Abstract
Cyclic GMP-AMP synthase (cGAS) is a key sensor critical for the recognition of DNA in the cytosol and catalyzes the synthesis of the second messenger cyclic GMP-AMP (cGAMP), which binds to the adapter protein MITA (also known as STING, MPYS, and ERIS) to initiate the innate immune response. How the binding of DNA to and the activation of cGAS are regulated remains poorly understood. Using a biochemical purification approach, we identified poly(rC)-binding protein 1 (PCBP1) as a cGAS-associated protein. PCBP1 was recruited to cGAS in a viral infection-dependent manner. PCBP1 directly bound to DNA and enhanced cGAS binding to its ligands, which was important for cGAS activation. Consistently, PCBP1 deficiency inhibited cytosolic DNA- and DNA virus-triggered transcription of downstream effector genes. These findings suggest that PCBP1 plays an important role in the cGAS-mediated innate immune response to DNA virus infection by promoting the binding of cGAS to viral DNA.
Keywords: cGAS, PCBP1, DNA, virus, Innate Immunity
Subject terms: Pattern recognition receptors, Cell signalling
Introduction
As the first line of host defense against viral infection, the innate immune system deploys various strategies to detect and respond to invading pathogens. Upon viral infection, host pattern recognition receptors (PRRs) detect nucleic acids derived from invaded viruses or necrotic cells to initiate a series of signaling events that lead to the induction of type I interferons (IFNs), proinflammatory cytokines, and other antiviral genes. These downstream effectors in combination elicit antiviral responses through various mechanisms.1–3
Viral nucleic acids, including RNA and DNA, represent key pathogen-associated molecular patterns that are detected by Toll-like receptors, retinoic acid-inducible gene-I (RIG-I)-like receptors (RLRs) and a variety of DNA sensors.1,4 Cyclic GMP-AMP synthase (cGAS) is a key cytosolic DNA sensor that detects mislocated self-DNA or DNA derived from pathogens in a DNA sequence-independent manner.5–10 The engagement of cGAS by DNA triggers a conformational change in the active site and catalyzes the synthesis of cyclic GMP-AMP (cGAMP) from ATP and GTP, which in turn acts as a second messenger to bind and activate the endoplasmic reticulum (ER)-membrane adapter protein MITA (also known as STING, MPYS, and ERIS).11–14 MITA is then trafficked via an ER-Golgi intermediate compartment and the Golgi apparatus to perinuclear punctate structures.15 During these processes, MITA recruits and activates the kinases TBK1 and IKKβ, which in turn phosphorylate the transcription factors IRF3 and NF-κB, leading to the ultimate induction of downstream effector genes.16
cGAS contains an N-terminal DNA-binding domain and a C-terminal nucleotidyltransferase and male abnormal 21 domains.5 In the absence of DNA, cGAS is in an autoinhibited state.17 Upon binding to double-stranded (ds) DNA, cGAS assembles into a 2:2 cGAS-dsDNA complex, and this binding induces a structural switch in cGAS in the active pocket, which carries out the cyclization of ATP and GTP to form cGAMP.18,19 Although dsRNA and single-stranded (ss) DNA also bind to cGAS, neither can rearrange the catalytic pocket, which might explain the specific activation of cGAS by dsDNA.20 Recent studies have also demonstrated that DNA binding to cGAS induces a robust phase transition to liquid-like droplets, which function as microreactors in which the enzyme and reactants are concentrated to greatly enhance the production of cGAMP. Long DNA is more potent than short DNA in driving cGAS phase separation, explaining why long DNA activates cGAS more efficiently than short DNA.21
Recently, it has been shown that polyglutamine binding protein 1 (PQBP1) directly binds to reverse-transcribed HIV-1 ssDNA and interacts with cGAS to initiate the innate immune response,22 while ZCCHC3 serves as a cosensor by directly binding to viral dsDNA and promoting the binding of cGAS to its ligands.23 To further understand how cGAS is regulated, we used tandem affinity purification followed by mass spectrometry (TAP-MS) to identify additional cGAS-associated proteins, which led to the identification of poly(rC)-binding protein 1 (PCBP1). PCBP1 belongs to the heterogeneous nuclear ribonucleoprotein family, which is ubiquitously expressed and plays multiple roles in transcriptional and posttranscriptional regulation.24 PCBP1 and PCBP2 have been reported to inhibit RLR-mediated signaling by mediating the degradation of the adapter protein VISA (also known as MAVS, IPS-1, and Cardif).25–27 In this study, we found that PCBP1 was associated with cGAS in a viral infection-dependent manner and promoted cGAS binding to DNA. PCBP1 deficiency inhibited cytosolic DNA- and DNA virus-triggered induction of downstream effector genes. Our findings suggest that PCBP1 is a critical regulator for cGAS sensing of DNA, which plays an important role in the innate antiviral response.
Materials and methods
Reagents, cells, and viruses
Phusion® high-fidelity DNA polymerase (New England Biolabs); Lipofectamine 2000, M-MLV reverse transcriptase and 2′3′-cGAMP (InvivoGen); digitonin and DNase I (Sigma-Aldrich); RiboLock RNase inhibitor (Thermo Scientific); polybrene (Millipore); RNAiso Plus (TaKaRa); SYBR Green mix (Bio-Rad); Dual-specific luciferase assay kit (Promega); 2′3′-cGAMP ELISA kit (Cayman Chemical); mouse antibodies against HA (OriGene), FLAG and β-actin (Sigma-Aldrich); rabbit antibodies against phospho-MITA (S366), MITA and cGAS (Cell Signaling Technology); against phospho-IRF3 (S396), phospho-TBK1 (S172), and TBK1 (Abcam); against phospho-STAT1 (Y701) and STAT1 (Santa Cruz Biotechnology); and against PCBP1 (ABclonal); and Alexa Fluor 488- and Alexa Fluor 594-conjugated goat anti-rabbit IgG antibodies (Invitrogen) were purchased from the indicated manufacturers. Mouse antisera against murine IRF3 was generated using recombinant protein murine IRF3 as an antigen, and mouse antisera against ZCCHC3 was generated using recombinant ZCCHC3 (133-404) as an antigen. HT1080 cells (China Center for Type Culture Collection); HeLa, HEK293, and THP-1 cells (American Type Culture Collection); SeV (Cantell strain, Charles River Laboratories); and HSV-1 (KOS strain, China Center for Type Culture Collection) were purchased from the indicated resources. Mouse lung fibroblasts (MLFs) were obtained from C57BL/6 mice and immortalized by SV40. The SV40 virus was obtained from Dr. Zhi-Ying Song (Wuhan University).
Constructs
Expression plasmids for HA-, FLAG-, and RFP-tagged PCBP1 and its truncated mutants, HA-, FLAG-, and GFP-tagged cGAS and its truncated mutants, FLAG-ZCCHC3, were constructed by standard molecular biology techniques. Guide-RNA plasmids targeting PCBP1 and cGAS were constructed into a lentiCRISPR V2 vector, which was provided by Dr. Shu-Wen Wu (Wuhan University).
