Abstract
A subset of families with co-dominant or recessive inheritance has been described in several genes previously associated with dominant inheritance. Those recessive families displayed similar, more severe, or even completely different phenotypes to their dominant counterparts. We report the first patients harboring homozygous disease-related variants in three genes that were previously associated with dominant inheritance: a loss-of-function variant in the CACNA1A gene and two missense variants in the RET and SLC20A2 genes, respectively. All patients presented with a more severe clinical phenotype than the corresponding typical dominant form. We suggest that co-dominant or recessive inheritance for these three genes could explain the phenotypic differences from those documented in their cognate dominant phenotypes. Our results reinforce that geneticists should be aware of the possible different forms of inheritance in genes when WES variant interpretation is performed. We also evidence the need to refine phenotypes and inheritance patterns associated with genes in order to avoid failures during WES analysis and thus, raising the WES diagnostic capacity in the benefit of patients.
Subject terms: Genetics research, Disease genetics
Introduction
Mendelian genetics has by far provided the most medically actionable data on human DNA variation, and thus, curating this data is a priority to move forward to personalized medicine. Several factors complicate this curation, such as the high incidence of rare benign variants or the erroneous disease-gene associations [1]. In addition, DNA variation expresses itself with a wide spectrum of phenotypes in human populations, favored by the presence of genetic modifiers and by the autozygosity and genetic variations in non-related disease genes, among others [2]. The possibility of different inheritance patterns associated with a particular gene also influences phenotype heterogeneity [3].
Whole exome sequencing (WES) has improved the genetic diagnosis of rare diseases, with a diagnostic yield that ranges from 25 to 50% depending on the disorder and the examined series [4–8]. This diagnostic yield of WES strongly relies on the strategy used for the variant’s prioritization and for the selection of genes to be analyzed [9]. Gaining insight into the different phenotypes and inheritance patterns associated with a gene will improve the curation of variants and genes linked to disease, and thus, should raise the WES diagnostics capacity.
Here, we report three consanguineous families with homozygous related disease variants in the CACNA1A, RET, and SLC20A2 genes, which had been only previously associated with dominant inheritance. We also discuss the clinical implications for each case and emphasize the importance of curating Mendelian genes to improve both variants interpretation and diagnostic yield of WES.
Methods
Subjects
Family 1 was derived for a second opinion to the Gynecology Department of the University Hospital 12 de Octubre. Probands from family 2 and 3 were full clinically evaluated by the Pediatry Department (Immunodeficiency Unit) and the Neurology Department of the Hospital, respectively. Since there was a suspicion of a genetic disorder, all patients were referred for analysis to the Clinical Genetics Department of the hospital.
Whole peripheral blood samples from the members of the three consanguineous families were collected. Written informed consent was obtained from each patient or their guardians. Genomic DNA extraction was performed following standard procedures, using the Maxwell 16 blood DNA purification (Promega).
The study was approved by the Ethics Committee of the University Hospital 12 Octubre, in accordance with the Declaration of Helsinki.
WES and segregation analysis
WES was performed on the probands using the kit xGen Exome Panel v1.0 (IDT –Integrated DNA technologies-). Paired-end sequencing (2x75bp) was carried out on a NextSeq 550 (-Illumina-) and the Bioinformatics analysis was done using the custom pipeline Karma, validated according to Somak Roy et al. [10]. Reads were aligned to the reference human genome (hg19) using BWA MEM (v0.7.17) [11] and Bowtie2 (v.2.4.1) [12]. The variant calling process was performed using GATK (Haplotype Caller from Genome Analysis Toolkit, v.4.1) [13] and VarDict (AstraZeneca, v1.7.0) [14]. Annovar (v2018Apr16) [15] was used for the annotation of variants. Copy number variations were not assessed.
WES data was analyzed using custom panels targeted to the clinical suspicion. Panels were developed based on public databases (OMIM, DisGeNet, GTR, PanelApp, HPOs) and by reviewing the medical specific literature (see the panel content used for each case in Supplementary Table 1)
An extended analysis of the clinical exome panel (5686 genes) was performed in each proband in order to rule out other possible related disease genetic alterations. Deep intronic, UTR, and variants with non-OMIM associated genes were not analyzed (the list of unfiltered identified variants in each case is shown in Supplementary Tables 2.1, 2.2, and 2.3). Guided for the consanguinity of the family, a special focus was made on homozygous variants. Trio analysis could not be performed and thus, a de novo causal variant could not be ruled out in any of the cases. However, taking into account the family history, the occurrence of a de novo variant in the probands seemed not to be the most likely genetic cause underlying the clinical suspicion.
All variants were prioritized by filtering through different clinical and population databases (gnomAD, dbSNP). The main requirements for a good-quality variant to be analyzed and interpreted were: (1) read mapping quality, (2) depth coverage ≥20X; (3) a variant fraction ≥20%, (4) a population frequency <3%, and (5) classification in the ClinVar database different from benign or likely benign by multiple suscriptors. According to these parameters, the list of all prioritized variants for each case are detailed in Supplementary Tables 2.1–2.3. Intronic, UTR variants and variants in genes not associated with OMIM are not shown.
All prioritized variants were classified following the ACMG criteria [16].
Family segregation analysis was carried out by Sanger sequencing using the kit BigDye Term v3.1 of Life Technologies, S.A (Applied Biosystems) and following standard procedures. The proband DNA was used as a positive control.
