Abstract
To understand the role of POL30 in mutation suppression, 11 Saccharomyces cerevisiae pol30 mutator mutants were characterized. These mutants were grouped based on their mutagenic defects. Many pol30 mutants harbor multiple mutagenic defects and were placed in more than one group. Group A mutations (pol30-52, -104, -108, and -126) caused defects in mismatch repair (MMR). These mutants exhibited mutation rates and spectra reminiscent of MMR-defective mutants and were defective in an in vivo MMR assay. The mutation rates of group A mutants were enhanced by a msh2 or a msh6 mutation, indicating that MMR deficiency is not the only mutagenic defect present. Group B mutants (pol30-45, -103, -105, -126, and -114) exhibited increased accumulation of either deletions alone or a combination of deletions and duplications (4 to 60 bp). All deletion and duplication breakpoints were flanked by 3 to 7 bp of imperfect direct repeats. Genetic analysis of one representative group B mutant, pol30-126, suggested polymerase slippage as the likely mutagenic mechanism. Group C mutants (pol30-100, -103, -105, -108, and -114) accumulated base substitutions and exhibited synergistic increases in mutation rate when combined with msh6 mutations, suggesting increased DNA polymerase misincorporation as a mutagenic defect. The synthetic lethality between a group A mutant, pol30-104, and rad52 was almost completely suppressed by the inactivation of MSH2. Moreover, pol30-104 caused a hyperrecombination phenotype that was partially suppressed by a msh2 mutation. These results suggest that pol30-104 strains accumulate DNA breaks in a MSH2-dependent manner.
Cancer can be viewed as a genetic disease whereby the accumulation of mutations leads to the eventual activation of cellular oncogenes or inactivation of tumor suppressor genes (53). These events, in turn, confer growth or survival advantages to tumor cells. This framework predicts that genetic defects causing increased mutation rates should predispose cells to carcinogenesis. Such a prediction has been fulfilled by the discovery that inherited defects in DNA mismatch repair genes underlie the etiology of hereditary nonpolyposis colon carcinoma, a cancer predisposition syndrome (22, 44). Given this example, it seems likely that other genetic defects giving rise to mutator phenotypes should also contribute to carcinogenesis. One potential class of such mutator genes is the group of genes encoding DNA replication accessory factors, since these factors modulate the fidelity of DNA replication as well as participate in various aspects of DNA repair.
Proliferating cell nuclear antigen (PCNA) is a replication accessory factor encoded by the essential gene POL30 in Saccharomyces cerevisiae (4). PCNA forms a homotrimeric ring around the DNA that serves as a binding platform for DNA polymerases. In doing so, PCNA enhances the processivity of DNA polymerases (26, 51). PCNA homologues from various eukaryotes share a high degree of sequence identity (19), and despite the lack of significant primary sequence homology between the Escherichia coli DNA polymerase III processivity factor and PCNA homologues, the three-dimensional crystal structures of these two proteins are superimposible (24, 26). This strong evolutionary conservation suggests that information obtained about PCNA in lower eukaryotes may be applicable to more complex organisms.
In addition to serving as a polymerase processivity factor, PCNA modulates the fidelity of DNA synthesis in vitro (41) as well as interacts with factors involved in nucleotide excision repair (9), mismatch repair (MMR) (18, 52), and base excision repair (42). PCNA also participates in the processing of branched DNA structures, including those formed during lagging-strand DNA synthesis (31, 56). The abundance of PCNA-interacting proteins led to the suggestion that PCNA may serve as a “docking bay” for many proteins and, in the process, coordinate various aspects of DNA synthesis and repair (19). Of the above-mentioned processes, DNA polymerase fidelity, MMR, and branch structure processing are mutation suppressing mechanisms pertinent to the results presented in this report.
Three major factors determine the ability of a polymerase to conduct faithful DNA replication, and all three are modulated by PCNA (5, 16, 30, 41): the selection of nucleotides that properly pair with the template (nucleotide discrimination), the 3′→5′ exonuclease activity that removes misincorporated nucleotides (proofreading), and adherence to the template during DNA replication (processivity). Defects in nucleotide discrimination or proofreading could lead to base substitution formation via nucleotide misincorporation. Failure of the DNA polymerase to adhere to the template properly could result in polymerase slippage, causing frameshifts, deletions, or duplications (27). Since specific types of mutations are associated with particular DNA polymerase defects, the defects of a mutant polymerase may be inferred by an analysis of the types of mutations that it caused. For instance, the pol2-4 allele in S. cerevisiae encodes a proofreading-deficient form of DNA polymerase ɛ that causes a roughly 10-fold increase in mutation rate. Not surprisingly, the mutation spectrum of pol2-4 is notable for an increased accumulation of base substitutions (40). Another example is the pol3-t allele, which encodes a temperature-sensitive (ts) form of DNA polymerase δ. This mutation facilitates the formation of deletions flanked by short repeats, suggesting that pol3-t increases polymerase slippage (48, 49).
In addition to modulating polymerase fidelity, PCNA is also required for MMR in a human crude extract system (52). MMR is an evolutionarily conserved process which corrects misincorporated bases that escape DNA polymerase proofreading. Cells completely defective in MMR exhibit a 10- to 1,000-fold increase in the rate of mutation accumulation. Most of these mutations occur as frameshift mutations in mononucleotide runs (13, 35, 47). In S. cerevisiae, MMR initiates with the binding of mismatches by the MSH2-MSH6 or the MSH2-MSH3 heterodimer. Subsequent to this binding, other factors are recruited to the mismatch, leading to the eventual nicking, degradation, and resynthesis of the DNA strand containing the mutagenic nucleotide (23). In E. coli, this strand discrimination is mediated by (i) transient undermethylation of newly synthesized GATC sites by the Dam methylase and (ii) activation of MutH, the MMR-initiating endonuclease, to cleave the unmethylated DNA strand at hemimethylated GATC sites (38). The mechanism of strand discrimination in eukaryotes, where DNA methylation is probably not involved, is less clear. The observation that PCNA functions in MMR at a step prior to strand excision (14, 52) raises the possibility that PCNA functions at the initiation stages of MMR and that this may involve coupling of mismatch repair proteins to the replication machinery by PCNA (11, 22, 28, 39).
In vitro studies suggest that PCNA may participate in the processing of branched DNA structures via its interaction with RAD27 (31, 56). RAD27 (also known as RTH1, FEN1, MF-1, and exonuclease IV) is an evolutionarily conserved protein that has both a 5′ flap endonuclease and a 5′-to-3′ exonuclease activity. These activities are crucial for the processing of Okazaki fragments during DNA replication (32, 55). S. cerevisiae rad27 mutants accumulate double-strand breaks (DSBs) and exhibit a strong mutator phenotype (47). The mutations that arise in rad27 mutants are predominately duplications where the region duplicated is flanked by short stretches of imperfect direct repeats. These duplications are believed to result from mutagenic repair of DNA strand breaks involving misannealing of short single-stranded DNA tails (21, 47). Similar duplications have been reported as inactivating somatic mutations in the adenomatous polyposis coli as well as the p53 gene (12, 47).
Since PCNA participates in many processes that normally contribute to mutation suppression, we screened a panel of previously isolated S. cerevisiae pol30 mutants for mutator phenotypes. Eleven pol30 mutator mutants were identified. Mutation rate, spectrum, and epistasis analyses suggest that PCNA functions in multiple mutation-suppressing processes, ranging from polymerase fidelity to MMR. Detailed characterization of one MMR-defective mutant (pol30-104) yielded genetic evidence consistent with the notion that PCNA plays an important role in strand discrimination during MMR.
MATERIALS AND METHODS
General genetic methods.
YPD (yeast extract-peptone-dextrose), sporulation, SD (synthetic dextrose), 5FOA (5-fluoroorotic acid), and canavanine media were prepared as previously described (6). Transformations were also performed as previously described (6). All strains were propagated at 37°C. Chromosomal DNA preparations were isolated by using glass bead lysis (18a) or Puregene kits (Gentra). PCR was performed in 50- to 100-μl reactions containing 0.5 U of Klentaq DNA polymerase (Ab Peptides), 0.08 U of Pfu DNA polymerase (Stratagene), 20 ng of genomic DNA, and 10 pmol of each primer in 1× PCII buffer (Ab Peptides). Primers were synthesized by the Dana-Farber Cancer Institute Molecular Biology Core Facility or Cybersyn Corp (Lenni, Pa.). Unless otherwise noted, standard three-temperature PCR was performed. Prior to DNA sequencing, PCR products were purified by using QIAquick Spin PCR purification kits (Qiagen). All DNA sequencing was performed with a Perkin Elmer/Applied Biosystems 377 DNA sequencer and dye terminator chemistry according to the manufacturer’s instructions.
Mutation rates and recombination rates were calculated by fluctuation analysis using the method of the median in experiments with sets of five independent cultures as previously described (29, 35). For the recombination/chromosomal loss experiment, independent colonies of approximately 1.5-mm diameter were inoculated into 5 ml of YPD and grown overnight at 30°C. Appropriate dilutions were then plated onto SD complete as well as canavanine-containing plates. The colonies were scored after 3 days. The colonies on canavanine plates were then replica plated onto SD −Thr (threonine-deficient SD) plates, and the SD −Thr plates were scored after further incubation at 30°C for 1 day. Statistical comparisons were done by the Mann-Whitney test, the chi-square test statistic, or the two-tailed t-test statistics (7). All rates shown represent the average of two or more independent fluctuation analyses. Mutation spectra were determined by PCR amplification of target regions followed by DNA sequence analysis of one mutant per independent culture as previously described (6, 35, 47).
Overexpression of RAD27 by pRDK762 in RDKY3578 was induced by inoculating isolates into SD −Ura containing 2% galactose and 2% raffinose. Appropriate dilutions of these cultures were grown to saturation at 30°C and then plated onto SD −Ura −Lys plates containing 2% galactose and 2% raffinose as well as SD −Ura plates containing 2% galactose and 2% raffinose. Colonies were scored after 5 days. Parallel experiments were performed with RDKY3588, which harbors the vector control. Mutation rates were calculated as described above.
Strain and plasmid construction.