DNA oligonucleotides
The following oligonucleotides were used to stimulate cells:
HSV60:
5′-TAAGACACGATGCGATAAAATCTGTTTGTAAAATTTATTAAGGGTACAAATTGCCCTAGC-3′;
VACV70:
5′-CCATCAGAAAGAGGTTTAATATTTTTGTGAGACCATGGAAGAGAGAAAGAGATAAAACTTTTTTACGACT-3′;
DNA90:
5′-TACAGATCTACTAGTGATCTATGACTGATCTGTACATGATCTACATACAGA CTACTAGTGATCTATGACTGATCTGTACATGATCTACA-3′;
HSV120:
5′-AGACGGTATATTTTTGCGTTATCACTGTCCCGGATTGGACACGGTCTTGTGGGATAGGCATGCCCAGAAGGCATATTGGGTTAACCCCTTTTTATTTGTGGCGGGTTTTTTGGAGGACTT-3′;
HIV-RT-DNA:
5′-ACAACAGACGGGCACACACTACTTGAAGCACTCAAGGCAAGCTTTATTGAGGCTTAAGCAGTGGGTTCCCTAGTTAGCCAGAGAGCTCCCAGGCTCAGATCTGGTCTAACCAGAGAGACC-3′.
The following oligonucleotides were used to construct the respective gRNA plasmids:
gPCBP1 #1-5′-GAGAAAGGGGAGTCGGTTAAG-3′, #2-5′-GATGCCGGTGTGACTGAAAG-3′,
gPcbp1 #1-5′-ACCAGCCGAAGTGTGACCGG-3′, #2-5′-AGATCGCTGCAGCGACGCGG-3′.
qPCR
Total RNA was isolated for a qPCR analysis to measure mRNA levels of the indicated genes using RNAiso Plus reagent (TaKaRa) according to the manufacturer’s protocol. The data report the relative abundance of the indicated mRNA derived from human or mouse cells normalized to that of GAPDH or Gapdh, respectively. The sequences of the qPCR primers were previously reported.23,28,29
Transfection and reporter assays
HEK293 cells were transfected by the standard calcium phosphate precipitation method. HeLa cells, THP-1 cells, and MLFs were transfected with Lipofectamine 2000. To ensure that each cell line was transfected with the same amount of total DNA, an empty control plasmid was added to each transfection culture. To normalize for transfection efficiency, a pRL-TK (Renilla luciferase) reporter plasmid (0.01 μg) was added to each transfection culture. Luciferase assays were performed using a dual-specific luciferase assay kit. Firefly luciferase activity levels were normalized based on the Renilla luciferase activity level.
Coimmunoprecipitation and immunoblot analysis
Cells were lysed in Triton X-100 lysis buffer (20 mM Tris-HCl, pH 7.4; 150 mM NaCl; 1 mM EDTA; 1% Triton X-100; 10 μg/mL aprotinin; 10 μg/mL leupeptin; and 1 mM phenylmethylsulfonyl fluoride). The lysates were centrifuged at 12,000 g for 10 min at 4 °C. For each immunoprecipitation experiment, the supernatant was incubated with the indicated antibody and 35 μL of 50% GammaBind G Plus-Sepharose slurry at 4 °C for 2 h. The beads were then washed three times with 1 mL of lysis buffer containing 500 mM NaCl. The bound proteins were separated by SDS-PAGE, and the associated proteins were analyzed by immunoblotting.
Confocal microscopy
HeLa cells, HT1080 cells, and MLFs were seeded on coverslips in 24-well plates. After transfection with Cy5-HSV60 and incubated for 4 h or with the indicated plasmids and incubated for 20 h, the cells were fixed with 4% paraformaldehyde for 15 min, permeabilized with Triton X-100 for 10 min and blocked in 1% BSA for 1 h. The cells were then incubated with primary antibodies overnight at room temperature. Alexa Fluor 488- and 546-conjugated secondary antibodies were incubated with the cells for 1 h. The nucleus were stained with DAPI for 2 min before images were acquired using a ZEISS confocal microscope.
In vitro pull-down assays
HEK293 cells transfected with the indicated plasmids or MLFs were lysed in Triton X-100 lysis buffer. The lysates were incubated with the indicated biotinylated DNA for 2 h at 4 °C and then incubated with streptavidin beads for 1 h at 4 °C. The beads were washed three times with lysis buffer and analyzed by immunoblotting with the indicated antibodies.
Semi-denaturing detergent agarose gel electrophoresis (SDD-AGE)
The MLFs were lysed in Triton X-100 lysis buffer, and the cell lysates were mixed in 1× sample buffer (0.5 × TBE, 10% glycerol, 2% SDS, and 0.0025% bromophenol blue) and loaded onto a vertical 2% agarose gel (Bio-Rad). After subjection to electrophoresis in running buffer (1 × TBE and 0.1% SDS) for ~1 h with a constant voltage of 100 V at 4 °C, the proteins were transferred to an immobile membrane (Millipore) for immunoblot analysis.
Measurement of cGAMP activity
The indicated MLFs (1 × 107) were untreated or transfected with HT-DNA (5 μg) for 6 h. The cells were then homogenized by a Dounce homogenizer in hypotonic buffer (10 mM Tris-HCl, pH 7.4; 10 mM KCl; and 1.5 mM MgCl2). After centrifugation at 13,000 rpm for 20 min, the supernatant was heated at 95 °C for 10 min and centrifuged at 13,000 rpm for another 10 min to remove denatured proteins. The heat-resistant supernatants containing cGAMP were delivered into digitonin-permeabilized MLFs at 37 °C for 30 min, and then, the cells were cultured with regular medium for another 4 h before qPCR analysis. The concentration of cGAMP in the cell extracts was also measured by a cGAMP ELISA kit.
Digitonin permeabilization
The protocol for digitonin permeabilization was previously described.28 The cells were treated with 2′3′-cGAMP in digitonin permeabilization solution (50 mM HEPES, pH 7.0; 100 mM KCl; 3 mM MgCl2; 0.1 mM DTT; 85 mM sucrose; 0.2% BSA; 1 mM ATP; 0.1 mM GTP; and 10 μg/mL digitonin) for 30 min at 37 °C. The cells were then incubated in regular medium for the indicated time before qPCR or immunoblot experiments were performed.
CRISPR-Cas9 knockout
The protocols for genome engineering using the CRISPR-Cas9 system were previously described.30,31 Briefly, ds-oligonucleotides corresponding to the target sequences were cloned into a lentiCRISPR V2 plasmid, which was transfected with packaging plasmids into HEK293 cells. Two days after transfection, the viruses were harvested and used to infect THP-1 cells, MLFs, and HEK293 cells. The infected cells were selected with puromycin (2 μg/mL for the MLFs and THP-1 cells and 1 μg/mL for the HEK293 cells) in culture for at least 5 days.
