Abstract
Sweet basil (Ocimum basilicum L.), a well-known medicinal and aromatic herb, rich in essential oils and antioxidants (contributed by phenolics), is widely used in traditional medicine. The biosynthesis of phytochemicals occurs via different biochemical pathways, and the expression of selected genes encoding enzymes involved in the formation of phenolic compounds is regulated in response to environmental factors. The synthesis of the compounds is closely interrelated: usually, the products formed in the first reaction steps are used as substrates for the next reactions. The current study attempted a comprehensive overview of the effect of aromatic amino acid composition (AAAs) in Ocimum basilicum in respect to the expression of genes related to the biosynthesis of phenolic compound and their content. The transcript expression levels of EOMT, PAL, CVOMT, HPPR, C4L, EGS, and FLS increased depending on the AAAs concentration compared to the control plants. The highest mRNA accumulation was obtained in EOMT, FLS, and HPPR in the leaves of sweet basil. The expression of the TAT gene in the leaves significantly reduced in response to all AAAs applications compared to untreated groups and it had the lowest transcript accumulation. Eleven individual phenolic compounds were determined in the basil leaves, and the contents of chicoric acid, methyl chavicol, caffeic acid, and vanillic acid increased depending on administered concentration to control (p < 0.05). Additionally, AAAs lead to an incremental change in the amount of chlorogenic acid at 50 and 100 mg kg−1 compared to control plants (p < 0.05). Rutin and rosmarinic acid were detected as the main phenolic compounds in all experimental groups of sweet basil in terms of quantity. However, their amount significantly decreased as compared to control plants based on the increase in AAAs concentrations (p < 0.05). Also, the accumulation of cinnamic acid, eugenol, and quercetin did not significantly change in the leaves of AAAs treated plants compared to control (p < 0.05). When AAAs was applied, total flavonoid content increased in all treatments compared to the control plants, but total phenolic content did not change significantly (p < 0.05). To the best of our knowledge, our work is the first detailed work to evaluate in detail the impact of AAAs on individual phenolic compounds at the phytochemistry and transcriptional levels in the O. basilicum plant. For a detailed understanding of the whole mechanism of phenolic compound regulation, further research is required to fill in some gaps and to provide further clarification.
Keywords: Aromatic amino acid, Ocimumbasilicum L., Phenolic compounds, Transcript expression
Introduction
Plants produce numerous phytochemicals, though these phytochemicals varies according to the plant family, genus, species, and variety (Koca and Karaman 2015; Wink 2015). The relevance of medicinal and aromatic plants (MAPs) has been well known since early times. Because they are a source of high and/or unique metabolites used for therapeutic purposes both in traditional and in modern medicine, these plants are being extracted heavily from their natural habitat. Of the different MAPs, the genus Ocimum (family-Lamiaceae) has drawn much attention due to the presence of a wide variety of essential oils and bioactive phytochemicals. This genus is comprised of more than 150 annual and perennial species which grow widely throughout the tropical and temperate regions of the world. Of all Ocimum spp., sweet basil (Ocimum basilicum L.), is the most important economically and have attracted a great deal of scientific interest due to their potential use as functional ingredients, and biologically active compounds which have been documented because of the high concentration of natural contents (Kwon et al. 2017; Purushothaman et al. 2018; Das et al. 2020). The fresh and dried herbs of the sweet basil plant are used as a seasoning, and its extracts are used in the pharmaceutical, cosmetic, food and perfume industries for the isolation of some secondary metabolites such as eugenol and methyl chavicol.
Synthesis of secondary metabolites in plants occurs through the shikimate pathway with a series of enzyme and non-enzyme reactions including deamination and hydroxylation. The first product of this pathway, chorismate, occurs as a result of a series of reactions through the phosphoenolpyruvate formed in the glycolysis metabolic pathway and erythrose-4-phosphate in the pentose phosphate pathway. In plants, the shikimate pathway is the entry pathway for phenylpropanoid synthesis, and AAAs are formed in several steps by branching through the chorismate. Tryptophan (Trp) is synthesized under the catalysis of the atranyl synthase (AS) enzyme, and phenylalanine (Phe), and tyrosine (Try) synthesis occur via the chorismate mutase (CM) enzyme. Then, plant phenolic compounds are formed as a result of a series of biochemical changes in the biosynthesis pathways, from the starting molecules of AAAs to the final phenolic compounds (Maeda and Dudareva 2012; Mandoulakani et al. 2017; Marchiosi et al. 2020). Phe, Try, and Trp are essential molecules of plant metabolism and act as the precursors of a series of natural plant products that play a vital role in plant growth, defense, development, environmental factors as well as in reproduction. Trp is an activator of the chorismate mutase, which plays a role in the secondary metabolite pathway, while Try is involved in gentisic acid, rosmarinic acid, and coumaric acid biosynthesis metabolism. Phe, on the other hand, serves as the primary molecule of the secondary metabolite series with various functions as the precursor of phenylpropanoid, flavonoids, lignin, and many other metabolites (Tzin and Galili 2010).
The regulation of AAAs biosynthesis in plants is coordinated with the activities of relevant sub-metabolic pathways. Transcript expression of related genes encoding enzymes in the AAAs pathway in plants is regulated in response to various environmental factors such as development, injury, pathogen infection, and elicitors. Several transcription factors regulate the biosynthesis of AAA-derived secondary products, and they regulate the expression of genes in the metabolic pathway (Maeda and Dudareva 2012; Rock 2017). The synthesis pathway of phenolic compounds is closely related to each other. For instance, CM enzyme plays a role in the conversion of chorismate to phenylalanine. Trp induces the activity of the CM enzyme and thus acts indirectly in the biosynthesis of phenolic compounds (Park and Lee 2010). One of the important enzymes in the regulation of phenylpropanoid formation in plants is the phenylalanine ammonium lyase (PAL) enzyme. Phenolic compounds are derived from phenylalanine to cinnamic acid (CA) by the deamination activity of PAL. After the synthesis of CA, which is the first phenolic compound synthesized in the phenylpropanoid pathway, various phenolic compounds are synthesized step by step with the role of many enzymes such as PAL, tyrosine aminotransferase (TAT), cinnamate 4-hydroxylase (C4H), cinnamyl-alcohol dehydrogenase (CAD), 4-coumarate CoA ligase (C4L), caffeic acid O-methyltransferase (COMT), cinnamoyl-CoA reductase (CCR), chavicol O-methyl transferase (CVOMT), eugenol/chavicol synthase (EGS), flavonol synthase (FLS), hydroxyphenylpyruvate reductase (HPPR), eugenol O-methyl transferase (EOMT), and p-coumaroyl shikimate 3′-hydroxylase (CS3′H) in the biosynthesis pathway according to KEGG Pathway Maps (Kyoto encyclopedia of genes and genomes, https://www.genome.jp/kegg-bin/show_pathway?map=map00940&show_description=show) (Petersen 2013; Rock 2017; Ru et al. 2017). Changes in the expression level of biosynthetic genes and the activity of their encoding enzymes involved in this metabolic pathway affect the regulation of phenylpropanoid production in plants (Rezaie et al. 2020).
It is well known that biotic and abiotic factors influence the content and composition of phenolic compounds, and this in turn determines the quality of MAPs. Furthermore, besides environmental factors, the genetic characteristics of the plant also influence the quantity of the phytochemicals (Amirkhiz et al. 2021; Zou et al. 2021). While studies on O. basilicum have been taken up in several fields including agriculture, food, ornamental plant, religious, and pharmacology, however, a little work has been carried out in the area of molecular biology. Moreover, there is no information on the role of AAAs in the phenylpropanoid pathway during biosynthesis of phenolic compounds in O. basilicum. Therefore, this investigation was undertaken to investigate the impact of AAAs treatment on the relative transcript accumulation levels of a set of 10 genes associated with secondary metabolites obtained from KEGG and phenolic content in O. basilicum with respect to industrial production. It is worth mentioning that basil is used as a culinary herb in Turkey, and approximately 6.8 tons of basil plants are produced annually; the production of MAPs has been encouraged in recent years to support agriculture and rural development in the country.
