Abstract
The multi-subunit bacterial RNA polymerase (RNAP) and its associated regulators carry out transcription and integrate myriad regulatory signals. Numerous studies have interrogated RNAP mechanism, and RNAP mutations drive Escherichia coli adaptation to many health- and industry-relevant environments, yet a paucity of systematic analyses hampers our understanding of the fitness trade-offs from altering RNAP function. Here, we conduct a chemical-genetic analysis of a library of RNAP mutants. We discover phenotypes for non-essential insertions, show that clustering mutant phenotypes increases their predictive power for drawing functional inferences, and demonstrate that some RNA polymerase mutants both decrease average cell length and prevent killing by cell-wall targeting antibiotics. Our findings demonstrate that RNAP chemical-genetic interactions provide a general platform for interrogating structure-function relationships in vivo and for identifying physiological trade-offs of mutations, including those relevant for disease and biotechnology. This strategy should have broad utility for illuminating the role of other important protein complexes.
Keywords: transcription, RNAP, chemical genetics, lineage-specific sequence insertion, SI2, stringent response, A22, mecillinam, FtsZ
Introduction
Multi-subunit RNA polymerases are responsible for transcription in all organisms. The core RNA polymerase (RNAP) enzyme (β’, β, α2, ω) is conserved across all domains of life (Jokerst et al., 1989; Lane and Darst, 2010; Sweetser et al., 1987). Bacterial-specific initiation factors, called sigmas (σs), transiently associate with the core complex to provide promoter recognition and assist in melting promoter DNA during initiation (Gruber and Gross, 2003). During elongation, RNAP associates with NusA, which enhances pausing and intrinsic termination at specific sequences (Artsimovitch and Landick, 2000), and NusG (Spt5 in archaea and eukaryotes), the only universally conserved elongation factor, which modulates elongation and ρ-dependent termination (Burova et al., 1995; Li et al., 1993). Termination in eubacteria is facilitated either by RNA structure (intrinsic termination) or by the termination factor ρ, which uses its helicase activity to release the transcript and recycle the RNAP complex (Figure 1A). Additionally, bacterial RNAPs differ from archaeal and eukaryotic RNAPs, for which the core enzymes acquired peripheral subunits (e.g., Rpb4,5,7–10,12 in RNAPII), by instead having acquired lineage-specific insertions in β’ and β (called sequence insertions 1–3 (SI1–3) in E. coli) whose functions remain largely unknown (Artsimovitch et al., 2003; Lane and Darst, 2010).
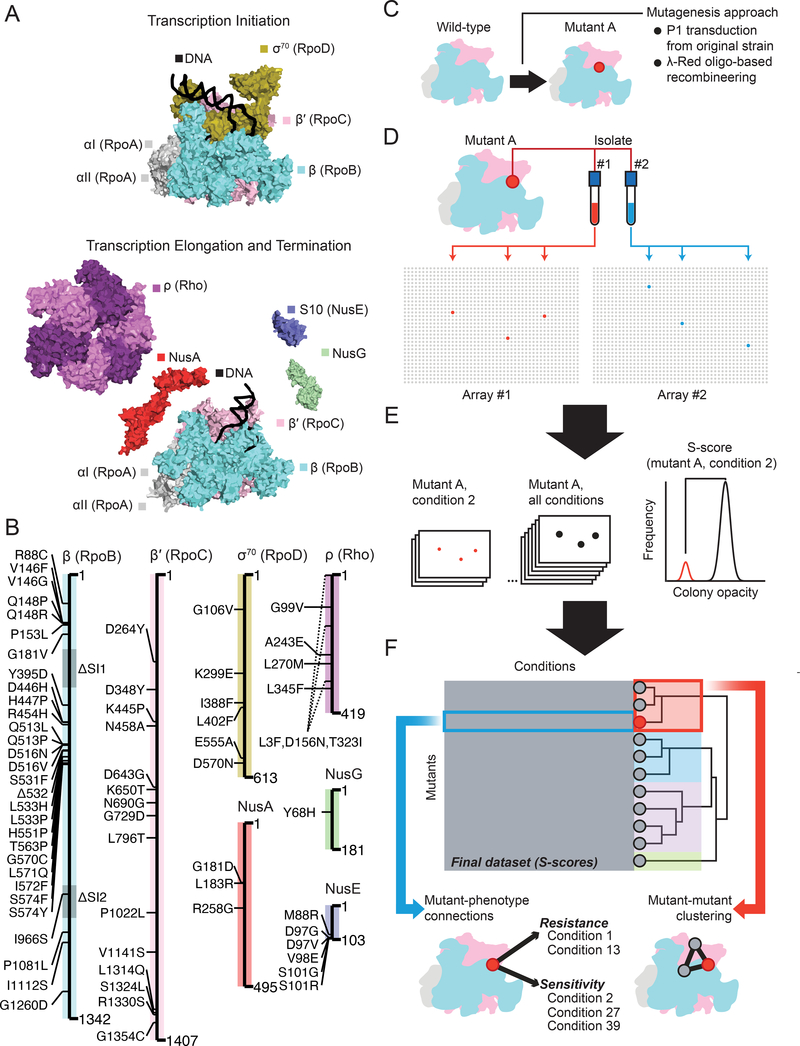
Figure 1: A chemical-genetic screen focused on the bacterial transcription machinery dissects structure-function relationships of RNA polymerase in vivo.
A) The core essential subunits of RNA polymerase (α2ββ′) form a complex with sigma factors such as σ70 during transcription initiation. During elongation, factors such as NusA, NusG, NusE, and ρ associate with RNAP to regulate pausing and termination.
B) A library of 68 mutations in RNAP was created in an isogenic background to dissect RNA polymerase function in vivo.
C) Mutations were introduced de novo using oligo-based recombineering or transduced from the original isolate using P1vir.
D) Independent isolates of the same mutation (red and blue) were split between parallel sets of colony arrays (Array #1 and Array #2). Each isolate was arrayed in triplicate and the position of each mutant was randomized between the two arrays. The randomized positions of the biological replicates are shown as red (Array #1) and blue (Array #2) colonies in a 32-row x 48-column array of colonies. The remaining mutants are gray.
E) At an appropriate time for each condition, a single image was taken of every plate and colony opacity was estimated using image analysis software. Following appropriate normalizations and filtering steps, the distribution of colony opacity measurements for a given mutant/condition pair were compared to the entire distribution of opacity measurements of the mutant across all conditions to generate an S-score. The S-score is a modified t-statistic that measures the significance of the difference between colony opacity (closely related to colony size) of a specific mutant/condition pair (red colonies, red distribution of colony opacity) and the control of the same mutant across all conditions (black colonies, black distribution of colony opacity). In this example, colony opacity is lower on the given condition, leading to negative S-scores that are interpreted as chemical sensitivity. Conversely, higher colony opacities would lead to positive S-scores that are interpreted as resistance. Importantly, S-scores are proportional to the statistical significance of an interaction, not the direct magnitude of the interaction itself.
F) The final dataset was a 68 × 83 matrix of mutant x condition S-scores. Individual S-scores were investigated to identify new mutant-phenotype connections, hierarchical clustering of mutants was used to assign new functions to mutations, and enrichments of chemical interactions were identified within mutant clusters. The colored boxes overlayed on the dendrogram represent a partitioning of the mutant set into discrete clusters (different colors) based on a distance cutoff (the extent that the colored boxes extend to the right) (see Supplemental Figure 1).
The central role played by this enzyme complex, both in orchestrating transcription and integrating diverse signals, is reflected in the pleiotropic phenotypes that arise from mutations in RNAP. Efforts to evolve E. coli in maladapted environments, such as growth on glycerol (Cheng et al., 2014), ethanol (Haft et al., 2014), or at elevated temperatures (Tenaillon et al., 2012), have all recovered mutations in RNAP as a predominant class, highlighting the intimate ties of RNAP to a wide range of cellular processes. Adaptation in these conditions is highly relevant for biotechnology applications, as directed mutagenesis of RNAP could serve as a rapid means of adapting bacteria to new production environments (Alper and Stephanopoulos, 2007). However, without a deeper understanding of how RNAP mutations affect cellular physiology, it will be difficult to predict the extent to which mutations in RNAP will have unintended physiological side effects.
Chemical-genetic screens measure the effect of stressful environments, such as the presence of an antibiotic, on growth across a large library of mutations (Brochado and Typas, 2013). By discovering novel growth phenotypes and identifying mutants with highly correlated growth phenotypes across conditions, such screens generate new hypotheses regarding biological pathways and gene functions (Nichols et al., 2011). Chemical screens can also be used to analyze a large collection of mutations in a single protein complex (Braberg et al., 2013), wherein discovery of new phenotypes and correlations between phenotypic profiles make possible in vivo structure-function analyses based on the effects of mutations on cellular physiology. By interrogating chemical-genetic interactions across a wide range of environments, these screens are also uniquely situated to identify the secondary effects of adaptive mutations.
In this work, we conducted a chemical-genetic screen focused on RNAP mutations in E. coli K-12, with the goal of interrogating connections between RNAP and cellular physiology and dissecting in vivo structure-function relationships within RNAP and its associated factors. We generated an isogenic library of 68 unique mutations in RNAP and essential transcription factors and screened the library in 83 unique conditions to generate a chemical-genetic dataset that we integrated with existing data from the Keio library (Baba et al., 2006) of all nonessential gene deletions. We confirmed that mutations in RNAP are highly pleiotropic, with altered sensitivities to antibiotics that target peptidoglycan synthesis, folate biosynthesis, DNA replication, and translation. We shed light on the effect of understudied features of RNAP like β-SI2 on transcription. Finally, we identified an antibiotic resistance phenotype of RNAP mutations that is associated with decreased average cell length, and showed that neither slow growth rate, altered UDP-glucose/OpgH regulation, nor increased FtsZ protein levels are sufficient to explain this phenomenon. Taken together, these data illustrate the power of chemical-genetic screens to illuminate in vivo structure-function landscapes.
Results
Construction of a library of strains with chromosomal mutations in the transcription machinery
Decades-long study of the E. coli transcriptional apparatus has generated a large set of mutations with diverse phenotypes, particularly in the two largest subunits of RNAP (β’ and β) and to a lesser extent in σ70, NusA, NusG, and ρ. Unfortunately, the phenotypes of these mutant strains are not immediately comparable, as RNAP mutations are in diverse genotype backgrounds and are often only found as episomal merodiploids with a wild-type chromosomal copy. Following a literature review to manually annotate and collate the existing mutants, we selected and successfully reconstructed 68 mutations (Figure 1B, Supplemental Table 1) at their endogenous locus in the BW25113 strain background, enabling comparison with published chemical genetics datasets (Nichols et al., 2011; Shiver et al., 2016). Some mutations were introduced via transduction using a closely linked antibiotic resistance cassette; others were reconstructed by λ-Red oligo-mediated recombineering into a strain containing that cassette (Figure 1C). RifR mutants that confer resistance to rifampicin and M+ mutants that restore growth in minimal media to strains that lack or are deficient in the mediators of the stringent response ppGpp and DksA (Murphy and Cashel, 2003) are overrepresented in this collection (32/68) because they could be identified by selection. Some M+ mutants have been shown to form innately unstable open promoter RNAP complexes in vitro, mimicking the effects of ppGpp and DksA binding to RNAP, likely explaining their phenotype in minimal media (Rutherford et al., 2009). The RifR and M+ mutants in our library have been isolated previously (Supplemental Table 1). Our library also included a wide variety of other mutants that ensured our capacity to detect diverse phenotypic profiles.
A chemical-genetic screen of the transcription library reveals residue-level phenotypes of transcription mutants in vivo
We performed a chemical-genetic screen of the arrayed mutant library using sub-inhibitory concentrations of 83 chemical stressors (Supplemental Table 2) that overlapped with previous screens (Nichols et al., 2011; Shiver et al., 2016). The screen was performed in duplicate with technical and biological replicates of the mutants (Figure 1D) as well as control strains with the antibiotic marker alone and a subset of deletion strains from the Keio library. Where possible, at least two independent isolates of the same mutant were distributed between two arrays as biological replicates. When only one strain was isolated, it was included in both arrays. We included a subset of the Keio deletion strains to facilitate direct comparison of our results to previous efforts (STAR Methods) (Nichols et al., 2011; Shiver et al., 2016). We quantified colony opacity from images of the colony arrays at a single time point to calculate S-scores (Figure 1E) (Collins et al., 2006; Kritikos et al., 2017). The S-score is a modified t-statistic that reflects the statistical significance of the deviation of colony opacity in a particular growth condition from the average behavior of the mutant across all growth conditions in the dataset. Positive S-scores in this study report chemical resistance and negative S-scores report chemical sensitivities (Collins et al., 2010; Collins et al., 2006). Our S-scores were internally reproducible (r=0.73 for the same mutant compared across the two arrays), and S-scores for the nonessential gene deletions were correlated with those determined in a previous screen (r=0.65) (Shiver et al., 2016). This final S-score dataset was used in subsequent analyses examining chemical sensitivities and mutant-mutant correlations (Figure 1F, Supplemental Dataset 1). The entire dataset is available in an interactive, searchable format on the Ontology of Microbial Phenotypes website at https://microbialphenotypes.org/wiki/index.php?title=Special:RNAPchemicalgenetics.
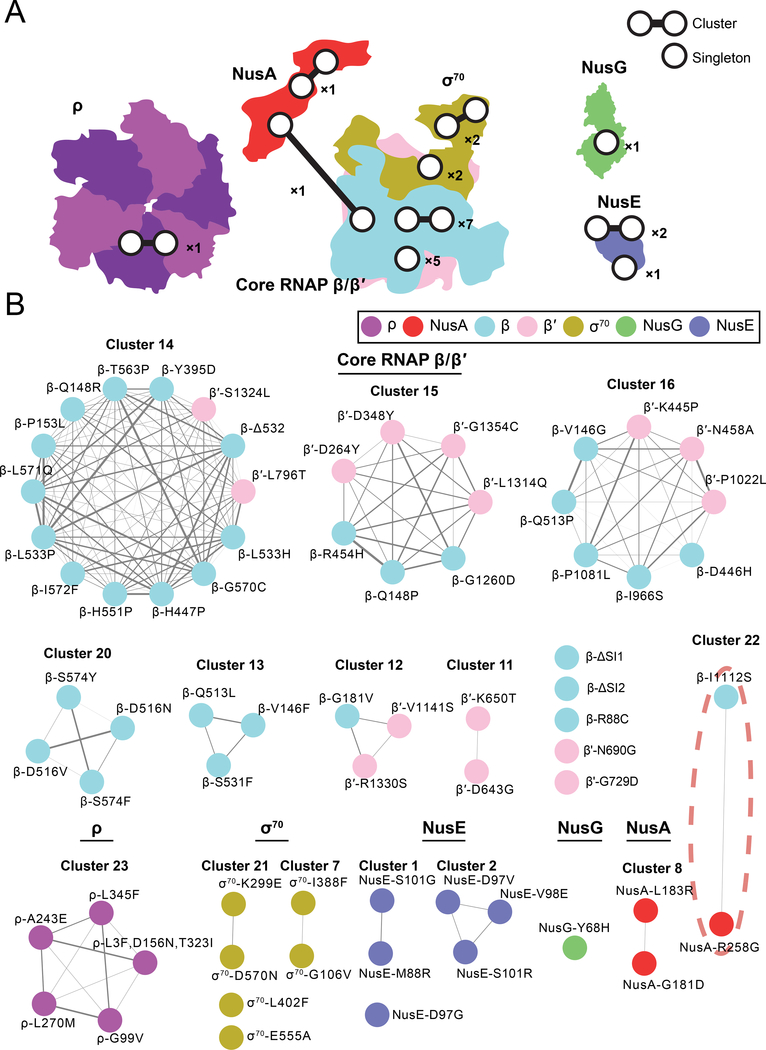
We used a cutoff based on hierarchical clustering of the S-scores to define 14 statistically significant clusters and 9 singletons of transcription mutants, which are numbered together from 1–23 (Supplemental Figure 1). Mutations largely clustered with others in the same polypeptide chain (Figure 2A), except for mutations in β and β′, which frequently clustered together. β and β′ are interwoven to form the core of the RNAP complex, and many of the mutations in these subunits are found on either side of the same DNA binding cleft. Clustering of mutations in these subunits likely reflects their tight functional coordination in the complex. Setting aside interactions between β and β′, only 1 of 171 co-clustering interactions was between mutations in different polypeptide chains (odds ratio=205, p=10−53). This interaction was between β-I1112S and NusA-R258G, which comprised cluster 22 (Figure 2A,B). β-I1112S and NusA-R258G were isolated from the same screen for ethanol tolerance (Haft et al., 2014).
