Abstract
Cardiovascular computed tomography (CT) is among the most active fields with ongoing technical innovation related to image acquisition and analysis. Artificial intelligence can be incorporated into various clinical applications of cardiovascular CT, including imaging of the heart valves and coronary arteries, as well as imaging to evaluate myocardial function and congenital heart disease. This review summarizes the latest research on the application of deep learning to cardiovascular CT. The areas covered range from image quality improvement to automatic analysis of CT images, including methods such as calcium scoring, image segmentation, and coronary artery evaluation.
Keywords: CT, Artificial intelligence, Deep learning, Heart
INTRODUCTION
Since artificial intelligence (AI) surpassed humans in the computerized version of the traditional board game Go, deep learning technology has become indispensable to technological innovation in medicine [1,2]. In cardiovascular imaging, where various technological innovations are rapidly applied, several papers on the applications of deep learning technology have been published [3]. Among cardiovascular imaging modalities, computed tomography (CT) is one of the most active fields with technical innovation. Cardiac CT has been used to evaluate coronary stenosis, identify hemodynamically significant stenosis, elucidate the pathology in structural heart disease, and measure cardiac function [4,5,6,7]. Some excellent reviews on the application of AI in cardiovascular imaging have been published recently [3,8,9,10,11]. Although these studies included cardiac CT, the subject of discussion was multimodality imaging, including echocardiography, nuclear imaging, and cardiac magnetic resonance imaging (MRI). These studies also addressed how clinicians could apply AI in the clinical workflow with multimodality imaging, such as patient screening, decision support, prognostication, and follow-up [3,9,11]. Although these discussions are worthwhile, a more focused review of the CT imaging workflow is also meaningful from the perspective of the radiologist. Litjens et al. [3] summarized deep learning research based on PubMed search results, including publications from inception until January 2019. In the present review, the results of the search using ‘deep learning’ OR ‘machine learning’ AND ‘Cardiac CT’ as keywords, with publication dates specified between January 2019 and August 2020, were added. AI can be applied to various tasks related to cardiovascular CT, such as the improvement of CT image quality, segmentation, and coronary stenosis evaluation. In this review, the latest studies have been divided into the following categories by topic: image quality improvement, segmentation of anatomic structures, automatic coronary calcium score, and coronary stenosis/plaque evaluation. The AI technologies that are currently available and those that require further research have also been discussed.
DEEP LEARNING ALGORITHMS
A detailed description of the deep learning networks is provided in the previous literature [1,3,12]. Radiologists need to understand that applied networks should be different depending on the deep learning task. The following four algorithm types have been used in the deep learning applications for cardiac CT: convolutional neural network (CNN), fully convolutional neural network (FCN), recurrent neural network (RNN), and generative adversarial network (GAN). Brief descriptions and application examples of these networks are summarized in Table 1. CNNs are the most widely used architectures, and they consist of a convolutional layer, pooling layer, and fully connected layers [3]. The convolutional layer extracts various features from an image and generates multiple feature maps. To apply CNNs to a large field of view, feature maps are progressively and spatially reduced by pooling the pixels together [1]. The pooling layer helps CNNs increase the receptive field through downsampling to facilitate an understanding of the contextual information. In the context of cardiovascular CT, CNNs can perform calcium scoring through the classification of specific voxels [13,14] or play a role in classification or slice selection as part of a segmentation algorithm [15,16]. FCNs are a modified form of CNNs that are specialized for image segmentation [3]. U-net is a type of FCN that is most widely used in organ segmentation studies [4,17,18]. After the downsampling path, which is similar to that of the CNNs, the FCNs contain an upsampling path in the architecture that produces an output image with the same resolution as the input images [1]. RNNs feed their output back as input through feedback loops, which is suitable for sequence data analysis [3]. Use cases of RNNs include electrocardiography (ECG), text, tracking of vessel centerline, cine MRI, and automatic labeling of anatomic structures [19]. GANs consist of two networks [3]: a generator and a discriminator. If the GAN network is sufficiently trained so that the discriminator cannot distinguish the image produced by the generator, a realistic image can be created [3]. Recently, GANs have become popular for image quality improvement [20] and the generation of virtual images [21].
Table 1. Deep Learning Algorithms for Cardiac CT.
| Method | Description | Example Applications |
|---|---|---|
| CNN | - The most common architecture in image analysis - CNN is composed of convolutional and pooling layers - Convolutional layer is to detect distinctive local motif-like edges and other visual elements. This operation mimics the extraction of visual features, such as edges and colors - Pooling layer downsamples the data, which helps CNNs incorporate more contextual information |
- Calcium scoring: CNN for voxel classification [13,14] - Segmentation of abdominal aortic thrombi in CT angiography [15] - Adipose tissue segmentation in a non-contrast CT: subsequent CNNs for slice selection and image segmentation [16] |
