Abstract
Peach is a typical climacteric fruit that releases ethylene during fruit ripening. Several studies have been conducted on the transcriptional regulation of ethylene biosynthesis in peach fruit. Herein, an ethylene response factor, PpERF.A16, which was induced by exogenous ethylene, could enhance ethylene biosynthesis by directly inducing the expression of 1-aminocyclopropane-1-carboxylic acid synthase (PpACS1) and 1-aminocyclopropane-1-carboxylic acid oxidase (PpACO1) genes. Moreover, the NAM/ATAF1/2/CUC2 (NAC) transcription factor (TF) PpNAC.A59 was coexpressed with PpERF.A16 in all tested peach cultivars. Interestingly, PpNAC.A59 can directly interact with the promoter of PpERF.A16 to induce its expression but not enhance LUC activity driven by any promoter of PpACS1 or PpACO1. Thus, PpNAC.A59 can indirectly mediate ethylene biosynthesis via the NAC-ERF signaling cascade to induce the expression of both PpACS1 and PpACO1. These results enrich the genetic network of fruit ripening in peach and provide new insight into the ripening mechanism of other perennial fruits.
Subject terms: Plant hormones, Transcription
Introduction
Fruit ripening is the last stage of the fruit developmental process and is accompanied by a series of physiological and metabolic changes in color, texture, flavor, and nutritional compounds1. Fleshy fruits are classically defined as climacteric or nonclimacteric; the former exhibit a burst of ethylene production and respiration during ripening, while the latter do not2. Ethylene is a major signaling molecule that controls fruit ripening in climacteric fruits3 and plays an important role in the fruit ripening of nonclimacteric fruits4,5.
Ethylene biosynthesis and signal transduction have been widely studied in angiosperms6–8. Ethylene biosynthesis is catalyzed by two enzymes, 1-aminocyclopropane-1-carboxylic acid synthase (ACS), and oxidase (ACO). Synthesized ethylene activates the expression of ripening-related genes through transcription factors (TFs), ETHYLENE INSENSITIVE-like factors and ethylene responsive factors (ERFs). In tomato, LeERF1 promotes fruit ripening9, while SlERF6 delays fruit ripening10. In perennial fruit trees, MaERF9, MdERF3, and PbERF24 induce ethylene biosynthesis during fruit ripening in banana, apple, and pear, respectively11–13. In contrast, MaERF11, MdERF2, and CpERF9 inhibit ethylene biosynthesis during fruit ripening in banana, apple, and papaya, respectively11,12,14. Notably, apple MdERF3 is the downstream target of MdERF2, MdMYC2, and MdARF512,15,16. These findings suggest that ERFs are important in ethylene biosynthesis during fruit ripening.
In addition to ERF, other TFs, such as NAM/ATAF1/2/CUC2 (NAC), MADS-box, homeobox, apetala 2, SQUAMOSA promoter binding protein (SBP), auxin response factor (ARF), and zinc-finger proteins, are also involved in mediating ethylene biosynthesis in climacteric fruits8,17–20. Among the NAC TFs, NOR, SlNAC4, and NAC-like 1 are positive regulators of fruit ripening21–23, while SlNAC1 is a transcriptional repressor of ripening-related genes in tomato24. AdNAC6 and AdNAC7 can bind to the AdACS1 and AdACO1 promoters to stimulate its expression and increase ethylene production in kiwifruit25. MaNAC1 and MaNAC2 can directly interact with ethylene signaling components, such as ETHYLENE INSENSITIVE, in banana26. CmNAC-NOR is located in a quantitative trait locus for climacteric ripening and is likely associated with ethylene biosynthesis and fruit ripening in melon27. Moreover, a heterodimer of two NACs can activate the transcription of an MYB TF28, indicating that the NAC TF is probably an upstream factor that regulates the expression of downstream TF(s). These findings suggest that NAC could be involved in regulating ethylene biosynthesis by directly and/or indirectly inducing ethylene biosynthesis genes. This suggestion is supported by previous reports, in which three MADS-box TFs, RIN, FUL1, and FUL2, can mediate the expression of ACS genes and their upstream TFs, including RIN, NOR, and CNR (an SBP TF)29,30.
Peach (Prunus persica) emits a large amount of ethylene during fruit ripening31,32. Exogenous ethylene accelerates fruit softening and induces the expression of softening-related genes33, such as endopolygalacturonase (PpendoPGM) controlling the melting flesh phenotype in the clingstone melting flesh (CMF) peach fruit34. Moreover, ethylene biosynthesis and signal transduction in peach can be interrupted by aminoethoxyvinylglycine and 1-methylcyclopropene (1-MCP), respectively, leading to the delay of fruit ripening35,36. Therefore, ethylene is crucial for catalyzing fruit ripening in peach. The genes PpACS1 and PpACO1, which encode the two catalytic enzymes for ethylene biosynthesis, have been identified in peach fruit37–39. Two TFs, PpHB.G7 and PpTCP.A2, were identified to be involved in peach fruit ripening40,41. PpHB.G7 can bind to the PpACS1 and PpACO1 promoters to induce the expression of both genes and increase ethylene production40, while PpTCP.A2 can inhibit ethylene biosynthesis by negatively affecting PpACS1 expression41. To broaden the transcription regulation network of ethylene biosynthesis in peach, two TFs, PpERF.A16 and PpNAC.A59 were isolated from peach fruit. Of these, PpERF.A16 could directly bind to the PpACS1 and PpACO1 promoters to positively regulate ethylene biosynthesis in peach fruit. PpNAC.A59 was coexpressed with PpERF.A16, and PpNAC.A59 could directly bind to the promoter of PpERF.A16 to indirectly promote ethylene biosynthesis in peach fruit. This study further elucidates the molecular network of ethylene-induced fruit ripening in peach and other perennial fruit trees.
Results
Identification of ACS, ACO, and ERF genes during fruit ripening
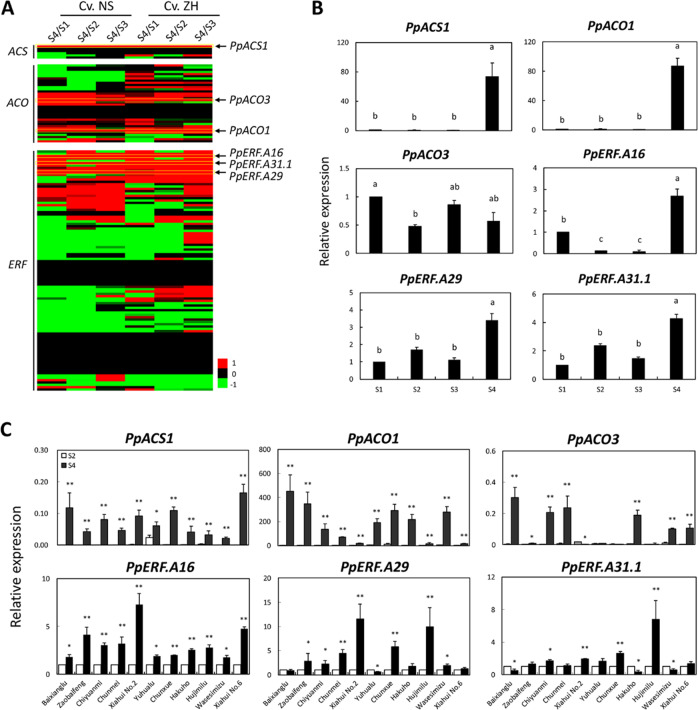
Members of the ACS, ACO, and ERF gene families were isolated from peach and other fruit trees. A total of 106 AP2/ERF genes (Table S1) were detected in the peach genome and could be divided into two groups, A and B (Fig. S1). Of the two groups, group A corresponded to subfamily ERF, which comprised 56 clusters (A1 → A56), and group B contained the AP2, RAV, and soloist subfamilies. Moreover, six members of the ACS gene family were detected in peach (Fig. S2) along with 32 possible members of the ACO (or ACO-like) gene family (Fig. S3). Based on the RNA-Seq data of fruit samples of the peach cultivars Nanshantiantao (NS) and Zhaohui (ZH) in a previous study34, the ACS, ACO, and AP2/ERF genes, which had higher levels of expression in ripening fruit than in developing fruit (Fig. 1A and Table S2), were detected. Based on previous reports37,40, the ACS gene Ppa004774m was identical to PpACS1, and the two ACO genes, Pp008791m and Ppa009228m, were identical to PpACO1 and PpACO3, respectively. The three ERF genes belonged to the A16, A29, and A31 clusters and were thus designated PpERF.A16 (Ppa008730m), PpERF.A29 (Ppa021711m), and PpERF.A31.1 (Ppa007193m), respectively (Table S1). However, the expression level of PpACO3 in ripening fruit was similar to that in developing fruit of cv. Xiahui No. 5 (Fig. 1B) and cvs. Yuhualu, Chumei, and Hujimilu (Fig. 1C). Moreover, PpERF.31.1 and PpERF.A29 had similar levels of expression between developing and ripening fruits of some cultivars (Fig. 1C), although these two genes were more highly expressed in ripening fruit than in developing fruit of cv. Xiahui No.5 (Fig. 1B). These results indicate that PpACO3, PpERF.31.1, and PpERF.A29 are not involved in peach fruit ripening. In contrast, PpACS1, PpACO1, and PpERF.A16 were more highly expressed in ripening fruit than in developing fruit of all 12 tested cultivars (Fig. 1B, C) and thus are probably associated with peach fruit ripening.
