Abstract
This bioinformatics study aimed to characterize and certify crucial anti-cancer targets, functional processes, and molecular mechanisms of Pachyman in treating hepatocellular carcinoma (HCC) by using pharmacology network and molecular docking analyses, by experimental validation. The crucial anti-HCC targets of Pachyman, including ALB, VEGFA, TNF, CASP3, SRC, EGF, CXCR4, STAT3, HRAS, HSP90AA1, MMP9, BCL2L1, FGF2, and PTPRC, were identified. In addition, the correlative networks of all crucial biotargets of Pachyman in treating HCC were created accordingly. Functionally, these crucial genes were correlated using angiogenesis and neoplastic metastasis of HCC. Interestingly, the molecular docking findings indicated that ALB and VEGFA in HCC might be potent pharmacological targets of Pachyman. In experimental validation, the clinical samples of HCC showed reduced ALB protein expression and increased VEGFA protein level. Following Pachyman treatments in vitro, the intracellular level of ALB protein was elevated, whereas the cellular content of VEGFA protein was downregulated. Taken together, current bioinformatics findings based on pharmacology network and molecular docking analyses elucidate the detailed molecular targets and signaling mechanisms of Pachyman in treating HCC. Interestingly, validated biotargets of ALB and VEGFA may be main potential biomarkers for detecting HCC medically.
Keywords: liver cancer, Pachyman, network pharmacology, molecular docking, biomarkers 3
Introduction
Hepatocellular carcinoma (HCC) refers to a malignant tumor, occurring in liver tissue in situ. Statistically, HCC cases account for a majority of cancer-causing deaths worldwide (McGlynn et al., 2021). As revealed in epidemiological assay, HCC is one of the deadly tumors in China, featured with elevated morbidity and mortality yearly (Fu and Wang, 2018). Notably, early symptoms of HCC may be inconspicuous during clinical characterization before the tumor develops as malignant and invasive. Accordingly, most of the HCC patients are diagnosed with advanced stages during initial examination, eventually leading to high mortality and undesired metastasis (Llovet et al., 2018; Llovet et al., 2021). For precision medicine, it is particularly necessary to identify more candidate anti-HCC targets for effective diagnosis, prognosis, and treatment (Anwanwan et al., 2020). In addition, some naturally occurring ingredients may be used for HCC treatment. Pachyman, Poria cocos polysaccharides, is a biologically active component that is isolated from the Poria cocos plant (Sun, 2014). Beneficially, Pachyman has been proven to exert potent pharmacological properties, such as antioxidation, anti-tumor action, hepatoprotection, enhancing immunity, and eliminating free radicals (Wu et al., 2018; Li et al., 2019; Liu et al., 2020). Although well-evidenced anti-cancer benefits have been found, there are still only few studies regarding the pharmacological effects of Pachyman in treating HCC. Intriguingly, an attractive strategy using network pharmacology and molecular docking analyses can function as an emerging and promising tool for revealing detailed biotargets and molecular mechanisms of candidate natural agents for treating medical diseases (Li et al., 2020a; Li et al., 2021a), including liver disease (Su et al., 2019), coronavirus disease 2019 (COVID-19) (Qin et al., 2021), and meningitis (Li et al., 2021b). Therefore, the present study was designed to optimize network pharmacology and molecular docking methods in identifying the bioinformatics findings of Pachyman in treating HCC, including detailed biological targets, functional processes, and signaling pathways. Experimentally, some bioinformatics data were validated for potential clinical use of Pachyman in treating HCC in future.
Materials and Methods
Screening of Anti-HCC Candidate Targets of Pachyman
All pharmacological targets of Pachyman were collected from databases of PharmMapper and Swiss Target Prediction databases. In addition, HCC-related targets were obtained from databases of OncoDB.HCC and Liverome. Furthermore, these identifiable targets of Pachyman and HCC were determined by using the FunRich software to construct an intersection map of Pachyman in treating HCC, as described by Li et al. (2021c).
Construction of an Integrated Network in Anti-HCC Targets of Pachyman
The available anti-HCC targets of Pachyman were imported to the STRING database for presenting protein–protein interaction (PPI) data with the confidence score greater than 0.9, and then introduced to the Cytoscape software for constructing a PPI map of Pachyman against HCC targets. The PPI network was modeled as a visualized graph, in which the nodes represented genes and the edges indicated the interaction proteins encoded by the related genes (Li et al., 2021d).
