Asians
Asian populations can be further subdivided into East Asians or South Asians based on geographical distribution and ethnicity. Although data in Asian groups are not as abundant, numerous studies have reported that East Asian populations exhibited higher iron stores and a higher risk of IO. This finding is evident despite the fact that the frequencies of the most common HFE mutations are lowest among Asians (44). Of note, HEIRS individuals that self-identified as Asians (predominantly East Asians: Chinese, Japanese, Vietnamese, and Filipino) exhibited the highest SF, TSAT, or both SF and TSAT compared with any other population group studied, even after excluding polymorphisms associated with IO (44, 191, 193). These differences remained significant after adjusting for diabetes or liver disease (193). In a further analysis of HEIRS data focused on women of reproductive age, Asian ancestry was associated with a decreased risk of ID and with increased iron stores compared with other ethnic groups independent of known HH mutation in HFE (C282Y) (191). Additional epidemiologic data from a study in Korean adults (n = 4904) found mean TSAT in both females and males was significantly higher than in Europeans as reported in the HEIRS study (194, 195). Few data exist on possible mechanisms explaining these ethnic differences. A recent functional study evaluated iron absorption in a group of young East Asian women (32) and found mean percentage of iron absorption was significantly higher in East Asian women compared with that reported in European women using the same methodology, even after correcting for a fixed amount of SF (32). This observation suggests that the storage threshold at which East Asians downregulate iron absorption is higher, supportive of increased risk of IO in East Asian populations at maturity. This may also explain the observation that East Asian populations have a greater risk of diabetes at a lower BMI (196, 197).
South Asians
Fewer data are available from South Asian populations, but existing data in this group suggest a lower iron status compared with Europeans and East Asians. Data from South Asian Surinamese adults (Hindustani Surinamese, n = 399) aged 35–60 y showed significantly lower SF concentrations than observed among a cohort of Dutch adults (n = 508) (174). South Asian pregnant women (primarily from Pakistan and Sri Lanka, n = 198) were reported to have the highest risk of ID and anemia when compared with pregnant European (n = 326) and East Asian (n = 43) women (177). Lower hemoglobin, SF, and TSAT have also been noted in a group of South Asian women of reproductive age (176).
Hispanics
At least 2 epidemiological studies have reported a significantly higher prevalence of ID in Hispanics (178, 190) compared with other ethnic groups, and Hispanic ethnicity has been associated with an increased risk of ID (191). In addition, evidence from NHANES found that Hispanic women of reproductive age had significantly lower SF and TSAT compared with European women (180).
Native Americans
Data on iron status among Native American populations are limited. The HEIRS study included data from 645 Native Americans. In this relatively small cohort, mean TSAT and SF concentrations in Native American men and women did not appreciably differ from the respective mean values reported in European men and women (198).
Statistical evaluation of population-based data on iron status
Multiple published reports of ethnicity and iron status exist but few attempts have been made to compile these data to explore statistical patterns of altered iron status. To address this gap, relevant population-based data were identified through PubMed, Web of Science, and Scopus using the following key words: ethnicity, race, genetic, Asian, European, Caucasian, African American, Chinese, Korean, genetics, iron, iron status, iron homeostasis, iron absorption, iron metabolism. Additional studies were identified through references cited within relevant articles. To systematically compare the data summarized above on iron status among different ethnical groups, we extracted iron status data from both population-based studies involving multiple ethnicities and other epidemiological or observational studies reporting data from 1 ethnic group only. We categorized studies by the number of ethnic groups involved and by the study type (Supplemental Table 3). For analytical purposes, we excluded studies designed to assess iron status in frequent blood donors or pregnant and/or breastfeeding women, as this would be expected to impact iron status. Studies were also excluded if there were no available iron status data to extract or if the iron status data were not reported by sex. As for different literature examining the same study cohort, data from the largest sample size were included in the analyses.
To explore possible differences in iron status indicators (SF, TSAT, and hemoglobin) between different ethnic groups, linear mixed-effects models were constructed with ethnicity (East Asian, European, African American, Hispanic, and South Asian) as a fixed-effect variable. Studies where the data were collected from were considered as a random-effect variable. The sample size of each ethnic group in these studies was used as weight for the fixed-effect variable and the mean age of each ethnic group was controlled for in the analysis. Mean age could not be controlled for models examining hemoglobin as this restricted the size of the dataset because many of the available studies did not report the mean age. Estimated marginal means of each iron status indicator were calculated for all ethnic groups using the package emmeans in R (R Foundation for Statistical Computing). All statistical analyses were performed using R version 3.4.3.
The estimated marginal means of iron status indicators evaluated are presented in the Table 1. A statistical evaluation of mean differences in each indicator between ethnic groups is presented in Table 2. With this approach, Asians (predominantly East Asians) were found to exhibit significantly higher SF concentrations and TSAT when compared with Europeans, African Americans, or South Asians, and this difference was significant for both males and females. While significant effects were evident in both sexes, the magnitude of the observed difference was significantly higher in males. This finding is expected as women of reproductive age have monthly losses of iron from menses, which may partially explain why risk of excess iron accumulation is greater in males. Evidence has shown that 1 μg SF/L is equivalent to 8 mg of iron stores (199); thus, the mean difference of 180 μg/L in SF would be translated into an additional 1440 mg of storage iron. If 1.5 mg of iron is absorbed daily (21), this additional 1440 mg of storage iron would be equivalent to the net amount of iron typically absorbed over 2.6 y.
TABLE 1.
Mean reported iron status indicators in different ethnic groups1
| Men | Women | |||||
|---|---|---|---|---|---|---|
| SF, μg/L | TSAT, % | Hb, g/dL | SF, μg/L | TSAT, % | Hb, g/dL | |
| Asian | 249 ± 17 [14] (40,874) | 37 ± 1 [6] (22,543) | 15.1 ± 0.1 [8] (20,265) | 113 ± 7 [13] (31,036) | 29 ± 1 [7] (13,075) | 13.1 ± 0.1 [9] (9706) |
| European | 68 ± 16 [15] (48,115) | 32 ± 1 [8] (38,532) | 14.8 ± 0.1 [10] (28,169) | 27 ± 7 [20] (62,736) | 25 ± 1 [10] (45,433) | 13.5 ± 0.1 [13] (33,352) |
| African American | 87 ± 16 [9] (14,025) | 30 ± 1 [8] (13,839) | 14.2 ± 0.1 [5] (2882) | 45 ± 7 [8] (20,413) | 22 ± 1 [7] (19,908) | 12.7 ± 0.2 [4] (1350) |
| South Asian | 57 ± 35 [1] (150) | 32 ± 5 [1] (150) | — | 35 ± 16 [2] (488) | 18 ± 2 [2] (242) | 12.6 ± 0.5 [1] (188) |
| Hispanic | 151 ± 83 [2] (5549) | 29 ± 4 [1] (5122) | 15.0 ± 0.2 [1] (427) | 54 ± 35 [2] (7784) | 25 ± 4 [1] (7241) | 13.3 ± 0.3 [1] (543) |
Values are estimated marginal means ± SEs [number of studies included in the analyses] (number of people included). The estimated marginal means were calculated from linear mixed-effects models using the lmer and emmeans functions in R (R Foundation for Statistical Computing). Hb, hemoglobin; SF, serum ferritin; TSAT, transferrin saturation.
TABLE 2.
Comparison of iron status indicators between comparison ethnic groups and reference ethnic groups1
| Reference ethnic group | Comparison ethnic groups | Men | Women | |||
|---|---|---|---|---|---|---|
| Iron indicator | Difference ± SE | n 2 | Difference ± SE | n 2 | ||
| SF, μg/L | Asian | [14] (40,874) | [13] (31,036) | |||
| European | −180 ± 53 | [15] (48,115) | −86 ± 33 | [20] (62,736) | ||
| African American | −161 ± 73 | [9] (14,025) | −68 ± 33 | [8] (20,413) | ||
| South Asian | −192 ± 303 | [1] (150) | −78 ± 163 | [2] (488) | ||
| European | [15] (48,115) | [20] (62,736) | ||||
| African American | 19 ± 44 | [9] (14,025) | 19 ± 23 | [8] (20,413) | ||
| TSAT, % | Asian | [6] (22,543) | [7] (13,075) | |||
| European | −5 ± 13 | [8] (38,532) | −3 ± 03 | [10] (45,433) | ||
| African American | −7 ± 13 | [8] (13,839) | −6 ± 03 | [7] (19,908) | ||
| South Asian | −5 ± 5 | [1] (150) | −11 ± 23 | [2] (242) | ||
| European | [8] (38,532) | [10] (45,433) | ||||
| South Asian | −1 ± 5 | [1] (150) | −7 ± 25 | [2] (242) | ||
| African American | −2 ± 16 | [8] (13,839) | −3 ± 03 | [7] (19,908) | ||
| Hb, g/dL | African American | [5] (2882) | [4] (1350) | |||
| Asian | 0.8 ± 0.13 | [8] (20,265) | 0.4 ± 0.2 | [9] (9760) | ||
| European | 0.6 ± 0.13 | [10] (28,169) | 0.8 ± 0.23 | [13] (33,352) | ||
| Hispanic | 0.8 ± 0.13 | [1] (427) | 0.6 ± 0.3 | [1] (543) | ||
| Asian | [8] (20,265) | [9] (9760) | ||||
| European | −0.2 ± 0.1 | [10] (28,169) | 0.4 ± 0.17 | [13] (33,352) | ||
The estimated differences were calculated from linear mixed-effects models using the lmer and emmeans functions in R (R Foundation for Statistical Computing). P values reported were corrected for multiple comparisons with Tukey's test. Hb, hemoglobin; SF, serum ferritin; TSAT, transferrin saturation.
n [number of studies included in the analyses] (number of people included).
