Abstract
Acute viral infection causes illness and death. In addition, an infection often results in increased susceptibility to a secondary infection, but the mechanisms behind this susceptibility are poorly understood. Since its initial identification as a marker for resident memory CD8+ T cells in barrier tissues, the function and regulation of CD103 integrin (encoded by ITGAE gene) have been extensively investigated. Nonetheless, the function and regulation of the resident CD103+CD8+ T cell response to acute viral infection remain unclear. Although TGFβ signaling is essential for CD103 expression, the precise molecular mechanism behind this regulation is elusive. Here, we reveal a TGFβ–SKI–Smad4 pathway that critically and specifically directs resident CD103+CD8+ T cell generation for protective immunity against primary and secondary viral infection. We found that resident CD103+CD8+ T cells are abundant in both lymphoid and nonlymphoid tissues from uninfected mice. CD103 acts as a costimulation signal to produce an optimal antigenic CD8+ T cell response to acute viral infection. There is a reduction in resident CD103+CD8+ T cells following primary infection that results in increased susceptibility of the host to secondary infection. Intriguingly, CD103 expression inversely and specifically correlates with SKI proto-oncogene (SKI) expression but not R-Smad2/3 activation. Ectopic expression of SKI restricts CD103 expression in CD8+ T cells in vitro and in vivo to hamper viral clearance. Mechanistically, SKI is recruited to the Itgae loci to directly suppress CD103 transcription by regulating histone acetylation in a Smad4-dependent manner. Our study therefore reveals that resident CD103+CD8+ T cells dictate protective immunity during primary and secondary infection. Interfering with SKI function may amplify the resident CD103+CD8+ T cell response to promote protective immunity.
Keywords: CD103, TGFβ, SKI, Smad4, LCMV infection, Histone acetylation
Subject terms: Immunology, Cell signalling
Introduction
Pathogenic infection has afflicted mankind throughout history. A common complication during acute viral infection is increased susceptibility to secondary infection morbidity and mortality.1–3 Multiple factors, including epithelial cell damage and innate immune dysfunction, contribute to the susceptibility to secondary infections.4,5 Nonetheless, whether and how adaptive immunity and its effectors, especially CD8+ T cells, are involved is poorly understood.
CD8+ T cells are critical for clearing infected and transformed cells because they mount effective primary and memory responses.6,7 Based on the expression of different surface markers, CD8+ T cells can be categorized into various subsets.8 In particular, CD103 was initially identified as a hallmark of tissue-resident memory (Trm) CD8+ T cells that contributed to Trm CD8+ T cell function.9 Encoded by the Itgae gene, CD103 is the αE subunit of the heterodimeric integrin αEβ7.10 CD103 binds to its receptor E-cadherin on target tissue cells to facilitate the tissue migration and retention of Trm CD8+ T cells in barrier tissues.11 Nonetheless, emerging evidence suggests that CD103 expression is not restricted to memory CD8+ T cells in nonlymphoid tissues12 and that CD103 may regulate diverse immune cell functions in a microenvironment-dependent fashion.5,13–17 In addition, CD103 can promote the cytotoxic killing activity of CD8+ T cells and increase TCR antigen sensitivity.18–22 This evidence suggests that CD103 plays a broad role in CD8+ T cell immunity beyond regulating the resident memory response. However, the regulation and function of resident CD103+CD8+ T cells during acute viral infection remain ill defined.
Therefore, the expression of CD103 is controlled is a question of significance that is under intense investigation.23,24 Transforming growth factor-beta (TGFβ) signaling is vital for CD103 expression.25–27 The binding of TGFβ to its receptors, including TGFβRI and TGFβRII, leads to the phosphorylation and activation of receptor-associated Smad2/3 (R-Smad2/3).28 TGFβ induces the degradation of the SKI pro-oncogene and, as such, it negatively regulates TGFβ signaling.29 Both R-Smad2/3 and SKI can bind to Smad4 to control target gene expression.28,30 Although TGFβR and R-Smad2/3 have been shown to be required for CD103 expression,25,27 whether and how SKI and Smad4 control CD103 expression remains unaddressed to date. More importantly, because interfering with TGFβR and R-Smad2/3 often leads to unwanted global effects beyond CD103 function because of their broad and pleiotropic functions, it is of interest to identify factors and pathways that specifically control CD103 expression and function.
Lymphocytic choriomeningitis virus (LCMV) is a prevalent human pathogen that infects a large number of people.31 Infection of mice with LCMV can be used to study the underlying principles of viral infection and immune response.32 Using mouse LCMV infection models, we discovered that the TGFβ–SKI–Smad4 pathway is critical in directing CD103+CD8+ T cell generation to fight against primary and secondary infection. We found that the resident CD103+CD8+ T cell population was abundant in nonpathogen-experienced mice, but it changed dynamically after viral infection. Mice with a reduced CD103+CD8+ T cell population were more susceptible to secondary infection. In addition, CD103 expression on CD8+ T cells was found to act as a costimulatory signal for the primary T cell response against acute viral infection. Interestingly, CD103 expression specifically and inversely correlated with SKI expression but not R-Smad activation. Ectopic expression of SKI restricted CD103+CD8+ T cells in vitro and in vivo to hamper viral clearance. In addition, we found that Smad4 was required for the suppression of CD103 expression because CD103 was constitutively expressed in the absence of Smad4 even when TGFβ signaling was abrogated. Furthermore, SKI co-opted Smad4 to restrain CD103 expression by directly binding to the Itgae loci and suppressing CD103 transcription through the inhibition of histone acetylation.
Results
Resident CD103+CD8+ T cells contribute to the control of primary and secondary LCMV infection
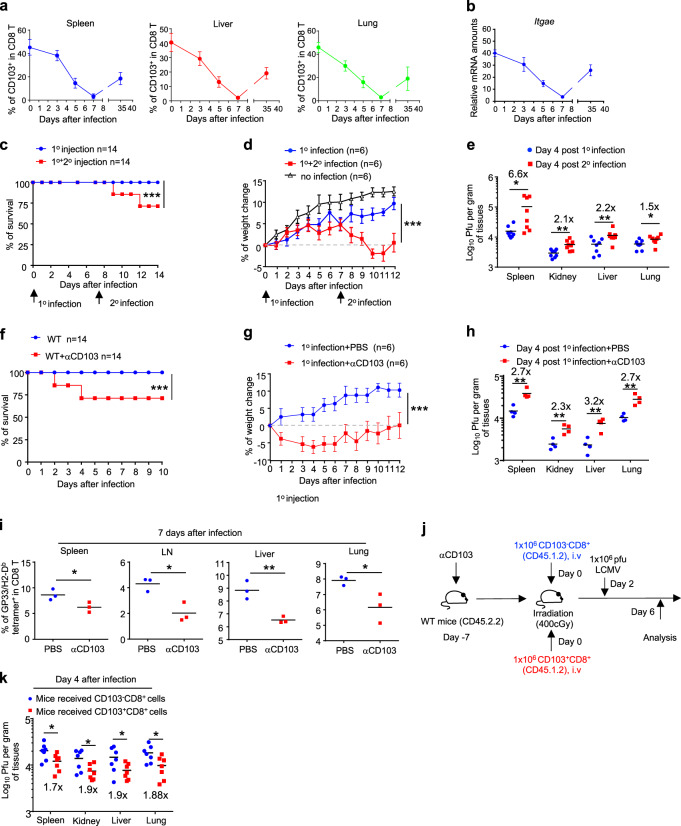
To understand the contribution of resident CD103+CD8+ T cells to immune defense, we examined the temporal change in CD103 expression in CD8+ T cells in different tissues before and after viral infection. Appreciable fractions of CD103+CD8+ T cells were identified in both lymphoid and nonlymphoid tissues in pathogen-naive mice (Figs. 1a and S1a–c), which was in agreement with previous reports and suggested a broad role of CD103 in immunity.33–35 Upon acute LCMV Armstrong strain infection,36 resident CD103+CD8+ T cell population declined dramatically and reached their lowest levels ~7 days after infection; this was followed by a gradual recovery of cell numbers, as indicated at both the protein and mRNA levels at day 35 post infection when the memory response was initiated (Figs. 1a, b and S1a–c). Such a dynamic change in the CD103+CD8+ T cell population suggests that these cells play a role in the acute antiviral response.
Fig. 1.
