Abstract
The COVID-19 pandemic caused by SARS-CoV-2 has made the development of safe and effective vaccines a critical priority. To date, four vaccines have been approved by European and American authorities for preventing COVID-19, but the development of additional vaccine platforms with improved supply and logistics profiles remains a pressing need. Here we report the preclinical evaluation of a novel COVID-19 vaccine candidate based on the electroporation of engineered, synthetic cDNA encoding a viral antigen in the skeletal muscle. We constructed a set of prototype DNA vaccines expressing various forms of the SARS-CoV-2 spike (S) protein and assessed their immunogenicity in animal models. Among them, COVID-eVax—a DNA plasmid encoding a secreted monomeric form of SARS-CoV-2 S protein receptor-binding domain (RBD)—induced the most potent anti-SARS-CoV-2 neutralizing antibody responses (including against the current most common variants of concern) and a robust T cell response. Upon challenge with SARS-CoV-2, immunized K18-hACE2 transgenic mice showed reduced weight loss, improved pulmonary function, and lower viral replication in the lungs and brain. COVID-eVax conferred significant protection to ferrets upon SARS-CoV-2 challenge. In summary, this study identifies COVID-eVax as an ideal COVID-19 vaccine candidate suitable for clinical development. Accordingly, a combined phase I-II trial has recently started.
Keywords: SARS-CoV-2, DNA vaccine, antiviral immunity, animal models, protection
Graphical abstract
We report the development, characterization, and preclinical evaluation of COVID-eVax, a novel COVID-19 vaccine candidate with improved supply and logistics profiles. The technology is based on the electroporation of engineered, synthetic cDNA encoding a secreted monomeric form of the receptor-binding domain of the SARS-CoV-2 spike protein.
Introduction
At the time of writing, SARS-CoV-2 has spread worldwide, causing over 220 million confirmed cases and more than 4.5 million confirmed deaths.
To date, the regulatory agencies European Medicines Agency (EMA) and Food and Drug Administration (FDA) have either approved or authorized the conditional or emergency use of four vaccines against SARS-CoV-2, two based on mRNA (produced by Pfizer and Moderna) and two based on adenoviral vectors (produced by AstraZeneca and Johnson & Johnson). Additional vaccine candidates are under development, and a continually updated list is available at https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines. Most COVID-19 vaccines and vaccine candidates target the SARS-CoV-2 full-length (FL) spike (S) glycoprotein, which mediates attachment and entry of the virus into host cells,1 and employ both traditional and novel vaccine platforms such as inactivated virus, protein-based preparations, and virus-vectored and nucleic acid-based formulations.1,2
Among the latter, DNA-based platforms show the greatest potential in terms of safety and ease of production.3 Prior work has demonstrated that a DNA-based vaccine approach for SARS- and MERS-CoV induces neutralizing antibody (nAb) responses and provides protection in challenge models.4,5 Moreover, in a phase I dose-escalation study subjects immunized with a DNA vaccine encoding the MERS-CoV S protein showed durable nAb and T cell responses and a seroconversion rate of 96%.5 The SARS-CoV-2 S protein is most similar in sequence and structure to SARS-CoV S and shares a global protein fold architecture with the MERS-CoV S protein.6 Of note, the receptor-binding site of the S protein is a vulnerable target for antibodies. In fact, anti-MERS antibodies targeting the receptor-binding domain (RBD) of the S protein tend to have greater neutralizing potency than those directed to other epitopes.7 More recently, a study by Piccoli et al.8 showed that depletion of anti-RBD antibodies in convalescent patient sera results in the loss of >90% neutralizing activity toward SARS-CoV-2, suggesting that the SARS-CoV-2 RBD represents a key target for vaccine development.
Here we describe the development of a DNA-based SARS-CoV-2 vaccine. Synthetic DNA is temperature stable and cold chain free, which are important advantages over approved RNA and vector vaccines for delivery to resource-limited settings. Furthermore, synthetic DNA vaccines are amenable to accelerated developmental timelines because of the relative simplicity with which multiple candidates can be designed, preclinically tested, manufactured in large quantities, and progressed through established regulatory pathways to the clinic. Injection of DNA plasmid into the skeletal muscle followed by a short electrical stimulation—referred to as electro-gene transfer (EGT) or electroporation (EP)—enhances DNA uptake and gene expression by several hundred-fold,9, 10, 11 leading to improved antigen expression and a local and transient tissue damage favoring inflammatory cell recruitment and cytokine production at the injection site.12
Exploiting our experience in the generation of vaccines based on the EP of plasmid DNA in the skeletal muscle,11 we produced and screened several constructs expressing different portions of the SARS-CoV-2 S protein and identified COVID-eVax—a DNA plasmid encoding a secreted monomeric form of SARS-CoV-2 S protein RBD—as a candidate for further clinical development. COVID-eVax has a favorable safety profile, it induces potent anti-SARS-CoV-2 nAb responses also against the current most common variants of concern (VOCs) as well as T cell responses, and it confers significant protection to human angiotensin-converting enzyme 2 (hACE2) transgenic mice and ferrets upon SARS-CoV-2 challenge.
Results
DNA vaccine constructs and immunogenicity
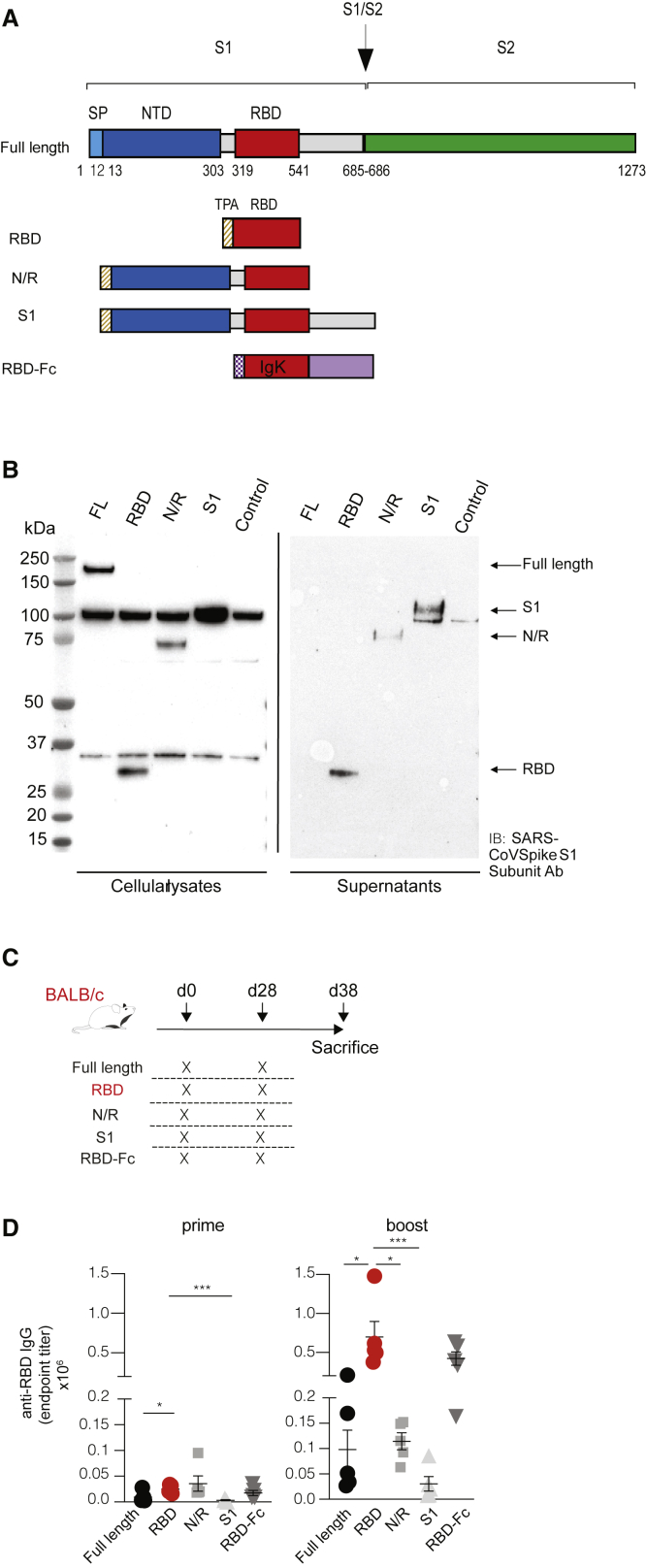
We designed five different DNA constructs (Figure 1A) encoding the following versions of the SARS-CoV-2 (Wuhan Hu-1, GenBank: MN_908947) S protein: (1) the FL protein; (2) the RBD; (3) the highly variable N-terminal domain (NTD) and the RBD domain (N/R); (4) the whole S1 subunit (S1); and (5) the RBD fused to a human immunoglobulin G (IgG)-Fc (RBD-Fc). To promote protein secretion, we introduced a tissue plasminogen activator (tPA) leader sequence in the RBD, N/R, and S1 constructs and an IgK leader sequence in the RBD-Fc construct. Western blot analyses confirmed expression of all constructs in cell lysates and of S1, N/R, and RBD in the culture supernatants (Figure 1B).
Figure 1.
DNA vaccine constructs and immunogenicity
(A) Schematic representation of SARS-CoV-2 DNA vaccine construct candidates, encoding (1) the full-length protein (FL); (2) the receptor-binding domain (RBD); (3) the highly variable N-terminal domain (NTD) and the RBD domain (N/R); (4) the whole S1 subunit (S1); and (5) the RBD fused to a human IgG-Fc (RBD-Fc). The RBD, N/R, and S1 constructs include a tPA leader sequence at the N terminus, whereas the RBD-Fc construct contains a IgK leader sequence. (B) Western blot analysis of SARS-CoV-2 DNA vaccine constructs after transfection in HEK293 cells. Forty-eight hours after transfection, both cell lysates and supernatants were resolved on a gel and blotted with a polyclonal SARS-CoV spike S1 subunit antibody. Cells transfected with empty plasmid vector were used as negative control (control). Non-specific bands were detected both in cell lysates and in supernatants, likely due to non-specific binding of primary antibody. (C) Schematic representation of the experimental setup. Each DNA construct was injected intramuscularly (20 μg total, 10 μg each quadriceps) into BALB/c mice (n = 5) at day 0 (prime) and day 28 (boost). Intramuscular injection was followed by electroporation (EP). Mice were euthanized and analyzed at day 38. (D) Sera of BALB/c mice (n = 5) were collected at day 14 (only prime) and day 38 (prime-boost), and anti-RBD IgG levels were measured through ELISA; each dot represents a mouse. ∗p value < 0.05, ∗∗∗p value < 0.001.
