Abstract
5-Methylcytosine (m5C) is a posttranscriptional RNA modification participating in many critical bioprocesses, but its functions in human cancer remain unclear. Here, by detecting the transcriptome-wide m5C profiling in esophageal squamous cell carcinoma (ESCC), we showed increased m5C methylation in ESCC tumors due to the overexpressed m5C methyltransferase NSUN2. Aberrant expression of NSUN2 was positively regulated by E2F Transcription Factor 1 (E2F1). High NSUN2 levels predicted poor survival of ESCC patients. Moreover, silencing NSUN2 suppressed ESCC tumorigenesis and progression in Nsun2 knockout mouse models. Mechanistically, NSUN2 induced m5C modification of growth factor receptor-bound protein 2 (GRB2) and stabilized its mRNA, which was mediated by a novel m5C mediator, protein lin-28 homolog B (LIN28B). Elevated GRB2 levels increased the activation of PI3K/AKT and ERK/MAPK signalling. These results demonstrate that NSUN2 enhances the initiation and progression of ESCC via m5C-LIN28B dependent stabilization of GRB2 transcript, providing a promising epitranscriptomic-targeted therapeutic strategy for ESCC.
Subject terms: Oesophageal cancer, Epigenetics, RNA metabolism
Introduction
Esophageal squamous cell carcinoma (ESCC) is one of the most malignant cancers with only ~19% of 5-year survival [1, 2]. Most ESCC patients eventually die of cancer progression due to lack of effective treatment modalities [3]. Therefore, further elucidation of the comprehensive molecular mechanisms underlying ESCC is urgently needed to develop more effective diagnostic and therapeutic interventions for ESCC.
Recent discoveries have demonstrated that aberrations in epigenetic regulation, such as RNA methylation, are crucial hallmarks of tumor initiation, progression, and recurrence [4]. 5-Methylcytosine (m5C) is one of the most well-known and conserved RNA modifications extensively occurring on various types of eukaryotic RNA, including rRNA, lncRNA, tRNA, and mRNA [5–10]. To date, the known m5C methyltransferases include the NOP2/Sun RNA methyltransferase family member 1−7 (NSUN1−7) and the DNA methyltransferase 2 [11]. Aly/REF export factor (ALYREF) and Y-box binding protein 1 (YBX1) are two m5C readers respectively mediating RNA nuclear export and enhancing RNA stability [9, 12, 13]. Accumulating evidence confirms that m5C modification regulates multiple RNA metabolic and biological processes, such as RNA stability [12, 13], RNA export [9], RNA translation [14, 15], and RNA processing [16, 17]. Recently, with the advance in detecting and mapping techniques, distribution profiles of m5C sites on mRNAs have been discovered, suggesting that m5C sites are distributed in 5′ untranslated regions (5′UTR), coding sequences (CDS) and 3′ untranslated regions (3′UTR) of mRNAs and are especially prominent near translation start sites [8, 9]. NSUN2 and NSUN6 are two major methyltransferase catalyzing m5C modification of mammalian mRNAs [7, 9, 11, 18] and NSUN2 is currently the best-studied one. Emerging evidence has shown that NSUN2-mediated RNA m5C methylation plays a critical role in cell proliferation, development, and differentiation [16,19–21]. Mutation or aberrant expression of NSUN2 is involved in various diseases, such as cancer and developmental disorders [12, 21−23]. However, very little is known about the precise regulatory mechanism of NSUN2-mediated mRNA m5C modification in human diseases, especially human cancer.
In this study, we indicate an oncogenic role of NSUN2-mediated RNA m5C modification in human ESCC. Specifically, E2F Transcription Factor 1 (E2F1) binds to the promoter of NSUN2 and enhances its expression, which significantly increases m5C formation in growth factor receptor-bound protein 2 (GRB2). An RNA-binding protein lin-28 homolog B (LIN28B) acts as an m5C mediator preferentially binding to the m5C-modified GRB2 RNA and enhancing its stability. Upregulation of GRB2 evokes the oncogenic PI3K/AKT and ERK/MAPK signaling. We propose that the NSUN2-m5C-GRB2-PI3K/AKT and ERK/MAPK signaling axes promote the initiation and the progression of ESCC.
Results
Aberrant upregulation of NSUN2 plays an oncogenic role in ESCC
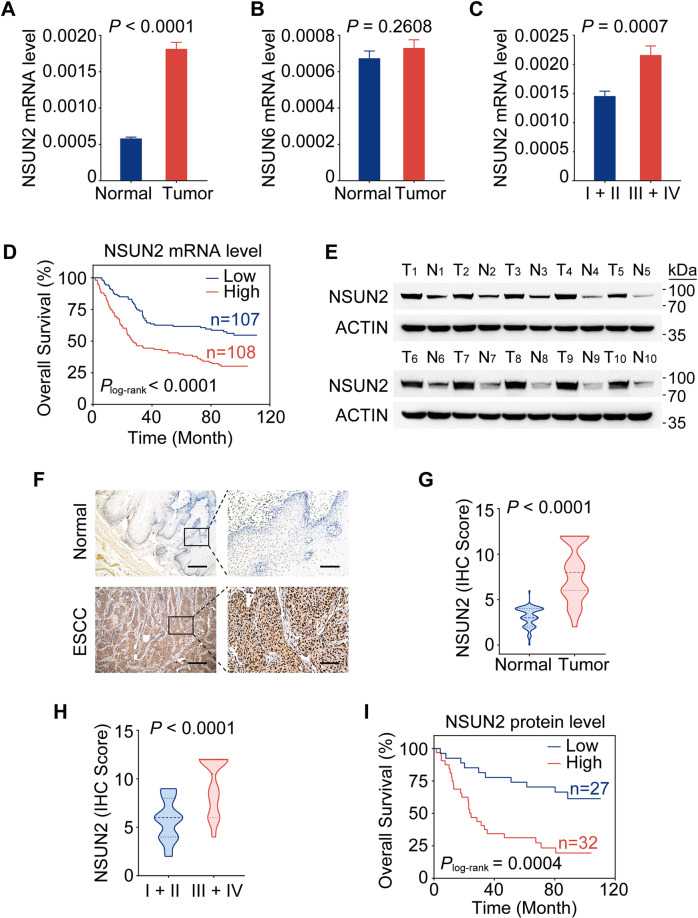
To determine the role of mRNA m5C modification in ESCC, we evaluated the expression levels of two major mRNA m5C methyltransferases NSUN2 and NSUN6 in an ESCC cohort (n = 215; Supplementary Table 1) from Sun Yat-sen University Cancer Center (SYSUCC) using qRT-PCR. We found aberrantly higher levels of NSUN2 RNA in tumors than in adjacent normal tissues (Fig. 1A). However, no obvious difference of NSUN6 RNA was observed (Fig. 1B). These findings were further verified in a public microarray dataset (GSE53625) consisting of 179 paired ESCC samples (Supplementary Fig. 1A, B). Furthermore, higher NSUN2 RNA was significantly associated with advanced ESCC tumor stage (Supplementary Table 2). Patients with stages III/IV ESCCs showed higher NSUN2 levels than those with stages I/II ESCCs (Fig. 1C). Kaplan–Meier analysis revealed that patients with high NSUN2 RNA levels had shorter overall survival times (OS) than those with low levels (Fig. 1D), with an adjusted HR of death for high level being 1.94 (95% CI, 1.32–2.85) (Supplementary Table 3).
Fig. 1. Elevated NSUN2 expression correlates with poor clinical outcomes in patients with ESCC.
A, BNSUN2 RNA levels were significantly higher in ESCC tumors than in paired normal tissues (n = 215; SYSUCC cohort) (A), but NSUN6 showed no obvious difference (B). C Increased NSUN2 RNA levels were observed in stage III/IV ESCC tumors (n = 109) than in stage I/II tumors (n = 106). D Kaplan–Meier estimates of survival time of patients in the SYSUCC cohort by different NSUN2 RNA levels in tumors. The median survival time for patients with high NSUN2 (≥median) was 26.9 months, significantly shorter than 87.7 months in patients with low NSUN2 (<median), with the adjusted HRs (95% CI) for death of high NSUN2 level being 1.94 (1.32–2.85). HR hazard ratio, CI confidence interval. E Western blotting analysis showing higher levels of NSUN2 protein in ESCC tumors than in paired normal tissues (n = 10). T tumor tissues, N paired normal tissues. F Representative immunohistochemical staining (IHC) images of NSUN2 protein levels in ESCC tumors and in paired normal tissues. Scale bar, 500 μm (left) and 100 μm (right). G, H NSUN2 protein levels were significantly higher in ESCC tumors than in paired normal tissues (G; n = 59; SYSUCC cohort), and in stage III/IV tumors (n = 34) than in stage I/II tumors (n = 25) (H). I Kaplan–Meier estimates of survival time of patients by different NSUN2 protein levels in tumors. The median survival time for patients with high NSUN2 (IHC score > 6; 24.2 months) was significantly shorter than those with low NSUN2 (IHC score ≤ 6; 88.4 months), with the adjusted HRs (95% CI) for death of high NSUN2 level being 3.52 (1.46–8.49). Data are represented as mean ± SEM in (A–C) and violin plots in (G) and (H). The centerlines of the violin plots represent median, while the upper and lower hinges indicate 25th and 75th percentiles, respectively. P values were calculated by two-sided paired Wilcoxon signed-rank test in (A), (B) and (G), and two-sided Mann-Whitney test in (C) and (H). Two-sided log-rank test was used in (D) and (I).