The distribution, dynamics, and function of resident CD103+CD8+ T cells during LCMV-Armstrong infection. a Flow cytometry analysis of CD103+CD8+ T cells in the spleen (blue), liver (red), and lung (green) from wild-type (WT) mice at different time points after infection with LCMV-Armstrong. (the mean ± s.d.; n = 3–4 mice of three experiments). b Itgae mRNA expression in splenic CD8+ T cells isolated from LCMV-Armstrong-infected WT mice at the indicated time points, assayed by qRT-PCR. (the mean ± s.d; n = 3 experiments). c–e WT mice were infected with LCMV-Armstrong once (1°) or twice (2°) as indicated. The survival (c), body weight change (d), and viral titers in different tissues (e) were determined at the indicated time points. Organ samples were collected either at day 4 post 1st injection or 4 days after the 2nd infection (11 days post 1st infection). (n = 14 mice of two experiments for c; n = 6 mice of three experiments for (d); the mean ± s.e.m. for (d); n = 8 from three experiments for e; ***p < 0.001 per two-way multiple-range ANOVA test for (c) and (d); *p < 0.05, and **p < 0.01 two-sided t-test for (e), centers indicate the mean values). f–h Mice were injected with 500 µg of anti-CD103 (αCD103) or with control PBS 3 days before they were infected once with LCMV-Armstrong. The survival rate (f), body weight change (g), and viral titers in different tissues (h) were monitored at the indicated time points. (n = 14 mice of two experiments for (h); n = 6 mice of three experiments for (g), n = 4 mice of two experiments for (h). The mean ± s.e.m. for (g); ***p < 0.001 per two-way multiple-range ANOVA test for (f) and (g); **p < 0.01 per two-sided t-test for (h); centers indicate the mean values). i The percentages of LCMV antigen-specific CD8+ T cells identified by LCMV-GP33/H2-Db tetramer staining in different tissues from LCMV-infected mice that were treated with αCD103 or PBS for 3 days prior to LCMV infection. (n = 3 mice of two experiments; *p < 0.05, and **p < 0.01 per two-sided t-test; centers indicate the mean values). j The experimental design of intravenous (i.v.) transfer of purified CD103-CD8+ T cells and CD103+CD8+ T cells (CD45.1+CD45.2+) into αCD103 pretreated and irradiated WT (CD45.2+) mice, which was followed by LCMV-Armstrong infection for (k) and Fig. S1g, h. k Viral titer in different tissues from recipient mice 4 days post LCMV-Armstrong infection as depicted in (j). (n = 7 mice of two experiments; *p < 0.05, per two-sided t-test; centers indicate the mean values)
It is noteworthy that re-exposure to LCMV infection 7 days after the first infection, when CD103+CD8+ T cells were at low levels, led to animal death (Fig. 1c), decline in body weight (Fig. 1d), and reduced pathogen clearance (Fig. 1e), suggesting a protective role of CD103+CD8+ T cells during secondary infection. Importantly, antibody-mediated blocking of CD103 (Fig. S1d) led to similar animal death (Fig. 1f), decline in body weight (Fig. 1g), and reduced viral clearance (Fig. 1h) upon LCMV infection. In addition, the virus-specific, GP33/H2-Db tetramer-positive37 CD8+ T cell population was substantially reduced in mice pretreated with an anti-CD103 antibody (Figs. 1i and S1e), although antibody treatment itself did not perturb the CD8+ T cell population (Fig. S1f). Thus, CD103 acts as a costimulation signal to ready CD8+ T cells for an optimal response to acute primary and secondary infection. Furthermore, we found that anti-CD103 antibody-treated mice that were also transplanted with CD103+CD8+ T cells (Fig. S1g, h) were more protected than those that were transplanted with CD103-CD8+ T cells (Fig. 1j, k). These results therefore suggest that reduced resident CD103+CD8+ T cell numbers result in increased susceptibility to secondary infection and that CD103 expression is important for CD8+ T cells to respond to and clear viral infection.
TGFβ signaling is critical for the induction and maintenance of CD103 expression
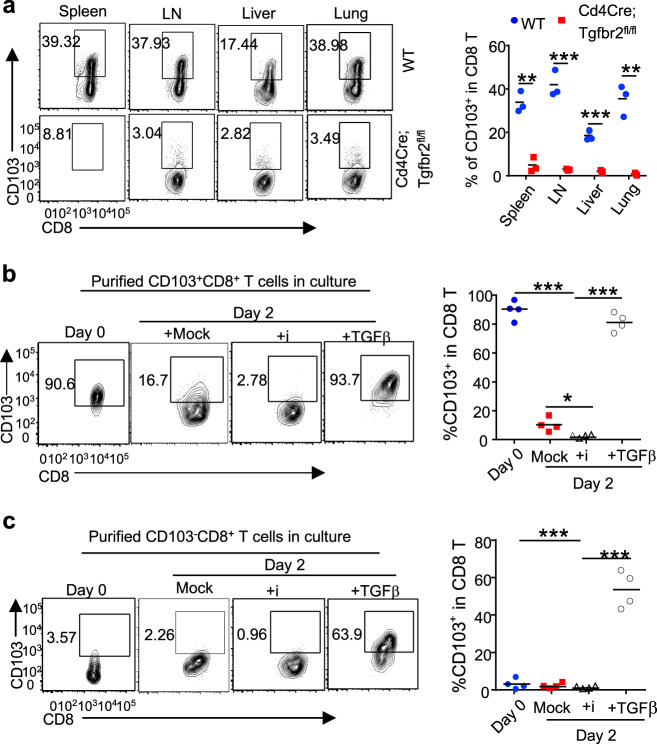
We next investigated how CD103 expression is regulated in CD8+ T cells. TGFβ signaling is required for the development of resident memory CD103+CD8+ cells in barrier tissues.25,38 Similarly, we found that the deletion of TGFβRII39 abrogated the CD103+CD8+ T cell population in both lymphoid and nonlymphoid tissues of Cd4Cre;Tgfbr2fl/fl mice, suggesting an essential role of TGFβR signaling in regulating CD103 expression during homeostasis (Figs. 2a and S2a). In addition, by culturing purified CD103+CD8+ T cells in the presence or absence of TGFβ, we found that TGFβ promoted the maintenance of CD103 expression on CD8+ T cells in vitro (Figs. 2b and S2b). Moreover, low doses of TGFβ induced de novo CD103 expression in activated CD103-CD8+ T cells in vitro (Figs. 2c and S2c). These findings underscore a central role for TGFβ signaling in inducing and maintaining CD103 expression in vivo and in vitro.
Fig. 2.
TGFβ is important for the induction and maintenance of CD103 expression in CD8+ T cells. a Flow cytometry of CD103+CD8+ T cells in different tissues from mice of the indicated genotypes. (n = 3 mice of three experiments; representative results are shown; **p < 0.01, and ***p < 0.001, per two-sided t-test; centers indicate the mean values). Flow cytometry of CD103 expression in purified CD103+CD8+ (b) or CD103-CD8+ (c) T cells after being activated in the absence (mock) or presence of TGFβR inhibitor (SB525334, i) or 0.25 ng/ml TGFβ for the indicated time. (n = 4 samples of three experiments; ***p < 0.001 per two-sided t-test; centers indicate the mean values)
SKI expression inversely correlates with CD103 expression in CD8+ T cells
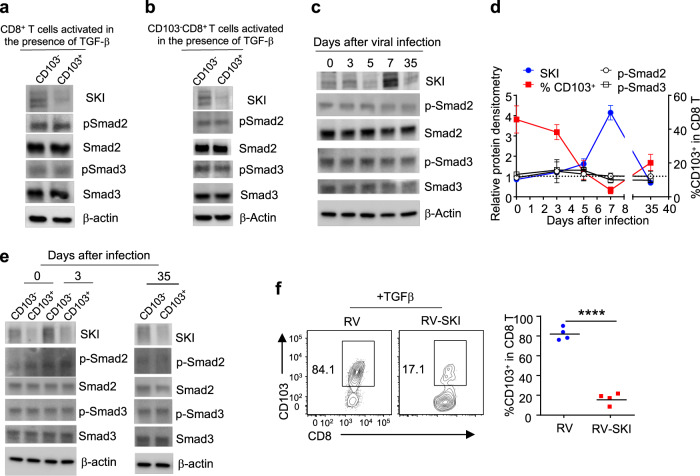
Intriguingly, we consistently observed that only a portion of CD8+ T cells expressed CD103 following TGFβ stimulation (Fig. 2c). Such observation could be attributed to a difference in the downstream signaling of TGFβR in CD103+ vs. CD103− CD8+ T cells. To address this, the phosphorylation levels of R-Smad2 and R-Smad3 were first determined in CD103+ vs. CD103− CD8+ T cells. Of note, TGFβ induced comparable phosphorylation of R-Smad2 and R-Smad3 in CD103+ and CD103− CD8+ T cells in vitro (Fig. 3a, b). Of interest, TGFβ-induced much more SKI degradation (Fig. S3a) in CD103+ than in CD103− CD8+ T cells in vitro (Fig. 3a, b). Similarly, SKI protein expression inversely correlated with CD103 expression in CD8+ T cells from mice during the course of LCMV infection, although the phosphorylation of R-Smad2 and R-Smad3 did not fluctuate (Fig. 3c, d). Mirroring what was observed in vitro, CD103+CD8+ T cells expressed much lower levels of SKI than their CD103−CD8+ counterparts in uninfected and LCMV-infected mice, while the phosphorylation of R-Smad2 and R-Smad3 appeared comparable (Fig. 3e). The consistent inverse correlation between SKI and CD103 expression strongly suggested a specific role for SKI in restricting CD103 expression. Indeed, retrovirus-mediated ectopic SKI expression potently suppressed the promotion of CD103 expression in CD8+ T cells by TGFβ at both the protein and mRNA levels (Figs. 3f and S3b). These findings suggest that TGFβ-induced SKI downregulation, but not R-Smad2/3 activation, specifically directs CD103 expression in CD8+ T cells.
