Abstract
The macronuclear genes coding for rRNA (ribosomal DNA [rDNA]) of Paramecium tetraurelia, stock 51, are arranged in polymers consisting of units made up of a transcribed coding region and a nontranscribed spacer region. The whole macronuclear polymer ends with a portion of the spacer on either end followed by a telomere. Six kinds of macronuclear units, or genes, were mapped. Spacers were different, and transcribed regions were the same. These genes are found in markedly different numbers in the macronucleus. The most common gene shows two regions in the spacer where a sequence is followed by a direct repeat. The next most common gene is similar but shows a deletion plus a number of base pair substitutions. Although most cosmid clones contain only a single kind of gene, many contain more than one. These are thought to be produced by somatic crossing over. The four micronuclear genes that have been isolated consist of a single central transcribed region and portions of the spacer on either end. Sequencing indicates that the two ends of the molecule are partially redundant. While the spacer region at the right end of the macronuclear polymer is derived from the micronuclear spacer on the right, the spacer at the left end of the macronuclear polymer is derived from regions of the micronuclear spacer on both the right and the left. To account for this situation, a rolling-circle model for generation of the macronuclear rDNA from the micronuclear DNA is proposed.
The genes for rRNAs (rDNAs) in most eucaryotic organisms are made up of units, each of which is a nontranscribed spacer and a transcribed region consisting of rDNAs coding for 18, 5.8, and 28S rRNAs. Many units are joined to form polymeric arrays. These arrays are normally chromosomal, but in some organisms, like Xenopus spp., Saccharomyces cerevisiae, Dictyostelium spp., and others, the rDNA can become extrachromosomal at certain stages of the life cycle.
Most ciliates have a micronucleus and a macronucleus. At conjugation and autogamy the macronucleus is destroyed and a new macronucleus is derived from the micronucleus. The DNA of the micronucleus is broken into fragments, internal sequences of DNA are eliminated, and telomeres are added to the ends of the fragments. Amplification of all genes then occurs.
In Tetrahymena spp., the only ciliate in which rDNA has been extensively investigated to date (14), the micronuclear copy of rDNA is a single gene. The gene, consisting of a transcribed portion between nontranscribed spacer regions, is in a micronuclear chromosome with flanking chromosomal regions. When a micronuclear gene gives rise to a macronuclear gene, the micronuclear gene is cut out from the micronuclear chromosome at chromosomal breakage sites (3) in the flanking region and made into a dimer, which is amplified.
Findly and Gall (4, 5) isolated a satellite band after treating whole-cell Paramecium tetraurelia stock 51 with actinomycin D and centrifuging it in cesium chloride for several cycles. This was demonstrated to be rDNA which was organized into repeating 9-kb units, portions of which hybridized to 17, 5.8, and 25S rRNAs as well as a nontranscribed spacer region. Based on differences in the spacers, they identified one major and one minor type (5). Electron microscopic analysis of partially denatured preparations showed that single molecules had spacers of different sizes. They mapped the two most common types of units and found a difference in the rDNAs of stocks 51 and 127.
We have investigated the rDNA from stock 51 of P. tetraurelia by studying clones isolated from libraries. In most instances, our work confirmed the findings of Findly and Gall. Macronuclear genes are organized into polymeric units, each consisting of a transcribed portion and a nontranscribed spacer region, as they described. The transcribed regions of all micronuclear and macronuclear genes in P. tetraurelia stock 51 have identical maps. Only the spacers differ. Six types of macronuclear genes have been delineated based on differences in the maps of their spacers. Maps of the two most common types of macronuclear genes are the same as those identified by Findly and Gall. Evidence that the macronuclear rDNAs are linear is presented.
Findly and Gall did not look at micronuclear rDNA; it was not possible to isolate micronuclei when they did their work. Upon screening, our micronuclear library produced 18 positive clones. On the basis of flanking sequences and maps of nontranscribed spacer sequences, four genes emerged. Each was a single unit. RIB2 appeared to be the source of the most common macronuclear gene, RIBX, and RIB3 appeared to be the source of the second most common macronuclear gene, RIBD. It should be noted that the micronuclear genes and the macronuclear genes differ from each other, part of the micronuclear gene being eliminated when the micronucleus forms the macronucleus. The recently adopted format for names of genes in ciliates makes no provision for differences in micronuclear and macronuclear genes (1). We adopt the names recommended for the micronuclear genes, RIB1, RIB2, RIB3, and RIB4. For the macronuclear genes we use the names RIBX, RIBD, RIBA, RIBB, RIBC, and RIBE.
This paper contains an account of the different genes that we have found in the micronucleus and the macronucleus of Paramecium. It includes sequences and an analysis of the physical conformation, as well as a discussion of data bearing on the conversion of the micronuclear genes into the macronuclear genes.
MATERIALS AND METHODS
Strains and culture of paramecia.
Stock 51.s of P. tetraurelia was taken from a culture derived from the Sonneborn collection in our laboratory in Bloomington, Ind. Stock 127 was from the American Type Culture Collection in Manassas, Va. (ATCC 30673). Paramecia were cultured as described previously (9).
Isolation of DNA.
Isolation of genomic DNA was performed according to the method of Steele et al. (12). Isolation of plasmid DNA for sequencing was done with a Qiagen (Chatsworth, Calif.) 100 kit.
Cosmid and lambda libraries.
See Table 1 for the cosmid and lambda clones used in this study. Most of our lambda clones came from a macronuclear library prepared from d48 whole-cell P. tetraurelia DNA in lambda EMBL 4 in our laboratory by Klaus Heckmann according to the method of Forney et al. (6). Lambda A3 and lambda C6r were isolated by Lloyd Epstein from a lambda library constructed by J. Preer. The micronuclear library was prepared from isolated micronuclei of P. tetraurelia in lambdaGEM-11 (12). The cosmid library was prepared from an Invitrogen (Carlsbad, Calif.) kit by placing segments of macronuclear DNA into the cos2 vector according to the manufacturer’s instructions. cosEM was isolated by Eric Meyer from a cosmid library he constructed.
TABLE 1.
