Abstract
Bacteria and their viruses (bacteriophages or phages) interact antagonistically and beneficially in polymicrobial communities such as the guts of animals. These interactions are multifaceted and are influenced by environmental conditions. In this review, we discuss phage-bacteria interactions as they relate to the complex environment of the gut. Within the mammalian and invertebrate guts, phages and bacteria engage in diverse interactions including genetic coexistence through lysogeny, and phages directly modulate microbiota composition and the immune system with consequences that are becoming recognized as potential drivers of health and disease. With greater depth of understanding of phage-bacteria interactions in the gut and the outcomes, future phage therapies become possible.
Keywords: bacteriophage, phage-bacteria interaction, intestinal virome, IBD, arthropod gut
1. INTRODUCTION
Bacteriophages, or phages, are viruses that infect bacteria and are the most abundant and genetically diverse biological entities on earth. Virtually all ecosystems that harbor bacteria contain phages that shape the composition of bacterial communities (1, 2). Complex interactions between phages, bacteria, and animal hosts are recognized as contributors to host-microbe interactions. In this review we focus on three areas: (a) an overview of phage biology that sets the stage for understanding phage-bacteria interactions studied in animals, (b) how phage-bacteria interactions affect health and disease in the gut, and (c) what effect these interactions have on animal hosts (Figure 1).
Figure 1.

Phage-bacteria interactions occur at the ecological and organismal levels in the gut and mucosal surfaces. (a) Abiotic and biotic factors, such as intestinal niches and mucus, influence phage-bacteria interactions. (b) Phage-bacteria interactions are associated with human disease and are being studied for their potential use as biomarkers and therapeutics. (c) Phages interact with bacteria in the guts and mucosal surfaces of diverse invertebrates. Understanding the outcomes of these interactions on microbial community structure and organismal biology is in its infancy.
Phages are proposed as targeted therapeutics against multidrug resistant bacteria and myriad other applications, including gut microbiota composition engineering and nucleic acid editing of bacteria in polymicrobial communities. The use of phages therapeutically should be considered within the broader context of phage-bacteria interactions and how these interactions shape the biology of phages, bacteria, and the animal hosts that harbor these microorganisms. A deeper knowledge of how phages modulate gut bacteria will refine efforts to design targeted and efficacious phage therapies.
2. A PRIMER OF BACTERIOPHAGE-BACTERIA INTERACTIONS
2.1. Phage Replication Strategies
Phages are temperate or nontemperate. Temperate phages lysogenize their bacterial hosts by integrating into bacterial chromosomes as prophages or are maintained as extrachromosomal episomes (3). Stably maintained chromosomal prophages are vertically transmitted in the host population. In response to host cell stress or other stimuli (4, 5), prophages are induced and enter the lytic replication cycle, where infectious virions are released following cell lysis (Figure 2a) (6).
Figure 2.
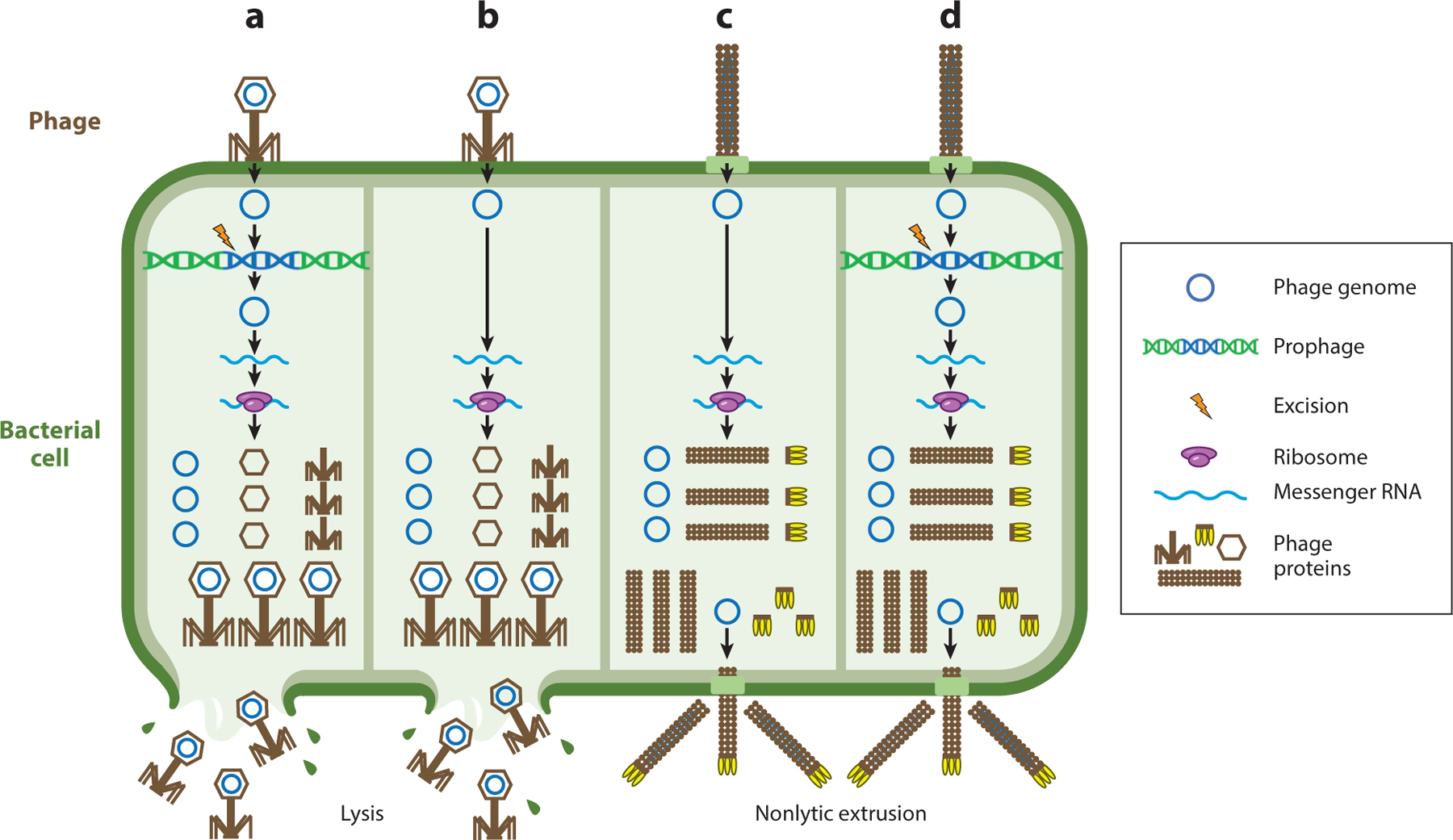
Phage replication strategies. Phage lifestyles can be divided into four categories: (a) lytic and temperate, (b) lytic and nontemperate, (c) chronic and nontemperate, and (d) chronic and temperate. All phages must adsorb to the bacterial surface and introduce their genome into the cell. Nontemperate phages replicate and assemble new virions (panels b and c) while temperate phages (panels a and d) may integrate into the bacterial chromosome as a prophage or be maintained as an episome. Early phage genes are expressed and typically include proteins involved in phage genome replication, followed by late gene expression where phage structural proteins are produced. Finally, phage virions are assembled and released via lysis (panels a and b) or by nonlytic extrusion (panels c and d).
Some phages do not form stable lysogens and replicate strictly via the lytic cycle. Lytic nontemperate phages (Figure 2b) are typically parasitic and are transmitted horizontally in the host bacterial population. Some phages, such as filamentous inoviruses, do not require host cell lysis to release infectious virions (7) and can establish chronic infections with continuous virion extrusion from the cell envelope (8, 9) (Figure 2c,d). Some inovirus species such as M13 do not form stable lysogens (8, 10), while others such as CTXϕ integrate into the bacterial chromosome as a prophage (11).