Fig. 3.
SKI expression inversely correlates with CD103 expression in CD8+ T cells both in vitro and in vivo. Immunoblotting of SKI, phospho-Smad2 (p-Smad2), Smad2, phospho-Smad3 (p-Smad3), and Smad3 in purified CD103+CD8+ and CD103-CD8+ T cells after total CD8+ (a) and CD103-CD8+ (b) T cells were activated for 2 days in the presence of 0.25 ng/ml TGFβ. The results are representative of three experiments. c Immunoblotting of SKI, phospho-Smad2 (p-Smad2), Smad2, phospho-Smad3 (p-Smad3), and Smad3 in splenic CD8+ T cells from LCMV-infected mice at the indicated time points. The results are representative of three experiments. d The comparison between the protein levels of SKI, p-Smad2 and p-Smad3 and CD103 expression in CD8+ T cells after LCMV-Armstrong infection at the indicated time points. (the mean ± s.d. of three experiments). e Immunoblotting of SKI, phospho-Smad2 (p-Smad2), Smad2, phospho-Smad3 (p-Smad3), and Smad3 in purified splenic CD103+CD8+ and CD103−CD8+ T cells at the indicated time points after LCMV-Armstrong infection. The results are representative of three experiments. f Purified CD8+ T cells were activated in the presence of TGFβ and then transduced with MSCV-IRES-Thy1.1 (RV) or MSCV-SKI-IRES-Thy1.1 (RV-SKI). CD103 expression in transduced (Thy1.1+) cells was detected by flow cytometry 6 days after activation. (n = 4 samples from three experiments; representative results are shown; ****p < 0.0001 per two-sided t-test; centers indicate the mean values)
SKI suppresses resident CD103+CD8+ T cells to hamper antiviral responses
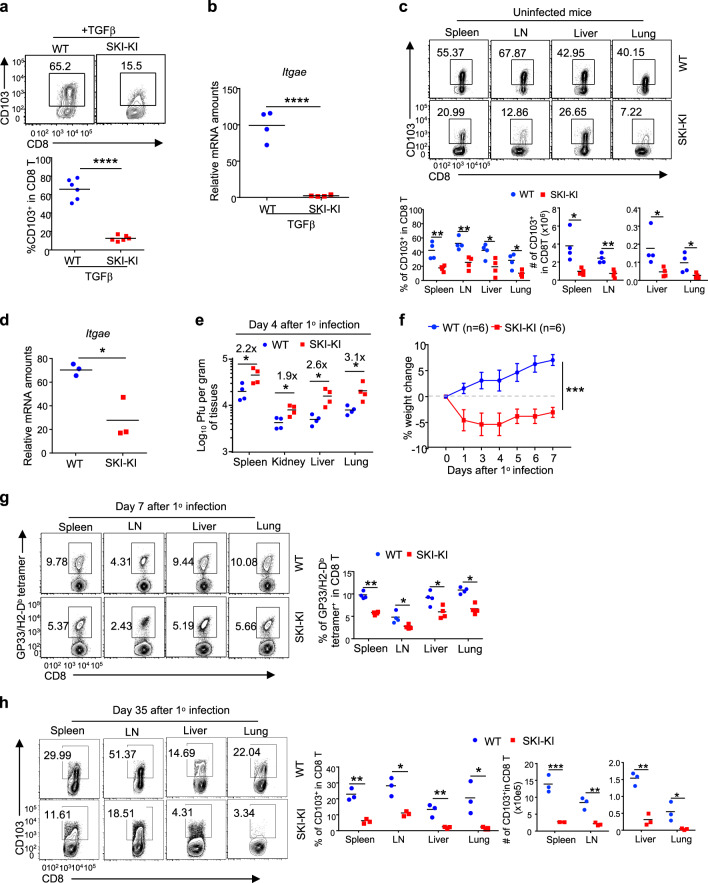
To further investigate the role of SKI in controlling resident CD103+CD8+ T cell-mediated antiviral function in vivo, we generated a mouse strain in which a Cre recombinase-inducible SKI expression cassette was knocked into the Rosa26 locus (Rosa26-iSki strain) (Fig. S4a). By crossing Rosa26-iSki with Cd4Cre40 mice, we ectopically expressed SKI in T cells in vivo (Fig. S4b). Although SKI has been suggested to inhibit TGFβ signaling,41 Cd4Cre;Rosa26-iSki (SKI-KI) mice were grossly normal, unlike mice whose T cells are deficient in TGFβR or R-Smad2/3.27,42–45 The homeostasis of CD4+ and CD8+ T cells in the periphery of these mice was largely unperturbed, although the CD8+ T cell population was slightly reduced in the spleen and lymph nodes (Fig. S4c, d). In addition, although SKI negatively regulates TGFβ signaling,30 the development of regulatory T cells remained undisturbed in SKI-KI mice (Fig. S4e). The normal phenotype of SKI-KI mice under steady state therefore allowed us to investigate how SKI specifically controls CD103 expression and function.
Consistent with the findings that retrovirus-mediated SKI expression suppressed CD103 expression in CD8+ T cells in vitro (Figs. 3f and S3b), CD8+ T cells from SKI-KI mice were defective in gaining CD103 expression upon TGFβ stimulation in vitro (Fig. 4a, b). Importantly, resident CD103+CD8+ T cell populations were greatly reduced in both lymphoid and nonlymphoid tissues from SKI-KI mice (Fig. 4c, d). By infecting SKI-KI mice with the LCMV-Armstrong virus, we assessed how SKI expression could impact host resistance to viral infection. We found that, unlike wild-type mice, SKI-KI mice cleared viruses poorly (Fig. 4e) and lost weight upon acute LCMV infection (Fig. 4f), which was associated with impaired expansion of LCMV-specific GP33/H2-Db tetramer-positive CD8+ T cells (Fig. 4g). In addition, the ability of SKI-KI mice to generate memory CD103+CD8+ T cells following viral clearance was also impaired, and this effect was most severe in the nonlymphoid tissues (Fig. 4h). These findings suggest that TGFβ-induced SKI downregulation is critical for enabling the generation of resident CD103+CD8+ T cells to fight against infection.
Fig. 4.