Significant difference between the comparison group and the reference group, P < 0.0001.
Significant difference between the comparison group and the reference group, P = 0.0002.
Significant difference between the comparison group and the reference group, P = 0.005.
Difference between the comparison group and the reference group approached significance, P = 0.05.
Significant difference between the comparison group and the reference group, P = 0.03.
Hemoglobin concentrations are impacted only when iron stores have been depleted (200). African-American men were found to have significantly lower hemoglobin compared with European, Asian, or Hispanic men. Similarly, African-American women had significantly lower hemoglobin when compared with European women. These findings are consistent with previous literature evaluating hemoglobin as a function of ethnicity (171, 176, 181–185).
Conclusions
This review highlighted the shared and unique genetic variants among ethnic groups that have been associated with iron status biomarkers, and possible differences in iron status as a function of ethnicity were explored using published data. To date, most genome-wide and candidate gene association studies on iron homeostasis have been conducted in European populations. Some of the associations found in Europeans have been replicated in other ethnic groups, but the clinical significance is unknown given the varying minor or effect allele frequencies among different populations. Interestingly, reported frequencies of key genetic variants associated with iron traits among different ethnic groups do not fully reflect epidemiological data on iron status in different populations. Because iron traits may be influenced by a combination of genetic, dietary, and lifestyle factors, methods taking into account several variants together, such as polygenic risk scores, may be better at predicting the risk of ID or IO in specific populations. Moreover, although recent studies have identified variants in non–iron-related genes that are associated with iron traits, how they vary as a function of ethnicity and the significance of these SNPs have yet to be determined. Finally, most genome-wide and candidate gene association studies have been focused on blood biomarkers of iron status. Although iron status biomarkers correlate with body iron stores, their concentrations are sensitive to diet and disease, which may have confounded current association studies. Iron homeostasis in humans depends predominantly on the tight regulation of dietary iron absorption by enterocytes. Therefore, attempts to characterize genetic determinants of interindividual variation in iron homeostasis using direct measures of nutrient utilization are needed in order to better understand the differences in iron homeostasis that exist among different populations. Additional data are needed to identify possible risks and benefits associated with universal iron supplementation policies, such as those currently recommended for pregnant North American women (201), to identify the genetic basis of population differences in iron metabolism and disease susceptibility, and to help inform population-specific dietary iron intake recommendations and surveillance in at-risk populations.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge Youri Jung for her assistance in creating the figure. The authors’ responsibilities were as follows—WK, AB, and KOO: designed and wrote the manuscript; AGC, YW, ZG, and XL: provided subject matter expertise, reviewed the manuscript, and provided feedback; and all authors: read and approved the final manuscript.
Notes
Supported by NIH grant R01 DK122216.
Author disclosures: The authors report no conflicts of interest.
Supplemental Tables 1–3 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/advances/.
WK and AB are co-first authors and contributed equally to this work.
Abbreviations used: ABO, ɑ1-3-N-acetylgalactosaminyltransferase and ɑ1-3-galactosyltransferase; BMP, bone morphogenetic protein; BMPR, bone morphogenetic protein receptor; DMT1, divalent metal transporter 1; FADS2, fatty acid desaturase 2; FeAST, Iron and Atherosclerosis Study; Fe3+, ferric iron; Fe2+, ferrous iron; FPN, ferroportin; GAB3, growth factor receptor bound protein 2–associated protein 3; GWAS, genome-wide association study; HAMP, hepcidin; HEIRS, Hemochromatosis and Iron Overload Screening; HFE, high iron protein; HH, hereditary hemochromatosis; HJV, hemojuvelin; ID, iron deficiency; IDA, iron deficiency anemia; IO, iron overload; IRIDA, iron-refractory iron deficiency anemia; JH, juvenile hemochromatosis; MAF, minor allele frequency; MCV, mean corpuscular volume; MT-2, matripase-2; NAT2, N-acetyltransferase 2; NGS, next-generation sequencing; NTBI, non–transferrin-bound iron; PMS1, postmeiotic segregation increased 1; SF, serum ferritin; SMAD, small mothers against decapentaplegic; SNP, single nucleotide polymorphism; sTfR, soluble transferrin receptor; TF, transferrin; TFR, transferrin receptor; TIBC, total iron binding capacity; TMPRSS6, transmembrane serine protease 6; TSAT, transferrin saturation.
Contributor Information
Wanhui Kang, Division of Nutritional Sciences, Cornell University, Ithaca, NY, USA.
Alexa Barad, Division of Nutritional Sciences, Cornell University, Ithaca, NY, USA.
Andrew G Clark, Department of Molecular Biology and Genetics, Cornell University, Ithaca, NY, USA; Department of Computational Biology, Cornell University, Ithaca, NY, USA.
Yiqin Wang, Division of Nutritional Sciences, Cornell University, Ithaca, NY, USA.
Xu Lin, Shanghai Institute of Nutrition and Health, Chinese Academy of Sciences, Shanghai, China.
Zhenglong Gu, Division of Nutritional Sciences, Cornell University, Ithaca, NY, USA.
Kimberly O O'Brien, Division of Nutritional Sciences, Cornell University, Ithaca, NY, USA.
References
- 1.Lynch S, Pfeiffer CM, Georgieff MK, Brittenham G, Fairweather-Tait S, Hurrell RF, McArdle HJ, Raiten DJ. Biomarkers of Nutrition for Development (BOND)—iron review. J Nutr. 2018;148(Suppl 1):1001S–67S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dev S, Babitt JL. Overview of iron metabolism in health and disease. Hemodial Int. 2017;21(Suppl 1):S6–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frey PA, Reed GH. The ubiquity of iron. ACS Chem Biol. 2012;7(9):1477–81. [DOI] [PubMed] [Google Scholar]
- 4.Camaschella C. Iron deficiency. Blood. 2019;133(1):30–9. [DOI] [PubMed] [Google Scholar]
- 5.Montonen J, Boeing H, Steffen A, Lehmann R, Fritsche A, Joost H-G, Schulze MB, Pischon T. Body iron stores and risk of type 2 diabetes: results from the European Prospective Investigation into Cancer and Nutrition (EPIC)-Potsdam study. Diabetologia. 2012;55(10):2613–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gan W, Guan Y, Wu Q, An P, Zhu J, Lu L, Jing L, Yu Y, Ruan S, Xie Det al. Association of TMPRSS6 polymorphisms with ferritin, hemoglobin, and type 2 diabetes risk in a Chinese Han population. Am J Clin Nutr. 2012;95(3):626–32. [DOI] [PubMed] [Google Scholar]
- 7.Gujja P, Rosing DR, Tripodi DJ, Shizukuda Y. Iron overload cardiomyopathy: better understanding of an increasing disorder. J Am Coll Cardiol. 2010;56(13):1001–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eftekhari MH, Mozaffari-Khosravi H, Shidfar F, Zamani A. Relation between body iron status and cardiovascular risk factors in patients with cardiovascular disease. Int J Prev Med. 2013;4(8):911–6. [PMC free article] [PubMed] [Google Scholar]
- 9.Basuli D, Stevens RG, Torti FM, Torti SV. Epidemiological associations between iron and cardiovascular disease and diabetes. Front Pharmacol. 2014;5:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torti SV, Torti FM. Iron and cancer: more ore to be mined. Nat Rev Cancer. 2013;13(5):342–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thévenod F. Iron and its role in cancer defense: a double-edged sword. Met Dev Action Anticancer Agents. 2018;18:437–68. [DOI] [PubMed] [Google Scholar]
