Abstract
Abscisic acid (ABA) is an important phytohormone involved in plant growth, plant development, and the protection of plants against abiotic stresses. PYL/RCAR (pyrabactin resistance/pyr1-like/regulatory components of ABA receptor) is the receptor protein of ABA and the core component of the ABA signal transduction network. The PYL gene family has been identified and analyzed in many species, however, there is no report about the research on the whole genome-wide identification of the alfalfa (Medicago sativa L.) PYL gene family. Therefore, to explore the function of alfalfa PYL genes, 39 MsPYL genes were identified by analyzing the recently published genome of alfalfa. Using bioinformatics methods, we systematically analyzed the chromosome location, protein physicochemical properties, evolutionary relationship, conserved motifs, and response to low-temperature stress of the MsPYL family of alfalfa. The results showed that 39 alfalfa MsPYL genes were distributed on 24 chromosomes, and the analysis of gene duplication events showed that fragment duplication was predominant duplication in alfalfa MsPYL family gene expansion. The phylogenetic tree of MsPYL protein of alfalfa and the phylogenetic tree of PYL genes of 3 species show that the MsPYL gene family can be divided into 3 subfamilies, and the structures of the same subfamilies are relatively similar. The 39 MsPYL gene family members of alfalfa contain 10 Motifs. Motif1, Motif2, Motif3, and Motif5 are the conserved motifs shared by these genes; cis-regulatory elements in promoter regions indicate that regulatory elements related to transcription, cell cycle, development, hormone, and stress response are abundantly present in the MsPYL promoter sequences; Real-time fluorescence quantitative PCR analysis showed that the expression of MsPYL genes can be induced by low-temperature treatment. This study provides a reference for further exploring the structural and functional characterization of the alfalfa PYL gene family.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12298-021-01066-3.
Keywords: Medicago sativa L., ABA receptor, PYL protein family, Cold stress, Phylogenetic tree, Gene expression
Introduction
Abscisic acid (ABA) is an important sesquiterpene plant hormone, which is involved in regulating all aspects of plant growth and development, including seed germination, dormancy, seedling growth, plant senescence, and leaf shedding (Pilet 1975; Smart et al. 1995). Many studies have shown that ABA can inhibit cell division, differentiation and elongation (Zhang et al. 2017). Zhang et al. (2010) observed that ABA inhibits Arabidopsis root growth by promoting the quiescent center (QC) inhibiting root tip stem cell differentiation.
The synthesis of ABA is carried out in the roots and transported to the leaves through the xylem, which promotes the flow of potassium ions, chloride ions and malate ions out of guard cells, thereby reducing the turgor pressure of guard cells and inducing stomatal closure (Schroeder et al. 2001; Levchenko et al. 2005; Vahisalu et al. 2008). As high affinity ABA binding receptors, the PYL gene family members interact with PP2Cs to form PYL-PP2C heterodimer that inhibits the phosphatase activity of key negative regulators PP2Cs, which leads to the activation of the serine/threonine-protein kinase SnRK2 (Santiago et al. 2009; Melcher et al. 2009; Miyakawa et al. 2012). The activated SnRK2 kinase thus regulates downstream transcription factors or proteins, which cause a physiological response to ABA (Nishimura et al. 2010a; Peterson et al. 2010).
ABA receptors (PYLs) are the core regulators of plant ABA signal transduction (Yadav et al. 2020). In 2009, Park et al. (2009) observed that PYR/PYL/RCAR proteins are ABA receptors in Arabidopsis thaliana through genetic screening and yeast two-hybrid methods. With the discovery of PYR/PYL/RCAR (Pyrabactin Resistance/PYR1-Like/Regulatory Components of ABA Receptor) as ABA receptors, PYR/PYL/RCAR has become the most widely recognized ABA receptors among various types of ABA receptor families (Wang et al. 2013). There are 14 members of this family in Arabidopsis, named PYR1 and PYL1 ~ 13. All of these are soluble proteins and contain START (STAR-RELATED LIPID-TRANSFER) characteristic regions, which are distributed in the cytoplasm and nucleus (Hu et al. 2012). Studies have established that the Arabidopsis single mutant (PYR1) is similar to the wild type in a series of responses such as seed germination, seedling growth, and ABA-induced downstream gene expression (Zhao et al. 2018), but mutants of three (PYR1/PYL1/PYL4) and four genes (PYR1/PYL1/PYL2/PYL4) showed that sensitivity of these transgenes to ABA is weakened (Raghavendra et al. 2010; Nishimura et al. 2010b). Six mutants (PYR1/PYL1/PYL2/PYL4/PYL5/PYL8) showed a phenotype that was completely insensitive to ABA were able germinate and grow under 100 µmol·L−1 ABA to environment conditions (Gonzalez-Guzman et al. 2012). Following Arabidopsis, other higher plants, such as rice (Oryza sativa L.), (Kim et al. 2012), soybean (Glycine max L.), (Bai et al. 2013), tomato (Solanum lycopersicum L.), (González-Guzmán et al. 2014), wheat (Triticum aestivum L.), (Gordon et al. 2016), corn (Zea mays L.), (Fan et al., 2018), tobacco (Nicotiana tabacum L.), (Bai et al. 2019), rubber tree (Hevea brasiliensis L.), (Guo et al. 2017), and other research on ABA receptors have also been gradually carried out. The results show that overexpression of PYR/PYL/RCAR family gene members in these crops can improve the sensitivity of transgenic crops to ABA, drought resistance, and promote fruit maturity.
The regulation of ABA in plants is accomplished through the ABA signal transduction pathway (Chen et al. 2020). Plant tend to accumulate ABA quickly when subjected to abiotic stress, such as drought, salinity, freezing damage, high temperature, etc., through a series of signal transmission (Cutler et al. 2010; Akhter et al. 2021), by controlling leaf stomatal openings, regulating cell ion balance and promoting old leaf shedding and other adaptive changes that respond to stress (Yang et al. 2019), thereby enhancing the stress resistance of plants (Zhao et al. 2017). Studies on a variety of ABA synthesis mutants have also established that ABA positively regulates seed dormancy. For example, Arabidopsis thaliana L. and tobacco (Nicotiana tabacum L.), mutants overexpressing ABA synthesis genes have increased ABA content, which can increase seed dormancy (Zhou et al. 2018). ABA can also regulate stomatal movements in stressed conditions by two signal transduction pathways i.e., promoting stoma closure and inhibiting stoma opening (Hewage et al. 2020). Exogenous ABA treatment can significantly reduce leaf stomata opening (Wang et al. 1998). In addition, it is also an anti-stress hormone that participates in the regulation of various biotic and abiotic stresses (Niyaz et al. 2017). When plants are under adverse conditions such as high temperature, freezing damage, drought, saline-alkaline conditions, etc., ABA can improve the plant's resistance to stress conditions, thereby reducing the degree of plant damage (Danquah et al. 2014). Ryynänen (1998) observed that ABA can promote the recovery of the vitrified shoot tip of silver-birch (Betula pendula L.) caused by cold injury, and enhance the ability of plants to adapt to cold stress by enhancing cold acclimation and callus formation. The application of exogenous ABA can reduce rice pollen sterility caused by high temperature stress. Because ABA enhances the transportation of sucrose, accelerates metabolism, maintains carbon balance and energy homeostasis, and reduces the surface temperature, thereby improving rice heat tolerance (Rezaul et al. 2019).
Alfalfa (Medicago sativa L.) is a plant of the leguminous alfalfa genus, which is naturally involved in biological nitrogen fixation, high protein content, accumulation, rich nutrition, and good quality (Sen et al. 1998). Simultaneously, alfalfa possesses the characteristics of strong stress resistance and can adapt to various environmental conditions. However, with the increasingly severe global climate and environmental situations, the growth and productivity of alfalfa is seriously threatened. Therefore, it is very important to have deep insights into the stress regulation mechanisms and to generate stress tolerant alfalfa varieties for cultivation. At present, the whole genome sequencing of alfalfa has been recently completed, which laid the foundation for studying the systematic evolution and functional analysis of the gene family of this species by using bioinformatics algorithms (Nian et al. 2021). As PYL genes play an important role in enhancing tolerance under abiotic stress, therefore, the identification and verification of PYL genes in alfalfa play a very important role in understanding their functions and ABA signal transduction pathways.
In this study, we identified PYL family genes in alfalfa based on whole-genome sequencing data. This study uses bioinformatics methods to identify the members of the PYL gene family in alfalfa, and subsequent analysis of gene structure, protein domains, conserved motifs, chromosome location, and other information. We further investigated the expression patterns of PYL gene family members using qRT-PCR techniques to study the differential expression patterns of PYL members in varying cold conditions. The relative expression level at the different time points elucidates the stress response behavior of the gene family members to cold stress, and provides some theoretical basis for the genetic improvement of frost tolerant alfalfa using molecular biology techniques.