We further assessed the protein levels of NSUN2 in ESCC and observed that NSUN2 protein was expressed at significantly higher levels in ESCC tumors than in paired normal tissues by western blotting (n = 10; Fig. 1E) and by immunohistochemical staining (IHC) (n = 59; Fig. 1F, G; Supplementary Table 4). Consistently, NSUN2 protein levels were also significantly correlated with tumor stages (Supplementary Table 5). Higher NSUN2 protein levels were found in stages III/IV ESCCs than in stages I/II ESCCs (Fig. 1H) and were correlated with worse prognosis (Fig. 1I; Supplementary Table 6). These results suggest that NSUN2 is frequently upregulated in ESCC and might serve as a prognostic indicator for ESCC patients.
NSUN2 expression is positively regulated by E2F1
To explore why NSUN2 is overexpressed in ESCC, we looked at genomic alterations including CNV and methylation status of NSUN2 in ESCC tissues derived from different datasets but the results were negative (Supplementary Fig. 2A, B). We then performed in silico analysis using five publicly available databases (ChIPBase, GTRD, AnimalTFDB, JASPAR and hTFtarget) to search for cis-element(s) in the NSUN2 promoter region (from −1000 bp to the transcription start site) and also looked at a public microarray dataset of 179 paired ESCC (GSE53625) to search for the transcription factors (TFs) positively correlated with NSUN2 levels (r > 0.3, P < 0.05). By overlapping the results from these approaches, we identified four potential TFs for NSUN2 (Fig. 2A; Supplementary Fig. 2C). Among the four TFs, only silencing E2F1 significantly downregulated NSUN2 at both RNA and protein levels in ESCC cells (Fig. 2B, C; Supplementary Fig. 2D). However, no significant alteration of NSUN6 level was observed when E2F1 was silenced in ESCC cells (Supplementary Fig. 2E). In silico analysis indicated that cis-element for E2F1 might be located between −623 and −613 bp upstream of the NSUN2 transcriptional start site (Fig. 2D), which was verified by chromatin immunoprecipitation (ChIP) assays (Fig. 2E). Moreover, luciferase reporter assays showed a significant increased transcriptional activity of NSUN2 wild-type promoter as compared to the empty vector or vector with putative E2F1 binding motif mutant NSUN2 promoter (Fig. 2F). This upregulation of luciferase expression was abrogated when E2F1 was silenced (Fig. 2F). Further analysis of the GSE53625 dataset and our in-house 215 paired ESCC cohort showed that E2F1 levels were significantly upregulated in ESCC tumors (Fig. 2G; Supplementary Fig. 2F). Consistently, NSUN2 levels were also positively correlated with E2F1 levels in our 215 paired ESCC samples (Fig. 2H). All these data suggest an E2F1-dependent positive regulation on NSUN2 expression in ESCC.
Fig. 2. Transcription factor E2F1 positively regulates NSUN2 expression in ESCC.
A In silico analysis of potential transcription factors in NSUN2 promoter region. B, C Relative NSUN2 RNA (B) and protein (C) levels in ESCC cells with or without knockdown of each of the four transcription factors indicated in (A). D Schema of the putative E2F1 binding site in NSUN2 promoter and the primers used for chromatin immunoprecipitation (ChIP) analysis. Highlighted are the consensus and mutant sequences for E2F1 binding. E ChIP-qPCR analysis of cells with anti-E2F1 antibody or IgG control. Left panel shows qPCR results and the right panel shows the images of agarose gel electrophoresis of the qPCR products. F Luciferase reporter assays in cells co-transfected with the indicated plasmids or siRNA for 48 h (upper panel). Knockdown efficiency of E2F1 was shown in the lower panel. pGL4-wt-promoter, pGL4-NSUN2-wt-promoter; pGL4-mut-promoter, pGL4-NSUN2-mut-promoter. G E2F1 RNA levels were significantly higher in ESCC tumors than in paired normal tissues (n = 215, SYSUCC cohort). H Spearman’s correlations between RNA levels of NSUN2 and E2F1 in ESCC tumors (n = 215). The center red line is the regression line. r, correlation coefficient. Data are shown as mean ± SEM in (B), (E), (F) and (G). All data are from at least three independent experiments. ACTIN was used as a control in (C) and (F). P values were calculated by two-sided Student’s t test (*P < 0.05, **P < 0.01 and ***P < 0.001) in (B), (E) and (F), and two-sided paired Wilcoxon signed-rank test in (G).
NSUN2 enhances tumorigenesis and progression of ESCC
We then investigate the role of NSUN2 by changing its expression in ESCC cells. Overexpressing NSUN2 substantially promoted cell proliferation, migration and invasion, while silencing NSUN2 showed opposite effects (Fig. 3A−D; Supplementary Fig. 3A, B, E, F). However, overexpressing NSUN2 catalytic mutants (MUT1 or MUT2) [9, 12] had no obvious effects on malignant phenotypes of ESCC cells (Fig. 3E, G; Supplementary Fig. 3C, G). Moreover, overexpressing wild-type but not mutant NSUN2 substantially reversed the inhibitory effects of NSUN2 knockdown on malignant phenotypes of ESCC cells (Fig. 3F, H; Supplementary Fig. 3D, H). These in-vitro results suggest an oncogenic role of NSUN2 in ESCC cells.
Fig. 3. NSUN2 promotes malignant phenotypes of ESCC cells and enhances the tumorigenesis and development of Nsun2 knockout mice.
A−D Effects of NSUN2 overexpression or knockdown on abilities of ESCC cell proliferation (A, B), migration and invasion (C, D). E, G Wild-type but not mutant NSUN2 enhanced proliferation (E), migration and invasion abilities (G) of ESCC cells. F, H Wild-type but not mutant NSUN2 reversed the decreased abilities of proliferation (F), migration and invasion (H) in NSUN2-depleted ESCC cells. WT, wild-type NSUN2 plasmids; MUT1, NSUN2 plasmid with a point mutation at catalytic site (C321A); MUT2, NSUN2 plasmid with point mutations at both catalytic site (C321A) and releasing site (C271A). All three plasmids were insensitive to shNSUN2 plasmid. I Schematic diagram of the timeline of establishing the 4-NQO-induced mouse model of ESCC. Colored arrows indicate the time when different events occurred. J Pathological features (upper panel) or NSUN2 levels (lower panel) of normal esophagus or esophagus tissues after 4-NQO withdrawal 4 or 8 weeks from Nsun2+/+ mice, as estimated by hematoxylin and eosin (H&E) staining or immunohistochemical staining (IHC). K−M Morphological images (K), tumor number (L) or tumor size (M) of esophagus collected from Nsun2+/+ mice (n = 10) and their Nsun2+/− (n = 10) littermate after 4-NQO withdraw for 12 weeks. N Pathological features (upper panel) or NSUN2 levels (lower panel) of esophagus collected from Nsun2+/+ mice and their Nsun2+/− littermate after 4-NQO withdraw for 12 weeks, as estimated by H&E or IHC. O Kaplan–Meier analysis showing significantly longer survival times in Nsun2+/− mice (n = 10) than in Nsun2+/+ mice (n = 10). Results of (A−H) are from at least three independent experiments. Data in (A−H) and (L, M) are displayed as mean ± SEM. Scale bars are 200 µm in (J) and (N). P values were calculated by two-sided Student’s t test in (A, H), two-sided Mann–Whitney test in (L, M), and two-sided log-rank test in (O). *P < 0.05, **P < 0.01 and ***P < 0.001.
We next generated Nsun2 knockout mice (Nsun2+/−, Supplementary Fig. 4A) to determine the oncogenic role of Nsun2 in vivo. A 14 bp shift due to the deletion mutation located on one allele of Nsun2 exon 4 was observed in the Nsun2+/− mice by Sanger sequencing (Supplementary Fig. 4B). Relative to those of Nsun2+/+ littermates, esophageal extracts of Nsun2+/− mice displayed significant reduction in NSUN2 protein levels (Supplementary Fig. 4C). We then used the Nsun2+/− mice and their wild-type (Nsun2+/+) littermates to induce ESCC by treating them with chemical carcinogen 4-nitroquilonine N-oxide (4-NQO) (Fig. 3I). We observed esophageal atypical hyperplasia lesions or squamous cell carcinoma in Nsun2+/+ mice after 4-NQO withdrawal for 4 or 8 weeks. IHC assays showed sequential upregulation of NSUN2 protein in normal esophageal epithelium, atypical hyperplasia lesions and ESCC tumor tissues (Fig. 3J). As expected, all (10/10) Nsun2+/+ mice developed esophageal masses after 4-NQO withdrawal for 12 weeks, but not all (8/10) Nsun2+/− mice at this timepoint (Fig. 3K). Both tumor number and tumor size of Nsun2+/− mice were smaller than those of Nsun2+/+ mice (Fig. 3L, M). Histopathological analysis showed advanced esophageal tumor stages of Nsun2+/+ mice than those of Nsun2+/− mice (Fig. 3N; Supplementary Fig. 4D). Consistently, Nsun2+/+ mice had worse prognosis than Nsun2+/− mice (Fig. 3O). These findings suggest that NSUN2 plays a critical role in the tumorigenicity and progression of ESCC.
