Abstract
Plant-specific BURP domain-containing proteins have an essential role in the plant's development and stress responses. Although BURP domain-containing proteins have been identified in several plant species, genome-wide analysis of the BURP gene family has not been investigated in the common bean. In the present study, we identified 11 BURP family members in the common bean (Phaseolus vulgaris) genome with a comprehensive in silico analysis. Pairwise alignment and phylogenetic analyses grouped PvBURP members into four subfamilies [RD-22 like (3), PG1β-like (4), BNM2-like (3), and USP-like (1)] according to their amino acid motifs, protein domains and intron–exon structure. The physical and biochemical characteristics of amino acids, motif and intron–exon structure, and cis-regulatory elements of BURPs members were determined. Promoter regions of BURP members included stress, light, and hormone response-related cis-elements. Therefore, expression profiles of PvBURP genes were identified with in silico tools and qRT-PCR analyses under stress (salt and drought) and hormone treatment (ABA, IAA) in the current study. While significant activity changes were not observed in BURP genes in RNA-seq data sets related to salt stress, it was determined that some BURP genes were expressed differently in those with drought stress. We identified 12 different miRNA, including miRNA395, miRNA156, miRNA169, miRNA171, miRNA319, and miRNA390, targeting the nine PvBURP genes using two different in silico tools based on perfect or near‐perfect complementarity to their targets. Here we present the first study to identify and characterize the BURP genes in common bean using whole-genome analysis, and the findings may serve as a reference for future functional research in common bean.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12298-021-01052-9.
Keywords: ABA, Abiotic stress, BURP, miRNA, IAA
Introduction
Common bean (Phaseolus vulgaris L.) is one of the most important foods used for direct human consumption providing essential proteins, complex carbohydrates, dietary fibers, minerals (Fe, Zn), and vitamins (Hayat et al. 2014). It also has disease-preventing and health-promoting effects on humans due to the existence of many phytochemicals such as polyphenolic compounds, fibers, lectins, and flavonoids in its seeds (McClean and Raatz 2017). As a legume species, it enhances soil fertility by fixing atmospheric nitrogen. All these qualities make common bean a good food of choice for more than 300 million people living in lower-income counties found in Asia, Eastern Africa, and Latin America (Cortés et al. 2013).
Besides its high demand and production, this crop is threatened by a series of abiotic stress. Common bean is known to be more susceptible to water shortages during the flowering and grain-filling stages. Even moderate levels of water deficit were reported to cause a reduction in common bean biomass, seed yield, and nitrogen fixation (Fageria et al. 2010). Common bean is primarily grown in drought-prone areas, and prolonged water deficiency is reported to create a global and endemic threat for most bean production areas (Caldas et al. 2016). Global warming-dependent climatic stresses became more widespread and intense in recent years. Effects of these stresses on the common bean production are most frequently handled in terms of water shortage and drought-dependent salinity (Pareek et al. 2020). Higher temperatures, combined with lower rainfall, are started to exacerbate evapotranspiration and drought in especially bean-producing areas of Asia, Latin America, and southern Africa (Darkwa et al. 2016). It is known that the accumulation of salts and ions in upper soil layers leads to osmotic stress and ion toxicity in plants during moderate or severe drought conditions. Therefore, global warming is estimated to change the adaptive altitudinal range of bean genotypes, accelerate the decomposition of soil organic matter (mineralization) and ion accumulation (salinization), combining the abiotic stresses even more acute in the near future. These estimates were verified with some studies that 73% and 40% of common bean production areas had been reported to be affected by drought and metal toxicity, respectively (Al Hassan et al. 2016; Arteaga et al. 2020; Beebe et al. 2009; Celmeli et al. 2018; Dipp et al. 2017; dos Santos Neto et al. 2020; Fageria et al. 2010; Lizana et al. 2006). Common bean can cope with these stress factors by activating tolerance genes and changing their cellular, biochemical and molecular mechanisms. Therefore, identifying the genes and transcription factors responsible for stress tolerance is highly important to develop resistant common bean cultivars and maintain their productivity.
Increasing evidence indicates that a gene family encoding BURP domain-containing proteins has important functions in plant development, metabolism, and stress tolerance. Members of the BURP protein family are distinguished from other proteins by the presence of a conserved amino acid motif located at the N-terminus. The BURP gene family derives its name from the four members of its family; the microsporogenesis-specific protein (BNM2) of Brassica napus (Boutilier et al. 1994), the unknown seed protein (USP) of Vicia faba (Bassüner et al. 1988), the responsive to dehydration 22 (RD22) in Arabidopsis thaliana (Yamaguchi-Shinozaki et al. 1993) and the non-catalytic β-subunit of the polygalacturonase isozyme 1 (PG1β) in Lycopersicon esculentum (Hattori et al. 1998; Zheng et al. 1992). The structure of BURP domain-containing proteins has three conserved modules: a conserved region containing four repeats of cysteine-histidine motifs following a single phenylalanine-glycine residue at the C-terminal region, a member-specific variable internal region, and a signal peptide at the hydrophobic N-terminal domain (Hattori et al. 1998). Depending on the variable region, the BURP domain-containing proteins are classified into seven subfamilies; BNM2-like, USP-like, RD22- like, PG1β-like, BURPV, BURPVII, and BURPVIII (Granger et al. 2002).
Several gene families encoding BURP domain-containing proteins have been identified and are found to be unique to plant species. Expression of these genes was associated with significant developmental and tolerance metabolisms in plants. For example, BNM2 expression was first linked to microspore embryogenesis in Brassica napus L. (Boutilier et al. 1994) and then realized to be related to seed formation due to its localization in seed protein storage vacuoles (Teerawanichpan et al. 2009). Another BURP domain-containing protein in Vicia faba L. called VfUSP is reportedly involved in regulating the early development of zygotic embryogenesis. In Panicum maximum, ASG1 was found to control the formation of apospory initial cells (Bassüner et al. 1988; Chen et al. 1999). A BURP protein in soybean (SCB1) was reported to be functional in the differentiation of seed coat parenchyma cells (Batchelor et al. 2002). In cotton, the BURP domain-containing proteins are expressed during fiber initiation, development, and elongation stages (Lee et al. 2007).
In addition to their contribution to plant development, some BURP domain-containing proteins (RD22-like and BNM2-like subfamilies) have been reported to show co-expression with stress conditions. Especially, the RD22 gene exhibited a strong molecular link between abscisic acid (ABA) and abiotic stress responses in plants. For instance, RD22-like genes of Arabidopsis and Vitis vinifera were significantly expressed in response to salt, drought, and ABA stress (Abe et al. 1997; Matus et al. 2014; Yamaguchi-Shinozaki et al. 1993). Similarly, RD22-like genes in Gossypium hirsutum (GhBURPs) were induced by ABA and salicylic acid exposure (Sun et al. 2019). Xun et al. (2019) identified some BURP domain-containing genes in soybean and indicated their upregulation in response to ABA exposure and soybean mosaic virus infection.
Some reports indicate the essential roles of BURP domain-containing proteins in metal toxicity tolerance in plants. SALI3-2, a BURP gene in soybean, was found to be induced by excess cadmium and copper exposure. This gene was estimated to have sequestering effects on metal ions in the soybean roots (Tang et al. 2014). Similarly, two other BURP genes, Sali5-4a and Sali3–2, found in soybean, were overexpressed by aluminum stress and Sali3–2 was also involved in the salt tolerance (Tang et al. 2014). In poplar, 18 BURP family genes, named PtBURPs, were identified and characterized according to their physical positions on the P. trichocarpa chromosomes (Shao et al. 2011). Genome-wide expression analysis of poplar verified the essential roles of PtBURPs on metal detoxification and drought tolerance (Yıldırım and Kaya 2017).
These studies represented that BURP domain-containing family genes had a significant role in plant development, abiotic stress response, and phytohormone signaling pathways. Although these plant-specific genes have been characterized in several plant species, a genome-wide analysis of BURP genes in common bean has not been reported to this date. The whole-genome sequence of P. vulgaris was released in 2014 (Schmutz et al. 2014), and now it is possible to analyze the entire family of common bean BURP domain-containing proteins. In the current study, 11 putative BURP genes were characterized with phylogenetic analysis, structural analysis, and expression profile analysis. The transcript levels of all 11 genes were determined under drought and salt stress as well as ABA and IAA hormone treatments. This study is the first report on genome-wide identification of PvBURP genes in common bean and characterization of their function on development and stress response of the common bean.
Material and methods
Identification of BURP members in common bean
We used two complementary methods to identify the genes encoding the BURP domain-containing proteins in common bean genome. In the first step, the Hidden Markov Model (HMM) profile of the BURP domain (PF03181) was obtained from Pfam30.0 (Finn et al. 2016). This profile information was used as a query to identify candidate BURPs from the bean genome using HMMER3.0 (Finn et al. 2015). In the second step, we made a keyword search in the Phytozome v12 database (https://phytozome.jgi.doe.gov/pz/portal.html) to find out other candidate BURP genes that may be overlooked from the first step. The presence of the BURP domain in a potential gene detected in Phaseolus vulgaris was further confirmed in Pfam (http://pfam.xfam.org/) and SMART databases (http://smart.embl-heidelberg.de/). We removed protein sequences that do not contain the BURP domain or have an uncertain domain belonging to other protein families. Finally, these remaining protein sequences were considered as members of the BURP family in P.vulgaris and used in subsequent analysis. Chromosome locations and protein-coding sequences (CDS) of all candidate PvBURP genes were downloaded from the Phytozome v12 database (https://phytozome.jgi.doe.gov/pz/portal.html#). We also determined the molecular and physicochemical properties of PvBURP genes by calculating the Mw, pI, instability index, and GRAVY using the ProtParam tool on the ExPASy server (https://web.expasy.org/protparam/). The PROSOII tool was used to predict the solubility of candidate BURP proteins based on their sequences (Smialowski et al. 2012).
Phylogenetic analysis
The amino acid sequences of BURPs from Arabidopsis thaliana, Brachypodium distachyon, Cucumis sativus, Glycine max, Medicago truncatula, Oryza sativa, Sorghum bicolor, and Zea mays were retrieved from the Phytozome v12 database to evaluate the expansion of BURP encoding genes within different species. The genes of BURPs were named based on numbering and their sequence homology with A. thaliana ortholog genes. Multiple sequence alignment was carried out by ClustalW 2.0 program. The neighbor-joining method-based phylogenetic tree was constructed with the bootstrap test (1000 replicates) and Jones-Taylor-Thornton (JTT) model in MEGAX. Phylogenetic trees were visualized with ITOL v3 (http://itol.embl.de/).