Materials and methods
Data acquisition and identification of MsPYLs in alfalfa genome
The genome data of alfalfa was obtained from the Alfalfa breeders toolbox (Alfalfa breeders toolbox, https://www.alfalfatoolbox.org/), whereas, the genomic and protein sequence information of Arabidopsis and rice PYL genes were obtained from the TAIR database (https://www.arabidopsis.org/index.jsp) and Phytozome database (https://phytozome.jgi.doe.gov/pz/portal.html). We performed local Blast method in the alfalfa genome using the Arabidopsis and rice PYL gene as query sequences with a threshold level of E-value < 0.00001 for the initial screening of candidate genes for the alfalfa PYL family. Further confirmation of the obtained genes was done by confirming the domain conserved in the PYL family using Pfam database (http://pfam.xfam.org) and the SMART database (http://smart.embl-heidelberg.de/). Using ExPASyProt Param (https://web.expasy.org/protparam/), we analyzed the physical and chemical properties of alfalfa PYL protein such as molecular weight, isoelectric point, and hydrophobicity. The subcellular localization was determined by Psort-Prediction (http://psort1.hgc.jp/form.html) forecast.
Chromosome location and gene duplication analysis
Using the annotation file (gff3 file) of the PYL genes in the alfalfa genome database, the distribution of alfalfa PYLs on the 32 chromosomes of alfalfa was analyzed. Duplicate gene pairs were detected from the plant genome duplication database server (http://chibba.agtec.uga.edu/duplication/index/locket). The amino acid sequence of the partially repeated MsPYL gene was predicted by Clustalw software. Synonymous substitution rate (Ks) and non-synonymous substitution rate (Ka) and Ka/Ks were calculated using online tools, Clustal Omega, and PAL2NAL (http://www.bork.embl.de/pal2nal/).
Sequence Alignment and Evolution Analysis of Alfalfa PYL Protein
We used 4 species to study the evolutionary relationship between alfalfa PYL genes and other plant PYL genes, including Arabidopsis, rice, tobacco, and identified alfalfa PYL genes. Among them, the protein sequences of tobacco were downloaded from the NCBI database. The multiple sequence alignment in evolutionary relationship analysis is a very important step. To obtain a more ideal multiple sequence alignment result, we first used the ClustalW tool to perform multiple sequence alignment on the PYL protein sequence, and then use the Gblocks online server to analyze the comparison results and selected the highly conserved protein segment sequences for the construction of the phylogenetic tree. Finally, Prot Test v3.4 an online software was used to evaluate the amino acid substitution model of the PYL conserved sequences used for phylogenetic tree construction. We selected the best substitution model based on the results and used the maximum likelihood method in the MEGA 7.0 software to construct a PYL evolutionary tree having bootstrap value as 1000 repeats.
Analysis of gene structure and conserved motif of Alfalfa PYL transcription factor family
According to the gff3 annotation file information of the whole genome of alfalfa, the online website Gene Structure Display Server (GSDS) (http://gsds.cbi.pku.edu.cn/) was used to obtain the visualization pictures of gene structure (exons-introns distributions) (Guo et al. 2007). The conserved motifs of alfalfa PYL proteins were analyzed by Multiple Expectation Maximization for Motif Elicitation (MEME Suite) (http://meme-suite.org/), and the number of Motifs was set to 10 (Bailey et al. 2006).
Promoter cis-acting elements and 3D structure analysis of PYL protein
The 1200 bp sequences upstream of the MsPYL genes were used as the promoters of the alfalfa genes and Plant CARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) database was used to predict and organize the cis-acting elements of the promoter i.e., in the form of a chart. At the same time, we used SWISS-MODEL (Schwede et al. 2003) (https://swissmodel.expasy.org/interactive) provided by the online software ExPaSy to model the three-dimensional structure homology of the protein spatial model of the MsPYL gene family of alfalfa. Additionally, the stereo-chemical quality of each gene model was evaluated by Ramachandran plot that gives a measure of structural error at each residue in the protein (Hooft et al. 1997).
Planting of plant materials and stress treatment
Seeds of alfalfa having identical shapes and sizes were selected and sterilized with 75% alcohol for 3 ~ 5 min. Sterilized seeds were rinsed with sterile deionized water 4 ~ 5 times, and spread in a pre-prepared petri dish with sterile filter paper. Alfalfa seeds were placed in the incubator for the optimum growth of seedlings. The environmental conditions of the incubator are set to temperature 20–22 °C, photoperiod set to 16/8 h (day/night), and relative humidity 55–60%. After the cotyledons were fully opened, the seedlings were transferred to the turf soil: vermiculite: perlite 3:2:1 nutrient soil to continue the cultivation. Four-week-old alfalfa seedlings with consistent growth status were subjected to low-temperature stress treatment at 4 °C, and the treatment time was 0 h, 3 h, 6 h, 9 h, 12 h, 24 h, and 48 h. The sampled tissues were alfalfa leaves, and the samples were immediately flash-frozen in liquid nitrogen, and then placed in -80 °C cryopreservation for subsequent quantitative experiments.
Alfalfa RNA extraction, reverse transcription, and q RT-PCR analysis
Using Shenggong’s UNIQ-10 column Trizol total RNA extraction kit, total RNA was extracted from each sample, and subjected to Nano-Drop 2000 UV spectrophotometer to detect RNA quality and concentration. We used M-Mu LV first-strand cDNA synthesis kit to reverse-transcribe RNA to obtain cDNA. After detecting the concentration, uniformly diluted to 100 ng/μl as the qRT-PCR reaction template. Specific primers were designed using Perl Primer 6 software, and the primers used in the experiments are listed in Table S1. Use Shenggong’s 2 × SG. Fast qPCR Master Mix kit, the reaction system is 20 μl, and the PCR reaction program was 95 °C pre-denaturation for 10 min, and then 40 cycles including 95 °C denaturation for 15 s and 60 °C annealing temperature for 1 min. The instrument used for qRT-PCR is Applied Biosystems 7500. In the q RT-PCR process, each sample was subjected to 3 technical repetitions to eliminate operational errors. In addition, we used the 2-△△Ct method to calculate the relative gene expression level (Livak et al. 2001). The SPSS statistical software was used to analyze the variance of the relative expression levels of each gene at different sampling points under low-temperature stress. Different letters in the figure indicate significant differences in the average of different sampling times, which are determined by Tukey's pairwise comparison test.
Result
Identification of Alfalfa MsPYL family genes
Using Arabidopsis and rice PYL protein sequences and alfalfa genome sequence for homology comparison analysis, a total of 39 alfalfa PYL genes were screened, and the predicted alfalfa PYL names were named according to the location on chromosomes (Fig. 1). The protein sequence information is shown in Table S2. As shown in Table 1, the coding sequences of 39 MsPYL transcription factors were between 528 and 696 nucleotides, and the lengths of their encoded protein sequences range from 175 to 231 aa. The molecular weight range of MsPYLs was 19.59 kDa to 25.31 kDa, and the average molecular weight was 22.46 kDa. The predicted isoelectric point of the encoded protein ranges from 5.66 to 8.50. Except for the GRAVY (Grand average of hydropathicity) of MsPYL2-3, the GRAVY value (Grand average of hydropathicity) of all other MsPYL proteins was less than 0, indicating that MsPYL2-3 was a neutral protein, and other proteins were hydrophilic proteins. The subcellular localization results showed that the MsPYL transcription factor family was mainly distributed in the cytoplasm of alfalfa cells, and partly distributed in the mitochondria and endoplasmic reticulum.