Identification and Construction Hub Network of Crucial Targets
Additionally, Network Analyzer in Cytoscape tool was used to determine topological parameters, such as average degree and maximum degree of PPI network of Pachyman in treating HCC, and then the crucial targets were screened and identified according to the contrivable degree value of the targets. Moreover, these crucial targets with interaction network were featured and visualized by using a software platform of Cytoscape for integrating complex networks, as described by Li et al. (2021e).
Enrichment and Function Analyses of Crucial Targets
Subsequently, Gene Ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment assays were generated by using the Metascape online platform. Additionally, the biological functions and pathway enrichment findings of all crucial targets were featured and visualized by using the OmicShare cloud platform before generating advanced bubble diagrams according to their p-values. Furthermore, the crucial target-signaling pathway network of Pachyman in treating HCC was constructed by using the Cytoscape software (Li et al., 2020b).
Molecular Docking Analysis
The molecular structure of the Pachyman compound was obtained from the PubChem database. The crucial target/protein structure in HCC was obtained from the Protein Data Bank database. Magnetometric minimum force field was obtained by using the three-dimensional structure of ChemBio3D Draw module in the ChemBioOffice software (version 2010). The PDBQT structure file necessary for virtual screening was created through the Raccoon software, and the docking active center was defined through the grid box function setting in the software. Data reliability was determined according to the size of the root-mean-square deviation of the docked and the original ligand molecules. Generally, it was designed in a way that the root-mean-square deviation that is ≤4 was the threshold for conforming the docked ligand to match the related original ligand (Nong et al., 2020).
Clinical Study
In a human study, 10 patients with HCC were detected, and clinical samples were collected for experimentative verification. All patients were medically diagnosed for HCC through clinical images and histological staining examination. HCC and HCC-free samples were harvested through surgical operation, followed by immunostaining and analyzing. These human protocols were approved by the Liuzhou Worker’s Hospital ethics committee, and correlative experiments were conducted based on the principles of the Declaration of Helsinki (Li et al., 2020c; Li et al., 2021f).
Cell Culture Study
A human liver cancer cell line of HepG2 was treated with Pachyman (Shanghai Yuanye Biotechnology Co., Ltd. China) at different doses of 0, 25, and 50 μM for 48 h. The cell proliferation was determined by using the commercially available reagents of Cell Counting Kit 8. In addition, the protein expressions of ALB and VEGFA were measured by using enzyme-linked immunosorbent assay (ELISA) kits. More experimental procedures have been described in previous reports (Wu et al., 2017; Wu et al., 2019).
Statistical Analyses
The statistical results were indicated as mean ± standard deviations (SD). Statistical assay was performed by using the Statistical Product and Service Solutions 19.0 software (IBM Corporation, Chicago, IL, United States). Two different comparison groups were determined through one-way analysis of variance, followed by Tukey's post hoc test. The statistical significance was set as p < 0.05.
Results
Potential Targets and Establishment of the PPI Network
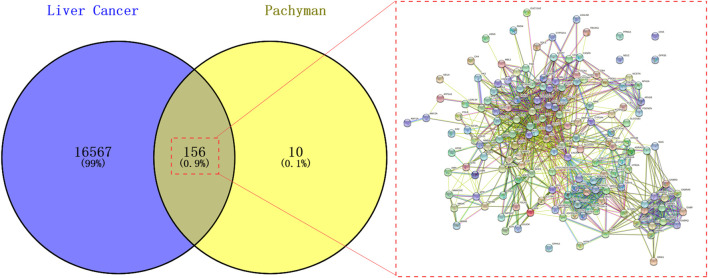
As shown in the Venn diagram (Figure 1), the data reported a total of 16,763 HCC-associated genes and 166 Pachyman-associated genes. As a result, 156 intersection genes of Pachyman and HCC were obtained when all duplicate genes were excluded. Visibly, the PPI network of the intersection genes showed 57 nodes and 139 edges (Figure 1).
FIGURE 1.
Candidate targets of a PPI network of Pachyman in treating HCC for identifying pharmacological and pathological biotargets.