Statistics
The data were analyzed using Student’s unpaired t-test with Prism GRAGHPAD 7. The number of asterisks represents the degree of significance with respect to P values, which are presented within each figure or figure legend.
Results
Identification of PCBP1 as a cGAS-associated protein
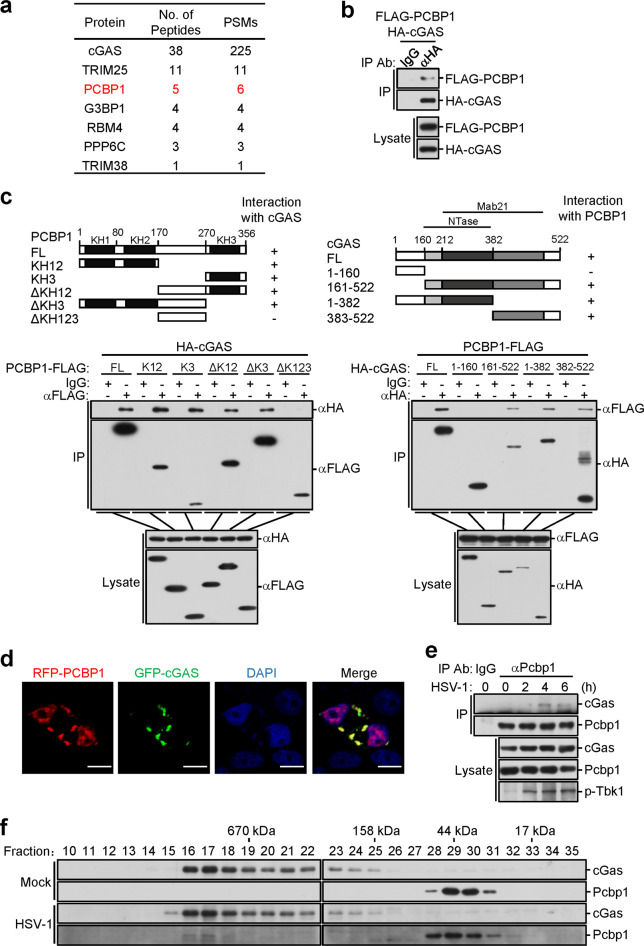
To identify candidate molecules involved in the cGAS-mediated innate immune response, we performed TAP-MS experiments. By comparing the samples with unrelated purified products obtained based on the same method, we identified PCBP1 as a candidate protein specifically associated with cGAS (Fig. 1a). Among the candidate proteins, G3BP1 and TRIM38 have been reported to be associated with cGAS.28,32 We focus on PCBP1 in this report, and the other candidate proteins will be validated in separate studies. To confirm that PCBP1 is associated with cGAS, we performed transient transfection and coimmunoprecipitation experiments. The results indicated that PCBP1 interacted with cGAS in the mammalian overexpression system (Fig. 1b). Domain mapping experiments indicated that the KH domains of PCBP1 interacted with cGAS (Fig. 1c). On the other hand, the C-terminal fragment of cGAS (aa161-522) interacted with PCBP1 (Fig. 1c). Confocal microscopy indicated that PCBP1 was colocalized with cGAS in the cytoplasm (Fig. 1d). Endogenous coimmunoprecipitation experiments indicated that PCBP1 was negligibly associated with cGAS in the uninfected cells, and its association was enhanced after infection with the DNA virus herpes simplex virus 1 (HSV-1) (Fig. 1e). Gel filtration experiments indicated that PCBP1 and cGAS were distributed in different fractions in the uninfected cells but could be detected in the overlapping high-molecular-weight fractions after HSV-1 infection (Fig. 1f). Taken together, these results suggest that PCBP1 is associated with cGAS after viral infection.
Fig. 1.
Identification of PCBP1 as a cGAS-associated protein. a HEK293 cells stably expressing SFB (S, FLAG and streptavidin-binding peptide)-tagged cGAS were subjected to TAP-MS experiments. Tables present summaries of the selected proteins identified by MS analysis. b PCBP1 interacts with cGAS. HEK293 cells were transfected with the indicated plasmids for 16 h before coimmunoprecipitation and immunoblot analysis with the indicated antibodies. c Domain mapping of the cGAS-PCBP1 interaction. HEK293 cells were transfected with the indicated plasmids before coimmunoprecipitation and immunoblot analysis with the indicated antibodies. Schematic presentations of the interactions are shown at the bottom. −, no interaction; +, positive interaction. d Colocalization of PCBP1 with cGAS. HeLa cells were transfected with GFP-cGAS and RFP-PCBP1 for 20 h before confocal microscopy. Scale bars, 10 μm. e Endogenous PCBP1 is associated with cGAS. MLFs (5 × 107) were untreated or infected with HSV-1 (MOI = 1) for the indicated times before coimmunoprecipitation and immunoblot analysis. f Analysis of the protein complexes containing PCBP1 and cGAS by size-exclusion chromatography. MLFs (5 × 107) were infected with HSV-1 (MOI = 1) for 4 h or were uninfected before lysis. Cell lysates were analyzed by size-exclusion chromatography on a Superdex 200 column. The individual fractions were analyzed by immunoblotting with anti-cGAS and anti-PCBP1 antibodies
PCBP1 positively regulates DNA virus-triggered signaling
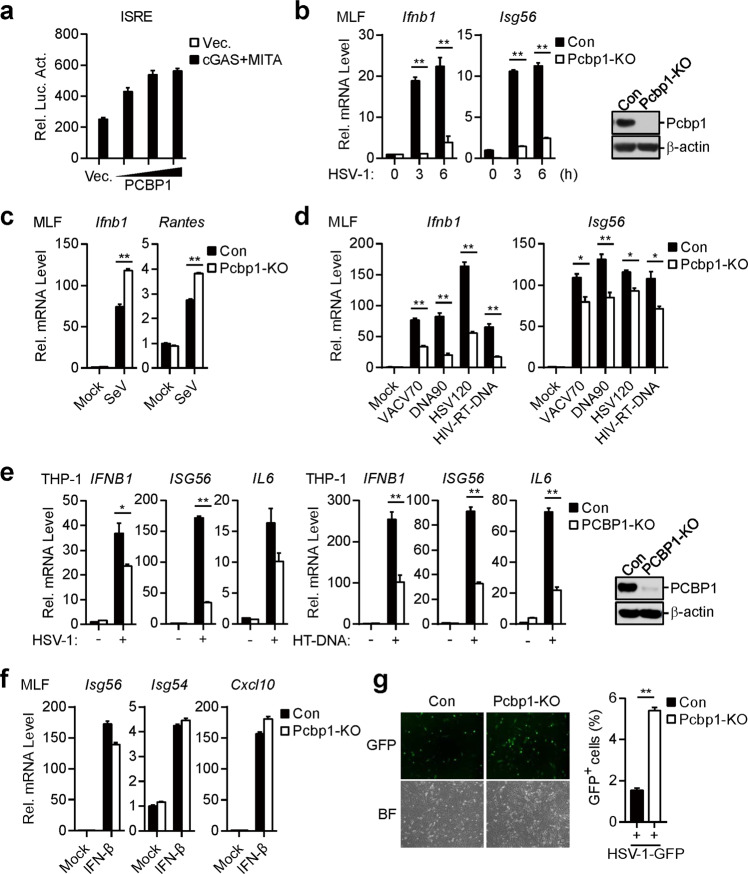
Because PCBP1 is associated with cGAS in a viral infection-dependent manner, we sought to determine whether PCBP1 regulates cGAS-mediated signaling. Reporter assays indicated that overexpressed PCBP1 potentiated the cGAS-MITA-mediated activation of IFN-stimulated response element (ISRE) in a dose-dependent manner in the HEK293 cells (Fig. 2a). To determine the roles of endogenous PCBP1 in cGAS-mediated signaling, we generated PCBP1-deficient MLFs by the CRISPR-Cas9 method. Quantitative PCR (qPCR) analysis indicated that the HSV-1-induced transcription of downstream genes, such as the Ifnb1 and Isg56 genes, was dramatically decreased in the PCBP1-deficient MLFs in comparison with that of the control cells (Fig. 2b). In contrast, the transcription of the Ifnb1 and Rantes genes induced by the Sendai virus (SeV) was increased in the PCBP1-deficient MLFs compared with that in the control cells (Fig. 2c), which is consistent with the previous publication.25 We next determined the effects of PCBP1 deficiency on the transcription of downstream genes induced by synthetic DNA, namely, a 120-mer dsDNA representing the respective genome of HSV-1 (HSV120), ~90 bp of dsDNA (DNA90) and a 70-mer dsDNA representing the vaccinia virus (VACV) genome (VACV70). As shown in Fig. 2d, PCBP1 deficiency dramatically inhibited the transfected dsDNA-induced transcription of the Ifnb1 and Isg56 genes. Since it has been reported that HIV-1 reverse-transcribed ssDNA can activate the cGAS-mediated innate immune response,33 we also determined the effects of PCBP1 deficiency on this process. PCBP1 deficiency inhibited the transcription of the Ifnb1 and Isg56 genes induced by the transfected 120-mer ssDNA representing SL2 and SL3 of HIV-sstDNA (HIV-RT-DNA) (Fig. 2d). In addition to inhibiting transcription in the MLFs, PCBP1 deficiency inhibited HSV-1- and herring testis DNA (HT-DNA)-induced transcription of the IFNB1, ISG56, and IL6 genes in the human monocytic THP-1 cells (Fig. 2e). In similar experiments, the transcription levels of the Isg56, Isg54, and Cxcl10 genes induced by IFN-β were comparable between the PCBP1-deficient and control MLFs (Fig. 2f). Because PCBP1 potentiated the DNA virus-triggered induction of downstream effector genes, we next determined whether it plays a role in the cellular antiviral response. We infected PCBP1-deficient and control cells with GFP-tagged HSV-1 and found that PCBP1 deficiency greatly increased the GFP signal intensity in the infected cells (Fig. 2g). These data suggest that PCBP1 is essential for the cellular defense against HSV-1 infection.
Fig. 2.
PCBP1 positively regulates DNA virus-triggered signaling. a PCBP1 potentiates cGAS-MITA-induced ISRE activation in a dose-dependent manner. HEK293 cells (1 × 105) were transfected with the ISRE luciferase reporter (50 ng), expression plasmids for cGAS (50 ng), MITA (50 ng), and increased amounts of PCBP1 and incubated for 20 h before luciferase assays. The graph shows the means ± SEM, n = 3. b Effects of PCBP1 deficiency on the HSV-1-triggered transcription of downstream genes in the MLFs. PCBP1-deficient MLFs were generated by the CRISPR-Cas9 method. PCBP1-deficient and control MLFs (1 × 106) were untreated or infected with HSV-1 (MOI = 1) for the indicated times before qPCR analysis. PCBP1 deficiency in the KO cells was confirmed by immunoblot analysis (right blots). The graph shows the means ± SEM, n = 3. **P < 0.01 (unpaired t-test). c Effects of PCBP1 deficiency on the transcription of downstream genes induced by SeV in the MLFs. PCBP1-deficient and control MLFs (1 × 106) were untreated or infected with SeV for 6 h before the qPCR analysis. The graph shows the means ± SEM, n = 3. **P < 0.01 (unpaired t-test). d Effects of PCBP1 deficiency on the transcription of downstream genes induced by cytosolic DNA in MLFs. PCBP1-deficient and control MLFs (1 × 106) were left untreated or transfected with the indicated DNA (1 μg) for 4 h before the qPCR analysis. The graph shows the mean ± SEM, n = 3. *P < 0.05, **P < 0.01 (unpaired t-test). e Effects of PCBP1 deficiency on the transcription of downstream genes induced by HSV-1 and cytosolic DNA in THP-1 cells. PCBP1-deficient THP-1 cells were generated by the CRISPR-Cas9 method. PCBP1-deficient and control THP-1 cells (1 × 106) were left untreated, infected with HSV-1 (MOI = 1) for 6 h or transfected with HT-DNA (0.5 μg) for 4 h before qPCR analysis. PCBP1 deficiency in KO cells was confirmed by immunoblotting analysis (right blots). The graph shows the mean ± SEM, n = 3. **P < 0.01 (unpaired t-test). f Deficiency of PCBP1 has no effect on transcription of downstream genes induced by IFN-β in MLFs. PCBP1-deficient and control MLFs (1 × 106) were left untreated or treated with IFN-β (100 ng/mL) for 4 h before the qPCR analysis. The graph shows the means ± SEM, n = 3. g Effects of PCBP1 deficiency on the replication of HSV-1 in the MLFs. PCBP1-deficient and control MLFs (1 × 106) were infected with HSV-1-GFP (MOI = 0.1) for 30 h followed by microscopy imaging (left) and flow cytometry (right). The graph shows the means ± SEM, n = 3. **P < 0.01 (unpaired t-test)
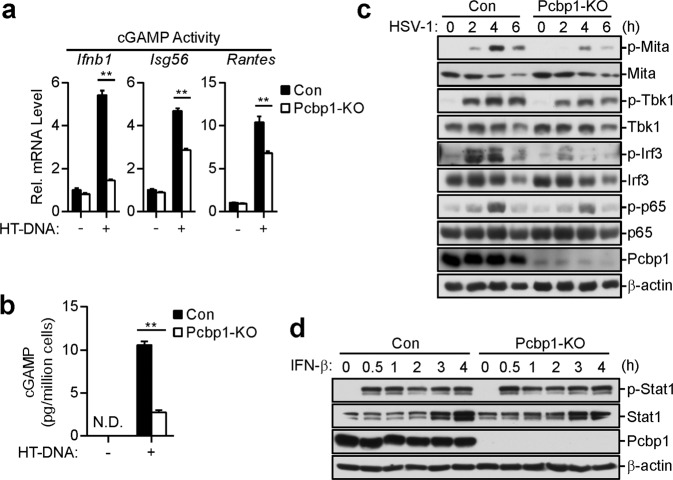
We also investigated whether PCBP1 deficiency affects the activation of downstream signaling components of cGAS. The measurement of cGAMP levels and activities indicated that the HT-DNA-induced cGAMP production was markedly reduced in the PCBP1-deficient MLFs compared with the level produced in the control cells (Fig. 3a, b). Consistently, the HSV-1-triggered phosphorylation of TBK1, IRF3, and p65, which are hallmarks of IRF3 and NF-κB activation, was impaired in the PCBP1-deficient cells compared with that in the control cells (Fig. 3c). In contrast, PCBP1 deficiency had no obvious effects on the phosphorylation of STAT1 (Y701) induced by IFN-β (Fig. 3d). These results suggest that PCBP1 is important for the activation of downstream components in the cGAS-mediated pathways.
