Abstract
Fucoidans are sulfated, fucose-rich marine polysaccharides primarily found in cell walls of brown seaweeds (macroalgae). Fucoidans are known to possess beneficial bioactivities depending on their structure and sulfation degree. Here, we report the first functional characterization and the first crystal structure of a prokaryotic sulfatase, PsFucS1, belonging to sulfatase subfamily S1_13, able to release sulfate from fucoidan oligosaccharides. PsFucS1 was identified in the genome of a Pseudoalteromonas sp. isolated from sea cucumber gut. PsFucS1 (57 kDa) is Ca2+ dependent and has an unusually high optimal temperature (68 °C) and thermostability. Further, the PsFucS1 displays a unique quaternary hexameric structure comprising a tight trimeric dimer complex. The structural data imply that this hexamer formation results from an uncommon interaction of each PsFucS1 monomer that is oriented perpendicular to the common dimer interface (~ 1500 Å2) that can be found in analogous sulfatases. The uncommon interaction involves interfacing (1246 Å2) through a bundle of α-helices in the N-terminal domain to form a trimeric ring structure. The high thermostability may be related to this unusual quaternary hexameric structure formation that is suggested to represent a novel protein thermostabilization mechanism.
Subject terms: Biochemistry, Biotechnology
Introduction
Brown seaweeds (brown macroalgae) contain fucoidan polysaccharides as part of their cell wall. The glycosidic backbone of fucoidan mainly consists of L-fucose residues linked by either α-1,3 glycosidic bonds or by alternating α-1,3 and α-1,4 bonds. The latter is a typical pattern in Fucus species, e.g. Fucus evanescens1. In other species, such as Turbinaria ornata and Sargassum mcclurei, the fucoidans are galactofucans, and S. mcclurei fucoidan can even have up to a 1:1 ratio of fucose to galactose in the backbone2. Fucoidans have attracted attention due to their manifold beneficial bioactivities that depend on the fucoidan configuration including the sulfation degree3–9. In fucoidans from e.g. F. evanescens and S. mcclurei the sulfate groups are mainly found on C2 and/or C4 of the fucosyl moieties1,2 (in S. mcclurei fucoidan also on some of the galactosyl moieties2), while C3 sulfation is rare1,2. The complexity of fucoidans requires that the organisms feeding on these polysaccharides possess an array of specific degradative enzymes, both glycosyl hydrolases and sulfatases10,11. Insight into these enzymes is a prerequisite for solving the puzzle of how bacteria degrade macroalgal polysaccharides and notably provide an important foundation for developing targeted, selective fucoidan modification processes to study and enhance specific fucoidan bioactivities. A few endo-acting fucoidanases that specifically catalyze cleavage of α-1,4 fucosyl12–14 or α-1,3 linkages15,16, have been described, while two fucoglucuronomannan specific endo-fucoidanase lyases have also been identified15.
Sulfatases represent a large family of enzymes that catalyze the hydrolytic cleavage of sulfate ester groups on carbohydrates (EC 3.1.6.-sulfuric ester hydrolases; EC 3.10.1.-sulfamidases) and are divided into four classes based on homology, crystal structures, and catalytic mechanism17. Type I sulfatases (S1) include the vast majority of the currently characterized sulfatases18,19. To function, these enzymes require a post-translational modification of the conserved catalytic site cysteine or serine into a formylglycine, usually catalyzed by a formylglycine-generating enzyme20,21.
A few prokaryotic polysaccharide-specific type S1 sulfatases have been characterized from marine bacteria such as Pseudoalteromonas carrageenovora22, Flammeovirga pacifica23, Pyrococcus furiosus24 and Thermotoga marina25 with activity on agar, while carrageenan active sulfatases have been found in Pseudoalteromonas atlantica26,27, Pseudoalteromonas sp. PS4718 and P. carrageenovora22,28. Likewise, a few fucoidan active sulfatases have been reported29–37, although only two have been functionally characterized, namely SWF1 and SWF4 from the marine bacterium Wenyingzhuangia fucanilytica CZ1127T36. SWF1 and SWF4 show activity on fuco-oligosaccharides; SWF1 appears to be exo-acting36.
Here we report the discovery, expression, characterization and structural elucidation of the recombinant fucoidan active sulfatase PsFucS1. PsFucS1 was isolated from Pseudoalteromonas sp. from the gut of sea cucumbers. The data represent the first characterized and crystallized subfamily S1_13 sulfatase as well as the first crystal structure of a fucoidan active sulfatase.
Results
Identification of putative sulfatases in a sea cucumber gut bacterium
To identify new fucoidan active sulfatases, 97 strains of aerobic bacteria (MB01-97) were isolated from sea cucumber guts and selected on fucoidan agar plates. Fucoidan modifying ability was observed for 16 strains (Supplementary Fig. 1), three of which, MB47, MB87 and MB104, showed distinct sulfatase activity (Supplementary Fig. 2). The sulfatase activity was highest in strain MB47 (Supplementary Figs. 2 and 3), which by 16S ribosomal DNA comparisons was determined to be a Pseudoalteromonas sp., closely related to Pseudoalteromonas shioyasakiensis (Supplementary Table 1). The strain was genome sequenced, resulting in a 100% complete genome draft (VFBD00000000; Supplementary Table 2).
A range of carbohydrate-active enzymes (CAZymes) were predicted from the genome (Supplementary Table 3), including putative enzymes active on seaweed carbohydrates, e.g. carrageenan (GH16), agar (GH16) and alginate (PL6, PL7 and PL17). The many predicted CAZymes suggests that this marine bacterium can degrade several complex seaweed carbohydrates.
Seven putative sulfatases, containing the sulfatase domain signature IPR000917, were also identified in the genome (Supplementary Table 4). Five of these proteins, named PsSUL1 to 4 and PsFucS1 contained the conserved sulfatase domain (CXXXRXXXXXG). BLASTp comparisons using the SulfAtlas database (v1.2; http://abims.sb-roscoff.fr/sulfatlas)17, grouped these putative sulfatases into subfamilies S1_4 (PsSUL2) and S1_13 (PsFucS1, PsSUL1, PsSUL3-4; Supplementary Table 4). While the subfamily S1_4 contains several characterized sulfatases (EC 3.1.6.1), the specificity of S1_13 sulfatases is unknown. Multiple alignment of protein sequences (Supplementary Fig. 4) revealed that PsSUL2 is slightly related to ARS (identity 30%) (previously AtsA; accession number: P51691) from Pseudomonas aeruginosa PAO1, which is active on small sulfated compounds, including pNCS 38,39, while PsSUL1, 3, 4 and PsFucS1 seem more related to the agar sulfatase Ary423 from F. pacifica (identity 51, 47, 46 and 29% respectively; Accession Number: AKL72071.1)23.
Functional characterization of the PsFucS1 sulfatase
The Pseudoalteromonas sp. MB47 sulfatases PsSUL1-4, PsFucS1 and the previously characterized sulfatases ARS and Ary423, used as benchmark sulfatases, were expressed in Escherichia coli and purified (Supplementary Fig. 5; Supplementary Table 5). ARS and PsFucS1 showed sulfatase activity, while the other sulfatases did not (Supplementary Fig. 6). Presumably, the heterologous expression in E. coli results in a mixture of unmodified and modified enzymes. Insufficient formylglycine formation by the heterologous formylglycine-generating enzyme in E. coli could therefore be the reason for the lack of activity of some of the sulfatases, while some of the ARS and PsFucS1 enzymes produced have been sufficiently transformed to result in measurable activity. All hitherto investigated enzymes in the S1 family of sulfatases require formylglycine modification for activity18. Hence, it is most likely that both modified and unmodified PsFucS1 molecules were present in the heterologously produced samples, and that a sufficient amount of PsFucS1 (and ARS) had been modified via the E. coli formylglycine-generating enzyme to elicit activity.