SKI suppresses CD103+CD8+ T cell generation and dampens antiviral responses. a Flow cytometry of CD103 expression on CD8+ T cells of the indicated genotypes after being activated in the presence of TGFβ for 2 days. (n = 5 samples from three experiments; representative results are shown; ***p < 0.001; per two-sided t-test; centers indicate the mean values). b qRT-PCR analysis of mRNA levels of Itgae gene-expressed CD8+ T cells of the indicated genotypes after being activated in the presence of TGFβ for 2 days. (n = 3 mice of three experiments; **p < 0.01 per two-sided t-test; centers indicate the mean values). c Flow cytometry of CD103 expression on CD8+ T cells from mice with the indicated genotypes to determine the percentages and numbers of CD103+CD8+ T cells in different tissues. (n = 4 mice of four experiments; representative results are shown; *p < 0.05, **p < 0.01 per two-sided t-test; centers indicate the mean values). d qRT-PCR analysis of mRNA levels of the Itgae gene expressed by splenic CD8+ T cells isolated from mice of the indicated genotypes. (n = 3 mice of three experiments; *p < 0.05 per two-sided t-test; centers indicate the mean values). e, f Mice with the indicated genotypes were infected once (1°) with LCMV-Armstrong. The viral titer in different tissues (e) and body weight changes (f) were assessed at the indicated time points. (n = 4 mice for (e) and n = 6 mice for (f) of two experiments; the mean ± s.e.m. for (f); *p < 0.05 per two-sided t-test for (e); ***p < 0.001 per two-way multiple-range ANOVA test for (f); centers indicate the mean values). g The percentages of LCMV-specific CD8+ T cells detected by GP33/H2-Db tetramer staining and flow cytometry in mice of the indicated genotypes 7 days after LCMV infection. (n = 4 mice of three experiments; *p < 0.05, **p < 0.01 per two-sided t-test; centers indicate the mean values). h The percentages and numbers of CD103+CD8+ T cells in different tissues from mice of the indicated genotypes 35 days post LCMV-Armstrong infection, assessed by flow cytometry. (n = 3 mice of three experiments; *p < 0.05, **p < 0.01, and ***p < 0.001 per two-sided t-test; centers indicate the mean values)
SKI interacts with Smad4, which is required to suppress CD103 expression in CD8+ T cells
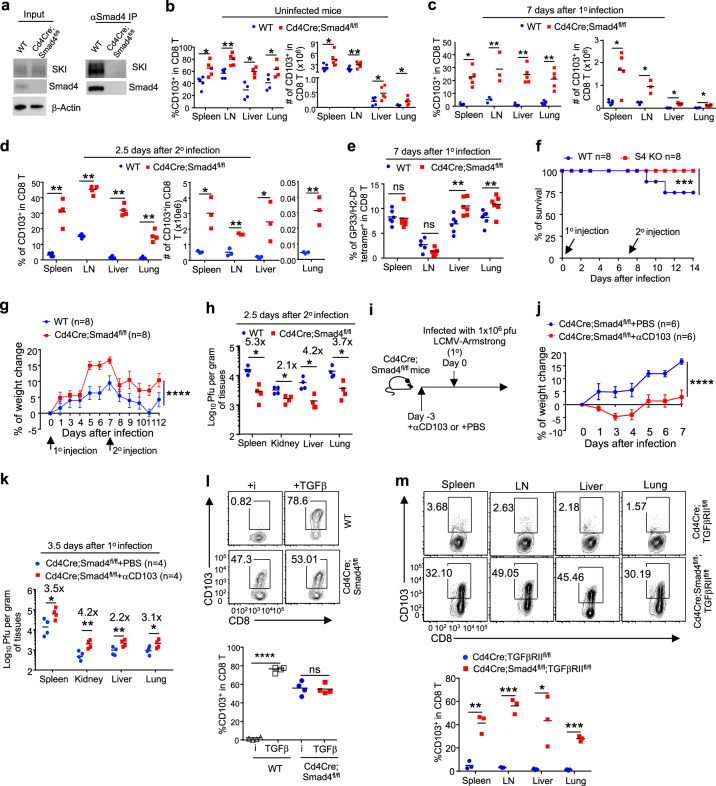
We found that CD103 mRNA (Itgae) expression virtually mirrored its protein expression under various in vivo and in vitro conditions (Figs. 1a, b, 2a–c, and S2a–c), and this expression was suppressed by SKI (Figs. 3f, 4a–d, and S3b), indicating that SKI regulates CD103 expression at the transcriptional level. Although SKI is not a transcription factor, it controls the expression of diverse genes by binding to transcription factors.30 SKI associates with the transcription factor Smad4 to control gene expression.46 To confirm this observation, we found that SKI bound to Smad4 in CD8+ T cells by immunoprecipitation (IP) (Fig. 5a), suggesting a role for Smad4 in controlling CD103 expression. To address whether and how Smad4 is required for the generation of resident CD103+CD8+ T cells, we investigated CD103 expression in CD8+ T cells of Cd4Cre;Smad4fl/fl mice.47 We found that Smad4 deficiency led to a consistent increase of the resident CD103+CD8+ T cell population in the lymphoid and nonlymphoid tissues of Cd4Cre;Smad4fl/fl mice when they remained uninfected or during the course of LCMV-Armstrong infection and reinfection (Figs. 5b–d and S5a–e). In addition, Smad4 deletion led to increased virus-specific GP33/H2-Db tetramer-positive CD8+ T cells following acute LCMV infection (Figs. 5e and S5f). Furthermore, compared with wild-type mice, Cd4Cre;Smad4fl/fl mice were more resistant to LCMV re-exposure-associated animal death (Fig. 5f), body weight decline (Fig. 5g), and unabated virus titer (Fig. 5h). Resident CD103+CD8+ T cells are important for the enhanced antiviral response observed in Cd4Cre;Smad4fl/fl mice because antibody-mediated CD103 inhibition (Fig. 5i) neutralized the enhanced ability of Cd4Cre;Smad4fl/fl mice to deal with LCMV infection (Fig. 5j, k).
Fig. 5.
Smad4 interacts with SKI and is required to suppress CD103 expression in CD8+ T cells. a The interaction between Smad4 and SKI in CD8+ T cells was detected by co-immunoprecipitation. The results are representative of three experiments. b The percentages and numbers of CD103+CD8+ T cells in different tissues of naïve mice with the indicated genotypes, as assessed by flow cytometry. (n = 4–5 mice of three experiments; *p < 0.05, **p < 0.01 per two-sided t-test; centers indicate the mean values). c The percentages and numbers of CD103+CD8+ T cells in different tissues of mice with the indicated genotypes after infection once (1°) with LCMV-Armstrong for 7 days, assessed by flow cytometry. (n = 3–5 mice of three experiments; *p < 0.05, **p < 0.01 per two-sided t-test; centers indicate the mean values). d The percentages and numbers of CD103+CD8+ T cells in different tissues of mice with the indicated genotypes after infection twice (2° as described in Fig. 1c) for 2.5 days, assessed by flow cytometry. (n = 3–4 mice of three experiments; *p < 0.05, and **p < 0.01 per two-sided t-test; centers indicate the mean values). e The percentages of LCMV-specific, GP33/H2-Db tetramer-positive CD8+ T cells in different tissues of mice of the indicated genotypes after infection once (1°) with LCMV-Armstrong for 7 days, assessed by flow cytometry. (n = 6 mice of three experiments; ns, not significant, **p < 0.01 per two-sided t-test; centers indicate the mean values) (f–h). Mice of the indicated genotypes were infected with LCMV-Armstrong twice (2°). The survival rate (f), change in body weight (g), and viral titers in different tissues (h) were monitored at the indicated time points. (n = 8 mice for (f, g) and n = 4 mice for (h) of two experiments; the mean ± s.e.m. for (g); ****p < 0.0001 per two-way multiple-range ANOVA test for (f) and (g); *p < 0.05 per two-sided t-test for (h); centers indicate the mean values). i The experimental design of anti-CD103 treatment followed by LCMV-Armstrong infection as shown in (j–k). The body weight change (j) and viral titers in different tissues (k) of LCMV-infected mice of the indicated genotypes as described in (i). (n = 6 mice of two experiments for (j); n = 4 mice of two experiments for (k); ****p < 0.0001 per two-way multiple-range ANOVA test for (j); *p < 0.05, **p < 0.01 per two-sided t-test for (k); centers indicate the mean values). l Flow cytometry of CD103 expression by CD8+ T cells of the indicated genotypes after being activated in the presence of TGFβ receptor inhibitor (i) or TGFβ. (Representative results are shown; n = 4 samples of three experiments; ns not significant, ****p < 0.0001 per two-sided t-test; centers indicate the mean values) m Flow cytometry of CD103 expression by CD8+ T cells in different tissues from mice of the indicated genotypes. (Representative results are shown; n = 3 mice of three experiments, *p < 0.05, **p < 0.01, and ***p < 0.001; per two-sided t-test; centers indicate the mean values)
We further assessed how Smad4 is involved in TGFβ-promoted CD103 expression. Interestingly, we found that Smad4 deletion enabled CD103 expression in CD8+ T cells even when TGFβ signaling was abrogated in vitro (Fig. 5l). Consistently, simultaneous deletion of Smad4 restored the CD103+CD8+ T cell population in both lymphoid and nonlymphoid organs from Cd4Cre;Tgfbr2fl/fl mice (Figs. 5m and S5g). Therefore, Smad4 is required to limit CD103 expression in CD8+ T cells through a mechanism that is downstream of TGFβR.
SKI co-opts Smad4 to directly suppress CD103 expression by inhibiting histone acetylation
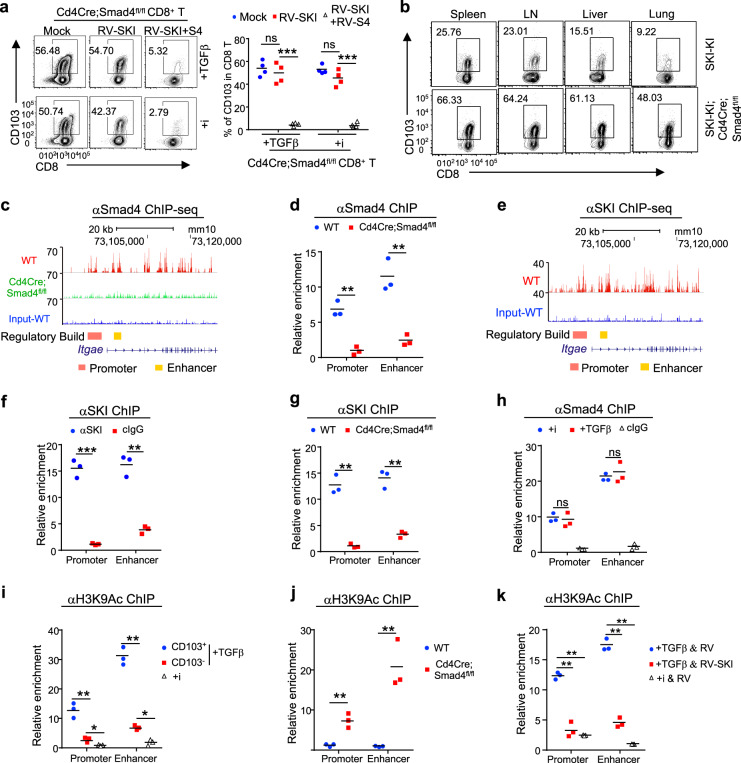
Intrigued by the findings described above, we further investigated the functional relationship of SKI and Smad4 in regulating CD103 expression. We found that ectopic SKI expression suppressed TGFβ-induced CD103 expression only when Smad4 was coexpressed in activated CD8+ T cells in vitro (Fig. 6a). In addition, Smad4 deletion restored the CD103+CD8+ T cell population in SKI-KI mice (Fig. 6b). Therefore, SKI suppresses CD103 expression through a Smad4-dependent mechanism. Further investigation revealed that such a mechanism involved the direct binding of the SKI–Smad4 complex to Itgae loci. Chromatin immunoprecipitation followed by sequencing (ChIP-seq) (Fig. 6c, e) as well as ChIP analysis (Fig. 6d, f) performed on CD8+ T cells revealed that both Smad4 and SKI bound to the Itgae loci containing Smad consensus sites. SKI failed to bind to Itgae loci when Smad4 was absent (Fig. 6g), although Smad4 bound to Itgae loci constitutively regardless of TGFβ signaling (Fig. 6h), suggesting that SKI is recruited to Itgae loci in a Smad4-dependent manner.