NSUN2-mediated m5C hypermethylation activates PI3K/AKT and ERK/MAPK signaling in ESCC
We next explore whether the oncogenic role of NSUN2 is m5C-dependent in ESCC. We performed RNA bisulfite sequencing (RNA-BisSeq) [9] on poly(A)-enriched RNAs purified from seven paired ESCC samples (Supplementary Table 7) to elucidate the transcriptomic m5C profile of ESCC. Conversion rates (C to T) of the methylation conversion control Dhfr were all >99%. We identified 115,187 m5C sites in 8263 transcripts. More than 90% of these m5Cs occurred in mRNAs (Supplementary Fig. 5A) and were enriched in 5’UTR, CDS and 3′UTR (Supplementary Fig. 5B). Moreover, the identified m5Cs were particularly accumulated in regions immediately downstream of translation initiation sites (Supplementary Fig. 5C) and were enriched in the CG-rich environments (Supplementary Fig. 5D).
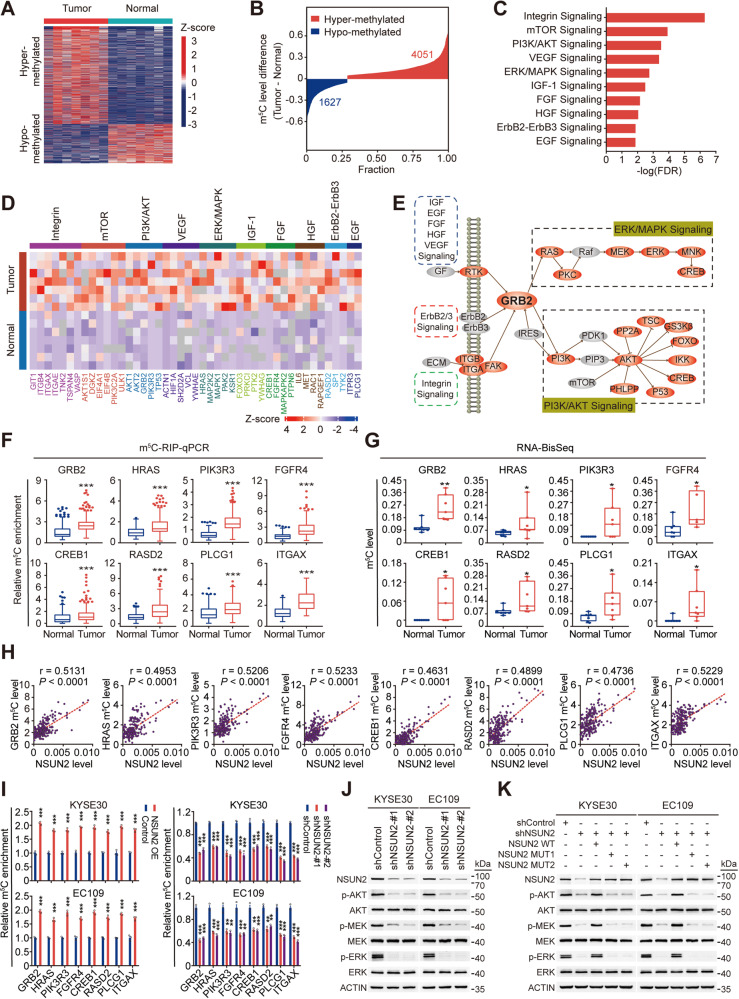
Among the 115,187 m5Cs, 4051 sites within 1362 transcripts were hypermethylated while 1627 sites within 626 transcripts were hypomethylated in tumors compared with those in normal tissues (Fig. 4A, B; Supplementary Data 1), indicating that m5C hypermethylation is a frequent event in ESCC. Furthermore, we found that RNA levels of NSUN2 but not NSUN6 were significantly upregulated in tumors from the seven paired ESCC samples (Supplementary Fig. 5E), suggesting that NSUN2 may be the main methyltransferase mediating the formation of aberrant mRNA m5C hypermethylation in ESCC. Then, pathway enrichment analysis using IPA software showed that the m5C-hypermethylated transcripts in tumors were mainly enriched in cancer-related pathways, such as PI3K/AKT, ERK/MAPK and cell-surface activated receptor-related pathways (Fig. 4C). Indeed, many oncogenes involved in these pathways exhibited m5C-hypermethylated in tumors (Fig. 4D, E). We randomly selected several transcripts involved in these pathways and verified their upregulated m5C levels in tumors (n = 215; Fig. 4F; Supplementary Table 1) using m5C-RIP-qPCR, which was consistent with our RNA-BisSeq results (Fig. 4G). Moreover, m5C levels of these transcripts were positively correlated with NSUN2 RNA levels in ESCC tumors (n = 215; Fig. 4H). Consistently, silencing NSUN2 decreased m5C levels of these transcripts in ESCC cells, while overexpressing NSUN2 produced opposite effects (Fig. 4I). In NSUN2-depleted cells, wild-type but not mutant NSUN2 restored m5C levels of these transcripts (Supplementary Fig. 5F).
Fig. 4. NSUN2 stimulates m5C hypermethylation in ESCC and activates PI3K/AKT and ERK/MAPK signaling.
A, B Heatmap (A, left) and distribution (B) of the differential methylation levels of m5C sites between ESCC tumors and paired normal tissues (n = 7). C Canonical pathway analysis of genes with m5C hypermethylated transcripts (n = 1362) in ESCC tumors than in paired normal tissues (n = 7) using IPA software. D Heatmap (upper) showing m5C hypermethylation of 45 representative genes involved in ten canonical cancer-related pathways in ESCC tumors. E Schematic diagram of genes involved in these cancer-related pathways. Genes with m5C hypermethylated transcripts in tumors are highlighted in red. F, G Substantially hypermethylated m5C of representative transcripts involved in these cancer-related pathways in ESCC tumors than in paired normal tissues by m5C-RIP-qPCR (F, n = 215) or RNA-BisSeq (G, n = 7). H Spearman’s correlation analysis between NSUN2 RNA levels and m5C levels of transcripts mentioned in (F) in ESCC tumors (n = 215). I Relative m5C levels of transcripts mentioned in (F) were increased or decreased in ESCC cells with NSUN2 overexpression or knockdown. J, K Western blotting analysis showing that NSUN2 depletion substantially suppressed AKT, MEK and ERK phosphorylation and activation in ESCC cells (J), while wild-type but not mutant NSUN2 reversed the decrease of AKT, MEK and ERK phosphorylation and activation caused by NSUN2 knockdown (K). Heatmaps in (A, right) and (D, lower) showing the z-score of m5C levels. Colors from blue to red indicate low to high. Data in (F, G) are displayed in boxplots. Data in (I) are shown as mean ± SEM from three independent experiments. The relative m5C levels in target transcripts in (F) and (I) were evaluated with input normalization. ACTIN was used as a control in (J, K). P values were calculated by two-sided paired Wilcoxon signed-rank test in (F), two-sided Mann–Whitney test in (G) and two-sided Student’s t test in (I) (*P < 0.05, **P < 0.01 and ***P < 0.001).
Previous studies have reported that PI3K/AKT and ERK/MAPK pathways are two major downstream pathways of multiple cell-surface activated receptor-related (VEGF, integrin, etc.) pathways [24, 25], and many transcripts in these pathways were hypermethylated in our results (Fig. 4E). We thus hypothesized that NSUN2-mediated m5C hypermethylation might promote ESCC tumor progression mainly by regulating PI3K/AKT and ERK/MAPK signaling. As expected, NSUN2 knockdown suppressed the activation of PI3K/AKT and ERK/MAPK pathways (Fig. 4J). This suppression could be reversed by overexpression of wild-type but not mutant NSUN2 (Fig. 4K). These results indicate that NSUN2-mediated m5C hypermethylation may trigger PI3K/AKT and ERK/MAPK pathways to promote ESCC malignancy.
GRB2 is a critical target via which NSUN2 stimulates PI3K/AKT and ERK/MAPK signaling
To identify downstream effectors involved in NSUN2-mediated activation of PI3K/AKT and ERK/MAPK signaling, we assessed potential targets with hypermethylated-m5Cs in ESCC tumors. We focused on the adaptor protein GRB2 for the following reasons. Firstly, GRB2 plays a central role in signal transduction between cell-surface receptors and PI3K/AKT and ERK/MAPK signalling, leading to simultaneous activation of both PI3K/AKT and ERK/MAPK pathways [26, 27] (Fig. 4E). Secondly, results above had showed that NSUN2 positively regulated m5C levels of GRB2 RNA via its m5C catalytic activity (Fig. 4I; Supplementary Fig. 5F), indicating that GRB2 was a substrate of NSUN2. Further experiments showed that GRB2 silencing substantially inhibited PI3K/AKT and ERK/MAPK signaling in ESCC cells (Fig. 5A). Overexpressing GRB2 partly restored the activation of PI3K/AKT and ERK/MAPK pathways in NSUN2-depleted cells (Fig. 5B), indicating that GRB2 is a key target of NSUN2 to activate both pathways.