Sequence analysis of PvBURP genes
The intron/exon, motif, and domain structure of PvBURPs were shown using Gene Structure View in TBtools software (https://github.com/CJ-Chen/TBtools) (Chen et al. 2018). Genomic DNA sequences and CDS of PvBURP genes were downloaded from the Phytozome v12 database (https://phytozome.jgi.doe.gov/pz/portal.html#) to determine their intron–exon structures. Conserved motifs in PvBURP proteins were analyzed by using the MEME suite (http://meme-suite.org/tools/meme) tool. Conserved motifs were analyzed by choosing 15 motifs and repeating any number of motifs. NCBI Batch-CD Search tool was used to show potential BURP domains (Lu et al. 2020). We visualized the distribution of PvBURP genes on chromosomes using TBtools software (https://github.com/CJ-Chen/TBtools). Locations of genes on chromosomes were determined with Gene on Genome from Fasta application using Blast. Potential cis-acting regulatory DNA elements (cis-elements) in the promotor sequences of PvBURP genes were analyzed using the place database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) with 2000 bp of upstream region of the predicted CDS. Putative miRNAs targeting PvBURP genes were estimated using psRNATarget server based on miRNA-target complementary match patterns (Dai et al. 2018). Arabidopsis thaliana, Glycine max, Populus trichocarpa, Vitis vinifera, and Brachypodium distachyon were selected for miRNA analysis. CLC Genomics Workbench software was used to assess the positions of conserved BURP domains and potential signal peptides of these proteins. In this study, the sequence of all candidate miRNA identified in Phaseolus vulgaris genome was retrieved from Plant miRNA Encyclopedia (PmiREN, http://www.pmiren.com/) (Guo et al. 2019). Potential miRNAs targeting the PvBURP genes are determined using the web-based psRNA Target Server (http://plantgrn.noble.org/psRNATarget) and Miranda with their default parameters (Enright et al. 2003).
Plant Genome Duplication Database (Lee et al. 2013) and BLASTP were used to identify gene duplications. Duplicated PvBURP genes found within the same chromosome were accepted as tandem duplication. For segmental duplications, the BLASTP search was performed against all identified peptide sequences of PvBURPs in the common bean. As potential anchors, the top five matches were taken into consideration according to their E-value (≤ 1e−05), and then, MCScan was utilized to determine their collinear blocks (Wang et al. 2012). In evolutionary analysis, ratio of the nonsynonymous mutation rate to the synonymous mutation rate (Ka/Ks) was calculated using TBtools software. First, BLAST analysis of PvBURP protein sequences was performed in the Phytozome v12 database. Sequences with a similarity of over 60% as a result of BLAST were obtained. At the end of this BLAST process, a tab-delimited text file was created, and Ka/Ks calculation was then performed in TBtools software.
Prediction of 3D protein homology, transmembrane helix, and sub-cellular localization
Subcellular localization of PvBURPs was predicted with the Plant-mPLoc server (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/) (Chou and Shen 2008). The Phyre2 server (Protein Homology / Analogy Recognition Engine) was used to estimate BURP proteins' 3D structure (Kelley et al. 2015). All PvBURP protein sequences have been downloaded from Phytozome v12 database (https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Pvulgaris). Then, PvBURPs protein sequences were analyzed in the Phyre2 server with an "intensive" mode to define the 3D structure. Trans-membrane helical domains were predicted using and TMHMM (http://www.cbs.dtu.dk/services/TMHMM/) (Krogh et al. 2001).
Plant material and stress treatments
A Turkish common bean genotype named 'İspir' was used in the study for gene expression analysis of PvBURPs under various stress conditions. İspir seeds were firstly surface sterilized in a 5% sodium hypochlorite solution and then planted in pots filled with vermiculite. Plants were grown in a fully-controlled growth chamber at 24 °C supplemented with 16 h light and 8 h dark photoperiod. After the plants were grown for 4 weeks, they were subjected to drought, salinity, and hormone treatments by applying polyethylene glycol 6000 (PEG), NaCl, ABA, and IAA. Salt and drought stress was subjected to the plants by adding 200 mM NaCl and PEG (20%) into the Hoagland's solution, respectively. Hormone treatment was achieved by spraying with 100 µM ABA and 100 µM IAA to the leaves. Stress and hormone-treated leaves and roots samples were collected at 6, 12, 24, 48, 60th hour and seventh day after stress treatment and stored at − 80 °C until their usage in RNA isolation.
RNA isolation and gene expression analysis
We isolated total RNA with the RNeasy Plant Mini Kit (Qiagen) following the manufacturer’s protocol. RNA quality was checked on NanoDrop ™ 2000/2000c spectrophotometer and on a 1.5% (w/v) agarose gel. The first strand of cDNA was synthesized with the iScript ™ cDNA Synthesis Kit. Tissue, hormone, and stress-related expression of 11 PvBURP genes were measured with qRT-PCR analysis performed on the Agilent Mx3000P device with Solis BioDyne 5 × HOT FIREPol® EvaGreen® qPCR Mix Plus (ROX). qRT-PCR conditions were carried out at 95 °C for 15 min, at 95 °C for 15 s, at 60 °C for 20 s, and at 72 °C for 20 s. Relative expression was calculated according to the 2−ΔΔCt method. Primers of 11 PvBURP genes used in this study are shown in Table S1.
In silico expression analysis
The expression profiles of PvBURP genes were also determined using eight RNA-seq data sets that were previously obtained from different genotypes in response to salt drought and pathogen stress. To evaluate the expression profiles of PvBURPs in different tissues under different conditions, the raw RNA-seq data sets were downloaded from NCBI Sequence Read Archive (SRA) under the accession number; PRJNA327176 and PRJNA508605 for drought treatment; PRJNA656794, PRJNA558376, PRJNA574280, PRJNA691982 for salt stress and PRJNA574280 for pathogen infection. The transcriptional analysis of downloaded files was done via CyVerse (https://www.cyverse.org/) and Galaxy (usegalaxy.eu), including virtual bioinformatics tools. The mapping of reads was done with the HISAT2 tool. Transcript assembly and differential expression analyses were performed with Stringtie 1.3.3 and Ballgown, respectively. The genes having fold change value log2 > 1 and a p value < 0.05 were accepted as differentially expressed genes (DEGs). Heatmap based on the log2 FC was prepared using TBTools. For this analysis, eight different comparisons were made. To determine the expression level of BURPs under salt stress, the first comparison was carried out with data previously obtained from salt-treated leaf explants of T43 (sensitive) and Ispir (resistant) common bean genotypes (PRJNA656794). The data belonging to root samples of the same genotypes were used for the second comparison. The third expression analysis was made by comparing the data obtained from the salt-tolerant Ispir's root tissues grown under both control and salt stress conditions (PRJNA656794). The transcription data obtained by applying salt stress to the salt-tolerant and the sensitive genotype at the bud stage were used for the fourth comparison to estimate the expression level of PvBURPS (PRJNA558376). In the fifth expression analysis, the data obtained by applying salt stress to the salt-tolerant bean genotype in the sprout stage were used (PRJNA691982). The sixth and seventh expression analyses were made to find the response of PvBURP genes under drought stress. The data used for these comparisons were obtained by RNA sequencing of drought-tolerant Pinto Saltillo (PRJNA508605) and Perola (PRJNA327176) genotypes grown under control and drought stress. The last comparison was made with data obtained from a library prepared with a common bean plant infected by the fungal pathogen Sclerotinia sclerotiorum (strain 1980) (PRJNA574280). Tissue-specific expression patterns of BURPs were retrieved from Phytozome v12.
Results
Genome-wide identification of BURP genes from the common bean genome
To identify the BURP family members in the common bean, we used different approaches included in the HMM search. After manually removing sequences containing a missing BURP domain, 11 putative PvBURP genes named PvBURP1-PvBURP11 were identified, depending on their chromosomal location (Table 1). We have found that PvBURP genes in the common bean genome vary widely in their length, MW, and pI value. In this context, gene lengths ranged from 1339 (PvBURP8) to 3379 bp (PvBURP7), MWs from 29.88 (PvBURP11) to 69.51 kDa (PvBURP8), and pI values from 5.75 (PvBURP6) to 9.03 (PvBURP8). The prediction of subcellular localization made by mGOASVM indicated that PvBURP proteins are active in golgi, chloroplast and cell walls. According to the instability index (II), most of the PvBURP proteins (8 out of 11) were determined to be stable in a test tube. Protein solubility prediction of PvBURPs based on amino acid sequence indicated that approximately 91% of those are insoluble in Escherichia coli. When the amino acid composition of these 11 BURP proteins is examined, it is seen that the most abundant amino acid is Ser (S).
Table 1.