Macronuclear subclones and clonesa
| Clone or subclone type | Clone(s) containing gene:
|
Mixed clone(s) (genes) | |||||
|---|---|---|---|---|---|---|---|
| RIBX | RIBD | RIBA | RIBB | RIBC | RIBE | ||
| Plasmid subclones | pXA3 from cosEM | pRD8.5 from λA3 | p38 from cos10a | pXA5 from cosEM | p110 from λ110 | p1 from cos1a | |
| Lambda clones | 4, 100, 101, 102, 104, 105, 106, 107, 108, 115, 116, 117, 119 | 109, 112, 114, 121, A3 | 110 | C6r (RIBX, RIBD) | |||
| Cosmid clones | 1b, 2a, 3a, 4b, 6a, 9a, 9b, 11a, 12a, 12b, 12c, 3, 11, 16, 19, 22, 23, 31, 34, 35, 36 | 8b, 2, 9, 15, 17, 25, 28, 33, 38 | 21 | 32 | 1a | 5a, 7a, 10a, 14 (RIBX, RIBA) EM (RIBX, RIBB) 3b, 6, 30 (RIBX, RIBD) 5b, 7, 20, 39 (RIBX, RIBE) 5 (RIBD, RIBB) 4 (RIBD, RIBC) 37 (RIBD, RIBE) 26 (RIBX, RIBB, RIBD) | |
Subclones are obtained by restricting the DNA from the lambda or cosmid clone at the single XbaI site in the rDNA unit, thus cutting out exactly one rDNA unit, or gene, of about 8.7 kb in length. Lambda clones are approximately 12 kb (1.5 rDNA units). Cosmid clones are approximately 35 kb (4 rDNA units). Clones listed under RIBX, RIBD, RIBA, RIBB, RIBC, and RIBE are pure clones, i.e., they consist of only one gene. Clones listed under “Mixed clone(s)” consist of at least two genes. cosEM was isolated by Eric Meyer and consisted not only of RIBX and RIBB but also of the gene for serotype antigen C; inclusion of a chromosomal serotype gene is thought to be a cloning artifact. Cosmid 26 is a mixture of RIBX, RIBB, and RIBD.
Plasmid strains.
Plasmid strains were subcloned in our laboratory at the single XbaI site into either pUC18 or pTZ18 (Table 1). pXA3 and pXA5 were derived by Eric Meyer from cosEM, which contained rDNA, as well as a section of the C immobilization antigen gene (thought to be an artifact of cloning). pRD8.5 was isolated by Jack Greenlee from lambda A3.
Southern hybridization.
Southern hybridization of blots was performed according to the method of Meyer (7).
PCR.
PCR amplification was carried out according to modifications of a method prescribed by Laurence Amar (1a). A lysate was prepared by centrifuging 40,000 living cells of stock 51, resuspending them in 60 μl of culture fluid, and adding 100 μl of 1% NP-40. The lysate was put at 65°C for 10 min and placed on ice until used. Four microliters was used in each reaction mixture and contained about 1,000 lysed cells (150 ng of DNA). Two 0.5-ml Ependorf tubes were prepared for the PCRs, one for the amplification of the left end of the gene and one for that of the right end. To the tube for the right end 1.2 μl of a 100-pmol/μl concentration of a primer beginning at base 3 (5′-GCTTATCCCTCTTCCTACGATCTGCTCATC-3′) was added. To the tube for the left end 1.2 μl of a 100-pmol/μl concentration of a primer beginning at base 677 (5′-CAAACTGTTCTTGAGAGTTTTCCTC-3′) was added. To each of the two tubes the following was added: 1.2 μl of a 100-pmol/μl concentration of degenerate 30-mer telomere primer, [5′-(C/A)AACCC-3′]5, 4 μl of the lysate, 10 μl of Tfl buffer (pH 8.6; Promega), 5 μl of 25 mM MgSO4, 2.8 μl of a mix of the four deoxyribonucleoside triphosphates at 2 mM each, and 0.8 μl of Tfl polymerase (5 U/μl; Promega). The volume was adjusted to 100 μl with water. The PCR was for 20 s at 92°C, 45 s at 63°C, and 1 min 30 s at 72°C for 45 cycles, and at the end of the run there was a 20-min extension at 72°C. The DNA was electrophoresed, removed from the gel with a Micropure-0.22 Separator and a Micron Concentrator (Amicon, Beverly, Mass.), and resuspended in 10 μl of Tris-EDTA. Cloning was carried out with a TOPA TA cloning kit (Invitrogen).
Contour-clamped homogeneous electric field (CHEF) gels.
Whole-cell DNA was prepared by lysing 200,000 cells in a mixture of 1% Sarkosyl, 0.5 M EDTA, 100 mM Tris HCl (pH 8.6), 1 μg of proteinase K per ml, and 1% low-melting-point agarose at 65°C, with the solution being kept overnight at 65°C. The lysate was then poured into a 1-ml syringe with the tip cut off and placed at 4°C for 30 min. The plug was pushed out into a small beaker of Tris-EDTA to dialyze overnight. Slices of the solidified lysate were put into the gel slots, and the slots were filled with agarose made up in 0.5× Tris-borate-EDTA, the running buffer. The gel was electrophoresed, blotted, and hybridized with 32P-labeled pXA3 and exposed to film.
Sequencing.
Sequencing was done with a sequencer from LI-COR (Lincoln, Nebr.) and dye-labeled forward and reverse M13 primers. A Sequitherm cycling kit (Epicentre, Madison, Wis.) was used for the reactions.
Dot blots.
Dot blots were carried out with a Schleicher and Schuell (Keene, N.H.) apparatus according to the directions of the manufacturer.
Nucleotide sequence accession numbers.
The complete sequence of RIBX has been given GenBank accession no. AF149979. The sequence of the spacer of RIBD has been given GenBank accession no. AF149980.
RESULTS
We decided first to determine the forms of rDNA which exist in both the macronucleus and the micronucleus of P. tetraurelia stock 51. Hence, we began by examining as many rDNA clones, derived from the macronucleus and micronucleus, as possible. Most of these clones were lambda clones, and most were from lambda libraries. We also found it desirable to look at larger pieces of DNA, and for this purpose we used cosmid libraries. Therefore, we begin with a listing of the clones that we used. Some were from earlier studies, but most were constructed specifically for this study.
Macronuclear clones.