EP of these DNA vaccines in the skeletal muscle of BALB/c mice was adopted to evaluate immunogenicity. The vaccination protocol consisted of the injection of 20 μg of DNA into both quadriceps (10 μg of DNA in each muscle) of 6-week-old mice (Figure 1C). A DNA plasmid expressing luciferase was used as a control for gene expression, whereas a group of mice injected with DNA but not electroporated served as additional controls (Figure S1). Mice received a second vaccination (boost) at day 28 and were sacrificed at day 38 (Figure 1C). The humoral response in the sera of vaccinated mice was evaluated by measuring anti-RBD IgG titers by ELISA at day 14 (prime) and at day 38 (boost) (Figure 1D). All mice showed detectable anti-RBD IgG antibodies at day 14, and their levels significantly increased at day 38 (Figure 1D). Notably, the most significant increase in antibody response was induced by the secreted RBD construct (Figure 1D), with a calculated geometric mean of IgG endpoint titers13 as high as 1:24,223 after prime and 1:617,648 after boost. Since these preliminary data showed the RBD construct to be the most immunogenic among the five DNA constructs, RBD was chosen as the main vaccine candidate for further development and was directly compared with the FL construct in subsequent experiments.
Detailed characterization of the humoral immune response elicited by the RBD vaccine candidate
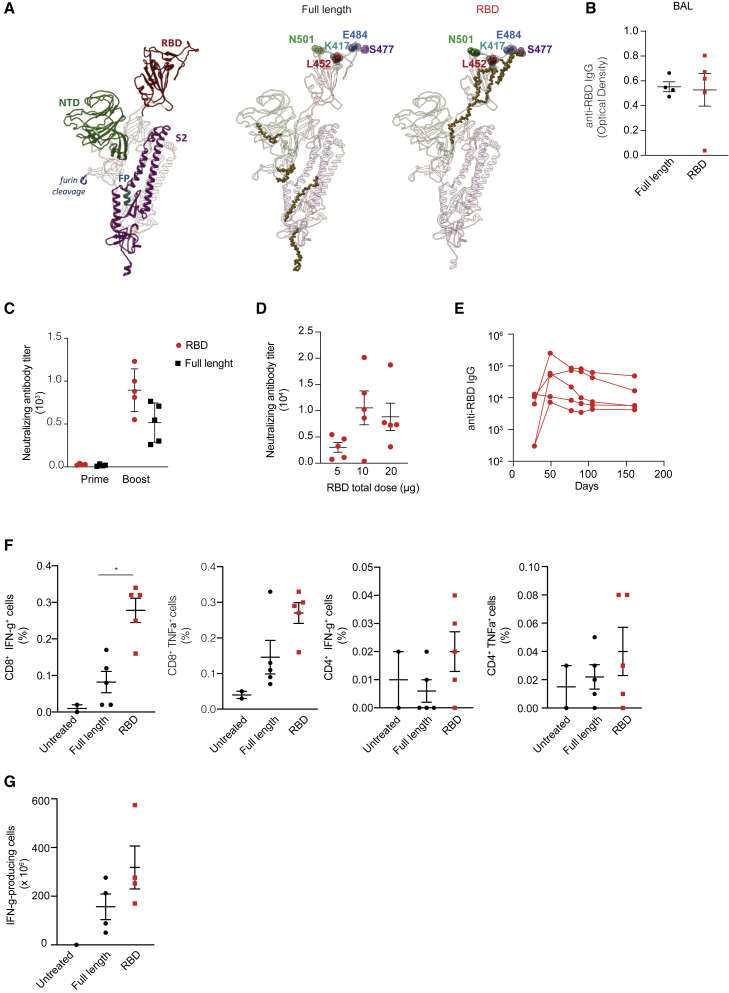
We next sought to characterize the humoral response to the RBD vaccine in depth, focusing on the specificity, duration, and neutralization capacity of the elicited antibodies. As per specificity, we carried out a B cell epitope mapping of the response elicited by the FL and RBD vaccines. To this end, a B cell ELISpot assay was performed by stimulating splenocytes collected from vaccinated BALB/c mice with 338 peptides covering the whole SARS-CoV-2 S protein. Sequences of positive hits (Table S1) were then mapped on the three-dimensional structure of the S protein,14 hence outlining the epitope domains (Figure 2A). Mice immunized with the RBD vaccine showed responses mapping mainly on conserved regions of the RBD, and not on regions most commonly affected by mutations in the current circulating VOCs (e.g., N501K, K417N, S477, E484K, and L452, Figure 2A). Despite the caveat that the abovementioned analysis detects linear and not conformational epitopes, this suggests that antibodies elicited by the RBD vaccine might be functional against the current most commonly circulating SARS-CoV-2 variants.
Figure 2.
Characterization of the immune response elicited by the RBD vaccine candidate
(A) Antibody linear epitopes mapped onto the structure of the FL spike protein. Each domain of the FL protein (NTD, RBD, furin cleavage, FP-fusion peptide, and S2) is outlined with a different color (left), and the linear epitopes are shown as gold spheres within the spike domains used for immunization (center and right). (B) Anti-RBD IgG levels measured in bronchoalveolar lavage (BAL) of FL- and RBD-vaccinated BALB/c mice at day 38. (C) Neutralizing antibody titers in sera collected from RBD- or FL-vaccinated BALB/c mice (n = 5) at day 14 (prime) and day 38 (boost), measured through a neutralization assay with infectious SARS-CoV-2-and Vero cells. (D) Neutralizing antibody titers in sera collected at day 38 from C57BL/6 mice (n = 5) vaccinated with increasing doses of RBD vaccine (5–10–20 μg) in a prime-boost regimen. (E) Serum anti-RBD IgG levels measured over time in sera of RBD-vaccinated C57BL/6 mice (prime-boost regimen, n = 5) up to 6 months starting from prime. (F) T cell immune response (IFN-γ+ and TNF-α+) in CD8+ and CD4+ cells measured by intracellular staining of splenocytes collected from FL- and RBD-vaccinated BALB/c mice (n = 5) at day 38 and restimulated with pool S1 peptides. (G) IFN-γ-producing T cells measured by ELISpot assay performed on BALs collected from BALB/c mice (n = 5) vaccinated with FL and RBD, intranasally challenged with 20 μg RBD protein at day 42, and culled the day after. ∗p value < 0.05.
SARS-CoV-2-specific antibodies elicited by both the FL and the RBD vaccine were present not only in the sera but also in the lungs of vaccinated mice, as shown by analysis of bronchoalveolar lavages (BALs) of mice 38 days after vaccination (Figure 2B).
We next sought to analyze whether the RBD-specific antibodies induced by the RBD vaccine were able to neutralize SARS-CoV-2. The neutralization capacity of antibodies induced by the RBD vaccine was comparable with FL after prime and superior after boost (Figure 2C), with 50% neutralization dose (ND50) at day 38 of 894 ± 249. A dose-response experiment indicated that the nAb titers plateaued at an RBD vaccine dose of 10 μg (injected in a single quadriceps muscle) in a prime-boost regimen serum (Figure 2D). Finally, total anti-RBD IgG antibodies persisted at high levels up to 6 months after vaccinations (Figure 2E).
Analysis of T cell responses elicited by the RBD vaccine candidate
Next we sought to evaluate the T cell response elicited by the RBD vaccine. To this end, we used peptide pools covering the S1 and S2 portions of the S protein (pools S1 and S2, respectively) to stimulate splenocytes collected from BALB/c mice at day 38 after vaccination (see Figure 1C for experimental setup). Interferon (IFN)-γ released by T cells upon peptide restimulation was evaluated by ELISpot assay. As expected, in the group vaccinated with the RBD vaccine we measured only T cell responses against pool S1 (which spans the RBD), whereas in the group vaccinated with the FL construct we measured T cell responses against both pools S1 and S2 (data not shown). In order to reveal immunodominant epitopes eliciting the T cell response, we performed epitope mapping using thirty-seven matrix mapping pools, covering the entire sequence of the S protein (Figure S2A), or twenty-four matrix pools, covering the RBD alone (Table S2 and Figure S2B). Most of the H2-Kd-restricted immunodominant epitopes were clustering in the RBD (Figure S2C). Cytokine production by antigen-specific T cells was also evaluated by intracellular staining of splenocytes collected from vaccinated BALB/c mice and restimulated with pool S1 (Figure 2F). Compared to FL, the RBD vaccine induced the highest frequency of CD8+ T cells producing either IFN-γ or tumor necrosis factor alpha (TNF-α) (Figure 2F).
To measure the potential recruitment of RBD-specific T cells to the lungs, 20 μg of the RBD protein was injected intranasally in a group of vaccinated BALB/c mice 2 weeks after the second immunization and IFN-γ production from lymphocytes recovered from BAL was measured by ELISpot assay 1 day later (Figure 2G). Mice vaccinated with RBD showed a higher recruitment of RBD-specific T cells than mice vaccinated with the FL vaccine (Figure 2G). A dose-response experiment conducted in C57BL/6 mice showed an even stronger specific T cell response than in BALB/c mice (data not shown) and a clear dose dependency (Figure S3A). A nonlinear fitting analysis of the curve (after pool S1 stimulation) revealed a median effective dose (ED50) of 2.06 ± 0.86 μg (Figure S3B). Cytokine analyses in vaccinated C57BL/6 mice revealed a predominant IFN-γ- and TNF-α-producing CD8+ T cell response, independent of the sex and age of the mice (Figure S3C and data not shown).
Safety and immunogenicity of the RBD vaccine candidate in rats
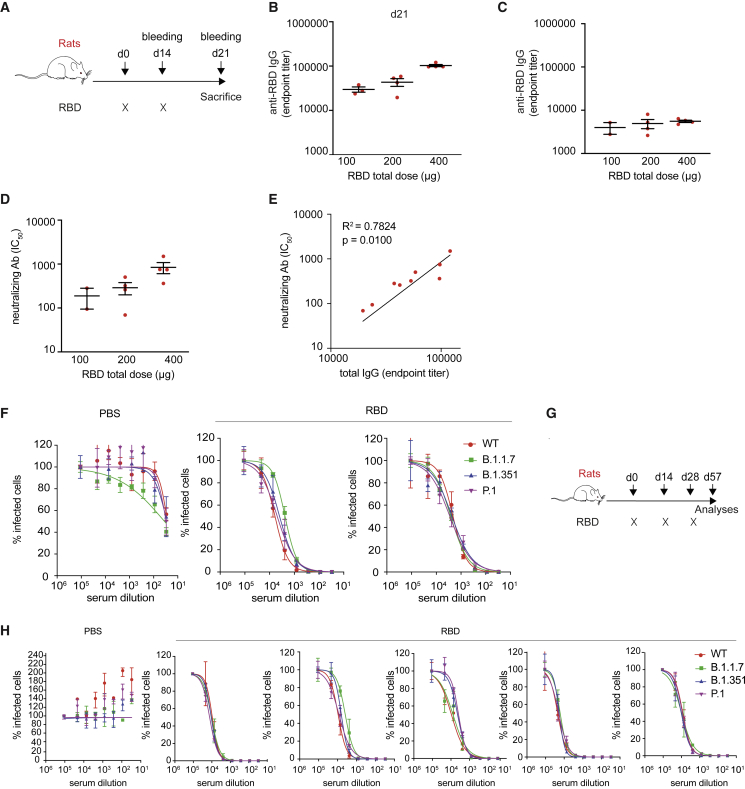
We next evaluated the safety and immunogenicity of the RBD vaccine candidate in rats, an animal model highly suited for toxicological studies. Seven-week-old female Sprague-Dawley rats were injected intramuscularly with phosphate-buffered saline (PBS) or 100, 200, or 400 μg of the RBD vaccine (divided equally in the 2 quadriceps) followed by EP at day 0 and day 14 (Figure 3A). The immunizations were well tolerated, with only mild to moderate lesions15 at the injection site that were almost fully recovered within 4 weeks (Figure S4) and an increased cellularity in the draining lymph nodes (data not shown).