Fig. 1. Transcriptome-based identification of the ACS, ACO, and AP2/ERF genes potentially associated with peach fruit ripening.
A Transcriptome-based analysis of the differentially expressed ACS, ACO, and AP2/ERF genes between developing (S1 to S3) and ripening (S4) fruits in two peach cvs. NS and ZH. The color bar is calculated by log2 fold-change value of the ratio. B The expression profiles of PpACS1, PpACO1, PpACO3, PpERF.A16, PpERF.A29, and PpERF.A31.1 were determined by quantitative real-time PCR (qRT-PCR) in cv. Xiahui No.5 fruits at S1, S2, S3, and S4. C The expression levels of PpACS1, PpACO1, PpACO3, PpERF.A16, PpERF.A29, and PpERF.A31.1 were determined in the fruits of 11 peach cultivars at S2 and S4
Interaction of PpERF.A16 with the promoters of the PpACS1, PpACO1, and PpendoPGM genes
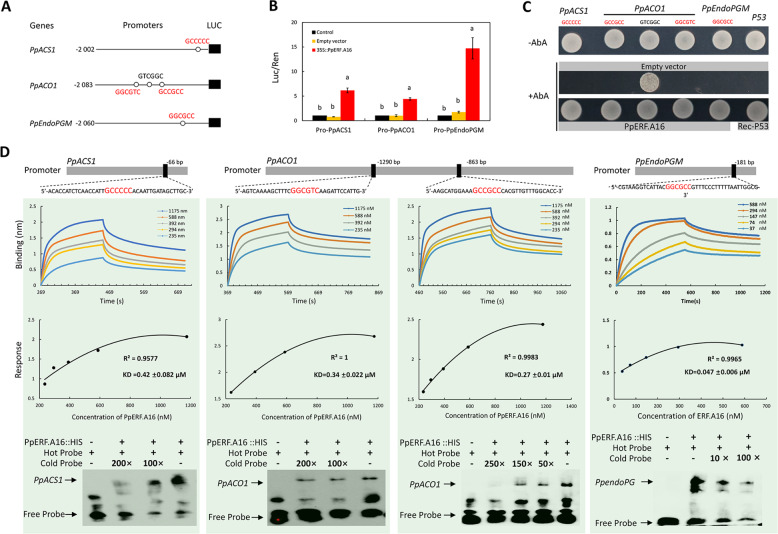
Similar to the previous study40, to test whether PpERF.A16 could bind to the promoters of ripening-related genes, PpACS1, PpACO1, and PpendoPGM, dual-luciferase assays were conducted and revealed that the LUC activities that were individually driven by the PpACS1, PpACO1, or PpendoPGM promoters were significantly increased in Arabidopsis protoplasts overexpressing PpERF.A16 (35S::PpERF.A16) compared to Arabidopsis protoplasts transformed with an empty vector or without any transformation (control; Fig. 2B). This result indicates that PpERF.A16 can interact with the promoters of PpACS1, PpACO1, and PpendoPGM. Subsequently, yeast one-hybrid (Y1H) assays showed that PpERF.A16 binds to the PpACS1, PpACO1, and PpendoPGM promoters (Fig. 2C). Moreover, a PpERF.A16::HIS fusion protein was generated by inserting the whole sequences of PpERF.A16 into the pCold-TF vector. Kinetic assay showed that binding values between PpERF.A16::HIS, and the promoters of the three ripening-related genes gradually increased with increasing concentrations of PpERF.A16::HIS (Fig. 2D). Furthermore, electrophoretic mobility shift assay (EMSA) showed that PpERF.A16::HIS could bind to the probes of the PpACS1, PpACO1, and PpendoPGM promoters, and the binding signals weakened with increasing concentrations of the cold probe (Fig. 2D). Taken together, PpERF.A16 directly binds to the PpACS1, PpACO1, and PpendoPGM promoters to enhance their activities.
Fig. 2. PpERF.A16 directly interacts with the promoters of PpACS1, PpACO1, and PpendoPGM.
A GCC-box element(s) were predicted in the promoters of PpACS1, PpACO1, and PpendoPGM. B PpERF.A16 activated LUC expression that was controlled by the promoters of PpACS1, PpACO1, or PpendoPGM. C Y1H assay of PpERF.A16 with the predicted GCC-box elements in the PpACS1, PpACO1, and PpendoPGM promoters. D Kinetic assay and EMSA of PpERF.A16 with the PpACS1, PpACO1, and PpendoPGM promoters. + and − indicate the presence and absence of recombinant PpERF.A16::HIS, biotin-labeled probe, or cold probe, respectively. Cold probe concentrations were tenfold (10×), 50-fold (50×), 100-fold (100×), 150-fold (150×), 200-fold (200×), or 250-fold (250×) those of the labeled probes
Effect of PpERF.A16 on ethylene biosynthesis
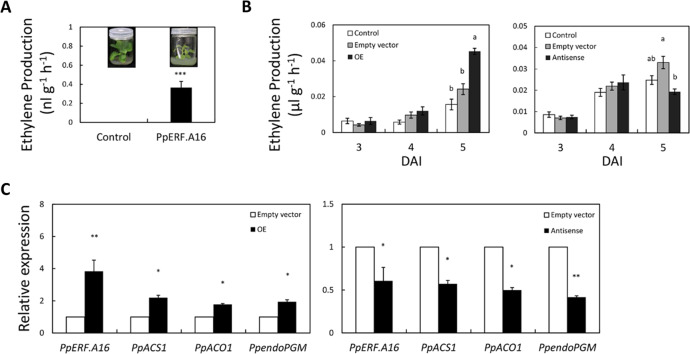
To unravel the role of PpERF.A16 in ethylene biosynthesis, overexpression and antisense vectors of PpERF.A16 were individually constructed and then introduced to Agrobacterium tumefaciens. After introducing the overexpression vector of PpERF.A16 into tobacco and identifying the positive lines (Fig. S4), the ethylene production rate was measured using a gas chromatography system. The results showed that the ethylene production rate was higher in the transgenic lines of PpERF.A16 (P value < 0.001) than in the nontransgenic regeneration lines (Fig. 3A), suggesting that PpERF.A16 can promote ethylene biosynthesis in transgenic tobacco. Furthermore, Agrobacterium tumefaciens containing overexpression and antisense vectors of PpERF.A16 was independently injected into the fruits harvested at ~10 days before the commercial harvesting date, and ethylene production rates were measured at 3, 4, and 5 days after injection (DAI). At 5 DAI, the ethylene production rate was increased in the fruits overexpressing PpERF.A16 but decreased in the fruits silencing PpERF.A16 compared to the fruits infiltrated with the empty vector (Fig. 3B). Thus, PpERF.A16 can positively mediate ethylene biosynthesis during peach fruit ripening.
Fig. 3. PpERF.A16 positively mediates ethylene biosynthesis.