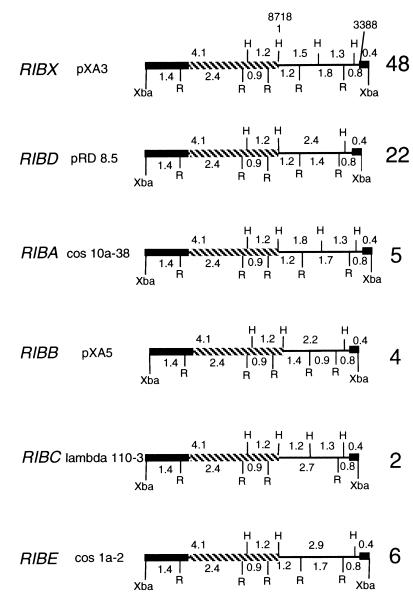
Two plasmid clones which had been in our laboratory for some time were pXA3, isolated by Eric Meyer from a cosmid clone, and pRD8.5, isolated by Jack Greenlee from a lambda clone. Maps of these subclones were identical to maps described by Findly and Gall (4, 5), and thus the subclones were presumed to be rDNA. These subclones were used to probe lambda and cosmid libraries; 69 positive clones were isolated and studied. Subclones of all macronuclear genes were obtained by restricting positive clones from the libraries with XbaI, which recognizes a single site within the gene. The six types of genes found, RIBX, RIBD, RIBA, RIBB, RIBC, and RIBE; plasmid subclones, representative of the six types of genes; lambda library clones; and cosmid clones are given in Table 1. Maps of representative plasmid subclones are also given (Fig. 1).
FIG. 1.
Maps of the six macronuclear genes found in P. tetraurelia stock 51. Since each gene contained only one XbaI site, this site was used to clone into the vector, pTZ18 (2.9 kb) or pUC18 (2.7 kb). Note that all differences are found in the spacer region (single line). Numbers to the right of each map indicate the numbers of clones in which the genes were found. The black bar represents DNA coding for 17S RNA, and the crosshatched bar represents DNA coding for 25S RNA. H, HindIII; R, EcoRI.
The libraries were usually made by partial restriction of genomic DNA with Sau3A, which approximated random cuts. Since the size of the DNA found in each lambda clone varied between 8 and 16 kb and the repeating unit was 8.7 kb, most lambda clones had different maps, depending on the locations of the restriction sites in the DNA used to make the library. Most of our lambda and cosmid clones consisted of one of these six genes or a mixture of two or three of them.
The most common type of gene, RIBX, named after clone pXA3, was found in 48 clones. The next most common type, RIBD, named after clone pRD8.5, was found in 22 clones. These are the two major types of genes mapped by Findly and Gall. Four other types of macronuclear genes were isolated: RIBA (5 clones), RIBB (4 clones), RIBC (2 clones), and RIBE (6 clones). The maps of all the types of genes have identical transcribed regions, differing only in their nontranscribed spacer regions.
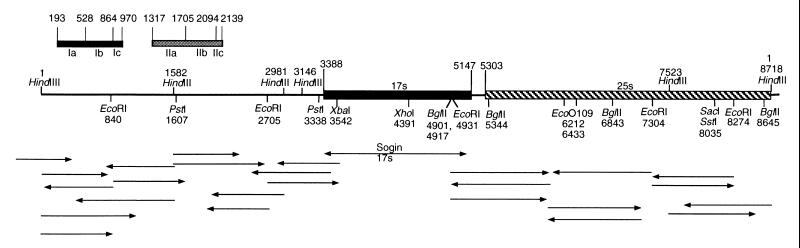
In order to facilitate comparisons, the whole of a RIBX unit (pXA3) was sequenced, with bases 1 to 3388 consisting of a nontranscribed spacer and bases 3389 to 8718 consisting of the transcribed region. The sequence of 17S rDNA is from the work of Sogin and Elwood (11). Base 1 is designated the start site at the HindIII site, but the true start site of the spacer region, based on homology with sequences in other organisms (3), is probably about 20 bases to the left of that point. The map and sequencing strategy are given in Fig. 2. The spacer of RIBX has two regions in which an extensive region is repeated once and then again partially. The first is a 336-bp sequence from bases 193 through 528 that is repeated at bases 529 through 864 and then repeated partially at bases 865 through 970. A second region of 389 bases from bases 1317 through 1705 is repeated at bases 1706 through 2094 and repeated partially at bases 2095 through 2139. The repeats were not identical but had a few base pair substitutions.
FIG. 2.
Sequencing strategy for pXA3. The spacer is represented by a single line, 17S RNA is represented by a black bar, and 25S RNA is represented by a crosshatched bar. The two repeat regions in the spacer are indicated at the top of the map by Ia to Ic and IIa to IIc. Regions were subcloned and sequenced as indicated below the map. The region labeled “Sogin 17s” represents sequences from the work of Sogin and Elwood (11).
The question arises as to whether the genes that map the same are actually identical. We sequenced parts of the spacers of five of the lambda clones of RIBX, clones 4, 102, 103, 104, and 107. Of about 650 bases, all were identical in all five clones. On the basis of this limited sequencing, there are no indications that the genes identified as RIBX by their maps differ from each other in sequence.
The deletion in RIBD.
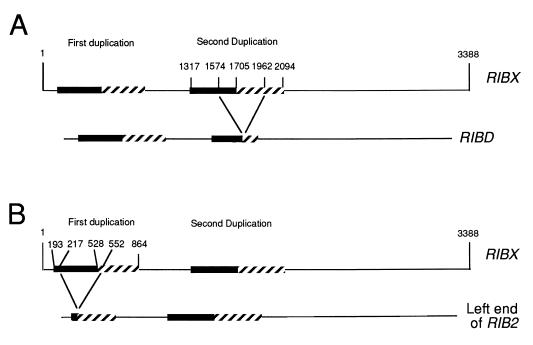
The spacer of RIBD was also sequenced. The sequencing strategy (not given) was similar to that used for RIBX (Fig. 2). The RIBD spacer had a total of 2,996 bases. Examination of RIBX compared with RIBD reveals the differences between the two genes. There were 25 base pair substitutions and four single-base deletions in RIBD compared to RIBX. RIBD also had a large deletion that removes the HindIII site at approximately position 1582 (Fig. 1 and 2). This deletion is 388 bases, almost exactly one repeat region (389 bases). It is impossible to tell the precise location of a deletion if it is of exactly the same length as the repeat region. The approximate location of the deletion can be obtained by comparing the sequence of RIBX (bases 1317 to 1705) and that of its repeat (bases 1706 to 2094) (between which there are a total of 49 differences) with the corresponding sequences in RIBD. When this was done, it was found that of the first 19 differences between the two sequences in RIBX ending at base 1574, 16 bases in the RIBD sequence fit the first sequence and 3 fit the repeat. With regard to the remaining 30 differences beginning at base 1582 (8 bases beyond 1574) and ending at base 2094, all bases in RIBD fit the bases in the repeat, except for one that fit neither. A deletion involving a portion of the first sequence and a portion of the repeat would account for the differences between RIBD and RIBX. Further, the point of shift of the deletion from the first sequence to the repeat can be fixed to a 9-bp region between bases 1574 and 1582 (Fig. 3A). Note that all numbers in this paper refer to RIBX.
