Abstract
Colitis is a common disease of the colon that is very difficult to treat. Probiotic bacteria could be an effective treatment. The probiotic Escherichia coli Nissle 1917 (EcN) was engineered to synthesize the ketone body (R)-3-hydroxybutyrate (3HB) for sustainable production in the gut lumen of mice suffering from colitis. Components of heterologous 3HB synthesis routes were constructed, expressed, optimized, and inserted into the EcN genome, combined with deletions in competitive branch pathways. The genome-engineered EcN produced the highest 3HB level of 0.6 g/L under microaerobic conditions. The live therapeutic was found to colonize the mouse gastrointestinal tract over 14 days, elevating gut 3HB and short-chain-length fatty acid (SCFA) levels 8.7- and 3.1-fold compared to those of wild-type EcN, respectively. The sustainable presence of 3HB in mouse guts promoted the growth of probiotic bacteria, especially Akkermansia spp., to over 31% from the initial 2% of all the microbiome. As a result, the engineered EcN termed EcNL4 ameliorated colitis induced via dextran sulfate sodium (DSS) in mice. Compared to wild-type EcN or oral administration of 3HB, oral EcNL4 uptake demonstrated better effects on mouse weights, colon lengths, occult blood levels, gut tissue myeloperoxidase activity and proinflammatory cytokine concentrations. Thus, a promising live bacterium was developed to improve colonic microenvironments and further treat colitis. This proof-of-concept design can be employed to treat other diseases of the colon.
Keywords: Escherichia coli Nissle 1917, Colitis, β-hydroxybutyrate, 3HB, Synthetic biology, Metabolic engineering, Microbiomes, Probiotics, Colonic microenvironment
Subject terms: Drug delivery, Mucosal immunology
Introduction
Engineered live therapeutics designed to deliver drug molecules or mediate diagnostic approaches through probiotic reconstruction have attracted much attention from researchers in the fields of biomedical engineering and synthetic biology due to their promising prospects [1–3]. It is desirable to reprogram microorganisms depending on actual medical and personalized demands. For next-generation probiotics, the engineering of microorganisms provides multiple advantages compared to their wild-type chassis, including in vivo animal diagnosis, site-specific drug administration, sustainable release, and mechanism-controllable therapy [4–6]. Successful reports of using genetically modified live bacteria to treat phenylketonuria, hyperammonemia, diabetes, colitis, obesity, alcoholic liver, and cancer have been published [1, 7–12]. Several biotherapeutic bacteria are under clinical trials, indicating their promising future pharmaceutical applications [13, 14].
Human commensal bacteria, especially species from Lactobacillus, Lactococcus, and Escherichia, are suitable chassis for medical engineering [15]. Among them, Escherichia coli Nissle 1917 (EcN) shows promise due to its easy genetic manipulation and probiotic characteristics [16]. The strain is nonpathogenic (serotype O6:K5:H1) with antagonistic activities against gut pathogens such as Shigella dysenteriae, Vibrio cholerae, Yersinia enterocolitica, Salmonella enteritidis, and invasive E. coli [17–19], and sufficient molecular biology toolboxes are available for EcN engineering. EcN was reported to be as effective as mesalazine for the remission of ulcerative colitis [17]. The bacterium is genetically stable and cannot be transformed by the tox- phages of enterohemorrhagic E. coli strains [20]. Production of defensins, cathelicidin, and calprotectin and anti-inflammatory effects, e.g., inhibition of IL-6 and TNF-α, have been demonstrated [21–23]. Live EcN bacteria exhibit robustness in the mammalian gastrointestinal tract [24]. Therefore, diverse biotherapeutic strategies based on EcN carriers are practicable and need to be further explored.
Biological drugs such as antitumor necrosis factors, interleukins, and small pharmaceutical molecules are likely easier to synthesize and permeate cell membranes, and they are interesting candidates for live therapeutics [25–27]. In addition, engineered probiotics overproducing chemicals such as butyrate or omega-3 fatty acids have been constructed for oral administration as reported previously [5, 28]. However, more therapeutic agents are needed to generate diversified live engineered probiotics for improved treatments.
(R)-β-Hydroxybutyrate (3HB) is the main component of animal ketone bodies, serving as an energy source during starvation or exercise [29, 30]. Despite its function in energy supply, 3HB is also regarded as a therapeutic agent [31, 32]. Beneficial effects of 3HB in the treatment of seizures, hypertension, NLRP3-mediated inflammation, or neurodegenerative diseases have been reported [33–36]. Additionally, bone formation promotion, improvements in learning and memory and glial cell protection were also observed after 3HB administration in mice [37–40].
The 3HB microbial synthesis pathway comprises three steps [41, 42]: two molecules of acetyl-CoA are first condensed to acetoacetyl-CoA by acetyl-CoA acetyltransferase; subsequently, acetoacetyl-CoA reductase converts acetoacetyl-CoA to (R)-β-hydroxybutyrate-CoA; and finally, the CoA moiety is removed to form 3HB. Recently, 3HB has been demonstrated to ameliorate dextran sulfate sodium (DSS)-induced colitis in animal models [43]. Since EcN is commonly used as a treatment for inflammatory bowel disease (IBD), its administration combined with therapeutic compounds and probiotics may exhibit synergy for enhanced therapeutic efficacy in colitis [44, 45]. Moreover, in situ synthesis of therapeutic substances can diminish the off-target effects of drugs and deliver the substances efficiently for extended periods of time [46, 47].
This study aims to develop a sustainable approach to treat chronic colitis using engineered EcN that can sustainably release 3HB.
Materials and methods
Strains, plasmids, chemicals, and culture conditions
Information on the strains and plasmids used in this study is listed in Supplementary Table 1 and Supplementary Data 1. E. coli strain Nissle 1917 served as the chassis for engineering in the study. Escherichia coli S17-1 was employed for plasmid construction and used as the conjugation donor for EcN derivatives. Q5 High-Fidelity DNA polymerase (New England Biolabs, Inc., USA) was used for amplification of DNA fragments. PCR products were purified using an E.Z.N.A. DNA/RNA Isolation Kit (Omega Bio-tek, Inc., USA). Plasmids were constructed via Gibson Assembly methods. Plasmids were extracted using a TIANprep Mini Plasmid Kit (Tiangen Biotech Co., Ltd., China). They were electroporated into E. coli S17-1 for further uses.
EcN and E. coli S17-1 were cultured at 37 °C in LB medium consisting of 10 g/L tryptone (Analytical Reagent, Oxoid, England), 10 g/L NaCl (Analytical Reagent, Sinopharm Chemical Reagent Co., Ltd., China) and 5 g/L yeast extract (Analytical Reagent, Oxoid, England). In fermentation studies, 10 g/L glucose (Analytical Reagent, Sinopharm Chemical Reagent Co., Ltd., China) was added in LB medium. Some essential experiments were repeated using M9 medium with 10 g/L glucose. In aerobic fermentations, the media were shaken at 200 rpm. In microaerobic fermentations, the culture medium was added to 15-mL tubes to full scale without shaking during the cultivation. In anaerobic fermentations, the cultures were incubated statically at 37 °C in an anaerobic chamber (Ruskinn Technology, England) under the following gas environments: 5% hydrogen, 5% carbon dioxide, and 90% nitrogen. An MM medium was used for selection of EcN strains from the mixture of S17-1 and EcN during the conjugation step. The MM medium consisted of 1% NaCl, 0.05% urea, 0.02% MgSO4, 1.0% Na2HPO4, 0.15% KH2PO4, 1.0% solution I and 0.1% solution II. Solution I consisted of 0.5% Fe(III)-NH4-citrate and 0.2% CaCl2, both dissolved in 1 M HCl. Solution II contained 0.01% ZnSO4, 0.003% MnCl2, 0.03% H3BO3, 0.02% CoCl2, 0.003% NaMoO4, 0.002% NiCl2 and 0.001% CuSO4 dissolved in 1 M HCl. Agar (15 g/L, BioDee Biotechnology Co., Ltd., Beijing, China) was added before sterilization. The chloramphenicol concentration was 25 μg/mL; it was purchased from BioDee Biotechnology Co., Ltd. (Beijing, China).
Construction of plasmid containing the 3HB synthesis pathway components in the EcN strain
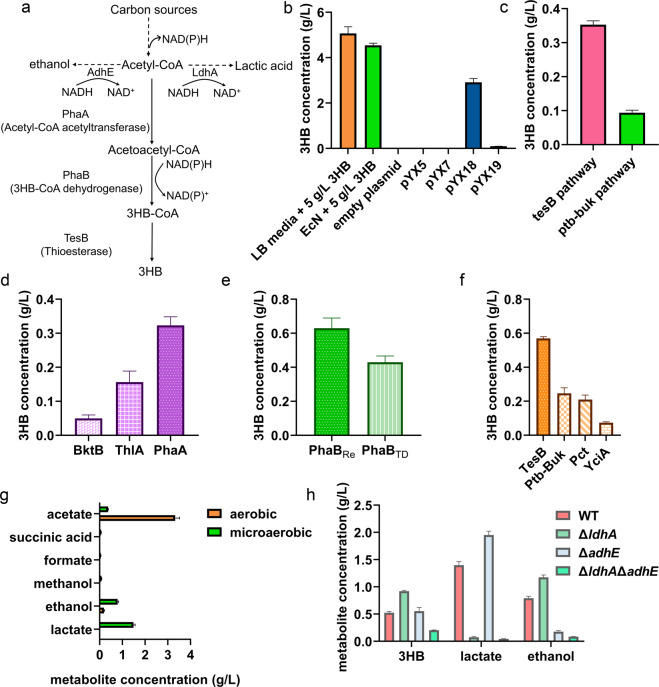
Possible degradation of 3HB in EcN was investigated by adding 5 g/L 3HB to the EcN cultures. EcN exhibited little 3HB degradation, as less than 0.5 g/L 3HB was utilized after 48 h of cultivation (Fig. 1a, b). Subsequently, a 3HB synthesis system comprising the phaA, phaBTD, and tesB genes encoding acetyl-CoA acetyltransferase from Cupriavidus necator H16 (GenBank accession no. CP039287.1), 3HB-CoA dehydrogenase from Halomonas bluephagenesis TD01 (GenBank accession no. WP_009724067.1) and thioesterase from E. coli MG1655 (GenBank accession no. CP032679.1) was cloned into the plasmids pYX5 and pYX18 under the weak promoter P32 and the strong promoter P23, respectively [48] (Fig. 1a). The rate-limiting enzyme TesB was replaced with its isozymes Ptb and Buk (GenBank accession no. AE001437.1), encoding phosphate acetyltransferase and butyrate kinase, respectively, to form the plasmids pYX7 and pYX19 (Supplementary Table 1). The EcN strain harboring the pYX18 plasmid produced 2.9 g/L 3HB in shake flasks under aerobic conditions compared to that of the other plasmids (Fig. 1b), indicating that the tesB gene was more favorable for 3HB production by the engineered EcN and that the stronger promoter P23 resulted in higher yields.