Molecular and physicochemical properties of PvBURP genes
| Gen ID | Phytozome Identfier | Chromosomes | Start and end positions (bp) | Length (bp) | CDS (bp) | Protein length (aa) | NCBI Accession number |
|---|---|---|---|---|---|---|---|
| PvBURP1 | Phvul.002G071500 | Chr02 | 9480419–9483312 | 2894 | 1881 | 626 | XM_007157400.1 |
| PvBURP2 | Phvul.003G052000 | Chr03 | 6643545–6646040 | 2496 | 1875 | 624 | XM_007153571.1 |
| PvBURP3 | Phvul.008G158600 | Chr08 | 43164301–43167172 | 2872 | 1023 | 340 | XM_007140936.1 |
| PvBURP4 | Phvul.008G210500 | Chr08 | 55885018–55887616 | 2599 | 1836 | 611 | XM_007141548.1 |
| PvBURP5 | Phvul.009G024400 | Chr09 | 5998841–6000194 | 1354 | 939 | 312 | XM_007136113.1 |
| PvBURP6 | Phvul.009G024700 | Chr09 | 6024647–6026346 | 1700 | 1110 | 369 | XM_007136116.1 |
| PvBURP7 | Phvul.009G105300 | Chr09 | 16335114–16338492 | 3379 | 1110 | 369 | XM_007137102.1 |
| PvBURP8 | Phvul.009G179500 | Chr09 | 26920688–26923132 | 2445 | 1884 | 627 | XM_007138028.1 |
| PvBURP9 | Phvul.009G180600 | Chr09 | 27156514–27159612 | 3099 | 1341 | 446 | XM_007138042.1 |
| PvBURP10 | Phvul.011G046300 | Chr11 | 4156692–4158030 | 1339 | 903 | 300 | XM_007131785.1 |
| PvBURP11 | Phvul.011G182200 | Chr11 | 49491661–49493447 | 1787 | 798 | 265 | XM_007133419.1 |
| Gen ID | pI | Molecular weight (Da) | Instability index | Stable or unstable | GRAVY | Solubility/Score | Subcellular location |
|---|---|---|---|---|---|---|---|
| PvBURP1 | 8.34 | 68435.07 | 29.21 | Stable | − 0.499 | Insoluble/0.353 | Cell wall |
| PvBURP2 | 6.44 | 68108.46 | 31.23 | Stable | − 0.500 | Insoluble/0.312 | Cell wall |
| PvBURP3 | 8.12 | 36925.07 | 32.18 | Stable | − 0.266 | Insoluble/0.336 | Cell wall/Chloroplast |
| PvBURP4 | 8.52 | 67265.46 | 25.42 | Stable | − 0.562 | Insoluble/0.354 | Cell wall |
| PvBURP5 | 6.99 | 35203.58 | 49.69 | Unstable | − 0.196 | Insoluble/0.224 | Cell wall |
| PvBURP6 | 5.75 | 42198.78 | 34.37 | Stable | − 0.588 | Soluble/0.526 | Nucleus |
| PvBURP7 | 8.56 | 40277.16 | 31.17 | Stable | − 0.107 | Insoluble/0.301 | Cell wall |
| PvBURP8 | 9.03 | 69512.12 | 34.71 | Stable | − 0.549 | Insoluble/0.330 | Cell wall |
| PvBURP9 | 6.23 | 51213.07 | 55.90 | Unstable | − 0.389 | Insoluble/0.336 | Cell wall/Chloroplast |
| PvBURP10 | 6.12 | 34038.78 | 46.29 | Unstable | − 0.331 | Insoluble/0.412 | Cell wall |
| PvBURP11 | 6.83 | 29885.33 | 39.92 | Stable | − 0.268 | Insoluble/0.351 | Golgi |
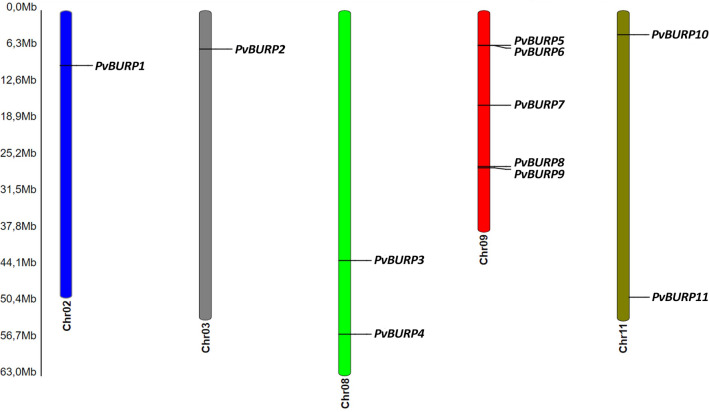
All genes encoding BURP domain-containing proteins have been successfully inserted into chromosomes and shown in Fig. 1. The chromosomal localizations indicated that one PvBURP gene was found on chromosomes 2 and 3 while two genes on chromosomes 8 and 11 and 5 genes on chromosome 9. There were no PvBURP genes found on other chromosomes. There is no common point regarding the positions of BURP genes on chromosomes. Few were seen located on the upper arm, some on the lower arm, and rest in the middle position.
Fig. 1.
Chromosomal location of common bean BURP domain-containing genes. The genes are located over the five linkage groups: Chr02, Chr03, Chr08, Chr09 and Chr11. The chromosome number is indicated at the bottom of each chromosome. The scale is in megabases (Mb)
Phylogenetic analysis of the BURP family
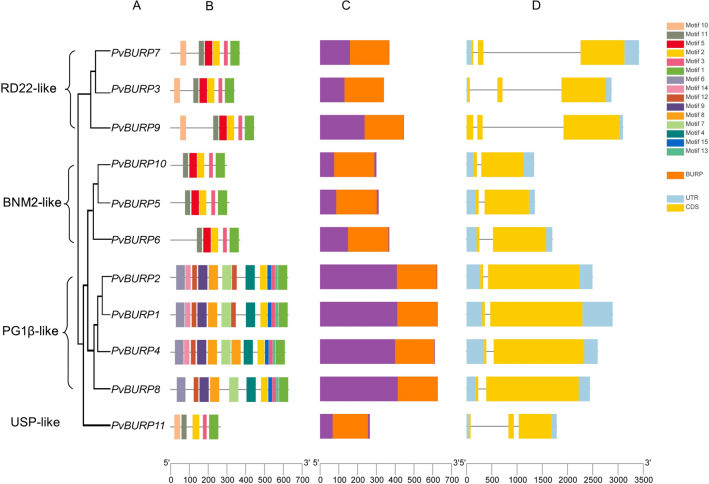
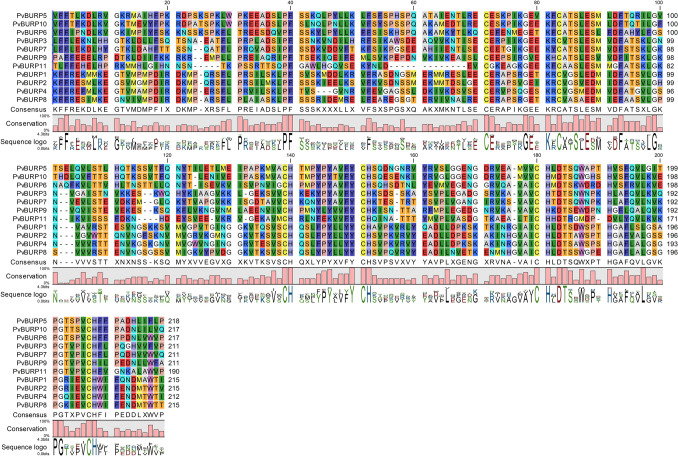
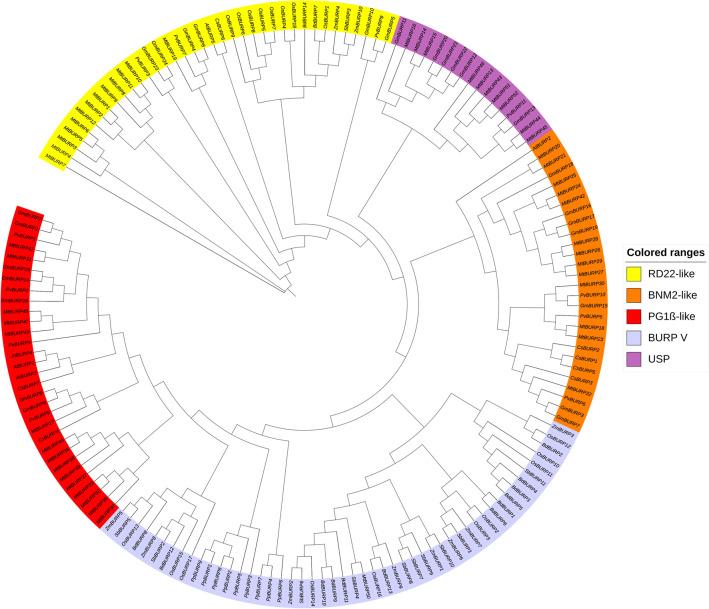
In this study, PvBURPs were divided into four groups according to the method of Hattori et al. (1998); BNM2A (PvBURP5, PvBURP6, and PvBURP10), USP (PvBURP11), RD22 (PvBURP3, PvBURP7, PvBURP9), and PG1β (PvBURP1, PvBURP2, PvBURP4, and PvBURP8) (Fig. 2). Alignment of BURP domains revealed that a total of 21 consensus amino acids (3F, 39P, 40F, 71C, 78G, 81K, 83C, 86S, 88E, 93F, 99G, 139C, 140H, 150Y, 151C, 152H, 180C, 181H, 183D, 184T, and 197L) were wholly conserved among the 11 proteins in the common bean BURP family members (Fig. 3). Additionally, this analysis of the BURP family members showed to have many extremely conserved amino acid sites and 4 CH motifs, indicating that these amino acids were essential for the basic functionality of these members of the BURP family. Members of the BURP family are generally characterized by the amino acid sequence located at the C-terminus, summarized as CHX3YX6CHX23-28CHXDX18-23CHX8W. However, PvBURP11, PvBURP1, PvBURP2, and PvBURP4 members, whose sixth amino acid is F, do not follow this rule (Fig. 3). We constructed a phylogenetic tree to investigate the evolutionary relationship of the BURP family in different species. For this purpose, members of the BURP family were identified in Arabidopsis thaliana (5 genes), Brachypodium distachyon (14 genes), Cucumis sativus (7 genes), Glycine max (26 genes), Medicago truncatula (52 genes), Oryza sativa (18 genes), Sorghum bicolor (11 genes), and Zea mays (10 genes) following the same workflow used in the identification of PvBURP genes. According to the phylogenetic tree, all BURPs were grouped into the five sub-families. These are. BNM2A, USP, RD22, PG1β, and BURP V. None of the BURP family members in Phaseolus vulgaris, Glycine max, Arabidopsis thaliana, and Cucumis sativus are in the BURP V group (Fig. 4).
Fig. 2.
Phylogenetic relationships, gene structure, conserved motifs and conserved domains in BURP genes from P. vulgaris. A Phylogenetic tree was constructed using the MEGA X software based on the full-length sequences of P. vulgaris BURP genes. PvBURP members are divided into four subfamilies RD-22 like, PG1β-like, BNM2-like and USP-like. B Conserved motifs were identified by MEME Suite and displayed in different colored boxes. C Distributions of the conserved domains in BURP proteins. Orange color boxes indicate cation BURP domains D Exon–intron structure of P. vulgaris BURP genes. Blue boxes indicated untranslated 5′- and 3′-regions; yellow boxes indicate exons
Fig. 3.
Multiple alignment of amino acid sequences of BURP proteins from P. vulgaris. BURP family members represent these to have many extremely conserved amino acid sites and 4 CH motifs
Fig. 4.
Phylogenetic relationship of BURP genes in P. vulgaris and other plant species. BURP genes are divided into five subfamilies named RD-22 like, PG1β-like, BNM2-like, USP-like, and BURP V. Each subfamily is shown in different colors. The tree was generated using the MEGA X software by the neighbor-joining method based on the Jones-Taylor-Thornton (JTT) model with bootstrap of 1000 replicates and visualized with ITOL v3
Analysis of gene structure, gene duplication, and conserved motifs of the PvBURPs
The analysis results of PvBURP proteins for identifying signal peptides found in the N-terminus revealed that all PvBURP proteins, excluding PvBURP7 and PvBURP9, have one signal peptide (Table S2). We also studied the exon/intron structure within the PvBURP family members to obtain more knowledge of the common bean BURPs' structural diversity. This structural analysis revealed that none of the BURP genes were without introns. Additionally, just one intron was present in seven of the BURP genes, while the others had two introns. The organization and number of introns of PVBURP genes included various patterns and distributions in the distinct subfamily. Accordingly, while two relatively long introns were found in the BURP genes belonging to the RD22 and USP sub-families, members of the other sub-families had a short and single intron. Another structural difference observed between proteins in subfamilies was the motifs. All members of PvBURPs have motif 1, 2, and 3 (Figure S1). Unlike other BURPs, those belonging to RD22 and BNM2 sub-family have motifs numbered 1, 2, 3, 5, and 11. Members of the PG1β sub-family with motifs numbered 1, 2, 3, 4, 6, 7, 8, 9, 12, 13, 14, and 15 were the most motif-bearing PvBURPs. The BURP member in the common bean with the least motifs is PvBURP11, annotated as USP (Fig. 2). Similar motifs and motif layouts were identified in closely related BURPs. Considering only the BURP domain located at the C terminal, it can be seen that Motif 5 belongs only to RD22 and BNM2 subfamily.