PsFucS1 exhibited activity between pH 6.0 to 7.0 with optimum at pH 6.5 and 68 °C (Supplementary Fig. 7). The kinetics analysis of PsFucS1 on pNCS showed that in the absence of NaCl, the apparent KM was 0.95 mM, kcat 1.5 min−1 and kcat/KM was 1.6 mM min−1. In the presence of 125 mM NaCl, the apparent KM was similar (0.88 mM) (Supplementary Fig. 8), but kcat was reduced to 1.1 min−1 and kcat/KM to 1.3 mM min−1.
The thermostability analysis verified an extraordinarily high thermal stability of the PsFucS1 at 68 °C with a slightly negative effect of the presence of NaCl. Hence, when pre-incubated without NaCl, kD of the enzyme was 9.5 × 10–4 min−1 at 68 °C (half-life 12.2 h); but when incubated with 125 mM NaCl the kD of the enzyme was higher, 13.4 × 10−4 min−1 (half-life 8.6 h) (Supplementary Figs. 9 and 10).
Crystal structure of PsFucS1
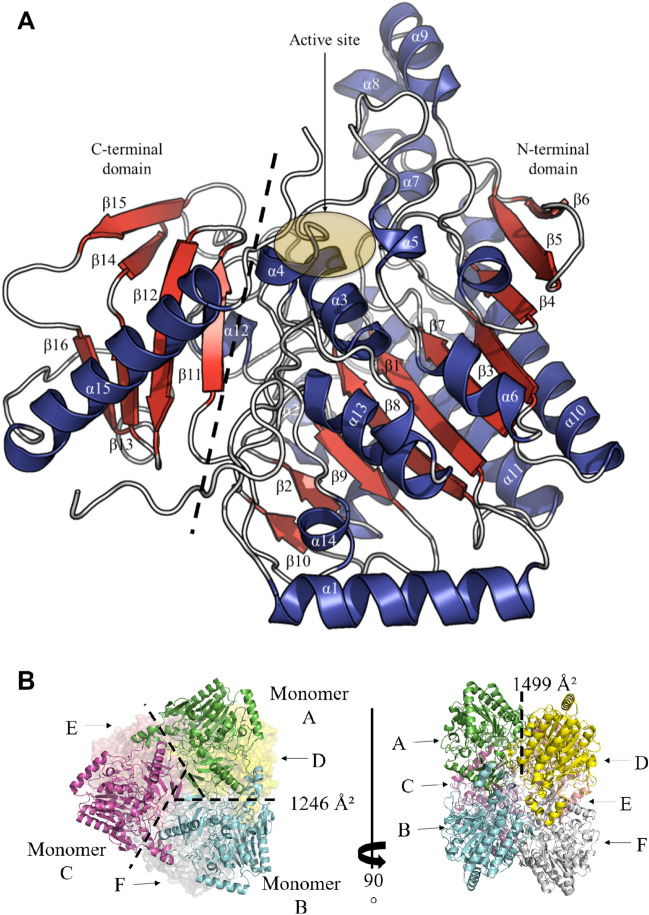
Although about forty crystal structures of sulfatases have been characterized to date, none are from the sulfatase subfamily S1_13 like PsFucS1 as defined by the SulfAtlas database17. The X-ray structure of PsFucS1 was solved by molecular replacement at 2.50 Å resolution (PDB entry 7AJ0), using the human N-acetylgalactosamine-6-sulfatase (GALNS; NM_000512.4; 4FDI; S1_5) as starting model. The crystals belonged to space group P212121 with six molecules in the asymmetric unit. The RMSD values of Cα atoms between the six copies were low, ranging from 0.10 to 0.15 Å, and the monomers can therefore be considered as identical (detailed data statistics are summarized in Supplementary Table 6).
The overall structure (Fig. 1A) revealed that each PsFucS1 monomer is organized in a catalytic N-terminal domain (Asn21-Leu419) region and a C-terminal domain (Val430-Gly521) typical of the formylglycine dependent sulfatase fold. The quaternary organization of the molecules in the asymmetric unit of the crystal can be described as a trimer of dimers (Fig. 1B), the dimer being similar to the closest homologous sulfatase 4FDI, while the trimer interface appears unique to PsFucS1.
Figure 1.
PsFucS1 fold and oligomeric state. (A) Cartoon representation of the PsFucS1 structure showing the domain organization and secondary structure elements. The N-terminal domain (Asn21-Leu419: α1-α14 helices and β1–β10 strands) and the C-terminal domain (Val430-Gly521: α15 helix and β11–β16 strands) are separated by a dashed line. The active site position is circled in yellow. Secondary structure elements are colored in red and blue for β-strands and α-helices, respectively. (B) Cartoon representation of the PsFucS1 hexamer (trimer of dimers) in the asymmetric unit. Monomers A, B, C, D, E and F are colored in green, cyan, purple, yellow, pink and white, respectively.
To further decipher the oligomeric state of PsFucS1 in solution, the enzyme was analyzed by size-exclusion chromatography. A single peak (Supplementary Fig. 11) with an estimated apparent molecular mass of approximately 291 kDa was observed, corroborating a hexameric structure of PsFucS1 in solution, the theoretical molecular mass of which is calculated to be 345.8 kDa (and 172.4 kDa for a trimer).
The common dimer interface, already observed in numerous sulfatases, is approximately 1500 Å2 (predicted by PISA, Fig. 1B) and involves 48 residues. Interfacing residues are mainly found in loops and very few are found in secondary structure elements. Surprisingly, interfacing residues are not that conserved within the sulfatase subfamily S1_13 (approximately 25%) and half of them are not conserved (present in less than 5% of the sequences). However, among the 48 interfacing residues, Gly80, Thr103, Arg363 and Asp482 residues are highly conserved (> 90%) and potentially play a key role in dimerization. The Gly80 residues of both monomers are face-to-face to avoid steric clashes. The Arg363 residue, stabilized by O-Thr103, Oγ-Ser104 and Oδ1-Asn381 atoms of the same monomer, is hydrogen-bonded to the O-Ala84 atom of the other monomer. The Asp482 residue participates in the construction of a π-turn structure and the formation of hydrogen bonds with N-Glu485, N-Met486 and N-Ala487 atoms. The π-turn located in the loop connecting the β16-strand to the α15-helix surely allows the α15-helix to cap β-strands of the C-terminal domain.
The trimer interface of each monomer is approximately 1246 Å2 (predicted by PISA, Fig. 1B) and involves 76 residues. The crystal structure of PsFucS1 is the first representative of the sulfatase subfamily S1_13, and interfacing residues, found both in loops and secondary structure elements in the trimer are poorly conserved (approximately 13%). This atypical interface is created through a bundle of α-helices (α7, α8 and α9) found in the N-terminal domain and composed of residues with very low conservation. Like the classic dimer interface, only four residues (His281, Lys354, Gly355 and Pro474) are highly conserved (> 90%) in the interface between each monomer forming the trimer.