Fig. 1.
Metabolic engineering of E. coli Nissle 1917 for 3HB overproduction. a Biosynthesis pathway for 3HB production utilizing glucose and acetyl-CoA as the substrate. Three enzymes, e.g., acetyl-CoA acetyltransferase, 3HB-CoA dehydrogenase, and thioesterase, were employed. Two intrinsic branch pathways are also indicated in the illustration. b Determination of the EcN characteristics for 3HB degradation and biosynthesis. LB media + 5 g/L 3HB was germfree medium supplemented with 5 g/L 3HB. The blank plasmid group was the EcN strain transformed with the pNZ8148 plasmid. The pYX5, pYX7, pYX18, and pYX19 groups consisted of EcN strains transformed with the appropriate plasmids. The Y-axis shows the residual 3HB concentration in media after fermentation. c The 3HB production under microaerobic conditions via different pathways. The groups with EcN overexpressing pathways introduced with corresponding plasmids. d The 3HB yields from various acetyl-CoA acetyltransferases. e The 3HB yields from various 3HB-CoA dehydrogenases. PhaBCn, PhaB from Cupriavidus necator H16; PhaBTD, PhaB from Halomonas bluephagenesis TD01. f The 3HB yields from various thioesterases. g Byproduct concentrations in 3HB fermentation processes under aerobic or microaerobic environments. The pYX50 plasmid was introduced for 3HB production. The monitored metabolites are listed on the Y-axis. h The 3HB, ethanol and lactate production levels under different EcN chassis. The pYX50 plasmid was transformed for 3HB production. Bacteria were grown in LB medium supplemented with 10 g/L glucose. All aerobic fermentation data were acquired after 48 h of cultivation at 200 rpm at 37 °C. Microaerobic fermentation results were obtained at 37 °C without shaking. All data are the mean value of 3 biologically independent experiments, and error bars represent standard deviations
Since shake flask conditions do not reflect the microaerobic/anaerobic circumstances of a gastrointestinal tract, microaerobic fermentations were conducted to imitate gut lumen environments under which 3HB should be produced. Similarly, the tesB pathway exerted better performance than the ptb-buk pathway under microaerobic conditions to form 0.3 g/L 3HB by engineered EcN (Fig. 1c). To increase 3HB production by the EcN strain, we studied various isozymes of thioesterases, acetyl-CoA acetyltransferases and 3HB-CoA dehydrogenases for better catalytic performance. The PhaA protein was replaced by different acetyl-CoA acetyltransferases, namely, BktB from C. necator H16 (GenBank accession no. AF026544.2) and ThlA from Clostridium acetobutylicum ATCC 824 (GenBank accession no. AE001437.1). The enzyme PhaA led to the highest level of 3HB, 0.3 g/L, among the other groups (Fig. 1d). The PhaB protein from C. necator H16 (GenBank accession no. FJ897462.1) was used to replace PhaBTD to obtain a better acetoacetyl-CoA reductase. PhaBCn was revealed to be better in the 3HB synthesis pathway, leading to 0.6 g/L 3HB (Fig. 1e). In addition to ptb-buk genes, the gene pct encoding propionate CoA-transferase from C. propionicum (GenBank accession no. KT199425.1) and yciA encoding acyl-CoA thioester hydrolase from E. coli MG1655 (GenBank accession no. CP032679.1) were also considered candidates for CoA removal genes. TesB overexpression resulted in the highest level of 3HB, 0.6 g/L, among the three alternatives by the engineered EcN (Fig. 1f and Table 1).
Table 1.
Enzyme activities in 3HB biosynthesis pathway encoded in EcN strains
| Enzymes | Activities (U/mg) for E. coli Nissle 1917 strainsa | ||||
|---|---|---|---|---|---|
| Acetyl-CoA acetyltransferase | Control | PhaA | BktB | ThlA | |
| <0.1 | 12.7 ± 2.2 | 2.6 ± 0.4 | 5.1 ± 0.9 | ||
| 3HB-CoA dehydrogenase | Control | PhaBCn | PhaBTD | ||
| NDb | 2.2 ± 0.3 | 1.9 ± 0.2 | |||
| Thioesterase | Control | TesB | Ptb-Buk | Pct | YciA |
| 1.8 ± 0.3 | 40.6 ± 5.7 | 10.4 ± 2.7 | 8.7 ± 1.2 | 2.8 ± 1.1 | |
aOne activity unit was defined as the conversion of 1 mole of substrate to product during 1 min at 37 °C. Data are the average values and the standard deviations are calculated from three independent cultures
bND not detected
To further enhance 3HB production by the EcN strains, we quantified other metabolites in cultures of EcN strains to find potential metabolic flux losses, including those of acetate, lactate, and ethanol, which were produced in amounts of 3.1, 1.5, and 0.8 g/L, respectively, under microaerobic conditions (Fig. 1g). The formation of these metabolites can be attributed to NADH accumulation during anaerobic processes [49]. Deleting the lactate and ethanol synthesis pathways should significantly increase 3HB formation, as the two pathways channeled their metabolic flux to themselves (Figs. 1h and 2a). The genes ldhA and adhE, encoding lactate dehydrogenase and aldehyde-alcohol dehydrogenase, respectively, were inactivated to reduce the production of lactate and ethanol. Moreover, 3HB was increased to 0.9 g/L in cultures with ldhA-deleted EcN. Furthermore, lactate was reduced to 0.1 from 1.5 g/L (Fig. 1h). Similarly, the adhE mutant also significantly reduced ethanol to 0.2 from 0.8 g/L, yet 3HB was not increased accordingly (Fig. 1h). However, the double-mutant EcNΔldhAΔadhE generated a reduced 3HB concentration compared to the ldhA mutant (Fig. 1h), possibly due to the unbalanced redox state generated from the disruption of two NADH-consuming enzymes. Impaired growth was observed with the EcNΔadhE and EcNΔldhAΔadhE strains, another possible reason for the low 3HB production (data not shown). Microaerobic and anaerobic fermentation studies based on the defined M9 media also exhibited similar results to those mentioned above (Supplementary Fig. 1). Generally, the EcNΔldhA strain harboring the pYX50 plasmid produced the highest titer and was employed as the bacterial chassis in the following experiments.
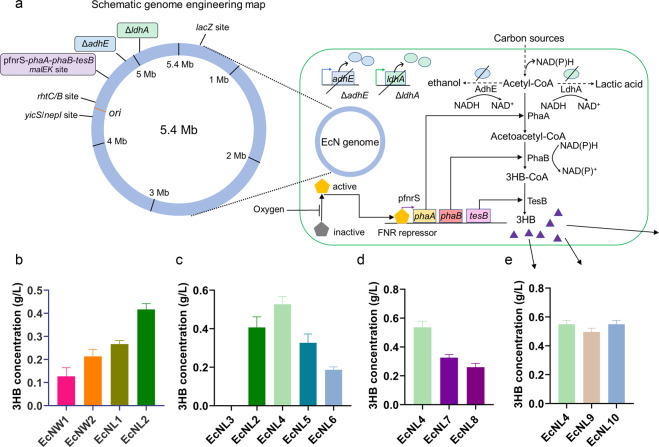
Fig. 2.
Genomic engineering to enhance 3HB production. a Schematic illustration of genome engineering in the EcN strains. The 3HB biosynthesis pathway is shown in the figure. The adhE or ldhA gene was knocked out. The 3HB biosynthesis route was integrated into different genomic sites. The 3HB molecules are presented as purple triangles. b Production of 3HB via different genome insertion strategies. EcNW strains were wild-type EcN integrated with the 3HB pathway. EcNL strains had EcNΔldhA inserted with the pathway. Group 1 had PhaBTD in the biosynthesis route. Group 2 had PhaBCn in the biosynthesis route. c The 3HB yields increased via promoter optimizations. EcNL3 was integrated with a promoterless 3HB biosynthesis pathway. EcNL4, EcNL5, and EcNL6 were inserted with the pathway driven by the pfnrS, J23119, and J23110 promoters. d The production of 3HB with different genomic integration sites. The biosynthesis route was integrated into the malEK, rhtCB and yicS/nepI sites for the EcNL4, EcNL7, and EcNL8 strains, respectively. e Determination of the 3HB yields in the EcN strains containing inserted selection markers. EcNL9 and EcNL10 were EcNL4 strains integrated with the ampR gene and sfGFP gene. Cells were grown in LB medium with 10 g/L glucose. Microaerobic fermentation results were acquired at 37 °C without shaking. All data represent the mean of 3 biologically independent parallel samples, and error bars indicate standard deviations
Conjugation
Conjugation was an effective way to introduce plasmids into EcN. E. coli S17-1 strains served as donors. E. coli S17-1 donors carrying plasmids and EcN strains were cultured at 37 °C for 4 h to OD600 = 0.5. Cells (1 mL) from the two strains were collected via centrifugation. Cell pellets were washed with LB medium to remove antibiotics. Then, the pellets were resuspended and mixed well at a ratio of 1:1 and cultured on selection MM medium plates at 37 °C for 12 h. The bacterial mixture was spread on MM solid plates containing selection antibiotics, followed by incubation at 37 °C for 24 h. Single colonies were verified via PCR.
Genome editing using homologous recombination
Gene integrations and deletions on the chromosome of EcN were performed using suicide plasmid pRE112-based homologous recombination strategies [50]. Antibiotics were employed as the selection marker of the single-crossover step. Colonies were verified via PCR. The sucrose concentration was 10 g/L to promote the double-crossover step on MM plates. Colonies were verified via PCR amplification and DNA sequencing. Descriptions of the modified EcN strains are listed in Supplementary Table 1. Strains were stocked in 25% glycerol solutions at −80 °C.
Fermentation studies
For 3HB production in shake flasks, LB medium with 10 g/L glucose was prepared for the experiments. Strains from glycerol stocks were resuscitated by streaking on LB plates. Single colonies were chosen and inoculated in LB liquid medium for 24 h of cultivation at 200 rpm and 37 °C, which served as the first seed culture. The second seed culture was acquired by inoculation of the first seed culture into fresh LB medium at 1% and continued growth for another 12 h. Bacteria were inoculated at 5% of the total volume and cultured for 48 h at 200 rpm at 37 °C. For microaerobic fermentations, the media and seed cultures were prepared the same way as in aerobic experiments. Bacteria were cultured in 15-mL tubes, fully filled in the tube, and then tightly sealed to prevent external oxygen uptake. Bacteria were also inoculated at 5% and cultured for 48 h at 37 °C without shaking.