FIG. 3.
Deletions. RIBX shows two regions where a sequence is duplicated. The first involves the sequence from bases 193 to 528, repeated at bases 529 to 864. The second is from bases 1317 to 1705, repeated at bases 1706 to 2094. (A) Deletion in RIBX to produce RIBD. The deletion to produce RIBD is shown from bases 1574 to 1963. The deletion could be shifted as far as 10 bases to the right. (B) Deletion in RIBX to produce the left end of RIB2. The deletion of 336 bases could start at base 217 or any base thereafter to 288. It is shown here at bases 217 to 552.
Mapping the linear ends of the macronuclear rDNA chromosome.
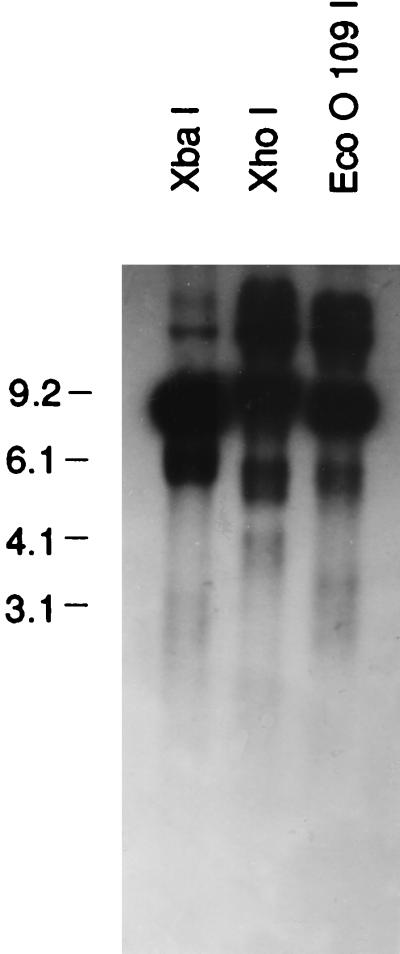
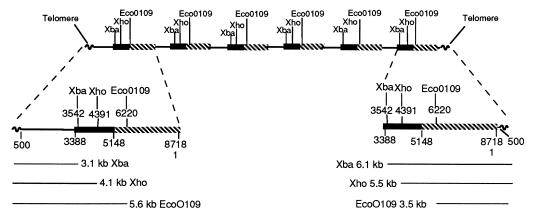
Samples of whole-cell DNA (equivalent to macronuclear DNA, since micronuclear DNA represents only 1/500 of whole-cell DNA) were each treated with a restriction enzyme which recognized a single site in the rDNA (XbaI, XhoI, or EcoO109I) and were electrophoresed (Fig. 4). (EcoO109I actually has two sites, but they are only 21 bases apart.) The gel was blotted and probed with pRD8.5. We expected and observed a major 9.0-kb band but also observed two smaller fuzzy bands. It was assumed that the two fuzzy bands arose from the ends of a linear DNA molecule. One cannot, of course, be sure which of the six genes gave rise to these bands, since whole-cell DNA was used. However, most should be RIBX, a few should be RIBD, and a very few should be the other four genes. Since the XhoI site is 1 kb to the right of the XbaI site, one fragment should be shorter with XbaI than with XhoI and one should be longer. This was observed. The precise positions were difficult to ascertain because the bands are broad (due to variation in the termination). EcoO190I is still farther to the right, and the band positions should readjust accordingly. They do. We could draw the map seen in Fig. 5.
FIG. 4.
P. tetraurelia stock 51 whole-cell DNA and aliquots cut with XbaI, XhoI, and EcoO109I and electrophoresed. The gel was blotted and probed with pRD8.5. The resulting blot is pictured here. Note that all digestions produced a very strong band at about 9 kb and two weaker bands. The 9-kb band represents the whole repeat region, while the two ends, including the telomeres, produced weaker fuzzy bands. The sizes of the bands and their interpretation are given in Fig. 5. Molecular size markers (in kilobases) are noted at the left.
FIG. 5.
Macronuclear chromosome containing RIBX. Shown are six repeating units terminating in bases 1 to 500 of the spacer on the right and in bases 500 to 3388 on the left. Note that XbaI produces an extra 6.1-kb unit on the right and a 3.1-kb unit on the left. When XhoI is used (1 kb to the right), the bands become 5.5 kb (5.1 kb expected) and 4.1 kb. Similar shifts are seen for EcoO109I. The number of repeating units is varied, so the size of the molecule is varied.
Definitive proof of the linear map ending in telomeres seen in Fig. 5 was obtained by analyses based on PCRs. One primer was degenerate and matched the telomere. It was directed toward the middle of the chromosome and was used for both the left and right ends. For the right end, a primer directed to the right starting at base 3 of RIBX was used. RIBX and RIBD are alike in this region. For the left end, a primer beginning at base 677 of RIBX and directed to the left was used. RIBX was like RIBD in this region also. The PCR products obtained were electrophoresed (Fig. 6), extracted from the gel, purified, subcloned, and sequenced. We found varied numbers of telomere sequences, along with the primer in each case. It was found that the point of attachment of the telomeres to the end of the chromosome was different for each subclone: the chromosomes ended on the right at bases 119, 342, 396, and 528 and on the left at bases 120, 139, 303, 341, and 475. Since RIBX and RIBD have occasional base pair substitutions in their sequences, it was usually possible to see whether the clones were like RIBX or RIBD. Inspection of the sequences revealed that seven of the nine clones were like RIBX and that two were too short to tell whether they were RIBX or RIBD. Thus, most of the clones were indeed like RIBX. The mean of these positions (119, 342, 396, and 528 and 120, 139, 303, 341, and 475) is not 500, as expected from Fig. 5, and is probably due to a failure to get a representative selection of subclones from the PCR and subsequent cloning steps.
FIG. 6.