Identification of Crucial Targets in Anti-HCC Targets of Pachyman
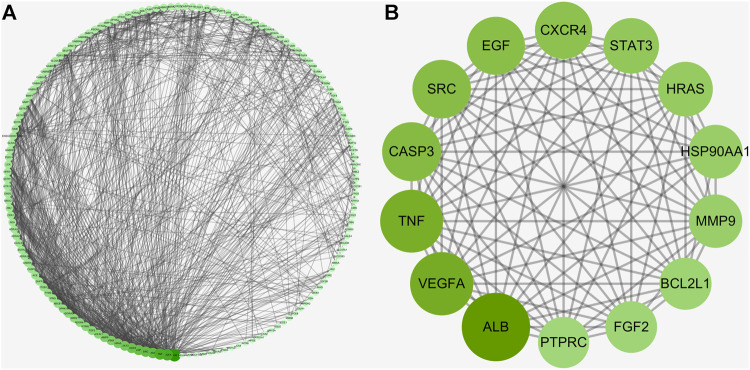
The topological data indicated a close relevance among the intersection genes, characterized in the PPI network map (Figure 2A). As revealed in Figure 2B, the top 14 crucial targets of ALB, VEGFA, TNF, CASP3, SRC, EGF, CXCR4, STAT3, HRAS, HSP90AA1, MMP9, BCL2L1, FGF2, and PTPRC were identified accordingly.
FIGURE 2.
Hub targets of a PPI network (A) for uncovering all crucial targets of Pachyman in treating HCC (B).
GO and KEGG Enrichment Findings of Crucial Targets
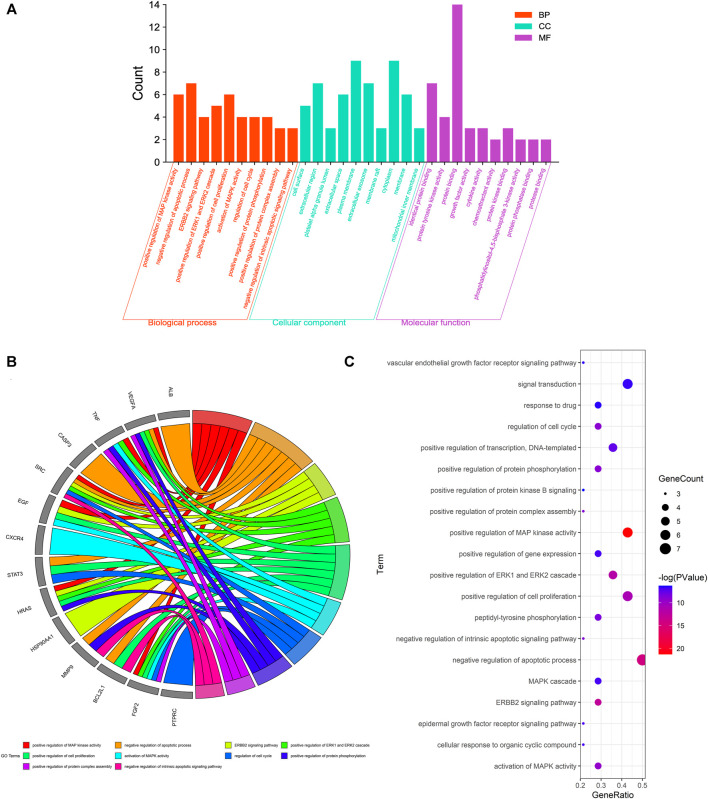
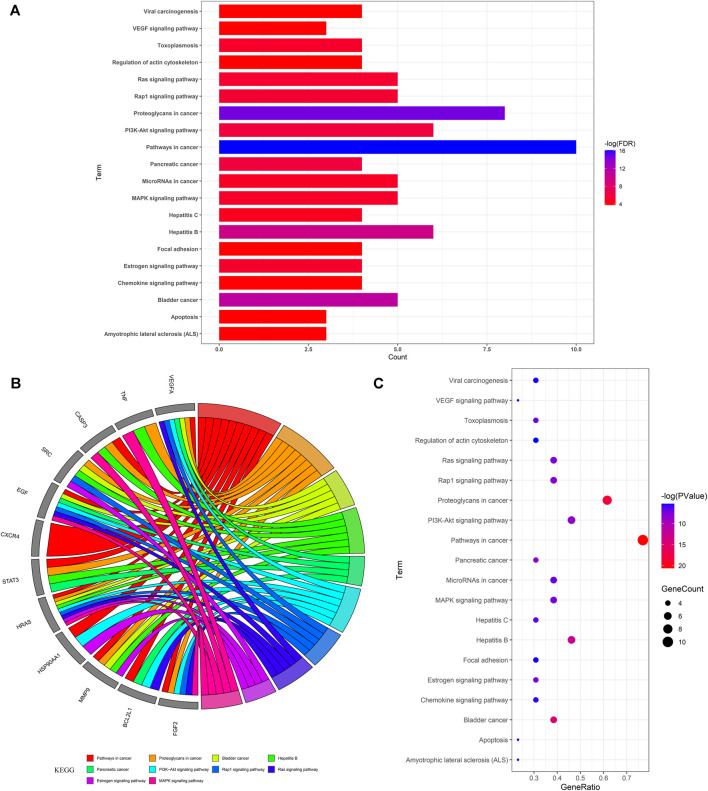
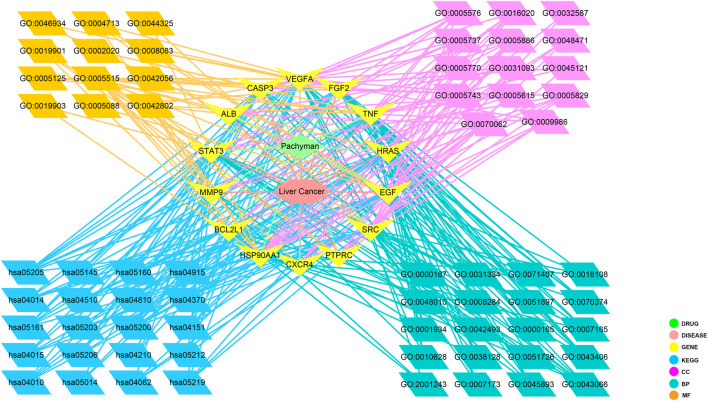
The biological functional processes and molecular pathways of the crucial targets in Pachyman for treating HCC were enriched and characterized respectively. The top 20 biological processes of Pachyman in treating HCC are illustrated in Figure 3A–C. The anti-cancer effects of Pachyman in treating HCC were mainly related to the regulation of the following: positive regulation of MAP kinase activity, negative regulation of apoptotic process, ERBB2 signaling pathway, positive regulation of extracellular signal-regulated kinase (ERK)—ERK1 and ERK2—cascades, positive regulation of cell proliferation, activation of mitogen-activated protein kinase (MAPK) activity, regulation of the cell cycle, positive regulation of protein phosphorylation, positive regulation of the protein complex assembly, negative regulation of the intrinsic apoptotic signaling pathway, peptidyl-tyrosine