All variants reported here have been submitted to the LOVD database and can be now accessed using the corresponding ID or the following URLs:
CACNA1A_000389: Variant #0000675114 (NC_000019.9:g.13409683G > A, CACNA1A(NM_001127221.1):c.2767C > T)—Global Variome shared LOVD
RET_000288: Variant #0000675117 (NC_000010.10:g.43608363G > A, RET(NM_020975.4):c.1711G > A)—Global Variome shared LOVD
SLC20A2_000056: Variant #0000675114 (NC_000019.9:g.13409683G > A, CACNA1A(NM_001127221.1):c.2767C > T)—Global Variome shared LOVD
Results and discussion
Family 1
Four newborns, all presenting with hypotonia, mild facial dysmorphic features, encephalopathy, and seizures were born from consanguineous parents of gipsy ethnicity. In all of them pregnancy and birth were normal, but they all died within the first 3–6 months of life. The couple had also two asymptomatic males and a spontaneous abortion in-between (Fig. 1A). Chromosome abnormalities in both parents were ruled out by conventional karyotype. In the proband (V:3), a mitochondrial disease was firstly suspected and thus, muscle mitochondrial DNA (mtDNA) copy number, whole mtDNA NGS and WES analysis of 305 nuclear-genes associated with mitochondrial disorders was performed. No genetic alterations were detected.
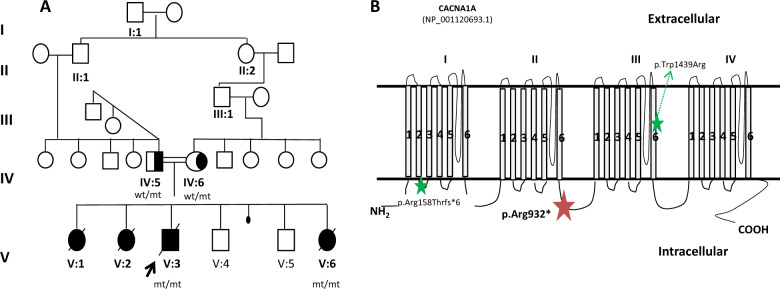
Fig. 1. Family 1: CACNA1A gene.
A Family 1 pedigree and genotypes; wild type (wt); mutant allele (mt). B Schematic structure of the Cav2.1 channel (adapted from Damaj et al. [17]). The red start illustrates the p.(Arg932*) variant detected in our family in the CACNA1A gene; the green starts indicate the variants described in Reinson et al. [31].
A second analysis of a panel containing 187 genes related to encephalopathies revealed the presence of the novel homozygous nonsense c.2767C>T p.(Arg932*) variant in the exon 19 of the CACNA1A gene (NM_001127221.1) where to our knowledge, only a heterozygous frameshift variant has been associated with Episodic ataxia type 2 (EA2) to date [17]. This variant is located in the intracytoplasmic loop between domain II and III of the protein, where most of the inherited loss-of-function (LOF) variants have been associated with encephalopathy epileptic and episodic ataxia [18] (Fig. 1B). The variant is not reported in the literature or in the clinical and population consulted databases (ClinVar, 1000 G, gnomAD, HGMD). Intriguingly and according to the gnomAD database, the CACNA1A gene is restricted for LOF variation and no homozygous LOF variants have been reported so far.
Segregation analysis showed that both parents were heterozygous carriers and the affected sister (V:2), who also died within the first 3 months of life with the same phenotype of the proband, was homozygous (Fig. 1A). Samples of the two affected sisters (V:1 and V:6) are not available and thus, segregation analysis could not be performed. The unaffected youngest and underage children (V:4 and V:5) were not genotyped but a clinical follow-up was recommended.
After genetic diagnosis, both apparently asymptomatic parents were referred to a Neurology consultation for clinical evaluation. The 44-year-old father referred frequent (2–3 per week) hours long ataxic episodes from the age of 17 to 30 that progressively decreased to completely disappear. The 39-year-old mother also referred to have similar episodes from the age of 30 to 37 years, that she defined as dizziness. Both parents had no clinical manifestations between episodes and remained asymptomatic since then. Therefore, they were diagnosed with EA2 due to the previous presence of ataxic episodes and the identification of the heterozygous apparently LOF variant in the CACNA1A gene. There were no other family members with similar background or phenotype to segregate the variant.
The extended analysis of the clinical exome panel, revealed the presence of the three following homozygous rare variants: (1) c.55-2A>C in the AMTN gene (NM_212557.3) associated with amelogenesis imperfecta IIIB (#617607); (2) c.1727C>T p.(Ser576Leu) in the ADGRE2 gene (NM_013447.3) associated vibratory urticaria (#606100), and (3) c.622G>A p.(Val208Leu) in the JAK3 gene (NM_000215.3) associated with severe combined immunodeficiency (#600173). These three genes are associated with phenotypes not related to that of this family. No other relevant homozygous, heterozygous, or hemizygous variants related to the clinical suspicion were detected. In addition, the occurrence of a de novo causing-disease variant is unlikely considering the family history. Taken together, all these data allowed us to categorize the variant c.2767C>T p.(Arg932*) in the CACNA1A gene as pathogenic following the ACMG criteria [16], being presumably the underlying cause of the clinical phenotype in this family.