Metabolite quantification via high-performance liquid chromatography (HPLC)
Concentrations of 3HB, acetic acid, succinic acid, formate, methanol, ethanol, and lactate were measured via high-performance liquid chromatography (HPLC). An LC-20 instrument (Shimadzu, Japan) equipped with an RID-10A refractive index detector (Shimadzu, Japan) was used for the experiments. The stationary phase was an Aminex HPX-87H column (Bio–Rad, U.S.A.). The mobile phase was 5 mM H2SO4 with a flow rate of 0.5 mL/min. The column temperature was set at 55 °C. Culture supernatants in the fermentation studies were collected after centrifugation at 12,000 g and 4 °C for 2 min and then subjected to a 0.22 μm polyethersulfone membrane syringe filter (Jinglong, China). The loading volume was 30 μL. Standards were purchased from Sinopharm Chemical Reagent and dissolved using five different concentrations to establish the standard curves. Sample concentrations were calculated based on the standard curve.
Enzyme activities
Bacteria were harvested via centrifugation at 5000 × g for 10 min after microaerobic growth, washed once with PBS, and then resuspended in a 10% volume of 0.1 M Tris-HCl buffer (pH = 7.5). Crude extract was acquired after the bacterial cells were disrupted by sonication at 450 W for 15 min with 3 s of pulse on and 2 s of pulse off time (SEIENTZ, China) on ice, followed by centrifugation at 12,000 × g for 10 min at 4 °C. The concentrations of total protein were determined via the Bradford method using Coomassie Plus Protein Assay reagent, and bovine serum albumin served as the standard [51]. Enzyme activities were assayed via a UV-visible light spectrophotometer (Shimadzu, Japan) at 25 °C. The activities of acetyl-CoA acetyltransferases were determined based on the decrease in acetoacetyl-CoA absorption at 303 nm [52]. One unit of acetyl-CoA acetyltransferase was defined as the amount of enzyme needed to consume 1 μmol of acetoacetyl-CoA in 1 min. Acetoacetyl-CoA reductase activity was determined via the decrease of NADPH absorption at 340 nm [53]. One unit of acetoacetyl-CoA reductase was defined as the amount of enzyme needed to consume 1 μmol of NADPH in 1 min. Thioesterase activity was assayed by monitoring the generation of CoA at 412 nm [54]. One unit of thioesterase activity was defined as the amount of enzyme needed to consume 1 μmol of 3-hydroxybutyryl-CoA per min.
Determination of microcins and siderophores
The microcin expression was determined as described previously [55]. In detail, bacteria were harvested after anaerobic fermentations. RNA was extracted using TRIzol® Reagent according to the manufacturer’s instructions. RNA concentrations were quantified via spectrophotometry at 260 nm. Genomic DNA removal and cDNA synthesis were performed using the PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa, Japan). qRT-PCR was performed using 2X RealStar Mixture with ROX II (GenStar, China) by using the 7500 Fast Real-Time PCR System (Applied Biosystems, USA). The constitutively transcribed gene gapA was used as a housekeeping gene control to normalize the total RNA quantity in different samples. The relative difference in mRNA levels was determined using the ΔΔCt method [56]. The genes mchB and mcmA encode microcin H47 and M, respectively. The primers used were reported previously [55].
Total siderophore concentrations were measured following the modified CAS assays [57, 58]. In detail, bacteria were grown in low-iron media, and the media were collected after anaerobic fermentation. CAS assay solution was freshly prepared. The concentration of siderophores was acquired by mixing 100 μl of supernatant and 100 μl of CAS reagent in a 96-well plate. Absorbance was measured at 630 nm using a VarioskanFlash microplate reader (Thermo Scientific, USA) after equilibration. The siderophore deferreoxamine E (Abcam, USA) was prepared at different concentrations as standards.
Animal experiments
The study was approved by the Laboratory Animal Research Center of Tsinghua University and by the Institutional Animal Care and Use Committee of Tsinghua University. Mice were maintained in a 12-h light-dark cycle environment and had full access to food and water. SPF male C57BL/6J mice (5 weeks) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. After 1 week of acclimation, mice were randomly allocated to different groups. For blood ketone dynamic assays, we used n = 4 in each group; for pharmacokinetics assays, we used n = 6 in each group. EcN strain groups were administered 5 × 1010 cells or other amounts mentioned in the main text. The 3HB group was orally administered doses of 100 mg/kg mouse weight. The bacteria were suspended in 0.2 ml of PBS, and other agents were also given at an equivalent volume. Blood ketone level was measured using the Freestyle Optium ketone monitoring system (Abbott, USA). Mouse feces (~0.2 g per mouse) were collected before drug administration and 3 days after administration or at other time points as indicated. Mice were sacrificed via euthanasia after the corresponding experiments.
For pharmacokinetics assays, mouse feces were collected 1, 3, 7, 14, and 21 days after oral administration of different bacteria. The weights of the feces were recorded, and the samples were spread on selection plates with gradient dilutions using PBS. Plates were placed in an incubator at 37 °C to quantify colonies. Colonies were randomly inspected via PCR.
For DSS-induced colitis mouse models, SPF male C57BL/6J mice (7 weeks) were used, and DSS (MP Biomedicals, USA) was given in drinking water at a concentration of 2.5%. Mice (n = 5) in each group were selected for each set of experiments. Treatments were continued for 7 days to establish the colitis mouse model. Mouse weights were measured every day. Feces were sampled on the 3rd day after oral administration. Blood and colon samples were collected at the end of experiments. Blood, colon, and feces were collected for biochemical and histological assays. Colonic myeloperoxidase activity experiments were conducted via MPO Assay Kits (Jiancheng Biotech, China). Occult blood was assayed by Hemoccult II Dispensapak Plus (Beckman Coulter, USA). The following scoring system was employed for comparative analysis of colonic bleeding. Grade 0: normal stool without hemoccult; Grade 1: normal stool with hemoccult; Grade 2: soft feces with positive hemoccult; Grade 3: very soft feces with blood traces; Grade 4: watery stools with visible bleeding.
Histological studies
Colonic samples were fixed and embedded in paraffin, sectioned (5 μm) and stained with hematoxylin and eosin (H&E). The slices were scored blindly by a pathologist with histological evidence of DSS colonic damage with the scoring system described [59].
Determination of serum cytokines
Serum cytokine concentrations were measured via the Mouse IL-1β ELISA Kit, Mouse IL-6 ELISA Kit and Mouse TNF-α ELISA Kit (Beyotime, China) according to the manufacturer’s instructions.
Bacterial 16S rRNA gene sequencing
Feces were collected on the 3rd day after administration, immediately frozen at −80 °C, and then transferred to Hangzhou Lianchuan Bio Technologies Co., Ltd., for further studies. Briefly, bacterial DNA was acquired using a QIAamp Fast DNA Stool Mini Kit (Qiagen, Germany) based on the manufacturer’s instructions. After PCR amplification using the primers 338F and 806R, sequencing was performed via Illumina MiSeq platforms. Linear discriminant analysis (LDA) was performed by online tools (http://huttenhower.sph.harvard.edu/galaxy/).
Liquid chromatography-mass spectrometry (LC-MS) quantification of SCFAs
Fecal samples were homogenized, and then, 1 ml of 50% acetonitrile was mixed with 100 mg feces by vortexing, followed by centrifugation at 4000 g and 10 °C for 10 min. Forty microliters of supernatant was mixed with 20 μL of 200 mM 3-nitrophenylhydrazine (Sigma, USA) and 20 μL of 120 mM N-(3-dimethylaminopropyl)methacrylamide (Sigma, USA) dissolved in 6% pyridine solution. The mixture was reacted at 40 °C for 30 min.
The Dionex Ultimate 3000 UPLC system was coupled with a TSQ Quantiva Ultra triple-quadrupole mass spectrometer (Thermo Fisher, USA) equipped with a heated electrospray ionization (HESI) probe in negative ion mode. Extracts were separated by a BEH C18 column (2.1 × 100 mm, 1.7 μm, Waters). A binary solvent system was employed: mobile phase A consisted of 100% H2O, and mobile phase B consisted of 100% acetonitrile. An 18-min gradient method with a flow rate of 350 μL/min was used as follows: 0–1.5 min at 5% B; 1.5–4 min, 5–15% B; 4–12 min, 15–55% B; 12–13 min, 55–98% B; 13–15 min, 98% B and 15.1–18 min, 5% B. The column chamber and sample tray were held at temperatures of 40 and 10 °C, respectively. Data acquired in selected reaction monitoring for 3HB, formate, acetate, propionate, butyrate and valerate. Both precursor and fragment ions were collected with a resolution of 0.7 FWHM. The source parameters were set as follows: spray voltage: 1000 V; ion transfer tube temperature: 350 °C; vaporizer temperature: 450 °C; sheath gas flow rate: 40 Arb; auxiliary gas flow rate: 20 Arb. CID gas: 2.0 mTorr. Data analysis and quantitation were performed by Xcalibur 3.0.63 software (Thermo Fisher, USA).
Statistical analysis
Data are shown as the mean ± SD. Statistical analysis was performed via GraphPad Prism 8 software. The details of the tests used are displayed in the figure legends. The threshold for statistical significance was set as P < 0.05.
Results
Construction, optimization, and genomic integration of the 3HB synthesis system for enhanced and sustainable 3HB production
First, we constructed and optimized the 3HB biosynthesis system using plasmids in EcN strains, and the details are accessible in the Materials and Methods section (Fig. 1 and Table 1). To prevent horizontal gene transfer and avoid using antibiotics to maintain plasmid stability [60], we inserted the whole 3HB synthesis pathway into the bacterial genome for in vivo applications. Importantly, 3HB should be maintained at a relatively high level to exert a therapeutic effect, and the pathway should be tuned concisely in the chromosome expression system.
The whole 3HB system consisting of the P23 promoter, phaA, phaBCn (or phaBTD) and tesB, was integrated into the intergenic region of malE and malK [1, 7], given the robustness of gene expression at the site. Moreover, two bacterial hosts, wild-type EcN or EcNΔldhA, were selected for genome engineering. EcNL2, which overexpressed phaBCn in the genome of the EcNΔldhA strain, produced 0.4 g/L 3HB, the highest 3HB concentration compared with those of the other genomic insertion candidates (Fig. 2b).
