Abstract
Most diploid organisms have polyploid ancestors. The evolutionary process of polyploidization is poorly understood but has frequently been conjectured to involve some form of ‘genome shock’, such as genome reorganization and subgenome expression dominance. Here we study polyploidization in Arabidopsis suecica, a post-glacial allopolyploid species formed via hybridization of Arabidopsis thaliana and Arabidopsis arenosa. We generated a chromosome-level genome assembly of A. suecica and complemented it with polymorphism and transcriptome data from all species. Despite a divergence around 6 million years ago (Ma) between the ancestral species and differences in their genome composition, we see no evidence of a genome shock: the A. suecica genome is colinear with the ancestral genomes; there is no subgenome dominance in expression; and transposon dynamics appear stable. However, we find changes suggesting gradual adaptation to polyploidy. In particular, the A. thaliana subgenome shows upregulation of meiosis-related genes, possibly to prevent aneuploidy and undesirable homeologous exchanges that are observed in synthetic A. suecica, and the A. arenosa subgenome shows upregulation of cyto-nuclear processes, possibly in response to the new cytoplasmic environment of A. suecica, with plastids maternally inherited from A. thaliana. These changes are not seen in synthetic hybrids, and thus are likely to represent subsequent evolution.
Subject terms: Speciation, Evolutionary genetics
Arabidopsis suecica is a natural allotetraploid species formed via hybridization of Arabidopsis thaliana and Arabidopsis arenosa. Comparative analysis of genome and transcriptome data shows no evidence for major genomic changes linked to structural and functional alterations in A. suecica but reveals changes to the meiotic machinery and cyto-nuclear processes.
Main
Ancient polyploidization or whole-genome duplication is a hallmark of most higher-organism genomes1,2, including our own3,4. While most of these organisms are now diploid and show only traces of polyploidy, there are many examples of recent polyploidization, especially among flowering plants5–9. These examples are important because they allow us to study the process of polyploidization.
Widespread naturally occurring polyploid hybrids (that is, allopolyploids) show that natural polyploid species can quickly become successful10–18 and even invasive19. However, new allopolyploid species face numerous challenges such as population bottlenecks13,20, competition with their diploid progenitors21, chromosome segregation22–24, changes to genome structure25 and genome regulation26,27—potentially leading to a ‘genome shock’28. In agreement with this, genomic and transcriptomic changes tied to the hybridization of diverged genomes have been reported in resynthesized polyploids of wheat29–35, Brassica napus36–38 and cotton39–42, although exceptions exist43.
The long-term importance of such changes is unclear. Evidence of the transcription and mobilization of transposable elements (TEs) in resynthesized wheat33,44–46 is not observed in cultivated wheat47. However, other cultivated crop genomes, like cotton, show evidence consistent with dramatic changes following allopolyploidy5,48–53. Strawberry6, peanut8 and the mesopolyploids Brassica rapa54 and maize55 show evidence of subgenome dominance, whereas wheat56, cotton51 and B. napus57 do not. The reasons for these differences are not understood.
Whether allopolyploid crops are representative of natural polyploidization is unclear. Domestication is frequently associated with very strong ‘artificial’ selection, which can markedly alter the fitness landscape58–62, and structural variants have been linked to favourable agronomic traits63–65. In addition, polyploid crops are generally evolutionarily recent.
Genomic changes have also been reported in natural allopolyploids such as Tragopogon miscellus66,67, Mimulus pergrinus17 and Spartina anglica68; however, these examples resemble resynthesized allopolyploids in being extremely recent (around 100 years). More-established allopolyploids generally do not show signs of genomic changes12–14,16,69–71.
Here, we focus on an allopolyploid comparable in age to these examples: the highly selfing72 A. suecica, which was formed through the hybridization of A. thaliana and A. arenosa around 16 thousand years ago, during the Last Glacial Maximum20, and which is currently found in northern Fennoscandia (Fig. 1a). The ancestral species diverged around 6 Ma (ref. 73), and based on organelle sequences, A. thaliana is the maternal and A. arenosa is the paternal parent74. This scenario is supported by A. arenosa being a ploidy-variable species, such that A. suecica could readily be generated by the fusion of an unreduced A. thaliana egg cell and an autotetraploid A. arenosa sperm cell20,75. Despite a severe genetic bottleneck20, most of the genetic variation in A. suecica is shared with the ancestral species, ruling out a unique origin. To study genomic change in A. suecica, we used long-read sequencing to generate a chromosome-level genome sequence, complemented by a partial assembly of a tetraploid A. arenosa, and by short-read genome and transcriptome sequencing data from populations of all three species—including ‘synthetic’ A. suecica. Our main goal was to look for evidence of genome changes (particularly the kind of dramatic changes discussed above) in A. suecica relative to the ancestral species.
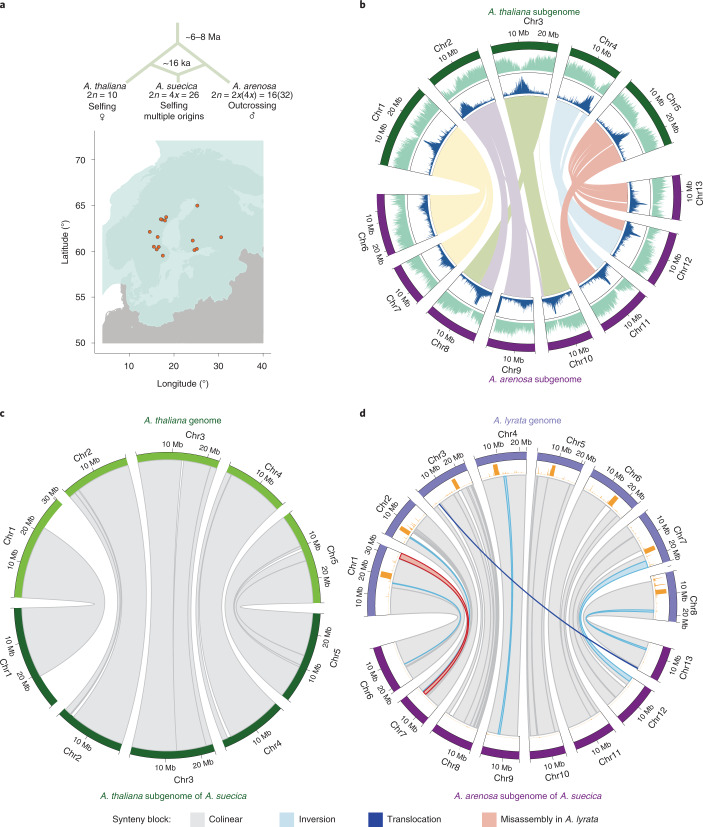
Fig. 1. The genome of A. suecica is largely colinear with the ancestral genomes.
a, Schematic depicting the origin of A. suecica and its current distribution in relation to the ice cover at the Last Glacial Maximum. ka, thousand years ago. Ice cover data are from Natural Resource Canada (https://open.canada.ca/data/en/dataset/a384bada-a787-5b49-9799-f5d589e97bd3). b, Chromosome-level assembly of the A. suecica genome with inner links depicting syntenic blocks between the A. thaliana and A. arenosa subgenomes of A. suecica. Histograms show the distribution of TEs (in blue) and protein-coding genes (in green) along the chromosomes. c, Synteny of the A. thaliana subgenome of A. suecica to the A. thaliana TAIR10 reference. In total 13 colinear synteny blocks were found. d, Synteny of the A. arenosa subgenome to A. lyrata. In total 40 synteny blocks were found, 33 of which were colinear. Of the remaining seven blocks, five represent inversions in the A. arenosa subgenome of A. suecica relative to A. lyrata, one is a translocation and one corresponds to a previously reported misassembly in the A. lyrata genome77. Orange bars show the density of missing regions (‘N’ bases) in the A. lyrata genome.
Results and analysis
The genome is conserved
We assembled a reference genome from a naturally inbred20,72 A. suecica accession (ASS3), using 50× long-read PacBio sequencing (PacBio RSII). The absence of heterozygosity and the substantial (around 11.6%) divergence between the subgenomes facilitated the assembly. By contrast, assembling A. arenosa is complicated by high heterozygosity (around 3.5% nucleotide diversity76) and high repeat content. Our assembly of a tetraploid A. arenosa individual, included in this study, is fragmented: 3,629 contigs; N50 of 331 kb. The A. suecica assembly, however, has an N50 contig size of 9.02 Mb. Contigs totalled 276 Mb (around 90% and around 88% of genome size estimated by flow cytometry and k-mer analysis, respectively; see Extended Data Fig. 1 and Methods). Contigs were placed into scaffolds using chromatin conformation capture (Hi-C) data and using the reference genomes of A. thaliana and Arabidopsis lyrata (as substitute for A. arenosa) as guides. This resulted in 13 chromosome-scale scaffolds (Extended Data Fig. 2). The placement and orientation of each contig within a scaffold was confirmed and corrected using a genetic map for A. suecica (see Methods and Extended Data Fig. 2). The final assembly (Fig. 1b) contains 262 Mb and has an N50 scaffold size of 19.59 Mb. The 5 + 8 chromosomes of the A. thaliana and A. arenosa subgenomes sum to 119 Mb and 143 Mb, respectively.
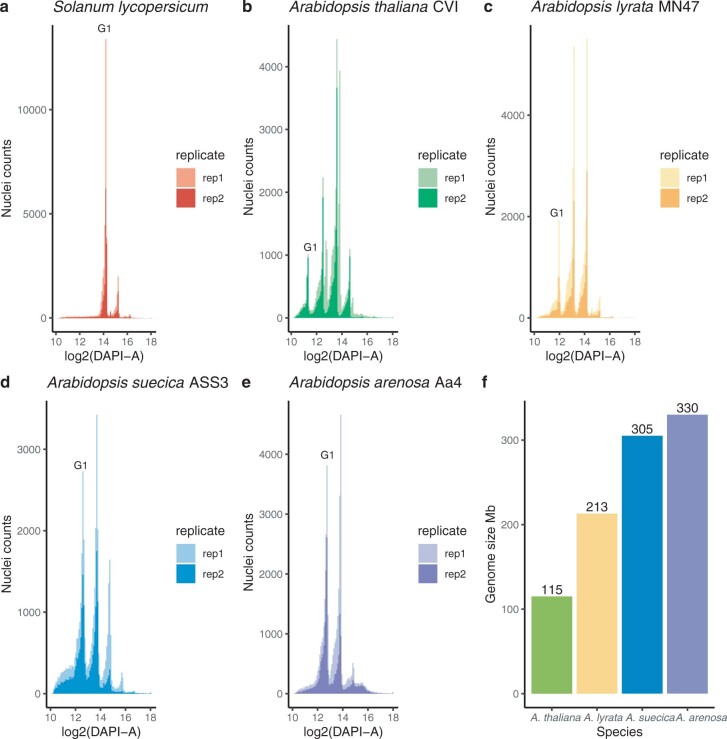
Extended Data Fig. 1. Measuring genome sizes of Arabidopsis species using flow cytometry.
a, FACS sorting of Solanum lycopersicum cells from 3-week-old leaf tissue for two replicates. G1 represents the peak denoting the G1 phase of the cell cycle. Cells in the G1 phase have 2 C DNA content (that is a 2 N genome). b, A. thaliana ‘CVI’ accession c, A. lyrata ‘MN47’ (the reference accession) d, A. suecica ‘ASS3’ (the reference accession) e autopolyploid A. arenosa accession ‘Aa4’ f, Bar chart shows calculated genome sizes (rounded to the nearest whole number) for each species using Solanum lycopersicum as the standard.
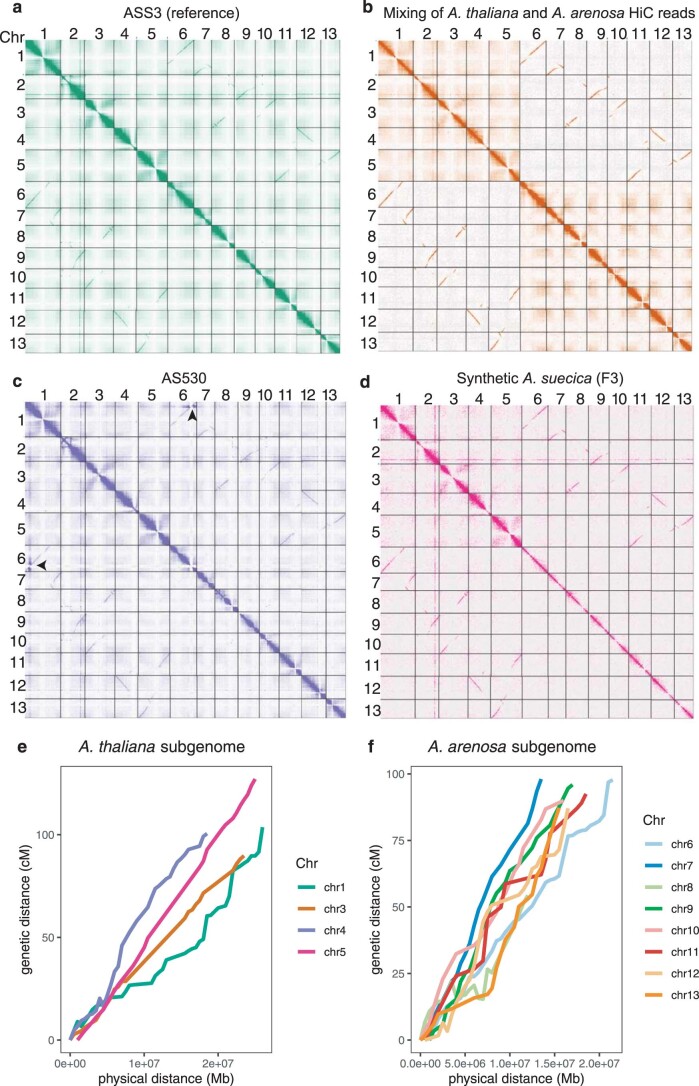
Extended Data Fig. 2. Hi-C and a genetic map analysis for the A. suecica genome.
a, Hi-C contact map for the genome of A. suecica. b, Mixing of A. thaliana and A. arenosa Hi-C reads suggest interchromosomal contacts between homeologous chromosomes is a result of mis-mapping for Hi-C reads. c, Accession ‘AS530’ with the region of HE highlighted with an arrow (Fig. 6), no other rearrangements were observed. d, Hi-C of synthetic A. suecica (third selfed generation). e and f physical distance (Mb) vs genetic distance (cM) is plotted for the A. thaliana and A. arenosa subgenome, respectively. Chromosome 2 is not plotted as there are too few SNPs on this chromosome in our cross, due to the recent bottleneck in A. suecica.
Of the A. thaliana and A. arenosa subgenomes of A. suecica, 108 Mb and 135 Mb are in large blocks syntenic to the genomes of the ancestral species: 13 and 40 blocks, respectively (Fig. 1c,d). The majority of these syntenic blocks are colinear, with the exception of five small-scale inversions (around 4.5 Mb) and one translocation (around 244 kb) on the A. arenosa subgenome—which may reflect differences between A. lyrata and A. arenosa, two highly polymorphic species separated by about a million years73,76. We also corrected the described77 misassembly in the A. lyrata reference genome using our genetic map. Overall we find that approximately 93% of the A. suecica genome is syntenic to the ancestral genomes (Fig. 1c,d). This highlights the conservation of the A. suecica genome and contrasts with the major rearrangements that have been observed in several resynthesized polyploids29,32,34,36 and some crops48,50,78. Notably, major rearrangements have also been observed in synthetic A. suecica79.
A total of 45,585 protein-coding genes were annotated for the A. suecica reference, of which 22,232 and 23,353 are located on the A. thaliana and A. arenosa subgenomes, respectively. We assessed completeness of the genome assembly and annotation with the BUSCO set for eudicots and found 2,088 (98.4%) complete genes for both the A. thaliana and A. arenosa subgenomes. Of the protein-coding genes, 18,023 had a one-to-one orthology between the subgenomes of A. suecica and 16,999 genes were conserved single-copy orthologues for each (sub-)genome of A. suecica and the ancestral species (Supplementary Data 2 and Extended Data Fig. 3). We annotated lineage-specific genes in A. suecica (that is, genes in A. suecica with no orthologue) using InterPro. We found significant enrichment in the A. thaliana subgenome for two gene ontology (GO) terms (GO:0008234 and GO:0015074) that are associated with repeat content (Supplementary Data 2). Ancestral genes missing in our annotation were overrepresented for functional categories of defence response. Examining DNA-sequencing coverage for these genes in the ancestral genomes did not confirm any gene loss, suggesting rather misassembly or misannotation, likely because of the repetitive and highly polymorphic nature of resistance genes (R-genes).