2.2. Ecological Forces Shape Bacteriophage-Bacteria Interactions
Phage-bacteria interactions drive coevolution that shapes microbial ecology. The Red Queen hypothesis when applied to phage-bacteria interactions supports parasitic relationships, characterized by cycles of bacterial resistance and phage counter resistance (12) (Figure 3a). Phage-bacteria interactions are also explained by kill-the-winner dynamics (Figure 3b). This model is analogous to the Lotka-Volterra equation describing predator-prey interactions (13). Kill-the-winner dynamics in the context of lytic nontemperate phages rely on bacterial population density. When host bacterial population densities are low, a phage interaction with a host is low, resulting in reduced infection. If the bacterial host population expands, the chances that a phage will encounter a susceptible host increase. Higher infection rates slow bacterial population expansion. Thus, phages can serve to stabilize bacterial community compositions by kill-the-winner infection biases that prevent fast-growing bacterial species from taking over the community (Figure 3b). However, when applied to virulent phages infecting diverse bacterial populations, kill-the-winner dynamics can cause abrupt population collapse (14). In bacterial populations containing multiple species, the dynamics of population expansion followed by abrupt collapse are cyclical for each species; so long as bacterial growth rates are high, the overall population diversity is maintained (14). Few examples of kill-the-winner dynamics have been described within the human intestine, with notable exceptions of phage-bacteria interactions in the infant intestine (15, 16).
Figure 3.

Phage-bacteria interactions within the intestine result in diverse ecological dynamics. (a) In Red Queen dynamics, the parasitic relationship between phage and bacteria results in the development of bacterial phage resistance. Over time, phage evolve counter resistance mechanisms allowing infection to continue. (b) In kill-the-winner dynamics, fast-growing and higher-density subpopulations are more susceptible to phage infection than slower-growing and lower-density populations. Lytic phages will thus infect and lyse higher-density subpopulations, ultimately restoring subpopulation density. (c) In piggyback-the-winner dynamics higher-density subpopulations are more likely to be infected by phage and lysogenized. Lysogenized subpopulations resistant to superinfection can then expand.
Piggyback-the-winner dynamics describe ecological parameters that can be applied to temperate phages in high-density microbial communities, as many ecological niches that harbor high-density bacterial communities (the gut, coral reefs, soil) display lower-than-expected phage-to-bacteria ratios (17). Reduced phage-to-bacteria ratios could be explained by temperate phages entering lysogeny. The majority of bacteria in the intestine are lysogenized by at least one temperate phage (18–21). Temperate phages protect their host from phage reinfection through a process called superinfection exclusion (22). Lysogenic conversion can also confer a variety of fitness advantages to the host bacterium (23). The piggyback-the-winner model postulates that high microbial densities favor lysogeny over lytic phage replication (Figure 3c).
3. BACTERIOPHAGE-BACTERIA INTERACTIONS OF THE INTESTINAL MICROBIOTA
3.1. Factors that Influence Composition and Abundances
The primary method used to study intestinal phages is next-generation sequencing of virus-like particles (VLPs) using Illumina short reads (24, 25). More recently, long-read sequencing methods, such as Oxford Nanopore and PacBio SMRT, are being used to provide full-length phage genomes from single reads and assess modifications such as DNA methylation patterns (26). A study of intestinal phages using Oxford Nanopore sequencing demonstrated that this method can detect known features of intestinal viral metagenomes (virome), such as highly abundant CrAss-phages, and novel methylation marks that were previously undescribed (27). Studies have determined that intestinal phages are largely composed of members of the Caudovirales, including the Myoviridae, Siphoviridae, and Podoviridae morphotypes, and the Petitvirales, represented by the Microviridae (19, 28, 29). Individuals have unique repertoires of intestinal phages (28, 30, 31), containing only a few shared core phages (21, 29). For instance, among 2,000 geographically dispersed viromes, 69% of the viral populations were present in less than 0.5% of the samples and 0.38% of viral populations were present in more than 20% of samples (31).
Factors affecting the composition of intestinal phages include diet, genetic factors, and disease state (Figure 4). Dietary intervention reduces the variation of phages among unrelated individuals and significantly alters virome composition compared to prediet (32). It is suggested that genetic factors play a role in phage composition, as monozygotic twins have more similar viromes than unrelated individuals (30, 33). An earlier study found that genetic relatedness did not lead to virome similarity (19), yet the results of this study were refuted in a more recent publication where a larger cohort of human samples was used (30).
Figure 4.

Virome composition is unique among humans, with a few shared viruses that numerically dominate the virome. Factors affecting virome composition are inflammation, genetics, diet, and maternal dissemination of the virome.
CrAss-phages, a family of intestinal phages, are the most abundant phages in human intestinal viromes and are ubiquitous worldwide (34). CrAss-phages are more prevalent in industrialized societies and are passed vertically from mother to child (35, 36). Different clades of CrAss-phages exist in humans, with each person having unique CrAss-phage clusters (37). Young children can have more than 1,400 different CrAss strains. CrAss-phages from similar geographic locations cluster together according to genome sequence, suggesting localized transmission. CrAss-phages may have evolved with humans because CrAss-like phages appear to be present in primates (37). While the effects and role of CrAss-phages are still unclear, CrAss-phage abundance does not correlate strongly with any diet, lifestyle, or health variable, suggesting these phages may not have a strong effect on human health (37). In a meta-analysis of individual viromes, one CrAss-phage population was found in 12% of samples analyzed from a variety of age groups worldwide (31). This study suggested that CrAss-phage abundance peaks in adults and declines in the elderly. Recently, evidence suggests that CrAss-phages infect Bacteroidetes bacteria such as Porpyromonas sp., Bacteroides thetaiotaomicron, and Bacteroides intestinalis (38–40). Bacteroides are dominant members of the intestinal microbiota of humans who consume a Western diet, further supporting these bacteria as a host for CrAss-phages (41).
The virome changes throughout all stages of human life (31, 42). Phage richness increases during infancy (15, 33) [although studies have posited the opposite (16, 43)], decrease during adolescence, reach high abundances during adulthood, and drop in abundance in the elderly. The infant virome is temporal and increases in the abundance of Microviridae and Siphoviridae and decreases in Myoviridae over time (15, 16, 33, 44). The adult virome is characterized by high levels of Podoviridae, Microviridae, and Siphoviridae. Podoviridae and Microviridae abundance remains high in the elderly.
Acquisition of phages in infants is thought to occur through bacterial-mediated dissemination of prophages. Neonates are devoid of intestinal phages up to 4 days after birth and show signs of phage intestinal occupation by one month postbirth, which indicates that intestinal phages are acquired after birth (45). Lack of in utero intestinal phages reflect the paucity of bacterial hosts, as the microbiota is widely considered to be acquired during and after birth (46). Factors such as diet and birthing method influence phage composition of the infant intestine. Bacterial species and their corresponding prophages are overabundant in breastfed infants compared to those fed formula (45). In addition, phages from infant stool associate with the milk microbiota from their corresponding mothers, and on average 30% of phages are present in both the infant stool and maternal milk (47). Spontaneous vaginal delivery of infants significantly increases intestinal virome diversity of one-year-old infants, compared to similarly aged infants delivered via caesarian section (48). The neonatal virome is mostly composed of temperate phages and becomes more virulent as time progresses (49). Prophages isolated from the intestinal bacteria of neonates show high similarity to phages of infants and have a strong positive correlation between a neonate’s intestinal bacterial abundance and the abundance of that bacteria’s prophages in the virome (45). In contrast, an inverse relationship between bacterial and phage richness and diversity in the infant gut has been observed, suggesting a primarily lytic phage composition (16).
The ratio between lysogenic and lytic phages in the adult intestine is disputed, likely because determining lysogeny in complex microbial samples is imprecise. Common methods for lysogenic measurements include treating bacterial populations with prophage inducers, such as mitomycin C or altering pH (50). However, a substantial number of intestinal bacteria resist cultivation and these methods are not absolute. Many studies use sequence-based methods to detect lysogeny (18, 28, 49). Several studies have found that when using the genomes of previously described phages as references (that account for a minority of phages detected in a complex virome) to identify intestinal phages, lysogenic phages dominate (19, 51). However, if database-independent methods are used for de novo identification of intestinal phages, lytic phages are more abundant (28). A study that utilized reporter strains found that the majority of intestinal phages that infect Escherichia coli are temperate, with the temperate phages having narrow host ranges and the virulent phages having broader host ranges (20, 52). A lack of temporal variability (<2.5 years) in intestinal phage diversity supports the rationale that phages are continually released from lysogenized bacteria (18, 19). A seminal study that sequenced intestinal viromes from ten individuals found that temperate phages dominated the virome of only one subject (28). Lysogenic phages have been shown to transition to the lytic state in the intestine. An Enterococcus faecalis prophage and genetically linked chromosomal island produce lytic phage particles that facilitate E. faecalis colonization of the mouse intestine (53). During mouse intestinal colonization Roseburia intestinalis prophages gain mutations in the cro antirepressor, immunity region, and other variable genes, producing hypervirulent lytic phages (54). This results in destruction of the R. intestinalis population, and phage-resistant bacteria survive. These studies demonstrate the highly variable nature of phage-bacteria interactions and the evolving perception of lytic-lysogenic dichotomy in the intestine.