Heterozygous variants in the CACNA1A gene (19p13.13, *601011) are associated with clinically heterogeneous and occasionally overlapping autosomal dominant disorders. In general, missense gain of function variants are mainly associated with familial hemiplegic migraine (FHM1, #141500), LOF variants with EA2 (#108500) and the gain of toxicity expansions of CAG repeats with spinocerebellar ataxia (SCA6, #183086) [19–22]. However, both gain and LOF heterozygous variants have also been associated with cognitive/behavioral impairments [17, 21, 23, 24], with epileptic encephalopathy [18, 25–27] and with congenital ataxia [28], suggesting that the neuropsychological disturbances are caused by the channel dysfunction. Indeed, no major cognitive deficits have been described in SCA6 patients, probably because polyglutamine expansions do not alter the calcium current of the channel [19].
To our knowledge, biallelic CACNA1A alterations have been described in a SCA6 patient homozygous for a 19-CAG repeats intermediate allele [29] and in two siblings with progressive myoclonus that were homozygous for a CAG insertion close to the C-terminal of the protein (c.6975_6976insCAG; p.(A2326delinsQA)) [30]. To date, the only previous association of biallelic CACNA1A variants with early-onset epileptic encephalopathy has been reported in two compound heterozygous siblings (c.[4315T>A];[472_478del], p.([Trp1439Arg];[Ala158Thrfs*6])) that presented with early-onset epileptic encephalopathy, progressive cerebral, cerebellar, and optic nerve atrophy. The elder sister suffered a significant clinical worsening that caused her death when she was 5 years old [31]. The authors described great intra-familial variability in the heterozygous reported relatives (parents and sister), showing a poor genotype-phenotype correlation that could be explained by the fact that the alteration of Cav2.1 channels function is differently compensated by the upregulation of other less efficient voltage-gated calcium channels (Cav2.2 and Cav1.2). In contrast to this reported genotype consisting of a missense and a presumably LOF frameshift variant in compound heterozygosis, the siblings we report were both homozygous for a nonsense presumably LOF variant, dying within the first months of life. We presume this genotype affects more severely the function of Cav2.1 channels causing a severe and lethal phenotype, as previously suggested in CACNA1A knock-out mice [32].
Herein, we describe the first lethal phenotype associated with a homozygous nonsense variant in the CACNA1A gene.
Family 2
The proband was a 15-year-old boy born from healthy and consanguineous parents (Fig. 2A). At birth, he was diagnosed with Hirschsprung disease (HSCR) that required several surgical interventions due to complications that included severe enterocolitis and abdominal sepsis. Complete colonic removal with preservation of anal sphincter, continent thereafter. He was on monthly iron i.v. supplementation until the age of 10 years, due to patchy gut inflammation as seen by radiolabeled leukocytes. Intravenous iron is no longer needed. Histopathological analysis revealed the absence of ganglion cells in myenteric and submucosal plexus of intestine. Duodenal biopsy was normal, including normal villous architecture. The couple had also two asymptomatic siblings and an HSCR affected 24-year-old boy (II:2) who, like his brother, was also totally ileostomized during his first year of life.
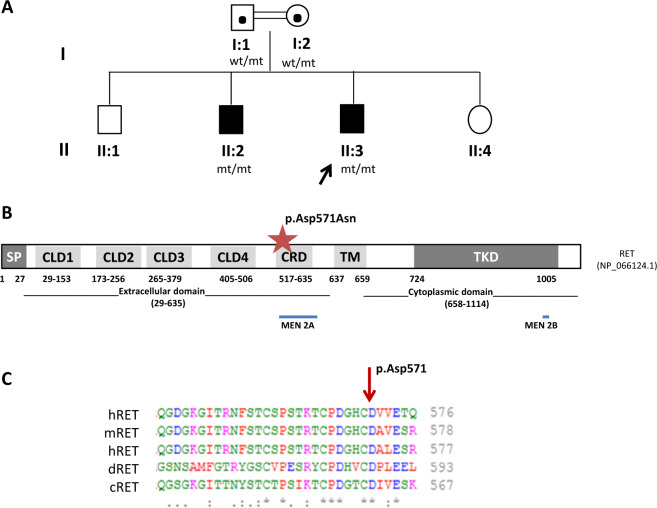
Fig. 2. Family 2: RET gene.
A Family 2 pedigree and genotypes; wild type (wt); mutant allele (mt). B Schematic domain structure of the RET protein (adapted from Wang et al. [49] and Wu et al. 2019 [50]). The red star indicates the position of the variant p.(Asp571Asn) detected in the RET gene in our family. The regions were MEN2A and MEN2B causal variants cluster are indicated with blue bars. C Multiple sequence alignment of residues adjacent to amino acid 571 of the RET protein from human (h), mouse (m), rat (r), drosophila (d), and chicken (c). The tool Clustal Omega was used for multiple alignments (https://www.ebi.ac.uk/Tools/msa/clustalo/). SP signal peptide, CLD cadherin-like domain, CRD Cysteine-rich domain, TM transmembrane, TKD tyrosine kinase domain.