phosphorylation, positive regulation of DNA-templated transcription, regulation of the epidermal growth factor receptor signaling pathway, regulation of cellular response to organic cyclic compound, MAPK cascade, positive regulation of gene expression, signal transduction, vascular endothelial growth factor receptor signaling pathway, response to drug, and positive regulation of protein kinase B signaling. As is revealed in pharmacological mechanisms, the KEGG pathways of Pachyman in treating HCC included the following: pathways in cancer, proteoglycans in cancer, bladder cancer, hepatitis B, pancreatic cancer, PI3K-Akt signaling pathway, Rap1 signaling pathway, Ras signaling pathway, estrogen signaling pathway, MAPK signaling pathway, toxoplasmosis, microRNAs in cancer, hepatitis C, amyotrophic lateral sclerosis (ALS), chemokine signaling pathway, viral carcinogenesis, focal adhesion, VEGF signaling pathway, apoptosis, and regulation of actin cytoskeleton (Figures 4A–C). Collectively, the integrated network diagram with detailed Pachyman–anti-HCC findings can be visualized in Figure 5.
FIGURE 3.
GO-based enrichment analysis for disclosing the top 20 biological processes of Pachyman in treating HCC, as shown in bar chart (A), circle diagram (B), bubble graph (C).
FIGURE 4.
KEGG-based enrichment analysis for revealing the top 20 molecular pathways of Pachyman in treating HCC as shown in bar chart (A), circle diagram (B), bubble graph (C).
FIGURE 5.
An integrative map of disease and ingredient-target-signaling pathway network of Pachyman in treating HCC.