- 12.Daugherty AM, Raz N. Appraising the role of iron in brain aging and cognition: promises and limitations of MRI methods. Neuropsychol Rev. 2015;25(3):272–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ward RJ, Dexter DT, Crichton RR. Neurodegenerative diseases and therapeutic strategies using iron chelators. J Trace Elem Med Biol. 2015;31:267–73. [DOI] [PubMed] [Google Scholar]
- 14.Simcox JA, McClain DA. Iron and diabetes risk. Cell Metab. 2013;17(3):329–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aregbesola A, Voutilainen S, Virtanen JK, Aregbesola A, Tuomainen T-P. Serum hepcidin concentrations and type 2 diabetes. World J Diabetes. 2015;6(7):978–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson GJ, Frazer DM. Current understanding of iron homeostasis. Am J Clin Nutr. 2017;106(Suppl 6):1559S–66S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gräsbeck R, Kouvonen I, Lundberg M, Tenhunen R. An intestinal receptor for heme. Scand J Haematol. 1979;23(1):5–9. [DOI] [PubMed] [Google Scholar]
- 18.Tenhunen R, Gräsbeck R, Kouvonen I, Lundberg M. An intestinal receptor for heme: its partial characterization. Int J Biochem. 1980;12(5–6):713–6. [DOI] [PubMed] [Google Scholar]
- 19.Knutson MD. Iron transport proteins: gateways of cellular and systemic iron homeostasis. J Biol Chem. 2017;292(31):12735–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishito Y, Kambe T. Absorption mechanisms of iron, copper, and zinc: an overview. J Nutr Sci Vitaminol. 2018;64(1):1–7. [DOI] [PubMed] [Google Scholar]
- 21.Camaschella C, Nai A, Silvestri L. Iron metabolism and iron disorders revisited in the hepcidin era. Haematologica. 2020;105(2):260–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vashchenko G, MacGillivray RTA. Multi-copper oxidases and human iron metabolism. Nutrients. 2013;5(7):2289–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen H, Attieh ZK, Syed BA, Kuo YM, Stevens V, Fuqua BK, Andersen HS, Naylor CE, Evans RW, Gambling Let al. Identification of zyklopen, a new member of the vertebrate multicopper ferroxidase family, and characterization in rodents and human cells. J Nutr. 2010;140(10):1728–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fleming RE, Ponka P. Iron overload in human disease. N Engl J Med. 2012;366(4):348–59. [DOI] [PubMed] [Google Scholar]
- 25.Jenkitkasemwong S, Wang C-Y, Coffey R, Zhang W, Chan A, Biel T, Kim J-S, Hojyo S, Fukada T, Knutson MD. SLC39A14 is required for the development of hepatocellular iron overload in murine models of hereditary hemochromatosis. Cell Metab. 2015;22(1):138–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen M, Cabantchik ZI, Chan S, Chan GC, Cheung Y. Iron overload and apoptosis of HL-1 cardiomyocytes: effects of calcium channel blockade. PLoS One. 2014;9(11):e112915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aschemeyer S, Qiao B, Stefanova D, Valore E V, Sek AC, Ruwe TA, Vieth KR, Jung G, Casu C, Rivella Set al. Structure-function analysis of ferroportin defines the binding site and an alternative mechanism of action of hepcidin. Blood. 2018;131(8):899–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sangkhae V, Nemeth E. Regulation of the iron homeostatic hormone hepcidin. Adv Nutr. 2017;8(1):126–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muckenthaler MU, Rivella S, Hentze MW, Galy B. A red carpet for iron metabolism. Cell. 2017;168(3):344–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ganz T. Erythropoietic regulators of iron metabolism. Free Radic Biol Med. 2019;133:69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.World Health Organization . Assessing the iron status of populations. Geneva (Switzerland): World Health Organization; 2012. [Google Scholar]
- 32.Ye K, Cao C, Lin X, O'Brien KO, Gu Z. Natural selection on HFE in Asian populations contributes to enhanced non-heme iron absorption. BMC Genet. 2015;16(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young MF, Glahn RP, Ariza-Nieto M, Inglis J, Olbina G, Westerman M, O'Brien KO. Serum hepcidin is significantly associated with iron absorption from food and supplemental sources in healthy young women. Am J Clin Nutr. 2009;89(2):533–8. [DOI] [PubMed] [Google Scholar]
- 34.Roe MA, Heath ALM, Oyston SL, Macrow C, Hoogewerff JA, Foxall R, Dainty JR, Majsak-Newman G, Willis G, Fairweather-Tait SJ. Iron absorption in male C282Y heterozygotes. Am J Clin Nutr. 2005;81(4):814–21. [DOI] [PubMed] [Google Scholar]
- 35.Zimmermann MB, Troesch B, Biebinger R, Egli I, Zeder C, Hurrell RF. Plasma hepcidin is a modest predictor of dietary iron bioavailability in humans, whereas oral iron loading, measured by stable-isotope appearance curves, increases plasma hepcidin. Am J Clin Nutr. 2009;90(5):1280–7. [DOI] [PubMed] [Google Scholar]
- 36.Roe MA, Collings R, Dainty JR, Swinkels DW, Fairweather-Tait SJ. Plasma hepcidin concentrations significantly predict interindividual variation in iron absorption in healthy men. Am J Clin Nutr. 2009;89(4):1088–91. [DOI] [PubMed] [Google Scholar]
- 37.CDC . Recommendations to prevent and control iron deficiency in the United States. Clin Nurse Spec. 1998;12(5):210. [Google Scholar]
- 38.Arezes J, Nemeth E. Hepcidin and iron disorders: new biology and clinical approaches. Int J Lab Hematol. 2015;37(S1):92–8. [DOI] [PubMed] [Google Scholar]
- 39.Brissot P, Pietrangelo A, Adams PC, De Graaff B, McLaren CE, Loreál O. Haemochromatosis. Nat Rev Dis Prim. 2018;4:18016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, Dormishian F, Domingo R, Ellis MC, Fullan Aet al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13(4):399–408. [DOI] [PubMed] [Google Scholar]
- 41.Wu XG, Wang Y, Wu Q, Cheng WH, Liu W, Zhao Y, Mayeur C, Schmidt PJ, Yu PB, Wang Fet al. HFE interacts with the BMP type I receptor ALK3 to regulate hepcidin expression. Blood. 2014;124(8):1335–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beutler E, Felitti VJ, Koziol JA, Ho NJ, Gelbart T. Penetrance of 845G → A (C282Y) HFE hereditary haemochromatosis mutation in the USA. Lancet. 2002;359(9302):211–8. [DOI] [PubMed] [Google Scholar]
- 43.Olynyk JK, Cullen DJ, Aquilia S, Rossi E, Summerville L, Powell LW. A population-based study of the clinical expression of the hemochromatosis gene. N Engl J Med. 1999;341(10):718–24. [DOI] [PubMed] [Google Scholar]
- 44.Adams PC, Reboussin DM, Barton JC, McLaren CE, Eckfeldt JH, McLaren GD, Dawkins FW, Acton RT, Harris EL, Gordeuk VRet al. Hemochromatosis and iron-overload screening in a racially diverse population. N Engl J Med. 2005;352(17):1769–78. [DOI] [PubMed] [Google Scholar]
- 45.Grosse SD, Gurrin LC, Bertalli NA, Allen KJ. Clinical penetrance in hereditary hemochromatosis: estimates of the cumulative incidence of severe liver disease among HFE C282Y homozygotes. Genet Med. 2018;20(4):383–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allen KJ, Gurrin LC, Constantine CC, Osborne NJ, Delatycki MB, Nicoll AJ, McLaren CE, Bahlo M, Nisselle AE, Vulpe CDet al. Iron-overload–related disease in HFE hereditary hemochromatosis. N Engl J Med. 2008;358(3):221–30. [DOI] [PubMed] [Google Scholar]
- 47.Camaschella C. Understanding iron homeostasis through genetic analysis of hemochromatosis and related disorders. Blood. 2005;106(12):3710–7. [DOI] [PubMed] [Google Scholar]
- 48.Merryweather-Clarke AT, Pointon JJ, Jouanolle AM, Rochette J, Robson KJH. Geography of HFE C282Y and H63D mutations. Genet Test. 2000;4(2):183–98. [DOI] [PubMed] [Google Scholar]
- 49.Pietrangelo A. Hereditary hemochromatosis—a new look at an old disease. N Engl J Med. 2004;350(23):2383–97. [DOI] [PubMed] [Google Scholar]
- 50.Steinberg KK, Cogswell ME, Chang JC, Caudill SP, McQuillan GM, Bowman BA, Grummer-Strawn LM, Sampson EJ, Khoury MJ, Gallagher ML. Prevalence of C282Y and H63D mutations in the hemochromatosis (HFE) gene in the United States. J Am Med Assoc. 2001;285(17):2216–22. [DOI] [PubMed] [Google Scholar]