Figure 3.
Immunogenicity of the RBD vaccine in rats
(A) Schematic representation of the experimental setup. Sprague-Dawley rats (n = 16) received two doses of RBD vaccine (day 0 and day 14) via intramuscular injection followed by EP. (B) Total IgG endpoint titer measured by ELISA assay performed on sera collected at day 14 (prime) from rats vaccinated with increasing doses of RBD (100–200–400 μg). (C) Total IgG endpoint titer measured by ELISA assay performed on sera collected at day 21 (prime-boost) from rats vaccinated with increasing doses of RBD vaccine. (D) Neutralizing antibody titer (IC50) of sera collected from the same rats as in (C). (E) Correlation between total IgG endpoint titers and neutralizing antibody IC50 values. (F) Dose-response curve representing neutralization activity of plasma against SARS-CoV-2 pseudovirus carrying the spike protein of wild-type (WT) virus or variants (B.1.1.7, B.1.351, and P.1). Plasma was collected at sacrifice from rats vaccinated with 400 μg of the RBD vaccine (two-dose vaccination regimen, day 0 and day 14) or with PBS, as negative control (G) Schematic representation of the experimental setup. Five Sprague-Dawley rats received three doses of RBD vaccine (days 0, 14, and 28) via intramuscular injection followed by EP. (H) Dose-response curve representing neutralization activity of plasma against SARS-CoV-2 pseudovirus carrying the SPIKE protein of WT virus or variants (B.1.1.7, B.1.351, and P.1). Plasma was collected at sacrifice from rats vaccinated with 400 μg of the RBD vaccine (three-dose vaccination regimen, days 0, 14, and 28) or with PBS, as negative control.
Quantification of the RBD-specific antibody titers showed a robust and dose-dependent antibody production, with ELISA endpoint titers up to 152,991 for the highest dose (Figures 3B and 3C). Immunization with the RBD vaccine induced high nAb titers (Figure 3D), which correlated with the total IgG endpoint titers (Figure 3E). Finally, sera from Sprague-Dawley rats that were immunized with 400 μg of the RBD vaccine at days 0 and 14, or that received a third dose at day 28, were assessed for neutralizing activity against three major SARS-CoV-2 variants (i.e., B.1.1.7, B.1.351, and P.1) with a lentiviral pseudotyped assay (Figure 3F–3H).
The RBD vaccine candidate elicits protective immune responses in K18-hACE2 transgenic mice and in ferrets
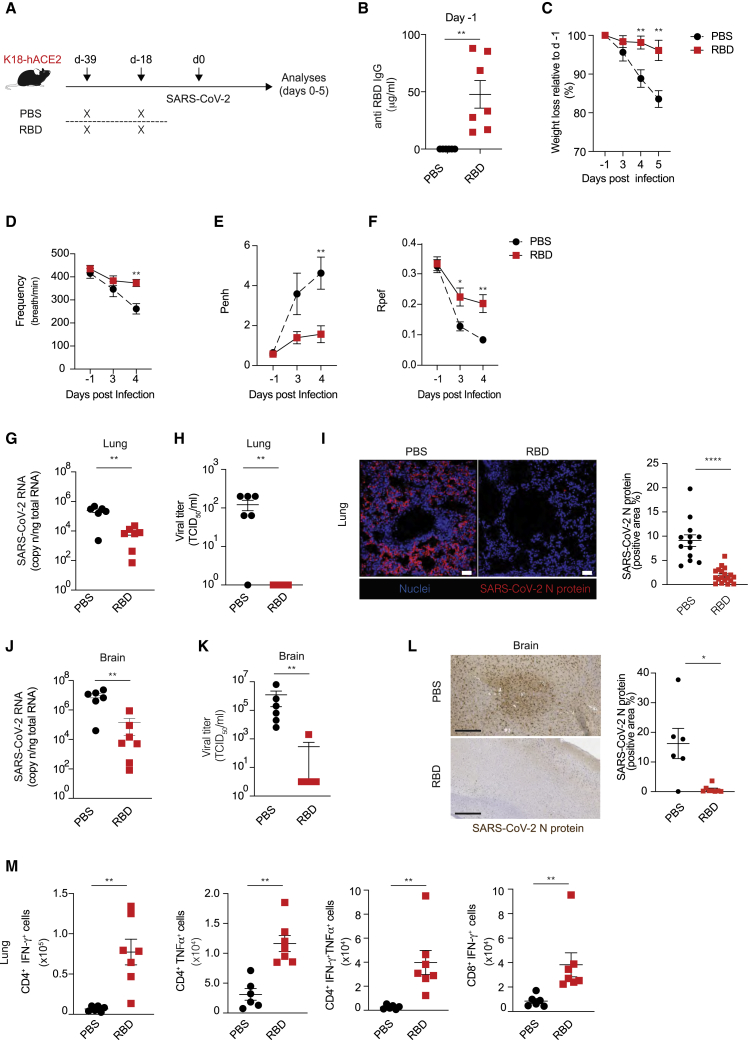
To explore the in vivo protection efficacy of RBD vaccine against SARS-CoV-2 challenge, K18-hACE2 transgenic mice16 received two intramuscular immunizations (at day −39 and at day −18) of 10 μg of the RBD vaccine (n = 7) or PBS (n = 6) followed by EP (Figure 4A). Pre-challenge sera collected 1 day prior to SARS-CoV-2 infection showed that the RBD vaccine induced robust RBD-specific IgG antibodies (average concentration of ∼50 μg/mL, Figure 4B). Eighteen days after the boost immunization, all mice were infected intranasally with 1 × 105 50% tissue culture infectious dose (TCID50) of SARS-CoV-2 (hCoV-19/Italy/LOM-UniSR-1/2020; GISAID Accession ID: EPI_ISL_413489) (Figure 4A). As expected,17 beginning 3–4 days post infection (p.i.) PBS-treated K18-hACE2 transgenic mice infected with SARS-CoV-2 exhibited a weight loss close to 20% of their body weight and a lethargic behavior (Figure 4C and data not shown). By contrast, K18-hACE2 transgenic mice immunized with RBD vaccine maintained stable body weight upon SARS-CoV-2 challenge and appeared more active (Figure 4C and data not shown). We used whole-body plethysmography (WBP) to evaluate several complementary metrics of pulmonary function, obstruction, and bronchoconstriction, including frequency, enhanced pause (PenH), and the fraction of expiration time at which the peak occurs (Rpef).18,19 PBS-treated mice infected with SARS-CoV-2 exhibited a decreased respiratory rate (Figure 4D), an increased PenH (Figure 4E), and a decreased Rpef (Figure 4F), indicative of pronounced loss of pulmonary function. By contrast, K18-hACE2 transgenic mice immunized with the RBD vaccine prior to infection maintained a relatively stable respiratory rate (Figure 4D) and had a much lower PenH (Figure 4E) and a higher Rpef (Figure 4F), indicative of better pulmonary function. Much higher amounts of viral RNA, infectious SARS-CoV-2, and viral N protein were detected in the lungs and brain of PBS-treated mice compared to mice immunized with the RBD vaccine (Figures 4G–4L). The lower viral titers in the lungs of immunized mice were associated with the detection of RBD-specific CD4+ T cells producing IFN-γ, TNF-α, or both as well as RBD-specific IFN-γ-producing CD8+ T cells (Figure 4M).
Figure 4.
In vivo protection efficacy of RBD vaccine against SARS-CoV-2 virus challenge in hACE2 transgenic mice
(A) Schematic representation of the experimental setup. K18-hACE2 (C57BL/6) mice received two immunizations (day −39, day −18) of 10 μg of RBD vaccine (n = 7) or PBS (n = 6) via intramuscular injection followed by EP before intranasal challenge with SARS-CoV-2. Lung and brain were collected and analyzed 5 days after SARS-CoV-2 infection. (B) Serum anti-RBD IgG levels of RBD vaccine- or PBS-challenged mice detected by ELISA assay; sera were collected right before SARS-CoV-2 infection. (C) Mouse body weights were monitored daily for up to 5 days. PBS-treated mice showed a rapid body weight decrease from day 4; in contrast, RBD vaccine-challenged mice demonstrated normal statuses. (D–F) Whole-body plethysmography assessing pulmonary function for frequency (D), PenH (E), and Rpef (F). (G) SARS-CoV-2 RNA in the lung was quantified by quantitative PCR with reverse transcriptase (qRT-PCR) 5 days after infection. (H) Viral titers in the lung 5 days after infection were determined by median tissue culture infectious dose (TCID50). (I) Representative confocal immunofluorescence micrographs of lung sections from PBS-treated mice (left) or RBD-treated mice (right) 5 days after SARS-CoV-2 infection. N-SARS-CoV-2 positive cells are depicted in red and nuclei in blue. Scale bars represent 30 μm. Right, quantification of N-SARS-CoV-2 signal; each dot represents a different section. (J) SARS-CoV-2 RNA in the brain was quantified by qRT-PCR 5 days after infection. (K) Viral titers in the brain 5 days after infection were determined by median tissue culture infectious dose (TCID50). (L) Representative immunohistochemical micrographs of brain sections from PBS-treated mice (top) or RBD-treated mice (bottom) 5 days after SARS-CoV-2 infection. N-SARS-CoV-2 expression is shown in brown. Scale bars, 300 μm. Right, quantification of N-SARS-CoV-2 signal; each dot represents a mouse. (M) Absolute numbers of CD4+ T cells producing IFN-γ, TNF-α, or both and of CD8+ T cells producing IFN-γ in the lung of the indicated mice 5 days after SARS-CoV-2 infection. ∗p value < 0.05, ∗∗p value < 0.01, ∗∗∗∗p value < 0.0001.