The analysis of the panel containing 23 genes related to HSCR revealed the presence of the homozygous rare variant c.1711G>A p.(Asp571Asn) (rs750958377) in the exon 9 of the RET gene (NM_020975.4). No homozygous, heterozygous, or hemizygous pathogenic or likely pathogenic neither candidate variants that could be related to the clinical suspicion of the patient and his brother were identified during the extended clinical exome analysis performed (see Supplementary Table 2.2).
Although there is not a mutational hot spot in HSCR, it seems to be a trend for HSCR-associated variants to cluster in the extracellular domain, particularly in the exon 3 of the gene [33]. The extracellular region of RET contains four atypical cadherin-like domains (CLD1–4) followed by a cysteine-rich domain (CRD) (Fig. 2), which are important for the RET receptor’s interaction with its cognate ligands and co-receptors and thus, for its essential dimerization and activation [34, 35]. Intriguingly, the variant c.1711G>A p.(Asp571Asn) in exon 9 of RET gene is predicted to affect the protein in the extracellular CDR domain (Fig. 2B), where only two heterozygous de novo disease-causing variants have been described so far associated with total and short-HSCR respectively: p.(Glu595Gln) and p.(Gly605Asp) [36, 37]. The 571 Aspartic amino acid appears not to be one of the residues coordinating the calcium ion or the backbone carbonyl of the protein, based on RET cryo-electron microscopy [35]. However, the Aspartate (negatively charged amino acid) is highly conserved among species (Fig. 2C) and thus, its substitution for an Asparagine (a polar uncharged amino acid) may somehow affect the stability and normal function of the RET protein, which has been demonstrated to be a versatile receptor that can generate distinct signaling outputs depending on the ligand [35]. Accordingly, all the in silico prediction programs tested (MutationTaster, PolyPhen, SIFT) consider the variant to be damaging. This variant had already been reported in three heterozygotes in the gnomAD population database (MAF < 0.001%) but it has not been described in Clinvar or previously reported in the literature.
The RET proto-oncogene (10q11.21, *164761) is the major susceptibility gene in HSCR (#142623) and encodes a transmembrane tyrosine kinase receptor that accounts for ~50% of familial and 15–20% of sporadic HSCR [38], a dominant congenital genetic disorder with reduced penetrance characterized by the absence of ganglion cells along with variable segments of the gut. Depending on the length of the affected segment, it is categorized as short-segment (up to 80%), long-segment (up to 20%) or total colonic aganglionosis (up to 8%). Most cases are sporadic (70–80%), though chromosomal abnormalities or congenital anomalies are associated in 12% and 18% of HSCR patients, respectively [38]. Germline LOF variants throughout the RET extracellular region have been associated with HSCR, but a specific hot spot has not been identified. Many of them are distributed in the co-receptor or the calcium-binding site, probably affecting the stability or co-receptor binding of RET, and thus impairing its activity [35]. It has been shown that causal variants located in the extracellular cadherin-like domain (CDL1-3) interfere with the RET maturation and its translocation to the membrane, while intracellular mutations alter the RET kinase activity and signaling ability (Fig. 2) [36, 38–41].
Heterozygous germline gain of function RET variants are also associated with autosomal dominant multiple endocrine neoplasia type 2 syndromes (MEN2A and MEN2B), which predispose to medullary thyroid carcinoma, pheochromocytoma, parathyroid hyperplasia (MEN2A only) and mucosal neuromas and thickened corneal nerves (MEN2B only). MEN2A is mainly caused by activating missense variants clustered in the CRD domain that not dramatically affect the folding or cell surface localization of RET, leading to RET dimerization and constitutive activation [35]. MEN2B is caused by the almost unique M918T variant that accelerate RET autophosphorylation (Fig. 2).
Segregation analysis in this family revealed the brother (II:2) affected with the same phenotype of the proband (total colonic aganglionosis) was homozygous while both asymptomatic parents were heterozygous carriers (I:1 and I:2) for the c.1711G>A variant in the RET gene (Fig. 2B). Unfortunately, the analysis in the apparently asymptomatic siblings (II:1 and II:4) could not be performed.
The absence of associated phenotype in the parents could be explained by the already known reduce penetrance of RET dominant causal variants. However, in this family, the detected variant seems to have low-penetrance in heterozygous (there was no family history of HSCR) but full-penetrance when presented in both alleles, suggesting a possible co-dominant inheritance. Considering the high heterogeneity of HSCR, there is no evidence enough to postulate a recessive inheritance associated with a more severe phenotype (both siblings had total colonic aganglionosis) though it cannot be completely discarded. Further functional studies are warranted to get insight into these issues, whose results will have an impact on the genetic counseling of this disorder.
Taking into account all the abovementioned information, the variant was classified as likely pathogenic according to the ACMG criteria [16] and thus, we describe the first family harboring a homozygous missense variant in the RET gene (c.1711G>A p.(Asp571Asn)), leading to a non-lethal but severe form of HSCR.
Family 3
The proband was a 55-year-old woman, the fourth of six siblings, born from healthy consanguineous parents (Fig. 3A). After an episode of vertigo, a computerized tomography scan (CT) that showed basal ganglia calcifications was performed when she was 20 years old. No endocrine or metabolic disorders related to brain calcifications were found. At the age of 28, she developed a psychotic disorder including delirium consistent with paranoia or delusional disorder that improved with aripiprazole. A CT scan showed extensive cortical bihemispheric calcifications in the basal ganglia, but also affecting the periventricular white matter, the posterior cortex and the cerebellum (Fig. 3B). In her forties, she started with motor clumsiness, balance disturbance, and severe dysarthria. In recent years, asymmetric Parkinsonism was observed. His father died at the age of 81 years, due to respiratory insufficiency.