3.2. Bacteriophage-Bacteria Interactions in the Mammalian Intestine
Coevolution of bacteria and phages occurs in the intestine, and these interactions shape both communities (55). Phages and their host bacteria evolve rapidly in culture, suggesting that the intestine is an ideal environment for phage evolution (56). Common intestinal bacteria, including bifidobacteria, contain variable phage resistance and infection genes (57–59), suggesting that intestinal bacteria are targets of predation and evolve to avoid phage infection (55). Intestinal bacteria frequently contain clustered regularly interspaced short palindromic repeats (CRISPR) spacers that align to phages, demonstrating that intestinal bacteria are actively evolving against phage infection (21).
Evidence of phage-induced genetic changes in intestinal bacteria is emerging. Studies focusing on E. coli and E. faecalis populations in the mouse intestine, following phage challenge, showed that phage-resistant bacteria can be recovered with mutations in phage receptors (60–62). Vibrio cholerae isolated from human stool carrying virulent phages were resistant to phage infection, indicating that bacterial adaptation that overcomes phage infection occurs in the intestine (63). All sequenced V. cholerae strains were isogenic except for mutations in the outer-membrane porin phage receptor, suggesting that phage predation selects for variants that can resist phage infection in the intestine. Bacterial adaptation to phages can also be transient. B. thetaiotaomicron regulates capsule polysaccharide expression through phase variation and downregulates the expression of capsule genes required for phage infection in the presence of phage, supporting phage-host coexistence (64).
Phage-bacteria coevolution occurs in the mammalian intestine. Mice colonized with E. coli and an E. coli–targeting phage resulted in loss of phage virulence over time due to bacterial resistance, and the phages regained infectivity following the acquisition of compensatory mutations (60). Phages gained new host ranges in the mouse intestine following continuous exposure to noncognate host cells (65). In this study, mice were orally inoculated with phages and both phage-sensitive and insensitive bacteria. Over time, these phages gained the ability to infect the phage-insensitive bacterium. A third intermediate host was required to reproduce these results in culture, suggesting that an endogenous intermediate host was present in the mouse intestine to aid in phage genome evolution and altered host range. Finally, longitudinal studies in humans have shown that genetic variation occurs over time in the healthy human virome, suggesting that phages evolve to selective pressures such as bacterial host density and CRISPR spacer acquisition (18).
The administration of phages to a defined intestinal microbiota in gnotobiotic mice has demonstrated that phages regulate microbial populations in the intestine. Gnotobiotic mice colonized with intestinal bacteria and administered VLPs from healthy human donors experienced a community-wide decrease in bacterial abundance (66). In separate research, gnotobiotic mice with defined microbial communities were treated with known lytic phages, which led to the loss of both phage-targeted and nontargeted bacteria (67). This suggests that phage predation can affect nonhost bacterial species. Furthermore, bacterial communities from young nonstunted children are modulated by phage communities from young stunted children (49). Bacteria isolated from young nonstunted children underwent a significant compositional change after treatment with phages from young stunted children, compared to a heat-killed phage treatment.
Phages actively transduce DNA in the intestine. Resident intestinal phages transduce E. coli in the mouse intestine (68). These acquired prophages transduce advantageous genes that aid in bacterial metabolism. Phage transduction can spread virulence factors, such as the Shiga toxin–encoding prophages of E. coli (69). Shiga toxin–encoding prophages lysogenize new E. coli hosts in the mouse intestine at a frequency of approximately 1:2,000 cells. Additionally, host inflammation promotes temperate phage transfer in Salmonella enterica serovar Typhimurium (S. Tm) and the spread of virulence genes (70). Mice infected with a donor S. Tm strain carrying an antibiotic-tagged prophage and a prophage-less recipient resulted in lysogenized recipient cells. Avirulent S. Tm donor strains, and thus a lack of intestinal inflammation, reduced temperate phage acquisition by recipients. Vaccination of mice before S. Tm colonization prevented lysogenic conversion, demonstrating that the host response increased prophage transfer to recipients. While these studies were the first to demonstrate that transduction and prophage acquisition occur in the intestine, more recently bioinformatics have been developed to monitor community-wide transduction and prophage excision events, showing that several modes of transduction including generalized and specialized transduction occur in the mouse intestine (71).
Intestinal cues such as bile salts, mucins, and intestinal spatial localization influence phage-bacteria interactions, indicating that the intestinal environment influences predator-prey relationships between phages and bacteria (72, 73). Phages and their host bacteria occupy different niches in the mouse intestine. Phage abundances relative to their target bacterium are higher in the intestinal lumen compared to the mucosa, indicating that the mucosa may serve as a hideaway for bacteria to escape phage predation, promoting phage-bacteria coexistence in the intestine (74). Phage infection efficiency of intestinal E. coli depends on the physical location of this interaction in the intestine, where E. coli cells are more permissive to phage infection in the ileum compared to the colon and phages multiply to a lesser degree in the cecum compared to the ileum or colon (75, 76). Other studies have shown that predatory phages fail to effectively eliminate pathogenic intestinal bacteria in the absence of phage-resistant bacteria, suggesting that phage-sensitive bacteria reside in niches in the intestine that are inaccessible by phages (73, 77). Bile salts can prevent adsorption and infection of a variety of E. coli phages (78–80). Finally, the production of the V. cholerae phage receptor, toxin coregulated pilus, is associated with proximity to the ileal epithelium, indicating that phage receptor expression is location dependent in the intestine (81).
Intestinal phages have an affinity for mucus and bind to a variety of mucosal surfaces in both vertebrates and invertebrates, which may play a role in the biogeography of phage predation (82). Phage adherence to the mucosa is suggested to protect these surfaces from bacterial invasion. Phage adherence to mucus is mediated by the immunoglobulin (Ig)-like domain capsid protein Hoc. This observation led to the proposed bacteriophage adherence to mucus (BAM) model, where mucus collects virulent phages as a defense against bacteria (82). Whether the BAM model applies to the intestinal mucosal surface in vivo remains to be determined. An in vitro intestinal epithelial model showed that Ig-like domain-containing phages and phages lacking the Ig-like domains are retained in mucus, but the presence of an Ig-like domain in the phage capsid more effectively reduced bacterial burden at the mucosa by slowing phage diffusion through the mucus and increasing phage-bacteria interactions (83). However, computational modeling suggests that the rate of phage and E. coli interactions in mucus is also dependent on E. coli motility (84). An alternative explanation is that mucins alter the bacterial host’s physiology to enhance phage infectivity because mucins can alter bacterial growth and increase phage production (85).
3.3. Phages and Intestinal Diseases
While our understanding of what constitutes a healthy microbial gut community is unclear, recent studies have identified differences in the phage community composition during intestinal disease. Inflammatory bowel disease (IBD), including ulcerative colitis (UC) and Crohn’s disease (CD), are influenced by the composition of the microbiota in the intestine. An observational study suggested that phage communities might also be involved in IBD (86). Using UC and CD patient cohorts, stool VLPs were isolated and metagenomic sequencing was performed to compare IBD to non-IBD individuals. Consistent with other studies, Caudovirales and Microviridae were the most abundant viral taxa detected in all three cohorts. Individuals with IBD harbored increased richness and abundances of Caudovirales and decreased abundances of Microviridae, indicating disease is associated with an expansion of Caudovirales phages. Phages identified to be uniquely associated with CD were closely related to Lactobacillus, Clostridium, Enterococcus, and Streptococcus phages, and these were not observed in individuals with UC, highlighting that specific phages associate with distinct diseases (86). In a related study, VLPs isolated from the rectal mucosa of 91 UC subjects were compared to VLPs isolated from 76 healthy controls (87). Similarly, an expansion of the order Caudovirales was observed in individuals with UC, including an enrichment in Escherichia and other Enterobacteriaceae phages. In contrast, while an overall increase in abundance of Caudovirales was observed, a decrease in the richness and diversity of Caudovirales was seen in the mucosa of UC patients, indicating that a specific subset of Caudovirales was expanding in these patients. In mice experiencing intestinal colitis, phages similar to those found in CD and UC patients dominate, suggesting colitic mice may serve as a useful tool for studying human IBD as it relates to phage composition (88). Finally, changes in phages were also seen in the more common intestinal disorder irritable bowel syndrome (89), demonstrating some commonalities in phage communities associated with intestinal disease. Using a database-independent method, Clooney and colleagues (90) found that in contrast to previous studies, there was no significant difference in viral richness during IBD. Instead, increased Caudovirales richness in individuals with IBD was seen only when the data were restricted to analyzing phages belonging to Caudovirales. While a healthy intestinal virome is dominated by a stable core of virulent phages, increased temperate phage abundances were linked to disease (90). Improvement of methods to characterize and identify phages from the microbiota will continue to refine our understanding of the changes that occur during IBD.