A variety of promoters were tested to increase the 3HB titer. The promoters pfnrS, J23119 and J23110 were cloned upstream of the functional genes in the EcNL4, EcNL5, and EcNL6 strains, respectively, taking EcNL3 as a control without the promoter preceding the genes (Fig. 2c). Strain EcNL4 exhibited better performance, as more than 0.5 g/L 3HB was produced under microaerobic conditions (Fig. 2c). In addition, both microaerobic and anaerobic fermentation studies based on the defined M9 media demonstrated that the EcNL4 strain could produce 0.5 g/L 3HB, the highest value among the candidates (Supplementary Fig. 2). In accordance with the literature, pfnrS is a low oxygen-induced promoter suitable for the anaerobic gut environment [61]. Other genomic insertion sites, e.g., the rhtCB and yicS/nepI intergenic areas, were integrated with the 3HB synthesis gene cluster to form strains EcNL7 and EcNL8. The malEK site was found to be preferable in this expression system, as over 0.5 g/L 3HB was obtained (Fig. 2d). Fermentations using M9 media under microaerobic or anaerobic conditions also proved that the malEK site was better than other sites for 3HB production (Supplementary Fig. 3). Multiple site insertions were also conducted for enhanced 3HB production, leading to a marginal 3HB increase and weaker cell growth when two sets of genes were expressed (data not shown). The genome-engineered bacteria produced over 3 g/L 3HB under aerobic conditions, the highest 3HB titer reported in shake flasks (Supplementary Fig. 4). EcNL4 with the ampicillin resistance gene ampR or the fluorescence marker sfGFP was constructed for subsequent in vivo detection. The resulting strains EcNL4, EcNL9, and EcNL10 produced at least 0.5 g/L 3HB under the same microaerobic conditions (Fig. 2e).
To investigate the possible influences generated by genetic modifications on the EcN probiotic capabilities, we determined the production of microcins and total siderophores. The results indicated that the H47 and M microcins as well as total siderophore concentrations changed little (<1.3-fold) in cultures of the EcNL4 strain compared with those of the wild-type EcN strain (Supplementary Fig. 5). Genetic modifications did not alter the intrinsic probiotic effects of EcN.
Pharmacokinetics of EcNL4 releasing 3HB in mouse models
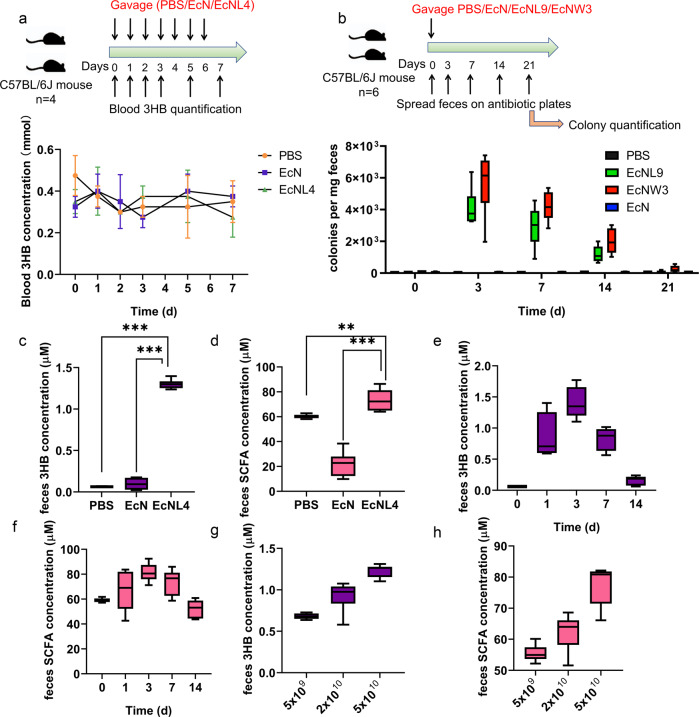
Mice were used to investigate the parameters of the engineered EcN bacteria in the colon environment for 3HB production. EcNL4 was orally administered to C57BL/6J mice once a day, and the blood 3HB levels were measured as described (Fig. 3a). The 3HB level was found to be between 0.3 and 0.5 mmol/L during the experiments (Fig. 3a), and administration of EcNL4 by gavage also generated little difference in blood ketone levels (Fig. 3a). The engineered probiotic bacteria did not change the blood ketone level for 24 h in the gut (Supplementary Fig. 6), which is reasonable due to the powerful buffering capacity of 3HB in mammalian blood systems [62].
Fig. 3.
In vivo pharmacokinetics of recombinant EcNL4 in mice. a Blood 3HB dynamics after oral administration of the EcNL4 strain. Cells were administered at 5 × 1010 per mouse. The mouse type, numbers, gavage, and quantification frequencies are indicated in the figure. b Engineered EcN gut colonization dynamics after cell administration. EcNL9 was an EcNL4 strain with an ampicillin resistance gene. The EcNW3 strain was EcNW2 with an ampicillin resistance gene. Cells were administered at 5 × 1010 per mouse. The mouse type, numbers, gavage, and detection frequencies are indicated in the figure. c Fecal 3HB concentrations after administration. Feces were sampled on the 3rd day after gavaging. d Fecal SCFA contents after the administration. Feces were sampled on the 3rd day after gavaging. SCFA titers were calculated by accumulating formate, acetate, propionate, butyrate and valerate concentrations. e The 3HB dynamics 14 days after EcNL4 administration. f The SCFA dynamics 14 days after EcNL4 administration. The SCFA titers were calculated by accumulating formate, acetate, propionate, butyrate and valerate concentrations. g The effects of cell amount on 3HB production. Feces were sampled on the 3rd day after gavaging. h The effects of cell amount on SCFA production. Feces were sampled on the 3rd day after gavaging. The SCFA titers were calculated by accumulating formate, acetate, propionate, butyrate and valerate concentrations. All data represent the mean value of 6 biologically independent samples except in a. Data in a were from 4 biologically independent samples. Error bars indicated standard deviations. Two-tailed Student’s t tests were conducted to suggest statistical significance for comparisons. **p < 0.01. ***p < 0.001
Antibiotic-marked EcNL4 (termed EcNL9), engineered EcN, EcNW3, and PBS buffer were administered to mice via gavage to examine the gut colonization ability of the engineered EcN (Fig. 3b). Strain EcNL9 had deletion of ldhA, while EcNW3 possesses an intact ldhA on the chromosome (Supplementary Table 1). Feces were sampled and spread on selection plates to quantify live engineered EcN cells (Fig. 3b). The engineered EcN could be retained in the gastrointestinal tract for more than 14 days, with the peak retention value appearing on the 3rd day (Fig. 3b). An approximate 30% reduction in gut colony number was observed 7 days after administration. Fluorescence-labeled engineered EcN exhibited similar results (Supplementary Fig. 7). EcNL9 was slightly inferior in colon colonization compared to EcNW3. However, EcNL9 and EcNW3 produced 0.6 and 0.3 g/L 3HB, respectively (Supplementary Fig. 8). Therefore, EcNL4 was chosen for subsequent animal experiments.
We studied 3HB produced in the colorectal tracts after uptake of the live bacteria using fecal samples for analysis of metabolites via LC-MS. The 3HB level was increased significantly in the gavage-treated EcNL4 group compared to the wild-type EcN group with barely altered gut lumen 3HB contents (Fig. 3c). Compared with that in the EcN group, the 3HB level in the EcNL4 group was elevated 8.7-fold (Fig. 3c). Moreover, the administration of engineered EcN increased the colonic short-chain fatty acid (SCFA) levels by 3.1-fold compared to those of the EcN group (Fig. 3d). The acetate and butyrate levels in the EcNL4 group were increased to 56.4 from 14.0 μM for acetate and 13.8 from 2.2 μM for butyrate compared to those of the EcN group. However, formate (0.4 μM vs. 0.5 μM), propionate (3.2 μM vs. 3.8 μM) and valerate (0.2 μM vs. 0.4 μM) showed weak changes (Supplementary Fig. 9). As SCFAs regulate gut immunity and benefit epithelial cells, the combination of 3HB and SCFAs indicates that EcNL4 can exert better probiotic functions than wild-type EcN does [63].
To obtain a dynamic profile of 3HB and SCFAs from intestinal colonization of EcNL4, we orally administered the bacteria to mice with subsequent fecal metabolites assayed at various time points. The 3HB level continued to increase within 3 days after administration from 0.1 to 1.3 μM before a slow decrease was observed over time, and it maintained a relatively high level of over 0.2 μM in 14 days (Fig. 3e). The kinetics of total SCFA concentrations were observed to be similar to those of 3HB; SCFA concentrations increased from 57.5 to 80.4 μM in the first 3 days before dropping to 52.4 μM after 14 days (Fig. 3f). Among the SCFAs, acetate, and butyrate were increased from 37.0 to 60.3 μM and 9.3 to 14.4 μM, respectively, during the initial 3–7 days and gradually dropped to the initial concentrations after 7 days of administration (Supplementary Fig. 10); the formate and propionate levels changed only from 0.4 to 0.5 μM and from 9.6 to 7.1 μM, respectively, over the experimental period of time (Supplementary Fig. 10); valerate exhibited an opposite pattern to 3HB as it decreased from 1.7 to 0.3 μM during the first 3 days, followed by an increase to 0.9 μM after 4th days. The results indicated probiotic-dependent concentration changes in metabolites.
Changes in the 3HB and SCFA contents were bacterial dose-dependent (Fig. 3g, h). The 3HB level was increased from 0.7 to 1.2 μM as the administered cell number increased from 5 × 109 to 5 × 1010 per mouse (Fig. 3g). Total SCFAs also increased from 55.5 to 80.6 μM after the cell number changed (Fig. 3h). Acetate (from 42.1 to 63.5 μM) and butyrate (from 7.9 to 12.0 μM) levels were positively correlated with EcNL4 numbers, while the levels of formate and propionate were independent of the probiotic number, and valerate (from 0.5 to 0.3 μM) showed a negative correlation with bacterial dosage (Supplementary Fig. 11). A total of 5 × 1010 cells were selected as the suitable dosage for the next step of the studies.
