Abstract
The comprehensive characterization of probiotic action has flourished during the past few decades, alongside the evolution of high-throughput, multiomics platforms. The integration of these platforms into probiotic animal and human studies has provided valuable insights into the holistic effects of probiotic supplementation on intestinal and extraintestinal diseases. Indeed, these methodologies have informed about global molecular changes induced in the host and residing commensals at multiple levels, providing a bulk of metagenomic, transcriptomic, proteomic, and metabolomic data. The meaningful interpretation of generated data remains a challenge; however, the maturation of the field of systems biology and artificial intelligence has supported analysis of results. In this review article, we present current literature on the use of multiomics approaches in probiotic studies, we discuss current trends in probiotic research, and examine the possibility of tailor-made probiotic supplementation. Lastly, we delve deeper into newer technologies that have been developed in the last few years, such as single-cell multiomics analyses, and provide future directions for the maximization of probiotic efficacy.
Keywords: probiotics, microbiome, metagenomics, transcriptomics, proteomics, metabolomics, lactic acid bacteria
High-throughput, multiomics platforms provide invaluable insights into the beneficial effects of probiotic supplementation on the host, setting the stage for personalized, tailor-made interventions.
Introduction
Probiotics are defined as “live microorganisms that when administered in adequate amounts confer a health benefit on the host” (1). Α plethora of studies has revealed the positive impact of probiotics on blood cholesterol concentrations (2), inflammation-related disorders (3), metabolic disorders such as obesity and diabetes (4), and even cancer (5). To evaluate the effectiveness of probiotics and elucidate their mechanisms of action, several preclinical and clinical models are commonly employed. For example, models of intestinal or systemic inflammation are used to study inflammation-related pathogenesis (6), models of high-fat diets have been developed to study metabolic disorders (7), and cancer models are used for the study of oncogenesis (8). Moreover, clinical trials that recruit healthy volunteers or patients, with the aim of correlating clinical outcomes to specific molecular changes caused by the probiotic supplementation, are also performed (9).
The evolution of high-throughput platforms and the development of robust bioinformatic tools have boosted our understanding of probiotic action, promoted the in-depth study of their biology and probiotic-induced cellular responses, and further validated their health-promoting properties. Genomic analyses have aided in the mining of probiotic features, such as acid tolerance or bacteriocin production (10), whereas metagenomic studies have contributed to the elucidation of probiotic–microbiota interactions (11). Transcriptomic, proteomic, and metabolomic platforms have supported the profiling of host–microbe crosstalk and have contextualized the holistic effects induced by the supplementation (12). Furthermore, recent comparative studies firmly support the species-, disease-, sex-, and host-specific probiotic actions, underlining the need for targeted interventions (13). The purpose of this review is to weigh the use of multiomics high-throughput platforms in preclinical and clinical studies, investigating the effects of probiotic supplementation on common pathophysiological conditions, such as immune and metabolic disorders. Furthermore, we critically discuss current trends in probiotic research, newly developed technologies, such as single-cell multiomics analysis, and provide future directions for the implementation of personalized probiotic interventions. To this aim, a comprehensive search of current literature on the PubMed database, using the term “probiotics” combined with “transcriptomics,” “proteomics,” “metabolomics,” “metagenomics,” “comparative genomics,” and “microbiome” was performed. Filters were applied to restrict results to animal and clinical studies. Finally, only articles published in English were included.
Current Status of Knowledge
Transcriptomic and proteomic studies of the immunomodulatory properties of probiotic bacteria
Characterization of the immunomodulatory effects of probiotic bacteria at the transcriptome level is performed either by RNA-sequencing (RNA-Seq) or microarray gene expression platforms. Accordingly, protein chips with antibodies, other proteins, or nucleic acids that bind specifically to protein targets are used to evaluate probiotic-induced changes at the proteome level. Animal studies are usually conducted on models of chemically induced colitis. The animals are treated with dinitrobenzene sulfonic acid (DNBS) or dextran sodium sulfate (DSS), which provoke strong inflammatory responses or induce colonic cell death, ultimately leading to the development of colitis (14). Preclinical and clinical studies employing these methodologies are listed in Table 1. For example, administration of Lactobacillus rhamnosus CNCM I-3690 to DNBS-induced chronic microinflammation mice, induced downregulation of systemic inflammation markers IL-6, IFN-β, and IFN-γ and restored gut permeability (15). Moreover, transcriptomic analysis with microarrays, targeting the sum of known mouse transcripts, demonstrated that the probiotic strain blocked the activation of the canonical NF-κB pathway and upregulated the expression of TNF receptor associated factor (TRAF) interaction protein with a forkhead-associated domain (TIFA). Validation of the microarray analysis for 7 representative genes was performed by qRT-PCR (15). In another study, colitis was induced by DSS in C57BL/6 mice that were also injected with the carcinogen azoxymethane (16). The simultaneous supplementation with Bifico, a commercially available probiotic cocktail, comprised of Bifidobacterium longum, L. acidophilus, and Enterococcus faecalis, managed to preserve the intestinal architecture (crypt morphology, infiltration of immunological populations). Notably, a reduction of tumor formation was also recorded. To analyze further the clinical outcomes, and elucidate the molecular pathways involved, global transcriptome analysis with cDNA microarrays, was performed. Two-dimensional hierarchical clustering revealed that 300 genes were differentially expressed; 166 genes were upregulated, and 134 genes were downregulated in the probiotic-treated group. The interpretation of the bulk of data derived from this analysis was performed by the Database for Annotation, Visualization, and Integrated Discovery (DAVID). It was found that Bifico influenced the expression of transcripts involved in tight junctions and cytokine–cytokine receptor interaction (16). Similarly, to investigate the global changes in the transcriptomic profile of BALB/c mice orally inoculated with Bifidobacterium bifidum PRL2010, the 90k CombiMatrix array was employed (17). It was demonstrated that specific genes coding for proinflammatory proteins, such as kallikrein B, plasma 1 protein (Klkb1), were downregulated in the probiotic group, whereas genes encoding tight junction proteins, including catenin α-like 1 (Ctnna1) and claudin 10 (Cldn10), were upregulated. Mechanistic studies in human colon adenocarcinoma cell lines revealed that these effects were likely attributed to the activation of the NF-κΒ pathway (17). Finally, to elucidate the holistic effect of L. plantarum JDFM LP11 consumption on weaned piglets, RNA-Seq was performed in ileum tissues. Comparative transcriptomic analysis revealed 25 differentially expressed genes (DEGs) involved in the downregulation of intestinal inflammatory responses (18). Similarly, RNA-Seq revealed that Saccharomyces boulardii administration to C57BL/6 mice stimulated systemic inflammatory responses (19) (Table 1).
TABLE 1.
Transcriptomic and proteomic preclinical and clinical studies of the immunomodulatory activities of probiotics1
| Type of study | Treatment | Sample | Omics platform | Molecular mechanisms | Clinical outcome | Reference |
|---|---|---|---|---|---|---|
| Animal study | Lactobacillus rhamnosus CNCM I-3690 (5 × 109 CFU) | Blood, MLN, spleen, colon, and ileum | Transcriptomics (microarrays) | Inhibition of ΝF-κB pathway | ↓Systemic inflammation, restored colon and ileum permeability | Martín et al. (15) |
| DNBS-induced chronic microinflammation (SPF male C57BL/6 mice) (n = 30) | 10-d gavage regimen | |||||
| Animal study | Bifico >1.2 × 107 CFU/d (Bifidobacterium longum, L. acidophilus, Enterococcus faecalis) | Colon | Transcriptomics (microarrays) | ↓Proinflammatory chemokines (Cxcl-1, -2, -3, and -5, Ccl-7) | ↓Tumor formation | Song et al. (16) |
| AOM/DSS C57BL/6J mice (n = 14) | Gavage, daily, 3 cycles of DSS treatments | ↓Colonic inflammation | ||||
| Animal study | B. bifidum PRL2010 (109 CFU) | Colon | Transcriptomics (microarrays) | Upregulation of tight-junction genes and β-defensin, downregulation of immune modulators | Modulation of innate immune responses | Turroni et al. (17) |
| Female BALB/c mice (n = 5) | 5-d oral inoculation | |||||
| Animal study | L. plantarum JDFM LP11 (2.5 × 107 CFU/ml) | Ileum | Transcriptomics (RNA-Seq) | ↓Bpi, ↓Rsad2, | Morphological changes in the epithelial layers of the intestines | Shin et al. (18) |
| Female crossbred piglets (n = 6) | Liquid and solid probiotics via drinking and feeds, daily for 4 wk | ↓Slpi, ↓Lum | ||||
| ↓Olfm4, ↓Dmbt1 | ||||||
| Animal study | Saccharomyces boulardii (108 CFU) | Draining mesenteric lymph nodes | Transcriptomics (RNA-Seq) | Modest transcriptional changes | Induced systemic immune response | Hudson et al. (19) |
| C57BL/6 mice (n = NS) | Gavage the day of analysis (single dose) | |||||
| Animal study | L. paracasei K5 (5 × 108 CFU) | Air pouch exudate | Proteomics (microarrays) | Modulation of chemokine expression | ↑Leukocyte recruitment (induced by all strains) | Chondrou et al. (20) |
| BALB/c mice (air pouch model) (n = 10) | L. casei ATCC 393 (5 × 108 CFU) | |||||
| LGG (5 × 108 CFU) | ||||||
| Subcutaneous injection in air pouch | ||||||
| Animal study | L. pentosus 281 (5 × 108 CFU) | Air pouch exudate | Proteomics (microarrays) | ↑sICAM | ↑Leukocyte recruitment (induced by all strains) | Saxami et al. (21) |
| BALB/c mice (air pouch model) (n = 10) | L. plantarum 282 (5 × 108 CFU) | ↑IL-16 | ||||
| L. casei ATCC 393 (5 × 108 CFU) | ↓CXCL-13 | |||||
| LGG (5 × 108 CFU) | ↓TIMP-1 | |||||
| Subcutaneous injection in air pouch | ↓M-CSF | |||||
| Animal study | L. casei ATCC 393 (109 CFU) | Tumor tissue | Proteomics (microarrays) | Modulation Th1 responses | ↓Mean tumor volume | Aindelis et al. (22) |
| Colon CT26 syngeneic tumor mice model (n = 10) | Oral administration, daily for 13 d | |||||
| Animal study | L. acidophilus (6 × 108 CFU) | Serum and colonic tissue | Proteomics (microarrays) | Elevation of TGF-β | ↓Mean tumor number | Mendes et al. (23) |
| AOM/DSS male C57BL/6 mice (n = 29) | L. rhamnosus (6 × 108 CFU) | |||||
| B. bifidum (6 × 108 CFU) | ||||||
| Oral gavage, daily for 3 cycles of DSS treatments | ||||||
| Randomized placebo-controlled, single-blind crossover study | L. casei Shirota (1.3 × 1010 CFU) | Venous blood, saliva | Proteomics (microarrays) | ↑NK cell activity | Immunomodulation | Dong et al. (24) |
| Healthy volunteers (55–80 y) (n = 16) | Per os, daily for 4 wk followed by a 4-wk washout period and 4 wk crossover to the other treatment | Increased | ||||
| IL-10/IL-12 ratio | ||||||
| Randomized, placebo-controlled, crossover study | L. acidophilus (5.2 × 1010 CFU) | Duodenum tissue | Transcriptomics (microarrays) | Modulation of the IFN and IL signaling | Species- and host-specific immunomodulation | van Baarlen et al. (25) |
| Healthy volunteers (n = 7) | L. casei (3.2 × 1010 CFU) | |||||
| L. rhamnosus (1.68 × 1010 CFU) | ||||||
| Per os, daily for 6 wk | ||||||
| Phase I open-label study | LGG (1010 CFU) | Venous blood | Transcriptomics (RNA-Seq) | Modulation of the expression of immune cell trafficking and inflammatory response gene-sets | Modulation of inflammation | Solano-Aguilar et al. (26) |
| Elderly healthy volunteers (65–80 y) (n = 11) | Per os, 2 capsules daily for 28 d |
AOM, azoxymethane; Bpi, bactericidal permeability increasing protein; Ccl, chemokine (C-C motif) ligand; CFU, colony-forming units; Cxcl, chemokine (C-X-C motif) ligand; Dmbt1, deleted in malignant brain tumors 1; DNBS, dinitrobenzene sulfonic acid; DSS, dextran sodium sulfate; LGG, Lactobacillus rhamnosus GG; Lum, lumican; M-CSF, macrophage colony-stimulating factor; MLN, mesenteric lymph node; NS, not specified; Olfm4, olfactomedin 4; Rsad2, radical S-adenosyl methionine domain-containing 2; sICAM, soluble intercellular adhesion molecule; Slpi, secretory leukocyte peptidase inhibitor; SPF, specific pathogen free; Th1, T helper cell type 1; TIMP-1, metallopeptidase inhibitor 1; ↓, statistically significant downregulation; ↑, statistically significant upregulation.