Fecal microbiota transplant (FMT) is emerging as a mechanism to restore healthy microbial communities and has proven clinically successful for the treatment of Clostridioides difficile infection. FMT for IBD has begun to be trialed to restore poor microbial diversity frequently associated with disease severity in both CD and UC patients. Several recent studies have demonstrated clinical success of FMT in patients with UC (91, 92). FMT increases microbial diversity in recipients irrespective of the clinical response, and it remains unclear if diversity or specific bacterial taxa are responsible for the clinical efficacy (91). A study published in 2017 (93) examined the efficacy of sterile fecal filtrate devoid of bacteria but retaining small particles such as phages and metabolites for treating C. difficile infection. Transfer of this filtrate to five individuals with C. difficile infection restored normal stool habits and removed the C. difficile infection in all five individuals. Virome analysis supported the presence of phages, thus suggesting that phages within FMTs can protect from intestinal disease. To determine whether phages are associated with successful FMT in UC patients, we examined differences in the intestinal phage community of individuals with UC that did or did not have successful clinical outcomes from FMT (94). Patients that had a successful clinical response to FMT had a lower relative abundance of Caudovirales bacteriophages at the time of transplant compared to patients that did not respond to therapy. Furthermore, the relative abundance of Caudovirales in nonresponders increased after FMT while no change was observed in responders. These data suggest that Caudovirales abundance might be indicative of FMT failure.
Stunting, a growth impairment that occurs in children under the age of five and is associated with malnutrition and/or environmental stress, has been associated with changes in the microbiota. In a recent study, Khan Mirzaei and colleagues (49) isolated and characterized phage communities in young children from Bangladesh. In these children, the Siphoviridae morphotype (within the Caudovirales order) dominated the phage sequencing reads from both stunted and nonstunted groups. However, stunted children had significantly increased reads assigned to Myoviridae and Microviridae. At the species level, Bacillus phages were the most abundant, indicating that an expansion in these phages is associated with stunting.
3.4. Functional Effects of the Virome During Disease
The majority of literature focused on phage communities in intestinal disease is observational; however, within the past few years researchers have performed functional analyses to demonstrate that phages themselves can influence disease. While it has long been speculated that phages can stimulate immune responses, most of these data have been garnered in vitro, whereby immune cells are incubated with phages, making it unclear whether phages could modulate immunity at the organismal level. Moreover, because phages are isolated from bacteria, phage preparations can contain bacterially derived antigens and must be highly purified to ensure effects on immune cells are truly in response to phages. Recently, highly purified E. coli phages were orally administered to germ-free mice, and phages alone were sufficient to induce T cell numbers to the same levels as those seen in specific pathogen-free (SPF) animals that possessed a complex microbiota (94). In vitro experiments confirmed that this effect occurred by dendritic cell–mediated phagocytosis of phages and presentation of phage antigens to activate T cells (94). Phage-mediated stimulation of immunity was dependent on Toll-like receptor 9 (TLR9) signaling by phage DNA (94). Another study demonstrated that the filamentous Pf phage that infects Pseudomonas aeruginosa can prevent phagocytosis and suppress tumor necrosis factor-α secretion dependent on TLR3 (95). These data demonstrate that phages can enhance and suppress host immunity through distinct mechanisms. As there are multiple receptors both outside and inside the cell that might detect phages, future studies are warranted to better understand this specificity. This could be particularly relevant in early development, as the neonatal intestine is being seeded by microorganisms, including phages, making it possible that phage stimulation could influence early immune system priming.
As discussed earlier, Caudovirales phages have been observed to expand in individuals with IBD. It is unclear whether this expansion has any consequences for this intestinal disease. To address this, SPF animals that did not harbor E. coli species, but contained an otherwise complex and normal microbiota, were orally treated with a cocktail of three Caudovirales phages that specifically targeted E. coli (94). Because the host targets of these phages were absent in these mice, this simulated an expansion of free Caudovirales phages that did not disrupt the commensal microbiota, mimicking the expansion of these phages in humans with IBD. Animals orally administered the phage cocktail were subsequently induced for IBD. Animals with the expanded phages developed worsened disease characterized by increased weight loss, enhanced inflammation, and greater epithelial damage when compared to untreated animals (94). Increased disease induced by phages was dependent on phage activation through TLR9, suggesting that the expanded population of phages detected in individuals with IBD may play a detrimental role during disease.
3.5. Harnessing Bacteriophages Therapeutically
Antibiotics have often been used to ameliorate IBD as well as treat intestinal infections. However, as antibiotics also kill the beneficial organisms in the body, the use of phage therapy for intestinal diseases would provide a more targeted approach to kill unwanted organisms, although recent reports show that some phage-bacteria interactions can have off-target effects (67, 96). This was recently tested in two distinct animal models. Genotoxin-producing adherent invasive E. coli (AIEC) is associated with both IBD and colorectal cancer (CRC) in humans and has been shown to increase tumor burden and number in a mouse model (97, 98). To determine whether phage therapy would be successful in a mouse model of CRC, a cocktail of three phages that specifically target AIEC was continuously administrated to animals that develop CRC due to a genetic mutation in the adenomatous polyposis coli (APC) gene (94). While a one-time dose was not sufficient to ameliorate disease, continuous treatment with this phage cocktail significantly reduced tumor burden and number, demonstrating for the first time that phage therapy could be successfully employed to treat CRC (94). A similar approach was used in a mouse model of alcoholic liver disease (99). The investigators observed that E. faecalis residing in the intestine secretes an exotoxin that causes hepatocyte death and liver injury. The abundance of intestinal E. faecalis is significantly higher in individuals with alcoholic liver disease. Treatment of animals with phages significantly reduced E. faecalis intestinal colonization and exotoxin production, which ameliorated liver injury. Thus, microbiota editing using phages provides a personalized approached to treat a multitude of disorders.
In addition to phages directly amplifying immune responses as discussed above, two recent studies have identified ways to utilize immune responses against phages as potential treatments for disease. The P. aeruginosa Pf phage promotes biofilm formation and enhances virulence of this bacterium. Immunization of animals with a Pf phage protein protected against P. aeruginosa wound infections (95). Immunized animals cleared the bacterial infection faster and developed milder disease. The cytotoxic T lymphocyte response to a prophage element, called TMP, found within the bacterium Enterococcus hirae is cross-reactive with tumor-associated antigens (100). T cell responses to E. hirae are associated with a positive clinical outcome in cancer patients, suggesting that commensal-specific T cell responses can contribute to antitumor immunity (100). The specific epitopes recognized by T cells and whether they can be harnessed to improve antitumor therapy are unclear. Interestingly, administration of bacterial strains engineered to express the prophage TMP epitope improved immune therapy in mice (100). In renal and lung patients, the presence of the prophage in stools and TMP cross-reactive antigens in tumors correlates with the benefit of PD-1 blockade (100). Thus, commensal-specific T cells can cross-react with tumor antigens and can be harnessed to improve cancer immunotherapy. Given that the intestinal microbiota contain numerous prophages, amplifying or vaccinating with phage antigens might be novel strategies for therapeutic intervention of intestinal disease.
4. BEYOND THE VERTEBRATE INTESTINE: BACTERIOPHAGE-BACTERIA INTERACTIONS IN ARTHROPODS AND OTHER NONVERTEBRATES
Arthropods are the largest group of terrestrial animals. Like vertebrates, arthropods display myriad relationships with bacteria (101). While many studies describe the functional diversity of bacterial communities in arthropods, the role of phages in arthropod microbiotas remains understudied. In this section, we discuss known and potential interactions between phages and bacteria in the guts of nonvertebrate hosts.