The 3HB-producing EcNL4 altered gut microbiota via the enrichment of Akkermansia spp
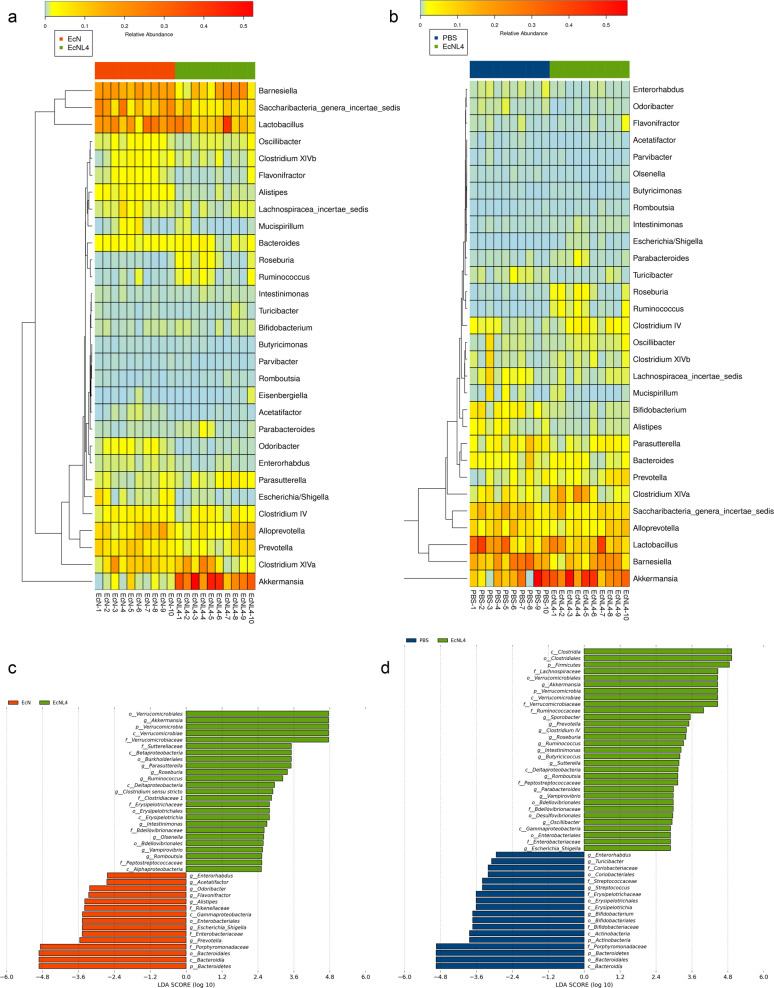
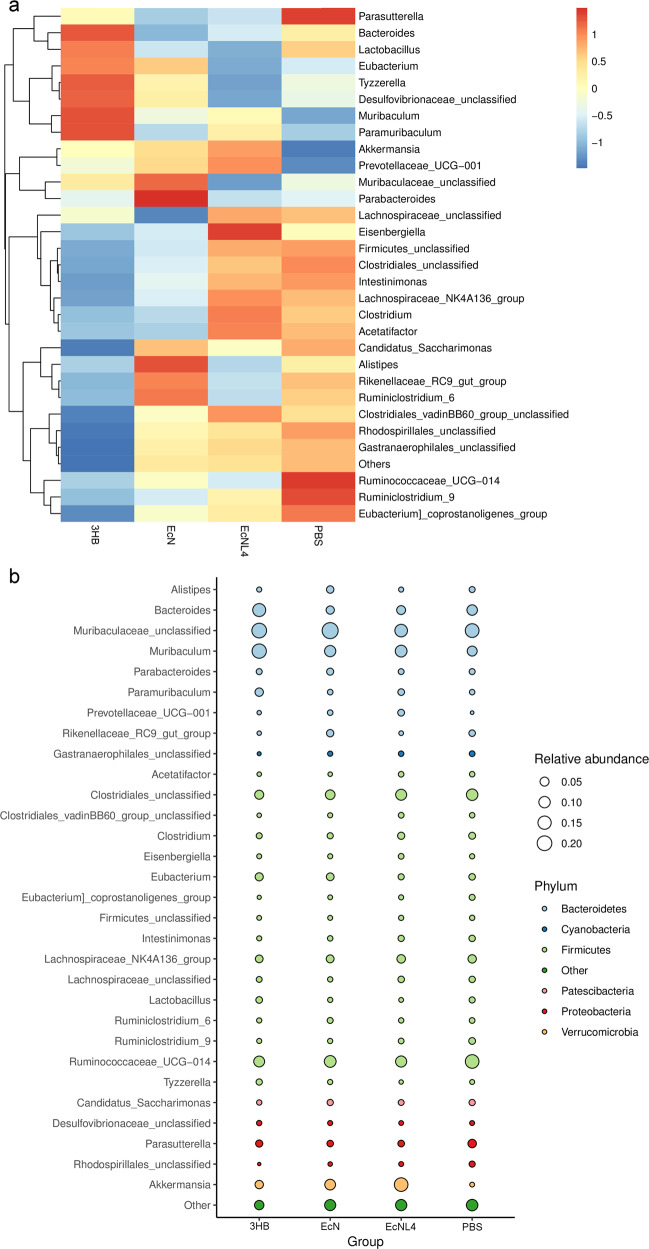
To further understand the microenvironments in the gastrointestinal tract, we analyzed colonic microflora compositions based on 16S sequencing. Three mouse groups gavaged with EcNL4, wild-type EcN and PBS buffer were established to determine their microbiome differences. Akkermansia, a mucin-degrading probiotic, was found to be enriched in 31% of the total gut bacteria in the EcNL4-administered mouse group compared to only 2% in the EcN group (Fig. 4a), indicating the strong positive effect of EcNL4 on the microbiome in the mouse gut lumen (Supplementary Fig. 12). Levels of Akkermansia or Verrucomicrobiae were found to distinguish the EcNL4 group from the EcN group via LDA (Fig. 4c). Some SCFA producers, such as Roseburia and Ruminococcus, were increased in number. The Roseburia abundance was 1.5% in the EcNL4 group compared with 0.2% in the EcN group (Supplementary Fig. 12). Ruminococcus was enriched to 1.2%, while it consisted of 0.3% in the EcN group (Supplementary Fig. 12). Since Akkermansia spp. produce SCFAs, the changing microbial populations change the SCFA levels [64]. The abundance of Alistipes, a genus highly relevant to diseases and dysbiosis [65], was decreased in the EcNL4 group to 0.5% compared to 2.6% in the EcN group. EcNL4 could increase probiotic abundance while reducing pathogens compared to wild-type EcN.
Fig. 4.
Gut microbiomes changed after EcNL4 administration in mice. a General gut microbiota genus differences between EcN and EcNL4 administration. b General gut microbiota genus differences between PBS and EcNL4 administration. Feces were collected 3 days after bacterial gavage. Data are presented in the heatmap. The color gradient represents the relative abundances in the samples. Vertical clustering indicates the similarity of all species between different samples. c LEfSe difference analysis between the EcN and EcNL4 groups. d LEfSe difference analysis between the PBS and EcNL4 groups. The figures represent the linear discriminant analysis (LDA) scores that suggested microorganisms with significant impacts on the differences of groups in microflora. The threshold was 2 in LDA analysis
Compared to those of the EcNL4- and PBS-administered mouse groups, the abundances of the probiotics Akkermansia, Roseburia, Clostridium subcluster XIVa and Ruminococcus were increased to 31.3% from 18.2%, 1.5% from 0.2%, 8.1% from 3.0%, and 1.2% from 0.1%, respectively. However, that of Alistipes was decreased to 0.5 from 1.8% (Fig. 4b and Supplementary Fig. 13). Furthermore, the abundance of Prevotella, also a probiotic in the colonic environment [66], was found to be 3.3% for the EcNL4 group and 1.7% for the PBS group. The LDA analysis revealed that Clostridia, certain Firmicutes, and Akkermansia showed the greatest discrimination between the EcNL4- and PBS-administered groups (Fig. 4d), demonstrating that the presence of EcNL4 in the gut enhanced probiotics and SCFAs in the gut, which have beneficial effects on host health.
Enhanced therapeutic effects of EcNL4 on ameliorating DSS-induced colitis
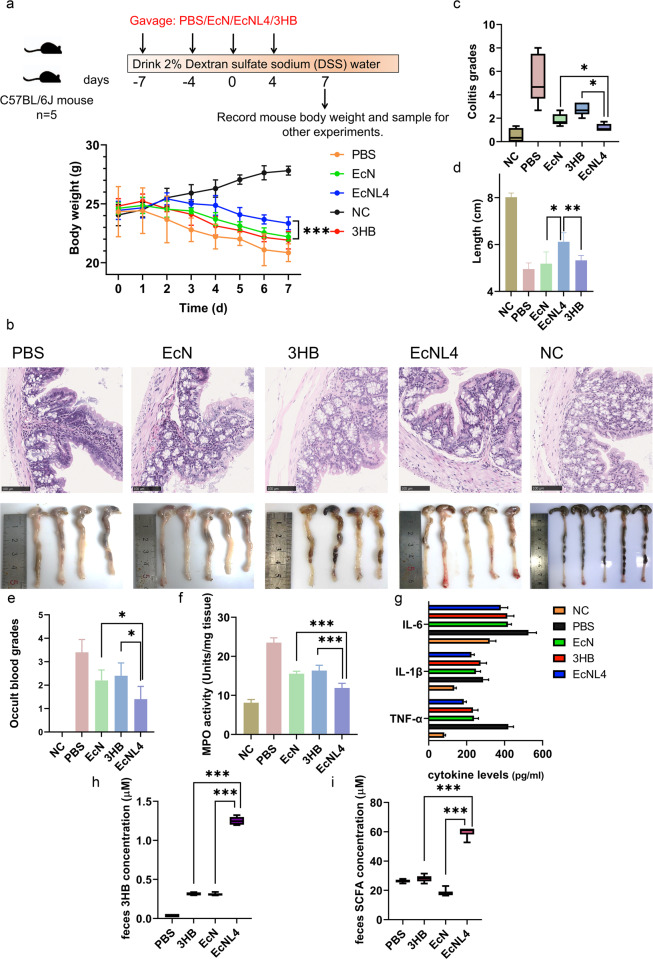
Since EcNL4 elevated gut 3HB and SCFA levels, increasing the abundances of Akkermansia and Clostridium subcluster XIVa, which ameliorates colitis by supporting Treg cell differentiation [67], we reasoned that EcNL4 could be used to treat IBD. Mice with DSS-induced colitis [68] were divided into four groups: groups orally administered EcNL4, EcN, PBS or 3HB solution, and the group of noncolitis mice gavaged with PBS was designated the negative control (NC) group.