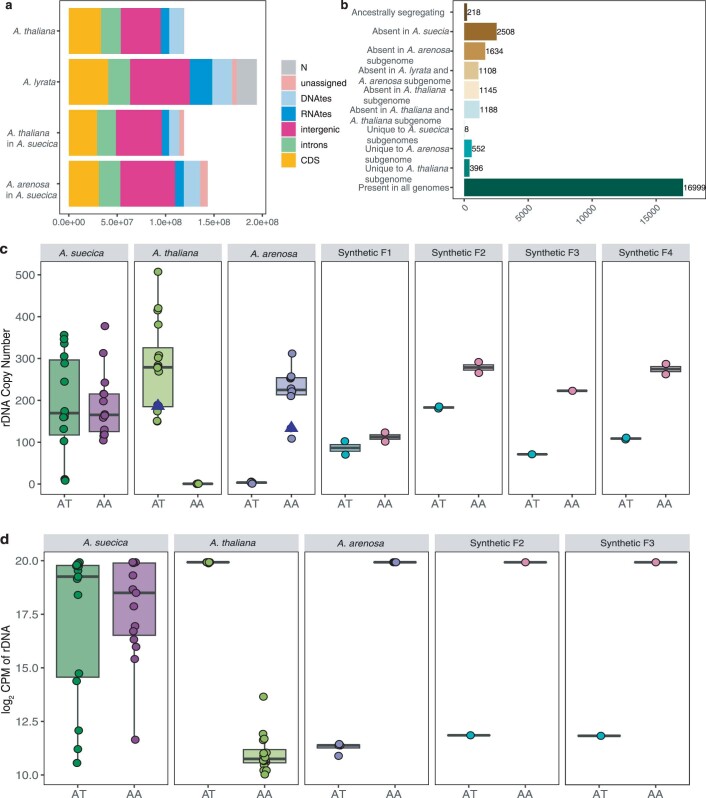
Extended Data Fig. 3. Genome composition and analysis of orthologues and the rDNA.
a, Genome composition of the A. suecica subgenomes and the ancestral genomes of A. thaliana and A. lyrata (here a substitute reference for A. arenosa because it is annotated). b, Counts of orthologous genes between the subgenomes of the reference A. suecica genome and the reference A. thaliana and A. lyrata genome. c, Copy number of A. thaliana and A. arenosa rDNA in natural A. suecica, ancestral species and synthetic lines. Blue triangles represent the A. thaliana and A. arenosa parent lines of the synthetic A. suecica cross. AT represents results when mapping to the A. thaliana consensus sequence and AA to the A. arenosa consensus sequences for the 45S rRNA b, Expression (log2(CPM)) of A. thaliana and A. arenosa rDNA in natural A. suecica, ancestral species and synthetic lines. Accessions with log2(CPM) of > =15 was taken as evidence for expression for the A. thaliana and A. arenosa 45S rRNA in A. suecica, as this CPM value was above the maximum level of mis-mapping observed in the ancestral species (A. thaliana mapping to the A. arenosa 45S rRNA).
The ribosomal DNA clusters are highly variable
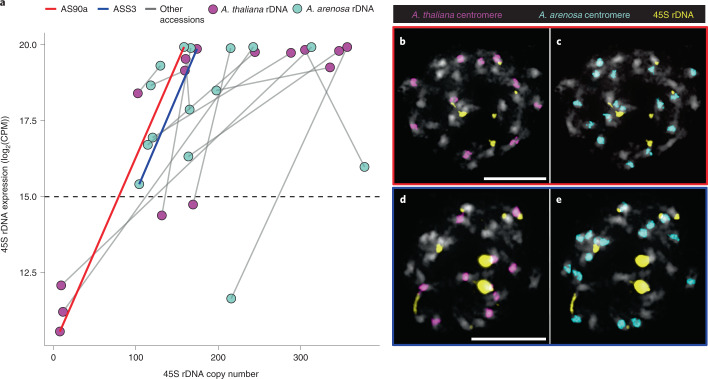
In eukaryotic genomes, genes encoding ribosomal RNA (rRNA) occur as tandem arrays in rDNA clusters. The 45S rDNA clusters are massive, containing hundreds or thousands of copies and spanning millions of base pairs80. The site of pre-ribosome assembly (nucleolus), forms at these clusters if they are actively transcribed. In inter-specific hybrids it was previously observed that the rDNA of only one parent tended to be involved in nucleolus formation, a phenomenon known as ‘nucleolar dominance’81–84. In A. suecica, it was observed that the rDNA clusters inherited from A. thaliana were silenced81–87, and structural changes associated with these clusters were also suggested88.
Given this, we examined the composition and transcription of 45S rDNA repeats. Although the large and highly repetitive 45S rDNA clusters are missing from the genome assembly, we can measure the copy number of A. thaliana and A. arenosa 45S rRNA genes using sequencing coverage (see Methods). We find that three accessions have experienced massive loss of the A. thaliana rDNA loci (Fig. 2a), which was confirmed for one of the accessions (AS90a) by fluorescence in situ hybridization (FISH) analysis (Fig. 2b,c). However, there is massive copy number variation for 45S rRNA genes in A. suecica (Fig. 2a), and some accessions (ASS3) have a higher 45S rRNA copy number in A. thaliana than in A. arenosa (Fig. 2d,e).
Fig. 2. Expression and copy number variation of 45S rDNA in A. suecica.
a, The relationship between expression levels (log2(CPM)) and copy number of 45S rDNA shows extensive variation of 45S rDNA copy number and varying direction of ‘nucleolar dominance’. Grey lines connect subgenomes of the same accession. Values above the dashed line are taken as evidence for expression of a particular 45S rDNA allele, as this is above the maximum level of mis-mapping seen in the ancestral species (see Extended Data Fig. 3). b,c, FISH results of a natural A. suecica accession AS90a that has largely lost the rDNA cluster of the A.thaliana subgenome (8 copies calculated for the A. thaliana 45S rDNA and 159 copies of the A. arenosa 45S rDNA). d,e, FISH results of a natural accession ASS3 that has maintained both ancestral rDNA loci (174 copies calculated for the A. thaliana 45S rDNA and 104 copies of the A. arenosa 45S rDNA). Scale bars, 10 μm (b, d).
We find nucleolar dominance to be variable in A. suecica (see Methods and Extended Data Fig. 3). The majority of accessions express both 45S rRNA alleles, five exclusively express A. arenosa 45S rRNA and one exclusively expresses A. thaliana 45S rRNA (Fig. 2a).
This extensive variation in 45S cluster size and expression is consistent with the intraspecific variation seen in A. thaliana89,90, and previous observations made in natural A. suecica91. This suggests that nucleolar dominance may partly be explained by retained ancestral variation. However, the large decrease in rDNA cluster size observed in some accessions may be a consequence of allopolyploidization, as synthetic A. suecica sometimes shows loss of 45S rDNA (even as early as the F1 stage) that varies between siblings and generations (Extended Data Fig. 3). Elimination of rDNA loci has also been previously observed in synthetic wheat92, and loss of rDNA sites has been reported in strawberry93.
No evidence for abnormal transposon activity
The possibility that allopolyploidization leads to a ‘genome shock’ in the form of increased transposon activity has been discussed27,28,94,95. Evidence for TE proliferation following hybridization has been found for Ty3/Gypsy retrotransposons in hybrid sunflower species96, although this may be due to environmental change97,98. Analysis of TE expression in F1 hybrids between A. thaliana and A. lyrata found strong correlation to the parent species, and little alteration of repressive chromatin marks99—although the F1 generation may be too early to study TE misregulation. Here we examine TE dynamics in A. suecica.
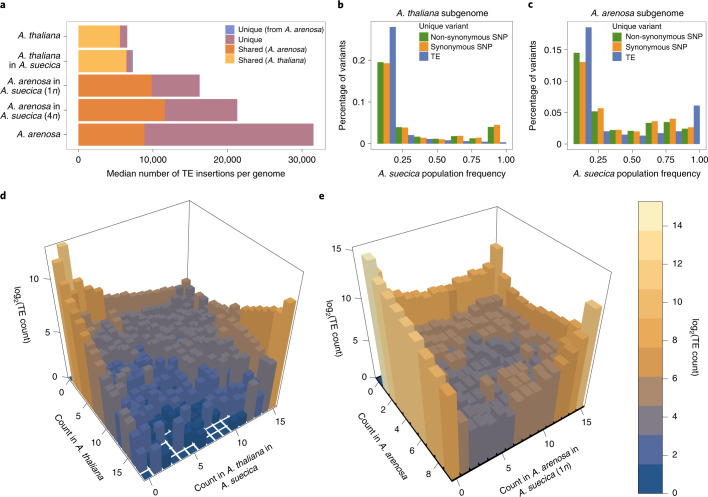
In A. suecica there are almost twice as many annotated transposons in the A. arenosa compared to the A. thaliana subgenome (66,722 versus 33,420). The difference is likely to be greater given that the A. arenosa subgenome assembly is less complete and TE annotation is biased towards A. thaliana. Whether the combination of these two genomes has led to increased transposon activity is unknown.
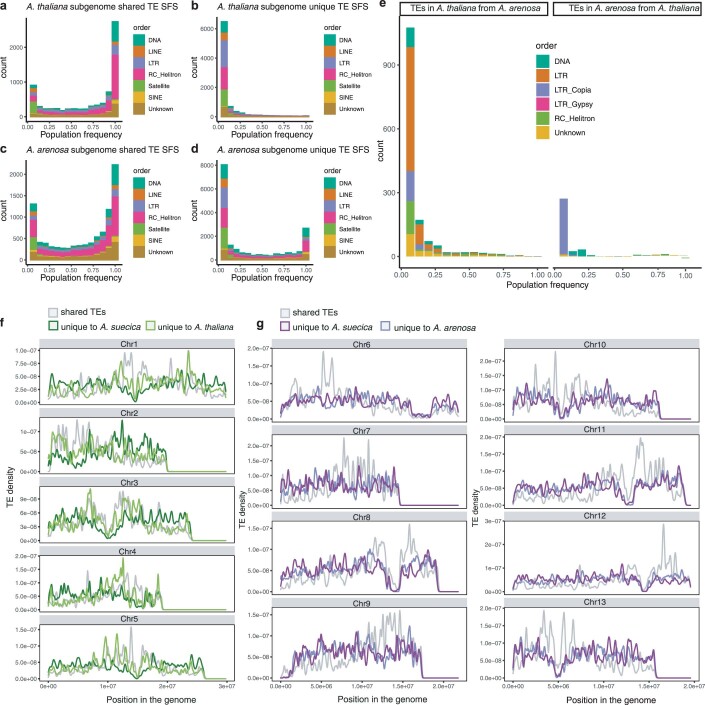
The A. thaliana subgenome contains around 3,000 more annotated transposons than the TAIR10 A. thaliana reference genome but could reflect a greater number of transposons in the A. thaliana ancestors rather than increased transposon activity in A. suecica. To investigate TE activity, unique TE jumps that occurred after the species separated are needed. We used the software PoPoolationTE2100 to call presence–absence variation on a population level using 15 natural A. suecica accessions, 18 A. thaliana accessions genetically close to A. suecica, and 9 A. arenosa lines. Of the 24,569 insertion polymorphisms in the A. thaliana subgenome, 8,767 were shared between A. thaliana and A. suecica, 7,196 were unique to A. thaliana and 8,606 were unique to A. suecica. Of the 115,336 insertion polymorphisms in the A. arenosa subgenome, 13,177 were shared with A. arenosa, 83,964 were unique to A. arenosa and 18,195 were unique to A. suecica (Supplementary Data 1a,b and Extended Data Fig. 4). Considering the number of transposons per individual genome (Fig. 3a), most transposon insertions in a typical A. thaliana subgenome are also found in A. thaliana. The slightly higher transposon load in the A. thaliana subgenome is probably due to the population bottleneck. Notably, the number of unique insertions is not higher in the A. thaliana subgenome, suggesting no increase in transposon activity.
Extended Data Fig. 4. Population frequency and genomic location of transposon polymorphisms.
Shared TE SFS for the a, A. thaliana and b, A. arenosa subgenome. Private TE SFS for the c, A. thaliana and d, A. arenosa subgenome. e, TEs ancestrally from A. arenosa that are present in the A. thaliana subgenome of A. suecica and TEs ancestrally from A. thaliana that are present in the A. arenosa subgenome of A. suecica. f, Shared TEs in the population between A. thaliana and the A. thaliana subgenome of A. suecica. Shared TEs are likely older than private TEs and are enriched around the pericentromeric regions in the A. thaliana subgenome. Private TEs are enriched in the chromosomal arms for both species, where protein-coding gene density is higher (Fig. 1b). g as in f but examining TEs in the population of A. arenosa and the A. arenosa part of A. suecica. Note the region between 5 and 10 on chromosome 2 was not included in the analysis as this region shows synteny with an unplaced contig.
Fig. 3. TE dynamics in A. suecica reveal no evidence for abnormal transposon activity.
a, Median TE insertions per genome. As the A. arenosa population is an autotetraploid outcrosser, four randomly chosen haploid A. arenosa subgenomes of A. suecica were combined to make a 4n A. suecica. A. suecica does not show an increase in private TE insertions compared with the ancestral species for either subgenome, and shared TEs constitute a higher fraction of TEs in A. suecica, reflecting the strong population bottleneck at its origin. b,c, Site-frequency spectra of non-synonymous SNPs, synonymous SNPs and TEs in the A. thaliana (b) and A. arenosa (c) subgenomes of A. suecica suggest that TEs are under purifying selection on both subgenomes. d, Three-dimensional histogram of a joint TE frequency spectrum for A. thaliana on the x axis and the A. thaliana subgenome of A. suecica on the y axis. e, Three-dimensional histogram of a joint TE frequency spectrum for A. arenosa on the x axis and the A. arenosa subgenome of A. suecica on the y axis. d and e show stable dynamics of private TEs in A. suecica and a bottleneck effect on the ancestral TEs (shared) at the origin of the A. suecica species.
Turning to the A. arenosa subgenome, we see that A. suecica contains only about half the number of transposons of A. arenosa on average (Fig. 3a). However, A. arenosa is an outcrossing tetraploid, therefore four randomly chosen A. arenosa subgenomes of A. suecica were used in the comparison (‘A. arenosa in A. suecica (4n)’ in Fig. 3a). This largely accounts for the observed difference, but there are still fewer transposons in A. suecica. A population bottleneck is likely to explain the difference, although decreased transposon activity in A. suecica, related to its transition to selfing101, cannot be ruled out.
In summary, we see no evidence for a burst of transposon activity in A. suecica, a conclusion supported by the analysis of transposon expression for A. suecica, which shows no upregulation relative to the ancestor species (Extended Data Fig. 5). The frequency distribution of polymorphic transposon insertions unique to A. suecica is heavily skewed towards zero, likely because of purifying selection as the distribution is more similar to that of non-synonymous than synonymous single-nucleotide polymorphisms (SNPs) (Fig. 3b,c). However, for both subgenomes, A. suecica also contains a large number of fixed or nearly fixed insertions that are present in the ancestral species at a lower frequency (Fig. 3d,e). These are likely to have reached high frequency as a result of a bottleneck. Shared transposons are enriched in the pericentromeric regions, while unique transposon insertions, generally at low frequency, are more uniformly distributed across the genome, which is consistent with strong selection against transposon insertions in the gene-dense chromosome arms102,103 (Extended Data Fig. 4).
Extended Data Fig. 5. Transposable element expression analysis.
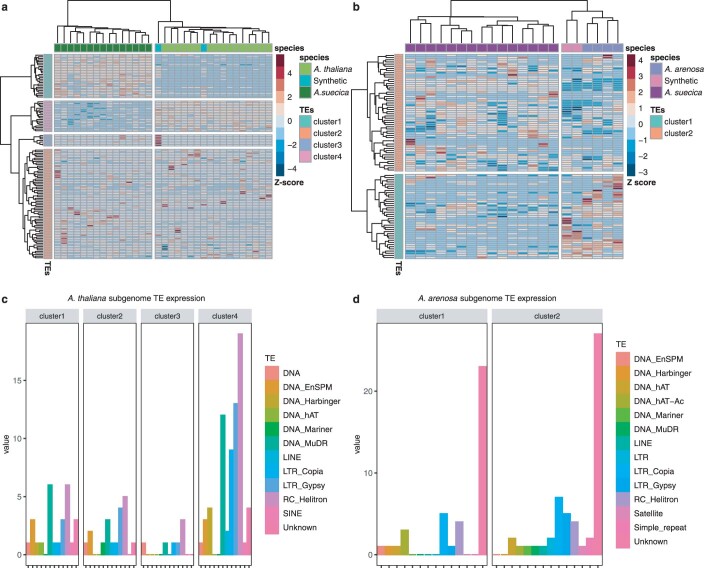
Patterns of TE expression in natural and synthetic A. suecica show that allopolyploidy is not accompanied by an overall upregulation in TE expression as predicted by the ‘genome shock’ hypothesis. a, Heat map of TE expression for the A. thaliana subgenome of A. suecica (dark green) synthetic A. suecica (cyan) and A. thaliana (light green). b, Heat map of TE expression for the A. arenosa subgenome of A. suecica (dark purple) synthetic A. suecica (pink) and A. arenosa (light purple). c and d the breakdown of TE families expressed in each cluster, with helitrons being the most abundant class on the A. thaliana subgenome and TEs of an unknown family being the most abundant in the A. arenosa subgenome.