Figure 2: Mutations in the transcription machinery cluster mainly according to the polypeptide chain in which they occur.
A) Summary statistics of clustering in the dataset are superimposed onto the structures of the 7 RNAP proteins with mutations included in the chemical-genetic screen. Two points connected by a line represent a cluster of ≥2 mutations as defined by the screen. A single point represents a singleton mutation with no significant correlation with any other transcription mutant. The number next to the points represent the number of each type (e.g., x2: either two clusters or two singletons). Mutations in ρ form a single cluster. Most NusA mutations form one intra-polypeptide chain cluster, but one NusA mutation clusters with a mutation from core RNAP. Mutations in σ70 form two clusters and two are singletons. Mutations in β/β′ form seven clusters, 5 mutations are singletons, and 1 mutation clusters with a mutation from NusA. The lone mutation in NusG is a singleton. Mutations in NusE form two clusters and one is a singleton.
B) The full clusters are shown color-coded and arranged according to polypeptide chain. The width of the edge connecting two mutations is proportional to their correlation in the dataset (Pearson’s r). Cluster 22, highlighted in red, is the only cluster that connects mutations from different polypeptide chains other than the β/β′ subunits of core RNAP.
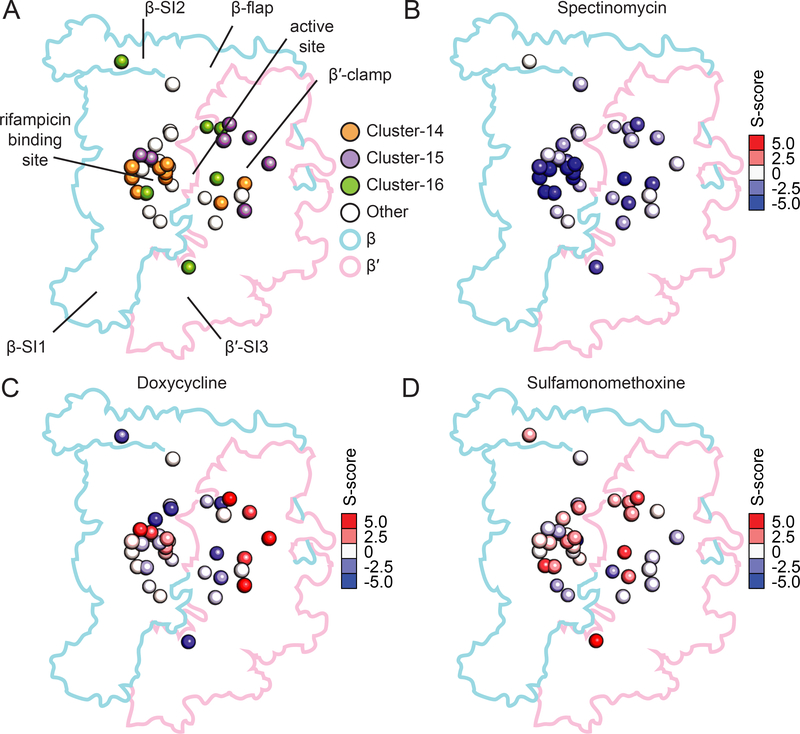
We calculated the enrichment of chemical-genetic interactions in clusters and mutant classes (Supplemental Table 3). Focusing on the three largest clusters of β and β’ mutations (out of 7 total) (Figure 3A), we found that each cluster could be associated with unique chemical-genetic interactions made by the mutants. The largest cluster (14) was enriched for sensitivities to aminoglycosides such as spectinomycin (Figure 3B). Cluster 15 was enriched for resistance to the tetracycline family of antibiotics (Figure 3C). Cluster 16 was enriched for sensitivity to tetracycline (Figure 3C) and resistance to sulfamonomethoxine (Figure 3D). Interestingly, this clustering did not necessarily follow the transcriptional classifications of the mutations: cluster 14 is comprised of both RifR and M+ mutations on the β-side of the RNAP cleft, cluster 15 is comprised predominantly of M+ mutations on the β’-side of the RNAP cleft, and cluster 16 contains mutations spread across the complex and with different known phenotypes (Figure 3A). Our identification of mutant clusters that are not aligned with previously defined classifications suggests that the chemical-genetic interactions in our dataset contain more detailed information regarding the effects of these diverse mutations on cellular physiology, a proposition we explore in the following sections as we investigate the phenotypes of specific RNAP mutations.
Figure 3: Mutations in core RNAP can be separated into clusters based on chemical sensitivities.
A) Three major clusters of point mutations within the core complex of β and β′ were identified. Cluster 14 (orange) is centered around the rifampicin binding site, cluster 15 (purple) is found mostly in the β′-clamp, and cluster 16 (green) is distributed throughout the complex, including β′-SI3, the active site, and β-SI2. Point mutations in RNAP not included in clusters 14–16 are shown in white. The alpha carbon of mutated residues is shown as a sphere in the structure.
B) Representation of S-scores indicating the extent of sensitivity to spectinomycin (enriched in Cluster 14). The alpha carbons of mutated residues are shown as spheres colored by the magnitude and direction of the mutant’s S-score in the dataset. Red indicates resistance and blue indicates sensitivity.
C) Representation of S-scores indicating the extent of resistance to doxycycline (enriched in Cluster 15) and sensitivity to doxycycline (enriched in Cluster 16). Mutations are presented in the structure as in (B).
D) Representation of S-scores indicating the extent of resistance to sulfamonomethoxine (enriched in Cluster 16). Mutations are presented as in (B).
Chemical-genetic profiling of the β subunit non-essential sequence insertions reveals environmental sensitivities
Strains with deletions of the large, non-essential sequence insertions β-SI1 and β-SI2 did not cluster with other transcription mutants (Figure 2B), suggesting that their impacts on RNAP function are unique within the library. β-ΔSI1 correlates with auxotrophic gene deletions, consistent with a role in the binding and function of the transcription factor DksA (Parshin et al., 2015). By contrast, β-ΔSI2 was not significantly correlated with any other transcription mutants or any mutants from the larger gene deletion library, making it difficult to ascertain its function by comparison to well-characterized mutants.
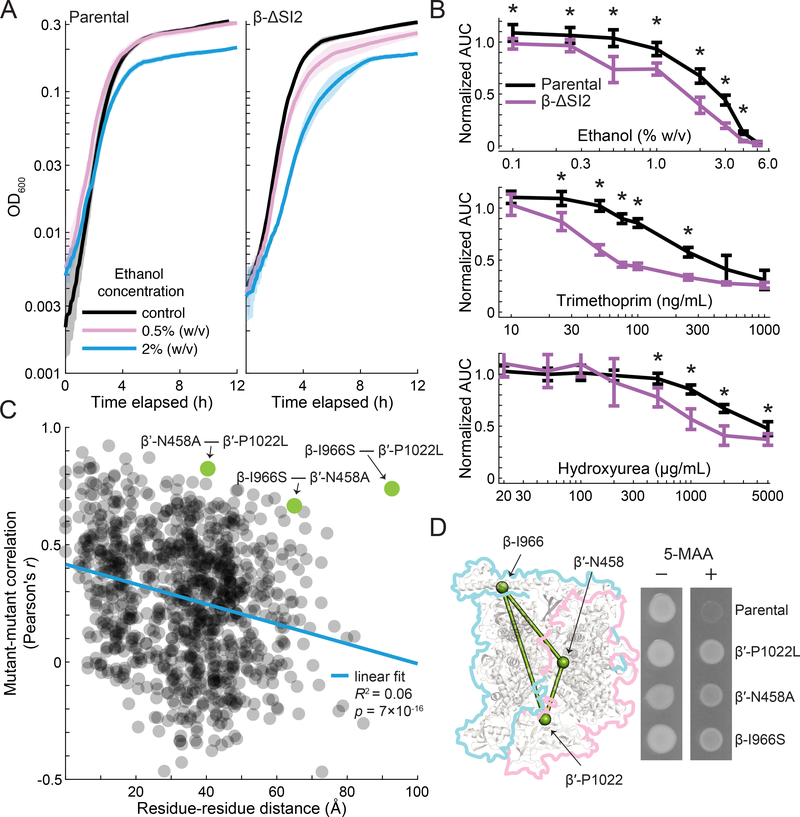
β-SI2 has alternatively been proposed to be dispensable for RNAP function (Borukhov et al., 1991; Nene and Glass, 1984; Severinov et al., 1992) or important for growth on M9 minimal media (Artsimovitch et al., 2003). In our dataset, we identified significantly negative S-scores for β-ΔSI2 in multiple treatments, including ethanol, trimethoprim, and hydroxyurea. To explore these potential sensitivities further, we monitored growth of β-ΔSI2 and its parental control in LB medium with increasing concentrations of all three compounds. At sub-lethal doses of ethanol, growth of the parental control slowed near the transition to stationary phase (Figure 4A). For β-ΔSI2, this phenotype was more pronounced and occurred at lower concentrations (Figure 4A). We found a similar chemical-genetic interaction with trimethoprim and hydroxyurea. In each case, growth of β-ΔSI2 was impacted by lower concentrations of the compounds but the mutant had no discernible effect on the MIC (Figure 4B). Thus, our screen revealed conditions under which deleting β-SI2 has an impact on growth, but mostly confirmed previous work that concluded the impact of deleting β-SI2 on cellular physiology is mild.
Figure 4: Mutations in β-SI2 have distinct phenotypes that include hyper-attenuation at the trp locus.
A) β-ΔSI2 has mild sensitivity to ethanol. Left: sublethal doses of ethanol hampered growth of the parental strain starting during the transition to stationary phase. Right: β-ΔSI2 has a more pronounced response to ethanol that begins at a lower concentration of ethanol.
B) β-ΔSI2 has mild sensitivity to ethanol, trimethoprim, and hydroxyurea. The normalized area under the curve (AUC) of growth curves as shown in (A) was calculated by integrating OD600 over time and normalizing by the AUC of the same strain without added drug. For ethanol (top), trimethoprim (middle), and hydroxyurea (bottom), growth of β-ΔSI2 was affected at lower (sub-inhibitory) concentrations, but the minimum inhibitory concentration remained the same. Error bars represent 95% confidence intervals. Drug concentrations for which the difference in relative AUC between β-ΔSI2 and its parental control is statistically significant are marked with asterisks (p<0.05).
C) Mutant-mutant correlations show a statistically significant association with distance between the residues in the RNA polymerase structure. Mutant-mutant correlations were calculated using Pearson’s r from the chemical-genetic dataset. Residue-residue distance was calculated based on the linear distance between alpha carbons of residues with mutations in the dataset as determined from the 3-dimensional structure. The PDB structure 4JKR was used for distance calculations. A three-mutant clique comprised of β-I966S, β′-N458A, and β′-P1022L was an exception to this rule, with high mutant-mutant correlations despite containing the largest inter-residue distance in the library (β-I966S to β′-P1022L).
D) Correlations among β-I966S, β′-N458A, and β′-P1022L were predictive of a shared hyper-attenuation phenotype that was originally identified for β′-P1022L (Weilbaecher et al., 1994). In a ΔtrpR background, expression of the trp locus is mainly controlled by attenuation. Hyper-attenuation reduces trp expression and makes cells resistant to a toxic analogue of a tryptophan biosynthesis intermediate, 5-methyl anthranilic acid (5-MAA) at 100 μg/mL.
Phenotypic clustering uncovers residue-level RNAP phenotypes
Similar to a previous study examining the genetic interactions of yeast RNA Pol II mutations (Braberg et al., 2013), we observed a statistical association between distance in the structure and pairwise phenotypic correlations between mutants in our dataset (Figure 4C) (Pearson’s r=−0.25, R2=0.06, p=7×10−16). In work published separately, this association provided sufficient information to use pairwise comparisons of mutant phenotypic profiles as a distance constraint to perform structural modeling of the core complex (Braberg et al., 2020).
Cluster 16 includes a high-correlation clique of three mutations: β-I966S, β′-N458A, and β′-P1022L. This clique was exceptional in that the phenotypic profiles of its members were highly correlated despite occurring in separated structural features of the RNAP complex (Figure 4C). Since β′-P1022L was isolated in a screen for increased transcription attenuation (Weilbaecher et al., 1994), we tested whether the other two mutants in the clique share this phenotype. As predicted by their high phenotypic correlation with β′-P1022L, both β-I966S and β′-N458A were resistant to 5-MAA (Figure 4D), indicative of increased transcription attenuation at the trp locus. To determine whether the 5-MAAR phenotype was widespread among the transcription mutant set, we tested three additional mutants in β (β-ΔSI2, β-P1081L, and β-I1112S), two additional mutants in the active site and secondary channel (β′-D643G and β′-V1141S) and two RifR mutations (β-S531F and β-I572F). Only the two RifR mutations were as resistant to 5-MAA as the members of the three-mutant clique (Supplemental Figure 2). These results indicate that the 5-MAAR phenotype is not universal among transcription mutants.
Resistance of β-P153L to mecillinam and A22 does not result from a classical stringent response
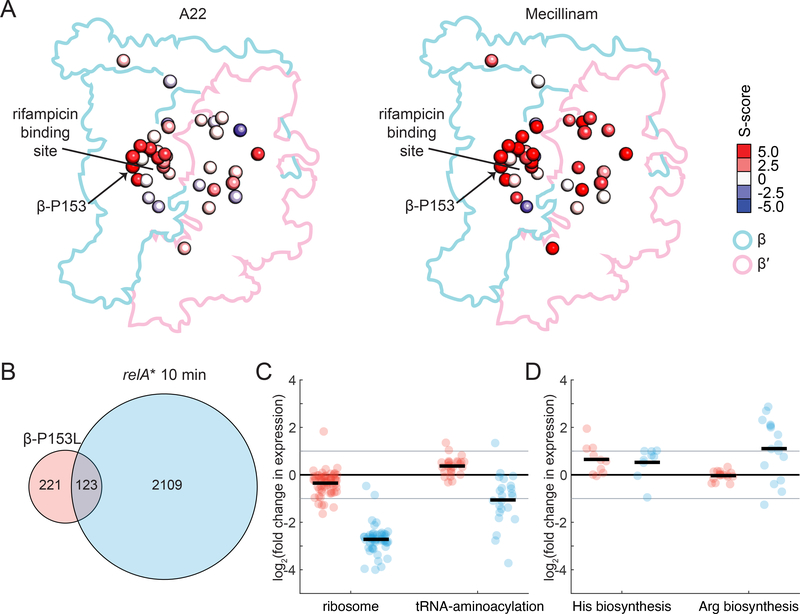
Both cluster 14 and the M+ class of mutations were enriched for resistance to the cell wall-targeting drugs mecillinam and A22 (Supplemental Table 3). At the sub-lethal doses used in our screen, A22 resistance was mostly restricted to a subset of cluster 14 mutants near the rifampin binding pocket, while mecillinam resistance was found throughout the complex (Figure 5A). To investigate these connections mechanistically, we focused on β-P153L, an M+ mutant in cluster 14 with the highest positive S-score for A22 and robust resistance to mecillinam (Figure 5A).