Molecular Docking Findings
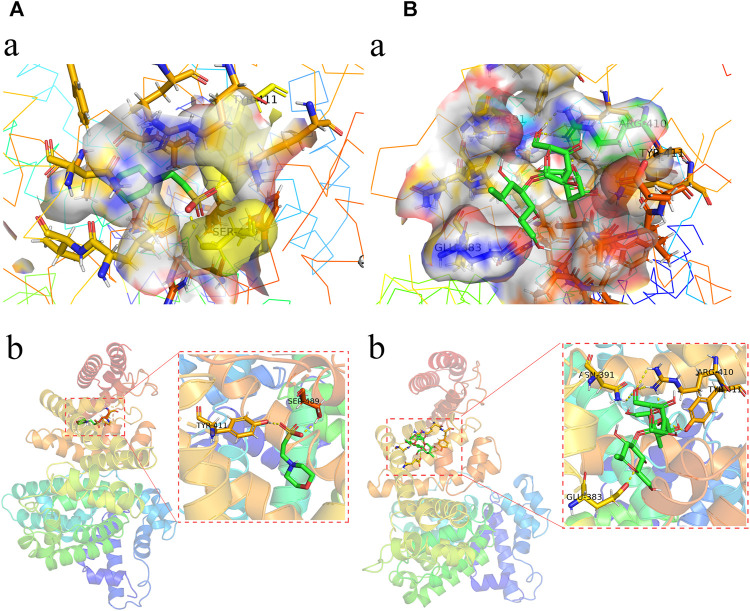
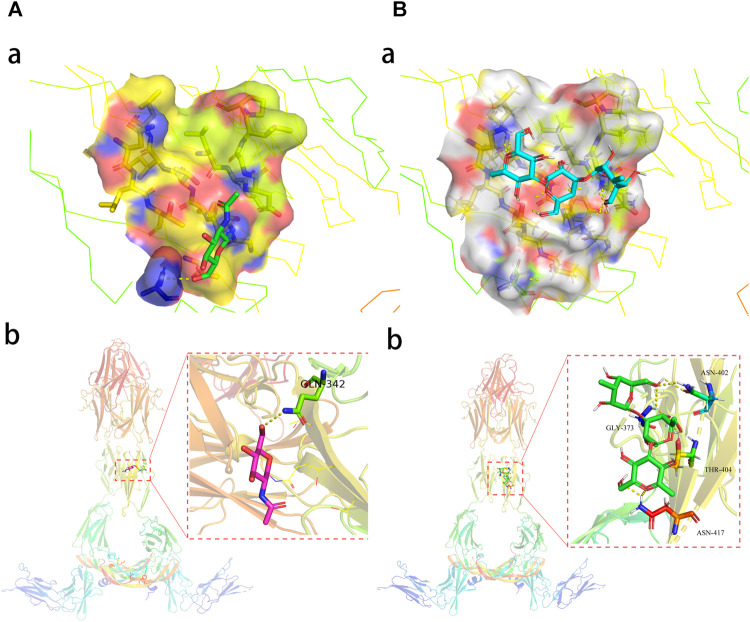
As shown in the ALB protein (Figure 6), the root-mean-square deviation of the original ligand is 2.713 Å. The hydrogen bonding of the pro-ligand MES to 6QIO protein acted on the amino acid residues TYR-411 (1.6Å) and SER-489 (2.0 Å). Pachyman formed bonds with the amino acid residues of TYR-411 (1.7 Å), ASN-391 (2.6 Å), ARG-410 (2.6 Å), and GLU-383 (2.2 Å) (Figure 6). As shown for VEGFA protein, the root-mean-square deviation of the original ligand was 2.586 Å. The hydrogen bonding of the pro-ligand NAG and 5T89 protein acted on the amino acid residues of GLN-342 (3.2 Å). Pachyman formed bonds with amino acid residues of ASN-402 (2.6 Å), THR-404 (2.5 Å), GLY-373 (2.8 Å), and ASN-417 (2.6 Å) (Figure 7).
FIGURE 6.
The molecular docking analysis shows potent binding activities of Pachyman with 6QIO in ALB as a crucial target in HCC.
FIGURE 7.
The molecular docking data–suggested potent binding activities of Pachyman with 5T89 in VEGFA as a crucial target in HCC.
Validated Data in Clinical HCC Samples
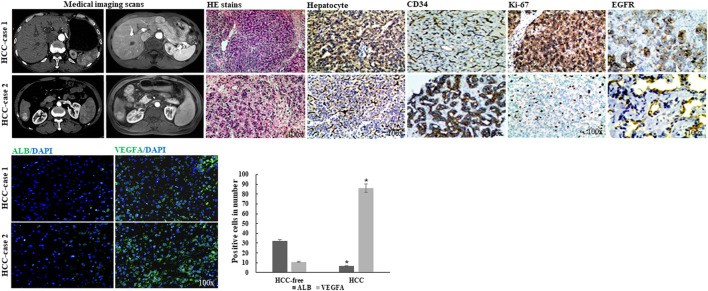
To further validate the molecular docking findings, the HCC and HCC-free clinical samples were collected for experimental tests. The HCC cases were well characterized medically with medical imaging scans and pathological diagnosis (hematoxylin and eosin stain (HE) and positive cells for hepatocyte, CD34, Ki-67, and EGFR expressions). Additionally, the data from the fluorescence immunostaining assay showed that the HCC samples resulted in significant downregulation of ALB expression and elevation of VEGFA expression when compared with those in HCC-free controls (p < 0.05) (Figure 8).
FIGURE 8.
Characterized imaging and immunostaining findings of clinical HCC samples, and validated data based on molecular docking analysis.