Fig. 3.
Effects of PCBP1 deficiency on the activation of downstream components of cGAS. a, b Effects of PCBP1 deficiency on the dsDNA-induced synthesis of cGAMP in the MLFs. PCBP1-deficient and control MLFs (1 × 107) were mock-transfected or transfected with HT-DNA (5 μg) for 6 h. Cell extracts containing cGAMP were delivered to digitonin-permeabilized MLFs and incubated for 4 h before qPCR (a), or cGAMP in the cell extracts was measured by ELISA (b). The graph shows the means ± SEM, n = 3. **P < 0.01 (unpaired t-test). c Effects of PCBP1 deficiency on the HSV-1-induced phosphorylation of downstream components in the MLFs. PCBP1-deficient and control MLFs (1 × 106) were left untreated or infected with HSV-1 (MOI = 1) for the indicated times before the immunoblot analysis was performed with the indicated antibodies. d Deficiency of PCBP1 has no effects on IFN-β-induced phosphorylation of STAT1 in the MLFs. PCBP1-deficient and control MLFs (1 × 106) were untreated or treated with IFN-β (100 ng/mL) for the indicated times before the immunoblot analysis was performed
PCBP1 promotes the binding of cGAS to DNA
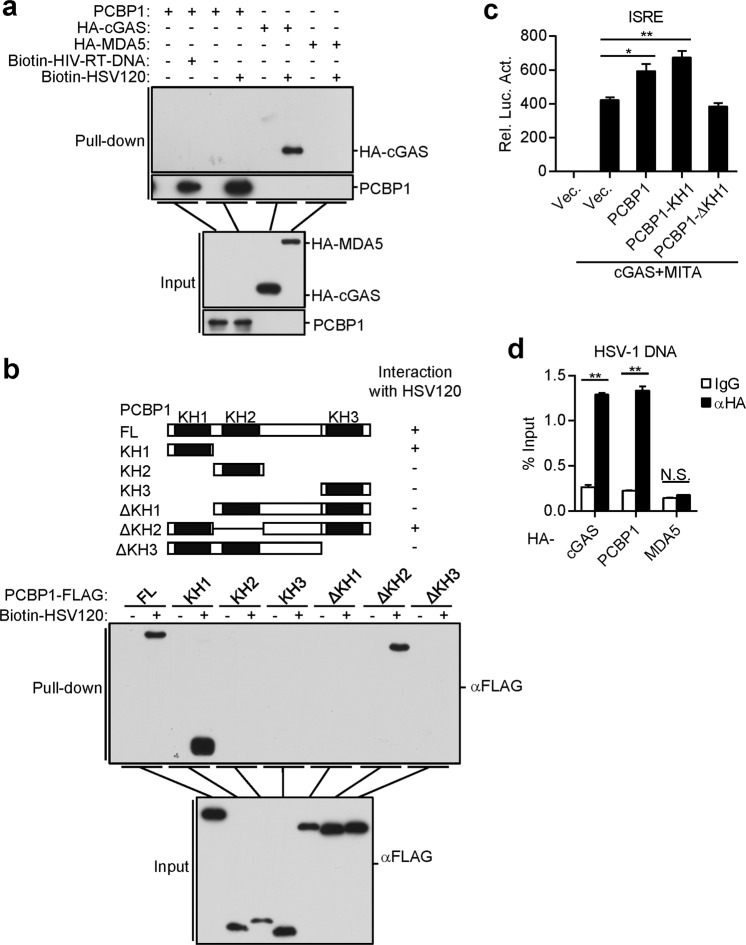
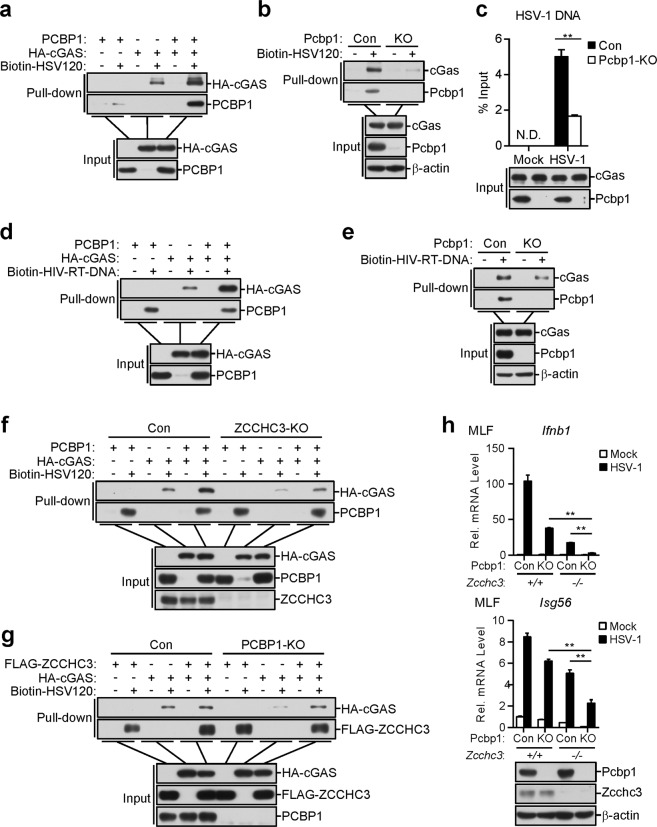
Since PCBP1 contains three KH domains that have been reported to bind to nucleic acids,34,35 we sought to determine whether PCBP1 binds to viral DNA. Pull-down assays showed that ectopically expressed PCBP1 could bind to HSV120 DNA and HIV-RT-DNA (Fig. 4a). In these experiments, the RNA sensor MDA5 did not bind to dsDNA. A deletion analysis indicated that the first KH domain (KH1) of PCBP1 was critical for its binding to the HSV120 DNA (Fig. 4b), a finding consistent with a previous publication.35 Consistently, the KH1 domain of PCBP1 potentiated the cGAS-MITA-induced activation of ISRE in the HEK293 cells (Fig. 4c). We also examined whether PCBP1 binds to viral DNA in infected cells. After cells were infected with HSV-1, we immunoprecipitated cGAS, PCBP1, and MDA5, and the protein-bound viral DNA was detected by qPCR with primers targeting HSV-1 DNA. As shown in Fig. 4d, cGAS and PCBP1, but not MDA5, could bind to HSV-1 DNA. These results suggest that PCBP1 binds to DNA through its KH1 domain.