Studies from our laboratory utilizing the dorsal air pouch model of inflammation showed that certain probiotic strains can also exert extraintestinal proinflammatory and immunostimulatory effects. Sterile air was subcutaneously injected to the back of BALB/c mice and after 6 d the intervention group received an injection with the probiotic strain L. paracasei K5. Targeted protein microarray analysis of cytokines and chemokines in the exudates revealed that L. paracasei K5 upregulated CC and CXC chemokines and cytokines of the IL-1 family, supporting the activation and migration of leukocytes on site. The expression of these factors, alongside the expression of Toll-like receptor (TLR)-2, TLR-4, TLR-6, and TLR-9, were validated using qPCR. These effects could be attributed to the probiotic-induced stimulation of p38 mitogen-activated protein kinase signaling pathway (20). Additional preclinical studies utilizing protein microarrays to investigate the immunoregulatory effects of probiotics are presented in Table 1 (21–23).
The employment of high-throughput platforms in clinical studies for the investigation of probiotic-induced immunomodulatory effects has been limited. L. casei Shirota or skim milk was administered to healthy elderly subjects in a randomized, placebo-controlled, single-blind crossover study, for a period of 4 wk, followed by a 4-wk washout, and a crossover to the other treatment. At the end of each treatment peripheral blood and saliva were analyzed in antibody microarrays for the presence of immunological markers. It was found that probiotic supplementation promoted the activity of NK cells and increased the ratio of IL-10 to IL-12. Overall, the intervention led to a reduction of the inflammatory status of the host, while supporting innate immunity (24). In a randomized, placebo-controlled, crossover study, 7 healthy volunteers were assigned to receive L. acidophilus, L. casei, L. rhamnosus, or a maltodextrin solution (placebo group), for 6 wk (25). At the day of intervention, the participants consumed the probiotics in maltodextrose solution every 30 min for 6 h, leading to duodenum biopsy. Affymetrix microarrays were utilized to analyze the transcriptomic profile of the duodenum. Results analysis was carried out by the generation of regulatory nodes (protein–protein and protein–DNA interactions) using the Bibliosphere software. The results demonstrated that L. casei induced the expression of TLR-3 and TLR-9, L. acidophilus upregulated various ILs and interfered with IL-23 signaling, whereas L. rhamnosus modulated IFN-signaling pathways. Interestingly, these transcriptomic changes were found to be highly variable among participants (25). The calculation of the CV revealed that genes encoding factors with predominant roles in cellular signaling, such as the transcriptional factor signal transducer and activator of transcription 3, demonstrated the lowest expression variability. In contrast, chemokines of the CC family exhibited the highest expression variability (25). Finally, RNA-Seq technology was used to investigate the effect of L. rhamnosus GG (LGG) on whole blood cell expression in elderly healthy volunteers (26). Differential expression and ingenuity pathway analysis showed that molecular networks involved in crucial processes, such as immune cell trafficking, inflammatory response, and cell-to-cell signaling, were modulated by the probiotic strain. These effects could be attributed to inhibition of NF-κB complex activation and CCL2 expression (26).
Transcriptomic and proteomic studies of the effect of probiotics on host metabolic pathways
Many studies have correlated mono- or multispecies probiotic interventions with distinct effects on the expression of genes associated with carbohydrate and lipid metabolic pathways. The introduction of multiomics approaches has supported the thorough investigation of these alterations in models of metabolic disease (Table 2). Transcriptomic microarray analysis of hepatic tissues extracted from high-fat-diet (HFD) mice supplemented with B. pseudocatenulatum CECT 7765, exhibited decreased expression of CD36 (Cd36), a fatty acid transporter that regulates lipid accumulation in the liver, and upregulated expression of phosphatase 1 regulatory subunit 3B (Ppp1r3b), early growth response 1 (Egr1), and insulin growth factor binding protein 2 (Igfbp2), counteracting the effects of the HFD (27). For in-depth analysis of the molecular events associated with L. acidophilus SJLH001 supplementation in HFD mice, RNA-Seq technology was used. It was shown that the intervention resulted in 844 DEGs, of which 275 DEGs were correlated with lipid metabolism and ion transport (28). Validation of the downregulated expression of glucose transporter type 4 (Glut4), scavenging receptor Cd36, and Tlr-2, and upregulated expression of cholesterol synthesis-related genes, such as apoA4 (ApoA4) and 3-hydroxy-3-methylglutaryl-CoA reductase (Hmgcr), was performed by qRT-PCR. Notably, the probiotic-treated mice exhibited significant reductions of total cholesterol and oral glucose concentrations (28). Similarly, RNA-Seq analysis was performed to study the modulation of lipid metabolism of broilers treated with L. johnsonii BS15 (29) and of weaned piglets fed with L. reuteri (30) (Table 2). Protein microarrays can also be used to investigate probiotic-induced alterations of host metabolic pathways. For example, it has been demonstrated that L. plantarum Ln4 administration in HFD obese mice significantly downregulated the expression of adipokines, leptin, lipocalin-2, and angiopoietin-like protein-L3, which are correlated with insulin resistance and obesity (31). Similarly, adipokine arrays showed a decrease in leptin concentration in mice supplemented with L. plantarum strains DSR M2 and DSR 920 (32).
TABLE 2.
Transcriptomic and proteomic preclinical and clinical studies of the effect of probiotics in metabolic signaling1
| Type of study | Treatment | Sample | Omics platform | Molecular mechanisms | Clinical outcome | Reference |
|---|---|---|---|---|---|---|
| Animal study | Bifidobacterium pseudocatenulatum CECT 7765 (109 CFU) oral gavage, daily for 7 wk | Liver tissue | Transcriptomics (microarrays) | ↓Lipid transport-related genes | Attenuated HFD-related hepatic steatosis and hyperlipidemia | Moya-Pérez et al. (27) |
| HFD C57BL/6J mice (n = 5) | ↑Carbohydrate metabolism-related genes | |||||
| Animal study | Lactobacillus acidophilus La-JLH001 (109 CFU), oral gavage, daily for 20 wk | Gastrointestinal and adipose tissue | Transcriptomics (RNA-seq) | Glucose metabolism-related genes: ↓Slc10a2, ↑Slc9a3 | Limited HFD-induced hypercholesterolemia and hyperglycemia | Sun et al. (28) |
| HFD C57BL/6J mice (n = 8) | Cholesterol synthesis-related genes: ↑Hmgcr | |||||
| Animal study | L. johnsonii BS15 (106 CFU), oral consumption for 15 d | Hepatic tissue | Transcriptomics (RNA-seq) | ↑Fabp2, ↑Acsbg1 | Altered hepatic metabolism | Qing et al. (29) |
| Male chicks (Cobb 500) (n = 100) | ↑Plin-1, Plin-2, ↓Pla2g4a | |||||
| Animal study | L. reuteri ZLR003 (2 × 109 CFU), oral consumption for 10 d | Jejunum tissues | Transcriptomics (RNA-seq) | Enrichment of GOs related to arachidonic and linoleic acid metabolism | Altered colonic metabolism | Zhang et al. (30) |
| Crossbred Landrace × Large White) (n = 9) | ||||||
| Animal study | L. plantarum Ln4 (5 × 108 CFU), oral gavage, daily for 5 wk | Plasma, epididymal fat tissue | Proteomics (microarrays) | Epididymal fat tissue: ↓CRP, ↓ANGPT-L | Attenuated diet-induced obesity and modulated the expression of T2D biomarkers | Lee et al. (31) |
| HFD C57BL/6J mice (n = 5–7) | ↓IGFBP-1, -3, -5, ↓leptin, ↓lipocalin-2 | |||||
| Animal study | L. plantarum DSR M2 (109 CFU), L. plantarum DSR 920 (109 CFU), oral gavage, daily for 12 wk | Serum, liver tissue, and epididymal fat | Proteomics (multiplex bead array) | Lipogenesis-related proteins: ↓PPARα | Improved obesity parameters | Lee et al. (32) |
| HFD C57BL/6J male mice (n = 5) | Inflammation markers: ↓MCP-1, ↓TNF-α | |||||
| Randomized, double-blind, crossover study design. Healthy young male volunteers (n = 7) | Probiotic yoghurt (Thermophilic Yoflex Culture and LGG) per os, daily for 2 wk | Blood | Transcriptomics (RNA-seq) | Differentially expressed KEGG insulin signaling gene-sets | ↑Postprandial insulin response | Burton et al. (33) |
| Randomized placebo-controlled crossover study. Healthy young volunteers (n = 7–8) | L. plantarum WCFS1 (1011–1012 CFU) pylorus infusion for 1 or 6 h | Pylorus tissue | Transcriptomics (microarrays) | ↓Degs1, ↑Fabp1, ↑Cd36, ↑Idh1 | Global transcriptomic changes | Troost et al. (35) |
Acsbg1, acyl-CoA synthetase bubblegum family member 1; ANGPT-L, angiopoietin-like protein-L; Cd36, clusters of differentiation 36; CFU, colony forming units; CRP, C-reactive protein; Degs1, Δ4-desaturase, sphingolipid 1; Fabp, fatty acid binding protein; GO, Gene Ontology; HFD, high-fat diet; Hmgcr, 3-hydroxy-3-methylglutaryl-CoA reductase; Idh1, isocitrate dehydrogenase (NADP(+)) 1; IGFBP, insulin-like growth factor-binding protein; KEGG, Kyoto Encyclopedia of Genes and Genomes; LGG, Lactobacillus rhamnosus GG; MCP, methyl-accepting chemotaxis protein; Pla2g4a, phospholipase A2 group IVA; Plin, perilipin; PPARα, peroxisome proliferator-activated receptor α; Slc, solute carrier family; T2D, type 2 diabetes; ↓, statistically significant downregulation; ↑, statistically significant upregulation.
Global transcriptomic analysis has also been employed in clinical studies focusing on the effects of probiotics on postprandial metabolism (Table 2). Healthy, young male volunteers participated in a randomized, double-blind, crossover study to investigate the effect of probiotic yoghurt (Thermophilic Yoflex Culture and LGG) on gene expression after meals (33). The volunteers received the probiotic yoghurt or acidified milk for 2 wk, followed by a 2-wk washout period and then a crossover to the other group. Distinct changes in the peripheral blood transcriptome between the probiotic and control groups were recorded. Specifically, RNA-Seq analysis revealed changes in the expression of gene sets involved in glycolysis, oxidative phosphorylation, and inflammation, such as aryl hydrocarbon receptor and epiregulin, with distinct kinetic characteristics in the probiotic group. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis indicated a significant positive enrichment of the insulin signaling pathway, after consumption of the probiotic yoghurt (33). The KEGG pathway database can facilitate the meaningful analysis of transcriptomic data, because it supports the visualization of results and determination of their biological significance (34).
In another study, cDNA and protein microarrays were used for the study of time-dependent effects of L. plantarum WCFS1 supplementation on healthy volunteers. To this aim, the probiotic was infused to the pylorus of the participants for 1 or 6 h and tissue samples were collected for transcriptomic (1 h) and proteomic (6 h) analysis. Transcriptomic analysis of samples from the duodenal mucosa after short-term stimulations revealed the abrupt inhibition in fatty acid metabolism and cell proliferation. Proteomic analysis showed that genes correlated to lipid and fatty acid metabolism, as well as genes involved in oxidative stress protection, were upregulated. Moreover, 1-h probiotic treatments led to modulation of genes encoding for innate immunity factors, whereas 6-h stimulations modulated the expression of members of the human leukocyte antigen family (35).
RNA-Seq and microarray technologies are employed to elucidate the molecular mechanisms of probiotic action. Both platforms can efficiently detect DEGs and provide a high-resolution snapshot of gene expression at a specific timepoint and under set conditions. Additionally, RNA-Seq can identify noncoding transcripts such as microRNAs and long noncoding RNAs, important epigenetic regulators of the cell, offering a more realistic picture of physiological conditions (36, 37). Recent comparative studies have shown that RNA-Seq exhibits higher sensitivity and wider dynamic range than microarrays (36). However, high cost, and complexity of data analysis correlated with this approach, must be taken into consideration. Conclusively, both platforms can contribute greatly to probiotic research by providing high-quality data.