- 51.Distante S, Berg JP, Lande K, Haug E, Bell H. High prevalence of the hemochromatosis-associated Cys282Tyr HFE gene mutation in a healthy Norwegian population in the City of Oslo, and its phenotypic expression. Scand J Gastroenterol. 1999;34(5):529–34. [DOI] [PubMed] [Google Scholar]
- 52.Hanson EH, Imperatore G, Burke W. HFE gene and hereditary hemochromatosis: a HuGE review. Am J Epidemiol. 2001;154(3):193–206. [DOI] [PubMed] [Google Scholar]
- 53.Simon M, Alexandre JL, Fauchet R, Genetet B, Bourel M. The genetics of hemochromatosis. Prog Med Genet. 1980;4:135–68. [PubMed] [Google Scholar]
- 54.Smith BN, Kantrowitz W, Grace ND, Greenberg MS, Patton TJ, Ookubo R, Sorger K, Semeraro JG, Dovle JR, Cooper AGet al. Prevalence of hereditary hemochromatosis in a Massachusetts corporation: is Celtic origin a risk factor?. Hepatology. 1997;25(6):1439–46. [DOI] [PubMed] [Google Scholar]
- 55.Lucotte G. Celtic origin of the C282Y mutation of hemochromatosis. Blood Cells Mol Dis. 1998;24(4):433–8. [DOI] [PubMed] [Google Scholar]
- 56.Milman N, Pedersen P. Evidence that the Cys282Tyr mutation of the HFE gene originated from a population in southern Scandinavia and spread with the Vikings. Clin Genet. 2003;64(1):36–47. [DOI] [PubMed] [Google Scholar]
- 57.Murphy S, Curran MD, McDougall N, Callender ME, O'Brien CJ, Middleton D. High incidence of the Cys 282 Tyr mutation in the HFE gene in the Irish population—implications for haemochromatosis. Tissue Antigens. 1998;52(5):484–8. [DOI] [PubMed] [Google Scholar]
- 58.Ryan E, O'Keane C, Crowe J. Hemochromatosis in Ireland and HFE. Blood Cells Mol Dis. 1998;24(4):428–32. [DOI] [PubMed] [Google Scholar]
- 59.Benyamin B, Esko T, Ried JS, Radhakrishnan A, Vermeulen SH, Traglia M, Gögele M, Anderson D, Broer L, Podmore Cet al. Novel loci affecting iron homeostasis and their effects in individuals at risk for hemochromatosis. Nat Commun. 2014;5(1):4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benyamin B, McRae AF, Zhu G, Gordon S, Henders AK, Palotie A, Peltonen L, Martin NG, Montgomery GW, Whitfield JBet al. Variants in TF and HFE explain ∼40% of genetic variation in serum-transferrin levels. Am J Hum Genet. 2009;84(1):60–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oexle K, Ried JS, Hicks AA, Tanaka T, Hayward C, Bruegel M, Gögele M, Lichtner P, Müller-Myhsok B, Döring Aet al. Novel association to the proprotein convertase PCSK7 gene locus revealed by analysing soluble transferrin receptor (sTfR) levels. Hum Mol Genet. 2011;20(5):1042–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Traglia M, Girelli D, Biino G, Campostrini N, Corbella M, Sala C, Masciullo C, Viganò F, Buetti I, Pistis Get al. Association of HFE and TMPRSS6 genetic variants with iron and erythrocyte parameters is only in part dependent on serum hepcidin concentrations. J Med Genet. 2011;48(9):629–34. [DOI] [PubMed] [Google Scholar]
- 63.McLaren CE, Garner CP, Constantine CC, McLachlan S, Vulpe CD, Snively BM, Gordeuk VR, Nickerson DA, Cook JD, Leiendecker-Foster Cet al. Genome-wide association study identifies genetic loci associated with iron deficiency. PLoS One. 2011;6(3):e17390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ganesh SK, Zakai NA, Van Rooij FJA, Soranzo N, Smith AV, Nalls MA, Chen MH, Kottgen A, Glazer NL, Dehghan Aet al. Multiple loci influence erythrocyte phenotypes in the CHARGE Consortium. Nat Genet. 2009;41(11):1191–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pichler I, Minelli C, Sanna S, Tanaka T, Schwienbacher C, Naitza S, Porcu E, Pattaro C, Busonero F, Zanon Aet al. Identification of a common variant in the TFR2 gene implicated in the physiological regulation of serum iron levels. Hum Mol Genet. 2011;20(6):1232–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Raffield LM, Louie T, Sofer T, Jain D, Ipp E, Taylor KD, Papanicolaou GJ, Avilés-Santa L, Lange LA, Laurie CCet al. Genome-wide association study of iron traits and relation to diabetes in the Hispanic Community Health Study/Study of Latinos (HCHS/SOL): potential genomic intersection of iron and glucose regulation?. Hum Mol Genet. 2017;26(10):1966–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li J, Lange LA, Duan Q, Lu Y, Singleton AB, Zonderman AB, Evans MK, Li Y, Taylor HA, Willis MSet al. Genome-wide admixture and association study of serum iron, ferritin, transferrin saturation and total iron binding capacity in African Americans. Hum Mol Genet. 2015;24(2):572–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kamatani Y, Matsuda K, Okada Y, Kubo M, Hosono N, Daigo Y, Nakamura Y, Kamatani N. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat Genet. 2010;42(3):210–5. [DOI] [PubMed] [Google Scholar]
- 69.De Gobbi M, Roetto A, Piperno A, Mariani R, Alberti F, Papanikolaou G, Politou M, Lockitch G, Girelli D, Fargion Set al. Natural history of juvenile haemochromatosis. Br J Haematol. 2002;117(4):973–9. [DOI] [PubMed] [Google Scholar]
- 70.Sandhu K, Flintoff K, Chatfield MD, Dixon JL, Ramm LE, Ramm GA, Powell LW, Nathan Subramaniam V, Wallace DF. Phenotypic analysis of hemochromatosis subtypes reveals variations in severity of iron overload and clinical disease. Blood. 2018;132(1):101–10. [DOI] [PubMed] [Google Scholar]
- 71.Koyama C, Hayashi H, Wakusawa S, Ueno T, Yano M, Katano Y, Goto H, Kidokoro R. Three patients with middle-age-onset hemochromatosis caused by novel mutations in the hemojuvelin gene. J Hepatol. 2005;43(4):740–2. [DOI] [PubMed] [Google Scholar]
- 72.Ravasi G, Pelucchi S, Mariani R, Silvestri L, Camaschella C, Piperno A. A severe hemojuvelin mutation leading to late onset of HFE2-hemochromatosis. Dig Liver Dis. 2018;50(8):859–62. [DOI] [PubMed] [Google Scholar]
- 73.Kong X, Xie L, Zhu H, Song L, Xing X, Yang W, Chen X. Genotypic and phenotypic spectra of hemojuvelin mutations in primary hemochromatosis patients: a systematic review. Orphanet J Rare Dis. 2019;14(1):171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wallace DF, Subramaniam VN. The global prevalence of HFE and non-HFE hemochromatosis estimated from analysis of next-generation sequencing data. Genet Med. 2016;18(6):618–26. [DOI] [PubMed] [Google Scholar]
- 75.Papanikolaou G, Samuels ME, Ludwig EH, MacDonald MLE, Franchini PL, Dubé MP, Andres L, MacFarlane J, Sakellaropoulos N, Politou Met al. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nat Genet. 2004;36(1):77–82. [DOI] [PubMed] [Google Scholar]
- 76.Gehrke S, Pietrangelo A, Kaščák M, Braner A, Eisold M, Kulaksiz H, Herrmann T, Hebling U, Bents K, Gugler Ret al. HJV gene mutations in European patients with juvenile hemochromatosis. Clin Genet. 2005;67(5):425–8. [DOI] [PubMed] [Google Scholar]
- 77.McDonald CJ, Wallace DF, Crawford DHG, Subramaniam VN. Iron storage disease in Asia-Pacific populations: the importance of non-HFE mutations. J Gastroenterol Hepatol. 2013;28(7):1087–94. [DOI] [PubMed] [Google Scholar]
- 78.Huang FW, Rubio-Aliaga I, Kushner JP, Andrews NC, Fleming MD. Identification of a novel mutation (C321X) in HJV. Blood. 2004;104(7):2176–7. [DOI] [PubMed] [Google Scholar]
- 79.Lv T, Zhang W, Xu A, Li Y, Zhou D, Zhang B, Li X, Zhao X, Wang Y, Wang Xet al. Non-HFE mutations in haemochromatosis in China: combination of heterozygous mutations involving HJV signal peptide variants. J Med Genet. 2018;55(10):650–60. [DOI] [PubMed] [Google Scholar]
- 80.Li S, Xue J, Chen B, Wang Q, Shi M, Xie X, Zhang L. Two middle-age-onset hemochromatosis patients with heterozygous mutations in the hemojuvelin gene in a Chinese family. Int J Hematol. 2014;99(4):487–92. [DOI] [PubMed] [Google Scholar]
- 81.Yuanfeng L, Hongxing Z, Haitao Z, Xiaobo P, Lili B, Fuchu H, Zewu Q, Gangqiao Z. Mutation analysis of the pathogenic gene in a Chinese family with hereditary hemochromatosis. Yi Chuan. 2014;36(11):1152–8. [DOI] [PubMed] [Google Scholar]