Materials and methods
Plant material and growth conditions
The seeds of Ocimum basilicum L. (sweet basil, green colored) obtained from Zengarden traditional seeds (Product no:141 H, İzmir, Turkey) were sterilized with 1% NaOCl and rinsed with tap water. Then, the seeds were sown in a plastic seedling tray (5 × 3.8 × 5 cm) containing a mixture of peat and perlite (3:1), one seed per hole. The seedlings with four true leaves were transferred to plastic pots (three per pot) containing an equal mixture of peat and sifted garden soil. The whole experiment was conducted in triplicate under the unheated greenhouse conditions with 16:8 photoperiods, at 25 ± 3 ºC in the periods of April to August. After the seedlings were acclimatized for approximately four weeks, they were treated with a composition of Phe, Try, and Trp under the three concentrations of 50, 100, and 200 mg kg−1 for each individual amino acid and the application of AAAs was performed three times with intervals of five days. Control plants were watered with tap water only. Leaf samples were taken in the early morning from approximately 90-days-old plants after ten days from the latest application. Whole samples were stored at − 80 °C until the laboratory work began.
Purification of total RNA and Real-Time qPCR analysis
The leaf material (70 mg) was ground into fine powder in liquid nitrogen (LN2) using a mortar and pestle, and the tissue powder was utilized for total RNA isolation utilizing the RNeasy Plant Mini Kit protocol (Qiagen, Sweden). To obtain total RNA, RNase-free water was added to the spin column, quantification and integrity of the RNA were determined using Nanodrop (MaestroGen Inc.) and agarose gel electrophoresis, respectively. The resultant RNA samples were kept at -80 °C for use in gene expression studies.
cDNA synthesis and qPCR studies were performed in one-step to determine gene expressions using the one-step SYBR Green RT-PCR Kit (QuantiTect_Qiagen, Germany) and according to the manufacturer’s recommendations with minor modifications. The kit procedure containing reverse transcriptase enzymes to obtain cDNA and fluorescent dye SYBR Green allows both reverse transcription and qPCR to take place in a single tube. OligoCalc web-based software (http://www.basic.northwestern.edu/biotools/oligocalc.html) was used to design and check gene-specific primers (Table 1). Ten genes were chosen based on their role in phenolic compound pathway submitted to NCBI, the KEGG data bank, and previous studies (Rastogi et al. 2014; Battini et al. 2016; Torre et al. 2016; Kwon et al. 2017; Mandoulakani et al. 2017; Rezaie et al. 2020). RT-qPCR was established in a volume of 25 µL reaction mixture comprising 12.5 µL of the master mix including HotStarTaq DNA polymerase, buffer, SYBR green dye, ROX dye, primers 2 × 0.5 µL, RT mix 0.3 µL, template-RNA 3 µL, and RNase-free water. The RT-qPCR conditions for cDNA synthesis and amplification were as follows: for the RT step 30 min at 50 °C; for the PCR initial activation stage, 15 min at 95 °C, 45 cycles of 30 s at 94 °C, 30 s at 56–60 °C, 30 s at 72 °C; and for final cooling, 20 s at 40 °C. PCR reactions were run in a Bio-Rad thermal cycler (CFX Connect System Real-Time PCR). For RT-PCR analysis, two biological copies of each sample were used. The expression level of selected genes was normalized with GAPDH as an internal control, and relative expression levels of mRNA transcript for the target genes were quantified by using the 2−ΔΔCT method (Livak and Schmittgen 2001).
Table 1.
Specific primers used for qPCR analysis of target genes in O. basilicum
| Genes | Forward | Reverse | Accession |
|---|---|---|---|
| C4H | GCCAACAACCCCGCTCAATG | CCAACGCCGAAGGGGAGGTAT | HM990150 |
| C4L | TCGTAGACAGGGTGAAGGAGC | CTTCACCAGCAGCCTCATCT | KC576841 |
| CAD | GTGAGGTGGTTGAGGTTGGT | ATATCTGAGTTGCAGGGGCG | AY879285 |
| CVOMT | AGTTGGCATTGGAGGGAAGG | TGTGAAGCCAGCATCCGAAA | AB530137 |
| EOMT | GTTGTGGCTGGCTTGGAAAG | CTTCCCTCCCATAACGACCG | AF435008 |
| EGS | AGAATAAACGCATTGCCGCC | TTGGATCATAAGGGCGGAGC | DQ372812 |
| FLS | GCTCGAAAAACTCCCTCCCA | GGGGGATCTTCCAAGTGGTG | Torre et al. 2016 |
| PAL | CCTCAACATCACTCCATGCC | CTCAAAGAAGGACGGGACGC | AB436791 |
| TAT | TACAGGCTGCAGTCCCAGAAA | ATGCCTTCGATGTCTTCGAGC | KJ004760 |
| HPPR | CGCCCTTACTCCAGAAACAA | CGAAGACATCAAGACCAGCA | KJ004761 |
| GAPDH | AACATTATCCCCAGCAGCAC | TAGGAACTCGGAATGCCATC | HM196352 |
Preparation of extracts for chemical analysis of plant samples
Basil leaves (1 g) were crushed into a fine powder in liquid N2 and 10 mL of methanol-chloroform (IsoLab, Germany) solution (4:1) was added to obtained powder for extraction. The homogenate was sonicated for 30 min at 37 °C and then centrifuged at 5000 × g for 10 min at ambient temperature. The obtained supernatants were used for the analysis of the total flavonoids and phenolic compounds.
Analysis of individual phenolic compounds
Quantitative analysis of the phenolic compounds in the leaf samples was carried out by using the HPLC (High Performance Liquid Chromatography) system (Shimadzu, Japan) coupled with an LC 20AT pump and SPD-M20A model. The samples extracted in the methanol-chloroform mentioned above were filtered (0.22 µm) and transferred to vials for the direct injection of 20 µL to the equipment. The mobile phase was composed of solvent A (methanol) and solvent B–acidified water (2% acetic acid, v/v), and the elution gradient was applied to reverse phase GL Sciences InertSustain C18 Analytical Columns (4.6 mm × 250 mm, 5 µm) as follows: 0–2 min, 13% A and 87% B; 2–7 min, 22% A and 68% B; 7–30 min, 40% A and 60% B; 30–50 min, 75% A and 25% B; 50–59 min, 90% A and 10% B; 59–67 min 95% A and 5% B and the equilibrate of the column (and back to the initial conditions). The flow rate of the solvents was 1 mL min−1, and the column temperature was kept to 25 °C. The detection of phenolic compounds was performed by scanning from 190 to 800 nm and read synchronously at 280 nm and 360 nm. The retention times and spectra of individual phenolic compounds were compared to those of known standards (SigmaAldrich, Germany) to identify them. The quantification was done based on external standard calibration curves for chicoric acid, chlorogenic acid, benzoic acid, caffeic acid, eugenol, gallic acid, ferulic acid, methyl chavicol, p-coumaric acid, rosmarinic acid, rutin, trans-cinnamic acid, vanillic acid, and quercetin; and the results were expressed as mg g−1 of leaves fw.
Determination of the total phenolic contents
The amount of total phenolic content (TPC) in the methanol–chloroform (4:1) extracts was determined by the Folin-Ciocalteu reagent in each sample (Singleton and Rosi 1965). Briefly, 100 µL of leaf extract was mixed with 100 µL of folin-ciocalteu’s reagent and 1.5 mL of distilled water and incubated at 25 °C for 3 min. Then, 300 µL of 2% aqueous sodium bicarbonate was added to the reaction tube, and the mixture was allowed to stand for two hours at 25 °C. The absorbance was read at 765 nm using a spectrophotometer (Cary 60, Agilent). The total phenolic concentration of the samples was expressed as gallic acid equivalents (GAEs) in mg per gram of plant extract.