We have also analyzed the gene duplication events in PvBURP genes. According to the analyses conducted with the plant genome duplication database and BLAST-P, we only identified segmentally duplicated PvBURP genes. Among the eleven genes in the PvBURP family, we identified four gene pairs (PvBURP1 /PvBURP2, PvBURP1/PvBURP8, PvBURP2/PvBURP8, and PvBURP5/PvBURP10) that had been duplicated in the evolution of the PvBURP family. Considering that synonymous silent substitutions per site (Ks) occur at a constant rate over time, it is possible to estimate the dates of large-scale duplications (Maher et al. 2006). The Ka, Ks, and the Ka/Ks ratios were calculated for each duplicated PvBURP gene pair to explore duplicated BURP genes' possible fate. If the Ka/Ks ratio is lower than one, it indicates functional constraints with the negative or purifying selection of the genes; if this ratio equals to zero, then it indicates neutral selection. A ratio higher than one shows accelerated evolution with positive selection (Juretic et al. 2005). Our research found that Ka/Ks ratios from four PvBURP duplicated gene pairs were less than 0.3 (Table 2). This finding indicates that the family of PvBURP genes has mainly evolved under strong purifying selection pressure, with a few functional variations following duplication. It was estimated that the duplication that created the four segmentally duplicated gene pairs occurred between 5.09 and 13.76 million years ago (Mya) (Table 2).
Table 2.
Ka/Ks ratios of PvBURP genes
| Group | Gene 1 | Gene 2 | Identity (%) | Ka | Ks | Ka/Ks | Purifying selection | Effective Length | Mya |
|---|---|---|---|---|---|---|---|---|---|
| PG1β | PvBURP1 | PvBURP2 | 65.15 | 0.267357054 | 1.68629 | 0.158547497 | Yes | 1851 | 12.97146 |
| PG1β | PvBURP1 | PvBURP8 | 51 | 0.438637093 | 1.464318 | 0.299550437 | Yes | 1833 | 11.26398 |
| PG1β | PvBURP2 | PvBURP8 | 51.75 | 0.426944018 | 1.789431 | 0.238592104 | Yes | 1833 | 13.76485 |
| BNM2 | PvBURP5 | PvBURP10 | 64.95 | 0.202641686 | 0.662749 | 0.305759265 | Yes | 876 | 5.09807 |
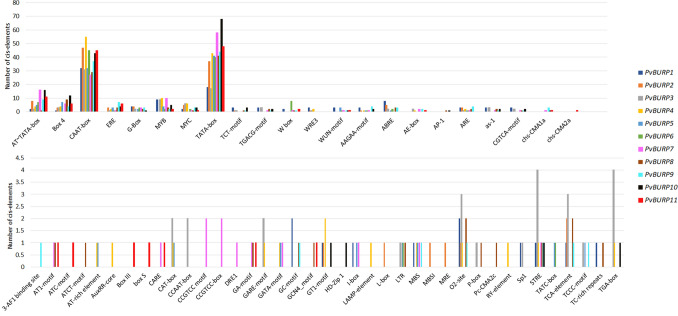
Analysis of cis-elements in putative PvBURP promoter regions
As recommended in all previous genome-wide studies, BURP genes have an important functional role in plants against abiotic stresses. We identified stress, light, and hormone response-related cis-elements in the promoter sequences of PvBURPs covering the upstream region of 2000 nucleotides from the gene start codon to fully understand and clarify the potential regulatory role of PvBURPs under various stresses. In this context, we identified 61 different cis-regulatory elements in the putative promoter regions. Ten types of elements related to plant hormones have been described. These are AuxRR-core (auxin), TGA-element (auxin), P-box (gibberellin), TATC-box (gibberellin), GARE-motif (gibberellin), CGTCA-motif (MeJA), TGACG-motif (MeJA), ERE (ethylene), TCA-element (SA), and ABRE (ABA). In these putative promoter regions, eight types of cis-regulatory elements were found to be related to various stress responses. Their names and potential functions are as follows; WUN (a wounding-responsive element), GC-motif (anoxic specific inducibility), W-box (defense and stress responsiveness), GT1 (drought), MBS (drought), TC-rich repeats (defense and stress responsiveness), and LTR (cold stress), ARE (anaerobic responsive elements). Another group of cis-elements found in the putative promoter regions of PvBURP genes those related to light response are Box4, G-box, I-box, Sp1, GT1-motif, TCT-motif, AT1-motif, MRE, L-box, AE-box, Gap-box, GATA-motif, chs-CMA1a, 3-AF1 binding site, Box3, GA-motif, chs-CMA2a, ATC-motif. The existence of CAAAGATATC-motif showed that most PvBURPs also have potential roles in regulating the circadian cycle in plants. (Fig. 5, Table S3). Likewise, 5UTR Py-rich (TTCTTCTAT) stretch found in the putative promoter region in PvBURP6, PvBURP8, and PvBURP11 provide them high transcription level.
Fig. 5.
Stress, light, and hormone response-related cis-elements in the promoter sequences of PvBURPs covering the upstream region of 2000 nucleotides from the gene start codon. PvBURPs are represented by different colors. The families of cis-elements were identified using place database
Identification of miRNAs targeting the genes plays a vital role in understanding both miRNAs and their target's functions. We identified 12 different miRNA targeting the nine PvBURP genes using two different in silico tools based on perfect or near perfect complementarity to their targets. We were unable to find a miRNA corresponding to the PvBURP5 and PvBURP6 during this study. The genes most targeted by the miRNAs in this analysis were PvBURP2 and PvBURP8. The most significant miRNA within this group is miRNA395, as it targets the highest number of genes: PvBURP9, PvBURP2, PvBURP11, and PvBURP8 (Table 3). The other important miRNAs targeting PvBURP genes were miRNA156, miRNA169, miRNA171, miRNA319, and miRNA390.
Table 3.
Prediction of potential miRNAs targeting PvBURPs
| miRNA_Acc | Target_Acc | Expectation | Unpaired energy | miRNA start | miRNA end | Target start | Target end |
|---|---|---|---|---|---|---|---|
| Pvu-miR156 | PvBURP3 | 5.5 | − 1 | 1 | 21 | 423 | 443 |
| Pvu-miR169 | PvBURP10 | 5.5 | − 1 | 1 | 21 | 498 | 518 |
| Pvu-miR171 | PvBURP8 | 5 | − 1 | 1 | 21 | 571 | 591 |
| Pvu-miR171 | PvBURP4 | 5.5 | − 1 | 1 | 21 | 1333 | 1354 |
| Pvu-miR319 | PvBURP2 | 6 | − 1 | 1 | 21 | 1141 | 1161 |
| Pvu-miR390 | PvBURP8 | 5 | − 1 | 1 | 21 | 717 | 737 |
| Pvu-miR390 | PvBURP10 | 6 | − 1 | 1 | 21 | 286 | 306 |
| Pvu-miR395 | PvBURP11 | 4 | − 1 | 1 | 21 | 91 | 111 |
| Pvu-miR395 | PvBURP9 | 5.5 | − 1 | 1 | 21 | 1251 | 1271 |
| Pvu-miR395 | PvBURP2 | 6 | − 1 | 1 | 21 | 98 | 118 |
| Pvu-miR395 | PvBURP11 | 4 | − 1 | 1 | 21 | 91 | 111 |
| Pvu-miR395 | PvBURP8 | 5.5 | − 1 | 1 | 21 | 585 | 605 |
| Pvu-miR397 | PvBURP2 | 5.5 | − 1 | 1 | 21 | 54 | 74 |
| Pvu-miR397 | PvBURP7 | 6 | − 1 | 1 | 21 | 21 | 41 |
| Pvu-miR408 | PvBURP3 | 6 | − 1 | 1 | 21 | 284 | 303 |
| Pvu-miR482 | PvBURP2 | 6 | − 1 | 1 | 22 | 1463 | 1484 |
| Pvu-miR1514 | PvBURP1 | 6 | − 1 | 1 | 22 | 1340 | 1361 |
| Pvu-miR4416 | PvBURP4 | 4.5 | − 1 | 1 | 21 | 924 | 944 |
| Pvu-miR4415 | PvBURP8 | 4 | − 1 | 1 | 21 | 1533 | 1553 |
| miRNA_Acc | miRNA_aligned_fragment | Alignment | Target_aligned_fragment | Inhibition |
|---|---|---|---|---|
| Pvu-miR156 | UGACAGAAGAGAGAGAGCACA | ::::.:::::::::: | AUUGGACUUGCUCUUCUCUCA | Cleavage |
| Pvu-miR169 | CAGCCAAGGGUGAUUUGCCGG | ::::::..::::::: | CACUCAAACCAUUCUUGGCUU | Cleavage |
| Pvu-miR171 | UGAUUGAGCCGUGCCAAUAUC | :.::::.:::::::::: | GGUUCUGCCGCGGCUCAAUCC | Cleavage |
| Pvu-miR171 | UUGAGCC-GCGCCAAUAUCACU | :::.::::.::::.:::: | UUUGAGGUUGGUGCCGGUUCAA | Cleavage |
| Pvu-miR319 | UGGACUGAAGGGAGCUCCUUC | ::.:::.:.:::::.:.: | GAGGGUGUUUCCUUCGCUUCU | Cleavage |
| Pvu-miR390 | AAGCUCAGGAGGGAUAGCACC | :::::::.:::::::: | UGCGCAAUCCUUCAUGAGCUA | Cleavage |
| Pvu-miR390 | AAGCUCAGGAGGGAUAGCACC | :::::.:::::::.: | CCUGCAACCUCUCCUAAGUUG | Cleavage |
| Pvu-miR395 | UGAAGUGUUUGGGGGAACUCU | :::…::::::::::: | UCUGUUUUUCCAAACACUACA | Cleavage |
| Pvu-miR395 | UGAAGUGUUUGGGGGAACUCU | :.::…:::.:.::::: | CGGCUUUUUACAAGCGCUUCA | Cleavage |
| Pvu-miR395 | CUGAAGUGUUUGGGGGAACUC | :::.:::::.::::: | CGUUUACUCCAAAGGCUUCUG | Cleavage |
| Pvu-miR395 | UGAAGUGUUUGGGGGAACUUU | :::…::::::::::: | UCUGUUUUUCCAAACACUACA | Cleavage |
| Pvu-miR395 | UUGAAGUGUUUGGAGGAACUC | :::::::.::::::: | UCAAUCCUCCAGCGACUUCAA | Cleavage |
| Pvu-miR397 | UCAUUGAGUGCAGCGUUGAUG | :::.:.:::.::..:: | ACUCACUGUUGCUUUCGGUGG | Cleavage |
| Pvu-miR397 | UCAUUGAGUGCAGCGUUGAUG | :::..:::.:::::::: | CAUUUUUGCUUUACUCAAUGU | Translation |
| Pvu-miR408 | AUGCACUGCCUCUUCCCUGGC | :::::::::.:::::: | CAAAGGGAA-AGCCGGUGCAU | Cleavage |
| Pvu-miR482 | UCUUCCCUACACCUCCCAUACC | ::.:::.::::.:::.:: | GUUGUGUGGGGUCUGUGGAGGA | Translation |
| Pvu-miR1514 | UUCAUUUUGAAAAUAGGCAUUG | :.:.:.::::::.:::.: | CGUUUUCCGUUUCCAAGAUGGA | Translation |
| Pvu-miR4416 | UACGGGUCGCUCUCACCUAGG | ::::.:::.:::.::: | CAACGGUGGGAGUGACUCGUU | Cleavage |
| Pvu-miR4415 | UUGAUUCUCAUCACAACAUGU | .:::::::.::::.::: | CAGUGUUGUGGUGAGGAGCAC | Cleavage |
Prediction of protein structure and subcellular localization
To estimate 3D structures of PvBURPs protein on the basis of homology modeling principles, Phyre2, a web-based bioinformatic server, was utilized (Figure S2). Although beta-sheets are very common in all PvBURP proteins, this situation is quite striking in PvBURP7 protein, containing only 10% alpha-helix (Table S4). The protein with the highest alpha-helix structure was estimated to be PvBURP3 (45.5%). Identifying where transmembrane (TM) segments are located can help narrow the potential conformations of the tertiary structures for the given protein and predict its function (Krogh et al. 2001). This analysis showed that there is one transmembrane helix in 6 PvBURP proteins (PvBURP1, PvBURP2, PvBURP3, PvBURP5, PvBURP6, and PvBURP7) (Figure S3). The most extended transmembrane helix consisting of 22 residues was observed in PvBURP3 and PvBURP7. The sizes of these segments in other proteins vary between 17 and 19 residues. When PvBURP proteins are compared structurally, another remarkable finding is the existence of signal peptides. In silico analysis indicated that all proteins except PvBURP7 and PvBURP9 contain Sec signal peptide (Sec / SPI) at the N-terminal (Table S2). Cell-PLoc2.0 predictions have shown that most PvBURPs except PvBURP7, active in the nucleus, were functional within the cell's membrane (Table 1).