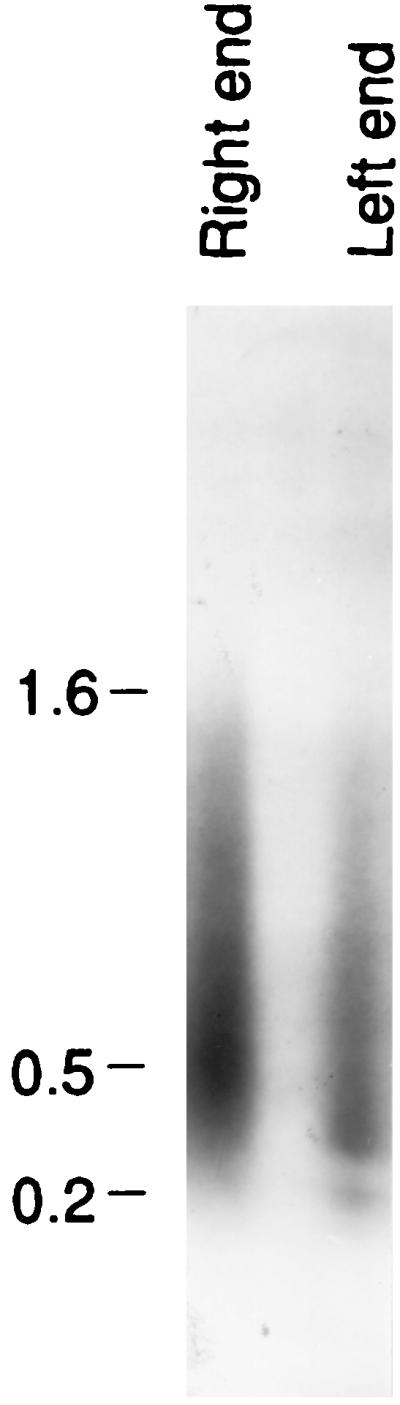
PCRs were carried out with one primer in the telomere for both the right and the left telomere. For the right region a second primer directed to the right, beginning at base 3, was used. For the left region a second primer directed to the left, beginning at base 677, was used. The material was electrophoresed, and the gel was blotted, hybridized with 32P-labeled pXA3, and exposed to film. The result is shown here. Clones were made from this material and sequenced. The sequences indicated that the material consisted of spacer plus telomere sequences as expected. Molecular size markers (in kilobases) are noted at the left.
To investigate further the size of the rDNA chromosome, CHEF gels were run (data not shown). We found a great spread in sizes, with a mean of about 450 kb. The size was close to that given by Phan et al. (8), who used pulsed-field gel electrophoresis. They also concluded, based on results of experiments using different switching times, that most of the molecules were probably circular. Findly and Gall (4, 5) detected many circular molecules in their electron micrographs. In our experiments there was no aberrant behavior on the gel at different switching times. Hence, we concluded that there was no evidence in our data for circular molecules, although the presence of some circular molecules could not be ruled out.
Cosmid libraries.
Because of our interest in the origin of the macronuclear genes from the micronuclear genes, we made a cosmid library, as already noted, and screened it for rDNAs. We were interested in knowing whether the different genes were combined at random or whether they tended to be alike in individual molecules (clones). Findly and Gall had examined partially denatured individual molecules of rDNA with an electron microscope and concluded that different kinds of units were found in the same molecule. The use of cosmid clones offered the possibility of an additional test. The library was screened with pXA3, and 48 positive clones were isolated. The inserts had an average length of 35 kb, enough DNA for approximately four genes per clone. Examination of these clones revealed that 33 were pure, i.e., that they contained four identical genes, and that 15 were mixed, i.e., that they were made up of different genes. The rare genes RIBA, RIBB, and RIBE were present as pure types. These represent far more pure clones than was expected on the basis of chance alone. For example, with reference to Fig. 1 and Table 1, the number of cosmid clones containing RIBE was 4 of 48 cosmid clones. Hence, a maximum estimate of the frequency of pure clones expected on the basis of a random drawing would be (4/48)4 times 48 or 0.03, whereas it is actually 1. Similar calculations for most of the other types show a wide deviation from randomness.
The number of genes per cell.
A dilution series of plasmid pXA3 was made, along with a dilution series of a known number of lysed paramecia, and dot blots were run. When the intensities of the spots in the two series were matched, it was found that there were about 20,000 molecules of rDNAs per cell (data not shown). The ploidy level in the macronucleus is generally assumed to be about 1,000 (1). Since the number of micronuclear genes that are amplified is unknown (see below), we cannot say how many times each is amplified on average.
Micronuclear genes.
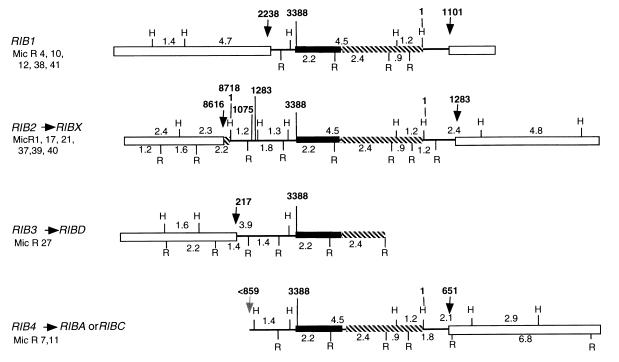
Nineteen micronuclear clones were isolated from a micronuclear library with pRD8.5 as a probe. Fourteen of these clones were different. They were mapped, and it was evident that some clones consisted of different parts of the same gene. Four different genes were distinguished (Fig. 7): RIB1, RIB2, RIB3, and RIB4. Base numbers indicated in this paper refer to homologous regions of the macronuclear gene RIBX.
FIG. 7.
Micronuclear genes. Four genes are drawn here. They are composites made from the individual clones indicated at the left. Clone Mic R 38, as noted in the text, has slight differences from the other clones and may be different from RIB1. Open bars are flanking sequences, black bars are 17S regions, and crosshatched bars are 26S regions. Single lines are spacers. Arrows indicate the junctions between flanking regions and the gene itself. Note that in RIB2 the sequences from bases 1076 to 1283 is repetitive on either end. H, HindIII; R, EcoRI.
The terminal regions of many of these genes were sequenced, and parts homologous to pRD8.5 and pXA3 were determined. The transcribed regions of the micronuclear genes all mapped alike and were like the transcribed regions of the macronuclear genes. Moreover, this identity extended to the regions adjacent to the transcribed regions as well. Immediately to the right of the transcribed region there was a HindIII site at base 1, and at the end of the spacer sequence there was always an EcoRI site at base 2705 followed closely by a HindIII site at base 2981. These relations are true for all micronuclear and macronuclear genes.