An interesting subset of recent transposon insertions unique to A. suecica are those that have jumped between the subgenomes. We searched for full-length transposon copies that are present in both subgenomes of A. suecica and assigned the transposon sequences to the A. thaliana or the A. arenosa ancestral genome (see Methods). We were able to assign 15 and 56 transposon sequences as belonging to the A. thaliana and A. arenosa ancestral genome, respectively. Using these sequences, we searched our transposon polymorphisms and identified 1,515 A. arenosa transposon polymorphisms on the A. thaliana subgenome, and 496 A. thaliana transposon polymorphisms on the A. arenosa subgenome. Like other private polymorphisms, these are skewed towards rare frequencies, and are uniformly distributed across the (sub-)genome. Most of the transposons that have jumped into the A. thaliana subgenome are helitron and long terminal repeat (LTR) elements (Extended Data Fig. 4). LTR elements also make up most of the A. thaliana transposons segregating in the A. arenosa subgenome. Three times as many transposon jumps from A. arenosa to A. thaliana than vice versa is notable, and suggests higher transposon activity in the A. arenosa subgenome, but we must consider differences in genome size and transposon number. If no differences in activity exist, we would expect the number of jumps to be proportional to the number of potential source elements and the size of the target genome. As the A. arenosa subgenome contains roughly twice as many transposons as the A. thaliana subgenome and is about 20% larger, we expect a 1.7-fold difference, not a three-fold one.
In conclusion, transposon activity in A. suecica appears to be governed by the same processes as in the ancestral species.
No global dominance in expression between the subgenomes
Over time the traces of polyploidy are erased through an evolutionary process referred to as fractionation or re-diploidization104–108. Analyses of retained homeologues in ancient allopolyploids such as A. thaliana109, maize55, B. rapa54 and Gossypium raimondii110 have revealed that one ‘dominant’ subgenome remains more intact, with more highly expressed homeologues compared to the ‘submissive’ genome(s)109. This pattern of biased fractionation has not been observed in ancient autopolyploids111,112, and is believed to be allopolyploid-specific.
Studying genome expression dominance in allopolyploids is useful for understanding or predicting which of the subgenomes will likely be refractory to, and which will likely experience this fractionation process more, over time55. Subgenome dominance in expression has been reported for a number of recent allopolyploids such as strawberry6, peanut8, Spartina68, T. miscellus113, monkeyflower17 and synthetic B. napus114. However, some allopolyploids display even subgenome expression: Capsella bursa-pastoris10,12, Trifolium repens13, Arabidopsis kamachatica70 and Brachypodium hybridum14.
Subgenome dominance is linked to differences in transposon content6 and/or large genetic differences between subgenomes115. This makes A. suecica, with 6 Ma divergence between the gene-dense A. thaliana and the transposon-rich A. arenosa, a promising candidate to study this phenomenon. Previous reports on subgenome dominance in A. suecica are conflicting, suggesting a bias to either the A. thaliana116 or the A. arenosa117 subgenome.
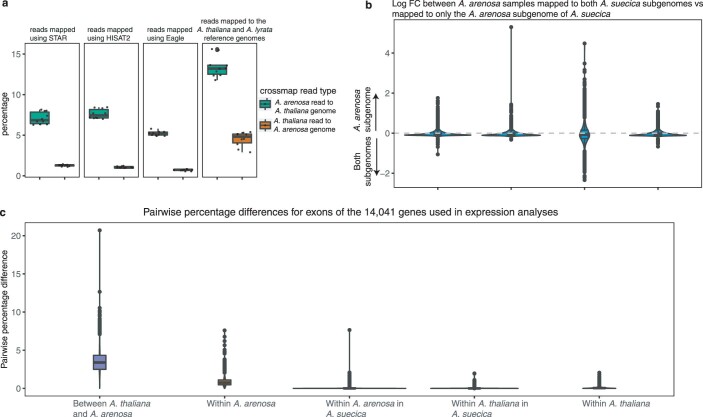
To investigate the evolution of gene expression in A. suecica, we generated RNA sequencing (RNA-seq) data for 15 natural A. suecica accessions, 15 closely related A. thaliana accessions, 4 A. arenosa individuals, a synthetically generated A. suecica from a lab cross (the second and third hybrid generations) and the parental lines of this cross. Each sample had 2–3 biological replicates (Supplementary Data 2). On average, we obtained 10.6 million raw reads per replicate, of which 7.6 million reads were uniquely mapped to the A. suecica reference genome and 14,041 homeologous gene pairs (see Methods). On average, around 1% of A. thaliana and around 6% of A. arenosa RNA reads cross-mapped between the subgenomes of A. suecica. The approximately 6% of cross-mapping in A. arenosa is likely to be because of the high level of polymorphism in this outcrossing species. However, diversity within A. suecica is massively lower, meaning that transcripts from the A. arenosa subgenome will probably map correctly (see Methods and Extended Data Fig. 6).
Extended Data Fig. 6. Cross-mapping of RNA-seq short reads.
a, Box plots of cross-mapping RNA short reads. This was examined by mixing reads in-silico between A. thaliana and A. arenosa. On average ~6% of A. arenosa reads map to A. thaliana subgenome instead of the A. arenosa subgenome, and ~1% vice versa. Mapping these reads to the combined reference genomes of A. thaliana and A. lyrata (box plot 4 in a) shows that reads map more precisely to the A. suecica reference and that cross-mapping is not due to unreported HE. b, LogFC of log2(CPM) read counts for A. arenosa (CPM of A. arenosa subgenome genes when reads are mapped only to A. arenosa subgenome of A. suecica/CPM of A. arenosa subgenome genes when reads are mapped to the full genome) show only a small effect of mapping strategy to estimate gene expression on the A. arenosa subgenome. c, Pairwise percentage differences (π) for each group measured for the exons of the 14,041 genes in the expression analysis. High levels of π in A. arenosa overlaps with the distribution of π between A. thaliana and A. arenosa. This explains why there is more cross-mapping for A. arenosa than for A. thaliana in a Importantly, lower π within A. suecica for both subgenomes means that measurements for subgenome dominance are not biased by cross-mapping, as we expect less cross-mapping since the distribution of π overlaps less with π between A. thaliana and A. arenosa.
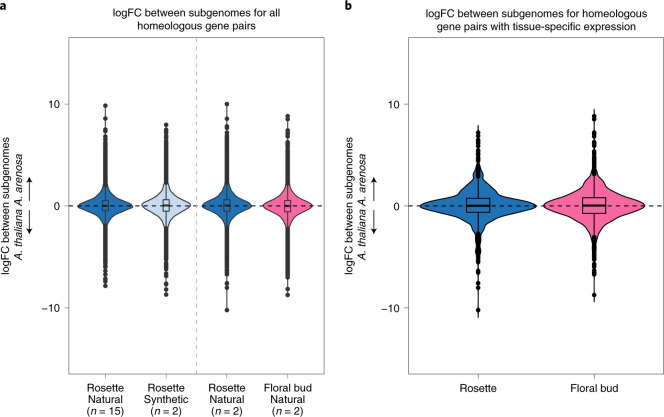
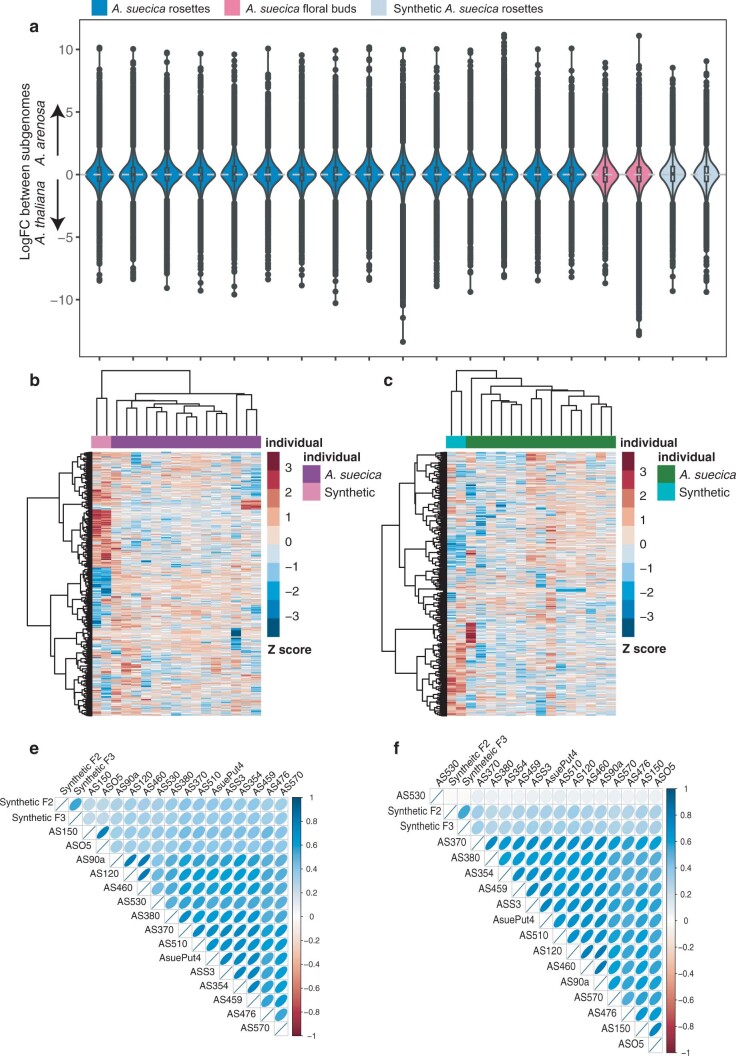
Examining expression differences between homeologous gene pairs, we found no general bias towards a subgenome of A. suecica (that is, the mean log fold change is 0), for any sample or tissue, including synthetic A. suecica (Fig. 4a and Extended Data Fig. 7). This suggests that the expression differences between the subgenomes have not changed systematically through polyploidization, and is in contrast to previous studies, which reported a bias towards the A. thaliana116 or the A. arenosa117 subgenome, likely because RNA-seq reads were not mapped to an appropriate reference genome.
Fig. 4. Patterns of gene expression between the subgenomes of A. suecica in rosettes and floral buds.
a, Violin plots of the mean log fold change (logFC) between the subgenomes for the 15 natural A. suecica accessions and 2 synthetic lines for whole rosettes. The centre line is the 50th percentile or median. The box limits represent the interquartile range. The whiskers represent the largest and smallest value within 1.5 times the interquartile range above and below the 75th and 25th percentile, respectively. Mean log fold change for the two accessions (ASS3 and AS530) for which transcriptome data for both whole rosettes and flower buds were available. All the distributions are centred around zero, suggesting even subgenome expression. b, Violin plots for the mean log fold change between the subgenomes for gene pairs with tissue-specific expression in at least one member of the pair.
Extended Data Fig. 7. Expression differences between subgenomes in natural and synthetic A. suecica.
The distribution of expression differences across homeologous gene pairs in natural and synthetic A. suecica. b, A heat map of expression for genes in the top 5% biased toward the A. arenosa subgenome. The gene must be in the 5% quantile for at least 1 accession. c, The same as in b but for the A. thaliana subgenome. Correlations of log fold change for genes in the tails of the distribution (top 5% quantile) for the A. arenosa subgenome d and the A. thaliana subgenome e.
The set of genes that show large expression differences between the subgenomes are not biased towards any particular GO category, and are not consistent between accessions and individuals (Fig. 4b and Extended Data Fig. 7). This suggests that many large subgenome expression differences are due to genetic polymorphisms within A. suecica rather than fixed differences between the ancestral species.
Levels of expression dominance were reported to vary across tissues in natural C. bursa-pastoris11 and resynthesized cotton118. To test whether expression dominance can vary for tissue-specific genes, we examined homeologous gene pairs in which at least one gene in the pair showed tissue-specific expression, in whole rosettes and floral buds. We do not find evidence for dominance between subgenomes in tissue-specific expression (Fig. 4b). A total of 897 genes with significant expression in whole rosettes for both homeologues showed GO overrepresentation that included both photosynthesis- and chloroplast-related functions (Supplementary Table 1). This result suggests that the A. arenosa subgenome has established important cyto-nuclear communication with the chloroplast inherited from A. thaliana, rather than being silenced. A total of 2,176 gene pairs with floral-bud-specific expression for both homeologues were overrepresented for GO terms related to responses to auxin and jasmonic acid that may reflect early developmental changes in this young tissue (Supplementary Table 1). Although flowers of selfing A. thaliana and A. suecica are scentless and are much smaller than those of the outcrosser A. arenosa72, this result suggests that the ‘selfing syndrome’119 has not hugely affected the transcriptome of floral buds in A. suecica at this stage of development.
In summary, we find no evidence that one subgenome is dominant and contributes more to the functioning of A. suecica. On the contrary, homeologous gene pairs are strongly correlated in expression across tissues.
Evolving gene expression in A. suecica
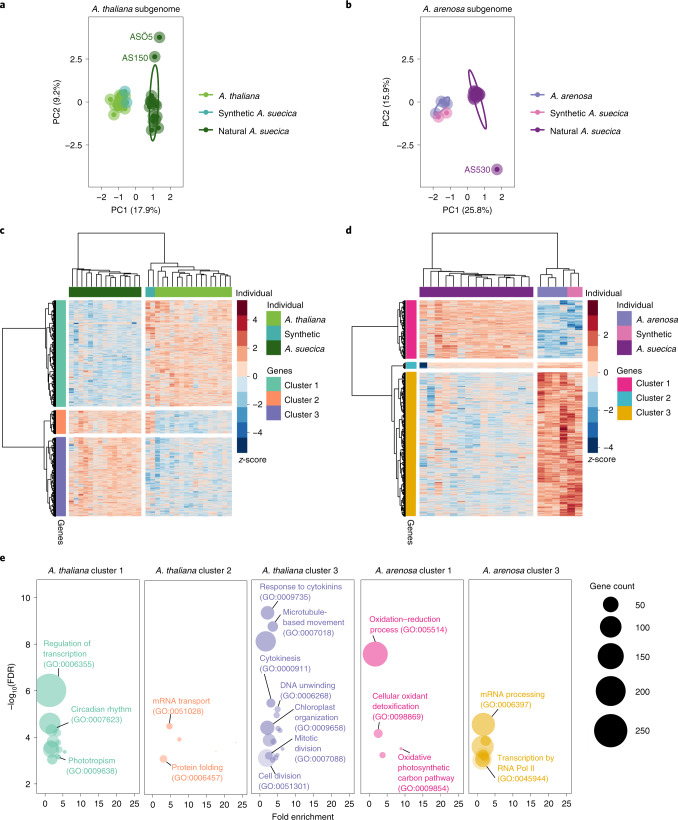
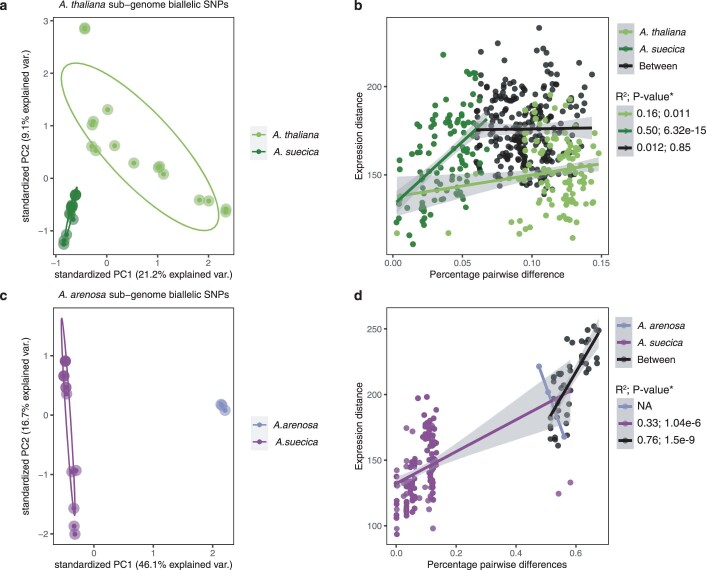
The previous section focused on differences in expression between the subgenomes within the same individual. This section will focus on differences between individuals. To provide an overview of expression differences between individuals we performed a principal component analysis (PCA) on gene expression separately for each (sub-)genome. For both subgenomes, the first principal component separates A. suecica from the ancestral species and the synthetic hybrid (Fig. 5a,b and Extended Data Fig. 8), suggesting that hybridization does not automatically result in large-scale transcriptional changes, and that gene expression changes in natural A. suecica have evolved over time. Given the limited time involved, and the fact that the genes that have changed expression are not random with respect to function (Fig. 5c), we suggest that the first principal component captures trans-regulated expression changes in A. suecica that are adaptive.