The application of metabolomics to probiotic interventions
Metabolomic approaches are utilized for the identification and characterization of the sum of metabolites in cells, tissues, organs, or organisms, produced under set conditions (38). GC-MS is the gold standard for metabolomic studies of volatile compounds with biological relevance (39). In this context GC-MS is widely applied for accurate detection and determination of SCFAs in probiotic animal studies (40–42) (Table 3). SCFAs are produced by gut microbiota and play a major role in host metabolism, inflammation, intestinal homeostasis, and microbiota-gut-brain communication (43, 44). For example, GC-MS recorded the enhanced production of SCFAs in C57BL/6J mice, after oral administration daily for 7 wk of the probiotic strain B. animalis ssp. lactis GCL2505. Notably, the probiotic-treated mice exhibited higher cecal and plasma concentrations of acetate and propionate that were positively correlated with improved glucose tolerance and reduced fat accumulation and adipocyte volume (40).
TABLE 3.
Metabolomic preclinical and clinical studies of the beneficial effects of probiotic supplementation1
| Type of study | Treatment | Sample | Omics platform | Molecular mechanisms | Clinical outcome | Reference |
|---|---|---|---|---|---|---|
| Animal study | Bifidobacterium animalis ssp. lactis GCL2505 (BlaG), | Blood, cecum | GC-MS | Cecum:↑SCFAs (acetate, propionate) | BlaG: improvement of clinical parameters of metabolic disorder BlaJ: NE | Aoki et al. (40) |
| Μale C57BL/6J mice (n = 23–26) | B. longum ssp. longum JCM1217T (BlaJ (109 CFU) oral administration, daily for 7 wk | Plasma: ↑acetate | ||||
| Animal study | B. bifidum TMC3115 (109 CFU) | Cecum | GC-MS | ↑SCFAs | Lower risk of IgE-mediated allergies in adulthood | Cheng et al. (41) |
| Pregnant (day 13 of gestation) SPF BALB/c mice and their offspring (OVA immunization) (n = 18) | Oral gavage, daily from birth to day 21 | |||||
| Animal study | SIMFORT- VITAFOR (107 CFU) | Cecum | GC-MS | Neonates and adults: ↑butyrate | Protection against experimental asthma in adulthood | Nunes et al. (42) |
| Neonatal and adult C57BL/6 mice (OVA immunization) (n = NS) | Oral gavage, 3 times a week for 21 d | |||||
| Animal study | B. animalis IPLA R1 (∼5 × 108 CFU) | Portal blood, liver, subcutaneous adipose tissues | GC-FID | ↓Triglycerides | Protection against DIO metabolic outcomes | Salazar et al. (46) |
| HFD male C57BL/6J mice (n = 8) | Oral intake of 10% skim milk containing the strain for 10 d | ↓Stearic acid | ||||
| ↓Arachidic acid | ||||||
| ↓Palmitoleic acid | ||||||
| Animal study | Lactobacillus fermentum CECT5716 (1010 CFU) oral gavage for 5 wk | Whole blood and gastrointestinal tissues | GC-FID | Dams: ↓EPA, ↑γ-linoleic acid | Immunomodulation | Azagra-Boronat et al. (47) |
| Wistar rat dams and offspring (n = 48) | (3 wk of pregnancy and 2 wk of lactation) | Pups: ↓total saturated fatty acids, ↓palmitic acid | ||||
| Double-blind, randomized, placebo-controlled, parallel trial in healthy volunteers (n = 35) | L. plantarum Q180 (4 × 109 CFU) | Feces | GC-FID | ↑SCFAs | ↓Postprandial TG | Park et al. (48) |
| Oral intake, daily for 12 wk | ↑Indole and phenol concentrations | ↓ApoB-48 | ||||
| ↓ApoB-100 concentrations | ||||||
| Double-blind, placebo-controlled, randomized intervention trial | L. paracasei ssp. paracasei F19 | Venous blood | GC-TOF | ↑Putrescine | NA | Chorell et al. (49) |
| Healthy term infants (n = 84) | (108 CFU) oral consumption, daily from 4 to 13 mo | ↓Palmitoleic acid | ||||
| Randomized, placebo controlled infant study. Infants with high risk of asthma (birth to 6 mo) (n = 10) | LGG (1010 CFU) Per os, daily from birth to 6 mo | Feces | UPLC-MS/MS | ↑PUFA, ↑DPA, ↑ETA | Immunological tolerance | Durack et al. (50) |
| Double-blind, randomized, placebo-controlled, early-life study. Newborns (birth to first year of life) (n = 49) | Polybiotic mix (107 CFU/g) (B. bifidum, B. breve, B. longum, B. longum ssp. infantis) per os, from birth to 1 y | Feces | UPLC/Q-TOF | Modulation of fecal fatty acids and sterol lipids | NA | Bazanella et al. (51) |
| Animal study | Lab4 mix (5 × 108 CFU) (L. acidophilis CUL21 and CUL60, B. bifidum CUL20, B. animalis ssp. lactis CUL34) and L. plantarum CUL66 | Fecal, blood, liver, and intestinal tissue samples | UPLC-MS | ↓Total cholesterol | Lowering of total plasma cholesterol, reduction of weight gain | Michael et al. (52) |
| Male C57BL/6J mice (n = 6) | Oral intake for 14 d | ↑Fecal unconjugated bile acids | ||||
| Randomized, double-blind crossover study | Bacillus subtilisR0179 (2.5 × 109 CFU) | Plasma | LC-MS/MS | ↑Deconjugated bile acids | NE | Culpepper et al. (53) |
| Healthy adults (n = 18–19) | L. plantarum HA-119 (2.5 × 109 CFU) | |||||
| Bifidobacterium animalis ssp. lactis B94 (2.5 × 109 CFU) oral intake, daily for 6 wk | ||||||
| Double-blind, randomized, parallel, placebo-controlled clinical trial. Overweight or obese individuals (n = 25) | B. animalis ssp. lactis 420 (1010 CFU) + Litesse Ultra polydextrose. Oral intake, daily for 6 mo | Plasma | UHPLC-MS/MS | ↓Deconjugated bile acids | Improved gut barrier and obesity-related markers | Hibberd et al. (54) |
| Animal study | Bifidobacterium longum BB536 (109 CFU) | Feces | NMR | ↑Pimelate | Modulation of gut luminal metabolism | Sugahara et al. (56) |
| GF female BALB/c mice (n = 6) | Oral intake, daily for 14 d | ↑Butyrate | ||||
| Double-blind, randomized, placebo-controlled trial | B. bifidum W23, B. animalis ssp. lactis W52 and L. lactis W58, Ecologic Panda (109 CFU per strain) | Feces | 1H-NMR | ↑Lactate | Temporary prevention of eczema onset | Kim et al. (57) |
| Infants with family history of allergic disease (n = 60) | Oral intake, daily for 12 mo | ↑SCFAs | ||||
| ↓Succinate | ||||||
| ↓Lactose | ||||||
| Animal study MS C57Bl/6J male mice (n = 18) | B. pseudocatenulatum CECT 7765 (108 CFU) gavage, daily for 21 d | Ileum tissue | HPLC-ECD | ↑Dopamine | Modulation of stress responses | Moya-Pérez et al. (59) |
| ↑Noradrenaline | ||||||
| Animal study | L. plantarum PS128 (109 CFU) Gavage, daily for 4 wk | Prefrontal cortex tissue | HPLC-ECD | ↑Dopamine | Amelioration of anxiety and depression-like behaviors | Liu et al. (60) |
| MS C57Bl/6J male mice (n = 10) |
BlaG, Bifidobacterium animalis ssp. lactis GCL2505; BlaJ, Bifidobacterium longum ssp. longum JCM1217T; CFU, colony-forming units; DIO, diet-induced obesity; DPA, docosapentaenoic acid; ECD, electrochemical detection; ETA, eicosatetraenoic acid; FID, flame ionization detector; GF, germ-free; HFD, high-fat diet; LGG, Lactobacillus rhamnosus GG; MS, maternally separated; NA, not available; NE, no effect; NS, not specified; OVA, ovalbumin; SPF, specific pathogen free; TOF, time of flight; UHPLC-MS/MS, ultra-high-performance liquid chromatography-tandem mass spectrometry; UPLC-MS, ultra-performance liquid chromatography-mass spectrometry; UPLC/Q-TOF, ultra-performance liquid chromatography/quadrupole time-of-flight mass spectrometry; ↓, statistically significant downregulation; ↑, statistically significant upregulation.
Variations of the GC-MS platform can be applied for targeted and untargeted metabolomic studies, and the identification of specific metabolites or the sum of metabolites in a sample (45). In this regard, GC coupled with a flame ionization detector is a reliable technique to assess changes in fatty acid profiles in animals following administration of probiotic bacteria (46–48) (Table 3). Furthermore, GC-time-of-flight-MS has been used to capture changes in fatty acid content of plasma samples, derived from infants who consumed cereal fortified with the probiotic strain L. paracasei LF19. Among the 288 differentially produced metabolites, concentrations of the MUFA palmitoleic acid were significantly lower in the probiotic group (49).
LC-MS is commonly preferred for studies investigating nonvolatile lipid compounds in complex samples (Table 3). In this respect, ultra-high-performance LC was employed to profile the fecal lipid content of infants who consumed LGG during the first months of life (50). It was demonstrated that probiotic treatment differentially altered the production of various fatty acids and led to the promotion of immunological tolerance, which could be beneficial in the context of allergy or asthma prevention (50). Similarly, newborns who were recruited to consume a polybiotic mix, consisting of Bifidobacterium bifidum, B. breve, B. longum, and B. longum ssp. infantis, showed significant changes in the concentrations of fecal fatty acids, sterol lipids, and glycerophospholipids compared with breastfed infants or newborns who consumed regular whey-based formula (51). Another important application of LC-MS in probiotic research is for the measurement of bile acids in plasma or feces of preclinical or clinical experimental models (52–54) (Table 3). Ultra-performance LC analysis showed an increase in fecal unconjugated bile acid content in mice fed the probiotic mix Lab4 (L. acidophilus CUL21 and CUL60, B. bifidum CUL20, and B. animalis ssp. lactis CUL34), and L. plantarum CUL66 (52). Moreover, alterations in amino acid metabolism and decreased plasma concentrations of primary and secondary conjugated bile acids were recorded in overweight adults who received B. animalis ssp. lactis 420 alone or in combination with Litesse Ultra polydextrose. These metabolic changes were accompanied by a clinical reduction of fat mass (54).
NMR spectroscopy is a commonly applied analytical method in metabolomics for the identification and quantification of compounds in nontargeted approaches. It can be used for the identification of low molecular weight compounds that are present in high concentrations in complex samples (55). NMR analysis showed that conventionalized gnotobiotic mice fed Bifidobacterium longum BB536 had significantly elevated fecal concentrations of butyrate, acetate, and pimelate, a biotin precursor, compared with control mice (56). In the same manner, children with a high risk of eczema who were enrolled in a double blind, randomized, placebo-controlled trial and consumed the polybiotic mix of strains B. bifidum W23, B. animalis ssp. lactis W52, and Lactococcus lactis W58, showed enhanced fecal total SCFA and acetate concentrations and temporary protection against the development of eczema (57).
Finally, metabolomic platforms are also used to investigate changes in the concentrations of neurotransmitters and hormones linked to probiotic supplementation (Table 3). For the study of monoamine neurotransmitters that could be present in nanomolar concentrations in samples, HPLC-electrochemical detection (HPLC-ECD) is commonly used (58). For example, intestinal tissues extracted from early-life-stressed maternally separated C57B1/6J male breastfed pups, gavaged with B. pseudocatenulatum, were analyzed with HPLC-ECD for the evaluation of monoamine content. It was found that probiotic supplementation normalized dopamine and adrenaline concentrations. These effects were accompanied by the amelioration of stress and depression-like behaviors (59). Similar findings were presented in an independent study investigating the psychotropic effects of L. plantrarum PS128 on maternally separated mice (60) (Table 3).
The use of metagenomics in probiotic research
Metagenomics are employed for the investigation of the structure (taxonomic metagenomics) and function (functional metagenomics) of complex microbial communities, present in any environment with specific conditions, without the need of isolation and propagation of the microbes in laboratory settings (61). Metagenomic studies utilize 2 approaches: amplicon-based detection of specific sequences, such as hypervariable regions of the 16S rRNA gene in bacteria; or whole-genome shotgun sequencing (WGS) (62). WGS provides higher sensitivity and higher yield in variant detection; however, in many cases amplicon sequencing is more accessible, due to lower cost and less complicated bioinformatic needs (63). Both platforms have been used for the investigation of probiotic-induced modulations in the gut microbiome, and the effects on inflammation, gut–brain communication and neurological disorders, and host metabolism (Table 4).