A The ethylene production rate was increased in tobacco transgenic plants overexpressing PpERF.A16 compared to the wild-type control. B The peach fruits harvested ~10 days before the commercial harvesting date were transiently transformed with an empty vector or a vector overexpressing PpERF.A16 (OE) or antisense PpERF.A16 (Antisense). Ethylene production rates were measured at 3, 4, and 5 days after injection (DAI) of Agrobacteria containing the vectors and showed a significant increase in the OE fruits but a decrease in the antisense fruits compared to the fruit injected with the pCAMBIA1301 empty vector and noninjected control fruit at 5 DAI. C The expression levels of PpERF.A16, PpACS1, PpACO1, and PpendoPGM genes were increased in the OE fruits but decreased in the antisense fruits compared to the empty vector fruits at 5 DAI
To test whether the expression of the PpACS1, PpACO1, and PpendoPGM genes was affected by PpERF.A16, qRT-PCR analyses were conducted on the transiently transformed peach fruits. As a result, both PpACS1 and PpACO1 were upregulated in fruits overexpressing PpERF.A16 but downregulated in the fruits silencing PpERF.A16 compared to the fruits infiltrated with the empty vector (Fig. 3C). Further qRT-PCR analysis showed that the transcript levels of PpERFA15.1/15.2/15.3 and PpERFA17.2/17.2 were most closely related to PpERF.A16 and were not downregulated in the fruits with silenced PpERF.A16 (Fig. S5). These results suggest that PpERF.A16 can positively regulate ethylene biosynthesis by inducing the expression of both PpACS1 and PpACO1. In addition, PpERF.A16 upregulated the expression of PpendoPGM (Fig. 3C) and ripening-related genes (such as PpCHI, PpPDS, PpZEP, and PpNECD) (Fig. S6). Together with the results of the promoter analyses described above, PpERF.A16 was shown to be able to directly affect the expression of the PpACS1, PpACO1, and PpendoPGM genes.
Identification of the NAC TF coexpressed with PpERF.A16
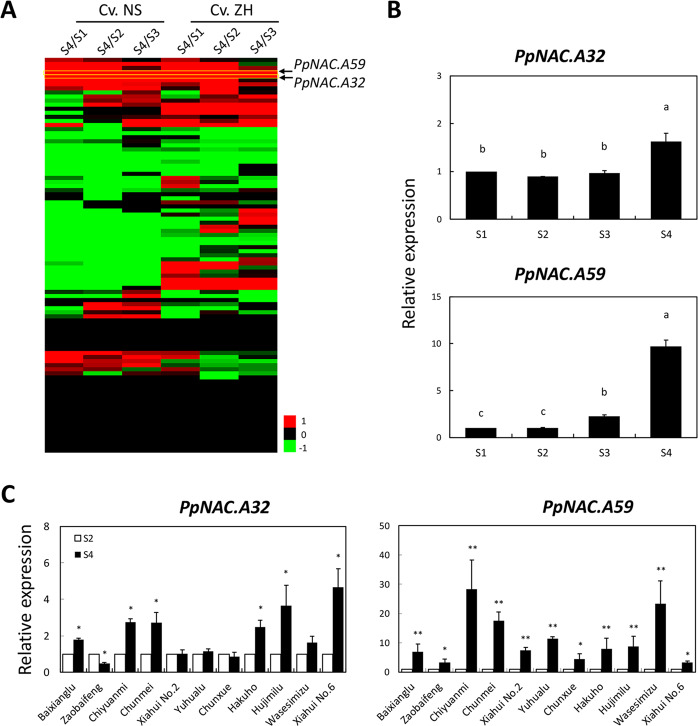
To mine the upstream TF(s) of PpERF.A16 expression and ethylene biosynthesis, the potential cis-regulatory elements in the PpERF.A16 promoter were predicted, revealing 12 elements (the sequences with red color) for the NAC TF (Fig. S7). A higher number of elements for the NAC TF indicated that this TF should play an important role in mediating PpERF.A16 expression and ethylene production. Subsequently, 103 members of the NAC gene family were isolated from the peach genome. Together with the NAC genes derived from other fruit trees, the NAC gene family can be divided into two groups, A and B (Fig. S8 and Table S3). Of the two groups, group A comprised 69 clusters (A1 → A69), while group B contained only three clusters (B1 → B3; Fig. S8). Based on the RNA-Seq data of fruit samples of cvs. NS and ZH in a previous study34, two NAC genes were more highly expressed in ripening fruit than in developing fruit of both cvs. NS and ZH (Fig. 4A and Table S2). These two genes, Ppa007445m and Ppa008828m, belong to clusters A32 and A59 and were thus designated PpNAC.A32 and PpNAC.A59, respectively (Table S3). Of the two genes, however, PpNAC.A32 had similar levels of expression between developing and ripening fruits of cv. Xiahui No.5 (Fig. 4B) and cvs. Xiahui No. 2, Yuhualu, Chunxue, and Wasesimizu (Fig. 4C), while PpNAC.A59 was more highly expressed in ripening fruit than in developing fruit of all 12 tested cultivars (Fig. 4B, C). Obviously, PpNAC.A59 had an identical expression profile to PpERF.A16 in all tested peach cultivars, indicating that this TF may induce the expression of PpERF.A16 and is probably associated with peach fruit ripening.
Fig. 4. Identification of the NAC genes probably associated with peach fruit ripening.
A Transcriptome analysis showed that PpNAC.A59 and PpNAC.A32 were differentially expressed between developing (S1 to S3) and ripening (S4) fruits in peach cvs. NS and ZH. The color bar was calculated by the log2 fold-change values of the ratio. B The expression profiles of PpNAC.A59 and PpNAC.A32 were tested by qRT-PCR in cv. Xiahui No. 5 fruits at S1, S2, S3, and S4. C The expression levels of PpNAC.A59 and PpNAC.A32 were tested in the fruits of 11 peach cultivars at S2 and S4
Interaction of PpNAC.A59 with the PpERF.A16 promoter
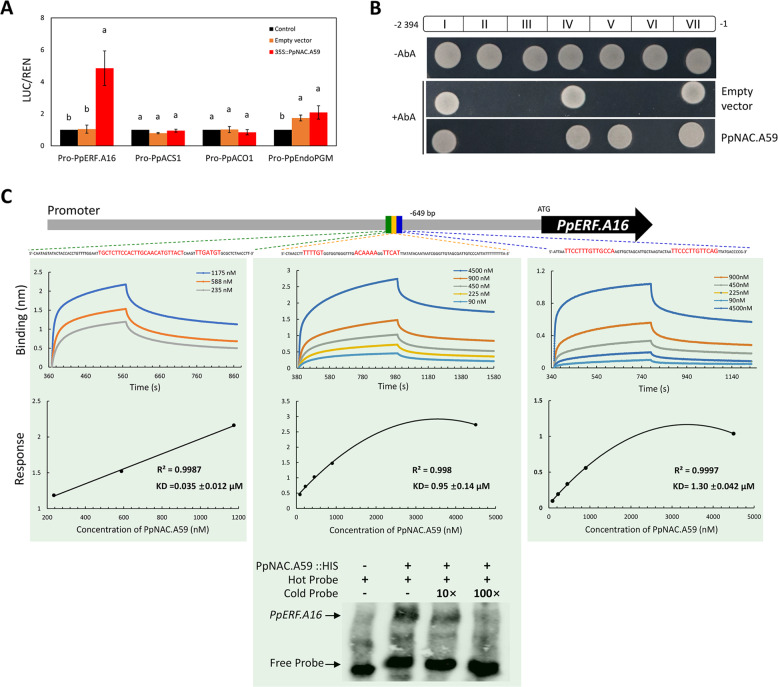
To test whether PpNAC.A59 interacts with the PpERF.A16 promoter, a dual-luciferase assay was conducted and showed an interesting result. The LUC gene driven by the PpERF.A16 promoter had higher activity in Arabidopsis protoplasts overexpressing PpNAC.A59 (35S::PpNAC.A59) than in Arabidopsis protoplasts transformed with an empty vector or without any transformation (control; Fig. 5A). In contrast, the LUC gene driven by any promoter of PpACS1, PpACO1, and PpendoPGM had similar activity in Arabidopsis protoplasts overexpressing PpNAC.A59 to that in Arabidopsis protoplasts transformed with an empty vector (Fig. 5A). Therefore, PpNAC.A59 interacts with the PpERF.A16 promoter.
Fig. 5. PpNAC.A59 directly interacts with the PpERF.A16 promoter.