All haploid strains used were derivatives of S288C parental strains provided by Fred Winston (Harvard Medical School). Five series of strains, all derived from RDKY3023 (MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8), were constructed. The first series (RDKY3546 to RDKY3548) was derived by transforming RDKY3023 with TRP1 ARS-CEN plasmids bearing pol30-41, -45, and -52 (pBL245-41, pBL230-45, pBL230-52; reagents kindly provided by Peter Burgers, Washington University, St. Louis, Mo. [3]). The POL30 gene in the first series was disrupted with a hisG-URA3-hisG cassette by transforming with MluI-KpnI-digested pBL243 (also provided by Peter Burgers) to generate the second series of strains, consisting of RDKY3549 to RDKY3551. Proper disruptions were verified by PCR amplification with 5′-TCA CAC TGA TGT AGT GGT GG and 5′-CCT GTT CCA CCG GCC CAG GG. The third series (RDKY3552 to RDKY3561) was derived by replacing POL30 in RDKY3023 with LEU2-tagged pol30 alleles by transformation with SacI-digested pCH1572 (POL30), pCH1573 (pol30-100), pCH1575 (pol30-102), pCH1576 (pol30-103), pCH1577 (pol30-104), pCH1579 (pol30-105), pCH1580 (pol30-106), pCH1578 (pol30-108), pCH1637 (pol30-126), and pCH1638 (pol30-114) (2, 37). Proper integrants were confirmed by PCR amplification with 5′-CGA ATT GAC CTT CTA CTG GGA and 5′-TGT CGC CGA AGA AGT TAA GA. The fourth series (RDKY3543 and RDKY3562 to RDKY3569) was derived by disrupting the MSH2 gene of the third series with a hisG-URA3-hisG cassette by transformation with AatII-PvuII-digested pRDK351 (35). Proper integrants were confirmed by PCR amplification with 5′-TCA AGT CAA TAC TTA AAC GCC and 5′-GAT TCG GTA ATC TCC GAA CAG AAG G. The fifth series (RDKY3544 and RDKY3570 to RDKY3577) was derived by disruption the MSH6 gene of the third series with a hisG-URA3-hisG cassette by transformation with EcoRI-SphI-digested pEAI108 (provided by Eric Alani, Cornell University). Proper integrants were confirmed by PCR amplification with 5′-CCA TTG ATG CGG ACG AAA ACT CGG and 5′-ATT TGC AAA GGG AAG GGA TG. RDKY3588 was constructed by transforming RDKY3560 with pCH1071 (URA3 GAL). RDKY3578 was constructed by transforming RDKY3560 with pRDK762 (URA3 GAL-RAD27). RDKY3668 was constructed by transforming RDKY3552 with pRDK762 (URA3 GAL-RAD27).
RDKY3545 was constructed by transforming RDKY3023 with pCH1511 (URA3 POL30). Likewise, RDKY3583 was constructed by transforming RDKY3556 with pCH1511 (URA3 POL30). 5′-GAA AAG ACG AAA AAT ATA GCG GCG GGC GGG TTA CGC GAC CGG TAT CGA GGC CTC CTC TAG TAC ACT C and 5′-TAA TAA ATA ATG ATG CAA ATT TTT TAT TTG TTT CGG CCA GGA AGC GTT GCG CGC CTC GTT CAG AAT G were used to amplify a HIS3-bearing RAD52 disruptor fragment. This fragment was used to disrupt the RAD52 gene in RDKY3583 to generate RDKY3585. The correct disruption was verified with 5′-ACG ACA CAT GGA GGA AAG AAA AAC T and 5′-CTC TCC CGT TAG TGA TTC TCG ATG. RDKY3584 was constructed by transforming RDKY2709, a strain that is isogenic to RDKY3023 except that the MSH2 gene is disrupted with hisG, with pCH1511 (URA3 POL30). POL30 in this strain was replaced with pol30-104.LEU2 by transformation with SacI-digested pCH1577 (2). Finally, the RAD52 gene in this strain was disrupted as described above to generate RDKY3586. RDKY3587 and RDKY3669 were constructed by transforming RDKY3586 and RDKY3585, respectively, with pRDK447 (TRP1 MSH2).
The diploid strain RDKY3579 was derived by crossing CH2165 (MATa ura3-52 leu2 LEU2.POL30) and CH2338 (MATα leu2 can1-51 hom3). RDKY3581 was derived by crossing CH2161 (MATa ura3-52 leu2 LEU2 pol30-104) and CH2340 (MATα leu2 can1-51 hom3 LEU2.pol30-104). 5′-ATG TCC TCC ACT AGG CCA GAG CTA AAA TTC TCT GAT GTA TCA GAG AGC AGC TGA AGC TTC GTA CGC and 5′-TTA TAA CAA CAA GGC TTT TAT ATA TTT CAG GTA ATT ATC GTT TTC CTT TTG CAT AGG CCA CTA GTG GAT CTG were used to PCR amplify a G418-bearing MSH2 disruptor fragment (54). The MSH2 gene in CH2165, CH2338, CH2161, and CH2340 was disrupted with the PCR fragment. Proper disruptions were confirmed by PCR amplification with 5′-CCA AAA ATC CAA TCA GAA CTC CAG and 5′-TGT ACC CAA TTC GTT CGG ACC TA. The resulting strains were crossed to yield RDKY3580 and RDKY3582. The genotypes of all diploids generated were further verified with POL30-specific primers (5′-CGA ATT GAC CTT CTA CTG GGA and 5′-TGT CGC CGA AGA AGT TAA GA) and MSH2-specific primers (5′-CCA AAA ATC CAA TCA GAA CTC CAG and 5′-TGT ACC CAA TTC GTT CGG ACC TA). All strains constructed for the purpose of this study are shown in Table 1.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype |
|---|---|
| RDKY3023a | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 |
| RDKY2706 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 MSH2::hisG |
| RDKY3545 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 [URA3 POL30] |
| RDKY3546 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 [TRP1 pol30-41] |
| RDKY3547 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 [TRP1 pol30-45] |
| RDKY3548 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 [TRP1 pol30-52] |
| RDKY3549 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 POL30::hisG-URA3-hisG [TRP1 pol30-41] |
| RDKY3550 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 POL30::hisG-URA3-hisG[TRP1 pol30-45] |
| RDKY3551 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 POL30::hisG-URA3-hisG[TRP1 pol30-52] |
| RDKY3552 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.POL30 |
| RDKY3553 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-100 |
| RDKY3554 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-102 |
| RDKY3555 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-103 |
| RDKY3556 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-104 |
| RDKY3557 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-105 |
| RDKY3558 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-106 |
| RDKY3559 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-108 |
| RDKY3560b | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-126 |
| RDKY3561 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-114 |
| RDKY3543 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.POL30 msh2::hisG-URA3-hisG |
| RDKY3562 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-100 msh2::hisG-URA3-hisG |
| RDKY3563 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-102 msh2::hisG-URA3-hisG |
| RDKY3564 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-103 msh2::hisG-URA3-hisG |
| RDKY3565 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-104 msh2::hisG-URA3-hisG |
| RDKY3566 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-105 msh2::hisG-URA3-hisG |
| RDKY3567 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-108 msh2::hisG-URA3-hisG |
| RDKY3568b | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-126 msh2::hisG-URA3-hisG |
| RDKY3569 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-114 msh2::hisG-URA3-hisG |
| RDKY3544 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.POL30 msh6::hisG-URA3-hisG |
| RDKY3570 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-100 msh6::hisG-URA3-hisG |
| RDKY3571 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-102 msh6::hisG-URA3-hisG |
| RDKY3572 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-103 msh6::hisG-URA3-hisG |
| RDKY3573 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-104 msh6::hisG-URA3-hisG |
| RDKY3574 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-105 msh6::hisG-URA3-hisG |
| RDKY3575 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-108 msh6::hisG-URA3-hisG |
| RDKY3576b | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-126 msh6::hisG-URA3-hisG |
| RDKY3577 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-114 msh6::hisG-URA3-hisG |
| RDKY3578b | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-126 [pRDK762 (URA3 GAL-RAD27)] |
| RDKY3588b | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-126 [pCH1071 (URA3 GAL)] |
| RDKY3668 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.POL30 [pRDK762 (URA3 GAL-RAD27)] |
| RDKY3579 | MATa ura3-52 leu2 LEU2.POL30 |
| MATα leu2 can1-51 hom3 | |
| RDKY3580 | MATa ura3-52 leu2 LEU2.POL30 MSH2::G418 |
| MATα leu2 can1-51 hom3 MSH2::G418 | |
| RDKY3581 | MATa ura3-52 leu2 LEU2 pol30-104 |
| MATα leu2 can1-51 hom3 LEU2 pol30-104 | |
| RDKY3582 | MATa ura3-52 leu2 LEU2 pol30-104 MSH2::G418 |
| MATα leu2 can1-51 hom3 LEU2 pol30-104 MSH2::G418 | |
| RDKY3583 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-104 [pCH1511 (URA3 POL30)] |
| RDKY3584 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-104 msh2::HisG [pCH1511 (URA3 POL30)] |
| RDKY3585 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-104 rad52::His3 [pCH1511 (URA3 POL30)] |
| RDKY3586 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-104 rad52::His3 msh2::hisG [pCH1511 (URA3 POL30)] |
| RDKY3587 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-104 rad52::his3 msh2::hisG [pCH1511(URA3 POL30)] [pRDK447 (TRP1 MSH2)] |
| RDKY3669 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 LEU2.pol30-104 msh2::hisG [pCH1511 (URA3 POL30)] [pRDK447 (TRP1 MSH2)] |
The nature of the ade8 mutation in RDKY3023 was previously undetermined. In the course of this study, we determined that the ade8 mutation resulted from the deletion of a CT dinucleotide at positions 104 and 105 (where the A of the initiating methionine codon is nucleotide +1).
pol30-112 was originally described as a mutation that changed nucleotide 719 from an A to a G (2). However, on resequencing the original pol30-112 plasmid, we found that the originally described mutation was present along with a second T→A change at nucleotide position 443, resulting in a V→D change at amino acid position 148. Thus, the original pol30-112 allele was a double mutant. When this double mutation was transferred to the strains constructed here by gene replacement, only the N-terminal T→A change at nucleotide position 443 was inherited and was responsible for the phenotypes observed. Because this resulted in the creation of a new allele, we have named this allele pol30-126 in accordance with the numbering used in reference 2.
A two-step method was used to construct the GAL-RAD27 overexpression plasmid (pRKY762). In the first step, the RAD27 gene was amplified from genomic DNA by using primers 5′-GAG CTC GAG ATG GGT ATT AAA GGT TTG AAT G and 5′-GCG CTC GAG CGA TGG TTC CGA TAT GCC AAA AGC. The resulting PCR fragment contains AvaI sites on both ends of RAD27. The fragment was gel purified, digested with AvaI and HindIII (cleaves at +1393 of RAD27), and ligated into SalI-HindIII-digested YEp352 (URA3 2μm). The second step involved excising the RAD27 coding region plus 12 bp of plasmid by digestion with BamHI-HindIII and ligating this fragment into BamHI-HindIII digested pCH1071 (pGAL YCp50). The resulting construct was sequenced with the following primers to verify that the RAD27 fragment cloned was free of mutations: 5′-ATG GGT ATT AAA GGT TTG AAT G, 5′-TAC CGT TAT CAA TCA TTC TCA GTG, 5′-TCA AGA AGG GTG GAA ACA GAA A, 5′-GGC GAC CAT TTC AAG TTT ATT TC, 5′-TCG CCA CCA AAG GAG AAG GAA CT, and CGA TGG TTC CGA TAT GCC AAA AGC. pRKY762 was shown to fully rescue the ts, methyl methanesulfonate-sensitive (MMSs), and mutator phenotypes of rad27 null mutants in both glucose- and galactose-containing media.
In vivo MMR assay.