Determination of the total flavonoid contents
The total flavonoid content (TFC) was evaluated by the aluminum chloride (AlCI3, SigmaAldrich, Germany) method with slight modifications (Pękal and Pyrzynska 2014). The reaction mixture of reactants was as follows: 20 µL of extract, 20 µL of AlCI3, and 20 µL of CH3COONa (Merck, Germany); and the final volume was completed to 1 ml with ethanol. All reagents were incubated for 30 min at room conditions, and the absorbance was measured at 425 nm in a spectrophotometer (Cary 60, Agilent). Total flavonoid concentration in O. basilicum extracts was expressed as mg of quercetin equivalents (QE).
Data analysis
Data analysis was performed using analysis of variance (ANOVA) and means comparison by the Duncan test (SPSS). The numerical data are presented as mean value ± standard deviation. Significant differences in each group were indicated with different letters at a p < 0.05 significance level.
Results
The transcript accumulation of associated genes in the biosynthesis pathway
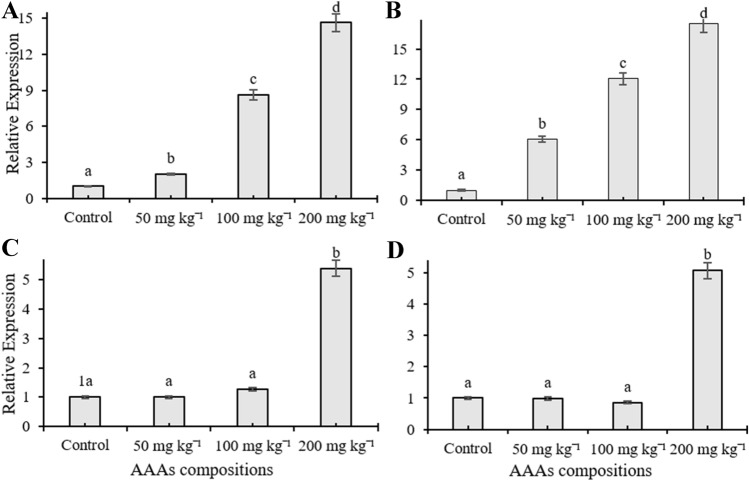
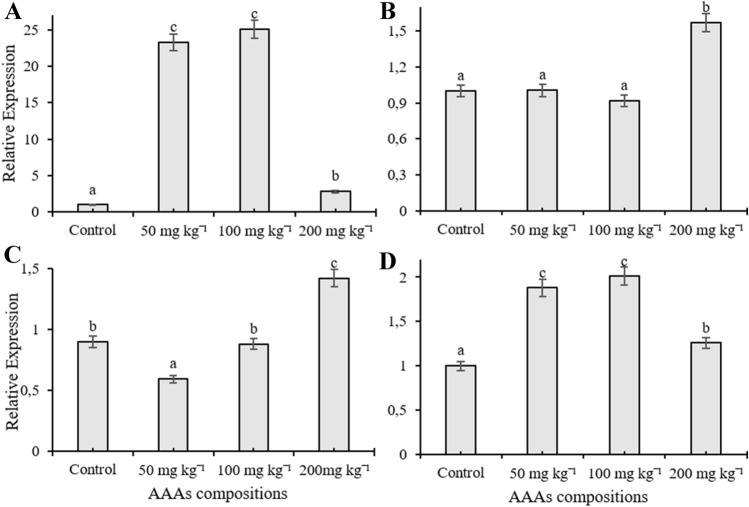
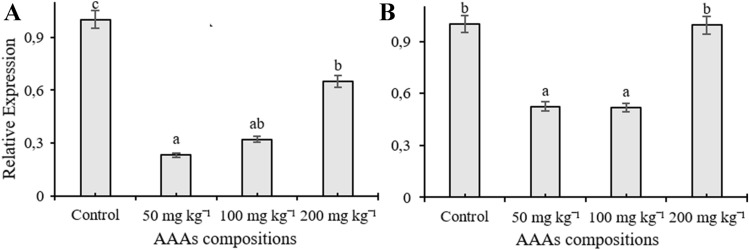
In the current study, RT-qPCR was used to monitor expression patterns of C4H, C4L, CAD, CVOMT, EOMT, EGS, FLS, PAL, TAT, and HPPR genes, and their transcription levels changed depending on the applied doses in the leaves of O. basilicum treated with AAAs. The mRNA transcription level of EOMT and HPPR genes increased compared to control plants depending on the increasing concentrations of AAAs and reached maximum transcription level at 200 mg kg−1 treatment (Fig. 1A, B). Application of 200 mg kg−1 AAAs elevated the transcription level of PAL and EGS genes about five times, but there were no significant changes in the expression level of the transcripts at 50 and 100 mg kg−1 of AAAs compared to control plants (Fig. 1C, D). The mRNA level of FLS were significantly induced in leaves of O. basilicum as compared to the control groups; it had the highest transcript accumulations at 50 and 100 mg kg−1 AAAs and the transcript level of FLS was increased by four times in plants subjected to 200 mg kg−1 AAAs treatments (Fig. 2A). The treatment of AAAs only increased the transcript level of CVOMT at 200 mg kg−1 whereas there was no significant change with other applications (Fig. 2B). AAAs resulted in 1.5-times upregulation of CAD mRNA transcript expression in growth medium treated with 200 mg kg−1 compared to control plants whereas there was no significant change when applied with 100 mg kg−1 AAAs (Fig. 2C). However, the transcript level of CAD gene decreased compared to the control group as a result of 50 mg kg−1 AAAs treatment. Applications of AAAs affected the C4L expression at the transcription level, and the mRNA accumulation increased compared to the control plants (Fig. 2D). The application of 50 and 100 mg kg−1 AAAs upregulated the gene expression level of C4L approximately two times while treatment of 200 mg kg−1 AAAs resulted in a slight increase in the mRNA transcript of C4L. The expression level of TAT gene in the leaves declined as a result of treatments compared to control groups, and TAT had the lowest transcript accumulation at 50 mg kg−1 applications (Fig. 3A). The treatment of 200 mg kg−1 AAAs did not alter the expression level of C4H gene compared to control plants while other treatments caused a decline in the mRNA transcription level. The expression level of C4H gene was downregulated in response to AAAs treatments (Fig. 3B).
Fig. 1.
The effect of AAAs on the expression level of EOMT (A), HPPR (B), PAL (C) and EGS (D). The relative quantities of genes in the basil leaves exposed to 0, 50, 100, 200 mg kg−1 AAAs treatment. Mean values with the different letters show significant differences at p ≤ 0.05 (Duncan test)
Fig. 2.
The effect of AAAs on the expression level of FLS (A), CVOMT (B), CAD (C) and C4L (D). The relative quantities of genes in the basil leaves exposed to 0, 50, 100, 200 mg kg−1 AAAs treatment. Mean values with the different letters show significant differences at p ≤ 0.05 (Duncan test)
Fig. 3.
