Abstract
Background
Coronavirus disease 2019 (COVID-19) is a pandemic that has caused a high number of deaths worldwide. Inflammatory factors may play important roles in COVID-19 progression. Yindan Jiedu granules (YDJDG) can inhibit the progression of COVID-19, but the associated mechanism is unclear.
Purpose
To evaluate the therapeutic effects of YDJDG on COVID-19 and explore its underlying mechanism.
Methods
We recruited 262 participants and randomly assigned 97 patients each to the YDJDG and control groups using one-to-one propensity score matching (PSM). Clinical effects were observed and serum inflammatory and immune indicators were measured. The target network model of YDJDG was established by predicting and determining the targets of identified compounds. The main constituents of the YDJDG extracts were identified and evaluated using high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) and molecular docking. Besides, the anti-inflammatory effects of YDJDG and its specific biological mechanism of action were studied.
Results
After PSM, the results showed that compared with the control group, the YDJDG group had a shorter time of dissipation of acute pulmonary exudative lesions (p < 0.0001), shorter time to negative conversion of viral nucleic acid (p < 0.01), more rapid decrease in serum amyloid A level and erythrocyte sedimentation rate (p < 0.0001), and a higher rate of increase in CD4+T cell count (p = 0.0155). By overlapping the genes of YDJDG and COVID-19, 213 co-targeted genes were identified. Metascape enrichment analysis showed that 25 genes were significantly enriched in the NF-κB pathway, which were mainly targets of luteolin, quercetin, and kaempferol as confirmed by MS analysis. Molecular docking revealed that the ligands of three compounds had strong interaction with NF-κB p65 and IκBα. In vivo, YDJDG significantly protected animals from lipopolysaccharide (LPS)-induced acute lung injury (ALI), decreasing the lung wet/dry weight ratio, ALI score, and lung histological damage. In LPS-treated RAW264.7 cells, YDJDG suppressed nuclear translocation of NF-κB p65. In vivo and in vitro, YDJDG exerted anti-inflammatory effects by inhibiting the production of inflammatory cytokines (IL-6, IL-1β, and TNF-α). These effects were accompanied by the inhibition of NF-ĸB activation and IκBα phosphorylation.
Conclusion
YDJDG may shorten the COVID-19 course and delay its progression by suppressing inflammation via targeting the NF-κB pathway.
Keywords: COVID-19, Yindan jiedugranules, Clinical research, Pharmacology analysis, Molecular docking, NF-κB signaling pathway
Abbreviations: ACE2, angiotensin-converting enzyme 2; ALI, acute lung injury; COVID-19, Coronavirus disease 2019; CRP, C-reactive protein; DL, drug-likeness property; ESR, erythrocyte sedimentation rate; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; LC, liquid chromatography; MS, mass spectrometry; NHC, National Health Commission; NLR, neutrophil-to-lymphocyte ratio; OB, satisfying oral bioavailability; PSM, propensity score matching; SAA, serum amyloid A; TCM, Traditional Chinese medicine; TCMSP, Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform; YDJDG, Yindan Jiedu granules.
Graphical abstract
Introduction
Since December 2019, the novel coronavirus disease (COVID-19) pandemic has been spreading rapidly worldwide. As of July 6, 2021, the number of confirmed infections exceeded 184,269,026 with 3986,902 deaths (https://coronavirus.jhu.edu/map.html). However, there are currently no effective therapeutic drugs against the virus.
Available data indicate that highly dysregulated exuberant inflammatory responses correlate with disease severity and lethality in COVID-19 patients. Epithelial-immune cell interactions and elevated levels of cytokines and chemokines trigger unchecked feed‐forward activation and amplification of the host immune system. This causes further massive release of a wide range of cytokines, such as interferon (IFN)‐γ, tumor necrosis factor (TNF), interleukin (IL)‐1, IL‐6, and IL‐18, which contributes to the formation of a “cytokine storm”, and plays a central role in disease severity and lethality (Gao et al., 2021; Kircheis et al., 2020). Therefore, it is very important to focus on the key factors associated with cytokine storm syndrome (CSS) in COVID-19 progression. In a recent study, a quantitative proteomic approach combined with bioinformatics analysis was used to detect proteomic changes in SARS-CoV-2-infected lung tissues. The result revealed the upregulation of several inflammatory factors, possibly caused by the activation of NF-κB signaling (Leng et al., 2020). The inhibition of the NF-κB pathway, such as immunomodulation at the level of NF-κB activation and inhibitors of NF-κB (IκB) degradation will potentially reduce cytokine storm and alleviate the severity of COVID-19. However, the NF-κB pathway is a complex cascade reaction system, and the simultaneous inhibition of multiple cytokines/chemokines is expected to have higher therapeutic potential than single target approaches to prevent cascade effects of multiple induced cytokines and chemokines at the critical stage of COVID-19 (Hariharan et al., 2021). Unfortunately, there is a lack of drugs with multi-target effects against the NF-κB cascade reaction system.
In China, traditional Chinese medicine (TCM) contains a variety of active compounds and may have the advantages of multi-factorial and multi-target effects on diseases. Some drugs, including QingFei Paidu decoction and Lianhua Qingwen capsule, significantly shortened the duration of fever and relieved symptoms in patients with severe COVID-19 (Yang et al., 2020; Zhang et al., 2020b). However, more drugs need to be evaluated. Yindan Jiedu granules (YDJDG), prescribed as a newly applied Chinese herbal formula at the Beijing Ditan Hospital, is a compounded Chinese herbal medicine produced through the combination of the Maxing Shigan decoction and Qingwen Baidu decoction, which have multiple therapeutic targets and comprehensive therapeutic action (Wen et al., 2020; Zhong et al., 2016). Between January and March 2020, YDJDG was used to treat COVID-19 patients at Beijing Ditan Hospital. YDJDG could hasten the recovery period in patients with mild/moderate COVID-19 or CD4+ T cell count < 660 cells/µl by shortening the mean duration of fever and pulmonary exudative lesions, compared with lopinavir-ritonavir treatment (Liu et al., 2020a). However, in this previous study, there was no significant difference in the time-to-negative conversion of SARS-CoV-2 nucleic acid of the two groups (p = 0.0729). Moreover, the study lacked a blank control (routine treatment, without any antiviral drugs). Furthermore, the mechanism of action of YDJDG in the treatment of COVID-19 has remained unclear.
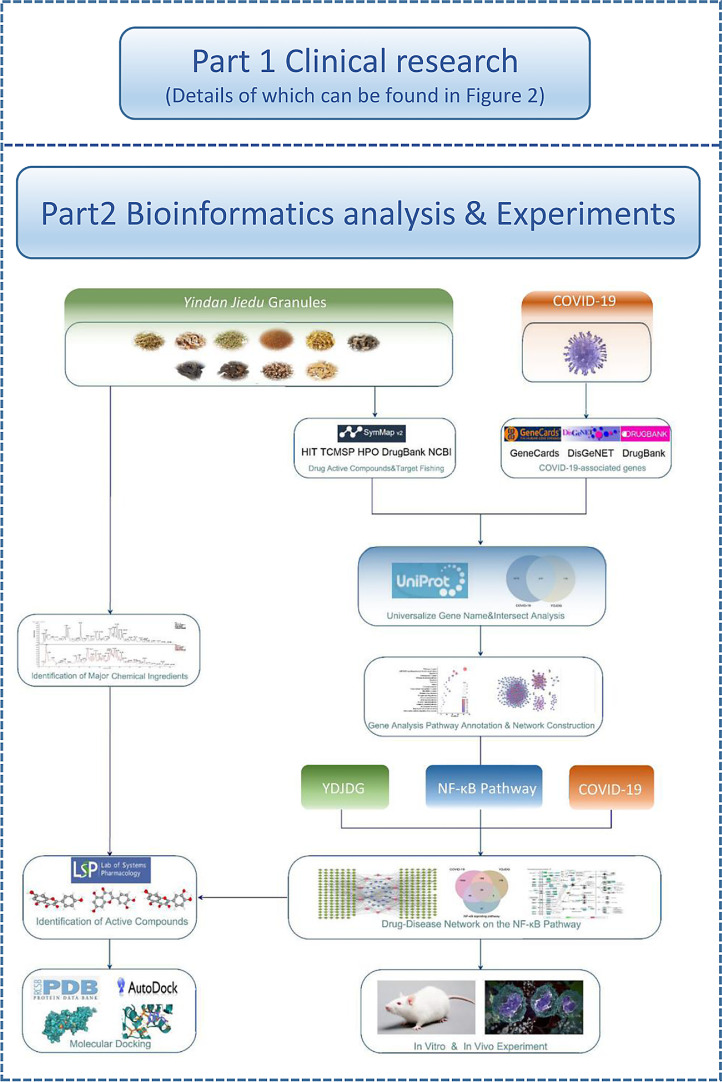
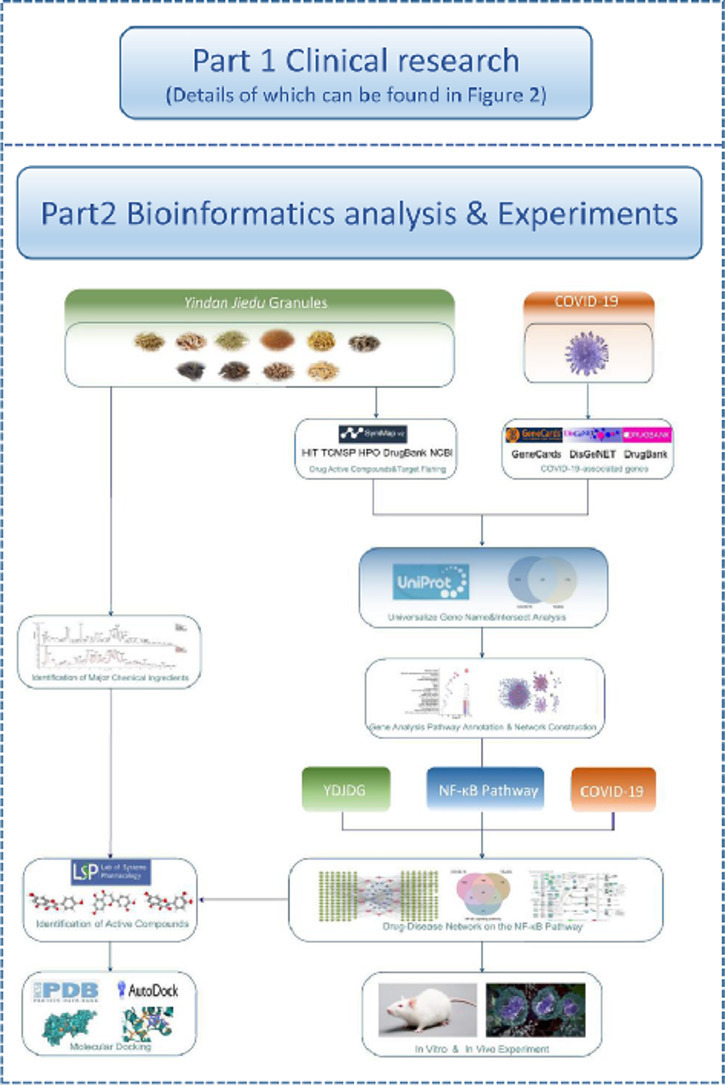
In this study, we first evaluated the effectiveness of YDJDG in COVID-19 patients. Furthermore, we established the target network model of YDJDG to predict and select the targets of identified compounds by conducting network pharmacology analysis and verified the anti-inflammatory effect in vivo and in vitro. The main constituents of the YDJDG extracts were identified and evaluated using high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS). Finally, based on the results of network pharmacology analysis and experiment, we conducted molecular docking to explore if some key components of YDJDG have potential inhibitory effect on the activation of the NF-κB pathway. In summary, this study provides clinical, bioinformatics and experiment basis for COVID-19 treatment with YDJDG. The flowchart of this research was shown in Fig. 1, Fig. 2 .