The junctions between the flanking regions and the genes were sequenced (Fig. 7). Each gene is discussed below.
The structure of RIB1.
It is seen in Fig. 7 that on the right, the RIB1 spacer begins at base 1 and ends at base 1101. On the left the RIB1 spacer begins at base 2238 and ends at 3388. Outside these regions there are flanking chromosomal regions which are unlike any rDNA sequence. The left region of RIB1 is characterized by an XbaI site, a SalI site (in the flanking region [not shown]), an EcoRI site, and a HindIII site (in the spacer), which are in order and all very close together in the region from bases 2238 to 3388. Clone Mic R 38 is almost identical in its sequence to the other clones in RIB1 but has a minor deviation upstream in the left flanking region, yielding a 3.5-kb fragment rather than a 4.7-fragment when it is digested with HindIII. It may therefore be a different gene from RIB1. No macronuclear gene which might have arisen from RIB1 has been found. RIB1 would have to produce a macronuclear gene with a spacer consisting of bases 1 to 1101 plus bases 2238 to 3388 for a total of 2.25 kb, smaller than any of the six spacers shown in Fig. 1. It would have only the HindIII sites at bases 1 and 2981 and the EcoRI site at base 2703. All of the six known spacers have additional EcoRI or HindIII sites or both, which are not found in the micronuclear RIB1 spacer. The product of RIB1 does not get into the macronucleus, presumably because it lacks the redundant ends required by the rolling-circle model. See below under “Conclusions.”
The structure of RIB2.
RIB2 appears to be the micronuclear gene from which RIBX is derived, for it contains sequences identical to those found in RIBX.
First, we note that the region from bases 8616 to 1 to 1283 is present as a duplication on both ends of RIB2. Comparison of the duplicated region on the left with that on the right shows that while the two regions are very similar, there are differences, the major one being a deletion of 336 bases from the region on the left (Fig. 3B, 7, and 8). This deletion comes within a 336-bp duplicated region, and as already noted, in these circumstances one cannot tell exactly where the deletion is. Proceeding as we did in the comparison of RIBX and RIBD, the deletion probably starts at any base between 217 and 288 and continues for 336 bases. Omitting the region from bases 217 to 552 and comparing the sequences on the left with those on the right, we found 13 individual base pair differences in the two regions.
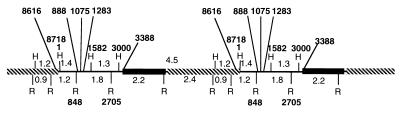
FIG. 8.
Macronuclear gene RIBX as it appears in a polymer. A complete gene is drawn with parts of adjacent RIBX genes. H, HindIII; R, EcoRI.
What are the regions in RIBX that are derived from RIB2? If we compare RIBX with these two left and right duplicated regions from bases 8616 to 1283 in RIB2, the region on the right is an exact match to RIBX. Only the region on the left contains discrepancies. These discrepancies on the left extend from bases 8616 to 1075. The region on the left from bases 1076 to 1283 is exactly like RIBX and like RIB2 on the right. In “Conclusions” we will consider a model that explains these relations.
The structure of RIB3.
RIB3 appears to be the micronuclear gene that gives rise to RIBD. The right end of RIB3 has never been found; it ends with the 2.4-kb EcoRI fragment in the transcribed portion of the gene (Fig. 7). The extreme left end of RIB3 was partially sequenced; it ends at position 217. The left end of RIB3 has remarkable similarities to RIBD (Fig. 1). While RIBX has a HindIII site present in a central region of its spacer that is removed by a deletion in RIBD, the same HindIII site is missing in RIB3. Moreover, the deletion in RIBD moves the two EcoRI sites on either side of the deletion closer together, producing a 1.4-kb EcoRI fragment rather than a 1.8-kb EcoRI fragment. This too is found in RIB3. Sequencing of the left and right portions of this left end of RIB3 reveals that the right portion is in perfect agreement with RIBD, even in a case where RIBX and RIBD disagree with each other. The left portion, however, beginning at base 217 shows at least eight base pair differences between RIB3 and RIBD. It should be recalled that this same region in RIB2 also shows differences between RIBX and RIB2.
The structure of RIB4.
Little is known about RIB4, except for the information given in Fig. 7. The map of RIB4, however, is consistent with its being the source of either RIBA or RIBC of the macronucleus.
Crosses.
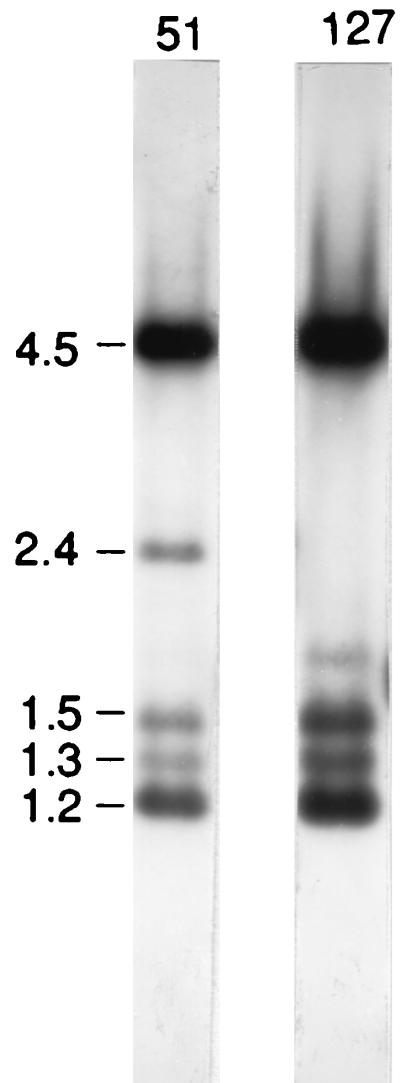
Findly and Gall (4, 5) reported a difference in the maps of the macronuclear rDNAs of stocks 51 and 127. We confirmed this observation with a blot of genomic DNAs from the two stocks cut with HindIII by probing with pRD8.5 (Fig. 9). The two major bands seen in the figure, the 4.5- and the 1.2-kb bands, are the transcribed portions of all the genes. The weaker bands, 1.3, 1.5, and 2.4 kb in size, represent portions of the individual spacer regions. The 2.4-kb spacer band, characteristic of RIBD, is missing from the blot of stock 127 and present in stock 51. Stock 127 also shows a band at 1.8 kb. This band is present in stock 51 but is too weak to show in this blot. It appears in RIBA as a major band (Fig. 1). A cross between stock 51 and stock 127 was made. Then F2 cells were obtained by inducing autogamy, which renders all loci homozygous and should yield a 1:1 segregation in the case of a single gene heterozygote. This 1:1 segregation was observed for the presence or absence of the 2.4-kb band. Hence, it is assumed that stock 127 lacks a micronuclear gene required for inclusion of RIBD in the macronucleus.