Roughly half of insects and other arthropods are infected by Wolbachia, a maternally transmitted endosymbiont (102–104). In some cases, Wolbachia are reproductive parasites, causing cytoplasmic incompatibility (CI) between sperm and egg, feminization, and male killing in a variety of arthropod species (105). In contrast to other obligate endosymbiotic bacteria, the Wolbachia genome contains a high level of mobile genetic elements, including the temperate phage WO, which is widely distributed among Wolbachia (106). Phage WO encodes ankyrin repeat domain proteins (107) that facilitate tripartite phage-bacteria-arthropod interactions (108, 109). Two ankyrin genes encoded by prophage WO (pk1 and pk2) are correlated with feminization and CI in different arthropod hosts (110, 111), and several other phage WO-encoded ankyrin domain-containing proteins are thought to interact with the arthropod host (109). Phage WO also encodes the nonankyrin genes cifA and cifB, which are responsible for causing CI in Drosophila melanogaster (112). CI is rescued if females are infected with the same strain of Wolbachia, which has the effect of driving CI-inducing Wolbachia, and thus phage WO, into host arthropod populations (113, 114). Wolbachia have been reported to be present in the midguts of various insect species (115); however, specific interactions between Wolbachia, phage WO, and the arthropod gut microbiota have not been described.
Compared to the vertebrate gut, arthropod guts contain relatively few microbial species (101). However, some arthropods harbor complex gut communities. For example, termites rely on gut symbionts for cellulose digestion and nutrient acquisition (116). Some termite lineages possess a purely prokaryotic gut microbiota, while other more primitive lineages digest cellulose with the help of a cellulolytic protist (117). Meta-analysis of the termite gut microbiota reveals phages are represented including several Caudovirales and Microviridae phages, and phages that infect endosymbiotic bacteria residing within the symbiotic protist (118, 119). Phages have been detected in other insect endosymbionts including Sodalis glossinidius from the tsetse fly gut (120), Proteus, Providencia, and Morganella symbionts in parasitoid wasps (Nasonia) (121), and gut bacteria in honey bees (Apis mellifera) (122). The presence of phages that infect endosymbionts throughout the stratified levels of the arthropod gut microbiota suggests phages play underappreciated roles in several aspects of arthropod biology.
Phages infect pathogenic bacteria transmitted by blood-feeding arthropod vectors. For example, Ixodes scapularis, the blacklegged deer tick, transmits numerous pathogens, including Lyme disease, the most common vector-borne disease in the Northern Hemisphere (123). Borrelia (Borreliella) burgdorferi, the bacterium that causes Lyme disease, has a complex genome that includes 32-kb circular plasmid (cp32) prophages (124). In B. burgdorferi, cp32 prophages undergo lytic replication where they are packaged into infectious virions designated ϕBB-1 (125, 126). cp32 prophages are likely induced during a blood meal because cp32 genes are transcriptionally active in the presence of blood and increased temperature (127). Putative ϕBB-1 structural genes are highly conserved and have not decayed among cp32 isoforms across the Borrelia genus (124), suggesting that lytic replication of ϕBB-1 occurs frequently during the natural tick-vertebrate life cycle. The cp32 prophages encode bacterial surface proteins that play critical roles in the B. burgdorferi arthropod-vertebrate life cycle (124, 128). What role if any cp32 prophages or ϕBB-1 virions play in the vertebrate host is not clear.
In addition to arthropod hosts, phages are important components of the microbiota in other invertebrate mucosal communities. Sponges harbor a complex but stable microbiota analogous to the gut microbiota of higher animals. Sponge bacterial symbionts are vertically transmitted and play critical roles in nutrient acquisition (129, 130). Filter-feeding sponges process several thousand liters of seawater each day (131). Because phages are abundant in seawater, sponges and their bacterial symbionts are exposed to billions of phages every day, and phage-defense mechanisms are enriched in sponge symbiont genomes (132, 133). A recent study found that phages that infect commensal bacteria in sponges encode the ankyrin repeat–containing protein ANKp. Phage-encoded ANKp suppresses inflammatory cytokine production and phagocytic bacterial uptake by the sponge immune system, which may facilitate sponge-commensal bacteria-phage coexistence (134). The mechanism underlying how ANKp suppresses host immunity is not known; however, the ankyrin repeat domain present in ANKp likely facilitates interactions with the animal host.
5. CONCLUSION
Knowledge of the interplay between bacteria and their phages has progressed greatly in recent years. This field has described numerous ways both bacteria and phages participate in a perpetual arms race to coexist. In the gut environment, where phage-bacteria interactions are affected by abiotic and biotic factors (i.e., spatial niches and mucus) and environmental conditions such as diet and host genetics, the interactions of bacteria and phages have broad implications for organismal gut biology. Perturbation of phage populations affect diverse biological systems ranging from disease states in humans to prophage-driven life cycle progression in blood-feeding ticks. Further research on all fronts to identify new features of phage-bacteria interactions in the gut will likely lead to more informed technologies for manipulating the microbiota. Additionally, new insights into how phage-bacteria and phage-animal interactions modulate health and disease will continue to place phages at the forefront of host-microbe interactions and next-generation therapeutics in the gut for years to come.
ACKNOWLEDGMENTS
This work was supported by NIH R01AI141479 (B.A.D.), NIH R01CA246368 (B.A.D.), NIH R01DK124317 (P.R.S. and J.L.R.), NIH R21AI151597 (P.R.S.), NIH P20GM103546 (P.R.S.), Helmsley award ID-G-1903-03786 (P.R.S.), and a medical grant from the W.M. Keck Foundation (P.R.S.).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Braga LPP, Spor A, Kot W, Breuil MC, Hansen LH, et al. 2020. Impact of phages on soil bacterial communities and nitrogen availability under different assembly scenarios. Microbiome 8:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Payet JP, McMinds R, Burkepile DE, Vega Thurber RL. 2014. Unprecedented evidence for high viral abundance and lytic activity in coral reef waters of the South Pacific Ocean. Front. Microbiol 5:493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wetzel KS, Aull HG, Zack KM, Garlena RA, Hatfull GF. 2020. Protein-mediated and RNA-based origins of replication of extrachromosomal mycobacterial prophages. mBio 11(2):e00385–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson AD, Poteete AR, Lauer G, Sauer RT, Ackers GK, Ptashne M. 1981. λ Repressor and cro–components of an efficient molecular switch. Nature 294:217–23 [DOI] [PubMed] [Google Scholar]
- 5.Little JW, Mount DW. 1982. The SOS regulatory system of Escherichia coli. Cell 29:11–22 [DOI] [PubMed] [Google Scholar]
- 6.Hobbs Z, Abedon ST. 2016. Diversity of phage infection types and associated terminology: the problem with ‘Lytic or lysogenic.’ FEMS Microbiol. Lett 363:fnw047. [DOI] [PubMed] [Google Scholar]
- 7.Calendar R 2006. The Bacteriophages. Oxford/New York: Oxford Univ. Press [Google Scholar]
- 8.Loh B, Kuhn A, Leptihn S. 2019. The fascinating biology behind phage display: filamentous phage assembly. Mol. Microbiol 111:1132–38 [DOI] [PubMed] [Google Scholar]
- 9.Rakonjac J, Bennett NJ, Spagnuolo J, Gagic D, Russel M. 2011. Filamentous bacteriophage: biology, phage display and nanotechnology applications. Curr. Issues Mol. Biol 13:51–76 [PubMed] [Google Scholar]
- 10.Stassen AP, Folmer RH, Hilbers CW, Konings RN. 1994. Single-stranded DNA binding protein encoded by the filamentous bacteriophage M13: structural and functional characteristics. Mol. Biol. Rep 20:109–27 [DOI] [PubMed] [Google Scholar]
- 11.McLeod SM, Kimsey HH, Davis BM, Waldor MK. 2005. CTXφ and Vibrio cholerae: exploring a newly recognized type of phage–host cell relationship. Mol. Microbiol 57:347–56 [DOI] [PubMed] [Google Scholar]