The effect of AAAs on the expression level of TAT (A) and C4H (B). The relative quantities of genes in the basil leaves exposed to 0, 50, 100, 200 mg kg−1 AAAs treatment. Mean values with the different letters show significant differences at p ≤ 0.05 (Duncan test)
Evaluation of individual phenolic compounds
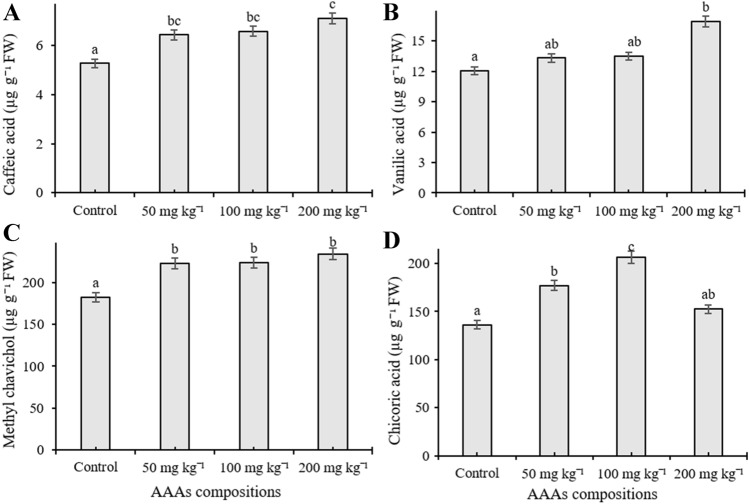
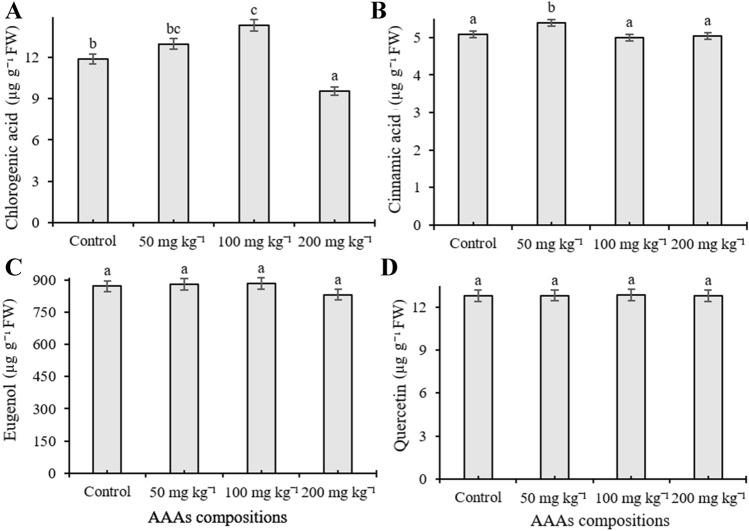
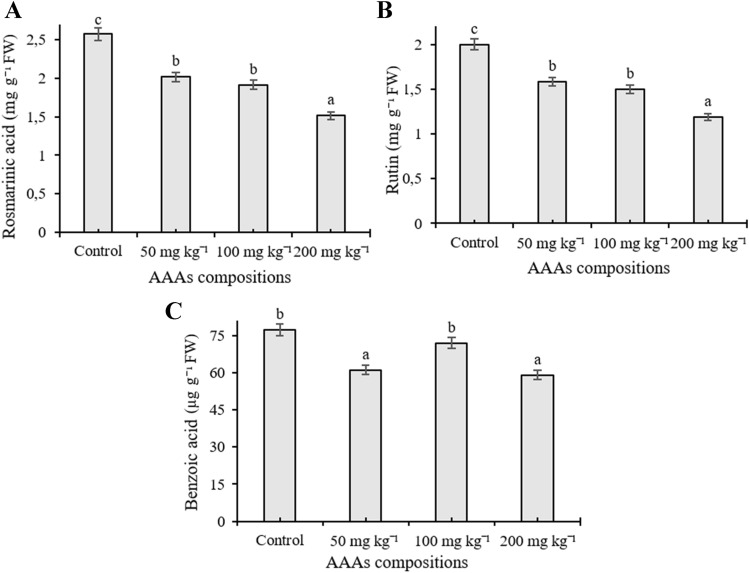
The phenolic compound content was quantified in O. basilicum exposed to different concentrations of AAA compositions by HPLC equipment. Eleven phenolic compounds including cinnamic acid, benzoic acid, chlorogenic acid, chicoric acid, caffeic acid, eugenol, methyl chavicol, quercetin, rosmarinic acid, vanillic acid, and rutin were investigated in the present work, and the number of individual phenolics changed depending on the applied AAAs, whereas gallic acid, p-coumaric acid, and ferulic acid remained below the detection limit. The content of caffeic acid increased in response to AAAs treatments and reached a high level (7.116 µg g−1) at 200 mg kg−1 treatments (p < 0.05) (Fig. 4A). Application of 200 mg kg−1 AAAs significantly elevated the amount of vanillic acid (16.92 µg g−1) whereas treatments did not statistically alter its content when plants were exposed to 50 and 100 mg kg−1 compared to control (p < 0.05) (Fig. 4B). It was observed that methyl chavicol content elevated as a response to applications, and the amount increased from 184.83 µg g−1 to 234.419 µg g−1 compared to the control groups, but there was no statistically significant change between administered doses (Fig. 4C). The level of chicoric acid significantly induced in basil leaves compared to control groups, the highest accumulation (206.47 µg g−1) was determined in plants treated with 100 mg kg−1 AAAs application, and its content was determined as 136.27, 177.21, and 152.04 µg g−1 at 0 (control), 50, and 200 mg kg−1 treatments, respectively (p < 0.05) (Fig. 4D). AAAs lead to an increase in the amount of chlorogenic acid at 100 mg kg−1 compared to control plants whereas a slight increase was observed when applied with 50 mg kg−1 application (p < 0.05). However, the level of chlorogenic acid was observed as decreased when applied with 200 mg kg−1 compared to untreated plants (Fig. 5A). The accumulation level of cinnamic acid, eugenol, and quercetin was not significantly changed in the leaves of sweet basil plants treated with AAAs compared to control (p < 0.05) (Fig. 5B, C and D). Eugenol is the third most abundant secondary metabolite in basil leaves. Quantitatively, rutin and rosmarinic acid were found to be the main phenolic compounds in all experimental groups. However, their amount significantly decreased compared to control samples with increasing concentrations of AAAs The level of rosmarinic acid decreased from 2.57 to 1.51 mg−1 g compared to control groups; it had low accumulation at 200 mg kg−1 AAAs treatment (p < 0.05) (Fig. 6A). It was seen that the quantity of rutin declined as a response to applications, and its amount declined by 1.99 and 1.18 mg g−1, respectively, from control samples to the 200 mg kg−1 AAAs administration, which was the lowest accumulation (p < 0.05) (Fig. 6B). The level of benzoic acid was observed as reduced as a result of all applications of AAAs compared to untreated plants grown in pots, and benzoic acid had low accumulation at 200 and 50 mg kg−1 treatments, respectively (p < 0.05) (Fig. 6C).
Fig. 4.
The content of caffeic acid (A), vanillic acid (B), methyl chavicol (C) and chicoric acid (D) of the leaves of O. basilicum exposed to 0, 50, 100, 200 mg kg−1 AAAs treatment. Mean values with the different letters show significant differences at p ≤ 0.05 (Duncan test). Data expressed is as µg g−1 FW
Fig. 5.
The content of chlorogenic acid (A), cinnamic acid (B), eugenol (C) and quercetin (D) of the leaves of O. basilicum exposed to 0, 50, 100, 200 mg kg−1 AAAs treatment. Mean values with the different letters indicate significant differences at p ≤ 0.05 (Duncan test). Data expressed is as µg g−1 FW
Fig. 6.
The content of rosmarinic acid (A), rutin (B) and benzoic acid of the leaves of O. basilicum exposed to 0, 50, 100, 200 mg kg−1 AAAs treatment. Mean values with the different letters indicate significant differences at p ≤ 0.05 (Duncan test). Data expressed is as µg g−1 FW
Total phenolic compounds and total flavonoid contents
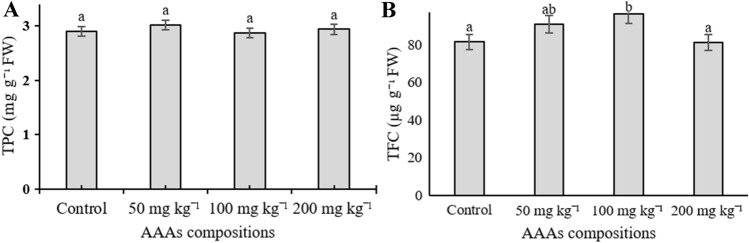
The contents of total phenolic compounds (TFC) and total flavonoid content were determined for the methanolic extract of sweet basil subjected to AAAs. It was observed that the evaluation of TPC did not reveal any significant difference between each treatment group in leaves of O. basilicum (p < 0.05). Total flavonoid content of samples changed depending on the applied concentrations. AAAs increased the accumulation of total flavonoids in all applications compared to control plants except for 200 mg kg−1, and it was not significantly changed (Fig. 7A and B).
Fig. 7.