Validated Data in Pachyman-Treated HCC Cells
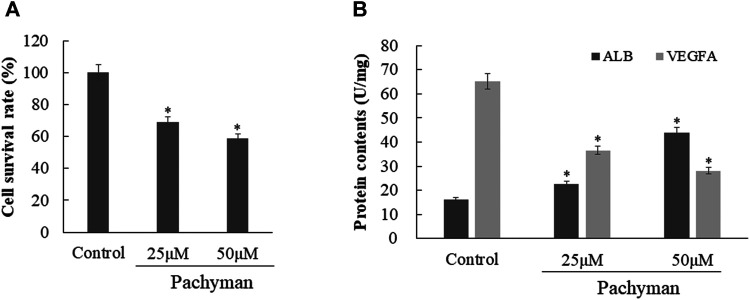
The experimental data in vitro suggested that Pachyman-treated HepG2 cells showed markedly reduced cell proliferation in comparison with the non-treated control (p < 0.05) (Figure 9A). In the quantitative test performed by using ELISA, the Pachyman-treated HepG2 cells exhibited increased ALB expression (p < 0.05) and reduced VEGFA content (p < 0.05) (Figure 9B).
FIGURE 9.
Validated data in vitro with Pachyman-treated liver cancer cells according to molecular docking findings, as shown in cell proliferation test (A), target protein determination (B).
Discussion
In the current bioinformatics findings of this study, network pharmacology–based analysis had identified individual and crucial targets, biological processes, and signaling pathways of Pachyman in treating HCC. The anti-HCC actions of Pachyman might be related to inducing cell death, enhancing immunologic function, and meliorating tumor microenvironmental regulation, as has been revealed in the GO-based analysis. Additionally, anti-proliferation pathways (such as apoptosis, and PI3K-Akt and MAPK signaling pathways), anti-invasiveness pathway (such as Ras and Rap1 signaling pathway), anti-angiogenesis pathway (such as VEGF signaling pathway), and other pathways (such as MicroRNAs in cancer) were all identified in Pachyman treating HCC actions, as revealed in the KEGG-based findings. During further screening of crucial targets by using molecular docking analysis, the Pachyman-activated anti-HCC core targets, including ALB and VEGFA, were identified accordingly, characterized with potent binding activities of Pachyman with 6QIO and 5T89 proteins in HCC. ALB, a monomeric protein, plays principally as a carrier protein for fatty acids, steroids, and thyroid hormones (Garcia-Martinez et al., 2013). Medically, serum ALB may be used for evaluating the nutritional condition in cancer patients, as malnutrition can be a function of prognostic significance in cancer cases (Gupta and Lis, 2010). It has been reported by using in situ hybridization test that endogenous albumin mRNA may serve as a sensitive marker for detecting primary hepatic carcinoma (Chen et al., 2021). In addition, the prognostic value of the albumin score in HCC cases has been identified clinically (Feng et al., 2020). VEGFA, a glycosylated mitogen, can induce vascular permeability, vasculogenesis, and endothelial cell proliferation or growth (Ballmer-Hofer, 2018). In tumor-related angiogenesis, VEGFA-based response mediates numerous functional effectors, including phosphoinositide 3 kinase (PI3K)/Akt, p38 MAPK, and ERKs (Claesson-Welsh and Welsh, 2013). In addition, VEGFA-induced HCC development and pathogenesis may be a potential pharmacological target for treating hepatocellular carcinoma (Morse et al., 2019). In the present validated experiments, clinical HCC samples showed decreased ALB expression and elevated VEGFA content. As shown in in vitro Pachyman treatments, intracellular protein of ALB was upregulated and VEGFA protein expression was reduced. These validated findings demonstrated that ALB and VEGFA may be the anti-HCC pharmacological targets activated by Pachyman. The limitation of the present study is that further in vivo experimental validation is needed for potential use of Pachyman in clinical application.
Conclusion
Collectively, the bioinformatics and validation findings revealed the pharmacological targets and mechanisms of Pachyman in treating HCC. Specially, computational identification of ALB and VEGFA proteins/genes may functionally serve as potential biomarkers in screening for and treating HCC.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
Author Contributions
HH contributed to the conception, design of the manuscript. LQ, DH, JH, and FQ contributed to the acquisition, analyses, and interpretation of the data in this manuscript. HH and LQ drafted this manuscript. HH revised this manuscript. All authors agree to be accountable for all aspects of work ensuring integrity and accuracy.
