Abstract
Increasing the vulnerability of plants especially crops to a wide range of cold stress reduces plant growth, development, yield production, and plant distribution. Cold stress induces physiological, morphological, biochemical, phenotypic, and molecular changes in plants. Transcription factor (TF) is one of the most important regulators that mediate gene expression. TF is activated by the signal transduction pathway, together with cis-acting element modulate the transcription of cold-responsive genes which contribute to increasing cold tolerance in plants. Here, AP2/ERF TF family is one of the most important cold stress-related TF families that along with other TF families, such as WRKY, bHLH, bZIP, MYB, NAC, and C2H2 interrelate to enhance cold stress tolerance. Over the past decade, significant progress has been found to solve the role of transcription factors (TFs) in improving cold tolerance in plants, such as omics analysis. Furthermore, numerous studies have identified and characterized the complexity of cold stress mechanisms among TFs or between TFs and other factors (endogenous and exogenous) including phytohormones, eugenol, and light. The role, function, and relationship among these TFs or between TFs and other factors to enhance cold tolerance still need to be clarified. Here, the current study analysed the role of AP2/ERF TF and the linkages among AP2/ERF with MYB, WRKY, bZIP, bHLH, C2H2, or NAC against cold stress tolerance.
Keywords: Abiotic stress, Cold stress, Cold tolerance, Low temperature, Transcription factors
Introduction
Global warming drives a drastic change in climate, which is accompanied by an increase in intensity and frequency of abiotic stresses including temperature, salinity, and drought stress (Watt et al. 2020; Zandalinas et al. 2021). Meanwhile, plants are sessile and acclimatize to these abiotic stresses (Beloiu et al. 2020). Cold stress (0–15 °C) and freezing stress (< 0 °C) are the major stresses in temperate and few subtropical areas that adversely influence plant growth and development, reduce yield production (Kang et al. 2020; Ritonga et al. 2021), and also cause worldwide economic losses in crop production. Cold stress generally alters all physio-chemical pathways of a living cell that influences enzyme activity, solute diffusion rates, membrane fluidity, and reverse the interactions of macromolecules like DNA, RNA, and proteins (Gualerzi et al. 2003).
Cell membrane and cell structure stability is the key point for plants survival under cold stress (Chen et al. 2018). Freezing stress leads to ice formation in plant tissues (Puhakainen et al. 2004). This phenomenon causes into the extracellular space of plant cells filled with ice crystals which leads to dehydration due to the water flowing (Ritonga and Chen 2020). Moreover, cold stress alters metabolic pathways of anthers to induce pollen sterility (Sharma and Nayyar 2016). Cold stress induces ovule infertility, flower abortion, fertilization breakdown and low quality of seed, and eventually lead to low grain yield in plants (Thakur et al. 2010; Alisoltani et al. 2019). Cold/freezing stress also leads to withered, dwarfism, and chlorosis in plants (Yadav 2010; Gu et al. 2019).
The ability of plants to survive under cold stress is referred to cold acclimation (CA) process (Kargiotidou et al. 2010). It was shown that non acclimated Arabidopsis thaliana is more sensitive to freezing stress (-20 °C) compared to 4 °C cold-acclimated A. thaliana under (Yu et al. 2021). The severity of the cold stress effect is related to species genotype, stress intensity, and the duration of cold exposure (Carvallo et al. 2011; Londo et al. 2018; Mehrotra et al. 2020). Prerostova et al. (2021) found that hormones such as salicylic acid (SA), jasmonic acid (JA), and abscisic acid (ABA) were elevated in the crowns, leaves, and plant roots under cold stress. Besides, antioxidant enzyme activities protect plants from higher H2O2 and O2− content under cold stress (Zhao et al. 2021). Over the past ten years (2010–2020), numerous studies had revealed that transcription factors (TFs) are primary regulators associated with cold stress (Mitsis et al. 2020). TFs play vital roles in regulating signal transduction, as well as gene expression under cold stress (Chen et al. 2015a). Several cold stress-responsive TF families have been analyzed and identified in numerous plant species (Mehrotra et al. 2020), including AP2/ERF (Byun et al. 2015; Lv et al. 2019), NAC (Nakashima et al. 2012), WRKY (Zhang et al. 2016), bZIP (Liu et al. 2018a), bHLH (Yao et al. 2018), and MYB (Su et al. 2014). Interestingly, TFs and stress-responsive genes regulate the plant's responses during and after cold/freezing stress (Mizoi et al. 2012).
Kashyap and Deswal (2017) reported the expression of C-repeat binding factor (CBF) gene from Hippophae rhamnoides (HrCBF) initially increased after 0.5 h of 4 °C exposure and continue to increase at 1 h, 3 h, 6 h, 24 h, and 1 week of cold exposure. Overexpression of CBF1 of Prunus persica (PpCBF) in Malus domestica was observed for three years in three growing seasons. It was revealed that PpCBF1 regulated anthocyanine and carotenoid content of transgenic apple during fall seasons, while PpCBF1 regulated plant height and lateral branches of transgenic apple during summer. These results illustrated that PpCBF1 functions during and after cold stress in transgenic M. domestica (Artlip et al. 2014). Some TFs also interact with other TFs to activate or repress the general transcriptional process (Eulgem and Somssich 2007). The current study summarized the role of AP2/ERF TF and the understanding of different TFs involved in cold stress to develop plant species resistant to low temperatures to achieve agricultural and forestry sustainability through TFs utilization.
Cold stress tolerance mechanism in plants
Plants utilize structural modifications such as alteration of membrane fluidity, protein structure transformation, and cytoskeleton movement, to respond and adapt to cold stress (Mehrotra et al. 2020). Previous studies have revealed that plants have different sensory levels under cold stress (Luo et al. 2020a). However, cold stress is initially sensed by receptors on plant membrane, which alter membrane fluidity and subsequently induce calcium cation (Ca2+), cyclic adenosine monophosphate (CAMP), and reactive oxygen species (ROS) production. Chloroplast also acts as a signal modulator to the nucleus through ROS production. Ca2+ signaling acts as a mediator of plant response to cold stress (Yuan et al. 2018). Meanwhile, Ca2+, CAMP, and ROS signaling mediate signal transduction via Calcineurin-B Like proteins (CBL), Ca2+-dependent protein kinases (CPKs/CDPKs) and CBL-interacting protein kinases (CIPKs) to the nucleus via a pathway interceded by ICE-CBF/DREB TFs.
In general, cold stress mechanism in plants involves the inducer of CBF expression 1 (ICE1), an inducer of CBF/ Dehydration-Responsive Element-Binding-Factor (DREB), which is interacted with cold regulated genes (COR) signaling pathway (Yang et al., 2019; Zhang et al., 2020). Thus, AP2/ERF, WRKY, bZIP, MYB, bHLH, C2H2, and NAC TFs regulate the expression of gene to activate cold stress-responsive genes (CORs), resulting in physiological responses to cold stress (Byun et al. 2015).
AP2/ERF, a key TF family in cold stress
The APETALA2/Ethylene responsive factor (AP2/ERF) is a large transcription factor (TF) family in plants involved in plant developmental processes and multiple environmental stimuli (Klay et al. 2018). The most famous family members of the AP2/ERF involved in cold stress are DREBs, also known as CBFs. CBFs act as pioneers of plant regulatory networks in response to cold stress and has homologs in many plants. Overexpression of CBF homologs from Oryza sativa, L. perenne, Zea mays, Hordeum vulgare, and T. aestivum in transgenic tobacco or A. thaliana have been found to increase the expression of cold-regulated genes belonging to the CBF regulon and cold/freezing tolerance (Mizuno et al. 2006; Medina et al. 2011; Rasmussen et al. 2013; Zhu et al. 2020).
Gene expression is an intricate mechanism, as well as CBFs regulatory network in the plant during cold stress (Shi et al. 2018). The role of CBFs in enhancing cold/freezing tolerance in plants has been well established in many species (Winfield et al. 2010; Artlip et al. 2014). CBFs activate cold stress-responsive genes through specific binding to the dehydration-responsive C-repeat (DRE/CRT) cis-acting element (A/GCCGAC) in RD29A promoters (Mizoi et al. 2012) to increase cold stress tolerance in plants (Hao et al. 2017). However, CBFs may have differential functions in cold stress response because different CBFs may activate disparate cis-acting elements. Transient transactivation tests have revealed that all Vitis riparia CBFs, except CBF5, can bind to DRE/CRT elements, whereas CBF3 and CBF4 prefer the CRT element (Carlow et al. 2017). In Zoysia japonica, ZjDREB1.4 demonstrated solid transactivation activity under -8 °C treatment, but weak binding to the DRE with ACCGAC as the core sequence. The ZjDREB1.4 protein preferentially binds to GCCGAC rather than ACCGAC (Feng et al. 2019). Using a TF-centered yeast one-hybrid (Y1H) experimental system, Lv et al. (2019) showed that BpERF13 activated the reporter gene by binding to LTRECOREATCOR15 and MYBCORE cis-elements under low temperature. Chromatin Immunoprecipitation-sequencing (ChIP-seq) and Chromatin Immunoprecipitation Polymerase Chain Reaction (ChIP-PCR) experiments further proved that BpERF13 binds to the promoter of CBF genes as well (Lv et al. 2019).
The up-regulated capacity of CBFs by low temperature is related to species genotype (Sakuma et al. 2002) and might be influenced by polymorphisms within promoter sequences of plants (Pan et al. 2013). For instance, the response of a cold-tolerant cultivar is slightly slower than the cold-sensitive cultivar under cold stress in Brassica rapa. Three OsCBFs genes (OsCBF1-3) showed a temporary induction in the CA process (10 °C) and were much more intense in Indica rice (93–11 variety) than Japonica rice (Nipponbare variety). OsLIP5 and OsLIP9 (the candidate downstream genes) were induced in Indica rice but not in Japonica rice. This result indicates that polymorphisms within promoter sequences caused differential expression of CBF regulon (Pan et al. 2013).