Fig. 5. Differential gene expression analysis in A. suecica.
Patterns of differential gene expression in A. suecica support adaptation to the whole-genome duplication for the A. thaliana subgenome and adaptation to the new plastid environment for the A. arenosa subgenome. a, PCA for A. thaliana and the A. thaliana subgenome of natural and synthetic A. suecica lines. Principal component 1 (PC1) separates natural A. suecica from the ancestral species and the synthetic lines. b, PCA for A. arenosa and the A. arenosa subgenome of natural and synthetic A. suecica lines. PC1 separates natural A. suecica from the ancestral species and the synthetic lines, whereas PC2 identifies outlier accessions discussed further below (see Fig. 6). c,d, Heat map of DEGs for the A. thaliana (c) and the A. arenosa (d) subgenome of A. suecica. Positive numbers (red colour) indicate higher expression. Genes and individuals have been clustered on the basis of similarity in expression, resulting in the clusters that are discussed in the text. e, GO enrichment for each cluster in c and d. Categories discussed in the text are highlighted. RNA Pol II, RNA polymerase II.
Extended Data Fig. 8. Comparison of genetic and expression distance.
a, PCA plot of biallelic SNPs in the population of A. thaliana and A. suecica for the A. thaliana subgenome of A. suecica (N=345,075 biallelic SNPs), of the analysed 13,647 genes in gene expression in addition to 500 bp up and downstream of each gene sequence b, Correlation of 𝜋 (pairwise genetic differences) and expression distance (that is, euclidean distance) for 14,041 genes (*=Bootstrapped 1000 times). c, PCA plot of biallelic SNPs in the population of A. arenosa (N.B. we had DNA sequencing for only 3 of the 4 accessions used in the expression analysis) and A. suecica for the A. arenosa subgenome of A. suecica (N= 1,761,708 biallelic SNPs), of the analysed 14,041 genes in gene expression in addition to 500 bp up and downstream of each gene sequence d, Correlation of Pi (pairwise genetic differences for mapped genomic regions) and expression distance (that is, euclidean distance) for 14,041 genes (*=Bootstrapped 1000 times). A. arenosa was too few samples to give reliable correlations and therefore is NA. Grey bars represent the 95 confidence intervals.
To further characterize expression changes in natural A. suecica, we analysed differentially expressed genes (DEGs) on each subgenome compared to the corresponding ancestral species. The total number of DEGs was 4,186 and 4,571 for the A. thaliana and A. arenosa subgenome, respectively (see Methods and Supplementary Data 2). These genes were clustered on the basis of the pattern of change across individuals (Fig. 5c,d) and GO enrichment analysis was performed for each cluster (Fig. 5e and Supplementary Table 2).
For the A. thaliana subgenome, we identified three clusters. Cluster 1 comprised 2,135 genes that showed decreased expression in A. suecica compared to A. thaliana. These genes are enriched for transcriptional regulation, which may be expected as we are examining DEGs between the species. Enrichments for circadian rhythm function and phototropism may be related to the ecology of A. suecica and its post-glacial migration to Fennoscandinavia (Fig. 1a).
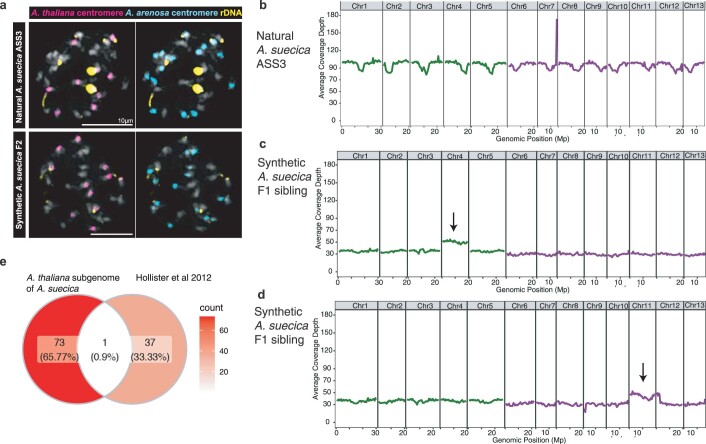
Cluster 2 consisted of 468 genes that are upregulated in both natural and synthetic A. suecica relative to A. thaliana. These expression changes most likely are an immediate consequence of trans-regulation in hybrids. Genes in this cluster are enriched for mRNA transport and protein folding. Adjustment of protein homeostasis has been reported previously in experimentally evolved stable polyploid yeast120. Notably, the synthetic lines used in the expression analysis did not show signs of aneuploidy (Extended Data Fig. 9).
Extended Data Fig. 9. Aneuploidy is frequently observed in synthetic A. suecica.
a, Comparison of FISH analyses of the reference natural A. suecica ‘ASS3’ and synthetic A. suecica. Synthetic A. suecica shows aneuploidy in both subgenomes in the F2 generation (gain of one chromosome on the A. thaliana subgenome (N=11) and loss of one chromosome on the A. arenosa subgenome (N=15)). Natural A. suecica shows a stable karyotype b, DNA-sequencing coverage in the reference natural A. suecica accession ‘ASS3’ c and d, DNA-sequencing coverage in siblings of F1 synthetic A. suecica show different cases of aneuploidy (indicated with arrow) in synthetic A. suecica, chromosome 4 in c and chromosome 11 in d e overlap of genes involved in cell division from Fig. 5e and genes previously shown to play a role in the adaptation to autopolyploidy in A. arenosa121. The little overlap in genes between A. suecica and A. arenosa highlights that successful meiosis in polyploids is likely a complex trait.
Cluster 3 consisted of 1,583 genes that are upregulated in A. suecica compared to A. thaliana, and several of the enriched GO categories, such as microtubule-based movement, cytokinesis, meiosis and cell division, suggest that the A. thaliana subgenome of A. suecica is adapting to polyploidy at a cellular level. Selection for this seems likely given that aneuploidy is frequent in synthetic A. suecica (Extended Data Fig. 9), while natural A. suecica has a stable conserved karyotype. Independent evidence for adaptation to polyploidy via modifications of the meiotic machinery in the other ancestor of A. suecica, A. arenosa, also exists23,121,122, although we see very little overlap in the genes involved (Extended Data Fig. 9). The nature of these changes in the A. thaliana subgenome of A. suecica will require further investigation, but we note the enrichment (see Methods and Supplementary Data 2) of MYB family transcription factor binding sites123 in cluster 3.
For the A. arenosa subgenome, we also found three clusters of DEGs (Fig. 5d) with GO enrichment for two clusters (Fig. 5e and Supplementary Table 2). Cluster 1 consisted of 1,278 genes that are upregulated in natural A. suecica compared to A. arenosa and synthetic A. suecica. We find enrichment for plastid-related functions that may be due to selection on the A. arenosa subgenome to restore communication with the maternally inherited plastids from A. thaliana. Twelve out of a total of 69 genes with structural evidence for direct plastid–nuclear interactions in A. thaliana overlap our genes in Cluster 1 using CyMIRA124 (P = 0.0072; one-sided Fisher’s exact test; Supplementary Data 2). Cluster 3 consists of 3,166 genes that are downregulated in A. suecica compared to A. arenosa and synthetic A. suecica. These genes were enriched for mRNA processing and epigenetic regulation of gene expression (Supplementary Table 2), and for positive regulation of transcription by RNA polymerase II, which suggests differences in the epigenetic regulation of expression between A. arenosa and A. suecica. Cluster 2 (127 genes), finally, did not have a GO overrepresentation and showed an intriguing pattern that will be discussed in the next section.
Homeologous exchange contributes to variation in gene expression
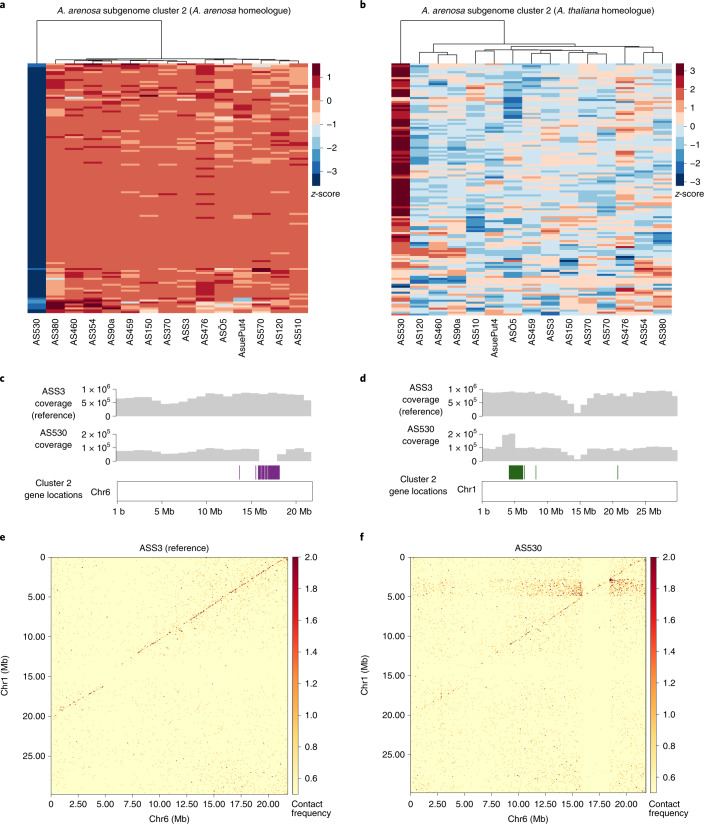
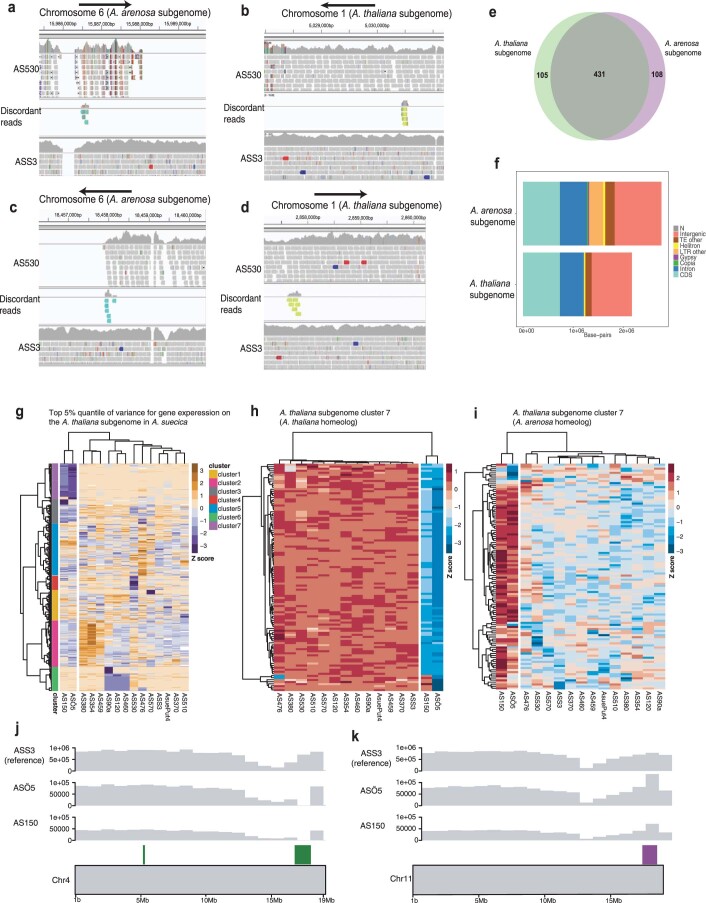
The second principal component for gene expression identified three outlier accessions of A. suecica: two for the A. thaliana subgenome (Fig. 5a) and one for the A. arenosa subgenome (Fig. 5b). While closely examining the latter accession, AS530, we realized that it is responsible for the cluster of genes with distinct expression patterns but no GO enrichment (Fig. 5d, Cluster 2). Genes from this cluster were significantly downregulated on the A. arenosa subgenome (Fig. 6a) and upregulated on the A. thaliana subgenome (Fig. 6b)—for AS530 only. The observation that 104 of the 127 genes (Extended Data Fig. 10) in the cluster are located in close proximity in the genome pointed to a structural rearrangement. The lack of DNA-sequencing coverage on the A. arenosa subgenome around these 104 genes, and the doubled coverage for their homeologues on the A. thaliana subgenome, suggested a homeologous exchange (HE) event resulting in AS530 carrying four copies of the A. thaliana subgenome and zero copies of the A. arenosa subgenome for this approximately 2.5 Mb region of the genome (Fig. 6c). This was further supported by Hi-C data, which showed clear evidence for interchromosomal contacts between A. thaliana subgenome chromosome 1 and A. arenosa subgenome chromosome 6 around the break points of the putative HE in AS530 (Fig. 6d,e), and by multiple discordant paired-end reads at the break points between the homeologous chromosomes, which independently support the HE event (Extended Data Fig. 10).
Fig. 6. Homeologous exchange contributes to expression variance within A. suecica.
a, Cluster 2 of Fig. 5d explains the outlier accession AS530, which is not expressing a cluster of genes on the A. arenosa subgenome. b, Homeologous genes of this cluster on the A. thaliana subgenome of A. suecica show the opposite pattern and are more highly expressed in AS530 compared to the rest of the population. c, Of the 122 genes from cluster 3, 97 are close to each other on the reference genome but appear to be deleted in AS530 on the basis of sequencing coverage. d, The A. thaliana subgenome homeologues have twice the DNA coverage, suggesting that they are duplicated. e,f, Hi-C data show (spurious) interchromosomal contacts at 25 kb resolution between chromosome 1 and chromosome 6 around the break point of the cluster of 97 genes in AS530 (f) but not in the reference accession ASS3 (e).
Extended Data Fig. 10. Evidence of HE in A. suecica.
Reads mapped to the beginning of the HE event in chromosome 6 (~ 15.9 Mb) in ‘AS530’. Arrows point to the direction of the break. Discordant reads map between the A. arenosa subgenome on chromosome 6 and the read pair maps to the homeologous chromosome 1 on the A. thaliana subgenome (~5 Mb) in b. The end of the HE event in chromosome 6 (~18.4 Mb). Discordant reads map between the A. arenosa subgenome in c and the read pair maps to chromosome 1 (~2.8 Mb) on the A. thaliana subgenome in d. e, Gene counts between the syntenic regions. 431 have a 1:1 relationship, 108 genes are specific to the A. arenosa subgenome and 105 genes are specific to the A. thaliana subgenome. f, Composition of the syntenic regions between the two subgenomes. g, The top 5% quantiles (N=702) for variation in gene expression for the A. thaliana subgenome shows in cluster 7 (N=111) the two outlier accessions (AS150 and ASÖ5) are expressing genes differently to the rest of the population. h, Homeologous genes of this cluster on the A. thaliana subgenome of A. suecica show that these genes are not expressed in these two accessions while i shows they are upregulated in ‘AS150’ and ‘ASÖ5’. j and k 101/111 genes in cluster 7 are located on chromosome 4 in close proximity to each other on the A. thaliana subgenome of the A. suecica reference genome and appear to be deleted in AS5Ö5 and AS150. The A. arenosa subgenome homeologues (located on chromosome 11) have twice the DNA coverage, suggesting they are duplicated, in agreement with expectations of HE event.