Fig. 5. NSUN2 stimulates oncogenic PI3K/AKT and ERK/MAPK signaling by enhancing GRB2 mRNA stability.
A Western blotting analysis showing substantial inhibition of AKT, MEK and ERK phosphorylation in GRB2-depleted ESCC cells. B Western blotting analysis showing that overexpression of GRB2 partially rescued the inhibitory effect of NSUN2 knockdown on phosphorylation of AKT, MEK and ERK in ESCC cells. C, D Significant increase or decrease in GRB2 mRNA (C) and protein (D) levels in NSUN2 overexpression or knockdown ESCC cells. E, F Wild-type but not mutant NSUN2 reversed the decrease of GRB2 mRNA (E) and protein (F) levels caused by NSUN2 depletion. G Expression levels of GRB2, p-AKT, p-MEK or p-ERK in esophagus tissues of different pathological stage from Nsun2+/+ mice. H Expression levels of GRB2, p-AKT, p-MEK or p-ERK in esophagus tissues from Nsun2+/+ mice and their Nsun2+/− littermate 12 weeks after 4-NQO withdraw. I, J Spearman’s correlation analysis between GRB2 and NSUN2 mRNA levels (I) or between GRB2 mRNA and m5C levels (J) in ESCC tumors (n = 215) from the SYSUCC cohort. K RNA stability assays displaying reduced GRB2 mRNA half-life in NSUN2 knockdown ESCC cells compared with the control cells by qRT-PCR at the indicated time points after treatment with 5 µg/mL actinomycin D. L Wild-type but not mutant NSUN2 reversed the decrease of GRB2 mRNA half-life induced by NSUN2 silencing. M Relative luciferase activity or mRNA levels of luciferase reporter gene with wild-type GRB2-m5C site (GRB2-WT) or mutant m5C site (GRB2-MUT) in control or NSUN2 knockdown ESCC cells. Data are displayed as mean ± SEM in (C), (E), (K–M). All data are from at least three independent experiments. Two-sided Student’s t tests were used in (C), (E), (K–M) (*P < 0.05, **P < 0.01 and ***P < 0.001. ns not significant). ACTIN served as a control in (A), (B), (D) and (F–H).
We then explored the effect of NSUN2-mediated m5C formation on GRB2. NSUN2 knockdown significantly reduced the GRB2 RNA and protein levels in ESCC cells, while NSUN2 overexpression exhibited opposite results (Fig. 5C, D). Moreover, overexpressing wild-type but not mutant NSUN2 recovered the GRB2 RNA (Fig. 5E) and protein (Fig. 5F) levels in NSUN2-knockdown cells. Consistently, we found both GRB2 levels and the activation levels of PI3K/AKT and ERK/MAPK pathways gradually increased in normal esophageal epithelium, atypical hyperplasia lesions and ESCC tumors in the 4-NQO-induced mouse model of ESCC (Fig. 5G). Higher levels of GRB2 and the activation levels of both pathways were also observed in esophageal tissues of Nsun2+/+ mice than in those of Nsun2+/− mice (Fig. 5H). Moreover, positive correlations were found between GRB2 RNA levels and NSUN2 RNA levels (Fig. 5I) or GRB2 m5C levels (Fig. 5J) in ESCCs (n = 215; Supplementary Table 1). Since the m5C site of GRB2 is in its 3′UTR, it is possible that NSUN2-mediated m5C modification maintains GRB2 expression by enhancing its mRNA stability. We treated ESCC cells with actinomycin D and found that NSUN2 silencing significantly reduced half-life of GRB2 RNA (Fig. 5K). This reduction could be restored by overexpressing wild-type but not mutant NSUN2 (Fig. 5L). Luciferase reporter assays showed substantial decreased luciferase expression of vector with wild-type GRB2 3′UTR (GRB2-WT), but not of that with m5C site mutant GRB2 3′UTR (GRB2-MUT) in NSUN2-depleted cells (Fig. 5M). These results suggest a central role of GRB2 in the NSUN2-enhanced activation of PI3K/AKT and ERK/MAPK signaling.
LIN28B recognizes m5C modification of GRB2 and stabilizes GRB2 mRNA
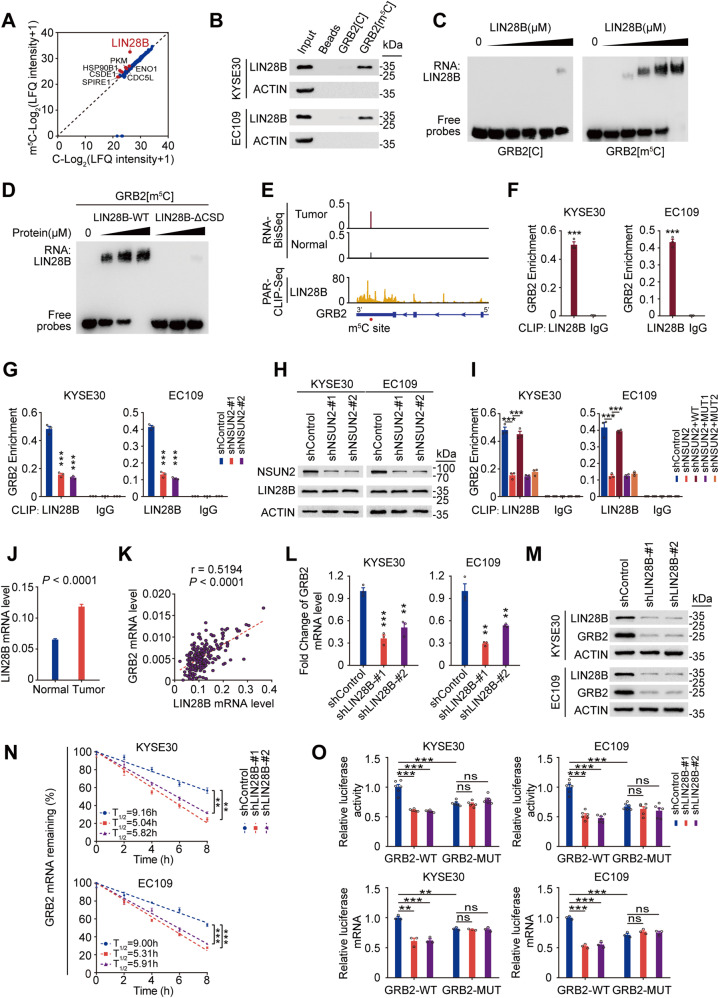
It is known that RNA methylation regulates its target RNA by reader proteins [28]. Since m5C modification could stabilize GRB2 mRNA, we first examined the effect of the known reader YBX1 on GRB2 and found that silencing YBX1 had no influence on GRB2 levels (Supplementary Fig. 6A). To identify the m5C mediators involved in GRB2 regulation, we performed mass spectrometry analysis of proteins obtained by RNA pulldown using biotin-labelled 50-bp GRB2 or m5C-GRB2 RNA probes (Supplementary Data 2). We identified seven proteins preferentially binding to m5C-GRB2 probes (Fig. 6A; Supplementary Fig. 6B), and among which, only LIN28B was verified by western blotting (Fig. 6B; Supplementary Fig. 6C) and REMSA assays (Fig. 6C). Since LIN28B has a conserved cold shock domain (CSD) similar to YBX1 [29], an m5C reader targeting m5C-modified RNAs through its CSD domain [12, 13], we hypothesis that LIN28B binds m5C-modified GRB2 through CSD domain. REMSA experiment confirmed this notion that truncation of LIN28B CSD domain (LIN28B-ΔCSD) led to reduction in binding affinity of LIN28B toward the m5C-containing GRB2 oligo compared with full-length LIN28B (LIN28B-WT) (Fig. 6D; Supplementary Fig. 6D). To further screen key residues for LIN28B to bind to m5C site of GRB2, we performed structural modeling for LIN28B in complex with the m5C containing GRB2 RNA hexamer oligo using YBX1-m5C RNA complex as a reference model [12]. According the modeled LIN28B-m5C RNA complex structure, W36, M41, F43, D61, H65, and K92 might be the key interacting residues (<3 angstrom), and N38 and D61 had polar contacts with m5C RNA fragment. Furthermore, the residue interaction network generated by RING software [30] showed that there was Van der Waals force between the W36 residue and m5C, which indicated that W36 was critical for the LIN28B-m5C RNA interaction (Supplementary Fig. 6E−G). REMSA assays further confirmed W36 as the key residue for LIN28B to distinguish and bind to m5C-modified GRB2 RNA (Supplementary Fig. 6D; Supplementary Fig. 6H). LIN28B-PAR-CLIP further showed direct binding of LIN28B to GRB2 m5C site (Fig. 6E, F). This interaction was reduced in NSUN2-silenced cells (Fig. 6G) where LIN28B protein level was not altered (Fig. 6H) and overexpressing wild-type but not mutant NSUN2 could reverse this reduced interaction (Fig. 6I). In addition, we found that LIN28B RNA levels were upregulated in ESCC tumors and were positively associated with GRB2 RNA levels (n = 215; Fig. 6J, K; Supplementary Table 1). Since LIN28B could bind and stabilize its target RNAs [31], we assumed that NSUN2-enhanced GRB2 stability depends on LIN28B. As expected, LIN28B silencing markedly decreased mRNA (Fig. 6L) and protein (Fig. 6M) levels of GRB2, and also reduced GRB2 mRNA stability (Fig. 6N). In addition, luciferase expression of GRB2-WT was strongly decreased in LIN28B-knockdown cells, whereas GRB2-MUT had no such effects (Fig. 6O). These findings suggest that LIN28B is a novel m5C mediator recognizing the m5C-modified GRB2 and stabilizing GRB2 mRNA.