| FCN/U-net | - Adapted CNN to perform image segmentation - FCN takes an image an input and directly predict an image-sized segmentation - The most common FCN in cardiovascular imaging is U-net |
- Segmentation of cardiac structures in CT angiography [4,17,18] |
| RNN | - RNNs feed their own output back as input, which is suitable for sequence data analysis, such as text, electrocardiography, or cine-MRI | - Automated anatomical labeling of the coronary a rtery tree [19] |
| GAN | - GANs consist of two networks: generator and discriminator - GANs are used for image noise reduction or generation (e.g., conversion of MRI to CT) |
- Image noise and artifact reduction [20,23] - Generation of virtual images [21] |
CNN = convolutional neural network, CT = computed tomography, FCN = fully convolutional neural network, GAN = generative adversarial network, MRI = magnetic resonance imaging, RNN = recurrent neural network
IMAGE QUALITY IMPROVEMENT
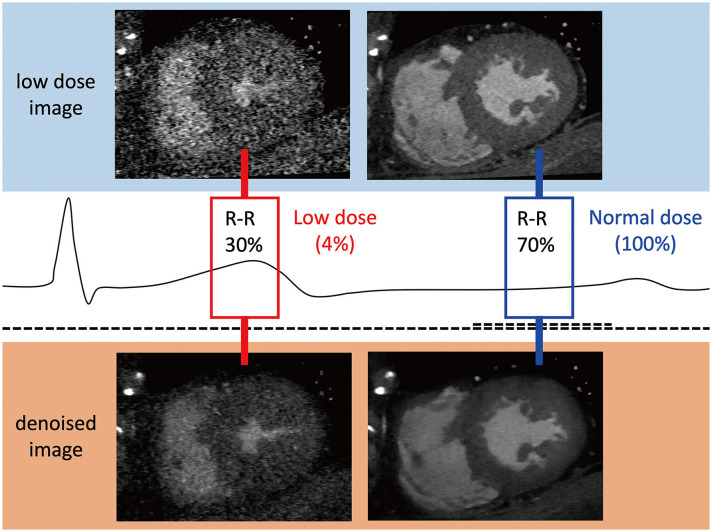
Iterative reconstruction (IR) is a classical method that has been widely used for image denoising [22]. However, IR requires huge computing resources, and IR performance highly depends on hyperparameters, which are often difficult to tune. AI-based CT denoising is rapidly emerging as an alternative to conventional IR [20,23,24]. For the AI-based method, denoised image reconstruction can be performed almost in real-time when the network has been trained, without concern about hyperparameter tuning. A ‘matched dataset’ of low-dose and standard-dose CT images is required for training a denoising network in supervised learning. Obtaining a matched dataset based on two different CT scans is a complex task, even in research settings. In contrast-enhanced CT scans, it is necessary to increase the contrast agent twice. Therefore, previous researchers have obtained low-dose datasets using anthropomorphic phantoms [23] or mathematical noise additions [24] from standard-dose images. Using paired datasets for training, AI networks learn noise patterns to predict noise maps. Denoised images can be obtained by subtracting the predicted noise maps from the low-dose images. In supervised learning, CNNs or FCNs have been used (Table 2) [24,25,26,27]. Green et al. [24] showed the efficiency of the FCN-based technique for reducing image noise in low-dose coronary computed tomography angiography (CCTA). Lossau et al. [25] applied a CNN technique to motion artifact detection and quantification in coronary arteries on CT, which is potentially useful for reducing motion artifacts. GANs are architectures for unsupervised learning [3,20]. Wolterink et al. [23] applied GANs to noise reduction of low-dose CT images obtained from an anthropomorphic phantom and nonenhanced cardiac CT. One of the important limitations of GANs for CT denoising is that potential mode-collapsing behavior means that some imaging features that are not present in the input images can be generated [28]. Kang et al. [20] introduced a cycle GAN as an alternative unsupervised learning method for denoising CCTA. They used a low-dose (20% of standard dose) image in early systole and late diastole and generated a retrospective ECG-gated scan with dose modulation. The standard-dose image was a mid-diastole (70–80% of the R-R interval) image. Using cycle GANs, they successfully obtained a denoised CT image with better performance than the state-of-the-art model-based IR technique [20]. Figure 1 and Supplementary Movie 1 show an example of denoised imaging using a cycle GAN-based image quality improvement algorithm with a very low-dose (4% of the standard dose in retrospective ECG-gated scan mode).
Table 2. Application of Deep Learning to Reduce Image Noise or Artifacts.
| Study | Year | CT Scans for Test (n) | Low Dose Data | Network | Summary |
|---|---|---|---|---|---|
| Wolterink et al. [23] | 2017 | 28 | Phantom | GAN | Noise reduction in low-dose CCTA; 3D GAN |
| Green et al. [24] | 2018 | 45 | Synthetic data | FCN | Noise reduction in low-dose CCTA; FCN for per voxel prediction |
| Lossau et al. [25] | 2019 | 4 | Synthetic data | CNN | Motion artifact recognition and quantification; CNN |
| Tatsugami et al. [26] | 2019 | 30 | Synthetic data | CNN | Deep learning–based imaging restoration; lower image noise and better CNR compared with hybrid IR images |
| Kang et al. [20] | 2019 | 50 | Patients data with different cardiac phase | GAN | Noise reduction in low-dose CCTA (multiple cardiac phase data); 2D cycle-consistent GAN (CycleGAN) |
| Benz et al. [52] | 2020 | 43 | Synthetic data | NA | Comparison between model-based IR and deep-learning image reconstruction in CCTA |
| Hong et al. [49] | 2020 | 82 | Synthetic data | FCN | Applying a deep learning–based denoising technique to CCTA along with IR for additional noise reduction |
CCTA = coronary computed tomography angiography, CNN = convolutional neural network, CNR = contrast-to-noise ratio, D = dimensional, FCN = fully convolutional neural network, GAN = generative adversarial network, IR = iterative reconstruction, NA = not available
Fig. 1. Image noise reduction by artificial intelligence in multiphase cardiac computed tomography obtained by retrospective electrocardiography-gated scanning.
SEGMENTATION OF ANATOMIC STRUCTURES
In cardiovascular CT research and quantitative reporting [6,17,29,30,31], image segmentation is the starting point and the most time-consuming step. Even with advanced cardiovascular CT applications, such as CT-derived fractional flow reserve (FFR) [5] and three-dimensional printing [7,32,33], accurate image segmentation is one of the most critical steps. If a segmentation algorithm with an accuracy similar to that of an expert is developed, radiological reporting will change, and advanced applications, such as CT-based simulation and printing, will be more widely used. Several researchers have published various studies that have applied AI to image segmentation and verified such techniques on this background (Table 3).
Table 3. Application of Deep Learning for Image Segmentation in Cardiovascular CT.