We examined the two outlier A. suecica accessions for the A. thaliana subgenome (Fig. 5a; AS150 and ASÖ5) and found that they probably share a single HE event in the opposite direction (four copies of the A. arenosa subgenome and no copies of the A. thaliana subgenome for a region of around 1.2 Mb in size; see Extended Data Fig. 10). This demonstrates that HE occurs in A. suecica and contributes to the intraspecific variation in gene expression (Fig. 5a,b). HE in allopolyploids is a main source of diversity, causing extensive phenotypic changes9,125. However, the majority of HEs are probably deleterious as they will lead to gene loss: although the A. thaliana and A. arenosa genomes are largely syntenic, AS530 is missing 108 genes (Extended Data Fig. 10) that are only present on the A. arenosa subgenome segment that has been replaced by the homeologous segment from the A. thaliana subgenome, and AS150 and ASÖ5 are missing 53 genes that were only present on the A. thaliana subgenome.
Discussion
This study has focused on the process of polyploidization in a natural allotetraploid species, A. suecica. Its ancestral species, A. thaliana and A. arenosa, differ substantially in genome size, chromosome number, ploidy, mating system and ecology.
Our main conclusion from this study is that the polyploid speciation leading to A. suecica appears to have been a gradual process rather than some kind of ‘event’. We confirmed previous results that genetic polymorphism is largely shared with the ancestral species, demonstrating that A. suecica did not originate through a single unique hybridization event, but rather through multiple crosses20. We also find no evidence for a ‘genome shock’, in the sense of major genomic changes linked to structural and functional alterations, which has often been suggested to accompany polyploidization and hybridization. Specifically, the genome has not been massively rearranged, transposable elements are not out of control and there is no subgenome dominance in expression. On the contrary, we find evidence of genetic adaptation to ‘stable’ life as a polyploid, in particular changes to the meiotic machinery and in interactions with the plastids. These findings made in natural (but not synthetic) A. suecica, together with the observation that experimentally generated A. suecica is often unviable and does exhibit evidence of genome rearrangements, similar to the young allopolyploid species in Tragopogon and monkeyflower, suggest that the most important bottleneck in polyploid speciation may be selective. If this is true, domesticated polyploids may not always be representative of natural polyploidization. Darwin famously argued that evolution is gradual126—we suggest that natural polyploids are no exception from this, but note that many more species will have to be studied before it is possible to draw general conclusions.
Methods
PacBio sequencing of A. suecica
We used genomic DNA from whole rosettes of one A. suecica (ASS3) accession to generate PacBio sequencing data. DNA was extracted using a modified PacBio protocol for preparing Arabidopsis genomic DNA for size-selected approximately 20-kb SMRTbell libraries. In brief, whole-genomic DNA was extracted from 32 g of 3–4-week-old plants, grown at 16 °C and subjected to a 2-day dark treatment. This generated 23 μg of purified genomic DNA with a fragment length of more than 40 kb for A. suecica. We assessed DNA quality with a Qubit fluorometer and a Nanodrop analysis and ran the DNA on a gel to visualize fragmentation. Genomic libraries and single-molecule real-time (SMRT) sequence data were generated at the Functional Genomics Center Zurich (FGCZ). The PacBio RSII instrument was used with P6-C4 chemistry and an average movie length of 6 hours. A total of 12 SMRT cells were processed, generating 16.3 Gb of DNA bases with an N50 read length of 20 kb and median read length of 14 kb. Using the same genomic library, an additional 3.3 Gb of data was generated by a PacBio Sequel instrument at the Vienna BioCenter Core Facilities (VBCF), with a median read length of 10 kbp.
A. suecica genome assembly
To generate the A. suecica assembly we first used FALCON127 (v.0.3.0) with a length cut-off for seed reads set to 1 kb in size. The assembly produced 828 contigs with an N50 of 5.81 Mb and a total assembly size of 271 Mb. In addition, we generated a Canu128 (v.1.3.0) assembly using default settings, which resulted in 260 contigs with an N50 of 6.65 Mb and a total assembly size of 267 Mb. Then we merged the two assemblies using the software quickmerge129. The resulting merged assembly consisted of 929 contigs with an N50 of 9.02 Mb and a total draft assembly size of 276 Mb. We polished the assembly using Arrow130 (smrtlink release 5.0.0.6792) and Pilon (v.1.22). For Pilon131, 100 bp (with PCR duplicates removed), and a second PCR-free 250 bp, Illumina paired-end reads were used that had been generated from the reference A. suecica accession ASS3.
PacBio sequencing of A. arenosa
A natural Swedish autotetraploid A. arenosa accession, Aa4, was inbred in a lab for two generations to reduce heterozygosity. We extracted whole-genomic DNA from 64 g of 3-week-old plants in the same way as described for A. suecica, generating 50 μg of purified genomic DNA with fragment sizes longer than 40 kb in length. The A. arenosa genomic libraries and SMRT sequence data were generated at the VBCF. A PacBio Sequel instrument was used to generate a total of 22 Gb of data from five SMRT cells, with an N50 of 13 kb and median read length of 10 kb. In addition, two runs of Oxford Nanopore sequencing were carried out at the VBCF, producing 750 Mb in 180,000 reads (median 5 kb and 2.6 kb; N50 8.7 and 6.7 kb, respectively).
Assembly of autotetraploid A. arenosa
We assembled a draft contig assembly for the autotetraploid A. arenosa accession Aa4 using FALCON (v.0.3.0) as for A. suecica. The assembly produced 3,629 contigs with an N50 of 331 kb, a maximum contig size of 2.5 Mb and a total assembly size of 461 Mb. The assembly size is greater than the calculated haploid size of 330 Mb using fluorescence-activated cell sorting (FACS) (see Extended Data Fig. 1), probably because of the high levels of heterozygosity in A. arenosa. The resulting assembly was polished as described for A. suecica.
Hi-C tissue fixation and library preparation
To generate physical scaffolds for the A. suecica assembly we generated proximity-ligation Hi-C sequencing data. We collected approximately 0.5 g of tissue from 3-week-old seedlings of the same reference A. suecica accession. Freshly collected plant tissue was fixed in 1% formaldehyde. Cross-linking was stopped by the addition of 0.15 M glycine. The fixed tissue was ground to a powder in liquid nitrogen and suspended in 10 ml nuclei isolation buffer. Nuclei were digested by adding 50 U DpnII and the digested chromatin was blunt-ended by incubation with 25 μl of 0.4 mM biotin–14-dCTP and 40 U of Klenow enzyme. T4 DNA ligase (20 U) was then added to start proximity ligation. The extracted DNA was sheared by sonication with a Covaris S220 to produce 250–500-bp fragments. This was followed by size fractionation using AMPure XP beads. Biotin was then removed from unligated ends. DNA fragments were blunt-end-repaired and adaptors were ligated to the DNA products following the NEBNext Ultra II RNA Library Prep Kit for Illumina.
To analyse structural rearrangements, we collected tissue for one other natural A. suecica (AS530), one A. thaliana accession (6978), one A. arenosa (Aa6) and one synthetic A. suecica (F3). Each sample had two replicates. We collected tissue and prepared libraries in the same manner as described above. Illumina reads (125-bp paired-end) were mapped using HiCUP132 (v.0.6.1).
Reference-guided scaffolding of the A. suecica genome with LACHESIS
We sequenced 207 million pairs of 125-bp paired-end Illumina reads from the Hi-C library of the reference accession ASS3. We mapped reads using HiCUP (v.0.6.1) to the draft A. suecica contig assembly. This resulted in around 137 million read pairs with a unique alignment.
Setting an assembly threshold of ≥1 kb in size, contigs of the draft A. suecica assembly were first assigned to the A. thaliana or A. arenosa subgenome. To do this, we used nucmer from the software MUMmer133 (v.3.23) to perform whole-genome alignments. We aligned the draft A. suecica assembly to the A. thaliana TAIR10 reference and to our A. arenosa draft contig assembly, simultaneously. We used the MUMer command dnadiff to produce 1-to-1 alignments. As the subgenomes are only around 86% identical, the majority of contigs could be conclusively assigned to either subgenome by examining how similar the alignments were. Contigs that could not be assigned to a subgenome on the basis of percentage identity were examined manually, and the length of the alignment was used to determine subgenome assignment.
Finally, we used the software LACHESIS134 (v.1.0.0) to scaffold our draft assembly, using the reference genomes of A. thaliana and A. lyrata as a guide to assist with scaffolding the contigs (we used A. lyrata here instead of our draft A. arenosa contig assembly, as A. lyrata is a chromosome-level assembly). This produced a 13-scaffold chromosome-level assembly for A. suecica.
Construction of the A. suecica genetic map
We crossed the natural A. suecica accession AS150 with the reference accession ASS3. The cross was uni-directional with AS150 as the maternal and ASS3 as the paternal plant. A total of 192 F2 plants were collected. We multiplexed the samples on 96-well plates using 75-bp paired-end reads and generated data of 1–2× coverage per sample. Samples were mapped to the repeat-masked scaffolds of the reference A. suecica genome using BWA-MEM135 (v.0.7.15). SAMtools136 (v.0.1.19) was used to filter reads for proper pairs and a minimum mapping quality of 5 (-F 256 -f 3 -q 5). We called variants directly from SAMtools mpileup giving a total of 590,537 SNPs. We required sites to have non-zero coverage in a minimum of 20 individuals and filtered SNPs to have frequency between 0.45-0.55 in our F2 population. We removed F2 individuals that did not have genotype calls for more than 90% of the data. This resulted in 183 individuals with genotype calls for 334,257 SNPs.
We applied a hidden Markov model implemented in the R package HMM137 to classify SNPs as homozygous or heterozygous for each of our F2 lines. We then divided the genome into 500-kb non-overlapping windows, and classified each window as homozygous or heterozygous. This was done per chromosome and the resulting file for each chromosome and their markers were processed in the R package qtl138, to generate a genetic map. Markers genotyped in fewer than 100 F2 individuals were excluded from the analysis. Linkage groups were assigned with a minimum log of the odds (LOD) score of 8 and a maximum recombination fraction of 0.35. We defined the final marker order by the best LOD score and the lowest number of crossover events.
We corrected the erroneous placement of a contig at the beginning of chromosome 1 of the A. arenosa subgenome. The misplaced contig was relocated from chromosome 1 to the pericentromeric region of chromosome 2 of the A. arenosa subgenome in A. suecica. Chromosome 2 of the A. thaliana subgenome of A. suecica was previously shown to be largely devoid of intraspecific variation, resulting in few markers for this chromosome. Therefore, this chromosome-scale scaffold was assembled by the manual inspection of 3D-proximity information based on our Hi-C sequencing using the software Juicebox139.
Gene prediction and annotation of the A. suecica genome
We combined de novo and evidence-based approaches to predict protein-coding genes. For de novo prediction, we trained AUGUSTUS140 on the set of conserved single-copy genes using BUSCO141 separately on A. thaliana and A. arenosa subgenomes of A. suecica. The evidence-based approach included both homology to the protein sequences of the ancestral species and the transcriptome of A. suecica. We aligned the peptide sequences from the TAIR10 A. thaliana assembly to the A. thaliana subgenome of A. suecica, while the peptides from A. lyrata annotation142 (Alyrata_384_v2.1) were aligned to the A. arenosa subgenome of A. suecica using GenomeThreader143 (v.1.7.0). We mapped the RNA-seq reads from the reference accession of A. suecica (ASS3) from the rosettes and flower bud tissues to the reference genome using TopHat144 and generated intron hints from the split reads using the bam2hints extension of AUGUSTUS. We split the alignment into A. thaliana and A. arenosa subgenomes and assembled the transcriptome of A. suecica for each subgenome separately in the genome-guided mode with Trinity145 (v.2.6.6). Separately for each of the subgenomes, we filtered the assembled transcripts using a transcripts per million (TPM) cut-off set to 1, collapsed similar transcripts using CD-HIT146,147 with sequence identity set to 90%, and chose the longest open reading frame from the six-frame translation. We then aligned the proteins from A. thaliana and A. arenosa parts of A. suecica to the corresponding subgenomes using GenomeThreader (v.1.7.0). We ran AUGUSTUS using retrained parameters from BUSCO and merged hints from all three sources, these being: (1) intron hints from A. suecica RNA-seq; (2) homology hints from ancestral proteins; and (3) hints from A. suecica proteins.
RepeatModeler148 (v.1.0.11) was used to build a de novo TE consensus library for A. suecica and identify repetitive elements based on the genome sequence. Genome locations for the identified TE repeats were determined by using RepeatMasker149 (v.4.0.7) and filtered for full-length matches using a code described previously150. Helitrons are the most abundant TE family in both subgenomes.
Synthetic A. suecica lines
To generate synthetic A. suecica we crossed a natural tetraploid A. thaliana accession (6978, also known as Wa-1) to a natural Swedish autotetraploid A. arenosa (Aa4) accession. Similar to the natural A. suecica, A. thaliana was the maternal and A. arenosa was the paternal plant in this cross. Crosses in the opposite direction were unsuccessful. We managed to obtain very few F1 hybrid plants, which after one round of selfing set higher levels of seed. The resulting synthetic line was able to self-fertilize. F2 seeds were descended from a common F1 and were similar to natural A. suecica in appearance. We further continued the synthetic line to F3 (selfed third generation).
Synteny analysis
We performed an all-against-all BLASTP search using CDS sequences for the reference A. suecica genome and the ancestral genomes, A. thaliana and A. lyrata. We used the SynMap tool151 from the online CoGe portal152. We examined synteny using the default parameters for DAGChainer (maximum distance between two matches = 20 genes; minimum number of aligned pairs = 5 genes).
Estimating the copy number of rDNA repeats using short DNA reads
To measure the copy number of 45S rRNA repeats in our populations of different species, we aligned short DNA reads to a single reference 45S consensus sequence of A. thaliana153. An A. arenosa 45S rRNA consensus sequence was constructed by finding the best hit using BLAST in our draft A. arenosa contig assembly. This hit matched position 1571–8232 bp of the A. thaliana consensus sequence, was 6,647 bp in length and is 97% identical to the A. thaliana 45S rRNA consensus sequence. The aligned regions of these two 45S rRNA consensus sequences, determined by BLAST, were used in copy number estimates, to ensure that the sizes of the sequences were equal. The relative increase in sequence coverage of these loci, when compared to the mean coverage for the reference genome, was used to estimate copy number.
Plant material for RNA-seq
Transcriptomic data generated in this study included 15 accessions of A. suecica, 16 accessions of A. thaliana, 4 accessions of A. arenosa and 2 generations of an artificial A. suecica line (the second and third selfed generation). The sibling of a paternal A. arenosa parent (Aa4) and the maternal tetraploid A. thaliana parent (6978, or Wa-1) of our artificial A. suecica line were included as part of our samples (Supplementary Data 1). Each accession was replicated three times. Seeds were stratified in the dark for 4 days at 4 °C in 1 ml of sterilized water. Seeds were then transferred to pots in a controlled growth chamber at 21 °C. Humidity was kept constant at 60%. Pots were thinned to two to three seedlings after one week. Pots were re-randomized each week in their trays. Whole rosettes were collected when plants reached the seven-to-nine true-leaf stage of development. Samples were collected between 14:00 and 17:00 and flash-frozen in liquid nitrogen.
RNA extraction and library preparation
For each accession, two to three whole rosettes in each pot were pooled and total RNA was extracted using the ZR Plant RNA MiniPrep kit. We treated the samples with DNAse and performed purification of mRNA and polyA selection using the AMPure XP magnetic beads and the Poly(A) RNA Selection Kit from Lexogen. RNA quality and degradation were assessed using the RNA Fragment Analyzer (DNF-471 stranded sensitivity RNA analysis kit, 15 nt). The concentration of RNA per sample was measured using the Qubit fluorometer. Library preparation was carried out following the NEBNext Ultra II RNA Library Prep Kit for Illumina. Barcoded adaptors were ligated using NEBNext Multiplex Oligos for Illumina (Index Primers Set 1 and 2). The libraries were PCR-amplified for seven cycles, and 125-bp paired-end sequencing was carried out at the VBCF on Illumina (HiSeq 2500) using multiplexing.
RNA-seq mapping and gene expression analysis
We mapped 125-bp paired-end reads to the de-novo-assembled A. suecica reference using STAR154 (v.2.7), and we filtered for primary and uniquely aligned reads using the parameters–outfilterMultimapNmax 1–outSamprimaryFlag OneBestScore. We quantified reads mapped to genes using–quantMode GeneCounts.
To reduce signals that are the result of cross-mapping between the subgenomes of A. suecica we used A. thaliana and A. arenosa as a control. For each gene in the A. thaliana subgenome we compared the log fold change of gene counts in our A. thaliana population to those in our A. arenosa population. We filtered for genes with a log2(A. thaliana/A. arenosa) below 0. We applied the same filters for genes on the A. arenosa subgenome. This reduced the number of genes analysed from 22,383 to 21,737 on the A. thaliana subgenome, and from 23,353 to 23,221 on the A. arenosa subgenome.