The effect of AAAs combination on TPC (A) and TFC (B) of the leaves of O. basilicum. Mean values with the different letters show significant differences at p ≤ 0.05 (Duncan test). Data for TPC and TFC expressed is as milligrams of gallic acids and quercetin equivalents (GAEs) per gram fresh weight
Discussion
One of the important features of AAAs is their ability to act as the precursor for secondary metabolites of the plant such as phenylpropanoids. Phe, Try, and Trp serves as a substrate for various secondary compounds such as rosmarinic acid, coumaric acid, alkaloid, lignin, phenylpropanoids, coumarins, flavonoids, etc. They may also act as activators for enzymes such as chorismate mutase. In fact, various molecules derived from these AAAs act through a series of interconnection points in the synthesis of phenolic compounds (Tzin and Galili 2010; Feduraev et al. 2020).
Genes selected for the present study are linked to phenolic compound metabolism in basil, and their expression levels and the content of phenolic compound changed when experimental plants were grown in AAAs-enriched growth medium. The level of methyl chavicol increased compared to control and reached the maximum level at 200 mg kg−1 treatments. EOMT and CVOMT enzymes catalyze the final step in the phenylpropanoid pathway to catalyze eugenol and chavicol to methyl eugenol and methyl chavicol. The gene expression level of EOMT and CVOMT was increased by the treatment with AAA compositions. The effect of AAAs on the expression of above mentioned genes has not been evaluated in O. basilicum so far, but it has been reported that drought stress increases the transcript expressions of EOMT and CVOMT, in agreement with our results (Mandoulakani et al. 2017). Research conducted on O. basilicum under cold stress has shown that moderate cold stress leads to upregulation of EOMT gene expression, but the transcription level of CVOMT remains relatively unchanged (Rezaie et al. 2020). The observed rise in caffeic acid content was accompanied by an increase in the transcript accumulation of PAL with the exception of the 50 mg kg−1 treatment. The increased amount of caffeic acid in our study is in agreement with those previously reported work on grown O. basilicum under iodine treatments where the application of KI or KIO3 induced the accumulation of caffeic acid (Kiferle et al. 2020). The increase in caffeic acid content may be associated with increased levels of expression of PAL gene. This gene encodes PAL, which is responsible for the synthesis of various phenolic compounds and the enzyme catalyzes phenylalanine to cinnamic acid. In a study in which the transcriptional expression of PAL was investigated, it was shown that the transcription level of PAL upregulated in sweet basil, supporting the findings of the current study (Rastogi et al. 2014).
The results showed that the application of AAAs significantly increased the content of chicoric acid compared to untreated plants. Chicoric acid is a derivative of tartaric acid and caffeic acid which are organic compounds related to the phenylpropanoids. Nazir et al. (2020) reported that stimulation of the synthesis of chicoric acid was enhanced by melatonin and UV-C treatments in callus cultures of purple basil (Nazir et al. 2020). The known genes related to chicoric acid biosynthesis are PAL, C3H, C4H, 4CL, and HTT (hydroxyl-cinnamoyl-CoA/tartaric acid hydroxycinnamoyl transferase). To date, no specific enzyme involved in chicoric acid biosynthesis has been reported, and there have been some efforts to identify the hydroxycinnamoyl-transferases involved in meso-chicoric acid biosynthesis. In the future, molecular analyses and hairy root culture techniques could help us learn more about chicoric acid biosynthesis (Lee and Scagel 2013; Sabet et al. 2017; Salmanzadeh et al. 2020).
Our analysis revealed that the level of vanillic acid significantly increased from 12.60 µg g−1 to 16.92 µg g−1 compared with 200 mg kg−1 AAAs application alone whereas there was a slight increase when applied with 50 and 100 mg kg−1 treatments. It was reported that the level of vanillic acid was higher in Ocimum basilicum than O. sanctum, and the concentration of vanillic acid was around 5.77 times higher than that of coumaric acid (Das et al. 2020). We found no published research comparing the vanillic acid level in basil plants subjected to different abiotic stress factors. Vanillic acid is widely accepted as a hydroxybenzoic acid derivative; it is the major product of ferulic acid and a precursor for the synthesis of vanillin (Li and Rosazza 2000; Ni et al. 2015; Delisi et al. 2016; Marchiosi et al. 2020). It is possible to posit that the low ferulic acid level is associated with the increased amount of vanillic acid.
The findings of the present research demonstrated that the chlorogenic acid content increased in plants as a response to 50 and 100 mg kg−1 AAAs, whereas there was a decrease when the plant was exposed to 200 mg kg−1 aromatic amino acid compositions. Researchers who study phenolic compounds have demonstrated that chlorogenic acid is one of the major phenolic acids after caftaric acid and gallic acid in O. basilicum (Irondi et al. 2016; Khatib et al. 2021). Chlorogenic acids (CGAs) and shikimic acid esters are synthesized through the phenylpropanoid pathway and enzymes involved in the early parts of the biosynthetic pathway such as PAL, C4H, C3H, and 4CL. The present findings indicated that the expression of C4L gene was close with an increase in the level of chlorogenic acid with the exception of 200 mg kg−1 AAAs treatment. Also, the transcript accumulation of PAL was consistent with an increase in the content of chlorogenic acid in the leaves of basil subjected to an application of 100 mg kg−1 AAAs. However, it was concluded that acyltransferases are necessary for the last steps in the CGAs biosynthesis, which was recently identified in some plants like coffee and eggplant. These enzymes involved in the CGAs pathway include hydroxyl-cinnamoyl-CoA shikimate/quinate hydroxyl-cinnamoil transferases (HCT), p-coumaroyl ester 3-hydroxylase (C3H), and hydroxyl-cinnamoyl-CoA quinate hydroxyl-cinnamoyl transferase (HQT). In earlier reported works, we did not observe any study on the gene expression involved in the last steps of the CGA biosynthesis in sweet basil (Koshiro et al. 2007; Lallemand et al. 2012; Gramazio et al. 2014; Šilarová et al. 2019).
In the current study, rosmarinic acid (RA) was a major phenolic compound of sweet basil, which is similar to previous studies (Majdi et al. 2020; Mousavi et al. 2020). The AAAs combination significantly decreased the content of rosmarinic acid with increasing treatments in the leaves compared to control. Findings from current work are in agreement with those reported in a previous study in which the treatment of Fe2+ significantly decreased the content of RA in Melissa officinalis seedlings exposed to various concentrations of FeSO4.7H2O compared to control (Salestani 2014). RA biosynthesis is related to both the phenylpropanoid and the tyrosine-derived pathways and involved enzymes in the RA biosynthesis from its amino acid precursor such as PAL, TAT, C4H, 4CL, HPPR, and RAS (rosmarinic acid synthase; 4-hydroxy-cinnamoyl-CoA:4-hydroxy-phenyllactate hydroxy-cinnamoyl-transferase) (Petersen 2013; Ru et al. 2017). Our results have shown that AAAs decreased the transcript level of genes playing a role in the RA biosynthesis such as TAT and C4H compared to the control plant with the exception of the 200 mg kg−1 treatment for C4H which remained constant. The decrease in rosmarinic acid content is generally in agreement with changes in the transcription level of these genes. However, HPPR and C4L gene expression elevated with increasing AAAs administration. Also, mRNA transcript accumulation of PAL similarly increased with the 200 mg kg−1 application. Mandoulakani et al. (2020) determined that the expression level of C4H and 4CL reduced under drought stress conditions in basil (Mandoulakani et al. 2017). While investigating the effect of cold, flood, drought, and salinity on Ocimum metabolomics in the leaf of Ocimum tenuiflorum it has also been observed that under the abiotic stress treatment there was a decrease in the transcript expression of 4CL, C4H, PAL, COMT (Caffeic acid O- methyltransferase), CS3’H (pCoumaroyl shikimate 3’-hydroxylase), C3H (Coumarate 3- hydroxylase), and CCR (Cinnamoyl-CoA reductase) with some exceptions under cold stress; however, there was an increase in the gene expression of CAD (Cinnamoyl alcohol dehydrogenase) and CCOMT (Caffeoyl CoA O-methyltransferase) in drought stress (Rastogi et al. 2019).