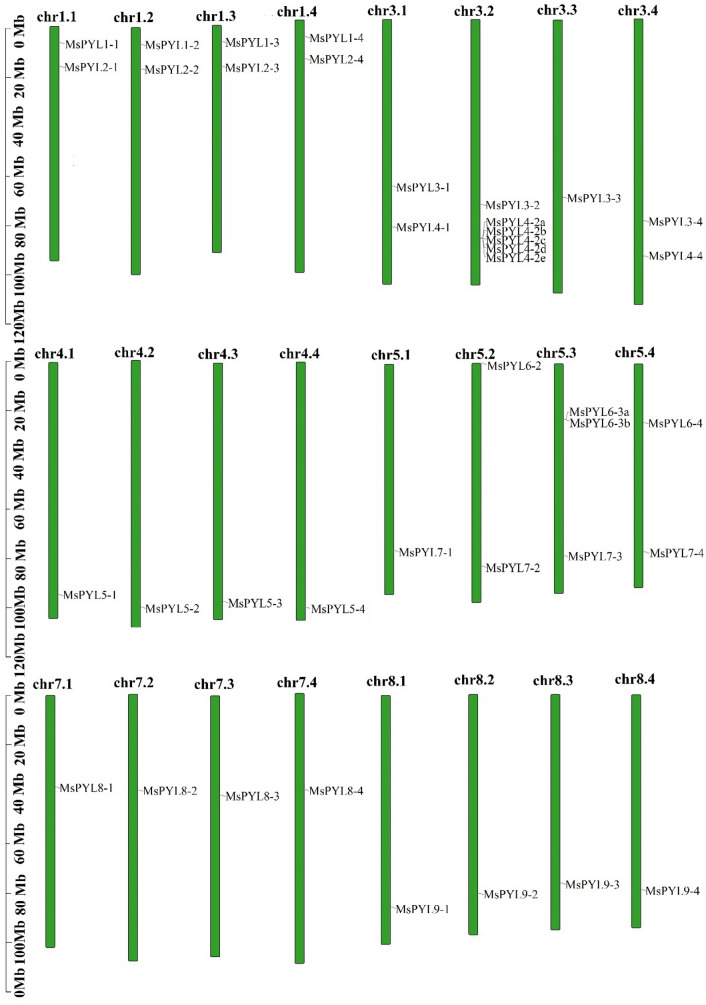
Fig. 1.
Chromosome localization of the MsPYL genes, distribution of MsPYL genes in the alfalfa genome. The MsPYL genes are located on chromosomes 1.1–1.4, 3.1–3.4, 4.1–4.4, 5.1–5.4, 7.1–7.4, and 8.1–8.4. Chromosomal distances are given in Mb
Table1.
Information about MsPYL gene family members
| Gene accession No | Gene | Size (aa) | Molecular weight (D) | Isoelectric point | GRAVY | Subcellular Localization |
|---|---|---|---|---|---|---|
| MS.gene32980.t1 | MsPYL1-1 | 189 | 21,265.22 | 5.81 | − 0.268 | Cytoplasm |
| MS.gene24547.t1 | MsPYL1-2 | 189 | 21,239.14 | 5.81 | − 0.292 | Cytoplasm |
| MS.gene051624.t1 | MsPYL1-3 | 189 | 21,239.14 | 5.81 | − 0.292 | Cytoplasm |
| MS.gene57223.t1 | MsPYL1-4 | 189 | 21,239.14 | 5.81 | − 0.292 | Cytoplasm |
| MS.gene31808.t1 | MsPYL2-1 | 186 | 21,016.97 | 6.38 | − 0.356 | Cytoplasm |
| MS.gene046813.t1 | MsPYL2-2 | 186 | 21,016.97 | 6.38 | − 0.353 | Cytoplasm |
| MS.gene054122.t1 | MsPYL2-3 | 175 | 19,594.60 | 5.66 | 0 | Mitochondrion |
| MS.gene32355.t1 | MsPYL2-4 | 186 | 21,016.97 | 6.38 | − 0.353 | Cytoplasm |
| MS.gene67442.t1 | MsPYL3-1 | 231 | 25,311.54 | 6.15 | − 0.197 | Cytoplasm |
| MS.gene040004.t1 | MsPYL3-2 | 231 | 25,311.54 | 6.15 | − 0.197 | Cytoplasm |
| MS.gene008006.t1 | MsPYL3-3 | 231 | 25,265.59 | 6.15 | − 0.142 | Endoplasmic reticulum |
| MS.gene031922.t1 | MsPYL3-4 | 231 | 25,311.54 | 6.15 | − 0.197 | Cytoplasm |
| MS.gene84133.t1 | MsPYL4-1 | 199 | 22,461.50 | 5.75 | − 0.397 | Cytoplasm |
| MS.gene06745.t1 | MsPYL4-2a | 199 | 22,403.46 | 5.90 | − 0.371 | Cytoplasm |
| MS.gene057109.t1 | MsPYL4-2b | 199 | 22,403.46 | 5.90 | − 0.371 | Cytoplasm |
| MS.gene057108.t1 | MsPYL4-2c | 199 | 22,403.46 | 5.90 | − 0.371 | Cytoplasm |
| MS.gene057110.t1 | MsPYL4-2d | 199 | 22,403.46 | 5.90 | − 0.371 | Cytoplasm |
| MS.gene06744.t1 | MsPYL4-2e | 199 | 22,403.46 | 5.90 | − 0.371 | Cytoplasm |
| MS.gene013622.t1 | MsPYL4-4 | 199 | 22,461.50 | 5.75 | − 0.397 | Cytoplasm |
| MS.gene93372.t1 | MsPYL5-1 | 218 | 23,957.71 | 5.90 | − 0.32 | Cytoplasm |
| MS.gene33451.t1 | MsPYL5-2 | 218 | 23,925.65 | 5.90 | − 0.332 | Cytoplasm |
| MS.gene33704.t1 | MsPYL5-3 | 218 | 23,911.62 | 5.90 | − 0.333 | Cytoplasm |
| MS.gene052795.t1 | MsPYL5-4 | 218 | 23,902.61 | 5.83 | − 0.333 | Cytoplasm |
| MS.gene054810.t1 | MsPYL6-2 | 219 | 24,334.06 | 5.78 | − 0.547 | Chloroplast |
| MS.gene054809.t1 | MsPYL6-3a | 219 | 24,334.06 | 5.78 | − 0.547 | Chloroplast |
| MS.gene069194.t1 | MsPYL6-3b | 215 | 23,932.64 | 5.78 | − 0.519 | Chloroplast |
| MS.gene027877.t1 | MsPYL6-4 | 219 | 24,342.08 | 5.98 | − 0.547 | Chloroplast |
| MS.gene51080.t1 | MsPYL7-1 | 205 | 22,465.57 | 8.50 | − 0.084 | Cytoplasm |
| MS.gene76095.t1 | MsPYL7-2 | 205 | 22,465.57 | 8.50 | − 0.084 | Cytoplasm |
| MS.gene29692.t1 | MsPYL7-3 | 205 | 22,465.57 | 8.5 | − 0.084 | Cytoplasm |
| MS.gene94210.t1 | MsPYL7-4 | 205 | 22,466.56 | 8.18 | − 0.084 | Cytoplasm |
| MS.gene99506.t1 | MsPYL8-1 | 186 | 20,340.9 | 6.14 | − 0.158 | Cytoplasm |
| MS.gene069000.t1 | MsPYL8-2 | 186 | 20,326.88 | 6.14 | − 0.158 | Cytoplasm |
| MS.gene071035.t1 | MsPYL8-3 | 186 | 20,358.93 | 6.28 | − 0.155 | Cytoplasm |
| MS.gene95677.t1 | MsPYL8-4 | 186 | 20,335.89 | 6.22 | − 0.156 | Cytoplasm |
| MS.gene95282.t1 | MsPYL9-1 | 190 | 21,564.47 | 6.23 | − 0.42 | Cytoplasm |
| MS.gene86009.t1 | MsPYL9-2 | 190 | 21,564.47 | 6.23 | − 0.42 | Cytoplasm |
| MS.gene59569.t1 | MsPYL9-3 | 190 | 21,564.47 | 6.23 | − 0.42 | Cytoplasm |
| MS.gene79764.t1 | MsPYL9-4 | 190 | 21,564.47 | 6.23 | − 0.42 | Cytoplasm |
The 39 PYL genes of alfalfa were distributed on 24 of the 32 chromosomes of the alfalfa genome (Fig. 1). MsPYL genes were distributed on four sets of chromosomes, of which, one set of chromosomes have 8 MsPYL genes located on chromosomes 1.1, 3.1, 4.1, 5.1, 7.1, 8.1; there were 13 MsPYL genes in the 2 groups of chromosomes, located on chromosomes 1.2, 3.2, 4.2, 5.2, 7.2, 8.2, and chromosomes; the 3 and 4 groups of chromosomes each have 9 MsPYL genes, located at 1.3, 3.3, 4.3, 5.3, On chromosomes 7.3, 8.3, 1.4, 3.4, 4.4, 5.4, 7.4, 8.4, the distribution of MsPYL gene was not found on other chromosomes. In addition, the maximum number of MsPYL genes distributed on chromosome 3.2 is 6, followed by 3 on chromosome 5.3, and there were two MsPYL genes on each of chromosomes 1.1, 1.2, 1.3, 1.4, 3.1, 3.4, 5.2, and 5.4. The other chromosomes had a MsPYL gene.