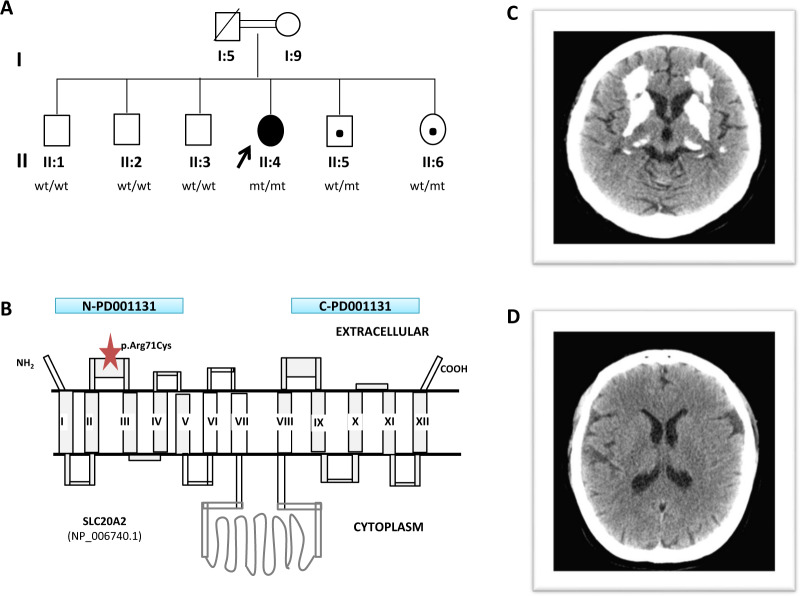
Fig. 3. Family 3: SLC20A2 gene.
A Family 3 pedigree and genotypes; wild type (wt); mutant allele (mt). B Schematic representation of the SLC20A2 protein (adapted from Hsu SC et al. 2013 [42]). Blue bars illustrate the N-and C-terminal ProDom domain (N-PD001131 and C-PD001131 respectively) of the protein. The red star indicates the position of the variant p.(Arg71Cys) in the SLC20A2 gene detected in our family. C Cranial computed tomography (CT) scan of the homozygous patient showing extensive areas of calcification in the frontal white matter and basal ganglia. D Normal cranial CT scan of her asymptomatic heterozygous sister (II:6).
The analysis of the panel containing 59 genes related to the presence of brain calcifications revealed the homozygous missense c.211C>T p.(Arg71Cys) variant (rs373139157; MAF < 0.0001%) in the exon 2 of the SLC20A2 gene (NM_006749.4) (Fig. 3C), where most of heterozygous missense disease-related variants have been described [42]. This variant has only been reported in 1 of 251,490 alleles in the gnomAD public population databases and, to our knowledge, it has not yet been associated with disease.
In the extended clinical exome analysis, no pathogenic or likely pathogenic neither candidate variants that could be related to the clinical suspicion of the patient were identified. Interestingly, we detected the rare homozygous c.476G>A p.(Arg159Lys) variant in the NDUFAF6 gene (NM_152416.3). Defects in this gene have been associated with autosomal recessive Mitochondrial complex I deficiency type 17 (#618239) with onset during infancy or early childhood. However, the related phenotype is not consistent with the patient’s phenotype, as she was diagnosed with Fahr’s disease after a vertigo episode in her second decade of life. In addition, in this family, even though a trio analysis could not be performed, the occurrence of a causal variant is unlikely.
The SLC20A2 gene encodes the type III sodium-dependent phosphate transporter 2, a Na/Pi transporter present in all kingdoms and ubiquitously expressed in various organs including the kidney, liver, and brain. The protein has 12 transmembrane regions and a large central intracytoplasmic domain (Fig. 3C). The two terminal transmembrane regions contain a unique sequence of ~150 amino acids with four highly conserved motifs known as ProDrom domain 001132 (PD001131). This domain is characteristic of all PiTs family members and is essential for human Pi transport function [43–46]. The Arg71 residue is located in the ProDom domain and it is highly conserved among species. All in silico predictors indicate the p.(Arg71Cys) substitution to be damaging (MutationTaster, PolyPhen, SIFT). Interestingly, the substitution of the Arg71 residue for a Histidine (p.Arg71His)—both positively charged amino amino acids—has been previously reported in a heterozygous woman who developed clumsiness of her hands and gait unsteadiness at the age of 71, being diagnosed with Parkinson disease. She had calcification in her brain CT and an asymptomatic daughter also with minimal intracranial calcification. However, Parkinson disease is a common disorder in aged people, and thus, a coincidental presence of IBGC in this patient cannot be discarded. Indeed, it is possible that IBGC would have been unnoticed if a CT were not performed in the patient [47, 48].
The SLC20A2 gene (8p11.2, *158378) is a main cause (41%) of familial idiopathic basal ganglia calcifications (IBGC; #213600) -also known as primary familial brain calcification or Fahr’s disease-, a rare progressive neurodegenerative dominant disorder characterized by bilateral calcium deposits in the basal ganglia and other brain regions. Frequent clinical manifestations include psychiatric conditions and neurological symptoms although the severity and the age of onset are very diverse. Many cases are subclinical and incidentally diagnosed during image studies. The penetrance of the disease is reduced and might be as low as 70% [47].