Fig. 6. LIN28B stabilizes GRB2 mRNA by recognizing its m5C site.
A Scatter plot of proteins binding to 50 bp GRB2 probe with or without m5C modification in KYSE30 cells. The filled red circles indicate proteins preferentially binding to GRB2[m5C] probes. B Western blotting analysis of potential GRB2[m5C] binding proteins obtained from RNA pull-down assays with 50 bp GRB2 probe with or without m5C modification shows specific association of LIN28B with GRB2[m5C] probes. C RNA Electrophoretic mobility shift (REMSA) assays of purified FLAG-tagged LIN28B with unmethylated or methylated GRB2 probes. The probes were maintained constantly while a gradient of 0–8 μM purified LIN28B was added to the reactions. D REMSA assays of GRB2[m5C] probes with purified FLAG-tagged LIN28B (wild-type or CSD domain truncation mutants). E Integrative-genomics-viewer (IGV) profiles showing the m5C levels of GRB2 in tumors and adjacent normal samples as well as the LIN28B-binding groups in PAR-CLIP-Seq data. The filled red circle represents the m5C site (chr17:75318971) in GRB2. F–G PAR-CLIP-qPCR assays showing direct in-vivo binding of LIN28B to GRB2 in ESCC cells (F), and significant reduction of LIN28B binding to GRB2 when NSUN2 was silenced (G). H NSUN2 depletion had no influence on LIN28B protein levels. I Wild-type but not mutant NSUN2 reversed the reduction of LIN28B binding to GRB2 caused by NSUN2 depletion. J LIN28B RNA levels were significantly higher in ESCC tumors than in paired normal tissues (n = 215, SYSUCC cohort). K Spearman’s correlation analysis between LIN28B and GRB2 RNA levels in ESCC tumors (n = 215). L–M LIN28B knockdown substantially decreased the RNA (L) and protein (M) levels of GRB2. N Effects of LIN28B knockdown on mRNA half-life of GRB2 by RNA stability assays. O Luciferase reporter assays of luciferase reporter gene with wild-type GRB2-m5C site (GRB2-WT) or mutant m5C site (GRB2-MUT) in the control or stable LIN28B knockdown ESCC cells. Data in (F), (G), (I), (J), (L), (N) and (O) are displayed as mean ± SEM. All data are from at least three independent experiments. P values are calculated by two-sided Student’s t test (*P < 0.05, **P < 0.01 and ***P < 0.001. ns not significant) in (F), (G), (I), (L), (N) and (O), and by two-sided paired Wilcoxon signed-rank test in (J). IgG served as an isotype control in (F), (G) and (I). ACTIN served as a control in (B), (H) and (M).
We then evaluate the LIN28B binding to other m5C-modified RNAs. A substantial decrease of LIN28B RNA-binding affinity was observed in ESCC cells when NSUN2 was silenced (Supplementary Fig. 6I). By further analyzing RNA-BisSeq and LIN28B-PAR-CLIP-Seq data, we observed a substantial overlap of m5C-modified RNAs and LIN28B-binding target RNAs (Supplementary Fig. 6J). Moreover, ~29% of the m5C sites were localized within the LIN28B peaks (Supplementary Fig. 6K). In addition, a significant enrichment of m5C modifications in LIN28B-bound RNAs was observed (Supplementary Fig. 6L). These findings indicate that LIN28B may also recognize m5C-modified RNAs other than GRB2.
GRB2 serves as an oncogene in ESCC, and the NSUN2-GRB2 axis is clinically relevant to ESCC
We further investigate the role of GRB2 in ESCC. Functionally, GRB2 knockdown dramatically suppressed cell proliferation, migration, and invasion (Supplementary Fig. 7A, B). Clinically, we found higher m5C and RNA levels of GRB2 in tumors than in adjacent normal tissues from our 215 paired ESCC cohort (Fig. 4F; Fig. 7A; Supplementary Table 1−2) and from the seven paired sequencing ESCC samples (Fig. 4G; Fig. 7B; Supplementary Table 7). Furthermore, patients with high GRB2 mRNA or m5C levels had shorter OS than those with low levels (Fig. 7C, D; Supplementary Table 3). Consistently, GRB2 protein levels were significantly higher in ESCC tumors than in paired normal tissues detecting by western blotting (n = 10; Fig. 7E) and IHC assays (n = 59; Fig. 7F, G; Supplementary Table 4−5). Patients with higher GRB2 protein level also had worse prognosis (Fig. 7H; Supplementary Table 6). These findings indicate that GRB2 was an independent prognostic factor in ESCC.
Fig. 7. Clinical significance of NSUN2-GRB2 axes in ESCC.
A, B Aberrant overexpression of GRB2 RNA in ESCC tumors than in paired normal tissues by qRT-PCR (n = 215; A) and by RNA-Seq (n = 7; B). C, D Kaplan–Meier estimates of survival time of patients in SYSUCC cohort (n = 215) by different GRB2 RNA levels (C) or m5C levels (D) in tumors. Median survival time for patients with high GRB2 RNA or m5C levels (≥median) was 33.6 months or 33.6 months compared with 85.9 or 88.4 months in patients with low GRB2 RNA (C) or m5C levels (D) (<median), with the adjusted HRs of 1.78 (95% CI, 1.22–2.61) and 1.70 (95% CI, 1.16–2.49), respectively. E Western blotting assays showing higher protein levels of GRB2 in ESCC tumors than in paired normal tissues (n = 10). F Representative immunohistochemical staining (IHC) images of GRB2 protein in ESCC tumors and in paired normal tissues. Scale bar, 500 μm (left panel) and 100 μm (right panel). G GRB2 protein levels were higher in ESCC tumors than in paired normal tissues (n = 59) as indicated by the IHC score. H Kaplan–Meier estimates of survival time of patients by different GRB2 protein levels in tumors. The median survival time for patients with high GRB2 (IHC score > 6) was 22.8 months compared with 85.4 months in patients with low GRB2 (IHC score ≤ 6), with the adjusted HRs of 3.64 (95% CI, 1.57–8.41). I Representative images showing positive correlations between protein levels of NSUN2 and GRB2 in ESCC specimens. Scale bar, 100 μm. J Statistical analysis of IHC staining showing the percentage of ESCC specimens displaying higher or lower NSUN2 levels and the corresponding GRB2 expression. K Spearman’s correlation analysis between protein levels of NSUN2 and GRB2 in ESCC tumors (n = 59). L A proposed model for the regulatory mechanism of E2F1-NSUN2-m5C/LIN28B-GRB2-PI3K/AKT and ERK/MAPK signaling axes in the tumorigenesis and progression of ESCC. Data represent as mean ± SEM in (A), boxplots in (B) and violin plots in (G). The centerlines of the boxplots and violin plots represent median, while the upper and lower hinges indicate 25th and 75th percentiles, respectively. P values were calculated using the two-sided paired Wilcoxon signed-rank test in (A) and (G), two-sided paired t-test in (B), two-sided Chi-square test in (J) and two-sided log-rank test in (C), (D) and (H). ACTIN served as a control in (E).
Finally, we explored the clinical importance of the NSUN2-GRB2 axis in ESCC. As expected, overexpressing GRB2 rescued the inhibition of malignant phenotypes in NSUN2-depleted cells (Supplementary Fig. 7C, D). Clinically, approximately 89% of the specimens with lower NSUN2 protein level presented weaker IHC staining of GRB2, while nearly 72% of the specimens with higher NSUN2 showed stronger IHC staining of GRB2 (Fig. 7I, J). Spearman’s correlation analysis also revealed a positive correlation between protein level of NSUN2 and GRB2 (Fig. 7K). These results demonstrate the clinical correlation between NSUN2 and GRB2, and reveal an oncogenic role of the NSUN2-GRB2 axis in ESCC.
Discussion
RNA m5C modification is a common posttranscriptional RNA modification participating in many cellular and physiological processes [32]. Abnormal m5C modification is associated with various diseases, including cancer [12, 33], inflammation [14], intellectual disabilities [22], neurodevelopmental disorders [34], infertility [35], and mitochondrial dysfunction [36]. However, the precise correlations between RNA m5C modification and tumor development remains largely unclear. In this study, we described a transcriptome-wide m5C profile in ESCC for the first time, which showed aberrantly increased levels of RNA m5C modification in ESCC tumors due to the overexpression of NSUN2. NSUN2 plays a critical role in ESCC by positively regulating GRB2 through the m5C-LIN28B-based posttranscriptional regulation, while its own transcription is positively regulated by E2F1. Increased expression of GRB2 promoted the development and progression of ESCC by triggering the abnormal activation of PI3K/AKT and ERK/MAPK signaling (Fig. 7L).