Tandem duplication and segmental duplications were two important processes in the evolution of gene families, so we investigated the duplication of MsPYL genes in alfalfa. A total of 93 replicated gene pairs (Table S3) were identified, and their Ka/Ks values ranged from 0.0243 to 0.9481. All the repetitive genes were fragment duplications, and there was no tandem duplication. Among them, 34 pairs of fragment duplication genes were partial homologous inter-chromosomal fragment duplications, and 59 pairs of genes are non-homologous inter-chromosomal fragment duplications. This shows that the in alfalfa MsPYL gene family, the main reason for the expansion was the integration of chromosomes 1, 2, 3, and 4 during the evolution of alfalfa (Fig. 1).
Evolutionary analysis of alfalfa MsPYL gene family
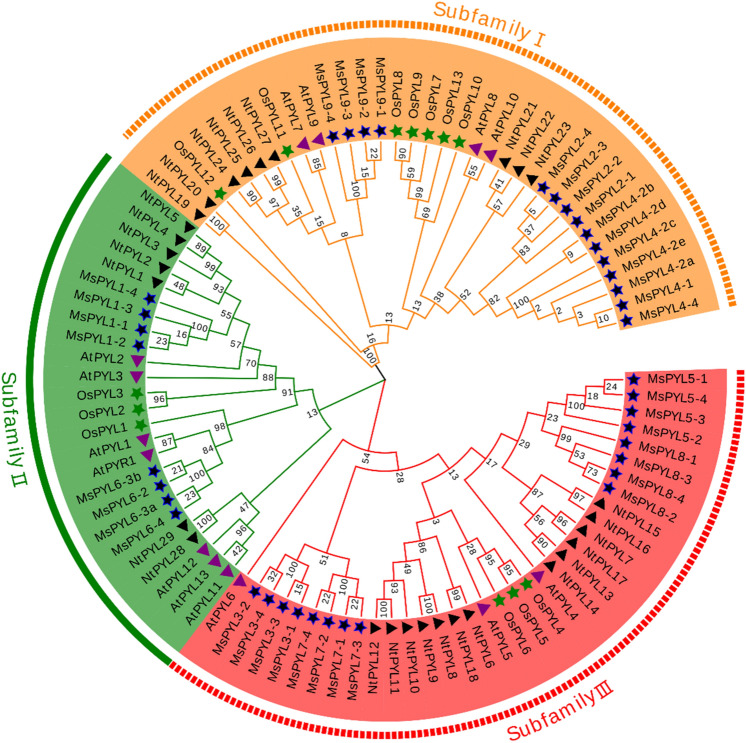
To explore the evolutionary relationship between the PYL gene family and predict its classification in alfalfa, 95 genes from different species (Arabidopsis, rice, tobacco, and alfalfa) of the PYL conserved protein sequence (Table S4) were used for the construction of the evolutionary tree, using MEGA7.0 according to the maximum likelihood method (Fig. 2). Although the phylogenetic tree of the members of a supergene family cannot clarify the biological functions of family members, the phylogenetic tree can clearly and intuitively predict the differentiation and functional diversity of family members. As shown in Fig. 2, the MsPYL gene family can be divided into three subfamilies, namely Subfamily I, Subfamily II, and Subfamily III. The number of members in each subgroup is unevenly distributed. Among them, the Subfamily III had the most members (16), and they were MsPYL3-1— MsPYL3-4, MsPYL5-1—MsPYL5-4, MsPYL7-1—MsPYL7-4 and MsPYL8-1—MsPYL8-4; Subfamily I had the second-highest number of members (15), and the members were MsPYL2-1—MsPYL2-4, MsPYL4-1—MsPYL4-4, and MsPYL9-1—MsPYL9-4. There were members of Subfamily II family, the members were MsPYL1-1—MsPYL1-4 and MsPYL6-2—MsPYL6-4. Alfalfa Subfamily I family members were closely related to AtPYL7-10, NtPYL19-27, and OsPYL7-13 and were properly grouped. Alfalfa Subfamily II family members were associated with AtPYR1, AtPYL1-3, AtPYL11-13, NtPYL1-5, NtPYL28, NtPYL1-29, and OsPYL1-29. Alfalfa Subfamily III members were closely related to AtPYL4-6, NtPYL6-18, and OsPYL4-6. These 3 closely related subfamilies form a family.
Fig. 2.
Phylogenetic tree analysis of PYL protein family members of alfalfa, Arabidopsis, rice, and tobacco. Note: Arabidopsis thaliana At (Arabidopsis thaliana L.), rice Os (Oryza sativa L.), tobacco Nt (Nicotiana tabacum L.), alfalfa Ms (Medicago sativa L.); three color areas represent three clades; blue stars mark common purple flowers Alfalfa PYL protein family members; green stars mark rice PYL protein family members; purple triangles mark Arabidopsis PYL protein family members; black triangles mark tobacco PYL protein family members
Alfalfa MsPYL gene structure and protein conserved domain analysis
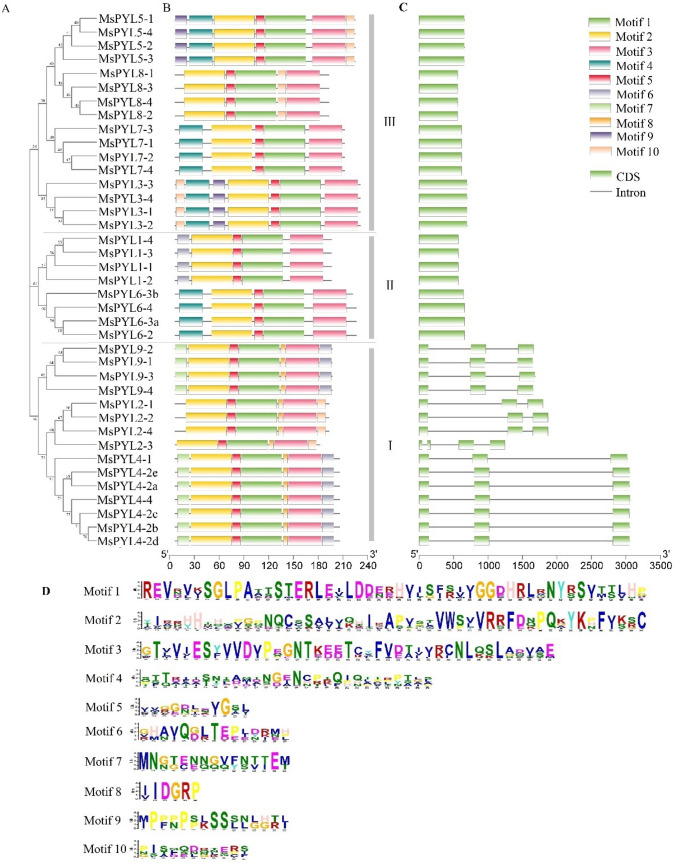
The structure of exons and introns of the MsPYL gene was analyzed by GSDS. We observed that the structure of the exons and introns of the MsPYL gene of alfalfa were different among different subfamilies, but relatively conserved within the same subfamily. As shown in Fig. 3C, the number and length of introns were significantly different among different subfamilies. The MsPYL genes of Subfamily III and Subfamily II had no introns. In Subfamily I, only MsPYL2-3 had 3 introns, while the other 14 MsPYL genes had only 2 introns. The conserved motifs of 39 alfalfa MsPYL genes were obtained by searching and comparing the MEME Suite database. A maximum of 10 conserved motifs (Motif) can be found in each OPR, and the length of these motifs ranges from 11 to 50 amino acids (Table S5). It can be seen from Fig. 3B that although the conserved motifs of the 39 alfalfa PYL family genes were different in composition, they all contain Motif 1, Motif 2, Motif 3 and Motif 5, and they were arranged in the same order, with Motif 2 first, then Motif 5 and Motif 1, Motif 3 was behind these three Motifs, and there was no duplication. MsPYL1-1—MsPYL1-4, MsPYL6-2—MsPYL6-4, MsPYL7-1—MsPYL7-4, and MsPYL8-1—MsPYL8-4 contains 5 Motifs; MsPYL2-1—MsPYL2-4, contains 6 Motifs; the remaining 19 MsPYL genes all contain 7 Motifs. Among them, only MsPYL3-1—MsPYL3-4 and MsPYL5-1—MsPYL5-4 contain Motif 9, and MsPYL4-1—MsPYL4-4 and MsPYL9-1—MsPYL9-4 contain Motif 7. The analysis results indicated that the MSLIM gene family should all contain Motif 1, Motif 2, Motif 3, and Motif 5, MsPYL3-1—MsPYL3-4, MsPYL5-1—MsPYL5-4, MsPYL4-1—MsPYL4-4, and MsPYL9-1— MsPYL9-4 was a gene with specific functions in the MSPYL gene family. This prediction helps to discover new members of the MSLIM gene family.
Fig. 3.