A PpNAC.A59-activated LUC expression, which was controlled by PpERF.A16 promoter. B Y1H assay of PpNAC.A59 with seven fragments (I to VII) in the PpERF.A16 promoter. C Kinetic assay and EMSA of PpNAC.A59 with the PpERF.A16 promoter. “+” and “−” indicate the presence and absence of recombinant PpNAC.A59::HIS, biotin-labeled probe, or cold probe, respectively. Cold probe concentrations were tenfold (10×) and 100-fold (100×) those of the labeled probes
To determine which fragment(s) in the promoter of PpERF.A16 could be bound by PpNAC.A59, the promoter sequences of PpERF.A16 were fragmented into seven sections (I → VII). Only fragment V was bound by PpNAC.A59 in yeast cells (Fig. 5B). In fragment V, we detected two possible cis-regulatory elements (WNNYBTNNNNNNNAMGNHW and TTRCGT) for NAC TF42–44, and these elements were used to synthesize the biotin-labeled probes (Fig. 5C). Subsequently, a recombinant PpNAC.A59::HIS fusion protein was generated by inserting the whole sequences of PpNAC.A59 into the pCold-TF vector. Kinetic assay showed that the binding value between PpNAC.A59::HIS and each of the three probes gradually increased with increasing concentrations of PpNAC.A59::HIS, and the binding response between PpNAC.A59::HIS and the probe boxed with a yellow color peaked when the concentration reached 4500 nM (KD <1 μM; Fig. 5C). Moreover, EMSA showed that PpNAC.A59::HIS could bind to the probe boxed with a yellow color, and the binding signal weakened as the concentration of the cold probe increased (Fig. 5C). Taken together, PpNAC.A59 can directly bind to the promoter of PpERF.A16.
Effect of PpNAC.A59 on ethylene biosynthesis
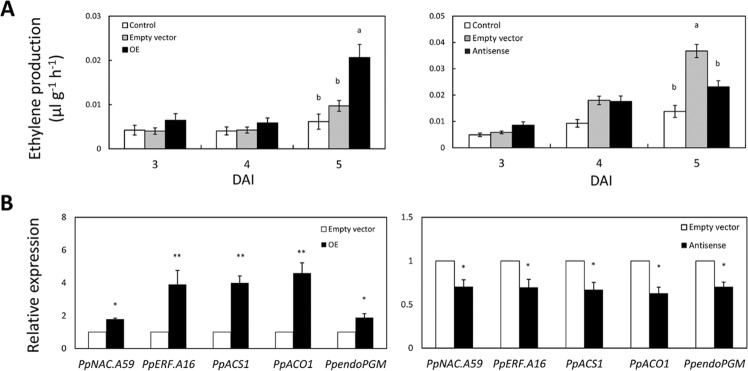
To unravel the role of PpNAC.A59 in ethylene biosynthesis, overexpression and antisense vectors of PpNAC.A59 were constructed and then introduced into A. tumefaciens. The resuspended Agrobacterium was injected into peach fruits, and ethylene production rates were measured at 3, 4, and 5 DAI. At 5 DAI, the ethylene production rate was increased in the fruits overexpressing PpNAC.A59 but reduced in the fruits with silenced PpNAC.A59 compared to the fruits injected with an empty vector (Fig. 6A). This result indicated that PpNAC.A59 can positively regulate ethylene biosynthesis during peach fruit ripening.
Fig. 6. PpNAC.A59 positively mediates ethylene biosynthesis.
A The peach fruits harvested ~10 days before the commercial harvesting date were transiently transformed with an empty vector or a vector overexpressing PpNAC.A59 (OE) or antisense PpNAC.A59 (Antisense). Ethylene production rates were measured at 3, 4, and 5 days after injection (DAI) of Agrobacteria containing the vectors and showed a significant increase in the OE fruits but a decrease in the antisense fruits compared to the fruits injected with the empty vector pCAMBIA1301 and noninjected control fruits at 5 DAI. B The expression levels of PpERF.A16, PpACS1, PpACO1, and PpendoPGM genes were increased in the OE fruits but decreased in the antisense fruits compared to the empty vector fruits at 5 DAI
To test whether the expression of ethylene biosynthesis and ripening-related genes was affected by PpNAC.A59, the transiently transformed peach fruits were analyzed using qRT-PCR. The results showed that the expression levels of PpACS1, PpACO1, and PpERF.A16 were upregulated in fruits overexpressing PpNAC.A59 and downregulated in the fruits with silenced PpNAC.A59 compared to the fruits infiltrated with the empty vector (Fig. 6B). Further qRT-PCR analysis showed that the transcript levels of PpNAC.A58/A50/A61, which were most closely related to PpNAC.A59, were not downregulated in the fruits with silenced PpNAC.A59 (Fig. S9). In addition, PpNAC.A59 upregulated the expression of PpendoPGM (Fig. 6B) and other ripening-related genes (such as PpCHI, PpZEP, and PpNECD) (Fig. S10). Due to no interaction being detected between PpNAC.A59 and the promoter sequences of PpACS1, PpACO1, and PpendoPGM (Fig. 5A), it is likely that PpNAC.A59 indirectly promotes the transcription of these genes via the PpERF.A16-ethylene pathway.
Discussion
PpERF.A16 directly regulates ethylene biosynthesis during fruit ripening
AP2/ERF TFs have been characterized in plant species, including peach45,46, and are involved in various biological processes, such as plant growth, development, senescence, and stress tolerance6,47. In fleshy fruits, AP2/ERF TFs play important roles in transmitting the ethylene signal and stimulating activities of downstream genes responsive to fruit ripening6,19. This type of AP2/ERF gene has been identified in climacteric fruits such as apple, banana, and pear11–13. This study identified a similar gene PpERF.A16 from previously reported RNA-Seq data from peach fruit. Overexpression of PpERF.A16 could enhance the expression of PpACS1 and PpACO1 and thus increase ethylene production during peach fruit ripening (Fig. 3). Furthermore, PpERF.A16 could also induce the expression of other ripening-related genes (Figs. 3C, S6) and is thus likely associated with peach fruit ripening. PpERF.A16 showed a similar function to the previously reported ERF genes MdERF3, PbERF24, and MaERF9 involved in regulating the ripening of climacteric fruits11–13. However, a difference is that PpERF.A16 binds to the promoters of both the PpACS1 and PpACO1 genes (Fig. 2), while MdERF3, PbERF24, and MaERF9 interact with the promoters of an ACS or ACO gene but not both genes11–13. Another difference is that these genes are clustered into different subgroups of the ERF family, i.e., PpERF.A16, PbERF24, MdERF3, and MaERF9 were clustered into the A16, A17, A39 and A13 subgroups, respectively (Table S1).
It has been reported that exogenous auxin can effectively induce the expression of PpACS1 and PpERF.A1637,48. Ethylene production is associated with the TC microsatellite genotypes of PpYUC11 in peach fruits with flesh melting and stony hard phenotypes49,50. Recently, it was confirmed that auxin can promote ethylene biosynthesis in apple fruit by activating MdARF516. MdARF5 can bind to the MdACS1, MdACS3a, MdACO1, and MdERF2 promoters to induce their expression in apple fruit. It is possible that auxin induces ethylene biosynthesis in peach fruit by an ARF TF. However, exogenous auxin treatment cannot upregulate ARF TFs in peach fruit37,48. Instead, the expression levels of auxin/indole-3-acetic acid (auxin/IAA) genes (IAA11, IAA13, and IAA9_2) were increased in the peach fruits with exogenous auxin treatment compared with those in the control fruits37,48. It is more possible that these auxin/IAA TFs directly or indirectly induce the expression of PpACS1 and PpERF.A16 to promote ethylene biosynthesis in peach fruit.
PpNAC.A59 indirectly mediates ethylene biosynthesis during fruit ripening
Plant-specific NAC TFs of woody and grassy plants have been phylogenetically analyzed51,52 and are reported to regulate various physiological traits53–56. NOR was the first NAC gene known to promote ethylene biosynthesis in a nonripening tomato mutant22. Subsequently, the SlNAC4 and NOR-like 1 genes were proven to be associated with fruit ripening in tomato21,23, similar to AdNAC6 and AdNAC7 in kiwifruit25. In the present study, PpNAC.A59 was shown to have a higher level of expression in ripening fruit than in developing fruit of all tested peach cultivars (Fig. 4). Interestingly, PpNAC.A59 cannot interact with the promoters of PpACS1 and PpACO1 (Fig. 5A) but could induce the expression of both PpACS1 and PpACO1 to promote ethylene biosynthesis in transiently transformed fruits (Fig. 6). This result is different from previous studies of the NACs associated with ethylene biosynthesis during fruit ripening. For example, NOR-like 1 of tomato interacts with the SlACS2 and SlACS4 promoters21, and AdNAC6/7 of kiwifruit interacts with the AdACS1 and AdACO1 promoters25. In contrast, PpNAC.A59 directly interacts with the PpERF.A16 promoter (Fig. 5) but not with the PpACS1 or PpACO1 promoter. Then, PpERF.A16 interacts with the promoters of PpACS1 and PpACO1. Obviously, PpNAC.A59 indirectly mediates ethylene biosynthesis during peach fruit ripening. It is noteworthy that PpNAC.A59 was not clustered together with any of the previously reported NACs associated with ethylene biosynthesis during fruit ripening. PpNAC.A59, AdNAC6/7, NOR/NOR-like 1, SlNAC1, and SlNAC4 were individually clustered into subgroups A59, A21, A35, A25, and A24 of the NAC family, respectively (Table S3).