Figure 5: Gene expression in β-P153L only weakly overlaps with the stringent response.
A) Resistance to A22 and mecillinam was enriched among M+ mutants and cluster 14. Left: In the screen, resistance to A22 was more concentrated in cluster 14 mutants around the rifampicin binding pocket (Figure 3A). Right: resistance to mecillinam was widespread throughout β and β′ at the sub-inhibitory concentration used in the screen. The mutation β-P153L had the highest level of resistance to A22 and had high resistance to mecillinam. Alpha carbons of residues with mutations are colored according to their S-score in the chemical-genetic dataset.
B) There was a small degree of overlap in the significantly differentially expressed genes in β-P153L (red, data collected in this study) and 10 min post-induction of a constitutively active relA* allele (blue, from (Sanchez-Vazquez et al., 2019)). 36% of differentially expressed genes in β-P153L overlapped with the much larger set from relA*.
C) Repression of ribosome gene expression was weak in β-P153L (red), but consistent with results from relA* induction (blue) and with the lower growth rate of the mutant in LB. The activation of tRNA-aminoacylation genes is opposite to the repression in the relA* dataset. Individual genes are plotted as circles. The set average is shown as a sold black line. Two-fold changes in expression are marked with a solid gray line.
D) Activation of genes involved in histidine biosynthesis in β-P153L (red) was consistent with the stringent response and comparable in magnitude to the relA* dataset (blue). However, histidine biosynthesis was the only biosynthetic pathway differentially expressed in the β-P153L mutant; for example, the lack of differential expression of the arginine biosynthetic pathways in β-P153L contrasted with the relA* dataset. Individual genes are plotted as circles. The set average is shown as a sold black line. Two-fold changes in expression are marked with a solid gray line.
M+ mutants have some stringent-like transcriptional responses (Rutherford et al., 2009; Zhou and Jin, 1998). Moreover, either lowering the nutritional content of the growth medium or artificially inducing the stringent response leads to high-level resistance to mecillinam and A22 in E. coli (Bendezú and de Boer, 2008). We therefore explored whether β-P153L resistance to these antibiotics arises from a gene expression program locked into a stringent-like state. We measured differential gene expression in β-P153L and its parental strain without and with induction of the stringent response. We achieved induction of the stringent response by expressing a constitutively active allele of RelA (relA*) and compared our results to a published dataset that used the same method (Sanchez-Vazquez et al., 2019) (Supplemental Dataset 2).
Steady-state gene expression in β-P153L without relA* showed limited similarity to stringent response gene expression. While induction of relA* in wild-type E. coli resulted in significant expression changes in >2,000 genes, only 344 genes were significantly differentially expressed in β-P153L, and only 36% of these overlapped with the changes associated with a stringent response (Figure 5B). Moreover, in the overlapping set, only inhibited genes were significantly enriched (p=0.02, odds ratio=1.6 for inhibited genes; p=0.09, odds ratio=1.3 for activated genes). Analysis by functional category reinforced the differences between β-P153L expression and the stringent response. Whereas ribosomal protein expression was uniformly repressed during the stringent response, in β-P153L only 8 genes for ribosomal proteins (14%) were significantly repressed and only 5 (8%) were repressed >2-fold (Figure 5C). Moreover, rather than repressing tRNA-aminoacylation genes, β-P153L significantly activated 12 (46%) (Figure 5C). Rather than prevalent activation of amino acid biosynthesis genes, only those for histidine biosynthesis were significantly upregulated in β-P153L (Figure 5D). Induction of relA* resulted in a response highly overlapping with the classic stringent response in both β-P153L and its parent (Supplemental Figure 3A, Supplemental Dataset 2), and the response of the mutant was greater than that of its parent (Supplemental Figure 3B), even though the fold induction of relA* was similar (Supplemental Figure 3C). Thus, despite in vitro behaviors of M+ mutants that mimic ppGpp binding (Rutherford et al., 2009; Zhou and Jin, 1998), the steady-state transcriptional program of β-P153L in vivo is largely distinct from the canonical stringent response.
β-P153L protects against death caused by loss of rod shape in rich media
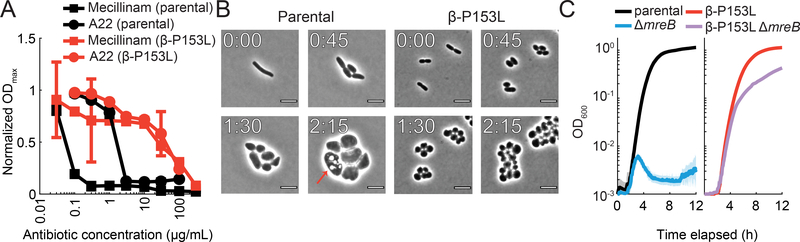
Mecillinam and A22 target PBP2 (Spratt, 1977) and MreB (Gitai et al., 2005), respectively, two components of the cell wall elongation machinery that directs lateral cell wall insertion and maintains rod shape in E. coli. They are essential during rapid growth (e.g. in LB), but dispensable in nutrient-poor environments (e.g. M9) (Bendezú and de Boer, 2008). As the stringent response is not obviously responsible for resistance in β-P153L, we sought to understand the origin of resistance by determining the full range of resistance responses and morphological changes associated with growth in the antibiotics.
We compared liquid growth curves in LB for β-P153L and its parental control over a range of A22 and mecillinam concentrations. β-P153L exhibited >10-fold higher MICs than the control (Figure 6A). To investigate how β-P153L protects E. coli under A22 or mecillinam treatment, we followed single-cell growth and morphology after exposure to supra-MIC concentrations of mecillinam using time-lapse microscopy. Wild-type cells stopped dividing and grew increasingly large, their membranes invaginated, and they eventually lysed (Figure 6B). By contrast, β-P153L cells morphed from small rods to small cocci that continued to grow and divide (Figure 6B). β-P153L cells displayed a similar morphological transition to small cocci in A22 (Supplemental Figure 3D), and fluorescent D-amino acid labeling (Kuru et al., 2012; Kuru et al., 2015) during growth with mecillinam revealed that β-P153L cocci retained a cell wall (Supplemental Figure 3E), as opposed to forming cell-wall-less spheroplasts.
Figure 6: β-P153L renders loss of rod shape non-lethal in rich media.
A) β-P153L is highly resistant to both mecillinam and A22, with MICs for both antibiotics that are higher by at least an order of magnitude. Maximum OD600 (ODmax) was extracted from growth curves of β-P153L (red curves) and its parental control (black curves) and normalized by the ODmax of each strain in the absence of antibiotic. Error bars represent 95% confidence intervals.
B) In the parental strain growing on agarose pads with LB+10 μg/mL mecillinam, division rapidly halted and cells expanded dramatically in volume, with the formation of apparent membrane invaginations (red arrows) and eventual lysis.
β-P153L cells growing on agarose pads with LB+10 μg/mL mecillinam lost rod-like shape but remained small and continued dividing without lysis. Elapsed time since imaging started is indicated in hours and minutes (H:MM) using white lettering at the top left of each panel. Scale bar: 5 μm.
C) β-P153L suppresses the lethality of ΔmreB in rich media. The ΔmreB deletion was introduced into the backgrounds of β-P153L and its parental control under permissive conditions (M9 minimal medium at 30 °C). Growth curves were measured after transitioning these strains to non-permissive conditions (LB at 37 °C). Left: ΔmreB halted bulk growth after a transition to non-permissive conditions. Right: β-P153L ΔmreB retained luxuriant growth in LB. The average OD600 from multiple growth curves is shown as an opaque line. The 95% confidence interval is shown is shown as a shading with the same color.
These results predicted that β-P153L should also render the genes encoding PBP2 and MreB (mrdA and mreB, respectively) non-essential during rapid growth conditions. We constructed ΔmreB and ΔmreB β-P153L mutants under permissive conditions (minimal medium, 30 °C), and tested growth of the double mutant after shifting to non-permissive conditions (LB, 37 °C). The ΔmreB β-P153L double mutant exhibited essentially normal growth, while the ΔmreB control quickly halted growth after the transfer (Figure 6C). Whole-genome resequencing confirmed that the strains did not contain second-site suppressors (Supplemental Table 4). We conclude that β-P153L renders mreB non-essential in rich media by preventing lysis after a loss of rod shape.
Decreased cell length is associated with A22 resistance in M+ mutants
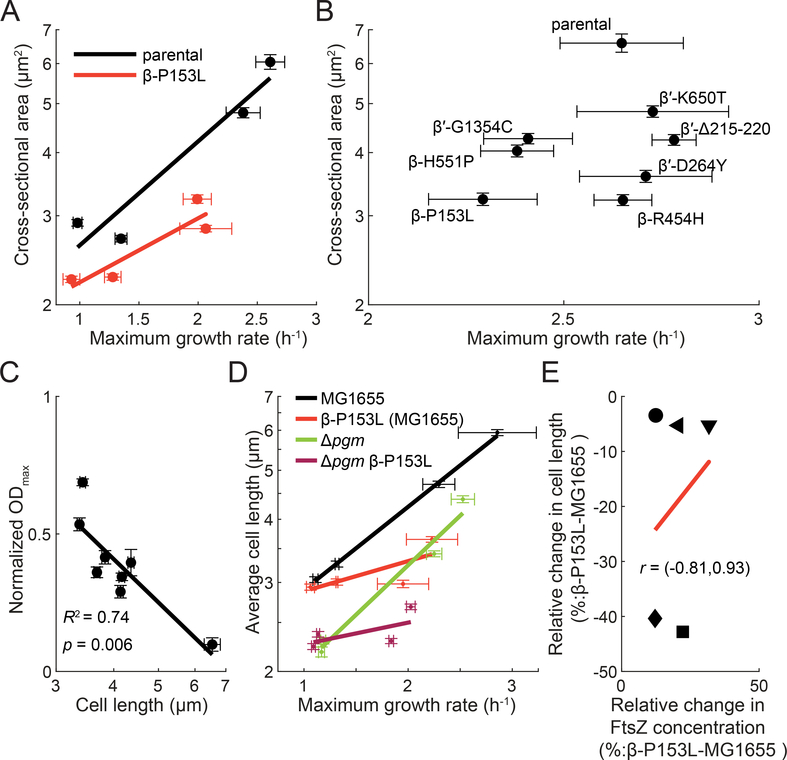
It has been proposed that the irreversible step toward death in A22-treated cells is the expansion of cell width beyond a limit at which division can no longer occur, leading to run-away cell widening and eventual lysis (Bendezú and de Boer, 2008). According to this model, the small size of β-P153L cells during treatment could keep the mutant below the non-reversible threshold and prevent death. However, the basis for the small size of β-P153L cells was not immediately clear. E. coli and many other rod-shaped bacteria have well-known a log-linear relationship between cell size and growth rate when the nutrient content of the medium is varied (Schaechter et al., 1958; Taheri-Araghi et al., 2015). This relationship, termed the Growth Law, suggested that the smaller size of β-P153L in LB might be simply due to its lower growth rate.
To test this idea, we measured cell size and growth rate across four media with different nutritional contents. If the small size of β-P153L were due to a growth rate defect alone, the overall relationship between growth rate and cell size would be indistinguishable between the two strains. Instead, we found that β-P153L was significantly smaller than its parental strain across all growth rates (Figure 7A). To determine whether our conclusions could be generalized to other M+ mutants, we chose 6 additional M+ mutants and measured the relationship between cell size and growth rate. We found that only a subset of M+ mutants had a slow growth phenotype in LB, but all M+ mutants had reduced size, with even the most subtle M+ mutant exhibiting a 27% reduction in cross-sectional area (Figure 7B). We conclude that M+ mutants exhibit a spectrum of reduced sizes and that their size reduction is not due solely to slower growth.
Figure 7: Decreased cell length in M+ mutants is associated with A22 resistance.
A) β-P153L cells are smaller than the parental control, even after controlling for growth rate. β-P153L (red) and its parental control (black) were grown for multiple generations in log phase in four media: MOPS minimal medium+0.2% glucose, MOPS minimal+0.2% glucose supplemented with 12 amino acids (see Methods), MOPS complete medium+0.2% glucose, and Tryptic Soy Broth. Maximum growth rate was extracted from growth curves started with log-phase cultures. Phase-contrast images of log-phase cells grown at steady state were acquired after spotting the cultures on PBS+1% (w/v) agarose pads, and cell area was computed from the segmented single-cell contours. Straight lines are linear regressions. Error bars on both axes are 95% confidence intervals for individual measurements.
B) M+ mutant cells are smaller than the parental control. Seven M+ mutants from different clusters were grown into log phase in lysogeny broth (LB) along with their parental control. Cultures were simultaneously spotted onto PBS+1% (w/v) agarose pads to measure cell size and used to inoculate growth curves to measure maximum growth rate. All M+ mutants were significantly smaller than their parental control, while only β-P153L, β-H551P, and β′-G1354C exhibited a statistically significant decrease in maximum growth rate.
C) A22 resistance is correlated with cell length. Maximum OD600 of the 7 M+ mutants and their parental control in LB with 13.5 μg/mL A22 was extracted from growth curves and normalized by growth curves in LB without antibiotic. Normalized ODmax values were strongly correlated with cell length from measurements in (B) (R2=0.74, p=0.006).
D) Δpgm is not epistatic to β-P153L. MG1655 (black), β-P153L (red), Δpgm (green), and β-P153L Δpgm (magenta) were grown for multiple generations in log phase in four media: M9 minimal medium with 0.2% (w/v) glucose, M9 minimal medium with 0.2% (w/v) glucose and supplemented with 12 amino acids (see Methods), LB, and LB with 0.2% (w/v) glucose. Growth curves were started in a plate reader with log-phase cultures. Phase-contrast images of log-phase cells grown in steady were acquired after spotting the cultures on LB + 1% (w/v) agarose pads, and cell length was calculated from the mesh computed for segmented single-cell contours. Straight lines are linear regressions. Error bars on both axes are 95% confidence intervals for individual measurements. If the length phenotype of Δpgm was epistatic to that of β-P153L, then the double mutant would have resembled Δpgm. Instead, Δpgm β-P153L exhibited a combination of the length phenotypes of both single mutants.