- 12.Forsberg KJ, Bhatt IV, Schmidtke DT, Javanmardi K, Dillard KE, et al. 2019. Functional metagenomics-guided discovery of potent Cas9 inhibitors in the human microbiome. eLife 8:e46540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winter C, Bouvier T, Weinbauer MG, Thingstad TF. 2010. Trade-offs between competition and defense specialists among unicellular planktonic organisms: the “killing the winner” hypothesis revisited. Microbiol. Mol. Biol. Rev 74:42–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maslov S, Sneppen K. 2017. Population cycles and species diversity in dynamic Kill-the-Winner model of microbial ecosystems. Sci. Rep 7:39642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breitbart M, Haynes M, Kelley S, Angly F, Edwards RA, et al. 2008. Viral diversity and dynamics in an infant gut. Res. Microbiol 159:367–73 [DOI] [PubMed] [Google Scholar]
- 16.Lim ES, Zhou Y, Zhao G, Bauer IK, Droit L, et al. 2015. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat. Med 21:1228–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knowles B, Silveira CB, Bailey BA, Barott K, Cantu VA, et al. 2016. Lytic to temperate switching of viral communities. Nature 531:466–70 [DOI] [PubMed] [Google Scholar]
- 18.Minot S, Bryson A, Chehoud C, Wu GD, Lewis JD, Bushman FD. 2013. Rapid evolution of the human gut virome. PNAS 110:12450–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, et al. 2010. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466:334–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathieu A, Dion M, Deng L, Tremblay D, Moncaut E, et al. 2020. Virulent coliphages in 1-year-old children fecal samples are fewer, but more infectious than temperate coliphages. Nat. Commun 11:378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stern A, Mick E, Tirosh I, Sagy O, Sorek R. 2012. CRISPR targeting reveals a reservoir of common phages associated with the human gut microbiome. Genome Res. 22:1985–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bondy-Denomy J, Qian J, Westra ER, Buckling A, Guttman DS, et al. 2016. Prophages mediate defense against phage infection through diverse mechanisms. ISME J. 10:2854–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Obeng N, Pratama AA, Elsas JDV. 2016. The significance of mutualistic phages for bacterial ecology and evolution. Trends Microbiol. 24:440–49 [DOI] [PubMed] [Google Scholar]
- 24.Shkoporov AN, Ryan FJ, Draper LA, Forde A, Stockdale SR, et al. 2018. Reproducible protocols for metagenomic analysis of human faecal phageomes. Microbiome 6:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleiner M, Hooper LV, Duerkop BA. 2015. Evaluation of methods to purify virus-like particles for metagenomic sequencing of intestinal viromes. BMC Genom. 16:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shkoporov AN, Hill C. 2019. Bacteriophages of the human gut: the “known unknown” of the microbiome. Cell Host Microbe 25:195–209 [DOI] [PubMed] [Google Scholar]
- 27.Cao J, Zhang Y, Dai M, Xu J, Chen L, et al. 2020. Profiling of human gut virome with Oxford Nanopore Technology. Med. Microecol 4:100012 [Google Scholar]
- 28.Shkoporov AN, Clooney AG, Sutton TDS, Ryan FJ, Daly KM, et al. 2019. The human gut virome is highly diverse, stable, and individual specific. Cell Host Microbe 26:527–41.e5 [DOI] [PubMed] [Google Scholar]
- 29.Manrique P, Bolduc B, Walk ST, van der Oost J, de Vos WM, Young MJ. 2016. Healthy human gut phageome. PNAS 113:10400–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreno-Gallego JL, Chou SP, Di Rienzi SC, Goodrich JK, Spector TD, et al. 2019. Virome diversity correlates with intestinal microbiome diversity in adult monozygotic twins. Cell Host Microbe 25:261–72.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gregory AC, Zablocki O, Zayed AA, Howell A, Bolduc B, Sullivan MB. 2020. The gut virome database reveals age-dependent patterns of virome diversity in the human gut. Cell Host Microbe 28:724–40.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, et al. 2011. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 21:1616–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reyes A, Blanton LV, Cao S, Zhao G, Manary M, et al. 2015. Gut DNA viromes of Malawian twins discordant for severe acute malnutrition. PNAS 112:11941–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dutilh BE, Cassman N, McNair K, Sanchez SE, Silva GG, et al. 2014. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat. Commun 5:4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Honap TP, Sankaranarayanan K, Schnorr SL, Ozga AT, Warinner C, Lewis CM Jr. 2020. Biogeographic study of human gut-associated crAssphage suggests impacts from industrialization and recent expansion. PLOS ONE 15:e0226930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siranosian BA, Tamburini FB, Sherlock G, Bhatt AS. 2020. Acquisition, transmission and strain diversity of human gut-colonizing crAss-like phages. Nat. Commun 11:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edwards RA, Vega AA, Norman HM, Ohaeri M, Levi K, et al. 2019. Global phylogeography and ancient evolution of the widespread human gut virus crAssphage. Nat. Microbiol 4:1727–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yutin N, Makarova KS, Gussow AB, Krupovic M, Segall A, et al. 2018. Discovery of an expansive bacteriophage family that includes the most abundant viruses from the human gut. Nat. Microbiol 3:38–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shkoporov AN, Khokhlova EV, Fitzgerald CB, Stockdale SR, Draper LA, et al. 2018. CrAss001 represents the most abundant bacteriophage family in the human gut and infects Bacteroides intestinalis. Nat. Commun 9:4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hryckowian AJ, Merrill BD, Porter NT, Van Treuren W, Nelson EJ, et al. 2020. Bacteroides thetaiotaomicron-infecting bacteriophage isolates inform sequence-based host range predictions. Cell Host Microbe 28:371–79.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gorvitovskaia A, Holmes SP, Huse SM. 2016. Interpreting Prevotella and Bacteroides as biomarkers of diet and lifestyle. Microbiome 4:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manrique P, Dills M, Young MJ. 2017. The human gut phage community and its implications for health and disease. Viruses 9(6):141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maqsood R, Rodgers R, Rodriguez C, Handley SA, Ndao IM, et al. 2019. Discordant transmission of bacteria and viruses from mothers to babies at birth. Microbiome 7:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan SK, Granados AC, Bouquet J, Hoy-Schulz YE, Green L, et al. 2020. Metagenomic sequencing of stool samples in Bangladeshi infants: virome association with poliovirus shedding after oral poliovirus vaccination. Sci. Rep 10:15392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liang G, Zhao C, Zhang H, Mattei L, Sherrill-Mix S, et al. 2020. The stepwise assembly of the neonatal virome is modulated by breastfeeding. Nature 581:470–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, et al. 2018. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 24:133–45.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pannaraj PS, Ly M, Cerini C, Saavedra M, Aldrovandi GM, et al. 2018. Shared and distinct features of human milk and infant stool viromes. Front. Microbiol 9:1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCann A, Ryan FJ, Stockdale SR, Dalmasso M, Blake T, et al. 2018. Viromes of one year old infants reveal the impact of birth mode on microbiome diversity. PeerJ 6:e4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khan Mirzaei M, Khan MAA, Ghosh P, Taranu ZE, Taguer M, et al. 2020. Bacteriophages isolated from stunted children can regulate gut bacterial communities in an age-specific manner. Cell Host Microbe 27:199–212.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller-Ensminger T, Garretto A, Stark N, Putonti C. 2020. Mimicking prophage induction in the body: induction in the lab with pH gradients. PeerJ 8:e9718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim MS, Bae JW. 2018. Lysogeny is prevalent and widely distributed in the murine gut microbiota. ISME J. 12:1127–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Furuse K, Osawa S, Kawashiro J, Tanaka R, Ozawa A, et al. 1983. Bacteriophage distribution in human faeces: continuous survey of healthy subjects and patients with internal and leukaemic diseases. J. Gen. Virol 64(9):2039–43 [DOI] [PubMed] [Google Scholar]