TABLE 4.
Metagenomic studies of the effects of probiotic supplementation on the gut microbiota and beneficial health outcomes1
| Type of study | Treatment | Sample | Omics platform | Microbiota composition changes | Clinical outcome | References |
|---|---|---|---|---|---|---|
| Animal study | Lactobacillus acidophilus (5 × 108 CFU) | Feces, rectum mucosa | 16S rDNA sequencing | ↑Bifidobacteria, ↑Lachnospiraceae_NK4A136 | ↓Colonic inflammation | Wang et al. (64) |
| DSS-treated C57BL/6J mice (n = 60) | L. rhamnosus (5 × 108 CFU) | ↑Lachnospiraceae_UCG-006 | ||||
| Bifidobacterium lactis (5 × 108 CFU) | ↓Alistipes | |||||
| Gavage, daily for 1 wk | ||||||
| Animal study | L. plantarum LP Onlly (109 CFU) | Feces | Shotgun sequencing | ↑Bacteroides | Prevention of gut inflammation | Chen et al. (65) |
| IL-10 knockout mice (n = 12) | Gavage, daily for 4 wks | WGS | ↑Akkermansia muciniphila | |||
| Single-center, prospective, randomized, controlled study | B. longum, L. acidophilus, and Enterococcus faecalis (1:1:1) (6 × 107 CFU) | Colorectal mucosa tissue | 16S rRNA sequencing | ↓Fusobacterium | NA | Gao et al. (69) |
| CRC patients (n = 11) | Oral intake, daily for 5 d | ↓Peptostreptococcus | ||||
| Prospective randomized study | B. lactis Bl-04 (1.4 × 1010 CFU) | Feces, tumor tissue | 16S rDNA sequencing | Fecal samples: ↑Firmicutes | NA | Hibberd et al. (70) |
| CRC patients (n = 28) | L. acidophilus NCFM (7 × 109 CFU) and inulin. Oral consumption for 31±28 d | Tumor microbiota: decrease of Fusobacterium and Peptostreptococcus | ||||
| Animal study | L. reuteri MM4-1A (108 organisms/mouse/d) | Feces | 16S rRNA sequencing | ↑L. reuteri | Restoration of social behavior | Buffington et al. (71) |
| C57Bl6/J mice with autism spectrum disorder (n = 10) | Oral administration, daily for 4 wk | |||||
| Animal study | L. reuteri ATG-F4 (107 CFU/mL) | Feces | 16S rRNA sequencing | ↑Bacteroidetes | ↑Serum dopamine concentrations | Beck et al. (72) |
| C57BL/6J male mice (n = 10) | Oral administration, daily for 4 wk | ↑Bacteroidales S24–7 | ||||
| ↑Prevotellaceae | ||||||
| Animal study | B. breve CCFM1025 (108 CFU/10g) oral gavage, daily for 6 wk | Feces | 16S rRNA sequencing | ↓Allobaculum spp. | Antidepressant effects | Tian et al. (73) |
| Chronically stressed male C57BL/6J mice (n = 10) | ↓Bifidobacterium spp. | |||||
| ↑Coprococcus spp. | ||||||
| ↑Oscillospira spp. | ||||||
| Animal study | L. plantarum LP3, L. rhamnosus LR5, B. lactis BL3, B. breve BR3, and Pediococcus pentosaceus PP1 (2 × 108 CFU/mL), oral administration, daily for 8 wk | Feces | 16S rRNA sequencing | ↑Bifidobacterium spp. | Antidepressant effects | Liu et al. (74) |
| ICR stressed mice (n = 10) | ||||||
| Randomized, double-blind, and placebo-controlled multicenter trial. Healthy elders (>65 y) | B. bifidum BGN4 (109 CFU/d) | Feces | 16S rRNA sequencing | ↓Eubacterium, ↓Clostridiales | ↑Cognitive function | Kim et al. (75) |
| B. longum BORI (109 CFU/d) | ↓Prevotellaceae, ↓Allisonella | |||||
| Oral intake, daily for 12 wk | ||||||
| Double-blind, placebo-controlled, parallel-group clinical trial. Healthy students (n = 29) | Heat-killed L. gasseri CP2305 (1010 CFU) per os, 2 tablets daily for 24 wk | Feces | 16S rRNA sequencing | ↑Bifidobacterium spp. | Ameliorated sleep disturbance and anxiety | Nishida et al. (76) |
| ↓Streptococcus spp. | ||||||
| Animal study | L. rhamnosus GG (107 CFU), oral administration, daily for 10 wk | Feces | 16S rRNA sequencing | ↓Oscillospira spp. | ↓Weight gain | Ji et al. (78) |
| HFD C57BL/6J mice (n = 7) | ↓Organ weight | |||||
| Animal study | B. pseudolongum (1011 CFU), gavage, daily for 6 wk | Feces | 16S rRNA sequencing | ↑Butyricimonas, ↑Bifidobacterium | ↓Body mass and visceral fat | Bo et al. (79) |
| HFD C57BL/6J mice (n = 6) | ↑Odoribacter | ↓Plasma triglycerides | ||||
| ↓Firmicutes/Bacteroidetes ratio | ||||||
| Animal study | Bacillus amyloliquefaciens SC06 (108 CFU), oral consumption, daily for 8 wk | Feces | 16S rRNA sequencing | ↓Firmicutes/Bacteroidetes ratio | ↓Subcutaneous fat weight, ↓insulin resistance | Wang et al. (80) |
| HFD C57BL/6J mice (n = 15) | ||||||
| Animal study | L. plantarum FZU3013 (109 CFU/mL), oral consumption, daily for 8 wk | Feces | 16S rRNA sequencing | ↑Alistipes | Potential role in prevention of NAFL and hyperlipidemia | Chen et al. (81) |
| HFD SPF Kunning mice (n = 8) |
CFU, colony forming units; CRC, colorectal cancer; DSS, dextran sodium sulfate; HFD, high-fat diet; ICR, Institute of Cancer Research; NA, not available; NAFL, nonalcoholic fatty liver; SPF, specific pathogen free; WGS, whole genome sequencing; ↓, statistically significant downregulation; ↑, statistically significant upregulation.
Metagenomics to study the effect of probiotic-induced microbiome modulation in intestinal inflammation
Gut dysbiosis has been implicated as a contributing factor in inflammation, because it supports the proliferation of inflammation-promoting bacteria, often leading to acute colitis. Several preclinical studies have aimed at evaluating the potential of probiotic and symbiotic treatments to reverse these structural microbiota changes (64, 65) (Table 4). To this end, administration of the probiotic mix L. acidophilus, L. rhamnosus, and B. lactis alone or in combination with inulin, to DSS-treated C57BL/6J mice, resulted in the increase of beneficial bacterial populations of Bifidobacterium, Lachnospiraceae_NK4A136, and Lachnospiraceae_UCG-006, and in the decrease of bacteria belonging to the Alistipes genus, as demonstrated by 16S rRNA sequencing. Additionally, these microbial alterations were accompanied by attenuation of gut inflammation, as evidenced by histological examinations (64). Accordingly, shotgun metagenomics reported that administration of L. plantarum LP-Onlly to IL-10−/− mice, an inflammatory bowel disease model, induced the proliferation of Bacteroides and Akkermansia muciniphila. These changes could be correlated with the improved inflammation profile observed (65). The dysbiotic gut is also characterized by epigenetic modifications in the colonic tissue that are associated with increased risk of carcinogenesis. These alterations mainly involve promoter hypermethylation of tumor suppressors, such as the Wnt inhibitory factor 1 (WIF1) and other genes involved in the Wnt signaling pathway (66). Recent metagenomic studies of human colon cancer tissues have correlated these epigenetic changes with high abundance of specific tumorigenic bacteria, such as Peptostreptococcus and Fusobacterium species (67). Furthermore, high abundance of these bacterial species has been linked with the onset and lower survival rate of patients with colorectal cancer (CRC) (68). Metagenomic analysis was employed in 2 clinical studies to investigate the potential inhibitory effect of probiotic supplementation against the proliferation of these bacterial species (69, 70) (Table 4). In the first study, CRC patients were assigned to receive a mixture of B. longum, L. acidophilus, and E. faecalis or placebo, for 5 d. Colonic tissue samples were collected, and their microbial load was analyzed by 16S rRNA pyrosequencing. Metagenomic analysis revealed a significant reduction in Peptostreptococcus and Comamonas populations and a decrease from 10% to 2% in Fusobacterium species (69). In the second study, 15 CRC patients were assigned to receive daily probiotic tablets containing B. lactis Bl-04 and L. acidophilus NCFM or placebo, from the day of diagnosis until surgery. The intervention resulted in elevated Clostridiales spp. and butyrate-producing Faecalibacterium, Roseburia, and Eubacterium bacteria, whereas the CRC-associated Fusobacterium and Peptostreptococcus species were decreased in the probiotic group, as evidenced by16S rRNA gene sequencing (70).
Metagenomics to study the effect of probiotic-directed microbiome modulation in neurological disorders
Probiotic administration can affect neurodevelopment, behavior, and mood disorders by modifying the gut microbiome (71–76) (Table 4). It has been demonstrated that Bifidobacterium breve CCFM1025 altered the gut microbiota composition of chronically stressed C57BL/6J male mice, after a 5-wk oral administration regimen (73). 16S rRNA sequencing of fecal samples revealed the increased abundance of SCFA-producing Allobaculum spp., Coprococcus spp., and Bifidobacterium spp. Higher stool SCFA concentrations are positively correlated with elevated colonic 5-hydroxytryptophan and 5-hydroxytryptamine hippocampal concentrations, resulting in antidepressant effects (73). Similarly, administration of a probiotic formulation containing L. plantarum LP3, L. rhamnosus LR5, B. lactis BL3, B. breve BR3, and Pediococcus pentosaceus PP1 for 8 wk to mice, reversed stress-induced microbiota structural changes by stimulating proliferation of Actinobacteria, Cyanobacteria, Lactobacillus, and Bifidobacterium populations. These compositional alterations could have supported attenuation of serum corticosterone concentrations and subsequent alleviation of depressive-like manifestations (74). Metagenomics was also employed in a randomized, double-blind, placebo-controlled, multicenter clinical trial to investigate the effect of probiotic capsules consisting of B. bifidum BGN4 and B. longum BORI to individuals aged >65 y for a 3-mo period (75). Stool samples were collected and their microbial load was examined with 16S rRNA gene sequencing. Significant reductions of Eubacterium, Clostridiales, Prevotellaceae, and Allisonella abundances in the gut were recorded. The intervention resulted in favorable outcomes in cognitive function and mental stress (75). Another clinical study indicated that administration of heat-killed L. gasseri CP2305 to medical students prior to national examination alleviated stress manifestations and positively influenced their sleep patterns, while it also reversed microbiota changes related to stress. Indeed, the probiotic managed to normalize the abundances of Bifidobacterium spp. and Streptococcus spp. populations that were disrupted due to stress. Furthermore, the authors postulated that this probiotic could directly signal to the brain via the vagus nerve and stimulate the gut–brain axis (76).
Metagenomics to study the impact of probiotics on host metabolic control
A disrupted gut microbiome negatively influences host metabolism, which can contribute to the development of metabolic syndrome (77). Metagenomics has revealed probiotic-directed modulation of gut microbiota for the management of metabolic disease (78–81) (Table 4). Amplicon sequencing of feces sampled from HFD mice gavaged with Bifidobacterium pseudolongum showed significant changes in the structure and function of the gut microbiota. The ratio of Firmicutes/Bacteroidetes was significantly reduced, whereas Butyricimonas and Bifidobacterium abundances increased. KEGG analysis for the identification of functional microbiota alterations showed that probiotics modulated pathways involved in lipid, carbohydrate, and amino acid metabolism. These findings were accompanied by the clinical decrease of body mass, visceral fat, and gross energy intake (79). Furthermore, hyperlipidemic HFD mice gavaged with L. plantarum FZU3013 had increased populations of Alistipes, a genus that contains several SCFA-producing species, and Ruminococcus. The administration of L. plantarum FZU3013 also minimized the gut population of Desulfovibrio, which is positively correlated with obesity and type 2 diabetes (81).