Funding
This study was supported by the Liuzhou Scientific Research and Technology Development Project (No. 2017BH20305).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Anwanwan D., Singh S. K., Singh S., Saikam V., Singh R. (2020). Challenges in Liver Cancer and Possible Treatment Approaches. Biochim. Biophys. Acta Rev. Cancer. 1873, 188314. 10.1016/j.bbcan.2019.188314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballmer-Hofer K. (2018). Vascular Endothelial Growth Factor, from Basic Research to Clinical Applications. Int. J. Mol. Sci. 19, 3750. 10.3390/ijms19123750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D. A., Koehne de Gonzalez A., Fazlollahi L., Coffey A., Remotti H. E., Lagana S. M. (2021). In Situ Hybridisation for Albumin RNA in Paediatric Liver Cancers Compared with Common Immunohistochemical Markers. J. Clin. Pathol. 74, 98–101. 10.1136/jclinpath-2020-206663 [DOI] [PubMed] [Google Scholar]
- Claesson-Welsh L., Welsh M. (2013). VEGFA and Tumour Angiogenesis. J. Intern. Med. 273, 114–127. 10.1111/joim.12019 [DOI] [PubMed] [Google Scholar]
- Feng D., Wang M., Hu J., Li S., Zhao S., Li H., et al. (2020). Prognostic Value of the Albumin-Bilirubin Grade in Patients with Hepatocellular Carcinoma and Other Liver Diseases. Ann. Transl Med. 8, 553. 10.21037/atm.2020.02.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Wang H. (2018). Precision Diagnosis and Treatment of Liver Cancer in China. Cancer Lett. 412, 283–288. 10.1016/j.canlet.2017.10.008 [DOI] [PubMed] [Google Scholar]
- Garcia-Martinez R., Caraceni P., Bernardi M., Gines P., Arroyo V., Jalan R. (2013). Albumin: Pathophysiologic Basis of Its Role in the Treatment of Cirrhosis and Its Complications. Hepatology 58, 1836–1846. 10.1002/hep.26338 [DOI] [PubMed] [Google Scholar]
- Gupta D., Lis C. G. (2010). Pretreatment Serum Albumin As a Predictor of Cancer Survival: a Systematic Review of the Epidemiological Literature. Nutr. J. 9, 69. 10.1186/1475-2891-9-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Guo C., Li Y., Liang X., Su M. (2021). Functional Benefit and Molecular Mechanism of Vitamin C against Perfluorooctanesulfonate-Associated Leukemia. Chemosphere 263, 128242. 10.1016/j.chemosphere.2020.128242 [DOI] [PubMed] [Google Scholar]
- Li R., Guo C., Li Y., Liang X., Yang L., Huang W. (2020). Therapeutic Target and Molecular Mechanism of Vitamin C-Treated Pneumonia: a Systematic Study of Network Pharmacology. Food Funct. 11, 4765–4772. 10.1039/d0fo00421a [DOI] [PubMed] [Google Scholar]
- Li R., Guo C., Li Y., Qin Z., Huang W. (2021). Therapeutic Targets and Signaling Mechanisms of Vitamin C Activity against Sepsis: a Bioinformatics Study. Brief Bioinform 22, bbaa079. 10.1093/bib/bbaa079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Guo C., Tse W. K. F., Su M., Zhang X., Lai K. P. (2020). Metabolomic Analysis Reveals Metabolic Alterations of Human Peripheral Blood Lymphocytes by Perfluorooctanoic Acid. Chemosphere 239, 124810. 10.1016/j.chemosphere.2019.124810 [DOI] [PubMed] [Google Scholar]
- Li R., Huang C., Ho J. C. H., Leung C. C. T., Kong R. Y. C., Li Y., et al. (2021). The Use of Glutathione to Reduce Oxidative Stress Status and Its Potential for Modifying the Extracellular Matrix Organization in Cleft Lip. Free Radic. Biol. Med. 164, 130–138. 10.1016/j.freeradbiomed.2020.12.455 [DOI] [PubMed] [Google Scholar]
- Li R., Huang X., Liang X., Su M., Lai K. P., Chen J. (2021). Integrated Omics Analysis Reveals the Alteration of Gut Microbe-Metabolites in Obese Adults. Brief Bioinform. 22 (3), bbaa165. 10.1093/bib/bbaa165 [DOI] [PubMed] [Google Scholar]