The mice in the EcNL4 group exhibited alleviated weight loss compared to those in the EcN and 3HB groups (Fig. 5a). The mice with colitis administered EcNL4, EcN, 3HB, and PBS and those in the NC group after 7 days weighed 23.7, 22.3, 21.9, 20.9, and 27.8 g, respectively. Hematoxylin-eosin (H&E) staining was conducted to study the colitis levels in the above mice. Architectural changes, inflammatory cell infiltrations, and epithelial injuries were relieved in the EcNL4 group (Fig. 5b). Based on colitis indices [59], the groups administered EcNL4, EcN, 3HB, PBS and the NC group scored 1.2, 1.9, 2.7, 4.6, and 0.3 points, respectively, indicating EcNL4 to be the most effective agent against colitis in mice (Fig. 5c). Colon lengths were found to be 6.1 cm in the EcNL4 group compared to 5.2 cm in the EcN group, 5.3 cm in the 3HB group, 5.0 cm in the PBS group and 8.0 cm in the NC group, demonstrating the obvious protective effects of EcNL4, as longer colons generally imply a healthier gut.
Fig. 5.
EcNL4 ameliorated colitis better than wild-type EcN in mouse models. a Body weight dynamics of the mice with colitis. The illustration above shows the mouse type, numbers, gavage, and model construction strategies. Mouse body weights were monitored every day. Feces were collected 3 days after administration. The NC group was normal mice gavaged with PBS. Statistical analysis was conducted between the EcNL4 group and the heaviest group (EcN group). b Physiological and histological results of the mice with colitis. The upper figure shows H&E staining of colon slices. The lower figure illustrates the colon length of the mice. c, d The figures show the statistical index results of the figures mentioned in b. e Occult blood level in mouse feces. f Mouse colon tissue MPO activities. g Blood cytokine levels. IL-6, IL-1β, and TNF-α levels were determined in the groups. h Fecal 3HB concentrations in the DSS-treated mice. i Gut SCFA concentrations in the DSS-treated mice. SCFA titers were calculated by accumulating formate, acetate, propionate, butyrate and valerate concentrations. All data represent the mean value of 5 biologically independent samples. Error bars indicate standard deviations. Two-tailed Student’s t tests were performed to determine statistical significance for comparisons. *p < 0.05, **p < 0.01. ***p < 0.001
Fecal occult blood levels indicate the degree of colon damage. The EcNL4 mice scored 1.4 points, the lowest occult blood level compared with 2.3 points for the EcN group, 2.4 points for the 3HB group, 3.4 points for the PBS group and 0 points for the NC group (Fig. 5e). Myeloperoxidase assays indicated that the EcNL4 group scored the lowest with 11.9 units and suffered from mild inflammation (Fig. 5f) compared to the EcN (15.6 units), 3HB (16.4 units), PBS (23.5 units) and NC groups (8.1 units). Furthermore, proinflammatory cytokines, including IL-6, IL-1β, and TNF-α, were depressed, as found in the EcNL4 group, suggesting reduced inflammation in the mice (Fig. 5g). IL-6 concentrations were 379.8, 417.4, 414.3, 525.7, and 321.8 pg/ml for the EcN, 3HB, PBS, and NC groups, respectively. The IL-1β levels were 225.1, 249.2, 273.0, 286.4, and 134.6 pg/ml for the EcN, 3HB, PBS, and NC groups, respectively. TNF-α concentrations reached 185.0, 238.5, 233.5, 419.6, and 82.1 pg/ml for the EcN, 3HB, PBS, and NC groups, respectively. All scores clearly indicated that EcNL4 suppressed DSS-induced colitis and gut inflammation better than EcN or 3HB solution alone.
Gut 3HB levels increased significantly in the mice with DSS-induced colitis administered EcNL4 (Fig. 5h), reaching a level 4.2-fold higher than that of the EcN group. The EcNL4 group produced 60.3 μM colonic SCFAs (Fig. 5i) compared to 18.0 μM for the EcN group, 27.3 μM for the 3HB group and 27.0 μM for the PBS group. Among the SCFAs, acetate and butyrate accumulated after gavaging EcNL4 to 45.1 μM vs. 8.9 μM in the EcN group and 12.1 μM vs. 3.6 μM in the EcN group, respectively (Supplementary Fig. 14). The abundances of Akkermansia and Prevotella were increased mostly in the EcNL4 group, similar to the previous results (Fig. 6a, b). The abundances of Akkermansia were found to be 0.35% for the PBS group, 3.85% for the 3HB group, 8.62% for the EcN group and 16.44% for the EcNL4 group. Similarly, Prevotella reached an abundance of 0.02% in the PBS group, 0.20% in the 3HB group, 0.79% in the EcN group and 1.92% in the EcNL4 group. The higher ratios of Akkermansia and Prevotella in the microbiome contributed by EcNL4 should provide remarkable benefits for the colon and thus for a healthy individual.
Fig. 6.
Gut microbiome changes after administration of bacteria in mice with colitis. a Gut microbiota differences at the genus level in the mice with colitis. Feces were collected 3 days after bacterial gavage. Data are presented in the heatmap. The color gradient represents the relative abundances in samples. Vertical clustering indicates the similarity of all species between different samples. b The relative abundance of different genera in the groups of mice with colitis. Data are exhibited in the bubble map format. The size of the bubbles represents the relative abundance of the corresponding genus. The different colors in the bubble indicate the phylum annotations of specific genera
Discussion
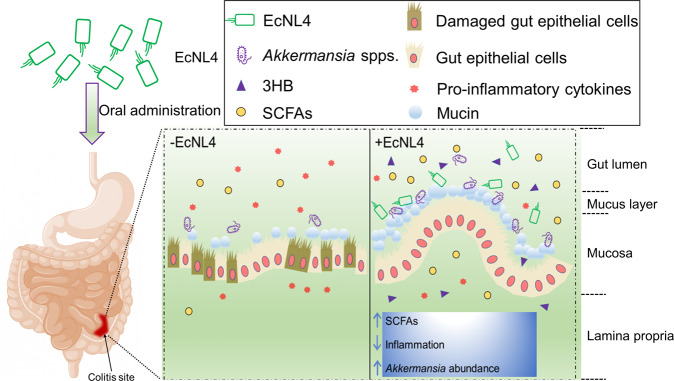
An engineered live therapeutic bacterium derived from EcN was successfully constructed to sustainably overproduce 3HB to alleviate colitis (Fig. 7). The resulting probiotic effects can originate from (i) the therapeutic agent 3HB, (ii) the probiotic nature of the EcN strain, (iii) elevated SCFA levels in gut lumen, and (iv) enhanced abundances of other probiotics. The effectiveness of 3HB disease treatment was demonstrated by systematic colonic niche improvements (Fig. 4). Engineered EcN colonization in colons can alter the SCFA concentration and enrich the probiotic abilities, reinforcing the amelioration of colitis. Rationally engineered EcNL4 overproduced 3HB (Fig. 3), and this bacterium can participate in the treatment of other conditions, including enteric diseases, serving as a dietary supplement to positively affect other body physiological statuses via the brain-gut axis and gut-liver axis [69, 70].
Fig. 7.
Schematic illustration of the EcNL4 therapeutic. Briefly, EcNL4 was orally administered, colonized the colitis sites, ameliorated colitis by upregulating SCFA concentrations, decreasing inflammation and enriching probiotics such as Akkermansia spp. Figure captions are exhibited above the figure
Initially, it was necessary to investigate whether the 3HB-producing chassis degrades 3HB, and the results showed this was not the case, indicating the ineffective intrinsic 3HB degradation pathway in EcN (Fig. 1b). Generally, 3HB is formed under microaerobic to anaerobic environments in gastrointestinal tracts (Fig. 1c–h), and it was increased from 0.3 to 0.9 g/L via rational metabolic flux tuning of plasmids, reaching the highest level reported (Fig. 1c–h and Table 1) [41]. After genomic integration of the 3HB production system, the 3HB level was increased to 3.2 and 0.6 g/L under aerobic and microaerobic circumstances, respectively (Fig. 2d and Supplementary Fig. 4), when the anoxia-induced promoter pfnrS and the malEK integration site were chosen for high expression requirements (Fig. 2c, d).
The engineered bacterium EcNL4 was applied orally for therapeutic investigation. However, the mouse blood 3HB level was not elevated after administration, indicating the vague ability of EcNL4 to generate sufficient 3HB levels for blood circulation (Fig. 3a and Supplementary Fig. 6). Similar to wild-type EcN, EcNL4 was able to grow and colonize mouse guts for at least 14 days (Fig. 3b), and gut 3HB and SCFA levels were increased 8.7- and 3.1-fold over the wild-type after administration, respectively (Fig. 3c, d), accompanied by increasing probiotic Akkermansia spp. from the initial 2–31% (Fig. 4).
As demonstrated above (Figs. 3 and 4), the engineered live therapeutic bacterium EcNL4 could have multiple beneficial effects colonic microenvironments, leading to alleviation of DSS-induced chronic colitis (Fig. 5). The therapeutic effects on colitis included increased body weights (23.7 g vs. 22.3 g in the EcN group) and colon lengths (6.1 cm vs. 5.2 cm in the EcN group) compared to those of the control groups and decreased immune cell invasion (colitis grades 1.2 vs. 1.9 in the EcN group), occult blood (occult blood grades 1.4 vs. 2.3 in the EcN group), proinflammatory cytokines and myeloperoxidase activities (activity units 12.8 vs. 16.2 in the EcN group) after the treatments (Fig. 5). These beneficial effects could be attributed to elevated 3HB (4.4-fold over the EcN group), SCFA levels (3.3-fold over the EcN group) and probiotic abundances, especially Akkermansia spp. (16.4% vs. 8.4% in the EcN group) (Figs. 5h, i and 6).
Engineered bacteria-releasing molecules offer multiple advantages over conventional drug administration. First, bacteria grown on biosubstances can produce therapeutic agents, are cost effective and sustain drug release. Second, therapeutic and probiotic bacteria have few side effects, as they mostly colonize the targets. Third, the method can be more effective than traditional drug delivery in regard to precision, duration, and control especially in the colon. This study provided a regular dosage of 100 mg/kg 3HB in mice under daily oral administrations [37]. In contrast, EcNL4 gavage every 3 days generated better preclinical effects. Moreover, the enhanced ketone bodies resulting from 3HB oral uptake were quickly consumed and thus not sustainable. Thus, engineered live therapeutics are an alternative to canonical dosing for sustainable treatments.
Diverse chemical agents could also be employed for live therapeutic engineering. As shown by recently successful examples, the general characteristics of candidate therapeutic molecules could be conveniently generated and optimized to produce low medical threshold concentrations [5]. Some metabolites with therapeutic effects, e.g., γ-aminobutyric acid (GABA) and serotonin, could be choices for healthy treatments [2, 3, 18]. Furthermore, multiple chemicals could be synthesized by the same engineered live bacteria to exert synergistic effects or target different diseases.