Besides inducing potent adaptive immune responses, the protection induced by the RBD vaccine might lie in the competitive inhibition of SARS-CoV-2 binding to ACE2 by the secreted RBD. Indeed, RBD is detectable in the sera and in the BAL of immunized BALB/c mice as early as 2 days after immunization (Figures S5A–S5C), a time point at which anti-RBD antibodies are not yet detectable (Figure S5D). To test whether this secreted RBD would compete with SARS-CoV-2 for ACE2 binding, we immunized K18-hACE2 transgenic mice with the RBD vaccine 2 days prior to intranasal inoculation with a luciferase-encoding lentiviral vector pseudotyped with the SARS-CoV-2 S protein. Two days later the lungs of treated mice were assessed for bioluminescence with an in vivo imaging system (IVIS). As shown in Figure S5E, compared to mice injected with PBS, mice immunized with the RBD vaccine exhibited a reduced bioluminescence, indicative of a significantly lower in vivo transduction. Further experiments should determine the extent to which the abovementioned mechanism confers protection by the RBD vaccine.
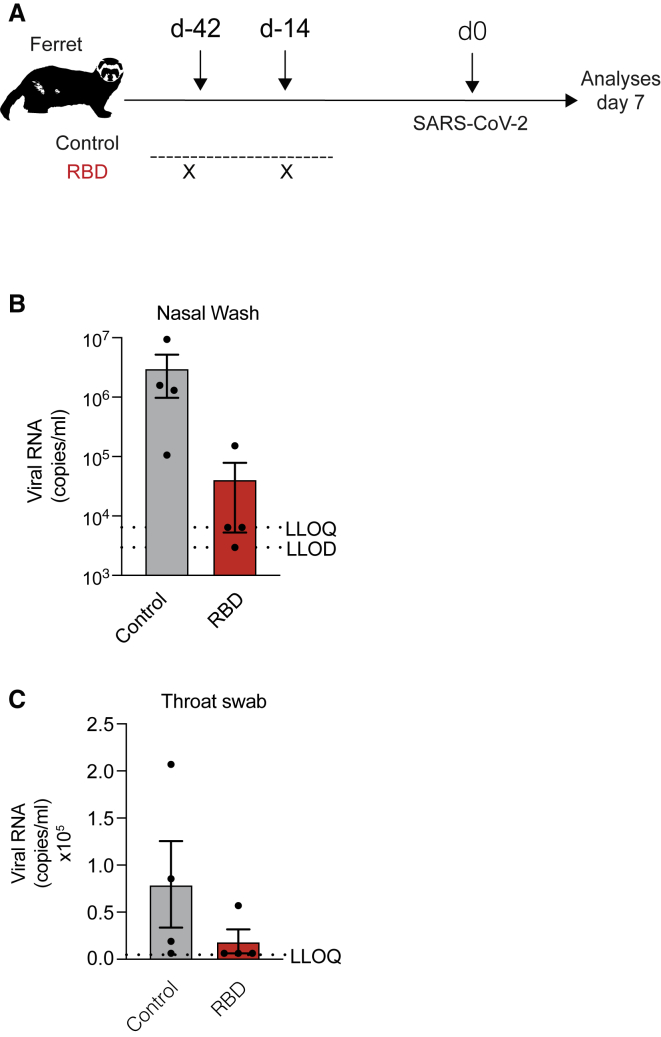
To confirm the immunogenicity and protective efficacy of the RBD vaccine against SARS-CoV-2 infection in a different and larger animal model, sixteen female ferrets weighing >750 g were either left untreated (control) or injected with 400 μg of the RBD vaccine followed by EP 42 and 14 days prior to intranasal infection with 5 × 106 plaque-forming units (PFU) of SARS-CoV-2 isolate Victoria/1/2020 (Figure 5A). Compared to control animals, viral subgenomic RNA detected in nasal washes and throat swabs at day 7 after challenge in immunized ferrets was significantly reduced (Figures 5B and 5C).
Figure 5.
Evaluation of RBD vaccine efficacy in a ferret infection model
(A) Schematic representation of the experimental setup. Female ferrets (n = 8) were either left untreated (control) or received two immunizations (day −42, day −14) of 400 μg of RBD vaccine via intramuscular injection followed by electroporation before intranasal challenge with 5 × 106 PFU/mL of SARS-CoV-2. Four animals from each group were euthanized at each time point (3 and 7 days after challenge). (B) Viral RNA detected in nasal wash from the control group or the vaccinated group after challenge. Results below the lower limit of detection (LLOD) have been assigned a value of 1,157 copies/mL, and results between the LLOD and the lower limit of quantification (LLOQ) have been assigned a value of 6,429 copies/mL. (C) Viral RNA detected in throat swabs from the control group or the vaccinated group after challenge. ∗p value < 0.05
Together, the results obtained in two distinct animal models of SARS-CoV-2 infection indicate that the RBD vaccine induces protective immune responses.
Discussion
The COVID-19 pandemic has stimulated the evaluation and the approval of novel genetic vaccination platform technologies in a very short time frame. COVID-eVax is based on DNA. Thus far, DNA vaccines have been approved only for veterinary applications and are being evaluated in oncology up to Phase III trial. A drawback of DNA-based vaccines has been the poor immunogenicity when moving from mice to larger species. However, the use of EP and other delivery technologies has greatly enhanced their potency. Similar to other DNA-based vaccines, such as those utilizing adenoviral vectors, the need for nuclear delivery, the risk for chromosomal integration, the potential activation of oncogenes, and induction of anti-DNA antibodies need to be taken into account. These safety concerns are carefully explored according to indications provided by the regulatory agencies (FDA Guidance for Industry—Considerations for Plasmid DNA Vaccines for Infectious Disease Indications, US Department of Health and Human Services, Food and Drug Administration, Center for Biologics Evaluation and Research, November 2007) and WHO (https://www.who.int/teams/health-product-policy-and-standards/standards-and-specifications/vaccines-quality/dna). Very recently (August 21, 2021), a DNA vaccine (ZyCoV-D) against SARS-CoV-2 developed by Zydus Cadila has achieved positive results in a Phase III clinical trial and was approved for emergency use authorization (EUA) with the office of the Drug Controller General of India (DCGI). We believe that this vaccine will pave the way for the approval of other DNA vaccines, such as COVID-eVax.
DNA-based vaccines are engineered for maximal gene expression and immunogenicity, they can be quickly designed from new genetic viral sequences, and they allow for fast and scalable manufacturing as well as long-term stability at room temperature. Moreover, DNA vaccines do not require complex formulations such as those based on nanoparticles (necessary for peptide- or RNA-based vaccines). An efficient DNA uptake can be obtained with different methods.20 Among others, EP increases the initial uptake of DNA plasmid by local cells by ∼500-fold.20, 21, 22 Here, we adopted an EP technology manufactured by the Italian company IGEA, a leader in tissue EP, and extensively tested both in mice and other animal species.11,23, 24, 25, 26 This platform technology has been referred to as X-eVax, where X represents the antigen (or the disease).
Multiple studies have reported that DNA vaccines allow for the generation of cellular and humoral responses against pathogens, making this platform ideal for rapid vaccine development against emerging infectious diseases.27 Among these, three DNA vaccines targeting coronavirus S protein have already been tested in humans. The first candidate DNA vaccine expressing SARS-CoV S protein was VRC-SRSDNS015-00-VP and was tested in 10 healthy adults, aged 21–49 yr, in 2004 and 2005.28 This vaccine was administered intramuscularly at a dose of 4 mg by a Biojector needle-free device and proved to be safe and immunogenic. Another candidate DNA vaccine expressing MERS S protein was GLS-5300, evaluated in 75 healthy subjects, aged 19–50 yr, in 2016.5 GLS-5300 DNA vaccine was also administered intramuscularly followed by EP, in a dose-escalation trial at 0.67, 2, or 6 mg. Overall, the vaccine was safe and immunogenic, as assessed by seroconversion and vaccine-induced T cell responses, in most vaccine recipients.5 The third candidate DNA vaccine, INO-4800, expressing the SARS-CoV-2 S protein, has been evaluated in 40 healthy subjects, aged 18–50 yr, at doses of 1 and 2 mg. Administration by intradermal injection followed by EP using Inovio’s Cellectra device has proven generally safe and immunogenic as assessed by humoral and cellular immune responses.29 INO-4800 is currently being investigated in a Phase II/III clinical trial (NCT04642638).
The RBD vaccine candidate reported here, referred to hereafter as COVID-eVax, was selected among 5 different candidates, encoding either the full-length S protein, portions of it, or engineered versions. Preclinical results have shown that all versions were capable of generating antibodies against the RBD region, key for viral entry. The RBD vaccine was chosen over the other candidates not only for its capacity to elicit potent nAbs but also in light of the capacity to induce a robust T cell response, the observation that anti-RBD antibodies represent >90% of the nAbs in convalescent patients,8,30 and the notion that an RBD vaccine might be devoid of potential antibody-dependent enhancement (ADE). Moreover, a recent report suggests that RBD vaccines could better promote the elicitation of high titers of broad sarbecovirus nAbs owing to enhanced accessibility of appropriate antigenic sites compared to the current full-length vaccines.31 Safety and immunogenicity of COVID-eVax was demonstrated in mice and rats. Antibodies binding to the RBD domain of SARS-CoV-2 were detected not only in sera but also in the lungs of vaccinated mice, and their functionality was assessed through neutralization of both wild-type (WT) virus and pseudovirus and by competitive inhibition of SARS-CoV-2 S protein binding to the ACE2 receptor in the presence of sera bled from immunized mice (data not shown). Besides the humoral response, a consistent T cell response, thought to contribute to preventing severe forms of COVID-19 in humans,32 was elicited by a prime-boost vaccination schedule of COVID-eVax. Moreover, since COVID-eVax targets a small region within the S protein, i.e., the RBD, the risk of inducing an ADE should be minimal.
The protective role played by the anti-RBD humoral and T cell response was demonstrated in both K18-hACE2 transgenic mice as well as ferrets. Besides inducing potent adaptive immune responses, a potential additional mechanism of action of COVID-eVax might be due to the competitive inhibition of SARS-CoV-2 binding to ACE2 by the secreted RBD. This is an intriguing potential mechanism that awaits confirmation and further characterization.
COVID-eVax is currently being evaluated in the COV-1/2-01 Phase I/II study (EudraCT 2020-003734-20) at four clinical sites in Italy. The trial is aimed at assessing the safety and immunogenicity of COVID-eVax in healthy subjects of both genders, 18–65 yr of age. In the Phase I Dose Escalation part, COVID-eVax is administered at 3 escalating doses (20 subjects/cohort), in a prime-boost setting (4 weeks apart), from 0.5 to 2 mg/dose. In addition, a cohort in a single 2-mg dose schedule will also be tested. The vaccine is administered by the intramuscular route followed by EP with the new IGEA ElectroPoration System (EPSGun) and the EGT technology for pulse generation (Cliniporator) commercially available in the EU. In both phases, subjects will be followed up for a total duration of 6 months after the first vaccination.