To evaluate the flavonoids contents in the present study, changes in the amount of rutin and quercetin were examined in the leaves of green basil. As the second most abundant phenolic compound in the plant, the amount of rutin decreased with the increasing concentration of AAAs whereas these applications did not statistically change the amount of quercetin. The main genes involved in the first steps for a biosynthetic pathway are PAL, C4H, 4CL (4-coumarate-CoA ligase), F3H (flavanone-3-hydroxylase), F3′H (flavonoid-3′-hydroxylase), CHS (chalcone synthase), CHI (chalcone isomerase), UFGT (flavonoid 3-O-glucosyltransferase), FLS (flavonol synthase), and RT (flavonol-3-O-glucoside L-rhamnosyltransferase). However, not all of these genes have yet been fully characterized in the basil plant (Huang et al. 2016; Kianersi et al. 2020). Rutin is a flavanol glycoside while quercetin is an aglycone produced by rutinase enzyme degradation (Luthar et al. 2020). A study investigating the effect of salicylic acid (SA) on flavonoid biosynthesis reported that SA treatments decreased the content of rutin in 7-day old plants while its concentration increased after 1 day in the leaves of wheat plants. The same study showed that the content of quercetin did not significantly change in the leaves of wheat grown by soaking the seed in SA solution (Gondor et al. 2016). However, another study declared that increasing UV-B irradiation increased the individual flavonoids such as catechin, quercetin, and kaempferol in the basil leaf extracts (Ghasemzadeh et al. 2016). When compared to our study, analogous findings may be observed in the quantity of rutin and quercetin in sweet basil. On the other hand, the transcript expression level of FLS significantly increased as compared to control basil plants here. This increase in the transcript accumulation of FLS is consistent with an increase in the total flavonoid content in the present study. Studies examining the effect of AAAs combination on flavonoids in O. basilicum are needed for a detailed comparison in the future.
Cinnamic acid (CA) and its derivatives (sinapic acids, caffeic, ferulic and p-coumaric) have been synthesized from Phe and Try by the action of phenylalanine ammonia-lyase (Shuab et al. 2016). The obtained results from the present study showed that the amount of cinnamic acid did not significantly change in the leaves of a basil growth AAAs-enriched medium. The CA results are consistent with the expression level of PAL at the 50 and 100 mg kg−1 applications. However, the mRNA transcript of PAL, whose protein product is in charge of the transformation of Phe to CA, turned out to be approximately five times higher at the 200 mg kg−1 compared to control. Kwon et al. (2020) demonstrated that the expression level of PAL increased at a high light regime in green basil, but it decreased at other light levels (Kwon et al. 2020). PAL is involved in the first step of phenylpropanoid biosynthesis metabolism, deaminating Phe to generate ammonia and trans-cinnamic acid. Khakdan et al. (2018) determined that the mRNA transcript level of the PAL gene elevated its transcription at different rates in basil subjected to drought stress. Their results demonstrated that PAL regulation is possibly a mechanism on basil cultivars and various drought stress (Khakdan et al. 2018). Another study that examined the accumulation of PAL mRNA transcript at different growth stages reported that the lowest and highest expression of PAL was in 10-leaves plant and the budding stage of basil, respectively, and fluctuation in PAL expression level (Ziaei et al. 2012).
Benzoic acids (BAs) are aromatic carboxylic acids, and their biosynthesis are formed through multiple routes that arise from the phenylpropanoid biosynthesis pathway. BA biosynthesis begins with the deamination of Phe to cinnamic acid by PAL activity, and then CA converts to BA with a shortening of the side-chain via CoA-dependent β-oxidative and non-oxidative pathways or a combination of both pathways (Qualley et al. 2012; Widhalm and Dudareva 2015). In the present study, the amount of benzoic acid decreased in basil leaves treated with AAAs. A study carried out on the chemical composition of O. sanctum revealed that p-hydroxybenzoic acid is one of the most prominent polyphenolic components (Hussain et al. 2017). Another study concluded that the gene expressions of phenylpropanoid pathways such as PAL, CAD, C4H, EOMT, and EGS varied depending on plant growth time (Rastogi et al. 2020).
TPC and TFC were assayed in basil plants treated with AAAs and TPC did not show a significant difference while TFC was elevated in the leaves of sweet basil. According to the results from the present work, it seems that the increase in TFC was consistent with the transcript accumulation of FLS which is significantly increased by the application of AAAs compared to control. A study conducted on the effect of light sources in the phytochemical composition of basil observed that basil leaves developed high TPC under the artificial sunlight (AS) bulb while sulfur plasma light (SPL) and high pressure-sodium (HPS) lamps caused a decrease in the TPC. No significant differences were determined in the TFC in the leaves of basil grown under the AS and SPL (Dörr et al. 2020). On the other hand, it was reported that melatonin and UV-C as elicitors led to an increase both in TPC and TFC in purple basil callus cultures (Nazir et al. 2020).
Conclusions
Phenolic compounds, generated products of the phenylpropanoid pathway, tend to be closely linked with the transcription levels of genes, an important regulatory step in secondary metabolite formation. In this study, the main goal was to get an overall view of the effect of AAAs on selected genes involved in the synthesis pathway and phenolic compounds of sweet basil; therefore, the combined analysis of phenolic profiles and transcriptomics was performed as a comparative study. In our study, plant nutritional value was positively affected by the AAAs treatment, especially in terms of the increasing amount of chicoric acid, caffeic acid, methyl chavicol, and the accumulation of TFC. For all that, although the content of rosmarinic acid and rutin decreased, it may not be stated that the decrease in these nutraceuticals reduce the nutritional value of the plant since their amount in sweet basil is high. Increase in the amount of caffeic acid, methyl chavicol, and TFC showed consistency with the elevation in the expression level of C4L, EOMT, and FLS genes associated with enzymes involved in their biosynthesis, respectively. The observed consistency among them suggests that AAAs probably increased the content of phenolic compounds partially via upregulating the expression of genes such as C4L, EOMT, PAL, and FLS. We studied several transcripts involved with the phenolic compound biosynthesis pathway and metabolite analysis of basil and concluded that AAAs treatment changed the expression of related biosynthesis genes. Secondary metabolites are first synthesized owing to metabolic deviation from the shikimate and mevalonate pathways. Further research is required for a detailed understanding of the effect of precursor molecules on the expression of genes related to biosynthesis of phenolic compounds and the accumulation of secondary metabolites because a number of genes, transcription factors, and their corresponding enzymes play a role in the regulatory mechanism of secondary metabolite production.
Acknowledgements
This study was financially supported by the BAP-Scientific Research Projects Commission, Bartın University, Bartın—TURKEY (Grand ID/Project No: 2019-FEN-A-002).
Author contributions
D.K. designed and performed the experiment, analyzed the data, and write the manuscript; R.İ did the experiments and conducted the real-time PCR; N.G. performed the HPLC. S.S cultivated the plants; M.A.Q write the manuscript and provided technical assistance; M.E. provides valuable suggestions and interpreted the data derived from HPLC.