Fig. 4.
PCBP1 binds to multiple types of nucleic acids. a PCBP1 binds to dsDNA and HIV-1-RT-DNA. HEK293 cells were transfected with the indicated plasmids. Twenty hours later, the cell lysates were incubated with the indicated biotinylated nucleic acids and streptavidin-Sepharose beads for in vitro pull-down assays. The bound proteins were then analyzed by immunoblotting with anti-HA or anti-PCBP1. b PCBP1 binds to dsDNA through its KH1 domain. HEK293 cells were transfected with the indicated plasmids. Twenty hours later, the cells were collected for in vitro pull-down assays as performed in a. c Effects of PCBP1 and its truncations on cGAS-MITA-induced ISRE activation. HEK293 cells (1 × 105) were transfected with ISRE luciferase reporter plasmid (50 ng) and the indicated plasmids for 20 h before luciferase assays were performed. The graph shows the mean ± SEM, n = 3. *P < 0.05, **P < 0.01 (unpaired t-test). d PCBP1 and cGAS, but not MDA5, bind to HSV-1 DNA of the infected cells. HEK293 cells were transfected with HA-tagged PCBP1, cGAS, and MDA5. Twenty hours after transfection, the cells were infected with HSV-1 and incubated for 3 h. Cell lysates were then immunoprecipitated with control IgG or anti-HA. The protein-bound DNAs were extracted and analyzed by qPCR with primers corresponding to the HSV-1 genome. The graph shows the means ± SEM, n = 3. **P < 0.01 (unpaired t-test)
We next investigated whether and how PCBP1 modulates the DNA-binding activity of cGAS. In virto pull-down analysis indicated that overexpression of PCBP1 enhanced the binding of cGAS to the HSV120 DNA (Fig. 5a). In contrast, deficiency of PCBP1 inhibited the binding of endogenous cGAS to the HSV120 DNA in the MLFs (Fig. 5b). We also examined whether deficiency of PCBP1 affects the binding of cGAS to viral DNA in the infected cells. Following cell infection with HSV-1, we immunoprecipitated cGAS, and the protein-bound viral DNA was detected by qPCR with primers targeting HSV-1 DNA. The results indicated that deficiency of PCBP1 decreased the abundance of the cGAS-bound HSV-1 DNA (Fig. 5c). Furthermore, PCBP1 was also bound to and increased the binding affinity of cGAS for HIV-RT-DNA (Fig. 5d). Consistently, PCBP1 deficiency had the opposite effects (Fig. 5e). These results suggest that PCBP1 acts as a cofactor for cGAS recognition of its ligands.
Fig. 5.
PCBP1 promotes the binding of cGAS to DNA. a PCBP1 enhances the binding of cGAS to dsDNA. HEK293 cells were transfected with the indicated plasmids. Twenty hours later, the cell lysates were incubated with biotinylated-HSV120 DNA and streptavidin-Sepharose beads for in vitro pull-down assays. The bound proteins were then analyzed by immunoblotting with anti-HA and anti-PCBP1 antibodies. b PCBP1 deficiency decreases the binding of cGAS to dsDNA in the MLFs. PCBP1-deficient and control MLFs were collected for in vitro pull-down assays as in a. c PCBP1 deficiency decreases the binding of cGAS to the HSV-1 DNA in the infected MLFs. PCBP1-deficient and control MLFs were infected with HSV-1, and 4 h later, the cell lysates were immunoprecipitated with anti-cGAS. The protein-bound DNAs were extracted and analyzed by qPCR analysis with primers corresponding to the HSV-1 genome. The graph shows the means ± SEM, n = 3. **P < 0.01 (unpaired t-test). d PCBP1 enhances the binding of cGAS to HIV-RT-DNA. HEK293 cells were transfected with the indicated plasmids. Twenty hours later, the cell lysates were incubated with biotinylated HIV-RT-DNA and streptavidin-Sepharose beads for in vitro pull-down assays. The bound proteins were then analyzed by immunoblotting with anti-HA and anti-PCBP1 antibodies. e PCBP1 deficiency decreases the binding of cGAS to HIV-RT-DNA in the MLFs. PCBP1-deficient and control MLFs were collected for in vitro pull-down assays as in d. f PCBP1 promotes cGAS binding to dsDNA in the ZCCHC3-deficient cells. ZCCHC3-deficient and control HEK293 cells were transfected with the indicated plasmids. Twenty hours later, the cell lysates were incubated with biotinylated-HSV120 DNA and streptavidin-Sepharose beads for in vitro pull-down assays. The bound proteins were then analyzed by immunoblotting with anti-HA and anti-PCBP1 antibodies. g ZCCHC3 promotes cGAS binding to dsDNA in the PCBP1-deficient cells. PCBP1-deficient and control HEK293 cells were transfected with the indicated plasmids. Twenty hours later, the cells were collected for in vitro pull-down assays as in f. h Effects of PCBP1/ZCCHC3 double deficiency on the HSV-1-triggered transcription of downstream genes in the MLFs. Deficiency of PCBP1 was generated by the CRISPR-Cas9 method in the Zcchc3+/+ and Zcchc3−/− MLFs. The indicated MLFs (1 × 106) were untreated or infected with HSV-1 (MOI = 1) for 6 h before the qPCR analysis. The graph shows the means ± SEM, n = 3. **P < 0.01 (unpaired t-test)
Since ZCCHC3 has been reported to serve as a coreceptor of cGAS by promoting its ligand binding,23 we investigated the relationship between PCBP1 and ZCCHC3 in regulating cGAS sensing of DNA. In vitro pull-down analysis indicated that the overexpression of PCBP1 enhanced the binding of cGAS to HSV120 DNA in the ZCCHC3-deficient cells (Fig. 5f). On the other hand, ZCCHC3 also markedly potentiated cGAS binding to the HSV120 DNA in the PCBP1-deficient cells (Fig. 5g). In addition, PCBP1 deficiency further inhibited the HSV-1-triggered transcription of the Ifnb1 and Isg56 genes in the ZCCHC3-deficient cells (Fig. 5h). These results suggest that PCBP1 and ZCCHC3 independently regulate cGAS sensing of DNA.