Gene structure and conserved motifs of alfalfa PYL family. A Phylogenetic tree of 39 PYL genes in alfalfa. B Schematic distribution of conserved motifs in the MsPYL proteins. C Intron–exon structures of MsPYL genes. D The amino acid composition of each motif (S5 Table)
Analysis of cis-acting elements of alfalfa MsPYLs gene promoter and prediction of protein 3D structure
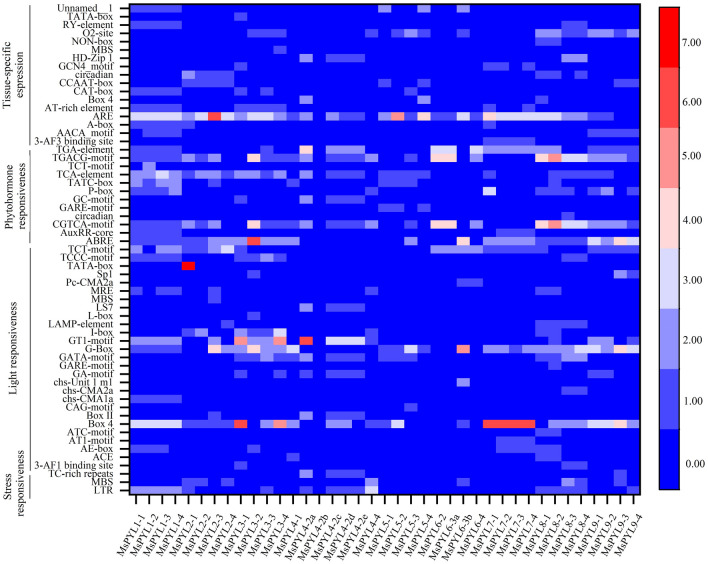
Promoter cis-acting elements were important binding regions of transcription initiation factors and played an important role in regulating gene expression. To further analyze the possible biological functions of MsPYL, the 1.2 kb sequence upstream of the alfalfa MsPYL gene promoter was used to predict the cis-elements using the PlantCARE database. It was predicted that there were many cis-elements related to transcription, cell cycle, development, hormone, and stress response in the promoter region of the alfalfa MsPYL gene, and some elements were closely related to seed specificity, meristem specificity, and endosperm specificity (Fig. 4, Table S6). In addition, we also found many elements related to hormone signaling pathways, such as methyl jasmonate (MeJA), abscisic acid (ABA), salicylic acid (SA), gibberellin (GA), and auxin (IAA). Among them, 29 MsPYL had methyl jasmonate response elements (TGACG-motif and CGTCA-motif), and 27 MsPYL had abscisic acid element ABRE, which indicates that most of MsPYL can participate in MeJA and ABA-mediated signaling pathways. The prediction also observed that these elements can participate in various abiotic stresses (mechanical damage, low-temperature, and drought). It was important to point out that all MsPYLs contain photo-responsive elements.
Fig. 4.
Promoter analysis of Medicago sativa L. PYL genes. The cis-acting elements in the promoter regions were obtained from the PlantCARE database. Stress responsiveness elements: LTR, MBS, TC-rich repeats; Light responsiveness elements:3-AF1 binding site, ACE, AE-box, AT1-motif, ATC-motif, Box 4, Box II, CAG-motif, chs-CMA1a, chs-CMA2a, chs-Unit 1 m1, GA-motif, GARE-motif, GATA-motif, G-Box, GT1-motif, I-box, LAMP-element, L-box, LS7, MBS, MRE, Pc-CMA2a, Sp1, TATA-box, TCCC-motif, TCT-motif; Phytohormone responsiveness elements: ABRE, AuxRR-core, CGTCA-motif, circadian, GARE-motif, GC-motif, P-box, TATC-box, TCA-element, TCT-motif, TGACG-motif, TGA-element; Tissue-specific expression elements: 3-AF3 binding site, AACA_motif, A-box, ARE,AT-rich element, Box 4, CAT-box, CCAAT-box, circadian, GCN4_motif, HD-Zip 1, MBS, NON-box, O2-site, RY-element, TATA-box, Unnamed_1.Different colors represent the number of cis-acting elements within the promoter, with red indicating a higher number and blue indicating a lower number
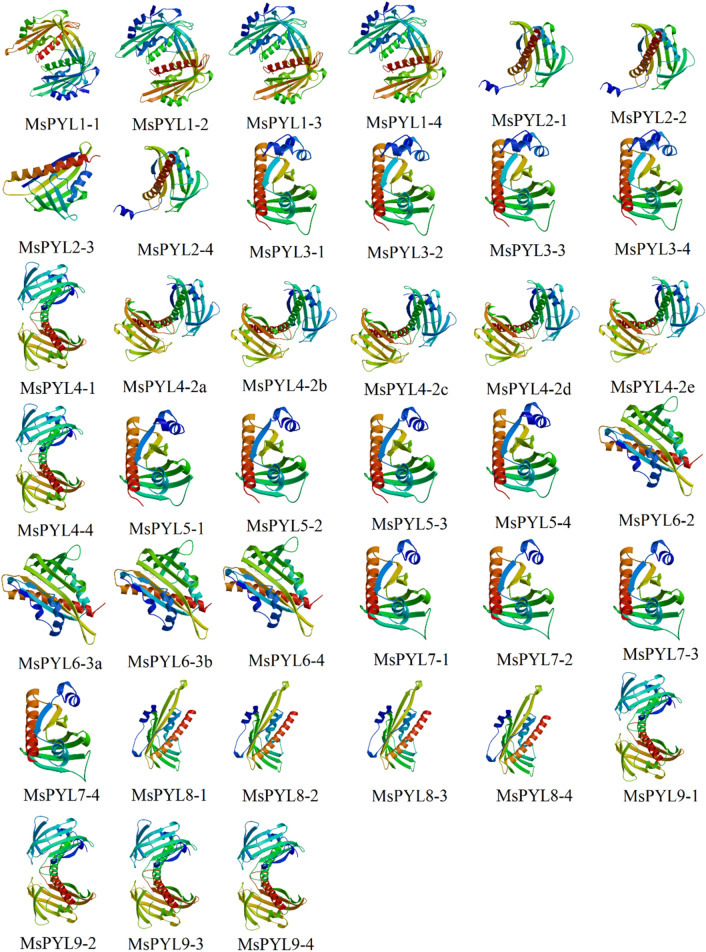
In this study, three-dimensional structural homology modeling was performed on the amino acid sequences of 39 MsPYL gene family members of alfalfa (Fig. 5). The online software Swiss-Model analysis showed that 39 MsPYL proteins had α-helix and β-sheet as their main structures. The validation of the MsPYL protein 3-D models obtained in this study was also carried out using Ramachandran plot, and the corresponding calculations were computed with the PROCHECK program. The Ramachandran plot results indicated that more than 90% of the residues of the MsPYL protein 3-D models were in favored and allowed regions (Supplementary file S7). The validation results using the Ramachandran plot revealed the stable conformation of constructed models with appropriate stereochemistry, indicating that these models can be used for further analyses. The tertiary structure of the amino acid sequence of its homologous genes was very similar, but there were exceptions. For example, the protein structure of MsPYL2-3 was significantly different from other genes. The tertiary structure of proteins of non-homologous genes had obvious differences, and these similarities or differences may be the reasons for their similar or different functions.
Fig. 5.