Active site of PsFucS1
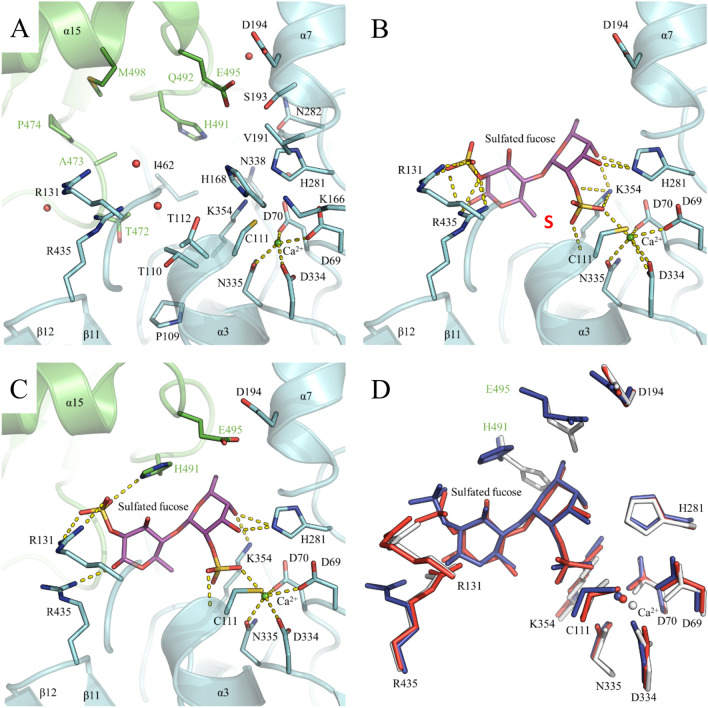
The active site of PsFucS1 (Fig. 2A) is delineated on one side by the presence of an extended β-hairpin loop and on the other side by a bundle of three α-helices, displaying a pocket topology. It is interesting to notice that the α15-helix coming from the other monomer (Fig. 2B) covers the active site. Mapping electrostatic potential onto the surface of PsFucS1 highlights a positively charged pocket (Fig. 2C), perfectly suited for the recognition of a sulfate group. The positive charge at the active site is similar to that of a 4S-ι-carrageenan active endo-sulfatase (6B0J; Fig. 2D), crystallized with a substrate molecule. The active site of sulfatases normally encompasses the catalytic nucleophile formylglycine residue but the Fo-Fc difference density of the X-ray structure of PsFucS1 showed no evidence of significant maturation of the catalytic Cys111, but undoubtedly showed the unmodified cysteine. This is common in crystal structures of sulfatases that are heterologously produced in the absence of the native formylglycine-generating enzyme, since the E. coli maturation machinery is often unable to post-translationally modify the sulfatase to a substantial extent18. If modified formylglycine residues are present in the crystal, they appear to be the minor fraction and are averaged out by the crystal structure.
Figure 2.
PsFucS1 active site topology. (A,B) Cartoon representation of PsFucS1. The N-terminal domain and the C-terminal domain are colored in blue and red, respectively. The active site pocket is circled in yellow. Structural elements delineating the protein active site are indicated by arrows. In (B) Part of the interfacing neighboring monomer is indicated in green including the α15 helix that partially covers the active site. (C) PsFucS1 crystal structure in electrostatic representation. The active site pocket is circled in yellow. (D) Electrostatic representation of the endo-4S-ι-carrageenan sulfatase from Pseudoalteromonas sp. PS47 (6B0J) in complex with κ-ι-κ-neocarrahexaose shown for comparison18. The active site groove is circled in yellow. Electrostatic potential is expressed as a spectrum ranging from − 5 kT e−1 (red) to + 5 kT e−1 (blue) and was calculated in APBS77. (E) Electron density around the calcium ion of PsFucS1. The map shown is a σA-weighted 2mFo-DFc map contoured at 1.2σ (0.67e− Å−3). Asp69, Asp70, Cys111, Asp334 and Asn335 residues are shown as sticks. The calcium atom is shown as a green sphere.
The catalytic residue Cys111 together with Asp69, Asp70, Asp334, and Asn335 coordinates a calcium ion (Fig. 2E) required for the binding and activation of the sulfate group of the substrate, which is highly conserved among S1 sulfatases. Analysis of the effect of presence of divalent cations on PsFucS1 activity showed profound activity retarding effects of the heavy metals Ni2+, Zn2+, Cu2+, and Fe2+, while addition of Mg2+, Mn2+ and Ca2+ produced increased activity of 127, 131 and 608%, respectively (Table 1), verifying that Ca2+ is indeed required for optimal activity of PsFucS1.
Table 1.
Effect of divalent cations on PsFucS1 activity.
| Divalent cations | Relative activity (%)1 | |||
|---|---|---|---|---|
| 0 mM | 2 mM | 5 mM | 10 mM | |
| Ca2+ | 100d | 258c ± 16 | 452b ± 16 | 608a ± 4 |
| Mg2+ | 100a | 110a ± 12 | 124a ± 12 | 127a ± 8 |
| Mn2+ | 100b | 131a ± 9 | nd | nd |
| Ni2+ | 100a | 16b ± 1 | 15b ± 1 | nd |
| Zn2+ | 100a | 6b ± 3 | nd | nd |
| Cu2+ | 100a | 35b ± 6 | nd | nd |
| Fe2+ | 100a | 8b ± 1 | nd | nd |
nd not determined.
1Data are given as averages of duplicate determinations in each row in this Table. Different roman superscript letters a–c indicate statistically different values at p < 0.05 for different addition levels of each cation.
The subsite nomenclature for carbohydrate sulfatases is such that the invariant sulfate binding site is denoted the S site. The sulfate of this site is appended to the 0 subsite sugar unit. Sugar subsites then increase in number (i.e. + 1, + 2) as the sugar moves toward the reducing end (free O1) and decreases in number as the sugar chain moves towards the non-reducing end (i.e. − 1, − 2)18. Molecular docking of C2 sulfated fucobiose into the active site (Fig. 3) allowed to position L-fucose units in subsites 0 and + 118 by replacing water molecules of the PsFucS1 crystal structure (Fig. 3A), and with the sulfate group correctly positioned at the S-site (defined in Hettle et al. 201818; Fig. 3B).
Figure 3.
Docking results of sugar-sulfatase complexes and PsFucS1 activity on fucoidan. (A) Active site pocket of PsFucS1 X-ray crystal structure. (B) Cartoon representation of C2 sulfated fucobiose-PsFucS1 complex obtained by docking. The red S indicates the position of the sulfate binding site as defined by Hettle et al. 201818. (C) Zoomed-in view of PsFucS1 active site with docked disaccharide at the dimer interface. (A–C) Monomers A and B are colored in green and cyan, respectively. Residues constituting the binding pocket are pointed as sticks and those from monomer A are labeled in green. Secondary structure elements are shown as cartoon. The calcium atom and water molecules are shown as green and red spheres, respectively. The bound disaccharide into the active site pocket is shown as purple sticks. H bonds between the ligand and protein are yellow dotted lines. (D) Superposition of the PsFucS1 X-ray crystal structure (white) to the monomer docked model (red) and the dimer docked model (blue).
The interactions observed in the docked ligand-enzyme structure (Fig. 3B–D) highlight the essential residues that possibly determine the observed substrate specificity. The best docking score was obtained in complex with a C2-sulfated fucobiose molecule as ligand. The C2-sulfate of fucose bound in subsite 0 is ideally positioned for catalysis, and the C2-sulfate of the adjacent fucose unit, in subsite + 1, is tightly bound by two arginine residues (namely Arg131 and Arg435, Fig. 3B,C) that are π-stacked to another. In this configuration the fucose unit at subsite 0 is further recognized by His281 (hydrogen bonded to O3 and O4) and Lys354 (hydrogen bonded to O3). Interestingly, one loop of a neighboring monomer (within the dimer) comes to lie right next to the + 1 site, possibly participating in the recognition of the sulfated-fucose oligosaccharides. Notably, this loop contains two residues, His491 and Glu495 that could be crucial for substrate specificity: His491 participates in binding of the fucose in subsite + 1, while the presence of Glu495 would sterically interfere with a sulfate at C3 of this fucose unit (Fig. 3C).