In addition, overexpression of CBFs improves cold tolerance through elevating antioxidant enzymes, including catalase (CAT), ascorbate peroxidase (APX), peroxidase (POD), superoxide (SOD), and proline, and also reduce EL, MDA, H2O2, and O2− contents under cold stress condition (4 °C) (Sun et al. 2019; Hu et al. 2020). Li et al. (2018) used Clustered Regularly Interspaced Short Palindromic Repeats Associated Protein 9 (CRISPR-Cas9) system to generate slcbf1 mutants. The mutants had lower proline and higher antioxidant enzyme activity compared to wild-type. The transgenic plants developed by the AP2/ERF TF family have been listed in Table 1.
Table 1.
AP2/ERF TF genes in the transgenic plant under cold stress
| No. | Gene | From | To | Other functions | Location | Homologs | Temperature and time points | Beneficial roles | References |
|---|---|---|---|---|---|---|---|---|---|
| 1 | AmCBF2 | Avicennia marina | A. marina | Salt, drought, heavy metals stress | NA | NA | 5 °C for 15 min, 2 h, 12 h, 24 h, 48 h, and 120 h | Involve in a cold signaling pathway | Peng et al. (2013b) |
| 2 | GmDREB1A;1GmDREB1A;2 | Glycine max | A. thaliana | NA | NA | NA | 4 °C for 0 h, 1 h, and 24 h | Responsive to cold stress culminating | Yamasaki and Randall (2016) |
| 3 | HvSHN1 | H. vulgare | Nicotiana tabacum | Salt, drought, and heat stress | Nucleus | TdSHN1AtSHN1, 2,3 | 4 °C for 5 d | Maintain chlorophyll content under multiple abiotic stresses | Djemal et al. (2018) |
| 4 | PpCBF1 | M. domestica | P. persica | NA | NA | NA | NA | A modest increase in cold hardiness and induces dormancy | Artlip et al. (2014) |
| 5 | TdSHN1 | Triticum durum | N. tabacum | Salt, and water stress | NA | NA | 4 °C for 5 d | Reduce the stomatal density, and increase the expression of osmotic stress pproteins, lipid transfer proteins (LTPs), defensive proteins, genes encoding oxidative stress-related proteins, and the wax biosynthesis gene (NtCER1)s | Djemal and Khoudi (2016) |
| 6 | DaCBF7 | Deschampsia antarctica | O. sativa | NA | Nucleus | NA | 4 °C for 8 d | Induces diverse sets of genes and confers cold tolerance | Byun et al. (2015) |
| ZjDREB1.4 | Z. japonica | A. thaliana | NA | Nucleus | DREB1 | 20, 15, 10, or 6 °C for 2 h | Induces the expression of multiple genes including a part of the CBF-regulon and | Feng et al. (2019) | |
| 7 | BpERF13 | B. platyphylla | B. platyphylla | NA | Nuclei | NA | 4 °C for 0, 2, 4, 6, or 12 h | Regulates physiological processes under cold stress in woody plants | Lv et al. (2019) |
| 8 | CdERF1 | Cynodon dactylon | A. thaliana | NA | Nucleus | NA | 4 °C, for 0, 1, 3,6, 12, and 24 h | Activate stress-related genes, PODs, CBF2 and LTPs | Hu et al. (2020) |
| 9 | VpERF2 | Vitis pseudoreticulata | A. thaliana | Drought, and heat stress | Nucleus | NA | 4 °C for 0, 2, 4, 6, 8, 10, 12, and 24 h | Involve in abiotic stress-responsive pathways | Zhu et al. (2013) |
| 10 | CsERF | Citrus sinensis | N. tabacum | NA | Nuclei | NA | 4 °C for 0 h, 2 h, 4 h, 8 h, 12 h and 24 h | Activate four indicator genes: two cold responsive transcription factor genes (NtCBF1 and NtCBF3), and two cold-induced genes(NtERD10B and NtERD10C) | Ma et al. (2014) |
| 11 | SmCBF1, SmCBF2, SmCBF3 | S. melongena | S. melongena | Drought, high salinity, and ABA stress | Nucleus and cytoplasm | NA | 4 °C for 0 h, 0.5 h, 1 h, 3 h, 6 h, 12 h, and 24 h | Involve in regulation of the response to abiotic stress | Zhou et al. (2018) |
| 12 | VaERF080, VaERF087 | Vitis amurensis | thaliana | NA | NA | NA | 4 °C for 0, 2, 4, 8, 24,and 48 h | Increase the antioxidant enzyme activities and nine representative cold-responsive Gene expressions (CBF1, CBF2, ICE1, ZAT12, KIN1, SIZ1, RD29A, COR15A, and COR47) | Sun et al. (2019) |
| 13 | SmCBF | S. melongena | S. melongena | NA | NA | NA | 4 ± 0.5 °C for 12 d | Increase cold tolerance and together with eugenol fumigation reduce chilling injury | Huang et al. (2019) |
| 14 | BjCBF | Brassica juncea | B. juncea | NA | Nucleus | NA | 4 °C for 0, 3, 6, 24 h | Binds to the DRE elements in the promoter of downstream cold-responsive genes resulting in increased cold tolerance | Kashyap and Deswal (2019) |
| 15 | AdERF2-AdERF14 | Actinidia deliciosa | A. deliciosa | NA | NA | NA | 0 °C for 12 weeks | Modulate cold stress response through changes in AP2/ERF key family expression | Yin et al. (2012) |
| 16 | EgCBF1 | Eucalyptus globulus | E. globulus | NA | NA | NA | 4 °C for 30 min | Participates in the cold-responsive pathway of E. globulus | Gamboa et al. (2007) |
| 17 | MfDREB1 and MfDREB1s | Medicago falcate | M. falcate | NA | NA | NA | 4 °C for 0 h, 0.5 h, 1 h, 3 h, 6 h and 12 h | Contribute to cold tolerance | Niu et al. (2010) |
| 18 | PhCBF4a and PhCBF4b | Populus hopeiensis | N. tabacum | Dehydration, and high salinity stress | NA | NA | 4 °C for 6 h | Induce elevated expression of the CBF/DREB1 regulons without prior stimulus | Wang et al. (2014) |
| 19 | AtCRAP2 | A. thaliana | A. thaliana | NA | NA | NA | 4 °C | Promote flowering under short-day conditions | Luo et al. (2020a) |
| 20 | BrcERF-B3 | B. rapa | B. rapa | Salt and plant hormone | NA | NA | 4 °C for 0, 1, 2, 4, 8, and 12 h | Involve in the formation of abnormal flower and BrcERF-B3 is more significant under cold stress in mutant plants | Xu et al. (2016) |
| 21 | GbCBF1 | G. hirsutum | N. tabacum | NA | NA | NA | 4 °C for 0, 0.25, 0.5, 1, 2, 4, 8, 16, or 24 h | Enhances cold tolerance in transgenic tobacco through reduce EL and increase proline and soluble sugar contents | Guo et al. (2011) |
| 22 | CRF2, CRF3 | A. thaliana | A. thaliana | NA | NA | NA | 1 °C for 8 h | Regulate Arabidopsis lateral root initiation under cold stress | Jeon et al. (2016) |
| 23 | OsCBF1, OsCBF2, OsCBF3 | O. sativa | O. sativa | NA | NA | NA | 5 °C for 3 d and 7 d | Transiently inducedin the process of cold acclimation to increase cold tolerance | Pan et al. (2013) |
| 24 | SlDREB3 | S. lycopersicum | S. lycopersicum | NA | NA | NA | 4 °C for 0, 3, 6, 9, 12 and 24 h | Improve the cold tolerance by upregulating SlLEAs expression | Wang et al. (2019a) |
| 25 | CRF4 | A. thaliana | A. thaliana | NA | NA | NA | 4 °C for 7 d | Contribute in short-term CA resulting in the enhancement of freezing tolerance | Zwack et al. (2016) |
| 26 | FTL1/DDF1 | A. thaliana | A. thaliana | Drought and heat stress | NA | NA | −5 °C for 24 h | Regulate response to freezing stress | Kang et al. (2011) |
| 27 | MgCBF6 | Miscanthus giganteus | M. giganteus | NA | NA | SbCBF3, SbDBF2, SbCBF6, ZmCBF3,ZmDBF2, and ZmCBF6 | 5 °C – (−3) °C for 3d | The dormancy strategy for overwintering | Rapacz et al. (2018) |
CBF genes are rapidly and transiently induced by low temperature and attenuated during the later stages of the cold stress response. In A. thaliana, the attenuation process of CBFs is mediated by protein kinase Brassinosteroid-Insensitive2 (BIN2) under freezing assay. The A. thaliana seedlings were moved to 4 °C conditions for 3 d and followed by − 9 °C treatment for 0.5 h. BIN2 associates with and phosphorylates ICE1 under prolonged low-temperature stress to facilitate the interaction between the E3 ubiquitin ligase High Expression of Osmotically Responsive Gene1 (HOS1) and ICE1, resulting in degradation of ICE1 in A. thaliana (Ye et al. 2019). On the contrary, cold-responsive protein kinase 1 (CRPK1) which is acts as a negative cold regulator phosphorylates 14–3-3 proteins. In the cytosol, the phosphorylated 14–3-3 proteins were translocated to nucleus and in association with CBFs disturbed the key cold-responsive CBF proteins under 4 °C for 3 h (Liu et al. 2017). Furthermore, when A. thaliana is imposed from 4 °C to -2 °C (2 °C per hour), the defense regulatory genes, such as PAD4, SAG101, and EDS1 increased the freezing tolerance by enhancing CBFs and their regulons (Chen et al. 2015b).
It has been shown that CBFs/DREBs play a key role in CORs induction to increase cold tolerance in plants. Recently, it was revealed that the MEKK1–MKK2–MPK4 cascade and six additional mitogen-activated protein kinases are involved in a gene regulatory network to regulate transcription factors and cold tolerance genes in Betula platyphylla (Chen et al. 2021). Meanwhile, proteins involved in reducing detrimental effects associated with cold stress have also been developed and divided into three categories: signaling molecules and regulatory proteins, degradative and defensive proteins, and protective proteins. It was proved that signaling, degradative, and defensive proteins function from upstream to downstream levels and, consequently, lead to cold tolerance improvement (Kazemi-Shahandashti and Maali-Amiri 2018).