EcN is a widely recognized engineering chassis for in vivo live therapies. Diversified properties of this bacterium include not only adhesion and antagonistic effects to pathogens but also convenient genetic manipulation and growth under aerobic, microaerobic and anaerobic conditions [16, 19]. Similarly, some gut probiotics and prebiotics, such as Bacteroides, Lactobacillus and Clostridia, can be developed as live therapeutics. These bacteria generally form a majority of gut consortia and possess strong a gastrointestinal colonization ability [66]. However, no mature molecular and synthetic biology technology has been developed for these bacteria. Thus, various live gut microorganisms should be developed to systemically target various diseases.
The study indicates that the bacteria EcN and 3HB could be combined to exert synergistic effects on IBD treatments. The molecule 3HB could be manufactured from poly(3-hydroxybutyrate) via hydrolysis reactions [71]. It can be mass produced because poly(3-hydroxybutyrate) and PHB can be acquired from large-scale industrial microbial fermentation [72]. As a component of human blood, 3HB is reasonably safe for therapeutic uses or food additives. The combination of EcN with 3HB could also be an interesting alternative that does not require the use of genetically engineered organisms.
In summary, an engineered live therapeutic was successfully constructed to target gastrointestinal tracts by sustainably releasing 3HB to improve the gut microbiome and to ameliorate colitis in mice. Interestingly, EcN was found to be closely related to uropathogenic E. coli (UPEC) strain CFT073 but proven safe with epithelial barrier protective abilities [73, 74]. However, some studies have indicated that mice are not very sensitive to enterohemorrhagic E. coli O157:H7 strains [73]. Subsequent studies on rats, rabbits or stem cell-derived human intestinal organoids should be carried out to further support the positive therapeutic results in mice. Further studies on primates and human clinical trials should be carried out to assess clinical applications.
Supplementary information
Acknowledgements
This research was financially supported by a grant from the Chunfeng Foundation (2020Z99CFG002) of Tsinghua University. Other support includes the National Natural Science Foundation of China (Grant Nos. 31870859, 21761132013, 31771886, and 31971170).
Source data
Author contributions
XY designed and conducted the study. XYL and DZ performed the experiments. YDZ and ZHL assisted with the animal experiments. XL helped draw the illustrations. XY wrote the manuscript. FQW and GQC supervised the project and helped write the manuscript. Source data are provided with this paper
Data availability
Data supporting the study are available from the corresponding authors by request. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-021-00760-2.
References
- 1.Kurtz CB, Millet YA, Puurunen MK, Perreault M, Charbonneau MR, Isabella VM, et al. An engineered E. coli Nissle improves hyperammonemia and survival in mice and shows dose-dependent exposure in healthy humans. Sci Transl Med. 2019;11:475. doi: 10.1126/scitranslmed.aau7975. [DOI] [PubMed] [Google Scholar]
- 2.Aggarwal N, Breedon AME, Davis CM, Hwang IY, Chang MW. Engineering probiotics for therapeutic applications: recent examples and translational outlook. Curr Opin Biotech. 2020;65:171–9. doi: 10.1016/j.copbio.2020.02.016. [DOI] [PubMed] [Google Scholar]
- 3.Pedrolli DB, Ribeiro NV, Squizato PN, de Jesus VN, Cozetto DA, Team AQA Unesp at iGEM. Engineering microbial living therapeutics: the synthetic biology toolbox. Trends Biotechnol. 2019;37:100–15. doi: 10.1016/j.tibtech.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Cao Z, Cheng S, Wang X, Pang Y, Liu J. Camouflaging bacteria by wrapping with cell membranes. Nat Commun. 2019;10:1–10. doi: 10.1038/s41467-018-07882-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai L, Gao M, Cheng X, Kang G, Cao X, Huang H. Engineered butyrate-producing bacteria prevents high fat diet-induced obesity in mice. Microb Cell Fact. 2020;19:1–13. doi: 10.1186/s12934-019-1269-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piñero-Lambea C, Ruano-Gallego D, Fernández LÁ. Engineered bacteria as therapeutic agents. Curr Opin Biotech. 2015;35:94–102. doi: 10.1016/j.copbio.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 7.Isabella VM, Ha BN, Castillo MJ, Lubkowicz DJ, Rowe SE, Millet YA, et al. Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat Biotechnol. 2018;36:857–64. doi: 10.1038/nbt.4222. [DOI] [PubMed] [Google Scholar]
- 8.Riglar DT, Silver PA. Engineering bacteria for diagnostic and therapeutic applications. Nat Rev Microbiol. 2018;16:214–25. doi: 10.1038/nrmicro.2017.172. [DOI] [PubMed] [Google Scholar]
- 9.Praveschotinunt P, Duraj-Thatte AM, Gelfat I, Bahl F, Chou DB, Joshi NS. Engineered E. coli Nissle 1917 for the delivery of matrix-tethered therapeutic domains to the gut. Nat Commun. 2019;10:1–14. doi: 10.1038/s41467-019-13336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho CL, Tan HQ, Chua KJ, Kang A, Lim KH, Ling KL, et al. Engineered commensal microbes for diet-mediated colorectal-cancer chemoprevention. Nat Biomed Eng. 2018;2:27–37. doi: 10.1038/s41551-017-0181-y. [DOI] [PubMed] [Google Scholar]
- 11.Chen Z, Guo L, Zhang Y, Walzem RL, Pendergast JS, Printz RL, et al. Incorporation of therapeutically modified bacteria into gut microbiota inhibits obesity. J Clin Investig. 2014;124:3391–406. doi: 10.1172/JCI72517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hendrikx T, Duan Y, Wang Y, Oh JH, Alexander LM, Huang W, et al. Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. Gut. 2019;68:1504–15. doi: 10.1136/gutjnl-2018-317232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ozdemir T, Fedorec AJ, Danino T, Barnes CP. Synthetic biology and engineered live biotherapeutics: toward increasing system complexity. Cell Syst. 2018;7:5–16. doi: 10.1016/j.cels.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 14.Charbonneau MR, Isabella VM, Li N, Kurtz CB. Developing a new class of engineered live bacterial therapeutics to treat human diseases. Nat Commun. 2020;11:1–11. doi: 10.1038/s41467-020-15508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rottinghaus AG, Amrofell MB, Moon TS. Biosensing in smart engineered probiotics. Biotechnol J. 2020;15:10. doi: 10.1002/biot.201900319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westendorf AM, Gunzer F, Deppenmeier S, Tapadar D, Hunger JK, Schmidt MA, et al. Intestinal immunity of Escherichia coli Nissle 1917: a safe carrier for therapeutic molecules. FEMS Immunol Med Mic. 2005;43:373–84. doi: 10.1016/j.femsim.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 17.Kruis W, Fric P, Pokrotnieks J, Lukás M, Fixa B, Kascák M, et al. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut. 2004;53:1617–23. doi: 10.1136/gut.2003.037747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan Y, Shen J, Si T, Ho CL, Li Y, Dai L. Engineered Live biotherapeutics: progress and challenges. Biotechnol J. 2020;15:2000155. doi: 10.1002/biot.202000155. [DOI] [PubMed] [Google Scholar]
- 19.Sonnenborn U. Escherichia coli strain Nissle 1917-from bench to bedside and back: history of a special Escherichia coli strain with probiotic properties. FEMS Microbiol Lett. 2016;363:1–6. [DOI] [PubMed]
- 20.Sonnenborn U, Schulze J. The non-pathogenic Escherichia coli strain Nissle 1917-features of a versatile probiotic. Microb Ecol Health Dis. 2009;21:122–58. [Google Scholar]
- 21.Wehkamp J, Harder J, Wehkamp K, Wehkamp-von Meissner B, Schlee M, Enders C, et al. NF-κB- and AP-1-mediated induction of human beta defensin-2 in intestinal epithelial cells by Escherichia coli Nissle 1917: a novel effect of a probiotic bacterium. Infect Immun. 2004;72:5750–8. doi: 10.1128/IAI.72.10.5750-5758.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grabig A, Paclik D, Guzy C, Dankof A, Baumgart DC, Erckenbrecht J, et al. Escherichia coli strain Nissle 1917 ameliorates experimental colitis via toll-like receptor 2-and toll-like receptor 4-dependent pathways. Infect Immun. 2006;74:4075–82. doi: 10.1128/IAI.01449-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kai-Larsen Y, Lüthje P, Chromek M, Peters V, Wang X, Holm A, et al. Uropathogenic Escherichia coli modulates immune responses and its curli fimbriae interact with the antimicrobial peptide LL-37. PLoS Pathog. 2010;6:e1001010. doi: 10.1371/journal.ppat.1001010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choudhary R, Mahadevan R. Toward a systematic design of smart probiotics. Curr Opin Biotech. 2020;64:199–209. doi: 10.1016/j.copbio.2020.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Fan J-X, Li ZH, Liu XH, Zheng DW, Chen Y, Zhang XZ. Bacteria-mediated tumor therapy utilizing photothermally-controlled TNF-α expression via oral administration. Nano Lett. 2018;18:2373–80. doi: 10.1021/acs.nanolett.7b05323. [DOI] [PubMed] [Google Scholar]
- 26.Huyghebaert N, Vermeire A, Neirynck S, Steidler L, Remaut E, Remon JP. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science. 2000;289:1352–5. doi: 10.1126/science.289.5483.1352. [DOI] [PubMed] [Google Scholar]
- 27.Oh J-H, Schueler KL, Stapleton DS, Alexander LM, Yen CLE, Keller MP, et al. Secretion of recombinant interleukin-22 by engineered Lactobacillus reuteri reduces fatty liver disease in a mouse model of diet-induced obesity. Msphere. 2020;5:3. doi: 10.1128/mSphere.00183-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amiri-Jami M, Abdelhamid AG, Hazaa M, Kakuda Y, Griffths MW. Recombinant production of omega-3 fatty acids by probiotic Escherichia coli Nissle 1917. FEMS Microbiol Lett. 2015;362:20. doi: 10.1093/femsle/fnv166. [DOI] [PubMed] [Google Scholar]