E) The relative change in FtsZ protein concentration is not correlated with decreases in average cell length in β-P153L as compared to MG1655. MG1655 FtsZ-msfGFP and MG1655 β-P153L FtsZ-msfGFP were grown in log phase for multiple generations in 5 media: M9 minimal medium with 0.2% (w/v) glycerol (circles), M9 minimal medium with 0.2% (w/v) glucose (leftwards-pointing triangles), M9 minimal medium with 0.2% (w/v) glucose and supplemented with 12 amino acids (downwards-pointing triangles), LB (squares), and LB with 0.2% (w/v) glucose (diamonds). Phase-contrast and fluorescence images of single cells were acquired after spotting log-phase cultures on PBS agarose pads with 1% (w/v) agarose. FtsZ-msGFP concentration was calculated from single-cell contours segmented from phase-contrast images by integrating the background-subtracted fluorescence within the contour area and normalizing by calculated cell volume. Relative cell length decreases the most in β-P153L in rich media like LB and LB 0.2% glucose, but this does not correspond to a proportional increase in FtsZ concentration. A linear fit to the data is shown as a red line. The correlation between the relative changes in FtsZ concentration and cell length is largely indeterminate as shown by the 95% confidence interval (Pearson’s r=0.25, 95% confidence interval: −0.81–0.93)
Finally, we asked whether cell length, cell width, or growth rate was correlated with A22 resistance of M+ mutants by determining the final OD600 in a supra-MIC concentration of A22 as a function of average cell width and length. We found that growth in A22 was strongly negatively correlated with cell length (R2=0.74, p=0.006), but not with cell width (R2=0.14, p=0.36) or growth rate (R2=0.17, p=0.31) (Figure 7C, Supplemental Figure 3F,G). This result is in stark contrast to mutations in MreB, for which cell width is the critical factor in A22 sensitivity (Shi et al., 2017), and suggests that altered division in M+ mutants both shortens the average cell length and protects against death.
The decreased cell length of β-P153L is independent of UDP-glucose signaling
The glucosyltransferase OpgH and the small molecule UDP-glucose link nutrient status to cell size in E. coli. OpgH localizes to the cell septum under nutrient rich conditions (high UDP-glucose), where it binds to and destabilizes FtsZ-rings to delay division (Hill et al., 2013). Deletion of opgH has a mild effect on cell size, while cells lacking pgm, which encodes a phosphoglucomutase necessary for UDP-glucose biosynthesis, have a pronounced decrease in cell length (Hill et al., 2013). Since OpgH sensing of UDP-glucose is the major known pathway for controlling cell length in response to nutrient availability, we investigated whether the decreased cell length of β-P153L was dependent on this pathway or reflected an independent mechanism of cell-size control.
Expression changes in genes related to the UDP-glucose sensing pathway were mild in β-P153L (Supplemental Table 5). However, as changes in protein levels are not always reflected in RNA levels, we performed a direct genetic test of the hypothesis. If the β-P153L small size phenotype resulted from a disturbance in OpgH/UDP-glucose signaling, then the small size phenotype of Δpgm cells would be epistatic to that of β-P153L. We compared cell length as a function of growth rate in the β-P153L Δpgm double mutant, single mutants, and wild-type strain. We found that the decrease in cell length due to the pgm deletion was independent of β-P153L, as the double mutant resulted in a combination of both the lowered slope of β-P153L and the negative length offset of Δpgm (Figure 7D). This result indicates that the cell size phenotype of β-P153L is independent of OpgH/UDP-glucose signaling. In this experiment and in the following experiment measuring FtsZ expression, we remade the β-P153L mutation in a MG1655 background without the rpoBC-cat marker to avoid potential complications from the cat cassette (see Microbial strains subsection in STAR Methods). However, we found that β-P153L had the same decrease in cell length regardless of strain background (Figure 7D). This, along with the observation that rpoBC-cat does not change the MIC of BW25113 for either A22 or mecillinam (Supplemental Figure 3 H,I) supports the conclusion that the M+ mutants do not simply correct the cell length phenotype of the parental strain (BW25113 rpoBC-cat).
Increased FtsZ expression does not explain the decreased length of β-P153L cells
Motivated by reports that the stringent response results in overexpression of FtsZ, which suppresses filamentation (Vinella and D’ari 1994), and other studies showing that FtsZ overexpression is sufficient to suppress mreB essentiality (Bendezú and de Boer, 2008), we next asked whether the lower cell length of β-P153L resulted from increased FtsZ levels. Using single-cell fluorescence microscopy, we quantified the concentration of an FtsZ-msfGFP translational fusion in β-P153L and MG1655 across media. FtsZ-msfGFP concentration was on average 19% higher in β-P153L (p=0.03, 95% confidence interval: 3–36%), consistent with the 68% increase in RNA levels of ftsZ in β-P153L (p-adj=0.0003) (Supplemental Table 5).
The cell length phenotype of β-P153L is exacerbated in richer nutrient environments (Figure 7 A,D). We reasoned that if increased FtsZ levels were the sole explanation for its decreased average cell length, then differences in FtsZ-msfGFP concentration between β-P153L and MG1655 would be negatively correlated with their respective differences in cell length across the media tested, with the highest concentration difference in rich media, where β-P153L cells are especially short. Instead, we found a non-significant positive correlation between the two variables (Figure 7E) (r=0.25, 95% confidence interval: −0.81–0.93). We conclude that β-P153L has somewhat elevated levels of FtsZ, but other factors are likely to underlie the decreased cell length of this mutant.
Discussion
As the enzyme responsible for bacterial transcription and the integrator of transcriptional control, RNAP has been the focus of an enormous amount of research. In addition to structural, biochemical, and evolutionary analyses, multiple studies have utilized RNAP-centric genetic approaches, including early work on resistance to RNAP-targeting drugs such as rifampicin (Jin and Gross, 1988) and streptolydigin (Heisler et al., 1993), temperature-sensitive (Saito et al., 1986) and dominant-negative mutations (Sagitov et al., 1993), mutations altering the function of RNAP (e.g. attenuation (Weilbaecher et al., 1994), mutations that increase survival in minimal media in sensitized genetic backgrounds (Rutherford et al., 2009)), and structure-guided mutational analysis (Wang et al., 2006). In addition to revealing the inner workings of RNAP, this top-down body of work has resulted in novel physiological discoveries. Furthermore, the repeated isolation of RNAP mutations during adaptation to biotechnology-related environments (Cheng et al., 2014; Haft et al., 2014; Tenaillon et al., 2012) has made evident the value and need for a deeper understanding of transcription-related pleiotropy (Alper and Stephanopoulos, 2007).
Here, we showed that a bottom-up approach based on unbiased, expansive screening and clustering of the phenotypes of large numbers of RNAP mutations is a powerful tool for functional discovery, illuminating structure-function relationships of RNAP at the single-residue level and systems-level connections between transcription and other cellular processes. That our strategy was successful even though our library is overrepresented in RifR and M+ mutants underscores the point that mutations isolated under the same selective pressure can have distinct, pleiotropic phenotypes.
Our analysis of the phenotypes of lineage-specific insertions in the β-subunit highlight both the strengths and challenges of chemical genetic screens. Although neither the β-ΔSI1 nor the β-ΔSI2 strain exhibited chemical sensitivities that clustered with other RNAP mutations, the strong chemical sensitivities of β-ΔSI1 were correlated with ΔdksA, providing a starting point for analysis. The proximity of β-SI1 to the known binding site of DksA on RNAP allowed us to predict a role for β-SI1 in the binding and function of DksA that was validated with biochemical and genetic evidence (Parshin et al., 2015). Further work by others has fleshed out this interaction by identifying the required conformational changes (Molodtsov et al., 2018), finding that additional secondary channel regulators bind to β-SI1 (Chen et al., 2019; Molodtsov et al., 2018), and identifying a role of ppGpp in the DksA–RNAP interaction (Molodtsov et al., 2018; Ross et al., 2016). By contrast, β-ΔSI2 displayed minor chemical sensitivities (Figure 4A,B) and the lack of phenotypic clustering information for β-ΔSI2 prevented hypothesis generation through traditional chemical-genetic inference. Our ability to successfully predict the attenuation-proficient phenotype of the β-I966S mutant in β-SI2 (Figure 4C,D), consistent with previous predictions of a hyper-termination phenotype (González-González et al., 2017), further highlights the higher predictive power of correlations between mutants for identifying the function of uncharacterized mutations compared with the interpretation of individual sensitivities, as has been the case for most high-throughput genetic screens to date (Schuldiner et al., 2005). The three mutants comprising the clique within cluster 16 that includes β-I966S are outliers to the negative correlation between phenotypic profile similarity and pairwise distance on the RNAP structure (Figure 4C), highlighting the potential for other long-distance interactions in RNAP that remain to be discovered.
Finally, our analysis of A22 and mecillinam resistance in M+ mutants demonstrates the power of this approach in discovering unexpected phenotypes. With prior knowledge that the stringent response confers resistance to A22 (Bendezú and de Boer, 2008) and mecillinam (Bendezú and de Boer, 2008; Vinella et al., 1992), and that M+ RNAP enzymes exhibit certain behaviors associated with the stringent response (Rutherford et al., 2009; Zhou and Jin, 1998), we first explored the possibility that A22 and mecillinam resistance reflected stringent-like transcription by M+ mutants. However, we found that the transcriptional program of β-P153L was largely dissimilar to the stringent response (Figure 5B-D). This finding was somewhat surprising given the clear effect of M+ mutants on transcriptional fusion reporters of rrn operon promoters (Zhou and Jin, 1998). One explanation worth investigating in future work is if long-term adaptation is capable of compensating for some of the transcription defects of M+ polymerases. Instead, we found that M+ mutants exhibited varying degrees of reduction in cell length relative to the parental strain (Figure 7B) and the extent of the reduction in average cell length was correlated with survival at high A22 concentration (Figure 7C). We presented evidence that none of the best-characterized mechanisms could explain this observation. Decreased growth rate wasn’t sufficient to explain the decrease in cell length in M+ mutants (Figure 7A,B,D). The gene expression program was largely dissimilar to that of the stringent response (Figure 5). Changes in FtsZ expression were not correlated with changes in cell length (Figure 7E), and the cell length of Δpgm is not epistatic to that of β-P153L (Figure 7D). These results suggest that an unknown mechanism underlies the decreased cell length of M+ mutants and motivate future work to uncover this mechanism.
The success of this proof-of-principle study motivates using chemical-genetics to explore structure-function relationships in additional protein complexes and machineries. The cell wall synthesis machinery of E. coli, for which extensive libraries of mreB and mrdA point mutations have recently been created (Shi et al., 2017) is an attractive first target. The simplicity of a chemical-genetic approach also motivates the study of RNAP function in a broader set of bacterial species, which could generate fascinating insights into the evolutionary conservation of structure-function relationships and physiological connections of this essential enzyme complex.
Limitations
The power of this proof-of-principle experiment highlights the value of a bottom-up chemical-genetic approach to interrogating structure-function relationships in vivo. However, the insight that we gained was limited by the modest number of mutations that we included in the study. New mutagenesis approaches such as CRISPR editing hold the promise of both increasing the scale of mutant libraries in essential genes and expanding the scope of mutations beyond those with easily selectable phenotypes, thereby enhancing the breadth and power of future screens. Moreover, a pooled deep sequencing approach could enable screening many more conditions simultaneously, increasing the capacity of screens such as ours to discover novel phenotypes.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Carol Gross (cgrossucsf@gmail.com).
Materials Availability
Strains, plasmids, and oligonucleotides generated in this study are available upon request from the Lead Contact.
Data and Code Availability
Raw images and Iris data files for the chemical-genetic screen along with two datasets generated in this work have been published on Dryad. Raw sequencing reads for whole genome resequencing and RNA sequencing have been deposited at NCBI. Reproducible compute capsules have been published on Code Ocean for the major findings and results of this study. Accession codes for deposited data are listed in the Key Resources Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| For the list of bacterial strains generated in this work, see Supplemental Table 1 | this study | Supplemental Table 1 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| For the list of chemicals used in the chemical-genetic screen, see Supplemental Table 2 | this study | Supplemental Table 2 |
| Trizol LS | Invitrogen | Catalog No. 10296028 |
| T4 RNA ligase buffer | NEB | Catalog No. B0216L |
| T4 RNA ligase 2 truncated KQ | NEB | Catalog No. M0373L |
| T4 Polynucleotide Kinase (PNK) | NEB | Catalog No. M0201L |
| T4 RNA Ligase 1 | NEB | Catalog No. M0204L |
| SuperScript IV | Thermo Fisher Scientific | Catalog No. 18090010 |
| HCC-amino-D-alanine (HADA) | Michael vanNieuwenhze lab | N/A |
| Teknova EZ rich defined media kit (EZ-RDM) | Teknova | Catalog No. M2105 |
| Critical Commercial Assays | ||
| Ribo-Zero rRNA removal kit (Gram-negative bacteria) | Illumina | MRZGN126 |
| Deposited Data | ||
| Images and data from the paper | this study | https://doi.org/10.5061/dryad.z612jm68q |
| Raw sequences and processed data for gene expression analysis of β-P153L. | this study | GSE151022 (NCBI GEO) |
| Raw sequences for whole genome resequencing of ΔmreB strains. | this study | PRJNA632897 (NCBI SRA) |
| Code Ocean compute capsule: Generating the chemical genetic interaction dataset (Supplemental Dataset 1) | this study | https://doi.org/10.24433/CO.9507705.v1 |
| Code Ocean compute capsule: Running DESeq2 on RNA-seq counts. | this study | https://doi.org/10.24433/CO.2313056.v1 |
| Code Ocean compute capsule: Generating Supplemental Dataset 2, figures, and statistics cited in the main text of the paper. | this study | https://doi.org/10.24433/CO.8886448.v3 |
| Code Ocean compute capsule: Generating the list of conditions and concentrations used in the screen. (Supplemental Table 2) | this study | https://doi.org/10.24433/CO.2239948.v1 |
| Code Ocean compute capsule: Generating the enrichment tables. (Supplemental Table 3) | this study | https://doi.org/10.24433/CO.1521219.v3 |
| Code Ocean compute capsule: Generating mutation reports using breseq. (Supplemental Table 4) | this study | https://doi.org/10.24433/CO.6559033.v1 |
| Code Ocean compute capsule: Collating the table of gene expression changes in genes of interest. (Supplemental Table 5) | this study | https://doi.org/10.24433/CO.3392553.v1 |
| Code Ocean compute capsule: Generating chemical genetic interaction datasets for the marker-only alleles. (Supplemental Dataset 3 and Supplemental Figure 4) | this study | https://doi.org/10.24433/CO.5034235.v1 |
| Code Ocean compute capsule: Comparing the results of hierarchical clustering with a cut-off to k-means clustering with k=23. (Supplemental Figure 5) | this study | https://doi.org/10.24433/CO.3570885.v2 |
| Oligonucleotides | ||
| For the list of oligonucleotides and plasmids used in this study, see Supplemental Table 6 | this study | Supplemental Table 6 |
| Software and Algorithms | ||
| MATLAB R2020b v. 9.9.0.1495850 | Mathworks | https://www.mathworks.com |
| Python v. 3.7.3 | Python Software Foundation | https://www.python.org/ |
| R v. 3.5.1 | R Foundation for Statistical Computing | https://www.r-project.org/ |
| Gene Set Enrichment Analysis v. 3.0 | (Subramanian et al., 2005) | https://www.gsea-msigdb.org/gsea/index.jsp |
| Cytoscape v. 3.7.2 | (Shannon et al., 2003) | https://cytoscape.org/ |
| Gen5 ™ v. 3.04 | BioTek ® | https://www.biotek.com/products/software-robotics-software/gen5-software-features-for-imaging-microscopy/ |
| Iris v. 0.9.4 | (Kritikos et al., 2017) | http://critichu.github.io/Iris |
| breseq v. 0.333.2 | (Deatherage and Barrick, 2014) | https://github.com/barricklab/breseq |
| DESeq2 v. 1.22.2 | (Love et al., 2014) | http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html |
| shake v. 5.0 | (Jos(10584), 2019) | https://www.mathworks.com/matlabcentral/fileexchange/10067-shake |
| ami v. 1.0.0.0 | (Nguyen, 2021) | https://www.mathworks.com/matlabcentral/fileexchange/33144-the-adjusted-mutual-information |
| venn v. 1.7.0.0 | (Darik, 2011) | https://www.mathworks.com/matlabcentral/fileexchange/22282-venn |
| Other | ||
| Data browser for chemical-genetic interaction dataset | this study | https://microbialphenotypes.org/wiki/index.php?title=Special:RNAPchemicalgenetics |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Microbial strains
Microbial strains used in this work are summarized in Supplemental Table 1 and in the Key Resources Table. Mutations used in the chemical-genetic screen were transduced into or generated in the E. coli K-12 BW25113 (CAG67001) genetic background. During subsequent work to confirm sensitivities found in the screen, we generated some strains in an MG1655 genetic background (KC001).