Fig. 6.
SKI co-opts Smad4 to directly suppress Itgae transcription by inhibiting histone acetylation. a Smad4-deficient CD8+ T cells were purified and activated in the presence of TGFβ or TGFβR inhibitor (i) and transduced with MSCV-SKI-IRES-Thy1.1 (RV-SKI) and/or MSCV-Smad4-IRES-EGFP (RV-S4), as indicated. The expression of CD103 by the transduced cells was detected by flow cytometry. (Representative results are shown. n = 4 samples from three experiments; ns not significant, ***p < 0.001 per two-sided t-test; centers indicate the mean values). b Flow cytometry of CD103 expression by CD8+ T cells in different tissues from mice of the indicated genotypes. (Results are representative of three experiments). c Chromatin immunoprecipitation followed by sequencing (ChIP-seq) analysis of Smad4 binding at the Itgae loci in CD8+ T cells cultured with TGFβR inhibitor for 2 days. d ChIP analysis of Smad4 binding at the Itgae promoter and enhancer regions in CD8+ T cells cultured with TGFβR inhibitor for 2 days. (n = 3 samples of three experiments, **p < 0.01 per two-sided t-test; centers indicate the mean values). e ChIP-seq analysis of SKI binding at the Itgae locus in CD8+ T cells cultured with TGFβR inhibitor for 2 days. f ChIP analysis of SKI binding at the Itgae promoter and enhancer regions in CD8+ T cells cultured with TGFβR inhibitor for 2 days. cIgG (control IgG). (n = 3 samples of three experiments, **p < 0.01, and ***p < 0.001, per two-sided t-test; centers indicate the mean values) g ChIP analysis of SKI binding at the Itgae promoter and enhancer regions in activated CD8+ T cells from mice with the indicated genotypes that were grown in the presence of TGFβR inhibitor for 2 days. (n = 3 samples of three experiments; **p < 0.01 per two-sided t-test; centers indicate the mean values). h ChIP analysis of Smad4 binding at the Itgae promoter and enhancer regions in activated CD8+ T cells cultured in the presence of TGFβR inhibitor (i) or TGFβ for 2 days. cIgG (control IgG). (n = 3 samples of three experiments; ns, not significant per two-sided t-test; centers indicate the mean values). i CD8+ T cells were purified and activated in the presence of TGFβR inhibitor (i) or TGFβ for 2 days. CD103+ and CD103− subsets were sorted from TGFβ-treated CD8+ T cells. H3K9 acetylation (H3K9Ac) at the Itgae promoter and enhancer regions was assessed by ChIP analysis. (n = 3 samples of three experiments, *p < 0.05, and **p < 0.01, per two-sided t-test; centers indicate the mean values). j ChIP analysis of H3K9Ac at the Itgae promoter and enhancer regions in CD8+ T cells of the indicated genotypes 2 days after activation in the presence of a TGFβR inhibitor. (n = 3 samples of three experiments; **p < 0.01 per two-sided t-test; centers indicate the mean values). k ChIP analysis of H3K9Ac at the Itgae promoter and enhancer regions in MSCV-SKI-IRES-Thy1.1 (RV-SKI)- or MSCV-IRES-Thy1.1 (RV)-transduced CD8+ T cells that were grown in the presence of TGFβ or TGFβR inhibitor (i). (n = 3 samples of three experiments, **p < 0.01 per two-sided t-test; centers indicate the mean values)
Since SKI may control target gene expression by modifying histone acetylation,30,46 we wondered how TGFβ–SKI–Smad4 signaling impacts the status of H3K9Ac, an epigenetic marker for transcriptional activation,48 of Itgae loci. We found that despite TGFβ treatment, very low levels of H3K9Ac were detected at Itgae loci in CD103−CD8+ T cells (Fig. 6i), which expressed high levels of SKI (Fig. 3a, b, e). In contrast, high levels of H3K9Ac were detected at Itgae loci in CD103+CD8+ T cells (Fig. 6i), which expressed low levels of SKI (Fig. 3a, b, e). In addition, in the absence of Smad4, SKI failed to be recruited to Itgae loci (Fig. 6g), and H3K9Ac was observed at high levels at Itgae loci to allow unconstrained CD103 expression in CD8+ T cells (Fig. 6j). Moreover, ectopic SKI expression suppressed H3K9Ac levels at the Itgae loci in CD8+ T cells (Fig. 6k). These findings therefore suggest that the SKI–Smad4 complex directly binds to Itgae loci to prevent H3K9Ac deposition and restrain CD103 expression.
In summary, this study reveals that CD103 is expressed by CD8+ T cells in lymphoid and nonlymphoid tissues. Such CD103 expression endows CD8+ T cells with an optimal ability to respond to and clear pathogens. In addition, CD103 expression is regulated specifically via the TGFβ–SKI–Smad4 pathway, shedding light on the long-sought mechanisms underlying TGFβ-controlled CD103 expression in CD8+ T cells. This work not only provides insights into the function and regulation of CD103 in CD8+ T cells but also unveils new molecular targets for specifically modulating resident CD103+CD8+ T cell functions for pathogen clearance.
Discussion
CD103+ Trm CD8+ cells have attracted great attention for their involvement in pathogen clearance, vaccination, and tumor immunity.38,49,50 As a hallmark of Trm CD8+ T cells, CD103 promotes the tissue migration and retention of CD8+ T cells in nonlymphoid tissues.23 Nevertheless, emerging evidence suggests that CD103 expression is not restricted to Trm CD8+ T cells.12 CD103+CD8+ T cells are easily detected in lymphoid and nonlymphoid tissues as well as in peripheral blood,35,51,52 implying that resident CD103+CD8+ T cells may have functions in addition to those in the memory response. In this study, we provided evidence to support the notion that CD103 is important for the CD8+ T cell-mediated effector response and that resident CD103+CD8+ T cells are poised to mount an optimal response towards pathogens. In light of the current finding that resident CD103+CD8+ T cells are important for controlling primary and secondary viral infection, it will be interesting to investigate how broadly this mechanism is involved in the clearance of various viral and bacterial pathogens as well as in effective vaccination.
We found that CD103 expression in CD8+ T cells was dynamically regulated during viral infection. CD103 expression declined aggressively and reached the lowest level at ~7 days post infection. Considering that CD103 is important for the tissue retention of T cells,12 the decline of its expression during the antiviral response is likely to facilitate the mobility of activated effector CD8+ T cells to survey the body and clear infected cells. In agreement with this notion, strong T cell receptor (TCR) activation induces the upregulation of T-bet and Eomes to potently suppress CD103 expression.53–56 Interestingly, we also noticed that CD8+ T cells regained CD103 expression 35 days post infection when the pathogens were largely cleared.57 Regaining CD103 expression in the absence of overt infection and strong T cell stimulation should allow pathogen-exposed migrating CD8+ T cells to locate to the tissues and facilitate the formation of Trm CD8+ T cells.
The expression of CD103 is controlled through multiple mechanisms,12,24 among which TGFβ signaling is critical for promoting its expression.25–27,58 However, the signaling pathways downstream of TGFβR in regulating CD103 expression remain poorly defined. R-Smads were activated in CD103+CD8+ T cells (Fig. 3) and are required for TGFβ-promoted CD103 expression.27 Mechanistically, Smad2/3 and NFAT are critical transcriptional factors that directly bind the promoter and enhancer regions of Itgae loci and promote CD103 transcription.58 Our data now suggest that SKI co-opts Smad4 to restrain CD103 expression by directly binding to Itgae loci. However, we found that R-Smads were comparably activated in CD103+CD8+ and CD103−CD8+ T cells, suggesting that an additional TGF-β-dependent mechanism beyond that of R-Smads directs CD103 expression in CD8+ T cells. Indeed, we found that SKI expression specifically and inversely correlated with CD103 expression and that SKI suppressed CD103 expression. These results uncover a unique role for SKI-dependent signaling in restricting CD103 expression. We thus propose a “dual role” model for TGFβ to promote CD103 expression1: TGFβ activates R-Smads to potentiate CD103 transcription and2 TGFβ triggers SKI downregulation to relieve SKI/Smad4-mediated suppression to permit CD103 transcription. Therefore, interfering with SKI function could be a viable approach to specifically target CD8+ T cell function for pathogen clearance.