The 3D structure modeling of MsPYL proteins. The structure image was generated using the SWISS-MODEL software. The validation of the models obtained in this study was also carried out using Ramachandran plot (Supplementary file S7)
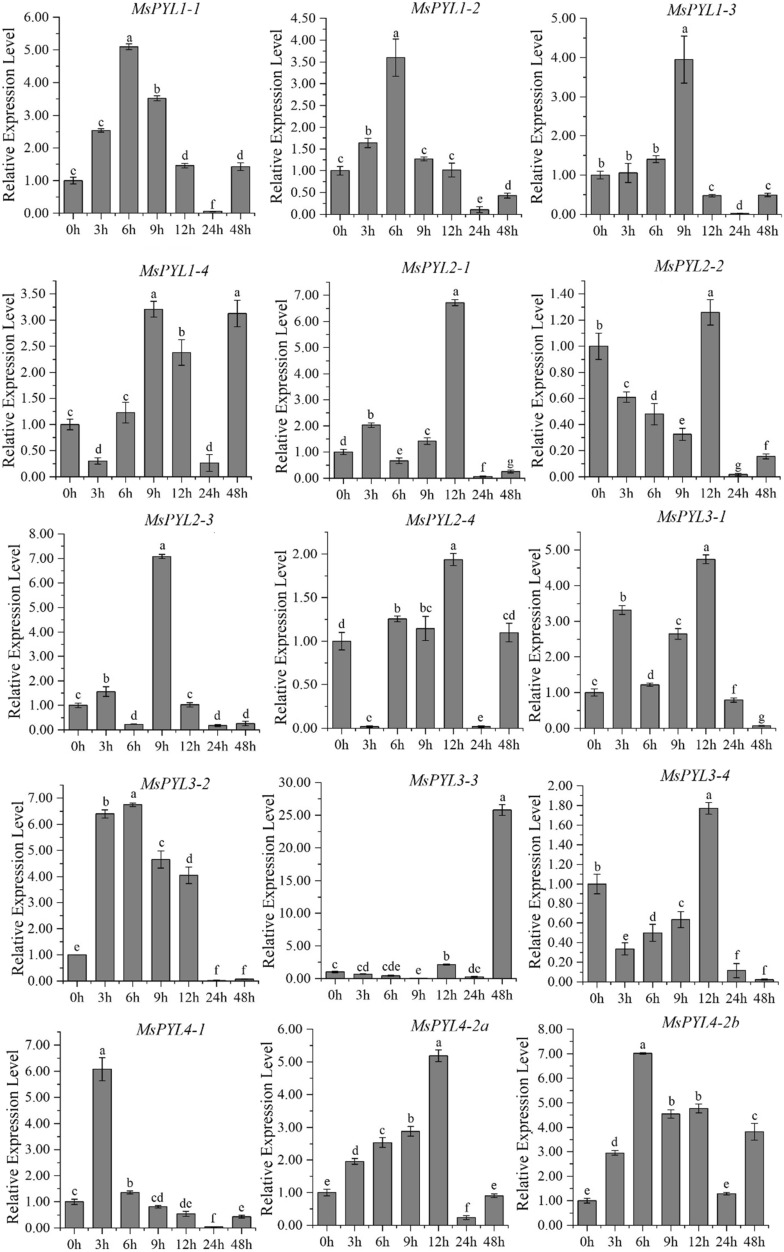
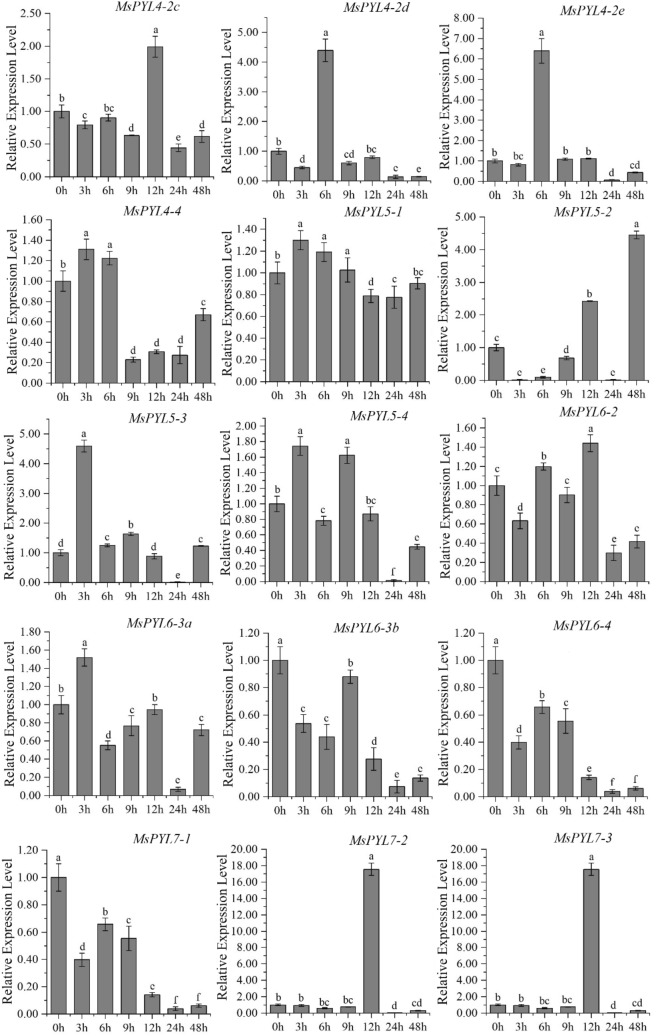
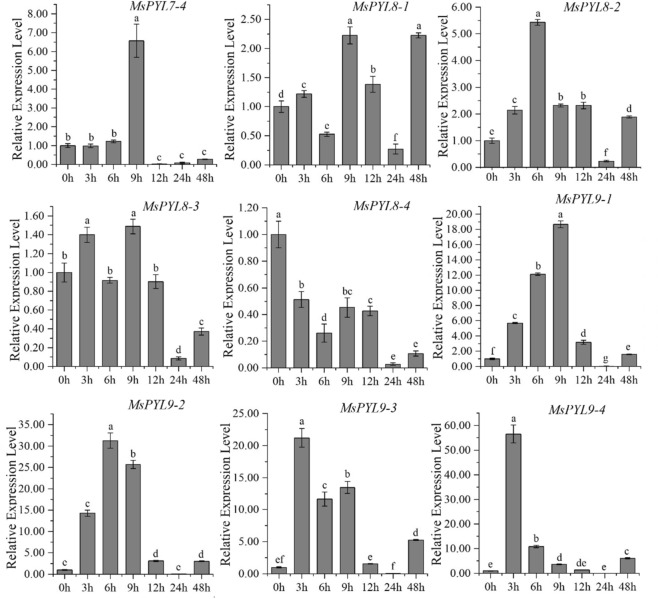
q-PCR analysis
To understand the possible role of alfalfa PYL genes under abiotic stress, we conducted a low-temperature (4 °C) induction expression analysis of MsPYL. In general, the genes of three sub-families of MsPYL family in alfalfa showed different expression patterns (Fig. 6, Table S8). These 39 genes can be induced to be expressed by low-temperature treatment, and most of the homologous genes showed the same expression pattern in response to stress. As shown in Fig. 6, as compared to the control (0 h), it has been observed that MsPYL4-1, MsPYL5-3, MsPYL5-4, MsPYL6-3a, MsPYL9-3, and MsPYL9-4 appeared with the highest value when low-temperature stress was induced for 3 h, indicating that these genes had a strong stress response. The expression levels of MsPYL1-1, MsPYL1-2, 3–2, MsPYL4-2b, MsPYL4-2d, MsPYL4-2e, MsPYL4-4, MsPYL5-1, MsPYL8-2, and MsPYL9-2 reached their maximum at 6 h of low-temperature induction. The expression levels of MsPYL6-3b, MsPYL6-4, MsPYL7-1, and MsPYL8–4 at each time point of low-temperature stress were lower compared to those of the control (0 h), indicating that these four genes had a significant response to low temperature stress. The expression of MsPYL1-3, MsPYL1-4, MsPYL2-3, MsPYL7-4, MsPYL8-1, MsPYL8-3, MsPYL9-1 peaked at 9 h, and the expression levels of MsPYL2-1, MsPYL2-2, MsPYL2-4, MsPYL3-1, MsPYL3-4, MsPYL4-2a, MsPYL4-2c, MsPYL6-2, MsPYL7-2, and MsPYL7-3 all reached their maximum at 12 h. The expression levels of MsPYL3-4, MsPYL4-2a, MsPYL4-2c, MsPYL6-2, MsPYL7-2, and MsPYL7-3 all reached their maximum at 12 h. It was important to note that MsPYL7-2 and MsPYL7-3; the expression level at 12 h was significantly higher compared to other time points. Only the expression of MsPYL3-3 and MsPYL5-2 reached the maximum after 48 h of stress treatment.
Fig. 6.
q-PCR expression analysis of 39 alfalfa PYL genes in response to low temperature treatment at 0, 3, 6, 9, 12, 24, 48 h of treatment. The vertical bars represent the standard error of the means of three independent replicates. Different letters indicate mean values at different sampling points after Tukey's pairwise comparison test
Discussion
PYL is a multi-gene family, which has been identified in Arabidopsis, corn, tomato, soybean, wheat, corn, poplar, rubber tree, strawberry, cotton, and other plants (Bai et al. 2019), but the systematic identification of PYL for alfalfa has not yet been reported. Tian et al. (2015) identified 13 PYL genes from rice and observed that some rice PYL members showed obvious tissue-specific expression patterns. At the same time, they also observed that these proteins can regulate ABA signals and improve tolerance to abiotic stress (Tian et al. 2015).