Expression analysis was then further restricted to 1:1 unique homeologous gene pairs between the subgenomes of A. suecica (17,881 gene pairs). Gene counts were normalized for gene size by calculating the TPM. The effective library sizes were calculated by computing a scaling factor based on the trimmed mean of M-values (TMM) in edgeR155, separately for each subgenome. Genes expressed at a low level were removed from the analysis by keeping genes that were expressed in at least three individuals of A. thaliana and A. suecica, at least one individual of A. arenosa and at least one individual of synthetic A. suecica. A total of 14,041 homeologous gene pairs satisfied our expression criteria. As A. suecica is expressing both subgenomes, to correctly normalize the effective library size in A. suecica accessions, the effective library size was calculated as a mean of TPM counts for both subgenomes. The effective library size of A. thaliana accessions was calculated for TPM counts using the A. thaliana subgenome of the reference genome, as genes from this subgenome will be expressed in A. thaliana, and the effective library size of A. arenosa lines was calculated using the A. arenosa subgenome of the reference A. suecica genome. Gene counts were transformed to counts per million (CPM) with a prior count of 1 and were log2-transformed. We used the mean of replicates per accession for downstream analyses.
To compare homeologous genes between the subgenomes in A. suecica we computed a log fold change using log2(A. arenosa homeologue/A. thaliana homeologue). For tissue-specific genes we took genes that showed a log fold change ≥ 2 in expression between two tissues.
For comparing homologous genes between the (sub-)genomes of A. suecica and the ancestral species A. thaliana and A. arenosa, we performed a Wilcoxon test independently for each of the 14,041 homeologous gene pairs. Using the normalized CPM values, we compared the relative expression level of a gene on the A. thaliana subgenome between our populations of A. thaliana and A. suecica. We performed the same test on the A. arenosa subgenome comparing the relative expression of a gene between our populations of A. arenosa and A. suecica. We filtered for genes with an adjusted P value of less than 0.05 (using false discovery rate (FDR) correction). This amounted to 4,186 and 4,571 DEGs for the A. thaliana and the A. arenosa subgenomes, respectively.
Cross-mapping of short reads
Cross-mapping of short RNA reads between the subgenomes of A. suecica was measured by mixing the RNA reads between A. thaliana and A. arenosa individuals to generate ‘in-silico’ A. suecica individuals. We mapped reads from 10 in-silico A. suecica individuals to the A. suecica genome. We compared different RNA-seq pipelines to determine cross-mapping error rates. We mapped reads using STAR154 (v.2.7), HISAT2156 (v.2.1) and EAGLE157. Around 1% of A. thaliana reads map to the A. arenosa subgenome and around 6% of the A. arenosa reads map to the A. thaliana subgenome, regardless of mapping strategy or pipeline (see Extended Data Fig. 6).
Expression analysis of rRNA
RNA reads were mapped in a similar manner to DNA reads for the analysis of rDNA copy number. Expression analysis was performed in a similar manner to protein-coding genes, in edgeR. We defined the exclusive expression of a particular 45S rRNA gene by taking a cut-off of 15 for log2(CPM) as this was the maximum level of cross-mapping we observed for the ancestral species (see Extended Data Fig. 3).
Expression analysis of transposable elements
To analyse the expression of transposable elements between species, the annotated TE consensus sequences in A. suecica were aligned using BLAST all vs all. Highly similar TE sequences (more than 85% similar for more than 85% of the TE sequence length), were removed, leaving 813 TE families out of 1,213. Filtered A. suecica TEs were aligned to annotated A. thaliana (TAIR10) and A. arenosa (the PacBio contig assembly presented in this study) TE sequences to assign each family to an ancestral species using BLAST. A total of 208 TE families were assigned to the A. thaliana parent and 171 TE families were assigned to the A. arenosa parent.
RNA reads were mapped to TE sequences using a similar approach as for gene expression analysis using edgeR. TEs that showed expression using a cut-off of log2(CPM) > 2 were kept. A total of 121 A. thaliana TE sequences and 93 A. arenosa TE sequences passed this threshold. We took the mean of replicates per accession for further downstream analyses.
GO enrichment analysis
We used the R package TopGO158 to conduct GO enrichment analysis. We used the ‘weight01’ algorithm when running TopGO, which accounts for the hierarchical structure of GO terms and thus implicitly corrects for multiple testing. GO annotations were based on the A. thaliana orthologue of A. suecica genes. Gene annotations for A. thaliana were obtained using the R package biomaRt159 from Ensembl ‘biomaRt::useMart(biomart = ’plants_mart’, dataset = ’athaliana_eg_gene’, host = 'plants.ensembl.org').
Genome size measurements
We measured genome size for the reference A. suecica accession ASS3 and the A. arenosa accession used for PacBio (Aa4), using Solanum lycopersicum cv. Stupicke (2C = 1.96 pg DNA) as the standard. The reference A. lyrata accession MN47 and the A. thaliana accession CVI were used as additional controls. Each sample had two replicates.
In brief, the leaves from three-week-old fresh tissue were chopped using a razor blade in 500 µl of UV Precise P extraction buffer with 10 µl mercaptoethanol per ml (kit PARTEC CyStain PI Absolute P no. 05- 5022) to isolate nuclei. Instead of the Partec UV Precise P staining buffer, however, 1 ml of a 5 mg DAPI solution was used, as DAPI provides DNA content histograms with high resolution. The suspension was then passed through a 30-µm filter (Partec CellTrics no. 04-0042-2316) and incubated for 15 minutes on ice before FACS.
Genome size was measured using flow cytometry and a FACS Aria III sorter with near UV 375-nm laser for DAPI. Debris was excluded by selecting peaks when plotting DAPI-W against DAPI-A for 20,000 events.
The data were analysed using the flowCore160 package in R. Genome size was estimated by comparing the mean G1 of the standard Solanum lycopersicum to that of each sample to calculate the 2C DNA content of that sample using the equation:
We also measured genome size for the reference A. suecica accession ASS3 using the software jellyfish161 and findGSE162 using k-mers (21mers). The genome size estimated was 312 Mb, compared to the 305 Mb estimated using FACS (see Extended Data Fig. 1).
Mapping of TE insertions
We used PoPoolationTE2100 (v.v1.10.04) to identify TE insertions. The advantage of this TE-calling software to others is that it avoids a reference bias by treating all TEs as de novo insertions. In brief, it works by using discordant read pairs to calculate the location and abundance of a TE in the genome for an accession of interest.
We mapped 100-bp Illumina DNA reads from previous studies20,76,163, in addition to our newly generated synthetic A. suecica using BWA-MEM135 (v.0.7.15) to a repeat-masked version of the A. suecica reference genome, concatenated with our annotated repeat sequences (see ‘Gene prediction and annotation of the A. suecica genome’), as this is the data format required by PoPoolationTE2. Reads were given an increased penalty of 15 for being unpaired. Reads were de-duplicated using SAMtools136 rmdup (v.1.9). The resulting bam files were then provided to PoPoolationTE2 to identify TE insertions in the genome of each of our A. suecica, A. thaliana and A. arenosa accessions. We used a mapping quality of 10 for the read in the discordant read pair mapping to the genome. We used the ‘separate’ mode in the ‘identify TE signatures’ step and a ‘–min-distance −200–max-distance 500’ in the ‘pairupsignatures’ step of the pipeline. TE counts within each accession were merged if they fell within 400 bp of each other and if they mapped to the same TE sequence. All TE counts (that is, the processed TE counts for each accession) were then combined to produce a population-wide count estimate. Population-wide TE insertions were merged if they mapped to the same TE sequence and fell within 400 bp of each other. Coverage of each TE insertion in the population was also calculated for each accession. The final file was a list of TE insertions present in the population and the presence or absence (or ‘NA’ if there was no coverage to support the presence or absence of a TE insertion) in each accession analysed (Supplementary Data 1).
Assigning ancestry to TE sequences
To examine TE consensus sequences that have mobilized between the subgenomes of A. suecica, we first examined which of our TE consensus sequences (n = 1,152) have at least the potential to mobilize (that is, have full-length TE copies in the genome of A. suecica). We filtered for TE consensus sequences that had TE copies in the genome of A. suecica that are more than 80% similar in identity for more than 80% of the consensus sequence length (n = 936). Of these, 188 consensus sequences were private to the A. thaliana subgenome, 460 were private to the A. arenosa subgenome and 288 TE consensus sequences were present in both subgenomes of A. suecica. To determine whether TEs have jumped from the A. thaliana subgenome to the A. arenosa subgenome and vice versa we next needed to assign ancestry to these 288 TE consensus sequences. To do this we used BLAST to search for these consensus sequences in the ancestral genomes of A. suecica, using the TAIR10 A. thaliana reference and our A. arenosa PacBio contig assembly. Using the same 80%-80% rule we assigned 55 TEs to A. arenosa and 15 TEs to A. thaliana ancestry.
Read mapping and SNP calling
To call biallelic SNPs we mapped reads to the A. suecica reference genome using the same filtering parameters described in ‘Mapping of TE insertions’. Biallelic SNPs were called using HaplotypeCaller from GATK164 (v.3.8) using default quality thresholds. SNPs were annotated using SnpEff165. Biallelic SNPs on the A. thaliana subgenome were polarized using 38 diploid A. lyrata lines76 and biallelic SNPS on the A. arenosa subgenome were polarized using 30 A. thaliana accessions163 closely related to A. suecica20.
Chromosome preparation and FISH
Whole inflorescences of A. arenosa, A. suecica and A. thaliana were fixed in freshly prepared ethanol:acetic acid fixative (3:1) overnight, transferred into 70% ethanol and stored at −20 °C until use. Selected inflorescences were rinsed in distilled water and citrate buffer (10 mM sodium citrate, pH 4.8), and digested by a 0.3% mix of pectolytic enzymes (cellulase, cytohelicase, pectolyase; all from Sigma-Aldrich) in citrate buffer for around 3 h. Mitotic chromosome spreads were prepared from pistils as previously described166 and suitable slides were pretreated by RNase (100 µg ml−1, AppliChem) and pepsin (0.1 mg ml−1, Sigma-Aldrich).
For identification of A. thaliana and A. arenosa subgenomes in the allotetraploid genome of A. suecica, FISH probes were made from plasmids pARR20–1 or pAaCEN containing 180 bp of A. thaliana (pAL) or around 250 bp of A. arenosa (pAa) pericentromeric repeats, respectively. The A. thaliana BAC clone T15P10 (AF167571) bearing 45S rRNA gene repeats was used for in situ localization of nucleolar organizer regions (NORs). Individual probes were labelled with biotin–dUTP, digoxigenin–dUTP and Cy3–dUTP by nick translation, pooled, precipitated and resuspended in 20 µl of hybridization mixture (50% formamide and 10% dextran sulfate in 2× saline sodium citrate (2× SSC)) per slide as previously described96.
Probes and chromosomes were denatured together on a hot plate at 80 °C for 2 min and incubated in a moist chamber at 37 °C overnight. Post-hybridization washing was performed in 20% formamide in 2× SSC at 42 °C. Fluorescent detection was as follows: biotin–dUTP was detected by avidin–Texas Red (Vector Laboratories) and amplified by goat anti-avidin–biotin (Vector Laboratories) and avidin–Texas Red; digoxigenin–dUTP was detected by mouse anti-digoxigenin (Jackson ImmunoResearch) and goat anti-mouse Alexa Fluor 488 (Molecular Probes). Chromosomes were counterstained with DAPI (4’,6-diamidino-2-phenylindole; 2 μg ml−1) in Vectashield (Vector Laboratories). Fluorescent signals were analysed and photographed using a Zeiss Axioimager epifluorescence microscope and a CoolCube camera (MetaSystems). Images were acquired separately for the four fluorochromes using appropriate excitation and emission filters (AHF Analysentechnik). The monochromatic images were pseudo-coloured and merged using Adobe Photoshop CS6 software (Adobe Systems).
DAP-seq enrichment analysis for transcription factor target genes
We downloaded the target genes of transcription factors from the plant cistrome database (http://neomorph.salk.edu/dap_web/pages/index.php), which is a collection of transcription-factor-binding sites and their target genes, in A. thaliana, based on DAP-seq167. To test for enrichment of a gene set (for example, the genes in A. thaliana cluster 2 on Fig. 5) for target genes of a particular transcription factor, we performed a hyper-geometric test in R. As a background we used the total 14,041 genes used in our gene expression analysis. We then performed FDR correction for multiple testing to calculate an accurate P value of the enrichment.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Supplementary Table 1. GO analysis for gene expression comparison between whole rosettes and floral buds in A. suecica. No significant GO was found for genes biased towards the A. thaliana subgenome of A. suecica for floral buds. Supplementary Table 2. List of overrepresented gene ontologies from Fig. 5e.
PoPoolationTE2 calls for A. suecica and the ancestral species for the subgenomes of A. suecica.
RNA-seq mapping statistics, DEGs, overlap with the CyMIRA and DAP-seq databases and gene evolutionary relationships in A. suecica.
Log fold change and CPM for genes on the A. thaliana and A. arenosa subgenome of A. suecica.
Gene annotation of the A. suecica genome.
TE sequences in A. suecica and TE hierarchy files.
Acknowledgements
This work was supported, in part, by DFG SPP 1529 to M.N. and D. Weigel. T.M. and M.A.L. were supported by the Czech Science Foundation (grant no. 19-03442S) and the CEITEC 2020 project (grant no. LQ1601). P.Y.N. acknowledges a postdoctoral fellowship of the Research Foundation–Flanders (12S9618N). We thank the Next Generation Sequencing Unit of the VBCF for assistance; S. Holm and T. Säll for material collections and discussions throughout; Y. Van de Peer for providing feedback on the manuscript; and J. Sharbrough for pointing us to the CyMIRA database
Extended data
Author contributions
R.B., T.M., J.G. and C.L. performed experiments. R.B., T.M., L.M.S.-J., M.A.L., P.Y.N. and M.N. collected and analysed data. R.B., P.Y.N. and M.N. wrote the manuscript. P.Y.N. and M.N. supervised the project.
Funding
Open access funding provided by Max Planck Institute for Plant Breeding Research (2).
Data availability
Genome assemblies and raw short reads can be found in the European Nucleotide Archive (ENA) (https://www.ebi.ac.uk/ena/browser/home). The genome assembly for A. suecica ASS3 can be found under the BioProject number PRJEB42198, assembly accession GCA_905175345. The raw reads for the A. suecica genome assembly generated by Pacbio RSII can be found under ERR5037702 and those from Sequel under ERR5031296. The Hi-C reads used for scaffolding the A. suecica assembly can be found under ERR5032369. The contig assembly for tetraploid A. arenosa (ssp. arenosa) can be found under the BioProject number PRJEB42276, assembly accession GCA_905175405. The raw reads for the A. arenosa Aa4 contig assembly generated by Sequel can be found under ERR5031542 and the reads generated by Nanopore under ERR5031541. Hi-C reads for the A. arenosa assembly can be found under ERR5032370. Hi-C sequencing data for the ancestral species, the outlier accession AS530 and synthetic A. suecica can be found under the BioProject number PRJEB42290. DNA resequencing data of synthetic A. suecica and parents generated in this study can be found under the BioProject number PRJEB42291. The RNA-seq reads are under the BioProject number PRJEB42277. TE presence or absence calls for A. suecica and the ancestral species can be found in Supplementary Data 1. A list of DEGs, orthologues, enriched DAP-seq transcription factors, CyMIRA gene overlaps and RNA-seq mapping statistics can be found in Supplementary Data 2. Log fold change and CPM for genes on the A. thaliana and A. arenosa subgenome can be found in Supplementary Data 3. The gene annotation (gff3 file) of the A. suecica genome can be found in Supplementary Data 4. TE consensus sequences and a hierarchy file of TE order for A. suecica can be found in Supplementary Data 5.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Ecology & Evolution thanks Z. Jeffrey Chen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
11/3/2021
In the version of this article initially published online, the following metadata was omitted and has now been included: Open access funding provided by Max Planck Institute for Plant Breeding Research (2).
Contributor Information
Polina Yu. Novikova, Email: pnovikova@mpipz.mpg.de
Magnus Nordborg, Email: magnus.nordborg@gmi.oeaw.ac.at.
Extended data
is available for this paper at 10.1038/s41559-021-01525-w.
Supplementary information
The online version contains supplementary material available at 10.1038/s41559-021-01525-w.