Fig. 1.
Graphical abstract. Part 1 is the clinical research, details of which can be found in Fig. 2. Part 2 is the flowchart of the network pharmacology research and experiments.
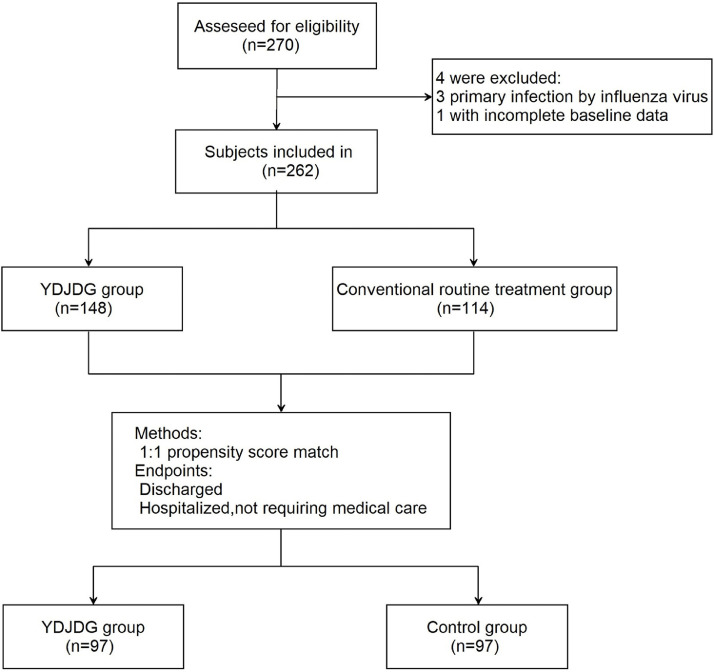
Fig. 2.
Flowchart of the clinical research. A total of 270 subjects were screened for eligibility, 262 patients were included in the research. According to propensity score matching (PSM), 194 participants were selected for each of the two groups: 97 in the YDJDG group and 97 in control group. COVID-19, Coronavirus disease-2019; YDJDG, Yindan Jiedu granules.
Patients and methods
Patients
A total of 270 patients with COVID-19 were recruited from the Beijing Ditan Hospital between January 29, 2020 and July 23, 2020. The diagnosis, clinical classification, inclusion, and exclusion criteria have been described in a paper previously published by our research group (Liu et al., 2020b). A total of 262 patients were included in the study; among these, 148 were receiving YDJDG therapy and 114 were receiving routine treatment (Fig. 2). This study was approved by the Institutional Research Ethics Committee of Beijing Ditan Hospital, Capital Medical University (Beijing, China). All the patients provided signed informed consent.
Drug preparation and treatment
YDJDG was approved by the Beijing Medical Products Administration (China) (No. Z20200012000) and was produced according to the Pharmacopeia of the People's Republic of China. YDJDG is composed of Ephedrae Herba, Gypsum Fibrosum, Mori Cortex, Scutellariae Radix, Lepidii Semen, Lonicerae Japonicae Flos, Scrophulariae Radix, Moutan Cortex, Rehmanniae Radix, Atractylodis Macrocephalae Rhizoma, and Cimicifugae Rhizoma (Beijing Medicinal Materials Company, Beijing, China). The production method was reported in our previous study (Liu et al., 2020a). Routine treatment generally consisted of supportive treatments, such as oxygen and symptomatic therapies, according to the Diagnosis and Treatment Protocol for Coronavirus Pneumonia (Trial version 8) at the discretion of the attending clinicians. The dose of YDJDG was 12 g or 24 g, which was administered orally three times per day (Fig. 2).
Propensity score matching (PSM)
To reduce the influence of confounding factors, we performed a greedy PSM at a 1:1 ratio to divide the participants into YDJDG and control groups (Fig. 2). The variables for this procedure were median age; sex; median days from onset of illness to admission time; number of coexisting comorbidities; disease severity; neutrophil, lymphocyte, CD4+ T cell, and platelet counts; neutrophil-to-lymphocyte ratio (NLR); potassium, sodium, hemoglobin, C-reactive protein (CRP), serum amyloid A (SAA), and lactic acid levels; and erythrocyte sedimentation rate (ESR). We confirmed the multicollinearity of all variables in PSM using the variance inflation factor. On the logit of the propensity score, we employed caliper widths equal to 0.010 of the standard deviation. The baseline characteristics of patients with COVID-19 before and after PSM are shown in Table 1 .
Table 1.
Baseline characteristics of COVID-19 patients before and after propensity score matching.
| Before propensity score matching | After propensity score matching | ||||||
|---|---|---|---|---|---|---|---|
| Variables | Control (n = 114) | YDJDG (n = 148) | p Value | Control (n = 97) | YDJDG (n = 97) | p Value | |
| Median Age (range) | 40 (18–86) | 42 (18–84) | 0.947 a) | 39 (18–86) | 42 (18–84) | 0.791 a) | |
| Gender (M/F) | 63/51 | 74/74 | 0.398 c) | 51/46 | 55/42 | 0.641 c) | |
| Median days from illness onset to admission time (range) | 4 (0–29) | 3 (1–38) | 0.004 b) | 3 (0–29) | 3 (1–38) | 0.108 b) | |
| No. of coexisting comorbidity | |||||||
| One | 42 (36.8) | 79 (53.4) | 0.103 c) | 38 (39.2) | 47 (48.5) | 0.193 c) | |
| Two | 9 (7.9) | 17 (11.5) | 0.382 c) | 8 (8.2) | 10 (10.3) | 0.621 c) | |
| Three | 2 (1.8) | 5 (3.4) | 0.431c) | 2 (2.1) | 2 (2.1) | 1.000 c) | |
| Disease severity | |||||||
| Mild | 8 (7.0) | 23 (15.5) | 0.059 c) | 8 (8.2) | 14 (14.4) | 0.174 c) | |
| Moderate | 94 (82.5) | 114 (77.0) | 0.716c) | 83 (85.6) | 74 (76.3) | 0.100 c) | |
| Severe | 10 (8.8) | 4 (2.7) | 0.041 c) | 5 (5.2) | 4 (4.1) | 0.733 c) | |
| Critical | 2 (1.8) | 7 (4.7) | 0.204 c) | 1 (1.0) | 5 (5.2) | 0.097 c) | |
| Potassium, mmol/l | 3.7 (3.4–4.0) | 3.6 (3.3–3.9) | 0.266 b) | 3.7 (3.4–4.0) | 3.7 (3.3–4.0) | 0.794 b) | |
| Sodium, mmol/l | 139.9 (137.9–141.0) | 140.0 (138.0–141.6) | 0.225 b) | 140.0 (138.1–141.2) | 139.9 (137.9–141.9) | 0.816 b) | |
| Hemoglobin, g/l | 143.5 (133.0–155.0) | 143.0 (129.3–153.8) | 0.388 b) | 144.0 (133.5–156.0) | 144.0 (130.5–153.0) | 0.418 b) | |
| Neutrophil count, × 10⁹/l | 2.9 (2.2–3.9) | 2.7 (2.1–3.9) | 0.438 b) | 2.8 (2.1–3.6) | 2.7 (2.1–3.9) | 0.559 b) | |
| Lymphocyte count, × 10⁹/l | 1.4 (1.0–1.9) | 1.4 (1.1–1.8) | 0.777 b) | 1.5 (1.1–1.9) | 1.4 (1.1–1.8) | 0.398 b) | |
| NLR | 2.1 (1.4–3.5) | 1.9 (1.4–2.7) | 0.425 b) | 1.9 (1.3–2.9) | 2.1 (1.4–2.8) | 0.263 b) | |
| Platelet count, × 10⁹/l | 206.5 (155.8–261.3) | 207.5 (168.0–249.8) | 0.925 b) | 208.0 (159.5–253.0) | 206.0 (161.5–249.0) | 0.548 b) | |
| CD4+T, cells/μl | 677.0 (442.0–788.0) | 645.5 (497.8–880.5) | 0.516 b) | 690.0 (467.5–783.5) | 624.0 (445.3–857.3) | 0.968 b) | |
| C-reactive protein, mg/l | 6.9 (1.6–27.6) | 2.8 (1.1–14.8) | 0.004 b) | 5.4 (1.1–17.9) | 4.9 (1.5–16.5) | 0.862 b) | |
| Serum amyloid A, mg/l | 31.4 (4.9–140.1) | 12.6 (3.7–48.4) | 0.049 b) | 17.0 (2.6–71.4) | 19.9 (4.3–64.0) | 0.801 b) | |
| Erythrocyte sedimentation rate, mm/h | 14.0 (9.0–31.0) | 10.0 (5.0–18.0) | 0.003 b) | 13.0 (8.0–23.0) | 11.0 (5.5–18.5) | 0.128 b) | |
| Lactic acid, mmol/l | 2.4 (1.9–2.8) | 2.7 (2.2–3.3) | 0.006 b) | 2.4 (1.9–2.9) | 2.3 (1.8–2.8) | 0.765 b) | |
Data are presented as median (range), n (%), median (interquartile range), or means ± SD.
p values comparing YDJD group and Control group are from t-test a), χ² test b), or Mann-Whitney U test c).