More importantly, DELLAs play a significant role in plant growth and development which is act as pivotal components of the GA signal transduction pathway in plants. DELLA genes contribute to inhibiting plant growth under cold stress (4 °C) in Glycine soja (Li et al. 2011). DELLAs work jointly with CBFs to retard plant growth. DELLAs contribute to the cold induction of CBF1, CBF2, and CBF3 through JA signaling. In addition, CBF3 encourages DELLAs accumulation by suppressing GA biosynthesis (Zhou et al. 2017). Taken together, JA plays a pivotal role in modulating multiple plant growth and development. It is an oxylipin compounds group that is ubiquitous in the plant kingdom (Hu et al. 2017). Low temperature-induced endogenous JA to activate ICE1 and ICE2, resulting in the activation of CBF/DREB1 transcriptional cascade (Hu et al. 2013).
Hu et al. (2017) also stated that JA positively modulates the transcriptional pathway of CBF to up-regulate COR genes, resulting in the cold tolerance improvement. In addition, JA associates with several hormones signaling pathways including ethylene, auxin, and gibberellin to regulate cold tolerance (Liu and Timko 2021). In specific circumstances, the outcome of hormone signaling may induce Brassinazole-Resistant 1 (BZR1) and consequently upregulates the expression of CBFs. BZR1 acts as an upstream of CBF1 and CBF2 and directly regulates CBF1 and CBF2 expression to increase cold stress tolerance without affecting plant growth (Barrero-Gil and Salinas 2017). Previous findings have also shown that BZR1 regulates other COR genes uncoupled with CBFs to regulate plants response to freezing stress (−4 °C – (−7) °C) (Li et al. 2017).
It has been reported that CBFs expression was also influenced by several factors such as exogenous ABA, circadian clock, eugenol, and light condition under cold stress (Yang et al. 2005; Jung and Seo 2019). In S. melongena, CBFs were strongly, rapidly, and transiently induced by exogenous ABA, indicating that SmCBFs might be affected plant response to ABA (Zhou et al. 2018). Light signaling components like phytochrome A (phyA), phytochrome B (phyB), and Phytochrome-Interacting Factors (PIFs) are involved in the expression of CBFs (Xu and Deng 2020). The interaction of CBFs with PIF3 was assumed to attenuate the mutually guaranteed destruction of PIF3–phyB. The interaction of phyB and CBFs positively regulates freezing tolerance (-5 °C and -9 °C) by degrading PIF1, PIF4, and PIF5 and eventually modulating the expression of COR genes in A. thaliana (Jiang et al. 2020).
Light and cold signals were integrated and transduced by molecular regulators to downstream signaling pathways. These molecular regulators also control the transcription process of numerous cold responsive and growth promoting genes, consequently balancing plant growth and development and increasing cold tolerance. Furthermore, investigations on the effect of eugenol fumigation on the chilling injury at 4 ± 0.5 °C to S. melongena have revealed that eugenol treatment increased SmCBF expression. This finding suggests that eugenol has a potential effect on alleviating cold injury in S. melongena (Huang et al. 2019). In addition to CBF genes, Cytokinin Response Factor2 (CRF2) and Cytokinin Response Factor3 (CRF3), members of AP2/ERF, contribute to cold-responsive genes and increase the lateral root adaptation of the plant to face cold stress (1 °C) (Jeon et al. 2016).
Other cold stress-related TFs
Basic helix-loop-helix (bHLH) is eukaryotes second-largest protein family, which has important functions in plant growth, survival, and the response to multiple abiotic stresses, especially chilling and freezing stress. Many studies have revealed that MYC-type bHLH activates the expression of CBF genes. Overexpression of SlICE1a, a member of MYC-type bHLH TF in N. tabacum, activated the expression of CBF3/DREB1A and their target genes, consequently increased proline levels, sugar contents, and late embryogenesis abundant (LEA)proteins under 4 °C treatment (Feng et al. 2013). In addition, expression of CBFs, such as AtCBF1-3, cold-responsive genes (AtCOR15A, AtCOR47, AtRD29A, and AtKIN1), and stress-responsive genes NtDREB1-3, NtLEA5, NtP5CS, and NtERD10C was also significantly increased in DlICE1 and RmICE1-overexpressing lines under cold stress (4 °C) (Yang et al. 2019; Zuo et al. 2019; Luo et al. 2020b).
Protein–protein interaction analysis in transgenic N. tabacum showed that MabHLH1, MabHLH2, and MabHLH4 interacted with each other to form heterodimers in the nucleus. Indeed, those genes also interacted with MaICE1, an important upstream component of cold signaling. The interaction of MabHLHs with MaICE1 might form a vast protein complex in the nucleus (Peng et al. 2013a). PuICE1 can physically interact with PuHHP1 protein to increase PuDREBa transcriptional levels under cold stress conditions (Huang et al. 2015). On the other hand, the interaction of MYC67 and MYC70 with ICE1 adversely affects cold tolerance in A. thaliana. Overexpression of MYC67 and MYC70 enhanced the cold sensitivity and down-regulated the cold-responsive gene expression. The cis-elements in the CBF3/DREB1A promoter bound by MYC proteins disrupt ICE1 interaction with the cis-elements (Ohta et al. 2018).
The overexpression of basic leucine zipper (bZIP) genes and CBFs significantly enhanced the resistance of chilling injury and cold storage in P. persica (Monteagudo et al. 2018). bZIP genes are involved in freezing stress and act as positive regulators (Cai et al. 2018). However, TabZIP6 has been found to negatively regulating freezing tolerance, where CBFs and several COR genes were down-regulated in TabZIP6-overexpressing lines by cold treatment.
Other studies also revealed that the overexpression of C2H2 Zinc finger proteins family gene (SlCZFP1) in transgenic O. sativa and A. thaliana induced the COR gene constitutive expression and increased freezing tolerance for non-acclimate transgenic plants under freezing stress conditions (Zhang et al. 2011). In banana, MaC2H2-2 and MaC2H2-3 were specifically induced by cold stress, which subsequently repressed MaICE1 expression (Han and Fu 2019). Sequence analyses revealed that a CRT/DRE element was found in ZFP245 promoter region and ZFP182 (Huang et al. 2005, 2012).
Another TF family that contributes to cold stress is Myeloblastosis (MYB) TF. The overexpressing of MdMYB15L in red-fleshed apple callus prevented the expression of MdCBF2 and resulted in decreased cold tolerance but did not give any effect on anthocyanin levels after cold treatment. ChIP-PCR and electrophoresis mobility shift assay (EMSA) analysis indicated that MdMYB15L binds to type II cis-acting element found in the promoter of MdCBF2 under 4 °C treatment (Xu et al. 2018). A study of MdMYB108L expression under cold stress showed that MdMYB108L upregulated MdCBF3 by binding its promoter region to increase cold tolerance in M. domestica. Conversely, the expression of MdHY5 was significantly downregulated by MdMYB108L (Wang et al. 2019d). EMSA and transient expression assay proved that MdHY5 positively regulated the transcription of MdCBF1 by binding to the G-Box motif of its promoter (An et al. 2017). Furthermore, CBF-independent cold-regulated genes expressions were also regulated by MdHY5 under 4 °C for ten days. Similar to MdMYB108L, MdMYB73 increased the expression of MdCBF2- MdCBF5 in transgenic M. domestica (Wang et al. 2019d), illustrating that MdMYB73 enhanced cold tolerance through CBF cold response pathway under 4 °C condition (Zhang et al. 2017).
The interactions of some cold stress-related genes in plants were shown in Fig. 1. The current study identified the interaction network using STRING version 11.0 (https://string-db.org/cgi/) (STRING 2021). AtACS2 showed co-expression patterns with other genes such as AtERF2, AtWRKY46, AtMYB73, AtABF2, and AtRHL41. Co-expression is the simultaneous expression of two or more genes which makes it impossible to determine whether AtERF2 activates AtACS2 and AtMYB73 activates AtRHL41, or AtMYB73 activates AtWRKY46, or whether another gene activates them in A. thaliana under cold stress. In addition, RHL41 and MYB73 were experimentally found that enhance CA and abiotic stress tolerance in plants (Iida et al. 2000; Rasmussen et al. 2013; Barrero‐Gil et al. 2016).
Fig. 1.
Interaction network analysis of TFs genes identified in A. thaliana by using STRING. The interaction network has significantly more interactions than expected. This means that 42 A. thaliana proteins have more interactions among themselves, indicating the proteins are at least partially biologically connected as a group. The line color is related to the type of interaction. The green line shows gene neighborhood, the pink line means experimentally determined, the black line means co-expression, the dark blue line means gene co-occurrence, and the blue line means protein homology. (For more interpretation of the color codes in this figure legend, the reader is referred to the web version of this network analysis (https://string-db.org/cgi/network?taskId=btbDj6SKVsTf&sessionId=b4DLtXYWAftY)
MdMYB88 and MdMYB124 are examples of genes that promote anthocyanin accumulation in response to cold stress. MdMYB88/MdMYB124 from R2R3-MYB TF acts as a key regulator of the MdCCA1, which enhances the expression of MdCBF3 under cold stress (4 °C) in M. domestica or A. thaliana (Xie et al. 2018). Interestingly, VcMYB4a expression in Vaccinium corymbosum was downregulated by cold, salt, and drought treatment, but it was induced by freezing and heating. Additionally, gene expression enhanced abiotic sensitivity under cold, freezing, heat, drought, and salt stress in V. corymbosum callus, illustrating that VcMYB4a might act as an important repressor of abiotic stress in this species (Zhang et al. 2020). OsMYB30 down-regulated a few β-amylase (BMY) genes during cold stress (4 °C), bonded to the promoters of BMY genes and interacted with OsJAZ9 to repress BMY gene expression (Lv et al. 2017).