The phagemids and most methods used to construct the mismatch-containing heteroduplexes were as previously reported (25). The heteroduplexes were purified by either benzoylated naphthoylated DEAE-cellulose chromatography followed by exonuclease V digestion (33) or high-pressure liquid chromatography (1). Comparable results were obtained in assays using substrates purified by using either protocol. The heteroduplex DNA was transformed into appropriate S. cerevisiae strains via lithium acetate or electroporation transformation. The transformants were plated onto SD −Ura plates supplemented with 6 mg of adenine per liter, incubated at 37°C, and scored after 5 days.
RESULTS
POL30 mutants exhibit variable increases in spontaneous mutation rates.
We screened a panel of previously isolated S. cerevisiae pol30 mutants (2, 3) for mutator phenotypes. Since many of the pol30 mutants were cold sensitive, mutation rates were assessed at 37°C. Three mutator assays were used: an assay that scores for arginine permease inactivation (CAN1), an assay that detects reversion of a +4 insertion in the LYS2 gene (lys2-Bgl), and an assay that detects reversion of a +1 insertion in the HOM3 gene (hom3-10) (13, 35, 47). Whereas the lys2-Bgl and the hom3-10 assays are particularly sensitive indicators of defective MMR, the CAN1 assay is less specific for defective MMR and more sensitive to other mutagenic pathways (6, 13, 35, 47, 50). Eleven of the twelve mutants examined exhibited elevated mutation rates in all three assays (Table 2, set A). Of these mutants, pol30-52 and -104 exhibited the most dramatic mutator phenotypes, particularly in assays sensitive to defective MMR (41- and 21-fold increases in lys2-Bgl, respectively; 243- and 324-fold increases in hom3-10, respectively). However, these increases were lower (Mann-Whitney test, P < 0.05) than those caused by an msh2 mutation, a mutation thought to completely inactivate MMR (82-fold in lys2-Bgl; 500-fold in hom3-10). These observations suggest that pol30-52 and -104 did not completely inactivate MMR. The comparable CAN1 mutation rates of the pol30-52, pol30-104, and msh2 mutants (15-, 26-, and 18-fold increases, respectively), then, suggests that pol30-52 and -104 caused mutagenic defects that are not related to MMR in addition to causing partial defects in MMR. The mutator phenotypes of the other pol30 mutants were more subtle. pol30-100, -108, -126, and -114 caused modest rate elevations that were most evident in the hom3-10 assay. pol30-41, -45, -102, -103, and -105 caused weak rate elevations in all three assays.
TABLE 2.
Mutation rate analysis
| Genotype | RDKY no. | Group(s) | Mutation ratea
|
||
|---|---|---|---|---|---|
| CAN1 | hom3-10 | lys2-bgl | |||
| Set a (pol30 mutants) | |||||
| POL30 | 3552 | 3.6 × 10−7 (1) | 1.0 × 10−8 (1) | 1.7 × 10−8 (1) | |
| POL30/POL30 | 3545 | 3.3 × 10−7 (1) | 1.2 × 10−8 (1) | 1.9 × 10−8 (1) | |
| pol30-41/POL30 | 3546 | 6.5 × 10−7 (2) | n.d. | 6.9 × 10−8 (3) | |
| pol30-45/POL30 | 3547 | 2.2 × 10−7 (1) | n.d. | 1.8 × 10−8 (1) | |
| pol30-52/POL30 | 3548 | 11 × 10−7 (3) | 57 × 10−8 (57) | 63 × 10−8 (37) | |
| pol30-41 | 3549 | 6.4 × 10−7 (2) | 2.7 × 10−8 (3) | 4.1 × 10−8 (2) | |
| pol30-45 | 3550 | B | 4.8 × 10−7 (1) | 1.5 × 10−8 (2) | 3.4 × 10−8 (2) |
| pol30-52 | 3551 | A | 55 × 10−7 (15) | 243 × 10−8 (243) | 70 × 10−8 (41) |
| pol30-100 | 3553 | C | 16 × 10−7 (4) | 16 × 10−8 (16) | 8.4 × 10−8 (5) |
| pol30-102 | 3554 | 6.8 × 10−7 (2) | 2.2 × 10−8 (2) | 2.8 × 10−8 (2) | |
| pol30-103 | 3555 | B, C | 13 × 10−7 (4) | 2.8 × 10−8 (3) | 6.6 × 10−8 (4) |
| pol30-104 | 3556 | A | 94 × 10−7 (26) | 324 × 10−8 (324) | 35 × 10−8 (21) |
| pol30-105 | 3557 | B, C | 6.8 × 10−7 (2) | 1.5 × 10−8 (2) | 3.3 × 10−8 (2) |
| pol30-106 | 3558 | 3.6 × 10−7 (1) | 0.9 × 10−8 (1) | 1.6 × 10−8 (1) | |
| pol30-108 | 3559 | A, C | 19 × 10−7 (5) | 15 × 10−8 (15) | 6.4 × 10−8 (4) |
| pol30-126 | 3560 | A, B | 22 × 10−7 (6) | 19 × 10−8 (19) | 8.6 × 10−8 (5) |
| pol30-114 | 3561 | B, C | 7.7 × 10−7 (2) | 6.0 × 10−8 (6) | 3.8 × 10−8 (2) |
| msh2 | 3543 | 64 × 10−7 (18) | 500 × 10−8 (500) | 140 × 10−8 (82) | |
| Set b (pol30 msh2 double mutants) | |||||
| msh2 | 3543 | 97 × 10−7 (27) | 800 × 10−8 (800) | 144 × 10−8 (85) | |
| msh2 pol30-100 | 3562 | C | 153 × 10−7 (43) | 742 × 10−8 (742) | 157 × 10−8 (92) |
| msh2 pol30-102 | 3563 | 75 × 10−7 (20) | 860 × 10−8 (860) | 134 × 10−8 (79) | |
| msh2 pol30-103 | 3564 | B, C | 200 × 10−7 (54) | 3,363 × 10−8 (3,363) | 663 × 10−8 (390) |
| msh2 pol30-104 | 3565 | A | 310 × 10−7 (84) | 1,700 × 10−8 (1,700) | 251 × 10−8 (148) |
| msh2 pol30-105 | 3566 | B, C | 127 × 10−7 (34) | 740 × 10−8 (740) | 155 × 10−8 (91) |
| msh2 pol30-108 | 3567 | A, C | 297 × 10−7 (80) | 1,567 × 10−8 (1,567) | 274 × 10−8 (161) |
| msh2 pol30-126 | 3568 | A, B | 97 × 10−7 (27) | 1,077 × 10−8 (877) | 140 × 10−8 (82) |
| msh2 pol30-114 | 3569 | B, C | 98 × 10−7 (27) | 985 × 10−8 (985) | 129 × 10−8 (76) |
| Set c (pol30 msh6 double mutants) | |||||
| msh6 | 3544 | 21 × 10−7 (6) | 5.8 × 10−8 (6) | 2.6 × 10−8 (2) | |
| msh6 pol30-100 | 3570 | C | 90 × 10−7 (25) | 44 × 10−8 (44) | 16 × 10−8 (9) |
| msh6 pol30-102 | 3571 | 20 × 10−7 (6) | 4.5 × 10−8 (5) | 2.2 × 10−8 (1) | |
| msh6 pol30-103 | 3572 | B, C | 84 × 10−7 (23) | 17 × 10−8 (17) | 15 × 10−8 (9) |
| msh6 pol30-104 | 3573 | A | 252 × 10−7 (70) | 879 × 10−8 (879) | 122 × 10−8 (72) |
| msh6 pol30-105 | 3574 | B, C | 48 × 10−7 (13) | 3.8 × 10−8 (4) | 2.8 × 10−8 (2) |
| msh6 pol30-108 | 3575 | A, C | 131 × 10−7 (36) | 56 × 10−8 (56) | 27 × 10−8 (16) |
| msh6 pol30-126 | 3576 | A, B | 55 × 10−7 (15) | 37 × 10−8 (37) | 11 × 10−8 (6) |
| msh6 pol30-114 | 3577 | B, C | 36 × 10−7 (10) | 32 × 10−8 (32) | 9 × 10−8 (5) |
| Set d (presence or absence of RAD27 overexpression) | |||||
| wt | 3552 | ND | ND | 1.0 × 10−8 (1) | |
| wt (GAL-RAD27) | 3668 | ND | ND | 1.8 × 10−8 (2) | |
| pol30-126 | 3588 | A, B | ND | ND | 6.5 × 10−8 (7) |
| pol30-126 (GAL-RAD27) | 3578 | A, B | ND | ND | 5.6 × 10−8 (6) |
Average of two or more experiments. Numbers in parentheses indicate fold induction relative to the wild-type (wt) mean value. pol30 msh2 or pol30 msh6 double mutants that are significantly higher than those for corresponding msh2, msh6, or pol30 single mutants (Mann-Whitney test, P < 0.05) are in boldface. ND, not determined.
Since pol30-52 exerts a dominant negative effect on cellular growth and sensitivity to DNA-damaging agents (3), we tested whether pol30-52 exerts a dominant negative effect with regard to its mutator phenotype. Although the mutation rates in a pol30-52/POL30 strain were lower than those of a pol30-52 strain (Table 2, set a), these rates were still significantly elevated relative to those of a wild-type strain. This dominant negative effect was not due to the presence of an additional copy of POL30 since a POL30/POL30 strain exhibited mutation rates comparable to those of a wild-type strain. Similar partial dominant mutator effects were observed in pol30-104/POL30 strains in patch assays (data not shown).
POL30 mutants can be divided into distinct categories based on CAN1 mutation spectra.
Although the assays used in this study are differentially sensitive to various mutagenic defects, they are not completely specific. Thus, it is difficult to determine the mutagenic defects of pol30 mutants based solely on mutation rate analyses. Because the analysis of mutations accumulated in mutator mutants have been instrumental in deciphering the mechanism of mutagenesis (6, 8, 13, 35, 47, 48), we determined the mutation spectra of the various pol30 mutants. The CAN1 assay was selected for this purpose because it is sensitive to a variety of mutational events, including base substitutions, frameshifts, deletions, duplications, inversions, and translocations (6, 35, 47). Given the large number of pol30 alleles studied and the absence of rationale for focusing on a specific allele, we sequenced 20 to 30 CAN1-inactivating mutations isolated from each pol30 mutant (Tables 3 and 4).
TABLE 3.