Sulfatase activity of PsFucS1 on fucoidan oligosaccharides
Examination of the sulfatase activity of PsFucS1 on fucoidans from S. mcclurei and F. evanescens revealed that PsFucS1 did not release detectable amounts of sulfate from native fucoidans (Table 2), but showed that the enzyme released sulfate from fucoidan oligosaccharides (Table 2) prepared by endo-fucoidanase treatment12,15,40. Of the total sulfate in the respective substrates, PsFucS1 released of 0.6% sulfate from F. evanescens fucoidan and 1.0 or 5.5% respectively from S. mcclurei fucoidan, dependent on the fucoidanase used. PsFucS1 activity on fucoidan oligosaccharides was, as expected, higher at 68 °C than 35 °C (Table 2).
Table 2.
Activity of PsFucS1 in combination with or without different fucoidanases on different fucoidan substrates at two different temperatures.
| Fucoidan origin | Total sulfate content (quantified as %SO42−) | Reaction temp. (°C) | %SO42− released by PsFucS11 | |||
|---|---|---|---|---|---|---|
| – | FdlA | FdlB | FcnAΔ229 | |||
| Fucus evanescens | 30.3 ± 1.9 | 35 | 0b,x | 0.2a,y ± 0.0 | 0.1a,y ± 0.0 | 0.2a,y ± 0.0 |
| 68 | 0b,x | 0.6a,x ± 0.2 | 0.6a,x ± 0.0 | 0.6a,x ± 0.1 | ||
| Sargassum mcclurei | 32.9 ± 2.3 | 35 | 0c,x | 1.3a,y ± 0.0 | 1.3a,y ± 0.0 | 0.7b,y ± 0.0 |
| 68 | 0c,x | 5.5a,x ± 0.0 | 5.5a,x ± 0.0 | 1.0b,x ± 0.0 | ||
– no fucoidanase.
1Sulfate-release data (%SO42− released) are given as averages of duplicate assays (data are relative to the theoretical maximum release); different roman superscript letters a–c indicate significantly different values (p < 0.05) in each row, i.e. in the PsFucS1 reactions together with the different fucoidanases (FdlA, FdlB, and FcnAΔ229), respectively, and different roman superscript letters x–y indicate significantly different values (p < 0.05) when comparing the two reaction temperatures for each type of fucoidan reaction.
Discussion
Considering its mesophilic marine bacterium origin, PsFucS1 had a surprisingly high thermal reaction optimum at 68 °C (at pH 6.5) and an unusually high thermostability. Three thermostable sulfatases have been described to date, the agar sulfatase from T. maritima with optimal temperature at 80 °C25, Atsa from P. aeruginosa with optimal temperature of 57 °C38 and Ary423 from F. pacifica, with optimal activity at 40 °C, but with a wide temperature range of 30–70 °C23. Ary423 retained more than 40% activity at 60 °C after 12 hours23, but the stability of PsFucS1 was even higher with a half-life at 68 °C of 12.2 h.
Interestingly, PsFucS1 formed hexamers in the crystal as well as in solution. Higher oligomerization has frequently been observed as a strategy to attain thermostability41, suggesting that the thermostability of PsFucS1 might be obtained through hexamerization. It is noteworthy that a recent report also links higher oligomerization states for cold adaptation of an enzyme42. While several oligomerization states, ranging from monomer to tetramer and even an octamer (1auk, family S1_1), have been observed for different S1 sulfatases so far, the hexameric organization of PsFucS1 observed here is unique. Among S1_13 sequences the crucial regions leading to the trimerization are not conserved. It is thus highly probable that not all S1_13 sulfatases occur as hexamers. The enzyme was Ca2+ dependent, in accord with the structural protein analysis that clearly indicated a negatively charged calcium cavity near the active site. Despite being of marine origin, PsFucS1 was both more active and more stable without NaCl than in the presence of NaCl with a 1.3 fold higher kcat without NaCl, but a similar KM value. Together with the thermal stability data these results might suggest that conditions with warmer temperatures and lower salt concentrations than found in the Vietnamese sea, could be the true origin of the PsFucS1 Pseudoalteromonas sp. MB47 bacterium.
PsFucS1 presumably removes sulfate from short fuco-oligosaccharides in an exo-acting manner. Only a small amount of sulfate was released as expected, largely due to the lack of complete degradation of the fucoidan by the fucoidanases, and due to the differentially positioned sulfate groups. Indeed, this hypothesis can also be inferred from the fucobiose-PsFucS1 complex that was obtained by molecular docking, where the complex with a C2-sulfated fucobiose molecule as ligand gave the best score. Moreover, the formylglycine post-translational modification might, as suggested above, not be fully obtained during the heterologous expression in E. coli, likely resulting in the lower apparent activity of the sulfatase. This interpretation is supported by the crystal structure, where no formylglycine was evident in the active site of the crystallized enzymes.
In summary we report the biochemical characterization and crystal structure of a hexameric sulfatase that releases sulfate from fucoidan oligosaccharides. Moreover, the PsFucS1 has a promising advantage of high thermostability, which may be attractive for application in production of partially desulfated fucoidan oligosaccharides in the future. Indeed, many bioactivities have been shown for fucoidans and fucoidan oligosaccharides3,5,43, while the exact underlying chemical structures responsible for the various bioactivities remain elusive. By the use of fucoidan sulfatases, including PsFucS1, the position-function relationship of sulfate groups on fucoidan for the bioactivity could potentially be elucidated.
Methods
Substrates
p-nitro catechol sulfate dipotassium salt (pNCS) was from Sigma-Aldrich (Steinheim, Germany). 5-Bromo-4-chloro-3-indolyl sulfate potassium salt (X-SO4) was from Carbosynth Ltd (Compton, UK). Fucoidans from S. mcclurei and T. ornata were obtained as described previously2. The F3 fraction of S. mcclurei fucoidan was obtained by anion exchange chromatography2,44 using DEAE-macroprep resin (Bio-Rad, Hercules, CA, USA). Crude fucoidans from F. evanescens were extracted using an enzyme-assisted method and further fractionated to obtain the F2 fraction by anion exchange chromatography using a DEAE-macroprep resin (as above), and characterized as previously described45.
Isolation of fucoidan-utilizing marine bacteria
Sea cucumbers were sampled at Hon Tre, Nha Trang Bay, Vietnam (12.23100 N, 109.24203 E). The sea cucumbers were stored on ice and transported to the laboratory within an hour of collection. After the surface of each of the sea cucumbers was sterilized with 70% ethanol, the ventral surface was dissected with a sterile scalpel and the digestive tract was removed and its contents were carefully squeezed into a sterile tube. All gut content and sediment samples were carefully mixed with sterile sea water and suspensions were spread on solid fucoidan MB media (F-MB) containing 15 g L−1 agar, 5 g L−1 Difco bactopeptone, 2 g L−1 Difco yeast extract, 1 g L−1 crude fucoidan from S. mcclurei, 0.2 g L−1 K2HPO4, and 0.05 g L−1 of MgSO4. Sampled seawater was used for media preparation (pH 7.5–7.8). After growth at 28–30 °C for 1–7 days, bacterial colonies were streaked on solid MB medium to obtain pure cultures.