Declarations
Conflict of interest
All of the authors declare no conflicts of interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Amirkhiz KF, Dehaghi MA, Modares Sanavy SAM, Rezazadeh A (2021) Evaluation of changes in fatty acid profile, grain, and oil yield of Carthamustinctorius L. in response to foliar application of polyamine compounds under eficit irrigation conditions. Ind Crops Prod 161:113231. doi: 10.1016/j.indcrop.2020.113231
- Battini F, Bernardi R, Turrini A, Agnolucci M, Giovannetti M. Rhizophagus intraradices or its associated bacteria affect gene expression of key enzymes involved in the rosmarinic acid biosynthetic pathway of basil. Mycorrhiza. 2016;26:699–707. doi: 10.1007/s00572-016-0707-2. [DOI] [PubMed] [Google Scholar]
- Clifford MN, Jaganath IB, Ludwig IA, Crozier A. Chlorogenic acids and the acyl-quinic acids: discovery, biosynthesis, bioavailability and bioactivity. Nat Prod Rep. 2017;34:1391–1421. doi: 10.1039/c7np00030h. [DOI] [PubMed] [Google Scholar]
- Das S, Barman S, Teron R, Bhattacharya SS, Kim KH (2020) Secondary metabolites and anti-microbial/anti-oxidant profiles in Ocimum spp.: role of soil physico-chemical characteristics as eliciting factors. Environ Res 188:109749. doi: 10.1016/j.envres.2020.109749 [DOI] [PubMed]
- Delisi R, Ciriminna R, Parrino F, Palmisano L, Xu YJ, Pagliaro M. One-pot, clean synthesis of vanillic acid from ferulic acid. ChemistrySelect. 2016;1:626–629. doi: 10.1002/slct.201600111. [DOI] [Google Scholar]
- Dörr OS, Brezina S, Rauhut D, Mibus H (2020) Plant architecture and phytochemical composition of basil (Ocimumbasilicum L.) under the influence of light from microwave plasma and high-pressure sodium lamps. J Photochem Photobiol B Biol 202:111678. doi: 10.1016/j.jphotobiol.2019.111678 [DOI] [PubMed]
- Feduraev P, Skrypnik L, Riabova A, Pungin A, Tokupova E, Maslennikov P, Chupakhina G. Phenylalanine and tyrosine as exogenous precursors of wheat (Triticumaestivum L.) secondary metabolism through PAL-associated pathways. Plants. 2020;9:39–64. doi: 10.3390/plants9040476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghasemzadeh A, Ashkani S, Baghdadi A, Pazoki A, Jaafar HZE, Rahmat A (2016) Improvement in flavonoids and phenolic acids production and pharmaceutical quality of sweet basil (Ocimumbasilicum L.) by ultraviolet-B irradiation. Molecules. doi: 10.3390/molecules21091203 [DOI] [PMC free article] [PubMed]
- Gondor OK, Janda T, Soós V, Pál M, Majláth I, Adak MK, Balázs E, Szalai G. Salicylic acid induction of flavonoid biosynthesis pathways in wheat varies by treatment. Front Plant Sci. 2016;7:1–12. doi: 10.3389/fpls.2016.01447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramazio P, Prohens J, Plazas M, Andjar I, Herraiz FJ, Castillo E, Knapp S, Meyer RS, Vilanova S. Location of chlorogenic acid biosynthesis pathway and polyphenol oxidase genes in a new interspecific anchored linkage map of eggplant. BMC Plant Biol. 2014;14:1–15. doi: 10.1186/s12870-014-0350-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou M, Zhang Y, Mu G, Cui S, Yang X, Liu L. Molecular cloning and expression characterization of flavonol synthase genes in peanut (Arachis hypogaea) Sci Rep. 2020;10:1–11. doi: 10.1038/s41598-020-74763-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Yao J, Zhao Y, Xie D, Jiang X, Xu Z. Efficient rutin and quercetin biosynthesis through flavonoids-related gene expression in Fagopyrum tataricum gaertn. Hairy root cultures with UV-B irradiation. Front Plant Sci. 2016;7:1–11. doi: 10.3389/fpls.2016.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain AI, Chatha SAS, Kamal GM, Ali MA, Hanif MA, Lazhari MI. Chemical composition and biological activities of essential oil and extracts from Ocimum sanctum. Int J Food Prop. 2017;20:1569–1581. doi: 10.1080/10942912.2016.1214145. [DOI] [Google Scholar]
- Irondi EA, Agboola SO, Oboh G, Boligon AA. Inhibitory effect of leaves extracts of ocimum basilicum and ocimum gratissimum on two key enzymes involved in obesity and hypertension in vitro. J Intercult Ethnopharmacol. 2016;5:396–402. doi: 10.5455/jice.20160814112756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakdan F, Alizadeh H, Ranjbar M. Molecular cloning, functional characterization and expression of a drought inducible phenylalanine ammonia-lyase gene (ObPAL) from Ocimumbasilicum L. Plant Physiol Biochem. 2018;130:464–472. doi: 10.1016/j.plaphy.2018.07.026. [DOI] [PubMed] [Google Scholar]
- Khatib S, Harnafi M, Touiss I, Bekkouch O, Milenkovic D, Amrani S. HPLC – DAD profiling of a phenolic extract from Moroccan sweet Basil and its application as oxidative stabilizer of sunflower oil. Chem Pap. 2021 doi: 10.1007/s11696-020-01472-z. [DOI] [Google Scholar]
- Kianersi F, Abdollahi MR, Mirzaie-asl A, Dastan D, Rasheed F. Identification and tissue-specific expression of rutin biosynthetic pathway genes in Capparis spinosa elicited with salicylic acid and methyl jasmonate. Sci Rep. 2020;10:1–15. doi: 10.1038/s41598-020-65815-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiferle C, Ascrizzi R, Martinelli M, Gonzali S, Mariotti L, Pistelli L, Flamini G, Perata P. Effect of Iodine treatments on Ocimumbasilicum L.: Biofortification, phenolics production and essential oil composition (PLoS ONE (2019) 14:12 (e0226559) DOI: 10.1371/journal.pone.0226559) PLoS ONE. 2020;15:1–23. doi: 10.1371/journal.pone.0229016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koca N, Karaman Ş. The effects of plant growth regulators and l-phenylalanine on phenolic compounds of sweet basil. Food Chem. 2015;166:515–521. doi: 10.1016/j.foodchem.2014.06.065. [DOI] [PubMed] [Google Scholar]
- Koshiro Y, Jackson MC, Katahira R, Wang ML, Nagai C, Ashihara H. Biosynthesis of chlorogenic acids in growing and ripening fruits of Coffea arabica and Coffea canephora plants. Zeitschrift Fur Naturforsch - Sect C J Biosci. 2007;62:731–742. doi: 10.1515/znc-2007-9-1017. [DOI] [PubMed] [Google Scholar]
- Kwon DY, Li X, Kim JK, Park SU. Molecular cloning and characterization of rosmarinic acid biosynthetic genes and rosmarinic acid accumulation in Ocimumbasilicum L. Saudi J Biol Sci. 2017 doi: 10.1016/j.sjbs.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon DY, Kim YB, Kim JK, Park SU. Production of rosmarinic acid and correlated gene expression in hairy root cultures of green and purple basil (Ocimumbasilicum L.) Prep Biochem Biotechnol. 2020;51:1–9. doi: 10.1080/10826068.2020.1789990. [DOI] [PubMed] [Google Scholar]
- Lallemand LA, Zubieta C, Lee SG, Wang Y, Acajjaoui S, Timmins J, McSweeney S, Jez JM, McCarthy JG, McCarthy AA. A structural basis for the biosynthesis of the major chlorogenic acids found in coffee. Plant Physiol. 2012;160:249–260. doi: 10.1104/pp.112.202051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Scagel CF. Chicoric acid: Chemistry, distribution, and production. Front Chem. 2013;1:1–17. doi: 10.3389/fchem.2013.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Rosazza JPN. Biocatalytic synthesis of vanillin. Appl Environ Microbiol. 2000;66:684–687. doi: 10.1128/AEM.66.2.684-687.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the method. Methods. 2001;25:02–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luthar Z, Germ M, Likar M, Golob A, Vogel-Mikuš K, Pongrac P, Kušar A, Pravst I, Kreft I. Breeding buckwheat for increased levels of rutin, quercetin and other bioactive compounds with potential antiviral effects. Plants. 2020;9:1–13. doi: 10.3390/plants9121638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Dudareva N. The Shikimate pathway and aromatic amino acid biosynthesis in plants. Annu Rev Plant Biol. 2012;63:73–105. doi: 10.1146/annurev-arplant-042811-105439. [DOI] [PubMed] [Google Scholar]