CAN1 mutation spectra for various pol30 mutants
| Genotype | Type of event | Frequency (%) | Mutationa |
|---|---|---|---|
| pol30-41 | |||
| Gross deletions | 0/22 (0) | ||
| Gross insertions | 0/22 (0) | ||
| Frameshifts | 7/22 (32) | ||
| 1/22 | G1→G0 | ||
| 1/22 | G2→G3 | ||
| 1/22 | G3→G2 | ||
| 1/22 | C1→C0 | ||
| 1/22 | T4→T3 | ||
| 1/22 | T6→T7 | ||
| 1/22 | T2→T1 | ||
| Base substitution | 15/22 (68) | ||
| 1/22 | G→T | ||
| 1/22 | G→A | ||
| 1/22 | G→C | ||
| 4/22 | C→T | ||
| 4/22 | C→G | ||
| 1/22 | C→A | ||
| 1/22 | T→G | ||
| 2/22 | A→T | ||
| Complex events | 0/21 (0) | ||
| pol30-45 | |||
| Gross deletions | 1/23 (4) | 39 bp1 | |
| Gross insertions | 1/23 (4) | 38 bp2 | |
| Frameshifts | 1/23 (4) | C1→C0 | |
| Base substitution | 20/23 (88) | ||
| 1/23 | C→T | ||
| 2/23 | C→A | ||
| 3/23 | C→G | ||
| 3/23 | G→A | ||
| 4/23 | G→C | ||
| 2/23 | G→T | ||
| 1/23 | T→A | ||
| 1/23 | T→G | ||
| 1/23 | T→C | ||
| 1/23 | A→T | ||
| 1/23 | A→G | ||
| Complex events | 0/23 (0) | ||
| pol30-52 | |||
| Gross deletions | 0/20 (0) | ||
| Gross insertions | 0/20 (0) | ||
| Frameshifts | 16/20 (80) | ||
| 2/20 | G4→G3 | ||
| 1/20 | G0→G1 | ||
| 1/20 | A3→A2 | ||
| 5/20 | A6→A5 | ||
| 1/20 | A2→A3 | ||
| 1/20 | A6→A7 | ||
| 1/20 | T5→T4 | ||
| 1/20 | T3→T2 | ||
| 1/20 | T4→T3 | ||
| 2/20 | T6→T7 | ||
| Base substitution | 4/20 (20) | ||
| 1/20 | C→T | ||
| 1/20 | T→C | ||
| 1/20 | A→T | ||
| 1/20 | A→G | ||
| Complex events | 0/20 (0) | ||
| pol30-100 | |||
| Gross deletions | 0/21 (0) | ||
| Gross insertions | 0/21 (0) | ||
| Frameshifts | 2/21 (10) | ||
| 1/21 | T6→T7 | ||
| 1/21 | G2→G1 | ||
| Base substitution | 19/21 (90) | ||
| 2/21 | G→T | ||
| 3/21 | G→A | ||
| 2/21 | G→C | ||
| 1/21 | C→T | ||
| 2/21 | C→G | ||
| 4/21 | C→A | ||
| 1/21 | T→G | ||
| 1/21 | T→A | ||
| 1/21 | A→G | ||
| 1/21 | A→C | ||
| Complex events | 0/21 (0) | ||
| pol30-102 | |||
| Gross deletions | 0/29 (0) | ||
| Gross insertions | 0/29 (0) | ||
| Frameshifts | 9/29 (31) | ||
| 3/29 | T2→T1 | ||
| 2/29 | T1→T0 | ||
| 1/29 | T3→T4 | ||
| 1/29 | T6→T5 | ||
| 2/29 | C3→C2 | ||
| Base substitution | 20/29 (69) | ||
| 5/29 | G→A | ||
| 3/29 | G→T | ||
| 6/29 | C→A | ||
| 3/29 | C→T | ||
| 1/29 | T→G | ||
| 1/29 | T→C | ||
| 1/29 | A→T | ||
| Complex events | 0/29 (0) | ||
| pol30-103 | |||
| Gross deletions | 4/20 (20) | 60 bp3 | |
| 40 bp4 | |||
| 49 bp5 | |||
| 41 bp6 | |||
| Gross insertions | 0/20 (0) | ||
| Frameshifts | 5/20 (25) | ||
| 1/20 | G3→G4 | ||
| 1/20 | A3→A4 | ||
| 1/20 | T6→T5 | ||
| 2/20 | T6→T7 | ||
| Base substitution | 10/20 (50) | ||
| 2/20 | C→A | ||
| 5/20 | G→T | ||
| 1/20 | T→G | ||
| 2/20 | C→G | ||
| Complex events | 1/20 (5) | ||
| 1/20 | TATgGGTTcTTT-GG→TATtGGTTtTTTtGG | ||
| pol30-104 | |||
| Gross deletions | 0/22 (0) | ||
| Gross insertions | 0/22 (0) | ||
| Frameshifts | 16/22 (73) | ||
| 1/22 | G1→G0 | ||
| 1/22 | G1→G2 | ||
| 1/22 | A6→A5 | ||
| 1/22 | C2→C1 | ||
| 4/22 | T4→T3 | ||
| 7/22 | T6→T7 | ||
| 1/22 | T1→T0 | ||
| Base substitution | 6/22 (27) | ||
| 2/22 | G→T | ||
| 2/22 | G→A | ||
| 1/22 | C→G | ||
| 1/22 | C→A | ||
| Complex events | 0/22 (0) | ||
| pol30-105 | |||
| Gross deletions | 1/28 (4) | 36 bp7 | |
| Gross insertions | 0/28 (0) | ||
| Frameshifts | 5/28 (18) | ||
| 3/28 | T7→T6 | ||
| 1/29 | T6→T5 | ||
| 2/28 | T2→T1 | ||
| 1/28 | A2→A1 | ||
| Base substitution | 21/28 (74) | ||
| 4/28 | G→A | ||
| 2/28 | G→T | ||
| 6/28 | C→A | ||
| 1/28 | C→T | ||
| 1/29 | C→G | ||
| 2/28 | T→G | ||
| 2/28 | T→C | ||
| 3/28 | T→A | ||
| Complex events | 1/28 (4) | ||
| pol30-108 | |||
| Gross deletions | 0/24 (0) | ||
| Gross insertions | 0/24 (0) | ||
| Frameshifts | 10/24 (42) | ||
| 1/24 | G3→G2 | ||
| 1/24 | G2→G1 | ||
| 1/24 | G1→G0 | ||
| 1/24 | A3→A2 | ||
| 1/24 | A5→A6 | ||
| 1/24 | T6→T5 | ||
| 4/24 | T6→T7 | ||
| Base substitution | 14/24 (58) | ||
| 3/24 | G→T | ||
| 4/24 | G→A | ||
| 1/24 | G→C | ||
| 2/24 | C→T | ||
| 4/24 | C→A | ||
| Complex events | 0/24 (0) | ||
| pol30-126 | |||
| Gross deletions | 2/22 (9) | 4 bp8 | |
| 10 bp9 | |||
| Gross insertions | 5/22 (23) | 38 bp insertion10 | |
| 13 bp insertion11 | |||
| 23 bp insertion12 | |||
| 27 bp insertion13 | |||
| 17 bp insertion14 | |||
| Frameshifts | 5/22 (23) | ||
| 2/22 | G1→G0 | ||
| 2/22 | T7→T8 | ||
| 1/22 | C2→C1 | ||
| Base substitution | 10/22 (45) | ||
| 5/22 | C→A | ||
| 1/22 | G→A | ||
| 1/22 | G→T | ||
| 2/22 | C→G | ||
| 1/22 | T→C | ||
| Complex events | 0/22 (0) | ||
| pol30-114 | |||
| Gross deletions | 1/28 (4) | 27 bp15 | |
| Gross insertions | 1/28 (4) | 34 bp16 | |
| Frameshifts | 4/28 (14) | ||
| 2/28 | C2→C1 | ||
| 1/28 | C1→C0 | ||
| 1/28 | G1→G0 | ||
| Base substitution | 19/28 (68) | ||
| 1/28 | C→T | ||
| 6/28 | C→A | ||
| 3/28 | C→G | ||
| 5/28 | G→A | ||
| 3/28 | G→C | ||
| 1/28 | G→T | ||
| Complex events | 3/28 (10) | ||
| 1/28 | GTTTTcTCACAA→GTTTTtTtCACAA | ||
| 1/28 | ATATcATATTTATttatGGG→ATATTTATTTATGCG | ||
| 1/28 | AAGAGAGaGcTTGAG→AAGAGAGGTTGAG |
Superscript numbers correspond to those preceding the sequences in Table 4. Altered nucleotides are in lowercase; relevant microhomologies are in boldface.
TABLE 4.
Sequences surrounding the junction of insertions and deletions
| Genotype | Sequencea |
|---|---|
| pol30-45 | 1GTTGGTTCC...GTTGGCTCCTAAATTCCT |
| 2AATGGTGTTAGCTttgctgccgcctatatctctattttcctgttcttagctTTGCTGCCGCCTATATCTCTATTTTCCTGTTCTTAGCTGTTTGGATC | |
| pol30-103 | 3GGCCGGCTT...TCACGGCTTTTGCACCAA |
| 4TTCTACATCATT...CCATACAATCCTAAACTA | |
| 5GGTGCTGGGGTT...GGTGCCTGGGGTCAAGGTATAATA | |
| 6GGTGCTGGGGTT...AAACCCAGGTGCCTGGGGTC | |
| pol30-105 | 7AGACATCTA...TATTAAATACACTGGTG |
| pol30-126 | 8ATATCATATTTATTTATGGGTTCTTT |
| 9AAGTACAGTAGTGCGCTGAAGTGAAGAGAGA | |
| 10CAATGGTGTTAGCTtttgctgccgcctatatctctattttccctgttcttagCTTTGCTGCCGCCTATATCTCTATTTTCCCTGTTCTTAGCTGTTTTG | |
| 11TCACAGTTTTCTacaaagattccttACAAAGATTCCTTCCAGCATT | |
| 12GTGTTAGCTTTGCTGccgcctatatctctattttcctgCCGCCTATATCTCTATTTTCCTGTTCTTAGCT | |
| 13TACCGGCCCAGTTGGattccgttattggagaaacccaggtggATTCCGTTATTGGAGAAACCCAGGTGGCTGGGGTCC | |
| 14ACGCTGAAGTGAAGAGAgagcttaagcaaagagaGAGCTTAAGCAAAGACATATTGGTAT | |
| pol30-114 | 15CATATTGGTACTATTGGTACAGGTCTTTT |
| 16CTCTATGGAAT̂attctgtca...aatggctacATTCTGTCA...AATGGCTAC |
Altered nucleotides are in lowercase; deleted sequences are underlined; inserted sequences are in italicized lowercase; ^ indicates that a space has been inserted. Relevant microhomologies are indicated in boldface.
To estimate the rate with which each type of mutations accumulated in the various pol30 mutants (Table 5), we multiplied the proportion of each type of mutation (Table 3) by the CAN1 mutation rate (Table 2, set a). Based on the rate of accumulating various types of mutations, the pol30 mutants were divided into groups. Some mutants were placed in more than one group; this reflects the likelihood that some of the pol30 mutations cause more than one defect due to the multifunctional nature of PCNA. Group A mutants (pol30-52, -104, -108, and -126) exhibited a greater tendency to accumulate frameshifts than base substitutions, an observation reminiscent of strains completely defective in MMR, such as msh2 (35, 47). The rates of accumulating frameshift mutations in msh2 and pol30-52, -104, -108, and -126 strains were elevated 60-, 48-, 76-, 8-, and 6-fold, respectively, relative to wild-type strains, whereas the rates of accumulating base substitution mutations were elevated only 4-, 5-, 11-, 4-, and 4-fold, respectively (Table 5).