In summary, COVID-eVax is a highly efficient vaccination platform capable of inducing robust, protective nAb and T cell responses in a variety of animal models. COVID-eVax can be administered multiple times, without the risk of inducing antibody responses to the vaccine itself, which may happen in the case of virus-based vector vaccines. We believe that the DNA vaccination platform described here offers unique advantages over other candidate vaccines, such as rapid manufacturing in response to sequence mutations (compared to protein- or viral vector-based vaccines) and greater stability at room temperature (compared to RNA-based platforms). With an increasing number of people having been immunized against SARS-CoV-2 with an RNA-, adenovirus-, or protein-based vaccine, COVID-eVax might be also considered as an additional platform for booster immunizations to extend the duration of protective immunity.
Materials and methods
Synthetic genes and constructs
The synthesis and codon optimization analysis of a cDNA encoding the SARS-CoV-2 protein S has been performed at GenScript (China). All constructs were completely synthetic and optimized for codon usage. Codon-optimized variants took into account codon usage bias, GC content, CpG dinucleotide content, mRNA secondary structure, cryptic splicing sites, premature poly(A) sites, internal chi sites and ribosomal binding sites, negative CpG islands, RNA instability motif (ARE), repeat sequences (direct repeat, reverse repeat, and dyad repeat), and restriction sites that may interfere with cloning. In addition, to improve translational initiation and performance, Kozak and Shine-Dalgarno sequences were inserted into the synthetic genes. To increase the efficiency of translational termination, two consecutive stop codons were inserted at the end of cDNAs. The codon usage bias in human was increased by upgrading the codon adaptation index (CAI) to 0.94. GC content and unfavorable peaks have been optimized to prolong the half-life of the mRNA. The stem-loop structures, which impact ribosomal binding and stability of mRNA, were broken. In addition, the optimization process screened and successfully modified those negative cis-acting sites. For the construction of RBD, N/R, and S1 constructs, the cDNA corresponding to each region was amplified via PCR by using sequence-specific primers and directionally cloned into the linearized pTK1A-TPA vector by PacI/NotI enzymatic restriction. FL expression vector was generated by the In-Fusion Cloning System (Takara), amplifying the cDNA by using specific primers overlapping both the synthetic gene and the acceptor empty vector pTK1A. The FL construct was then cloned into the BglII restriction site of pTK1A.
Transient expression of recombinant SARS-CoV-2 spike proteins and western blotting
HEK293 cells were transiently transfected with SARS-CoV-2-S fragment expression vectors with Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific). Two days later, the supernatants were collected and concentrated by Amicon Ultra centrifugal filters (Sigma) and cells were pelleted and lysed in radioimmunoprecipitation assay (RIPA) buffer (Thermo Fisher Scientific). Cell lysates and supernatants were separated by SDS-PAGE and transferred to nitrocellulose membranes. Immunoblotting was performed by using SARS-CoV2 Spike S1 subunit primary antibody (Sino Biological) diluted 1:1,000 in 5% milk-0.05% PBS-Tween 20. Chemiluminescence detection was performed with the ECL Prime Western Blotting System (Cytiva, Merck) and acquired by the ChemiDoc Imaging System (Bio-Rad).
Recombinant proteins and peptides
RBD-Fc and RBD-6xHis proteins were produced by transient transfection of Expi293F high-density cells with ExpiFectamine 293 Lipid Cation Transfection Reagent (Thermo Fisher) according to the manufacturer’s instructions. The supernatant containing the proteins was collected 1 week later and subjected to clarification by centrifugation and filtration for the subsequent purification steps. The RBD-Fc protein was purified by affinity chromatography with the AktaPure system with a protein A column (TOYOSCREEN AF-RPROTEIN A HC-650F; Tosoh Bioscience). Briefly, the column was equilibrated with binding buffer (buffer phosphate 0.1 M pH 8) and loaded with the supernatant diluted 1:1 in the same buffer. After the column was washed, the protein was recovered by acid elution in 0.1 M pH 3 citrate buffer, neutralized in Tris-HCl pH 9, and subjected to dialysis in 1× PBS with Slide-A-Lyzers (Thermo Fisher) as indicated in the product datasheet. The RBD-6xHIS protein was purified by affinity chromatography of His tag residues for metals immobilized on the AktaPure system with HisPur Ni-NTA Chromatography Cartridges (Thermo Fisher) according to the manufacturer’s instructions. Briefly, the column was equilibrated in 5 mM 1× PBS-imidazole and loaded with the supernatant diluted 1:1 in the same buffer. After washing, the protein was eluted with 1× PBS-imidazole 0.3 M, pH 7.4 and dialyzed in 1× PBS with Slide-A-Lyzers (Thermo Fisher) as indicated in the product datasheet. Once recovered from dialysis, the RBD-Fc and RBD-6xHis proteins were quantified on the spectrophotometer by absorbance at 280 nm. Protein purity was evaluated by SDS-PAGE and western blot analysis, carried out in both reduced and non-reduced conditions and by standard methods. Lyophilized S protein peptides were purchased from JPT (Berlin, Germany) and resuspended in DMSO at 40 mg/mL. Pools of peptides of 15 aa overlapping by 11 residues were assembled in two pools: pool S1 (residue 1 to 635) and pool S2 (residue 625 to 1273). Peptides and pools were stored at −80°C.
Production of SARS-CoV2S pseudoparticles based on VSV
Viral pseudoparticles (PPs) based on vesicular stomatitis virus (VSV) bearing SARS-CoV-2 S were produced as previously described.33 In brief, HEK293T cells were seeded and, the next day, transfected with 44 μg of plasmid encoding SARS-CoV-2 Spike (pCG1-SARS-2-S, Wuhan Hu-1) using TransIT-LT1 (Mirus). The next day, medium was removed and 15 mL of fresh medium and then VSV(Fluc-eGFP)-VSV-G added to deliver the defective viral reporter genome (ΔVSV-G, kindly provided by Gert Zimmer, Institute of Virology and Immunology, Mittelhäusern, Switzerland).34 After 2 h, inoculum was removed, cells washed, and fresh medium added. PPs were harvested after 16–24 h: the supernatant was centrifuged at 1,200 rpm for 5 min to pellet debris, and the supernatant of anti-VSV-G-expressing hybridoma cells (Anti-VSV-G antibody I1, produced from CRL-2700 mouse hybridoma cells, ATCC) was added to virus stocks at 1:10 (v/v) to block residual VSV-G-containing particles. Supernatants were then concentrated by Vivaspin 20 100 kDa ultrafiltration devices, immediately aliquoted, and frozen at −80°C until use.
Production of SARS-CoV2S pseudoparticles based on lentiviral vectors
To generate SARS-CoV-2 lentiviral pseudotype particles, HEK293TN (System Bioscience) cells were plated in 15-cm dishes with complete DMEM medium. The following day, 32 μg of reporter plasmid pLenti CMV-GFP-TAV2A-LUC Hygro, 12.5 μg of pMDLg/pRRE (Addgene #12251), 6.25 μg of pRSV-Rev (Addgene #12253), and 9 μg of pcDNA3.1_Spike_del19 were co-transfected after a calcium phosphate transfection. pcDNA3.1_Spike_del19 (Addgene #155297) was generated by deletion of the last 19 aa of S starting from pcDNA3.1-SARS2-Spike (a gift from Fang Li, Addgene plasmid #145032). pLenti CMV-GFP-TAV2A-LUC Hygro was generated from pLenti CMV GFP Hygro (Addgene #17446) by addition of T2A-luciferase by PCR cloning. Twelve hours after transfection, the medium was replaced with complete Iscove. Thirty hours after transfection, the supernatant was collected, clarified by filtration (0.45-μm pore -size), and concentrated by centrifugation for 2 h at 20,000 rpm. Viral PP suspensions were aliquoted and stored at −80°C.
Animals
BALB/c (H-2d) and C57BL/6 mice (H-2b) were purchased from Envigo (Italy). B6.Cg-Tg(K18-ACE2)2Prlmn/J mice were purchased from The Jackson Laboratory. Mice were housed under specific pathogen-free conditions, and heterozygous mice were used at 6–10 weeks of age. All experimental animal procedures were approved by the Institutional Animal Committee of the San Raffaele Scientific Institute, and all infectious work was performed in designed BSL-3 workspaces.
Sixteen 7-week-old female Sprague-Dawley rats, with a body weight range of 140–155 g, were purchased from Envigo (Italy).
Sixteen 7-month-old female ferrets (Mustela putorius furo) were obtained from a UK Home Office-accredited supplier (Highgate Farm, UK). Animals were housed in pairs at Advisory Committee on Dangerous Pathogens (ACDP) containment level 3. Cages met with the UK Home Office Code of Practice for the Housing and Care of Animals Bred, Supplied or Used for Scientific Procedures (December 2014). All experimental work was conducted under the authority of a UK Home Office-approved project license that had been subject to local ethical review at PHE Porton Down by the Animal Welfare and Ethical Review Body (AWERB) as required by the Home Office Animals (Scientific Procedures) Act 1986.
Vaccination
Mice
DNA-EGT was performed in mice quadriceps injected with doses ranging from 0.1 μg to 20 μg and electrically stimulated as previously described.10 The DNA was formulated in PBS at a concentration of 0.2 mg/mL. DNA-EP was performed with an IGEA Cliniporator (Carpi, Italy) using a needle electrode (electrode A-15-4B). At different time points, antibody and cell-mediated immune response were analyzed.
Rats
After a suitable quarantine period, animals were divided in three different experimental groups (4 females/group) and immunized by intramuscular EP, alternating quadriceps at each vaccine administration.
Ferrets
Eight ferrets were immunized twice at days −42 and −14 with 400 μg of RBD intramuscularly in the quadriceps muscle of the right leg, followed by EP. An additional eight ferrets remained unvaccinated.
Viruses and in vivo treatments
The hCoV-19/Italy/LOM-UniSR-1/2020 (GISAID Accession ID: EPI_ISL_413489) isolate of SARS-CoV-2 was used in this study. Virus isolation studies were carried out in a BSL-3 workspace and performed in Vero E6 cells, which were cultured at 37°C, 5% CO2 in complete medium (DMEM supplemented with 10% fetal bovine serum [FBS], 1% penicillin plus streptomycin, 1% l-glutamine). Virus stocks were titrated with both plaque reduction assay (PRA, PFU/mL) and endpoint dilutions assay (EDA, TCID50/mL). In PRA, confluent monolayers of Vero E6 cells were infected with eight 10-fold dilutions of virus stock. After 1 h of adsorption at 37°C, the cell-free virus was removed. Cells were then incubated for 48 h in DMEM containing 2% FBS and 0.5% agarose. Cells were fixed and stained, and viral plaques were counted. In EDA, Vero E6 cells were seeded into 96-well plates and infected at 95% of confluence with base 10 dilutions of virus stock. After 1 h of adsorption at 37°C, the cell-free virus was removed, cells were washed with 1× PBS, and complete medium was added to cells. After 48 h, cells were observed to evaluate the presence of a cytopathic effect (CPE). TCID50/mL of viral stocks was then determined by applying the Reed-Muench formula.