| Study | Year | Development Dataset | CT Scans for Test (n) | Performance, Dice Coefficient | Network | Target of Segmentation/Modality for Prediction |
|---|---|---|---|---|---|---|
| Trullo et al. [34] | 2017 | Nonenhanced chest CT | 30 | 0.91 | FCN | Heart, thoracic aorta/nonenhanced chest CT |
| Commandeur et al. [16] | 2018 | Nonenhanced cardiac CT | 250 | 0.82 | Multi-task CNN (ConvNet) | Epicardial adipose tissue, paracardial adipose tissue, thoracic adipose tissue/ nonenhanced cardiac CT |
| Jin et al. [18] | 2017 | CCTA | 150 | 0.94 | FCN | Left atrial appendage/CCTA |
| López-Linares et al. [15] | 2018 | Aorta CT angiography | 13 | 0.82 | FCN | Postoperative thrombus after EVAR/aorta CT angiography |
| Cao et al. [38] | 2019 | Aorta CT angiography | 30 | 0.93 | FCN (3D U-Net) | Whole aorta, true lumen, false lumen/aorta CT angiography |
| Morris et al. [35] | 2020 | Paired CT/MRI data | 11 | 0.88 | FCN (3D U-Net) | Cardiac chambers, great vessels, coronary artery/nonenhanced CT for simulation |
| Baskaran et al. [4] | 2020 | CCTA | 17 | 0.92 | FCN (3D U-Net) | Cardiac chambers, LV myocardium/CCTA |
| Bruns et al. [36] | 2020 | CCTA (dual-energy set) | 290 | 0.89 | 3D CNN | Cardiac chambers, LV myocardium/nonenhanced cardiac CT |
| Chen et al. [31] | 2020 | CCTA | 518 | - | CNN | Left atrium/CCTA |
| Koo et al. [17] | 2020 | CCTA | 1000 | 0.88 | FCN | LV myocardium/CCTA |
CCTA = coronary computed tomography angiography, CNN = convolutional neural network, CT = computed tomography, D = dimensional, DC = dice coefficient, EVAR = endovascular aneurysm repair, FCN = fully convolutional neural network, LV = left ventricle
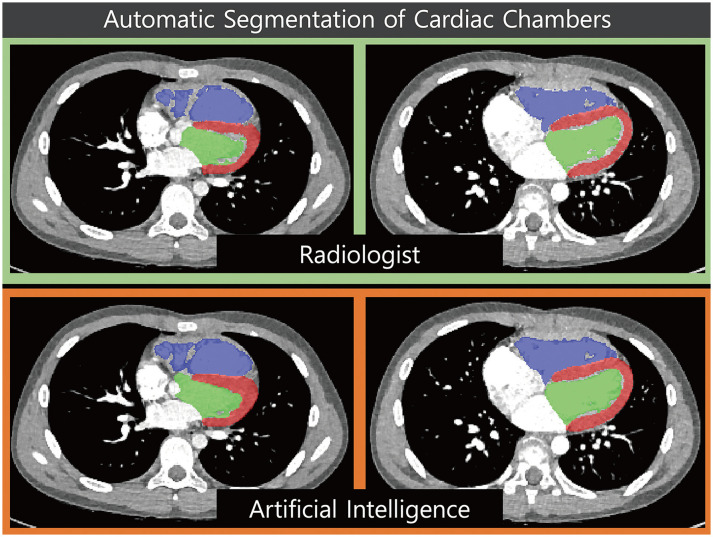
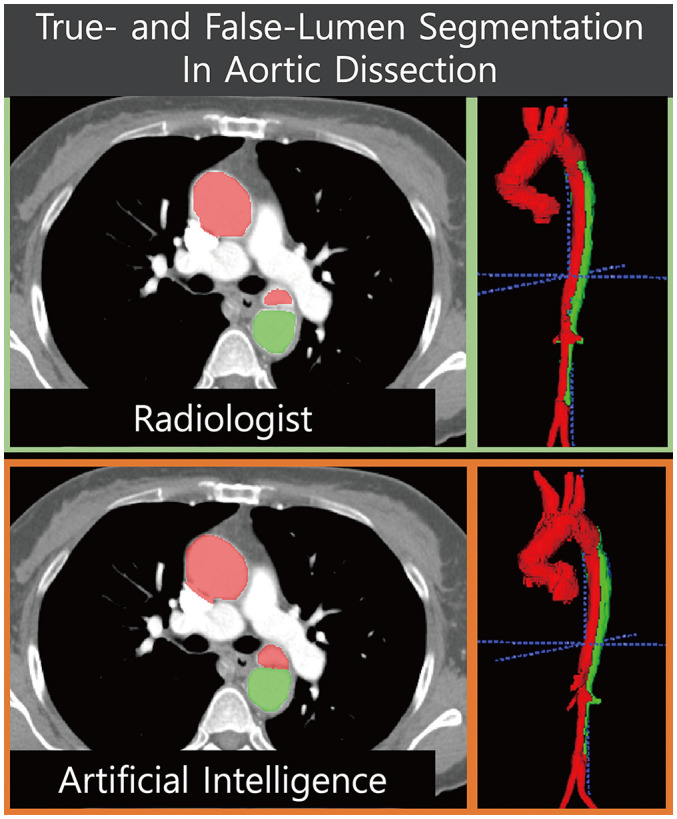
The most popular and essential targets for image segmentation are the cardiac chambers [4,31,34,35,36,37] (Fig. 2). Researchers have reported AI-based automatic segmentation of the four cardiac chambers using cardiac CT [4,17]. Koo et al. [17] reported the accuracy of AI-based segmentation of the left ventricular (LV) myocardium and contrast-filled LV chamber in 1000 CT scans. The sensitivity and specificity of the automatic segmentation for each LV segment (1–16 segments) were high (85.5–100.0%) [17]. Some researchers have studied specific areas, other than cardiac chambers, that are suitable for clinical purposes [18]. Commandeur et al. [16] reported the results of segmentation of epicardial and paracardial fat tissue on nonenhanced CT. Automatic quantification of adipose tissue showed good agreement with manual segmentation in epicardial locations (median Dice score coefficient 0.82) [16]. Cao et al. [38] split false and true lumen automatically in 276 patients with type B aortic dissection (Fig. 3). Although they showed good agreement, as defined by the Dice score coefficient (whole aorta, 0.93; true lumen, 0.93; false lumen, 0.91), the patients were relatively few (training set n = 246; testing set n = 30). Automatic quantification of a peri-graft thrombus after endovascular repair of an aortic aneurysm can be performed automatically using an AI-empowered algorithm [15]. For segmentation tasks, most researchers have applied FCNs or U-Nets (Table 3).
Fig. 2. Example of ventricular segmentation by artificial intelligence.
Fig. 3. Fully automated segmentation of true and false lumina in a patient with aortic dissection.