COVID-19, 2019 coronavirus disease. YDJDG, Yindan Jiedu granule; NLR, neutrophil-to-lymphocyte ratio.
Collection and screening of compounds and targets
The original compounds and the related targets of 10 herbs of YDJDG (except gypsum) were searched in the SymMap database, which integrated information on the ingredients from the TCMID (version 2015), TCMSP (version 2.3), and TCM-ID (version 1.0) databases and information on these targets was obtained from the HIT, TCMSP, HPO, DrugBank, and NCBI databases. Active compounds with oral bioavailability (OB) ≥ 30% and their related targets were selected.
Screening of targets for COVID-19
COVID-19-associated genes were screened from the GeneCards (www.genecards.org) database, DisGeNET platform (http://www.disgenet.org/), and DrugBank (www.drugbank.ca) using the keyword “COVID-19″. Proteins were transformed to the gene symbols with “Homo sapiens” in the UniProt (https://www.uniprot.org/) database.
Gene analysis and pathway annotation
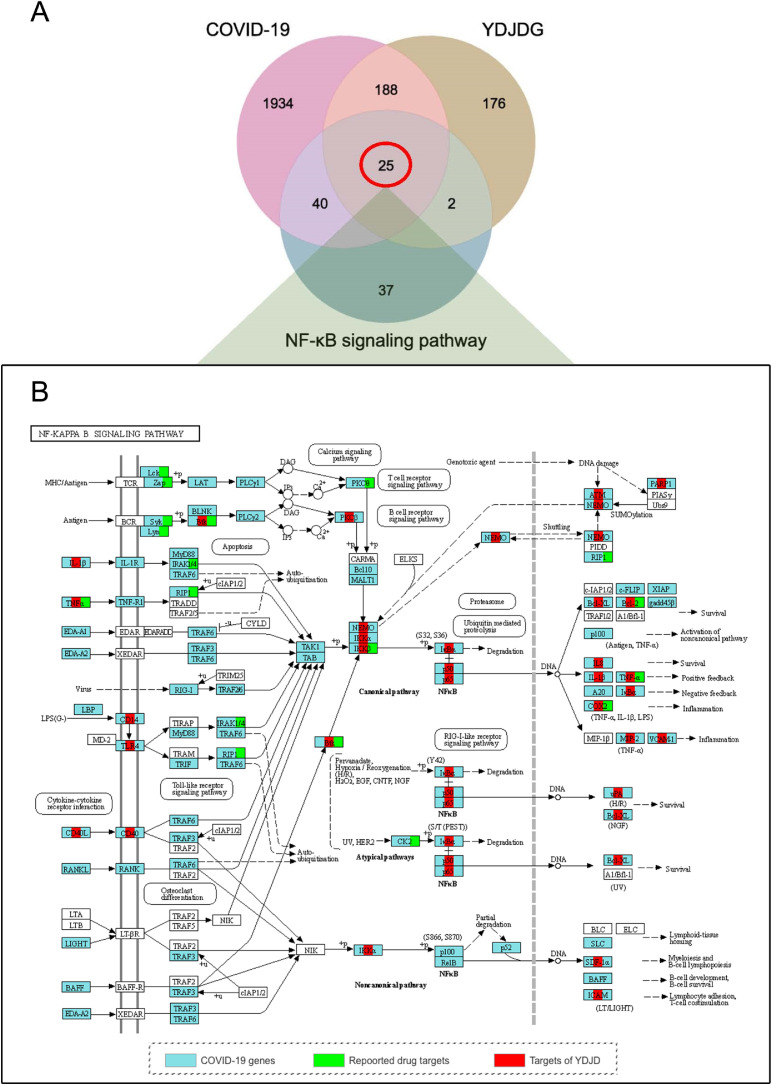
The overlaps between COVID-19 and YDJDG targets were analyzed using the online tool Draw Venn Diagram (http://bioinformatics.psb.ugent.be/Webtools/Venn/). These gene targets were supposed to be the active targets of YDJDG for the treatment of COVID-19. The targets of antiviral action were entered into the Metascape platform (http://metascape.org) for enrichment analysis of the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. The targets were analyzed, and the results were saved and sorted based on the number of targets involved in each entry to screen the top 20 pathways. The NF-κB signaling pathway, as a classic inflammation-related pathway, ranked number 5, is considered one of the critical pathways associated with the therapeutic effect of YDJDG against COVID-19; hence, the drug-disease network of YDJDG and COVID-19 in relation to the NF-κB signaling pathway was constructed. The results were visualized using the Pathview website (https://pathview.uncc.edu/).
Network construction
We established an intervention network between the potential therapeutic targets of YDJDG and the target genes of COVID-19. In order to further focus on the effect of YDJDG on inflammation, we mapped the herb-ingredient-gene network of YDJDG, COVID-19, and the NF-κB signaling pathway. The protein-protein interaction (PPI) network was constructed using STRING (https://www.string-db.org/) and Cytoscape 3.8.0.
Identification of major chemical ingredients
Three bags of YDJDG were randomly selected, and approximately 200 mg of sample was collected. The method was as reported in our previous study (Liu et al., 2020a). The data were initially collated and then searched in three databases (mzCloud, mzV ault, and ChemSpider) for comparison.
Serum collection from YDJDG-treated Sprague Dawley (SD) rats
Serum from YDJDG-treated SD rats was collected according to a previously described method (Yang et al., 2021). Healthy adult SD rats (male, weighing 200 g, 8-week-old) were purchased and maintained in Beijing Vital River Laboratory Animal Technologies Co. Ltd (Beijing, China), starved for 4 h and then intragastrically administered YDJDG (1.26 ml, 504 mg) once every 12 h. An hour after the fifth administration, blood samples were collected from the rats using aseptic methods.
Establishment of acute Lung injury (ALI) model
BALB/c mice (male, weighing 18–22 g, 5-week-old) were purchased from Beijing Vital River Laboratory Animal Technologies Co. Ltd (Beijing, China) and raised under specific-pathogen-free conditions. The study was approved by the Vital River Institutional Animal Care and Use Committee (permit number: RSD-SOP-002–01). A total of 24 mice were randomly divided into four groups: control, lipopolysaccharide (LPS), LPS + YDJDG, and LPS + Methylprednisolone (MP). MP is a common clinical oral drug to treat inflammation, and it was intragastrically administered to the mice at a dose of 6 mg/kg, based on the recommended dose for the treatment of COVID-19 patients and the effective dose for ALI mice model in previous studies (Pinzón et al., 2021; Shang et al., 2020; Silva et al., 2009). ALI was induced by intratracheal instillation of LPS (E. coli O111:B4, 5 mg/kg, lot no. L2630, Sigma, USA). The mice in the LPS + YDJDG group received 0.182 ml (72.8 mg) YDJDG, which was administered 4 h and 10 h after LPS instillation. The mice in the LPS + MP group received 0.12 ml (0.12 mg) MP, 4 h after LPS instillation. The mice in the LPS+saline group received saline solution (0.9% NaCl) 4 h and 10 h after LPS instillation, while those in the control group received only saline solution. The animals were euthanized 24 h after LPS administration.
Processing of bronchoalveolar lavage fluid (BALF)
The right main bronchus was tied with a string at the hilum. BALF from all the groups was obtained from the left lung by connecting a syringe to the cannula placed in the trachea and then gently flushing through 1.5 ml sterile 0.9% NaCl solution five times with 1 mM of ethylenediaminetetraacetic acid. BALF recovery was always greater than 85%. BALF was spun at 800 × g for 10 min, and the supernatant was stored at −80 °C for subsequent ELISA analysis. Next, the RIPA lysis buffer (1% phenylmethanesulfonyl fluoride, PMSF, and 1% cocktail) was used to extract the total protein of the left lung for subsequent western blot analysis.
Histopathological evaluation
The right lower lobe lungs of the mice were immersed in normal 10% paraformaldehyde, cut into 5 µm sections, stained with hematoxylin and eosin and analyzed by two independent experts blinded to treatment. The entire surface of the lung was analyzed for inflammation and damage, and was scored on a 5-point scale as follows: 0 = minimal damage, 1+ = mild damage, 2+ = moderate damage, 3+ = severe damage, and 4+ = maximal damage (Mikawa et al., 2003).
Lung wet/dry weight ratio
To assess edema, the remaining right lung was dissected immediately after exsanguination and the wet weight was recorded. The lung was then placed in an incubator at 80 °C for 48 h, after which the dry weight was recorded and the lung wet-to-dry weight ratio was determined.