References
- 1.Van de Peer Y, Mizrachi E, Marchal K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017;18:411–424. doi: 10.1038/nrg.2017.26. [DOI] [PubMed] [Google Scholar]
- 2.Soltis PS, Soltis DE. Ancient WGD events as drivers of key innovations in angiosperms. Curr. Opin. Plant Biol. 2016;30:159–165. doi: 10.1016/j.pbi.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 3.Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Z, et al. Multiple large-scale gene and genome duplications during the evolution of hexapods. Proc. Natl Acad. Sci. USA. 2018;115:4713–4718. doi: 10.1073/pnas.1710791115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen ZJ, et al. Genomic diversifications of five Gossypium allopolyploid species and their impact on cotton improvement. Nat. Genet. 2020;52:525–533. doi: 10.1038/s41588-020-0614-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edger PP, et al. Origin and evolution of the octoploid strawberry genome. Nat. Genet. 2019;51:541–547. doi: 10.1038/s41588-019-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramírez-González RH, et al. The transcriptional landscape of polyploid wheat. Science. 2018;361:eaar6089. doi: 10.1126/science.aar6089. [DOI] [PubMed] [Google Scholar]
- 8.Zhuang W, et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019;51:865–876. doi: 10.1038/s41588-019-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertioli DJ, et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 2019;51:877–884. doi: 10.1038/s41588-019-0405-z. [DOI] [PubMed] [Google Scholar]
- 10.Kasianov AS, et al. High-quality genome assembly of Capsella bursa-pastoris reveals asymmetry of regulatory elements at early stages of polyploid genome evolution. Plant J. 2017;91:278–291. doi: 10.1111/tpj.13563. [DOI] [PubMed] [Google Scholar]
- 11.Kryvokhyzha D, et al. Towards the new normal: transcriptomic convergence and genomic legacy of the two subgenomes of an allopolyploid weed (Capsella bursa-pastoris) PLoS Genet. 2019;15:e1008131. doi: 10.1371/journal.pgen.1008131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Douglas GM, et al. Hybrid origins and the earliest stages of diploidization in the highly successful recent polyploid Capsella bursa-pastoris. Proc. Natl Acad. Sci. USA. 2015;112:2806–2811. doi: 10.1073/pnas.1412277112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffiths AG, et al. Breaking free: the genomics of allopolyploidy-facilitated niche expansion in white clover. Plant Cell. 2019;31:1466–1487. doi: 10.1105/tpc.18.00606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon SP, et al. Gradual polyploid genome evolution revealed by pan-genomic analysis of Brachypodium hybridum and its diploid progenitors. Nat. Commun. 2020;11:3670. doi: 10.1038/s41467-020-17302-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Catalán P, López-Álvarez D, Bellosta C, Villar L. Updated taxonomic descriptions, iconography, and habitat preferences of Brachypodium distachyon, B. stacei, and B. hybridum (Poaceae) An. Jard. Bot. Madr. 2016;73:e028. doi: 10.3989/ajbm.2428. [DOI] [Google Scholar]
- 16.Paape T, et al. Patterns of polymorphism and selection in the subgenomes of the allopolyploid Arabidopsis kamchatica. Nat. Commun. 2018;9:3909. doi: 10.1038/s41467-018-06108-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edger PP, et al. Subgenome dominance in an interspecific hybrid, synthetic allopolyploid, and a 140-year-old naturally established neo-allopolyploid monkeyflower. Plant Cell. 2017;29:2150–2167. doi: 10.1105/tpc.17.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soltis DE, et al. Recent and recurrent polyploidy in Tragopogon (Asteraceae): cytogenetic, genomic and genetic comparisons. Biol. J. Linn. Soc. 2004;82:485–501. doi: 10.1111/j.1095-8312.2004.00335.x. [DOI] [Google Scholar]
- 19.te Beest M, et al. The more the better? The role of polyploidy in facilitating plant invasions. Ann. Bot. 2012;109:19–45. doi: 10.1093/aob/mcr277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novikova PY, et al. Genome sequencing reveals the origin of the allotetraploid Arabidopsis suecica. Mol. Biol. Evol. 2017;34:957–968. doi: 10.1093/molbev/msw299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fowler NL, Levin DA. Ecological constraints on the establishment of a novel polyploid in competition with its diploid progenitor. Am. Nat. 1984;124:703–711. doi: 10.1086/284307. [DOI] [Google Scholar]
- 22.Bomblies K, Madlung A. Polyploidy in the Arabidopsis genus. Chromosome Res. 2014;22:117–134. doi: 10.1007/s10577-014-9416-x. [DOI] [PubMed] [Google Scholar]
- 23.Hollister JD, et al. Genetic adaptation associated with genome-doubling in autotetraploid Arabidopsis arenosa. PLoS Genet. 2012;8:e1003093. doi: 10.1371/journal.pgen.1003093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bomblies K, Jones G, Franklin C, Zickler D, Kleckner N. The challenge of evolving stable polyploidy: could an increase in ‘crossover interference distance’ play a central role? Chromosoma. 2016;125:287–300. doi: 10.1007/s00412-015-0571-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leitch AR, Leitch IJ. Genomic plasticity and the diversity of polyploid plants. Science. 2008;320:481–483. doi: 10.1126/science.1153585. [DOI] [PubMed] [Google Scholar]
- 26.Bottani S, Zabet NR, Wendel JF, Veitia RA. Gene expression dominance in allopolyploids: hypotheses and models. Trends Plant Sci. 2018;23:393–402. doi: 10.1016/j.tplants.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Parisod C, et al. Impact of transposable elements on the organization and function of allopolyploid genomes. New Phytol. 2010;186:37–45. doi: 10.1111/j.1469-8137.2009.03096.x. [DOI] [PubMed] [Google Scholar]
- 28.McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 29.Feldman M, et al. Rapid elimination of low-copy DNA sequences in polyploid wheat: a possible mechanism for differentiation of homoeologous chromosomes. Genetics. 1997;147:1381–1387. doi: 10.1093/genetics/147.3.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H, et al. Transcriptome shock invokes disruption of parental expression-conserved genes in tetraploid wheat. Sci. Rep. 2016;6:26363. doi: 10.1038/srep26363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, et al. Transcriptome asymmetry in synthetic and natural allotetraploid wheats, revealed by RNA-sequencing. New Phytol. 2016;209:1264–1277. doi: 10.1111/nph.13678. [DOI] [PubMed] [Google Scholar]
- 32.Zhang H, et al. Persistent whole-chromosome aneuploidy is generally associated with nascent allohexaploid wheat. Proc. Natl Acad. Sci. USA. 2013;110:3447–3452. doi: 10.1073/pnas.1300153110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kashkush K, Feldman M, Levy AA. Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics. 2002;160:1651–1659. doi: 10.1093/genetics/160.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaked H, Kashkush K, Ozkan H, Feldman M, Levy AA. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell. 2001;13:1749–1759. doi: 10.1105/TPC.010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ozkan H, Levy AA, Feldman M. Allopolyploidy-Induced rapid genome evolution in the wheat (Aegilops–Triticum) group. Plant Cell. 2001;13:1735–1747. doi: 10.1105/TPC.010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiong Z, Gaeta RT, Pires JC. Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc. Natl Acad. Sci. USA. 2011;108:7908–7913. doi: 10.1073/pnas.1014138108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu J, et al. Homoeolog expression bias and expression level dominance in resynthesized allopolyploid Brassica napus. BMC Genomics. 2018;19:586. doi: 10.1186/s12864-018-4966-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szadkowski E, et al. The first meiosis of resynthesized Brassica napus, a genome blender. New Phytol. 2010;186:102–112. doi: 10.1111/j.1469-8137.2010.03182.x. [DOI] [PubMed] [Google Scholar]
- 39.Zhao T, et al. LncRNAs in polyploid cotton interspecific hybrids are derived from transposon neofunctionalization. Genome Biol. 2018;19:195. doi: 10.1186/s13059-018-1574-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoo M-J, Szadkowski E, Wendel JF. Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity. 2013;110:171–180. doi: 10.1038/hdy.2012.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li A, et al. mRNA and small RNA transcriptomes reveal insights into dynamic homoeolog regulation of allopolyploid heterosis in nascent hexaploid wheat. Plant Cell. 2014;26:1878–1900. doi: 10.1105/tpc.114.124388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flagel LE, Wendel JF. Evolutionary rate variation, genomic dominance and duplicate gene expression evolution during allotetraploid cotton speciation. New Phytol. 2010;186:184–193. doi: 10.1111/j.1469-8137.2009.03107.x. [DOI] [PubMed] [Google Scholar]
- 43.Liu B, Brubaker CL, Mergeai G, Cronn RC, Wendel JF. Polyploid formation in cotton is not accompanied by rapid genomic changes. Genome. 2001;44:321–330. doi: 10.1139/g01-011. [DOI] [PubMed] [Google Scholar]
- 44.Kashkush K, Feldman M, Levy AA. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 2003;33:102–106. doi: 10.1038/ng1063. [DOI] [PubMed] [Google Scholar]
- 45.Kraitshtein Z, Yaakov B, Khasdan V, Kashkush K. Genetic and epigenetic dynamics of a retrotransposon after allopolyploidization of wheat. Genetics. 2010;186:801–812. doi: 10.1534/genetics.110.120790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yaakov B, Kashkush K. Mobilization of Stowaway-like MITEs in newly formed allohexaploid wheat species. Plant Mol. Biol. 2012;80:419–427. doi: 10.1007/s11103-012-9957-3. [DOI] [PubMed] [Google Scholar]
- 47.International Wheat Genome Sequencing Consortium (IWGSC) et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science361, eaar7191 (2018). [DOI] [PubMed]
- 48.Wang M, et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 2019;51:224–229. doi: 10.1038/s41588-018-0282-x. [DOI] [PubMed] [Google Scholar]
- 49.Yang Z, et al. Extensive intraspecific gene order and gene structural variations in upland cotton cultivars. Nat. Commun. 2019;10:2989. doi: 10.1038/s41467-019-10820-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang G, et al. Genome sequence of gossypium herbaceum and genome updates of Gossypium arboreum and Gossypium hirsutum provide insights into cotton A-genome evolution. Nat. Genet. 2020;52:516–524. doi: 10.1038/s41588-020-0607-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang T, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015;33:531–537. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- 52.Han J, et al. Rapid proliferation and nucleolar organizer targeting centromeric retrotransposons in cotton. Plant J. 2016;88:992–1005. doi: 10.1111/tpj.13309. [DOI] [PubMed] [Google Scholar]
- 53.Wang M, et al. Evolutionary dynamics of 3D genome architecture following polyploidization in cotton. Nat. Plants. 2018;4:90–97. doi: 10.1038/s41477-017-0096-3. [DOI] [PubMed] [Google Scholar]
- 54.Cheng F, et al. Biased gene fractionation and dominant gene expression among the subgenomes of Brassica rapa. PLoS One. 2012;7:e36442. doi: 10.1371/journal.pone.0036442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schnable JC, Springer NM, Freeling M. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc. Natl Acad. Sci. USA. 2011;108:4069–4074. doi: 10.1073/pnas.1101368108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.International Wheat Genome Sequencing Consortium (IWGSC). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science345, 1251788 (2014). [DOI] [PubMed]
- 57.Chalhoub B, et al. Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014;345:950–953. doi: 10.1126/science.1253435. [DOI] [PubMed] [Google Scholar]
- 58.Wang M, et al. Asymmetric subgenome selection and cis-regulatory divergence during cotton domestication. Nat. Genet. 2017;49:579–587. doi: 10.1038/ng.3807. [DOI] [PubMed] [Google Scholar]
- 59.Gaut BS, Seymour DK, Liu Q, Zhou Y. Demography and its effects on genomic variation in crop domestication. Nat. Plants. 2018;4:512–520. doi: 10.1038/s41477-018-0210-1. [DOI] [PubMed] [Google Scholar]
- 60.Kremling KAG, et al. Dysregulation of expression correlates with rare-allele burden and fitness loss in maize. Nature. 2018;555:520–523. doi: 10.1038/nature25966. [DOI] [PubMed] [Google Scholar]
- 61.Qian L, Qian W, Snowdon RJ. Sub-genomic selection patterns as a signature of breeding in the allopolyploid Brassica napus genome. BMC Genomics. 2014;15:1170. doi: 10.1186/1471-2164-15-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang L, et al. The interplay of demography and selection during maize domestication and expansion. Genome Biol. 2017;18:215. doi: 10.1186/s13059-017-1346-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alonge M, et al. Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell. 2020;182:145–161. doi: 10.1016/j.cell.2020.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Y, et al. Pan-genome of wild and cultivated soybeans. Cell. 2020;182:162–176. doi: 10.1016/j.cell.2020.05.023. [DOI] [PubMed] [Google Scholar]
- 65.Zhou Y, et al. The population genetics of structural variants in grapevine domestication. Nat. Plants. 2019;5:965–979. doi: 10.1038/s41477-019-0507-8. [DOI] [PubMed] [Google Scholar]
- 66.Buggs RJA, et al. Transcriptomic shock generates evolutionary novelty in a newly formed, natural allopolyploid plant. Curr. Biol. 2011;21:551–556. doi: 10.1016/j.cub.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 67.Chester M, et al. Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae) Proc. Natl Acad. Sci. USA. 2012;109:1176–1181. doi: 10.1073/pnas.1112041109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chelaifa H, Monnier A, Ainouche M. Transcriptomic changes following recent natural hybridization and allopolyploidy in the salt marsh species Spartina × townsendii and Spartina anglica (Poaceae) New Phytol. 2010;186:161–174. doi: 10.1111/j.1469-8137.2010.03179.x. [DOI] [PubMed] [Google Scholar]
- 69.Kryvokhyzha D, et al. Parental legacy, demography, and admixture influenced the evolution of the two subgenomes of the tetraploid Capsella bursa-pastoris (Brassicaceae) PLoS Genet. 2019;15:e1007949. doi: 10.1371/journal.pgen.1007949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Akama S, Shimizu-Inatsugi R, Shimizu KK, Sese J. Genome-wide quantification of homeolog expression ratio revealed nonstochastic gene regulation in synthetic allopolyploid Arabidopsis. Nucleic Acids Res. 2014;42:e46. doi: 10.1093/nar/gkt1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu H, Yu Q, Ran J-H, Wang X-Q. Unbiased subgenome evolution in allotetraploid species of Ephedra and its implications for the evolution of large genomes in gymnosperms. Genome Biol. Evol. 2020;13:evaa236. doi: 10.1093/gbe/evaa236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Säll T, Lind-Halldén C, Jakobsson M, Halldén C. Mode of reproduction in Arabidopsis suecica. Hereditas. 2004;141:313–317. doi: 10.1111/j.1601-5223.2004.01833.x. [DOI] [PubMed] [Google Scholar]
- 73.Hohmann N, Wolf EM, Lysak MA, Koch MA. A time-calibrated road map of Brassicaceae species radiation and evolutionary history. Plant Cell. 2015;27:2770–2784. doi: 10.1105/tpc.15.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.O’Kane SL, Schaal BA, Al-Shehbaz IA. The origins of Arabidopsis suecica (Brassicaceae) as indicated by nuclear rDNA sequences. Syst. Bot. 1996;21:559–566. doi: 10.2307/2419615. [DOI] [Google Scholar]
- 75.Jakobsson M, et al. A unique recent origin of the allotetraploid species Arabidopsis suecica: evidence from nuclear DNA markers. Mol. Biol. Evol. 2006;23:1217–1231. doi: 10.1093/molbev/msk006. [DOI] [PubMed] [Google Scholar]
- 76.Novikova PY, et al. Sequencing of the genus Arabidopsis identifies a complex history of nonbifurcating speciation and abundant trans-specific polymorphism. Nat. Genet. 2016;48:1077–1082. doi: 10.1038/ng.3617. [DOI] [PubMed] [Google Scholar]
- 77.Slotte T, et al. The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat. Genet. 2013;45:831–835. doi: 10.1038/ng.2669. [DOI] [PubMed] [Google Scholar]
- 78.Liu S, et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014;5:3930. doi: 10.1038/ncomms4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Madlung A, et al. Genomic changes in synthetic Arabidopsis polyploids. Plant J. 2005;41:221–230. doi: 10.1111/j.1365-313X.2004.02297.x. [DOI] [PubMed] [Google Scholar]
- 80.Copenhaver GP, Pikaard CS. Two-dimensional RFLP analyses reveal megabase-sized clusters of rRNA gene variants in Arabidopsis thaliana, suggesting local spreading of variants as the mode for gene homogenization during concerted evolution. Plant J. 1996;9:273–282. doi: 10.1046/j.1365-313X.1996.09020273.x. [DOI] [PubMed] [Google Scholar]
- 81.Navashin M. Chromosome alterations caused by hybridization and their bearing upon certain general genetic problems. Cytologia. 1934;5:169–203. doi: 10.1508/cytologia.5.169. [DOI] [Google Scholar]
- 82.Tucker S, Vitins A, Pikaard CS. Nucleolar dominance and ribosomal RNA gene silencing. Curr. Opin. Cell Biol. 2010;22:351–356. doi: 10.1016/j.ceb.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Maciak S, Michalak K, Kale SD, Michalak P. Nucleolar dominance and repression of 45S ribosomal RNA genes in hybrids between Xenopus borealis and X. muelleri (2n = 36) Cytogenetic Genome Res. 2016;149:290–296. doi: 10.1159/000450665. [DOI] [PubMed] [Google Scholar]