When developing an AI segmentation algorithm, preparing and labeling the dataset for development is a fundamental and critical step [1,3]. Most studies use the same imaging technique for labeling and prediction (e.g., left atrial labeling on CCTA for AI training [labeling] and automatic segmentation of the left atrium on CCTA [prediction]) [4,17,31,38]. Some researchers have developed an AI algorithm that divides the four cardiac chambers into nonenhanced cardiac CT images [35,36]. With nonenhanced CT, it is challenging, even for experts, to prepare a labeled dataset for AI training. Researchers have solved this problem in a quite creative way. Morris et al. [35] used a paired CT/MRI dataset to label nonenhanced CT images. Bruns et al. [36] prepared enhanced CT and virtual nonenhanced CT datasets using dual-energy cardiac CT and developed a segmentation algorithm that works for nonenhanced CT images. Lee et al. [39] developed an algorithm that works for nonenhanced CT images using matched CCTA and nonenhanced CT training datasets, which were used for automatic coronary calcium (CAC) scoring. The use of these unmatched datasets to solve technically challenging issues is an excellent example of how radiologists can contribute to AI algorithm development.
CORONARY CALCIUM SCORING
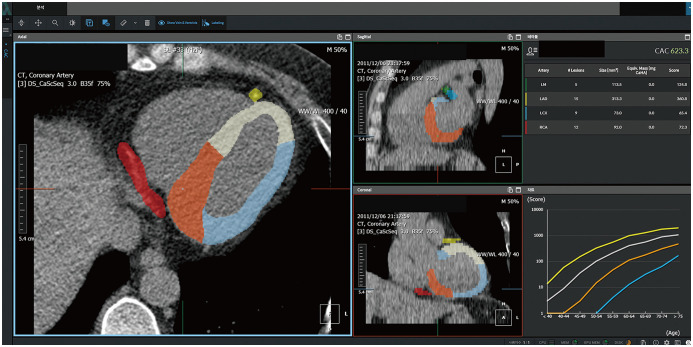
The CAC scoring workflow can be described as follows: 1) in a nonenhanced scan, determine whether calcium over 130 Hounsfield unit is present in the coronary artery, and 2) if calcium is in a coronary artery, identify the coronary artery (e.g., left main, right coronary artery, left anterior descending artery). Several researchers attempted automatic CAC scoring using feature extraction or multi-atlas methods even in the pre-AI era, but the results were unsatisfactory based on the high false-positive lesions and long calculation durations [40,41,42]. Recently, automatic CAC scoring using AI has been reported, showing promising results in clinical practice (Table 4). Lessmann et al. [13] employed a CNN to classify calcium candidate objects (sensitivity of 91.2% per lesion). Martin et al. [14] presented a multi-step deep learning model and tested it in 511 patients. The first step was used to identify and segment the regions, such as the coronary artery, aorta, aortic valve, and mitral valve. The second step classified the voxels as coronary calcium. They achieved a good sensitivity of 93.2% and an intraclass correlation coefficient of 0.985. Recently, Lee et al. [39] proposed an atlas-based fully automated CAC scoring system that uses AI (Fig. 4). The novelty of the system is that it can precisely detect coronary artery regions using a deep learning model based on semantic segmentation in a single step. This method can also provide regional information about the coronary artery and surrounding structures, such as the aorta, ventricular chambers, and myocardium. Therefore, this method can be easily extended to the segmentation of the aortic and mitral valves. This atlas-based automatic algorithm showed good agreement with manual segmentation (sensitivity, 93.3%; intraclass correlation coefficient, 0.99) as well as a low false-positive rate (0.11 calcium lesion per CT scan).
Table 4. Application of Automatic Coronary Calcium Scoring.
| Study | Year | Sensitivity (%), per Lesion | False Positives per CT Scan | ICC | CT Scans for Test (n) | Protocol of CT Scan | Method of Detection |
|---|---|---|---|---|---|---|---|
| Wolterink et al. [53] | 2016 | 79 | 0.2 | 0.96 | 530 | ECG-gated CT | 2.5D patch-based CNN (15 or 25 sizes from axial, coronal, sagittal planes) |
| Lessmann et al. [13] | 2018 | 91.2 | 40.7 mm3/scan | NA | 506 | Non-ECG-gated chest CT | Cascaded two 2.5D CNNs (CNN1 with large receptive field and CNN2 with smaller receptive field) |
| Cano-Espinosa et al. [54] | 2018 | NA | NA | 0.93* | 1000 | Non-ECG-gated chest CT | CNN based regression model, which directly predicts CAC score |
| Martin et al. [14] | 2020 | 93.2† | NA | 0.985 | 511 | ECG-gated CT | Two-fold deep-learning models, first one to exclude aorta, aortic valve, mitral valve regions; second one to classify coronary calcium voxels |
| van Velzen et al. [55] | 2020 | 93 | 4 mm3/scan | 0.99 | 529 | ECG-gated CT Non-EGC-gated chest CT (n = 3811) |
Same as Lessmann et al. 2018 [13] |
| Lee et al. [39] | 2021 | 93.3 | 0.11 | 0.99 | 2985 | ECG-gated CT | 3D patch-based CNN for semantic segmentation |
*Pearson correlation coefficient, †Per-patient sensitivity. CAC = coronary artery calcium score, CNN = convolutional neural network, CT = computed tomography, D = dimensional, ECG = electrocardiography, ICC = intraclass correlation coefficient, NA = not applicable
Fig. 4. Fully automated coronary artery calcium scoring software.