Our parental strain for mutations in β and β′ (BW25113 rpoBC-cat) exhibited an increase in mean cell length, sometimes resulting in filamentous cells >10 μm in length. This phenotype likely resulted from insertion of the chloramphenicol resistance cassette (cat) between rpoB and rpoC, as the strain had no additional mutations (Supplemental Table 4). The rpoBC-cat marker did not change the MIC of BW25113 in A22 or mecillinam (Supplemental Figure 3 H,I).
To test if the decreased cell length of M+ mutants (Figure 7B) was due to suppression of the rpoBC-cat phenotype, we remade β-P153L in a MG1655 background without the cat cassette (used in Figure 7D,E). The β-P153L mutation had the same cell length phenotype regardless of strain background (Figure 7D), suggesting the M+ mutant phenotype is not just due to a genetic interaction with rpoBC-cat.
Growth conditions are described in the Method Details section of the STAR Methods.
METHOD DETAILS
Oligonucleotide design
All oligonucleotides used in this study are listed in Supplemental Table 6. Oligonucleotides were synthesized by Integrated DNA Technologies (Skokie, IL) with standard desalting purification. Oligonucleotides used for recombineering were designed to anneal to the lagging strand to increase efficiency (Ellis et al., 2001). Recombineering oligonucleotides were 79 nucleotides long, unless high efficiency of mutagenesis was required in which case the length was extended to 89 or 90 nucleotides. For all recombineering oligonucleotides, mismatch(es) were located in the center of the sequence. For recombineering oligonucleotides that required highly efficient mutagenesis, four phosphorothioation modifications were included between the five bases closest to the 5’ end of the oligonucleotide to prevent 5’ degradation (Wang et al., 2009).
Oligonucleotide recombineering mutagenesis
We generated some mutations reported in this study de novo using oligonucleotide recombineering. We transformed strains of the appropriate genetic background with pSIM6 to introduce the λ-Red system (Datta et al., 2006), made electrocompetent cells using published methods (Thomason et al., 2014), and electroporated the cells with mutagenic oligonucleotides. To prepare recombineering-competent cells, an overnight culture was grown at 30 °C in LB with 100 μg/mL ampicillin and 500 μL of this culture was diluted into 35 mL of fresh LB with 100 μg/mL ampicillin in a 250-mL baffled flask and grown in a shaking water bath (Gyrorotory® Water Bath Model G76, New Brunswick Scientific Co., Incorporated) at 32 °C. The culture was shaken at 330 rpm throughout early log phase until it reached an OD600 of 0.4–0.6 as measured on a Genesys 20 spectrophotometer (Thermo-Scientific). Fifteen milliliters of culture were then transferred to a 125-mL baffled flask in an orbital shaking water bath set to 42 °C and 200 rpm for 15 min. After heat shock to induce the λ-Red system from pSIM6, cultures were chilled on ice for 10 min.
Chilled cells were transferred to a 50-mL conical tube, pelleted at 3,709g for 4 min (Allegra X-30R, C0650 adaptor, Beckman Coulter), and resuspended in 50 mL of ice-cold deionized water (MilliQ Biocel A10, Millipore). Cells were pelleted again at 3,709g for 4 min and resuspended in 800 μL of ice-cold deionized water. Finally, cells were transferred to a 1.5-mL Eppendorf tube, pelleted in a tabletop centrifuge (Centrifuge 5417 C, Eppendorf) at 10,000g for 30 s, and resuspended in 200 μL of ice-cold deionized water. These competent cells were kept on ice until they were electroporated. Before electroporation, 50 μL of competent cells were mixed with 2 μL of 100 μM oligonucleotide solution before being transferred to an electroporation cuvette. Transformation mixtures were electroporated at 18 kV/cm (Gene Pulser Cuvettes (0.1-cm gap), E. coli Pulser, Gene Pulser Attachment, Bio-Rad).
Electroporated cells were immediately resuspended in 500 μL of LB, transferred to a 5-mL test tube, and incubated on a roller drum at 37 °C for 1 h. Recovered cells were plated according to the selection scheme necessary to isolate the intended mutations.
Selection of RifR mutants
RifR mutations were selected in rpoB in BW25113 rpoBC-cat. Two hundred microliters of recovered transformants were plated on LB with 10–50 μg/mL rifampicin. Resistant colonies were struck on rifampin plates a second time to purify the colonies and confirm the RifR phenotype. Mutations were confirmed using Sanger sequencing.
Selection of M+ mutants
We selected for M+ mutations in rpoB and rpoC in BW25113 rpoBC-cat ΔdksA::kan using a standard genetic selection (Rutherford et al., 2009). After recovery in LB, cells were pelleted at 10,000g for 30 s in a tabletop centrifuge and resuspended in 1 mL of M9 minimal medium with 0.2% glucose. Two hundred microliters were then plated on M9 minimal medium plates with 0.2% glucose. Colonies that grew in the first 48 h were struck again on M9 minimal medium plates to confirm growth. Mutations were transduced into BW25113 using the genetically linked cat gene. Separation of the rpoBC locus from ΔdksA::kan was confirmed by testing for a Kans phenotype. Co-transduction of the mutations with cat was confirmed with Sanger sequencing.
Screening for attenuation mutants
We screened for attenuation-enhancing mutations in a BW25113 rpoBC-cat ΔtrpR::kan genetic background. Recovered cells were plated in a dilution series on LB agar, and 200 μL of the 10−3, 10−4, and 10−5 dilutions were plated on LB agar plates and grown at 37 °C overnight. Single colonies were patched onto a grid on an LB plate and replica-plated onto M9 minimal glucose plates supplemented with L-cysteine (400 μg/mL), L-methionine (400 μg/mL), L-leucine (400 μg/mL), indole (5 μg/mL), and 5-methyl anthranilic acid (5-MAA, 100 μg/mL) and grown at 30 °C. Patches with robust growth in the presence of 5-MAA were considered to be potential mutants (Weilbaecher et al., 1994; Yanofsky and Horn, 1981). Single colonies were isolated by streaking from the non-selective patch, and the mutation was confirmed with Sanger sequencing. Finally, the mutant was transduced into BW25113 using the genetically linked cat gene. Separation of the mutation from ΔtrpR::kan was confirmed by testing for a Kans phenotype. Co-transduction of the mutation with cat was confirmed with Sanger sequencing.
Screening for mutants with PCR
Some of the mutations that we generated using oligonucleotide recombineering had no available selective phenotype, so we directly screened for these mutations using a PCR-based assay. For point mutations, we used the BW25113 rpoBC-cat genetic background. We designed oligonucleotides that introduced silent mutations in the codons surrounding the mutation of interest to increase the efficiency of mutagenesis (Thomason et al., 2014) and improve our ability to distinguish between the mutant and wild-type alleles through primer annealing. After transformation with the mutagenic oligonucleotide, recovered cells were plated in a dilution series on LB agar. Single colonies were isolated and suspended in 50 μL of PCR buffer. One microliter of this suspension was used for colony PCR (KAPA2G Robust HotStart PCR Kit, Roche), wherein a ~500-bp amplicon was amplified by a primer pair in which the 3’ end of one primer was complementary to the mutant allele. A colony that led to amplification with the mutation-specific primers was considered to be a positive hit. To further purify the putative mutant, the colony suspension was struck on LB agar plates, grown overnight at 37 °C, and the PCR screen was repeated. We then verified the mutation of interest using Sanger sequencing.
To generate deletions of the lineage-specific insertions SI1 and SI2, we used the strain BW25113 rpoBC-cat ΔmutS::kan. Primers were designed to detect the deletions through a shift in amplicon size. After transformation with the mutagenic oligonucleotide, recovered cells were plated in a dilution series on LB agar. Single colonies were isolated and suspended in 50 μL of PCR buffer. One microliter of this suspension was used for colony PCR (KAPA2G Robust HotStart PCR Kit, Roche). Samples with a small amplicon size were considered positive hits. The colony suspension was struck on LB, grown overnight at 37 °C, and the PCR screen was repeated on single colonies. After confirmation of the deletion with PCR, the suspensions were used to inoculate 5 mL of LB and grown to an OD600 of ~0.6. This culture was used to create a P1vir lysate for transduction into BW25113. Co-transduction of the deletions with the cat antibiotic resistance gene was confirmed with PCR and Sanger sequencing. Separation from the ΔmutS::kan locus was confirmed by testing for a Kans phenotype.
P1vir transduction
Some of the mutants used in our study were collected from the scientific community and transduced into a BW25113 background. We first introduced the relevant antibiotic markers into the original strain using λ-Red recombineering with the pSIM6 plasmid. We then transduced the mutations into BW25113 using P1vir and selected for the antibiotic resistance cassette that we introduced in the previous step. Co-transduction of the genetically linked mutations was confirmed with Sanger sequencing.
Assembly of the 1536-colony arrays
We split biological replicates for each transcription mutation or gene deletion into two sets (Array #1 and Array #2). We then arrayed the mutants within each set in triplicate with randomized positions in a 32×48 array of 1536 colonies. To minimize edge effects (French et al., 2016), we filled the outermost two columns and rows of the 1536-colony array with wild-type controls and only analyzed the inner positions. Mutants were split according to antibiotic resistance phenotype (Camr and Kanr) into 16 groups that corresponded to each of the 16 96-well plates that would comprise the 1536 array. Based on the final position in the 1536-well array, spaces in each 96-well plate were devoted to wild-type (either BW25113 or BW25113 rpoBC-cat) and used as “dummy” colonies that would grow in all conditions.
For storage, plates were grown overnight at 37 °C with shaking at 900 rpm in a humidified platform shaker (Infors HT). Glycerol was added to a final concentration of 12.5%, and aliquots of each plate were stored at −80 °C in a 96-well format. The two 1536-colony arrays were assembled by thawing copies of the 2×16 96-well plates and using a Rotor pinning robot (Singer Instruments) to spot the plates, first into 2×4 384-colony plates, and finally into two 1536-colony format plates (Array #1 and Array #2).
Screening the mutant library
We pinned Array #1 and Array #2 in parallel onto agar plates with antibiotics and other chemical stressors added to the agar. Screens were performed in four batches with 30 or more conditions in each batch to allow for normalization within each batch (Collins et al., 2010). Chemical perturbations were chosen to overlap with existing chemical-genetics datasets (Nichols et al., 2011; Shiver et al., 2016). For the dataset from (Nichols et al., 2011), which used three concentrations per chemical, the concentration with the highest number of significant interactions was chosen. For the dataset from (Shiver et al., 2016), Array #1 and Array #2 were screened at the same time using the same batch of chemicals as used for the gene deletion library.
For each condition, drug was added to melted LB agar and 45 mL was poured into PlusPlates (Singer Instruments). Source plates were generated by pinning the colony arrays onto LB agar plates, and each source plate was used to pin the array onto multiple drug plates. Plates were incubated at 37 °C for a time interval over which colonies had grown appreciably but had not overgrown to the point that colony edges overlapped. Images were taken with a Powershot G10 camera (Canon) and a custom illumination configuration. Colony opacity was estimated using the software Iris v. 0.9.4 (Kritikos et al., 2017).
Liquid growth curves
Growth curves were measured in a Synergy H1 (BioTek ® Instruments) or an Epoch 2 (BioTek ® Instruments) plate reader using Gen5™ v. 3.04 (BioTek Instruments). Data were collected for approximately 24 h at 37 °C using a 2-min discontinuous loop comprised of a read step at 600 nm and 1 min of double-orbital shaking at slow orbital speed and an orbital frequency of 237 cycles per minute.
All experiments used clear, flat-bottom, polystyrene 96-well plates (Greiner Bio-one) covered with a clear polystyrene lid (E&K Scientific). All conditions other than ethanol used a final culture volume of 200 μL per well. Ethanol experiments used a final culture volume of 150 μL per well and 50 μL of mineral oil was overlaid on the culture to reduce ethanol evaporation.
All cultures to measure chemical sensitivities were inoculated at an OD600 of 0.01. Growth curves of ΔmreB strains were inoculated at an OD600 of 10−4 because we found that a lower inoculation density clarified the growth defects of MG1655 ΔmreB. For growth in mecillinam, A22, and minimal medium, the inoculum was log-phase culture that had been kept below an OD600 of 0.3 using sequential dilutions in the same medium as the growth curve for 6–8 h. For transition experiments of ΔmreB strains, the inoculum was log-phase culture that had been kept at a low OD600 using sequential dilutions in M9 minimal medium with 0.2% glucose at 30 °C. For sensitivity to ethanol, hydroxyurea, and trimethoprim, the initial inoculum was a stationary-phase culture that had grown for 16–24 h in LB. Stationary-phase cultures were used in these measurements to enhance sensitivity of β-ΔSI2 to the compounds.
trp-locus attenuation assay
We used the trp-locus attenuation assay from (Weilbaecher et al., 1994; Yanofsky and Horn, 1981) to test for a hyper-attenuation phenotype. We transduced mutations and controls from the BW25113 background to an MG1655 ΔtrpR genetic background using P1vir transduction and selected for the linked rpoBC-cat antibiotic resistance cassette. Sequences of all transductants were verified with Sanger sequencing.
We used resistance to 5-MAA to test for hyper-attenuation at the trp locus. Strains were grown overnight in M9 minimal medium with 0.2% glucose at 30 °C, pelleted using centrifugation, and resuspended at a normalized OD600 of 1.0 in M9 minimal salts. We then spotted 2.5 μL of the resuspended cultures onto M9 minimal medium agar plates supplemented with 0.2% glucose, L-cysteine (400 μg/mL), L-methionine (400 μg/mL), L-leucine (400 μg/mL), and indole (5 μg/mL) to which 5-MAA had either been excluded (−) or added at a concentration of 100 μg/mL (+). The spots were allowed to grow at 30 °C for 2 days before pictures were taken with an EOS Rebel T5i (Canon).