In previous studies, NACs were reported to be involved in abscisic acid (ABA) biosynthesis57,58. Both Oryza sativa OsNAC2 and Solanum lycopersicum SlNAP2 directly control the expression of the ABA biosynthesis gene 9-cis-epoxycarotenoid dioxygenase and the ABA catabolic gene ABA 8’-hydroxylase to affect ABA production59,60. It is noteworthy that ABA indirectly promotes fruit ripening by triggering ethylene biosynthesis in climacteric fruits61,62. In this study, we found that PpNAC.A59 positively regulated the expression of ABA biosynthesis genes (Zeaxanthin epoxidase and 9-cis-epoxycarotenoid dioxygenase; Fig. S10), indicating that PpNAC.A59 is likely involved in ABA biosynthesis in peach fruit. Therefore, ethylene biosynthesis may also be influenced by the PpNAC.A59-ABA-ethylene signal cascade during peach fruit ripening.
Molecular network of fruit ripening in peach
Controlling the ripening process of climacteric fruits is important to the commercialization of these fruit crops and has been widely studied at the molecular genetics level19,63. In tomato, a genetic network regulating fruit ripening has been established and consists of TFs of different families, including RIN, FUL1, FUL2, HB-1, AP2a, ERF6, TAGL1, NOR, CNR, and TDR429,30. This network was further enriched by the most recently identified TFs, SlNAC4, SlNAC1, SlZFP2, and NOR-like 120,21,23,24, and epigenetic modifiers, such as DNA methylation64,65, microRNA66,67, and long noncoding RNA68,69. Obviously, the understanding of the genetic network regulating fruit ripening in tomato is much advanced compared to our understanding of the network in tree fruits. During apple fruit ripening, the regulatory network of ethylene biosynthesis was constructed to include the TFs MYC2, ARF5, ERF2, and ERF3 and ethylene biosynthesis genes12,15,16. However, in other perennial fruit trees, few TFs were concatenated into a network. In peach fruit, two TFs, PpHB.G7 and PpTCP.A2, were identified in the previous studies40,41. Of these two TFs, the transcription activator PpHB.G7 can directly bind to the PpACS1 and PpACO1 promoters to increase ethylene production40, while the transcription repressor PpTCP.A2 can inhibit the expression of PpACS1 to hinder ethylene biosynthesis41.
In the present study, two TFs, PpERF.A16 and PpNAC.A59, were shown to be involved in ethylene biosynthesis during peach fruit ripening. These results could be integrated into a primary network of fruit ripening in peach (Fig. 7). First, PpNAC.A59 can induce the expression of PpERF.A16 by directly binding to its promoter. Second, PpERF.A16 and PpHB.G7 enhances the expression of both PpACS1 and PpACO1 by directly interacting with their promoters, contributing to an increase in ethylene production. Finally, ethylene accelerates fruit ripening by mediating fruit metabolism, such as flavonoid and carotenoid metabolism. It is noteworthy that this conclusion is supported by a previous study in which the NAC TF was predicted to control ethylene biosynthesis during peach fruit ripening70. The only difference is that the predicted NAC TF can induce the expression of ethylene biosynthesis genes by directly interacting with their promoters70, while PpNAC.A59 indirectly induces the expression of both PpACS1 and PpACO1 by directly mediating the expression of PpERF.A16. Further work will focus on the enrichment of this network by identifying more TFs associated with fruit ripening.
Fig. 7. A regulatory model of ethylene biosynthesis induced by PpNAC.A59-activated PpERF.A16.
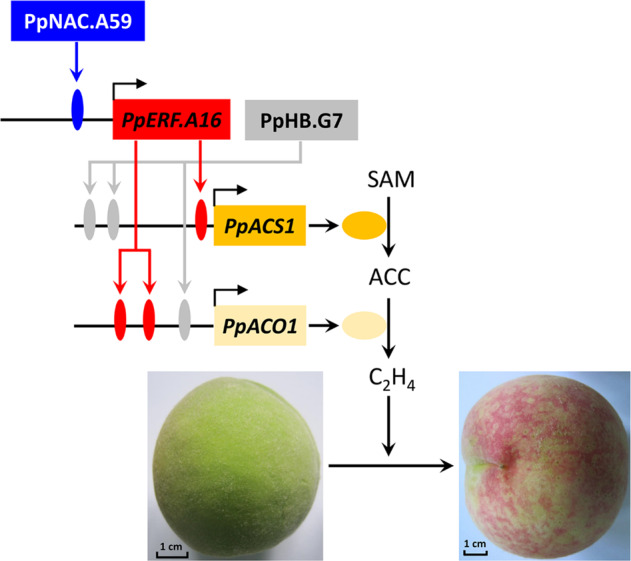
PpNAC.A59 activates the expression of PpERF.A16 by directly binding to its promoter. PpERF.A16 and PpHB.G7 enhance the expression of both PpACS1 and PpACO1 by directly interacting with their promoters, resulting in an increase in ethylene (C2H2) production in peach fruit
Materials and methods
Plant materials
A total of 12 peach cultivars were used in this study. Of these cultivars, Wasesimizu and Xiahui No. 5 were maintained at the Jiangsu Academy of Agricultural Sciences (Nanjing, Jiangsu Province, China). The other ten cultivars (Chunxue, Hakuho, Hujinmilu, Xiahui No. 2, Yuhualu, Chiyuanmi, Chunmei, Xiahui No. 6, Baixianglu, and Zaobaifeng) were identical to those in a previous study40. The cv. Baixianglu fruit presents a semifreestone melting flesh phenotype, and the fruits of the other ten peach cultivars present a clingstone melting flesh phenotype. Based on the dynamic investigation of fruit development and ripening71, the fruits of cv. Xiahui No. 5 were collected at the fruitlet (S1), stone hardening (S2), preripening (S3), and ripening (S4) stages, and the fruits of the other 11 cultivars were collected at the rapid enlargement (roughly corresponding to S2) and ripening stages (S4; Table S4). Grouping, biological replication, and storage of fruit samples were as described in a previous study40.
Sequence and phylogenetic analyses of the ACS, ACO, AP2/ERF, and NAC genes
To isolate the ACS, ACO, AP2/ERF, and NAC genes in peach and other fruit trees, the nucleotide and amino acid sequences of the ACS, ACO, AP2/ERF, and NAC genes in peach were extracted from the peach genome version 1.0 (Genome Database for Rosaceae; http://www.rosaceae.org/). Then, these downloaded sequences were used as indexes to find their homologs in papaya (Carica papaya ASGPBv0.4), strawberry (Fragaria vesca v 1.1), orange (Citrus sinensis v1.1), grape (Vitis vinifera Genoscope.12X), apple (Malus domestica v1.0) (https://phytozome.jgi.doe.gov/), and pear (Pyrus bretschneideri; http://peargenome.njau.edu.cn/) genomes (Tables S1, S3). Sequence alignment and phylogenetic analyses were as described in a previous study40.
Gene expression analysis in developing and ripening fruits
In a previous study, RNA-Seq was performed in the developing and ripening fruits of three peach cultivars, Nanshantiantao (NS), Zhaohui (ZH), and Myojo34. Of these three cultivars, NS and ZH fruits presented a melting flesh phenotype, and the fruit hardness gradually decreased during ripening, while fruits of the Myojo cultivar did not soften during fruit ripening. For this reason, the transcriptome data fruits from cvs. NS and ZH were used to isolate the ACS, ACO, AP2/ERF, and NAC genes that may be associated with fruit ripening. The hierarchical clustering of transcriptome data was performed according to a previous study40. By comparing the gene expression levels of the ripening fruit (S4) to those of the developing fruits (S1 to S3), we isolated the target genes PpACS1, PpACO1, PpACO3, PpERF.A16, PpERF.A29, PpERF.A31.1, PpNAC.A32, and PpNAC.A59. Thus, these eight genes were further tested by qRT-PCR analysis. The details for qRT-PCR and data analysis were identical to those of a previous study40.