RNA m5C modification is catalyzed by several methyltransferases [11], among which, NSUN2 has attracted increasing attentions because of its oncogenic role in various types of cancers [12, 23, 37–39]. Our results in this study are consistent with previous findings and extend the oncogenic role of NSUN2 to ESCC. We found aberrant overexpression of NSUN2 in ESCCs, which was highly associated with poor prognosis and advanced tumor stages, suggesting a prognostic value for NSUN2. In-vitro experiments showed that NSUN2 promoted malignant phenotypes of ESCC cells dependent on its methyltransferase activity, suggesting an m5C-dependent oncogenic role of NSUN2. In according with in-vitro data, NSUN2 silencing markedly suppressed ESCC tumor initiation and progression in the 4-NQO-induced ESCC model in transgenic mice, indicating that NSUN2 overexpression may be an early molecular event of ESCC. Our findings demonstrate that NSUN2 is essential for the malignancy of ESCC in an m5C-dependent manner. As a previous study [21] has indicated that NSUN2 is not expressed in quiescent stem and undifferentiated progenitor cells but highest expressed in committed progenitors in mouse skin squamous cell carcinoma model, it would be interesting to investigate differences in tumor cell population ratios between different tumor models.
Another interesting finding is the identification of E2F1 as an NSUN2 transcriptional activator. In this study, we found that overexpression of NSUN2 in ESCC was not caused by genomic changes as indicated by no significant alterations of mutations, CNV and DNA methylation status at NSUN2. Integrated analysis suggests that E2F1 was a potential regulator of NSUN2. E2F1 is a well-known TF that has been shown to be aberrant expressed in ESCC tumors [40]. Consistently, in our study, we demonstrated that E2F1 was upregulated in ESCC tumors and was positively correlated to NSUN2 expression, supporting a positive regulation of NSUN2 expression by E2F1. Moreover, we demonstrated that E2F1 could bind to the promoter region of NSUN2, stimulating NSUN2 transcription. These findings reveal for the first time that overexpression of NSUN2 in ESCC may be partially mediated by trans-element(s).
To further address the oncogenic function of NSUN2-mediated m5C methylation in ESCC, we provided an RNA m5C landscape in ESCC and demonstrated an oncogenic role of RNA m5C-hypermethylation in ESCC. Notably, genes with hypermethylated-m5Cs in ESCC tumors were significantly enriched in PI3K/AKT and ERK/MAPK pathways. It is known that both PI3K/AKT and ERK/MAPK pathways play vital roles in various cellular processes [25, 41]. Dysfunction of both pathways is involved in many diseases such as malignant tumors [25, 41]. Several studies have reported that ESCC patients show abnormal activation of PI3K/AKT and ERK/MAPK signaling, which may provide useful therapeutic targets of ESCC [42, 43]. However, how these two pathways are activated in ESCC and by what remains largely unclear. In the present study, we demonstrated that aberrant expression of NSUN2 could trigger abnormal activation of PI3K/AKT and ERK/MAPK signaling via stabilizing GRB2 mRNA. It has been reported that GRB2, as a growth factor receptor-bound protein, is involved in various biological processes, such as cell growth, proliferation, and metabolism [26]. GRB2 plays a central role between cell-surface activation receptors (RTK, integrin, etc.) and other downstream signaling pathways, especially PI3K/AKT and ERK/MAPK signalling [26, 27, 44]. Abnormal expression of GRB2 promotes tumor malignancies by activating both PI3K/AKT and ERK/MAPK pathways [26, 27, 44]. Here, we have linked aberrant m5C modifications in GRB2 to the development and progression of ESCC via PI3K/AKT and ERK/MAPK pathways. NSUN2 silencing attenuated PI3K/AKT and ERK/MAPK signaling via decreasing both m5C and mRNA levels of GRB2, while GRB2 overexpression reversed the inhibition of malignant cellular phenotypes in NSUN2-depleted cells, suggesting that GRB2 is a key mediator of malignancy induced by abnormal NSUN2-meidated m5C modification in ESCC. Our results suggest the central role of NSUN2-m5C-GRB2-PI3K/AKT and ERK/MAPK axes to the pro-tumorigenic effect of NSUN2 in ESCC and provide a new acting model for NSUN2-mediated regulation of ESCC progression. Moreover, a number of reports have indicated that GRB2 overexpression is associated with poor prognosis in cancers [26] including ESCC [45]. Our results are in accordance with these studies showing that both mRNA and protein level of GRB2 were upregulated in ESCC tumors and high level of GRB2 predicted poor prognosis. Furthermore, GRB2 overexpression was previously indicated to be related to lymph node metastasis in ESCC [45], indicating GRB2 would play critical roles in metastasis in ESCC. This observation supports the results of our study showing that GRB2 promotes invasion and metastasis in ESCC cells. Until now, mechanisms controlling the expression of GRB2 remain unclear. Regulation of GRB2 by miRNAs binding to its 3′UTR such as miR-433-3p was previously shown in ESCC [46], suggesting that targeting GRB2-asscocited miRNAs may have therapeutic benefits in ESCC. In addition, a potential antitumor drug F806 was previously reported to downregulate GRB2 level to inhibit cellular proliferation signaling network in ESCC [47]. All these findings suggest that reducing the GRB2 level could arrest the malignant growth of cancers, which may provide a potential treatment strategy for ESCC. In our current research, we have shown that the m5C site in the 3′UTR of GRB2 RNA plays critical roles in GRB2 upregulation through enhancing GRB2 mRNA stability. Our findings have extended the regulatory mechanism of GRB2 expression from an epigenetic perspective and revealed a new mechanism for regulating GRB2 expression through m5C modification, indicating that downregulation of m5C modification in GRB2 may also have therapeutic benefits in ESCC via decreasing GRB2 expression. As GRB2 has been suggested to be a therapeutic target for cancers by targeting its SH2/SH3 domains [48, 49], it may be interesting to further study the synergistic anticancer effect of GRB2 and NSUN2-mediated m5C modification in ESCC. Since other oncogenic transcripts, such as PIK3R3, which has been previously shown to trigger the activation of the PI3K/AKT pathway in ESCC cells [50], also exhibit abnormal m5C modification, it would be interesting to further explore whether m5C modification has the same regulatory function as GRB2 on various transcripts or cancer-related signaling.
Another interesting finding is a new RNA m5C mediator. The biological importance of RNA methylation relies on reader proteins [28]. Previous studies have reported ALYREF and YBX1 as m5C readers [9, 12, 13]. In our study, we showed an m5C-mediated RNA stabilization of GRB2, which was YBX1-independent. Then, by mass spectrometry analysis, we have showed that LIN28B preferentially interacts with m5C-modified GRB2. This notion is further confirmed by several experiments showing that the binding of LIN28B to GRB2 is m5C-dependent. LIN28B is a known oncoprotein aberrantly expressed in a subset of human cancers [51], including ESCC [52], which is consistent with our results. It contains a conserved CSD domain [29], which is necessary for YBX1 binding to m5C-modified RNAs [12]. In our study, we have identified a critical role of CSD domain for LIN28B binding to m5C-carrying GRB2. By performing structure modeling analysis and in-vitro RNA-protein interaction assays, we have identified the LIN28B as an m5C mediator preferentially recognizing m5C-carrying GRB2 RNA through the W36 residue in its CSD domain. The mode of LIN28B binding to m5C-modified GRB2 RNA is extremely similar to YBX1, which binds to m5C-modified RNAs through the indole ring of W65 in its CSD [12]. However, this binding mechanism is different from that of ALYREF, which recognizes and interacts with m5C-modified RNAs mainly through a conserved positively charged residue, K171 [9]. It has been shown that LIN28B regulates RNA stability mainly through inhibiting let-7 microRNAs biogenesis [53] or through directly binding to its target RNAs [31, 54]. As our observations suggest, LIN28B could indeed stabilize GRB2 transcripts. However, this stabilization is m5C-dependent, which might be a novel mechanism of LIN28B-dependent regulation to its target RNAs. Our results strongly indicate that LIN28B is likely an m5C mediator stabilizing m5C-modified GRB2. This function is similar to YBX1 that plays an essential role in maintaining m5C-carrying RNA stability [12] but not ALYREF that exerts an RNA-export-promoting function through recognizing m5C-modified RNAs [9]. Since the m5C modification was located in 3′UTR of GRB2 RNA, which was previously shown to be bound by miR-433-3p in ESCC [46], it would be worth exploring the association between m5C modification and miRNA binding in GRB2 3′UTR. Previous study has suggested that presence of RNA m6A methylation on some transcripts could affect the RBP-RNA interactions and the miRNA targeting in 3′UTR region of RNAs [55], whether m5C modification exerts similar regulatory function remains to be further clarified. Although we have also shown the overlap of LIN28B-binding targets and m5C-modified RNAs, whether LIN28B is a common reader and the exact molecular mechanism for the association of LIN28B and other m5C-modified RNAs are warranted to further investigation.