Sample preparation for RNA-seq
The parental rpoBC-cat and β-P153L strains were first transformed with the pALS13 (Ptrc::relA*) and pALS14 (Ptrc::relA-) plasmids. Cells were grown overnight in Teknova Rich Defined Media (EZ-RDM) with 100 μg/mL ampicillin to maintain plasmid selection. Overnight cultures were inoculated into fresh EZ-RDM with 100 μg/mL ampicillin to maintain plasmid selection. After strains had grown to mid-log phase, samples were taken for the uninduced control, 10 mg/mL isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to induce expression of the relA alleles, and samples were taken 5 min after induction. All samples were immediately stored on ice with a 1:8 volume of 5% phenol in ethanol as a stop solution. Samples were transferred to a −80 °C freezer for storage before further processing.
RNA isolation and library prep
RNA was isolated from frozen cell pellets using Trizol (Invitrogen) extraction according to the manufacturer’s protocol. One microgram of purified RNA was fragmented at 95 °C for 7 min in 1X T4 RNA Ligase buffer (NEB, B0216L) with an equal volume of 2X alkaline fragmentation buffer (0.6 volumes of 100 mM Na2CO3 plus 4.4 volumes of 100 mM NaHCO3). After 3’-end healing with T4 Polynucleotide Kinase (NEB, M0201L) in T4 RNA ligase buffer for 1 h, 3’ ligation to a pre-adenylated, barcoded TruSeq R1 adapter with 5 random bases at its 5’ end was performed overnight using T4 RNA Ligase 2 truncated KQ (NEB, M0373L). The barcoded samples were then pooled and run on a 6% TBE-Urea gel for size selection (>15 nucleotide insert size), eluted, and ethanol precipitated before performing ribosomal RNA subtraction (Ribo-Zero rRNA Removal Kit, Illumina, MRZGN126). Reverse transcription with SuperScript IV (Thermo Fisher Scientific, 18090010) was performed using a TruSeq R1 RT primer, followed by ligation of the TruSeq R2 adapter to the 3’ end of the cDNA overnight using T4 RNA ligase 1 (NEB, M0204L), prior to another gel size selection as described above. A final PCR of the library was performed with indexed TruSeq PCR primers to add the index and P5/P7 flowcell adapters, followed by gel extraction, precipitation, and a BioAnalyzer (Agilent) run for quality control before sequencing on a HiSeq4000 platform (Illumina).
Fluorescent D-amino acid incorporation
Fluorescent D-amino acid labeling of the cell wall was performed according to published protocols (Kuru et al., 2015). β-P153L was grown into log phase in LB broth and transferred to LB broth with 30 μg/mL mecillinam. The strain was grown for two doublings in the presence of mecillinam, diluted to an OD600 of 0.05 in LB broth with 30 μg/mL mecillinam and 500 μM HCC-amino-D-alanine (HADA), and grown for 1.5 h. The culture was then washed three times in phosphate buffer saline, and 1 μL was spotted onto phosphate buffer saline 1% (w/v) agarose pads. Fluorescence microscopy images were collected using a Ti-E microscope (Nikon) with a 100X (NA: 1.4) objective and a Zyla 5.5 sCMOS camera (Andor).
Quantifying the relationship between growth rate and cell size
For the data in Figure 7A, we individually inoculated the parental control (CAG67202) and β-P153L (CAG68095) into 5 mL test tubes filled with one of four media (8 total cultures). The media used were MOPS minimal medium+0.2% glucose, MOPS minimal medium+0.2% glucose supplemented with 12 amino acids (L-methionine, L-histidine, L-arginine, L-proline, L-threonine, and L-tryptophan, L-serine, L-leucine, L-tyrosine, L-alanine, and L-asparagine at 500 μg/mL each and L-aspartate at 12.5 μg/mL), EZ-RDM (Teknova), and Tryptic Soy Broth (Taheri-Araghi et al., 2015). For the data in Figure 7B,C, we individually inoculated 8 strains into 5 mL test tubes filled with lysogeny broth (8 total cultures). For the data in Figure 7D, we individually inoculated the four strains (MG1655, MG1655 β-P153L, MG1655 Δpgm, and MG1655 Δpgm β-P153L) into one of four media (M9 minimal medium with 0.2% (w/v) glucose, M9 minimal medium with 0.2% (w/v) glucose and supplemented with 12 amino acids (L-methionine, L-histidine, L-arginine, L-proline, L-threonine, and L-tryptophan, L-serine, L-leucine, L-tyrosine, L-alanine, and L-asparagine at 500 μg/mL each and L-aspartate at 12.5 μg/mL), lysogeny broth (LB), and LB with 0.2% (w/v) glucose). For Figures 7A,B, after incubating for ~16 h at 37 °C on a roller drum, each culture was diluted 1:200 into 3 mL of pre-warmed (37 °C) medium of the same type in 5 mL tubes and incubated in a roller drum at 37 °C. All cultures were continuously monitored and repeatedly diluted into pre-warmed media over 6 h to ensure that even the slowest growing cultures had grown in log phase long enough for cell size to stabilize. For Figure 7D, overnight and log-phase cultures were treated similarly except that cultures were agitated on a platform shaker instead of a roller drum. All subsequent steps were equivalent for the four figure panels.
After growing all cultures into log phase, each culture was split into two experiments. In the first, culture densities were normalized to an OD600 of 0.1, then used to inoculate the same media in a 96-well plate at a final volume of 200 μL and an initial inoculum with an OD600 of 0.01. Growth curves were measured as described above, and maximum growth rates were computationally extracted from the growth curves. For the data in Figure 7C and Supplemental Figure 3 F,G, in addition to growing the M+ mutants in LB, we also generated liquid growth curves in LB with 13.5 μg/mL of A22 or mecillinam. Maximum OD600 was computationally extracted from the growth curves in the presence of drug and normalized against maximum OD600 of the same strain in LB without antibiotic.
In the second experiment, cultures were directly spotted onto either a phosphate buffer saline 1% (w/v) agarose pad (Figure 7A,B,C) or a lysogeny broth 1% (w/v) agarose pad (Figure 7D) and phase-contrast images were acquired using a Ti-E microscope (Nikon) with a 100X (NA: 1.4) objective and a Zyla 5.5 sCMOS camera (Andor). Phase-contrast images were segmented and meshed using Morphometrics (Ursell et al., 2017) and shape parameters were computationally extracted from the mesh.
Single-cell quantification of FtsZ concentration
P1vir transduction was used to introduce the ftsZ-msfgfp::kan allele into the MG1655 and MG1655 β-P153L strain backgrounds. To quantify FtsZ-msfGFP, overnight cultures grown in one of five media (M9 minimal medium with 0.2% (w/v) glycerol, M9 minimal medium with 0.2% (w/v) glucose, M9 minimal medium with 0.2% (w/v) glucose and supplemented with 12 amino acids (L-methionine, L-histidine, L-arginine, L-proline, L-threonine, and L-tryptophan, L-serine, L-leucine, L-tyrosine, L-alanine, and L-asparagine at 500 μg/mL each and L-aspartate at 12.5 μg/mL), LB, and LB with 0.2% (w/v) glucose) were inoculated into tubes with fresh media. Strains were kept in log phase via repeated dilution for 3–8 h. After growth in log phase, cells were spotted on phosphate buffer saline agarose pads with 1% (w/v) agarose and images were collected. Phase and fluorescence microscopy images were collected using a Ti-E microscope (Nikon) with a 100X (NA: 1.4) objective and a Zyla 5.5 sCMOS camera (Andor).
Cells were segmented from phase-contrast images using Morphometrics (Ursell et al., 2017). We background-corrected the fluorescence images by subtracting the median value of pixels that were not contained within a cell contour from the entire image and calculated the fluorescence concentration as the sum of background-subtracted fluorescence within each contour divided by the calculated volume for each cell. Cell volume was estimated as series of cylinders with dimensions defined by the pill mesh of Morphometrics. The dataset was filtered to eliminate poorly fit contours.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis to generate S-scores
Data were analyzed using similar algorithms to the gene deletion library (Collins et al., 2006; Nichols et al., 2011; Shiver et al., 2016), with several specialized steps added to or modifying the analysis. Data in the outer rows and columns were immediately discarded. After normalizing average colony size on each plate and the spatial bias in colony size due to the amount of colony transferred to the uneven agar surface during pinning (Collins et al., 2006), we performed an additional normalization to account for effects in the transcription mutants that were due to the genetically linked antibiotic marker alone. For every control strain (antibiotic marker alone), we computed the multiplicative factor required to make its average colony size equal to the average of the entire plate. We then multiplied the control strain and every associated mutant by this factor to normalize for marker-specific effects.
S-scores were computed for every colony position in the library, and S-scores of the same strain were averaged within Array #1 and Array #2 separately. S-scores for each mutant were then averaged between Array #1 and Array #2. Measurements of mutation-condition combinations for which only one of the two plates passed quality control were transformed by a pseudo-averaging mapping (Collins et al., 2006).
Finally, we leveraged the Keio deletion mutants included in our screen to compare our results to a previously published genome-wide deletion dataset (Shiver et al., 2016) on a condition-by-condition basis. There were two goals: first, to decide whether our results sufficiently matched previous efforts in order to warrant inclusion in the final dataset; second, for chemicals that were screened previously at multiple concentrations, to identify the concentration from previous work that best matched results from our current screen. To achieve these goals, we used S-scores for Keio gene deletions that overlapped between our dataset and the previous study to calculate a pairwise correlation between each condition in the current study and those in the previously reported gene deletion dataset. We labeled these cross-dataset comparisons as either matched (same stress condition label, any concentration) or unmatched (all other comparisons). We expected correlations between matched comparisons to be high and correlations between unmatched comparisons to be relatively low. To achieve our first goal, we threw out any condition in our screen where the strength of the correlations for all matched comparisons were less than the 95th percentile of all non-matched correlations. If the correlation of at least one matched condition was in the 95th percentile of non-matched correlations, we achieved our second goal by choosing the most correlated concentration within the matched set.
Because of the early normalizations, S-scores of marker-only alleles of the transcription genes were close to zero (Supplemental Dataset 3). Including the marker-only alleles from analysis would lead to their co-clustering as a distinct group in the hierarchical clustering analysis (Supplemental Figure 4).
Defining and visualizing clusters
We used hierarchical clustering to cluster the dataset, which is standard for chemical-genetic (Nichols et al., 2011) and genetic-interaction (Schuldiner et al., 2005) datasets. To separate the dendrogram that results from hierarchical clustering into discrete clusters, we developed an approach for defining a statistically significant distance cut-off based on randomization of the S-score dataset, as has been done previously to identify statistically significant pairwise correlations in chemical-genomic screens of the Keio deletion library (Nichols et al., 2011; Shiver et al., 2016).
First, we hierarchically clustered a randomized copy of the dataset. We then calculated the smallest cophenetic distance in the randomized dendrogram. After repeating these steps 30,000 times, we used the 5th percentile of the distribution of cophenetic distances as a cutoff to define significant clusters in the original dataset. This cutoff represents the cophenetic distance that is closer than the closest pairwise distance in 95% of randomized matrices. Linkage was calculated using average correlation and distance was calculated using correlation. To speed up iteration over randomized matrices, missing values were imputed with a zero value. The matrix was randomized in one dimension only using shake v. 5.0 from the MATLAB File Exchange (Jos(10584), 2019).
Notably, the cophenetic distance between cluster 14 and 15 was very close to the cutoff used to define statistically significant clusters. Estimating the cutoff using fewer iterations (~1,000) led to run-to-run variation in the determination of whether cluster 14 and cluster 15 were separated; increasing the number of iterations stabilized their separation into two distinct clusters.
The full dendrogram along with statistically significant clusters is shown in Supplemental Figure 1. The undirected graphs in Figure 2 were generated by exporting mutant-mutant correlations that exceeded the cutoff into Cytoscape v. 3.7.2 (Shannon et al., 2003).
This analytical approach was taken because of the difficulty in identifying a priori the optimal number of clusters to use in k-means clustering. We then used the 23 clusters defined with our approach (14 clusters + 9 singletons, k=23) as input to k-means clustering and compared the performance of the two approaches. Overall, the clusters defined by the two methods are similar but not equivalent, with an adjusted mutual information score of 0.70 (Supplemental Figure 5 A,B). Clusters defined by hierarchical clustering are less dispersed, with an average silhouette score 0.046 higher than clusters defined by k-means clustering (Supplemental Figure 5C). The k-means approach also resulted in 4 co-clustering interactions between strains with mutations in different polypeptide chains of the transcription complex (excluding the common co-clustering of mutations in β and β′), as compared to the single intra-chain co-clustering interaction defined by hierarchical clustering (Supplemental Figure 5D).
Interestingly, enrichment analysis between the clusters and conditions revealed a larger difference between the two approaches. Analysis of the hierarchical clustering involved enrichment analysis using the software Gene Set Enrichment Analysis (GSEA) v. 3.0 (Subramanian et al., 2005) to identify enrichment of chemical interactions between clusters and conditions from the screen. Using multiple hypothesis-corrected significance from GSEA as the only criterion for significant enrichments, clusters defined by hierarchical clustering had 70 significant enrichments with stress conditions while clusters defined by k-means clustering had only 41 (Supplemental Figure 5E). Of the significant enrichments, those from hierarchical clustering also had 0.054 higher average normalized enrichment score than those from k-means clustering (Supplemental Figure 5E). Notably, neither the difference in average silhouette score nor the difference in average normalized enrichment score were significant at the p=0.05 level. Therefore, we conclude that hierarchical clustering with a statistically defined cut-off performs as well as, if not better than, k-means clustering in defining groups of similar mutations with similar chemical sensitivities, at least at the level of clustering (k=23) examined in this work.
The undirected graphs in Supplemental Figure 5 were generated with the plot and graph functions of MATLAB. Adjusted mutual information score was calculating using ami v.1.0.0.0 from MATLAB File Exchange (Nguyen, 2021).
Generating enrichment tables
Enrichment tables were created by combining two metrics for the significance of chemical genetic interactions. First, we calculated the significance of individual S-scores based on the distribution of S-scores in each condition (Nichols et al., 2011). Second, we determined the significantly enriched chemical perturbations in groups of genes defined either by clustering or by previous knowledge. We then combined this information.
We calculated the enrichment of S-scores within mutant groups using the software Gene Set Enrichment Analysis (GSEA) v. 3.0 (Subramanian et al., 2005). Ranked lists of S-scores for all transcription mutants were exported for every individual screen condition. Groups were defined from hierarchical clusters and from predefined classes such as ethanol-tolerant mutations or M+ mutations. GSEA was run on every condition individually to identify enrichment of mutant groups in either positive or negative S-scores. Options for GSEA included running the analysis for pre-ranked lists and the flags “-norm meandiv”, “-scoring_scheme weighted”,“-create_svgs false”,“-make_sets true”, “-plot_top_x 20”, “-rnd_seed 081889”,“-set_max 500”,”-set_min 3”,”-zip_report true”, and “-gui false”. The output for each condition was collected into a single table and the combined set of nominal p-values was corrected using the Benjamini-Hochberg FDR correction.
Separately, the significance of a chemical-genetic interaction was calculating using the overall distribution of S-scores in the same condition to calculate a null hypothesis (Nichols et al., 2011). Mutant x condition interactions that were significant above a cut-off (FDR<0.05) were considered individually significant.