TABLE 5.
Summary of CAN1 mutation spectra for pol30 mutantsa
| Genotype | Group(s) | Substitution (%) | Frameshifts (%) | Deletions (%) | Insertions (%) | Complex events (%) |
|---|---|---|---|---|---|---|
| Wild typeb | 65 (1) | 25 (1) | 0 | 0 | 10 | |
| msh2b | 15 (4) | 85 (60) | 0 | 0 | 0 | |
| pol30-41 | 68 (1) | 32 (1) | 0 | 0 | 0 | |
| pol30-45 | B | 88 (1) | 4 (0.3) | 4 | 4 | 0 |
| pol30-52 | A | 20 (5) | 80 (48) | 0 | 0 | 0 |
| pol30-100 | C | 90 (6) | 10 (2) | 0 | 0 | 0 |
| pol30-102 | 69 (2) | 31 (2) | 0 | 0 | 0 | |
| pol30-103 | B, C | 50 (4) | 25 (4) | 20 | 0 | 0 |
| pol30-104 | A | 27 (11) | 73 (76) | 0 | 0 | 0 |
| pol30-105 | B, C | 74 (2) | 18 (1) | 4 | 0 | 4 |
| pol30-108 | A, C | 58 (4) | 42 (8) | 0 | 0 | 0 |
| pol30-126 | A, B | 45 (4) | 23 (6) | 9 | 23 | 0 |
| pol30-114 | B, C | 68 (2) | 14 (1) | 4 | 4 | 10 |
Indicated in parentheses is the fold increase in the estimated rate of frameshift and base substitution accumulation in various mutants over the wild-type rate. The estimated rates were calculated by multiplying the proportion of frameshifts and base substitutions (Table 2) by the respective CAN1 mutation rates (Table 1). Group A mutants exhibit mutation rates and spectra reminiscent of mutants defective in MMR. Group B mutants accumulated either deletions or a combination of deletions and duplications in addition to base substitutions and frameshifts. Group C mutants exhibit mutation rates which were synergistic in the presence of a msh6 mutation. Many pol30 mutants harbor multiple mutagenic defects and were placed into more than one group.
From reference 47.
Group B mutants (pol30-45, -103, -105, -126, and -114) accumulated either deletions or a combination of deletions and duplications in addition to base substitutions and frameshifts. pol30-103 and -105 strains accumulated only deletions (36 to 60 bp), whereas pol30-45, -126, and -114 accumulated both deletions (4 to 39 bp) and duplications (13 to 38 bp). The percentage of deletions and duplications was small in some pol30 mutants. However, the detection of one deletion or duplication in a sample size of 30 translates to approximately 10−8 deletion/duplication events per cell per generation. This rate represents a significant increase over the rate of accumulating such mutations in wild-type cells, which is approximately 10−10 per cell per generation (5a). The deletion breakpoints in pol30 mutants were flanked by direct imperfect repeats (3 to 7 bp), similar to those observed in rad27 and pol3-t mutants. The duplications in pol30 mutants were also similar to those seen in rad27 mutants in that the region duplicated was flanked by direct imperfect repeats (5 to 7 bp) and that the duplication always included one of the flanking repeats. However, the duplications in pol30 mutants were, on average, smaller than those seen in rad27 mutants (27 bp versus 39 bp; chi-square test, P < 0.05). This difference was mainly due to the absence of duplications of >50 bp in pol30 mutants.
The two remaining pol30 mutator mutants could not be easily classified based on their mutation spectra. pol30-100 caused a moderate increase in the rate of accumulating base substitution mutations (sixfold) and a small increase in the rate of accumulating frameshift mutations (twofold). pol30-102 caused roughly equal increases (twofold) in the rate of accumulating base substitution and frameshift mutations.
The lys2-Bgl reversion spectrum of pol30-104 suggests a defect in DNA MMR.
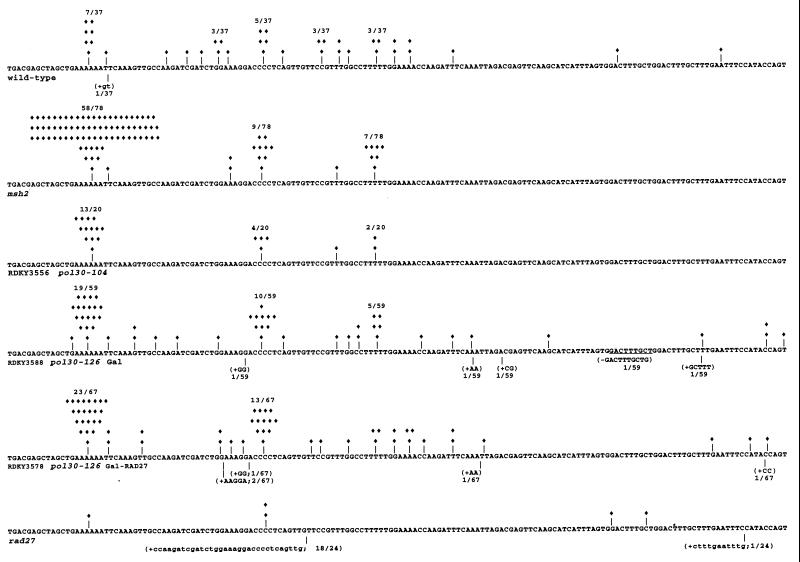
Since mutants that are completely defective in MMR (e.g., msh2, mlh1, and pms1) exhibit a characteristic lys2-Bgl reversion spectrum (8, 13, 35), we wished to determine whether such a spectrum could be seen in any of the group A mutants. pol30-104 was selected as a representative allele for this purpose. As shown in Fig. 1, all of the pol30-104 Lys+ revertants resulted from −1 frameshifts; 65% of these frameshifts occurred in a run of 6 A’s (nucleotides 664 to 669), 20% occurred in a run of four C’s (nucleotides 697 to 700), and 10% occurred in a run of five T’s (nucleotides 689 to 693). The distribution of these frameshift mutations is similar to those observed in msh2, mlh1, and pms1 strains, further supporting the notion that pol30-104 causes defective MMR.
FIG. 1.
Lys2-Bgl reversion spectra from wild-type, msh2, rad27, and pol30 mutants. The sequence shown represents the reversion window (nucleotides 650 to 798) of the lys2-Bgl assay. The location of the nucleotide alteration resulting in functional restoration of LYS2 is indicated by |. Each independent −1 frameshift is indicated by a ⧫ above the deleted nucleotide. Duplications are indicated by (+) followed by the nucleotides inserted. Deletions of greater than one base pair are marked by (−) followed by the nucleotides deleted. The ratio indicated represents the proportion of each particular class of events. The wild-type and msh2 spectra are from reference 8; the rad27 spectra is from reference 47.
An in vivo MMR assay indicates that pol30-104, -108, and -126 cause defects in MMR.
To directly assess the defects of group A mutants in MMR, various pol30 ura3-52 ade2 ade8 strains were transformed with either an A/C or a G/T mismatch-containing URA3 ADE8 plasmid (25). The mismatch resides in the ADE8 gene such that only one strand of the plasmid encodes a functional protein. If the mismatch is corrected prior to DNA replication, the resulting colony will be nonsectored and either red or white. However, if the mismatch is not corrected, then the two ADE8 alleles will segregate after replication, resulting in a red/white sectored colony. Thus, the efficiency of MMR in various pol30 mutants can be assessed by scoring for Ura+ sectored colonies. For comparison, we selected pol30-103 from group B and pol30-100.
Comparable results were obtained using an A/C or a G/T substrate (Table 6). For the wild-type strain, 3% of all transformants were sectored. This percentage increased to approximately 60% in a MMR-defective strain (msh2). No significant percentage increase in sectored colonies was observed in pol30-100 or -103 mutants, indicating that these mutants are not defective in MMR. The group A mutants (pol30-104, -108, and -126) showed significant increases in the percentage of sectored colonies (to approximately 40, 25, and 14%, respectively), indicating that they are defective in MMR. Of these mutants, pol30-104 mutants exhibited the largest increase in the sectoring phenotype. However, this increase is still significantly lower than that observed for a msh2 mutant (P < 0.005), suggesting that pol30-104, -108, and -126 all caused only partial defects in MMR. The same partial MMR defect was observed for pol30-104 strains in two chromosome-based mutator assays (Table 2, set a). Overall, these results suggest that group A mutants are partially defective in MMR.
TABLE 6.
In vivo MMR assay
| Substrate | Genotype | RDKY no. | No. of sectored coloniesa | Total no. of coloniesa | % Sectoringa |
|---|---|---|---|---|---|
| A/C | |||||
| Wild type | 3023 | 57 | 1,783 | 3.2 | |
| msh2 | 2706 | 843 | 1,400 | 60.2 | |
| pol30-100 | 3553 | 116 | 2,723 | 4.3 | |
| pol30-103 | 3555 | 71 | 2,192 | 3.2 | |
| pol30-104 | 3556 | 439 | 1,025 | 42.8 | |
| pol30-108 | 3559 | 83 | 293 | 28.3 | |
| pol30-126 | 3560 | 401 | 2,513 | 15.9 | |
| G/T | |||||
| Wild type | 3023 | 85 | 3,142 | 2.7 | |
| msh2 | 2706 | 943 | 1,455 | 64.8 | |
| pol30-100 | 3553 | 99 | 2,255 | 4.4 | |
| pol30-103 | 3555 | 66 | 1,801 | 3.7 | |
| pol30-104 | 3556 | 238 | 704 | 33.8 | |
| pol30-108 | 3559 | 249 | 1,140 | 21.8 | |
| pol30-126 | 3560 | 183 | 1,701 | 10.7 |
Results of a typical experiment, each repeated at least twice.
lys2-Bgl duplications in pol30-126 mutants differ from those observed in rad27 mutants.
The duplication mutations that arose in group B mutants were similar to those observed in rad27 mutants (47). Since PCNA is known to stimulate the endo/exonucleolytic activity of RAD27 in vitro (31, 56), one interpretation of these observations is that group B mutants are defective in stimulating RAD27 activity. To explore this possibility, we tested whether galactose-induced RAD27 expression in a pol30-126 strain would suppress duplication formation. pol30-126 was selected because it exhibited the highest rate of duplication accumulation in group B. For this analysis we used the lys2-Bgl assay because the small mutation window in this system (0.5 kb versus 2.1 kb for CAN1) minimizes the sequencing effort necessary to obtain a large sample size (13, 35).