In conclusion, our current work elucidates a vicious role of the NSUN2-m5C-GRB2-PI3K/AKT and ERK/MAPK signaling axes in the initiation and the progression of ESCC. These findings illustrate an m5C-mediated epigenetic regulation mechanism of ESCC and highlight the opportunity for epitranscriptomic-targeted therapy for ESCC.
Materials and methods
Patient sample collection
Written informed consent was obtained from each patient, and this study was approved by the Institutional Review Board of the SYSUCC.
Cell lines and cell culture
Human ESCC cell lines KYSE30 and EC109 were kind gifts from Dr. Xinyuan Guan at SYSUCC.
Cell proliferation, migration and invasion assays
Details of in-vitro functional experimental procedures could see in Supplementary Materials and methods.
RNA extraction and quantitative real-time PCR (qRT-PCR)
The primer sequences are shown in Supplementary Table 8.
Western blotting analysis
Antibodies against the interest proteins are shown in Supplementary Table 9.
Chromatin immunoprecipitation (ChIP) assays
Specific primers used are listed in Supplementary Table 8. qPCR products were used for agarose gel electrophoresis.
4-NQO-induced ESCC model in Nsun2 knockout transgenic mice
Nsun2 knockout (Nsun2+/−) or Nsun2 wild-type (Nsun2+/+) C57BL/6J mice were donated from Nanjing Medical University. Animal experiments were carried out with protocols and guidelines approved by the Institutional Animal Care and Use Committee of SYSUCC.
Construction of RNA-BisSeq and RNA-Seq libraries
The procedure was performed according to a previous study with some modifications [9]. Analysis of the Dhfr spike-in showed C to T conversion rates >99%.
Differential m5C methylation analysis
M5C sites with P ≤ 0.05 and a mean m5C level difference ≥0.05 (|mean m5C level tumor − mean m5C level normal|) were considered to contain statistically significantly different m5C methylation.
Pathway analysis via Ingenuity Pathway Analysis (IPA)
Genes with m5C-hypermethylated transcripts were uploaded into IPA software for core analysis to identify canonical pathways (FDR ≤ 0.1) [56].
M5C RNA immunoprecipitation followed by qRT-PCR (m5C-RIP-qPCR)
The m5C-RIP-qPCR procedure was performed according to a previous study with some modifications [13]. Gene-specific primers are shown in Supplementary Table 8. Relative m5C levels of the indicated transcripts were evaluated with input normalization.
Photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP)
Cells were cultured in medium supplemented with 4-thiouridine for 14 h, followed by UV light crosslinking. The relative enrichment of the interest transcripts was calculated with input normalization.
RNA interference
Small interfering RNA (siRNA) targeting TFAP2C, SP1, NRF1, E2F1 or YBX1 genes are listed in Supplementary Table 10.
Plasmids, lentivirus production and transduction
Short hairpin RNA specifically targeting NSUN2, LIN28B or GRB2 are listed in Supplementary Table 10.
RNA stability assay
Cells were treated with actinomycin D and the mRNA half-life time was calculated as previously described [57].
Luciferase reporter gene assays
The luciferase activity or RNA level was examined using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) or qRT-PCR, respectively.
RNA pulldown and mass spectrometry analysis
Biotin-labeled RNA fragment containing 50 bp GRB2 RNA sequences with (GRB2[m5C]) or without (GRB2[C]) m5C modification at m5C site (chr17:75318971) were listed in Supplementary Table 10.
RNA electrophoretic mobility shift assays (REMSA)
Assays were performed using the LightShift Chemiluminescent RNA EMSA Kit (Thermo Fisher Scientific, Waltham, MA, USA).
Public data processing
Details are described in Supplementary Materials and methods.
Statistical analysis
All the statistical analyses were performed using SPSS version 20.0 software (SPSS Inc., Chicago, IL, USA) or GraphPad Prism 8.0 software (GraphPad Software, La Jolla, CA, USA). P < 0.05 was considered statistically significant.
Supplementary materials and methods
For the details of other experimental methods, see the Supplementary Materials and methods.
Supplementary information
Acknowledgements
We thank Dr. Bin Shen (Department of Histology and Embryology, Nanjing Medical University, Nanjing, China) for his generous donation of Nsun2 +/− C57BL/6J mice. This study was supported by the Program for Guangdong Introducing Innovative and Entrepreneurial Teams (2017ZT07S096 to DL), Natural Science Foundation of China (U1601229 to DL), National Young Top-notch Talent Support Program (to JZ) and Sun Yat-sen University Intramural Funds (to DL and to JZ).
Author contributions
JZheng and DL conceptualized and supervised the research. JS, XH, YZ, and RB designed and performed most experiments. GW, JZhang and LZhuang performed animal experiments. YY, RL, SD, ZL and ZZ were engaged in biostatistics and bioinformatics analysis. JL, ML, LP, JD, LZeng and SZ were responsible for patient recruitment, biospecimen and clinical data collection. JZheng, DL and JS drafted the paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jiachun Su, Guandi Wu, Ying Ye, Jialiang Zhang.
Contributor Information
Junzhong Lin, Email: linjzh@sysucc.org.cn.
Dongxin Lin, Email: lindx@sysucc.org.cn.
Jian Zheng, Email: zhengjian@sysucc.org.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41388-021-01978-0.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Smyth EC, Lagergren J, Fitzgerald RC, Lordick F, Shah MA, Lagergren P, et al. Oesophageal cancer. Nat Rev Dis Prim. 2017;3:17048. doi: 10.1038/nrdp.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delaunay S, Frye M. RNA modifications regulating cell fate in cancer. Nat Cell Biol. 2019;21:552–9. doi: 10.1038/s41556-019-0319-0. [DOI] [PubMed] [Google Scholar]
- 5.Bourgeois G, Ney M, Gaspar I, Aigueperse C, Schaefer M, Kellner S, et al. Eukaryotic rRNA modification by yeast 5-methylcytosine-methyltransferases and human proliferation-associated antigen p120. PLoS ONE. 2015;10:e0133321. doi: 10.1371/journal.pone.0133321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.David R, Burgess A, Parker B, Li J, Pulsford K, Sibbritt T, et al. Transcriptome-wide mapping of RNA 5-methylcytosine in arabidopsis mRNAs and noncoding RNAs. Plant Cell. 2017;29:445–60. doi: 10.1105/tpc.16.00751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–33. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amort T, Rieder D, Wille A, Khokhlova-Cubberley D, Riml C, Trixl L, et al. Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genom Biol. 2017;18:1. doi: 10.1186/s13059-016-1139-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY, et al. 5-methylcytosine promotes mRNA export-NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. 2017;27:606–25. doi: 10.1038/cr.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khoddami V, Cairns BR. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat Biotechnol. 2013;31:458–64. doi: 10.1038/nbt.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Motorin Y, Lyko F, Helm M. 5-methylcytosine in RNA: detection, enzymatic formation and biological functions. Nucleic Acids Res. 2010;38:1415–30. doi: 10.1093/nar/gkp1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, Li A, Sun BF, Yang Y, Han YN, Yuan X, et al. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat Cell Biol. 2019;21:978–90. doi: 10.1038/s41556-019-0361-y. [DOI] [PubMed] [Google Scholar]
- 13.Yang Y, Wang L, Han X, Yang WL, Zhang M, Ma HL, et al. RNA 5-methylcytosine facilitates the maternal-to-zygotic transition by preventing maternal mRNA decay. Mol Cell. 2019;75:1188–202. doi: 10.1016/j.molcel.2019.06.033. [DOI] [PubMed] [Google Scholar]
- 14.Luo Y, Feng J, Xu Q, Wang W, Wang X. NSun2 deficiency protects endothelium from inflammation via mRNA methylation of ICAM-1. Circ Res. 2016;118:944–56. doi: 10.1161/CIRCRESAHA.115.307674. [DOI] [PubMed] [Google Scholar]
- 15.Schumann U, Zhang HN, Sibbritt T, Pan A, Horvath A, Gross S, et al. Multiple links between 5-methylcytosine content of mRNA and translation. BMC Biol. 2020;18:40. doi: 10.1186/s12915-020-00769-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sajini AA, Choudhury NR, Wagner RE, Bornelov S, Selmi T, Spanos C, et al. Loss of 5-methylcytosine alters the biogenesis of vault-derived small RNAs to coordinate epidermal differentiation. Nat Commun. 2019;10:2550. doi: 10.1038/s41467-019-10020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y, et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013;4:255–61. doi: 10.1016/j.celrep.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Selmi T, Hussain S, Dietmann S, Heiss M, Borland K, Flad S, et al. Sequence- and structure-specific cytosine-5 mRNA methylation by NSUN6. Nucleic Acids Res. 2021;49:1006–22. doi: 10.1093/nar/gkaa1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flores JV, Cordero-Espinoza L, Oeztuerk-Winder F, Andersson-Rolf A, Selmi T, Blanco S, et al. Cytosine-5 RNA methylation regulates neural stem cell differentiation and motility. Stem. Cell Rep. 2017;8:112–24. doi: 10.1016/j.stemcr.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang W. mRNA methylation by NSUN2 in cell proliferation. Wiley Interdiscip Rev RNA. 2016;7:838–42. doi: 10.1002/wrna.1380. [DOI] [PubMed] [Google Scholar]