Stress-responsive expression profiles of BURPs
To evaluate the expression pattern of BURPs in different stress conditions, available RNA-seq data sets were retrieved from the NCBI SRA database. In this context, six BioProject was analyzed conducting in common bean under drought, salt, and pathogen stress. As shown in Fig. 6 prepared based on log2 fold change, BURP genes' expression differs according to stressors. It was observed that in all experiments, at least one BURP gene was expressed differentially. The data set in which the least number of BURP genes were differentially expressed were first and fourth. In the first data set, the expression levels of BURP genes in salt-treated leaves of the T43 (salt-sensitive) and ISPIR (salt resistant) genotype were studied. In this first comparison, PvBURP6 was significantly up-regulated with salt stress (log2FC = 1.58), while PvBURP4 was down-regulated (log2FC = 1.50) with the same stressor. In the second data set obtained by comparing the data retrieved from the roots of T43 and Ispir genotypes under salt stress, only the PvBURP10 gene was significantly induced in both genotypes (log2FC = 1.94).
Fig. 6 .
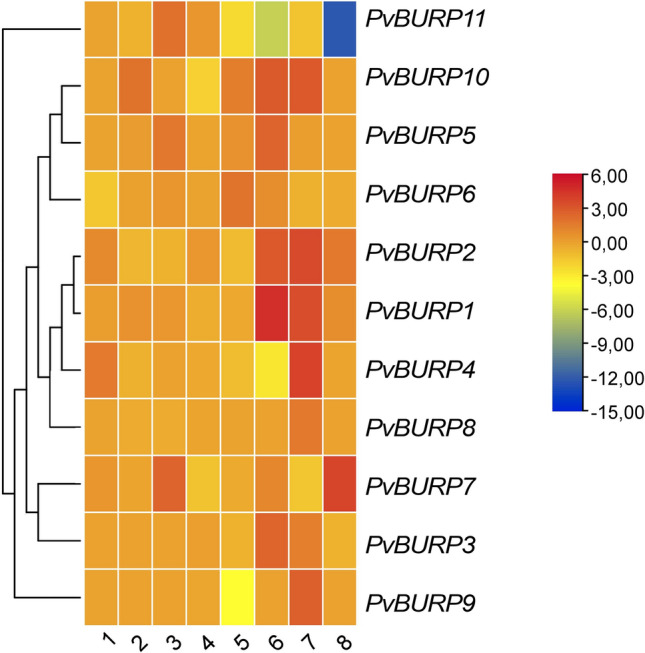
A Heat map illustrating BURP expression in various RNA-Seq datasets. Red, positive log fold-change (log FC) indicates higher expression in the first genotypes compared with the second; blue, negative log FC. The raw RNA-seq datasets were downloaded from NCBI Sequence Read Archive (SRA) under the accession number; 1 and 2 represent salt-treated leaf explants of T43 (sensitive) and Ispir (resistant) common bean genotypes (PRJNA656794), 3 represents salt-tolerant Ispir's root tissues grown under both control and salt stress conditions (PRJNA656794), 4 indicates salt stress to the salt-tolerant and the sensitive genotype at the bud stage (PRJNA558376), 5 represents salt stress to the salt-tolerant bean genotype in the sprout stage (PRJNA691982), 6 and 7 represent RNA sequencing of drought-tolerant Pinto saltillo (PRJNA508605) and Perola (PRJNA327176) genotypes grown under control and drought stress and 8 indicates a common bean plant infected by the fungal pathogen Sclerotinia sclerotiorum (strain 1980) (PRJNA574280). The transcriptional analysis of downloaded files was done via CyVerse and Galaxy (usegalaxy.eu) with in silico tools. The mapping of reads was done with the HISAT2 tool. Transcript assembly and differential expression analyses were performed with Stringtie 1.3.3 and Ballgown, respectively. The genes having fold change value log2 > 1 and a p value < 0.05 were accepted as differentially expressed genes (DEGs). Heatmap based on the log2FC was prepared using TBTools
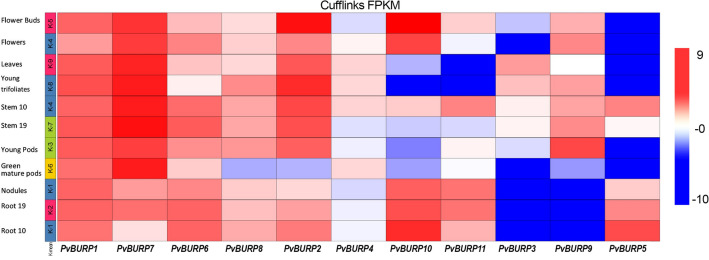
Additionally, PvBURP5, PvBURP7, PvBURP5, and PvBURP11 were differentially up-regulated in the salt-treated Ispir roots (third data set) compared to control conditions. In the fourth data set, when the salt-stressed sensitive and resistant genotypes were compared, it was found that only two genes, PvBURP7 (log2 FC = − 1.35) and PvBURP10 (log2 FC = − 1.96), differentially suppressed with salt stress. In the last data set (fifth data set), four genes (PvBURP2, PvBURP4, PvBURP9, and PvBURP11) were differentially up-regulated and PvBURP10 was down-regulated in response to salt stress. In the sixth and seventh data sets expression level of PvBURP genes under drought indicated similar regulations to salt stress. All these in silico gene expression analyses indicated that PvBURP1, PvBURP2, PvBURP3, and PvBURP10 were differentially up-regulated in both data sets, while the PvBURP11 gene was down-regulated. When compared with other stress conditions, it was observed that more PvBURP genes were expressed differentially in drought stress applications. In this context, the data set having the highest number of BURP genes (9 genes) with different expression levels was the seventh. In this study, drought stress was applied when the plants reached the true three leaves stage, and after two weeks of stress application, all aboveground organs were collected for RNA isolation. During the in silico expression analysis of these data, expression profiles of control and drought-treated Pinto Saltillo plants were compared. According to this comparison, PvBURP1, PvBURP2, PvBURP3, PvBURP4, PvBURP8, PvBURP9, and PvBURP10 were found to be differentially up-regulated (Fig. 6 and Table S5). However, PvBURP7 and PvBURP11 genes were found to be down-regulated. The maximum log2 fold change value with a − 12.3 was calculated for the PvBURP11 gene using the RNA-seq data obtained from a library (eighth data set) prepared with a common bean plant infected by the fungal pathogen Sclerotinia sclerotiorum (strain 1980) (PRJNA574280) (Table S5). In addition to this differentially down-regulated gene, PvBURP2 and PvBURP7 were also differentially up-regulated in this library. Tissue-specific expression profiles of PvBURPs were retrieved from Phytozome v12 (Fig. 7). In the light of this data, we observed that PvBURP1, PvBURP2, and PvBURP7 were differentially up-regulated in almost all tissues, including flower, flower buds, leaves, young trifoliates, young pods, stem, root, and nodules. However, the expression levels of PvBURP8 and PvBURP4 did not change significantly between tissues of the common bean. The expression levels of other PvBURP genes increased in some tissues but decreased in some other tissues. For instance, while the PvBURP10 gene increased significantly in the tissue of flowers, flower buds, and nodules, it decreased in the tissue of leaves, young three leaves, young pods, and green mature pods (Fig. 7). Likewise, while less PvBURP11 activity was observed in leaves and young leaves, it was seen that this gene was expressed at a higher rate in nodule and root tissue. It was determined that PvBURP3 and PvBURP9 genes' activity did not change in other tissues, but expression levels were increased in green mature pods, nodules, and root tissues. It was seen that the PvBURP5 gene is less expressed in other aboveground organs other than the stem but has a significantly increased activity in the root tissue (Fig. 7).
Fig. 7.