FIG. 9.
Whole-cell DNA from stock 51 and stock 127 was restricted with HindIII and electrophoresed. The gel was then blotted and probed with 32P-labeled pXA3 DNA. Stock 51 shows the expected bands for RIBX: 4.5 and 1.2 kb for the transcribed portion of the gene and 1.3 and 1.5 kb for the nontranscribed portion. The 2.4-kb band is derived from RIBD. Longer exposure also shows a weak band at 1.8 kb from RIBA. Stock 127 shows the absence of the 2.4-kb band of RIBD and also a slightly stronger 1.8-kb RIBA band. Molecular size markers (in kilobases) are noted at the left.
DISCUSSION
Conclusions.
RIB2 is the apparent progenitor of the macronuclear RIBX gene (Fig. 7 and 8). From bases 1 to 1075 on the left of RIB2 there are approximately 12 base pair substitutions and a 336-bp deletion when the region is compared to the RIBX spacer. However, from bases 1076 to 3388 on the left of RIB2 and from bases 1 to 1283 on the right of RIB2, there is identity with the RIBX spacer. Thus, there is redundancy from bases 1076 to 1283. What is the situation with the left and right ends of the macronuclear polymer sequenced in the PCR experiments? Both the left and right ends of the polymer are like the spacer of RIBX and unlike bases 1 to 1075 on the left of RIB2. Therefore, the sequence from bases 1 to 1075 on the left of RIB2, which is unlike RIBX, having a deletion and base substitutions, cannot be the template when the gene produces RIBX. The only places from which the sequences on the left side of the polymer from bases 1 to 1075 could have come are the sequences on the right side of RIB2. The remaining sequences on the left side of the polymer, from bases 1076 to 3388, must have come from the left side of RIB2. The sequences on the right end of the polymer are like the sequences on the right end of RIB2.
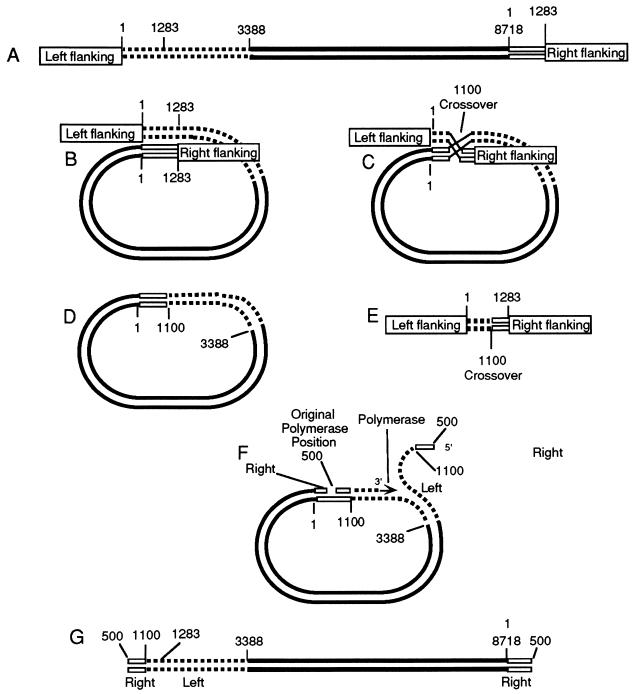
How could these relations come about? Since the two ends of the micronuclear RIB2 are identical to bases 1076 to 1283, they might pair to form a circle (Fig. 10B). There is also partial identity in the region from bases 1 to 1075, and pairing might also occur there. Crossing over in the paired region of bases 1076 to 1283 (at base 1100 in our diagram [Fig. 10C]) could cut out a circular piece of DNA (Fig. 10D) and eliminate the original defective micronuclear left end (Fig. 10E). Thereafter, a rolling circle of replication might occur. The data suggest that the polymerase can begin at a multiplicity of sites within the nontranscribed spacer at bases 100 to 600, the initial position establishing the left end of the final linear macronuclear chromosome. Note that, as represented in the diagram, the left end of the polymer would consist of bases 500 to 1100 from the right end of the micronuclear gene and the region from bases 1100 to 3388 from the left end of the initial micronuclear gene. After several times around the circle the polymerase falls off at a point near where it started, establishing the right end of the molecule. Note that the right end of the polymer is a copy of the right end of the micronuclear chromosome, just as observed. Finally, the single strand becomes duplexed and telomeres are added.
FIG. 10.
Model for the formation of macronuclear DNA from micronuclear DNA. (A) Linear micronuclear RIB2. (B) The molecule bends back on itself, and homologous regions pair. (C) Next there is a crossover at about base 1100, which cuts out the region with the deletion of the micronuclear gene. (D) Resultant circular form of the micronuclear gene. (E) Defective region that was cut out. (F) A polymerase molecule now attaches at about base 500 and begins to transcribe in a typical rolling-circle replication. The left part of the molecule peeling off the template begins at about base 500, and the polymerase continues to transcribe, going around the circle to the position where replication started. (G) Molecule produced by one cycle of replication. The polymerase continues around the circle numerous times to produce a continuous sequence of transcribed and nontranscribed regions. Finally, replication stops near the point where it started, producing the right end of the molecule. After a complementary strand has been synthesized, telomeres are then added to the free ends of the linear molecule to produce the macronuclear polymer of RIBX. Alternatively, the two free ends, instead of adding telomeres, may combine to form a circle.
Are all the spacer regions identical within a single molecule in the macronucleus? Findly and Gall (4) measured the distances between units of partially denatured molecules spread and shadowed on an electron microscope grid. They concluded that individual molecules contained units of different lengths randomly arranged. Our data on cosmid clones, which show many different clones containing more than one kind of molecule, corroborate their conclusion that different types of units are associated. However, our data show that the arrangements are not random but that they tend to be alike on individual molecules. The rolling-circle model described above could not account for mixed clones. One must assume somatic crossing over in the macronucleus to account for these arrangements.