Family segregation analysis in the five clinically asymptomatic siblings of the proband (both asymptomatic parents were not available) showed that only the two younger brothers (II:5 and II:6) were heterozygous for the c.211C>T variant (Fig. 3A). The CT performed in the elder asymptomatic sibling (II:6) was normal (Fig. 3D). Therefore, relying on all these findings, the variant was classified as likely pathogenic, according to the ACMG criteria.
Here, we describe the first patient harboring a homozygous variant in the SLC20A2 gene associated with a severe IBGC phenotype. Based on these results, a co-dominant pattern for the variant in the SLC20A2 gene cannot be excluded in this family.
Concluding remarks
Biallelic disease-causing variants in genes in which only dominant alleles have been are reported so far, as well as in genes with incomplete phenotypes, could be easily lost during variant analysis, and consequently lead to a failure in the genetic diagnosis. Identifying new forms of inheritance for particular genes and their associated phenotype is essential to provide a reliable diagnosis through WES.
In this study, we expand the number of biallelic disease-causing variants reported in the CACNA1A, RET, and SLC20A2 genes which were previously only associated with a dominant phenotype. We postulate that distinct inheritance forms in the same gene can contribute to phenotypic variability. Our results suggest that genetic analysts should be aware of this possibility when WES variant analysis is performed.
Supplementary information
Acknowledgements
The authors would like to thank all the clinical staff from the University Hospital 12 de Octubre, who made this study possible. Grant number PI17/1067 to MIAB from the Spanish “Instituto de Salud Carlos III” (ISCIII). Grant number PI18/01374 to MAM from the Spanish “Instituto de Salud Carlos III” (ISCIII) and European Regional Development Fund (ERDF; FEDER).
Competing interests
All authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41431-021-00919-5.
References
- 1.Alkuraya FS. Discovery of mutations for Mendelian disorders. Hum Genet. 2016;135:615–23. doi: 10.1007/s00439-016-1664-8. [DOI] [PubMed] [Google Scholar]
- 2.Alsalem AB, Halees AS, Anazi S, Alshamekh S, Alkuraya FS. Autozygome sequencing expands the horizon of human knockout research and provides novel insights into human phenotypic variation. PLoS Genet. 2013;9:e1004030. [DOI] [PMC free article] [PubMed]
- 3.Monies D, Maddirevula S, Kurdi W, Alanazy MH, Alkhalidi H, Al-Owain M, et al. Autozygosity reveals recessive mutations and novel mechanisms in dominant genes: implications in variant interpretation. Genet Med. 2017;19:1144–50. doi: 10.1038/gim.2017.22. [DOI] [PubMed] [Google Scholar]
- 4.Dillon OJ, Lunke S, Stark Z, Yeung A, Thorne N, Gaff C, et al. Exome sequencing has higher diagnostic yield compared to simulated disease-specific panels in children with suspected monogenic disorders. Eur J Hum Genet. 2018;26:644–51. doi: 10.1038/s41431-018-0099-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee H, Deignan JL, Dorrani N, Strom SP, Kantarci S, Quintero-Rivera F, et al. Clinical exome sequencing for genetic identification of rare mendelian disorders. JAMA. 2014;312:1880–7. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seo GH, Kim T, Choi IH, Park Jyoung, Lee J, Kim S, et al. Diagnostic yield and clinical utility of whole exome sequencing using an automated variant prioritization system, EVIDENCE. Clin Genet. 2020;98:562–70. doi: 10.1111/cge.13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu X, Guo R, Guo J, Qi Z, Li W, Hao C. Parallel tests of whole exome sequencing and copy number variant sequencing increase the diagnosis yields of rare pediatric disorders. Front Genet. 2020;11:473. doi: 10.3389/fgene.2020.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stefanski A, Calle-López Y, Leu C, Pérez-Palma E, Pestana-Knight E, Lal D. Clinical sequencing yield in epilepsy, autism spectrum disorder, and intellectual disability: a systematic review and meta-analysis. Epilepsia. 2021;62:143–51. doi: 10.1111/epi.16755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shamseldin HE, Maddirevula S, Faqeih E, Ibrahim N, Hashem M, Shaheen R, et al. Increasing the sensitivity of clinical exome sequencing through improved filtration strategy. Genet Med. 2017;19:593–8. doi: 10.1038/gim.2016.155. [DOI] [PubMed] [Google Scholar]
- 10.Roy S, Coldren C, Karunamurthy A, Kip NS, Klee EW, Lincoln SE, et al. Standards and guidelines for validating next-generation sequencing bioinformatics pipelines: a joint recommendation of the association for molecular pathology and the college of american pathologists. J Mol Diagn. 2018;20:4–27. [DOI] [PubMed]
- 11.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ArXiv. 2013; p. 1303.