- 82.Kaneko Y, Miyajima H, Piperno A, Tomosugi N, Hayashi H, Morotomi N, Tsuchida KI, Ikeda T, Ishikawa A, Ota Yet al. Measurement of serum hepcidin-25 levels as a potential test for diagnosing hemochromatosis and related disorders. J Gastroenterol. 2010;45(11):1163–71. [DOI] [PubMed] [Google Scholar]
- 83.Hattori A, Miyajima H, Tomosugi N, Tatsumi Y, Hayashi H, Wakusawa S. Clinicopathological study of Japanese patients with genetic iron overload syndromes. Pathol Int. 2012;62(9):612–8. [DOI] [PubMed] [Google Scholar]
- 84.Ikuta K, Hatayama M, Addo L, Toki Y, Sasaki K, Tatsumi Y, Hattori A, Kato A, Kato K, Hayashi Het al. Iron overload patients with unknown etiology from national survey in Japan. Int J Hematol. 2017;105(3):353–60. [DOI] [PubMed] [Google Scholar]
- 85.Nagayoshi Y, Nakayama M, Suzuki S, Hokamaki J, Shimomura H, Tsujita K, Fukuda M, Yamashita T, Nakamura Y, Sugiyama Set al. A Q312X mutation in the hemojuvelin gene is associated with cardiomyopathy due to juvenile haemochromatosis. Eur J Heart Fail. 2008;10(10):1001–6. [DOI] [PubMed] [Google Scholar]
- 86.Dhillon BK, Chopra G, Jamwal M, Chandak GR, Duseja A, Malhotra P, Chawla YK, Garewal G, Das R. Adult onset hereditary hemochromatosis is associated with a novel recurrent hemojuvelin (HJV) gene mutation in north Indians. Blood Cells Mol Dis. 2018;73:14–21. [DOI] [PubMed] [Google Scholar]
- 87.Chun YL, Merryweather-Clarke AT, Viprakasit V, Chinthammitr Y, Srichairatanakool S, Limwongse C, Oleesky D, Robins AJ, Hudson J, Wai Pet al. Iron overload in the Asian community. Blood. 2009;114(1):20–5. [DOI] [PubMed] [Google Scholar]
- 88.Barton JC, LaFreniere SA, Leiendecker-Foster C, Li H, Acton RT, Press RD, Eckfeldt JH. HFE, SLC40A1, HAMP, HJV, TFR2, and FTL mutations detected by denaturing high-performance liquid chromatography after iron phenotyping and HFE C282Y and H63D genotyping in 785 HEIRS study participants. Am J Hematol. 2009;84(11):710–4. [DOI] [PubMed] [Google Scholar]
- 89.Lee PL, Barton JC, Brandhagen D, Beutler E. Hemojuvelin (HJV) mutations in persons of European, African-American and Asian ancestry with adult onset haemochromatosis. Br J Haematol. 2004;127(2):224–9. [DOI] [PubMed] [Google Scholar]
- 90.Hamdi-Rozé H, Ben Ali Z, Ropert M, Detivaud L, Aggoune S, Simon D, Pelletier G, Deugnier Y, David V, Bardou-Jacquet E. Variable expressivity of HJV related hemochromatosis: “juvenile” hemochromatosis?. Blood Cells Mol Dis. 2019;74:30–3. [DOI] [PubMed] [Google Scholar]
- 91.Lanzara C, Roetto A, Daraio F, Rivard S, Ficarella R, Simard H, Cox TM, Cazzola M, Piperno A, Gimenez-Roqueplo APet al. Spectrum of hemojuvelin gene mutations in 1q-linked juvenile hemochromatosis. Blood. 2004;103(11):4317–21. [DOI] [PubMed] [Google Scholar]
- 92.Santos PCJL, Cançado RD, Pereira AC, Schettert IT, Soares RAG, Pagliusi RA, Hirata RDC, Hirata MH, Teixeira AC, Figueiredo MSet al. Hereditary hemochromatosis: mutations in genes involved in iron homeostasis in Brazilian patients. Blood Cells Mol Dis. 2011;46(4):302–7. [DOI] [PubMed] [Google Scholar]
- 93.Mendes AI, Ferro A, Martins R, Picanço I, Gomes S, Cerqueira R, Correia M, Nunes AR, Esteves J, Fleming Ret al. Non-classical hereditary hemochromatosis in Portugal: novel mutations identified in iron metabolism-related genes. Ann Hematol. 2009;88(3):229–34. [DOI] [PubMed] [Google Scholar]
- 94.Roettol A, Papanikolaou G, Politou M, Alberti F, Girelli D, Christakis J, Loukopoulos D, Camaschella C. Mutant antimicrobial peptide hepcidin is associated with severe juvenile hemochromatosis. Nat Genet. 2003;33(1):21–2. [DOI] [PubMed] [Google Scholar]
- 95.Matthes T, Aguilar-Martinez P, Pizzi-Bosman L, Darbellay R, Rubbia-Brandt L, Giostra E, Michel M, Ganz T, Beris P. Severe hemochromatosis in a Portuguese family associated with a new mutation in the 5′-UTR of the HAMP gene. Blood. 2004;104(7):2181–3. [DOI] [PubMed] [Google Scholar]
- 96.Porto G, Roetto A, Daraio F, Pinto JP, Almeida S, Bacelar C, Nemeth E, Ganz T, Camaschella C. A Portuguese patient homozygous for the -25G+>A mutation of the HAMP promoter shows evidence of steady-state transcription but fails to up-regulate hepcidin levels by iron. Blood. 2005;106(8):2922–3. [DOI] [PubMed] [Google Scholar]
- 97.Fonseca PFS, Cançado RD, Uellendahl Lopes MM, Correia E, Lescano MA, Santos PCJL. HAMP gene mutation associated with juvenile hemochromatosis in Brazilian patients. Acta Haematol. 2016;135(4):228–31. [DOI] [PubMed] [Google Scholar]
- 98.Lescano MA, Tavares LC, Santos PCJL. Juvenile hemochromatosis: HAMP mutation and severe iron overload treated with phlebotomies and deferasirox. World J Clin Cases. 2017;5(10):381–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Roetto A, Daraio F, Porporato P, Caruso R, Cox TM, Cazzola M, Gasparini P, Piperno A, Camaschella C. Screening hepcidin for mutations in juvenile hemochromatosis: identification of a new mutation (C70R). Blood. 2004;103(6):2407–9. [DOI] [PubMed] [Google Scholar]
- 100.Majore S, Binni F, Pennese A, De Santis A, Crisi A, Grammatico P. HAMP gene mutation c.208T>C (p.C70R) identified in an Italian patient with severe hereditary hemochromatosis. Hum Mutat. 2004;23(4):400. [DOI] [PubMed] [Google Scholar]
- 101.Delatycki M, Allen K, Gow P, MacFarlane J, Radomski C, Thompson J, Hayden M, Goldberg Y, Samuels M. A homozygous HAMP mutation in a multiply consanguineous family with pseudo-dominant juvenile hemochromatosis. Clin Genet. 2004;65(5):378–83. [DOI] [PubMed] [Google Scholar]
- 102.Hattori A, Tomosugi N, Tatsumi Y, Suzuki A, Hayashi K, Katano Y, Inagaki Y, Ishikawa T, Hayashi H, Goto Het al. Identification of a novel mutation in the HAMP gene that causes non-detectable hepcidin molecules in a Japanese male patient with juvenile hemochromatosis. Blood Cells Mol Dis. 2012;48(3):179–82. [DOI] [PubMed] [Google Scholar]
- 103.Gichohi-Wainaina WN, Tanaka T, Towers GW, Verhoef H, Veenemans J, Talsma EF, Harryvan J, Boekschoten MV, Feskens EJ, Melse-Boonstra A. Associations between common variants in iron-related genes with haematological traits in populations of African ancestry. PLoS One. 2016;11(6):e0157996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ravasi G, Rausa M, Pelucchi S, Arosio C, Greni F, Mariani R, Pelloni I, Silvestri L, Pineda P, Camaschella Cet al. Transferrin receptor 2 mutations in patients with juvenile hemochromatosis phenotype. Am J Hematol. 2015;90(12):E226–7. [DOI] [PubMed] [Google Scholar]
- 105.Piperno A, Roetto A, Mariani R, Pelucchi S, Corengia C, Daraio F, Piga A, Garozzo G, Camaschella C. Homozygosity for transferrin receptor-2 Y250X mutation induces early iron overload. Haematologica. 2004;89(3):359–60. [PubMed] [Google Scholar]
- 106.Le Gac G, Mons F, Jacolot S, Scotet V, Ferec C, Frebourg T. Early onset hereditary hemochromatosis resulting from a novel TFR2 gene nonsense mutation (R105X) in two siblings of north French descent. Br J Haematol. 2004;125(5):674–8. [DOI] [PubMed] [Google Scholar]
- 107.Joshi R, Shvartsman M, Moran E, Lois S, Aranda J, Barque A, de la Cruz X, Bruguera M, Vagace JM, Gervasini Get al. Functional consequences of transferrin receptor-2 mutations causing hereditary hemochromatosis type 3. Mol Genet Genomic Med. 2015;3(3):221–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Girelli D, Bozzini C, Roetto A, Alberti F, Daraio F, Colombari R, Olivieri O, Corrocher R, Camaschella C. Clinical and pathologic findings in hemochromatosis type 3 due to a novel mutation in transferrin receptor 2 gene. Gastroenterology. 2002;122(5):1295–302. [DOI] [PubMed] [Google Scholar]
- 109.Hattori A, Wakusawa S, Hayashi H, Harashima A, Sanae F, Kawanaka M, Yamada G, Yano M, Yoshioka K. AVAQ 594–597 deletion of the TfR2 gene in a Japanese family with hemochromatosis. Hepatol Res. 2003;26(2):154–6. [DOI] [PubMed] [Google Scholar]