Cell culture
The RAW 264.7 mouse macrophage cell line was preserved in our laboratory, cultured in DMEM containing 10% FBS, and incubated at 37 °C with 5% CO2. In all the experiments, the cells were allowed to acclimatize for 24 h before any treatment.
Cytotoxicity assay
According to the manufacturer's instructions, cell viability was evaluated using Cell Counting Kit-8 (CCK-8, lot no. CA1210, Solarbio, China). Briefly, the cells were seeded in 96-well plates at a density of 2 × 104 cells/well in the presence or absence of 0.1–10 μg/ml LPS (E. coli O111:B4, lot no. L4391, Sigma, USA) and YDJDG (final concentration of 10–30%) for 24 h. After incubating the cells with CCK-8 solution for 2 h, optical density (OD) was measured at 450 nm using a microplate reader (Sigma Aldrich, St. Louis, MO, USA).
NO production and cytokine analysis
RAW264.7 cells were plated at a density of 2 × 104 cells/ml in 96-well plates and incubated with or without LPS (10 μg/ml) in the absence or presence of 20% YDJDG for 6–24 h. Dexamethasone (Dex, 100 μg/ml; lot no. D1756–25MG, Merck, USA) was used as a positive control. The concentration of nitric oxide (NO) in the culture medium was measured using the Griess reagent method (lot no. S0021S; Beyotime, China), according to the manufacturer's protocol. Absorbance at 540 nm was measured using a microplate reader (Sigma Aldrich, St. Louis, MO, USA).
The amount of cytokine expression was measured by a commercial ELISA kit: TNF-α (lot no. ADI-900–047; Neobioscience, China), IL-6 (lot no.88–7064–86; Neobioscience, China), and IL-1β (lot no. 88–8014–22; Neobioscience, China), following the instructions provided by the manufacturer. The absorbance of the resultant solutions were read at 450 nm on a microplate reader (Sigma Aldrich, St. Louis, MO, USA).
Analysis of NF-κB nuclear translocation by immunofluorescent staining
RAW264.7 cells (1.2 × 106 cells/ml) were cultured in confocal dishes for 24 h, pretreated with YDJDG (20%) for 2 h, and LPS-stimulated (10 μg/ml) for 1 h. Using the NF-κB nuclear translocation assay kit (antibodies against NF-κB p65, 1:400; lot no.8242T; CST, USA), according to the manufacturer's instructions. Finally, coverslips were mounted on slides, and fluorescence signals were analyzed by confocal laser scan microscopy (Leica, LSM3, Germany).
Western blot analysis
Protein levels in the cells or lung tissues were detected by western blotting. RIPA buffer was used to extract the total protein, and the protein concentration was determined using the bicinchoninic acid method. Equal amounts (10–20 μg) of the protein were added to the sample wells. Sodium dodecyl sulfate-polyacrylamide gels (8–15%) were used to separate proteins, and the gels were then transferred onto polyvinylidene difluoride membranes. Western blotting was performed with antibodies against NF-κB p65 (1:1000; lot no.8242T; CST, USA), Phospho-NF-κB p65 (Ser536) (1:1000; lot no. 3033T; CST, USA), IκBα (44D4) (1:1000; lot no. 4812S; CST, USA), and Phospho-IκBα (1:1000; lot no. 2859T; CST, USA). GAPDH antibody (1:1000; lot no.5174; CST, USA) was used as a loading control. Image J software (National Institutes of Health, Bethesda, MD, USA) was used to quantify the integrated density of each band.
Molecular docking
The 2D structure of the main active ingredients was downloaded from TCMSP, and the 3D structure of the target proteins was obtained from the RCSB database (https://www.rcsb.org/). The Pymol software was used to remove solvent molecules and ligands, and the AutoDock 4.2 software was used for hydrogenation, electron addition, and ROOT addition. After the completion of molecular docking, the protein structure was set to rigid macromolecules, and the algorithm was used as a local search parameter. The binding energy was defined as ≤ −5.0 kJ∙mol−1, which was the screening criterion, and Pymol was used to construct the hydrogen bonds between the active ingredients and the target proteins.
Statistical analysis
Statistical analyses were conducted using SPSS (version 22.0; IBM, NY, USA), GraphPad (GraphPad Software, CA, USA), and R (version 4.0.4, R Foundation for Statistical Computing, Vienna, Austria) software packages. Categorical data were recorded as the number of observations. The variables that conformed to a normal distribution are presented as mean ± standard deviation. Non-normal distribution variables are presented as medians (Q1 and Q3). The clinical and demographic characteristics in this study are summarized as median, range, and number. Baseline characteristics, before and after matching, were evaluated using standardized differences. Student's t-test was used to compare samples in terms of continuous variables, and Pearson's chi-square test or Mann-Whitney U test was applied to compare the relationship between two or more categorical variables. The markers at different time points were analyzed using two-way repeated-measures ANOVA. Statistical significance was set at p < 0.05, and all experiments were repeated at least three times.
Results
Clinical characteristics of participants
A total of 270 subjects were screened for eligibility and 262 patients were included in this study. After PSM, 194 participants were selected: 97 each in the YDJDG and control groups. There was no significant difference between the two groups with regard to age; gender; median days from onset of illness to admission time; number of coexisting comorbidities; disease severity; neutrophil, lymphocyte, platelet, and CD4+ T cell counts; CRP, SAA, potassium, sodium, hemoglobin, and lactic acid levels; NLR; and ESR (p > 0.05) (Table 1).
Recovery time after treatment
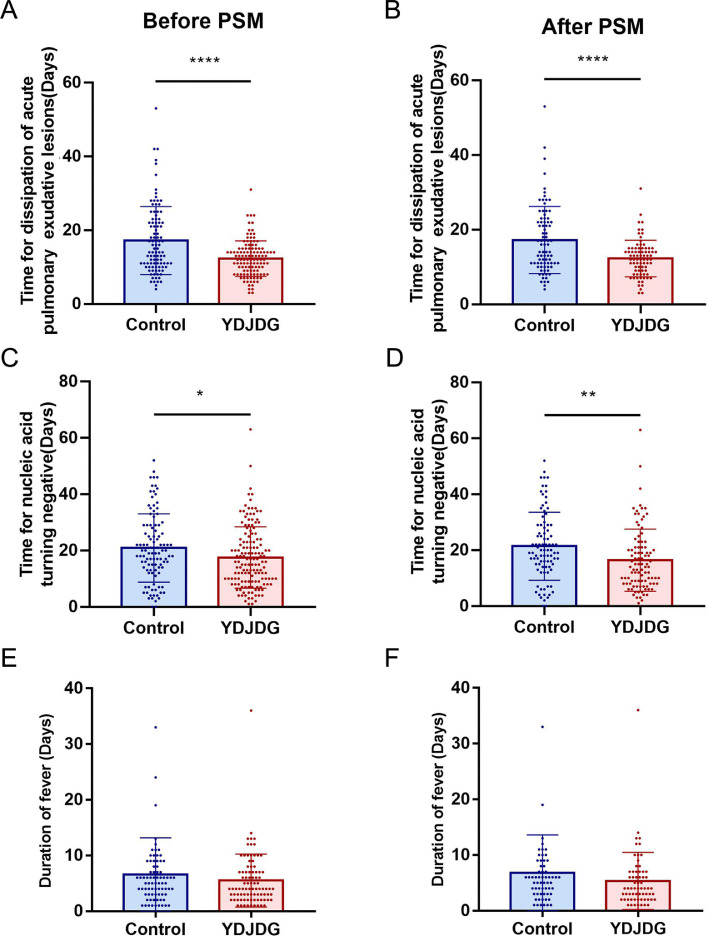
Compared with the control group, the YDJDG group had a significantly shorter time of dissipation of acute pulmonary exudative lesions (p < 0.0001) and a shorter time to negative conversion of viral nucleic acid (p < 0.05) in 262 subjects (Fig. 3 A, C). After PSM, the time of dissipation of acute pulmonary exudative lesions in the YDJDG group was still shorter than that in the control group (p < 0.0001) (Fig. 3B), and the time to negative conversion of viral nucleic acid in the YDJDG group was shorter than that in the control group (p < 0.01) (Fig. 3D). There were no significant differences in the duration of fever between the two groups (Fig. 3E, F).
Fig. 3.
Recovery time after treatment. Comparison of the time of dissipation of acute pulmonary exudative lesions of YDJDG and the control group before (A) and after PSM (B). Comparison of the times of negative conversion of nucleic acid of the YDJDG and control groups before (C) and after PSM (D). Comparison of fever duration of the YDJDG and control groups before (E) and after PSM (F). **** p < 0.0001, *** p < 0.001, ** p < 0.01, and * p < 0.05. YDJDG, Yindan Jiedu granules; PSM, propensity score matching.