- 12.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinforma. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai Z, Markovets A, Ahdesmaki M, Chapman B, Hofmann O, McEwen R, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016;44:e108. doi: 10.1093/nar/gkw227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–23. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Damaj L, Lupien-Meilleur A, Lortie A, Riou É, Ospina LH, Gagnon L, et al. CACNA1A haploinsufficiency causes cognitive impairment, autism and epileptic encephalopathy with mild cerebellar symptoms. Eur J Hum Genet. 2015;23:1505–12. doi: 10.1038/ejhg.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang X, Raju PK, D’Avanzo N, Lachance M, Pepin J, Dubeau F, et al. Both gain-of-function and loss-of-function de novo CACNA1A mutations cause severe developmental epileptic encephalopathies in the spectrum of Lennox-Gastaut syndrome. Epilepsia. 2019;60:1881–94. doi: 10.1111/epi.16316. [DOI] [PubMed] [Google Scholar]
- 19.Indelicato E, Nachbauer W, Karner E, Eigentler A, Wagner M, Unterberger I, et al. The neuropsychiatric phenotype in CACNA1A mutations: a retrospective single center study and review of the literature. Eur J Neurol. 2019;26:66. doi: 10.1111/ene.13765. [DOI] [PubMed] [Google Scholar]
- 20.Tottene A, Fellin T, Pagnutti S, Luvisetto S, Striessnig J, Fletcher C, et al. Familial hemiplegic migraine mutations increase Ca2+ influx through single human Cav2.1 channels and decrease maximal Cav2.1 current density in neurons. Proc Natl Acad Sci USA. 2002;99(Oct):13284–9. doi: 10.1073/pnas.192242399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Travaglini L, Nardella M, Bellacchio E, D’Amico A, Capuano A, Frusciante R, et al. Missense mutations of CACNA1A are a frequent cause of autosomal dominant nonprogressive congenital ataxia. Eur J Paediatr Neurol. 2017;21:450–6. doi: 10.1016/j.ejpn.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 22.Zhuchenko O, Bailey J, Bonnen P, Ashizawa T, Stockton DW, Amos C, et al. Autosomal dominant cerebellar ataxia (SCA6) associated with small polyglutamine expansions in the α(1A)-voltage-dependent calcium channel. Nat Genet. 1997;15:62–9. [DOI] [PubMed]
- 23.Blumkin L, Michelson M, Leshinsky-Silver E, Kivity S, Lev D, Lerman-Sagie T. Congenital ataxia, mental retardation, and dyskinesia associated with a novel CACNA1A mutation. J Child Neurol. 2010;25:892–7. doi: 10.1177/0883073809351316. [DOI] [PubMed] [Google Scholar]
- 24.García Segarra N, Gautschi I, Mittaz-Crettol L, Kallay Zetchi C, Al-Qusairi L, Van Bemmelen MX, et al. Congenital ataxia and hemiplegic migraine with cerebral edema associated with a novel gain of function mutation in the calcium channel CACNA1A. J Neurol Sci. 2014;342:69–78. doi: 10.1016/j.jns.2014.04.027. [DOI] [PubMed] [Google Scholar]
- 25.Mammadova D, Kraus C, Leis T, Trollmann R. Severe Epileptic Encephalopathy in Siblings due to a Novel Heterozygous CACNA1A Gene Mutation. Neuropediatrics 2019;50(S 02):S1–S55
- 26.Hayashida T, Saito Y, Ishii A, Yamada H, Itakura A, Minato T, et al. CACNA1A-related early-onset encephalopathy with myoclonic epilepsy: a case report. Brain Dev. 2018;40:130–3. doi: 10.1016/j.braindev.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 27.Myers CTT, McMahon JMM, Schneider ALL, Petrovski S, Allen ASS, Carvill GLL, et al. De novo mutations in SLC1A2 and CACNA1A are important causes of epileptic encephalopathies. Am J Hum Genet. 2016;99:287–98. doi: 10.1016/j.ajhg.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Izquierdo-Serra M, Fernández-Fernández JM, Serrano M. Rare CACNA1A mutations leading to congenital ataxia. Pflugers Arch. 2020;472:791–809. [DOI] [PubMed]
- 29.Mariotti C, Gellera C, Grisoli M, Mineri R, Castucci A, Di, Donato S. Pathogenic effect of an intermediate-size SCA-6 allele (CAG)19 in a homozygous patient. Neurology. 2001;57:1502–4. doi: 10.1212/WNL.57.8.1502. [DOI] [PubMed] [Google Scholar]
- 30.Lv Y, Wang Z, Liu C, Cui L. Identification of a novel CACNA1A mutation in a Chinese family with autosomal recessive progressive myoclonic epilepsy. Neuropsychiatr Dis Treat. 2017;13:2631. doi: 10.2147/NDT.S145774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reinson K, Õiglane-Shlik E, Talvik I, Vaher U, Õunapuu A, Ennok M, et al. Biallelic CACNA1A mutations cause early onset epileptic encephalopathy with progressive cerebral, cerebellar, and optic nerve atrophy. Am J Med Genet Part A. 2016;170:2173–6. doi: 10.1002/ajmg.a.37678. [DOI] [PubMed] [Google Scholar]