Hierarchical clustering of FPKM values of PvBURP genes in different tissues. Red to blue frames represent positive to negative expressions. The raw data was normalized and retrieved from the Phytozome v12
Determination of expression pattern of BURPs under various stress conditions
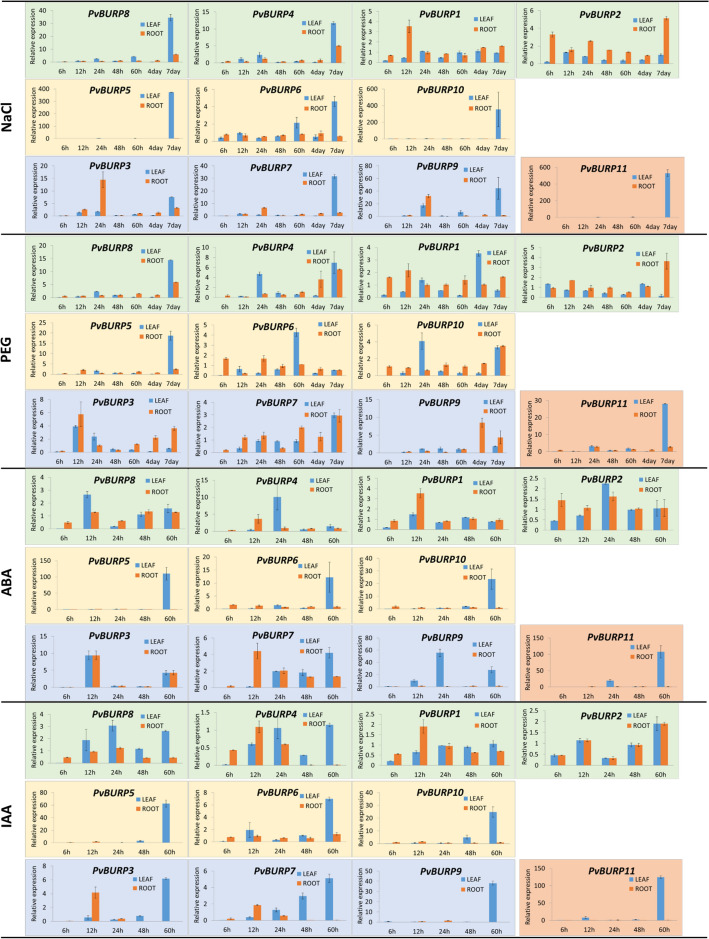
The expression levels of the BURP genes in roots and leaves of common bean plants subjected to salt, drought, IAA, and ABA stress were determined by quantitative real-time PCR (Fig. 8). Under salt stress conditions, it was observed that PvBURP3, 4, 5, 6, 7, 8, 9, 10, and PvBURP11 were most significantly up-regulated at seven days in the leaf tissue. These all were up-regulated at different levels, but it was clearly seen that PvBURP5, 10, 11 represent a significant correlation at the highest response level in leaf tissue and only at seven days. Although no clear correlation appears in root tissue, it was understood that PvBURP1 at 12 h, PvBURP2 at six hours and seven days, and PvBURP3 at 24 h were up-regulated in the root tissue (Fig. 8).
Fig. 8.
qRT-PCR analysis of PvBURP genes in roots and leaves of common bean plants under salt, drought, IAA and ABA treatments. Expression levels were measured at different time points. Relative expression was calculated according to the 2−ΔΔCt method
Under PEG-mediated drought stress condition, almost all BURPs in that experiment showed different positive responses at different time points in leaf tissue, but the significant ones that show up-regulation were; PvBURP3 at 12 h, PvBURP4, and PvBURP10 at 24 h, and PvBURP4, 5, 7, 8, 10 and 11 at seven days. One of the clear positive correlations between BURP4 and BURP10 appear at 24 h in leaf and seven days both in root and leaf tissues. The second correlation at seven days in the leaf tissue between BURP4, 5, 7, 8, 10, and 11 looks similar to PvBURPs under salt treatment except for PvBURP3, 6, 9, 10 was no significant up-regulation under PEG condition. PvBUPR3 at 12 h, PvBURP4 and PvBURP9 at four days, and PvBURP2, 3, 4, 7, 8, 9, 10 at seven days exhibited up-regulation in the root tissue (Fig. 8).
After ABA treatment, some BURP members represented positive responses at different time-point in the leaf tissue like PvBURP1, 3, 8, 9 at 12 h, PvBURP2, 4, 7, 9 at 24 h, and PvBURP3, 7, 9, 11 at 60 h. The exciting correlation results have appeared from PvBURP9, and PvBURP11 respond at the highest level at 24 h and 60 h to ABA only in leaf tissue. The PvBURP1, 3, 4, 7 at 12 h, PvBURP7 at 24 h, and PvBURP3 at 60 h showed up-regulation in root tissue. The general idea about the response of BURPs to ABA was that BURPs were much more active in leaf tissue than the root tissue under ABA treatment (Fig. 8).
Under IAA treatment, almost all PvBURPs except for PvBURP1 and PvBURP4 are highly active at 60 h, and PvBURP8 is found highly active at all time-point except for six hours in leaf tissue. Moreover, only PvBURP7 shows up-regulation at 48 h. In the root tissue, PvBURPs are much less active compared to the result in leaf tissue. Only PvBURP3 and PvBURP7 at 12 h and PvBURP2 at 60 h represent significant up-regulation in root tissue (Fig. 8).
Overall, as it is evident from these results, BURPs are not regulated at the onset of the stress response in both leaf and root tissues. There is a robust correlation indicating the up-regulation of BURPs towards the end of all stress conditions in leaf tissue. Moreover, BURPs are much more active in leaf tissue than root tissue for all conditions.
Discussion
There are studies in the literature which aims to understand the roles of BURP genes in development and stress responses in plants, but it mainly covers the RD22 and USP-like subfamily only (Bassüner et al. 1988; Batchelor et al. 2002; Harshavardhan et al. 2014; Hattori et al. 1998; Yamaguchi-Shinozaki et al. 1993). In parallel with the development of new generation sequencing systems, although a considerable increase has been observed in the genome-wide analysis of different genes and transcription factors, the same cannot be said for BURPs. Until now, genome-wide analysis studies of BURP genes have been carried out on very few plants, including rice, maize, grapevine, soybean, cotton, sorghum, and poplar (Ding et al. 2009; Gan et al. 2011; Matus et al. 2014; Shao et al. 2011; Sun et al. 2019; Xu et al. 2010). When these previous studies were examined, it was found that the richest genome in terms of BURP genes was in cotton with 30 genes, and the least found was in sorghum with 11 genes. Consistent with previous studies, we also found 11 putative BURP genes distributed to the five chromosomes in the common bean genome using various in-silico tools. We observed that most of the BURP encoding genes, 5 out of 11, were located on chromosome 9. During the evolution and expansion of gene families in plants, gene duplication such as segmental and tandem play a key role (Cannon et al. 2004). Duplication events allow the quantity of genetic material to evolve during evolution and natural selection. When a gene duplication event takes place, some duplicated genes keep their functions, while others show partial or complete divergence from another (Pickett and Meeks-Wagner 1995). It can be speculated that the evolutionary origin of the BURP gene family in the common bean genome by searching the location of these genes in the genome. A gene cluster, located on chromosome 9, consists of five genes (PvBURP5–9), of which three genes (PvBURP5, PvBURP6, and PvBURP8) share common parent genes. Five out of 11 BURP genes (PvBURP1, PvBURP2, PvBURP8, PvBURP5, and PvBURP10) were found located in duplicated regions (Table 2). The analysis of gene duplication events indicated that segmental gene duplication played a significant role in forming the PvBURP gene family. The same is reported for poplar (Shao et al. 2011), grapevine (Matus et al. 2014), and soybean (Xu et al. 2010) alike. Another similarity of our study with previous studies was related to the distribution of BURP genes on chromosomes. When previous studies carried out in poplar, grapevine, sorghum, and maize were examined, it was seen that BURP genes were unevenly distributed to the chromosomes, such as seven genes in the seventh chromosome at poplar, 13 genes in the fourth chromosome at grapevine, four genes in the eighth chromosome at sorghum and six genes in the seventh chromosome at maize. This data suggests that, even though the number of BURP genes between species is similar, the BURP superfamily may have undergone a specific expansion of a particular group within these chromosomes (Matus et al. 2014).
To gain insight into the relationships between the common bean BURP genes, we first constructed a phylogenetic tree including the 11 common bean proteins identified in this work based on their deduced amino acid sequences. As reported in previous studies, the common bean BURP proteins are subdivided into four different subfamilies, including USP-like, BNM2- like, PG1β-like and RD22-like. We identified at least one member for each subfamily. However, the classification of the BURP genes varies in different plant species. For instance, Xu et al. (2010) reported that there were five subfamilies, including BURP V, in addition to the previously mentioned subfamilies in the soybean genome. We confirmed this phylogenetic relationship using the motif patterns retrieved from MEME Suite. As in our study, Ding et al. (2009) found that PG1βs show a range of unique motif patterns with a minimal degree of divergence amongst species. For this reason, these sequences were not included in the further analysis. We observed that USP-like, BNM2-like, and PG1β-like members were all detected in dicotyledons only, whereas RD22-like members were identified in both dicotyledon and monocotyledon plants, based on the phylogenetic relationship between PvBURPs and other BURP proteins identified in other analyzed plants. Additionally, the BURPV subfamily consists of the members belong to the monocotyledons, indicating that these genes might evolve separately and perform different functions between monocots and dicots (Ding et al. 2009; Gan et al. 2011; Sun et al. 2019). Our findings related to the classification of BURPs is consistent with previous studies, as no Arabidopsis gene was present in the USP subfamily (Matus et al. 2014). Actually, it was seen that this subfamily included BURP genes belonging to barrelclover, soybean, and bean plants; that is, it contained only the leguminous family.
In previous studies, it has been shown that at least one of the BURP genes in different plants, including maize and rice, is without introns and there are BURP members, which generally carry 2 or 3 introns (Ding et al. 2009; Gan et al. 2011). In contrast, in the present study, we found that there was no intronless PvBURP gene and the maximum number of introns in the PvBURP genes was two. The intron–exon distribution pattern can be used as an independent criterion for testing the reliability of the phylogenetic analysis (Du et al. 2012). The intron–exon pattern of PvBURP genes of a particular subfamily displayed remarkable consistency with the location of the intron almost fully conserved, which highlighted how related the individual members of the subfamily are.
miRNA can play various roles in plant growth and development, reproductive processes, response to different stresses, and cellular signaling (Zhao et al. 2015). Predicting the miRNAs that target genes allows us to obtain information about the possible functions of those genes. The best example of this assumption is miRNA156. Because miRNA156, one of the first discovered miRNAs, plays a role in regulating many developmental events in plants (Xing et al. 2010). The most important micro-RNA found in our study was mirRNA395, which is involved in sulfur metabolism. Sulfur actively participates in numerous biological processes and plays a vital role in plant development (Ai et al. 2016). Since micro RNAs such as miRNA169, mirNA171, and miRNA319, which are among the other miRNAs found in our study, play a key role in the response of plants to various stresses and the formation of plant organs, this confirms the previously defined functions of the BURP genes they target.
Another proof that BURP genes have roles in response to various stress factors and regulating many different developmental processes in plants is the different cis-elements found in the promoter regions of PvBURPs. Since they regulate a variety of stress responses, cis-acting regulatory elements are essential transcriptional gene regulatory units (Sheshadri et al. 2016). In the present study, we identified ten types of plant hormone-related and eight types of stress response-related cis-acting regulatory elements in the promoter region of PvBURP genes inconsistent with their potential functions. Some of the cis-acting regulatory elements identified in this study are not frequently observed in other plants. These are GARE and GT1 motifs, thymine- and cytosine-rich repeats. GARE element, located between 139 and 145 bp downstream of the TSS, is involved in the gibberellin response. One of the light response elements is the G1 motif (ATGGTGGTTGG), which can be located 168–178 bp downstream of the TSS. (Biłas et al. 2016).