PCBP1 is critical for DNA-induced aggregation of cGAS
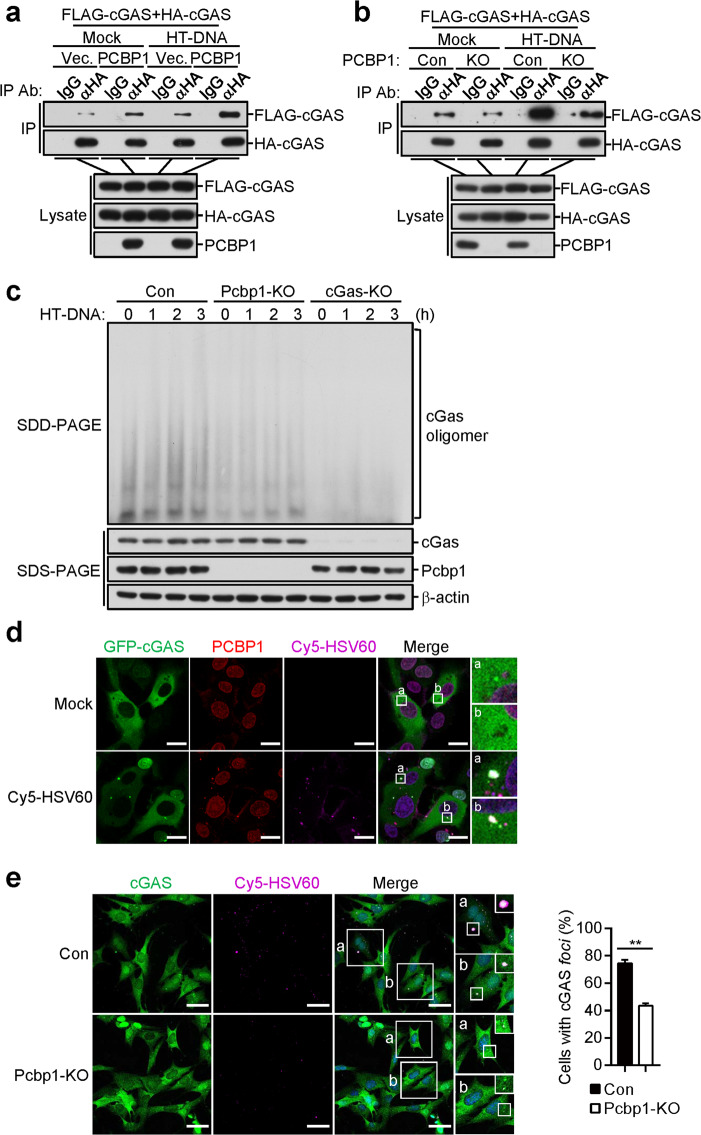
Previously, it has been shown that the binding of cGAS to dsDNA induces its oligomerization, which is required for its activation.18 Coimmunoprecipitation experiments indicated that the overexpression of PCBP1 potentiated the self-association of cGAS, which was enhanced following stimulation with transfected HT-DNA (Fig. 6a). In contrast, PCBP1 deficiency inhibited cGAS self-association in the HEK293 cells (Fig. 6b). In addition, oligomerization of cGAS induced by the transfected HT-DNA was inhibited in the PCBP1-deficient MLFs in comparison with control cells (Fig. 6c). These results suggest that PCBP1 plays an important role in cGAS oligomerization.
Fig. 6.
PCBP1 is critical for DNA-induced liquid phase separation of cGAS. a PCBP1 promotes the HT-DNA-induced self-association of cGAS in the HEK293 cells. HEK293 cells (1 × 107) were transfected with the indicated plasmids for 20 h. The cells were then mock-transfected or transfected with HT-DNA (10 μg) for 4 h before the coimmunoprecipitation and immunoblot analyses were performed with the indicated antibodies. b Effects of PCBP1 deficiency on the HT-DNA-induced self-association of cGAS in the HEK293 cells. PCBP1-deficient and control HEK293 cells (1 × 107) were transfected with the indicated plasmids for 20 h. The cells were then mock-transfected or transfected with HT-DNA (10 μg) for 4 h before the coimmunoprecipitation and immunoblot analyses were performed with the indicated antibodies. c Effects of PCBP1 deficiency on the HT-DNA-induced oligomerization of cGAS in the MLFs. PCBP1-deficient and control MLFs (1 × 107) were mock-transfected or transfected with HT-DNA (10 μg) for 4 h. Cell lysates were then fractionated by SDD-AGE and SDS-PAGE and analyzed by immunoblotting with the indicated antibodies. d PCBP1 and cGAS form puncta with Cy5-HSV60. HT1080 cells stably expressing GFP-tagged cGAS (2 × 105) were mock-transfected or transfected with Cy5-HSV60 (0.5 μg) for 4 h followed by immunofluorescence staining and analysis. Insets show enlarged images of cGAS-DNA-PCBP1 puncta. Scale bars, 20 μm. e Effects of PCBP1 deficiency on the formation of cGAS-DNA foci in the MLFs. PCBP1-deficient and control MLFs (2 × 105) were mock-transfected or transfected with Cy5-HSV60 (0.5 μg) for 4 h before immunofluorescence staining and analysis. Insets show enlarged images of the foci (left). The percentage of cells with cGAS foci was quantified (right), and at least 100 cells from each group were analyzed. Scale bars, 50 μm. The graph shows the means ± SEM, n = 3. **P < 0.01 (unpaired t-test)
Recent studies have shown that the liquid phase separation of cGAS is important for its activation after binding to DNA.21 After cell transfection with fluorescence-labeled DNA, we observed that PCBP1, cGAS, and DNA formed punctate liquid-like structures (Fig. 6d). We also investigated the effects of PCBP1 on cytosolic DNA-induced formation of cGAS-DNA punctate structures in cells. As shown in Fig. 6e, PCBP1 deficiency reduced the formation of cGAS-DNA foci in the MLFs. These data suggest that PCBP1 is important for DNA-induced aggregation and the liquid phase condensation of cGAS.
Discussion
cGAS is a key intracellular PRR that recognizes viral DNA or mislocated self-DNA in the cytosol.5,6 However, the components of the cGAS-associated complex and their roles in the innate immune response have not been fully defined. In this study, we identified PCBP1 as a cofactor of cGAS, which is important for the DNA virus-induced innate immune response.
Using a TAP purification approach, we found that PCBP1 was associated with cGAS. Endogenous coimmunoprecipitation experiments indicated that PCBP1 did not interact with cGAS in uninfected cells but associated with cGAS after HSV-1 infection. Gel filtration experiments indicated that PCBP1 and cGAS existed in overlapping high-molecular-weight complexes after HSV-1 infection. Overexpression of PCBP1 promoted the self-association of cGAS, while deficiency of PCBP1 impaired DNA-induced aggregation and the liquid phase condensation of endogenous cGAS, which is important for cGAS activation after its binding to dsDNA.
