Abstract
Coronavirus disease 2019 (COVID-19), caused by SARS-CoV-2 pathogen, has led to waves of global pandemic claiming lives and posing a serious threat to public health and social cum physical interactions. To evaluate the mutational landscape and conserved regions in the genome of the causative pathogen, we analysed 7213 complete SARS-CoV-2 protein sequences mined from the Global Initiative on Sharing All Influenza Data (GISAID) repository from infected patients across all regions on the EpiCov web interface. Regions of origin and the corresponding number of sequences mined are as follows: Asia – 2487; Oceania – 2027; Europe – 1240; Africa – 717; South America – 391; and North America – 351. High recurrent mutations, namely: T265I in non-structural protein 2 (nsp2), L3606F in nsp6, P4715L in RNA-dependent RNA polymerase (RdRp), D614G in spike glycoprotein, R203K and G204R in nucleocapsid phosphoprotein and Q57H in ORF3a with well-conserved envelope and membrane proteins, 3CLpro and spike S2 domains across regions were observed. Comparative analyses of the viral sequences reveal the prevalence P4715L and D614G mutations as the most recurrent and concurrent in Africa (97.20%), Europe (89.83%) and moderately in Asia (61.60%). Mutation rates are central to viral transmissibility, evolution and virulence, which help them to invade host immunity and develop drug resistance. Based on the foregoing, it is important to understand the mutational spectra of SARS-CoV-2 genome across regions. This will help in identifying specific genomic sites as potential targets for drug design and vaccine development, monitoring the spread of the virus and unraveling its evolution, virulence and transmissibility.
Keywords: Coronavirus, Conserved regions, Transmissibility, Mutations, Genome, SARS-CoV-2
1. Introduction
The novel coronavirus was first reported on December 30, 2019 in Wuhan of Hubei Province, China (Wang et al., 2020). With the use of reverse-transcription polymerase chain reaction (RT-PCR) technique, the first whole-genome sequence of the virus was determined and published; this information subsequently helped in identifying the virus in patients (Wang et al., 2020). The virus was initially named 2019 novel coronavirus (2019-nCoV), or “Wuhan seafood market pneumonia virus” (GenBank, 2020) before the disease was officially named coronavirus disease 2019 (COVID-19) by the World Health Organisation (WHO) on February 12, 2020. Its causative agent is severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Wang et al., 2020). As of 5:36 p.m. CEST, September 13, 2021, global fatality due to COVID-19 infections had risen to 4,627,540 with 224,511,226 confirmed cases (WHO Coronavirus (COVID-19) Dashboard | WHO Coronavirus (COVID-19) Dashboard With Vaccination Data [WWW Document], 2021).
The SARS-CoV-2 is classified thus: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirinae; Coronaviridae; Orthocoronavirinae; Betacoronavirus; Sarbecovirus (GenBank, 2020; Malta et al., 2020). In general, coronaviruses are large, enveloped, single-stranded RNA viruses with the largest genome (ranging from 26 to 32 kb) among all RNA viruses (Wang et al., 2020; Woo et al., 2010). The genomic RNA consists of: 3′-, 5′-untranslated regions (UTRs), ORF1ab, ORF3a, ORF6, ORF7a/b, ORF8, ORF10 and regions for four structural proteins, namely: nucleocapsid phosphoprotein (N), membrane protein (M), envelope protein (E), and the spike protein (S). The spike proteins facilitate viral entry and membrane fusion, and they form large protrusions in the form of crowns (Latin, corona) (Banerjee et al., 2020; Li, 2017; Shi et al., 2003). The ORF1ab region coding sequence encodes very important non-structural proteins (nsp), among which the following have essential functions: nsp2, which interacts with host protein (prohibitin 1 and 2) post-infection; nsp3 (papain-like proteinase), which promotes cytokine expression by blocking host's innate immune response; 3-chymothrypsin-like proteinase (3CLpro), which is crucial for RNA replication and mediates cleavages downstream of nsp4 that are important for viral replication; nsp12, the RNA-dependent RNA polymerase (RdRp); and nsp13, the NTPase/helicase or zinc-binding domain. The ORF1ab polyprotein (7096 amino acids) plays a crucial role in altering host cell environment and viral replication (GenBank, 2020; Woo et al., 2010; Yin, 2020).