If a chemical condition was both significantly enriched within a group (adjusted p<0.05 from GSEA) and had at least one individually significant chemical interaction within the group, it was recorded as a significant enrichment. Supplemental Table 3 contains enrichment tables for hierarchical clusters and previously defined classes.
Quantification of β-ΔSI2 chemical sensitivities
The β-ΔSI2 strain did not have an appreciably different minimum inhibitory concentration, maximum OD600, or maximum growth rate from the parental control in any of the conditions measured. Instead, growth of β-ΔSI2 slowed at an earlier OD600 value at lower concentrations of the compounds. To quantify this effect, we measured area under the curve (AUC). To define a range for the AUC, we used the growth curve of each strain with no drug as a reference point. We extracted maximum growth rate from the curve, computed the time at which the growth rate first dropped below 10% of this value, and added two hours to define the time t2 that determines the upper limit of the area to be measured for every drug concentration for that strain. This calculation sets the upper limit for the area of integration to two hours into the transition phase of the strain when grown without stressors. The lower limit of the area to be integrated was the initial time t1 that measurements started. The area integrated was the blanked OD600 between times t1 and t2. OD600 was not log-transformed for this calculation. We then normalized mean AUC for every combination of strain and drug concentration to the no-drug control. The no-drug control was included in every plate and every measurement was compared to the control on the same plate.
Differential gene expression analysis
Indexed raw sequencing data were demultiplexed according to their R1 barcodes and the degenerate linker sequence was clipped using a custom script. Mapping of individual reads to the E. coli genome (GenBank ID U00096.3) was performed with STAR (Dobin et al., 2013), followed by read counting for individual genome regions according to gene annotations from assembly ASM584v2.
Raw read counts for all samples were used as input for DESeq2 v. 1.22.2 (Love et al., 2014). Information on the strain (parental or mutant), plasmid (pALS13 or pALS14), and induction (yes or no) was used to group samples for statistical analysis.
Estimates for the differential expression of genes in response to induction of RelA* were obtained using the full factorial linear model y ~ strain + plasmid + induction + strain:plasmid + strain:induction + plasmid:induction + strain:plasmid:induction.
To estimate the response of the parental strain with pALS14 to induction of RelA−, reference levels for the three factors were set to (strain: parental), (plasmid: pALS14) and (induction: no). The (induced: yes) versus (induced: no) contrast was used as output.
To estimate the response of the parental strain with pALS13 to induction of RelA*, the same model and reference factors were used as above, but the interaction term of plasmid:induction was used as output.
To estimate the response of the mutant strain with pALS14 to induction of RelA−, reference levels for the three factors were set to (strain: mutant), (plasmid: pALS14) and (induction: no). The (induced: yes) versus (induced: no) contrast was used as output.
To estimate the response of the mutant strain with pALS13 to induction of RelA*, the same model and reference factors were used as above, but the interaction term of plasmid:induction was used as output.
Finally, to estimate the effect of the mutant strain alone, we used the average contrast between mutant and wild-type for strains with pALS14 across induced and uninduced conditions. The linear model used was y ~ strain + plasmid + induction + strain:plasmid + strain:induction. The reference factors were set to (strain: parental), (plasmid: pALS14), and (induction: no). The (strain: mutant) versus (strain: parental) contrast was combined with 50% of the strain:induction interaction term as output.
Before counting the overlap between β-P153L and the relA* condition, we filtered both datasets so that they only included genes that were measured in both experiments. The list of significant genes was combined with a flag indicating the direction of change (activated or repressed) and this modified gene set was used as input for generating the Venn diagram in Figure 5B. Intersections between sets were calculated and used as input to the venn package in MATLAB File Exchange (Darik, 2011).
Quantification of A22 and mecillinam sensitivity
Certain combinations of mutants and growth conditions led to a clear shift in the minimum inhibitory concentration of both A22 and mecillinam. To quantify this shift, maximum OD600 (ODmax) of each culture was computationally extracted from growth curves. To account for plate-to-plate, reader-to-reader, and day-to-day variability in OD600 measurements, as well as idiosyncratic growth curves of the strains and growth conditions, ODmax of every combination of strain and drug concentration was normalized by a no-drug control that was run within the same plate.
Whole genome resequencing of ΔmreB strains
Raw reads from whole genome resequencing were used as input into breseq v.0.33.2 (Deatherage and Barrick, 2014) to elucidate potential mutations.
ADDITIONAL RESOURCES
The entire dataset is available in an interactive, searchable format on the Ontology of Microbial Phenotypes website https://microbialphenotypes.org/wiki/index.php?title=Special:RNAPchemicalgenetics.
Supplementary Material
Supplemental Table 1: Strains used in this study, Related to Experimental Model and Subject Details in STAR Methods.
Supplemental Table 6: Oligos and plasmids used in this study, Related to Method Details in STAR Methods, subsection, Oligonucleotide design.
Data S2: Gene expression changes in β-P153L with and without induction of the stringent response, Related to Figure 5.
Data S3: The S-score values of parental strains, Related to quantification and statistical analysis in STAR Methods.
Acknowledgements
We thank Seth Darst for advice on the design of lineage-specific insertion/deletion mutations, Ann Hochschild for sharing mutants in σ70 that were used in the screen, Wilma Ross and Rick Gourse for sharing the pALS13 and pALS14 plasmids, and Curtis Ross contributions to the data browser. Funding was provided by the Department of Energy Office of Science Graduate Fellowship Program (to A.L.S.), NSF REU grant 1659873 (to K.K.H.), NIH grants R01 GM084448 and P50 AI150476 (to N.J.K.), the Allen Discovery Center at Stanford on Systems Modeling of Infection (to K.C.H), and MIRA grant R35GM118061 (to C.A.G.). K.C.H. is a Chan Zuckerberg Investigator.
Footnotes
Declaration of interests
The authors declare no competing interests.
References
- Alper H, and Stephanopoulos G (2007). Global transcription machinery engineering: a new approach for improving cellular phenotype. Metab Eng 9, 258–267. [DOI] [PubMed] [Google Scholar]
- Artsimovitch I, and Landick R (2000). Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proceedings of the National Academy of Sciences 97, 7090–7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Svetlov V, Murakami KS, and Landick R (2003). Co-overexpression of E. coli RNA polymerase subunits allows isolation and analysis of mutant enzymes lacking lineage-specific sequence insertions. Journal of Biological Chemistry. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, and Mori H (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Molecular systems biology 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett MS, Gaal T, Ross W, and Gourse RL (1998). RNA polymerase mutants that destabilize RNA Polymerase-Promoter complexes alter NTP-sensing by rrn P1 promoters. Journal of Molecular Biology 279, 331–345. [DOI] [PubMed] [Google Scholar]
- Bendezú FO, and de Boer PA (2008). Conditional lethality, division defects, membrane involution, and endocytosis in mre and mrd shape mutants of Escherichia coli. Journal of bacteriology 190, 1792–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borukhov S, Severinov K, Kashlev M, Lebedev A, Bass I, Rowland GC, Lim PP, Glass RE, Nikiforov V, and Goldfarb A (1991). Mapping of trypsin cleavage and antibody-binding sites and delineation of a dispensable domain in the beta subunit of Escherichia coli RNA polymerase. J Biol Chem 266, 23921–23926. [PubMed] [Google Scholar]
- Braberg H, Echeverria I, Bohn S, Cimermancic P, Shiver A, Alexander R, Xu J, Shales M, Dronamraju R, and Jiang S (2020). Genetic interaction mapping informs integrative structure determination of protein complexes. Science 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braberg H, Jin H, Moehle EA, Chan YA, Wang S, Shales M, Benschop JJ, Morris JH, Qiu C, Hu F, et al. (2013). From structure to systems: high-resolution, quantitative genetic analysis of RNA polymerase II. Cell 154, 775–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochado AR, and Typas A (2013). High-throughput approaches to understanding gene function and mapping network architecture in bacteria. Current opinion in microbiology 16, 199–206. [DOI] [PubMed] [Google Scholar]
- Burova E, Hung S, Sagitov V, Stitt B, and Gottesman M (1995). Escherichia coli NusG protein stimulates transcription elongation rates in vivo and in vitro. Journal of bacteriology 177, 1388–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Gopalkrishnan S, Chiu C, Chen AY, Campbell EA, Gourse RL, Ross W, and Darst SA (2019). E. coli TraR allosterically regulates transcription initiation by altering RNA polymerase conformation. Elife 8, e49375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K-K, Lee B-S, Masuda T, Ito T, Ikeda K, Hirayama A, Deng L, Dong J, Shimizu K, and Soga T (2014). Global metabolic network reorganization by adaptive mutations allows fast growth of Escherichia coli on glycerol. Nature communications 5, 3233. [DOI] [PubMed] [Google Scholar]
- Collins SR, Roguev A, and Krogan NJ (2010). Quantitative genetic interaction mapping using the E-MAP approach. In Methods in enzymology (Elsevier; ), pp. 205–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Schuldiner M, Krogan NJ, and Weissman JS (2006). A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome biology 7, R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darik (2011). venn (https://www.mathworks.com/matlabcentral/fileexchange/22282-venn), MATLAB Central File Exchange. Retrieved February 14th 2011.
- Das A, Merril C, and Adhya S (1978). Interaction of RNA polymerase and rho in transcription termination: coupled ATPase. Proceedings of the National Academy of Sciences of the United States of America 75, 4828–4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, and Wanner BL (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Costantino N, and Court DL (2006). A set of recombineering plasmids for gram-negative bacteria. Gene 379, 109–115. [DOI] [PubMed] [Google Scholar]
- Deatherage DE, and Barrick JE (2014). Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. In Engineering and analyzing multicellular systems (Springer; ), pp. 165–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ederth J, Mooney RA, Isaksson LA, and Landick R (2006). Functional interplay between the jaw domain of bacterial RNA polymerase and allele-specific residues in the product RNA-binding pocket. J Mol Biol 356, 1163–1179. [DOI] [PubMed] [Google Scholar]
- Ellis HM, Yu D, DiTizio T, and Court DL (2001). High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc Natl Acad Sci U S A 98, 6742–6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French S, Mangat C, Bharat A, Côté J-P, Mori H, and Brown ED (2016). A robust platform for chemical genomics in bacterial systems. Molecular biology of the cell 27, 1015–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman DI (1971). A bacterial mutant affecting lambda development. In The Bacteriophage Lambda, Hershey AD, ed. (Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory; ), pp. 733–738. [Google Scholar]
- Garibyan L, Huang T, Kim M, Wolff E, Nguyen A, Nguyen T, Diep A, Hu K, Iverson A, Hanjing Y, et al. (2003). Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair 2, 593–608. [DOI] [PubMed] [Google Scholar]
- Gitai Z, Dye NA, Reisenauer A, Wachi M, and Shapiro L (2005). MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120, 329–341. [DOI] [PubMed] [Google Scholar]
- González-González A, Hug SM, Rodríguez-Verdugo A, Patel JS, and Gaut BS (2017). Adaptive mutations in RNA polymerase and the transcriptional terminator Rho have similar effects on Escherichia coli gene expression. Molecular biology and evolution 34, 2839–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber TM, and Gross CA (2003). Multiple sigma subunits and the partitioning of bacterial transcription space. Annual Reviews in Microbiology 57, 441–466. [DOI] [PubMed] [Google Scholar]
- Haft RJ, Keating DH, Schwaegler T, Schwalbach MS, Vinokur J, Tremaine M, Peters JM, Kotlajich MV, Pohlmann EL, Ong IM, et al. (2014). Correcting direct effects of ethanol on translation and transcription machinery confers ethanol tolerance in bacteria. Proc Natl Acad Sci U S A 111, E2576–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer K, Jensen KF, Poulsen P, Oppenheim AB, and Gottesman M (1987). Isolation of Escherichia coli rpoB mutants resistant to killing by λ cII protein and altered in pyrE gene attenuation. Journal of Bacteriology 169, 5289–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler L, Suzuki H, Landick R, and Gross C (1993). Four contiguous amino acids define the target for streptolydigin resistance in the beta subunit of Escherichia coli RNA polymerase. Journal of Biological Chemistry 268, 25369–25375. [PubMed] [Google Scholar]
- Hill NS, Buske PJ, Shi Y, and Levin PA (2013). A moonlighting enzyme links Escherichia coli cell size with central metabolism. PLoS genetics 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin DJ, and Gross CA (1988). Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. Journal of Molecular Biology 202, 45–58. [DOI] [PubMed] [Google Scholar]
- Jokerst R, Weeks J, Zehring W, and Greenleaf A (1989). Analysis of the gene encoding the largest subunit of RNA polymerase II in Drosophila. Molecular and General Genetics MGG 215, 266–275. [DOI] [PubMed] [Google Scholar]
- Jos (10584) (2019). shake (https://www.mathworks.com/matlabcentral/fileexchange/10067-shake), MATLAB Central File Exchange. Retrieved December 6th, 2019.
- Kirschbaum JB, and Konrad EB (1973). Isolation of a Specialized Lambda Transducing Bacteriophage Carrying the Beta Subunit Gene for Escherichia coli Ribonucleic Acid Polymerase. Journal of Bacteriology 116, 517–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko DC, Marr MT, Guo J, and Roberts JW (1998). A surface of Escherichia coli σ70 required for promoter function and antitermination by phage λ Q protein. Genes & Development 12, 3276–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kritikos G, Banzhaf M, Herrera-Dominguez L, Koumoutsi A, Wartel M, Zietek M, and Typas A (2017). A tool named Iris for versatile high-throughput phenotyping in microorganisms. Nature microbiology 2, 17014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S, Cava F, de Pedro MA, Brun YV, and VanNieuwenhze MS (2012). In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angewandte Chemie 124, 12687–12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuru E, Tekkam S, Hall E, Brun YV, and Van Nieuwenhze MS (2015). Synthesis of fluorescent D-amino acids and their use for probing peptidoglycan synthesis and bacterial growth in situ. Nature protocols 10, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane WJ, and Darst SA (2010). Molecular evolution of multisubunit RNA polymerases: sequence analysis. Journal of molecular biology 395, 671–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibman M, and Hochschild A (2007). A σ-core interaction of the RNA polymerase holoenzyme that enhances promoter escape. The EMBO journal 26, 1579–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Mason S, and Greenblatt J (1993). Elongation factor NusG interacts with termination factor rho to regulate termination and antitermination of transcription. Genes & development 7, 161–172. [DOI] [PubMed] [Google Scholar]
- Lisitsyn NA, Sverdlov ED, Moiseyeva EP, Danilevskaya ON, and Nikiforov VG (1984). Mutation to rifampicin resistance at the beginning of the RNA polymerase β subunit gene in Escherichia coli. Molecular & General Genetics 196, 173–174. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molodtsov V, Sineva E, Zhang L, Huang X, Cashel M, Ades SE, and Murakami KS (2018). Allosteric effector ppGpp potentiates the inhibition of transcript initiation by DksA. Molecular cell 69, 828–839. e825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney RA, Schweimer K, Rosch P, Gottesman M, and Landick R (2009). Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J Mol Biol 391, 341–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Imai M, and Shigesada K (1989). Mutant rho factors with increased transcription termination activities. II: Identification and functional dissection of amino acid changes. Journal of Molecular Biology 210, 39–49. [DOI] [PubMed] [Google Scholar]
- Murphy H, and Cashel M (2003). Isolation of RNA polymerase suppressors of a (p) ppGpp deficiency. In Methods in enzymology (Elsevier; ), pp. 596–601. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Mizusawa S, Court DL, and Tsugawa A (1986). Regulatory defects of a conditionally lethal nusAts mutant of Escherichia coli: Positive and negative modulator roles of NusA protein in vivo. Journal of Molecular Biology 189, 103–111. [DOI] [PubMed] [Google Scholar]
- Nene V, and Glass RE (1984). Genetic studies on the beta subunit of Escherichia coli RNA polymerase. VI. A redundant region in the beta polypeptide. Mol Gen Genet 196, 64–67. [DOI] [PubMed] [Google Scholar]
- Nguyen XV (2021). The Adjusted Mutual Information (https://www.mathworks.com/matlabcentral/fileexchange/33144-the-adjusted-mutual-information), MATLAB Central File Exchange. Retrieved Jan 1st 2021.