Rolling-circle replication is well known, occurring in plasmids, viruses, mitochondria, bacteria, and rDNA. Rolling-circle replication in Xenopus laevis has been amply documented (13). Since the genes are arranged in tandem on the chromosome of Xenopus, individual repetitive units can pair with each other and amplify copies. The units (which vary in their spacer regions) are arranged at random on the chromosomes. Typical rolling-circle intermediates are found. Moreover, amplified repeated copies originate from single chromosomal units and often contain only a single type, providing additional evidence for rolling circles. The situation is much like we postulate here for Paramecium, except that the final product is circular in Xenopus rather than linear as in Paramecium.
Could there be other models that explain the data? Suppose that the micronuclear gene were able to get out of the chromosome either by an enzyme that simply cuts it out with terminal redundancies or by “onion skin” replication (2, 10), as occurs with simian virus 40. If the portion cut out were still redundant, then during or after amplification crossing over might produce the polymers. However, in order to obtain the observed structure of the left end of the polymer, a special cut that removes one or a few units from the left end of the polymer once the polymer was formed would have to be made. How such a cut could be achieved is not apparent. Although these events might occur, this hypothesis is not as appealing as the rolling-circle model, which neatly explains all the facts and for which there is a precedent in Xenopus.
In Tetrahymena, of course, amplification proceeds by a totally different mechanism. The DNA is cut out at chromosome breakage sequences with no terminal redundancies and processed into a dimer, in which form it is replicated.
Do some of the different macronuclear genes arise from alternate processing of a single micronucleus gene, or are there undiscovered micronuclear genes for each of these types seen in the macronucleus? If there are many undiscovered genes, we would have to assume that the micronuclear genes that we have found in our micronuclear library represent a rather skewed sample. We have no way at present of resolving this problem.
Are the molecules circular? Findly and Gall (4, 5) found with an electron microscope that about one-third of the molecules were circular. It was not possible to tell whether the remainder were broken circles or linear. Our data prove that a substantial number of the rDNA molecules are linear with telomeres. The results of Phan et al. (8) using pulsed-field gels suggest that a large portion of the molecules are circular. When we used CHEF gels, we did not find the same sort of evidence for circular molecules that they found. It is not clear how these data are to be reconciled. Perhaps the material was different in our two cases. In any event, the rolling-circle model may easily produce circular molecules as well as linear.
The absence of macronuclear RIBD in stock 127 may be due simply to a lack of the micronuclear gene coding for it, or it could be the result of a gene that influences processing of the micronuclear rDNA.
ACKNOWLEDGMENTS
This work was supported by a joint grant from the Rockefeller Foundation and Indiana University.
Footnotes
This paper is dedicated to Geoffrey H. Beale, who began the study in our laboratory.
REFERENCES
- 1.Allen S L, Altschuler M I, Bruns P J, Cohen J, Doerder F P, Gaertig J, Gorovsky M, Orias E, Turkewitz A. Proposed genetic nomenclature rules for Tetrahymena thermophila, Paramecium primaurelia and Paramecium tetraurelia. Genetics. 1998;149:459–462. doi: 10.1093/genetics/149.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Amar, L. Personal communication.
- 2.Botchan M, Todd W, Sambrook J. Studies on simian virus 40 excision from cellular chromosomes. Cold Spring Harbor Symp Quant Biol. 1978;43:709–719. doi: 10.1101/sqb.1979.043.01.079. [DOI] [PubMed] [Google Scholar]
- 3.Din N, Engberg J, Gall J G. The nucleotide sequence of the transcription termination site of the ribosomal RNA gene in Tetrahymena thermophila. Nucleic Acids Res. 1982;10:1503–1513. doi: 10.1093/nar/10.5.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Findly R C, Gall J G. Free ribosomal RNA genes in Paramecium are tandemly repeated. Proc Natl Acad Sci USA. 1978;75:3312–3316. doi: 10.1073/pnas.75.7.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Findly R C, Gall J G. Organization of ribosomal genes in Paramecium tetraurelia. J Cell Biol. 1980;84:547–549. doi: 10.1083/jcb.84.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forney J D, Epstein L M, Preer L B, Rudman B M, Widmayer D J, Klein W H, Preer J R., Jr Structure and expression of genes for surface proteins in Paramecium. Mol Cell Biol. 1983;3:466–474. doi: 10.1128/mcb.3.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer E. Induction of specific macronuclear developmental mutations by microinjection of a cloned telomeric gene in Paramecium primaurelia. Genes Dev. 1992;6:211–222. doi: 10.1101/gad.6.2.211. [DOI] [PubMed] [Google Scholar]
- 8.Phan H L, Forney J, Blackburn E H. Analysis of Paramecium macronuclear DNA using pulsed field gel electrophoresis. J Protozool. 1989;36:402–408. doi: 10.1111/j.1550-7408.1989.tb05535.x. [DOI] [PubMed] [Google Scholar]
- 9.Rudman B M, Preer J R., Jr Non-Mendelian inheritance of revertants of paranoiac in Paramecium. Eur J Protozool. 1996;32:141–146. [Google Scholar]
- 10.Sambrook J, Botchan M, Gallimore P, Ozanne B, Pettersson V, Williams J, Sharp P A. Viral DNA sequences in cells transformed by simian virus 40, adenovirus type 2 and adenovirus type 5. Cold Spring Harbor Symp Quant Biol. 1974;39:615–632. doi: 10.1101/sqb.1974.039.01.075. [DOI] [PubMed] [Google Scholar]
- 11.Sogin M L, Elwood H J. Primary structure of the Paramecium tetraurelia small sub-unit rRNA coding region: phylogenetic relationships within the Ciliophora. J Mol Evol. 1986;23:53–60. doi: 10.1007/BF02100998. [DOI] [PubMed] [Google Scholar]
- 12.Steele C J, Barkocy-Gallagher G A, Preer L B, Preer J R., Jr Developmentally excised sequences in micronuclear DNA of Paramecium. Proc Natl Acad Sci USA. 1994;91:2255–2259. doi: 10.1073/pnas.91.6.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wellauer P K, Reeder R H, Dawid I B, Brown D D. The arrangement of length heterogeneity in repeating units of amplified and chromosomal ribosomal DNA from Xenopus laevis. J Mol Biol. 1976;105:487–505. doi: 10.1016/0022-2836(76)90230-8. [DOI] [PubMed] [Google Scholar]
- 14.Yao M-C. Amplification of ribosomal RNA genes. In: Gall J G, editor. The molecular biology of ciliated protozoa. New York, N.Y: Academic Press; 1986. pp. 179–201. [Google Scholar]