- Li R., Li Y., Liang X., Yang L., Su M., Lai K. P. (2021). Network Pharmacology and Bioinformatics Analyses Identify Intersection Genes of Niacin and COVID-19 as Potential Therapeutic Targets. Brief Bioinform. 22, 1279–1290. 10.1093/bib/bbaa300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Wu K., Li Y., Liang X., Lai K. P., Chen J. (2021). Integrative Pharmacological Mechanism of Vitamin C Combined with Glycyrrhizic Acid against COVID-19: Findings of Bioinformatics Analyses. Brief Bioinform. 22, 1161–1174. 10.1093/bib/bbaa141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Wu K., Li Y., Liang X., Tse W. K. F., Yang L., et al. (2020). Revealing the Targets and Mechanisms of Vitamin A in the Treatment of COVID-19. Aging (Albany NY). 12, 15784–15796. 10.18632/aging.103888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., He Y., Zeng P., Liu Y., Zhang M., Hao C., et al. (2019). Molecular Basis for Poria Cocos Mushroom Polysaccharide Used As an Antitumour Drug in China. J. Cell Mol Med. 23, 4–20. 10.1111/jcmm.13564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Yu J., Peng X. (2020). Poria Cocos Polysaccharides Alleviates Chronic Nonbacterial Prostatitis by Preventing Oxidative Stress, Regulating Hormone Production, Modifying Gut Microbiota, and Remodeling the DNA Methylome. J. Agric. Food Chem. 68, 12661–12670. 10.1021/acs.jafc.0c05943 [DOI] [PubMed] [Google Scholar]
- Llovet J. M., Kelley R. K., Villanueva A., Singal A. G., Pikarsky E., Roayaie S., et al. (2021). Hepatocellular Carcinoma. Nat. Rev. Dis. Primers. 7, 6. 10.1038/s41572-020-00240-3 [DOI] [PubMed] [Google Scholar]
- Llovet J. M., Montal R., Sia D., Finn R. S. (2018). Molecular Therapies and Precision Medicine for Hepatocellular Carcinoma. Nat. Rev. Clin. Oncol. 15, 599–616. 10.1038/s41571-018-0073-4 [DOI] [PubMed] [Google Scholar]
- McGlynn K. A., Petrick J. L., El-Serag H. B. (2021). Epidemiology of Hepatocellular Carcinoma. Hepatology 73, 4–13. 10.1002/hep.31288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse M. A., Sun W., Kim R., He A. R., Abada P. B., Mynderse M., et al. (2019). The Role of Angiogenesis in Hepatocellular Carcinoma. Clin. Cancer Res. 25, 912–920. 10.1158/1078-0432.CCR-18-1254 [DOI] [PubMed] [Google Scholar]
- Nong Y., Liang Y., Liang X., Li Y., Yang B. (2020). Pharmacological Targets and Mechanisms of Calycosin against Meningitis. Aging (Albany NY). 12, 19468–19492. 10.18632/aging.103886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X., Huang C., Wu K., Li Y., Liang X., Su M., et al. (2021). Anti-coronavirus Disease 2019 (COVID-19) Targets and Mechanisms of Puerarin. J. Cell Mol Med. 25, 677–685. 10.1111/jcmm.16117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M., Guo C., Liu M., Liang X., Yang B. (2019). Therapeutic Targets of Vitamin C on Liver Injury and Associated Biological Mechanisms: A Study of Network Pharmacology. Int. Immunopharmacol 66, 383–387. 10.1016/j.intimp.2018.11.048 [DOI] [PubMed] [Google Scholar]
- Sun Y. (2014). Biological Activities and Potential Health Benefits of Polysaccharides from Poria Cocos and Their Derivatives. Int. J. Biol. Macromol. 68, 131–134. 10.1016/j.ijbiomac.2014.04.010 [DOI] [PubMed] [Google Scholar]
- Wu K., Fan J., Huang X., Wu X., Guo C. (2018). Hepatoprotective Effects Exerted by Poria Cocos Polysaccharides against Acetaminophen-Induced Liver Injury in Mice. Int. J. Biol. Macromol 114, 137–142. 10.1016/j.ijbiomac.2018.03.107 [DOI] [PubMed] [Google Scholar]
- Wu K., Guo C., Yang B., Wu X., Wang W. (2019). Antihepatotoxic Benefits of Poria Cocos Polysaccharides on Acetaminophen-Lesioned Livers In Vivo and In Vitro . J. Cell Biochem. 120, 7482–7488. 10.1002/jcb.28022 [DOI] [PubMed] [Google Scholar]
- Wu X., Liang M., Yang Z., Su M., Yang B. (2017). Effect of Acute Exposure to PFOA on Mouse Liver Cells In Vivo and In Vitro . Environ. Sci. Pollut. Res. Int. 24, 24201–24206. 10.1007/s11356-017-0072-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.