Viral genome variability and evolution is centered on mutation rates, which help the virus to invade the host, evade host's immunity, develop drug resistance (Pachetti et al., 2020) and the possible emergence of more virulent strains with a high mortality rate (Koyama et al., 2020). Alterations of host range and tissue tropism through mutations and genetic recombination aid the adaptability of coronaviruses to new environments, which results in constant and long-term health threats (Li, 2017). Understanding the mutational pattern of coronaviruses and monitoring their spread can be crucial for drug/vaccine development, economic stability, survival and global health (Li, 2017). More so, genomic sequence comparisons using single nucleotide polymorphisms (SNPs) and mutational studies have often been used for evolutionary research in recognising mutations in the genomes (Yin, 2020). Therefore, the present study was carried out to gain insight into the genomic variabilities and conserved domains in the SARS-CoV-2 genome across different continents.
2. Material and methods
2.1. Data acquisition
SARS-CoV-2 genomic sequences (7213, Asia – 2487, Oceania – 2027, Europe – 1240, Africa – 717, South America – 391, and North America – 351) submitted between December 2019 and September 2020, filtered as complete (>29,000 bp), human samples, low coverage excluded, high coverage only, all clades, all geographic regions and with patient status were obtained from the GISAID database (epicov.org/epi3/frontend). Detailed information on the regions infected by SARS-CoV-2 and from which complete genome sequences, used in this study, have been shared on the GISAID database is available as Supplementary File S1.
2.2. Sequence and mutational analysis
The mined 7213 SARS-CoV-2 genomic sequences were analysed with respect to the reference strain (accession number: hCoV-19/Wuhan/WIV04/2019) on the EpiCov web interface (epicov.org/epi3/frontend) to evaluate the global population variability in the SARS-CoV-2 genome since the onset of COVID-19 pandemic in order to identify conserved domains and mutational patterns. Mutations that occurred multiple times (≥1%) independently were focused on as they are likely candidates for the ongoing adaptive strategies of SARS-CoV-2 to human host. However, sites with few (<1%) sequences and undetermined residues (labelled as X), which might have resulted due to putative artefactual single nucleotide polymorphism, were excluded from the study.
3. Results
Mutational analysis of the 7213 SARS-CoV-2 genome sequences revealed the following high rate recurrent mutations in the ORF1ab polyprotein sequence: T265I and D448del (nsp2 coding region), T2016K (nsp3), G3278S (nsp5), L3606F (nsp6), A4489V and P4715L (RNA dependent RNA polymerase), and P5828L, Y5865C (RNA helicase region). In addition, very high rate recurrent mutations exist at the following locations: spike protein; D614G, nucleocapsid phosphoprotein; S194L, R203K and G204R, ORF3a; Q57H and G251V, and ORF8; L84S. The recurrent mutations observed from this study flags these three (3) regions (S, N and RdRp regions) as SARS-CoV-2 genome hotspots. All observed high-frequency mutations are shown in Table 1 .
Table 1.
High frequency mutations in the 7213 SARS-CoV-2 genome.a
| SARS-CoV-2 region | Mutation | Africa | Asia | Europe | Oceania | North America | South America | Total | % incidence |
|---|---|---|---|---|---|---|---|---|---|
| S protein | D614G | 696 (97.1%) | 1452 (58.4%) | 1113 (89.8%) | 1689 (83.3%) | 321 (91.5%) | 361 (92.3%) | 5632 | 78% |
| N protein | P13L | 2 (0.3%) | 248 (10%) | 116 (5.7%) | 366 | 5% | |||
| S194L | 2 (0.3%) | 314 (12.6%) | 11 (0.9%) | 14 (0.7%) | 10 (2.8%) | 1 (0.3%) | 352 | 5% | |
| R203K | 489 (68.2%) | 502 (20.2%) | 435 (35.1%) | 1348 (66.5%) | 47 (13.4%) | 252 (64.5%) | 3073 | 43% | |