- 32.Fletcher CF, Tottene A, Lennon VA, Wilson SM, Dubel SJ, Paylor R, et al. Dystonia and cerebellar atrophy in Cacna1a null mice lacking P/Q calcium channel activity. FASEB J. 2001;15:1288–90. doi: 10.1096/fj.00-0562fje. [DOI] [PubMed] [Google Scholar]
- 33.Ngo DN, So MT, Gui H, Tran AQ, Bui DH, Cherny S, et al. Screening of the RET gene of Vietnamese Hirschsprung patients identifies 2 novel missense mutations. J Pediatr Surg. 2012;47:1859–64. doi: 10.1016/j.jpedsurg.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 34.Bigalke JM, Aibara S, Roth R, Dahl G, Gordon E, Dorbéus S, et al. Cryo-EM structure of the activated RET signaling complex reveals the importance of its cysteine-rich domain. Sci Adv. 2019;5(7):eaau4202. [DOI] [PMC free article] [PubMed]
- 35.Li J, Shang G, Chen Y-J, Brautigam CA, Liou J, Zhang X, et al. Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands. Elife. 2019;8:e47650. [DOI] [PMC free article] [PubMed]
- 36.So M-T, Leon TY-Y, Cheng G, Tang CS-M, Miao X-P, Cornes BK, et al. RET mutational spectrum in Hirschsprung disease: evaluation of 601 Chinese patients. PLoS One. 2011;6:e28986. doi: 10.1371/journal.pone.0028986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang Q, Wang Y, Li Q, Zhang Z, Xiao P, Wang H, et al. Sequence characterization of RET in 117 Chinese Hirschsprung disease families identifies a large burden of de novo and parental mosaic mutations. Orphanet J Rare Dis. 2019;14:237. doi: 10.1186/s13023-019-1194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amiel J, Sproat-Emison E, Garcia-Barcelo M, Lantieri F, Burzynski G, Borrego S, et al. Hirschsprung disease, associated syndromes and genetics: a review. J Med Genet. 2008;45:1–14. doi: 10.1136/jmg.2007.053959. [DOI] [PubMed] [Google Scholar]
- 39.De Falco V, Carlomagno F, Li H yu, Santoro M. The molecular basis for RET tyrosine-kinase inhibitors in thyroid cancer. Vol. 31, Best Prac Res Clin Endocrino Metabol. 2017;31:307–18. [DOI] [PMC free article] [PubMed]
- 40.Iwashita T, Murakarni H, Asai N, Takahashi M. Mechanism of Ret dysfunction by Hirschsprung mutations affecting its extracellular domain. Hum Mol Genet. 1996;5:1577–80. doi: 10.1093/hmg/5.10.1577. [DOI] [PubMed] [Google Scholar]
- 41.Iwashita T, Kurokawa K, Qiao S, Murakami H, Asai N, Kawai K, et al. Functional analysis of RET with Hirschsprung mutations affecting its kinase domain. Gastroenterology. 2001;121:24–33. doi: 10.1053/gast.2001.25515. [DOI] [PubMed] [Google Scholar]
- 42.Hsu SC, Sears RL, Lemos RR, Quintáns B, Huang A, Spiteri E, et al. Mutations in SLC20A2 are a major cause of familial idiopathic basal ganglia calcification. Neurogenetics. 2013;14:11–22. doi: 10.1007/s10048-012-0349-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai JY, Chu CH, Lin MG, Chou YH, Hong RY, Yen CY, et al. Structure of the sodium-dependent phosphate transporter reveals insights into human solute carrier SLC20. Sci Adv. 2020;6(32):eabb4024. [DOI] [PMC free article] [PubMed]
- 44.Salaün C, Maréchal V, Heard JM. Transport-deficient Pit2 phosphate transporters still modify cell surface oligomers structure in response to inorganic phosphate. J Mol Biol. 2004;340:39–47. doi: 10.1016/j.jmb.2004.04.050. [DOI] [PubMed] [Google Scholar]
- 45.Bøttger P, Pedersen L Mapping of the minimal inorganic phosphate transporting unit of human PiT2 suggests a structure universal to PiT-related proteins from all kingdoms of life. BMC Biochem. 2011;12(1):21. [DOI] [PMC free article] [PubMed]
- 46.Travaglini L, Nardella M, Bellacchio E, D’Amico A, Capuano A, Frusciante R, et al. Evolutionary and experimental analyses of inorganic phosphate transporter PiT family reveals two related signature sequences harboring highly conserved aspartic acids critical for sodium-dependent phosphate transport function of human PiT2. FEBS J. 2005;272:3060–74. doi: 10.1111/j.1742-4658.2005.04720.x. [DOI] [PubMed] [Google Scholar]
- 47.Westenberger A, Klein C. The genetics of primary familial brain calcifications. Curr Neurol Neurosci Rep. 2014;14:490. doi: 10.1007/s11910-014-0490-4. [DOI] [PubMed] [Google Scholar]
- 48.Yamada M, Tanaka M, Takagi M, Kobayashi S, Taguchi Y, Takashima S, et al. Evaluation of SLC20A2 mutations that cause idiopathic basal ganglia calcification in Japan. Neurology. 2014;82:705–12. doi: 10.1212/WNL.0000000000000143. [DOI] [PubMed] [Google Scholar]
- 49.Wang XJ, Camilleri M. Hirschsprung disease: Insights on genes, penetrance, and prenatal diagnosis. Neurogastroenterol Motil. 2019;31(11):e13732. [DOI] [PubMed]
- 50.Wu W, Lu L, Xu W, Liu J, Sun J, Zheng L, et al. Whole exome sequencing identifies a novel pathogenic RET variant in Hirschsprung disease. Front Genet. 2019;14(9):752. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.