Changes in indices after treatment
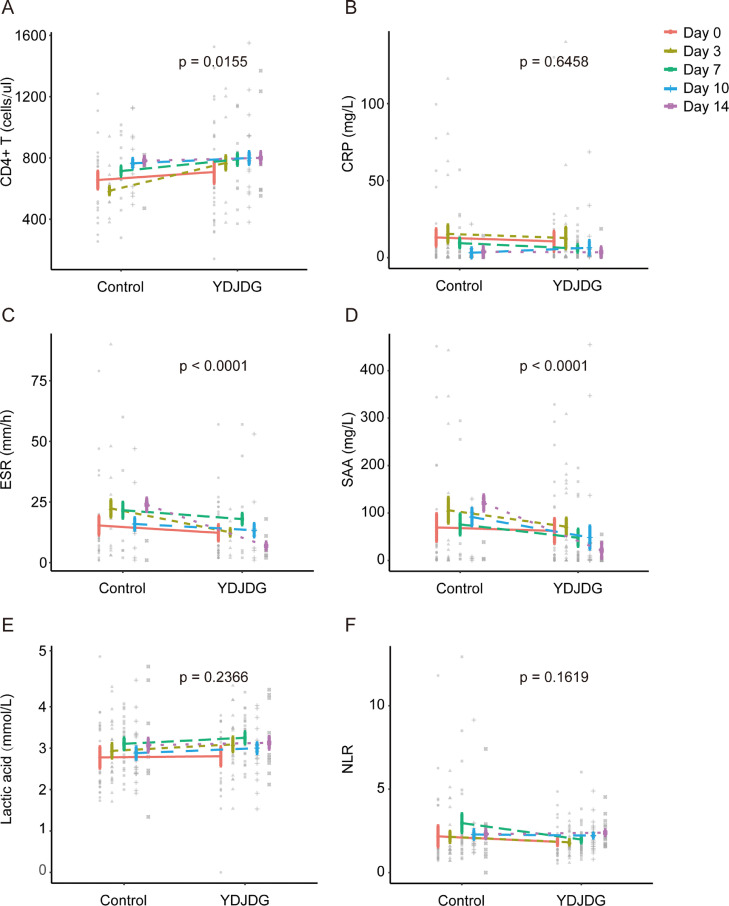
Because not all patients were reviewed regularly, we observed dynamic changes in CD4+T cell count, CRP, SAA, and lactic acid levels, ESR, and NLR in 35 pairs of patients from the YDJDG and control groups, which were checked regularly on day 0, day 3, day 7, day 10, and day 14 after PSM. The baseline values of the two groups were not significantly different (p > 0.05, Supplementary Table 1). A significant difference in the rate of increase of CD4+ T cell count was observed between the two groups (p = 0.0155) (Fig. 4 A). A more rapid reduction in ESR and SAA level was observed in the YDJDG group than in the control group (p < 0.0001) (Fig. 4C, D). However, there were no significant differences in CRP and lactic acid levels and NLR between the two groups (Fig. 4B, E, F).
Fig. 4.
Dynamic changes in CD4+T cell count (A), CRP (B), ESR (C), SAA (D), lactic acid levels (E), and NLR (F) at different times in the control and YDJDG groups after PSM. CRP, C-reactive protein; ESR, erythrocyte sedimentation rate; SAA, serum amyloid A; NLR, neutrophil-to-lymphocyte ratio; YDJDG, Yindan Jiedu granules; PSM, propensity score matching.
Identification of bioactive compounds of YDJDG
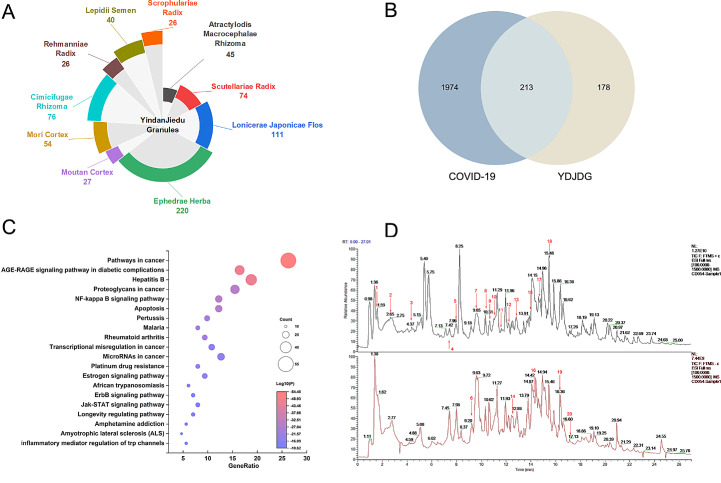
A total of 387 active compounds were retrieved from the SymMap database (Fig. 5 A). These active compounds primarily originated from Ephedrae Herba (MaHuang, 220 compounds), Lonicerae Japonicae Flos (JinYinHua, 111 compounds), Scutellariae Radix (HuangQin, 74 compounds), Mori Cortex (SangBaiPi, 54 compounds), Lepidii Semen (TingLiZi, 40 compounds), Cimicifugae Rhizoma (ShengMa, 76 compounds), Moutan Cortex (MuDanPi, 27 compounds), Scrophulariae Radix (XuanShen, 26 compounds), Atractylodis Macrocephalae Rhizoma (BaiZhu, 45 compounds), and Rehmanniae Radix (DiHuang, 26 compounds).
Fig. 5.
Interaction network of YDJDG compounds with YDJDG and anti-COVID-19 drug targets. (A) Interaction network of the herbs and compounds of YDJDG. A total of 387 active compounds were retrieved from 10 herbs. (B) Venn diagram for comparative analysis of targets of anti-COVID19 drugs and YDJDG. (C) Bubble chart of enriched terms across input gene lists, area determined by count, colored by p values. (D) Mass spectrum chromatograms of YDJDG. Black depicts the Positive mode and Red the Negative mode. Red numbers represent 20 identified compounds. YDJDG, Yindan Jiedu granules.
Gene function and pathway enrichment analysis
Based on the aforementioned results, we further searched for the targets of these ingredients. After eliminating duplicate targets, 390 targets remained. Next, 2187 COVID-19-associated genes were obtained from the GeneCards database, DisGeNET platform, and DrugBank. These targets were considered the active targets of YDJDG in COVID-19 treatment. We screened 213 overlapping targets of YDJDG and COVID-19-associated genes (Fig. 5B). Next, we used the Metascape platform to conduct enrichment analysis of the KEGG pathway of 213 potential targets, with p < 0.01 set as the threshold. The top 20 enrichment pathways with high coincidence are shown in Fig. 5C. As shown in the legend, the size of the bubbles represented the number of genes enriched to this pathway, and the colors from blue to red represented the large to small LogP. Pathway enrichment analysis showed that most of the genes were associated with cancer-related pathways, AGE-RAGE signaling pathway in diabetic complications, hepatitis B, proteoglycans in cancer, NF-κB signaling pathway, apoptosis, etc. The NF-κB signaling pathway, ranged number 5, regulate genes involved in immunity, inflammation, and cell survival, and hence, it is considered one of the critical pathways associated with the therapeutic effect of YDJDG against COVID-19.
Establishment of “herb-compound-target”, YDJDG therapeutic network for COVID-19 and identification of the constituents in the YDJDG
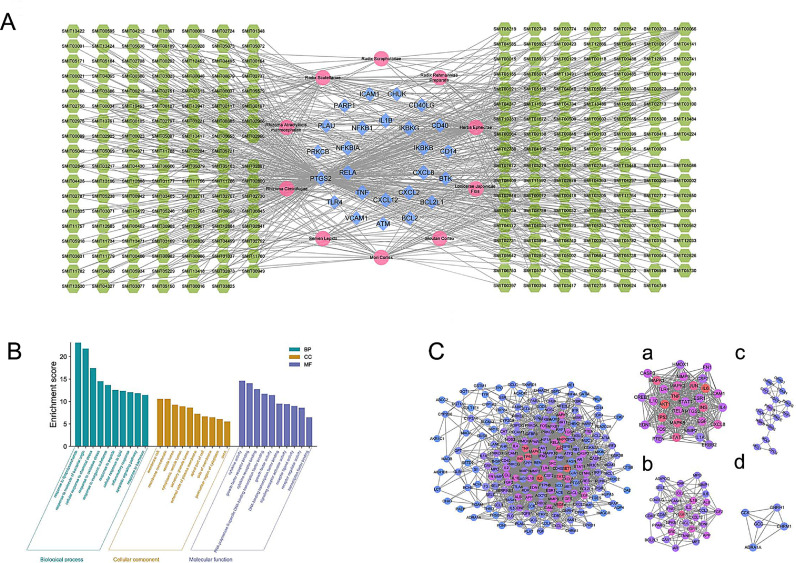
We further screened 25 overlapping targets of YDJDG, COVID-19, and the NF-κB signaling pathway (Fig. 6 A,). Among the genes interfered by YDJDG, 18 genes are both disease-related genes and drug therapy related genes (Fig. 6B). Next, we constructed a network of active compound targets. The 25 overlapping genes corresponded to 248 compounds, which corresponded to 10 herbs. (Fig. 7 A, Supplementary Table 2–3). We subsequently ranked these ingredients according to their degrees. The top five compounds were as follows: quercetin, 19; oleic acid, 12; beta-sitosterol, 12; luteolin, 10; kaempferol, 9. Using MS, 1280 compounds were identified in YDJDG (Fig. 5D). A total of 20 compounds overlapped with the YDJDG components in network pharmacology (Supplementary Table 4). For the top five compounds with the highest scores, luteolin, quercetin and kaempferol were confirmed by chromatography (Table 2 ). Peaks representing luteolin, quercetin, and kaempferol were detected, and the peak outflow times were 12.243, 10.669 and 16.359 min, respectively (Fig. 10A-C). The peak intensities were 5.24e7, 5.68e4, and 1.44e6 cps, respectively. Moreover, we enriched the results of the biological process from GO and the results of pathways from Reactome database, and we discovered that cytokine activation was the core physiological and pathological process of COVID-19 (Fig. 7B).
Fig. 6.
Regulatory effect of YDJDG on the NF-κB signaling pathway.
Fig. 7.
Establishment of YDJDG therapeutic network for COVID-19 and functional attributes of target proteins. (A) The “herb-component-target” network. The pink dots represent the herbs of YDJDG, the green pentagons represent the compounds of the herbs, and the blue diamonds represent the targets of YDJDG. (B) The enrichment of genes with GO method. (C) PPI network and MCODE components identified in the gene lists. Four gene clusters are clustered according to their main biological functions. YDJDG, Yindan Jiedu granules; GO, gene ontology; PPI, protein-protein interaction.
Table 2.