Screening of marine bacteria for enzymatic activity
The bacterial isolates were screened for fucoidan-modifying enzymes by the fucoidan agar plate cetavlon method46. In brief, isolates were cultivated for 3 days at 28 °C on fucoidan agar medium containing 0.5% (w/v) S. mcclurei or T. ornata fucoidan. Bacterial cells were removed from the agar plate surface and a 1% aqueous solution of the cationic hexadecyltrimethylammonium bromide salt (cetavlon; Sigma-Aldrich, Steinheim, Germany) was added to each plate. After incubation for 30 min at 25 °C, the agar plates were thoroughly washed with water. Presence of the cationic cetavlon salt forms a water-insoluble white precipitate in the presence of fucoidan: hence transparent zones indicate that the bacteria secrete enzymes with fucoidan-modifying activity since enzymatically degraded fucoidan has a lowered charge density and will not precipitate with cetavlon. Sulfatase activity was detected using MB solid media supplemented with 100 μg ml−1 X-SO4. Enzymatic activity was detected by blue color formation around the colonies38.
Strain identification by 16S ribosomal RNA analysis
Bacterial cells from 5 ml overnight culture in MB liquid media were collected by centrifugation and genomic DNA of selected strains was isolated by DNeasy Blood and Tissue DNA kit (Qiagen, Hilden, Germany), following the manufacturer’s recommendations. The 16S rRNA gene fragment was amplified using Phusion High-Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA, USA) with the universal bacterial primers (533F: 5’-GTGCCAGCAGCCGCGGTAA3’ and 1392R: 5’-GGTTACCTTGTTACGACTT-3’), blunt-end cloned into pJET1.2 vector using CloneJET PCR Cloning Kit (ThermoFisher Scientific, Waltham, MA, USA) and propagated in the E. coli DH5α strain (Invitrogen Life Technologies, Thermo Fisher Scientific, Waltham, MA, USA). After plasmid preparation, inserts were sequenced at Macrogen Inc. (Seoul, Korea) using the Sanger capillary sequencing method. Sequences were aligned against 16S ribosomal RNA entries of NCBI RefSeq database to identify the closest taxonomic bacterial identity.
Genome sequencing of and sequence assembly
Genomic DNA of the Pseudoalteromonas sp. MB47 strain was isolated as described for the 16S ribosomal analysis. The genomic DNA was submitted to Macrogen Inc. (Seoul, Korea) for Illumina TruSeq DNA PCR-free shotgun library preparation and sequencing using Illumina Miseq (300 bp paired-end). Residual sequencing adapters from resulting reads were removed with Cutadapt47 and BBDuk48. After quality filtering, draft genomes were assembled with Ray using a number of different k-mer lengths49. Best assemblies were selected based on the assessment of the constructed deBrujin graphs and quality of the resulting contigs. Reads were mapped back to contigs using the Bowtie2 algorithm50. To further clean assemblies abundance/GC binning was performed with MetaBat51. The completeness of the resulting draft genome was evaluated using Checkm52. Draft genome annotation and CAZymes identification protein-coding features and ribosomal RNA features were predicted with Prodigal and barrnap as part of the prokka annotation pipeline. Additionally, all predicted protein sequences were subjected to annotation using the Interproscan pipeline (v5.27-66.0), using all available databases and analyses. Taxonomy was investigated by aligning all predicted protein sequences against the NR database. Analysis of selected phylogenetic protein markers was done using AMPHORA253 and analysis of identified sequences encoding 16S rRNA genes. dbCAN models were used to predict CAZymes54.
Identification of sulfatases in the Pseudoalteromonas sp. genome and sequence alignment
Protein-coding features in the Pseudoalteromonas sp. MB47 genome were predicted with Prodigal (used as part of the prokka annotation pipeline)55 and subjected to annotation using Interproscan pipeline56, v5.27-66.0), using all available databases and analyses. All signatures corresponding to sulfatase families and domains have been retrieved from the InterPro database57 and used for extraction of sulfatase protein sequences. Signal peptides were predicted using SignalP 4.058. SulfAtlas (http://abims.sb-roscoff.fr/sulfatlas) was used to identify the sulfatase families and subfamilies.
Multiple sequence alignments were performed using ClustalW59 via the NPS@ server (https://npsa-prabi.ibcp.fr/)60. Sequences included previously characterized polysaccharide-active sulfatases as well as the human GALNS sequence. The alignment was rendered using ESPript 3.061.
Cloning, expression and purification of PsFucS1
Enzymes and gene constructs
Constructs containing the genes encoding the sulfatases were designed without the predicted N-terminal signal peptide but with an N-terminal 6xHis tag. The synthetic codon-optimized genes (optimized for E. coli expression), all devoid of their original signal peptide, were synthesized by GenScript (Piscataway, NJ, USA) and delivered inserted into the pET-45b(+) vector between the KpnI and PacI restriction sites. The E. coli strain DH5α (Invitrogen, Waltham, MA, USA), was used as plasmid propagation host.
Production of recombinant sulfatases and fucoidanases
Expression of the sulfatase and endo-fucoidanase FcnAΔ229 was performed in E. coli BL21 (DE3; Invitrogen, Waltham, MA, USA) harbouring the Pch2 (pGro7) plasmid, as previously described40, and expression of FdlA, FdlB was performed in E. coli C41 (DE3) as previously described40. The enzymes were desalted using a PD10 desalting column (GE Healthcare, Chicago, IL, USA) and the protein content was determined by Bradford assay62 with bovine serum albumin as standard.
SDS–PAGE and western blot analysis
The homogeneity and molecular weight of the recombinantly expressed proteins were estimated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) in 12% acrylamide gels, according to the Laemmli protocol63 and western blot analysis using poly-histidine antibodies as previously described40. 7.5 µl E. coli culture, 40 µg crude protein and 5 µg purified protein was loaded on the gels. The Protein Plus molecular weight marker (Bio-Rad Laboratories, Hercules, CA, USA) was used as standard.
Enzyme activity assays
X-SO4 plate screening
Whole E. coli cells harboring the different sulfatase gene-constructs were tested for sulfatase activity by streaking the cells onto X-SO4 agar plates (15 g l−1 agar, 20 mM Tris–HCl, 125 mM NaCl, pH 6.5 and 100 µg ml−1 X-SO4). Crude E. coli cell lysates containing recombinant sulfatases were tested on the X-SO4 agar plates by punching a hole in the plates and adding 50 µl of the cell lysate into the hole. All plates were incubated at 37 °C for 30 min to 24 h.
pNCS sulfatase assay
Sulfatase activity was measured by incubating enzyme samples (0.5 mg ml−1 sulfatase) in a reaction mixture of 40 mM Tris–HCl pH 6.5, 10 mM CaCl2, 125 mM NaCl, and 2.5 mM pNCS at 37 °C or 68 °C. The reaction was stopped by addition of NaOH (final concentration 0.8 M NaOH) before recording absorbance at 515 nm. One unit of sulfatase activity was defined as the amount of enzyme capable of hydrolyzing 1 µmol of pNCS per minute at the assay conditions. Kinetics experiments were performed with different pNCS concentrations (0.5, 2.5 and 4 mM) at pH 6.5, 68 °C and KM and kcat values of the sulfatase were calculated according to Michaelis–Menten kinetics64.
The influence of cations on PsFucS1 sulfatase activity was determined after pre-incubation with 0–10 mM CaCl2, MgCl2, MnCl2, NiCl2, CuCl2, ZnCl2, or FeCl2 for 5 min followed by assay on 2.5 mM pNCS at 37 °C as described above. After the calcium dependence was discovered 10 mM CaCl2 was included in all assays.