K18-hACE2 mice were immunized with 10 μg of COVID-eVax or saline solution intramuscularly twice 21 days apart followed by EP as described above.
Virus infection was performed via intranasal administration of 1 × 105 TCID50 per mouse under isoflurane 2% (#IsoVet250) anesthesia. Mice were monitored to record body weight and clinical and respiratory parameters.
Eight ferrets were immunized twice at days −42 and −14 with 400 μg of RBD intramuscularly in the quadriceps muscle of the right leg followed by EP as described above. An additional eight ferrets remained unvaccinated. All ferrets were challenged with SARS-CoV-2 Victoria/01/202035 6 weeks after first vaccination. Challenge virus (5 × 106 PFU/mL) was delivered by intranasal instillation (1.0 mL total, 0.5 mL per nostril) diluted in PBS. Nasal washes and throat swabs were taken from all ferrets at 2, 4, 6, and 7 days after challenge. Nasal washes were obtained by flushing the nasal cavity with 2 mL of PBS. For throat swabs, a flocked swab (MWE Medical Wire, Corsham, UK) was gently stroked across the back of the pharynx in the tonsillar area. Four animals from each group were euthanized at day 3, and the remaining animals from each group were euthanized at day 7.
Luciferase assay
BALB/c mice (5 mice/group) were anesthetized with 97% oxygen and 3% isoflurane (Isoba, MSD Animal Health, Walton, UK) and then injected by DNA EP with a DNA plasmid encoding luciferase (pcDNA3-Hygro-Luc 1 μg/mouse) in a 50-μL volume in quadriceps muscle. Mice were electroporated by means of Cliniporator Device EPS01 N-10-4B electrodes with the following electrical conditions in EGT modality: 8 pulses 20 ms each at 110 V, 8 Hz, 120-ms interval. Imaging was performed under gas anesthesia at Xenogen IVIS 200 at 48 h after injection, 8 min after injection of a luciferin solution subcutaneously (s.c.) (15 mg/mL, PerkinElmer) at 10 μL/g of body weight.
ELISpot assays
For the B cell ELISpot assay, pools of sera collected from mice vaccinated with RBD or FL constructs were tested against each of the 338 peptides covering the entire S protein, pre-coated on 96-well plates, in order to identify the linear epitopes. Sequences of positive hits were then mapped on three-dimensional structure of S protein, hence outlining the epitope domains. The T cell ELISpot for mouse IFN-γ was performed as previously described.36 RBD peptides are 132 out of the 338 peptides covering the whole S protein (from peptide nr.4 to peptide nr.136). In order to identify immunodominant RBD epitopes (here highlighted in yellow), ELISpot assay was performed by stimulating splenocytes from RBD-vaccinated BALB/c mice for 20 h with RBD peptide pools. Pools (from 1 to 24) were distributed as a matrix (the intersection of two pools identifies one RBD peptide), with each pool comprising up to 12 RBD peptides. Immunodominant RBD peptides were identified at the intersection of pools showing >50 spot-forming cells (SFCs).
Antibody detection assays
Antibody titration was performed both on sera, obtained by retro-orbital bleeding, and on BALs, obtained by flushing 1 mL of PBS in the lungs. The ELISA plates were functionalized by coating with the RBD-6xHis protein at a concentration of 1 μg/mL and incubated ∼18 h at 4°C. Subsequently, the plates were blocked with 3% BSA-0.05% Tween 20-PBS for 1 h at room temperature, and then the excess solution was eliminated. The sera of the immunized mice were then added at a dilution of 1/300 and diluted 1:3 up to 1/218,700, in duplicate, and the plates were incubated for 2 h at room temperature. After a double wash with 0.05% Tween 20-PBS, the secondary anti-murine IgG or anti-murine immunoglobulin M (IgM) conjugated with alkaline phosphatase was added and the plates were incubated for 1 h at room temperature. After a double wash with 0.05% Tween 20-PBS, the binding of the secondary was detected by adding the substrate for alkaline phosphatase and measuring the absorbance at 405 nm by means of an ELISA reader after incubation for 2 h. IgG antibody titers against the S protein RBD were evaluated at several time points. Regarding antibody titration on sera of vaccinated K18-hACE2 mice, the sera were added at a dilution of 1/300 to 1/21,8700, in duplicate, and the plates were incubated overnight (O/N) at 4°C. After three washes with 0.05% Tween 20-PBS, the secondary anti-murine IgG horseradish peroxidase (HRP) (1:2,000) was added and the plates were incubated for 1 h at room temperature. After a wash with 0.05% Tween 20-PBS, the binding of the secondary was detected by adding 3,3′,5,5′-tetramethylbenzidine (TMB) substrate reagent (BD Biosciences). The reaction was blocked with 0.5 M H2SO4, and the absorbance at 450 nm and reference 630 nm was measured.
For neutralization experiments with SARS-CoV-2 VSV-based PPs, Caco-2 cells were seeded in 96-well plates in 180 μL of medium at 10,000 cells/well. The next day, serially diluted heat-inactivated sera in PBS or diluted BALs were added to CoV2S-PPs at 1:1 dilution with max. 10% serum or BAL on PPs. Sera/BAL-PP mixtures were incubated 1 h at 37°C and then added to Caco-2 cells in duplicates at 1:10 dilution (max. 1% serum on cells), and cells were incubated at 37°C. Sixteen hours after transduction, firefly luciferase activity was measured with the Promega Luciferase Assay System (E1501) and values normalized to PPs treated with PBS only. Inhibition of 50% of CPE (IC50) was fit by Prism Inhibitor versus Normalized response (variable slope); samples with <50% inhibition at 10% serum were excluded. These assays have been performed in a BSL-2 facility at Ulm University.
For lentiviral pseudotype neutralization assay, HEK293TN-hACE2 were plated at 104 cells/well in white 96-well plates (100 μL/well of complete DMEM medium). The next day, cells were infected with 0.1 MOI of SARS-CoV-2-GFP/luciferase PPs that were subjected to preincubation with serially diluted sera. In detail, sera samples were serially diluted 3-fold in PBS in order to obtain a 7-point dose-response curve (plus PBS alone as untreated control). Thereafter, aliquots of undiluted or 3-fold serially diluted sera were further diluted 1:10 in aliquots of SARS-CoV-2 PPs adjusted to contain 0.1 MOI/50 μL of complete culture medium. After incubation for 1 h at 37°C, 50 μL of serum-SARS-CoV-2 PP mixture was added to each well and plates were incubated for 24 h at 37°C. Thus, the starting serum dilutions was 1:30. Each dilution was tested in triplicate. After 24 h of incubation, cell infection was measured by luciferase assay with the Bright-Glo Luciferase System (Promega). The Infinite F200 plate reader (Tecan) was used to read luminescence. Obtained relative light units (RLUs) were normalized to controls, and dose-response curves were generated by nonlinear regression curve fitting with GraphPad Prism to calculate ND50.
HEK293TN-hACE2 were generated by transduction of HEK293TN with a lentiviral vector engineered to stably express hACE2 (unpublished data).
To test the ability of elicited antibodies to neutralize the virus in vitro, Vero E6 cells (20,000 cells/well) were seeded 24 h prior to infection in 96-well plates (Costar). Serum samples from mice were incubated at 56°C for 30 min and then serially 2-fold diluted in cell culture medium (10- to 10,240-fold). Serum dilutions were then mixed to 100 TCID50 of SARS-CoV-2 (virus isolated in January 2020 at the National Institute for Infectious Diseases “L. Spallanzani” in Rome, 2019-nCoV/Italy-INMI1, clade V strain; GISAID accession number: EPI_ISL_410545) in 96-well plates and incubated at 37°C for 30 min in 5% (v/v) CO2. The virus-serum mixture was then added to the cells and incubated at 37°C for microscopic examination of the cytopathic effect after 48 h. Cell supernatants were then removed, and cells were fixed/stained for 30 min with a solution of 2% formaldehyde (AppliChem, Darmstadt, Germany) in crystal violet (Diapath, Martinengo, Bergamo, Italy). The fixing solution was removed and cell viability measured by photometer at 595 nm (Synergy HTX; BioTek Instruments, Winooski, VT, USA). The serum IC50 was calculated with Prism 7 (GraphPad Software, San Diego, CA, USA) as described previously.37 Tests were performed in duplicate with negative control samples from unvaccinated mice and positive control samples from a COVID-19 patient with known neutralizing titer. The same assay was repeated with the SARS-CoV-2 strain belonging to the G clade (SARS-CoV-2/Human/ITA/PAVIA10734/2020, clade G, D614G (S), in GISAID EPI_ISL_568579; isolated at Policlinico San Matteo in the Laboratory of Prof. Fausto Baldanti). These assays have been performed in a BSL-3 facility at the Spallanzani Institute in Rome.
Cell isolation and flow cytometry
In experiments performed with vaccinated WT mice, the intracellular staining was performed according to the procedure described in Giannetti et al.38 Briefly, peripheral blood mononuclear cells (PBMCs) or splenocytes were treated with ACK Lysing Buffer (Life Technologies) for red blood cell lysis and resuspended in 0.6 mL RPMI, 10% fetal calf serum (FCS) and incubated with the indicated pool of peptides (5 μg/mL final concentration of each peptide) and brefeldin A (1 μg/mL; BD Pharmingen) at 37°C for 12–16 h. Cells were then washed and stained with surface antibodies. After washing, cells were fixed, permeabilized, and incubated with anti-IFN-γ (XMG1.2) and -TNF-α (MP6-XT22; all from eBioscience, San Diego, CA, USA), fixed with 1% formaldehyde in PBS, and analyzed on a CytoFLEX flow cytometer. DMSO and phorbol myristate acetate/ionomycin (PMA/IONO) (Sigma) at 10 μg/mL were used as internal negative and positive control of the assay, respectively.
In experiments performed with SARS-CoV-2-infected mice, lung was perfused through the right ventricle with PBS at the time of autopsy and after the brain was removed from the skull. Lung tissue was digested in RPMI 1640 containing 3.2 mg/mL collagenase IV (Sigma) and 25 U/mL DNase I (Sigma) for 30 min at 37°C. Brain was digested in RPMI 1640 containing 1 mg/mL collagenase D (Sigma) and 6.3 μg/mL DNase I (Sigma) for 30 min at 37°C. Homogenized lung and brain were passed through 70-μm nylon mesh to obtain a single-cell suspension. Cells were resuspended in 36% Percoll solution (Sigma) and centrifuged for 20 min at 2,000 rpm (light acceleration and low brake). The remaining red blood cells were removed with ACK lysis.