- 110.Zamani F, Bagheri Z, Bayat M, Fereshtehnejad SM, Basi A, Najmabadi H, Ajdarkosh H. Iranian hereditary hemochromatosis patients: baseline characteristics, laboratory data and gene mutations. Med Sci Monit. 2012;18(10):CR622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Camaschella C, Roetto A, Calì A, De Gobbi M, Garozzo G, Carella M, Majorano N, Totaro A, Gasparini P. The gene TFR2 is mutated in a new type of haemochromatosis mapping to 7q22. Nat Genet. 2000;25(1):14–5. [DOI] [PubMed] [Google Scholar]
- 112.Lee PL, Halloran C, West C, Beutler E. Mutation analysis of the transferrin receptor-2 gene in patients with iron overload. Blood Cells Mol Dis. 2001;27(1):285–9. [DOI] [PubMed] [Google Scholar]
- 113.Hayashi H, Wakusawa S, Motonishi S, Miyamoto KI, Okada H, Inagaki Y, Ikeda T. Genetic background of primary iron overload syndromes in Japan. Intern Med. 2006;45(20):1107–11. [DOI] [PubMed] [Google Scholar]
- 114.Soranzo N, Spector TD, Mangino M, Kühnel B, Rendon A, Teumer A, Willenborg C, Wright B, Chen L, Li Met al. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen Consortium. Nat Genet. 2009;41(11):1182–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.An P, Wu Q, Wang H, Guan Y, Mu M, Liao Y, Zhou D, Song P, Wang C, Meng Let al. TMPRSS6, but not TF, TFR2 or BMP2 variants are associated with increased risk of iron-deficiency anemia. Hum Mol Genet. 2012;21(9):2124–31. [DOI] [PubMed] [Google Scholar]
- 116.Nemeth E, Tuttle MS, Powelson J, Vaughn MD, Donovan A, Ward DMV, Ganz T, Kaplan J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306(5704):2090–3. [DOI] [PubMed] [Google Scholar]
- 117.Drakesmith H, Nemeth E, Ganz T. Ironing out ferroportin. Cell Metab. 2015;22(5):777–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pietrangelo A. The ferroportin disease. Blood Cells Mol Dis. 2004;32(1):131–8. [DOI] [PubMed] [Google Scholar]
- 119.Mayr R, Janecke AR, Schranz M, Griffiths WJH, Vogel W, Pietrangelo A, Zoller H. Ferroportin disease: a systematic meta-analysis of clinical and molecular findings. J Hepatol. 2010;53(5):941–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vlasveld LT, Janssen R, Bardou-Jacquet E, Venselaar H, Hamdi-Roze H, Drakesmith H, Swinkels DW. Twenty years of ferroportin disease: a review or an update of published clinical, biochemical, molecular, and functional features. Pharmaceuticals. 2019;12(3):132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Roetto A, Merryweather-Clarke AT, Daraio F, Livesey K, Pointon JJ, Barbabietola G, Piga A, Mackie PH, Robson KJH, Camaschella C. A valine deletion of ferroportin 1: a common mutation in hemochromatosis type 4. Blood. 2002;100(2):733–4. [DOI] [PubMed] [Google Scholar]
- 122.Callebaut I, Joubrel R, Pissard S, Kannengiesser C, Gérolami V, Ged C, Cadet E, Cartault F, Ka C, Gourlaouen Iet al. Comprehensive functional annotation of 18 missense mutations found in suspected hemochromatosis type 4 patients. Hum Mol Genet. 2014;23(17):4479–90. [DOI] [PubMed] [Google Scholar]
- 123.Cazzola M, Cremonesi L, Papaioannou M, Soriani N, Kioumi A, Charalambidou A, Paroni R, Romtsou K, Levi S, Ferrari Met al. Genetic hyperferritinaemia and reticuloendothelial iron overload associated with a three base pair deletion in the coding region of the ferroportin gene (SLC11A3). Br J Haematol. 2002;119(2):539–46. [DOI] [PubMed] [Google Scholar]
- 124.Speletas M, Kioumi A, Loules G, Hytiroglou P, Tsitouridis J, Christakis J, Germenis AE. Analysis of SLC40A1 gene at the mRNA level reveals rapidly the causative mutations in patients with hereditary hemochromatosis type IV. Blood Cells Mol Dis. 2008;40(3):353–9. [DOI] [PubMed] [Google Scholar]
- 125.Devalia V, Carter K, Walker AP, Perkins SJ, Worwood M, May A, Dooley JS. Autosomal dominant reticuloendothelial iron overload associated with a 3-base pair deletion in the ferroportin 1 gene (SLC11A3). Blood. 2002;100(2):695–7. [DOI] [PubMed] [Google Scholar]
- 126.Wallace DF, Browett P, Wong P, Kua H, Ameratunga R, Subramaniam VN. Identification of ferroportin disease in the Indian subcontinent. Gut. 2005;54(4):567–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Beutler E, Barton JC, Felitti VJ, Gelbart T, West C, Lee PL, Waalen J, Vulpe C. Ferroportin 1 (SCL40A1) variant associated with iron overload in African-Americans. Blood Cells Mol Dis. 2003;31(3):305–9. [DOI] [PubMed] [Google Scholar]
- 128.Kasvosve I, Gomo ZAR, Nathoo KJ, Matibe P, Mudenge B, Loyevsky M, Gordeuk VR. Effect of ferroportin Q248H polymorphism on iron status in African children. Am J Clin Nutr. 2005;82(5):1102–6. [DOI] [PubMed] [Google Scholar]
- 129.McNamara L, Gordeuk VR, MacPhail AP. Ferroportin (Q248H) mutations in African families with dietary iron overload. J Gastroenterol Hepatol. 2005;20(12):1855–8. [DOI] [PubMed] [Google Scholar]
- 130.Albuquerque D, Manco L, Loua KM, Arez AP, De Jesus Trovoada M, Relvas L, Millimono TS, Rath SL, Lopes D, Nogueira Fet al. SLC40A1 Q248H allele frequencies and associated SLC40A1 haplotypes in three West African population samples. Ann Hum Biol. 2011;38(3):378–81. [DOI] [PubMed] [Google Scholar]
- 131.Gordeuk VR, Caleffi A, Corradini E, Ferrara F, Jones RA, Castro O, Onyekwere O, Kittles R, Pignatti E, Montosi Get al. Iron overload in Africans and African-Americans and a common mutation in the SCL40A1 (ferroportin 1) gene. Blood Cells Mol Dis. 2003;31(3):299–304. [DOI] [PubMed] [Google Scholar]
- 132.Kasvosve I, Tshwenyego U, Phuthego T, Koto G, Zachariah M, Nyepetsi NG, Motswaledi MS. Serum ferritin concentration is affected by ferroportin Q248H mutation in Africans. Clin Chim Acta. 2015;444:257–9. [DOI] [PubMed] [Google Scholar]
- 133.Rivers CA, Barton JC, Gordeuk VR, Acton RT, Speechley MR, Snively BM, Leiendecker-Foster C, Press RD, Adams PC, McLaren GDet al. Association of ferroportin Q248H polymorphism with elevated levels of serum ferritin in African Americans in the Hemochromatosis and Iron Overload Screening (HEIRS) Study. Blood Cells Mol Dis. 2007;38(3):247–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Muriuki JM, Mentzer AJ, Band G, Gilchrist JJ, Carstensen T, Lule SA, Goheen MM, Joof F, Kimita W, Mogire Ret al. The ferroportin Q248H mutation protects from anemia, but not malaria or bacteremia. Sci Adv. 2019;5(9):eaaw0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Meidtner K, Podmore C, Kröger J, Van Der Schouw YT, Bendinelli B, Agnoli C, Arriola L, Barricarte A, Boeing H, Cross AJet al. Interaction of dietary and genetic factors influencing body iron status and risk of type 2 diabetes within the EPIC-InterAct study. Diabetes Care. 2018;41(2):277–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Liao M, Shi J, Huang L, Gao Y, Tan A, Wu C, Lu Z, Yang X, Zhang S, Hu Yet al. Genome-wide association study identifies variants in PMS1 associated with serum ferritin in a Chinese population. PLoS One. 2014;9(8):e105844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang CY, Meynard D, Lin HY. The role of TMPRSS6/matriptase-2 in iron regulation and anemia. Front Pharmacol. 2014;5:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Finberg KE, Heeney MM, Campagna DR, Aydinok Y, Pearson HA, Hartman KR, Mayo MM, Samuel SM, Strouse JJ, Markianos Ket al. Mutations in TMPRSS6 cause iron-refractory iron deficiency anemia (IRIDA). Nat Genet. 2008;40(5):569–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Du X, She E, Gelbart T, Truksa J, Lee P, Xia Y, Khovananth K, Mudd S, Mann N, Moresco EMYet al. The serine protease TMPRSS6 is required to sense iron deficiency. Science. 2008;320(5879):1088–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Silvestri L, Guillem F, Pagani A, Nai A, Oudin C, Silva M, Toutain F, Kannengiesser C, Beaumont C, Camaschella Cet al. Molecular mechanisms of the defective hepcidin inhibition in TMPRSS6 mutations associated with iron-refractory iron deficiency anemia. Blood. 2009;113(22):5605–8. [DOI] [PubMed] [Google Scholar]
- 141.Donker AE, Raymakers RAP, Thom Vlasveld L, Van Barneveld T, Terink R, Dors N, Brons PPT, Knoers NVAM, Swinkels DW. Practice guidelines for the diagnosis and management of microcytic anemias due to genetic disorders of iron metabolism or heme synthesis. Blood. 2014;123(25):3873–86. [DOI] [PubMed] [Google Scholar]