Regarding the OsMYB30, the use of CRISPR–Cas9 significantly increases yield production and cold stress resistance in O. sativa. Zeng et al. (2020) used the CRISPR–Cas9 system to edit two target sites of OsMYB30 with high efficiency 63% for OsMYB30-site1, and 58% for OsMYB30-site2. The results showed that the osmyb30 mutants exhibited enhanced cold tolerance. The study proved that cold stress resistance and high performance rice varieties can be resulted via gene-editing techniques. However, there is still little information concerning how PLANT U-BOX 25 and 26 (PUB25 and PUB26) contribute the enhancement of CBF gene expression by degrading MYB15 to improve cold tolerance in A. thaliana under 4 °C treatment (Wang et al. 2019c).
Meanwhile, cold signaling pathway was positively regulated by CsWRKY46 in an ABA-dependent manner. Numerous cis-regulatory elements were found in the upstream region of WRKY genes in C. sativus. Phytohormones (MeJa, ABA, SA, GA, auxin, and zeatin) were associated with WRKY expression to obtain cold tolerance enhancement (Zhao et al. 2015; Govardhana and Kumudini 2020). CsWRKY46 associates with the W-box (TTGACC/T) in the ABA-responsive transcription factor (ABI5) to increase the expression of RD29A and COR47, the member of stress-inducible genes. Consequently, C. sativus over-expressing CsWRKY46 and A. thaliana over-expressing CsWRKY46 had higher survival rates, higher proline accumulation, and less EL and MDA levels under 4 °C cold stress (Zhang et al. 2016). An increase in cold stress tolerance can be seen by a higher survival rate (Ju et al. 2020).
Overexpression of SlNAM1, a member of NAC TF family in transgenic N. tabacum increased minor wilting, photosynthetic rates (Pn), germination rates, and osmolytes contents under chilling stress. Reduction in H2O2 contents under cold stress has a pivotal role in minimizing the cell membrane's oxidative damage in transgenic N. tabacum overexpressed SlNAM1. The increased cold stress tolerance in transgenic N. tabacum was also assumed to increase transcripts of NtDREB1, NtP5CS, and NtERD10s (Li et al. 2016). Activation of the DREB/CBF–COR pathway caused by over-expression of GmNAC20 under freezing stress may promote and control lateral root formation through auxin signaling-related genes alteration (Hao et al. 2011). Likewise, PbeNAC1 protein associates with PbeDREB1 and PbeDREB2A to enhance mRNA levels in some stress-associated genes in P. betulifolia under cold stress (Jin et al. 2017).
Other studies in NAC genes from M. domestica found that MdNAC029 acts as a negative regulator of cold stress. Over-expression of MdNAC029 decreased cold tolerance in calli of M. domestica and transgenic A. thaliana. These findings were supported by EMSA and transient expression assays which illustrated that MdNAC029 suppressed the MdCBF1 and MdCBF4 expression by binding to their promoters (An et al. 2018). Moreover, Liu et al. (2018b) found that ShNAC1 has a negative role in cold stress tolerance in plants. ShNAC1 acts as a negative regulator in cold tolerance in S. lycopersicum by regulating the ethylene biosynthesis and signal transduction pathways.
A potential transcriptional regulatory network, including AP2/ERF, MYB, bZIP, bHLH, and NAC, is directly associated with other gene expressions (Wang et al. 2019b). CsLEA (a novel gene encoding a late embryogenesis abundant protein from LEA_3 subfamily protein) was significantly induced by cold stress, illustrating that CsLEA associated with TFs increases cold tolerance in plants. Furthermore, it was assumed that low molecular weight and high hydrophilicity of CsLEA1 might be related to the heterologous expression of CsLEA1 which increased the cold stress tolerance of Escherichia coli and yeast (Wang et al. 2019b). MfNAC3, an NAC TF from M. falcate increased MtICE3, MtCAS15, and MtCAS31 expression under cold treatment in M. truncatula. ICE1 is an important TF in regulating the expression of CBFs, while CASs are important regulons of CBFs. Consequently, MfNAC3 binds to the CATGTG and CACG motifs in the promoter region of MtCBF4 to increase cold tolerance in M. truncatula (Qu et al. 2016). In transgenic A. thaliana overexpressed LlNAC2 from L. lancifolium, various stress-related cis-acting regulatory elements were demonstrated in the LlNAC2 promoter and this promoter was able to enhance GUS activity under cold stress. LlDREB1 and LlZFHD4 bind and interact with the LlNAC2 promoter to enhance stress tolerance (Yong et al. 2019). Besides the CBF-dependent pathway, NAC genes improve freezing stress tolerance through ABA-dependent pathway, a phenomenon that might be related to the enhancement of expression level of stress-responsive genes and scavenging capability of ROS (Zhao et al. 2016).
Conclusion and future perspective
Molecular approaches have been used to identify the role, function, regulation, interaction, and changes of AP2/ERF TF and other TFs under cold stress, such as transgenic breeding (overexpression and gene silencing), Y2H, EMSA, ChIP Seq, omics analysis, and CRISPR-Cas9 along with bioinformatics tools and web-based genetic database as well to improve cold stress resistance in plants (Fig. 2). Though transgenic technique promises to be a good source of cold stress resistance plant (Shahzad et al. 2021), but this technique still has several shortcomings such as unexplored metabolic pathways. Therefore, the omics analysis oupled with CRISPR-Cas9 and bioinformatics tools have been used to reveal several functional features in the plant genome to provide the best plant characteristics (Raza et al. 2021; Razzaq et al. 2021).
Fig. 2.
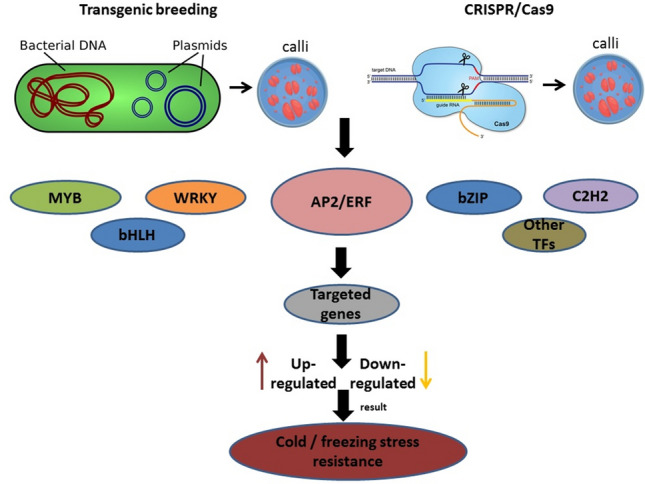
Schematic illustration of improvement techniques particularly targeted modifications in TFs via transgenic breeding and CRISPR/Cas9. Different TFs that can be used for incorporation of cold or freezing stress tolerance in plants. TFs activate or modify different signal transduction pathways such as up-regulate or down-regulate targeted genes. TFs alleviate cold/freezing stress and consequently increase cold stress resistance
As shown in Fig. 3, CBFs (a member of AP2/ERF TF family) were activated or upregulated by the bHLH, bZIP, C2H2, MYB, WRKY, and NAC TFs. CBFs bind to the promoter of COR genes and induce the cold stress genes expression to improve cold/freezing tolerance. Contrary, TCP TF contributed to repressing CBFs expression resulted in reducing cold tolerance in plants. In addition, other novel genes and factors such as light, hormones, and other exogenous treatments related to cold stress are needed to improve our understanding of cold stress mechanisms in plants. The intricate physiological and molecular mechanism in plants under cold/freezing stress calls for researchers to identify other possible factors related to the cold stress mechanism. An understanding of such factors could allow scientists to identify the most suitable molecular breeding technique that can provide the best cold stress tolerance plants.
Fig. 3.
The mechanism of cold tolerance in plants. Cold is sensed by receptors in the plant membrane and followed by the increase of Ca2+, CAMP, ROS, and ABA as well. The chloroplast modulates the signal to the nucleus through ROS production while Ca2+ signaling mediates plant response to cold stress. Besides, Ca2+, CAMP, and ROS signaling mediate signal transduction via CBL, CPKs/CDPKs, and CIPKs to the nucleus through a pathway mediated by ICE-CBF TFs. The MEKK1–MKK2–MPK4 cascade and mitogen-activated protein kinases also involve in the gene regulatory network to regulate TFs and cold tolerance genes. ICE1 and CBFs (member of AP2/ERF TF family) are associated with the COR signaling pathway to increase cold tolerance in plants. Furthermore, WRKY, bZIP, MYB, bHLH, C2H2, NAC, and other AP2/ERF transcription factor members regulate gene expression for the activation of COR genes. In addition, the CBFs were induced by other TF families, such as WRKY, bHLH, bZIP, MYB, NAC, and C2H2. The up-regulated CBFs regulate and activate the expression of COR genes and resulted in improving cold tolerance in plants
Acknowledgements
The authors appreciate the reviewers for their comments and suggestions.
Abbreviations
- CA
Cold acclimation
- TF
Transcription factor
- TFs
Transcription factors
- ABA
Abscisic acid
- JA
Jasmonic acid
- SA
Salicylic acid
- CAMP
Cyclic adenosine monophosphate
- ROS
Reactive oxygen species
- CBL
Calcineurin-B Like proteins
- CPKs/CDPKs
Ca2+-dependent protein kinases
- CIPKs
CBL-interacting protein kinases
- CBF
C-repeat Binding Factor
- DREB
Dehydration-Responsive Element-Binding-Factor
- ICE1
CBF expression 1
- COR
Cold regulated genes
- AP2/ERF
APETALA2/Ethylene responsive factor
- DRE/CRT
Dehydration-responsive C-repeat
- ChIP-seq
Chromatin Immunoprecipitation-sequencing
- ChIP-PCR
Chromatin Immunoprecipitation Polymerase Chain Reaction
- SOD
Superoxide
- POD
Peroxide
- CAT
Catalase
- APX
Ascorbate peroxidase
- BIN2
Brassinosteroid-Insensitive2
- HOS1
Osmotically Responsive Gene1
- GA
Gibberellic Acid
- phyA
Phytochrome A
- phyB
Phytochrome B
- PIFs
Phytochrome-Interacting Factors
- bHLH
Basic helix-loop-helix
- LEA
Late embryogenesis abundant
- bZIP
Basic leucine zipper
- MYB
Myeloblastosis
Authors' contributions
FNR had contributed to writing, editing, and original draft preparation. JNN, YRW, MAK, and UF contributed to editing the manuscript. SC had contributed to supervision, project administration, funding acquisition, review, and editing manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This review was funded by the Fundamental Research Funds for the Central Universities, grant number 2572019CG08, the National Natural Science Foundation of China, Grant Number 31870659 and Heilongjiang Touyan Innovation Team Program (Tree Genetics and Breeding Innovation Team).
Declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Alisoltani A, Karimi M, Ravash R, Fallahi H, Shiran B (2019). Molecular responses to cold stress in temperate fruit crops with focus on rosaceae family," in Genomics Assisted Breeding of Crops for Abiotic Stress Tolerance, Vol. II. Springer), 105–130
- An JP, Yao JF, Wang XN, You CX, Wang XF, Hao YJ. MdHY5 positively regulates cold tolerance via CBF-dependent and CBF-independent pathways in apple. J Plant Physiol. 2017;218:275–281. doi: 10.1016/j.jplph.2017.09.001. [DOI] [PubMed] [Google Scholar]
- An JP, Li R, Qu FJ, You CX, Wang XF, Hao YJ. An apple NAC transcription factor negatively regulates cold tolerance via CBF-dependent pathway. J Plant Physiol. 2018;221:74–80. doi: 10.1016/j.jplph.2017.12.009. [DOI] [PubMed] [Google Scholar]
- Artlip TS, Wisniewski ME, Norelli JL. Field evaluation of apple overexpressing a peach CBF gene confirms its effect on cold hardiness, dormancy, and growth. Environ Exp Bot. 2014;106:79–86. doi: 10.1016/j.envexpbot.2013.12.008. [DOI] [Google Scholar]
- Barrero-Gil J, Salinas J. CBFs at the crossroads of plant hormone signaling in cold stress response. Mol Plant. 2017;10(4):542–544. doi: 10.1016/j.molp.2017.03.004. [DOI] [PubMed] [Google Scholar]
- Barrero-Gil J, Huertas R, Rambla JL, Granell A, Salinas J. Tomato plants increase their tolerance to low temperature in a chilling acclimation process entailing comprehensive transcriptional and metabolic adjustments. Plant Cell Environ. 2016;39(10):2303–2318. doi: 10.1111/pce.12799. [DOI] [PubMed] [Google Scholar]
- Beloiu M, Stahlmann R, Beierkuhnlein C. High recovery of saplings after severe drought in temperate deciduous forests. Forests. 2020;11(5):546. doi: 10.3390/f11050546. [DOI] [Google Scholar]
- Byun MY, Lee J, Cui LH, Kang Y, Oh TK, Park H, Kim WT, et al. Constitutive expression of DaCBF7, an Antarctic vascular plant Deschampsia antarctica CBF homolog, resulted in improved cold tolerance in transgenic rice plants. Plant Sci. 2015;236:61–74. doi: 10.1016/j.plantsci.2015.03.020. [DOI] [PubMed] [Google Scholar]
- Cai WT, Yang YL, Wang WW, Guo GY, Liu W, Bi CL. Overexpression of a wheat (Triticum aestivum L.) bZIP transcription factor gene, TabZIP6, decreased the freezing tolerance of transgenic Arabidopsis seedlings by down-regulating the expression of CBFs. Plant Physiol Biochem. 2018;124:100–111. doi: 10.1016/j.plaphy.2018.01.008. [DOI] [PubMed] [Google Scholar]
- Carlow CE, Faultless JT, Lee C, Siddiqua M, Edge A, Nassuth A. Nuclear localization and transactivation by Vitis CBF transcription factors are regulated by combinations of conserved amino acid domains. Plant Physiol Biochem. 2017;118:306–319. doi: 10.1016/j.plaphy.2017.06.027. [DOI] [PubMed] [Google Scholar]
- Carvallo MA, Pino M-T, Jeknić Z, Zou C, Doherty CJ, Shiu S-H, Thomashow MF, et al. A comparison of the low temperature transcriptomes and CBF regulons of three plant species that differ in freezing tolerance: Solanum commersonii, Solanum tuberosum, and Arabidopsis thaliana. J Exp Bot. 2011;62(11):3807–3819. doi: 10.1093/jxb/err066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HY, Chen XL, Chai XF, Qiu YW, Gong C, Zhang ZZ, Wang AX, et al. Effects of low temperature on mRNA and small RNA transcriptomes in Solanum lycopersicoides leaf revealed by RNA-Seq. Biochem Biophys Res Commun. 2015;464(3):768–773. doi: 10.1016/j.bbrc.2015.07.029. [DOI] [PubMed] [Google Scholar]
- Chen QF, Xu L, Tan WJ, Chen L, Qi H, Xie LJ, Yao N, et al. Disruption of the Arabidopsis defense regulator genes SAG101, EDS1, and PAD4 confers enhanced freezing tolerance. Mol Plant. 2015;8(10):1536–1549. doi: 10.1016/j.molp.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhao Y, Xu S, Zhang Z, Xu Y, Zhang J, Chong K. OsMADS57 together with OsTB1 coordinates transcription of its target OsWRKY94 and D14 to switch its organogenesis to defense for cold adaptation in rice. New Phytol. 2018;218(1):219–231. doi: 10.1111/nph.14977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wang Y, Yu L, Zheng T, Wang S, Yue Z, Yang C, et al. Genome sequence and evolution of Betula platyphylla. Hortic Res. 2021;8(1):37. doi: 10.1038/s41438-021-00481-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djemal R, Mila I, Bouzayen M, Pirrello J, Khoudi H. Molecular cloning and characterization of novel WIN1/SHN1 ethylene responsive transcription factor HvSHN1 in barley (Hordeum vulgare L.) J Plant Physiol. 2018;228:39–46. doi: 10.1016/j.jplph.2018.04.019. [DOI] [PubMed] [Google Scholar]
- Djemal R, Khoudi H. TdSHN1, a WIN1/SHN1-type transcription factor, imparts multiple abiotic stress tolerance in transgenic tobacco. Environ Exp Bot. 2016;131:89–100. doi: 10.1016/j.envexpbot.2016.07.005. [DOI] [Google Scholar]
- Eulgem T, Somssich IE. Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol. 2007;10(4):366–371. doi: 10.1016/j.pbi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Feng HL, Ma NN, Meng X, Zhang S, Wang JR, Chai S, Meng QW. A novel tomato MYC-type ICE1-like transcription factor, SlICE1a, confers cold, osmotic and salt tolerance in transgenic tobacco. Plant Physiol Biochem. 2013;73:309–320. doi: 10.1016/j.plaphy.2013.09.014. [DOI] [PubMed] [Google Scholar]
- Feng WQ, Li J, Long SX, Wei SJ. A DREB1 gene from zoysiagrass enhances Arabidopsis tolerance to temperature stresses without growth inhibition. Plant Sci. 2019;278:20–31. doi: 10.1016/j.plantsci.2018.10.009. [DOI] [PubMed] [Google Scholar]
- Gamboa MC, Rasmussen-Poblete S, Valenzuela PD, Krauskopf E. Isolation and characterization of a cDNA encoding a CBF transcription factor from E. globulus. Plant Physiol Biochem. 2007;45(1):1–5. doi: 10.1016/j.plaphy.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Govardhana M, Kumudini BS. In-silico analysis of cucumber (Cucumis sativus L) Genome for WRKY transcription factors and cis-acting elements. Comput Biol Chem. 2020;85:107212. doi: 10.1016/j.compbiolchem.2020.107212. [DOI] [PubMed] [Google Scholar]
- Gu H, Yang Y, Xing M, Yue C, Wei F, Zhang Y, Huang J, et al. Physiological and transcriptome analyses of Opisthopappus taihangensis in response to drought stress. Cell & Biosci. 2019;9(1):56. doi: 10.1186/s13578-019-0318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualerzi CO, Giuliodori AM, Pon CL. Transcriptional and post-transcriptional control of cold-shock genes. J Mol Biol. 2003;331(3):527–539. doi: 10.1016/s0022-2836(03)00732-0. [DOI] [PubMed] [Google Scholar]
- Guo H, Li Z, Han Z, Xin Y, Cheng H. Cloning of cotton CBF gene for cold tolerance and its expression in transgenic tobacco. Acta Agro Sin. 2011;37(2):286–293. doi: 10.1016/S1875-2780(11)60009-6. [DOI] [Google Scholar]
- Han YC, Fu CC. Cold-inducible MaC2H2s are associated with cold stress response of banana fruit via regulating MaICE1. Plant Cell Rep. 2019;38(5):673–680. doi: 10.1007/s00299-019-02399-w. [DOI] [PubMed] [Google Scholar]
- Hao YJ, Wei W, Song QX, Chen HW, Zhang YQ, Wang F, Zhang WK, et al. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 2011;68(2):302–313. doi: 10.1111/j.1365-313X.2011.04687.x. [DOI] [PubMed] [Google Scholar]
- Hao JJ, Yang JL, Dong JL, Fei SZ. Characterization of BdCBF genes and genome-wide transcriptome profiling of BdCBF3-dependent and-independent cold stress responses in Brachypodium distachyon. Plant Sci. 2017;262:52–61. doi: 10.1016/j.plantsci.2017.06.001. [DOI] [PubMed] [Google Scholar]
- Hu Y, Jiang Y, Han X, Wang H, Pan J, Yu D. Jasmonate regulates leaf senescence and tolerance to cold stress: crosstalk with other phytohormones. J Exp Bot. 2017;68(6):1361–1369. doi: 10.1093/jxb/erx004. [DOI] [PubMed] [Google Scholar]