For analysis of ex vivo intracellular cytokine production, 1 mg/mL of brefeldin A (Sigma) was included in the digestion buffer. All flow cytometry stainings of surface-expressed and intracellular molecules were performed as described previously.39 Briefly, cells were stimulated for 4 h at 37°C with 15-mer peptides overlapping by 11 amino acids (5 μg/mL) covering the RBD of SARS-CoV-2. Cell viability was assessed by staining with Viobility 405/520 fixable dye (Miltenyi). Antibodies (Abs) used are indicated in Table 1.
Table 1.
Antibodies used in the study
| Name | Clone | Source and catalog number |
|---|---|---|
| CD8α | 53-6.7 | BioLegend 100723 |
| CD3 | 145-2C11 | Pharmingen 552774 |
| CD44 | IM7 | BioLegend 103028 |
| IFNγ | XMG1.2 | BioLegend 505830 |
| TNFα | MP6-XT22 | BioLegend 506329 |
| CD62L | MEL-14 | BioLegend 104453 |
| CD4 | RM4-5 | BD Biosciences 740208 |
| IL-17A | TC11-18H10 | BD Pharmingen 562542 |
| IL-5 | TRFK5 | BD 554395 |
| IL-4 | 11B11 | BD Pharmingen 554436 |
Flow cytometry analysis was performed on a Beckman Coulter CytoFLEX LX cytometer and analyzed with FlowJo software (Treestar).
Tissue homogenates and viral titers
Tissues homogenates were prepared by homogenizing perfused lung with a gentleMACS Octo Dissociator (Miltenyi) in M tubes containing 1 mL of DMEM 0%. Samples were homogenized three times with the program m_Lung_01_02 (34 s, 164 rpm). The homogenates were centrifuged at 3,500 rpm for 5 min at 4°C. The supernatant was collected and stored at −80°C for viral isolation and viral load detection. Viral titer was calculated by TCID50. Briefly, Vero E6 cells were seeded at a density of 1.5 × 104 cells per well in flat-bottom 96-well tissue culture plates. The following day, 2-fold dilutions of the homogenized tissue were applied to confluent cells and incubated 1 h at 37°C. Then, cells were washed with PBS and incubated for 72 h at 37°C in DMEM 2% FBS. Cells were fixed with 4% paraformaldehyde (PFA) for 20 min and stained with 0.05% (w/v) crystal violet in 20% methanol.
RNA extraction and qPCR
Tissue homogenates were prepared by homogenizing perfused lung with the gentleMACS dissociator (Miltenyi) with the RNA_02 program in M tubes in 1 mL of TRIzol (Invitrogen). The homogenates were centrifuged at 2,000 × g for 1 min at 4°C, and the supernatant was collected. RNA extraction was performed by combining phenol/guanidine-based lysis with silica membrane-based purification. Briefly, 100 μL of chloroform was added to 500 μL of homogenized sample; after centrifugation, the aqueous phase was added to 1 volume of 70% ethanol and loaded on a ReliaPrep RNA Tissue Miniprep column (Promega, Cat #Z6111). Total RNA was isolated according to the manufacturer’s instructions. qPCR was performed with TaqMan Fast Virus 1-Step PCR Master Mix (Life Technologies); standard curve was drawn with 2019_nCOV_N Positive control (IDT). The primers used were 2019-nCoV_N1-Forward Primer (5′-GAC CCC AAA ATC AGC GAA AT-3′), 2019-nCoV_N1-Reverse Primer (5′-TCT GGT TAC TGC CAG TTG AAT CTG-3′), and 2019-nCoV_N1-Probe (5′-FAM-ACC CCG CAT TAC GTT TGG ACC-BHQ1-3′) (Centers for Disease Control and Prevention [CDC] Atlanta, GA, USA). All experiments were performed in duplicate.
Whole-body plethysmography
WBP was performed with a WBP chamber (DSI Buxco respiratory solutions, DSI). First, mice were allowed to acclimate inside the chamber for 10 min, and then respiratory parameters were acquired for 15 min with FinePointe software.
Confocal immunofluorescence histology and histochemistry and N-SARS-CoV-2 signal quantification
Lungs of infected mice were collected and fixed in 4% PFA. Samples were then dehydrated in 30% sucrose prior to embedding in OCT freezing medium (Bio-Optica). Twenty-micrometer sections were cut on a CM1520 cryostat (Leica) and adhered to Superfrost Plus slides (Thermo Scientific). Sections were then permeabilized and blocked in PBS containing 0.3% Triton X-100 (Sigma-Aldrich) and 5% FBS followed by staining in PBS containing 0.3% Triton X-100 and 1% FBS. Slides were stained for SARS-CoV-2 nucleocapsid (GeneTex) for 1 h at room temperature (RT). Then, slides were stained with Alexa Fluor 568 Goat Anti-Rabbit antibody for 2 h at RT. All slides were analyzed by confocal fluorescence microscopy (Leica TCS SP5 Laser Scanning Confocal). For SARS-CoV-2 N protein immunohistochemistry, mice were perfused with PBS and brains were collected in Zn-formalin and transferred into 70% ethanol 24 h later. Tissue was then processed, embedded in paraffin, and automatically stained for SARS-CoV-2 (2019-nCoV) Nucleocapsid Antibody (Sino Biological, 40143-R019) through Leica Bond RX for 1 h at RT and developed with Bond Polymer Refine Detection (Leica, DS9800). Brightfield images were acquired through an Aperio Scanscope System CS2 microscope and an ImageScope program (Leica Biosystems) according to the manufacturer’s instructions. In both immunofluorescence and histochemistry, SARS-CoV-2 N protein percentage of positive area was determined by QuPath (Quantitative Pathology & Bioimage Analysis) software.
Statistical analyses and software
Detailed information concerning the statistical methods used is provided in the figure legends. Flow cytometry data were collected with FlowJo version 10.5.3 (Treestar). Statistical analyses were performed with GraphPad Prism software version 8 (GraphPad). Immunofluorescence and histochemical imaging quantifications were performed with the Aperio Scanscope System and QuPath software (Quantitative Pathology & Bioimage Analysis). n represents individual mice analyzed per experiment. Error bars indicate the standard error of the mean (SEM). We used Mann-Whitney U-tests to compare two groups with non-normally distributed continuous variables. We used two-way ANOVA followed by Sidak’s multiple comparisons tests to analyze experiments with multiple groups and two independent variables. Significance is indicated as follows: ∗p < 0.05; ∗∗p < 0.01. Comparisons are not statistically significant unless indicated.
Data and materials availability
All data are available in the main text or the Supplemental information.
Acknowledgments
We thank the entire Rottapharm Biotech and IGEA Teams for useful scientific and regulatory discussions and for setting up the EPSGun technology in a short time. We also thank M. Mainetti, M. Freschi, and A. Fiocchi for technical support; M. Silva for secretarial assistance; and the members of the Iannacone laboratory for helpful discussions. Flow cytometry was carried out at FRACTAL, a flow cytometry resource and advanced cytometry technical applications laboratory established by the San Raffaele Scientific Institute. Confocal immunofluorescence histology was carried out at Alembic, an advanced microscopy laboratory established by the San Raffaele Scientific Institute and the Vita-Salute San Raffaele University. We would like to acknowledge the PhD program in Basic and Applied Immunology and Oncology at Vita-Salute San Raffaele University, as D.M. and E. Sala conducted this study as partial fulfillment of their PhD in Molecular Medicine within that program. M.I. is supported by European Research Council (ERC) Consolidator Grant 725038, ERC Proof of Concept Grant 957502, Italian Association for Cancer Research (AIRC) Grants 19891 and 22737, Italian Ministry of Health Grants RF-2018-12365801 and COVID-2020-12371617, Lombardy Foundation for Biomedical Research (FRRB) Grant 2015-0010, the European Molecular Biology Organization Young Investigator Program, and Funded Research Agreements from Gilead Sciences, Takis Biotech, Toscana Life Sciences, and Asher Bio. L.G.G. is supported by AIRC Grant 22737, Lombardy Open Innovation Grant 229452, PRIN Grant 2017MPCWPY from the Italian Ministry of Education, University and Research, Funded Research Agreements from Gilead Sciences, Avalia Therapeutics, and CNCCS SCARL, and donations from FONDAZIONE SAME and FONDAZIONE PROSSIMO MIO for COVID-19-related research. M.K. is supported by Italian Ministry of Education, University and Research grant PRIN-2017ZXT5WR. J.M. is supported by funding from the Deutsche Forschungsgemeinschaft through Fokus-Förderung COVID-19 and the CRC1279, the European Union Horizon 2020 Framework Programme for Innovation and Research (Fight-nCoV), the Ministry for Science, Research and Arts of Baden-Württemberg, and the BMWi–Federal Ministry for Economic Affairs and Energy (COMBI-COV-2). R.G., A.S., and L.W. are part of the International Graduate School in Molecular Medicine Ulm. Takis Research activities are supported in part by the Italian Ministry of Economic Development through grants F/050298/02/X32, F/090033/01-04/X36, and F/190180/01/X44. We are also grateful to Lazio Innova for the funding provided through grant A0376-2020-0700050 Prog. T0002E0001 “Emergenza Coronavirus ed oltre” to Vitares no profit organization, and Fondazione Melanoma for providing support to develop the assays used in clinical trials. PHE activities (Covivax project) are supported by the European Network of vaccine research and development (TRANSVAC2).
Author contributions
A.C., E.M., F.P., G.R., M.R., V.F., A.M., M.M., L. Luberto, L. Lione, E. Salvatori, M. Compagnone, E. Pinto, E. Pavoni, F.B., G.V., D.S., M.L.P., M. Cappalletti, F.F.F., E.D., V.C, R.A., A.N., P.D.L., D.M., E.B., L.G., E. Sala, C.P., J.P., K.A.R., A.R.C., G.M., F.C., G. Caselli, E.C., N.C., N.M., R.G., A.S., L.W., L.D., and M. Conti performed experiments; A.C., M.R., V.F., E. Sala, G. Caselli, J.M., L.D., M. Compagnone, and R.D.F. analyzed and interpreted data; A.C. and M.R. prepared the figures and edited the manuscript; J.M., R.D.F., M.K., G. Ciliberto, C.C., M.R.C., G.I., L.G.G., and L.R. provided funding and conceptual advice and edited the manuscript; M.I. and L.A. coordinated the study, provided funding, and wrote the paper.