The current study highlights an important role for SKI in controlling CD103 expression and CD8+ T cell function for viral clearance. How SKI expression and function are regulated is thus of interest. We found that SKI expression was increased in activated CD8+ T cells, facilitating CD103 downregulation in these cells. Because the SKI expression level is inversely correlated with TGFβ stimulation, the increased SKI expression in activated T cells could be due to their reduced sensitivity towards TGFβ stimulation because TGFβRI expression is drastically downregulated in T cells upon activation.59 SKI expression is regulated predominantly at a posttranslational level through modulation of protein stability. Polyubiquitination and proteasome-mediated protein degradation are important for SKI degradation.30 One of the E3 ubiquitin ligases, Arkadia, has been shown to facilitate TGFβ-induced SKI protein degradation in nonlymphoid cells.60 Nonetheless, whether Arkadia, other E3 ubiquitin ligase(s), and proteasomal-independent mechanisms contribute to the regulation of SKI expression in T cells warrants further investigation.
SKI functions by regulating gene transcription through epigenetic modification of gene loci.30 Because SKI is neither a transcription factor nor an enzyme that regulates epigenetic modifications, it is imperative for SKI to co-opt-related factors to carry out its functions.61 Indeed, we found that SKI interacted with the transcription factor Smad4, which is critical for the recruitment of SKI to the Itgae loci to control CD103 expression. In addition to Smad4, SKI can interact with other transcription factors, including PU.1, GATA1, and Gli3, in a cellular context-dependent manner.30 If and how SKI functions with any of these other transcription factors to control Itgae expression remain to be addressed. In addition, we found that the recruitment of SKI to Itgae loci resulted in transcriptional repression associated with decreased histone acetylation, which can be achieved by either recruiting histone deacetylase (HDAC) or excluding histone acetyl transferase.48 Because SKI has been shown to bind to HDAC1 and HDAC3,62,63 it is likely that SKI suppresses Itgae expression by recruiting HDACs. Further investigation is therefore warranted to shed light on the precise mechanisms underlying SKI-controlled Itgae expression.
Materials and methods
Animal
Cd4Cre, Smad4fl/fl, Tgfbr2fl/fl, Cd4Cre;Rosa26-iSki (SKI-KI), and CD45.1 congenic wild-type mice were on a C57BL/6 background. Two- to four-month-old, age- and sex-matched male and female mice were used for experiments. Littermates were used unless stated otherwise. All mice were housed and bred in specific pathogen-free conditions in the animal facility at the University of North Carolina at Chapel Hill. All mouse experiments were approved by the Institutional Animal Care and Use Committee of the University of North Carolina. We have complied with all relevant ethical regulations.
Generation of Cd4Cre;Rosa26-iSki (SKI-KI) mice
The synthesized mSki-flag cDNA was ligated into the mROSA-KI-12 vector to generate the mROSA26-mSki-flag targeting vector. This cassette was inserted between the first and second exons of the safe harbor locus of Rosa26 loci by sgRosa26/Cas9-introduced double-strand break. Microinjection of the targeting vector was performed in zygotes from C57BL/6 female mice. F0 mice were bred with wild-type C57BL/6 mice to obtain F1 germline transmission heterozygous mice after validation by PCR and southern blotting. F1 germline transmitted mice were bred with Cd4Cre mice to generate Cd4Cre;Rosa26-iSki (SKI-KI) mice.
LCMV-Armstrong infection and in vivo antibody treatment
Mice at 8–12 weeks of age were infected intraperitoneally with LCMV-Armstrong (1 × 106 plaque-forming units, pfu). For the repeated infection model, mice were rechallenged intraperitoneally with 1 × 106 pfu LCMV-Armstrong on day 7 after the 1st. infection. InVivoMAb anti-mouse CD103 (M209; BioXcell) was administered intraperitoneally at a dose of 0.5 mg per mouse 3 days before LCMV-Armstrong infection.
LCMV-Armstrong propagation and titering
Viral stocks of LCMV-Armstrong were propagated from infected BHK21 cell monolayers. Virus titers in the spleen, liver, lung and kidney were determined by plaque assay on Vero cell monolayers.
Flow cytometry
T cells from various experiments were stained with the following fluorescence-labeled antibodies: anti-CD4 (RM4-5), anti-CD8 (53–6.7), anti-CD25 (PC61), anti-CD103 (2E7) (Biolegend), and anti-Foxp3 (FJK-16s) from eBioscience. The MHC class I tetramer of H-2Db complexed with the LCMV GP33-41 peptide was obtained from the NIH tetramer core facility at Emory University. The data were acquired by FACSCanto (BD Bioscience) and were analyzed by FlowJo software (Treestar).
T cell isolation and culturing in vitro
CD8+ T cells were isolated from the peripheral lymph nodes and spleens from mice by CD8 magnetic beads (Miltenyi Biotec) per the manufacturer’s protocols. To isolate CD103+ cells, cells were stained with a biotin-conjugated anti-mouse CD103 antibody (2E7, Biolegend) and then were purified by streptavidin magnetic beads (Miltenyi Biotec). The flow-through CD8+ T cells were collected by CD8 magnetic beads (Miltenyi Biotec) to obtain CD103−CD8+ cells. Sorted cells were then subjected to subsequent culture and analysis. To activate purified T cells, they were cultured on plates precoated with 5 μg/ml anti-CD3 (145-2C11, BioXCell) and 2 μg/ml anti-CD28 (37.51, BioXCell) in RPMI medium supplemented with 10% FBS. TGFβ (0.25 ng/ml, Biolegend) or 10 μM TGFβR inhibitor SB525334 (Selleckchem) was added as required.
Lymphocyte isolation from nonlymphoid tissues
Lung tissues were cut into pieces and digested for 45 min with 1 mg/ml collagenase D (Sigma) in PBS at 37 °C with shaking every 15 min. Digested tissues were filtered through a 70 μm nylon cell strainer (Falcon). Lymphocytes were then separated by using a 38% Percoll (Sigma) solution with centrifugation. Liver tissues were excised and homogenized. Lymphocyte suspensions were then separated using a 40/70% Percoll density gradient with centrifugation. Lymphocytes were then extracted and subjected to subsequent analysis.
Quantitative RT-PCR (qRT-PCR) analysis
Total RNA was extracted from lymphocytes using TRIsure reagent (Bioline), and it was reverse-transcribed to generate cDNA with Superscript III reverse transcriptase (Bio-Rad) per the manufacturer’s protocols. Quantitative PCR (qPCR) was performed on a QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific). The housekeeping gene actin was used as an internal control for normalization of all qPCR results. Normalized qPCR results are presented as relative mRNA amounts with arbitrary units to show different levels of gene expression.
Immunoblotting and IP
For immunoblotting, protein extracts were resolved by 4–15% Mini-PROTEAN TGX Precast Protein Gels (Bio-Rad), transferred to a polyvinylidene fluoride membrane (Bio-Rad) and analyzed by immunoblotting with the following antibodies: anti-SKI (G8, Santa Cruz), anti-Smad4 (D3M6U, CST), anti-β-actin (C4, Santa Cruz), anti-phospho-Smad2 (S465/467, CST), anti-phospho-Smad3 (C25A9, CST), anti-Smad2 (D43B4, CST), and anti-Smad3 (C67H9, CST).
For IP, cells were lysed with IP lysis buffer (10 mM HEPES, pH 7.5, 1.5 mM MgCl2, 0.2 mM EDTA, and 150 mM NaCl containing 1% NP40) containing a protease inhibitor mixture (Roche Applied Science), and then they were sonicated with Bioruptor PICO. Cell lysates were incubated with 50 μl of magnetic protein A/G beads (Bio-Rad) that had been conjugated to the indicated antibodies and treated with dimethyl pimelimidate. After overnight incubation, the associated protein was eluted with 2 × Laemmli sample buffer (Bio-Rad) after five washes and incubated at 95 °C for 5 min. Then, the eluted samples were separated by SDS-PAGE and analyzed by immunoblotting.
Retroviral transduction
For retroviral transduction, CD8+ T cells were isolated and cultured under various conditions and then spin-inoculated (at 1500 × g) with the indicated MSCV retroviruses in the presence of 8 μg/ml polybrene (Sigma-Aldrich) and HEPES buffer (Gibco) at 30 °C for 90 min 24 h after activation. Transduced cells were harvested and analyzed by flow cytometry.
ChIP assay
Isolated CD8+ T cells were cross-linked with 1% formaldehyde (Sigma) and lysed in lysis buffer. Lysates were sonicated with Bioruptor PICO to shear genomic DNA. Chromatin was then subjected to immunoprecipitation overnight at 4 °C with anti-Smad4 (EP618Y, Abcam), anti-Ski (G8, Santa Cruz), anti-H3K9Ac (ab4441, Abcam), normal mouse IgG (sc-2025, Santa Cruz), and normal rabbit IgG (sc-2027, Santa Cruz). qPCR was performed to detect the relative abundance of the target genomic DNA. Specific PCR primers to detect the Itgae promoter and enhancer region (predicted by ENCODE) are as follows: promoter: CTCCTAGGGAGCAGGTGTCT and TAGAAAAGCCTGCACGGGAT; enhancer: GGTACATGGAAGCCTGAGCA and AGACAGAGGGCCTAAGAGCA.