PCBP1 deficiency inhibited the HSV-1- and transfected DNA-triggered induction of downstream genes in murine and human cells. Moreover, the replication of HSV-1 was increased in the PCBP1-deficient cells compared with that in the control cells. Unfortunately, PCBP1-deficient mice exhibited early embryonic lethality,36 which ended our investigation of PCBP1 in vivo. Nevertheless, our results suggest that PCBP1 plays an important role in the cGAS-mediated innate immune response to cytosolic DNA and DNA viruses.
Mechanistic studies suggest that PCBP1 acts as a cofactor for the recognition of cytosolic DNA by cGAS. PCBP1 deficiency inhibited the DNA-triggered induction of cGAMP in the MLFs, which suggests that PCBP1 functions through cGAS. PCBP1 bound to synthetic DNA and naturally infected HSV-1 DNA through its KH1 domain. Overexpression of PCBP1 promoted the binding of cGAS to dsDNA, whereas PCBP1 deficiency had the opposite effect. Consistently, overexpression of KH1 in PCBP1 was sufficient to potentiate cGAS-MITA-mediated activation of the ISRE reporter in the HEK293 cells. These results suggest that PCBP1 acts as a cofactor for cGAS binding to its ligands.
It has been reported that PQBP1 functions as a specific coreceptor of cGAS in the recognition of reverse-transcribed HIV-1 ssDNA, but it is not required for the recognition of transfected dsDNA or other viral DNA. In contrast, we found that PCBP1 directly interacted with ssDNA and dsDNA and was required for ssDNA- and dsDNA-triggered innate immune responses, suggesting that PCBP1 acts as a general cofactor of cGAS for the regulation of both ssDNA and dsDNA. ZCCHC3, another DNA-binding protein, acts as a coreceptor of cGAS by promoting its ligand binding. Both ZCCHC3 and PCBP1 are associated with cGAS after HSV-1 infection. Our results suggest that PCBP1 and ZCCHC3 act independently in regulating cGAS sensing of DNA. The identification of PCBP1 as a cofactor for cGAS provides additional insights into the molecular mechanisms of the innate immune response to viral or mislocated self-DNA.
Acknowledgements
We thank members of our laboratory for technical help and discussion. This work was supported by grants from the State Key R&D Program of China (2017YFA0505800 and 2016YFA0502102) and the National Natural Science Foundation of China (31830024, 31630045, and 31870870).
Author contributions
H.-B.S., C.-Y.L., and C.-Q.L. designed the research; C.-Y.L. performed the research; H.-B.S., C.-Y.L., and C.-Q.L. analyzed the data; and H.-B.S., C.-Q.L., and C.-Y.L. wrote the paper.
Competing interests
The authors declare no competing interests.
References
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Hu MM, Shu HB. Cytoplasmic mechanisms of recognition and defense of microbial nucleic acids. Annu Rev. Cell Dev. Biol. 2018;34:357–379. doi: 10.1146/annurev-cellbio-100617-062903. [DOI] [PubMed] [Google Scholar]
- 3.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 4.Paludan SR, Bowie AG. Immune sensing of DNA. Immunity. 2013;38:870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li XD, et al. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.West AP, et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520:553–557. doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao D, et al. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lahaye X, et al. The capsids of HIV-1 and HIV-2 determine immune detection of the viral cDNA by the innate sensor cGAS in dendritic cells. Immunity. 2013;39:1132–1142. doi: 10.1016/j.immuni.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Lam E, Stein S, Falck-Pedersen E. Adenovirus detection by the cGAS/STING/TBK1 DNA sensing cascade. J. Virol. 2014;88:974–981. doi: 10.1128/JVI.02702-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ablasser A, et al. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao P, et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong B, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 14.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo WW, et al. iRhom2 is essential for innate immunity to DNA viruses by mediating trafficking and stability of the adaptor STING. Nat. Immunol. 2016;17:1057–1066. doi: 10.1038/ni.3510. [DOI] [PubMed] [Google Scholar]
- 16.Shu HB, Wang YY. Adding to the STING. Immunity. 2014;41:871–873. doi: 10.1016/j.immuni.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 17.Civril F, et al. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498:332–337. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, et al. Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity. 2013;39:1019–1031. doi: 10.1016/j.immuni.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, et al. The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Rep. 2014;6:421–430. doi: 10.1016/j.celrep.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mankan AK, et al. Cytosolic RNA:DNA hybrids activate the cGAS-STING axis. EMBO J. 2014;33:2937–2946. doi: 10.15252/embj.201488726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du M, Chen ZJ. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science. 2018;361:704–709. doi: 10.1126/science.aat1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoh SM, et al. PQBP1 is a proximal sensor of the cGAS-dependent innate response to HIV-1. Cell. 2015;161:1293–1305. doi: 10.1016/j.cell.2015.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lian H, et al. ZCCHC3 is a co-sensor of cGAS for dsDNA recognition in innate immune response. Nat. Commun. 2018;9:3349. doi: 10.1038/s41467-018-05559-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim SS, et al. Poly(C) binding protein family is a transcription factor in mu-opioid receptor gene expression. Mol. Pharm. 2005;68:729–736. doi: 10.1124/mol.105.012245. [DOI] [PubMed] [Google Scholar]
- 25.Zhou X, You F, Chen H, Jiang Z. Poly(C)-binding protein 1 (PCBP1) mediates housekeeping degradation of mitochondrial antiviral signaling (MAVS) Cell Res. 2012;22:717–727. doi: 10.1038/cr.2011.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.You F, et al. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat. Immunol. 2009;10:1300–1308. doi: 10.1038/ni.1815. [DOI] [PubMed] [Google Scholar]
- 27.Xu LG, et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 28.Hu MM, et al. Sumoylation promotes the stability of the DNA sensor cGAS and the adaptor STING to regulate the kinetics of response to DNA virus. Immunity. 2016;45:555–569. doi: 10.1016/j.immuni.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 29.Wei J, et al. SNX8 modulates innate immune response to DNA virus by mediating trafficking and activation of MITA. PLoS Pathog. 2018;14:e1007336. doi: 10.1371/journal.ppat.1007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu ZS, et al. G3BP1 promotes DNA binding and activation of cGAS. Nat. Immunol. 2019;20:18–28. doi: 10.1038/s41590-018-0262-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herzner AM, et al. Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA. Nat. Immunol. 2015;16:1025–1033. doi: 10.1038/ni.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siomi H, Matunis MJ, Michael WM, Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoga YM, et al. Contribution of the first K-homology domain of poly(C)-binding protein 1 to its affinity and specificity for C-rich oligonucleotides. Nucleic Acids Res. 2012;40:5101–5114. doi: 10.1093/nar/gks058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghanem LR, et al. The poly(C) binding protein Pcbp2 and its retrotransposed derivative Pcbp1 are independently essential to mouse development. Mol. Cell Biol. 2016;36:304–319. doi: 10.1128/MCB.00936-15. [DOI] [PMC free article] [PubMed] [Google Scholar]