CORONARY STENOSIS AND PLAQUE EVALUATION
Among the techniques for predicting hemodynamically significant coronary stenosis with CCTA, CT-FFR has attracted the most attention [29,43]. However, this technique requires extensive coronary lumen segmentation and complex simulations of computational fluid dynamics [29]. Recently, machine learning-based CT-derived FFR technology was reported and is expected to increase efficiency [44]. This machine learning application was trained using 12000 synthetic coronary trees with various degrees of coronary stenosis, for which the CT-FFR values were computed using the computational fluid dynamics method [4]. It should be noted that machine learning shortens the computational simulation time and not the time required for coronary lumen segmentation [44]. The diameters of the coronary artery and plaque are relatively small, and research to accurately distinguish them is still in the early stages (Table 5). Other alternatives for predicting FFR using CCTA have also been reported [45,46,47]. van Hamersvelt et al. [46] applied feature extraction from the LV myocardium to classify patients with hemodynamically significant stenosis. The deep learning algorithm that characterized the LV myocardium improved the diagnosis of hemodynamically significant coronary stenosis (area under the receiver operating characteristic curve [AUC] = 0.76) as compared with the diameter stenosis (AUC = 0.68) [4]. Zreik et al. [45] successfully identified patients requiring invasive angiography in the stretched multiplanar reformatted image of CCTA using an autoencoder and a support vector machine. Using invasive FFR as a reference standard, the AUC for detecting coronary stenosis requiring invasive evaluation was 0.81 at the per-vessel level and 0.87 at the per-patient level [45]. Deep learning applications for relatively basic technologies, such as centerline extraction of the coronary tree [48] and annotation of coronary segments [19], have also been recently reported. The tree labeling network, a type of RNN, can annotate main coronary branches and major side branches with high accuracy (main branch 97%, side branch 90%) [19]. This automated anatomical labeling technology can streamline the diagnostic workflow in daily practice for radiologists and radiology technologists. Hong et al. [27] reported deep learning-based coronary stenosis quantification using CCTA. All quantitative measurements showed a good correlation between an expert reader and a deep learning algorithm (minimal lumen area r = 0.984, diameter stenosis r = 0.975, and percent contrast density difference r = 0.975) [27]. To date, one study reported by Kumamaru et al. [21] predicted FFR using only CT images without any human input. They overcame the complex process of coronary lumen segmentation using GAN. The deep-learning CT-FFR model achieved 76% accuracy in detecting abnormal FFR [21]. Coronary stenosis and plaque analysis require continuous technological development in the future for clinical applications.
Table 5. Application of Coronary Stenosis and Plaque.
| Study | Year | Role of Artificial Intelligence | Dataset for Test | Performance | Algorithm |
|---|---|---|---|---|---|
| Coenen et al. [44] | 2018 | Prediction of FFR based on synthetic data (manual segmentation of coronary tree) in CCTA | 351 | 0.78 accuracy | Machine learning using synthetic stenosis model and computational fluid dynamics results |
| Zreik et al. [47] | 2019 | Automatic plaque and stenosis characterization in stretched MPR image of CCTA | 65 | 0.77 accuracy | 3D recurrent CNN |
| van Hamersvelt et al. [46] | 2019 | Identification of patients with functionally significant coronary stenosis in CCTA | 101 | 0.76 AUC | CNN for LV myocardial segmentation; SVM for patient classification |
| Wolterink et al. [48] | 2019 | Coronary centerline extraction in CCTA | 24 | 93.7% overlap | 3D CNN for prediction of vessel orientation and radius to guide iterative tracker |
| Wu et al. [19] | 2019 | Coronary artery tree segment labeling in CCTA | 436 | 0.87 F1 | Bidirectional LSTM in three graph representation |
| Hong et al. [27] | 2019 | Quantification of coronary stenosis automatically in CCTA | 156 | 0.95 correlation coefficient | U-Net |
| Zreik et al. [45] | 2020 | Identification of a patient requiring invasive coronary angiography in stretched MPR image of CCTA | 137 | 0.81 AUC | Autoencoder and SVM |
| Kumamaru et al. [21] | 2020 | Fully automatic estimation of minimum FFR from CCTA (i.e., free from human input) | 131 | 0.76 accuracy | Lumen extraction block using GAN; residual extraction block; prediction block for minimum FFR estimation |
AUC = area under the receiver operating characteristic curve, CCTA = coronary computed tomography angiography, CNN = convolutional neural network, CT = computed tomography, D = dimensional, ECG = electrocardiography, FFR = fractional flow reserve, GAN = generative adversarial network, LSTM = long short-term memory, LV = left ventricle, MPR = multiplanar reformatted, SVM = support vector machine
FUTURE PERSPECTIVES
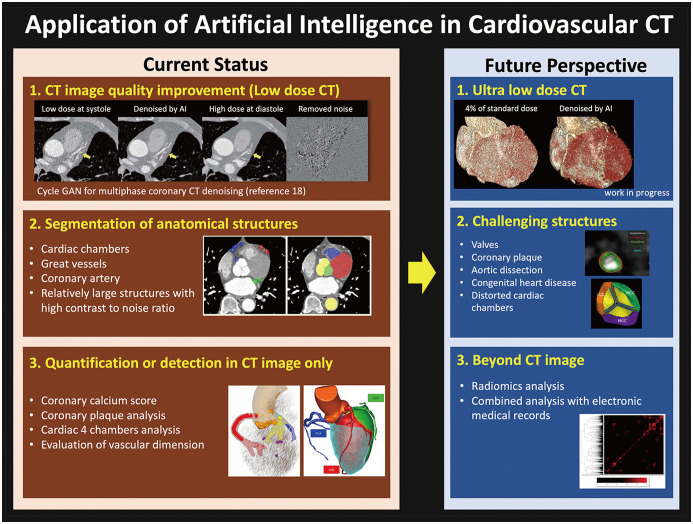
The application of AI in cardiovascular CT is being developed in various ways, from image improvement to quantitative diagnosis. Some technologies are ready to be used in routine workflows or are already being used (Fig. 5); for example, low-dose CT denoising [49] or automatic quantification of relatively easy to distinguish structures (e.g., coronary calcification, cardiac chambers) [17]. Some technologies have promising results [27], but they require more time to be applied in routine practice. Typical examples include the improvement of ultra-low-dose CT and the segmentation of very small or delicate structures (e.g., coronary plaques and valves) [21,27]. Recently, electronic medical records with large data have been prepared for various AI research [50]. Cardiovascular CT powered by AI or radiomic analysis [51] can be combined with other imaging modalities or clinical information (e.g., ECG and blood laboratory tests) to guide decision-making or prognostication.
Fig. 5. Current status and future perspectives of artificial intelligence in cardiovascular CT.
CT = computed tomography, GAN = generative adversarial network
Cardiovascular CT requires quantitative reporting for several scenarios, such as CAC scoring, ventricular function assessment, and coronary stenosis evaluation [29,33,43]. For quantitative reporting, a process that lasts for a shorter duration is essential, and AI can significantly help shorten the duration of the quantification process [17,39]. To date, one of the best techniques for shortening the actual workflow is automatic CAC scoring [14,39]. At Asan Medical Center, Seoul, Korea, from October 2020, manual primary analysis of calcium scoring by an experienced radiological technologist was replaced by AI software-guided analysis. After primary analysis by AI, the CAC results are automatically transferred to a picture archiving system, and the radiologist carries out the final confirmation of CAC. The time saved by AI will be used for higher-level image analysis processes (e.g., ventricular function analysis) to improve the quality of CT reporting. Cardiac CT can be applied to the evaluation of the coronary arteries and other indications, such as the evaluation of the cardiac valves, myocardium, and congenital heart disease. Therefore, it is necessary to develop specific AI applications to promote research and quantitative reporting in related fields.