- Hu Y, Jiang L, Wang F, Yu D. Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell. 2013;25(8):2907–2924. doi: 10.1105/tpc.113.112631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu ZR, Huang XB, Amombo E, Liu A, Fan JB, Bi AY, Fu JM, et al. The ethylene responsive factor CdERF1 from bermudagrass (Cynodon dactylon) positively regulates cold tolerance. Plant Sci. 2020;294:110432. doi: 10.1016/j.plantsci.2020.110432. [DOI] [PubMed] [Google Scholar]
- Huang J, Wang JF, Wang QH, Zhang HS. Identification of a rice zinc finger protein whose expression is transiently induced by drought, cold but not by salinity and abscisic acid. DNA Seq. 2005;16(2):130–136. doi: 10.1080/10425170500061590. [DOI] [PubMed] [Google Scholar]
- Huang J, Sun SJ, Xu DQ, Lan HX, Sun H, Wang ZF, Zhang HS, et al. A TFIIIA-type zinc finger protein confers multiple abiotic stress tolerances in transgenic rice (Oryza sativa L) Plant Mol Biol. 2012;80(3):337–350. doi: 10.1007/s11103-012-9955-5. [DOI] [PubMed] [Google Scholar]
- Huang XS, Li KQ, Jin C, Zhang SL. ICE1 of Pyrus ussuriensis functions in cold tolerance by enhancing PuDREBa transcriptional levels through interacting with PuHHP1. Sci Rep. 2015;5:17620. doi: 10.1038/srep17620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang QH, Qian XC, Jiang TJ, Zheng XL. Effect of eugenol fumigation treatment on chilling injury and CBF gene expression in eggplant fruit during cold storage. Food Chem. 2019;292:143–150. doi: 10.1016/j.foodchem.2019.04.048. [DOI] [PubMed] [Google Scholar]
- Iida A, Kazuoka T, Torikai S, Kikuchi H, Oeda K. A zinc finger protein RHL41 mediates the light acclimatization response in Arabidopsis. Plant J. 2000;24(2):191–203. doi: 10.1046/j.1365-313x.2000.00864.x. [DOI] [PubMed] [Google Scholar]
- Jeon J, Cho C, Lee MR, Van Binh N, Kim J. Cytokinin Response Factor2 (CRF2) and CRF3 regulate lateral root development in response to cold stress in Arabidopsis. Plant Cell. 2016;28(8):1828–1843. doi: 10.1105/tpc.15.00909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang BC, Shi YT, Peng Y, Jia Y, Yan Y, Dong XJ, Gong ZZ, et al. Cold-Induced CBF-PIF3 Interaction Enhances Freezing Tolerance by Stabilizing the phyB Thermosensor in Arabidopsis. Mol Plant. 2020 doi: 10.1016/j.molp.2020.04.006. [DOI] [PubMed] [Google Scholar]
- Jin C, Li KQ, Xu XY, Zhang HP, Chen HX, Chen YH, Zhang SL, et al. A novel NAC transcription factor, PbeNAC1, of Pyrus betulifolia confers cold and drought tolerance via interacting with PbeDREBs and activating the expression of stress-responsive genes. Front Plant Sci. 2017;8:1049. doi: 10.3389/fpls.2017.01049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju YL, Yue XF, Min Z, Wang XH, Fang YL, Zhang JX. VvNAC17, a novel stress-responsive grapevine (Vitis vinifera L.) NAC transcription factor, increases sensitivity to abscisic acid and enhances salinity, freezing, and drought tolerance in transgenic Arabidopsis. Plant Physiol Biochem. 2020;146:98–111. doi: 10.1016/j.plaphy.2019.11.002. [DOI] [PubMed] [Google Scholar]
- Jung WJ, Seo YW. Identification of novel C-repeat binding factor (CBF) genes in rye (Secale cereale L.) and expression studies. Gene. 2019;684:82–94. doi: 10.1016/j.gene.2018.10.055. [DOI] [PubMed] [Google Scholar]
- Kang HG, Kim JK, Kim BH, Jeong HN, Choi SH, Kim EK, Lim PO, et al. Overexpression of FTL1/DDF1, an AP2 transcription factor, enhances tolerance to cold, drought, and heat stresses in Arabidopsis thaliana. Plant Sci. 2011;180(4):634–641. doi: 10.1016/j.plantsci.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Kang WH, Sim YM, Koo NJ, Nam JY, Js L, Kim NY, Yeom SI, et al. Transcriptome profiling of abiotic responses to heat, cold, salt, and osmotic stress of Capsicum annuum L. Sci Data. 2020;7(1):1–7. doi: 10.1038/s41597-020-0352-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kargiotidou A, Kappas I, Tsaftaris A, Galanopoulou D, Farmaki T. Cold acclimation and low temperature resistance in cotton: Gossypium hirsutum phospholipase Dα isoforms are differentially regulated by temperature and light. J Exp Bot. 2010;61(11):2991–3002. doi: 10.1093/jxb/erq124. [DOI] [PubMed] [Google Scholar]
- Kashyap P, Deswal R. A novel class I Chitinase from Hippophae rhamnoides: Indications for participating in ICE-CBF cold stress signaling pathway. Plant Sci. 2017;259:62–70. doi: 10.1016/j.plantsci.2017.03.004. [DOI] [PubMed] [Google Scholar]
- Kashyap P, Deswal R. Two ICE isoforms showing differential transcriptional regulation by cold and hormones participate in Brassica juncea cold stress signaling. Gene. 2019;695:32–41. doi: 10.1016/j.gene.2019.02.005. [DOI] [PubMed] [Google Scholar]
- Kazemi-Shahandashti SS, Maali-Amiri R. Global insights of protein responses to cold stress in plants: signaling, defence, and degradation. J Plant Physiol. 2018;226:123–135. doi: 10.1016/j.jplph.2018.03.022. [DOI] [PubMed] [Google Scholar]
- Klay I, Gouia S, Liu M, Mila I, Khoudi H, Bernadac A, Pirrello J, et al. Ethylene Response Factors (ERF) are differentially regulated by different abiotic stress types in tomato plants. Plant Sci. 2018;274:137–145. doi: 10.1016/j.plantsci.2018.05.023. [DOI] [PubMed] [Google Scholar]
- Li KL, Bai X, Li Y, Cai H, Ji W, Tang LL, Zhu YM, et al. GsGASA1 mediated root growth inhibition in response to chronic cold stress is marked by the accumulation of DELLAs. J Plant Physiol. 2011;168(18):2153–2160. doi: 10.1016/j.jplph.2011.07.006. [DOI] [PubMed] [Google Scholar]
- Li XD, Zhuang KY, Liu ZM, Yang DY, Ma NN, Meng QW. Overexpression of a novel NAC-type tomato transcription factor, SlNAM1, enhances the chilling stress tolerance of transgenic tobacco. J Plant Physiol. 2016;204:54–65. doi: 10.1016/j.jplph.2016.06.024. [DOI] [PubMed] [Google Scholar]
- Li H, Ye KY, Shi YT, Cheng JK, Zhang XY, Yang SH. BZR1 positively regulates freezing tolerance via CBF-dependent and CBF-independent pathways in Arabidopsis. Mol Plant. 2017;10(4):545–559. doi: 10.1016/j.molp.2017.01.004. [DOI] [PubMed] [Google Scholar]
- Li R, Zhang LX, Wang L, Chen L, Zhao RR, Sheng JP, Shen L. Reduction of tomato-plant chilling tolerance by CRISPR–Cas9-mediated SlCBF1 mutagenesis. J Agric Food Chem. 2018;66(34):9042–9051. doi: 10.1021/acs.jafc.8b02177. [DOI] [PubMed] [Google Scholar]
- Liu H, Timko MP. Jasmonic acid signaling and molecular crosstalk with other phytohormones. Int J Mol Sci. 2021 doi: 10.3390/ijms22062914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZY, Jia YX, Ding YL, Shi YT, Li Z, Guo Y, Yang SH, et al. Plasma membrane CRPK1-mediated phosphorylation of 14–3–3 proteins induces their nuclear import to fine-tune CBF signaling during cold response. Mol Cell. 2017;66(1):117–128. doi: 10.1016/j.molcel.2017.02.016. [DOI] [PubMed] [Google Scholar]
- Liu C, Ou S, Mao B, Tang J, Wang W, Wang H, Xiao G, et al. Early selection of bZIP73 facilitated adaptation of japonica rice to cold climates. Nat Commun. 2018;9(1):1–12. doi: 10.1038/s41467-018-05753-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhou Y, Li H, Wang T, Zhang J, Ouyang B, Ye Z. Molecular and functional characterization of ShNAC1, an NAC transcription factor from Solanum habrochaites. Plant Sci. 2018;271:9–19. doi: 10.1016/j.plantsci.2018.03.005. [DOI] [PubMed] [Google Scholar]
- Londo JP, Kovaleski AP, Lillis JA. Divergence in the transcriptional landscape between low temperature and freeze shock in cultivated grapevine (Vitis vinifera) Hortic Res. 2018;5(1):1–14. doi: 10.1038/s41438-018-0020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Liu H, Ren J, Chen D, Cheng X, Sun W, Huang C, et al. Cold-inducible expression of an Arabidopsis thaliana AP2 transcription factor gene, AtCRAP2, promotes flowering under unsuitable low-temperatures in chrysanthemum. Plant Physiol Biochem. 2020;146:220–230. doi: 10.1016/j.plaphy.2019.11.022. [DOI] [PubMed] [Google Scholar]