Transformations in peach fruit and tobacco
The full length CDSs of PpERF.A16 and PpNAC.A59 were amplified with the PCR primers listed in Table S5 and inserted in a sense orientation downstream of the CaMV 35S promoter in the pCAMBIA1301 vector to generate gene overexpression vectors. An approximately 400-bp fragment of the CDS of PpERF.A16 or PpNAC.A59 was amplified using the primers listed in Table S5 and inserted in an antisense orientation downstream of the CaMV 35 S promoter in the pSAK277 vector to create a gene silencing construct. The 400 bp fragments were located in the nonconserved region and showed no significant similarity (defined as no >21 bp in length with 100% identity) to the sequences of the most closely related genes within the same gene family (Figs. S11–S20). pCAMBIA1301 contains a hygromycin resistance gene, and pSAK277 contains a kanamycin resistance gene as a selection marker in plant transformation. These vectors were transferred into Agrobacterium tumefaciens EHA 105 cells that were used to transiently transform peach fruit tissues with a previously reported Agrobacterium infiltration protocol40. The fruit used for the transformation experiments was harvested from the peach cultivar Xiahui No. 5 at ~10 days before the commercial harvesting date. At three, four and five DAI, at least five fruits were sealed in a 5-L airtight jar, and six jars were used for each construct. Three DAI, ethylene production was measured every day. Fruit flesh tissues were sampled at 5 DAI for qRT-PCR analysis. All of the PCR primer sequences are listed in Supplementary Table S5.
Overexpression vectors of PpERF.A16 were also introduced into tobacco (Nicotiana tabacum) based on Agrobacterium-mediated transformation as described previously72. The regenerated tobacco plants were tested by PCR for amplification of the hygromycin gene from the pCAMBIA1301 vector. PCR-positive plants and wild-type control plants were used for measuring ethylene production.
Ethylene measurements
The ethylene production rate was measured using a gas chromatography system (GC2010, Shimadzu, Japan) with a 1000 μl rheodyne injector (Gaoge, Shangshai, China) according to a previously reported procotol73 with a few modifications. In brief, at room temperature, at least four fruits were weighed as a replicate and then placed into a 3-l airtight chamber. For tobacco, each plant was a replicate and individually weighed after collection from tissue culture medium and placed into a 15 ml centrifuge tube. Six replicates were used for each gene construct and each control. A 0.5-ml gas sample was collected from the chambers and tubes and injected into a gas chromatograph fitted with a 30-cm glass column (3.2 mm ID). Gas flows for N2, H2, and air were 40, 40, and 500 ml/min, respectively. The oven, injector, and FID temperatures were 300, 60, and 260 °C, respectively.
Protoplast isolation and dual-luciferase assay
Protoplast isolation was as described in a previous report40. Approximately 2000-bp sequences upstream of the initiation codons of PpACS1, PpACO1, PpendoPGM, and PpERF.A16 were inserted into a pGreenII 0800-LUC vector for a dual-luciferase assay that was performed according to a previous study40. At least six biological replicates were used in each assay. All of the primer sequences are listed in Table S5.
Yeast one-hybrid assay (Y1H)
It is well known that ERF TFs can bind to the GCC-box element74,75. Thus, the possible GCC-box elements were surveyed from the promoters of PpACS1, PpACO1, and PpEndoPGM (Fig. 2A). The fragments containing the possible GCC-box element were amplified from the genomic DNA of cv. Xiahui No.5 and then inserted into the pAbAi vector. The whole sequences of PpERF.A16 and PpNAC.A59 genes were individually inserted into the pGADT7 vector. The Y1H assay was performed using the Matchmaker Gold Yeast One-Hybrid Library Screening System (Clontech, Palo Alto, CA). All of the primer sequences are listed in Table S5.
Recombinant proteins of both PpERF.A16 and PpNAC.A59 in Escherichia coli
The whole sequences of PpERF.A16 and PpNAC.A59 were individually inserted into the pCold-TF expression vector (including a HIS tag) to create recombinant proteins. Recombinant plasmids were introduced into Escherichia coli cells and incubated at 37 °C until an OD660 of 0.6 was reached. Protein expression was induced at 16 °C for 24 h, and the recombinant protein was extracted and purified at 4 °C76. SDS-PAGE was conducted to analyze the extracted proteins. All of the primer sequences are listed in Table S5.
Kinetic assay
Fragments containing possible protein-binding elements were used to synthesize probes biotinylated at the ends of both the N- and C-termini (Sangon, Shanghai, China). Protein-DNA interactions were determined by biolayer interferometry using a ForteBio Octet 96 Red Instrument (Menlo Park, CA, USA). The kinetic assay comprised the following five steps: (1) baseline acquisition was conducted by loading PBS buffer (1.4 mM KH2PO4, 2.7 mM KCl, 4.3 mM Na2HPO4, 137 mM NaCl, 0.02% (w/v) Tween-20; pH = 7.2) onto streptavidin-coated biosensors, (2) the biotin-labeled probe was loaded onto the sensor, (3) a second baseline acquisition was conducted with the same PBS buffer, and steps (4) and (5) were the respective association and dissociation between the recombinant protein and the biotin-labeled probe(s). Equilibrium binding affinities (KD) were measured using nanomole quantities of samples as well as rates of association and dissociation (KA/KD)77.
Electrophoretic mobility shift assay (EMSA)
The recombinant proteins and biotin-labeled probes were also used for EMSAs. In brief, the binding reaction was performed in a 0.2 ml centrifuge tube for 20 min at room temperature, and the reaction mixtures contained 2 μl loading buffer, 1 mg poly(dI·dC), 5 mM MgCl2, 2.5% glycerol, 0.05% NP-40, 200 nM probe, and 1 μg recombinant protein, with ultrapure water added for a total volume of 20 μl. For probe competition, 2, 10, 20, 30, 40, or 50 μM nonlabeled probe was individually mixed into the reaction mixtures. After running in a 6% polyacrylamide gel, the protein-DNA complexes were electrophoretically transferred to a nylon membrane (GE Healthcare, Danderyd, Sweden). SDS-PAGE imaging was performed by a Tanon 5200 chemiluminescence imaging system (Tanon, Shanghai, China) and scanned using a ChemiDoc™ XRS+ System (BioRad, Hercules, CA).
Statistical analysis
Means and standard errors were calculated from at least three biological replicates for qRT-PCR analysis, five biological replicates for ethylene measurement in tobacco, and at least six replicates for ethylene measurement in peach fruit. Analysis of variance was calculated by Student’s t test. The lowercase letters a, b, and c indicate the level of significance at P < 0.05. Single, double and triple asterisks represent the levels of significance at P < 0.05, <0.01, and <0.001, respectively.
Supplementary information
Acknowledgements
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this paper. This work was supported by the Natural Science Foundations of China (31672118 and 31471856) and the Natural Science Foundations of Jiangsu Province (BK20140756).
Author contributions
C.G. and S.L.Z. conceived this study; C.G. conducted transcriptome-based data analyses, isolated the candidate genes by qRT-PCR, and drafted the paper. Z.H.G. contributed to most experiments with help from Y.J.Z. and Z.H.X. Y.Y.Z. collected peach samples at the developing and ripening stages. J.L.Y. and S.L.Z. revised this paper.
Conflict of interest
The authors declare no competing interests.
Supplementary information
The online version contains supplementary material available at 10.1038/s41438-021-00644-6.