CONCLUSION
AI can be implemented and applied in various ways to cardiovascular CT-from image reconstruction to quantitative analysis. The interpretation room environment of radiologists changes as AI technology develops. As imaging specialists, radiologists should be actively involved in the entire process of AI development: from conception to clinical validation.
Footnotes
This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HR20C0026020021); and a grant of the KHIDI, funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI18C0022).
Conflicts of Interest: The authors have no potential conflicts of interest to disclose.
Supplement
The Supplement is available with this article at https://doi.org/10.3348/kjr.2020.1314.
Image noise reduction by AI in multiphase cardiac CT obtained by retrospective ECG-gated scanning.
References
- 1.Lee JG, Jun S, Cho YW, Lee H, Kim GB, Seo JB, et al. Deep learning in medical imaging: general overview. Korean J Radiol. 2017;18:570–584. doi: 10.3348/kjr.2017.18.4.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silver D, Schrittwieser J, Simonyan K, Antonoglou I, Huang A, Guez A, et al. Mastering the game of go without human knowledge. Nature. 2017;550:354–359. doi: 10.1038/nature24270. [DOI] [PubMed] [Google Scholar]
- 3.Litjens G, Ciompi F, Wolterink JM, de Vos BD, Leiner T, Teuwen J, et al. State-of-the-art deep learning in cardiovascular image analysis. JACC Cardiovasc Imaging. 2019;12:1549–1565. doi: 10.1016/j.jcmg.2019.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Baskaran L, Maliakal G, Al'Aref SJ, Singh G, Xu Z, Michalak K, et al. Identification and quantification of cardiovascular structures from CCTA: an end-to-end, rapid, pixel-wise, deep-learning method. JACC Cardiovasc Imaging. 2020;13:1163–1171. doi: 10.1016/j.jcmg.2019.08.025. [DOI] [PubMed] [Google Scholar]
- 5.Yang DH, Kim YH, Roh JH, Kang JW, Ahn JM, Kweon J, et al. Diagnostic performance of on-site CT-derived fractional flow reserve versus CT perfusion. Eur Heart J Cardiovasc Imaging. 2017;18:432–440. doi: 10.1093/ehjci/jew094. [DOI] [PubMed] [Google Scholar]
- 6.Yang DH, Kim DH, Handschumacher MD, Levine RA, Kim JB, Sun BJ, et al. In vivo assessment of aortic root geometry in normal controls using 3D analysis of computed tomography. Eur Heart J Cardiovasc Imaging. 2017;18:780–786. doi: 10.1093/ehjci/jew146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim W, Lim M, Jang YJ, Koo HJ, Kang JW, Jung SH, et al. Novel resectable myocardial model using hybrid three-dimensional printing and silicone molding for mock myectomy for apical hypertrophic cardiomyopathy. Korean J Radiol. 2021;22:1054–1065. doi: 10.3348/kjr.2020.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen C, Qin C, Qiu H, Tarroni G, Duan J, Bai W, et al. Deep learning for cardiac image segmentation: a review. Front Cardiovasc Med. 2020;7:25. doi: 10.3389/fcvm.2020.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dey D, Slomka PJ, Leeson P, Comaniciu D, Shrestha S, Sengupta PP, et al. Artificial intelligence in cardiovascular imaging: JACC state-of-the-art review. J Am Coll Cardiol. 2019;73:1317–1335. doi: 10.1016/j.jacc.2018.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siegersma KR, Leiner T, Chew DP, Appelman Y, Hofstra L, Verjans JW. Artificial intelligence in cardiovascular imaging: state of the art and implications for the imaging cardiologist. Neth Heart J. 2019;27:403–413. doi: 10.1007/s12471-019-01311-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sermesant M, Delingette H, Cochet H, Jaïs P, Ayache N. Applications of artificial intelligence in cardiovascular imaging. Nat Rev Cardiol. 2021;18:600–609. doi: 10.1038/s41569-021-00527-2. [DOI] [PubMed] [Google Scholar]
- 12.Do S, Song KD, Chung JW. Basics of deep learning: a radiologist's guide to understanding published radiology articles on deep learning. Korean J Radiol. 2020;21:33–41. doi: 10.3348/kjr.2019.0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lessmann N, van Ginneken B, Zreik M, de Vos PA, de Vos BD, Viergever MA, et al. Automatic calcium scoring in low-dose chest CT using deep neural networks with dilated convolutions. IEEE Trans Med Imaging. 2018;37:615–625. doi: 10.1109/TMI.2017.2769839. [DOI] [PubMed] [Google Scholar]
- 14.Martin SS, van Assen M, Rapaka S, Hudson HT, Jr, Fischer AM, Varga-Szemes A, et al. Evaluation of a deep learning–based automated CT coronary artery calcium scoring algorithm. JACC Cardiovasc Imaging. 2020;13:524–526. doi: 10.1016/j.jcmg.2019.09.015. [DOI] [PubMed] [Google Scholar]
- 15.López-Linares K, Aranjuelo N, Kabongo L, Maclair G, Lete N, Ceresa M, et al. Fully automatic detection and segmentation of abdominal aortic thrombus in post-operative CTA images using deep convolutional neural networks. Med Image Anal. 2018;46:202–214. doi: 10.1016/j.media.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 16.Commandeur F, Goeller M, Betancur J, Cadet S, Doris M, Chen X, et al. Deep learning for quantification of epicardial and thoracic adipose tissue from non-contrast CT. IEEE Trans Med Imaging. 2018;37:1835–1846. doi: 10.1109/TMI.2018.2804799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koo HJ, Lee JG, Ko JY, Lee G, Kang JW, Kim YH, et al. Automated segmentation of left ventricular myocardium on cardiac computed tomography using deep learning. Korean J Radiol. 2020;21:660–669. doi: 10.3348/kjr.2019.0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin C, Feng J, Wang L, Yu H, Liu J, Lu J, et al. Left atrial appendage segmentation using fully convolutional neural networks and modified three-dimensional conditional random fields. IEEE J Biomed Health Inform. 2018;22:1906–1916. doi: 10.1109/JBHI.2018.2794552. [DOI] [PubMed] [Google Scholar]