Galactose-induced RAD27 expression did not alter the lys2-Bgl reversion rate or spectrum of pol30-126 mutants (Table 2, set d; Fig. 1). The RAD27-expressing plasmid was shown to fully rescue the ts, MMSs, and mutator phenotypes of rad27 null mutants in both glucose- and galactose-containing media (data not shown). When galactose induction was carried out in cells harboring the vector control plasmid or the RAD27 expression plasmid, approximately 90% of the Lys+ revertants were −1 frameshifts; 10% were duplications or deletions, indicating that RAD27 function is not likely to be limiting in pol30-126 mutants. Furthermore, the Lys+ duplications that occurred in the lys2-Bgl assay in pol30-126 mutants were different from those that occurred in rad27 mutants. The rad27 lys2-Bgl revertants consisted of 21% −1 frameshifts and 79% duplications which involved duplication of unique segment of DNA bounded by short repeat sequences (47). In contrast, unlike the pol30-126 CAN1 duplications (Table 3), the pol30-126 Lys+ duplications consisted of simple duplications of two or five nucleotides and did not involve duplication of a unique sequence bounded by repeated sequences. The rad27 Lys+ duplications were identical to the rad27 CAN1 duplications, as both types of duplications involved duplication of unique sequences located between short imperfect repeats. Finally, the frameshift mutations in pol30-126 mutants were different from those that occurred in rad27 mutants. Approximately 50% of the −1 frameshifts in pol30-126 were clustered at the same hot spots as those seen in mutants completely defective in MMR, such as msh2, mlh1, and pms1 (8, 13, 35). This result is consistent with the partial defect that pol30-126 mutants exhibited in the plasmid-based MMR assay. Such clustering was not observed for the rad27 −1 frameshifts (47).
Epistasis analyses with msh2 and msh6 mutations suggest that pol30 mutations also cause mutagenic defects that are distinct from defects in MMR.
Except for group A mutants, the mutation spectra of pol30 mutants differed from that of a msh2 mutant, suggesting that pol30 mutations cause defects in processes distinct from MMR. Additionally, mutation rate analyses of some group A mutants (pol30-52 and -104) suggest that defective MMR constitutes only one of the mutagenic defects in these mutants. To explore these possibilities, we conducted an epistasis analysis between the various pol30 mutations and a msh2 mutation. In at least one mutator assay, double mutants containing a combination of msh2 and pol30-100, -103, -104, or -108 exhibited mutation rates significantly higher than that of a msh2 or a pol30 single mutant (Mann-Whitney test, P < 0.05) (Table 2, set b). This result indicates that pol30-100, -103, -104, and -108 caused defects in processes distinct from MMR.
The disparity between the mutation rates of a msh2 mutant and those of many pol30 mutants makes the detection of synergistic effects difficult (Table 2, set b), especially if the pol30 mutants are confined to accumulating base substitution mutations. We therefore compared the mutation rates of various pol30 msh6 double mutants to those of msh6 and pol30 single mutants (Table 2, set c). Recall that MMR initiates with mismatch binding by either the MSH2-MSH6 heterodimer, which recognizes primarily base-base mispairs, or the MSH2-MSH3 heterodimer, which recognizes only insertion/deletion mispairs (10, 35). Since MSH2 is required for the formation of both complexes, msh2 mutations completely inactivate MMR and cause a strong mutator phenotype. The mutation rates of msh6 mutants are lower than those of msh2 mutants but comparable to those of most pol30 mutants. Because base-base mispairs are recognized by the MSH2-MSH6 heterodimer and not by the MSH2-MSH3 heterodimer (10, 35), msh6 mutations completely inactivate MMR of base-base mispairs (23). Thus, the mutation rates in msh6 strains primarily reflect increased accumulation of base substitution mutations. Of all of the mutations examined (Table 2, set c), pol30-102 was the only one whose mutation rate was not enhanced by a msh6 mutation. Of the remaining mutations, five (pol30-100, -103, -105, -108, and -114) exhibited synergistic mutator effects in the CAN1 assay when combined with a msh6 mutation. Because this synergy suggests that pol30-100, -103, -105, -108, and -114 caused defects that increased polymerase misincorporation (see Discussion), these mutants were placed into group C. Many of the group C mutants also belonged to group A or B (Table 5).
A msh2 mutation rescues the synthetic lethality between pol30-104 and rad52.
A series of genetic observations in E. coli raised an interesting hypothesis regarding the defect of group A mutants in MMR. The initial proposal of methyl-directed MMR in E. coli was based on the genetic observation that dam mutations were lethal in combination with recA mutations and that this lethality was suppressed by MMR-inactivating mutations, such as mutS (36). These observations were interpreted to mean that in the absence of a strand discriminating signal, the MMR machinery (e.g., MutH) aberrantly nicks both the parental and the daughter DNA strands, leading to DSBs which require RecA-mediated recombinational repair. When MMR is inactivated by a mutS mutation, DNA breaks are not generated, and RecA is no longer required for viability. The mechanism of daughter strand discrimination has yet to be described in eukaryotes. However, the recent observation that PCNA functions in MMR at a step prior to strand excision raises the possibility that it can facilitate strand discrimination (14, 52). If PCNA does facilitate strand discrimination in MMR, then defects leading to aberrantly initiated MMR may cause DNA breaks. Interestingly, a number of pol30 mutations, including the group A mutations, were reported to be synthetically lethal with mutations in the RAD52 series of genes—genes required for recombinational repair of DNA breaks (37). Thus, by analogy to the genetics of MMR in E. coli, a msh2 mutation might rescue the synthetic lethality between rad52 and group A mutations. Moreover, if the above analogy is appropriate, then msh2 should not rescue the synthetic lethality between rad52 and other pol30 mutations.
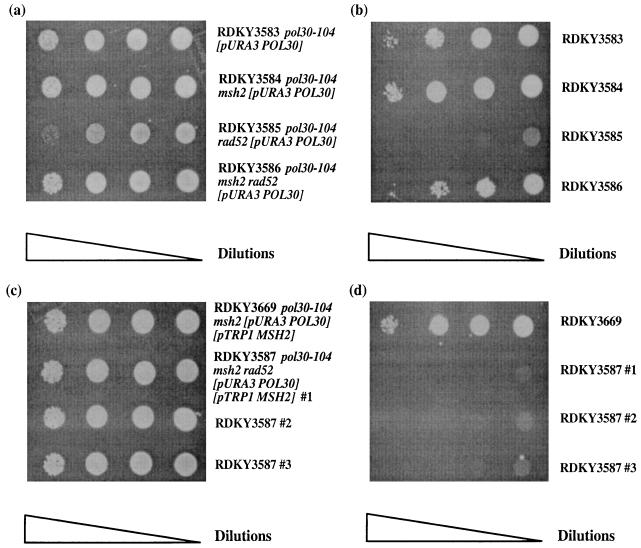
To test these possibilities, the RAD52 gene was disrupted in pol30-104, pol30-100, pol30-103, pol30-104 msh2, pol30-100 msh2, and pol30-103 msh2 strains containing a Pol30+ (POL30 URA3) plasmid. pol30-104 was selected because it caused the strongest MMR defect among group A mutations. pol30-100 and -103 were selected because they exhibit synthetic lethality with rad52 but do not cause MMR defects. The pol30-104 rad52 [pPOL30 URA3] strain did not grow on 5FOA plates, whereas the pol30-104 msh2 rad52 [pPOL30 URA3] strain grew, albeit with slightly reduced viability (Fig. 2a and b). These results indicate that the synthetic lethality between pol30-104 and rad52 was almost completely rescued by a msh2 mutation. This suppression, as predicted, was abolished by the introduction of a MSH2-containing plasmid (Fig. 2c and d). The effect of the msh2 mutation appeared specific for defects in POL30 since msh2 mutations did not suppress the MMS or the hydroxyurea sensitivity of rad52 mutants (data not shown). pol30-103 rad52, pol30-100 rad52, pol30-103 msh2 rad52, and pol30-100 msh2 rad52 strains containing a Pol30+ plasmid were sensitive to killing by 5FOA (data not shown), suggesting that MSH2 inactivation did not rescue the synthetic lethality between pol30-100 or pol30-103 and rad52. Similar to the behavior of pol30-100 and pol30-103, a RFC allele exhibiting synthetic lethality with rad52 that is not suppressed by MSH2 inactivation has been reported (57). Together these studies suggest that the suppression of the synthetic lethality between pol30-104 and rad52 by msh2 is allele specific.
FIG. 2.
A msh2 mutation rescues the synthetic lethality between pol30-104 and rad52. (a and b) pol30-104 (RDKY3583), pol30-104 msh2 (RDKY3584), pol30-104 rad52 (RDKY3585) and pol30-104 msh2 rad52 (RDKY3586) strains bearing a URA3 POL30 plasmid were grown at 30°C in liquid SD −Ura to saturation; 10-fold serial dilutions of each culture were spotted onto an SD −Ura plate to determine viability (a) and an SD +5FOA plate to determine whether the URA3 POL30 plasmid was required for viability (b). (c and d) pol30-104 msh2 (RDKY3669) and three independent isolates of pol30-104 msh2 rad52 (RDKY3587), all bearing URA3 POL30 and TRP1 MSH2 plasmids, were grown at 30°C in liquid SD −Ura −Trp media to saturation; 10-fold serial dilutions of each culture were spotted onto an SD −Ura −Trp plate to determine viability (c) and an SD −Trp +5FOA plate to determine whether the URA3 POL30 plasmid was required for viability in the presence of the TRP1 MSH2 plasmid (d).
A msh2 mutation decreases the hyper-rec phenotype caused by pol30-104.
Since DNA breaks are known to be recombinagenic (43) and since pol30-104 strains accumulate DNA breaks (37), pol30-104 might be expected to cause a hyperrecombination (hyper-rec) phenotype. Moreover, if the DNA strand breaks in pol30-104 mutants occurred as a result of aberrantly initiated MMR, then the hyper-rec phenotype should be suppressed by a msh2 mutation. To test this, we used a diploid strain carrying two recessive alleles (can1-51 and hom3) on one copy of chromosome V; the other copy of chromosome V is wild type for both alleles (15, 17). Phenotypically, the strain is canavanine sensitive and prototrophic. However, if gene conversion at CAN1 occurs or if there is a crossover between CAN1 and the centromere followed by proper segregation, canavanine-resistant and homoserine prototrophic progeny will be obtained. On the other hand, loss of the wild-type chromosome V will lead to canavanine-resistant but homoserine auxotrophic progeny.
As shown in Table 7, a pol30-104 diploid strain had a 50-fold increase in mitotic recombination and a 72-fold increase in chromosome loss. A msh2 mutation caused a twofold increase in mitotic recombination but had essentially no effect on chromosome loss. In the pol30-104 msh2 double mutant, the rates of mitotic recombination and chromosome loss were higher than wild-type rates (17- and 22-fold, respectively). However, these rates were threefold lower (P < 0.05, Mann-Whitney) than those observed in pol30-104 mutants. While the suppression of the pol30-104 hyper-rec phenotype by msh2 was modest in absolute terms, it was striking considering that pol30-104 and msh2 each independently caused a hyper-rec phenotype.
TABLE 7.