Chemical formulation of bioactive components identified from YDJDG.
| Ingredient-ID | Name | Molecular formula | CAS | Molecular Weight | RT [min] | Herbs of YDJDG |
|---|---|---|---|---|---|---|
| SMIT00002 | Luteolin | C15H10O6 | 491–70–3 | 286.25 | 12.243 |
Ephedrae Herba Lonicerae Japonicae Flos |
| SMIT00013 | Quercetin | C15H10O7 | 117–39–5 | 302.25 | 10.669 |
Ephedrae Herba Lonicerae Japonicae Flos Mori Cortex Lepidii Semen Moutan Cortex |
| SMIT00041 | Kaempferol | C15H10O6 | 520–18–3 | 286.25 | 16.359 |
Ephedrae Herba Lonicerae Japonicae Flos Mori Cortex Lepidii Semen Moutan Cortex |
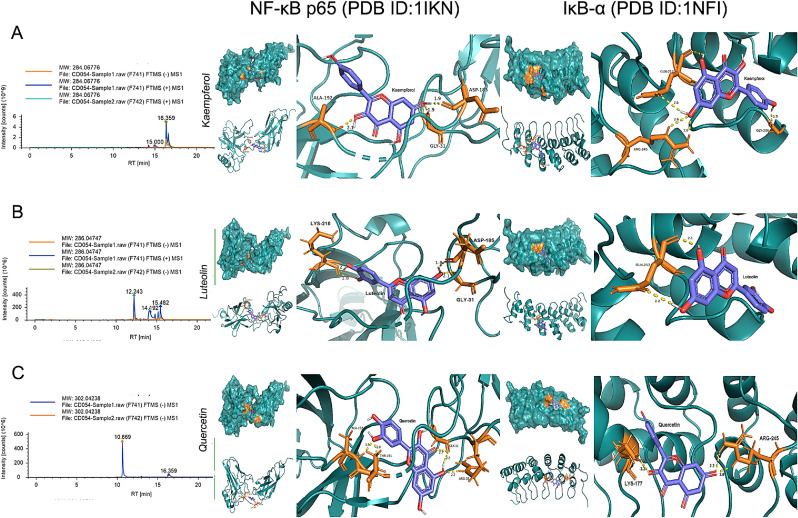
Fig. 10.
Molecular docking analysis. Docking of NF-κB p65 (PDB ID:1IKN) and IκBα (PDB ID:1NFI) with luteolin, quercetin, and kaempferol. All of these amino acids are near the active site of NF-κB p65 and IκBα, and forms hydrogen bonds, thus promoting binding.
Functional attributes of target proteins
Using the STRING platform (https://www.string-db.org/), PPI network and MCODE were generated, as shown in Fig. 7C. The four most significant MCODE components were extracted from the PPI network, and independent enrichment analysis was applied to each MCODE component. The results showed that the biological functions of cluster1 (a) and cluster2 (b) were mainly related to signaling by interleukins as well as response to biotic stimulus and lipopolysaccharide. In cluster3 (c) and cluster4 (d), the protein function annotations were apoptosis, antimicrobial humoral immune response mediated by antimicrobial peptides, and chemokine-mediated signaling pathways.
YDJDG decreases inflammatory response and the expression of inflammatory cytokines in LPS-induced ALI by inhibiting the phosphorylation of NF-κB and i-κb in vivo
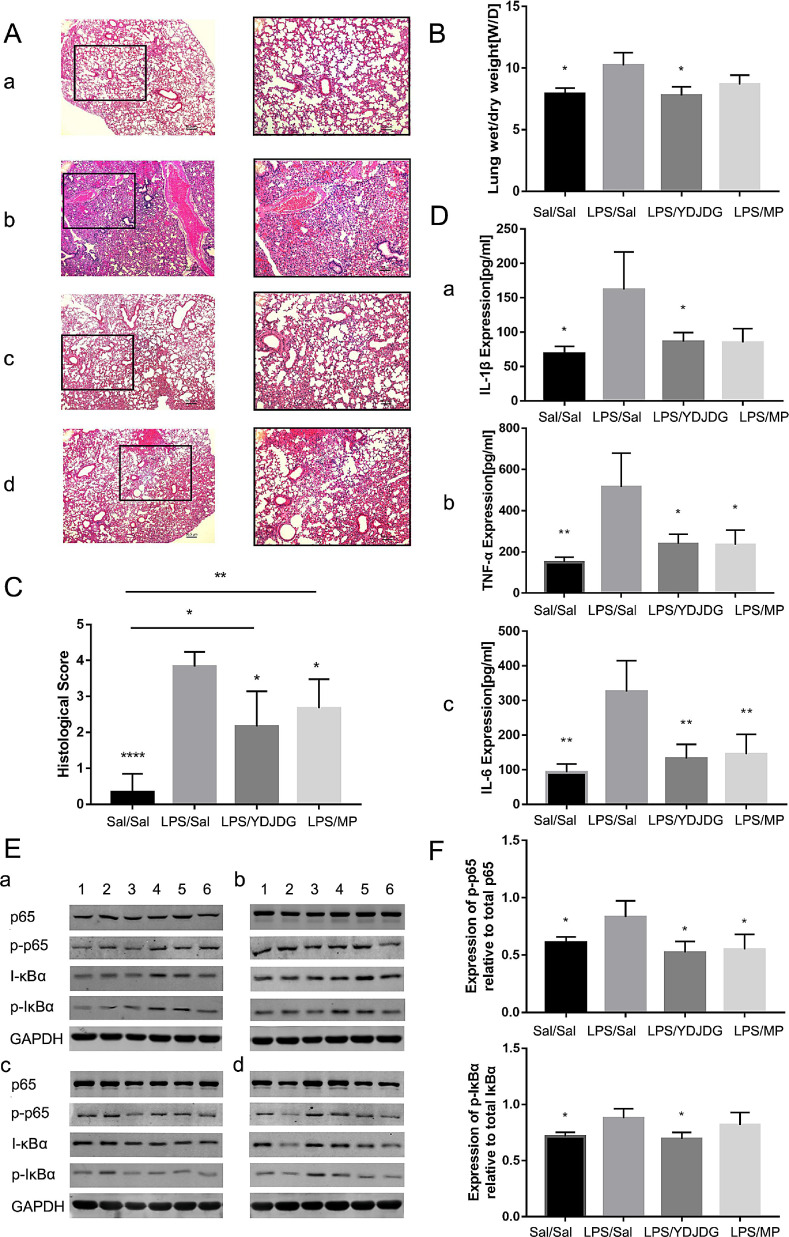
Histological analysis of lung tissue detected more severe lung injury (hemorrhage, interstitial edema, peribronchial and intra-alveolar infiltration, and alveolar pneumocyte hyperplasia) in LPS-instilled mice than in the control group (p < 0.0001) (Fig. 8 A, C). Lung injury was considerably reduced in the LPS + YDJDG and LPS + MP mice, especially in the former group (p < 0.05) (Fig.8A, C). The wet-to-dry ratio were significantly greater in LPS-instilled mice than in the control group (p < 0.05), suggesting increased permeability of the alveolocapillary barrier. However, YDJDG markedly reduced the wet-to-dry ratio (p < 0.05) (Fig. 8B). Moreover, in lung homogenates, the concentrations of the pro-inflammatory cytokines IL-6, TNF-α, and IL-1β reduced significantly in groups of mice treated with YDJDG, compared with the LPS group (Fig. 8D, a-c).
Fig. 8.
In vivo experiment. (A) Lung tissue analysis using hematoxylin and eosin staining. Representative images of (a) Control group; (b) LPS group; (c) LPS + YDJDG group; (d) LPS + MP group. (B) Wet/dry weight ratio and (C) Histological score, (D) The levels of cytokines in bronchoalveolar lavage fluid, (E) Expression of NF-κB p65, phosphorylated NF-κB, IκBα, and phosphorylated IκBα in the lung tissue of mice in different groups, as determined by western blot 24 h after the induction of injury. The letters a-d represents different groups: (a) Control group; (b) LPS group; (c) LPS + YDJDG group; (d) LPS + MP group. The numbers 1–6 represents the reference number of each mouse in one group. Original magnification × 200. Data are presented as mean ± standard error of mean. ANOVA followed by the post hoc Fisher's PLSD test were used. * p < 0.05; ** p < 0.001. LPS, lipopolysaccharide; YDJDG, Yindan Jiedu granules.
In order to evaluate the activation status of NF-κB p65 and IκBα in the lung tissue, we determined the expression levels of NF-κB p65, p-NF-κB, IκBα, and p-IκBα in the lung tissues of mice in each group by using western blot analysis. As shown in Fig. 8E, F, the P/T levels of NF-κB and IκBα in YDJDG group was significantly lower than that in the ALI model group (p < 0.05), suggesting that the activation of the NF-κB pathway was inhibited, thus alleviating inflammatory injury in the lung.