References
- 1.Klee HJ, Giovannoni JJ. Genetics and control of tomato fruit ripening and quality attributes. Annu. Rev. Genet. 2011;45:41–59. doi: 10.1146/annurev-genet-110410-132507. [DOI] [PubMed] [Google Scholar]
- 2.Vrebalov J, et al. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (Rin) locus. Science. 2002;296:343–346. doi: 10.1126/science.1068181. [DOI] [PubMed] [Google Scholar]
- 3.Pech JC, Purgatto E, Bouzayen M, Latché A. Ethylene and fruit ripening. Annu. Plant Rev. 2012;44:275–304. doi: 10.1002/9781118223086.ch11. [DOI] [Google Scholar]
- 4.Merchante C, et al. Ethylene is involved in strawberry fruit ripening in an organ-specific manner. J. Exp. Bot. 2013;64:4421–4439. doi: 10.1093/jxb/ert257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Villarreal NM, Bustamante CA, Civello PM, Martínez GA. Effect of ethylene and 1-MCP treatments on strawberry fruit ripening. J. Sci. Food Agric. 2010;90:683–689. doi: 10.1002/jsfa.3868. [DOI] [PubMed] [Google Scholar]
- 6.Gu C, et al. Multiple regulatory roles of AP2/ERF transcription factor in angiosperm. Bot. Stud. 2017;58:6. doi: 10.1186/s40529-016-0159-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo HW, Ecker JR. The ethylene signaling pathway: new insights. Curr. Opin. Plant Biol. 2004;7:40–49. doi: 10.1016/j.pbi.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 8.Seymour GB, Ostergaard L, Chapman NH, Knapp S, Martin C. Fruit development and ripening. Annu. Rev. Plant Biol. 2013;64:219–241. doi: 10.1146/annurev-arplant-050312-120057. [DOI] [PubMed] [Google Scholar]
- 9.Li YC, et al. LeERF1 positively modulated ethylene triple response on etiolated seedling, plant development and fruit ripening and softening in tomato. Plant Cell Rep. 2007;26:1999–2008. doi: 10.1007/s00299-007-0394-8. [DOI] [PubMed] [Google Scholar]
- 10.Lee JM, et al. Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant J. 2012;70:191–204. doi: 10.1111/j.1365-313X.2011.04863.x. [DOI] [PubMed] [Google Scholar]
- 11.Xiao YY, et al. Banana ethylene response factors are involved in fruit ripening through their interactions with ethylene biosynthesis genes. J. Exp. Bot. 2013;64:2499–2510. doi: 10.1093/jxb/ert108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li T, et al. Apple (Malus domestica) MdERF2 negatively affects ethylene biosynthesis during fruit ripening by suppressing MdACS1 transcription. Plant J. 2016;88:735–748. doi: 10.1111/tpj.13289. [DOI] [PubMed] [Google Scholar]
- 13.Hao PP, et al. Transcriptome analysis unravels an ethylene response factor involved in regulating fruit ripening in pear. Physiol. Plant. 2018;163:124–135. doi: 10.1111/ppl.12671. [DOI] [PubMed] [Google Scholar]
- 14.Fu CC, et al. Papaya CpERF9 acts as a transcriptional repressor of cell-wall-modifying genes CpPME1/2 and CpPG5 involved in fruit ripening. Plant Cell Rep. 2016;35:2341–2352. doi: 10.1007/s00299-016-2038-3. [DOI] [PubMed] [Google Scholar]
- 15.Li T, et al. The jasmonate-activated transcription factor MdMYC2 regulates ETHYLENE RESPONSE FACTOR and ethylene biosynthetic genes to promote ethylene biosynthesis during apple fruit ripening. Plant Cell. 2017;29:1316–1334. doi: 10.1105/tpc.17.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yue P, et al. Auxin-activated MdARF5 induces the expression of ethylene biosynthesis genes to initiate apple fruit ripening. N. Phytol. 2020;226:1781–1795. doi: 10.1111/nph.16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breitel DA, et al. AUXIN RESPONSE FACTOR 2 intersects hormonal signals in the regulation of tomato fruit ripening. PLoS Genet. 2016;12:e1005903. doi: 10.1371/journal.pgen.1005903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hao YW, et al. Auxin response factor SlARF2 is an essential component of the regulatory mechanism controlling fruit ripening in tomato. PLoS Genet. 2015;11:e1005649. doi: 10.1371/journal.pgen.1005649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karlova R, et al. Transcriptional control of fleshy fruit development and ripening. J. Exp. Bot. 2014;65:4527–4541. doi: 10.1093/jxb/eru316. [DOI] [PubMed] [Google Scholar]
- 20.Weng L, et al. The zinc finger transcription factor SlZFP2 negatively regulates abscisic acid biosynthesis and fruit ripening in tomato. Plant Physiol. 2015;167:931–949. doi: 10.1104/pp.114.255174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao Y, et al. A NAC transcription factor, NOR-like1, is a new positive regulator of tomato fruit ripening. Hortic. Res. 2018;5:75. doi: 10.1038/s41438-018-0111-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osorio S, et al. Systems Biology of tomato fruit development: combined transcript, protein, and metabolite analysis of tomato transcription factor (nor, rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiol. 2011;157:405–425. doi: 10.1104/pp.111.175463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu M, et al. A new tomato NAC (NAM/ATAF1/2/CUC2) transcription factor, SlNAC4, functions as a positive regulator of fruit ripening and carotenoid accumulation. Plant Cell Physiol. 2014;55:119–135. doi: 10.1093/pcp/pct162. [DOI] [PubMed] [Google Scholar]
- 24.Meng C, et al. Suppression of tomato SlNAC1 transcription factor delays fruit ripening. J. Plant Physiol. 2016;193:88–96. doi: 10.1016/j.jplph.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 25.Wang WQ, et al. Genome-wide analysis of coding and non-coding RNA reveals a conserved miR164-NAC regulatory pathway for fruit ripening. N. Phytol. 2019;225:1618–1634. doi: 10.1111/nph.16233. [DOI] [PubMed] [Google Scholar]
- 26.Shan W, et al. Molecular characterization of banana NAC transcription factors and their interactions with ethylene signalling component EIL during fruit ripening. J. Exp. Bot. 2012;63:5171–5187. doi: 10.1093/jxb/ers178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rios P, et al. ETHQV6.3 is involved in melon climacteric fruit ripening and is encoded by a NAC domain transcription factor. Plant J. 2017;91:671–683. doi: 10.1111/tpj.13596. [DOI] [PubMed] [Google Scholar]
- 28.Zhou H, et al. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 2015;82:105–121. doi: 10.1111/tpj.12792. [DOI] [PubMed] [Google Scholar]
- 29.Fujisawa M, Nakano T, Shima Y, Ito Y. A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell. 2013;25:371–386. doi: 10.1105/tpc.112.108118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujisawa M, et al. Transcriptional regulation of fruit ripening by tomato FRUITFULL homologs and associated MADS box proteins. Plant Cell. 2014;26:89–101. doi: 10.1105/tpc.113.119453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayama H, Tatsuki M, Ito A, Kashimura Y. Ethylene and fruit softening in the stony hard mutation in peach. Postharvest Biol. Technol. 2006;41:16–21. doi: 10.1016/j.postharvbio.2006.03.006. [DOI] [Google Scholar]
- 32.Tonutti P, Casson P, Ramina A. Ethylene biosynthesis during peach fruit development. J. Am. Soc. Hortic. Sci. 1991;116:274–279. doi: 10.21273/JASHS.116.2.274. [DOI] [Google Scholar]
- 33.Hayama H, Shimada T, Fujii H, Ito A, Kashimura Y. Ethylene-regulation of fruit softening and softening-related genes in peach. J. Exp. Bot. 2006;57:4071–4077. doi: 10.1093/jxb/erl178. [DOI] [PubMed] [Google Scholar]
- 34.Gu C, et al. Copy number variation of a gene cluster encoding endopolygalacturonase mediates flesh texture and stone adhesion in peach. J. Exp. Bot. 2016;67:1993–2005. doi: 10.1093/jxb/erw021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bregoli AM, et al. Peach (Prunus persica) fruit ripening: aminoethoxyvinylglycine (AVG) and exogenous polyamines affect ethylene emission and flesh firmness. Physiol. Plant. 2002;114:472–481. doi: 10.1034/j.1399-3054.2002.1140317.x. [DOI] [PubMed] [Google Scholar]
- 36.Hayama H, Tatsuki M, Ito A, Kashimura Y. Combined treatment of aminoethoxyvinylglycine (AVG) and 1-methylcyclopropene (1-MCP) reduces melting-flesh peach fruit softening. Postharvest Biol. Technol. 2008;50:228–230. doi: 10.1016/j.postharvbio.2008.05.003. [DOI] [Google Scholar]
- 37.Tadiello A, et al. On the role of ethylene, auxin and a GOLVEN-like peptide hormone in the regulation of peach ripening. BMC Plant Biol. 2016;16:44. doi: 10.1186/s12870-016-0730-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tatsuki M, Haji T, Yamaguchi M. The involvement of 1-aminocyclopropane-1- carboxylic acid synthase isogene, Pp-ACS1, in peach fruit softening. J. Exp. Bot. 2006;57:1281–1289. doi: 10.1093/jxb/erj097. [DOI] [PubMed] [Google Scholar]