- 53.Duerkop BA, Clements CV, Rollins D, Rodrigues JLM, Hooper LV. 2012. A composite bacteriophage alters colonization by an intestinal commensal bacterium. PNAS 109:17621–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cornuault JK, Moncaut E, Loux V, Mathieu A, Sokol H, et al. 2020. The enemy from within: A prophage of Roseburia intestinalis systematically turns lytic in the mouse gut, driving bacterial adaptation by CRISPR spacer acquisition. ISME J. 14:771–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scanlan PD. 2017. Bacteria-bacteriophage coevolution in the human gut: implications for microbial diversity and functionality. Trends Microbiol. 25:614–23 [DOI] [PubMed] [Google Scholar]
- 56.Paterson S, Vogwill T, Buckling A, Benmayor R, Spiers AJ, et al. 2010. Antagonistic coevolution accelerates molecular evolution. Nature 464:275–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O’Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, et al. 2011. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. PNAS 108:11217–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu A, Sunagawa S, Mende DR, Bork P. 2015. Inter-individual differences in the gene content of human gut bacterial species. Genome Biol. 16:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patrick S, Blakely GW, Houston S, Moore J, Abratt VR, et al. 2010. Twenty-eight divergent polysaccharide loci specifying within- and amongst-strain capsule diversity in three strains of Bacteroides fragilis. Microbiology 156:3255–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.De Sordi L, Lourenço M, Debarbieux L. 2019. “I will survive”: a tale of bacteriophage-bacteria coevolution in the gut. Gut Microbes 10:92–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Duerkop BA, Huo W, Bhardwaj P, Palmer KL, Hooper LV. 2016. Molecular basis for lytic bacteriophage resistance in enterococci. mBio 7(4):e01304–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chatterjee A, Johnson CN, Luong P, Hullahalli K, McBride SW, et al. 2019. Bacteriophage resistance alters antibiotic-mediated intestinal expansion of enterococci. Infect. Immun 87(6):e00085–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seed KD, Yen M, Shapiro BJ, Hilaire IJ, Charles RC, et al. 2014. Evolutionary consequences of intra-patient phage predation on microbial populations. eLife 3:e03497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Porter NT, Hryckowian AJ, Merrill BD, Fuentes JJ, Gardner JO, et al. 2020. Phase-variable capsular polysaccharides and lipoproteins modify bacteriophage susceptibility in Bacteroides thetaiotaomicron. Nat. Microbiol 5:1170–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.De Sordi L, Khanna V, Debarbieux L. 2017. The gut microbiota facilitates drifts in the genetic diversity and infectivity of bacterial viruses. Cell Host Microbe 22:801–8.e3 [DOI] [PubMed] [Google Scholar]
- 66.Reyes A, Wu M, McNulty NP, Rohwer FL, Gordon JI. 2013. Gnotobiotic mouse model of phage-bacterial host dynamics in the human gut. PNAS 110:20236–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hsu BB, Gibson TE, Yeliseyev V, Liu Q, Lyon L, et al. 2019. Dynamic modulation of the gut microbiota and metabolome by bacteriophages in a mouse model. Cell Host Microbe 25:803–14.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frazão N, Sousa A, Lässig M, Gordo I. 2019. Horizontal gene transfer overrides mutation in Escherichia coli colonizing the mammalian gut. PNAS 116:17906–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Acheson DW, Reidl J, Zhang X, Keusch GT, Mekalanos JJ, Waldor MK. 1998. In vivo transduction with Shiga toxin 1-encoding phage. Infect. Immun 66:4496–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Diard M, Bakkeren E, Cornuault JK, Moor K, Hausmann A, et al. 2017. Inflammation boosts bacteriophage transfer between Salmonella spp. Science 355:1211–15 [DOI] [PubMed] [Google Scholar]
- 71.Kleiner M, Bushnell B, Sanderson KE, Hooper LV, Duerkop BA. 2020. Transductomics: sequencing-based detection and analysis of transduced DNA in pure cultures and microbial communities. Microbiome 8:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.De Sordi L, Lourenço M, Debarbieux L. 2019. The battle within: interactions of bacteriophages and bacteria in the gastrointestinal tract. Cell Host Microbe 25:210–18 [DOI] [PubMed] [Google Scholar]
- 73.Sausset R, Petit MA, Gaboriau-Routhiau V, De Paepe M. 2020. New insights into intestinal phages. Mucosal Immunol. 13:205–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lourenço M, Chaffringeon L, Lamy-Besnier Q, Pédron T, Campagne P, et al. 2020. The spatial heterogeneity of the gut limits predation and fosters coexistence of bacteria and bacteriophages. Cell Host Microbe 28:390–401.e5 [DOI] [PubMed] [Google Scholar]
- 75.Galtier M, De Sordi L, Sivignon A, de Vallée A, Maura D, et al. 2017. Bacteriophages targeting adherent invasive Escherichia coli strains as a promising new treatment for Crohn’s disease. J. Crohn’s Colitis 11:840–47 [DOI] [PubMed] [Google Scholar]
- 76.Maura D, Galtier M, Le Bouguénec C, Debarbieux L. 2012. Virulent bacteriophages can target O104:H4 enteroaggregative Escherichia coli in the mouse intestine. Antimicrob. Agents Chemother 56:6235–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weiss M, Denou E, Bruttin A, Serra-Moreno R, Dillmann ML, Brüssow H. 2009. In vivo replication of T4 and T7 bacteriophages in germ-free mice colonized with Escherichia coli. Virology 393:16–23 [DOI] [PubMed] [Google Scholar]
- 78.Gabig M, Herman-Antosiewicz A, Kwiatkowska M, Los M, Thomas MS, Wegrzyn G. 2002. The cell surface protein Ag43 facilitates phage infection of Escherichia coli in the presence of bile salts and carbohydrates. Microbiology 148:1533–42 [DOI] [PubMed] [Google Scholar]
- 79.Scanlan JG, Hall AR, Scanlan PD. 2019. Impact of bile salts on coevolutionary dynamics between the gut bacterium Escherichia coli and its lytic phage PP01. Infect. Genet. Evol 73:425–32 [DOI] [PubMed] [Google Scholar]
- 80.Scanlan PD, Bischofberger AM, Hall AR. 2017. Modification of Escherichia coli–bacteriophage interactions by surfactants and antibiotics in vitro. FEMS Microbiol. Ecol 93:fiw211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nielsen AT, Dolganov NA, Rasmussen T, Otto G, Miller MC, et al. 2010. A bistable switch and anatomical site control Vibrio cholerae virulence gene expression in the intestine. PLOS Pathog. 6:e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Barr JJ, Auro R, Furlan M, Whiteson KL, Erb ML, et al. 2013. Bacteriophage adhering to mucus provide a non-host-derived immunity. PNAS 110:10771–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Barr JJ, Auro R, Sam-Soon N, Kassegne S, Peters G, et al. 2015. Subdiffusive motion of bacteriophage in mucosal surfaces increases the frequency of bacterial encounters. PNAS 112:13675–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Joiner KL, Baljon A, Barr J, Rohwer F, Luque A. 2019. Impact of bacteria motility in the encounter rates with bacteriophage in mucus. Sci. Rep 9:16427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Almeida GMF, Laanto E, Ashrafi R, Sundberg LR. 2019. Bacteriophage adherence to mucus mediates preventive protection against pathogenic bacteria. mBio 10(6):e01984–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, et al. 2015. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 160:447–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zuo T, Lu XJ, Zhang Y, Cheung CP, Lam S, et al. 2019. Gut mucosal virome alterations in ulcerative colitis. Gut 68:1169–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Duerkop BA, Kleiner M, Paez-Espino D, Zhu W, Bushnell B, et al. 2018. Murine colitis reveals a disease-associated bacteriophage community. Nat. Microbiol 3:1023–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ansari MH, Ebrahimi M, Fattahi MR, Gardner MG, Safarpour AR, et al. 2020. Viral metagenomic analysis of fecal samples reveals an enteric virome signature in irritable bowel syndrome. BMC Microbiol. 20:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Clooney AG, Sutton TDS, Shkoporov AN, Holohan RK, Daly KM, et al. 2019. Whole-virome analysis sheds light on viral dark matter in inflammatory bowel disease. Cell Host Microbe 26:764–78.e5 [DOI] [PubMed] [Google Scholar]
- 91.Jacob V, Crawford C, Cohen-Mekelburg S, Viladomiu M, Putzel GG, et al. 2017. Single delivery of high-diversity fecal microbiota preparation by colonoscopy is safe and effective in increasing microbial diversity in active ulcerative colitis. Inflamm. Bowel Dis 23:903–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moayyedi P, Surette MG, Kim PT, Libertucci J, Wolfe M, et al. 2015. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology 149:102–9.e6 [DOI] [PubMed] [Google Scholar]