- 19.Wu D, Wang X, Bai J, Xu X, Ouyang B, Li Y, et al. Automated anatomical labeling of coronary arteries via bidirectional tree LSTMs. Int J Comput Assist Radiol Surg. 2019;14:271–280. doi: 10.1007/s11548-018-1884-6. [DOI] [PubMed] [Google Scholar]
- 20.Kang E, Koo HJ, Yang DH, Seo JB, Ye JC. Cycle-consistent adversarial denoising network for multiphase coronary CT angiography. Med Phys. 2019;46:550–562. doi: 10.1002/mp.13284. [DOI] [PubMed] [Google Scholar]
- 21.Kumamaru KK, Fujimoto S, Otsuka Y, Kawasaki T, Kawaguchi Y, Kato E, et al. Diagnostic accuracy of 3D deep-learning-based fully automated estimation of patient-level minimum fractional flow reserve from coronary computed tomography angiography. Eur Heart J Cardiovasc Imaging. 2020;21:437–445. doi: 10.1093/ehjci/jez160. [DOI] [PubMed] [Google Scholar]
- 22.Naoum C, Blanke P, Leipsic J. Iterative reconstruction in cardiac CT. J Cardiovasc Comput Tomogr. 2015;9:255–263. doi: 10.1016/j.jcct.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 23.Wolterink JM, Leiner T, Viergever MA, Isgum I. Generative adversarial networks for noise reduction in low-dose CT. IEEE Trans Med Imaging. 2017;36:2536–2545. doi: 10.1109/TMI.2017.2708987. [DOI] [PubMed] [Google Scholar]
- 24.Green M, Marom EM, Konen E, Kiryati N, Mayer A. 3-D Neural denoising for low-dose Coronary CT Angiography (CCTA) Comput Med Imaging Graph. 2018;70:185–191. doi: 10.1016/j.compmedimag.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Lossau T, Nickisch H, Wissel T, Bippus R, Schmitt H, Morlock M, et al. Motion artifact recognition and quantification in coronary CT angiography using convolutional neural networks. Med Image Anal. 2019;52:68–79. doi: 10.1016/j.media.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 26.Tatsugami F, Higaki T, Nakamura Y, Yu Z, Zhou J, Lu Y, et al. Deep learning-based image restoration algorithm for coronary CT angiography. Eur Radiol. 2019;29:5322–5329. doi: 10.1007/s00330-019-06183-y. [DOI] [PubMed] [Google Scholar]
- 27.Hong Y, Commandeur F, Cadet S, Goeller M, Doris MK, Chen X, et al. Deep learning-based stenosis quantification from coronary CT Angiography. Proc SPIE Int Soc Opt Eng. 2019;10949:109492I. doi: 10.1117/12.2512168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu JY, Park T, Isola P, Efros AA. Unpaired image-to-image translation using cycle-consistent adversarial networks; Proceedings of the IEEE International Conference on Computer Vision (ICCV); 2017 Oct 22–29; Venice, Italy. IEEE; 2017. pp. 2223–2232. [Google Scholar]
- 29.Yang DH, Kang SJ, Koo HJ, Kweon J, Kang JW, Lim TH, et al. Incremental value of subtended myocardial mass for identifying FFR-verified ischemia using quantitative CT angiography: comparison with quantitative coronary angiography and CT-FFR. JACC Cardiovasc Imaging. 2019;12:707–717. doi: 10.1016/j.jcmg.2017.10.027. [DOI] [PubMed] [Google Scholar]
- 30.Yang DH, Kang SJ, Koo HJ, Chang M, Kang JW, Lim TH, et al. Coronary CT angiography characteristics of OCT-defined thin-cap fibroatheroma: a section-to-section comparison study. Eur Radiol. 2018;28:833–843. doi: 10.1007/s00330-017-4992-8. [DOI] [PubMed] [Google Scholar]
- 31.Chen HH, Liu CM, Chang SL, Chang PY, Chen WS, Pan YM, et al. Automated extraction of left atrial volumes from two-dimensional computer tomography images using a deep learning technique. Int J Cardiol. 2020;316:272–278. doi: 10.1016/j.ijcard.2020.03.075. [DOI] [PubMed] [Google Scholar]
- 32.Yang DH, Park SH, Lee K, Kim T, Kim JB, Yun TJ, et al. Applications of three-dimensional printing in cardiovascular surgery: a case-based review. Cardiovasc Imaging Asia. 2018;2:166–175. [Google Scholar]
- 33.Yang DH, Park SH, Kim N, Choi ES, Kwon BS, Park CS, et al. Incremental value of 3D printing in the preoperative planning of complex congenital heart disease surgery. JACC Cardiovasc Imaging. 2021;14:1265–1270. doi: 10.1016/j.jcmg.2020.06.024. [DOI] [PubMed] [Google Scholar]
- 34.Trullo R, Petitjean C, Nie D, Shen D, Ruan S. Joint segmentation of multiple thoracic organs in CT images with two collaborative deep architectures. Deep Learn Med Image Anal Multimodal Learn Clin Decis Support (2017) 2017;10553:21–29. doi: 10.1007/978-3-319-67558-9_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morris ED, Ghanem AI, Dong M, Pantelic MV, Walker EM, Glide-Hurst CK. Cardiac substructure segmentation with deep learning for improved cardiac sparing. Med Phys. 2020;47:576–586. doi: 10.1002/mp.13940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bruns S, Wolterink JM, Takx RAP, van Hamersvelt RW, Suchá D, Viergever MA, et al. Deep learning from dual-energy information for whole-heart segmentation in dual-energy and single-energy non-contrast-enhanced cardiac CT. Med Phys. 2020;47:5048–5060. doi: 10.1002/mp.14451. [DOI] [PubMed] [Google Scholar]
- 37.Yu Y, Gao Y, Wei J, Liao F, Xiao Q, Zhang J, et al. A three-dimensional deep convolutional neural network for automatic segmentation and diameter measurement of type B aortic dissection. Korean J Radiol. 2021;22:168–178. doi: 10.3348/kjr.2020.0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao L, Shi R, Ge Y, Xing L, Zuo P, Jia Y, et al. Fully automatic segmentation of type B aortic dissection from CTA images enabled by deep learning. Eur J Radiol. 2019;121:108713. doi: 10.1016/j.ejrad.2019.108713. [DOI] [PubMed] [Google Scholar]