Genomics and comparative genomics of probiotics
The genome of novel potentially probiotic isolates should be fully sequenced and available in public databases to facilitate accurate taxonomic identification, investigation of functional traits, and evaluation of the safety profile (82). The advancement of next-generation sequencing (NGS) technologies has made WGS of probiotic strains more affordable and accessible. Furthermore, the numerous available bioinformatics tools support assembly, annotation, and phylogenetic analysis of the genomic sequences (83, 84). Computational mining of whole genomes is used to construct phylogenetic trees, provide more accurate insights into the evolutionary relations between strains, and facilitate molecular taxonomy. In this light, the genus Lactobacillus has recently been reclassified as 25 genera, based on shared physiological and metabolic properties. The new taxonomic classification could enhance our understanding of common probiotic mechanisms of action (85, 86).
Comparative genomics is also employed to investigate the range of phenotypic variability between strains, as well as to identify the entire set of strain-specific genes in their pangenome (the entire set of genes of all strains within a monophyletic group) and to highlight shared properties (87). Moreover, it can elucidate conserved sequences in the genome of probiotics that code for essential cellular functions, and regulatory elements that modulate their expression. Cluster of orthologous genes analysis can classify these findings into distinct categories, such as clusters involved in transcription, metabolism, cell motility, and signal transduction, among others (88). Furthermore, genomic comparison of novel isolates with already characterized probiotics can reveal functional properties, such as adhesion to epithelial cells, autoaggregation, stress response mechanisms, and defense mechanisms, including the presence of virulence factors and resistance to antibiotics (89).
The rapid spread of antibiotic resistance is a major threat to global health. Probiotic bacteria with intrinsic resistance to antibiotics are generally recognized as safe because they present a minimal risk of spreading drug-resistant genes to other more harmful species. However, in probiotic stains with acquired resistance genes, mainly carried on mobile genetic elements, such as plasmids, transposons, and integrons, the possibility of horizontal transmission is considered to be high, thus representing a serious safety issue (90). Indeed, the transfer of a vancomycin resistance gene from enterococci to the probiotic strain L. acidophilus has been recorded in vitro and, more importantly, in the gut of mice at high frequencies (91). Assessment of antibiotic resistance of novel probiotic strains is performed phenotypically by the determination of minimal inhibitory concentrations (92), and genotypically by PCR-based techniques and sequencing (93, 94). For the identification and localization of previously undetected antibiotic resistance genes, DNA microarrays, as well as WGS platforms, might also be used (94, 95). Furthermore, multiomics technologies and other advanced molecular tools, such as live imaging, can accurately represent probiotics–microbiome interactions and evaluate the risk of antibiotic resistance gene transfer, as well as risks linked to long-term consumption (96). To date, horizontal gene transfer from probiotics to other bacteria has not been recorded in the human host; however, further research is needed.
Future probiotics and the road to success
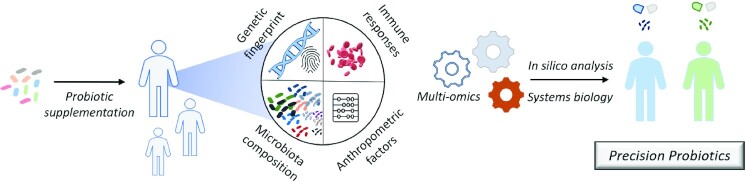
Recent breakthroughs in the study of the microbiome, supported by multiomics and systems biology approaches, have highlighted the role of the gut microbiota in the host's health. It has also been demonstrated that the gut microbiota, alongside other host- and sex-specific traits, contributes significantly to the efficacy of probiotic supplementation. Consequently, the idea of personalized, tailor-made probiotics has been developed, setting the stage for a new era of probiotic research (97). Evidently, personalized probiotic supplementation should consider anthropometric and immunological features, as well as the microbiome and genetic fingerprint of the host (Figure 1). Indeed, several studies have associated genetic diversity with specific gut microbial populations (98, 99). The observed variation was pinpointed in loci coding for factors involved in host–microbial interactions, such as the pattern recognition receptor and chemokine signaling, and in metabolic pathways. These data could allude to the fact that the genetic makeup of the host could affect probiotic action. Furthermore, a number of studies have shown that the human gut microbiota exerts a host-specific resistance to probiotic colonization. In a recent elegant study, individuals who consumed the commercial probiotic mix Supherb Bio-25 (B. bifidum, L. rhamnosus, L. lactis, L. casei ssp. casei, B. breve, S. thermophilus, B. longum ssp. longum, L. casei ssp. paracasei, L. plantarum, and B. longum ssp. infantis) exhibited host- and site-specific patterns of colonization. Interestingly, germ-free mice that were conventionalized with fecal microbiota from individuals resistant to probiotic colonization, were less receptive to probiotic supplementation, whereas mice that were fed with fecal microbiota of permissive individuals showed a better response. These findings underline the fundamental role that the gut microbiota plays in the efficiency of probiotic colonization (13). Similarly, it has been shown that probiotic resistance to colonization could be associated with the diversity of the residing microbial populations. In this respect, L. helveticus MTCC 5463 colonization was more efficient in healthy individuals, who had a higher gut microbiome α-diversity (100). It has also been demonstrated recently that antibiotic treatments can alleviate this innate resistance to probiotic colonization. Indeed, individuals who were treated with wide-spectrum antibiotics prior to supplementation with Supherb Bio-25, were more permissive than naïve individuals (101). Comparative analysis of metagenomic data from permissive and nonpermissive individuals could reveal species that could act as predictive markers for the success of probiotic supplementation. For example, the microbiome of the responders to L. helveticus MTCC 5463 supplementation had decreased abundance of Clostridium and increased Eubacterium populations (100).
FIGURE 1.
The road to precision probiotics. The unique host genetic and microbiota signatures, alongside anthropological and immune parameters, can determine the efficiency of probiotic supplementation. The identification of these factors using high-throughput multiomics analysis and systems biology approaches can set the basis for tailor-made probiotic interventions.
Probiotic research needs to gravitate toward the understanding of strain-specific mechanisms of action, by delving deeper into their specific genetic and metabolic signatures. The advancement of the field of comparative genomics has facilitated the prediction of probiotic characters and attributes. Additionally, global transcriptome and proteome analyses are being employed to investigate the production of proteins and other small molecules that implement probiotic action, and posttranslational modifications that can modulate their activity (102). Metabolomic studies have aided in the characterization of produced metabolites, termed “postbiotics.” Postbiotics are soluble, bioactive compounds secreted by probiotic microorganisms in their growth medium (103), that can originate from both intracellular and extracellular compartments and can be cell surface proteins, secreted proteins and peptides, exopolysaccharides, teichoic acids, organic acids such as SCFAs, enzymes, neurotransmitters such as γ-aminobutyric acid, or vitamins (104). They can act alone or synergistically and induce immunomodulatory or epigenetic effects in the host, whereas some of them can exert antimicrobial activities against pathogenic bacteria (105). The fractionation of cell-free supernatants, and enzymatic (proteinase K, trypsin, catalase, α-chymotrypsin), heat, and pH treatments, can provide valuable information about their molecular weight and chemical identity. However, only high-throughput analyses can provide definitive answers about the identity and physicochemical properties of these compounds. Such analyses can be carried out in platforms like LC-MS or GC-MS and NMR. Importantly, metabolomics can decipher the context in which these molecules can be produced and secreted. Several studies have shown that probiotics can produce a wide array of metabolites, based on the environmental stimuli that they receive. More specifically, it has been shown that gastrointestinal tract conditions (106), available nutrients (107), as well as proinflammatory microenvironments (108), can determine the production and secretion of certain metabolites. Apart from host-related interactions, bacteria grown in food matrices show different metabolic signatures compared with those cultured in common laboratory media (109). These platforms can also be used to investigate the interactions between different probiotic strains of multispecies supplements, because interbacterial communication can also modulate gene expression and consequent metabolite production. It is known that bacteria utilize an array of language signals, such as quorum sensing, to communicate with other bacteria of the same or different species. The exchanged signals can determine important behaviors such as biofilm formation, production of virulence factors, metabolism, and response to stress (110). These language signals and other metabolites could also determine the probiotic–microbiome interactions. For instance, a recent study supported that lactobacilli contained in the mix Supherb Bio-25 delayed microbiome reconstitution after antibiotic treatments, because they produced uncharacterized antibacterial compound(s) (101). Conclusively, comparative metabolomic studies between different strains of probiotics, or strains grown in different media or simulated host conditions, could be informative about their species- and host-specific actions.
The current trend in probiotic research is the identification of microorganisms with probiotic potential that are not originated from dairy products. These microorganisms, termed “next-generation probiotics” (NGPs), are usually isolated from the gastrointestinal tract of mammals and are solely intended for pharmaceutical use (111). Because they require stringent survival conditions, such as complex growth media and reduced oxygen availability, their propagation in laboratory conditions is quite challenging. The advent of culture-independent platforms has facilitated the study of their probiotic attributes and safety profile in silico. In this context, metagenomics has supported the discovery of novel NGPs, whereas comparative genomics has provided a basis for the characterization of their biological functions, virulence, and antibiotic susceptibility (112). Furthermore, proteomics and metabolomics can be employed for the investigation of temporal and spatial actions on the host and accurately describe their holistic effects. Indeed, several studies have already associated NGPs, such as Prevotella copri, Bacteroides thetaiotaomicron, Akkermansia muciniphila, and Faecalibacterium prausnitzii, with favorable health outcomes in intestinal inflammation and metabolic disorders (113). For the commercialization of these strains, it is important to ensure maximum viability during production and shelf life, taking into consideration their unique survival requirements (111). Furthermore, survival during gastric passage should be guaranteed, because these bacteria have evolved to withstand the conditions of the lower, but not the upper, gastrointestinal tract (114). Lastly, it is important to note that appropriate regulatory frameworks should be put in place for the evaluation, characterization, and commercialization of products containing these bacteria (111).
Multiomics analysis has, indeed, broadened our understanding about the global effects of probiotic supplementation. Although these platforms have been extensively used to explore the interaction of probiotics with immune and metabolic signaling, as well as with the residing gut populations, the meaningful interpretation of the generated data remains challenging. The evolving field of systems biology has provided the mathematical and computational tools to create probabilistic models of biological function at the single-cell, tissue, or organ-system scale based on the data derived from these platforms (115). More specifically, systems biology utilizes statistical methods, such as gene set enrichment analysis, and computational units, such as biological networks and dynamic models, to decipher the information flow from gene to transcript, to protein and metabolites. However, the rapid production of experimental data demands more robust tools than mechanistic models. To this aim, artificial intelligence (AI) approaches, such as machine learning, deep learning, and artificial neural networks, can be utilized to manage big numerical datasets. The predictive models produced by AI could be invaluable for precision medicine (115). Systems biology has, indeed, been proven useful in probiotic and microbiome research. More specifically, the compilation of data extracted by high-throughput platforms has aided in the understanding of the genomic and functional properties of probiotics and has supported the strain specificity of their actions (116), whereas systems medicine and microbiome-wide association studies have provided insights about the role of the microbiome in health and disease, paving the way to targeted therapeutics (117). In a representative study, Bisanz et al. (118) characterized the global molecular and cellular events triggered by vaginal probiotic treatments for postmenopausal women (118). Microbial profiling by NGS, microarray analysis of chemokine and cytokine production, and metabolomic GC-MS analysis of the vaginal fluid suggested that L. rhamnosus GR-1 and L. reuteri RC-14 did induce mild changes that were, however, not reflected in clinical outcomes. The authors postulated that host-specific factors and the small sample size contributed to the lack of statistically significant clinical results and proposed that further studies are needed. Thus, this multifaceted analysis managed to reveal fine molecular changes that would otherwise be overlooked.
Novel technologies that have recently been introduced could complement our understanding about host–microbe interactions. Single-cell multiomics analyses can realize the simultaneous profiling of nucleic acids and proteins at single-cell resolution. Different platforms allow for combined genome and transcriptome sequencing (single-cell genome and transcriptome sequencing), genome, epigenome, and transcriptome sequencing (single-cell triple omics sequencing), or transcriptome and proteome sequencing (RNA expression and protein sequencing) (119). These cutting-edge technologies are highly suitable to study dynamic processes, such as time-dependent host–microbe interactions (120). The 2 major limitations on the implementation of such techniques are cost and data analysis. Data analysis of these platforms includes the integration of a bulk of data generated from different platforms (121, 122), which complicates the meaningful interpretation of the results (123). However, as the use of these technologies and the field of systems biology mature, these obstacles are expected to be overcome.