| G204R | 489 (68.2%) | 495 (19.9%) | 432 (34.8%) | 1348 (66.5%) | 45 (12.8%) | 252 (64.5%) | 3061 | 42% | |
| I292T | 2 (0.2%) | 4 (0.2%) | 113 (28.9%) | 119 | 2% | ||||
| ORF3a | Q57H | 49 (6.8%) | 774 (31.1%) | 295 (23.8%) | 192 (9.5%) | 238 (67.8%) | 63 (16.1%) | 1611 | 22% |
| G251V | 3 (0.4%) | 61 (2.5%) | 29 (2.3%) | 67 (3.3%) | 3 (0.9%) | 4 (1%) | 167 | 2% | |
| ORF8 | S24L | 64 (18.2%) | 64 | 1% | |||||
| L84S | 12 (1.7%) | 259 (10.4%) | 60 (4.8%) | 171 (8.4%) | 24 (6.8%) | 12 (3.1%) | 538 | 7% | |
| ORF1ab; nsp2 | T265I | 20 (2.8%) | 79 (3.2%) | 188 (15.2%) | 148 (7.3%) | 207 (59%) | 28 (7.2%) | 670 | 9% |
| D448del | 5 (0.7%) | 4 (0.2%) | 4 (0.3%) | 13 | 0% | ||||
| ORF1ab; nsp3 | T2016K | 2 (0.3%) | 254 (10.2%) | 24 (1.2%) | 280 | 4% | |||
| ORF1ab; nsp5 | G3278S | 67 (9.3%) | 1 (0.04%) | 13 (1%) | 2 (0.6%) | 1 (0.3%) | 84 | 1% | |
| ORF1ab; nsp6 | L3606F | 35 (4.9%) | 425 (17.1%) | 56 (4.5%) | 152 (7.5%) | 11 (3.1%) | 6 (1.5%) | 685 | 9% |
| ORF1ab; nsp12 | A4489V | 2 (0.3%) | 256 (10.3%) | 25 (1.2%) | 1 (0.3%) | 1 (0.3%) | 285 | 4% | |
| P4715L | 697 (28%) | 1532 (61.6%) | 1088 (87.7%) | 1689 (83.3%) | 316 (90%) | 357 (91.3%) | 5679 | 79% | |
| ORF1ab; nsp13 | P5828L | 7 (0.3%) | 50 (2.5%) | 14 (4%) | 2 (0.5%) | 73 | 1% | ||
| Y5865C | 7 (0.3%) | 51 (2.5%) | 14 (4%) | 2 (0.5%) | 74 | 1% |
Key: One letter code for corresponding amino acid: A – Alanine, R – Arginine, N – Asparagine, D – Aspartic acid, C – Cysteine, E – Glutamate, Q – Glutamine, G – Glycine, H – Histidine, I – Isoleucine, L – Leucine, K – Lysine, M – Methionine, F – Phenylalanine, P – Proline, S – Serine, T – Threonine, W – Tryptophan, Y – Tyrosine, V – Valine.
The mutation positions are with respect to the reference (hCoV-19/Wuhan/WIV04/2019). The percentage (%) incidence is calculated as number of observed mutations across regions from the total 7213 viral samples. Highly recurrent mutations were observed in nsp12; P4715L, ORF3a; Q57H, N protein; R203K and G204R, and S protein; D614G.
It is worth noting that the D614G mutation in the S protein coincides with P4715L variants in the ORF1ab RdRp region. The R203K and G204R variants ran concurrently almost in same viral sequences. The 78% viral samples bearing the D614G and P4715L mutations also bears either the concurrent N protein R203K and G204R or the ORF3a Q57H mutations (Table 2 ). The following mutations: N protein (P13L and S194L), ORF8 (L84S), nsp3 (T2016K), and nsp6 (L3606F) are prevalent in Asian population. Mutations P5828L and Y5865C in nsp13 are prevalent in Oceania samples and they are concurrent in same viral sequences. ORF8 variants having S24L mutations are only observed in viral samples from the USA. More so, the N protein variants having I292T mutation are prevalent in viral samples from South America. The SARS-CoV-2 viral sequences were classified into clades and the corresponding pangolin lineage (Table 3 ); clade GR (40.22%), GH (20.37%), G (16.87%), O (10.69%), S (6.49%), L (3.88%) and V (1.48%).
Table 2.
Most prevalent combination of mutations.a
| Region (s) | Mutations | Frequency of occurrence |
|---|---|---|
| S protein and ORF1ab | D614G and P4715L | 78% |
| N protein | R203K and G204R | 42% |
| S protein, ORF1ab and N protein | D614G, P4715L, R203K and G204R | 40% |
| S protein, ORF1ab and ORF3a | D614G, P4715L and Q57H | 22% |
The D614G coincides with the leader protein P4715L variant alongside either of the Q57H in ORF3a or the concurrent R203K and G204R in the N protein.
Table 3.
| Pangolin lineage | Clades | Mutation(s) |
|---|---|---|
| B.1.1 | GR | D614G and G204R |
| B.1. | GH | D614G and Q57H |
| B.1. | G | D614G |
| B.2 | V | L3606F and G251V |
| A | S | L84S |
| L | SARS-CoV-2 genomes with reference alleles | |
| O |
The Clade O (depicted as “others”), a general group that do not belong to any of the SARS-CoV-2 major clades, while the SARS-CoV-2 reference strain belongs to the Clade L.