In this study, 39 MsPYL genes were identified in alfalfa, and divided into 3 subfamilies (Subfamily I, Subfamily II, and Subfamily III). Through phylogenetic and gene structure analysis, most genes in the same subfamily have similar exon or intron structures. We have observed a diversity of protein conserved motifs in alfalfa MsPYLs. MsPYLs in Subfamily I lack Motif 4, and the missing motifs are crucial to the structure of the protein. Due to the selective pressure during the evolution of the alfalfa genome, MsPYL genes in the same subfamily gradually formed different gene structures or protein motifs to adapt to complex environmental conditions. By comparing the PYL of alfalfa and other plants (rice, tobacco, and Arabidopsis), we explored the evolutionary relationship of the PYL gene family. Thirteen PYLs were observed in monocotyledonous rice, 14 and 29 PYLs were observed in dicotyledonous plants Arabidopsis and tobacco, respectively, and 39 PYLs were observed in alfalfa. The number of PYLs in alfalfa is far more than that in rice and Arabidopsis, the reason may be related to the auto-tetraploid genome and complicated evolution of alfalfa.
Gene duplication events are the outcome of polyploidization or tandem and fragment replication, which contribute to gene family expansion and genome evolution (Moore and Purugganan, 2005). In this study, there were 93 pairs of duplicated MsPYL genes in all subfamilies, and there were no tandem duplicated genes. In addition, 34 of the 93 MsPYL genes replicated are homologous genes, but not all MsPYL genes have homologous genes on homologous chromosomes 1, 2, 3, and 4. Studies have shown that some homologous genes may be lost during the process of genome polyploidization (Lynch et al. 2000). Therefore, gene duplication plays an important role in the expansion of the PYL gene family, and these new genes help to produce new biological functions (Conant and Wolfe, 2008).
The analysis of promoter homeopathic elements indicated that MsPYLs may be involved in the regulation of a variety of biological processes. In this study, alfalfa MsPYLs are closely related to plant hormones (MeJA, ABA, SA, GA, IAA) and various stresses (mechanical damage, low-temperature, and drought). It is speculated that the MsPYL genes may participate in the regulation of alfalfa growth and development and defense response. Studies have shown that over-expression of the PYL gene in Arabidopsis not only significantly increased the plant's sensitivity to ABA during seed germination, seedling growth, and stomatal opening and closing, but also increased the drought resistance of plants (Ma et al. 2009; Saavedra et al. 2010; Santiago et al. 2009). The rice ABA receptor OsPYL/RCAR5 has been reported to be able to positively regulate ABA signaling, including increasing the sensitivity of seeds to ABA during germination and seedling growth and improving the drought resistance and salt-alkali stress resistance of rice (Kim et al. 2012, 2014), that is consistent with our research conclusions. Therefore, the PYL gene can participate in the regulation of the growth and development of alfalfa and the stress defense response through specific hormone signaling pathways.
PYLs play an important role in many biological processes of plants. Alfalfa is often affected by low temperature and chilling damage and doesn’t show winter hardiness during frost and cold. If cold damage occurs and persists for a longer time during the seedling stage, the alfalfa seedlings might die. Plant cold tolerance is a measure of plant stress tolerance. To further illustrate that expression of alfalfa MsPYL gene family members can improve alfalfa’s ability to withstand adversity stress, and to explore whether MsPYL genes function in cold stress, we observed the gene expression patterns of MsPYL gene family of cold subjected alfalfa. The qRT-PCR data under low-temperature stress showed (Fig. 6) that the MsPYL genes can participate in the low-temperature stress response in alfalfa. When alfalfa suffers low-temperature stress, a large number of MsPYL genes are up-regulated. Some MsPYL genes showed a strong stress response after 3 h of stress, after which these MsPYL genes were all down-regulated. Most of the homologous genes showed similar expression patterns under low-temperature stress, indicating that these genes may have similar physiological functions. Certain alfalfa MsPYLs exhibit different expression patterns under low-temperature stress, which may be because they participate in different defense mechanisms against this stress, and more experiments, are needed to confirm.
The results of this study fill the gaps regarding the alfalfa ABA receptor MsPYL transcription factor family, laying a foundation for further research on family members. In short, this study provides basis for the future use of genetic engineering techniques to improve the agronomic traits of alfalfa, but the molecular mechanism of the role of alfalfa PYL gene family members in other stress related response needs to be further elucidated.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This research was financially supported by The Scientific Research Start-up Funds for Openly Recruited Doctors of Gansu Agriculture University (2017RCZX32); Research on the Coordinated Relationship between Land Urbanization and Population Cities (GSAU-ZL-2015-046) and The Youth Science and Technology Fund of Gansu Province (20JR5RA014).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xuelu Liu, Email: liuxl@gsau.edu.cn.
Yingbo Yang, Email: yyb_17929@163.com.
References
- Akhter Z, Bi Z, Ali K, Sun C, Fiaz S, Haider FU, Bai J. In response to abiotic stress, DNA methylation confers epigenetic changes in plants. Plants (basel) 2021;10(6):1096. doi: 10.3390/plants10061096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai G, Yang DH, Zhao Y, Ha S, Yang F, Ma J, Gao XS, Wang ZM, Zhu JK. Interactions between soybean ABA receptors and type 2C protein phosphatases. Plant Mol Biol. 2013;83:651–664. doi: 10.1007/s11103-013-0114-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai G, Xie H, Yao H, Li F, Chen X, Zhang Y, Xiao B, Yang J, Li Y, Yang DH. Genome-wide identification and characterization of ABA receptor PYL/RCAR gene family reveals evolution and roles in drought stress in Nicotiana tabacum. BMC Genomics. 2019;20:575. doi: 10.1186/s12864-019-5839-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Li GJ, Bressan RA, Song CP, Zhu JK, Zhao Y. Abscisic acid dynamics, signaling, and functions in plants. J Integr Plant Biol. 2020;62:25–54. doi: 10.1111/jipb.12899. [DOI] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet. 2008;9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- Danquah A, de Zelicourt A, Colcombet J, Hirt H. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol Adv. 2014;32(1):40–52. doi: 10.1016/j.biotechadv. [DOI] [PubMed] [Google Scholar]
- Fan W, Zhao M, Li S, Bai X, Li J, Meng H, Mu Z. Contrasting transcriptional responses of PYR1/PYL/RCAR ABA receptors to ABA or dehydration stress between maize seedling leaves and roots. BMC Plant Biol. 2018;16:99. doi: 10.1186/s12870-016-0764-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guzman M, Pizzio GA, Antoni R, Vera-Sirera F, Merilo E, Bassel GW, Fernández MA, Holdsworth MJ, Perez-Amador MA, Kollist H, Rodriguez PL. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell. 2012;24:2483–2496. doi: 10.1105/tpc.112.098574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Guzmán M, Rodríguez L, Lorenzo-Orts L, Pons C, Sarrión-Perdigones A, Fernández MA, Peirats-Llobet M, Forment J, Moreno-Alvero M, Cutler SR, Albert A, Granell A, Rodríguez PL. Tomato PYR/PYL/RCAR abscisic acid receptors show high expression in root, differential sensitivity to the abscisic acid agonist quinabactin, and the capability to enhance plant drought resistance. J Exp Bot. 2014;65:4451–4464. doi: 10.1093/jxb/eru219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon CS, Rajagopalan N, Risseeuw EP, Surpin M, Ball FJ, Barber CJ, Buhrow LM, Clark SM, Page JE, Todd CD, Abrams SR, Loewen MC. Characterization of Triticum aestivum abscisic acid receptors and a possible role for these in mediating fusairum head blight susceptibility in wheat. PLoS ONE. 2016;11:e0164996. doi: 10.1371/journal.pone.0164996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo AY, Zhu QH, Chen X, Luo JC. GSDS: a gene structure display server. Yi Chuan. 2007;29(8):1023–1026. doi: 10.1360/yc-007-1023. [DOI] [PubMed] [Google Scholar]