- Luo P, Li Z, Chen W, Xing W, Yang J, Cui Y. Overexpression of RmICE1, a bHLH transcription factor from Rosa multiflora, enhances cold tolerance via modulating ROS levels and activating the expression of stress-responsive genes. Environ Exp Bot. 2020;178:104160. doi: 10.1016/j.envexpbot.2020.104160. [DOI] [Google Scholar]
- Lv Y, Yang M, Hu D, Yang Z, Ma S, Li X, Xiong L. The OsMYB30 transcription factor suppresses cold tolerance by interacting with a JAZ protein and suppressing β-amylase expression. Plant Physiol. 2017;173(2):1475–1491. doi: 10.1104/pp.16.01725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv K, Li J, Zhao K, Chen S, Nie J, Zhang W, Wei H, et al. Overexpression of an AP2/ERF family gene, BpERF13, in birch enhances cold tolerance through upregulating CBF genes and mitigating reactive oxygen species. Plant Sci. 2019 doi: 10.1016/j.plantsci.2019.110375. [DOI] [PubMed] [Google Scholar]
- Ma Y, Zhang L, Zhang J, Chen J, Wu T, Zhu S, Zhong G, et al. Expressing a citrus ortholog of arabidopsis ERF1 enhanced cold-tolerance in tobacco. Sci Hortic. 2014;174:65–76. doi: 10.1016/j.scienta.2014.05.009. [DOI] [Google Scholar]
- Medina J, Catalá R, Salinas J. The CBFs: three Arabidopsis transcription factors to cold acclimate. Plant Sci. 2011;180(1):3–11. doi: 10.1016/j.plantsci.2010.06.019. [DOI] [PubMed] [Google Scholar]
- Mehrotra S, Verma S, Kumar S, Kumari S, Mishra BN. Transcriptional regulation and signalling of cold stress response in plants: an overview of current understanding. Environ Exp Bot. 2020;180:104243. doi: 10.1016/j.envexpbot.2020.104243. [DOI] [Google Scholar]
- Mitsis T, Efthimiadou A, Bacopoulou F, Vlachakis D, Chrousos GP, Eliopoulos E. Transcription factors and evolution: An integral part of gene expression. World Acad Sci Eng Technol. 2020;2(1):3–8. doi: 10.3892/wasj.2020.32. [DOI] [Google Scholar]
- Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) AP2/ERF family transcription factors in plant abiotic stress responses. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms 1819(2): 86–96 10.1016/j.bbagrm.2011.08.004 [DOI] [PubMed]
- Mizuno S, Hirasawa Y, Sonoda M, Nakagawa H, Sato T. Isolation and characterization of three DREB/ERF-type transcription factors from melon (Cucumis melo) Plant Sci. 2006;170(6):1156–1163. doi: 10.1016/j.plantsci.2006.02.005. [DOI] [Google Scholar]
- Monteagudo A, Forcada CF, Estopañán G, Dodd RS, Alonso JM, Rubio-Cabetas MJ, Marti ÁF. Biochemical analyses and expression of cold transcription factors of the late PDO ‘Calanda’peach under different post-harvest conditions. Sci Hortic. 2018;238:116–125. doi: 10.1016/j.scienta.2018.04.043. [DOI] [Google Scholar]
- Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) NAC transcription factors in plant abiotic stress responses. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms 1819(2): 97–103. 10.1016/j.bbagrm.2011.10.005 [DOI] [PubMed]
- Niu Y, Hu T, Zhou Y, Hasi A. Isolation and characterization of two Medicago falcate AP2/EREBP family transcription factor cDNA, MfDREB1 and MfDREB1s. Plant Phys Biochem. 2010;48(12):971–976. doi: 10.1016/j.plaphy.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Ohta M, Sato A, Renhu N, Yamamoto T, Oka N, Zhu JK, Miura K, et al. MYC-type transcription factors, MYC67 and MYC70, interact with ICE1 and negatively regulate cold tolerance in Arabidopsis. Sci Rep. 2018;8(1):1–12. doi: 10.1038/s41598-018-29722-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan XW, Li YC, Li XX, Liu WQ, Jun M, Lu TT, Sheng XN, et al. Differential regulatory mechanisms of CBF regulon between Nipponbare (Japonica) and 93–11 (Indica) during cold acclimation. Rice Sci. 2013;20(3):165–172. doi: 10.1016/S1672-6308(13)60121-3. [DOI] [Google Scholar]
- Peng HH, Shan W, Kuang JF, Lu WJ, Chen JY. Molecular characterization of cold-responsive basic helix-loop-helix transcription factors MabHLHs that interact with MaICE1 in banana fruit. Planta. 2013;238(5):937–953. doi: 10.1007/s00425-013-1944-7. [DOI] [PubMed] [Google Scholar]
- Peng YL, Wang YS, Cheng H, Sun CC, Wu P, Wang LY, Fei J. Characterization and expression analysis of three CBF/DREB1 transcriptional factor genes from mangrove Avicennia marina. Aquat Toxicol. 2013;140:68–76. doi: 10.1016/j.aquatox.2013.05.014. [DOI] [PubMed] [Google Scholar]
- Prerostova S, Zupkova B, Petrik I, Simura J, Nasinec I, Kopecky D, Vankova R, et al. Hormonal responses associated with acclimation to freezing stress in Lolium perenne. Environ Exp Bot. 2021;182:104295. doi: 10.1016/j.envexpbot.2020.104295. [DOI] [Google Scholar]
- Puhakainen T, Li C, Boije-Malm M, Kangasjärvi J, Heino P, Palva ET. Short-day potentiation of low temperature-induced gene expression of a C-repeat-binding factor-controlled gene during cold acclimation in silver birch. Plant Physiol. 2004;136(4):4299–4307. doi: 10.1104/pp.104.047258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu YT, Duan M, Zhang ZQ, Dong JL, Wang T. Overexpression of the Medicago falcata NAC transcription factor MfNAC3 enhances cold tolerance in Medicago truncatula. Environ Exp Bot. 2016;129:67–76. doi: 10.1016/j.envexpbot.2015.12.012. [DOI] [Google Scholar]
- Rapacz M, Jurczyk B, Krępski T, Płażek A. C-repeat binding transcription factors from Miscanthus× giganteus and their expression at a low temperature. Ind Crops Prod. 2018;113:283–287. doi: 10.1016/j.indcrop.2018.01.058. [DOI] [Google Scholar]
- Rasmussen S, Barah P, Suarez-Rodriguez MC, Bressendorff S, Friis P, Costantino P, Mundy J, et al. Transcriptome responses to combinations of stresses in Arabidopsis. Plant Physiol. 2013;161(4):1783–1794. doi: 10.1104/pp.112.210773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raza A, Tabassum J, Kudapa H, Varshney RK (2021) Can omics deliver temperature resilient ready-to-grow crops? Crit Rev Biotechnol 10.1080/07388551.2021.1898332 [DOI] [PubMed]
- Razzaq MK, Aleem M, Mansoor S, Khan MA, Rauf S, Iqbal S, Siddique KHM. Omics and CRISPR-Cas9 approaches for molecular insight, functional gene analysis, and stress tolerance development in crops. Int J Mol Sci. 2021 doi: 10.3390/ijms22031292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritonga FN, Chen S. Physiological and molecular mechanism involved in cold stress tolerance in plants. Plants. 2020;9(560):13. doi: 10.3390/plants9050560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritonga FN, Ngatia JN, Song RX, Farooq U, Somadona S, Andi TL, Chen S (2021) Abiotic stresses induced physiological, biochemical, and molecular changes in Betula platyphylla a review. Silva Fenn. 10.14214/sf.10516
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration-and cold-inducible gene expression. Biochem Biophys Res Commun. 2002;290(3):998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- Shahzad R, Jamil S, Ahmad S, Nisar A, Amina Z, Saleem S, Wang X, et al. Harnessing the potential of plant transcription factors in developing climate resilient crops to improve global food security: Current and future perspectives. Saudi J Biol Sci. 2021;28(4):2323–2341. doi: 10.1016/j.sjbs.2021.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma KD, Nayyar H. Regulatory networks in pollen development under cold stress. Front Plant Sci. 2016;7:402–402. doi: 10.3389/fpls.2016.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YT, Ding YL, Yang SH. Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 2018;23(7):623–637. doi: 10.1016/j.tplants.2018.04.002. [DOI] [PubMed] [Google Scholar]
- STRING (2021). Protein-protein Interaction networks, Functional enrichment analysis [Online]. Available: (https://string-db.org/cgi/) [Accessed 21 June, 2021].