Conclusions
Τhere is a growing body of evidence that demonstrates the prophylactic and therapeutic potential of probiotics against acute or chronic diseases. Nevertheless, probiotics have not yet been introduced in clinical practice, except for the management of a limited number of gastrointestinal disorders, because several questions related to probiotic production, efficacy, and health benefits remain unanswered. The great variability of probiotic mechanisms of action and host-specific traits has contributed greatly to discrepancies in outcomes presented in preclinical and clinical studies. Clinical protocols differ a lot between probiotic studies, thus meta-analysis often leads to contradictory conclusions. Furthermore, the dynamics of probiotic–probiotic interactions are insufficiently researched; consequently it is difficult to assess the effectiveness of multistrain supplementation because it is unclear whether inhibitory or proliferative relations exist among strains. To this end, multiomics approaches can be applied to systematically characterize and predict host–microbe and microbe–microbe interactions and evaluate probiotic efficacy. The identification of probiotic genes and the context in which they are expressed, as well as the immense range of metabolites that can be produced by each strain, can contribute to the elucidation of strain-specific actions and interactions with the host and its microbiota. Accordingly, high-throughput analyses of the unique genomic, proteomic, metabolomic, and metagenomic signature of the host can shed light on factors that influence the efficiency of probiotic action. Adding to that, the thorough investigation of NGP and postbiotic attributes can make available an arsenal of bioactive compounds with potential health-promoting effects to people who are allergic to dairy products or to other vulnerable populations, such as the immunocompromised, and are advised not to consume traditional probiotic foods or supplements. In future, routine use of multiomics platforms, single-cell technologies, and the integration of systems biology in probiotic research will contribute to the careful design of tailor-made interventions that would take into consideration species-, host-, and disease-specific factors and hopefully bring probiotic supplementation from bench to bedside.
ACKNOWLEDGEMENTS
The authors’ responsibilities were as follows—DEK, MR, MT, PC: searched, collected, and analyzed the literature and drafted the manuscript; DEK: discussed and revised all drafts; AG: was responsible for conception, design, writing, reviewing, and editing; and all authors: read and approved the final manuscript.
Notes
This work was supported by the project ELIXIR-GR: Hellenic Research Infrastructure for the Management and Analysis of Data from the Biological Sciences (MIS 5002780), which is implemented under the Action Reinforcement of the Research and Innovation Infrastructure, funded by the Operational Program Competitiveness, Entrepreneurship and Innovation (NSRF 2014-2020) and co-financed by Greece and the European Union (European Regional Development Fund).
Author disclosures: The authors report no conflicts of interest.
Abbreviations used: AI, artificial intelligence; CRC, colorectal cancer; DEG, differentially expressed gene; DNBS, dinitrobenzene sulfonic acid; DSS, dextran sodium sulfate; HFD, high-fat diet; HPLC-ECD, high-pressure liquid chromatography-electrochemical detection; KEGG, Kyoto Encyclopedia of Genes and Genomes; LGG, Lactobacillus rhamnosus GG; NGP, next-generation probiotic; NGS, next-generation sequencing; RNA-Seq, RNA-sequencing; TIFA, TNF receptor-associated factor (TRAF) interaction protein with a forkhead-associated domain; TLR, Toll-like receptor; WGS, whole genome sequencing.
Contributor Information
Despoina Eugenia Kiousi, Department of Molecular Biology and Genetics, Faculty of Health Sciences, Democritus University of Thrace, Alexandroupolis, Greece.
Marina Rathosi, Department of Molecular Biology and Genetics, Faculty of Health Sciences, Democritus University of Thrace, Alexandroupolis, Greece.
Margaritis Tsifintaris, Department of Molecular Biology and Genetics, Faculty of Health Sciences, Democritus University of Thrace, Alexandroupolis, Greece.
Pelagia Chondrou, Department of Molecular Biology and Genetics, Faculty of Health Sciences, Democritus University of Thrace, Alexandroupolis, Greece.
Alex Galanis, Department of Molecular Biology and Genetics, Faculty of Health Sciences, Democritus University of Thrace, Alexandroupolis, Greece.
References
- 1.Food and Agriculture Organization of the United Nations/World Health Organization (FAO/WHO) . Guidelines for the evaluation of probiotics in food. 2002; [Internet]. [cited 2020 May 22]. Available from: https://www.who.int/foodsafety/fs_management/en/probiotic_guidelines.pdf [Google Scholar]
- 2.Kumar M, Nagpal R, Kumar R, Hemalatha R, Verma V, Kumar A, Chakraborty C, Singh B, Marotta F, Jain Set al. Cholesterol-lowering probiotics as potential biotherapeutics for metabolic diseases. Exp Diabetes Res. 2012;2012:902917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plaza-Díaz J, Ruiz-Ojeda FJ, Vilchez-Padial LM, Gil A. Evidence of the anti-inflammatory effects of probiotics and synbiotics in intestinal chronic diseases. Nutrients. 2017;9:555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alard J, Lehrter V, Rhimi M, Mangin I, Peucelle V, Abraham AL, Mariadassou M, Maguin E, Waligora-Dupriet AJ, Pot Bet al. Beneficial metabolic effects of selected probiotics on diet-induced obesity and insulin resistance in mice are associated with improvement of dysbiotic gut microbiota. Environ Microbiol. 2016;18:1484–97. [DOI] [PubMed] [Google Scholar]
- 5.So SS, Wan ML, El-Nezami H. Probiotics-mediated suppression of cancer. Curr Opin Oncol. 2017;29:62–72. [DOI] [PubMed] [Google Scholar]
- 6.Abraham BP, Quigley EMM. Probiotics in inflammatory bowel disease. Gastroenterol Clin North Am. 2017;46:769–82. [DOI] [PubMed] [Google Scholar]
- 7.Le Barz M, Daniel N, Varin TV, Naimi S, Demers-Mathieu V, Pilon G, Audy J, É Laurin, Roy D, Urdaci MCet al. In vivo screening of multiple bacterial strains identifies Lactobacillus rhamnosus Lb102 and Bifidobacterium animalis ssp. lactis Bf141 as probiotics that improve metabolic disorders in a mouse model of obesity. FASEB J. 2019;33:4921–35. [DOI] [PubMed] [Google Scholar]
- 8.Casas-Solís J, Huizar-López MDR, Irecta-Nájera CA, Pita-López ML, Santerre A. Immunomodulatory effect of Lactobacillus casei in a murine model of colon carcinogenesis. Probiotics Antimicrob Proteins. 2020;12:1012–24. [DOI] [PubMed] [Google Scholar]
- 9.Kim SK, Guevarra RB, Kim YT, Kwon J, Kim H, Cho JH, Kim HB, Lee JH. Role of probiotics in human gut microbiome-associated diseases. J Microbiol Biotechnol. 2019;29:1335–40. [DOI] [PubMed] [Google Scholar]
- 10.Salvetti E, O'Toole PW. The genomic basis of Lactobacilli as health-promoting organisms. Microbiol Spectr[Internet] 2017;5(3). doi:10.1128/microbiolspec.BAD-0011-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gueimonde M, Collado MC. Metagenomics and probiotics. Clin Microbiol Infect. 2012;18:32–4. [DOI] [PubMed] [Google Scholar]
- 12.Bottacini F, van Sinderen D, Ventura M. Omics of Β ifidobacteria: research and insights into their health-promoting activities. Biochem J. 2017;474:4137–52. [DOI] [PubMed] [Google Scholar]
- 13.Zmora N, Zilberman-Schapira G, Suez J, Mor U, Dori-Bachash M, Bashiardes S, Kotler E, Zur M, Regev-Lehavi D, Brik RBet al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell. 2018;174:1388–405. [DOI] [PubMed] [Google Scholar]
- 14.Wirtz S, Popp V, Kindermann M, Gerlach K, Weigmann B, Fichtner-Feigl S, Neurath MF. Chemically induced mouse models of acute and chronic intestinal inflammation. Nat Protoc. 2017;12:1295–309. [DOI] [PubMed] [Google Scholar]
- 15.Martín R, Chamignon C, Mhedbi-Hajri N, Chain F, Derrien M, Escribano-Vázquez U, Garault P, Cotillard A, Pham HP, Chervaux Cet al. The potential probiotic Lactobacillus rhamnosus CNCM I-3690 strain protects the intestinal barrier by stimulating both mucus production and cytoprotective response. Sci Rep. 2019;9:5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song H, Wang W, Shen B, Jia H, Hou Z, Chen P, Sun Y. Pretreatment with probiotic Bifico ameliorates colitis-associated cancer in mice: transcriptome and gut flora profiling. Cancer Sci. 2018;109:666–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turroni F, Taverniti V, Ruas-Madiedo P, Duranti S, Guglielmetti S, Lugli GA, Gioiosa L, Palanza P, Margolles A, van Sinderen Det al. Bifidobacterium bifidum PRL2010 modulates the host innate immune response. Appl Environ Microbiol. 2014;80:730–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shin D, Chang SY, Bogere P, Won K, Choi JY, Choi YJ, Lee HK, Hur J, Park BYet al. Beneficial roles of probiotics on the modulation of gut microbiota and immune response in pigs. PLoS One. 2019;14:e0220843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hudson LE, McDermott CD, Stewart TP, Hudson WH, Rios D, Fasken MB, Corbett AH, Lamb TJ. Characterization of the probiotic yeast Saccharomyces boulardii in the healthy mucosal immune system. PLoS One. 2016;11:e0153351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chondrou P, Karapetsas A, Kiousi DE, Vasileiadis S, Ypsilantis P, Botaitis S, Alexopoulos A, Plessas S, Bezirtzoglou E, Galanis A. Assessment of the immunomodulatory properties of the probiotic strain Lactobacillus paracasei K5 in vitro and in vivo. Microorganisms. 2020;8:709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saxami G, Karapetsas A, Chondrou P, Vasiliadis S, Lamprianidou E, Kotsianidis I, Ypsilantis P, Botaitis S, Simopoulos C, Galanis A. Potentially probiotic Lactobacillus strains with anti-proliferative activity induce cytokine/chemokine production and neutrophil recruitment in mice. Beneficial Microbes. 2017;8:615–23. [DOI] [PubMed] [Google Scholar]
- 22.Aindelis G, Tiptiri-Kourpeti A, Lampri E, Spyridopoulou K, Lamprianidou E, Kotsianidis I, Ypsilantis P, Pappa A, Chlichlia K. Immune responses raised in an experimental colon carcinoma model following oral administration of Lactobacillus casei. Cancers. 2020;12:368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mendes MCS, Paulino DS, Brambilla SR, Camargo JA, Persinoti GF, Carvalheira JBC. Microbiota modification by probiotic supplementation reduces colitis associated colon cancer in mice. World J Gastroenterol. 2018;24:1995–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong H, Rowland I, Thomas LV, Yaqoob P. Immunomodulatory effects of a probiotic drink containing Lactobacillus c asei Shirota in healthy older volunteers. Eur J Nutr. 2013;52:1853–63. [DOI] [PubMed] [Google Scholar]
- 25.van Baarlen P, Troost F, van der Meer C, Hooiveld G, Boekschoten M, Brummer RJ, Kleerebezem M. Human mucosal in vivo transcriptome responses to three lactobacilli indicate how probiotics may modulate human cellular pathways. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4562–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solano-Aguilar G, Molokin A, Botelho C, Fiorino AM, Vinyard B, Li R, Chen C, Urban J Jr, Dawson H, Andreyeva Iet al. Transcriptomic profile of whole blood cells from elderly subjects fed probiotic bacteria Lactobacillus rhamnosus GG ATCC 53103 (LGG) in a phase I open label study. PLoS One. 2016;11:e0147426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moya-Pérez A, Romo-Vaquero M, Tomás-Barberán F, Sanz Y, García-Conesa MT. Hepatic molecular responses to Bifidobacterium p seudocatenulatum CECT 7765 in a mouse model of diet-induced obesity. Nutr Metab Cardiovasc Dis. 2014;24:57–64. [DOI] [PubMed] [Google Scholar]