Rates of heteroallelic recombination and chromosome loss in msh2, pol30-104, and pol30-104 msh2 mutants
| Strain | Ratea
|
|
|---|---|---|
| Heteroallelic recombination | Chromosome loss | |
| RKY3579 (wild type/wild type) | 3.6 × 10−5 (1) | 3.2 × 10−6 (1) |
| RKY3580 (msh2/msh2) | 8.0 × 10−5 (2) | 4.2 × 10−6 (1) |
| RKY3581 (pol30-104/pol30-104) | 1.8 × 10−3 (50) | 2.3 × 10−4 (72) |
| RKY3582 (pol30-104 msh2/pol30-104 msh2) | 0.6 × 10−3 (17) | 0.7 × 10−4 (22) |
Average of two more more experiments. Numbers in parentheses indicate fold induction relative to the wild-type rate. The Mann-Whitney test indicates that the recombination rate and the chromosomal loss rate in pol30-104/pol30-104 mutants were significantly higher than those of pol30-104 msh2/pol30-104 msh2 diploids (P < 0.05).
DISCUSSION
In this report, we described the effects of 12 pol30 alleles on the rate of mutation accumulation. Eleven of the twelve pol30 mutations studied caused increased accumulation of base substitutions, frameshifts, deletions, duplications, or some combination of these types of mutations. Although the pol30 alleles studied here were previously shown to cause defects in DNA replication and to cause sensitivity to various DNA-damaging agents (2, 3, 37), we could not find any correlation between these phenotypes and the mutator phenotype of pol30 mutants. The lack of such correlation suggests that the pol30 mutants are simultaneously defective in multiple cellular processes. Consistent with this interpretation, many pol30 mutations caused more than one mutagenic defect.
Based on their mutator phenotypes, the pol30 mutants were divided into three groups. Because many pol30 mutants harbor multiple mutagenic defects, they were placed in more than one group (Table 5). Group A mutations (pol30-52, -104, -108, and -126) caused MMR defects. Two of the group A mutations (pol30-52 and -104) were previously reported to cause defects in MMR because they induce mutator phenotypes that were epistatic to MMR-inactivating mutations (18, 52). Our results extend these observations by demonstrating that pol30-52 and -104 strains exhibited mutation spectra indistinguishable from mutants completely inactivated for MMR, such as msh2 (35, 47). Moreover, we showed that pol30-104 strains are defective in the in vivo repair of plasmids containing mispaired bases. Using mutation spectra analysis and in vivo MMR defect as criteria, two additional MMR-defective POL30 alleles, pol30-108 and -126, were identified. pol30-104 and -108 were mutated at the same amino acid (A251V for pol30-104 and A251T for pol30-108). Although these two alleles were similarly defective in terms of cold sensitivity and cell cycle arrest, pol30-104 caused a significantly more severe MMSs phenotype (2, 37) and a stronger mutator phenotype.
Although the mutator phenotypes of the pol30-52 and -104 strains were initially thought to result solely from defective MMR, two recent studies indicate that polymerase slippage also contributes to the pol30-52 mutator phenotype (20, 57). Our mutation spectra analyses as well as the msh2 and msh6 epistasis analyses of pol30-104, -108, and -126 suggest that these MMR-defective pol30 alleles, like pol30-52, are additionally defective in some aspect of replicative fidelity. Contrary to the results presented here, Johnson et al. (18) reported that pol30-104 caused a mutator phenotype that is epistatic to msh2, mlh1, and pms1. This discrepancy may be explained if the plasmid based dinucleotide instability assay used by Johnson et al. is more specific to MMR-related defects than our assays. The disagreement between our results and the reported epistatic relationship between pol30-104 and mlh1 in the CAN1 assay (18) may have resulted from differences in strain background or from differences between a mlh1 and a msh2 mutation.
Group B mutants (pol30-45, -103, -105, -126, and -114) exhibited increased accumulation of either deletions alone or a combination of deletions and duplications. All of the CAN1 duplications in pol30 mutants, like those seen in rad27 mutants, involved short repeated sequences at the breakpoint. Since PCNA stimulates the activity of RAD27 in vitro (31, 56), one interpretation of these observations is that group B mutants are defective in stimulating RAD27 activity. This interpretation suggests that group B mutants, like rad27 mutants, accumulate DSBs and that these breaks are repaired by misannealing of single-stranded DNA tails (21, 47). However, two lines of evidence are inconsistent with this idea. First, the lys2-Bgl duplications in pol30-126 mutants differed from those observed in rad27 mutants. Second, the CAN1 duplications in pol30-126 mutants were, on average, shorter than those observed in rad27 mutants. Finally, the duplications in pol30-126 could not be suppressed by the expression of RAD27 under a GAL10 promoter. Although the third observation could be explained by insufficient RAD27 expression, this explanation fails to account for the first two observations. One explanation that takes into account all three observations is that defective PCNA-DNA polymerase or PCNA-DNA interactions decreases the adherence of the polymerase to the template, thereby facilitating polymerase slippage. Consistent with this explanation, recent studies suggest that pol30-52, an allele defective in MMR, also induced replication slippage in the context of simple repeated sequences such as dinucleotide repeats (20, 57). Also, DNA polymerase mutants have been shown to accumulate repeat-mediated deletions and duplications (34, 48). While the above hypothesis is appealing, we cannot exclude the possibility that in the absence of wild-type PCNA, RAD27 may mediate the removal of some but not all of a flap structure. In vitro, however, RAD27 removes the entire flap structure in the absence of PCNA (32). We also cannot exclude that possibility that the pol30 mutants are defective in a yet to be identified repair system that specifically prevents deletion/duplication formation.
Group C mutants (pol30-100, -103, -105, -108, and -114) exhibited increased accumulation of base substitutions (Table 5) and synergistic mutator effects in the CAN1 assay when combined with msh6 mutations (Table 2, set c). The synergistic effect with msh6, a mutation thought to completely inactivate the repair of base-base mispairs, suggests that group C mutations increased polymerase misincorporation. Another interpretation of this synergy with msh6 is that the pol30 mutants are defective in MSH3-dependent MMR. This interpretation is unlikely because the mutator phenotype of msh3 msh6 double mutants differed from those of group C msh6 double mutants; msh3 msh6 double mutants were potent mutators in the hom3-10 and the lys2-bgl assay but were modest mutators in the CAN1 assay (increases of roughly 500-, 100-, and 30-fold, respectively, over wild-type rates) (35). The group C msh6 double mutants exhibited CAN1 mutation rates comparable to that of msh3 msh6 double mutants. However, the hom3-10 and lys2-bgl reversion rates of group C msh6 double mutants (ranging from increases of 6- to 15-fold over wild-type rates) were significantly lower than those of msh3 msh6 double mutants.
Since the crystal structure of PCNA has been solved (26), we wished to determine whether group A, B, and C mutations were localized to any particular region of PCNA. The amino acids altered by group A mutations (pol30-52, -104, -108, and -126) were located in three distinct regions. pol30-52 changed a residue located in the monomer-monomer interface region. pol30-104(-108) changed a residue in the beta sheets connecting the two monomer domains (i.e., the interdomain region). pol30-126 changed a residue in one of the alpha helices contacting DNA. Therefore, we could not define a single region of PCNA where amino acid substitutions caused MMR defects. Likewise, group B and C mutations were not localized to any particular region of the PCNA structure. We were also unable to find any correlation between the known biochemical defects of pol30 mutants and the severity of their defects in MMR. For instance, pol30-52 trimers exhibited a notable decrease in stability relative to pol30-104 trimers (7a). However, the two mutants were equally defective in MMR by the criteria established here.
Suppression of the pol30-104 rad52 synthetic lethality and the pol30-104-induced hyper-rec phenotype by MSH2 inactivation provides some insights into the mechanistic details of MMR. One interpretation of these results is that PCNA mediates a signal that targets newly synthesized DNA strands for excision during MMR. By analogy to the genetic interaction between mutations in dam, recA, and mutS in E. coli (36), in the absence of such a signal (as may be the case in a pol30-104 mutant), the MMR machinery might nick both the template and the daughter DNA strands. Alternatively, in the absence of a strand discrimination signal, the MMR machinery can nick the template strand. In the presence of the many nascent strand breaks induced by pol30-104 (37), these template strand nicks lead to the formation of DSBs. Another explanation is that PCNA couples MMR proteins to replication proteins. In this scenario, pol30-104 causes replication fork stalling when MMR is initiated such that the stalled replication fork is converted to a DSB (46). These models are not mutually exclusive. We are currently testing these possibilities by isolating additional suppressors of the pol30-104 rad52 synthetic lethality.
The observations reported here have a number of implications regarding the genetics of cancer susceptibility. First, the observation that many pol30 mutants are mutators suggests that missense mutations in the human PCNA could increase cancer susceptibility. This possibility is particularly intriguing when one takes into consideration the partial dominant mutator effect of some pol30 alleles, such as pol30-52 and -104. Second, though many pol30 mutants were by themselves weak mutators, their mutator effects were dramatically elevated in the presence of a msh6 mutation. This observation suggests that weak pol30 mutator alleles, which might exist as natural polymorphic variants in a population, could act as modifiers of partially defective MMR alleles, such as partial loss of function MSH2 alleles or MSH6 null alleles. Finally, pol30-induced duplications may lead to trinucleotide expansion, a process underlying the pathogenesis of many human neurological disorders (45). All of these possibilities await further examination.
ACKNOWLEDGMENTS
We thank John Weger and Pamela Hunt for DNA sequence analysis. We also thank Takuro Nakagawa, Abhijit Datta, Kyung Jae Myun, Alex Shoemaker, Richura Das Gupta, and Neelam Amin for helpful discussions and comments on the manuscript and Alex Shoemaker and Neelam Amin for resequencing the pol30-126 allele.
This work was supported by NIH grants GM50006 to R.D.K. and GM36510 to C.H.
REFERENCES
- 1.Alani E, Lee S, Kane M F, Griffith J, Kolodner R D. Saccharomyces cerevisiae MSH2, a mispaired base recognition protein, also recognizes Holliday junctions in DNA. J Mol Biol. 1997;265:289–301. doi: 10.1006/jmbi.1996.0743. [DOI] [PubMed] [Google Scholar]
- 2.Amin N S, Holm C H. In vivo analysis reveals that the interdomain region of the yeast proliferating cell nuclear antigen is important for DNA replication and DNA repair. Genetics. 1996;144:479–493. doi: 10.1093/genetics/144.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayyagari R, Impellizzeri K J, Yoder B L, Gary S L, Burgers P M J. A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and DNA repair. Mol Cell Biol. 1995;15:4420–4429. doi: 10.1128/mcb.15.8.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer G A, Burgers P M. Molecular cloning, structure and expression of the yeast proliferating cell nuclear antigen gene. Nucleic Acids Res. 1990;18:261–265. doi: 10.1093/nar/18.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauer G A, Burgers P M. Protein-protein interactions of yeast DNA polymerase III with mammalian and yeast proliferating cell nuclear antigen (PCNA)/cyclin. Biochim Biophy Acta. 1988;951:274–279. doi: 10.1016/0167-4781(88)90097-8. [DOI] [PubMed] [Google Scholar]
- 5a.Chen, C., and R. D. Kolodner. Unpublished data.