ChIP-seq
The sequencing libraries were prepared with an MGI Easy DNAseq kit (MGI), and sequence data were collected using 50 nt single-end reads with a BGISEQ-500 system (MGI) at the BGI Genomics company. Raw reads (24–25 million reads per sample) were first processed to remove adapter sequences using Trim Galore (version 0.4) with default parameters. The resulting reads were aligned to mm10 using Bowtie (version 1.2) with the parameters -m 1, -v 2, and -X 1500. These parameters ensured that fragments that were up to 1500 bp (-X 1500) with mismatches of up to 2 (-v 2) were allowed to align and that only unique aligning reads were collected (-m 1). For all the data files, duplicated alignments were removed using Picard (version 1.115). The ChIP-seq data are available in the Gene Expression Omnibus repository at the National Center for Biotechnology Information under accession number GSE135533.
The findPeaks program from Homer (version 4.8.3) was applied to call all reported ChIP-seq peaks. The basic goal was to identify regions in the genome where more sequencing reads could be found than would be expected by chance. Peak calling for all samples used the paired input sample as noise control. The findPeaks program was run using default parameters of “-F 4 -L 0 -C 2 -fdr 0.001”. “-F” required each putative peak to have fourfold more normalized tags in the target experiment than the control. “-L 0” disabled the local signal. “-C 2” was used to remove peaks near repeat elements that contain odd tag distributions. “-fdr 0.001” indicated that FDR-adjusted p value < 0.05 was used as cutoff. All significant peaks were associated with genes using GREAT (Version 3).
Statistical analysis
A Student’s t test or two-way ANOVA was used for two-group comparisons. p values of < 0.05 were considered significant. In the figures, *, **, *** and **** were used to indicate p < 0.05, p < 0.01, p < 0.001 and p < 0.0001, respectively. All results shown are the mean ± s.d. unless otherwise stated.
Supplementary information
Acknowledgements
We thank E. Robertson and E. Bikoff for providing Smad4fl/fl mice, H. Moses for providing TgfbrIIfl/fl mice, and the NIH tetramer core facility at Emory University for providing the tetramers. This work was supported by NIH funding (R01AI143894; R01AI138337) for J.K.W.; the NIH (AI123193); the National Multiple Sclerosis Society (RG-1802-30483); and the Yang Family Biomedical Scholars Award for Y.Y.W.
Author contributions
B.W. and G.Z. contributed equally to the design and implementation of the cellular, molecular, biochemical, and animal experiments; B.W. contributed to the writing of the manuscript; Z.G., G.W., and J. Z. contributed to the generation and characterization of SKI-KI mice; J.L. and X.X. contributed to the bioinformatic analysis of ChIP data; J.K.W. contributed to the LCMV infection model; and Y.Y.W. conceived the project, designed the experiments, and wrote the manuscript.
Data availability
ChIP-seq data are deposited in the GEO database under ID code GSE135533.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Bing Wu, Ge Zhang
Contributor Information
Junnian Zheng, Email: jnzheng@xzhmu.edu.cn.
Yisong Y. Wan, wany@email.unc.edu
Supplementary information
The online version of this article (10.1038/s41423-020-0495-7) contains supplementary material.
References
- 1.Rynda-Apple A, Robinson KM, Alcorn JF. Influenza and bacterial superinfection: illuminating the immunologic mechanisms of disease. Infect. Immun. 2015;83:3764–3770. doi: 10.1128/IAI.00298-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Sluijs KF, et al. IL-10 is an important mediator of the enhanced susceptibility to pneumococcal pneumonia after influenza infection. J. Immunol. 2004;172:7603–7609. doi: 10.4049/jimmunol.172.12.7603. [DOI] [PubMed] [Google Scholar]
- 3.Brundage JF. Interactions between influenza and bacterial respiratory pathogens: implications for pandemic preparedness. Lancet Infect. Dis. 2006;6:303–312. doi: 10.1016/S1473-3099(06)70466-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paget C, Trottein F. Mechanisms of bacterial superinfection post-influenza: a role for unconventional T cells. Front. Immunol. 2019;10:336. doi: 10.3389/fimmu.2019.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ivanov S, et al. Key role for respiratory CD103(+) dendritic cells, IFN-gamma, and IL-17 in protection against Streptococcus pneumoniae infection in response to alpha-galactosylceramide. J. Infect. Dis. 2012;206:723–734. doi: 10.1093/infdis/jis413. [DOI] [PubMed] [Google Scholar]
- 6.Wong P, Pamer EG. CD8 T cell responses to infectious pathogens. Annu. Rev. Immunol. 2003;21:29–70. doi: 10.1146/annurev.immunol.21.120601.141114. [DOI] [PubMed] [Google Scholar]
- 7.Restifo NP, Dudley ME, Rosenberg SA. Adoptive immunotherapy for cancer: harnessing the T cell response. Nat. Rev. Immunol. 2012;12:269–281. doi: 10.1038/nri3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang JT, Wherry EJ, Goldrath AW. Molecular regulation of effector and memory T cell differentiation. Nat. Immunol. 2014;15:1104–1115. doi: 10.1038/ni.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gebhardt T, et al. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat. Immunol. 2009;10:524–530. doi: 10.1038/ni.1718. [DOI] [PubMed] [Google Scholar]
- 10.Cepek KL, et al. Adhesion between epithelial cells and T lymphocytes mediated by E-cadherin and the alpha E beta 7 integrin. Nature. 1994;372:190–193. doi: 10.1038/372190a0. [DOI] [PubMed] [Google Scholar]
- 11.Iijima N, Iwasaki A. Tissue instruction for migration and retention of TRM cells. Trends Immunol. 2015;36:556–564. doi: 10.1016/j.it.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardenberg JB, Braun A, Schon MP. A Yin and Yang in epithelial immunology: the roles of the alphaE(CD103)beta7 Integrin in T Cells. J. investig. Dermatol. 2018;138:23–31. doi: 10.1016/j.jid.2017.05.026. [DOI] [PubMed] [Google Scholar]
- 13.Schlickum S, et al. Integrin alpha E(CD103)beta 7 influences cellular shape and motility in a ligand-dependent fashion. Blood. 2008;112:619–625. doi: 10.1182/blood-2008-01-134833. [DOI] [PubMed] [Google Scholar]
- 14.Smazynski J, Webb JR. Resident memory-like tumor-infiltrating lymphocytes (TILRM): latest players in the immuno-oncology repertoire. Front. Immunol. 2018;9:1741. doi: 10.3389/fimmu.2018.01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Annacker O, et al. Essential role for CD103 in the T cell-mediated regulation of experimental colitis. J. Exp. Med. 2005;202:1051–1061. doi: 10.1084/jem.20040662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duhen T, et al. Co-expression of CD39 and CD103 identifies tumor-reactive CD8 T cells in human solid tumors. Nat. Commun. 2018;9:2724. doi: 10.1038/s41467-018-05072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gabriely G, et al. Targeting latency-associated peptide promotes antitumor immunity. Sci. Immunol. 2017;2:eaaj1738. doi: 10.1126/sciimmunol.aaj1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shields BD, et al. Loss of E-cadherin inhibits CD103 antitumor activity and reduces checkpoint blockade responsiveness in melanoma. Cancer Res. 2019;79:1113–1123. doi: 10.1158/0008-5472.CAN-18-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boutet M, et al. TGFbeta signaling intersects with CD103 integrin signaling to promote T-lymphocyte accumulation and antitumor activity in the lung tumor microenvironment. Cancer Res. 2016;76:1757–1769. doi: 10.1158/0008-5472.CAN-15-1545. [DOI] [PubMed] [Google Scholar]
- 20.Abd Hamid M, et al. Self-maintaining CD103(+) cancer-specific T cells are highly energetic with rapid cytotoxic and effector responses. Cancer Immunol. Res. 2020;8:203–216. doi: 10.1158/2326-6066.CIR-19-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franciszkiewicz K, et al. CD103 or LFA-1 engagement at the immune synapse between cytotoxic T cells and tumor cells promotes maturation and regulates T-cell effector functions. Cancer Res. 2013;73:617–628. doi: 10.1158/0008-5472.CAN-12-2569. [DOI] [PubMed] [Google Scholar]
- 22.Xue D, Liu P, Chen W, Zhang C, Zhang L. An anti-CD103 antibody-drug conjugate prolongs the survival of pancreatic islet allografts in mice. Cell Death Dis. 2019;10:735. doi: 10.1038/s41419-019-1980-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hadley GA, Higgins JM. Integrin alphaEbeta7: molecular features and functional significance in the immune system. Adv. Exp. Med. Biol. 2014;819:97–110. doi: 10.1007/978-94-017-9153-3_7. [DOI] [PubMed] [Google Scholar]