- Nichols RJ, Sen S, Choo YJ, Beltrao P, Zietek M, Chaba R, Lee S, Kazmierczak KM, Lee KJ, and Wong A (2011). Phenotypic landscape of a bacterial cell. Cell 144, 143–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov YA, Monastyrskaya GS, Gubanov VV, Lipkin VM, Sverdlov ED, Kiver IF, Bass IA, Mindlin SZ, Danilevskaya ON, and Khesin RB (1981). Primary structure of Escherichia coli RNA polymerase nucleotide substitution in the β subunit gene of the rifampicin resistant rpoB255 mutant. Molecular & General Genetics 184, 536–538. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov YA, Monastyrskaya GS, Guriev SO, Kalinina NF, Sverdlov ED, Gragerov AI, Bass IA, Kiver IF, Moiseyeva EP, Igumnov VN, et al. (1983). RNA polymerase rifampicin resistance mutations in Escherichia coli: Sequence changes and dominance. Molecular & General Genetics 190, 344–348. [DOI] [PubMed] [Google Scholar]
- Parshin A, Shiver AL, Lee J, Ozerova M, Schneidman-Duhovny D, Gross CA, and Borukhov S (2015). DksA regulates RNA polymerase in Escherichia coli through a network of interactions in the secondary channel that includes Sequence Insertion 1. Proceedings of the National Academy of Sciences 112, E6862–E6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira S, and Platt T (1995). Analysis of E. coli Rho Factor: Mutations Affecting Secondary-site Interactions. Journal of Molecular Biology 251, 30–40. [DOI] [PubMed] [Google Scholar]
- Ratner D (1976). Evidence that mutations in the suA polarity suppressing gene directly affect termination factor rho. Nature 259, 151–153. [DOI] [PubMed] [Google Scholar]
- Ross W, Sanchez-Vazquez P, Chen AY, Lee J-H, Burgos HL, and Gourse RL (2016). ppGpp binding to a site at the RNAP-DksA interface accounts for its dramatic effects on transcription initiation during the stringent response. Molecular cell 62, 811–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford ST, Villers CL, Lee JH, Ross W, and Gourse RL (2009). Allosteric control of Escherichia coli rRNA promoter complexes by DksA. Genes Dev 23, 236–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagitov V, Nikiforov V, and Goldfarb A (1993). Dominant lethal mutations near the 5’ substrate binding site affect RNA polymerase propagation. Journal of Biological Chemistry 268, 2195–2202. [PubMed] [Google Scholar]
- Saito M, Tsugawa A, Egawa K, and Nakamura Y (1986). Revised sequence of the nusA gene of Escherichia coli and identification of nusA11 (ts) and nusA1 mutations which cause changes in a hydrophobic amino acid cluster. Molecular & General Genetics 205, 380–382. [DOI] [PubMed] [Google Scholar]
- Sanchez-Vazquez P, Dewey CN, Kitten N, Ross W, and Gourse RL (2019). Genome-wide effects on Escherichia coli transcription from ppGpp binding to its two sites on RNA polymerase. Proceedings of the National Academy of Sciences 116, 8310–8319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos DA, Shi L, Tu BP, and Weissman JS (2019). Cycloheximide can distort measurements of mRNA levels and translation efficiency. Nucleic acids research 47, 4974–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaechter M, Maaløe O, and Kjeldgaard NO (1958). Dependency on medium and temperature of cell size and chemical composition during balanced growth of Salmonella typhimurium. Microbiology 19, 592–606. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, and Greenblatt JF (2005). Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 123, 507–519. [DOI] [PubMed] [Google Scholar]
- Severinov K, Mustaev A, Kashlev M, Borukhov S, Nikiforov V, and Goldfarb A (1992). Dissection of the beta subunit in the Escherichia coli RNA polymerase into domains by proteolytic cleavage. Journal of Biological Chemistry 267, 12813–12819. [PubMed] [Google Scholar]
- Severinov K, Soushko M, Goldfarb A, and Nikiforov V (1993). Rifampicin region revisited. New rifampicin-resistant and streptolydigin-resistant mutants in the β subunit of Escherichia coli RNA polymerase. Journal of Biological Chemistry 268, 14820–14825. [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, and Ideker T (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp MM, Chan CL, Lu CZ, Marr MT, Nechaev S, Merritt EW, Severinov K, Roberts JW, and Gross CA (1999). The interface of σ with core RNA polymerase is extensive, conserved, and functionally specialized. Genes Dev 13, 3015–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Colavin A, Bigos M, Tropini C, Monds RD, and Huang KC (2017). Deep phenotypic mapping of bacterial cytoskeletal mutants reveals physiological robustness to cell size. Current Biology 27, 3419–3429. e3414. [DOI] [PubMed] [Google Scholar]
- Shiver AL, Osadnik H, Kritikos G, Li B, Krogan N, Typas A, and Gross CA (2016). A chemical-genomic screen of neglected antibiotics reveals illicit transport of kasugamycin and blasticidin S. PLoS genetics 12, e1006124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegele DA, Hu JC, and Gross CA (1988). Mutations in rpoD, the gene encoding the σ70 subunit of Escherichia coli RNA polymerase, that increase expression of the lac operon in the absence of CAP-cAMP. Journal of Molecular Biology 203, 29–37. [DOI] [PubMed] [Google Scholar]
- Spratt BG (1977). The mechanism of action of mecillinam. Journal of antimicrobial chemotherapy 3, 13–19. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, and Lander ES (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitil A, Cashel M, and Zyskind J (1993). Guanosine tetraphosphate inhibits protein synthesis in vivo. A possible protective mechanism for starvation stress in Escherichia coli. Journal of Biological Chemistry 268, 2307–2311. [PubMed] [Google Scholar]
- Sweetser D, Nonet M, and Young RA (1987). Prokaryotic and eukaryotic RNA polymerases have homologous core subunits. Proceedings of the National Academy of Sciences 84, 1192–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalewska-Palasz A, Johansson LU, Bernardo LM, Skarfstad E, Stec E, Brannstrom K, and Shingler V (2007). Properties of RNA polymerase bypass mutants: implications for the role of ppGpp and its co-factor DksA in controlling transcription dependent on sigma54. J Biol Chem 282, 18046–18056. [DOI] [PubMed] [Google Scholar]
- Taheri-Araghi S, Bradde S, Sauls JT, Hill NS, Levin PA, Paulsson J, Vergassola M, and Jun S (2015). Cell-size control and homeostasis in bacteria. Current biology 25, 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O, Rodriguez-Verdugo A, Gaut RL, McDonald P, Bennett AF, Long AD, and Gaut BS (2012). The Molecular Diversity of Adaptive Convergence. Science 335, 457–461. [DOI] [PubMed] [Google Scholar]
- Thomason LC, Sawitzke JA, Li X, Constantino N, and Court DL (2014). Recombineering: Genetic Engineering in Bacteria Using Homologous Recombination. Current Protocols in Molecular Biology, 1.16.11–11.16.39. [DOI] [PubMed] [Google Scholar]
- Trautinger BW, and Lloyd RG (2002). Modulation of DNA repair by mutations flanking the DNA channel through RNA polymerase. The EMBO Journal 21, 6944–6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurushita N, and Shigesada K (1989). Mutant rho factors with increased transcription termination activities. I. Functional correlations of the primary and secondary polynucleotide binding sites with the efficiency and site-selectivity of rho-dependent termination. Journal of Molecular Biology 210, 23–37. [DOI] [PubMed] [Google Scholar]
- Ursell T, Lee TK, Shiomi D, Shi H, Tropini C, Monds RD, Colavin A, Billings G, Bhaya-Grossman I, and Broxton M (2017). Rapid, precise quantification of bacterial cellular dimensions across a genomic-scale knockout library. BMC biology 15, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinella D, D’Ari R, Jaffe A, and Bouloc P (1992). Penicillin binding protein 2 is dispensable in Escherichia coli when ppGpp synthesis is induced. The EMBO journal 11, 1493–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Bushnell DA, Westover KD, Kaplan CD, and Kornberg RD (2006). Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell 127, 941–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, and Church GM (2009). Programming cells by multiplex genome engineering and accelerated evolution. Nature 460, 894–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weilbaecher R, Hebron C, Feng G, and Landick R (1994). Termination-altering amino acid substitutions in the β’ subunit of Escherichia coli RNA polymerase identify regions involved in RNA chain elongation. Genes & Development 8, 2913–2927. [DOI] [PubMed] [Google Scholar]
- Yanofsky C, and Horn V (1981). Rifampin resistance mutations that alter the efficiency of transcription termination at the tryptophan operon attenuator. Journal Of Bacteriology 145, 1334–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan AH, Gregory BD, Sharp JS, McCleary KD, Dove SL, and Hochschild A (2008). Rsd family proteins make simultaneous interactions with regions 2 and 4 of the primary sigma factor. Mol Microbiol 70, 1136–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YN, and Jin DJ (1998). The rpoB mutants destabilizing initiation complexes at stringently controlled promoters behave like “stringent” RNA polymerases in Escherichia coli. Proceedings of the National Academy of Sciences 95, 2908–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1: Strains used in this study, Related to Experimental Model and Subject Details in STAR Methods.
Supplemental Table 6: Oligos and plasmids used in this study, Related to Method Details in STAR Methods, subsection, Oligonucleotide design.
Data S2: Gene expression changes in β-P153L with and without induction of the stringent response, Related to Figure 5.
Data S3: The S-score values of parental strains, Related to quantification and statistical analysis in STAR Methods.
Data Availability Statement
Raw images and Iris data files for the chemical-genetic screen along with two datasets generated in this work have been published on Dryad. Raw sequencing reads for whole genome resequencing and RNA sequencing have been deposited at NCBI. Reproducible compute capsules have been published on Code Ocean for the major findings and results of this study. Accession codes for deposited data are listed in the Key Resources Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| For the list of bacterial strains generated in this work, see Supplemental Table 1 | this study | Supplemental Table 1 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| For the list of chemicals used in the chemical-genetic screen, see Supplemental Table 2 | this study | Supplemental Table 2 |
| Trizol LS | Invitrogen | Catalog No. 10296028 |
| T4 RNA ligase buffer | NEB | Catalog No. B0216L |
| T4 RNA ligase 2 truncated KQ | NEB | Catalog No. M0373L |
| T4 Polynucleotide Kinase (PNK) | NEB | Catalog No. M0201L |
| T4 RNA Ligase 1 | NEB | Catalog No. M0204L |
| SuperScript IV | Thermo Fisher Scientific | Catalog No. 18090010 |
| HCC-amino-D-alanine (HADA) | Michael vanNieuwenhze lab | N/A |
| Teknova EZ rich defined media kit (EZ-RDM) | Teknova | Catalog No. M2105 |
| Critical Commercial Assays | ||
| Ribo-Zero rRNA removal kit (Gram-negative bacteria) | Illumina | MRZGN126 |
| Deposited Data | ||
| Images and data from the paper | this study | https://doi.org/10.5061/dryad.z612jm68q |
| Raw sequences and processed data for gene expression analysis of β-P153L. | this study | GSE151022 (NCBI GEO) |
| Raw sequences for whole genome resequencing of ΔmreB strains. | this study | PRJNA632897 (NCBI SRA) |
| Code Ocean compute capsule: Generating the chemical genetic interaction dataset (Supplemental Dataset 1) | this study | https://doi.org/10.24433/CO.9507705.v1 |
| Code Ocean compute capsule: Running DESeq2 on RNA-seq counts. | this study | https://doi.org/10.24433/CO.2313056.v1 |
| Code Ocean compute capsule: Generating Supplemental Dataset 2, figures, and statistics cited in the main text of the paper. | this study | https://doi.org/10.24433/CO.8886448.v3 |
| Code Ocean compute capsule: Generating the list of conditions and concentrations used in the screen. (Supplemental Table 2) | this study | https://doi.org/10.24433/CO.2239948.v1 |
| Code Ocean compute capsule: Generating the enrichment tables. (Supplemental Table 3) | this study | https://doi.org/10.24433/CO.1521219.v3 |
| Code Ocean compute capsule: Generating mutation reports using breseq. (Supplemental Table 4) | this study | https://doi.org/10.24433/CO.6559033.v1 |
| Code Ocean compute capsule: Collating the table of gene expression changes in genes of interest. (Supplemental Table 5) | this study | https://doi.org/10.24433/CO.3392553.v1 |
| Code Ocean compute capsule: Generating chemical genetic interaction datasets for the marker-only alleles. (Supplemental Dataset 3 and Supplemental Figure 4) | this study | https://doi.org/10.24433/CO.5034235.v1 |
| Code Ocean compute capsule: Comparing the results of hierarchical clustering with a cut-off to k-means clustering with k=23. (Supplemental Figure 5) | this study | https://doi.org/10.24433/CO.3570885.v2 |
| Oligonucleotides | ||
| For the list of oligonucleotides and plasmids used in this study, see Supplemental Table 6 | this study | Supplemental Table 6 |
| Software and Algorithms | ||
| MATLAB R2020b v. 9.9.0.1495850 | Mathworks | https://www.mathworks.com |
| Python v. 3.7.3 | Python Software Foundation | https://www.python.org/ |
| R v. 3.5.1 | R Foundation for Statistical Computing | https://www.r-project.org/ |
| Gene Set Enrichment Analysis v. 3.0 | (Subramanian et al., 2005) | https://www.gsea-msigdb.org/gsea/index.jsp |
| Cytoscape v. 3.7.2 | (Shannon et al., 2003) | https://cytoscape.org/ |
| Gen5 ™ v. 3.04 | BioTek ® | https://www.biotek.com/products/software-robotics-software/gen5-software-features-for-imaging-microscopy/ |
| Iris v. 0.9.4 | (Kritikos et al., 2017) | http://critichu.github.io/Iris |
| breseq v. 0.333.2 | (Deatherage and Barrick, 2014) | https://github.com/barricklab/breseq |
| DESeq2 v. 1.22.2 | (Love et al., 2014) | http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html |
| shake v. 5.0 | (Jos(10584), 2019) | https://www.mathworks.com/matlabcentral/fileexchange/10067-shake |
| ami v. 1.0.0.0 | (Nguyen, 2021) | https://www.mathworks.com/matlabcentral/fileexchange/33144-the-adjusted-mutual-information |
| venn v. 1.7.0.0 | (Darik, 2011) | https://www.mathworks.com/matlabcentral/fileexchange/22282-venn |
| Other | ||
| Data browser for chemical-genetic interaction dataset | this study | https://microbialphenotypes.org/wiki/index.php?title=Special:RNAPchemicalgenetics |