Effect of pH and temperature on sulfatase activity was assessed in a statistical factorial design (CCF) using two overlapping buffer systems ranging from pH 5.0 to 8.0 (200 mM acetate buffer pH 5.0 and 200 mM Tris–HCl buffer, center point pH 6.5) and reaction temperatures from 60 to 75 °C, with 67.5 °C as center point.
Thermostability of recombinant PsFucS1 was evaluated by measuring the residual enzyme activity after pre-incubation of the enzyme in 40 mM Tris–HCl, 10 mM CaCl2, pH 6.5 at 68 °C with various levels of NaCl 0–125 mM for up to 12 h. First-order rate constants of the thermal denaturation (KD) and half-life at 68 °C was calculated from the activity decay (Supplementary Figs. 9 and 10).
Fucoidan desulfation
Desulfation of fucoidans was measured after fucoidan hydrolysis catalysed by different fucoidanases in reaction mixtures containing 0.5 µg µl−1 fucoidanase (FcnA, FdlA and FdlB) in 20 mM Tris–HCl buffer pH 7.4, 250 mM NaCl, 10 mM CaCl2 and 1% (w/v) fucoidans from either S. mcclurei or F. evanescens at 35 °C and 68 °C for 24 h. The PsFucS1 (1 mg ml−1) was then added to the reaction resulting in the following reaction mixture: 125 mM NaCl, 10 mM CaCl2, 60 mM Tris–HCl pH 6.5, 0.5% (w/v) fucoidan. Desulfation data were compared by one-way analysis of variance (ANOVA) using Tukey–Kramer’s Significant Differences at p ≤ 0.05 using JMP 15 Pro (SAS Institute Inc., Cary, NC, USA).
Sulfate detection by ion-exchange chromatography
The sulfate content of fucoidans substrates as well as sulfate release after sulfatase treatment was analyzed by high performance anion exchange chromatography (HPAEC) using a Dionex ICS-5000 chromatography system equipped with a GP40 gradient pump and an ED40 electrochemical detector. The ions were separated on an HC-AS11 anion-exchange column (4 × 250 mm; Dionex) with accompanying HC-AG11 guard column (4 × 50 mm; Dionex). Elution was performed with 35 mM NaOH using an isocratic flow rate of 1.5 ml min−1. Background signal and noise originating from the eluent was reduced using an anion self-regenerating suppressor (AERS-500, Dionex) with a current of 180 mA. Sulfate concentration was deduced from the signal intensity and calculated from a standard sulfate calibration curve using Na2SO4. The release of sulfate was related to the amount of sulfate present in the polysaccharide substrate, 33 ± 3% in S. mcclurei2 and 30 ± 2% in F. evanescens45.
Crystallization and structure determination
PsFucS1 crystals were grown in a hanging drop set up at 20 °C using 2 µl protein solution (6 mg ml−1) and 1 µl mother liquor solution containing 0.1 M sodium acetate pH 4.6, 0.1 M sodium chloride and 20% (w/v) PEG 3350. Crystals were flash-frozen and stored in mother liquor supplemented with 14% (v/v) glycerol prior to diffraction experiments. Diffraction data were collected on the PROXIMA-2A beamline at the SOLEIL Synchrotron. Data were processed using XDS65 and scaled and merged with Aimless66 in CCP467. An initial homology model was generated from the SWISS-MODEL server68 based on the N-acetylgalactosamine-6-sulfatase X-ray structure from human (4fdi)69, with which PsFuc1 shares 25% sequence identity. Only residues with a predicted local similarity to target > 0.6 were kept for molecular replacement. The PsFucS1 crystal structure was then solved by molecular replacement using phenix.phaser70. Density modification and model building was carried out with phenix.autobuild71. Refinement was carried out using phenix.refine72 interspersed with manual model revisions using the program Coot73. All statistics are summarized in Table S6.
Analytical size-exclusion chromatography (SEC)
Analytical SEC was performed using a HiLoad 16/600 Superdex 200 pg column connected to an ÄKTA-Purifier at room temperature. The column was calibrated with molecular mass standards ranging from 13.7 to 440 kDa. The molar mass of each protein and their elution volume are as follows: Ferritin (440 kDa—58.08 ml); Aldolase (158 kDa—69.71 ml); Ovalbumin (44 kDa—83.31 ml); Carbonic anhydrase (29 kDa—91.10 ml); Ribonuclease A (13.7 kDa—99.31 ml). 200 µl of His-tagged PsFucS1 (6 mg ml−1) were loaded on the column equilibrated with 50 mM Tris–HCl buffer pH 8.0 and 100 mM NaCl at a flow rate of 0.5 ml min−1. The absorbance of the eluted fractions was recorded at 280 nm.
Molecular docking with a sulfated fucose molecule
Molecular docking was performed on a single PsFucS1 monomer. Only protein atoms were kept (hydrogen, water molecules, and chloride atoms were removed). The initial coordinates of a C2-sulfated fucobiose molecule was generated from the smiles string using phenix.elbow74. Then, AutoDock vina75 was used to obtain a docking model of the PsFucS1/C2-sulfated fucobiose complex. Before docking, the side chains of residues in the active site pocket were set as flexible. Docking was done over a search space of 20 × 20 × 20 Å that covers the entire active site pocket. The calculation yielded 10 possible models, of which the one with the highest ranking was selected as the most likely. Finally, the PsFucS1 structure in complex with the C2-sulfated fucobiose was energy minimized using UCSF Chimera76 with standard steepest descent and conjugate gradient steps. In addition, after the first energy minimization, the C2-sulfated fucobiose molecule binding site at the dimer interface was generated by superposition of the structures followed by energy minimization of the dimer in complex with the substrate using UCSF Chimera76.
Supplementary Information
Acknowledgements
This work was supported by grants from Vietnam National Foundation for Science and Technology Development (NAFOSTED-106.02-2018.353). This study was part of the FucoSan-Health from the Sea Project, supported by EU InterReg-Deutschland-Denmark and the European Fund of Regional Development. This project was also funded by MARIKAT JPI Cofund Blue BioEconomy Project Grant No. 9082-00021B and by the Technical University of Denmark. M.C. and T.R. acknowledge support from ANR under grant ANR-10-BTBR-04 (investment expenditure program IDEALG). The authors appreciate the access to the CristalO platform (FR2424, Station Biologique de Roscoff), which is part of the Biogenouest core facility network and supported by the Conseil Regional Bretagne (Grant No: 20000943). The authors thank the staff of PROXIMA-2A beamline at SOLEIL for its assistance in data collection. We also thank the French synchrotron at SOLEIL (St Aubin) for regular access to the MX beamline Proxima-2A, and especially William Shepard, Serena Sirigu and Martin Savko for valuable help during the X-ray data collection and data treatment.
Author contributions
M.D.M., H.T.T.C., and A.S.M. conceived and planned the study. Genome mining was done by M.L., J.M., and M.D.M., molecular cloning, recombinant expression, protein purification were performed by M.D.M., H.T.T.C., and N.R.-K., fucoidan purification, enzyme assays and characterizations were done by V.T.T.T., T.T.N., V.T.N.H., and T.D.P., chromatographic sulfate analyses were done by N.R.-K., J.H., and crystallography and crystallography interpretation were done by T.R. and M.C.. The paper was written and edited by M.D.M., H.T.T.C., M.C., and A.S.M. All authors reviewed the manuscript.
Data availability
The crystal structure presented has been deposited with PDB entry 7AJ0.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Maria Dalgaard Mikkelsen and Hang Thi Thuy Cao.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-98588-3.