- 28.Sun Q, Zhang Y, Li Z, Yan H, Li J, Wan X. Mechanism analysis of improved glucose homeostasis and cholesterol metabolism in high-fat-induced obese mice treated with La-SJLH001 via transcriptomics and culturomics. Food Funct. 2019;10:3556–66. [DOI] [PubMed] [Google Scholar]
- 29.Qing X, Zeng D, Wang H, Ni X, Lai J, Liu L, Khalique A, Pan K, Jing B. Analysis of hepatic transcriptome demonstrates altered lipid metabolism following Lactobacillus johnsonii BS15 prevention in chickens with subclinical necrotic enteritis. Lipids Health Dis. 2018;17:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang D, Shang T, Huang Y, Wang S, Liu H, Wang J, Wang Y, Ji H, Zhang R. Gene expression profile changes in the jejunum of weaned piglets after oral administration of Lactobacillus or an antibiotic. Sci Rep. 2017;7:15816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee E, Jung SR, Lee SY, Lee NK, Paik HD, Lim SI. Lactobacillus plantarum strain Ln4 attenuates diet-induced obesity, insulin resistance, and changes in hepatic mRNA levels associated with glucose and lipid metabolism. Nutrients. 2018;10:643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee J, Jang JY, Kwon MS, Lim SK, Kim N, Lee J, Park HK, Yun M, Shin MY, Jo HEet al. Mixture of two Lactobacillus plantarum strains modulates the gut microbiota structure and regulatory T cell response in diet-induced obese mice. Mol Nutr Food Res. 2018;62:1800329. [DOI] [PubMed] [Google Scholar]
- 33.Burton KJ, Pimentel G, Zangger N, Vionnet N, Drai J, McTernan PG, Pralong FP, Delorenzi M, Vergères G. Modulation of the peripheral blood transcriptome by the ingestion of probiotic yoghurt and acidified milk in healthy, young men. PLoS One. 2018;13:e0192947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40(Database issue):D109–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Troost FJ, van Baarlen P, Lindsey P, Kodde A, de Vos WM, Kleerebezem M, Brummer RJ. Identification of the transcriptional response of human intestinal mucosa to Lactobacillus plantarum WCFS1 in vivo. BMC Genomics. 2008;9:374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rao MS, Van Vleet TR, Ciurlionis R, Buck WR, Mittelstadt SW, Blomme EAG, Liguori MJ. Comparison of RNA-Seq and microarray gene expression platforms for the toxicogenomic evaluation of liver from short-term rat toxicity studies. Front Genet. 2019;9:636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao S, Fung-Leung WP, Bittner A, Ngo K, Liu X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One. 2014;9:e78644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klassen A, Faccio AT, Canuto GA, da Cruz PL, Ribeiro HC, Tavares MF, Sussulini A. Metabolomics: definitions and significance in systems biology. Adv Exp Med Biol. 2017;965:3–17. [DOI] [PubMed] [Google Scholar]
- 39.Vernocchi P, Del Chierico F, Putignani L. Gut microbiota profiling: metabolomics based approach to unravel compounds affecting human health. Front Microbiol. 2016;7:1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aoki R, Kamikado K, Suda W, Takii H, Mikami Y, Suganuma N, Hattori M, Koga Y. A proliferative probiotic Bifidobacterium strain in the gut ameliorates progression of metabolic disorders via microbiota modulation and acetate elevation. Sci Rep. 2017;7:43522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng RY, Yao JR, Wan Q, Guo JW, Pu FF, Shi L, Hu W, Yang YH, Li L, Li Met al. Oral administration of Bifidobacterium bifidum TMC3115 to neonatal mice may alleviate IgE-mediated allergic risk in adulthood. Beneficial Microbes. 2018;9:815–28. [DOI] [PubMed] [Google Scholar]
- 42.Nunes CF, Nogueira JS, Vianna PHO, Ciambarella BT, Rodrigues PM, Miranda KR, Lobo LA, Domingues R, Busch M, Atella GCet al. Probiotic treatment during neonatal age provides optimal protection against experimental asthma through the modulation of microbiota and T cells. Int Immunol. 2018;30:155–69. [DOI] [PubMed] [Google Scholar]
- 43.Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L. The role of short-chain fatty acids in health and disease. Adv Immunol. 2014;121:91–119. [DOI] [PubMed] [Google Scholar]
- 44.Dalile B, Van Oudenhove L, Vervliet B, Verbeke K. The role of short-chain fatty acids in microbiota-gut-brain communication. Nat Rev Gastroenterol Hepatol. 2019;16:461–78. [DOI] [PubMed] [Google Scholar]
- 45.Chen L, Zhong F, Zhu J. Bridging targeted and untargeted mass spectrometry-based metabolomics via hybrid approaches. Metabolites. 2020;10:348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salazar N, Neyrinck AM, Bindels LB, Druart C, Ruas-Madiedo P, Cani PD, de Los Reyes-Gavilán CG, Delzenne NM. Functional effects of EPS-producing Bifidobacterium administration on energy metabolic alterations of diet-induced obese mice. Front Microbiol. 2019;10:1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Azagra-Boronat I, Tres A, Massot-Cladera M, Franch À, Castell M, Guardiola F, Pérez-Cano FJ, Rodríguez-Lagunas MJ. Lactobacillus fermentum CECT5716 supplementation in rats during pregnancy and lactation impacts maternal and offspring lipid profile, immune system and microbiota. Cells. 2020;9:575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park YE, Kim MS, Shim KW, Kim YI, Chu J, Kim BK, Choi IS, Kim JY. Effects of Lactobacillus plantarum Q180 on postprandial lipid levels and intestinal environment: a double-blind, randomized, placebo-controlled, parallel trial. Nutrients. 2020;12:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chorell E, Karlsson Videhult F, Hernell O, Antti H, West CE. Impact of probiotic feeding during weaning on the serum lipid profile and plasma metabolome in infants. Br J Nutr. 2013;110:116–26. [DOI] [PubMed] [Google Scholar]
- 50.Durack J, Kimes NE, Lin DL, Rauch M, McKean M, McCauley K, Panzer AR, Mar JS, Cabana MD, Lynch SV. Delayed gut microbiota development in high-risk for asthma infants is temporarily modifiable by Lactobacillus supplementation. Nat Commun. 2018;9:707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bazanella M, Maier TV, Clavel T, Lagkouvardos I, Lucio M, Maldonado-Gòmez MX, Autran C, Walter J, Bode L, Schmitt-Kopplin Pet al. Randomized controlled trial on the impact of early-life intervention with Β ifidobacteria on the healthy infant fecal microbiota and metabolome. Am J Clin Nutr. 2017;106:1274–86. [DOI] [PubMed] [Google Scholar]
- 52.Michael DR, Davies TS, Moss JWE, Calvente DL, Ramji DP, Marchesi JR, Pechlivanis A, Plummer SF, Hughes TR. The anti-cholesterolaemic effect of a consortium of probiotics: an acute study in C57BL/6J mice. Sci Rep. 2017;7:2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Culpepper T, Rowe CC, Rusch CT, Burns AM, Federico AP, Girard SA, Tompkins TA, Nieves C Jr, Dennis-Wall JC, Christman MCet al. Three probiotic strains exert different effects on plasma bile acid profiles in healthy obese adults: randomised, double-blind placebo-controlled crossover study. Beneficial Microbes. 2019;10:497–509. [DOI] [PubMed] [Google Scholar]
- 54.Hibberd AA, Yde CC, Ziegler ML, Honoré AH, Saarinen MT, Lahtinen S, Stahl B, Jensen HM, Stenman LK. Probiotic or synbiotic alters the gut microbiota and metabolism in a randomised controlled trial of weight management in overweight adults. Beneficial Microbes. 2019;10:121–35. [DOI] [PubMed] [Google Scholar]
- 55.Zhao L, Hartung T. Metabonomics and toxicology. Methods Mol Biol. 2015;1277:209–31. [DOI] [PubMed] [Google Scholar]
- 56.Sugahara H, Odamaki T, Fukuda S, Kato T, Xiao JZ, Abe F, Kikuchi J, Ohno H. Probiotic Bifidobacterium longum alters gut luminal metabolism through modification of the gut microbial community. Sci Rep. 2015;5:13548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim HK, Rutten NB, Besseling-van der Vaart I, Niers LE, Choi YH, Rijkers GT, van Hemert S. Probiotic supplementation influences faecal short chain fatty acids in infants at high risk for eczema. Beneficial Microbes. 2015;6:783–90. [DOI] [PubMed] [Google Scholar]
- 58.Zapata A, Chefer VI, Shippenberg TS, Denoroy L. Detection and quantification of neurotransmitters in dialysates. Curr Protoc Neurosci. 2009;Chapter 7:Unit 7.4.1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moya-Pérez A, Perez-Villalba A, Benítez-Páez A, Campillo I, Sanz Y. Bifidobacterium CECT 7765 modulates early stress-induced immune, neuroendocrine and behavioral alterations in mice. Brain Behav Immun. 2017;65:43–56. [DOI] [PubMed] [Google Scholar]
- 60.Liu YW, Liu WH, Wu CC, Juan YC, Wu YC, Tsai HP, Wang S, Tsai YC. Psychotropic effects of Lactobacillus plantarum PS128 in early life-stressed and naïve adult mice. Brain Res. 2016;1631:1–12. [DOI] [PubMed] [Google Scholar]
- 61.Ghosh A, Mehta A, Khan AM. Metagenomic analysis and its applications. In: Ranganathan S, Gribskov M, Schönbach C, editors. Encyclopedia of bioinformatics and computational biology. Academic Press;2019; p. 184–93. [Google Scholar]
- 62.Rausch P, Rühlemann M, Hermes BM, Doms S, Dagan T, Dierking K, Domin H, Fraune S, von Frieling J, Hentschel Uet al. Comparative analysis of amplicon and metagenomic sequencing methods reveals key features in the evolution of animal metaorganisms. Microbiome. 2019;7:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ranjan R, Rani A, Metwally A, McGee HS, Perkins DL. Analysis of the microbiome: advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem Biophys Res Commun. 2016;469:967–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang YN, Meng XC, Dong YF, Zhao XH, Qian JM, Wang HY, Li JN. Effects of probiotics and prebiotics on intestinal microbiota in mice with acute colitis based on 16S rRNA gene sequencing. Chin Med J. 2019;132:1833–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen H, Xia Y, Zhu S, Yang J, Yao J, Di J, Liang Y, Gao R, Wu W, Yang Yet al. Lactobacillus plantarum LPOnlly alters the gut flora and attenuates colitis by inducing microbiome alteration in interleukin10 knockout mice. Mol Med Rep. 2017;16:5979–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sobhani I, Bergsten E, Couffin S, Amiot A, Nebbad B, Barau C, de'Angelis N, Rabot S, Canoui-Poitrine F, Mestivier Det al. Colorectal cancer-associated microbiota contributes to oncogenic epigenetic signatures. Proc Natl Acad Sci U S A. 2019;116:24285–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xia X, Wu WKK, Wong SH, Liu D, Kwong TNY, Nakatsu G, Yan PS, Chuang YM, Chan MW, Coker OOet al. Bacteria pathogens drive host colonic epithelial cell promoter hypermethylation of tumor suppressor genes in colorectal cancer. Microbiome. 2020;8:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song Met al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65:1973–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gao Z, Guo B, Gao R, Zhu Q, Wu W, Qin H. Probiotics modify human intestinal mucosa-associated microbiota in patients with colorectal cancer. Mol Med Rep. 2015;12:6119–27. [DOI] [PubMed] [Google Scholar]