Both clade GR and GH are the offshoot of the clade G.
4. Discussion
Since its emergence and identification in December 2019 at Wuhan, China, the globally infectious coronavirus disease, caused by novel SARS-CoV-2, has presented untold hardship and critical challenge to global economies, social integration and public health (Omotoso, 2020; Wang et al., 2020). An earlier report (Dorp et al., 2020) suggested an accumulation of moderate mutations in the SARS-CoV-2 global population with an average pairwise variance of 9.6 SNPs at th phase of the pandemic and an estimated mutation rate of ~6 × 10−4 nucleotides/genome/year (CI: 4 × 10−4 - 7 × 10−4). Since the first human transmission in December 2019 (Wang et al., 2020), SARS-CoV-2 transmission is estimated to have reached 14 generations (Yin, 2020).
The ORF1ab nsp2 maintains the mitochondria functional integrity (Shuvam et al., 2020); in relation to nsp4, nsp2 is involved in viral replication. It modulates signalling in host cell environment by interacting with two host proteins, prohibitin 1 and prohibitin 2, which have been implicated in cell cycle progression, cell migration, mitochondrial biogenesis, and apoptosis (Lu, 2020; Shuvam et al., 2020). Mutations at position 265 in nsp2 induce structural alteration (Shuvam et al., 2020) and could serve as one of the mechanisms through which SARS-CoV-2 aid survival in the host cell environment. We observed this mutation to be predominant in viral sequences from Europe, Oceania and the USA, which is corroborated by an earlier report (Shuvam et al., 2020) and with very few occurrence in regions with low fatality due to COVID-19.
The mutation, L3606F, in ORF1ab nsp6 is prevalent in Asia. The protein, nsp6, intersects a presumed immunogenic peptide that could result in CD4+ and CD8+ T-cell reactivities. This missense mutation (position 3606 aa; 11083G > T or L3606F) in this report is consistent with other findings (Dorp et al., 2020; Khailany et al., 2020; Yin, 2020) as one of the strongest observed homoplasies in their studies. A leucine residue is present in wild-type position 3606 of the ORF1ab-encoded polyprotein in SARS-CoV-2 and MERS-CoV, while a valine residue in SARS-CoV. At present, there is little or no literature to substantiate the mechanism of immune response of humans to SARS-CoV-2 infection. However, CD4+ T cells induce B cells to produce antibodies, and CD8+ T-cells eliminate viral-infected cells. These cells play key roles in clearing respiratory viral infections (Dorp et al., 2020).
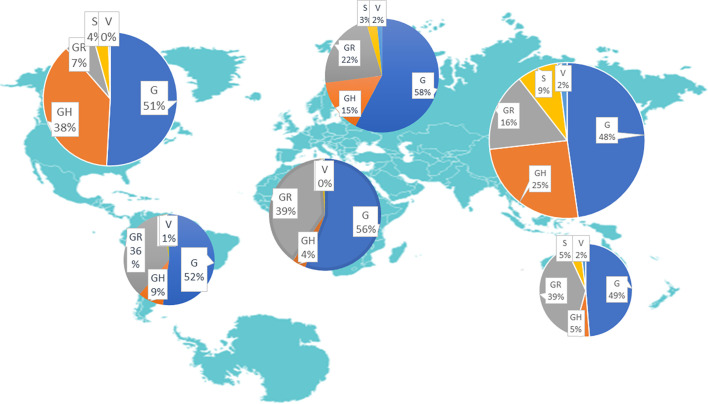
Furthermore, the most observed mutations: P4715L in RdRp and D614G in S protein are observed in same viral sequences across all regions. The polymerase is one of the primary targets for antiviral drug development (Pachetti et al., 2020; Shuvam et al., 2020) and genomic sequencing (Paul, 2020) while the S protein facilitates receptor binding and membrane fusion (Korber et al., 2020; Li et al., 2020). Recurrent mutations at these important sites can possibly be a mechanism through which SARS-CoV-2 promotes evolution, viral transmissibility and establish successful host-entry, as earlier reported (Pachetti et al., 2020). The P4715L mutation was first observed in Italy during the sporadic increase in incidence and fatality of COVID-19 in Europe (Pachetti et al., 2020). This finding is consistent with an earlier study (Yin, 2020) in a genotyping analysis of 588 SARS-CoV-2 strains where P4715L mutation (14408C > T) was reported to be located in a critical protein necessary for RNA replication. Our study discovered the mutation (P4715L) as being prevalent in Africa (97.20%), South America (91.30%), North America (90.03%), and Europe (89.83%); this was corroborated by other studies (Pachetti et al., 2020; Shuvam et al., 2020; Yin, 2020) which observed the prevalence of the mutation in viral isolates from Europe (especially France, Spain and Greece) and the USA where COVID-19 pandemic transmission is very severe. Mutations observed in this study in the nsp13 (P5828L and Y5865C) of ORF1ab polyprotein of SARS-CoV-2 which functions as a replicase or helicase corroborate other findings (Shuvam et al., 2020; Yin, 2020). These mutations were only observed simultaneously in the same viral sequence strains from Australia, South America and North America. This further buttresses previous studies (Pachetti et al., 2020; Shuvam et al., 2020) where the mutation was observed to be predominant in the USA and with very few or no occurrence in African, European or Asian population. In line with an earlier finding (Pachetti et al., 2020), we observed that most of the recurrent mutations in the viral genomes have little prevalence in the Asian population where the first case was reported (Wang et al., 2020). The intercontinent distribution showed the high prevalence of the clades G and GR in Africa, Asia, Australia and South America. While, clade GH were also prevalent in Europe and North America (Fig. 1 ). This corroborates a recent report (Hamed et al., 2021) which also reports same trend of clade predominace across the continents. Furthermore, a recent report (Omotoso, 2020) on the SARS-CoV-2 mutational landscape in Africa likewise showed the prevalence of clades G and GR in Africa.
Fig. 1.
Intercontinent distribution of the most predominant SARS-CoV-2 clades.
The diagnostic test for COVID-19, Reverse Transcriptase – Polymerase Chain Reaction (RT-PCR) assays, targets the RdRp region of ORF1ab sequence as being the region with the highest analytical sensitivity for screening via next-generation sequencing or RT-PCR (Wang et al., 2020). An earlier report (Yin, 2020) has attributed the basis of mutations (which might impact pathogenicity, survival or transmissibility) to lack of proof-reading activity by RdRp encoded in RNA viruses. The 3CLpro proteinase (which processes nsp4 – nsp16 in all coronaviruses), encoded in the ORF1ab sequence, plays a crucial role in RNA replication. Supported by other findings (Muhammad et al., 2020; Yin, 2020), our study observed sequence conservation in the 3CLpro region which serves an important role in viral replication and has been targeted in SARS-CoV and MERS-CoV for drug design and development. Based on the importance of this region and the observed genomic conservation, 3CLpro catalytic sites and conserved assembly may serve as attractive targets for the design of vaccine and antiviral drugs (Muhammad et al., 2020).
The avalanche of data on SARS-CoV-2 genome deposited in public repositories gives an advantage of abundant viral sequences for researchers to work with. One major limitation we observed, which might also limit future studies is the availability of patients' ages and disease status for most of the viral sequences. Therefore, we encourage the continual release of comprehensive data from every region which include sociobiological informations, most especially the disease outcome of the infected patients. This can help future studies to correlate the observed mutations to disease severity and chances of survival. Likewise, further studies, such as molecular docking studies, are required to determine the impact of the aforementioned mutations on protein structures, functions and viral interactions with the host in order to facilitate drug design and vaccine development.