- Guo D, Zhou Y, Li HL, Zhu JH, Wang Y, Chen XT, Peng SQ. Identification and characterization of the abscisic acid (ABA) receptor gene family and its expression in response to hormones in the rubber tree. Sci Rep. 2017;7:45157. doi: 10.1038/srep45157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewage KAH, Yang JF, Wang D, Hao GF, Yang GF, Zhu JK. Chemical Manipulation of Abscisic Acid Signaling: A New Approach to Abiotic and Biotic Stress Management in Agriculture. Adv Sci (weinh) 2020;7(18):2001265. doi: 10.1002/advs.202001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooft RW, Sander C, Vriend G. Objectively judging the quality of a protein structure from a Ramachandran plot. Comput Appl Biosci. 1997;13(4):425–430. doi: 10.1093/bioinformatics/13.4.425. [DOI] [PubMed] [Google Scholar]
- Hu S, Wang FZ, Liu ZN, Liu YP, Yu XL. ABA signaling mediated by PYR/PYL/RCAR in plants. Yi Chuan. 2012;34:560–572. doi: 10.3724/sp.j.1005.2012.00560. [DOI] [PubMed] [Google Scholar]
- Kim H, Hwang H, Hong JW, Lee YN, Ahn IP, Yoon IS, Yoo SD, Lee S, Lee SC, Kim BG. A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J Exp Bot. 2012;63:1013–1024. doi: 10.1093/jxb/err338. [DOI] [PubMed] [Google Scholar]
- Kim H, Lee K, Hwang H, Bhatnagar N, Kim DY, Yoon IS, Byun MO, Kim ST, Jung KH, Kim BG. Overexpression of PYL5 in rice enhances drought tolerance, inhibits growth, and modulates gene expression. J Exp Bot. 2014;65:453–464. doi: 10.1093/jxb/ert397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko V, Konrad KR, Dietrich P, Roelfsema MR, Hedrich R. Cytosolic abscisic acid activates guard cell anion channels without preceding Ca2+ signals. Proc Natl Acad Sci U S A. 2005;102(11):4203–4208. doi: 10.1073/pnas.0500146102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lynch M, Force A (2000) The probability of duplicate gene preservation by subfunctionalization. Genetics.154:459–73. PMID: 10629003; PMCID: PMC1460895. [DOI] [PMC free article] [PubMed]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- Melcher K, Ng LM, Zhou XE, Soon FF, Xu Y, Suino-Powell KM, Park SY, Weiner JJ, Fujii H, Chinnusamy V, Kovach A, Li J, Wang Y, Li J, Peterson FC, Jensen DR, Yong EL, Volkman BF, Cutler SR, Zhu JK, Xu HE. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature. 2009;462:602–608. doi: 10.1038/nature08613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa T, Fujita Y, Yamaguchi-Shinozaki K, Tanokura M. Structure and function of abscisic acid receptors. Trends Plant Sci. 2012;18:259–266. doi: 10.1016/j.tplants.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Moore RC, Purugganan MD. The evolutionary dynamics of plant duplicate genes. CurrOpin Plant Biol. 2005;8:122–128. doi: 10.1016/j.pbi.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Nian L, Liu X, Yang Y, Zhu X, Yi X, Haider FU. Genome-wide identification, phylogenetic, and expression analysis under abiotic stress conditions of LIM gene family in Medicago sativa L. PLoS ONE. 2021;16:e0252213. doi: 10.1371/journal.pone.0252213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, Schroeder JI, Getzoff ED. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science. 2010;326:1373–1379. doi: 10.1126/science.1181829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC, Lee S, Caddell DF, Cutler SR, Chory J, Yates JR, Schroeder JI. PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 2010;61:290–299. doi: 10.1111/j.1365-313X.2009.04054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyaz AD, Insha A, Wasia W, Shafiq A, Wani AB, Shikari SHW, Khalid ZM. Abscisic acid: A key regulator of abiotic stress tolerance in plants. Plant Gene. 2017;11:106–111. doi: 10.1016/j.plgene.2017.07.003. [DOI] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, Alfred SE, Bonetta D, Finkelstein R, Provart NJ, Desveaux D, Rodriguez PL, McCourt P, Zhu JK, Schroeder JI, Volkman BF, Cutler SR. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson FC, Burgie ES, Park SY, Jensen DR, Weiner JJ, Bingman CA, Chang CE, Cutler SR, Phillips GNJ, Volkman BF. Structural basis for selective activation of ABA receptors. Nat Struct Mol Biol. 2010;17:1109–1113. doi: 10.1038/nsmb.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilet PE. Abscisic acid as a root growth inhibitor: Physiological analyses. Planta. 1975;122:299–302. doi: 10.1007/BF00385279. [DOI] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E. ABA perception and signalling. Trends Plant Sci. 2010;15(7):395–401. doi: 10.1016/j.tplants.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Rezaul IM, Baohua F, Tingting C, Weimeng F, Caixia Z, Longxing T, Guanfu F. Abscisic acid prevents pollen abortion under high-temperature stress by mediating sugar metabolism in rice spikelets. Physiol Plant. 2019;165(3):644–663. doi: 10.1111/ppl.12759. [DOI] [PubMed] [Google Scholar]
- Ryynänen L. Effect of Abscisic Acid, Cold Hardening, and Photoperiod on Recovery of Cryopreserved in Vitro Shoot Tips of Silver Birch. Cryobiology. 1998;36(1):32–39. doi: 10.1006/cryo.1997.2067. [DOI] [PubMed] [Google Scholar]
- Saavedra X, Modrego A, Rodríguez D, González-García MP, Sanz L, Nicolás G, Lorenzo O. The nuclear interactor PYL8/RCAR3 of Fagus sylvatica FsPP2C1 is a positive regulator of abscisic acid signaling in seeds and stress. Plant Physiol. 2010;152:133–150. doi: 10.1104/pp.109.146381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, Cutler SR, Rodriguez PL, Márquez JA. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature. 2009;462:665–668. doi: 10.1038/nature08591. [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D. GUARD CELL SIGNAL TRANSDUCTION. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:627–658. doi: 10.1146/annurev.arplant.52.1.627. [DOI] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S, Makkar HP, Becker K. Alfalfa saponins and their implication in animal nutrition. J Agric Food Chem. 1998;46:131–140. doi: 10.1021/jf970389i. [DOI] [PubMed] [Google Scholar]
- Smart CC, Fleming AJ, Chaloupkova K, Hanke DE. The Physiological role of abscisic acid in eliciting Turion Morphogenesis. Plant Physiol. 1995;108:623–632. doi: 10.1104/pp.108.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Wang Z, Li X, Lv T, Liu H, Wang L, Niu H, Bu Q. Characterization and functional analysis of pyrabactin resistance-like abscisic acid receptor family in rice. Rice (n Y) 2015;8:28. doi: 10.1186/s12284-015-0061-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahisalu T, Kollist H, Wang YF, Nishimura N, Chan WY, Valerio G, Lamminmäki A, Brosché M, Moldau H, Desikan R, Schroeder JI, Kangasjärvi J. SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature. 2008;452(7186):487–491. doi: 10.1038/nature06608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XQ, Wu WH, Assmann SM. Differential responses of abaxial and adaxial guard cells of broad bean to abscisic acid and calcium. Plant Physiol. 1998;118(4):1421–1429. doi: 10.1104/pp.118.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen ZH, Zhang B, Hills A, Blatt MR. PYR/PYL/RCAR abscisic acid receptors regulate K+ and Cl- channels through reactive oxygen species-mediated activation of Ca2+ channels at the plasma membrane of intact Arabidopsis guard cells. Plant Physiol. 2013 doi: 10.1104/pp.113.219758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav SK, Kumar VVS, Verma RK, Yadav P, Saroha A, Wankhede DP, Chinnusamy V. Genome-wide identification and characterization of ABA receptor PYL gene family in rice. BMC Genomics. 2020;21(1):1–27. doi: 10.1186/s12864-020-07083-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Zhao T, Rao P, Gao K, Yang X, Chen Z, An X. Transcriptome profiling of Populus tomentosa under cold stress. Ind Crops Prod. 2019;135:283–293. doi: 10.1016/j.indcrop.2019.04.056. [DOI] [Google Scholar]
- Zhang H, Han W, De Smet I, Talboys P, Loya R, Hassan A, Rong H, Jürgens G, Paul Knox J, Wang MH. ABA promotes quiescence of the quiescent centre and suppresses stem cell differentiation in the Arabidopsis primary root meristem. Plant J. 2010;64(5):764–774. doi: 10.1111/j.1365-313X.2010.04367.x. [DOI] [PubMed] [Google Scholar]
- Zhang G, Lu T, Miao W, Sun L, Tian M, Wang J, Hao F. Genome-wide identification of ABA receptor PYL family and expression analysis of PYLs in response to ABA and osmotic stress in Gossypium. PeerJ. 2017;5:e4126. doi: 10.7717/peerj.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Gao J, Im KJ, Chen K, Bressan RA, Zhu JK. Control of plant water use by ABA induction of senescence and dormancy: An overlooked lesson from evolution. Plant Cell Physiol. 2017;58:1319–1327. doi: 10.1093/pcp/pcx086. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Zhang Z, Gao J, Wang P, Hu T, Wang Z, Hou YJ, Wan Y, Liu W, Xie S, Lu T, Xue L, Liu Y, Macho AP, Tao WA, Bressan RA, Zhu JK. Arabidopsis Duodecuple Mutant of PYL ABA Receptors Reveals PYL Repression of ABA-Independent SnRK2 Activity. Cell Rep. 2018;23:3340–3351.e5. doi: 10.1016/j.celrep.2018.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YP, Wu JH, Xiao WH, Chen W, Chen QH, Fan T, Xie CP. Tian CE (2018) Arabidopsis IQM4, a Novel Calmodulin-Binding Protein, Is Involved With Seed Dormancy and Germination in Arabidopsis. Front Plant Sci. 2018;5(9):721. doi: 10.3389/fpls.2018.00721. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.