Gene expression patterns are considered to be important clues used to make inferences about the functions of those genes (Kavas et al. 2016). To determine the potential functions of PvBURP genes, RNA-seq data belong to different tissues exposed to various stresses were evaluated. The RNA-seq data from Perola and Pinto Saltillo genotypes have been analyzed to determine expression profiles of PvBURP genes under drought stress. It was reported that the total RNAs from drought tolerant Pinto Saltillo were isolated from all aboveground tissues after two weeks of stress treatment to the 45 days-old plants (Gregorio Jorge et al. 2020). However, in the other drought-tolerant Perola genotype, drought stress was applied to the two-week-old plants for just 150 min (Pereira et al. 2020). Although the age of plants when stress treatment was started and the duration of stress were different in these two experiments, the expression pattern of BURP genes was generally consistent in both genotypes. In this context, the expression level of members of RD22 (PvBURP3), BNM2 (PvBURP10), and PG1β (PvBURP1 and PvBURP2) were significantly increased in both treatments. As a similar result, the increase in the expression of some BURP genes in response to drought stress and the decrease in some of them were observed in the genome-wide analysis carried out in the Medicago plants (Li et al. 2016). A similar change in expression of BURP genes occurred under salt stress. As a result of the comparison of the transcriptome data obtained from the roots of the salt-tolerant Ispir and sensitive T43 genotypes exposed to salt stress, it was determined that only the PvBURP10 gene was expressed differently. However, in the same study conducted with the roots of the ISPIR genotype exposed and not exposed to salt stress, more genes (PvBURP11, PvBURP7, and PvBURP5) were found to be up-regulated. Banzai et al. (2002) cloned and evaluated the expression of BgBDC genes, RD22 homolog in Bruguiera gymnorrhiza. They reported that the expression of these genes could be changed between different leaves depending on ABA levels under salt stress. A similar result to previous studies was that the USP-like gene, PvBURP11, is down-regulated under drought stress. In their study, Harshavardhan et al. (2014) found that plants with the mutant AtUSPL1 gene have higher drought tolerance than wild-type plants. We also determined the expression pattern of PvBURPs under IAA, ABA, salt, and drought stress conditions with qRT-PCR. We observed that PvBURPs were not regulated at the onset of the stress response in both leaf and root tissues. There is a solid correlation indicating the upregulation of PvBURPs towards the end of all stress conditions in leaf tissue. Moreover, PvBURPs are much more active in leaf tissue than root tissue for all conditions.
In conclusion, for the first time, a complete analysis of the BURP family in common bean was analyzed, and the relationship between this family and drought, salt, IAA, and ABA stress response was evaluated. Additionally, we showed gene structures, chromosomal locations and sequence homologies of these genes. This study provided a useful resource for future studies on the structure and function of BURP proteins in the regulation of drought, salt, IAA, and ABA stress response in plants.
Supplementary Information
Below is the link to the electronic supplementary material.
Table S1. Primer sequences of PvBURP genes for qRT-PCR analysis (XLSX 11 kb)
Table S2. Location of the BURP domain and signal peptide in BURP proteins (XLSX 12 kb)
Table S3. A-biotic stress response-related cis-elements in the promoter sequences of PvBURPs covering the upstream region of 2000 nucleotides from the gene start codon (XLSX 11 kb)
Table S4. Secondary structures of PvBURP proteins (XLSX 25 kb)
Table S5. Expression profiles of PvBURP genes in different RNA-seq dataset based on log2FC value (XLSX 11 kb)
Figure S1. Conserved motifs in PvBURP proteins were obtained through the MEME Suit. Conserved motifs were analyzed by choosing 15 motifs and repeating any number of motifs. (PNG 1192 kb)
Figure S2. 3D structure of PvBURP proteins illustrated using Phyre2 server with an “intensive” mode. (PNG 640 kb)
Figure S3. Prediction of transmembrane helices in PvBURP proteins. (PNG 534 kb)
Funding
This research was supported by Research Fund of Ondokuz Mayıs University PYO.ZRT.1901.17.010.
Declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell. 1997;9:1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai Q, Liang G, Zhang H, Yu D. Control of sulfate concentration by miR395-targeted APS genes in Arabidopsis thaliana. Plant Divers. 2016;38:92–100. doi: 10.1016/j.pld.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Hassan M, Morosan M, López-Gresa MD, Prohens J, Vicente O, Boscaiu M. Salinity-induced variation in biochemical markers provides insight into the mechanisms of salt tolerance in common (Phaseolus vulgaris) and runner (P. coccineus) beans. Int J Mol Sci. 2016 doi: 10.3390/ijms17091582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arteaga S, Yabor L, Díez MJ, Prohens J, Boscaiu M, Vicente O. The use of proline in screening for tolerance to drought and salinity in common bean (Phaseolus vulgaris L.) genotypes. Agronomy. 2020;10:817. doi: 10.3390/agronomy10060817. [DOI] [Google Scholar]
- Banzai T, Sumiya K, Hanagata N, Dubinsky Z, Karube I. Molecular cloning and characterization of genes encoding BURP domain-containing protein in the mangrove, Bruguiera gymnorrhiza. Trees. 2002;16:87–93. doi: 10.1007/s00468-001-0144-4. [DOI] [Google Scholar]
- Bassüner R, Bäumlein H, Huth A, Jung R, Wobus U, Rapoport TA, Saalbach G, Müntz K. Abundant embryonic mRNA in field bean (Vicia faba L.) codes for a new class of seed proteins: cDNA cloning and characterization of the primary translation product. Plant Mol Biol. 1988;11:321–334. doi: 10.1007/BF00027389. [DOI] [PubMed] [Google Scholar]
- Batchelor AK, Boutilier K, Miller SS, Hattori J, Bowman L, Hu M, Lantin S, Johnson DA, Miki BL. SCB1, a BURP-domain protein gene, from developing soybean seed coats. Planta. 2002;215:523–532. doi: 10.1007/s00425-002-0798-1. [DOI] [PubMed] [Google Scholar]
- Beebe SE, Rao IM, Blair MW, Butare L (2009) Breeding for abiotic stress tolerance in common bean: Present and future challenges. In: Australasian Plant Breeding; SABRAO Conference (14; 11; 2009, Cairns, Queensland, Australia). Proceedings. Global Partnership Initiative for Plant Breeding Capacity Building (GIPB), Queensland, AU, p 11
- Biłas R, Szafran K, Hnatuszko-Konka K, Kononowicz AK. Cis-regulatory elements used to control gene expression in plants. Plant Cell Tissue Organ Cult (PCTOC) 2016;127:269–287. doi: 10.1007/s11240-016-1057-7. [DOI] [Google Scholar]
- Boutilier KA, Ginés M-J, DeMoor JM, Huang B, Baszczynski CL, Iyer V, Miki BL. Expression of the BnmNAP subfamily of napin genes coincides with the induction of Brassica microspore embryogenesis. Plant Mol Biol. 1994;26:1711–1723. doi: 10.1007/BF00019486. [DOI] [PubMed] [Google Scholar]
- Caldas DGG, Konzen ER, Recchia GH, Pereira ACVZ, Tsai SM (2016) Functional genomics of biotic and abiotic stresses in Phaseolus vulgaris. In: Shanker ASaC (ed) Abiotic and biotic stress in plants-recent advances and future perspectives. London: IntechOpen, pp 121–150
- Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celmeli T, Sari H, Canci H, Sari D, Adak A, Eker T, Toker C. The nutritional content of common bean (Phaseolus vulgaris L.) landraces in comparison to modern varieties. Agronomy. 2018;8:166. doi: 10.3390/agronomy8090166. [DOI] [Google Scholar]
- Chen L, Miyazaki C, Kojimai A, Saito A, Adachi T. Isolation and characterization of a gene expressed during early embryo sac development in apomictic guinea grass (Panicum maximum) J Plant Physiol. 1999;154:55–62. doi: 10.1016/S0176-1617(99)80318-6. [DOI] [Google Scholar]
- Chen C, Chen H, He Y, Xia RJB. TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. bioRxiv. 2018 doi: 10.1101/289660. [DOI] [Google Scholar]
- Chou KC, Shen HB. Cell-PLoc: a package of Web servers for predicting subcellular localization of proteins in various organisms. Nat Protoc. 2008;3:153–162. doi: 10.1038/nprot.2007.494. [DOI] [PubMed] [Google Scholar]
- Cortés AJ, Monserrate FA, Ramírez-Villegas J, Madriñán S, Blair MW. Drought tolerance in wild plant populations: the case of common beans (Phaseolus vulgaris L.) PLoS ONE. 2013;8:e62898. doi: 10.1371/journal.pone.0062898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhuang Z, Zhao PX. psRNATarget: a plant small RNA target analysis server (2017 release) Nucleic Acids Res. 2018;46:W49–W54. doi: 10.1093/nar/gky316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darkwa K, Ambachew D, Mohammed H, Asfaw A, Blair MW. Evaluation of common bean (Phaseolus vulgaris L.) genotypes for drought stress adaptation in Ethiopia. Crop J. 2016;4:367–376. doi: 10.1016/j.cj.2016.06.007. [DOI] [Google Scholar]
- Ding X, Hou X, Xie K, Xiong LJP. Genome-wide identification of burp domain-containing genes in rice reveals a gene family with diverse structures and responses to abiotic stresses. Planta. 2009;230:149–163. doi: 10.1007/s00425-009-0929-z. [DOI] [PubMed] [Google Scholar]
- Dipp CC, Marchese JA, Woyann LG, Bosse MA, Roman MH, Gobatto DR, Paludo F, Fedrigo K, Kovali KK, Finatto T. Drought stress tolerance in common bean: what about highly cultivated Brazilian genotypes? Euphytica. 2017;213:102. doi: 10.1007/s10681-017-1893-5. [DOI] [Google Scholar]
- dos Santos Neto J, Delfini J, Willian ST, Akihide HA, Marcos NJ, Simões Azeredo Gonçalves L, Moda-Cirino V. Response of common bean cultivars and lines to aluminum toxicity. Agronomy. 2020;10:296. doi: 10.3390/agronomy10020296. [DOI] [Google Scholar]
- Du H, Yang SS, Liang Z, Feng BR, Liu L, Huang YB, Tang YX. Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 2012;12:106. doi: 10.1186/1471-2229-12-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright A, John B, Gaul U, Tuschl T, Sander C, Marks D. MicroRNA targets in drosophila. Genome Biol. 2003;4:P8. doi: 10.1186/gb-2003-4-11-p8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fageria NK, Baligar VC, Moreira A, Portes TA. Dry bean genotypes evaluation for growth, yield components and phosphorus use efficiency. J Plant Nutr. 2010;33:2167–2181. doi: 10.1080/01904167.2010.519089. [DOI] [Google Scholar]