- 6.Chen C, Umezu K, Kolodner R D. Chromosomal rearrangements occur in S. cerevisiae rfa1 mutator mutants due to mutagenic lesions processed by double-strand-break repair. Mol Cell. 1998;2:9–22. doi: 10.1016/s1097-2765(00)80109-4. [DOI] [PubMed] [Google Scholar]
- 7.Daniel W W. Biostatistics: a foundation for analysis in the health sciences. 5th ed. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 7a.Flores-Rozas, H. Personal communication.
- 8.Flores-Rozas H, Kolodner R D. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc Natl Acad Sci USA. 1998;96:12404–12409. doi: 10.1073/pnas.95.21.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gary R, Ludwig D L, Cornelius H L, MacInnes M A, Park M S. The DNA repair endonuclease XPG binds to proliferating cell nuclear antigen (PCNA) and shares sequence elements with the PCNA-binding regions of FEN-1 and cyclin-dependent kinase inhibitor p21. J Biol Chem. 1997;272:24522–24529. doi: 10.1074/jbc.272.39.24522. [DOI] [PubMed] [Google Scholar]
- 10.Genschel J, Littman S J, Drummond J T, Modrich P. Isolation of MutSbeta from human cells and comparison of the mismatch repair specificities of MutSbeta and MutSalpha. J Biol Chem. 1998;273:19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- 11.Gradia S, Subramanian D, Wilson T, Acharya S, Makhov A, Griffith J, Fishel R. hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol Cell. 1999;3:255–261. doi: 10.1016/s1097-2765(00)80316-0. [DOI] [PubMed] [Google Scholar]
- 12.Greenblatt M S, Grollman A P, Harris C C. Deletions and insertions in the p53 tumor suppressor gene in human cancers: confirmation of the DNA polymerase slippage/misalignment model. Cancer Res. 1996;56:2130–2136. [PubMed] [Google Scholar]
- 13.Greene C N, Jinks-Robertson S. Frameshift intermediates in homopolymer runs are removed efficiently by yeast mismatch repair proteins. Mol Cell Biol. 1997;17:2844–2850. doi: 10.1128/mcb.17.5.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu L, Hong Y, McCulloch S, Watanabe H, Li G M. ATP-dependent interaction of human mismatch repair proteins and dual role of PCNA in mismatch repair. Nucleic Acids Res. 1998;25:1173–1178. doi: 10.1093/nar/26.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartwell L H, Smith D. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics. 1985;110:381–395. doi: 10.1093/genetics/110.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hindges R, Hubscher U. DNA polymerase delta, an essential enzyme for DNA transactions. Biol Chem. 1997;378:345–362. doi: 10.1515/bchm.1997.378.5.345. [DOI] [PubMed] [Google Scholar]
- 17.Holm C, Stearns T, Botstein D. DNA topoisomerase II must act at mitosis to prevent nondisjunction and chromosome breakage. Mol Cell Biol. 1989;9:159–168. doi: 10.1128/mcb.9.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson R E, Kovvali G K, Guzder S N, Amin N S, Holm C, Habraken Y, Sung P, Prakash L, Prakash S. Evidence for involvement of yeast proliferating cell nuclear antigen in DNA mismatch repair. J Biol Chem. 1996;271:27987–27990. doi: 10.1074/jbc.271.45.27987. [DOI] [PubMed] [Google Scholar]
- 18a.Kaiser C, Michaelis S, Mitchel A. A ten minute DNA prep from yeast. Methods Yeast Genet. 1994;1994:141–143. [Google Scholar]
- 19.Kelman Z, Hurwitz J. Protein-PCNA interactions: a DNA-scanning mechanism. Trends Biochem Sci. 1998;1998:236–238. doi: 10.1016/s0968-0004(98)01223-7. [DOI] [PubMed] [Google Scholar]
- 20.Kokoska R J, Stefanovic L, Buermeyer A B, Liskay R M, Petes T D. A mutation of the yeast gene encoding PCNA destabilizes both microsatellite and minisatellite DNA sequences. Genetics. 1999;151:511–519. doi: 10.1093/genetics/151.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kokoska R J, Stefanovic L, Tran H T, Resnick M A, Gordenin D A, Petes T D. Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase δ (pol3-t) Mol Cell Biol. 1998;18:2779–2788. doi: 10.1128/mcb.18.5.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolodner R. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 23.Kolodner R D, Marsischky G T. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 24.Kong X P, Onrust R, O’Donnell M, Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme, a sliding DNA clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 25.Kramer B, Kramer W, Williamson M S, Fogel S. Heteroduplex DNA correction in Saccharomyces cerevisiae is mismatch specific and requires functional PMS genes. Mol Cell Biol. 1989;9:4432–4440. doi: 10.1128/mcb.9.10.4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krishna T S R, Kong X P, Gary S, Burgers P, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 27.Kunkel T A. DNA replication fidelity. J Biol Chem. 1992;267:18251–18254. [PubMed] [Google Scholar]
- 28.Kunkel T A. DNA-mismatch repair. The intricacies of eukaryotic spell-checking. Curr Biol. 1995;5:1091–1094. doi: 10.1016/s0960-9822(95)00218-1. [DOI] [PubMed] [Google Scholar]
- 29.Lea D E, Coulson C A. The distribution of the numbers of mutants in bacterial populations. J Genet. 1948;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 30.Lee S H. The 3′-5′ exonuclease of human DNA polymerase delta (pol delta) is regulated by pol delta accessory factors and deoxyribonucleoside triphosphates. Nucleic Acids Res. 1993;21:1935–1939. doi: 10.1093/nar/21.8.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X, Li J, Harrington J, Lieber M R, Burgers P M. Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen. J Biol Chem. 1995;270:22109–22112. doi: 10.1074/jbc.270.38.22109. [DOI] [PubMed] [Google Scholar]
- 32.Lieber M R. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination, and repair. Bioessays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- 33.Lin Y L, Shivji M K, Chen C, Kolodner R, Wood R D, Dutta A. The evolutionarily conserved zinc finger motif in the largest subunit of human replication protein A is required for DNA replication and mismatch repair but not nucleotide excision repair. J Biol Chem. 1998;273:1453–1461. doi: 10.1074/jbc.273.3.1453. [DOI] [PubMed] [Google Scholar]
- 34.Liu V, Bhaumik D, Wang T S-F. Mutator phenotype induced by aberrant replication. Mol Cell Biol. 1999;19:1126–1135. doi: 10.1128/mcb.19.2.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marsischky G T, Filosi N, Kane M F, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 36.McGraw B R, Marinus M G. Isolation and characterization of Dam+ revertants and suppressor mutations that modify secondary phenotypes of dam-3 strains of Escherichia coli K-12. Mol Gen Genet. 1980;178:309–315. doi: 10.1007/BF00270477. [DOI] [PubMed] [Google Scholar]
- 37.Merrill B J, Holm C. The RAD52 recombinational repair pathway is essential in pol30 (PCNA) mutants that accumulate small single-stranded DNA fragments during DNA synthesis. Genetics. 1998;148:611–624. doi: 10.1093/genetics/148.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Modrich P. DNA mismatch correction. Annu Rev Biochem. 1987;56:435–466. doi: 10.1146/annurev.bi.56.070187.002251. [DOI] [PubMed] [Google Scholar]
- 39.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 40.Morrison A, Sugino A. The 3′→5′ exonucleases of both DNA polymerase delta and epsilon participate in correcting errors of DNA replication in Saccharomyces cerevisiae. Mol Gen Genet. 1994;242:289–296. doi: 10.1007/BF00280418. [DOI] [PubMed] [Google Scholar]
- 41.Mozzherin D J, McConnell M, Jasko M V, Krayevsky A A, Tan C K, Downey K M, Fisher P A. Proliferating cell nuclear antigen promotes misincorporation catalyzed by calf thymus DNA polymerase delta. J Biol Chem. 1996;271:31711–31717. doi: 10.1074/jbc.271.49.31711. [DOI] [PubMed] [Google Scholar]
- 42.Muller-Weeks S J, Caradonna S. Specific association of cyclin-like uracil-DNA glycosylase with the proliferating cell nuclear antigen. Exp Cell Res. 1996;226:346–355. doi: 10.1006/excr.1996.0235. [DOI] [PubMed] [Google Scholar]
- 43.Osman F, Subramani S. Double-strand break-induced recombination in eukaryotes. Prog Nucleic Acid Res Mol Biol. 1998;58:263–299. doi: 10.1016/s0079-6603(08)60039-2. [DOI] [PubMed] [Google Scholar]
- 44.Peltomaki P, Vasen H F A. Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. Gastroenterology. 1997;113:1146–1158. doi: 10.1053/gast.1997.v113.pm9322509. [DOI] [PubMed] [Google Scholar]
- 45.Ross C A, Margolis R L, Becher M W, Wood J D, Engelender S, Cooper J K, Sharp A H. Pathogenesis of neurodegenerative diseases associated with expanded glutamine repeats: new answers, new questions. Prog Brain Res. 1998;117:397–419. doi: 10.1016/s0079-6123(08)64029-7. [DOI] [PubMed] [Google Scholar]
- 46.Seigneur M, Bidnenko V, Ehrlich S D, Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 47.Tishkoff D X, Filosi N, Gaida G M, Kolodner R D. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 48.Tran H T, Degtyareva N P, Koloteva N N, Sugino A, Masumoto H, Gordenin D A, Resnick M A. Replication slippage between distant short repeats in Saccharomyces cerevisiae depends on the direction of replication and the RAD50 and RAD52 genes. Mol Cell Biol. 1995;15:5607–5617. doi: 10.1128/mcb.15.10.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tran H T, Gordenin D A, Resnick M A. The prevention of repeat-associated deletions in Saccharomyces cerevisiae by mismatch repair depends on size and origin of deletions. Genetics. 1996;143:1579–1587. doi: 10.1093/genetics/143.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tran H T, Keen J D, Kricker M, Resnick M A, Gordenin D A. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol Cell Biol. 1997;17:2859–2865. doi: 10.1128/mcb.17.5.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsurimoto T. PCNA, a multifunctional ring on DNA. Biochim Biophys Acta. 1998;1443:23–39. doi: 10.1016/s0167-4781(98)00204-8. [DOI] [PubMed] [Google Scholar]
- 52.Umar A, Buermeyer A B, Simon J A, Thomas D C, Clark A B, Liskay R M, Kunkel T A. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- 53.Vogelstein B, Kinzler K W. The multistep nature of cancer. Trends Genet. 1993;9:138–141. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- 54.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 55.Waga S, Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 56.Wu X, Li J, Li X, Hsieh C L, Burgers P M, Lieber M R. Processing of branched DNA intermediates by a complex of human FEN-1 and PCNA. Nucleic Acids Res. 1996;24:2036–2043. doi: 10.1093/nar/24.11.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xie Y, Counter C, Alani E. Characterization of the repeat-tract instability and mutator phenotypes conferred by a Tn3 insertion in RFC1, the large subunit of the yeast clamp loader. Genetics. 1999;151:499–509. doi: 10.1093/genetics/151.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]