5. Conclusion
Our study revealed the mutational spectra present in the SARS-CoV-2 genome and the possible implications on viral structure-function, evolution, transmissibility and virulence. Our result showed the important conserved domains as important regions for new antiviral vaccines or therapeutics in targeting the SARS-CoV-2. Certain recurrent signature mutations observed to be linked to specific regions could be used to monitor the spread, evolution and transmissibility of the virus in humans. This could play an important role in understanding SARS-CoV-2 genetic epidemiology. The mutational analysis revealed high recurrent mutations that aid viral replication and survival in the host. Considering the current trend of COVID-19 incidence and fatality across the globe (covid19.who.int), we can deduce that these mutations confer a selective advantage of higher virulence and transmissibility of the virus, but might not really be the major player in fatality and higher severity of the disease considering the low fatality due to COVID-19 in Africa despite having the highest prevalence of the most recurrent mutations (P4715L and D614G) observed.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgements
The authors would like to thank those who sequenced SARS-CoV-2 genomes and deposited in public domains.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2021.105097.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
Supplementary material 3
Supplementary material 4
Supplementary material 5
Supplementary material 6
References
- Banerjee A.K., Begum F., Ray U. 2020. Mutation Hot Spots in Spike Protein of COVID-19. [DOI] [Google Scholar]
- GenBank . NCBI GenBank; 2020. Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1, complete genome [WWW Document]https://www.ncbi.nlm.nih.gov/genbank/sars-cov-2-seqs/ [Google Scholar]
- Hamed S.M., Elkhatib W.F., Khairalla S.A., Noreddin M.A. Global dynamics of SARS - CoV - 2 clades and their relation to COVID - 19 epidemiology. Sci. Rep. 2021;11:8435. doi: 10.1038/s41598-021-87713-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khailany R.A., Safdar M., Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Reports. 2020;19:100682. doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Labranche C.C., Saphire E.O., Montefiori D.C., Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Partridge D.G., Evans C.M., Freeman T.M., De Silva T.I. Tracking changes in SARS-CoV-2 spike : evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Platt D., Parida L. Variant analysis of COVID-19 genomes. Bull. World Health Organ. 2020 doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, function and evolution of coronavirus spike proteins. Ann. Rev. Virol. 2017;3:237–261. doi: 10.1146/annurev-virology-110615-042301.Structure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Wang L., Ren L. Antiviral mechanisms of candidate chemical medicines and traditional Chinese medicines for SARS-CoV-2 infection. Virus Res. 2020;286:198073. doi: 10.1016/j.virusres.2020.198073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acid Res. 2020;48(D1):D265–D268. doi: 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malta F., Amgarten D., de Oliveira D.B.L., Araujo D.B., Machado R.R.G., Santana R.A.F., Mangueira C.L.P., Durigon E.L., Pinho J.R.R. GenBank; 2020. ORF1ab Polyprotein [ Severe Acute Respiratory Syndrome Coronavirus 2] [WWW Document] (URL SARS COV/Journal/ORF1ab polyprotein [Severe acute respiratory syndrome coronavirus 2] - Protein - NCBI.mhtml) [Google Scholar]
- Muhammad T.Q., Safar A.M., Alamri L.-L.C. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Anal. 2020 doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omotoso O.E. Contributory role of SARS-CoV-2 genomic variations and life expectancy in COVID-19 transmission and low fatality rate in Africa. Egypt. J. Med. Hum. Genet. 2020;21:1–6. doi: 10.1186/s43042-020-00116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., Masciovecchio C., Angeletti S., Ciccozzi M., Gallo R.C., Zella D., Ippodrino R. Emerging SARS - CoV - 2 mutation hot spots include a novel RNA - dependent - RNA polymerase variant. J. Transl. Med. 2020;18:179. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul O. First African SARS-CoV-2 genome sequence from Nigerian COVID-19 case. Genome Reports. 2020 https://virological.org/t/first-african-sars-cov-2-genome-sequence-from-nigerian-covid-19-case/421 (accessed 4.20.20) [Google Scholar]
- Shi D., Zhou H.-J., Wang B.-B., Gu Y.-H., Wang Y.-F. Multiple sequence alignment of the M protein in SARS-associated and other known coronaviruses. J. Shanghai Univ. 2003;7:118–123. doi: 10.1007/s11741-003-0078-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuvam B., Sohan S., Riju D., Kousik M., Pritha B. Mutational spectra of SARS-CoV-2 ORF1ab Polyprotein and signature mutations in the United States of America. bioRxiv. 2020 doi: 10.1101/2020.05.01.071654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dorp L., Acman M., Richard D., Shaw L.P., Ford C.E., Ormond L., Owen C.J., Pang J., Tan C.C.S., Boshier F.A.T., Torres A., Balloux F. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect. Genet. Evol. 2020;83:104351. doi: 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Li X., Li T., Zhang S., Wang L., Wu X., Liu J. The genetic sequence, origin, and diagnosis of SARS-CoV-2. Eur. J. Clin. Microbiol. Infect. Dis. 2020 doi: 10.1007/s10096-020-03899-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Coronavirus (COVID-19) Dashboard | WHO Coronavirus (COVID-19) Dashboard With Vaccination Data [WWW Document] 2021. https://covid19.who.int/ n.d. (accessed 9.14.21)
- Woo P.C.Y., Huang Y., Lau S.K.P., Yuen K. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C. Genotyping coronavirus SARS-CoV-2: methods and implications. Genomics. 2020:1–9. doi: 10.1016/j.ygeno.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2
Supplementary material 3
Supplementary material 4
Supplementary material 5
Supplementary material 6