YDJDG decreases pro-inflammatory cytokine and increases anti-inflammatory cytokine levels in LPS-treated RAW264.7 cells by blocking LPS-induced activation of the NF-κB signaling pathway in vitro
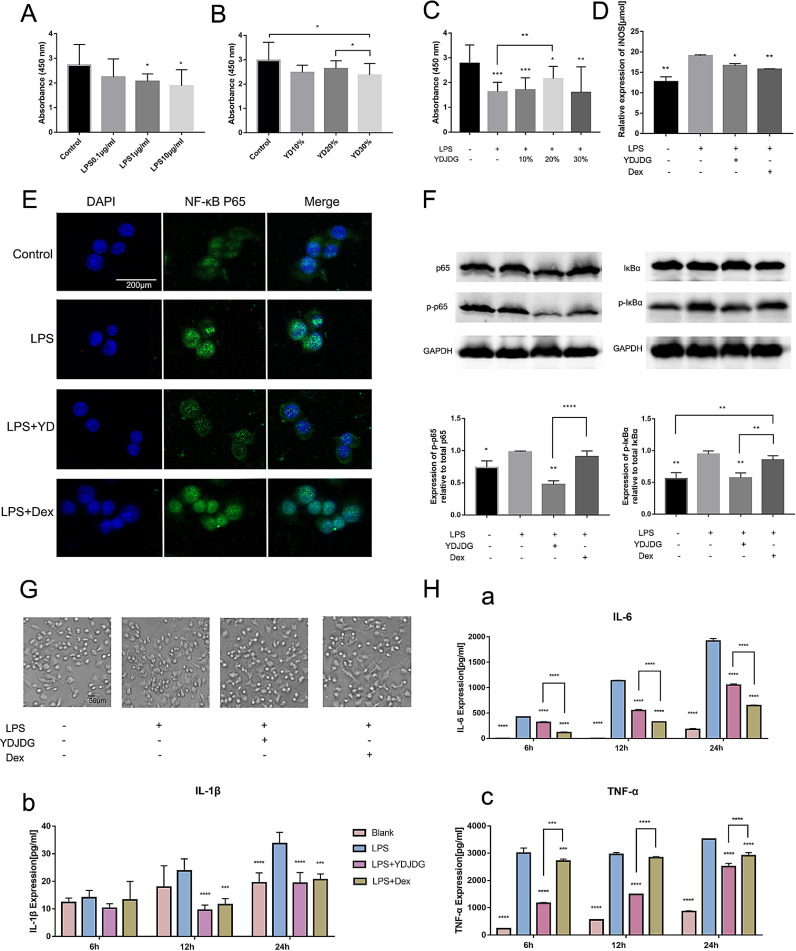
In order to ascertain if YDJDG serum exerted cytotoxicity on RAW264.7 cells and determine the working concentration of both LPS and YDJDG, a CCK-8 assay was conducted. We added 10, 20, or 30% serum from YDJDG-treated rats or LPS (0.1–10 μg/ml) to DMEM, as described in our previous study (Yang et al., 2021). The results demonstrated that cell viability was significantly inhibited at a concentration of 30% YDJDG serum or 1 μg/ml LPS (p < 0.05). Therefore, we selected 20% as the treatment concentration of serum for further experiments, as well as 1 μg/ml LPS (Fig. 9 A,B). However, in our pre-experiment, we found that 10 μg/ml LPS stimulated higher cytokine production while the cell viability did not decrease significantly, and hence, we eventually chose 10 μg/ml LPS as the working concentration. Interestingly, 20% YDJDG-treated serum effectively reversed the inhibitory effect of LPS on cell viability (p < 0.01 vs. LPS group) (Fig. 9C).
Fig. 9.
In vitro experiment. (A-C). Effect of different concentrations of LPS and YDJDG serum on the viability of RAW264.7 cells. * p < 0.05; ** p < 0.01 vs. control group. (D) The levels of nitric oxide in the different groups as determined by Griess reagent method. * p < 0.05; ** p < 0.01 vs. LPS group. (E) Inhibitory effects of YDJDG on LPS-induced nuclear translocation of p65 in RAW264.7 cells. Dexamethasone was used as positive control. (F) Expression of NF-κB p65, phosphorylated NF-κB, IκBα, and phosphorylated IκBα in RAW264.7 cells as determined by western blot. (G) Inhibition of macrophage activation by YDJDG in LPS-stimulated macrophages. (H) Inhibitory effect of YDJDG serum on cytokine release at different time points in LPS-treated RAW264.9 cells. **** p < 0.0001, *** p < 0.001, ** p < 0.01, and * p < 0.05 vs. LPS group. LPS, lipopolysaccharide; Dex, Dexamethasone; YDJDG, Yindan Jiedu granules.
NO is one of the overexpressed inflammatory mediators triggered by LPS. The amount of NO released by RAW264.7 cells increased significantly upon exposure to LPS (10 μg/ml). However, YDJDG serum significantly decreased LPS-induced NO production in RAW264.7 cells (p < 0.05 vs. LPS group). The NO release results are shown in Fig. 9D.
As shown in Fig. 9E/F, the results of immunofluorescence staining and western blotting indicated that LPS stimulation markedly promoted the expression of NF-κB p65, p-NF-κB, and p-IκBα, accelerated the degradation of IκBα, and promoted the translocation of cytosolic NF-κB p65 into the nucleus. However, western blotting showed that the trend was reversed by YDJDG. Immunofluorescence staining of NF-κB also revealed that YDJDG effectively blocked the translocation of cytosolic NF-κB p65 into the nucleus in LPS-stimulated RAW264.7 cells.
As LPS-induced morphological change is a hallmark of macrophage activation, we investigated the effects of YDJDG on macrophage activation by light microscopy. The RAW 264.7 cells (1.2 × 106 cells/ml) were cultured in the presence of YDJDG and LPS for 12 h on cover slides in 6-well plates. Unstimulated cells hardly showed morphological changes, whereas LPS stimulation induced lamellipodia extension and spreading of cells. However, treatment of RAW 264.7 cells with serum from YDJDG-treated rats significantly prevented LPS-induced morphological changes, compared with Dex treatment (Fig. 9G). These data suggested that YDJDG inhibited the activation of LPS-stimulated RAW 264.7 cells.
The ELISA assay results also indicated that YDJDG serum attenuated IL-6, IL-1β and TNF-α levels in the supernatant of cells cultured for 6, 12, or 24 h (Fig. 9H a-c), indicating that YDJDG sustained diminished inflammatory responses in LPS-stimulated RAW264.7 cells.
Results of molecular docking
We also performed molecular docking to verify if the three key compounds of YDJDG played a significant role in the regulation of NF-κB p65, and IκBα. We selected the proteins from the PDB database for further analysis. The results showed that the three key compounds had strong affinity for the proteins and could enter the active pocket of two proteins (Fig. 10 and Table 3 ).
Table 3.
Molecular docking among NF-κB, IκB-α and bioactive components from YDJDG.
| Targets | PDB ID | Compounds | PubChem CID | Docking Score | H-bounds |
|---|---|---|---|---|---|
| NF-κB | 1IKN | Luteolin | 5,280,445 | −6.17 | Gly31 Asp185 Lys218 |
| NF-κB | 1IKN | Quercetin | 5,280,343 | −5.62 | Gly31 Arg33 Thr191 Ala192 |
| NF-κB | 1IKN | Kaempferol | 5,280,863 | −6.42 | Gly31 Alh192 Asp185 |
| IκB-α | 1NFI | Luteolin | 5,280,445 | −5.20 | Gln 212 |
| IκB-α | 1NFI | Quercetin | 5,280,343 | −6.67 | Lys177 Arg245 Asp208 |
| IκB-α | 1NFI | Kaempferol | 5,280,863 | −5.86 | Gln212 Gly206 Arg245 |
Discussion
Highly dysregulated inflammatory responses are associated with disease severity and lethality in patients infected with SARS-CoV-2. Corticosteroids, which are common anti-inflammatory drugs for COVID-19, may increase secondary opportunistic infections and delay viral clearance due to their inherent immunosuppressive effect; hence, their role is controversial (Russell et al., 2020). In this study, through dynamic observation of computed tomography images of the chest and serum inflammatory markers, we found that YDJDG significantly promoted the absorption of inflammatory exudates, reduced the time of negative conversion of viral nucleic acid, and improved the CD4+ T cell count in the peripheral blood. Therefore, we believe that the clinical efficacy of YDJDG against COVID-19 is mainly associated with its anti-inflammatory effect as well as the ability to promote negative conversion of nucleic acid of SARS-CoV-2 and regulate the immune system.
In order to explore the main anti-inflammatory mechanism of YDJDG, network pharmacology analysis was performed. Through gene enrichment analysis, we found that the intersection between COVID-19 and YDJDG targets were concentrated in the NF-κB pathway. Through the analysis of the KEGG pathway, we found that a total of 25 genes are promising target genes of YDJDG in COVID-19 treatment. Molecular docking showed that luteolin, quercetin, and kaempferol could strongly interact with NF-κB p65 residues, which may inhibit the phosphorylation of p65 by changing the spatial conformation of the protein. Generally, bioinformatics analysis showed that YDJDG may inhibit the body's inflammatory response by regulating the NF-κB pathway.
As a classic inflammation-related pathway, NF-κB pathway can be activated by many factors, such as LPS, cytokines, and viruses (Kircheis et al., 2020). In normal conditions, NF-κB interacts with IκB, with no effect on the downstream genes. Viral proteins, such as nsp1, nsp3a, nsp7a, spike, and nucleocapsid protein can cause excessive activation of NF-κB (Hariharan et al., 2021). Previous studies have also shown that NF-κB activation is central to the acute respiratory RNA virus-induced cytokine storm (Gao et al., 2021; Kircheis et al., 2020). SARS-CoV-2 binds to its receptors, angiotensin converting enzyme 2 (ACE2) and serine protease TMPRSS2, which triggers endocytosis into host cells, activates toll like receptors TLR7 and TLR8, induces transcription activation of interferon regulatory factor family and antiviral response. However, the activation of TLR can also lead to the degradation of the cytoplasmic inhibitor, IκBα, through phosphorylation and ubiquitination. This causes the NF-κB derived from IκBα, then transferred into the nucleus to initiate gene transcription and control the expression of pro-inflammatory proteins, such as cytokines, chemokines, adhesion molecules, and growth factors (de Martin et al., 1993). Our in vivo experiments verified that YDJDG could significantly reduce lung inflammatory and immune responses, pulmonary edema, and release of inflammatory factors related to the NF-κB pathway, such as IL-6, IL-1β, and TNF- α in LPS-induced ALI models. In vitro experiments results showed that YDJDG serum could maintain most of the original morphology of macrophages by reducing the release of inflammatory factors, significantly inhibiting p65 translocation to the nucleus, and reducing NO concentrations in LPS-treated RAW264.7 cells. Western blot results showed that the phosphorylation of NF-κB and IκB could be expressed both in vitro and in vivo. These results were consistent with our network pharmacology analysis and molecular docking results, which supported and confirmed our hypothesis.