- 21.Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J, et al. Stem cell function and stress response are controlled by protein synthesis. Nature. 2016;534:335–40. doi: 10.1038/nature18282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khan MA, Rafiq MA, Noor A, Hussain S, Flores JV, Rupp V, et al. Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am J Hum Genet. 2012;90:856–63. doi: 10.1016/j.ajhg.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frye M, Watt FM. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr Bio.l. 2006;16:971–81. doi: 10.1016/j.cub.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 24.Caunt CJ, Sale MJ, Smith PD, Cook SJ. MEK1 and MEK2 inhibitors and cancer therapy: the long and winding road. Nat Rev Cancer. 2015;15:577–92. doi: 10.1038/nrc4000. [DOI] [PubMed] [Google Scholar]
- 25.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 26.Ijaz M, Shahbaz M, Jiang W, Fathy A, Nesa E, Wang F. The role of Grb2 in cancer and peptides as Grb2 antagonists. Protein Pept Lett. 2017;24:1084–95. doi: 10.2174/0929866525666171123213148. [DOI] [PubMed] [Google Scholar]
- 27.Sattler M, Mohi MG, Pride YB, Quinnan LR, Malouf NA, Podar K, et al. Critical role for Gab2 in transformation by BCR/ABL. Cancer Cell. 2002;1:479–92. doi: 10.1016/S1535-6108(02)00074-0. [DOI] [PubMed] [Google Scholar]
- 28.Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Budkina KS, Zlobin NE, Kononova SV, Ovchinnikov LP, Babakov AV. Cold shock domain proteins: structure and interaction with nucleic acids. Biochemistry. 2020;85:1–19. doi: 10.1134/S0006297920140011. [DOI] [PubMed] [Google Scholar]
- 30.Piovesan D, Minervini G, Tosatto SC. The RING 2.0 web server for high quality residue interaction networks. Nucleic Acids Res. 2016;44:W367–74. doi: 10.1093/nar/gkw315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keskin T, Bakaric A, Waszyk P, Boulay G, Torsello M, Cornaz-Buros S, et al. LIN28B underlies the pathogenesis of a subclass of Ewing sarcoma. Cell Rep. 2020;30:4567–83. doi: 10.1016/j.celrep.2019.12.053. [DOI] [PubMed] [Google Scholar]
- 32.Trixl L, Lusser A. The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark. Wiley Interdiscip Rev RNA. 2019;10:e1510. doi: 10.1002/wrna.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng JX, Chen L, Li Y, Cloe A, Yue M, Wei J, et al. RNA cytosine methylation and methyltransferases mediate chromatin organization and 5-azacytidine response and resistance in leukaemia. Nat Commun. 2018;9:1163. doi: 10.1038/s41467-018-03513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33:2020–39. doi: 10.15252/embj.201489282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khosronezhad N, Hosseinzadeh Colagar A, Mortazavi SM. The Nsun7 (A11337)-deletion mutation, causes reduction of its protein rate and associated with sperm motility defect in infertile men. J Assist Reprod Genet. 2015;32:807–15. doi: 10.1007/s10815-015-0443-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Haute L, Dietmann S, Kremer L, Hussain S, Pearce SF, Powell CA, et al. Deficient methylation and formylation of mt-tRNA(Met) wobble cytosine in a patient carrying mutations in NSUN3. Nat Commun. 2016;7:12039. doi: 10.1038/ncomms12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frye M, Dragoni I, Chin SF, Spiteri I, Kurowski A, Provenzano E, et al. Genomic gain of 5p15 leads to over-expression of Misu (NSUN2) in breast cancer. Cancer Lett. 2010;289:71–80. doi: 10.1016/j.canlet.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 38.Lu L, Zhu G, Zeng H, Xu Q, Holzmann K. High tRNA transferase NSUN2 gene expression is associated with poor prognosis in head and neck squamous carcinoma. Cancer Investig. 2018;36:246–53. doi: 10.1080/07357907.2018.1466896. [DOI] [PubMed] [Google Scholar]
- 39.Yang JC, Risch E, Zhang M, Huang C, Huang H, Lu L. Association of tRNA methyltransferase NSUN2/IGF-II molecular signature with ovarian cancer survival. Future Oncol. 2017;13:1981–90. doi: 10.2217/fon-2017-0084. [DOI] [PubMed] [Google Scholar]
- 40.Li B, Xu WW, Guan XY, Qin YR, Law S, Lee NPY, et al. Competitive binding between Id1 and E2F1 to Cdc20 regulates E2F1 degradation and thymidylate synthase expression to promote esophageal cancer chemoresistance. Clin Cancer Res. 2016;22:1243–55. doi: 10.1158/1078-0432.CCR-15-1196. [DOI] [PubMed] [Google Scholar]
- 41.Samatar AA, Poulikakos PI. Targeting RAS-ERK signalling in cancer: promises and challenges. Nat Rev Drug Disco. 2014;13:928–42. doi: 10.1038/nrd4281. [DOI] [PubMed] [Google Scholar]
- 42.Song M, Liu X, Liu K, Zhao R, Huang H, Shi Y, et al. Targeting AKT with oridonin inhibits growth of esophageal squamous cell carcinoma in vitro and patient-derived xenografts in vivo. Mol Cancer Ther. 2018;17:1540–53. doi: 10.1158/1535-7163.MCT-17-0823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nan P, Wang T, Li C, Li H, Wang J, Zhang J, et al. MTA1 promotes tumorigenesis and development of esophageal squamous cell carcinoma via activating the MEK/ERK/p90RSK signaling pathway. Carcinogenesis. 2019;41:1263–72. doi: 10.1093/carcin/bgz200. [DOI] [PubMed] [Google Scholar]
- 44.Zhang JP, Song Z, Wang HB, Lang L, Yang YZ, Xiao W, et al. A novel model of controlling PD-L1 expression in ALK(+) anaplastic large cell lymphoma revealed by CRISPR screening. Blood. 2019;134:171–85. doi: 10.1182/blood.2019001043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li LY, Li EM, Wu ZY, Cao HH, Shen JH, Xu XE, et al. Overexpression of GRB2 is correlated with lymph node metastasis and poor prognosis in esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7:3132–40. [PMC free article] [PubMed] [Google Scholar]
- 46.Shi Q, Wang Y, Mu Y, Wang X, Fan Q. MiR-433-3p inhibits proliferation and invasion of esophageal squamous cell carcinoma by targeting GRB2. Cell Physiol Biochem. 2018;46:2187–96. doi: 10.1159/000489548. [DOI] [PubMed] [Google Scholar]
- 47.Li LY, Zhang K, Jiang H, Xie YM, Liao LD, Chen B, et al. Quantitative proteomics reveals the downregulation of GRB2 as a prominent node of F806-targeted cell proliferation network. J Proteom. 2015;117:145–55. doi: 10.1016/j.jprot.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 48.Ohanian M, Tari Ashizawa A, Garcia-Manero G, Pemmaraju N, Kadia T, Jabbour E, et al. Liposomal Grb2 antisense oligodeoxynucleotide (BP1001) in patients with refractory or relapsed haematological malignancies: a single-centre, open-label, dose-escalation, phase 1/1b trial. Lancet Haematol. 2018;5:e136–46. doi: 10.1016/S2352-3026(18)30021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Giubellino A, Gao Y, Lee S, Lee MJ, Vasselli JR, Medepalli S, et al. Inhibition of tumor metastasis by a growth factor receptor bound protein 2 Src homology 2 domain-binding antagonist. Cancer Res. 2007;67:6012–6. doi: 10.1158/0008-5472.CAN-07-0022. [DOI] [PubMed] [Google Scholar]
- 50.Nicolau-Neto P, Da Costa NM, de Souza Santos PT, Gonzaga IM, Ferreira MA, Guaraldi S, et al. Esophageal squamous cell carcinoma transcriptome reveals the effect of FOXM1 on patient outcome through novel PIK3R3 mediated activation of PI3K signaling pathway. Oncotarget. 2018;9:16634–47. doi: 10.18632/oncotarget.24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–8. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamano R, Miyata H, Yamasaki M, Sugimura K, Tanaka K, Kurokawa Y, et al. High expression of Lin28 is associated with tumour aggressiveness and poor prognosis of patients in oesophagus cancer. Br J Cancer. 2012;106:1415–23. doi: 10.1038/bjc.2012.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Madison BB, Liu Q, Zhong X, Hahn CM, Lin N, Emmett MJ, et al. LIN28B promotes growth and tumorigenesis of the intestinal epithelium via Let-7. Genes Dev. 2013;27:2233–45. doi: 10.1101/gad.224659.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X, Hu H, Liu H. RNA binding protein Lin28B confers gastric cancer cells stemness via directly binding to NRP-1. Biomed Pharmacother. 2018;104:383–9. doi: 10.1016/j.biopha.2018.05.064. [DOI] [PubMed] [Google Scholar]
- 55.Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16:191–8. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Veschi V, Liu Z, Voss T, Ozbun L, Gryder B, Yan C, et al. Epigenetic siRNA and chemical screens identify SETD8 inhibition as a therapeutic strategy for p53 activation in high-risk neuroblastoma. Cancer Cell. 2017;31:50–63. doi: 10.1016/j.ccell.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–95. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.