- 84.Książczyk T, et al. Immediate unidirectional epigenetic reprogramming of NORs occurs independently of rDNA rearrangements in synthetic and natural forms of a polyploid species Brassica napus. Chromosoma. 2011;120:557–571. doi: 10.1007/s00412-011-0331-z. [DOI] [PubMed] [Google Scholar]
- 85.Chen ZJ, Comai L, Pikaard CS. Gene dosage and stochastic effects determine the severity and direction of uniparental ribosomal RNA gene silencing (nucleolar dominance) in Arabidopsis allopolyploids. Proc. Natl Acad. Sci. USA. 1998;95:14891–14896. doi: 10.1073/pnas.95.25.14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pontes O, et al. Postembryonic establishment of megabase-scale gene silencing in nucleolar dominance. PLoS One. 2007;2:e1157. doi: 10.1371/journal.pone.0001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lewis MS, Pikaard CS. Restricted chromosomal silencing in nucleolar dominance. Proc. Natl Acad. Sci. USA. 2001;98:14536–14540. doi: 10.1073/pnas.251424098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pontes O, et al. Chromosomal locus rearrangements are a rapid response to formation of the allotetraploid Arabidopsis suecica genome. Proc. Natl Acad. Sci. USA. 2004;101:18240–18245. doi: 10.1073/pnas.0407258102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Long Q, et al. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat. Genet. 2013;45:884–890. doi: 10.1038/ng.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rabanal FA, et al. Epistatic and allelic interactions control expression of ribosomal RNA gene clusters in Arabidopsis thaliana. Genome Biol. 2017;18:75. doi: 10.1186/s13059-017-1209-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pontes O, et al. Natural variation in nucleolar dominance reveals the relationship between nucleolus organizer chromatin topology and rRNA gene transcription in Arabidopsis. Proc. Natl Acad. Sci. USA. 2003;100:11418–11423. doi: 10.1073/pnas.1932522100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guo X, Han F. Asymmetric epigenetic modification and elimination of rDNA sequences by polyploidization in wheat. Plant Cell. 2014;26:4311–4327. doi: 10.1105/tpc.114.129841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu B, Davis TM. Conservation and loss of ribosomal RNA gene sites in diploid and polyploid Fragaria (Rosaceae) BMC Plant Biol. 2011;11:157. doi: 10.1186/1471-2229-11-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Steige KA, Slotte T. Genomic legacies of the progenitors and the evolutionary consequences of allopolyploidy. Curr. Opin. Plant Biol. 2016;30:88–93. doi: 10.1016/j.pbi.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 95.Vicient CM, Casacuberta JM. Impact of transposable elements on polyploid plant genomes. Ann. Bot. 2017;120:195–207. doi: 10.1093/aob/mcx078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ungerer MC, Strakosh SC, Zhen Y. Genome expansion in three hybrid sunflower species is associated with retrotransposon proliferation. Curr. Biol. 2006;16:R872–R873. doi: 10.1016/j.cub.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 97.Rieseberg LH, et al. Major ecological transitions in wild sunflowers facilitated by hybridization. Science. 2003;301:1211–1216. doi: 10.1126/science.1086949. [DOI] [PubMed] [Google Scholar]
- 98.Cavrak VV, et al. How a retrotransposon exploits the plant’s heat stress response for its activation. PLoS Genet. 2014;10:e1004115. doi: 10.1371/journal.pgen.1004115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Göbel U, et al. Robustness of transposable element regulation but no genomic shock observed in interspecific Arabidopsis hybrids. Genome Biol. Evol. 2018;10:1403–1415. doi: 10.1093/gbe/evy095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kofler R, Gomez-Sanchez D, Schlotterer C. PoPoolationTE2: Comparative population genomics of transposable elements using Pool-Seq. Mol. Biol. Evol. 2016;33:2759–2764. doi: 10.1093/molbev/msw137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lockton S, Gaut BS. The evolution of transposable elements in natural populations of self-fertilizing Arabidopsis thaliana and its outcrossing relative Arabidopsis lyrata. BMC Evol. Biol. 2010;10:10. doi: 10.1186/1471-2148-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Quadrana L, et al. The Arabidopsis thaliana mobilome and its impact at the species level. eLife. 2016;5:e15716. doi: 10.7554/eLife.15716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stuart T, et al. Population scale mapping of transposable element diversity reveals links to gene regulation and epigenomic variation. eLife. 2016;5:e20777. doi: 10.7554/eLife.20777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wolfe KH. Yesterday’s polyploids and the mystery of diploidization. Nat. Rev. Genet. 2001;2:333–341. doi: 10.1038/35072009. [DOI] [PubMed] [Google Scholar]
- 105.Conant GC, Birchler JA, Pires JC. Dosage, duplication, and diploidization: clarifying the interplay of multiple models for duplicate gene evolution over time. Curr. Opin. Plant Biol. 2014;19:91–98. doi: 10.1016/j.pbi.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 106.Aköz G, Nordborg M. The Aquilegia genome reveals a hybrid origin of core eudicots. Genome Biol. 2019;20:256. doi: 10.1186/s13059-019-1888-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jiao Y, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- 108.Soltis PS, Marchant DB, Van de Peer Y, Soltis DE. Polyploidy and genome evolution in plants. Curr. Opin. Genet. Dev. 2015;35:119–125. doi: 10.1016/j.gde.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 109.Thomas BC, Pedersen B, Freeling M. Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 2006;16:934–946. doi: 10.1101/gr.4708406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Renny-Byfield S, Gong L, Gallagher JP, Wendel JF. Persistence of subgenomes in paleopolyploid cotton after 60 my of evolution. Mol. Biol. Evol. 2015;32:1063–1071. doi: 10.1093/molbev/msv001. [DOI] [PubMed] [Google Scholar]
- 111.Garsmeur O, et al. Two evolutionarily distinct classes of paleopolyploidy. Mol. Biol. Evol. 2014;31:448–454. doi: 10.1093/molbev/mst230. [DOI] [PubMed] [Google Scholar]
- 112.Li Q, et al. Unbiased subgenome evolution following a recent whole-genome duplication in pear (Pyrus bretschneideri Rehd.) Hortic. Res. 2019;6:34. doi: 10.1038/s41438-018-0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shan S, et al. Transcriptome dynamics of the inflorescence in reciprocally formed allopolyploid Tragopogon miscellus (Asteraceae) Front. Genet. 2020;11:888. doi: 10.3389/fgene.2020.00888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bird KA, et al. Replaying the evolutionary tape to investigate subgenome dominance in allopolyploid Brassica napus. New Phytol. 2021;230:354–371. doi: 10.1111/nph.17137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Alger EI, Edger PP. One subgenome to rule them all: underlying mechanisms of subgenome dominance. Curr. Opin. Plant Biol. 2020;54:108–113. doi: 10.1016/j.pbi.2020.03.004. [DOI] [PubMed] [Google Scholar]
- 116.Carlson KD, et al. Natural variation in stress response gene activity in the allopolyploid Arabidopsis suecica. BMC Genomics. 2017;18:653. doi: 10.1186/s12864-017-4067-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chang PL, Dilkes BP, McMahon M, Comai L, Nuzhdin SV. Homoeolog-specific retention and use in allotetraploid Arabidopsis suecica depends on parent of origin and network partners. Genome Biol. 2010;11:R125. doi: 10.1186/gb-2010-11-12-r125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Adams KL, Percifield R, Wendel JF. Organ-specific silencing of duplicated genes in a newly synthesized cotton allotetraploid. Genetics. 2004;168:2217–2226. doi: 10.1534/genetics.104.033522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sicard A, Lenhard M. The selfing syndrome: a model for studying the genetic and evolutionary basis of morphological adaptation in plants. Ann. Bot. 2011;107:1433–1443. doi: 10.1093/aob/mcr023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lu Y-J, Swamy KBS, Leu J-Y. Experimental evolution Reveals Interplay between Sch9 and polyploid stability in yeast. PLoS Genet. 2016;12:e1006409. doi: 10.1371/journal.pgen.1006409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yant L, et al. Meiotic adaptation to genome duplication in Arabidopsis arenosa. Curr. Biol. 2013;23:2151–2156. doi: 10.1016/j.cub.2013.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Morgan C, Zhang H, Henry CE, Franklin FCH, Bomblies K. Derived alleles of two axis proteins affect meiotic traits in autotetraploid Arabidopsis arenosa. Proc. Natl Acad. Sci. USA. 2020;117:8980–8988. doi: 10.1073/pnas.1919459117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Haga N, et al. Mutations in MYB3R1 and MYB3R4 cause pleiotropic developmental defects and preferential down-regulation of multiple G2/M-specific genes in Arabidopsis. Plant Physiol. 2011;157:706–717. doi: 10.1104/pp.111.180836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Forsythe ES, Sharbrough J, Havird JC, Warren JM, Sloan DB. CyMIRA: the cytonuclear molecular interactions reference for Arabidopsis. Genome Biol. Evol. 2019;11:2194–2202. doi: 10.1093/gbe/evz144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wu Y, et al. Genomic mosaicism due to homoeologous exchange generates extensive phenotypic diversity in nascent allopolyploids. Natl Sci. Rev. 2020;8:nwaa277. doi: 10.1093/nsr/nwaa277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Darwin, C. The Origin of Species by Means of Natural Selection, or The Preservation of Favored Races in the Struggle for Life (Hurst, 1872).
- 127.Chin C-S, et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods. 2016;13:1050–1054. doi: 10.1038/nmeth.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Koren S, et al. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chakraborty M, Baldwin-Brown JG, Long AD, Emerson JJ. Contiguous and accurate de novo assembly of metazoan genomes with modest long read coverage. Nucleic Acids Res. 2016;44:e147. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chin C-S, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 131.Walker BJ, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wingett S, et al. HiCUP: Pipeline for mapping and processing Hi-C data. F1000Res. 2015;4:1310. doi: 10.12688/f1000research.7334.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Marçais G, et al. MUMmer4: a fast and versatile genome alignment system. PLoS Comput. Biol. 2018;14:e1005944. doi: 10.1371/journal.pcbi.1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Burton JN, et al. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 2013;31:1119–1125. doi: 10.1038/nbt.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Himmelmann, L. HMM: Hidden Markov Models. R package version 1.0 (2010); https://cran.r-project.org/web/packages/HMM/index.html
- 138.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 139.Durand NC, et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 2016;3:95–98. doi: 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Stanke M, Morgenstern B. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005;33:W465–W467. doi: 10.1093/nar/gki458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Seppey, M., Manni, M. & Zdobnov E. M. in Gene Prediction. Methods in Molecular Biology (ed. Kollmar, M.) Vol. 1962, 227–245 (Springer, 2019). [DOI] [PubMed]
- 142.Rawat V, et al. Improving the annotation of Arabidopsis lyrata using RNA-seq data. PLoS One. 2015;10:e0137391. doi: 10.1371/journal.pone.0137391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gremme G, Brendel V, Sparks ME, Kurtz S. Engineering a software tool for gene structure prediction in higher organisms. Inf. Softw. Technol. 2005;47:965–978. doi: 10.1016/j.infsof.2005.09.005. [DOI] [Google Scholar]
- 144.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Grabherr MG, et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 148.Smit, A. F. A. & Hubley, R. RepeatModeler Open-1.0 (2008–2015); http://www.repeatmasker.org
- 149.Smit, A. F. A., Hubley, R. & Green, P. RepeatMasker Open-4.0 (2013–2015); http://www.repeatmasker.org
- 150.Bailly-Bechet M, Haudry A, Lerat E. ‘One code to find them all’: a perl tool to conveniently parse RepeatMasker output files. Mob. DNA. 2014;5:13. doi: 10.1186/1759-8753-5-13. [DOI] [Google Scholar]
- 151.Lyons E, Pedersen B, Kane J, Freeling M. The value of nonmodel genomes and an example using SynMap within CoGe to dissect the hexaploidy that predates the rosids. Trop. Plant Biol. 2008;1:181–190. doi: 10.1007/s12042-008-9017-y. [DOI] [Google Scholar]
- 152.Lyons E, Freeling M. How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 2008;53:661–673. doi: 10.1111/j.1365-313X.2007.03326.x. [DOI] [PubMed] [Google Scholar]
- 153.Rabanal FA, et al. Unstable Inheritance of 45S rRNA genes in Arabidopsis thaliana. G3. 2017;7:1201–1209. doi: 10.1534/g3.117.040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kuo T, Frith MC, Sese J, Horton P. EAGLE: Explicit alternative genome likelihood evaluator. BMC Med. Genomics. 2018;11(Suppl. 2):28. doi: 10.1186/s12920-018-0342-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Alexa, A. & Rahnenführer, J. Gene set enrichment analysis with topGO. Bioconductor Improv (2009); https://bioconductor.org/packages/release/bioc/html/topGO.html
- 159.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hahne F, et al. flowCore: a Bioconductor package for high throughput flow cytometry. BMC Bioinformatics. 2009;10:106. doi: 10.1186/1471-2105-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sun H, Ding J, Piednoël M, Schneeberger K. findGSE: estimating genome size variation within human and Arabidopsis using k-mer frequencies. Bioinformatics. 2018;34:550–557. doi: 10.1093/bioinformatics/btx637. [DOI] [PubMed] [Google Scholar]
- 163.The 1001 Genomes Consortium. 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell166, 481–491 (2016). [DOI] [PMC free article] [PubMed]
- 164.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mandáková T, Lysak MA. Chromosome preparation for cytogenetic analyses in Arabidopsis. Curr. Protoc. Plant Biol. 2016;1:43–51. doi: 10.1002/cppb.20009. [DOI] [PubMed] [Google Scholar]
- 167.O’Malley RC, et al. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell. 2016;165:1280–1292. doi: 10.1016/j.cell.2016.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. GO analysis for gene expression comparison between whole rosettes and floral buds in A. suecica. No significant GO was found for genes biased towards the A. thaliana subgenome of A. suecica for floral buds. Supplementary Table 2. List of overrepresented gene ontologies from Fig. 5e.
PoPoolationTE2 calls for A. suecica and the ancestral species for the subgenomes of A. suecica.
RNA-seq mapping statistics, DEGs, overlap with the CyMIRA and DAP-seq databases and gene evolutionary relationships in A. suecica.
Log fold change and CPM for genes on the A. thaliana and A. arenosa subgenome of A. suecica.
Gene annotation of the A. suecica genome.
TE sequences in A. suecica and TE hierarchy files.
Data Availability Statement
Genome assemblies and raw short reads can be found in the European Nucleotide Archive (ENA) (https://www.ebi.ac.uk/ena/browser/home). The genome assembly for A. suecica ASS3 can be found under the BioProject number PRJEB42198, assembly accession GCA_905175345. The raw reads for the A. suecica genome assembly generated by Pacbio RSII can be found under ERR5037702 and those from Sequel under ERR5031296. The Hi-C reads used for scaffolding the A. suecica assembly can be found under ERR5032369. The contig assembly for tetraploid A. arenosa (ssp. arenosa) can be found under the BioProject number PRJEB42276, assembly accession GCA_905175405. The raw reads for the A. arenosa Aa4 contig assembly generated by Sequel can be found under ERR5031542 and the reads generated by Nanopore under ERR5031541. Hi-C reads for the A. arenosa assembly can be found under ERR5032370. Hi-C sequencing data for the ancestral species, the outlier accession AS530 and synthetic A. suecica can be found under the BioProject number PRJEB42290. DNA resequencing data of synthetic A. suecica and parents generated in this study can be found under the BioProject number PRJEB42291. The RNA-seq reads are under the BioProject number PRJEB42277. TE presence or absence calls for A. suecica and the ancestral species can be found in Supplementary Data 1. A list of DEGs, orthologues, enriched DAP-seq transcription factors, CyMIRA gene overlaps and RNA-seq mapping statistics can be found in Supplementary Data 2. Log fold change and CPM for genes on the A. thaliana and A. arenosa subgenome can be found in Supplementary Data 3. The gene annotation (gff3 file) of the A. suecica genome can be found in Supplementary Data 4. TE consensus sequences and a hierarchy file of TE order for A. suecica can be found in Supplementary Data 5.