- 142.Donker AE, Schaap CCM, Novotny VMJ, Smeets R, Peters TMA, van den Heuvel BLP, Raphael MF, Rijneveld AW, Appel IM, Vlot AJet al. Iron refractory iron deficiency anemia: a heterogeneous disease that is not always iron refractory. Am J Hematol. 2016;91(12):E482–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.De Falco L, Sanchez M, Silvestri L, Kannengiesser C, Muckenthaler MU, Iolascon A, Gouya L, Camaschella C, Beaumont C. Iron refractory iron deficiency anemia. Haematologica. 2013;98(6):845–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.de Falco L, Silvestri L, Kannengiesser C, Morán E, Oudin C, Rausa M, Bruno M, Aranda J, Argiles B, Yenicesu Iet al. Functional and clinical impact of novel Tmprss6 variants in iron-refractory iron-deficiency Anemia patients and genotype-phenotype studies. Hum Mutat. 2014;35(11):1321–9. [DOI] [PubMed] [Google Scholar]
- 145.Benyamin B, Ferreira MAR, Willemsen G, Gordon S, Middelberg RPS, McEvoy BP, Hottenga JJ, Henders AK, Campbell MJ, Wallace Let al. Common variants in TMPRSS6 are associated with iron status and erythrocyte volume. Nat Genet. 2009;41(11):1173–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tanaka T, Roy CN, Yao W, Matteini A, Semba RD, Arking D, Walston JD, Fried LP, Singleton A, Guralnik Jet al. A genome-wide association analysis of serum iron concentrations. Blood. 2010;115(1):94–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chambers JC, Zhang W, Li Y, Sehmi J, Wass MN, Zabaneh D, Hoggart C, Bayele H, McCarthy MI, Peltonen Let al. Genome-wide association study identifies variants in TMPRSS6 associated with hemoglobin levels. Nat Genet. 2009;41(11):1170–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wilman HR, Parisinos CA, Atabaki-Pasdar N, Kelly M, Thomas EL, Neubauer S, Jennison C, Ehrhardt B, Baum P, Schoelsch Cet al. Genetic studies of abdominal MRI data identify genes regulating hepcidin as major determinants of liver iron concentration. J Hepatol. 2019;71(3):594–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kloss-Brandstätter A, Erhart G, Lamina C, Meister B, Haun M, Coassin S, Seifert M, Klein-Franke A, Paulweber B, Kedenko Let al. Candidate gene sequencing of SLC11A2 and TMPRSS6 in a family with severe anaemia: common SNPs, rare haplotypes, no causative mutation. PLoS One. 2012;7(4):e35015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.He M, Workalemahu T, Manson JAE, Hu FB, Qi L. Genetic determinants for body iron store and type 2 diabetes risk in US men and women. PLoS One. 2012;7(7):e40919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gichohi-Wainaina WN, Towers GW, Swinkels DW, Zimmermann MB, Feskens EJ, Melse-Boonstra A. Inter-ethnic differences in genetic variants within the transmembrane protease, serine 6 (TMPRSS6) gene associated with iron status indicators: a systematic review with meta-analyses. Genes Nutr. 2015;10(1):442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jallow MW, Cerami C, Clark TG, Prentice AM, Campino S. Differences in the frequency of genetic variants associated with iron imbalance among global populations. PLoS One. 2020;15(7):e0235141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Goldwurm S, Casati C, Venturi N, Strada S, Santambrogio P, Indraccolo S, Arosio P, Cazzola M, Piperno A, Masera Get al. Biochemical and genetic defects underlying human congenital hypotransferrinemia. Hematol J. 2000;1(6):390–8. [DOI] [PubMed] [Google Scholar]
- 154.Trombini P, Coliva T, Nemeth E, Mariani R, Ganz T, Biondi A, Piperno A. Effects of plasma transfusion on hepcidin production in human congenital hypotransferrinemia. Haematologica. 2007;92(10):1407–10. [DOI] [PubMed] [Google Scholar]
- 155.Nazir H, Al-Khabori MK, Kohistani N, Puchalka J, Klein C. A rare case of congenital atransferrinemia: international collaboration for genetic diagnosis. Blood. 2014;124(21):4883. [Google Scholar]
- 156.Dabboubi R, Amri Y, Yahyaoui S, Mahjoub R, Sahli CA, Sahli C, Hadj Fredj S, Bibi A, Sammoud A, Messaoud T. A new case of congenital atransferrinemia with a novel splice site mutation: c.293-63del. Eur J Med Genet. 2020;63(5):103874. [DOI] [PubMed] [Google Scholar]
- 157.McLaren CE, McLachlan S, Garner CP, Vulpe CD, Gordeuk VR, Eckfeldt JH, Adams PC, Acton RT, Murray JA, Leiendecker-Foster Cet al. Associations between single nucleotide polymorphisms in iron-related genes and iron status in multiethnic populations. PLoS One. 2012;7(6):e38339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kayaalti Z, Akyüzlü DK, Söylemezoğlu T. Evaluation of the effect of divalent metal transporter 1 gene polymorphism on blood iron, lead and cadmium levels. Environ Res. 2015;137:8–13. [DOI] [PubMed] [Google Scholar]
- 159.Tolone C, Bellini G, Punzo F, Papparella A, Miele E, Vitale A, Nobili B, Strisciuglio C, Rossi F. The DMT1 IVS4+44C>A polymorphism and the risk of iron deficiency anemia in children with celiac disease. PLoS One. 2017;12(10):e0185822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Constantine CC, Anderson GJ, Vulpe CD, McLaren CE, Bahlo M, Yeap HL, Gertig DM, Osborne NJ, Bertalli NA, Beckman KBet al. A novel association between a SNP in CYBRD1 and serum ferritin levels in a cohort study of HFE hereditary haemochromatosis. Br J Haematol. 2009;147(1):140–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pelucchi S, Mariani R, Calza S, Fracanzani AL, Modignani GL, Bertola F, Busti F, Trombini P, Fraquelli M, Forni GLet al. CYBRD1 as a modifier gene that modulates iron phenotype in HFE p.C282Y homozygous patients. Haematologica. 2012;97(12):1818–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Milet J, Déhais V, Bourgain C, Jouanolle AM, Mosser A, Perrin M, Morcet J, Brissot P, David V, Deugnier Yet al. Common variants in the BMP2, BMP4, and HJV genes of the hepcidin regulation pathway modulate HFE hemochromatosis penetrance. Am J Hum Genet. 2007;81(4):799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gill D, Benyamin B, Moore LSP, Monori G, Zhou A, Koskeridis F, Evangelou E, Laffan M, Walker AP, Tsilidis KKet al. Associations of genetically determined iron status across the phenome: a mendelian randomization study. PLoS Med. 2019;16(6):e1002833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wolf I, Jenkins BJ, Liu Y, Seiffert M, Custodio JM, Young P, Rohrschneider LR. Gab3, a new DOS/Gab family member, facilitates macrophage differentiation. Mol Cell Biol. 2002;22(1):231–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wöhrle FU, Daly RJ, Brummer T. Function, regulation and pathological roles of the Gab/DOS docking proteins. Cell Commun Signal. 2009;7(1):1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hirano N, Butler MO, Guinan EC, Nadler LM, Kojima S. Presence of anti-kinectin and anti-PMS1 antibodies in Japanese aplastic anaemia patients. Br J Haematol. 2005;128(2):221–3. [DOI] [PubMed] [Google Scholar]
- 167.James WPT, Johnson RJ, Speakman JR, Wallace DC, Frühbeck G, Iversen PO, Stover PJ. Nutrition and its role in human evolution. J Intern Med. 2019;285(5):533–49. [DOI] [PubMed] [Google Scholar]
- 168.McLaren CE, Barton JC, Adams PC, Harris EL, Acton RT, Press N, Reboussin DM, McLaren GD, Sholinsky P, Walker APet al. Hemochromatosis and iron overload screening (HEIRS) Study design for an evaluation of 100,000 primary care-based adults. Am J Med Sci. 2003;325(2):53–62. [DOI] [PubMed] [Google Scholar]
- 169.Endres-Dighe SM, Guo Y, Kanias T, Lanteri M, Stone M, Spencer B, Cable RG, Kiss JE, Kleinman S, Gladwin MTet al. Blood, sweat, and tears: Red Blood Cell-Omics study objectives, design, and recruitment activities. Transfusion. 2019;59(1):46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zacharski LR, Shamayeva G, Chow BK, Depalma RG. Racial differences in iron measures and outcomes observed during an iron reduction trial in peripheral arterial disease. J Health Care Poor Underserved. 2015;26(1):243–59. [DOI] [PubMed] [Google Scholar]