- 70.Hibberd AA, Lyra A, Ouwehand AC, Rolny P, Lindegren H, Cedgård L, Wettergren Y. Intestinal microbiota is altered in patients with colon cancer and modified by probiotic intervention. BMJ Open Gastroenterol. 2017;4:e000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buffington SA, Di Prisco GV, Auchtung TA, Ajami NJ, Petrosino JF, Costa-Mattioli M. Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell. 2016;165:1762–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beck BR, Park GS, Jeong DY, Lee YH, Im S, Song WH, Kang J. Multidisciplinary and comparative investigations of potential psychobiotic effects of Lactobacillus strains isolated from newborns and their impact on gut microbiota and ileal transcriptome in a healthy murine model. Front Cell Infect Microbiol. 2019;9:269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tian P, O'Riordan KJ, Lee YK, Wang G, Zhao J, Zhang H, Cryan JF, Chen W. Towards a psychobiotic therapy for depression: Bifidobacterium breve CCFM1025 reverses chronic stress-induced depressive symptoms and gut microbial abnormalities in mice. Neurobiol Stress. 2020;12:100216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu QF, Kim HM, Lim S, Chung MJ, Lim CY, Koo BS, Kang SS. Effect of probiotic administration on gut microbiota and depressive behaviors in mice. Daru. 2020;28:181–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim CS, Cha L, Sim M, Jung S, Chun WY, Baik HW, Shin DM. Probiotic supplementation improves cognitive function and mood with changes in gut microbiota in community-dwelling elderly: a randomized, double-blind, placebo-controlled, multicenter trial. J Gerontol A Biol Sci Med Sci. 2020;17:glaa090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nishida K, Sawada D, Kuwano Y, Tanaka H, Rokutan K. Health benefits of Lactobacillus gasseri CP2305 tablets in young adults exposed to chronic stress: a randomized, double-blind, placebo-controlled study. Nutrients. 2019;11:1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sanz Y, Olivares M, Moya-Pérez Á, Agostoni C. Understanding the role of gut microbiome in metabolic disease risk. Pediatr Res. 2015;77:236–44. [DOI] [PubMed] [Google Scholar]
- 78.Ji Y, Park S, Park H, Hwang E, Shin H, Pot B, Holzapfel WH. Modulation of active gut microbiota by Lactobacillus rhamnosus GG in a diet induced obesity murine model. Front Microbiol. 2018;9:710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bo TB, Wen J, Zhao YC, Tian SJ, Zhang XY, Wang DH. Bifidobacterium pseudolongum reduces triglycerides by modulating gut microbiota in mice fed high-fat food. J Steroid Biochem Mol Biol. 2020;198:105602. [DOI] [PubMed] [Google Scholar]
- 80.Wang Y, Wu Y, Wang B, Xu H, Mei X, Xu X, Zhang X, Ni J, Li W. Bacillus amyloliquefaciens SC06 protects mice against high-fat diet-induced obesity and liver injury via regulating host metabolism and gut microbiota. Front Microbiol. 2019;10:1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen M, Guo WL, Li QY, Xu JX, Cao YJ, Liu B, Yu XD, Rao PF, Ni L, Lv XC. The protective mechanism of Lactobacillus plantarum FZU3013 against non-alcoholic fatty liver associated with hyperlipidemia in mice fed a high-fat diet. Food Funct. 2020;11:3316–31. [DOI] [PubMed] [Google Scholar]
- 82.EFSA FEEDAP Panel . Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 2018;16:5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Koren S, Schatz MC, Walenz BP, Martin J, Howard JT, Ganapathy G, Wang Z, Rasko DA, McCombie WR, Jarvis EDet al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol. 2012;30:693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EEet al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–9. [DOI] [PubMed] [Google Scholar]
- 85.Zheng J, Wittouck S, Salvetti E, Franz C, Harris HMB, Mattarelli P, O'Toole PW, Pot B, Vandamme P, Walter Jet al. A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus b eijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int J Syst Evol Microbiol. 2020;70:2782–858. [DOI] [PubMed] [Google Scholar]
- 86.Ghosh S, Sarangi AN, Mukherjee M, Bhowmick S, Tripathy S. Reanalysis of Lactobacillus paracasei Lbs2 strain and large-scale comparative genomics places many strains into their correct taxonomic position. Microorganisms. 2019;7:487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Botthoulath V, Upaichit A, Thumarat U. Identification and in vitro assessment of potential probiotic characteristics and antibacterial effects of Lactobacillus plantarum subsp. plantarum SKI19, a bacteriocinogenic strain isolated from Thai fermented pork sausage. J Food Sci Technol. 2018;55:2774–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kazou M, Alexandraki V, Blom J, Pot B, Tsakalidou E, Papadimitriou K. Comparative genomics of Lactobacillus acidipiscis ACA-DC 1533 isolated from traditional Greek kopanisti cheese against species within the Lactobacillus salivarius clade. Front Microbiol. 2018;9:1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fontana A, Falasconi I, Molinari P, Treu L, Basile A, Vezzi A, Campanaro S, Morelli L. Genomic comparison of Lactobacillus helveticus strains highlights probiotic potential. Front Microbiol. 2019;10:1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gueimonde M, Sánchez B, G de Los Reyes-Gavilán C, Margolles A. Antibiotic resistance in probiotic bacteria. Front Microbiol. 2013;4:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mater DD, Langella P, Corthier G, Flores MJ. A probiotic Lactobacillus strain can acquire vancomycin resistance during digestive transit in mice. J Mol Microbiol Biotechnol. 2008;14:123–7. [DOI] [PubMed] [Google Scholar]
- 92.Argyri AA, Zoumpopoulou G, Karatzas KA, Tsakalidou E, Nychas GJ, Panagou EZ, Tassou CC. Selection of potential probiotic lactic acid bacteria from fermented olives by in vitro tests. Food Microbiol. 2013;33:282–91. [DOI] [PubMed] [Google Scholar]
- 93.Ouoba LI, Lei V, Jensen LB. Resistance of potential probiotic lactic acid bacteria and Bifidobacteria of African and European origin to antimicrobials: determination and transferability of the resistance genes to other bacteria. Int J Food Microbiol. 2008;121:217–24. [DOI] [PubMed] [Google Scholar]
- 94.Ammor MS, Flórez AB, van Hoek AH, de Los Reyes-Gavilán CG, Aarts HJ, Margolles A, Mayo B. Molecular characterization of intrinsic and acquired antibiotic resistance in lactic acid bacteria and Bifidobacteria. J Mol Microbiol Biotechnol. 2008;14:6–15. [DOI] [PubMed] [Google Scholar]
- 95.Bennedsen M, Stuer-Lauridsen B, Danielsen M, Johansen E. Screening for antimicrobial resistance genes and virulence factors via genome sequencing. Appl Environ Microbiol. 2011;77:2785–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zheng M, Zhang R, Tian X, Zhou X, Pan X, Wong A. Assessing the risk of probiotic dietary supplements in the context of antibiotic resistance. Front Microbiol. 2017;8:908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Veiga P, Suez J, Derrien M, Elinav E. Moving from probiotics to precision probiotics. Nat Microbiol. 2020;5:878–80. [DOI] [PubMed] [Google Scholar]
- 98.Turpin W, Espin-Garcia O, Xu W, Silverberg MS, Kevans D, Smith MI, Guttman DS, Griffiths A, Panaccione R, Otley Aet al. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat Genet. 2016;48:1413–7. [DOI] [PubMed] [Google Scholar]
- 99.Bonder MJ, Kurilshikov A, Tigchelaar EF, Mujagic Z, Imhann F, Vila AV, Deelen P, Vatanen T, Schirmer M, Smeekens SPet al. The effect of host genetics on the gut microbiome. Nat Genet. 2016;48:1407–12. [DOI] [PubMed] [Google Scholar]
- 100.Senan S, Prajapati JB, Joshi CG, Sreeja V, Gohel MK, Trivedi S, Patel RM, Pandya H, Singh US, Phatak Aet al. Geriatric respondents and non-respondents to probiotic intervention can be differentiated by inherent gut microbiome composition. Front Microbiol. 2015;6:944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Suez J, Zmora N, Zilberman-Schapira G, Mor U, Dori-Bachash M, Bashiardes S, Zur M, Regev-Lehavi D, Ben-Zeev Brik Ret al. Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell. 2018;174:1406–23..e16. [DOI] [PubMed] [Google Scholar]
- 102.Ruiz L, Hidalgo C, Blanco-Míguez A, Lourenço A, Sánchez B, Margolles A. Tackling probiotic and gut microbiota functionality through proteomics. J Proteomics. 2016;147:28–39. [DOI] [PubMed] [Google Scholar]
- 103.Aguilar-Toalá JE, Garcia-Varela R, Garcia HS, Mata-Haro V, González-Córdova AF, Vallejo-Cordoba B, Hernández-Mendoza. Postbiotics: an evolving term within the functional foods field. Trends Food Sci Technol. 2018;75:105–14. [Google Scholar]
- 104.Pyclik M, Srutkova D, Schwarzer M, Górska S. Bifidobacteria cell wall-derived exopolysaccharides, lipoteichoic acids, peptidoglycans, polar lipids and proteins – their chemical structure and biological attributes. Int J Biol Macromol. 2020;147:333–49. [DOI] [PubMed] [Google Scholar]
- 105.Chen MJ, Tang HY, Chiang ML. Effects of heat, cold, acid and bile salt adaptations on the stress tolerance and protein expression of kefir-isolated probiotic Lactobacillus kefiranofaciens M1. Food Microbiol. 2017;66:20–7. [DOI] [PubMed] [Google Scholar]
- 106.Yuan J, Wang B, Sun Z, Bo X, Yuan X, He X, Zhao H, Du X, Wang F, Jiang Zet al. Analysis of host-inducing proteome changes in Β ifidobacterium longum NCC2705 grown in vivo. J Proteome Res. 2008;7:375–85. [DOI] [PubMed] [Google Scholar]
- 107.Zhao J, Cheung PC. Comparative proteome analysis of Bifidobacterium longum subsp. infantis grown on β-glucans from different sources and a model for their utilization. J Agric Food Chem. 2013;61:4360–70. [DOI] [PubMed] [Google Scholar]
- 108.Schumann S, Alpert C, Engst W, Klopfleisch R, Loh G, Bleich A, Blaut M. Mild gut inflammation modulates the proteome of intestinal Escherichia coli. Environ Microbiol. 2014;16:2966–79. [DOI] [PubMed] [Google Scholar]
- 109.Koskenniemi K, Koponen J, Kankainen M, Savijoki K, Tynkkynen S, de Vos WM, Kalkkinen N, Varmanen P. Proteome analysis of Lactobacillus rhamnosus GG using 2-D DIGE and mass spectrometry shows differential protein production in laboratory and industrial-type growth media. J Proteome Res. 2009;8:4993–507. [DOI] [PubMed] [Google Scholar]
- 110.Di Cagno R, De Angelis M, Calasso M, Gobbetti M. Proteomics of the bacterial cross-talk by quorum sensing. J Proteomics. 2011;74:19–34. [DOI] [PubMed] [Google Scholar]
- 111.O'Toole PW, Marchesi JR, Hill C. Next-generation probiotics: the spectrum from probiotics to live biotherapeutics. Nat Microbiol. 2017;2:17057. [DOI] [PubMed] [Google Scholar]
- 112.Lin TZ, Shu CC, Lai WF, Tzeng CM, Lai HC, Lu CC. Investiture of next generation probiotics on amelioration of diseases – strains do matter. Medicine in Microecology. 2019;1–2:100002. [Google Scholar]
- 113.Chang CJ, Lin TL, Tsai YL, Wu TR, Lai WF, Lu CC, Lai HC. Next generation probiotics in disease amelioration. J Food Drug Anal. 2019;27:615–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.El Hage R, Hernandez-Sanabria E, Van de Wiele T. Emerging trends in “smart probiotics”: functional consideration for the development of novel health and industrial applications. Front Microbiol. 2017;8:1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tavassoly I, Goldfarb J, Iyengar R. Systems biology primer: the basic methods and approaches. Essays Biochem. 2018;62:487–500. [DOI] [PubMed] [Google Scholar]
- 116.Yadav M, Shukla P. Recent systems biology approaches for probiotics use in health aspects: a review. 3 Biotech. 2019;9:448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Peñalver Bernabé B, Cralle L, Gilbert JA. Systems biology of the human microbiome. Curr Opin Biotechnol. 2018;51:146–53. [DOI] [PubMed] [Google Scholar]
- 118.Bisanz JE, Seney S, McMillan A, Vongsa R, Koenig D, Wong L, Dvoracek B, Gloor GB, Sumarah M, Ford Bet al. A systems biology approach investigating the effect of probiotics on the vaginal microbiome and host responses in a double blind, placebo-controlled clinical trial of post-menopausal women. PLoS One. 2014;9:e104511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hu Y, An Q, Sheu K, Trejo B, Fan S, Guo Y. Single cell multi-omics technology: methodology and application. Front Cell Dev Biol. 2018;6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Song Y, Xu X, Wang W, Tian T, Zhu Z, Yang C. Single cell transcriptomics: moving towards multi-omics. Analyst. 2019;144:3172–89. [DOI] [PubMed] [Google Scholar]
- 121.Leonavicius K, Nainys J, Kuciauskas D, Mazutis L. Multi-omics at single-cell resolution: comparison of experimental and data fusion approaches. Curr Opin Biotechnol. 2019;55:159–66. [DOI] [PubMed] [Google Scholar]
- 122.Chappell L, Russell AJC, Voet T. Single-cell (multi)omics technologies. Annu Rev Genom Hum Genet. 2018;19:15–41. [DOI] [PubMed] [Google Scholar]
- 123.Sandhu C, Qureshi A, Emili A. Panomics for precision medicine. Trends Mol Med. 2018;24:85–101. [DOI] [PMC free article] [PubMed] [Google Scholar]