- 39.Tonutti P, Bonghi C, Ruperti B, Tornielli GB, Ramina A. Ethylene evolution and 1-aminocyclopropane-1-carboxylate oxidase gene expression during early development and ripening of peach fruit. J. Am. Soc. Hortic. Sci. 1997;122:642–647. doi: 10.21273/JASHS.122.5.642. [DOI] [Google Scholar]
- 40.Gu C, et al. A HD-ZIP II HOMEOBOX transcription factor, PpHB.G7, mediates ethylene biosynthesis during fruit ripening in peach. Plant Sci. 2019;278:12–19. doi: 10.1016/j.plantsci.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 41.Guo ZH, et al. Expression analysis of TCP genes in peach reveals an involvement of PpTCP.A2 in ethylene biosynthesis during fruit ripening. Plant Mol. Biol. Report. 2018;36:588–595. doi: 10.1007/s11105-018-1105-z. [DOI] [Google Scholar]
- 42.Ge H, et al. EjNAC3 transcriptionally regulates chilling-induced lignification of loquat fruit via physical interaction with an atypical CAD-like gene. J. Exp. Bot. 2017;68:5129–5136. doi: 10.1093/jxb/erx330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nieuwenhuizen NJ, et al. Natural variation in monoterpene synthesis in kiwifruit: transcriptional regulation of terpene synthases by NAC and ETHYLENE-INSENSITIVE3-like transcription factors. Plant Physiol. 2015;167:1243–1258. doi: 10.1104/pp.114.254367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhong RQ, Lee CH, Ye ZH. Global analysis of direct targets of secondary wall NAC master switches in arabidopsis. Mol. Plant. 2010;3:1087–1103. doi: 10.1093/mp/ssq062. [DOI] [PubMed] [Google Scholar]
- 45.Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006;140:411–432. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang CH, et al. Genome-wide analysis of the AP2/ERF superfamily in peach (Prunus persica) Genet. Mol. Res. 2012;11:4789–4809. doi: 10.4238/2012.October.17.6. [DOI] [PubMed] [Google Scholar]
- 47.Licausi F, Ohme-Takagi M, Perata P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. N. Phytol. 2013;199:639–649. doi: 10.1111/nph.12291. [DOI] [PubMed] [Google Scholar]
- 48.Trainotti L, Tadiello A, Casadoro G. The involvement of auxin in the ripening of climacteric fruits comes of age: the hormone plays a role of its own and has an intense interplay with ethylene in ripening peaches. J. Exp. Bot. 2007;58:3299–3308. doi: 10.1093/jxb/erm178. [DOI] [PubMed] [Google Scholar]
- 49.Pan L, et al. PpYUC11, a strong candidate gene for the stony hard phenotype in peach (Prunus persica L. Batsch), participates in IAA biosynthesis during fruit ripening. J. Exp. Bot. 2015;66:7031–7044. doi: 10.1093/jxb/erv400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cirilli M, et al. Integrative genomics approaches validate PpYUC11-like as candidate gene for the stony hard trait in peach (P. persica L. Batsch) BMC Plant Biol. 2018;18:88. doi: 10.1186/s12870-018-1293-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu R, et al. Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol. 2010;10:145. doi: 10.1186/1471-2229-10-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nuruzzaman M, et al. Genome-wide analysis of NAC transcription factor family in rice. Gene. 2010;465:30–44. doi: 10.1016/j.gene.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 53.Jensen MK, Skriver K. NAC transcription factor gene regulatory and protein-protein interaction networks in plant stress responses and senescence. IUBMB Life. 2014;66:156–166. doi: 10.1002/iub.1256. [DOI] [PubMed] [Google Scholar]
- 54.Kim HJ, Nam HG, Lim PO. Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 2016;33:48–56. doi: 10.1016/j.pbi.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 55.Olsen AN, Ernst HA, Lo Leggio L, Skriver K. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 2005;10:79–87. doi: 10.1016/j.tplants.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 56.Zhong RQ, Demura T, Ye ZH. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell. 2006;18:3158–3170. doi: 10.1105/tpc.106.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shang XG, et al. A cotton NAC transcription factor GhirNAC2 plays positive roles in drought tolerance via regulating ABA biosynthesis. Plant Sci. 2020;296:110498. doi: 10.1016/j.plantsci.2020.110498. [DOI] [PubMed] [Google Scholar]
- 58.Ren TT, et al. Involvement of NAC transcription factor SiNAC1 in a positive feedback loop via ABA biosynthesis and leaf senescence in foxtail millet. Planta. 2018;247:53–68. doi: 10.1007/s00425-017-2770-0. [DOI] [PubMed] [Google Scholar]
- 59.Mao CJ, et al. A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol. 2017;174:1747–1763. doi: 10.1104/pp.17.00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma XM, et al. The NAC Transcription Factor SlNAP2 Regulates Leaf Senescence and Fruit Yield in Tomato. Plant Physiol. 2018;177:1286–1302. doi: 10.1104/pp.18.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang M, Yuan B, Leng P. The role of ABA in triggering ethylene biosynthesis and ripening of tomato fruit. J. Exp. Bot. 2009;60:1579–1588. doi: 10.1093/jxb/erp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kou XH, et al. The interplay between ABA/ethylene and NAC TFs in tomato fruit ripening: a review. Plant Mol. Biol. 2021;106:223–238. doi: 10.1007/s11103-021-01128-w. [DOI] [PubMed] [Google Scholar]
- 63.Shinozaki Y, et al. High-resolution spatiotemporal transcriptome mapping of tomato fruit development and ripening. Nat. Commun. 2018;9:364. doi: 10.1038/s41467-017-02782-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lang Z, et al. Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc. Natl Acad. Sci. 2017;114:E4511–E4519. doi: 10.1073/pnas.1705233114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu R, et al. A DEMETER-like DNA demethylase governs tomato fruit ripening. Proc. Natl Acad. Sci. 2015;112:10804–10809. doi: 10.1073/pnas.1503362112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gao C, et al. MicroRNA profiling analysis throughout tomato fruit development and ripening reveals potential regulatory role of RIN on microRNAs accumulation. Plant Biotechnol. J. 2015;13:370–382. doi: 10.1111/pbi.12297. [DOI] [PubMed] [Google Scholar]
- 67.Moxon S, et al. Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res. 2008;18:1602–1609. doi: 10.1101/gr.080127.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu B, et al. RNA sequencing and functional analysis implicate the regulatory role of long non-coding RNAs in tomato fruit ripening. J. Exp. Bot. 2015;66:4483–4495. doi: 10.1093/jxb/erv203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li R, Fu D, Zhu B, Luo Y, Zhu H. CRISPR/Cas9-mediated mutagenesis of lncRNA1459 alters tomato fruit ripening. Plant J. 2018;94:513–524. doi: 10.1111/tpj.13872. [DOI] [PubMed] [Google Scholar]
- 70.Lu P, et al. Genome encode analyses reveal the basis of convergent evolution of fleshy fruit ripening. Nat. Plants. 2018;4:784–791. doi: 10.1038/s41477-018-0249-z. [DOI] [PubMed] [Google Scholar]
- 71.Gu C, et al. RNA-Seq analysis unveils gene regulation of fruit size cooperatively determined by velocity and duration of fruit swelling in peach. Physiol. Plant. 2018;164:320–336. doi: 10.1111/ppl.12736. [DOI] [PubMed] [Google Scholar]
- 72.Liu Y, et al. A WRKY transcription factor PbrWRKY53 from Pyrus betulaefolia is involved in drought tolerance and AsA accumulation. Plant Biotechnol. J. 2019;17:1770–1787. doi: 10.1111/pbi.13099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fan X, Argenta L, Mattheis JP. Inhibition of ethylene action by 1-methylcyclopropene prolongs storage life of apricots. Postharvest Biol. Technol. 2000;20:135–142. doi: 10.1016/S0925-5214(00)00121-6. [DOI] [Google Scholar]
- 74.Han YC, et al. Banana transcription factor MaERF11 recruits histone deacetylase MaHDA1 and represses the expression of MaACO1 and expansins during fruit ripening. Plant Physiol. 2016;171:1070–1084. doi: 10.1104/pp.16.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yin XR, et al. Involvement of an ethylene response factor in chlorophyll degradation during citrus fruit degreening. Plant J. 2016;86:403–412. doi: 10.1111/tpj.13178. [DOI] [PubMed] [Google Scholar]
- 76.Chen J, et al. Phosphatidic Acid Counteracts S-RNase Signaling in Pollen by Stabilizing the Actin Cytoskeleton. Plant Cell. 2018;30:1023–1039. doi: 10.1105/tpc.18.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sultana A, Lee JE. Measuring protein-protein and protein-nucleic acid interactions by biolayer interferometry. Curr. Protoc. Protein Sci. 2015;79:19.25.1–19.25.26. doi: 10.1002/0471140864.ps1925s79. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.