In COVID-19, the final sequence of NF-κB activation is shared with a variety of cytokine receptor and toll like receptor-mediated signal cascades, resulting in “waterfall effect”, which eventually leads to progressive exacerbation of pulmonary edema and pneumonia and increases the risk of death. Moreover, patients with severe COVID-19 often suffer from secondary bacterial infections, with LPS inducing the activation of the NF-κB pathway. Therefore, inhibiting a single inflammatory factor or protein in this pathway is not significant enough to control COVID-19. However, multiple interference on the later stages (i.e. IκBα phosphorylation, ubiquitination and/or proteasome degradation) can effectively inhibit continuous activation of the NF-κB pathway. The main active compounds of YDJDG include quercetin, luteolin, and kaempferol. Quercetin has antioxidant, antiviral, and antibacterial effects, and it can inhibit the activation of inflammatory cells (Cheng et al., 2019). Luteolin reduces the production of inflammatory factors by inhibiting the activation of the (PI3K)/Akt and NF-κB pathways (Lin et al., 2008). Kaempferol can reduce the expression of iNOS and NO, block the NF-κB pathway, and reduce the production of downstream inflammatory factors (Kadioglu et al., 2015). Therefore, YDJDG may inhibit inflammatory cascades, exerting multi-factorial and multi-target effects on NF-κB-related pathways.
Our study has several limitations. First, this study was not randomized, controlled, and double-blinded; however, PSM was utilized to reduce this bias. Besides, COVID-19 could not be directly induced in this study, as there are strict restrictions on the use of SARS-CoV-2 in experiments. However, viral infections were mimicked by LPS-induced ALI and cell model. Finally, the anti-inflammatory mechanisms of the active constituents of YDJDG were not fully explored in this study.
Conclusion
This study revealed that YDJDG significantly shortened the COVID-19 course and delayed its progression by suppressing inflammation. Network pharmacology showed that the NF-κB pathway was the main target of YDJDG. HPLC-MS analysis identified 1280 active components of YDJDG, and verified 20 compounds identified by network pharmacology. Molecular docking revealed that three active compounds specifically inhibited the activation of the NF-κB pathway by modifying NF-κB p65 and IκBα. YDJDG suppressed inflammatory cytokine release, alleviated LPS-induced damage, and blocked the phosphorylation of NF-κB p65 and IκBα through the NF-κB pathway in vitro and in vivo. YDJDG demonstrated significant clinical efficacy against inflammation, indicating its potential as a new effective drug for COVID-19.
Authors’ contributions
XBW, YYJ, and YF designed and supervised the entire study. YF and BBZ performed the in vitro and in vivo experiments, pharmacology analysis, and molecular docking, and they wrote the manuscript. YL analyzed the clinical data. GQZ, YL, LY, and LL collected the clinical data. RJ and HY performed the HPLC-MS/MS assay. YXH and PPM provided support for the analysis of clinical data. All authors agree to be responsible for all aspects of their work, ensuring completeness and accuracy.
Declaration of competing interest
The authors have no conflicts of interest.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.phymed.2021.153784.
Appendix. Supplementary materials
Baseline characteristics of 131 COVID-19 patients before and after propensity score matching.
List of 25 overlapping genes of the “Herb-Compound-Target” network.
List of 248 compounds corresponding to 25 overlapping genes and 10 herbs.
Overlapping components of YDJDG and mass spectrometry analysis.
Reference
- Cheng S.C., Huang W.C., JH S.P., Wu Y.H., Cheng C.Y. Quercetin inhibits the production of IL-1β-induced inflammatory cytokines and chemokines in ARPE-19 cells via the MAPK and NF-κB signaling pathways. Int. J. Mol. Sci. 2019:20. doi: 10.3390/ijms20122957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Martin R., Vanhove B., Cheng Q., Hofer E., Csizmadia V., Winkler H., Bach F.H. Cytokine-inducible expression in endothelial cells of an I kappa B alpha-like gene is regulated by NF kappa B. EMBO J. 1993;12:2773–2779. doi: 10.1002/j.1460-2075.1993.tb05938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y.M., Xu G., Wang B., Liu B.C. Cytokine storm syndrome in coronavirus disease 2019: a narrative review. J. Intern. Med. 2021;289:147–161. doi: 10.1111/joim.13144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan A., Hakeem A.R., Radhakrishnan S., Reddy M.S., Rela M. The Role and Therapeutic Potential of NF-kappa-B Pathway in Severe COVID-19 Patients. Inflammopharmacology. 2021;29:91–100. doi: 10.1007/s10787-020-00773-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadioglu O., Nass J., Saeed M.E., Schuler B., Efferth T. Kaempferol is an anti-inflammatory compound with activity towards NF-κB pathway proteins. Anticancer Res. 2015;35:2645–2650. [PubMed] [Google Scholar]
- Kircheis R., Haasbach E., Lueftenegger D., Heyken W.T., Ocker M., Planz O. NF-κB pathway as a potential target for treatment of critical stage COVID-19 patients. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.598444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng L., Cao R., Ma J., Mou D., Zhu Y., Li W., Lv L., Gao D., Zhang S., Gong F., Zhao L., Qiu B., Xiang H., Hu Z., Feng Y., Dai Y., Zhao J., Wu Z., Li H., Zhong W. Pathological features of COVID-19-associated lung injury: a preliminary proteomics report based on clinical samples. Signal Transduct. Target Ther. 2020;5:240. doi: 10.1038/s41392-020-00355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Shi R., Wang X., Shen H.M. Luteolin, a flavonoid with potential for cancer prevention and therapy. Curr. Cancer Drug Targets. 2008;8:634–646. doi: 10.2174/156800908786241050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Jiang Y., Liu Y., Pu L., Du C., Li Y., Wang X., Ren J., Liu W., Yang Z., Chen Z., Song R., Xie W., Wang X. Yindan Jiedu Granules, a traditional chinese medicinal formulation, as a potential treatment for coronavirus disease 2019. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.634266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Liu Y., Xiang P., Pu L., Xiong H., Li C., Zhang M., Tan J., Xu Y., Song R., Song M., Wang L., Zhang W., Han B., Yang L., Wang X., Zhou G., Zhang T., Li B., Wang Y., Chen Z., Wang X. Neutrophil-to-lymphocyte ratio predicts critical illness patients with 2019 coronavirus disease in the early stage. J. Transl. Med. 2020;18:206. doi: 10.1186/s12967-020-02374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikawa K., Nishina K., Takao Y., Obara H. ONO-1714, a nitric oxide synthase inhibitor, attenuates endotoxin-induced acute lung injury in rabbits. Anesth. Analg. 2003;97:1751–1755. doi: 10.1213/01.ANE.0000086896.90343.13. [DOI] [PubMed] [Google Scholar]
- Pinzón M.A., Ortiz S., Holguín H., Betancur J.F., Cardona Arango D., Laniado H., Arias Arias C., Muñoz B., Quiceno J., Jaramillo D., Ramirez Z. Dexamethasone vs methylprednisolone high dose for Covid-19 pneumonia. PLoS ONE. 2021;16 doi: 10.1371/journal.pone.0252057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell C.D., Millar J.E., Baillie J.K. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet (London, England) 2020;395:473–475. doi: 10.1016/S0140-6736(20)30317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang L., Zhao J., Hu Y., Du R., Cao B. On the use of corticosteroids for 2019-nCoV pneumonia. Lancet (London, England) 2020;395:683–684. doi: 10.1016/S0140-6736(20)30361-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva P.L., Garcia C.S., Maronas P.A., Cagido V.R., Negri E.M., Damaceno-Rodrigues N.R., Ventura G.M., Bozza P.T., Zin W.A., Capelozzi V.L., Pelosi P., Rocco P.R. Early short-term versus prolonged low-dose methylprednisolone therapy in acute lung injury. Eur. Respir. J. 2009;33:634–645. doi: 10.1183/09031936.00052408. [DOI] [PubMed] [Google Scholar]
- Wen J., Wang R., Liu H., Tong Y., Wei S., Zhou X., Li H., Jing M., Wang M., Zhao Y. Potential therapeutic effect of Qingwen Baidu decoction against corona virus disease 2019: a mini review. Chin Med. 2020;15:48. doi: 10.1186/s13020-020-00332-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Liu H., Bai C., Wang Y., Zhang X., Guo R., Wu S., Wang J., Leung E., Chang H., Li P., Liu T., Wang Y. Chemical composition and pharmacological mechanism of Qingfei Paidu Decoction and Ma Xing Shi Gan Decoction against Coronavirus Disease 2019 (COVID-19): in silico and experimental study. Pharmacol. Res. 2020;157 doi: 10.1016/j.phrs.2020.104820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Feng Y., Liu Y., Ye X., Ji X., Sun L., Gao F., Zhang Q., Li Y., Zhu B., Wang X. Fuzheng Jiedu Xiaoji formulation inhibits hepatocellular carcinoma progression in patients by targeting the AKT/CyclinD1/p21/p27 pathway. Phytomedicine. 2021;87 doi: 10.1016/j.phymed.2021.153575. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Cao F., Ji G., Xu X., Sun Y., Li J., Qi X., Sun S., Wang Y., Song B. The efficacy and safety of Lianhua Qingwen (LHQW) for coronavirus disease 2019 (COVID-19): a protocol for systematic review and meta analysis. Medicine (Baltimore). 2020;99:e20979. doi: 10.1097/MD.0000000000020979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y., Zhou J., Liang N., Liu B., Lu R., He Y., Liang C., Wu J., Zhou Y., Hu M., Zhou J. Effect of maxing Shigan Tang on H1N1 influenza a virus-associated acute lung injury in Mice. Intervirology. 2016;59:267–274. doi: 10.1159/000458726. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Baseline characteristics of 131 COVID-19 patients before and after propensity score matching.
List of 25 overlapping genes of the “Herb-Compound-Target” network.
List of 248 compounds corresponding to 25 overlapping genes and 10 herbs.
Overlapping components of YDJDG and mass spectrometry analysis.