- 39.Lee JG, Kim HS, Kang H, Koo HJ, Kang JW, Kim YH, et al. Fully automatic coronary calcium score software empowered by artificial intelligence technology: validation study using three CT cohorts. Korean J Radiol. 2021 Aug; doi: 10.3348/kjr.2021.0148. [Epub] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Isgum I, Rutten A, Prokop M, van Ginneken B. Detection of coronary calcifications from computed tomography scans for automated risk assessment of coronary artery disease. Med Phys. 2007;34:1450–1461. doi: 10.1118/1.2710548. [DOI] [PubMed] [Google Scholar]
- 41.Brunner G, Chittajallu DR, Kurkure U, Kakadiaris IA. Toward the automatic detection of coronary artery calcification in non-contrast computed tomography data. Int J Cardiovasc Imaging. 2010;26:829–838. doi: 10.1007/s10554-010-9608-1. [DOI] [PubMed] [Google Scholar]
- 42.Shahzad R, van Walsum T, Schaap M, Rossi A, Klein S, Weustink AC, et al. Vessel specific coronary artery calcium scoring: an automatic system. Acad Radiol. 2013;20:1–9. doi: 10.1016/j.acra.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 43.Nørgaard BL, Leipsic J, Gaur S, Seneviratne S, Ko BS, Ito H, et al. Diagnostic performance of noninvasive fractional flow reserve derived from coronary computed tomography angiography in suspected coronary artery disease: the NXT trial (analysis of coronary blood flow using CT angiography: next steps) J Am Coll Cardiol. 2014;63:1145–1155. doi: 10.1016/j.jacc.2013.11.043. [DOI] [PubMed] [Google Scholar]
- 44.Coenen A, Kim YH, Kruk M, Tesche C, De Geer J, Kurata A, et al. Diagnostic accuracy of a machine-learning approach to coronary computed tomographic angiography–based fractional flow reserve: result from the MACHINE consortium. Circ Cardiovasc Imaging. 2018;11:e007217. doi: 10.1161/CIRCIMAGING.117.007217. [DOI] [PubMed] [Google Scholar]
- 45.Zreik M, van Hamersvelt RW, Khalili N, Wolterink JM, Voskuil M, Viergever MA, et al. Deep learning analysis of coronary arteries in cardiac CT angiography for detection of patients requiring invasive coronary angiography. IEEE Trans Med Imaging. 2020;39:1545–1557. doi: 10.1109/TMI.2019.2953054. [DOI] [PubMed] [Google Scholar]
- 46.van Hamersvelt RW, Zreik M, Voskuil M, Viergever MA, Išgum I, Leiner T. Deep learning analysis of left ventricular myocardium in CT angiographic intermediate-degree coronary stenosis improves the diagnostic accuracy for identification of functionally significant stenosis. Eur Radiol. 2019;29:2350–2359. doi: 10.1007/s00330-018-5822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zreik M, van Hamersvelt RW, Wolterink JM, Leiner T, Viergever MA, Isgum I. A recurrent CNN for automatic detection and classification of coronary artery plaque and stenosis in coronary CT angiography. IEEE Trans Med Imaging. 2019;38:1588–1598. doi: 10.1109/TMI.2018.2883807. [DOI] [PubMed] [Google Scholar]
- 48.Wolterink JM, van Hamersvelt RW, Viergever MA, Leiner T, Išgum I. Coronary artery centerline extraction in cardiac CT angiography using a CNN-based orientation classifier. Med Image Anal. 2019;51:46–60. doi: 10.1016/j.media.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 49.Hong JH, Park EA, Lee W, Ahn C, Kim JH. Incremental image noise reduction in coronary CT angiography using a deep learning-based technique with iterative reconstruction. Korean J Radiol. 2020;21:1165–1177. doi: 10.3348/kjr.2020.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahn I, Na W, Kwon O, Yang DH, Park GM, Gwon H, et al. CardioNet: a manually curated database for artificial intelligence-based research on cardiovascular diseases. BMC Med Inform Decis Mak. 2021;21:29. doi: 10.1186/s12911-021-01392-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kolossváry M, Park J, Bang JI, Zhang J, Lee JM, Paeng JC, et al. Identification of invasive and radionuclide imaging markers of coronary plaque vulnerability using radiomic analysis of coronary computed tomography angiography. Eur Heart J Cardiovasc Imaging. 2019;20:1250–1258. doi: 10.1093/ehjci/jez033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benz DC, Benetos G, Rampidis G, von Felten E, Bakula A, Sustar A, et al. Validation of deep-learning image reconstruction for coronary computed tomography angiography: impact on noise, image quality and diagnostic accuracy. J Cardiovasc Comput Tomogr. 2020;14:444–451. doi: 10.1016/j.jcct.2020.01.002. [DOI] [PubMed] [Google Scholar]
- 53.Wolterink JM, Leiner T, de Vos BD, van Hamersvelt RW, Viergever MA, Išgum I. Automatic coronary artery calcium scoring in cardiac CT angiography using paired convolutional neural networks. Med Image Anal. 2016;34:123–136. doi: 10.1016/j.media.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 54.Cano-Espinosa C, González G, Washko GR, Cazorla M, Estépar RSJ. Automated Agatston score computation in non-ECG gated CT scans using deep learning. Proc SPIE Int Soc Opt Eng. 2018;10574:105742K. doi: 10.1117/12.2293681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Velzen SGM, Lessmann N, Velthuis BK, Bank IEM, van den Bongard DHJG, Leiner T, et al. Deep learning for automatic calcium scoring in CT: validation using multiple cardiac CT and chest CT protocols. Radiology. 2020;295:66–79. doi: 10.1148/radiol.2020191621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Image noise reduction by AI in multiphase cardiac CT obtained by retrospective ECG-gated scanning.