- 171.Beutler E, West C. Hematologic differences between African-Americans and whites: the roles of iron deficiency and α-thalassemia on hemoglobin levels and mean corpuscular volume. Blood. 2005;106(2):740–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Beutler E, Felitti V, Gelbart T, Ho N. The effect of HFE genotypes on measurements of iron overload in patients attending a health appraisal clinic. Ann Intern Med. 2000;133(5):329–37. [DOI] [PubMed] [Google Scholar]
- 173.Zacharski LR, Ornstein DL, Woloshin S, Schwartz LM. Association of age, sex, and race with body iron stores in adults: analysis of NHANES III data. Am Heart J. 2000;40(1):98–104. [DOI] [PubMed] [Google Scholar]
- 174.Dekker LH, Nicolaou M, Van Der A DL, Busschers WB, Brewster LM, Snijder MB, Stronks K, Van Valkengoed IGM. Sex differences in the association between serum ferritin and fasting glucose in type 2 diabetes among South Asian Surinamese, African Surinamese, and ethnic Dutch: the population-based SUNSET study. Diabetes Care. 2013;36(4):965–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Jenum AK, Sletner L, Voldner N, Vangen S, Mørkrid K, Andersen LF, Nakstad B, Skrivarhaug T, Rognerud-jensen O, Roald Bet al. The STORK Groruddalen research programme: a population-based cohort study of gestational diabetes, physical activity, and obesity in pregnancy in a multiethnic population. Rationale, methods, study population, and participation rates. Scand J Public Health. 2010;38(5):60–70. [DOI] [PubMed] [Google Scholar]
- 176.Godsland IF, Seed M, Simpson R, Broom G, Wynn V. Comparison of haematological indices between women of four ethnic groups and the effect of oral contraceptives. J Clin Pathol. 1983;36(2):184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Næss-Andresen ML, Eggemoen ÅR, Berg JP, Falk RS, Jenum AK. Serum ferritin, soluble transferrin receptor, and total body iron for the detection of iron deficiency in early pregnancy: a multiethnic population-based study with low use of iron supplements. Am J Clin Nutr. 2019;109(3):566–75. [DOI] [PubMed] [Google Scholar]
- 178.Spencer BR, Guo Y, Cable RG, Kiss JE, Busch MP, Page GP, Endres-Dighe SM, Kleinman S, Glynn SA, Mast AE. Iron status and risk factors for iron depletion in a racially/ethnically diverse blood donor population. Transfusion. 2019;59(10):3146–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Barton JC, Acton RT, Dawkins FW, Adams PC, Lovato L, Leiendecker-Foster C, McLaren CE, Reboussin DM, Speechley MR, Gordeuk VRet al. Initial screening transferrin saturation values, serum ferritin concentrations, and HFE genotypes in whites and blacks in the Hemochromatosis and Iron Overload Screening Study. Genet Test. 2005;9(3):231–41. [DOI] [PubMed] [Google Scholar]
- 180.Miller EM. Iron status and reproduction in US women: National Health and Nutrition Examination Survey, 1999–2006. PLoS One. 2014;9(11):e112216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Dallman PR, Barr GD, Allen CM, Shinefield HR. Hemoglobin concentration in white, black, and Oriental children: is there a need for separate criteria in screening for anemia?. Am J Clin Nutr. 1978;31(3):377–80. [DOI] [PubMed] [Google Scholar]
- 182.Perry GS, Byers T, Yip R, Margen S. Iron nutrition does not account for the hemoglobin differences between blacks and whites. J Nutr. 1992;122(7):1417–24. [DOI] [PubMed] [Google Scholar]
- 183.McLaren CE, Li KT, Gordeuk VR, Hasselblad V, McLaren GD. Relationship between transferrin saturation and iron stores in the African American and US Caucasian populations: analysis of data from the third National Health and Nutrition Examination Survey. Blood. 2001;98(8):2345–51. [DOI] [PubMed] [Google Scholar]
- 184.Johnson-Spear MA, Yip R. Hemoglobin difference between black and white women with comparable iron status: justification for race-specific anemia criteria. Am J Clin Nutr. 1994;60(1):117–21. [DOI] [PubMed] [Google Scholar]
- 185.Meyers LD, Habicht JP, Johnson CL. Components of the difference in hemoglobin concentrations in blood between black and white women in the United States. Am J Epidemiol. 1979;109(5):539–49. [DOI] [PubMed] [Google Scholar]
- 186.World Health Organization; Centers for Disease Control and Prevention . Assessing the iron status of populations. Second edition including literature reviews. Geneva (Switzerland): World Health Organization; 2007. [Google Scholar]
- 187.Pan Y, Jackson RT. Ethnic difference in the relationship between acute inflammation and serum ferritin in US adult males. Epidemiol Infect. 2008;136(3):421–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zacharski LR, Shamayeva G, Chow BK, DePalma RG. Racial health disparities, and variant red cell and iron homeostasis. J Health Care Poor Underserved. 2016;27(2):741–61. [DOI] [PubMed] [Google Scholar]
- 189.Pan Y, Jackson RT. Insights into the ethnic differences in serum ferritin between black and white US adult men. Am J Hum Biol. 2008;20(4):406–16. [DOI] [PubMed] [Google Scholar]
- 190.Barton JC, Wiener HH, Acton RT, Adams PC, Eckfeldt JH, Gordeuk VR, Harris EL, McLaren CE, Harrison H, McLaren GDet al. Prevalence of iron deficiency in 62,685 women of seven race/ethnicity groups: the HEIRS study. PLoS One. 2020;15(4):e0232125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Gordeuk VR, Brannon PM. Ethnic and genetic factors of iron status in women of reproductive age. Am J Clin Nutr. 2017;106(Suppl 6):1594S–9S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Cossrow N, Falkner B. Race/ethnic issues in obesity and obesity-related comorbidities. J Clin Endocrinol Metab. 2004;89(6):2590–4. [DOI] [PubMed] [Google Scholar]
- 193.Harris EL, McLaren CE, Reboussin DM, Gordeuk VR, Barton JC, Acton RT, McLaren GD, Vogt TM, Snively BM, Leiendecker-Foster Cet al. Serum ferritin and transferrin saturation in Asians and Pacific Islanders. Arch Intern Med. 2007;167(7):722. [DOI] [PubMed] [Google Scholar]
- 194.Oh SH, Jeong TD, Lee W, Chun S, Min WK. Analysis of familial tendencies in transferrin saturation in a Korean population. Dig Dis Sci. 2015;60(10):3136–41. [DOI] [PubMed] [Google Scholar]
- 195.Lee SH, Kim JW, Shin SH, Kang KP, Choi HC, Choi SH, Park KU, Kim HY, Kang W, Jeong SH. HFE gene mutations, serum ferritin level, transferrin saturation, and their clinical correlates in a Korean population. Dig Dis Sci. 2009;54(4):879–86. [DOI] [PubMed] [Google Scholar]
- 196.Hsu WC, Araneta MRG, Kanaya AM, Chiang JL, Fujimoto W. BMI cut points to identify at-risk Asian Americans for type 2 diabetes screening. Diabetes Care. 2015;38(1):150–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.WHO Expert Consultation . Appropriate body-mass index for Asian populations and its implications for policy and intervention strategies. Lancet. 2004;363(9403):157–63. [DOI] [PubMed] [Google Scholar]
- 198.Barton JC, Acton RT, Lovato L, Speechley MR, McLaren CE, Harris EL, Reboussin DM, Adams PC, Dawkins FW, Gordeuk VRet al. Initial screening transferrin saturation values, serum ferritin concentrations, and HFE genotypes in Native Americans and whites in the Hemochromatosis and Iron Overload Screening Study. Clin Genet. 2006;69(1):48–57. [DOI] [PubMed] [Google Scholar]
- 199.Walters GO, Miller FM, Worwood M. Serum ferritin concentration and iron stores in normal subjects. J Clin Pathol. 1973;26(10):770–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lynch S. The rationale for selecting and standardizing iron status indicators. Geneva (Switzerland): World Health Organization; 2012. [Google Scholar]
- 201.O'Brien KO, Ru Y. Iron status of North American pregnant women: an update on longitudinal data and gaps in knowledge from the United States and Canada. Am J Clin Nutr. 2017;106(Suppl 6):1647S–54S. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.