- 24.Milner JJ, Goldrath AW. Transcriptional programming of tissue-resident memory CD8(+) T cells. Curr. Opin. Immunol. 2018;51:162–169. doi: 10.1016/j.coi.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang N, Bevan MJ. Transforming growth factor-beta signaling controls the formation and maintenance of gut-resident memory T cells by regulating migration and retention. Immunity. 2013;39:687–696. doi: 10.1016/j.immuni.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.El-Asady R, et al. TGF-{beta}-dependent CD103 expression by CD8(+) T cells promotes selective destruction of the host intestinal epithelium during graft-versus-host disease. J. Exp. Med. 2005;201:1647–1657. doi: 10.1084/jem.20041044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takimoto T, et al. Smad2 and Smad3 are redundantly essential for the TGF-beta-mediated regulation of regulatory T plasticity and Th1 development. J. Immunol. 2010;185:842–855. doi: 10.4049/jimmunol.0904100. [DOI] [PubMed] [Google Scholar]
- 28.Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Transforming growth factor-beta regulation of immune responses. Annu. Rev. Immunol. 2006;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- 29.Sun Y, Liu X, Ng-Eaton E, Lodish HF, Weinberg RA. SnoN and Ski protooncoproteins are rapidly degraded in response to transforming growth factor beta signaling. Proc. Natl Acad. Sci. USA. 1999;96:12442–12447. doi: 10.1073/pnas.96.22.12442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deheuninck J, Luo K. Ski and SnoN, potent negative regulators of TGF-beta signaling. Cell Res. 2009;19:47–57. doi: 10.1038/cr.2008.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonthius DJ. Lymphocytic choriomeningitis virus: an underrecognized cause of neurologic disease in the fetus, child, and adult. Semin Pediatr. Neurol. 2012;19:89–95. doi: 10.1016/j.spen.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou X, Ramachandran S, Mann M, Popkin DL. Role of lymphocytic choriomeningitis virus (LCMV) in understanding viral immunology: past, present and future. Viruses. 2012;4:2650–2669. doi: 10.3390/v4112650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grueter B, et al. Runx3 regulates integrin alpha E/CD103 and CD4 expression during development of CD4-/CD8+ T cells. J. Immunol. 2005;175:1694–1705. doi: 10.4049/jimmunol.175.3.1694. [DOI] [PubMed] [Google Scholar]
- 34.Andrew DP, Rott LS, Kilshaw PJ, Butcher EC. Distribution of alpha 4 beta 7 and alpha E beta 7 integrins on thymocytes, intestinal epithelial lymphocytes and peripheral lymphocytes. Eur. J. Immunol. 1996;26:897–905. doi: 10.1002/eji.1830260427. [DOI] [PubMed] [Google Scholar]
- 35.Fousteri G, et al. Minimal effect of CD103 expression on the control of a chronic antiviral immune response. Viral Immunol. 2010;23:285–294. doi: 10.1089/vim.2009.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dutko FJ, Oldstone MB. Genomic and biological variation among commonly used lymphocytic choriomeningitis virus strains. J. Gen. Virol. 1983;64(Pt 8):1689–1698. doi: 10.1099/0022-1317-64-8-1689. [DOI] [PubMed] [Google Scholar]
- 37.Murali-Krishna K, et al. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity. 1998;8:177–187. doi: 10.1016/S1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- 38.Mackay LK, et al. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat. Immunol. 2013;14:1294–1301. doi: 10.1038/ni.2744. [DOI] [PubMed] [Google Scholar]
- 39.Chytil A, Magnuson MA, Wright CV, Moses HL. Conditional inactivation of the TGF-beta type II receptor using Cre:Lox. Genesis. 2002;32:73–75. doi: 10.1002/gene.10046. [DOI] [PubMed] [Google Scholar]
- 40.Lee PP, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/S1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 41.Akiyoshi S, et al. c-Ski acts as a transcriptional co-repressor in transforming growth factor-beta signaling through interaction with smads. J. Biol. Chem. 1999;274:35269–35277. doi: 10.1074/jbc.274.49.35269. [DOI] [PubMed] [Google Scholar]
- 42.Li MO, Sanjabi S, Flavell RA. Transforming growth factor-beta controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms. Immunity. 2006;25:455–471. doi: 10.1016/j.immuni.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 43.Marie JC, Liggitt D, Rudensky AY. Cellular mechanisms of fatal early-onset autoimmunity in mice with the T cell-specific targeting of transforming growth factor-beta receptor. Immunity. 2006;25:441–454. doi: 10.1016/j.immuni.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 44.Liu Y, et al. A critical function for TGF-beta signaling in the development of natural CD4+CD25+Foxp3+ regulatory T cells. Nat. Immunol. 2008;9:632–640. doi: 10.1038/ni.1607. [DOI] [PubMed] [Google Scholar]
- 45.Gu AD, Wang Y, Lin L, Zhang SS, Wan YY. Requirements of transcription factor Smad-dependent and -independent TGF-beta signaling to control discrete T-cell functions. Proc. Natl Acad. Sci. USA. 2012;109:905–910. doi: 10.1073/pnas.1108352109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang S, et al. Reversing SKI-SMAD4-mediated suppression is essential for TH17 cell differentiation. Nature. 2017;551:105–109. doi: 10.1038/nature24283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chu GC, Dunn NR, Anderson DC, Oxburgh L, Robertson EJ. Differential requirements for Smad4 in TGFbeta-dependent patterning of the early mouse embryo. Development. 2004;131:3501–3512. doi: 10.1242/dev.01248. [DOI] [PubMed] [Google Scholar]
- 48.Wang Z, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amsen D, van Gisbergen K, Hombrink P, van Lier RAW. Tissue-resident memory T cells at the center of immunity to solid tumors. Nat. Immunol. 2018;19:538–546. doi: 10.1038/s41590-018-0114-2. [DOI] [PubMed] [Google Scholar]
- 50.Park CO, Kupper TS. The emerging role of resident memory T cells in protective immunity and inflammatory disease. Nat. Med. 2015;21:688–697. doi: 10.1038/nm.3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woon HG, et al. Compartmentalization of total and virus-specific tissue-resident memory CD8+ T cells in human lymphoid organs. PLoS Pathog. 2016;12:e1005799. doi: 10.1371/journal.ppat.1005799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sathaliyawala T, et al. Distribution and compartmentalization of human circulating and tissue-resident memory T cell subsets. Immunity. 2013;38:187–197. doi: 10.1016/j.immuni.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laidlaw BJ, et al. CD4+ T cell help guides formation of CD103+ lung-resident memory CD8+ T cells during influenza viral infection. Immunity. 2014;41:633–645. doi: 10.1016/j.immuni.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mackay LK, et al. T-box transcription factors combine with the cytokines TGF-beta and IL-15 to control tissue-resident memory T cell fate. Immunity. 2015;43:1101–1111. doi: 10.1016/j.immuni.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 55.Pearce EL, et al. Control of effector CD8+ T cell function by the transcription factor eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 56.Sullivan BM, Juedes A, Szabo SJ, von Herrath M, Glimcher LH. Antigen-driven effector CD8 T cell function regulated by T-bet. Proc. Natl Acad. Sci. USA. 2003;100:15818–15823. doi: 10.1073/pnas.2636938100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 2012;12:749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mokrani M, Klibi J, Bluteau D, Bismuth G, Mami-Chouaib F. Smad and NFAT pathways cooperate to induce CD103 expression in human CD8 T lymphocytes. J. Immunol. 2014;192:2471–2479. doi: 10.4049/jimmunol.1302192. [DOI] [PubMed] [Google Scholar]
- 59.Tu E, et al. T cell receptor-regulated TGF-beta type I receptor expression determines T cell quiescence and activation. Immunity. 2018;48:e746. doi: 10.1016/j.immuni.2018.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagano Y, et al. Arkadia induces degradation of SnoN and c-Ski to enhance transforming growth factor-beta signaling. J. Biol. Chem. 2007;282:20492–20501. doi: 10.1074/jbc.M701294200. [DOI] [PubMed] [Google Scholar]
- 61.Tecalco-Cruz AC, Rios-Lopez DG, Vazquez-Victorio G, Rosales-Alvarez RE, Macias-Silva M. Transcriptional cofactors Ski and SnoN are major regulators of the TGF-beta/Smad signaling pathway in health and disease. Signal Transduct. Target. Ther. 2018;3:15. doi: 10.1038/s41392-018-0015-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nomura T, et al. Ski is a component of the histone deacetylase complex required for transcriptional repression by Mad and thyroid hormone receptor. Genes Dev. 1999;13:412–423. doi: 10.1101/gad.13.4.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tabata T, Kokura K, Ten Dijke P, Ishii S. Ski co-repressor complexes maintain the basal repressed state of the TGF-beta target gene, SMAD7, via HDAC3 and PRMT5. Genes Cells. 2009;14:17–28. doi: 10.1111/j.1365-2443.2008.01246.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-seq data are deposited in the GEO database under ID code GSE135533.