- 93.Ott SJ, Waetzig GH, Rehman A, Moltzau-Anderson J, Bharti R, et al. 2017. Efficacy of sterile fecal filtrate transfer for treating patients with Clostridium difficile infection. Gastroenterology 152(4):671–74 [DOI] [PubMed] [Google Scholar]
- 94.Gogokhia L, Buhrke K, Bell R, Hoffman B, Brown DG, et al. 2019. Expansion of bacteriophages is linked to aggravated intestinal inflammation and colitis. Cell Host Microbe 25:285–99.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sweere JM, Van Belleghem JD, Ishak H, Bach MS, Popescu M, et al. 2019. Bacteriophage trigger antiviral immunity and prevent clearance of bacterial infection. Science 363(6434):eaat9691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chatterjee A, Willett JLE, Dunny GM, Duerkop BA. 2021. Phage infection and sub-lethal antibiotic exposure mediate Enterococcus faecalis type VII secretion system dependent inhibition of bystander bacteria. PLOS Genet. 17:e1009204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, et al. 2012. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338:120–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Palmela C, Chevarin C, Xu Z, Torres J, Sevrin G, et al. 2018. Adherent-invasive Escherichia coli in inflammatory bowel disease. Gut 67:574–87 [DOI] [PubMed] [Google Scholar]
- 99.Duan Y, Llorente C, Lang S, Brandl K, Chu H, et al. 2019. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature 575:505–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fluckiger A, Daillere R, Sassi M, Sixt BS, Liu P, et al. 2020. Cross-reactivity between tumor MHC class I–restricted antigens and an enterococcal bacteriophage. Science 369:936–42 [DOI] [PubMed] [Google Scholar]
- 101.Engel P, Moran NA. 2013. The gut microbiota of insects—diversity in structure and function. FEMS Microbiol. Rev 37:699–735 [DOI] [PubMed] [Google Scholar]
- 102.Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH. 2008. How many species are infected with Wolbachia?—A statistical analysis of current data. FEMS Microbiol. Lett 281:215–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Werren JH, Zhang W, Guo LR. 1995. Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc. Biol. Sci 261:55–63 [DOI] [PubMed] [Google Scholar]
- 104.Jeyaprakash A, Hoy MA. 2000. Long PCR improves Wolbachia DNA amplification: wsp sequences found in 76% of sixty-three arthropod species. Insect Mol. Biol 9:393–405 [DOI] [PubMed] [Google Scholar]
- 105.Werren JH, Baldo L, Clark ME. 2008. Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol 6:741–51 [DOI] [PubMed] [Google Scholar]
- 106.Gavotte L, Henri H, Stouthamer R, Charif D, Charlat S, et al. 2007. A survey of the bacteriophage WO in the endosymbiotic bacteria Wolbachia. Mol. Biol. Evol 24:427–35 [DOI] [PubMed] [Google Scholar]
- 107.Bork P 1993. Hundreds of ankyrin-like repeats in functionally diverse proteins: mobile modules that cross phyla horizontally? Proteins 17:363–74 [DOI] [PubMed] [Google Scholar]
- 108.Tanaka K, Furukawa S, Nikoh N, Sasaki T, Fukatsu T. 2009. Complete WO phage sequences reveal their dynamic evolutionary trajectories and putative functional elements required for integration into the Wolbachia genome. Appl. Environ. Microbiol 75:5676–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bordenstein SR, Bordenstein SR. 2016. Eukaryotic association module in phage WO genomes from Wolbachia. Nat. Commun 7:13155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pichon S, Bouchon D, Liu C, Chen L, Garrett RA, Greve P. 2012. The expression of one ankyrin pk2 allele of the WO prophage is correlated with the Wolbachia feminizing effect in isopods. BMC Microbiol. 12:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Walker T, Klasson L, Sebaihia M, Sanders MJ, Thomson NR, et al. 2007. Ankyrin repeat domain-encoding genes in the wPip strain of Wolbachia from the Culex pipiens group. BMC Biol. 5:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.LePage DP, Metcalf JA, Bordenstein SR, On J, Perlmutter JI, et al. 2017. Prophage WO genes recapitulate and enhance Wolbachia-induced cytoplasmic incompatibility. Nature 543:243–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Turelli M, Cooper BS, Richardson KM, Ginsberg PS, Peckenpaugh B, et al. 2018. Rapid global spread of wRi-like Wolbachia across multiple Drosophila. Curr. Biol 28:963–71.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hancock PA, Sinkins SP, Godfray HC. 2011. Population dynamic models of the spread of Wolbachia. Am. Nat 177:323–33 [DOI] [PubMed] [Google Scholar]
- 115.Pietri JE, DeBruhl H, Sullivan W. 2016. The rich somatic life of Wolbachia. MicrobiologyOpen 5:923–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Brune A 2014. Symbiotic digestion of lignocellulose in termite guts. Nat. Rev. Microbiol 12:168–80 [DOI] [PubMed] [Google Scholar]
- 117.Hongoh Y, Sharma VK, Prakash T, Noda S, Toh H, et al. 2008. Genome of an endosymbiont coupling N2 fixation to cellulolysis within protist cells in termite gut. Science 322:1108–9 [DOI] [PubMed] [Google Scholar]
- 118.Pramono AK, Kuwahara H, Itoh T, Toyoda A, Yamada A, Hongoh Y. 2017. Discovery and complete genome sequence of a bacteriophage from an obligate intracellular symbiont of a cellulolytic protist in the termite gut. Microbes Environ. 32:112–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tikhe CV, Husseneder C. 2017. Metavirome sequencing of the termite gut reveals the presence of an unexplored bacteriophage community. Front. Microbiol 8:2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Clark AJ, Pontes M, Jones T, Dale C. 2007. A possible heterodimeric prophage-like element in the genome of the insect endosymbiont Sodalis glossinidius. J. Bacteriol 189:2949–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Leigh BA, Bordenstein SR, Brooks AW, Mikaelyan A, Bordenstein SR. 2018. Finer-scale phylosymbiosis: insights from insect viromes. mSystems 3(6):e00131–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Deboutte W, Beller L, Yinda CK, Maes P, de Graaf DC, Matthijnssens J. 2020. Honey-bee–associated prokaryotic viral communities reveal wide viral diversity and a profound metabolic coding potential. PNAS 117:10511–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mead PS. 2015. Epidemiology of Lyme disease. Infect. Dis. Clin. North Am 29:187–210 [DOI] [PubMed] [Google Scholar]
- 124.Stevenson B, Zuckert WR, Akins DR. 2000. Repetition, conservation, and variation: the multiple cp32 plasmids of Borrelia species. J. Mol. Microbiol. Biotechnol 2:411–22 [PubMed] [Google Scholar]
- 125.Eggers CH, Kimmel BJ, Bono JL, Elias AF, Rosa P, Samuels DS. 2001. Transduction by φBB-1, a bacteriophage of Borrelia burgdorferi. J. Bacteriol 183:4771–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Eggers CH, Samuels DS. 1999. Molecular evidence for a new bacteriophage of Borrelia burgdorferi. J. Bacteriol 181:7308–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tokarz R, Anderton JM, Katona LI, Benach JL. 2004. Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole genome DNA array. Infect. Immun 72:5419–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hefty PS, Brooks CS, Jett AM, White GL, Wikel SK, et al. 2002. OspE-related, OspF-related, and Elp lipoproteins are immunogenic in baboons experimentally infected with Borrelia burgdorferi and in human Lyme disease patients. J. Clin. Microbiol 40:4256–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Freeman CJ, Thacker RW, Baker DM, Fogel ML. 2013. Quality or quantity: Is nutrient transfer driven more by symbiont identity and productivity than by symbiont abundance? ISME J. 7:1116–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Webster NS, Taylor MW, Behnam F, Lucker S, Rattei T, et al. 2010. Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ. Microbiol 12:2070–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Vogel S 1977. Current-induced flow through living sponges in nature. PNAS 74:2069–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Horn H, Slaby BM, Jahn MT, Bayer K, Moitinho-Silva L, et al. 2016. An enrichment of CRISPR and other defense-related features in marine sponge-associated microbial metagenomes. Front. Microbiol 7:1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bergh O, Borsheim KY, Bratbak G, Heldal M. 1989. High abundance of viruses found in aquatic environments. Nature 340:467–68 [DOI] [PubMed] [Google Scholar]
- 134.Jahn MT, Arkhipova K, Markert SM, Stigloher C, Lachnit T, et al. 2019. A phage protein aids bacterial symbionts in eukaryote immune evasion. Cell Host Microbe 26:542–50.e5 [DOI] [PubMed] [Google Scholar]