- Su LT, Li JW, Liu DQ, Zhai Y, Zhang HJ, Li XW, Wang QY, et al. A novel MYB transcription factor, GmMYBJ1, from soybean confers drought and cold tolerance in Arabidopsis thaliana. Gene. 2014;538(1):46–55. doi: 10.1016/j.gene.2014.01.024. [DOI] [PubMed] [Google Scholar]
- Sun XM, Zhu ZF, Zhang LL, Fang LC, Zhang JS, Wang QF, Xin HP, et al. Overexpression of ethylene response factors VaERF080 and VaERF087 from Vitis amurensis enhances cold tolerance in Arabidopsis. Sci Hortic. 2019;243:320–326. doi: 10.1016/j.scienta.2018.08.055. [DOI] [Google Scholar]
- Thakur P, Kumar S, Malik JA, Berger JD, Nayyar H. Cold stress effects on reproductive development in grain crops: an overview. Environ Exp Bot. 2010;67(3):429–443. doi: 10.1016/j.envexpbot.2009.09.004. [DOI] [Google Scholar]
- Wang Z, Liu J, Guo H, He X, Wu W, Du J, An X, et al. Characterization of two highly similar CBF/DREB1-like genes, PhCBF4a and PhCBF4b, in Populus hopeiensis. Plant Physiol Biochem. 2014;83:107–116. doi: 10.1016/j.plaphy.2014.07.012. [DOI] [PubMed] [Google Scholar]
- Wang GD, Xu XP, Wang H, Liu Q, Yang XT, Liao LX, Cai GH. A tomato transcription factor, SlDREB3 enhances the tolerance to chilling in transgenic tomato. Plant Physiol Biochem. 2019;142:254–262. doi: 10.1016/j.plaphy.2019.07.017. [DOI] [PubMed] [Google Scholar]
- Wang WD, Gao T, Chen JF, Yang JK, Huang HY, Yu YB. The late embryogenesis abundant gene family in tea plant (Camellia sinensis): Genome-wide characterization and expression analysis in response to cold and dehydration stress. Plant Physiol Biochem. 2019;135:277–286. doi: 10.1016/j.plaphy.2018.12.009. [DOI] [PubMed] [Google Scholar]
- Wang X, Ding Y, Li Z, Shi Y, Wang J, Hua J, Yang S, et al. PUB25 and PUB26 promote plant freezing tolerance by degrading the cold signaling negative regulator MYB15. Dev Cell. 2019;51(2):222–235. doi: 10.1016/j.devcel.2019.08.008. [DOI] [PubMed] [Google Scholar]
- Wang Y, Mao Z, Jiang H, Zhang Z, Chen X. A feedback loop involving MdMYB108L and MdHY5 controls apple cold tolerance. Biochem Biophys Res Commun. 2019;512(2):381–386. doi: 10.1016/j.bbrc.2019.03.101. [DOI] [PubMed] [Google Scholar]
- Watt C, Zhou G, Li C (2020) Harnessing transcription factors as potential tools to enhance grain size under stressful abiotic conditions in cereal crops. Front Plant Sci 11(1273). 10.3389/fpls.2020.01273 [DOI] [PMC free article] [PubMed]
- Winfield MO, Lu C, Wilson ID, Coghill JA, Edwards KJ. Plant responses to cold: transcriptome analysis of wheat. Plant Biotechnol J. 2010;8(7):749–771. doi: 10.1111/j.1467-7652.2010.00536.x. [DOI] [PubMed] [Google Scholar]
- Xie Y, Chen P, Yan Y, Bao C, Li X, Wang L, Niu C, et al. An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBF-independent pathways in apple. New Phytol. 2018;218(1):201–218. doi: 10.1111/nph.14952. [DOI] [PubMed] [Google Scholar]
- Xu D, Deng XW. CBF-phyB-PIF module links light and low temperature signaling. Trends Plant Sci. 2020 doi: 10.1016/j.tplants.2020.06.010. [DOI] [PubMed] [Google Scholar]
- Xu YC, Hou XL, Xu WW, Shen LL, Lü SW, Zhang SL, Hu CM. Isolation and characterization of an ERF-B3 gene associated with flower abnormalities in non-heading Chinese cabbage. J Integr Agric. 2016;15(3):528–536. doi: 10.1016/S2095-3119(15)61203-5. [DOI] [Google Scholar]
- Xu H, Yang G, Zhang J, Wang Y, Zhang T, Wang N, Chen X, et al. Overexpression of a repressor MdMYB15L negatively regulates anthocyanin and cold tolerance in red-fleshed callus. Biochem Biophys Res Commun. 2018;500(2):405–410. doi: 10.1016/j.bbrc.2018.04.088. [DOI] [PubMed] [Google Scholar]
- Yadav SK. Cold stress tolerance mechanisms in plants A review. Agronomy Sustain Develop. 2010;30(3):515–527. doi: 10.1051/agro/2009050. [DOI] [Google Scholar]
- Yamasaki Y, Randall SK. Functionality of soybean CBF/DREB1 transcription factors. Plant Sci. 2016;246:80–90. doi: 10.1016/j.plantsci.2016.02.007. [DOI] [PubMed] [Google Scholar]
- Yao PF, Sun ZX, Li CL, Zhao XR, Li MF, Deng RY, Wu Q, et al. Overexpression of Fagopyrum tataricum FtbHLH2 enhances : tolerance to cold stress in transgenic Arabidopsis. Plant Physiol Biochem. 2018;125:85–94. doi: 10.1016/j.plaphy.2018.01.028. [DOI] [PubMed] [Google Scholar]
- Yang T, Zhang L, Zhang T, Zhang H, Xu S, An L. Transcriptional regulation network of cold-responsive genes in higher plants. Plant Sci. 2005;169(6):987–995. doi: 10.1016/j.plantsci.2005.07.005. [DOI] [Google Scholar]
- Xy Yang, Wang R, Ql Hu, Li SL, Mao XD, Jing HH, Liu CM, et al. DlICE1, a stress-responsive gene from Dimocarpus longan, enhances cold tolerance in transgenic Arabidopsis. Plant Physiol Biochem. 2019;142:490–499. doi: 10.1016/j.plaphy.2019.08.007. [DOI] [PubMed] [Google Scholar]
- Ye K, Li H, Ding Y, Shi Y, Song C, Gong Z, Yang S (2019) BRASSINOSTEROID-INSENSITIVE2 negatively regulates the stability of transcription factor ICE1 in response to cold stress in Arabidopsis. The Plant Cell 31(11), 2682-2696 [DOI] [PMC free article] [PubMed]
- Yin XR, Allan AC, Xu Q, Burdon J, Dejnoprat S, Ks C, Ferguson IB. Differential expression of kiwifruit ERF genes in response to postharvest abiotic stress. Postharvest Biol Technol. 2012;66:1–7. doi: 10.1016/j.postharvbio.2011.11.009. [DOI] [Google Scholar]
- Yong YB, Zhang Y, Lyu YM. A Stress-Responsive NAC transcription factor from tiger lily (LlNAC2) interacts with LlDREB1 and LlZHFD4 and enhances various abiotic stress tolerance in Arabidopsis. Int J Mol Sci. 2019;20(13):3225. doi: 10.3390/ijms20133225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ZC, Wang TQ, Luo YN, Zheng XT, He W, Chen LB, Peng CL. Overexpression of the V-ATPase c subunit gene from Antarctic notothenioid fishes enhances freezing tolerance in transgenic Arabidopsis plants. Plant Physiol Biochem. 2021;160:365–376. doi: 10.1016/j.plaphy.2021.01.038. [DOI] [PubMed] [Google Scholar]
- Yuan P, Yang T, Poovaiah BW. Calcium signaling-mediated plant response to cold stress. Int J Mol Sci. 2018;19(12):3896. doi: 10.3390/ijms19123896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandalinas SI, Fritschi FB, Mittler R. Global warming, climate change, and environmental pollution: recipe for a multifactorial stress combination disaster. Trends Plant Sci. 2021;26(6):588–599. doi: 10.1016/j.tplants.2021.02.011. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Wen J, Zhao W, Wang Q, Huang W (2020) Rational Improvement of Rice Yield and Cold Tolerance by Editing the Three Genes OsPIN5b, GS3, and OsMYB30 With the CRISPR–Cas9 System. Front Plant Sci 10(1663). 10.3389/fpls.2019.01663 [DOI] [PMC free article] [PubMed]
- Zhang X, Guo XP, Lei CL, Cheng ZJ, Lin QB, Wang JL, Wan JM, et al. Overexpression of SlCZFP1, a novel TFIIIA-type zinc finger protein from tomato, confers enhanced cold tolerance in transgenic Arabidopsis and rice. Plant Mol Biol Rep. 2011;29(1):185–196. doi: 10.1007/s11105-010-0223-z. [DOI] [Google Scholar]
- Zhang Y, Yu HJ, Yang XY, Li Q, Ling J, Wang H, Jiang WJ, et al. CsWRKY46, a WRKY transcription factor from cucumber, confers cold resistance in transgenic-plant by regulating a set of cold-stress responsive genes in an ABA-dependent manner. Plant Physiol Biochem. 2016;108:478–487. doi: 10.1016/j.plaphy.2016.08.013. [DOI] [PubMed] [Google Scholar]
- Zhang QY, Yu JQ, Wang JH, Hu DG, Hao YJ. Functional characterization of MdMYB73 reveals its involvement in cold stress response in apple calli and Arabidopsis. J Integr Agric. 2017 doi: 10.1007/s11103-019-00846-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CY, Liu HC, Zhang XS, Guo QX, Bian SM, Wang JY, Zhai LL. VcMYB4a, an R2R3-MYB transcription factor from Vaccinium corymbosum, negatively regulates salt, drought, and temperature stress. Gene. 2020;757:144935. doi: 10.1016/j.gene.2020.144935. [DOI] [PubMed] [Google Scholar]
- Zhao H, Wang S, Chen S, Jiang J, Liu GF. Phylogenetic and stress-responsive expression analysis of 20 WRKY genes in Populus simonii× Populus nigra. Gene. 2015;565(1):130–139. doi: 10.1016/j.gene.2015.04.002. [DOI] [PubMed] [Google Scholar]
- Zhao X, Yang XW, Pei SQ, He G, Wang XY, Tang Q, Zhou GK, et al. The Miscanthus NAC transcription factor MlNAC9 enhances abiotic stress tolerance in transgenic Arabidopsis. Gene. 2016;586(1):158–169. doi: 10.1016/j.gene.2016.04.028. [DOI] [PubMed] [Google Scholar]
- Zhao C, Liu XF, He JQ, Xie YP, Xu Y, Ma FW, Guan QM. Apple TIME FOR COFFEE contributes to freezing tolerance by promoting unsaturation of fatty acids. Plant Sci. 2021;302:110695. doi: 10.1016/j.plantsci.2020.110695. [DOI] [PubMed] [Google Scholar]
- Zhou MQ, Chen H, Wei DH, Ma H, Lin J. Arabidopsis CBF3 and DELLAs positively regulate each other in response to low temperature. Sci Rep. 2017;7(1):1–13. doi: 10.1038/srep39819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Li J, He YJ, Liu Y, Chen HY. Functional characterization of SmCBF genes involved in abiotic stress response in eggplant (Solanum melongena) Sci Hortic. 2018;233:14–21. doi: 10.1016/j.scienta.2018.01.043. [DOI] [Google Scholar]
- Zhu Z, Shi J, Xu W, Li H, He M, Xu Y, Wang Y, et al. Three ERF transcription factors from Chinese wild grapevine Vitis pseudoreticulata participate in different biotic and abiotic stress-responsive pathways. J Plant Physiol. 2013;170(10):923–933. doi: 10.1016/j.jplph.2013.01.017. [DOI] [PubMed] [Google Scholar]
- Zhu YY, Liu XL, Gao YD, Li K, Guo WD (2020) Transcriptome-based identification of AP2/ERF family genes and their cold-regulated expression during the dormancy phase transition of Chinese cherry flower buds. Sci Hortic, 109666. 10.1016/j.scienta.2020.109666
- Zuo ZF, Kang HG, Park MY, Jeong H, Sun HJ, Song PS, Lee HY. Zoysia japonica MYC type transcription factor ZjICE1 regulates cold tolerance in transgenic Arabidopsis. Plant Sci. 2019;289:110254. doi: 10.1016/j.plantsci.2019.110254. [DOI] [PubMed] [Google Scholar]
- Zwack PJ, Compton MA, Adams CI, Rashotte AM. Cytokinin response factor 4 (CRF4) is induced by cold and involved in freezing tolerance. Plant Cell Rep. 2016;35(3):573–584. doi: 10.1007/s00299-015-1904-8. [DOI] [PubMed] [Google Scholar]