References
- 1.Bilan MI, et al. Structure of a fucoidan from the brown seaweed Fucus evanescens C. Ag. Carbohydr. Res. 2002;37:719–730. doi: 10.1016/S0008-6215(02)00053-8. [DOI] [PubMed] [Google Scholar]
- 2.Thinh PD, et al. Structural characteristics and anticancer activity of fucoidan from the brown alga Sargassum mcclurei. Mar. Drugs. 2013;11:1456–1476. doi: 10.3390/md11051456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ale MT, Mikkelsen JD, Meyer AS. Important determinants for fucoidan bioactivity: A critical review of structure-function relations and extraction methods for fucose-containing sulfated polysaccharides from brown seaweeds. Mar. Drugs. 2011;9:2106–2130. doi: 10.3390/md9102106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torres MD, et al. Fucoidans: The importance of processing on their anti-tumoral properties. Algal Res. 2020;45:101748. doi: 10.1016/j.algal.2019.101748. [DOI] [Google Scholar]
- 5.Silchenko AS, et al. Structure, enzymatic transformation, anticancer activity of fucoidan and sulphated fucooligosaccharides from Sargassum horneri. Carbohydr. Polym. 2017;175:654–660. doi: 10.1016/j.carbpol.2017.08.043. [DOI] [PubMed] [Google Scholar]
- 6.Dörschmann P, et al. Effects of a newly developed enzyme-assisted extraction method on the biological activities of fucoidans in ocular cells. Mar. Drugs. 2020;18:282. doi: 10.3390/md18060282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blondin C, Fischer E, Boisson-Vidal C, Kazatchkine MD, Jozefonvicz J. Inhibition of complement activation by natural sulfated polysaccharides (fucans) from brown seaweed. Mol. Immunol. 1994;31:247–253. doi: 10.1016/0161-5890(94)90121-X. [DOI] [PubMed] [Google Scholar]
- 8.Nardella A, et al. Anticoagulant low molecular weight fucans produced by radical process and ion exchange chromatography of high molecular weight fucans extracted from the brown seaweed Ascophyllum nodosum. Carbohydr. Res. 1996;19:201–208. doi: 10.1016/0008-6215(96)00110-3. [DOI] [PubMed] [Google Scholar]
- 9.Koyanagi S, Tanigawa N, Nakagawa H, Soeda S, Shimeno H. Oversulfation of fucoidan enhances its anti-angiogenic and antitumor activities. Biochem. Pharmacol. 2003;15:173–179. doi: 10.1016/S0006-2952(02)01478-8. [DOI] [PubMed] [Google Scholar]
- 10.Sichert A, et al. Verrucomicrobia use hundreds of enzymes to digest the algal polysaccharide fucoidan. Nat. Microbiol. 2020;5:1026–1039. doi: 10.1038/s41564-020-0720-2. [DOI] [PubMed] [Google Scholar]
- 11.Michel G, Czjzek M. Polysaccharide-degrading enzymes from marine bacteria. In: Trincone A, editor. Marine Enzymes for Biocatalysis: Sources, Biocatalytic Characteristics and Bioprocesses of Marine Enzymes. Elsevier; 2013. pp. 429–464. [Google Scholar]
- 12.Colin S, et al. Cloning and biochemical characterization of the fucanase FcnA: Definition of a novel glycoside hydrolase family specific for sulfated fucans. Glycobiology. 2006;16:1021–1032. doi: 10.1093/glycob/cwl029. [DOI] [PubMed] [Google Scholar]
- 13.Silchenko AS, et al. Expression and biochemical characterization and substrate specificity of the fucoidanase from Formosa algae. Glycobiology. 2017;27:254–263. doi: 10.1093/glycob/cww138. [DOI] [PubMed] [Google Scholar]
- 14.Vuillemin M, et al. Functional characterization of a new GH107 endo-α-(1,4)-fucoidanase from the marine bacterium Formosa haliotis. Mar. Drugs. 2020;18:562. doi: 10.3390/md18110562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takayama, M., Koyama, N., Sakai, T. & Kato, I. Enzymes capable of degrading a sulfated-fucose-containing polysaccharide and their encoding genes. US Patent 6489155 B1 (2002).
- 16.Shen J, Chang Y, Zhang Y, Mei X, Xue C. Discovery and characterization of an endo-1,3-fucanase from marine bacterium Wenyingzhuangia fucanilytica: A novel glycoside hydrolase family. Front. Microbiol. 2020;11:1674. doi: 10.3389/fmicb.2020.01674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barbeyron T, et al. Matching the diversity of sulfated biomolecules: Creation of a classification database for sulfatases reflecting their substrate specificity. PLoS ONE. 2016;11(10):e0164846. doi: 10.1371/journal.pone.0164846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hettle AG, et al. The molecular basis of polysaccharide sulfatase activity and a nomenclature for catalytic subsites in this class of enzyme. Structure. 2018;26:747–758. doi: 10.1016/j.str.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 19.Hanson SR, Best MD, Wong CH. Sulfatases: Structure, mechanism, biological activity, inhibition, and synthetic utility. Angew. Chem. Int. Ed. 2004;43:5736–5763. doi: 10.1002/anie.200300632. [DOI] [PubMed] [Google Scholar]
- 20.Cosma MP, et al. The multiple sulfatase deficiency gene encodes an essential and limiting factor for the activity of sulfatases. Cell. 2003;113:445–456. doi: 10.1016/S0092-8674(03)00348-9. [DOI] [PubMed] [Google Scholar]
- 21.Dierks T, et al. Posttranslational formation of formylglycine in prokaryotic sulfatases by modification of either cysteine or serine. J. Biol. Chem. 1998;273:25560–25564. doi: 10.1074/jbc.273.40.25560. [DOI] [PubMed] [Google Scholar]
- 22.Kim DE, et al. Purification and characterization of the recombinant arylsulfatase cloned from Pseudoalteromonas carrageenovora. Protein Expr. Purif. 2005;39:107–115. doi: 10.1016/j.pep.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 23.Gao C, Jin M, Yi Z, Zeng R. Characterization of a recombinant thermostable arylsulfatase from deep-sea bacterium Flammeovirga pacifica. J. Microbiol. Biotechnol. 2015;25:1894–1901. doi: 10.4014/jmb.1504.04028. [DOI] [PubMed] [Google Scholar]
- 24.Jung KT, et al. Identification of the first archaeal arylsulfatase from Pyrococcus furiosus and its application to desulfatation of agar. Biotechnol. Bioprocess Eng. 2012;17:1140–1146. doi: 10.1007/s12257-012-0228-6. [DOI] [Google Scholar]
- 25.Lee D-G, Shin JG, Jeon MJ, Lee S-H. Heterologous expression and characterization of a recombinant thermophilic arylsulfatase from Thermotoga maritima. Biotechnol. Bioprocess Eng. 2013;18:897–902. doi: 10.1007/s12257-013-0094-x. [DOI] [Google Scholar]
- 26.Préchoux A, Genicot S, Rogniaux H, Helbert W. Controlling carrageenan structure using a novel formylglycine-dependent sulfatase, an endo-4S-iota-carrageenan sulfatase. Mar. Biotechnol. 2013;15:265–274. doi: 10.1007/s10126-012-9483-y. [DOI] [PubMed] [Google Scholar]
- 27.Préchoux A, Helbert W. Preparation and detailed NMR analyses of a series of oligo-α- carrageenans. Carbohydr. Polym. 2014;101:864–870. doi: 10.1016/j.carbpol.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 28.Genicot SM, et al. Discovery of a novel iota carrageenan sulfatase isolated from the marine bacterium Pseudoalteromonas carrageenovora. Front. Chem. 2014;2:67. doi: 10.3389/fchem.2014.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thanassi NM, Nakada HI. Enzymic degradation of fucoidan by enzymes from the hepatopancreas of abalone, Haliotus species. Arch. Biochem. Biophys. 1967;118:172–177. doi: 10.1016/0003-9861(67)90294-9. [DOI] [Google Scholar]
- 30.Sasaki K, et al. Partial purification and characterization of an enzyme releasing 2-sulfo-α-l- fucopyranose from 2-sulfo-α-l-fucopyranosyl-(1→2) pyridylaminated fucose from a sea urchin, Strongylocentrotus nudus. Biosci. Biotechnol. Biochem. 1996;60:666–668. doi: 10.1271/bbb.60.666. [DOI] [Google Scholar]