- 29.Newman JC, Verdin E. β-hydroxybutyrate: much more than a metabolite. Diabetes Res Clin Pr. 2014;106:173–81. doi: 10.1016/j.diabres.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kashiwaya Y, Takeshima T, Mori N, Nakashima K, Clarke K, Veech RL. d-β-Hydroxybutyrate protects neurons in models of Alzheimer’s and Parkinson’s disease. P Natl Acad Sci USA. 2000;97:5440–4. doi: 10.1073/pnas.97.10.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, Cao Q, Li S, Lu X, Zhao Y, Guan JS, et al. 3-Hydroxybutyrate methyl ester as a potential drug against Alzheimer’s disease via mitochondria protection mechanism. Biomaterials. 2013;34:7552–62. doi: 10.1016/j.biomaterials.2013.06.043. [DOI] [PubMed] [Google Scholar]
- 32.Olson CA, Vuong HE, Yano JM, Liang QY, Nusbaum DJ, Hsiao EY. The gut microbiota mediates the anti-seizure effects of the ketogenic diet. Cell. 2018;173:1728–41. doi: 10.1016/j.cell.2018.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kossoff EH. More fat and fewer seizures: dietary therapies for epilepsy. Lancet Neuro. 2004;3:415–20. doi: 10.1016/S1474-4422(04)00807-5. [DOI] [PubMed] [Google Scholar]
- 34.Chakraborty S, Galla S, Cheng X, Yeo JY, Mell B, Singh V, et al. Salt-responsive metabolite, β-hydroxybutyrate, attenuates hypertension. Cell Rep. 2018;25:677–89. doi: 10.1016/j.celrep.2018.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Youm Y-H, Nguyen KY, Grant RW, Goldberg EL, Bodogai M, Kim D, et al. The ketone metabolite β-hydroxybutyrate blocks NLRP3 inflammasome–mediated inflammatory disease. Nat Med. 2015;21:263–9. doi: 10.1038/nm.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rahman M, Muhammad S, Khan MA, Chen H, Ridder DA, Müller-Fielitz H, et al. The β-hydroxybutyrate receptor HCA 2 activates a neuroprotective subset of macrophages. Nat Commun. 2014;5:1–11. doi: 10.1038/ncomms4944. [DOI] [PubMed] [Google Scholar]
- 37.Cao Q, Zhang J, Liu H, Wu Q, Chen J, Chen GQ. The mechanism of anti-osteoporosis effects of 3-hydroxybutyrate and derivatives under simulated microgravity. Biomaterials. 2014;35:8273–83. doi: 10.1016/j.biomaterials.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, Jiang XL, Peng SW, Guo XY, Shang GG, Chen JC, et al. Induced apoptosis of osteoblasts proliferating on polyhydroxyalkanoates. Biomaterials. 2013;34:3737–46. doi: 10.1016/j.biomaterials.2013.01.088. [DOI] [PubMed] [Google Scholar]
- 39.Xiao XQ, Zhao Y, Chen GQ. The effect of 3-hydroxybutyrate and its derivatives on the growth of glial cells. Biomaterials. 2007;28:3608–16. doi: 10.1016/j.biomaterials.2007.04.046. [DOI] [PubMed] [Google Scholar]
- 40.Obruca S, Sedlacek P, Mravec F, Samek O, Marova I. Evaluation of 3-hydroxybutyrate as an enzyme-protective agent against heating and oxidative damage and its potential role in stress response of poly (3-hydroxybutyrate) accumulating cells. Appl Microbiol Biot. 2016;100:1365–76. doi: 10.1007/s00253-015-7162-4. [DOI] [PubMed] [Google Scholar]
- 41.Liu Q, Ouyang S-P, Chung A, Wu Q, Chen G-Q. Microbial production of R-3-hydroxybutyric acid by recombinant E. coli harboring genes of phbA, phbB, and tesB. Appl Microbiol Biot. 2007;76:811–8. doi: 10.1007/s00253-007-1063-0. [DOI] [PubMed] [Google Scholar]
- 42.Tseng H-C, Martin CH, Nielsen DR, Prather KLJ. Metabolic engineering of Escherichia coli for enhanced production of (R)-and (S)-3-hydroxybutyrate. Appl Environ Microb. 2009;75:3137–45. doi: 10.1128/AEM.02667-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh V, Yeoh BS, Walker RE, Xiao X, Saha P, Golonka RM, et al. Microbiota fermentation-NLRP3 axis shapes the impact of dietary fibres on intestinal inflammation. Gut. 2019;68:1801–12. doi: 10.1136/gutjnl-2018-316250. [DOI] [PubMed] [Google Scholar]
- 44.Riglar DT, Giessen TW, Baym M, Kerns SJ, Niederhuber MJ, Bronson RT, et al. Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation. Nat Biotechnol. 2017;35:653–8. doi: 10.1038/nbt.3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rembacken B, Snelling A, Hawkey P, Chalmers D, Axon A. Non-pathogenic Escherichia coli versus mesalazine for the treatment of ulcerative colitis: a randomised trial. Lancet. 1999;354:635–9. doi: 10.1016/S0140-6736(98)06343-0. [DOI] [PubMed] [Google Scholar]
- 46.Leventhal DS, Sokolovska A, Li N, Plescia C, Kolodziej SA, Gallant CW, et al. Immunotherapy with engineered bacteria by targeting the STING pathway for anti-tumor immunity. Nat Commun. 2020;11:1–15. doi: 10.1038/s41467-020-16602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Toole PW, Marchesi JR, Hill C. Next-generation probiotics: the spectrum from probiotics to live biotherapeutics. Nat Microbiol. 2017;2:1–6. doi: 10.1038/nmicrobiol.2017.57. [DOI] [PubMed] [Google Scholar]
- 48.Van der Vossen J, van der Lelie D, Venema G. Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl Environ Microb. 1987;53:2452–7. doi: 10.1128/aem.53.10.2452-2457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Graef MR, Alexeeva S, Snoep JL, de Mattos MJT. The steady-state internal redox state (NADH/NAD) reflects the external redox state and is correlated with catabolic adaptation in Escherichia coli. J Bacteriol. 1999;181:2351–7. doi: 10.1128/JB.181.8.2351-2357.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat Biotechnol. 1983;1:784–91. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 51.Kruger NJ. The Bradford Method For Protein Quantitation. In: Walker J.M. editor. The Protein Protocols Handbook. Totowa, NJ: Springer Protocols Handbooks. Humana Press; 2009.
- 52.Nishimura T, Saito T, Tomita K. Purification and properties of β-ketothiolase from Zoogloea ramigera. Arch Microbiol. 1978;116:21–27. doi: 10.1007/BF00408729. [DOI] [PubMed] [Google Scholar]
- 53.Senior PJ, Dawes EA. The regulation of poly-β-hydroxybutyrate metabolism in Azotobacter beijerinckii. Biochem J. 1973;134:225–38. doi: 10.1042/bj1340225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng Z, Gong Q, Liu T, Deng Y, Chen JC, Chen GQ. Thioesterase II of Escherichia coli plays an important role in 3-hydroxydecanoic acid production. Appl Environ Microb. 2004;70:3807–13. doi: 10.1128/AEM.70.7.3807-3813.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sassone-Corsi M, Nuccio SP, Liu H, Hernandez D, Vu CT, Takahashi AA, et al. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature. 2016;540:280–3. doi: 10.1038/nature20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 57.Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 58.Alexander DB, Zuberer DA. Use of chrome azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol Fertil Soils. 1991;12:39–45. doi: 10.1007/BF00369386. [DOI] [Google Scholar]
- 59.Smith P, Mangan NE, Walsh CM, Fallon RE, McKenzie AN, van Rooijen N, et al. Infection with a helminth parasite prevents experimental colitis via a macrophage-mediated mechanism. J Immunol. 2007;178:4557–66. doi: 10.4049/jimmunol.178.7.4557. [DOI] [PubMed] [Google Scholar]
- 60.Amrofell MB, Rottinghaus AG, Moon TS. Engineering microbial diagnostics and therapeutics with smart control. Curr Opin Biotech. 2020;66:11–17. doi: 10.1016/j.copbio.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Volbeda A, Darnault C, Renoux O, Nicolet Y, Fontecilla-Camps JC. The crystal structure of the global anaerobic transcriptional regulator FNR explains its extremely fine-tuned monomer-dimer equilibrium. Sci Adv. 2015;1:e1501086. doi: 10.1126/sciadv.1501086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Puchalska P, Crawford PA. Multi-dimensional roles of ketone bodies in fuel metabolism, signaling, and therapeutics. Cell Metab. 2017;25:262–84. doi: 10.1016/j.cmet.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–73. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bian X, Yang L, Wu W, Lv L, Jiang X, Wang Q, et al. Pediococcus pentosaceus LI05 alleviates DSS‐induced colitis by modulating immunological profiles, the gut microbiota and short‐chain fatty acid levels in a mouse model. Microb Biotechnol. 2020;13:1228–44. doi: 10.1111/1751-7915.13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parker BJ, Wearsch PA, Veloo AC, Rodriguez-Palacios A. The genus Alistipes: Gut bacteria with emerging implications to inflammation, cancer, and mental health. Front Immunol. 2020;11:906. doi: 10.3389/fimmu.2020.00906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, et al. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab. 2015;22:971–82. doi: 10.1016/j.cmet.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 67.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–6. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 68.Chassaing B, Aitken JD, Malleshappa M, Vijay‐Kumar M. Dextran sulfate sodium (DSS)‐induced colitis in mice. Curr Protoc Immunol. 2014;104:11–15. doi: 10.1002/0471142735.im1525s104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rhee SH, Pothoulakis C, Mayer EA. Principles and clinical implications of the brain-gut-enteric microbiota axis. Nat Rev Gastro Hepat. 2009;6:306–14. doi: 10.1038/nrgastro.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tripathi A, Debelius J, Brenner DA, Karin M, Loomba R, Schnabl B, et al. The gut-liver axis and the intersection with the microbiome. Nat Rev Gastro Hepat. 2018;15:397–411. doi: 10.1038/s41575-018-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu J, Plackett D, Chen LX. Kinetics and mechanism of the monomeric products from abiotic hydrolysis of poly [(R)-3-hydroxybutyrate] under acidic and alkaline conditions. Polym Degrad Stab. 2005;89:289–99. doi: 10.1016/j.polymdegradstab.2004.12.026. [DOI] [Google Scholar]
- 72.Tan D, Xue YS, Aibaidula G, Chen GQ. Unsterile and continuous production of polyhydroxybutyrate by Halomonas TD01. Bioresour Technol. 2011;102:8130–6. doi: 10.1016/j.biortech.2011.05.068. [DOI] [PubMed] [Google Scholar]
- 73.Pradhan S, Weiss AA. Probiotic properties of Escherichia coli Nissle in human intestinal organoids. Mbio. 2020;11:e01470–20. doi: 10.1128/mBio.01470-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hancock V, Vejborg RM, Klemm P. Functional genomics of probiotic Escherichia coli Nissle 1917 and 83972, and UPEC strain CFT073: comparison of transcriptomes, growth and biofilm formation. Mol Genet Genomics. 2010;284:437–54. doi: 10.1007/s00438-010-0578-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the study are available from the corresponding authors by request. Source data are provided with this paper.