Declaration of interests
A.C. and M.M. are Evvivax employees. E.M., F.P., G.R., A.M., L.L., L.L., E. Salvatori, M. Cappalletti, F.F.F., E.D., V.C., and L.A. are Takis employees. G. Caselli and L.R. are Rottapharm Biotech employees. Takis and Rottapharm Biotech are jointly developing COVID-eVax. M.I. participates in advisory boards/consultancies for or receives funding from Gilead Sciences, Roche, Third Rock Ventures, Amgen, Allovir, Asher Bio. L.G.G is a member of the board of directors at Genenta Science and Epsilon Bio and participates in advisory boards/consultancies for Gilead Sciences, Roche, and Arbutus Biopharma.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2021.09.011.
Contributor Information
Matteo Iannacone, Email: iannacone.matteo@hsr.it.
Luigi Aurisicchio, Email: aurisicchio@takisbiotech.it.
Supplemental information
References
- 1.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 2.Gebre M.S., Brito L.A., Tostanoski L.H., Edwards D.K., Carfi A., Barouch D.H. Novel approaches for vaccine development. Cell. 2021;184:1589–1603. doi: 10.1016/j.cell.2021.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conforti A., Marra E., Roscilli G., Palombo F., Ciliberto G., Aurisicchio L. Are Genetic Vaccines the Right Weapon against COVID-19? Mol. Ther. 2020;28:1555–1556. doi: 10.1016/j.ymthe.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Z.Y., Kong W.P., Huang Y., Roberts A., Murphy B.R., Subbarao K., Nabel G.J. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428:561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Modjarrad K., Roberts C.C., Mills K.T., Castellano A.R., Paolino K., Muthumani K., Reuschel E.L., Robb M.L., Racine T., Oh M.D., et al. Safety and immunogenicity of an anti-Middle East respiratory syndrome coronavirus DNA vaccine: a phase 1, open-label, single-arm, dose-escalation trial. Lancet Infect. Dis. 2019;19:1013–1022. doi: 10.1016/S1473-3099(19)30266-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L., Shi W., Chappell J.D., Joyce M.G., Zhang Y., Kanekiyo M., Becker M.M., van Doremalen N., Fischer R., Wang N., et al. Importance of Neutralizing Monoclonal Antibodies Targeting Multiple Antigenic Sites on the Middle East Respiratory Syndrome Coronavirus Spike Glycoprotein To Avoid Neutralization Escape. J. Virol. 2018;92:e02002–e02017. doi: 10.1128/JVI.02002-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piccoli L., Park Y.-J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fattori E., La Monica N., Ciliberto G., Toniatti C. Electro-gene-transfer: a new approach for muscle gene delivery. Somat. Cell Mol. Genet. 2002;27:75–83. doi: 10.1023/a:1022927822244. [DOI] [PubMed] [Google Scholar]
- 10.Cappelletti M., Zampaglione I., Rizzuto G., Ciliberto G., La Monica N., Fattori E. Gene electro-transfer improves transduction by modifying the fate of intramuscular DNA. J. Gene Med. 2003;5:324–332. doi: 10.1002/jgm.352. [DOI] [PubMed] [Google Scholar]
- 11.Aurisicchio L., Mancini R., Ciliberto G. Cancer vaccination by electro-gene-transfer. Expert Rev. Vaccines. 2013;12:1127–1137. doi: 10.1586/14760584.2013.836903. [DOI] [PubMed] [Google Scholar]
- 12.Hojman P. Basic principles and clinical advancements of muscle electrotransfer. Curr. Gene Ther. 2010;10:128–138. doi: 10.2174/156652310791110994. [DOI] [PubMed] [Google Scholar]
- 13.van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J.N., Port J.R., Avanzato V.A., Bushmaker T., Flaxman A., Ulaszewska M., et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature. 2020;586:578–582. doi: 10.1038/s41586-020-2608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huo J., Le Bas A., Ruza R.R., Duyvesteyn H.M.E., Mikolajek H., Malinauskas T., Tan T.K., Rijal P., Dumoux M., Ward P.N., et al. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 2020;27:846–854. doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- 15.Schafer K.A., Eighmy J., Fikes J.D., Halpern W.G., Hukkanen R.R., Long G.G., Meseck E.K., Patrick D.J., Thibodeau M.S., Wood C.E., Francke S. Use of Severity Grades to Characterize Histopathologic Changes. Toxicol. Pathol. 2018;46:256–265. doi: 10.1177/0192623318761348. [DOI] [PubMed] [Google Scholar]
- 16.McCray P.B., Jr., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H.P., Halabi C., Sigmund C.D., et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng J., Wong L.R., Li K., Verma A.K., Ortiz M.E., Wohlford-Lenane C., Leidinger M.R., Knudson C.M., Meyerholz D.K., McCray P.B., Jr., Perlman S. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature. 2021;589:603–607. doi: 10.1038/s41586-020-2943-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menachery V.D., Gralinski L.E., Baric R.S., Ferris M.T. New Metrics for Evaluating Viral Respiratory Pathogenesis. PLoS ONE. 2015;10:e0131451. doi: 10.1371/journal.pone.0131451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dinnon K.H., 3rd, Leist S.R., Schäfer A., Edwards C.E., Martinez D.R., Montgomery S.A., West A., Yount B.L., Jr., Hou Y.J., Adams L.E., et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020;586:560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jorritsma S.H.T., Gowans E.J., Grubor-Bauk B., Wijesundara D.K. Delivery methods to increase cellular uptake and immunogenicity of DNA vaccines. Vaccine. 2016;34:5488–5494. doi: 10.1016/j.vaccine.2016.09.062. [DOI] [PubMed] [Google Scholar]
- 21.Diaz C.M., Chiappori A., Aurisicchio L., Bagchi A., Clark J., Dubey S., Fridman A., Fabregas J.C., Marshall J., Scarselli E., et al. Phase 1 studies of the safety and immunogenicity of electroporated HER2/CEA DNA vaccine followed by adenoviral boost immunization in patients with solid tumors. J. Transl. Med. 2013;11:62. doi: 10.1186/1479-5876-11-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aurisicchio L., Fridman A., Mauro D., Sheloditna R., Chiappori A., Bagchi A., Ciliberto G. Safety, tolerability and immunogenicity of V934/V935 hTERT vaccination in cancer patients with selected solid tumors: a phase I study. J. Transl. Med. 2020;18:39. doi: 10.1186/s12967-020-02228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peruzzi D., Gavazza A., Mesiti G., Lubas G., Scarselli E., Conforti A., Bendtsen C., Ciliberto G., La Monica N., Aurisicchio L. A vaccine targeting telomerase enhances survival of dogs affected by B-cell lymphoma. Mol. Ther. 2010;18:1559–1567. doi: 10.1038/mt.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Impellizeri J.A., Gavazza A., Greissworth E., Crispo A., Montella M., Ciliberto G., Lubas G., Aurisicchio L. Tel-eVax: a genetic vaccine targeting telomerase for treatment of canine lymphoma. J. Transl. Med. 2018;16:349. doi: 10.1186/s12967-018-1738-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aurisicchio L., Salvatori E., Lione L., Bandini S., Pallocca M., Maggio R., Fanciulli M., De Nicola F., Goeman F., Ciliberto G., et al. Poly-specific neoantigen-targeted cancer vaccines delay patient derived tumor growth. J. Exp. Clin. Cancer Res. 2019;38:78. doi: 10.1186/s13046-019-1084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Impellizeri J.A., Ciliberto G., Aurisicchio L. Electro-gene-transfer as a new tool for cancer immunotherapy in animals. Vet. Comp. Oncol. 2014;12:310–318. doi: 10.1111/vco.12006. [DOI] [PubMed] [Google Scholar]
- 27.Gary E.N., Weiner D.B. DNA vaccines: prime time is now. Curr. Opin. Immunol. 2020;65:21–27. doi: 10.1016/j.coi.2020.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin J.E., Louder M.K., Holman L.A., Gordon I.J., Enama M.E., Larkin B.D., Andrews C.A., Vogel L., Koup R.A., Roederer M., et al. VRC 301 Study Team A SARS DNA vaccine induces neutralizing antibody and cellular immune responses in healthy adults in a Phase I clinical trial. Vaccine. 2008;26:6338–6343. doi: 10.1016/j.vaccine.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith T.R.F., Patel A., Ramos S., Elwood D., Zhu X., Yan J., Gary E.N., Walker S.N., Schultheis K., Purwar M., et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 2020;11:2601. doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29:463–476.e6. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tortorici M.A., Czudnochowski N., Starr T.N., Marzi R., Walls A.C., Zatta F., Bowen J.E., Jaconi S., Di Iulio J., Wang Z., et al. Broad sarbecovirus neutralization by a human monoclonal antibody. Nature. 2021;597:103–108. doi: 10.1038/s41586-021-03817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jarjour N.N., Masopust D., Jameson S.C. T cell memory: Understanding COVID-19. Immunity. 2021;54:14–18. doi: 10.1016/j.immuni.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffmann M., Arora P., Groß R., Seidel A., Hörnich B.F., Hahn A.S., Krüger N., Graichen L., Hofmann-Winkler H., Kempf A., et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393.e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berger Rentsch M., Zimmer G. A vesicular stomatitis virus replicon-based bioassay for the rapid and sensitive determination of multi-species type I interferon. PLoS ONE. 2011;6:e25858. doi: 10.1371/journal.pone.0025858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryan K.A., Bewley K.R., Fotheringham S.A., Slack G.S., Brown P., Hall Y., Wand N.I., Marriott A.C., Cavell B.E., Tree J.A., et al. Dose-dependent response to infection with SARS-CoV-2 in the ferret model and evidence of protective immunity. Nat. Commun. 2021;12:81. doi: 10.1038/s41467-020-20439-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conforti A., Peruzzi D., Giannetti P., Biondo A., Ciliberto G., La Monica N., Aurisicchio L. A novel mouse model for evaluation and prediction of HLA-A2-restricted CEA cancer vaccine responses. J. Immunother. 2009;32:744–754. doi: 10.1097/CJI.0b013e3181aee1b6. [DOI] [PubMed] [Google Scholar]
- 37.Ferrara F., Temperton N. Pseudotype Neutralization Assays: From Laboratory Bench to Data Analysis. Methods Protoc. 2018;1:8. doi: 10.3390/mps1010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giannetti P., Facciabene A., La Monica N., Aurisicchio L. Individual mouse analysis of the cellular immune response to tumor antigens in peripheral blood by intracellular staining for cytokines. J. Immunol. Methods. 2006;316:84–96. doi: 10.1016/j.jim.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 39.Bénéchet A.P., De Simone G., Di Lucia P., Cilenti F., Barbiera G., Le Bert N., Fumagalli V., Lusito E., Moalli F., Bianchessi V., et al. Dynamics and genomic landscape of CD8+ T cells undergoing hepatic priming. Nature. 2019;574:200–205. doi: 10.1038/s41586-019-1620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.