- 31.Daniel R, et al. Regioselective desulfation of sulfated L-fucopyranoside by a new sulfoesterase from the marine mollusk Pecten maximus: Application to the structural study of algal fucoidan (Ascophyllum nodosum) Eur. J. Biochem. 2001;268:5617–5626. doi: 10.1046/j.1432-1033.2001.02497.x. [DOI] [PubMed] [Google Scholar]
- 32.Lloyd PF, Forrester PF. Desulphation of L-fucose monosulphates by an enzyme from Patella vulgata. Biochem. J. 1971;124:21. doi: 10.1042/bj1240021Pa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Furukawa S, Fujikawa T, Koga D, Ide A. Production of fucoidan-degrading enzymes, fucoidanase, and fucoidan sulfatase by Vibrio sp. N-5. Nippon suisan Gakk. 1992;58:1499–1503. doi: 10.2331/suisan.58.1499. [DOI] [Google Scholar]
- 34.Wegner CE, et al. Expression of sulfatases in Rhodopirellula baltica and the diversity of sulfatases in the genus Rhodopirellula. Mar. Genom. 2013;9:51–61. doi: 10.1016/j.margen.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 35.Sakai T, Ishizuka K, Shimanaka K, Ikai K, Kato I. Structures of oligosaccharides derived from Cladosiphon okamuranus fucoidan by digestion with marine bacterial enzymes. Mar. Biotechnol. 2003;5:536–544. doi: 10.1007/s10126-002-0107-9. [DOI] [PubMed] [Google Scholar]
- 36.Silchenko AS, et al. Fucoidan sulfatases from marine bacterium Wenyingzhuangia fucanilytica CZ1127T. Biomolecules. 2018;8:98. doi: 10.3390/biom8040098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang Y, et al. Isolation and characterization of a sea cucumber fucoidan-utilizing marine bacterium. Lett. Appl. Microbiol. 2010;50:301–307. doi: 10.1111/j.1472-765X.2009.02792.x. [DOI] [PubMed] [Google Scholar]
- 38.Beil S, et al. Purification and characterization of the arylsulfatase synthesized by Pseudomonas aeruginosa PAO during growth in sulfate-free medium and cloning of the arylsulfatase gene (atsA) Eur. J. Biochem. 1995;229:385–394. doi: 10.1111/j.1432-1033.1995.0385k.x. [DOI] [PubMed] [Google Scholar]
- 39.Boltes I, et al. 1.3 Å structure of arylsulfatase from Pseudomonas aeruginosa establishes the catalytic mechanism of sulfate ester cleavage in the sulfatase family. Structure. 2001;9:483–491. doi: 10.1016/S0969-2126(01)00609-8. [DOI] [PubMed] [Google Scholar]
- 40.Cao HTT, et al. Novel enzyme actions for sulphated galactofucan depolymerisation and a new engineering strategy for molecular stabilisation of fucoidan degrading enzymes. Mar. Drugs. 2018;16:422. doi: 10.3390/md16110422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li WF, Zhou XX, Lu P. Structural features of thermozymes. Biotechnol. Adv. 2005;23:271–281. doi: 10.1016/j.biotechadv.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 42.Mangiagalli M, et al. The co-existence of cold activity and thermal stability in an Antarctic GH42 β-galactosidase relies on its hexameric quaternary arrangement. FEBS J. 2020;228:546–565. doi: 10.1111/febs.15354. [DOI] [PubMed] [Google Scholar]
- 43.Chen MC, Hsu WL, Hwang PA, Chou TC. Low molecular weight fucoidan inhibits tumor angiogenesis through downregulation of HIF-1/VEGF signaling under hypoxia. Mar. Drugs. 2015;13:4436–4451. doi: 10.3390/md13074436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ermakova SP, et al. Structure, chemical and enzymatic modification, and anticancer activity of polysaccharides from the brown alga Turbinaria ornata. J. Appl. Phycol. 2016;28:2495–2505. doi: 10.1007/s10811-015-0742-y. [DOI] [Google Scholar]
- 45.Nguyen TT, et al. Enzyme-assisted fucoidan extraction from brown macroalgae Fucus distichus subsp. evanescens and Saccharina latissima. Mar. Drugs. 2020;18:246–263. doi: 10.3390/md18050246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silchenko AS, et al. A simple plate method for the screening and detection of fucoidanases. Achiev. Life Sci. 2015;9:104–106. [Google Scholar]
- 47.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 48.Bushnell, B. BBMap: A fast, accurate, splice-aware aligner. Joint Genome Instritute, Department of Energy (2014).
- 49.Boisvert S, Raymond F, Godzaridis É, Laviolette F, Corbeil J. Ray Meta: Scalable de novo metagenome assembly and profiling. Genome Biol. 2012;13:R122. doi: 10.1186/gb-2012-13-12-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kang DD, Froula J, Egan R, Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ. 2015;3:e1165. doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu M, Scott AJ. Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics. 2012;28:1033–1034. doi: 10.1093/bioinformatics/bts079. [DOI] [PubMed] [Google Scholar]
- 54.Yin Y, et al. DbCAN: A web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40:445–451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 56.Jones P, et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mitchell AL, et al. InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019;47:351–360. doi: 10.1093/nar/gky1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petersen TN, Brunak S, Von Heijne G, Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 59.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Combet C, Blanchet C, Geourjon C, Deléage G. NPS@: Network protein sequence analysis. Trends Biochem. Sci. 2000;25:147–150. doi: 10.1016/S0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 61.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:320–324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 63.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 64.Cornish-Bowden A. Robust estimation in enzyme kinetics. In: Endrenyi L, editor. Kinetic Data Analysis: Design and Analysis of Enzyme and Pharmaco-Kinetic Experiments. Plenum Press; 1981. pp. 105–119. [Google Scholar]
- 65.Kabsch W, et al. XDS. Acta Cryst. D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Evans P. Scaling and assessment of data quality. Acta Cryst. D. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 67.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Cryst. D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Waterhouse A, et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:296–303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rivera-Colon Y, Schutsky EK, Kita AZ, Garman SC. The structure of human GALNS reveals the molecular basis for mucopolysaccharidosis IV A. J. Mol. Biol. 2012;423:736–751. doi: 10.1016/j.jmb.2012.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Cryst. D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Terwilliger TC, Adams PD, Afonine PV, Sobolev OV. A fully automatic method yielding initial models from high-resolution cryo-electron microscopy maps. Nat. Methods. 2018;15:905–908. doi: 10.1038/s41592-018-0173-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liebschner D, et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Cryst. D. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Cryst. D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moriarty NW, Grosse-Kunstleve RW, Adams PD. Electronic ligand builder and optimization workbench (eLBOW): A tool for ligand coordinate and restraint generation. Acta Cryst. D. 2009;65:1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Oleg T, Arthur OJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pettersen EF, et al. UCSF Chimera: A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 77.Jurrus E, et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018;27:112–128. doi: 10.1002/pro.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The crystal structure presented has been deposited with PDB entry 7AJ0.