- Finn RD, Clements J, Arndt W, Miller BL, Wheeler TJ, Schreiber F, Bateman A, Eddy SR. HMMER web server: 2015 update. Nucleic Acids Res. 2015;43:W30–W38. doi: 10.1093/nar/gkv397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan D, Jiang H, Zhang J, Zhao Y, Zhu S, Cheng B. Genome-wide analysis of BURP domain-containing genes in Maize and Sorghum. Mol Biol Rep. 2011;38:4553–4563. doi: 10.1007/s11033-010-0587-z. [DOI] [PubMed] [Google Scholar]
- Granger C, Coryell V, Khanna A, Keim P, Vodkin L, Shoemaker RC. Identification, structure, and differential expression of members of a BURP domain containing protein family in soybean. Genome. 2002;45:693–701. doi: 10.1139/g02-032. [DOI] [PubMed] [Google Scholar]
- Gregorio JJ, Villalobos-López MA, Chavarría-Alvarado KL, Ríos-Meléndez S, López-Meyer M, Arroyo-Becerra A. Genome-wide transcriptional changes triggered by water deficit on a drought-tolerant common bean cultivar. BMC Plant Biol. 2020;20:525. doi: 10.1186/s12870-020-02664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Kuang Z, Wang Y, Zhao Y, Tao Y, Cheng C, Yang J, Lu X, Hao C, Wang T, Cao X, Wei J, Li L, Yang X. PmiREN: a comprehensive encyclopedia of plant miRNAs. Nucleic Acids Res. 2019;48:D1114–D1121. doi: 10.1093/nar/gkz894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshavardhan VT, Van Son L, Seiler C, Junker A, Weigelt-Fischer K, Klukas C, Altmann T, Sreenivasulu N, Bäumlein H, Kuhlmann M. AtRD22 and AtUSPL1, members of the plant-specific BURP domain family involved in Arabidopsis thaliana drought tolerance. PLoS ONE. 2014;9:e110065. doi: 10.1371/journal.pone.0110065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori J, Boutilier KA, Campagne MMV, Miki BL. A conserved BURP domain defines a novel group of plant proteins with unusual primary structures. Mol Gen Genet. 1998;259:424–428. doi: 10.1007/s004380050832. [DOI] [PubMed] [Google Scholar]
- Hayat I, Ahmad A, Masud T, Ahmed A, Bashir S. Nutritional and health perspectives of beans (Phaseolus vulgaris L.): an overview. Cri Rev Food Sci Nutr. 2014;54(5):580–592. doi: 10.1080/10408398.2011.596639. [DOI] [PubMed] [Google Scholar]
- Juretic N, Hoen DR, Huynh ML, Harrison PM, Bureau TE. The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res. 2005;15:1292–1297. doi: 10.1101/gr.4064205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavas M, Baloğlu MC, Atabay ES, Ziplar UT, Daşgan HY, Ünver T. Genome-wide characterization and expression analysis of common bean bHLH transcription factors in response to excess salt concentration. Mol Genet Genomics. 2016;291:129–143. doi: 10.1007/s00438-015-1095-6. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Lee JJ, Woodward AW, Chen ZJ. Gene expression changes and early events in cotton fibre development. Ann Bot. 2007;100:1391–1401. doi: 10.1093/aob/mcm232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Tang H, Wang X, Paterson AH. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 2013;41:D1152–D1158. doi: 10.1093/nar/gks1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chen X, Chen Z, Cai R, Zhang H, Xiang Y. Identification and expression analysis of BURP domain-containing genes in Medicago truncatula. Front Plant Sci. 2016 doi: 10.3389/fpls.2016.00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizana C, Wentworth M, Martinez JP, Villegas D, Meneses R, Murchie EH, Pastenes C, Lercari B, Vernieri P, Horton P, Pinto M. Differential adaptation of two varieties of common bean to abiotic stress: I. Effects of drought on yield and photosynthesis. J Exp Bot. 2006;57:685–697. doi: 10.1093/jxb/erj062. [DOI] [PubMed] [Google Scholar]
- Lu SN, Wang JY, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Marchler GH, Song JS, Thanki N, Yamashita RA, Yang MZ, Zhang DC, Zheng CJ, Lanczycki CJ, Marchler-Bauer A. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2020;48:D265–D268. doi: 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher C, Stein L, Ware D. Evolution of Arabidopsis microRNA families through duplication events. Genome Res. 2006;16:510–519. doi: 10.1101/gr.4680506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus JT, Aquea F, Espinoza C, Vega A, Cavallini E, Dal Santo S, Cañón P, de la Guardia AR-H, Serrano J, Tornielli GB, Arce-Johnson P. Inspection of the grapevine BURP superfamily highlights an expansion of RD22 genes with distinctive expression features in berry development and ABA-mediated stress responses. PLoS ONE. 2014;9:e110372. doi: 10.1371/journal.pone.0110372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClean PE, Raatz B. Common bean genomes: mining new knowledge of a major societal crop. The common bean genome. Berlin: Springer; 2017. pp. 129–145. [Google Scholar]
- Pareek A, Dhankher OP, Foyer CH. Mitigating the impact of climate change on plant productivity and ecosystem sustainability. J Exp Bot. 2020;71:451–456. doi: 10.1093/jxb/erz518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira WJ, Melo ADTO, Coelho ASG, Rodrigues FA, Mamidi S, Alencar SAD, Lanna AC, Valdisser PAMR, Brondani C, Nascimento-Júnior IRD, Borba TCDO, Vianello RP. Genome-wide analysis of the transcriptional response to drought stress in root and leaf of common bean. Genet Mol Biol. 2020 doi: 10.1590/1678-4685-GMB-2018-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett FB, Meeks-Wagner DR. Seeing double: appreciating genetic redundancy. Plant Cell. 1995;7:1347–1356. doi: 10.1105/tpc.7.9.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, McClean PE, Mamidi S, Wu GA, Cannon SB, Grimwood J, Jenkins J, Shu S, Song Q, Chavarro C, et al. A reference genome for common bean and genome-wide analysis of dual domestications. Nat Genet. 2014;46:707–713. doi: 10.1038/ng.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Wei G, Wang L, Dong Q, Zhao Y, Chen B, Xiang Y. Genome-wide analysis of BURP domain-containing genes in Populus trichocarpa. J Integr Plant Biol. 2011;53:743–755. doi: 10.1111/j.1744-7909.2011.01068.x. [DOI] [PubMed] [Google Scholar]
- Sheshadri SA, Nishanth MJ, Simon B. Stress-mediated cis-element transcription factor interactions interconnecting primary and specialized metabolism in planta. Front Plant Sci. 2016 doi: 10.3389/fpls.2016.01725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smialowski P, Doose G, Torkler P, Kaufmann S, Frishman D. PROSO II–a new method for protein solubility prediction. FEBS J. 2012;279:2192–2200. doi: 10.1111/j.1742-4658.2012.08603.x. [DOI] [PubMed] [Google Scholar]
- Sun H, Wei H, Wang H, Hao P, Gu L, Liu G, Ma L, Su Z, Yu S. Genome-wide identification and expression analysis of the BURP domain-containing genes in Gossypium hirsutum. BMC Genomics. 2019;20:558. doi: 10.1186/s12864-019-5948-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Cao Y, Qiu J, Gao Z, Ou Z, Wang Y, Zheng Y. Expression of a vacuole-localized BURP-domain protein from soybean (SALI3–2) enhances tolerance to cadmium and copper stresses. PLoS ONE. 2014;9:e98830. doi: 10.1371/journal.pone.0098830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teerawanichpan P, Xia Q, Caldwell SJ, Datla R, Selvaraj G. Protein storage vacuoles of Brassica napus zygotic embryos accumulate a BURP domain protein and perturbation of its production distorts the PSV. Plant Mol Biol. 2009;71:331. doi: 10.1007/s11103-009-9541-7. [DOI] [PubMed] [Google Scholar]
- Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, Kissinger JC, Paterson AH. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S, Salinas M, Höhmann S, Berndtgen R, Huijser P. miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell. 2010;22:3935–3950. doi: 10.1105/tpc.110.079343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Li Y, Yan Y, Wang K, Gao Y, Hu Y. Genome-scale identification of soybean BURP domain-containing genes and their expression under stress treatments. BMC Plant Biol. 2010;10:197. doi: 10.1186/1471-2229-10-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xun H, Yang X, He H, Wang M, Guo P, Wang Y, Pang J, Dong Y, Feng X, Wang S, Liu B. Over-expression of GmKR3, a TIR–NBS–LRR type R gene, confers resistance to multiple viruses in soybean. Plant Mol Biol. 2019;99:95–111. doi: 10.1007/s11103-018-0804-z. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana. Mol Gen Genet MGG. 1993;238:17–25. doi: 10.1007/BF00279525. [DOI] [PubMed] [Google Scholar]
- Yıldırım K, Kaya Z. Gene regulation network behind drought escape, avoidance and tolerance strategies in black poplar (Populus nigra L.) Plant Physiol Biochem. 2017;115:183–199. doi: 10.1016/j.plaphy.2017.03.020. [DOI] [PubMed] [Google Scholar]
- Zhao M, Meyers BC, Cai C, Xu W, Ma J. Evolutionary patterns and coevolutionary consequences of MIRNA genes and microRNA targets triggered by multiple mechanisms of genomic duplications in soybean. Plant Cell. 2015;27:546–562. doi: 10.1105/tpc.15.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Heupel RC, DellaPenna D. The beta subunit of tomato fruit polygalacturonase isoenzyme 1: isolation, characterization, and identification of unique structural features. Plant Cell. 1992;4:1147–1156. doi: 10.1105/tpc.4.9.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primer sequences of PvBURP genes for qRT-PCR analysis (XLSX 11 kb)
Table S2. Location of the BURP domain and signal peptide in BURP proteins (XLSX 12 kb)
Table S3. A-biotic stress response-related cis-elements in the promoter sequences of PvBURPs covering the upstream region of 2000 nucleotides from the gene start codon (XLSX 11 kb)
Table S4. Secondary structures of PvBURP proteins (XLSX 25 kb)
Table S5. Expression profiles of PvBURP genes in different RNA-seq dataset based on log2FC value (XLSX 11 kb)
Figure S1. Conserved motifs in PvBURP proteins were obtained through the MEME Suit. Conserved motifs were analyzed by choosing 15 motifs and repeating any number of motifs. (PNG 1192 kb)
Figure S2. 3D structure of PvBURP proteins illustrated using Phyre2 server with an “intensive” mode. (PNG 640 kb)
Figure S3. Prediction of transmembrane helices in PvBURP proteins. (PNG 534 kb)