- Majdi C, Pereira C, Dias MI, Calhelha RC, Alves MJ, Rhourri-Frih B, Charrouf Z, Barros L, Amaral JS, Ferreira ICFR. Phytochemical Characterization and Bioactive Properties of Cinnamon Basil (Ocimum basilicum cv. ‘Cinnamon’) and Lemon Basil (Ocimum × citriodorum) Antioxidants. 2020;2:1–17. doi: 10.3390/antiox9050369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandoulakani AB, Eyvazpour E, Ghadimzadeh M. The effect of drought stress on the expression of key genes involved in the biosynthesis of phenylpropanoids and essential oil components in basil (Ocimumbasilicum L.) Phytochemistry. 2017;139:1–7. doi: 10.1016/j.phytochem.2017.03.006. [DOI] [PubMed] [Google Scholar]
- Marchiosi R, dos Santos WD, Constantin RP, de Lima RB, Soares AR, Finger-Teixeira A, Mota TR, de Oliveira DM, Foletto-Felipe M de P, Abrahão J, Ferrarese-Filho O (2020) Biosynthesis and metabolic actions of simple phenolic acids in plants
- Mousavi M, Zaiter A, Becker L, Modarressi A, Baudelaire E, Dicko A. Optimisation of phytochemical characteristics and antioxidative properties of Foeniculum vulgare Mill. seeds and Ocimumbasilicum L. leaves superfine powders using new parting process. Phytochem Anal. 2020;31:154–163. doi: 10.1002/pca.2875. [DOI] [PubMed] [Google Scholar]
- Muhlemann JK, Woodworth BD, Morgan JA, Dudareva N. The monolignol pathway contributes to the biosynthesis of volatile phenylpropenes in flowers. New Phytol. 2014;204:661–670. doi: 10.1111/nph.12913. [DOI] [PubMed] [Google Scholar]
- Nazir M, Ullah MA, Mumtaz S, Siddiquah A, Shah M, Drouet S, Hano C, Abbasi BH (2020) Interactive effect of melatonin and UV-C on phenylpropanoid metabolite production and antioxidant potential in callus cultures of purple Basil (Ocimumbasilicum L. var purpurascens). Molecules. doi: 10.3390/molecules25051072 [DOI] [PMC free article] [PubMed]
- Ni J, Tao F, Du H, Xu P. Mimicking a natural pathway for de novo biosynthesis: natural vanillin production from accessible carbon sources. Sci Rep. 2015;5:1–12. doi: 10.1038/srep13670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EJ, Lee WY. Tryptophan enhanced accumulation of phenolic compounds via chorismate mutase activation in the Ganoderma neo-japonicum mycelia. J Appl Biol Chem. 2010 doi: 10.3839/jksabc.2010.056. [DOI] [Google Scholar]
- Pękal A, Pyrzynska K. Evaluation of aluminium complexation reaction for flavonoid content assay. Food Anal Methods. 2014;7:1776–1782. doi: 10.1007/s12161-014-9814-x. [DOI] [Google Scholar]
- Petersen M. Rosmarinic Acid: New Aspects Phytochem Rev. 2013;12:207–227. doi: 10.1007/s11101-013-9282-8. [DOI] [Google Scholar]
- Purushothaman B, Srinivasan RP, Suganthi P, Ranganathan B, Gimbun J, Shanmugam K. A Comprehensive Review on Ocimum basilicum. J Nat Remedies. 2018;18:71–85. doi: 10.18311/jnr/2018/21324. [DOI] [Google Scholar]
- Qualley AV, Widhalm JR, Adebesin F, Kish CM, Dudareva N. Completion of the core β-oxidative pathway of benzoic acid biosynthesis in plants. Proc Natl Acad Sci U S A. 2012;109:16383–16388. doi: 10.1073/pnas.1211001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi S, Meena S, Bhattacharya A, Ghosh S, Shukla R, Sangwan N, Lal R, Gupta M, Lavania U, Gupta V, Nagegowda DA, Shasany A. De novo sequencing and comparative analysis of holy and sweet basil transcriptomes. BMC Genomics. 2014;15:588. doi: 10.1186/1471-2164-15-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi S, Shah S, Kumar R, Vashisth D, Akhtar MQ, Kumar A, Dwivedi UN, Shasany AK. Ocimum metabolomics in response to abiotic stresses: Cold, flood, drought and salinity. PLoS ONE. 2019;14:1–26. doi: 10.1371/journal.pone.0210903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi S, Shah S, Kumar R, Kumar A, Shasany AK. Comparative temporal metabolomics studies to investigate interspecies variation in three Ocimum species. Sci Rep. 2020;10:1–15. doi: 10.1038/s41598-020-61957-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaie R, AbdollahiMandoulakani B, Fattahi M. Cold stress changes antioxidant defense system, phenylpropanoid contents and expression of genes involved in their biosynthesis in Ocimumbasilicum L. Sci Rep. 2020;10:1–10. doi: 10.1038/s41598-020-62090-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock CD (2017) Phenylpropanoid metabolism. In: Plant Science. p 17
- Ru M, Wang K, Bai Z, Peng L, He S, Wang Y, Liang Z. A tyrosine aminotransferase involved in rosmarinic acid biosynthesis in Prunella vulgaris L. Sci Rep. 2017;7:1–12. doi: 10.1038/s41598-017-05290-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabet MS, Salmanzadeh M, Moieni A. Identification of gene sequences involved in chicoric acid biosynthesis pathway in Echinaceapurpurea through RNA-SEQ transcriptome analysis. J Plant Physiol Pathol. 2017;05:4172. doi: 10.4172/2329-955x-c1-012. [DOI] [Google Scholar]
- Salestani K. Effects of iron ions on rosmarinic acid production and antioxidant system in Melissaofficinalis L. Seed Annu Res Rev Biol. 2014;4:3359–3372. doi: 10.9734/arrb/2014/9300. [DOI] [Google Scholar]
- Salmanzadeh M, Sabet MS, Moieni A, Homaee M. Heterologous expression of an acid phosphatase gene and phosphate limitation leads to substantial production of chicoric acid in Echinaceapurpurea transgenic hairy roots. Planta. 2020;251:1–14. doi: 10.1007/s00425-019-03317-w. [DOI] [PubMed] [Google Scholar]
- Shuab R, Lone R, Koul KK. Cinnamate and cinnamate derivatives in plants. Acta Physiol Plant. 2016;38:1–9. doi: 10.1007/s11738-016-2076-z. [DOI] [Google Scholar]
- Šilarová P, Boulekbache-Makhlouf L, Pellati F, Česlová L. Monitoring of chlorogenic acid and antioxidant capacity of Solanummelongena L. (eggplant) under different heat and storage treatments. Antioxidants. 2019;8:1–11. doi: 10.3390/antiox8070234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton VL, Rosi JA. Colorimetry of total phenolics with phosphomolybdic-phosphotungstic acid reagents. Am J Oenol Vitic. 1965;16:144–158. [Google Scholar]
- Torre S, Tattini M, Brunetti C, Guidi L, Gori A, Marzano C, Landi M, Sebastiani F. De Novo assembly and comparative transcriptome analyses of red and green morphs of sweet basil grown in full sunlight. PLoS ONE. 2016;11:1–19. doi: 10.1371/journal.pone.0160370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzin V, Galili G. New Insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol Plant. 2010;3:956–972. doi: 10.1093/mp/ssq048. [DOI] [PubMed] [Google Scholar]
- Widhalm JR, Dudareva N. A familiar ring to it: Biosynthesis of plant benzoic acids. Mol Plant. 2015;8:83–97. doi: 10.1016/j.molp.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Wink M. Modes of action of herbal medicines and plant secondary metabolites. Medicines. 2015;2:251–286. doi: 10.3390/medicines2030251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziaei M, Sharifi M, Behmanesh M, Razavi K. Gene expression and activity of phenyl alanine amonia- lyase and essential oil composition of Ocimumbasilicum L. at different growth stages. Iran J Biotechnol. 2012;10:32–39. [Google Scholar]
- Zou Q, Wang T, Guo Q, Yang F, Chen J, Zhang W (2021) Combined metabolomic and transcriptomic analysis reveals redirection of the phenylpropanoid metabolic flux in different colored medicinal Chrysanthemum morifolium. Ind Crop Prod 164